
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 05 October 2022
Sec. Plant Breeding
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.1012368
 Manoj Kumar Patel1
Manoj Kumar Patel1 Rajat Chaudhary1
Rajat Chaudhary1 Yashpal Taak1
Yashpal Taak1 Priya Pardeshi1
Priya Pardeshi1 Joghee Nanjundan2
Joghee Nanjundan2 K. K. Vinod1
K. K. Vinod1 Navinder Saini1
Navinder Saini1 Sujata Vasudev1
Sujata Vasudev1 D. K. Yadava1*
D. K. Yadava1*Seed coat colour is an important trait in Indian mustard. Breeding for seed coat colour needs precise knowledge of mode of inheritance and markers linked to it. The present study was focussed on genetics and development of functional markers for seed coat colour. F1s (direct and reciprocal) and F2 populations were developed by crossing two contrasting parents for seed coat colour (DRMRIJ-31, brown seeded and RLC-3, yellow seeded). Phenotypic results have shown that the seed coat colour trait was under the influence of maternal effect and controlled by digenic-duplicate gene action. Further, Bju.TT8 homologs of both parents (DRMRIJ-31 and RLC-3) were cloned and sequenced. Sequencing results of Bju.TT8 homologs revealed that in RLC-3, gene Bju.ATT8 had an insertion of 1279bp in the 7th exon; whereas, gene Bju.BTT8 had an SNP (C→T) in the 7th exon. These two mutations were found to be associated with yellow seed coat colour. Using sequence information, functional markers were developed for both Bju.TT8 homologs, validated on F2 population and were found highly reliable with no recombination between the markers and the phenotype. Further, these markers were subjected to a germplasm assembly of Indian mustard, and their allelic combination for the seed coat colour genes has been elucidated. The comparative genomics of TT8 genes revealed high degree of similarity between and across the Brassica species, and the respective diploid progenitors in tetraploid Brassica species are the possible donors of TT8 homologs. This study will help in the marker-assisted breeding for seed coat colour, and aid in understanding seed coat colour genetics more precisely.
Indian mustard [Brassica juncea (L.) Czern. and Coss., AABB; 2n=36] is one of the major oilseed crops and is globally cultivated in tropical and subtropical regions including the Indian sub-continent. In India, it is a predominant species among the rapeseed-mustard group and accounts for more than 90% of its total acreage (Chand et al., 2021; Yadava et al., 2022). Based on seed coat colour, Indian mustard is broadly categorized into two classes viz., brown and yellow seeded. However, most Indian cultivars come under the brown seeded types (Padmaja et al., 2005). Seed coat colour in B. juncea is an important trait related to oil and seed meal quality. Yellow seeded Brassicas are known to contain more oil, protein and low fibre content than the brown seeded ones (Woods, 1980; Rahman and McVetty, 2011). Seed coat colour also affects the earliness in seed germination (Zhang et al., 2015). There are reports about poor storability of yellow seeded mustard than brown seeded due to thin testa leading to imbibitional damage (Zhang et al., 2006; Swami et al., 2016; Yadav et al., 2020). Therefore, there is an urgent need to improve the storability of Indian mustard which could be met by developing brown seeded cultivars.
The seed coat is developed from the differentiation of the integuments and chalaza tissues of the ovule after fertilization. In crucifers, accumulation of proanthocyanidin (oxidized form of flavonoids) in inner integuments of seed coat gives rise to brown pigmentation (Lepiniec et al., 2006) and mutations in the pathway genes lead to failure of proanthocyanidin production. In B. juncea, the brown seed coat colour is dominant over the yellow type, however, environmental conditions, maternal effect, and epistasis make its expression complex and varying (Mahmood et al., 2005). Further, several previous studies on the inheritance pattern and the number of genes governing the seed coat colour in B. juncea, indicate ambiguous results, suggesting a complex inheritance pattern. However, reports on monogenic (2016; Xu et al., 2010; Huang et al., 2012), digenic (Padmaja et al., 2005; Yan et al., 2009) and multigenic (Mahmood et al., 2005) inheritance are available in the literature. By mapping, QTLs controlling seed coat colour in B. rapa are reported later (Rahman et al., 2014; Zhang et al., 2019). Earlier, seed coat phenotypes of a series of Arabidopsis thaliana mutants, transparent testa (TT), having varying seed colour has been reported by Debeaujon et al. (2000). Among these mutants, Nesi et al. (2000) identified that the mutant gene of TT8 produces an aberrant protein affecting the expression of two other genes, dihydroflavonol 4-reductase (DFR) and Banyuls (BAN), involved in flavonoid metabolism in interaction with genes such as TT2, and TTG1. Subsequently, Baudry et al. (2004) identified that these three synergistically acting genes regulate proanthocyanidin biosynthesis in Arabidopsis. The role of TT8 in determining seed coat colour in B. rapa was later reported (Li et al., 2012; Niu et al., 2020), and also in B. juncea (Padmaja et al., 2014). They reported two homologs of the TT8 gene are to be responsible for seed coat colour development. Recently, Zhai et al. (2020) reported the role of BnTT8 homologs in seed coat colour development in B. napus, through targeted mutagenesis of BnTT8 homologs that leads to production of yellow seed coat colour coupled with increased seed oil and protein content. In addition, Shen et al. (2021) have also suggested the role of TT8 gene governing seed coat colour in different Brassica species through metabolite profiling and transcriptome analysis. However, TT2 gene is also reported to be involved in flavonoid biosynthesis that leads to seed coat colour formation in B. napus (Zhou et al., 2016; Xie et al., 2020).
DNA-based markers are indispensable for marker-assisted breeding as they facilitate the selection of the trait, irrespective of plant growth stages, environments, and even in the absence of phenotypic expression. In B. juncea, Negi et al. (2000) found linked AFLP markers for seed coat colour and were converted into SCAR markers using the bulked segregant analysis (BSA) approach in the F4 generation. Subsequently, using RFLP markers, three QTLs for coat colour variation viz. SC-B04, SC-A10, and SC-A06 were reported in a doubled haploid (DH) population derived from brown-seeded cultivar ‘RLM-514’ and a yellow-seeded breeding line (Mahmood et al., 2005). At the same time, Padmaja et al. (2005), identified two genes, BjSC1 and BjSC2 linked with SSR markers Na10-A08 and Ni4-F11, respectively. Although, Padmaja et al. (2014) has reported two independent markers targeting both the homologs of TT8 genes in B. juncea, but required more operational time that are poorly suitable in marker assisted selection. Recently, Huang et al. (2016) used intron polymorphism (IP) and sequence characterised amplified region (SCAR) markers to map linked genomic regions to seed coat colour using backcross inbred lines, and found that marker IP4 and Y1 were located on either side of yellow seed colour gene. In addition, seed coat colour gene, Brsc1fine-mapped employing whole genome re-sequencing coupled with BSA was reported by Wang et al. (2016). Besides, two co-localized major QTLs, qSC9.1 and qSCb9.1, and one minor QTL, qSC3.1 were also reported for seed coat colour variation in B. rapa (Zhang et al., 2019). Despite these limited reports, functional markers for seed coat colour that work across the genetic backgrounds and are useable in marker-assisted crop improvement in B. juncea have not yet been developed. To address this, we have developed functional markers targeting the Bju.TT8 homologs and attempted to validate the same in a segregating and a germplasm assembly. The same set of germplasm assembly was evaluated using developed functional markers to deduce allelic pattern for Bju.TT8 genes.
For studying the variability for seed coat colour, a set of 127 diverse genotypes were screened and two contrasting varieties viz., DRMRIJ-31 and RLC-3 were selected and crossed. DRMRIJ-31 was a conventional, high yielding, brown seeded cultivar developed from the directorate of Rapeseed and Mustard, Bharatpur, India and RLC-3 was a double zero canola quality, having yellow seeded and originated from Punjab Agricultural University, Ludhiana, Punjab. The F1s (both direct and reciprocal) were developed during rabi 2018-19. Grown during rabi 2019-20, the F1s were selfed to develop F2 seeds. Two rows of each parent and 15 rows of the F2 population were raised rabi 2020-21. Each row was 5m in length, with a row-to-row spacing of 45 cm and plant to plant distance of 15 cm. All the recommended agricultural operations such as thinning, weeding, irrigation, and fertilizer application were done as per recommended package of practices for better crop establishment. The seed coat colour of 526 F2 individual plants (F3 seeds) was scored visually and broadly categorized into two classes i.e., brown and yellow.
The maternal-effect was examined by observing the seed coat colour differences between direct and reciprocal F1 crosses. Inheritance pattern was determined by studying the segregation of F3 seeds for coat colour. The gene action and marker-phenotype association were analysed using the χ2-test (Pearson, 1916). The χ2-calculated value was compared with χ2-tabulated value at a 5% level of significance with (n-1) degrees of freedom. If χ2-calculated value is less than the χ2-tabulated value then it concludes that markers and phenotype are following expected segregation and vice-versa. The χ2-test can be done using the following formula:
where, Oi and Ei represent the observed and expected frequency of each class, respectively.
Approximately 300 mg of fresh tender leaves from each 127 genotypes, including individual F2 plants were used for DNA isolation using the CTAB method (Doyle and Doyle, 1987). DNA was quantified employing nanodrop (NanoDrop 2000, Thermo Scientific, USA) and quality was evaluated using 0.8% of agarose on gel electrophoresis. The DNA was stored at -20°C. For one PCR sample, a reaction mixture of volume 15µl was used in a thermal cycler (MiniAmp Plus, Applied biosystem) using ~50ng DNA template, 1.5µl 10x buffer S, 750 µM dNTP, 1U Taq-polymerase (Vivantis) and DNA primers 10 pM each (forward and reverse) were used. The PCR profile contained 40 cycles with customised profiles for each marker. The amplicons were resolved in 2.5% Agarose gel in a gel electrophoresis system; visualised and pictographed in a gel documentation system (SYNGENE), under ultra-violet transillumination.
Full-length Bju.TT8 genes were amplified in DRMRIJ-31 (brown seeded parent) and RLC-3 (yellow seeded parent) using primers TT8-RsF1 (5’-ggaaagtgatcggggctgagaaag-3’) and TT8-RsR4 (5’-ctcaaaatgattgtcgttcaaggca-3’) for Bju.ATT8 gene, TT8-NsF1 (5’-aggtatggaaagtgatcggagctgagg-3’) and TT8-NsMPR (5’-catgttcttctctaaagttggcatcag-3’) for Bju.BTT8 gene (Padmaja et al., 2014). Amplified products were cloned in pGEM®-T Easy vector (www.promega.com) and sequenced using Sanger method (Applied Biosystems). Using this sequencing information, the PCR-based markers were developed for both the homologs of Bju.TT8 gene using online tool PrimerQuest™ (https://www.idtdna.com/PrimerQuest).
526 F2 plants were used to validate functional markers for seed coat colour by studying segregation pattern with the phenotype. In addition, a germplasm assembly of 130 genotypes, which includes a diverse panel of 127 genotypes of B. juncea, two genotypes of B. rapa (2n=20; AA) i.e., TL-15 (brown seeded), Pusa gold (yellow seeded), and one brown seeded B. nigra (2n=16; BB) genotype (IC-281862), were screened for seed coat colour alleles. Further, these genotypes were categorised into four different allelic combinations of Bju.TT8 genes i.e., AABB, AAbb, aaBB, and aabb among B. juncea genotypes.
Available coding (CDS) and protein sequences of TT8 homologs and orthologs in different Brassica species were downloaded from NCBI Genbank (https://www.ncbi.nlm.nih.gov). TT8 gene sequences of B. juncea cv. Varuna and Heera were taken from Padmaja et al. (2014). In some Brassica species, where the TT8 gene has not been reported, predicted CDS and protein sequences were obtained through NCBI blast (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and an online software Fgenesh+ (Solovyev, 2007), using the TT8 protein sequence of Arabidopsis thaliana (NM_117050.3, Mayer et al., 1999) as reference. The CDS and protein sequences of TT8 genes of different Brassicas i.e., B. rapa (XM_009115326.3; GU255866.1 and HQ337791.1, Wang et al., 2012), B. nigra (cv. YZ12151 and Sangam), B. oleracea (GU255867.1 and GU219990.1, Chiu et al., 2010), B. napus (NM_001315974.2, Zhai et al., 2020; GU255864.1; GU255865.1; cv. Da-Ae), B. juncea (KJ942581.1, Xie et al., 2014; cv. Varuna; DRMRIJ-31; RLC-3; Heera; AU-213; Sichuan Huangzi and T84-66), B. carinata (cv. sxm20200214) and A. thaliana (NM_117050.3, Mayer et al., 1999) were aligned using the online tool, Clustal Omega (Sievers et al., 2011).
Parents DRMRIJ-31 and RLC-3 were used to produce reciprocal F1 crosses. The F1 seeds harvested from DRMRIJ-31 × RLC3 cross were brown in colour, whereas the yellow seeds were obtained from RLC-3 × DRMRIJ-31 cross (Figure 1A). Since the seed coat colour exhibited maternal effect, the F3 seeds harvested from 526 F2 plants segregated into 500 plants producing brown seeds, and 26 plants producing yellow seeds (Figure 1B, Supplementary Figure 1, Table 1A). The observed values well agreed with 15:1 ratio for duplicate dominant gene action, involving two genes. The χ2-test showed that was a non-significant difference at 5% level of significance with a probability of 0.22 (Table 1A).
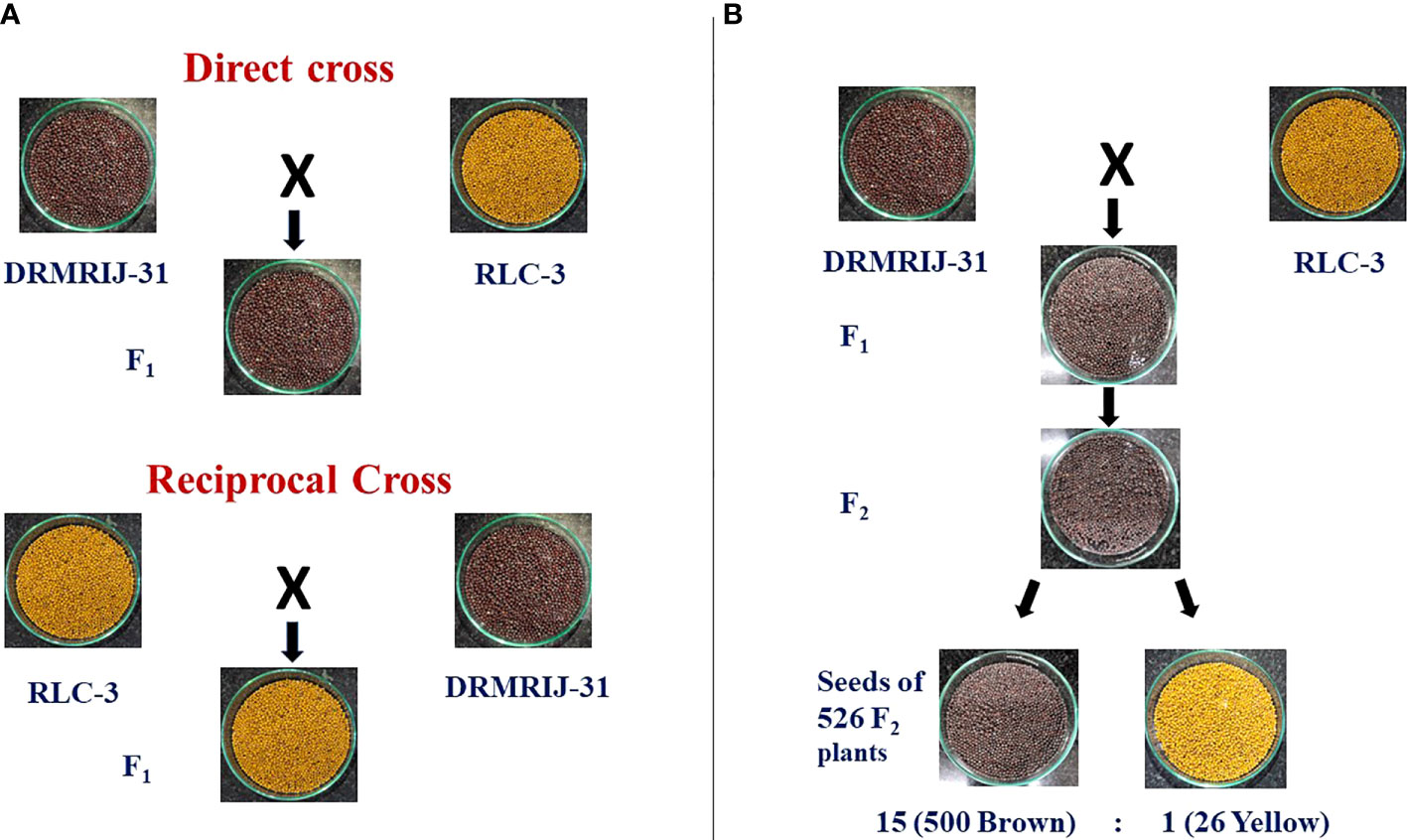
Figure 1 Inheritance pattern of seed coat colour (A) Seed coat colour differences in direct and reciprocal crosses of DRMRIJ-31 and RLC-3 (B) Duplicate dominant type segregation of seed coat colour in seeds of F2 plants.
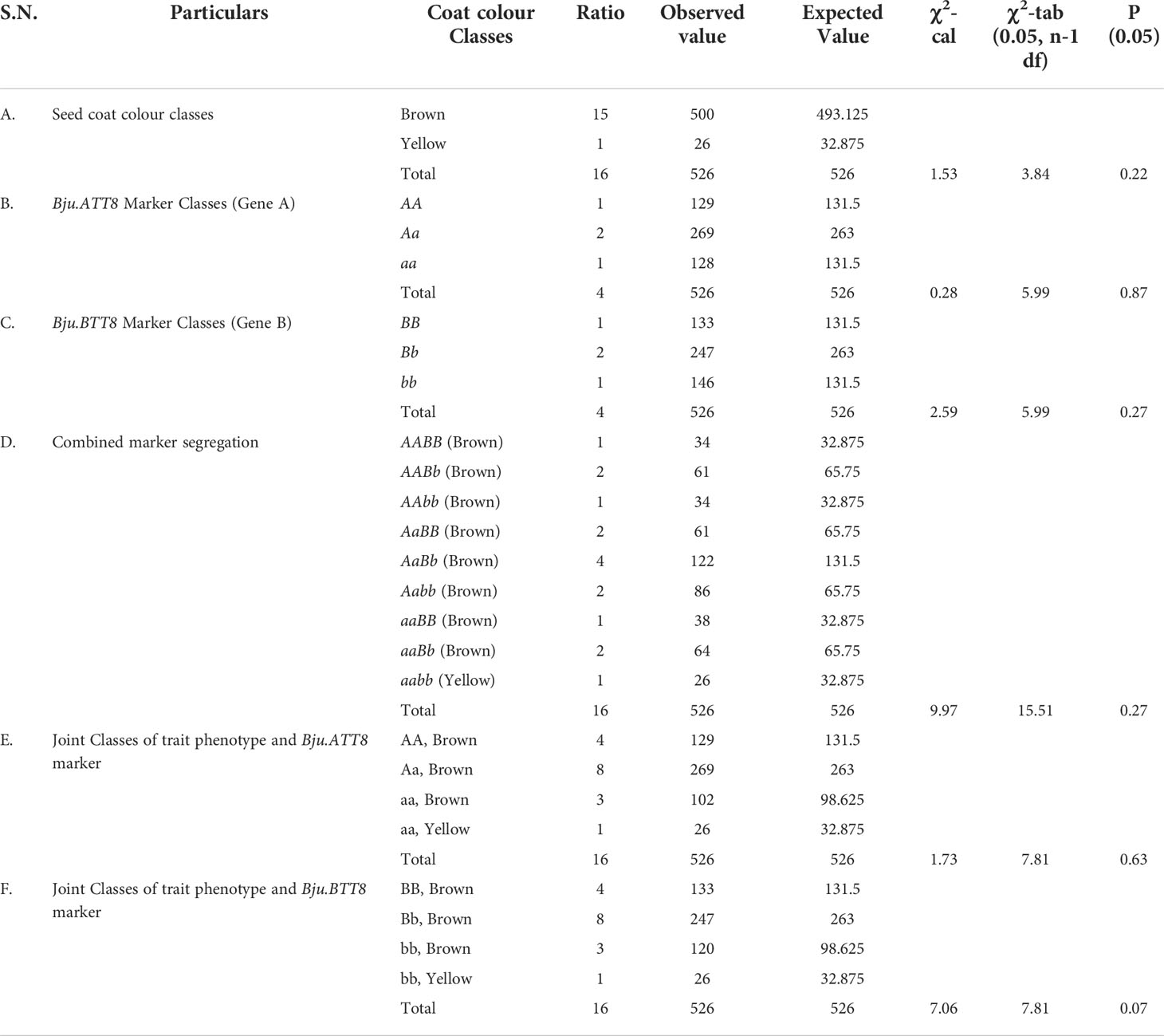
Table 1 Segregation analysis of seed coat colour and functional markers in F2-population using χ2-test.
The Bju.TT8 homologs amplified using respective primers were named as Bju.ATT8 and Bju.BTT8, referring to their location at chromosome A09 and B03, respectively. The in-silico sequence analysis showed that Bju.TT8 genes were consisting of 7 exons and 6 introns. The length of the first five exons of Bju.TT8 homologs were well conserved; however, 6th and 7th exons showed variation in the sequence length (Figure 2). Bju.TT8 homolog sequences when compared between two parents, DRMRIJ-31 and RLC-3, showed that RLC-3 has long insertion of 1279 bp in the 7th exon (from 3047bp to 4325bp) of Bju.ATT8 gene resulting an additional intron of 1297 bp and a single point mutation (C/T) was observed at position 2742bp from start point, in the 7th exon of Bju.BTT8 gene (Figure 2).
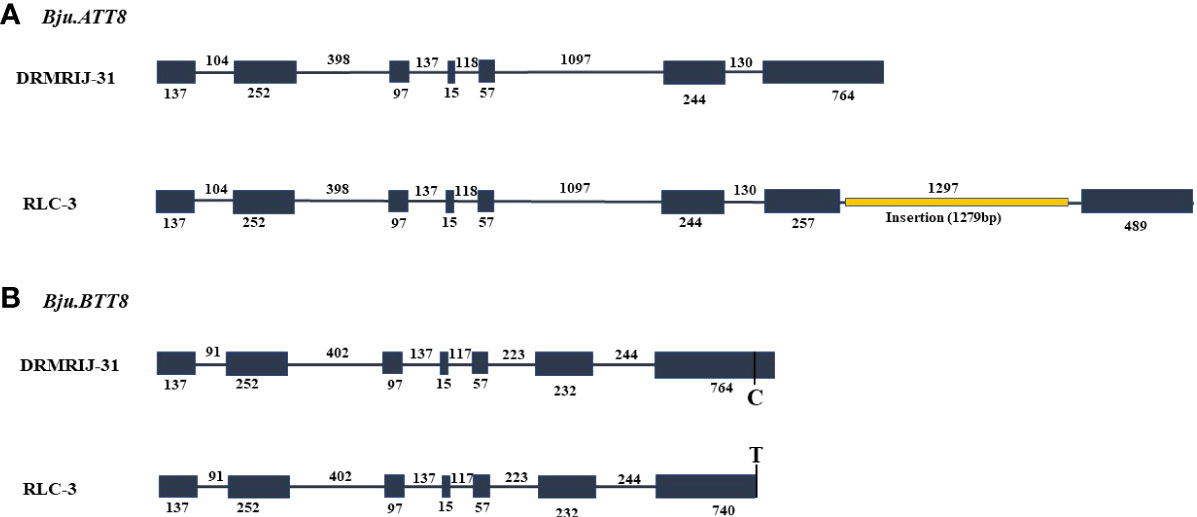
Figure 2 Gene structure of Bju.TT8 homologs, exonic region represented by grey boxes and grey line denoting the intronic region (A) Diagrammatic representation of Gene differences observed in DRMRIJ-31 and RLC-3 for Bju.ATT8 gene, RLC-3 had large insertion of 1279 bp (depicted by orange line) in the seventh exon (B) Gene differences observed in DRMIJ-31 and RLC-3 for Bju.BTT8 gene, RLC-3 had a SNP (C→T) in the seventh exon.
Located on A09 chromosome, Bju.ATT8 gene had an insertion rendering it non-functional in RLC-3. Exploiting this insertion of 1279 bp in the RLC-3, we have developed a functional sequence-tagged sites (STS) marker, that comprised of a forward primer, COLBA-F1, designed outside the insertion site (88bp from the insertion site), and the reverse primer TT8RsR4, from an earlier study (Padmaja et al., 2014) were developed. The amplified product of this primer set was 499bp in DRMRIJ-31 (brown seeded) and 1778 bp in RLC-3 (yellow seeded). However, the larger fragment of 1778 bp in RLC-3 seemed less user friendly due to its large size. Hence, an additional reverse primer, COLYA-R1 was designed using the sequences within the insertion (263bp inside the insertion site) to specifically target the yellow seeded parent. When put together in a multiplex PCR, COLBA-F1 and COLYA-R1 could amplify a 351bp fragment in RLC-3, while a 499 bp fragment was generated in DRMRIJ-31 with the help of the reverse primer, TT8RsR4 (Figures 3A, B; Supplementary Figure 2A). Interestingly, a large-sized amplicon (1778 bp) in RLC-3 generated by TT8RsR4 could be suppressed by changing the PCR conditions i.e., annealing temperature was kept at 62°C for 30s and extended for one minute at 72°C. The primer concentration of COLBA-F1, COLYA-R1, and TT8RsR4 were kept 10, 5 and 5 pM, respectively (Table 2A).
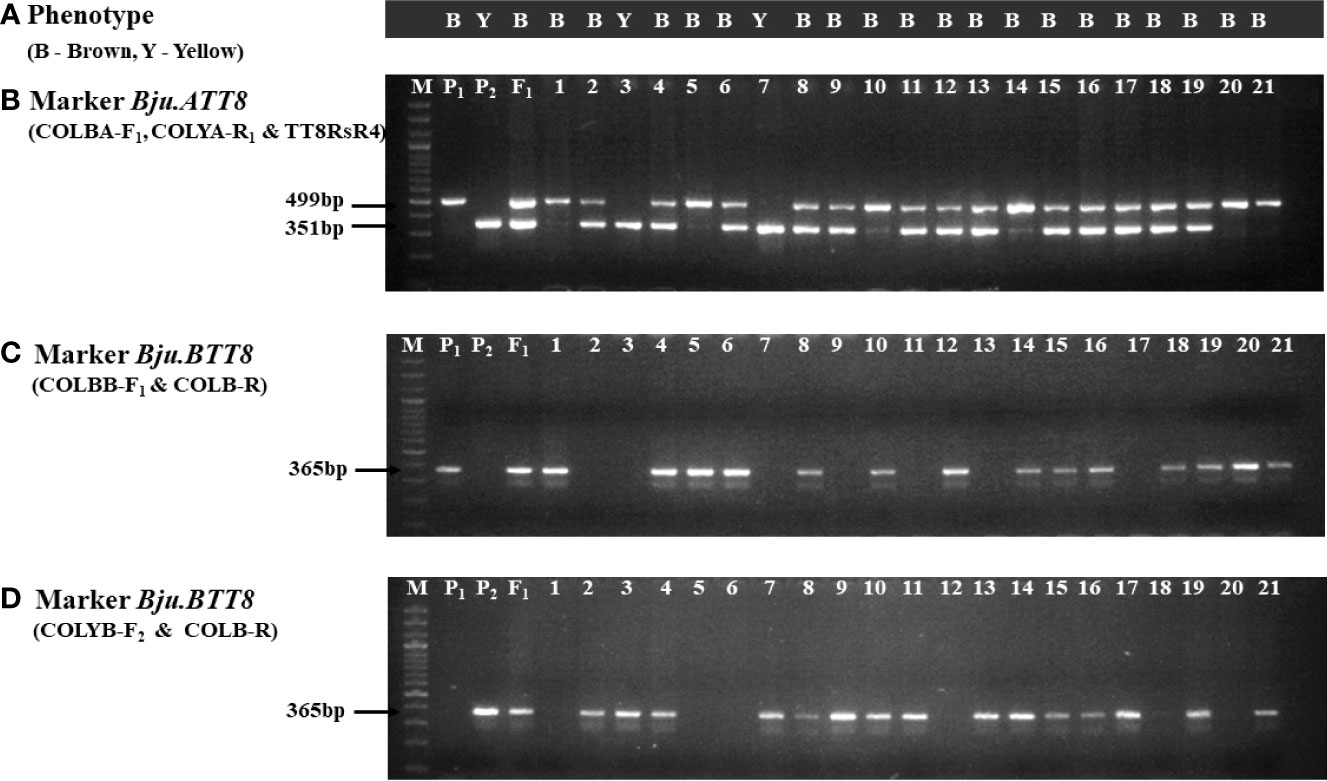
Figure 3 Markers segregation in F2 population (A) Phenotype of individual F2 plants (brown and yellow); (B) Gel image of Bju.ATT8 marker; (C, D) Gel image of Bju.BTT8 markers using two different combinations of forward primers; M-Ladder, P1-DRMRIJ-31, P2-RLC-3, F1-Hybrid, 1-21-F2 plants.
The functional markers developed for Bju.BTT8 gene located on B03 chromosome targeted the SNP (C/T) present in the seventh exon. Two forward (COLBB-F1, COLYB-F2) and one reverse (COLB-R) primers were developed. COLBB-F1 targeted the SNP nucleotide ‘C’ with an additional mismatch of ‘G’ at the third nucleotide position from the SNP at the 3’ end as suggested by Liu et al. (2012). COLYB-F1 was similar to COLBB-F1, but targeted the SNP nucleotide ‘T’. Although both the primers amplified a band size of 365bp, allele specificity of these markers were remarkable, since COLBB-F1 could amplify only the brown allele, while COLYB-F1 amplified the yellow allele. However, two separate PCR reactions having different primer combinations, COLBB-F1+COLB-R and COLYB-F2 + COLB-R were required to differentiate homozygotes and heterozygotes, because of their dominant nature (Figures 3A, C, D, Supplementary Figure 2B). Further, the PCR reaction for COLBB-F1 + COLB-R required annealing for the 20s at 60°C and extension at 72°C for 30s, while COLYB-F2 + COLB-R required annealing at 64°C for 20s and extension at 72°C for 30s (Table 2B). The concentration of forward and reverse primers was kept 10 pM each for PCR reaction.
Functional markers validated over a F2 population of 526 plants, derived from the cross DRMRIJ-31/RLC3, the Bju.ATT8 marker revealed a segregation of 129 AA, 269 Aa and 128 aa plants. ‘A’ represented the brown allele from DRMRIJ-31 and ‘a’ represented the yellow allele from RLC3. The phenotype of the F2 plants perfectly associated to the allelic status. Based on the goodness of fit, the marker segregation followed the expected ratio of 1:2:1 with a probability of 0.87. Similarly, the segregation of the Bju.BTT8 gene has shown 133 BB, 247 Bb and 146 bb plants, having a probability of 0.27 (Supplementary Figures 1B, C, Tables 1B, C).
By combined marker analysis, 526 F2 plants were categorized into 9 different genotypic classes based on the marker allelic pattern as AABB (34), AABb (61), AAbb (34), AaBB (61), AaBb (122), Aabb (86), aaBB (38), aaBb (64) and aabb (26). The segregation of both markers was found to follow the expected genotypic ratio for mendelian dihybrids (1:2:1:2:4:2:1:2:1) with a probability of 0.27 (Table 1D). When F2 individual plants were categorized based on the presence of dominant alleles on either or both the loci, there were 34 plants had all four dominant alleles (AABB) identical to DRMRIJ-31. 122 plants had three dominant alleles (AABb or AaBB), 194 plant with two dominant alleles and 150 plants with one dominant allele. All the plants that carried dominant allele were having brown seed coat. Altogether, 26 plants carried all recessive alleles (aabb) identical to RLC-3 and having yellow seed coat colour (Supplementary Figure 1D).
Since both the genes (Bju.ATT8 and Bju.BTT8) assorted independently and showed duplicate type gene action, a separate goodness of fit test was run for each marker to confirm the significance of their influence on the seed coat colour. Based on the allelic pattern, the segregation of Bju.ATT8 and Bju.BTT8 gene showed perfect agreement with the expected pattern having a χ2-value of 1.73 and 7.06 having a probability of 0.63 and 0.07, respectively (Tables 1E, F).
Out of 130 genotypes used for validation, 96, 10, 9, and 12 B. juncea genotypes were having the genetic constitution of AABB, AAbb, aaBB, and aabb, respectively. Genotypes of the first three genotypic classes (AABB, AAbb, aaBB) had brown seed coat colour, whereas the fourth class (aabb) had yellow seed coat colour (Table 3). Of the remaining, contrary to the expectations, both the yellow and brown seeded genotypes of B. rapa produced wild type (brown) allele (AA; 499bp) with Bju.ATT8 gene. As expected, there was no amplification of Bju.BTT8 gene in B. rapa. Similarly, Bju.ATT8 gene was not amplified in B. nigra genotype IC-281862; but Bju.BTT8 did amplify with its wild type (brown) allele (BB; 365bp) (Table 3).
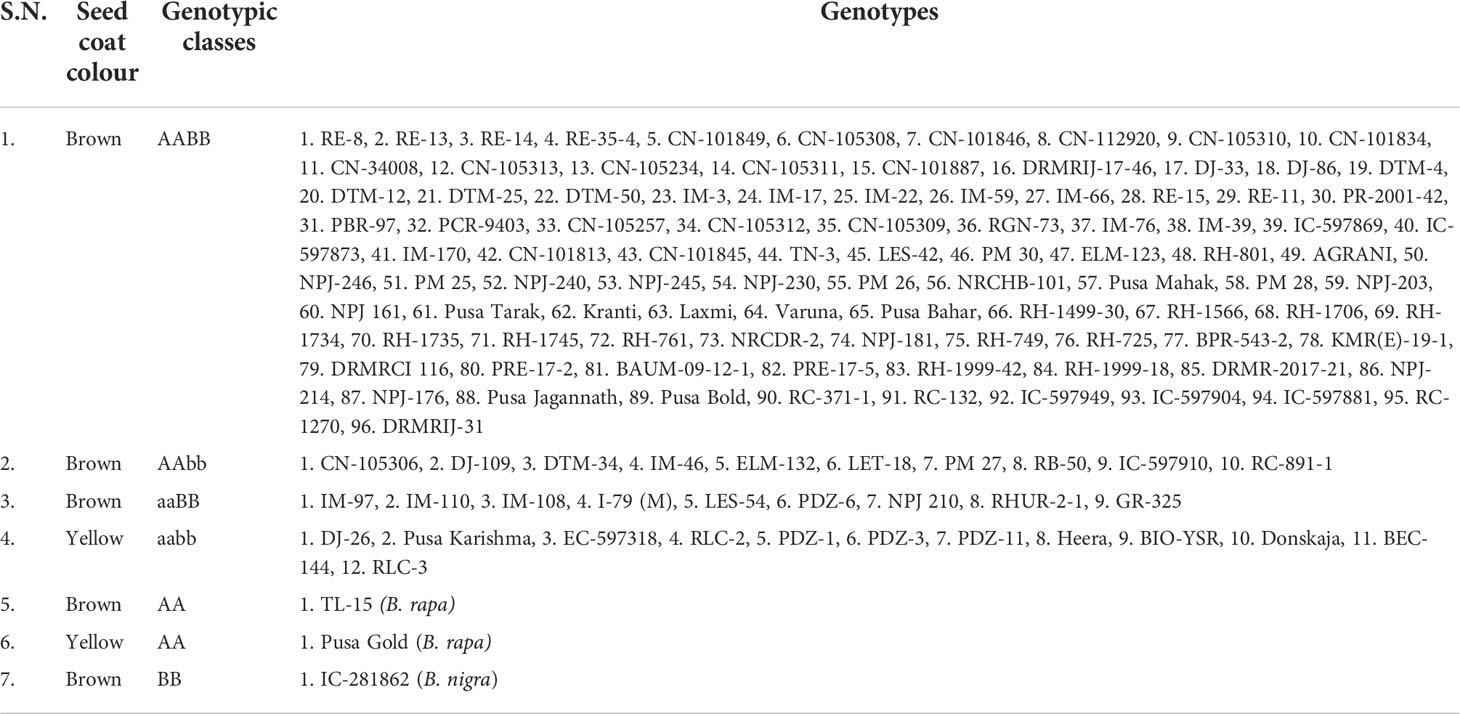
Table 3 Classification of mustard genotypes based on seed coat colour and their genotypic classes using Bju.ATT8 (AA/aa) and Bju.BTT8 (BB/bb) markers.
Multiple sequence alignment of available and predicted TT8 sequences of different Brassica species i.e., B. rapa (XM_009115326.3, HQ337791.1, GU255866.1), B. nigra (cv. YZ12151, Sangam), B. oleracea (GU255867.1, GU219990.1), B. napus (NM_001315974.2, GU255864.1, GU255865.1, cv. Da-Ae), B. juncea (KJ942581.1, cv. Varuna, DRMRIJ-31, RLC-3, Heera, AU-213, Sichuan Huangzi, T84-66), B. carinata (cv. sxm20200214) with A. thaliana (NM_117050.3) showed >83% homology for coding sequences (CDS) and >77% homology for protein sequences across the B. species (Supplementary Tables 1, 2). Comparison of Bju.ATT8 and Bju.BTT8 in DRMRIJ-31 showed 95.2% CDS similarity and 92.3% protein identity. These sequences were 100% similar to that of published sequences (Padmaja et al., 2014; Xie et al., 2014). Further, CDS and protein sequences of Bju.ATT8 showed high similarity (99.5% for CDS and 99.42% for protein) with TT8 gene of B. rapa (cv. Chiffu, Chiifu-401-42), whereas Bju.BTT8 showed high sequence similarity (99.9% for CDS and 99.8% for protein) with TT8 gene of B. nigra (cv. YZ12151, Sangam). In the case of RLC-3, the CDS for both the Bju.TT8 homologs showed 95.2% similarity and 92% protein identity. These sequences were 100% similar to that of Heera, a yellow seeded cultivar (Padmaja et al., 2014). Similarly, the TT8 homologs of other two tetraploid species i.e., B. napus and B. carinata had shown greater homology with their progenitors at CDS as well as protein level (Supplementary Tables 1, 2). In general, the CDS region of TT8 comprised of at least 1530 bp that codes for protein of 509 amino acids. However, number of base pairs and amino acids for TT8 gene varied across the B. species with a range of CDS sequence of 1530bp to 1566bp and protein sequence of 509 amino acid to 521 amino acids (Supplementary Table 3). Interestingly, CDS and amino acid length of TT8 in different cultivars within a diploid species was identical, whereas the length varies among the species. B. rapa (cv. Chiffu, Chiifu-401-42, Tsuda), B. nigra (cv. YZ12151, Sangam), B. oleracea (cv. C10, Stovepipe) have 1566, 1554, 1551 bp CDS and 521, 517, 516 amino acids, respectively. However, in B. juncea differences were observed in CDS and amino acid length of TT8 homologs among different cultivars. The black seeded cultivars (Tumida, DRMRIJ-31, Varuna, T84-66) have 1566 bp CDS and 521 amino acid sequence for Bju.ATT8 gene, while yellow seeded cultivars (RLC-3, Heera, AU-213) have 1548 bp CDS and 515 amino acids. Likewise, for Bju.BTT8 gene, black seeded cultivars have 1554 bp CDS and 517 amino acids, but yellow seeded cultivars have 1548 bp CDS and 515 amino acids. In addition, other tetraploid brassicas i.e., B. napus and B. carinata, have CDS and amino acid length of TT8 homologs identical to their respective diploid genomes (Supplementary Table 3).
Brown seed coat colour in Brassicas is a result of proanthocyanidin accumulation in the inner integuments of seed coat (Lepiniec et al., 2006). Working out the genetics of this trait, several previous studies have suggested inheritance pattern ranging from monogenic to multigenic in species of Brassica triangle. Although a variation in the seed coat colour intensity was also observed where brown seeded plants had more variation than yellow seeded plants, but these seeds were broadly classified into two categories i.e., brown and yellow. Further, a difference in seed coat colour phenotyping of direct and reciprocal F1 crosses was also reported suggesting the trait is influenced by maternal effect. McGugan (1948) while describing the botany of Brassica genus observes that seed coat tissues contain the inner integument (pigment layer) that provides the colour to the seeds. Since the pigment layer of the seed coat is developed from integuments of the ovule, coat colour shows the maternal effect (Lepiniec et al., 2006), and therefore shows segregation only in F2 sporophyte. Also, the colour of F1 seeds is identical to the seeds of the female parent, irrespective of the nature of nuclear genes governing the trait (Padmaja et al., 2005). The first available report of inheritance of seed coat colour in B. juncea, suggests involvement of two duplicate genes with purple/brown being dominant over yellow (Sun, 1945). Several later studies, also confirmed a duplicate dominant type gene interaction, wherein the presence of a single dominant allele of any gene would be sufficient enough to produce brown seed coat colour, whereas individuals having all the recessive alleles will produce yellow seed coat colour (Vera et al., 1979; Vera and Woods, 1982; Padmaja et al., 2005; Yan et al., 2009). However, monogenic inheritance was also reported (Li et al., 2012; Ren et al., 2017) including in B. juncea (2016; Xu et al., 2010; Huang et al., 2012). Under a digenic dominant system with duplicate effect, like that is observed in this study, monogenic inheritance is possible when the brown seeded parent is having single homozygous dominant gene (either AA or BB) at one locus and the other gene in homozygous recessive state. In this study too, we have observed that some of the brown seeded genotypes having AAbb and aaBB genetic constitution for seed coat colour genes, supported the monogenic inheritance. The variation in seed coat colour intensity noticed among the brown seeded types was later attributed to prevailing environmental conditions (Deynze et al., 1993), and/or genetic factors such as minor genes/QTLs (Xu et al., 2010). In support of this, recently, a minor QTL, qSC3.1 with two major QTLs, qSC9.1 and qSCb9.1 controlling the seed coat colour was reported in the RILs and successfully validated in CSSLs of B. rapa (Zhang et al., 2019).
Flavonoid metabolism is an integral process in the seed coat pigmentation in Brassica seeds through proanthocyanidin accumulation in the inner integuments (Shirley, 1998; Lepiniec et al., 2006). Several mutants collectively known as transparent testa (tt) (Koornneef, 1990) identified in the cruciferous model species, Arabidopsis thaliana and there are 11 genes reported (Shirley et al., 1995). Most important among these genes, such as TT2, TT8, TTG1 and TTG2 encoded transcription factors that regulated the flavonoid metabolism in developing siliquae (Coen et al., 2020). Located on different genomic regions across Brassica species, these loci drive failed accumulation of proanthocyanidin (PA), leading to pigment less layer, resulting in yellow seeds. However, different TT loci are found to be involved in the different molecular and biochemical mechanisms (Baudry et al., 2004; Lepiniec et al., 2006; Chai et al., 2009). TT1 gene was found to positively regulate the PA biosynthetic genes (Appelhagen et al., 2011), whereas, the TT8 gene was found to have a role in seed coat colour development (Li et al., 2012; Padmaja et al., 2014; Zhai et al., 2020).
Investigated in this study, using two contrasting B. juncea cultivars, DRMRIJ-31 (brown seeded) and RLC-3 (yellow seeded), the nucleotide sequence of the Bju.TT8 genes revealed two homologs produced similar results as earlier reported by Padmaja et al. (2014), using two other cultivars, Varuna (brown seeded) and Heera (yellow seeded). Their corresponding mutant alleles, Bju.att8 showed an insertion of 1279 bp in the seventh exon while the Bju.btt8 allele showed a C/T substitution on the seventh exon leading to a stop codon, TAA (Padmaja et al., 2014). Both the mutations yielded aberrant non-functional proteins, leading to yellow seeded phenotype. In this study, having found similar alleles, across a set of 127 B. juncea germplasm lines, we conclude that the coat colour variation in Indian mustard is governed by two duplicate genes, Bju.ATT8 and Bju.BTT8, possibly indicating a common source of origin.
Padmaja et al. (2014) have developed primers flanking Bju.ATT8 and Bju.BTT8 to amplify the entire genes and suggested that the polymorphism in these genes could be used in marker assisted crop improvement. Accordingly, targeting the insertion site in the mutant gene (Bju.att8), we have developed an STS marker which amplified a 499 bp fragment with the wild type allele (Bju.ATT8), while it amplified a very long fragment (1.78kb) in the yellow seeded mutant. Although the absence of 499 bp allele can indicate the presence of the mutant allele, we have designed an additional marker to target an intermediate region of the insertion site, that could amplify a specific band of 351 bp in the mutant alone for confirming the presence of the Bju.att8 allele. The amplified products of this primer set (499bp in brown seeded and 351bp in yellow seeded parent) were more user convenient than that of Padmaja et al. (2014) (3550 bp in brown seeded parent and 4708 bp in the yellow seeded parent). This multiplex marker system can be used for selecting the target allele for the Bju.ATT8. In earlier studies also, a similar approach was used for developing markers for BjuBCYP79F1 gene-regulating synthesis of aliphatic glucosinolates in B. juncea (Sharma et al., 2016). However, the use of these marker may be limited to B. juncea, because a yellow seeded genotype of B. rapa (Pusa Gold) failed to produce the desired amplicon of 351 bp, and instead produced the 499 bp allele typical to that of the wild type allele. This indicates that a different mutation is the cause of the yellow allele in B. rapa cv. Pusa Gold. Two different and independent insertional mutations in the TT8 gene were earlier reported in B. rapa (BrTT8; Li et al., 2012) and B. juncea (Bju.ATT8; Padmaja et al., 2014) that cause yellow seed coat. It evidently suggests that insertional event in Bju.ATT8 gene of B. juncea had occurred after its evolution from B. rapa.
Targeting the Bju.BTT8 gene for marker-based selection was a challenge, because a SNP was responsible for the mutation. Moreover, Bju.BTT8 gene was found to be more conserved than the Bju.ATT8 gene. The wild type nucleotide in the SNP was ‘C’ that produced the brown seeded plants, which when changed to T produced the yellow seeded plants. Although SNPs are the most common type of DNA polymorphism (Brookes, 1999), the sequencing techniques targeting a specific SNP in molecular breeding programs is still a cumbersome process. Development of markers targeting functional polymorphism in the form of PCR based markers therefore still remains the choice of the breeders. There are several methods in use to target an SNP in marker development, such as the use of cleaved amplified polymorphic sequence (CAPS), derived CAPS (dCAPS) and allele specific primers. Allele-specific PCR based markers are cost-effective and require less operational time, the use of CAPS and dCAPS markers depends on restriction sites by endonuclease and are expensive (Liu et al., 2012). However, precision of the allele specific markers depends on the PCR conditions. We have developed two separate allele specific primers for Bju.BTT8 gene, one targeting ‘C’ nucleotide and the other targeting ‘T’. A common reverse primer was used. To improve the allele specificity a mismatch was added at the third nucleotide from the SNP at the 3’ end of the primer (Liu et al., 2012). Under the PCR conditions described in this study, both the primers could perfectly differentiate both the alleles producing an amplicon of 365 bp. Two independent PCRs with independent resolution of amplicons in separate electrophoretic gels were required to test the allele specificity and a comparative evaluation of both the gels were required to identify the heterozygotes.
Linked markers for seed coat colour in B. juncea were reported in several studies, such as AFLP markers (Negi et al., 2000), three QTLs viz. SC-B04, SC-A10, and SC-A06 (Mahmood et al., 2005), gene linked markers Na10-A08 and Ni4-F11 (Padmaja et al., 2005), IP and SCAR markers (Huang et al., 2016). However, none of these linked markers were prove to be effective for marker assisted selection. The functional markers (Gupta and Rustgi, 2004) developed in this study targeting Bju.TT8 homologs showed unambiguous Mendelian segregation with no recombination between marker and trait, assuring their high reliability in marker-assisted breeding. Efforts to develop gene-based and functional markers in Brassica species, have been made in the past to select FAE genes governing erucic acid content (Saini et al., 2016, Saini et al., 2019), seed coat colour (Padmaja et al., 2014), erucic acid genes (Gill et al., 2021), and BnFAD2 gene controlling high oleic acid content (Fu et al., 2021).
Validation of the newly developed markers in a panel of 127 genotypes of B. juncea proved their perfect utility in selection, particularly in B. juncea, which could differentiate the population into four classes based on allelic combinations. All these combinations, interestingly, did not deviate from the expected seed coat colour. Saini et al. (2016) have conducted a similar study to categorize different genotypes of B. juncea based on allelic combinations of FAE genes. As explained earlier, one exception we noticed pertains to a B. rapa genotype, Pusa Gold. Recently, it was reported that a major QTL for yellow seed coat, cqSC-A09 located on chromosome A09 (same chromosome as that of Bju.ATT8) increases the oil and fibre content of the seed in B. napus (Chao et al., 2022). If, a similar effect exists with Bju.ATT8, breeding B. juncea carrying this mutant allele can definitely have advantage towards Indian mustard improvement.
Comparison of homolog sequences of the TT8 gene within and between the Brassica genomes indicates high level of conservation, suggesting a descent from a common progenitor during evolution. Similar observations were also made for TT12 gene (related again to seed coat colour) of different Brassica species (Chai et al., 2009). In the present study, consanguineous relationships indicated by the sequence homology among the TT8 genes, suggest that in tetraploid Brassicas the homologs might have come from their respective diploid progenitors before the speciation, as more proximity was observed with the progenitor genes than among the homologs themselves present within the same species. Based on this hypothesis, it is apparent that Bju.TT8 homologs descended directly from B. rapa and B. nigra and evolved subsequently and independently. The similar predictions about progenitors of seed coat colour genes in B. juncea were documented earlier (Negi et al., 2000).
In this study, we confirm that the duplicate dominant genes for seed coat colour in B. juncea, are the homologs of TT8 gene originating from the two diploid progenitor species, B. rapa and B. nigra. Both the genes affect the nucellar pigmentation in the testa, thereby exhibiting maternal effect. They code for transcription factors affecting proanthocyanidin metabolism, and mutation in both the genes results in yellow seeds in place of wild type brown seeds. In Indian mustard, two major mutations exist in both the homologs, both affecting the seventh exon of the TT8 gene resulting in yellow seed coat colour. These genes are the major genes that control seed coat colour inheritance; however, the role of environmental factors and minor genes could not be ignored. In the breeding perspective, there is a need to improve seed storability which is associated with brown seed, while higher oil and protein contents are attributed to yellow seed. Therefore, depending on the breeding objective, it is essential to have functional marker systems to target the desired choice of seed coat colour. We have developed functional markers in this study targeting both wild and mutant alleles, which have been validated in a segregating as well as a germplasm assembly. We conclude that these markers are highly reliable, cost-effective and could be used in marker-assisted breeding. The results from this study also indicate that the marker systems developed herein are for Indian mustard alone, as different genetic systems governing seed coat colour may be operational in other Brassica species.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Conceptualization- NS and DY. Investigation- MP and YT. Methodology- MP. Validation- RC and MP. Formal analysis- MP and KV. Resources- SV and NS. Software- MP and RC. Data curation- PP and JN. Writing- MP. Writing, review and editing- NS, KV, YT, SV and DY. Visualization- NS. Supervision- NS. All authors contributed to the article and approved the submitted version.
The authors are grateful to Director, ICAR-IARI, New Delhi for providing fellowship to MP and CRP-MB, ICAR, New Delhi for providing required facility during research programme.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1012368/full#supplementary-material
Appelhagen, I., Lu, G. H., Huep, G., Schmelzer, E., Weisshaar, B., Sagasser, M. (2011). TRANSPARENT TESTA1 interacts with R2R3-MYB factors and affects early and late steps of flavonoid biosynthesis in the endothelium of Arabidopsis thaliana seeds. Plant J. 67, 406–419. doi: 10.1111/j.1365-313X.2011.04603.x
Baudry, A., Heim, M. A., Dubreucq, B., Caboche, M., Weisshaar, B., Lepiniec, L. (2004). TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 39, 366–380. doi: 10.1111/j.1365-313X.2004.02138.x
Chai, Y. R., Lei, B., Huang, H. L., Li, J. N., Yin, J. M., Tang, Z. L., et al. (2009). TRANSPARENT TESTA 12 genes from Brassica napus and parental species: Cloning, evolution, and differential involvement in yellow seed trait. Mol. Genet. Genomics 281, 109–123. doi: 10.1007/s00438-008-0399-1
Chand, S., Patidar, O. P., Chaudhary, R., Saroj, R., Chandra, K., Meena, V. K., et al. (2021). “Rapeseed-mustard breeding in India: Scenario, achievements and research needs,” in Brassica breeding and biotechnology. (London, UK: IntechOpen). doi: 10.5772/intechopen.96319
Chao, H., Guo, L., Zhao, W., Li, H., Li, M. (2022). A major yellow − seed QTL on chromosome A09 significantly increases the oil content and reduces the fiber content of seed in Brassica napus. Theor. Appl. Genet. 135, 1293–1305. doi: 10.1007/s00122-022-04031-0
Chiu, L. W., Zhou, X., Burke, S., Wu, X., Prior, R. L., Li, L. (2010). The purple cauliflower arises from activation of a MYB transcription factor. Plant Physiol. 154, 1470–1480. doi: 10.1104/pp.110.164160
Coen, O., Lu, J., Xu, W., Pateyron, S., Grain, D., Péchoux, C., et al. (2020). A TRANSPARENT TESTA transcriptional module regulates endothelium polarity. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01801
Debeaujon, I., Léon-Kloosterziel, K. M., Koornneef, M. (2000). Influence of the testa on seed dormancy, germination, and longevity in arabidopsis. Plant Physiol. 122, 403–413. doi: 10.1104/pp.122.2.403
Deynze, A. E., Beversdorf, W. D., Pauls, K. P. (1993). Temperature effects on seed color in black- and yellow-seeded rapeseed. Can. J. Plant Sci. 73, 383–387. doi: 10.4141/cjps93-057
Doyle, J. J., Doyle, J. L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19, 11–15.
Fu, Y., Mason, A. S., Zhang, Y., Yu, H. (2021). Identification and development of KASP markers for novel mutant BnFAD2 alleles associated with elevated oleic acid in Brassica napus. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.715633
Gill, K. S., Kaur, G., Kaur, G., Kaur, J., Kaur, S., Sra, Kaur, K., et al. (2021). Development and validation of kompetitive allele-specific PCR assays for erucic acid content in Indian mustard [Brassica juncea (L.) czern and coss.]. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.738805
Gupta, P. K., Rustgi, S. (2004). Molecular markers from the transcribed/expressed region of the genome in higher plants. Funct. Integr. Genomics 4, 139–162. doi: 10.1007/s10142-004-0107-0
Huang, Z., Ban, Y., Yang, L., Zhang, Y., Li, H., Xiao, E., et al. (2012). Fine mapping of the yellow seed locus in Brassica juncea l. Genome 55, 8–14. doi: 10.1139/g11-072
Huang, Z., Liu, L., Lu, H., Lang, L., Zhao, N., Ding, J., et al. (2016). Development of IP and SCAR markers linked to the yellow seed color gene in Brassica juncea l. Breed. Sci. 66, 175–180. doi: 10.1270/jsbbs.66.175
Koornneef, M. (1990). Mutations affecting the testa color in arabidopsis. Arabidopsis. Inf. Serv. 27, 1–4.
Lepiniec, L., Debeaujon, I., Routaboul, J. M., Baudry, A., Pourcel, L., Nesi, N., et al. (2006). Genetics and biochemistry of seed flavonoids. Annu. Rev. Plant Biol. 57, 405–430. doi: 10.1146/annurev.arplant.57.032905.105252
Li, X., Chen, L., Hong, M., Zhang, Y., Zu, F., Wen, J., et al. (2012). A Large insertion in bHLH transcription factor BrTT8 resulting in yellow seed coat in Brassica rapa. PloS One 7, 1–9. doi: 10.1371/journal.pone.0044145
Liu, J., Huang, S., Sun, M., Liu, S., Liu, Y., Wang, W., et al. (2012). An improved allele-specific PCR primer design method for SNP marker analysis and its application. Plant Methods 8, 1–9. doi: 10.1186/1746-4811-8-34
Mahmood, T., Rahman, M. H., Stringam, G. R., Raney, J. P., Good, A. G. (2005). Molecular markers for seed colour in Brassica juncea. Genome 48, 755–760. doi: 10.1139/G04-122
Mayer, K., Schüller, C., Wambutt, R., Murphy, G., Volckaert, G., Pohl, T., et al. (1999). Sequence and analysis of chromosome 4 of the plant arabidopsis thaliana. Nature 402 (6763), 769–777. doi: 10.1038/47134
McGugan, J. M. (1948). Seeds and seedlings of the genus brassica. Can. J. Res. 26, 520–587. doi: 10.1139/cjr48c-037
Negi, M. S., Devic, M., Delseny, M., Lakshmikumaran, M. (2000). Identification of AFLP fragments linked to seed coat colour in Brassica juncea and conversion to a SCAR marker for rapid selection. Theor. Appl. Genet. 101, 146–152. doi: 10.1007/s001220051463
Nesi, N., Debeaujon, I., Jond, C., Pelletier, G., Caboche, M., Lepiniec, L. (2000). The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in arabidopsis siliques. Plant Cell 12, 1863–1878. doi: 10.1105/tpc.12.10.1863
Niu, Y., Wu, L., Li, Y., Huang, H., Qian, M., Sun, W., et al. (2020). Deciphering the transcriptional regulatory networks that control size, color, and oil content in Brassica rapa seeds. Biotechnol. Biofuels 13, 1–20. doi: 10.1186/s13068-020-01728-6
Padmaja, L. K., Agarwal, P., Gupta, V., Mukhopadhyay, A., Sodhi, Y. S., Pental, D., et al. (2014). Natural mutations in two homoeologous TT8 genes control yellow seed coat trait in allotetraploid Brassica juncea (AABB). Theor. Appl. Genet. 127, 339–347. doi: 10.1007/s00122-013-2222-6
Padmaja, K. L., Arumugam, N., Gupta, V., Mukhopadhyay, A., Sodhi, Y. S., Pental, D., et al. (2005). Mapping and tagging of seed coat colour and the identification of microsatellite markers for marker-assisted manipulation of the trait in Brassica juncea. Theor. Appl. Genet. 111, 8–14. doi: 10.1007/s00122-005-1933-8
Pearson, K. (1916). XLII. on a brief proof of the fundamental formula for testing the goodness of fit of frequency distributions, and on the probable error of “P.” London. Edinburgh. Dublin. Philos. Magazine. J. Sci. 31, 369–378. doi: 10.1080/14786440408635509
Rahman, M., Mamidi, S., Mcclean, P. (2014). Quantitative trait loci mapping of seed colour, hairy leaf, seedling anthocyanin, leaf chlorosis and days to flowering in F2 population of Brassica rapa l. Plant Breed. 133, 381–389. doi: 10.1111/pbr.12165
Rahman, M., McVetty, P. B. E. (2011). A review of brassica seed color. Can. J. Plant Sci. 91, 437–446. doi: 10.4141/cjps10124
Ren, Y., Wu, J., Zhao, J., Hao, L., Zhang, L. (2017). Identification of SSR markers closely linked to the yellow seed coat color gene in heading Chinese cabbage (Brassica rapa l. ssp. pekinensis). Biol. Open 6, 278–282. doi: 10.1242/bio.021592
Saini, N., Koramutla, M. K., Singh, N., Singh, S., Singh, R., Yadav, S., et al. (2019). Promoter polymorphism in FAE1. 1 and FAE1. 2 genes associated with erucic acid content in Brassica juncea. Mol. Breed. 39, 1–9. doi: 10.1007/s11032-019-0971-x
Saini, N., Singh, N., Kumar, A., Vihan, N., Yadav, S., Vasudev, S., et al. (2016). Development and validation of functional CAPS markers for the FAE genes in Brassica juncea and their use in marker-assisted selection. Breed. Sci. 66, 831–837. doi: 10.1270/jsbbs.16132
Sharma, M., Mukhopadhyay, A., Gupta, V., Pental, D., Pradhan, A. K. (2016). BjuB.CYP79F1 regulates synthesis of propyl fraction of aliphatic glucosinolates in oilseed mustard Brassica juncea: Functional validation through genetic and transgenic approaches. PloS One 11, 1–17. doi: 10.1371/journal.pone.0150060
Shen, S., Tang, Y., Zhang, C., Yin, N., Mao, Y., Sun, F., et al. (2021). Metabolite profiling and transcriptome analysis provide insight into seed coat color in Brassica juncea. Int. J. Mol. Sci. 22, 7215. doi: 10.3390/ijms22137215
Shirley, B. W. (1998). Flavonoids in seeds and grains: physiological function, agronomic importance and the genetics of biosynthesis. Seed. Sci. Res. 8, 415–422. doi: 10.1017/S0960258500004372
Shirley, B. W., Kubasek, W. L., Storz, G., Bruggemann, E., Koornneef, M., Ausubel, F. M., et al. (1995). Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J. 8, 659–671. doi: 10.1046/j.1365-313X.1995.08050659.x
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., et al. (2011). Fast , scalable generation of high-quality protein multiple sequence alignments using clustal omega. Molecular Systems Biology 7, 539–539. doi: 10.1038/msb.2011.75
Solovyev, V. V. (2007). “Statistical approaches in eukaryotic gene prediction,” in In handbook of statistical genetics. Eds. Balding D., B. M., Cannings, C. (Hoboken, NJ, USA: Wiley-Interscience) 1616.
Sun, P. C. (1945). Genetic studies on Brassica juncea coss. flower colour, leaf shape, seed colour and branching habit. J. Agr. Assoc. China 50, 12–13.
Swami, S., Yadav, S., Yadav, S. K., Dahuja, A., Yadav, D. K. (2016). Imbibition behaviour and germination response in conventional and quality of Indian mustard (Brassica juncea) seeds. Indian J. Agric. Sci. 86, 1625–1629.
Vera, C. L., Woods, D. L. (1982). Isolation of independent gene pairs at two loci for seed coat color in Brassica juncea. Can. J. Plant Sci. 62, 47–50. doi: 10.4141/cjps82-006
Vera, C. L., Woods, D. L., Downey, R. K. (1979). Inheritance of seed coat color in Brassica juncea. Can. J. Plant Sci. 59, 635–637. doi: 10.4141/cjps79-100
Wang, Y., Xiao, L., Guo, S., An, F., Du, D. (2016). Fine mapping and whole-genome resequencing identify the seed coat color gene in Brassica rapa. PloS One 11, 1–14. doi: 10.1371/journal.pone.0166464
Wang, Y., Zhou, B., Sun, M., Li, Y., Kawabata, S. (2012). UV-A light induces anthocyanin biosynthesis in a manner distinct from synergistic blue+ UV-b light and UV-a/blue light responses in different parts of the hypocotyls in turnip seedlings. Plant Cell Physiol. 53, 1470–1480. doi: 10.1093/pcp/pcs088
Woods, D. L. (1980). The association of yellow seed coat with other characters in mustard B. juncea. Cruciferae. Newslett. 5, 23–24.
Xie, T., Chen, X., Guo, T., Rong, H., Chen, Z., Sun, Q., et al. (2020). Targeted knockout of BnTT2 homologues for yellow-seeded Brassica napus with reduced flavonoids and improved fatty acid composition. J. Agric. Food Chem. 68, 5676–5690. doi: 10.1021/acs.jafc.0c01126
Xie, Q., Hu, Z., Zhang, Y., Tian, S., Wang, Z., Zhao, Z., et al. (2014). Accumulation and molecular regulation of anthocyanin in purple tumorous stem mustard (Brassica juncea var. tumida tsen et Lee). J. Agric. Food Chem. 62, 7813–7821. doi: 10.1021/jf501790a
Xu, A., Huang, Z., Ma, C., Xiao, E., Tian, G., Zhang, X., et al. (2010). Inheritance of seed color and molecular markers linked to the seed color gene in Brassica juncea. Mol. Breed. 25, 57–65. doi: 10.1007/s11032-009-9308-5
Yadava, D. K., Yadav, R., Vishwakarma, H., Yadav, S., Saini, N., Vasudev, S., et al. (2022). “Genetic diversity characterization and population structure in brassica juncea,” in The brassica juncea genome, compendium of plant genomes. Eds. Mohapatra, T., Kole, C. (Switzerland: Springer) 73–84. doi: 10.1007/978-3-030-91507-0_5
Yadav, S., Yadav, S. K., Swami, S., Saini, N., Hussain, Z., Vasudev, S., et al. (2020). Seed quality differences in diverse seed colored Indian mustard (Brassica juncea) genotypes. Indian J. Agric. Sci. 90, 401–406.
Yan, M., Liu, Z., Guan, C., Chen, S., Yuan, M., Liu, X. (2009). Inheritance and molecular markers for the seed coat color in Brassica juncea. Front. Agric. China 3, 1–6. doi: 10.1007/s11703-009-0019-9
Zhai, Y., Yu, K., Cai, S., Hu, L., Amoo, O., Xu, L., et al. (2020). Targeted mutagenesis of BnTT8 homologs controls yellow seed coat development for effective oil production in Brassica napus l. Plant Biotechnol. J. 18, 1153–1168. doi: 10.1111/pbi.13281
Zhang, J., Hu, L., Redden, B., Yan, G. (2015). Identification of fast and slow germination accessions of Brassica napus l. for genetic studies and breeding for early vigour. Crop Pasture Sci. 66, 481–491. doi: 10.1071/CP14269
Zhang, Y., Sun, Y., Sun, J., Feng, H., Wang, Y. (2019). Identification and validation of major and minor QTLs controlling seed coat coloin Brassica rapa l. Breed. Sci. 69, 47–54. doi: 10.1270/jsbbs.18108
Zhang, X. K., Yang, G. T., Chen, L., Yin, J. M., Tang, Z. L., Li, J. N. (2006). Physiological differences between yellow-seeded and black-seeded rapeseed (Brassica napus l.) with different testa characteristics during artificial ageing. Seed. Sci. Technol. 34, 373–381. doi: 10.15258/sst.2006.34.2.13
Keywords: Bju.ATT8, Bju.BTT8, duplicate gene action, functional markers, Indian mustard, maternal-effect, seed coat colour
Citation: Patel MK, Chaudhary R, Taak Y, Pardeshi P, Nanjundan J, Vinod KK, Saini N, Vasudev S and Yadava DK (2022) Seed coat colour of Indian mustard [Brassica juncea (L.) Czern. and Coss.] is associated with Bju.TT8 homologs identifiable by targeted functional markers. Front. Plant Sci. 13:1012368. doi: 10.3389/fpls.2022.1012368
Received: 05 August 2022; Accepted: 14 September 2022;
Published: 05 October 2022.
Edited by:
Leonardo Velasco, Institute for Sustainable Agriculture, (CSIC), SpainReviewed by:
Dandan Hu, Henan Agricultural University, ChinaCopyright © 2022 Patel, Chaudhary, Taak, Pardeshi, Nanjundan, Vinod, Saini, Vasudev and Yadava. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: D. K. Yadava, ZGt5Z2VuZXRAZ21haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.