
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 26 September 2022
Sec. Plant Bioinformatics
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.1009723
This article is part of the Research TopicOmics-Driven Crop Improvement for Stress ToleranceView all 21 articles
Post-translational histone modifications play important roles in regulating chromatin structure and transcriptional regulation. Histone 3 lysine 4 trimethylation (H3K4me3) is a prominent histone modification mainly associated with gene activation. Here we showed that a histone demethylase, JMJ15, belonging to KDM5/JARID group, is involved in salt stress response in Arabidopsis thaliana. Jmj15 loss-of-function mutants displayed increased sensitivity to salt stress. Moreover, knockout of JMJ15 impaired the salt responsive gene expression program and affected H3K4me3 levels of many stress-related genes under salt-stressed condition. Importantly, we demonstrated that JMJ15 regulated the expression level of two WRKY transcription factors, WRKY46 and WRKY70, which were negatively involved in abiotic stress tolerance. Furthermore, JMJ15 directly bound to and demethylated H3K4me3 mark in the promoter and coding regions of WRKY46 and WRKY70, thereby repressing these two WRKY gene expression under salt stress. Overall, our study revealed a novel molecular function of the histone demethylase JMJ15 under salt stress in plants.
Higher plants can adapt to changing environmental conditions in different ways via adjustments in gene expression. Epigenetic regulation of gene expression involving chromatin modification such as histone 3 lysine 4 trimethylation (H3K4me3) plays essential roles in plant response to environmental conditions (van Dijk et al., 2010; Foroozani et al., 2021). H3K4me3 has long been known as an active mark for gene transcription (Santos-Rosa et al., 2002; Li et al., 2007). Genome-wide analysis revealed that a large number of genes were marked by H3K4me3 which predominantly found at the promoter and 5′ end of genes in plants (Zhang et al., 2009). H3K4me3 was often associated with stress-induced gene expression by plant internal and external signals (Sokol et al., 2007; Kim et al., 2008; van Dijk et al., 2010), suggesting an active transcription role for H3K4me3. However, in some cases the increase of H3K4me3 was found to be lagged behind stressed gene activation (Kim et al., 2008; Hu et al., 2011), indicating that H3K4me3 might have a function to mark active gene status. In addition, H3K4me3 has also been found to play a role in transcriptional memory of stress-responsive genes in plants (Jaskiewicz et al., 2011; Ding et al., 2012; Kim et al., 2012, 2015). H3K4 methylation is mediated by the Trithorax group proteins (TRX), including five Trithorax-like (ATX) proteins and two ATX-related (ATXR) proteins in Arabidopsis (Ng et al., 2007). Arabidopsis ATX1, ATX4, and ATX5 were found to be necessary for abscisic acid (ABA) and dehydration stress responses (Ding et al., 2011; Liu et al., 2018).
Histone methylation could be reversed by histone demethylases. Lysine Specific Demethylase 1 (LSD1) was the first identified histone demethylase to remove mono- and di-methyl groups from H3K4 (Shi et al., 2004). Jumonji C (jmjC) domain-containing proteins were the second known class of histone demethylases. JmjC proteins, conserved in plants, animals and fungi, catalyze histone lysine demethylation through a ferrous ion [Fe(II)] and a-ketoglutaric acid (a-KG)-dependent oxidative reaction (Tsukada et al., 2006). JmjC domain containing demethylases are divided into distinct groups including KDM2A/JHDM1A, KDM5/JARID, KDM4/JMJD2, KDM6/JMJD3, and KDM3/JMJD1/JHDM2, depending on their binding and catalytic specificities. In humans, the KDM5/JARID group demethylases specially catalyze H3K4me2/3 demethylation (Mosammaparast and Shi, 2010). The Arabidopsis genome encodes ∼20 jmjC domain-containing histone demethylases (JMJ11–JMJ31), among which six jmjC proteins (JMJ14, JMJ15, JMJ16, JMJ17, JMJ18, and JMJ19) belonging to KDM5/JARID group demethylate H3K4me1/2/3 (Lu et al., 2008; Sun and Zhou, 2008; Chen et al., 2011). JMJ14 demethylates H3K4me3 in vitro and represses floral integrator genes Flowering Locus T (FT) and Suppressor of overexpression of CO1 (SOC1) by removing H3K4me3 (Jeong et al., 2009; Lu et al., 2010; Yang et al., 2010, 2018). JMJ16 negatively regulates leaf senescence through repressing the positive regulators of WRKY53 and SAG201 by reducing H3K4me3 (Liu et al., 2019). JMJ15 and JMJ18 regulate flowering time by demethylating H3K4me3 at the locus of the floral repressor gene Flowering Locus C (FLC) (Yang et al., 2012a,b). In addition to the essential roles in plant development, JMJ15 and JMJ17 are also involved in stress response. The jmj17 loss-of-function mutants display dehydration stress tolerance and ABA hypersensitivity (Huang et al., 2019; Wang et al., 2021). In detail, JMJ17 regulates the stomatal closure and ABA response through modulating OPEN STOMATA1 (OST1) and ABI5 genes by demethylating H3K4me3 (Huang et al., 2019; Wang et al., 2021). Our previous results showed that JMJ15 participated in salinity stress response (Shen et al., 2014). Jmj15 gain-of-function mutants increased tolerance to salinity stress and down-regulated many genes encoding transcription regulators involved in stress responses (Shen et al., 2014). Although the overexpression of JMJ15 confers tolerance to salt stress, the molecular mechanism of JMJ15 mediated salt tolerance remains elusive.
In the present study, we showed that two jmj15 loss-of-function mutant alleles (jmj15-3 and jmj15-4) revealed more sensitive phenotypes to salinity stress. Importantly, the jmj15 loss-of-function mutations led to up-regulation of many stress-related genes under salt condition rather than under normal condition. In addition, chromatin immunoprecipitation sequencing (ChIP-seq) showed that knockout of JMJ15 regulated H3K4me3 levels of many salt and water deprivation stress-related genes under salt-stressed condition. Furthermore, our results showed that JMJ15 directly bound to and repressed the genes of WRKY46 and WRKY70 that played negative roles in salt stress tolerance. Taken together, our data unravel a novel molecular function of JMJ15 in salt stress responsiveness, which is distinct from the function of other characterized jmjC genes in Arabidopsis.
In this study, Arabidopsis thaliana wild type Col-0 and mutant lines jmj15-1 (GABI_257F10), jmj15-2 (GABI_876B01), jmj15-3 (GABI-663C11), and jmj15-4 (GABI-229F03) in the Col-0 background were used. The insertional knock-out lines jmj15-3 (Shen et al., 2014) and jmj15-4 (Yang et al., 2012b) have been previously characterized. The gain-of-function T-DNA mutant lines jmj15-1, jmj15-2, and the over-expressing tagged line 35s-JMJ15-HA have been reported (Shen et al., 2014). The Arabidopsis seeds were surface-sterilized by 5% (w/v) sodium hypochlorite for 7 min, washed with 95% (v/v) ethanol twice, and then sown on 1/2 Murashige and Skoog (MS) medium. After stratification in darkness at 4°C for 2 days, seeds were transferred into a growth chamber (20°C) under white light (120 μmol m–2 s–1) in 16 h light photoperiods. Wild-type and mutant plants were grown together and their mature seeds were harvested at the same time to avoid differences in post-maturation that can affect seed germination. Seeds of each genotype were harvested as a single bulk consisted of at least five plants. Seeds were stored in open tubes inside a closed box and maintained in darkness with silica gel at 4°C until the experiments were performed.
For measuring seed germination, more than 100 seeds of each genotype (Col-0, jmj15-3, and jmj15-4) were sown on NaCl-infused media (1/2 MS medium with 0, 100, 150, and 200 mM NaCl, respectively). After stratification in darkness at 4°C for 2 days, the plates were transferred into a growth chamber under white light in 16 h light photoperiods. The germination (fully emerged radicle) rates were measured per day for a duration of 5 days. Each test was performed with more than three biological replicates.
Measure root elongation under salt stress mainly according to the previous protocol (Lee and Zhu, 2009). More than 50 seedlings of Col-0 and jmj15 mutation lines were sown on 1/2 MS medium. After stratification in darkness at 4°C for 2 days, the plates were moved to a vertical position in the growth chamber (16 h white light photoperiod, 20°C) for germination. The 4-day-old germinated seedlings were transferred onto vertical 1/2 MS agar plates supplemented with 0 m M NaCl, 50 m M NaCl, 100 mM NaCl and 150 mM NaCl, respectively. After 5 days with the salt treatment, the primary root length was measured and statistical significance was determined by two-sided t-test. Three replicates were performed for each line and treatment.
Ten-day-old wild type (Col-0) and jmj15-4 mutants Arabidopsis plants treated with NaCl (0 mM or 150 mM NaCl solution in 1/2 MS liquid medium) for 5 h were pooled for RNA extraction and transcriptomic analysis. Plants harvested from three independent cultures were used as the biological replicates. Total RNA was extracted with Trizol (Invitrogen, Carlsbad, CA, United States), and treated with DNase I (Promega, Madison, WI, United States) to remove the genomic DNA. Then the mRNA was enriched by using oligo(dT) magnetic beads and broken into short fragments (200 bp) using the fragmentation buffer. The first strand cDNA was synthesized by using a random primer. RNase H, DNA polymerase I and dNTPs were used to synthesize the second strand. The double-strand cDNA was purified with magnetic beads. cDNA ends were repaired and a nucleotide A (adenine) was added at the 3′-end. Finally, PCR amplification was performed with fragments ligated by sequencing adaptors. The Agilent 2100 Bioanalyzer and the ABI Step One Plus real-time PCR system were used to qualify and quantify the QC library. The library products were sequenced with the Illumina HiSeqX-ten platform, and the library construction and sequencing were completed at Novogene Corporation (Tianjin, China). Gene expression was defined by RSEM (Li and Dewey, 2011) and estimated by FPKM. Differential expression analysis between two conditions/groups was performed using DESeq R package. The resulting P-values were adjusted using the Benjamini and Hochberg’s approach for controlling the false discovery rate (Anders and Huber, 2010). The RNA-seq data were deposited to NCBI-SRA databases under the accession PEJNA822702.
Total RNA was extracted from plant seedlings using Trizol (Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s instructions. RNA concentration was measured by using a Nanodrop. For RT-qPCR analysis, 2 μg of total RNA treated with DNaseI (RQ1, Promega, M6101, Madison, WI, United States) were used to synthesize first-strand cDNA with Oligo(dT)15 primers using ImPromII reverse transcriptase (M3104A, Promega, M3104A, Madison, WI, United States). RT-qPCR was performed with LightCycler 480 SYBR Green I Master mix on the LightCycler 480 (Roche, Mannheim, Germany). The reactions were performed in triplicate for each run and at least three biological replicates were carried out for each reaction. Transcript levels were calculated using the comparative Ct (threshold cycle) method and utilizing ACTIN2 as an internal control for data normalization. Primer sequences used in this study were summarized in Supplementary Table 1.
Chromatin immunoprecipitation (ChIP) experiments were carried out as described previously with minor modifications (Shen et al., 2015). Ten-day-old plants were treated with NaCl (0 mM or 150 mM NaCl solution in 1/2 MS liquid medium) for 5 h before harvest. About 4 g of plant seedlings were harvested, which were then cross-linked with 1% formaldehyde for 10 min under vacuum and ground into fine powder in liquid nitrogen. The chromatin DNA was isolated and sonicated in the 200–1000 bp range with Diagenode Bioruptor UCD-300. The sonicated chromatin was pre-cleared and incubated with anti-H3K4me3 (Millipore, 07-473, Darmstadt, Germany), anti-RNAPII (Abcam, ab5408, Cambridge, United Kingdom) or anti-HA (Sigma, H6908, St. Louis, MO, United States) antibodies loaded Dynabeads™ protein A (Invitrogen, #10002D, Carlsbad, CA, United States) at 4°C overnight. Subsequently, the immunoprecipitated DNA was decrosslinked and purified by using the MinElute PCR Purification Kit (Qiagen, #28004, Hilden, Germany) according to the manufacturer’s instruction. ChIP DNA was used for sequencing or qPCR. ChIP-qPCR was performed with three biological replicates, and results were calculated as the percentage of input DNA. Sequences of the primers used for ChIP-qPCR were listed in Supplementary Table 1.
At least 5 ng of each ChIP DNA was used to construct ChIP-seq library, and two biological replicates for each sample. The Illumina libraries were constructed using the ChIP-seq DNA Sample Prep Kit (Illumina) according to manufacturer’s protocol. The ChIP-seq library was sequenced by Novogene Corporation (Tianjin, China) on the Illumina HiSeq2500 sequencing system. Base calling and read quality control were performed following the standard Illumina protocol. Reads passing quality control were aligned to the Arabidopsis genome (TAIR101) using BWA (Burrows Wheeler Aligner) with default parameters and only uniquely mapped reads were kept (Li and Durbin, 2009). MACS2 (version 2.1.0) peak calling software was used to identify regions of enriched intervals over the background. A q-value threshold of 0.05 was used for all data sets. Differential enrichments were assessed by MACS2 (bdgdiff), with an FDR cutoff < 0.001. The differential histone modification regions were intersected with annotated genes to obtain the target genes using BED tools2. The alignments were converted to wiggle (WIG) files. The data were then imported into the Integrated Genome Browser for visualization. The ChIP-seq data were deposited to NCBI-SRA databases under the accession PRJNA823378.
To explore the transcriptional level of genes involved in plant response to salt stress, we conducted RNA-seq analysis using wild type seedlings under normal or salt-stressed conditions (0 mM or 150 mM NaCl solution in 1/2 MS liquid medium for 5 h), each with three biological replicates. The RNA-seq results showed that the three biological replicates were all highly correlated (R = 0.91–0.99). Analysis of differentially expressed genes (DEGs) revealed that 5470 up-regulated genes (URGs) and 5393 down-regulated genes (DRGs) in wild type seedlings as a result of the salt stress (P < 0.05) (Figure 1A and Supplementary Table 2). Using the strict criteria (fold change > 4.0, P < 0.001), 619 URGs and 833 DRGs were found in salt treated versus normal growth seedlings (Figure 1A and Supplementary Table 2). Among the genes up-regulated by salt stress, inducible genes implicated in salt stress response in other studies were present, including COR15A (AT2G42540), COR15B (AT2G42530), RD20 (AT2G33380), RD29A (AT5G52310), RD29B (AT5G52300) (Figure 1B; Chen et al., 2010; Zheng et al., 2016; Yang et al., 2019).
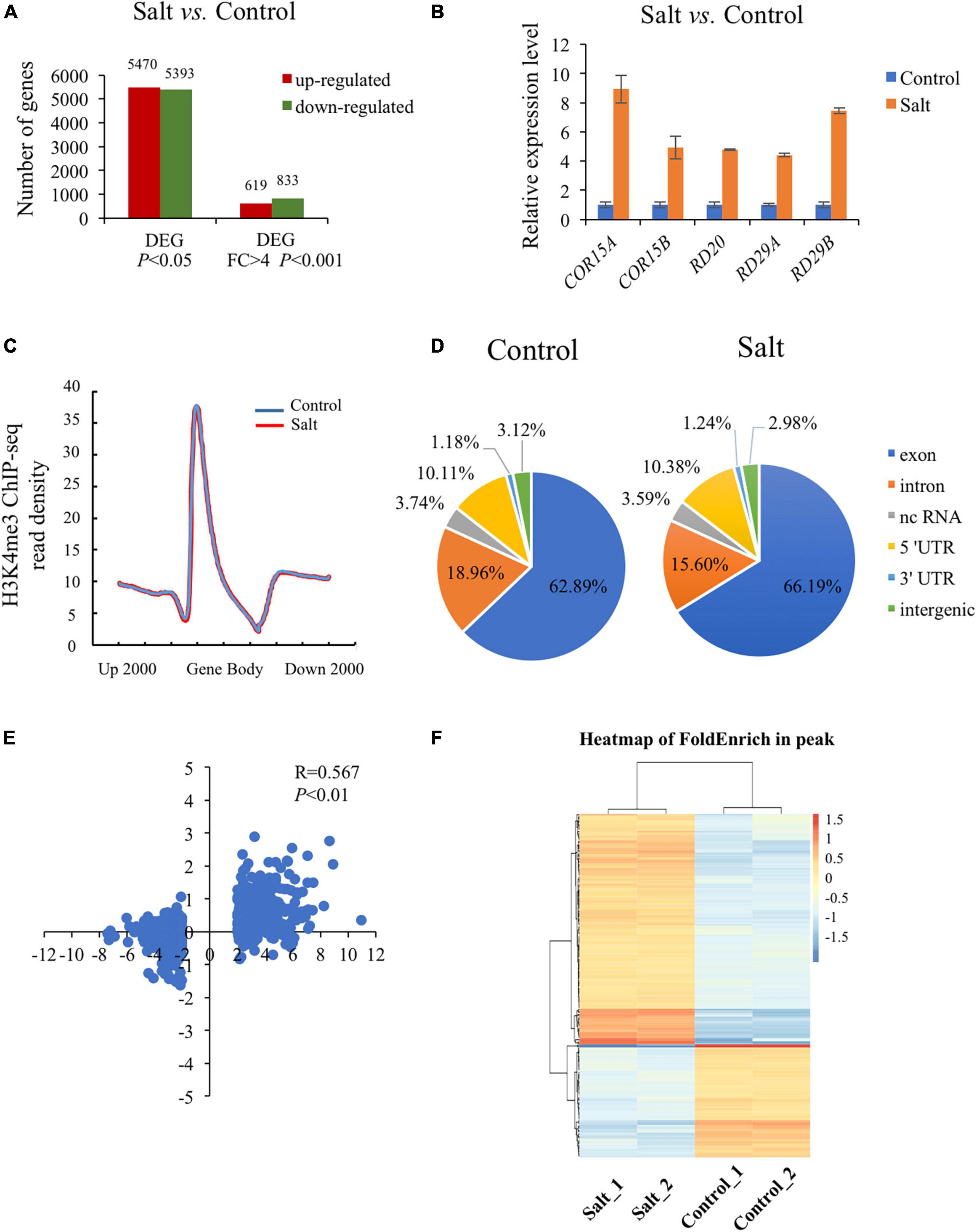
Figure 1. Global analysis of dynamic changes of H3K4me3 and mRNA levels during the plant response to salt stress. (A) Differently expressed genes in the wild type under salt condition compared with normal condition. The Y-axis represents the number of upregulated genes or downregulated genes by using the criteria P < 0.05 or fold change > 4, P < 0.001. (B) The mRNA level of 5 well known up-regulated genes which were strongly induced by salt stress. The data was calculated by log2(FPKM) and wild type under normal condition were set to 1. (C) Metaplot of the H3K4me3 distribution on genes in Col-0 under normal and salt stress conditions. Gene body in the X-axis represents all genes in the genome. Up 2000 represents 2000 bp upstream of Transcriptional Start Site (TSS) and down 2000 represents 2000 bp downstream of Transcriptional Terminal Site (TTS), respectively. The Y-axis represents read density of H3K4me3. (D) Genomic distribution of H3K4me3 peaks in Col-0 under normal and salt conditions. The definition of each region is described on the right. (E) Scatter plot of H3K4me3 and transcription changes of dysregulated genes in salt vs. normal (fold change > 4, P < 0.001). The horizontal axis represents the log2(fold change) of mRNA expression level. The vertical axis represents the log2(fold change) of H3K4me3 level. Pearson correlation indicates a correlation between the H3K4me3 and transcript dynamic changes under salt stress (R = 0.567, P < 0.01). (F) Heatmap shows the H3K4me3 enrichment of different peaks associated with genes under normal and salt condition (P < 0.05). The genes (rows) were subjected to hierarchical clustering. The color bar on the right indicates the Z-score.
Increasing evidence has shown that transcriptional regulation directed by H3K4me3 is very important for environmental responses in plants (van Dijk et al., 2010; Huang et al., 2019; Foroozani et al., 2021). To reveal the possible regulatory role of H3K4me3 methylation in the plant response to salt stress, ChIP-seq was performed by using the wild type seedlings under normal and salt stress (the same conditions as the RNA-seq), each with two biological replicates. The results revealed that 21894 H3K4me3 methylation peaks in Col-0 samples under normal condition (Supplementary Table 3), while 24141 H3K4me3 peaks under salt stress treatment (Supplementary Table 4). The distribution of H3K4me3 was similar between normal and salt-treated seedlings, with the most enrichment occurring at the gene body (Figure 1C). In detail, most H3K4me3 amounts were found at exons (63–66%), followed by introns (16–19%) and 5′ untranslated regions (5′ UTR) (10%). Lower amounts of H3K4me3 were found at non-coding RNA (3.4%), intergenic region (3%) and 3′ UTR (1%) (Figure 1D), which was consistent with the previous data (Zhang et al., 2009; Huang et al., 2019). To determine whether/how salt stress-altered gene expression correlated with changes in H3K4me3, we examined the H3K4me3 changes for the 1452 genes with altered transcript abundance (|fold change| > 4.0, P < 0.001) (Supplementary Table 5). The plot analysis revealed a correlation between the H3K4me3 and transcript dynamic changes under salt stress (Pearson correlation coefficient R = 0.567, P < 0.01) (Figure 1E), indicating that H3K4me3 dynamics is associated with the transcript level of many genes involved in salt stress response. Totals of 2591 and 1905 genes were differentially hypermethylated and hypomethylated between salt and normal conditions (P < 0.05), respectively (Figure 1F and Supplementary Table 6). Gene ontology (GO) enrichment analysis revealed that the H3K4me3 changed genes under salt stress were categorized into ‘response to water deprivation,’ ‘response to abiotic stimulus,’ ‘response to abscisic acid,’ and ‘response to chemical’ (Figure 2A). In addition, the up-regulated salt-responsive genes (i.e., COR15A, COR15B, RD20, RD29A, RD29B) were found hypermethylated under salt stress condition by using the Integrated Genome Browser (Figure 2B). These results suggest that the H3K4me3 change is associated with the transcriptional change of the salt responsive genes during the stress process.
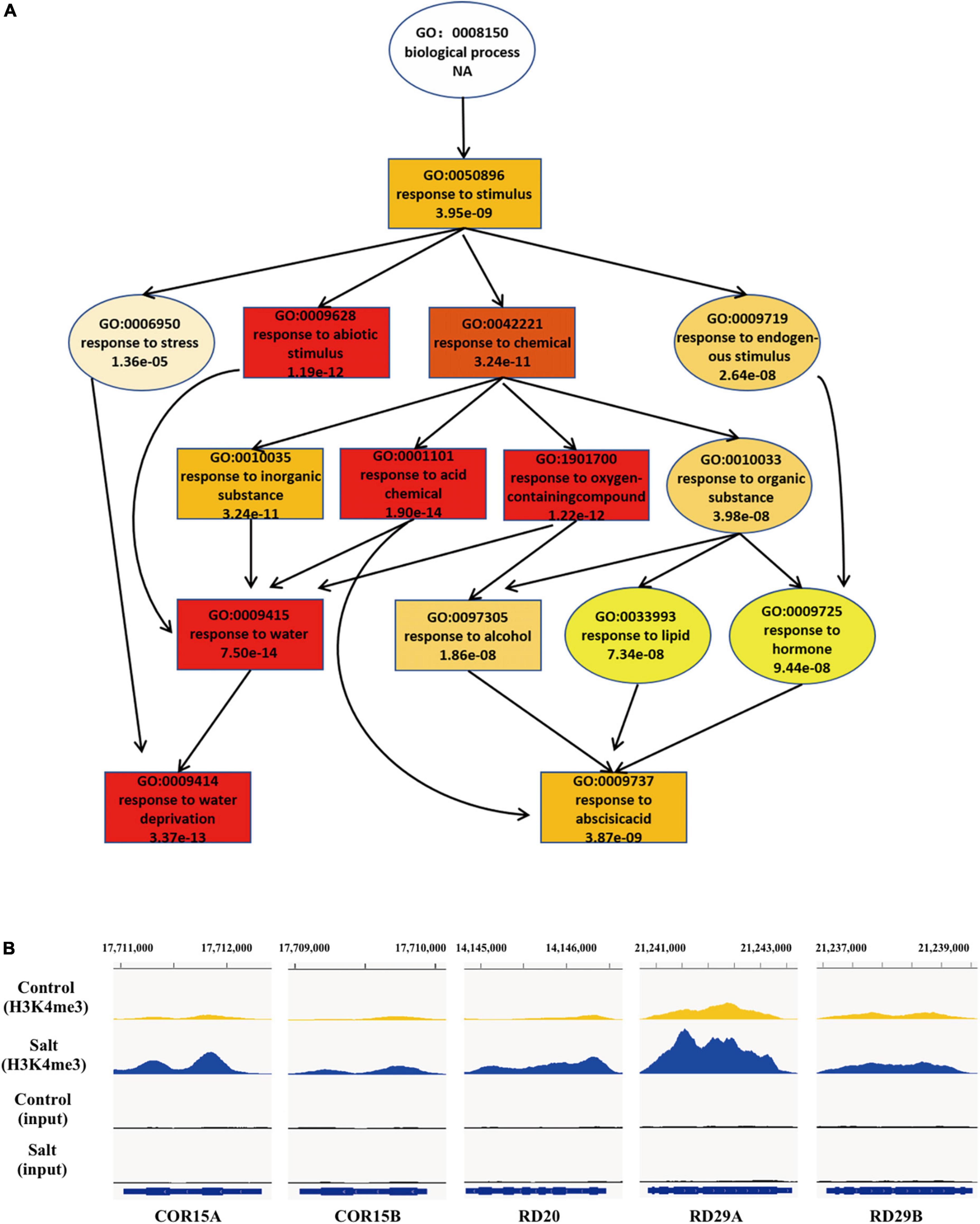
Figure 2. During the response to salt stress the H3K4me3 changes are associated with many of the genes involved in stress response. (A) A tree structure graph of the major significantly enriched GO terms of genes associated with different peaks (P < 0.05) of H3K4me3 between normal and salt conditions. GO terms are represented as boxes containing detailed description, including GO accession, description and P-value. (B) Integrative Genome Viewer (IGV) of H3K4me3 modifications at five representative salt-induced genes in Figure 1B under normal and salt-stressed conditions.
We have shown that gain-of-function mutants (jmj15-1 and jmj15-2) exhibited increased tolerance to salinity stress, whereas one allele of loss-of-function mutant (jmj15-3) exhibited increased sensitivity to salinity stress (Shen et al., 2014). To further confirm the response of jmj15 loss-of-function mutant to salt stress, we obtained another mutant allele jmj15-4. In jmj15-4, T-DNA was inserted in the 5th intron of JMJ15 (Figure 3A). RT-qPCR analysis revealed that the JMJ15 transcripts were abolished in both jmj15-3 and jmj15-4 alleles (Figure 3B). Both loss-of-function lines together with the wild type (Col-0) were tested for germination rates on normal MS media and MS supplemented with different salt concentrations. The germination rates were scored at different time points after 2 days. Seeds from three various harvests were tested. Under normal conditions, the germination rates of wild type and jmj15 loss-of-function mutants were more than 95%. When treated with salt, the germination rate of jmj15 loss-of-function mutants were lower than the wild type at different indicated salt concentration and time points (Figures 3C,D). Root elongation lengths were also measured to analyze the sensitivity of plants to the salt stress. The results revealed that after 5 days under normal condition, the root lengths of loss-of-function jmj15 and wild type plants were similar. However, the root lengths of jmj15 mutants were shorter than that of wild type after 5 days under various concentration of NaCl treatment (Figures 3E,F). Together, these results suggest that JMJ15 regulates the response to salt stress in Arabidopsis.
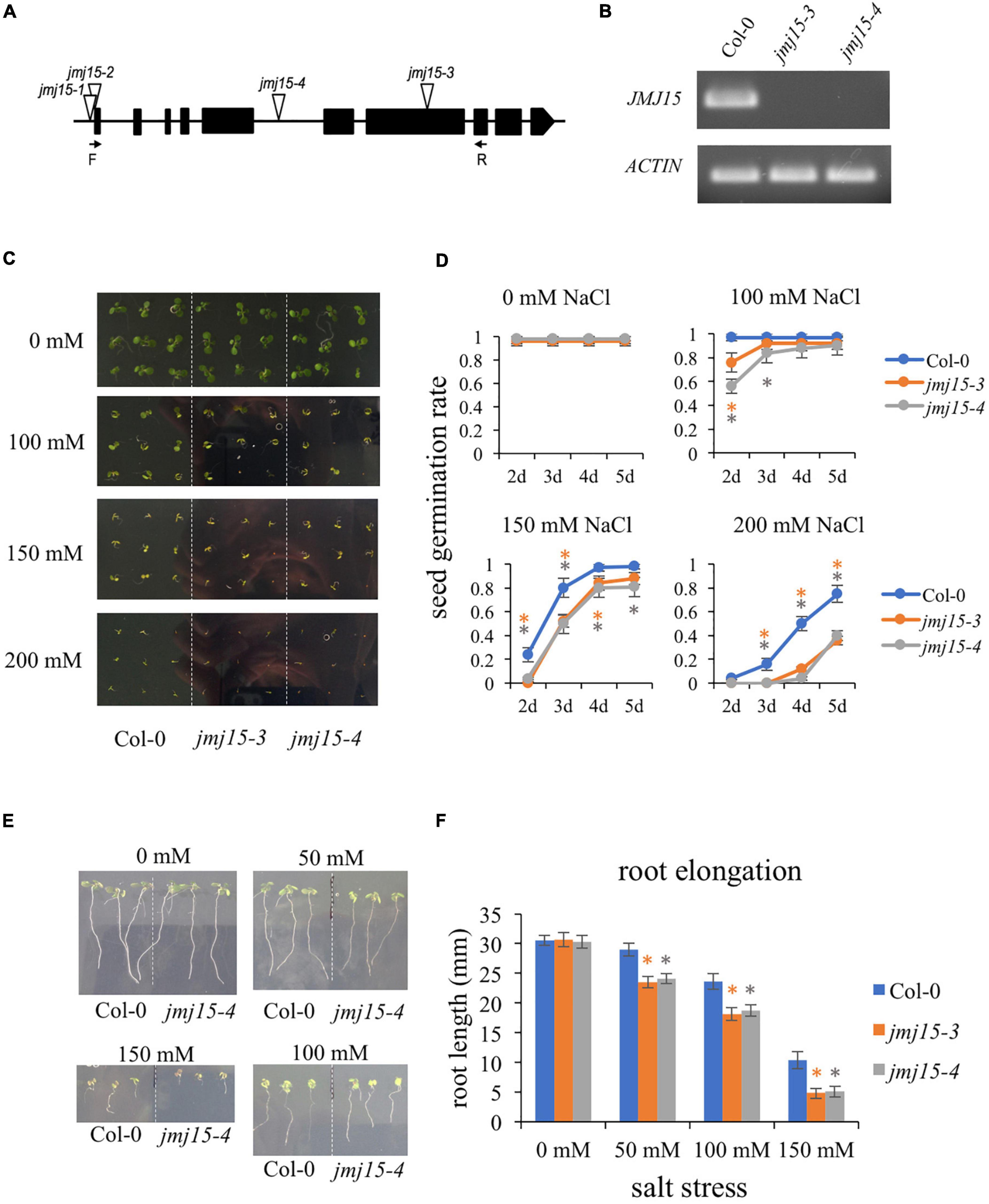
Figure 3. Loss-of-function of JMJ15 in Arabidopsis thaliana are hypersensitive to salt stress. (A) Schematic diagram of the JMJ15 gene. Exons are represented by black boxes. Triangles indicate T-DNA insertions. The primer positions of forward (F) and reverse (R) are indicated by arrows. (B) Reverse transcription polymerase chain reaction (RT-PCR) analysis of transcripts of Col-0, jmj15-3 and jmj15-4 by using the indicated primers. ACTIN2 was used as an internal control. (C,D) Measurement of seed germination rates. (C) Images of the wild type, jmj15-3 and jmj15-4 seeds germinated on 1/2 MS medium supplemented with or without indicated concentrations of NaCl for 5 days. (D) Quantification of seed germination rates of genotypes shown in (C) at different time points. Data represent means of ± SD (n = 3, three biological replicates were performed with at least 100 seeds per genotype in each replicate). (E,F) Measurement of root elongation. (E) Wild-type and jmj15 seedlings were grown on vertical 1/2 MS plate for 4 days and then transfered onto vertical 1/2 MS medium supplemented with or without different concentration of NaCl for 5 days. (F) The primary root length was measured after 5 days with different indicated concentrations of NaCl. Data represent means of ± SD (n = 3, three biological replicates were performed with at least 50 seedlings per genotype in each replicate). Asterisks indicate where the difference between Col-0 and mutant is statistically significant (Student’s t-test, *P < 0.05).
The developmental phenotype between jmj15-4 and wild type was highly significant different under salt stress (Figure 3). To explore the genome-wide transcriptional landscape directed by JMJ15, we conducted RNA-seq analysis of jmj15 loss-of-mutation (jmj15-4) and Col-0 under normal and salt stress conditions, each with three replicates. The total of 897 and 1852 DEGs were identified in jmj15-4 compared with Col-0 under normal and salt stress conditions, respectively (P < 0.05) (Figure 4A). Among 897 DEGs in jmj15-4 under normal condition, 393 were upregulated (URGs) and 504 were downregulated (DRGs) in jmj15-4 (Figure 4A and Supplementary Table 7), while among 1852 DEGs under salt stress, 1020 were URGs and 832 were DRGs in jmj15-4 (Figure 4A and Supplementary Table 8), suggesting that JMJ15 may mainly play a role as a transcriptional repressor during the salt responsive process. GO enrichment revealed that the URGs in jmj15/Col-0 under salt stress were enriched in response to stress, peptide metabolic process, flavonoid metabolic process, pigment synthetic process, and the DRGs were enriched in response to stress, response to organic substance, response to chemical stimulus, systematic acquired resistance (Figure 4B). URGs in jmj15/Col-0 under normal condition were enriched in response to stress, but no GO enrichment for DRGs (Figure 4B). These results suggest that loss of JMJ15 may mostly affect metabolism and the responsiveness to environmental stimuli of seedlings which is consistent with the previous result that overexpression of JMJ15 preferentially represses the stress regulatory genes (Shen et al., 2014).
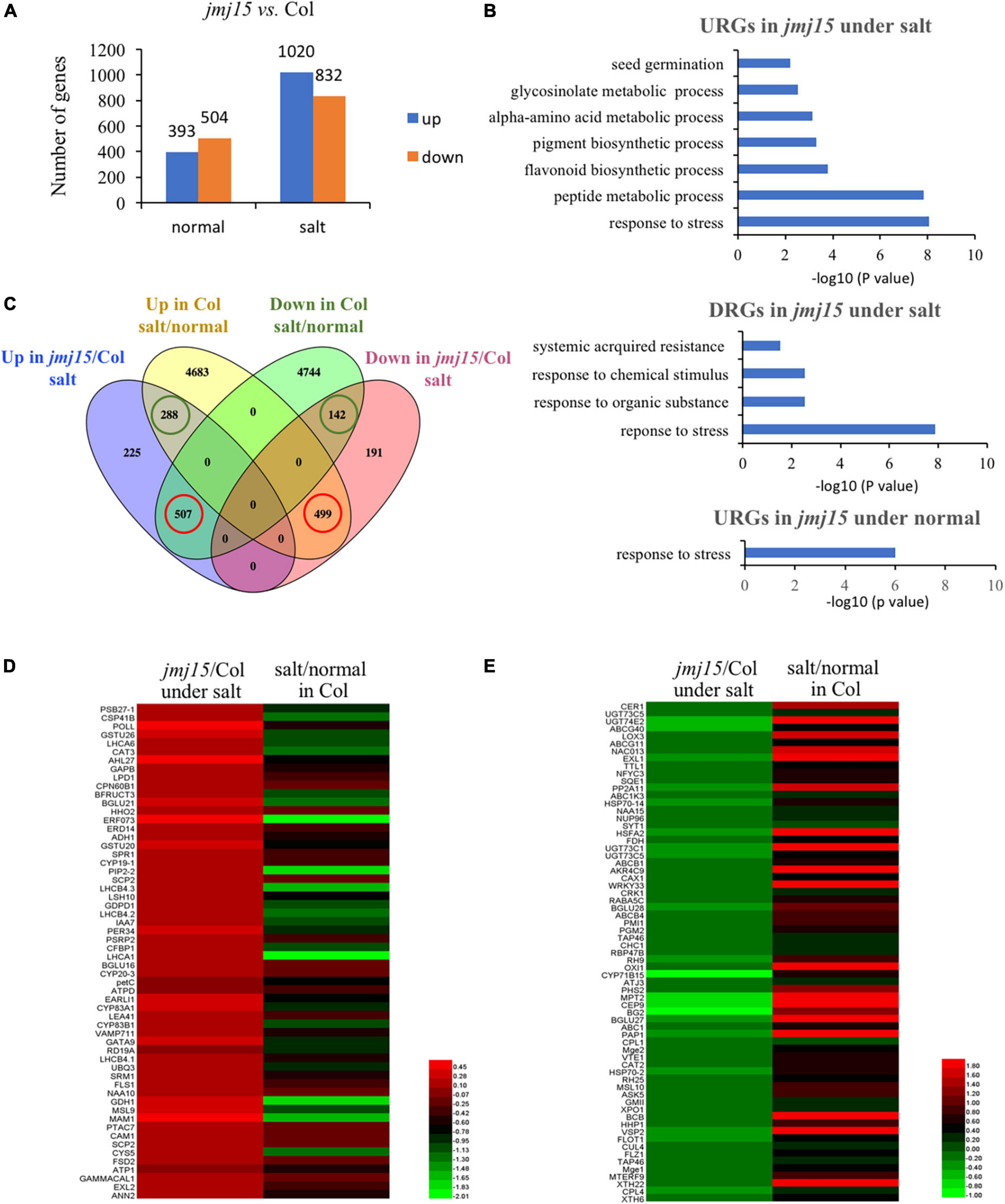
Figure 4. Transcriptomic analysis of jmj15 loss-of-function mutants compared to Col-0 under normal and salt-stressed conditions. (A) Differently expressed genes in the wild type and jmj15 mutations under normal and salt conditions. The Y-axis represents the number of upregulated genes or downregulated genes by using the criteria P < 0.05. (B) GO term enrichment of the upregulated genes (URGs) and downregulated genes (DRGs) in jmj15 mutants at the salt and normal conditions. (C) Venn diagram of comparison of DEGs in jmj15-4 (relative to Col-0 under salt condition) and DEGs in Col-0 (under salt relative to normal condition). The green cycles represent the number of genes that were up-regulated or down-regulated both in jmj15/Col-0 under salt condition and Col-0 under salt/normal. The red cycles represent the number of genes that were up-regulated in jmj15/Col-0 under salt condition but down-regulated in Col-0 under salt/normal, or down-regulated in jmj15/Col-0 under salt condition but up-regulated in Col-0 under salt/normal. (D,E) Heatmaps show the expression of the misregulated stress-related genes (GO: 0006950) in jmj15/Col-0 and salt/normal (red cycles in C). (D) Shows URGs in jmj15/Col-0 and DRGs under salt/normal; (E) shows DRGs in jmj15/Col-0 and URGs under salt/normal.
To investigate how the jmj15 mutation affects the salt responsive gene expression program, we compared DEGs in jmj15 mutation versus Col-0 under salt treatment (jmj15/Col-0 under salt) with that of Col-0 under salt versus normal condition (salt/normal in Col-0). Interestingly, we found that (507/1020, 49.7%) upregulated and (499/832, 55.6%) downregulated DEGs in jmj15/Col-0 under salt stress overlapped with the downregulated and upregulated DEGs in Col-0 under salt/normal respectively (Figure 4C red cycle and Supplementary Table 9). However, the overlap was much less between the upregulated (288/1020, 28.2%) and downregulated (142/832, 17.1%) DEGs in jmj15/Col-0 under salt stress with the upregulated and downregulated DEGs in Col-0 under salt/normal (Figure 4C green cycle). These data indicated that jmj15 mutation impaired the salt responsive gene expression program, that may explain the salt-stress-sensitive phenotype in jmj15 loss-of-function mutation. Further investigation of these 507 genes that were upregulated in jmj15/Col-0 under salt but downregulated in Col-0 under salt/normal found some key components of plant tolerance to stress, such as PRXIIF (AT3G06050), ERF73 (AT1G72360), PIP2B (AT2G37170), CAM1 (AT5G37780), LTP5 (AT3G51600), PDF1.2 (AT5G44420), PDF1.2B (AT2G26020), PDF1.3 (AT2G26010), LEA41/DI21 (AT4G15910), SAP18 (AT2G45640), ERD14 (AT1G76180), SESA (AT4G27170), EARLI1 (AT4G12480), GSTF9 (AT2G30860), GSTU26 (AT1G17190), GSTU20 (AT1G78370) (Figure 4D and Supplementary Table 9). The result suggested that loss of JMJ15 derepressed the expression of some stress-responsive genes, which was inhibited during the salt treatment. Similarly, we found some stress related genes such as CAT2 (AT1G58030), XTH6 (AT5G65730), XTH22/TCH4 (AT5G57560), HSFA2 (AT2G26150), NRT2.6 (AT3G45060), HSP70-2 (AT5G02490), COR15A (AT2G42540), COR15B (AT2G42530), NAC13 (AT1G32870), VSP2 (AT5G24770) that were upregulated in Col-0 under salt/normal but downregulated in jmj15/Col-0 under salt stress (Figure 4E and Supplementary Table 9). Together, the analysis suggested that JMJ15 was required for both gene activation and repression of the salt-responsive gene expression program.
To investigate whether JMJ15 is involved in the salt stress response by demethylating H3K4me3, we examined H3K4me3 patterns in the jmj15 loss-of-function mutants under normal and salt-stressed conditions by ChIP-seq. The result revealed that 21964 H3K4me3 methylation peaks in jmj15 mutation under normal condition (Supplementary Table 10), while 24273 H3K4me3 methylation peaks under salt stress treatment (Supplementary Table 11). The distribution pattern is similar in jmj15 to WT under both conditions (Figures 1D, 5A). However, totals of 722 and 1763 genes were differentially hypermethylated and hypomethylated in the jmj15 loss-of-function mutants under salt stress, respectively (P < 0.05) (Supplementary Table 12). Scatterplot analysis of increased expressed genes in jmj15/Col-0 under salt showed that most transcriptionally URGs were hypermethylated (Figure 5B and Supplementary Table 13). GO enrichment analysis of these hypermethylated and URGs in jmj15 under salt was the most enriched in response to stimulus (Figure 5C). By using Integrated Genome Browser, the H3K4me3 level was increased for some stimulus responsive genes such as EARLI (AT4G12480), DI21 (AT4G15910), ERD14 (AT1G76180) and LTP2 (AT2G38530) (Figure 5D and Supplementary Table 13), suggesting that the increased transcription of these stress related genes may be due to decreased demethylation activity of H3K4me3 in the jmj15 mutants. Of note, we found some transcriptional factors that both the transcription and H3K4me3 levels increased in jmj15 knockout mutations under salt condition such as some stress-responsive WRKY genes and ethylene-responsive-element binding factor genes (Supplementary Table 14), consisting with our previous data that overexpression of JMJ15 preferentially represses the stress regulatory genes, especially some transcription factors.
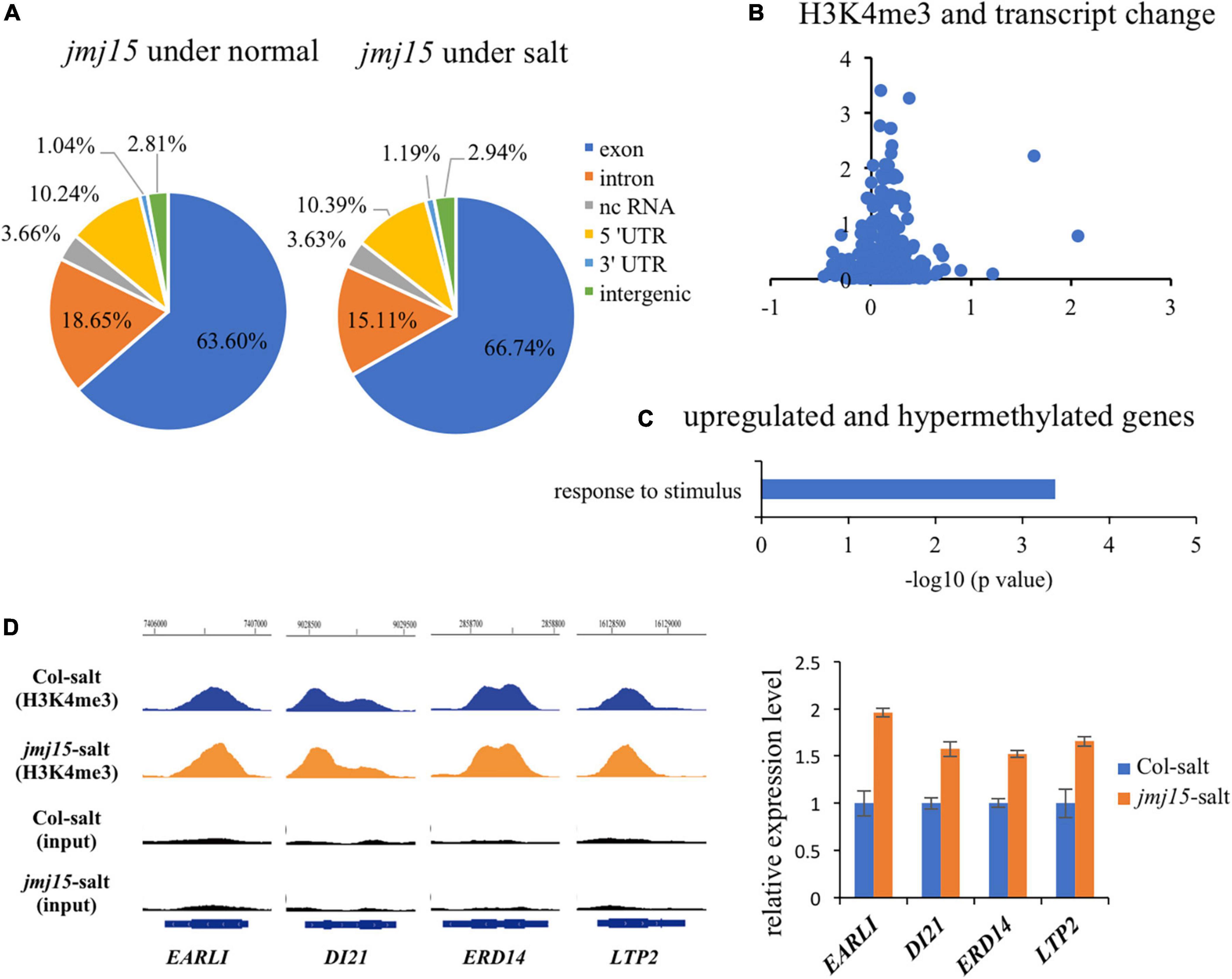
Figure 5. JMJ15 affects H3K4me3 levels at many stress-related genes under salt stress. (A) Genomic distribution of H3K4me3 peaks in jmj15-4 under normal and salt conditions. (B) Scatterplot shows that most of up-regulated genes of jmj15 mutation under salt stress were H3K4me3 hypermethylated. The x-axis represents the log2(fold change) of H3K4me3 level in jmj15 compared to Col-0 under salt stress. The y-axis represents the log2(fold change) of mRNA level in jmj15 compared to Col-0 under salt stress. (C) GO term enrichment of the genes upregulated and H3K4me3 hypermethylated in jmj15 mutants compared to Col-0 under salt stress. (D) H3K4me3 and mRNA level of selective stress-related genes in jmj15 mutants under salt stress. Left, IGV showing H3K4me3 levels of stress-related genes in jmj15 mutants under salt stress. Right, the transcript level in jmj15 mutants under salt stress. The data was calculated by log2(FPKM) and wild type were set to 1.
Interestingly, among these hypermethylated and URGs in jmj15 under salt stress, we found two known transcription factors, WRKY46 and WRKY70, that function as negative regulators in abiotic stress signaling in plants (Li et al., 2013; Ding et al., 2015; Chen et al., 2017). WRKY46 modulates the lateral root development in salt stress condition via regulation of ABA signaling and auxin homeostasis (Ding et al., 2015). WRKY70⋅modulates osmotic stress response by regulating stomatal aperture in Arabidopsis (Li et al., 2013). Meanwhile, WRKY70 was shown to be a direct target of ATX1 and positively regulated by ATX1-generated H3K4me3 (Alvarez-Venegas et al., 2007). Therefore, we hypothesized that JMJ15 might be involved in salt stress responses by modulating the H3K4me3 level of WRKY46 and WRKY70 genes. First, we examined the transcript level of these two WRKY genes in the jmj15 loss-of-function mutants (jmj15-3 and jmj15-4), jmj15 gain-of-function mutants (jmj15-1 and jmj15-2) and wild type (Col-0) under normal and salt-stressed conditions. The result showed that the transcript level of WRKY46 and WRKY70 was induced in wild type by salt stress. In jmj15 loss-of-function mutants, the transcript level of these two WRKY genes was enhanced compared to wild type under salt treatment, whereas the transcript level was obviously decreased in jmj15 gain-of-function mutations compared to wild type under salt treatment (Figure 6A). However, no discernible alterations were detected in jmj15 loss-of-function mutants under normal condition, but the transcript level of two WRKY genes were declined in jmj15 gain-of-function mutants under normal condition (Figure 6A), which was consistent with our previous result that overexpression of JMJ15 inhibited the expression of some transcription factors including some WRKY genes under normal condition (Shen et al., 2014). To confirm the role of JMJ15 in regulating chromatin status at the loci of WRKY46 and WRKY70 during the salt responsive process, we performed ChIP-qPCR analysis of the jmj15 loss-of-function mutants and jmj15 gain-of-function mutants (Figure 6B). The results showed that H3K4me3 level of WRKY46 and WRKY70 were obviously increased in jmj15-3 and jmj15-4 compared with wild type under salt stress, but decreased in jmj15-1 and jmj15-2 compared to wild type under normal and salt stress conditions (Figure 6C), suggesting that JMJ15 regulates the transcript levels of WRKY46 and WRKY70 genes by modulating their H3K4me3 levels.
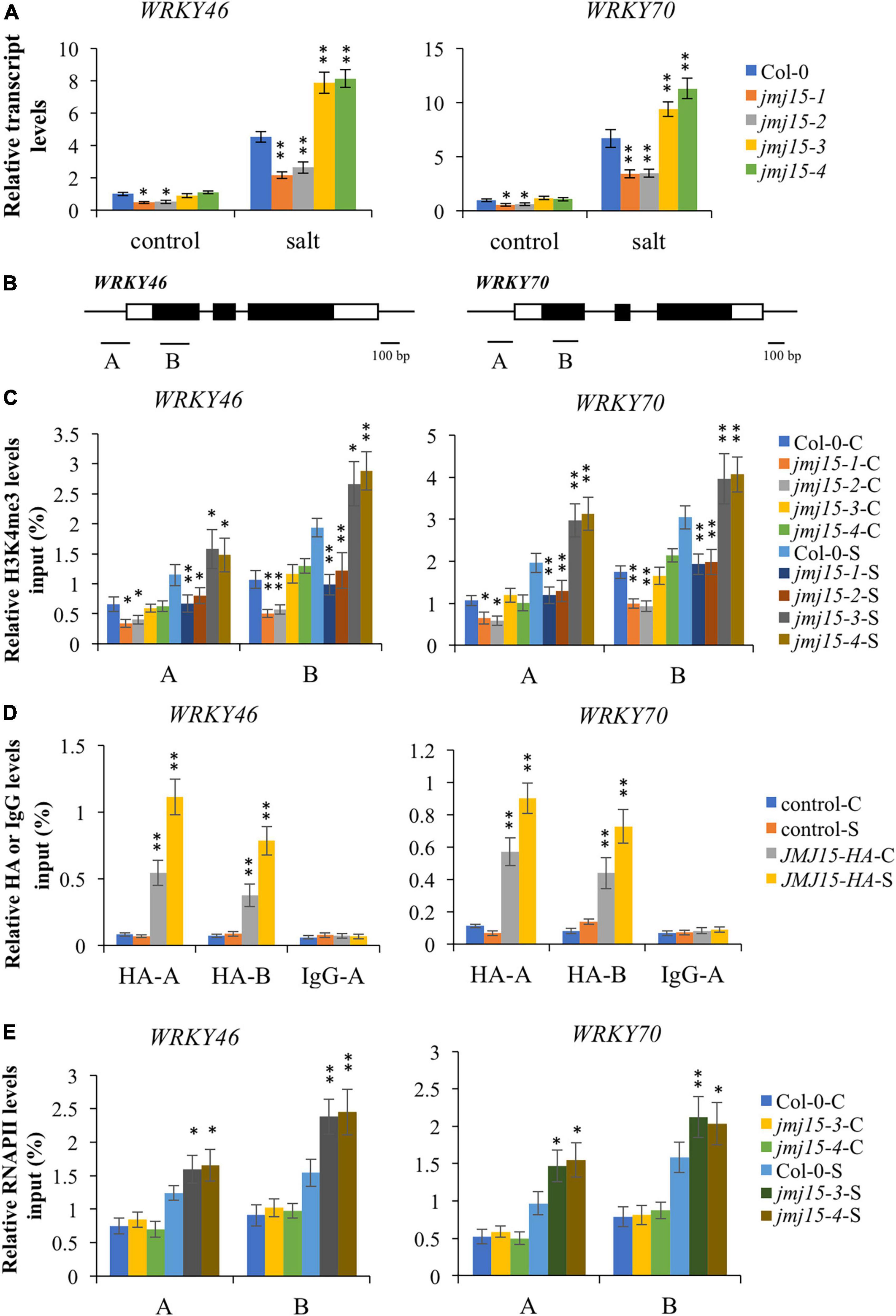
Figure 6. Transcript and chromatin changes at the WRKY46 and WRKY70 loci in jmj15 mutations. (A) RT-qPCR analysis of the transcript level of WRKY46 and WRKY70 genes in jmj15 mutants compared to Col-0 under normal and salt conditions. ACTIN2 was used as an internal control to normalize the gene expression level. The wild type under normal condition were set to 1. Bars are means ± SD of data from three biological replicates. *P < 0.05 and **P < 0.001, Student’s t-test. (B) Schematic diagram of WRKY46 and WRKY70. Regions A and B of WRKY genes were used for ChIP-qPCR analysis. Black bar represents 100 bp. (C) ChIP-qPCR analysis of H3K4me3 of WRKY46 and WRKY70 genes in Col-0 and jmj15 mutants under control condition (C) and salt condition (S). Results are presented as percentage of input. Bars are means ± SD of data from three biological replicates. *P < 0.05 and **P < 0.001, Student’s t-test. (D) JMJ15-HA ChIP-qPCR was performed by using HA antibody to determine the occupancy of JMJ15-HA at the WRKY46 and WRKY70 loci under control condition (C) and salt condition (S). IgG was used as a negative control. Results are presented as percentage of input. Bars are means ± SD from three biological replicates. *P < 0.05 and **P < 0.001, Student’s t-test. (E) ChIP-qPCR analysis of occupancy of RNA polymerase II (RNAPII) at the WRKY46 and WRKY70 loci in Col-0 and jmj15 loss-of-function mutants under control condition (C) and salt stress condition (S). Results are presented as percentage of input. Bars are means ± SD from three biological replicates. *P < 0.05 and **P < 0.001, Student’s t-test.
To investigate whether JMJ15 is directly associated with WRKY46 and WRKY70 genes, we analyzed the 35S-JMJ15-HA plants by ChIP-qPCR using anti-HA antibody (Shen et al., 2014). The analysis revealed that JMJ15-HA was enriched in WRKY46 and WRKY70 relative to the negative control (IgG), and JMJ15-HA binding levels were higher at the promoter and gene body under the salt stress compared to normal condition (Figure 6D). The results suggest that JMJ15 is recruited to the WRKY46 and WRKY70 genes and regulates their expression during the salt stress process. It has been established that H3K4me3 levels have a positive correlation with transcription rates and occupancy of activated RNA polymerase II (RNAPII) (Ardehali et al., 2011). We then tested whether the RNAPII occupancy at WRKY46 and WRKY70 was affected by JMJ15 following salt treatment. The results revealed that binding of RNAPII to WRKY46 and WRKY70 were increased in jmj15 loss-of-function mutants compared to wild type under salt stress (Figure 6E). Taken together, these results indicate that JMJ15 modulates H3K4me3 levels at the WRKY46 and WRKY70 loci and thus in turn affects RNAPII occupancy, which is essential for gene expression.
H3K4me3 has been implicated in the regulation of a number of environmental responses, such as drought stress, heat stress, submergence stress, and salt stress. For example, H3K4me3 abundance along with transcript levels of the inducible drought marker genes (RD29A, RD29B, RD20, and RAP2.4) were increased in response to drought (Kim et al., 2008). The genome-wide studies also showed that many genes up- or down-regulated by dehydration exhibited the similar increased or decreased H3K4me3 abundance, respectively (van Dijk et al., 2010; Zong et al., 2013). ATX1, H3K4me3 methyltransferase, was reported to involve in drought stress by affecting biosynthesis of ABA resulting from regulating H3K4me3 and transcript of NCED3 (Ding et al., 2011). Investigation of a number of stress-responsive genes in Arabidopsis has shown that several chromatin marks impact the responsiveness of salt-induced genes. For instance, salt stress led to enrichment of H3K4me3 and H3K9K14 acetylation, but decreased H3K9me2 at a subset of salt stress-responsive loci (Chen et al., 2010). Histone deacetylase 6 (HDA6) and HDA9 led to increased H3 acetylation at salt-inducible genes in hda6 mutants compared to wild type (Chen et al., 2010; Zheng et al., 2016). Interestingly, mutation of HDA6 also abolished H3K4me3 enrichment of ABA-induced loci (Chen et al., 2010). A recent study showed that salt stress altered the distribution of H3K4me3 in a tissue specific manner. In response to salt, rice seedlings exhibited H3K4me3 decreases and increases in exons and introns respectively, whereas only exons decrease of H3K4me3 in roots (Zheng et al., 2019). In our study, the distribution pattern of H3K4me3 was similar between normal and salt-treated seedlings. However, when compared the H3K4me3 level and transcript dynamic abundance for some transcript changed genes (| fold change| > 4.0), there was a correlation between the H3K4me3 and transcript dynamic changes under salt stress (Figure 1E). In addition, the H3K4me3 peak difference associated genes under salt stress were enriched in response to stress (Figure 2). These results indicate that H3K4me3 dynamics is associated with the transcript level of many genes involved in salt stress response.
In humans, KDM5/JARID group proteins are active histone H3K4me1/2/3 demethylases (Lu et al., 2008; Sun and Zhou, 2008). In Arabidopsis, six proteins (JMJ14-19) have been identified as members of this group based on phylogenetic analysis. Among these, JMJ14, JMJ15, JMJ16, JMJ18 harbor JmjN, JmjC, C5HC2, FY-rich N-terminus (FYRN), and FY-rich C-terminus (FYRC) domains. By contrast, JMJ19 harbors only JmjN, JmjC, and C5HC2 domains, whereas JMJ17 harbors JmjC, zf-C5HC2, PLU-1, and PHD domains (Lu et al., 2008). JMJ14, JMJ15, and JMJ18 play important roles in regulating flowering by reducing H3K4me3 abundance at various flowering regulatory genes. In addition to regulating the transition to flowering, Arabidopsis demethylases also play roles in RNA silencing (Searle et al., 2010); circadian clock (Song et al., 2019); defense (Li et al., 2020); senescence (Liu et al., 2019); de-etiolation process (Islam et al., 2021), and dehydration stress (Huang et al., 2019). JMJ17 played a role in dehydration stress by directly binding to OST1 and demethylating H3K4me3 levels (Huang et al., 2019). JMJ17 also regulates ABA responsive genes by interacting with WRKY40 (Wang et al., 2021). Our data showed that JMJ15 involved in salt stress by directly binding to the chromatin and regulating the expression of WRKY46 and WRKY70 genes. It seems that JMJ17 did not function in salt stress response as jmj17/jmj15 double mutations showed the similar salt stress response phenotype as jmj15 single mutant while jmj17 single mutant did not show discernible phenotype of salt stress response compared to wild type (Huang et al., 2019). These results indicate that these two KDM5/JARID histone demethylases function in different physiological processes without functional redundancy, which might be explained by their existing different domains and various target genes.
WRKY transcription factors, as a large family of plant transcription factors, participate in a variety of biological processes, including root growth, the quality of blossom clusters, senescence of leaf, fruit maturation, resistance to pathogens, abiotic stress response (Chen et al., 2017; Viana et al., 2018; Wang et al., 2020; de Bont et al., 2022; Goyal et al., 2022). WRKY gene family members were categorized into three groups (Rushton et al., 2010). WRKY46 and WRKY70 belonged to the group III with one WRKY domain and CCHC zinc finger motif, which played critical roles in the regulation of biotic stress response (Li et al., 2006; Hu et al., 2012). Furthermore, WRKY46 and WRKY70 acted as essential regulators in abiotic stress response which had also been demonstrated (Li et al., 2013; Ding et al., 2014; Chen et al., 2017). WRKY46 was found to negatively regulate plant responses to abiotic stress as the overexpression of WRKY46 resulted in hypersensitivity to drought and salt stress with a higher rate of water loss (Ding et al., 2014). Similarly, WRKY70 functions as negative regulator in drought stress and osmotic stress response by affecting water retention and stomatal conductance (Li et al., 2013; Chen et al., 2017). In our study, we found that the expression of WRKY46 and WRKY70 were negatively regulated by JMJ15 under salt stress (Figure 6A), which was consistent with the hypersensitivity of JMJ15 loss-of-function mutants to salt stress response. Furthermore, we found that JMJ15 bound to the chromatin of WRKY46 and WRKY70, and the enrichment was slightly induced under salt stress treatment (Figure 6D). Notably, overexpression of JMJ15 led to decrease the transcript levels of WRKY genes as well as H3K4me3 levels at these loci. According to publicly available microarray data3 (Winter et al., 2007), the expression of JMJ15 was low but rapidly induced by salt treatment (0.5–1 h), while WRKY46 and WRKY70 were induced at later time (3–6 h) (Supplementary Figure 1). These data suggest that JMJ15 may mainly repress the gene expression of WRKY46 and WRKY70 at the early stage of salt stress. Our results showed that H3K4me3 levels increased substantially at the WRKY46 and WRKY70 genes after 5 h salt treatment. This result suggests the existence of H3K4me3 methyltransferases and other chromatin remodelers that may also modulate the chromatin of WRKY46 and WRKY70 genes during the salt stress process. WRKY70 was previously reported as one target gene of H3K4me3 histone methyltransferase ATX1 (Ding et al., 2011, 2012). The SUMO E3 ligase, SIZ1 was found to modulate transcript and H3K4me3 level at the WRKY70 gene (Miura et al., 2020). However, it is not excluded other chromatin remodelers regulate H3K4me3 abundance and the transcript levels of WRKY46 and WRKY70 genes. Further studies should be conducted to understand how JMJ15 cooperates with other chromatin remodelers to regulate target genes.
The data presented in the study are deposited in the NCBI-SRA repository, accession number PRJNA822702 https://www.ncbi.nlm.nih.gov/bioproject/PRJNA822702 and PRJNA823378 https://www.ncbi.nlm.nih.gov/bioproject/PRJNA823378.
YS, YC, and LS designed the research and analyzed the data. YS, YC, SL, HL, and LS performed the experiments. YS and LS wrote the manuscript. All authors have read and approved the final manuscript.
This work was supported by National Natural Science Foundation of China (No. 31500982) and the doctoral starting funds from Xinxiang Medical University (No. 505197).
The authors thank Dao-xiu Zhou (Institute of Plant Sciences Paris-Saclay, France) for constructive discussion.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1009723/full#supplementary-material
Supplementary Figure 1 | Expression patterns of JMJ15, WRKY46, and WRKY70 under 150 mM NaCl treatment for 24 h. The expression data were obtained from a public database, eFP browser (http://bar.utoronto.ca/efp_arabidopsis/cgi-bin/efpWeb.cgi). The color bar on the right indicates the absolute expression values.
Alvarez-Venegas, R., Abdallat, A. A., Guo, M., Alfano, J. R., and Avramova, Z. (2007). Epigenetic control of a transcription factor at the cross section of two antagonistic pathways. Epigenetics 2, 106–113. doi: 10.4161/epi.2.2.4404
Anders, S., and Huber, W. (2010). Differential expression analysis for sequence count data. Genome. Biol. 11:R106. doi: 10.1186/gb-2010-11-10-r106
Ardehali, M. B., Mei, A., Zobeck, K. L., Caron, M., Lis, J. T., and Kusch, T. (2011). Drosophila Set1 is the major histone H3 lysine 4 trimethyltransferase with role in transcription. EMBO J. 30, 2817–2828. doi: 10.1038/emboj.2011.194
Chen, J., Nolan, T. M., Ye, H., Zhang, M., Tong, H., Xin, P., et al. (2017). Arabidopsis WRKY46, WRKY54, and WRKY70 transcription factors are involved in brassinosteroid-regulated plant growth and drought responses. Plant Cell 29, 1425–1439. doi: 10.1105/tpc.17.00364
Chen, L. T., Luo, M., Wang, Y. Y., and Wu, K. (2010). Involvement of arabidopsis histone deacetylase HDA6 in ABA and salt stress response. J. Exp. Bot. 61, 3345–3353. doi: 10.1093/jxb/erq154
Chen, X., Hu, Y., and Zhou, D. X. (2011). Epigenetic gene regulation by plant Jumonji group of histone demethylase. Biochim. Biophys. Acta 1809, 421–426. doi: 10.1016/j.bbagrm.2011.03.004
de Bont, L., Mu, X., Wei, B., and Han, Y. (2022). Abiotic stress-triggered oxidative challenges: Where does H2S act? J. Genet. Genom. 49, 748–755. doi: 10.1016/j.jgg.2022.02.019
Ding, Y., Avramova, Z., and Fromm, M. (2011). The Arabidopsis trithorax-like factor ATX1 functions in dehydration stress responses via ABA-dependent and ABA-independent pathways. Plant J. 66, 735–744. doi: 10.1111/j.1365-313X.2011.04534.x
Ding, Y., Fromm, M., and Avramova, Z. (2012). Multiple exposures to drought ‘train’ transcriptional responses in Arabidopsis. Nat. Commun. 3:740. doi: 10.1038/ncomms1732
Ding, Z. J., Yan, J. Y., Li, C. X., Li, G. X., Wu, Y. R., and Zheng, S. J. (2015). Transcription factor WRKY46 modulates the development of Arabidopsis lateral roots in osmotic/salt stress conditions via regulation of ABA signaling and auxin homeostasis. Plant J. 84, 56–69. doi: 10.1111/tpj.12958
Ding, Z. J., Yan, J. Y., Xu, X. Y., Yu, D. Q., Li, G. X., Zhang, S. Q., et al. (2014). Transcription factor WRKY46 regulates osmotic stress responses and stomatal movement independently in Arabidopsis. Plant J. 79, 13–27. doi: 10.1111/tpj.12538
Foroozani, M., Vandal, M. P., and Smith, A. P. (2021). H3K4 trimethylation dynamics impact diverse developmental and environmental responses in plants. Planta 253:4. doi: 10.1007/s00425-020-03520-0
Goyal, P., Devi, R., Verma, B., Hussain, S., Arora, P., Tabassum, R., et al. (2022). WRKY transcription factors: Evolution, regulation, and functional diversity in plants. Protoplasma [Epub ahead of print]. doi: 10.1007/s00709-022-01794-7
Hu, Y., Dong, Q., and Yu, D. (2012). Arabidopsis WRKY46 coordinates with WRKY70 and WRKY53 in basal resistance against pathogen Pseudomonas syringae. Plant Sci. 185-186, 288–297. doi: 10.1016/j.plantsci.2011.12.003
Hu, Y., Shen, Y., Conde, E. S. N., and Zhou, D. X. (2011). The role of histone methylation and H2A.Z occupancy during rapid activation of ethylene responsive genes. PLoS One 6:e28224. doi: 10.1371/journal.pone.0028224
Huang, S., Zhang, A., Jin, J. B., Zhao, B., Wang, T. J., Wu, Y., et al. (2019). Arabidopsis histone H3K4 demethylase JMJ17 functions in dehydration stress response. New Phytol. 223, 1372–1387. doi: 10.1111/nph.15874
Islam, M. T., Wang, L. C., Chen, I. J., Lo, K. L., and Lo, W. S. (2021). Arabidopsis JMJ17 promotes cotyledon greening during de-etiolation by repressing genes involved in tetrapyrrole biosynthesis in etiolated seedlings. New Phytol. 231, 1023–1039. doi: 10.1111/nph.17327
Jaskiewicz, M., Conrath, U., and Peterhansel, C. (2011). Chromatin modification acts as a memory for systemic acquired resistance in the plant stress response. EMBO Rep. 12, 50–55. doi: 10.1038/embor.2010.186
Jeong, J. H., Song, H. R., Ko, J. H., Jeong, Y. M., Kwon, Y. E., Seol, J. H., et al. (2009). Repression of flowering locus T chromatin by functionally redundant histone H3 lysine 4 demethylases in Arabidopsis. PLoS One 4:e8033. doi: 10.1371/journal.pone.0008033
Kim, J. M., Sasaki, T., Ueda, M., Sako, K., and Seki, M. (2015). Chromatin changes in response to drought, salinity, heat, and cold stresses in plants. Front. Plant Sci. 6:114. doi: 10.3389/fpls.2015.00114
Kim, J. M., To, T. K., Ishida, J., Matsui, A., Kimura, H., and Seki, M. (2012). Transition of chromatin status during the process of recovery from drought stress in Arabidopsis thaliana. Plant Cell Physiol. 53, 847–856. doi: 10.1093/pcp/pcs053
Kim, J. M., To, T. K., Ishida, J., Morosawa, T., Kawashima, M., Matsui, A., et al. (2008). Alterations of lysine modifications on the histone H3 N-tail under drought stress conditions in Arabidopsis thaliana. Plant Cell Physiol. 49, 1580–1588. doi: 10.1093/pcp/pcn133
Lee, B. H., and Zhu, J. K. (2009). Phenotypic analysis of Arabidopsis mutants: Root elongation under salt/hormone-induced stress. Cold Spring Harb. Protoc. 2009:dbrot4968. doi: 10.1101/pdb.prot4968
Li, B., Carey, M., and Workman, J. L. (2007). The role of chromatin during transcription. Cell 128, 707–719. doi: 10.1016/j.cell.2007.01.015
Li, B., and Dewey, C. N. (2011). RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 12:323. doi: 10.1186/1471-2105-12-323
Li, D., Liu, R., Singh, D., Yuan, X., Kachroo, P., and Raina, R. (2020). JMJ14 encoded H3K4 demethylase modulates immune responses by regulating defence gene expression and pipecolic acid levels. New Phytol. 225, 2108–2121. doi: 10.1111/nph.16270
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Li, J., Besseau, S., Toronen, P., Sipari, N., Kollist, H., Holm, L., et al. (2013). Defense-related transcription factors WRKY70 and WRKY54 modulate osmotic stress tolerance by regulating stomatal aperture in Arabidopsis. New Phytol. 200, 457–472. doi: 10.1111/nph.12378
Li, J., Brader, G., Kariola, T., and Palva, E. T. (2006). WRKY70 modulates the selection of signaling pathways in plant defense. Plant J. 46, 477–491. doi: 10.1111/j.1365-313X.2006.02712.x
Liu, P., Zhang, S., Zhou, B., Luo, X., Zhou, X. F., Cai, B., et al. (2019). The histone H3K4 demethylase JMJ16 represses leaf senescence in Arabidopsis. Plant Cell 31, 430–443. doi: 10.1105/tpc.18.00693
Liu, Y., Zhang, A., Yin, H., Meng, Q., Yu, X., Huang, S., et al. (2018). Trithorax-group proteins Arabidopsis Trithorax4 (ATX4) and ATX5 function in abscisic acid and dehydration stress responses. New Phytol. 217, 1582–1597. doi: 10.1111/nph.14933
Lu, F., Cui, X., Zhang, S., Liu, C., and Cao, X. (2010). JMJ14 is an H3K4 demethylase regulating flowering time in Arabidopsis. Cell Res. 20, 387–390. doi: 10.1038/cr.2010.27
Lu, F., Li, G., Cui, X., Liu, C., Wang, X. J., and Cao, X. (2008). Comparative analysis of JmjC domain-containing proteins reveals the potential histone demethylases in Arabidopsis and rice. J Integr. Plant Biol. 50, 886–896. doi: 10.1111/j.1744-7909.2008.00692.x
Miura, K., Renhu, N., and Suzaki, T. (2020). The PHD finger of Arabidopsis SIZ1 recognizes trimethylated histone H3K4 mediating SIZ1 function and abiotic stress response. Commun. Biol. 3:23. doi: 10.1038/s42003-019-0746-2
Mosammaparast, N., and Shi, Y. (2010). Reversal of histone methylation: Biochemical and molecular mechanisms of histone demethylases. Annu. Rev. Biochem. 79, 155–179. doi: 10.1146/annurev.biochem.78.070907.103946
Ng, D. W., Wang, T., Chandrasekharan, M. B., Aramayo, R., Kertbundit, S., and Hall, T. C. (2007). Plant SET domain-containing proteins: Structure, function and regulation. Biochim. Biophys. Acta 1769, 316–329. doi: 10.1016/j.bbaexp.2007.04.003
Rushton, P. J., Somssich, I. E., Ringler, P., and Shen, Q. J. (2010). WRKY transcription factors. Trends Plant Sci. 15, 247–258. doi: 10.1016/j.tplants.2010.02.006
Santos-Rosa, H., Schneider, R., Bannister, A. J., Sherriff, J., Bernstein, B. E., Emre, N. C., et al. (2002). Active genes are tri-methylated at K4 of histone H3. Nature 419, 407–411. doi: 10.1038/nature01080
Searle, I. R., Pontes, O., Melnyk, C. W., Smith, L. M., and Baulcombe, D. C. (2010). JMJ14, a JmjC domain protein, is required for RNA silencing and cell-to-cell movement of an RNA silencing signal in Arabidopsis. Genes Dev. 24, 986–991. doi: 10.1101/gad.579910
Shen, Y., Conde, E. S. N., Audonnet, L., Servet, C., Wei, W., and Zhou, D. X. (2014). Over-expression of histone H3K4 demethylase gene JMJ15 enhances salt tolerance in Arabidopsis. Front. Plant Sci. 5:290. doi: 10.3389/fpls.2014.00290
Shen, Y., Devic, M., Lepiniec, L., and Zhou, D. X. (2015). Chromodomain, Helicase and DNA-binding CHD1 protein, CHR5, are involved in establishing active chromatin state of seed maturation genes. Plant Biotechnol. J. 13, 811–820. doi: 10.1111/pbi.12315
Shi, Y., Lan, F., Matson, C., Mulligan, P., Whetstine, J. R., Cole, P. A., et al. (2004). Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119, 941–953. doi: 10.1016/j.cell.2004.12.012
Sokol, A., Kwiatkowska, A., Jerzmanowski, A., and Prymakowska-Bosak, M. (2007). Up-regulation of stress-inducible genes in tobacco and Arabidopsis cells in response to abiotic stresses and ABA treatment correlates with dynamic changes in histone H3 and H4 modifications. Planta 227, 245–254. doi: 10.1007/s00425-007-0612-1
Song, Q., Huang, T. Y., Yu, H. H., Ando, A., Mas, P., Ha, M., et al. (2019). Diurnal regulation of SDG2 and JMJ14 by circadian clock oscillators orchestrates histone modification rhythms in Arabidopsis. Genome Biol. 20:170. doi: 10.1186/s13059-019-1777-1
Sun, Q., and Zhou, D. X. (2008). Rice JmjC domain-containing gene JMJ706 encodes H3K9 demethylase required for floral organ development. Proc. Natl. Acad. Sci. U. S. A. 105, 13679–13684. doi: 10.1073/pnas.0805901105
Tsukada, Y., Fang, J., Erdjument-Bromage, H., Warren, M. E., Borchers, C. H., Tempst, P., et al. (2006). Histone demethylation by a family of JmjC domain-containing proteins. Nature 439, 811–816. doi: 10.1038/nature04433
van Dijk, K., Ding, Y., Malkaram, S., Riethoven, J. J., Liu, R., Yang, J., et al. (2010). Dynamic changes in genome-wide histone H3 lysine 4 methylation patterns in response to dehydration stress in Arabidopsis thaliana. BMC Plant Biol. 10:238. doi: 10.1186/1471-2229-10-238
Viana, V. E., Busanello, C., da Maia, L. C., Pegoraro, C., and Costa de Oliveira, A. (2018). Activation of rice WRKY transcription factors: An army of stress fighting soldiers? Curr. Opin. Plant Biol. 45, 268–275. doi: 10.1016/j.pbi.2018.07.007
Wang, T. J., Huang, S., Zhang, A., Guo, P., Liu, Y., Xu, C., et al. (2021). JMJ17-WRKY40 and HY5-ABI5 modules regulate the expression of ABA-responsive genes in Arabidopsis. New Phytol. 230, 567–584. doi: 10.1111/nph.17177
Wang, Y., Cui, X., Yang, B., Xu, S., Wei, X., Zhao, P., et al. (2020). WRKY55 transcription factor positively regulates leaf senescence and the defense response by modulating the transcription of genes implicated in the biosynthesis of reactive oxygen species and salicylic acid in Arabidopsis. Development 147:dev189647. doi: 10.1242/dev.189647
Winter, D., Vinegar, B., Nahal, H., Ammar, R., Wilson, G. V., and Provart, N. J. (2007). An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2:e718. doi: 10.1371/journal.pone.0000718
Yang, H., Han, Z., Cao, Y., Fan, D., Li, H., Mo, H., et al. (2012a). A companion cell-dominant and developmentally regulated H3K4 demethylase controls flowering time in Arabidopsis via the repression of FLC expression. PLoS Genet. 8:e1002664. doi: 10.1371/journal.pgen.1002664
Yang, H., Mo, H., Fan, D., Cao, Y., Cui, S., and Ma, L. (2012b). Overexpression of a histone H3K4 demethylase, JMJ15, accelerates flowering time in Arabidopsis. Plant Cell Rep. 31, 1297–1308. doi: 10.1007/s00299-012-1249-5
Yang, R., Hong, Y., Ren, Z., Tang, K., Zhang, H., Zhu, J. K., et al. (2019). A role for pickle in the regulation of cold and salt stress tolerance in Arabidopsis. Front. Plant Sci. 10:900. doi: 10.3389/fpls.2019.00900
Yang, W., Jiang, D., Jiang, J., and He, Y. (2010). A plant-specific histone H3 lysine 4 demethylase represses the floral transition in Arabidopsis. Plant J. 62, 663–673. doi: 10.1111/j.1365-313X.2010.04182.x
Yang, Z., Qiu, Q., Chen, W., Jia, B., Chen, X., Hu, H., et al. (2018). Structure of the Arabidopsis JMJ14-H3K4me3 complex provides insight into the substrate specificity of KDM5 subfamily histone demethylases. Plant Cell 30, 167–177. doi: 10.1105/tpc.17.00666
Zhang, X., Bernatavichute, Y. V., Cokus, S., Pellegrini, M., and Jacobsen, S. E. (2009). Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol. 10:R62. doi: 10.1186/gb-2009-10-6-r62
Zheng, D., Wang, L., Chen, L., Pan, X., Lin, K., Fang, Y., et al. (2019). Salt-responsive genes are differentially regulated at the chromatin levels between seedlings and roots in rice. Plant Cell Physiol. 60, 1790–1803. doi: 10.1093/pcp/pcz095
Zheng, Y., Ding, Y., Sun, X., Xie, S., Wang, D., Liu, X., et al. (2016). Histone deacetylase HDA9 negatively regulates salt and drought stress responsiveness in Arabidopsis. J. Exp. Bot. 67, 1703–1713. doi: 10.1093/jxb/erv562
Keywords: jumonji demethylase, H3K4me3, JMJ15, WRKY46, WRKY70, salt stress
Citation: Shen Y, Chi Y, Lu S, Lu H and Shi L (2022) Involvement of JMJ15 in the dynamic change of genome-wide H3K4me3 in response to salt stress. Front. Plant Sci. 13:1009723. doi: 10.3389/fpls.2022.1009723
Received: 02 August 2022; Accepted: 29 August 2022;
Published: 26 September 2022.
Edited by:
Yi Han, Anhui Agricultural University, ChinaReviewed by:
Yongfeng Hu, China Three Gorges University, ChinaCopyright © 2022 Shen, Chi, Lu, Lu and Shi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuan Shen, eXVhbi5zaGVuQHlhaG9vLmNvbQ==; Lei Shi, ZG91ZG91MjQ2QDE2My5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.