
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 17 January 2022
Sec. Plant Physiology
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.813691
This article is part of the Research Topic Plant Oxidative Processes Honoring James Nash Siedow View all 8 articles
Alternative oxidase (AOX) is an important component of the plant respiratory pathway, enabling a route for electrons that bypasses the energy-conserving, ROS-producing complexes of the mitochondrial electron transport chain. Plants contain numerous isoforms of AOX, classified as either AOX1 or AOX2. AOX1 isoforms have received the most attention due to their importance in stress responses across a wide range of species. However, the propensity for at least one isoform of AOX2 to accumulate to very high levels in photosynthetic tissues of all legumes studied to date, suggests that this isoform has specialized roles, but we know little of its properties. Previous studies with sub-mitochondrial particles of soybean cotyledons and roots indicated that differential expression of GmAOX1, GmAOX2A, and GmAOX2D across tissues might confer different activation kinetics with pyruvate. We have investigated this using recombinantly expressed isoforms of soybean AOX in a previously described bacterial system (Selinski et al., 2016, Physiologia Plantarum 157, 264-279). Pyruvate activation kinetics were similar between the two GmAOX2 isoforms but differed substantially from those of GmAOX1, suggesting that selective expression of AOX1 and 2 could determine the level of AOX activity. However, this alone cannot completely explain the differences seen in sub-mitochondrial particles isolated from different legume tissues and possible reasons for this are discussed.
It is well documented that the plant mitochondrial electron transport chain (mETC) is branched, with phosphorylating and non-phosphorylating pathways coexisting (see Siedow and Day, 2000 for an overview). In addition to the classical mETC linked to proton translocation and ATP synthesis, plant mitochondria possess an alternative pathway that is non-proton translocating and consists of external and internal NAD(P)H dehydrogenases (NDBs and NDAs, respectively) and the so-called alternative oxidase (AOX), which accepts electrons from ubiquinol and provides an alternative route for electrons to reduce oxygen. It has been proposed based on co-expression analysis that NDB2, the major external NADH dehydrogenase, and AOX could potentially form a complete respiratory chain (Clifton et al., 2005). A functional link has been recently confirmed between the two proteins, which together help the plant to cope with the oxidative stress associated with photo inhibitory conditions (Sweetman et al., 2019b). An important role for AOX in minimizing oxidative stress under many different conditions has been shown by numerous studies (Maxwell et al., 1999; Pastore et al., 2001; Umbach et al., 2005; Smith et al., 2009; Cvetkovska and Vanlerberghe, 2013; Li et al., 2013; Mhadhbi et al., 2013; Vishwakarma et al., 2015; Dahal and Vanlerberghe, 2017; Demircan et al., 2020; Jayawardhane et al., 2020). However, operation of the alternative pathway has the potential to dramatically decrease the yield of ATP during respiration (Ribas-Carbo et al., 2005; Soole and Sweetman, 2021) and it is important that it is regulated carefully.
The in vivo activity, or engagement, of alternative oxidase (AOX) in plants reflects many levels of regulation, including transcriptional and post-transcriptional control. AOX capacity may be limited by protein abundance (e.g., Elthon and McIntosh, 1987; Rhoads and McIntosh, 1992; Vanlerberghe and McIntosh, 1994; McCabe et al., 1998), but increasing protein abundance does not always result in increased capacity nor activity (Obenland et al., 1990; Guy and Vanlerberghe, 2005; Florez-Sarasa et al., 2011). Protein abundance may or may not be related to gene expression (Rhoads and McIntosh, 1992; Vanlerberghe and McIntosh, 1994, 1996; Cruz-Hernandez and Gomez-Lim, 1995; Aubert et al., 1997; McCabe et al., 1998), with discrepancies potentially due to the half-life of AOX proteins, some of which can remain long after their transcripts subside. Activity is dependent on dimerization of the 30–40 kDa monomers, which must be in the reduced (i.e., non-covalently bound) form (Umbach and Siedow, 1993), with the reduction state of AOX in vivo probably regulated by thioredoxin (but see Florez-Sarasa et al., 2019; Moller et al., 2020). Another layer of regulation is the provision of substrate to the mitochondria, since the reduction state of ubiquinol (Q) to a particular threshold (i.e., high QH2:Q), at least partly determines AOX activity (but see Millenaar and Lambers, 2003), depending on the activation status of the protein (Hoefnagel et al., 1995), which in turn is dependent on various metabolites. In particular, activity of AOX in the reduced dimer state is regulated by certain organic acids, most notably pyruvate and other 2-oxo acids (Millar et al., 1993, 1996; Selinski et al., 2017), which interact with key cysteine residues of AOX within the mitochondrial matrix (Umbach and Siedow, 1996; Rhoads et al., 1998; Vanlerberghe et al., 1998; Djajanegara et al., 1999), forming a thiohemiacetal group (Umbach and Siedow, 1996). The presence of such activators decreases the threshold of QH2:Q required for AOX engagement (Umbach et al., 1994; Hoefnagel et al., 1995). The in vivo relevance of these various layers of regulation have been debated in the literature (e.g., Day and Wiskich, 1995; Del-Saz et al., 2018; Schwarzlander and Fuchs, 2019).
Plant AOX proteins are classified into two subfamilies, AOX1 and AOX2 (Considine et al., 2002) and four phylogenetic clades: AOX1a-c/1e, AOX1d, AOX2a-c, and AOX2d (Costa et al., 2014). While most angiosperms possess an expanded AOX1 subfamily alongside a single AOX2, legumes generally exhibit a single AOX1 and an expanded AOX2 subfamily (Costa et al., 2014; Sweetman et al., 2019a). Across all plants studied to date, there is at least one stress-inducible gene, typically an Aox1a, b and/or Aox1d (Considine et al., 2002; Clifton et al., 2005; Polidoros et al., 2005; Feng et al., 2013; Wanniarachchi et al., 2018), although Aox2d in chickpea and other legumes is also responsive to cold, drought and salinity stresses (Costa et al., 2014; Sweetman et al., 2019a,2020). The genome of soybean (Glycine max) contains three AOX genes GmAox1, GmAox2a, and GmAox2d and these are preferentially expressed in different tissues. The most abundant AOX protein of soybean photosynthetic tissues, GmAOX2a, is absent in roots and this pattern of expression is conserved for orthologs in other legumes (Kearns et al., 1992; Finnegan et al., 1997; Sweetman et al., 2019a). Importantly, in contrast to non-legumes in which AOX expression and activity are strictly controlled by environmental (stress) conditions, the AOX2 isoforms of legumes are constitutively expressed (Sweetman et al., 2019a) and can contribute substantially to overall respiration, at least in soybean (Millar et al., 1998).
Recently it was shown that specific AOX1 isoforms of Arabidopsis thaliana respond to different sets of organic acids (Selinski et al., 2018), but it is not known whether the same is true of AOX2 isoforms, nor whether there are major differences in activation kinetics between AOX1 and AOX2 subfamilies. Activation kinetics of AOX are very different in sub-mitochondrial particles from different soybean tissues, with cotyledon and root particles exhibiting a tenfold difference in the K1/2PYRUVATE (Finnegan et al., 1997). While soybean roots only express AOX2D, cotyledons have high levels of both AOX2A and 2D, but it is unclear whether this explains the different activation kinetics of AOX in two tissues. It is important that we understand the activation kinetics of the different AOX isoforms in legumes, to facilitate the development of more targeted biotechnological approaches for improvement of plant growth and stress tolerance. For example, predicting situations where plant health might be improved via AOX engagement, e.g., during abiotic stress (Del-Saz et al., 2016; Vanlerberghe et al., 2016; Watanabe et al., 2016; Dahal and Vanlerberghe, 2018; Sweetman et al., 2019b), or controlling AOX when energy conservation is necessary for improvement of plant productivity (Amthor et al., 2019). Using a system developed specifically for measuring enzyme kinetics of recombinantly expressed AOX proteins (Selinski et al., 2016), we herein describe the differences between pyruvate activation kinetics of soybean AOX1 and AOX2 proteins.
Mitochondrial target peptide sequences of the G. max AOX proteins, GmAOX1, GmAOX2A and GmAOX2D (Whelan et al., 1996) were predicted using Mitoprot (Claros and Vincens, 1996). Primers were designed to amplify the full-length open reading frames, immediately downstream of the mitochondrial transit peptide coding region. Primers also contained restriction sites compatible with pET22b(+): GmAox1 (F: 5′-CATATGGAGAGCACTTTGGCTTTGTC-3′, R: 5′-GAATTC TTGGCATCATGAGACATAAC-3′), GmAox2a (F: 5′-CATATG ATGGTTTCGCCGGCGGA-3′, R: 5′-GTCGACTCATATCCGT GGCAATAAAGACTA-3′), GmAox2d (F: 5′-CATATGTCCAC TCTTCCAGAGGTAAA-3′, R: 5′-GAATTCAGCAAAGGCT AGATATCGTT-3′). PCR products were amplified from G. max leaf cDNA, inserted into pGEMtEasy via A-T ligation (Promega, WI, United States) then digested and cloned into pET22b(+) using standard restriction enzymes (NEB, MA, United States) and T4 ligase (Promega, WI, United States). Resultant constructs: pET22b(+)GmAox1, pET22b(+)GmAox2a and pET22b(+)GmAox2d were confirmed via Sanger sequencing (AGRF, SA, Australia), then transformed into E.coli strain BHH8 (Selinski et al., 2016) via heat-shock. Due to removal of the start codon (as part of the N-terminal target peptide), a new start codon was introduced via the NdeI restriction site within the F primers. Consequently, the translated peptide sequences contained a methionine that would not typically occur in the mature GmAOX proteins.
Methods for bacterial culture, protein synthesis, cell harvesting, and membrane vesicle isolations were carried out as previously described (Selinski et al., 2016). Protein concentrations were determined in a 96-well plate using the Pierce™ BCA Protein Assay Kit (Thermo Fisher Scientific, MA, United States).
Assays were carried out with a Clark type oxygen electrode (Hansatech, Norfolk, United Kingdom), essentially as described by Selinski et al. (2016) but with some modifications. Two electrode chambers were first calibrated and pre-equilibrated with 1 ml NKM buffer (150 mM NaCl, 50 mM potassium phosphate buffer (pH 7.0), 10 mM KCl and 5 mM MgCl2). Before beginning the assay, 5 mM DTT, 1 mM KCN and a 10 μl aliquot of inverted membrane vesicles (IMVs; approximately 100 μg) were added, followed by 10 μl of the “effector.” Final pyruvate concentrations ranged between 0.5 and 5,000 μM. After inserting the lids and equilibrating for 3 min, 2.5 mM NADH was used to initiate the reaction, which was inhibited by 1 mM SHAM after 4 min and the residual rate was allowed to proceed for a further 4 min. Each pyruvate concentration was measured in a separate assay; incremental additions were not possible because the activity of membrane vesicles subsided over time. A control reaction containing no pyruvate was run alongside each assay for the same reason, and results were presented as a percentage activation relative to the control. Prior to each set of assays, both chambers were tested with 5 mM pyruvate to ensure the electrodes yielded equivalent rates.
Aliquots of each BHH8 IMV sample were snap-frozen in liquid nitrogen immediately after isolation and stored at −80°C until use for Western blots. Samples were thawed gently, then suspended in loading buffer [62.5 mM Tris (pH 6.8), 30% (v/v) glycerol, 5% (v/v) β-mercaptoethanol, 2% (w/v) SDS, and 0.002% (w/v) bromophenol blue] containing 25 mM DTT. After boiling at 95°C for 2 min and centrifuging briefly, a further 25 mM DTT was added to the supernatant. Samples of 2.5 μg equivalent protein were loaded directly onto 10% SDS-PAGE gels and resolved at 60 V for approximately 60 min, before being transferred to a 0.45 μm nitrocellulose membrane (Trans-Blot Transfer Medium, BioRad, CA, United States). The membrane was incubated with “AOA” antibody raised against Sauromatum guttatum AOX in mouse (Elthon et al., 1989) then a peroxidase-conjugated goat anti-mouse IgG (BioRad, CA, United States) and visualized using a chemiluminescent stain (Clarity ECL, BioRad, CA, United States).
Data were analyzed using SPSS version 25 (IBM). Michaelis Menten constants were determined by non-linear regression. Specific activities of different AOX isoforms were compared using one-way ANOVA with post hoc Tukey test.
E. coli BHH8 contains a stable, highly reduced ubiquinone pool due to the lack of cytochrome bo quinol oxidase and cytochrome bd-1 quinol oxidase, making it ideal for the investigation of heterologously expressed AOX protein kinetics (Selinski et al., 2016, 2017, 2018). Inverted membrane vesicles (IMVs) were prepared from BHH8 cells expressing one of GmAOX1, GmAOX2A or GmAOX2D, and used to measure activation kinetics at different concentrations of pyruvate. Pyruvate activated all three isoforms (Figure 1), but the K1/2PYR for GmAOX2A and GmAOX2D (99 and 40 μM, respectively) were much higher than that for GmAOX1 (4 μM) (Table 1). With saturating concentrations of pyruvate, GmAOX1 and GmAOX2A activity was stimulated by about 30%, while GmAOX2D was stimulated by 50%. These maximal activation levels were considerably lower than previous measurements with membrane vesicles containing Arabidopsis AOX isoforms (sixfold increase; Selinski et al., 2016), and with soybean sub-mitochondrial particles (1.85-fold increase in cotyledon SMPs and fourfold increase in root SMPs; Finnegan et al., 1997), suggesting some variability based on expression system and isoform. All of the GmAOX proteins were also activated by 5 mM 2-oxoglutarate (2-OG) (Supplementary Table 1).
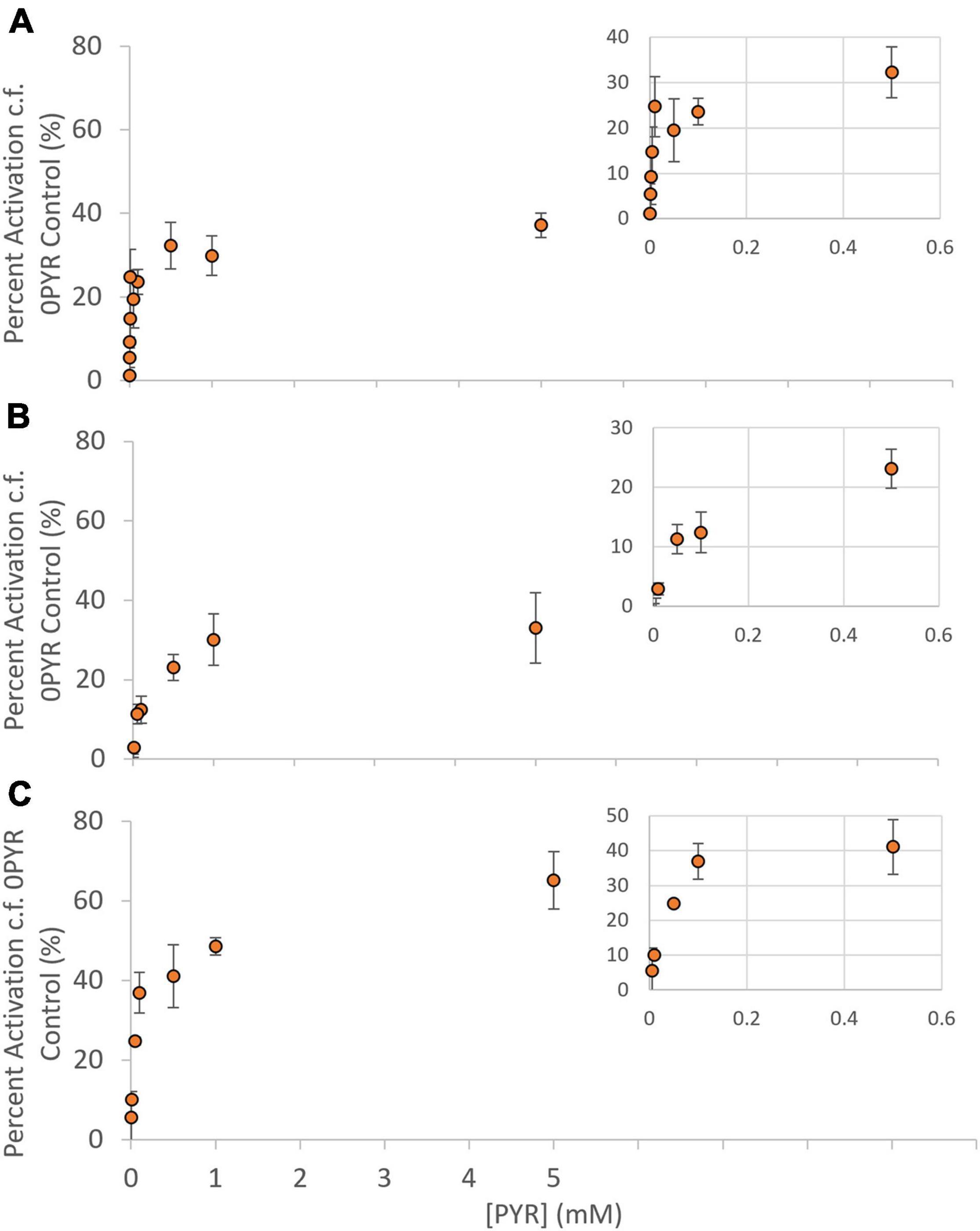
Figure 1. Activation kinetics of AOX1, AOX2A, and AOX2D. Activator saturation curves for (A) AOX1, (B) AOX2A, and (C) AOX2D. Rates presented as Percent Activation; at each pyruvate concentration a simultaneous assay containing no pyruvate (0PYR) was used to calculate activation levels. Each data point represents the mean of measurements taken from three separate IMV preparations (n = 3 ± SEM). Michaelis Menten constants were determined by non-linear regression (IBM SPSS v25), and kinetic data summarized in Table 1. Inset: Expanded graph for pyruvate concentrations ≤ 0.5 mM.
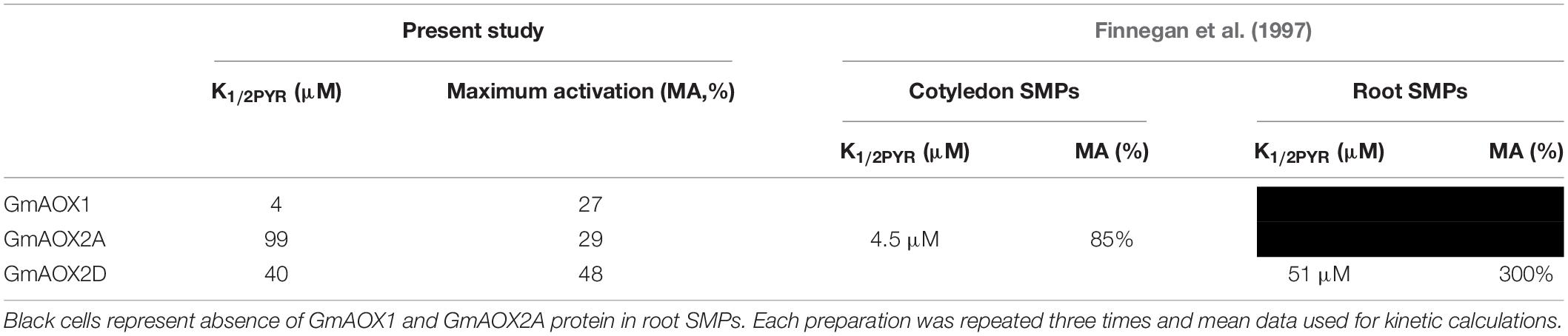
Table 1. Summary of pyruvate activation kinetics for individual soybean AOX isoforms of inverted membrane vesicles and for combinations of AOX isoforms within submitochondrial particles.
To ensure that enzyme activities were not influenced by reduction state, DTT was included in all isolation solutions and assay mixtures. Monomeric AOX subunits were confirmed using immunoblots from non-reducing SDS-PAGE (Supplementary Figure 1). GmAOX1, GmAOX2A, and GmAOX2D resolved to different apparent molecular weights: 28, 29, and 31 kDa, respectively (Figure 2), in agreement with apparent Mr measured in soybean mitochondria (Kearns et al., 1992; Finnegan et al., 1997).
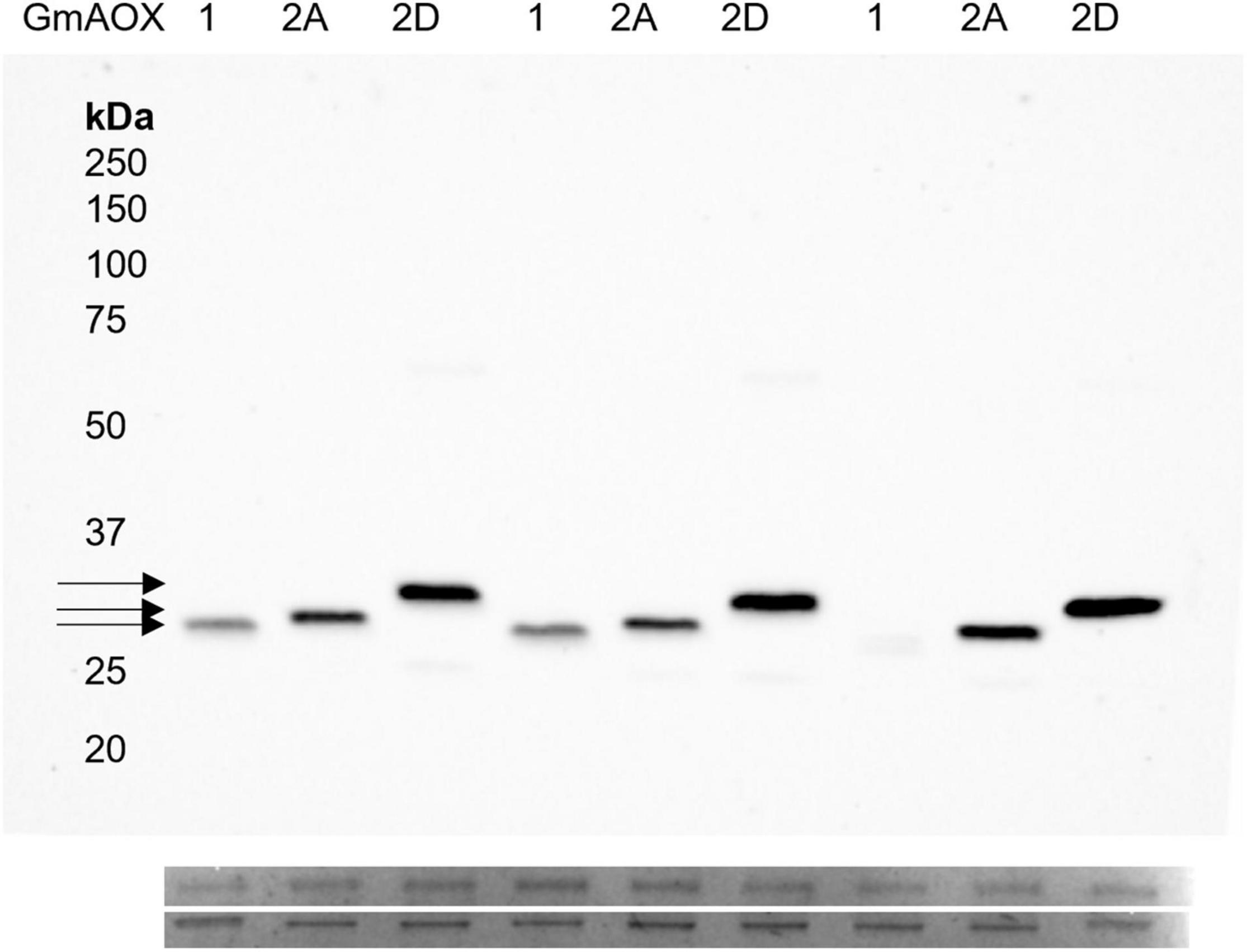
Figure 2. Expression of AOX proteins in BHH8 inverted membrane vesicles. Each lane was loaded with 2.5 μg protein sample, treated with 50 mM DTT. The membrane was probed with AOA antibody, which is non-selective for AOX isoforms (Elthon et al., 1989). Apparent molecular weights were: 28, 29, and 31 kDa for AOX1, AOX2A, and AOX2D, respectively, each lacking the predicted mitochondrial target peptide. Below, two prominent bands from Coomassie stain indicate protein loading.
The immunoblots showed that GmAOX1 protein was less abundant in membrane vesicle preparations than were the GmAOX2 isoforms (Figure 2). Such differences have been observed previously for Arabidopsis AOX isoforms (Selinski, J. unpublished results). Consequently, specific activities were calculated for the three isoforms based on their relative band intensities on immunoblots of IMV proteins separated by reducing SDS-PAGE (i.e., in the presence of 50 mM DTT; Figure 2). On this basis, it is particularly noteworthy that the specific activity of GmAOX1 (relative level of respiration per unit of AOX protein) was threefold higher than GmAOX2A and fivefold higher than GmAOX2D (Figure 3).
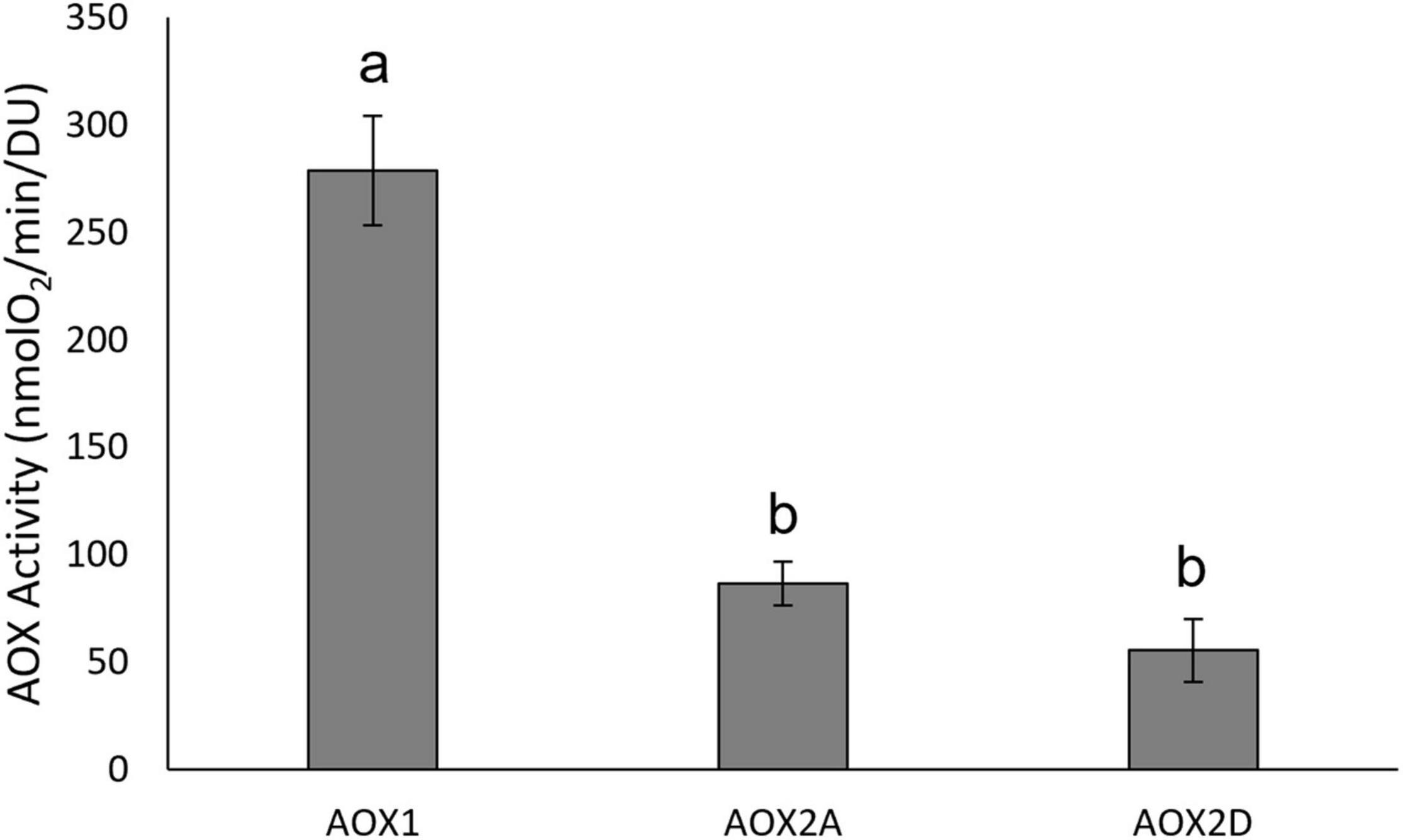
Figure 3. Specific activities of AOX isoforms. Oxygen consumption rates of AOX from inverted membrane vesicles. Rate determined as nmolO2/min/μg membrane vesicle protein in the presence of potassium cyanide, DTT and 5 mM pyruvate and initiated with the addition of NADH. Rates were normalized to AOX protein content (density unit reflects the intensity of protein bands in immunoblots, see Selinski et al., 2016) (n = 3 ± SEM). Significant differences between isoforms are indicated by different letters, based on one-way ANOVA with post hoc Tukey tests (IBM SPSS v25).
It is always difficult to compare data obtained with a heterologously expressed enzyme to those obtained with native membranes. However, the K1/2PYR for GmAOX2D measured here in bacterial membrane vesicles (40 μM) was strikingly similar to that measured previously in sub-mitochondrial particles from soybean root (51 μM; Finnegan et al., 1997), in which only AOX2D is expressed. This gives us some confidence that our results reflect AOX properties in vivo. The situation in sub-mitochondrial particles from cotyledons, however, is more complicated since they express multiple AOX isoforms (Finnegan et al., 1997). Interestingly, while the K1/2PYR of GmAOX2A (90 μM) was only moderately higher than that of AOX2D (40 μM) in our vesicle system, the K1/2PYR for AOX activation in cotyledon sub-mitochondrial particles was much less (4.5 μM; Finnegan et al., 1997), closer to that measured for GmAOX1 in the vesicle system (6 μM). Yet cotyledons of soybean, like young shoots from other legumes, display AOX2A at much higher levels than AOX2D and AOX1 (Finnegan et al., 1997; Sweetman et al., 2019a).
Two possible explanations for this discrepancy come to mind. Firstly, the higher specific activity of GmAOX1 (Figure 3) led it to contribute more strongly to overall AOX activity in cotyledon sub-mitochondrial particles, even though GmAOX2A protein was substantially more abundant, thereby masking the activation kinetics of the GmAOX2 isoforms. However, if this were the case, then a double sigmoid curve of pyruvate activation kinetics would be expected from soybean cotyledon sub-mitochondrial particles, which was not reported by Finnegan et al. (1997). We consider this option as unlikely.
Alternatively, the formation of AOX heterodimers in mitochondria may affect pyruvate activation kinetics. Within the inner mitochondrial membrane, it is generally accepted that active AOX proteins exist as homodimers (Vanlerberghe, 2013). However, this has never been confirmed experimentally and there are hints of heterodimer formation in early work with AOX antibodies (Elthon et al., 1989; Umbach and Siedow, 1993), which have been echoed throughout the literature (Finnegan et al., 1997; Sluse and Jarmuszkiewicz, 1998; Vanlerberghe et al., 2020). It is possible, therefore, that the formation of heterodimers in tissues and cells where multiple isoforms of AOX coexist might affect activation kinetics, with, in the case of soybean, heterodimers being much more sensitive to pyruvate. This would explain the differences observed between the values of K1/2PYR values in cotyledon sub-mitochondrial particles and those obtained with AOX proteins expressed singly in the vesicle system. Further work is required to resolve these possibilities.
We also need to consider whether all AOX isoforms reside within the same mitochondrion in vivo, since isolated mitochondria originate from multiple cell types when purified from complex tissues such as cotyledons. It is not known whether GmAOX1, GmAOX2A and GmAOX2D co-localize within the same cell types of soybean. However, in Arabidopsis thaliana shoots there is indirect evidence for AOX transcript co-localization in some cell types, based on laser microdissection transcriptomics that suggested both AtAox1a and AtAox1b were present in vasculature cells, while AtAox1a, AtAox1d and AtAox2 transcripts were all found in mesophyll (Berkowitz et al., 2021). In addition, interactions between different AOX peptides have been suggested, based on Blue Native (BN)-PAGE experiments with isolated A. thaliana leaf mitochondrial proteins; specifically AtAOX1A, AtAOX1B, and AtAOX1D could be found in a complex of approximately 140 kDa (Senkler et al., 2017). The simultaneous expression of multiple AOX isoforms using the experimental system described in this paper could help to confirm, or refute, the formation of heterodimers.
The observation that GmAOX2D was more highly activated by pyruvate than GmAOX2A, at saturating levels of pyruvate, has precedence in the literature. During soybean seedling development, McCabe et al. (1998) observed a shift in transcript and protein level, from predominantly GmAOX2A to predominantly GmAOX2D (AOX1 transcript and protein were low or undetectable), at which point AOX in isolated mitochondria became more highly activated by 2 mM pyruvate. Clearly, the spatiotemporal profiles of specific AOX isoforms can influence regulatory properties of the enzyme in soybean and probably in other plant species. Further research in this area is warranted.
The fact that GmAOX1 has higher specific activity and requires less pyruvate for its activation, compared to its more highly expressed AOX2 counterparts in soybean, may be significant in planta because AOX1 is the most stress-inducible isoform in legumes (Sweetman et al., 2019a). GmAOX2D is also stress-responsive and, compared to GmAOX2A, requires about half the amount of pyruvate and is activated to a higher Vmax in the presence of pyruvate. This suggests that stress-induced AOX isoforms respond more quickly to pyruvate (and presumably other organic acid activators) and have a higher inherent capacity than constitutively expressed isoforms such as GmAOX2A that may carry out more ‘house-keeping’ functions in the tissues in which they are expressed.
While it is clear that different AOX isoforms have tissue-specific expression (eg Finnegan et al., 1997), different kinetics of pyruvate activation (this report), and also respond differently to other organic acid activators (Selinski et al., 2018), we still lack precise knowledge of the concentrations of activators in planta and how they vary with tissue type and growth conditions. Organic acid concentrations have been estimated in whole tissue extracts from numerous plant species, but for AOX activation it is the intramitochondrial concentrations that are important, and these are very difficult to measure, especially pyruvate given that its import and generation within mitochondria can occur via multiple pathways (Le et al., 2021a). The advent of a pyruvate sensor for in vivo estimation of pyruvate levels (Arce-Molina et al., 2020) may be of help in this context, although very recent data suggest that even within mitochondria distinct pools of pyruvate exist, and the fate of the different pyruvate pools may be predetermined by source, e.g., import vs in situ biosynthesis (Le et al., 2021b).
This article is dedicated to the memory of Jim Siedow, a pioneer of the field of plant respiration, a highly valued colleague, and a very good friend: DD.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
DD conceived the project. CS, JS, and TM designed and carried out all experimental procedures. JW contributed to experimental design and provided facilities and guidance. CS drafted the manuscript. JS, JW, and DD edited the manuscript. All authors approved the final manuscript.
This research was funded by the Australian Research Council, Grains Research and Development Corporation, and Industrial Transformation Research Hub (IH140100013).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We are grateful to Kathleen Soole and Colin Jenkins for valuable discussions on this topic.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.813691/full#supplementary-material
Supplementary Figure 1 | AOX proteins from non-reducing SDS-PAGE.
Supplementary Table 1 | Activation of individual soybean AOX isoforms by 5 mM 2-oxoglutarate.
Amthor, J. S., Bar-Even, A., Hanson, A. D., Millar, A. H., Stitt, M., Sweetlove, L. J., et al. (2019). Engineering strategies to boost crop productivity by cutting respiratory carbon loss. Plant Cell 31, 297–314. doi: 10.1105/tpc.18.00743
Arce-Molina, R., Cortes-Molina, F., Sandoval, P. Y., Galaz, A., Alegria, K., Schirmeier, S., et al. (2020). A highly responsive pyruvate sensor reveals pathway-regulatory role of the mitochondrial pyruvate carrier MPC. Elife 9:e53917. doi: 10.7554/eLife.53917
Aubert, S., Bligny, R., Day, D. A., Whelan, J., and Douce, R. (1997). Induction of alternative oxidase synthesis by herbicides inhibiting branched-chain amino acid synthesis. Plant J. 11, 649–657. doi: 10.1046/j.1365-313x.1997.11040649.x
Berkowitz, O., Xu, Y., Liew, L. C., Wang, Y., Zhu, Y. Q., Hurgobin, B., et al. (2021). RNA-seq analysis of laser microdissected Arabidopsis thaliana leaf epidermis, mesophyll and vasculature defines tissue-specific transcriptional responses to multiple stress treatments. Plant J. 107, 938–955. doi: 10.1111/tpj.15314
Claros, M. G., and Vincens, P. (1996). Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 241, 779–786. doi: 10.1111/j.1432-1033.1996.00779.x
Clifton, R., Lister, R., Parker, K. L., Sappl, P. G., Elhafez, D., Millar, A. H., et al. (2005). Stress-induced co-expression of alternative respiratory chain components in Arabidopsis thaliana. Plant Mol. Biol. 58, 193–212.
Considine, M. J., Holtzapffel, R. C., Day, D. A., Whelan, J., and Millar, A. H. (2002). Molecular distinction between alternative oxidase from monocots and dicots. Plant Physiol. 129, 949–953. doi: 10.1104/pp.004150
Costa, J. H., McDonald, A. E., Arnholdt-Schmitt, B., and de Melo, D. F. (2014). A classification scheme for alternative oxidases reveals the taxonomic distribution and evolutionary history of the enzyme in angiosperms. Mitochondrion 19, 172–183. doi: 10.1016/j.mito.2014.04.007
Cruz-Hernandez, A., and Gomez-Lim, M. A. (1995). Alternative oxidase from mango (Mangifera-indica, L) is differentially regulated during fruit ripening. Planta 197, 569–576.
Cvetkovska, M., and Vanlerberghe, G. C. (2013). Alternative oxidase impacts the plant response to biotic stress by influencing the mitochondrial generation of reactive oxygen species. Plant Cell Environ. 36, 721–732. doi: 10.1111/pce.12009
Dahal, K., and Vanlerberghe, G. C. (2017). Alternative oxidase respiration maintains both mitochondrial and chloroplast function during drought. New Phytol. 213, 560–571. doi: 10.1111/nph.14169
Dahal, K., and Vanlerberghe, G. C. (2018). Improved chloroplast energy balance during water deficit enhances plant growth: more crop per drop. J. Exp. Bot. 69, 1183–1197. doi: 10.1093/jxb/erx474
Day, D. A., and Wiskich, J. T. (1995). Regulation of alternative oxidase activity in higher plants. J. Bioenerget. Biomembr. 27, 379–385. doi: 10.1007/bf02110000
Del-Saz, N. F., Florez-Sarasa, I., Jose Clemente-Moreno, M., Mhadhbi, H., Flexas, J., Fernie, A. R., et al. (2016). Salinity tolerance is related to cyanide-resistant alternative respiration in Medicago truncatula under sudden severe stress. Plant Cell Environ. 39, 2361–2369. doi: 10.1111/pce.12776
Del-Saz, N. F., Ribas-Carbo, M., McDonald, A. E., Lambers, H., Fernie, A. R., and Florez-Sarasa, I. (2018). An in vivo perspective of the role(s) of the alternative oxidase pathway. Trends Plant Sci. 23:3. doi: 10.1016/j.tplants.2017.11.006
Demircan, N., Cucun, G., and Uzilday, B. (2020). Mitochondrial alternative oxidase (AOX1a) is required for the mitigation of arsenic-induced oxidative stress in Arabidopsis thaliana. Plant Biotechnol. Rep. 14, 235–245. doi: 10.1007/s11816-020-00595-9
Djajanegara, I., Holtzapffel, R., Finnegan, P. M., Hoefnagel, M. H. N., Berthold, D. A., Wiskich, J. T., et al. (1999). A single amino acid change in the plant alternative oxidase alters the specificity of organic acid activation. FEBS Lett. 454, 220–224. doi: 10.1016/s0014-5793(99)00808-x
Elthon, T. E., and McIntosh, L. (1987). Identification of the alternative terminal oxidase of higher-plant mitochondria. Proc. Natl. Acad. Sci. U.S.A. 84, 8399–8403. doi: 10.1073/pnas.84.23.8399
Elthon, T. E., Nickels, R. L., and McIntosh, L. (1989). Monoclonal-antibodies to the alternative oxidase of higher-plant mitochondria. Plant Physiol. 89, 1311–1317. doi: 10.1104/pp.89.4.1311
Feng, H. Q., Guan, D. D., Sun, K., Wang, Y. F., Zhang, T. G., and Wang, R. F. (2013). Expression and signal regulation of the alternative oxidase genes under abiotic stresses. Acta Biochim. Biophys. Sin. 45, 985–994. doi: 10.1093/abbs/gmt094
Finnegan, P. M., Whelan, J., Millar, A. H., Zhang, Q., Smith, M. K., Wiskich, J. T., et al. (1997). Differential expression of the multigene family encoding the soybean mitochondrial alternative oxidase. Plant Physiol. 114, 455–466.
Florez-Sarasa, I., Flexas, J., Rasmusson, A. G., Umbach, A. L., Siedow, J. N., and Ribas-Carbo, M. (2011). In vivo cytochrome and alternative pathway respiration in leaves of Arabidopsis thaliana plants with altered alternative oxidase under different light conditions. Plant Cell Environ. 34, 1373–1383. doi: 10.1111/j.1365-3040.2011.02337.x
Florez-Sarasa, I., Obata, T., Del-Saz, N. F., Reichheld, J. P., Meyer, E. H., Rodriguez-Concepcion, M., et al. (2019). The lack of mitochondrial thioredoxin TRXo1 affects in vivo alternative oxidase activity and carbon metabolism under different light conditions. Plant Cell Physiol. 60, 2369–2381. doi: 10.1093/pcp/pcz123
Guy, R. D., and Vanlerberghe, G. C. (2005). Partitioning of respiratory electrons in the dark in leaves of transgenic tobacco with modified levels of alternative oxidase. Pyhsiol. Plant. 125, 171–180. doi: 10.1111/j.1399-3054.2005.00557.x
Hoefnagel, M. H. N., Millar, A. H., Wiskich, J. T., and Day, D. A. (1995). Cytochrome and alternative respiratory pathways compete for electrons in the presence of pyruvate in soybean mitochondria. Arch. Biochem. Biophys. 318, 394–400. doi: 10.1006/abbi.1995.1245
Jayawardhane, J., Cochrane, D. W., Vyas, P., Bykova, N. V., Vanlerberghe, G. C., and Igamberdiev, A. U. (2020). Roles for plant mitochondrial alternative oxidase under normoxia, hypoxia, and reoxygenation conditions. Front. Plant Sci. 11:566. doi: 10.3389/fpls.2020.00566
Kearns, A., Whelan, J., Young, S., Elthon, T. E., and Day, D. A. (1992). Tissue-specific expression of the alternative oxidase in soybean and siratro. Plant Physiol. 99, 712–717. doi: 10.1104/pp.99.2.712
Le, X. H., Lee, C. P., and Millar, A. H. (2021a). The mitochondrial pyruvate carrier (MPC) complex mediates one of three pyruvate-supplying pathways that sustain Arabidopsis respiratory metabolism. Plant Cell 33, 2776–2793. doi: 10.1093/plcell/koab148
Le, X. H., Lee, C. P., Monachello, D., and Millar, A. H. (2021b). Metabolic evidence for distinct pyruvate pools inside plant mitochondria. bioRxiv [Preprint] doi: 10.1101/2021.10.30.466557
Li, C. R., Liang, D. D., Xu, R. F., Li, H., Zhang, Y. P., Qin, R. Y., et al. (2013). Overexpression of an alternative oxidase gene, OsAOX1a, improves cold tolerance in Oryza sativa L. Genet. Mol. Res. 12, 5424–5432. doi: 10.4238/2013.November.11.4
Maxwell, D. P., Wang, Y., and McIntosh, L. (1999). The alternative oxidase lowers mitochondrial reactive oxygen production in plant cells. Proc. Natl. Acad. Sci. U.S.A. 96, 8271–8276. doi: 10.1073/pnas.96.14.8271
McCabe, T. C., Finnegan, P. M., Millar, A. H., Day, D. A., and Whelan, J. (1998). Differential expression of alternative oxidase genes in soybean cotyledons during postgerminative development. Plant Physiol. 118, 675–682. doi: 10.1104/pp.118.2.675
Mhadhbi, H., Fotopoulos, V., Mylona, P. V., Jebara, M., Aouani, M. E., and Polidoros, A. N. (2013). Alternative oxidase 1 (Aox1) gene expression in roots of Medicago truncatula is a genotype-specific component of salt stress tolerance. J. Plant Physiol. 170, 111–114. doi: 10.1016/j.jplph.2012.08.017
Millar, A. H., Atkin, O. K., Menz, R. I., Henry, B., Farquhar, G., and Day, D. A. (1998). Analysis of respiratory chain regulation in roots of soybean seedlings. Plant Physiol. 117, 1083–1093. doi: 10.1104/pp.117.3.1083
Millar, A. H., Hoefnagel, M. H. N., Day, D. A., and Wiskich, J. T. (1996). Specificity of the organic acid activation of alternative oxidase in plant mitochondria. Plant Physiol. 111, 613–618.
Millar, A. H., Wiskich, J. T., Whelan, J., and Day, D. A. (1993). Organic-acid activation of the alternative oxidase of plant-mitochondria. FEBS Lett. 329, 259–262. doi: 10.1016/0014-5793(93)80233-k
Millenaar, F. F., and Lambers, H. (2003). The alternative oxidase: in vivo regulation and function. Plant Biol. 5, 2–15.
Moller, I. M., Igamberdiev, A. U., Bykova, N. V., Finkemeier, I., Rasmusson, A. G., and Schwarzlander, M. (2020). Matrix redox physiology governs the regulation of plant mitochondrial metabolism through posttranslational protein modifications. Plant Cell 32, 573–594. doi: 10.1105/tpc.19.00535
Obenland, D., Diethelm, R., Shibles, R., and Stewart, C. (1990). Relationship of alternative respiratory capacity and alternative oxidase amount during soybean seedling growth. Plant Cell Physiol. 31, 897–901.
Pastore, D., Trono, D., Laus, M. N., Di Fonzo, N., and Passarella, S. (2001). Alternative oxidase in durum wheat mitochondria. Activation by pyruvate, hydroxypyruvate and glyoxylate and physiological role. Plant Cell Physiol. 42, 1373–1382.
Polidoros, A. N., Mylona, P. V., Pasentsis, K., Scandalios, J. G., and Tsaftaris, A. S. (2005). The maize alternative oxidase 1a (Aox1a) gene is regulated by signals related to oxidative stress. Redox Rep. 10, 71–78. doi: 10.1179/135100005x21688
Rhoads, D. M., and McIntosh, L. (1992). Salicylic acid regulation of respiration in higher plants: alternative oxidase expression. Plant Cell 4, 1131–1139.
Rhoads, D. M., Umbach, A. L., Sweet, C. R., Lennon, A. M., Rauch, G. S., and Siedow, J. N. (1998). Regulation of the cyanide-resistant alternative oxidase of plant mitochondria – identification of the cysteine residue involved in alpha-keto acid stimulation and intersubunit disulfide bond formation. J. Biol. Chem. 273, 30750–30756.
Ribas-Carbo, M., Taylor, N. L., Giles, L., Busquets, S., Finnegan, P. M., Day, D. A., et al. (2005). Effects of water stress on respiration in soybean leaves. Plant Physiol. 139, 466–473. doi: 10.1104/pp.105.065565
Schwarzlander, M., and Fuchs, P. (2019). Keeping mitochondrial alternative oxidase reduced and active in vivo does not require Thioredoxin o1. Plant Cell Physiol. 60, 2357–2359. doi: 10.1093/pcp/pcz173
Selinski, J., Hartmann, A., Deckers-Hebestreit, G., Day, D. A., Whelan, J., and Scheibe, R. (2018). Alternative oxidase isoforms are differentially activated by tricarboxylic acid cycle intermediates. Plant Physiol. 176, 1423–1432. doi: 10.1104/pp.17.01331
Selinski, J., Hartmann, A., Hofler, S., Deckers-Hebestreit, G., and Scheibe, R. (2016). Refined method to study the posttranslational regulation of alternative oxidases from Arabidopsis thaliana in vitro. Physiol. Plant. 157, 264–279. doi: 10.1111/ppl.12418
Selinski, J., Hartmann, A., Kordes, A., Deckers-Hebestreit, G., Whelan, J., and Scheibe, R. (2017). Analysis of posttranslational activation of alternative oxidase isoforms. Plant Physiol. 174, 2113–2127. doi: 10.1104/pp.17.00681
Senkler, J., Senkler, M., Eubel, H., Hildebrandt, T., Lengwenus, C., Schertl, P., et al. (2017). The mitochondrial complexome of Arabidopsis thaliana. Plant J. 89, 1079–1092. doi: 10.1111/tpj.13448
Siedow, J. N., and Day, D. A. (2000). “Respiration and photorespiration,” in Biochemistry and Molecular Biology of Plants, eds B. B. Buchanan, W. Gruissem, and R. L. Jones (Rockville, MD: American Society of Plant Physiologists), 676–728.
Sluse, F. E., and Jarmuszkiewicz, W. (1998). Alternative oxidase in the branched mitochondrial respiratory network: an overview on structure, function, regulation, and role. Braz. J. Med. Biol. Res. 31, 733–747. doi: 10.1590/s0100-879x1998000600003
Smith, C. A., Melino, V. J., Sweetman, C., and Soole, K. L. (2009). Manipulation of alternative oxidase can influence salt tolerance in Arabidopsis thaliana. Physiol. Plant. 137, 459–472. doi: 10.1111/j.1399-3054.2009.01305.x
Soole, K. L., and Sweetman, C. (2021). “Respiration | plant mitochondria – substrates, inhibitors, uncouplers,” in Encyclopedia of Biological Chemistry III, 3rd Edn, ed. J. Jez (Oxford: Elsevier), 536–544.
Sweetman, C., Miller, T. K., Booth, N. J., Shavrukov, Y., Jenkins, C. L. D., Soole, K. L., et al. (2020). Identification of alternative mitochondrial electron transport pathway components in chickpea indicates a differential response to salinity stress between cultivars. Int. J. Mol. Sci. 21:3844. doi: 10.3390/ijms21113844
Sweetman, C., Soole, K. L., Jenkins, C. L. D., and Day, D. A. (2019a). Genomic structure and expression of alternative oxidase genes in legumes. Plant Cell Environ. 42, 71–84. doi: 10.1111/pce.13161
Sweetman, C., Waterman, C. D., Rainbird, B. M., Smith, P. M. C., Jenkins, C. L. D., Day, D. A., et al. (2019b). AtNDB2 is the main external NADH dehydrogenase in mitochondria and is important for tolerance to environmental stress. Plant Physiol. 181, 774–788. doi: 10.1104/pp.19.00877
Umbach, A. L., and Siedow, J. N. (1993). Covalent and noncovalent dimers of the cyanide-resistant alternative oxidase protein in higher plant mitochondria and their relationship to enzyme activity. Plant Physiol. 103, 845–854.
Umbach, A. L., and Siedow, J. N. (1996). The reaction of the soybean cotyledon mitochondrial cyanide-resistant oxidase with sulfhydryl reagents suggests that alpha-keto acid activation involves the formation of a thiohemiacetal. J. Biol. Chem. 271, 25019–25026.
Umbach, A. L., Fiorani, F., and Siedow, J. N. (2005). Characterization of transformed Arabidopsis with altered alternative oxidase levels and analysis of effects on reactive oxygen species in tissue. Plant Physiol. 139, 1806–1820. doi: 10.1104/pp.105.070763
Umbach, A. L., Wiskich, J. T., and Siedow, J. N. (1994). Regulation of alternative oxidase kinetics by pyruvate and intermolecular disulfide bond redox status in soybean seedling mitochondria. FEBS Lett. 348, 181–184.
Vanlerberghe, G. C. (2013). Alternative oxidase: a mitochondrial respiratory pathway to maintain metabolic and signaling homeostasis during abiotic and biotic stress in plants. Int. J. Mol. Sci. 14, 6805–6847. doi: 10.3390/ijms14046805
Vanlerberghe, G. C., and McIntosh, L. (1994). Mitochondrial electron-transport regulation of nuclear gene-expression – studies with the alternative oxidase gene of tobacco. Plant Physiol. 105, 867–874.
Vanlerberghe, G. C., and McIntosh, L. (1996). Signals regulating the expression of the nuclear gene encoding alternative oxidase of plant mitochondria. Plant Physiol. 111, 589–595.
Vanlerberghe, G. C., Dahal, K., Alber, N. A., and Chadee, A. (2020). Photosynthesis, respiration and growth: a carbon and energy balancing act for alternative oxidase. Mitochondrion 52, 197–211. doi: 10.1016/j.mito.2020.04.001
Vanlerberghe, G. C., Martyn, G. D., and Dahal, K. (2016). Alternative oxidase: a respiratory electron transport chain pathway essential for maintaining photosynthetic performance during drought stress. Physiol. Plant. 157, 322–337. doi: 10.1111/ppl.12451
Vanlerberghe, G. C., McIntosh, L., and Yip, J. Y. H. (1998). Molecular localization of a redox-modulated process regulating plant mitochondrial electron transport. Plant Cell 10, 1551–1560.
Vishwakarma, A., Tetali, S. D., Selinski, J., Scheibe, R., and Padmasree, K. (2015). Importance of the alternative oxidase (AOX) pathway in regulating cellular redox and ROS homeostasis to optimize photosynthesis during restriction of the cytochrome oxidase pathway in Arabidopsis thaliana. Ann. Bot. 116, 555–569. doi: 10.1093/aob/mcv122
Wanniarachchi, V. R., Dametto, L., Sweetman, C., Shavrukov, Y., Day, D. A., Jenkins, C. L. D., et al. (2018). Alternative respiratory pathway component genes (AOX and ND) in rice and barley and their response to stress. Int. J. Mol. Sci. 19:915. doi: 10.3390/ijms19030915
Watanabe, C. K. A., Yamori, W., Takahashi, S., Terashima, I., and Noguchi, K. (2016). Mitochondrial alternative pathway-associated photoprotection of photosystem II is related to the photorespiratory pathway. Plant Cell Physiol. 57, 1426–1431. doi: 10.1093/pcp/pcw036
Keywords: alternative oxidase, isoform, recombinant, kinetics, pyruvate, activation, soybean, legume
Citation: Sweetman C, Selinski J, Miller TK, Whelan J and Day DA (2022) Legume Alternative Oxidase Isoforms Show Differential Sensitivity to Pyruvate Activation. Front. Plant Sci. 12:813691. doi: 10.3389/fpls.2021.813691
Received: 12 November 2021; Accepted: 27 December 2021;
Published: 17 January 2022.
Edited by:
Paula Mulo, University of Turku, FinlandReviewed by:
Igor Florez-Sarasa, Centre for Research in Agricultural Genomics, Spanish National Research Council (CSIC), SpainCopyright © 2022 Sweetman, Selinski, Miller, Whelan and Day. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: David A. Day, ZGF2aWQuZGF5QGZsaW5kZXJzLmVkdS5hdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.