- 1Department of Ecological Plant Protection, University of Kassel, Witzenhausen, Germany
- 2Section of Organic Plant Breeding and Agrobiodiversity, University of Kassel, Witzenhausen, Germany
- 3Foundation Ecology & Agriculture (SOEL), Ahrweiler, Germany
A survey across Germany was undertaken from 2016-2019 to evaluate effects of management system (organic vs conventional), pedo-climatic conditions and crop rotation history on faba bean root health status, diversity of major root rot pathogens and yield. Root rot incidence was generally low and there was no effect of the management system on the spectrum of pathogens isolated. Among the most common fungal species identified, frequencies of Fusarium redolens and Didymella pinodella were significantly higher in roots from organic fields compared with conventional and lower was observed for F. avenaceum, F. tricinctum and F. culmorum. Faba bean roots were colonized at similar rates by F. equiseti and the members of the F. oxysporum (FOSC) and F. solani (FSSC) species complexes in both management systems. Almost no legumes had been grown in the 5-11 years preceding the conventional faba beans surveyed while legumes had almost always been present during this period in the organic fields. This difference in rotational histories between the farming systems led to apparent cropping systems effects on the isolation frequencies of several species. For example, D. pinodella was ubiquitous in organic fields with a high frequency of legumes in the rotations but much rarer and often absent in conventional fields. Pedo-climatic conditions, particularly cool conditions at sowing and plant emergence and/or during the vegetative season favored most of the most prevalent Fusarium species identified in this study. In organic systems, yields correlated negatively with D. pinodella and F. redolens frequencies whereas higher levels of F. tricintum in faba bean roots had a positive correlation with yield. In conventional systems, faba bean yields depended more on the total precipitation before sowing and during the main growing season but were also negatively correlated with the frequencies of FOSC and F. culmorum. Phylogenetic analysis based on the TEF1 alpha locus indicated that the FSSC isolates mainly belonged to the F. pisi lineage. In contrast, the FOSC isolates were placed in 9 different lineages, with a conspicuous dominance of F. libertatis that has until now not been associated with any leguminous host.
Introduction
Foot and root rots, caused by a complex of soil-borne pathogens are among the most widespread and important grain legume diseases and one of the major constrains in grain legume production worldwide (Wille et al., 2019). Several Fusarium species, Aphanomyces euteiches and the Ascochyta complex pathogens, Didymella pinodella and D. pinodes, are the most commonly associated pathogens with the disease complex (Baćanović-Šišić et al., 2018; Wille et al., 2019). Plants under field conditions are usually colonized by multiple pathogens simultaneously, and the importance of each species varies depending on the geographical region, pedo-climatic conditions and the crop management strategy (Esmaeili Taheri et al., 2016; Naseri and Ansari Hamadani, 2017; Baćanović-Šišić et al., 2018; Chatterton et al., 2019; Williamson-Benavides and Dhingra, 2021). In northern USA and Canada for example, F. avenaceum together with Aphanomyces eutheiches is a major threat to pea and lentil production (Chittem et al., 2015; Chatterton et al., 2019), whereas in France and northern European countries including Denmark and Sweden, in addition to A. euteiches and F. avenaceum, F. solani, F. redolens and Didymella pinodella (syn. Phoma medicaginis var. pinodella) play an important role in pea growing areas (Persson et al., 1997; Hossain et al., 2012; Gibert et al., 2022). Other pathogens such as, Rhizoctonia solani, Pythium spp., Thielaviopsis basicola and Macrophomina phaseolina have also been implicated as important parts of the grain legume root rot complex (Naseri and Mousavi, 2015; Wille et al., 2019; Williamson-Benavides and Dhingra, 2021; Wohor et al., 2022).
In the past 15 years, two large root rot surveys have been conducted on grain legumes in Germany. The first (2005-2007) focused on root health assessments of conventional peas, including detailed identification of the pathogens involved based on morphology (Pflughöft et al., 2012). This survey indicated declining importance of F. solani and F. oxysporum while D. pinodella together with F. redolens and F. avenaceum were identified as the primary pathogens in the pea root rot complex in Germany. The second survey (2008-2012) was conducted in four regions of Germany and, in addition to pea, also included faba beans but covered organic fields only (Wilbois et al., 2013). This survey focused solely on rating of the plants in the fields and, besides pointing to the importance of Fusarium spp. did not identify the major Fusarium species involved in the root rot complex of organic pea and faba bean. Although faba bean usually appeared healthier than pea they frequently harbored the same pathogens as peas (Pflughöft et al., 2012; Wilbois et al., 2013). No information is available on pathogens associated with conventional faba bean in Germany. Since the pathogen complex as a whole has a wide host range among legumes and, in addition, especially Fusarium spp. often affect cereals (Bainard et al., 2017; Walder et al., 2017), farmers are forced to grow pulses in wide rotations in order to avoid disease problems.
As a result of the new protein crop strategy adopted in 2012 by the German Federal Ministry of Food and Agriculture, and the EU Common Agricultural Policy (CAP) greening measures in 2015, faba bean production area in Germany has quadrupled since 2014 to about 60.000 ha in 2020. To avoid inoculum build-up and maintain or increase the area grown to faba bean and pulses in general, there is a need to evaluate current pathological risks in order to plan rotations. The overall objective of this study was to determine the root health status of organic and conventional faba beans in Germany and to characterize diversity and frequency of root associated pathogens as affected by farming system, pedo-climatic conditions and crop rotation management. We further report on the effects of root health status and root infections with major fungal species on faba bean yield and provide insights into the genetic variability of the F. oxysporum and F. solani isolates recovered.
Material and methods
Weather data, cropping history and site characteristics
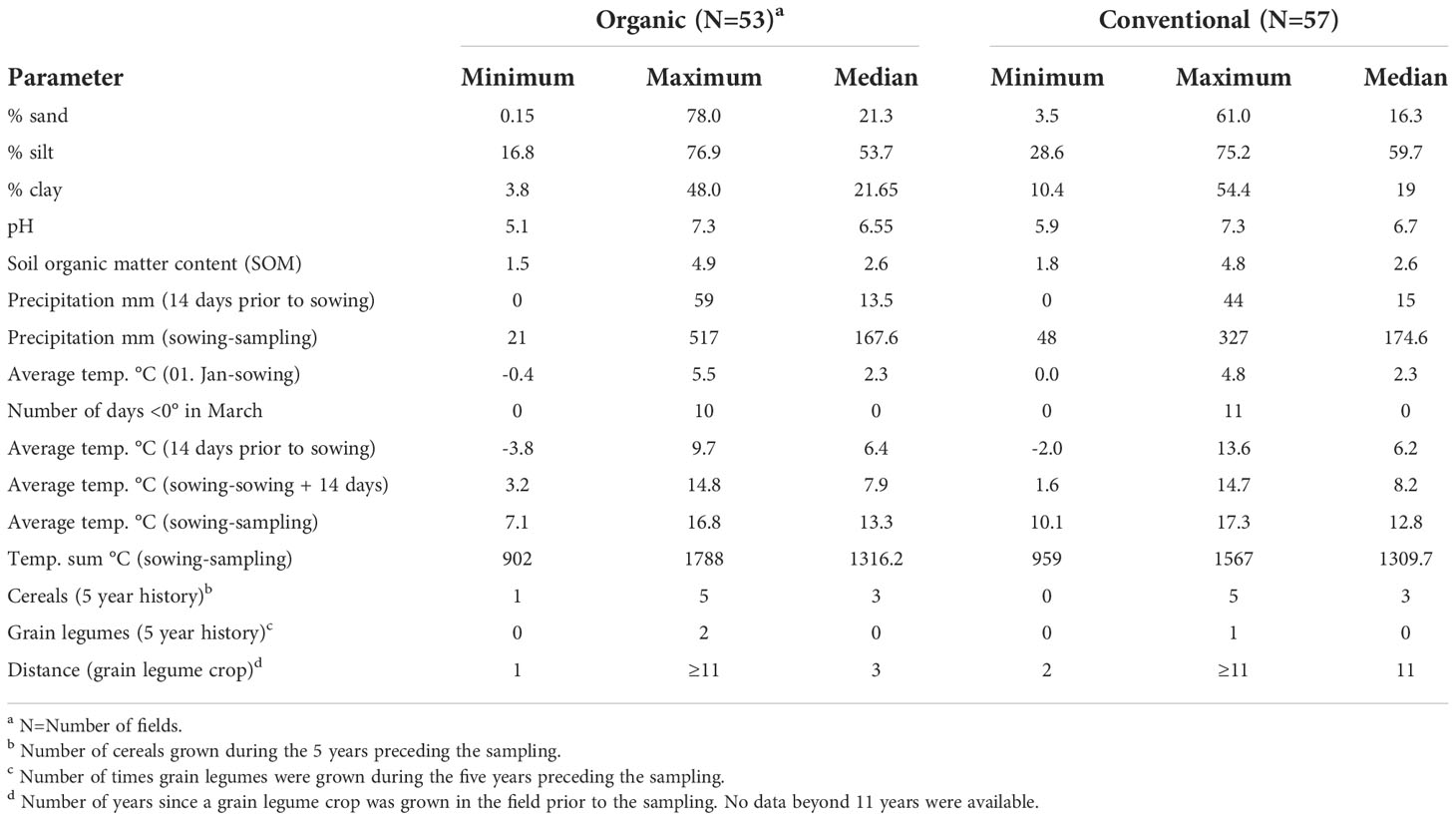
Between 2016 and 2019, a total of 110 faba bean fields were sampled throughout Germany (Figure 1). Of these, 53 surveyed fields were managed organically according to the European Union and national standards and 57 fields conventionally. The meteorological data were obtained from the closest weather stations (<10 km) to the fields. Sand, silt, clay, and soil organic matter content as well as pH were determined in accordance with the standard DIN EN ISO/IEC 17025:2018-03. Data on cropping history were obtained from the farmers directly. These included the number of years fields were planted to different leguminous species (i.e. clover species and alfalfa, pea, faba bean, lentil, lupin, soybean, vetch and the unspecified group of ‘other grain’ or ‘small seeded legumes’) and the number of years of cereals (bulked data for all cereal crops) for a 5- and 11-year period prior to the sampling of faba bean. The complete data set is given in Supplementary Table 1.
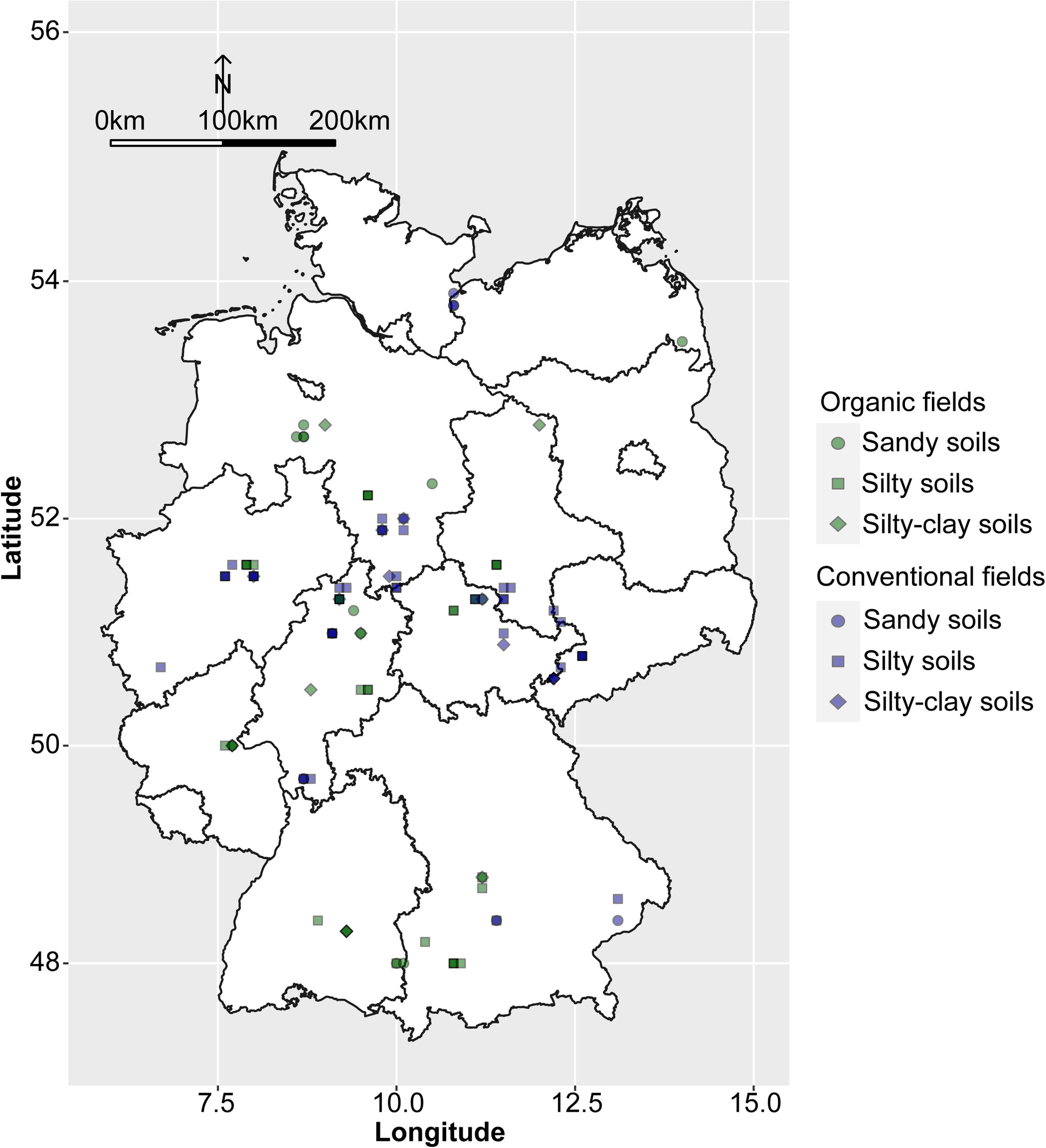
Figure 1 Map of Germany showing locations and the soil types of the surveyed organic and conventional faba bean fields.
Sampling, disease assessments, morphological characterization of the isolates and yield estimation
Thirty-six to 40 faba bean plants were uprooted at full flowering from two areas per field, each 5 m2 in size with the distance between the two areas of 10 to 20 meters, depending on the field. Half of the roots were immediately washed to remove adhering soil and individual plants were evaluated for the severity of root rot symptoms using a visual 1-9 score (1=healthy, 9=dying plant) based on external root tissue discoloration levels (Figure 2) according to Pflughöft et al. (2012). The remaining half of the sampled roots were shipped to the University of Kassel and stored at -18°C until fungal isolations were performed as described previously (Šišić et al., 2018b). Isolations were targeted at the species belonging to the genus Fusarium and those sharing Didymella (Phoma) like morphology as these had been identified previously as the most common pathogens associated with field peas and faba beans in Germany (Pflughöft et al., 2012; Wilbois et al., 2013; Baćanović-Šišić et al., 2018). Briefly, roots were thoroughly washed under running tap water, surface sterilized with 3% sodium hypochlorite for 10 s, rinsed in distilled water and placed on filter paper under a laminar flow hood for ≥1 h. Three approximately 1-cm-long pieces per plant, representing root, crown, and the transition zone, were placed on COONs (Coons, 1916) media and incubated at 20˚C under 12 h cycles of UV light and dark. Roots included both lateral and tap roots up to the point of seed attachment, the crown was considered as the point of seed attachment up to ca. 0.5 cm below the soil surface and the transition zone covered about 1.5 cm between crown and stem.

Figure 2 Varying root discoloration levels and the assigned root rot disease severity ratings (1=healthy plant, 9=dying plant). Plants with disease severity ratings of 8 and 9 (i.e. severe rots) were not observed during the survey.
After 1 to 2 weeks incubation, fungal colonies developing from the root pieces were sub-cultured separately in Petri dishes containing half-strength potato dextrose agar (19 g/l Difco PDA and 10 g/l agar). Pure cultures were generated either through hyphal tipping (Fusarium morphology) or transfer of single pycnida (Didymella morphology). Each isolate was examined microscopically and identified to the species level based on cultural appearance (colony color and pigmentation) and morphology of conidiogenous cells following the protocols of Leslie and Summerell (2006) for Fusarium spp. and Boerema et al. (2004) for Didymella spp.
Grain yield was estimated by hand harvesting the plants in five 0.5 m² plots within each of the 5 m2 area used for root collection. The yield was adjusted to 86% dry matter.
Molecular confirmation of fungal species identity and phylogenetic analyses
The identity of 120 Fusarium and 8 Didymella randomly selected isolates obtained in this study representing 11 different fungal species (Supplementary Table 2) was confirmed by sequencing the portion of the translation elongation factor 1 (TEF1) alpha for Fusarium spp. (O’Donnell et al., 1998) and the beta tubulin gene region for Didymella spp. (Chen et al., 2015). DNA extraction, PCR amplification, sequencing and raw sequence data analysis were performed as described previously (Šišić et al., 2018a). Briefly, genomic DNA was extracted from pure cultures growing on half strength PDA agar plates (Fusarium spp; ½ strength PDA; 19 g/l Difco PDA and 10 g/l agar) or Coons (Didymella spp.; Coons, 1916) medium using the protocol described by Doyle and Doyle (1987). A portion of the translation-elongation factor 1 alpha (tef1) gene was amplified using primer pairs EF1 and EF2 (O’Donnell et al., 1998). The β tubulin (tub2) gene region was amplified with the primers Btub2Fd and Btub4Rd (Chen et al., 2015). Amplicons were visualized via electrophoresis on a 1% agarose gel and purified using the DNA Clean & Concentrator kit (Zymo Research, Freiburg, Germany) according to the manufacturer’s instructions. Sanger sequencing in both directions was performed by Macrogen Europe Laboratories (Amsterdam, Netherlands). Obtained row sequence data were assembled and errors identified and corrected manually in SeqMan Lasergene software (DNAStar, Madison, WI, U.S.A.). To confirm the taxonomic identity of the isolates, these sequences were used as queries for the Fusarium-ID v. 1.0 (Geiser et al., 2004) and NCBI (Madden, 2002) databases.
Phylogenetic analyses
Single locus phylogenetic analyses based on the TEF1 alpha gene sequences were performed for the 35 Fusarium oxysporum species complex (FOSC) and 33 Fusarium solani species complex (FSSC) isolates. Reference sequences for the analysis were selected based on the previously published phylogenetic relationship within the FOSC (O’Donnell et al., 2009; Lombard et al., 2018) and the FSSC (O’Donnell et al., 2020; Geiser et al., 2021) (Supplementary Tables 3 and 4). The sequence alignments were generated using MAFFT v.7 (Katoh and Standley, 2013) and further adjusted manually with MEGA v6 (Tamura et al., 2013). A bootstrapped Maximum-Likelihood (ML) analysis was performed using the RAxML-VI-HPC v. 7.0.3 with non-parametric bootstrapping and 1000 replicates implemented on the Cipres portal (Stamatakis et al., 2008). For the FOSC we first performed the phylogenetic analysis on the data set which included 174 representative isolates from the study of O’Donnel et al. (2009) and 91 isolates from Lombard et al. (2018). The resulting TEF1 alpha tree topology revealed several isolates which belonged to the same forma specialis, shared identical or had similar TEF1 alpha gene sequences. These were excluded from subsequent analysis resulting in a data set which comprised 181 FOSC sequences. The FSSC data set consisted of 96 TEF1 sequences (Supplementary Tables 2–4). For outgroup purposes, F. udum (CBS 177.31) and F. thapsinum (H05-557S-1 DCPA) were used to generate the phylogenetic trees.
Statistical analyses
All statistical analyses were performed in R (R Core Team, 2013). Prior to the analysis, the abundance of individual fungal species was used to calculate isolation frequencies (i.e., percent colonized roots) by dividing the number of roots in which the species occurred by the total number of roots processed. The fields were further scored as positive or negative for presence of a particular fungal species and these data were used to calculate the prevalence for each pathogen in each management system (organic and conventional) by dividing the number of fields in which each species was present by the total number of fields sampled. In addition, the root rot incidence for each management system was calculated as the percentage of fields with mean disease severity score greater than 3 i.e., roots with clearly visible symptoms (Figure 2).
In the analysis of the isolation frequencies of individual fungal species associated with faba bean roots, rare species (i.e., <2% of total isolations) were not considered. To determine if the isolation frequencies were affected by the management system or sampling year, a generalized linear mixed model analysis was performed on proportional data with a binomial distribution and logit link function (Brooks et al., 2017). Fields were used as random effects, and to account for the two sampling areas within each field, sampling replicates were nested within. In data analyzed across sampling years, year was also used as random effect. Prevalence of fungal species and root rot incidence (the proportion of fields with mean disease severity ratings > 3; Chatterton et al., 2019) were treated as binary data (presence/absence) and were analyzed with Bayesian generalized linear model and logit link function (package ‘arm’, Su et al., 2018). The goodness of fit of the models was assessed using Pearson chi-square residual tests and further verified by the Kolmogorov–Smirnov test of normality and by checking if the data contain potentially significant outliers (package ‘DHARMa’, Hartig, 2021). Data were also visually inspected for normality by plotting the Pearson residuals against the expected values (package ‘ggplot2’, Wickham, 2016). The significance of the main effects in generalized linear models was assessed using an ANOVA function with the type III margin sum of squares (package ‘arm’). If significant treatment effects were observed, comparisons of least squares means with Tukey’s correction across the effect levels were performed (P < 0.05) (package ‘lsmeans’, Lenth, 2016).
Root rot severity data were analyzed with the non-parametric ranking procedure of the Kruskal-Wallis test (Conover, 1999) using the package ‘agricolae’ (Mendiburu, 2014). Management system and/or year were included as main explanatory variables. If significant treatment effects were observed (P<0.05), mean rank values were separated with the Kruskal multiple comparison test. For both, Kruskal-Wallis test and Kruskal multiple comparison test, the Benjamini and Hochberg (Benjamini and Hochberg, 1995) stepwise adjustment of P-values was used to control false discovery rate (FDR) and reduce type I errors.
The relationship between frequencies of individual fungal species and root rot incidence data including pedo-climatic, crop rotation and yield effects were examined using path analysis (package ‘lavaan’, Rosseel, 2012). Prior to the analysis, highly correlated environmental variables (Pearson r ≥ ± 0.7) were removed. Data were then subjected to the stepwise forward selection procedure (package ‘stats’, R Core Team, 2013) and only significant variables were retained in the path analysis. In addition, distinct soil clusters of the sampled fields were used as entries clustered based on their similarities in soil abiotic properties (sand, silt, clay, soil organic matter content and pH) employing the hierarchical clustering on principle components (HCPC) (package ‘FactoMineR’, Lê et al., 2008).
Results
Environmental conditions of the fields sampled
The 110 organic and conventional faba bean fields sampled represented a wide range of environments with respect to soil and climatic conditions (Table 1). Organic and conventional fields were placed in soil types ranging from sandy to loamy; large variation in soil organic matter (SOM) contents and moderate ranges in pH were present in organic and conventional fields.
Sowing conditions ranged from very wet (up to 59 mm of rain in the 2 weeks before sowing) to no rain during the same period and, from very cold soils (minimal mean temperature two weeks before sowing -3.8°C) to very warm (maximal mean temperature before sowing 13.8°C). The driest conditions were observed in the year 2018 with a field receiving as little as 21 mm, the wettest in 2016 with a field that received 517 mm of rain between sowing and sampling (Table 1 and Supplementary Table 1).
The hierarchical clustering of faba bean fields on principal components (HCPC) based on their similarities in soil parameters i.e. soil pH, sand, silt, clay and organic matter content (SOM) grouped the 110 faba bean fields into three clusters (cluster I, II and III) (Figure 3). The first two dimensions of the PCA summarized 77% of the variability in the data. Dimension 1 explained 50.6% of the variance and separated fields in cluster I from fields in clusters II and III based on their differences in soil pH values, sand and silt content. Dimension 2 explained 26.8% of the variance and separated the clusters mainly based on their differences in SOM content, which was also positively correlated with the soil clay content (e.g. cluster III) (Figure 3 and Table 2). The PCA dimension 3 explained an additional 14.9% of the variability in the data and was most strongly related to the pH (not shown).
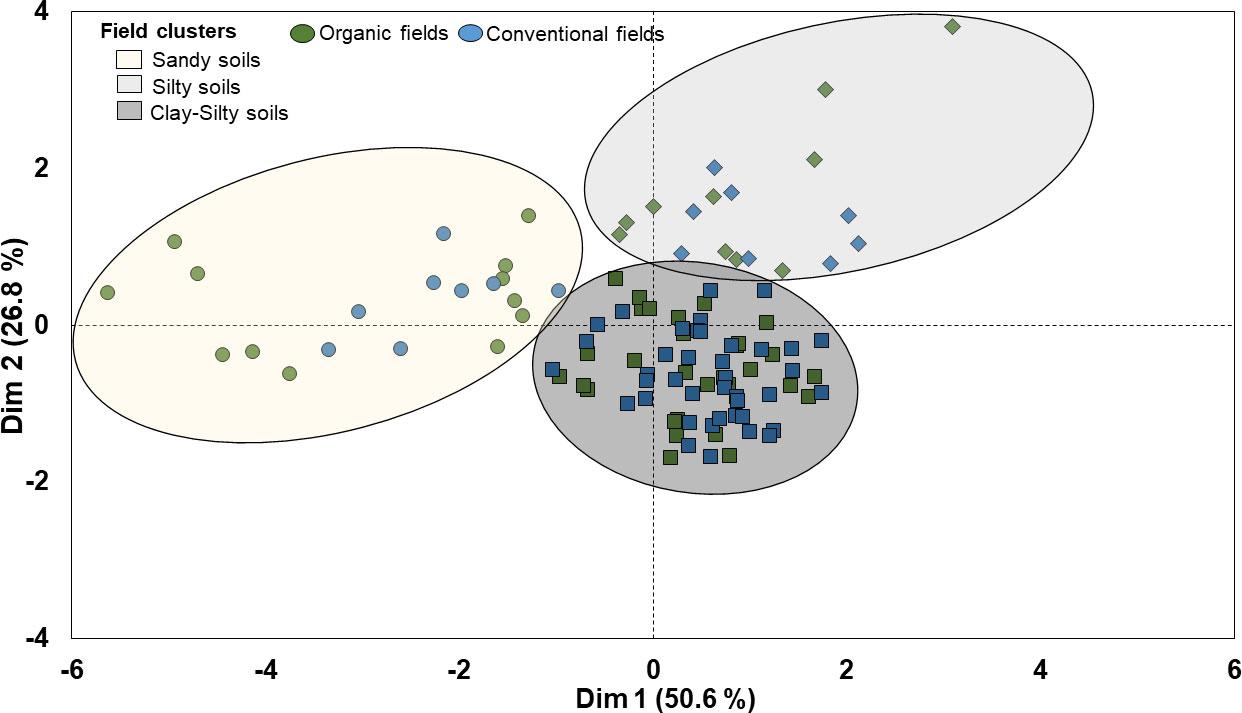
Figure 3 Cluster analysis for 110 organic and conventional faba bean fields based on their similarities in soil abiotic properties.
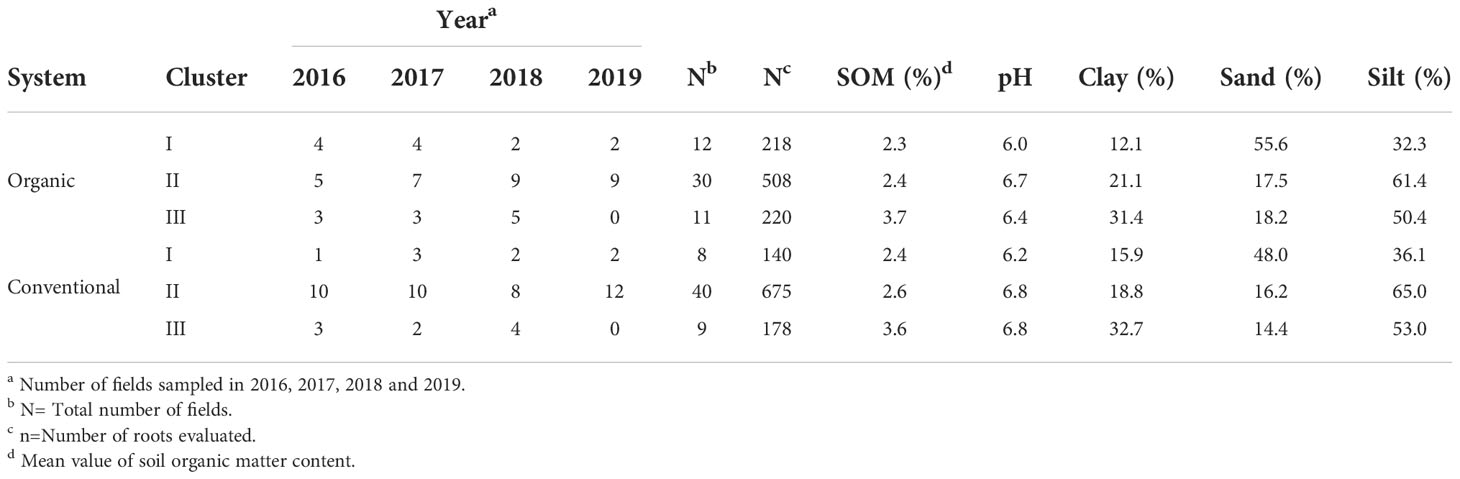
Table 2 The number of organic and conventional fields in each year, grouped by their similarities according to soil abiotic properties.
Cluster 1 included 12 organic and 8 conventional fields classified as sandy loam and characterized by lower pH (around 6.0) compared with the clusters II and III (around 6.7), and mean SOM content of 2.3% and 2.4% in organic and conventional fields, respectively (Figure 3 and Table 2). Cluster II included 30 organic and 40 conventional fields associated with silty soils with higher pH (around 6.7) but similar SOM content as cluster I (Figure 3 and Table 2). Cluster III included 11 organic and 9 conventional fields. It included silty clay soils but with higher clay and SOM (around 3.6%) contents than cluster II (SOM ca. 2.4%) (Figure 3 and Table 2).
Cropping histories
Across all fields for the 5-year period prior to the faba bean sampling, approximately 26% of the crops in rotations under organic management were legumes. Of these, approximately 70% were clover and alfalfa and about 30% grain legumes (mainly peas and faba beans - see below; hereafter grain legumes). In contrast, legumes in conventional fields constituted only about 5% of the crops in the 5-year rotation plan. The ratio of cereals in organic and conventional crops rotations was similar and accounted for 57% (organic) and 68% (conventional) of all crops. Overall frequencies of legumes and cereals for the 11-year period in both management systems was similar to the 5 year rotation plan (Supplementary Table 5).
In the organic fields, 37 out of the 52 (71%) fields for which data were available had been cropped with legumes either as main or cover crops during the past five years (Figure 4A). Clover and alfalfa had been grown in 32 (62%) fields usually for one to two years and grain legumes in 17 (32%) fields for one or two seasons. Also, cereals were part of the rotation during the preceding 5-years usually for two to three seasons (73% of the organic fields; Supplementary Table 6). In contrast, 43 (75%) of the conventional fields had not been planted to any legume in the preceding 5 years (Figure 4A). Twelve of the remaining 14 fields (25%) had been planted once with faba bean, one with pea during that period and one with small seeded legumes. Most rotations (48 out of 57 fields; 84%) included three or four years of cereals and five fields even 5 years while one conventional field had not been grown to cereals (Figure 4B and Supplementary Table 6). When considering the past 11 years prior to faba bean sampling, almost all organic farmers had grown legumes at least once while 32 of the 57 conventional farmers had not grown legumes during the least 11 years or longer (Figure 4B).
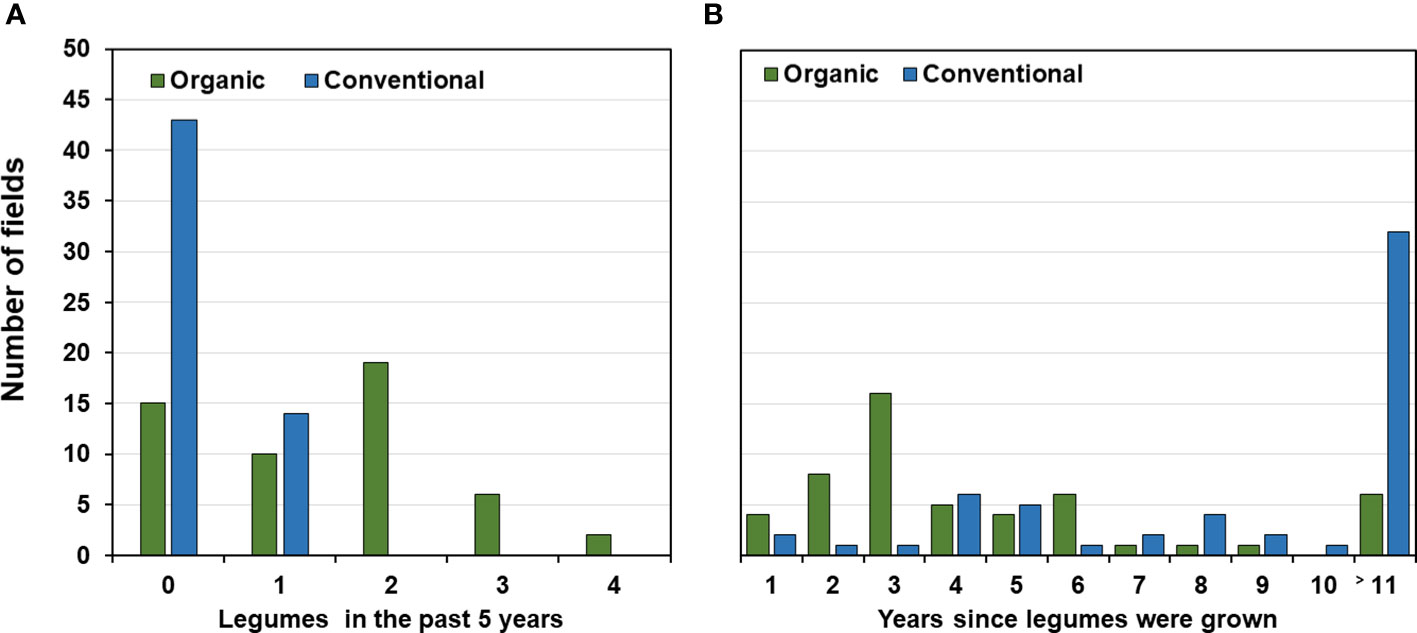
Figure 4 Ratio of legumes in organic and conventional crop rotations. (A) Number of fields and years since legumes were grown before the faba bean sampling. No information available for 11 or more years. (B) Number of fields depending on the frequency of legumes planted for the five-year period preceding the faba bean sampling.
Root rot incidence and root health status
Overall disease severity ratings (DSR) did not significantly differ between management systems (mean DSR of 2.6 and 2.0 for organic and conventional system, respectively) or among years (P≥0.28). However, there was a difference in the proportion of organic and conventional fields with clearly visible symptoms of root rot (i.e. root rot incidence, mean DSR>3). Faba bean plants from 18 out of 53 organic fields (34%) had mean DSR>3, significantly (P=0.03) more than in the conventional fields (9/57, 16%) with some variation among years (Table 3). Nevertheless, even in the organic fields, mean root rot severity was usually just above the threshold level of 3 with no significant difference in overall root rot symptom severity between the management systems (fields with mean DSR>3; Table 3).

Table 3 Mean root rot incidence (%) and root rot severity ratings (DSR) of faba bean fields in Germany, 2016-2019.
Among the years, the highest root rot incidence in both management systems was recorded in 2016 (58% organic and 36% conventional fields), followed by 2018 (50% organic and 29% conventional fields). In 2017, all roots collected from conventional fields showed no visible symptoms of rot (mean DSR<3) whereas roots collected from 21% of the organic fields were symptomatic (mean DSR>3). In 2019, all roots collected from both management systems appeared healthy (mean DSR<3).
Fungal species associated with root infections
Out of a total of 2175 roots analyzed over the four years, 1939 yielded fungal isolates. A total of 4213 Fusarium and 490 Didymella- like isolates were obtained from the 110 fields. Of these, 49.7% (n=2093) of the Fusarium and 91.8% (n=450) of the Didymella isolates originated from organically managed fields (N = 53 fields; n = 946 roots), whereas the remaining 50.3% (n=2093) Fusarium and 8.2% (n=40) Didymella isolates were obtained from conventional fields (N=57; n=993).
Combined over years and management systems, members of the Fusarium oxysporum species complex (FOSC; 36% colonized roots), F. redolens (34%), members of the F. solani species complex (FSSC; 27%) and F. avenaceum (23%) were the most frequent. Together, they constituted 74% of all isolates recovered, and occurred in 82 to 94% of the fields. The next most frequent Fusarium species were F. tricinctum, F. culmorum and F. equiseti which were isolated from 6% (F. equiseti) to 11% (F. tricinctum) of the roots. Didymella pinodella was recovered from 17% of all roots, but occurred mainly in organic fields with overall isolation frequency of 31%. The latter four species together represented 23% of all isolates recovered and were found in 44 to 54% of the fields over the years. Species isolated at frequencies ≤ 2% included seven Fusarium and one Didymella spp.: F. acuminatum, F. graminearum, F. crookwalance, F. torulosum, F. sporotrichioides, F. sambucinum, F. flocciferum (Šišić et al., 2020) and D. eupyrena (syn. Juxtiphoma eupyrena).
Across years, frequencies of F. redolens and D. pinodella were significantly higher in roots from organic fields than from conventional fields. In contrast, F. avenaceum, F. tricinctum and F. culmorum more frequently colonized roots in conventionally managed fields (for all comparisons P < 0.01). Overall mean isolation frequencies of the species within the F. oxysporum and the F. solani species complexes and the mean isolation frequencies of F. equiseti did not differ significantly between the management systems (Figure 5A).
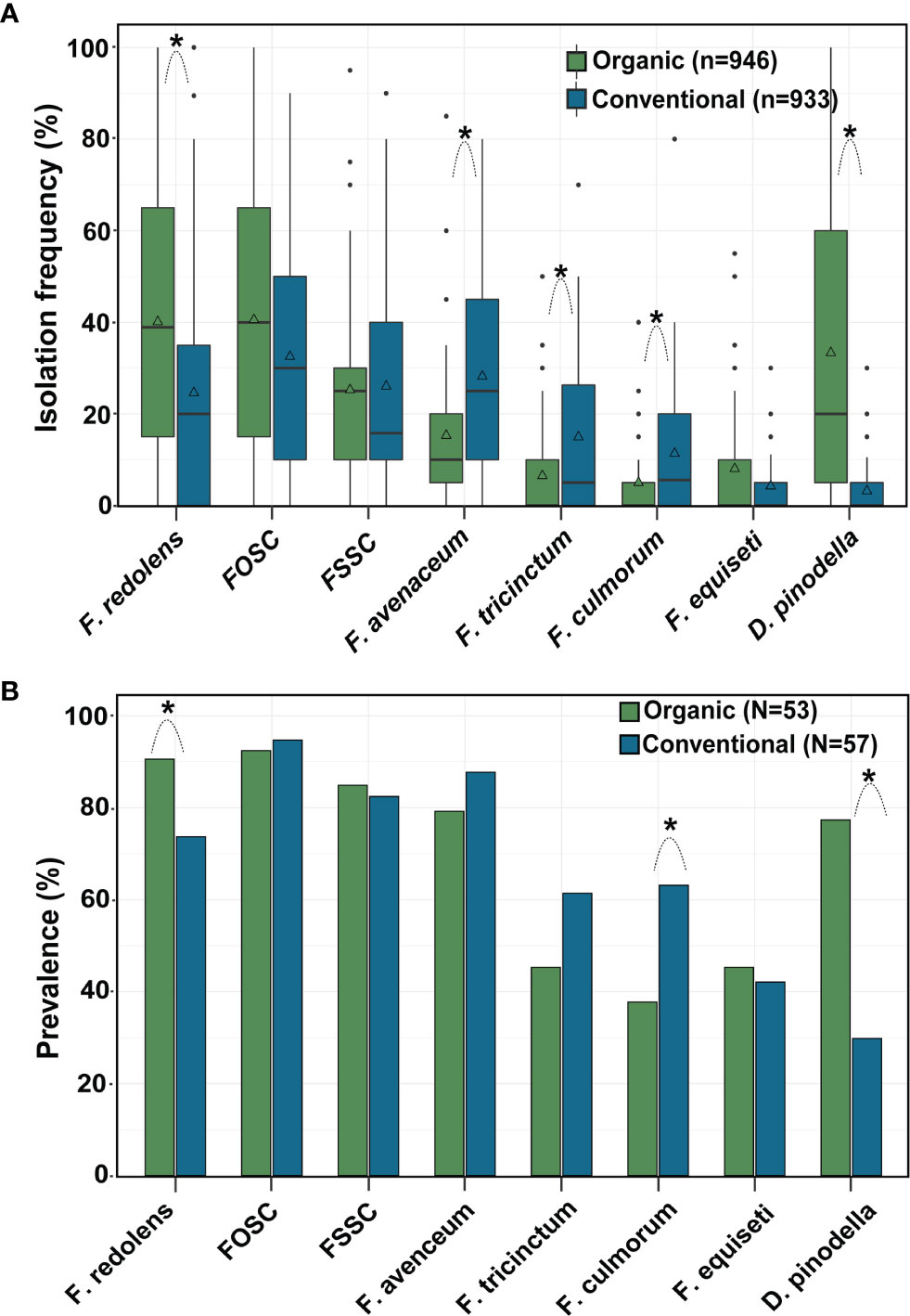
Figure 5 Effect of management system on (A) isolation frequency (%) and (B) prevalence (% fields yielding a given species) of the eight most common fungal species recovered from faba bean roots. Asterisks indicate significant differences (P < 0.05) between organic and conventional fields for each fungal species separately (Tukey-adjusted pairwise LSMeans comparisons). n = number of roots, N = number of fields evaluated. The horizontal line in the boxplot shows the median value, the bottom and tops of the box the 25th and 75th percentiles and the vertical lines the minimum and maximum values, outliers as single points. Mean values are marked with triangles.
With few exceptions, the trends observed over the years were similar within years (Table 4). For example, during 2016-2018, F. redolens was more common in organically grown faba beans roots (31 to 66%) compared with conventionally grown faba beans (17 to 44%) while in 2019, the species occurred only rarely in either management system. Didymella pinodella varied considerably in frequency among organic fields depending on the year (15 to 51%) but was consistently low in conventional fields (≤ 6%) (Table 4).
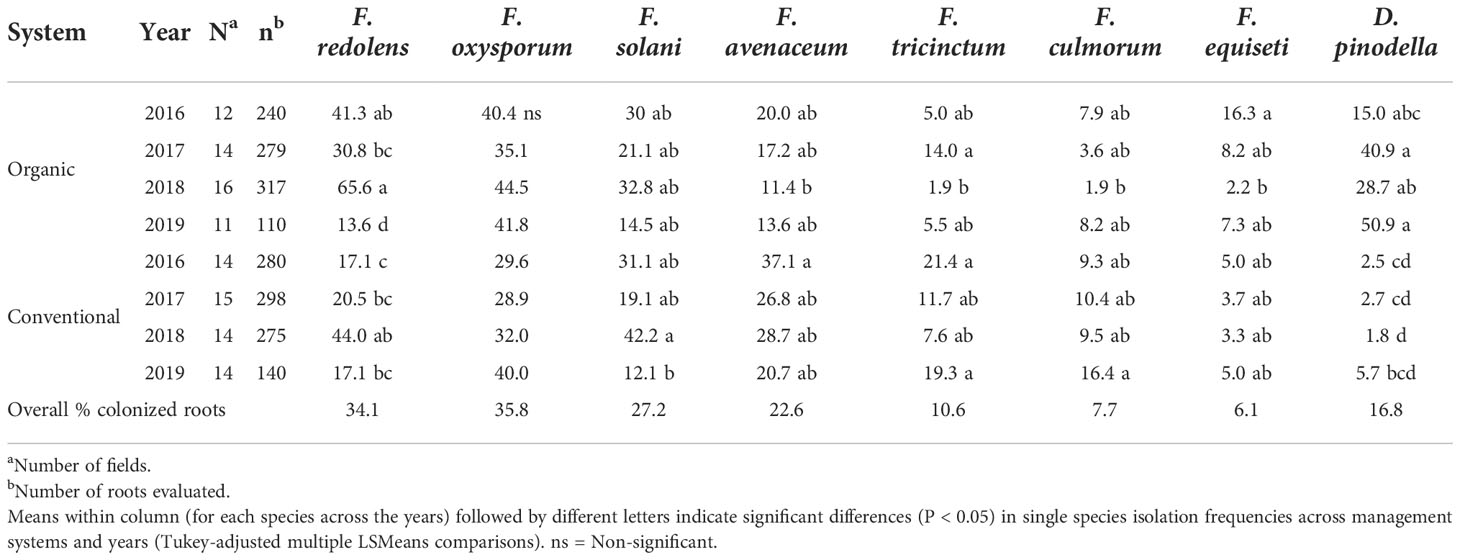
Table 4 Variation in isolation frequencies (%) of the eight most common fungal species recovered from faba bean roots separated based on sampling year and management system.
Fusarium avenaceum, F. tricinctum and F. culmorum isolation frequencies varied greatly among years with the latter two species occurring generally less frequently in both management systems (11 to 37% for F. avenaceum vs 2 to 21% for F. tricintum, and 2 to 16% colonized roots for F. culmorum). Fusarium equiseti was recovered at low frequencies in both growing systems (2-16%) (Table 4) while members of the FOSC and the FSSC were common in isolation frequencies among years and between management systems (Table 4).
Pathogen prevalence, i.e. the percentage of fields in which the pathogens occurred, followed a similar pattern to the isolation frequencies (Figure 5B). Across years, F. redolens and D. pinodella were more prevalent (P=0.0018) in organic fields (91% for F. redolens and 77% for D. pinodella) compared with conventional fields (74% for F. redolens and 30% for D. pinodella) (Figure 5B and Table 5). Presence of F. redolens was considerably lower in conventional fields in 2016 and 2019, while in 2017 and 2018, the prevalence rates of F. redolens in organic and conventional fields were more similar (Table 5). The prevalence patterns of D. pinodella were more variable. Predominantly occurring in organic systems, this species showed moderate to high prevalence rates over the years ranging from 63% (2018) to 93% (2017). In conventional systems, the highest D. pinodella prevalence rate was observed in 2017 (47%), the lowest also in 2018 (14%) (Table 5).
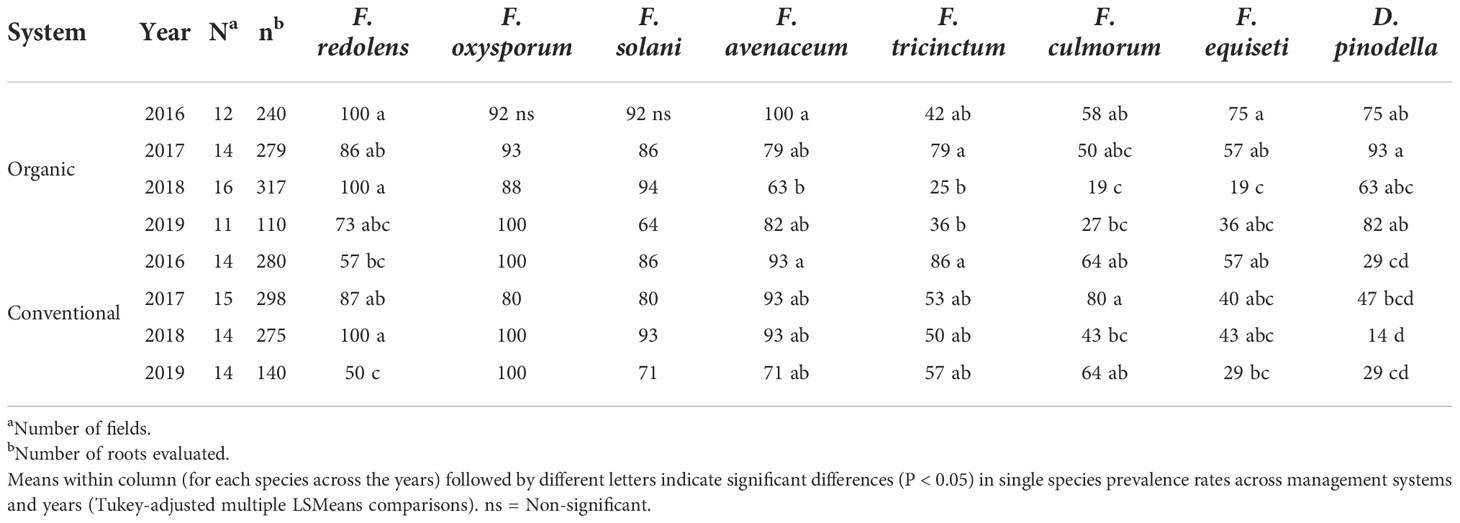
Table 5 Variation in prevalence (% of fields) of the eight most common fungal species recovered from faba bean roots separated based on sampling year and management system.
Although isolated more frequently from roots collected from conventional fields, the overall prevalence rates of F. avenaceum and F. tricinctum did not differ significantly between the management systems (Figure 5B). The prevalence of F. culmorum was significantly higher in conventional fields (63%) than in organic (38%) as observed for isolation frequencies.
For the members of the FOSC and FSSC, some variability in the proportion of positive fields was observed similar to the isolation frequencies, but these differences were not significant between the management systems over the years (Figure 5B) or during any single year (Table 5). Members of the FOSC were consistently found in more than 80% of the fields. The proportion of FSSC positive fields ranged from 64 to 93%. Fusarium equiseti prevalence rates varied between 19 and 75% depending on year and growing system (Table 5).
Phylogeny
Phylogenetic analyses inferred from the TEF1 alpha gene sequences resolved the phylogenetic positions of the 35 FOSC and 33 FSSC isolates studied in relation to currently recognized species in both species complexes (Figures 6, 7). Both the members of the FOSC and the FSSC varied greatly in morphology, which was reflected in high genetic variability, particularly for the members of the FOSC. Based on the single locus phylogeny, the 35 FOSC isolates were distributed throughout the FOSC clade and belonged to 9 different lineages. The most abundant group comprising 17 isolates was placed in the F. libertatis lineage. The second most abundant group, represented by 6 isolates, did not cluster clearly with any of the recently described species within the FOSC. These were most closely related to the previously assigned F. oxysporum special form conglutinans (NRRL 36364). The results of the TEF1 alpha tree topology further revealed 4 isolates matching recently erected epitype specimen (Lombard et al., 2018) and 3 isolates were placed in F. odoratissimum linages. In addition, single isolates were placed in the F. nirenbergie, F. hodiae, F. fabacearum/F. calistephi, F. curvatum and F. commune lineages (Figure 6).
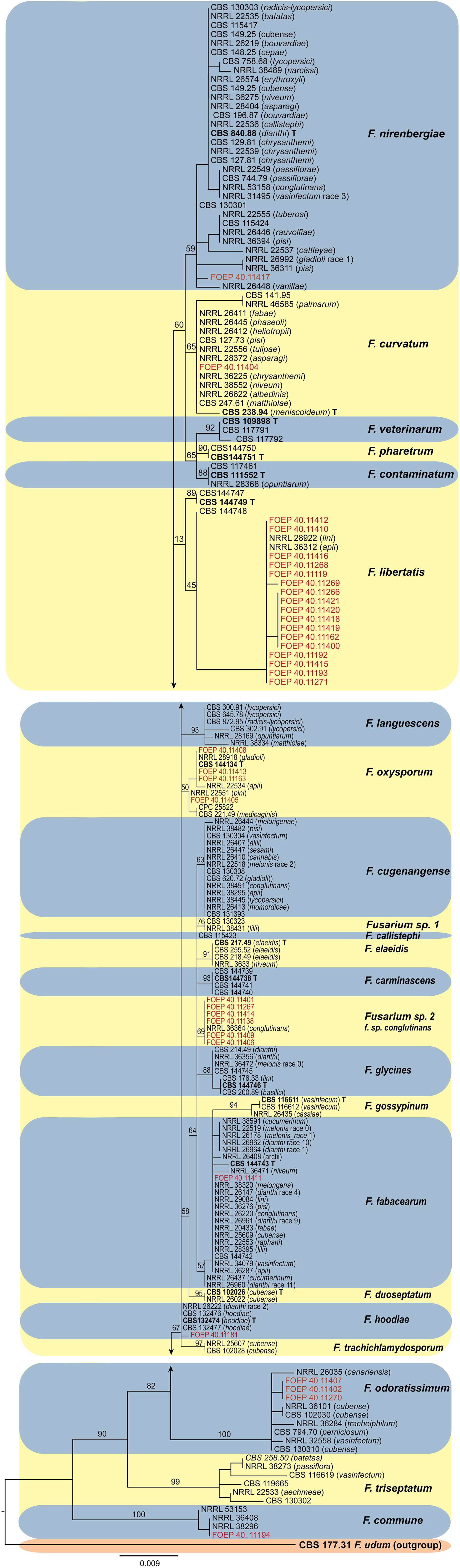
Figure 6 The maximum likelihood (RAxML) tree inferred from the partial TEF1 alpha gene sequence alignments of the Fusarium oxysporum species complex isolates (FOSC) used in this study. Isolates (i.e. FOEP) are indicated in red. Epi- and ex-type strains are indicated in bold and superscript ‘T’ (Lombard et al., 2018). The scale bar indicates 0.009 expected changes per site. The tree is rooted to F. udum (CBS 177.31).
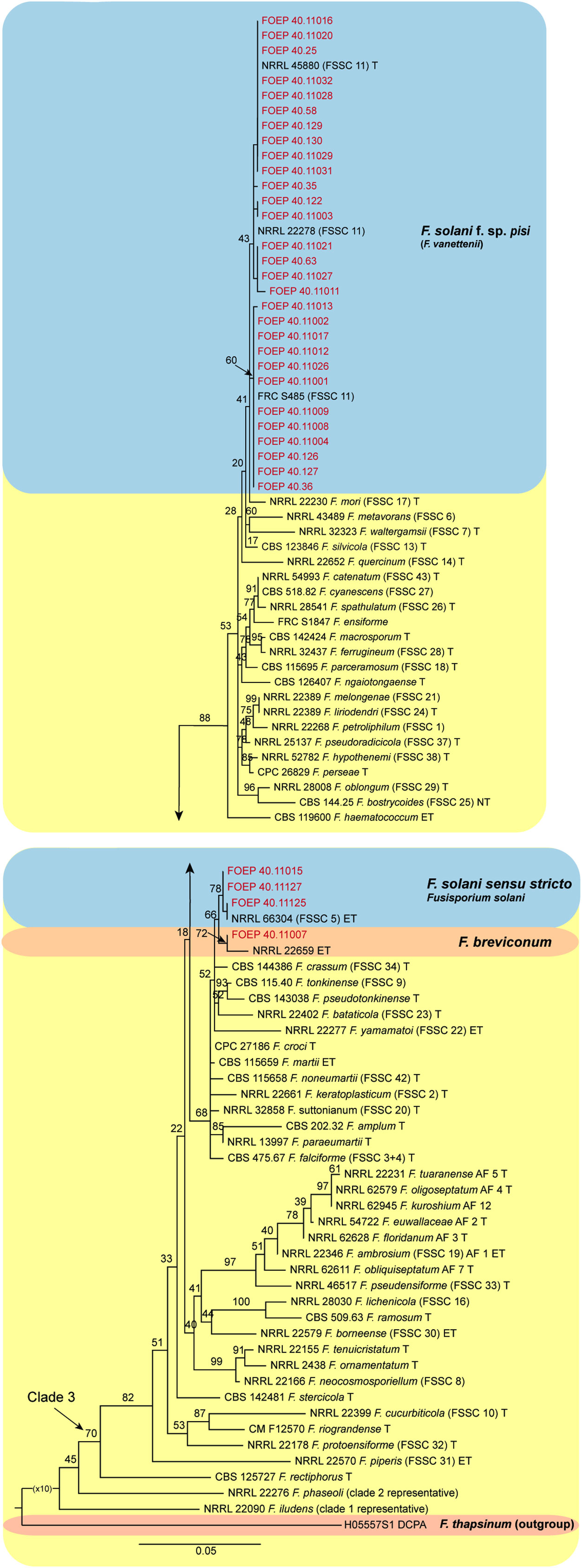
Figure 7 The maximum likelihood (RAxML) tree inferred from the partial TEF1 alpha gene sequence alignments of the Fusarium solani species complex isolates (FSSC) used in this study. Isolates (i.e. FOEP) are indicated in red. Epi- and ex-type strains are indicated in bold and superscript ‘T’ (O’Donnell et al., 2020; Geiser et al., 2021). The scale bar indicates 0.05 expected changes per site. The tree is rooted to F. thapsinum (H05557S1 DCPA).
The 33 FSSC isolates were placed into three different lineages, all nested within clade 3. Most isolates (n=29) matched F. pisi (syn. F. solani f. sp pisi), the lineage recently renamed to Fusarium vanettenii (Geiser et al., 2021). A group of 3 isolates were placed in the Fusarium solani sensu stricto lineage, and one isolate matched F. breviconum (Figure 7).
Major factors influencing pathogen frequency and faba bean yield
The overall path model from stepwise regression showed a good fit with the data for both organic and conventional fields (Supplementary Table 8). In organic fields, yield correlated negatively with D. pinodella (β = -0.42) and F. redolens (β = -0.40) frequencies and positively with F. tricintum in faba bean roots (β = 0.33). In conventional fields, weak but significant negative correlations between yield and FOSC (β = -0.20) and F. culmorum (β = -0.21) were found. Additionally, yield correlated positively with the total precipitation before sowing (β = 0.27) and in particular the total precipitation from sowing to root sampling (β = 0.48) for conventional faba bean. Overall, these variables accounted for approximately 40% of the yield variation in both management systems (Supplementary Table 7).
The frequencies of F. redolens in both management systems were positively correlated with the number of days below zero in March (i.e. colder conditions at sowing and plant emergence: path coefficients for organic/conventional system, β = 0.44/0.50) and the average temperature measured for the period from sowing to root sampling (β = 0.62/0.42). Following this pattern, in organic fields F. redolens frequencies was correlated negatively with the average temperature two weeks before sowing (β = -0.30). In addition, the frequency of cereals in the preceding five years before sampling (β = 0.24) and silty soils (β = 0.22) was associated with this pathogen. Cumulatively, these variables explained 55% and 25% of variation in F. redolens frequencies in organic and conventional systems, respectively (Table 6 and Supplementary Table 7).
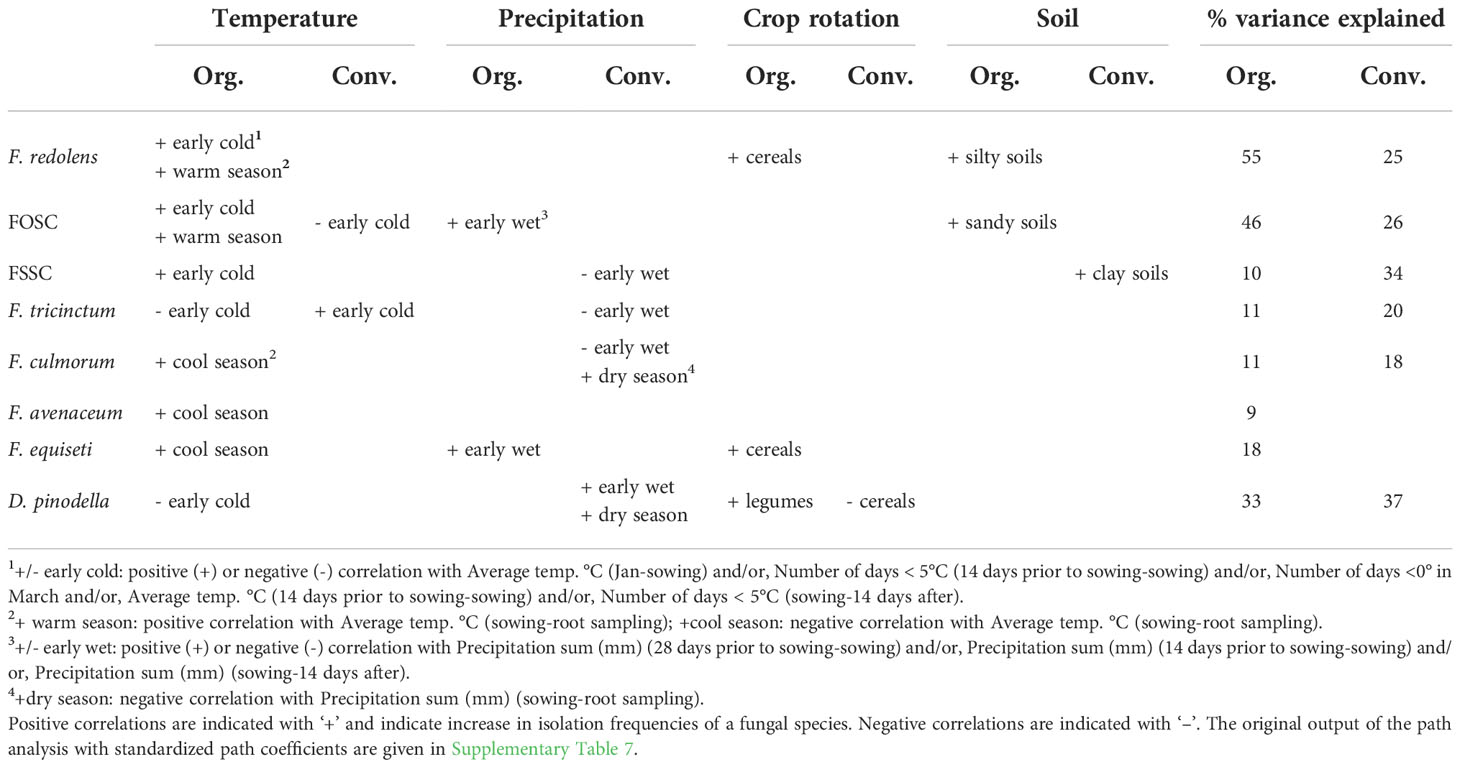
Table 6 Summary of the path analysis results showing the main environmental and cropping history factors affecting abundance (isolation frequencies) of major fungal species in roots of organically (Org.) and conventionally (Conv.) grown faba beans.
The FOSC in both management systems was positively correlated with sandy, slightly acidic soils (β = 0.52/0.43). In organic fields, FOSC frequencies correlated positively with the number of days below 5°C two weeks after sowing (β = 0.46), the total precipitation from sowing to two weeks after (β = 0.21) and the average temperature from sowing to root sampling (β = 0.24). Thus, wet and colder conditions up to plant emergence followed by higher temperatures during the main growing season favored this pathogen complex in organic fields similar to what was observed for F. redolens. In conventional fields, in addition to the observed correlation with sandy soils, the FOSC frequencies were favored by higher temperatures early in the year (January to sowing; β = 0.30). Together, these variables explained 46% and 26% of the FOSC total variance in organic and conventional fields, respectively (Table 6 and Supplementary Table 7).
Similar to F. redolens and to the FOSC in organic systems, colder conditions at sowing and/or plant emergence correlated positively with the FSSC frequencies (in conventional system: positive correlation with the number of days below zero in March, β = 0.32; in organic system: negative correlation with the average temperatures from January to sowing, β = -0.32). The analysis also indicated that low precipitation in the period of plant emergence (from sowing to 2 weeks after; β = -0.28) and growing faba bean in clay soils (β = 0.31) enhanced FSSC root colonization rates in conventional but not organic fields. Together, these variables explained 10% and 34% of the FSSC total variance in organic and conventional fields, respectively (Table 6 and Supplementary Table 7).
For F. tricinctum, 11% of the variance in organic fields was explained by the negative correlation with number of days below 5°C two weeks before sowing (β = -0.33), i.e. warmer conditions before sowing favored it. In conventional systems this effect was opposite (β = 0.34). In addition, the lower precipitation two weeks before sowing (β = 0.23) explained 20% variation in the species root colonization rates data (Table 6 and Supplementary Table 7).
The frequencies of F. culmorum in both management systems correlated negatively correlated with the average temperature measured for the period from sowing to root sampling (β = -0.28/-0.49). In conventional fields only, there was also a negative correlation of F. culmorum frequencies with the total precipitation 4 weeks prior to sowing (β = -0.27) and from sowing to root sampling (β = -0.38) i.e. colder and drier conditions favored this pathogen. Together, these variables explained 11% and 18% of the F. culmorum total variance in organic and conventional fields, respectively (Table 6 and Supplementary Table 7).
The frequencies of F. avenaceum and F. equiseti could not be related to any of the environmental factors or the cropping history in conventional systems. In organic systems, isolation frequencies of both species correlated negatively with total temperature between sowing and root sampling (F. avenaceum/F. equiseti; β = -0.31/-0.27). In addition, F. equiseti was correlated negatively with the total precipitation two weeks after sowing (β = -0.27; as the FSSC) and positively with the frequency of cereals in the preceding five years (β = 0.30) similar to F. redolens. These variables together explained 9% and 18% of the variation in the F. avenaceum and F. equiseti data, respectively (Table 6 and Supplementary Table 7).
Didymella pinodella in organic fields was positively correlated with frequencies of cool season grain and small seeded legumes in the preceding five years before sampling (β = 0.47) and clover and alfalfa for the same period (β = 0.31). Conversely, in conventional systems, the frequencies of cereals in the preceding five years before sampling negatively affected D. pinodella root colonization rates (β = -0.45). Also, D. pinodella frequencies were negatively associated with the number of days below zero in March (conventional system: β = -0.47) and the number of days below zero 2 weeks prior to sowing (organic system: β = -0.27) i.e. opposite to F. redolens and FOSC. In conventional systems, D. pinodella frequencies were also positively correlated with the total precipitation during the 14 days after sowing (β = 0.44) but negatively with the total precipitation from sowing to root sampling (β = -0.42) i.e. warmer and wetter conditions at sowing/plant emergence followed by drier growing seasons favored this pathogen in conventional fields. The model explained 33% and 37% of the species variation in organic and conventional system, respectively (Table 6 and Supplementary Table 7).
Discussion
Root rot incidence was generally low in both management systems especially in 2017 and 2019 when precipitation prior to sowing was low followed by dry growing seasons. Among the 14 Fusarium and two Didymella species identified, the F. oxysporum (FOSC) and F. solani (FSSC) species complexes, F. redolens, and F. avenaceum were present in 82 to 94% of the fields and were most abundant, accounting for 74% of all isolates recovered. Less frequently found Fusarium spp. included F. tricinctum, F. culmorum and F. equiseti. The species D. pinodella occurred in moderate abundance in organic fields but was much less frequent in conventional fields. Fusarium redolens was also more common in roots from organic fields compared with conventional fields. In contrast, F. avenaceum, F. tricinctum and F. culmorum occurred more frequently in conventionally managed fields. Faba bean roots were colonized at similar rates by FOSC, FSSC and F. equiseti in both management systems.
There was no difference in the composition nor the frequencies (% colonized roots) of the fungal species in symptomatic and asymptomatic faba bean roots. There were also no effects of the management system (organic vs conventional) on the spectrum of pathogens isolated. Cropping system effects were intricately connected with differences in cropping histories as conventional systems had been devoid of legumes for at least five years in most cases. Cropping history affected the isolation frequencies of a number of species.
The low levels of root rot symptoms observed in some fields may have been due to other factors not assessed in this study. This could have included the presence of other pathogens such as Pythium or Rhizoctonia spp. (Esmaeili Taheri et al., 2017; Chatterton et al., 2019; Wille et al., 2019) and/or adverse abiotic soil properties such as elevated levels of Fe and Mn contents in the soil which are contributing factors in development of root rot disease (Nayyar et al., 2009). Soil-borne pathogens need to reach a threshold population within the root before causing visible disease symptoms (Gu et al., 2022) and cultural methods fail to reflect pathogen densities in roots. Also, a given root is often colonized by multiple fungal species simultaneously and competitive interactions may have played a role in the generally low symptom expression. For example, we have previously shown that co-occurrence of F. equiseti and highly aggressive strains of F. avenacem and D. pinodella in pea roots can almost completely neutralize detrimental effects of these pathogens (Šišić et al., 2017). Similarly, other beneficial members of root associated microbial communities such as arbuscular mycorrhizal fungi are also known for their ability to reduce biotic stresses (Wille et al., 2019). It is unknown to what extent such interactions played a role in symptom expression. This could be evaluated using, for example, quantitative real-time PCR (qPCR). While qPCR assays targeting all major Fusarium species identified in this study including the qPCR for detection and quantification of D. pinodella have been developed recently (Zitnick-Anderson et al., 2018; Šišić et al., 2022), these assays were not available when this study was initiated. This study provides solid foundation to further analyze the nature of interactions among the major fungal species isolated as the success of qPCR depends on prior knowledge of the pathogen population targeted. Despite the limitations resulting from the culture based identifications used in our study, taken together, our results indicate a generally high tolerance of faba beans to major root rot pathogens of grain legumes which are a common part of the root microflora of this crop.
Yield effects
The lack of a clear association between root rot incidence and the major pathogens identified in this study and between root rot incidence and faba bean yield is in line with previous reports pointing to the ability of legume associated D. pinodella and Fusarium spp. (Rodriguez et al., 2009; Šišić et al., 2018b; Šišić et al., 2022) to infect various hosts without causing visible root rot disease symptoms. Nevertheless, path analysis indicated significant negative correlations of D. pinodella and F. redolens with yield in organic systems, and significant negative yield effects of FOSC and F. culmorum in conventional systems. These results are in line with the hypothesis that there are rarely neutral biological interactions (Schulz and Boyle, 2005). Thus, asymptomatic plant infections likely result from mutually balanced antagonisms between a plants defense system and pathogen virulence factors (Schulz and Boyle, 2005). The positive association of pathogen frequencies with yield reductions in this study could reflect a need for higher investment of the faba bean to maintain a balanced antagonism with the aforementioned pathogens (i.e. absence to low levels of root rot), resulting in lower yields. We have recently demonstrated that D. pinodella is highly aggressive on pea causing symptoms and biomass reductions. In contrast, it can colonize wheat roots without causing visible disease symptoms while reducing wheat biomass (Šišić et al., 2022). This is the first instance where predominantly asymptomatic root infections by this pathogen appear to be negatively associated with faba bean yield. The fact that this negative correlation occurred only in organic systems is a result of the differences in rotational history between the management systems as this pathogen was inseparably connected with the frequency of legumes in rotation which constituted only about 5% of the crops in conventional fields compared to 26% of the crops in organic fields (Figure 4 and Supplementary Table 5).
Fusarium redolens is commonly isolated from diseased roots of different grain legumes, however its role in the root rot complex is not fully understood. For example, Booth (1971) reported that this pathogen was wide spread in temperate regions causing damping-off, wilts and cortical rots on a variety of non-legume and legume crops including pea and faba bean. A more recent study established F. redolens as an important and very aggressive root rot pathogen of pea in northern France causing similar damage as F. solani f. sp. pisi (Gibert et al., 2022). In contrast, Persson et al. (1997) and more recently Safarieskandari et al. (2020) found F. redolens to be a generally weak root rot pathogen based on the in vitro pathogenicity assays despite its frequent isolation from symptomatic field pea and lentil roots (Chatterton et al., 2019). In our study, it is possible, however, that asymptomatic infections with F. redolens could have affected yields opportunistically when the plants were stressed by cold conditions in early crop growth stages followed by warm and dry conditions. There is a need to more precisely characterize the interactions of this pathogen, faba bean yield and environmental variables including other fungal species. This may be particularly important as F. redolens is also pathogenic on cereals (Esmaeili Taheri et al., 2011; Yegin et al., 2017; Gebremariam et al., 2018) which are common part of crop rotations.
In contrast to negative yield effects of D. pinodella and F. redolens, higher abundance of F. tricintum in faba bean roots was positively associated with faba bean yield in organic systems. Although frequently isolated from legume crops (Šišić et al., 2018b; Chatterton et al., 2019), F. tricinctum is mainly associated with the Fusarium Head Blight (FHB) complex of small grain cereals in Europe and North America (Uhlig et al., 2007). Our previous research (Šišić et al., 2018b) indicated that this species was only a weak pathogen on pea where root colonization even resulted in increased pea biomass. In contrast, Yan and Nelson (2020) recently reported that F. tricinctum is an important soybean root rot pathogen. Due to apparent positive yield effects observed, however, further studies are recommended to better understand the role of this species in the faba bean root rot complex.
In contrast to organic systems, higher abundance of FOSC and F. culmorum in faba bean roots led to yield depressions in conventional fields, possibly reflecting cropping system-driven differences in overall soil properties. Fusarium root rot of faba bean caused by multiple Fusarium species including F. culmorum, and Fusarium wilt in particular caused by F. oxysporum, are among the most destructive diseases of this crop worldwide (Sillero et al., 2010; Lv et al., 2020). However, it is important to note that, in contrast to organic systems, faba bean yields in conventional systems were less affected by the pathogens and depended more on the total precipitation.
Cropping history effects on main root associated fungal species
Almost no legumes had been grown in the 5-11 years preceding the conventional faba beans surveyed in this study while grain legumes (mostly pea and faba bean) and also clover and alfalfa had almost always been present during this period in the organic fields sampled. This difference in rotational histories was strongly correlated with the occurrence of D. pinodella, which was ubiquitous in organic fields but much rarer or even absent in conventional fields. (Bainard et al., 2017) reported similar results with pea intensified rotations leading to substantial increase in abundance of this pathogen in soil and pea roots.
It is likely that the higher frequencies of legumes in organic rotations also contributed to higher abundance of F. redolens in organic systems compared to conventional as abundance of this pathogen in soil has been shown previously to increase substantially following intensified grain legume rotations (Bainard et al., 2017). The positive correlation of this pathogen with the frequency of cereals in organic systems is not surprising, however, as F. redolens is also a wheat (Esmaeili Taheri et al., 2011; Gebremariam et al., 2018) and barley (Yegin et al., 2017) pathogen.
The higher relative abundance of F. avenaceum, F. tricinctum and F. culmorum in conventionally grown faba beans is likely related to the generally higher ratio of cereals in conventional rotations despite the absence of clear correlations. In most conventional fields, cereals were grown three to five times in the five years preceding faba beans. The three species are major small grain cereal and maize pathogens frequently associated with ear, stem and root rots and responsible for pre-harvest mycotoxin contaminations (Bottalico and Perrone, 2002; Pfordt et al., 2020). Among the three species, F. avenaceum is the most important and wide-spread. It is an opportunistic pathogen especially in the absence of organic matter (Baćanović-Šišić et al., 2018) and the major causal agents of pea and lentil root rot in Canada (Esmaeili Taheri et al., 2016; Chatterton et al., 2019) and pea root rot in the USA (Chittem et al., 2015) in the past 20 years. In Europe, F. avenacem is frequently isolated from diseased pea roots but usually at moderate frequencies (Persson et al., 1997; Pflughöft et al., 2012; Baćanović-Šišić et al., 2018; Šišić et al., 2019).
Effects of environmental conditions on major fungal species identified
Pedo-climatic conditions appeared to be the main drivers for the occurrence of most of the Fusarium species identified in this study. Cold conditions at sowing and plant emergence and/or during the vegetative season in particular were found to favor most of the dominant Fusarium species identified in this study. These conditions were associated with increased root colonization rates by F. redolens, F. solani and F. culmorum in both management systems, and the FOSC and F. equiseti in organic systems. Sowing into cold soils prolongs seedling emergence favoring early plant infections by these pathogens (Naseri and Marefat, 2011). Although Fusarium spp. can infect their hosts at all growth stages, previous research has shown that the timing of infection plays a crucial role in the extent of root rot severity and yield reduction (Papavizas, 1974; Šišić et al., 2017; Navas-Cortés et al., 2020). For example, Šišić et al. (2017) showed that the detrimental effect of F. avenaceum on pea root health and biomass depended strongly on the timing of pathogen inoculation. When inoculated at sowing, F. avenaceum caused severe wilting resulting in 83% pea biomass loss compared to a non-inoculated control. In contrast, inoculation five days after pea sowing resulted in moderate root rot disease severity with greatly reduced negative effects on pea biomass (-14%). Therefore, vigorous seeds are likely to rapidly outgrow the highly susceptible seedling growth stage, reducing the overall risks of Fusarium damage. But also higher temperatures e.g. hot weather following sowing and/or in the period from sowing to root sampling can favor root infections e.g. by F. redolens which was more severe in both management systems in hot and dry seasons. The opposite was observed for F. culmorum in both management systems and F. avenaceum and F. equiseti in organic systems. Their root colonization rates were favored by cooler growing season. The precipitation effects were highly variable and mostly management system and Fusarium species specific. While Fusarium spp. are able to adapt to various ranges of environmental conditions, the competitive advantage of each species in the faba bean root rot complex will vary depending on the site specific pedo-climatic conditions (Yergeau et al., 2009). Furthermore, abiotic plant stress (i.e. the crop defense response) seems to have an important impact on the susceptibility of faba beans to Fusarium infections.
In contrast to Fusarium spp., warmer conditions at sowing/plant emergence favored D. pinodella in both management systems. In addition, although this species occurred rarely in conventional systems, D. pinodella frequencies were positively correlated with warmer and wetter conditions at sowing/plant emergence followed by drier growing seasons. The positive effect of drier conditions on D. pinodella root colonization rates and especially the high incidence of this pathogen in 2019, the driest sampling year in this study, are in contrast with what has been reported in the D. pinodella-pea system where infections are primarily favored by wet and humid conditions during the main growing season (Bretag et al., 2006; Esmaeili Taheri et al., 2016). These results suggest that short periods of wet conditions are sufficient for infections by this pathogen and also point to its opportunistic nature where abiotic plant stress (e.g. lack of precipitations) can enhance colonization process once the primary infections occur.
Our observations that the frequency of the FOSC was correlated positively with sandy soils characterized by lower pH supports previous reports that sandy soils and lower pH are more conducive for this pathogen and contribute to increased root rot and wilt incidence in a range of different crops, including chickpea, banana, flax, carnations, watermelon, tomato and marigold (Scher and Baker, 1980; Amir and Alabouvette, 1993; Singh et al., 2017; Orr and Nelson, 2018; Saeedi and Jamali, 2021). Some of these studies also showed that silty and clay soils and/or increasing soil pH were often suppressive to Fusarium wilt development. We also found a weak but statistically significant association of F. redolens with organic silty soils in this study. (Saeedi and Jamali, 2021) recently reported that F. redolens is a very aggressive pathogen of chickpea in Iran where F. oxysporum dominated on sandy soils while F. redolens was highly correlated with low sand and organic matter among others. In addition, the positive correlation between the FSSC frequencies in conventional system and silty-clay soils with generally high SOM content (i.e. fields in soil cluster 3) suggest that the FSSC members are highly competitive saprophytes.
Genetic diversity among F. oxysporum and F. solani isolates
The observation that the members of the FOSC and the FSSC are important components of the faba bean root rot complex both in organic and conventional fields is consistent with many previous findings (Chittem et al., 2015; Esmaeili Taheri et al., 2016; Šišić et al., 2018a; Šišić et al., 2018b; Chatterton et al., 2019). The wide-spread occurrence of the FOSC and FSSC over a range of soil and environmental conditions observed in this study indicates a high adaptability of both species complexes to a range of pedo-climatic and environmental conditions, which may in turn indicate high genetic diversity. The high genetic variability among the 35 F. oxysporum isolates from a single host observed in this study came as a surprise, however. With the exception of F. curvatum, F. nirenbergie, F. oxysporum and F. fabacearum lineages which have been previously associated either with faba bean or other legumes, the remaining five lineages including the most abundant F. libertatis (17/35 isolates) have not been associated with any legume host previously. Further analysis is required to determine their role in the faba bean root rot complex.
In contrast to FOSC, 29 of the 33 FSSC isolates analyzed matched F. pisi (syn. F. solani f. sp. pisi; Fusarium vanettenii), and a group of 3 isolates were placed in Fusarium solani sensu stricto lineage, and one isolate matched F. breviconum. These results confirm the common association of F. pisi with various legumes and its ability to occupy diverse ecological niches (Šišić et al., 2018a). More recently, Safarieskandari et al. (2020) demonstrated a high level of aggressiveness of F. pisi to faba beans. It is also important to note that, while the phylogenetic analysis generally confirmed a good resolution power of the TEF1 alpha locus in Fusarium, poor bootstrap support for some lineages within the FOSC was observed. These results were expected however, due to the single locus analysis and were consistent with the results of the TEF1 alpha tree topology reported for the FOSC (Lombard et al., 2018) and for the FSSC (Šišić et al., 2018a; Geiser et al., 2021). Work is on-going to obtain additional sequence data from more loci and also aggressiveness tests to better understand genetic variation and the role of the collected isolates in the faba bean root rot complex.
Conclusions
This four-year survey provides the first comparative documentation on the prevalence and frequency of Fusarium and Didymella species associated with faba bean roots in organic and conventional fields in Germany. By covering a wide range of pedoclimatic and field history conditions under conventional and organic production systems, it was possible to develop inferences about the main drivers currently influencing the pathobiome community on faba beans. Pedoclimatic conditions were the main drivers of the Fusarium communities, whereas D. pinodella was primarily influenced by the presence of grain legumes in the recent cropping history. This led to the dominance of this pathogen in organic systems, almost certainly because of the higher frequency of legumes in organic rotations. The role of F. libertatis and the other FOSC members identified that had not previously been associated with root rot of grain legumes needs to be examined. This study indicated that several major root rot pathogens of grain legumes may asymptomatically colonize faba bean roots as had been reported previously for other crops (Šišić et al., 2018b; Šišić et al., 2022). The study indicated that faba bean yields in organic systems apparently were affected by asymptomatic root infections by D. pinodella and F. redolens whereas yields in conventional systems depended more on the total precipitation during the main growing season. As described in the introduction, the new protein strategy of the EU has encouraged many conventional farmers to adopt grain legumes since 2012 and our survey shows that most conventionally grown faba beans were grown for the first time in many years. An overall increase of grain legume production under conventional conditions will likely change their health status, however, as has already been observed in Canada (Bainard et al., 2017; Niu et al., 2018).
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Files. Further inquiries can be directed to the corresponding author.
Author contributions
AŠ, HS, JŠ, MF study design/methodology/investigation. AŠ data analysis/drafted the manuscript. AŠ, MF resources/funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
This work was carried out within the framework of the research projects PATHO-ID (2814EPS40) and APSOLU (2814EPS035), funded by the Federal Ministry of Food and Agriculture within the framework of the BMEL protein plant strategy, in cooperation with the German Demonstration network Pea/Bean.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1009906/full#supplementary-material
References
Amir, H., Alabouvette, C. (1993). Involvement of soil abiotic factors in the mechanisms of soil suppressiveness to Fusarium wilts. Soil Biol. Biochem. 25, 157–164. doi: 10.1016/0038-0717(93)90022-4
Baćanović-Šišić, J., Šišić, A., Schmidt, J. H., Finckh, M. R. (2018). Identification and characterization of pathogens associated with root rot of winter peas grown under organic management in Germany. Eur. J. Plant Pathol. 151, 745–755. doi: 10.1007/s10658-017-1409-0
Bainard, L. D., Navarro-Borrell, A., Hamel, C., Braun, K., Hanson, K., Gan, Y. (2017). Increasing the frequency of pulses in crop rotations reduces soil fungal diversity and increases the proportion of fungal pathotrophs in a semiarid agroecosystem. Agricult. Ecosyst. Environ. 240, 206–214. doi: 10.1016/j.agee.2017.02.020
Benjamini, Y., Hochberg, Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. 57, 289–300. doi: 0035-9246/95/57289
Boerema, G. H., Gruyter, J., Noordeloos, M. E., Hamers, M. E. C. (2004). Phoma identification manual: Differentiation of specific and infra-specific taxa in culture (Wallingford, Oxfordshire, UK: CABI publishing).
Bottalico, A., Perrone, G. (2002). Toxigenic Fusarium species and mycotoxins associated with head blight in small-grain cereals in Europe. Eur. J. Plant Pathol. 108, 611–624. doi: 10.1023/A:1020635214971
Bretag, T. W., Keane, P. J., Price, T. V. (2006). The epidemiology and control of ascochyta blight in field peas: A review. Aust. J. Agric. Res. 57, 883. doi: 10.1071/AR05222
Brooks, M. E., Kristensen, K., Benthem, K. J., Magnusson, A., Berg, C. W., Nielsen, A., et al. (2017). glmmTMB balances speed and flexibility among packages for zero-inflated generalized linear mixed modeling. R J. 9, 378–400. doi: 10.32614/RJ-2017-066
Chatterton, S., Harding, M. W., Bowness, R., Mclaren, D. L., Banniza, S., Gossen, B. D. (2019). Importance and causal agents of root rot on field pea and lentil on the Canadian prairies 2014–2017. Can. J. Plant Pathol. 41, 98–114. doi: 10.1080/07060661.2018.1547792
Chen, Q., Jiang, J. R., Zhang, G. Z., Cai, L., Crous, P. W. (2015). Resolving the Phoma enigma. Stud. Mycology 82, 137–217. doi: 10.1016/j.simyco.2015.10.003
Chittem, K., Mathew, F. M., Gregoire, M., Lamppa, R. S., Chang, Y. W., Markell, S. G., et al. (2015). Identification and characterization of fusarium spp. associated with root rots of field pea in north Dakota. Eur. J. Plant Pathol. 143, 641–649. doi: 10.1007/s10658-015-0714-8
Conover, W. J. (1999). Practical nonparametric statistics. 3rd ed (New York, USA: John Wiley & Sons, Inc).
Coons, G. H. (1916). Factors involved in the growth and the pycnidium formation of Plenodomus fuscomaculans. J. Agric. Res. 5, 713–769.
Doyle, J. J., Doyle, J. L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bull. 19, 11–15.
Esmaeili Taheri, A., Chatterton, S., Foroud, N. A., Gossen, B. D., McLaren, D. L. (2016). Identification and community dynamics of fungi associated with root, crown, and foot rot of field pea in western Canada. Eur. J. Plant Pathol. 147, 489–500. doi: 10.1007/s10658-016-1017-4
Esmaeili Taheri, A., Chatterton, S., Gossen, B. D., McLaren, D. L. (2017). Metagenomic analysis of oomycete communities from the rhizosphere of field pea on the Canadian prairies. Can. J. Microbiol. 63, 758–768. doi: 10.1139/cjm-2017-0099
Esmaeili Taheri, A., Hamel, C., Gan, Y., Vujanovic, V. (2011). First report of Fusarium redolens from Saskatchewan and its comparative pathogenicity. Can. J. Plant Pathol. 33, 559–564. doi: 10.1080/07060661.2011.620631
Gebremariam, E. S., Sharma-Poudyal, D., Paulitz, T. C., Erginbas-Orakci, G., Karakaya, A., Dababat, A. A. (2018). Identity and pathogenicity of Fusarium species associated with crown rot on wheat (Triticum spp.) in Turkey. Eur. J. Plant Pathol. 150, 387–399. doi: 10.1007/s10658-017-1285-7
Geiser, D. M., Al-Hatmi, A. M. S., Aoki, T., Arie, T., Balmas, V., Barnes, I., et al. (2021). Phylogenomic analysis of a 55.1-kb 19-gene dataset resolves a monophyletic Fusarium that includes the Fusarium solani species complex. Phytopathology 111, 1064–1079. doi: 10.1094/PHYTO-08-20-0330-LE
Geiser, D. M., Jiménez-Gasco, M. M. D., Kang, S., Makalowska, I., Veeraraghavan, N., Ward, T. J., et al. (2004). FUSARIUM-ID v. 1.0: A DNA sequence database for identifying Fusarium. Eur. J. Plant Pathol. 110, 473–479. doi: 10.1007/978-1-4020-2285-2_2
Gibert, S., Edel-Hermann, V., Gautheron, E., Gautheron, N., Bernaud, E., Sol, J. M., et al. (2022). Identification, pathogenicity and community dynamics of fungi and oomycetes associated with pea root rot in northern France. Plant Pathol. 71, 1550–1569. doi: 10.1111/ppa.13583
Gu, Y., Banerjee, S., Dini-Andreote, F., Xu, Y., Shen, Q., Jousset, A., et al. (2022). Small changes in rhizosphere microbiome composition predict disease outcomes earlier than pathogen density variations. ISME J. 14, 2448–2456. doi: 10.1038/s41396-022-01290-z
Hartig, F. (2021) DHARMa: Residual diagnostics for hierarchical (multi-level / mixed) regression models. Available at: http://florianhartig.github.io/DHARMa/.
Hossain, S., Bergkvist, G., Berglund, K., Mårtensson, A., Persson, P. (2012). Aphanomyces pea root rot disease and control with special reference to impact of brassicaceae cover crops. Acta Agriculturae Scandinavica Section B - Soil Plant Sci. 62, 1–11. doi: 10.1080/09064710.2012.668218
Katoh, K., Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Lê, S., Josse, J., Husson, F. (2008). FactoMineR: An R package for multivariate analysis. J. Stat. Software 25, 1–18. doi: 10.18637/jss.v025.i01
Lenth, R. V. (2016). Least-squares means: The R package lsmeans. J. Stat. Soft. 69, 1–33. doi: 10.18637/jss.v069.i01
Leslie, J. F., Summerell, B. A. (2006). The fusarium laboratory manual. 1st ed (Ames, Iowa, USA: Blackwell publishing).
Lombard, L., Sandoval-Denis, M., Lamprecht, S. C., Crous, P. W. (2018). Epitypification of fusarium oxysporum – clearing the taxonomic chaos. persoonia. 43, 1–47. doi: 10.3767/persoonia.2019.43.01
Lv, J., Dong, Y., Dong, K., Zhao, Q., Yang, Z., Chen, L. (2020). Intercropping with wheat suppressed fusarium wilt in faba bean and modulated the composition of root exudates. Plant Soil 448, 153–164. doi: 10.1007/s11104-019-04413-2
Madden, T. (2002). “Chapter 16 “The BLAST sequence analysis tool,” in The NCBI handbook. Eds. McEntyre, J., Ostell, J. (Bethesda (MD: National Center for Biotechnology Information (US) 2002). Available at: http://www.ncbi.nlm.nih.gov/books/NBK21097/.
Mendiburu, D. F. (2014). Agricolae: Statistical procedures for agricultural research 1, 2. R package version.
Naseri, B., Ansari Hamadani, S. (2017). Characteristic agro-ecological features of soil populations of bean root rot pathogens. Rhizosphere 3, 203–208. doi: 10.1016/j.rhisph.2017.05.005
Naseri, B., Marefat, A. (2011). Large-Scale assessment of agricultural practices affecting fusarium root rot and common bean yield. Eur. J. Plant Pathol. 131, 179–195. doi: 10.1007/s10658-011-9798-y
Naseri, B., Mousavi, S. S. (2015). Root rot pathogens in field soil, roots and seeds in relation to common bean ( Phaseolus vulgaris ), disease and seed production. Int. J. Pest Manage. 61, 60–67. doi: 10.1080/09670874.2014.993001
Navas-Cortés, J. A., Hau, B., iménez-Díaz, R. M. (2020). Yield loss in chickpeas in relation to development of Fusarium wilt epidemics. Phytopathology 11, 1269–1278. doi: 10.1094/PHYTO.2000.90.11.1269
Nayyar, A., Hamel, C., Lafond, G., Gossen, B. D., Hanson, K., Germida, J. (2009). Soil microbial quality associated with yield reduction in continuous-pea. Appl. Soil Ecol. 43, 115–121. doi: 10.1016/j.apsoil.2009.06.008
Niu, Y., Bainard, L. D., May, W. E., Hossain, Z., Hamel, C., Gan, Y. (2018). Intensified pulse rotations buildup pea rhizosphere pathogens in cereal and pulse based cropping systems. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.01909
O’Donnell, K., Al-Hatmi, A. M. S., Aoki, T., Brankovics, B., Cano-Lira, J. F., Coleman, J. J., et al. (2020). No to Neocosmospora: phylogenomic and practical reasons for continued inclusion of the Fusarium solani species complex in the genus. Fusarium 5, 7. doi: 10.1128/mSphere.00810-20
O’Donnell, K., Gueidan, C., Sink, S., Johnston, P. R., Crous, P. W., Glenn, A., et al. (2009). A two-locus DNA sequence database for typing plant and human pathogens within the fusarium oxysporum species complex. Fungal Genet. Biol. 46, 936–948. doi: 10.1016/j.fgb.2009.08.006
O’Donnell, K., Kistler, H. C., Cigelnik, E., Ploetz, R. C. (1998). Multiple evolutionary origins of the fungus causing Panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proc. Natl. Acad. Sci. 95, 2044–2049. doi: 10.1073/pnas.95.5.2044
Orr, R., Nelson, P. N. (2018). Impacts of soil abiotic attributes on Fusarium wilt, focusing on bananas. Appl. Soil Ecol. 132, 20–33. doi: 10.1016/j.apsoil.2018.06.019
Papavizas, G. C. (1974). Aphanomyces species and their root diseases in pea and sugarbeet: A review (US Department of Agriculture: Agricultural Research Service).
Persson, L., Bødker, L., Larsson-Wikström, M. (1997). Prevalence and pathogenicity of foot and root rot pathogens of pea in southern Scandinavia. Plant Dis. 81, 171–174. doi: 10.1094/PDIS.1997.81.2.171
Pflughöft, O., Merker, C., von Tiedemann, A., Schäfer, B. C. (2012). Zur verbreitung und bedeutung von pilzkrankheiten in körnerfuttererbsen (Pisum sativum l.) in deutschland. Gesunde Pflanzen 64, 39–48. doi: 10.1007/s10343-011-0270-x
Pfordt, A., Ramos Romero, L., Schiwek, S., Karlovsky, P., von Tiedemann, A. (2020). Impact of environmental conditions and agronomic practices on the prevalence of Fusarium species associated with ear- and stalk rot in maize. Pathogens 9, 236. doi: 10.3390/pathogens9030236
R Core Team (2013). R: A language and environment for statistical computing (Vienna, Austria: R Foundation for Statistical Computing). Available at: http://www.R-project.org/.
Rodriguez, R. J., White, J. F., Jr., Arnold, A. E., Redman, R. S. (2009). Fungal endophytes: diversity and functional roles. New Phytol. 182, 314–330. doi: 10.1111/j.1469-8137.2009.02773.x
Rosseel, Y. (2012). Lavaan: An R package for structural equation modeling. J. Stat. Software 48, 1–36. doi: 10.18637/jss.v048.i02
Saeedi, S., Jamali, S. (2021). Mol. characterization distribution Fusarium isolates uncultivated soils chickpea Plants Iran special reference to Fusarium redolens. J. Plant Pathol. 103, 167–183. doi: 10.1007/s42161-020-00698-w
Safarieskandari, S., Chatterton, S., Hall, L. M. (2020). Pathogenicity and host range of Fusarium species associated with pea root rot in Alberta, Canada. Can. J. Plant Pathol. 43, 162–171. doi: 10.1080/07060661.2020.1730442
Scher, F. M., Baker, R. (1980). Mechanism of biological control in a Fusarium-suppressive soil. Phytopathology 70, 412–417. doi: 10.1094/Phyto-70-412
Schulz, B., Boyle, C. (2005). The endophytic continuum. Mycological Res. 109, 661–686. doi: 10.1017/S095375620500273X
Sillero, J. C., Villegas-Fernández, A. M., Thomas, J., Rojas-Molina, M. M., Emeran, A. A., Fernández-Aparicio, M., et al. (2010). Faba bean breeding for disease resistance. Field Crops Res. 115, 297–307. doi: 10.1016/j.fcr.2009.09.012
Singh, B., Singh, V., Srivastava, S., Pandey, A., Shukla, D. (2017). Influence of soil properties on wilt incidence of water melon, tomato and marigold. ARRB 19, 1–6. doi: 10.9734/ARRB/2017/34350
Šišić, A., Baćanović, J., Finckh, M. R. (2017). Endophytic Fusarium equiseti stimulates plant growth and reduces root rot disease of pea (Pisum sativum l.) caused by Fusarium avenaceum and Peyronellaea pinodella. Eur. J. Plant Pathol. 148, 271–282. doi: 10.1007/s10658-016-1086-4
Šišić, A., Baćanović-Šišić, J., Al-Hatmi, A. M. S., Karlovsky, P., Ahmed, S. A., Maier, W., et al. (2018a). The ‘forma specialis’ issue in Fusarium: A case study in Fusarium solani f. sp. pisi. Sci. Rep. 8, 1252. doi: 10.1038/s41598-018-19779-z
Šišić, A., Baćanović-Šišić, J., Karlovsky, P., Wittwer, R., Walder, F., Campiglia, E., et al. (2018b). Roots of symptom-free leguminous cover crop and living mulch species harbor diverse Fusarium communities that show highly variable aggressiveness on pea (Pisum sativum). PloS One 13, e0191969. doi: 10.1371/journal.pone.0191969
Šišić, A., Baćanović-Šišić, J., Schmidt, H., Finckh, M. R. (2019). Root pathogens occurring on pea (Pisum sativum) and faba bean (Vicia faba) in Germany (Sarajevo: Springer International Publishing). Sept. 26-27, 2019.
Šišić, A., Baćanović-Šišić, J., Schmidt, H., Finckh, M. R. (2020). First report of Fusarium flocciferum causing root rot of pea (Pisum sativum) and faba bean (Vicia faba) in Germany. Plant Dis. 104, 283–283. doi: 10.1094/PDIS-06-19-1302-PDN
Šišić, A., Oberhänsli, T., Baćanović-Šišić, J., Hohmann, P., Finckh, M. R. (2022). A novel real time PCR method for the detection and quantification of Didymella pinodella in symptomatic and asymptomatic plant hosts. J. Fungi 8, 41. doi: 10.3390/jof8010041
Stamatakis, A., Hoover, P., Rougemont, J. (2008). A rapid bootstrap algorithm for the RAxML web servers. Systematic Biol. 57, 758–771. doi: 10.1080/10635150802429642
Su, Y. S., Yajima, M., Hill, J., Pittau, M. G., Kerman, J., Zheng, T., et al. (2018) Package a’rm’: Data analysis using regression and multilevel/hierarchical models. Available at: https://cran.r-project.org/web/packages/car/car.pdf (Accessed February 17, 2020).
Tamura, K., Stecher, G., Peterson, D., Filipski, A., Kumar, S. (2013). MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Uhlig, S., Jestoi, M., Parikka, P. (2007). Fusarium avenaceum — the north European situation. Int. J. Food Microbiol. 119, 17–24. doi: 10.1016/j.ijfoodmicro.2007.07.021
Walder, F., Schlaeppi, K., Wittwer, R., Held, A. Y., Vogelgsang, S., van der Heijden, M. G. A. (2017). Community profiling of Fusarium in combination with other plant-associated fungi in different crop species using SMRT sequencing. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.02019
Wickham, H. (2016). ggplot2: Elegant graphics for data analysis. 2nd ed (New York: Springer International Publishing). Available at: https://ggplot2.tidyverse.org.
Wilbois, K. P., Böhm, H., Bohne, B., Brandhuber, R., Bruns, C., Demmel, M., et al. (2013). “Steigerung der wertschöpfung ökologisch angebauter marktfrüchte durch optimierung des managements der bodenfruchtbarkeit,” in Gesamtprojekt abschlussbericht. Available at: www.orgprints.org/28973.
Wille, L., Messmer, M. M., Studer, B., Hohmann, P. (2019). Insights to plant–microbe interactions provide opportunities to improve resistance breeding against root diseases in grain legumes. Plant Cell Environ. 42, 20–40. doi: 10.1111/pce.13214
Williamson-Benavides, B. A., Dhingra, A. (2021). Understanding root rot disease in agricultural crops. Horticulturae 7, 33. doi: 10.3390/horticulturae7020033
Wohor, O. Z., Rispail, N., Ojiewo, C. O., Rubiales, D. (2022). Pea breeding for resistance to rhizospheric pathogens. Plants 11, 2664. doi: 10.3390/plants11192664
Yan, H., Nelson, B. (2020). Effect of temperature on Fusarium solani and F. tricinctum growth and disease development in soybean. Can. J. Plant Pathol. 42, 527–537. doi: 10.1080/07060661.2020.1745893
Yegin, N. Z., Ünal, F., Tekiner, N., Dolar, F. S. (2017). First report of Fusarium redolens causing root and crown rot of barley (Hordeum vulgare) in Turkey. J. Turkish Phytopathol. 46, 101–105.
Yergeau, E., Labour, K., Hamel, C., Vujanovic, V., Nakano-Hylander, A., Jeannotte, R., et al. (2009). Patterns of fusarium community structure and abundance in relation to spatial, abiotic and biotic factors in soil. FEMS Microbiol. Ecol. 71, 34–42. doi: 10.1111/j.1574-6941.2009.00777.x
Keywords: fusarium, didymella, root rot, faba bean, grain legumes, organic agriculture, conventional agriculture
Citation: Šišić A, Baćanović-Šišić J, Schmidt H and Finckh MR (2022) Farming system effects on root rot pathogen complex and yield of faba bean (vicia faba) in Germany. Front. Plant Sci. 13:1009906. doi: 10.3389/fpls.2022.1009906
Received: 02 August 2022; Accepted: 29 November 2022;
Published: 21 December 2022.
Edited by:
Sabine Banniza, University of Saskatchewan, CanadaReviewed by:
Bita Naseri, Education and Extension Organization (AREEO), IranBruce D. Gossen, Agriculture and Agri-Food Canada (AAFC), Canada
Copyright © 2022 Šišić, Baćanović-Šišić, Schmidt and Finckh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Adnan Šišić, YWRuYW5fc2lzaWNAdW5pLWthc3NlbC5kZQ==
 Adnan Šišić
Adnan Šišić Jelena Baćanović-Šišić2
Jelena Baćanović-Šišić2 Maria R. Finckh
Maria R. Finckh