- 1Division of Agricultural Bioinformatics, ICAR-Indian Agricultural Statistics Research Institute, New Delhi, India
- 2Division of Computer Application, ICAR-Indian Agricultural Statistics Research Institute, New Delhi, India
- 3Department of Biotechnology, School of Interdisciplinary and Applied Sciences, Central University of Haryana, Mahendergarh, Haryana, India
The impact of climate change has been alarming for the crop growth. The extreme weather conditions can stress the crops and reduce the yield of major crops belonging to Poaceae family too, that sustains 50% of the world’s food calorie and 20% of protein intake. Computational approaches, such as artificial intelligence-based techniques have become the forefront of prediction-based data interpretation and plant stress responses. In this study, we proposed a novel activation function, namely, Gaussian Error Linear Unit with Sigmoid (SIELU) which was implemented in the development of a Deep Learning (DL) model along with other hyper parameters for classification of unknown abiotic stress protein sequences from crops of Poaceae family. To develop this models, data pertaining to four different abiotic stress (namely, cold, drought, heat and salinity) responsive proteins of the crops belonging to poaceae family were retrieved from public domain. It was observed that efficiency of the DL models with our proposed novel SIELU activation function outperformed the models as compared to GeLU activation function, SVM and RF with 95.11%, 80.78%, 94.97%, and 81.69% accuracy for cold, drought, heat and salinity, respectively. Also, a web-based tool, named DeepAProt (http://login1.cabgrid.res.in:5500/) was developed using flask API, along with its mobile app. This server/App will provide researchers a convenient tool, which is rapid and economical in identification of proteins for abiotic stress management in crops Poaceae family, in endeavour of higher production for food security and combating hunger, ensuring UN SDG goal 2.0.
1. Introduction
The drastic climatic changes due to global warming after the 1980s lead to significant yield loss in various crops (Lobell et al., 2011). The Poaceae family of crops, especially rice, wheat, and maize, which account for ~50% of the world’s food calories and 20% of its protein intake (Erenstein et al., 2022), are highly susceptible to abiotic stress like heat, salinity, drought, and cold (Landi et al., 2017). On the other hand, due increasing global population, which may be around 9.5 billion by 2050, the current food availability gap requires a dramatic increase in food by 2050 (Cobb et al., 2013). It is already well known that environmental stressors negatively regulate the growth and development of plants leading to substantial yield and quality losses (Boyer, 1982; Palanog et al., 2014, Gupta et al., 2021). A recent study suggests that climate change could reduce global crop yields by 3–12% by mid-century, and by 11–25% by the century’s end, under a vigorous warming scenario (Sue Wing et al., 2021).
Stresses in plants, like drought, salinity, cold, etc. are their defensive states which result from deviations from their optimal growth conditions (Jansen and Potters, 2017). These stresses lead to a loss in yield, thus affecting food security, especially in the current scenario of climate change (Rico-Chávez et al., 2022). Therefore, there is a need to conceive comprehensive strategies for trait improvement of important crops, especially of the Poaceae family, under adverse climatic conditions. Artificial intelligence (AI)- based machine learning techniques have become the forefront of prediction-based data interpretation and plant stress responses (Gill et al., 2022). Analyses of high-throughput genomic data in recent years, like, genes, transcripts, proteins, metabolites, etc., require advanced analytical methods for proper associations and interactions. The promising computational power in terms of artificial intelligence (AI) based methodologies had been a promising means for analyzing various plant stress mechanisms (Fenu and Malloci, 2021). Also, machine learning (ML) based methodologies for identifying DNA N6-methyladenine sites of plant genomes (Hasan et al., 2021), a deep-learning-based hybrid framework for identifying human RNA N5-methylcytosine sites (Hasan et al., 2022), solving classification problems in molecular data like amino acid sequence, protein sequences and structures (Cai et al., 2020; Xu et al., 2020; Gelman et al., 2021; Sridevi and Kanimozhi, 2021; Wang, 2022; Ding et al., 2022) proves the versatility of ML methodologies. The use of ML-based studies to identify, classify, and predict various stresses in plants are well reported, namely, in basil, coriander, parsley, baby-leaf, coffee, pea, and maize for water stress (Niu et al., 2021; Zahid et al., 2022), in Arabidopsis thaliana for heat, cold, salt, and drought (Kang et al., 2018), salt stress in rice (Das et al., 2020) and wheat (Moghimi et al., 2018), drought stress in Bromus inermis (Dao et al., 2021), and biotic stresses in soybean (Venal et al., 2019), etc.
Various studies have been done using ML/Deep Learning techniques to classify stress-responsive varieties in corn using deep convolutional neural networks (Ghosal et al., 2018; Khaki et al., 2019), neural networks (Etminan et al., 2019), linear mixed model (Chen et al, 2012) and CNN (An et al., 2019), etc. However, there are limited resources of deep-learning-based prediction models for the abiotic stress protein sequence of the Poaceae crop family. Therefore, we developed a deep learning approach for the classification of the abiotic stress protein sequence of this family. In addition, we developed a novel activation function, namely, sielu that has increased accuracy as compared to the existing models. The same has been applied to the stress datasets. Most of the data under study were benchmark data collected from Uniprot. Although, the DL model works well in the structure, unstructured, and complex features of the dataset, however, it requires a large dataset to train the model (Elaraby and Elmogy, 2016). It also uses different optimization techniques, weight functions, loss functions, and activation functions during model development (Wen et al., 2018; Salman and Liu, 2019). During model building, an activation function plays an important role in boosting the performance of the model as this helps in the activation or deactivation of neurons (Benvenuto and Piazza, 1992; Sarker, 2021). DL model without an activation function converges to linear regression model. Several activation functions like sigmoid, ReLU, LeakyReLU, Tanh, and Softmax have been reported in the literature (Xu et al., 2015; Hendrycks and Gimpel, 2016; Agarap, 2018; Pratiwi et al., 2020) are being used in building DL for the classification and prediction (Li et al., 2018; Armenteros et al., 2019; Bileschi et al., 2022). Some of the major limitations of these activation functions are the vanishing gradient, loss of neurons, and problems in training small datasets (Srinivasan et al., 2019).
In this study, we proposed a novel activation function, named Gaussian Error Linear Unit with Sigmoid (SIELU) to overcome issues related to the activation function. Further, we have built a DL model using the proposed activation function for the prediction of abiotic stresses, i.e. heat, drought, cold, and salinity responsive protein sequences from the crops of the Poaceae family. Also, a Web server has been developed, which can be extensively used by researchers/breeders for the development of abiotic stress resistance varieties of the crops of the Poaceae family for increasing agricultural production and productivity. In the future, there is a scope for developing different weight initialization techniques, activation functions, optimizers, etc. for more efficient classification using deep learning models.
2. Materials and methodology
2.1. Activation function
A series of studies have been carried out related to various activation functions and their performance in DL network building. The extensively used activation functions in DL models are Sigmoid, Tanh, ReLU, LeakyReLU, SoftMax, etc. (Dunn et al., 2011).
Sigmoid function: For any given input of data, the sigmoid maps to 0 or 1. If a given input goes above the predetermined threshold value, it will give output as 1, otherwise, 0, i.e., the neuron will remain deactivated. Scientifically, it has been proven that the human brain functions like the sigmoid function for differentiating and classifying objects (Pratiwi et al., 2020). Mathematically, it is expressed as:
Tanh function: It is similar to the Sigmoid function with little modification for the output and expressed mathematically as (LeCun et al., 2012):
Rectified Linear Unit (ReLU): This activation function uses stochastic gradient descent for back-propagation by adjusting the learning rate and minimizing the errors during training a model. Also, it provides a better solution without decaying the hidden layers by adjusting the learning rate and minimizing the error differentiation by removing all the negative values in back-propagation. Mathematically, ReLU can be expressed as (Agarap, 2018):
Leaky Rectified Linear Unit (LaekyReLU): It is an extension of ReLU i.e., by using some value, say σ=0.01 that makes the neuron active instead of deactivating for zero values. Mathematically, the LeakyReLU function is expressed as (Xu et al., 2015):
Softmax function: It gives the probability of each true class and is expressed as (Kanai et al., 2018):
Many other activation functions have been developed which are mainly derived from the above activation functions such as Gaussian Error Linear Unit (gelu) (Hendrycks and Gimpel, 2016), a multi-layer perceptron model with a sigmoid, tanh, conic section, and radial bases function (RBF), etc. (Karlik and Olgac, 2011; Cai et al., 2015).
2.2. Proposed Gaussian error linear unit with sigmoid activation function (SIELU) activation function
It may be noted that the Tanh activation is used in the Cumulative Distribution Function of GELU. Also, Tanh activation function is reported to perform better than sigmoid (Szandała, 2021; Ingole and Patil, 2020; Jiang et al., 2020) but takes more time. However, in the prediction of high-dimension datasets, computational time is one of the crucial factors. It has been pointed out that the sigmoid function requires less time and is computationally inexpensive by approximating its polynomial for positive outputs (Wang et al., 2020). Further, the sigmoid function is computationally easy to perform. Therefore, a thorough investigation was done to derive a novel activation function i.e., SIELU from the GELU function.
An approximation of normal distribution (q) was carried out in 1955 for the first time by (Hastings, 1955; Brophy, 1985) which was expressed as:
Hence ;
where , α0=2.30753 , α1=0.27061 , b1=0.99229 , b2=0.04481 or ,
were, q→ normal distribution , t→ time, α0=2.515517 , α1=0.802853 , α1=0.010328 , b1=1.432788 , b2=0.189269 , b2=0.001308 (Hastings., 1955).
With the advancement of technology, a more accurate approximation was introduced by estimating the standard normal deviated distribution z by (Zelen and Severo, 1964) followed by Emerson, 1979.
where, and |e(p)|<4,5×10−4 , C0=2.515517 , C1=0.802853 , C2=0.010328 , d1=1.43288 , d2=0.189269 , d3=0.001308 .
Later, in 2008, standard normal deviated distribution to approximate the function was given by Kiani and co-workers (Kiani et al., 2008) as follows:
; −∞<z<∞ where ; −∞<z<∞ .
Moreover, the approximation of Φ(x)−0.5 with absolute error< 3×10−5 (Bagby, 1995) is estimated from:
Our proposed Gaussian Error Linear Unit with Sigmoid (SiELU) was constructed by modifying the GELU function as follows:
Let where, On simplification of the equation (1):
Tanh and Sigmoid functions are mathematically defined as:
On further simplification of the equation (2),
By dividing numerator and denominator by e-x, equation (4) is changes to:
From equation (1) , f(x)=0.5x[1+tanhy] Now, equating sigmoid with tanh function and simplifying, we get:
Finally, the SiELU can be expressed as:
On simplification, we got the Gaussian Error Linear Unit with Sigmoid activation function, termed SIELU as follows:
2.3. Deep learning model with proposed activation function
2.3.1. Data collection and pre-processing:
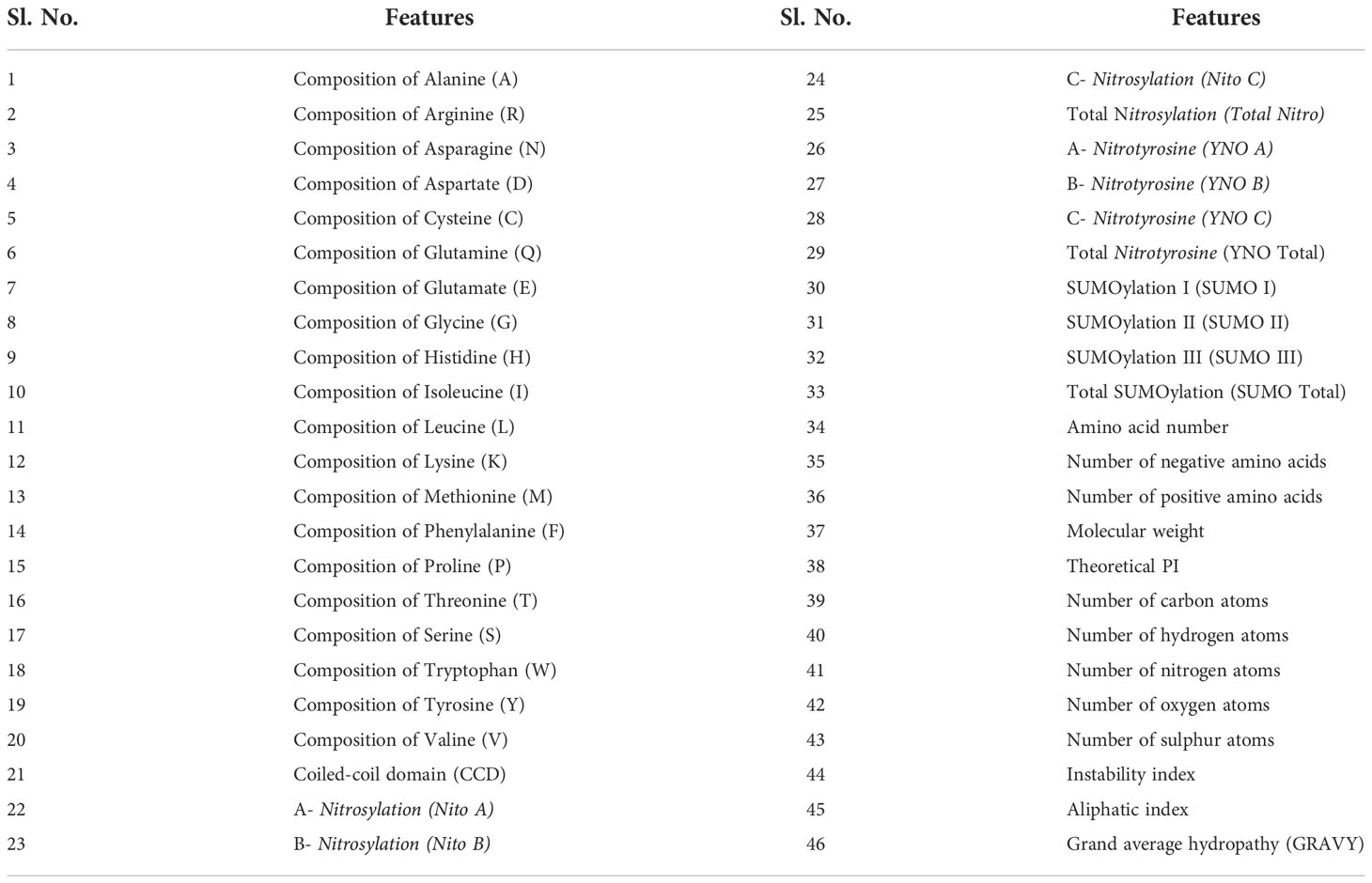
Abiotic stress responsive protein sequence data, namely, “salt stress”, “drought stress”, “heat stress” and “cold stress” of the Poaceae family were retrieved using Boolean operator from the public domain (Uniprot database: https://www.uniprot.org/). Also, the negative dataset of the corresponding stress conditions has been downloaded with the NOT operator. A total of 46 features were extracted from each of these sequences using the bio-python package, (Cock et al., 2009) (Table 1). All the redundant sequences were removed with a similarity of 80% or more using the CD-Hit suite (Huang et al., 2010). For pre-processing the dataset, StandardScaler was used to transform these datasets into Standard Normal Distribution (SND) of the data having zero mean and unit variance, which reduces the biases of the models (Ahsan et al., 2021; Karlaš et al., 2022; Cha and Bae, 2022).
This data pertains to various features that were scaled down and standardized as follows to achieve consistency in the varying range of datasets:
where, Z is standard normalization with x variables, μ mean, and σ2 variance (Tauber and Sánchez, 2002).
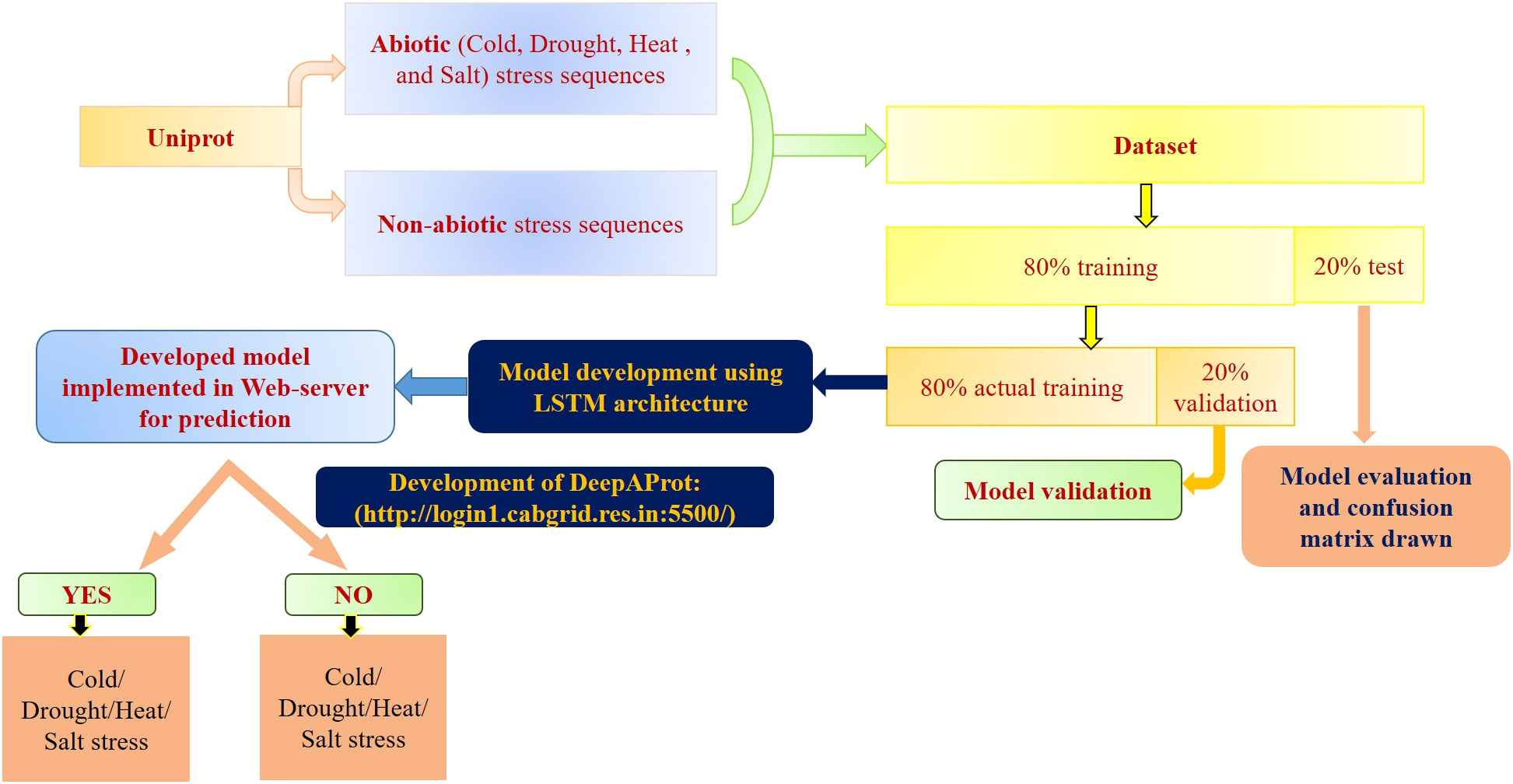
For different layers and epochs, first, stratified sampling was performed, followed by random selection of the training dataset using python script, sklearn library. Different combinations of training:test sets, like, 70:30, 80:20, and 90:10 were made, and finally we proceeded with 80:20 based on the accuracy parameter (Gholamy et al., 2018; Akarsh et al., 2019; Pham et al., 2020; Nguyen et al., 2021; Gu et al., 2022). From this training data, actual training data and drop-out prediction data were retained at 80:20. Fine tuning of weight initializer, layers, epochs, and activation function was carried out in the model to assess the model performance in each epoch. For the given datasets of four stresses, different machine learning algorithms such as SVM, RF, LSTM models were applied using GeLU. For SVM models, polynomial kernel function, 0.01 coeff, and 5-fold StratifiedKFold were used in SVM models for maximum efficiency. In the case of Random Forest, we used a minimum of 0.1 leaf weight with 5-fold StratifiedKFold. For the deep learning model, 150 units, He normal kernel initializers, gelu activation function, and the proposed activation function i.e., sielu were used for comparative analysis in input layers. In the case of the hidden layer, 50 units, 0.02 dropout, and sigmoid activation with 1 unit for binary classification (in the output layer) were employed. During the model compilation, an Adam optimizer and mean square error loss function were used with 500 epochs. The schematic diagram of the methodology is represented in Figure 1.
2.4. Model evaluation indicators
For model evaluation, measures such as accuracy, precision, recall, F1 Score, specificity, and MCC were applied. These parameters were calculated for all four abiotic stresses for SVM, RF, LSTM with GeLU, and LSTM with SieLU activation functions. These are expressed as follows:
where, TP = True Positive, TN = True Negative, FP = False Positive, FN = False Negative.
3. Results and discussion
A thorough screening of “salt stress”, “drought stress”, “heat stress” and “cold stress” associated protein sequences from the Poaceae family retrieved from the public domain resulted in a total of 739 positive and 1305 negative protein sequences of cold stress, 642 positive and 1284 negative protein sequences of drought stress, 977 positive and 1305 negative protein sequences of drought stress, and 473 positive and 946 negative protein sequences of salt stress. For these datasets, 46 protein sequence features were extracted (Table 1) using the bio-python package. These features were scaled down and standardized. The scaling method was used followed by the transformation of feature information into 0 to 1 to reduce the dominance of one feature over others (Beljkas et al., 2020).
The DL models were built using Sigmoid, Tanh, ReLU, LeakyReLU, SoftMax using the above data set and their performance was evaluated with respect model using the proposed SIELU activation function. Also, models were built based on these stress-associated datasets with different machine learning algorithms, namely, SVM, RF, and DL with GeLU activation function were also evaluated with the model using the proposed SEILU activation function. Off course, the proposed SIELU activation function was used in LSTM along with other fine-tuning hyper-parameters for the model development of four different abiotic stress protein sequence datasets of the Poaceae family. All these developed models were subjected to five-fold cross-validation.
The performance of these models was recorded from the test dataset in the form of a confusion matrix for calculating the various evaluation measures, namely, accuracy, precision, recall, F1 Score, specificity, and MCC. The following points emerged from this analysis:
It was observed that, for the cold stress dataset, accuracy and MCC were highest for LSTM with the proposed activation function, SieLU, i.e., 95.11% and 0.90, respectively for testing and 99.20% and 0.98 for the training dataset. LSTM with GeLU activation function gave an accuracy of 94.62% and MCC of 0.89 for the testing dataset and 100% accuracy and MCC of 0.89 in the training dataset. The performance of RF was lowest, i.e., 87.53% accuracy and 0.74 MCC for the testing dataset, accuracy of 88.43% and MCC of 0.75 for the training dataset (Table 2).
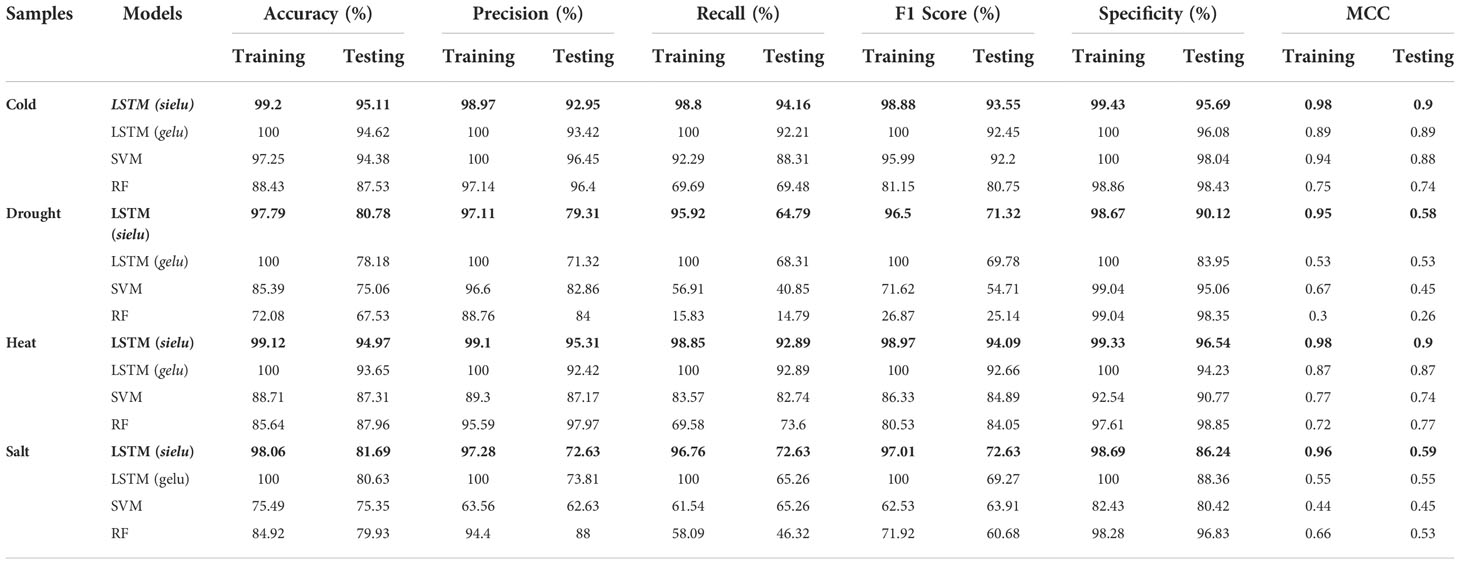
Table 2 Comparison of LSTM with sielu and gelu, SVM, and RF for different abiotic stress-associated protein sequences. The figures in bold denote the evaluation parameters of the best fit model for given stress.
For the drought-responsive protein sequences, the performance of LSTM with SieLU activation function was best with accuracy and MCC as 80.78% and 0.58, respectively for the testing dataset and 97.79% accuracy and MCC 0.95 for the training dataset. This was followed by LSTM with GeLU activation function (Accuracy 78.18%, MCC 0.53 for testing dataset and Accuracy of 100% and MCC 0.53 for training dataset), SVM (Accuracy 75.06%, MCC 0.45 for testing and Accuracy 85.39% and MCC 0.67 for training dataset) and RF (Accuracy 67.53, MCC 0.26 for testing dataset and Accuracy 72.03% and MCC 0.30 for training dataset).
In the case of heat stress also, we found LSTM with a novel activation function, SieLU to perform best with 94.97% accuracy and 0.90 MCC for the testing dataset while an Accuracy of 99.12% and MCC 0.98 for the training dataset. The accuracies for LSTM (GeLU), SVM, and RF were 94.97%, 93.65%, and 87.31%, and 87.96% respectively for the testing dataset whereas for the training dataset, it was found as 99.12%, 100%, 88.71%, and 85.64% respectively, while MCCs were 0.90, 0.87, 0.74, and 0.77 respectively for testing dataset whereas for training it was 0.98, 0.87, 0.77, and 72 respectively. A similar trend was observed in performance for the salt stress dataset also. Accuracy of LSTM (SieLU), LSTM (GeLU), SVM, and RF were 81.69%, 80.63%, 75.35, and 79.93 respectively for the testing dataset, whereas for the training dataset, it was 98.06%, 100%, and 75.49%, and 84.92% respectively. Table 2 delineates the performance of models in detail.
Training accuracy vs. validation accuracy was captured for each epoch in which performance LSTM (SieLU) was found to be superior for all four abiotic stress datasets (Figure 2). For the binary classification of four different abiotic datasets, we used a precision-Recall graph (Supplementary Figure 1) for measurement of the performance of our developed models (Flach and Kull, 2015; Boyd et al., 2012). Analogously, the ROC (Receiver Operating Characteristics) curve shows the comparison of the performance of the developed ML/DL models for all the abiotic stress datasets (Supplementary Figure 2) (Majnik and Bosnić, 2013). Therefore, it can be concluded that the LSTM model with the proposed SIELU activation function outperformed in all datasets as compared to the other competitive models used in this study for classifying protein sequences. Further, these models were also cross-validated with the benchmark heart disease dataset available in the UCI machine learning repository which consists of 303 samples with the 13 most significant features (Otoom et al., 2015). The results showed LSTM (SiELU) to have the highest accuracy (94.74%) and MCC (0.89) as compared to other machine learning models, namely, LSTM (GELU), SVM and RF which showed MCC of 0.86, 0.57 and 0.53, respectively.
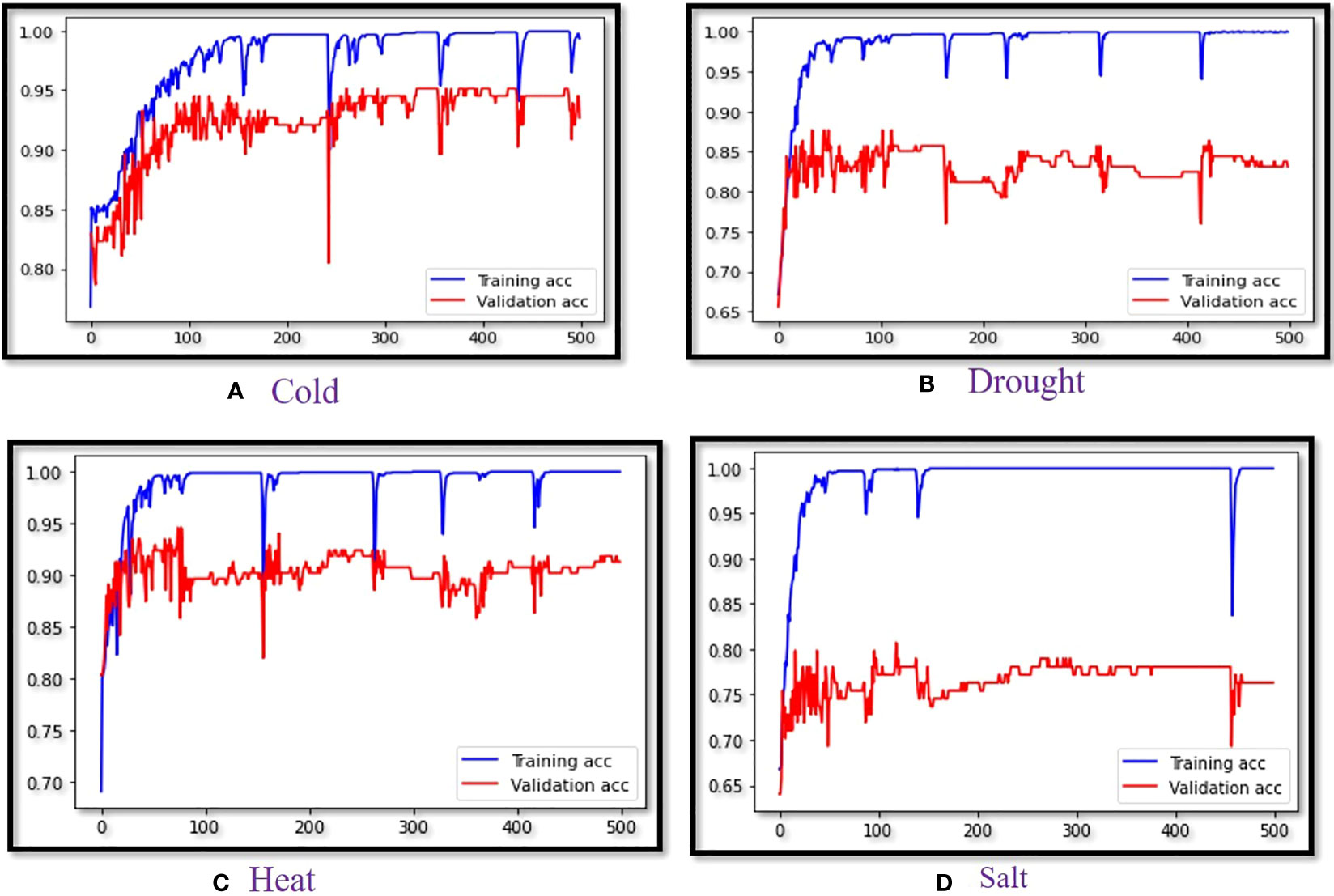
Figure 2 Validation curve of LSTM (SiELU) for (A) Cold stress , (B) Drought stress, (C) Heat stress and (D) Salt stress.
3.1. DeepAProt: Web implementation
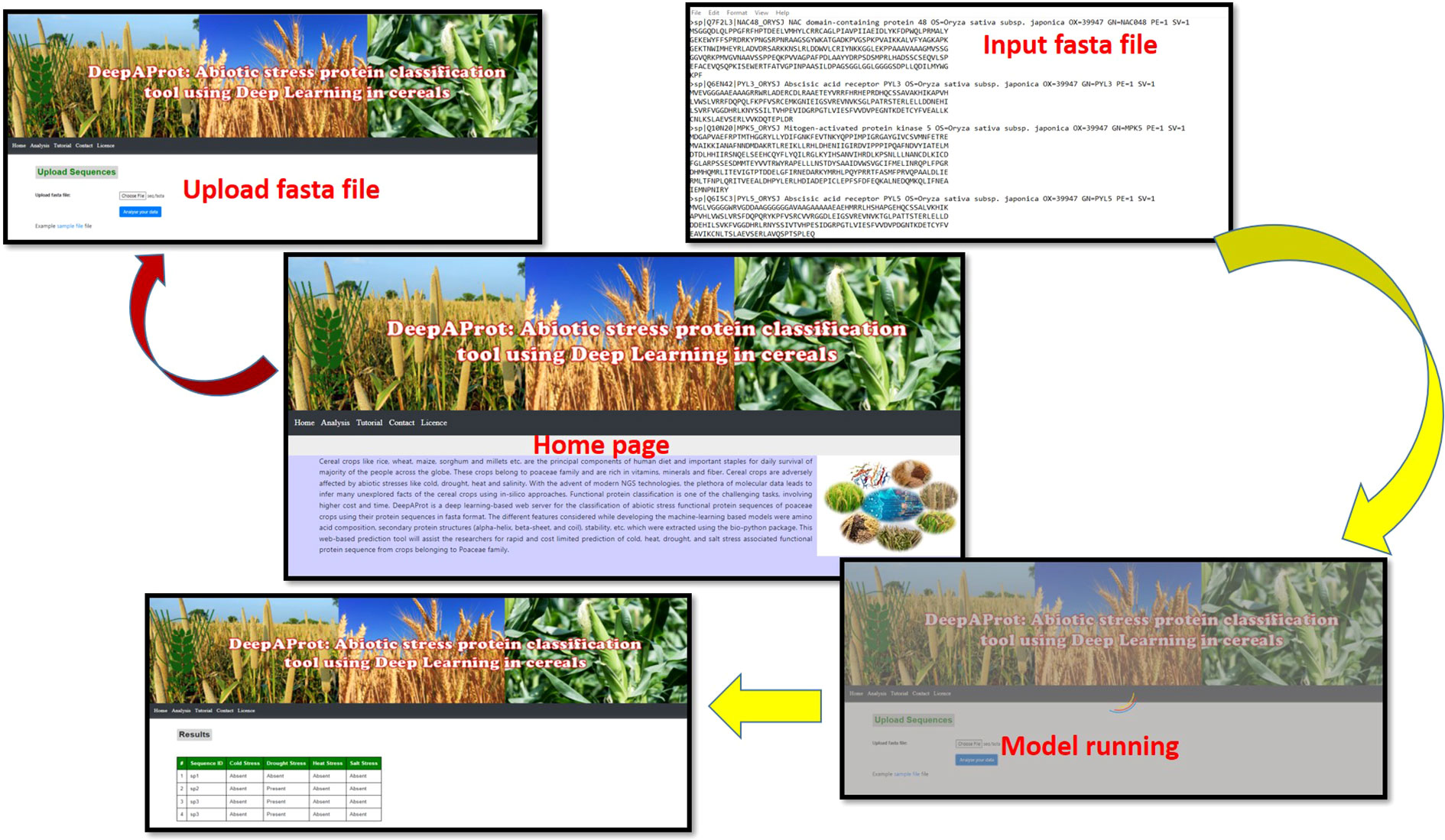
A web-based tool, named as DeepAProt, was developed using the Application Programming Interface (API) flask for the deployment of these DL models. In this web server, the best model for each of the stress-responsive datasets was implemented at the backend to develop a web server for the prediction of related stress-responsive proteins. The architecture of a web-based tool followed the standard three-tier architecture, namely, presentation, web-API, and application layer. The presentation layer is the user interface of the tool which was implemented using HTML and CSS languages. In web-API, a REST API was developed for deploying the model in the server. This layer was implemented using the Python programming language. Finally, the application layer contains the models for the end users, making it more user-friendly for easy use and access. For its application at remote locations, a mobile app “DeepAProt app” was also developed. “DeepAProt app” is developed using Java and XML as a front-end mobile app using android studio. For the interface of the web tool, the Python Flask framework has been used. The Back-end web tool is developed on a python framework using a deep learning module i.e., TensorFlow. This app has the provision to upload protein sequence data in fasta format for analysis and the result will be presented in a tabular form regarding the given protein sequences association with abiotic stresses such as cold, drought, heat, and salt. In this app, a provision was also made to download and help document and sample data. It makes use of HTML (Peroni et al., 2017), javascript (Delcev and Draskovic, 2018), and CSS (Genevès et al., 2012) at the back-end and front-end to classify any protein sequence (in fasta format) that has to be upload as input by biologists.
The user can select either of the abiotic stresses, (i.e., heat/cold/salt/drought) followed by uploading the sequence. Once the raw protein sequence is uploaded in fasta format, the output classifies the sequences to the predicted category. This web server is user-friendly and freely accessible at http://login1.cabgrid.res.in:5500/. Figure 3 shows the interface of this web-implemented server and its usage. This web-based tool helps the biologist to classify the unknown protein sequence to the respective class of abiotic stress. Also, the developed mobile app can be popularized for easy and quick handling of data for the identification of stress. It can be downloaded from the Homepage.
As classification and prediction of proper abiotic stress protein sequences help the biologist to implement it in crop improvement. Machine learning and deep learning models help to find out the abiotic stress protein sequences in a cost and effective manner. However, most biologists do not have enough knowledge about machine learning and deep learning to predict the proper abiotic stress protein sequences. Therefore, our models help them to distinguish between the abiotic stress and non-abiotic stress protein sequence that comes from the sequencing laboratory directly.
4. Conclusion
In this study, we proposed a novel activation function name SIELU which was used to build the DL model along with other hyperparameters. The performance of this novel activation function has been studied using public domain data to predict stress-responsive proteins under four abiotic stresses, namely, cold, heat, salinity, and drought from the major crops of the Poaceae family. Further, a comparative analysis was carried out between SVM, RF, and LSTM with GELU, and SIELU activation functions. It has been observed that LSTM with SIELU activation function outperformed as compared to other competitive models used in this study. Hence, LSTM with SIELU models was implemented in the form of web servers for the classification of unknown protein sequences into different abiotic stresses of crops from the Poaceae family. This work can be of immense use for plant breeders for in silico identification of the stress-responsive proteins in crops of the Poaceae family, leading to the rapid development of abiotic stress-resistant varieties.
Resource used: The research was carried out using python programming packages, version 3.7.8. Also, for the graphical user interface (GUI), Anaconda Repository was used for coding these models in a Jupyter notebook with necessary python libraries. All these model buildings have been carried out in HP-Z400-Workstation dual booting system where Linux - Ubuntu version with 16.04 LTS is used with the memory of 99.3 GB. The RAM of the system was 16 BGB with a processor of Intel® Xeon(R) CPU W3565 at 3.20GHz × 4 having NVC1 graphics.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found here: Python library: PyPi (https://pypi.org/project/sielu/). Web-based application: http://login1.cabgrid.res.in:5500/ Mobile Application: download from http://login1.cabgrid.res.in:5500/.
Author contributions
SJ, AR, and DK conceived the theme of the study. BA, MI, SJ, and AR developed the methodology, BA collected the data. BA, MH, SJ, MI, and UA were involved in the computational analysis and development of web resources and mobile applications. SJ, MI, and AR supervised the study. BA wrote the original draft. DK and AR reviewed and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The authors are thankful to the CABin grant, Indian Council of Agricultural Research, Ministry of Agriculture and Farmers’ Welfare, Govt. of India (F. no. Agril. Edn. 4–1/2013-AandP) for providing financial support. The grant of the IARI Merit scholarship to BA is duly acknowledged.
Acknowledgments
The financial grants, ICAR- CABin and IARI Merit scholarship to BA are duly acknowledged. The authors further acknowledge the supportive role of the Director, ICAR-IASRI, New Delhi.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1008756/full#supplementary-material
Supplementary Figure 1 | Precision-Recall curve of different abiotic stress data.
Supplementary Figure 2 | Receiver Operating Characteristics curve of different abiotic stress data.
References
Ahsan, M., Mahmud, M., Saha, P., Gupta, K., Siddique, Z. (2021). Effect of data scaling methods on machine learning algorithms and model performance. Technologies 9 (3), 52. doi: 10.3390/technologies9030052
Akarsh, S., Poornachandran, P., Menon, V. K., Soman, K. P. (2019). “A detailed investigation and analysis of deep learning architectures and visualization techniques for malware family identification,” in Advanced sciences and technologies for security applications (Springer International Publishing). doi: 10.1007/978-3-030-16837-7_12
An, J., Li, W., Li, M., Cui, S., Yue, H. (2019). Identification and classification of maize drought stress using deep convolutional neural network. Symmetry 11, 256. doi: 10.3390/sym11020256
Armenteros, J. J. A., Salvatore, M., Emanuelsson, O., Winther, O., Von Heijne, G., Elofsson, A., et al. (2019). Detecting sequence signals in targeting peptides using deep learning. Life Sci. Alliance. 2 (5), 1–14. doi: 10.26508/lsa.201900429
Bagby, R. J. (1995). Calculating normal probabilities. Am. Math. monthly. 102, 46–49. doi: 10.1080/00029890.1995.11990532
Beljkas, Z., Knezevic, M., Rutesic, S., Ivanisevic, N. (2020). Application of artificial intelligence for the estimation of concrete and reinforcement consumption in the construction of integral bridges. Adv. Civil Eng. 2020, 1–8. doi: 10.1155/2020/8645031
Benvenuto, N., Piazza, F. (1992). On the complex back-propagation algorithm. IEEE Trans. Signal Process. 40 (4), 967–969. doi: 10.1109/78.127967
Bileschi, M., Belanger, D., Bryant, D., Sanderson, T., Carter, B., DePristo, M., et al. (2022). Using deep learning to annotate the protein universe. Nat Biotechnol 40, 932–937. doi: 10.1038/s41587-021-01179-w
Boyd, K., Costa, V. S., Davis, J., Page, C. D. (2012). “Unachievable region in precision-recall space and its effect on empirical evaluation,” in Proceedings of the 29th International Conference on Machine Learning, ICML 2012, (Edinburgh, Scotland, UK: The International Conference on Machine Learning (ICML)) Vol. 1639–646.
Brophy, A. L. (1985). Approximation of the inverse normal distribution function. Behav. Res. Methods Instrum. Comput. 17 (3), 415–417. doi: 10.3758/bf03200956
Cai, Y., Wang, J., Deng, L. (2020). SDN2GO : An integrated deep learning model for protein function prediction 8, April, 1–11. doi: 10.3389/fbioe.2020.00391
Cai, C., Xu, Y., Ke, D., Su, K. (2015). Deep neural networks with multistate activation functions. Comput. Intell. Neurosci. 721367, 1–10. doi: 10.1155/2015/721367
Cha, J., Bae, G. (2022). Deep learning based infant cry analysis utilizing computer vision 17, 1, 30–35.
Chen, J., Xu, W., Velten, J., Xin, Z., Stout, J. (2012). Characterization of maize inbred lines for drought and heat tolerance. J. Soil Water Conserv. 67 (5), 354–364. doi: 10.2489/jswc.67.5.354
Cobb, J. N., DeClerck, G., Greenberg, A., Clark, R., McCouch, S. (2013). Next-generation phenotyping: requirements and strategies for enhancing our understanding of genotype–phenotype relationships and its relevance to crop improvement. Theor. Appl. Genet. 126 (4), 867–887. doi: 10.1007/s00122-013-2066-0
Cock, P. J. A., Antao, T., Chang, J. T., Chapman, B. A., Cox, C. J., Dalke, A., et al. (2009). Biopython: Freely available python tools for computational molecular biology and bioinformatics. Bioinformatics 25 (11), 1422–1423. doi: 10.1093/bioinformatics/btp163
Dao, P. D., He, Y., Proctor, C. (2021). Plant drought impact detection using ultra-high spatial resolution hyperspectral images and machine learning. Int. J. Appl. Earth Obs. Geoinformation 102, 102364. doi: 10.1016/j.jag.2021.102364
Das, B., Manohara, K. K., Mahajan, G. R., Sahoo, R. N. (2020). Spectroscopy based novel spectral indices, PCA- and PLSR-coupled machine learning models for salinity stress phenotyping of rice. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 229, 117983. doi: 10.1016/j.saa.2019.117983
Delcev, S., Draskovic, D. (2018). “Modern JavaScript frameworks: A survey study,” in 2018 Zooming Innovation in Consumer Technologies Conference (ZINC). (Novi Sad, Serbia) 106–109 (IEEE). doi: 10.1109/ZINC.2018.8448444
Ding, W., Nakai, K., Gong, H. (2022). Protein design via deep learning. Briefings Bioinf. 23 (3), 1–16. doi: 10.1093/bib/bbac102
Dunn, A. M., Hofmann, O. S., Waters, B., Witchel, E. (2011). “Cloaking malware with the trusted platform module,” in Proceedings of the 20th USENIX Security Symposium. (San Francisco, CA: USENIX Security Symposium) 395–410.
Elaraby, N. M., Elmogy, M. (2016). Deep Learning : Effective tool for big data analytics. Int. J. Comput. Sci. Eng. 5 (05), 254–262.
Emerson, P. L. (1979). Computer approximation of the inverse of the normal distribution function. Behav. Res. Methods Instrum. 11, 397–398. doi: 10.3758/BF03205685
Erenstein, O., Jaleta, M., Mottaleb, K. A., Sonder, K., Donovan, J., Braun, H. J. (2022). “Global trends in wheat production, consumption and trade,” in Wheat improvement. Eds. Reynolds, M. P., Braun, H. J. (Springer, Cham). doi: 10.1007/978-3-030-90673-3_4
Etminan, A., Pour-Aboughadareh, A., Mohammadi, R., Shooshtari, L., Yousefiazarkhanian, M., Moradkhani, H. (2019). Determining the best drought tolerance indices using artificial neural network (ANN): Insight into application of intelligent agriculture in agronomy and plant breeding. Cereal Res. Commun. 47 (1), 170–181. doi: 10.1556/0806.46.2018.057
Fenu, G., Malloci, F. M. (2021). Review forecasting plant and crop disease: An explorative study on current algorithms. Big Data Cogn. Computing 5 (1), 1–24. doi: 10.3390/bdcc5010002
Flach, P. A., Kull, M. (2015). Precision-Recall-Gain curves: PR analysis done right. Adv. Neural Inf. Process. Syst., 1838–846.
Gelman, S., Fahlberg, S. A., Heinzelman, P., Romero, P. A., Gitter, A. (2021). Neural networks to learn protein sequence-function relationships from deep mutational scanning data. Proc. Natl. Acad. Sci. United States America 118 (48), e2104878118. doi: 10.1073/pnas.2104878118
Genevès, P., Layaïda, N., Quint, V. (2012). “On the analysis of cascading style sheets,” in WWW’12 - Proceedings of the 21st Annual Conference on World Wide Web, (New York, United States: Springer Link) 809–818.
Gholamy, A., Kreinovich, V., Kosheleva, O. (2018). Why 70/30 or 80/20 relation between training and testing Sets : A pedagogical explanation. Departmental Tech. Rep. (CS) 1209, 1–6.
Ghosal, S., Blystone, D., Singh, A. K., Ganapathysubramanian, B., Singh, A., Sarkar, S. (2018). An explainable deep machine vision framework for plant stress phenotyping. Proc Natl Acad Sci U S A 115 (18), 4613–4618. doi: 10.1073/pnas.1716999115
Gill, M., Anderson, R., Hu, H., Bennamoun, M., Petereit, J., Valliyodan, B., et al. (2022). Machine learning models outperform deep learning models, provide interpretation and facilitate feature selection for soybean trait prediction. BMC Plant Biol. 22, 180. doi: 10.1186/s12870-022-03559-z
Gupta, C., Ramegowda, V., Basu, S., Pereira, A. (2021). Using network-based machine learning to predict transcription factors involved in drought resistance. Front. Genet. 943. doi: 10.3389/fgene.2021.652189
Gu, Z., Sharma, S., Riley, D., Pantawane, M., Joshi, S., Fu, S., et al. (2022). A universal predictor-based machine learning model for optimal process maps in laser powder bed fusion process. J. Intell. Manuf., 1–23. doi: 10.1007/s10845-022-02004-0
Hasan, M. M., Basith, S., Khatun, M. S., Lee, G., Manavalan, B., Kurata, H. (2021). Meta-i6mA: Deepm5C N6-methyladenine sites of plant genomes by exploiting informative features in an integrative machine-learning framework. Briefings Bioinf. 22 (3), bbaa202. doi: 10.1093/bib/bbaa202
Hasan, M. M., Tsukiyama, S., Cho, J. Y., Kurata, H., Alam, M. A., Liu, X., et al. (2022). Deepm5C: A deep-learning-based hybrid framework for identifying human RNA N5-methylcytosine sites using a stacking strategy. Mol. Ther. 30 (8), 2856–2867. doi: 10.1016/j.ymthe.2022.05.001
Hastings, C. J. (1955). Approximations for digital computers (Princeton, NJ: Princeton University Press). Available at: https://books.google.co.in/books?hl=enandlr=andid=IRTWCgAAQBAJandoi=fndandpg=PP1andots=UKUd8hGL3Landsig=-cTtzuSYIcORnRC1Co6f1vkMesandredir_esc=y#v=onepageandqandf=false.
Huang, Y., Niu, B., Gao, Y., Fu, L., Li, W.. (2010). CD-HIT suite: A web server for clustering and comparing biological sequences. Bioinformatics 26, 680–682. doi: 10.1093/bioinformatics/btq003
Ingole, K., Patil, N. (2020). Performance analysis of various activation function on a shallow neural network. Int. J. Emerging Technol. Innovative Res. 7 (6), 269–276. doi: 10.1729/Journal.24670
Jansen, M. A., Potters, G. (2017). “Plant stress physiology,” in Stress: The way of life, 2nd ed (London, UK: CABI), ix–xiv.
Jiang, X., Zhu, Z., Chen, L. (2020). An intelligent deep feature learning method with improved activation functions for machine fault diagnosis. IEEE Access. 8, 1975–1985. doi: 10.1109/ACCESS.2019.2962734
Kanai, S., Fujiwara, Y., Yamanaka, Y., Adachi, S. (2018). “Sigsoftmax: Reanalysis of the softmax bottleneck,” in NIPS'18: Proceedings of the 32nd International Conference on Neural Information Processing Systems. (CA, USA: Neural Information Processing Systems Foundation, Inc. (NeurIPS)) 284–294. doi: 10.5555/3326943.3326970
Kang, D., Ahn, H., Lee, S., Lee, C. J., Hur, J., Jung, W., et al. (2018). “Identifying stress-related genes and predicting stress types in arabidopsis using logical correlation layer and CMCL loss through time-series data,” in Proceedings - 2018 IEEE International Conference on Bioinformatics and Biomedicine, BIBM 2018, (Madrid, Spain: Institute of Electrical and Electronics Engineers (IEEE)) December 2018. 399–404. doi: 10.1109/BIBM.2018.8621581
Karlaš, B., Dao, D., Interlandi, M., Li, B., Schelter, S., Wu, W., et al. (2022). Data debugging with shapley importance over end-to-End machine learning pipelines. arXiv:2204.11131 [cs.LG], 1–43.
Karlik, B., Olgac, A. V. (2011). Performance analysis of various activation functions in generalized MLP architectures of neural networks. Int. J. Artif. Intell. Expert Syst. 1 (4), 111–122.
Khaki, S., Khalilzadeh, Z., Wang, L. (2019). Classification of crop tolerance to heat and drought–a deep convolutional neural networks approach. Agronomy 9 (12), 833. doi: 10.3390/agronomy9120833
Kiani, M., Panaretos, J., Psarakis, S., Saleem, M. (2008). Approximations to the normal distribution function and an extended table for the mean range of the normal variables. J. Iranian Stat. Soc. (Jirss) 7 (12), 57–72.
Landi, S., Hausman, J. F., Guerriero, G., Esposito, S. (2017). Poaceae vs. abiotic stress: focus on drought and salt stress, recent insights and perspectives. Front. Plant science. 8, 1214.
LeCun, Y. A., Bottou, L., Orr, G. B., Müller, K. R. (2012). “Efficient backprop,” in Neural networks:Tricks of the trade, lecture notes in computer science, vol. 7700 . Eds. Montavon, G., Orr, G. B., Müller, K. R. (Berlin, Heidelberg: Springer), 375–405. doi: 10.1007/978-3-642-35289-8_3
Li, Y., Wang, S., Umarov, R., Xie, B., Fan, M., Li, L., et al. (2018). DEEPre: Sequence-based enzyme EC number prediction by deep learning. Bioinformatics 34 (5), 760–769. doi: 10.1093/bioinformatics/btx680
Lobell, D. B., Schlenker, W., Costa-Roberts, J. (2011). Climate trends and global crop production since 1980. Science 333, 616–620. doi: 10.1126/science.1204531
Majnik, M., Bosnić, Z. (2013). ROC analysis of classifiers in machine learning: A survey. Intell. Data Anal. 17 (3), 531–558. doi: 10.3233/IDA-130592
Moghimi, A., Yang, C., Marchetto, P. M. (2018). Ensemble feature selection for plant phenotyping: A journey from hyperspectral to multispectral imaging. IEEE Access 6, 56870–56884. doi: 10.1109/ACCESS.2018.2872801
Nguyen, Q. H., Ly, H. B., Ho, L. S., Al-Ansari, N., Van Le, H., Tran, V. Q., et al. (2021). Influence of data splitting on performance of machine learning models in prediction of shear strength of soil. Math. Problems Eng. 2021. doi: 10.1155/2021/4832864
Niu, Y., Han, W., Zhang, H., Zhang, L., Chen, H. (2021). Estimating fractional vegetation cover of maize under water stress from UAV multispectral imagery using machine learning algorithms. Comput. Electron. Agric. 189 (106414). doi: 10.1016/j.compag.2021.106414
Otoom, A. F., Abdallah, E. E., Kilani, Y., Kefaye, A., Ashour, M. (2015). Effective diagnosis and monitoring of heart disease. Int. J. software Eng. its Appl. 9 (1), 143–156. doi: 10.14257/ijseia.2015.9.1.12
Palanog, A. D., Swamy, B. P. M., Shamsudin, N. A. A., Dixit, S., Hernandez, J. E., Boromeo, T. H., Cruz, P. C. S., et al (2014). Grain yield QTLs with consistent-effect under reproductive-stage drought stress in rice. Field Crops Res 161, 46–54.
Peroni, S., Osborne, F., Di Iorio, A., Nuzzolese, A. G., Poggi, F., Vitali, et al. (2017). Research articles in simplified HTML: a web-first format for HTML-based scholarly articles. PeerJ Comput. Sci. 3, e132. doi: 10.7717/peerj-cs.132
Pham, B. T., Qi, C., Ho, L. S., Nguyen-Thoi, T., Al-Ansari, N., Nguyen, M. D., et al. (2020). A novel hybrid soft computing model using random forest and particle swarm optimization for estimation of undrained shear strength of soil. Sustain. (Switzerland) 12 (6), 1–16. doi: 10.3390/su12062218
Pratiwi, H., Windarto, A. P., Susliansyah, S., Aria, R. R., Susilowati, S., Rahayu, L. K., et al. (2020). Sigmoid activation function in selecting the best model of artificial neural networks. J. Physics: Conf. Ser. 1471 (1), 12010. doi: 10.1088/1742-6596/1471/1/012010
Rico-Chávez, A. K., Franco, J. A., Fernandez-Jaramillo, A. A., Contreras-Medina, L. M., Guevara-González, R. G., Hernandez-Escobedo, Q. (2022). Machine learning for plant stress modeling: A perspective towards hormesis management. Plants 11 (7), 1–22. doi: 10.3390/plants11070970
Salman, S., Liu, X. (2019). Overfitting mechanism and avoidance in deep neural networks. arXiv:1901.06566 [cs.LG].
Sarker, I. H. (2021). Deep learning: A comprehensive overview on techniques, taxonomy, applications and research directions. SN Comput. Sci. 2 (6), 1–20. doi: 10.1007/s42979-021-00815-1
Sridevi, S., Kanimozhi, T. (2021). “Classification of protein sequences using hybrid recurrent deep learning models,” in IEEE International Conference on Technology, Research, and Innovation for Betterment of Society (TRIBES), 2021. (Raipur, India: Institute of Electrical and Electronics Engineers (IEEE)) 1–4. doi: 10.1109/TRIBES52498.2021.9751666
Srinivasan, K., Cherukuri, A. K., Vincent, D. R., Garg, A., Chen, B. Y. (2019). An efficient implementation of artificial neural networks with K-fold cross-validation for process optimization. J. Internet Technol. 20 (4), 1213–1225. doi: 10.3966/160792642019072004020
Sue Wing, I., De Cian, E., Mistry, M. N. (2021). Global vulnerability of crop yields to climate change. J. Environ. Econ. Manage. 109, 102462. doi: 10.1016/j.jeem.2021.102462
Szandała, T. (2021). Review and comparison of commonly used activation functions for deep neural networks. In: Bhoi, A., Mallick, P., Liu, C. M., Balas, V. eds. Bio-inspired Neurocomputing. Studies in Computational Intelligence Singapore: Springer, 903. doi: 10.1007/978-981-15-5495-7_11
Tauber, L., Sánchez, V. (2002). “Introducing the normal distribution in a data analysis course: specific meaning contributed by the use of computers,” in Proceedings of Seventh International Congress for Teaching Statistics, Citeseer. (Brazil: IEEE) 1–6.
Venal, M. C. A., Fajardo, A. C., Hernandez, A. A. (2019). “Plant stress classification for smart agriculture utilizing convolutional neural network-support vector machine,” in Proceeding - 2019 International Conference on ICT for Smart Society: Innovation and Transformation Toward Smart Region, ICISS 2019, (Indonesia: School of Electrical Engineering and Informatics ITB) February 2020. doi: 10.1109/ICISS48059.2019.8969799
Wang, A. (2022). Deep learning methods for protein family classification on PDB sequencing data. ArXiv, abs/1505.00853.
Wang, Y., Li, Y., Song, Y., Rong, X. (2020). The influence of the activation function in a convolution neural network model of facial expression recognition. Appl. Sci. 10 (5). doi: 10.3390/app10051897
Wen, M., Cong, P., Zhang, Z., Lu, H., Li, T. (2018). DeepMirTar: a deep learning approach for predicting human miRNA targets. Bioinformatics 34 (22), 3781–3787. doi: 10.1093/bioinformatics/bty424
Xu, Y., Verma, D., Sheridan, R. P., Liaw, A., Ma, J., Marshall, N. M., et al. (2020). Deep dive into machine learning models for protein engineering. J. Chem. Inf. Modeling 60 (6), 2773–2790. doi: 10.1021/acs.jcim.0c00073
Xu, B., Wang, N., Chen, T., Li, M. (2015). Empirical evaluation of rectified activations in convolutional network. arXiv, abs/1505.00853.
Zahid, A., Dashtipour, K., Abbas, H. T., Mabrouk, I.B., Al-Hasan, M., Ren, A., et al. (2022). Machine learning enabled identification and real-time prediction of living plants’ stress using terahertz waves. Defence Technol. 18 (8), 1330–1339. doi: 10.1016/j.dt.2022.01.003
Keywords: abiotic stress, activation function, deep learning, web-server, mobile application
Citation: Ahmed B, Haque MA, Iquebal MA, Jaiswal S, Angadi UB, Kumar D and Rai A (2023) DeepAProt: Deep learning based abiotic stress protein sequence classification and identification tool in cereals. Front. Plant Sci. 13:1008756. doi: 10.3389/fpls.2022.1008756
Received: 01 August 2022; Accepted: 14 November 2022;
Published: 12 January 2023.
Edited by:
Nabin Bhusal, Agriculture and Forestry University, NepalReviewed by:
Md Mehedi Hasan, Tulane University, United StatesPiyush Priya, National Institute of Plant Genome Research (NIPGR), India
Copyright © 2023 Ahmed, Haque, Iquebal, Jaiswal, Angadi, Kumar and Rai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sarika Jaiswal, c2FyaWthQGljYXIuZ292Lmlu
 Bulbul Ahmed
Bulbul Ahmed Md Ashraful Haque
Md Ashraful Haque Mir Asif Iquebal
Mir Asif Iquebal Sarika Jaiswal
Sarika Jaiswal U. B. Angadi
U. B. Angadi Dinesh Kumar
Dinesh Kumar Anil Rai
Anil Rai