
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 20 December 2021
Sec. Plant Symbiotic Interactions
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.804527
This article is part of the Research Topic Role of Endophytic Bacteria in Improving Plant Stress Resistance View all 8 articles
 Tao Chen1,2†
Tao Chen1,2† Ruiwen Hu1†
Ruiwen Hu1† Zhongyi Zheng1
Zhongyi Zheng1 Jiayi Yang1
Jiayi Yang1 Huan Fan1
Huan Fan1 Xiaoqiang Deng2
Xiaoqiang Deng2 Wang Yao3
Wang Yao3 Qiming Wang4
Qiming Wang4 Shuguang Peng5*
Shuguang Peng5* Juan Li1*
Juan Li1*The shortage of land resources restricts the sustainable development of agricultural production. Multiple cropping has been widely used in Southern China, but whether the continuous planting will cause a decline in soil quality and crop yield is unclear. To test whether multiple cropping could increase grain yield, we investigated the farmlands with different cultivation years (10–20 years, 20–40 years, and >40 years). Results showed that tobacco-rice multiple cropping rotation significantly increased soil pH, nitrogen nutrient content, and grain yield, and it increased the richness of the bacterial community. The farmland with 20–40 years of cultivation has the highest soil organic carbon (SOC), ammonium nitrogen, and grain yield, but there is no significant difference in the diversity and structure of the bacterial community in farmlands with different cultivation years. The molecular ecological network indicated that the stability of the bacterial community decreased across the cultivation years, which may result in a decline of farmland yields in multiple cropping system> 40 years. The Acidobacteria members as the keystone taxa (Zi ≥ 2.5 or Pi ≥ 0.62) appeared in the tobacco-rice multiple cropping rotation farmlands, and the highest abundance of Acidobacteria was found in the farmland with the highest SOC and ammonium nitrogen content, suggesting Acidobacteria Gp4, GP7, GP12, and GP17 are important taxa involved in the soil carbon and nitrogen cycle. Therefore, in this study, the multiple cropping systems for 20 years will not reduce the crop production potential, but they cannot last for more than 40 years. This study provides insights for ensuring soil quality and enhancing sustainable agricultural production capacity.
- Multiple cropping can improve soil carbon and nitrogen, and increase crop yield for 20–40 years.
- Decreased bacterial community stability in multiple cropping system for long cultivation years (>40 years) could result in decrease in yield of farmland.
- Acidobacteria subgroups (e.g., Gp4 and GP17) respond sensitively to the changes of soil carbon and nitrogen.
The multiple cropping system increases crop diversity, makes full use of limited soil resources, to a certain extent alleviates the shortage of cultivated land resources in agricultural production, and also guarantees food security (Yang et al., 2015; Xu et al., 2019b). Tobacco-rice multiple cropping rotation, that is, planting tobacco in spring and rice in autumn on the same farmland, is one of the main double-cropping systems in southern China (Hu et al., 2021). It not only ensures crop security production but also improves the income of farmers and effectively alleviates the social problems of rural laborers moving to cities and no production activities in cultivated land due to insufficient income (Xu et al., 2019a). Different cropping systems not only lead to differences in soil physical, chemical, and biological properties (Viaud et al., 2018; Wu et al., 2021) but also affect crop yield and quality (Caviglia et al., 2011), as well as have important implications for greenhouse gas emissions (Zhang, 2019). Under multiple cropping system, different fertilization methods, crop residue management, and rhizosphere microecosystem mediated by root exudates lead to differences in soil nutrient cycle and microbial community (Arcand et al., 2016; Li et al., 2017; Guyonnet et al., 2018; Zhao et al., 2021).
The paddy field needs flooding, and rice planting on the same farmland for many years will cause the occurrence of soil gleization (Liu et al., 2015). Studies have pointed out that the gleization of rice fields has low effective nutrient content, poor biological activity, and serious diseases, which are not conducive to the growth of rice, and the yield is severely reduced (Yuan et al., 2020). Continuous agricultural planting will reduce the diversity of soil bacterial community, change the community composition, find the proliferation of harmful bacteria in the soil with serious diseases in continuous cropping farm systems, and reduce the beneficial bacteria (Wu et al., 2017; Hu et al., 2019). Increasing cultivation years changed the structure and diversity of soil bacterial and fungal communities in farmland, and the long-term fertilization increased soil organic carbon (SOC) storage and macroaggregate content (Zhang et al., 2019; Li et al., 2020), but these studies focused on farmland whose cultivation years generally spanned about 20 years, and there were fewer studies on the changing trends of soil bacterial community in multiple cropping farmlands with more than 40 years of cultivation.
As an important part of the soil, microorganisms are very sensitive to soil environmental changes, such as pH, SOC, nutrient, water, and temperature (Griffiths and Philippot, 2013; Hartmann et al., 2015; Barcenas-Moreno et al., 2016; Frac et al., 2018). Soil microorganisms play an important role in the decomposition of soil organic matter, material transformation, energy transfer, and other ecological processes (Frey et al., 2013; Viggi et al., 2014). The interaction between rhizosphere microorganisms and plants can promote plant growth, enhance plant stress resistance, and help plants resist diseases and abiotic stress, which is called “the second genome of plants” (Mendes et al., 2013; Rodriguez et al., 2019). The energy and nutrients required by microorganisms mainly come from soil carbon and nitrogen, so the change of soil carbon and nitrogen concentration will affect the structure and composition of the microbial community (Yao et al., 2021b), leading to the transformation of its function in the soil ecosystem. Ammonium and nitrate nitrogen are the two main inorganic nitrogen that plants can directly absorb and utilize, both of which affect the diversity and structure of the microbial community (Li et al., 2019b). However, different forms of soil nitrogen have different effects on bacterial and fungal communities: nitrate drives bacterial communities, and ammonium nitrogen affects fungal communities (Li et al., 2021b).
A large number of different bacterial taxa were present in the soil, yet <1% of bacteria from the natural environment could be cultured by the conventional culture techniques (Chaudhary et al., 2019). In recent years, researchers have paid increasing attention to the role played by Acidobacteria taxa in participating in soil nutrient cycling and interactions with plants (Kielak et al., 2016). However, due to the difficulty of isolation and culture of Acidobacteria, the understanding of its physiological characteristics and function is limited (Eichorst et al., 2011). Acidobacteria exists in various habitats, and the quantity of Acidobacteria in soil accounts for about 20–50% of the total bacterial community, which plays an important role in the construction of soil ecosystem (Jones et al., 2009). Studies have reported that Acidobacteria is sensitive to soil pH and nitrogen deposition levels, and different Acidobacteria subgroups have different responses to pH and nitrogen levels (Liu et al., 2017; Yao et al., 2021a). At the same time, different soil habitats will also lead to differences in the distribution of Acidobacteria (Naether et al., 2012).
The purpose of this study was to clarify the differences in soil properties and crop yields in multiple cropping farmlands under different cultivation years and to evaluate the relationship between the changes of bacterial community diversity and composition and soil properties. We hypothesized that the differences in soil physicochemical properties and bacterial community in multiple cropping farmlands with different cultivation years caused the changes in crop yields. Our research attempts to explain the change of crop yield from the perspective of bacterial interaction and community stability, which will help to understand the importance of the interaction between soil-crop-microbial in the farmland ecosystem.
On November 5, 2020, after the harvest of rice in the tobacco-rice multiple cropping rotation farmlands, soil samples were collected from farmlands with different cultivation years (Table 1). All samples were collected from Guiyang County, Chenzhou City, Hunan Province, China. The average annual temperature in Guiyang County from 2018 to 2020 is 17.6°C, the annual average precipitation is 1,568 mm, the annual average frost-free period is about 281 days, and the annual sunshine hours are 1,475 h. According to the principle of random, multisite mixed soil sample collection, a shovel was used to collect 6 soil samples at each sampling site, and 3 of them were completely mixed into 1 soil sample, so 2 soil samples per sampling site, a total of 16 sampling sites, and 32 soil samples. The impurities, such as roots, stones, and residual fertilizer, were removed from the soil. Then, we divided each soil sample into two parts and put them into sterile ziplock bags. One of the bags of soil was frozen in liquid nitrogen, then put in an icebox and transported back to the laboratory, and stored in an ultralow temperature refrigerator at −80°C for DNA extraction. Another bag of soil was stored at room temperature to determine the physical and chemical properties of the soil.
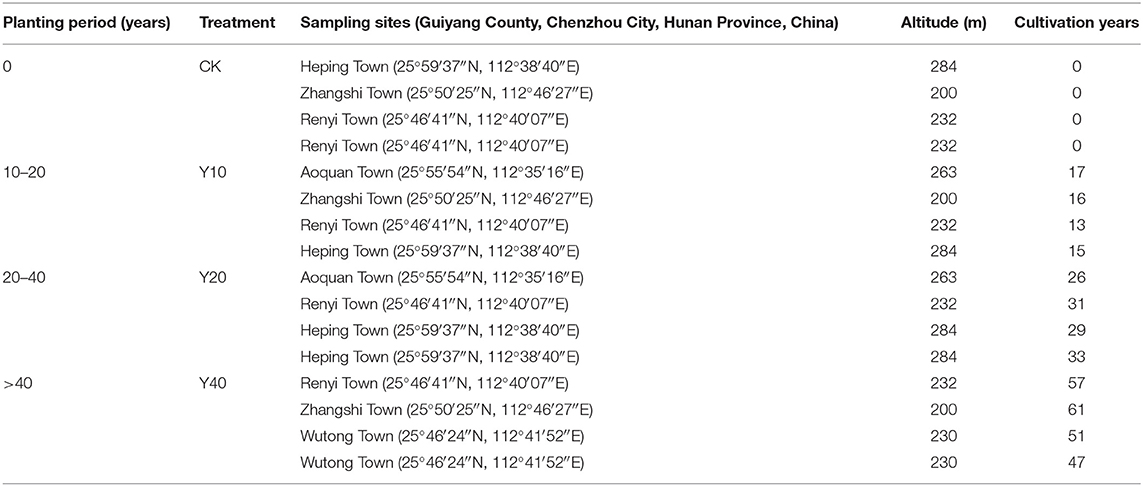
Table 1. Sampling sites, altitude, and cultivation years of tobacco-rice multiple cropping rotation farmlands.
In rice monoculture farmland, rice is planted in April each year and then fallow. The tobacco-rice multiple cropping rotation farmlands is planted with flue-cured tobacco in March and rice in August each year. All sampling sites (farmlands) were fertilized according to the same local fertilization scheme: the total fertilization amount used for flue-cured tobacco planting was 165 kg N ha−1, 165 kg P2O5 ha−1, and 420 kg K2O ha−1, and the total fertilization amount used for rice planting was 150 kg N ha−1, 75 kg P2O5 ha−1, and 120 kg K2O ha−1. During the planting period of flue-cured tobacco, nitrogen fertilizer is applied according to base fertilizer: topdressing = 5:5, phosphorus fertilizer is applied as base fertilizer at one time, and potassium fertilizer is applied according to base fertilizer: topdressing = 3:7. In the rice planting period, nitrogen fertilizer is applied according to base fertilizer: tiller fertilizer: panicle fertilizer = 5:3:2, phosphorus fertilizer is applied as base fertilizer at one time, and potassium fertilizer is applied according to base fertilizer: tiller fertilizer = 5:5. The tobacco planted variety was Yunyan 87, and the rice planted variety was Qianfengyou 877 (hybrid rice). All other farmland management measures were consistent.
The measurement methods of soil physical and chemical properties followed the protocols detailed in the study by Bao (2000). Soil pH was measured in 1:2.5 mixtures of soil and deionized water with a pH meter (PHS-3C, Lei-ci, Shanghai, China). SOC was determined by potassium dichromate (K2Cr2O7) and sulfuric acid (H2SO4) oxidation and titration. Ammonium-N (-N) and nitrate-N (-N) were extracted with the ratio of 5 g fresh soil to 50 ml 2 M potassium chloride. The contents of -N and -N were analyzed by a continuous flow analytical system (San++ system, Skalar, Holland).
The grain yield measurement of the tobacco-rice multiple cropping rotation farmlands was completed on October 29, and the grain yield measurement of the rice monoculture farmland was on July 20. Grain yield was determined using a 5 m2 area in the center of each sampling site and adjusted to 14% of the standard water content (Huang et al., 2020).
Genomic DNA was extracted from fresh samples of 1.0 g with the Fast DNA® SPIN Kit for Soil (MP Biomedicals, Santa Ana, CA, USA). The V4–V5 region of bacterial 16S rRNA was amplified together with a specific Illumina adapter and barcode sequence primer pair 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) (Gu et al., 2019). PCR was conducted in 25 μl mixtures containing 1.0 μl (~25 ng DNA) template, 12.5 μl PCR Premix, 2.5 μl of forward/reverse primers, and DNase-RNase-free deionized water to adjust the volume. The PCR procedure consisted of 98°C denaturing for 30 s, followed by 32 cycles of 98°C for 10 s, 54°C for 30 s, 72°C for 45 s, and a final step of 72°C extensions for 10 min. After purification and quantification, the PCR product constructed that the library sequencing was performed on the Illumina NovaSeq PE250 platform (LC-Bio Technology Co., Ltd., Hang Zhou, Zhejiang Province, China).
The sequence was processed, the primers on the Galaxy pipeline were removed (http://mem.rcees.ac.cn:8080/), Btrim was used for low-quality sequence filtering, the low-quality reads with a quality control score of <20 and a length of <200 bp were removed, and then Flash was used to splice the forward and reverse sequences together and overlap by 20–250 bp to obtain high-quality sequences (Gu et al., 2019). Through the UPARSE classification operation, operational taxonomic unit (OTU) clustering is performed at a similar level of 97%, and the ribosomal database project (RDP) classifier is used to classify and annotate each sequence (Bacci et al., 2015). We obtained 45,637–73,902 clean sequences per sample and then randomly rarefied to 45,637 sequences per sample.
The bacterial community analysis includes alpha diversity analysis (Shannon, Simpson, Pielou evenness, and Chao1) and beta diversity analysis, including non-metric multidimensional scaling (NMDS) analysis, principal component analysis (PCA), and community dissimilarity test based on the Bray–Curtis distance matrix (ANOSIM, MRPP, and ADONIS), and all the above analyses were calculated using “vegan” and “stats” packages and plotted using the “ggplot2” package (Oksanen et al., 2006; Gómez-Rubio, 2017). A Venn diagram was obtained using the “VennDiagram” package. Pearson's correlation between bacteria and soil properties was calculated using the Statistical Package for Social Science (SPSS, IBM, New York, USA) 19.0 for Windows. To determine the specific differential taxa of bacterial community composition in different treatments, the linear discriminant analysis (LDA) Effect Size (LEfSe) was completed on the Galaxy pipeline (http://mem.rcees.ac.cn:8080/), and the threshold on the logarithmic LDA score was 3.0. Pairwise Pearson's correlation matrix of the edaphic factors and Acidobacteria subgroups was completed with the “corrplot” package (Yang et al., 2021). At the p < 0.05 level, the least significant difference (LSD) method was used for ANOVA.
Based on the random matrix theory (RMT), a phylogenetic molecular ecological network (pMEN) was constructed by using the 16S rRNA sequencing data (Deng et al., 2012). The network was constructed using at least 8 detected OTUs out of the 10 replicates in this study. Network construction and analysis were executed on the MENA pipeline of the Oklahoma University IEG (http://ieg4.rccc.ou.edu/mena/). The first step of network construction is to upload the standardized OTU sequence data. Second, the relative abundance and the OTU of different samples based on Pearson's correlation coefficient are analyzed and converted into a similarity matrix, which can measure the degree of agreement. Third, using the RMT-based network method, the adjacency matrix is obtained by applying an appropriate threshold to the similarity matrix, thereby defining the distance between each pair of nodes (Zhou et al., 2010). Finally, a network containing nodes and links is obtained, and some network topology characteristics are generated. The constructed network is visualized by Gephi 0.9.2-beta. To determine the topological role of each node in the network, a Zi-Pi chart display based on the within-module connectivity (Zi) and among-module connectivity (Pi) values was used. Module hubs, connectors, and network hubs have been proposed as potential keystone taxa due to their important roles in network topology.
Compared with rice monoculture (CK treatment), the tobacco-rice multiple cropping rotation (Y10, Y20, and Y40 treatments) significantly improved soil pH value, SOC content, ammonium nitrogen content, and grain yield (p < 0.05, Table 2). Comparing tobacco-rice multiple cropping rotation farmlands with different cultivation years, the soil pH value, SOC content, ammonium nitrogen content, and grain yield were the highest in the Y20 treatment, and the nitrate nitrogen content was the highest in the Y40 treatment. The content of SOC and ammonium nitrogen affected the grain yield, while nitrate nitrogen had no effect.
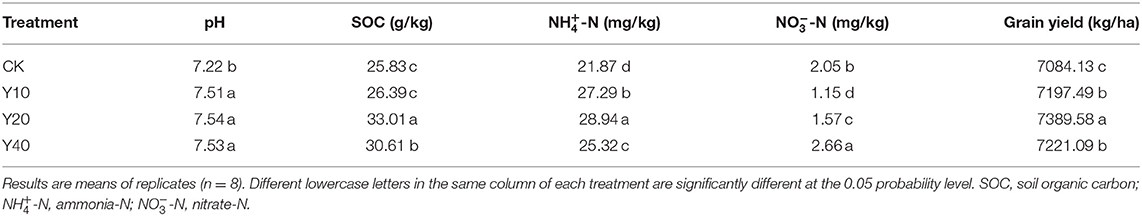
Table 2. Comparison of soil physical and chemical properties and grain yield in different cultivation years of tobacco-rice multiple cropping rotation farmlands.
The alpha diversity of soil bacterial communities in different treatments generally showed that there is no significant difference among treatments (Figures 1A–D), only Chao1 value was significantly lower in CK treatment than in Y10, Y20, and Y40 treatments (p < 0.05). Compared with rice monoculture farmland, a higher abundance of the soil bacterial community was observed in tobacco-rice multiple cropping rotation farmlands, but there is no significant difference in the alpha diversity of soil bacterial community in tobacco-rice multiple cropping rotation farmlands with different cultivation years.
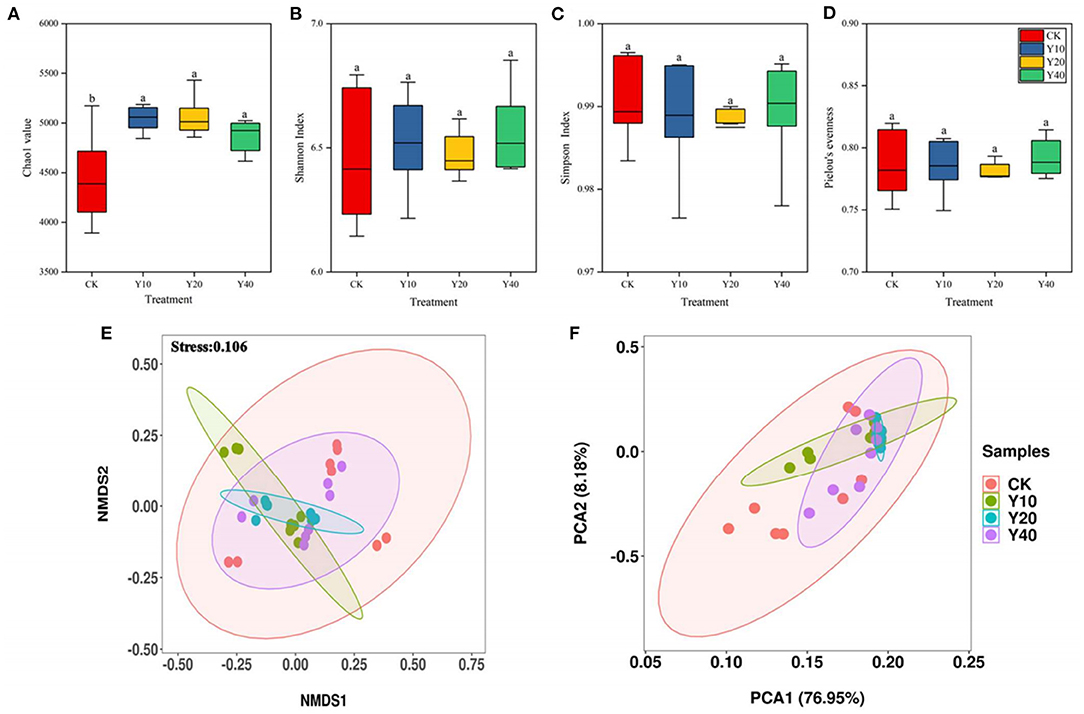
Figure 1. Alpha diversity of 16S rRNA gene sequencing data among groups CK, Y10, Y20 and Y40. (A) Chao1 value, (B) Shannon Index, (C) Simpson Index, (D) Pielou's evenness. Results are means of eight replicates. Different letter means the significant difference (P < 0.05). The non-metric multidimensional scaling (NMDS) (E) and principal component analysis (PCA) (F) plots of soil bacterial community beta diversity.
The NMDS (Figure 1E) and PCA (Figure 1F) further confirmed that the bacterial community structure of the CK treatment and Y10, Y20, and Y40 treatments was separated clearly. The two main axes of PCA explained 72.08% of the variation, indicating that it can better represent the characteristics of microbial community composition, of which 61.61% of the variation is explained by PC1, and 10.47% of the variation is explained by PC2. The dissimilarity analysis (i.e., ADNOIS, ANOSIM, and MRPP) based on the Bray–Curtis distance further confirmed that there are significant differences in the soil bacterial community structure between rice monoculture farmland and tobacco-rice multiple cropping rotation farmlands (p < 0.05, Supplementary Table S1), while there were no significant differences in soil bacterial community structure between tobacco-rice multiple cropping rotation farmlands with different cultivation years.
In terms of the taxonomic composition of soil bacterial communities at the phylum level, there were 57, 57, 54, and 59 different phyla in CK, Y10, Y20, and Y40 treatments, respectively. The bacterial communities in all treatments were mainly composed of Chloroflexi, Proteobacteria, and Acidobacteria, and their relative abundance accounted for more than half of the entire community (Figure 2A). It should be noted that the relative abundance of Proteobacteria in CK treatment was higher than that in multiple cropping treatments, while the relative abundance of Chloroflexi and Acidobacteria was lower than that in multiple cropping treatments. Tobacco-rice multiple cropping rotation showed a stronger symbiotic relationship between Chloroflexi and Acidobacteria. At the genus level, there are 720, 729, 694, and 751 different genera in CK, Y10, Y20, and Y40 treatments, respectively. UTCFX1 is the genus with the highest relative abundance among all identified genera (Figure 2B). The mean relative abundance of UTCFX1 in CK, Y10, Y20, and Y40 treatments were 7.99, 10.27, 12.43, and 9.39%, respectively. From the relative abundance of the heat map and the scale of the color change, the differences in the composition of the soil bacterial community at the genus level among the treatments can be observed more clearly (Supplementary Figure S1). The relative abundance of MND1 and Anaeromyxobacter was higher in CK; Pseudolabrys, Anaerolinea, Sideroxydans, Geobacter, and Haliangium were higher in Y10; Nitrospira, Gemmatimonas, RB41, and UTCFX1 were higher in Y20; and Thiobacillus was higher in Y40. In addition, according to the cluster analysis of the heat map, the treatments were divided into two groups. All tobacco-rice multiple cropping rotation farmlands (Y10, Y20, and Y40) were clustered together, and rice monoculture farmlands (CK) were grouped into one group separately, which is consistent with the results of NMDS.
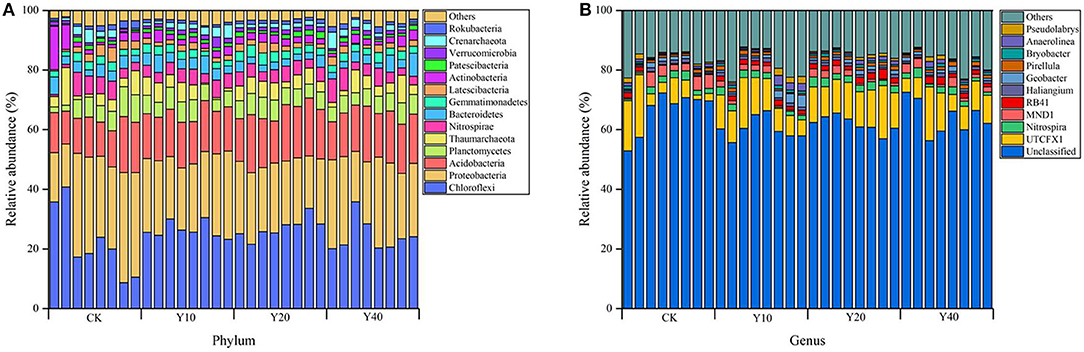
Figure 2. Soil bacterial community composition: (A) Relative abundance at phylum level; (B) Relative abundance at genus level. Among the bacterial communities in all samples, phylum and genera with an abundance of <1% were classified as “others.”
The Venn diagram showed that the CK treatment had more unique OTUs (Figure 3A), and with the increase in the cultivation years of tobacco-rice multiple cropping rotation farmlands, the number of unique OTUs in the Y20 and Y40 treatments gradually decreased. The environmental factors driving the change of bacterial community were revealed by the redundancy analysis (RDA) analysis and the envfit analysis. The results of the RDA plot (Figure 3B) and the envfit analysis (Supplementary Table S2) show that soil pH, ammonium nitrogen, and nitrate nitrogen were the environmental variables that significantly affected the changes of soil bacterial communities (p < 0.05), and the four environmental variables cumulatively explain 14.98% of the variation of the bacterial community (Supplementary Table S3).
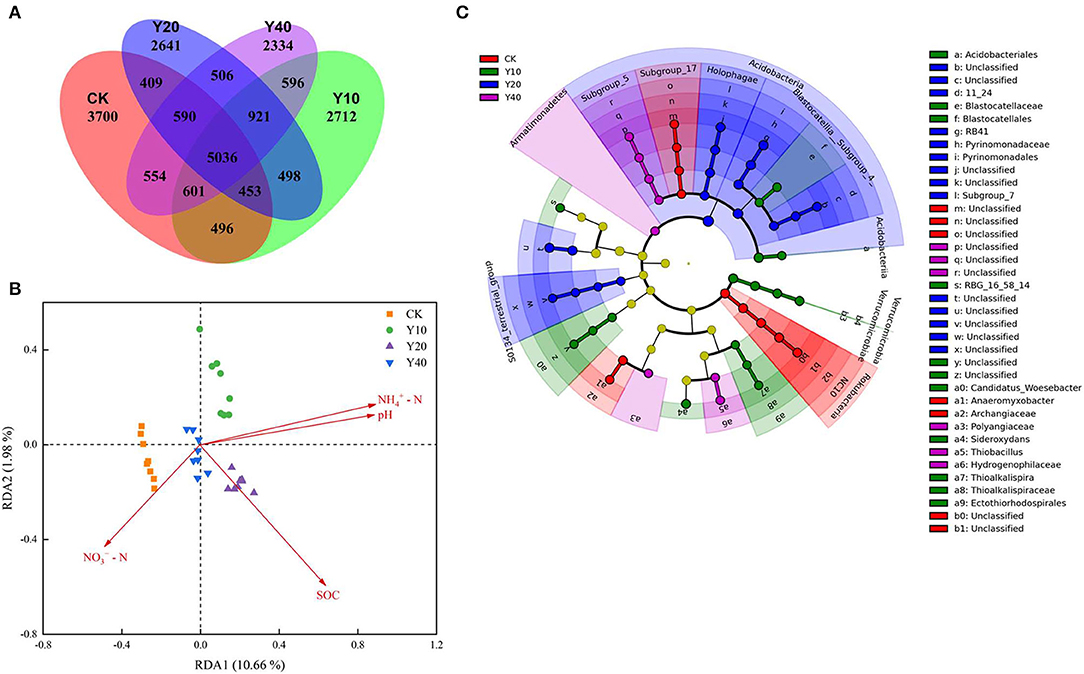
Figure 3. Venn diagrams showing the shared and unique OTUs (A) and redundancy analysis shows soil properties drive the bacterial community (B). Linear discriminant analysis Effect Size (LEfSe) identified the size of differentiation in bacterial abundances between different treatments with a threshold value of 3.0 (C). SOC, Soil organic carbon; -N, Ammonia-N; -N, Nitrate-N.
Through the LEfSe analysis (threshold value of 3.0), the specificity in bacterial community composition among different treatments was analyzed, and 53 bacterial taxa (Supplementary Table S4) showed significant differences (p < 0.05). The cladogram (Figure 3C) showed that at the phylum level, Rokubacteria was highly enriched in the CK treatment, Verrucomicrobia was highly enriched in the Y10 treatment, Acidobacteria was highly enriched in the Y20 treatment, and Armatimonadetes was highly enriched in the Y40 treatment. Pearson's correlation analysis (Table 3) of the bacterial phyla that were significantly enriched in each treatment and soil properties showed that Rokubacteria was significantly positively correlated with soil nitrate nitrogen; Verrucomicrobia was significantly positively correlated with soil ammonium nitrogen and significantly negatively correlated with nitrate nitrogen; and Acidobacteria and Armatimonadetes were significantly positively correlated with soil pH, SOC, and ammonium nitrogen. The soil characteristics of the highest pH, SOC, and ammonium nitrogen in tobacco-rice multiple cropping rotation farmlands with a cultivation period of 20–40 years drive more Acidobacteria to survive in the soil.
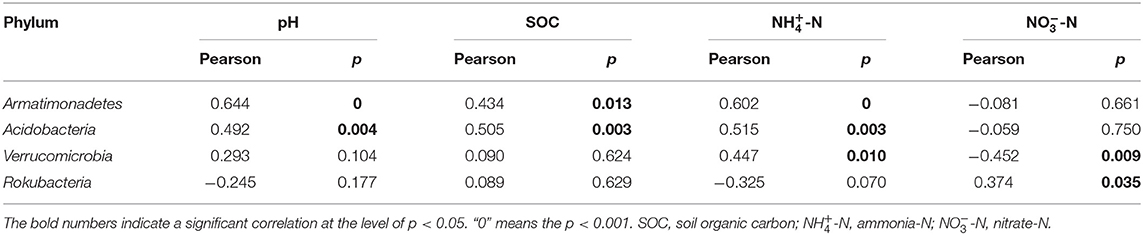
Table 3. Pearson's correlation between soil properties and phylum abundance in the bacterial community.
Since the farmland (Y20) with the highest SOC and soil ammonium nitrogen content had the most enrichment of Acidobacteria (Supplementary Table S4), we speculated that Acidobacteria plays an important role in the soil carbon and nitrogen cycle. To further understand the distribution of Acidobacteria in farmland with different cultivation years, we compared the relative abundance of 21 different Acidobacteria subgroups (Table 4). In the CK treatment, the relative abundance of Acidobacteria GP17 was significantly higher than other treatments (p < 0.05). In the Y10 treatment, the relative abundance of Acidobacteria GP1 was significantly higher than other treatments (p < 0.05). In the Y20 treatment, the relative abundance of Acidobacteria GP4 and Acidobacteria GP7 was significantly the highest (p < 0.05). The relative abundance of Acidobacteria GP15 was significantly highest in the Y40 treatment (p < 0.05). The heat map of the pairwise Pearson's correlation matrix (Figure 4) of edaphic factors and Acidobacteria subgroups shows that Acidobacteria GP1 is significantly negatively correlated with soil nitrate nitrogen, Acidobacteria GP3, GP4, GP5, GP7, and GP12 are significantly positively correlated with soil pH and ammonium nitrogen, and Acidobacteria GP17 and GP23 are significantly negatively correlated with soil pH, SOC, and ammonium nitrogen (p < 0.01). Noticeably, compared with rice monoculture, tobacco-rice multiple cropping rotation significantly increased soil pH (Table 2), and the Acidobacteria GP17 abundance decreased significantly (p < 0.01), with a maximum decrease of 54.65%. The abundance of Acidobacteria GP4 and GP5 increased significantly (p < 0.01), with the highest increases of 152.05 and 101.08%, respectively.
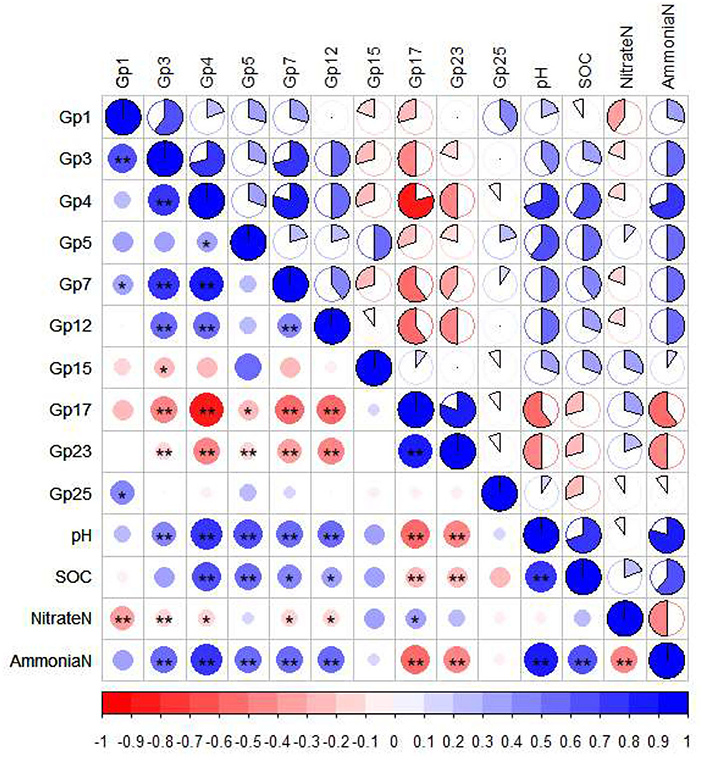
Figure 4. Pairwise Pearson's correlation matrix of the edaphic factors and Acidobacteria subgroups was shown with heat map, * Significant at the P = 0.05 level, ** Significant at the P = 0.01 level. SOC, Soil organic carbon.
To investigate the differences in bacterial interaction between rice monoculture farmland and tobacco-rice multiple cropping rotation farmlands with different cultivation years, four molecular ecological networks based on RMT were constructed using the 16s rRNA sequencing data of each treatment (Figure 5). The topological features of the molecular ecological networks (Table 5) showed that the connectivity distribution curves of all four networks fit well with the power-law model, with the R2 values of 0.818, 0.915, 0.913, and 0.839, respectively. Y10, Y20, and Y40 have more nodes than CK, but Y10 and Y20 have fewer links than CK, and Y40 has the most nodes and links (913 nodes and 3,680 links). Compared with other treatments, Y40 has the highest node, link, and average connectivity, as well as the lowest average path distance and harmonic geodetic distance, so its network is more complex, and the connection between bacteria was closer. However, Y40 has the lowest modularity, indicating that its network stability was poor, and it was more difficult to resist the interference of external environmental changes. The percentages of positive correlations in CK (80.03%), Y10 (70.96%), Y20 (67.25%), and Y40 (73.80%) were greater than the negative correlations, but the percentages of positive correlations in Y10, Y20, and Y40 treatments were lower than those of CK, indicating that tobacco-rice multiple cropping rotation would strengthen the competitive relationship between bacteria. We tried to clarify the relationship between edaphic factors and bacterial positive correlation by using the linear regression equation. The results showed that there is a significant negative correlation (p < 0.01) between soil ammonium nitrogen and the percentages of positive links (Supplementary Figure S2). The cooperative relationship between bacteria will weaken, and the competitive relationship will become stronger with the increase of soil ammonium nitrogen content.
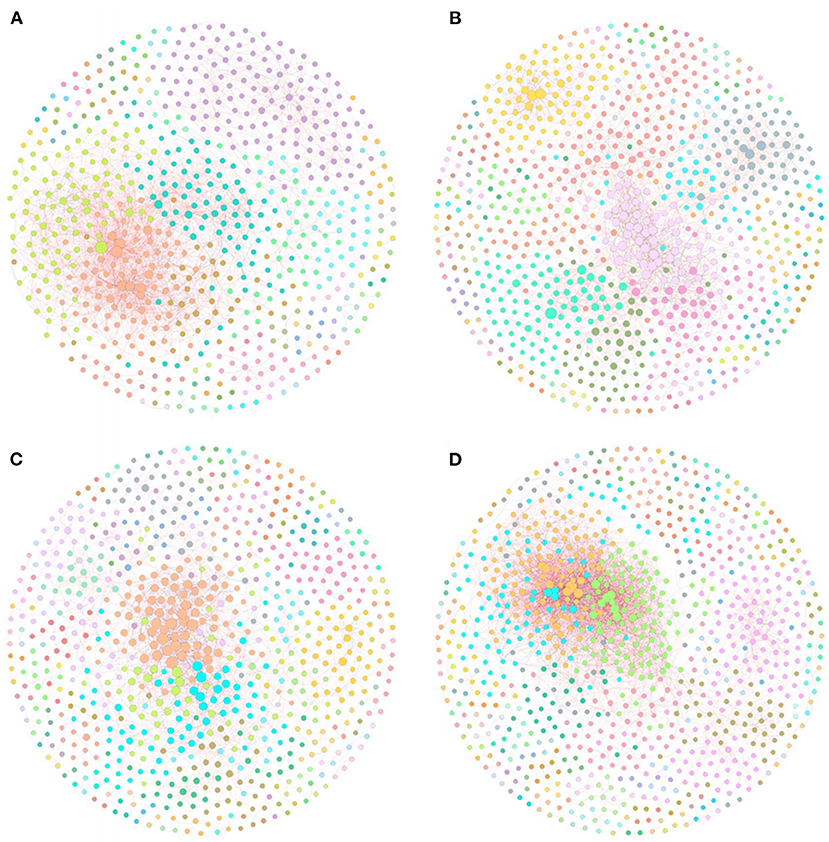
Figure 5. The RMT-based molecular ecological networks of CK (A), Y10 (B), Y20 (C), Y40 (D). In the networks, each node represents an OUT, and edges between nodes correspond to either positive (red) or negative (black) correlations. Modules were randomly colored.
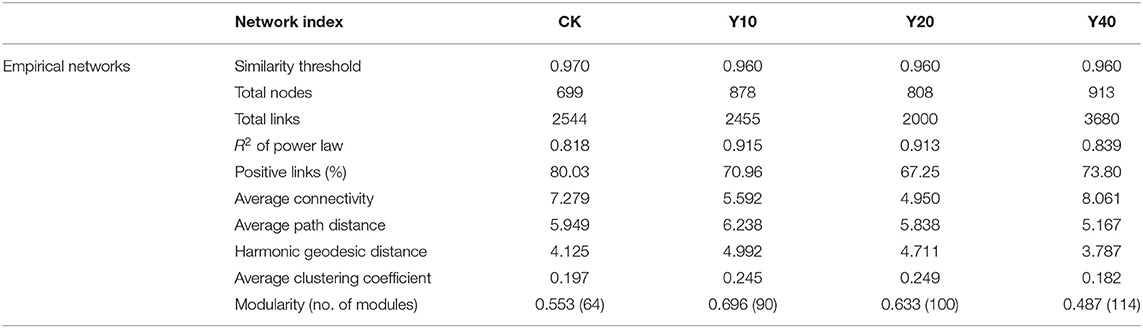
Table 5. Major topological features of the molecular ecological networks of bacterial communities in CK, Y10, Y20, and Y40.
To ascertain the possible topological role of taxa in the network, the nodes are divided into four categories (i.e., peripherals, connectors, module hubs, and network hubs) according to the within-module connectivity (Zi) and among-module connectivity (Pi) values of taxa (Figure 6). From the Zi-Pi plot, it can be observed that 3.29, 2.39, 1.98, and 2.63% of the nodes in CK, Y10, Y20, and Y40 were classified as module hubs, and 1.14, 0.80, 1.11, and 0.77% of the nodes were classified as connectors, respectively. None of the nodes meet the classification of network hubs. The taxonomy information of keystone taxa (Supplementary Table S5) showed that the members of some phyla such as Proteobacteria, Acidobacteria, Chloroflexi, and Planctomycetes were the highlighted keystone taxa in networks. A total of 8 keystone OTUs appeared in the module hubs and connectors of different networks, namely, OTU102, OTU4, OTU11, OTU2548, OTU358, OTU1329, OTU409, and OTU2943, of which OTU102, OTU4, and OTU11 appeared in the module hubs of CK and Y40 networks, OTU2548 in the module hubs of Y10 networks, OTU409 in the module hubs of Y20 network also appeared in the module hubs of Y40 network, OTU358 in Y10 network connector, and OTU2943 in Y20 network connector appeared in Y40 network connector. According to the taxonomic information of these 8 OTUs (Supplementary Table S6), the members of the Acidobacteria accounted for half, indicating that Acidobacteria played an important role in multiple different networks. While different Acidobacteria members in the connector and module hubs of Y10 network and the module hubs of Y20 network appeared in the module hubs of Y40 network (OTU2548, OTU358, and OTU409), and they did not appear in the module hubs of CK network (Table 6), indicating that tobacco-rice multiple cropping rotation can change the role of Acidobacteria members in the molecular ecological networks.
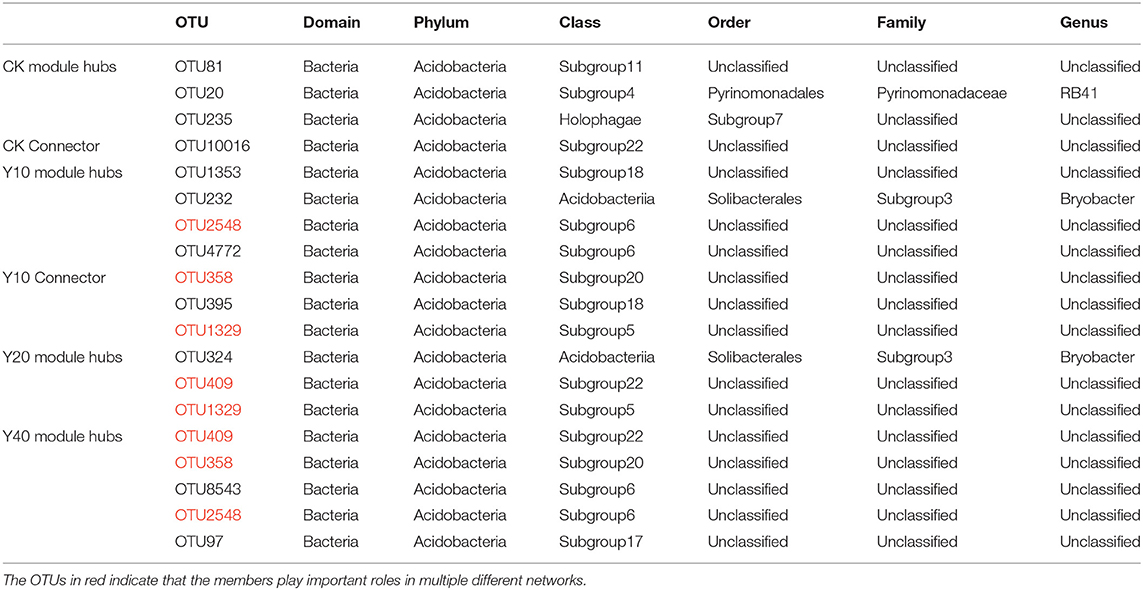
Table 6. Taxonomy information of keystone operational taxonomic units (OTUs) of Acidobacteria members.
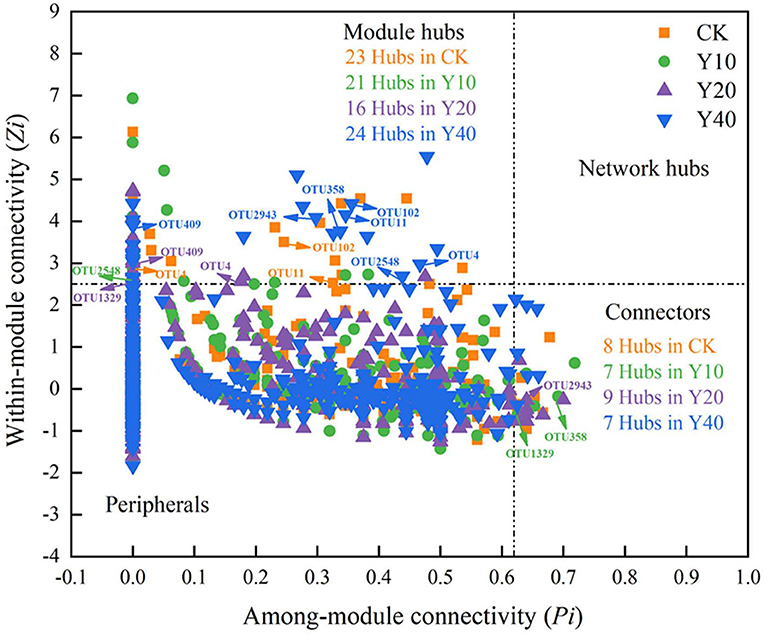
Figure 6. Each node was identification to classify the keystone taxa in molecular ecological networks of bacterial communities. Each symbol represents an OTU from the network of CK, Y10, Y20 and Y40. The value of Zi and Pi confirmed each OTU's topological role. Nodes were identified into four models depended on their Zi and Pi values: module hubs (Zi > 2.5 and Pi < 0.62), network hubs (Zi > 2.5 and Pi > 0.62), connectors (Zi < 2.5 and Pi > 0.62) and peripherals (Zi < 2.5 and Pi < 0.62). Besides, the number of module hubs and connectors as well as the OTUs that repeatedly appear in different networks were shown in the corresponding area of the plot.
Paddy-upland multiple cropping rotation can accelerate the soil carbon cycle and the decomposition rate of soil organic matter in the farmland and, therefore, improve soil fertility (Zheng et al., 2016). A previous study has shown that under the condition of planting green manure in the winter fallow period, compared with the double rice cropping system, the paddy-upland multiple cropping rotation significantly increased the soil pH, SOC content, and nitrate nitrogen content and decreased the ammonium nitrogen content (Zhong et al., 2021). Although soil ammonium nitrogen and nitrate nitrogen are the two main forms of nitrogen that can be absorbed by crops, different crops have different preferences for ammonium nitrogen and nitrate nitrogen (Alpha et al., 2009; Wang et al., 2015). In this study, the farmland with the highest soil ammonium nitrogen content (Y20) has a higher grain yield than the farmland with the highest soil nitrate nitrogen content (Y40), suggesting that ammonium nitrogen is more conducive to the increase of grain yield. Understanding the effects of different soil nitrogen nutrient forms on grain yield is crucial for us to develop sustainable agriculture.
The structure and diversity of the soil microbial community are easily affected by factors such as climate change, vegetation types, and soil properties (Liang et al., 2015; Tang et al., 2020; Tan et al., 2021). Previous studies showed that the soil bacterial community structure of paddy fields with different cultivation years of continuous fertilization has differentiated, and the soil bacterial community of paddy fields with 25 years of cultivation is significantly different from that of 9 and 15 years of cultivation (Li et al., 2019a). In this study, we unexpectedly found that there was no significant difference in the diversity and structure of soil bacterial community in tobacco-rice multiple cropping rotation farmlands with different cultivation years. The reason is that tobacco-rice multiple cropping rotation farmlands with different cultivation years has no significant change in soil pH, resulting in no difference in the diversity and structure of bacterial community. Furthermore, since our sampling sites were all in Guiyang County, the homogeneity of geographical location and climatic environment may also cause no difference in the diversity and structure of the bacterial community.
Research has reported that compared with rice monoculture, the composition and diversity of soil bacterial community in paddy-upland multiple cropping rotation have changed (Do Thi et al., 2012), and the taxa of the community, especially the core taxa, play an important role in maintaining the functions of agricultural ecosystems (Shi et al., 2020; Li et al., 2021a). Our results showed that the differences in the nutrient characteristics of farmlands with different cultivation years shaped the differentiation of dominant bacterial genus, and the LEfSe analysis showed that the specific taxa significantly enriched in each treatment were different. Recent studies have shown that Acidobacteria RB41 plays a key role in control over soil carbon cycle (Stone et al., 2021), and Acidobacteria is involved in the metabolism of inorganic and organic nitrogen sources (Eichorst et al., 2018), and Acidobacteria members can also act as plant growth-promoting rhizobacteria (PGPR) to improve plant growth (Kalam et al., 2020). Our research found that Acidobacteria were highly enriched in farmland with high SOC and ammonium nitrogen content, and the relative abundance of Acidobacteria was significantly positively correlated with pH, SOC, and ammonium nitrogen. Different Acidobacteria subgroups respond differently to soil properties. Some studies have reported that Acidobacteria subgroups 1, 2, and 3 were negatively correlated with soil pH, total C, and N, and Acidobacteria subgroups 4, 6, 7, and 25 were positively correlated with soil pH, total C, and N (Navarrete et al., 2015). Liu et al. (2016) also supported that Acidobacteria subgroups 1, 3, and 13 were negatively correlated with soil pH, while subgroups 4, 6, 7, 11, 17, 18, and 25 were positively correlated with soil pH. In this study, Acidobacteria subgroup 3 was positively correlated with soil pH, and subgroup 17 was negatively correlated with soil pH, which was different from the previous research results. The reason may be the differences in land use and status, covered crops, and geographical regions (Ge et al., 2008; Lu et al., 2019). Our study demonstrated the involvement of Acidobacteria in soil carbon and nitrogen cycle, but we were unable to determine the role of Acidobacteria in soil carbon and nitrogen cycle, and we may need to verify the role of Acidobacteria by isolation and experimental phenotype characterizations in the future.
Studies have proved that the interaction and coexistence patterns between microorganisms have a certain feedback and regulation effect on global climate change, biogeochemical processes, plant growth, and stress resistance (Wang et al., 2019; Mohapatra et al., 2021; Yuan et al., 2021). Our research proves that compared with rice monoculture, tobacco-rice multiple cropping rotation can increase the nodes of the network, and Y40 with the longest cultivation years has the highest network complexity, with most nodes, links, and modules. However, due to the lowest modularity of Y40, its network stability was poor, the interaction between bacteria was more vulnerable to the interference of external environmental stress (Hernandez et al., 2021), and the weaker bacterial community stability and resistance may be the reason for the decline of grain yield in Y40 farmland (Horner et al., 2019). Moreover, we found that the increase of soil ammonium nitrogen content strengthened the competitive relationship between bacterial interactions, and more competitive relationships between bacterial interactions will enhance the stability of the community (Coyte et al., 2015; Ghoul and Mitri, 2016). The better the soil nutrient conditions, the stronger the competitive relationship between bacterial interactions has also been confirmed (Wang et al., 2018). By classifying the keystone taxa in the molecular ecological network, we found that there were 8 repeated OTUs in different networks, of which 4 OTUs were Acidobacteria. We hold the opinion that Acidobacteria plays an important role in all networks. In addition, tobacco-rice multiple cropping rotation farmlands with different cultivation years have the same keystone OTUs of Acidobacteria members, and these OTUs are inconsistent with rice monoculture farmland, which indicates that tobacco-rice multiple cropping rotation has changed the Acidobacteria members with important functions in the network.
The cultivation years of the farmland affected the richness of the bacterial community, and the increase of cultivation age reduced the diversity of the bacterial community and changed the community structure and composition (Chang et al., 2021; Tong et al., 2021). Continuous cropping has caused the aggravation of soil-borne diseases in farmland, resulting in a decline in crop quality and yield, and severely restricted the productivity and sustainable development of farmland ecosystems (Arafat et al., 2019; Gao et al., 2020; Tian et al., 2020). However, recent studies have shown that continuous cultivation of farmland has no significant negative impact on crop yield, which is attributed to the use of organic fertilizers and crop straw mulching return (Rannestad and Gessesse, 2020). With the increase of cultivation years, the diversity and structure of bacterial communities did not change significantly in tobacco-rice multiple cropping rotation farmlands in this study. Compared with the farmland with 10–20 years of cultivation, the grain yield of farmland with 20–40 years of cultivation has increased significantly. These studies prove that under the tobacco-rice multiple cropping rotation system in this study, there are no continuous cropping obstacles due to reasonable nutrient planning, agronomic management, and crop straw returning (Chen et al., 2018). However, the decline of soil bacterial community stability and grain yield in farmland with cultivation years greater than 40 years indicates that excessive cultivation years are not conducive to the sustainable use of soil and the stable production of crops. Our research provides a theoretical basis for rationally developing crop planting patterns, resisting continuous cropping obstacles, and stabilizing crop production and has important practical significance for guiding sustainable agricultural production.
Tobacco-rice multiple cropping rotation improved soil fertility and the diversity of the bacterial community and finally increased crop yields. The farmland with the highest SOC and ammonium nitrogen content enriched Acidobacteria members. Different Acidobacteria subgroups respond differently to changes in soil properties, and Acidobacteria members play a key role in maintaining biodiversity and the local ecosystem. The decrease in the stability of bacterial communities in farmland where the cultivation period is too long may be the reason for the decrease in crop yield. Thus, choosing reasonable crop planting patterns according to local conditions and reducing the years of planting crops in the same farmland are more conducive to the sustainable development and production of farmland and crops.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: the raw sequencing data can be found at the National Centre for Biotechnology Information (NCBI) Sequence Read Archive (SRA) (Accession Number: PRJNA775899).
TC: carried out the experiments and wrote the article. RH: formal analysis and wrote the article. ZZ, JY, HF, XD, and WY: carried out the experiments. QW: methodology. SP: methodology and formal analysis. JL: methodology, formal analysis, and wrote the article. All authors contributed to the article and approved the submitted version.
This study was supported by the Natural Science Foundation of Hunan Province (2020JJ4038 and 2020JJ4374) and the Key Project of Science and Technology of Hunan Tobacco Company Chenzhou Branch (CYKJ2018-05 and CZYC2021JS10).
TC, XD and WY were employed by the Hunan Tobacco Company.
This study received funding from Key Project of Science and Technology of Hunan Tobacco Company Chenzhou Branch and Natural Science Foundation of Hunan Province. The funder had involvement with the study design and data collection. All authors declare no other competing interests.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.804527/full#supplementary-material
Alpha, J. M., Chen, J., and Zhang, G. (2009). Effect of nitrogen fertilizer forms on growth, photosynthesis, and yield of rice under cadmium stress. J. Plant Nutr. 32, 306–317. doi: 10.1080/01904160802608635
Arafat, Y., Tayyab, M., Khan, M. U., Chen, T., Amjad, H., Awais, S., et al. (2019). Long-term monoculture negatively regulates fungal community composition and abundance of tea orchards. Agronomy 9:466. doi: 10.3390/agronomy9080466
Arcand, M. M., Helgason, B. L., and Lemke, R. L. (2016). Microbial crop residue decomposition dynamics in organic and conventionally managed soils. Appl. Soil Ecol. 107, 347–359. doi: 10.1016/j.apsoil.2016.07.001
Bacci, G., Bani, A., Bazzicalupo, M., Ceccherini, M. T., Galardini, M., Nannipieri, P., et al. (2015). Evaluation of the performances of ribosomal database project (RDP) classifier for taxonomic assignment of 16S rRNA metabarcoding sequences generated from illumina-solexa NGS. J Genom. 3, 36–39. doi: 10.7150/jgen.9204
Bao, S. D. (2000). Soil and Agricultural Chemistry Analysis. Beijing, China: Agriculture Publication.
Barcenas-Moreno, G., Baath, E., and Rousk, J. (2016). Functional implications of the pH-trait distribution of the microbial community in a re-inoculation experiment across a pH gradient. Soil Biol. Biochem. 93, 69–78. doi: 10.1016/j.soilbio.2015.10.024
Caviglia, O. P., Sadras, V. O., and Andrade, F. H. (2011). Yield and quality of wheat and soybean in sole- and double-cropping. Agron. J. 103, 1081–1089. doi: 10.2134/agronj2011.0019
Chang, F., Jia, F., Lv, R., Li, Y., Wang, Y., Jia, Q., et al. (2021). Soil bacterial communities reflect changes in soil properties during the tillage years of newly created farmland on the Loess Plateau. Appl. Soil Ecol. 161:103853. doi: 10.1016/j.apsoil.2020.103853
Chaudhary, D. K., Khulan, A., and Kim, J. (2019). Development of a novel cultivation technique for uncultured soil bacteria. Sci. Reports. 9:1–11. doi: 10.1038/s41598-019-43182-x
Chen, G., Qiao, J., Zhao, G., Zhang, H., Shen, Y., and Cheng, W. (2018). Rice-straw biochar regulating effect on Chrysanthemum morifolium Ramat. cv. 'Hangbaiju'. Agron. J. 110, 1996–2003. doi: 10.2134/agronj2017.12.0710
Coyte, K. Z., Schluter, J., and Foster, K. R. (2015). The ecology of the microbiome: networks, competition, and stability. Science 350, 663–666. doi: 10.1126/science.aad2602
Deng, Y., Jiang, Y.-H., Yang, Y., He, Z., Luo, F., and Zhou, J. (2012). Molecular ecological network analyses. BMC Bioinform. 13:113. doi: 10.1186/1471-2105-13-113
Do Thi, X., Vo Thi, G., Rosling, A., Alstrom, S., Chai, B., and Hogberg, N. (2012). Different crop rotation systems as drivers of change in soil bacterial community structure and yield of rice, Oryza sativa. Biol. Fertil. Soils. 48, 217–225. doi: 10.1007/s00374-011-0618-5
Eichorst, S. A., Kuske, C. R., and Schmidt, T. M. (2011). Influence of plant polymers on the distribution and cultivation of bacteria in the phylum Acidobacteria. Appl. Environ. Microbiol. 77, 586–596. doi: 10.1128/aem.01080-10
Eichorst, S. A., Trojan, D., Roux, S., Herbold, C., Rattei, T., and Woebken, D. (2018). Genomic insights into the Acidobacteria reveal strategies for their success in terrestrial environments. Environ. Microbiol. 20, 1041–1063. doi: 10.1111/1462-2920.14043
Frac, M., Hannula, S. E., Belka, M., and Jedryczka, M. (2018). Fungal biodiversity and their role in soil health. Front. Microbiol. 9:707. doi: 10.3389/fmicb.2018.00707
Frey, S. D., Lee, J., Melillo, J. M., and Six, J. (2013). The temperature response of soil microbial efficiency and its feedback to climate. Nat. Clim. Chang. 3, 395–398. doi: 10.1038/nclimate1796
Gao, J., Pei, H., and Xie, H. (2020). Synergistic effects of organic fertilizer and corn straw on microorganisms of pepper continuous cropping soil in China. Bioengineered 11, 1258–1268. doi: 10.1080/21655979.2020.1840753
Ge, Y., He, J.-,z., Zhu, Y.-,g., Zhang, J.-,b., Xu, Z., Zhang, L.-, m., et al. (2008). Differences in soil bacterial diversity: driven by contemporary disturbances or historical contingencies? Isme J. 2, 254–264. doi: 10.1038/ismej.2008.2
Ghoul, M., and Mitri, S. (2016). The ecology and evolution of microbial competition. Trends Microbiol. 24, 833–845. doi: 10.1016/j.tim.2016.06.011
Gómez-Rubio, V. (2017). ggplot2—Elegant Graphics for Data Analysis (2nd Edition). J. Stat. Softw. 77, 1–3. doi: 10.18637/jss.v077.b02
Griffiths, B. S., and Philippot, L. (2013). Insights into the resistance and resilience of the soil microbial community. FEMS Microbiol. Rev. 37, 112–129. doi: 10.1111/j.1574-6976.2012.00343.x
Gu, Y., Meng, D., Yang, S., Xiao, N., Li, Z., Liu, Z., et al. (2019). Invader-resident community similarity contribute to the invasion process and regulate biofertilizer effectiveness. J. Clean. Prod. 241:118278. doi: 10.1016/j.jclepro.2019.118278
Guyonnet, J. P., Guillemet, M., Dubost, A., Simon, L., Ortet, P., Barrakat, M., et al. (2018). Plant nutrient resource use strategies shape active rhizosphere microbiota through root exudation. Front. Plant Sci. 9:1662. doi: 10.3389/fpls.2018.01662
Hartmann, M., Frey, B., Mayer, J., Maeder, P., and Widmer, F. (2015). Distinct soil microbial diversity under long-term organic and conventional farming. Isme J. 9, 1177–1194. doi: 10.1038/ismej.2014.210
Hernandez, D. J., David, A. S., Menges, E. S., Searcy, C. A., and Afkhami, M. E. (2021). Environmental stress destabilizes microbial networks. Isme J. 15, 1722–1734. doi: 10.1038/s41396-020-00882-x
Horner, A., Browett, S. S., and Antwis, R. E. (2019). Mixed-cropping between field pea varieties alters root bacterial and fungal communities. Sci. Rep. 9, 1–10. doi: 10.1038/s41598-019-53342-8
Hu, L., Zi, H., Wu, P., Wang, Y., Lerdau, M., Wu, X., et al. (2019). Soil bacterial communities in grasslands revegetated using Elymus nutans are largely influenced by soil pH and total phosphorus across restoration time. Land Degrad. Dev. 30, 2243–2256. doi: 10.1002/ldr.3414
Hu, R., Liu, Y., Chen, T., Zheng, Z., Peng, G., Zou, Y., et al. (2021). Responses of soil aggregates, organic carbon, and crop yield to short-term intermittent deep tillage in Southern China. J. Clean. Prod. 298:126767. doi: 10.1016/j.jclepro.2021.126767
Huang, M., Fang, S., Cao, F., Chen, J., Shan, S., Liu, Y., et al. (2020). Early sowing increases grain yield of machine-transplanted late-season rice under single-seed sowing. Field Crops Res. 253:107832. doi: 10.1016/j.fcr.2020.107832
Jones, R. T., Robeson, M. S., Lauber, C. L., Hamady, M., Knight, R., and Fierer, N. (2009). A comprehensive survey of soil acidobacterial diversity using pyrosequencing and clone library analyses. Isme J. 3, 442–453. doi: 10.1038/ismej.2008.127
Kalam, S., Basu, A., Ahmad, I., Sayyed, R. Z., El-Enshasy, H. A., Dailin, D. J., et al. (2020). Recent understanding of soil acidobacteria and their ecological significance: a critical review. Front. Microbiol. 11:580024. doi: 10.3389/fmicb.2020.580024
Kielak, A. M., Cipriano, M. A. P., and Kuramae, E. E. (2016). Acidobacteria strains from subdivision 1 act as plant growth-promoting bacteria. Arch. Microbiol. 198, 987–993. doi: 10.1007/s00203-016-1260-2
Li, F., Chen, L., Zhang, J., Yin, J., and Huang, S. (2017). Bacterial Community structure after long-term organic and inorganic fertilization reveals important associations between soil nutrients and specific taxa involved in nutrient transformations. Front. Microbiol. 8:187. doi: 10.3389/fmicb.2017.00187
Li, M., Guo, J., Ren, T., Luo, G., Shen, Q., Lu, J., et al. (2021a). Crop rotation history constrains soil biodiversity and multifunctionality relationships. Agric. Ecosyst. Environ. 319:107550. doi: 10.1016/j.agee.2021.107550
Li, M., Wang, G., Kang, X., Hu, H., Wang, Y., Zhang, X., et al. (2020). Long-term fertilization alters microbial community but fails to reclaim soil organic carbon stocks in a land-use changed soil of the Tibetan Plateau. Land Degrad. Dev. 31, 531–542. doi: 10.1002/ldr.3469
Li, W., Jiang, L., Zhang, Y., Teng, D., Wang, H., Wang, J., et al. (2021b). Structure and driving factors of the soil microbial community associated with Alhagi sparsifolia in an arid desert. PLoS ONE. 16:e0254065. doi: 10.1371/journal.pone.0254065
Li, W., Liu, M., Wu, M., Jiang, C., Kuzyakov, Y., Gavrichkova, O., et al. (2019a). Bacterial community succession in paddy soil depending on rice fertilization. Appl. Soil Ecol. 144, 92–97. doi: 10.1016/j.apsoil.2019.07.014
Li, W., Niu, S., Liu, X., and Wang, J. (2019b). Short-term response of the soil bacterial community to differing wildfire severity in Pinus tabulaeformis stands. Sci. Rep. 9, 1–10. doi: 10.1038/s41598-019-38541-7
Liang, Y., Jiang, Y., Wang, F., Wen, C., Deng, Y., Xue, K., et al. (2015). Long-term soil transplant simulating climate change with latitude significantly alters microbial temporal turnover. Isme J. 9, 2561–2572. doi: 10.1038/ismej.2015.78
Liu, C., Dong, Y., Hou, L., Deng, N., and Jiao, R. (2017). Acidobacteria community responses to nitrogen dose and form in Chinese fir plantations in Southern China. Curr. Microbiol. 74, 396–403. doi: 10.1007/s00284-016-1192-8
Liu, J., Sui, Y., Yu, Z., Yao, Q., Shi, Y., Chu, H., et al. (2016). Diversity and distribution patterns of acidobacterial communities in the black soil zone of northeast China. Soil Biol. Biochem. 95, 212–222. doi: 10.1016/j.soilbio.2015.12.021
Liu, Z., Zhou, W., Li, S., He, P., Liang, G., Lv, J., et al. (2015). Assessing soil quality of gleyed paddy soils with different productivities in subtropical China. Catena 133, 293–302. doi: 10.1016/j.catena.2015.05.029
Lu, M., Ren, Y., Wang, S., Tian, K., Sun, X., and Peng, S. (2019). Contribution of soil variables to bacterial community composition following land use change in Napahai plateau wetlands. J. Environ. Manage. 246, 77–84. doi: 10.1016/j.jenvman.2019.05.149
Mendes, R., Garbeva, P., and Raaijmakers, J. M. (2013). The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 37, 634–663. doi: 10.1111/1574-6976.12028
Mohapatra, M., Yadav, R., Rajput, V., Dharne, M. S., and Rastogi, G. (2021). Metagenomic analysis reveals genetic insights on biogeochemical cycling, xenobiotic degradation, and stress resistance in mudflat microbiome. J. Environ. Manage. 292. doi: 10.1016/j.jenvman.2021.112738
Naether, A., Foesel, B. U., Naegele, V., Wuest, P. K., Weinert, J., Bonkowski, M., et al. (2012). Environmental factors affect acidobacterial communities below the subgroup level in grassland and forest soils. Appl. Environ. Microbiol. 78, 7398–7406. doi: 10.1128/aem.01325-12
Navarrete, A. A., Venturini, A. M., Meyer, K. M., Klein, A. M., Tiedje, J. M., Bohannan, B. J. M., et al. (2015). Differential response of acidobacteria subgroups to forest-to-pasture conversion and their biogeographic patterns in the Western Brazilian Amazon. Front. Microbiol. 6:1443. doi: 10.3389/fmicb.2015.01443
Oksanen, J., Guillaume, B. F., Kindt, R., Legendre, P., Minchin, P., O'Hara, R., et al. (2006). vegan: Community Ecology Package version 2.0-10. J. Stat. Softw. 48, 103–132.
Rannestad, M. M., and Gessesse, T. A. (2020). Deforestation and subsequent cultivation of nutrient poor soils of miombo woodlands of tanzania: long term effect on maize yield and soil nutrients. Sustainability 12:4113. doi: 10.3390/su12104113
Rodriguez, P. A., Rothballer, M., Chowdhury, S. P., Nussbaumer, T., Gutjahr, C., and Falter-Braun, P. (2019). Systems biology of plant-microbiome interactions. Mol. Plant. 12, 804–821. doi: 10.1016/j.molp.2019.05.006
Shi, Y., Delgado-Baquerizo, M., Li, Y., Yang, Y., Zhu, Y.-G., Penuelas, J., et al. (2020). Abundance of kinless hubs within soil microbial networks are associated with high functional potential in agricultural ecosystems. Environ. Int. 142:105869. doi: 10.1016/j.envint.2020.105869
Stone, B. W., Li, J., Koch, B. J., Blazewicz, S. J., Dijkstra, P., Hayer, M., et al. (2021). Nutrients cause consolidation of soil carbon flux to small proportion of bacterial community. Nat. Commun. 12, 1–9. doi: 10.1038/s41467-021-23676-x
Tan, G., Liu, Y., Peng, S., Yin, H., Meng, D., Tao, J., et al. (2021). Soil potentials to resist continuous cropping obstacle: three field cases. Environ. Res. 200:111319. doi: 10.1016/j.envres.2021.111319
Tang, M., Li, L., Wang, X., You, J., Li, J., and Chen, X. (2020). Elevational is the main factor controlling the soil microbial community structure in alpine tundra of the Changbai Mountain. Sci. Rep. 10, 1–15. doi: 10.1038/s41598-020-69441-w
Tian, L., Shi, S., Ma, L., Lam-Son Phan, T., and Tian, C. (2020). Community structures of the rhizomicrobiomes of cultivated and wild soybeans in their continuous cropping. Microbiol. Res. 232:126390. doi: 10.1016/j.micres.2019.126390
Tong, A.-Z., Liu, W., Liu, Q., Xia, G.-Q., and Zhu, J.-Y. (2021). Diversity and composition of the Panax ginseng rhizosphere microbiome in various cultivation modesand ages. BMC Microbiol. 21, 1–13. doi: 10.1186/s12866-020-02081-2
Viaud, V., Santillan-Carvantes, P., Akkal-Corfini, N., Le Guillou, C., Prevost-Boure, N. C., Ranjard, L., et al. (2018). Landscape-scale analysis of cropping system effects on soil quality in a context of crop-livestock farming. Agric. Ecosyst. Environ. 265, 166–177. doi: 10.1016/j.agee.2018.06.018
Viggi, C. C., Rossetti, S., Fazi, S., Paiano, P., Majone, M., and Aulenta, F. (2014). Magnetite particles triggering a faster and more robust syntrophic pathway of methanogenic propionate degradation. Environ. Sci. Technol. 48, 7536–7543. doi: 10.1021/es5016789
Wang, Q., Liu, X., Jiang, L., Cao, Y., Zhan, X., Griffin, C. H., et al. (2019). Interrogation of internal workings in microbial community assembly: play a game through a behavioral network? Msystems. 4:e00550–19. doi: 10.1128/mSystems.00550-19
Wang, S., Wang, X., Han, X., and Deng, Y. (2018). Higher precipitation strengthens the microbial interactions in semi-arid grassland soils. Glob. Ecol. Biogeogr. 27, 570–580. doi: 10.1111/geb.12718
Wang, Z.-h., Miao, Y.-f., and Li, S.-x. (2015). Effect of ammonium and nitrate nitrogen fertilizers on wheat yield in relation to accumulated nitrate at different depths of soil in drylands of China. Field Crops Res. 183, 211–224. doi: 10.1016/j.fcr.2015.07.019
Wu, J., Jiao, Z., Zhou, J., Guo, F., Ding, Z., and Qiu, Z. (2017). Analysis of bacterial communities in rhizosphere soil of continuously cropped healthy and diseased konjac. World J. Microbiol. Biotechnol. 33, 1–8. doi: 10.1007/s11274-017-2287-5
Wu, Z., Liu, Y., Han, Y., Zhou, J., Liu, J., and Wu, J. (2021). Mapping farmland soil organic carbon density in plains with combined cropping system extracted from NDVI time-series data. Sci. Total Environ. 754:142120. doi: 10.1016/j.scitotenv.2020.142120
Xu, D., Deng, X., Guo, S., and Liu, S. (2019a). Labor migration and farmland abandonment in rural China: empirical results and policy implications. J. Environ. Manage. 232, 738–750. doi: 10.1016/j.jenvman.2018.11.136
Xu, W., Jin, J., Jin, X., Xiao, Y., Ren, J., Liu, J., et al. (2019b). Analysis of changes and potential characteristics of cultivated land productivity based on MODIS EVI: a case study of Jiangsu Province, China. Remote Sens 11:2041. doi: 10.3390/rs11172041
Yang, D., Lyu, W., Hu, Z., Gao, J., Zheng, Z., Wang, W., et al. (2021). Probiotic effects of Lactobacillus fermentum ZJUIDS06 and Lactobacillus plantarum ZY08 on hypercholesteremic golden hamsters. Front. Nutr. 8:705763. doi: 10.3389/fnut.2021.705763
Yang, X., Chen, F., Lin, X., Liu, Z., Zhang, H., Zhao, J., et al. (2015). Potential benefits of climate change for crop productivity in China. Agric. For. Meteorol. 208, 76–84. doi: 10.1016/j.agrformet.2015.04.024
Yao, Q., Liu, J., Yu, Z., Li, Y., Jin, J., Liu, X., et al. (2021a). Response of acidobacterial communities to 3 years of biochar addition in a black soil of northeast China. Arch. Agron. Soil Sci. 67, 889–902. doi: 10.1080/03650340.2020.1766679
Yao, R., Yang, J., Wang, X., Xie, W., Zheng, F., Li, H., et al. (2021b). Response of soil characteristics and bacterial communities to nitrogen fertilization gradients in a coastal salt-affected agroecosystem. Land Degrad. Dev. 32, 338–353. doi: 10.1002/ldr.3705
Yuan, M. M., Guo, X., Wu, L., Zhang, Y., Xiao, N., Ning, D., et al. (2021). Climate warming enhances microbial network complexity and stability. Nat. Clim. Chang. 11, 343–U100. doi: 10.1038/s41558-021-00989-9
Yuan, P., Wang, J., Li, C., and Cao, C. (2020). Long-term rice-crayfish farming aggravates soil gleying and induced changes of soil iron morphology. Soil Use Manag. doi: 10.1111/sum.12688. [Epub ahead of print].
Zhang, X. (2019). Multiple cropping system expansion: increasing agricultural greenhouse gas emissions in the north China plain and neighboring regions. Sustainability. 11:3941. doi: 10.3390/su11143941
Zhang, Y., Li, Q., Chen, Y., Dai, Q., and Hu, J. (2019). Mudflat reclamation causes change in the composition of fungal communities under long-term rice cultivation. Can. J. Microbiol. 65, 530–537. doi: 10.1139/cjm-2019-0005
Zhao, M., Zhao, J., Yuan, J., Hale, L., Wen, T., Huang, Q., et al. (2021). Root exudates drive soil–microbe–nutrient feedbacks in response to plant growth. Plant Cell Environ. 44, 613–628. doi: 10.1111/pce.13928
Zheng, H., Huang, H., Zhang, C., and Li, J. (2016). National-scale paddy-upland rotation in Northern China promotes sustainable development of cultivated land. Agric. Water Manag. 170, 20–25. doi: 10.1016/j.agwat.2016.01.009
Zhong, C., Liu, Y., Xu, X., Yang, B., Aamer, M., Zhang, P., et al. (2021). Paddy-upland rotation with Chinese milk vetch incorporation reduced the global warming potential and greenhouse gas emissions intensity of double rice cropping system. Environ. Pollut. 276:116696. doi: 10.1016/j.envpol.2021.116696
Keywords: multiple cropping system, grain yield, soil properties, soil bacterial community, Acidobacteria, molecular ecology networks
Citation: Chen T, Hu R, Zheng Z, Yang J, Fan H, Deng X, Yao W, Wang Q, Peng S and Li J (2021) Soil Bacterial Community in the Multiple Cropping System Increased Grain Yield Within 40 Cultivation Years. Front. Plant Sci. 12:804527. doi: 10.3389/fpls.2021.804527
Received: 29 October 2021; Accepted: 22 November 2021;
Published: 20 December 2021.
Edited by:
Xiao-Xia Zhang, Institute of Agricultural Resources and Regional Planning (CAAS), ChinaReviewed by:
Yuanyuan Qu, Dalian University of Technology, ChinaCopyright © 2021 Chen, Hu, Zheng, Yang, Fan, Deng, Yao, Wang, Peng and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuguang Peng, a3lzcHNnQHNpbmEuY29t; Juan Li, YWRhbGVlNjE5QDE2My5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.