- 1College of Life Sciences, Jilin Agricultural University, Changchun, China
- 2Joint International Research Laboratory of Modern Agricultural Technology, Ministry of Education, Changchun, China
- 3College of Agronomy, Jilin Agricultural University, Changchun, China
Maize (Zea mays L.) is an annual gramineous herb and is among the world’s most important crop species. Drought is the main factor contributing to maize yield reduction. The lateral organ boundaries domain (LBD) proteins belong to a class of higher-plant-specific transcription factors. LBD proteins usually include the highly conserved lateral organ boundaries (LOB) domains that play essential roles in plant growth and response to biotic stresses. However, few studies have addressed the biological functions of LBD genes associated with maize response to drought. Here we cloned the ZmLBD2 gene from maize and described its role in combating drought. Investigating ZmLBD2 subcellular localization, we show that it localizes to the cell nucleus and can specifically bind with inverted repeats of “GCGGCG”. Under drought stress, Arabidopsis thaliana overexpressing ZmLBD2 performed better than the wild-type plants in terms of seed germination rates, root length, relative water content, fresh weight, chlorophyll content, proline content, and antioxidant enzyme content. Arabidopsis overexpressing ZmLBD2 contained less MDA, H2O2, and than the wild-type plants. Our protein-protein interaction results indicate an interaction between the ZmLBD2 and ZmIAA5 genes. In conclusion, the ZmLBD2 gene positively regulates H2O2 homeostasis in plants, strengthening drought resistance.
Introduction
Maize (Zea mays L.) is the world’s number one food crop, with strong adaptability and wide cultivation areas across the globe. Drought stress is one of the common factors reducing crop yield. Agricultural economic losses increase year on year as drought rages worldwide. In recent years, a series of genes crucial to combat drought stress has been described in plants (Sahebi et al., 2018; Liu et al., 2020; Wei et al., 2021; Chen et al., 2021; Wang B. et al., 2022). Reactive oxygen species (ROS), mainly including superoxide anion radicals (), hydroxyl ions (OH-), hydroxyl radicals (·OH), and hydrogen peroxide (H2O2), accumulate under drought stress and work in various ways to influence the growth of plants and their responses to environmental stresses (Wang et al., 2018; Xiong et al., 2022). Yuan et al. (2019) found that the overexpression of the NAC066 gene could significantly enhance the tolerance of rice (Oryza sativa L.) against drought and oxidative stress by reducing the accumulation of ROS. Hu et al. (2020) reported that TaCDPK13 could be considered the upstream regulatory factor of TaNOX7 to regulate the generation of ROS in wheat (Triticum aestivum L.). Xiong et al. (2018) reported that the natural mutation of the OsLG3 gene strengthened the resistance of rice against drought by inducing the removal of ROS.
The lateral organ boundaries domain (LBD) proteins belong to a class of plant-specific transcription factors, including the conserved lateral organ boundaries (LOB) domain. This domain consists of the highly conserved C motif (C-block), Gly-Ala-Ser motif (GAS-block), and leucine zipper-like motif (leucine zipper-like block). Most LBD family members attribute to the class I, which is mainly responsible for several biological reactions in higher plants, such as growth and responses to adversities (drought, salt, cold, etc.).There are still many unanswered questions about the LBD gene. For example, does the LBD gene function in other species of plants, and what function(s) does it play? Xiong et al. (2022) found that the ZmLBD5 of maize could increase the tolerance of Arabidopsis thaliana against drought by inhibiting the accumulation of ROS. Liu et al. (2020) discovered that the knockout of the SlLBD40 gene could promote the drought tolerance of tomatoes (Solanum lycopersicum L.). Liu et al. (2019) illustrated that the overexpression of StLBD2-6 and StLBD3-5 genes might help maintain a normal metabolism for potato (Solanum tuberosum L.) to improve its ability to resist drought. Research published by Guo et al. (2020) stated that drought stress induced the ABA(abscisic acid) signal to promote the expression of the LBD15 gene in A. thaliana, which enhanced its tolerance against water stress. Ariel et al. (2010) found that overexpression of Mt LBD1 could control the structure of Medicago truncatula roots under salt stress. Wang Z. et al. (2021) found that the lateral organ boundaries domain (LBD) genes, a plant-specific transcription factor family, play crucial roles in controlling plant architecture and stress tolerance. Wang J. et al. (2021) found that RrLBD12c, RrLBD25, RrLBD39, and RrLBD40 of Rosa may be potential regulators of salt stress signaling. Ba et al. (2016) reported that MaLBD5 and MaJAZ1 might act antagonistically concerning MeJA-induced cold tolerance of banana fruit.
In previous studies, 44 LBD transcription factors have been identified in the maize genome (Zhang et al., 2014). However, only the ZmLBD5 gene (Zm00001d032286) has been functionally verified regarding the response to drought. The functions of other ZmLBD genes remain unclear. In this study, we analyzed how the ZmLBD2 gene responded to drought. The overexpression of ZmLBD2 in the transgenic A. thaliana could improve its drought tolerance by reducing the accumulation of ROS. Our results show that ZmLBD2 can mediate the response of maize seedlings to drought by regulating the homeostasis of hydrogen peroxide, which may positively affect plants’ drought tolerance.
Materials and methods
Plant materials and growth conditions
The wild-type (ecotype: Col-0) and transgenic A. thaliana (OE2 and OE3) seeds were disinfected with 75% alcohol for 1 min. After surface sterilization with 1% NaClO3 for 10 min, they were washed four times with sterile water. Following the disinfection treatment, all seeds were cultivated in 1/2 Murashige and Skoog (MS) media at 4°C in the dark for 3 days. Next, the culture media were moved to a plant incubator (at 22°C with a 16 h light/8 h dark cycle) for the seeds to grow. Two weeks later, the seedlings were transplanted into the soil and grown in a phytotron (at 22°C with a 16 h light/8 h dark cycle). They were treated with 8% PEG6000 (polyethylene glycol 6000) at the three-leaf stage, and the physiological data were measured. We collected their leaves at 0, 6, 12, 24, and 48 h and froze them in liquid nitrogen immediately after the collection to extract their RNA.
Gene isolation and sequence analysis
The maize LBD2 gene was obtained from MaizeGDB (https://www.maizegdb.org/) (accessed on 2 June 2022). Homologous sequences of ZmLBD2 were retrieved from the Phytozome database (https://phytozome-next.jgi.doe.gov/) (accessed on 2 June 2022). Amino and multiple sequence alignments were constructed with ClustalX. Our laboratory completed the transcriptome data of maize under different drought treatment times, and the transcriptome data has been uploaded to the NCBI (National Center for Biotechnology Information) database (PRJNA793522).
The subcellular localization of ZmLBD2
To explore the possible subcellular location of ZmLBD2, the full-length gene coding region of ZmLBD2 was obtained from the TA cloning vector containing the ZmLBD2 gene fragment. The amplified product was then fused to the CaMV35S promoter and inserted upstream of the coding region of the pCAMBIA1302-eGFP subcellular localization vector. The constructed vector pCAMBIA1302-ZmLBD2-eGFP was introduced into tobacco leaves using agrobacterium-mediated methods. The leaves were then examined at 45 h after infiltration using an Olympus confocal laser scanning microscope (DAPY Olympus, Tokyo, Japan). The primers used here are listed in Supplementary Table S1.
Induced expression of ZmLBD2 protein and detection by Western blotting
Full-length cDNA of ZmLBD2 was amplified and inserted into a pET28a vector driven by a Cauliflower Mosaic Virus 35S (CaMV35S) promoter. The constructed vector pET28a-ZmLBD2-His was transformed into BL21 E. coli competent cells. Then, the protein expression of the positive recombinant plasmid was performed under isopropyl β-D-thiogalactoside (IPTG) induction conditions, and finally, the protein content was detected by SDS-PAGE (polyacrylamide gel electrophoresis) electrophoresis and Western blotting.
Analysis of DNA binding characteristics of ZmLBD2
First, the cDNA of the target gene (ZmLBD2) was constructed on the pGADT7 empty vector to form the recombinant vector pGAD-ZmLBD2. Three tandem repeats of GCGGCG were synthesized by artificial synthesis to construct the yeast monohybrid reporter vector pHIS2-TCS. To verify the specificity of ZmLBD2 for this target recognition, we synthesized three tandem repeated mutant targets, GAGGAG. EcoR I and Sac I restriction sites was added at both ends to construct the yeast monohybrid reporter vector pHIS2-mTCS. Finally, the binding site of the ZmLBD2 gene was verified by yeast one-hybrid (the yeast one-hybrid strain used in this study was Y187).
Analysis of the drought resistance of the ZmLBD2 gene in yeast
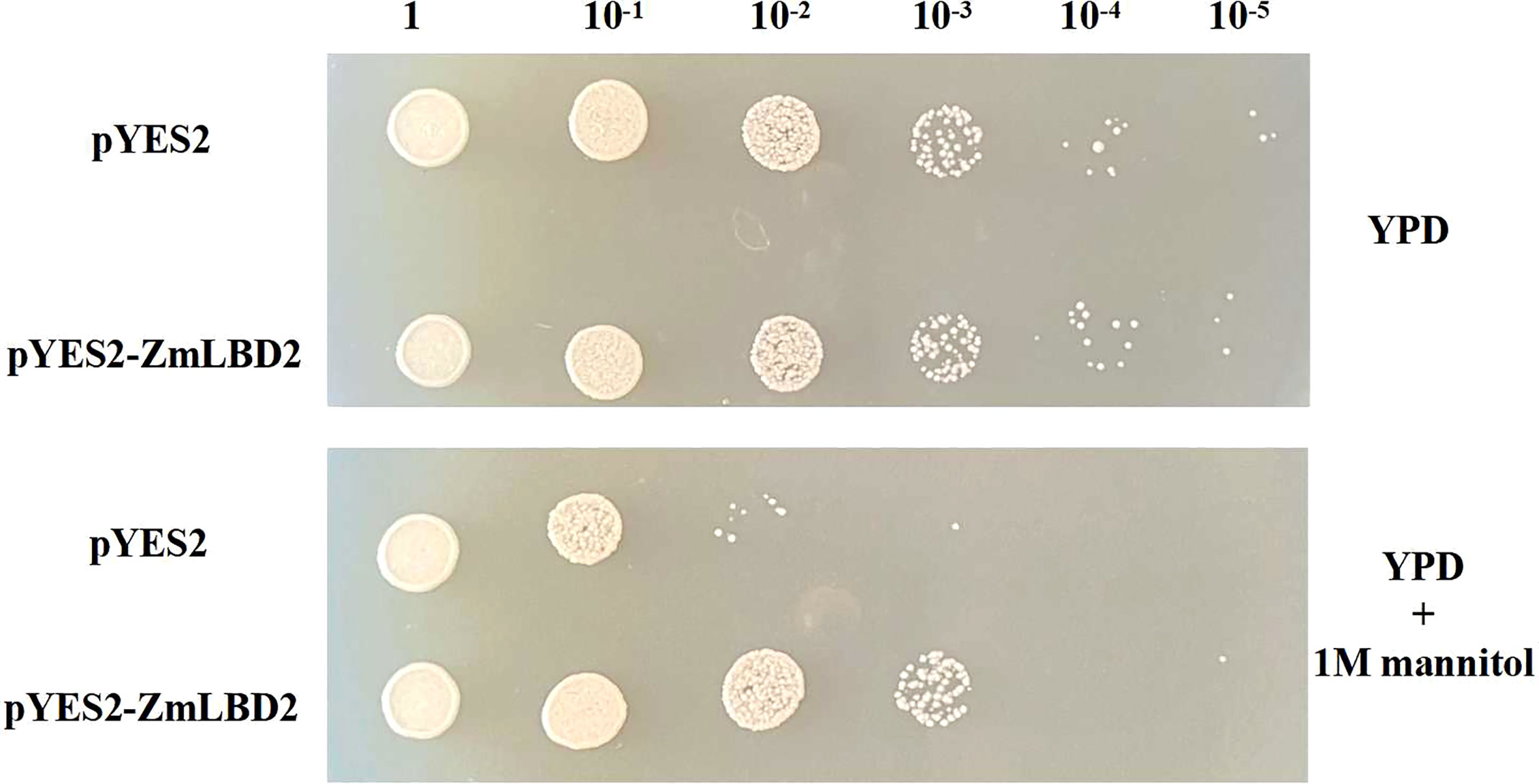
We designed the yeast plasmid vectors pYES2-ZmLBD2 and transformed them into competent INVSc1 yeast strains by the yeast transformation method. We cultivated the transformed strain in YPD (Yeast Extract Peptone Dextrose) liquid media. When the OD600 (optical density 600) concentration reached 0.8 after shake culture, we collected the strain bodies by centrifugation and dissolved them with physiological saline. After diluting the solution into six gradients, we added 5uL of each gradient into YPD culture media with and without mannitol.
RNA extraction and quantitative RT-qPCR analysis
TRIzol reagent (TAKARA BIO INC. Dalian, China) was used to extract total RNA from the maize (maize inbred lines H8186) treated under different stresses and from the A. thaliana seedlings. After being identified, the RNA was used to generate cDNA with TaKaRa reverse transcription kit. And SYBR GreenFastqPCRMix kit (ABclonal Biotechnology Co., Ltd, RM21203) was used for qPCR amplification. The reaction procedure was as follows: denaturation at 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 10s, at 55°C for the 40s, at 72°C for 20s, and 72°C for 4 min. Zm18S and AtACTIN8 were selected as the internal reference genes for maize and A. thaliana, respectively. The 2−ΔΔCT method was used to analyze the relative expression levels of relevant genes (Jiao et al., 2022; Wang C. et al., 2022). All primers used in this study are listed in Supplementary Table S1.
The genetic transformation of A. thaliana and phenotypic analysis
In this study, we constructed the plant overexpression vectors pCAMBIA3301-ZmLBD2-bar and transformed the recombinant plasmids into Agrobacterium tumefaciens strain GV3101; and then we infected A. thaliana with the strain by floral dip method (Noman et al., 2019). We cultivated the A. thaliana in 1/2 Murashige and Skoog (MS) media, screened out the transgenic plants of T0 generation and collected their leaves for PCR and test strip assay. Our research used the seeds of representative homozygous A. thaliana OE2 and OE3 of T3 generation (Transgenic generation 3).
To examine the responses of the transgenic and wild A. thaliana to drought, we disinfected both the seeds of the wild type and the transgenic one with the ZmLBD2 gene (OE2 and OE3) with 75% alcohol for 1 min. After conducting surface sterilization with 1% NaClO3 for 10 min, we washed the seeds four times with sterile water. We cultivated the seeds in 1/2MS media and performed drought experiments by adding 200 and 300 mM mannitol. After cultivation at 4°C for 3 days, they were moved to another environment and grew at 24°C and 80% relative humidity with a 16 h light/8 h dark cycle for 5 days. Then, we determined their germination rates and root length. The main root length was measured using the DJ-GXG02 root image analysis system (DianJiang Technology; http://www.dianjiangtech.com/). Each sample contains three biological replicates. One-way ANOVA method was used for statistical analysis. To identify the survival rates, we transplanted the A. thaliana seedlings, which sprouted in the 1/2MS media into the soil, and cultivated them at 22°C and 50% relative humidity with a 16 h light/8 h dark cycle for 3 weeks. After that, we stopped watering them for 14 days and took some pictures. Fourteen days later, we re-watered them for 3 days and calculated their survival rates (Ju et al., 2020).
Tetranitroblue tetrazolium chloride (NBT) staining, diaminobenzidine (DAB) staining, and oxidative stress analyses
Tetranitroblue tetrazolium chloride (NBT) and diaminobenzidine (DAB) were used to stain the seedling leaves of the wild-type and transgenic A. thaliana (OE2 and OE3) after drought treatment to examine the accumulation of superoxide anion () and Hydrogen peroxide (H2O2) (Zhao J. Y. et al., 2020). The spectrophotography was used to test the activity of catalase (CAT), superoxide dismutase (SOD), malondialdehyde (MDA), and peroxidase (POD) (Zhao Q. et al., 2020; Ju et al., 2020; Liu et al., 2021; Li et al., 2021). The weighing method was used to determine relative water content (Sapes and Sala, 2021). The methods mentioned above were used to evaluate the concentration of hydrogen peroxide and the amount of .
Yeast two-hybrid system
The CDS of the ZmLBD2 gene was cloned into the c-terminus of the GAL4 DNA-binding domain of the pGBKT7 vector. We used the recombinant plasmid ZmLBD2-BD as a decoy to search for every potential interaction between the encoded proteins on the STRING database (https://string-db.org/), and we cloned the CDS of interactive candidate gene ZmIAA5 into the pGADT7 vector. The recombinant plasmids ZmLBD2-BD and ZmIAA5-AD were then transformed into yeast cells AH109 using the lithium acetate method (Xu et al., 2007). The transformation efficiency was examined on DDO culture medium SD (synthetic-defined)/-Trp/-Leu, and the protein-protein interaction was verified on QDO medium SD/-Leu/-Trp/-His/-Ade. pGADT7-T and pGBKT7-53 were used as positive controls, and pGADT7-T and pGBKT7-lam were used as negative controls. The yeast transformation procedure was completed concerning the yeast transformation system produced by Beijing Kulaibo Technology Co., Ltd.
Bimolecular fluorescence complementation
The ORFs (Open Reading Frames) of ZmLBD2 and ZmIAA5 were cloned to the n-terminus and c-terminus of the coding regions of yellow fluorescent proteins (YFP) to construct plasmids nYFP-ZmLBD2 and cYFP-ZmIAA5. Five microliters of each of the two recombinant plasmids were co-transformed into tobacco mesophyll cells by agroinfiltration (Yu et al., 2021). The leaves were then examined at 45 h after infiltration using an Olympus confocal laser scanning microscope (DAPY Olympus, Tokyo, Japan).
Pull-down assay
We constructed Flag-ZmIAA5 and Myc-ZmLBD2 vectors and purified the fusion proteins by immunoprecipitation using Anti-Flag M1 Affinity Gel. And we purified the Myc-ZmLBD2 recombinant proteins using amylose resin (Park et al., 1998). Flag or Flag-ZmIAA5 was added with equal amounts of Myc-ZmLBD2 protein beads into the buffer (50 mmol/L Tris-HCl, pH 8.0, 100 mmol/L sodium chloride, 0.2% glycerol and 0.5% TritonX-100) and incubated at 4°C for 6 h. The protein complexes were eluted by boiling the beads with electrophoresis loading buffer. Ultimately, the crude extracts and eluted products were determined on 10% SDS-PAGE, and the chemiluminescence signal was detected by Western Blot using Flag-tagged antibodies.
Statistical analysis
Data were tested by analysis of variance using SPSS software (SPSS USA). The data are the mean ± standard deviation (SD) of three biological replicates. The significance was analyzed using Student’s t-tests. The * and ** represents p < 0.05 and p < 0.01, respectively. The figures were prepared with GraphPad Prism 7.
Results
Analysis of ZmLBD2 sequence and expression pattern
In this study, we cloned the ZmLBD2 gene from maize from the H8186 selfing line. ZmLBD2 has a full-length CDS of 831bp, which encodes a polypeptide consisting of 244 amino acid residues with a predicted molecular weight of 25.47kD and a pI value of 5.66 (Table 1). The results of sequence alignment show that the ZmLBD2 contains CX2CX6CX3C, a typical conserved DNA-binding domain. GAS and leucine zipper-like motifs (LX6LX3LX6L) are primarily used to distinguish the members of the maize LBD family into class I and class II (Figure 1A).
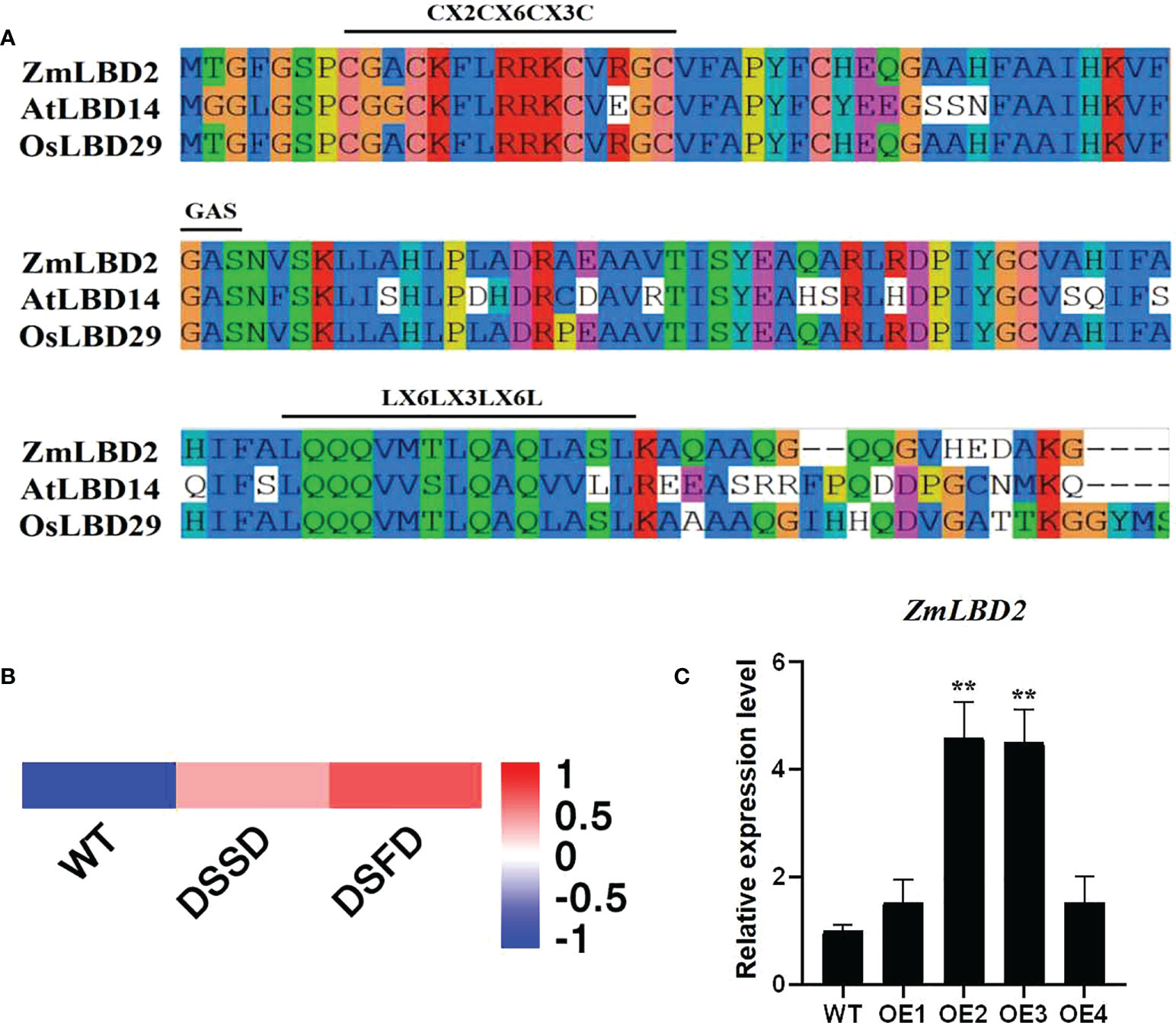
Figure 1 Sequence and expression pattern analysis of ZmLBD2. (A) LOB domain sequence alignment of ZmLBD2 and LBD members from Arabidopsis and Oryza sativa. The class-I LBD members had typical CX2CX6CX3C, GAS, and LX6LX3LX6L domains. The alignment was performed using DNAMAN software. (B, C) The expression of ZmLBD2 upon drought treatment in maize. Total RNA was isolated from 3-leaf seedlings grown without (0 h) or with 8% PEG6000 treatment. Transcript levels of ZmLBD2 were determined by qPCR, using Zm18S as reference genes. The ** represents p < 0.01.
We drew a heatmap (Figure 1B) based on the transcriptome data of maize roots (PRJNA793522) with different drought treatment processing times. The response of ZmLBD2 to drought stress was studied by RT-qPCR using maize plants under 8% PEG6000 drought treatment. The expression of the ZmLBD2 gene was highly induced as the time of suffering drought stress increased (Figure 1C), indicating that the ZmLBD2 gene plays a crucial role in responding to drought.
Subcellular localization of ZmLBD2 protein in tobacco
We designed subcellular localization vectors for the ZmLBD2 gene and transformed them into tobacco mesophyll cells via Agrobacterium tumefaciens. By confocal microscope (Olympus, Japan), we observed that the proteins transcribed from the ZmLBD2 gene localized to the cell nucleus (Figure 2).
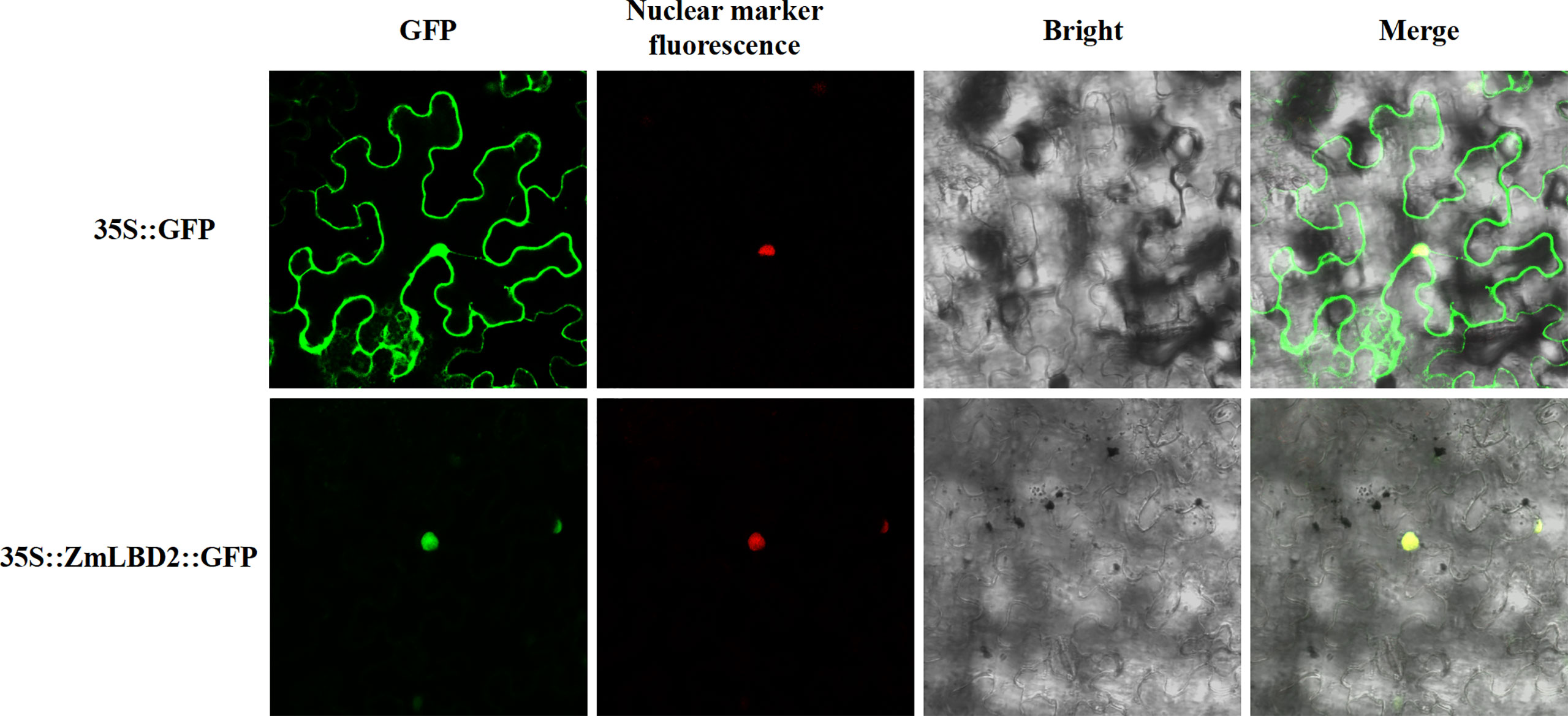
Figure 2 Subcellular localization analysis of ZmLBD2 proteins in tobacco cells. The scale bar represents 50 μm.
Efficient expression of ZmLBD2 in a prokaryotic system
BL21 strain with pET28a-ZmLBD2 recombinant plasmids was selected for single-colony shake cultivation. When the OD600 concentration of the cultures was between 0.6 and 1.0, IPTG with a final concentration of 0.1 mM was added to induce protein expression. After that, BL21 strain with transformed pET28a-ZmLBD2 recombinant plasmids was obtained. We induced the expression at 26°C for 4 h and observed the changes in protein expression. The SDS-PAGE results showed that a clear band around 25.47 kDa was spotted for the recombinant protein of the ZmLBD2, which indicated that the ZmLBD2 recombinant protein could be expressed stably and effectively in the BL21 strain with the induction of IPTG (Figure 3A). The Western blot results showed that a specific band could be seen clear between 25 kDa and 35 kDa (Figure 3B), indicating that the ZmLBD2 protein of maize was successfully induced and expressed in vitro.
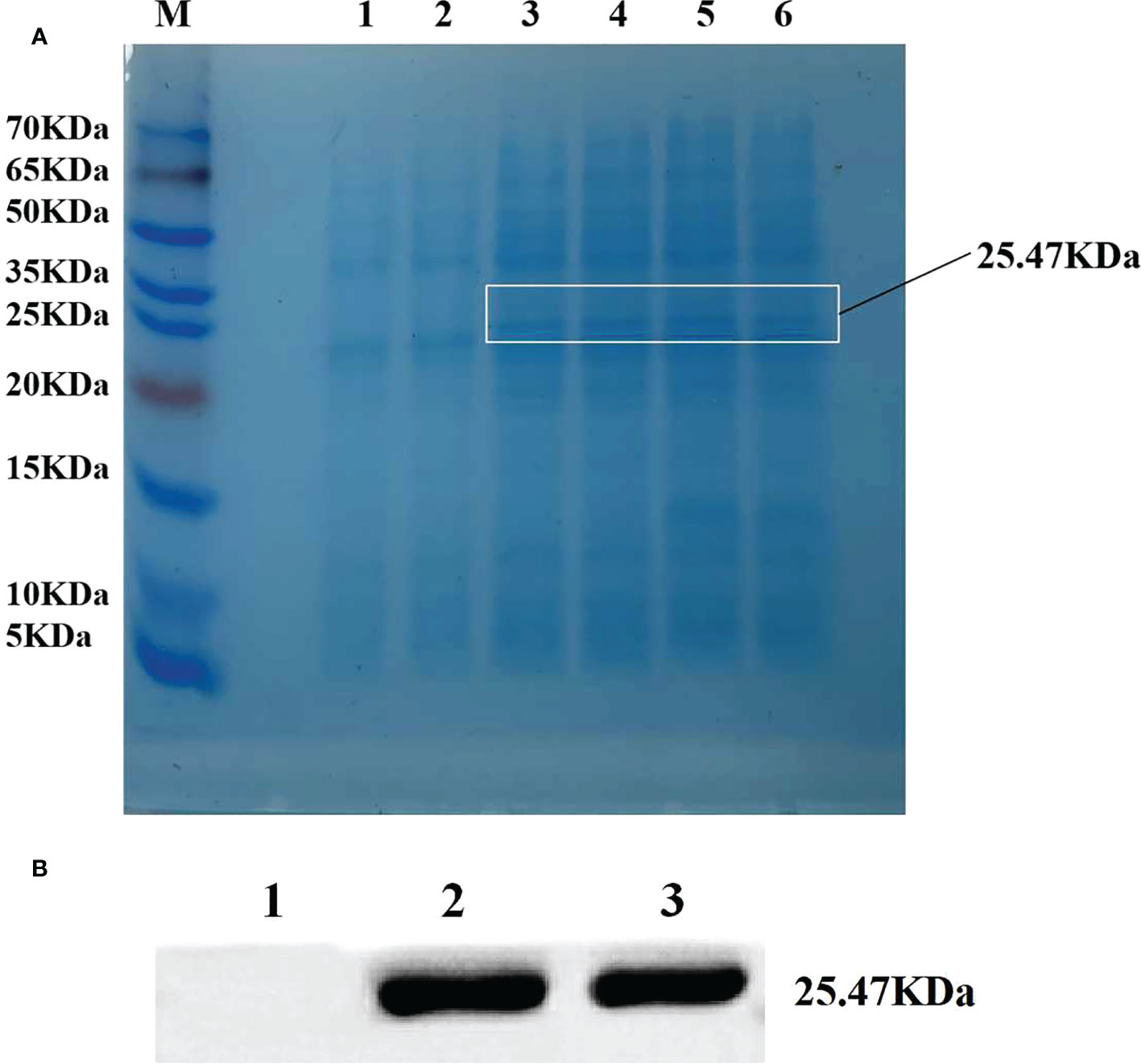
Figure 3 Efficient expression of ZmLBD2 in a prokaryotic system. (A) SDS-PAGE electrophoretic analysis of ZmLBD2 recombinant protein. M: Protein marker; 1: pET22b empty vector was not induced by IPTG; 2: pET22b-ZmLBD2 non-induced bacteria; 3-6: pET22b-ZmLBD2 was produced by IPTG for 4 h. (B) Western blotting detection of ZmLBD2 protein; 1: pET22b empty vector; 2-3: pET22b-ZmLBD2 after inducing.
The DNA binding characteristics of ZmLBD2
On the 20 mM 3-AT SD/Trp-/Leu-/His triple deficiency culture plate, only the yeast strains co-transformed with pGADT7-ZmLBD2 and pHIS2-TCS and the positive control p53HIS2+pGAD-Rec2-p53 can grow normally (Figure 4), indicating that ZmLBD2 can activate the expression of the reporter gene HIS by binding to the “GCGGCG” inverted repeat sequence. To verify the specificity of the DNA binding method, the pGADT7-ZmLBD2 and pHIS2-mTCS vectors were co-transformed into yeast cells on the SD/Trp-/Leu-/His- triple lack culture plate containing 20 mM 3-AT. Only the positive control could grow normally. The binding of the ZmLBD2 gene to the “GCGGCG” inverted repeat is specific.
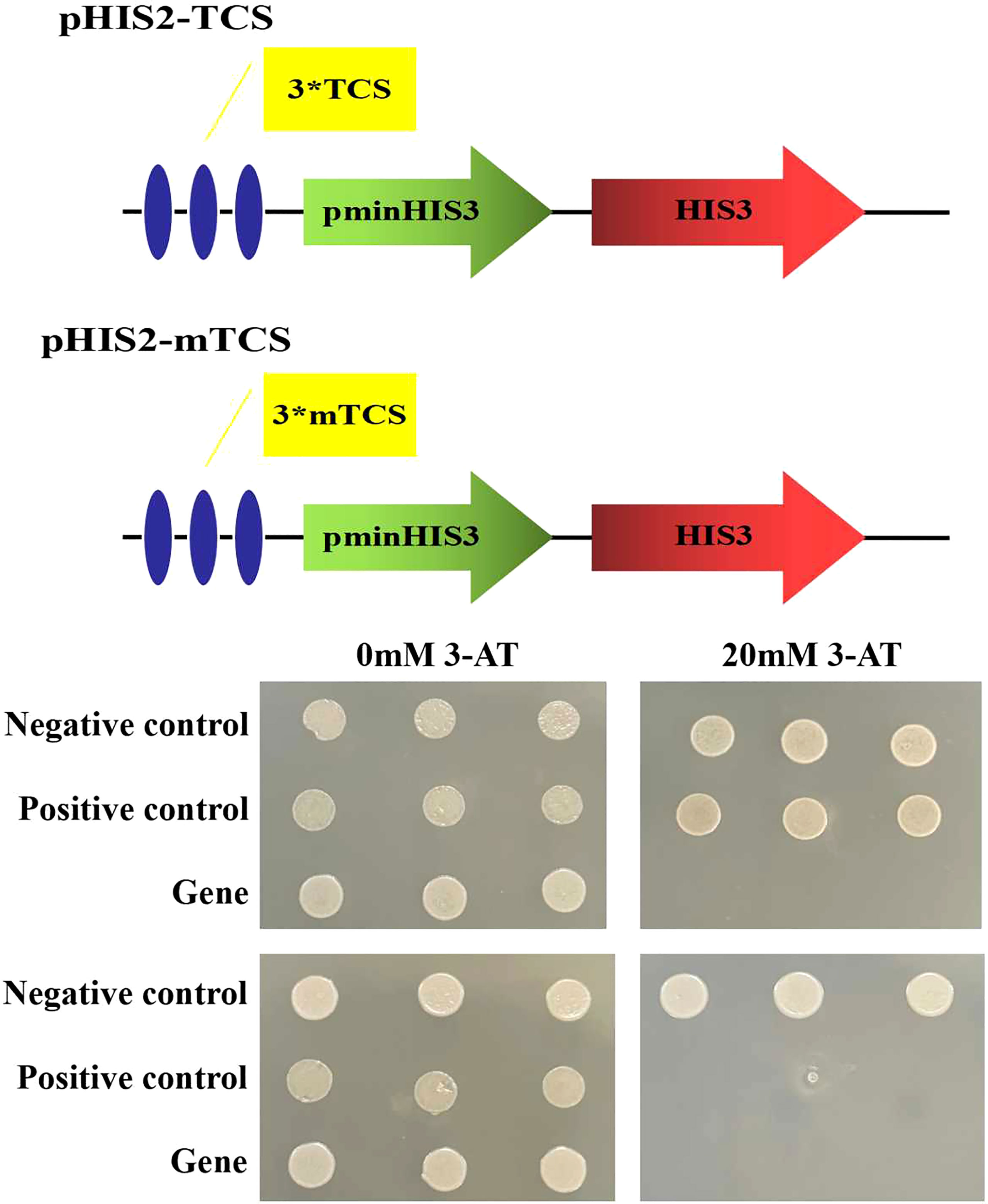
Figure 4 DNA-binding assays of ZmLBD2 in Y1HGold yeast cells. Yeast cells were assayed on SD/Trp-/Leu-/His- medium; Negative control: p53HIS2+p GAD-ZmLBD2; Positive control: p53HIS2+pGAD-Rec2-53p53; Gene: different positive colonies; TCS: GCGGCG; mTCS: GAGGAG; 3*: The sequence was repeated three times; 3-AT: 3-amino-1,2,4-triazole.
Dimer-forming ability of ZmLBD2
To test the dimerization ability of ZmLBD2, six truncated peptide fragments (a, b, c, ab, bc, and abc) of ZmLBD2 were tested for interaction with ZmLBD2. Fragments A, B, and C represent the N-terminal CX2CX6CX3C, the GAS and LX6LX3LX6L coiled-coil motifs, and the C-terminal domain, respectively (Figure 5A). To further define the necessary regions for ZmLBD2 interaction, we screened for homo- and heterodimerization by yeast two-hybridization (Figure 5B).
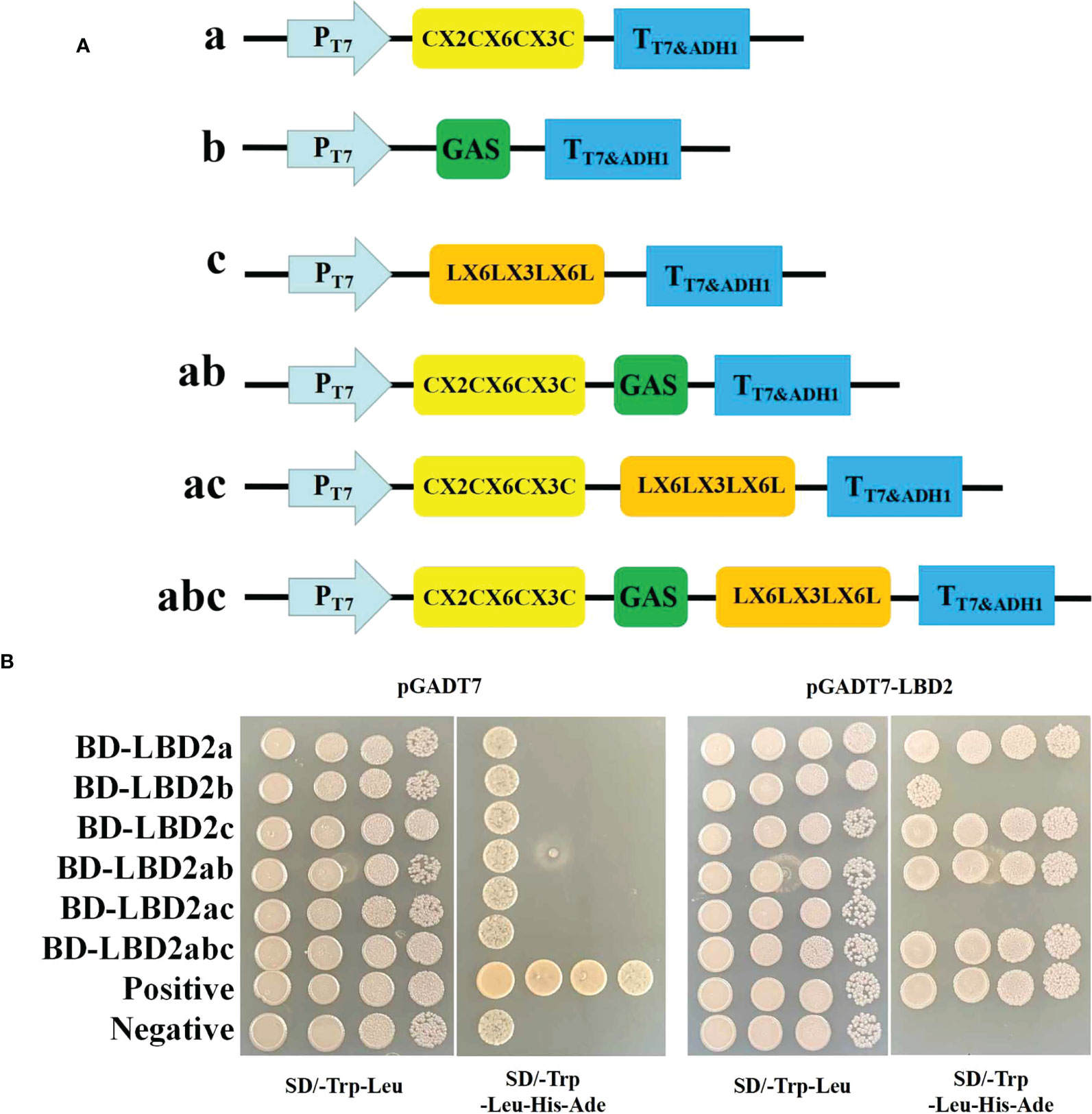
Figure 5 Analysis of the dimer-forming ability of ZmLBD2. (A) Strategy for yeast two-hybrid vector construction of the maize ZmLBD2 sequence. (B) The ability of ZmLBD2 to form dimers in the yeast strain Y2H Gold. The Y2H Gold strains containing target plasmids were diluted and cultured on SD/-Trp-Leu media or SD/-Trp-Leu-His-Ade media. Fragments a, b, and c represent CX2CX6CX3C, GAS, and LX6LX3LX6L.
Identification of the drought tolerance of transgenic yeast with ZmLBD2
We detected no differences between transgenic yeast with pYES2 empty vector and that with ZmLBD2 gene in regular YPD culture media (Figure 6). But, in the YPD culture media containing 1M of mannitol, the transgenic yeast with ZmLBD2 grew better than that the pYES2 empty vector. This tentatively indicates that the ZmLBD2 gene has the power to resist drought.
Genetic transformation and molecular identification of the ZmLBD2 gene in A. thaliana
In this study, we transformed the plasmid DNA of constructed plant overexpression vector pCAMBIA3301-ZmLBD2-bar (Figure 7A) into the EHA105 chemically competent cells and finally, obtained four transgenic positive A. thaliana plants of T3 generation by using floral dip method (Figures 7B-D). By qRT-PCR, we found that the ZmLBD2 gene was expressed in all transgenic plants, but a higher expression was observed in OE2 and OE3 (Figure 7E). In conclusion, the transgenic plants OE2 and OE3 could be used to further identify the ability of ZmLBD2 to combat drought.
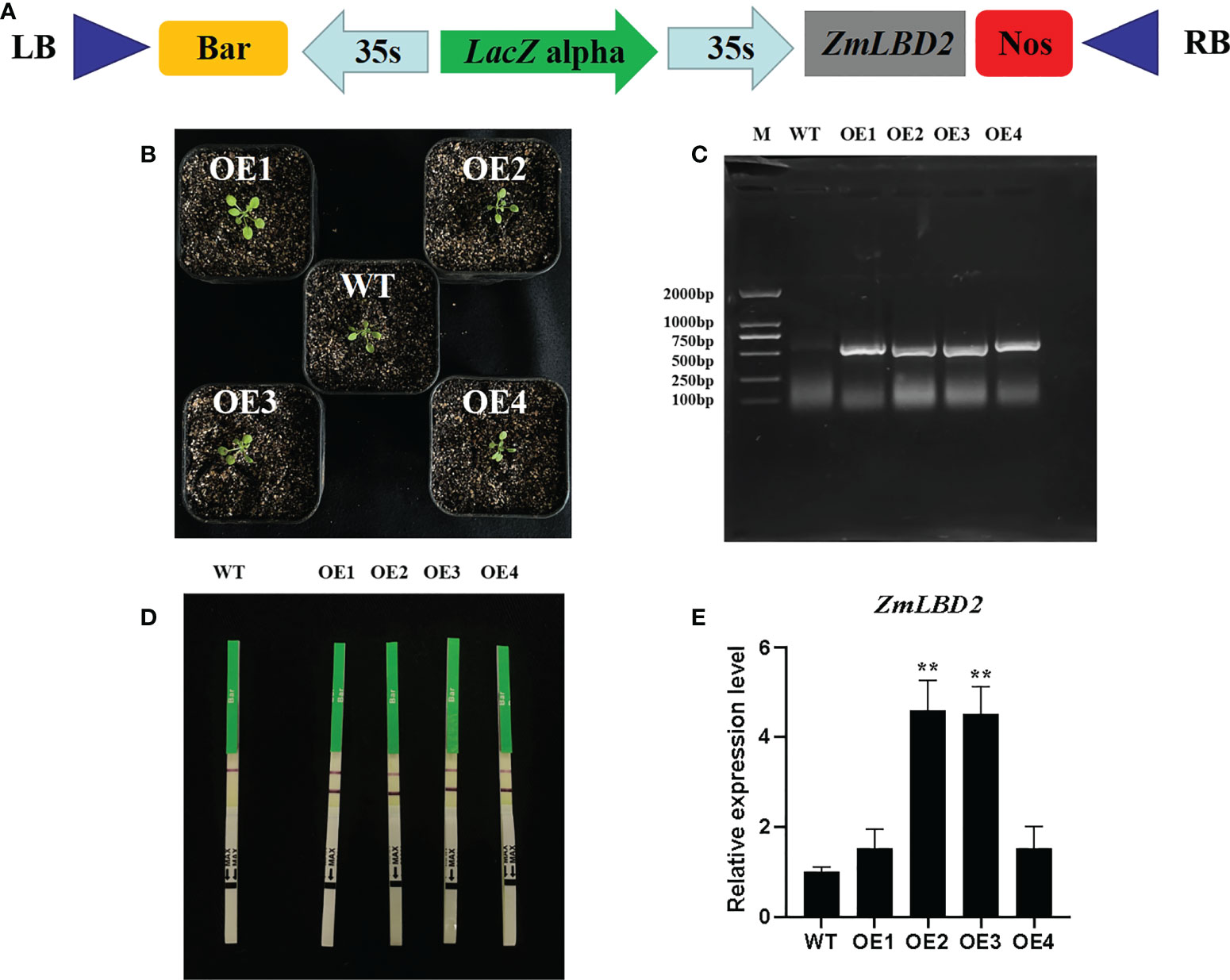
Figure 7 Identification of Arabidopsis overexpressing the ZmLBD2 gene. (A) Schematic of expression vectors pCAMBIA3301-ZmLBD2-Bar. (B) Transgenic Arabidopsis lines. (C) The PCR detection of the Bar gene in leaves of wild type (WT) and transgenic lines. (D) Rapid test strip analysis of the Bar gene in wild-type (WT) leaves and transgenic lines. (E) qRT-PCR analysis of the ZmLBD2 gene in leaves of wild type (WT) and transgenic lines. The expression level was normalized to that of Arabidopsis AtACTIN8. Data were expressed as the mean of triplicate values, and error represented the SD. The * and ** represents p < 0.05 and p < 0.01, respectively.
Overexpression of ZmLBD2 enhanced drought tolerance in transgenic A. thaliana
To investigate its biological function, we generated ZmLBD2 overexpressing A. thaliana. The seeds of the transgenic plants OE2 and OE3 of T3 generation were cultivated in 1/2 MS media with 0, 200, or 300 mM of mannitol. After cultivation for 5 days, the results of comparing their germination status showed no differences between the wild type and the transgenic A. thaliana in the 1/2 MS media with 0 mannitol in terms of germination. As the concentration of mannitol increased, however, the OE2 and OE3 grew better than the wild type did (Figure 8A). Moreover, the germination rates (Figure 8B) and fresh weight (Figure 8C) and root length (Figure 8D) of the OE2 and OE3 were much higher than that of the wild type as the concentration of mannitol increased.
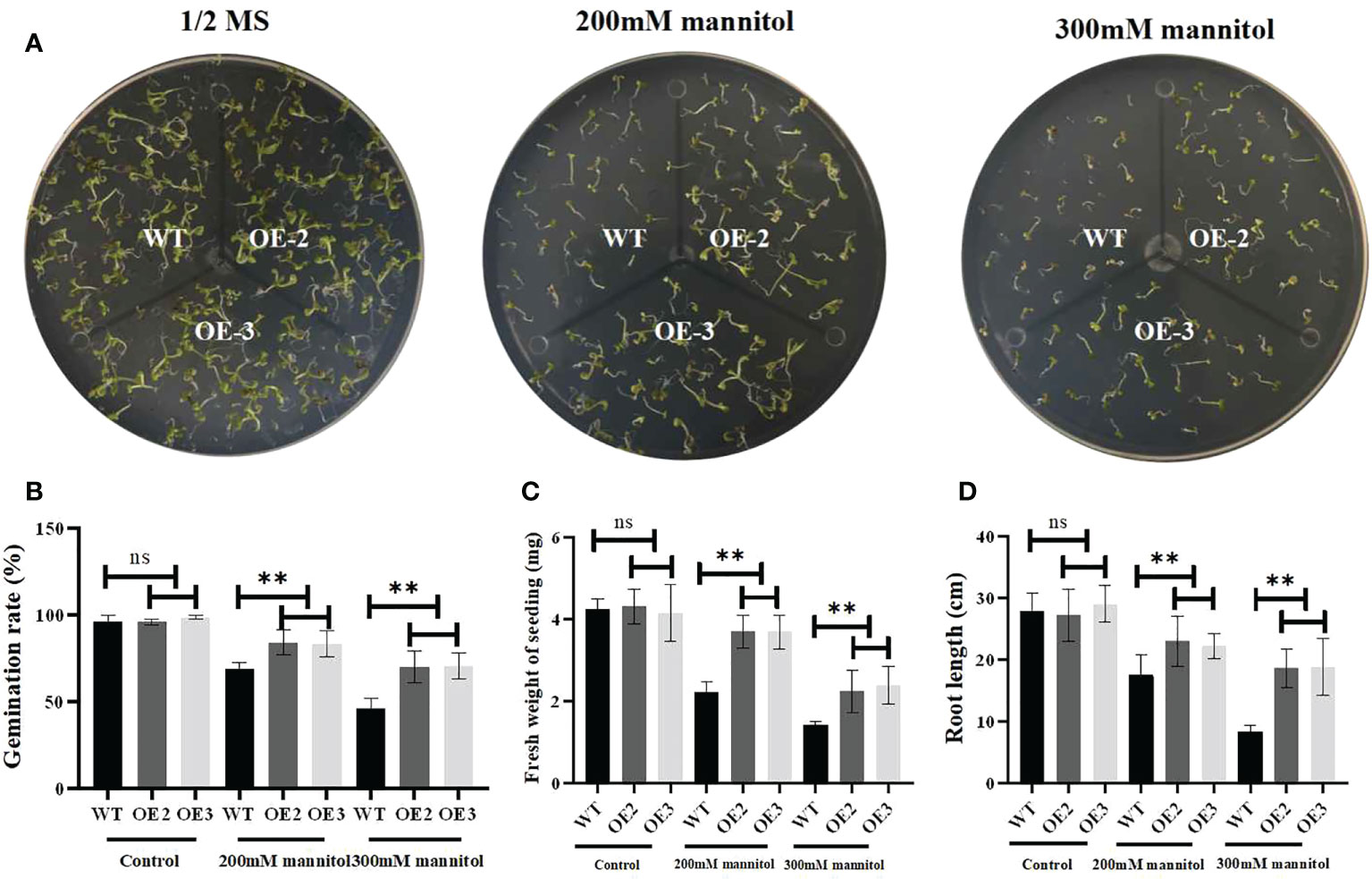
Figure 8 Drought stress responses of ZmLBD2-overexpressing transgenic Arabidopsis and wild-type plants. (A) Seedling growth by wild-type and ZmLBD2-overexpressing transgenic Arabidopsis plants under treatment with 0, 200, and 300 mM of mannitol for 5 days. (B) Statistical analysis of ZmLBD2-overexpressing transgenic lines and wild-type seed germination rates under 0, 200, and 300 mM mannitol, respectively. (C) Statistical analysis of ZmLBD2-overexpressing transgenic lines’ and wild-type’s fresh weight under 0, 200, and 300 mM mannitol, respectively. (D) Statistical analysis of ZmLBD2-overexpressing transgenic lines’ and wild-type’s root length under 0, 200, and 300 mM mannitol, respectively. Data were expressed as the mean of triplicate values, and error represented the SD. The ** represents p < 0.01, respectively. Non-significant (ns).
To further study the functions of ZmLBD2 in response to drought stress, we transplanted the 5-day-old cultivated plants into the soil. Then, we conducted drought treatment on 30-day-old A. thaliana for 10 days. After re-watering for 5 days, we found that the OE2 and OE3 performed much better than the wild type in terms of survival rates (Figures 9A, B), fresh weight (Figure 9C), relative water content (Figure 9D), chlorophyll content (Figures 9E, F), and proline content (Figure 9G). OE2 and OE3 also had less MDA than the wild-type plants (Figure 9H). These results indicate that the overexpression of ZmLBD2 can significantly boost the drought resistance of A. thaliana.
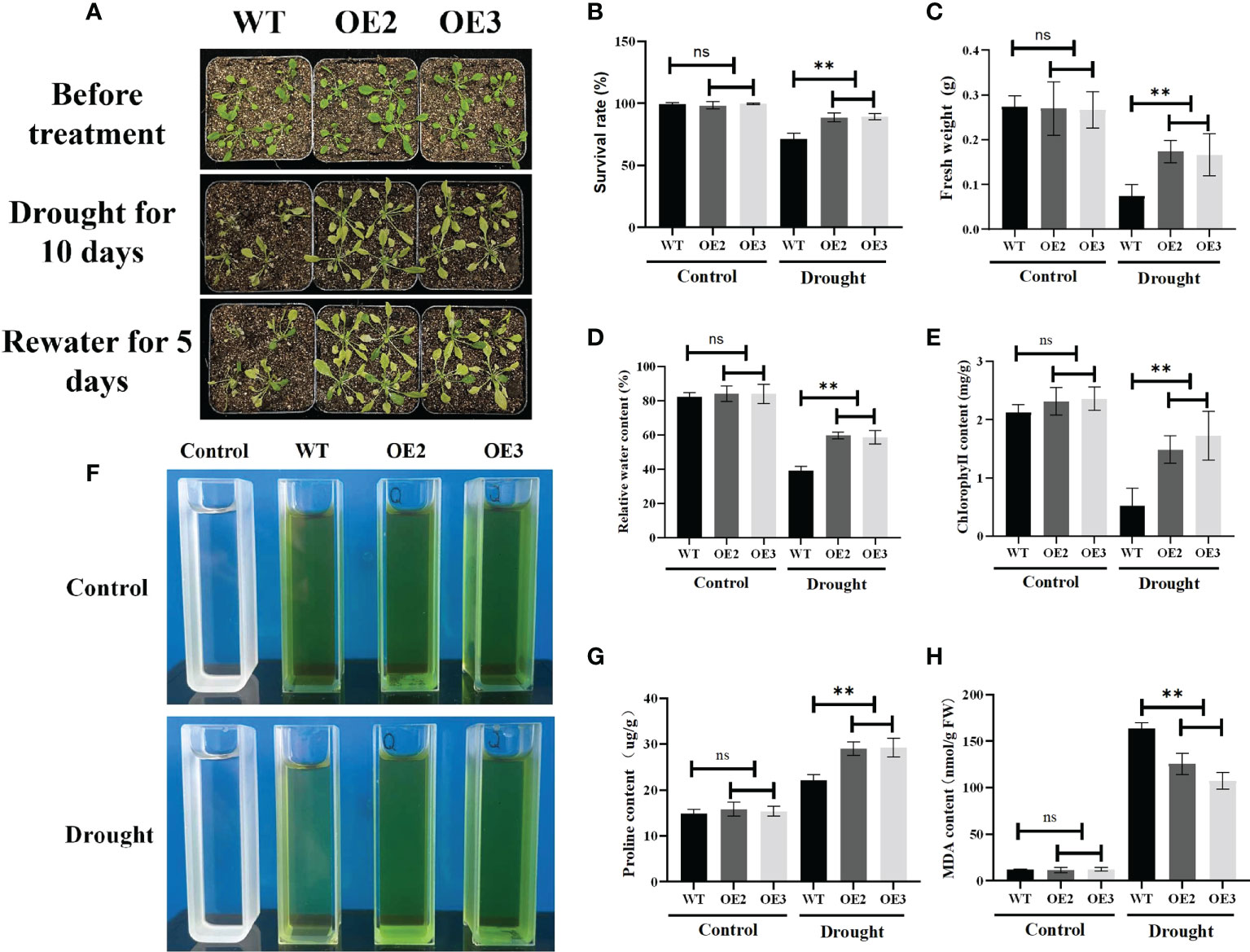
Figure 9 Phenotypes of the ZmLBD2-overexpressing transgenic plants and wild-type controls in A. thaliana under drought conditions. (A) Drought treatment using 30 days plants. The wild-type controls and transgenic plants were grown in pots for 30 days and then subjected to drought treatment for 10 days. The plants were watered again 5 days after the treatment. Survival rate (B), fresh weight (C), relative water content (D), chlorophyll content (E, F), proline content (G), and MDA content (H) in transgenic Arabidopsis plants and wild-type controls under normal conditions and drought stress. Data were expressed as the mean of triplicate values, and error represented the SD. The ** represents p < 0.01, respectively. Non-significant (ns).
Reducing the accumulation of reactive oxygen species by the overexpression of the ZmLBD2 gene under drought stress
Reactive oxygen species (ROS) are essential signaling molecules for plant growth, which play a crucial role in adapting to environments. Therefore, we analyzed the ROS levels of wild and transgenic plants by evaluating the accumulated amount of hydrogen peroxide and superoxide under drought stress. After staining with Nitrotetrazolium Blue chloride (NBT) and Diaminobenzidine (DAB), we found that the accumulation of hydrogen peroxide and superoxide of the wild type was more than that of the OE2 and OE3 (Figures 10A, B). Meanwhile, we compared the accumulation and production of hydrogen peroxide and O2− in the leaves of the wide type OE2 and OE3 maize seedlings. Hydrogen peroxide and superoxide anion accumulated less in the leaves of seedlings with the overexpression of ZmLBD2 than in wild-type seedlings (Figures 10C, D). SOD, CAT, and POD enzymes directly eliminate ROS to regulate ROS levels in plant cells. They turn the ROS into less active and less toxic hazardous substances. We found that, with the threat of drought, SOD, CAT, and POD enzymes were more active in the A. thaliana with the overexpression of ZmLBD2 than those in the wild type (Figures 10E-G).
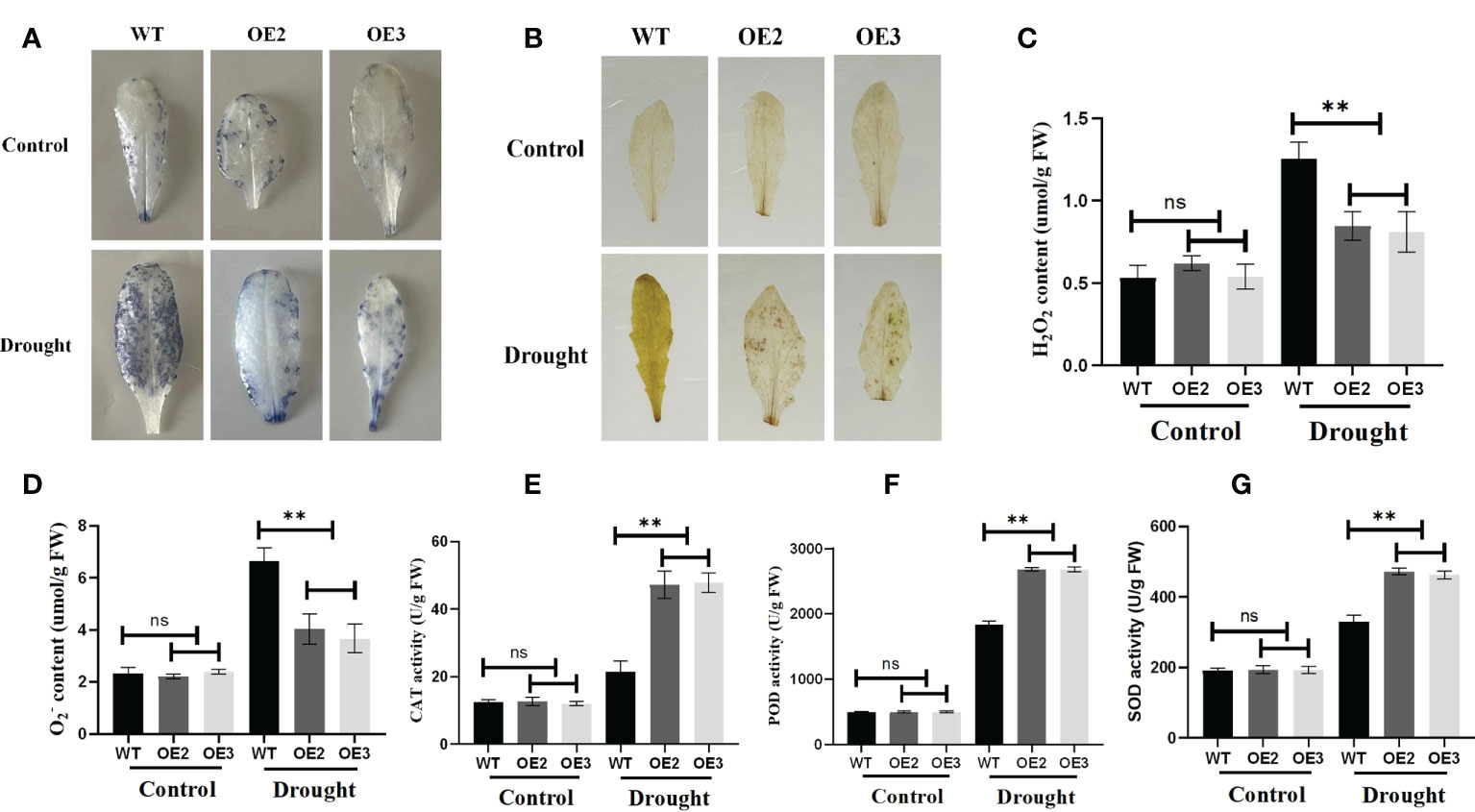
Figure 10 Reactive oxygen species staining and physiological indices in ZmLBD2 transgenic Arabidopsis. (A) NBT staining of leaves for H2O2 from ZmLBD2 transgenic seedlings and wild-type plants under normal conditions and drought stress (30 days seedlings were subjected to drought for 10 days). (B) DAB staining of leaves for H2O2 from ZmLBD2 transgenic seedlings and wild-type plants under normal conditions and drought stress (30 days seedlings were subjected to drought for 10 days). (C, D) H2O2 and content in leaves from ZmLBD2 transgenic seedlings and wild-type plants under normal conditions and drought stress. CAT activity (E), POD activity (F), and SOD activity (G) in leaves from ZmLBD2 transgenic seedlings and wild-type plants under normal and drought conditions. Data were expressed as the mean of triplicate values, and error represented the SD. The ** represents p < 0.01, respectively. Non-significant (ns).
The effects of ZmLBD2 on the expression of drought-stress-responsive genes
To investigate the molecular mechanism of ZmLBD2 regulation under drought conditions, we performed a quantitative expression analysis of six stress-related genes in transgenic Arabidopsis and wild-type plants. Under normal growth conditions, the transcript expression levels of P5CS1, RD29A, COR15A, NCED3, DREB2A, and ABI4 were not significantly different between wild-type control and overexpressing lines. However, under drought conditions, the expression of these genes was rapidly induced and at higher levels, and the expression levels of these genes in ZmLBD2-overexpressing seedlings were lower than in wild-type controls (Figure 11). Therefore, ZmLBD2 may positively regulate the expression of drought stress-related genes in plants under drought stress.
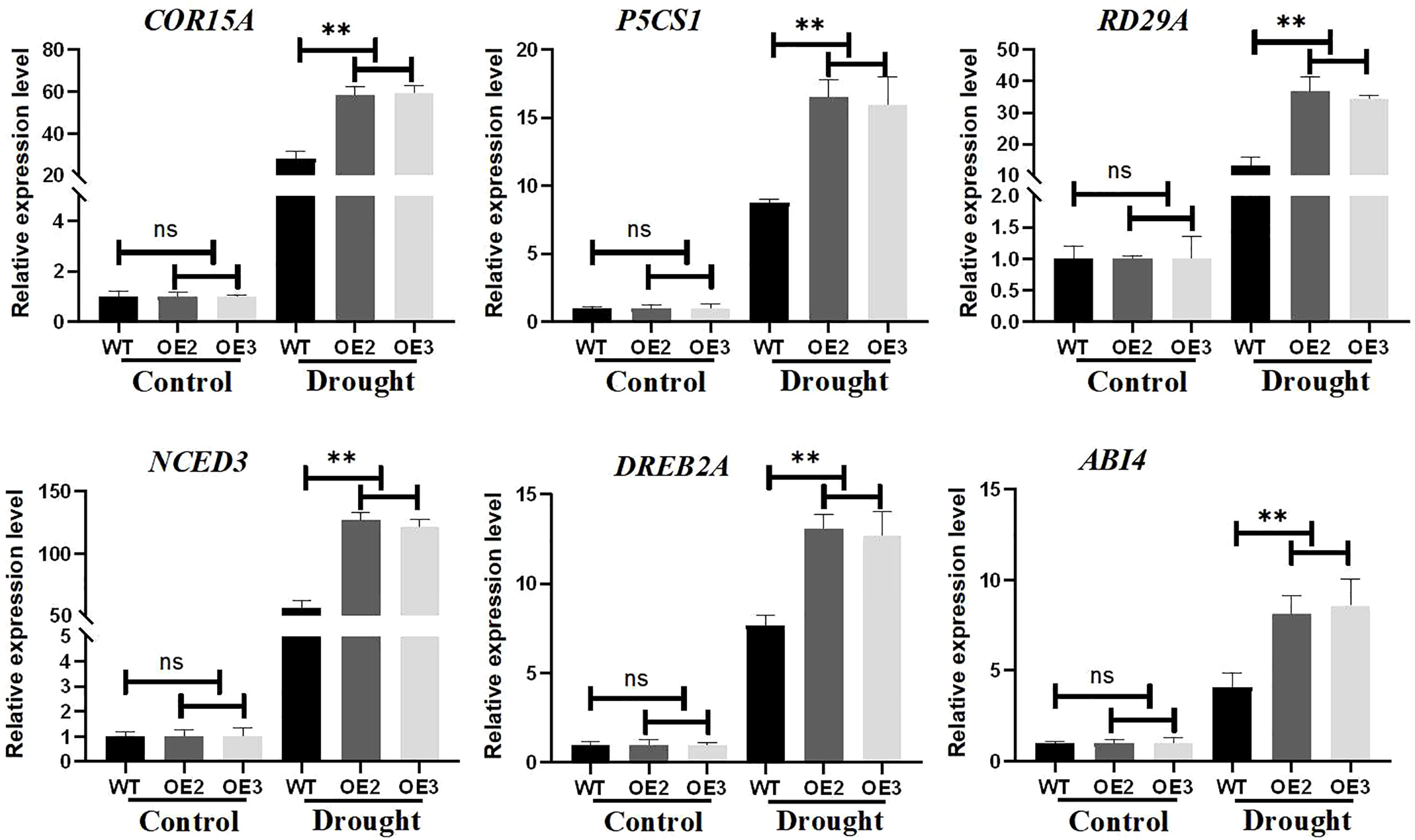
Figure 11 Expression levels of drought-stress-related genes in ZmLBD2 transgenic Arabidopsis. Total RNA was isolated from 15-day-old seedlings grown without (CK) or drought treatment for 10 days. Transcript levels of COR15A, P5CS1, RD29A, NCED3, DREB2A, and ABI4 in the transgenic lines and wild type were determined by qPCR AtACTIN8 as reference genes. Fold change was calculated by 2−ΔΔCT. The ** represents p < 0.01, respectively. Non-significant (ns).
ZmIAA5, a candidate ZmLBD2 interacting gene
To study how the ZmLBD2 gene influences the drought resistance of the transgenic A. thaliana, we screened a cDNA library for maize in a yeast two-hybrid system (with ZmLBD2 as a decoy) and identified an interactive candidate gene ZmIAA5 (Zm00001d043878). Figure 12A shows that all yeast grew normally in DDD culture media (SD/-Trp/-Leu), whereas only that with the coexpression of AD-ZmIAA5 and BD-ZmLBD2 grew well in QDO selective media (SD/-Trp/-Leu/-His/-Ade). We also conducted a BiFC assay to corroborate the interaction between ZmLBD2 and ZmIAA5. When nYFP-ZmLBD2 and cYFP-ZmIAA5 were coexpressed, we detected the fluorescence from yellow fluorescent proteins (YFP) in the nuclei of tobacco mesophyll cells. However, no fluorescence was detected in terms of the coexpression of nYFP-ZmLBD2 and cYFP, or nYFP and cYFP-ZmIAA5 (Figure 12B). Pull-down assay was performed to test the purified recombinants Myc-ZmLBD2 and Flag-ZmIAA5 in vitro. The Myc-ZmLBD2 protein interacted with Flag-ZmIAA5 protein, but it did not bind to Myc (Figure 12C). These results indicate a direct gene interaction between ZmLBD2 and ZmIAA5.
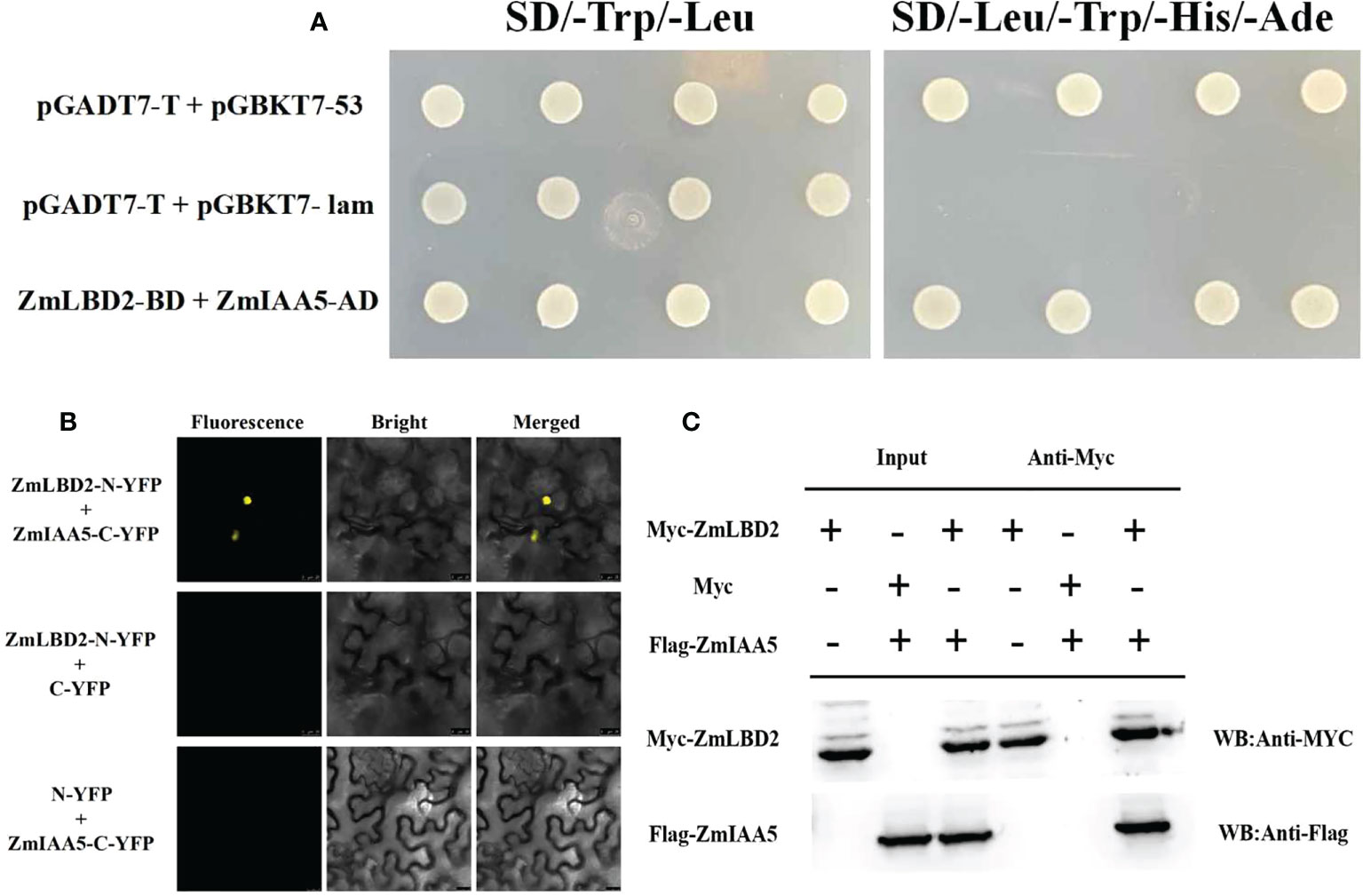
Figure 12 Interaction between ZmLBD2 and ZmIAA5. (A) Interaction analysis of ZmLBD2 and ZmIAA5 using a yeast two-hybrid system. (B) Interaction of ZmLBD2 and ZmIAA5 as determined with a BiFC assay. (C) Pull-down assay ofZmLBD2 and ZmIAA5.
Discussion
LBD genes are specific plant transcription factors. They are divided into class I and class II following the differences in the LOB domain. LBD transcription factors from the class I contain the complete LOB domain, and they are mainly responsible for plant growth, such as developing lateral organs, extending leaves, and regeneration (Bell et al., 2012; Liu et al., 2018; Zhang et al., 2020; Huang et al., 2020). Class II LBD proteins hold an incomplete leucine zipper-like domain, so they cannot form the coiled-coil structure. They are primarily responsible for secondary metabolite production in plants, such as the synthesis of anthocyanidin and the growth of roots. They also play an active role in responding to pathogenesis, osmotic stress, salt stress, and drought stress (Lim et al., 2015; Liu et al., 2016; Zhang et al., 2017; Fernando Gil et al., 2018; Liu et al., 2019; Liu et al., 2020; Guo et al., 2020). Our study found that the ZmLBD2 gene was significantly induced to express under drought.
The growth of plants is not only regulated by internal hormones and transcription factors but also influenced by external environments. When the living environment changes or hazardous factors occur, plants need to respond to the environmental signals from the outside to better adapt to the environment. Liu et al. (2020) reported that the SlLBD40 gene in tomato (Solanum lycopersicum L.) was overexpressed under drought stress, which caused the plant to wither. In addition, the expression of this gene was affected by the signal transmission pathway of jasmonic acid. Huang et al. (2020) found that most Physcomitrium patens mosses increased the expression of the PpLBDs gene after they were processed by mannitol. They extrapolated that the expression of PpLBDs might strengthen the drought resistance of the mosses. The expression of the VvLBD1 gene in grape (Vitis vinifera L.) increased considerably under the threats of mannitol, salt stress, and heat stress, indicating the significance of VvLBD1 in the responses to environmental signals (Cao et al., 2016). Liu et al. (2019) adopted qRT-PCR assay to potato (Solanum tuberosum L.) with drought treatment and found that the expression of StLBD1-5 decreased while that of the StLBD2-6 and StLBD3-5 increased. This finding reveals the connection between these three genes and the response of potatoes to drought stress. The research published by Guo et al. (2020) showed that the A. thaliana AtLBD15 gene could directly bind to the abscisic acid signaling pathway factor ABI14’s promoter to close the stomas and enhance drought resistance. Our study shows that the overexpression of the maize ZmLBD2 gene can promote A. thaliana seeds to burgeon and grow roots under drought, showing that the ZmLBD2 can strengthen the drought tolerance of plants.
Drought stress is to blame for the production of ROS. To protect cells from oxidative damage caused by oxidative stress when accumulating too much ROS, plants have developed various antioxidant mechanisms to remove toxins caused by excess ROS (McGrann et al., 2015). Xiong et al. (2022) found that the ROS content in Arabidopsis overexpressing zmLBD5 was reduced, and the SOD and POD activities were higher than those in wild-type Arabidopsis. Xing et al. (2020) found that A. thaliana overexpressing the CmLOX10 gene under drought conditions had lower MDA and hydrogen peroxide content than the wild type and had strong drought resistance. In our research, the activity of CAT, SOD, and POD increased in the A. thaliana overexpressing ZmLBD2, and the levels of H2O2 and were higher in the wild type. This finding indicates that the ZmLBD2 can enhance drought resistance by reducing ROS accumulation.
Protein interaction has become a necessary link to further explore the role of maize LBD transcription factor genes in plant growth and development and abiotic stress response. Xu et al. (2018) found that Arabidopsis LBD16 interacted with bZIP59 protein to jointly initiate the expression of the downstream gene FAD-BD, thereby promoting callus formation. Their study found that the AtbZIP59-LBD complex is an auxin-induced callus. Key regulators of cell fate change during wound tissue formation, which may open the door to further exploration of the relationship between the remarkable regenerative capacity of plants and developmental plasticity. In this study, through yeast two-hybrid, BiFC, and Pull-down assays, we identified IAA5 as an interacting protein of ZmLBD2. The Aux/IAA proteins are a large family of auxin coreceptors and transcriptional repressors, which are involved in auxin signaling. Aux/IAA proteins also play an important role in plant responses to abiotic stresses (Cheol Park et al., 2013; Wang F. et al., 2021). We found that the auxin content decreased under drought conditions, and the transcriptional expression levels of IAA-related genes were also affected. Jung et al. (2015) found that the rice Aux/IAA gene OsIAA6 was highly induced by drought stress, thereby improving drought tolerance through the regulation of auxin biosynthesis genes. Therefore, we speculate that ZmLBD2 interacts with ZmIAA5 to regulate the expression of downstream auxin synthesis-related genes to improve the drought resistance of plants. We plan to further verify our hypothesis in future work.
Conclusion
In conclusion, we show that the ZmLBD2 gene is a positive regulatory factor for drought response. Subcellular localization results showed that the ZmLBD2 transcriptional protein was located in the nucleus. The ZmLBD2 gene specifically binds with the inverted repeats of “GCGGCG”. Under drought stress, overexpression of ZmLBD2 improved drought resistance by increasing plant germination rate, root length, fresh weight, relative water content, chlorophyll, and proline content. In addition, it can also be overexpressed to promote the activity of CAT, SOD, and POD in A. thaliana to strengthen drought resistance by lowering ROS levels. Meanwhile, we also screened out the ZmIAA5 gene interaction with ZmLBD2. This study provides a theoretical basis for the future analysis of the biological function of the interaction between maize LBD2 and IAA5 in auxin-regulated plant responses to drought stress.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
SYG and YYM conceived research plans and designed experiments. PJ, XTW and ZZJ conducted experiments. PJ wrote the draft. SYL and ZZJ analyzed the data. PJ, SYG and YYM reviewed and edited this article and provided helpful comments and discussions. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Jilin Province Science and Technology Development Plan Project [20220202008NC].
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1000149/full#supplementary-material
References
Ariel, F. D., Diet, A., Crespi, M., Chan, R. L. (2010). The LOB-like transcription factor Mt LBD1 controls medicago truncatula root architecture under salt stress. Plant Signaling Behav. 5 (12), 1666–1668. doi: 10.4161/psb.5.12.14020
Ba, L. J., Kuang, J. F., Chen, J. Y., Lu, W. J. (2016). MaJAZ1 attenuates the MaLBD5-mediated transcriptional activation of jasmonate biosynthesis gene MaAOC2 in regulating cold tolerance of banana fruit. J. Agric. Food Chem. 64 (4), 738–745. doi: 10.1021/acs.jafc.5b05005
Bell, E. M., Lin, W. C., Husbands, A. Y., Yu, L. F., Jaganatha, V., Jablonska, B., et al. (2012). Arabidopsis LATERAL ORGAN BOUNDARIES negatively regulates brassinosteroid accumulation to limit growth in organ boundaries, proc. Natl. Acad. Sci. U.S.A. 109 (51), 21146–21151. doi: 10.1073/pnas.1210789109
Cao, H., Liu, C. Y., Liu, C. X., Zhao, Y. L., Xu, R. R. (2016). Genomewide analysis of the lateral organ boundaries domain gene family in vitis vinifera. J. Genet. 95 (3), 515–526. doi: 10.1007/s12041-016-0660-z
Chen, K., Su, C., Tang, W., Zhou, Y., Xu, Z., Chen, J., et al. (2021). Nuclear transport factor GmNTF2B-1 enhances soybean drought tolerance by interacting with oxidoreductase GmOXR17 to reduce reactive oxygen species content. Plant J. Cell Mol. Biol. 107 (3), 740–759. doi: 10.1111/tpj.15319
Cheol Park, H., Cha, J. Y., Yun, D. J. (2013). Roles of YUCCAs in auxin biosynthesis and drought stress responses in plants. Plant Signaling Behav. 8 (6), e24495. doi: 10.4161/psb.24495
Fernando Gil, J, Liebe, S., Thiel, H., Lennefors, B. L., Kraft, T., Gilmer, D., et al. (2018). Massive up-regulation of LBD transcription factors and EXPANSINs highlights the regulatory programs of rhizomania disease. Mol. Plant Pathol. 19 (10), 2333–2348. doi: 10.1111/mpp.12702
Guo, Z., Xu, H., Lei, Q., Du, J., Li, C., Wang, C., et al. (2020). The arabidopsis transcription factor LBD15 mediates ABA signaling and tolerance of water-deficit stress by regulating ABI4 expression. Plant J. Cell Mol. Biol. 104 (2), 510–521. doi: 10.1111/tpj.14942
Huang, X. L., Yan, H. Q., Liu, Y. J., Yi, Y. (2020). Genome-wide analysis of LATERAL ORGAN BOUNDARIES DOMAIN-in physcomitrella patens and stress responses. Genes Genomics 42 (6), 651–662. doi: 10.1007/s13258-020-00931-x
Hu, C. H., Zeng, Q. D., Tai, L., Li, B. B., Zhang, P. P., Nie, X. M., et al. (2020). Interaction between TaNOX7 and TaCDPK13 contributes to plant fertility and drought tolerance by regulating ROS production. J. Agric. Food Chem. 68 (28), 7333–7347. doi: 10.1021/acs.jafc.0c02146
Jiao, P., Jiang, Z., Wei, X., Liu, S., Qu, J., Guan, S., et al. (2022). Overexpression of the homeobox-leucine zipper protein ATHB-6 improves the drought tolerance of maize (Zea mays l.). Plant Sci. an Int. J. Exp. Plant Biol. 316, 111159. doi: 10.1016/j.plantsci.2021.111159
Jung, H., Lee, D. K., Choi, Y. D., Kim, J. K. (2015). OsIAA6, a member of the rice Aux/IAA gene family, is involved in drought tolerance and tiller outgrowth. Plant Sci. an Int. J. Exp. Plant Biol. 236, 304–312. doi: 10.1016/j.plantsci.2015.04.018
Ju, Y. L., Yue, X. F., Min, Z., Wang, X. H., Fang, Y. L., Zhang, J. X. (2020). VvNAC17, a novel stress-responsive grapevine (Vitis vinifera l.) NAC transcription factor, increases sensitivity to abscisic acid and enhances salinity, freezing, and drought tolerance in transgenic arabidopsis. Plant Physiol. Biochem. PPB 146, 98–111. doi: 10.1016/j.plaphy.2019.11.002
Lim, C. W., Han, S. W., Hwang, I. S., Kim, D. S., Hwang, B. K., Lee, S. C., et al. (2015). Te Pepper lipoxygenase CaLOX1 plays a role in osmotic, drought and high salinity stress response. Plant Cell Physiol. 56(5), 930–942. doi: 10.1093/pcp/pcv020
Liu, H., Cao, M., Chen, X., Ye, M., Zhao, P., Nan, Y., et al. (2019). Genome-wide analysis of the lateral organ boundaries domain (LBD) gene family in solanum tuberosum. Int. J. Mol. Sci. 20 (21), 5360. doi: 10.3390/ijms20215360
Liu, J., Hu, X. M., Qin, P., Prasad, K., Hu, Y. X., Xu, L. (2018). The WOX11-LBD16 pathway promotes pluripotency acquisition in callus cells during de novo shoot regeneration in tissue culture. Plant Cell Physiol. 59 (4), 739–748. doi: 10.1093/pcp/pcy010
Liu, J., Xu, L., Shang, J., Hu, X., Yu, H., Wu, H., et al. (2021). Genome-wide analysis of the maize superoxide dismutase (SOD) gene family reveals important roles in drought and salt responses. Genet. Mol. Biol. 44 (3), e20210035. doi: 10.1590/1678-4685-gmb-2021-0035
Liu, J. Y., Zhang, C., Shao, Q., Tang, Y. F., Cao, S. X., Guo, X. O., et al. (2016). Efects of abiotic stress and hormones on the expressions of fve 13-CmLOXs and enzyme activity in oriental melon (Cucumis melo var. makuwa makino). J. Integr. Agric. 15, 326–338. doi: 10.1016/S2095-3119(15)61135-2
Liu, L., Zhang, J., Xu, J., Li, Y., Guo, L., Wang, Z., et al. (2020). CRISPR/Cas9 targeted mutagenesis of SlLBD40, a lateral organ boundaries domain transcription factor, enhances drought tolerance in tomato. Plant Sci. an Int. J. Exp. Plant Biol. 301, 110683. doi: 10.1016/j.plantsci.2020.110683
Li, L. H., Yi, H. L., Liu, X.-P., Qi, H. X. (2021). Sulfur dioxide enhance drought tolerance of wheat seedlings through H2S signaling. Ecotoxicol. Environ. Saf. 207, 111248. doi: 10.1016/j.ecoenv.2020.111248
McGrann, G. R., Steed, A., Burt, C., Goddard, R., Lachaux, C., Bansal, A., et al. (2015). Contribution of the drought tolerance-related stress-responsive NAC1 transcription factor to resistance of barley to ramularia leaf spot. Mol. Plant Pathol. 16 (2), 201–209. doi: 10.1111/mpp.12173
Noman, M., Jameel, A., Qiang, W. D., Ahmad, N., Liu, W. C., Wang, F. W., et al. (2019). Overexpression of GmCAMTA12 enhanced drought tolerance in arabidopsis and soybean. Int. J. Mol. Sci. 20 (19), 4849. doi: 10.3390/ijms20194849
Park, J. H., Choi, E. A., Cho, E. W., Hahm, K. S., Kim, K. L. (1998). Maltose binding protein (MBP) fusion proteins with low or no affinity to amylose resins can be single-step purified using a novel anti-MBP monoclonal antibody. Molecules Cells 8 (6), 709–716. doi: 10.1016/S0022-0981(98)00091-4
Sahebi, M., Hanafi, M. M., Rafii, M. Y., Mahmud, T., Azizi, P., Osman, M., et al. (2018). Improvement of drought tolerance in rice (Oryza sativa l.): Genetics, genomic tools, and the WRKY gene family. BioMed. Res. Int. 2018, 3158474. doi: 10.1155/2018/3158474
Sapes, G., Sala, A. (2021). Relative water content consistently predicts drought mortality risk in seedling populations with different morphology, physiology and times to death. Plant Cell Environ. 44 (10), 3322–3335. doi: 10.1111/pce.14149
Wang, C., Chen, N., Liu, J., Jiao, P., Liu, S., Qu, J., et al. (2022). Overexpression of ZmSAG39 in maize accelerates leaf senescence in arabidopsis thaliana. Plant Growth Regul. doi: 10.1007/s10725-022-00874-1
Wang, F., Niu, H., Xin, D., Long, Y., Wang, G., Liu, Z., et al. (2021). OsIAA18, an Aux/IAA transcription factor gene, is involved in salt and drought tolerance in rice. Front. Plant Sci. 12, 738660. doi: 10.3389/fpls.2021.738660
Wang, C. T., Ru, J. N., Liu, Y. W., Li, M., Zhao, D., Yang, J. F., et al. (2018). Maize WRKY transcription factor ZmWRKY106 confers drought and heat tolerance in transgenic plants. Int. J. Mol. Sci. 19 (10), 3046. doi: 10.3390/ijms19103046
Wang, Y., Yu, Y., Wan, H., Tang, J., Ni, Z. (2022). The sea-island cotton GbTCP4 transcription factor positively regulates drought and salt stress responses. Plant Sci. an Int. J. Exp. Plant Biol. 322, 111329. doi: 10.1016/j.plantsci.2022.111329
Wang, J., Zhang, W., Cheng, Y., Feng, L. (2021). Genome-wide identification of LATERAL ORGAN BOUNDARIES DOMAIN (LBD) transcription factors and screening of salt stress candidates of Rosa rugosa thunb. Biology 10 (10), 992. doi: 10.3390/biology10100992
Wang, Z., Zhang, R., Cheng, Y., Lei, P., Song, W., Zheng, W., et al. (2021). Genome-wide identification, evolution, and expression analysis of LBD transcription factor family in bread wheat (Triticum aestivum l.). Front. Plant Sci. 12, 721253. doi: 10.3389/fpls.2021.721253
Wei, S., Xia, R., Chen, C., Shang, X., Ge, F., Wei, H., et al. (2021). ZmbHLH124 identified in maize recombinant inbred lines contributes to drought tolerance in crops. Plant Biotechnol. J. 19 (10), 2069–2081. doi: 10.1111/pbi.13637
Xing, Q., Liao, J., Cao, S., Li, M., Lv, T., Qi, H. (2020). CmLOX10 positively regulates drought tolerance through jasmonic acid -mediated stomatal closure in oriental melon (Cucumis melo var. makuwa makino). Sci. Rep. 10 (1), 17452. doi: 10.1038/s41598-020-74550-7
Xiong, H., Yu, J., Miao, J., Li, J., Zhang, H., Wang, X., et al. (2018). Natural variation in OsLG3 increases drought tolerance in rice by inducing ROS scavenging. Plant Physiol. 178 (1), 451–467. doi: 10.1104/pp.17.01492
Xiong, J., Zhang, W., Zheng, D., Xiong, H., Feng, X., Zhang, X., et al. (2022). ZmLBD5 increases drought sensitivity by suppressing ROS accumulation in arabidopsis. Plants (Basel Switzerland) 11 (10), 1382. doi: 10.3390/plants11101382
Xu, C., Cao, H., Zhang, Q., Wang, H., Xin, W., Xu, E., et al. (2018). Control of auxin-induced callus formation by bZIP59-LBD complex in arabidopsis regeneration. Nat. Plants 4 (2), 108–115. doi: 10.1038/s41477-017-0095-4
Xu, Z. S., Xia, L. Q., Chen, M., Cheng, X. G., Zhang, R. Y., Li, L. C., et al. (2007). Isolation and molecular characterization of the triticum aestivum l. ethylene-responsive factor 1 (TaERF1) that increases multiple stress tolerance. Plant Mol. Biol. 65 (6), 719–732. doi: 10.1007/s11103-007-9237-9
Yuan, X., Wang, H., Cai, J., Bi, Y., Li, D., Song, F. (2019). Rice NAC transcription factor ONAC066 functions as a positive regulator of drought and oxidative stress response. BMC Plant Biol. 19 (1), 278. doi: 10.1186/s12870-019-1883-y
Yu, T. F., Liu, Y., Fu, J. D., Ma, J., Fang, Z. W., Chen, J., et al. (2021). The NF-Y-PYR module integrates the abscisic acid signal pathway to regulate plant stress tolerance. Plant Biotechnol. J. 19 (12), 2589–2605. doi: 10.1111/pbi.13684
Zhang, J., Huguet-Tapia, J. C., Hu, Y., Jones, J., Wang, N., Liu, S., et al. (2017). Homologues of CsLOB1 in citrus function as disease susceptibility genes in citrus canker. Mol. Plant Pathol. 18 (6), 798–810. doi: 10.1111/mpp.12441
Zhang, Y. W., Li, Z. W., Ma, B., Hou, Q. C., Wan, X. Y. (2020). Phylogeny and functions of LOB domain proteins in plants. Int. J. Mol. Sci. 21 (7), 2278. doi: 10.3390/ijms21072278
Zhang, Y. M., Zhang, S. Z., Zheng, C. C. (2014). Genomewide analysis of LATERAL ORGAN BOUNDARIES domain gene family in zea mays. J. Genet. 93 (1), 79–91. doi: 10.1007/s12041-014-0342-7
Zhao, Q., Hu, R. S., Liu, D., Liu, X., Wang, J., Xiang, X. H., et al. (2020). The AP2 transcription factor NtERF172 confers drought resistance by modifying NtCAT. Plant Biotechnol. J. 18 (12), 2444–2455. doi: 10.1111/pbi.13419
Keywords: lateral organ boundaries domain, LBD2, transcription factor, drought tolerance, maize
Citation: Jiao P, Wei X, Jiang Z, Liu S, Guan S and Ma Y (2022) ZmLBD2 a maize (Zea mays L.) lateral organ boundaries domain (LBD) transcription factor enhances drought tolerance in transgenic Arabidopsis thaliana. Front. Plant Sci. 13:1000149. doi: 10.3389/fpls.2022.1000149
Received: 21 July 2022; Accepted: 26 September 2022;
Published: 13 October 2022.
Edited by:
Ibrahim Al-Ashkar, Al-Azhar University, EgyptReviewed by:
Hayssam M. Ali, King Saud University, Saudi ArabiaZongliang Xia, Henan Agricultural University, China
Copyright © 2022 Jiao, Wei, Jiang, Liu, Guan and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuyan Guan, Z3VhbnNodXlhbkBqbGF1LmVkdS5jbg==; Yiyong Ma, bTE4NDA0MzE5MjAyXzFAMTI2LmNvbQ==
 Peng Jiao
Peng Jiao Xiaotong Wei2,3
Xiaotong Wei2,3 Shuyan Guan
Shuyan Guan Yiyong Ma
Yiyong Ma