
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 29 September 2022
Sec. Plant Bioinformatics
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.1012231
This article is part of the Research Topic Omics-Driven Crop Improvement for Stress Tolerance View all 21 articles
 Longsheng Chen1,2
Longsheng Chen1,2 Shuwen Xu2
Shuwen Xu2 Yujun Liu2
Yujun Liu2 Yanhong Zu2
Yanhong Zu2 Fuyuan Zhang2
Fuyuan Zhang2 Liji Du2
Liji Du2 Jun Chen2
Jun Chen2 Lei Li3
Lei Li3 Kai Wang2
Kai Wang2 Yating Wang2
Yating Wang2 Shijin Chen2
Shijin Chen2 Ziping Chen2*
Ziping Chen2* Xianfeng Du1*
Xianfeng Du1*Plant polysaccharides, a type of important bioactive compound, are involved in multiple plant defense mechanisms, and in particular polysaccharide-alleviated abiotic stress has been well studied. Polygonatum cyrtonema Hua (P. cyrtonema Hua) is a medicinal and edible perennial plant that is used in traditional Chinese medicine and is rich in polysaccharides. Previous studies suggested that sucrose might act as a precursor for polysaccharide biosynthesis. However, the role of sucrose metabolism and transport in mediating polysaccharide biosynthesis remains largely unknown in P. cyrtonema Hua. In this study, we investigated the contents of polysaccharides, sucrose, glucose, and fructose in the rhizome, stem, leaf, and flower tissues of P. cyrtonema Hua, and systemically identified the genes associated with the sucrose metabolism and transport and polysaccharide biosynthesis pathways. Our results showed that polysaccharides were mainly accumulated in rhizomes, leaves, and flowers. Besides, there was a positive correlation between sucrose and polysaccharide content, and a negative correlation between glucose and polysaccharide content in rhizome, stem, leaf, and flower tissues. Then, the transcriptomic analyses of different tissues were performed, and differentially expressed genes related to sucrose metabolism and transport, polysaccharide biosynthesis, and transcription factors were identified. The analyses of the gene expression patterns provided novel regulatory networks for the molecular basis of high accumulation of polysaccharides, especially in the rhizome tissue. Furthermore, our findings explored that polysaccharide accumulation was highly correlated with the expression levels of SUS, INV, SWEET, and PLST, which are mediated by bHLH, bZIP, ERF, ARF, C2H2, and other genes in different tissues of P. cyrtonema Hua. Herein, this study contributes to a comprehensive understanding of the transcriptional regulation of polysaccharide accumulation and provides information regarding valuable genes involved in the tolerance to abiotic stresses in P. cyrtonema Hua.
Polygonati rhizoma, commonly known as Huangjing, is a vital traditional and classic Chinese herb and is widely distributed in the world (Luo et al., 2016). There are various species, including Polygonatum cyrtonema Hua, Polygonatum kingianum Collett and Hemsley, and Polygonatum sibiricum Red et al in China (Chinese Pharmacopoeia Commission, 2020). Among these, Polygonatum cyrtonema Hua (P. cyrtonema Hua) is a key constituent of herbal medicines in Anhui Province. Indeed, it usually grows on forested mountains, bushes, and shade hillsides at an altitude of 500–1,200 m above sea level (Figures 1A,B). P. cyrtonema Hua often blooms from late April to early June, and the perianth shows the yellow-green color (Figures 1B,C). Its fruit ripens around the middle of August. Moreover, the rhizomes are harvested in late October and are used to manufacture the Huangjing slice, nine cycles of steam-sun drying processing products, functional beverages, fruit wine, and well-received sweetmeats (Figure 1C; Chinese Pharmacopoeia Commission, 2020; Li et al., 2021; Wu et al., 2022). They have a high abundance of health benefit-promoting metabolites and are among the widely consumed medicines and functional foods (Cheng et al., 2021; Li et al., 2021; Hu et al., 2022; Zhu et al., 2022).

Figure 1. Growth environment, phenotype, and processing of Polygonatum cyrtonema (P. cyrtonema) Hua. (A) P. cyrtonema Hua is often grown on the mountainside by underwood planting. (B,C) The phenotype of the whole and different parts of P. cyrtonema Hua. Bar = 5 cm. To date, steaming and drying are the most used methods in the processing of rhizome of P. cyrtonema Hua. Polysaccharide (PCP) is one of the most crucial compounds of processed product and contributes to flavor and has health benefits.
Polysaccharides of P. cyrtonema Hua are important bioactive compounds (Li et al., 2021). Numerous pharmaceutical results revealed that polysaccharides of rhizomes have a large set of functions, such as anti-oxidation, anti-herpes, antibacterial, anti-cancer, lowering of blood sugar levels, and lowering of blood lipids, as well as the lung-protective and anti-viral effects (Liu et al., 2004; Li et al., 2018a,b, 2020, 2021; Xie et al., 2020; Gan et al., 2022). For example, polysaccharides that are sequentially extracted from P. cyrtonema Hua exhibited anti-cancerous function against cervical cancer Hela cells via regulating the cell cycle and the genes related to different apoptosis pathways which are involved in Hela cell cycle arrest and apoptosis (Li et al., 2020). Previous studies also indicated that polysaccharides can act as compatible solutes and enhance abiotic stress tolerance, such as drought and salt stress in plants (Yu et al., 2017, 2021). In recent years, intermediate metabolites and genes related to polysaccharide biosynthesis have been explored in various plants, including Arabidopsis, wheat, rice, Cyclocarya paliurus, Cynomorium songaricum, Dendrobium officinale, and Polygonatum odoratum (Northcote and Pickett-Heaps, 1966; Meng et al., 2009; Oh et al., 2013; Zhang et al., 2016; He et al., 2017; Zhang et al., 2020; Lin et al., 2021; Wang et al., 2021a). Studies have discovered that polysaccharides are biosynthesized via continuous enzymatic reactions from sucrose, requiring invertase (INV), hexokinase (HK), fructokinase (scrK), mannose-6-phosphate isomerase (MPI), phosphomannomutase (PMM), mannose-1-phosphate guanylyltransferase (GMPP), GDP-mannose 4,6-dehydratase (GMDS), GDP-L-fucose synthase (TSTA3), UDP-glucose 4-epimerase (GALE), UDP-glucose 6-dehydrogenase (UGDH), UDP-apiose/xylose synthase (AXS), UDP-arabinose 4-epimerase (UXE), UDP-glucose 4,6-dehydratase (RHM), 3,5-epimerase/4-reductase (UER1), and glycosyltransferase (GT) (Figure 2; Zhang et al., 2020). First, sucrose is converted to glucose (Glc) and fructose (Fru) by INV. Then, HK and scrK convert Glc and Fru to glucose-6-phosphate (Glc-6P) and fructose-6-phosphate (Fru-6P), respectively. Later, they are used to synthesize UDP-glucose (UDP-Glc) and guanosine di phosphomannose (GDP-Man). A series of glycosyltransferase (GT) reactions result in the production of polysaccharides.
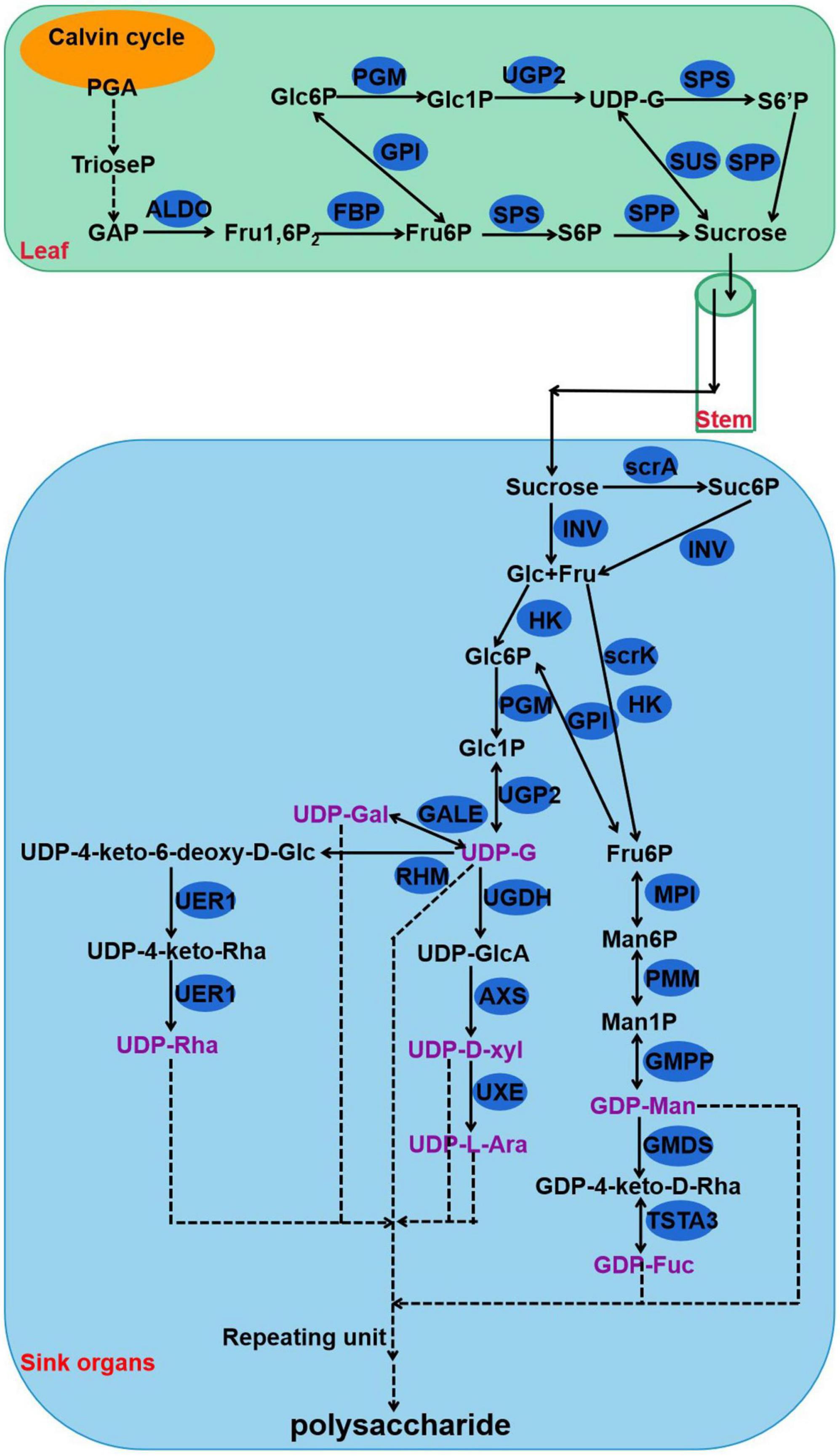
Figure 2. The proposed pathway for the biosynthesis of polysaccharides in Polygonatum cyrtonema. 3-phosphoglycerate, PGA; Triose phosphate, TrioseP; Glyceraldehyde-3-phosphate, GAP; fructose-1,6-diphosphatase, Fru1,6P; Fructose 6-phosophate, Fru-6P; Sucrose 6-phosophate, S6P; Glucose, Glc; Fructose, Fru; Glucose 6-phosophate, Glc-6P; Glucose 1-phosophate, Glc1P; Sucrose 6′-phosophate, S6′P; Uridine diphosphate glucose, UDP-G; Mannose-6-phosphate, Man6P; Mannose-1-phosphate, Man1P; Guanosine diphosphate mannose, GDP-Man; UDP-4keto-D-rhamnose, GDP-4keto-D-Rha; Guanosine diphosphate fucose, GDP-Fuc; UDP-glucuronic acid, UDP-GlcA; UDP-D-xylose, UDP-D-xyl; UDP-L-arabinose, UDP-L-Ara; UDP-galactose, UDP-Gal; UDP-4-keto-6-deoxy-D-Glucose, UDP-4-keto-6-deoxy-D-Glc; UDP-4-keto-rhamnose, UDP-4-keto-Rha; UDP-rhamnose, UDP-Rha; fructose-bisphosphate aldolase, ALDO; fructose-1,6-bisphosphatase, FBP; glucose-6-phosphate isomerase, GPI; phosphoglucomutase, PGM; UTP-glucose-1-phosphate uridylyltransferase, UGP2; sucrose synthase, SUS; sucrose PTS system EIIBCA or EIIBC component, scrA; sucrose-phosphate synthase, SPS; sucrose-6-phosphatase, SPP; invertase, INV; sucrose synthase, SUS; hexokinase, HK; fructokinase, scrK; mannose-6-phosphate isomerase, MPI; phosphomannomutase, PMM; mannose-1-phosphate guanylyltransferase, GMPP; GDPmannose 4,6-dehydratase, GMDS; GDP-L-fucose synthase, TSTA3; UDP-glucose 4-epimerase, GALE; UDP-glucose 6-dehydrogenase, UGDH; UDP-apiose/xylose synthase, AXS; UDP-arabinose 4-epimerase, UXE; UDP-glucose 4,6-dehydratase, RHM; 3,5-epimerase/4-reductase, UER1.
Recently, some molecular studies on the genes related to polysaccharide biosynthesis of P. cyrtonema Hua have been published (Wang et al., 2019; Li et al., 2022). For example, Wang et al. (2019) speculated the multiple genes encoding important enzymes, such as UDP glycosyltransferases (UGTs), and the transcription factors through comprehensive analyses of the transcriptome in leaf, root, and rhizome tissues. Besides, a positively correlated link between polysaccharide accumulation and the MPI, TSTA3, AXS, and UGDH transcript levels was also suggested via the transcriptome sequencing for rhizomes at different ages (Li et al., 2022).
Plant leaves capture light energy and convert this to carbohydrates, such as 3-phosphoglycerate (PGA) and glyceraldehyde-3-phosphate (GAP), during the process of photosynthesis (Osorio et al., 2014). Then, sucrose is synthesized by sucrose synthase (SUS), sucrose-phosphate synthase (SPS), and sucrose-6-phosphatase (SPP) in the chloroplast during the daytime (Figure 2; Osorio et al., 2014). Other enzymes, such as fructose-bisphosphate aldolase (ALDO), fructose-1,6-bisphosphatase (FBP), glucose-6-phosphate isomerase (GPI), phosphoglucomutase (PGM), and UTP-glucose-1-phosphate uridylyltransferase (UGP2), are also involved in the pathway of sucrose synthesis (Figure 2). Plants have evolved a complex system to balance the distribution and allocation of carbon (C) (Raines and Paul, 2006; Kircher and Schopfer, 2012). For example, sucrose metabolism is vital for both the allocation of crucial carbon resources and the initiation of hexose-based sugar signals (Koch, 2004). As a pivotal energy substance, sucrose is transported through the vasculature, type, and sink tissues by SUTs/SUCs (sucrose transporters or sucrose carriers) and/or SWEETs (Sugars Will Eventually be Exported Transporters) and serves as a metabolite for plant growth and development (Feng et al., 2019; Mathan et al., 2021a). These sugar metabolism and transport pathways are mainly controlled by transcription factors, such as bZIP, WRKY, bHLH, AP2/ERF, NAC, and MYB (Sun et al., 2003; Zhu et al., 2011; Phukan et al., 2018; Yang et al., 2021; Wang et al., 2021b; Zhang et al., 2022). Besides, this transporter-regulated sucrose accumulation played a specific role in plant abiotic stress tolerance and adaptation (Chandran, 2015; Eggert et al., 2016). Interestingly, in the rhizome of P. cyrtonema Hua, sucrose might act as a precursor for polysaccharide biosynthesis (Wang et al., 2019; Li et al., 2022). However, whether or not, or how polysaccharide accumulation in rhizomes is modulated by sucrose synthesis and transported in leaf and/or stem remains unknown in P. cyrtonema Hua.
In this study, we investigated the polysaccharides, sucrose, glucose, and fructose content in different parts (rhizome, stem, leaf, and flower) of P. cyrtonema Hua. By the whole-transcriptome analysis, we performed a systematic analysis of polysaccharide biosynthetic genes and estimated the transcript levels of these genes in rhizome, stem, leaf, and flower tissues. Furthermore, several sugar transporters, such as SWEETs, ERDs, and SUTs, were involved in sucrose transport from leaves to stem, rhizome, and flower tissues, thereby coordinately the regulation of the polysaccharide biosynthesis, especially in the rhizome. Finally, we filtrated some key transcription factors, such as bZIP, bHLH, and ERF, which might regulate the above genes related to sucrose metabolism and transport. Briefly, our findings would provide the potential key regulatory points of polysaccharide accumulation in P. cyrtonema Hua.
Plants of P. cyrtonema Hua were collected on May 20 from the P. cyrtonema Hua germplasm conservation and breeding base at Jinzhai County in Anhui Province, China. They were identified by Associate Professor Yujun Liu (Anhui Promotion Center for Technology Achievements Transfer, Anhui Academy of Science and Technology). Plants were cleaned and dried on filter paper, and then imaged using a Canon IXUS 130 camera. Meanwhile, rhizomes, leaves, stems, and flower parts were sampled, and immediately frozen in liquid nitrogen and stored at −80°C.
Rhizomes, leaves, stems, and flower parts of P. cyrtonema Hua were oven-dried at 70°C to constant weight. Then, the dried samples were ground into powder using the grinding mill and kept in a glassware dryer, and further used for the determination of polysaccharide, glucose, fructose, and sucrose content.
Total polysaccharides were extracted and detected from different dried tissues of P. cyrtonema Hua by the anthrone-sulfuric acid method in the Chinese Pharmacopoeia Commission (2020). Briefly, 0.2 g of samples was weighed into a round-bottom flask with 120 mL of 80% ethanol, and boiled for 1 h to remove the small organic compounds and pigments. Afterward, 120 mL of distilled water was added and extracted at 80°C for another 1 h. The supernatant was transferred and diluted in a 250 mL volumetric flask. Polysaccharide content was determined spectrophotometrically by measuring the absorbance at 582 nm with 1 mL of extract, 1 mL of distilled water, and 8 mL of 0.2% anthrone-sulfuric acid solution. The concentration was calculated according to the standard curve of glucose.
Sucrose in different tissues of P. cyrtonema Hua was extracted and determined as previously described (Feng et al., 2019). Samples were homogenized with 1 mL of extracting solution, heated to 80°C in a water bath for 10 min, and then centrifuged (10,000 × g; 25°C) for 15 min. About 5 mg of activated carbon was added to the supernatant for decoloring at 80°C for 30 min. After the solution was cooled to room temperature, 60 μL of 0.1 M NaOH solution was added to 100 μL of the supernatant in a 100°C water bath for 5 min. Then, 700 μL of 10 M HCl and 200 μL of 0.1% resorcinol were added to a 100°C water bath for 10 min. After cooling, the sucrose content was determined and calculated by measuring the absorbance at 480 nm.
Glucose in different tissues of P. cyrtonema Hua was extracted and determined using the Glucose Assay Kit (Solarbio Life Sciences, Beijing, China) according to the manufacturer’s instructions. Samples were homogenized with 1 mL of H2O and heated to 80°C in a boiling water bath for 10 min, and then centrifuged (10,000 × g; 25°C) for another 10 min. Glucose concentration was determined by a glucose oxidase enzyme system and calculated by measuring the absorbance at 505 nm.
Fructose in different tissues of P. cyrtonema Hua was extracted and determined using the Fructose Assay Kit (Solarbio Life Sciences, Beijing, China) according to the manufacturer’s instructions. Samples were homogenized with 1 mL of extracting solution, heated to 80°C in a water bath for 10 min, and then centrifuged (10,000 × g; 25°C) for 10 min. About 5 mg of activated carbon was added to the supernatant for decoloring at 80°C for 30 min. After the solution cooled to room temperature, 1.4 mL of 10 M HCl and 400 μL of 0.1% resorcinol were added to 200 μL of supernatant in a 100°C water bath for 10 min. After cooling, the fructose content was determined and calculated by measuring the absorbance at 480 nm.
Total RNA of different tissues (rhizome, stem, leaf, and flower) of P. cyrtonema Hua was extracted using the Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. RNA concentration and quality were checked using the NanoDrop 2000 (Thermo Fisher Scientific, Inc., Waltham, MA, USA). Sequencing libraries were generated using the TruSeq RNA Sample Preparation Kit (Illumina Inc., San Diego, CA, USA) by using three biological replicates according to the manufacturer’s instructions. Then, the libraries were sequenced using the next-generation sequencing (NGS) and Illumina HiSeq. Enrichment of mRNA, fragment interruption, addition of adapters, size selection, CR amplification, and RNA-Seq were performed by staff at Personal Biotechnology Co., Ltd., Shanghai, China.
After the data filtration, the unigene set of sequences was constructed by the clean reads with high-quality samples using Trinity v2.10.0. The fragments per kilobase million (FPKM) of transcript sequence presented the expression level of each gene in each library. To determine the transcriptional changes among different parts of P. cyrtonema Hua, differentially expressed genes (DEGs) were identified by comparing the expression levels in rhizome vs. stem, rhizome vs. leaf, and rhizome vs. flower, respectively. Transcripts with | log2FoldChange| > 1 and P-value < 0.05 were considered as the thresholds for significant differences, which was performed by DEGseq 2.
All unigenes were annotated against the Kyoto Encyclopedia of Genes and Genome (KEGG) and Gene ontology (GO). KEGG annotation1 was performed to identify the metabolic pathways of genes. GO classifications2 were obtained according to molecular function, biological process, and cellular component. Over-represented KEGG and GO terms were detected via Fisher’s exact test, and multi-test adjustment was made using the Yekutieli (FDR under dependency) method with a cutoff of FDR < 0.05.
Statistical analysis was performed using SPSS 18.0 software. Values were mean values and SD of three independent experiments with three replicates for each. For statistical analysis, data were analyzed by one-way analysis of variance (ANOVA) followed by Duncan’s multiple range test. P-values < 0.05 were considered statistically significant. The correlation coefficient between polysaccharide, sucrose, glucose, and fructose content and gene expression levels was analyzed via Pearson’s correlation coefficient and Student’s t-test (*P < 0.05, **P < 0.01, ***P < 0.001).
Since sucrose is the major transportable form of photosynthate from source leaves to sink organs, we tested the polysaccharides, sucrose, glucose, and fructose contents in rhizome, stem, leaf, and flower tissues to unravel the mechanism underlying the polysaccharide biosynthesis in P. cyrtonema Hua (Figures 1, 3). Among them, the polysaccharide content of rhizome was the highest (93.87 mg⋅g–1DW), followed by flower (59.03 mg⋅g–1DW), and lowest in stem (14.51 mg⋅g–1DW) (Figure 3A). We also found that sucrose and fructose levels were higher in the rhizome than in the stem, leaf, and flower (Figures 3B,D). In contrast, the glucose content of the stem was the highest, followed by the leaf (Figure 3C). It was lowest in both rhizome and flower of P. cyrtonema Hua (Figure 3C). The above results clearly suggested that the polysaccharides are synthesized in rhizome, stem, leaf, and flower tissues. Meanwhile, the biosynthesis and transport of sucrose, glucose, and fructose in the leaf and stem significantly impacted the polysaccharide accumulation in the rhizome and flower of P. cyrtonema Hua.
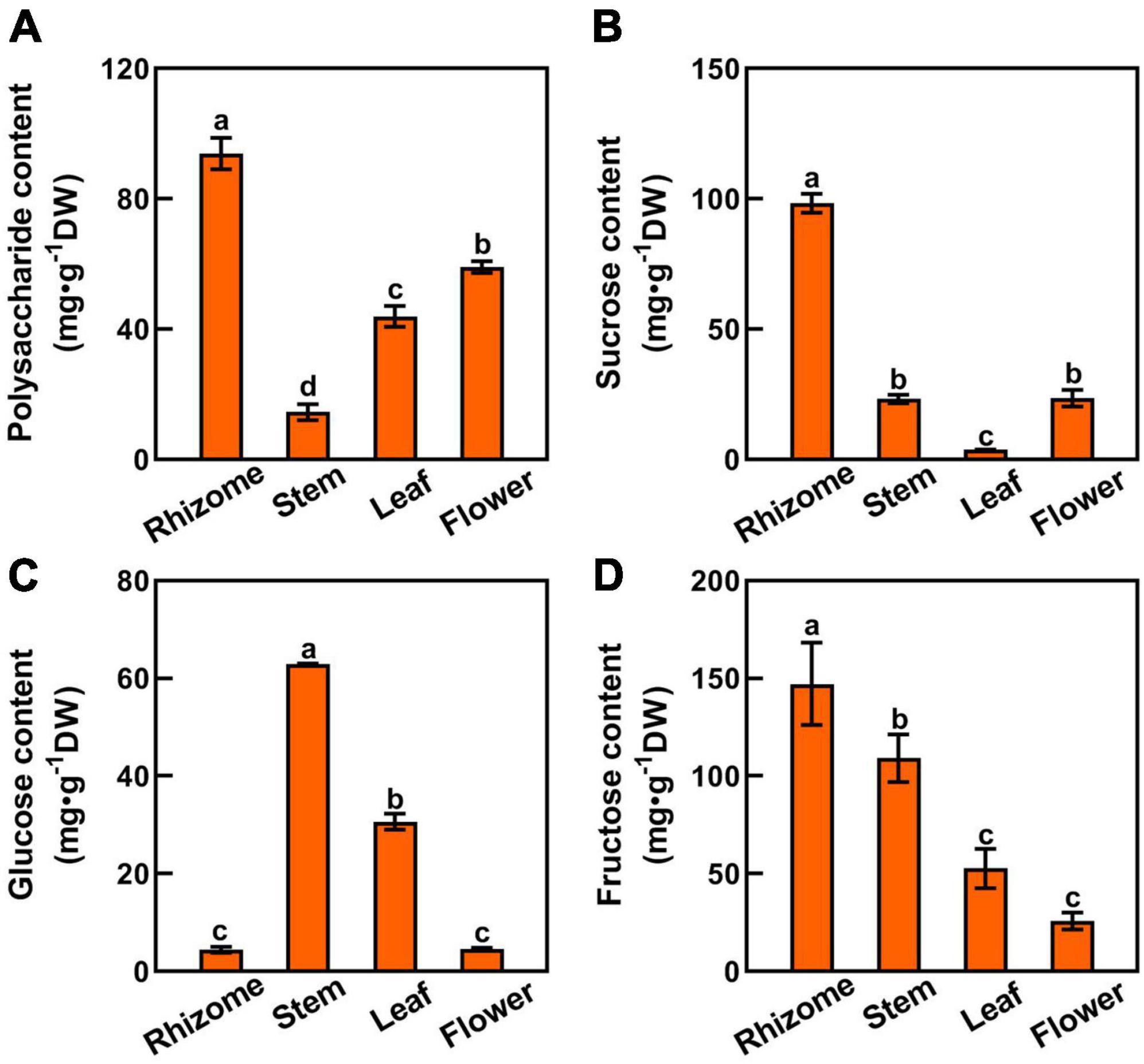
Figure 3. Sugar composition in different parts of P. cyrtonema. (A–D) Polysaccharide, sucrose, glucose, and fuctose content of rhizome, stem, leaf, and flower tissues of P. cyrtonema. Data represent mean ± SD (n = 3). Bars with different letters denote significant differences at P < 0.05 according to Duncan’s multiple comparison test.
To elucidate the molecular mechanisms of polysaccharide biosynthesis, a genome-wide transcriptional analysis was performed in the rhizome, stem, leaf, and flower tissues of P. cyrtonema Hua. For transcriptomic analysis, total RNA was extracted and used to prepare cDNA libraries by using three biological replicates. After processing, the raw sequence reads (between 42,336,126 and 52,068,538 clean reads in total) were aligned, and the Q20% (the percentage of bases with a Phred value >20) value for the clean reads was >97% and the Q30% (the percentage of bases with a Phred value >30) value of the clean reads was >92% and were found to map (Supplementary Table 1). A total of 182,700 unigenes were assembled. With regard to the unigenes, the total length and mean length of the unigenes were 157,174,631 and 860.29 bp, respectively, and their GC% was 44.78% (Supplementary Table 2). These results implied that high-quality sequencing data were obtained for the following analyses.
In order to construct a detailed regulatory network for polysaccharide biosynthesis in different tissues, we compared differentially expressed genes (DEGs) between stem and rhizome, leaf and rhizome, and flower and rhizome, respectively. The DEGs were filtered by expression levels | log 2 (fold-change)| > 1 and FDR < 0.05 in each pairwise comparison, thus revealing 7,054 (3,963 up- and 3,091 downregulated) DEGs in stem/rhizome, 13,468 (7,440 up- and 6,028 downregulated) DEGs in leaf/rhizome, and 19,437 (18,010 up- and 1,427 downregulated) DEGs in flower/rhizome (Figure 4A). Then, we constructed a Venn diagram to investigate the numbers of co-expressed and uniquely expressed DEGs in each pairwise comparison. A total of 953 co-expressed DEGs were obtained among these (Figure 4B). Additionally, hierarchical clustering analysis showed that strong changes in DEG expression levels were observed in the different tissues (Figure 4C).
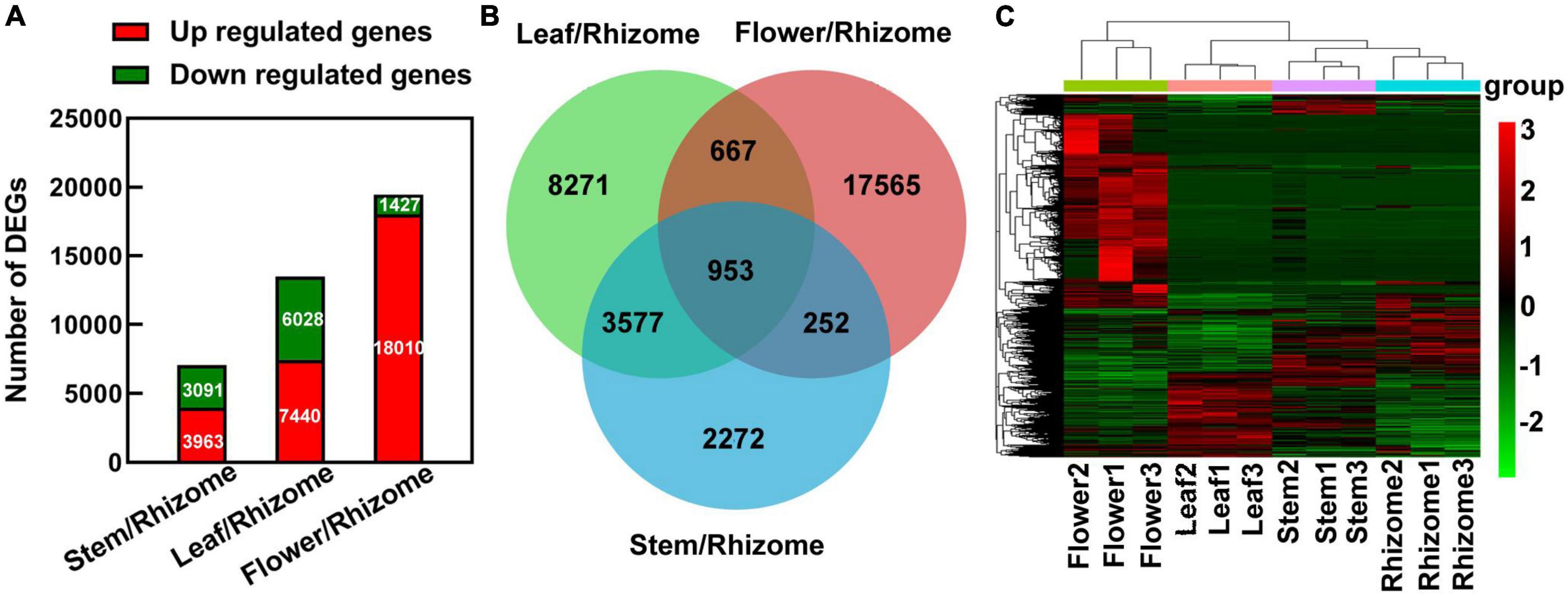
Figure 4. An overview of differentially expressed genes (DEGs) responsive to different parts of Polygonatum cyrtonema. (A) The number of DEGs was examined in the comparison between rhizome and stem, leaf, or flower. (B) Venn diagram showing the distribution of DEGs in the comparison between rhizome and stem, leaf, or flower. (C) Hierarchical clustering represents relative expression levels of DEGs in comparison between rhizome and stem, leaf, or flower. Multiple of differential expression [foldChange (Case/Control)]: foldChange (stem/rhizome), foldChange (leaf/rhizome), and foldChange (flower/rhizome). log2FoldChange > 1, P-value < 0.05. Red indicates upregulated genes, and green indicates downregulated genes.
The above-assembled unigenes were searched and annotated in NR, GO, KEGG, PFAM, eggNOG, and Swiss prot databases using the BLAST algorithm. Our results showed that 49.47, 21.77, 20.66, 30.53, 40.79, and 37.27% unigenes were annotated as significant hits in NR, GO, KEGG, PFAM, eggNOG, and Swiss prot databases, respectively (Supplementary Table 3).
To understand the biological processes and pathways involved in polysaccharide biosynthesis, Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) pathway-based enrichment analyses were performed (Figures 5, 6). The results showed that the top 20 pathway classifications with the smallest FDR value were selected by KEGG pathway-based enrichment analysis due to the DEGs between the rhizome and stem, leaf, or flower group (Figure 5A). Among these, genes related to photosynthesis, carbon fixation in photosynthetic organisms, galactose metabolism, and starch and sucrose metabolism processes were significantly enriched. Then, we screened 15 pathway classifications from the DEGs related to carbohydrate metabolism, which are most likely associated with polysaccharide biosynthesis, such as starch and sucrose metabolism, fructose and mannose metabolism, glycolysis/gluconeogenesis, and pentose and glucuronate interconversions (Figure 5B and Supplementary Table 4). For example, there were 52, 103, and 206 DEGs involved in starch and sucrose metabolism among stem/rhizome, leaf/rhizome, and flower/rhizome, respectively. Moreover, GO pathway analysis revealed that there were numerous DEGs in the polysaccharide metabolic process, including cellular/cell wall polysaccharide biosynthetic and catabolic processes, directly related to polysaccharide biosynthesis in P. cyrtonema Hua (Figure 6 and Supplementary Table 5).
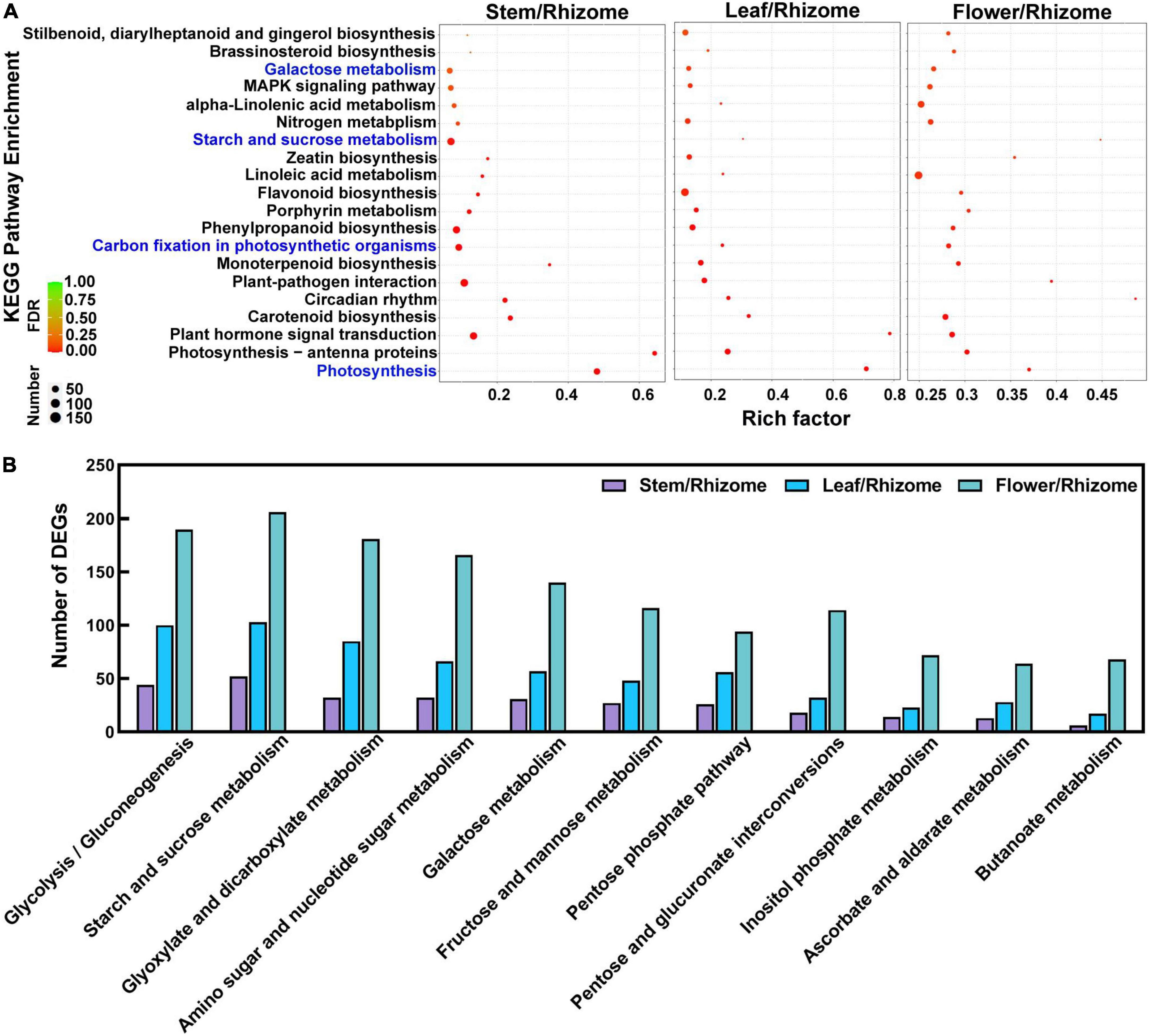
Figure 5. Functional enrichment analysis of differentially expressed genes (DEGs) by Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway-based enrichment analysis. (A) The top 20 pathway classifications with the smallest FDR value were selected by KEGG pathway-based enrichment analysis due to the DEGs between rhizome and stem, leaf, or flower group. (B) The 11 pathway classifications for carbohydrate metabolism. Rich factor indicates the matched gene ratio, the circle size is proportional to the target gene number, and 0 < FDR (false discovery rate) <1.
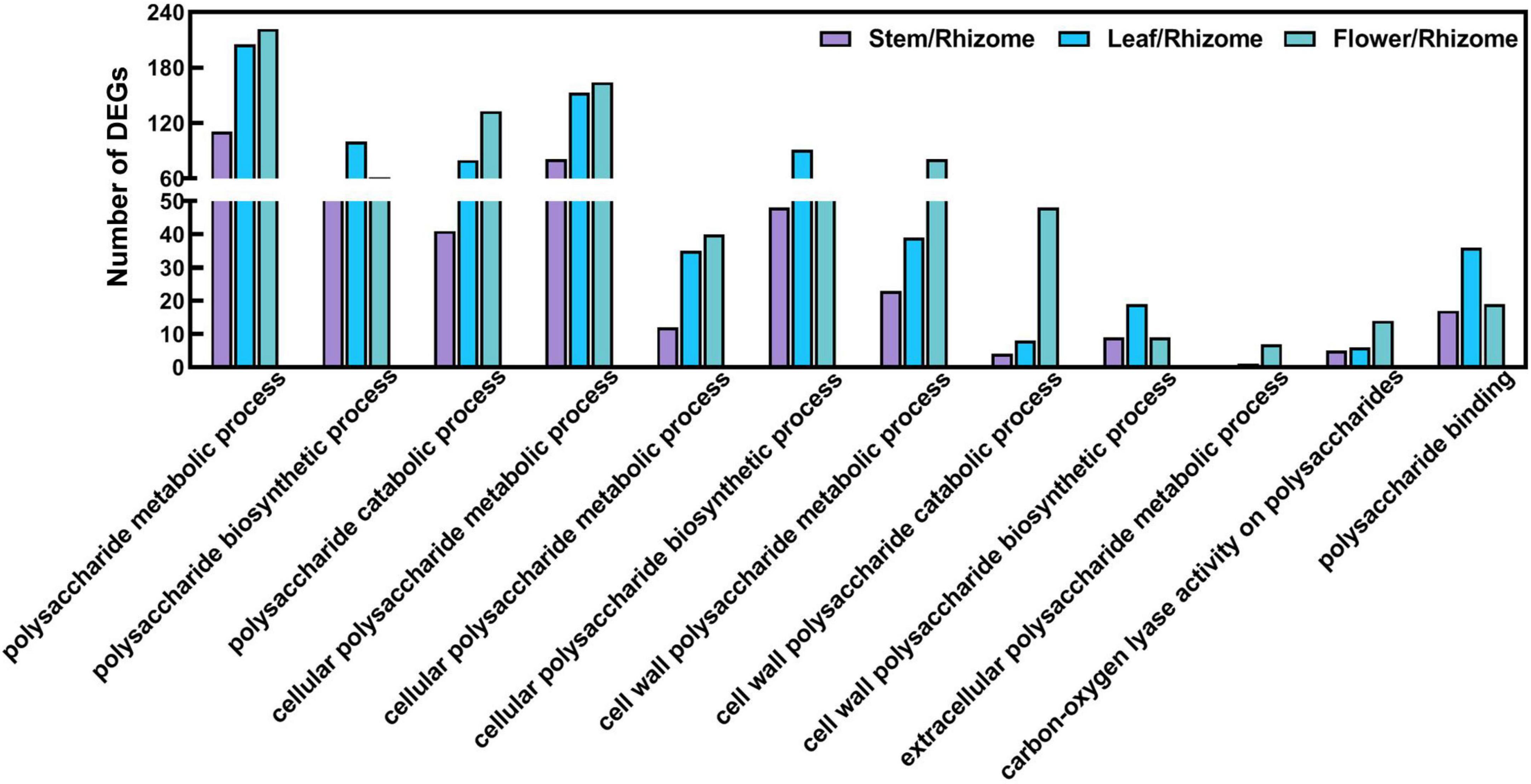
Figure 6. Functional enrichment analysis of differentially expressed genes (DEGs) by Gene Ontology (GO) pathway-based enrichment analysis. The 12 pathway classifications for the polysaccharide metabolism were selected by GO pathway-based enrichment analysis due to the DEGs between rhizome and stem, leaf, or flower group.
To further comprehend the polysaccharide biosynthesis in different tissues, we compared DEGs between stem and rhizome, leaf and rhizome, and flower and rhizome, respectively (Figure 7 and Supplementary Table 6). We found that 67 genes involved in polysaccharide biosynthesis showed significant differences in expression, such as MPI, GMPP, GMDS, GALE, and RHM (Figure 7). Interestingly, the transcript levels of GMPP (DN3245_c1_g2), GMDS (DN608_c0_g1), and RHM (DN155122_c0_g1) were lower in stem, leaf, and flower compared to these of rhizome, respectively. Moreover, multiple genes involved in sucrose metabolism, including sucrose synthesis and degradation, were also discovered, such as SPS, SUS, and INV. As expected, the transcript levels of SPS (DN19938_c0_g1) and SUS (DN15329_c0_g1) were higher in the leaf than in other tissues. Similarly, most of the genes involved in the glycolysis/gluconeogenesis pathway, such as ALDO, FBP, GPI, PGM, and UGP2 transcript levels, showed a similar tendency. Besides, PGM (DN27690_c0_g1 and DN5147_c0_g1), GPI (DN12212_c0_g1), FBP (DN17750_c0_g1), and UGP2 (DN2305_c0_g1 and DN2305_c0_g2) were mainly expressed in the flower tissues of P. cyrtonema Hua.
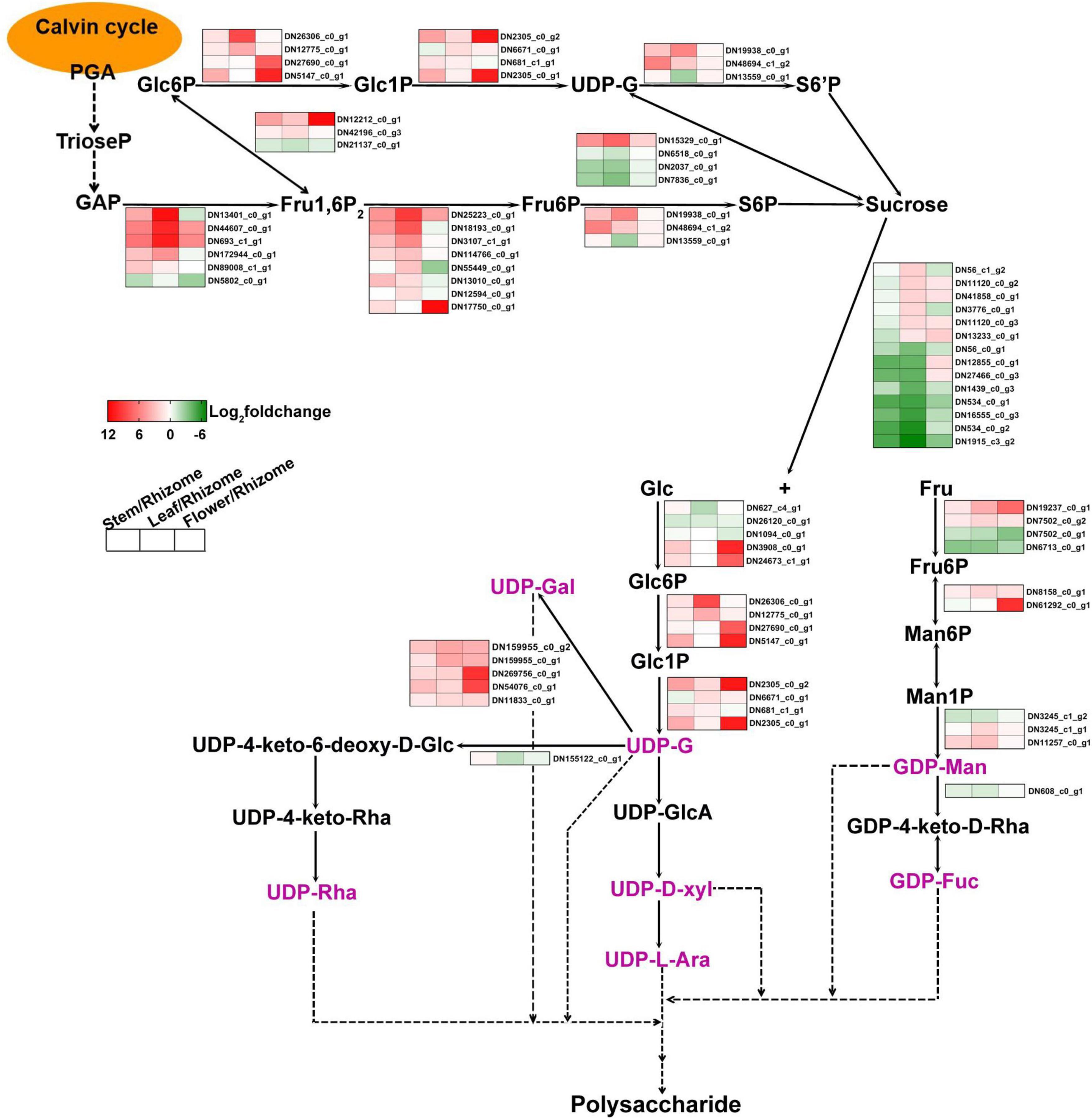
Figure 7. Heat map illustrating major expression profiles of differentially expressed genes (DEGs) involved in polysaccharide biosynthesis. Red indicates high expression, white indicates intermediate expression, and green indicates low expression.
Sugar transporters play key roles in source-sink dynamics, which is important for carbon distribution and plant growth (Hennion et al., 2019). Thus, we analyzed the expression levels of SWEETs, SUTs, ERDs, MSTs, PLSTs, and STPs in the different tissues of P. cyrtonema Hua (Figure 8 and Supplementary Table 7). As shown in Figure 8, SWEET (DN60284_c0_g1) was mainly expressed in both leaf and stem, SWEET (DN6616_c0_g1) was mainly expressed in both stem and flower, and other SWEETs and ERDs were mainly expressed in stem and/or rhizome. SUT (DN6616_c0_g1) was mainly expressed in both rhizome and flower, and SUT (DN2552_c0_g1) was mainly expressed in both leaf and stem. Additionally, most of the monosaccharide transporters (MSTs), plastidic glucose transporters (PLSTs), and sugar transport proteins (STPs) were mainly expressed in the leaf. Combined with the content of sugar compositions, the above results indicated that these transporters might play important roles in the source-sink dynamics, especially in the accumulation of polysaccharides.
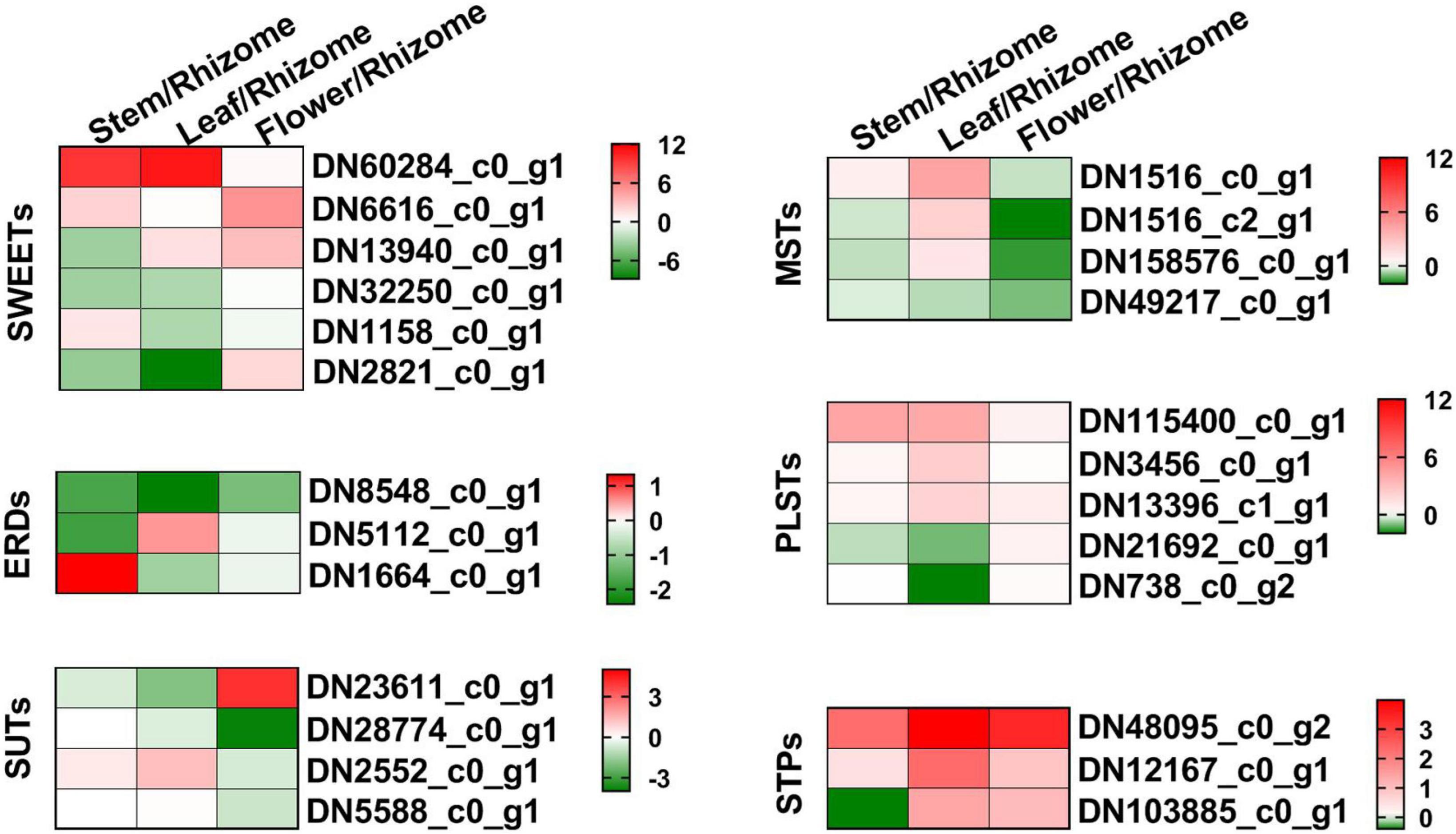
Figure 8. Heat map illustrating major expression profiles of differentially expressed genes (DEGs) related to the sugar transport. Sugar will eventually be exported transporters, SWEET; early response to dehydration, ERD; H + /sucrose transporters, SUT/SUC; monosaccharide transporters, MST; plastidic glucose transporter, PLST; sugar transport protein, STP. Red indicates high expression, white indicates intermediate expression, and green indicates low expression.
To get further insight into the regulation of polysaccharide accumulation, we next analyzed the correlation between polysaccharide, sucrose, glucose, and fructose contents and the expression levels of genes related to polysaccharide biosynthesis and sugar transporters in the rhizome, stem, leaf, and flower tissues (Figure 9). Here, we found sucrose content was generally and positively correlated with polysaccharide content (r = 0.79), while glucose content showed a significant negative correlation (r = −0.90). There was a highly significant correlation between the expression levels of GPI (DN21137_c0_g1), SUS (DN7836_c0_g1), INV (DN56_c0_g1, DN1439_c0_g3, DN534_c0_g1, DN16555_c0_g3, DN534_c0_g2, and DN1915_c3_g2), HK (DN26120_c0_g1), scrK (DN7502_c0_g2, DN7502_c0_g1, and DN6713_c0_g1), MPI (DN8158_c0_g1), GMPP (DN3245_c1_g2), GMDS (DN608_c0_g1), GALE (DN159955_c0_g2 and DN11833_c0_g1), ERD (DN8548_c0_g1), and sucrose content. We also observed that the expression of ALDO (DN89008_c1_g1), FBP (DN13010_c0_g1), UGP2 (DN681_c1_g1), and SPS (DN48694_c1_g2) showed high positive correlation with the glucose content. Besides, the expression of scrK (DN7502_c0_g1) and GALE (DN159955_c0_g2) was positively and negatively correlated with fructose content, respectively. Moreover, the expression levels of SUS (DN2037_c0_g1 and DN7836_c0_g1), INV (DN534_c0_g2), SWEET (DN32250_c0_g1), and PLST (DN32250_c0_g1) genes significantly correlated with polysaccharide content. These results suggested that the above genes were involved in the polysaccharide biosynthesis in P. cyrtonema Hua.
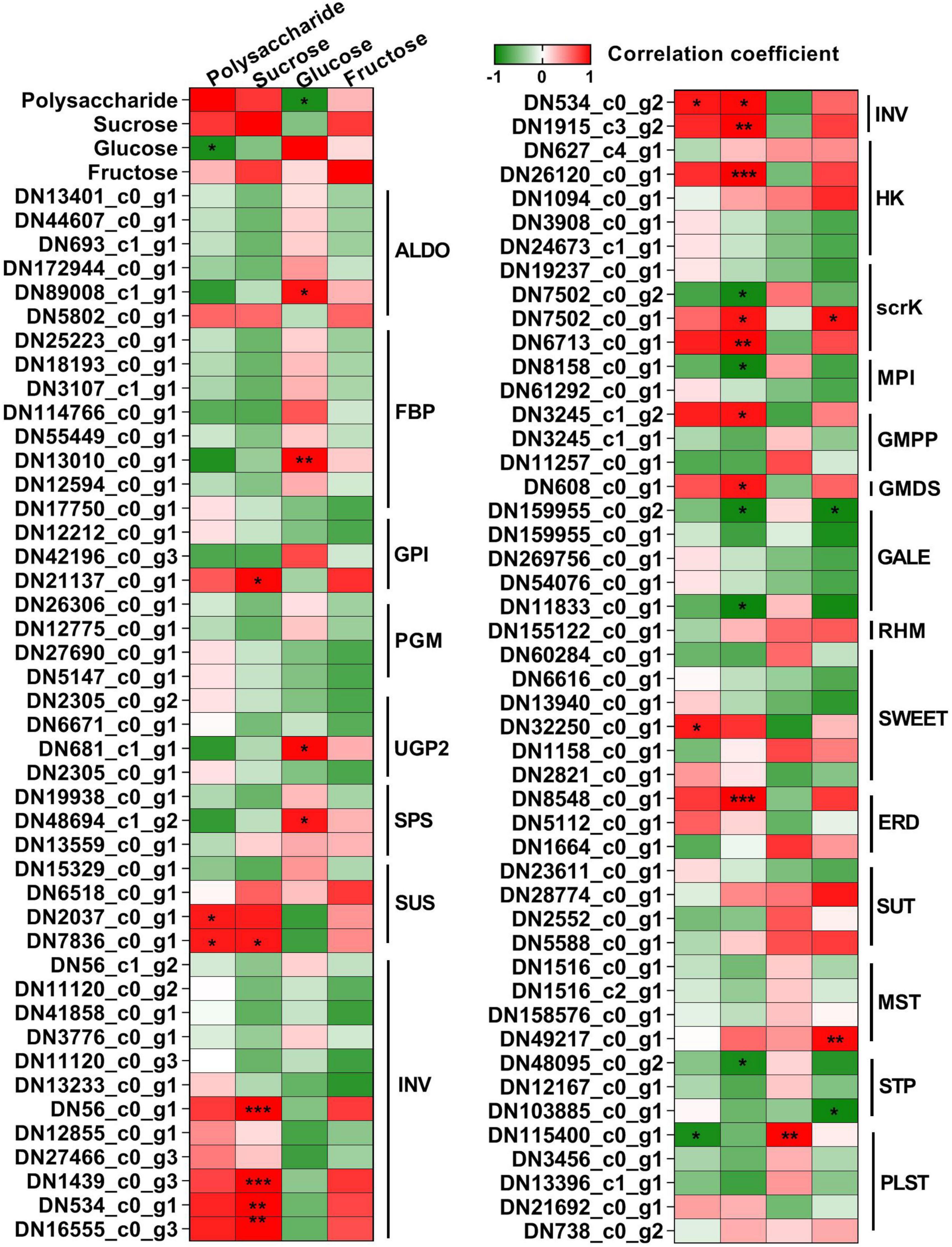
Figure 9. Heat map of the correlation coefficients between polysaccharide, sucrose, glucose, and fructose and the expression levels of genes related to sugar metabolism and transport in different tissues. Asterisks represent statistical significance determined by Student’s t-test (*P < 0.05, **P < 0.01, and ***P < 0.001).
To explore the transcriptional regulation of genes related to polysaccharide accumulation, we next analyzed the transcription factors (TFs) in the rhizome, stem, leaf, and flower tissues of P. cyrtonema Hua. We found that 2,728 TFs were significantly up/downregulated (Figure 10). The major TF families were screened, including bHLH (296 unigenes), ERF (163 unigenes), MYB_related (158 unigenes), C2H2 (157 unigenes), NAC (147 unigenes), B3 (135 unigenes), FAR (131 unigenes), bZIP (119 unigenes), MYB (109 unigenes) groups (Figure 10A and Supplementary Table 8). Then, we constructed the co-expression network according to the differentially expressed transcription factors and sugar metabolism and transport genes (Figure 10B). There were 87 genes related to sugar metabolism and transport genes that presented very high correlations. For example, the UGP2 (DN681_c1_g1) expression was highly and positively correlated with bZIP (DN16926_c0_g1), bHLH (DN918_c1_g1), and ERF (DN25257_c0_g1) expression (r = 0.96, 0.95, 0.98, p < 0.001). These results suggested that transcription factors might be involved in polysaccharide accumulation by regulating the expression of sugar metabolism and transport genes.
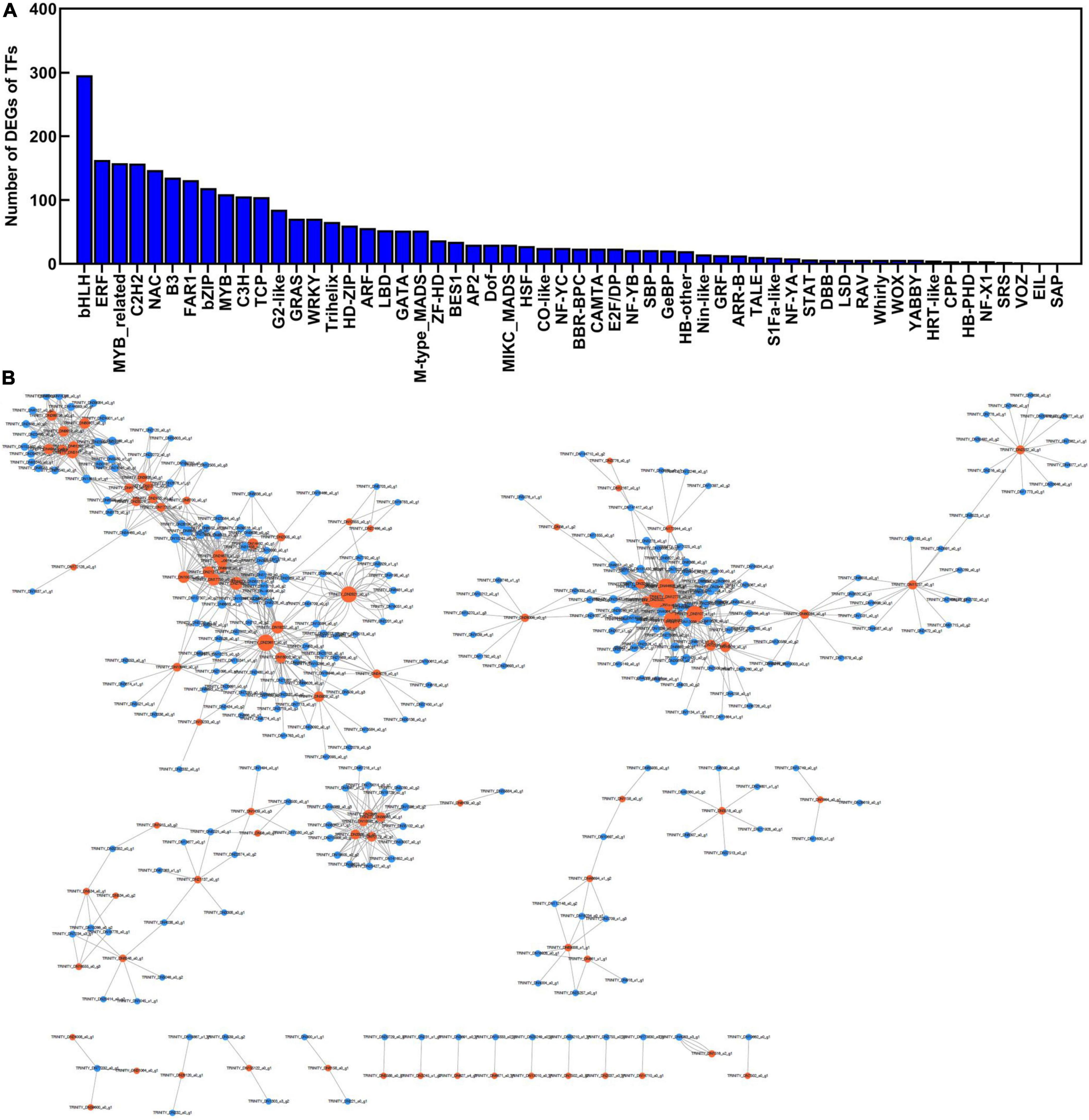
Figure 10. The co-expression network was constructed according to the differentially expressed transcription factors and sugar metabolism and transport genes in different tissues of Polygonatum cyrtonema Hua. (A) Type and number of transcription factors (TFs) of P. cyrtonema Hua. (B) The correlation coefficients of expression levels between transcription factors and sugar metabolism and transport in different tissues.
As bioactive ingredients, polysaccharides are of great value in the merits of medicinal plants (Li et al., 2021). Because rhizomes are the harvested targets, some studies have shown that rhizomes of P. cyrtonema Hua accumulated high content of polysaccharides, and unearthed several key genes involved in polysaccharide biosynthesis pathway from rhizomes at different ages by transcriptome sequencing (Li et al., 2022). Besides, polysaccharide accumulation is also highly cultivar- and tissue-dependent (Wang et al., 2019; Hu et al., 2022). For instance, Wang et al found that polysaccharide content was low in the leaf (1.62%) (Wang et al., 2019).
Most of the studies on polysaccharide accumulation were performed by examining the gene expression involved in the proposed pathway from sucrose to polysaccharide (Wang et al., 2019; Li et al., 2022). Furthermore, sucrose is the main form that is transported from source leaves to different sinks via SWEETs, SUTs, and/or ERDs transporters (Julius et al., 2017). Previous developments indicated that the role of sucrose synthase involved in sucrose import may play a role in directing carbon toward polysaccharide biosynthesis (Koch, 2004). Therefore, polysaccharide accumulation is dynamic and highly regulated. However, whether or how sucrose metabolism is involved in polysaccharide accumulation of shoots and rhizomes remains to be investigated in P. cyrtonema Hua.
In this report, we clearly observed that polysaccharides highly accumulated in rhizome tissue, which is consistent with the previously published results (Figure 3A; Wang et al., 2019). In addition, our results also indicated that sucrose and fructose levels were higher in the rhizome than in stem, leaf, and flower tissues (Figures 3B,D). In contrast, glucose content was lower in both the rhizome and flower of P. cyrtonema Hua (Figure 3C). Then, we analyzed the correlation between polysaccharide content and sucrose, glucose, or fructose in different tissues, respectively (Figure 9). These results demonstrated that the contents of sucrose and glucose were generally correlated with polysaccharide content. This was consistent with the regulation of sugar metabolism in rhizomes of P. cyrtonema Hua and P. sibiricum Red (Hu et al., 2022). Sugars were involved in most of the metabolic and signaling pathways controlling plant growth and development, as well as the responses to abiotic stresses. For example, leaf-derived sucrose accumulated in large amounts in the hypocotyls of tomato plants under flooding stress conditions (Mignolli et al., 2021). The above results suggested that sucrose metabolism might be involved in regulating the polysaccharide accumulation, which might play a critical role in tolerance to abiotic stresses in P. cyrtonema Hua.
The previous studies showed that a few hundred unigenes were involved in the sugar metabolism pathway. They found that the expression levels of unigenes encoding sacA, RHM, MPI, GMDS, TSTA3, UER1, and UGE enzymes were higher in rhizomes compared to leaves, while the expression levels of unigenes encoding HK, scrK, UGP2, GPI, GMPP, GALE, UXE, and UGDH were higher in leaves (Wang et al., 2019; Li et al., 2022). Combined with the sugar accumulation pattern in the above results, these findings led us to explore the regulation of the expression of polysaccharide biosynthesis genes in response to polysaccharide accumulation in rhizome, stem, leaf, and flower during the growth of P. cyrtonema Hua.
In this report, we analyzed the correlation between the expression of polysaccharide and sucrose biosynthesis pathway genes and polysaccharide, sucrose, or glucose content in rhizome, stem, leaf, and flower tissues, respectively. These results demonstrated that the expression of polysaccharide biosynthesis pathway genes is generally correlated with polysaccharide content in rhizomes (Figures 3, 7). Additionally, there were several unigenes, such as HK (DN3908_c0_g1 and DN24673_c1_g1), MPI (DN61292_c0_g1), and GALE (DN259756_c0_g1 and DN54076_c0_g1), specifically expressed in flowers, which are beneficial to polysaccharide biosynthesis in flowers (Figure 7). These results suggested that polysaccharide accumulation is generally regulated at the transcription level of polysaccharide biosynthesis genes.
In plants, sucrose, as an important form, is delivered to different tissues for metabolism and storage by the phloem (Hennion et al., 2019). This is apoplastically mediated through SWEET, SUT/SUC, and EDR6 transporters (Stadler et al., 1995; Kiyosue et al., 1998; Milne et al., 2018; Chen et al., 2010, 2012). The molecular mechanisms of sugar transport, especially sucrose transport, are largely unknown in P. cyrtonema Hua. In this study, we found that there were several DEGs involved in sugar transport among rhizome, stem, leaf, and flower tissues (Figure 8). For example, SWEET (DN60284_c0_g1) was specifically expressed in both leaf and stem, while SWEET (DN2821_c0_g1) was specifically expressed in the rhizome. SUT (DN23611_c0_g1) was specifically expressed in the flower. These results suggested that the above genes showed different roles in sugar efflux and/or uptake, thereby participating in polysaccharide biosynthesis. Previous results showed that OsSWEET13 and OsSWEET15 genes modulated sucrose transport and distribution to maintain sugar homeostasis in response to drought and salinity stresses (Mathan et al., 2021b). It was also found that AtSWEET17 was predicted to act as a vacuolar fructose importer, with a function in sequestering excess cytosolic fructose into vacuoles for storage in roots (Chardon et al., 2013). AtSWEET16 overexpression increased the vacuolar sucrose and glucose accumulation to improve the freezing tolerance in Arabidopsis (Klemens et al., 2013). Furthermore, AtSWEET15 was strongly induced by osmotic stresses, including cold, high salinity, and drought (Seo et al., 2011). It is also worth noting that both glucose content and plastidic glucose transporter (PLST and DN115400_c0_g1) expression showed a significantly negative correlation with polysaccharide content. Thus, the biological functions of these transporters should be further elucidated in P. cyrtonema Hua in the future.
In this study, we observed that the expression of sucrose synthesis and decomposition genes correlated differently with sucrose and polysaccharide contents among the different tissues (Figure 9). For example, SUS (DN2037_c0_g1 and DN7836_c0_g1) and INV (DN534_c0_g2) transcripts positively correlated with the polysaccharide content. Emerging evidence had shed light on the potential role of INVs in the response to abiotic stress in various plant species (Dahro et al., 2016). Therefore, the vital role of INV in alleviating the abiotic stress in P. cyrtonema Hua needs to be explored in the future. Furthermore, the correlation between sucrose contents and the expression of polysaccharide biosynthesis genes was also discovered, such as MPI (DN8158_c0_g1), GMPP (DN3245_c0_g2), GMDS (DN608_c0_g1), and GALE (DN159955_c0_g2 and DN11833_c0_g1) (Figure 9). Correlations between polysaccharide content and the expression of MPI, GMPP, GMDS, and GALE genes were observed by other reports in P. cyrtonema Hua (Wang et al., 2019; Li et al., 2022). Therefore, these results suggested that sucrose might be transported by SWEET (DN32250_c0_g1) to rhizomes, and then decomposed by INV to promote polysaccharide biosynthesis. Furthermore, several studies revealed that various transcription factors are involved in the regulation of sugar metabolism pathways which overcome the negative effects of abiotic stress (Kumar et al., 2022). For example, MaRAP2-4, an ERF transcription factor, activated SWEET10 that directly or indirectly assists in sugar accumulation, thus enhancing tolerance to waterlogging, drought, and salinity (Phukan et al., 2018). Future research will focus on the functional role of these key genes involved in polysaccharide accumulation and their roles in responses to abiotic stress in P. cyrtonema Hua. Other studies suggested that gas signaling molecules and hormones also enhanced plant tolerance to abiotic stress by regulating sugar metabolism (Wei et al., 2015; Yang et al., 2016; Chen et al., 2020; Rangani et al., 2020; Gu et al., 2021, 2022; de Bont et al., 2022). For example, exogenous melatonin application improved the tolerance to salinity and drought stresses via modulating the starch/sucrose metabolism in soybean (Wei et al., 2015). Therefore, the roles of gas signaling molecules and hormones involved in polysaccharide accumulation are also worth in-depth study in P. cyrtonema Hua.
In this study, we explored a detailed regulatory network for polysaccharide accumulation in P. cyrtonema Hua. Our results showed that polysaccharides generally accumulated in rhizome, stem, leaf, and flower tissues. Moreover, the polysaccharide biosynthesis in rhizome and flower is closely related to sucrose metabolism and transport in the leaf and stem (Figure 11). Sucrose is mainly synthesized by the upregulation of ALDO, GPI, UGP2, SPS, and SUS genes in the leaf. Then, a high amount of sucrose and several monosaccharides are transported to the stem, and then get unloaded into the rhizome and flower tissues due to the functions of SWEETs, SUTs, MSTs, ERDs, PLSTs, and/or STP genes. Afterward, sucrose is catalyzed by INV, and their products glucose and fructose are used to synthesize polysaccharides. Furthermore, these key genes are possibly regulated by transcription factors, such as bZIP, bHLH, ERF, ARF, and C2H2, which are involved in the regulatory processes of abiotic stresses. To our knowledge, this is the first report to link sucrose metabolism and transport with polysaccharide accumulation in P. cyrtonema Hua. This study provided new insights into the molecular mechanism of polysaccharide biosynthesis and valuable genes involved in abiotic stress tolerance in P. cyrtonema Hua. Our results also suggested that as polysaccharides have various health effects, not only rhizome, but leaves and flowers can also be used to process the foods to meet the market demand.
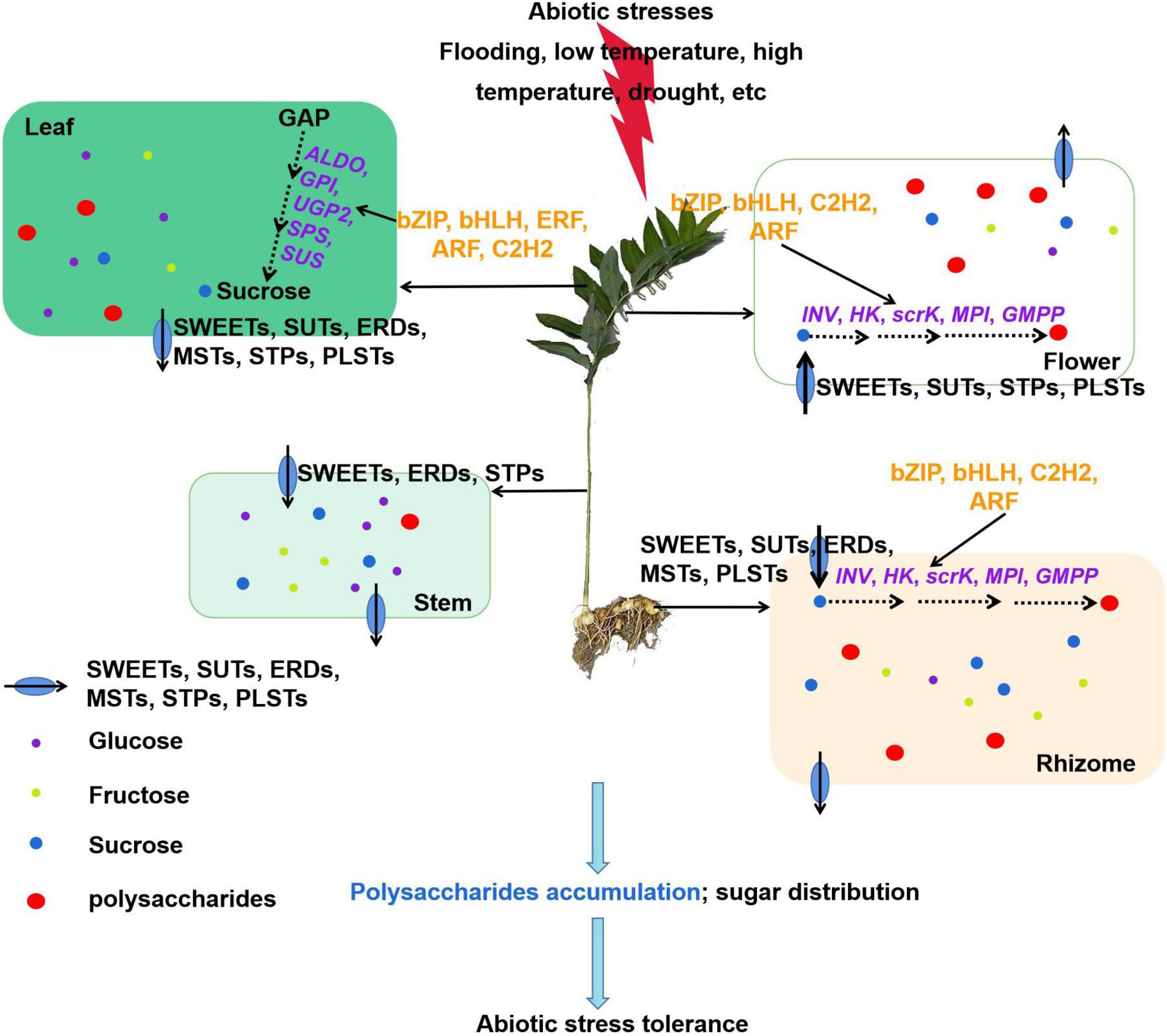
Figure 11. A model explaining the networks of transcriptional regulation of polysaccharide accumulation and their possible mechanisms in response to abiotic stress in Polygonatum cyrtonema Hua. Sucrose is mainly synthesized by the upregulation of ALDO, GPI, UGP2, SPS, and SUS genes in the leaf. Then, a high amount of sucrose and several monosaccharides is transported to the stem by SWEETs, ERDs, and/or STPs genes, and then get unloaded into the rhizome and flower tissues due to the functions of SWEETs, SUTs, MSTs, ERDs, PLSTs, and/or STPs genes. Afterward, sucrose is catalyzed by INV, and their products glucose and fructose are used to synthesize polysaccharides. Furthermore, these key genes are possibly regulated by transcription factors, such as bZIP, bHLH, ERF, ARF, and C2H2, which are involved in the regulatory processes of abiotic stresses. In general, our study contributes to a comprehensive understanding of the transcriptional regulation of polysaccharide accumulation.
Publicly available datasets were analyzed in this study. RNA-seq data of the tissues can be found in the publicly accessible NCBI under BioProject PRJNA878799.
XD, ZC, and LC conceived the study and designed the experiments. LC, SX, YL, YZ, FZ, LD, JC, KW, and SC performed the experiments. LL and YW participated in the preparation of plant materials. LC wrote the manuscript. XD and ZC revised and finalized the manuscript. All authors read and approved the final version of the manuscript.
This work was supported by the Anhui Province Key Research and Development Program (Grant No. 202204c06020010) and Postdoctoral Projects of Anhui Province (Grant No. 2020A396).
LL was employed by Jinzhai Senfeng Agricultural Technology Development Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1012231/full#supplementary-material
Chandran, D. (2015). Co-option of developmentally regulated plant SWEET transporters for pathogen nutrition and abiotic stress tolerance. IUBMB Life 67, 461–471. doi: 10.1002/iub.1394
Chardon, F., Bedu, M., Calenge, F., Klemens, P. A., Spinner, L., Clement, G., et al. (2013). Leaf fructose content is controlled by the vacuolar transporter SWEET17 in Arabidopsis. Curr. Biol. 23, 697–702. doi: 10.1016/j.cub.2013.03.021
Chen, L. Q., Hou, B. H., Lalonde, S., Takanaga, H., Hartung, M. L., Qu, X. Q., et al. (2010). Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 468, 527–532. doi: 10.1038/nature09606
Chen, L. Q., Qu, X. Q., Hou, B. H., Sosso, D., Osorio, S., Fernie, A. R., et al. (2012). Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science 335, 207–211. doi: 10.1126/science.1213351
Chen, T., Tian, M., and Han, Y. (2020). Hydrogen sulfide: A multi-tasking signal molecule in the regulation of oxidative stress responses. J. Exp. Bot. 71, 2862–2869. doi: 10.1093/jxb/eraa093
Cheng, X., Ji, H., Cheng, X., Wang, D., Li, T., Ren, K., et al. (2021). Characterization, classification, and authentication of Polygonatum sibiricum samples by volatile profiles and flavor properties. Molecules 27:25. doi: 10.3390/molecules27010025
Chinese Pharmacopoeia Commission (2020). Chinese Pharmacopoeia Commission. Beijing: China Medical Science Press, 319–320.
Dahro, B., Wang, F., Peng, T., and Liu, J. H. (2016). PtrA/NINV, an alkaline/neutral invertase gene of Poncirus trifoliata, confers enhanced tolerance to multiple abiotic stresses by modulating ROS levels and maintaining photosynthetic efficiency. BMC Plant Biol. 16:76. doi: 10.1186/s12870-016-0761-0
de Bont, L., Mu, X., Wei, B., and Han, Y. (2022). Abiotic stress-triggered oxidative challenges: Where does H2S act? J. Genet. Genom. 49, 748–755. doi: 10.1016/j.jgg.2022.02.019
Eggert, E., Obata, T., Gerstenberger, A., Gier, K., Brandt, T., Fernie, A. R., et al. (2016). A sucrose transporter-interacting protein disulphide isomerase affects redox homeostasis and links sucrose partitioning with abiotic stress tolerance. Plant Cell Environ. 39, 1366–1380. doi: 10.1111/pce.12694
Feng, Y. X., Yu, X. Z., Mo, C. H., and Lu, C. J. (2019). Regulation network of sucrose metabolism in response to trivalent and hexavalent chromium in Oryza sativa. J. Agric. Food Chem. 67, 9738–9748. doi: 10.1021/acs.jafc.9b01720
Gan, Q., Wang, X., Cao, M., Zheng, S., Ma, Y., and Huang, Q. (2022). NF-κB and AMPK-Nrf2 pathways support the protective effect of polysaccharides from Polygonatum cyrtonema Hua in lipopolysaccharide-induced acute lung injury. J. Ethnopharmacol. 291:115153. doi: 10.1016/j.jep.2022.115153
Gu, Q., Wang, C., Xiao, Q., Chen, Z., and Han, Y. (2021). Melatonin confers plant cadmium tolerance: An update. Int. J. Mol. Sci. 22:11704. doi: 10.3390/ijms222111704
Gu, Q., Xiao, Q., Chen, Z., and Han, Y. (2022). Crosstalk between melatonin and reactive oxygen species in plant abiotic stress responses: An update. Int. J. Mol. Sci. 23:5666. doi: 10.3390/ijms23105666
He, C., Wu, K., Zhang, J., Liu, X., Zeng, S., Yu, Z., et al. (2017). Cytochemical localization of polysaccharides in Dendrobium officinale and the involvement of DoCSLA6 in the synthesis of mannan polysaccharides. Front. Plant Sci. 8:173. doi: 10.3389/fpls.2017.00173
Hennion, N., Durand, M., Vriet, C., Doidy, J., Maurousset, L., Lemoine, R., et al. (2019). Sugars en route to the roots. Transport, metabolism and storage within plant roots and towards microorganisms of the rhizosphere. Physiol. Plant 165, 44–57. doi: 10.1111/ppl.12751
Hu, Y., Yin, M., Bai, Y., Chu, S., Zhang, L., Yang, M., et al. (2022). An evaluation of traits, nutritional, and medicinal component quality of Polygonatum cyrtonema Hua and P. sibiricum Red. Front. Plant Sci. 13:891775. doi: 10.3389/fpls.2022.891775
Julius, B. T., Leach, K. A., Tran, T. M., Mertz, R. A., and Braun, D. M. (2017). Sugar transporters in plants: New insights and discoveries. Plant Cell Physiol. 58, 1442–1460. doi: 10.1093/pcp/pcx090
Kircher, S., and Schopfer, P. (2012). Photosynthetic sucrose acts as cotyledon-derived long-distance signal to control root growth during early seedling development in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 109, 11217–11221. doi: 10.1073/pnas.1203746109
Kiyosue, T., Abe, H., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1998). ERD6, a cDNA clone for an early dehydration-induced gene of Arabidopsis, encodes a putative sugar transporter. Biochim. Biophys. Acta 1370, 187–191. doi: 10.1016/s0005-2736(98)00007-8
Klemens, P. A., Patzke, K., Deitmer, J., Spinner, L., Le Hir, R., Bellini, C., et al. (2013). Overexpression of the vacuolar sugar carrier AtSWEET16 modifies germination, growth, and stress tolerance in Arabidopsis. Plant Physiol. 163, 1338–1352. doi: 10.1104/pp.113.224972
Koch, K. (2004). Sucrose metabolism: Regulatory mechanisms and pivotal roles in sugar sensing and plant development. Curr. Opin. Plant Biol. 7, 235–246. doi: 10.1016/j.pbi.2004.03.014
Kumar, S., Thakur, M., Mitra, R., Basu, S., and Anand, A. (2022). Sugar metabolism during pre- and post-fertilization events in plants under high temperature stress. Plant Cell Rep. 41, 655–673. doi: 10.1007/s00299-021-02795-1
Li, D., Wang, Q., Chen, S., Liu, H., Pan, K., Li, J., et al. (2022). De novo assembly and analysis of Polygonatum cyrtonema Hua and identification of genes involved in polysaccharide and saponin biosynthesis. BMC Genom. 23:195. doi: 10.1186/s12864-022-08421-y
Li, L., Liao, B. Y., Thakur, K., Zhang, J. G., and Wei, Z. J. (2018a). The rheological behavior of polysaccharides sequential extracted from Polygonatum cyrtonema Hua. Int. J. Biol. Macromol. 109, 761–771. doi: 10.1016/j.ijbiomac.2017.11.063
Li, L., Thakur, K., Cao, Y. Y., Liao, B. Y., Zhang, J. G., and Wei, Z. J. (2020). Anticancerous potential of polysaccharides sequentially extracted from Polygonatum cyrtonema hua in Human cervical cancer Hela cells. Int. J. Biol. Macromol. 148, 843–850. doi: 10.1016/j.ijbiomac.2020.01.223
Li, L., Thakur, K., Liao, B. Y., Zhang, J. G., and Wei, Z. J. (2018b). Antioxidant and antimicrobial potential of polysaccharides sequentially extracted from Polygonatum cyrtonema Hua. Int. J. Biol. Macromol. 114, 317–323. doi: 10.1016/j.ijbiomac.2018.03.121
Li, X. L., Ma, R. H., Zhang, F., Ni, Z. J., Thakur, K., Wang, S. Y., et al. (2021). Evolutionary research trend of Polygonatum species: A comprehensive account of their transformation from traditional medicines to functional foods. Crit. Rev. Food Sci. Nutr. of print]. [Epub ahead doi: 10.1080/10408398.2021.1993783
Lin, W., Chen, H., Wang, J., Zheng, Y., Lu, Q., Zhu, Z., et al. (2021). Transcriptome analysis associated with polysaccharide synthesis and their antioxidant activity in Cyclocarya paliurus leaves of different developmental stages. PeerJ 9:e11615. doi: 10.7717/peerj.11615
Liu, F., Liu, Y., Meng, Y., Yang, M., and He, K. (2004). Structure of polysaccharide from Polygonatum cyrtonema Hua and the antiherpetic activity of its hydrolyzed fragments. Antivir. Res. 63, 183–189. doi: 10.1016/j.antiviral.2004.04.006
Luo, M., Zhang, W. W., Deng, C. F., Tan, Q. S., Luo, C., and Luo, S. (2016). Advances in studies of medicinal crop Polygonatum cyrtonema Hua. Lishizhen Med. Mater. Med. Res. 27, 1467–1469.
Mathan, J., Singh, A., and Ranjan, A. (2021a). Sucrose transport and metabolism control carbon partitioning between stem and grain in rice. J. Exp. Bot. 72, 4355–4372. doi: 10.1093/jxb/erab066
Mathan, J., Singh, A., and Ranjan, A. (2021b). Sucrose transport in response to drought and salt stress involves ABA-mediated induction of OsSWEET13 and OsSWEET15 in rice. Physiol. Plant 171, 620–637. doi: 10.1111/ppl.13210
Meng, M., Geisler, M., Johansson, H., Harholt, J., Scheller, H. V., Mellerowicz, E. J., et al. (2009). UDP-glucose pyrophosphorylase is not rate limiting, but is essential in Arabidopsis. Plant Cell Physiol. 50, 998–1011. doi: 10.1093/pcp/pcp052
Mignolli, F., Barone, J. O., and Vidoz, M. L. (2021). Root submergence enhances respiration and sugar accumulation in the stem of flooded tomato plants. Plant Cell Environ. 44, 3643–3654. doi: 10.1111/pce.14152
Milne, R. J., Grof, C. P. L., and Patrick, J. W. (2018). Mechanisms of phloem unloading: Shaped by cellular pathways, their conductances and sink function. Curr. Opin. Plant Biol. 43, 8–15. doi: 10.1016/j.pbi.2017.11.003
Northcote, D. H., and Pickett-Heaps, J. D. (1966). A function of the Golgi apparatus in polysaccharide synthesis and transport in the root-cap cells of wheat. Biochem. J. 98, 159–167. doi: 10.1042/bj0980159
Oh, C. S., Kim, H., and Lee, C. (2013). Rice cell wall polysaccharides: Structure and biosynthesis. J. Plant Biol. 56, 274–282. doi: 10.1007/s12374-013-0236-x
Osorio, S., Ruan, Y. L., and Fernie, A. R. (2014). An update on source-to-sink carbon partitioning in tomato. Front. Plant Sci. 5:516. doi: 10.3389/fpls.2014.00516
Phukan, U. J., Jeena, G. S., Tripathi, V., and Shukla, R. K. (2018). MaRAP2-4, a waterlogging-responsive ERF from Mentha, regulates bidirectional sugar transporter AtSWEET10 to modulate stress response in Arabidopsis. Plant Biotechnol. J. 16, 221–233. doi: 10.1111/pbi.12762
Raines, C. A., and Paul, M. J. (2006). Products of leaf primary carbon metabolism modulate the developmental programme determining plant morphology. J. Exp. Bot. 57, 1857–1862. doi: 10.1093/jxb/erl011
Rangani, J., Panda, A., and Parida, A. K. (2020). Metabolomic study reveals key metabolic adjustments in the xerohalophyte Salvadora persica L. during adaptation to water deficit and subsequent recovery conditions. Plant Physiol. Biochem. 150, 180–195. doi: 10.1016/j.plaphy.2020.02.036
Seo, P. J., Park, J. M., Kang, S. K., Kim, S. G., and Park, C. M. (2011). An Arabidopsis senescence-associated protein SAG29 regulates cell viability under high salinity. Planta 233, 189–200. doi: 10.1007/s00425-010-1293-8
Stadler, R., Brandner, J., Schulz, A., Gahrtz, M., and Sauer, N. (1995). Phloem loading by the PmSUC2 sucrose carrier from Plantago major occurs into companion cells. Plant Cell 7, 1545–1554. doi: 10.1105/tpc.7.10.1545
Sun, C. X., Palmqvist, S., Olsson, H., Boren, M., Ahlandsberg, S., and Jansson, C. (2003). A novel WRKY transcription factor, SUSIBA2, participates in sugar signaling in barley by binding to the sugar-responsive elements of the iso1 promoter. Plant Cell 15, 2076–2092. doi: 10.1105/tpc.014597
Wang, C., Peng, D., Zhu, J., Zhao, D., Shi, Y., Zhang, S., et al. (2019). Transcriptome analysis of Polygonatum cyrtonema Hua: Identification of genes involved in polysaccharide biosynthesis. Plant Methods 15:65. doi: 10.1186/s13007-019-0441-9
Wang, J., Su, H., Han, H., Wang, W., Li, M., Zhou, Y., et al. (2021a). Transcriptomics reveals host-dependent differences of polysaccharides biosynthesis in Cynomorium songaricum. Molecules 27:44. doi: 10.3390/molecules27010044
Wang, J., Wang, Y., Zhang, J., Ren, Y., Li, M., Tian, S., et al. (2021b). The NAC transcription factor ClNAC68 positively regulates sugar content and seed development in watermelon by repressing ClINV and ClGH3.6. Hortic. Res. 8:214. doi: 10.1038/s41438-021-00649-1
Wei, W., Li, Q. T., Chu, Y. N., Reiter, R. J., Yu, X. M., Zhu, D. H., et al. (2015). Melatonin enhances plant growth and abiotic stress tolerance in soybean plants. J. Exp. Bot. 66, 695–707. doi: 10.1093/jxb/eru392
Wu, W., Huang, N., Huang, J., Wang, L., Wu, L., Wang, Q., et al. (2022). Effects of the steaming process on the structural properties and immunological activities of polysaccharides from Polygonatum cyrtonema. J. Funct. Foods 88:104866. doi: 10.1016/j.jff.2021.104866
Xie, S. Z., Yang, G., Jiang, X. M., Qin, D. Y., Li, Q. M., Zha, X. Q., et al. (2020). Polygonatum cyrtonema Hua polysaccharide promotes GLP-1 secretion from enteroendocrine L-Cells through sweet taste receptor-mediated cAMP signaling. J. Agric. Food Chem. 68, 6864–6872. doi: 10.1021/acs.jafc.0c02058
Yang, M., Qin, B. P., Ma, X. L., Wang, P., Li, M. L., Chen, L. L., et al. (2016). Foliar application of sodium hydrosulfide (NaHS), a hydrogen sulfide (H2S) donor, can protect seedlings against heat stress in wheat (Triticum aestivum L.). J. Integr. Agric. 15, 2745–2758. doi: 10.1016/S2095-3119(16)61358-8
Yang, Y., Jiang, M., Feng, J., Wu, C., Shan, W., Kuang, J., et al. (2021). Transcriptome analysis of low-temperature-affected ripening revealed MYB transcription factors-mediated regulatory network in banana fruit. Food Res. Int. 148:110616. doi: 10.1016/j.foodres.2021.110616
Yu, Z., He, C., Teixeira da Silva, J. A., Zhang, G., Dong, W., Luo, J., et al. (2017). Molecular cloning and functional analysis of DoUGE related to water-soluble polysaccharides from Dendrobium officinale with enhanced abiotic stress tolerance. Plant Cell Tiss. Organ. 131, 579–599. doi: 10.1007/s11240-017-1308-2
Yu, Z., Zhang, G., Teixeira da Silva, J. A., Li, M., Zhao, C., He, C., et al. (2021). Genome-wide identification and analysis of DNA methyltransferase and demethylase gene families in Dendrobium officinale reveal their potential functions in polysaccharide accumulation. BMC Plant Biol. 21:21. doi: 10.1186/s12870-020-02811-8
Zhang, H., Tao, X., Fan, X., Zhang, S., and Qin, G. (2022). PpybZIP43 contributes to sucrose synthesis in pear fruits by activating PpySPS3 expression and interacts with PpySTOP1. Physiol. Plant 174:e13732. doi: 10.1111/ppl.13732
Zhang, J., He, C., Wu, K., Teixeira da Silva, J. A., Zeng, S., Zhang, X., et al. (2016). Transcriptome analysis of Dendrobium officinale and its application to the identification of genes associated with polysaccharide synthesis. Front. Plant Sci. 7:5. doi: 10.3389/fpls.2016.00005
Zhang, S., Shi, Y., Huang, L., Wang, C., Zhao, D., Ma, K., et al. (2020). Comparative transcriptomic analysis of rhizomes, stems, and leaves of Polygonatum odoratum (Mill.) Druce reveals candidate genes associated with polysaccharide synthesis. Gene 744:144626. doi: 10.1016/j.gene.2020.144626
Zhu, A., Li, W., Ye, J., Sun, X., Ding, Y., Cheng, Y., et al. (2011). Microarray expression profiling of postharvest Ponkan mandarin (Citrus reticulata) fruit under cold storage reveals regulatory gene candidates and implications on soluble sugars metabolism. J. Integr. Plant Biol. 53, 358–374. doi: 10.1111/j.1744-7909
Zhu, S., Liu, P., Wu, W., Li, D., Shang, E. X., Guo, S., et al. (2022). Multi-constituents variation in medicinal crops processing: Investigation of nine cycles of steam-sun drying as the processing method for the rhizome of Polygonatum cyrtonema. J. Pharm. Biomed. Anal. 209:114497. doi: 10.1016/j.jpba.2021.114497
Keywords: polysaccharides, sucrose metabolism and transport, transcription factors, gene expression, Polygonatum cyrtonema Hua
Citation: Chen L, Xu S, Liu Y, Zu Y, Zhang F, Du L, Chen J, Li L, Wang K, Wang Y, Chen S, Chen Z and Du X (2022) Identification of key gene networks controlling polysaccharide accumulation in different tissues of Polygonatum cyrtonema Hua by integrating metabolic phenotypes and gene expression profiles. Front. Plant Sci. 13:1012231. doi: 10.3389/fpls.2022.1012231
Received: 05 August 2022; Accepted: 29 August 2022;
Published: 29 September 2022.
Edited by:
Weicong Qi, Jiangsu Academy of Agricultural Sciences (JAAS), ChinaReviewed by:
Zhaojun Wei, Hefei University of Technology, ChinaCopyright © 2022 Chen, Xu, Liu, Zu, Zhang, Du, Chen, Li, Wang, Wang, Chen, Chen and Du. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ziping Chen, enBjaGVuQGFoYXUuZWR1LmNu; Xianfeng Du, ZHhmQGFoYXUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.