- College of Agronomy, Shandong Agricultural University/State Key Laboratory of Crop Biology, Tai’an, Shandong, China
Wheat has a specific preference for NO3- and shows toxicity symptoms under high NH4+ concentrations. Increasing the nitrate supply may alleviate ammonium stress. Nevertheless, the mechanisms underlying the nitrate regulation of wheat root growth to alleviate ammonium toxicity remain unclear. In this study, we integrated physiological and weighted gene co-expression network analysis (WGCNA) to identify the hub genes involved in nitrate alleviation of ammonium toxicity at the wheat seedling stage. Five NH4+/NO3- ratio treatments, including 100/0 (Na), 75/25 (Nr1), 50/50 (Nr2), 25/75 (Nr3), and 0/100 (Nn) were tested in this study. The results showed that sole ammonium treatment (Na) increased the lateral root number but reduced root biomass. Increasing the nitrate supply significantly increased the root biomass. Increasing nitrate levels decreased abscisic acid (ABA) content and increased auxin (IAA) content. Furthermore, we identified two modules (blue and turquoise) using transcriptome data that were significantly related to root physiological growth indicators. TraesCS6A02G178000 and TraesCS2B02G056300 were identified as hub genes in the two modules which coded for plastidic ATP/ADP-transporter and WRKY62 transcription factors, respectively. Additionally, network analysis showed that in the blue module, TraesCS6A02G178000 interacts with downregulated genes that coded for indolin-2-one monooxygenase, SRG1, DETOXIFICATION, and wall-associated receptor kinase. In the turquoise module, TraesCS2B02G056300 was highly related to the genes that encoded ERD4, ERF109, CIGR2, and WD40 proteins, and transcription factors including WRKY24, WRKY22, MYB30, and JAMYB, which were all upregulated by increasing nitrate supply. These studies suggest that increasing the nitrate supply could improve root growth and alleviate ammonium toxicity through physiological and molecular regulation networks, including ROS, hormonal crosstalk, and transcription factors.
Introduction
Wheat (Triticum aestivum L.) is an important cereal crop worldwide. Nitrogen (N) is a crucial limiting nutrient that has a central effect on crop growth and yield formation. NO3- and NH4+ are the most prominent forms of inorganic N taken up by plant roots (Feng et al., 2020). In agricultural fields, the use of nitrification inhibitors together with ammonium fertilizers or organic fertilizers stabilizes ammonium at high concentrations in the soil for several weeks (Marino and Moran, 2019). Nevertheless, wheat has a specific preference for NO3- and shows toxicity symptoms under high NH4+ concentrations, including reduced plant growth, decreased root length, and leaf chlorosis (Imran et al., 2019).
Previous studies have demonstrated a few physiological and molecular mechanisms underlying the toxicity of NH4+ stress. Firstly, endogenous hormones can be imbalanced by an excess supply of ammonium. A previous study reported that high ammonium concentrations increased ethylene production (Lynch and Brown, 1997). Yang et al. (2015) found that the expression of auxin-regulated genes is repressed in NH4+ -stressed plants. Furthermore, abscisic acid (ABA) plays a positive role in regulating the OsSAPK9–OsbZIP20 pathway in rice to increase tolerance to high-NH4+ stress (Sun et al., 2020). Britto et al. (2001) proposed that the transportation of NH4+ across membranes was energy-intensive when high concentrations of ammonium were present in the medium, resulting in massive ATP consumption and waste. On the other hand, NH4+ excess causes inhibition cation uptake, such as K+ and Mg2+, which consequently change plant ion balance (Esteban et al., 2016). A recent study found that excessive assimilation of ammonium by plastidic glutamine synthetase produces high levels of protons and aggravates the acidic burden that leads to plant toxicity (Hachiya et al., 2021). NO3- has positive effects on hormone synthesis and transport to recover from the negative effects of ammonium stress (Liu et al., 2019). Sucrose non-fermenting-1-related protein kinase (SnRK1.1) negatively regulates the nitrate channel SLOW ANION CHANNEL Homologue 3 (SLAH3), which is involved in nitrate-dependent alleviation of ammonium toxicity (Sun et al., 2021). Xiao et al. (2022) revealed that NRT1.1 and SLAH3 could form a functional unit to regulate nitrate-dependent alleviation of ammonium toxicity by regulating NO3- transport and balancing rhizosphere acidification. In our previous study, we reported that a high NH4+/NO3- ratio enhances the expression of genes and proteins involved in lignin biosynthesis, leading to root lignification, thereby resulting in increased root oxidative tolerance at the cost of reducing nitrate transport and utilization (Yang et al., 2022). These results suggest that increasing nitrate levels may alleviate ammonium toxicity. These studies have succeeded in determining the physiological and molecular components associated with ammonium toxicity. Nevertheless, the molecular mechanisms underlying the nitrate regulation of wheat root growth to alleviate ammonium toxicity remain unclear.
In the post-genomic era, omics and bioinformatics are essential for understanding the complex regulatory networks in plants associated with stress adaptation and tolerance (Urano et al., 2010; Mochida and Shinozaki, 2011). For example, a previous study integrated transcriptome and metabolome analyses to reveal the physiological and molecular responses of rapeseed to ammonium toxicity (Li et al., 2021). Weighted gene co-expression network analysis (WGCNA) is widely used to identify hubs in biological systems, including plant responses to abiotic stresses (Randhawa and Pathania, 2020; Di Silvestre et al., 2021). WGCNA was used to explore candidate hub genes involved in drought adaptation in wheat (Luo et al., 2019). A study by Chen et al. (2021) showed that WGCNA could be performed to identify hub genes, networks, and pathways relevant to whole leaf responses to waterlogging stress. Integrated WGCNA and physiological analysis may provide new insights to uncover hub genes involved in nitrate alleviation of ammonium toxicity. The objectives of the present study were to (i) investigate the wheat root transcriptome profiles in response to different NH4+/NO3- ratios treatments, (ii) identify functional gene module networks that are involved in the root response to ammonium stress by WGCNA analysis, and (iii) provide new insights into nitrate alleviation of ammonium toxicity.
Materials and methods
Plant materials and culture conditions
The experiments were conducted at the experimental station of Shandong Agricultural University, Tai’ an, China. The Jimai 22 winter wheat cultivar was grown under controlled conditions. Variations in the temperature, illumination intensity, and relative humidity are shown in Supplementary Figure S1. Seeds were surface-sterilized with 1% NaClO3 for 30 min, rinsed five× with sterile water, and germinated on wet filter paper in the dark at 25°C for two days. Germinated seeds were sown in plastic pots (10 × 10 × 10 cm; 10 plants/pot) filled with perlite. The pots were placed on plastic trays (55 × 45 × 5 cm; 20 pots/tray).
Treatments and experimental design
Five NH4+/NO3- ratio treatments with a total N concentration of 6 mM were used to investigate how nitrate alleviated ammonium toxicity in a controlled climate chamber experiment. Treatments included 100/0 (Na), 75/25 (Nr1), 50/50 (Nr2), 25/75 (Nr3), and 0/100 (Nn). Ca(NO3)2, NH4Cl, and NH4NO3 were used to set these ratios. The NH4+/NO3- ratios were determined according to our previous study (Yang et al., 2022). The NH4+/NO3- ratios preparation according to the following method: 6 mmol L-1 NH4Cl and 4 mmol L-1 CaCl2 for Na; 4.5 mmol L-1NH4Cl, 0.75 mmol L-1 Ca(NO3)2 and 3.25 mmol L-1 CaCl2 for Nr1; 3 mmol L-1 NH4NO3 and 4 mmol L-1 CaCl2 for Nr2; 1.5 mmol L-1NH4Cl, 2.25 mmol L-1 Ca(NO3)2 and 1.75 mmol L-1 CaCl2 for Nr3; 3 mmol L-1 Ca(NO3)2 and 1 mmol L-1 CaCl2 for Nn. The concentration of all other nutrient elements was referred to a modified Hoagland’s nutrient solution with the following chemical composition: 5 mmol·L-1 KCl, 4 mmol·L-1 MgSO4, 1 mmol·L-1 KH2PO4, 50 μmol·L-1 Fe-EDTA, 0.5 μmol·L-1 H3BO3, 0.74 μmol·L-1 MnSO4, 0.27 μmol·L-1 ZnSO4, 0.001 μmol·L-1 CuSO4, 0.001 μmol·L-1 CoCl2, and 0.005 μmol·L-1 Na2MoO4, 0.025 μmol·L-1 KI; and pH 6.5.
Assays of root number and biomass
Wheat root number, including the primary and lateral roots, was evaluated 42 days after sowing (DAS). Ten wheat plants from each treatment group were sampled. The roots were washed with water, and the primary and lateral root numbers were counted. Finally, root samples were dried at 60°C for biomass weight measurements.
Assays of root O2-, malondialdehyde content, and superoxide dismutase activity
O2- content was determined according to the method described by Bai et al. (2015). Root samples for each treatment were homogenized in 3 mL of 65 mM phosphate buffer (pH 7.8) and the homogenate was centrifuged at 10,000 × g for 10 min. The supernatant (2 mL) was added to 50 mM phosphate buffer (pH 7.8) and 0.1 mL of 10 mM hydroxylamine hydrochloride (0.5 mL). After 20 min at 25°C, the mixture was added to 1 mL of 58 mM sulfanilamide and 1 mL of 7 mM α-naphthyl-amine and incubated at 30°C for 30 min. After this period, the absorbance was measured at 530 nm.
Root samples (0.5 g) were homogenized using a mortar and pestle at 4°C in 5 mL of 50 mmol L−1 phosphate buffer (pH 7.8). The homogenate was filtered through muslin cloth and centrifuged at 15000×g for 20 min at 4°C, and the supernatant was used to assay the enzyme activities of SOD and MDA content according to Yang et al. (2018). One unit of SOD activity was defined as the amount of crude enzyme extract required to inhibit the reduction of NBT by 50%. The extract (1 ml) and 2 ml of 0.6% thiobarbituric acid were boiled for 20 min and cooled to room temperature. The absorbance of MDA was measured at 600, 532, and 450 nm using a spectrophotometer after centrifugation at 3000 g for 15 min. MDA concentrations were calculated by the equation: MDA (μmol g-1 FW) = [6.45 × (OD532 – OD600) – 0.56 × OD450] × V/W, where OD532, OD600, and OD450 are the absorbance at 532, 600, and 450 nm, respectively; V is the volume of extraction, W is the fresh mass of sample.
Assays of root IAA and ABA content
IAA and ABA were extracted, purified, and measured by high-performance liquid chromatography (HPLC) using the method described by Yang et al. (2018). Root samples (0.1 g) from each treatment were ground to a powder in liquid nitrogen, 4 mL acetonitrile was added, and the homogenate was incubated in the dark at 4°C for 12 h. The extract was centrifuged at 10800 rpm for 10 min at 4°C. The residue was extracted twice with the same solvent. The supernatant was combined and concentrated to residue at 37°C by rotatory evaporation, and re-dissolved in 8.0 mL 0.4 mol L-1 phosphate buffer and then added 6.0 mL chloroform and to remove pigment. The pH of the aqueous phase was adjusted to pH3 using pure formic acid. The aqueous phase was extracted thrice with ethyl acetate (3.0 mL). The ethyl acetate phase was concentrated by rotary evaporation and redissolved in 1 mL of the mobile phase. Phytohormone extracts were filtered through 0.2-μm hydrophobic membranes and 10-μL aliquots were injected into a Waters Symmetry C18 column (4.6 mm × 150.0 mm; 5 μm; Waters Corp., Milford, MA, USA) using acetonitrile: methanol: 0.6% acetic acid (5:50:45, v:v:v) as the mobile phase. The flow rate was held at 0.6 mL min-1 and the peaks were identified with a photodiode array detector (Waters 2998; Waters Corp., Milford, MA, USA) at an absorbance of 254 nm.
Co‐expression network analysis for construction of modules of transcriptomics genes
Root RNA extraction, cDNA library construction, Illumina sequencing, read mapping, and differential gene expression analyses have been described previously (Yang et al., 2022). Gene co-expression networks were constructed using the WGCNA package in R software. Network construction and module detection were implemented using the blockwiseModule function. The soft thresholding power was 14. The ‘maxBlockSize’ is ‘nGenes’. The networkType parameter is set to ‘unsigned’. The ‘mergeCutHeight’ was 0.25, and ‘minModuleSize’ was 30. The other parameters were kept at their default values. Modules significantly associated with root physiological traits (root weight, O2-, MDA, and ABA content) were identified using Pearson’s correlation between eigengene expression profiles and physiological traits. The gene significance (GS) of a gene is defined as the absolute value of the correlation between a gene and a certain clinical parameter, and the module membership (MM) of a gene is defined as the correlation between the module eigengene and the gene expression profile. Genes with higher GS and MM were defined as hub genes in the module. Module networks were visualized using Cytoscape (v3.9.1).
Statistical analysis and processing
Data for root number and biomass, O2-, MDA, IAA, ABA content, and SOD activity were processed using DPS v. 7.05. Multiple comparisons were performed using a preliminary F-test. The means were tested using the least significant difference test, and the significance was set at a probability level of 0.05. Graphs were plotted using OriginPro 2017 software (OriginLab Inc., USA).
Results
Effect of increasing nitrate supply on root biomass and number
In the present study, we investigated the effects of different NH4+/NO3- ratios on root biomass and primary and lateral root numbers. The results showed that the sole ammonium supply treatment (Na) significantly decreased root biomass, but increased nitrate increased biomass (Figure 1A). There was no significant difference in the number of primary roots among treatments (Figure 1B). But, 42-d of treatment of wheat roots with ammonium significantly increased the lateral root number and total root number. Increasing the nitrate supply decreased the lateral and total root numbers. These results suggest that ammonium can elicit root emergence but inhibit root dry matter accumulation. Increasing the nitrate supply could improve root dry matter accumulation.
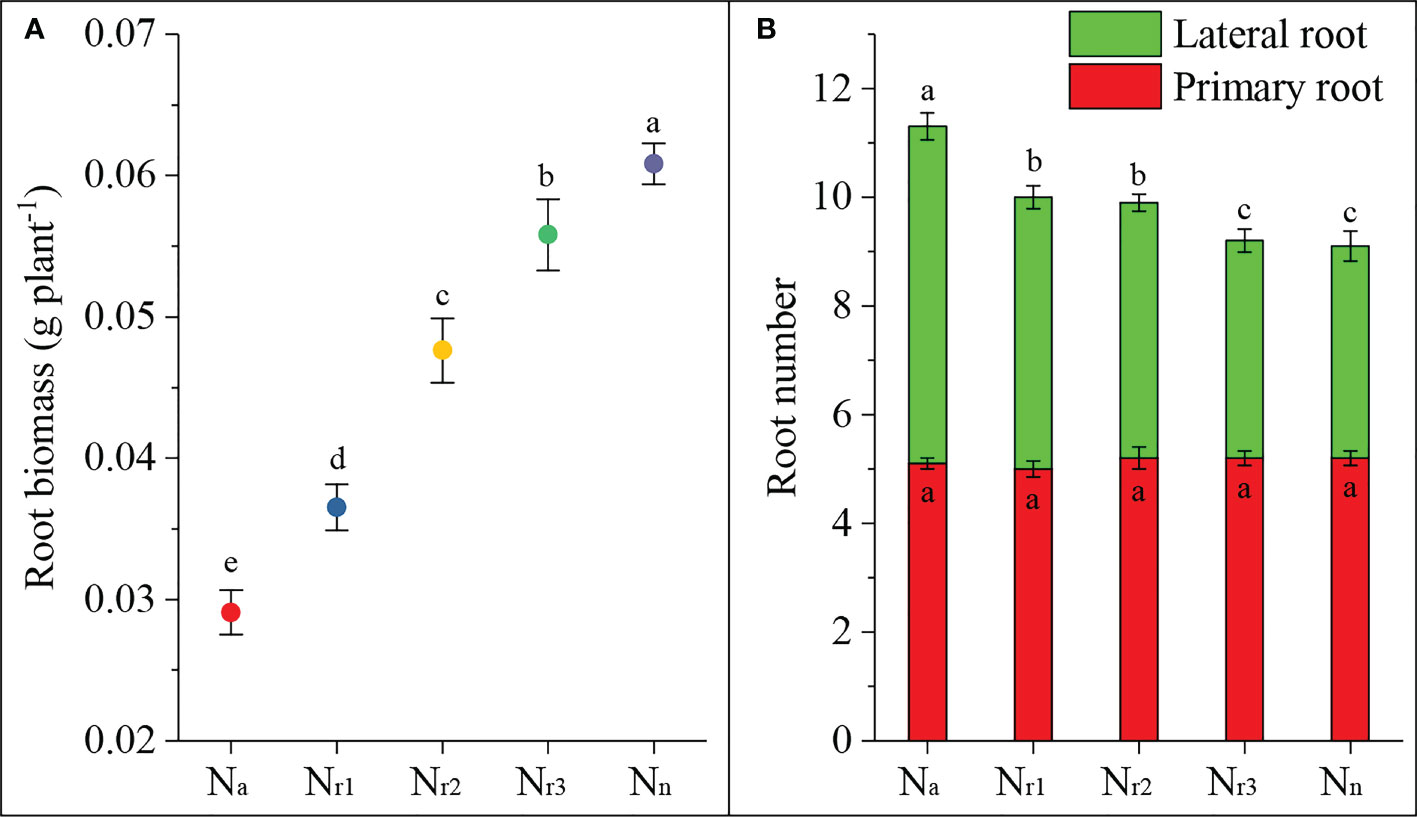
Figure 1 The effects of different NH4+/NO3- ratios on root biomass (A) and number (B) of wheat root at 42 days after sowing. Segments represent the standard error of the mean (mean ± SE, n = 3). Different letters indicate significant differences among each treatment, P < 0.05.
Changes in ROS, MDA content, and antioxidant enzyme activity
The results showed that root O2- and MDA contents gradually and significantly decreased with increasing nitrate supply (Figures 2A, B). The Na treatment had the highest O2- and MDA content, while the sole nitrate supply treatment (Nn) yielded the lowest value. Compared with the Nr1 treatment, root O2- and MDA contents under the Nr3 treatment decreased by 33.12% and 22.21%, respectively. These results suggest that increasing the NO3- -N ratio or sole nitrate supply could alleviate ammonium-induced oxidative stress. Roots of ammonium-treated wheat plants had much higher SOD activity, whereas Nn treatment had the lowest antioxidant enzyme activities (Figure 2C). SOD activity was significantly decreased by increasing nitrate supply. For example, compared to Nr1 treatment, SOD activity under Nr3 treatment decreased by 57.72%.
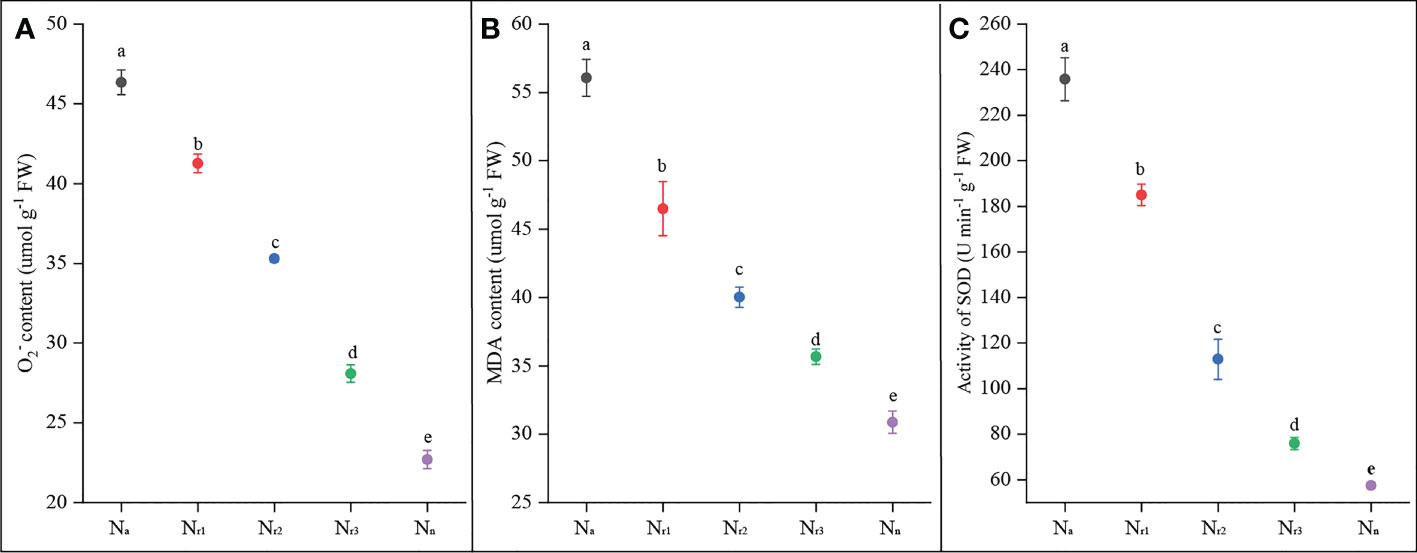
Figure 2 The effects of different NH4+/NO3- ratios on root O2- (A), MDA content (B), and SOD(C) of wheat root at 42 days after sowing. Segments represent the standard error of the mean (mean ± SE, n = 3). Different letters indicate significant differences among each treatment, P < 0.05.
Transcriptomic genes expression and gene ontology enrichment
Next, we analyzed the number of differentially expressed genes (DEGs) under different NH4+/NO3- ratios. For example, 449 upregulated and 293 downregulated genes were identified in the Na/Nr1 group, whereas 4570 upregulated and 4474 downregulated genes were identified in the Na/Nr3 group (Figure 3, Table S1). A total of 2066 upregulated and 768 downregulated genes were identified in the Nr3/Nn group, whereas 5618 upregulated and 4634 downregulated genes were identified in the Nr1/Nn group.
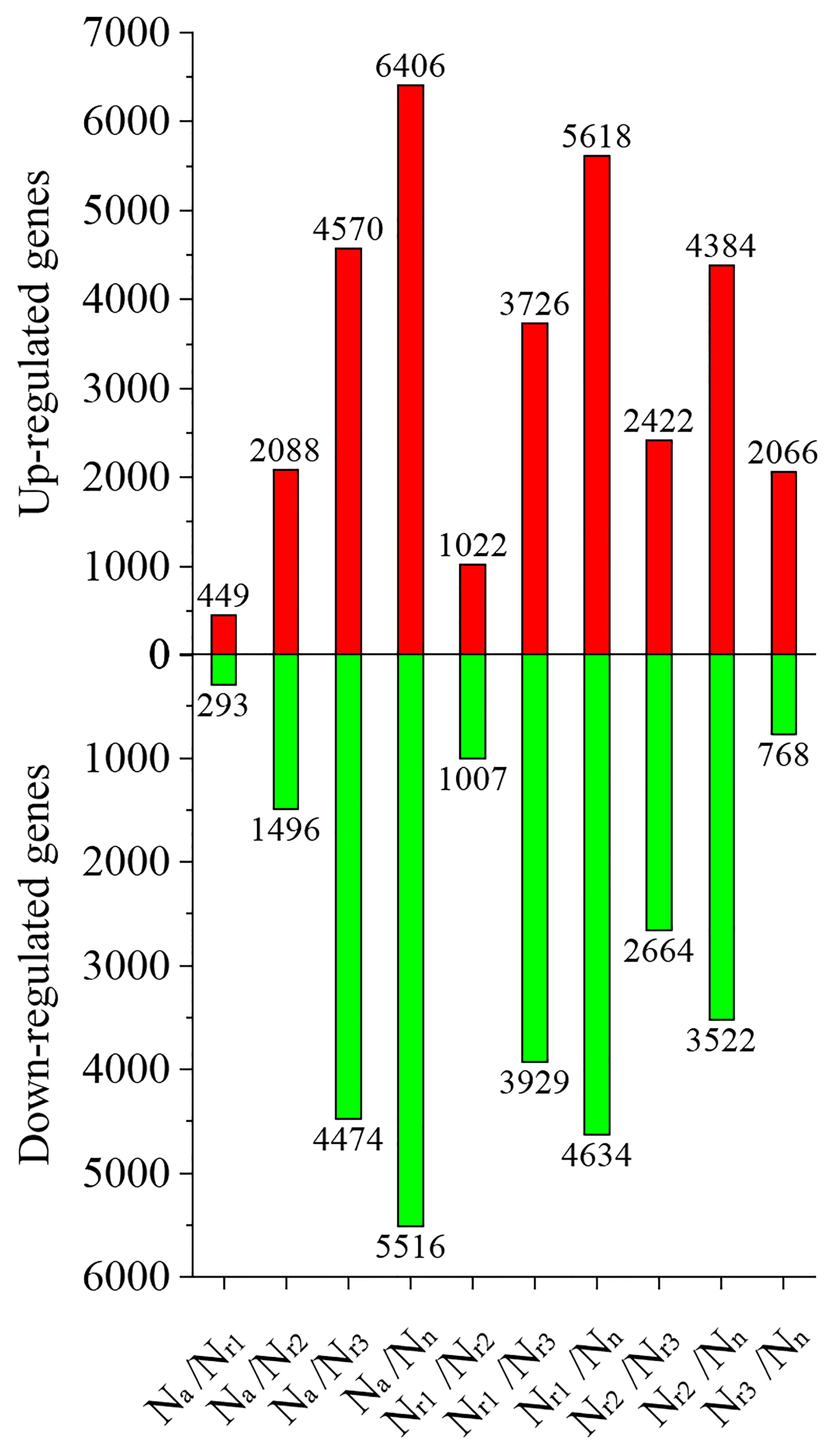
Figure 3 The effects of different NH4+/NO3- ratios on the differentially expressed genes (DEGs) in the wheat root samples.
GO enrichment analysis revealed that 558 significant GO terms were identified in the Na/Nn transcriptome comparison group, which was the highest group (Figure 4A, Table S2). Only 177 GO terms were identified for the Na/Nr1 group. 31 significant GO terms were unique to Na/Nn, including 16 GO terms enriched in molecular functions, such as glutamate-cysteine ligase activity, and 15 enriched in biological processes, such as the glutathione biosynthetic process (Table S3). Additionally, 21 GO terms were shared among all groups, including plant-type cell wall (GO:0009505), peroxidase activity (GO:0004601), nitrate transport (GO:0015706), and phenylpropanoid biosynthetic process (GO:0009699) (Figure 4B; Table S3).
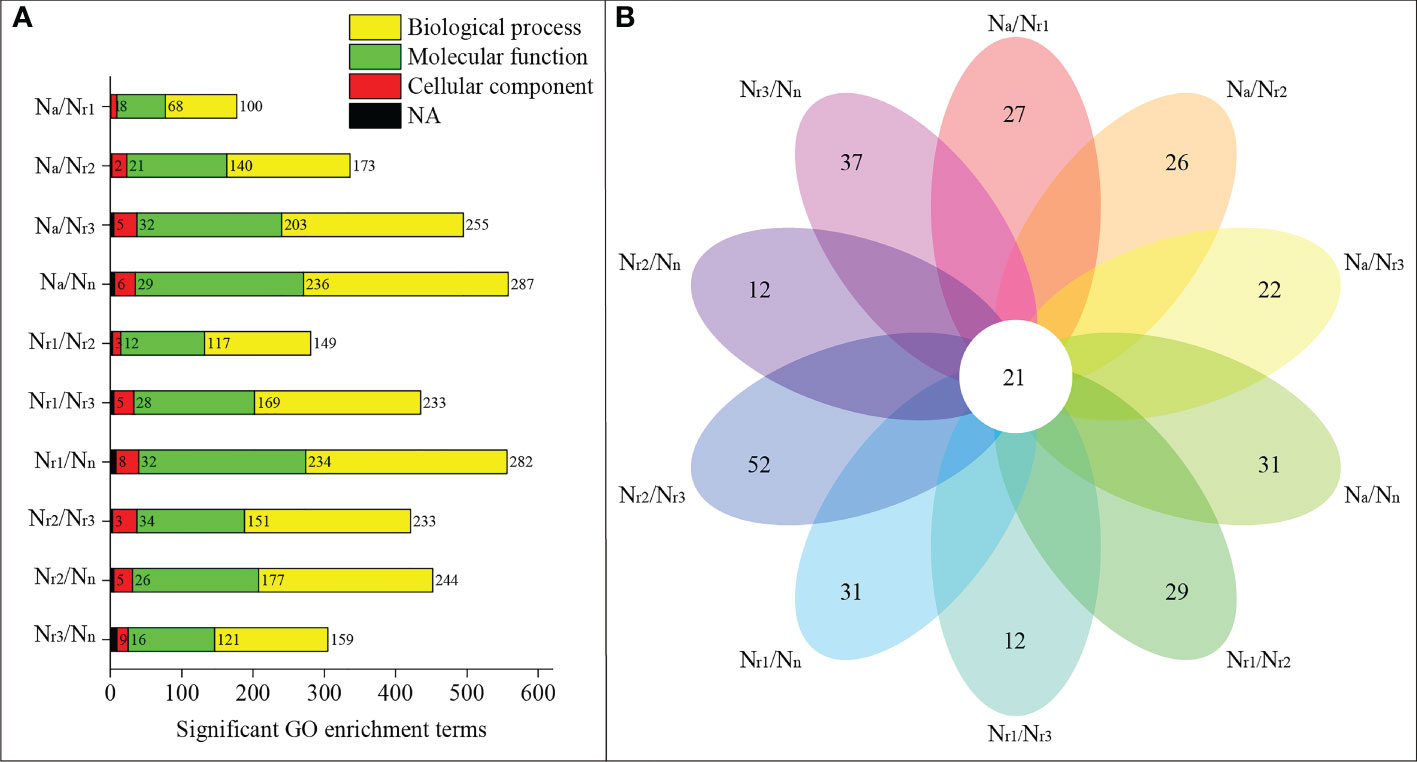
Figure 4 (A) Functional enrichment analysis of differentially expressed genes (DEGs) in the wheat root samples of different NH4+/NO3- ratio treatments. (B) Venn diagrams represent the number of shared and unique GO terms in each comparisons.
Weighted co-expression network analysis
To understand the regulatory network of increasing nitrate under ammonium stress, we performed WGCNA using the expression data of 85570 genes from the transcriptome data (Table S4). The top 75% of the most varying genes selected by a robust covariation estimator relative to median absolute deviation were used as inputs to construct the weighted network. 47861 genes were considered for downstream analysis (Table S5). Results showed that a total of 23 distinct modules were identified for the five NH4+/NO3- ratio treatments (Figure 5). A total of 23 genes were identified and subsequently defined as hub genes based on the principle of the highest KME (Tables 1 and S6). For example, TraesCS7B02G271400 was coded as a 40S ribosomal protein in the brown module. TraesCS6A02G178000 is a plastidic ATP/ADP transporter in the blue module. TraesCS2B02G056300 was annotated as a WRKY62 transcription factor in the turquoise module. TraesCS7A02G549000 was annotated as an NAC domain-containing protein 22 in the yellow module.
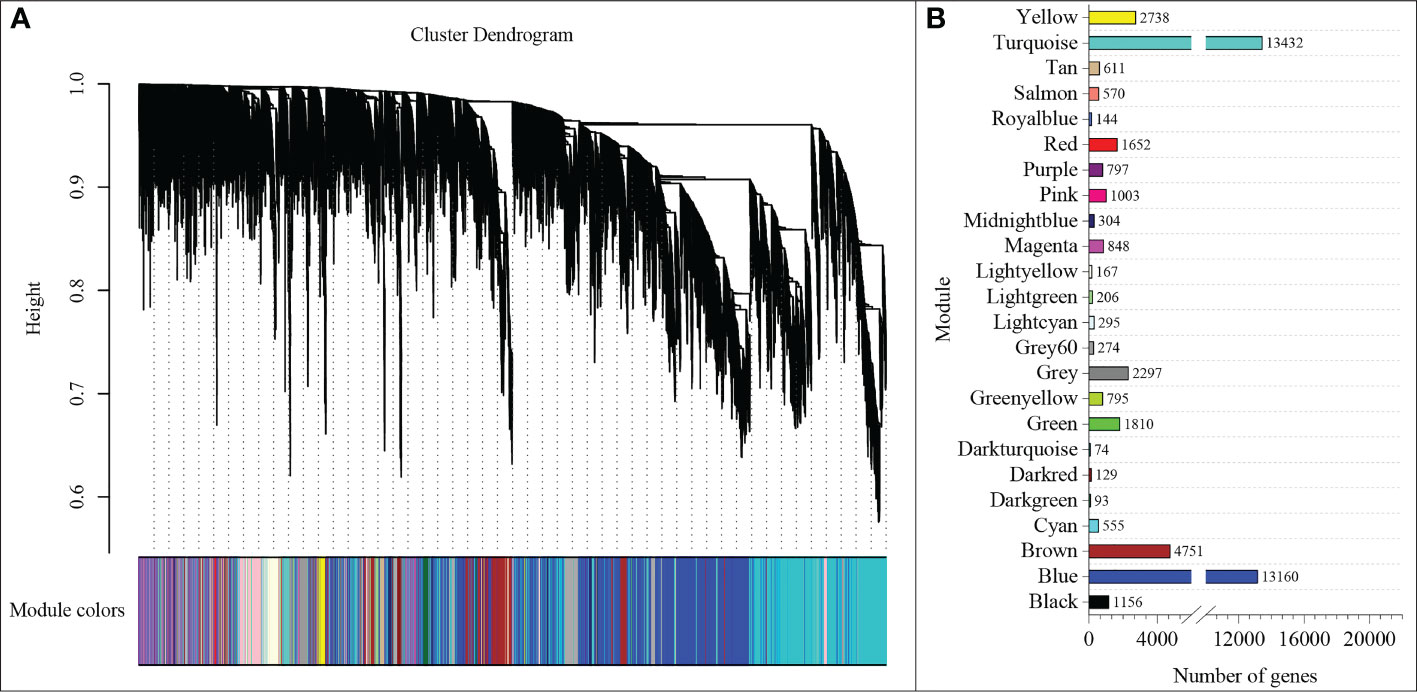
Figure 5 WGCNA of genes (A) and modules (B) were identified under different NH4+/NO3- ratios treatments.
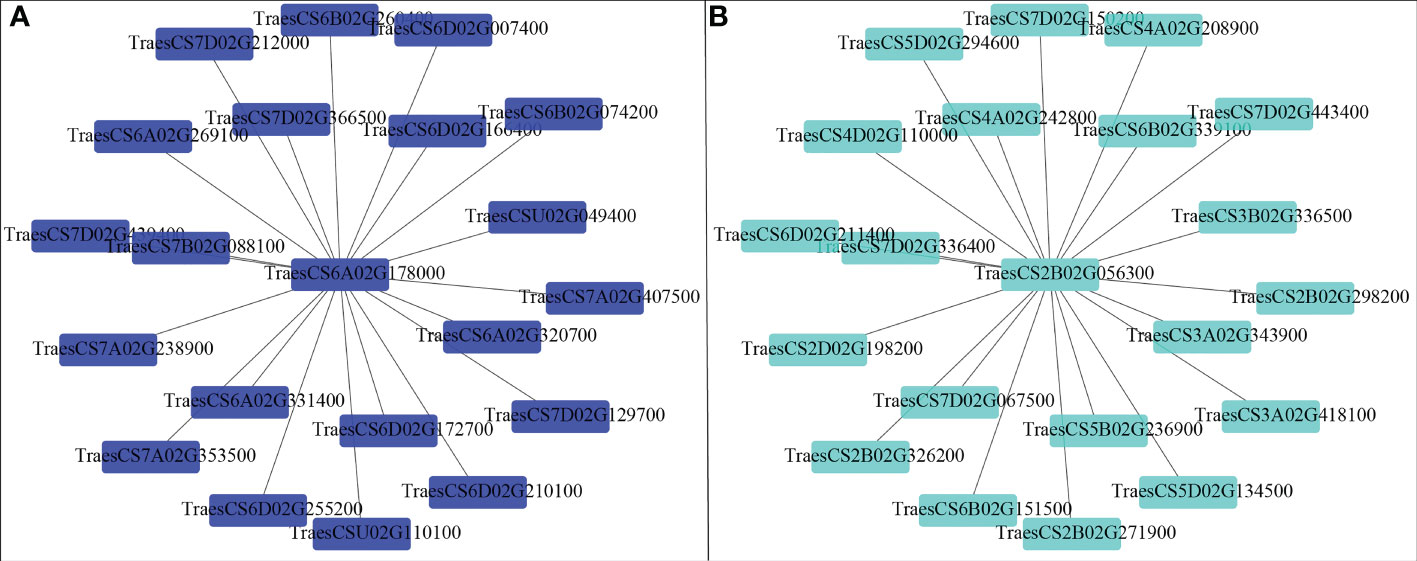
Correlation between the modules and root physiological traits, including root biomass, O2-, MDA, SOD activity, IAA, and ABA content (Figure 6). The results showed that the turquoise module was highly related to root biomass. The blue module (with 13160 genes) was highly related to O2-, MDA, SOD activity, and ABA content. Based on the correlation coefficients, we selected the blue and turquoise modules to perform functional analysis and construct gene networks. According to the TOM values calculated by WGCNA, we constructed a gene co-expression network for the blue and turquoise modules. The top 20 interacting genes were mapped using the weight values (Figure 7). The results showed that there were downregulated genes with increasing nitrate supply, which encoded indolin-2-one monooxygenase, Stress responsive gene 1 (SRG1), laccase, ATP/ADP-transporter, Beta3GalT1, DETOXIFICATION, and wall-associated receptor kinase interacting with hub genes in the blue module (Figure 8A; Table 2). Four upregulated gene transcription factors were identified under nitrate supply conditions: WRKY24, WRKY22, JAMYB, and MYB30 in the turquoise module (Figure 8B; Table 2). Genes encoding the CSC1-like protein ERD4, xylan arabinosyl transferase, WD40 protein, NRT1/PTR FAMILY 4.3 were up-regulated by nitrate supply and interacted with hub genes in the turquoise module. In addition, nitrate induced the upregulation of TraesCS5B02G236900 and TraesCS2D02G198200 in the turquoise module, which were related to ethylene and gibberellin signal response, respectively.
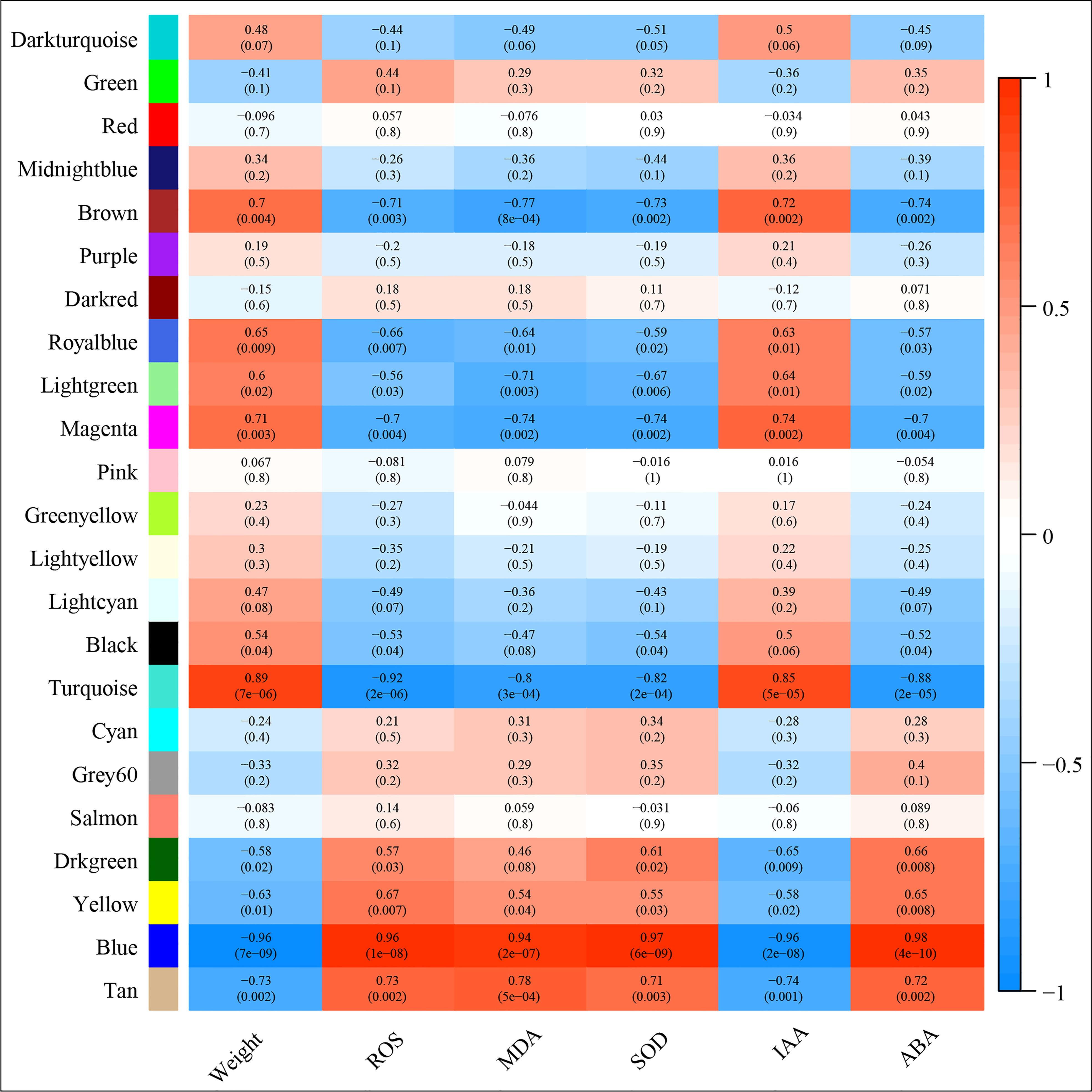
Figure 6 Module‐trait associations. Each row corresponds to a module eigengene, column to a trait. Each frame contains the corresponding correlation and p‐value. The table is color‐coded by correlation according to the color legend.
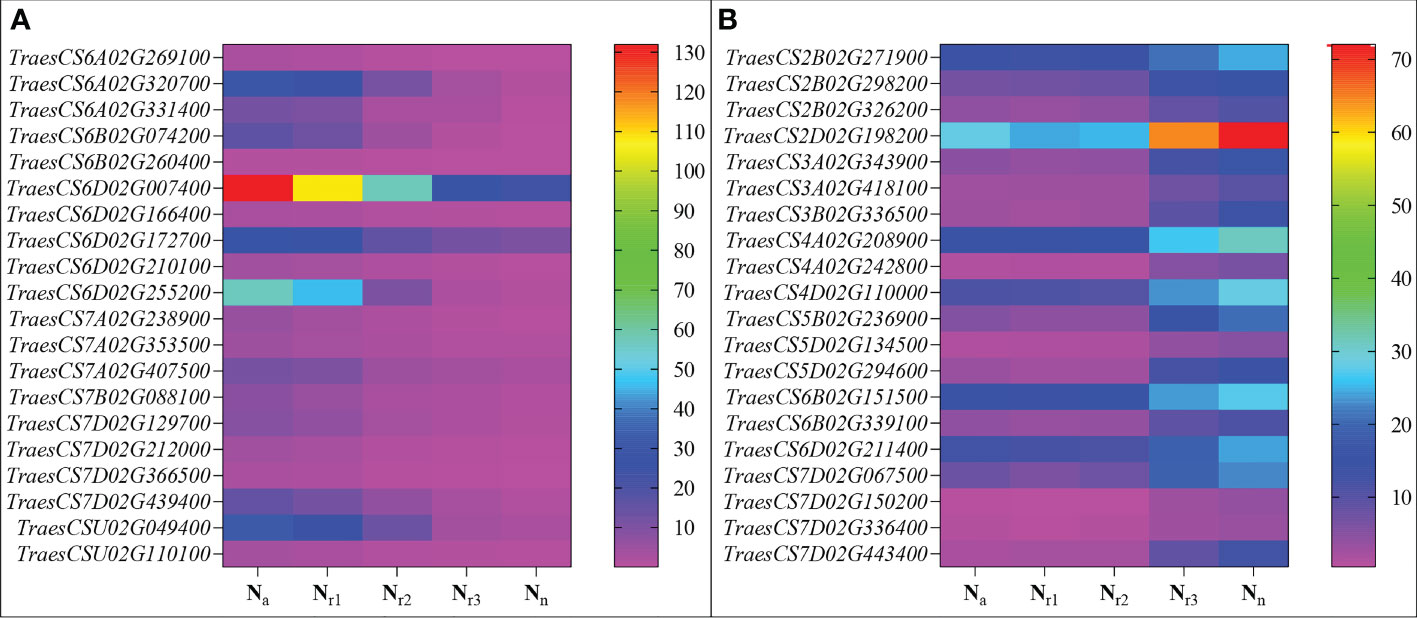
Figure 8 Heatmaps represent the candidate genes expression abundance in the blue (A) and turquoise (B) module under different NH4+/NO3- ratios treatments. The gene expression was scaled using the fragment per kilobase of transcript per million mapped reads (FPKM) based on a mean value of three biological replicates. Different colors represent the upregulation and downregulation difference of genes among treatments.
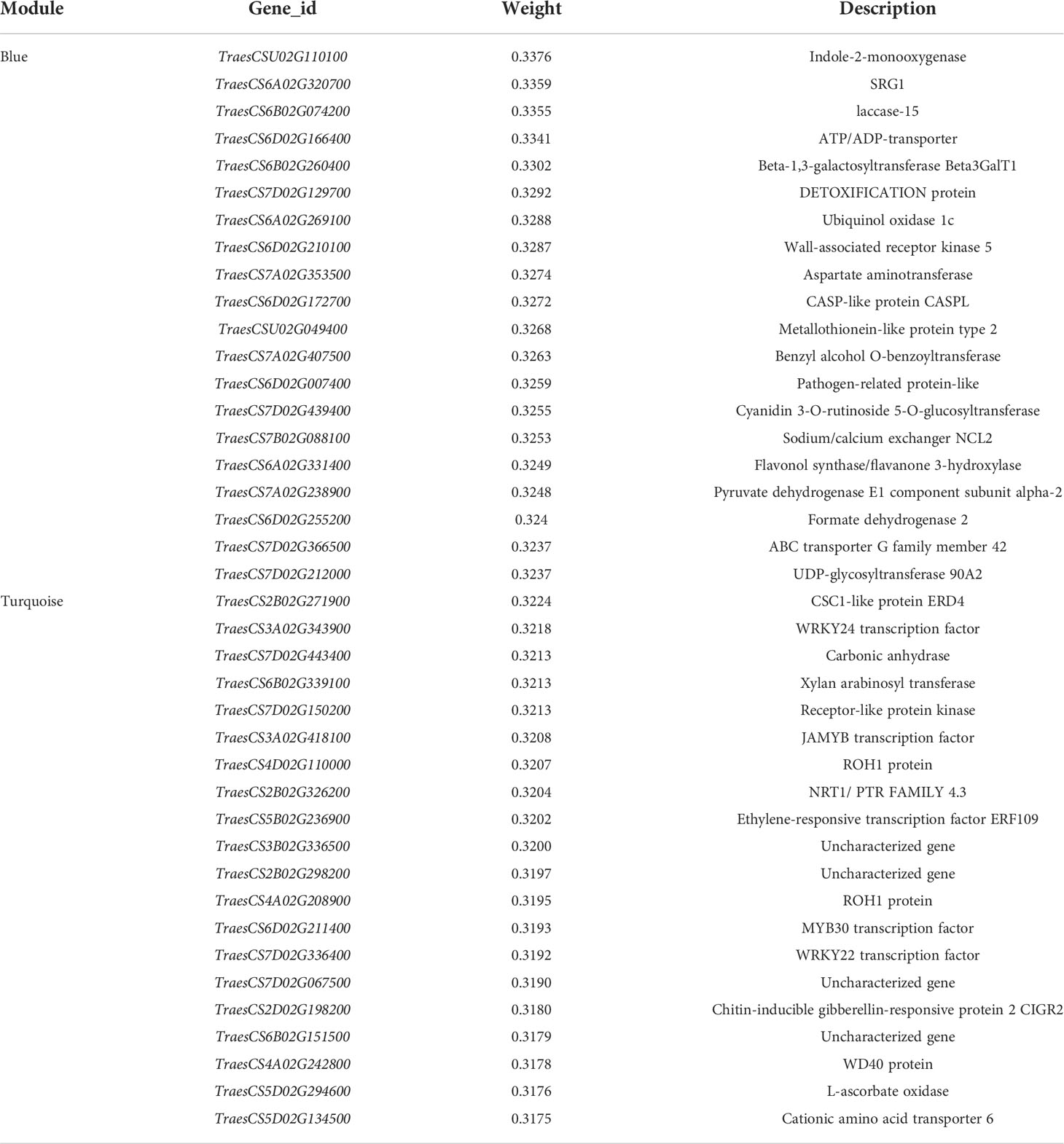
Table 2 Genes associated with hub gene in blue and turquoise modules and their function annotations.
Discussion
The physiological effects of increasing nitrate supply on alleviation of ammonium toxicity
Nitrate and ammonium are the main inorganic forms of nitrogen absorbed by plants, but ammonium nutrition often represents an important growth-limiting stressor (Sarasketa et al., 2016). In a previous study, we found that ammonium supply resulted in lower wheat leaf photochemical efficiency and root uptake capacity, thereby reducing plant biomass and leading to stunted growth (Yang et al., 2022). Investigating agronomy management practices and underlining their regulation and signaling processes may contribute to alleviating ammonium inhibition. In the present study, we found that increasing nitrate supply and reducing the NH4+/NO3- ratio inhibited lateral root number, but increased root biomass (Figure 1). On the one hand, this may be due to the lower O2- content under sole nitrate supply and the low NH4+/NO3- ratio condition (Figure 2A). Reactive oxygen species (ROS) are toxic molecules that can cause protein, membrane, and DNA damage. Malondialdehyde (MDA) is a widely used marker of oxidative lipid injury (Davey et al., 2005). Our study showed that the lower NH4+/NO3- ratio treatment significantly decreased MDA content in wheat roots (Figure 2B). Furthermore, in the current study, ammonium treatment (Na) induced the highest SOD activity, while SOD activity decreased with decreasing NH4+/NO3- ratio (Figure 2C). SOD catalyzes the conversion of O2- to H2O2 (Mittler, 2002), which accumulates under ammonium supply and inhibits primary root elongation (Liu et al., 2022). These results indicate that increasing the nitrate supply could alleviate the oxidative stress elicited by ammonium in wheat roots.
On the other hand, nitrate and ammonium supply regulate root system architecture through related hormones, such as auxin and ABA (Patterson et al., 2010; Li et al., 2014; Meier et al., 2020; Liu and von Wirén, 2022). Di et al. (2018) found that high NH4+ decreased free IAA content in rice roots. Consistent with this result, we found that sole ammonium or high NH4+/NO3- ratio treatment decreased the IAA content of wheat roots (Figure 9A). In contrast, increasing the nitrate supply could increase the IAA content. Nevertheless, a previous study stated that high-NH4+ stress decreased free IAA in roots by increasing IAA inactivation but not by decreasing IAA biosynthesis (Di et al., 2021). Elevated NH4+ levels can significantly accelerate ABA accumulation in rice tissue ABA accumulation (Sun et al., 2020). ABA plays a direct role in mediating the inhibitory effects of nitrate on lateral root development (Signora et al., 2002). In our study, both sole ammonium and high NH4+/NO3- ratio treatments increased wheat root ABA content, while increasing nitrate supply reduced root ABA content (Figure 9B). These results suggest that increasing nitrate supply alleviated ammonium toxicity by regulating root physiological indicators, including SOD activity, ROS, and hormone content.
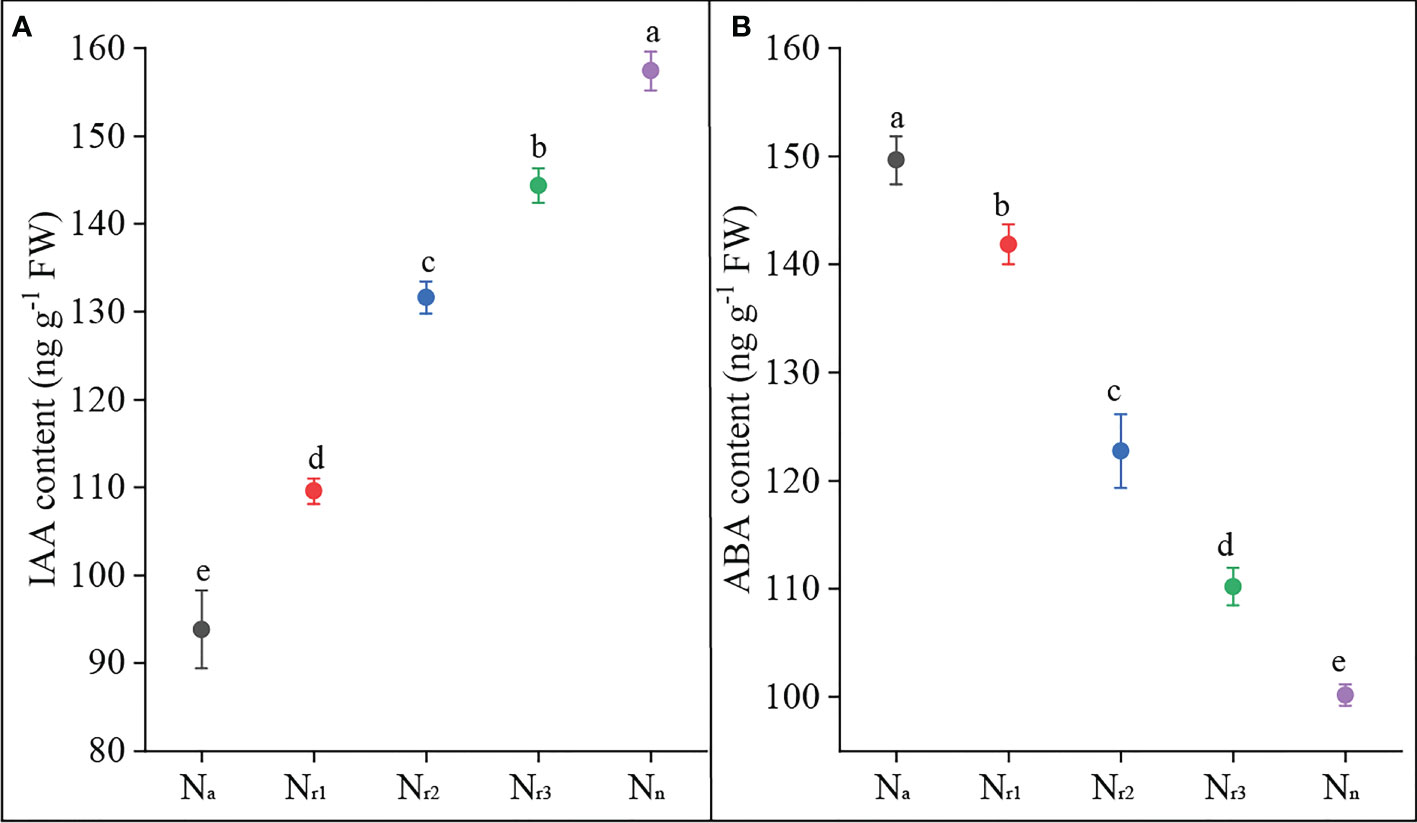
Figure 9 The effects of different NH4+/NO3- ratios on IAA (A) and ABA content (B) of wheat root at 42 days after sowing. Segments represent the standard error of the mean (mean ± SE, n = 3). Different letters indicate significant differences among each treatment, P < 0.05.
The molecular regulation networks of increasing nitrate supply on alleviation of ammonium toxicity
The wheat root is the first organ to be exposed to high NH4+ concentrations. Previous studies have demonstrated some physiological response mechanisms of ammonium stress from the aspect of transamination ability and oxidative metabolism and identified techniques to alleviate NH4+ toxicity (Wang et al., 2016; Liu et al., 2021). Molecular adjustment of roots to excess ammonium nutrition may determine the capacity of plants to cope with ammonium stress. In the present study, we combined the transcriptome with physiological responses and used WGCNA to explore functional hub genes and detect the key regulatory factor of nitrate that alleviates NH4+ toxicity and contributes to root development under ammonium stress. We found that the Na/Nn comparison group had the most DEGs (Figure 5). The results of GO enrichment analysis showed that DEGs were mainly related to plant-type cell wall, peroxidase activity, nitrate transport, and phenylpropanoid biosynthetic processes among all treatments. Further, conjoint analysis of physiological indicators as well as transcriptome data was used to identify root growth-specific gene modules, such as blue and turquoise modules (Figure 6). The blue module was strongly related to the ROS and MDA content, SOD activity, and ABA content. The gene encoding the plastidic ATP/ADP-transporter was the hub gene in the blue module (Table 1), which was upregulated by the sole ammonium treatment but downregulated with increasing nitrate supply (Figure 8A). The main function of plastidic ATP/ADP transporters is to supply ATP to storage plastids (Reiser et al., 2004). Previous studies have suggested that high energy costs of carbohydrate transport and metabolism or futile transmembrane NH4+ cycling are induced by high ammonium stress (Britto et al., 2001; Ren et al., 2020). In this study, we selected genes that were highly associated with hub genes via weight values. For example, TraesCS6A02G178000 was highly related to the downregulated genes encoding indole-2-monooxygenase, SRG1, DETOXIFICATION 27, CASPL protein, and ABC transporter G family member 42 (Figure 7A; Table 2). The molecular function of indole-2-monooxygenase shows that it has oxidoreductase activity and can incorporate or reduce molecular oxygen. SRG1 appears to act as a transcriptional repressor and contributes to the engagement of plant defense response (Cui et al., 2018). DETOXIFICATION is also known as a multidrug and toxic compound extrusion protein that is involved in a wide variety of physiological functions throughout plant development. It transports a broad range of substrates, such as organic acids, plant hormones, and secondary metabolites (Takanashi et al., 2014). A previous study found that overexpression of CASPL in Arabidopsis significantly decreased primary root growth, indicating that CASPL negatively regulates plant growth (Yang et al., 2015). In our study, TraesCS6D02G172700 encoded the CASPL protein, which was upregulated by ammonium treatment alone and downregulated by increasing nitrate supply (Figure 8A). ABC transporters might be a detoxifying mechanism to protect against ammonium stress through the accumulation of N compounds in the root vacuole (Vega-Mas et al., 2017).
In addition, the gene encoding the transcription factor WRKY62 was the hub gene in the turquoise module and was upregulated by increasing the nitrate supply. Previous studies have reported that WRKY transcription factors are among the largest families of transcriptional regulators and play a pivotal role in modulating various signal transduction pathways during biotic and abiotic stresses (Wani et al., 2021). For example, Gao et al. (2018) found that TaWRKY2 enhances drought tolerance and increases grain yield in wheat. WRKY62 has been previously reported to be related to disease resistance in rice and Arabidopsis (Kim et al., 2008; Fukushima et al., 2016). In the present study, we found that the turquoise module was highly correlated with root biomass and IAA content (Figure 6). As its hub gene, this suggest that WRKY62 may positively regulate wheat root growth under increasing nitrate supply condition. Previous studies have suggested that hormones and transcription factors cooperate to regulate root growth (Vega et al., 2019; Ortigosa et al., 2021). Ding et al. (2015) reported that WRKY46 contributes to the feedforward inhibition of osmotic/salt stress-dependent lateral root inhibition via the regulation of ABA signaling and auxin homeostasis.
In this study, we constructed correlation networks for WRKY62 gene (Figure 7B). The results showed that TraesCS2B02G056300 was highly related to the genes that encoded ERD4, ERF109, CIGR2, WD40 proteins, and transcription factors that include WRKY24, WRKY22, MYB30, and JAMYB, which were all up-regulated by increasing nitrate supply (Figure 8B). Overexpression of the ERD4 gene could mitigate tobacco plant physiology by enduring stress tolerance (Jha and Mishra, 2021). Cai et al. (2014) reported that ERF109 integrates JA signalling into auxin pathways to regulate root architecture. CIGR2 is correlated with the bioactivities of GA and has been reported to play a role in stress responses (Yuan et al., 2016). WD40 interacts with MYB and bHLH to form a ternary regulatory complex (MYB-bHLH-WD40) and could improve tolerance to abiotic stress by regulating root growth and development (Ramsay and Glover, 2005; Kong et al., 2015; Tan et al., 2021). WRKY24 and WRKY22 have been identified in the response to salt stress in rice at the seedling stage (Zhou et al., 2016). JAMYB overexpression in transgenic Arabidopsis improves tolerance to high-salinity stress during root elongation (Yokotani et al., 2013). MYB30 is downstream of ROS signalling and regulates root development (Mabuchi et al., 2018). Sakaoka et al. (2018) showed that MYB30-regulated root cell elongation is mediated by ROS production under ABA signaling. These studies suggest that increasing the nitrate supply could improve root growth and alleviate ammonium toxicity through molecular regulation networks, including ROS, hormonal crosstalk, and transcription factors.
Conclusion
In the present study, wheat seedling roots responded to ammonium stress by increasing ROS, MDA, and ABA contents and decreasing root biomass. WGCNA revealed two-hub genes, TraesCS6A02G178000 and TraesCS2B02G056300 encode the plastidic ATP/ADP-transporter and transcription factor WRKY62, which is related to root growth and ammonium stress, respectively. Increasing nitrate supply alleviated the physiological effects of ammonium toxicity such as a decrease in ROS and ABA content and an increase in IAA content. Further, an increase in nitrate supply down-regulated genes that encoded the CASPL protein and the ABC transporter, and up-regulated genes that encoded ERD4, ERF109, CIGR2, WD40 proteins, and transcription factors including WRKY24, WRKY22, MYB30, and JAMYB, which enhanced ammonium tolerance.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found here: https://figshare.com/articles/dataset/WGCNA/20409792.
Author contributions
DY conceived and designed the experiments, analyzed the data, prepared figures and tables, authored and reviewed drafts of the manuscript, and approved the final draft. LL, XZ, JL, and JR performed the experiments. All authors contributed to the article and approved the submitted version.
Funding
The research report here was supported by the National Natural Science Foundation of China (31801295), the Shandong Province Natural Science Foundation (ZR2017BC106), China Postdoctoral Science Foundation funded Project (2018M632701), and supported by China Postdoctoral Science Special Foundation (2019T120600).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1012966/full#supplementary-material
References
Bai, X. Y., Dong, Y. J., Wang, Q. H., Xu, L. L., Kong, J., Liu, S. (2015). Effects of lead and nitric oxide on photosynthesis, antioxidative ability, and mineral element content of perennial ryegrass. Biol. Plantarum. 59, 163–170. doi: 10.1007/s10535-014-0476-8
Britto, D. T., Siddiqi, M. Y., Glass, A. D. M., Kronzucker, H. J. (2001). Futile transmembrane NH4+ cycling: A cellular hypothesis to explain ammonium toxicity in plants. PNAS 98, 4255–4258. doi: 10.1073/pnas.061034698
Cai, X. T., Xu, P., Zhao, P. X., Liu, R., Yu, L. H., Xiang, C. B. (2014). Arabidopsis ERF109 mediates cross-talk between jasmonic acid and auxin biosynthesis during lateral root formation. Nat. Commun. 5, 5833. doi: 10.1038/ncomms6833
Chen, S., Xu, Z. Y., Adil, M. F., Zhang, G. P. (2021). Cultivar-, stress duration- and leaf age-specific hub genes and co-expression networks responding to waterlogging in barley. Environ. Exp. Bot. 191, 104599. doi: 10.1016/j.envexpbot.2021.104599
Cui, B. M., Pan, Q. N., Clarke, D., Villarreal, M. O., Umbreen, S., Yuan, B., et al. (2018). S-nitrosylation of the zinc finger protein SRG1 regulates plant immunity. Nat. Commun. 9, 4226. doi: 10.1038/s41467-018-06578-3
Davey, M. W., Stals, E., Panis, B., Keulemans, J., Swennen, R. L. (2005). High-throughput determination of malondialdehyde in plant tissues. Anal. Biochem. 347, 201–207. doi: 10.1016/j.ab.2005.09.041
Di, D. W., Li, G. J., Sun, L., Wu, J. J., Wang, M., Kronzucker, H. J., et al. (2021). High ammonium inhibits root growth in Arabidopsis thaliana by promoting auxin conjugation rather than inhibiting auxin biosynthesis. J. Plant Physiol. 261, 153415. doi: 10.1016/j.jplph.2021.153415
Ding, Z. J., Yan, J. Y., Li, C. X., Li, G. X., Wu, Y. R., Zheng, S. J. (2015). Transcription factor WRKY46 modulates the development of arabidopsis lateral roots in osmotic/salt stress conditions via regulation of ABA signaling and auxin homeostasis. Plant J. 84, 56–69. doi: 10.1111/tpj.12958
Di Silvestre, D., Vigani, G., Mauri, P., Hammadi, S., Morandini, P., Murgia, I. (2021). Network topological analysis for the identification of novel hubs in plant nutrition. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.629013
Di, D. W., Sun, L., Zhang, X. N., Li, G. J., Kronzucker, H. J., Shi, W. M. (2018). Involvement of auxin in the regulation of ammonium tolerance in rice (Oryza sativa l.). Plant Soil. 432, 373–387. doi: 10.1007/s11104-018-3813-4
Esteban, R., Ariz, I., Cruz, C., Moran, J. F. (2016). Mechanisms of ammonium toxicity and the quest for tolerance. Plant. Sci. 248, 92–101. doi: 10.1016/j.plantsci.2016.04.008
Feng, H. M., Fan, X. R., Miller, A. J., Xu, G. H. (2020). Plant nitrogen uptake and assimilation: regulation of cellular pH homeostasis. J. Exp. Bot. 71, 4380–4392. doi: 10.1093/jxb/eraa150
Fukushima, S., Mori, M., Sugano, S., Takatsuji, H. (2016). Transcriptionfactor WRKY62 plays a role in pathogen defense and hypoxia-responsive gene expression in rice. Plant Cell. Physiol. 57, 2541–2551. doi: 10.1093/pcp/pcw185
Gao, H. M., Wang, Y. F., Xu, P., Zhang, Z. B. (2018). Overexpression of a WRKY transcription factor TaWRKY2 enhances drought stress tolerance in transgenic wheat. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00997
Hachiya, T., Inaba, J., Wakazaki, M., Sato, M., Toyooka, K., Miyagi, A., et al. (2021). Excessive ammonium assimilation by plastidic glutamine synthetase causes ammonium toxicity in Arabidopsis thaliana. Nat. Commun. 12, 4944. doi: 10.1038/s41467-021-25238-7
Imran, M., Sun, X. C., Hussain, S., Ali, U., Rana, M. S., Rasul, F., et al. (2019). Molybdenum-induced effects on nitrogen metabolism enzymes and elemental profile of winter wheat (Triticum aestivum l.) under different nitrogen sources. Int. J. Mol. Sci. 20, 3009. doi: 10.3390/ijms20123009
Jha, R. K., Mishra, A. (2021). Introgression of SbERD4 gene encodes an early-responsive dehydration-stress protein that confers tolerance against different types of abiotic stresses in transgenic tobacco. Cells-Basel. 11, 62. doi: 10.3390/cells11010062
Kim, K. C., Lai, Z. B., Fan, B. F., Chen, Z. X. (2008). Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. Plant Cell. 20, 2357–2371. doi: 10.1105/tpc.107.055566
Kong, D. J., Li, M. J., Dong, Z. H., Ji, H. T., Li, X. (2015). Identification of TaWD40D, a wheat WD40 repeat-containing protein that is associated with plant tolerance to abiotic stresses. Plant Cell. Rep. 34, 395–410. doi: 10.1007/s00299-014-1717-1
Li, B. H., Li, G. J., Kronzucker, H. J., Baluška, F., Shi, W. M. (2014). Ammonium stress in Arabidopsis: signaling, genetic loci, and physiological targets. Trends. Plant Sci. 19, 107–114. doi: 10.1016/j.tplants.2013.09.004
Liu, Y., Li, Y. X., Li, Y. X., Tian, Z. W., Hu, J. L., Steve, A., et al. (2021). Changes of oxidative metabolism in the roots of wheat (Triticum aestivum l.) seedlings in response to elevated ammonium concentrations. J. Integr. Agr. 20, 1216–1228. doi: 10.1016/S2095-3119(20)63216-6
Liu, Y., Maniero, R. A., Giehl, R. F. H., Melzer, M., Steensma, P., Krouk, G., et al. (2022). PDX1.1-dependent biosynthesis of vitamin B6 protects roots from ammonium-induced oxidative stress. Mol. Plant. 15, 820–839. doi: 10.1016/j.molp.2022.01.012
Liu, Y., Sun, S. Z., Lei, K. Q., Tian, Z. W., Dai, T. B. (2019). Effect of exogenous substances on seed germination and seedling growth under elevated ammonium stress in wheat. J. Triti. Crop. 39, 1477–1485. doi: 10.7606/j.issn.1009-1041.2019.12.11
Liu, Y., von Wirén, N. (2022). Integration of nutrient and water availabilities via auxin into the root developmental program. Curr. Opin. Plant. Biol. 65, 102117. doi: 10.1016/j.pbi.2021.102117
Li, S., Yan, L., Riaz, M., White, P. J., Yi, C., Wang, S. L., et al. (2021). Integrated transcriptome and metabolome analysis reveals the physiological and molecular responses of allotetraploid rapeseed to ammonium toxicity. Environ. Exp. Bot. 189, 104550. doi: 10.1016/j.envexpbot.2021.104550
Luo, Y. L., Pang, D. W., Jin, M., Chen, J., Kong, X., Li, W. Q., et al. (2019). Identification of plant hormones and candidate hub genes regulating flag leaf senescence in wheat response to water deficit stress at the grain-filling stage. Plant Direct. 3, 1–23. doi: 10.1002/pld3.152
Lynch, J., Brown, K. M. (1997). Ethylene and plant responses to nutritional stress. Physiol. Plantarum. 100, 613–619. doi: 10.1111/j.1399-3054.1997.tb03067.x
Mabuchi, K., Maki, H., Itaya, T., Suzuki, T., Nomoto, M., Sakaoka, S., et al. (2018). MYB30 links ROS signaling, root cell elongation,and plant immune responses. PNAS. 115, 4710–4719. doi: 10.1073/pnas.1804233115
Marino, D., Moran, J. F. (2019). Can ammonium stress be positive for plant performance? Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01103
Meier, M., Liu, Y., Lay-Pruitt, K. S., Takahashi, H., von Wirén, N. (2020). Auxin-mediated root branching is determined by the form of available nitrogen. Nat. Plants. 6, 1136–1145. doi: 10.1038/s41477-020-00756-2
Mittler, R. (2002). Oxidative stress, antioxidants and stress tolerance. Trends. Plant Sci. 7, 405–410. doi: 10.1016/S1360-1385(02)02312-9
Mochida, K., Shinozaki, K. (2011). Advances in omics and bioinformatics tools for systems analyses of plant functions. Plant Cell Physiol. 52, 2017–2038. doi: 10.1093/pcp/pcr153
Ortigosa, F., Lobato-Fernández, C., Shikano, H., Ávila, C., Taira, S., Cánovas, F. M., et al. (2021). Ammonium regulates the development of pine roots throughhormonal crosstalk and differential expression of transcription factors in the apex. Plant Cell. Environ. 45, 915–935. doi: 10.1111/pce.14214
Patterson, K., Cakmak, T., Cooper, A., Lager, I., Rasmusson, A. G., Escobar, M. A. (2010). Distinct signalling pathways and transcriptome response signatures differentiate ammonium- and nitrate-supplied plants. Plant Cell. Environ. 33, 1486–1501. doi: 10.1111/j.1365-3040.2010.02158.x
Ramsay, N. A., Glover, B. J. (2005). MYB–bHLH–WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 10, 63–70. doi: 10.1016/j.tplants.2004.12.011
Randhawa, V., Pathania, S. (2020). Advancing from protein interactomes and gene co-expression networks towards multi-omics-based composite networks: approaches for predicting and extracting biological knowledge. Brief. Funct. Genomics 19, 364–376. doi: 10.1093/bfgp/elaa015
Reiser, J., Linka, N., Lemke, L., Jeblick, W., Neuhaus, H. K. (2004). Molecular physiological analysis of the two plastidic ATP/ADP transporters from Arabidopsis. Plant Physiol. 136, 3524–3536. doi: 10.1104/pp.104.049502
Ren, Q. F., Zhou, Y. C., Zhou, X. W. (2020). Combined transcriptome and proteome analysis of masson pine (Pinus massoniana lamb.) seedling root in response to nitrate and ammonium supplementations. Int. J. Mol. Sci. 21, 7548. doi: 10.3390/ijms21207548
Sakaoka, S., Mabuchi, K., Morikami, A., Tsukagoshi, H. (2018). MYB30 regulates root cell elongation under abscisic acid signaling. Commun. Integr. Biol. 11, e1526604. doi: 10.1080/19420889.2018.1526604
Sarasketa, A., González-Moro, M. B., González-Murua, C., Marino, D. (2016). Nitrogen source and external medium pH interaction differentially affects root and shoot metabolism in arabidopsis. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00029
Signora, L., De Smet, I., Foyer, C. H., Zhang, H. M. (2002). ABA plays a central role in mediating the regulatory effects of nitrate on root branching in Arabidopsis. Plant J. 28, 655–662. doi: 10.1046/j.1365-313x.2001.01185.x
Sun, L., Di, D. W., Li, G. J., Kronzucker, H. J., Wu, X. Y., Shi, W. M. (2020). Endogenous ABA alleviates rice ammonium toxicity by reducing ROS and free ammonium via regulation of the SAPK9–bZIP20 pathway. J. Exp. Bot. 71, 4562–4577. doi: 10.1093/jxb/eraa076
Sun, D. D., Fang, X. M., Xiao, C. B., Ma, Z., Huang, X. M., Su, J. R., et al. (2021). Kinase SnRK1.1 regulates nitrate channel SLAH3 engaged in nitrate-dependent alleviation of ammonium toxicity. Plant Physiol. 186, 731–749. doi: 10.1093/plphys/kiab057
Takanashi, K., Shitan, N., Yazaki, K. (2014). The multidrug and toxic compound extrusion (MATE) family in plants. Plant Biotechnol. 31, 417–430. doi: 10.5511/plantbiotechnology.14.0904a
Tan, L., Salih, H., Htet, N. N. W., Azeem, F., Zhan, R. L. (2021). Genomic analysis of WD40 protein family in the mango reveals a TTG1 protein enhances root growth and abiotic tolerance in Arabidopsis. Sci. Rep. 11, 2266. doi: 10.1038/s41598-021-81969-z
Urano, K., Kurihara, Y., Seki, M., Shinozaki, K. (2010). ‘Omics’ analyses of regulatory networks in plant abiotic stress responses. Curr. Opin. Plant Biol. 13, 132–138. doi: 10.1016/j.pbi.2009.12.006
Vega-Mas, I., Pérez-Delgado, C. M., Marino, D., Fuertes-Mendizábal, T., González-Murua, C., Márquez, A. J., et al. (2017). Elevated CO2 induces root defensive mechanisms in tomato plants when dealing with ammonium toxicity. Plant Cell. Physiol. 58, 2112–2125. doi: 10.1093/pcp/pcx146
Vega, A., O’Brien, J. A., Gutiérrez, R. A. (2019). Nitrate and hormonal signaling crosstalk for plant growth and development. Curr. Opin. Plant Biol. 52, 155–163. doi: 10.1016/j.pbi.2019.10.001
Wang, F., Gao, J. W., Liu, Y., Tian, Z. W., Muhammad, A., Zhang, Y. X., et al. (2016). Higher ammonium transamination capacity can alleviate glutamate inhibition on winter wheat (Triticum aestivum l.) root growth under high ammonium stress. PloS One 11, e0160997. doi: 10.1371/journal.pone.0160997
Wani, S. H., Anand, S., Singh, B., Bohra, A., Joshi, R. (2021). WRKY transcription factors and plant defense responses: latest discoveries and future prospects. Plant Cell. Rep. 40, 1071–1085. doi: 10.1007/s00299-021-02691-8
Xiao, C. B., Sun, D. D., Liu, B. B., Fang, X. M., Li, P. C., Jiang, Y., et al. (2022). Nitrate transporter NRT1.1 and anion channel SLAH3 form a functional unit to regulate nitrate-dependent alleviation of ammonium toxicity. J. Integr. Plant Biol. 64, 942–957. doi: 10.1111/jipb.13239
Yang, J. H., Ding, C. Q., Xu, B. C., Chen, C., Narsai, R., Whelan, J., et al. (2015). A casparian strip domain-like gene, CASPL, negatively alters growth and cold tolerance. Sci. Rep. 5, 14299. doi: 10.1038/srep14299
Yang, D. Q., Luo, Y. L., Dong, W. H., Yin, Y. P., Li, Y., Wang, Z. L. (2018). Response of photosystem II performance and antioxidant enzyme activities in stay-green wheat to cytokinin. Photosynthetica 56, 567–577. doi: 10.1007/s11099-017-0708-1
Yang, H. Y., von der Fecht-Bartenbach, J., Friml, J., Lohmann, J. U., Neuhäuser, B., Ludewig, U. (2015). Auxin-modulated root growth inhibition in Arabidopsis thaliana seedlings with ammonium as the sole nitrogen source. Funct. Plant Biol. 42, 239–251. doi: 10.1071/FP14171
Yang, D. Q., Zhao, J. H., Bi, C., Li, L. Y., Wang, Z. L. (2022). Transcriptome and proteomics analysis of wheat seedling roots reveals that increasing NH4+/NO3- ratio induced root lignification and reduced nitrogen utilization. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.797260
Yokotani, N., Ichikawa, T., Koudou, Y., Iwabuchi, M., MFatsui, M., Hirochika, H., et al. (2013). Role of the rice transcription factor JAmyb in abiotic stress response. J. Plant Res. 126, 131–139. doi: 10.1007/s10265-012-0501-y
Yuan, Y. Y., Fang, L. C., Karungo, S. K., Zhang, L. L., Gao, Y. Y., Li, S. H., et al. (2016). Overexpression of VaPAT1, a GRAS transcription factor from Vitis amurensis, confers abiotic stress tolerance in arabidopsis. Plant Cell. Rep. 35, 655–666. doi: 10.1007/s00299-015-1910-x
Keywords: wheat seedlings, transcriptome, weighted gene co-expression network, nitrate, ammonium toxicity
Citation: Li L, Zang X, Liu J, Ren J, Wang Z and Yang D (2022) Integrated physiological and weighted gene co-expression network analysis reveals the hub genes engaged in nitrate-regulated alleviation of ammonium toxicity at the seedling stage in wheat (Triticum aestivum L.). Front. Plant Sci. 13:1012966. doi: 10.3389/fpls.2022.1012966
Received: 06 August 2022; Accepted: 02 November 2022;
Published: 17 November 2022.
Edited by:
Hamada AbdElgawad, Beni-Suef University, EgyptReviewed by:
Kunal Mukhopadhyay, Birla Institute of Technology, IndiaTie Cai, Northwest A&F University, China
Copyright © 2022 Li, Zang, Liu, Ren, Wang and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dongqing Yang, Y2hlbmd5YW5nMjM2NEAxMjYuY29t
†These authors have contributed equally to this work
 Liuyin Li†
Liuyin Li† Zhenlin Wang
Zhenlin Wang Dongqing Yang
Dongqing Yang