- 1Key Laboratory of Plant Resources Conservation and Germplasm Innovation in Mountainous Region (Ministry of Education), College of Life Sciences/Institute of Agro-bioengineering, Guizhou University, Guiyang, China
- 2Guizhou Academy of Tobacco Science, Molecular Genetics Key Laboratory of China Tobacco, Guiyang, China
- 3Department of Microbiology, Baluchistan University of Information Technology and Managemnet Sciences, Quetta, Pakistan
- 4Plant Conservation Technology Center, Guizhou Key Laboratory of Agricultural Biotechnology, Guizhou Academy of Agricultural Sciences, Guiyang, China
Trichomes are specialized hair-like organs found on epidermal cells of many terrestrial plants, which protect plant from excessive transpiration and numerous abiotic and biotic stresses. However, the genetic basis and underlying mechanisms are largely unknown in Nicotiana tabacum (common tobacco), an established model system for genetic engineering and plant breeding. In present study, we identified, cloned and characterized an unknown function transcription factor NtMYB306a from tobacco cultivar K326 trichomes. Results obtained from sequence phylogenetic tree analysis showed that NtMYB306a-encoded protein belonged to S1 subgroup of the plants’ R2R3-MYB transcription factors (TFs). Observation of the green fluorescent signals from NtMYB306a-GFP fusion protein construct exhibited that NtMYB306a was localized in nucleus. In yeast transactivation assays, the transformed yeast containing pGBKT7-NtMYB306a construct was able to grow on SD/-Trp-Ade+X-α-gal selection media, signifying that NtMYB306a exhibits transcriptional activation activity. Results from qRT-PCR, in-situ hybridization and GUS staining of transgenic tobacco plants revealed that NtMYB306a is primarily expressed in tobacco trichomes, especially tall glandular trichomes (TGTs) and short glandular trichomes (SGTs). RNA sequencing (RNA-seq) and qRT-PCR analysis of the NtMYB306a-overexpressing transgenic tobacco line revealed that NtMYB306a activated the expression of a set of key target genes which were associated with wax alkane biosynthesis. Gas Chromatography–Mass Spectrometry (GC-MS) exhibited that the total alkane contents and the contents of n-C28, n-C29, n-C31, and ai-C31 alkanes in leaf exudates of NtMYB306a-OE lines (OE-3, OE-13, and OE-20) were significantly greater when compared to WT. Besides, the promoter region of NtMYB306a contained numerous stress-responsive cis-acting elements, and their differential expression towards salicylic acid and cold stress treatments reflected their roles in signal transduction and cold-stress tolerance. Together, these results suggest that NtMYB306a is necessarily a positive regulator of alkane metabolism in tobacco trichomes that does not affect the number and morphology of tobacco trichomes, and that it can be used as a candidate gene for improving stress resistance and the quality of tobacco.
Introduction
A well-integrated epidermis is critical for several important processes in plant development, shoot growth and plant defense (Javelle et al., 2011). The outmost epidermal layer of the plant leaf is composed of pavement cells, followed by specialized guard cells and trichomes, respectively (Bird and Gray, 2003). Trichomes are uni- or multi-cellular appendages resulting from divisions of epidermal cells, which primarily help in reducing evaporation by protecting the plant from wind and heat. Research has shown that trichomes found on leaves and stems accumulate and produce secondary metabolites in a species- and cultivar-specific manner, which provides forefront protection against herbivores, pathogens, and insects. Numerous studies have documented the important roles of trichomes in plant resistance to biotic and abiotic stresses, including drought and ultraviolet radiation (Wagner, 1991; Serna and Martin, 2006; McDowell et al., 2011; Glas et al., 2012). The trichomes of Tobacco (Nicotiana tabacum L.), an emerging model system for plant biology over the past several decades, are morphologically divided into three types: tall glandular trichome (TGT), short glandular trichome (SGT), and non-glandular trichome (NGT) (Wang et al., 2004; Huang et al., 2018; Wang et al., 2021b). In tobacco, many compounds such as cembranoids, sucrose esters, alkanes, and fatty acids are synthesized and secreted by trichomes, which are considered to be the main components of tobacco leaf exudates. In addition to inducing stress resistance in tobacco, they are also reported to be the precursors of important compounds produced in the process of leaf mellowing and are associated with the aroma of tobacco (Heemann et al., 1983; Johnson et al., 1985; Wagner, 1991; Sallets et al., 2014; Huang et al., 2018). Additionally, in flavology, alkanes produced from plant been shown to have a significant impact on flavor release and odor switch (Fu et al., 2021b). However, little is known about the key enzymes and associated genetic pathways involved in the biosynthesis of wax-alkanes in tobacco trichomes.
Plant epidermal waxes are complex mixtures of very-long-chain fatty acids (VLCFAs, with chain lengths of C20 to C34), alkanes, ketones, aldehydes, esters, and alcohols (Yeats and Rose, 2013; Jetter and Riederer, 2016). In Arabidopsis thaliana, the synthesis of these compounds occurs in endoplasmic reticulum, where four types of fatty acid elongase (FAE) complexes, including ketoacyl-CoA synthase (KCS), enoyl-CoA reductase (ECR), ketoacyl-CoA reductase (KCR), and hydroxyacyl-CoA dehydratase (HCD), catalyze the precursors of C16/C18 fatty acyls to form VLCFAs (Beaudoin et al., 2009; Javelle et al., 2011; Bernard and Joubès, 2013; Lee and Suh, 2015a; Xue et al., 2017). The first reaction catalyzed by KCS is the rate-limiting step of the FAE complex (Bird and Gray, 2003; Lee and Suh, 2015a). It is believed that KCS can specifically control the chain length of different substrates or products, while this specialty is not observed in the other three enzymes involved in chain extension (Hegebarth et al., 2016). VLCFAs serve as precursors for wax synthesis in plants, and the associated pathway split-up into two sub-pathways: the alkane synthesis pathway and alcohol synthesis pathway. The alkane synthesis pathway is also called the decarbonylation pathway, which mainly produces alkanes, aldehydes, ketones, and secondary alcohols (Samuels et al., 2008; Lee and Suh, 2015a; Pascal et al., 2019). So far, only a few genes involved in the wax-alkane synthesis pathway have been reported. Elongase 3-ketoacyl-CoA synthase encoded by KCS1, as one of the FAE complexes, is responsible for the extension of VLCFA chains (Todd et al., 1999). Two genes, CER1 and CER3, encoding alkane synthetase and VLCFA reductase enzymes respectively, play a vital role in the biosynthesis of VLC-alkanes by controlling the reduction and decarbonylation of VLCFA-CoA to produce alkanes. Among these, the VLCFA reductase produces fatty aldehydes and catalyzes the conversion of very long-chain acyl-CoA to very long-chain alkanes, while alkane synthetase catalyzes the decarbonylation of fatty aldehydes to n-alkanes (Bourdenx et al., 2011; Bernard et al., 2012; Yeats and Rose, 2013; Xue et al., 2017; Pascal et al., 2019). Overexpression of CER1 in Poa pratensis (Wang et al., 2021a) and cucumber (Wang et al., 2015) further confirmed the significant role of CER1 in alkane biosynthesis. In addition, GDSL, ABCG, and HOTHEAD, along with CYP77A and CYP85A belonging to the cytochrome P450 (CYP450) family, are all involved in the synthesis or transport of waxes and cutins to varying degrees (Panikashvili et al., 2011; Girard et al., 2012; Fabre et al., 2015; Lashbrooke et al., 2015; Nguyen et al., 2018; Shi et al., 2018; Tang et al., 2020). Research has shown that wax components in Arabidopsis trichomes are different from those of other epidermal cells, with higher concentrations of alkanes in trichomes. A study exhibited that KCS genes, such as KCS1, KCS5, and KCS10, were highly expressed in trichome cells, and may participate in the biosynthesis of wax components in trichomes (Hegebarth et al., 2016).
MYB transcription factors (TFs) are one of the largest TF families in plants and can function as gene activators. According to the number of MYB domains, the MYB TFs can be divided into four subfamilies including 1R-MYB, R2R3-MYB, 3R-MYB (R1R2R3-MYB), and 4R-MYB (Dubos et al., 2010; Liu et al., 2015). R2R3-MYB is the largest subfamily of MYB TFs, usually containing two MYB domains near the N-terminus and C-terminus. Based on sequence variation at the, R2R3-MYB members are further divided into 23 subgroups (Dubos et al., 2010), and members of the same subgroup show similar functions (Dubos et al., 2010; Jiang and Rao, 2020). R2R3-MYB TFs are involved in many plants physiological and biochemical activities, such as growth and development, primary and secondary metabolism, cell fate determination, and biotic and abiotic stress responses (Dubos et al., 2010; Liu et al., 2015; Jiang and Rao, 2020). It is known that members of the S1 and S9 subgroups of R2R3-MYB can regulate the synthesis of VLCFAs including waxes and cutins. For example, overexpression of MYB94, a member of the S1 subgroup, increased the total amount of waxes in Arabidopsis by activating the expression of genes (e.g., KCS2 and CER2) related to the wax synthesis pathway (Lee and Suh, 2015b). MYB96, a transcriptional activator of wax biosynthesis in Arabidopsis, up-regulated the expression of KCS1, KCS2, CER1, and CER3 (Seo et al., 2011). MdMYB30 bound to the MdKCS1 promoter to activate its expression and regulate wax biosynthesis in apple (Zhang et al., 2019). In wheat, TaMYB31 has been reported to play a key role in drought stress tolerance by up-regulating the expression of wax biosynthesis genes (Zhao et al., 2018). The S9 subgroup members (i.e., MIXTA-MYB, AaMIXTA1 and MYB16) play an important role in controlling plant cell shape, epidermal permeability, and trichome development by regulating the expression of the structure and transport genes in the wax/cuticle biosynthesis pathway (Oshima and Mitsuda, 2013; Oshima et al., 2013; Shi et al., 2018; Jiang and Rao, 2020; Xu et al., 2021). Although, the members of the S1 subgroup have been well characterized in plants such as Arabidopsis, their roles in tobacco require further exploration.
In this study, we isolated and characterized a MYB TF, NtMYB306a, which was predominantly expressed in tobacco trichomes. Analyses of phylogenetic tree, protein sequence alignment, subcellular localization, and yeast transactivation assays indicated that NtMYB306a-encoded protein belongs to the S1 subgroup of MYB-R2R3 TFs and has transcriptional activation activity. The high expression of NtMYB306a in tobacco trichomes was further verified by quantitative real-time PCR (qRT-PCR), in situ hybridization, and GUS staining. In addition, the expression patterns of NtMYB306a under salicylic acid (SA), abscisic acid (ABA), and cold stress conditions were examined in tobacco. Finally, the target genes possibly activated by NtMYB306a were analyzed by RNA sequencing (RNA-seq) and qRT-PCR in tobacco plants overexpressing NtMYB306a. Gas chromatography-mass spectrometry (GC-MS) further detected the increase of alkane content in NtMYB306a-overexpression (NtMYB306a-OE) plants. Overall, our study provides a valuable reference and new insight for the regulatory network and candidate genes of the alkane biosynthesis pathway in tobacco trichomes.
Materials and methods
Plant materials and growth conditions
Tobacco (N. tabacum L.) cultivar K326 was used in this study. Tobacco plants were grown under 25 ± 2°C and 60% humidity conditions, with a photoperiod of 16/8 h light/dark.
Isolation and characterization of NtMYB306a
In order to amplify NtMYB306a gene, PCR was performed using specific primers (Supplementary Table S1) and using tobacco trichomes that were obtained from frozen leaves according to a previous report (Zhang et al., 2018). The amino acid sequences of 126 R2R3-MYB proteins in A. thaliana were obtained from the TAIR database (http://www.arabidopsis.org/). A phylogenetic tree based on full-length amino acid sequences of R2R3-MYB proteins from tobacco and A. thaliana was constructed using MAGE v6.0. The neighbor-joining (NJ) method was performed with 1,000 bootstrap replications. Evolview v3.0 was used to further visualize the phylogenetic tree (Subramanian et al., 2019).
Sample collection
To investigate the spatial and temporal expression patterns of NtMYB306a, we collected different tissues (roots, stems, flowers, leaves, leaves without trichomes, and trichomes) and leaves of different positions (the 1st, 4th, 7th, 11th, 15th, and 19th leaf, from the top to the bottom of the 5-month-old plant). All samples were frozen immediately in liquid nitrogen and stored at -80°C until RNA extraction, and three biological replicates were conducted for each sample.
Hormone and abiotic stress treatments
Base leaves of 40-day-old tobacco plants were subjected to hormonal and cold stress treatments. The criteria of selection for the phytohormones and different treatment type was based on the in-silico detection of cis-acting elements in upstream regions of NtMYB306a gene. For hormone treatments, 10 μmol·L-1 ABA, 50 mg·L-1 SA, and 100 μmol·L-1 MeJA were sprayed on the leaves, respectively. For abiotic stress treatments, samples were subjected cold stress by transferring the plants to 4°C conditions. Samples were taken at 0, 6, 12, 24, 48, and 72 h post-treatment and immediately frozen in liquid nitrogen and stored at -80°C until further use. All treatments were performed with three biological replicates.
RNA extraction and qRT-PCR analysis
Total RNA was extracted by using a plant RNA extraction kit (TransGen Biotech, Beijing, China) according to the manufacturer’s instructions. For the first-strand cDNA synthesis, a TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix Kit (TransGen Biotech, Beijing, China) was used. qRT-PCR analysis was performed with TB Green Premix DimerEraser (TaKaRa, Dalian, China) on a ViiA7 Real-Time PCR Detection System (Applied Biosystems, USA), β-actin was used as the internal reference for normalization, and data were analyzed by using the 2-ΔΔCT method (Livak and Schmittgen, 2001). The qRT-PCR program was as follows: 95°C for 30 s; followed by 40 cycles of 95°C for 5 s, 60°C for 30 s, and 72°C for 34 s. Then, a melting curve analysis was performed. Primers used for qRT-PCR are listed in Supplementary Table S1.
Yeast transactivation assay
For the yeast transactivation assay, full-length open reading frame (ORF) of NtMYB306a was cloned and fused in the GAL4 DNA-binding domain of the pGBKT7 vector through Nde I and Not I restriction sites (primers are listed in Supplementary Table S1). The pGBKT7-NtMYB306a construct was transformed into yeast strain AH109 and grown on SD/-Trp, SD/-Trp-Ade, and SD/-Trp-Ade+X-α-gal selection media at 30°C for 3 d. Transactivation ability was determined by evaluating the growth of yeast cells on the selection medium.
Subcellular localization
To determine the subcellular location of the NtMYB306a protein, the coding sequence (CDS) without the stop codon was ligated into the pBWA(V)HS-GLosgfp vector, which contains the CaMV 35S promoter and green fluorescent protein (GFP) gene, resulting in a fusion gene driven by the 35S promoter. Subsequently, this fusion vector and the control vector (NLS-mCherry vector that contains a red fluorescent protein) were electroporated into A. tumefaciens strain GV3101 and simultaneously infiltrated into the N. benthamiana leaves. After 48 h of infection, a Nikon C2+ confocal microscope (Nikon, Japan) was used to observe the green and red fluorescent signals. Primers used for plasmid construction are listed in Supplementary Table S1.
In situ hybridization
Leaves of 60-day-old plants were used for in situ hybridization assay. Probes (5’-DIG-GUGUAAAAUUACCACCCUCAUCGACCGACAUG-DIG-3’) were designed and labeled with DIG-UTP. Tobacco leaves were fixed with FAA fixative, and the following procedures including embedding, section, hybridization, and development were performed as previously described (Choi et al., 2011). Leaf sections were then observed and photographed by a bright field microscopy.
Molecular cloning of the NtMYB306a promoter and promoter-GUS fusion in transgenic tobacco plants
The 2,077 bp sequence upstream of the NtMYB306a translation start site was considered as the promoter region and amplified from the K326 genome, primers are listed in Supplemental Table S1. The cis-acting elements of the NtMYB306a promoter were analyzed using the PlantCARE website (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/). The NtMYB306a promoter was constructed into the pCAMBIA1391Z vector and transformed into tobacco plants. Positive plants were identified by PCR, and GUS staining was conducted according to the method described by Jefferson et al. (1987). Tobacco tissues were immersed in the GUS staining buffer (0.5 mg/mL X-Gluc, 50 mmol/L sodium phosphate buffer [pH 7.0], 0.5 mol/L EDTA, 1% Triton X-100, and 40 mL/L methanol) at 37°C for 16-20 h, and then the material was dehydrated through an ethanol gradient and then photographed under a stereomicroscope.
Tobacco transformation
Tobacco plants were transformed by the Agrobacterium-mediated method (Horsch et al., 1985), and the selection marker gene of the genetic transformation vector was hygromycin phosphotransferase (HPT). After three rounds of hygromycin (20 mg/L) selection on Murashige and Skoog (MS) medium, the regenerated buds were excised and rooted, and the T0 plants were self-crossed and harvested. The positive transgenic lines were screened on MS plates containing hygromycin (20 mg/L), and the expression level of NtMYB306a was detected by qRT-PCR. The T2 or T3 plants were used for molecular and metabolic analyses.
RNA-seq and data analysis
Leaves were collected from 5-mouth-old plants including OE-20 T1 and WT plants, with three biological replicates per sample. Total RNA was extracted as described above. The following procedures were performed by Novogene Bioinformatics Technology Co., Ltd. (Beijing, China). Libraries were constructed and sequenced using an Illumina HiSeq 2500 platform. The RNA-seq raw data were transformed, filtered, and aligned to the N. tabacum cv.TN90 reference genome (https://www.ncbi.nlm.nih.gov/assembly/GCF_000715135.1/) (Sierro et al., 2014). FPKM of each gene was calculated based on the length of the gene and read counts mapped to this gene. Differential expression level, GO, and KEGG enrichment analyses of DEGs were performed on the Novomagic cloud platform (Novogene Bioinformatics Technology Co., Ltd., Beijing, China). DEGs were determined according to the following thresholds: |log2(fold-change)| > 1 and adjusted padj < 0.05.
Analysis of alkanes in tobacco trichomes using GC-MS
According to Huang et al. (2018), leaves of 5-mouth-old tobacco plants were immersed in dichloromethane (CH2Cl2) solution three times for 2 s each time, which were combined and added with n-tetracosane (C24 alkane) as an internal reference. The extracted leaf exudate was vacuum-dried until most of the solvent was evaporated and the final volume reached 1 mL. The leaf exudate was passed through an organic filter membrane and analyzed by a GC-MS (7890B/5977B, Agilent, USA). Helium was used as the carrier gas, the flow rate was 1.0 mL/min; the temperature of the injection port was 250°C, and the injection volume was 1 µL; split injection was performed, and the split ratio was 10:1; the initial temperature was 50°C, hold for 1 min; from 50°C to 280°C at a rate of 5°C/min, hold for 1 min; then from 280°C to 300°C at a rate of 20°C/min, hold for 15 min; the total running time was 64 min. For the MS, the following parameters were used: MS detector; solvent delay: 3 min; ionization voltage: 70 eV; ion source temperature: 230°C; transmission line temperature: 280°C; scanning ion range: 35-450 amu; MS scanning mode: SCAN. The spectrum of each peak was analyzed by GC-MS using the mass spectrum database and the relative retention time of each peak, to determine the substance corresponding to each peak.
Trichome count and SEM observation
Leaves of four randomly selected 5-month-old tobacco plants were collected for trichome count, and 5-8 mm2 pieces between the margin and middle rib were taken from leaves of the 10th to 12th positions, avoiding the midrib. Leaf pieces were fixed with 0.1 M phosphate buffer (pH 7.4) containing 2.5% glutaraldehyde for 24 h and 1% osmic acid for 2 h. After dehydration, replacement, drying, mounting, and gold spraying, a HITACHI Regulus 8100 SEM was used to observe the trichomes with an accelerating voltage of 3 kV, and the number of trichomes in the 1 mm2 area was calculated.
Results
Identification and characterization of NtMYB306a in tobacco
To identify the TFs that regulate important metabolites in tobacco trichomes, our research focused on the TFs that are predominantly expressed in trichomes. Therefore, the transcriptomic data (data not shown) of tobacco cultivar K326 trichomes was compared to those of leaves without trichomes. As a result, we identified 18 MYB genes that were predominantly expressed in tobacco trichomes, among which eight were annotated as MYB306 TFs but with unknown function. It is reported that tobacco leaf exudates mainly include cembranoids, sucrose esters, alkanes, and fatty acids (Johnson et al., 1985; Sallets et al., 2014). Given that the trichomes are tiny specialized organs derived from epidermal cells, it was initially speculated that these MYB306 genes, expressed predominantly in trichomes, may be related to the occurrence and development of trichomes, and the synthesis or transport of secondary metabolites.
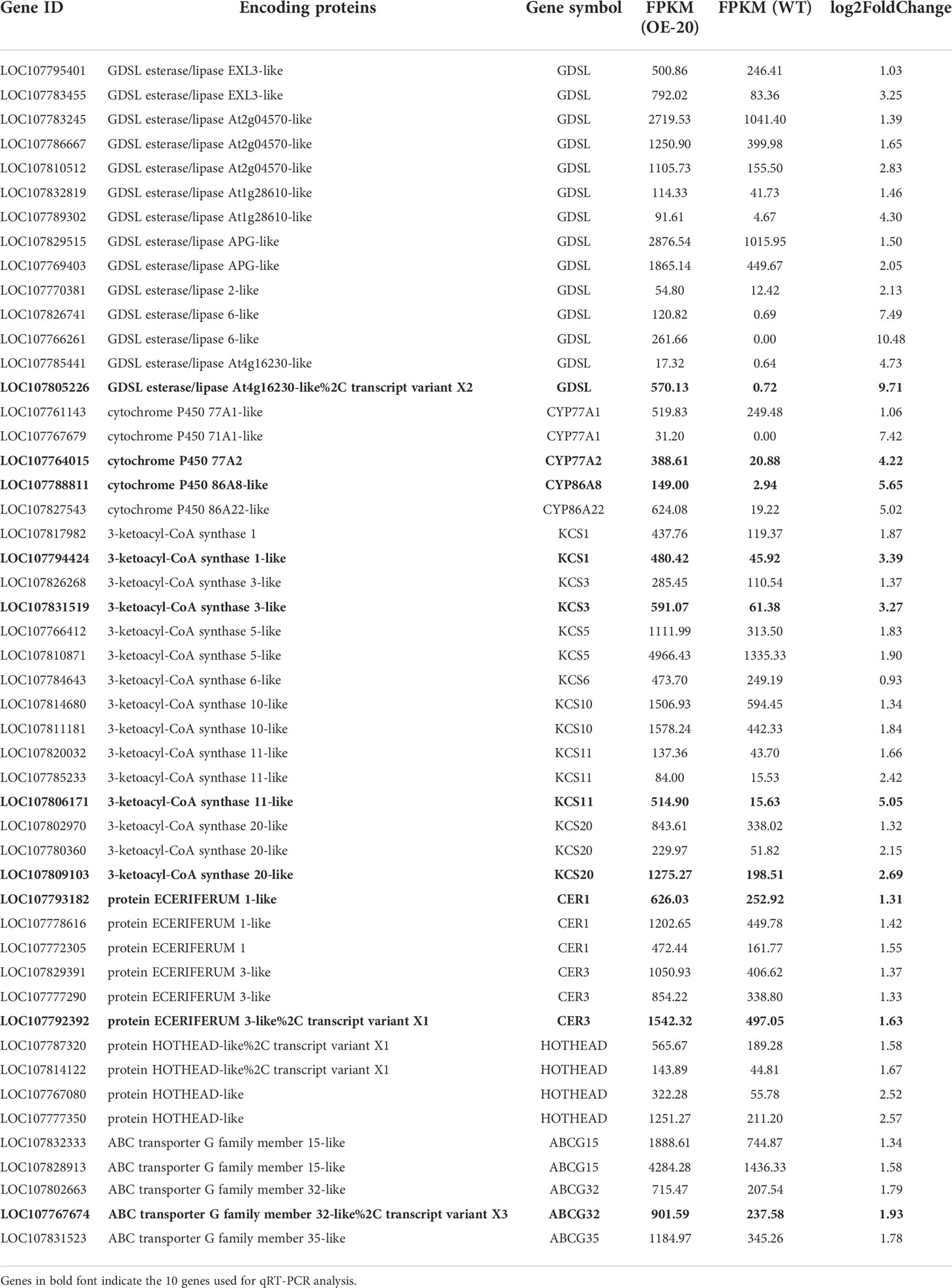
To identify the biological function of the MYB306 gene, one of these genes from the cDNA of tobacco trichomes (accession number: LOC107818868) was cloned and named NtMYB306a (Supplementary Figure S1). Sequence analysis showed that the cDNA of NtMYB306a contained 1,095 nucleotides, encoding 364 amino acids (aa); the predicted molecular weight (MW) of the encoded protein was 40.46 kDa, and the theoretical isoelectric point (pI) was 6.29. Ten homologous genes of NtMYB306a from 10 other species were retrieved from the NCBI database using homology-based search. These homologous genes, together with NtMYB306a, were used to perform amino acid sequence alignment analysis and construct a phylogenetic tree (Figures 1A, B). Phylogenetic analysis of these 10 MYB306s revealed that NtMYB306a clustered with NtomMYB306 (Figure 1B), and the similarity between the amino acid sequences of NtMYB306a and those of NtomMYB306 and NsMYB306 were 99.73% and 92.16%, respectively (Figure 1A). It is reported that N. tabacum has evolved from chromosome duplication after an interspecific hybridization of two diploid ancestors (i.e., N. sylvestris and N. tomentosiformis) (Leitch et al., 2008). The clustering pattern and close homology between NtMYB306a and NtomMYB306 indicated that NtMYB306a may have originated from the diploid parent N. tomentosiformis. Moreover, NtMYB306a was found to share close homology to the MYB306s of other plants such as pepper, potato, and tomato, ascribing to the fact that these species belong to the Solanaceae family. The amino acid sequences of these MYB306s were highly conserved at the N-terminus that contained two DNA-binding domains, R2 and R3, with amino acids located at 13-63aa and 66-114aa, respectively (Figure 1A; Supplementary Figure S2). On the contrary, the C-terminal regions of the MYB306s were found to be more diverged. These results suggest that the MYB306 genes are conserved during evolution, and NtMYB306a belongs to the R2R3-MYB subfamily. Members of the same subgroup of R2R3-MYB proteins often harbor similar biological functions (Dubos et al., 2010; Jiang and Rao, 2020). The phylogenetic tree constructed based on NtMYB306a and 126 Arabidopsis R2R3-MYBs showed that NtMYB306a belonged to the S1 subgroup of R2R3-MYB, and clustered with AtMYB30 (Figure 1C), with amino acid similarity of 45.53%. It has been shown that members of the S1 subgroup of plant R2R3-MYB proteins are regulators of wax and cutin biosynthesis (Seo et al., 2011; Lee and Suh, 2015b; Xue et al., 2017; Zhao et al., 2018; Zhang et al., 2019). Therefore, we speculated that NtMYB306a-encoded protein is likely to be involved in tobacco wax or cutin biosynthesis.
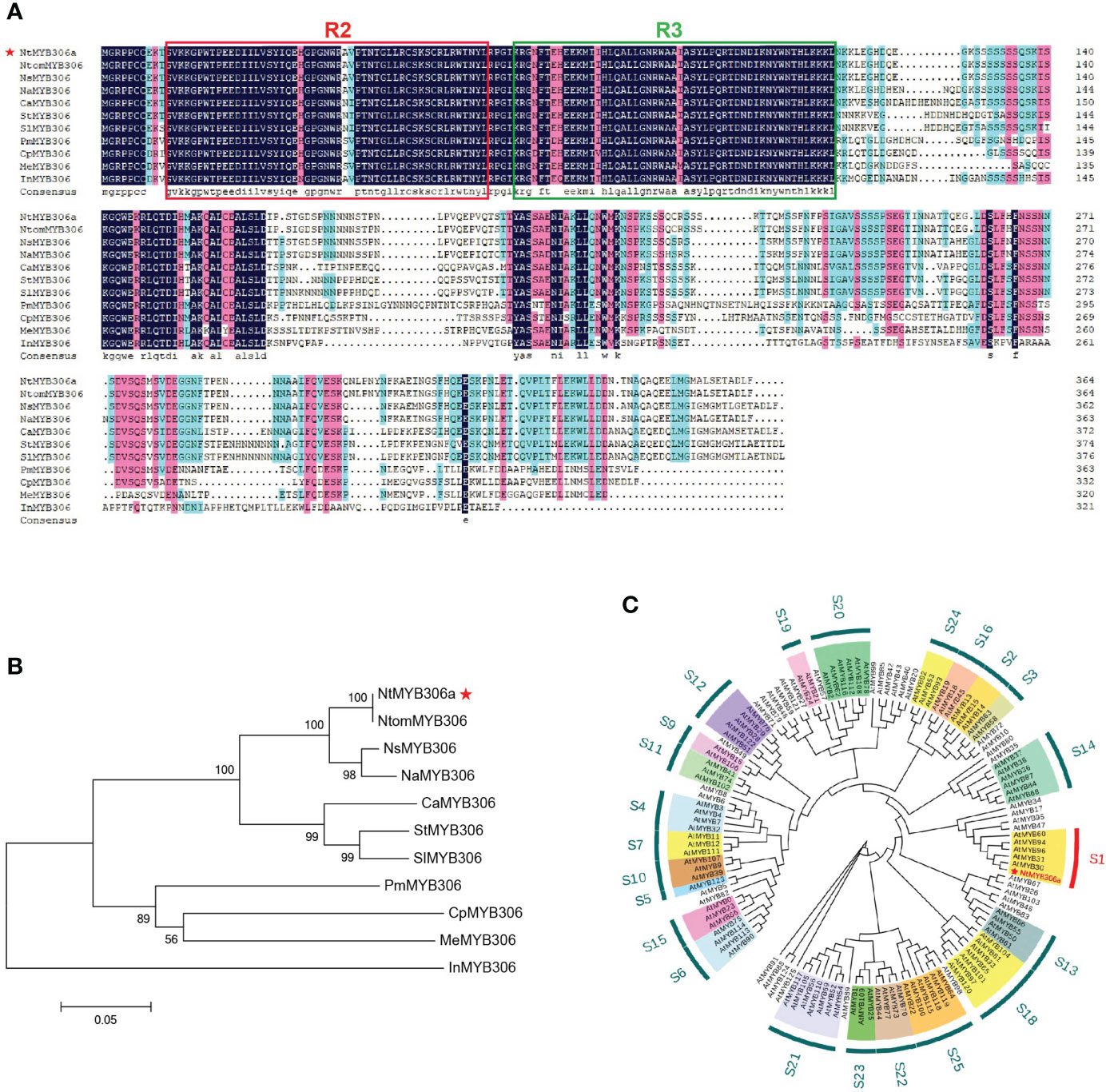
Figure 1 Multiple sequence alignment and phylogenetic analysis of the NtMYB306a protein and its homologs from various plant species. (A) Protein sequence alignment of MYB306 homologs. The R2 and R3 MYB DNA binding domains are indicated by red and green boxes, respectively. (B) Phylogram of NtMYB306a and related R2R3-MYBs from other plant species, including Nicotiana tomentosiformis (NtomMYB306: XP_009602669.1), Nicotiana sylvestris (NsMYB306: XP_009760429.1), Nicotiana attenuata (NaMYB306: XP_019239267.1), Capsicum annuum (CaMYB306: XP_016578359.1), Solanum tuberosum (StMYB306: XP_006354829.1), Ipomoea nil (InMYB306: XP_019198062.1), Solanum lycopersicum (SlMYB306: XP_004241580.1), Prunus mume (PmMYB306: XP_008239949.1), Manihot esculenta (MeMYB306: XP_021614785.1), and Carica papaya (CpMYB306: XP_021891841.1). NtMYB306a is highlighted with a red star. (C) Phylogenetic tree of NtMYB306a and Arabidopsis thaliana R2R3-MYBs. Individual subgroups are indicated with different colors. NtMYB306a, belonging to the S1 subgroup and being most closely related to AtMYB30, is highlighted with a red asterisk.
Subcellular localization and transcriptional activation activity of NtMYB306a
Our above results revealed that NtMYB306a encodes a typical R2R3-MYB TF with transcriptional regulation activity (Figure 1), and suggested that NtMYB306a may function inside the nucleus. To investigate the subcellular localization of the NtMYB306a protein, a open reading frame (ORF) of the NtMYB306a without the terminator codon was fused to the GFP. The fusion protein construct was transiently expressed in N. benthamiana leaves and the GFP fluorescence of 35S: NtMYB306a-GFP was observed. These results demonstrated that the green fluorescence of NtMYB306a::GFP fusion protein completely coincided with the nuclear localized control vector (NLS-mCherry vector that contains a red fluorescent protein), indicating that NtMYB306a was localized in the nucleus (Figure 2A), which is also consistent with its role as a TF.
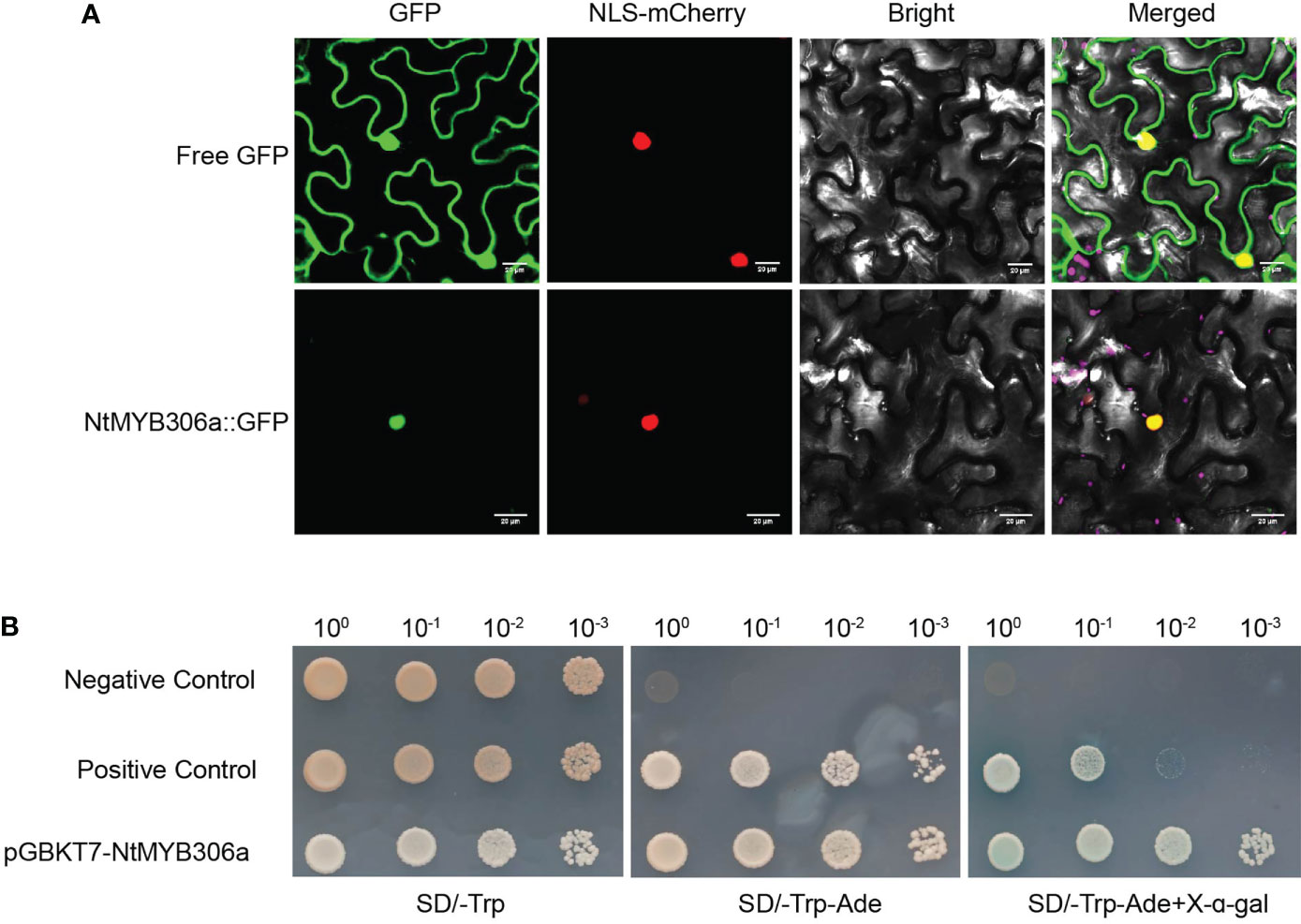
Figure 2 Subcellular localization and transcriptional activation of NtMYB306a. (A) NtMYB306a is located in the nucleus. GFP: green fluorescent protein signal; NLS-mCherry: red fluorescence indicates the nucleus-localized signal (NLS); Bright: bright field; Merged: GFP and NLS-mCherry overlay. Scale bars = 20 µm. (B) Transcription activation analysis of NtMYB306a protein in yeast. The pGBKT7 and pCL1 plasmids were used as negative and positive controls, respectively. 100, 10-1, 10-2 and 10-3 indicate the yeast dilutions (1:1, 1:10, 1:100 and 1:1000).
In addition to nuclear localization, transcriptional activation is another important feature of TFs. In the yeast transactivation assays (Figure 2B), the cells containing the pGBKT7-NtMYB306a plasmid was able to grow on SD/-Trp medium, signifying successful transformation. Furthermore, the yeast cells could grow on the SD/-Trp-Ade+X-α-gal selective medium and turned blue after 3 days. Simultaneously, the yeast cells transformed with the positive control pCL1 plasmid were able to grow on SD/-Trp-Ade medium, while those transformed with the negative control pGBKT7 were only able to grow on SD/-Trp medium. The above results indicate that NtMYB306a is a TF with transcriptional activation activity.
NtMYB306a is predominantly expressed in tobacco trichomes
To elucidate the expression pattern of NtMYB306a, various tobacco tissues were subjected to qRT-PCR analysis. As the data shown in Figure 3A, NtMYB306a was the highest expression level in trichomes, followed by leaves, flowers, and stems, respectively. Almost no expression was found in roots. The expression of NtMYB306a had no significant difference between leaves with or without trichomes. Additionally, we measured the relative expression levels of NtMYB306a in leaf tissues from different positions (i.e., L1-L6) in 5-months-old tobacco plants. The obtained data showed that the expression level of NtMYB306a in L3 leaves was higher than the leaves at other positions (Figure 3B). The expression level of NtMYB306a increasingly up-regulated from L1 to L2, peaked at L3, and started to decline at L4 and L5, respectively. At L6 position, the gene expression level was up-regulated to the level of L2 and L3. The in-situ hybridization analysis of NtMYB306a mRNA in 60-day-old tobacco leaves exhibited strong signal accumulation in tall glandular trichomes (TGTs) and short glandular trichomes (SGTs) (Figures 3C, D), which further indicated that NtMYB306a plays a role in tobacco glandular trichomes. In addition, strong hybridization signals were also observed in epidermis of some leaves (Supplementary Figure S3), which may be ascribed to the fact that trichomes are derived from epidermal cells. Next, we cloned the 2,077 bp long promoter sequence of NtMYB306a (Supplementary Figure S4) and built the pNtMYB306a:GUS construct to transform tobacco plants. In pNtMYB306a:GUS transgenic tobacco plants, GUS staining was clearly observed in TGTs and SGTs (Figures 3E, F). The above results indicated that NtMYB306a plays a key role in tobacco stems, leaves, and flowers, especially in trichomes.
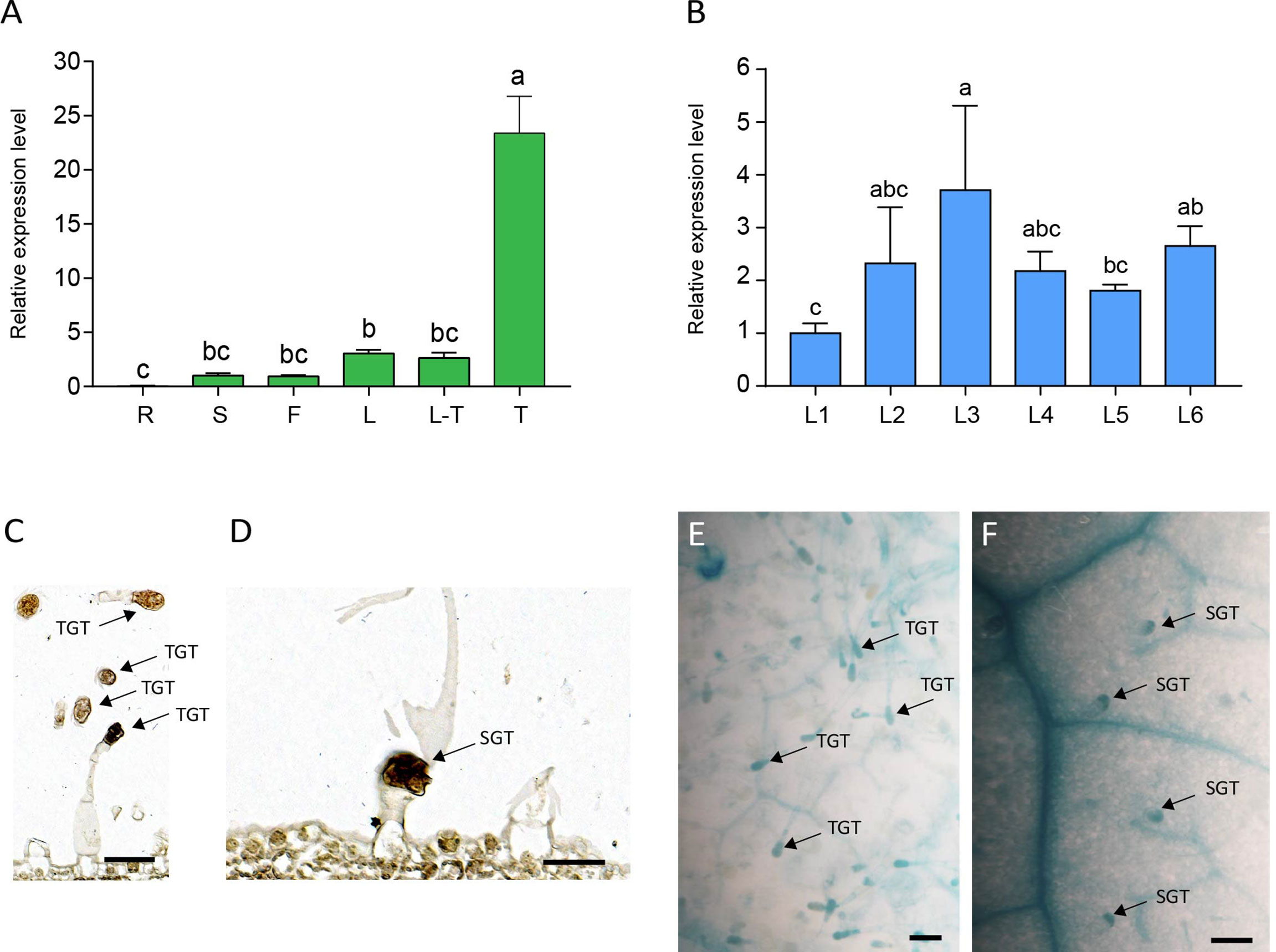
Figure 3 Spatial and temporal expression patterns of NtMYB306a in tobacco plants. (A) Relative expression levels of NtMYB306a in different tissues of tobacco plants. R, roots; S, stems; F, flowers; L, leaves; L-T, leaves without trichomes; T, trichomes. Error bars indicate the standard deviation (SD) of three biological replicates. Different letters above the bars indicate that those values are significantly different (P < 0.05) by one-way ANOVA. (B) Relative expression levels of NtMYB306a in leaves of different positions. L1-L6: the 1st, 4th, 7th, 11th, 15th, and 19th leaf position, respectively, from the top to the bottom of a plant. (C, D) mRNA in situ hybridization analysis of NtMYB306a in 60-day-old tobacco leaves. Strong signals were detected in tall glandular trichomes (TGTs) and short glandular trichomes (SGTs), which are indicated by black arrows. Scale bars = 50 µm. (E, F) GUS staining of pNtMYB306a:GUS transgenic tobacco plants. Black arrows indicate TGTs and SGTs, respectively. Scale bars = 100 µm.
Promoter region of NtMYB306a harbour multiple stress-related cis-elements
To understand the mechanism of transcriptional regulation and involvement of different regulatory components, the cis-acting elements of the NtMYB306a promoter were analyzed using the PlantCARE online tool. The in-silico analysis predicted numerous regulatory sequences present in the 5′ upstream regions of NtMYB306a, including ABA- (Abscisic acid), MeJA- (Methyle-jasmonate), SA- (Salicylic acid), light-, and cold-stress-responsive element (Supplementary Table S2). To compliment in-silico analysis of promoter regions, qRT-PCR experiment was designed to assess the expression of NtMYB306a in response to treatment with exogenous hormones (i.e., ABA, MeJA and SA) and cold stress (4°C) at different time points (0h, 6h, 12h, 24h, 48h and 72h). The results are shown in Figure 4. After ABA or MeJA treatment, no significant changes in the transcript abundance level of NtMYB306a were detected, while the NtMYB306a expression was significantly up-regulated after 6 h of SA treatment. At 12, 48, and 72 h of cold stress, the expression level of NtMYB306a was significantly down-regulated, showing negative co-regulation. The differential expression of NtMYB306a towards SA and cold stresses reflects complex regulatory mechanism during plant-environment interaction.
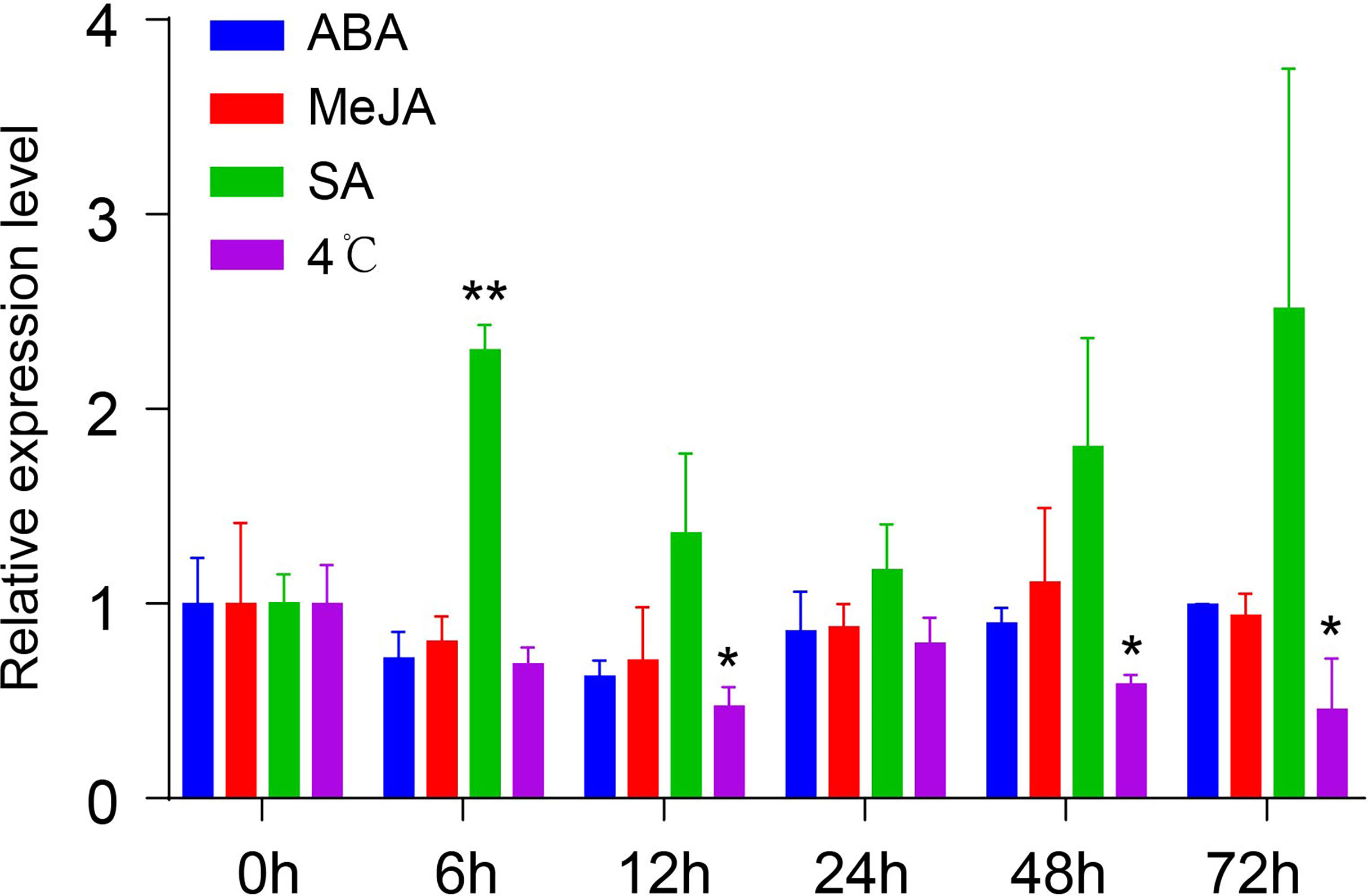
Figure 4 Relative expression levels of NtMYB306a in the leaves of tobacco plants under ABA, MeJA, SA, and 4°C treatments, respectively. Student’s t-test was used to analyze the difference between 0 h and other time points (*P < 0.05; **P < 0.01). Error bars indicate the SD of three biological replicates.
Overexpression of NtMYB306a upregulates the transcription of wax/cutin synthesis-related genes
To identify the key genes that may be regulated by NtMYB306a and explore the molecular mechanism, NtMYB306a was overexpressed in tobacco plants driven by the cauliflower mosaic virus (CMV) 35S promoter, and 11 NtMYB306a-OE transgenic lines (T0) were generated (Supplementary Figures S5, S6). To analyze the differential gene expression, RNA-seq analysis was performed on the leaves of OE-20 T1 transgenic and wild-type (WT) plants. The parameters for screening differently expressed genes (DEGs) were set to adjusted padj <0.05 and |log2(fold-change)|>1, which yielded a total of 1,036 DEGs. Compared with WT, 700 (67.57%) genes were up-regulated and 336 genes (32.43%) were down-regulated in OE-20 line (Figure 5A). Since NtMYB306a was predicted as TF with transcriptional activation activity, to have a further understanding on their function in biological system, the obtained 700 up-regulated DEGs were mapped to the Kyoto encyclopedia of genes and genomes (KEGG) database. As shown in Figure 5B, these DEGs were heavily enriched in biosynthesis of cutin, suberin, and wax. These results are consistent with our previous predictions based on the evolutionary tree, suggesting that NtMYB306a was related to wax and cutin biosynthesis. Genes that may be involved in wax and/or cutin biosynthesis are shown in Table 1. Among these, GDSL belonging to the lipase family, ABCG32 belonging to the ABCG family, and P450 family members CYP86A and CYP77A are involved in cutin deposition or transport; KCS is involved in VLCFA biosynthesis; and CER1 and CER3 can convert VLCFAs into alkanes. In addition, genes involved in metabolic and/or biosynthetic processes (including glycerolipid, fatty acid, and unsaturated fatty acids), fatty acid degradation processes, and fatty acid elongation were also found to be enriched (Figure 5B). Valine and isoleucine are the precursors for generating alkanes (Grice et al., 2008), and genes related to the biosynthesis and degradation of valine and isoleucine were also enriched by KEGG analysis. In higher plants, alkanes are the decarboxylation products of fatty acids (Grice et al., 2008), indicating that NtMYB306a is involved in wax accumulation by regulating the transcription of lipid-related genes. In addition to genes related to lipid metabolism, plant-pathogen interaction pathways were also identified in KEGG enrichment analysis (Figure 5B), suggesting that NtMYB306a is also involved in plant defense.
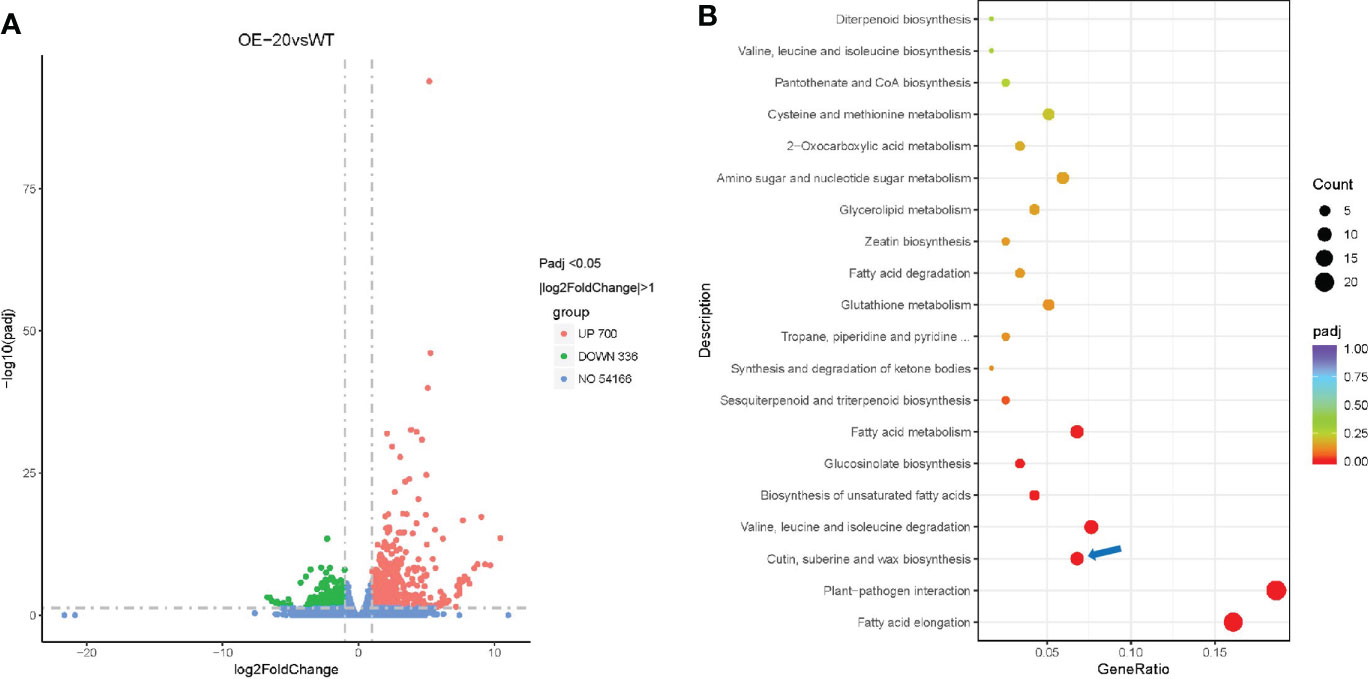
Figure 5 RNA-seq analysis of OE-20 and WT plants. (A) Volcano plot of DEGs between OE-20 and WT plants. Abundance of each gene was normalized as FPKM and DEGs were filtered for adjusted padj < 0.05 and |log2(fold-change)| > 1. DEGs shown in red color represent up-regulated genes (700), green showing down-regulated genes (336), and blue color indicates genes that were not differentially expressed (no-DEGs, 54,166). (B) KEGG enrichment analysis of up-regulated genes in OE-20 plants. The coloring of the padj-values represents the significance of the enrichment factor. Circles indicate the involved target genes, and the size is proportional to the gene number. The blue arrow indicates the cutin, suberine, and wax biosynthesis pathway.
To further confirm the possible target genes regulated by NtMYB306a, we performed qRT-PCR assay to investigate the expression patterns of ten putative target wax/cutin metabolism-related genes in the T2 plants of OE-3, OE-13, and OE-20 lines. The chosen target genes included GDSL, CYP77A2, CYP86A8, ABCG32, KCS1, KCS3, KCS11, KCS20, CER1, and CER3. As expected, their expression was induced in these transgenic lines (Figure 6), which was consistent with the data obtained from RNA-seq (Table 1). Nearly all lines had small increases in expression of target genes relative to wild-type. Comparison between over-expression lines, OE-3, OE-13 and OE-20, eight genes were significantly expressed in OE-20 and OE-13, while seven genes were expressed in OE-3 as compared to WT lines. The expression of NtMYB306a showed 40 - 60-fold increase in all three homozygous lines compared to WT. Among ten target genes, the highest level of expression was noted for GDSL, which was ~20 fold increased in OE-20, followed by CYP86A8 (~7fold) and KCS3 (~3fold) respectively. Other genes, including CER1, CER3, KCS1, KCS11, KCS20, CYP77A2, ABCG32 showed lower yet significant level of expression in over-expression lines relative segregating wild-type plants. In addition, the expression level of these ten genes was examined in NtMYB306a-OE tobacco plants under the control of tobacco glandular trichome-specific promoter pCBTS (Ennajdaoui et al., 2010). In the pCBTS : NtMYB306a-T1 plants, the expression of the target set of genes showed similar significantly up-regulation trends (Supplementary Figure S7). These results suggest that NtMYB306a may be an activator of a set of genes related to wax and cutin biosynthesis in tobacco trichomes.
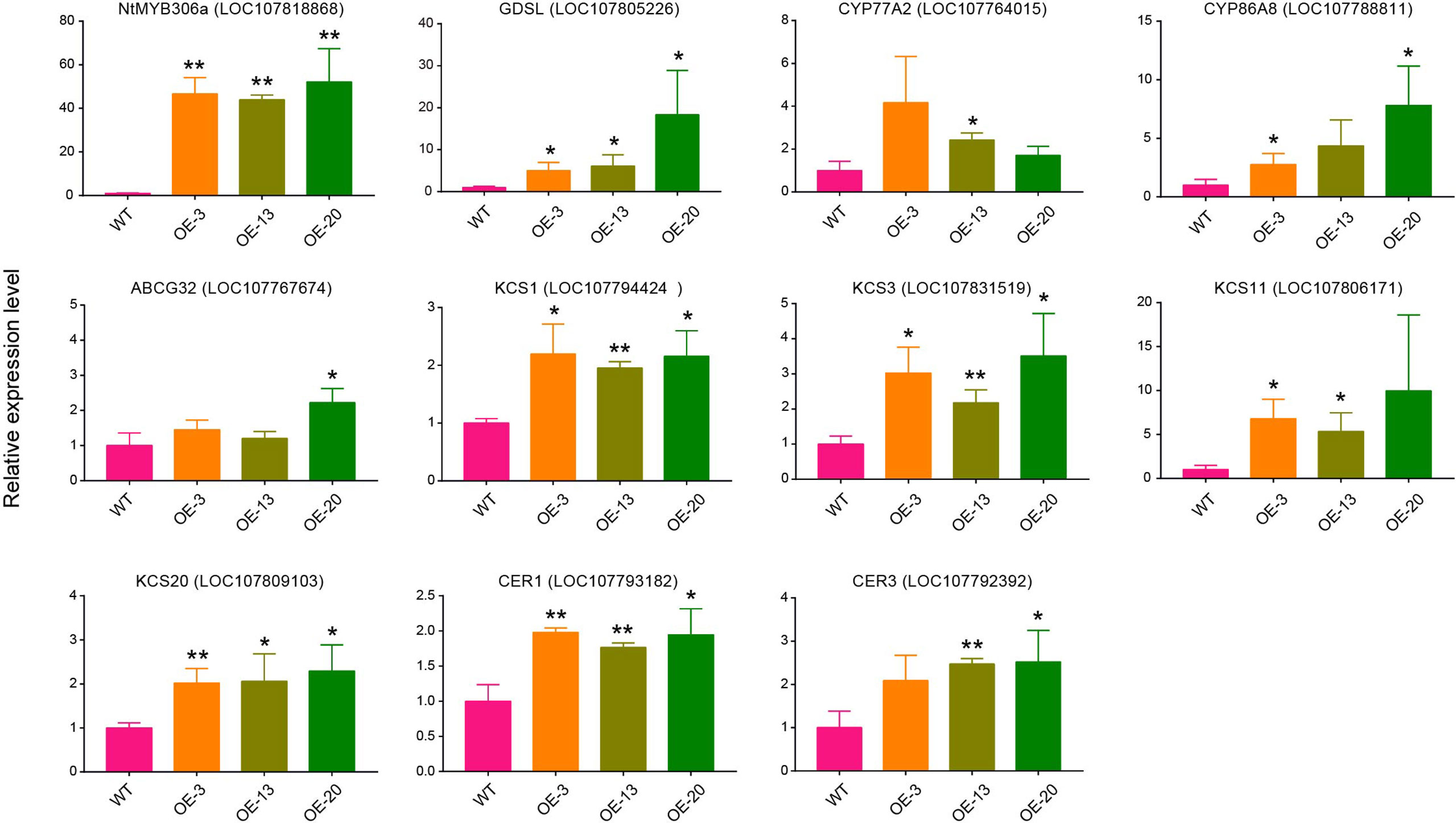
Figure 6 Relative expression levels of genes involved in the wax/cutin biosynthesis pathway in NtMYB306a-OE and WT plants. Ten putative target wax/cutin metabolism-related genes, including GDSL, CYP77A2, CYP86A8, ABCG32, KCS1, KCS3, KCS11, KCS20, CER1, and CER3, were selected. Error bars indicate the SD of three biological replicates. Student’s t-test: *P < 0.05; **P < 0.01.
Overexpression of NtMYB306a enhances alkane content in leaf exudates
In Arabidopsis, the composition of surface wax on trichomes is different from other epidermal cells, and alkanes are the central components of trichome waxes (Hegebarth et al., 2016). In this study, NtMYB306a belonging to the S1 subgroup of R2R3-MYB was predominantly expressed in trichomes, indicating that NtMYB306a may be involved in the synthesis or secretion of alkanes in tobacco trichomes. Components of tobacco leaf exudates were surveyed by Gas Chromatography–Mass Spectrometry (GC-MS) that exhibited that the total alkane contents in leaf exudates of NtMYB306a-OE lines (OE-3, OE-13, and OE-20) were significantly greater than those of the WT (Figure 7A), and the contents of n-C28, n-C29, n-C31, and ai-C31 alkanes were significantly higher than those in WT (Figure 7B). In WT plants, n-C31 and n-C33 alkanes accounted for the majority of n-alkanes ranging between n-C25 and n-C33, and there was a small amount of n-C25 and n-C26, alkanes (Figure 7B). The contents of i-C31 and i-C33 contents among iso-alkanes and the ai-C30 and ai-C32 contents in anteiso-alkanes was relatively high in all samples. A similar result was reported by Grice et al. (2008), where the n-alkanes were found to be predominant in WT tobacco leaf exudates among the three homologs (i.e., n-alkanes, iso-alkanes, and anteiso-alkanes). Moreover, the trend for alkane contents presented in tobacco trichome samples is similar to that reported in Arabidopsis trichomes, where the n-C29 and n-C31 homologs were predominant in WT Arabidopsis samples (Hegebarth et al., 2016). These results further suggest that NtMYB306a is a positive regulator of alkane metabolism in tobacco trichomes.
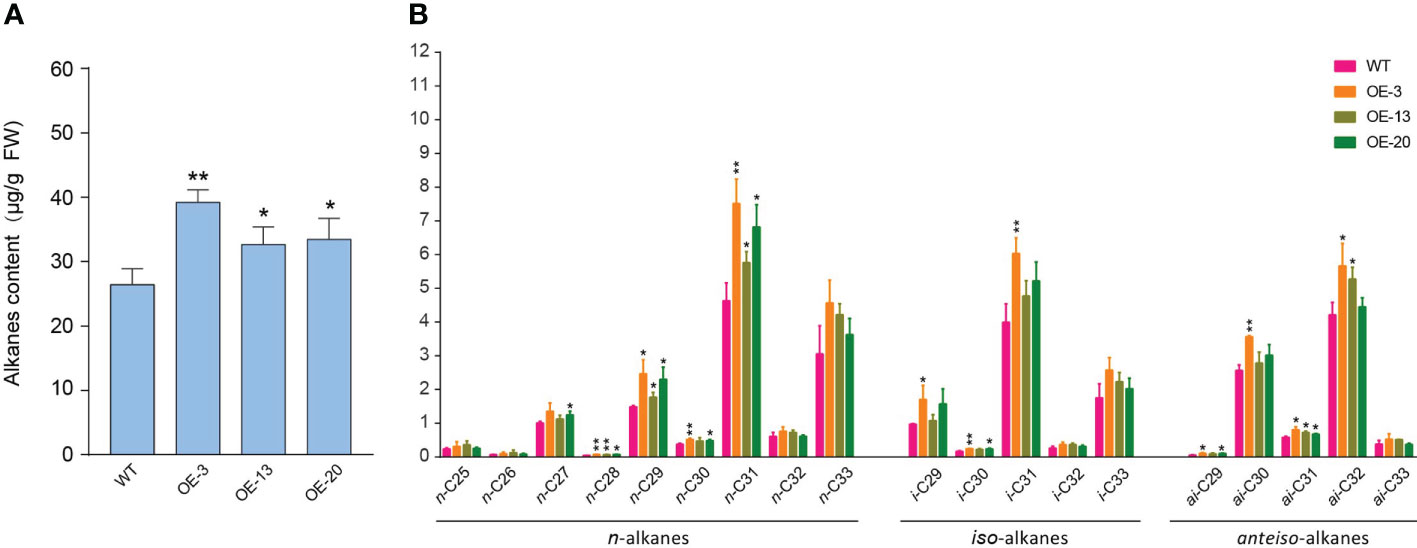
Figure 7 Alkane contents in NtMYB306a-OE and WT leaf exudates. (A) Total alkane content. (B) The content of n-alkanes, iso-alkanes, and anteiso-alkanes, respectively. Error bars indicate the SD of three biological replicates. Student’s t-test: *P < 0.05; **P < 0.01.
Overexpression of NtMYB306a does not influence tobacco trichome density
Trichomes are specialized organs of epidermal cells, and wax and cuticle biosynthesis is reportedly associated with the differentiation of epidermal cells, and the initiation and development of trichomes (Shi et al., 2018). Our scanning electron microscopy (SEM) images showed that there were no obvious differences in leaf surface structures between the TGTs, SGTs, NGTs, and pavement cells of the overexpression NtMYB306a-OE line (OE-20) and WT plants (Figure 8A). We observed that the numbers of leaf trichomes was not significantly different between NtMYB306a-OE lines (OE-3, OE-13, and OE-20) and WT (Figures 8B, C). Additionally, 2-month-old NtMYB306a-OE lines showed no significant difference in macroscopic phenotype compared with WT plants (Figure 8D). The above results indicate that overexpression of NtMYB306a in tobacco fails to alter the tobacco trichome density, and thus had no significant effect on the morphology of trichomes.
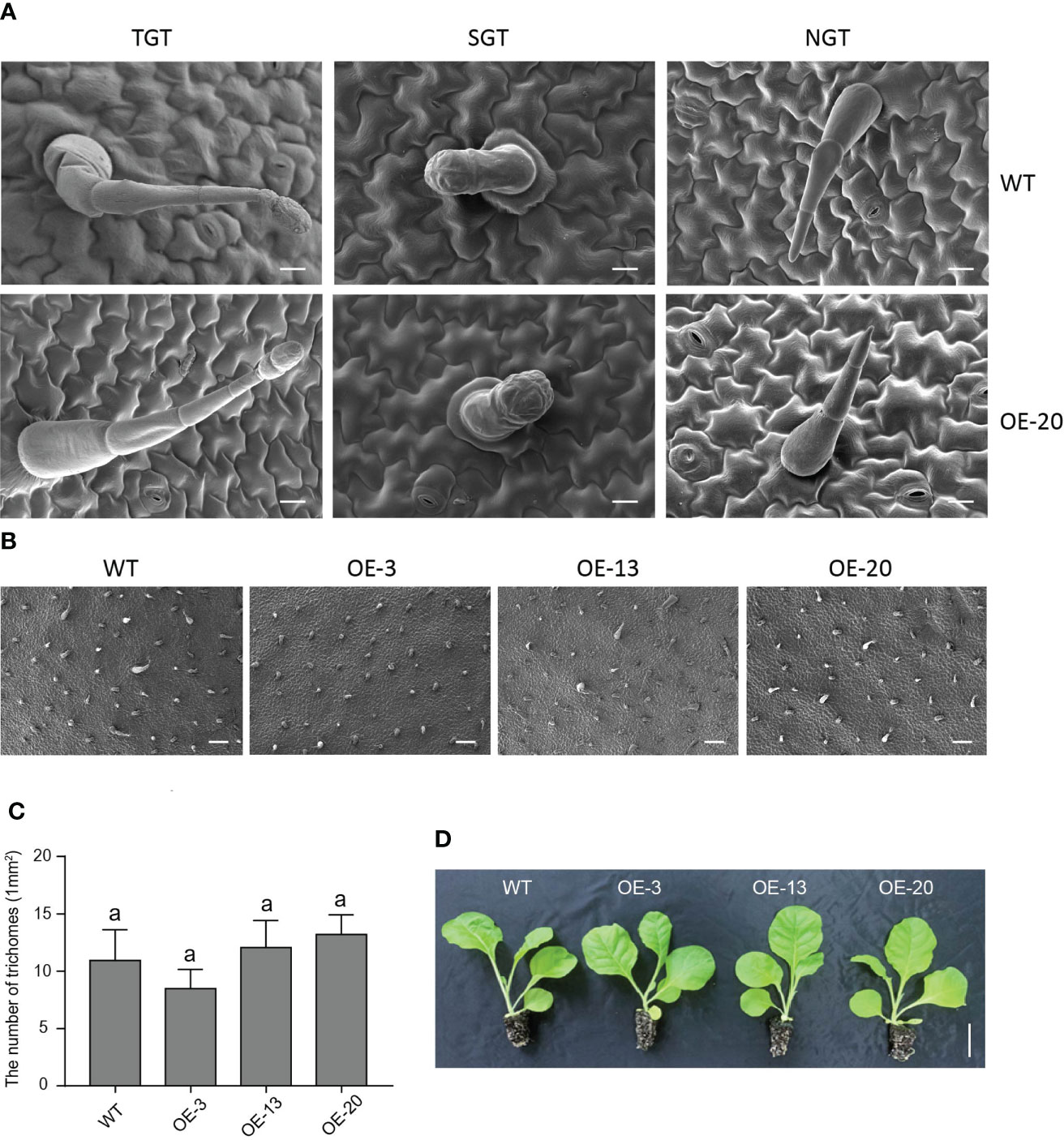
Figure 8 Phenotypes of NtMYB306a-OE and WT leaves. (A) The morphologies of tall glandular trichome (TGT), short glandular trichome (SGT), and non-glandular trichome (NGT) in OE-20 and WT plants. Scale bars = 20 µm. (B) Trichome abundance on WT, OE-3, OE-13, and OE-20 leaves, respectively. Scale bars = 20 µm. (C) The number of trichomes on WT, OE-3, OE-13, and OE-20 leaves. Error bars indicate the SD of four biological replicates. Different letters above the bars indicate that those values are significantly different (P < 0.05) by one-way ANOVA. (D) Phenotype observation of WT and three NtMYB306a-OE transgenic lines (OE-3, OE-13, and OE-20). Scale bars = 5 cm.
Discussion
NtMYB306a responds to SA induction and is predominantly expressed in tobacco trichomes
Trichomes are specialized tissues located on the epidermis of many plant species, that protect plant from excessive transpiration, and participate in resistance mechanisms to numerous abiotic stresses including high temperature, radiation and UV light, and biotic stresses such as pathogens and herbivore attack (Wagner, 1991; Serna and Martin, 2006; Glas et al., 2012). In tobacco, glandular trichomes act as sites for the biosynthesis and secretion of cembranoids and several secondary metabolites (Johnson et al., 1985; Wang et al., 2004; Ennajdaoui et al., 2010), hence they serve as important tool of genetic engineering for quality improvement of varieties (Glas et al., 2012; Tissier, 2012; Wu et al., 2020). However, limited studies to date have investigated the development of tobacco trichomes, and the detailed mechanism has not been fully understood. The present study was conducted to identify regulatory factors that may play a role in the metabolism of leaf trichomes in tobacco. Previous studies have shown that some TFs, which are predominantly expressed in plant glandular trichomes, are involved in the secondary metabolite synthesis of glandular trichome and trichome development. For example, artemisinin of Artemisia annua and menthol of spearmint are specifically synthesized and secreted in glandular trichomes. Among different genes discovered, AaGSW1 (Chen et al., 2017), AaWRKY9 (Fu et al., 2021a), AaORA (Lu et al., 2013), and AaMYB15 (Wu et al., 2021) of A. annua regulate the artemisinin synthesis pathway, AaTAR2 (Zhou et al., 2020) is involved in the regulation of trichome development, while, MsMYB is involved in the production of monoterpenes in spearmint (Reddy et al., 2017). In our current study, we isolated, characterized, and subsequently cloned the NtMYB306a (Supplementary Figure S1) gene from tobacco by comparing the transcriptomic data between leaves with and without trichomes. NtMYB306a was localized in the nucleus and showed transcriptional activation activity. Scores obtained from protein sequence alignment and phylogenetic tree analysis showed that NtMYB306a belonged to the S1 subgroup of R2R3-MYB TFs, which are involved in wax and cutin biosynthesis of plants (Seo et al., 2011; Lee and Suh, 2015b; Zhang et al., 2019; Wen et al., 2021). We further confirmed the predominant expression of NtMYB306a in tobacco trichomes, especially glandular trichomes, by employing qRT-PCR, in situ hybridization, and GUS staining, and speculated that NtMYB306a was involved in the biosynthesis or transport of wax and cutin in tobacco trichomes. In addition, it has been reported that the S1 subgroup members, such as AtMYB30, regulate the transduction of cell death-related lipid signals by enhancing the synthesis of VLCFAs or their derivatives (Raffaele and Rivas, 2013). Moreover, AtMYB30 plays an important role in plant defense and stress response (Marino et al., 2013; Raffaele and Rivas, 2013; Liao et al., 2017; Mabuchi et al., 2018), while SA is an important signaling molecule in plant defense (Ding and Ding, 2020). We found that NtMYB306a expression was induced by SA, and the RNA-seq data strongly supported the results, i.e., several stress-related DEGs were enriched in NtMYB306a-OE transgenic lines. These findings suggest that NtMYB306a can enhance VLCFA synthesis and participate in SA signal transduction.
Up-regulation of wax/cutin-related genes increases the alkane content in NtMYB306a-OE lines
Through genome-wide transcriptome analysis, we revealed that many protein-coding genes involved in wax biosynthesis were regulated by the NtMYB306a. Considering that NtMYB306a showed transcriptional activation activity, we found that the expression levels of wax and cutin synthesis-related genes were up-regulated in NtMYB306a-OE lines. Among these, KCS1, KCS3, KCS5, KCS10, KCS11, and KCS20 may be related to the elongation of VLCFAs (Todd et al., 1999; Lee et al., 2009; Bernard and Joubès, 2013; Xiong et al., 2020; Batsale et al., 2021), while CER3 and CER1 convert VLCFAs into alkanes (Bourdenx et al., 2011; Bernard et al., 2012; Pascal et al., 2019). In addition, CYP77A and CYP86A of the CYP450 family, which are involved in the biosynthesis of cutin monomers (Li-Beisson et al., 2009); and GDSL lipase (Girard et al., 2012; Tang et al., 2020) and ABCG protein (Panikashvili et al., 2011; Nguyen et al., 2018; Philippe et al., 2022), which are related to wax and cutin synthesis, transport, and secretion, were also up-regulated in NtMYB306a-OE plants. However, the functions of most of the above genes have not been characterized due to the lack of research on the wax and cutin synthesis pathways in tobacco. Therefore, we also performed an RNAi experiment on NtMYB306a, but we did not obtain NtMYB306a-RNAi lines with silenced NtMYB306a (data not shown), which might be due to the fact that cultivated tobacco is allotetraploid and has multiple structurally similar MYB306 homologs. The predominant wax components of trichomes are mainly alkanes in Arabidopsis (Hegebarth et al., 2016); therefore, we next focused on detecting the alkane content in tobacco leaf exudates and observed an increase in the alkane content in NtMYB306a-OE plants. Combining with the results of RNA-seq and qRT-PCR, we speculate that NtMYB306a can activate the expression of several genes of wax and cutin biosynthesis pathways. These genes include those involved in VLC-alkane synthesis, thereby increasing the content of alkanes.
NtMYB306a positively regulates genes in trichomes to synthesize and transport trichome-specific wax alkane components
We detected alkanes in tobacco leaf exudates, but whether these substances were specifically synthesized in trichomes or secreted by glandular trichomes after being synthesized by epidermal cells is unclear and requires detailed validation with further experiments. It is challenging to isolate the required amount of trichome metabolites from tobacco leaves, which limited accurate examination of trichome-specific wax components. However, results of some studies support the synthesis of specific wax alkanes in plant trichomes. For example, in Arabidopsis, all FAE complexes share the same KCR, HCD, and ECR compounds but different KCSs, which determine the specificity of substrates and product chain lengths of the entire complex (Hegebarth et al., 2017). Some cell-specific KCS genes, which are only expressed in trichomes or guard cells but not in the pavement cells of the epidermis (Samuels et al., 2008), may be involved in different pathways of trichomes (Hegebarth et al., 2017). For example, KCS1, KCS5/CER60, KCS8, and KCS10 were mainly expressed during trichome development, which may be involved in the formation of trichome waxes (Hegebarth et al., 2016). Furthermore, Arabidopsis trichomes have higher concentrations of alkanes, which is consistent with the relatively high expression of two enzyme genes (CER3 and CER1) involved in the alkane synthesis pathway in trichomes (Hegebarth et al., 2016). In our study, some genes were up-regulated after overexpression of NtMYB306a, such as KCS1 (LOC107794424), KCS3 (LOC107831519), KCS20 (LOC107809103), CER1 (LOC107793182), CER3 (LOC1077492392), and GDSL (LOC1078052767). The expression levels of these genes were much higher in trichomes than in other epidermal cells (Supplementary Figure S8), further supporting that NtMYB306a positively regulates these genes in trichomes to synthesize trichome-specific wax alkane components. Collectively, these genes that are predominantly expressed in tobacco trichomes have not been characterized and are candidate genes related to wax alkane biosynthesis and transport in tobacco trichomes.
Overexpression of NtMYB306a does not influence the number and morphology of tobacco trichomes
Some studies have shown that the R2R3-MYB TFs can affect epidermis development by activating the expression of genes related to wax and cutin biosynthesis (Shi et al., 2018), thus regulating the occurrence and development of trichomes. Most of these genes are members of the S9 subgroup, for example, in Arabidopsis, a MIXTA-like TF MYB106 regulates trichome branching by inducing cutin and wax biosynthesis (Oshima et al., 2013); in A. annua, AaMIXTA1 promotes GST initiation by regulating cuticle biosynthesis (Shi et al., 2018). These genes are closely related to the initiation and development of trichomes, which may be because the plant leaf epidermis is a mixture composed of cutins and waxes, and trichomes protrude from the epidermal cell layer (Bird and Gray, 2003). Some correlational studies have demonstrated the relationship among the S1 subgroup members of MYB, trichome development and wax synthesis. For instance, SlMYB31 interacted with the HD-Zip IV TF Woolly to regulate the expression of SlCER6, a KCS-encoding gene, to synergistically regulate cuticle wax biosynthesis in tomato (Xiong et al., 2020); deposition of wax crystals was observed outside the epidermis after CsMYB30 was expressed in Arabidopsis (Wen et al., 2021). Therefore, we observed the leaf surface morphology of 5-month-old NtMYB306a-OE lines with SEM; however, we did not find significant differences in the number and morphology of trichomes between NtMYB306a-OE and WT plants. These results indicated that the increased alkanes content in the NtMYB306a-OE lines is not due to an increase in trichome density. In addition, we did not observe obvious wax accumulation on the epidermis, and no significant differences was found in cross-section leaves (Supplementary Figure S9).
Conclusion
In this report, we identified NtMYB306a as a member of the S1 subgroup of the R2R3-MYB transcription factor family, and provided evidence to show that NtMYB306a function in wax alkane biosynthesis in trichomes of N. tabacum. We showed that NtMYB306a function as activator to regulate the expression of their target genes, including GDSL, CYP77A2, CYP86A8, ABCG32, KCS1, KCS3, KCS11, KCS20, CER1, and CER3. Besides, the promoter region of NtMYB306a harbored multiple stress-related cis-elements, which positively responded to SA and cold stress during qRT-PCR analysis. Finally, we found that the number and morphology of glandular trichomes were not influenced by NtMYB306a. The NtMYB306a can be used as a candidate gene for stress resistance and quality improvement of tobacco.
Data availability statement
The raw sequencing data generated from this study have been deposited in NCBI Sequence Read Archive (SRA) (http://www.ncbi.nlm.nih.gov/sra) with the accession number PRJNA883934.
Author Contributions
JY and BL planned and designed research. BW, YG, XZ, MJ and HY performed the experiments. HZ and KK analyzed and discussed the data. JY and DZ wrote the manuscript. JY and KK revised the manuscript. All authors approved the manuscript.
Funding
This work was supported by the Program of China National Tobacco Corporation Project [110202101005(JY-05)], the Science and Technology project of Guizhou Company of China Tobacco Corporation (2021XM02), the Guizhou Province High-level Innovative Talent Training Program Project ([(2016)4003]), the Guizhou Province Science and Technology Project ([2021]5642), and the National Natural Science Foundation (31860491).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1005811/full#supplementary-material
References
Batsale, M., Bahammou, D., Fouillen, L., Mongrand, S., Joubès, J., Domergue, F. (2021). Biosynthesis and functions of very-Long-Chain fatty acids in the responses of plants to abiotic and biotic stresses. Cells 10, 1284. doi: 10.3390/cells10061284
Beaudoin, F. D. R., Wu, X., Li, F., Haslam, R. P., Markham, J. E., Zheng, H., et al. (2009). Functional characterization of the arabidopsis beta-ketoacyl-coenzyme a reductase candidates of the fatty acid elongase. Plant Physiol. 150, 1174–1191. doi: 10.1104/pp.109.137497
Bernard, A., Domergue, F., Pascal, S., Jetter, R., Renne, C., Faure, J. D., et al. (2012). Reconstitution of plant alkane biosynthesis in yeast demonstrates that arabidopsis ECERIFERUM1 and ECERIFERUM3 are core components of a very-long-chain alkane synthesis complex. Plant Cell 24, 3106–3118. doi: 10.1105/tpc.112.099796
Bernard, A., Joubès, J. (2013). Arabidopsis cuticular waxes: advances in synthesis, export and regulation. Prog. Lipid Res. 52, 110–129. doi: 10.1016/j.plipres.2012.10.002
Bird, S. M., Gray, J. E. (2003). Signals from the cuticle affect epidermal cell differentiation. New Phytol. 157, 9–23. doi: 10.1046/j.1469-8137.2003.00543.x
Bourdenx, B., Bernard, A. L., Domergue, F. D. R., Pascal, S. P., Le´Ger, A., Roby, D., et al. (2011). Overexpression of arabidopsis ECERIFERUM1 promotes wax very-long-chain alkane biosynthesis and influences plant response to biotic and abiotic stresses. Plant Physiol. 156, 29–45. doi: 10.1104/pp.111.172320
Chen, M. H., Yan, T. X., Shen, Q., Lu, X., Pan, Q. F., Huang, Y. R., et al. (2017). GLANDULAR TRICHOME-SPECIFIC WRKY 1 promotes artemisinin biosynthesis in artemisia annua. New Phytol. 214, 304–316. doi: 10.1111/nph.14373
Choi, Y. E., Lim, S., Kim, H. J., Han, J. Y., Lee, M. H., Yang, Y., et al. (2011). Tobacco NtLTP1, a glandular-specific lipid transfer protein, is required for lipid secretion from glandular trichomes. Plant J. 70, 480–491. doi: 10.1111/j.1365-313X.2011.04886.x
Ding, P. T., Ding, Y. L. (2020). Stories of salicylic acid: A plant defense hormone. Trends Plant Sci. 25, 549–565. doi: 10.1016/j.tplants.2020.01.004
Dubos, C., Stracke, R., Grotewold, E., Weisshaar, B., Martin, C., Lepiniec, L. (2010). MYB transcription factors in arabidopsis. Trends Plant Sci. 15, 573–581. doi: 10.1016/j.tplants.2010.06.005
Ennajdaoui, H., Vachon, G., Giacalone, C., Besse, I., Sallaud, C., Herzog, M., et al. (2010). Trichome specific expression of the tobacco (Nicotiana sylvestris) cembratrien-ol synthase genes is controlled by both activating and repressing cis-regions. Plant Mol. Biol. 73, 673–685. doi: 10.1007/s11103-010-9648-x
Fabre, G., Garroum, I., Mazurek, S., Daraspe, J., Mucciolo, A., Sankar, M., et al. (2015). The ABCG transporter PEC1/ABCG32 is required for the formation of the developing leaf cuticle in arabidopsis. New Phytol. 209, 192–201. doi: 10.1111/nph.13608
Fu, X. Q., Peng, B. W., Hassani, D., Xie, L. H., Liu, H., Li, Y. P., et al. (2021a). AaWRKY9 contributes to light- and jasmonate-mediated to regulate the biosynthesis of artemisinin in artemisia annua. New Phytol. 231, 1858–1874. doi: 10.1111/nph.17453
Fu, Y. J., Zhang, Y. P., Zeng, S. T., Luo, L. W., Xi, H., Li, P., et al. (2021b). The effect of long-chain alkanes on flavour release and olfactory characteristics of rose essential oil. Flavour. Fragrance. J. 37, 72–80. doi: 10.1002/ffj.3681
Girard, A. L., Mounet, F., Chamley, M. L., Gaillard, C., Elmorjani, K., Vivancos, J., et al. (2012). Tomato GDSL1 is required for cutin deposition in the fruit cuticle. Plant Cell 24, 3119–3134. doi: 10.1105/tpc.112.101055
Glas, J. J., Schimmel, B. C. J., Alba, J. M., Escobar-Bravo, R., Schuurink, R. C., Kant, M. R. (2012). Plant glandular trichomes as targets for breeding or engineering of resistance to herbivores. Int. J. Mol. Sci. 13, 17077–17103. doi: 10.3390/ijms131217077
Grice, K., Lu, H., Zhou, Y. P., Williams, H. S., Farquhar, G. D. (2008). Biosynthetic and environmental effects on the stable carbon isotopic compositions of anteiso-(3-methyl) and iso-(2-methyl) alkanes in tobacco leaves. Phytochemistry 69, 2807–2814. doi: 10.1016/j.phytochem.2008.08.024
Heemann, V., Brümmer, U., Paulsen, C., Seehofer, F. (1983). Composition of the leaf surface gum of some nicotiana species and nicotiana tabacum cultivars. Phytochemistry 22, 133–135. doi: 10.1016/S0031-9422(00)80073-4
Hegebarth, D., Buschhaus, C., Jérômejoubès, Thoraval, D., Bird, D., Jetter, R. (2017). Arabidopsis ketoacyl-CoA synthase 16 (KCS16) forms C36/C38 acyl precursors for leaf trichome and pavement surface wax. Plant. Cell Environ. 40, 1761–1776. doi: 10.1111/pce.12981
Hegebarth, D., Buschhaus, C., Wu, M., Bird, D., Jetter, R. (2016). The composition of surface wax on trichomes of arabidopsis thaliana differs from wax on other epidermal cells. Plant J. 88, 762–774. doi: 10.1111/tpj.13294
Horsch, R. B., Rogers, S. G., Fraley, R. T. (1985). Transgenic plants. Cold Spring Harb. Symp. Quant. Biol. 50, 433–437. doi: 10.1101/SQB.1985.050.01.054
Huang, M. Y., Zhang, H. Y., Wang, Z. J., Niu, D. X., Li, Y. H., Cui, H. (2018). Comparative studies of leaf surface chemical biosynthesis in different tobacco cultivars. Acta Physiol. Plantarum. 40, 67. doi: 10.1007/s11738-018-2642-7
Javelle, M., Vernoud, V., Rogowsky, P. M., Ingram, G. C. (2011). Epidermis: the formation and functions of a fundamental plant tissue. New Phytol. 189, 17–39. doi: 10.1111/j.1469-8137.2010.03514.x
Jefferson, R. A., Kavanagh, T. A., Bevan, M. W. (1987). GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x
Jetter, R., Riederer, M. (2016). Localization of the transpiration barrier in the epi- and intracuticular waxes of eight plant species: Water transport resistances are associated with fatty acyl rather than alicyclic components. Plant Physiol. 170, 921–934. doi: 10.1104/pp.15.01699
Jiang, C. K., Rao, G. Y. (2020). Insights into the diversification and evolution of R2R3-MYB transcription factors in plants. Plant Physiol. 183, 637–655. doi: 10.1104/pp.19.01082
Johnson, A. W., Severson, R. F., Hudson, J. (1985). Tobacco leaf trichomes and their exudates. Tobacco. Control. 27, 67–72.
Lashbrooke, J., Adato, A., Lotan, O., Alkan, N., Tsimbalist, T., Rechav, K., et al. (2015). The tomato MIXTA-like transcription factor coordinates fruit epidermis conical cell development and cuticular lipid biosynthesis and assembly. Plant Physiol. 169, 2553–2571. doi: 10.1104/pp.15.01145
Lee, S. B., Jung, S. J., Go, Y. S., Kim, H. U., Kim, J. K., Cho, H.-J., et al. (2009). Two arabidopsis 3-ketoacyl CoA synthase genes, KCS20 and KCS2/DAISY, are functionally redundant in cuticular wax and root suberin biosynthesis, but differentially controlled by osmotic stress. Plant J. 60, 462–475. doi: 10.1111/j.1365-313X.2009.03973.x
Lee, S. B., Suh, M. C. (2015a). Advances in the understanding of cuticular waxes in arabidopsis thaliana and crop species. Plant Cell Rep. 34, 557–572. doi: 10.1007/s00299-015-1772-2
Lee, S. B., Suh, M. C. (2015b). Cuticular wax biosynthesis is up-regulated by the MYB94 transcription factor in arabidopsis. Plant Cell Physiol. 56, 48–60. doi: 10.1093/pcp/pcu142
Leitch, I. J., Hanson, L., Lim, K. Y., Kovarik, A., Chase, M. W., Clarkson, J. J., et al. (2008). The ups and downs of genome size evolution in polyploid species of nicotiana (Solanaceae). Ann. Bot. 101, 805–814. doi: 10.1093/aob/mcm326
Liao, C. C., Zheng, Y., Guo, Y. (2017). MYB30 transcription factor regulates oxidative and heat stress responses through ANNEXIN-mediated cytosolic calcium signaling in arabidopsis. New Phytol. 216, 163–177. doi: 10.1111/nph.14679
Li-Beisson, Y., Pollard, M., Sauveplane, V., Pinot, F., Ohlrogge, J., Beisson, F. (2009). Nanoridges that characterize the surface morphology of flowers require the synthesis of cutin polyester. PNAS 106, 22008–22013. doi: 10.1073/pnas.0909090106
Liu, J. Y., Osbourn, A., Ma, P. (2015). MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Mol. Plant 8, 689–708. doi: 10.1016/j.molp.2015.03.012
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2 -ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, X., Zhang, L., Zhang, F. Y., Jiang, W. M., Shen, Q., Zhang, L., et al. (2013). AaORA, a trichome-specific AP2/ERF transcription factor ofArtemisia annua, is a positive regulator in the artemisinin biosynthetic pathway and in disease resistance to botrytis cinerea. New Phytol. 198, 1191–1202. doi: 10.1111/nph.12207
Mabuchi, K., Maki, H., Itaya, T., Suzuki, T., Nomoto, M., Sakaoka, S., et al. (2018). MYB30 links ROS signaling, root cell elongation, and plant immune responses. PNAS 115, E4710–E4719. doi: 10.1073/pnas.1804233115
Marino, D., Froidure, S. N., Canonne, J., Khaled, S. B., Khafif, M., Pouzet, C. C., et al. (2013). Arabidopsis ubiquitin ligase MIEL1 mediates degradation of the transcription factor MYB30 weakening plant defence. Nat. Commun. 4, 1476. doi: 10.1038/ncomms2479
McDowell, E. T., Kapteyn, J., Adamschmidt, C., Kang, J. H., Descour, A., Shi, F., et al. (2011). Comparative functional genomic analysis of solanum glandular trichome types. Plant Physiol. 155, 524–539. doi: 10.1104/pp.110.167114
Nguyen, V. N. T., Lee, S. B., Suh, M. C., An, G., Jung, K. H. (2018). OsABCG9 is an important ABC transporter of cuticular wax deposition in rice. Front. Plant Sci. 9, 960. doi: 10.3389/fpls.2018.00960
Oshima, Y., Mitsuda, N. (2013). The MIXTA-like transcription factor MYB16 is a major regulator of cuticle formation in vegetative organs. Plant Signaling Behav. 8, e26826. doi: 10.4161/psb.26826
Oshima, Y., Shikata, M., Koyama, T., Ohtsubo, N., Mitsuda, N., Ohme-Takagia, M. (2013). MIXTA-like transcription factors and WAX INDUCER1/SHINE1 coordinately regulate cuticle development in arabidopsis and torenia fournieri. Plant Cell 25, 1609–1624. doi: 10.1105/tpc.113.110783
Panikashvili, D., Shi, J. X., Schreiber, L., Aharoni, A. (2011). The arabidopsis ABCG13 transporter is required for flower cuticle secretion and patterning of the petal epidermis. New Phytol. 190, 113–124. doi: 10.1111/j.1469-8137.2010.03608.x
Pascal, S., Bernard, A., Deslous, P., Gronnier, J., Goss, A. F., Domergue, F., et al. (2019). Arabidopsis CER1-LIKE1 functions in a cuticular very-Long-Chain alkane-forming complex. Plant Physiol. 179, 415–432. doi: 10.1104/pp.18.01075
Philippe, G., Bellis, D. D., Rose, J. K. C., Nawrath, C. (2022). Trafficking processes and secretion pathways underlying the formation of plant cuticles. Front. Plant Sci. 12, 786874. doi: 10.3389/fpls.2021.786874
Raffaele, S., Rivas, S. (2013). Regulate and be regulated: integration of defense and other signals by the AtMYB30 transcription factor. Front. Plant Sci. 4, 98. doi: 10.3389/fpls.2013.00098
Reddy, V. A., Wang, Q., Dhar, N., Kumar, N., Venkatesh, P. N., Rajan, C., et al. (2017). Spearmint R2R3-MYB transcription factor MsMYB negatively regulates monoterpene production and suppresses the expression of geranyl diphosphate synthase large subunit (MsGPPS.LSU). Plant Biotechnol. J. 15, 1105–1119. doi: 10.1111/pbi.12701
Sallets, A., Beyaert, M., Boutry, M., Champagne, A. (2014). Comparative proteomics of short and tall glandular trichomes of nicotiana tabacum reveals differential metabolic activities. J. Proteome Res. 13, 3386–3396. doi: 10.1021/pr5002548
Samuels, L., Kunst, L., Jetter, R. (2008). Sealing plant surfaces: Cuticular wax formation by epidermal cells. Annu. Rev. Plant Biol. 59, 683–707. doi: 10.1146/annurev.arplant.59.103006.093219
Seo, P. J., Lee, S. B., Suh, M. C., Park, M. J., Go, Y. S., Park, C. M. (2011). The MYB96 transcription factor regulates cuticular wax biosynthesis under drought conditions in arabidopsis. Plant Cell 23, 1138–1152. doi: 10.1105/tpc.111.083485
Serna, L., Martin, C. (2006). Trichomes: Different regulatory networks lead to convergent structures. Trends Plant Sci. 11, 274–280. doi: 10.1016/j.tplants.2006.04.008
Shi, P., Fu, X. Q., Shen, Q., Liu, M., Pan, Q. F., Tang, Y. L., et al. (2018). The roles of AaMIXTA1 in regulating the initiation of glandular trichomes and cuticle biosynthesis in artemisia annua. New Phytol. 217, 261–276. doi: 10.1111/nph.14789
Sierro, N., Battey, J. N., Ouadi, S., Bakaher, N., Bovet, L., Willig, A., et al. (2014). The tobacco genome sequence and its comparison with those of tomato and potato. Nat. Commun. 5, 3833. doi: 10.1038/ncomms4833
Subramanian, B., Gao, S. H., Lercher, M. J., Hu, S., Chen, W. H. (2019). Evolview v3: A webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 47, W270–W275. doi: 10.1093/nar/gkz357
Tang, J., Yang, X. P., Xiao, C. L., Li, J. Y., Chen, Y. Q., Li, R. Y., et al. (2020). GDSL lipase occluded stomatal pore 1 is required for wax biosynthesis and stomatal cuticular ledge formation. New Phytol. 228, 1880–1896. doi: 10.1111/nph.16741
Tissier, A. (2012). Glandular trichomes: What comes after expressed sequence tags? Plant J. 70, 51–68. doi: 10.1111/j.1365-313X.2012.04913.x
Todd, J., Post-Beittenmiller, D., Jaworski, J. G. (1999). KCS1 encodes a fatty acid elongase 3-ketoacyl-CoA synthase affecting wax biosynthesis in arabidopsis thaliana. Plant J. 17, 119–130. doi: 10.1046/j.1365-313X.1999.00352.x
Wagner, G. J. (1991). Secreting glandular trichomes: More than just hairs. Plant Physiol. 96, 675–679. doi: 10.1104/pp.96.3.675
Wang, E., Hall, J. T., Wagner, G. J. (2004). Transgenic nicotiana tabacum l. with enhanced trichome exudate cembratrieneols has reduced aphid infestation in the field. Mol. Breed. 13, 49–57. doi: 10.1023/B:MOLB.0000012328.04974.fb
Wang, Z. J., Li, Y. H., Zhang, H. Y., Yan, X. X., Cui, H. (2021b). Methyl jasmonate treatment, aphid resistance assay, and transcriptomic analysis revealed different herbivore defensive roles between tobacco glandular and non-glandular trichomes. Plant Cell Rep. 41, 195–208. doi: 10.1007/s00299-021-02801-6
Wang, D. J., Ni, Y., Liao, L. X., Xiao, Y., Guo, Y. J. (2021a). Poa pratensis ECERIFERUM1 (PpCER1) is involved in wax alkane biosynthesis and plant drought tolerance. Plant Physiol. Biochem. 159, 312–321. doi: 10.1016/j.plaphy.2020.12.032
Wang, W. J., Zhang, Y., Xu, C., Ren, J. J., Liu, X. F., Black, K., et al. (2015). Cucumber ECERIFERUM1 (CsCER1), which influences the cuticle properties and drought tolerance of cucumber, plays a key role in VLC alkanes biosynthesis. Plant Mol. Biol. 87, 219–233. doi: 10.1007/s11103-014-0271-0
Wen, X. F., Geng, F., Cheng, Y. J., Wang, J. Q. (2021). Ectopic expression of CsMYB30 from citrus sinensis enhances salt and drought tolerance by regulating wax synthesis in arabidopsis thaliana. Plant Physiol. Biochem. 166, 777–788. doi: 10.1016/j.plaphy.2021.06.045
Wu, M. L., Cui, Y. C., Ge, L., Cui, L. P., Xu, Z. C., Zhang, H. Y., et al. (2020). NbCycB2 represses nbwo activity via a negative feedback loop in tobacco trichome development. J. Exp. Bot. 71, 1815–1827. doi: 10.1093/jxb/erz542
Wu, Z. K. Y., Li, L., Liu, H., Yan, X., Ma, Y. N., Li, Y. P., et al. (2021). AaMYB15, an R2R3-MYB TF in artemisia annua, acts as a negative regulator of artemisinin biosynthesis. Plant Sci. 308, 110920. doi: 10.1016/j.plantsci.2021.110920
Xiong, C., Xie, Q. M., Yang, Q. H., Sun, P. Y., Gao, S. H., Li, H. X., et al. (2020). WOOLLY, interacting with MYB transcription factor MYB31, regulates cuticular wax biosynthesis by modulating CER6 expression in tomato. Plant J. 103, 323–337. doi: 10.1111/tpj.14733
Xue, D. W., Zhang, X. Q., Lu, X. L., Chen, G., Chen, Z. H. (2017). Molecular and evolutionary mechanisms of cuticular wax for plant drought tolerance. Front. Plant Sci. 8, 621. doi: 10.3389/fpls.2017.00621
Xu, B., Taylor, L., Pucker, B., Feng, T., Glover, B. J., Brockington, S. F. (2021). The land plant-specific MIXTA-MYB lineage is implicated in the early evolution of the plant cuticle and the colonization of land. New Phytol. 229, 2324–2338. doi: 10.1111/nph.16997
Yeats, T. H., Rose, J. K. C. (2013). The formation and function of plant cuticles. Plant Physiol. 163, 5–20. doi: 10.1104/pp.113.222737
Zhang, Y. L., Zhang, C. L., Wang, G. L., Wang, Y. X., Qi, C. H., Zhao, Q., et al. (2019). The R2R3 MYB transcription factor MdMYB30 modulates plant resistance against pathogens by regulating cuticular wax biosynthesis. BMC Plant Biol. 19, 362. doi: 10.1186/s12870-019-1918-4
Zhang, H. Y., Zhang, S. T., Yang, Y. X., Jia, H. F., Cui, H. (2018). Metabolic flux engineering of cembratrien-ol production in both the glandular trichome and leaf mesophyll in nicotiana tabacum. Plant Cell Physiol. 59, 566–574. doi: 10.1093/pcp/pcy004
Zhao, Y., Cheng, X. Y., Liu, X. D., Wu, H. F., Bi, H. H., Xu, H. X. (2018). The wheat MYB transcription factor TaMYB31 is involved in drought stress responses in arabidopsis. Front. Plant Sci. 9, 1426. doi: 10.3389/fpls.2018.01426
Keywords: NtMYB306a, transcription factors, trichomes, Nicotiana tabacum, alkane biosynthesis
Citation: Yu J, Lei B, Zhao H, Wang B, Kakar KU, Guo Y, Zhang X, Jia M, Yang H and Zhao D (2022) Cloning, characterization and functional analysis of NtMYB306a gene reveals its role in wax alkane biosynthesis of tobacco trichomes and stress tolerance. Front. Plant Sci. 13:1005811. doi: 10.3389/fpls.2022.1005811
Received: 28 July 2022; Accepted: 14 September 2022;
Published: 06 October 2022.
Edited by:
Kun Lu, Southwest University, ChinaReviewed by:
Qinlong Zhu, South China Agricultural University, ChinaYing Fu, Zhejiang Academy of Agricultural Sciences, China
Copyright © 2022 Yu, Lei, Zhao, Wang, Kakar, Guo, Zhang, Jia, Yang and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Degang Zhao, ZGd6aGFvQGd6dS5lZHUuY24=
 Jing Yu
Jing Yu Bo Lei
Bo Lei Huina Zhao2
Huina Zhao2 Bing Wang
Bing Wang Yushuang Guo
Yushuang Guo Xiaolian Zhang
Xiaolian Zhang Mengao Jia
Mengao Jia Degang Zhao
Degang Zhao