
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 30 August 2022
Sec. Plant Pathogen Interactions
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.1006409
This article is part of the Research Topic Genomics and disease resistance in wheat and maize View all 16 articles
 Lijing Sun†
Lijing Sun† Liangjie Lv†
Liangjie Lv† Jie Zhao
Jie Zhao Mengyun Hu
Mengyun Hu Yelun Zhang
Yelun Zhang Yun Zhao
Yun Zhao Xiaodong Tang
Xiaodong Tang Peinan Wang
Peinan Wang Qianying Li
Qianying Li Xiyong Chen
Xiyong Chen Hui Li*
Hui Li* Yingjun Zhang*
Yingjun Zhang*Cytokinin is an important endogenous hormone in plants performing a wide spectrum of biological roles. The type-A response regulators (RRAs) are primary cytokinin response genes, which are important components of the cytokinin signaling pathway and are involved in the regulation of plant growth and development. By analysis of the whole genome sequence of wheat, we identified 20 genes encoding RRAs which were clustered into eight homologous groups. The gene structure, conserved motifs, chromosomal location, and cis-acting regulatory elements of the TaRRAs were analyzed. Quantitative real-time polymerase chain reaction (qRT-PCR) results showed that the expression levels of most of the TaRRAs increased rapidly on exogenous cytokinin application. Moreover, the TaRRA family members displayed different expression profiles under the stress treatments of drought, salt, cold, and heat. This study provides valuable insights into the RRA gene family in wheat and promotes the potential application of these genes in wheat genetic improvement.
Cytokinin is a vital phytohormone responsible for regulating numerous aspects of plant growth and development, including cell division and differentiation, apical dominance, leaf senescence, photomorphogenesis, fertility, and seed development (Werner and Schmülling, 2009; Hwang et al., 2012; Schaller et al., 2015; Wybouw and De Rybel, 2019). In addition, cytokinin plays an important role in plant response to many environmental stresses such as drought, salt, cold, and heat (Ha et al., 2012; Pavlů et al., 2018; Liu et al., 2020). Cytokinin signal transduction is mediated by a multistep two-component system (TCS) involving a His-Asp-His-Asp phosphorelay from histidine kinase receptors (HKs) and histidine-containing phosphotransfer proteins (HPs) to downstream response regulators (RRs) (Argueso et al., 2010; El-Showk et al., 2013; Kieber and Schaller, 2018).
The response regulators are classified into four distinct groups based on phylogenetic and conserved domain analysis, namely, type-A response regulators (RRAs), type-B response regulators (RRBs), type-C response regulators (RRCs), and circadian clock-related pseudo response regulators (PRRs) (Heyl et al., 2013), all with a conserved N-terminal receiver domain and a variable-length C-terminal. The C-terminal of RRAs is short and has yet to be assigned any specific function. RRBs have a large C-terminal extension containing Myb-like DNA binding, nuclear localization, and transcription activation domains (Sakai et al., 2000; Hosoda et al., 2002). The C-terminal of RRCs is also short; however, they are not grouped in the same class as RRAs. PRRs lack the highly conserved Asp residue required for phosphorylation and have a C-terminal including a CCT domain (CO, CO-LIKE, TOC1) (Makino et al., 2000; Schaller et al., 2008). RRBs are transcription factors that mediate the transcriptional response to cytokinin (Argyros et al., 2008). RRAs are transcriptionally induced in response to cytokinin via direct activation by RRBs and are responsible for repressing cytokinin signaling via a negative feedback loop (Hwang et al., 2012).
Many reports have established that RRAs play a critical role in plant growth and development. In Arabidopsis, 10 RRAs have been reported (ARR3-ARR9 and ARR15-ARR17) (D’Agostino et al., 2000), of which ARR3 and ARR4 act redundantly in the determination of the circadian rhythm (Salomé et al., 2006). ARR4 interacts with phytochrome B, modulating red light signaling by stabilizing the active Pfr form of phytochrome B (Sweere et al., 2001; Mira-Rodado et al., 2007). ARR7 negatively influences meristem size through regulation of its expression by WUSCHEL (Leibfried et al., 2005). Moreover, ARR7, together with ARR15, also participates in the cytokinin–auxin hormonal control of the shoot stem-cell niche (Zhao et al., 2010). In addition, ARR16 has been reported to be involved in Arabidopsis seedling development via regulation of its expression by MYC2 (Srivastava et al., 2019).
Accumulating evidence indicates that RRAs are involved in abiotic stress responses. Dehydration stress transiently induces the expression of ARR5, ARR7, and ARR15, but reduces the expression of ARR8 and ARR17 (Kang et al., 2012). Also, the phosphorylation of ARR5 Ser residues by SnRK2s enhances its stability and plant drought tolerance (Huang et al., 2018). The expression of a variety of RRAs, including ARR5, ARR6, ARR7, and ARR15, are induced by cold (Jeon et al., 2010). However, ARR-OE plants (ARR5-OE, ARR7-OE, and ARR15-OE) and arr mutants (arr5, arr6, and arr7) show similar enhanced freezing tolerance, indicating that RRAs play a complex role in regulating cold stress response (Jeon et al., 2010; Shi et al., 2012). In rice, the expression of type-A OsRR6 is induced by salt, dehydration, and low-temperature treatments (Jain et al., 2006), and overexpression of OsRR6 enhances seedling drought and salinity tolerance (Bhaskar et al., 2021); whereas, OsRR9 and OsRR10 negatively regulate rice salinity tolerance (Wang et al., 2019). Recently, ZmRR1, a maize type-A RR, has been demonstrated to positively regulate maize chilling tolerance by modulating the expression of ZmDREB1s and ZmCesAs. The phosphorylation of ZmRR1 Ser residues by ZmMPK8 accelerates its degradation, thereby reducing the chilling tolerance (Zeng et al., 2021).
RRAs have been widely studied in Arabidopsis and rice, however, limited information is available for RRAs in wheat. The completion of the wheat whole genome sequence and further improvements of the wheat genome database have immensely contributed to decoding the wheat genome at the molecular level. In the present study, we systematically performed a genome-wide analysis of the wheat RRA gene family and investigated their gene structures, conserved motifs, chromosomal locations, cis-acting regulatory elements, and expression patterns in response to cytokinin treatment and various stresses. This work provides valuable information on the wheat RRA gene family and lays a foundation for further functional analysis of this gene family.
Whole-genome and protein sequence data of wheat (IWGSC Assembly GCA_900519105.1) and the hidden Markov model (HMM) file for the response regulator receiver domain (PF00072) were downloaded from the EnsemblPlants database1 and Pfam database,2 respectively. A wheat-specific HMM file was established by the alignment of the response regulator receiver domain HMM file with the wheat protein sequences (E-value < 1e–20). The wheat-specific HMM file was then used as bait to search against the local reference genome database to identify candidate wheat RRs (E-value < 0.01). Redundant genes were removed, and the longest representative transcripts were selected for further analysis. The identified proteins were then submitted to Pfam (see text footnote 2), SMART,3 and NCBI conserved domains search tool4 to further check the receiver domain as well as other conserved domains. The protein sequence of RRs in wheat, Arabidopsis, and rice was used to carry out multiple sequence alignment using ClustalX 2.1 software (Larkin et al., 2007). The phylogenetic tree was established using MEGA 7.0 based on the neighbor-joining (NJ) method with 1,000 bootstrap replicates (Kumar et al., 2016), and RRs were named according to the standard nomenclature for the plant TCS elements (Heyl et al., 2013).
Information about the TaRRA gene family, such as chromosomal localization, number of exons, and cDNA and protein length, was obtained from the EnsemblPlants. The protein sequence of TaRRAs was analyzed in the Expasy server5 to obtain the theoretical isoelectric point (PI) and molecular weight (MW).
The gene structure of TaRRAs was constructed by the gene structure display server (GSDS 2.06) using the coding sequence (CDS) and corresponding genomic sequence retrieved from the EnsemblPlants database. Conserved motifs of TaRRAs were predicted using the Multiple Em for Motif Elicitation (MEME 5.4.17), with the following parameters: maximum number of 10 motifs and optimum motif widths of 6-50 residues.
The wheat genomic sequence and genome annotation files downloaded from the EnsemblPlants database were used to generate a graph of chromosomal location and collinearity relationship of TaRRAs by TBtools software (Chen et al., 2020). The synteny relationship of RRAs between wheat and rice was constructed using the Dual Systeny Plot for MCscanX.8
The promoter region, 1,500 bp upstream of the initiation code (ATG), of all of the TaRRAs, was obtained from the EnsemblPlants database and the cis-acting regulatory elements were predicted by PlantCARE.9
The expression data in various tissues were downloaded from the WheatOmics 1.0 (Ma et al., 2021). The transcripts per million (TPM) values were used to create a heat map by using Heatmap (see text footnote 8).
Jimai325, a high-yielding and water-saving wheat variety cultivated by our lab, was used for qRT-PCR analysis. Ten-day-old hydroponically grown seedlings were exposed to 50 μM 6-BA, 20% PEG-6000 (drought stress), 200 mM NaCl (salt stress), 4°C (cold stress), or 40°C (heat stress) for 0, 1, 3, 6, 12, and 24 h, and samples were then collected. Total RNA was isolated using TRNzol Universal reagent (TIANGEN) according to the manufacturer’s instructions, and 1 μg of total RNA was used as the template for cDNA synthesis. Real-time PCR was subsequently performed to quantify the cDNA using SYBR Premix Ex Taq (TaKaRa) in a CFX96™ real-time PCR detection system (BIO-RAD). TaActin was used as an internal control to normalize all data. The primers used were listed in Supplementary Table 1.
The wheat-specific HMM file for the response regulator receiver domain was aligned with the whole protein sequences in wheat, and 151 non-redundant TaRR genes in wheat were identified after receiver domain confirmation. An unrooted phylogenetic tree was generated by using the conserved receiver domain and incorporating the well-established family members from Arabidopsis and rice for the subfamily classification of TaRRs (Figure 1; Heyl et al., 2013). To confirm the subfamily classification, the protein sequences of all TaRRs were further analyzed for conserved domains, including the receiver domain, Myb-like DNA binding domain of RRBs, and CCT domain of PRRs. Among the 151 TaRR genes, there were 20 TaRRAs, 71 TaRRBs, 43 TaRRCs, and 17 TaPRRs (Supplementary Table 2), and each type of TaRR gene in wheat was found to be more abundant than the corresponding type of RR genes in Arabidopsis and rice (Table 1).
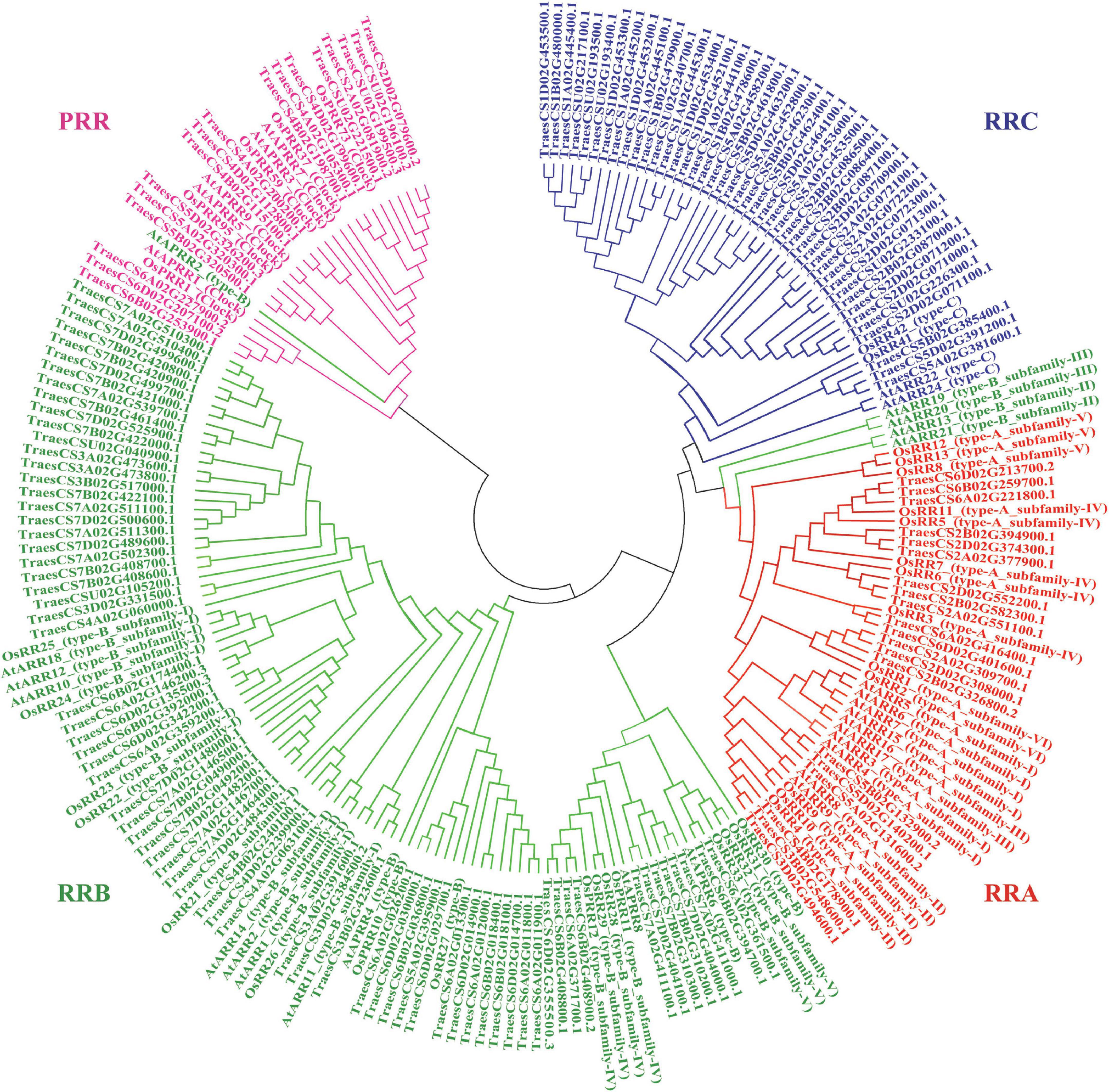
Figure 1. Phylogenetic analysis of RR proteins among the wheat, Arabidopsis, and rice. All RR proteins were allocated into four clades. Red, green, blue, and pink represent RRA, RRB, RRC, and PRR, respectively. The unrooted tree was established by the neighbor-joining method using the MEGA7.0 software with 1,000 bootstrap replicates.
Further phylogenetic analysis using the full-length protein sequence of type-A RRs in wheat clustered the 20 TaRRA genes into eight homologous groups (Figure 2A), which were named TaRRA1 to TaRRA8. TaRRA1, 2, 3, 6, and 7 had three orthologous genes (TaRRA1-A/B/D, TaRRA2-A/B/D, TaRRA3-A/B/D, TaRRA6-A/B/D, and TaRRA7-A/B/D), while TaRRA4 and 8 contained two orthologous genes (TaRRA4-B/D and TaRRA8-A/D) and TaRRA5 possessed only one gene copy (TaRRA5-B). All the proteins encoded by TaRRA genes varied from 108 to 269 amino acids with predicted molecular weights (MW) ranging from 12.22 to 28.89 kDa and the isoelectric points (PI) ranging from 5.05 to 8.48 (Table 2).
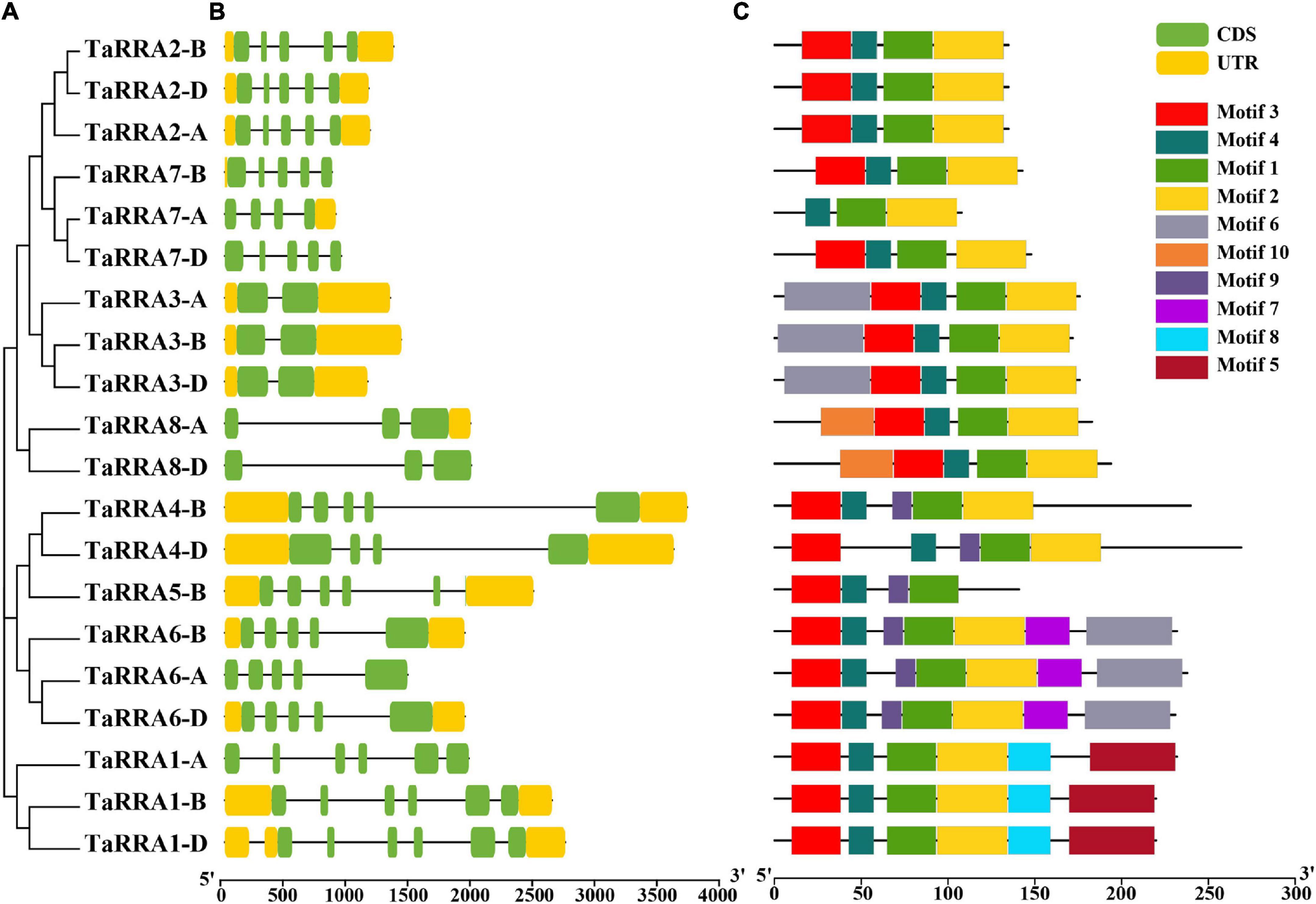
Figure 2. Phylogenetic analysis, gene structure, and conserved motifs of TaRRAs. (A) The phylogenetic tree was constructed based on the full-length amino acid sequence of TaRRAs. (B) Gene structure analysis of TaRRA genes. Yellow boxes represent UTR regions; green boxes indicate exons; black lines represent introns. (C) Conserved domain structures identified in the TaRRAs. Different color boxes show different motifs.
To gain further insights into the TaRRA gene members, we surveyed the gene structure and conserved motifs of each TaRRA. Although the lengths of genomic DNA varied from 859 to 3,709 bp in different TaRRAs, orthologous genes at each TaRRA locus usually had similar genomic DNA lengths. For instance, genomic DNA length of TaRRA7-A, TaRRA7-B, and TaRRA7-D was 888, 859, and 932 bp, respectively, while genomic DNA length of TaRRA4-B and TaRRA4-D was 3,709 and 3,604 bp, respectively (Figure 2B). Additionally, the number of exons, ranging from 2 to 6 in different TaRRAs, was generally the same in orthologous genes at each TaRRA locus. For example, TaRRA3-A, TaRRA3-B, and TaRRA3-D all harbored 2 exons, while TaRRA1-A, TaRRA1-B, and TaRRA1-D all contained 6 exons (Figure 2B and Table 2).
Using the MEME tool to predict the conserved protein motifs of the TaRRA family, 10 conversed motifs were identified (Figure 2C and Supplementary Table 3). Except for TaRRA5-B and TaRRA7-A, all TaRRAs contained motif 1, motif 2, motif 3, and motif 4. Additionally, TaRRA1-A, TaRRA1-B, and TaRRA1-D also possessed motif 5 and motif 8 (both unique in this group). TaRRA3-A, TaRRA3-B, and TaRRA3-D also had motif 6, while TaRRA4-B and TaRRA4-D had an added motif 9. TaRRA6-A, TaRRA6-B, and TaRRA6-D additionally contained motif 6, motif 7 (unique in this group), and motif 9. Likewise, TaRRA8-A and TaRRA8-D had motif 10 (unique in this group). In addition, all TaRRAs contained the highly conserved Lys and two Asp residues (D-D-K) in the receiver domain, except for TaRRA5-B and TaRRA7-A, which lacked the Lys and the first Asp respectively (Supplementary Figure 1). It is worth mentioning that all the 20 TaRRAs contained the predicted Asp phosphorylation site (the second Asp in the conserved D-D-K motif), which was embedded in a conserved TDY sequence. The above results indicate that orthologous TaRRA genes in the A, B, and D wheat subgenomes were usually similar in gene structure and encoding protein motifs, suggesting that TaRRA genes were conserved during evolution.
Chromosomal localization analysis showed that the 20 TaRRA genes were unevenly distributed on 12 of the 21 wheat chromosomes (Figure 3), with the number of TaRRA genes on each chromosome ranging from 1 (3B, 3D, 4B, 5A, 5B, 5D, and 6B) to 3 (2A, 2B, and 2D). Chromosomal group II harbored 9 (45.0%) TaRRA genes, the largest number, followed by chromosomal groups VI, V, III, and IV, which contained 5 (25.0%), 3 (15.0%), 2 (10.0%), and 1 (5.0%) TaRRA genes, respectively (Figure 3). Whereas, there was no TaRRA gene located in the rest two chromosomal groups (I and VII). Collinear relationship displayed the homology between TaRRAs (Figure 3), which was consistent with the phylogenetic analysis (Figure 2).
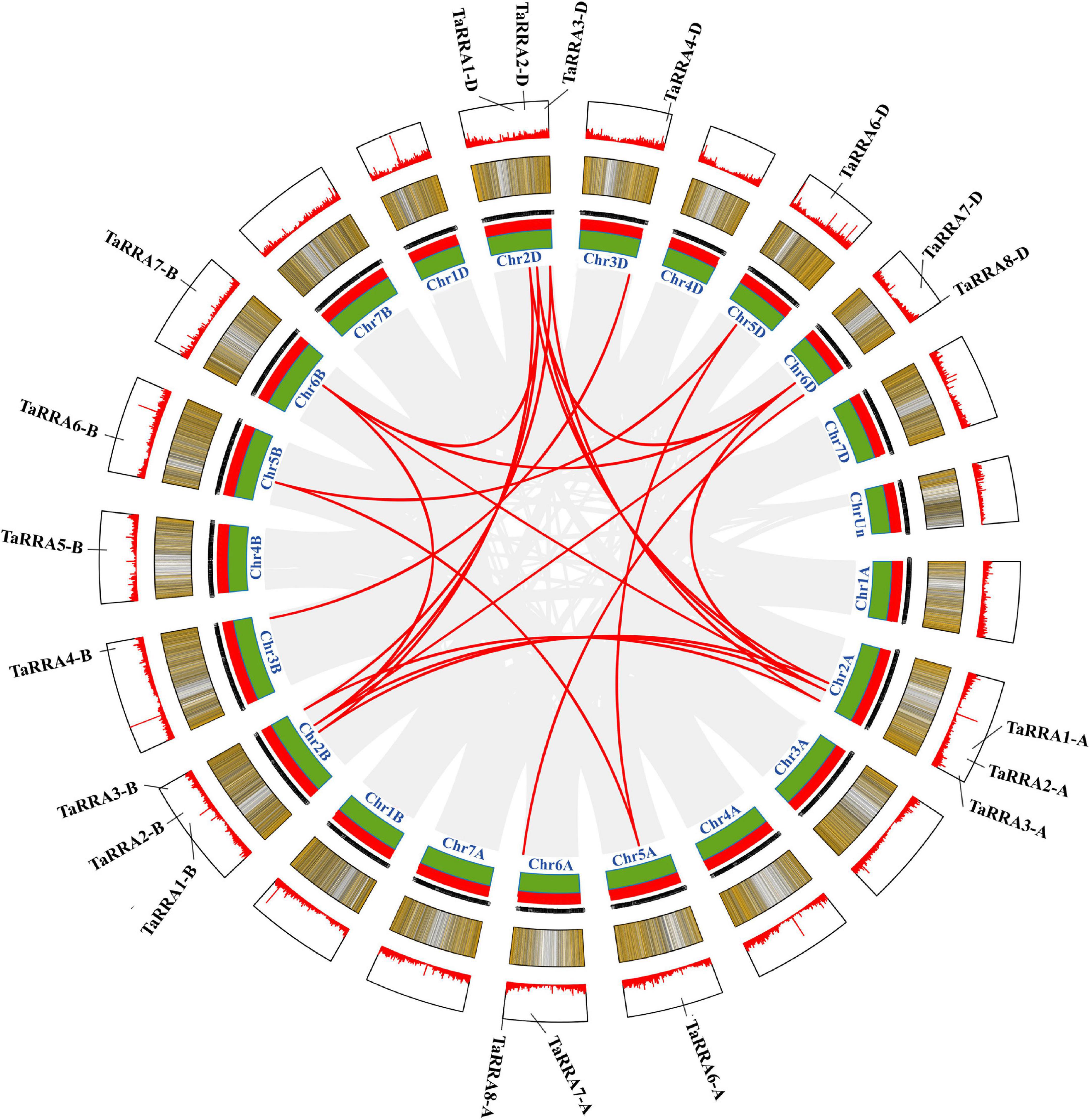
Figure 3. Schematic diagram of the inter-chromosomal relationships of TaRRA genes. Gray lines indicate all syntenic blocks in the wheat genome, and red lines indicate the presence of TaRRA genes.
Synteny analysis between the wheat and rice was conducted and 30 orthologous RRA gene pairs were found (Figure 4 and Supplementary Table 4), with a close similarity to the phylogenetic analysis (Figure 1), indicating that these syntenic gene pairs were relatively conserved during the evolution of gramineous species.
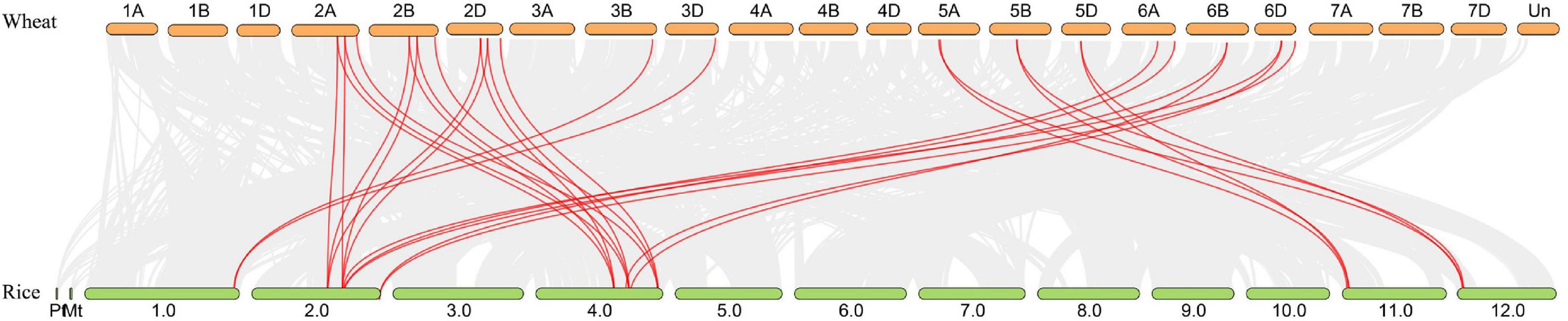
Figure 4. Synteny analysis of RRA genes in wheat and rice. Gray lines in the background indicate all syntenic blocks within the two genomes, while red lines highlight the syntenic RRA gene pairs.
The cis-acting elements in the promoter region play important roles in gene transcription regulation. The RRAs are cytokinin response genes that are targets of RRB transcription factors in Arabidopsis (To et al., 2004). Therefore, the 1,500 bp DNA sequence upstream of TaRRAs was analyzed for BA-dependent (+BA) and BA-independent (-BA) RRB binding cis-acting elements (Figure 5A and Supplementary Table 5; Xie et al., 2018). Both the BA-dependent and BA-independent RRB binding cis-acting elements were distributed widely throughout all of the TaRRA genes, suggesting that transcription of TaRRAs was probably regulated in part by RRBs.
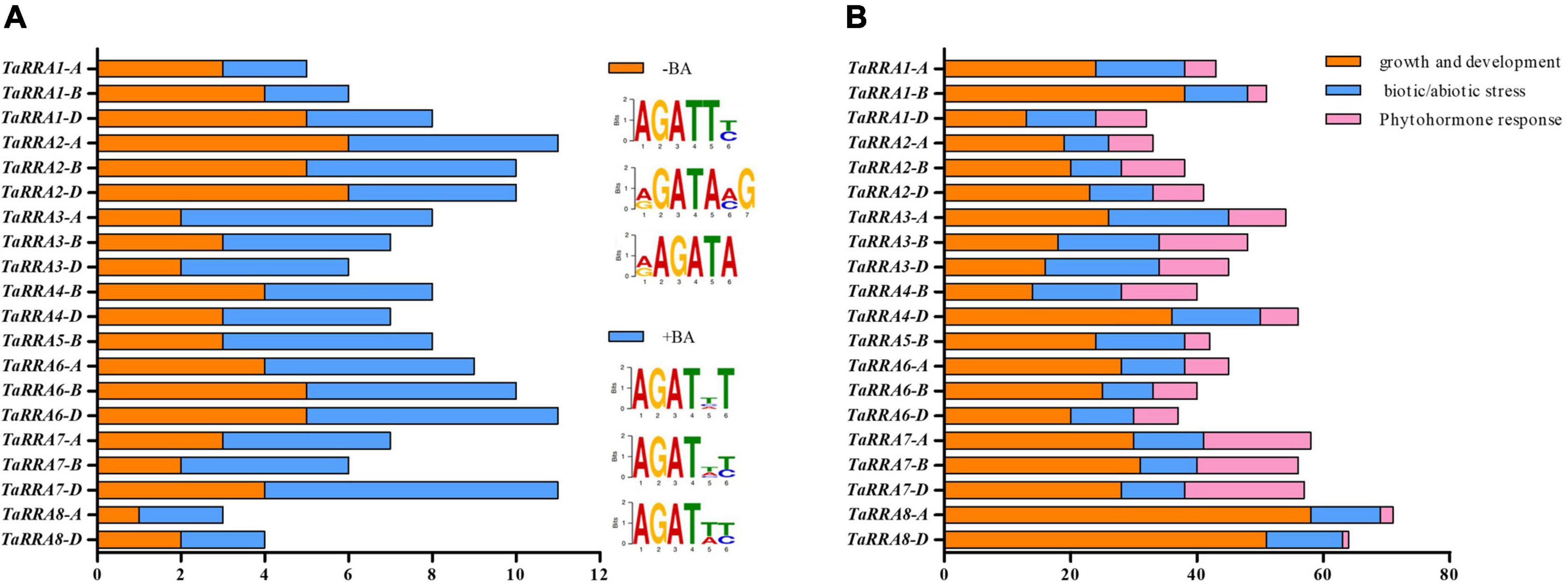
Figure 5. Prediction of cis-acting elements in the promoter region of TaRRAs. (A) BA-dependent (+BA) and BA-independent (-BA) RRB binding cis-acting elements in the promoter of TaRRAs. (B) Growth and development, biotic/abiotic stress, and phytohormone responses cis-acting elements in the promoter of TaRRAs.
To fully understand the potential role of TaRRAs, the 1,500 bp promoter region of TaRRAs was further analyzed in the PlantCARE database for more cis-acting elements. A total of 43 types of cis-acting elements with known functions were identified in the TaRRAs promoter region, which were divided into three different categories, i.e., growth and development response elements, biotic/abiotic stress response elements, and phytohormone response elements (Figure 5B and Supplementary Tables 6, 7). Among the growth and development response cis-elements, CAAT-box (common cis-acting element in promoter and enhancer regions) and TATA-box (core promoter element around -30 of transcription start) were highly enriched in all the TaRRA promoters. In addition, O2-site involved in zein metabolism regulation, RY-element involved in seed-specific regulation, and CAT-box related to meristem expression were identified in some of the TaRRA promoters. Among the biotic/abiotic stress response cis-elements, the proportion of light-response elements was large, including 19 cis-regulatory factors, such as G-box, Sp1, and TCCC-motif. Additionally, ARE and GC-motif involved in anaerobic induction and LTR and MBS elements involved in low-temperature and drought responsiveness, respectively, were found in several TaRRA promoters. In the phytohormone response category, ABRE in ABA response and CGTCA-motif and TGACG-motif in MeJA response were distributed in most of the TaRRA promoters. Moreover, GARE-motif, P-box, and TATC-box implicated in gibberellin response, AuxRR-core and TGA-element in auxin response, and TCA-element in salicylic acid response were also distributed in several TaRRA promoters. The presence of different numbers and types of cis-acting elements in TaRRA promoters indicates that these genes may be involved in different regulatory mechanisms.
To characterize the expression profiles of the TaRRA gene family, we analyzed the RNA-seq data downloaded from the WheatOmics 1.0 (Ma et al., 2021). Notably, TaRRA2-A/B/D and TaRRA7-A/B/D were hardly detected in any of the tissues tested, while other TaRRAs showed relatively high expression in the root, except for TaRRA1-A/B/D with higher expression in the stem (Figure 6A).
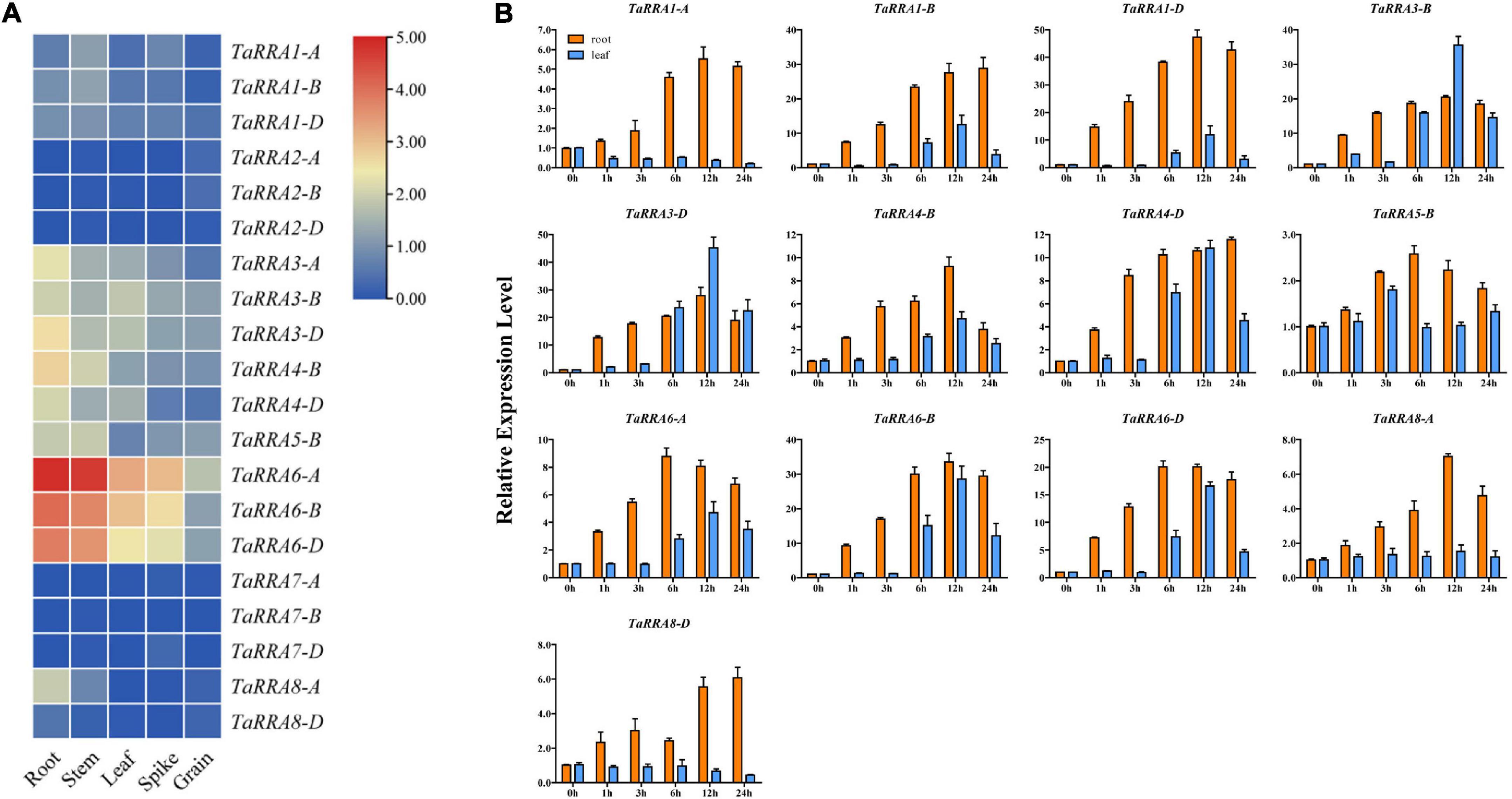
Figure 6. Expression analysis of TaRRA genes. (A) Expression profiles of TaRRA genes in various tissues. The RNA-sequence data was obtained from leaf, root, stem, spike, and grain of Chinese Spring (International Wheat Genome Sequencing Consortium [IWGSC], 2014). (B) Quantitative RT-PCR analysis of TaRRA genes in response to cytokinin treatment. Leaves and roots of 10-day-old seedlings were sampled after 50 μM 6-BA treatment for 0, 1, 3, 6, 12, and 24 h. The data are given as means ± SE of three biological replicates.
Since most of the RRAs are rapidly induced by exogenous cytokinin in monocots and dicots (D’Agostino et al., 2000; Jain et al., 2006), we investigated the expression profiles of TaRRAs in response to cytokinin treatment by qRT-PCR (Figure 6B). In the root, 10 out of 13 detectable TaRRAs were up-regulated after 1 h BA treatment and displayed an increasing expression trend afterward. Most of the TaRRAs reached maximal induction at 12 h, except for TaRRA5-B and TaRRA6-A, which were maximally induced at 6 h, and TaRRA1-B, TaRRA4-D, and TaRRA8-D, which were maximally induced at 24 h. Although all the 13 detectable TaRRAs were cytokinin-induced, their fold-change varied greatly after BA treatment, ranging from 2.6 (TaRRA5-B, 6 h) to 47.3 (TaRRA1-D, 12 h) times compared with the control (0 h). It is worth noting that expression levels of orthologous genes at each TaRRA locus could differ significantly after BA treatment. For instance, TaRRA1-D was up-regulated by 47.3 times at 12 h, whereas TaRRA1-A was up-regulated by 5.5 times. In the leaf, 9 out of 13 detectable TaRRAs displayed an obvious increase in the transcription level after 6 h BA treatment and showed maximal induction at 12 h. However, there was no increase in the transcription levels of TaRRA5-B and TaRRA8-A, and a decrease was observed in the transcription levels of TaRRA1-A and TaRRA8-D. The above results showed that most TaRRAs responded more rapidly and strongly to BA treatment in the root than that in the leaf. However, we could hardly detect the expression of TaRRA2-A/B/D and TaRRA7-A/B/D by qRT-PCR, which was consistent with previous RNA-seq data (Figure 6A).
To further evaluate the potential function of TaRRAs in response to abiotic stress, the TaRRA gene expression patterns were analyzed by qRT-PCR under drought, salt, cold, and heat stress treatments (Figures 7, 8). The results showed that under drought stress, the expression levels of TaRRA1-B, TaRRA3-B, TaRRA3-D, TaRRA4-D, and TaRRA8-A decreased continuously from 0 to 6 h and later increased marginally at 12 and 24 h (Figure 7A). TaRRA8-D was exclusively up-regulated by drought stress, while the expression of the rest of the TaRRAs showed insignificant changes under drought stress. Under salt stress, the expression levels of most TaRRAs were up-regulated at least at one time point (Figure 7B). Notably, TaRRA1-D, TaRRA3-D, TaRRA6-B, and TaRRA6-D were induced at each time point compared with the control (0 h). Inversely, TaRRA5-B was gradually down-regulated by salt stress. Under cold stress, the expression of 8 TaRRAs increased after 6 h treatment, among which the expression of TaRRA1-D, TaRRA4-B, TaRRA6-A, TaRRA6-B, and TaRRA6-D continued to increase till 12 h, whereas the expression of TaRRA3-B, TaRRA3-D, and TaRRA8-D was reduced subsequently (Figure 8A). Heat significantly inhibited the expression of most TaRRAs after 3 h treatment, except for that of TaRRA8-A and TaRRA8-D, which increased after 1 h treatment (Figure 8B).
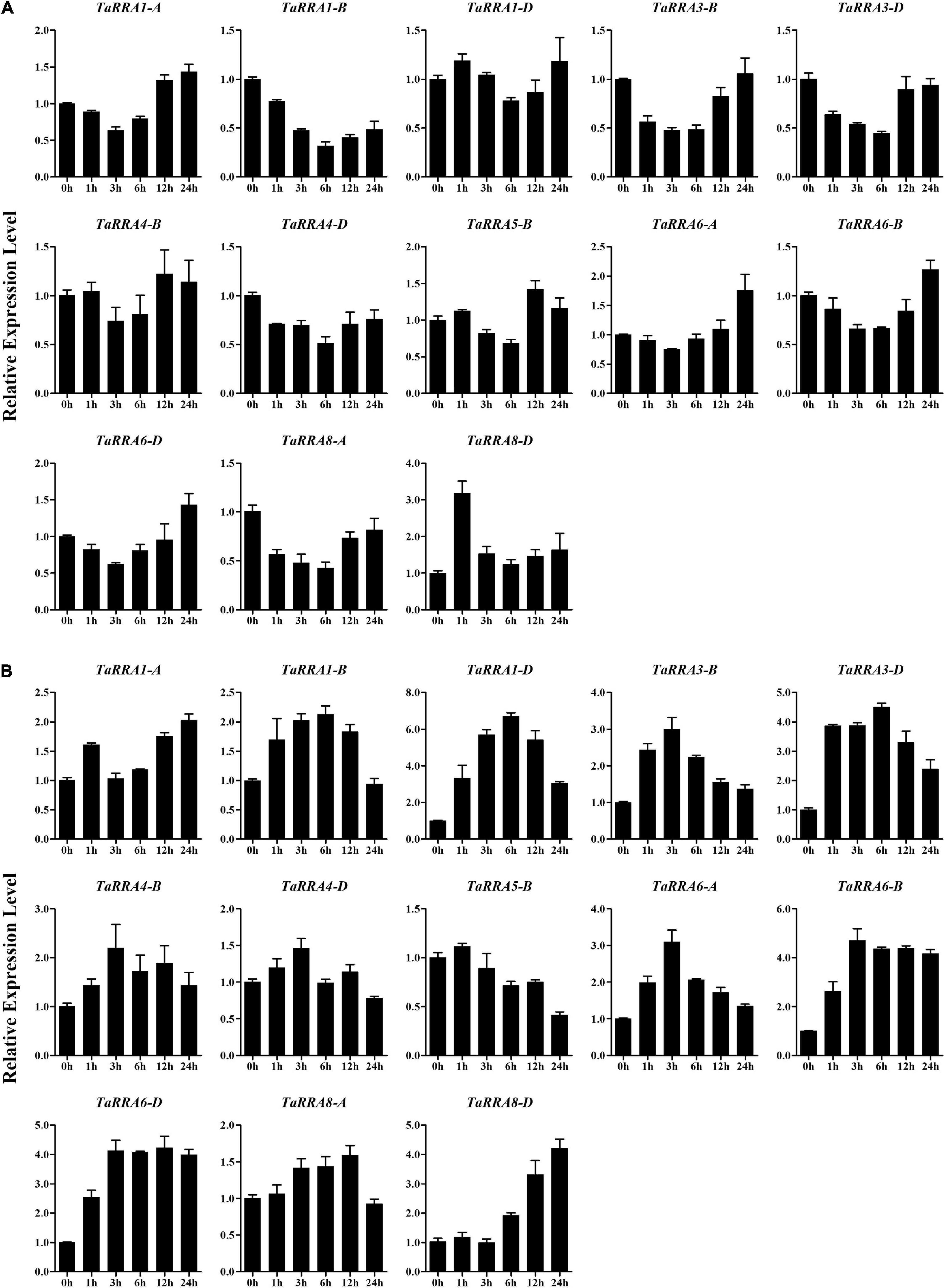
Figure 7. Quantitative RT-PCR analysis of TaRRA genes under drought and salt stress. Roots of 10-day-old seedlings were sampled after 0, 1, 3, 6, 12, and 24 h drought stress (A) and salt stress (B). The data are given as means ± SE of three biological replicates.
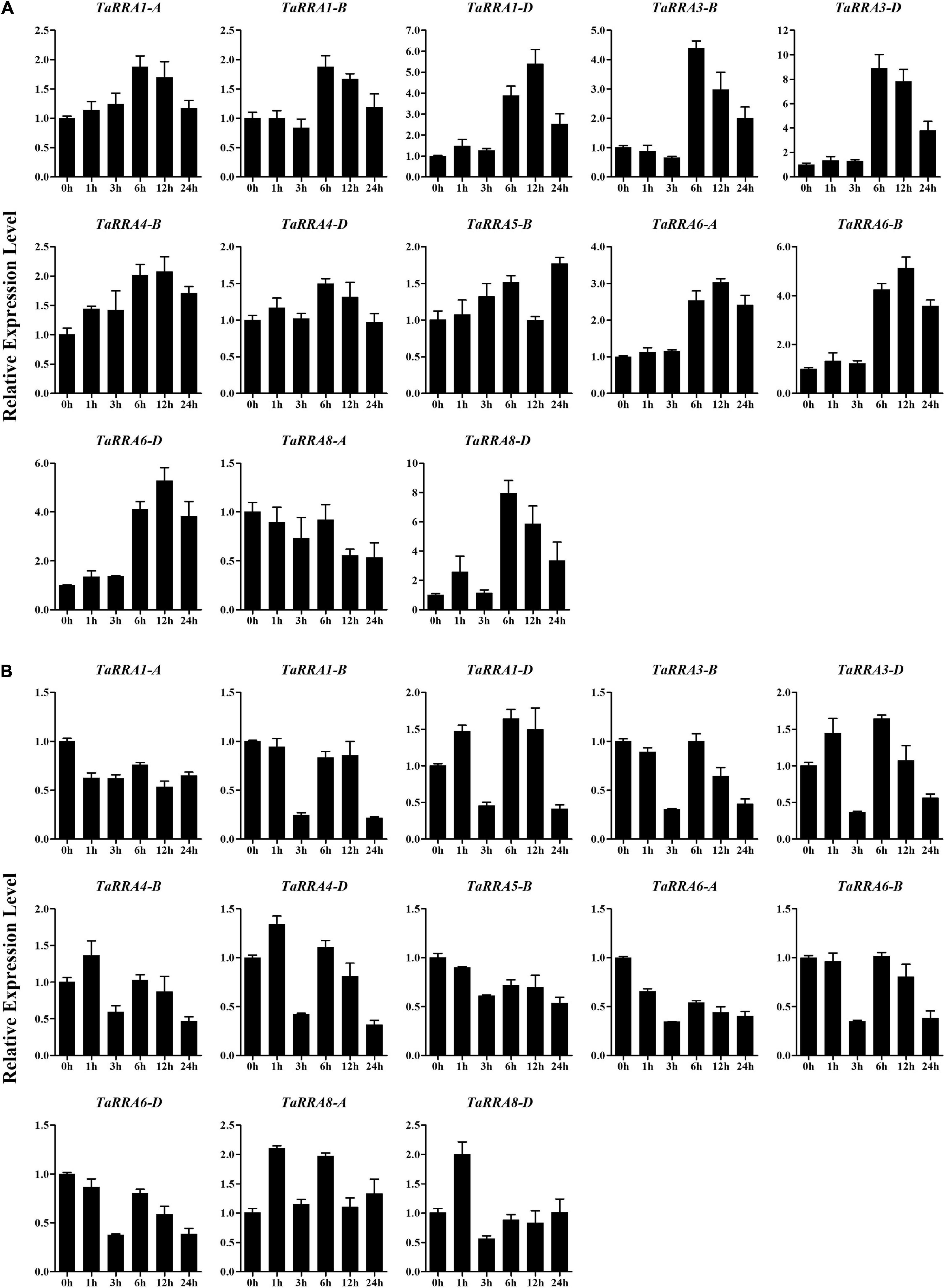
Figure 8. Quantitative RT-PCR analysis of TaRRA genes under cold and heat stress. Roots of 10-day-old seedlings were sampled after 0, 1, 3, 6, 12, and 24 h cold stress (A) and heat stress (B). The data are given as means ± SE of three biological replicates.
It is worth noting that some orthologous genes at each TaRRA locus (TaRRA3-B/D and TaRRA6-A/B/D) exhibited similar expression patterns in response to abiotic stress, indicating that they may have similar biological functions under stress conditions. On the contrary, TaRRA8-A and TaRRA8-D showed different expression patterns under drought and cold stress, suggesting that they could play different roles under some abiotic stresses. In general, all the TaRRAs showed significant changes in response to at least one abiotic stress.
RRAs are rapidly induced by exogenous cytokinin and are thus considered to be primary cytokinin response genes (D’Agostino et al., 2000; Jain et al., 2006). The RRA gene family, a subfamily of the RR gene family, is relatively small in higher plants, with only 10, 13, 21, and 18 members in Arabidopsis, rice, maize, and soybean, respectively (Mochida et al., 2010; Chu et al., 2011; Heyl et al., 2013). In this study, we performed a systematic genome-wide analysis of the wheat RRA gene family by combining two different approaches. First, we performed phylogenetic analysis by using the conserved receiver domain of 151 TaRRs, and simultaneously adding the well-established family members from Arabidopsis and rice into the analysis (Figure 1; Heyl et al., 2013). Second, we analyzed the conserved domains of all the TaRRs to further confirm whether they contain a Myb-like DNA binding domain or a CCT domain in addition to the receiver domain. We used the protein sequence of the longest transcript of each TaRR for domain analysis to avoid domain missing of the shorter transcript. According to the above method, we identified 20 RRAs, 71 RRBs, 43 RRCs, and 17 PRRs from the wheat reference genome (Table 1). However, in a previous study, 41 RRAs, 2 RRBs, and 2 PRRs have been identified in wheat (Gahlaut et al., 2014). Given that RRAs only carry a receiver domain, while RRBs contain an additional Myb-like DNA binding domain and PRRs have an extra CCT domain, some truncated RRBs and PRRs may be identified as RRAs due to the earlier incomplete wheat reference genome. In recent years, great progress has been made in genome sequencing, assembly, and annotation of wheat (International Wheat Genome Sequencing Consortium [IWGSC], 2018; Alonge et al., 2020; Zhu et al., 2021), which provides a high-quality reference genome for the study of the TaRRA gene family. The 20 TaRRA genes belonging to 8 homologous groups were unequally distributed on 12 wheat chromosomes (Figure 3). As wheat is an allohexaploid (AABBDD), most TaRRA genes had homologs in the A, B, and D subgenomes due to polyploidization, sharing similar gene structures and protein motifs within the homologous group (Figure 2).
Most of the RRAs are rapidly induced by exogenous cytokinin in plants (D’Agostino et al., 2000; Asakura et al., 2003; Jain et al., 2006). Consistent with previous results, the transcription levels of most detectable TaRRAs displayed an obvious increase after BA treatment in both the root and leaf (Figure 6B). However, the transcripts of most TaRRAs reached a maximal induction at 12 h cytokinin treatment, while most RRAs in Arabidopsis and rice showed maximal induction within 1 h cytokinin treatment and then gradually declined (D’Agostino et al., 2000; Jain et al., 2006). Although TaRRA1-A, TaRRA5-B, TaRRA8-A, and TaRRA8-D were induced by cytokinin in the root, their transcription levels were unchanged or even decreased in the leaf after cytokinin treatment (Figure 6B), indicating their potential function differentiation between root and leaf. Similarly, there is no significant change in the transcript abundance of a few RRAs in Arabidopsis and rice following cytokinin treatment, including ARR8, ARR9, OsRR3, and OsRR8 (D’Agostino et al., 2000; Jain et al., 2006). The rapid induction of RRAs by exogenous cytokinin has been shown to mediate a feedback mechanism, probably by competing with RRBs for the phosphotransfer from HPs. Phosphorylation of RRAs increases their protein stability (To et al., 2007), whereas phosphorylation of RRBs enables them to bind to DNA, thus initiating transcription of downstream targets (Kim et al., 2006; Zubo et al., 2017). Double and higher-order type-A arr mutants, rather than the single arr mutants, show increasing sensitivity to cytokinin, indicating that RRAs act as negative regulators of cytokinin signaling with partially redundant functions (To et al., 2007).
Cytokinin signaling components are widely involved in plant response to abiotic stress. A series of Arabidopsis mutants, including ahk2,3, ahp2,3,5, and arr1,10,12, show significantly increased drought and salt tolerance, indicating that HKs, HPs, and RRBs are negative regulators of these stress responses (Tran et al., 2007; Nishiyama et al., 2013; Nguyen et al., 2016; Abdelrahman et al., 2021). In contrast, plants overexpressing ARR5 exhibit enhanced drought tolerance, suggesting that type-A ARR5 is a positive regulator of drought tolerance (Huang et al., 2018). Functional analysis of the single and double ahk mutants indicates that HKs function partially redundantly as negative regulators of the cold stress adaptation response (Jeon et al., 2010). However, type-B ARR1 is a positive factor in cold signaling, because arr1 shows reduced cold resistance, whereas ARR1 overexpression increases plant cold resistance (Jeon and Kim, 2013). Additionally, RRAs also play important roles in cold stress signaling (Jeon et al., 2010; Shi et al., 2012).
Given that the expression pattern of genes has a correlation with its function, we monitored the expression profiles of TaRRA family members under multiple stresses. Drought stress reduced the expression of TaRRA1-B, TaRRA3-B, TaRRA3-D, TaRRA4-D, and TaRRA8-A, but induced the expression of TaRRA8-D (Figure 7A). Similarly, drought stress reduces the expression of ARR8 and ARR17, whereas induces the expression of ARR5, ARR7, and ARR15 in Arabidopsis (Kang et al., 2012). We identified 8 TaRRAs with significantly up-regulated expression under cold stress (Figure 8A), which is consistent with the cold-induced changes in ARR5, ARR6, ARR7, and ARR15 expression in Arabidopsis (Jeon et al., 2010). Furthermore, overexpression of ARR5, ARR7, and ARR15 enhances the freezing tolerance of plants, while arr5, arr6, and arr7 also lead to higher freezing tolerance (Jeon et al., 2010; Shi et al., 2012). These different results suggest the complexity of the molecular mechanism involved and hence further research is warranted. In addition, a considerable number of TaRRAs were shown to respond to salt and heat stress (Figures 7B, 8B), suggesting that they are promising regulators for salt and heat response in wheat. TaRRA3-B/D, orthologs of drought and salt positive regulator OsRR6, and TaRRA6-A/B/D, orthologs of salt negative regulator OsRR9 and OsRR10 (Supplementary Table 4), showed responses to salt, cold, and heat stress, which may be important candidate genes for genetic improvement of stress tolerance in wheat.
In conclusion, this study provided comprehensive insights into the TaRRA gene family in wheat. The systematical identification and investigation of the TaRRA gene family will inevitably contribute to further elucidation of the biological function and genetic improvement application of TaRRAs in wheat.
The original contributions presented in this study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
HL and YJZ designed the experiments. LS, LL, JZ, MH, YLZ, YZ, XT, PW, QL, and XC performed the experiments and analyzed the data. LS and LL wrote the manuscript. All authors reviewed and approved the manuscript.
This work was funded by the S&T Program of Hebei (C2019301089 and 20326345D), the HAAFS Science and Technology Innovation Special Project (2022KJCXZX-LYS-1), the Basic Research Funds of Hebei Academy of Agriculture and Forestry Sciences (2018060304), the Hebei Modern Agricultural Industrial Technology System (HBCT2018010201), and the Project for Hebei Scientific and Technological Innovation Team of Modern Wheat Seed Industry (21326318D).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1006409/full#supplementary-material
Abdelrahman, M., Nishiyama, R., Tran, C. D., Kusano, M., Nakabayashi, R., Okazaki, Y., et al. (2021). Defective cytokinin signaling reprograms lipid and flavonoid gene-to-metabolite networks to mitigate high salinity in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 118:e2105021118. doi: 10.1073/pnas.2105021118
Alonge, M., Shumate, A., Puiu, D., Zimin, A. V., and Salzberg, S. L. (2020). Chromosome-scale assembly of the bread wheat genome reveals thousands of additional gene copies. Genetics 216, 599–608. doi: 10.1534/genetics.120.303501
Argueso, C. T., Raines, T., and Kieber, J. J. (2010). Cytokinin signaling and transcriptional networks. Curr. Opin. Plant Biol. 13, 533–539. doi: 10.1016/j.pbi.2010.08.006
Argyros, R. D., Mathews, D. E., Chiang, Y. H., Palmer, C. M., Thibault, D. M., Etheridge, N., et al. (2008). Type B response regulators of Arabidopsis play key roles in cytokinin signaling and plant development. Plant Cell 20, 2102–2116. doi: 10.1105/tpc.108.059584
Asakura, Y., Hagino, T., Ohta, Y., Aoki, K., Yonekura-Sakakibara, K., Deji, A., et al. (2003). Molecular characterization of His-Asp phosphorelay signaling factors in maize leaves: Implications of the signal divergence by cytokinin-inducible response regulators in the cytosol and the nuclei. Plant Mol. Biol. 52, 331–341. doi: 10.1023/a:1023971315108
Bhaskar, A., Paul, L. K., Sharma, E., Jha, S., Jain, M., and Khurana, J. P. (2021). OsRR6, a type-A response regulator in rice, mediates cytokinin, light and stress responses when over-expressed in Arabidopsis. Plant Physiol. Biochem. 161, 98–112. doi: 10.1016/j.plaphy.2021.01.047
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chu, Z. X., Ma, Q., Lin, Y. X., Tang, X. L., Zhou, Y. Q., Zhu, S. W., et al. (2011). Genome-wide identification, classification, and analysis of two-component signal system genes in maize. Genet. Mol. Res. 10, 3316–3330. doi: 10.4238/2011.December.8.3
D’Agostino, I. B., Deruère, J., and Kieber, J. J. (2000). Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol. 124, 1706–1717. doi: 10.1104/pp.124.4.1706
El-Showk, S., Ruonala, R., and Helariutta, Y. (2013). Crossing paths: Cytokinin signalling and crosstalk. Development 140, 1373–1383. doi: 10.1242/dev.086371
Gahlaut, V., Mathur, S., Dhariwal, R., Khurana, J. P., Tyagi, A. K., Balyan, H. S., et al. (2014). A multi-step phosphorelay two-component system impacts on tolerance against dehydration stress in common wheat. Funct. Integr. Genomics 14, 707–716. doi: 10.1007/s10142-014-0398-8
Ha, S., Vankova, R., Yamaguchi-Shinozaki, K., Shinozaki, K., and Tran, L. S. (2012). Cytokinins: Metabolism and function in plant adaptation to environmental stresses. Trends Plant Sci. 17, 172–179. doi: 10.1016/j.tplants.2011.12.005
Heyl, A., Brault, M., Frugier, F., Kuderova, A., Lindner, A. C., Motyka, V., et al. (2013). Nomenclature for members of the two-component signaling pathway of plants. Plant Physiol. 161, 1063–1065. doi: 10.1104/pp.112.213207
Hosoda, K., Imamura, A., Katoh, E., Hatta, T., Tachiki, M., Yamada, H., et al. (2002). Molecular structure of the GARP family of plant Myb-related DNA binding motifs of the Arabidopsis response regulators. Plant Cell 14, 2015–2029. doi: 10.1105/tpc.002733
Huang, X., Hou, L., Meng, J., You, H., Li, Z., Gong, Z., et al. (2018). The antagonistic action of abscisic acid and cytokinin signaling mediates drought stress response in Arabidopsis. Mol. Plant 11, 970–982. doi: 10.1016/j.molp.2018.05.001
Hwang, I., Sheen, J., and Müller, B. (2012). Cytokinin signaling networks. Annu. Rev. Plant Biol. 63, 353–380. doi: 10.1146/annurev-arplant-042811-105503
International Wheat Genome Sequencing Consortium [IWGSC] (2014). A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345:1251788. doi: 10.1126/science.1251788
International Wheat Genome Sequencing Consortium [IWGSC] (2018). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361:eaar7191. doi: 10.1126/science.aar7191
Jain, M., Tyagi, A. K., and Khurana, J. P. (2006). Molecular characterization and differential expression of cytokinin-responsive type-A response regulators in rice (Oryza sativa). BMC Plant Biol. 6:1. doi: 10.1186/1471-2229-6-1
Jeon, J., and Kim, J. (2013). Arabidopsis response regulator1 and Arabidopsis histidine phosphotransfer protein2 (AHP2), AHP3, and AHP5 function in cold signaling. Plant Physiol. 161, 408–424. doi: 10.1104/pp.112.207621
Jeon, J., Kim, N. Y., Kim, S., Kang, N. Y., Novák, O., Ku, S. J., et al. (2010). A subset of cytokinin two-component signaling system plays a role in cold temperature stress response in Arabidopsis. J. Biol. Chem. 285, 23371–23386. doi: 10.1074/jbc.M109.096644
Kang, N. Y., Cho, C., Kim, N. Y., and Kim, J. (2012). Cytokinin receptor-dependent and receptor-independent pathways in the dehydration response of Arabidopsis thaliana. J. Plant Physiol. 169, 1382–1391. doi: 10.1016/j.jplph.2012.05.007
Kieber, J. J., and Schaller, G. E. (2018). Cytokinin signaling in plant development. Development 145:dev149344. doi: 10.1242/dev.149344
Kim, H. J., Ryu, H., Hong, S. H., Woo, H. R., Lim, P. O., Lee, I. C., et al. (2006). Cytokinin-mediated control of leaf longevity by AHK3 through phosphorylation of ARR2 in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 103, 814–819. doi: 10.1073/pnas.0505150103
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Leibfried, A., To, J. P., Busch, W., Stehling, S., Kehle, A., Demar, M., et al. (2005). WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 438, 1172–1175. doi: 10.1038/nature04270
Liu, Y., Zhang, M., Meng, Z., Wang, B., and Chen, M. (2020). Research progress on the roles of cytokinin in plant response to stress. Int. J. Mol. Sci. 21:6574. doi: 10.3390/ijms21186574
Ma, S., Wang, M., Wu, J., Guo, W., Chen, Y., Li, G., et al. (2021). WheatOmics: A platform combining multiple omics data to accelerate functional genomics studies in wheat. Mol. Plant 14, 1965–1968. doi: 10.1016/j.molp.2021.10.006
Makino, S., Kiba, T., Imamura, A., Hanaki, N., Nakamura, A., Suzuki, T., et al. (2000). Genes encoding pseudo-response regulators: Insight into His-to-Asp phosphorelay and circadian rhythm in Arabidopsis thaliana. Plant Cell Physiol. 41, 791–803. doi: 10.1093/pcp/41.6.791
Mira-Rodado, V., Sweere, U., Grefen, C., Kunkel, T., Fejes, E., Nagy, F., et al. (2007). Functional cross-talk between two-component and phytochrome B signal transduction in Arabidopsis. J. Exp. Bot. 58, 2595–2607. doi: 10.1093/jxb/erm087
Mochida, K., Yoshida, T., Sakurai, T., Yamaguchi-Shinozaki, K., Shinozaki, K., and Tran, L. S. (2010). Genome-wide analysis of two-component systems and prediction of stress-responsive two-component system members in soybean. DNA Res. 17, 303–324. doi: 10.1093/dnares/dsq021
Nguyen, K. H., Ha, C. V., Nishiyama, R., Watanabe, Y., Leyva-González, M. A., Fujita, Y., et al. (2016). Arabidopsis type B cytokinin response regulators ARR1, ARR10, and ARR12 negatively regulate plant responses to drought. Proc. Natl. Acad. Sci. U.S.A. 113, 3090–3095. doi: 10.1073/pnas.1600399113
Nishiyama, R., Watanabe, Y., Leyva-Gonzalez, M. A., Van Ha, C., Fujita, Y., Tanaka, M., et al. (2013). Arabidopsis AHP2, AHP3, and AHP5 histidine phosphotransfer proteins function as redundant negative regulators of drought stress response. Proc. Natl. Acad. Sci. U.S.A. 110, 4840–4845. doi: 10.1073/pnas.1302265110
Pavlů, J., Novák, J., Koukalová, V., Luklová, M., Brzobohatý, B., and Černý, M. (2018). Cytokinin at the crossroads of abiotic stress signalling pathways. Int. J. Mol. Sci. 19:2450. doi: 10.3390/ijms19082450
Sakai, H., Aoyama, T., and Oka, A. (2000). Arabidopsis ARR1 and ARR2 response regulators operate as transcriptional activators. Plant J. 24, 703–711. doi: 10.1111/j.1365-313X.2000.00909.x
Salomé, P. A., To, J. P., Kieber, J. J., and McClung, C. R. (2006). Arabidopsis response regulators ARR3 and ARR4 play cytokinin-independent roles in the control of circadian period. Plant Cell 18, 55–69. doi: 10.1105/tpc.105.037994
Schaller, G. E., Bishopp, A., and Kieber, J. J. (2015). The yin-yang of hormones: Cytokinin and auxin interactions in plant development. Plant Cell 27, 44–63. doi: 10.1105/tpc.114.133595
Schaller, G. E., Kieber, J. J., and Shiu, S. H. (2008). Two-component signaling elements and histidyl-aspartyl phosphorelays. Arabidopsis Book 2008:e0112. doi: 10.1199/tab.0112
Shi, Y., Tian, S., Hou, L., Huang, X., Zhang, X., Guo, H., et al. (2012). Ethylene signaling negatively regulates freezing tolerance by repressing expression of CBF and type-A ARR genes in Arabidopsis. Plant Cell 24, 2578–2595. doi: 10.1105/tpc.112.098640
Srivastava, A. K., Dutta, S., and Chattopadhyay, S. (2019). MYC2 regulates ARR16, a component of cytokinin signaling pathways, in Arabidopsis seedling development. Plant Direct 3:e00177. doi: 10.1002/pld3.177
Sweere, U., Eichenberg, K., Lohrmann, J., Mira-Rodado, V., Bäurle, I., Kudla, J., et al. (2001). Interaction of the response regulator ARR4 with phytochrome B in modulating red light signaling. Science 294, 1108–1111. doi: 10.1126/science.1065022
To, J. P., Deruère, J., Maxwell, B. B., Morris, V. F., Hutchison, C. E., Ferreira, F. J., et al. (2007). Cytokinin regulates type-A Arabidopsis response regulator activity and protein stability via two-component phosphorelay. Plant Cell 19, 3901–3914. doi: 10.1105/tpc.107.052662
To, J. P., Haberer, G., Ferreira, F. J., Deruère, J., Mason, M. G., Schaller, G. E., et al. (2004). Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell 16, 658–671. doi: 10.1105/tpc.018978
Tran, L. S., Urao, T., Qin, F., Maruyama, K., Kakimoto, T., Shinozaki, K., et al. (2007). Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 104, 20623–20628. doi: 10.1073/pnas.0706547105
Wang, W. C., Lin, T. C., Kieber, J., and Tsai, Y. C. (2019). Response Regulators 9 and 10 negatively regulate salinity tolerance in rice. Plant Cell Physiol. 60, 2549–2563. doi: 10.1093/pcp/pcz149
Werner, T., and Schmülling, T. (2009). Cytokinin action in plant development. Curr. Opin. Plant Biol. 12, 527–538. doi: 10.1016/j.pbi.2009.07.002
Wybouw, B., and De Rybel, B. (2019). Cytokinin – a developing story. Trends Plant Sci. 24, 177–185. doi: 10.1016/j.tplants.2018.10.012
Xie, M., Chen, H., Huang, L., O’Neil, R. C., Shokhirev, M. N., and Ecker, J. R. (2018). A B-ARR-mediated cytokinin transcriptional network directs hormone cross-regulation and shoot development. Nat. Commun. 9:1604. doi: 10.1038/s41467-018-03921-6
Zeng, R., Li, Z., Shi, Y., Fu, D., Yin, P., Cheng, J., et al. (2021). Natural variation in a type-A response regulator confers maize chilling tolerance. Nat. Commun. 12:4713. doi: 10.1038/s41467-021-25001-y
Zhao, Z., Andersen, S. U., Ljung, K., Dolezal, K., Miotk, A., Schultheiss, S. J., et al. (2010). Hormonal control of the shoot stem-cell niche. Nature 465, 1089–1092. doi: 10.1038/nature09126
Zhu, T., Wang, L., Rimbert, H., Rodriguez, J. C., Deal, K. R., De Oliveira, R., et al. (2021). Optical maps refine the bread wheat Triticum aestivum cv. Chinese Spring genome assembly. Plant J. 107, 303–314. doi: 10.1111/tpj.15289
Zubo, Y. O., Blakley, I. C., Yamburenko, M. V., Worthen, J. M., Street, I. H., Franco-Zorrilla, J. M., et al. (2017). Cytokinin induces genome-wide binding of the type-B response regulator ARR10 to regulate growth and development in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 114, E5995–E6004. doi: 10.1073/pnas.1620749114
Keywords: type-A response regulators, gene family, wheat (Triticum aestivum L.), expression pattern, cytokinin, abiotic stress
Citation: Sun L, Lv L, Zhao J, Hu M, Zhang Y, Zhao Y, Tang X, Wang P, Li Q, Chen X, Li H and Zhang Y (2022) Genome-wide identification and expression analysis of the TaRRA gene family in wheat (Triticum aestivum L.). Front. Plant Sci. 13:1006409. doi: 10.3389/fpls.2022.1006409
Received: 29 July 2022; Accepted: 12 August 2022;
Published: 30 August 2022.
Edited by:
Cheng Liu, Shandong Academy of Agricultural Sciences, ChinaReviewed by:
Shuanghe Cao, Institute of Crop Sciences (CAAS), ChinaCopyright © 2022 Sun, Lv, Zhao, Hu, Zhang, Zhao, Tang, Wang, Li, Chen, Li and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hui Li, endzbGlodWlAMTYzLmNvbQ==; Yingjun Zhang, emhhbmd5aW5nanVuMTk3N0AxNjMuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.