
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 28 September 2022
Sec. Plant Symbiotic Interactions
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.1005303
This article is part of the Research Topic Rhizosphere Interactions: Root Exudates and the Rhizosphere Microbiome View all 23 articles
To improve tobacco leaf quality, excessive K2SO4 fertilizers were applied to soils in major tobacco-planting areas in China. However, the effects of K2SO4 application on soil microbial community and functions are still unclear. An eight-year field experiment with three kinds of K2SO4 amounts (low amount, K2O 82.57 kg hm-2, LK; moderate amount, K2O 165.07 kg hm-2, MK; high amount, K2O 247.58 kg hm-2, HK) was established to assess the effects of K2SO4 application on the chemical and bacterial characteristics of tobacco-planting soil using 16S rRNA gene and metagenomic sequencing approaches. Results showed that HK led to lower pH and higher nitrogen (N), potassium (K), sulfur(S) and organic matter contents of the soil than LK. The bacterial community composition of HK was significantly different from those of MK and LK, while these of MK and LK were similar. Compared to LK, HK increased the relative abundance of predicted copiotrophic groups (e.g. Burkholderiaceae, Rhodospirillaceae families and Ellin6067 genus) and potentially beneficial bacteria (e.g. Gemmatimonadetes phylum and Bacillus genus) associated with pathogens and heavy metal resistance, N fixation, dissolution of phosphorus and K. While some oligotrophic taxa (e.g. Acidobacteria phylum) related to carbon, N metabolism exhibited adverse responses to HK. Metagenomic analysis suggested that the improvement of pathways related to carbohydrate metabolism and genetic information processing by HK might be the self-protection mechanism of microorganisms against environmental stress. Besides, the redundancy analysis and variation partitioning analysis showed that soil pH, available K and S were the primary soil factors in shifting the bacterial community and KEGG pathways. This study provides a clear understanding of the responses of soil microbial communities and potential functions to excessive application of K2SO4 in tobacco-planting soil.
Tobacco (Nicotiana tobacum L.) is an economically important crop that is widely planted worldwide. As a main ingredient in tobacco, K is recognized as an important indicator of quality by the cigarette industry. A higher K content in tobacco leaves can improve the flammability, aroma and processability of cigarettes (Zhang and Kong, 2014). K plays a crucial role in cell expansion, the transportation of compounds, stomatal opening and closing and the activation of enzymes, which could promote the growth of tobacco (Hu et al., 2019). China is a main producing country of flue-cured tobacco in the world, while K deficiency of soil is a common problem in major tobacco-producing areas. As a result, the K content in tobacco is generally lower than the global standard for high-quality tobacco (Ding et al., 2017). K2SO4 is a high-quality and efficient K fertilizer with good solubility and a lack of chlorine, and the application of K2SO4 is currently the most effective measure to improve soil K content and tobacco quality. However, to pursue higher tobacco quality, a large amount of K2SO4 fertilizer (270-360 kg hm-2) is often applied to tobacco-planting soil in China. The application rate of K fertilizer is far beyond the K requirement of tobacco plants, leading to a large amount of K+ and left in the soil. Moreover, long-term application of K2SO4 possibly increased acidification in acidic and neutral soils, mainly due to the H+ released by tobacco roots when it absorbed excess K+ from the soil (Dai et al., 2021). These changes would lead to loss of base ions and inhibition of microbial activity and tobacco root growth (Shen et al., 2018). Therefore, environmental degradation caused by long-term excessive application of K2SO4 greatly limits the sustainable development of tobacco industry, which should be of concern.
Soil microbes are vital for maintaining soil quality and ecosystem, including the turnover of organic matter (OM), the degradation of toxic substances, the acceleration of nutrient availability and the improvement of stress tolerance to pathogens (Jin et al., 2022a). At the same time, soil microbes are also closely related to nutrient uptake, disease occurrence, growth and quality of tobacco (Zheng et al., 2021; Jin et al., 2022b). The variety and quantity of microbes in tobacco-planting soil are abundant. For example, Proteobacteria, Actinobacteria, Acidobacteria, Firmicutes and Bacteroidetes were always the dominant phyla in tobacco-planting soil (Zheng et al., 2021). Arthrobacter and Lysobacter were reported to be significant negative correlated with tobacco bacterial wilt disease (She et al., 2017). The increase of Codinaea acaciae and Saitozyma podzolica species were adverse to tobacco nicotine (Wang et al., 2022). Fertilization is an important means of shaping soil microorganisms. While excessive fertilization leads to the deterioration of soil physicochemical properties and microbial diversity and communities, which in turn leads to a decrease in tobacco yield and quality (Xin et al., 2012). For instance, excessive K fertilizer delays ripening and limits nicotine production (Henry et al., 2019).
Previously, pH and contents of OM, AK and water were considered to be key environmental factors, which significantly shaped the microbial community and diversity of tobacco-planting soils (Wang Z. B. et al., 2019; Shen et al., 2022). Excessive application of K2SO4 reduced the pH, which is widely recognized as the strongest predictor of microbial activity and composition (Rath et al., 2019; Zeng et al., 2019) and explain approximately 70% of species changes (Liu et al., 2022). Previous studies showed that soil pH had significantly positive relationships with soil bacterial α-diversity and bacterial operational taxonomic unit abundance and influenced ecological functions and biogeographic distribution (Wang C. et al., 2019). Wan et al. (2020) demonstrated that enzymes and proteins related to carbon (C), N, phosphorus (P) and S were downregulated in more acidic soils (pH< 5.5) compared to those in soils with pH values higher than 5.5. Wang W. et al. (2019) found that with decreasing pH, the functions of nitrification, ammonia oxidation, N fixation, nitrite respiration, and denitrification were restrained, while the functions of chemoheterotrophy, nitrate reduction and aromatic compound degradation were enriched.
A large amount of residual in soil not only increases soil acidification but also activates Fe3+ and Al3+ in soil and forms precipitation with Ca2+ and Mg2+ (Xu and Ji, 2001). Under anaerobic conditions, excess in soil may form H2S, which destroys aerobic beneficial microorganisms and promotes anaerobic harmful microorganisms (Liu et al., 2021). addition has been shown to significantly affect soil microbial communities by enriching specific microbial taxa associated with the bioavailability and transformation of metals such cadmium, arsenic and iron (Fe) (Li et al., 2019; Wang et al., 2021). Moreover, the addition of Na2SO4 altered the bacterial composition of the dominant phyla by increasing the relative abundance of Proteobacteria and Acidobacteria and depleting Firmicutes (Tang et al., 2020).
Abundant K+ in soil has an antagonistic effect with Ca2+, and so on (Nieder et al., 2011), leading to nutrient imbalance in tobacco soils and plants. Twenty-one consecutive years of KCl application alone significantly decreased the Shannon, Simpson and McIntosh indices of the functional diversity of microbial communities compared to no fertilizer treatment in maize soil (Zhong et al., 2010).
However, studies on the regulation of soil chemical and biological characteristics by K and S fertilizers mostly focus on fertilizers such as KCl and Na2SO4. Moreover, many previous studies have shown that a single application of N fertilizer causes soil acidification and indirectly affects soil microbial communities (Bai et al., 2020; Yang et al., 2020). However, the effects of K2SO4 application and its resulting acidification on soil biological properties are still unclear, especially in tobacco fields. Therefore, in this study, we applied 16S rRNA gene and metagenomic sequencing technologies to analyze changes in the microbial community structure and function of tobacco-planting soil treated with three K2SO4 rates (LK, MK and HK) based on an 8-year experiment. We hypothesized that, HK had a negative and strong impact on the soil bacterial community and functions, when compared to LK. And MK had less effect on soil biological properties than HK. The variations of biological properties were due to the altered soil physicochemical properties, especially soil pH. The objectives of this study were to investigate (1) the effects of 8-year application of K2SO4 on soil physicochemical properties and nutrient uptake of tobacco plants; (2) the effects of 8-year application of K2SO4 on soil bacterial diversity and community composition of tobacco-planting soil; (3) impacts of 8-year application of K2SO4 on potential functional pathways of tobacco-planting soil; and (4) the key soil environmental variables that strongly affected soil microbial community and function.
This study was established in tobacco resources and environment field scientific observation and experiment station of the Chinese Academy of Agricultural Sciences (36°26’54″N, 120°34’38″E, 75 m a.s.l.) in 2010 in Qingdao city, Shandong province, China. This region has a temperate monsoon climate with a mean annual rainfall of 708.9 mm, average daily air temperature of 12.1°C, frost-free period of 200 d and annual accumulated temperature of 4410°C. The typical soil in this region is Alfisols (FAO Soil Taxonomic System). Before the experiment began in May 2010, the soil at 0-20 cm depth had a pH of 5.56, OM of 11.66 g kg-1, AN, available phosphorus (AP) and AK of 52.69 mg kg-1, 10.60 mg kg-1 and 105.25 mg kg-1, respectively.
Three treatments were included in the experiment: (1) applying compound fertilizer (N 15%, P2O5 15%, K2O 15%) 550.5 kg hm-2 (LK); (2) applying compound fertilizer 550.5 kg hm-2 and K2SO4 (K2O 50%, S 18%) 165 kg hm-2 (MK); and (3) applying compound fertilizer 550.5 kg hm-2 and K2SO4 330 kg hm-2 (HK). The nutrient application amount of each treatment is shown in Table 1. The experiment used a randomized block design. Each plot (5 m long and 4.4 m wide) was separated by concrete walls from others. Each treatment had 3 replicates, giving a total of 9 pots. The variety used in the experiment was NC89, and 40 tobacco plants were planted in each plot with a line spacing of 1.1 m and row of 0.5 m. All fertilizers were applied to the surface in a certain amount, and then mixing with 0-10 cm soil manually, ridging and planting tobacco. The tobaccos were transplanted in the first ten days of June every year. Irrigation was carried out according to the water requirement of tobacco and rainfall during different growth periods. The other field management practices were in accordance with local farming practices.
Soil and plant samplings were conducted in the mature season of flue-cured tobacco on August 30, 2017. One composite rhizosphere soil sample was taken from each plot consisting of roots of 5 randomly selected tobacco plants. The roots were uprooted by shaking the roots, removing the loose soil at the roots, and collecting the soil at the roots with a sterile brush. Then, the fresh soil was passed through a 2 mm sieve and divided into two fractions. One part of the fresh soil was used for the analysis of bacterial community and functions. The other part was air dried for the determination of soil physicochemical properties. After collecting soil, the 5 tobacco plants taken from each plot were washed and then were mixed into one sample for determining dry matter weight and nutrient content. Three composite samples of both soil and plant were conducted for each treatment.
Soil physicochemical properties and plant nutrients were determined according to Bao (2011). Soil pH was measured with a soil-to-water ratio of 1:2.5 using a pH meter (Meter3100C the US). The OM content was analyzed using dichromate oxidation. The total nitrogen (TN) content was digested by H2SO4-K2Cr2O7 and measured by Kjeldahl digestion with automatic N analyzer (KjeltecTM 8400 Denmark). The AN content was determined by the alkali-diffusion method. Total potassium (TK) and AK were digested by HClO4 and CH3COONH4, respectively, and both were measured by flame atomic absorption spectrophotometry with flame spectrometry (Sherwood M410 Britain). Total sulfur (TS) and AS were digested by Mg(NO3)2 and extracting agent of Ca(H2PO4)2 and CH3COOH, respectively, and both were determinated by BaSO4 turbidimetry with UV-visible spectrophotometer (SHIMADZU UV-2700 Japan).
The plant dry matter was weighed after drying at 105°C for 30 mins and subsequently at 80°C to a constant weight. Then, the dry plant was ground and sieved to< 2 mm. The contents of plant N and K were digested by H2SO4-H2O2 digestion and then N content was determined by the Kjeldahl method with automatic N analyzer and K content was measured by flame photometry with flame spectrometry. The contents of plant S were digested by HNO3-HClO4 and then measured by the BaSO4 turbidimetric method with UV-visible spectrophotometer. N (K, S) accumulation was calculated as the product of dry matter and the N (K, S) content of the plant.
Soil DNA was extracted using a PowerSoil DNA Isolation Kit (MoBio Laboratories, Carlsbad, CA) following the manual. The purity and quality of the genomic DNA were checked on 0.8% agarose gels.
The V3-4 hypervariable region of the bacterial 16S rRNA gene was amplified with the primers 338F (5’-ACTCCTACGGGAGGCAGCAG-3’) and 806R (5’-GGACTACNNGGGTATCTAAT-3’) (Caporaso et al., 2012). For each soil sample, an 8-digit barcode sequence was added to the 5’ end of the forward and reverse primers (provided by Allwegene Company, Beijing). PCR was carried out on a Mastercycler Gradient (Eppendorf, Germany) using 25 μl reaction volumes, containing 12.5 μl 2× Taq PCR MasterMix, 3 μl BSA (2 ng μl-1), 1 μl forward primer (5 μM), 1 μL reverse primer (5 μM), 2 μl template DNA, and 5.5 μl ddH2O. The cycling parameters were 95°C for 5 min, followed by 28 cycles of 95°C for 45 s, 55°C for 50 s and 72°C for 45 s with a final extension at 72°C for 10 min. The PCR products were purified using an Agencourt AMPure XP Kit.
The raw data were first screened, and sequences were removed from consideration if they were shorter than 230 bp, had a low-quality score (≤ 20), contained ambiguous bases or did not exactly match to primer sequences and barcode tags and separated using the sample-specific barcode sequences. Qualified reads were clustered into operational taxonomic units (OTUs) at a similarity level of 97% using the Uparse algorithm of Vsearch (v2.7.1) software (Edgar, 2013). The Ribosomal Database Project (RDP) Classifier tool was used to classify all sequences into different taxonomic groups against the SILVA128 database (Cole et al., 2009). The sequencing was performed on Illumina Miseq PE300 platform. The raw sequences of the 16S rRNA gene were deposited into the NCBI database under the accession number PRJNA805374.
Total microbial genomic DNA samples were extracted using the OMEGA Soil DNA Kit (D5625-01), following the manufacturer’s instructions, and stored at -20°C prior to further assessment. The quantity and quality of extracted DNAs were measured using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA,USA) and agarose gel electrophoresis, respectively. The extracted microbial DNA was processed to construct metagenome shotgun sequencing libraries with insert sizes of 400 bp by using Illumina TruSeq Nano DNA LT Library Preparation Kit. Each library was sequenced by Illumina HiSeq X-ten platform (Illumina, USA) with PE150 strategy.
Raw sequencing reads were processed to obtain quality-filtered reads for further analysis. Firstly, sequencing adapters were removed from sequencing reads using Cutadapt (v1.2.1) (Martin, 2011). Secondly, low-quality reads were trimmed using a sliding-window algorithm in fastp (Chen et al., 2018). Megahit (v1.1.2) (Li et al., 2015) was used to assemble each sample using the meta-large preset parameters. The generated contigs (longer than 200 bp) were then pooled together and clustered using mmseqs2 (Steinegger and Soding, 2017) with “easy-Linclust” mode, setting the sequence identity threshold to 0.95 and covering residues of the shorter contig to 90%. MetaGeneMark (Zhu et al., 2010) was used to predict the genes in the contigs. The CDSs of all samples were clustered by mmseqs2 (Steinegger and Soding, 2017) with “easy-cluster” mode, setting the protein sequence identity threshold to 0.90 and covering residues of the shorter contig to 90%. To assess the abundances of these genes, the high-quality reads from each sample were mapped onto the predicted gene sequences using salmon (Patro et al., 2015) in the quasi-mapping-based mode with “–meta –minScoreFraction = 0.55”, and the CPM (copy per kilobase per million mapped reads) was used to normalize abundance values in metagenomes. The functionality of the nonredundant genes was obtained by annotation using mmseqs2 (Steinegger and Soding, 2017) with the “search” mode against the protein databases of KEGG.
Analysis of variance was conducted to determine significant differences in indices of soil physicochemical characteristics, α-bacterial diversity, plant dry matter and nutrient accumulation. OTUs and pathways (Level 3) were analyzed by principal coordinate analyses (PCoA) based on Bray–Curtis distance by CANOCO 5.0. Permutational multivariate analysis of variance (PERMANOVA) was conducted using the adonis function (Vegan package, Rstudio) to evaluate similarities in the OTUs and pathways. Venn diagrams were constructed to show the number of shared OTUs by MOTHUR software (Schloss et al., 2011). Post hoc analysis by Stamp was used to compare the differences in the top 20 phyla and genera between different soil samples. Differentially abundant OTUs between groups were calculated using a moderate t-test, and the obtained P values were adjusted using the Benjamini–Hochberg correction method. Enriched OTUs were further visualized in volcano and heatmap plots using the Limma R package and heatmap.2 function R package. Linear discriminant analysis effect size (LEfSe) for detecting significant differences in KEGG pathways was performed on the online Galaxy platform (http://huttenhower.sph.harvard.edu/galaxy/) (Segata et al., 2011). Redundancy analysis (RDA) and variation partitioning analysis (VPA) were applied to clarify the influence of environmental factors on the microbial community and functional composition. RDA and VPA were implemented using R project Vegan package (version 2.5.3) and CANOCO 5.0, respectively. Co-occurrence network analysis was used to explore environmental factor-OTU and environmental factor-pathway interactions. Only top100 OTUs and pathways with coefficients>0.85 (or<-0.85) and FDR corrected P values<0.05 (Spearman’s correlation) were identified and established into a network. All statistical analyses were performed using igraph package in R (Csardi and Nepusz, 2006) and networks were constructed and visualized in Cytoscape v.3.8.0 (Shannon et al., 2003). Structural equation modeling (SEM) was performed by SPSS-AMOS to analyze hypothetical pathways to explain soil physicochemical characteristics, bacterial community and KEGG pathways effects on plant biomass. The model fit was assessed by a χ2-test, the comparative fit index (CFI) and the root square mean error of approximation (RMSEA). Mean values ± SE were reported here.
The results of a comparative analysis of soil physicochemical properties among different K2SO4 treatments are presented in Figure 1. Applying medium and high amounts of K2SO4 (MK and HK) promoted N (AN and TN), K (AK and TK) and S (AS and TS) contents compared to LK, and the differences between HK and LK was always remarkable. The OM contents were also higher in MK and HK than in LK and there was significant difference between MK and LK. Unlike other soil nutrients, pH dropped with the increasing K2SO4 rate, and the difference between LK and HK was significant.
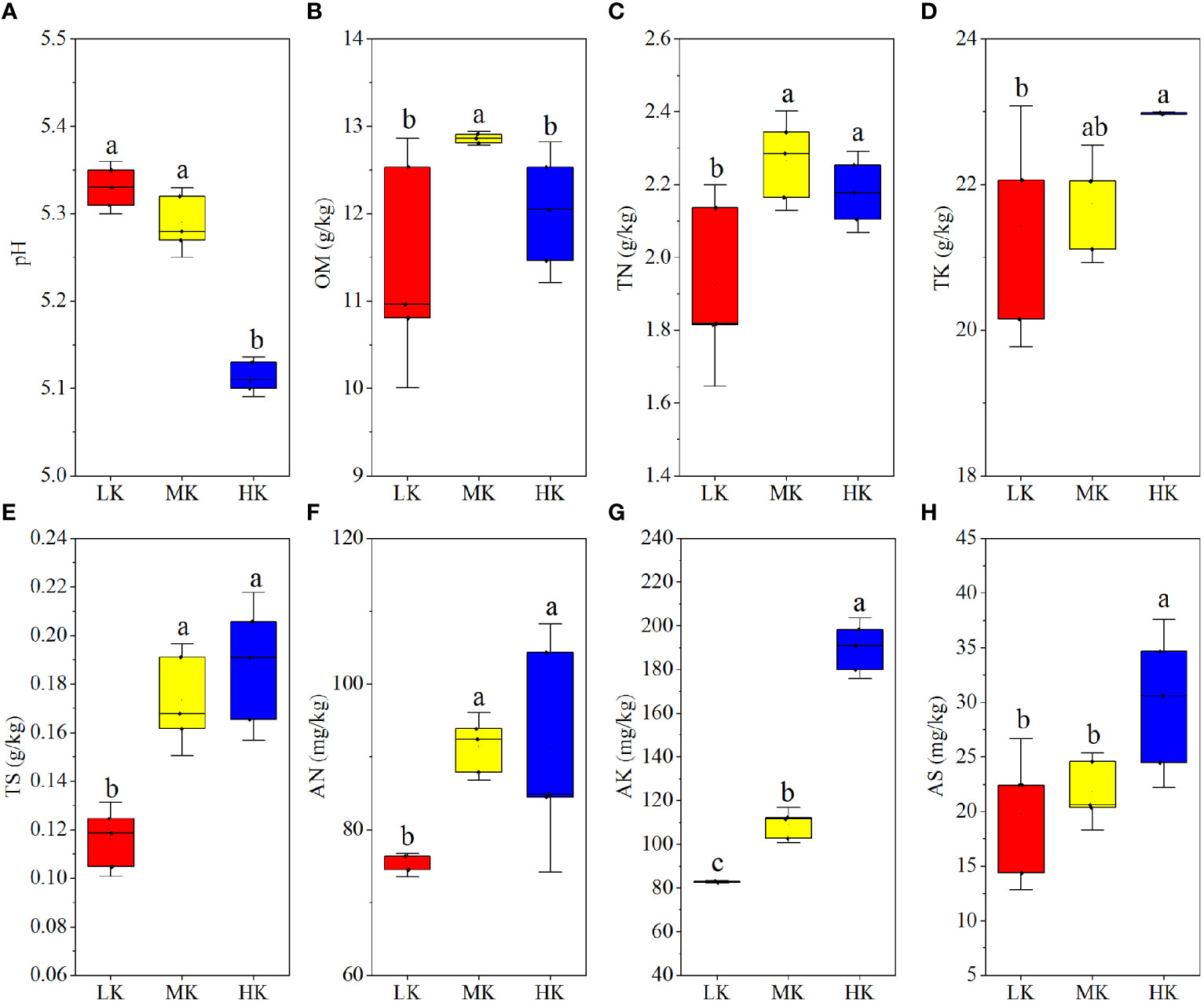
Figure 1 Soil physicochemical properties of plots with different fertilization regimes (A-H). Different lowercase letters above the boxes indicate significant differences (P < 0.05) among the different treatments, as determined by ANOVA followed by the LSD test. LK, Low amount of K2SO4; MK, Middle amount of K2SO4; HK, High amount of K2SO4. OM, organic matter; TN, total nitrogen; TK, total potassium; TS, total sulfur; AN, available nitrogen; AK, available potassium; AS, available sulfur.
The N, K, S nutrient and dry matter accumulation of different plots are displayed in Figure 2. The N and dry matter accumulation of whole plant decreased with increasing K2SO4 amount, while the K accumulation showed the opposite trend. Compared to LK, the dry matter accumulation was decreased by 10.50% and 26.05% in MK and HK, respectively. The differences in accumulations of N, K and dry matter between LK and HK were all remarkable.
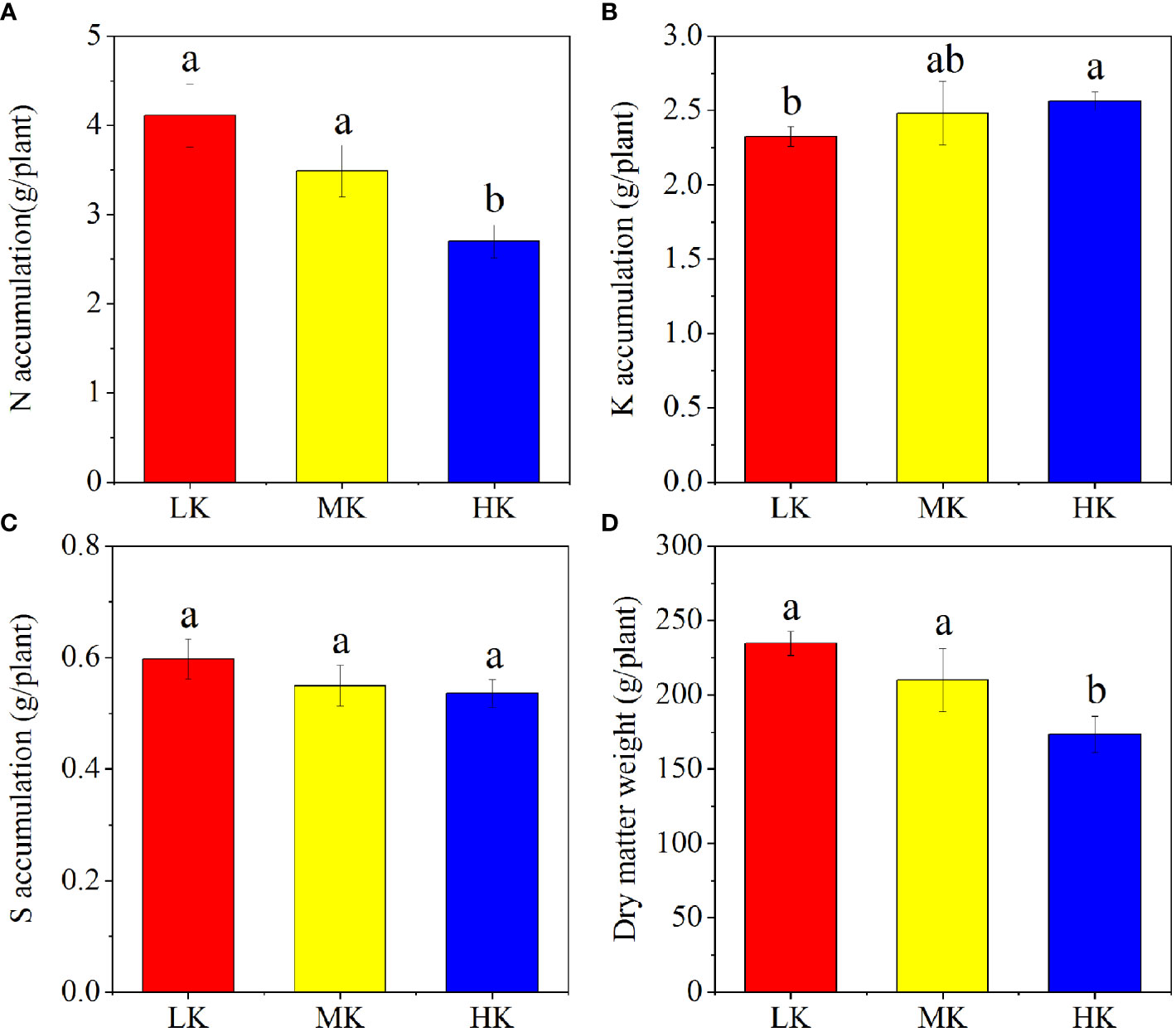
Figure 2 N (A), K (B), S (C) and dry matter (D) accumulation in plants under different treatments. Different lowercase letters above the bars indicate significant differences (P< 0.05) among the different treatments, as determined by ANOVA followed by the LSD test. LK, Low amount of K2SO4; MK, Middle amount of K2SO4; HK, High amount of K2SO4.
Through the 16S rRNA gene sequencing analysis of 9 soil samples, a total of 36020-40641 raw tags and 34116-337892 clean tags were obtained (Table 2). The sequencing coverage rate was 0.9760-0.9778 with 97% similarity. The results basically covered all the species in the tested samples, and further analysis of the bacterial community structure could be carried out.
α-diversity analysis based on OTU showed that Chao1, Observed_species, PD_whole_tree and Shannon were not significantly different among the three treatments, signifying that the addition of K2SO4 had little effect on bacterial α-diversity (Figure 3).
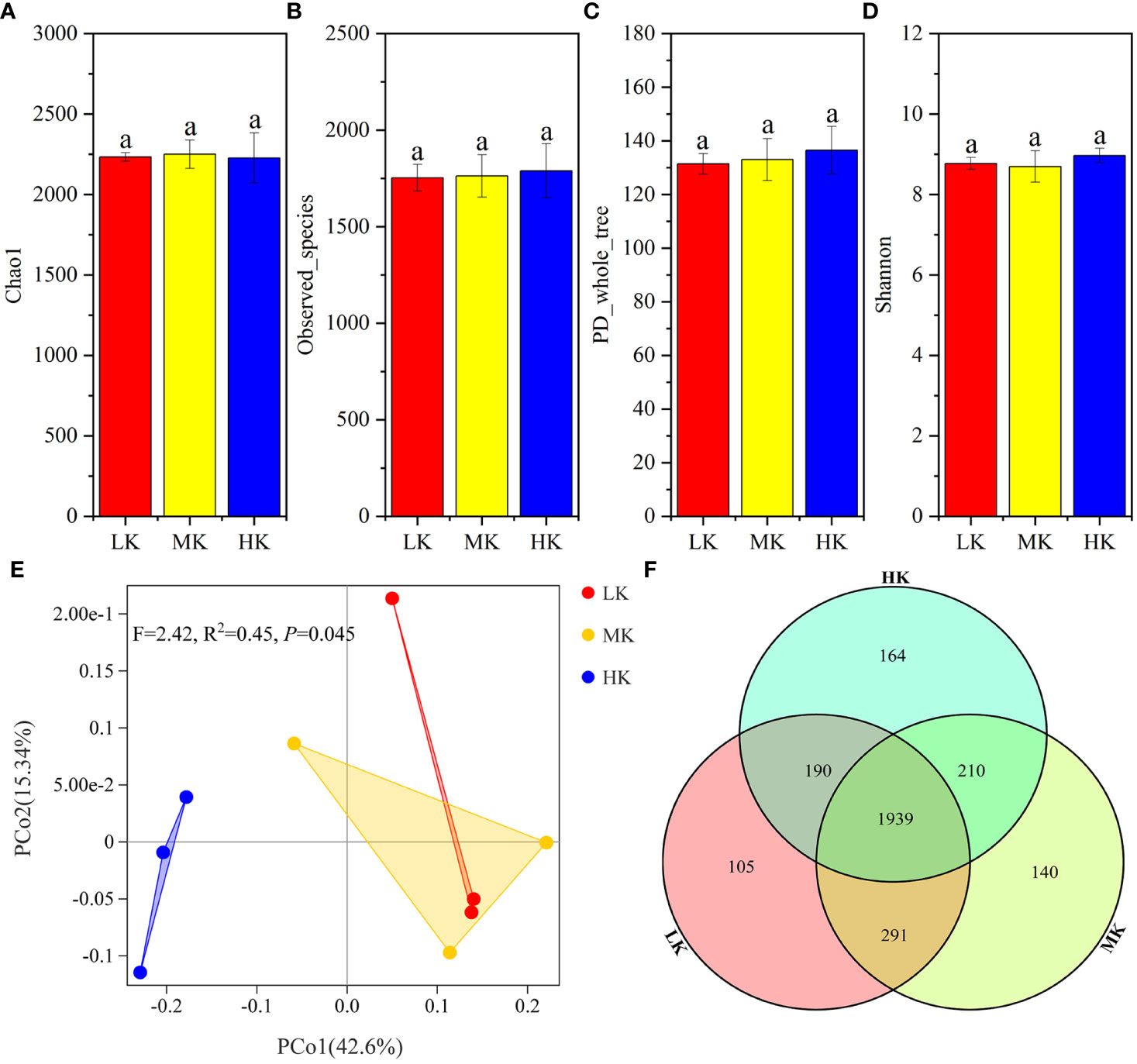
Figure 3 Similarity and differentiation of the bacterial community with different fertilization treatments. (A-D). α-diversity of soils treated with different fertilization treatments; (E). PCoA plot of bacterial communities at the OTU level; (F). Venn diagram of exclusive and shared bacterial taxa at the OTU level. Values with the same lowercase letters are not significantly different among the different treatments (LSD test). LK, Low amount of K2SO4; MK, Middle amount of K2SO4; HK, High amount of K2SO4.
PCoA is performed to determine the OTU compositions in soils with different treatments (Figure 3). PERMANOVA results showed that application of K2SO4 explained the variation in OTU compositions significantly (P =0.045). The variance contribution rates of the first and second principal components were 42.6% and 15.34%, respectively. The three treatments could be divided into two groups along the PCoA1 axis. The three samples for the HK gathered together on the left side of the abscissa PCoA1, well separated from those of MK and LK. Most samples of MK and LK were clustered together on the right half of the plot. It was indicated that the bacterial composition of MK and LK was similar, and HK significantly changed the bacterial community.
The Venn diagram (Figure 3) showed that the number of OTUs unique to each treatment increased from 105 to 164 with the increasing K2SO4 rate, and the number of OTUs shared by LK and MK (2230) was greater than that shared by HK and LK (2129) and HK and MK (2149). These sequencing data indicated that applying a high amount of K2SO4 had a stronger stimulatory effect on the soil bacterial community, but MK had little effect.
Post hoc tests are used to detect significant differences in the relative abundance of the top 20 phyla and genera (Figure 4). Addition of K2SO4 significantly altered the bacterial composition at the phylum and genus levels. HK enriched the relative abundances of Proteobacteria, Gemmatimonadetes, Bacteroidetes, Patescibacteria phyla and Sphingomonas, Bacillus, Ellin6067 genera, while it reduced the proportions of Acidobacteria, Chloroflexi phyla and uncultured_Acidobacteria_bacterium, Bryobacter genera compared with LK. Only one genus, Pseudolabrys, showed significant difference between MK and LK, and there was no significant difference at the phylum level between the two treatments. This result indicated that HK greatly shaped the bacterial composition at the phylum and genus levels, while MK had little effect on the bacterial community.
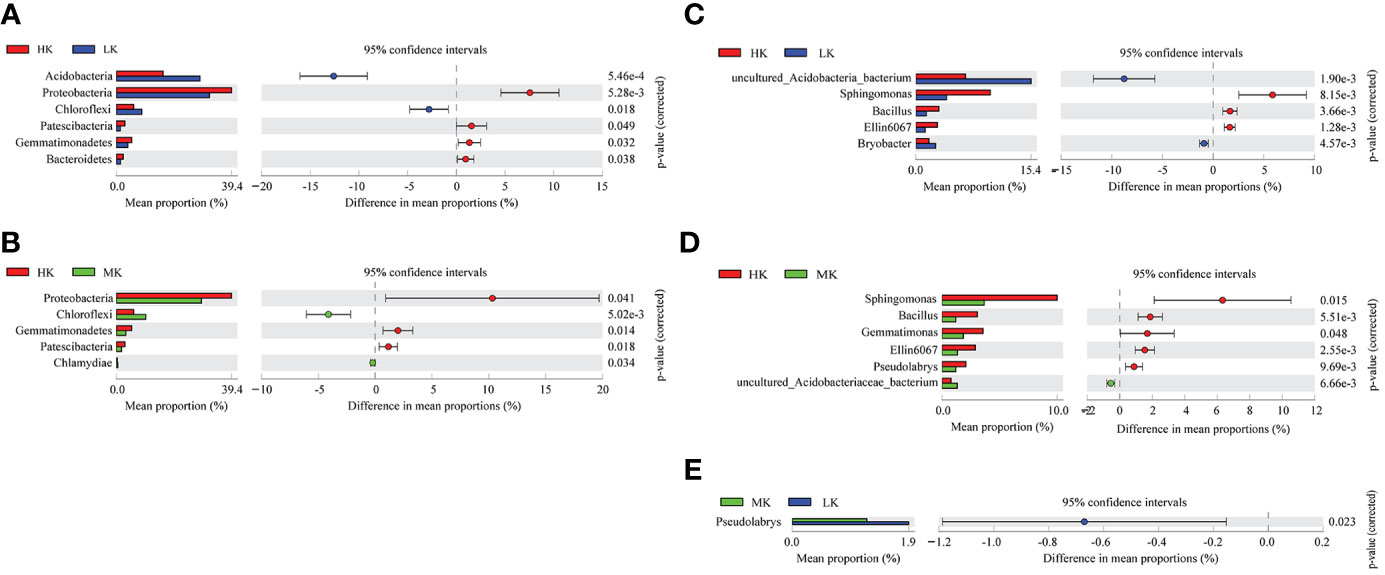
Figure 4 Comparative analysis of the top 20 species phyla and genera with significant differences between different treatments. (A) Differences at phylum level between LK and HK. (B) Differences at phylum level between LK and MK. (C) Differences at genus level between LK and HK. (D) Differences at genus level between MK and HK. (E) Differences at genus level between MK and LK. Only significant differences are shown (P< 0.05); LK, Low amount of K2SO4; MK, Middle amount of K2SO4; HK, High amount of K2SO4.
We further identified OTUs correlated with the differences between different treatments to explore the enrichment or exclusion of different bacterial taxa by fertilization (Figure 5). The number of OTUs with significant differential relative abundance between the HK and LK group was greater than those between the HK and MK group and the MK and LK group, indicating that HK had a greater effect on the composition of OTUs than MK.
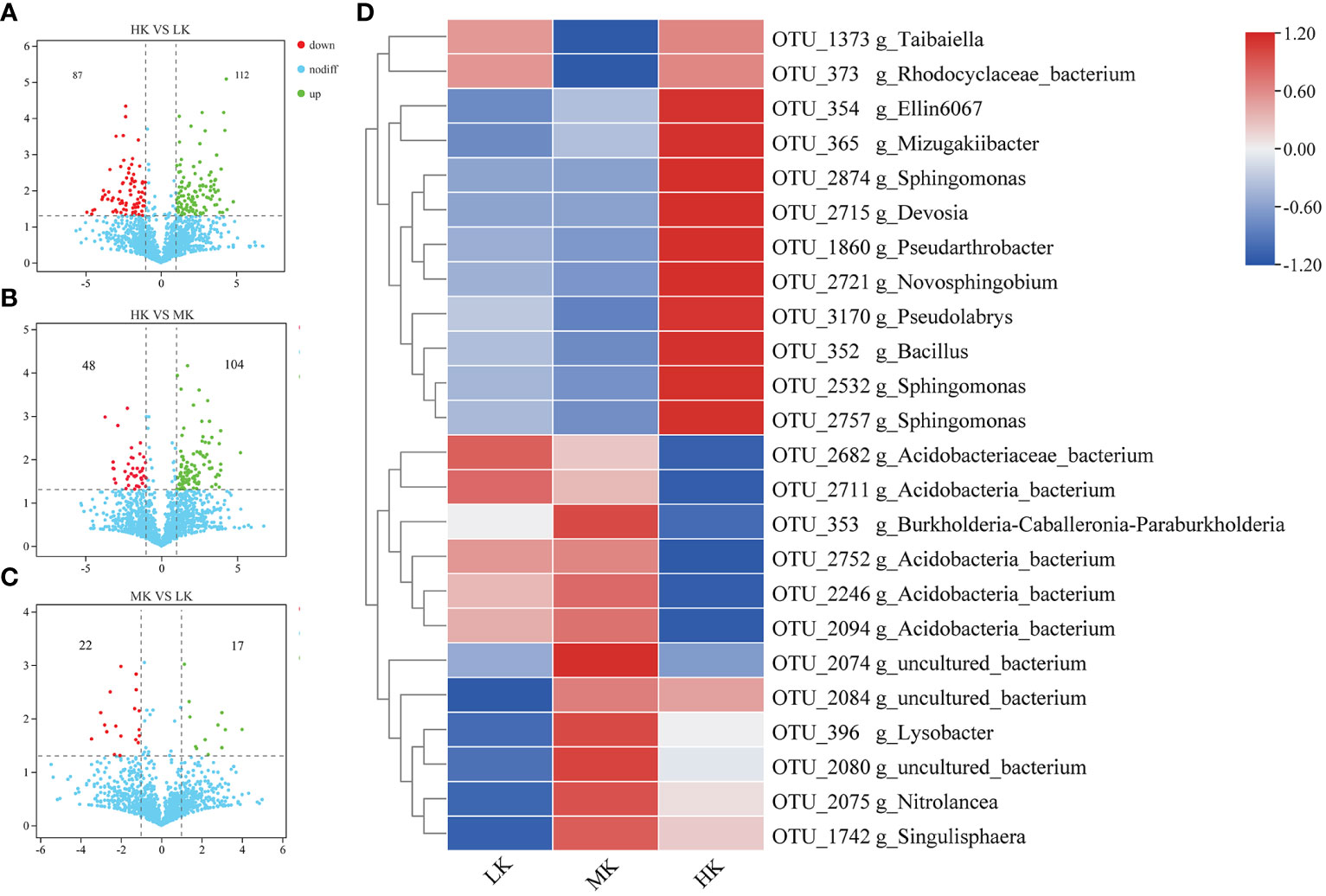
Figure 5 The relative abundance of enriched or depleted OTUs in different fertilized soils after pairwise comparison. (A-C) Volcano plot showing the differentially abundant OTUs between HK and LK, HK and MK, MK and LK. The position along the y-axis represents the log2 of average abundance of each OTU, and the x-axis represents the log2 of fold change between two groups. (D) Heatmap of the relative abundance of significantly enriched OTUs (top 30) among the different treatments.
To display the differences in the relative abundance of bacterial operational taxonomic units (OTUs), a heatmap is constructed based on the top 30 enriched OTUs (Figure 5). Compared with LK, MK enriched 7 OTUs, and 4 of them belonged to uncultured_bacterium, and the other three were affiliated with Lysobacter, Nitrolancea and Singulisphaera genera, respectively. MK also depleted 3 OTUs belonging to Taibaiella, uncultured_bacterium and uncultured_Rhodocyclaceae. HK improved 5 OTUs compared to LK, and 3 of them were affiliated with Sphingomonas genus and the other two belonged to Ellin6067 and Pseudarthrobacter genera, respectively. There were 5 OTUs downregulated in HK compared to LK. And 4 of them belonged to uncultured_Acidobacteria_bacterium and the other belonged to uncultured_bacterium.
Phylogenetic investigation of communities by reconstruction of unobserved states (PICRUSt) analysis predicts KEGG functional pathways (Level 3) associated with the metagenomes of the three treatments. PCoA analysis is carried out to investigate the effects of the treatments on the composition of KEGG pathways (Figure 6). PERMANOVA results confirmed that application of K2SO4 had no significant effect on the composition of KEGG pathways (P =0.163). The variance contribution rates of the first and second principal components were 31.54% and 25.81%, respectively. The three samples of HK could be completely separated from those of the LK treatment. In comparison, the samples of MK were closer to those of LK. This result indicated that HK had a greater effect on the potential functional composition of microorganisms than MK. A Venn diagram showed small differences in the number of shared and exclusive pathways across the three treatments (Figure 6).
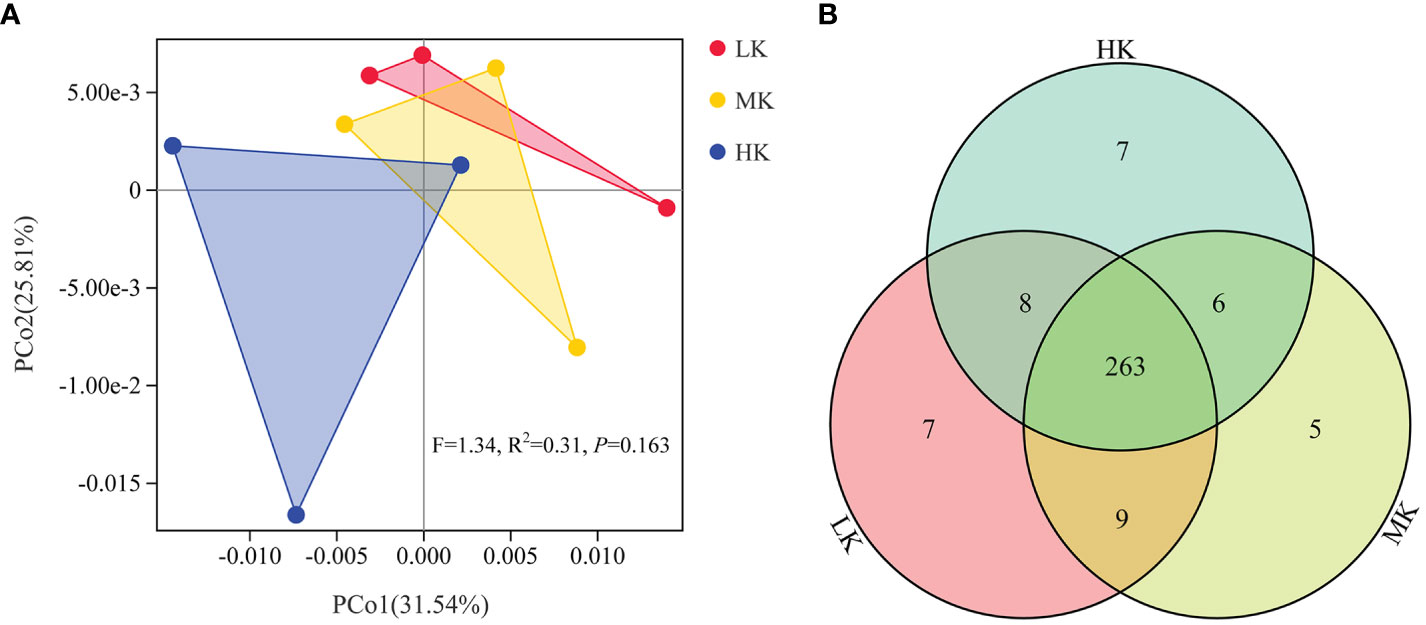
Figure 6 Similarity and differentiation of KEGG pathways with different fertilization treatments. (A) PCoA plot of KEGG pathways at level 3; (B) Venn diagram of exclusive and shared KEGG pathways at level 3. LK, Low amount of K2SO4; MK, Middle amount of K2SO4; HK, High amount of K2SO4.
LEfSe analysis is conducted on the top 100 level 3 KEGG pathways to determine pathways at level 3 KEGG gene annotation with significant differences in abundance across the three treatments. When comparing HK and LK (Figure 7), HK was primarily associated with pathways of homologous recombination, starch and sucrose metabolism, DNA replication, glycolysis/gluconeogenesis, base excision repair, aminoacyl-tRNA biosynthesis and the TCA cycle, while LK was primarily associated with valine, leucine and isoleucine biosynthesis, pantothenate and CoA biosynthesis and C5-branched dibasic acid metabolism. In the MK and LK group (Figure 7), MK was mainly associated with the functions of one carbon pool by folate, streptomycin biosynthesis, monobactam biosynthesis and homologous recombination, while LK was mainly associated with glyoxylate and dicarboxylate metabolism, alanine, aspartate and glutamate metabolism, RNA degradation and pantothenate and CoA biosynthesis.
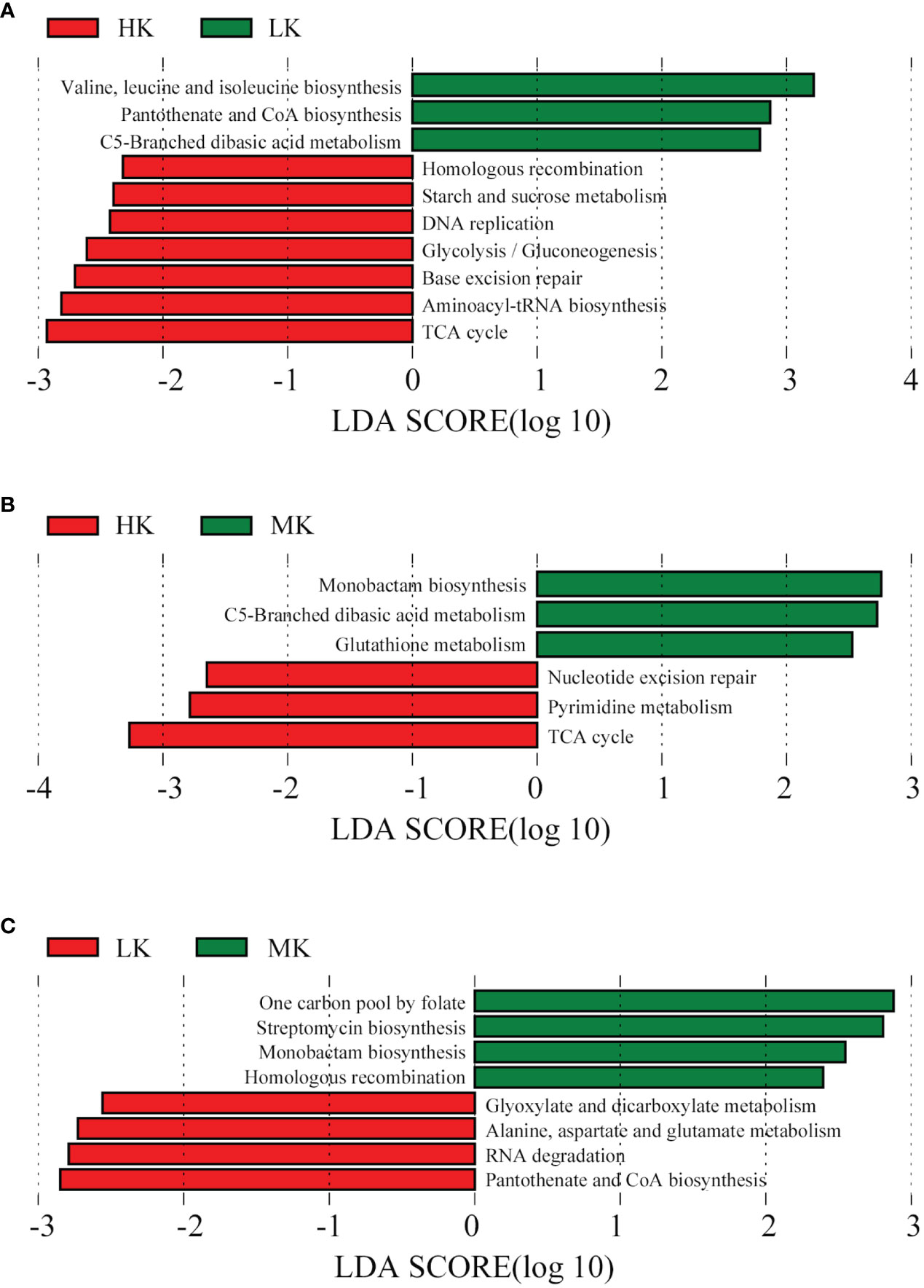
Figure 7 Histogram of the LDA effect value of differentially enriched KEGG pathways at level 3. Lineages with LDA values higher than 2.5 are displayed. (A) HK vs. LK; (B) HK vs. MK; (C). MK vs. LK.
The results of RDA analysis for the bacterial community and functional pathways are visualized in Figure 8. The first two axes explained 56.17% and 17.29% of the total variance in the bacterial community, respectively, and 40.23% and 26.95% in pathways. Among the 8 environmental variables, a remarkable impact of pH (r2 = 0.636, P = 0.029) was found in the bacterial community, as well as pH (r2 = 0.757, P = 0.025), AK (r2 = 0.74, P = 0.023) and AS (r2 = 0.878, P = 0.004) on pathway composition (Table 3). VPA analysis showed that pH, AK, AS and TK explained 29.23%, 26.03%, 24.84% and 4.95% of bacterial community variation, respectively (Figure 8). AS, AK, pH, TS and TK accounted for 14.61%, 13.09%, 11.42%, 7.53% and 4.57% of pathway variation, respectively (Figure 8). In summary, pH, AK and AS were the critical factors in shaping bacterial community and functional composition.
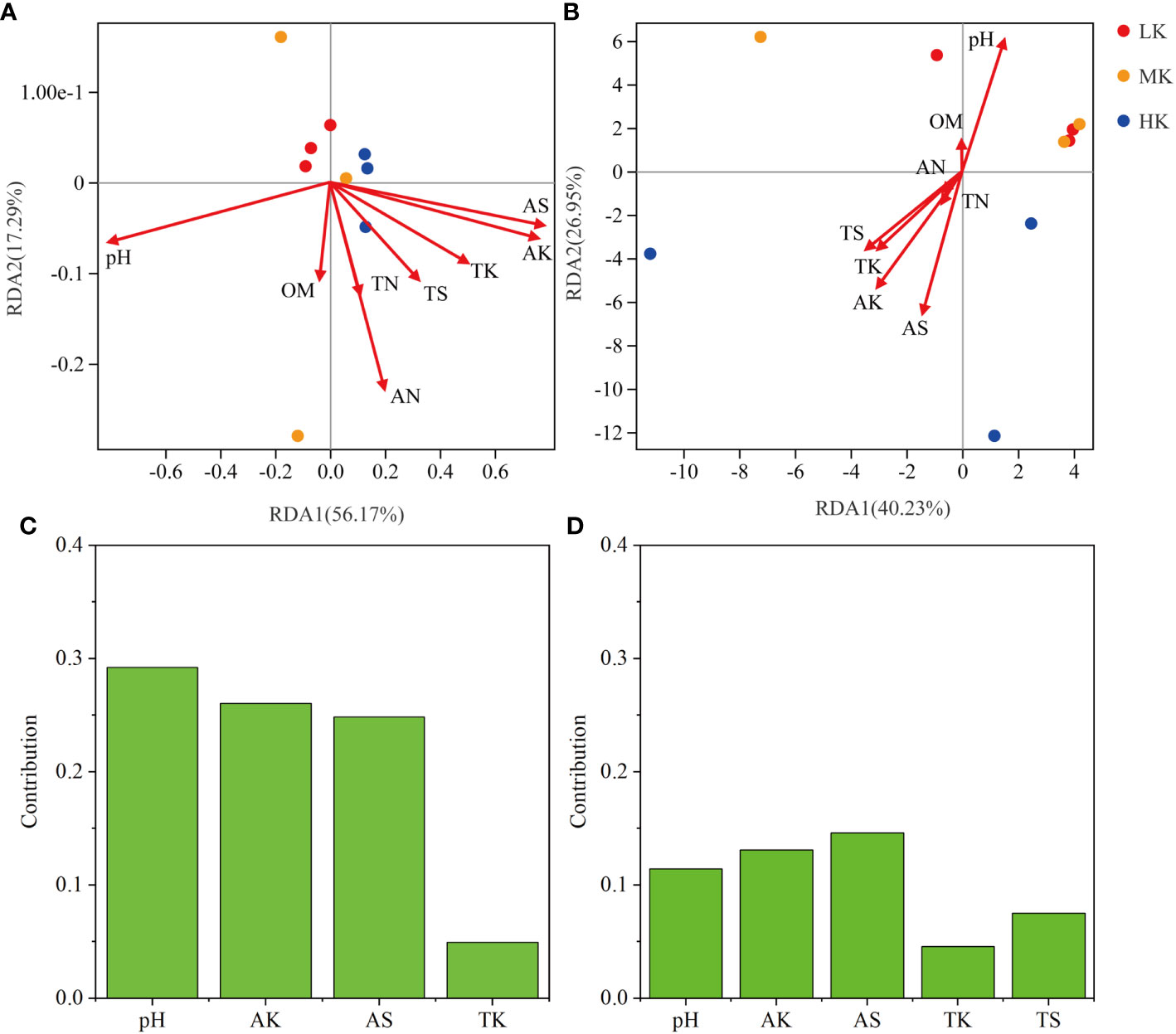
Figure 8 RDA analysis among soil samples based on all OTUs (A) and pathways_L3 (B), and VPA analysis of the effects of soil physicochemical properties on OTUs (C) and pathways_L3 (D). LK, Low amount of K2SO4; MK, Middle amount of K2SO4; HK, High amount of K2SO4. OM, organic matter; TN, total nitrogen; TK, total potassium; TS, total sulfur; AN, available nitrogen; AK, available potassium; AS, available sulfur.
Further, the interactions between environmental factors and microbial communities and functions were investigated using person’s correlation and were visualized by co-occurrence networks (Figure 9). In the network of OTU, pH recorded the highest node connectivity (34), followed by AK (31) and AS (21). In most cases, pH had significantly or extremely significantly negative correlations with OTUs belonged to Proteobacteria and had positive correlations with OTUs belonged to Acidobacteria (Table 1S). AS and AK showed the opposite trends. In the network of pathways, AS recorded the highest node connectivity, and was significantly negatively correlated with 21 pathways, which belonged to pathways (L1) of Metabolism, Environmental Information Processing, Human Diseases, Genetic Information Processing, Cellular Processes, Organismal Systems (Table 2S). And the node connectivities of AN, TS and pH were 11, 10, and 8, respectively.
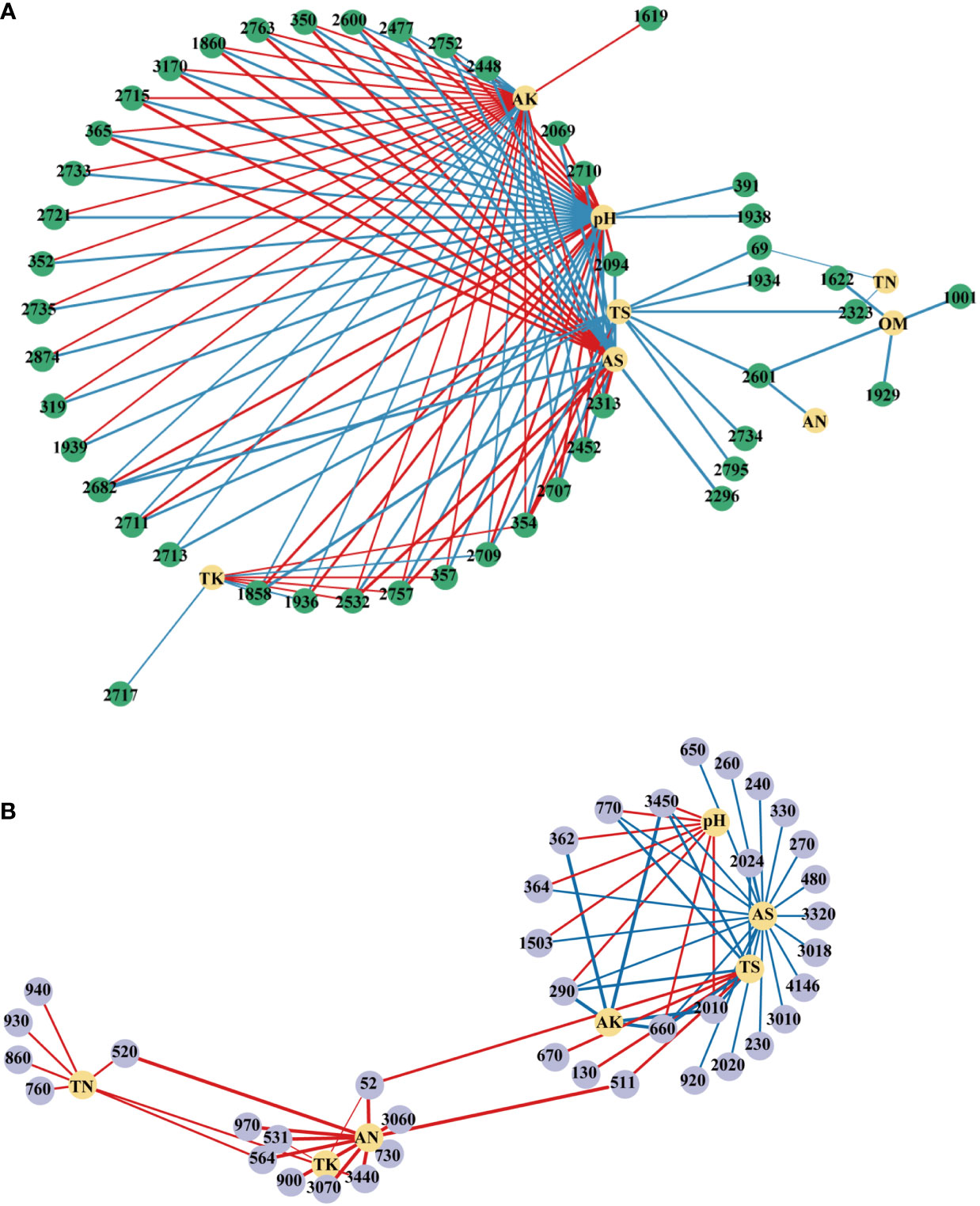
Figure 9 Network analysis revealing associations between soil physicochemical properties and top100 OTUs (A) and pathways (B). A blue line indicates a negative interaction, while a red line indicates a positive interaction. The numbers in the figure represent the numbers of the OTU or pathway. LK, Low amount of K2SO4; MK, Middle amount of K2SO4; HK, High amount of K2SO4. OM, organic matter; TN, total nitrogen; TK, total potassium; TS, total sulfur; AN, available nitrogen; AK, available potassium; AS, available sulfur.
SEM results showed that AK and pH significantly affected abundance of KEGG pathway (SPC = -2.364, P< 0.001; SPC = -1.622, P< 0.05), while no significant effect on abundance of bacterial community (Figure 10). And all the three physicochemical indicators and abundance of bacterial community and KEGG pathway had no significant effect on plant biomass.
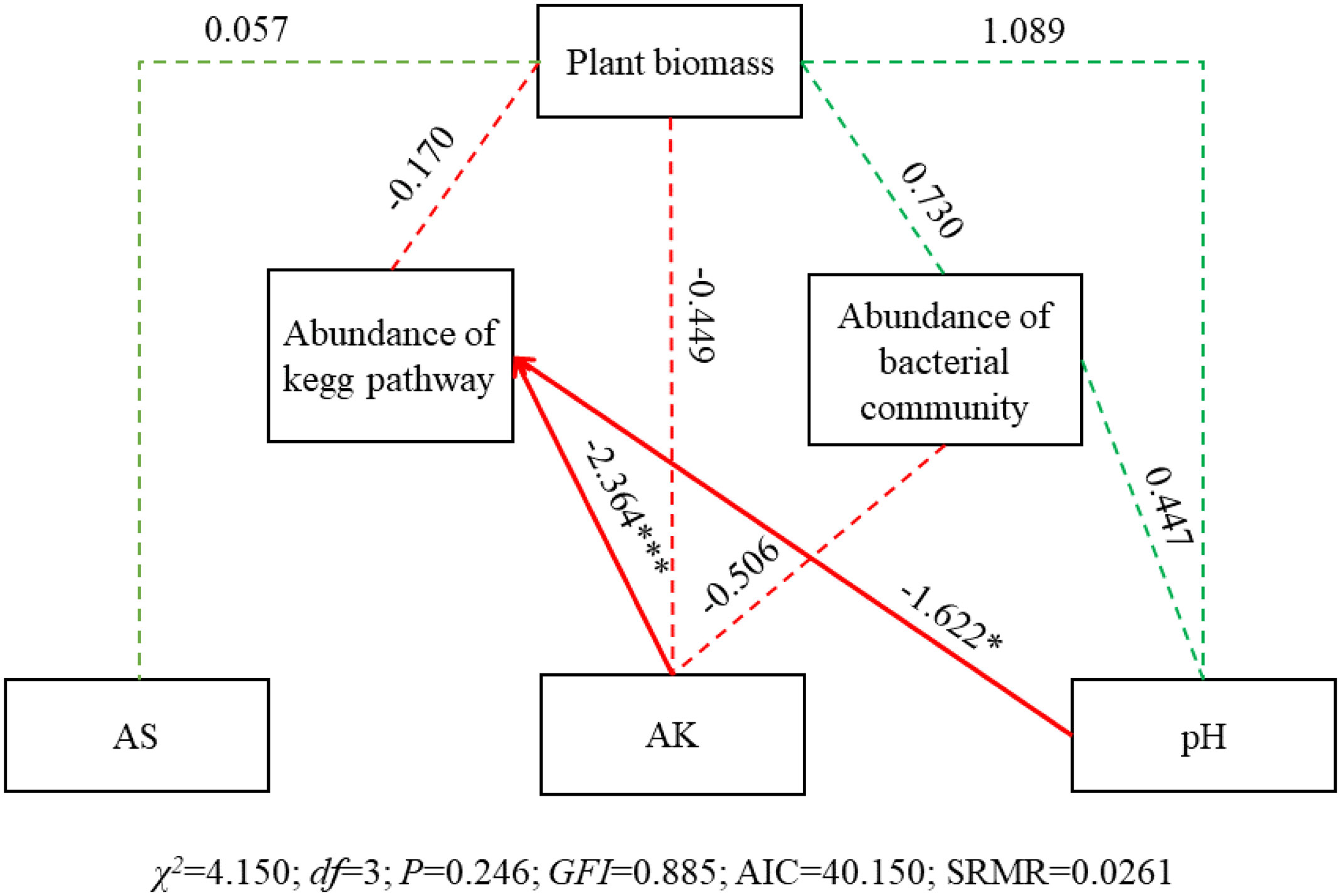
Figure 10 SEM of the effects of soil physicochemical characteristics, bacterial community and KEGG pathways on plant biomass. Square boxes denote variables included in the models. Values associated with solid arrows represent standardized path coefficients (SPCs) and asterisks mark their significance: *P< 0.05; ***P< 0.001. Solid arrows denote the directions and effects that were significant (P< 0.05). Dashed arrows represent the directions and effects that were non-significant (P< 0.05). Green arrows indicate a positive relationship (P< 0.05), while red indicates a negative correlation. CFI = 0.885 is result from the chi-squared value is less than the degrees of freedom in this SEM. Abundance of bacterial community, PCA1 of the abundance of the top 100 OTUs; Abundance of the abundance of the top 100 pathways; AK, available potassium; AS, available sulfur.
The results of the present study demonstrated that after the 8-year application of K2SO4 fertilizer significantly changed the physicochemical properties of tobacco-planting soil. MK and HK led to increase of soil acidification, when compared to LK (Figure 1). The main reasons for this phenomenon may be as follows. First, tobacco plant absorbed much more K+ than , and H+ was released by root to maintain the charge balance in its body (Wallace, 1994). Second, almost all the base ions (mainly K+) absorbed by tobacco plant were taken away from soil at the harvest time. When 1 mol of base ions were removed from the soil, the acid buffer capacity of the soil decreased by 1 mol (Dong et al., 2022). Third, to maintain the ion balance, when 1 mol of were leached from soil, the same amount of base ions would also be washed to maintain charge balance (Xu and Ji, 2001). Our results also showed that the AN contents were higher in MK and HK than that in LK (Figure 1 C and F). It might be attributed to the following reasons. First, K application reduced soil N (mainly N2O emissions) loss by reducing denitrification (Li et al., 2021). Second, tobacco roots were damaged by the excess H+ and Al3+ brought by MK and HK (Dai et al., 2021), which inhibited the N uptake of plants (Figure 2) and left more N in the soil. Additionally, MK and LK improved OM contents (Figure 1), mainly because the increasing Fe and Al oxides of soil, caused by low pH, could protect OM by adsorbing organic biomolecules (Berhe et al., 2012). This finding was in accordance with previous studies (Wen et al., 2019; Huang et al., 2020), which reported that long-term application of only N fertilizer or combined application of NPK fertilizer improved soil OM content by reducing soil pH.
In our study, dry matter weight and N and S accumulation were restrained in the soil treated with HK (Figure 2). The main reason was that excessive application of K2SO4 intensified soil acidification, and the roots were harmed by the abundant H+ and Al3+ (Yang et al., 2018; Dai et al., 2021).
In our study, the percentages of Proteobacteria, Gemmatimonadetes, Bacteroidetes, Patescibacteria phyla and Sphingomonas, Bacillus, Ellin6067 genera were significantly higher in HK than in LK (Figure 4). And except for Bacillus, most members of all these taxa are considered to be predicted copiotrophic bacteria (Xia et al., 2005; Spain et al., 2009; Albertsen et al., 2013; Mowlick et al., 2014; Naas et al., 2014; Brown et al., 2015; Banerjee et al., 2016), which grow fastly in nutrient-rich conditions (Fierer et al., 2007), such as Burkholderiaceae, Rhodospirillaceae and Rhizobiaceae families in Proteobacteria phylum (Table 3S; Finn et al., 2021). And all taxa (Acidobacteria, Chloroflexi phyla and uncultured_Acidobacteria_bacterium, Bryobacter genera) significantly reduced in HK were reported to be predicted oligotrophic bacteria (Sorokin et al., 2012; Dedysh et al., 2017; Kalam et al., 2020), which grow slowly and are able to metabolize nutrient poor and recalcitrant C substrates (Fierer et al., 2007). These shifts in community structure between HK and LK could be mainly ascribed to the more OM supplied by HK (Figure 1B). Interestingly, Gemmatimonadetes phyla, Bacillus and Sphingomonas genera enriched in the HK treatment were considered to be potential beneficial bacteria. Gemmatimonadetes facilitates P dissolution and suppress diseases (Zeng et al., 2020). Bacillus (belongs to Firmicutes phylum) has the abilities of resistance to pathogens, N fixation, dissolution of P and K (Radhakrishnan et al., 2017). Sphingomonas is reported to degrade recalcitrant compounds and fix N (Xu et al., 2018). It was mainly because soil acidification of HK increased harmful metals (e.g. Al3+ and Cd2+) (Dai et al., 2021) and pathogenic bacteria (e.g. bacterial wilt) (Shen et al., 2018), resulting in the improvement of resistant taxa. This was also a self-protection mechanism of microorganisms against environmental stress. These were in accordance with results of Chen et al. (2022), in which probiotics Bacteroidetes and Firmicutes phyla were higher in acid soil than in non-acid soil. Additionally, Acidobacteria played an important role in the degradation of various organic materials and in the biogeosmic cycling of C, H and Fe (Kalam et al., 2020). And Chloroflexi is associated with nitrification and degradation of cellulose and polysaccharide (Sorokin et al., 2012). Both two phyla involve in C and N metabolism and play a key role in microbial community formation and stability under adverse environmental conditions. In brief, compared with LK, HK favored the growth of predicted copiotrophic groups and beneficial groups involved in pathogens and heavy metal resistance and N fixation, dissolution of P and K, while had negative with some oligotrophic taxa related to C, N metabolism. And the specific functions of potential beneficial bacteria (Gemmatimonadetes phyla and Bacillus and Sphingomonas genera) under environmental stress in tobacco-planting soils need to be investigated in the further research.
Different functional pathways could lead to different physiological consequences. In contrast to LK, HK had higher abundances of functional pathways mainly involved in carbohydrate metabolism (L2), such as starch and sucrose metabolism, glycolysis/gluconeogenesis and the TCA cycle (Figure 7). All the three pathways can release a large amount of energy for microbial life activities (Wang et al., 2015; Zhang et al., 2018). The TCA cycle is also the final metabolic pathway and hub of the three major nutrients (sugars, lipids, and amino acids). Some intermediate products formed during the decomposition of sucrose and starch can also be used as raw materials for the synthesis of biological macromolecules such as lipids, proteins and nucleic acids (Wang et al., 2015). In our study, pathways of homologous recombination, DNA replication, base excision repair and aminoacyl-tRNA biosynthesis, belonging to genetic information processing (L1) were also up-regulated in HK compared to LK (Figure 7). Homologous recombination plays an important role in the processing, integration and transformation of genes and is an important factor in maintaining gene frequency and gene diversity (Wielgoss et al., 2016). DNA replication enables genetic information to be passed from parent to offspring, thus ensuring the continuity of genetic information (Aklilu et al., 2014). Base excision repair is an important DNA oxidative damage defense response (Bauer et al., 2015). The primary function of aminoacyl-tRNA biosynthesis is protein synthesis, but they also play a role in gene expression, cell wall formation, protein labeling for degradation, and antibiotic biogenesis (Ling et al., 2009). Additionally, only three pathways of valine, leucine and isoleucine biosynthesis, pantothenate and CoA biosynthesis and C5-branched dibasic acid metabolism were enhanced in the soil of LK. They belonged to amino acid metabolism, metabolism of cofactors and vitamin and carbohydrate metabolism, respectively (Figure 7). These results indicated that, compared with LK, the addition of HK could provide more energy, promote microbial metabolism, and improve the functions of gene replication, recombination and repair. Our study also showed that MK promoted the functional pathways of one carbon pool by folate, streptomycin biosynthesis, monobactam biosynthesis and homologous recombination compared to LK (Figure 7). Streptomycin and monobactam are both antibiotics that effectively prevent disease (Li et al., 2017; Westhoff et al., 2019). One pool of carbon by folate is involved in protein synthesis and cell division (Ducker and Rabinowitz, 2017). Homologous recombination plays an important role in DNA damage repair and mutation processes (Wielgoss et al., 2016). The pathways of glyoxylate and dicarboxylate metabolism, alanine, aspartate and glutamate metabolism, RNA degradation and pantothenate and CoA biosynthesis were depleted in MK (Figure 7), which were related to sugar, fat and amino acid metabolism and cell regulation (Hjorth et al., 2006; Rubio et al., 2008). It was indicated that MK had both positive and negative effects on soil functions. All in all, we speculate that these pathways in microbes may have implications for plant survival and acid tolerance to some extent, which need further research.
Previous studies reported that soil physicochemical properties played an important role in regulating microbial communities and functions (Lu et al., 2020; Kang et al., 2021). Our study showed that pH significantly shaped soil microbial community and functional composition. This result was consistent with many previous studies showing that pH was the most important indicator for determining bacterial composition, which was due to the relatively narrow growth tolerances of most bacterial taxa (Zhou et al., 2015). In this study, AK also significantly affected microbial community and KEGG pathways. The probable reason was that, excessive AK in the soil can affect the availability of other elements (e.g. Ca and N) (Nieder et al., 2011), thereby indirectly affecting the microbial community and function. Additionally, AS was another key factor in soil, which was significantly negatively correlated with multiple taxa and pathways (Figure 9). This was mainly attributable to damage to microorganisms caused by excess S in soil (Ma et al., 2020).
In summary, these results are broadly consistent with our hypothesis and advance our understanding of the impacts of K2SO4 application on the physicochemical and microbial properties of soils in typical tobacco fields, with implications for the scientific and reasonable application of K fertilizer in tobacco production. The common K2SO4 rate of 495 kg hm-2 in tobacco planting areas with Alfisols risks exacerbating soil acidification and adversely affecting plant growth. Considering yield, efficiency and environment, the optimal K2SO4 rate was 165-330 kg hm-2. Moreover, calcium magnesium phosphate fertilizer or organic fertilizer should also be used to prevent soil acidification.
The present data showed that, compared with LK, HK promoted the N, K, S and OM contents of soil, while increased soil acidification. Due to this change, addition of HK resulted in higher percentages of predicted copiotrophic groups and beneficial bacterium and lower percentages of some oligotrophic taxa. According to the changes of KEGG pathways, carbohydrate metabolism and genetic information processing might improve in soils treated with HK. Additionally, the responses of soil physicochemical properties, composition of microbial community and functions to MK were less sensitive than HK. These results provide critical information to support the rational application of K fertilization in tobacco planting. However, further studies are required to investigate the variations of soil basic cations, heavy metal element, pathogenic bacteria and functional genes (C, N, P and S) to comprehensively and deeply evaluate the effect of excessive application of K2SO4 on ecological environment of tobacco-planting soil.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA805374.
JD and WS conceived and designed the experiments. SK carried out the experiments. LT analyzed the data. YLi finalized the figures and tables. YLu and PC wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was financially supported by the Central Public-interest Scientific Institution Basal Research Fund (1610232022007), the Natural Science Foundation of Shandong Province (ZR2021QD036), Qingdao postdoctoral Foundation (2021).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1005303/full#supplementary-material
Aklilu, B. B., Soderquist, R. S., Culligan, K. M. (2014). Genetic analysis of the replication protein a large subunit family in arabidopsis reveals unique and overlapping roles in DNA repair, meiosis and DNA replication. Nucleic Acids Res. 42, 3104–3118. doi: 10.1093/nar/gkt1292
Albertsen, M., Hugenholtz, P., Skarshewski, A., Nielsen, K. L., Tyson, G. W., Nielsen, P. H. (2013). Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat. Biotechnol. 31, 533–538. doi: 10.1038/nbt.2579
Bai, Y., Chang, Y., Hussain, M., Lu, B., Zhang, J., Song, X., et al. (2020). Soil chemical and microbiological properties are changed by long-term chemical fertilizers that limit ecosystem functioning. Microorganisms 8, 694. doi: 10.3390/microorganisms8050694
Banerjee, S., Baah-Acheamfour, M., Carlyle, C. N., Bissett, A., Richardson, A. E., Siddique, T., et al. (2016). Determinants of bacterial communities in Canadian agroforestry systems. Environ. Microbiol. 18, 1805–1816. doi: 10.1111/1462-2920.12986
Bauer, N. C., Corbett, A. H., Doetsch, P. W. (2015). The current state of eukaryotic DNA base damage and repair. Nucleic Acids Res. 21, 10083–10101. doi: 10.1093/nar/gkv1136
Berhe, A. A., Suttle, K. B., Burton, S. D., Banfield, J. F. (2012). Contingency in the direction and mechanics of soil organic matter responses to increased rainfall. Plant Soil. 358, 371–383. doi: 10.1007/s11104-012-1156-0
Brown, C. T., Hug, L. A., Thomas, B. C., Sharon, I., Castelle, C. J., Singh, A., et al. (2015). Unusual biology across a group comprising more than 15% of domain bacteria. Nature 523, 208–211. doi: 10.1038/nature14486
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Huntley, J., Fierer, N., et al (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6, 1621–1624. doi: 10.1038/ismej.2012.8
Chen, H., Ren, H. Y., Liu, J. J., Tian, Y., Lu, S. G. (2022). Soil acidification induced decline disease of myrica rubra: aluminum toxicity and bacterial community response analyses. Environ. Sci. Pollut. R. 29, 45435–45448. doi: 10.1007/s11356-022-19165-3
Chen, S., Zhou, Y., Chen, Y., Gu, J. (2018). Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Cole, J. R., Wang, Q., Cardenas, E., Fish, J., Chai, B., Farris, R. J., et al. (2009). The ribosomal database project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37, D141–D145. doi: 10.1093/nar/gkn879
Csardi, G., Nepusz, T. (2006). The igraph software package for complex network research. Int. J. Complex Syst. 1695, 1–9.
Dai, P., Cong, P., Wang, P., Dong, J., Dong, Z., Song, W. (2021). Alleviating soil acidification and increasing the organic carbon pool by long-term organic fertilizer on tobacco planting soil. Agron. J. 11, 2135. doi: 10.3390/agronomy11112135
Dedysh, S. N., Kulichevskaya, I. S., Huber, K. J., Overmann, J. (2017). Defining the taxonomic status of described subdivision 3 Acidobacteria: proposal of Bryobacteraceae fam. nov. Int. J. Syst. Evol. Micr. 67, 498–501. doi: 10.1099/ijsem.0.001687
Ding, M., Wei, B., Zhang, Z., She, S., Huang, L., Ge, S., et al. (2017). Effect of potassium organic and inorganic salts on thermal decomposition of reconstituted tobacco sheet. J. Therm. Anal. Calorim. 129, 975–984. doi: 10.1007/s10973-017-6214-7
Dong, Y., Yang, J., Zhao, X., Yang, S., Mulder, J., Dörsch, P., et al. (2022). Soil acidification and loss of base cations in a subtropical agricultural watershed. Sci. Total Environ. 827, 154338. doi: 10.1016/j.scitotenv.2022.154338
Ducker, G. S., Rabinowitz, J. D. (2017). One-carbon metabolism in health and disease. Cell. Metab. 25, 27–42. doi: 10.1016/j.cmet.2016.08.009
Edgar, R. C. (2013). PARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Fierer, N., Bradford, M. A., Jackson, R. B. (2007). Toward an ecological classification of soil bacteria. Ecology 88, 1354–1364. doi: 10.1890/05-1839
Finn, D. R., Bergk-Pinto, B., Hazard, C., Nicol, G. W., Tebbe, C. C., Vogel, T. M. (2021). Functional trait relationships demonstrate life strategies in terrestrial prokaryotes. FEMS Microbiol. Ecol. 97, finb068. doi: 10.1093/femsec/fiab068
Henry, J. B., Vann, M. C., Lewis, R. S. (2019). Agronomic practices affecting nicotine concentration in flue-cured tobacco: A review. Agron. J. 111, 3067–3075. doi: 10.2134/agronj2019.04.0268
Hjorth, M., Mathiassen, S. K., Kudsk, P., Ravn, H. W. (2006). Amino acids in loose silky-bent (Apera spica-venti (L.) Beauv.) responding to prosulfocarb exposure and the correlation with physiological effects. Pestic. Biochem. Phys. 86, 138–145. doi: 10.1016/j.pestbp.2006.02.006
Huang, X., Kang, W., Guo, J., Wang, L., Tang, H., Li, T., et al. (2020). Highly reactive nanomineral assembly in soil colloids: Implications for paddy soil carbon storage. Sci. Total. Environ. 703, 134728. doi: 10.1016/j.scitotenv.2019.134728
Hu, W., Di, Q., Wang, Z., Zhang, Y., Zhang, J., Liu, J., et al. (2019). Grafting alleviates potassium stress and improves growth in tobacco. BMC. Plant Biol. 19, 130. doi: 10.1186/s12870-019-1706-1
Jin, X., Bai, Y., Rahman, M. K. U., Kang, X., Pan, K., Wu, F., et al. (2022b). Biochar stimulates tomato roots to recruit a bacterial assemblage contributing to disease resistance against fusarium wilt. iMeta 1, e37. doi: 10.1002/imt2.37
Jin, X., Wang, Z. L., Wu, F. Z., Li, X. G., Zhou, X. G. (2022a). Litter mixing alters microbial decomposer community to accelerate tomato root litter decomposition. Microbiol. Spectr. 10, e00186-22. doi: 10.1128/spectrum.00186-22
Kalam, S., Basu, A., Ahmad, I., Sayyed, R. Z., El-Enshasy, H. A., Dailin, D. J., et al. (2020). Recent understanding of soil Acidobacteria and their ecological significance: A critical review. Front. Microbiol. 11, 580024. doi: 10.3389/fmicb.2020.580024
Kang, H., Yu, W., Dutta, S., Gao, H. (2021). Soil microbial community composition and function are closely associated with soil organic matter chemistry along a latitudinal gradient. Geoderma 383, 114744. doi: 10.1016/j.geoderma.2020.114744
Li, D., Liu, C., Luo, R., Sadakane, K., Lam, T. (2015). MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de bruijn graph. Bioinformatics 31, 1674–1676. doi: 10.1093/bioinformatics/btv033
Li, Z., Li, L., Xia, S., Zhang, R., Zhang, R., Chen, P., et al. (2021). K Fertilizer alleviates N2O emissions by regulating the abundance of nitrifying and denitrifying microbial communities in the soil-plant system. J. Environ. Manage. 291, 112579. doi: 10.1016/j.jenvman.2021.112579
Ling, J., Reynolds, N., Ibba, M. (2009). Aminoacyl-tRNA synthesis and translational quality control. Annu. Rev. Microbiol. 63, 61–78. doi: 10.1146/annurev.micro.091208.073210
Li, R., Oliver, R. A., Townsend, C. A. (2017). Identification and characterization of the sulfazecin Monobactam biosynthetic gene cluster. Cell. Chem. Biol. 24, 24–34. doi: 10.1016/j.chembiol.2016.11.010
Liu, J., Wang, Q. Q., Ku, Y. L, Zhang, W. W, Zhu, H. L., Zhao, Z. (2022). Precipitation and soil pH drive the soil microbial spatial patterns in the Robinia pseudoacacia forests at the regional scale. Catena 212, 106120. doi: 10.1016/j.catena.2022.106120
Liu, Z., Zhuang, Z., Yu, Y., Wang, Q., Wan, Y., Li, H. (2021). Arsenic transfer and accumulation in the soil-rice system with sulfur application and different water managements. Chemosphere 269, 128772. doi: 10.1016/j.chemosphere.2020.128772
Li, X., Yu, H., Sun, X., Yang, J., Wang, D., Shen, L., et al. (2019). Effects of sulfur application on cadmium bioaccumulation in tobacco and its possible mechanisms of rhizospheric microorganisms. J. Hazard. Mater. 368, 308–315. doi: 10.1016/j.jhazmat.2018.12.099
Lu, H., Yan, M. X., Wong, M. H., Mo, W. Y., Wang, Y. H., Chen, X. W., et al. (2020). Effects of biochar on soil microbial community and functional genes of a landfill cover three years after ecological restoration. Sci. Total Environ. 717, 137133. doi: 10.1016/j.scitotenv.2020.137133
Ma, J., Quan, Z., Sun, Y., Du, J., Liu, B. (2020). Excess sulfur and fe elements drive changes in soil and vegetation at abandoned coal gangues, guizhou China. Sci. Rep-UK. 10, 10456. doi: 10.1038/s41598-020-67311-z
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. Embnet J. 17, 1. doi: 10.14806/ej.17.1.200
Mowlick, S., Inoue, T., Takehara, T., Tonouchi, A., Kaku, N., Ueki, K., et al. (2014). Usefulness of Japanese-radish residue in biological soil disinfestation to suppress spinach wilt disease accompanying with proliferation of soil bacteria in the firmicutes. Crop Prot. 61, 64–73. doi: 10.1016/j.cropro.2014.03.010
Naas, A. E., Mackenzie, A. K., Mravec, J., Schückel, J., Willats, W. G. T., Eijsink, V. G. H., et al. (2014). Do RumenBacteroidetes utilize an alternative mechanism for cellulose degradation? mBio 5, e01401–e01414. doi: 10.1128/mBio.01401-14
Nieder, R., Benbi, D. K., Scherer, H. W. (2011). Fixation and defixation of ammonium in soils: a review. Biol. Fert. Soils 47, 1–14. doi: 10.1007/s00374-010-0506-4
Patro, R., Duggal, G., Kingsford, C. (2015). Salmon: accurate, versatile and ultrafast quantification from RNA-seq data using lightweight-alignment. BioRxiv 021592. doi: 10.1101/021592
Radhakrishnan, R., Hashem, A., Abd Allah, E. F. (2017). Bacillus: A biological tool for crop improvement through bio-molecular changes in adverse environments. Front. Physiol. 8, 667. doi: 10.3389/fphys.2017.00667
Rath, K. M., Fierer, N., Murphy, D. V., Rousk, J. (2019). Linking bacterial community composition to soil salinity along environmental gradients. ISME J. 13, 836–846. doi: 10.1038/s41396-018-0313-8
Rubio, S., Whitehead, L., Larson, T. R., Graham, I. A., Rodriguez, P. L. (2008). The coenzyme a biosynthetic enzyme phosphopantetheine adenylyltransferase plays a crucial role in plant growth, salt/osmotic stress resistance, and seed lipid storage. Plant Physiol. 148, 546–556. doi: 10.1104/pp.108.124057
Schloss, P. D., Gevers, D., Westcott, S. L. (2011). Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS. One 6, e27310. doi: 10.1371/journal.pone.0027310
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12, R60. doi: 10.1186/gb-2011-12-6-r60
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
She, S., Niu, J., Zhang, C., Xiao, Y., Chen, W., Dai, L., et al. (2017). Significant relationship between soil bacterial community structure and incidence of bacterial wilt disease under continuous cropping system. Arch. Microbiol. 199, 267–275. doi: 10.1007/s00203-016-1301-x
Shen, M. C., Zhang, Y. Z., Bo, G. D., Yang, B., Wang, P., Ding, Z. Y., et al. (2022). Microbial responses to the reduction of chemical fertilizers in the rhizosphere soil of flue-cured tobacco. Front. Bioeng Biotech. 9, 812316. doi: 10.3389/fbioe.2021.812316
Shen, G., Zhang, S., Liu, X., Jiang, Q., Ding, W. (2018). Soil acidification amendments change the rhizosphere bacterial community of tobacco in a bacterial wilt affected field. Appl. Microbiol. Biot. 102, 9781–9791. doi: 10.1007/s00253-018-9347-0
Sorokin, D. Y., Lucker, S., Vejmelkova, D., Kostrikina, N. A., Kleerebezem., R., Rijpstra, W. I., et al. (2012). Nitrification expanded: discovery, physiology and genomics of a nitrite-oxidizing bacterium from the phylum Chloroflexi. ISME. J 6, 2245–2256. doi: 10.1038/ismej.2012.70
Spain, A. M., Krumholz, L. R., Elshahed, M. S. (2009). Abundance, composition, diversity and novelty of soil Proteobacteria. ISME J. 3, 992–1000. doi: 10.1038/ismej.2009.43
Steinegger, M., Soding, J. (2017). MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat. Biotechnol. 35, 1026–1028. doi: 10.1038/nbt.3988
Tang, X., Li, L., Wu, C., Khan, M. I., Manzoor, M., Zou, L., et al. (2020). The response of arsenic bioavailability and microbial community in paddy soil with the application of sulfur fertilizers. Environ. Pollut. 264, 114679. doi: 10.1016/j.envpol.2020.114679
Wallace, A. (1994). Soil acidification from use of too much fertilizer. Commun. Soil. Sci. Plan. 25, 87–92. doi: 10.1080/00103629409369010
Wang, W., Luo, X., Chen, Y., Ye, X., Wang, H., Cao, Z., et al. (2019). Succession of composition and function of soil bacterial communities during key rice growth stages. Front. Microbiol. 10, 421. doi: 10.3389/fmicb.2019.00421
Wang, M., Wang, L., Shi, H., Liu, Y., Chen, S. (2021). Soil bacteria, genes, and metabolites stimulated during sulfur cycling and cadmium mobilization under sodium sulfate stress. Environ. Res. 201, 111599. doi: 10.1016/j.envres.2021.111599
Wang, Z., Xu, Y., Chen, T., Zhang, H., Yang, J., Zhang, J. (2015). Abscisic acid and the key enzymes and genes in sucrose-to-starch conversion in rice spikelets in response to soil drying during grain filling. Planta 241, 1091–1107. doi: 10.1007/s00425-015-2245-0
Wang, Z. B., Yang, Y., Xia, Y. Z., Wu, T., Zhu, J., Yang, J. M., et al. (2019). Time-course relationship between environmental factors and microbial diversity in tobacco soil. Sci. Rep-UK. 9, 19969. doi: 10.1038/s41598-019-55859-4
Wang, M., Zhang, L., He, Y., Huang, L., Liu, L., Chen, D., et al. (2022). Soil fungal communities affect the chemical quality of flue-cured tobacco leaves in bijie, southwest China. Sci. Rep-UK. 22, 2815. doi: 10.1038/s41598-022-06593-x
Wang, C., Zhou, X., Guo, D., Zhao, J., Yan, L., Feng, G., et al. (2019). Soil pH is the primary factor driving the distribution and function of microorganisms in farmland soils in northeastern China. Ann. Microbiol. 69, 1461–1473. doi: 10.1007/s13213-019-01529-9
Wan, W., Tan, J., Wang, Y., Qin, Y., He, H., Wu, H., et al. (2020). Responses of the rhizosphere bacterial community in acidic crop soil to pH: Changes in diversity, composition, interaction, and function. Sci. Total. Environ. 700, 134418. doi: 10.1016/j.scitotenv.2019.134418
Wen, Y., Liu, W., Deng, W., He, X., Yu, G. (2019). Impact of agricultural fertilization practices on organo-mineral associations in four long-term field experiments: Implications for soil c sequestration. Sci. Total Environ. 651, 591–600. doi: 10.1016/j.scitotenv.2018.09.233
Westhoff, S., Otto, S. B., Swinkels, A., Bode, B., Wezel, G. P., Rozen, D. E. (2019). Spatial structure increases the benefits of antibiotic production in Streptomyces griseus. bioRxiv 74, 179–187. doi: 10.1101/482786
Wielgoss, S., Didelot, X., Chaudhuri, R. R., Liu, X., Weedall, G. D., Velicer, G. J., et al. (2016). A barrier to homologous recombination between sympatric strains of the cooperative soil bacterium Myxococcus xanthus. ISME J. 10, 2468–2477. doi: 10.1038/ismej.2016.34
Xia, S., Shi, Y., Fu, Y., Ma, X. (2005). DGGE analysis of 16S rDNA of ammonia-oxidizing bacteria in chemical-biological flocculation and chemical coagulation systems. Appl. Microbiol. Biot. 69, 99–105. doi: 10.1007/s00253-005-0035-5
Xin, L., Li, X., Tan, M. (2012). Temporal and regional variations of china’s fertilizer consumption by crops during 1998-2008. J. Geogr. Sci. 22, 643–652. doi: 10.1007/s11442-012-0953-y
Xu, R. K., Ji, G. L. (2001). Effect of H2SO4 and HNO3 on soil acidification and alumunum speciation in variable and constant charge soils. Water Air Soil Pollut. 129, 33–43. doi: 10.1023/A:1010315011341
Xu, J., Kloepper, J. W., Huang, P., McInroy, J. A., Hu, C. H. (2018). Isolation and characterization of N2-fixing bacteria from giant reed and switchgrass for plant growth promotion and nutrient uptake. J. Basic. Microb. 58, 459–471. doi: 10.1002/jobm.201700535
Yang, X., Ni, K., Shi, Y., Yi, X., Ji, L., Ma, L., et al. (2020). Heavy nitrogen application increases soil nitrification through ammonia-oxidizing bacteria rather than archaea in acidic tea (Camellia sinensis l.) plantation soil. Sci. Total Environ. 717, 137248. doi: 10.1016/j.scitotenv.2020.137248
Yang, X., Ni, K., Shi, Y., Yi, X., Zhang, Q., Fang, L., et al. (2018). Effects of long-term nitrogen application on soil acidification and solution chemistry of a tea plantation in China. Agr. Ecosyst. Environ. 252, 74–82. doi: 10.1016/j.agee.2017.10.004
Zeng, Q., An, S., Liu, Y., Wang, H., Wang, Y. (2019). Biogeography and the driving factors affecting forest soil bacteria in an arid area. Sci. Total Environ. 680, 124–131. doi: 10.1016/j.scitotenv.2019.04.184
Zeng, Y. H., Nupur, W., Madsen, A. M., Chen, X. H., Gardiner, A. T., Koblížek, M. (2020). Gemmatimonas groenlandica sp. nov. is an aerobic anoxygenic phototroph in the phylum Gemmatimonadetes. Front. Microbiol. 11, 606612. doi: 10.3389/fmicb.2020.606612
Zhang, C., Kong, F. (2014). Isolation and identification of potassium-solubilizing bacteria from tobacco rhizospheric soil and their effect on tobacco plants. Appl. Soil. Ecol. 82, 18–25. doi: 10.1016/j.apsoil.2014.05.002
Zhang, Y., Swart, C., Alseekh, S., Scossa, F., Jiang, L., Obata, T., et al. (2018). The extra-pathway interactome of the TCA cycle: expected and unexpected metabolic interactions. Plant Physiol. 177, 966–979. doi: 10.1104/pp.17.01687
Zheng, J. Y., Zhang, J. X., Gao, L., Wang, R., Gao, J., Dai, Y., et al. (2021). Effect of straw biochar amendment on tobacco growth, soil properties, and rhizosphere bacterial communities. Sci. Rep-UK. 11, 20727. doi: 10.1038/s41598-021-00168-y
Zhong, W., Gu, T., Wang, W., Zhang, B., Lin, X., Huang, Q., et al. (2010). The effects of mineral fertilizer and organic manure on soil microbial community and diversity. Plant Soil. 326, 511–522. doi: 10.1007/s11104-009-9988-y
Zhou, J., Guan, D., Zhou, B., Zhao, B., Ma, M., Qin, J., et al. (2015). Influence of 34-years of fertilization on bacterial communities in an intensively cultivated black soil in northeast China. Soil. Biol. Biochem. 90, 42–51. doi: 10.1016/j.soilbio.2015.07.005
Keywords: tobacco-planting soil, K2SO4 fertilizer, soil physicochemical properties, bacterial community, pathway
Citation: Lu Y, Cong P, Kuang S, Tang L, Li Y, Dong J and Song W (2022) Long-term excessive application of K2SO4 fertilizer alters bacterial community and functional pathway of tobacco-planting soil. Front. Plant Sci. 13:1005303. doi: 10.3389/fpls.2022.1005303
Received: 28 July 2022; Accepted: 05 September 2022;
Published: 28 September 2022.
Edited by:
Xingang Zhou, Northeast Agricultural University, ChinaReviewed by:
Henry Joseph Oduor Ogola, Jaramogi Oginga Odinga University of Science and Technology, KenyaCopyright © 2022 Lu, Cong, Kuang, Tang, Li, Dong and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianxin Dong, ZG9uZ2ppYW54aW5AY2Fhcy5jbg==; Wenjing Song, c29uZ3dlbmppbmdAY2Fhcy5jbg==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.