
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 04 January 2022
Sec. Plant Breeding
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.815589
Fruit shape is one of the most important quality traits of pepper (Capsicum spp.) and is used as a major attribute for the classification of fruit types. Wide natural variation in fruit shape exists among the major cultivated species Capsicum annuum, allowing the identification of several QTLs controlling the trait. However, to date, no genes underlying fruit shape QTLs have been conclusively identified, nor has their function been verified in pepper. We constructed a mapping population from a cross of round- and elongated-fruited C. annuum parents and identified a single major QTL on chromosome 10, termed fs10, explaining 68 and 70% of the phenotypic variation for fruit shape index and for distal fruit end angle, respectively. The QTL was mapped in several generations and was localized to a 5 Mbp region containing the ortholog of SlOFP20 that suppresses fruit elongation in tomato. Virus-induced gene silencing of the pepper ortholog CaOFP20 resulted in increased fruit elongation on two independent backgrounds. Furthermore, CaOFP20 exhibited differential expression in fs10 near-isogenic lines, as well as in an association panel of elongated- and round-fruited accessions. A 42-bp deletion in the upstream region of CaOFP20 was most strongly associated with fruit shape variation within the locus. Histological observations in ovaries and fruit pericarps indicated that fs10 exerts its effect on fruit elongation by controlling cell expansion and replication. Our results indicate that CaOFP20 functions as a suppressor of fruit elongation in C. annuum and is the most likely candidate gene underlying fs10.
Fruit shape is one of the most important quality traits affecting consumer preference and breeder goals in vegetable crops. Typically, wild fruit is small and has a round shape, whereas intensive breeding following domestication has resulted in a wide diversity of sizes and shapes (van der Knaap and Ostergaard, 2017).
Tomato (Solanum lycopersicum) is the most important model plant species for studying fleshy fruit development and morphology (Seymour et al., 2013), and for which numerous quantitative trait loci (QTLs) controlling fruit shape variation have been identified (reviewed by van der Knaap et al., 2014; Wang et al., 2015a). OVATE was the first isolated gene underlying a tomato fruit shape QTL, and a null mutation in the gene resulted in pear-shaped fruit (Liu et al., 2002). OVATE was determined as a negative regulator of cell division and a suppressor of plant growth, because its overexpression resulted in smaller organs (Liu et al., 2002). Recently, another member of the OVATE FAMILY PROTEINs (OFP), SlOFP20, was identified as underlying the tomato fruit shape QTL suppressor of ovate1 (sov1; Wu et al., 2018). OFP homologs have been identified in diverse plant species (Liu et al., 2014; Snouffer et al., 2020) and have been associated with regulation of fruit shape in melon and tuber shape in potato (Wu et al., 2018). More recently, flat fruit of peach (Prunus persica) was also found to be associated with an inversion containing a member of the OFP family, PpOFP1, which is closely related to Arabidopsis thaliana OFP1 and SlOFP20, where its expression was activated by the inversion (Zhou et al., 2020). Furthermore, OFP members have been associated with regulation of organ growth in rice (Xiao et al., 2017; Yang et al., 2018) and Arabidopsis (Li et al., 2011; Zhang et al., 2016), as well as with variation in plant architecture and response to abiotic stresses and hormones (Snouffer et al., 2020). Using a yeast two-hybrid screen, TONNEAU1-recruiting motif (TRM) proteins were identified as interacting with OVATE and SlOFP20 (Wu et al., 2018). TRM LONGIFOLIA homologs have been associated with variation in leaf and grain morphology in Arabidopsis and rice, respectively (Lee et al., 2006; Wang et al., 2015b). Collectively, these results indicate the involvement of the OFP–TRM module in controlling diverse aspects of plant organ morphology in multiple plant species.
A second gene family associated with control of fruit shape is IQ67 domain (IQD). The tomato fruit shape QTL SUN belongs to this family and mediates fruit elongation primarily by controlling cell-division patterns (Xiao et al., 2008; Wu et al., 2011). Interestingly, sun is a gain-of-function mutation mediated by inserting a retrotransposon near the locus. Association of IQD homologs and fruit shape has also been demonstrated in the cucurbits (Pan et al., 2017; Dou et al., 2018). In addition to SUN, OVATE, and SlOFP20, which were identified as underlying tomato fruit elongation QTLs, a fourth locus, fs8.1, has been fine-mapped to tomato chromosome 8 but the underlying gene has not yet been identified (Sun et al., 2015). The involvement of FRUITFULL-like MADS-box gene in regulating fruit elongation was demonstrated in cucumber (Zhao et al., 2019), but its function in regulating Solanaceae fruit shape is not known. LOCULE NUMBER (LC) and FASCIATED (FAS) synergistically control flat fruit shape and locule number, and have a pleiotropic effect on fruit weight and shape (Cong et al., 2008; Huang and van der Knaap, 2011; Munos et al., 2011). WUSCHEL, which is required for stem cell identity in meristems, underlies the lc mutation, whereas CLAVATA3 underlies the fas mutation (Munos et al., 2011; Xu et al., 2015). Together, these genes control various developmental aspects, such as cell division, number of carpels, meristem size, and organization, which contribute to the morphological diversity of the fruit (van der Knaap and Ostergaard, 2017).
Pepper (Capsicum spp.), a member of the Solanaceae family, exhibits vast natural variation in fruit shape that is utilized for breeding of diverse fruit types. The genetic and molecular bases of the natural variation in pepper fruit shape have been analyzed mainly by QTL mapping studies (Ben Chaim et al., 2001, 2003a,b; Rao et al., 2003; Zygier et al., 2005; Barchi et al., 2009; Borovsky and Paran, 2011; Yarnes et al., 2013; Han et al., 2016; Chunthawodtiporn et al., 2018; Du et al., 2019; Lee et al., 2020). These studies detected two major fruit elongation QTLs, fs3.1 and fs10.1, on chromosomes 3 and 10, respectively, and additional, more minor QTLs on multiple chromosomes. Analysis of genomic population structure in C. annuum revealed multiple fixed regions that are conserved in non-pungent varieties and partly overlap with known QTLs for fruit weight and shape in tomato (Hill et al., 2017). Furthermore, based on expression and QTL colocalization, candidate genes putatively associated with fruit morphology were identified (Hill et al., 2017). Genome-wide association study (GWAS) of 220 C. annuum accessions revealed four loci associated with fruit shape attributes, including a non-synonymous mutation in the gene LONGIFOLIA1-like on chromosome 3 (Colonna et al., 2019). Furthermore, a pepper homolog of OVATE was shown to be associated with fruit shape variation by downregulation using virus-induced gene silencing (VIGS; Tsaballa et al., 2011). However, it is not known whether this gene is associated with the natural variation in pepper fruit shape. Thus, to date, no genes underlying fruit weight and shape QTLs have been conclusively identified or their function verified in pepper. The objective of the present study was to dissect the phenotypic, genetic, and molecular bases for the extreme natural variation in C. annuum fruit shape. QTL mapping enabled the identification of a major fruit shape QTL, fs10. We provide evidence for the pepper ortholog of tomato SlOFP20 underlying fs10 and describe the cellular mechanism by which fs10 exerts its effect on regulation of fruit shape.
For QTL mapping of fruit shape, we constructed an intraspecific C. annuum F2 population from the cross of two parents that exhibit extreme variation in fruit shape. Inbred line 1901 (long sweet CGN23289, kindly provided by Dr. A. Bovy, Wageningen University, Netherlands) has very long and narrow fruit and inbred line 5226 has small round fruit. For initial mapping, an F2 population of 240 progenies was grown in the greenhouse and measured by digital caliper for fruit maximal length and width, fruit shape index (length/width), and the angle of the distal end at 5% from the fruit tip by Tomato Analyzer software version 3.0.1 For fine mapping of the QTL, selected F2 plants were self-pollinated until the F4 generation in which markers in the vicinity of the QTL were fixed for the parental alleles. For construction of QTL near-isogenic lines (QTL-NILs), selected F4 plants that were heterozygous at the most closely linked marker to the QTL (130 Mbp) were self-pollinated and fixed for alternate parental alleles in the F5 generation. All other chromosome 10 markers were monomorphic in the QTL-NILs.
We employed bulked segregant analysis using RNA sequencing (BSAseq) for QTL mapping by constructing bulks based on the phenotypic distribution of the F2 population (Borovsky et al., 2019). Three bulks composed of 13–15 plants per bulk for each of the phenotypic extremes were constructed and RNA was extracted from ovaries at anthesis for each of the six bulks. Total RNA was extracted using the GeneElute™ Mammalian Total RNA Extraction Miniprep Kit (Sigma). Genomic DNA was removed by DNaseI (Sigma) treatment. RNA sequencing was performed on a lane of 60-bp single-end reads in an Illumina HiSeq 2500 System at the Weizmann Institute of Science, Israel.
Putative QTL peaks were identified by plotting the number of homozygous single-nucleotide polymorphisms (SNPs) for each phenotypic extreme across the genome. Then, specific SNPs derived from the RNAseq data were genotyped in the entire F2 population (Supplementary Table S1) and QTL mapping was performed using MapQTL software version 52 with the interval mapping and Multiple QTL Model (MQM) functions. To study the interaction between CaOFP20 and CaOVATE, we used an F3 population of 154 individuals and two-way ANOVA in JMP V.14.
To study the expression pattern of the QTL candidate gene in a wide pepper germplasm, we made use of the pepper G2P-SOL core collection that consists of 450 accessions representing the global diversity of this species.3 Based on preliminary visual assessment of the fruit shape index, we chose a subset of 30 C. annuum accessions representing elongated and round fruit. We further employed association mapping of fruit shape index and shape categories in a panel of 286 accessions from the G2P-SOL core collection. Fruit shape categories were determined by visual assessment of the fruits. For GWAS, we used the genotyping-by-sequencing (GBS) markers described in Tripodi et al. (2021), as well as specific DNA polymorphisms at the QTL candidate gene (Supplementary Table S1).
SNP calling for an association mapping population of 286 pepper accessions was carried out by Tassel 5.0 GBS v2 pipeline4 using the genome reference of Zunla-1 C. annuum L.5 and BWA software. The proportion of taxa with a genotype (minimum locus coverage) was set to 0.1 and minor allele frequency (MAF) was 0.01. The GWAS was performed using the mixed linear model by including kinship matrix in TASSEL 5.2.57 (Bradbury et al., 2007).
Raw reads from ovary RNA sequencing of the bulks were subjected to a filtering and cleaning procedure as described by Borovsky et al. (2019). Gene abundance was estimated using Cufflinks (Trapnell et al., 2010, v. 2.2) combined with gene annotations from the Pepper Genome Platform.6 Gene-expression values were computed as FPKM. Differential expression analysis was performed using the DESeq2 R package (Love et al., 2014). Genes with an adjusted p-value of no more than 0.05 were considered differentially expressed. The gene sequences were used as a query term for a search of the NCBI non-redundant (nr) protein database that was carried out with the DIAMOND program (Buchfink et al., 2015). Homologous sequences were also identified by searching against the Heinz 1706 S. lycopersicum genome v.37 with the BLAST tool and an E-value threshold of 10−5. The search results were imported into Blast2GO version 4.0 for gene ontology (GO) assignments. The GO-enrichment analysis was carried out using the Blast2GO program based on Fisher’s Exact Test with multiple testing correction of false discovery rate (FDR). KOBAS 3.0 tool8 was used to detect the statistical enrichment of differentially expressed genes in the KEGG pathway and GO analysis.
For transcriptome SNP analysis, mapped bam files were preprocessed prior to performing SNP identification with Samtools/Picard Tools (Li et al., 2009)9 to mark duplicates, sort and add read groups. We performed local realignment to minimize false positives during the SNP-calling procedure (McKenna et al., 2010). Then, we performed genotype calling using Genome Analysis Toolkit, (GATK v.3.7) with the HaplotypeCaller option.
Expression levels of specific genes were determined by quantitative (q) RT-PCR in a Rotor-Gene 6000™ thermal cycler (Corbett Research, Australia) with at least five biological replications. Expression levels were normalized relative to CaUbiquitin (DQ975458.1). The PCR primers are listed in Supplementary Table S1.
A 296-bp fragment of CA10g10680 amplified by primers OFP-Kpn-F and OFP-Xba-R (Supplementary Table S1) was cloned into the pTRV2 vector previously digested with the restriction enzymes KpnI and XbaI. Empty vector pTRV2 served as a negative control. As a positive control, we used pTRV2:CaPDS, which contains the phytoene desaturase sequence and induces a photobleaching phenotype. The vectors were used to transform Escherichia coli DH5α competent cells, which were then plated on selective media (with 50 μg/ml kanamycin). After incubation at 37°C for 18 h, colonies were PCR-screened for the presence of the modified vectors using pTRV2-specific primers (PYL156-F and PYL156-R, Supplementary Table S1). Plasmid DNA from positive transformant colonies was isolated, digested with Kpn1 and XbaI, and analyzed on an agarose gel to check for fragment release from the pTRV2 vector. Plasmids from positive colonies were sequenced to verify successful vector construction.
For Agrobacterium infiltration, pTRV1, pTRV2, and the recombinant plasmids (pTRV2:CaOFP20 and pTRV2:CaPDS) were transformed into Agrobacterium tumefaciens strain GV3101 using the freeze–thaw method. A 4-ml culture of each construct was grown at 28°C for 2 days in YEP medium containing 50 μg/ml kanamycin and 30 μg/ml gentamicin. The cultures were then diluted with YEP medium containing acetosyringone (0.01%, 200 mm), MES (1.0%, 1 M) and kanamycin and incubated at 28°C with shaking at 200 rpm overnight. On day of treatment, the cultures were centrifuged at 4000 rpm for 8 min, the supernatants were discarded and the pellets were resuspended in MMA medium [2.0% sucrose, 0.5% MS salts without vitamins, 1.0% MES (1 M), 0.1% acetosyringone (200 mm)] depending on the cell density to ensure OD600 = 0.8. Prior to the treatment, cell solutions were incubated in the dark for 4 h at room temperature.
Pepper seedlings were germinated in a speedling tray for 20 days before Agrobacterium infiltration. At the time of infiltration, the seedlings (20 each for negative and positive controls and 40 for gene silencing) had fully developed cotyledons while true leaves had only initiated. One day prior to infection, the seedlings were moved into a growing room with long-day conditions (16 light, 8 dark) at 16°C. The leaf infiltration media contained the pTRV1 plasmid and each of the pTRV2 plasmids mixed in a 1:1 ratio. For each seedling, both cotyledons were infiltrated with the prepared solutions using a 1-mL needleless syringe. Two weeks after inoculation, the seedlings were transplanted into 500-mL plastic pots and continued to grow under long-day conditions at 25°C, for a total of 7–8 weeks. Flowers were labeled at anthesis; 3 weeks after anthesis, fruit were removed and measured for fruit shape, and the tissues were harvested for RNA extraction to determine gene-expression level.
Fresh samples of ovaries at anthesis and fruit pericarp at 3 weeks after anthesis were collected from the QTL-NILs and stored in FAA solution for 24 h. For histological analyses, samples were processed as described by Singh et al. (2019). Briefly, samples were dehydrated in an ethanol dilution series, embedded in paraffin wax, and sectioned with a microtome (Leica RM2245, Leica Biosystems, Germany). For ovaries and pericarps, 12 μm- and 20 μm-thick longitudinal and transverse sections, respectively, were made. The sections were then deparaffinized in Histo-Clear solution, rehydrated, and used for histochemical staining with Safranin Fast Green. Sections were examined under a light microscope (Leica DMLB) equipped with a Nikon DS-Fi1 digital camera. Image analyses for calculations of cell number, size, shape, and cross-sectional distances were done by the software ImageJ (NIH, United States) and NIS-Elements (Nikon).
Cell area, length, width, and shape index in the ovary and pericarp were measured in both the longitudinal and transverse sections. For longitudinal sections, the ovaries were divided into four zones, indicated by red lines in Supplementary Figures S1A,B. All cell parameters were measured in the four zones along the red lines in the ovary wall on one side of the ovary. Because the cell attributes were similar in zones 1, 2, and 3, the values for these three zones were averaged. For calculation of cell size in each zone, cells were counted in four squares of 100 μm × 100 μm in a grid in the middle of the ovary wall in areas without vascular tissue. Cell area was defined as the square area divided by the number of cells within the square. A total of 20 measurements were done for each zone in each NIL (five fruit per NIL, four squares per zone). Cell length, width, and shape index (length/width) were measured for 20 random cells per fruit in each zone in five fruit (a total of 100 cells per zone for each NIL). Cell attributes in the transverse sections were measured in zone 2 in the middle of the ovary. Cell number and distance across the ovary wall along the medio-lateral axis were measured along the red lines in the four zones. Cell number and distance along the proximal-distal axis of the entire longitudinal sections of the ovaries (black line in Supplementary Figures S1AB), as well as ovary maximum length, ovary maximum width, and ovary shape index were measured for intact ovaries. For the fruit pericarp, the same cell attributes were measured in three zones marked by red lines at 1/3, 2/3, and 3/3 positions (Supplementary Figure S1C). In each zone, a slice of 10 mm was excised from the pericarp and sectioning was done in the longitudinal direction on one side of the slice and in the transverse direction on the second side of the slice. For calculation of cell size in each zone, cells were counted in four squares of 500 μm × 500 μm in a grid in the middle of the pericarp in areas without vascular tissue.
To construct a QTL mapping population, we crossed line 1901 with an elongated fruit shape, and line 5226 with a small round fruit (Figure 1A; Table 1). Fruit shape index of the F2 progeny ranged from 0.9 to 6.4, with a mean of 2.5 indicating partial dominance of the round fruit shape. For initial QTL mapping, we employed BSA (Michelmore et al., 1991) using RNAseq of ovaries at anthesis from the two phenotypic extremes of fruit shape index in the F2 population. The mean fruit shape indices of the two bulks were 4.5 ± 0.9 and 1.3 ± 0.2 for the elongated- and round-fruited bulks, respectively. Anthesis was chosen for RNA extraction because it is a well-defined developmental stage at which differences in ovary shape are clearly observed (Figure 1B). In addition to fruit shape index which reflects the degree of fruit elongation, we measured the angle at the distal end of the fruit, which reflects the difference between elongated and round shape at that site (Table 1). The angles of the distal end of the F2 progeny ranged from 17.3o to 160.2o (Table 1). Values for fruit shape index and the angle of the distal end were strongly correlated (r = −0.83), suggesting common genetic control underlying these traits. Fruit shape index was highly correlated with fruit length (r = 0.93) and moderately correlated with fruit width (r = −0.5), indicating that fruit length is the predominant factor determining fruit shape index in the population.

Figure 1. Fruit and ovaries of parents and near-isogenic lines (NILs) used in this study. (A) Ripe fruit of the mapping parents 5226 and 1901. (B) Ovaries at anthesis of the mapping parents 5226 and 1901. Ovary photos were taken by a Leica MZFLIII binocular. (C) Ripe fruit of the fs10-NILs 34-18R and 34-2 L. Scale bar = 5 cm.
A total of 2,517 homozygous SNPs were identified between the bulks (Supplementary Table S2). Plotting the number of homozygous SNPs differentiating the bulks across the genome indicated a major peak in a 27-Mbp region between 142 and 169 Mbp on chromosome 10 (Zunla genome, Qin et al., 2014; Figures 2A,B). Several additional minor peaks were identified at the beginning of chromosome 10 and across the genome which may represent minor QTLs affecting the trait (Figure 2A). We genotyped the F2 population with SNP markers derived from the RNAseq data flanking the putative QTL (Supplementary Table S1), and use of MapQTL software and MQM mapping enabled the identification of a single major QTL in the interval of 127–144 Mbp termed fs10, 130–132 Mbp being the most significant markers for fruit length, fruit shape index, and angle of the distal fruit end (Figures 3A,B; Table 2).
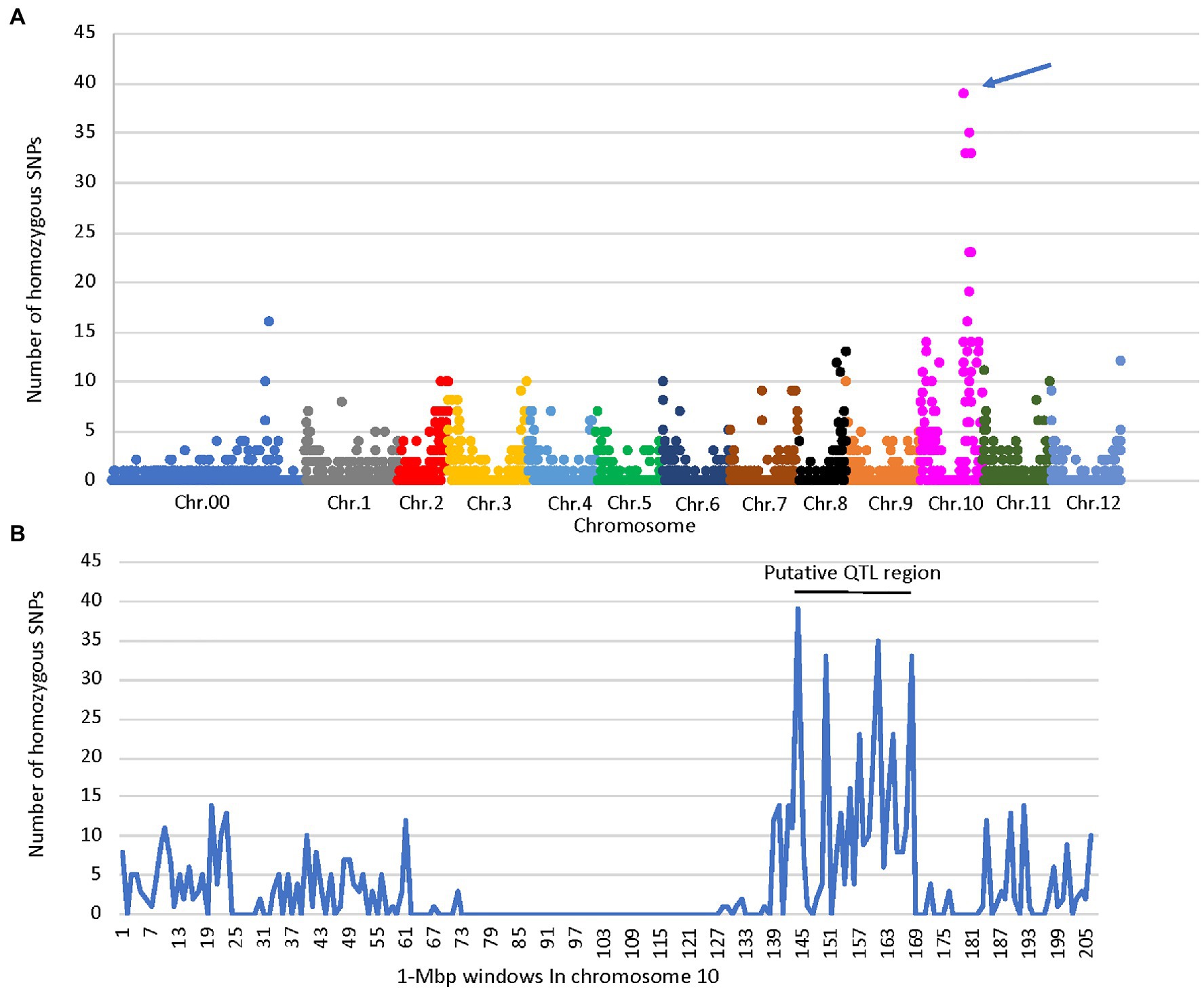
Figure 2. Distribution of homozygous SNPs identified by RNAseq of the elongated- and round-fruited bulks. (A) Distribution of SNPs on each chromosome in 1 Mbp windows, indicating a QTL peak marked by an arrow on chromosome 10. (B) Distribution of SNPs on chromosome 10 in 1 Mbp windows, indicating a putative QTL region at 142–169 Mbp. Chr.00 – SNPs that were not assigned to chromosomes.
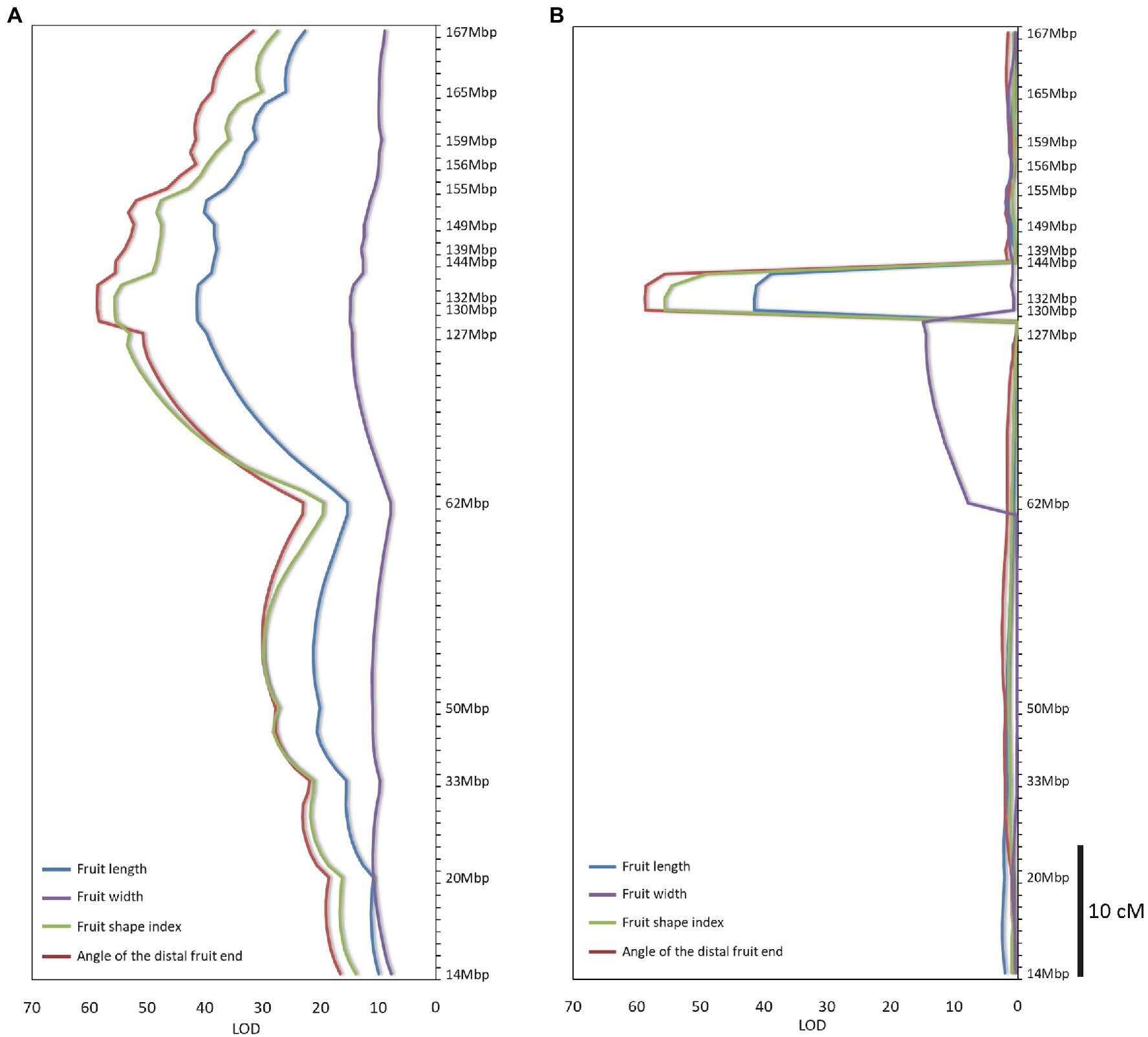
Figure 3. QTL mapping of fruit shape parameters in the F2 population. (A) Interval mapping. (B) MQM mapping. Significance threshold of LOD = 2.2 (p = 0.05) was determined for QTL detection by permutation test. Mapping analyses were performed by MapQTL software version 5 (Kyazma B.V., Netherlands).
To verify the mapping results based on the F2 generation and to further fine map the QTL, we fixed recombinant progeny in the F4 generation and phenotyped them for fruit shape index (Table 3). The data consistently indicated that the marker at 130 Mbp is the most tightly linked to the QTL because all of the lines possessing the 1901 allele at this marker had elongated fruit and all of the lines possessing the 5226 allele at this marker had round fruit. Line 16 allowed delineating the QTL to a 5-Mbp region between 127 and 132 Mbp.
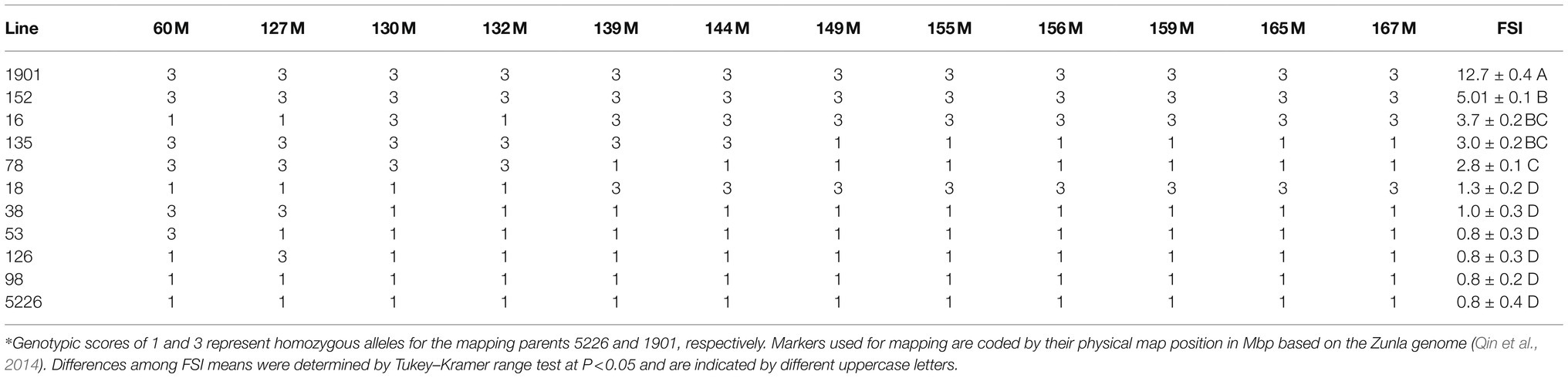
Table 3. Marker genotypes and fruit shape index (FSI) of fixed recombinant lines and the mapping parents in the fs10 region*.
For further characterization of fs10’s effect, we constructed NILs in the F5 generation that were fixed for contrasting alleles at the 130 Mbp marker (Figure 1C; Table 1). Flower organs and leaf shape of the NILs were measured at anthesis, and exhibited significant differences in the shape indices of the ovary, anther, style, and petals (Supplementary Table S3). These indicated that fs10 affects the shape of multiple flower organs and that organ shape differences are determined at early stages of flower differentiation prior to anthesis. No difference in leaf shape was observed between the NILs.
To identify candidate genes underlying the QTL, the 5-Mbp region flanking fs10 was examined for gene content (Zunla reference genome, Qin et al., 2014) as well as for differentially expressed genes based on transcriptome analysis of the F2 bulks. The QTL region contains 14 genes (Table 4), and the most likely candidate is Capana10g001230, the ortholog of SlOFP20 that controls fruit elongation in tomato (Wu et al., 2018). Because only a partial sequence of Capana10g001230 was available, we obtained the full-length sequence from the homologous gene CA10g10680 of the reference genome CM334 V.1.55 (Kim et al., 2014). Two genes were differentially expressed (Padj ≤ 0.05, Table 4) between the F2 bulks: Capana10g001225 coding for a non-specific lipid transfer protein (nsLTP) and Capana10g001244 coding for a transmembrane protein with unknown function. Neither gene is known for its involvement in fruit shape control in plants. Interestingly, Capana10g001230 was not differentially expressed between the bulks.
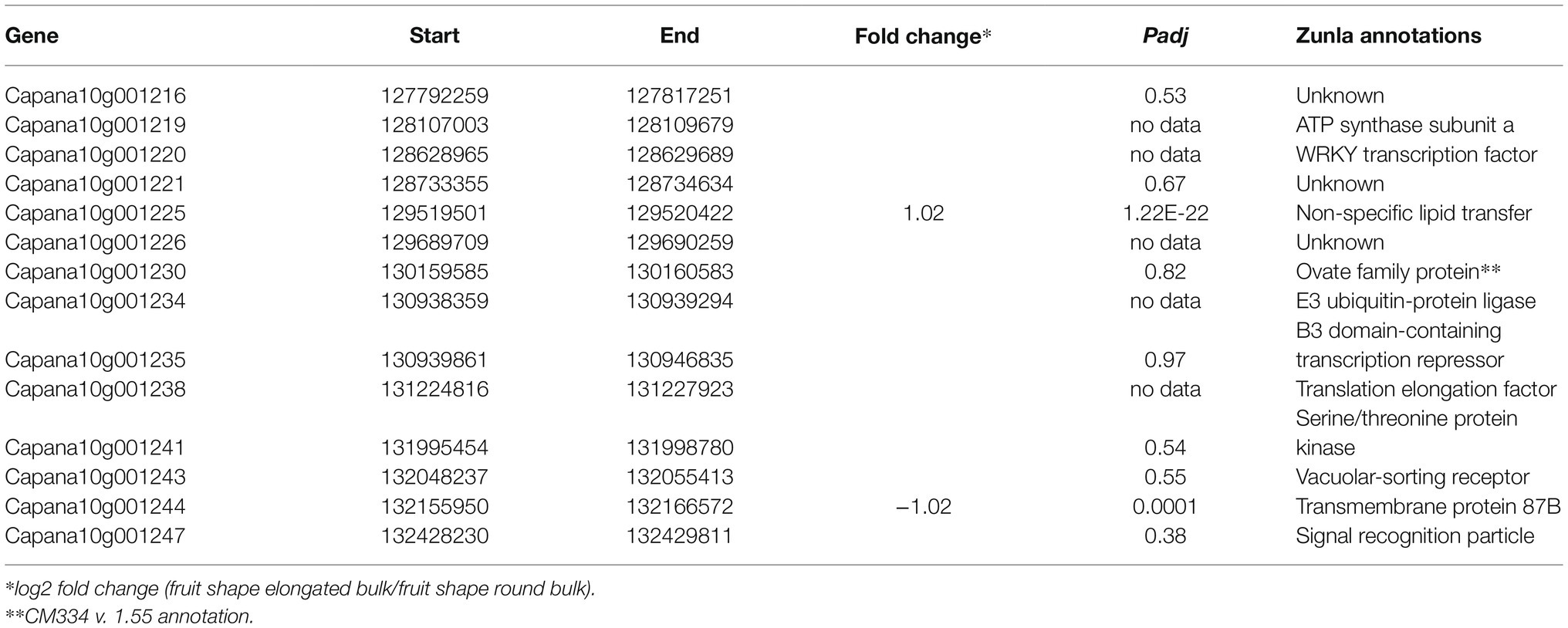
Table 4. Genes contained within the 5-Mbp fs10 region and their expression in RNAseq of the F2 fruit shape index bulks.
Because we knew that SlOFP20 is involved in controlling tomato fruit shape, we further focused on the role of its ortholog Capana10g001230 (termed CaOFP20) in determining fruit shape variation in pepper. Sequencing the open reading frame (ORF) of the gene in the parents of the mapping population revealed a single non-synonymous K131E amino acid substitution leading to a change of lysine (basic amino acid) in 1901 to glutamic acid (acidic amino acid) in 5226. This substitution is not included within the conserved OVATE domain at the C terminus of the protein (Liu et al., 2002).
Examination of the expression pattern of CaOFP20 throughout flower and fruit development in line 642110 (Liu et al., 2017) indicated that the gene is expressed at its highest level during flower development at anthesis but is also highly expressed at later stages of fruit pericarp and placenta development (Supplementary Table S4). We therefore measured its expression in three tissues of the QTL-NILs: ovary at anthesis, and fruit pericarp and placenta at 3 weeks after anthesis. In all three tissues, CaOFP20 exhibited significantly higher expression in the round-fruited NIL compared to the elongated-fruited one (Figure 4A), consistent with the role of SlOFP20 as a suppressor of fruit elongation in tomato (Wu et al., 2018).
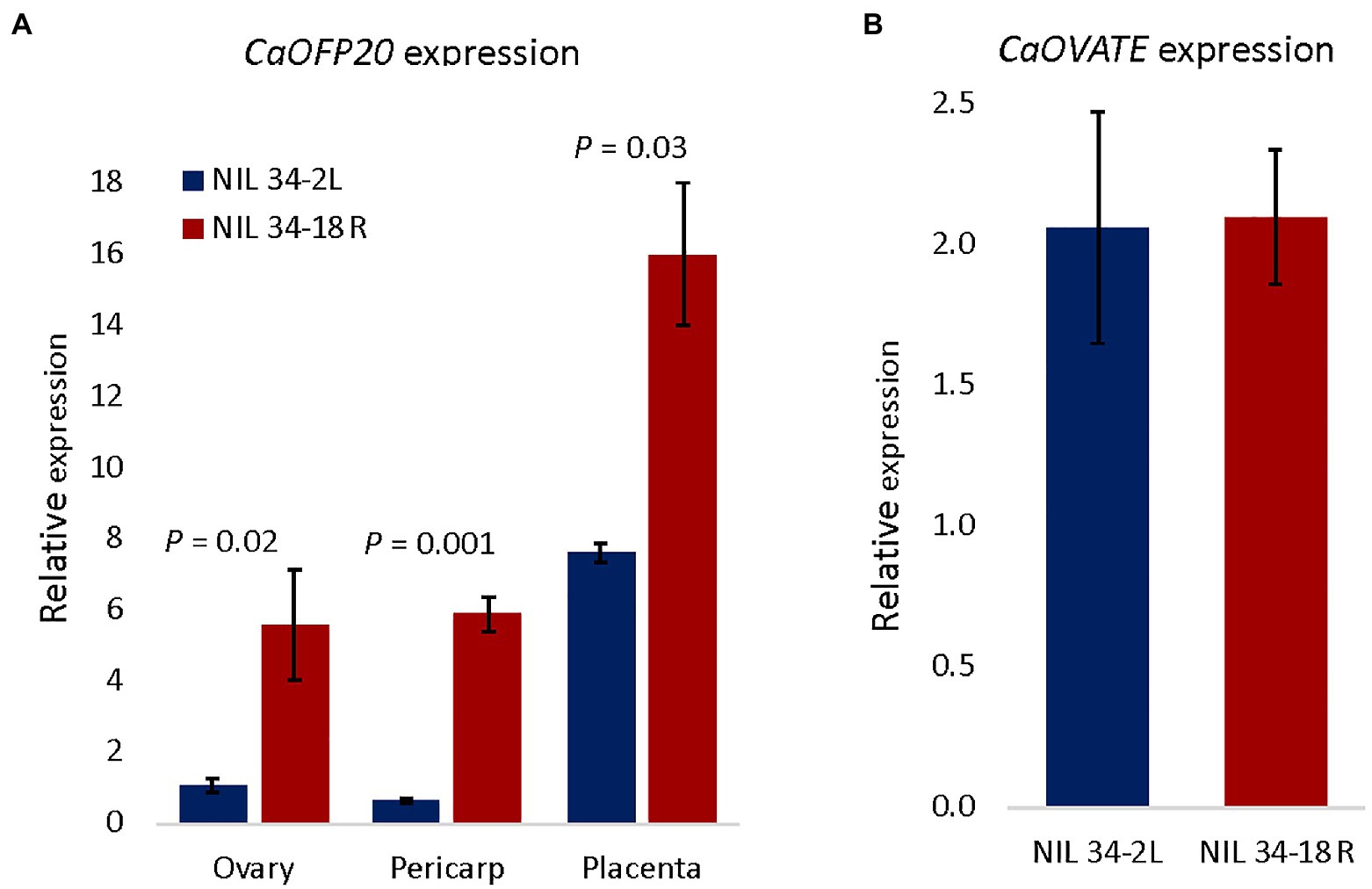
Figure 4. Expression of CaOFP20 is downregulated in the elongated fruit fs10-NIL 34-2L, whereas no difference was observed for CaOVATE. (A) Expression of CaOFP20 in ovary at anthesis and in pericarp and placenta of the fruit 3 weeks after anthesis. (B) Expression of CaOVATE in ovary at anthesis. Significant differences between the NILs were determined by Student’s t-test.
Because SlOFP20 had been identified as underlying the suppressor of ovate (sov1) mutation in tomato, we determined whether the expression of the pepper OVATE ortholog (CaOVATE, Capana02g002672) is altered in the NILs; no difference in expression level of this gene was observed at anthesis (Figure 4B). Very low levels of expression of CaOVATE were observed in the pericarp and placenta. To further assess the relationship between CaOFP20 and CaOVATE in determining fruit shape index, we scored both markers in the F3 generation and tested their possible interaction by two-way ANOVA. While CaOFP20 had the expected major effect on fruit shape index (p < 0.0001), CaOVATE had a minor effect on the trait (p = 0.02), and the interaction was not significant (p = 0.06).
To determine whether CaOFP20 underlies fruit shape variation, we employed VIGS to knock down its expression in the blocky fruited cv. Maor and changes in fruit morphology were examined 3 weeks after anthesis. Out of 20 fruit examined, 7 exhibited an elongated shape (Figures 5A,B). Silenced elongated fruits were longer and narrower than the control fruit (inoculated with empty vector), resulting in a mean fruit shape index of 1.8 ± 0.3 and 1.0 ± 0.1 for the elongated and control fruit, respectively. Expression of CaOFP20 was examined by qRT-PCR in ovaries at anthesis and in fruit pericarp and placenta tissue 3 weeks after anthesis. In all three tissues, CaOFP20 exhibited significantly lower expression levels in the elongated genotypes (Figure 5C).
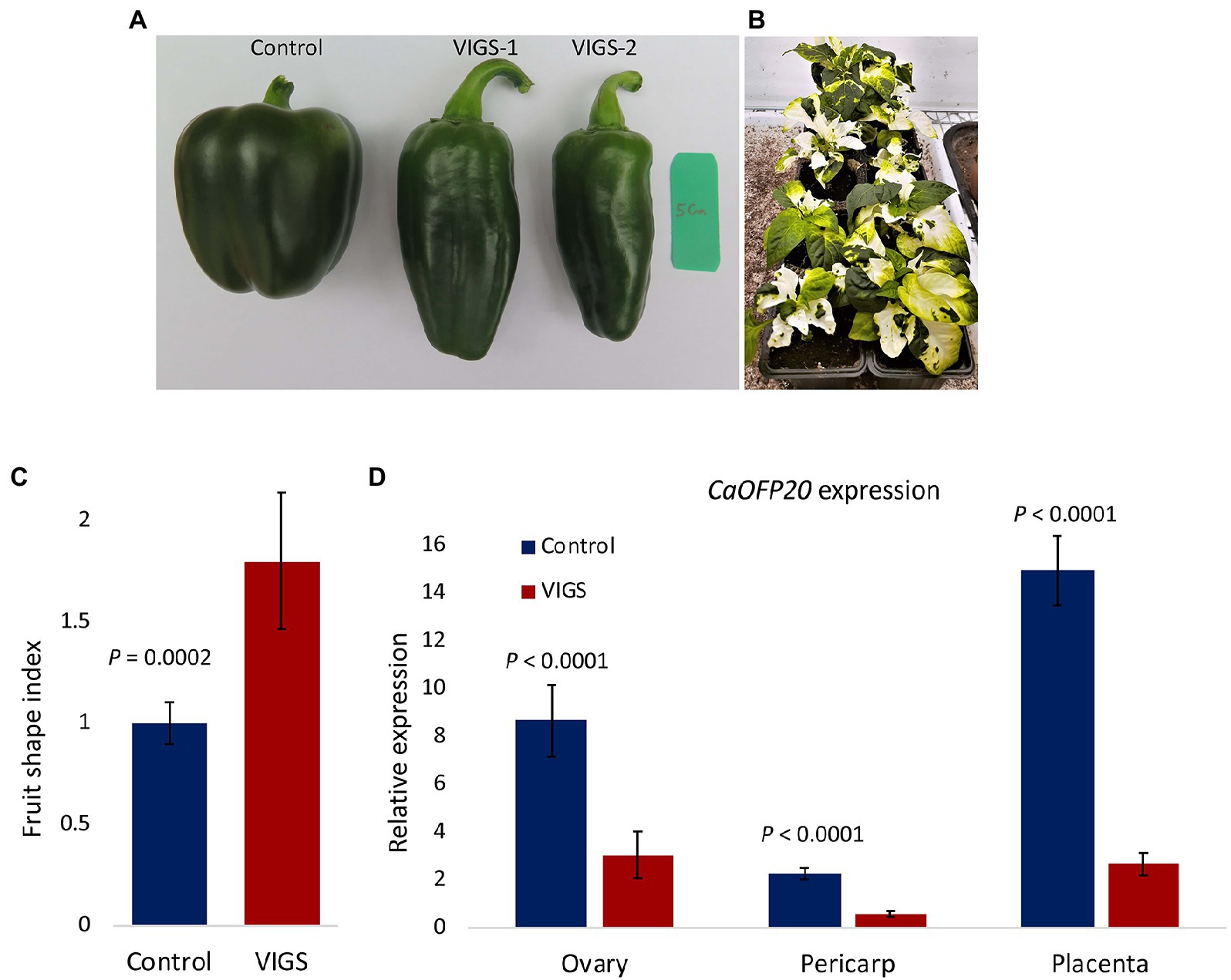
Figure 5. Silencing of CaOFP20 in cv. Maor by VIGS. (A,C) Silencing of CaOFP20 results in fruit elongation. Control – inoculation with empty vector. VIGS-1 and VIGS-2 are examples of silenced fruit 3 weeks after anthesis. (B) Silencing CaPDS as a positive control. Silenced plants did not set fruits. (D) Expression of CaOFP20 is downregulated in silenced elongated fruit. Significant differences between the silenced and control fruit were determined by Student’s t-test.
To test the effect of silencing CaOFP20 on the background of a round fruit, we employed VIGS on the round-fruited line 4590. Out of 20 fruit examined, 7 had an elongated shape (Figure 6A). Silenced elongated fruits were longer and narrower than the control fruits (inoculated with empty vector), resulting in a fruit shape index of 1.59 ± 0.3 in the silenced fruit compared to 0.8 ± 0.06 in the control fruit (Figure 6B). Pericarp thickness was significantly thinner in the elongated fruit (Figure 6C), but no difference in fruit weight was observed (Figure 6D). Expression of CaOFP20 in the pericarp 3 weeks after anthesis was significantly reduced in the elongated fruit (Figure 6E), similar to the silenced “Maor” fruit. Taken together, silencing of CaOFP20 by VIGS provided evidence for this gene’s function as a suppressor of fruit elongation in blocky and round pepper.
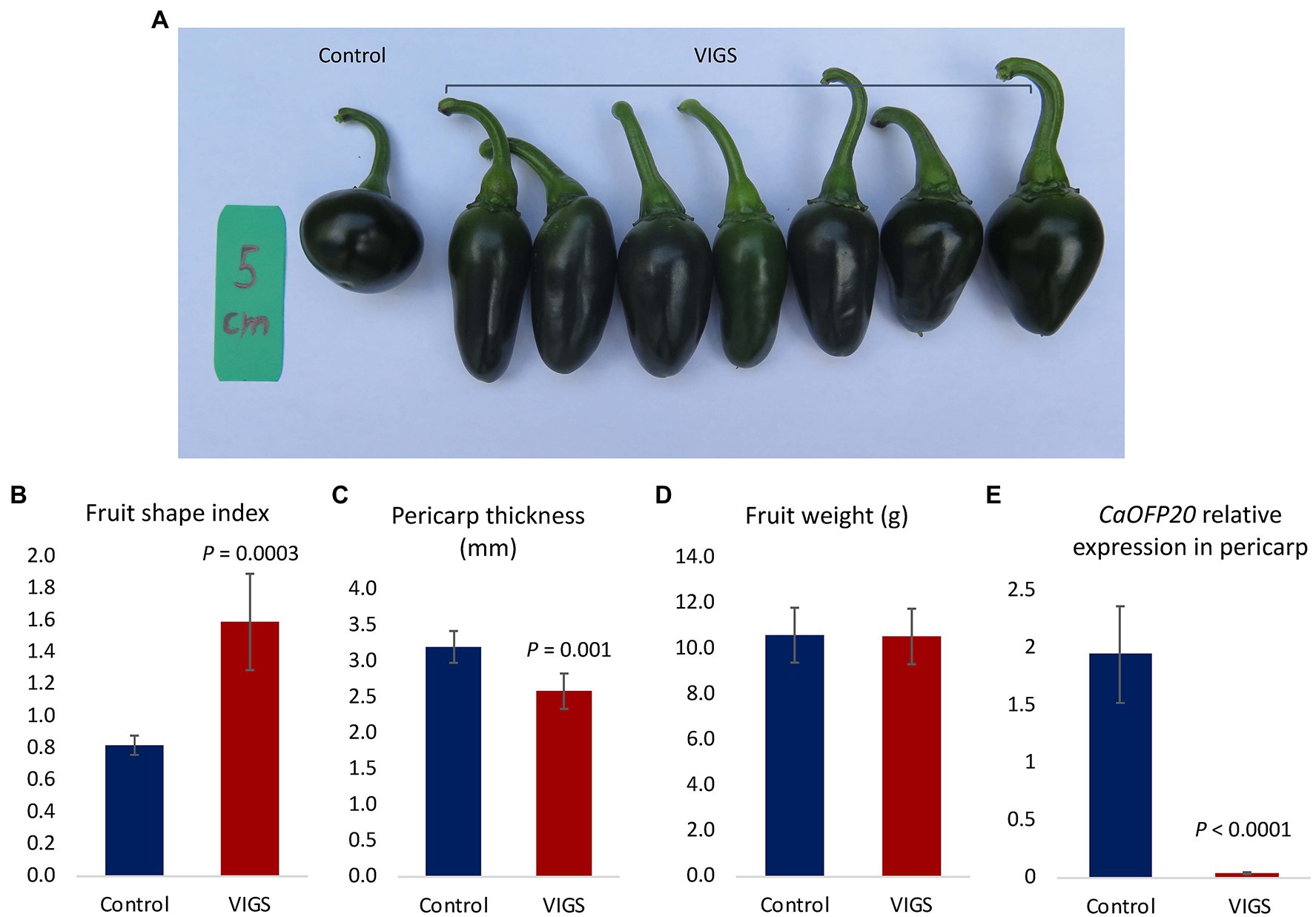
Figure 6. Silencing of CaOFP20 in line 4590 by VIGS. (A) Silencing of CaOFP20 results in fruit elongation. Control – inoculation with empty vector. VIGS – examples of silenced fruit 3 weeks after anthesis. (B–D) Fruit shape index, pericarp thickness, and fruit weight of silenced and control fruit. (E) Expression of CaOFP20 is downregulated in silenced elongated fruit. Significant differences between the silenced and control fruit were determined by Student’s t-test.
To test whether the expression pattern of CaOFP20 is associated with natural variation in fruit shape, we selected 30 C. annuum accessions representing elongated and round fruit from the pepper G2P-SOL core collection. The mean fruit shape indices of the elongated- and round- fruited accessions were 11.09 ± 3.6 and 0.92 ± 0.13, respectively (Figure 7A). We performed qRT-PCR of CaOFP20 using RNA from fruit pericarp tissues 3 weeks after anthesis. The expression level of the gene was significantly higher in the round-fruited accessions than in the elongated ones (Figure 7B), and expression level was strongly negatively correlated with fruit shape index (r = −0.82), indicating that CaOFP20 is widely associated with fruit shape variation in C. annuum.
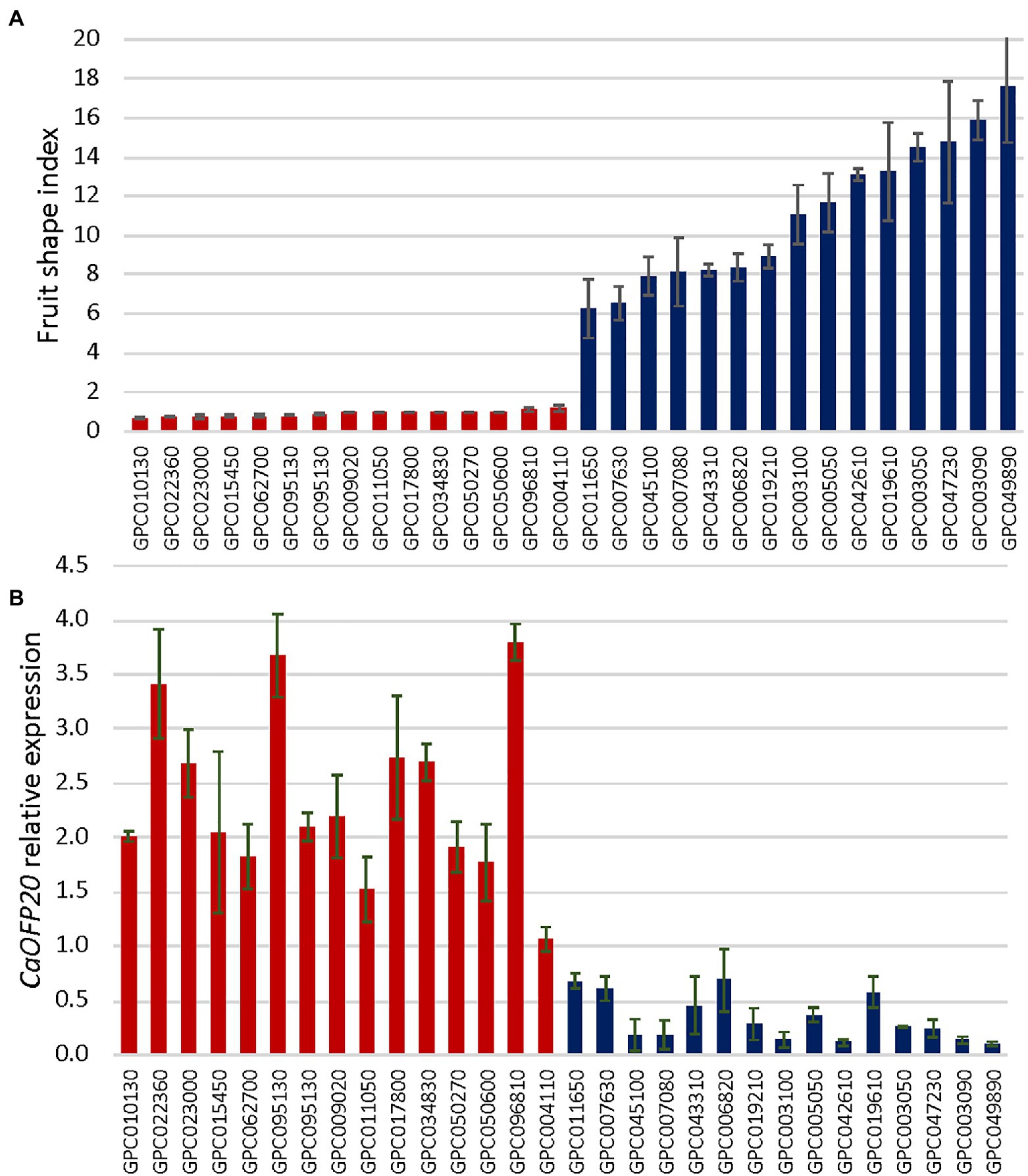
Figure 7. Fruit shape index is associated with expression level of CaOFP20 in a panel of round- and elongated-fruited accessions. (A) Ripe fruit shape index of accessions in the panel. (B) Expression of CaOFP20 in fruit pericarp 3 weeks after anthesis. Error bars represent standard deviation of three fruit per accession. Columns of round- and elongated-fruited accessions are colored in red and blue, respectively.
To identify additional polymorphisms at the CaOFP20 locus, we exploited the 30X resequencing genomes of the two parental lines of the mapping population as part of the G2P-SOL project. Within 50 kbp of the intergenic region upstream of CaOFP20, we detected several SNPs and a deletion of 42 bp at position 130121672 bp, 38381 bp upstream of the start codon of the gene in line 1901. Putative cis-acting elements could be found in the SNP regions as well as within the 42 bp InDel (Supplementary Tables S5, S6). Furthermore, sequencing the ORF of CaOFP20 in the 30 elongated- and round-fruited accessions revealed five non-synonymous substitutions (Table 5). We developed molecular markers based on the promoter InDel and the ORF substitutions (Supplementary Table S1) and genotyped all 286 accessions. These genotypic data allowed us to identify six haplotypes consisting of at least five accessions per haplotype based on the CaOFP20 locus (Table 6). Haplotypes a and b were clearly distinguished from the others in their effect on fruit shape, associated with circular and oblate shapes, respectively (Figures 8B,C). Haplotypes a and b had the same amino acid substitutions and differed in presence/absence of the promoter InDel (Table 6). Most of the accessions with circular fruit shape did not carry the 42-bp deletion, while all other accessions with various fruit shape categories, carried the 42-bp deletion. GWAS revealed a highly significant association of fruit shape categories with CaOFP20 markers on chromosome 10 (Table 5; Figure 8A), whereas no association of CaOFP20 markers was observed for fruit shape index (data not shown). Among the CaOFP20 markers, the InDel marker was most strongly associated with fruit shape (p = 9.86 × 10−11, Table 5). Surprisingly, the most significantly associated marker with the trait in the panel was found at position 142898783 bp on chromosome 10 (p = 7.34 × 10−13), about 12 kb downstream of CaOFP20.
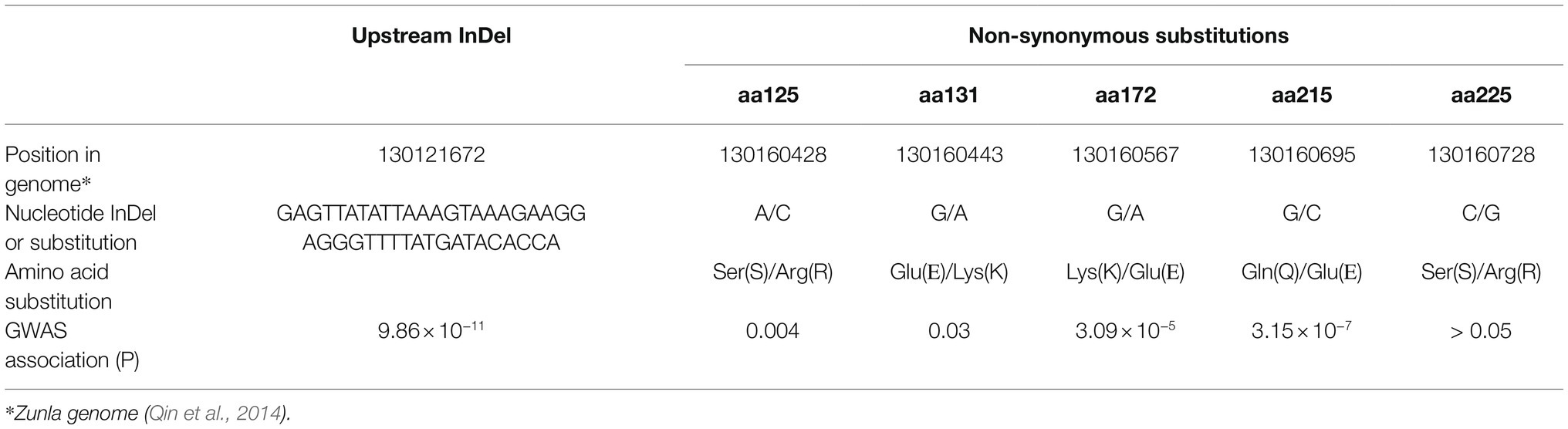
Table 5. InDel in the promoter of CaOFP20 and non-synonymous substitutions in its coding region, and their significance level for fruit shape categories in the association panel.
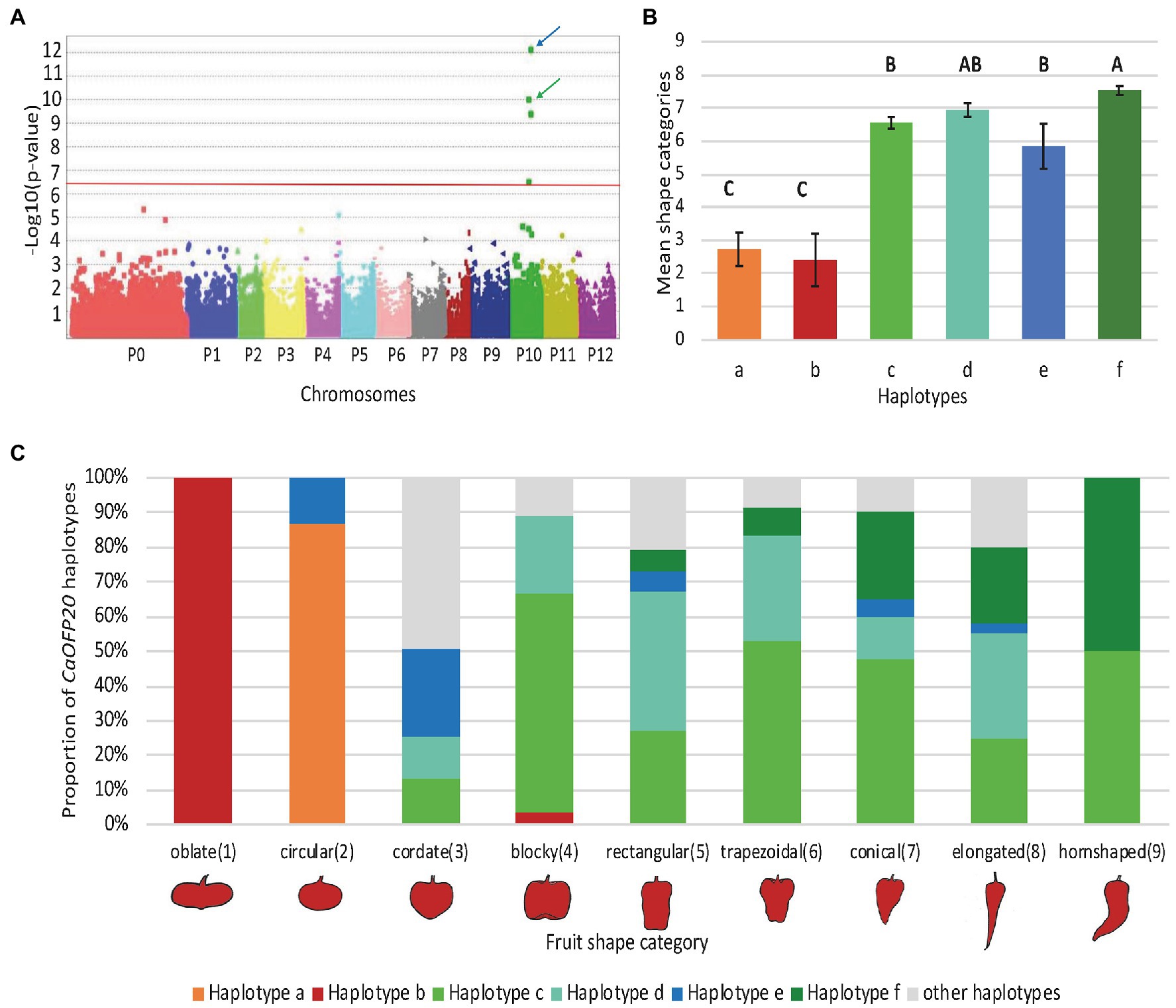
Figure 8. GWAS plot and haplotypes based on allelic variation at CaOFP20 and their association with fruit shape categories in the association panel. (A) GWAS plot for fruit shape categories in the association panel. Blue arrow indicates the SNP at position 142898783 bp, downstream of CaOFP20; green arrow indicates the most significant marker at the CaOFP20 locus at 130121672 bp. Red line represents the FDR-derived genome-wide significance threshold. P0 – SNPs that were not assigned to chromosomes. (B) Means of fruit shape categories of the different haplotypes. The six haplotypes in the association panel had at least five accessions per haplotype. Differences among means were determined by Tukey–Kramer range test at p < 0.05 and are indicated by different uppercase letters. (C) Haplotype composition of fruit shape categories.
To decipher the cellular mechanisms underlying fruit shape variation conferred by fs10, we analyzed cellular parameters of fs10-NIL ovaries at anthesis and fruit pericarp tissue 3 weeks after anthesis. Longitudinal and transverse sections were performed in four and three zones in ovaries and pericarps, respectively (Supplementary Figures S1A–C). The total distance and total number of cells between the proximal and distal ends of the entire ovary in longitudinal section, marked along the black line (Supplementary Figures S1A,B), did not change significantly (Supplementary Table S7). In contrast, maximum ovary width and ovary shape index were greater in NIL 34-18R and NIL 34-2 L, respectively. Average cell area, cell length, cell width, cell shape index, number of cell layers between internal and external epidermis, and distance between internal and external epidermis in the ovary wall in longitudinal sections were significantly increased in NIL 34-18R. Similarly, these parameters, except for cell shape index, were significantly increased in transverse sections of the NIL 34-18R ovaries. These results indicate that at anthesis, ovary cells in the round-fruited NIL were larger and more numerous than in the elongated-fruited NIL. In pericarp longitudinal sections (Supplementary Figure S1C), the predominant factors were cell length and cell shape index, which were significantly increased in NIL 34-2 L in accordance with the difference in fruit elongation (Supplementary Table S8). In transverse sections, the thicker pericarp of NIL 34-18R had a larger cell area and greater number of cell layers between the internal and external epidermis tissues. Taken together, fs10 exerts its effect on fruit shape by coordinately affecting multiple cellular mechanisms, including cell size, cell number, and cell shape.
Transcriptome analysis of the RNA bulks from ovaries at anthesis revealed 278 and 332 differentially upregulated and downregulated genes in the elongated-fruited vs. round-fruited bulk, respectively (Supplementary Table S9). GO analysis revealed significant enrichment of processes associated with photosynthesis, oxidation reduction, hormone metabolism, flavonoid synthesis, cell wall organization, protein binding, circadian rhythm, defense response, and fatty acid and lipid metabolism (Supplementary Figure S2). Hormone-enriched categories included response to auxin, auxin binding, auxin metabolism, cytokinin signaling, gibberellin (GA) signaling, response to GA, response to abscisic acid, response to jasmonic acid (JA), JA signaling and biosynthesis, and response to ethylene.
Thirty-five genes associated with cell wall organization were differentially expressed between the bulks, mostly upregulated in the round-fruited vs. elongated-fruited bulk and in accordance with the round-fruited NIL that had larger cells in the ovary (Supplementary Table S9). These genes coded for enzymes associated with biosynthesis, modification, and degradation of cell wall components and their involvement in cell wall extension, such as pectinesterase, beta-galactosidase, and polygalacturonase, and were either up- or downregulated in the round-fruited bulk. Two genes encoding extensin, a non-enzymatic protein associated with cell wall loosening, were upregulated in the round-fruited bulk. Additional genes associated with cell wall loosening were xyloglucan endotransglucosylase and expansin, which were downregulated in the round-fruited bulk.
GAs are plant hormones that control organ growth (Wang et al., 2017). Four genes associated with GA metabolism were differentially expressed between the bulks. Gibberellic acid 2-oxidase (GA2ox), associated with deactivation of GA, exhibited marked upregulation in the round-fruited bulk. Overexpression of GA2ox results in organ growth inhibition in several species, e.g., Arabidopsis and Jatropha, due to a decrease in endogenous bioactive GA (Hu et al., 2017). Similarly, 2-oxoglutarate and Fe(II)-dependent oxygenase, associated with structural modification of GA, were upregulated in the round-fruited bulk. In contrast, gibberellin 3-beta-hydroxylase 3, associated with GA activation, was upregulated in the elongated vs. round-fruited bulk.
Auxin is involved in the regulation of cell wall loosening and cell elongation (Majda and Robert, 2018; Wang et al., 2019). Eight genes associated with auxin signaling were differentially expressed between the bulks, mostly upregulated in the round-fruited bulk. These included several auxin response factors (ARFs), such as ARF2, ARF6, and ARF8. Mutations in ARF2 and ARF8 in Arabidopsis are associated with an elongated hypocotyl (Tian et al., 2004; Okushima et al., 2005). Other differentially expressed auxin-signaling genes were TIR1 and LAX1, which function in auxin reception and transport, respectively. Additional genes associated with auxin biosynthesis (indole-3-acetamide hydrolase) and conjugation (indole-3-acetic acid-amido synthetase GH3.5) were upregulated in the elongated-fruited bulk.
Cytokinin is involved in the control of cell division and organ growth, among other developmental processes (Wybouw and De Rybel, 2019). Cytokinin hydroxylase catalyzes the biosynthesis of trans-zeatin and was upregulated in the round-fruited bulk vs. elongated-fruited bulk in this study. Trans-zeatin functions in shoot growth in Arabidopsis (Kiba et al., 2013), and in early tomato fruit development (Matsuo et al., 2012); however, its precise mode of action is not known.
JA is involved in the control of plant defense as well as plant growth under stress conditions. JA further interacts with other hormones involved in controlling plant growth, such as cytokinin, GA, and auxin (Jang et al., 2020). Six upregulated genes involved in JA biosynthesis and signaling were differentially expressed in the round-fruited vs. elongated-fruited bulk.
Ethylene acts as a regulator of plant growth, promoting cell expansion, and division (Dubois et al., 2018). Ethylene’s effect on cell expansion is mediated by the cell wall loosening expansins. Eleven genes in the round-fruited bulk, involved in ethylene biosynthesis and signaling, were differentially expressed, mostly upregulated.
To decipher the control of natural variation in fruit shape in C. annuum, we crossed two parents that exhibit diverse fruit types and extreme variation in fruit elongation. By combining BSAseq and map-based approaches, we detected a single major QTL, fs10, on chromosome 10 explaining 69% of the phenotypic variation for fruit shape index in the population. Additional minor QTLs may be present throughout the genome, as implied from the genome-wide homozygous SNP distribution; however, MapQTL analysis revealed only one significant QTL on chromosome 10. fs10 likely corresponds to fs10.1 which has been mapped in interspecific crosses of C. annuum×C. chinense (Ben Chaim et al., 2003a; Borovsky and Paran, 2011). The most closely linked marker to fs10.1 was CT11 (Borovsky and Paran, 2011), corresponding to Capana10g001248 at position 132521846 in the Zunla genome, which is tightly linked to CaOFP20. Sequencing the ORF of CaOFP20 in the EMS elongated-fruited mutant E-1654 (Borovsky and Paran, 2011) did not reveal any mutation in the coding region of the gene. Thus, it is likely that another as yet unknown fruit shape gene is tightly linked to CaOFP20.
Interestingly, CaOFP20 at 130 Mbp mapped outside the major peak identified by BSAseq in the 142–169 Mbp region of chromosome 10 (Figure 2B). This discrepancy likely results from a bias of SNP frequency along chromosome 10; the CaOFP20 region had a very low content of polymorphisms compared to the high content of polymorphisms in the major peak area detected by BSAseq. Fine mapping using fixed recombinants clearly excluded the 142–169 Mbp region from containing the QTL (Table 3). It may be, however, that the peak discovered by the GWAS in the 142 Mbp region (Figure 8A) corresponds to an unknown QTL. Mining the 142 Mbp region for putative candidates did not reveal genes with known association to organ growth.
The parents of the mapping population and NILs differed not only in the degree of fruit elongation but also in the shape of the distal fruit tip; elongated fruit had more pointed fruit tips than the short and round fruit. QTL mapping and the high correlation between these two traits indicate that they are likely controlled by the same locus, suggesting that fruit elongation is associated with reduced angle of the distal end of the fruit. Compared to the parents of the mapping population that differed 13-fold in their fruit shape index, the QTL-NILs differed only 4-fold, and the maximal fruit shape index in the F2 generation differed only 2-fold. This indicates that additional unknown QTLs with low effects, as well as epistasis, likely contribute to the genetic control of the trait, as was demonstrated by the interaction of SUN, ovate, and fs8.1 (Wu et al., 2015), as well as of ovate and sov, in tomato (Wu et al., 2018).
Fine mapping localized fs10 to a 5-Mbp region that contains 14 genes, including CaOFP20. Due to prior identification of the tomato and potato CaOFP20 orthologs underlying tomato fruit shape and potato tuber shape (Wu et al., 2018), as well as its differential expression between the QTL-NILs, we chose this gene as a primary target for gene silencing to test its function in pepper. VIGS knockdown of CaOFP20 expression in both blocky and round-fruited backgrounds was associated with fruit elongation (Figures 5, 6). Unexpectedly, CaOFP20 did not exhibit differential expression in the ovary transcriptome analysis of the bulks, likely due to masking of its expression by other unknown genes in the genome. However, qRT-PCR of the QTL-NILs using three tissues (ovary, pericarp, and placenta), as well as pericarp tissue from 30 diverse accessions, showed its higher expression level in the round-fruited accessions (Figures 4, 7).
Resequencing the genomes of the two parental lines revealed a 42 bp InDel about 38 Mbp upstream of CaOFP20. The 42-bp deletion was very strongly associated with fruit shape variation in the GWAS and is the most likely factor affecting reduction in the gene’s expression in elongated accessions, as no other structural variations were observed in the promoter region. Among the putative cis-acting regulatory elements found within the 42 bp InDel, the NTBBF1ARROLB motif (Supplementary Table S6) associated with auxin regulated expression may be affecting the expression of CaOFP20. Furthermore, sequencing the ORF of CaOFP20 in 30 round- and elongated-fruited accessions revealed five SNPs causing non-synonymous amino acid substitutions. None of these substitutions was included in the conserved OVATE domain in the protein and it is not known whether they affect the protein’s function. Nevertheless, two of these substitutions, at aa 172 and aa 215, were strongly associated with fruit shape variation in the association panel (Table 5). Taken together, these results indicate that CaOFP20 functions as a suppressor of fruit elongation in diverse C. annuum backgrounds, and it likely underlies fs10. Shape measurements of additional flower organs in the QTL-NILs indicated that the QTL suppresses growth of additional flower organs but has no effect on leaf growth. GWAS of the G2P-SOL panel revealed a significant association of CaOFP20 markers with fruit shape categories, but not with fruit shape index. This indicates that the main effect of CaOFP20 on fruit shape variation is the differentiation between elongated and round shapes, but not the magnitude of the fruit elongation.
Among the 14 genes located within the fs10 QTL interval, two genes exhibited differential expression between the bulks. Capana10g001225 was upregulated in the elongated-fruited bulk and codes for nsLTP, which belongs to a multigene family in plants that is involved in many physiological processes, such as response to biotic and abiotic stress, cutin and wax metabolism, and seed and pollen development (Liu et al., 2015). The potential involvement of nsLTP in cell wall organization and cell shape determination was shown in tobacco and Arabidopsis (Nieuwland et al., 2005; Ambrose et al., 2013). In tobacco, a nsLTP was identified that functions as a cell wall loosening protein in the pistil, likely associated with pollen tube growth. The tobacco gene (NCBI accession no. BAA03044) is highly homologous to Capana10g001225, as well as to an additional nine genes from the nsLTP family, seven of which are located in different regions of chromosome 10. Therefore, it is not possible to determine whether Capana10g001225 and BAA03044 are orthologous. Moreover, due to the high redundancy of the nsLTP family, it is not possible to downregulate Capana10g001225 in a gene-specific manner by VIGS. Therefore, we cannot exclude this gene’s possible involvement in fruit shape regulation by cell shape determination.
Variation in fruit shapes of the QTL-NILs is already manifested at anthesis. At this stage, the ovary wall of NIL 34-18R is thicker due to increased cell size and number. The decrease in ovary shape index in NIL 34-18R results from expansion of the ovary wall along the medio-lateral axis. This effect is most pronounced at the distal end of the ovary, where the two ovary walls fuse together. Similarly, at 3 weeks after anthesis, the fruit pericarp of NIL 34-18R is thicker because of increased cell size and number in the medio-lateral axis of the pericarp. Furthermore, fruit elongation in NIL 34-2 L is associated with a significant increase in cell elongation along the proximal-distal axis. Therefore, multiple processes, including cell expansion, cell division, and cell elongation, during the period of ovary and fruit growth, are affected by fs10.
The involvement of OFPs in regulating cell division and elongation has been demonstrated in multiple species. In tomato ovate, fruit elongation at the proximal end was associated with increased cell number along the proximal-distal axis (Wu et al., 2018). Whereas in tomato, the activity of SlOFP20 was dependent on a mutation in OVATE, we did not observe such an interaction in pepper. This indicates that while the main effect of OFP members is mostly conserved in pepper and tomato, the interaction among OFPs may be species-specific. In rice (Oryza sativa), GS9 acts as a transcriptional activator and regulator of grain elongation by altering cell division (Zhao et al., 2018). GS9 interacts with two OFPs, OsOFP14, and OsOFP8, and the latter interacts with OsGSK2 as a negative regulator of the brassinosteroid signaling pathway. Knockdown of OsOFP6 by RNAi resulted in altered grain shape and reduced plant growth associated with a reduction in cell number, but with no effect on cell elongation (Ma et al., 2017). In Arabidopsis, a gain-of-function mutation in AtOFP1 resulted in reduced length of the shoot and flower organs, and was associated with reduced cell elongation (Wang et al., 2007).
The fs10 transcriptome-enrichment analysis revealed a large number of genes putatively associated with cell expansion; however, no enrichment was associated with cell division, despite the change in cell number in the ovaries of the QTL-NILs. This may be the result of gene activation related to cell division occurring at earlier stages of ovary development, i.e., before anthesis. Multiple hormone metabolism processes that are putatively associated with organ growth were enriched in the fs10 transcriptome; however, there is no direct evidence linking the activity of specific differentially expressed genes with fruit shape determination. The major limitation of the transcriptome analysis in the present study was that RNA was sampled from a single narrow developmental stage, which precludes a more comprehensive assessment of gene-expression patterns during ovary and fruit development. Therefore, to study gene networks associated with CaOFP20, transcriptome analyses of multiple ovary and fruit developmental stages will be necessary.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: NCBI BioProject - PRJNA783486.
YB, AR, AD-F, and HZ designed and conducted the experiments and analyzed the data. EK produced the plants. IP wrote the manuscript. All authors contributed to the article and approved the submitted version.
Support for this research was provided by the G2P-SOL project and funded by the European Union Horizon 2020 research and innovation program under grant agreement no. 677379.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Dr. Arnaud Bovy, Wageningen University, Netherlands for providing the VIGS protocol.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.815589/full#supplementary-material
1. ^https://vanderknaaplab.uga.edu/tomato_analyzer.html
2. ^https://www.kyazma.nl/index.php/MapQTL/
4. ^https://bitbucket.org/tasseladmin/tassel-5-source/wiki/Tassel5GBSv2Pipeline
5. ^https://plantgarden.jp/en/list/t4072/genome/t4072.G002
6. ^http://peppergenome.snu.ac.kr/download.php
8. ^http://kobas.cbi.pku.edu.cn/kobas3/?t=1
Ambrose, C., DeBono, A., and Wasteneys, G. (2013). Cell geometry guides the dynamic targeting of apoplastic GPI-linked lipid transfer protein to cell wall elements and cell borders in Arabidopsis thaliana. PLoS One 8:e81215. doi: 10.1371/journal.pone.0081215
Barchi, L., Lefebvre, V., Sage-Palloix, A.-M., Lantery, S., and Palloix, A. (2009). QTL analysis of plant development and fruit traits in pepper and performance of selective phenotyping. Theor. Appl. Genet. 118, 1157–1171. doi: 10.1007/s00122-009-0970-0
Ben Chaim, A., Borovsky, Y., De Jong, W., and Paran, I. (2003a). Linkage of the A locus for the presence of anthocyanin and fs10.1, a major fruit shape QTL in pepper. Theor. Appl. Genet. 106, 889–894. doi: 10.1007/s00122-002-1132-9
Ben Chaim, A., Borovsky, E., Rao, G. U., Tanyolac, B., and Paran, I. (2003b). fs3.1: a major fruit shape QTL conserved in capsicum. Genome 46, 1–9. doi: 10.1139/g02-096
Ben Chaim, A., Paran, I., Grube, R., Jahn, M., van Wijk, R., and Peleman, J. (2001). QTL mapping of fruit related traits in pepper (Capsicum annuum). Theor. Appl. Genet. 102, 1016–1028. doi: 10.1007/s001220000461
Borovsky, Y., Monsonego, N., Mohan, V., Shabtai, S., Kamara, I., Faigenboim, A., et al. (2019). The zinc-finger transcription factor CcLOL1 controls chloroplast development and immature pepper fruit color in Capsicum chinense and its function is conserved in tomato. Plant J. 99, 41–55. doi: 10.1111/tpj.14305
Borovsky, Y., and Paran, I. (2011). Characterization of fs10.1, a major QTL controlling fruit elongation in capsicum. Theor. Appl. Genet. 123, 657–665. doi: 10.1007/s00122-011-1615-7
Bradbury, P. J., Zhang, Z., Kroon, D. E., Casstevens, T. M., Ramdoss, Y., and Buckler, E. S. (2007). TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23, 2633–2635. doi: 10.1093/bioinformatics/btm308
Buchfink, B., Xie, C., and Huson, D. H. (2015). Fast and sensitive protein alignment using DIAMOND. Methods 12, 59–60. doi: 10.1038/nmeth.3176
Chunthawodtiporn, J., Hill, T., Stoffel, K., and Van Deynze, A. (2018). Quantitative trait loci controlling fruit size and other horticultural traits in bell pepper (Capsicum annuum). Plant Genome. 11:160125. doi: 10.3835/plantgenome2016.12.0125
Colonna, V., D'Agostino, N., Garrison, E., Albrechten, A., Meisner, J., Facchiano, A., et al. (2019). Genomic diversity and novel genome-wide association with fruit morphology in capsicum, from 746k polymorphic sites. Sci. Rep. 9:10067. doi: 10.1038/s41598-019-46136-5
Cong, B., Barrero, L. S., and Tanksley, S. D. (2008). Regulatory change in YABBY-like transcription factor led to evolution of extreme fruit size during tomato domestication. Nat. Genet. 40, 800–804. doi: 10.1038/ng.144
Dou, J., Zhao, S., Lu, X., He, N., Zhang, L., Ali, A., et al. (2018). Genetic mapping reveals a candidate gene (ClFS1) for fruit shape in watermelon (Citrullus lanatus L.). Theor. Appl. Genet. 131, 947–958. doi: 10.1007/s00122-018-3050-5
Du, H., Yang, J., Chen, B., Zhang, X., Zhang, J., and Yang, K. (2019). Target sequencing reveals genetic diversity, population structure, core-SNP markers, and fruit shape-associated loci in pepper. BMC Plant Biol. 19:578. doi: 10.1186/s12870-019-2122-2
Dubois, M., Van den Broeck, L., and Inze, D. (2018). The pivotal role of ethylene in plant growth. Trends Plant Sci. 23, 311–323. doi: 10.1016/j.tplants.2018.01.003
Han, K., Jeong, H.-J., Yang, H.-B., Kang, S.-M., Kwon, J.-K., Kim, S., et al. (2016). An ultra-high-density bin map facilitates high-throughput QTL mapping of horticultural traits in pepper (Capsicum annuum). DNA Res. 23, 81–91. doi: 10.1093/dnares/dsv038
Hill, A. T., Chunthawodtiporn, J., Ashrafi, H., Stoffel, K., Weir, A., and van Deynze, A. (2017). Regions underlying population structure and the genomics of organ size determination in Capsicum annuum. Plant Genome. 10, 1–14. doi: 10.3835/plantgenome2017.03.0026
Hu, Y.-X., Tao, Y.-B., and Xu, Z.-F. (2017). Overexpression of Jatropha Gibberellin2-oxidase 6 (JcGA2ox6) induces dwarfism and smaller leaves, flowers, and fruits in Arabidopsis and Jatropha. Front. Plant Sci. 8:2103. doi: 10.3389/fpls.2017.02103
Huang, Z., and van der Knaap, E. (2011). Tomato fruit weight 11.3 maps close to fasciated on the bottom of chromosome 11. Theor. Appl. Genet. 123, 465–474. doi: 10.1007/s00122-011-1599-3
Jang, G., Yoon, Y., and Choi, Y. D. (2020). Crosstalk with Jasmonic acid integrates multiple responses in plant development. Int. J. Mol. Sci. 21:305. doi: 10.3390/ijms21010305
Kiba, T., Takei, K., Kojima, M., and Sakakibara, H. (2013). Side-chain modification of cytokinins controls shoot growth in Arabidopsis. Dev. Cell 27, 452–461. doi: 10.1016/j.devcel.2013.10.004
Kim, S., Park, M., Yeom, S.-I., Kim, Y.-M., Lee, J. M., et al. (2014). Genome sequence of the hot pepper provides insights into the evolution of pungency in capsicum species. Nature Genet. 46, 270–278. doi: 10.1038/ng.2877
Lee, Y. K., Kim, G.-T., Kim, I.-J., Park, J., Kwak, S.-S., Choi, G., et al. (2006). LONGIFOLIA1 and LONGIFOLIA2, two homologous genes, regulate longitudinal cell elongation in Arabidopsis. Development 133, 4305–4314. doi: 10.1242/dev.02604
Lee, H.-Y., Ro, N.-Y., Patil, A., Lee, J.-H., Kwon, J.-K., and Kang, B.-C. (2020). Uncovering Candidate Genes Controlling Major Fruit-Related Traits in Pepper via Genotype-by-Sequencing Based QTL Mapping and Genome-Wide Association Study. Front. Plant Sci. 11:1100. doi: 10.3389/fpls.2020.01100
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Li, E., Wang, S., Liu, Y., Chen, J. G., and Douglas, C. J. (2011). OVATE FAMILY PROTEIN4 (OFP4) interaction with KNAT7 regulates secondary cell wall formation in Arabidopsis thaliana. Plant J. 67, 328–341. doi: 10.1111/j.1365-313X.2011.04595.x
Liu, D., Sun, W., Yuan, Y., Zhang, N., Hayward, A., Liu, Y., et al. (2014). Phylogenetic analyses provide the first insights into the evolution of OVATE family proteins in land plants. Ann. Bot. 113, 1219–1233. doi: 10.1093/aob/mcu061
Liu, J., Van Eck, J., Cong, B., and Tanksley, S. D. (2002). A new class of regulatory genes underlying the cause of pear-shaped tomato fruit. Proc. Natl. Acad. Sci. U. S. A. 99, 13302–13306. doi: 10.1073/pnas.162485999
Liu, F., Yu, H., Deng, Y., Zheng, J., Liu, M., Ou, L., et al. (2017). PepperHub, an informatics hub for the chili pepper research community. Mol. Plant 10, 1129–1132. doi: 10.1016/j.molp.2017.03.005
Liu, F., Zhang, X., Lu, C., Zeng, X., Li, Y., Fu, D., et al. (2015). Non-specific lipid transfer proteins in plants: presenting new advances and an integrated functional analysis. J. Exp. Bot. 66, 5663–5681. doi: 10.1093/jxb/erv313
Love, M., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Ma, Y., Yang, C., He, Y., Tian, Z., and Li, J. (2017). Rice OVATE family protein6 6 regulates plant development and confers resistance to drought and cold stresses. J. Exp. Bot. 68, 4885–4898. doi: 10.1093/jxb/erx309
Majda, M., and Robert, S. (2018). The role of auxin in cell wall expansion. Int. J. Mol. Sci. 19:951. doi: 10.3390/ijms19040951
Matsuo, S., Kikuchi, K., Fukuda, M., Honda, I., and Imanishi, S. (2012). Roles and regulation of cytokinins in tomato fruit development. J. Exp. Bot. 63, 5569–5579. doi: 10.1093/jxb/ers207
McKenna, A., Hanna, M., Banks, E., Sivachenko, A., Cibulskis, K., Kernytsky, A., et al. (2010). The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303. doi: 10.1101/gr.107524.110
Michelmore, R. W., Paran, I., and Kesseli, R. V. (1991). Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. U. S. A. 88, 9828–9832. doi: 10.1073/pnas.88.21.9828
Munos, S., Ranc, N., and Botton, E. (2011). Increase in tomato locule number is controlled by two single-nucleotide polymorphisms located near WUSCHEL. Plant Physiol. 156, 2244–2254. doi: 10.1104/pp.111.173997
Nieuwland, J., Feron, R., Huisman, B. A., Fasolino, A., Hilbers, C. W., Derksen, J., et al. (2005). Lipid transfer proteins enhance cell wall extension in tobacco. Plant Cell 17, 2009–2019. doi: 10.1105/tpc.105.032094
Okushima, Y., Mitina, I., Quach, H. L., and Theologis, A. (2005). AUXIN RESPONSE FACTOR 2 (ARF2): a pleiotropic developmental regulator. Plant J. 43, 29–46. doi: 10.1111/j.1365-313X.2005.02426.x
Pan, Y., Liang, X., Gao, M., Liu, H., Meng, H., Weng, Y., et al. (2017). Round fruit shape in WI7239 cucumber is controlled by two interacting quantitative trait loci with one putatively encoding a tomato SUN homolog. Theor. Appl. Genet. 130, 573–586. doi: 10.1007/s00122-016-2836-6
Qin, C., Yu, C., Shen, Y., Fang, X., Chen, L., Min, J., et al. (2014). Whole- genome sequencing of cultivated and wild peppers provides insights into capsicum domestication and specialization. Proc. Natl. Acad. Sci. U. S. A. 111, 5135–5140. doi: 10.1073/pnas.1400975111
Rao, G. U., Ben Chaim, A., Borovsky, E., and Paran, I. (2003). Mapping of yield related QTLs in pepper in an inter-specific cross of Capsicum annuum and C. frutescens. Theor. Appl. Genet. 106, 1457–1466. doi: 10.1007/s00122-003-1204-5
Seymour, G. B., Ostergaard, L., Chapman, N. H., Knapp, S., and Martin, C. (2013). Fruit development and ripening. Ann. Rev. Plant Biol. 64, 219–241. doi: 10.1146/annurev-arplant-050312-120057
Singh, V., Sergeeva, L., Ligterink, W., Aloni, R., Zemach, H., Doron-Faigenboim, A., et al. (2019). Gibberellin promotes sweetpotato root vascular lignification and reduces storage-root formation. Front. Plant Sci. 10:1320. doi: 10.3389/fpls.2019.01320
Snouffer, A., Kraus, C., and van der Knapp, E. (2020). The shape of things to come: ovate family proteins regulate plant organ shape. Curr. Opin. Plant Biol. 53, 98–105. doi: 10.1016/j.pbi.2019.10.005
Sun, L., Rodriguez, G. R., Clevenger, J. P., Illa-Berenguer, E., Lin, J., Blakeslee, J. J., et al. (2015). Candidate gene selection and detailed morphological evaluations of fs8.1, a quantitative trait locus controlling tomato fruit shape. J. Exp. Bot. 66, 6471–6482. doi: 10.1093/jxb/erv361
Tian, C. E., Muto, H., Higuchi, K., Matamura, T., Tatematsu, K., Koshiba, T., et al. (2004). Disruption and overexpression of auxin response factor 8 gene of Arabidopsis affect hypocotyl elongation and root growth habit, indicating its possible involvement in auxin homeostasis in light condition. Plant J. 40, 333–343. doi: 10.1111/j.1365-313X.2004.02220.x
Trapnell, C., Williams, B. A., Pertea, G., Mortazavi, A., Kwan, G., van Baren, M. J., et al. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515. doi: 10.1038/nbt.1621
Tripodi, P., Rabanus-Wallace, M., Barchi, L., Kale, S., Esposito, S., Acquadro, A., et al. (2021). Global range expansion history of pepper (capsicum spp.) revealed by over 10,000 genebank accessions. Proc. Natl. Acad. Sci. U. S. A. 118:e2104315118. doi: 10.1073/pnas.2104315118
Tsaballa, A., Pasentsis, K., Darzentas, N., and Tsaftaris, A. S. (2011). Multiple evidence for the role of an ovate-like gene in determining fruit shape in pepper. BMC Plant Biol. 11:46. doi: 10.1186/1471-2229-11-46
van der Knaap, E., Chakrabarti, M., Chu, Y. H., Clevenger, J., Illa-Berenguer, E., Huang, Z., et al. (2014). What lies beyond the eye: the molecular mechanisms regulating tomato fruit weight and shape. Front. Plant Sci. 5:227. doi: 10.3389/fpls.2014.00227
van der Knaap, E., and Ostergaard, L. (2017). Shaping a fruit: developmental pathways that impact growth patterns. Semin. Cell Dev. Biol. 79, 27–36. doi: 10.1016/j.semcdb.2017.10.028
Wang, S., Chang, Y., Guo, J., and Chen, J.-G. (2007). Arabidopsis ovate family protein 1 is a transcriptional repressor that suppresses cell elongation. Plant J. 50, 858–872. doi: 10.1111/j.1365-313X.2007.03096.x
Wang, Y., Clevenger, J. P., Illa-Berenguer, E., Meulia, T., van der Knaap, E., and Sun, L. (2019). A comparison of sun, ovate, fs8.1 and auxin application on tomato fruit shape and gene expression. Plant Cell Physiol. 60, 1067–1081. doi: 10.1093/pcp/pcz024
Wang, L., Li, J., Zhao, J., and He, C. (2015a). Evolutionary developmental genetics of fruit morphological variation within the Solanaceae. Front. Plant Sci. 6:248. doi: 10.3389/fpls.2015.00248
Wang, Y., Xiong, G., Hu, J., Jiang, L., Yu, H., Xu, J., et al. (2015b). Copy number variation at the GL7 locus contributes to grain size diversity in rice. Nat. Genet. 47, 944–948. doi: 10.1038/ng.3346
Wang, Y., Zhao, J., Lu, W., and Deng, D. (2017). Gibberellin in plant height control: old player, new story. Plant Cell Rep. 36, 391–398. doi: 10.1007/s00299-017-2104-5
Wu, S., Clevenger, J. P., Sun, L., Visa, S., Kamiya, Y., Jikumaru, Y., et al. (2015). The control of tomato fruit elongation orchestrated by sun, ovate and fs8.1 in a wild relative of tomato. Plant Sci. 238, 95–104. doi: 10.1016/j.plantsci.2015.05.019
Wu, S., Xiao, H., Cabrera, A., Meulia, T., and van der Knaap, E. (2011). SUN regulates vegetative and reproductive organ shape by changing cell division patterns. Plant Physiol. 157, 1175–1186. doi: 10.1104/pp.111.181065
Wu, S., Zhang, B., Keyhaninejad, N., Rodriguez, G. R., Kim, H. J., Chakrabarti, M., et al. (2018). A common genetic mechanism underlies morphological diversity in fruits and other plant organs. Nature Cummun. 9:4734. doi: 10.1038/s41467-018-07216-8
Wybouw, B., and De Rybel, B. (2019). Cytokinin-A developing story. Trends Plant Sci. 24, 177–185. doi: 10.1016/j.tplants.2018.10.012
Xiao, H., Jiang, N., Schaffner, E. K., Stockinger, E. J., and Van der Knaap, E. (2008). A retrotransposon-mediated gene duplication underlies morphological variation of tomato fruit. Science 319, 1527–1530. doi: 10.1126/science.1153040
Xiao, Y., Liu, D., Zhang, G., Tong, H., and Chu, C. (2017). Brassinosteroids regulate OFP1, a DLT interacting protein, to modulate plant architecture and grain morphology in rice. Front. Plant Sci. 8:1698. doi: 10.3389/fpls.2017.01698
Xu, C., Liberatore, K. L., MacAlister, C. A., Huang, Z., Chu, Y.-H., Jiang, K., et al. (2015). A cascade of arabinosyltansferases controls shoot meristem size in tomato. Nat. Genet. 47, 784–792. doi: 10.1038/ng.3309
Yang, C., Ma, Y., He, Y., Tian, Z., and Li, J. (2018). OsOFP19 modulates plant architecture by integrating the cell division pattern and brassinosteroid signaling. Plant J. 93, 489–501. doi: 10.1111/tpj.13793
Yarnes, S. C., Ashrafi, A., Reyes-Chin-Wo, S., Hill, T. A., Stoffel, K. M., and Van Deynze, A. (2013). Identification of QTLs for capsaicinoids, fruit quality, and plant architecture-related traits in an interspecific capsicum RIL population. Genome 56, 61–74. doi: 10.1139/gen-2012-0083
Zhang, L., Zhang, X., Ju, H., Chen, J., Wang, S., Wang, H., et al. (2016). Ovate family protein1 interaction with BLH3 regulates transition timing from vegetative to reproductive phase in Arabidopsis. Biochem. Biophs. Res. Commun. 470, 492–497. doi: 10.1016/j.bbrc.2016.01.135
Zhao, J., Jiang, L., Che, G., Pan, Y., Li, Y., Hou, Y., et al. (2019). A functional allele of CsFUL1 regulates fruit length through repressing CsSUP and inhibiting auxin transport in cucumber. Plant Cell 31, 1289–1307. doi: 10.1105/tpc.18.00905
Zhao, D. S., Li, Q. F., Zhang, C. Q., Zhang, C., Yang, Q. Q., Pan, L. X., et al. (2018). GS9 acts as a transciptional activator to regulate rice grain shape and appearance quality. Nat. Commun. 9:1240. doi: 10.1038/s41467-018-03616-y
Zhou, H., Ma, R., Gao, L., Zhang, J., Zhang, A., Zhang, X., et al. (2020). A 1.7-Mb chromosomal inversion downstream of a PpOFP1 gene is responsible for flat fruit shape in peach. Plant Biotech. J. 19, 192–205. doi: 10.1111/pbi.13455
Keywords: fruit shape, pepper (Capsicum spp.), ovate family protein, QTL mapping, natural variation in plants
Citation: Borovsky Y, Raz A, Doron-Faigenboim A, Zemach H, Karavani E and Paran I (2022) Pepper Fruit Elongation Is Controlled by Capsicum annuum Ovate Family Protein 20. Front. Plant Sci. 12:815589. doi: 10.3389/fpls.2021.815589
Received: 15 November 2021; Accepted: 02 December 2021;
Published: 04 January 2022.
Edited by:
Qian-Hao Zhu, Commonwealth Scientific and Industrial Research Organisation (CSIRO), AustraliaReviewed by:
Zhangsheng Zhu, South China Agricultural University, ChinaCopyright © 2022 Borovsky, Raz, Doron-Faigenboim, Zemach, Karavani and Paran. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ilan Paran, aXBhcmFuQHZvbGNhbmkuYWdyaS5nb3YuaWw=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.