- 1National Key Facility for Crop Gene Resources and Genetic Improvement, Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing, China
- 2College of Agriculture, Fisheries and Forestry, Fiji National University, Nausori, Fiji
Rice inflorescence is one of the major organs in determining grain yield. The genetic and molecular regulation on rice inflorescence architecture has been well investigated over the past years. In the present review, we described genes regulating rice inflorescence architecture based on their roles in meristem activity maintenance, meristem identity conversion and branch elongation. We also introduced the emerging regulatory pathways of phytohormones involved in rice inflorescence development. These studies show the intricacies and challenges of manipulating inflorescence architecture for rice yield improvement.
Introduction
Rice (Oryza sativa L.) is one of the major crops in the world, providing energy for over half of the population on earth (Sreenivasulu et al., 2021). Rice grain yield is influenced by a variety of factors, one of which is the rice inflorescence, also known as the panicle. A rice inflorescence is typically composed of a main rachis, primary branches (PB), secondary branches (SB), and spikelets. The spikelets are divided into two types: lateral spikelets (LS) and terminal spikelets (TS) which grow on the tips of branches (Figure 1A). Based on the quantity and length of these organs, the inflorescence architecture can be categorized into different types, such as short or long, erect or dense.
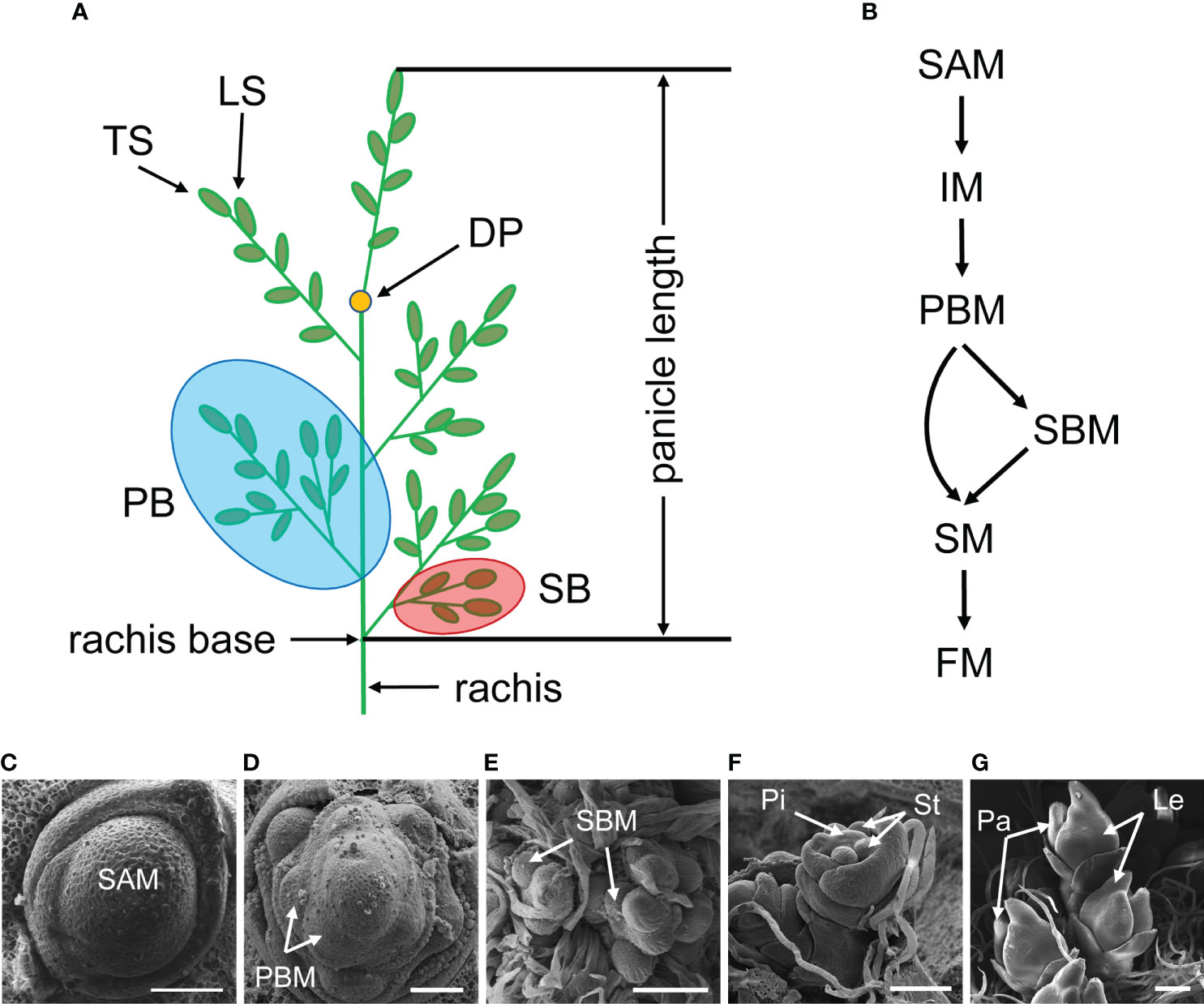
Figure 1 Rice inflorescence architecture and development process. (A) Schematic diagram of the rice inflorescence. The rice inflorescence is also called “panicle” that primarily contains a rachis, branches and spikelets. DP, degenerate point of rachis; PB, primary branch; SB, secondary branch; LS, lateral spikelet; TS, terminal spikelet. (B) A hypothesis of meristem identity conversion. SAM, shoot apical meristem; IM, inflorescence meristem; PBM, primary branch meristem; SBM, secondary branch meristem; SM, spikelet meristem; FM, floral meristem. (C-G) Scanning electron micrographs (SEM) showing early stages of rice inflorescence development. Pi, pistil; St, stamen; Pa, palea; Le, lemma. Bars = 50 μm in (C) and (D); Bars = 100 μm in (E, F); Bar = 200 μm in (G).
Rice inflorescence, like most other aboveground organs, is generated from a group of cells referred to as the shoot apical meristem (SAM). SAM initiates in the embryo and continues to generate leaves throughout the vegetative stage by maintaining its activity. During the reproductive stage, SAM converts to inflorescence meristem (IM) and produces two types of lateral meristem: primary branch meristem (PBM) and secondary branch meristem (SBM). Both meristems are undetermined until they acquire the terminated identity, at which point branch meristem (BM) differentiates into spikelet meristem (SM). Subsequently, floral organs, such as pistil, stamen, lemma and palea are developed, eventually forming a complete spikelet (Figures 1B–G). The branches begin to elongate during the differentiation stage of SM, which is another event crucial for inflorescence morphology.
In the past years, our understanding of genetic and molecular mechanisms that underpin rice inflorescence architecture has advanced significantly. These studies support breeders to create ideal inflorescence architecture of rice. Here, we summarize current advancements in elucidating the mechanisms of rice inflorescence development, with an emphasis on meristem activity maintenance, meristem identity conversion, branch elongation, and phytohormone regulation. We also explore the flaws of present researches and the challenges for future studies.
Maintenance of SAM activity
SAM is formed during embryogenesis and keeps itself from differentiating through continuous cell division and vegetative development (Laux and Schoof, 1997). Rice IM directly develops from SAM, thus SAM activity has a significant impact on inflorescence architecture and yield. Despite several genes have been identified (Figure 2), the mechanism of SAM establishment and maintenance in rice is not as clear as that in Arabidopsis (Guan and Jiao, 2020; Wang et al., 2020; Eshed, 2021; Li et al., 2022).
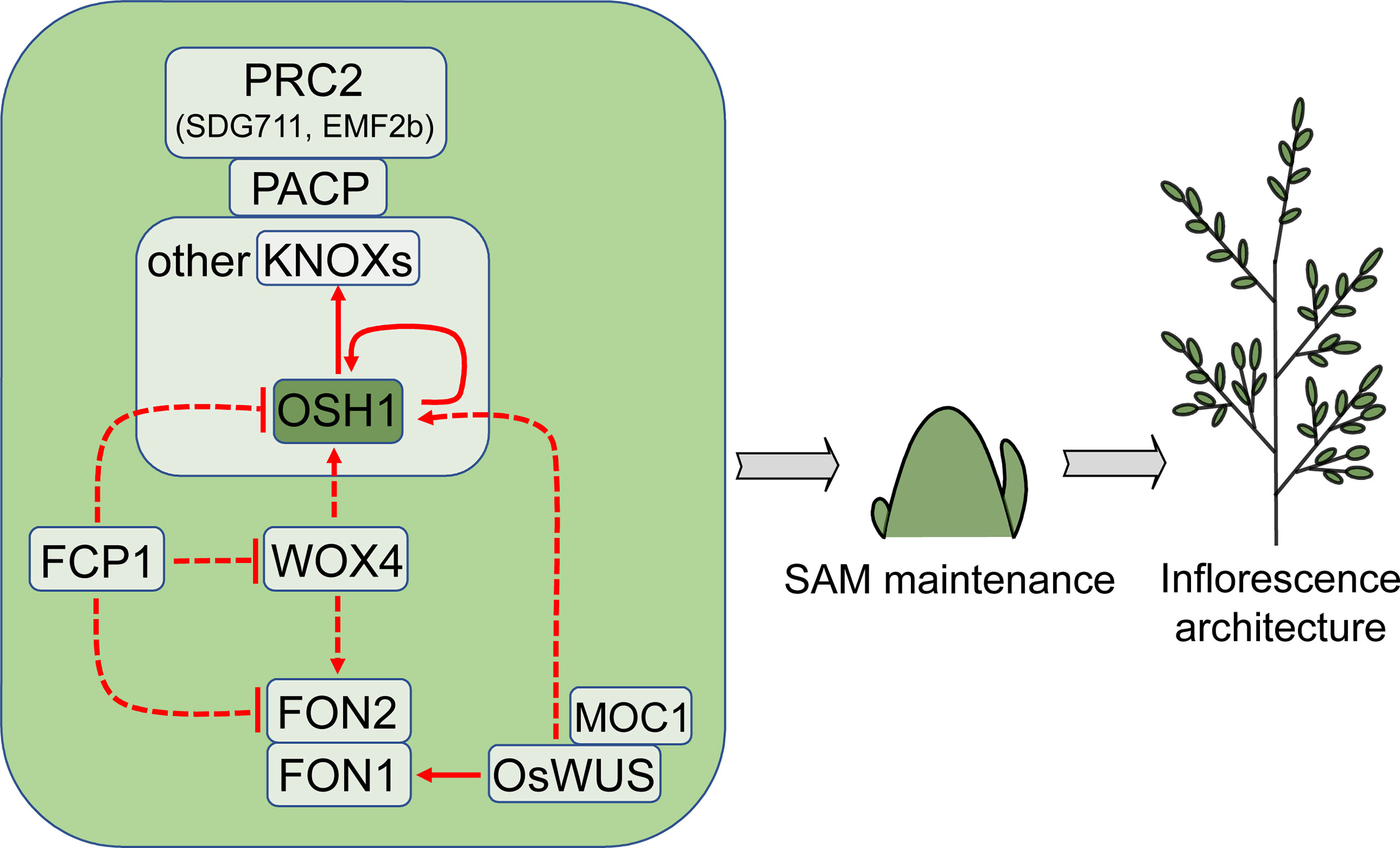
Figure 2 Regulators that control SAM or IM activity and affect inflorescence architecture. OSH1 is a critical factor that is commonly used as a marker gene for rice SAM maintenance. Arrows and blocked arrows represent positive and negative regulation, respectively. Solid and dashed lines represent direct and indirect regulation, respectively. Red lines represent regulation at transcription level. SAM, shoot apical meristem; IM, inflorescence meristem.
Rice SAM establishment and maintenance are dependent on CLASS 1 KNOTTED1-LIKE HOMEOBOX (KNOX) family, among which ORYZA SATIVA HOMEOBOX1 (OSH1) is expressed throughout SAM but not in leaf primordia (Sentoku et al., 1999). The expression of OSH1 precedes organ differentiation and continues until floral organs formation (Sato et al., 1996). OSH1 deficiency results in undersized rice inflorescence and fewer spikelets. As a transcription factor, OSH1 directly regulates itself and other KNOX genes (Tsuda et al., 2011).
OSH1 and several other KNOX proteins physically interact with PRC2-associated coiled-coil protein (PACP) in SAM (Tan et al., 2022). PACP is involved in the maintenance of H3K27me3 and inhibition of cell differentiation-promoting genes targeted by KNOXs in SAM through recruiting PRC2 complex proteins, including SDG711 and EMF2b. The growth of the pacp mutant is severely repressed due to the underdevelopment of SAM, leading to a much smaller inflorescence compared with wild type (Tan et al., 2022). These phenotypes are similar to those of the SDG711 and EMF2b RNAi plants (Liu et al., 2015b).
In Arabidopsis, the negative feedback regulatory module of WUSCHEL-CLAVATA (WUS-CLV) mediates the maintenance of stem cells in SAM (Mayer et al., 1998; Schoof et al., 2000; Aichinger et al., 2012; Somssich et al., 2016). Interestingly, OsWUS in rice is involved in the formation of axillary meristems rather than SAM, suggesting that the function of these two genes may be distinct (Lu et al., 2015b; Tanaka et al., 2015). One explanation for this difference is that OsWUS is specifically expressed in lateral precursor meristem instead of SAM (Tanaka et al., 2015). OSH1 expression is down-regulated in the loss-of-function mutant of OsWUS, suggesting that OsWUS maintains axillary meristem activity by promoting OSH1 expression (Tanaka et al., 2015). The Arabidopsis WUS directly interacts with STM, the ortholog of OSH1, coordinately maintaining SAM activity (Su et al., 2020), whereas it is unknown whether this mechanism is conserved in rice.
FLORAL ORGAN NUMBER1 (FON1) and FON2 (also known as FON4) in rice are the homologs of CLV1 and CLV3, respectively. The loss-of-function mutants fon2/fon4 and fon1 produce inflorescences with more primary branches due to the increased size of SAM or IM (Suzaki et al., 2004; Chu et al., 2006; Moon et al., 2006; Suzaki et al., 2006). CLV1 is mainly expressed in the central zone of SAM in Arabidopsis (Clark et al., 1997), but FON1 is expressed throughout the SAM and BM in rice (Suzaki et al., 2004). These studies show that the maintenance of meristem activity mediated by CLV signaling is conserved in rice, but the precise regulation may differ from that in Arabidopsis.
Current studies show that OsWUS can directly bind and enhance FON1 expression, and this binding is reliant on another meristem activity regulator, MONOCULM1 (MOC1), which has been identified as a QTL controlling rice inflorescence architecture (Shao et al., 2019; Zhang et al., 2020). MOC1 physically interacts with and activates OsWUS to modulate FON1 expression (Shao et al., 2019). Although MOC1 is expressed in axillary meristem rather than SAM, its loss-of-function mutant showed reduced number of panicle branches and spikelets (Li et al., 2003; Zhang et al., 2020).
Rice SAM maintenance is also dependent on another plant-specific homeobox transcription factor, WUSCHEL-RELATED HOMEOBOX4 (WOX4), which belongs to the same WOX subfamily as OsWUS (Ohmori et al., 2013). Unlike OsWUS, the expression of WOX4 is distributed throughout the SAM. At the reproductive stage, WOX4 is expressed in BM and SM. Interference of WOX4 expression results in a smaller SAM and down-regulation of OSH1 and FON2 (Ohmori et al., 2013). FON2-LIKE CLE PROTEIN1 (FCP1), a FON2-related CLE domain-containing protein, inhibits WOX4 expression. Overexpression of FCP1 prevents SAM formation and decreases the expression of OSH1, FON2, and WOX4, showing that FCP1 plays a negative regulatory role in SAM maintenance (Ohmori et al., 2013).
Conversion of meristem identity
The conversion of SAM to IM and BM to SM are two main events in the establishment of rice inflorescence architecture. Delayed conversion may result in a larger inflorescence with more branches and spikelets. Many genes associated with meristem identity conversion have been identified via studies using Arabidopsis as a model, including LEAFY (LFY), APETALA1 (AP1), and CAULIFLOWER (CAL). These genes enable the lateral meristem to acquire a floral identity and differentiate into flower organs (Mandel et al., 1992; Weigel et al., 1992; Kempin et al., 1995). In recent years, quite a few genes involved in the meristem identity conversion have also been identified in rice (Figure 3).
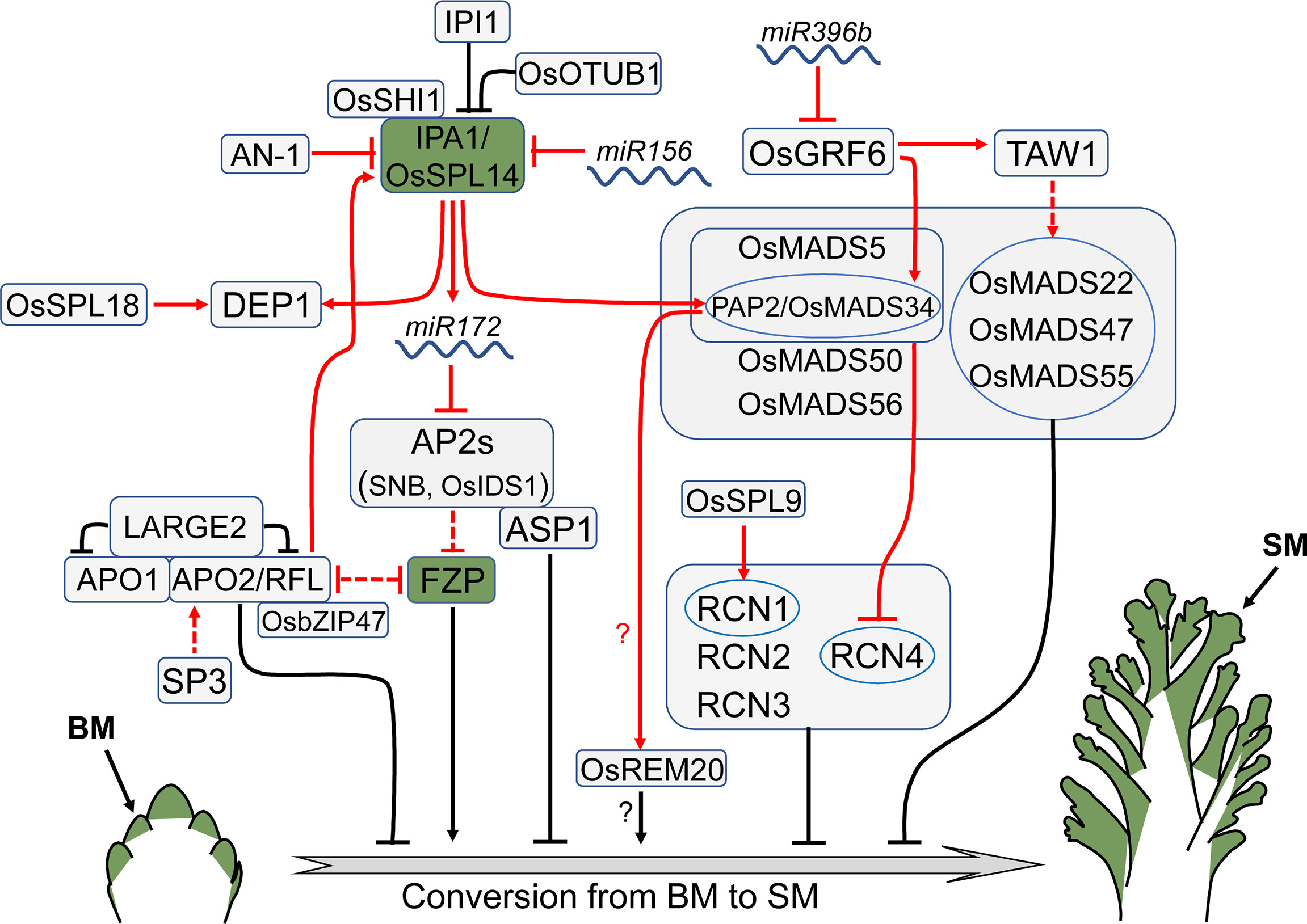
Figure 3 Regulators that control meristem identity conversion. FZP promotes the conversion of BM to SM, and its expression indicates that the meristem has acquired terminal identity. IPA1, also known as WFP/OsSPL14, is an important rice yield regulator that affects both meristem activity and identity conversion. Arrows and blocked arrows represent positive and negative regulation, respectively. Solid and dashed lines represent direct and indirect regulation, respectively. Red and black lines represent regulation between factors at transcriptional and protein level, respectively. BM, branch meristem; SM, spikelet meristem.
FRIZZY PANICLE (FZP), encoding an ERF (ethylene response factor) family transcription factor, promotes the conversion of BM identity to SM identity. The expression of FZP is restricted in a half-ring domain of SM before and during the formation of rudimentary glume meristem (Komatsu et al., 2003). In the loss-of-function fzp mutant, the generation of spikelets is prevented, leading to the formation of higher order branches instead of spikelets (Komatsu et al., 2003). On the contrary, overexpression of FZP severely inhibits the development of secondary branches, resulting in significantly reduced length of inflorescence and number of secondary branches and spikelets (Bai et al., 2016).
FZP is a major negative regulator of ABERRANT PANICLE ORGANIZATION2 (APO2)/RICE FLORICAULA LEAFY (RFL), the rice ortholog of Arabidopsis LFY encoding a plant-specific transcription factor. The precise expression pattern of APO2/RFL in the vegetative stage remains controversial, since two studies on whether APO2 is expressed in SAM reported opposite results (Kyozuka et al., 1998; Ikeda-Kawakatsu et al., 2012). In the reproductive stage, APO2/RFL is expressed in both PBM and SBM, but its expression is downregulated after the branch formation (Kyozuka et al., 1998; Ikeda-Kawakatsu et al., 2012). Expression of APO2/RFL is increased in young panicles of the fzp mutant, but reduced in the FZP overexpression plants (Bai et al., 2016). Similar to the FZP overexpression plants, the apo2 loss-of-function mutant or transgenic lines with reduced APO2/RFL expression generate small inflorescence with decreased primary branches, due to the premature termination of BM (Rao et al., 2008; Ikeda-Kawakatsu et al., 2012). Interestingly, FZP is up-regulated in the RFL knock-down plants (Rao et al., 2008), implying mutual repression between FZP and APO2/RFL. The transcription factor OsbZIP47 interacts with APO2/RFL, suppressing BM identity conversion to SM. Knocking down OsbZIP47 leads to a reduction in inflorescence axis length, primary branch number and spikelet number (Prakash et al., 2022).
SHORT PANICLE3 (SP3), which encodes a Dof transcription factor, also regulates APO2/RFL. The expression of APO2/RFL is down-regulated in the young panicle of the sp3 mutant, resulting in a reduction in panicle length, secondary branch number, and spikelet number (Huang et al., 2019). Another IM identity regulator APO1 directly interacts with APO2, coordinately controlling rice IM identity (Ikeda-Kawakatsu et al., 2012). APO1 is an F-box protein that positively controls panicle branch number and spikelet number by inhibiting the conversion of IM to SM. The expression of APO1 is detectable in SAM and BM, particularly in the outer layers of the rachis meristem and PBM (Ikeda et al., 2007). The apo1 mutant has fewer panicle branches and spikelets, whereas APO1 overexpression results in opposite effects (Ikeda et al., 2007). Recent studies reveal that APO1 and APO2 physically interact with LARGE2, a HECT-domain E3 ubiquitin ligase OsUPL2. The APO1-APO2 complex is accumulated in the large2 mutant, contributing to a larger inflorescence and increased grain number in comparison with wild type (Huang et al., 2021).
ABERRANT SPIKELET AND PANICLE1 (ASP1) encodes a TOPLESS-related transcription corepressor, the rice homolog of Arabidopsis TOPLESS (TPL) which determines SAM fate (Long et al., 2006). Similarly, ASP1 in rice is also involved in determination of meristem identity. The expression of ASP1 is strong during the initiation of IM and BM. Similar to APO1, ASP1 exhibits higher expression in the outer region than the inner region of BM (Yoshida et al., 2012). The asp1 mutant shows shortening of primary branch and the reduction of spikelet number, as the result of early conversion of BM into SM (Yoshida et al., 2012). ASP1 interacts with several APETALA2 (AP2) family transcription factors, such as SUPERNUMERARY BRACT (SNB) and ORYZA SATIVA INDETERMINATE SPIKELET1 (OsIDS1), which positively regulate panicle branch number through inhibiting the acquisition of SM identity (Wang et al., 2015). The expression of FZP in the snb osids1 double mutant precedes that in wild type, indicating that the BM of the snb osids1 double mutant is transformed into SM in advance, contributing to fewer branches and spikelets (Lee and An, 2012). As the targets of miR172, SNB and OsIDS1 are down-regulated in the miR172 overexpression plants, which display a phenotype identical to the snb osids1 double mutant (Lee and An, 2012).
IDEAL PLANT ARCHITECTURE1 (IPA1), also known as WEALTHY FARMER’S PANICLE (WFP), simultaneously regulates tiller number and inflorescence size, shaping rice plant with ideal architecture (Jiao et al., 2010; Miura et al., 2010). IPA1, encoding a plant-specific transcription factor SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 14 (OsSPL14), is a target of miR156. IPA1 is expressed in both SAM and IM, especially highly in PBM and SBM (Jiao et al., 2010; Miura et al., 2010). Appropriately elevating IPA1 expression, such as by interfering with miR156 expression, altering the miR156 target site in IPA1 or reducing epigenetic repression of IPA1, can produce larger inflorescences and control the number of tillers at an optimal level, resulting in enhanced yield (Jiao et al., 2010; Miura et al., 2010; Wang et al., 2015; Zhang et al., 2017). However, overexpression of IPA1 at high levels results in a small inflorescence with fewer branches and spikelets, particularly the secondary branches (Wang et al., 2015; Du et al., 2017). These morphological alterations may be achieved by regulating meristem identity conversion, as evidenced by the ectopic expression of FZP in BM of the IPA1 overexpression plants (Wang et al., 2015; Du et al., 2017). IPA1 is a directly target of APO2 in regulating inflorescence architecture. Overexpression of IPA1 recovers the panicle defects of the apo2 mutant, indicating that APO2 acts upstream of IPA1 (Miao et al., 2022).
Being a transcription factor, IPA1 binds directly to the promoter of miR172 and activates its expression, thus inhibiting the AP2 genes (Wang et al., 2015). IPA1 also directly targets and activates DENSE AND ERECT PANICLE 1 (DEP1) (Lu et al., 2013). Another plant-specific transcription factor, ORYZA SATIVA SHORT INTERNODES1 (OsSHI1), is involved in this regulation (Duan et al., 2019a). OsSHI1 interacts with IPA1 and inhibits its transcriptional activity by affecting the binding of IPA1 to the DEP1 promoter (Duan et al., 2019a). The level of IPA1 protein is regulated by the RING-finger ubiquitin E3 ligase, IPA1 INTERACTING PROTEIN1 (IPI1). IPI1 stimulates IPA1 degradation by adding K48-linked polyubiquitin chains in panicles but stabilizes IPA1 by adding K63-linked polyubiquitin chains in SAM. As a result, IPI1 controls panicle branching and tillering by differentially regulating IPA1 levels in various rice tissues (Wang et al., 2017a). IPA1 also interacts with the deubiquitinating enzyme OsOTUB1, which restricts the K63-linked ubiquitination of IPA1, in turn promoting IPA1 degradation through the K48Ub dependent-proteasome pathway. OsOTUB1 deficiency or reduced expression leads to the accumulation of IPA1, resulting in a large inflorescence (Wang et al., 2017b).
In Arabidopsis, several MADS-box genes determine floral meristem identity by directly inhibiting IM transition factor TERMINAL FLOWER1 (TFL1) (Liu et al., 2013). This genetic mechanism seems conserved in rice. Loss-of-function mutation of PANICLE PHYTOMER2 (PAP2)/OsMADS34 prevents newly generated meristems from developing into SM, resulting in more branches and spikelets. However, a large number of branches and spikelets are aborted at basal nodes of the panicle (Gao et al., 2010; Kobayashi et al., 2010; Zhu et al., 2022). Inactivation of other MADS-box genes, such as OsMADS5, SHORT VEGETATIVE PHASE (SVP) (OsMADS22, OsMADS47 and OsMADS55), SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1) (OsMADS50 and OsMADS56) in the pap2/osmads34 mutant gives rise to further increase of inflorescence branches, even generates tertiary branches, but the numerous branches are severely aborted (Liu et al., 2013; Zhu et al., 2022). This phenotype resembles the rice plants overexpressing RICE CENTRORADIALISs (RCNs) which are the rice orthologs of Arabidopsis TFL1 (Nakagawa et al., 2002; Zhu et al., 2022). Particularly, RCN4 is highly and ectopically expressed in the PBM of OsMADSs multiple knock-down plants, demonstrating that these MADS-box genes promote inflorescence meristem identity conversion by suppressing RCN4 expression (Liu et al., 2013). A recent study further confirms that OsMADS5 and OsMADS34 directly target RCN4 (Zhu et al., 2022). However, overexpression of two SVP genes, OsMADS22 and OsMADS55 separately, delays conversion of BM to SM, leading to increased secondary branch number (Yoshida et al., 2013). These studies suggest that SVP genes and other MADS-box genes may play opposite roles in determining rice inflorescence architecture, and SVPs could directly suppress meristem identity conversion instead of promoting conversion through suppressing RCNs expression.
The MADS-TFL1 module in controlling meristem identity conversion and inflorescence architecture is regulated by SPL genes, such as SPL3 and SPL9, in Arabidopsis (Wang et al., 2009; Yamaguchi et al., 2009). This mechanism seems also conserved in rice, evidenced by that IPA1/OsSPL14 directly binds the PAP2/OsMADS34 promoter and up-regulates its expression (Wang et al., 2015). Meanwhile, overexpression of RCN1 in the OsSPL14 knock-down plants can rescue the secondary branch defects, suggesting that OsSPL14 acts upstream of RCN1 in controlling conversion of BM to SM (Wang et al., 2015). Recently, OsSPL9 has been identified as a positive regulator of secondary branch number, which directly activates RCN1 expression (Hu et al., 2021a), implying that SPLs mediate meristem identity conversion may not always through MADS-box genes.
REPRODUCTIVE MERISTEM20 (OsREM20), encoding a B3 domain transcription factor, is expressed in the reproductive meristems, including PBM, SBM and SM. The loss-of-function mutation of OsREM20 generates small inflorescence with reduced grain number. OsMADS34 directly binds the CArG box-containing inverted repeat sequence in the OsREM20 promoter and activates OsREM20 expression (Wu et al., 2021). However, whether OsREM20 regulates rice inflorescence architecture through controlling rice meristem identity conversion and how OsMADS34 activates OsREM20 expression as a transcriptional repressor is unknown.
TAWAWA1 (TAW1) encodes a protein with an unknown function that influences rice inflorescence architecture by blocking the transition from BM to SM (Yoshida et al., 2013). TAW1 is highly expressed in SAM and BM but disappears after BM formation. In the gain-of-function mutant tawawa1-d, the IM activity is prolonged and spikelet formation is delayed due to the continuously high expression of TAW1 in BM and SM, resulting in branch extension and spikelet number increase. On the contrary, reduced TAW1 expression causes IM prematuration and early formation of spikelets, producing small inflorescence (Yoshida et al., 2013). TAW1 acts upstream of the SVP genes and positively regulates their expression to suppress SM identity (Yoshida et al., 2013). GROWTH-REGULATING FACTOR6 (OsGRF6) directly binds the promoter of TAW1 and OsMADS34, positively controlling secondary branch number. Overexpression of OsGRF6 leads to the increase of both primary and secondary branches, while knocking down OsGRF6 generates abnormal inflorescences with no secondary branch (Gao et al., 2015). miR396b targets OsGRF6 and suppresses its expression. Consistent with the role of OsGRF6, plants overexpressing the target mimicry of miR396b generate more secondary branches, but the miR396b overexpression plants have no secondary branch. Both TAW1 and OsMADS34 are up-regulated in the miR396b mimicry plants, suggesting that OsGRF6 could activate those two genes (Gao et al., 2015). Because TAW1 and OsMADS34 play opposing roles in determining rice inflorescence architecture, the function of OsGRF6 may be mediated predominantly by TAW1, at the very least by balancing TAW1 and OsMADS34 expression.
Control of branch elongation
Panicle branch elongation is another crucial event during rice inflorescence development. Although the development of BM has been extensively studied, the research on branch elongation in rice is still quite limited.
SHORT PANICLE1 (SP1)/PANICLE LENGTH3 (PAL3) encodes a polypeptide transporter located on the plasma membrane and is involved in the regulation of rice panicle branch elongation. Development of IM is normal in the loss-of-function mutants sp1/pal3, but in the later stage, the branches of the sp1/pal3 mutants could not extend properly, resulting in delayed elongation or even degeneration of branches and subsequently short inflorescences (Li et al., 2009; Shang et al., 2021). SP1 is highly expressed in the branch phloem of the inflorescence, consistent with its role in branch elongation (Li et al., 2009).
DEP2 (DENSE AND ERECT PANICLE2) influences the elongation of the main axis and inflorescence branches. The dep2 mutant generates the dense and erect inflorescence. DEP2 encodes a plant-specific protein with unknown function and is highly expressed in rachis and branches of the young panicle. Several cell cycle genes are down-regulated in the dep2 mutant, suggesting that cell proliferation is affected (Li et al., 2010).
Gibberellins (GAs) play a vital role in controlling organ elongation including rice inflorescence. This topic will be discussed in the ‘Gibberellins’ section below.
Roles of phytohormones in controlling inflorescence architecture
Phytohormones are tiny regulatory molecules that affect almost all aspects of plant growth and development. Rice inflorescence formation and development are regulated by a range of hormones, such as cytokinins, auxin and gibberellins. These hormones frequently communicate with and affect one another, making them essential components of rice yield attributes.
Cytokinins
Cytokinins (CKs) are adenine-derived compounds that are mainly involved in plant cell division (Werner et al., 2001; Schaller et al., 2014; Yang et al., 2021), playing a crucial role in many plant development processes, such as promoting the initiation and maintenance of SAM, regulating the development of flower organs, and determining the root meristem size (Wybouw and De Rybel, 2019). Many genes have recently been discovered to regulate CK homeostasis and signaling, which affect meristem activity and inflorescence development in rice (Figure 4).
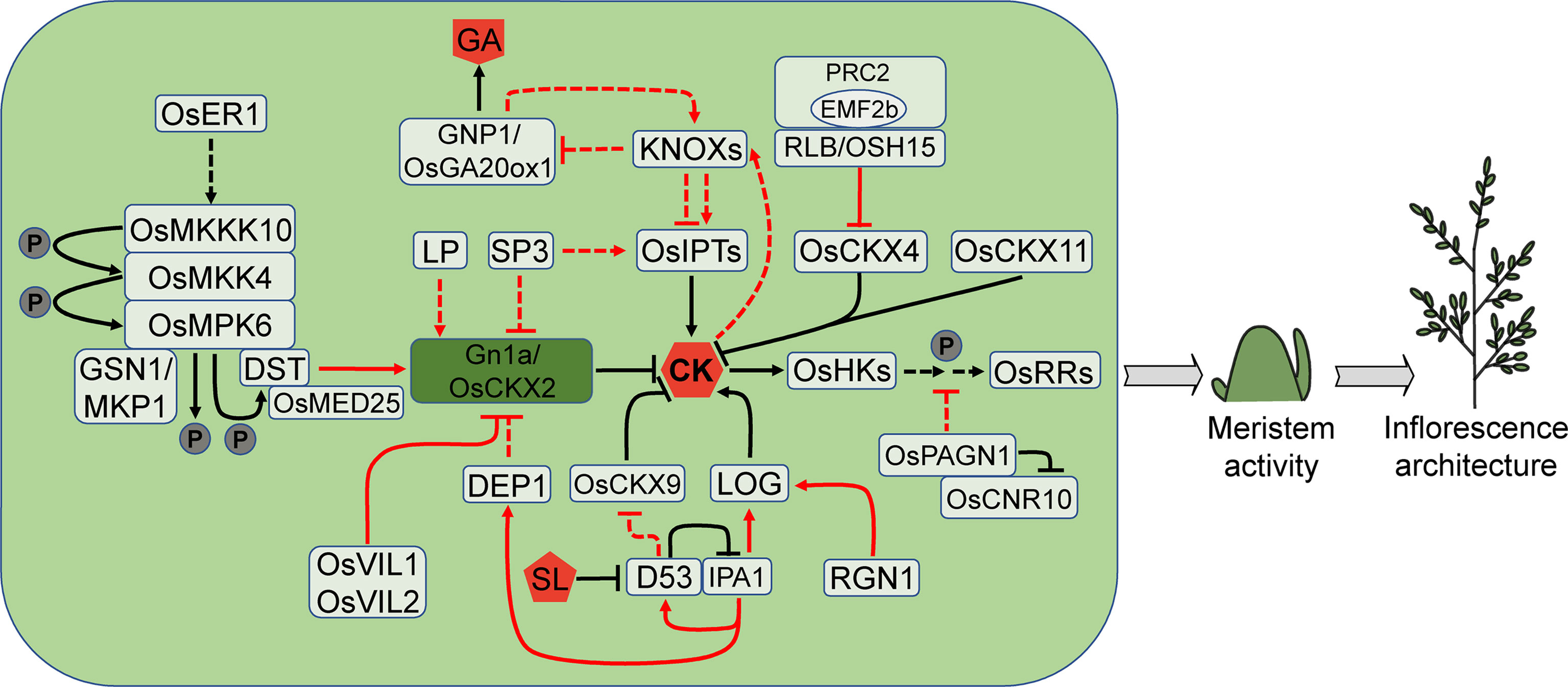
Figure 4 CK pathways that are involved in rice meristem activity. CK stimulates meristem activity and has the potential to increase yield. The CK levels are modulated by several enzymes, such as OsCKXs, LOGs and OsIPTs. Noticeably, Gn1a/OsCKX2, as a master regulator of rice inflorescence size and yield, is controlled by numerous other factors. CK also collaborates with other hormones such as GA and SL, in controlling rice inflorescence architecture. Arrows and blocked arrows represent positive and negative regulation, respectively. Solid and dashed lines represent direct and indirect regulation, respectively. Red and black lines represent regulation between factors at transcriptional and protein level, respectively. P, phosphorylation; CK, cytokinin; GA, gibberellin; SL, strigolactone.
The CK signaling transduction is mediated by the two-component system (TCS) that includes receptor histidine kinases (HKs), histidine phosphotransfer proteins (HPts) and response regulators (RRs). Knockout of rice CK receptors OsHK5/OHK3 or OsHK6/OHK5 impairs various aspects of rice development, including root and shoot growth (Burr et al., 2020). The panicle length, branch number and spikelet number are all reduced in the hk5 and hk6 single mutants, and the hk5 hk6 double mutant displays more severe defects. These abnormalities in the hk mutants probably arise from the poor establishment of IM (Burr et al., 2020). Type-B RRs, as transcription factors, function downstream of HKs (Sakai et al., 2001; Mason et al., 2005; Zubo et al., 2017; Xie et al., 2018). The rice rr21 rr22 rr23 triple mutant produces shorter inflorescences and fewer branches, leading to the reduction in spikelet number (Worthen et al., 2019). Type-A RRs are primary responsors to CK signaling and regulated by type-B RRs (Brandstatter and Kieber, 1998; Rashotte et al., 2003; Brenner et al., 2005). Overexpression of type-A OsRR6 generates small inflorescences with fewer branches and spikelets (Hirose et al., 2007).
PLANT ARCHITECTURE AND GRAIN NUMBER1 (OsPAGN1) encodes a RING U-box protein, likely an E3 ubiquitin ligase. Knockout of OsPAGN1 results in an increase in primary branch number and grain number. The expression of several CK signaling genes including type-A OsRR9/10, HPt gene OsAHP1 and OsAHP2 is elevated in the pagn1 mutant, implying that OsPAGN1 may negatively regulate CK signaling. OsPAGN1 interacts with CELL NUMBER REGULATOR10 (OsCNR10) and probably ubiquitinates OsCNR10 for degradation. However, it is very perplexing that the cnr10 mutant shows similar changes to the pagn1 mutant in inflorescence architecture (Yan et al., 2022). Other regulators may be involved in the interaction of the two proteins.
GRAIN NUMBER 1a (Gn1a), one major QTL controlling rice grain number per panicle, encodes CK oxidase/dehydrogenase OsCKX2 which can irreversibly degrade the active CK into adenine or adenosine and side chain. Gn1a is predominantly expressed in the vascular tissue of young panicles and culms. The decreased Gn1a expression leads to the accumulation of CKs in IM and the increased meristem activity, resulting in the increased grain number and yield (Ashikari et al., 2005).
Numerous genes so far have been discovered to influence CK contents in IM through modulating Gn1a expression, which consequently affects inflorescence architecture and yield. DEP1 is another major QTL that controls rice inflorescence architecture. The dominant allele dep1 leads to truncation of the phosphatidylethanolamine-binding protein-like domain protein, resulting in increased meristem activity, short and erect inflorescence. In the NIL-dep1 plants, Gn1a expression is decreased significantly, indicating that DEP1 may affect meristem activity by regulating CKs accumulation (Huang et al., 2009). SP3 can affect CK contents in young inflorescence. The expression of CK synthesis gene ISOPENTENYL TRANSFERASEs (OsIPTs) is decreased in the sp3 mutant, but the degradation genes, including OsCKX2 and other OsCKXs, are up-regulated, resulting in a drop in trans-zeatin (tZ) and a reduction in inflorescence size (Huang et al., 2019). LARGER PANICLE (LP) encodes a kelch repeat-containing F-box protein that negatively regulates inflorescence size by affecting OsCKX2 expression. The loss of function of LP leads to an increase in branch number and grain number, which is most likely attributed to up-regulation of CK levels (Li et al., 2011). The chromatin interacting factor VIN3-LIKE1 (OsVIL1) and OsVIL2 directly binds to the promoter of OsCKX2 and regulates H3K27 methylation, resulting in decreased OsCKX2 expression, increased CK levels and an increase in the number of branches and grains (Yang et al., 2019; Yoon et al., 2021).
OsCKX2 promoter can also be bound by the zinc finger transcription factor DROUGHT AND SALT TOLERANCE (DST). REGULATOR OF Gn1a (REG), the semi-dominant allele of DST, disrupts OsCKX2 expression mediated by DST, leading to higher CK levels in the IM, increased meristem activity, and more branches and grains (Li et al., 2013). DST physically interacts with the Mediator complex subunit 25 (OsMED25) which is probably a coactivator of DST. The inflorescence phenotype of OsMED25 knock-down plants and the osmed25 mutant is similar to the dst/reg mutant. The DST-OsMED25 complex recruits RNA polymerase II (Pol II) to activate OsCKX2 expression (Lin et al., 2022). The DST-OsCKX2 module for inflorescence development is also regulated by the OsMKKK10-OsMKK4-OsMPK6 cascade signal which adversely affects spikelet formation (Guo et al., 2018; Guo et al., 2020). OsMPK6 interacts with and phosphorylates DST, enhancing the transcriptional activation capacity of DST on OsCKX2, boosting CK degradation and maintaining normal CK levels during inflorescence development (Guo et al., 2020). The receptor-like kinase ERECTA1 (OsER1) acts upstream of the OsMKKK10-OsMKK4-OsMPK6 cascade signal and modulates OsMPK6 phosphorylation level. The oser1 loss-of-function mutant has a lower level of OsMPK6 phosphorylation and produces more spikelets. OsMPK6 interacts with and is dephosphorylated by the mitogen-activated protein kinase phosphatase GRAIN SIZE AND NUMBER1 (GSN1)/OsMKP1, resulting in deactivation which positively regulates spikelet number (Guo et al., 2018). Suppressing GSN1 expression causes decreased spikelet number, while overexpression leads to increased spikelet number. The gsn1 osmpk6 double mutant produces much more spikelets than the gsn1 mutant, but fewer than the osmpk6 mutant (Guo et al., 2018; Guo et al., 2020).
Overexpression of OsCKX4 also leads to changes in inflorescence architecture as demonstrated by sparser and smaller panicles (Wang et al., 2022). The KNOX protein RICE LATERAL BRANCH (RLB)/OSH15 epigenetically suppresses OsCKX4 expression through recruiting OsEMF2b, a component of the polycomb repressive complex 2 (PRC2) which mediates H3K27 tri-methylation on target genes. The loss-of-function mutant rlb has shorter inflorescences with reduced spikelets, due to the decrease of secondary branches (Wang et al., 2022).
It is intriguing that either knockout or overexpression of another CK oxidase/dehydrogenase gene OsCKX9 leads to the decrease of panicle length, number of branches and grains (Duan et al., 2019b). This reveals that the steady-state levels of the OsCKX9 expression may play a key role in the regulation of rice inflorescence architecture. However, OsCKX9 may not directly regulate inflorescence size but through regulating tillering. The smaller panicle is perhaps a tradeoff effect of increased tiller number. OsCKX9 is a primary responsor to the strigolactone (SL) signaling as its expression is up-regulated within 1 h by SL treatment. This induction is dependent on the SL signaling repressor DWARF53 (D53) which inhibits the IPA1 transcriptional activation activity by interacting with it. In turn, IPA1 directly regulates D53 expression, forming a negative feedback loop (Song et al., 2017; Duan et al., 2019b). In the shoot base of the gain-of-function d53 mutant, the OsCKX9 expression is decreased, leading to the significant increase of CK contents (Duan et al., 2019b).
Another CK oxidase/dehydrogenase gene OsCKX11 simultaneously mediates leaf senescence and grain number. The osckx11 mutant shows delayed leaf senescence under dark treatment and produces a larger inflorescence with more primary branches and high grain yield. Expression pattern analysis shows that OsCKX11 is highly expressed in PBM, SBM and SM (Zhang et al., 2021).
CK synthesis involves two families of key genes, OsIPTs and LONELY GUYs (LOGs). LOG encodes CK-activating enzyme, which directly converts inactive CK nucleosides into free active CKs. LOG is expressed at a low level in a small region of upper SAM but highly in BM and SM. In the log mutant, the IM ceases soon after generating a modest amount of lateral meristem in the reproductive stage, resulting in a reduction of inflorescence size and fewer branches and spikelets (Kurakawa et al., 2007). The LOG expression is directly regulated by the R2R3 MYB transcription factor REGULATOR OF GRAIN NUMBER1 (RGN1). The mutation of RGN1 leads to the absence of lateral spikelets on secondary branches (Li et al., 2022). The promoter of LOG is also bound by IPA1 (Lu et al., 2013; Du et al., 2017), implying that IPA1 mediates inflorescence architecture by altering CK levels directly or indirectly.
CKs may maintain SAM activity in rice through controlling the expression of KNOXs, which are induced by CKs (Tsuda et al., 2011). Overexpression of CK signalling genes, such as OHK3, OHP2 and type-B RRs, promotes CK-mediated induction of OSH1. However, the response of OSH1 to exogenous CKs lags behind that of the CK primary response genes, type-A RRs, suggesting that type-B RRs may not directly regulate OSH1 (Naruse et al., 2018). In the formation and maintenance of rice SAM, there appears to be a complicated connection between the CK synthesis genes OsIPTs and KNOXs. Increasing KNOXs expression induces OsIPT2 and OsIPT3, but inhibits other OsIPTs. In the aboveground tissues, overexpression of OSH1 and OsIPT3 results in comparable phenotypes. Apart from upregulating the CK synthesis genes, KNOXs inhibit expression of the gibberellins (GAs) synthesis genes OsGA20oxs, permitting the meristem to maintain a state of high-level CKs and low-level GAs, which is required for meristem establishment and maintenance (Sakamoto et al., 2006).
Auxin
Auxin is the first plant hormone identified, and it is involved in many aspects of plant growth and development, such as embryogenesis, root structure, geotropism, phototropism, and the formation of plant lateral organs (Rashotte et al., 2000; Reinhardt et al., 2000; Benková et al., 2003; Blakeslee et al., 2004; Möller and Weijers, 2009). Auxin is mainly synthesized in the shoot tip and transported downward by polar transport, indirectly suppressing the formation of axillary buds (Sieberer and Leyser, 2006; Zwiewka et al., 2019). During the formation of the rice inflorescence, auxin is involved in the initiation and maintenance of the axillary meristem, which influences the development of panicle branches (Deveshwar et al., 2020) (Figure 5).
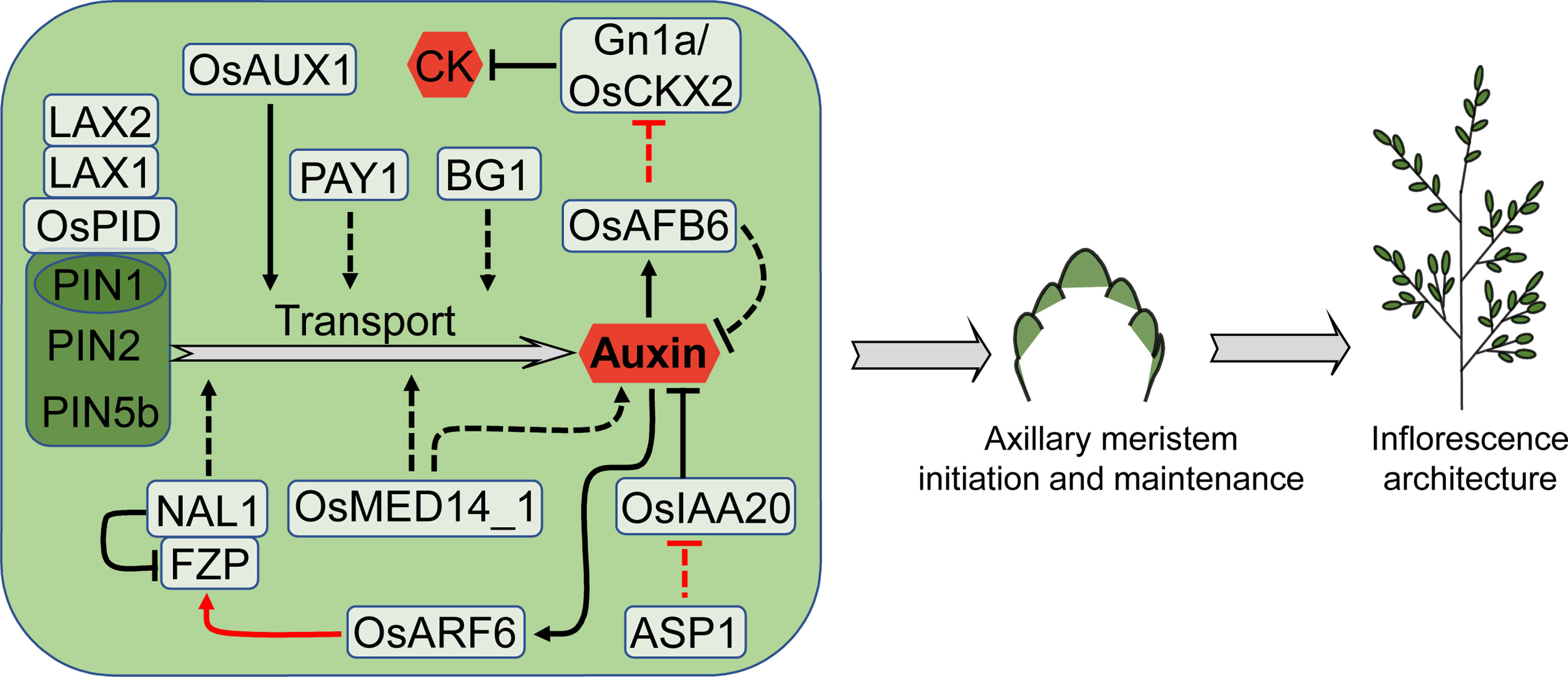
Figure 5 Factors involved in auxin-mediated regulation of rice inflorescence architecture. Auxin plays a vital role in axillary meristem initiation and maintenance at both vegetative and reproductive stage. PINs-mediated auxin transport is a crucial process for auxin function. Auxin also interacts with CK in determining rice inflorescence architecture. Arrows and blocked arrows represent positive and negative regulation, respectively. Solid and dashed lines represent direct and indirect regulation, respectively. Red and black lines represent regulation between factors at transcriptional and protein level, respectively. CK, cytokinin.
Auxin transport through tissues is essential for many aspects of plant development. This movement is regulated by auxin transporters, among which the PIN-FORMED (PIN) protein family plays a vital role in accelerating the outward transport of auxin from cells (Zazímalová et al., 2010). PIN1 is the first auxin efflux carrier discovered in Arabidopsis, having polarity in the plasma membranes of root, stem, inflorescence axis, and embryo cells, and its loss-of-function mutant has a substantial impact on organ initiation (Okada et al., 1991; Gälweiler et al., 1998; Friml et al., 2003; Blilou et al., 2005; Adamowski and Friml et al., 2015). There are four PIN1 homologues (PIN1a-PIN1d) in rice. No evident phenotypic change is observed in any single pin1 mutant, but the pin1a pin1b double mutation changes root architecture and results in a wider panicle branching angle; the pin1c pin1d double mutant has fewer branches and no spikelet (Li et al., 2019; Liu et al., 2022). Overexpression of OsPIN2, another member of PIN family, promotes auxin transport from the shoot to the root-shoot junction, causing a higher non-tissue-specific concentration of free auxin at the root-shoot junction. This non-specific auxin accumulation gives rise to reduced plant height, increased tillering, shorter panicles and fewer grains (Chen et al., 2012). Another member of PIN family OsPIN5b participates in auxin homeostasis, transportation and distribution, thereby regulating rice plant architecture and yield (Mravec et al., 2009; Barbez et al., 2012; Lu et al., 2015a). OsPIN5b overexpression causes diverse morphologies, including decreased plant height, fewer tillers, lower seed-setting rate and shorter panicles. On the contrary, knockdown of OsPIN5b produces more tillers, a better developed root system and longer panicles (Lu et al., 2015a).
Serine/threonine protein kinase OsPINOID (OsPID) interacts with PIN1a and PIN1b to govern polar transport and distribution of auxin, and modulates the formation and development of rice flower organs. OsPID expression is high in young panicles and its overexpression plants have more panicle branches and grains (Wu et al., 2020). The bHLH transcription factor LAX PANICLE1 (LAX1) also interacts with OsPID, probably controlling inflorescence architecture through affecting auxin polar transport (Wu et al., 2020), and this mechanism is conserved in maize (Gallavotti et al., 2004). The expression of LAX1 is not detected in SAM but restricted to the boundary between the inflorescence rachis and the region of new meristem formation. With the elongation of new meristem, LAX1 expression is gradually diminished. The lax1 mutant lacks lateral spikelets in favor of sole terminal spikelets (Komatsu et al., 2001). LAX1 interacts with LAX2, and their double mutations enhance the phenotype of the lax1 mutant, indicating that LAX1 governs the inflorescence meristem initiation by controlling auxin signal transduction and transport, either independently or in collaboration with LAX2. Unlike LAX1, LAX2 expression is observed in PBM, SBM and SM, covering the expression domain of LAX1 (Tabuchi et al., 2011).
By high-throughput single-cell RNA sequencing technology, auxin influx transporter gene OsAUX1 is identified to have enriched expression in BM. Consistent with its expression pattern, the loss-of-function mutant of OsAUX1 produces smaller inflorescence with reduced branch and spikelet number (Zong et al., 2022).
A few other genes also affect inflorescence architecture through modulating auxin transport. PLANT ARCHITECTURE AND YIELD1 (PAY1) encodes a nuclear-localized peptidase, controlling rice plant architecture by influencing the auxin polar transport and the level of endogenous indole 3-acetic acid (IAA). Overexpression of PAY1 produces a significant increase in grain number and yield (Zhao et al., 2015). BIG GRAIN1 (BG1) encodes a membrane-localized protein that, when overexpressed, can increase grain and inflorescence size as well as yield through altering auxin response and transport (Liu et al., 2015a). OsMED14_1 is a subunit of the Mediator complex that regulates diverse biological processes. In the OsMED14_1 knock-down plants, auxin level and PINs expression decreased, giving rise to fewer primary and secondary branches as well as spikelet (Malik et al., 2020). NARROW LEAF1 (NAL1) encodes a trypsin-like serine/cysteine protease, which controls rice leaf width by regulating auxin polar transport activity (Qi et al., 2008; Jiang et al., 2015). Overexpression of the NAL1 alleles LSCHL4 and SPIKELET NUMBER (SPIKE) results in larger inflorescences and higher yields (Fujita et al., 2013; Zhang et al., 2014). NAL1 interacts with FZP and promotes the degradation of FZP. Down-regulation of FZP or up-regulation of NAL1 increases the number of secondary branches, grains per panicle and yield per plant (Huang et al., 2018).
Auxin receptor TRANSPORT INHIBITOR RESPONSES (TIRs)/AUXIN SIGNALING F-box (AFBs) are a class of F-box protein. Overexpression of OsAFB6 in rice causes a rise in primary branches and spikelets due to reduced IAA levels and Gn1a expression, resulting in an increase in CK contents and IM activity (He et al., 2018). AUXIN RESPONSE FACTORs (ARFs) are auxin signal response factors, of which OsARF6 can directly bind to the promoter of FZP and activate its expression. A 4-bp tandem repeat deletion near the binding element affects the capacity of OsARF6 to regulate FZP, improving secondary branch number and grain yield (Huang et al., 2018). Similar to TPL in Arabidopsis, the meristem fate regulator ASP1 is also related to auxin signaling (Szemenyei et al., 2008). In the asp1 mutant, expression of the auxin signaling negative regulatory gene OsIAA20 is up-regulated, indicating that auxin signal is interrupted (Yoshida et al., 2012).
Gibberellins
Gibberellins (GAs) are a kind of tetracyclic diterpene hormones that regulate many aspects of plant development including organ elongation, seed germination and flowering (Cheng et al., 2004; Tyler et al., 2004; Kuroha et al., 2018). GA signaling is received by the receptor GIBBERELLIN INSENSITIVE DWARF1 (GID1) (Ueguchi-Tanaka et al., 2005). Loss-of-function mutations of GID1 cause multiple phenotype changes including dwarfism, small inflorescence and reduced fertility (Ueguchi-Tanaka et al., 2005; Ayano et al., 2014; Wang et al., 2018). In recent years, several additional genes involved in GA synthesis and signaling have been discovered to influence rice inflorescence architecture by regulating IM activity or inflorescence branch elongation (Figure 6).
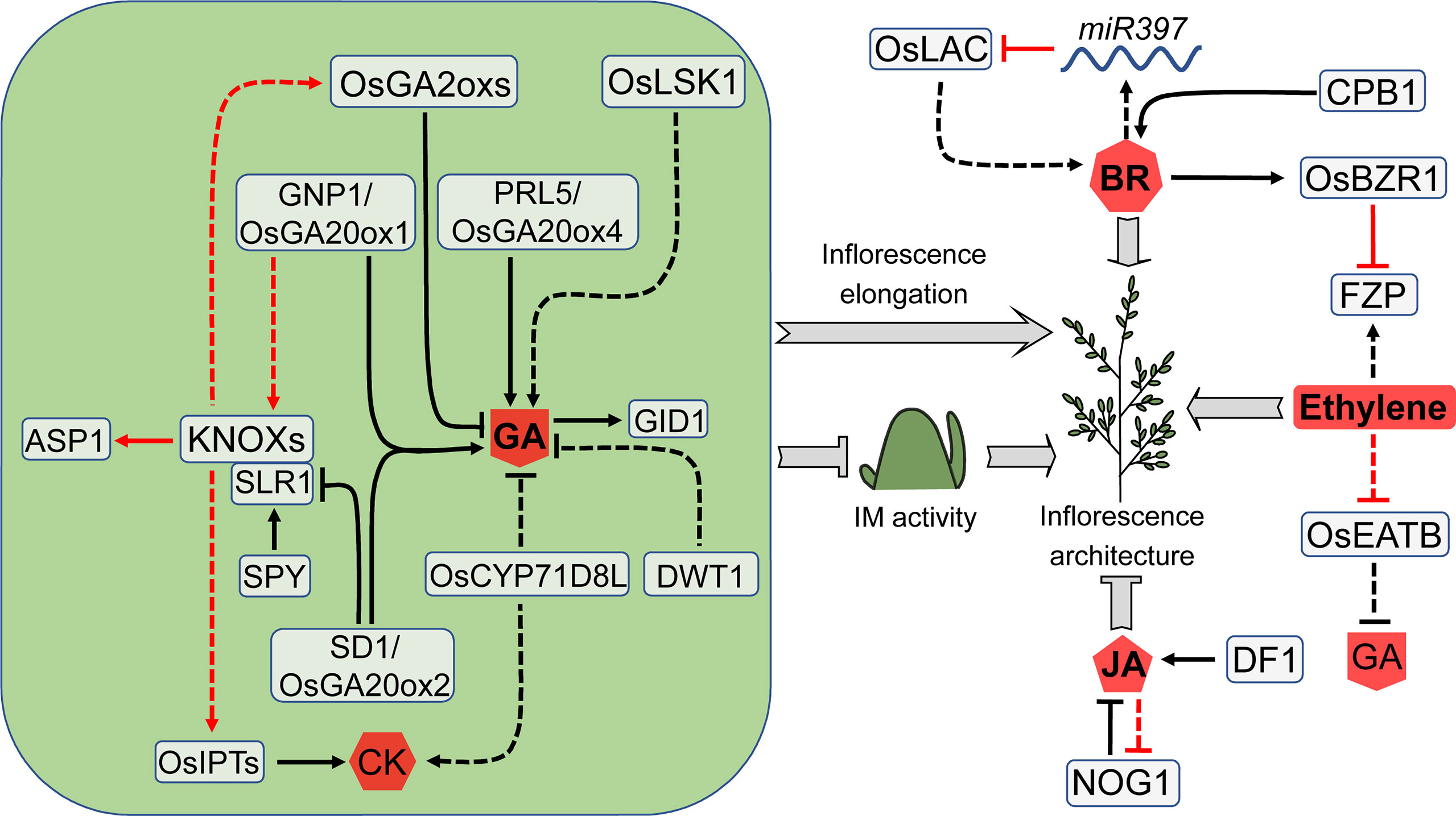
Figure 6 Factors involved in GA- and other hormone-mediated regulation of rice inflorescence architecture. Through an opposite way, GA promotes branch elongation or inhibits meristem activity to influence the rice inflorescences architecture. GA antagonizes CK in maintaining meristem activity, and KNOX genes act as coordinators between GA and CK. Several additional hormones also have a substantial impact on rice inflorescence architecture. These hormone regulators form a complicated network that influences one another. Arrows and blocked arrows represent positive and negative regulation, respectively. Solid and dashed lines represent direct and indirect regulation, respectively. Red and black lines represent regulation between factors at transcriptional and protein level, respectively. BR, brassinosteroid; CK, cytokinin; GA, gibberellin; JA, jasmonic acid.
Studies in Arabidopsis and rice show that GAs antagonize CKs in maintaining SAM activity, and KNOXs act as coordinators between these two hormones (Hay et al., 2002; Jasinski et al., 2005; Sakamoto et al., 2006). GRAIN NUMBER PER PANICLE1 (GNP1), a major QTL controlling the grain number in rice, encodes the GAs synthesis enzyme OsGA20ox1 and is expressed in SAM and BM (Wu et al., 2016b). GNP1 upregulation in IM leads to an increase of KNOXs in a feedback manner, which activates OsIPTs. Subsequently, elevated CK levels and KNOXs trigger expression of the GA catabolic genes GA2oxs, leading to enhanced active GAs catabolism. Therefore, the unfavorable effect of GA production on meristem activity is decreased because GA1 and GA3 are not accumulated in IM. This equilibrium mechanism improves yield by enhancing IM activity through boosting CKs activity and decreasing GAs activity (Wu et al., 2016b).
Overexpression or gain-of-function mutation of the cytochrome P450 monooxygenase gene OsCYP71D8L leads to the reduction in panicle length and grain number (Zhou et al., 2020), probably due to decreased GA levels. Surprisingly, CK levels increase in OsCYP71D8L overexpression plants (Zhou et al., 2020), but these enhanced CK levels do not compensate panicle defects induced by decreased GA levels. This study indicates that OsCYP71D8L functions mainly through GA rather than CK in regulating inflorescence architecture.
The rice ‘Green Revolution’ gene SEMI-DWARF 1 (SD1) encodes GA20ox2, which has been used in breeding for many years. Recent studies reveal that SD1 also participates in inflorescence development. The loss of function of SD1 causes a decrease in panicle length, branch number and grain number. Consistent with its function, SD1 is expressed in elongated PBM, SBM, and SM. The level of DELLA protein SLR1, a negative regulator of GA signaling, is increased dramatically in the sd1 mutant. SLR1 interacts with KNOXs to inhibit KNOXs-mediated activation of downstream genes, such as ASP1 (Su et al., 2021). Although both SD1 and GNP1 belong to GA20 oxidases, they may work independently to regulate inflorescence architecture due to their diverse spatiotemporal expression patterns (Su et al., 2021). PANICLE RACHIS LENGTH5 (PRL5) is a major QTL for rice panicle length, encoding another GA oxidases, OsGA20ox4. PRL5 overexpression increases GA levels in the inflorescence, resulting in elongation of the panicle axis (Agata et al., 2020).
SPINDLY (OsSPY) encodes N-acetyl glucosamine transferase, a negative regulator of GA signaling by enhancing the SLR1 activity (Shimada et al., 2006; Olszewski et al., 2010). The R833L substitution of OsSPY at the conserved C-terminus of the enzymatic domain leads to the decreased O-fucosyltransferase activity of OsSPY to SRL1, consequently increased panicle length, primary and secondary branch number and spikelet number (Yano et al., 2019).
LARGE SPIKE S-DOMAIN RECEPTOR LIKE KINASE 1 (OsLSK1) is an s-domain receptor kinase. Overexpression of truncated OsLSK1 increases the number of primary branches and grains, which is probably attributable to the upregulation of several critical genes involved in GAs synthesis and signaling (Zou et al., 2015). DWARF TILLER1 (DWT1) encodes a WUS-like homeobox transcription factor. The dwt1 mutant produces main culm with normal height and dwarf tillers with unelongated internodes. Aa a result, those two types of tillers generate large and small inflorescences, respectively. It seems that DWT1 is a direct regulator of tiller growth with indirect effect on panicle size. Interestingly, the expression of DWT1 is detectable in PBM and SBM but not in the elongating internode. The expression of GA20 oxidase genes is up-regulated in the dwt1 mutant, and their responsiveness to GAs is weakened, suggesting that DWT1 is directly or indirectly related to GA signaling pathway (Wang et al., 2014).
Other hormones
In addition to the three vital hormones mentioned above, several additional hormones are also involved in the regulation of rice inflorescence architecture (Figure 6).
Ethylene is a gaseous hormone that regulates a variety of plant growth processes. In rice, ethylene influences several essential agronomic traits, including flowering, grain size, and grain filling (Yin et al., 2017). Ethylene response factors (ERFs) are transcription factors that regulate ethylene signal transduction and response, among which OsEATB mediates the crosstalk between GA and ethylene. OsEATB expression is inhibited by ethylene, and its overexpression reduces GA contents, shortens the panicle length but increases grain number per panicle. OsEATB inhibits the ethylene-induced GA response by downregulating GA synthase, ent-kaurene synthase A (Qi et al., 2011). FZP is also an ERF domain-containing protein. As described above, FZP mediates the transformation of meristem identity, which affects inflorescence architecture and yield (Komatsu et al., 2003).
Brassinosteroids (BRs) are steroid hormones that regulate a wide range of biological processes, including plant development and stress response (Yang et al., 2011; Zhao et al., 2013; Gao et al., 2014; Saini et al., 2015; Divi and Krishna, 2009). DWARF11 (D11) encodes a cytochrome P450 protein that is involved in the BR biosynthesis. CLUSTERED PRIMARY BRANCH 1 (CPB1) is a D11 allele that controls the clustering of rice inflorescence branches and is highly expressed in young panicles. The cpb1 mutant has clustered primary branches and elongated internodes at the base of the main axis (Wu et al., 2016a). OsBZR1 is a primary regulator of BR signaling in rice, and its overexpression results in phenotypic alterations in anther and grain size, as well as an increase in grain number per panicle (Zhu et al., 2015). OsBZR1 inhibits FZP expression through binding to the CGTG motif which is located in an 18-bp fragment inserted 5.3 kb upstream of FZP, resulting in the increased number of branches and grains (Bai et al., 2017).
OsLAC encodes a laccase protein that modulates plant response to BRs. This protein may influence multiple rice development processes by controlling BR signals. The panicle length, branch number, and grain number are all decreased in the OsLAC overexpression plants (Zhang et al., 2013). OsLAC is a target of miR397. In contrast to OsLAC overexpression, miR397 overexpression increases the number of branches and grains. Plants with high levels of miR397 are more sensitive to BR treatment. Additionally, BR levels declined somewhat in miR397 overexpression plants but increased dramatically in OsLAC overexpression plants (Zhang et al., 2013).
Jasmonic acids (JAs) and derivatives are lipid-derived hormones that control plant defense responses and development processes, such as seed germination, root growth, tuber formation, tendril curling, trichome initiation, reproduction and aging (Kessler et al., 2004; Browse and Howe, 2008; Browse, 2009; Acosta and Farmer, 2010; Wasternack and Hause, 2013). In rice, JA suppresses spikelet growth and reduces yield through adversely regulating spikelet development (Kim et al., 2009; Cai et al., 2014). DOUBLE FLORET1 (DF1) is an allele of EXTRA GLUME1 (EG1) which controls spikelet development, encoding a plastid lipase involved in JA biosynthesis (Cai et al., 2014; Ren et al., 2018). The df1 mutant develops two complete florets with normal grain in one pair of glumes (Ren et al., 2018). NUMBER OF GRAINS 1 (NOG1) encodes an enoyl-CoA hydratase/isomerase that is involved in the regulation of JA synthesis and β-oxidation of fatty acid. A 12-bp insertion in the NOG1 promoter enhances NOG1 expression, resulting in higher levels of enoyl-CoA hydratase/isomerase, lower total fatty acid and linolenic acid levels, as well as lower JA levels. Changes in these components lead to more grains per panicle and higher yield. Excessively applying JAs reduces NOG1 expression and yield, suggesting that JAs have a negative impact on NOG1 expression and rice yield (Huo et al., 2017).
Conclusions and perspectives
Owing to the importance of rice inflorescence architecture for grain yield, studies on inflorescence development have risen remarkedly in recent years, and multiple relevant genes have been identified. These genes mainly function in meristem activity maintenance, meristem identity conversion, and phytohormones regulation. Nevertheless, several other crucial genes involved in rice inflorescence architecture, such as OsPDCD5 (Dong et al., 2021), OsTPR (Pasion et al., 2021), OsKNR2 (Chen et al., 2022) etc. are not discussed here due to their unclear or unrelated genetic mechanism. Although so many genes related to rice inflorescence development have been identified, how can the revealed genetic pathways be fine-tuned to create an ideal inflorescence architecture to maximize the yield? It is not easy to answer this question.
The first challenge is environment cues, such as light, temperature, humidity and nutrients. The ideal inflorescence architecture should be appropriate for local ecological niche. For example, the inflorescence of high-yielding japonica varieties grown in northern China is generally dense and erect, due to the gain-of-function mutation of DEP1 (Huang et al., 2009). Despite of the potential of improving grain yield, this allele is rarely used in indica varieties grown in southern China (Huang et al., 2009). One of the possible reasons could be that dense and erect panicle is prone to diseases, such as false smut, under the high temperature and humidity in southern China (Sun et al., 2020). On the contrary, the indica rice generates long and drooping inflorescences that provide more space for spikelets, establishing a microenvironment to prevent pathogen attacks.
Meanwhile, the tradeoff between the inflorescence architecture and other traits severely restricts breeding. In many cases, this tradeoff is controlled by common genes. FUWA encodes a protein that contains an NHL domain. The mutation of FUWA causes short and erect panicles, while the grains become smaller and thicker (Chen et al., 2015). DEP1, DEP2 and DEP3 also influence grain size apart from inflorescence architecture (Huang et al., 2009; Li et al., 2010; Qiao et al., 2011). Knockout of OsSPL4 or overexpression of OsSPL13 enlarges both inflorescences and grains (Si et al., 2016; Hu et al., 2021b). The loss of function of MOC1, LAX1, and LAX2 simultaneously reduces the number of tillers and panicle branches (Komatsu et al., 2001; Li et al., 2003; Tabuchi et al., 2011). Because the impacts of these genes on inflorescence architecture and other traits are not always beneficial to yield, resolving the tradeoff between inflorescence size and other morphologies to enhance yield is an issue worth investigating. Recently, a study addressed tradeoff effects between inflorescence size and tiller number. Using tilling-deletion-based screen for the IPA1 promoter by CRISPR-Cas9, a 54-bp deletion that contains an AN-1 binding site in the IPA1 promoter simultaneously increases tiller number and inflorescence size (Song et al., 2022). This research reveals a possibility for breeders that the association between multiple traits can be dissected, and genetic effects can be precisely modified.
Phytohormone is another challenge for creating ideal inflorescence architecture, owing to its intricacy and cross-talk at various regulatory levels. Auxin promotes axillary meristem initiation, and its efflux determines both inflorescence branch number and branch angle (Li et al., 2019; Liu et al., 2022). CK positively regulates inflorescence meristem activity, while GA is detrimental for meristem activity. This antagonism is linked by KNOXs and GNP1 (Jasinski et al., 2005; Sakamoto et al., 2006; Wu et al., 2016b). Apart from inflorescence architecture, ethylene, brassinosteroids, and JA usually act on other biological processes, such as grain filling, leaf angle and stress response (Yin et al, 2017; Tong and Chu, 2018). Pyramiding the pleiotropic positive effects of hormones by optimizing their levels or balancing their connections will be a challenging task in the future.
Author contributions
YC and AK wrote the manuscript. XL made valuable suggestions and revised the manuscript. All authors read and approved the final manuscript.
Funding
Research in our lab was funded by the Agricultural Variety Improvement Project of Shandong Province, China (grant number 2021LZGC020), and the Agricultural Science and Technology Innovation Program of Chinese Academy of Agricultural Sciences.
Acknowledgments
We thank China Scholarship Council (CSC) for providing full scholarship to AK for his Master and Ph.D. study, respectively.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adamowski, M., Friml, J. (2015). PIN-dependent auxin transport: Action, regulation, and evolution. Plant Cell 27, 20–32. doi: 10.1105/tpc.114.134874
Agata, A., Ando, K., Ota, S., Kojima, M., Takebayashi, Y., Takehara, S., et al. (2020). Diverse panicle architecture results from various combinations of Prl5/GA20ox4 and Pbl6/APO1 alleles. Commun. Biol. 3, 302. doi: 10.1038/s42003-020-1036-8
Aichinger, E., Kornet, N., Friedrich, T., Laux, T. (2012). Plant stem cell niches. Annu. Rev. Plant Biol. 63, 615–636. doi: 10.1146/annurev-arplant-042811-105555
Ashikari, M., Sakakibara, H., Lin, S., Yamamoto, T., Takashi, T., Nishimura, A., et al. (2005). Cytokinin oxidase regulates rice grain production. Science 309, 741–745. doi: 10.1126/science.1113373
Ayano, M., Kani, T., Kojima, M., Sakakibara, H., Kitaoka, T., Kuroha, T., et al. (2014). Gibberellin biosynthesis and signal transduction is essential for internode elongation in deepwater rice. Plant Cell Environ. 37, 2313–2324. doi: 10.1111/pce.12377
Bai, X., Huang, Y., Hu, Y., Liu, H., Zhang, B., Smaczniak, C., et al. (2017). Duplication of an upstream silencer of FZP increases grain yield in rice. Nat. Plants 3, 885–893. doi: 10.1038/s41477-017-0042-4
Bai, X., Huang, Y., Mao, D., Wen, M., Zhang, L., Xing, Y. (2016). Regulatory role of FZP in the determination of panicle branching and spikelet formation in rice. Sci. Rep. 6, 19022. doi: 10.1038/srep19022
Barbez, E., Kubeš, M., Rolčík, J., Béziat, C., Pěnčík, A., Wang, B., et al. (2012). A novel putative auxin carrier family regulates intracellular auxin homeostasis in plants. Nature 485, 119–122. doi: 10.1038/nature11001
Benková, E., Michniewicz, M., Sauer, M., Teichmann, T., Seifertová, D., Jürgens, G., et al. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115, 591–602. doi: 10.1016/s0092-8674(03)00924-3
Blakeslee, J. J., Bandyopadhyay, A., Peer, W. A., Makam, S. N., Murphy, A. S. (2004). Relocalization of the PIN1 auxin efflux facilitator plays a role in phototropic responses. Plant Physiol. 134, 28–31. doi: 10.1104/pp.103.031690
Blilou, I., Xu, J., Wildwater, M., Willemsen, V., Paponov, I., Friml, J., et al. (2005). The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433, 39–44. doi: 10.1038/nature03184
Brandstatter, I., Kieber, J. J. (1998). Two genes with similarity to bacterial response regulators are rapidly and specifically induced by cytokinin in Arabidopsis. Plant Cell 10, 1009–1019. doi: 10.1105/tpc.10.6.1009
Brenner, W. G., Romanov, G. A., Köllmer, I., Bürkle, L., Schmülling, T. (2005). Immediate-early and delayed cytokinin response genes of Arabidopsis thaliana identified by genome-wide expression profiling reveal novel cytokinin-sensitive processes and suggest cytokinin action through transcriptional cascades. Plant J. 4, 314–333. doi: 10.1111/j.1365-313X.2005.02530.x
Browse, J. (2009). The power of mutants for investigating jasmonate biosynthesis and signaling. Phytochemistry 70, 1539–1546. doi: 10.1016/j.phytochem.2009.08.004
Browse, J., Howe, G. A. (2008). New weapons and a rapid response against insect attack. Plant Physiol. 146, 832–838. doi: 10.1104/pp.107.115683
Burr, C. A., Sun, J., Yamburenko, M. V., Willoughby, A., Hodgens, C., Boeshore, S. L., et al. (2020). The HK5 and HK6 cytokinin receptors mediate diverse developmental pathways in rice. Development 147, dev191734. doi: 10.1242/dev.191734
Cai, Q., Yuan, Z., Chen, M., Yin, C., Luo, Z., Zhao, X., et al. (2014). Jasmonic acid regulates spikelet development in rice. Nat. Commun. 5, 3476. doi: 10.1038/ncomms4476
Chen, W., Chen, L., Zhang, X., Yang, N., Guo, J., Wang, M., et al. (2022). Convergent selection of a WD40 protein that enhances grain yield in maize and rice. Science 375, eabg7985. doi: 10.1126/science.abg7985
Chen, Y., Fan, X., Song, W., Zhang, Y., Xu, G. (2012). Over-expression of OsPIN2 leads to increased tiller numbers, angle and shorter plant height through suppression of OsLAZY1. Plant Biotechnol. J. 10, 139–149. doi: 10.1111/j.1467-7652.2011.00637.x
Chen, J., Gao, H., Zheng, X. M., Jin, M., Weng, J. F., Ma, J., et al. (2015). An evolutionarily conserved gene, FUWA, plays a role in determining panicle architecture, grain shape and grain weight in rice. Plant J. 83, 427–438. doi: 10.1111/tpj.12895
Cheng, H., Qin, L., Lee, S., Fu, X., Richards, D. E., Cao, D., et al. (2004). Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development 131, 1055–1064. doi: 10.1242/dev.00992
Chu, H., Qian, Q., Liang, W., Yin, C., Tan, H., Yao, X., et al. (2006). The FLORAL ORGAN NUMBER4 gene encoding a putative ortholog of Arabidopsis CLAVATA3 regulates apical meristem size in rice. Plant Physiol. 142, 1039–1052. doi: 10.1104/pp.106.086736
Clark, S. E., Williams, R. W., Meyerowitz, E. M. (1997). The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89, 575–585. doi: 10.1016/s0092-8674(00)80239-1
Deveshwar, P., Prusty, A., Sharma, S., Tyagi, A. K. (2020). Phytohormone-mediated molecular mechanisms involving multiple genes and QTL govern grain number in rice. Front. Genet. 11. doi: 10.3389/fgene.2020.586462
Divi, U. K., Krishna, P. (2009). Brassinosteroid: a biotechnological target for enhancing crop yield and stress tolerance. N. Biotechnol. 26, 131–136. doi: 10.1016/j.nbt.2009.07.006.
Dong, S., Dong, X., Han, X., Zhang, F., Zhu, Y., Xin, X., et al. (2021). OsPDCD5 negatively regulates plant architecture and grain yield in rice. Proc. Natl. Acad. Sci. U. S. A. 118, e2018799118. doi: 10.1073/pnas.2018799118
Du, Y., Liu, L., Li, M., Fang, S., Shen, X., Chu, J., et al. (2017). UNBRANCHED3 regulates branching by modulating cytokinin biosynthesis and signaling in maize and rice. New Phytol. 214, 721–733. doi: 10.1111/nph.14391
Duan, E., Wang, Y., Li, X., Lin, Q., Zhang, T., Wang, Y., et al. (2019a). OsSHI1 regulates plant architecture through modulating the transcriptional activity of IPA1 in rice. Plant Cell 31, 1026–1042. doi: 10.1105/tpc.19.00023
Duan, J., Yu, H., Yuan, K., Liao, Z., Meng, X., Jing, Y., et al. (2019b). Strigolactone promotes cytokinin degradation through transcriptional activation of CYTOKININ OXIDASE/DEHYDROGENASE 9 in rice. Proc. Natl. Acad. Sci. U. S. A. 116, 14319–14324. doi: 10.1073/pnas.1810980116
Eshed, W. (2021). Genetics of shoot meristem and shoot regeneration. Annu. Rev. Genet. 55, 661–681. doi: 10.1146/annurev-genet-071719-020439
Friml, J., Vieten, A., Sauer, M., Weijers, D., Schwarz, H., Hamann, T., et al. (2003). Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426, 147–153. doi: 10.1038/nature02085
Fujita, D., Trijatmiko, K. R., Tagle, A. G., Sapasap, M. V., Koide, Y., Sasaki, K., et al. (2013). NAL1 allele from a rice landrace greatly increases yield in modern indica cultivars. Proc. Natl. Acad. Sci. U. S. A. 110, 20431–20436. doi: 10.1073/pnas.1310790110
Gallavotti, A., Zhao, Q., Kyozuka, J., Meeley, R. B., Ritter, M. K., Doebley, J. F., et al. (2004). The role of barren stalk1 in the architecture of maize. Nature 432, 630–635. doi: 10.1038/nature03148
Gälweiler, L., Guan, C., Müller, A., Wisman, E., Mendgen, K., Yephremov, A., et al. (1998). Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282, 2226–2230. doi: 10.1126/science.282.5397.2226
Gao, X., Liang, W., Yin, C., Ji, S., Wang, H., Su, X., et al. (2010). The SEPALLATA-like gene OsMADS34 is required for rice inflorescence and spikelet development. Plant Physiol. 153, 728–740. doi: 10.1104/pp.110.156711
Gao, F., Wang, K., Liu, Y., Chen, Y., Chen, P., Shi, Z., et al. (2015). Blocking miR396 increases rice yield by shaping inflorescence architecture. Nat. Plants 2, 15196. doi: 10.1038/nplants.2015.196
Gao, Y., Wang, G., Yuan, S., Qin, Y., Zhao, J., Zhang, Y., et al. (2014). Phenotypic analysis and molecular characterization of an allelic mutant of the D61 gene in rice. Crop J. 2, 175–182. doi: 10.1016/j.cj.2014.04.003
Guan, C., Jiao, Y. (2020). Interplay between the shoot apical meristem and lateral organs. aBIOTECH 1, 178–184. doi: 10.1007/s42994-020-00021-2
Guo, T., Chen, K., Dong, N. Q., Shi, C. L., Ye, W. W., Gao, J. P., et al. (2018). GRAIN SIZE AND NUMBER1 negatively regulates the OsMKKK10-OsMKK4-OsMPK6 cascade to coordinate the trade-off between grain number per panicle and grain size in rice. Plant Cell 30, 871–888. doi: 10.1105/tpc.17.00959
Guo, T., Lu, Z. Q., Shan, J. X., Ye, W. W., Dong, N. Q., Lin, H. X. (2020). ERECTA1 acts upstream of the OsMKKK10-OsMKK4-OsMPK6 cascade to control spikelet number by regulating cytokinin metabolism in rice. Plant Cell 32, 2763–2779. doi: 10.1105/tpc.20.00351
Hay, A., Kaur, H., Phillips, A., Hedden, P., Hake, S., Tsiantis, M. (2002). The gibberellin pathway mediates KNOTTED1-type homeobox function in plants with different body plans. Curr. Biol. 12, 1557–1565. doi: 10.1016/s0960-9822(02)01125-9
He, Q., Yang, L., Hu, W., Zhang, J., Xing, Y. (2018). Overexpression of an auxin receptor OsAFB6 significantly enhanced grain yield by increasing cytokinin and decreasing auxin concentrations in rice panicle. Sci. Rep. 8, 14051. doi: 10.1038/s41598-018-32450-x
Hirose, N., Makita, N., Kojima, M., Kamada-Nobusada, T., Sakakibara, H. (2007). Overexpression of a type-a response regulator alters rice morphology and cytokinin metabolism. Plant Cell Physiol. 48, 523–539. doi: 10.1093/pcp/pcm022
Hu, L., Chen, W., Yang, W., Li, X., Zhang, C., Zhang, X., et al. (2021a). OsSPL9 regulates grain number and grain yield in rice. Front. Plant Sci. 12, 682018. doi: 10.3389/fpls.2021.682018
Hu, J., Huang, L., Chen, G., Liu, H., Zhang, Y., Zhang, R., et al. (2021b). The elite alleles of OsSPL4 regulate grain size and increase grain yield in rice. Rice (N Y) 14, 90. doi: 10.1186/s12284-021-00531-7
Huang, Y., Bai, X., Luo, M., Xing, Y. (2019). Short panicle 3 controls panicle architecture by upregulating APO2/RFL and increasing cytokinin content in rice. J. Integr. Plant Biol. 61, 987–999. doi: 10.1111/jipb.12729
Huang, L., Hua, K., Xu, R., Zeng, D., Wang, R., Dong, G., et al. (2021). The LARGE2-APO1/APO2 regulatory module controls panicle size and grain number in rice. Plant Cell 33, 1212–1228. doi: 10.1093/plcell/koab041
Huang, X., Qian, Q., Liu, Z., Sun, H., He, S., Luo, D., et al. (2009). Natural variation at the DEP1 locus enhances grain yield in rice. Nat. Genet. 41, 494–497. doi: 10.1038/ng.352
Huang, Y., Zhao, S., Fu, Y., Sun, H., Ma, X., Tan, L., et al. (2018). Variation in the regulatory region of FZP causes increases in secondary inflorescence branching and grain yield in rice domestication. Plant J. 96, 716–733. doi: 10.1111/tpj.14062
Huo, X., Wu, S., Zhu, Z., Liu, F., Fu, Y., Cai, H., et al. (2017). NOG1 increases grain production in rice. Nat. Commun. 8, 1497. doi: 10.1038/s41467-017-01501-8
Ikeda, K., Ito, M., Nagasawa, N., Kyozuka, J., Nagato, Y. (2007). Rice ABERRANT PANICLE ORGANIZATION 1, encoding an F-box protein, regulates meristem fate. Plant J. 51, 1030–1040. doi: 10.1111/j.1365-313X.2007.03200.x
Ikeda-Kawakatsu, K., Maekawa, M., Izawa, T., Itoh, J., Nagato, Y. (2012). ABERRANT PANICLE ORGANIZATION 2/RFL, the rice ortholog of Arabidopsis LEAFY, suppresses the transition from inflorescence meristem to floral meristem through interaction with APO1. Plant J. 69, 168–180. doi: 10.1111/j.1365-313X.2011.04781.x
Jasinski, S., Piazza, P., Craft, J., Hay, A., Woolley, L., Rieu, I., et al. (2005). KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr. Biol. 15, 1560–1565. doi: 10.1016/j.cub.2005.07.023
Jiang, D., Fang, J., Lou, L., Zhao, J., Yuan, S., Yin, L., et al. (2015). Characterization of a null allelic mutant of the rice NAL1 gene reveals its role in regulating cell division. PloS One 10, e0118169. doi: 10.1371/journal.pone.0118169
Jiao, Y., Wang, Y., Xue, D., Wang, J., Yan, M., Liu, G., et al. (2010). Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 42, 541–544. doi: 10.1038/ng.591
Kempin, S. A., Savidge, B., Yanofsky, M. F. (1995). Molecular basis of the cauliflower phenotype in Arabidopsis. Science 267, 522–525. doi: 10.1126/science.7824951
Kessler, A., Halitschke, R., Baldwin, I. T. (2004). Silencing the jasmonate cascade: induced plant defenses and insect populations. Science 305, 665–668. doi: 10.1126/science.1096931
Kim, E. H., Kim, Y. S., Park, S. H., Koo, Y. J., Choi, Y. D., Chung, Y. Y., et al. (2009). Methyl jasmonate reduces grain yield by mediating stress signals to alter spikelet development in rice. Plant Physiol. 149, 1751–1760. doi: 10.1104/pp.108.134684
Kobayashi, K., Maekawa, M., Miyao, A., Hirochika, H., Kyozuka, J. (2010). PANICLE PHYTOMER2 (PAP2), encoding a SEPALLATA subfamily MADS-box protein, positively controls spikelet meristem identity in rice. Plant Cell Physiol. 51, 47–57. doi: 10.1093/pcp/pcp166
Komatsu, M., Chujo, A., Nagato, Y., Shimamoto, K., Kyozuka, J. (2003). FRIZZY PANICLE is required to prevent the formation of axillary meristems and to establish floral meristem identity in rice spikelets. Development 130, 3841–3850. doi: 10.1242/dev.00564
Komatsu, M., Maekawa, M., Shimamoto, K., Kyozuka, J. (2001). The LAX1 and FRIZZY PANICLE 2 genes determine the inflorescence architecture of rice by controlling rachis-branch and spikelet development. Dev. Biol. 231, 364–373. doi: 10.1006/dbio.2000.9988
Kurakawa, T., Ueda, N., Maekawa, M., Kobayashi, K., Kojima, M., Nagato, Y., et al. (2007). Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445, 652–655. doi: 10.1038/nature05504
Kuroha, T., Nagai, K., Gamuyao, R., Wang, D. R., Furuta, T., Nakamori, M., et al. (2018). Ethylene-gibberellin signaling underlies adaptation of rice to periodic flooding. Science 361, 181–186. doi: 10.1126/science.aat1577
Kyozuka, J., Konishi, S., Nemoto, K., Izawa, T., Shimamoto, K. (1998). Down-regulation of RFL, the FLO/LFY homolog of rice, accompanied with panicle branch initiation. Proc. Natl. Acad. Sci. U. S. A. 95, 1979–1982. doi: 10.1073/pnas.95.5.1979
Laux, T., Schoof, H. (1997). Maintaining the shoot meristem-the role of CLAVATA1. Trends Plant Sci. 2, 325–327. doi: 10.1016/s1360-1385(97)84613-4
Lee, D. Y., An, G. (2012). Two AP2 family genes, SUPERNUMERARY BRACT (SNB) and OsINDETERMINATE SPIKELET 1 (OsIDS1), synergistically control inflorescence architecture and floral meristem establishment in rice. Plant J. 69, 445–461. doi: 10.1111/j.1365-313X.2011.04804.x
Li, F., Liu, W., Tang, J., Chen, J., Tong, H., Hu, B., et al. (2010). Rice DENSE AND ERECT PANICLE 2 is essential for determining panicle outgrowth and elongation. Cell Res. 20, 838–849. doi: 10.1038/cr.2010.69
Li, G., Xu, B., Zhang, Y., Xu, Y., Khan, N. U., Xie, J., et al. (2022). RGN1 controls grain number and shapes panicle architecture in rice. Plant Biotechnol. J. 20, 158–167. doi: 10.1111/pbi.13702
Li, M., Tang, D., Wang, K., Wu, X., Lu, L., Yu, H., et al. (2011). Mutations in the F-box gene LARGER PANICLE improve the panicle architecture and enhance the grain yield in rice. Plant Biotechnol. J. 9, 1002–1013. doi: 10.1111/j.1467-7652.2011.00610.x
Li, S., Meng, S., Weng, J., Wu, Q. (2022). Fine-tuning shoot meristem size to feed the world. Trends Plant Sci. 27, 355–363. doi: 10.1016/j.tplants.2021.10.004
Li, S., Qian, Q., Fu, Z., Zeng, D., Meng, X., Kyozuka, J., et al. (2009). Short panicle1 encodes a putative PTR family transporter and determines rice panicle size. Plant J. 58, 592–605. doi: 10.1111/j.1365-313X.2009.03799.x
Li, S., Zhao, B., Yuan, D., Duan, M., Qian, Q., Tang, L., et al. (2013). Rice zinc finger protein DST enhances grain production through controlling Gn1a/OsCKX2 expression. Proc. Natl. Acad. Sci. U. S. A. 110, 3167–3172. doi: 10.1073/pnas.1300359110
Li, X., Qian, Q., Fu, Z., Wang, Y., Xiong, G., Zeng, D., et al (2003). Control of tillering in rice. Nature 422, 618–621. doi: 10.1038/nature01518.
Li, Y., Zhu, J., Wu, L., Shao, Y., Wu, Y., Mao, C. (2019). Functional divergence of PIN1 paralogous genes in rice. Plant Cell Physiol. 60, 2720–2732. doi: 10.1093/pcp/pcz159
Lin, L., Du, M., Li, S., Sun, C., Wu, F., Deng, L., et al. (2022). Mediator complex subunit MED25 physically interacts with DST to regulate spikelet number in rice. J. Integr. Plant Biol. 64, 871–883. doi: 10.1111/jipb.13238
Liu, J., Shi, X., Chang, Z., Ding, Y., Ding, C. (2022). Auxin efflux transporters OsPIN1c and OsPIN1d function redundantly in regulating rice (Oryza sativa l.) panicle development. Plant Cell Physiol. 63, 305–316. doi: 10.1093/pcp/pcab172
Liu, C., Teo, Z. W., Bi, Y., Song, S., Xi, W., Yang, X., et al. (2013). A conserved genetic pathway determines inflorescence architecture in Arabidopsis and rice. Dev. Cell 24, 612–622. doi: 10.1016/j.devcel.2013.02.013
Liu, L., Tong, H., Xiao, Y., Che, R., Xu, F., Hu, B., et al. (2015a). Activation of Big Grain1 significantly improves grain size by regulating auxin transport in rice. Proc. Natl. Acad. Sci. U. S. A. 112, 11102–11107. doi: 10.1073/pnas.1512748112
Liu, X., Zhou, S., Wang, W., Ye, Y., Zhao, Y., Xu, Q., et al. (2015b). Regulation of histone methylation and reprogramming of gene expression in the rice inflorescence meristem. Plant Cell 27, 1428–1444. doi: 10.1105/tpc.15.00201
Long, J. A., Ohno, C., Smith, Z. R., Meyerowitz, E. M. (2006). TOPLESS regulates apical embryonic fate in Arabidopsis. Science 312, 1520–1523. doi: 10.1126/science.1123841
Lu, G., Coneva, V., Casaretto, J. A., Ying, S., Mahmood, K., Liu, F., et al. (2015a). OsPIN5b modulates rice (Oryza sativa) plant architecture and yield by changing auxin homeostasis, transport and distribution. Plant J. 83, 913–925. doi: 10.1111/tpj.12939
Lu, Z., Shao, G., Xiong, J., Jiao, Y., Wang, J., Liu, G., et al. (2015b). MONOCULM 3, an ortholog of WUSCHEL in rice, is required for tiller bud formation. J. Genet. Genomics 42, 71–78. doi: 10.1016/j.jgg.2014.12.005
Lu, Z., Yu, H., Xiong, G., Wang, J., Jiao, Y., Liu, G., et al. (2013). Genome-wide binding analysis of the transcription activator IDEAL PLANT ARCHITECTURE1 reveals a complex network regulating rice plant architecture. Plant Cell 25, 3743–3759. doi: 10.1105/tpc.113.113639
Malik, N., Ranjan, R., Parida, S. K., Agarwal, P., Tyagi, A. K. (2020). Mediator subunit OsMED14_1 plays an important role in rice development. Plant J. 101, 1411–1429. doi: 10.1111/tpj.14605
Mandel, M. A., Gustafson-brown, C., Savidge, B., Yanofsky, M. F. (1992). Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 360, 273–277. doi: 10.1038/360273a0
Mason, M. G., Mathews, D. E., Argyros, D. A., Maxwell, B. B., Kieber, J. J., Alonso, J. M., et al. (2005). Multiple type-b response regulators mediate cytokinin signal transduction in Arabidopsis. Plant Cell 17, 3007–3018. doi: 10.1105/tpc.105.035451
Mayer, K. F., Schoof, H., Haecker, A., Lenhard, M., Jürgens, G., Laux, T. (1998). Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95, 805–815. doi: 10.1016/s0092-8674(00)81703-1
Miao, Y., Xun, Q., Taji, T., Tanaka, K., Yasuno, N., Ding, C., et al. (2022). ABERRANT PANICLE ORGANIZATION2 controls multiple steps in panicle formation through common direct-target genes. Plant Physiol. 189, 2210–2226. doi: 10.1093/plphys/kiac216
Miura, K., Ikeda, M., Matsubara, A., Song, X. J., Ito, M., Asano, K., et al. (2010). OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet. 42, 545–549. doi: 10.1038/ng.592
Möller, B., Weijers, D. (2009). Auxin control of embryo patterning. Cold Spring Harb. Perspect. Biol. 1, a001545. doi: 10.1101/cshperspect.a001545
Moon, S., Jung, K. H., Lee, D. E., Lee, D. Y., Lee, J., An, K., et al. (2006). The rice FON1 gene controls vegetative and reproductive development by regulating shoot apical meristem size. Mol. Cells 21, 147–152. doi: 10.14348/.1970.0.0.
Mravec, J., Skůpa, P., Bailly, A., Hoyerová, K., Krecek, P., Bielach, A., et al. (2009). Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter. Nature 459, 1136–1140. doi: 10.1038/nature08066
Nakagawa, M., Shimamoto, K., Kyozuka, J. (2002). Overexpression of RCN1 and RCN2, rice TERMINAL FLOWER 1/CENTRORADIALIS homologs, confers delay of phase transition and altered panicle morphology in rice. Plant J. 29, 743–750. doi: 10.1046/j.1365-313x.2002.01255.x
Naruse, M., Takahashi, H., Kurata, N., Ito, Y. (2018). Cytokinin-induced expression of OSH1 in a shoot-regenerating rice callus. Plant Biotechnol. (Tokyo) 35, 267–272. doi: 10.5511/plantbiotechnology.18.0614a
Ohmori, Y., Tanaka, W., Kojima, M., Sakakibara, H., Hirano, H. Y. (2013). WUSCHEL-RELATED HOMEOBOX4 is involved in meristem maintenance and is negatively regulated by the CLE gene FCP1 in rice. Plant Cell 25, 229–241. doi: 10.1105/tpc.112.103432
Okada, K., Ueda, J., Komaki, M. K., Bell, C. J., Shimura, Y. (1991). Requirement of the auxin polar transport system in early stages of Arabidopsis floral bud formation. Plant Cell 3, 677–684. doi: 10.1105/tpc.3.7.677
Olszewski, N. E., West, C. M., Sassi, S. O., Hartweck, L. M. (2010). O-GlcNAc protein modification in plants: Evolution and function. Biochim. Biophys. Acta 1800, 49–56. doi: 10.1016/j.bbagen.2009.11.016
Pasion, E. A., Badoni, S., Misra, G., Anacleto, R., Parween, S., Kohli, A., et al. (2021). OsTPR boosts the superior grains through increase in upper secondary rachis branches without incurring a grain quality penalty. Plant Biotechnol. J. 19, 1396–1411. doi: 10.1111/pbi.13560
Prakash, S., Rai, R., Zamzam, M., Ahmad, O., Peesapati, R., Vijayraghavan, U. (2022). OsbZIP47 is an integrator for meristem regulators during rice plant growth and development. Front. Plant Sci. 13, 865928. doi: 10.3389/fpls.2022.865928
Qi, J., Qian, Q., Bu, Q., Li, S., Chen, Q., Sun, J., et al. (2008). Mutation of the rice Narrow leaf1 gene, which encodes a novel protein, affects vein patterning and polar auxin transport. Plant Physiol. 147, 1947–1959. doi: 10.1104/pp.108.118778
Qi, W., Sun, F., Wang, Q., Chen, M., Huang, Y., Feng, Y. Q., et al. (2011). Rice ethylene-response AP2/ERF factor OsEATB restricts internode elongation by down-regulating a gibberellin biosynthetic gene. Plant Physiol. 157, 216–228. doi: 10.1104/pp.111.179945
Qiao, Y., Piao, R., Shi, J., Lee, S. I., Jiang, W., Kim, B. K., et al. (2011). Fine mapping and candidate gene analysis of dense and erect panicle 3, DEP3, which confers high grain yield in rice (Oryza sativa l.). Theor. Appl. Genet. 122, 1439–1449. doi: 10.1007/s00122-011-1543-6
Rao, N. N., Prasad, K., Kumar, P. R., Vijayraghavan, U. (2008). Distinct regulatory role for RFL, the rice LFY homolog, in determining flowering time and plant architecture. Proc. Natl. Acad. Sci. U. S. A. 105, 3646–3451. doi: 10.1073/pnas.0709059105
Rashotte, A. M., Brady, S. R., Reed, R. C., Ante, S. J., Muday, G. K. (2000). Basipetal auxin transport is required for gravitropism in roots of Arabidopsis. Plant Physiol. 122, 481–490. doi: 10.1104/pp.122.2.481
Rashotte, A. M., Carson, S. D., To, J. P., Kieber, J. J. (2003). Expression profiling of cytokinin action in Arabidopsis. Plant Physiol. 132, 1998–2011. doi: 10.1104/pp.103.021436
Reinhardt, D., Mandel, T., Kuhlemeier, C. (2000). Auxin regulates the initiation and radial position of plant lateral organs. Plant Cell 12, 507–518. doi: 10.1105/tpc.12.4.507
Ren, D., Yu, H., Rao, Y., Xu, Q., Zhou, T., Hu, J., et al. (2018). 'Two-floret spikelet' as a novel resource has the potential to increase rice yield. Plant Biotechnol. J. 16, 351–353. doi: 10.1111/pbi.12849
Saini, S., Sharma, I., Pati, P. K. (2015). Versatile roles of brassinosteroid in plants in the context of its homoeostasis, signaling and crosstalks. Front. Plant Sci. 6, 950. doi: 10.3389/fpls.2015.00950
Sakai, H., Honma, T., Aoyama, T., Sato, S., Kato, T., Tabata, S., et al. (2001). ARR1, a transcription factor for genes immediately responsive to cytokinins. Science 294, 1519–1521. doi: 10.1126/science.1065201
Sakamoto, T., Sakakibara, H., Kojima, M., Yamamoto, Y., Nagasaki, H., Inukai, Y., et al. (2006). Ectopic expression of KNOTTED1-like homeobox protein induces expression of cytokinin biosynthesis genes in rice. Plant Physiol. 142, 54–62. doi: 10.1104/pp.106.085811
Sato, Y., Hong, S. K., Tagiri, A., Kitano, H., Yamamoto, N., Nagato, Y., et al. (1996). A rice homeobox gene, OSH1, is expressed before organ differentiation in a specific region during early embryogenesis. Proc. Natl. Acad. Sci. U. S. A. 93, 8117–8122. doi: 10.1073/pnas.93.15.8117
Schaller, G. E., Street, I. H., Kieber, J. J. (2014). Cytokinin and the cell cycle. Curr. Opin. Plant Biol. 21, 7–15. doi: 10.1016/j.pbi.2014.05.015
Schoof, H., Lenhard, M., Haecker, A., Mayer, K. F., Jürgens, G., Laux, T. (2000). The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100, 635–644. doi: 10.1016/s0092-8674(00)80700-x
Sentoku, N., Sato, Y., Kurata, N., Ito, Y., Kitano, H., Matsuoka, M. (1999). Regional expression of the rice KN1-type homeobox gene family during embryo, shoot, and flower development. Plant Cell 11, 1651–1664. doi: 10.1105/tpc.11.9.1651
Shang, J. Y., Chun, Y., Li, X. Y. (2021). Map-based cloning and natural variation analysis of the PAL3 gene controlling panicle length in rice. Chin. Bull. Bot. 56, 520–532. doi: 10.11983/CBB21119
Shao, G., Lu, Z., Xiong, J., Wang, B., Jing, Y., Meng, X., et al. (2019). Tiller bud formation regulators MOC1 and MOC3 cooperatively promote tiller bud outgrowth by activating FON1 expression in rice. Mol. Plant 12, 1090–1102. doi: 10.1016/j.molp.2019.04.008
Shimada, A., Ueguchi-Tanaka, M., Sakamoto, T., Fujioka, S., Takatsuto, S., Yoshida, S., et al. (2006). The rice SPINDLY gene functions as a negative regulator of gibberellin signaling by controlling the suppressive function of the DELLA protein, SLR1, and modulating brassinosteroid synthesis. Plant J. 48, 390–402. doi: 10.1111/j.1365-313X.2006.02875.x
Si, L., Chen, J., Huang, X., Gong, H., Luo, J., Hou, Q., et al. (2016). OsSPL13 controls grain size in cultivated rice. Nat. Genet. 48, 447–456. doi: 10.1038/ng.3518
Sieberer, T., Leyser, O. (2006). Plant science. auxin transport, but in which direction? Science 312, 858–860. doi: 10.1126/science.1127659
Somssich, M., Je, B. I., Simon, R., Jackson, D. (2016). CLAVATA-WUSCHEL signaling in the shoot meristem. Development 143, 3238–3248. doi: 10.1242/dev.133645
Song, X., Lu, Z., Yu, H., Shao, G., Xiong, J., Meng, X., et al. (2017). IPA1 functions as a downstream transcription factor repressed by D53 in strigolactone signaling in rice. Cell Res. 27, 1128–1141. doi: 10.1038/cr.2017.102
Song, X., Meng, X., Guo, H., Cheng, Q., Jing, Y., Chen, M., et al. (2022). Targeting a gene regulatory element enhances rice grain yield by decoupling panicle number and size. Nat. Biotechnol 40, 1403–1411. doi: 10.1038/s41587-022-01281-7
Sreenivasulu, N., Pasion, E., Kohli, A. (2021). Idealizing inflorescence architecture to enhance rice yield potential for feeding nine billion people in 2050. Mol. Plant 14, 861–863. doi: 10.1016/j.molp.2021.05.003
Su, S., Hong, J., Chen, X., Zhang, C., Chen, M., Luo, Z., et al. (2021). Gibberellins orchestrate panicle architecture mediated by DELLA-KNOX signalling in rice. Plant Biotechnol. J. 19, 2304–2318. doi: 10.1111/pbi.13661
Su, Y., Zhou, C., Li, Y., Yu, Y., Tang, L., Zhang, W., et al. (2020). Integration of pluripotency pathways regulates stem cell maintenance in the Arabidopsis shoot meristem. Proc. Natl. Acad. Sci. U. S. A. 117, 22561–22571. doi: 10.1073/pnas.2015248117
Sun, W., Fan, J., Fang, A., Li, Y., Tariqjaveed, M., Li, D., et al. (2020). Ustilaginoidea virens: Insights into an emerging rice pathogen. Annu. Rev. Phytopathol. 58, 363–385. doi: 10.1146/annurev-phyto-010820-012908
Suzaki, T., Sato, M., Ashikari, M., Miyoshi, M., Nagato, Y., Hirano, H. Y. (2004). The gene FLORAL ORGAN NUMBER1 regulates floral meristem size in rice and encodes a leucine-rich repeat receptor kinase orthologous to Arabidopsis CLAVATA1. Development 131, 5649–5657. doi: 10.1242/dev.01441
Suzaki, T., Toriba, T., Fujimoto, M., Tsutsumi, N., Kitano, H., Hirano, H. Y. (2006). Conservation and diversification of meristem maintenance mechanism in Oryza sativa: Function of the FLORAL ORGAN NUMBER2 gene. Plant Cell Physiol. 47, 1591–1602. doi: 10.1093/pcp/pcl025
Szemenyei, H., Hannon, M., Long, J. A. (2008). TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319, 1384–1386. doi: 10.1126/science.1151461
Tabuchi, H., Zhang, Y., Hattori, S., Omae, M., Shimizu-Sato, S., Oikawa, T., et al. (2011). LAX PANICLE2 of rice encodes a novel nuclear protein and regulates the formation of axillary meristems. Plant Cell 23, 3276–3287. doi: 10.1105/tpc.111.088765
Tan, F. Q., Wang, W., Li, J., Lu, Y., Zhu, B., Hu, F., et al. (2022). A coiled-coil protein associates polycomb repressive complex 2 with KNOX/BELL transcription factors to maintain silencing of cell differentiation-promoting genes in the shoot apex. Plant Cell 34, 2969–2988. doi: 10.1093/plcell/koac133
Tanaka, W., Ohmori, Y., Ushijima, T., Matsusaka, H., Matsushita, T., Kumamaru, T., et al. (2015). Axillary meristem formation in rice requires the WUSCHEL ortholog TILLERS ABSENT1. Plant Cell 27, 1173–1184. doi: 10.1105/tpc.15.00074
Tong, H., Chu, C. (2018). Functional specificities of brassinosteroid and potential utilization for crop improvement. Trends Plant Sci. 23, 1016–1028. doi: 10.1016/j.tplants.2018.08.007
Tsuda, K., Ito, Y., Sato, Y., Kurata, N. (2011). Positive autoregulation of a KNOX gene is essential for shoot apical meristem maintenance in rice. Plant Cell 23, 4368–4381. doi: 10.1105/tpc.111.090050
Tyler, L., Thomas, S. G., Hu, J., Dill, A., Alonso, J. M., Ecker, J. R., et al. (2004). Della Proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol. 135, 1008–1019. doi: 10.1104/pp.104.039578
Ueguchi-Tanaka, M., Ashikari, M., Nakajima, M., Itoh, H., Katoh, E., Kobayashi, M., et al. (2005). GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437, 693–698. doi: 10.1038/nature04028
Wang, J. W., Czech, B., Weigel, D. (2009). miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138, 738–749. doi: 10.1016/j.cell.2009.06.014
Wang, W., Li, G., Zhao, J., Chu, H., Lin, W., Zhang, D., et al. (2014). DWARF TILLER1, a WUCHEL-related homeobox transcription factor, is required for tiller growth in rice. PloS Genet. 10, e1004154. doi: 10.1371/journal.pgen.1004154
Wang, M., Qin, Y., Chun, Y., Zhao, J., Fang, J., Zafar, S. A., et al. (2018). Characterization of a novel semi-dwarf GID1 allele identifies an amino acid required for its interaction with SLR1 in rice. J. Plant Growth Regul. 37, 840–848. doi: 10.1007/s00344-018-9790-2
Wang, J., Su, Y., Kong, X., Ding, Z., Zhang, X. (2020). Initiation and maintenance of plant stem cells in root and shoot apical meristems. aBIOTECH 1, 194–204. doi: 10.1007/s42994-020-00020-3
Wang, L., Sun, S., Jin, J., Fu, D., Yang, X., Weng, X., et al. (2015). Coordinated regulation of vegetative and reproductive branching in rice. Proc. Natl. Acad. Sci. U. S. A. 112, 15504–15509. doi: 10.1073/pnas.1521949112
Wang, H., Tong, X., Tang, L., Wang, Y., Zhao, J., Li, Z., et al. (2022). RLB (RICE LATERAL BRANCH) recruits PRC2-mediated H3K27 tri-methylation on OsCKX4 to regulate lateral branching. Plant Physiol. 188, 460–476. doi: 10.1093/plphys/kiab494
Wang, S., Wu, K., Qian, Q., Liu, Q., Li, Q., Pan, Y., et al. (2017b). Non-canonical regulation of SPL transcription factors by a human OTUB1-like deubiquitinase defines a new plant type rice associated with higher grain yield. Cell Res. 27, 1142–1156. doi: 10.1038/cr.2017.98
Wang, J., Yu, H., Xiong, G., Lu, Z., Jiao, Y., Meng, X., et al. (2017a). Tissue-specific ubiquitination by IPA1 INTERACTING PROTEIN1 modulates IPA1 protein levels to regulate plant architecture in rice. Plant Cell 29, 697–707. doi: 10.1105/tpc.16.00879
Wasternack, C., Hause, B. (2013). Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. an update to the 2007 review in annals of botany. Ann. Bot. 111, 1021–1058. doi: 10.1093/aob/mct067
Weigel, D., Alvarez, J., Smyth, D. R., Yanofsky, M. F., Meyerowitz, E. M. (1992). LEAFY controls floral meristem identity in Arabidopsis. Cell 69, 843–859. doi: 10.1016/0092-8674(92)90295-n
Werner, T., Motyka, V., Strnad, M., Schmülling, T. (2001). Regulation of plant growth by cytokinin. Proc. Natl. Acad. Sci. U. S. A. 98, 10487–10492. doi: 10.1073/pnas.171304098
Worthen, J. M., Yamburenko, M. V., Lim, J., Nimchuk, Z. L., Kieber, J. J., Schaller, G. E. (2019). Type-b response regulators of rice play key roles in growth, development and cytokinin signaling. Development 146, dev174870. doi: 10.1242/dev.174870
Wu, Y., Fu, Y., Zhao, S., Gu, P., Zhu, Z., Sun, C., et al. (2016a). CLUSTERED PRIMARY BRANCH 1, a new allele of DWARF11, controls panicle architecture and seed size in rice. Plant Biotechnol. J. 14, 377–386. doi: 10.1111/pbi.12391
Wu, X., Liang, Y., Gao, H., Wang, J., Zhao, Y., Hua, L., et al. (2021). Enhancing rice grain production by manipulating the naturally evolved cis-regulatory element-containing inverted repeat sequence of OsREM20. Mol. Plant 14, 997–1011. doi: 10.1016/j.molp.2021.03.016
Wu, Y., Wang, Y., Mi, X. F., Shan, J. X., Li, X. M., Xu, J. L., et al. (2016b). GNP1 encodes GA20ox1, which increases grain number and yield by increasing cytokinin activity in rice panicle meristems. PloS Genet. 12, e1006386. doi: 10.1371/journal.pgen.1006386
Wu, H. M., Xie, D. J., Tang, Z. S., Shi, D. Q., Yang, W. C. (2020). PINOID regulates floral organ development by modulating auxin transport and interacts with MADS16 in rice. Plant Biotechnol. J. 18, 1778–1795. doi: 10.1111/pbi.13340
Wybouw, B., De Rybel, B. (2019). Cytokinin - a developing story. Trends Plant Sci. 24, 177–185. doi: 10.1016/j.tplants.2018.10.012
Xie, M., Chen, H., Huang, L., O'Neil, R. C., Shokhirev, M. N., Ecker, J. R. (2018). A b-ARR-mediated cytokinin transcriptional network directs hormone cross-regulation and shoot development. Nat. Commun. 9, 1604. doi: 10.1038/s41467-018-03921-6
Yamaguchi, A., Wu, M. F., Yang, L., Wu, G., Poethig, R. S., Wagner, D. (2009). The microRNA-regulated SBP-box transcription factor SPL3 is a direct upstream activator of LEAFY, FRUITFULL, and APETALA1. Dev. Cell 17, 268–278. doi: 10.1016/j.devcel.2009.06.007
Yan, P., Zhu, Y., Wang, Y., Ma, F., Lan, D., Niu, F., et al. (2022). A new RING finger protein And GRAIN NUMBER 1, affects plant architecture and grain yield in rice. Int. J. Mol. Sci. 23, 824. doi: 10.3390/ijms23020824
Yang, J., Cho, L. H., Yoon, J., Yoon, H., Wai, A. H., Hong, W. J., et al. (2019). Chromatin interacting factor OsVIL2 increases biomass and rice grain yield. Plant Biotechnol. J. 17, 178–187. doi: 10.1111/pbi.12956
Yang, C. J., Zhang, C., Lu, Y. N., Jin, J. Q., Wang, X. L. (2011). The mechanisms of brassinosteroids' action: from signal transduction to plant development. Mol. Plant 4, 588–600. doi: 10.1093/mp/ssr020
Yang, W., Cortijo, S., Korsbo, N., Roszak, P., Schiessl, K., Gurzadyan, A. (2021). Molecular mechanism of cytokinin-activated cell division in Arabidopsis. Science 371, 1350–5. doi: 10.1126/science.abe2305
Yano, K., Morinaka, Y., Wang, F., Huang, P., Takehara, S., Hirai, T., et al (2019). GWAS with principal component analysis identifies a gene comprehensively controlling rice architecture. Proc. Natl. Acad. Sci. U. S. A. 116, 21262–7. doi: 10.1073/pnas.1904964116
Yin, C. C., Zhao, H., Ma, B., Chen, S. Y., Zhang, J. S. (2017). Diverse roles of ethylene in regulating agronomic traits in rice. Front. Plant Sci. 8, 1676. doi: 10.3389/fpls.2017.01676
Yoon, J., Jeong, H. J., Baek, G., Yang, J., Peng, X., Tun, W., et al. (2021). A VIN3-like protein OsVIL1 is involved in grain yield and biomass in rice. Plants (Basel) 11, 83. doi: 10.3390/plants11010083
Yoshida, A., Ohmori, Y., Kitano, H., Taguchi-Shiobara, F., Hirano, H. Y. (2012). ABERRANT SPIKELET AND PANICLE1, encoding a TOPLESS-related transcriptional co-repressor, is involved in the regulation of meristem fate in rice. Plant J. 70, 327–339. doi: 10.1111/j.1365-313X.2011.04872.x
Yoshida, A., Sasao, M., Yasuno, N., Takagi, K., Daimon, Y., Chen, R., et al. (2013). TAWAWA1, a regulator of rice inflorescence architecture, functions through the suppression of meristem phase transition. Proc. Natl. Acad. Sci. U. S. A. 110, 767–772. doi: 10.1073/pnas.1216151110
Zazímalová, E., Murphy, A. S., Yang, H., Hoyerová, K., Hosek, P. (2010). Auxin transporters–why so many? Cold spring harb. Perspect. Biol. 2, a001552. doi: 10.1101/cshperspect.a001552
Zhang, G. H., Li, S. Y., Wang, L., Ye, W. J., Zeng, D. L., Rao, Y. C., et al. (2014). LSCHL4 from japonica cultivar, which is allelic to NAL1, increases yield of indica super rice 93-11. Mol. Plant 7, 1350–1364. doi: 10.1093/mp/ssu055
Zhang, L., Yu, H., Ma, B., Liu, G., Wang, J., Wang, J., et al (2017). A natural tandem array alleviates epigenetic repression of IPA1 and leads to superior yielding rice. Nat. Commun. 8, 14789. doi: 10.1038/ncomms14789.
Zhang, W., Peng, K., Cui, F., Wang, D., Zhao, J., Zhang, Y., et al. (2021). Cytokinin oxidase/dehydrogenase OsCKX11 coordinates source and sink relationship in rice by simultaneous regulation of leaf senescence and grain number. Plant Biotechnol. J. 19, 335–350. doi: 10.1111/pbi.13467
Zhang, Z., Sun, X., Ma, X., Xu, B., Zhao, Y., Ma, Z., et al. (2020). GNP6, a novel allele of MOC1, regulates panicle and tiller development in rice. Crop J. 9, 57–67. doi: 10.1016/j.cj.2020.04.011
Zhang, L., Yu, H., Ma, B., Liu, G., Wang, J., Wang, J., et al. (2017). A natural tandem array alleviates epigenetic repression of IPA1 and leads to superior yielding rice. Nat. Commun. 8, 14789. doi: 10.1038/ncomms14789
Zhang, Y. C., Yu, Y., Wang, C. Y., Li, Z. Y., Liu, Q., Xu, J., et al. (2013). Overexpression of microRNA OsmiR397 improves rice yield by increasing grain size and promoting panicle branching. Nat. Biotechnol. 31, 848–852. doi: 10.1038/nbt.2646
Zhao, L., Tan, L., Zhu, Z., Xiao, L., Xie, D., Sun, C. (2015). PAY1 improves plant architecture and enhances grain yield in rice. Plant J. 83, 528–536. doi: 10.1111/tpj.12905
Zhao, J., Wu, C., Yuan, S., Yin, L., Sun, W., Zhao, Q., et al. (2013). Kinase activity of OsBRI1 is essential for brassinosteroids to regulate rice growth and development. Plant Sci. 199-200, 113–120. doi: 10.1016/j.plantsci.2012.10.011
Zhou, J., Li, Z., Xiao, G., Zhai, M., Pan, X., Huang, R., et al. (2020). CYP71D8L is a key regulator involved in growth and stress responses by mediating gibberellin homeostasis in rice. J. Exp. Bot. 71, 1160–1170. doi: 10.1093/jxb/erz491
Zhu, X., Liang, W., Cui, X., Chen, M., Yin, C., Luo, Z., et al. (2015). Brassinosteroids promote development of rice pollen grains and seeds by triggering expression of carbon starved anther, a MYB domain protein. Plant J. 82, 570–581. doi: 10.1111/tpj.12820
Zhu, W., Yang, L., Wu, D., Meng, Q., Deng, X., Huang, G., et al. (2022). Rice SEPALLATA genes OsMADS5 and OsMADS34 cooperate to limit inflorescence branching by repressing the TERMINAL FLOWER1-like gene RCN4. New Phytol. 233, 1682–1700. doi: 10.1111/nph.17855
Zong, J., Wang, L., Zhu, L., Bian, L., Zhang, B., Chen, X., et al. (2022). A rice single cell transcriptomic atlas defines the developmental trajectories of rice floret and inflorescence meristems. New Phytol. 234, 494–512. doi: 10.1111/nph.18008
Zou, X., Qin, Z., Zhang, C., Liu, B., Liu, J., Zhang, C., et al. (2015). Over-expression of an s-domain receptor-like kinase extracellular domain improves panicle architecture and grain yield in rice. J. Exp. Bot. 66, 7197–7209. doi: 10.1093/jxb/erv417
Zubo, Y. O., Blakley, I. C., Yamburenko, M. V., Worthen, J. M., Street, I. H., Franco-Zorrilla, J. M., et al. (2017). Cytokinin induces genome-wide binding of the type-b response regulator ARR10 to regulate growth and development in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 114, E5995–E6004. doi: 10.1073/pnas.1620749114
Keywords: rice (Oryza sativa L.), inflorescence, meristem, identity conversion, panicle branch, phytohormone
Citation: Chun Y, Kumar A and Li X (2022) Genetic and molecular pathways controlling rice inflorescence architecture. Front. Plant Sci. 13:1010138. doi: 10.3389/fpls.2022.1010138
Received: 02 August 2022; Accepted: 13 September 2022;
Published: 28 September 2022.
Edited by:
Qian-Hao Zhu, Commonwealth Scientific and Industrial Research Organisation (CSIRO), AustraliaReviewed by:
Longbiao Guo, China National Rice Research Institute (CAAS), ChinaXiujuan Yang, University of Adelaide, Australia
Copyright © 2022 Chun, Kumar and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xueyong Li, bGl4dWV5b25nQGNhYXMuY24=
†These authors have contributed equally to this work and share first authorship
 Yan Chun
Yan Chun Ashmit Kumar
Ashmit Kumar Xueyong Li
Xueyong Li