
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 14 December 2022
Sec. Plant Breeding
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.1006264
 Karina Y. Morales1,2†
Karina Y. Morales1,2† Aya H. Bridgeland1†
Aya H. Bridgeland1† Kater D. Hake3
Kater D. Hake3 Joshua A. Udall1
Joshua A. Udall1 Michael J. Thomson2
Michael J. Thomson2 John Z. Yu1*
John Z. Yu1*Upland cotton (Gossypium hirsutum L.) accounts for more than 90% of the world’s cotton production, providing natural material for the textile and oilseed industries worldwide. One strategy for improving upland cotton yields is through increased adoption of hybrids; however, emasculation of cotton flowers is incredibly time-consuming and genetic sources of cotton male sterility are limited. Here we review the known biochemical modes of plant nuclear male sterility (NMS), often known as plant genetic male sterility (GMS), and characterized them into four groups: transcriptional regulation, splicing, fatty acid transport and processing, and sugar transport and processing. We have explored protein sequence homology from 30 GMS genes of three monocots (maize, rice, and wheat) and three dicots (Arabidopsis, soybean, and tomato). We have analyzed evolutionary relationships between monocot and dicot GMS genes to describe the relative similarity and relatedness of these genes identified. Five were lowly conserved to their source species, four unique to monocots, five unique to dicots, 14 highly conserved among all species, and two in the other category. Using this source, we have identified 23 potential candidate genes within the upland cotton genome for the development of new male sterile germplasm to be used in hybrid cotton breeding. Combining homology-based studies with genome editing may allow for the discovery and validation of GMS genes that previously had no diversity observed in cotton and may allow for development of a desirable male sterile mutant to be used in hybrid cotton production.
Heterosis, or hybrid vigor, is defined as the superior performance of a hybrid offspring over its inbred parents. Many plant species exhibit heterosis, including upland cotton (Gossypium hirsutum L.), the leading crop for the production of natural fibers used in the textile industry (Voora et al., 2020) and an important oilseed crop for human food and animal feed (Kohel and Yu, 2007). To produce hybrid seeds, an inbred male plant is crossed with an inbred female plant that has been prevented from self-pollinating, thus producing only hybrid progeny. In most plants, pollen control in the female parent can be accomplished through hand emasculation, but this is a time and labor-consuming task that cannot be scaled to the demands of commercial agriculture. As an alternative to emasculation, genetic male sterility can be used for hybrid seed production, but sources of genetic male sterility in upland cotton are limited. At this time, only a small number of genes associated with male reproductive function have been identified in cotton (summarized in Supplementary Table 1), despite studies in other species that have established that large numbers of genes are involved in the process of anther formation and microsporogenesis (Richmond and Kohel, 1961; Allison and Fisher, 1964; Ferrario et al., 2004; Ma, 2005; Wang and Li, 2009; Chang et al., 2011; Li et al., 2013; Wei et al., 2013; Wei et al., 2013; Wu et al., 2015; Wang et al., 2019; Li et al., 2020; Ma et al., 2021, Zhang et al., 2021a). Of these previously identified cotton genes potentially for male reproductivity and/or sterility, many are not mapped or have no known protein sequence available, thus making it difficult to manipulate them via methods such as genetic engineering or genome editing. For example, between the 1960s to 1990s, over 10 genes associated with genetic male sterility were identified in upland cotton (Supplementary Table 1), but only recently have three of these genes, ms15, ms5, and ms6, been mapped (Justus and Leinwebr, 1960; Richmond and Kohel, 1961; Justus et al., 1963; Allison and Fisher, 1964; Weaver, 1968; Weaver and Ashley, 1971; Bowman and Weaver, 1979; Huang and Shi, 1988; Zhang et al., 1992; Chen et al., 2009). These factors hinder the development and utilization of hybrid cotton in mechanized agriculture.
Male sterility arises from the abortion or malformation of male gametes, but usually does not affect female gametes, effectively creating a female parent that lacks the ability to self-pollinate (Kun et al., 2013). Classically, pollen has been described as containing four microspores, or pollen sacs, with each of these containing cells which will develop into male gametophytes, surrounded by a tapetal layer, layer(s) of parietal tissue, an epidermal layer, and a connective tissue which will hold these four pollen sacs together as a grouping within the anther (Horner and Palmer, 1995). Formation of the tapetum is a critical part of anther development, as the tapetum will collect resources from vascular tissue to redistribute to developing microspores (Pacini, 2010). Premature cell death of tapetal cells prevents the viability of pollen cells and is controlled by a variety of genes including Baymax1, Carbon Starved Anther, Aborted Microspores, and AtMs1 (Sorensen et al., 2003; Zhang et al., 2010; Zhang et al., 2013; Li et al., 2016; Zhang et al., 2018; An et al., 2020; Xiang et al., 2021; Li et al., 2022). Traditionally, anther development is divided into 10 stages with meiosis occurring in stage 1 and 2, followed by mitotic divisions to form tetrads in stages 3-4. Microspore development then occurs in stages 5-7 with pollen development completing in stages 8-10 (Horner and Palmer, 1995). Abnormal development at any of these stages may lead to male sterility with mutations earlier in the developmental stages often having a larger impact.
The genetic control of male sterility can occur via cytoplasmic male sterility (CMS) or nuclear male sterility (NMS), also known as genetic male sterility (GMS), systems. CMS is controlled by the interaction of gene products between mitochondrial and nuclear genomes. In hybrid breeding, a three-line system is used that involves the use of a CMS line, a male-fertile restorer line, and a fertile sterility-maintainer line that is isogenic to the CMS line. The three-line system includes two crosses. To produce fertile F1 hybrid planting seed, the restorer line (used as the male parent) is crossed with the CMS line (female parent). To continue to produce CMS seed, the isogenic maintainer line (male parent) is crossed with the CMS line (female parent), which ensures that the seeds from the cross are CMS while preserving agronomically beneficial traits. CMS is stable across environments, leading to widespread historical use in some species such as rice and maize, but some types of cytoplasm are known to spontaneously revert and become fertile to varying degrees under specific environmental conditions, and fertility restoration is not always reliable (Havey, 2004; Weider et al., 2009; Chen and Liu, 2014; Abbas et al., 2021; Zhang et al., 2022). As of a 2014 review by Chen and Liu, 28 CMS genes have been identified across 13 crops (Chen and Liu, 2014). Within upland cotton, some CMS markers have been developed for breeding purposes, while transcriptomic and metabolomic studies have identified some interesting differentially expressed genes for further study of this trait (Supplementary Table 1) (Jiang et al., 2007; Wang et al., 2010; Wu et al., 2011; Chen and Liu, 2014; Yang et al., 2014; Kong et al., 2017; Li et al., 2018; Nie et al., 2018; Li et al., 2019; Li et al., 2021; You et al., 2022). At this time, identifying the specific mechanisms behind CMS in cotton is still an area that is under development; most recently, Zhang, et al. identified and mapped orf610a, a mitochondrial chimeric gene, as one causal gene of CMS in cotton (Supplementary Table 1) (Zhang et al., 2022).
Environment-sensitive genetic male sterility (EGMS) contrasts with CMS as its fertility is controlled by known environmental conditions such as temperature (TGMS), photoperiod (PGMS), humidity (HGMS), or a combination of temperature and photoperiod (TPGMS) (Min et al., 2013; Chen and Liu, 2014; Chang et al., 2016; Ding et al., 2017; Zhang et al., 2020; Ma et al., 2021, Zhang et al., 2021b, Li et al., 2022). Under EGMS, a male sterile line can revert to fully fertile depending on the environment. For example, the same line can be male sterile at high temperature while having full fertility at low temperature (Fan and Zhang, 2018). Essentially, male sterility of EGMS lines is stable within an environment (barring extreme weather events), but not across environments. Using this reversible sterile/fertile trait, EGMS lines can be produced in a two-line system. The EGMS line is grown in the sterility-inducing environment, where it is crossed with the inbred male parent to produce hybrid planting seed. To obtain new male-sterile seed, the EGMS line is grown in the fertility-permissive environment for seed increase (Chen and Liu, 2014). Because specific maintainer and restorer lines are not required for EGMS seed increase, there is a broader range of germplasm available for developing hybrids in the EGMS system than for CMS (Abbas et al., 2021). Examples of genes contributing to EGMS include ZmTms5, OCL4, and dcl5 in maize and Tms5 and Tms10 in rice (Supplementary Table 1) (Zhou et al., 2016; Li et al., 2017; Yu et al., 2017; Teng et al., 2020; Yadava et al., 2021).
In rice and maize, genes associated with GMS are well-characterized, and hybrid production using male sterile lines is common (Li et al., 2007; Qi et al., 2020; Rout et al., 2020). In these crops, as well as in model organisms like Arabidopsis, substantial genetic resources exist regarding GMS that can be used for the discovery of homologous genes in other species. In this review, we summarized the current methods in genome editing that can be used for the generation of male sterile traits. We assessed the known biochemical modes of plant GMS and characterized them into four general groups: transcriptional regulation, splicing, fatty acid transport and processing, and sugar transport and processing. In addition, we explored protein sequence homology from EGMS genes of three monocots (maize, rice, and wheat) and three dicots (Arabidopsis, soybean, and tomato) and have identified 23 candidate genes within the upland cotton genome for the development of new male sterile germplasm to be used in cotton breeding. We also examined evolutionary relationships between monocot and dicot GMS genes to describe the relative similarity and relatedness of these genes.
Various strategies can be used to introduce male sterility in a crop of interest. Historically, male sterile traits have been introduced via cross pollination to elite germplasm after observing naturally occurring male sterility within a species (Yamagishi and Bhat, 2014; Bruns, 2017; Zheng et al., 2020). This strategy was widely used before the advent of plant transformation; many of the well-known historical examples of crop male sterility such as cytoplasmic Texas male sterile maize and two-line hybrid rice were developed in this manner (Virmani, 2003; Bruns, 2017). A common drawback of this method is the long timeline needed for backcrossing the trait into elite germplasm – a process that takes multiple generations. Additionally, the discovery of natural male sterility in a breeding population or wild relative is oftentimes a chance event, and sufficient diversity may not exist or may not be visually observable in the species of interest. Traditional plant transformation (using Agrobacterium or the gene gun) has also been used to confer male sterility in crop plants. The development and adoption of genetic engineering techniques furthered the potential of developing male sterile lines and hybrid systems as breeders were no longer confined to the genetic variation available within the species of interest. Genetic engineering of male sterility meant that the gene of interest could be inserted directly into the elite line, but a reliance on tissue culture to recover mutants still makes this process time and labor-consuming. Additionally, plants engineered using Agrobacterium t-DNA systems are under strict regulations and the regulatory process adds considerable additional time to varietal release.
In recent years, CRISPR/Cas9 genome editing has revolutionized genetic engineering with its ability to precisely alter DNA using sequence-specific nucleases (Doudna and Charpentier, 2014). Briefly, the Cas9 protein is led to the target sequence in DNA using a homologous segment of RNA referred to as single-guide RNA (sgRNA) (Doudna and Charpentier, 2014). At the target sequence, the Cas9’s endonuclease activity causes double-stranded breaks (DSBs) in target DNA, predominantly triggering the error-prone non-homologous end joining (NHEJ) DNA repair pathway (Jiang and Doudna, 2017). NHEJ’s repair process joins the two broken ends of DNA together without using a template for repair, commonly causing random insertions and deletions (indels), leading to a knockout or knockdown of the target gene (Jiang and Doudna, 2017). Less frequently, the homology-directed repair (HDR) pathway is engaged, where a homologous segment of DNA is used as a repair template and precise insertions are made into the target sequence (Jiang and Doudna, 2017). The sequence-specific targeting of DNA is an improvement on Agrobacterium t-DNA and gene gun methods as the insertion will occur in a specific, predetermined location rather than randomly within the genome. In the United States, regulatory oversight is also less stringent for crops that are genome edited but do not contain transgenes when compared to crops genetically engineered with Agrobacterium (Entine et al., 2021).
Currently, the most well-defined and common use for CRISPR/Cas9 in plants is for performing gene knockouts with NHEJ. This makes the CRISPR/Cas system particularly useful in male sterility-related studies as the male sterile phenotype is frequently due to loss-of-function mutations in genes related to reproduction and fertility. For example, Jiang et al. (2021) identified 12 new GMS genes in maize by knocking out selected transcription factor genes using the CRISPR/Cas9 system. Although knock-ins with HDR are less often used due to the low-efficiency nature of this pathway in plant cells, HDR has been used in a few studies to introduce or entirely replace promoters for a gene of interest, referred to as “promoter swapping”; this method could be used to target promoters associated with male reproductive organs to induce male sterility (Čermák et al., 2015; Shi et al., 2017; Hummel et al., 2018; Li et al., 2020; Lu et al., 2021). This is a still developing area of study with great potential; however, no instances of promoter swapping have been reported in upland cotton.
Other CRISPR/Cas system capabilities include multiplex editing, a commonly used strategy where multiple sites within a genome can be targeted at the same time by one CRISPR/Cas9 construct. Multiplex editing is typically used when attempting knockouts of multiple genes predicted to affect a trait, members of gene families, or simply to increase the chances of achieving the desired phenotype by targeting multiple sites within a single gene (Xie et al., 2015; Iqbal et al., 2016; Chilcoat et al., 2017; Abdelrahman et al., 2021). Base editing is another application of the CRISPR/Cas system which uses a Cas protein variant fused to a cytosine deaminase or adenine deaminase (Komor et al., 2016; Nishida et al., 2016; Gaudelli et al., 2017). In this system, specific substitutions (C to A, C to T, C to G, and A to G) can be directly induced in a target site without generating a DSB; this can be used to correct previous mutations, introduce premature stop codons to generate a knockout, or to produce a different amino acid than the one currently coded for in the DNA (Komor et al., 2016; Nishida et al., 2016; Gaudelli et al., 2017; Kim, 2018; Evanoff and Komor, 2019; Guyon-Debast et al., 2021). In upland cotton, successful base editing has been established for A to G and C to T substitutions, but at the present time other substitution types have not been reported (Qin et al., 2020; Wang et al., 2022). Prime editing, a more recent innovation in the CRISPR/Cas system originally developed in human cells, uses a modified Cas9 nickase-reverse transcriptase fused protein, paired with a prime editing guide RNA (pegRNA) for editing (Anzalone et al., 2019). Among the variations of the CRISPR/Cas system, prime editing is one of the most versatile and promising. Similar to base editing, prime editing creates mutations without the generation of a DSB, but, unlike base editing, prime editing can write new DNA sequences directly into the target genomic DNA and can create targeted insertions, deletions, and all twelve types of substitutions; this function has been dubbed “search and replace” (Anzalone et al., 2019; Lin et al., 2020; Marzec and Hensel, 2020). Prime editing has been tested in multiple crop and model species but has not yet been established in upland cotton (Jiang et al., 2020; Lin et al., 2020; Lu et al., 2021; Molla et al., 2021; Perroud et al., 2022). Some studies indicate that targeted insertions with prime editing are more efficient or comparable to HDR, but further optimizations will be necessary to fully adapt this technology to plants as editing efficiencies are still low (Lin et al., 2020; Molla et al., 2021). A summary of the pros and cons of each of these methods for developing cotton male sterility can be found in Figure 1.
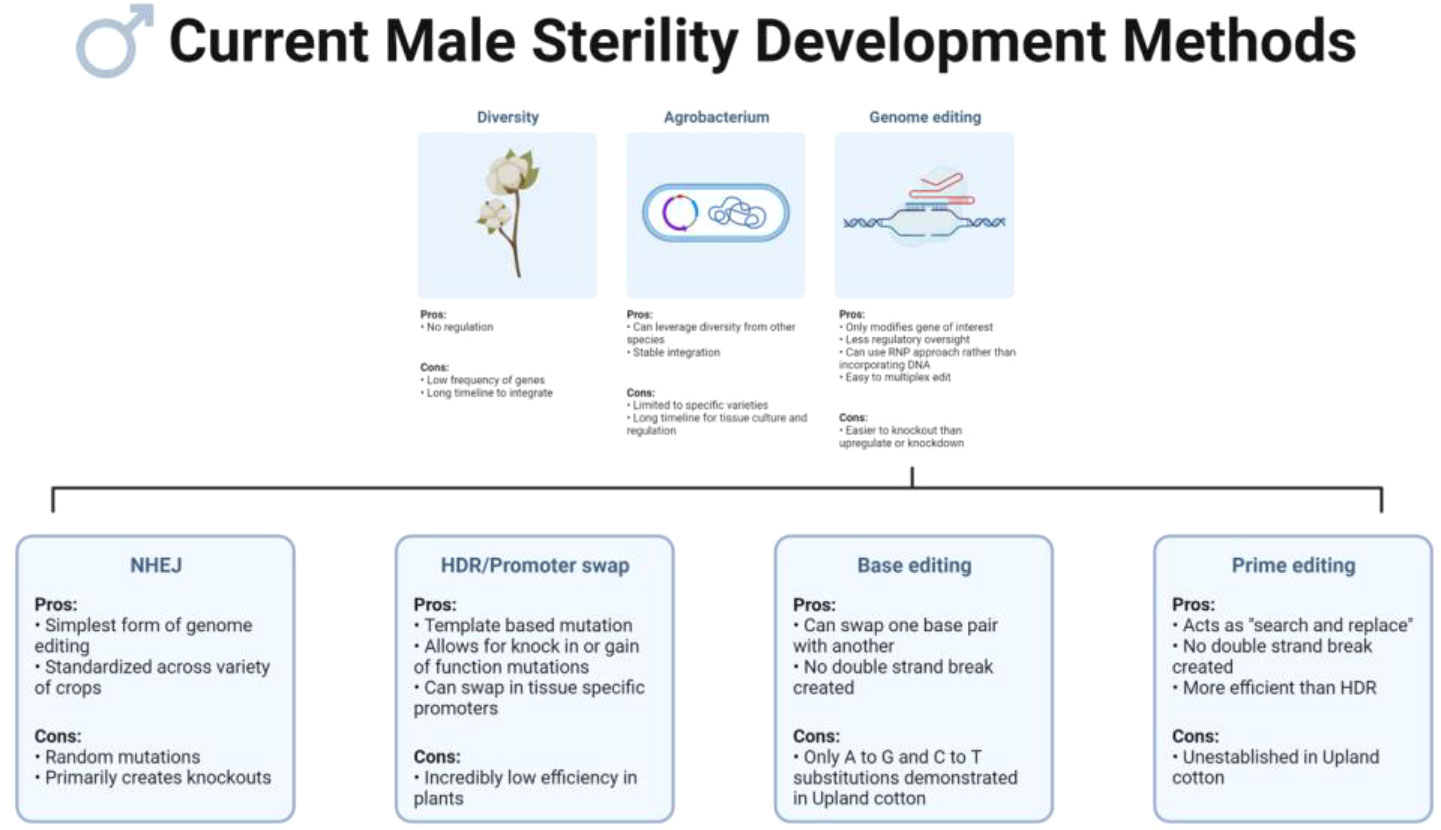
Figure 1 Current methods to develop male sterility in crops include making use of natural diversity, genetic engineering with Agrobacterium, and genome editing. Promising genome editing technologies include non-homologous end joining (NHEJ) for gene knockouts, homology dependent repair (HDR) for promoter swaps and gain-of-function mutations, base editing for amino acid substitutions, and prime editing for precise allele modifications. Created on biorender.com.
One of the most important issues that must be accounted for when starting a CRISPR/Cas9 experiment is the recognition of possible off-target editing. Off-target editing often occurs when sgRNAs incorrectly target DNA sequences that are highly similar, but not identical, to the target sequence, leading to Cas9 cleavage and mutations in off-target locations (Modrzejewski et al., 2020). Compared to animal or human systems, Cas9 has highly specific activity in plants due to its the overall lower expression level, which reduces the occurrence of off-target editing (Peterson et al., 2016). Additionally, undesirable mutations can be eliminated through backcrossing in plants (Hahn and Nekrasov, 2019; Hajiahmadi et al., 2019). In addition to highly specific sgRNA design, various factors such as GC content of the target region, length of the sgRNA, number of mismatches proximal to the PAM site, plant tissue culture time, the type of Cas protein variant used and the associated promoter, and the fidelity of the available reference sequence, among others, have been shown to have effects on off-target activity, and are described in depth in recent reviews by Hajiahmadi et al. (2019) and Modrzejewski et al. (2020). However, generally speaking, the most important step in of avoiding off-target effects is first designing highly specific sgRNAs; if done well, off-target effects can be undetectable in the edited plant (Hajiahmadi et al., 2019). At present, multiple in silico prediction methods have been developed to streamline sgRNA design, and, while accounting for these factors, and suggest and rank the ‘best’ sgRNAs to use for a particular target (Haeussler et al., 2016; Alkan et al., 2018).
Although the control of male sterility can be divided between genes within the cytoplasm and the nucleus, the way these genes cause the sterility can broadly differ. To best review the broad classification of pathways these genes act in, four categories of functions were identified as common mechanisms: transcriptional regulation, splicing and long non-coding RNAs, fatty acid processing and transport, and sugar transport and processing. It is important to note that inclusion of a gene in one category does not mean that it will not impact other classes. Instead, genes were classified based on their direct mode of action while noting that they may cause downstream effects in other areas. These four categories were chosen with transcriptional regulators influencing the expression of other genes and pathways; splicing and long non-coding RNAs indicating genes that do not get translated into proteins but still have an impact on male sterility; and the final two categories indicating a gene preventing the proper formation or transport of a sugar or fatty acid.
When considering the central dogma of biology, one of the most vital steps in the expression of a trait is the transcriptional regulation of the gene(s) controlling the trait. Many of the known genes controlling male sterility in crop species are classified as transcriptional regulators that up- or downregulate the expression of other genes. Often, a single gene will work to impact the expression of one or multiple genes, resulting in the change of phenotype. Examples of this include Carbon Starved Anther from rice which upregulates the expression of MST8 and impacts sugar partitioning (Zhang et al., 2010); Aborted Microspores from Arabidopsis which causes premature tapetum and microspore degeneration (Sorensen et al., 2003); ZmGAMYB/ZmGAMYB2 from maize which causes brown and shrunken pollen grains (Li et al., 2021); and DYT1 from Arabidopsis which transcriptionally regulates 22 genes, with the knockout of DYT1 preventing normal formation of the tapetum (Feng et al., 2012). Homology among plant species has been demonstrated for some of these genes including ZmMs7, where premature expression with an anther-specific promoter affects downstream genes, disrupting tapetum and pollen exine development. ZmMs7 was originally discovered in maize but is homologous with AtMs1 from Arabidopsis and OsMs1 from rice (Zhang et al., 2018; An et al., 2020). Baymax1 is a MYB transcription factor in rice which is conserved amongst many other species and causes male sterility by changing the expression of genes controlling tapetum and microspore development (Xiang et al., 2021). Although the focus of this paper is on male sterility, it is important to highlight that genes involved in transcriptional regulation can often show pleiotropic effects by impacting other traits, including biomass accumulation, a key trait contributing to crop yield (Mukhtar et al., 2018). Furthermore, it is possible that transcriptional regulators controlling male sterility have previously been unobserved in traditional mutation studies due to the quantitative nature of this trait. Recently, the advancement of genome editing technologies has aided in identifying novel genes impacting male sterility in maize through multiplexing the editing of several genes simultaneously. Among the 14 genes edited in a maize study by Jiang, et al., there was no male sterility displayed among single mutations, while some double and triple mutations caused sterility (Jiang et al., 2021). Furthermore, it is possible that the transcriptional regulation of a gene may only be observed in specific growing conditions, as shown with TMS1 from Arabidopsis, which is overexpressed under heat stress to cause male sterility, or Carbon Starved Anther from rice, which causes photoperiod dependent male sterility (Yang et al., 2009; Li et al., 2022). Transcriptional factors are a key target for genome editing with many previously described genes in this classification preventing the proper development of pollen cells; however, when modifying transcriptional factors, it is important to recognize that they often interact with other genes and may cause undesired effects to traits that are unrelated to sterility and that the impact of these genes may change under varying environments.
Although the actual expression of a gene is incredibly important in impacting a phenotype, it is equally important that the gene is properly modified post-transcription. In plants, phased secondary small interfering RNAs (phasiRNAs) act as signals for cleaving specific mRNA sequences. PhasiRNAs can be separated based on their length as either 21 or 24 nucleotides (nt) in length. While 21-nt phasiRNAs were characterized in rice, 24-nt phasiRNAs were discovered in a variety of crops including rice, maize, wheat, litchi, orange, strawberry, and grape (Xia et al., 2019; Bélanger et al., 2020). Although 24-nt phasiRNAs were found in both monocots and dicots, each group underwent a different processing pathway for their development. Monocots have a unique version of Dicer, DCL5, which specifically contributes towards making 24-nt phasiRNAs, while dicots were thought to have 24-nt phasiRNAs produced by DCL3 (Xia et al., 2019). Modification of DCL5 resulted in temperature-dependent male sterility in maize, a beneficial trait to have as this removes the need for having a restorer line (Teng et al., 2020). Along with Dicer-like genes, MEL1 in Arabidopsis and Argonaute 18 are also involved in the creation of phased RNAs, with Argonaute 18 causing a decrease in the stature of plants (Das et al., 2020; Lian et al., 2021). Mutations in the splicing signal of a gene can also cause significant changes in male sterility as observed with PL1 from rice. The splicing mutation in PL1 prevents proper formation of pollen exine, causing irregular shape to pollen cells and a lack of protection from stresses (Zou et al., 2020). Long non-coding RNAs may also contribute to male sterility with LDMAR shown to cause premature cell death in anthers under long day conditions (Ding et al., 2012). Long non-coding RNAs can be found in a broad range of species, acting in the modification of transcriptional messages. While these genes can impact sterility, they may also interact with alternative traits (Xia et al., 2019). As a result, when considering the editing of long non-coding RNAs, it is important to recognize that even if two genes appear to fall in the same family, they may not have the same impact across species.
Another vital process for the stability of pollen cells is the processing and transport of fatty acids. Fatty acid chain length can be a critical component in the fertility of pollen cells as demonstrated by ZmMs25, AtCUT1, OsHMS1, and OsGLI-4. Expression of ZmMs25 primarily occurs in the anther and it is thought this gene is involved in Acyl Co-A metabolism (Zhang et al., 2021c). Very long-chain fatty acids (VLCFAs) and their derivatives help maintain water content in pollen, an important component of pollen vitality. Mutations in these genes disrupt VLCFA biosynthesis or processing, leading to pollen desiccation and male sterility in dry environments. This produces a unique phenomenon in which male sterility is regulated by the humidity levels the plant is exposed to, as plants can still maintain pollen fertility under sufficiently humid environments (Millar et al., 1999; Yu et al., 2019; Chen et al., 2020). Although this trait does not allow for practical use in field-based conditions, it could be highly useful in controlled environment conditions to create a new source of controlling male fertility. Often, genes related to fatty acid transport and processing are vital in maintaining the integrity of the cell wall or membrane in pollen. Examples of this gene group include OsDGD2β (Basnet et al., 2019), AtMS2 (Aarts et al., 1997), NEF1 Arabidopsis (Ariizumi et al., 2004), and OsNP1 (Chang et al., 2016). OsNP1 has been further used in developing a transgenic sterility system in which a red fluorescent protein is used to separate transgenic fertile seeds from non-transgenic male sterile seeds (Chang et al., 2016). Ms10 from rice is less studied; however, it likely functions in a similar manner to the genes described above as it causes a decrease in the exine waxy layer. In Arabidopsis, AtFAX1 functions as a fatty acid exporter vital in ROS signaling and maintaining homeostasis throughout pollen wall development with a mutation in this gene causing sterility (Zhu et al., 2020). Acos5 from Arabidopsis has proven to be another vital gene with its knockout preventing the formation of sporopollenin (de Azevedo Souza et al., 2009). When knocked out using genome editing, OsACOS12, a homolog of ACOS5 in Arabidopsis found in rice, the pollen wall was unable to form correctly and caused male sterility (Zou et al., 2017). Homology between sterility genes can further be found in defective pollen wall from rice which is homologous with ms2 from Arabidopsis and shifts fatty acid and alcohol content in pollen cells (Shi et al., 2011). CYP703A3 works similarly in rice with mutants displaying small, pale anthers (Yang et al., 2018). Alongside genetics, environmental conditions could play a critical role in fatty acid regulation as CRISPR/Cas9 knockdown of ACO2 in cotton resulted in male sterility when plants were grown in normal day and night temperatures, but plants grown in high day and night temperatures were fertile (Khan et al., 2020). Oftentimes genes involved in the processing and transport of fatty acids impact the stability of the pollen cell wall, with changes in function preventing pollen cells from retaining the appropriate amount of water. As a result, modifications in this class of genes can allow for an interesting range of effects depending on the environment, with humidity often being the largest controlling factor.
Although transcriptional regulation, splicing, and fatty acid processing play a much larger part in the regulation of male sterility in plants, genes related to the processing and transport of sugar have also been shown an impact on the fertility of pollen. One example of this is UGPase 1 in rice, where a SNP in this gene causes pleiotropic effects by increasing rice chalkiness and causing male sterility (Chen et al., 2007; Woo et al., 2008). Loss of HXK5 in rice causes male sterility as pollen cells experience a decrease in starch content (Lee et al., 2019). This is further demonstrated in cucumber, where the downregulation of CsSUT1, a sucrose-proton transporter, leads to male sterility by altering sucrose, hexose, and starch supply available in pollen development (Sun et al., 2019). In Arabidopsis, Thin Exine 2 (TEX2 or ROCK1) functions as a nucleotide-sugar transporter, which when knocked out leads to the formation of an incredibly thin exine (Wang and Dobritsa, 2021). Ms8 from corn is an example of a ß-1,3-galactosyltransferase which has been genome edited to create a transgene free, male sterile variety of corn (Chen et al., 2018). ZmMs45 is another gene in maize which is thought to function as a strictosamide synthase and causes complete sterility when both copies are knocked out (Wu et al., 2016). When ms39 or callose synthase12 in maize is knocked out, the tapetum develops irregularly causing aborted microspores (Zhu et al., 2018). Finally, the regulation of genes surrounding carbohydrate metabolism are suggested to impact cytoplasmic male sterility in wheat; however, this has not been pinned to single gene impacts and thus remains largely hypothetical (Geng et al., 2018). Although the majority of genes contributing to male sterility can fit within the above categories, one which does not is OsDEX1 which acts in calcium transport and homeostasis (Yu et al., 2016). Similarly, OsFIGNL1 is a AAA-ATPase which causes problems in separation of chromosomes in pollen cells when mutated (Zhang et al., 2017). Elucidation of biochemical function of some male sterility genes still has not been completed including GmMs1 from soybean and ms2 from wheat (Ni et al., 2017; Jiang et al., 2021). Carbohydrate metabolism functions in a broad range of roles in the creation of male sterile lines, with some genes causing thin exines, others impacting the tapetum formation, and still others changing the carbohydrate availability in developing cells. Despite the broad range of knowledge known about male sterility, a significant amount of work is still needed in determining the biochemical functions of genes and how they may overlap across species.
The sources of male sterility in a crop are practically limited with only a few genes in each species known to contribute to the fertility of pollen in a crop. In the past, methods for identifying genes related to male sterility have included genome-wide association studies (GWAS) and mutation analyses (Chen et al., 2007; Lafarge et al., 2017; Ma et al., 2021; Han et al., 2022). However, both methods require the phenotyping of hundreds of lines to identify genes controlling a trait for which variation may not exist in the population being studied. As a result, homology-based identification of genes controlling male sterility may offer an alternative approach to shorten the time necessary to create novel male sterile lines in economically important crops. A homology-based approach can be pursued through identification of known male sterility genes in a variety of crop species. As these genes are identified, a BLAST sequence similarity search is used to screen a variety of monocot and dicot species with the aim of finding genes with a high level of conservation amongst either monocots, dicots, or both.
The advent of genome editing in crops has allowed for rapid integration of gene knockouts in a variety of crops including rice, cotton, soybean, tomato, wheat, lettuce, and many other plant species (Shan et al., 2014; Belhaj et al., 2015; Long et al., 2018). In tomato, CRISPR-based genome editing has already been used to create novel sources of male sterility with the knockout of SlSTR1. The project took a transgenic approach when developing lines as a restorer gene was transformed into the tomato plants with genome editing elements (Du et al., 2020). Additionally, genome editing offers the capability to target specific genes and to differentiate amongst members of large gene families, an important consideration as genes like Dcl5 can be members of gene families with a broad diversity of functions beyond male sterility (Teng et al., 2020). Furthermore, as genes controlling environmentally dependent male sterility show a range of responses depending on the variety, it is possible this trait may be quantitatively inherited (Teng et al., 2020). In order to overcome this complexity, genome editing allows for multiplexed knockout of many genes simultaneously allowing investigation of how genes impact sterility individually and in combination with one another (Minkenberg et al., 2017). Genome editing offers the opportunity to quickly mutate and screen dozens of genes for insertion into novel varieties and breeding schemes for use in crop improvement programs. Strategies utilizing genome editing can also be designed in such a way that the scheme will work across multiple species as demonstrated with the editing of ms26 in corn, rice, sorghum, and wheat (Cigan et al., 2017). This homology based approach has already proven successful in rice where TMS5 was mutated using CRISPR/Cas9 based on homology with maize TMS5, suggesting that our approach should be successful in cotton (Barman et al., 2019).
Upon an extensive search for male sterility genes from diverse sources (Supplementary Table 2), sequences for these genes were obtained from databases including CottonGen (Yu et al., 2021), Sol Genomics Network (Fernandez-Pozo et al., 2015), RAP-DB (Sakai et al., 2013), TAIR (Berardini et al., 2015), SoyBase (Grant et al., 2010), and the Maize Genetics and Genomics Database (Woodhouse et al., 2021). A total of 38 genes were identified which contributed to male sterility; however, eight of these genes were excluded from subsequent analyses as they had not been finely mapped or had no corresponding protein. Sequences similar with each gene were searched in the cotton, soybean, rice, Arabidopsis, corn, wheat, and tomato genome databases to identify the level of sequence similarity for each gene (Table 1) as well as the protein it codes (Figure 2); this information is summarized in Supplementary Table 2.
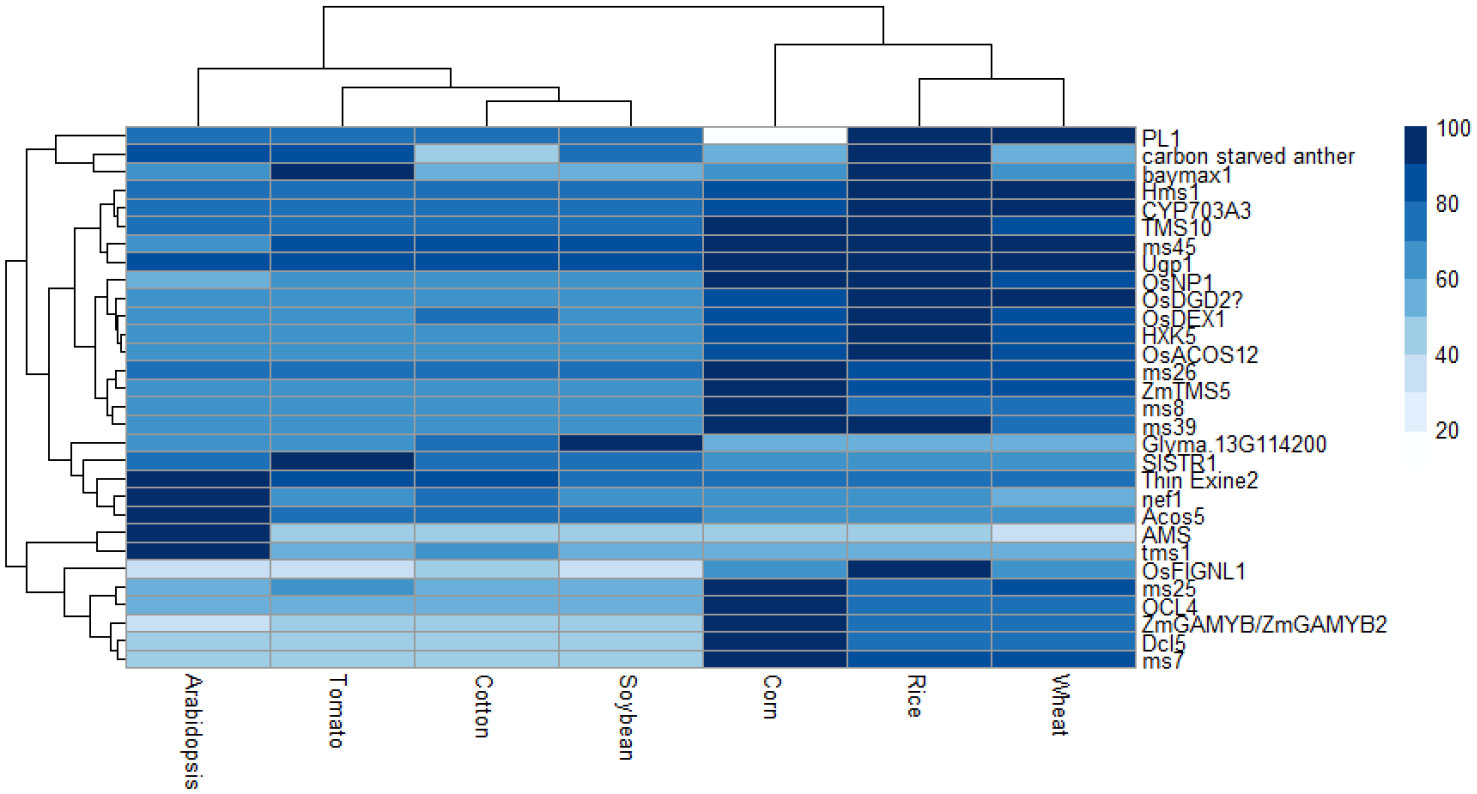
Figure 2 Heat map displaying the percentage of protein similarity between species. Dark blue is most similar and white is least similar.
By use of this method, genes identified can be classified into one of five categories: low conserved, in which only one species has the gene of interest, monocot-specific, dicot-specific, conserved among all species, or other. Among the queried genes, five are classified as low conserved, four as monocot-specific, five as dicot-specific, and 14 highly conserved among all species. As carbon starved anther, a gene discovered in rice, is conserved among dicot species except cotton but not among other monocot species, it is placed in the “Other” category. Additionally, the gene PL1 is conserved among all species except maize, and thus is in the “Other” category (Table 1; Figure 2).
Utilizing candidate gene analysis from a homology perspective, the number of genes recommended for validation of their function within a novel species can be quickly narrowed down to make a starter list of candidate genes. For example, genes which are unique to one species may not be beneficial to pursue incorporation of them into another species as it is unlikely will have any impact on the new species of interest. On the other hand, genes conserved across a wide variety of species are likely to maintain the same function and cause similar phenotypes, so these types of genes may be good candidates. A final consideration to make when using homology-based candidate gene analysis is whether a gene is pleiotropic or not. In general, when trying to create a novel phenotype for incorporation into breeding programs, it would be most beneficial to modify only the trait of interest and, as a result, genes with pleiotropic effects should be avoided. In summary, the original list of 30 candidate genes was developed following the criteria: (1) reported genes that had already been mapped, (2) they had a protein sequence available, and (3) they did not have known pleiotropic effects. Among these, we prioritized genes that were conserved across all species based on our homology analysis.
To specifically search for candidate genes in upland cotton after the homology-based analysis, the 30-gene list acquired in the previous step (Figure 2) was narrowed down in a series of steps to a list of 23 genes (Table 2). Sequences from each gene were BLASTed against the TM-1 UTX v2.1 genome (Chen et al., 2020; Zhang, et al., 2015), and any genes that did not return a hit (ms7, OsFIGNL1) were discarded. Some genes from different species, for example, ACOS5 from Arabidopsis and OsACOS12, identified a single ortholog in the TM-1 genome. Ms25 from maize and baymax1 from rice were another set of genes that were orthologous for a single gene in TM-1. Orthologous genes of this nature were consolidated to one candidate, further reducing the number of candidates. Next, we found orthologues for these genes in the TM-1 NAU-NBI genome to assess gene expression across tissue types using RNA-seq data (Dai et al., 2022); we presented these orthologues in Supplementary Table 3. Some genes that were present in the TM-1 UTX v2.1 genome were not present in the TM-1 NAU-NBI genome, so a few genes (ZmGAMYB/ZmGAMYB2, Dcl5, and Glyma.13G114200) were also discarded here, as we could not find expression data for these genes. Of this list of 23 candidates, tms1, Zmtms5, OCL4, and TMS10 were TGMS genes, while Hms1 and carbon starved anther were the only HGMS and PGMS genes, respectively. The remaining 17 candidate genes are not known to be environmentally sensitive or reversible male-sterile. This list of GMS genes in Table 2, as well as the expression data in Supplementary Table 3, provides a good starting point for researchers interested in hybrid cotton development, as well as basic research into cotton male sterility and reproduction in general.

Table 2 Twenty-three candidate genes identified for male sterility in upland cotton through homology-based analysis.
A homology-based analysis for the identification of target genes has clear benefits for the researcher, reducing the time and financial commitment required to perform large genetic screens, as is needed to perform GWAS. One of the major drawbacks of traditional genetic screens is their reliance on existing genetic variation to identify genes of interest. When using DNA or protein sequence homology as the basis for interspecific gene discovery, natural variation within a species is not required as we take advantage of the evolutionary conservation of genetic material. Targets can be identified even in species where sterile allele frequency is incredibly rare or phenotypic differences cannot be easily discerned. This is particularly relevant for male sterile traits as common phenotypes, such as differences in pollen shape or pollen shedding, can be subtle. Because this approach takes advantage of pre-existing resources and can be performed entirely in silico, the pipeline from identifying a trait of interest to generating multiple target genes is efficient.
In this study, we have demonstrated that protein sequence homology can be a basis for the discovery of many new target genes in upland cotton. Prior to this report, only a few confirmed sources of male sterility were previously known in this Gossypium species. These candidate genes now present promising targets for future genome editing efforts. Moreover, in crops where genetic resources are limited, such as orphan crops, this method can also be a cost-effective and time-efficient way to quickly identify new target genes and rapidly expand knowledge of crops’ genomes. In plant breeding, where the development of novel germplasm is an unabating need, identifying highly homologous genes among crop plants may facilitate new breeding strategies to capture hybrid vigor for improved crop productivity.
KM, KH, JU, and JY conceived the study. KM and JY designed the study. KM and AB collected the data. KM, AB, MT, and JY analyzed the data. KM and AB drafted the manuscript. KH, JU, MT, and JY revised the manuscript. All authors contributed to the article and approved the submitted version.
This study was supported in part by a grant from the Cotton Incorporated project #20-229.
Author KH is employed by Cotton Incorporated.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1006264/full#supplementary-material
Aarts, M. G., Hodge, R., Kalantidis, K., Florack, D., Wilson, Z. A., Mulligan, B. J., et al. (1997). The arabidopsis MALE STERILITY 2 protein shares similarity with reductases in elongation/condensation complexes. Plant J. Cell Mol. Biol. 12 (3), 615–623. doi: 10.1046/j.1365-313X.1997.00615.x
Abbas, A., Yu, P., Sun, L., Yang, Z., Chen, D., Cheng, S., et al. (2021). Exploiting genic Male sterility in rice: From molecular dissection to breeding applications. Front. Plant Sci. 12, 220. doi: 10.3389/fpls.2021.629314
Abdelrahman, M., Wei, Z., Rohila, J. S., Zhao, K. (2021). Multiplex genome-editing technologies for revolutionizing plant biology and crop improvement. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.721203
Alkan, F., Wenzel, A., Anthon, C., Havgaard, J. H., Gorodkin, J. (2018). CRISPR-Cas9 off-targeting assessment with nucleic acid duplex energy parameters. Genome Biol. 19 (1), 1–13. doi: 10.1186/s13059-018-1534-x
Allison, D. C., Fisher, W. D. (1964). A dominant gene for Male-sterility in upland cotton 1. Crop Sci. 4 (5), 548–549. doi: 10.2135/cropsci1964.0011183X000400050035x
An, X., Ma, B., Duan, M., Dong, Z., Liu, R., Yuan, D., et al. (2020). Molecular regulation of ZmMs7 required for maize male fertility and development of a dominant male-sterility system in multiple species. Proc. Natl. Acad. Sci. 117 (38), 23499–23509. doi: 10.1073/pnas.2010255117
Anzalone, A. V., Randolph, P. B., Davis, J. R., Sousa, A. A., Koblan, L. W., Levy, J. M., et al. (2019). Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576 (7785), 149–157. doi: 10.1038/s41586-019-1711-4
Ariizumi, T., Hatakeyama, K., Hinata, K., Inatsugi, R., Nishida, I., Sato, S., et al. (2004). Disruption of the novel plant protein NEF1 affects lipid accumulation in the plastids of the tapetum and exine formation of pollen, resulting in male sterility in arabidopsis thaliana. Plant J. 39 (2), 170–181. doi: 10.1111/j.1365-313X.2004.02118.x
Barman, H. N., Sheng, Z., Fiaz, S., Zhong, M., Wu, Y., Cai, Y., et al. (2019). Generation of a new thermo-sensitive genic male sterile rice line by targeted mutagenesis of TMS5 gene through CRISPR/Cas9 system. BMC Plant Biol. 19 (1), 109. doi: 10.1186/s12870-019-1715-0
Basnet, R., Hussain, N., Shu, Q. (2019). OsDGD2β is the sole digalactosyldiacylglycerol synthase gene highly expressed in anther, and its mutation confers Male sterility in rice. Rice 12 (1), 66. doi: 10.1186/s12284-019-0320-z
Bélanger, S., Pokhrel, S., Czymmek, K., Meyers, B. C. (2020). Premeiotic, 24-nucleotide reproductive phasiRNAs are abundant in anthers of wheat and barley but not rice and maize. Plant Physiol. 184 (3), 1407–1423. doi: 10.1104/pp.20.00816
Belhaj, K., Chaparro-Garcia, A., Kamoun, S., Patron, N. J., Nekrasov, V. (2015). Editing plant genomes with CRISPR/Cas9. Curr. Opin. Biotechnol. 32, 76–84. doi: 10.1016/j.copbio.2014.11.007
Berardini, T. Z., Reiser, L., Li, D., Mezheritsky, Y., Muller, R., Strait, E., et al. (2015). The arabidopsis information resource: Making and mining the “gold standard” annotated reference plant genome. Genesis (New York N.Y. 2000) 53 (8), 474–485. doi: 10.1002/dvg.22877
Bowman, D. T., Weaver, J. B. (1979). Analyses of a dominant Male-sterile character in upland cotton. II. genetic studies 1. Crop Sci. 19 (5), 628–630. doi: 10.2135/cropsci1979.0011183X001900050020x
Bruns, H. A. (2017). Southern corn leaf blight: A story worth retelling. Agron. J. 109 (4), 1218–1224. doi: 10.2134/agronj2017.01.0006
Čermák, T., Baltes, N. J., Čegan, R., Zhang, Y., Voytas, D. F. (2015). High-frequency, precise modification of the tomato genome. Genome Biol. 16 (1), 1–15. doi: 10.1186/s13059-015-0796-9
Chang, Z., Chen, Z., Wang, N., Xie, G., Lu, J., Yan, W., et al. (2016). Construction of a male sterility system for hybrid rice breeding and seed production using a nuclear male sterility gene. Proc. Natl. Acad. Sci. 113 (49), 14145–14150. doi: 10.1073/pnas.1613792113
Chang, F., Wang, Y., Wang, S., Ma, H. (2011). Molecular control of microsporogenesis in arabidopsis. Curr. Opin. Plant Biol. 14 (1), 66–73. doi: 10.1016/j.pbi.2010.11.001
Chen, D., Ding, Y., Guo, W., Zhang, T. (2009). Molecular mapping of genic male-sterile genes ms15, ms5 and ms6 in tetraploid cotton. Plant Breed. 128 (2), 193–198. doi: 10.1111/j.1439-0523.2008.01562.x
Chen, L., Liu, Y.-G. (2014). Male Sterility and fertility restoration in crops. Annu. Rev. Plant Biol. 65, 579–606. doi: 10.1146/annurev-arplant-050213-040119
Chen, R., Xu, Q., Liu, Y., Zhang, J., Ren, D., Wang, G., et al. (2018). Generation of transgene-free maize Male sterile lines using the CRISPR/Cas9 system. Front. Plant Sci. 9 (1180). doi: 10.3389/fpls.2018.01180
Chen, H., Zhang, Z., Ni, E., Lin, J., Peng, G., Huang, J., et al. (2020). HMS1 interacts with HMS1I to regulate very-long-chain fatty acid biosynthesis and the humidity-sensitive genic male sterility in rice (Oryza sativa). New Phytol. 225 (5), 2077–2093. doi: 10.1111/nph.16288
Chen, R., Zhao, X., Shao, Z., Wei, Z., Wang, Y., Zhu, L., et al. (2007). Rice UDP-glucose Pyrophosphorylase1 is essential for pollen callose deposition and its cosuppression results in a new type of thermosensitive genic Male sterility. Plant Cell 19 (3), 847–861. doi: 10.1105/tpc.106.044123
Chilcoat, D., Liu, Z.-B., Sander, J. (2017). “Use of CRISPR/Cas9 for crop improvement in maize and soybean,” Progress in Molecular Biology and Translational Science (San Diego, CA, United States: Elsevier) 149, 27–46.
Cigan, A. M., Singh, M., Benn, G., Feigenbutz, L., Kumar, M., Cho, M.-J., et al. (2017). Targeted mutagenesis of a conserved anther-expressed P450 gene confers male sterility in monocots. Plant Biotechnol. J. 15 (3), 379–389. doi: 10.1111/pbi.12633
Dai, F., Chen, J., Zhang, Z., Liu, F., Li, J., Zhao, T., et al. (2022). COTTONOMICS: a comprehensive cotton multi-omics database. Database 2022, 1–8. doi: 10.1093/database/baac080
Das, S., Swetha, C., Pachamuthu, K., Nair, A., Shivaprasad, P. (2020). Loss of function of oryza sativa argonaute 18 induces male sterility and reduction in phased small RNAs. Plant Reprod. 33 (1), 59–73. doi: 10.1007/s00497-020-00386-w
de Azevedo Souza, C., Kim, S. S., Koch, S., Kienow, L., Schneider, K., McKim, S. M., et al. (2009). A novel fatty acyl-CoA synthetase is required for pollen development and sporopollenin biosynthesis in arabidopsis. Plant Cell 21 (2), 507–525. doi: 10.1105/tpc.108.062513
Ding, J., Lu, Q., Ouyang, Y., Mao, H., Zhang, P., Yao, J., et al. (2012). A long noncoding RNA regulates photoperiod-sensitive male sterility, an essential component of hybrid rice. Proc. Natl. Acad. Sci. 109 (7), 2654–2659. doi: 10.1073/pnas.1121374109
Ding, Y., Ma, Y., Liu, N., Xu, J., Hu, Q., Li, Y., et al. (2017). Micro RNA s involved in auxin signalling modulate male sterility under high-temperature stress in cotton (Gossypium hirsutum). Plant J. 91 (6), 977–994. doi: 10.1111/tpj.13620
Doudna, J. A., Charpentier, E. (2014). The new frontier of genome engineering with CRISPR-Cas9. Science 346 (6213), 1258096. doi: 10.1126/science.1258096
Du, M., Zhou, K., Liu, Y., Deng, L., Zhang, X., Lin, L., et al. (2020). A biotechnology-based male-sterility system for hybrid seed production in tomato. Plant J. 102 (5), 1090–1100. doi: 10.1111/tpj.14678
Entine, J., Felipe, M. S. S., Groenewald, J.-H., Kershen, D. L., Lema, M., McHughen, A., et al. (2021). Regulatory approaches for genome edited agricultural plants in select countries and jurisdictions around the world. Transgenic Res. 30 (4), 551–584. doi: 10.1007/s11248-021-00257-8
Evanoff, M., Komor, A. C. (2019). Base editors: modular tools for the introduction of point mutations in living cells. Emerging Topics Life Sci. 3 (5), 483–491. doi: 10.1042/ETLS20190088
Fan, Y., Zhang, Q. (2018). Genetic and molecular characterization of photoperiod and thermo-sensitive male sterility in rice. Plant Reprod. 31 (1), 3–14. doi: 10.1007/s00497-017-0310-5
Feng, B., Lu, D., Ma, X., Peng, Y., Sun, Y., Ning, G., et al. (2012). Regulation of the arabidopsis anther transcriptome by DYT1 for pollen development. Plant J. 72 (4), 612–624. doi: 10.1111/j.1365-313X.2012.05104.x
Fernandez-Pozo, N., Menda, N., Edwards, J. D., Saha, S., Tecle, I. Y., Strickler, S. R., et al. (2015). The sol genomics network (SGN)–from genotype to phenotype to breeding. Nucleic Acids Res. 43 (Database issue), D1036–D1041. doi: 10.1093/nar/gku1195
Ferrario, S., Immink, R. G., Angenent, G. C. (2004). Conservation and diversity in flower land. Curr. Opin. Plant Biol. 7 (1), 84–91. doi: 10.1016/j.pbi.2003.11.003
Gaudelli, N. M., Komor, A. C., Rees, H. A., Packer, M. S., Badran, A. H., Bryson, D. I., et al. (2017). Programmable base editing of a• T to g• c in genomic DNA without DNA cleavage. Nature 551 (7681), 464–471. doi: 10.1038/nature24644
Geng, X., Ye, J., Yang, X., Li, S., Zhang, L., Song, X. (2018). Identification of proteins involved in carbohydrate metabolism and energy metabolism pathways and their regulation of cytoplasmic Male sterility in wheat. Int. J. Mol. Sci. 19 (2), 324. doi: 10.3390/ijms19020324
Grant, D., Nelson, R. T., Cannon, S. B., Shoemaker, R. C. (2010). SoyBase, the USDA-ARS soybean genetics and genomics database. Nucleic Acids Res. 38 (Database issue), D843–D846. doi: 10.1093/nar/gkp798
Guyon-Debast, A., Alboresi, A., Terret, Z., Charlot, F., Berthier, F., Vendrell-Mir, P., et al. (2021). A blueprint for gene function analysis through base editing in the model plant physcomitrium (Physcomitrella) patens. New Phytol. 230 (3), 1258–1272. doi: 10.1111/nph.17171
Haeussler, M., Schönig, K., Eckert, H., Eschstruth, A., Mianné, J., Renaud, J.-B., et al. (2016). Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 17 (1), 1–12. doi: 10.1186/s13059-016-1012-2
Hahn, F., Nekrasov, V. (2019). CRISPR/Cas precision: do we need to worry about off-targeting in plants? Plant Cell Rep. 38 (4), 437–441. doi: 10.1007/s00299-018-2355-9
Hajiahmadi, Z., Movahedi, A., Wei, H., Li, D., Orooji, Y., Ruan, H., et al. (2019). Strategies to increase on-target and reduce off-target effects of the CRISPR/Cas9 system in plants. Int. J. Mol. Sci. 20 (15), 3719. doi: 10.3390/ijms20153719
Han, X., Gao, C., Liu, L., Zhang, Y., Jin, Y., Yan, Q., et al. (2022). Integration of eQTL analysis and GWAS highlights regulation networks in cotton under stress condition. Int. J. Mol. Sci. 23 (14), 7564. doi: 10.3390/ijms23147564
Havey, M. J. (2004). The use of cytoplasmic male sterility for hybrid seed production. Mol. Biol. Biotechnol. Plant organelles 1, 623–634. doi: 10.1007/978-1-4020-3166-3_23
Horner, H. T., Palmer, R. G. (1995). Mechanisms of genic male sterility. Crop Sci. 35 (6), 1527–1535. doi: 10.2135/cropsci1995.0011183X003500060002x
Huang, G., Shi, S. (1988). Chuanza 4, a cotton hybrid by genic male sterile line. China Cotton 15 (3), 19.
Hummel, A. W., Chauhan, R. D., Cermak, T., Mutka, A. M., Vijayaraghavan, A., Boyher, A., et al. (2018). Allele exchange at the EPSPS locus confers glyphosate tolerance in cassava. Plant Biotechnol. J. 16 (7), 1275–1282. doi: 10.1111/pbi.12868
Iqbal, Z., Sattar, M. N., Shafiq, M. (2016). CRISPR/Cas9: a tool to circumscribe cotton leaf curl disease. Front. Plant Sci. 7, 475. doi: 10.3389/fpls.2016.00475
Jiang, Y., An, X., Li, Z., Yan, T., Zhu, T., Xie, K., et al. (2021). CRISPR/Cas9-based discovery of maize transcription factors regulating male sterility and their functional conservation in plants. Plant Biotechnol. J 19, 1769–84. doi: 10.1111/pbi.13590
Jiang, Y.-Y., Chai, Y.-P., Lu, M.-H., Han, X.-L., Lin, Q., Zhang, Y., et al. (2020). Prime editing efficiently generates W542L and S621I double mutations in two ALS genes in maize. Genome Biol. 21 (1), 1–10. doi: 10.1186/s13059-020-02170-5
Jiang, B., Chen, L., Yang, C., Wu, T., Yuan, S., Wu, C., et al. (2021). The cloning and CRISPR/Cas9-mediated mutagenesis of a male sterility gene MS1 of soybean. Plant Biotechnol. J. 19 (6), 1098–1100. doi: 10.1111/pbi.13601
Jiang, F., Doudna, J. A. (2017). CRISPR–Cas9 structures and mechanisms. Annu. Rev. Biophys. 46 (1), 505–529. doi: 10.1146/annurev-biophys-062215-010822
Jiang, P., Zhang, X., Zhu, Y., Zhu, W., Xie, H., Wang, X. (2007). Metabolism of reactive oxygen species in cotton cytoplasmic male sterility and its restoration. Plant Cell Rep. 26 (9), 1627–1634. doi: 10.1007/s00299-007-0351-6
Justus, N., Leinwebr, C. (1960). A heritable partially male-sterile character in cotton. J. Hered. 51, 191–192. doi: 10.1093/oxfordjournals.jhered.a106987
Justus, N., Meyer, J. R., Roux, J. B. (1963). A partially Male-sterile character in upland cotton 1. Crop Sci. 3 (5), 428–429. doi: 10.2135/cropsci1963.0011183X000300050018x
Khan, A. H., Min, L., Ma, Y., Wu, Y., Ding, Y., Li, Y., et al. (2020). High day and night temperatures distinctively disrupt fatty acid and jasmonic acid metabolism, inducing male sterility in cotton. J. Exp. Bot. 71 (19), 6128–6141. doi: 10.1093/jxb/eraa319
Kim, J.-S. (2018). Precision genome engineering through adenine and cytosine base editing. Nat. Plants 4 (3), 148–151. doi: 10.1038/s41477-018-0115-z
Kohel, R. J., Yu, J. Z. (2007). Cottonseed, pp. 89-102. In: Singh, R. J. (eds) Genetic Resources, Chromosome Engineering and Crop Improvement: Oilseed Crops. (Boca Raton, FL.: CRC Press), 4.
Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A., Liu, D. R. (2016). Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533 (7603), 420–424. doi: 10.1038/nature17946
Kong, X., Liu, D., Liao, X., Zheng, J., Diao, Y., Liu, Y., et al. (2017). Comparative analysis of the cytology and transcriptomes of the cytoplasmic male sterility line H276A and its maintainer line H276B of cotton (Gossypium barbadense l.). Int. J. Mol. Sci. 18 (11), 2240. doi: 10.3390/ijms18112240
Kun, W., Xiaojue, P., Yanxiao, J., Yang, P., Yingguo, Z., Li, S. (2013). Gene, protein, and network of male sterility in rice. Front. Plant Sci. 4, 92. doi: 10.3389/fpls.2013.00092
Lafarge, T., Bueno, C., Frouin, J., Jacquin, L., Courtois, B., Ahmadi, N. (2017). Genome-wide association analysis for heat tolerance at flowering detected a large set of genes involved in adaptation to thermal and other stresses. PloS One 12 (2), e0171254. doi: 10.1371/journal.pone.0171254
Lee, S.-K., Kim, H., Cho, J.-I., Nguyen, C. D., Moon, S., Park, J. E., et al. (2019). Deficiency of rice hexokinase HXK5 impairs synthesis and utilization of starch in pollen grains and causes male sterility. J. Exp. Bot. 71 (1), 116–125. doi: 10.1093/jxb/erz436
Lian, J.-P., Yang, Y.-W., He, R.-R., Yang, L., Zhou, Y.-F., Lei, M.-Q., et al. (2021). Ubiquitin-dependent argonauteprotein MEL1 degradation is essential for rice sporogenesis and phasiRNA target regulation. Plant Cell 33, 2685–2700. doi: 10.1093/plcell/koab138
Li, M., Chen, L., Khan, A., Kong, X., Khan, M. R., Rao, M. J., et al. (2021). Transcriptome and MiRNAomics analyses identify genes associated with cytoplasmic male sterility in cotton (Gossypium hirsutum l.). Int. J. Mol. Sci. 22 (9), 4684. doi: 10.3390/ijms22094684
Li, S., Chen, Z., Zhao, N., Wang, Y., Nie, H., Hua, J. (2018). The comparison of four mitochondrial genomes reveals cytoplasmic male sterility candidate genes in cotton. BMC Genomics 19 (1), 1–15. doi: 10.1186/s12864-018-5122-y
Li, Y., Jiang, J., Du, M.-L., Li, L., Wang, X.-L., Li, X.-B. (2013). A cotton gene encoding MYB-like transcription factor is specifically expressed in pollen and is involved in regulation of late anther/pollen development. Plant Cell Physiol. 54 (6), 893–906. doi: 10.1093/pcp/pct038
Li, Y., Li, Y., Su, Q., Wu, Y., Zhang, R., Li, Y., et al. (2022). High temperature induces male sterility via MYB66–MYB4–Casein kinase I signaling in cotton. Plant Physiol 189, 2091–2109. doi: 10.1093/plphys/kiac213
Li, Y., Li, L., Wang, Y., Wang, Y.-C., Wang, N.-N., Lu, R., et al. (2020). Pollen-specific protein PSP231 activates callose synthesis to govern male gametogenesis and pollen germination. Plant Physiol. 184 (2), 1024–1041. doi: 10.1104/pp.20.00297
Lin, Q., Zong, Y., Xue, C., Wang, S., Jin, S., Zhu, Z., et al. (2020). Prime genome editing in rice and wheat. Nat. Biotechnol. 38 (5), 582–585. doi: 10.1038/s41587-020-0455-x
Li, Y., Qin, T., Wei, C., Sun, J., Dong, T., Zhou, R., et al. (2019). Using transcriptome analysis to screen for key genes and pathways related to cytoplasmic male sterility in cotton (Gossypium hirsutum l.). Int. J. Mol. Sci. 20 (20), 5120. doi: 10.3390/ijms20205120
Li, Q., Sapkota, M., van der Knaap, E. (2020). Perspectives of CRISPR/Cas-mediated cis-engineering in horticulture: unlocking the neglected potential for crop improvement. Horticul. Res. 7, 1–11. doi: 10.1038/s41438-020-0258-8
Li, J., Wang, D., Sun, S., Sun, L., Zong, J., Lei, Y., et al. (2022). The regulatory role of CARBON STARVED ANTHER-mediated photoperiod-dependent male fertility in rice. Plant Physiol. 189 (2), 955–971. doi: 10.1093/plphys/kiac076
Li, S., Yang, D., Zhu, Y. (2007). Characterization and use of male sterility in hybrid rice breeding. J. Integr. Plant Biol. 49 (6), 791–804. doi: 10.1111/j.1744-7909.2007.00513.x
Li, Q., Zhang, D., Chen, M., Liang, W., Wei, J., Qi, Y., et al. (2016). Development of japonica photo-sensitive genic Male sterile rice lines by editing carbon starved anther using CRISPR/Cas9. J. Genet. Genomics 43 (6), 415–419. doi: 10.1016/j.jgg.2016.04.011
Li, J., Zhang, H., Si, X., Tian, Y., Chen, K., Liu, J., et al. (2017). Generation of thermosensitive male-sterile maize by targeted knockout of the ZmTMS5 gene. J. Genet. Genomics 44 (9), 465–468. doi: 10.1016/j.jgg.2017.02.002
Li, Z., Zhu, T., Liu, S., Jiang, Y., Liu, H., Zhang, Y., et al. (2021). Genome-wide analyses on transcription factors and their potential microRNA regulators involved in maize male fertility. Crop J. 9 (6), 1248–1262. doi: 10.1016/j.cj.2021.03.016
Long, L., Guo, D.-D., Gao, W., Yang, W.-W., Hou, L.-P., Ma, X.-N., et al. (2018). Optimization of CRISPR/Cas9 genome editing in cotton by improved sgRNA expression. Plant Methods 14 (1), 85. doi: 10.1186/s13007-018-0353-0
Lu, Y., Tian, Y., Shen, R., Yao, Q., Zhong, D., Zhang, X., et al. (2021). Precise genome modification in tomato using an improved prime editing system. Plant Biotechnol. J. 19 (3), 415. doi: 10.1111/pbi.13497
Lu, Y., Wang, J., Chen, B., Mo, S., Lian, L., Luo, Y., et al. (2021). A donor-DNA-free CRISPR/Cas-based approach to gene knock-up in rice. Nat. Plants 7 (11), 1445–1452. doi: 10.1038/s41477-021-01019-4
Ma, H. (2005). Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annu. Rev. Plant Biol. 56 (1), 393–434. doi: 10.1146/annurev.arplant.55.031903.141717
Ma, Y., Min, L., Wang, J., Li, Y., Wu, Y., Hu, Q., et al. (2021). A combination of genome-wide and transcriptome-wide association studies reveals genetic elements leading to male sterility during high temperature stress in cotton. New Phytol. 231 (1), 165–181. doi: 10.1111/nph.17325
Marzec, M., Hensel, G. (2020). Prime editing: game changer for modifying plant genomes. Trends Plant Sci. 25 (8), 722–724. doi: 10.1016/j.tplants.2020.05.008
Millar, A. A., Clemens, S., Zachgo, S., Giblin, E. M., Taylor, D. C., Kunst, L. (1999). CUT1, an arabidopsis gene required for cuticular wax biosynthesis and pollen fertility, encodes a very-long-chain fatty acid condensing enzyme. Plant Cell 11 (5), 825–838. doi: 10.1105/tpc.11.5.825
Minkenberg, B., Wheatley, M., Yang, Y. (2017). CRISPR/Cas9-enabled multiplex genome editing and its application. Prog. Mol. Biol. Trans. Sci 149, 111–132. doi: 10.1016/bs.pmbts.2017.05.003
Min, L., Zhu, L., Tu, L., Deng, F., Yuan, D., Zhang, X. (2013). Cotton gh CKI disrupts normal male reproduction by delaying tapetum programmed cell death via inactivating starch synthase. Plant J. 75 (5), 823–835. doi: 10.1111/tpj.12245
Modrzejewski, D., Hartung, F., Lehnert, H., Sprink, T., Kohl, C., Keilwagen, J., et al. (2020). Which factors affect the occurrence of off-target effects caused by the use of CRISPR/Cas: a systematic review in plants. Front. Plant Sci. 11, 574959. doi: 10.3389/fpls.2020.574959
Molla, K. A., Sretenovic, S., Bansal, K. C., Qi, Y. (2021). Precise plant genome editing using base editors and prime editors. Nat. Plants 7 (9), 1166–1187. doi: 10.1038/s41477-021-00991-1
Mukhtar, M. S., Liu, X., Somssich, I. E. (2018). Elucidating the role of WRKY27 in male sterility in arabidopsis. Plant Signaling Behav. 13 (11), e1363945. doi: 10.1080/15592324.2017.1363945
Nie, H., Wang, Y., Su, Y., Hua, J. (2018). Exploration of miRNAs and target genes of cytoplasmic male sterility line in cotton during flower bud development. Funct. Integr. Genomics 18 (4), 457–476. doi: 10.1007/s10142-018-0606-z
Ni, F., Qi, J., Hao, Q., Lyu, B., Luo, M.-C., Wang, Y., et al. (2017). Wheat Ms2 encodes for an orphan protein that confers male sterility in grass species. Nat. Commun. 8 (1), 15121. doi: 10.1038/ncomms15121
Nishida, K., Arazoe, T., Yachie, N., Banno, S., Kakimoto, M., Tabata, M., et al. (2016). Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 353 (6305), aaf8729. doi: 10.1126/science.aaf8729
Pacini, E. (2010). Relationships between tapetum, loculus, and pollen during development. Int. J. Plant Sci. 171 (1), 1–11. doi: 10.1086/647923
Perroud, P.-F., Guyon-Debast, A., Veillet, F., Kermarrec, M.-P., Chauvin, L., Chauvin, J.-E., et al. (2022). Prime editing in the model plant physcomitrium patens and its potential in the tetraploid potato. Plant Sci. 316, 111162. doi: 10.1016/j.plantsci.2021.111162
Peterson, B. A., Haak, D. C., Nishimura, M. T., Teixeira, P. J., James, S. R., Dangl, J. L., et al. (2016). Genome-wide assessment of efficiency and specificity in CRISPR/Cas9 mediated multiple site targeting in arabidopsis. PloS One 11 (9), e0162169. doi: 10.1371/journal.pone.0162169
Qin, L., Li, J., Wang, Q., Xu, Z., Sun, L., Alariqi, M., et al. (2020). High-efficient and precise base editing of c• G to t• a in the allotetraploid cotton (Gossypium hirsutum) genome using a modified CRISPR/Cas9 system. Plant Biotechnol. J. 18 (1), 45–56. doi: 10.1111/pbi.13168
Qi, X., Zhang, C., Zhu, J., Liu, C., Huang, C., Li, X., et al. (2020). Genome editing enables next-generation hybrid seed production technology. Mol. Plant 13 (9), 1262–1269. doi: 10.1016/j.molp.2020.06.003
Richmond, T. R., Kohel, R. J. (1961). Analysis of a completely Male-sterile character in American upland cotton 1. Crop Sci. 1 (6), 397–401. doi: 10.2135/cropsci1961.0011183X000100060005x
Rout, D., Jena, D., Singh, V., Kumar, M., Arsode, P., Singh, P., et al. (2020). Hybrid rice research: Current status and prospects. Recent Adv. Rice Res, 23–59. doi: 10.5772/intechopen.87472
Sakai, H., Lee, S. S., Tanaka, T., Numa, H., Kim, J., Kawahara, Y., et al. (2013). Rice annotation project database (RAP-DB): An integrative and interactive database for rice genomics. Plant Cell Physiol. 54 (2), e6–e6. doi: 10.1093/pcp/pcs183
Shan, Q., Wang, Y., Li, J., Gao, C. (2014). "Genome editing in rice and wheat using the CRISPR/Cas system." nat. Protocols 9 (10), 2395–2410. doi: 10.1038/nprot.2014.157
Shi, J., Gao, H., Wang, H., Lafitte, H. R., Archibald, R. L., Yang, M., et al. (2017). ARGOS8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol. J. 15 (2), 207–216. doi: 10.1111/pbi.12603
Shi, J., Tan, H., Yu, X.-H., Liu, Y., Liang, W., Ranathunge, K., et al. (2011). Defective pollen wall is required for anther and microspore development in rice and encodes a fatty acyl carrier protein reductase. Plant Cell 23 (6), 2225–2246. doi: 10.1105/tpc.111.087528
Sorensen, A.-M., Kröber, S., Unte, U. S., Huijser, P., Dekker, K., Saedler, H. (2003). The arabidopsis ABORTED MICROSPORES (AMS) gene encodes a MYC class transcription factor. Plant J. 33 (2), 413–423. doi: 10.1046/j.1365-313X.2003.01644.x
Sun, L., Sui, X., Lucas, W. J., Li, Y., Feng, S., Ma, S., et al. (2019). Down-regulation of the sucrose transporter CsSUT1 causes male sterility by altering carbohydrate supply. Plant Physiol. 180 (2), 986–997. doi: 10.1104/pp.19.00317
Teng, C., Zhang, H., Hammond, R., Huang, K., Meyers, B. C., Walbot, V. (2020). Dicer-like 5 deficiency confers temperature-sensitive male sterility in maize. Nat. Commun. 11 (1), 2912. doi: 10.1038/s41467-020-16634-6
Virmani, S. S. (2003). Two-line hybrid rice breeding manual. Int. Rice Res. Inst (Los Baños, Philippines: International Rice Research Institute) 1, 1–24. Available at: http://www.knowledgebank.irri.org/ricebreedingcourse/documents/2LineHybridRiceBreeding.pdf.
Voora, V., Larrea, C., Bermudez, S. (2020). Global market report: Cotton (Winnepeg, Canada: International Institute for Sustainable Development (IISD), JSTOR).
Wang, R., Dobritsa, A. A. (2021). Loss of THIN EXINE2 disrupts multiple processes in the mechanism of pollen exine formation. Plant Physiol. 187 (1), 133–157. doi: 10.1093/plphys/kiab244
Wang, F., Feng, C., O’Connell, M. A., Stewart, J. M., Zhang, J. (2010). RFLP analysis of mitochondrial DNA in two cytoplasmic male sterility systems (CMS-D2 and CMS-D8) of cotton. Euphytica 172 (1), 93–99. doi: 10.1007/s10681-009-0055-9
Wang, X. L., Li, X. B. (2009). The GhACS1 gene encodes an acyl-CoA synthetase which is essential for normal microsporogenesis in early anther development of cotton. Plant J. 57 (3), 473–486. doi: 10.1111/j.1365-313X.2008.03700.x
Wang, Y., Li, Y., He, S.-P., Gao, Y., Wang, N.-N., Lu, R., et al. (2019). A cotton (Gossypium hirsutum) WRKY transcription factor (GhWRKY22) participates in regulating anther/pollen development. Plant Physiol. Biochem. 141, 231–239. doi: 10.1016/j.plaphy.2019.06.005
Wang, G., Xu, Z., Wang, F., Huang, Y., Xin, Y., Liang, S., et al. (2022). Development of an efficient and precise adenine base editor (ABE) with expanded target range in allotetraploid cotton (Gossypium hirsutum). BMC Biol. 20 (1), 1–15. doi: 10.1186/s12915-022-01232-3
Weaver, J., Jr. (1968). Analysis of a genetic double recessive completely Male-sterile cotton 1. Crop Sci. 8 (5), 597–600. doi: 10.2135/cropsci1968.0011183X000800050027x
Weaver, J. B., Ashley, T. (1971). Analysis of a dominant gene for Male-sterility in upland cotton, gossypium hirsutum l. 1. Crop Sci. 11 (4), 596–598. doi: 10.2135/cropsci1971.0011183X001100040048x
Weider, C., Stamp, P., Christov, N., Hüsken, A., Foueillassar, X., Camp, K. H., et al. (2009). Stability of cytoplasmic male sterility in maize under different environmental conditions. Crop Sci. 49 (1), 77–84. doi: 10.2135/cropsci2007.12.0694
Wei, M., Song, M., Fan, S., Yu, S. (2013). Transcriptomic analysis of differentially expressed genes during anther development in genetic male sterile and wild type cotton by digital gene-expression profiling. BMC Genomics 14 (1), 1–16. doi: 10.1186/1471-2164-14-97
Wei, M., Wei, H., Wu, M., Song, M., Zhang, J., Yu, J., et al. (2013). Comparative expression profiling of miRNA during anther development in genetic male sterile and wild type cotton. BMC Plant Biol. 13 (1), 1–14. doi: 10.1186/1471-2229-13-66
Woodhouse, M. R., Cannon, E. K., Portwood, J. L., Harper, L. C., Gardiner, J. M., Schaeffer, M. L., et al. (2021). A pan-genomic approach to genome databases using maize as a model system. BMC Plant Biol. 21 (1), 385. doi: 10.1186/s12870-021-03173-5
Woo, M. O., Ham, T. H., Ji, H. S., Choi, M. S., Jiang, W., Chu, S. H., et al. (2008). Inactivation of the UGPase1 gene causes genic male sterility and endosperm chalkiness in rice (Oryza sativa l.). Plant J. 54 (2), 190–204. doi: 10.1111/j.1365-313X.2008.03405.x
Wu, Y., Fox, T. W., Trimnell, M. R., Wang, L., Xu, R.-j., Cigan, A. M., et al. (2016). Development of a novel recessive genetic male sterility system for hybrid seed production in maize and other cross-pollinating crops. Plant Biotechnol. J. 14 (3), 1046–1054. doi: 10.1111/pbi.12477
Wu, J., Gong, Y., Cui, M., Qi, T., Guo, L., Zhang, J., et al. (2011). Molecular characterization of cytoplasmic male sterility conditioned by gossypium harknessii cytoplasm (CMS-D2) in upland cotton. Euphytica 181 (1), 17–29. doi: 10.1007/s10681-011-0357-6
Wu, Y., Min, L., Wu, Z., Yang, L., Zhu, L., Yang, X., et al. (2015). Defective pollen wall contributes to male sterility in the male sterile line 1355A of cotton. Sci. Rep. 5 (1), 1–8. doi: 10.1038/srep09608
Xia, R., Chen, C., Pokhrel, S., Ma, W., Huang, K., Patel, P., et al. (2019). 24-nt reproductive phasiRNAs are broadly present in angiosperms. Nat. Commun. 10 (1), 627. doi: 10.1038/s41467-019-08543-0
Xiang, X. J., Sun, L. P., Yu, P., Yang, Z. F., Zhang, P. P., Zhang, Y. X., et al. (2021). The MYB transcription factor Baymax1 plays a critical role in rice male fertility. Theor. Appl. Genet. 134 (2), 453–471. doi: 10.1007/s00122-020-03706-w
Xie, K., Minkenberg, B., Yang, Y. (2015). Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc. Natl. Acad. Sci. 112 (11), 3570–3575. doi: 10.1073/pnas.1420294112
Yadava, P., Tamim, S., Zhang, H., Teng, C., Zhou, X., Meyers, B. C., et al. (2021). Transgenerational conditioned male fertility of HD-ZIP IV transcription factor mutant ocl4: impact on 21-nt phasiRNA accumulation in pre-meiotic maize anthers. Plant Reprod. 34 (2), 117–129. doi: 10.1007/s00497-021-00406-3
Yamagishi, H., Bhat, S. R. (2014). Cytoplasmic male sterility in brassicaceae crops. Breed. Sci. 64 (1), 38–47. doi: 10.1270/jsbbs.64.38
Yang, P., Han, J., Huang, J. (2014). Transcriptome sequencing and de novo analysis of cytoplasmic male sterility and maintenance in JA-CMS cotton. PloS One 9 (11), e112320. doi: 10.1371/journal.pone.0112320
Yang, K.-Z., Xia, C., Liu, X.-L., Dou, X.-Y., Wang, W., Chen, L.-Q., et al. (2009). A mutation in THERMOSENSITIVE MALE STERILE 1, encoding a heat shock protein with DnaJ and PDI domains, leads to thermosensitive gametophytic male sterility in arabidopsis. Plant J. 57 (5), 870–882. doi: 10.1111/j.1365-313X.2008.03732.x
Yang, Z., Zhang, Y., Sun, L., Zhang, P., Liu, L., Yu, P., et al. (2018). Identification of cyp703a3-3 and analysis of regulatory role of CYP703A3 in rice anther cuticle and pollen exine development. Gene 649, 63–73. doi: 10.1016/j.gene.2018.01.058
You, J., Li, M., Li, H., Bai, Y., Zhu, X., Kong, X., et al. (2022). Integrated methylome and transcriptome analysis widen the knowledge of cytoplasmic Male sterility in cotton (Gossypium barbadense l.). Front. Plant Sci. 13. doi: 10.3389/fpls.2022.770098
Yu, J., Han, J., Kim, Y.-J., Song, M., Yang, Z., He, Y., et al. (2017). Two rice receptor-like kinases maintain male fertility under changing temperatures. Proc. Natl. Acad. Sci. 114 (46), 12327–12332. doi: 10.1073/pnas.1705189114
Yu, J., Jung, S., Cheng, C.-H., Lee, T., Zheng, P., Buble, K., et al. (2021). CottonGen: The community database for cotton genomics, genetics, and breeding research. Plants 10 (12), 2805. doi: 10.3390/plants10122805
Yu, B., Liu, L., Wang, T. (2019). Deficiency of very long chain alkanes biosynthesis causes humidity-sensitive male sterility via affecting pollen adhesion and hydration in rice. Plant Cell Environ. 42 (12), 3340–3354. doi: 10.1111/pce.13637
Yu, J., Meng, Z., Liang, W., Behera, S., Kudla, J., Tucker, M. R., et al. (2016). A rice Ca2+ binding protein is required for tapetum function and pollen formation. Plant Physiol. 172 (3), 1772–1786. doi: 10.1104/pp.16.01261
Zhang, T., Feng, Y., Pan, J. (1992). Genetic analysis of four genetic male-sterile lines found in upland cottons in China. Acta Gossypii Sin. (China) 4, 1–8.
Zhang, Y., Han, Y., Zhang, M., Zhang, X., Guo, L., Qi, T., et al. (2022). The cotton mitochondrial chimeric gene orf610a causes male sterility by disturbing the dynamic balance of ATP synthesis and ROS burst. Crop J 10. doi: 10.1016/j.cj.2022.02.008
Zhang, T., Hu, Y., Jiang, W., Fang, L., Guan, X., Chen, J., et al. (2015). Sequencing of allotetraploid cotton (Gossypium hirsutum l. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 33 (5), 531–537. doi: 10.1038/nbt.3207
Zhang, H., Liang, W., Yang, X., Luo, X., Jiang, N., Ma, H., et al. (2010). Carbon starved anther encodes a MYB domain protein that regulates sugar partitioning required for rice pollen development. Plant Cell 22 (3), 672–689. doi: 10.1105/tpc.109.073668
Zhang, M., Wei, H., Hao, P., Wu, A., Ma, Q., Zhang, J., et al. (2021a). GhGPAT12/25 are essential for the formation of anther cuticle and pollen exine in cotton (Gossypium hirsutum l.). Front. Plant Sci. 12, 720. doi: 10.3389/fpls.2021.667739
Zhang, M., Wei, H., Liu, J., Bian, Y., Ma, Q., Mao, G., et al. (2021b). Non-functional GoFLA19s are responsible for the male sterility caused by hybrid breakdown in cotton (Gossypium spp.). Plant J. 107 (4), 1198–1212. doi: 10.1111/tpj.15378
Zhang, D., Wu, S., An, X., Xie, K., Dong, Z., Zhou, Y., et al. (2018). Construction of a multicontrol sterility system for a maize male-sterile line and hybrid seed production based on the ZmMs7 gene encoding a PHD-finger transcription factor. Plant Biotechnol. J. 16 (2), 459–471. doi: 10.1111/pbi.12786
Zhang, S., Wu, S., Niu, C., Liu, D., Yan, T., Tian, Y., et al. (2021c). ZmMs25 encoding a plastid-localized fatty acyl reductase is critical for anther and pollen development in maize. J. Exp. Bot. 72 (12), 4298–4318. doi: 10.1093/jxb/erab142
Zhang, H., Xu, C., He, Y., Zong, J., Yang, X., Si, H., et al. (2013). Mutation in CSA creates a new photoperiod-sensitive genic male sterile line applicable for hybrid rice seed production. Proc. Natl. Acad. Sci. 110 (1), 76–81. doi: 10.1073/pnas.1213041110
Zhang, M., Zhang, X., Guo, L., Qi, T., Liu, G., Feng, J., et al. (2020). Single-base resolution methylome of cotton cytoplasmic male sterility system reveals epigenomic changes in response to high-temperature stress during anther development. J. Exp. Bot. 71 (3), 951–969. doi: 10.1093/jxb/erz470
Zhang, P., Zhang, Y., Sun, L., Sinumporn, S., Yang, Z., Sun, B., et al. (2017). The rice AAA-ATPase OsFIGNL1 is essential for Male meiosis. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01639
Zhang, T. Y., Hu, W., Fang, X., Guan, J., Chen, J., Zhang, C. A., et al. (2015). Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nature Biotechnol 33 (5), 531–537.
Zheng, W., Ma, Z., Zhao, M., Xiao, M., Zhao, J., Wang, C., et al. (2020). Research and development strategies for hybrid japonica rice. Rice 13 (1), 1–22. doi: 10.1186/s12284-020-00398-0
Zhou, H., He, M., Li, J., Chen, L., Huang, Z., Zheng, S., et al. (2016). Development of commercial thermo-sensitive genic Male sterile rice accelerates hybrid rice breeding using the CRISPR/Cas9-mediated TMS5 editing system. Sci. Rep. 6, 1–12. doi: 10.1038/srep37395
Zhu, L., He, S., Liu, Y., Shi, J., Xu, J. (2020). Arabidopsis FAX1 mediated fatty acid export is required for the transcriptional regulation of anther development and pollen wall formation. Plant Mol. Biol. 104 (1), 187–201. doi: 10.1007/s11103-020-01036-5
Zhu, Y., Shi, Z., Li, S., Liu, H., Liu, F., Niu, Q., et al. (2018). Fine mapping of the novel male-sterile mutant gene ms39 in maize originated from outer space flight. Mol. Breed. 38 (10), 125. doi: 10.1007/s11032-018-0878-y
Zou, T., He, Z., Qu, L., Liu, M., Zeng, J., Liang, Y., et al. (2017). Knockout of OsACOS12 caused male sterility in rice. Mol. Breed. 37 (10), 126. doi: 10.1007/s11032-017-0722-9
Keywords: upland cotton, candidate genes, heterosis, homology, genetic male sterility, genome editing
Citation: Morales KY, Bridgeland AH, Hake KD, Udall JA, Thomson MJ and Yu JZ (2022) Homology-based identification of candidate genes for male sterility editing in upland cotton (Gossypium hirsutum L.). Front. Plant Sci. 13:1006264. doi: 10.3389/fpls.2022.1006264
Received: 29 July 2022; Accepted: 18 November 2022;
Published: 14 December 2022.
Edited by:
Qian-Hao Zhu, Commonwealth Scientific and Industrial Research Organisation (CSIRO), AustraliaReviewed by:
Qiuming Yao, University of Nebraska-Lincoln, United StatesCopyright © 2022 Morales, Bridgeland, Hake, Udall, Thomson and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: John Z. Yu, john.yu@usda.gov
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.