
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 13 October 2022
Sec. Plant Development and EvoDevo
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.1009998
This article is part of the Research TopicThe Evolution, Diversity and Functions of Alternative Splicing (AS) in Plant Responses to Abiotic StressView all 5 articles
 Lingli Yang1,2
Lingli Yang1,2 Li Yang2,3
Li Yang2,3 Chuanji Zhao2
Chuanji Zhao2 Jie Liu2
Jie Liu2 Chaobo Tong2
Chaobo Tong2 Yuanyuan Zhang2
Yuanyuan Zhang2 Xiaohui Cheng2
Xiaohui Cheng2 Huifang Jiang2
Huifang Jiang2 Jinxiong Shen1
Jinxiong Shen1 Meili Xie2*
Meili Xie2* Shengyi Liu2
Shengyi Liu2Alternative splicing (AS) is an important regulatory process that affects plant development and stress responses by greatly increasing the complexity of transcriptome and proteome. To understand how the AS landscape of B. napus changes in response to abiotic stresses, we investigated 26 RNA-seq libraries, including control and treatments with cold, dehydration, salt, and abscisic acid (ABA) at two different time points, to perform comparative alternative splicing analysis. Apparently, AS events increased under all stresses except dehydration for 1 h, and intron retention was the most common AS mode. In addition, a total of 357 differential alternative splicing (DAS) genes were identified under four abiotic stresses, among which 81 DAS genes existed in at least two stresses, and 276 DAS genes were presented under only one stress. A weighted gene co-expression network analysis (WGCNA) based on the splicing isoforms, rather than the genes, pinpointed out 23 co-expression modules associated with different abiotic stresses. Among them, a number of significant hub genes were also found to be DAS genes, which encode key isoforms involved in responses to single stress or multiple stresses, including RNA-binding proteins, transcription factors, and other important genes, such as RBP45C, LHY, MYB59, SCL30A, RS40, MAJ23.10, and DWF4. The splicing isoforms of candidate genes identified in this study could be a valuable resource for improving tolerance of B. napus against multiple abiotic stresses.
Alternative splicing (AS) is a common phenomenon in eukaryotes, which can use different splicing sites to produce multiple mRNA isoforms from the same pre-mRNA, thus increasing transcriptomic and proteomic diversity (Reddy et al., 2013; Staiger and Brown, 2013). More than 95% of intron-containing genes undergo AS in humans (Wang et al., 2008), and the proportion is as high as 70% in plants (Shen et al., 2014b; Thatcher et al., 2014; Zhang et al., 2017; Ruan et al., 2018). The selection of alternative splicing sites is determined by both cis-regulatory elements and trans-splicing factors. The cis-regulatory elements contain splice sites, polypyrimidine tract, branch point, and exonic and intronic splicing enhancer/silencer, which are the binding sites of splicing factors. Trans-splicing factors control the selection of splice sites and instruct splice assembly, including serine-/arginine-rich (SR) proteins and heterogeneous nuclear ribonucleoproteins (hnRNPs), in which SR proteins promote splicing, while hnRNPs do the opposite, both belonging to the important RNA-binding proteins (Syed et al., 2012; Erkelenz et al., 2013; Geuens et al., 2016). Splicing factors are widely alternatively spliced in plant development and responses to abiotic stresses, thus affecting the AS patterns and expression of downstream target genes (Staiger and Brown, 2013; Thatcher et al., 2014; Gu et al., 2019).
There are five basic types of AS events, namely, exon skipping (ES), intron retention (IR), alternative 5′ splice sites (A5SS), alternative 3′ splice sites (A3SS), and mutually exclusive exons (MEs). The proportion of AS events varies in different species: IR is the most common AS event in plants, while ES is the most prevalent one in animals (Shen et al., 2014b). AS can augment proteome diversity by generating diverse protein isoforms (Syed et al., 2012; Kruse et al., 2020), which is also coupled to mRNA stability and translation through nonsense-mediated decay (NMD) and microRNAs (miRNAs) regulation (Reddy et al., 2013; Chen et al., 2018). It was reported that 35% of AS events in polysome-associated mRNAs were translated into proteins, most of which resulted in diverse proteins in Arabidopsis (Yu et al., 2016). About 11%–18% of intron-containing genes were alternatively spliced to generate transcripts containing premature termination codons (PTCs) and subsequently coupled with the NMD pathway in Arabidopsis, such as CCA1, SR34a, GRP8, and GRP7 (Kalyna et al., 2012; Drechsel et al., 2013). Similarly, 32% of AS genes were subjected to NMD in Physcomitrella patens (Lloyd et al., 2018). In addition, a slice of transcripts produced by AS could obtain or lose miRNA binding sites, thus varying their expression by the regulation of miRNA (Iwakawa and Tomari, 2015; Chen et al., 2018).
AS is an important regulatory process for plants to respond to various stresses (Laloum et al., 2018). It has been proven that many vital genes, including those encoding transcription factors (TFs) and RNA-binding proteins (RBPs), underwent alternative splicing due to environmental stresses. For instance, in Arabidopsis, the transcription factor IDD14 produced a truncated protein variant lacking a functional DNA binding domain through intron retention, which was unable to activate the expression of starch-degrading enzymes, thereby resulting in starch accumulation under low temperatures (Seo et al., 2011). In Arabidopsis and rice, the heat shock transcription factor HsfA2 produced HsfA2-III isoform, instead of the PTC-containing HsfA2-II isoform, leading to the generation of truncated proteins under heat stress (Sugio et al., 2009; Liu et al., 2013; Cheng et al., 2015). Another crucial abiotic stress regulator, DREB2B, was also reported to be alternatively spliced in wheat and rice. Under drought and salt stresses, wheat WDREB2 was prone to produce WDREB2α and WDREB2γ isoforms, rather than WDREB2β encoding a non-functional polypeptide (Terashima and Takumi, 2009). Similarly, the functional isoform OsDREB2B2, but not the PTC-containing non-functional isoform OsDREB2B1, was generated to enhance stress tolerance in rice under high temperature and drought stresses (Matsukura et al., 2010), highlighting the conservation of AS regulation in plants. As to RBPs, multiple studies have also focused on the splicing pattern in response to stresses. For example, one SR gene, atSR30, could generate five different isoforms atSR30 mRNA1–5 via AS, while it tended to produce atSR30 mRNA1 and atSR30 mRNA4 under high-light irradiation and atSR30 mRNA1 with the treatment of paraquat in Arabidopsis (Tanabe et al., 2007). In addition, other SR genes, such as SR34, SR34b, RS31, RS40, RSZ32, and SR33/SCL33, in Arabidopsis, showed prominent changes in AS patterns under various abiotic treatments, including hormones and cold or heat stresses (Palusa et al., 2007). For example, SR45, SR30, and SR34, along with nuclear ribonucleic protein U1A, were more likely to produce AS events under high temperatures in grape (Jiang et al., 2017). A splicing factor RBM25 underwent AS activated by ABA, which was responsible for splicing many pre-mRNAs participated in ABA signal transduction pathways in Arabidopsis (Zhan et al., 2015; Cheng et al., 2017). Meanwhile, other types of genes were also reported recently. For example, the cyclin-D3-1-like gene may respond to environmental stresses via alternative splicing in Brassica napus (Guo et al., 2019), and the gene encoding cyclophilin 38 was induced to increase IR events nearly 30-fold under heat stress in P. patens (Chang et al., 2014). In the past, the AS events of most genes in response to stresses were identified by wet-lab experiments, while recent studies have developed the methodology for genome-wide identification of AS events and AS networks by transcriptomics, so that more splicing variations can be detected. For example, the AS landscapes ascribable to individual stress such as Sclerotinia sclerotiorum, Leptosphaeria maculans, and boron deficiency in B. napus have been illustrated (Gu et al., 2019; Ma et al., 2019; Ma et al., 2020). The AS dynamics of Arabidopsis, maize, and wheat, caused by salt, drought, and cold and heat stresses, alone or in combination, were also elaborated in detail (Ding et al., 2014; Thatcher et al., 2016; Liu et al., 2018).
Global agriculture is confronted with tremendous environmental changes, such as extreme temperatures, drought, and salt stresses, which are critical limiting factors influencing the growth and yield of crops such as rapeseed (Brassica napus L.), the worldwide second largest oilseed crop. Therefore, identifying genes with broad-spectrum effects on different stress responses is one of the long-term goals of genetic improvement in rapeseed. With the development of sequencing technology, many mature research methods have been widely used to identify potential genes involved in response to abiotic stresses, including genome-wide association studies (Zhang et al., 2015; Liu et al., 2021; Wassan et al., 2021) and comparative transcriptome analyses that focus primarily on differential expression analyses (Du et al., 2016; Pu et al., 2019; Raza et al., 2021). Nevertheless, these methodologies rarely consider the widespread AS events in plants, which, as described above, play an important role in response and adaptation to various abiotic stresses. Hence, identifying genes that alter AS modes in response to stresses could also provide resources for improving stress tolerance. However, comprehensive analysis of AS responsive to multiple abiotic stresses such as heat, drought, salt, and ABA in B. napus is still rarely available.
In this study, to identify candidate resistance genes in response to multiple abiotic stresses, comparative differential AS analyses were performed between control and treatments subjected to cold, dehydration, salt, and ABA at two different time points in B. napus. Weighted gene co-expression network analysis (WGCNA) was implemented to determine the regulatory network of B. napus in reaction to different abiotic stresses based on isoform expression level. This research described the global dynamics of AS isoforms, which would deepen our understanding of the response of B. napus to various abiotic stresses. In addition, the findings of the candidate stress responsive genes could assist in rapeseed variety breeding with extensive abiotic stress tolerance.
The RNA-seq data of B. napus leaves before and after cold, dehydration, salt, and ABA treatments at two different time points were downloaded from the National Genomics Data Center with project number CRA001775 (https://bigd.big.ac.cn/), of which data for replicate 1 of 1 h of dehydration was removed from subsequent analysis due to poor biological reproducibility (Zhang et al., 2019). Raw reads were first filtered by removing reads containing adapter, ploy-N, and low-quality reads using Trimmomatic v0.36 (Bolger et al., 2014). The generated paired-end clean reads were then aligned to the reference genome using Hisat2 v2.2.0 (Chalhoub et al., 2014; Kim et al., 2015). Furthermore, the in-house perl scripts were used to exclude reads aligned to multiple chromosome positions to minimize pseudomorphisms caused by ambiguous mapping. Finally, StringTie v2.1.5 was used to construct and identify both known and novel transcripts at the base of alignments and to quantify transcripts per million (TPM) of each isoform across all samples (Pertea et al., 2015).
Based on the gtf annotations generated by StringTie, Astanavista v4.0 was adopted to analyze the AS patterns of each sample (Foissac and Sammeth, 2007), including intron retention, exon skipping, alternative acceptors, and alternative donors events. Differential alternative splicing (DAS) genes are defined as genes that undergo differential AS events and produce different types of isoforms under different stress conditions compared with the control. DAS events between control and different treatments were respectively quantified using rMATS. rMATS is a software for differential alternative splicing analysis from RNA-Seq data with biological replicates. rMATS uses likelihood-ratio test to calculate the p-value and false discovery rate (FDR) to denote the difference in inclusion level (IncLevel, also designated as ψ) between the two groups of samples. First, the IncLevel1 and IncLevel2 from the two groups of samples were calculated, respectively, as follows: ψ = (I/LI)/(I/LI + S/LS). For example, ψ = Inclusion Level (IncLevel1), where I = number of reads mapped to the exon inclusion isoform (IC_SAMPLE_1), S = number of reads mapped to the exon skipping isoform (SC_SAMPLE_1), LI = effective length of the exon inclusion isoform (IncFormLen), and LS = effective length of the exon skipping isoform (SkipFormLen). Each biological replicate for one group had an IncLevel value, and IncLevelDifference (ΔIncLevel) = average(IncLevel1) − average(IncLevel2). rMATS uses a likelihood-ratio test to calculate the p-value for the difference in the mean IncLevel values between two groups of samples. Benjamini–Hochberg method (FDR) was used to correct p-value, and the thresholds of |ΔIncLevel| ≥ 0.1 and FDR < 0.05 were used to identify DAS. The command “python rmats.py –b1 control.txt –b2 treatment.txt -t paired –readLength 144 –gtf stringtie_merge.gtf –od treatment.dir –nthread 20 –cstat 0.1 –libType fr-unstranded” was used for this process (Shen et al., 2014a). Moreover, rmats2sashimiplot was used to visualize DAS events (https://github.com/Xinglab/rmats2sashimiplot).
In order to investigate the regulatory relationships between isoforms, WGCNA was performed on the top 50% transcripts selected based on their TPM variances using R v3.6.2. This analysis consisted of a series of steps, including sample clustering, outlier detection, soft threshold selection, one-step network construction, module identification, and module-treatment relationship analysis (Langfelder and Horvath, 2008). Subsequently, in-house perl scripts were used to filter out pair-wise co-expressed transcripts with weighted value <0.15 to obtain significant co-expression networks, which were finally visualized in Cytoscape v3.8.2 (Kohl et al., 2011). Isoforms whose absolute value of kME (eigengene connectivity) was not <0.8 (|kME|≥0.8) were designated as hub isoforms within each module. The average TPM values of different biological replicates were converted to log2 (TPM+1) for isoform heatmap visualization using R package pheatmap.
To gain functional annotation of the differentially spliced genes in B. napus, all genes were aligned against Arabidopsis protein database using local Blast similarity search, and the best hits with e‐value <1e−10 were retained. The functional information of Arabidopsis genes was extracted from TAIR database (http://www.arabidopsis.org/). Moreover, Gene Ontology (GO) enrichment analyses were performed using software TBtools (Chen et al., 2020).
To identify the AS landscape of B. napus in response to multiple abiotic stresses, RNA-seq data of 26 samples from leaves of cultivar Zhongshuang 11 (ZS11) were downloaded from NGDC database to perform comparative alternative splicing analyses. These samples included control and treatments with 1 and 8 h of dehydration, 4 and 24 h of cold, NaCl and ABA (Zhang et al., 2019). The downloaded data were then subjected to quality trimming, reads mapping, transcript assembly, followed by transcript merging to quantify TPM for each isoform (Supplementary Table 1). These analyses resulted into a total of 28,720 new transcripts across all these conditions (Supplementary Table 2).
The frequency of four different types of AS events was identified in B. napus under different abiotic stress conditions using Astalavista v4.0 (Foissac and Sammeth, 2007). As shown in Figure 1 and Supplementary Table 1, IR was the most common among the four AS events, accounting for 45.94% of all AS events identified under different stress conditions on average, while ES events accounted for the lowest proportion, with an average of just 5.67%. The average ratios of A5SS and A3SS events were very similar, 23.96% and 23.88%, respectively. These results were consistent with the frequency distribution of alternative splicing in plant genomes reported in previous studies (Zhang et al., 2017; Ruan et al., 2018). In general, the AS events of most treatments increased with the extension of stress treatment time compared with the control (Figure 1).
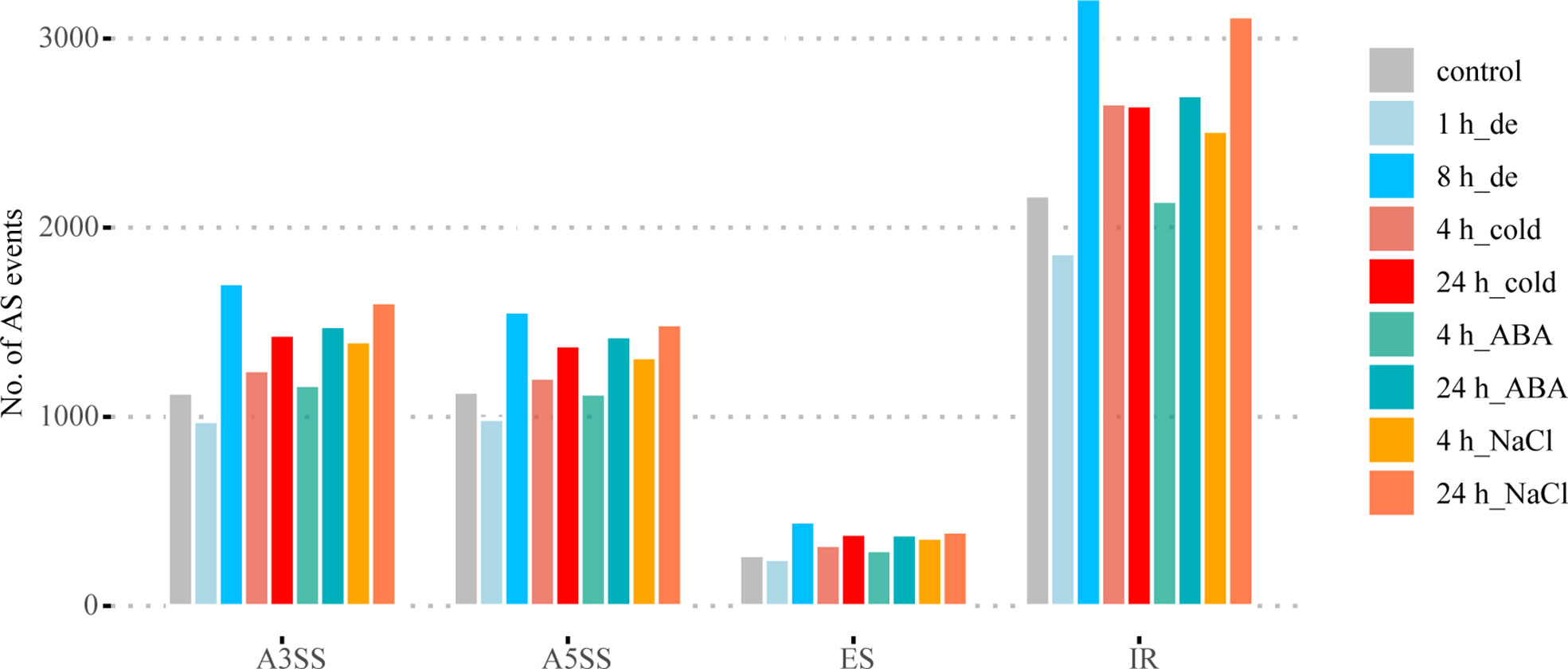
Figure 1 AS landscapes of control and treatments with dehydration, cold, ABA, and NaCl at two different time points in B. napus. A3SS, alternative 3′ splice site; A5SS, alternative 5′ splice site; ES, exon skip; IR, intron retain.
In order to study the AS transcriptional dynamics at different time points upon the treatments of four stresses, DAS genes were identified and systematically investigated between control and each of the treatments. Overall, this analysis resulted into a total of 357 DAS genes by integrating all comparative data sets. More DAS genes under treatments of dehydration and cold were observed than that of ABA and NaCl stresses, implying a stronger response of AS evoked by the former in B. napus (Figure 2). Specifically, the number of DAS genes was significantly variable across different treatments. A striking increase in the number of DAS genes (63 to 125) was observed with the time course of dehydration stress treatment, while the number of DAS genes enhanced slightly under ABA and cold stresses, from 26 to 37 and from 98 to 107 at 4 and 24 h, respectively (Figure 2). However, with the treatment course of NaCl stress, the number of DAS genes was found to show a moderate reduction from 56 to 33 (Figure 2). Comparative analysis of DAS genes between different stress treatments revealed that 81 DAS genes were detected under at least two stresses (Supplementary Table 3). For example, exon skipping of MSTRG.1012 was identified between control and several stresses including ABA, dehydration, and NaCl (Supplementary Figure 1A). In addition, the number of DAS genes specifically induced by individual stress was largest under cold stress (137), followed by dehydration (94), NaCl (25), and ABA (20) (Supplementary Table 3). For example, differential ES event of MSTRG.17416 was identified only at 8 h of dehydration compared with control (Supplementary Figure 1B). Overall, these results implied that ~23% of DAS genes could be induced by multiple stresses, and most DAS genes were independently induced by individual stress.
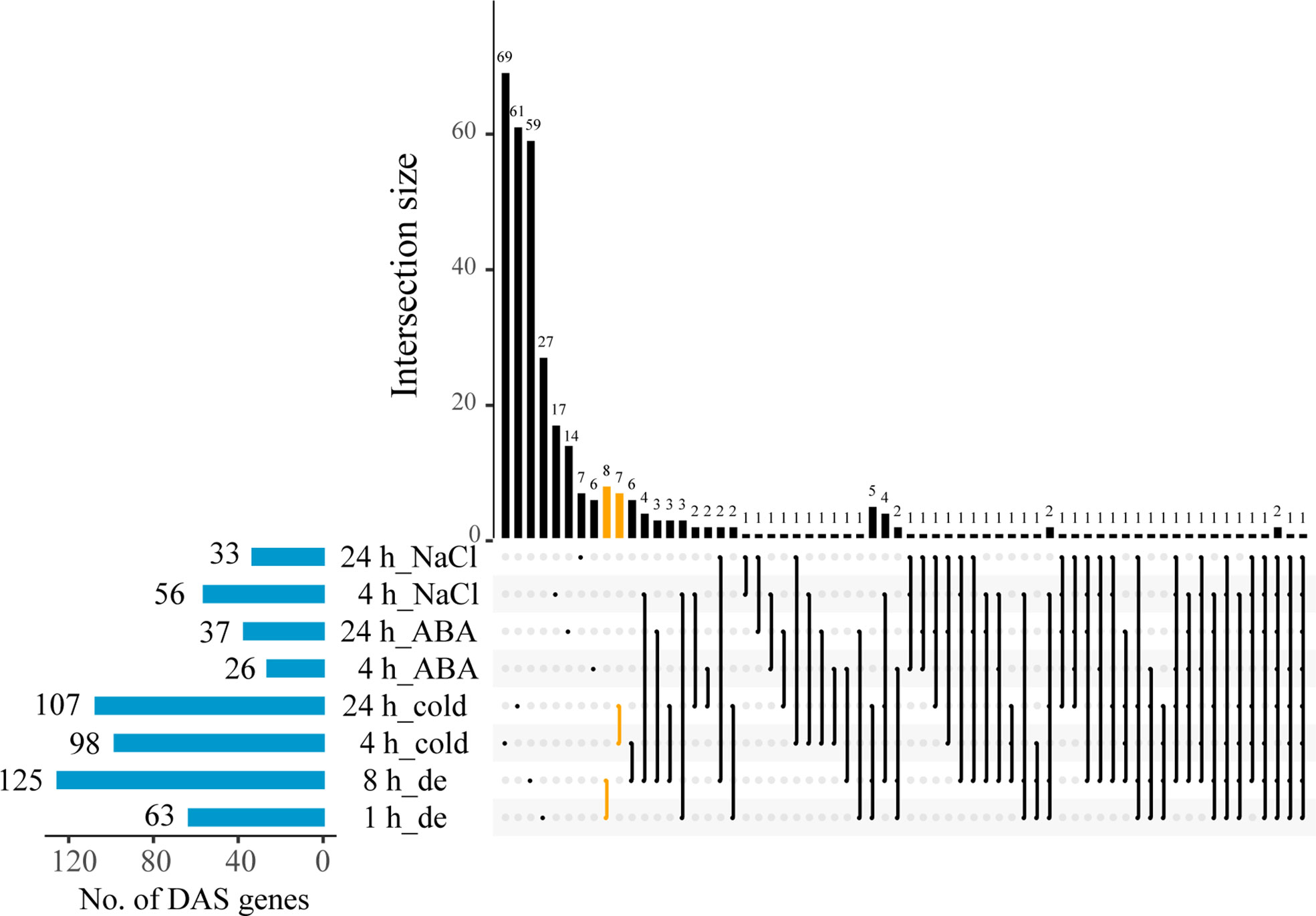
Figure 2 The numbers of differentially alternative splicing (DAS) genes between control and treatments with dehydration, cold, ABA, and NaCl at two different time points in B. napus.
In total, 286 homologs of 357 B. napus DAS genes were identified in A. thaliana (Supplementary Table 3), of which 45 (corresponding to 53 DAS genes in B. napus) were reported to be associated with abiotic stress (Naika et al., 2013). Moreover, 125 out of the 286 homologous genes have been previously shown to be AS genes according to the ASIP database (Wang and Brendel, 2006). These DAS genes included RBPs, TFs, and other important genes, such as RBP45C, LHY, MYB59, SCL30A, RS40, MAJ23.10, and DWF4. By GO enrichment analysis, all 357 DAS genes were found to be mainly involved in splicing and metabolic processes, including mRNA metabolism, mRNA splicing via spliceosome, RNA splicing via transesterification reactions, photoperiodism, and regulation of transcription (Supplementary Table 4).
To explore the potential genes/isoforms in response to various stress treatments, WGCNA was performed at the base of 35,527 identified isoforms with high expression level and top 50% TPM variances, to generate isoform co-expression network. A scale-free network was constructed based on weight value β of 6, and 23 co-expression modules were obtained accordingly (Figure 3). The generated modules were color-coded, and of note was that the gray module contained isoforms not assigned to any other modules. Remarkably, the isoform numbers of different modules varied greatly, ranging from 85 to 8,241, among which the turquoise module possessed most isoforms (8,241), while the dark turquoise module had the least number of isoforms (85) (Supplementary Figure 2).
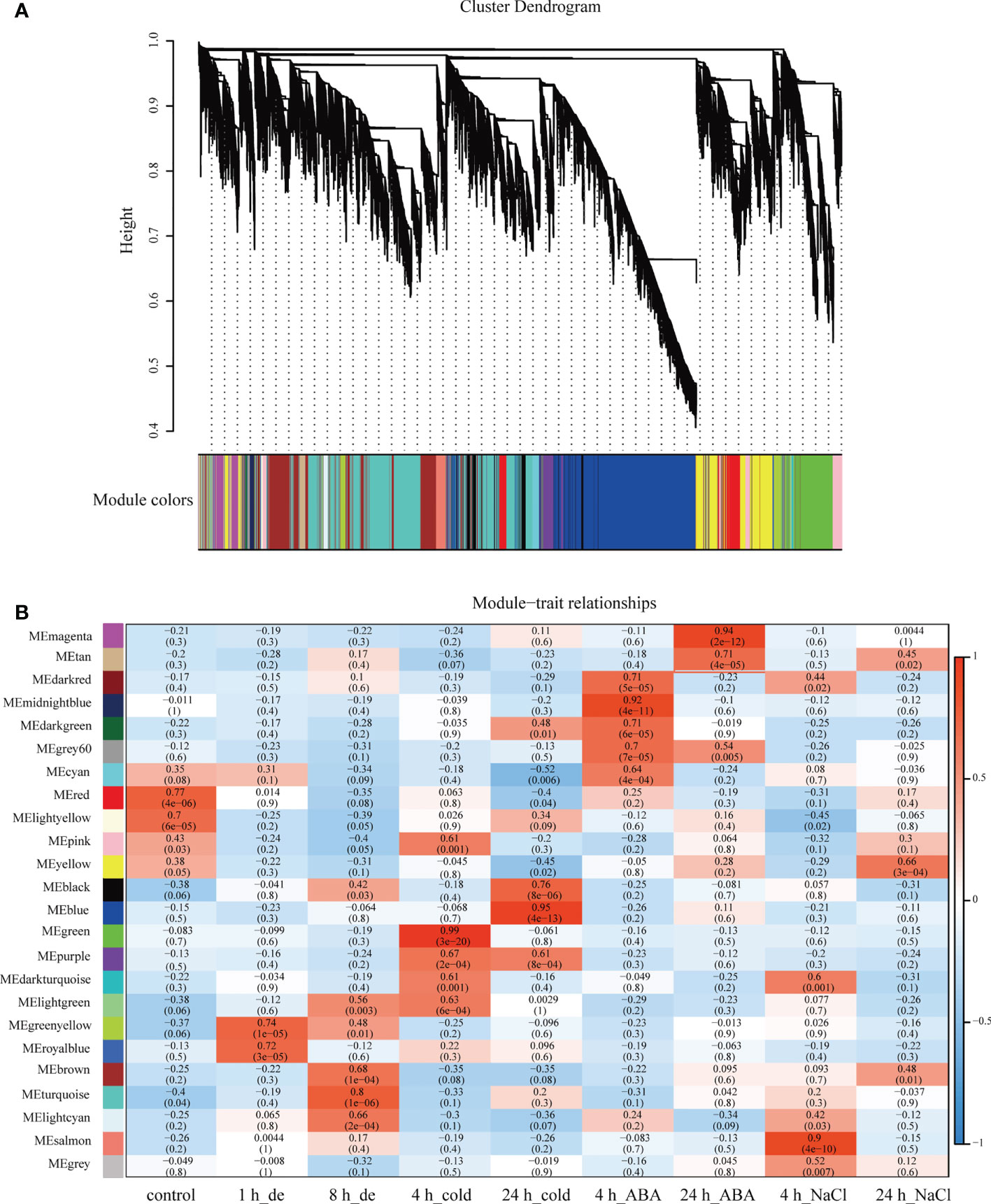
Figure 3 Co-expression network generation on isoform level by WGCNA. (A) Hierarchical cluster tree showing co-expression modules identified by WGCNA. Each leaf in the tree represents one isoform. The major tree branches constitute 23 modules, labeled with different colors. (B) Heatmap of correlation between modules and treatments. Each row corresponds to a module, labeled with a color as in panel (A).
Here, we identified 576 isoforms whose precursor genes were identified as 257 DAS genes, of which approximately 50% genes (131) produced hub isoforms in 16 stress-related modules, such as brown-8 h_de, turquoise-8 h_de, green-4 h_cold, pink-4 h_cold, blue-24 h_cold, magenta-24 h_ABA, and yellow-24 h_NaCl, suggesting that alternative splicing may play a central role in co-expression regulation of B. napus in response to abiotic stresses (Figure 4 and Supplementary Table 5). To explore gene functions within stress-related modules, GO enrichment analysis was performed, and the top 20 significantly enriched terms are shown in Supplementary Figure 3. For genes belonging to brown and turquoise modules associated with 8 h of dehydration, the significantly enriched GO terms (p < 0.05) were various processes of carbohydrate metabolism, water-soluble vitamin biosynthetic process, coenzyme biosynthetic/metabolic process, ubiquitin-dependent protein catabolic process, and splicing-related processes such as RNA splicing, nucleosome assembly, and DNA conformation change. As for 4 h cold-stress-related pink and green modules, the genes were mainly involved in the molecule metabolism/biosynthesis, transport, and the regulation of these processes. With the extension of cold stress, the blue module was identified to be related, and various processes mediating proteins synthesis and transport were detected, such as translation, translational elongation, ribosome biogenesis, cellular protein metabolic process, and protein transport. Additionally, the magenta and yellow modules were found to be associated with the later stages of ABA and NaCl treatments, respectively. The genes were mainly enriched in ion transports, nucleotide metabolic process, and fatty acid biosynthetic process for the former, while the genes were mostly responsible for carboxylic acid, cellular amino acid, and small molecule metabolic/biosynthetic/catabolic processes for the latter.
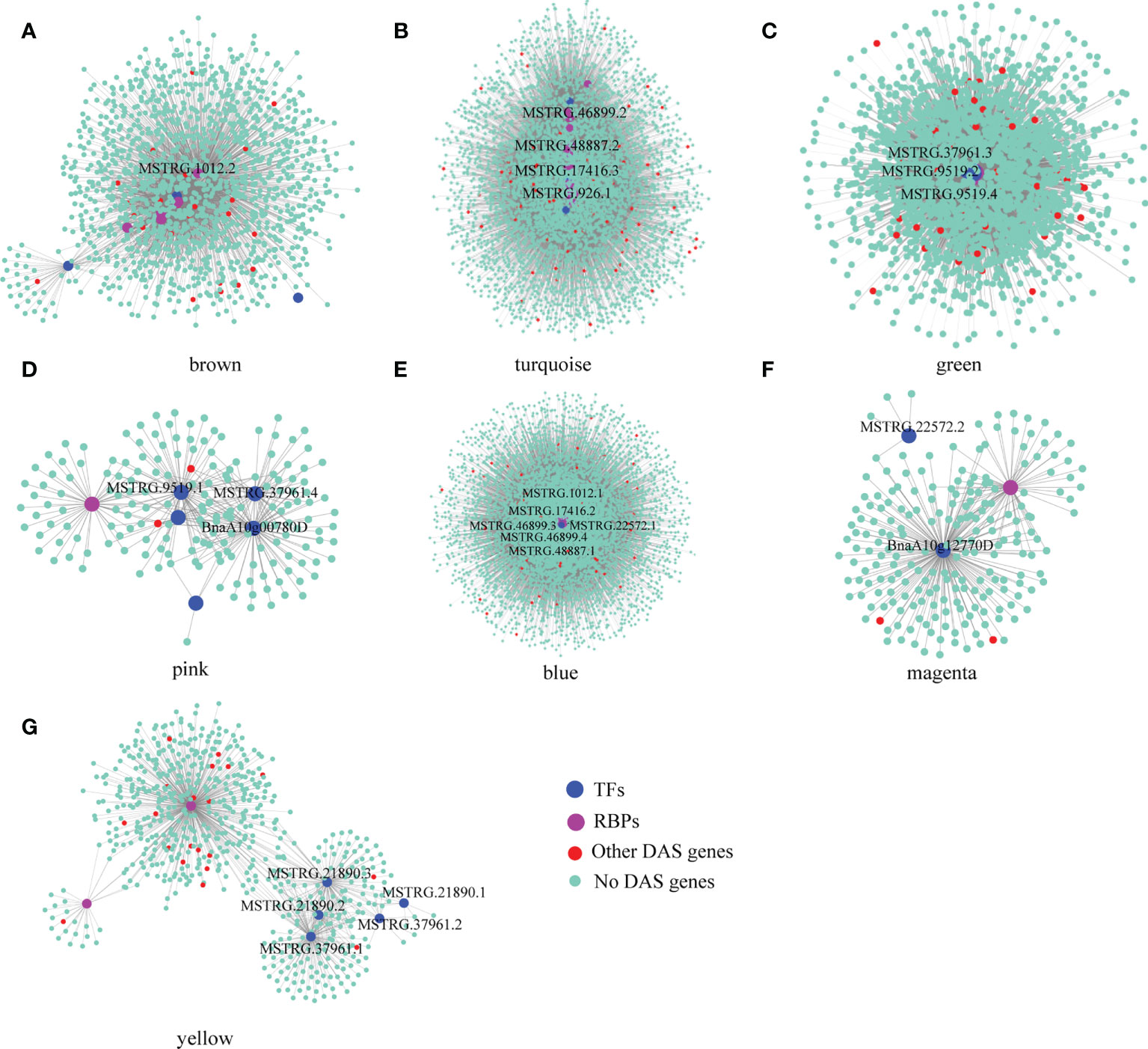
Figure 4 Visualization of the stress-related co-expression networks by Cytoscape. Panels (A–G), respectively, represented brown, turquoise, green, pink, blue, magenta, and yellow module. Using Cytoscape, the purple nodes represent the isoforms of the RBPs of DAS genes, the blue nodes represent the isoforms of TFs of DAS genes, the red nodes represent the isoforms of other DAS genes, and the light blue nodes represent other connected isoforms.
A total of 32 genes producing hub isoforms within stress-related modules were differentially alternatively spliced only under dehydration stress, including those encoding RNA-binding (RRM/RBD/RNP motifs) family protein, glycine-rich protein, tyrosine kinase family protein, and RING/U-box superfamily protein (Supplementary Table 5). For example, MSTRG.17416, encoding an RRM motif containing protein, was DAS through ES events under dehydration for 8 h. Based on the structure analysis of gene transcripts, ES events were found to occur between BnaA08g14210D/MSTRG.17416.2 and MSTRG.17416.3 (Figure 5A). Notably, the IncLevels of MSTRG.17416 were much higher under dehydration for 8 h than those of the mock control (Figure 5A), which indicated that this gene was prone to reduce ES events and retain exon 3 as much as possible, resulting into the generation of MSTRG.17416.3. As for the transcript levels of these divergent isoforms, MSTRG.17416.3 had the highest expression at 8 h of dehydration, which belonged to the turquoise module closely related to 8 h of dehydration (Figure 8). The protein encoded by MSTRG.17416.3 contained only two RRM_1 domains, while those encoded by other isoforms contained one more RRM_1 domain. Moreover, this gene was not differentially expressed, suggesting that alternative splicing provides an additional layer of transcriptional regulation in response to abiotic stress.
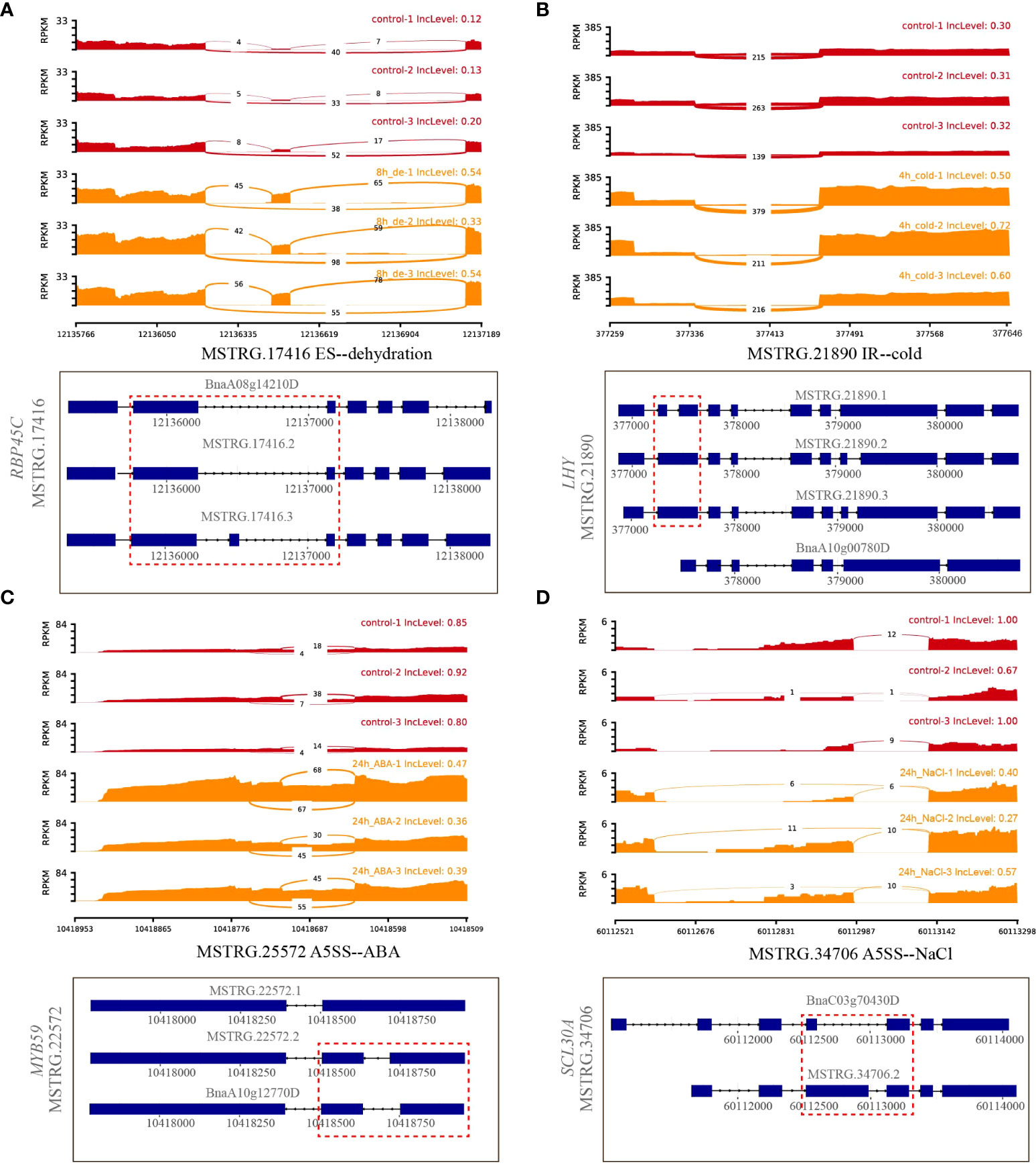
Figure 5 Quantitative visualization (sashimiplot) and transcripts structures of DAS genes under single abiotic stress. Panels (A–D) represented differential alternative splicing of different genes under dehydration, cold, ABA, and salt stress, respectively. Red dashed box indicated exons and introns showed in sashimiplots corresponding to gene structures below. The IncLevel value represented the normalized proportion of AS event. The red color denote control, and orange color denote treatments. The blue rectangular boxes denote the exons, the lines denote the introns, and the arrows denote the transcriptional direction.
In total, 56 hub DAS genes were found only under cold stress. These genes mainly encode homeodomain-like superfamily protein, WRKY DNA-binding protein 11, SC35-like splicing factor 33, DEAD/DEAH box RNA helicase, tyrosine kinase family protein, and RING/U-box superfamily protein (Supplementary Table 5). Among them, the MYB_related family TF MSTRG.21890 (LHY) encoding a homeodomain-like superfamily protein is homologous to AT1G01060, which was involved in circadian rhythm along with another myb transcription factor CCA1 in Arabidopsis (Adams et al., 2018). This gene experienced multiple AS events under cold stress for 4 h, including IR, ES, and A3SS, and also underwent IR event at 24 h of cold (Supplementary Table 5). Accordingly, IR events occurred between MSTRG.21890.1 and MSTRG.21890.2/MSTRG.21890.3 (Figure 5B). Remarkably, the IncLevels of IR events of MSTRG.21890 were much higher than those of control (Figure 5B). In addition, the transcript level of MSTRG.21890.1 was higher than that of the control at 4 h of cold (Figure 8). These results denoted that MSTRG.21890 tended to produce more MSTRG.21890.1 isoform that is responsive to cold stress.
Next, eight hub DAS genes were specifically found in response to ABA stress, including genes encoding cytochrome C1 family, cyclic nucleotide-binding transporter 1, and myb domain protein 59 MSTRG.22572 (MYB59) (Supplementary Table 5). Remarkably, AT4G31550, the homolog of MYB59 in Arabidopsis, has been reported to respond to ABA and cold stresses (Naika et al., 2013). This gene was identified as a DAS gene through A5SS event at 24 h of ABA (Supplementary Table 5), and the IncLevels were notably lower than the control, suggesting a tendency to increase the transcript of BnaA10g12770D (Figure 5C). Moreover, BnaA10g12770D displayed the highest expression level at 24 h of ABA, which was designated to the magenta module linked to 24 h of ABA (Figure 8). All of these findings indicated that MSTRG.22572 was likely to generate BnaA10g12770D isoform in response to ABA stress.
However, five hub DAS genes were identified only in NaCl stress, including SCL30A, SR30, HD2B, SLP1, and MQB2.8 (Supplementary Table 5). One splicing factor MSTRG.34706 (SCL30A) was differentially spliced via A5SS event at 24 h of NaCl (Supplementary Table 5). From the structures of gene transcripts, the A5SS event occurred between BnaC03g70430D and MSTRG.34706.2 (Figure 5D). The IncLevels of MSTRG.34706 were considerably lower than control at 24 h of NaCl stress (Figure 5D), suggesting that this gene enhanced the expression of BnaC03g70430D. Interestingly, this gene was not a differentially expressed gene (DEG), indicating that alternative splicing is of crucial importance for abiotic stress tolerance in B. napus.
A total of 30 DAS genes generating hub isoforms in stress-related modules were detected under at least two stresses. These genes mainly encode RRM/RBD/RNP motif protein, DEAD/DEAH box RNA helicase, ser-/arg-rich protein 34A, arginine-/serine-rich splicing factor 35, homeodomain-like superfamily protein, circadian clock associated 1, glycine-rich protein, cytochrome P450 protein, and RING/U-box superfamily protein (Supplementary Table 5). Among them, MSTRG.926 (RS40), encoding arginine/serine-rich splicing factor 35, was differentially alternatively spliced through IR event under multiple stress treatments, such as dehydration (8 h), cold (24h), ABA (4 h), and NaCl (4 h) (Figures 6A–D). Structure analyses revealed that IR event occurred between MSTRG.926.1 and MSTRG.926.3 (Figure 6E), and all IncLevels of MSTRG.926 showed much lower than those of the control under aforementioned treatments. MSTRG.926.1 had the highest expression level at 8 h of dehydration, which existed in the turquoise module closely related to 8 h of dehydration (Figure 8). These findings implied that this splicing factor encoding gene showed the same AS pattern in response to different stress conditions. Furthermore, one RBP MSTRG.1012, encoding RRM/RBD/RNP motif protein, was also shown as DAS by ES event under several stress treatments (Supplementary Figure 4). However, MSTRG.1012 was detected as a DEG only under dehydration and NaCl, but not ABA, which indicated that differential alternative splicing was partially independent of differential expression.
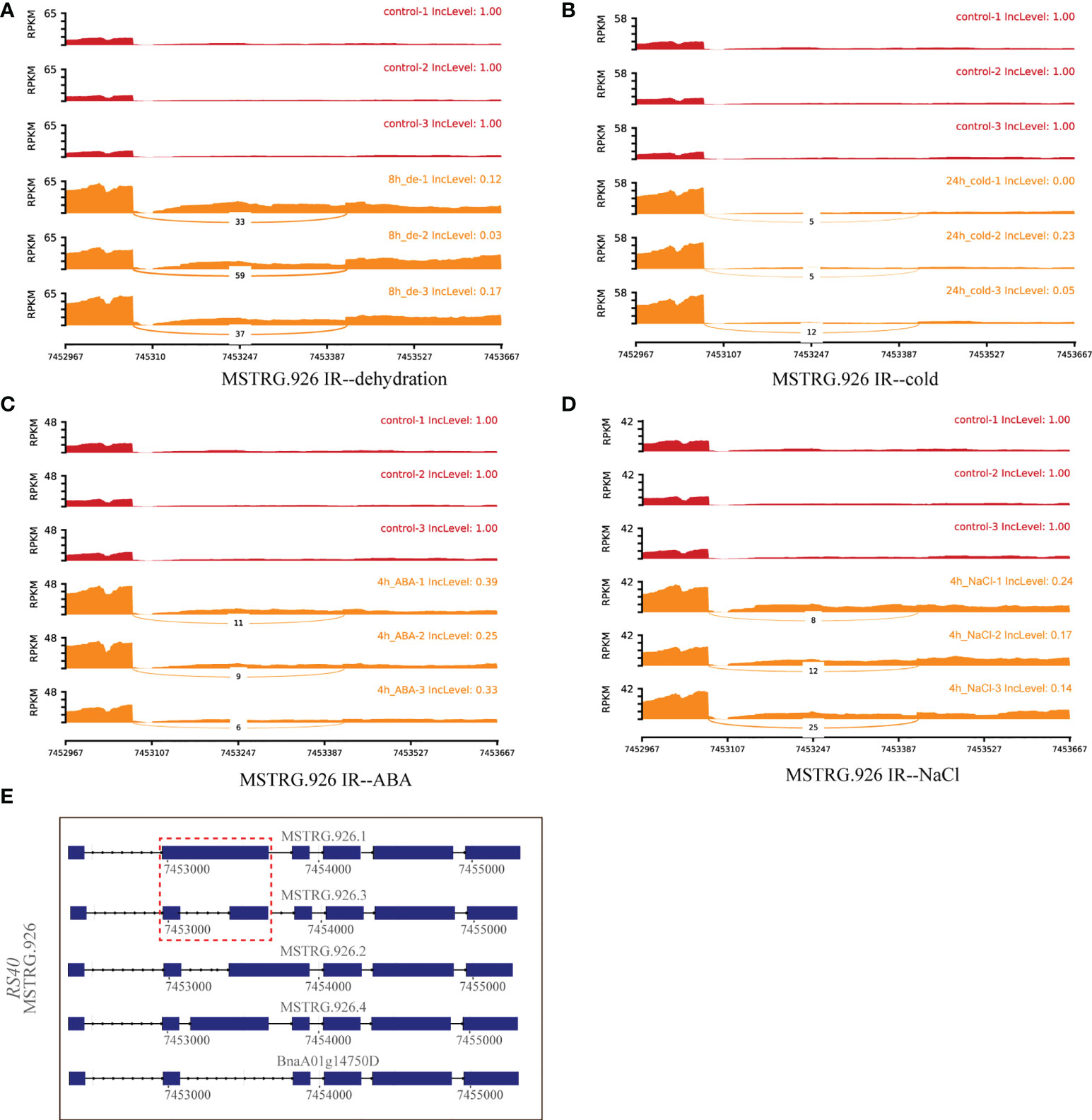
Figure 6 Quantitative visualization (sashimiplot) and transcripts structures of the DAS gene (MSTRG.926, RBP) under multiple abiotic stresses. Panels (A–D) represent dehydration, cold, ABA, and NaCl stress, respectively. The IncLevel value represents the normalized proportion of AS event. The red color denotes control, and orange color denotes treatments. Panel (E) displays the structures of all transcripts of this gene. Red dashed box indicates exons and introns shown in sashimiplots corresponding to gene structures below. The blue rectangular boxes denote the exons, the lines denote the introns, and the arrows denote the transcriptional direction.
Additionally, the myb-related transcription factor MSTRG.37961 (LHY), encoding homeodomain-like superfamily protein, experienced different IR events between MSTRG.37961.1 and MSTRG.37961.2/BnaC05g00840D under dehydration for 8 h and cold stress for 4 h (Figure 7). Remarkably, the IncLevels of IR event of MSTRG.37961 were significantly higher than what was found for the control under dehydration for 8 h (Figure 7A), whereas they were much lower under cold stress for 4 h (Figure 7B). Meanwhile, the transcript BnaC05g00840D displayed the highest expression at 4 h of cold, which was present in the green module associated with 4 h of cold (Figure 8). These results suggested that this TF encoding gene underwent different alternative splicing patterns in response to different stresses. Taken together, one gene may experience same or different AS patterns in response to diverse stresses.
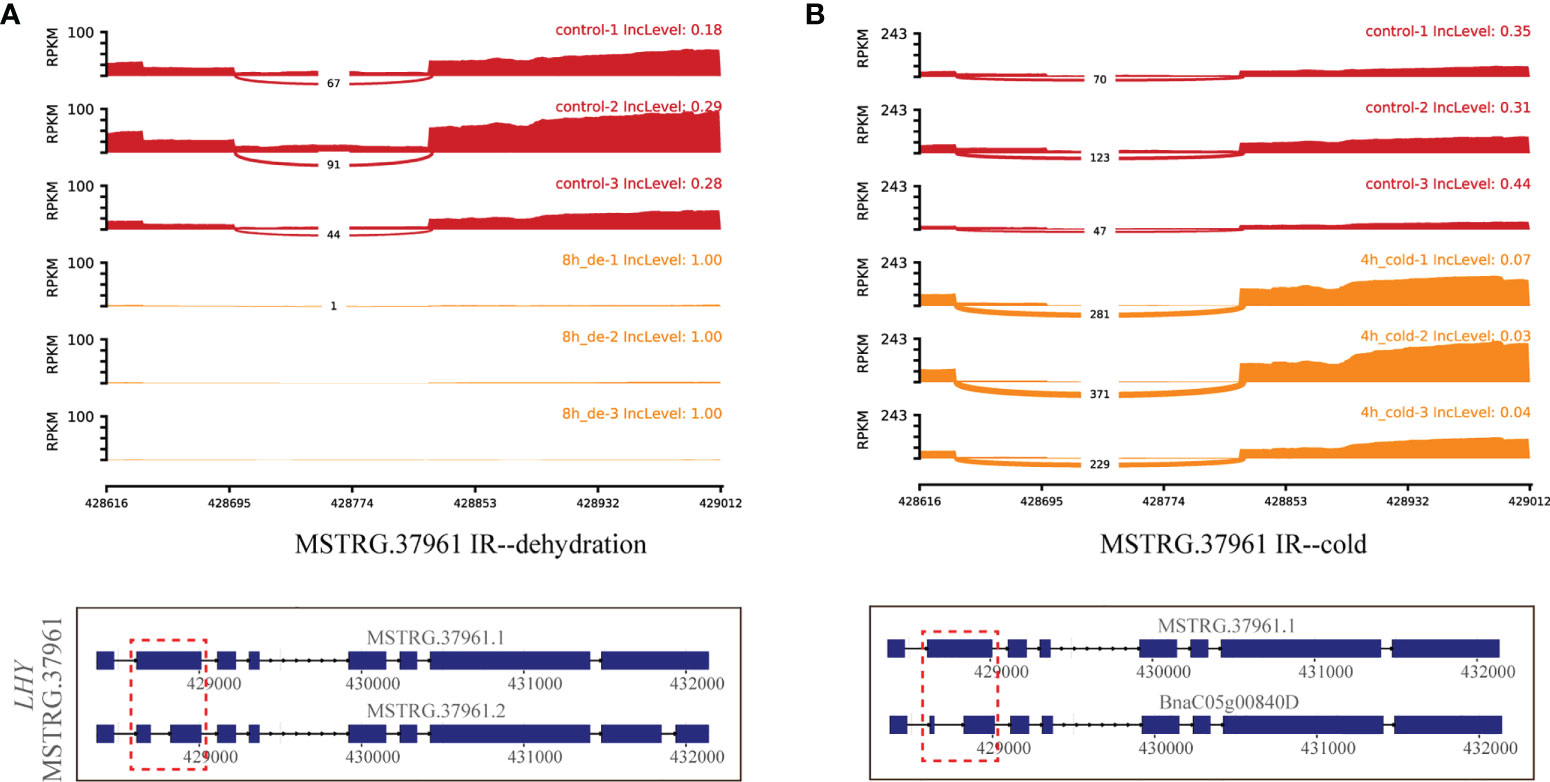
Figure 7 Quantitative visualization (sashimiplot) and transcripts structures of the DAS gene (MSTRG.37961, TF) under multiple abiotic stresses. Panels (A, B) represent dehydration and cold stress, respectively. Red dashed box indicates exons and introns shown in sashimiplots corresponding to gene structures below. The IncLevel value represents the normalized proportion of AS event. The red color denotes control, and orange color denotes treatments. The blue rectangular boxes denote the exons, the lines denote the introns, and the arrows denote the transcriptional direction.
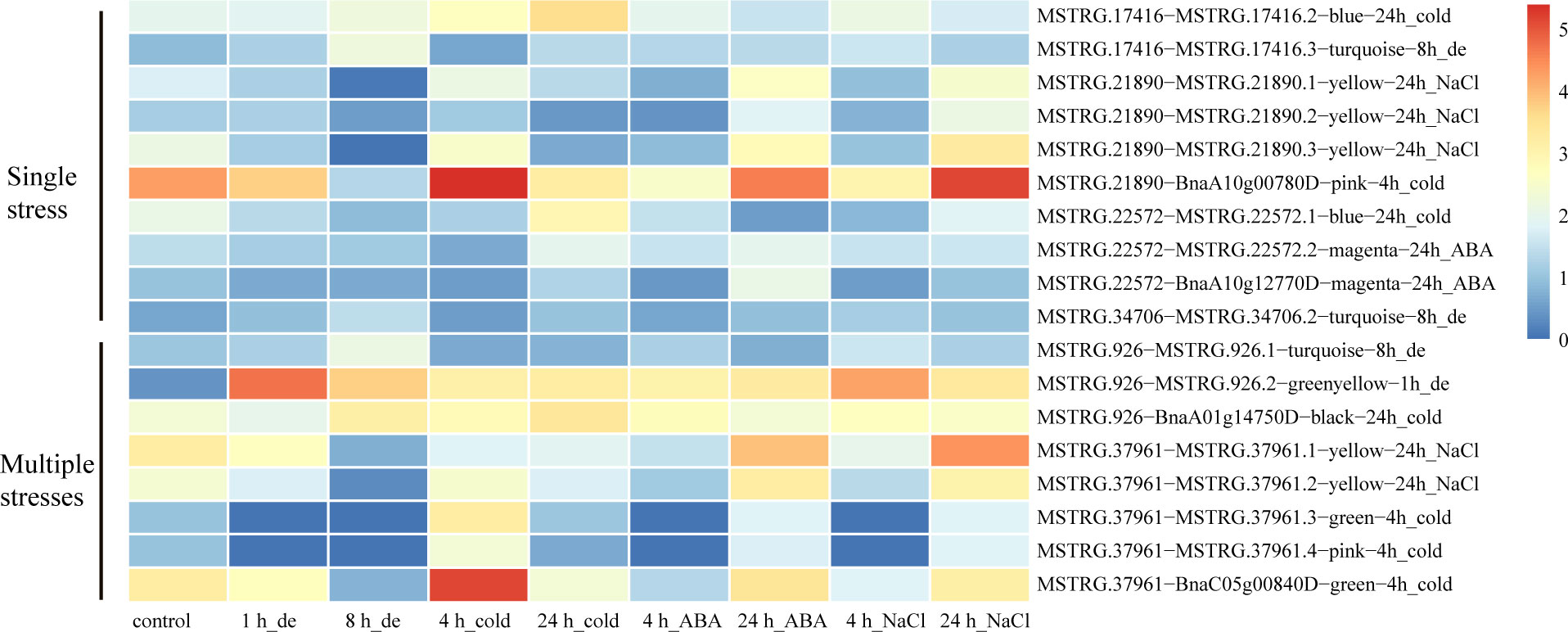
Figure 8 Expression heatmaps of each isoform of the candidate DAS genes under single stress or multiple stresses. The texts written behind the isoforms signifies their contained co-expression module that related to corresponding stress.
In this study, we aimed to determine the role of AS in response to multiple abiotic stresses by performing comparative transcriptome analyses on B. napus accession ZS11 at two different time points after treatments of dehydration, cold, ABA, and NaCl. Identification of DAS and analysis of co-expression networks at isoform level were conducted to elucidate plant internal response of AS to abiotic stresses. A total of 357 DAS genes were identified between control and treatments of the aforementioned four stresses, among which 131 DAS genes produced hub isoforms in the corresponding stress-related modules by WGCNA. These hub DAS genes included those encoding RNA-binding proteins, TFs, RING/U-box superfamily proteins, tyrosine kinase superfamily proteins, cytochrome P450 superfamily protein, and Transducin/WD40 repeat-like superfamily protein.
In the original analysis of Zhang et al. (2019), a total of 30,908 DEGs were identified under these four abiotic stresses, which were involved in small molecules and cellular macromolecule metabolism, ribosome biogenesis, and translation (Zhang et al., 2019). These two different analyses originating from the same transcriptome data led us to consider whether there is a potential link between AS and expression level. The subsequent comparative analysis revealed that a total of nearly 53% DAS genes were differentially expressed under all stress conditions (Supplementary Table 6). With the time course of different treatments, the ratios of DEGs from DAS genes showed an increasing trend under dehydration and low temperature, ranging from 33% to 67% and from 35% to 61%, respectively, while it showed an opposite trend under NaCl treatment, with the ratio of 62% to 39% (Supplementary Figure 5). Moreover, the proportions of differentially expressed DAS genes were similar at two time points after ABA treatment, about 54% and 51%, respectively (Supplementary Figure 5). Remarkably, a previous study also pinpointed out that approximately 42% DAS genes were identified as DEGs under combined heat and drought stresses in wheat (Liu et al., 2018). These results suggested that AS can work together with expression regulation to contribute to abiotic stress responses. Nevertheless, some studies indicated that AS might function independently to gene expression under abiotic and biotic stresses (Gu et al., 2019; Ma et al., 2019; Ma et al., 2020; Li et al., 2022a). Thereafter, the exact relationship between AS and expression level in response to stress needs to be further investigated in detail.
RBPs play an important role in the post-transcriptional gene regulation, such as modulating the activity and fate of RNA transcripts by binding to the specific target RNAs (Marondedze, 2020). A multitude of RBP-encoding genes were found to be induced under various abiotic stress conditions and played a comprehensive role in the process of response (Yan et al., 2022). In this study, we identified 16 RBP-encoding genes, such as CCR1, SR34a, SCL30A, SCL33, SR30, RS40, U1-70K, U1A, and U2AF65A, by integrating the DAS genes and hub genes identified from co-expression modules. One CCR1 gene MSTRG.34246 (GRP8), encoding a glycine-rich protein with RNA binding domain, was identified to be a DAS gene in reaction to dehydration, ABA, and NaCl stresses. AtGRP8 was previously reported to affect AS of downstream targets in Arabidopsis (Streitner et al., 2012). Furthermore, SR protein, a type of RBP, functions in pre-mRNA splicing and subsequent steps of post-transcriptional gene expression. It is composed of one or two RNA recognition motifs and a ser-/arg-rich domain responsible for recognizing pre-mRNA and recruiting spliceosomal components, respectively. It cannot only regulate the splicing of downstream target genes but also can be alternatively spliced itself (Joris et al., 2017; Xie et al., 2022). A growing body of evidence have reported that SR protein encoding genes underwent AS events in response to environmental stresses in plants. For example, SR45a and RS2Z33 produced different isoforms to respond to drought stress in G. uralensis (Li et al., 2022b). A few SR genes were certificated to exhibit responsive AS patterns under heat, drought, and salt stress conditions in wheat (Liu et al., 2018). In B. napus, the AS modes of SR splicing factors have also been reported to change under different stress conditions, such as boron deficiency, S. sclerotiorum, and L. maculans (Gu et al., 2019; Ma et al., 2019; Ma et al., 2020). In this study, two SR34a (MSTRG.46899 and MSTRG.27677) produced different isoforms in response to dehydration and cold stresses, suggesting that there was a certain responsive relationship between dehydration and cold. RS40 (MSTRG.926) also experienced more IR events under four abiotic stresses, indicating that it was a common splicing factor responding to multiple stresses. SCL30A (MSTRG.34706) and SR30 (MSTRG.38321) were differentially alternatively spliced under NaCl stress, while SCL33 (MSTRG.11046) was a DAS gene under cold stress. These results indicated that RBP-encoding genes play an important role in response to various abiotic stresses via alternative splicing.
Moreover, U1-70K, a U1 snRNP-specific protein encoding gene, plays a crucial role in the early steps of spliceosome formation via binding to 5′ splice sites and is involved in basic and alternative splicing of nuclear pre-mRNAs. U1-70K is important in a lot of biological processes, such as the development of sepals and petals and stress resistance response in Arabidopsis (Golovkin and Reddy, 2003; Amorim et al., 2018). In this study, U1-70K (MSTRG.48887) was found to undergo DAS to produce different isoforms belonging to two distinct modules (turquoise and blue) related to 8 h of dehydration and 24 h of cold, respectively. It was reported that U1 snRNP protein can interact with serine-/arginine-rich proteins to regulate splicing of pre-mRNAs in Arabidopsis (Golovkin and Reddy, 2003). A similar phenomenon was also found in our work, according to the co-expression of U1-70K and SR34a in the turquoise and blue modules. Spliceosomal protein encoding gene U1A (MSTRG.42942) was also a DAS gene identified in this study under stresses of dehydration and NaCl. Notably, a previous study demonstrated that U1A was involved in pre-mRNA processing and salt tolerance in A. thaliana (Gu et al., 2018). In addition, we also found that U2 snRNP auxiliary factor encoding gene, U2AF65A (MSTRG.17488), was DAS by A3SS event under 4 h of cold. OsU2AF65A was certificated to mediate response to various stresses, such as drought, cold, high salt, and heavy metal exposure in rice (Lu et al., 2021).
Transcription factors are proteins that bind to specific cis-acting response elements in the promoters of stress-inducible genes, which can modulate the transcription of target genes and control key downstream responses, thus enhancing the stress tolerance in plants. In this study, two LHY genes (MSTRG.37961 and MSTRG.21890) and one CCA1 gene (MSTRG.9519) were identified to be DAS genes in reaction to dehydration and/or cold stresses. LHY played a crucial role in promoting the expression of ABA-responsive genes, which improved Arabidopsis tolerance to drought and osmotic stresses (Yamaura et al., 2020). Furthermore, AS of LHY was also proved to influence circadian rhythm, and CCA1 can produce two isoforms in Arabidopsis, a full-length isoform CCA1α and a truncated isoform CCA1β resulting from an intron retention, which exhibited enhancive tolerance and sensitivity to cold, respectively (Seo et al., 2012). AS of CCA1 was also reported to regulate the circadian clock in response to low temperature in B. napus (Lee and Adams, 2020).
In our work, HSFA1E (MSTRG.11411) and WRKY11 (MSTRG.26624) were differentially alternatively spliced owing to cold stress. The heat shock transcription factors, HSFA1D and HSFA6B, were induced to produce different isoforms by AS under drought and heat stresses in wheat (Liu et al., 2018). WRKY19 and WRKY20 were also prone to generate corresponding isoform under drought stress in G. uralensis (Li et al., 2022a). A DAS gene (MSTRG.22572) encoding myb domain protein 59 was identified in responding to ABA stress. MYB59 was recently reported to respond to low K+ stress in Arabidopsis (Du et al., 2019), and its transcriptional level was also influenced by alternative splicing in Arabidopsis and rice (Li et al., 2006). These results revealed that transcription factors cannot only activate or inhibit gene transcription, but also undergo constitutive and inducible AS under abiotic stresses. Interestingly, we found that TFs tended to co-express with RBPs in the modules related to various abiotic stresses, and both of them were hub genes/isoforms, which was consistent with previous reports (He and Hu, 2021).
Several other important DAS genes were also pinpointed in response to abiotic stress. For example, RING/U-box superfamily protein encoding gene MSTRG.50841 was DAS by A5SS event under cold, ABA, and NaCl stresses. It is homologous to AT5G10650, which was involved in ABA-mediated microtubule depolymerization and tolerance response to drought stress (Yu et al., 2020). A DAS gene MSTRG.27581 (DWF4), encoding P450 protein, was identified in reaction to dehydration, cold, and NaCl stresses. DWF4 played a key role in brassinosteroid (BR) homeostasis and was related to root elongation in BR-auxin crosstalk in Arabidopsis (Rahman et al., 2011). Transducin/WD40 repeat-like superfamily protein encoding gene MSTRG.42815 was also a DAS gene identified in this study under stresses of dehydration and cold. Its homolog in Arabidopsis, AT1G78070, was involved in drought, cold, ABA, and NaCl stresses (Naika et al., 2013). In addition, MSTRG.16072 and MSTRG.42565, encoding tyrosine kinase family proteins, were DAS genes under dehydration and cold stress, respectively. Both of these two genes were homologous to AT1G73660, which may function as a negative regulator of salt tolerance (Gao and Xiang, 2008).
In this study, a total of 357 DAS genes were identified by comparative transcriptome analyses under abiotic stress conditions, such as dehydration, cold, ABA, and salt stress. Through isoform-based WGCNA, 23 co-expression modules related to different stresses were determined, and a number of crucial genes, such as RBPs and TFs, changed AS modes in responding to single abiotic stress or multiple stresses in B. napus. The identified potential candidate isoforms could be useful targets for further functional studies to explore the molecular mechanism of AS in response to abiotic stresses. Taken together, this study provides novel insights of AS dynamics associated with abiotic stresses in B. napus and forms a beneficial resource for molecular breeding to create cultivars tolerant to multiple stresses not only in B. napus but also in other crops.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
LLY, MX, and SL designed the research. LLY performed the research. JL, CZ, YZ, CT and XC supplied assistance in data analysis. LLY wrote the draft manuscript. MX, LY, HJ, and JS revised the manuscript. CT and SL applied the funding. All authors contributed to the article and approved the submitted version.
This research was funded by the Yong Top-notch Talent Cultivation Program of Hubei Province for Dr. Chaobo Tong, the National Natural Science Foundation of China (No. 31770250), the Agricultural Science and Technology Innovation Program of Chinese Academy of Agricultural Sciences (CAAS-ASTIP-2013-OCRI), and the Earmarked Fund for China Agriculture Research System (CARS-12).
We sincerely thank Prof. Liang Guo (Huazhong Agricultural University) for publishing and uploading the sequencing data to the public database. We sincerely thank Zhixian Qiao of the Analysis and Testing Center at IHB for technical supports in RNA-seq analysis.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1009998/full#supplementary-material
Supplementary Figure 1 | IGV showing the RNA-seq reads aligned to each transcript of DAS gene under control and stress conditions. (A, B) represented MSTRG.1012 and MSTRG.17416, respectively. Green box indicated the position of the skipped exons and the reads spanning the junction. The transcripts structures were presented as dark blue exon-plots at the bottom: boxes represented the exons, lines represented the introns, and arrows represented the transcriptional direction.
Supplementary Figure 2 | Distribution of the number of genes/isoforms contained in each module.
Supplementary Figure 3 | Top20 GO terms enrichment of genes in the seven stress-related modules. (A-G) respectively represented brown, turquoise, green, pink, blue, magenta, and yellow module. The x-axis represented the -log10(Pvalue) which shows the significant level. The size of the bubble represented the enriched gene number.
Supplementary Figure 4 | Quantitative visualization (sashimiplot) and transcripts structures of the DAS gene (MSTRG.1012, RBP) under multiple abiotic stresses. (A-C) represented dehydration, ABA and NaCl stress, respectively. The IncLevel value represented the normalized proportion of AS event. The red color denoted control and orange color denoted treatments. (D) displayed the structures of all transcripts of this gene. Red dashed box indicated exons and introns showed in sashimiplots corresponding to gene structures below. The blue rectangular boxes denoted the exons, the lines denoted the introns, and the arrows denoted the transcriptional direction.
Supplementary Figure 5 | The percentage of DAS genes overlapping with DEGs under each stress condition.
Adams, S., Grundy, J., Veflingstad, S. R., Dyer, N. P., Hannah, M. A., Ott, S., et al. (2018). Circadian control of abscisic acid biosynthesis and signalling pathways revealed by genome-wide analysis of LHY binding targets. New Phytol. 220 (3), 893–907. doi: 10.1111/nph.15415
Amorim, M. D., Willing, E. M., Szabo, E. X., Francisco-Mangilet, A. G., Droste-Borel, I., Macek, B., et al. (2018). The U1 snRNP subunit LUC7 modulates plant development and stress responses via regulation of alternative splicing. Plant Cell 30 (11), 2838–2854. doi: 10.1105/tpc.18.00244
Bolger, A. M., Lohse, M., Usadel, B. (2014). Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 30 (15), 2114–2120. doi: 10.1093/bioinformatics/btu170
Chalhoub, B., Denoeud, F., Liu, S., Parkin, I. A., Tang, H., Wang, X., et al. (2014). Early allopolyploid evolution in the post-neolithic Brassica napus oilseed genome. Science 345 (6199), 950–953. doi: 10.1126/science.1253435
Chang, C. Y., Lin, W. D., Tu, S. L. (2014). Genome-wide analysis of heat-sensitive alternative splicing in Physcomitrella patens. Plant Physiol. 165 (2), 826–840. doi: 10.1104/pp.113.230540
Chen, C. J., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y. H., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13 (8), 1194–1202. doi: 10.1016/j.molp.2020.06.009
Cheng, C. H., Wang, Z. J., Yuan, B. J., Li, X. (2017). RBM25 mediates abiotic responses in plants. Front. Plant Sci. 8: 292. doi: 10.3389/fpls.2017.00292
Cheng, Q., Zhou, Y., Liu, Z., Zhang, L., Song, G., Guo, Z., et al. (2015). An alternatively spliced heat shock transcription factor, OsHSFA2dI, functions in the heat stress-induced unfolded protein response in rice. Plant Biol. 17 (2), 419–429. doi: 10.1111/plb.12267
Chen, Q., Han, Y., Liu, H., Wang, X., Sun, J., Zhao, B., et al. (2018). Genome-wide association analyses reveal the importance of alternative splicing in diversifying gene function and regulating phenotypic variation in maize. Plant Cell 30 (7), 1404–1423. doi: 10.1105/tpc.18.00109
Ding, F., Cui, P., Wang, Z. Y., Zhang, S. D., Ali, S., Xiong, L. M. (2014). Genome-wide analysis of alternative splicing of pre-mRNA under salt stress in Arabidopsis. BMC Genom. 15 (1), 431. doi: 10.1186/1471-2164-15-431
Drechsel, G., Kahles, A., Kesarwani, A. K., Stauffer, E., Behr, J., Drewe, P., et al. (2013). Nonsense-mediated decay of alternative precursor mRNA splicing variants is a major determinant of the Arabidopsis steady state transcriptome. Plant Cell 25 (10), 3726–3742. doi: 10.1105/tpc.113.115485
Du, C. F., Hu, K. N., Xian, S. S., Liu, C. Q., Fan, J. C., Tu, J. X., et al. (2016). Dynamic transcriptome analysis reveals AP2/ERF transcription factors responsible for cold stress in rapeseed (Brassica napus l.). Mol. Genet. Genom. 291 (3), 1053–1067. doi: 10.1007/s00438-015-1161-0
Du, X. Q., Wang, F. L., Li, H., Jing, S., Yu, M., Li, J., et al. (2019). The transcription factor MYB59 regulates K+/NO3- translocation in the Arabidopsis response to low k+ stress. Plant Cell 31 (3), 699–714. doi: 10.1105/tpc.18.00674
Erkelenz, S., Mueller, W. F., Evans, M. S., Busch, A., Schoneweis, K., Hertel, K. J., et al. (2013). Position-dependent splicing activation and repression by SR and hnRNP proteins rely on common mechanisms. RNA 19 (1), 96–102. doi: 10.1261/rna.037044.112
Foissac, S., Sammeth, M. (2007). ASTALAVISTA: dynamic and flexible analysis of alternative splicing events in custom gene datasets. Nucleic Acids Res. 35, W297–W299. doi: 10.1093/nar/gkm311
Gao, L., Xiang, C. B. (2008). The genetic locus At1g73660 encodes a putative MAPKKK and negatively regulates salt tolerance in Arabidopsis. Plant Mol. Biol. 67 (1-2), 125–134. doi: 10.1007/s11103-008-9306-8
Geuens, T., Bouhy, D., Timmerman, V. (2016). The hnRNP family: insights into their role in health and disease. Hum. Genet. 135 (8), 851–867. doi: 10.1007/s00439-016-1683-5
Golovkin, M., Reddy, A. S. (2003). Expression of U1 small nuclear ribonucleoprotein 70K antisense transcript using APETALA3 promoter suppresses the development of sepals and petals. Plant Physiol. 132 (4), 1884–1891. doi: 10.1104/pp.103.023192
Gu, J., Li, W., Wang, S., Zhang, X., Coules, A., Ding, G., et al. (2019). Differential alternative splicing genes in response to boron deficiency in Brassica napus. Genes 10 (3), 224. doi: 10.3390/genes10030224
Guo, Y., Li, J., Fang, Y., Wan, Y., Tang, J., Wei, T., et al. (2019). An event of alternative splicing affects the expression of two BnCYCD3-1-like genes in. Brassica. napus. Gene 694, 33–41. doi: 10.1016/j.gene.2018.12.085
Gu, J., Xia, Z., Luo, Y., Jiang, X., Qian, B., Xie, H., et al. (2018). Spliceosomal protein U1A is involved in alternative splicing and salt stress tolerance in Arabidopsis thaliana. Nucleic Acids Res. 46 (4), 1777–1792. doi: 10.1093/nar/gkx1229
He, M., Hu, F. (2021). TF-RBP-AS triplet analysis reveals the mechanisms of aberrant alternative splicing events in kidney cancer: Implications for their possible clinical use as prognostic and therapeutic biomarkers. Int. J. Mol. Sci. 22 (16), 8789. doi: 10.3390/ijms22168789
Iwakawa, H. O., Tomari, Y. (2015). The functions of MicroRNAs: mRNA decay and translational repression. Trends Cell Biol. 25 (11), 651–665. doi: 10.1016/j.tcb.2015.07.011
Jiang, J. F., Liu, X. N., Liu, C. H., Liu, G. T., Li, S. H., Wang, L. J. (2017). Integrating omics and alternative splicing reveals insights into grape response to high temperature. Plant Physiol. 173 (2), 1502–1518. doi: 10.1104/pp.16.01305
Joris, M., Schloesser, M., Baurain, D., Hanikenne, M., Muller, M., Motte, P. (2017). Number of inadvertent RNA targets for morpholino knockdown in danio rerio is largely underestimated: evidence from the study of Ser/Arg-rich splicing factors. Nucleic Acids Res. 45 (16), 9547–9557. doi: 10.1093/nar/gkx638
Kalyna, M., Simpson, C. G., Syed, N. H., Lewandowska, D., Marquez, Y., Kusenda, B., et al. (2012). Alternative splicing and nonsense-mediated decay modulate expression of important regulatory genes in Arabidopsis. Nucleic Acids Res. 40 (6), 2454–2469. doi: 10.1093/nar/gkr932
Kim, D., Landmead, B., Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12 (4), 357–360. doi: 10.1038/Nmeth.3317
Kohl, M., Wiese, S., Warscheid, B. (2011). Cytoscape: Software for visualization and analysis of biological networks. Methods Mol. Biol. 696, 291–303. doi: 10.1007/978-1-60761-987-1_18
Kruse, C. P. S., Meyers, A. D., Basu, P., Hutchinson, S., Luesse, D. R., Wyatt, S. E. (2020). Spaceflight induces novel regulatory responses in Arabidopsis seedling as revealed by combined proteomic and transcriptomic analyses. BMC Plant Biol. 20 (1), 237. doi: 10.1186/s12870-020-02392-6
Laloum, T., Martin, G., Duque, P. (2018). Alternative splicing control of abiotic stress responses. Trends Plant Sci. 23 (2), 140–150. doi: 10.1016/j.tplants.2017.09.019
Langfelder, P., Horvath, S. (2008). WGCNA: an r package for weighted correlation network analysis. BMC Bioinform. 9, 559. doi: 10.1186/1471-2105-9-559
Lee, J. S., Adams, K. L. (2020). Global insights into duplicated gene expression and alternative splicing in polyploid Brassica napus under heat, cold, and drought stress. Plant Genome 13 (3), e20057. doi: 10.1002/tpg2.20057
Li, M., Hu, M., Xiao, Y., Wu, X., Wang, J. (2022b). The activation of gene expression and alternative splicing in the formation and evolution of allopolyploid Brassica napus. Hortic. Res. 9, uhab075. doi: 10.1093/hr/uhab075
Li, J., Li, X., Guo, L., Lu, F., Feng, X., He, K., et al. (2006). A subgroup of MYB transcription factor genes undergoes highly conserved alternative splicing in Arabidopsis and rice. J. Exp. Bot. 57 (6), 1263–1273. doi: 10.1093/jxb/erj094
Liu, Z., Qin, J., Tian, X., Xu, S., Wang, Y., Li, H., et al. (2018). Global profiling of alternative splicing landscape responsive to drought, heat and their combination in wheat (Triticum aestivum l.). Plant Biotechnol. J. 16 (3), 714–726. doi: 10.1111/pbi.12822
Liu, J. J., Sun, N., Liu, M., Liu, J. C., Du, B. J., Wang, X. J., et al. (2013). An autoregulatory loop controlling Arabidopsis HsfA2 expression: Role of heat shock-induced alternative splicing. Plant Physiol. 162 (1), 512–521. doi: 10.1104/pp.112.205864
Liu, H. J., Wang, J. C., Zhang, B. B., Yang, X. Y., Hammond, J. P., Ding, G. D., et al. (2021). Genome-wide association study dissects the genetic control of plant height and branch number in response to low-phosphorus stress in Brassica napus. Ann. Bot. 128 (7), 919–929. doi: 10.1093/aob/mcab115
Li, G. Z., Xu, D. X., Huang, G., Bi, Q., Yang, M., Shen, H. T., et al. (2022a). Analysis of whole-transcriptome RNA-seq data reveals the involvement of alternative splicing in the drought response of Glycyrrhiza uralensis. Front. Genet. 13, 885651. doi: 10.3389/fgene.2022.885651
Lloyd, J. P. B., Lang, D., Zimmer, A. D., Causier, B., Reski, R., Davies, B. (2018). The loss of SMG1 causes defects in quality control pathways in Physcomitrella patens. Nucleic Acids Res. 46 (11), 5822–5836. doi: 10.1093/nar/gky225
Lu, S., Gao, C., Wang, Y., He, Y., Du, J., Chen, M., et al. (2021). Phylogenetic analysis of the plant U2 snRNP auxiliary factor Large subunit a gene family in response to developmental cues and environmental stimuli. Front. Plant Sci. 12, 739671. doi: 10.3389/fpls.2021.739671
Marondedze, C. (2020). The increasing diversity and complexity of the RNA-binding protein repertoire in plants. Proc. R. Soc B. 287 (1935), 20201397. doi: 10.1098/rspb.2020.1397
Matsukura, S., Mizoi, J., Yoshida, T., Todaka, D., Ito, Y., Maruyama, K., et al. (2010). Comprehensive analysis of rice DREB2-type genes that encode transcription factors involved in the expression of abiotic stress-responsive genes. Mol. Genet. Genom. 283 (2), 185–196. doi: 10.1007/s00438-009-0506-y
Ma, J. Q., Wei, L. J., Lin, A., Zhang, C., Sun, W., Yang, B., et al. (2019). Brassica napus infected with Leptosphaeria maculans. Genes 10 (4), 296. doi: 10.3390/genes10040296
Ma, J. Q., Xu, W., Xu, F., Lin, A., Sun, W., Jiang, H. H., et al. (2020). Differential alternative splicing genes and isoform regulation networks of rapeseed (Brassica napus l.) infected with Sclerotinia sclerotiorum. Genes 11 (7), 784. doi: 10.3390/genes11070784
Naika, M., Shameer, K., Mathew, O. K., Gowda, R., Sowdhamini, R. (2013). STIFDB2: An updated version of plant stress-responsive transcription factor DataBase with additional stress signals, stress-responsive transcription factor binding sites and stress-responsive genes in Arabidopsis and rice. Plant Cell Physiol. 54 (2), e8. doi: 10.1093/pcp/pcs185
Palusa, S. G., Ali, G. S., Reddy, A. S. N. (2007). Alternative splicing of pre-mRNAs of Arabidopsis serine/arginine-rich proteins: regulation by hormones and stresses. Plant J. 49 (6), 1091–1107. doi: 10.1111/j.1365-313X.2006.03020.x
Pertea, M., Pertea, G. M., Antonescu, C. M., Chang, T. C., Mendell, J. T., Salzberg, S. L. (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33 (3), 290–295. doi: 10.1038/nbt.3122
Pu, Y. Y., Liu, L. J., Wu, J. Y., Zhao, Y. H., Bai, J., Ma, L., et al. (2019). Transcriptome profile analysis of winter rapeseed (Brassica napus l.) in response to freezing stress, reveal potentially connected events to freezing stress. Int. J. Mol. Sci. 20 (11), 2771. doi: 10.3390/ijms20112771
Rahman, A., Yoshimitsu, Y., Tanaka, K., Fukuda, W., Asami, T., Yoshida, S., et al. (2011). Transcription of DWARF4 plays a crucial role in auxin-regulated root elongation in addition to brassinosteroid homeostasis in Arabidopsis thaliana. PloS One 6 (8), 23851. doi: 10.1371/journal.pone.0023851
Raza, A., Su, W., Hussain, M. A., Mehmood, S. S., Zhang, X. K., Cheng, Y., et al. (2021). Integrated analysis of metabolome and transcriptome reveals insights for cold tolerance in rapeseed (Brassica napus l.). Front. Plant Sci. 12, 721681. doi: 10.3389/fpls.2021.721681
Reddy, A. S., Marquez, Y., Kalyna, M., Barta, A. (2013). Complexity of the alternative splicing landscape in plants. Plant Cell 25 (10), 3657–3683. doi: 10.1105/tpc.113.117523
Ruan, J., Guo, F., Wang, Y. Y., Li, X. G., Wan, S. B., Shan, L., et al. (2018). Transcriptome analysis of alternative splicing in peanut (Arachis hypogaea l.). BMC Plant Biol. 18(1): 139. doi: 10.1186/s12870-018-1339-9
Seo, P. J., Kim, M. J., Ryu, J. Y., Jeong, E. Y., Park, C. M. (2011). Two splice variants of the IDD14 transcription factor competitively form nonfunctional heterodimers which may regulate starch metabolism. Nat. Commun. 2, 303. doi: 10.1038/ncomms1303
Seo, P. J., Park, M. J., Lim, M. H., Kim, S. G., Lee, M., Baldwin, I. T., et al. (2012). A self-regulatory circuit of CIRCADIAN CLOCK-ASSOCIATED1 underlies the circadian clock regulation of temperature responses in Arabidopsis. Plant Cell 24 (6), 2427–2442. doi: 10.1105/tpc.112.098723
Shen, S., Park, J. W., Lu, Z. X., Lin, L., Henry, M. D., Wu, Y. N., et al. (2014a). rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-seq data. Proc. Natl. Acad. Sci. U.S.A. 111 (51), E5593–E5601. doi: 10.1073/pnas.1419161111
Shen, Y. T., Zhou, Z. K., Wang, Z., Li, W. Y., Fang, C., Wu, M., et al. (2014b). Global dissection of alternative splicing in paleopolyploid soybean. Plant Cell 26 (3), 996–1008. doi: 10.1105/tpc.114.122739
Staiger, D., Brown, J. W. (2013). Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell 25 (10), 3640–3656. doi: 10.1105/tpc.113.113803
Streitner, C., Koster, T., Simpson, C. G., Shaw, P., Danisman, S., Brown, J. W., et al. (2012). An hnRNP-like RNA-binding protein affects alternative splicing by in vivo interaction with transcripts in Arabidopsis thaliana. Nucleic Acids Res. 40 (22), 11240–11255. doi: 10.1093/nar/gks873
Sugio, A., Dreos, R., Aparicio, F., Maule, A. J. (2009). The cytosolic protein response as a subcomponent of the wider heat shock response in Arabidopsis. Plant Cell 21 (2), 642–654. doi: 10.1105/tpc.108.062596
Syed, N. H., Kalyna, M., Marquez, Y., Barta, A., Brown, J. W. S. (2012). Alternative splicing in plants-coming of age. Trends Plant Sci. 17 (10), 616–623. doi: 10.1016/j.tplants.2012.06.001
Tanabe, N., Yoshimura, K., Kimura, A., Yabuta, Y., Shigeoka, S. (2007). Differential expression of alternatively spliced mRNAs of Arabidopsis SR protein homologs, atSR30 and atSR45a, in response to environmental stress. Plant Cell Physiol. 48 (12), 1826–1826. doi: 10.1093/pcp/pcm156
Terashima, A., Takumi, S. (2009). Allopolyploidization reduces alternative splicing efficiency for transcripts of the wheat DREB2 homolog, WDREB2. Genome 52 (1), 100–105. doi: 10.1139/G08-101
Thatcher, S. R., Danilevskaya, O. N., Meng, X., Beatty, M., Zastrow-Hayes, G., Harris, C., et al. (2016). Genome-wide analysis of alternative splicing during development and drought stress in maize. Plant Physiol. 170 (1), 586–599. doi: 10.1104/pp.15.01267
Thatcher, S. R., Zhou, W. G., Leonard, A., Wang, B. B., Beatty, M., Zastrow-Hayes, G., et al. (2014). Genome-wide analysis of alternative splicing in Zea mays: Landscape and genetic regulation. Plant Cell 26 (9), 3472–3487. doi: 10.1105/tpc.114.130773
Wang, B. B., Brendel, V. (2006). Genome-wide comparative analysis of alternative splicing in plants. Proc. Natl. Acad. Sci. U.S.A. 103 (18), 7175–7180. doi: 10.1073/pnas.0602039103
Wang, E. T., Sandberg, R., Luo, S. J., Khrebtukova, I., Zhang, L., Mayr, C., et al. (2008). Alternative isoform regulation in human tissue transcriptomes. Nature 456 (7221), 470–476. doi: 10.1038/nature07509
Wassan, G. M., Khanzada, H., Zhou, Q. H., Mason, A. S., Keerio, A. A., Khanzada, S., et al. (2021). Identification of genetic variation for salt tolerance in Brassica napus using genome-wide association mapping. Mol. Genet. Genom. 296 (2), 391–408. doi: 10.1007/s00438-020-01749-8
Xie, M., Zuo, R., Bai, Z., Yang, L., Zhao, C., Gao, F., et al. (2022). Genome-wide characterization of Serine/Arginine-rich gene family and its genetic effects on agronomic traits of Brassica napus. Front. Plant Sci. 13, 829668. doi: 10.3389/fpls.2022.829668
Yamaura, S., Yamauchi, Y., Makihara, M., Yamashino, T., Ishikawa, A. (2020). CCA1 and LHY contribute to nonhost resistance to Pyricularia oryzae (syn. Magnaporthe oryzae) in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 84 (1), 76–84. doi: 10.1080/09168451.2019.1660612
Yan, Y., Gan, J., Tao, Y., Okita, T. W., Tian, L. (2022). RNA-Binding proteins: The key modulator in stress granule formation and abiotic stress response. Front. Plant Sci. 13, 882596. doi: 10.3389/fpls.2022.882596
Yu, S. G., Kim, J. H., Cho, N. H., Oh, T. R., Kim, W. T. (2020). Arabidopsis RING E3 ubiquitin ligase JUL1 participates in ABA-mediated microtubule depolymerization, stomatal closure, and tolerance response to drought stress. Plant J. 103 (2), 824–842. doi: 10.1111/tpj.14775
Yu, H. P., Tian, C. H., Yu, Y., Jiao, Y. L. (2016). Transcriptome survey of the contribution of alternative splicing to proteome diversity in Arabidopsis thaliana. Mol. Plant 9 (5), 749–752. doi: 10.1016/j.molp.2015.12.018
Zhang, Y., Ali, U., Zhang, G., Yu, L., Fang, S., Iqbal, S., et al. (2019). Transcriptome analysis reveals genes commonly responding to multiple abiotic stresses in rapeseed. Mol. Breed. 39 (10-11), 158. doi: 10.1007/s11032-019-1052-x
Zhang, R. X., Calixto, C. P. G., Marquez, Y., Venhuizen, P., Tzioutziou, N. A., Guo, W. B., et al. (2017). A high quality Arabidopsis transcriptome for accurate transcript-level analysis of alternative splicing. Nucleic Acids Res. 45 (9), 5061–5073. doi: 10.1093/nar/gkx267
Zhang, J., Mason, A. S., Wu, J., Liu, S., Zhang, X. C., Luo, T., et al. (2015). Identification of putative candidate genes for water stress tolerance in canola (Brassica napus). Front. Plant Sci. 6, 1058. doi: 10.3389/fpls.2015.01058
Keywords: Brassica napus, abiotic stress, RNA-seq, alternative splicing, WGCNA, RBPs, TFs
Citation: Yang L, Yang L, Zhao C, Liu J, Tong C, Zhang Y, Cheng X, Jiang H, Shen J, Xie M and Liu S (2022) Differential alternative splicing genes and isoform co-expression networks of Brassica napus under multiple abiotic stresses. Front. Plant Sci. 13:1009998. doi: 10.3389/fpls.2022.1009998
Received: 02 August 2022; Accepted: 23 September 2022;
Published: 13 October 2022.
Edited by:
Fang Bai, University of Florida, United StatesReviewed by:
Lijuan Wei, Southwest University, ChinaCopyright © 2022 Yang, Yang, Zhao, Liu, Tong, Zhang, Cheng, Jiang, Shen, Xie and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Meili Xie, eGllbWVpbGkwMTAxQDE2My5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.