- 1School of Grassland Science, Beijing Forestry University, Beijing, China
- 2College of Resources and Environment, International Magnesium Institute, Fujian Agriculture and Forestry University, Fuzhou, China
Plants and arbuscular mycorrhizal fungi (AMF) can form complex symbiotic networks based on functional trait selection, contributing to the maintenance of ecosystem biodiversity and stability. However, the selectivity of host plants on AMF and the characteristics of plant-AMF networks remain unclear in Tibetan alpine meadows. In this study, we studied the AMF communities in 69 root samples from 23 plant species in a Tibetan alpine meadow using Illumina-MiSeq sequencing of the 18S rRNA gene. The results showed a significant positive correlation between the phylogenetic distances of plant species and the taxonomic dissimilarity of their AMF community. The plant-AMF network was characterized by high connectance, high nestedness, anti-modularity, and anti-specialization, and the phylogenetic signal from plants was stronger than that from AMF. The high connected and nested plant-AMF network potentially promoted the interdependence and stability of the plant-AMF symbioses in Tibetan alpine meadows. This study emphasizes that plant phylogeny and plant-AMF networks play an important role in the coevolution of host plants and their mycorrhizal partners and enhance our understanding of the interactions between aboveground and belowground communities.
Introduction
Arbuscular mycorrhizal fungi (AMF) are an ancient root symbiotic group, whose origin is consistent with the first appearance of terrestrial plants, and generally considered to be the result of coevolution of fungi and plants (Bonfante and Genre, 2008; Walder and van der Heijden, 2015; Genre et al., 2020). As a key component of the underground biological community, AMF can form a symbiotic relationship with most terrestrial plant species and can increase the absorption of phosphorus and nitrogen for plants (Wang and Qiu, 2006; Smith and Read, 2008; Zhang et al., 2021). At the same time, plants provide photosynthetic carbon products to AMF (Bever et al., 2009; Kiers et al., 2011; Wipf et al., 2019). The combination of AMF and plants improves the adaptability of plants and AMF symbionts (Blackwell, 2000; Chen et al., 2018; Tedersoo et al., 2020). Furthermore, the plant-AMF symbioses are not formed stochastically but are dependent on their functional traits (Bever et al., 2009; Kiers et al., 2011). Both plant hosts and AMF have been shown to preferentially allocate resources to higher-quality partners (Ji and Bever, 2016), inducing reciprocal shifts in each assemblage, ultimately affecting the partner selection in the mycorrhizal symbiosis (Werner and Kiers, 2015). Due to the simultaneous processes of coevolution, niche differentiation, and niche conservatism, the closely related plants tend to interact with similar AMF species (i.e., phylogenetic signal in the interaction) (Gomez et al., 2010; Sargent et al., 2011). However, negative correlations between the phylogenetic distance of plants and AMF community distance were also reported in three locations of the northern Great Plains of the United States (Reinhart and Anacker, 2014). In addition, the specificity of the interactions between the plant and AMF changed with community succession (Montesinos-Navarro et al., 2012a). These contrasting findings indicate the complexity of plant and AMF interactions.
Many studies have shown that phylogenetic relatedness affects the mycorrhizal symbiotic network structure, which could answer the question of how coevolution explained species assemblages (Barberan et al., 2012; Chagnon et al., 2012; Montesinos-Navarro et al., 2012b; Chen et al., 2017; Goberna et al., 2019). The network is increasingly used to study co-occurrence patterns in plant and AMF symbioses, providing insights for biological interactions (Barberan et al., 2012; Chagnon et al., 2012; Chen et al., 2017; Gao et al., 2019). The AMF communities of 166 root samples from 17 woody plants in subtropical forests of China showed that the woody plant-AMF network was highly interconnected and nested but in anti-modular and anti-specialized manners (Chen et al., 2017). Most of the studies on the plant-fungus symbiotic network focused on temperate and tropical ecosystems, and the research objects were mainly woody plant-AMF symbiotic interactions. However, herbaceous plant-AMF network characteristics in the alpine ecosystems remain poorly understood.
As the Third Pole on the earth, the Tibetan Plateau is a unique geographic unit with great biodiversity and critical ecosystem functions and is experiencing striking climate warming (Bai-ping et al., 2002; Duan et al., 2006; Qiu, 2008). Alpine meadow is a typical vegetation type, accounting for approximately 40% of the total grassland area on the Tibetan Plateau (Miehe et al., 2008; Wang et al., 2016). The importance of AMF in plant succession on the Tibetan plateau has been well documented (Liu et al., 2011; Gai et al., 2012; Li et al., 2015). However, most of the studies focused on the effects of altitude gradients (Liu et al., 2011; Gai et al., 2012; Li et al., 2014), precipitation (Zhang et al., 2016), and vegetation types (Gai et al., 2009; Gao and Guo, 2010; Xu et al., 2017) on AMF community composition. The selectivity of the interactions between the plant and AMF and their network properties in Tibetan alpine meadows remains unclear.
In this study, to explore the selectivity in the plant-AMF interactions and clarify the maintenance mechanism of symbiosis, 23 species of common herbs in a Tibetan alpine meadow were selected. Illumina-MiSeq sequencing was used to characterize AMF communities living in each plant root to address the following questions: (1) How is the selectivity in the plant-AMF interactions in Tibetan alpine meadow? and (2) What are the characteristics of the plant-AMF network in Tibetan alpine meadows?
Materials and Methods
Site and Sampling
The study site is located at the Naqu Alpine Grassland Ecosystem National Field Scientific Observation and Research Station (Kaima Station) of Tibetan Plateau Research of Chinese Academy of Sciences (31°16′N, 92°06′E, 4453 m a.s.l). This region is a typical alpine meadow ecosystem, where the mean annual temperature is −2.1°C, the mean annual rainfall is 406 mm, and most of the precipitation was concentrated from June to September. Summers are short and cool, and winters are long and severely cold (Cui et al., 2017). The plant community is dominated by Kobresia Pygmaea and Kobresia Humilis, and the Carex moorcroftii, Stipa purpurea, Poa tibetica, Potentilla bifurca, and Heteropappus semiprostratus (Hs) are companion species.
In mid-August 2018, 23 representative plant species (Figure 1) were selected based on a vegetation survey in the alpine meadow of Naqu county. Notably, 23 sampling sites were randomly chosen for plant sampling around the Kaima Station, which was at least 50 m away from each other to avoid interactions among plant species. Then, three 0.5 × 0.5 m2 quadrats were randomly selected in each sampling site, and three plant individuals in each quadrat were collected and pooled as one composite sample. Finally, 207 plant individuals of 69 samples (i.e., 23 plant species × 3 replicates) belonging to 11 families and 20 genera (Figure 1, according to the genetic relationship of species) were collected in this study. Plant roots were carefully separated from aboveground parts and soil, packaged in plastic bags, refrigerated and transported to the laboratory, and stored at −2°C for colonization determination and DNA extraction.
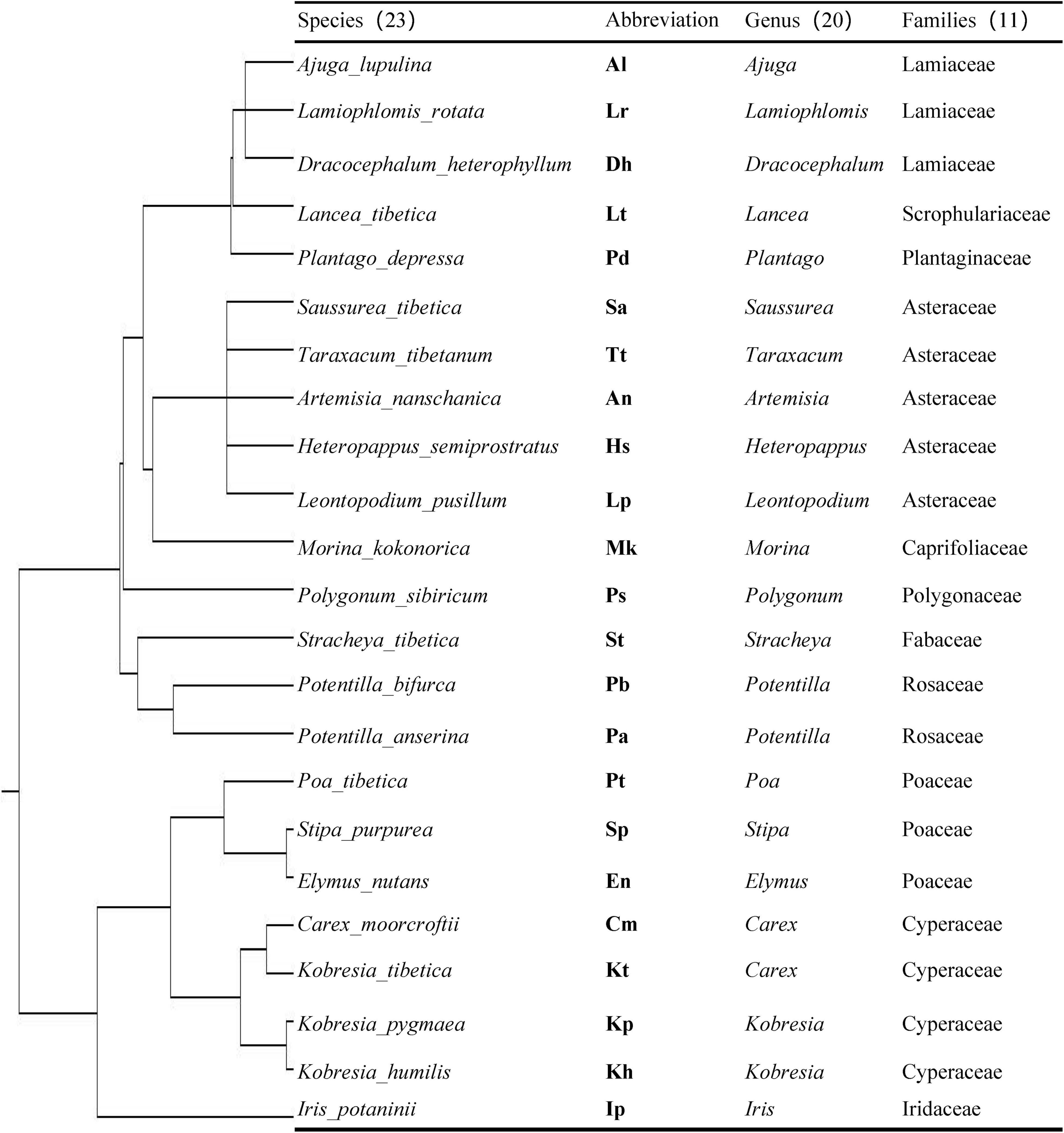
Figure 1. Phylogenetic tree of 23 plants species constructed using Phylomatic. The detailed information of the 23 plant species is shown including their abbreviation, genus, and families.
Plant Phylogenetic Tree
Based on Zanne phylogeny as the backbone (Zanne et al., 2014), the phylogenetic tree of plant species was constructed using Phylomatic (V.3)1. The phylogenetic distance of plant species was calculated using R software 3.1.1 with the package “picante” (V.1.8.2) (Kembel et al., 2010).
Arbuscular Mycorrhizal Fungi Colonization
The plant roots were washed carefully and cut into 1 cm length. Approximately, 100 root segments of each sample were cleared in 10% KOH at 90°C for 90 min, then acidified in 2% HCl at room temperature for 10 min, and stained with 0.05% Trypan blue at 90°C for 10 min. Finally, the dyed root segments were randomly selected and measured for the mycorrhizal colonization by the magnified intersections method at 200 × microscopes (McGonigle et al., 1990).
DNA Extraction and Polymerase Chain Reaction
Total DNA was extracted from a 0.05 g (freeze-dried) subsample of fine roots using a PowerPlant DNA Isolation Kit (MoBio Laboratories, United States) following the instructions of manufacturer. A NanoDrop 8000 Ultramicro UV spectrophotometer (NanoDrop Technologies, Wilmington, DE, United States) was used to determine the quantity and purity of DNA samples. A two-step polymerase chain reaction (PCR) was performed to amplify the 18S ribosomal RNA (rRNA) gene. The primers used in the first PCR were NS31 (TTGGAGGGCAAGTCTGGTGCC) and AML2 (GAACCCAAACACTTTGGTTTCC) and the second PCR amplification with the primers AMV4.5NF (AAGCTCGTAGTTGAATTTCG) and AMDGR (CCCAA CTATCCCTATTAATCAT) (Lumini et al., 2010; Van Geel et al., 2014). The PCR products were purified with an agarose gel DNA purification kit (AP-GX-250G; Axygen, United States) and quantified using a NanoDrop 8000. The purified products were pooled in equimolar amounts and then sequenced on the Illumina-MiSeq platform (Shanghai Magigene Inc., Shanghai, China).
Bioinformatics Analysis
We used the Quantitative Insights Into Microbial Ecology (QIIME) and the UPARSE pipeline to treat raw sequences as described previously (Caporaso et al., 2010; Edgar, 2013). Sequence quality control was conducted with QIIME, and the obtained high-quality sequences were imported into a Usearch (vision 11). Then, the Usearch was used for dereplication and merging of paired-end reads. The singletons were removed, and the chimeras were detected and removed using the de novo approach. The operational taxonomic units (OTUs) were clustered at 97% similarity. The representative sequences of OTUs were uploaded to the National Center for Biotechnology Information (NCBI) and MarrjAM2 database for taxonomy identification. Non-AMF reads were removed from the OTU table. Then, the OTU table was rarefied to a sequencing depth of 5,000 reads per sample, resulting in 345,000 AMF reads for all samples (i.e., 69 samples × 5,000 reads = 345,000 AMF reads). The rarefaction curves of each sample tended to be flat, indicating adequate sequencing depth (Supplementary Figure 1). Alpha diversity metrics (richness and Shannon-Wiener) were calculated using the rarefied OTU table. The phylogenetic tree of AMF OTUs was constructed according to Neighbor-Joining using MEGA 7.0 software (Supplementary Figure 2). The sequences obtained in this study have been submitted to the GenBank database (PRJNA 748864).
Statistical Analysis
Analysis of variance (ANOVA) test and Tukey’s honestly (HSD) significant difference test (P < 0.05) were used for the pairwise comparisons of the diversity of AMF in different plant species. We tested for host-plant species effects on AMF community dissimilarity using Permutational ANOVA (PERMANOVA) using the “adonis” function in the R package “vegan” (V.2.5.7) with 999 permutations. Mantel tests were used to examine the correlation between AMF community dissimilarity matrix (Bray-Curtis and Unifrac distance) and plant phylogenetic distance matrix, and the relationship between pairwise AMF community dissimilarity distances and plant phylogenetic distances was tested by linear regressions. To better explain the plant-AMF symbiotic network, the plant-AMF visualization matrix at the species level was established. For a given paired plant-OTU sample, the association frequency (i.e., 0, 1, 2, or 3) was defined as the number of plant root samples with AMF OTUs observed (Toju et al., 2016), which reflected the selectivity of plant-AMF association. The association frequency matrix was represented by the heat map using TBtools software (v.0.6655) (Chen et al., 2020).
The species-level matrix was used to calculate the network indexes, such as connectance, nestedness, modularity, specialization, and checkerboard score. The ecological meaning of these indexes can be found in the Supplementary Material.
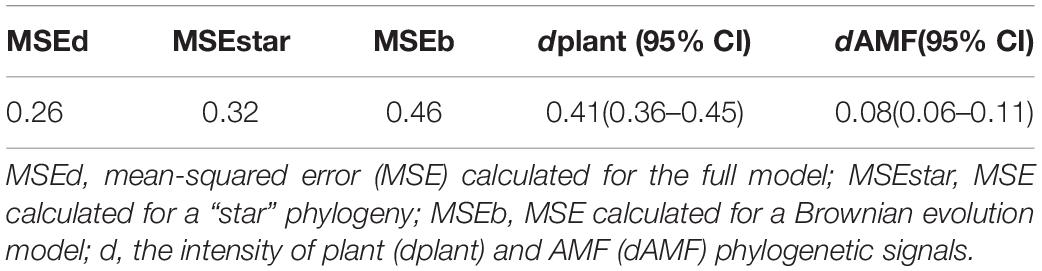
With network analysis and null model comparison, we could infer the non-randomness constructional laws of biological communities (Ulrich and Gotelli, 2007). The result of the network structure index was stable using the random test. First, random values of 1,000 randomly arranged samples of the null model matrix were generated, and then the t-test (P < 0.05) was used to test the significant difference between the observed values and the random values based on the null model. Based on the association frequency matrix and random matrix, weighted connectance, H2′, weighted nestedness metric based on the overlap and decreasing fill (WNODF), and checkerboard score index were calculated using “bipartite” package “networklevel” command in R (Dormann et al., 2013). For the modular analysis, the association frequency matrix and random matrix were the outputs from R, and then the data were analyzed using the MODULAR program. The estimated generalized least squares method (EGLS) was used to assess the phylogenetic signals of plant-AMF. The analysis fitted the phylogenetic variance-covariance matrix to the plant-AMF association frequency matrix (Ives and Godfray, 2006; Montesinos-Navarro et al., 2012b). Using the EGLS method, we calculated the independent phylogenetic signals of the plant (dplant) and AMF (dAMF) on association frequency matrix, assessed the overall intensity of the two phylogenetic signals combined with the mean square error (MSE) value, and regarded the model of minimizing MSE as the best fit. The MSE full model (MSEd), the MSE derived under the assumption of no phylogenetic signal (i.e., a star phylogeny, MSEstar), and maximum phylogenetic signal (i.e., Brownian motion evolution, MSEb) were calculated according to the description of Chen et al. (2017).
Results
Arbuscular Mycorrhizal Fungi Colonization, Diversity, and Community Composition of Different Plant Species
The sequences annotated as AMF were grouped into 121 OTUs, covering 3 orders, 6 families, and 9 genera (Supplementary Figure 3 and Supplementary Table 1). Glomus was the dominant genus (181013 sequences corresponding to 52.5% of the total sequences and 84 OTUs corresponding to 69.4% of the total OTUs), followed by Claroideoglomus and Rhizophagus (Supplementary Table 1).
Arbuscular mycorrhizal fungi colonization was significantly different (F = 9.14, P < 0.001) among the 23 plant species (Figure 2A). The colonization of Hs was the lowest (18.3 ± 3.0%) and Lancea tibetica (Lt) was the highest (90.0 ± 2.7%). Significant differences in the Shannon-Wiener index were found among the AMF community hosted by different plant species (Figure 2B; F = 3.07, P = 0.001), but no significant differences were found in OTU richness (Figure 2C; F = 1.55, P = 0.10). Elymus nutans (En) of Poace had the highest AMF richness and Shannon-Wiener index, while Leontopodium Pusillum (Lp) of Asteraceae showed the lowest diversity. For example, Glomus and Rhizophagus were more enriched in Potentilla bifurca (Pb) and Lp root separately relative to other species, respectively (Figure 3). PERMANOVA showed that taxonomic composition was significantly different (Figure 4A; F = 2.81, P = 0.001), but not phylogenetic composition (Figure 4B; F = 1.38, P = 0.18) of AMF communities varied among plant species.
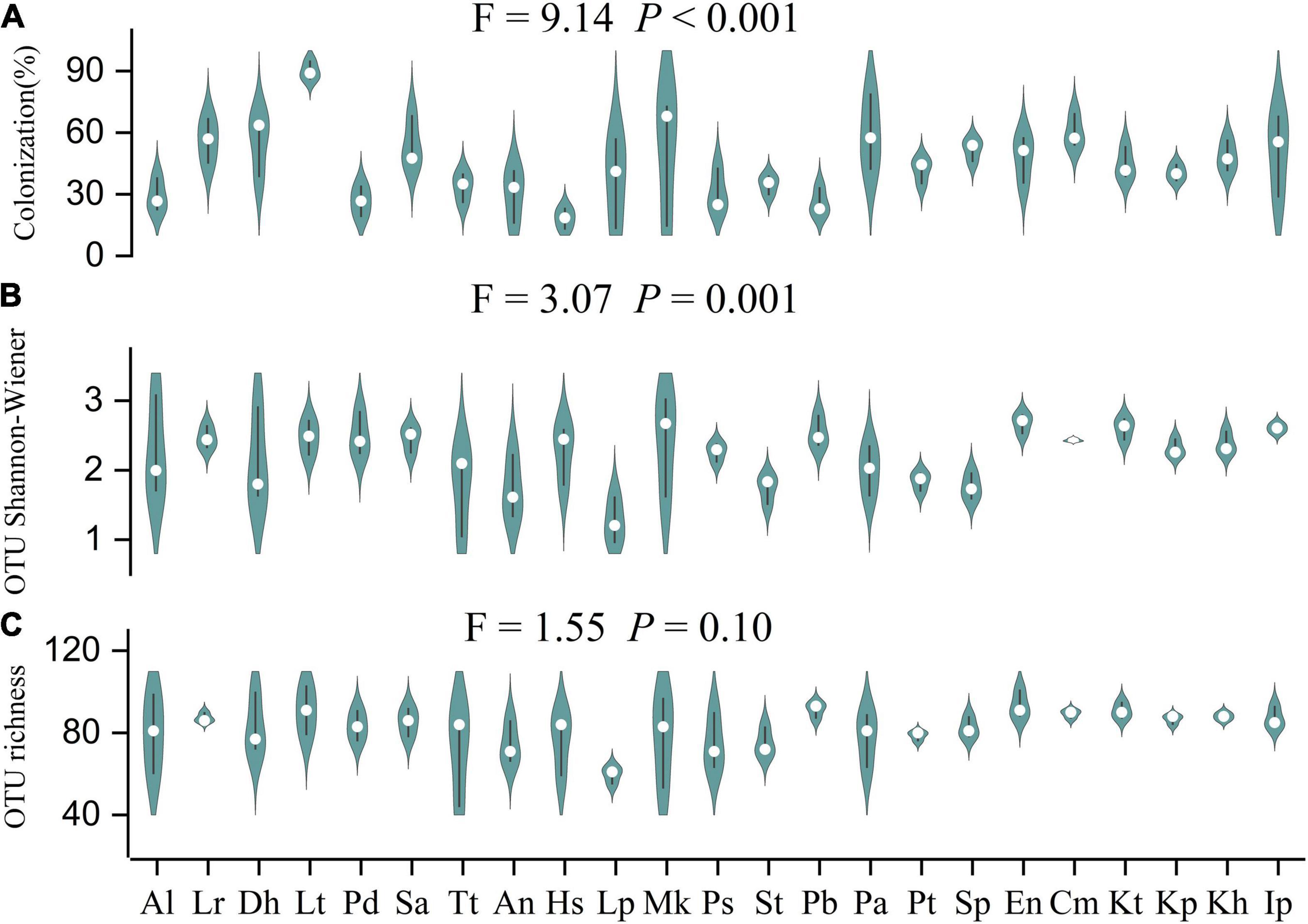
Figure 2. Differences in Arbuscular mycorrhizal fungi (AMF) colonization (A); OTU Shannon-Wiener diversity index (B); and OTU richness (C) of 23 plant roots. Al: Ajuga lupulina; Lr: Lamiophlomis rotata; Dh: Dracocephalum heterophyllum; Lt: Lancea tibetica; Pd: Plantago depressa; Sa: Saussurea tibetica; Tt: Taraxacum tibetanum; An: Artemisia nanschanica; Hs: Heteropappus semiprostratus; Lp: Leontopodium pusillum; Mk: Morina kokonorica; Ps: Polygonum sibiricum; St: Stracheya tibetica; Pb: Potentilla bifurca; Pa: Potentilla anserina; Pt: Poa tibetica; Sp: Stipa purpurea; En: Elymus nutans; Cm: Carex moorcroftii; Kt: Kobresia tibetica; Kp: Kobresia pygmaea; Kh: Kobresia humilis; Ip: Iris potaninii.
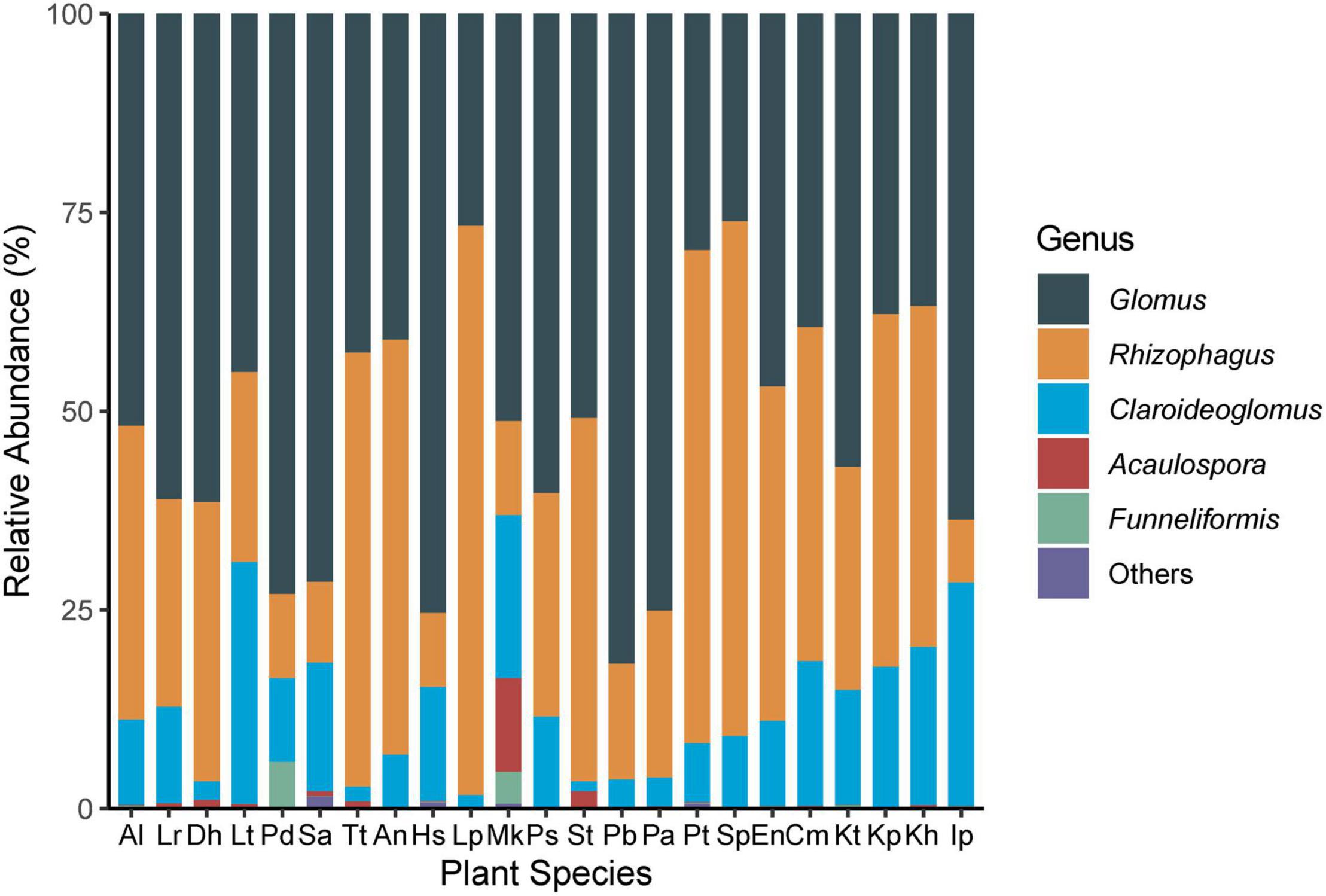
Figure 3. Composition of AMF genera in roots of 23 plant roots. Al: Ajuga lupulina; Lr: Lamiophlomis rotata; Dh: Dracocephalum heterophyllum; Lt: Lancea tibetica; Pd: Plantago depressa; Sa: Saussurea tibetica; Tt: Taraxacum tibetanum; An: Artemisia nanschanica; Hs: Heteropappus semiprostratus; Lp: Leontopodium pusillum; Mk: Morina kokonorica; Ps: Polygonum sibiricum; St: Stracheya tibetica; Pb: Potentilla bifurca; Pa: Potentilla anserina; Pt: Poa tibetica; Sp: Stipa purpurea; En: Elymus nutans; Cm: Carex moorcroftii; Kt: Kobresia tibetica; Kp: Kobresia pygmaea; Kh: Kobresia humilis; Ip: Iris potaninii.
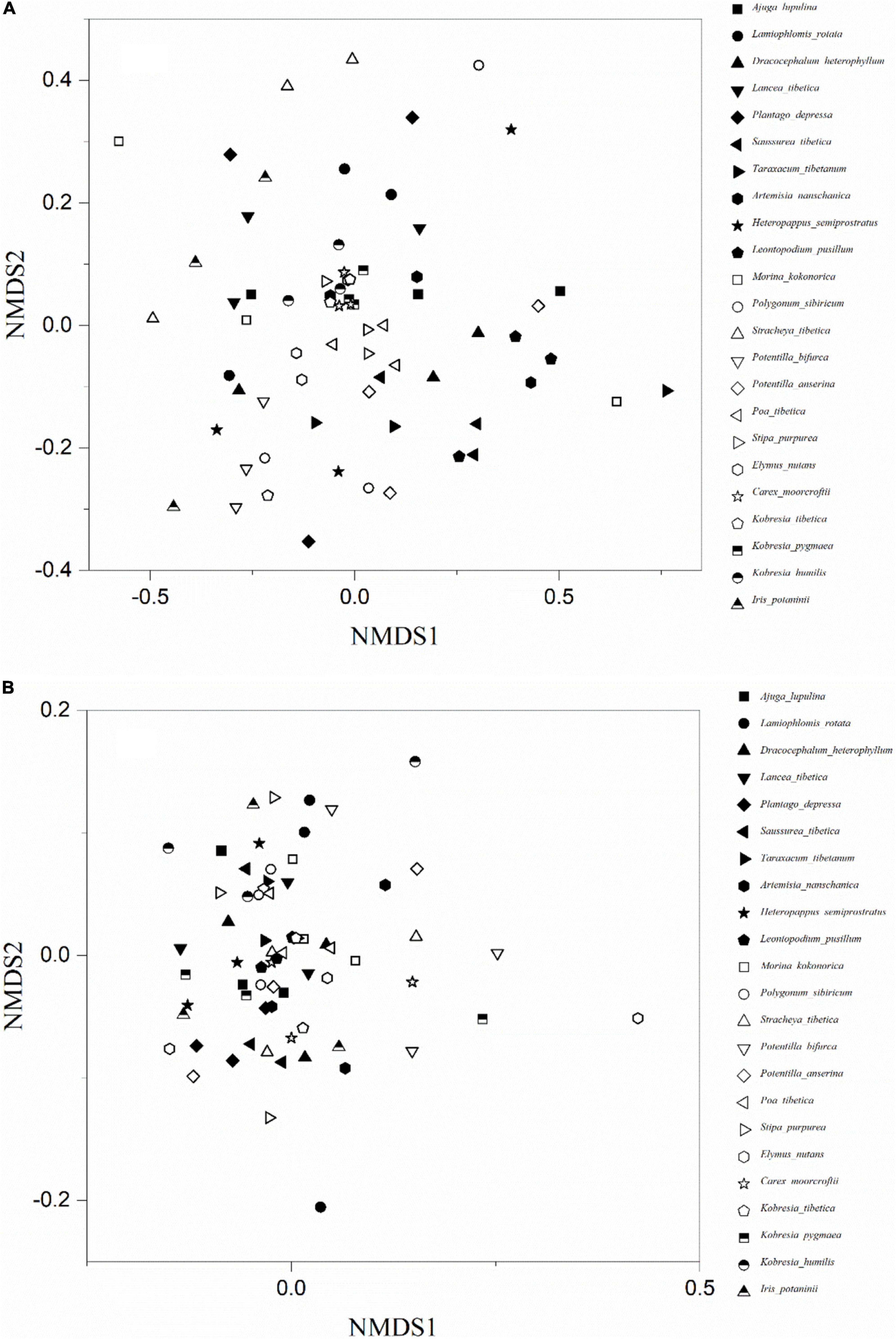
Figure 4. The non-metric multidimensional scaling (NMDS) ordinations of AMF taxonomic (A) and phylogenetic (B) community composition of 23 plant roots.
Phylogenetic Correlations Between Plants and Arbuscular Mycorrhizal Fungi
The result of linear fitting and mantel test showed that there was a significant positive correlation between the plant interspecies phylogenetic distance and the corresponding AMF taxonomic community dissimilarity (Figure 5A; R2 = 0.13, P < 0.001; r = 0.36, P = 0.001), meaning that closely related plant species in phylogenetic trees tend to have more similar AMF community compositions. The result of linear fitting between the plant interspecies phylogenetic distance and corresponding AMF phylogenetic community dissimilarity was the same as above, but the mantel test was not significant (Figure 5B; R2 = 0.01, P = 0.03; r = 0.03, P = 0.26).
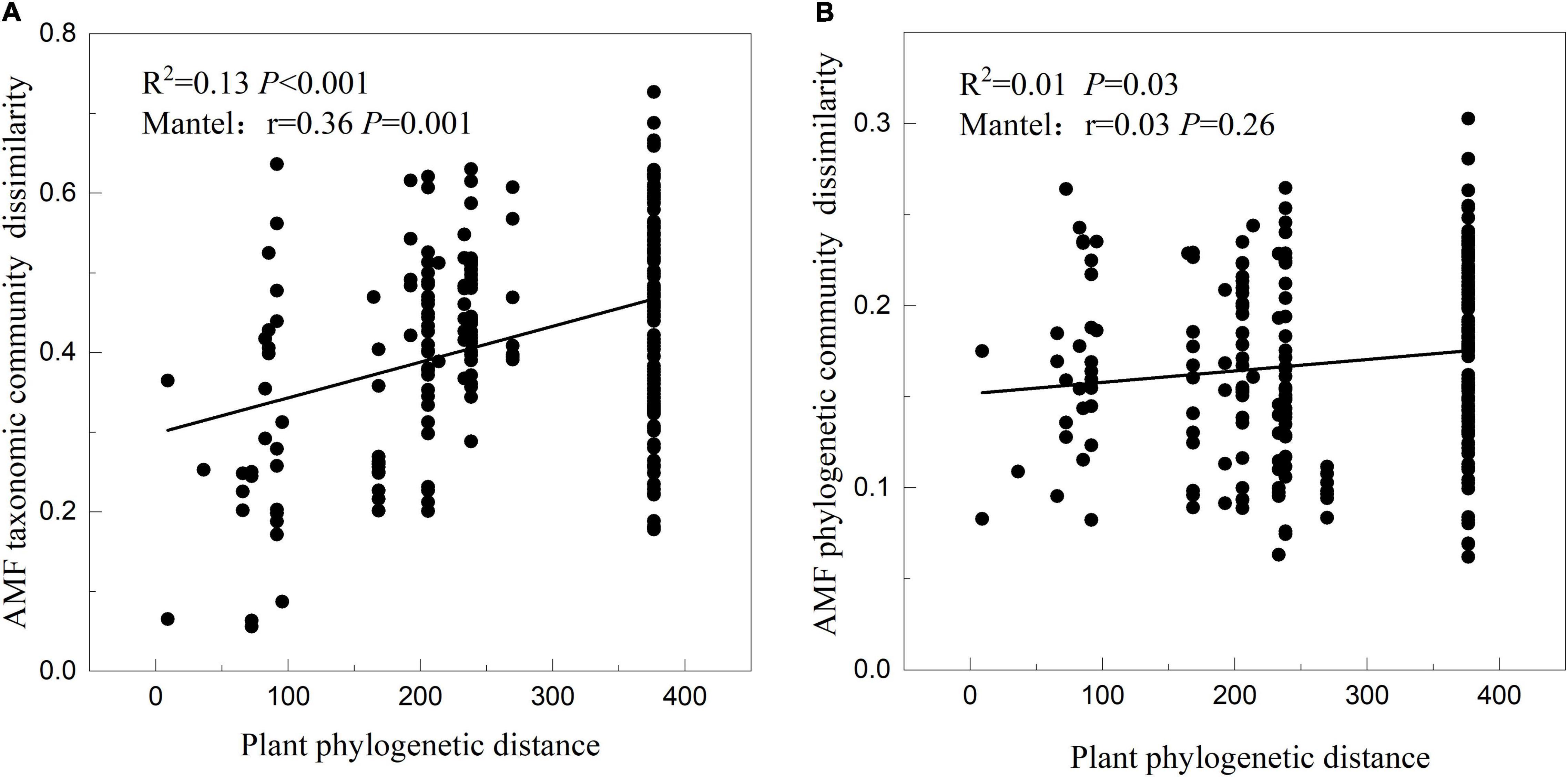
Figure 5. Relationships between the phylogenetic distance of 23 plant species and their corresponding AMF taxonomic (A) and phylogenetic (B) community dissimilarity tested by linear regression and mantel test.
Characteristics of Plant-AMF Network
The heat map of the plant-AMF incidence matrix showed that the symbiotic network relationship between the plant and AMF was established (Figure 6). Compared with the predicted value of the null model, the characteristics of the plant-AMF network, such as high connectance, high nestedness, anti-modularity, and anti-specialization, were obtained (Figures 7A–D). The observed values of weighted connectance and WNODF were significantly higher than the predicted values (P < 0.001). The values of H2′ and modularity were both significantly lower than the observed values (P < 0.001). The results of the checkerboard score analysis showed that the observed values of plants and AMF communities were significantly lower than the expected values based on the null model (P < 0.001), but the checkerboard score (Figures 7E,F; fungus: 0.51, plant: 0.37) showed that the competition intensity of AMF was higher than that of plant species. The plant-AMF association frequency matrix showed that the phylogenetic signals of the plant species and AMF OTUs had significant effects on the plant-AMF network structure, and the phylogenetic signals from the plant were stronger than that from AMF (Table 1).
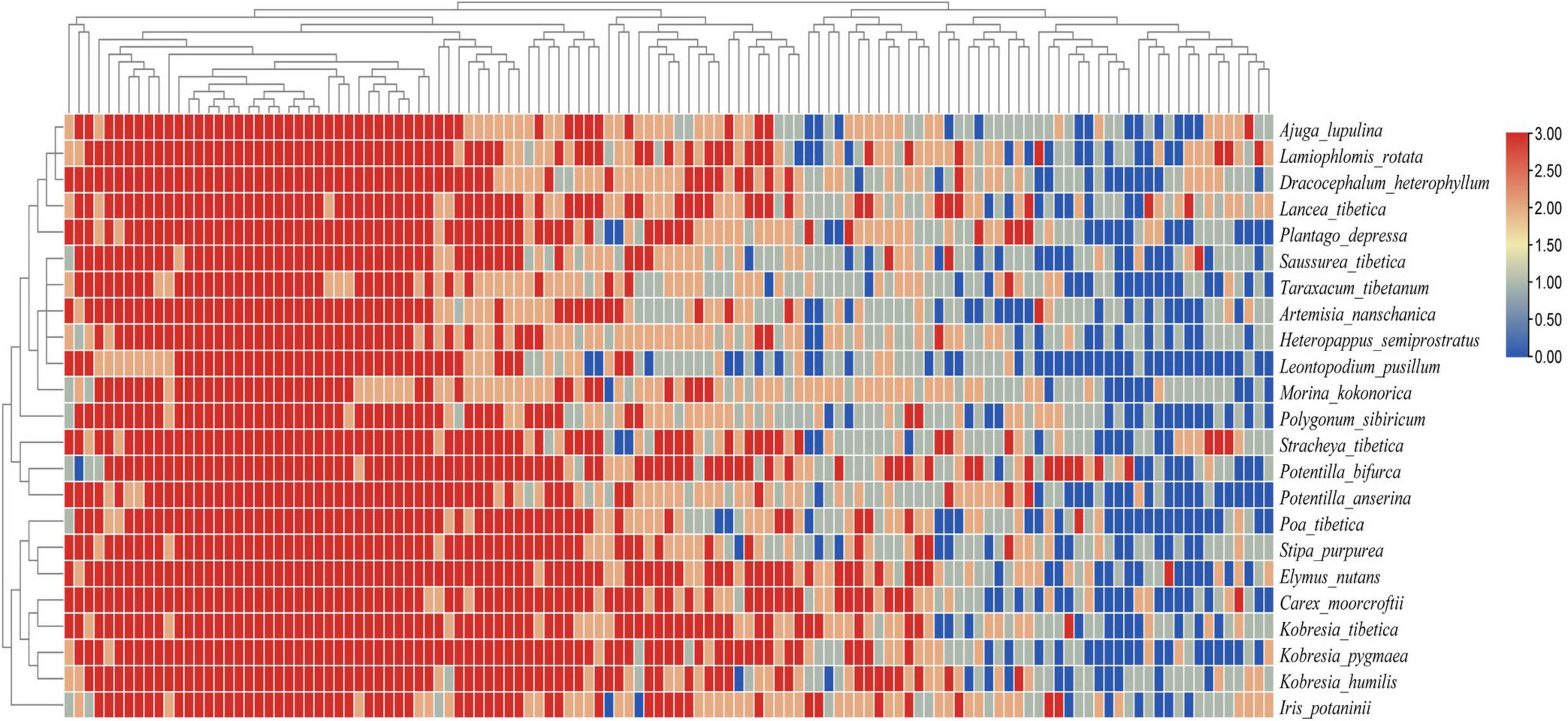
Figure 6. Heat map of plant-AMF association frequency matrix combined with the phylogenetic tree. Different shades of color denote the association frequency between different plant species and AMF OTUs, and blue denotes an absence of association.
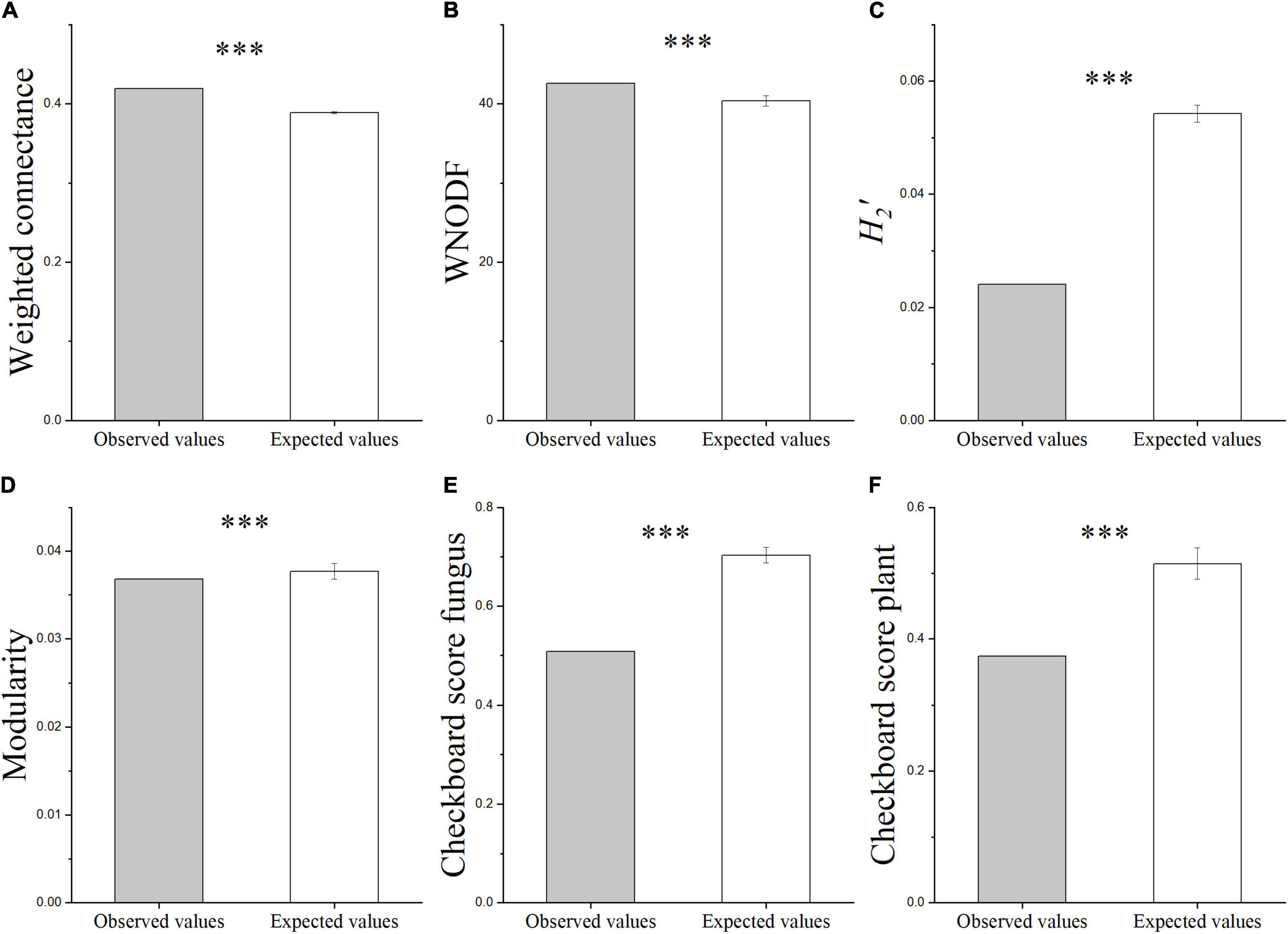
Figure 7. The characteristics of different plant-AMF networks. Weight connectance (A), WNODF (B), H2′ (C), modularity (D), checkerboard score fungus [(E), degree of avoiding overlapping of plants in the fungal community], and checkerboard score plant [(F), degree of avoiding overlapping of fungi in the plant community]. Asterisks indicate significant differences between the observed and expected values according to the t-test (***P < 0.001; bars indicate SE).
Discussion
There were 121 AMF molecular species detected from plant roots in this study, indicating diverse AMF species in Tibetan alpine meadow, in concordance with the previous studies (Gai et al., 2009, 2012; Liu et al., 2011; Zhang et al., 2016). At the same time, it was consistent with previous studies (Gai et al., 2012; Li et al., 2014) that Cyperaceae plants which were considered unable or not easy to form mycorrhizal fungi also had high AMF colonization (Figure 2A; 40.4–60.2%) and diversity (Figures 2B,C; Shannon-Wiener: 2.31–2.60; richness: 86.67–90.67). These findings suggest the potential ecological importance of AMF for plant growth in the alpine meadow. To survive in the harsh environment of the alpine meadow, plants might choose AMF species to improve their ability to absorb nitrogen and phosphorus or the stress-resistance ability (Bever, 2002). It has been found that some AMF species with strong cold resistance ability could survive, thus affecting the growth of host plants and community dynamics (Deepika and Kothamasi, 2015; Bauer et al., 2017).
Furthermore, this study found a significant positive correlation between plant phylogenetic distance and AMF community dissimilarity. Thus, the selectivity existed in the plant-AMF interactions, so that closely phylogenetic-related plant species select similar AMF communities (Figure 5). This plant-AMF selectivity has been well documented in previous studies (Vandenkoornhuyse et al., 2003; Horn et al., 2014; Veresoglou and Rillig, 2014; Chagnon et al., 2015; Deepika and Kothamasi, 2015). The different AMF communities in the rhizosphere of different plant species may be achieved by preferentially allocating plant carbon to the most beneficial fungal partners (Bever et al., 2009; Kiers et al., 2011). The construction of the plant-AMF community was to maximize the functional matching between partners (Thompson, 2005). Plants with similar traits preferentially hosted similar AMF and, likewise, phylogenetically related AMF (assumed to have similar functional traits) interacted with similar plants. No doubt that the plant phylogeny was used to map the mycorrhizal information to examine evolutionary patterns (Wang and Qiu, 2006), but the complex symbiotic relationships between plants and AMF cannot be fully explained by plant phylogeny alone. The phylogenetic correlations between the interacting plant and AMF were modified by the competitive intransitivity (Laird and Schamp, 2006) and AMF species competition within a plant host (Roger et al., 2013). Therefore, future studies should involve factors, such as plant and AMF functional traits, species competition, climates, and soil nutrients, which can well explain the AMF community dynamics.
The network structure of 23 common plants and AMF was characterized by high connectance, high nestedness, anti-modularity, and anti-specialization, which was in line with the characteristics of the woody plant-AMF network in a subtropical forest in China (Chen et al., 2017). Generally, with the increase of connectance, the nestedness increased and the modularity decreased (Põlme et al., 2018), while the nestedness decreased, the modularity and the specialization increased (van der Heijden et al., 2015), as found in our network. High network connectance and nestedness and low network modularity indicated less segregation or sparse connections within subnetworks and dense connections between subnetworks for the plant-AMF network. These network properties enhanced the stability of the communities in the mutually beneficial network, which was considered to be an important property to promote the coexistence of species in the mutual system (Thebault and Fontaine, 2010). Theoretical studies suggested that the common nestedness patterns in the species network can determine the feasibility, resilience, durability, and structural stability of ecological communities (Toju et al., 2014), and the higher the nestedness of the interaction network, the better it can adapt to the effect of environmental disturbance.
In the analysis of plant-AMF phylogenetic signals based on the association frequency matrix, the results showed that the phylogenetic signals of plants and AMF had significant effects on the structure of the plant-AMF network. The phylogenetic signal from plants was stronger than that from AMF (Table 1). Close relatives of plants often interacted with AMF OTU species in the same group, but the interaction between AMF and plant species had a low correlation with AMF phylogeny, which was consistent with the study by Chen et al. (2017). In contrast, findings by Montesinos-Navarro et al. (2012a,b) showed that AMF phylogenetic diversity might also explain that there were different plant-AMF network structures in different ecosystems. In the AMF community with a more diverse phylogeny, there were more opportunities for specific AMF to coexist with common host plants, thus forming a highly nested symbiotic network structure (Chen et al., 2017). The phylogeny of plants and AMF played an important role in the non-random pattern of the AMF network in subtropical forest sites (Chen et al., 2017). In addition, the plant root traits in subtropical forests also showed strong interspecific phylogenetic signals (Kong et al., 2014). The modularity and nestedness in the plant-AMF network were caused by a variety of ecological processes, such as habitat heterogeneity (Olesen et al., 2007), specific selection of plant and AMF combination, relative species abundance and phylogenetic diversity, or AMF competition within the root (Verdu and Valiente-Banuet, 2011). The complex symbiotic interaction between plants and AMF has a potential effect on community assembly. Our study found that the characteristics of the plant-AMF network with high connectance and nestedness promote the stable symbiosis and interdependence between plants and AMF in Tibetan alpine meadows. Plants established network relationships with underground AMF through selection, which may affect the nutrient cycle of symbionts and even the succession of grassland community, so as to improve the adaptation to alpine ecological environment and stress. Therefore, further studies are required to elucidate the reciprocal symbiotic selection strategy, the coexistence pattern, community construction mechanism, and community dynamic rules of plant and AMF species. Furthermore, to enhance our understanding of the influence factors in the coevolution of plant-AMF is of great significance to study community structure, species evolution, and biodiversity.
Conclusion
This study assesses the relationship between the common plant phylogeny and AMF and the characteristics of the plant-AMF symbiotic network in a Tibetan alpine meadow. There was a significant positive correlation between the phylogenetic distance of the 23 herbs and their AMF community dissimilarity, indicating the selectivity of plant species on their AMF community. The network structure of the plant and AMF was characterized by high connectance, high nestedness, anti-modularity, and anti-specialization, which potentially promoted the stable symbiosis and interdependence between plants and AMF in Tibetan alpine meadows. The plant-AMF network was affected by the plant-AMF phylogenies, and the phylogenetic signal from plants was stronger than that from AMF. This study emphasizes that plant phylogeny and plant-AMF network play an important role in the complex plant-AMF community mechanism and enhance our understanding of the interaction between aboveground and belowground communities.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA748864.
Author Contributions
QD, BJ, and YL designed the experiments, which were then carried out by QD, KC, and SR. QD, XG, and YL collected the data and conducted the statistical analyses. QD wrote the first draft of the manuscript. QD, MM, JZ, YL, and BJ reviewed and edited the manuscript before submission. All authors made substantial contributions to the discussion of the content.
Funding
This study was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA2005010402), the Second Tibetan Plateau Scientific Expedition and Research (2019QZKK0302), the National Natural Science Foundation of China (31800380 and 41871067), and the National Key Research and Development Program of China (2016YFC0501802).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Ping Wang for his help in providing data analysis methods and Congcong Li, Yinhang Luo, and Zhen Fang for their help in field sampling and lab analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.804861/full#supplementary-material
Footnotes
References
Bai-ping, Z., Xiao-dong, C., Bao-lin, L., and Yong-hui, Y. (2002). Biodiversity and conservation in the Tibetan Plateau. J. Geogr. Sci. 12, 135–143. doi: 10.1007/BF02837467
Barberan, A., Bates, S. T., Casamayor, E. O., and Fierer, N. (2012). Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J. 6, 343–351. doi: 10.1038/ismej.2011.119
Bauer, J. T., Blumenthal, N., Miller, A. J., Ferguson, J. K., Reynolds, H. L., and Brudvig, L. (2017). Effects of between-site variation in soil microbial communities and plant-soil feedbacks on the productivity and composition of plant communities. J. Appl. Ecol. 54, 1028–1039. doi: 10.1111/1365-2664.12937
Bever, J. D. (2002). Negative feedback within a mutualism: host-specific growth of mycorrhizal fungi reduces plant benefit. Proc. Biol. Sci. 269, 2595–2601. doi: 10.1098/rspb.2002.2162
Bever, J. D., Richardson, S. C., Lawrence, B. M., Holmes, J., and Watson, M. (2009). Preferential allocation to beneficial symbiont with spatial structure maintains mycorrhizal mutualism. Ecol. Lett. 12, 13–21. doi: 10.1111/j.1461-0248.2008.01254.x
Blackwell, M. (2000). Terrestrial life-fungal from the start? Science 289, 1884–1885. doi: 10.1126/science.289.5486.1884
Bonfante, P., and Genre, A. (2008). Plants and arbuscular mycorrhizal fungi: an evolutionary-developmental perspective. Trends Plant Sci. 13, 492–498. doi: 10.1016/j.tplants.2008.07.001
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Chagnon, P. L., Bradley, R. L., and Klironomos, J. N. (2012). Using ecological network theory to evaluate the causes and consequences of arbuscular mycorrhizal community structure. New Phytol. 194, 307–312. doi: 10.1111/j.1469-8137.2011.04044.x
Chagnon, P.-L., Bradley, R. L., and Klironomos, J. N. (2015). Trait-based partner selection drives mycorrhizal network assembly. Oikos 124, 1609–1616. doi: 10.1111/oik.01987
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, E. C. H., Morin, E., Beaudet, D., Noel, J., Yildirir, G., Ndikumana, S., et al. (2018). High intraspecific genome diversity in the model arbuscular mycorrhizal symbiont Rhizophagus irregularis. New Phytol. 220, 1161–1171. doi: 10.1111/nph.14989
Chen, L., Zheng, Y., Gao, C., Mi, X. C., Ma, K. P., Wubet, T., et al. (2017). Phylogenetic relatedness explains highly interconnected and nested symbiotic networks of woody plants and arbuscular mycorrhizal fungi in a Chinese subtropical forest. Mol. Ecol. 26, 2563–2575. doi: 10.1111/mec.14061
Cui, S., Meng, F., Suonan, J., Wang, Q., Li, B., Liu, P., et al. (2017). Responses of phenology and seed production of annual Koenigia islandica to warming in a desertified alpine meadow. Agric. Meteorol. 247, 376–384. doi: 10.1016/j.agrformet.2017.08.034
Deepika, S., and Kothamasi, D. (2015). Soil moisture–a regulator of arbuscular mycorrhizal fungal community assembly and symbiotic phosphorus uptake. Mycorrhiza 25, 67–75. doi: 10.1007/s00572-014-0596-1
Dormann, C. F., Fründ, J., Blüthgen, N., and Gruber, B. (2013). Open access indices, graphs and null models: analyzing bipartite ecological networks. Open Ecol. J. 2, 7–24. doi: 10.2174/1874213000902010007
Duan, A. M., Wu, G. X., Zhang, Q., and Liu, Y. M. (2006). New proofs of the recent climate warming over the Tibetan Plateau as a result of the increasing greenhouse gases emissions. Chin. Sci. Bull. 51, 1396–1400. doi: 10.1007/s11434-006-1396-6
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Gai, J. P., Christie, P., Cai, X. B., Fan, J. Q., Zhang, J. L., Feng, G., et al. (2009). Occurrence and distribution of arbuscular mycorrhizal fungal species in three types of grassland community of the Tibetan Plateau. Ecol. Res. 24, 1345–1350. doi: 10.1007/s11284-009-0618-1
Gai, J. P., Tian, H., Yang, F. Y., Christie, P., Li, X. L., and Klironomos, J. N. (2012). Arbuscular mycorrhizal fungal diversity along a Tibetan elevation gradient. Pedobiologia 55, 145–151. doi: 10.1016/j.pedobi.2011.12.004
Gao, C., Montoya, L., Xu, L., Madera, M., Hollingsworth, J., Purdom, E., et al. (2019). Strong succession in arbuscular mycorrhizal fungal communities. ISME J. 13, 214–226. doi: 10.1038/s41396-018-0264-0
Gao, Q.-M., and Guo, L.-D. (2010). A comparative study of arbuscular mycorrhizal fungi in forest, grassland and cropland in the Tibetan Plateau, China. Mycology 1, 163–170. doi: 10.1080/21501203.2010.510123
Genre, A., Lanfranco, L., Perotto, S., and Bonfante, P. (2020). Unique and common traits in mycorrhizal symbioses. Nat. Rev. Microbiol. 18, 649–660. doi: 10.1038/s41579-020-0402-3
Goberna, M., Montesinos-Navarro, A., Valiente-Banuet, A., Colin, Y., Gomez-Fernandez, A., Donat, S., et al. (2019). Incorporating phylogenetic metrics to microbial co-occurrence networks based on amplicon sequences to discern community assembly processes. Mol. Ecol. Resour. 19, 1552–1564. doi: 10.1111/1755-0998.13079
Gomez, J. M., Verdu, M., and Perfectti, F. (2010). Ecological interactions are evolutionarily conserved across the entire tree of life. Nature 465, 918–U916. doi: 10.1038/nature09113
Horn, S., Caruso, T., Verbruggen, E., Rillig, M. C., and Hempel, S. (2014). Arbuscular mycorrhizal fungal communities are phylogenetically clustered at small scales. ISME J. 8, 2231–2242. doi: 10.1038/ismej.2014.72
Ives, A. R., and Godfray, H. C. J. (2006). Phylogenetic analysis of trophic associations. Am. Nat. 168, E1–E14. doi: 10.1086/505157
Ji, B. M., and Bever, J. D. (2016). Plant preferential allocation and fungal reward decline with soil phosphorus: implications for mycorrhizal mutualism. Ecosphere 7:e01256. doi: 10.1002/ecs2.1256
Kembel, S. W., Cowan, P. D., Helmus, M. R., Cornwell, W. K., Morlon, H., Ackerly, D. D., et al. (2010). Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26, 1463–1464. doi: 10.1093/bioinformatics/btq166
Kiers, E. T., Duhamel, M., Beesetty, Y., Mensah, J. A., Franken, O., Verbruggen, E., et al. (2011). Reciprocal rewards stabilize cooperation in the mycorrhizal symbiosis. Science 333, 880–882. doi: 10.1126/science.1208473
Kong, D., Ma, C., Zhang, Q., Li, L., Chen, X., Zeng, H., et al. (2014). Leading dimensions in absorptive root trait variation across 96 subtropical forest species. New Phytol. 203, 863–872. doi: 10.1111/nph.12842
Laird, R. A., and Schamp, B. S. (2006). Competitive intransitivity promotes species coexistence. Am. Nat. 168, 182–193. doi: 10.1086/506259
Li, X., Gai, J., Cai, X., Li, X., Christie, P., Zhang, F., et al. (2014). Molecular diversity of arbuscular mycorrhizal fungi associated with two co-occurring perennial plant species on a Tibetan altitudinal gradient. Mycorrhiza 24, 95–107. doi: 10.1007/s00572-013-0518-7
Li, X., Zhang, J., Gai, J., Cai, X., Christie, P., and Li, X. (2015). Contribution of arbuscular mycorrhizal fungi of sedges to soil aggregation along an altitudinal alpine grassland gradient on the Tibetan Plateau. Environ. Microbiol. 17, 2841–2857. doi: 10.1111/1462-2920.12792
Liu, Y., He, J., Shi, G., An, L., Opik, M., and Feng, H. (2011). Diverse communities of arbuscular mycorrhizal fungi inhabit sites with very high altitude in Tibet Plateau. FEMS Microbiol. Ecol. 78, 355–365. doi: 10.1111/j.1574-6941.2011.01163.x
Lumini, E., Orgiazzi, A., Borriello, R., Bonfante, P., and Bianciotto, V. (2010). Disclosing arbuscular mycorrhizal fungal biodiversity in soil through a land-use gradient using a pyrosequencing approach. Environ. Microbiol. 12, 2165–2179. doi: 10.1111/j.1462-2920.2009.02099.x
McGonigle, T. P., Miller, M. H., Evans, D. G., Fairchild, G. L., and Swan, J. A. (1990). A new method which gives an objective measure of colonization of roots by vesicular-arbuscular mycorrhizal fungi. New Phytol. 115, 495–501. doi: 10.1111/j.1469-8137.1990.tb00476.x
Miehe, G., Mlehe, S., Kaiser, K., Liu, J. Q., and Zhao, X. Q. (2008). Status and dynamics of Kobresia pygmaea ecosystem on the Tibetan plateau. Ambio 37, 272–279. doi: 10.1579/0044-7447(2008)37[272:sadotk]2.0.co;2
Montesinos-Navarro, A., Segarra-Moragues, J. G., Valiente-Banuet, A., and Verdu, M. (2012a). The network structure of plant-arbuscular mycorrhizal fungi. New Phytol. 194, 536–547. doi: 10.1111/j.1469-8137.2011.04045.x
Montesinos-Navarro, A., Segarra-Moragues, J. G., Valiente-Banuet, A., and Verdu, M. (2012b). Plant facilitation occurs between species differing in their associated arbuscular mycorrhizal fungi. New Phytol. 196, 835–844. doi: 10.1111/j.1469-8137.2012.04290.x
Olesen, J. M., Bascompte, J., Dupont, Y. L., and Jordano, P. (2007). The modularity of pollination networks. Proc. Natl. Acad. Sci. U.S.A. 104, 19891–19896. doi: 10.1073/pnas.0706375104
Põlme, S., Bahram, M., Jacquemyn, H., Kennedy, P., Kohout, P., Moora, M., et al. (2018). Host preference and network properties in biotrophic plant-fungal associations. New Phytol. 217, 1230–1239. doi: 10.1111/nph.14895
Reinhart, K. O., and Anacker, B. L. (2014). More closely related plants have more distinct mycorrhizal communities. AoB Plants 6:lu051. doi: 10.1093/aobpla/plu051
Roger, A., Colard, A., Angelard, C., and Sanders, I. R. (2013). Relatedness among arbuscular mycorrhizal fungi drives plant growth and intraspecific fungal coexistence. ISME J. 7, 2137–2146. doi: 10.1038/ismej.2013.112
Sargent, R. D., Kembel, S. W., Emery, N. C., Forrestel, E. J., and Ackerly, D. D. (2011). Effect of local community phylogenetic structure on pollen limitation in an obligately insect-pollinated plant. Am. J. Bot. 98, 283–289. doi: 10.3732/ajb.1000329
Smith, S. E., and Read, D. J. (2008). Mycorrhizal Symbiosis. London: Academic Press. doi: 10.1016/B978-0-12-370526-6.X5001-6
Tedersoo, L., Bahram, M., and Zobel, M. (2020). How mycorrhizal associations drive plant population and community biology. Science 367:eaba1223. doi: 10.1126/science.aba1223
Thebault, E., and Fontaine, C. (2010). Stability of ecological communities and the architecture of mutualistic and trophic networks. Science 329, 853–856. doi: 10.1126/science.1188321
Thompson, J. N. (2005). Coevolution: the geographic mosaic of coevolutionary arms races. Curr. Biol. 15, R992–R994. doi: 10.1016/j.cub.2005.11.046
Toju, H., Guimaraes, P. R., Olesen, J. M., and Thompson, J. N. (2014). Assembly of complex plant-fungus networks. Nat. Commun. 5:5273. doi: 10.1038/ncomms6273
Toju, H., Tanabe, A. S., and Ishii, H. S. (2016). Ericaceous plant-fungus network in a harsh alpine-subalpine environment. Mol. Plant 25, 3242–3257. doi: 10.1111/mec.13680
Ulrich, W., and Gotelli, N. J. (2007). Null model analysis of species nestedness patterns. Ecology 88, 1824–1831. doi: 10.1890/06-1208.1
van der Heijden, M. G. A., Martin, F. M., Selosse, M.-A., and Sanders, I. R. (2015). Mycorrhizal ecology and evolution: the past, the present, and the future. New Phytol. 205, 1406–1423. doi: 10.1111/nph.13288
Van Geel, M., Busschaert, P., Honnay, O., and Lievens, B. (2014). Evaluation of six primer pairs targeting the nuclear rRNA operon for characterization of arbuscular mycorrhizal fungal (AMF) communities using 454 pyrosequencing. J. Microbiol. Methods 106, 93–100. doi: 10.1016/j.mimet.2014.08.006
Vandenkoornhuyse, P., Ridgway, K. P., Watson, I. J., Fitter, A. H., and Young, J. P. W. (2003). Co-existing grass species have distinctive arbuscular mycorrhizal communities. Mol. Plant 12, 3085–3095. doi: 10.1046/j.1365-294X.2003.01967.x
Verdu, M., and Valiente-Banuet, A. (2011). The relative contribution of abundance and phylogeny to the structure of plant facilitation networks. Oikos 120, 1351–1356. doi: 10.1111/j.1600-0706.2011.19477.x
Veresoglou, S. D., and Rillig, M. C. (2014). Do closely related plants host similar arbuscular mycorrhizal fungal communities? A meta-analysis. Plant Soil 377, 395–406. doi: 10.1007/s11104-013-2008-2
Walder, F., and van der Heijden, M. G. (2015). Regulation of resource exchange in the arbuscular mycorrhizal symbiosis. Nat. Plants 1:15159. doi: 10.1038/nplants.2015.159
Wang, B., and Qiu, Y. L. (2006). Phylogenetic distribution and evolution of mycorrhizas in land plants. Mycorrhiza 16, 299–363. doi: 10.1007/s00572-005-0033-6
Wang, Z. Q., Zhang, Y. Z., Yang, Y., Zhou, W., Gang, C. C., Zhang, Y., et al. (2016). Quantitative assess the driving forces on the grassland degradation in the Qinghai-Tibet Plateau, in China. Ecol. Inform. 33, 32–44. doi: 10.1016/j.ecoinf.2016.03.006
Werner, G. D. A., and Kiers, E. T. (2015). Partner selection in the mycorrhizal mutualism. New Phytol. 205, 1437–1442. doi: 10.1111/nph.13113
Wipf, D., Krajinski, F., van Tuinen, D., Recorbet, G., and Courty, P. E. (2019). Trading on the arbuscular mycorrhiza market: from arbuscules to common mycorrhizal networks. New Phytol. 223, 1127–1142. doi: 10.1111/nph.15775
Xu, M., Li, X., Cai, X., Li, X., Christie, P., and Zhang, J. (2017). Land use alters arbuscular mycorrhizal fungal communities and their potential role in carbon sequestration on the Tibetan Plateau. Sci. Rep. 7:3067. doi: 10.1038/s41598-017-03248-0
Zanne, A. E., Tank, D. C., Cornwell, W. K., Eastman, J. M., Smith, S. A., FitzJohn, R. G., et al. (2014). Three keys to the radiation of angiosperms into freezing environments. Nature 506, 89–92. doi: 10.1038/nature12872
Zhang, J., Wang, F., Che, R. X., Wang, P., Liu, H. K., Ji, B. M., et al. (2016). Precipitation shapes communities of arbuscular mycorrhizal fungi in Tibetan alpine steppe. Sci. Rep. 6:23488. doi: 10.1038/srep23488
Keywords: arbuscular mycorrhizal fungi, plant species, host selectivity, symbiotic network, phylogenetic distance, community dissimilarity, Tibetan alpine meadow
Citation: Dong Q, Guo X, Chen K, Ren S, Muneer MA, Zhang J, Li Y and Ji B (2021) Phylogenetic Correlation and Symbiotic Network Explain the Interdependence Between Plants and Arbuscular Mycorrhizal Fungi in a Tibetan Alpine Meadow. Front. Plant Sci. 12:804861. doi: 10.3389/fpls.2021.804861
Received: 29 October 2021; Accepted: 24 November 2021;
Published: 17 December 2021.
Edited by:
Jian Sun, Institute of Tibetan Plateau Research, Chinese Academy of Sciences (CAS), ChinaCopyright © 2021 Dong, Guo, Chen, Ren, Muneer, Zhang, Li and Ji. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yaoming Li, eWFvbWluZ2xpQGJqZnUuZWR1LmNu; Baoming Ji, YmFvbWluZ2ppQGJqZnUuZWR1LmNu
 Qiang Dong
Qiang Dong Xin Guo
Xin Guo Keyu Chen1
Keyu Chen1 Shijie Ren
Shijie Ren Muhammad Atif Muneer
Muhammad Atif Muneer