- 1Zhejiang Key Laboratory of Super Rice Research, State Key Laboratory of Rice Biology, China National Rice Research Institute, Hangzhou, China
- 2Department of Resource and Environment, Moutai Institute, Renhuai, China
Grain size is a key constituent of grain weight and appearance in rice. However, insufficient attention has been paid to the small-effect quantitative trait loci (QTLs) on the grain size. In the present study, residual heterozygous populations were developed for mapping two genetically linked small-effect QTLs for grain size. After the genotyping and the phenotyping of five successive generations, qGS7.1 was dissected into three QTLs and two were selected for further analysis. The qTGW7.2a was finally mapped into a 21.10 kb interval containing four annotated candidate genes. Transcript levels assay showed that the expression of the candidates LOC_Os07g39490 and the LOC_Os07g39500 were significantly reduced in the NIL-qTGW7.2aBG1. The cytological observation indicated that qTGW7.2a regulated the grain width through controlling the cell expansion. Using the same strategy, qTGW7.2b was fine-mapped into a 52.71 kb interval containing eight annotated candidate genes, showing a significant effect on the grain length and width with opposite allelic directions, but little on the grain weight. Our study provides new genetic resources for yield improvement and for fine-tuning of grain size in rice.
Introduction
Rice (Oryza sativa L.) is one of the most important staple crops which feeds half of the population of the world. Therefore, grain yield became a prime target for breeders. Grain yield is characterized by three components: panicle number, filled grain number per panicle, and grain weight. Grain weight is mainly determined by grain size, which simultaneously affects appearance (Zuo and Li, 2014). Thus, grain size is a primary target for yield improvement.
Grain length (GL) and grain width (GW) determine grain size, and both are complex traits controlled by the quantitative trait locus (QTL). To date, 20 grain size-related QTLs with large effects have been cloned and characterized. Several signals and regulatory pathways, controlling grain size and thousand-grain weight (TGW), have been identified in rice, such as the G-protein signaling pathway, the ubiquitin-proteasome pathway, the mitogen-activated protein kinase (MAPK) signaling pathway, the phytohormone signaling, and transcriptional regulators (Li and Li, 2016; Fan and Li, 2019). The GS3 and DEP1 encode G-protein γ-subunits and regulate the grain size and weight (Fan et al., 2006; Huang et al., 2009). The OsLG3b encodes a MADS-domain transcription factor OsMADS1, which acts as a key downstream effector of G-protein βγ dimers in controlling grain size and appearance (Yu et al., 2018). The HGW, GW2, WTG1, and OsUBP15 regulate the grain size and weight via the ubiquitin-proteasome pathway (Song et al., 2007; Li et al., 2012; Huang et al., 2017; Shi et al., 2019). The OsMKKK10, OsMKK4/SMG1, and OsMAPK6 are involved in the MAPK signaling pathway (Duan et al., 2014; Liu S. Y. et al., 2015; Guo et al., 2018). The OsRac1 and GSN1 directly interact with OsMAPK6, and also inactivate and activate the OsMAPK6 via dephosphorylation and phosphorylation (Guo et al., 2018; Zhang et al., 2019). Furthermore, ERECTA1 acts upstream of the OsMKKK10-OsMKK4-OsMPK6 cascade to control the spikelet number by regulating cytokinin metabolism in rice (Guo et al., 2020). Some proteins participate in the brassinosteroids (BR) signal pathway. Particularly, GW5 encodes a calmodulin-binding protein, GS5 encodes a putative serine carboxypeptidase, GL3.1 encodes a protein phosphatase kelch (PPKL), GS2 encodes transcription factor OsGRF4, and GSK2 kinase has multiple substrates that carry out various BR responses (Li et al., 2011; Qi et al., 2012; Hu et al., 2015; Liu et al., 2017). In addition, TGW6, BG1, GL3.3/TGW3/qTGW3, GSA1, and RBG1 are involved in the auxins signaling pathway (Ishimaru et al., 2013; Liu L. C. et al., 2015; Hu et al., 2018; Xia et al., 2018; Ying et al., 2018; Dong et al., 2020; Lo et al., 2020). The GNP1 encodes the GA20ox1, and at the same time, increases the grain number and yield by increasing cytokinin activity. The GW6 encodes a GA-regulated GAST family protein and positively regulates the grain width and weight through the gibberellins pathway (Wu et al., 2016; Shi et al., 2020). Additionally, many other major QTLs regulate the grain size and weight through the transcriptional levels, such as GW8, GL7/GW7, GW6a, GLW7, GL4, OsLG3, GS9, GL6, TGW2, and SG3 (Wang et al., 2012; Song et al., 2015; Wang S. K. et al., 2015; Wang Y. X. et al., 2015; Si et al., 2016; Wu et al., 2017; Yu et al., 2017; Xiong et al., 2018; Zhao et al., 2018; Wang A. H. et al., 2019; Li et al., 2020; Ruan et al., 2020).
Small-effect QTLs also play important roles in regulating the grain size and are widely utilized in commercial rice varieties (Kinoshita et al., 2017). Many QTLs with small-effects are responsible for quantitative genetic variation. These QTLs are often unexpected based on prior knowledge of the trait or correspond to computationally predicted genes (Mackay et al., 2009). Therefore, it is beneficial to validate these small-effect QTLs for breeding. In recent years, more than 400 small-effect QTLs for grain size and weight were reported (Huang et al., 2013). However, only a few were fine-mapped or cloned. The DTH2 encodes a CONSTANS-like protein that promotes heading by inducing the florigen genes Hd3a and RFLT1 (Wu et al., 2013). Notably, qTGW1.2b regulates grain weight which encodes a VQ-motif protein OsVQ4 (Chan et al., 2020). A naturally varying QTL, the qTGW12a, which encodes the multidrug and the toxic compound extrusion (MATE) transporter, regulates grain weight in rice (Du et al., 2021). Validation and dissection of more small-effect QTLs could provide a large number of the candidate genes and would be beneficial for establishing the network-controlling grain weight and grain size in rice.
The residual heterozygous method (Du et al., 2008) was mainly used for QTL mapping in this study. Residual heterozygote populations, the genotypic compositions are showing heterozygosity of the target region and high homozygosity in the background. The progeny population obtained by selfing is equal to the natural near-isogenic line (NIL)-F2 population, which applies to the QTL validating, resolving, and fine-mapping. To date, a series of small-effect QTLs have been fine mapped using this method (Dong et al., 2018; Wang W. H. et al., 2019; Zhu et al., 2019; Zhang et al., 2020).
In a previous study, a set of backcross recombinant inbred lines, between the indica rice variety BG1 (Big Grain 1) and the japonica rice variety XLJ (Xiaolijing), were used for QTL mapping. There were significant differences in a wide range of traits, including the heading date (HD), plant height (PH), flag leaf length (FLL), flag leaf width (FLW), GL, GW, and TGW, between the two parents. A small-effect grain size QTL, the qGS7.1, which had considerable effect on GL, GW, and TGW has been identified on chromosome 7 (Xue et al., 2019). In the present study, the qGS7.1 was dissected into three QTLs, named qTGW7.1, qTGW7.2a, and qTGW7.2b. Finally, the qTGW7.2a was located into a 21.10 kb region which controls the grain width and weight, while the qTGW7.2b was mapped to a 52.71 kb interval, which inversely affects the grain length and width, not grain weight.
Materials and Methods
Plant Materials
Five runs and a total of 23 residual heterozygous populations were used to map the target QTL in this study. The populations were derived from two BC4F6 individuals, which showed heterozygosity of the target region and high homozygosity in the background from the cross of XLJ/////XLJ////XLJ///XLJ//XLJ/BG1 (Supplementary Figure 1).
In the first run, two single plants with heterozygous regions of the qGS7.1 were selected. These plants were then developed into two BC4F7 populations consisting of 137 plants (R7) and 142 plants (R8) used for QTL validation and mapping. New polymorphic markers were designed and used to test genotypes of these populations.
In the second run, six resultant BC4F8 populations, R9 to R14, consisting of 189, 193, 198, 151, 116, and 213 plants, respectively, were developed from six residual heterozygous BC4F7 single plants with updated target regions. Then, the BC4F9 population containing 3,989 individuals derived from the R9 population was constructed and was used for selecting the recombinants.
In the third run for the QTL validation and mapping, eleven single plants were selected from the BC4F9 generation to develop eleven BC4F10 populations, to be named R15 to R25, for a total of 794 plants.
In the fourth run, three homozygous NIL populations in the segregating region, namely, N1 to N3, were developed to validate the QTL. Two single plants without the qTGW7.2b target region were selected and selfed to develop two populations, named R26 and R27, which were made up of 209 and 223 plants, respectively. Meanwhile, a BC4F11 population, including 6,128 individuals derived from the R23 population, was constructed and used for further mapping.
In the fifth run, two single plants were selected from BC4F11 plants in the XP7-12-XP7-23 interval to develop progeny populations consisting of 233 (R28) and 98 (R29) plants for validation and fine-mapping of the qTGW7.2a. Later, two non-recombinants were selected from the R28 population and selfed to develop two NILs of the NIL-qTGW7.2aBG1 and the NIL-qTGW7.2aXLJ for traits measurement.
Field Experiments and Traits Measurement
Plants were grown at the field stations of the China National Rice Research Institute in Lingshui, Hainan province, and Fuyang, Zhejiang province. After harvesting, 300 dry seeds were randomly selected for measuring TGW (g), GL (mm), GW (mm), and the ratio of the grain length to width (RLW) using an automatic seed counting and analyzing instrument (Model SC-G, Wanshen Ltd., Hangzhou, China).
DNA Extraction and Molecular Markers Development
Total DNA was extracted from fresh leaf samples by the cetyltrimethylammonium bromide (CTAB) method (Murray and Thompson, 1980). The PCR products were visualized on 8% non-denaturing polyacrylamide gels by silver staining. A total of 31 polymorphic DNA markers were used (Supplementary Table 1).
RNA Extraction and Quantitative Reverse-Transcriptase Polymerase Chain Reaction
Total RNA was extracted from rice panicles using RNAprep pure Plant Kit (TIANGEN, Beijing, China). Quantitative reverse-transcriptase polymerase chain reaction (qRT-PCR) was performed using SYBR Premix Ex Taq II (TAKARA, Dalian, China). Data analysis used the 2–ΔΔCt method, while the UBQ10 was used as the internal reference to normalize the gene expression (Livak and Schmittgen, 2001). The qRT-PCR primers used in this study are listed in Supplementary Table 1.
Cytological Observation
During the heading stage, young spikelet hulls of NIL-qTGW7.2aBG1 and NIL-qTGW7.2aXLJ were fixed in 2.5% glutaraldehyde for 12 h at 4°C, and then dehydrated in serial graded ethanol (30, 50, 70, 80, 90, 95, and 100%) before finally preserved in 100% ethanol. The samples were dried in a Hitachi HCP-2 critical point drier, and the cell length and width of the inner glumes were observed by scanning electron microscopy (Hitachi SU-8010, Hitachi, Japan). ImageJ software was used to measure cell numbers and cell size.
Data Analysis
Three genotypes were obtained after genotyping this population. Two homozygous genotype plants which carried alleles from XLJ and BG1 were used to detect the phenotypic differences by Student’s t-test. We deduce there was a QTL when p < 0.05. Subsequently, the heterozygous individual harboring the target QTL was used for developing a new residual heterozygous population.
All the analysis data, including the additive effect (A) and the proportion of phenotypic variance explained by the QTL (R2), were obtained from the Windows QTL Cartographer Version 2.5 software to estimate the genetic effects.
Results
Validation and Mapping of qGS7.1
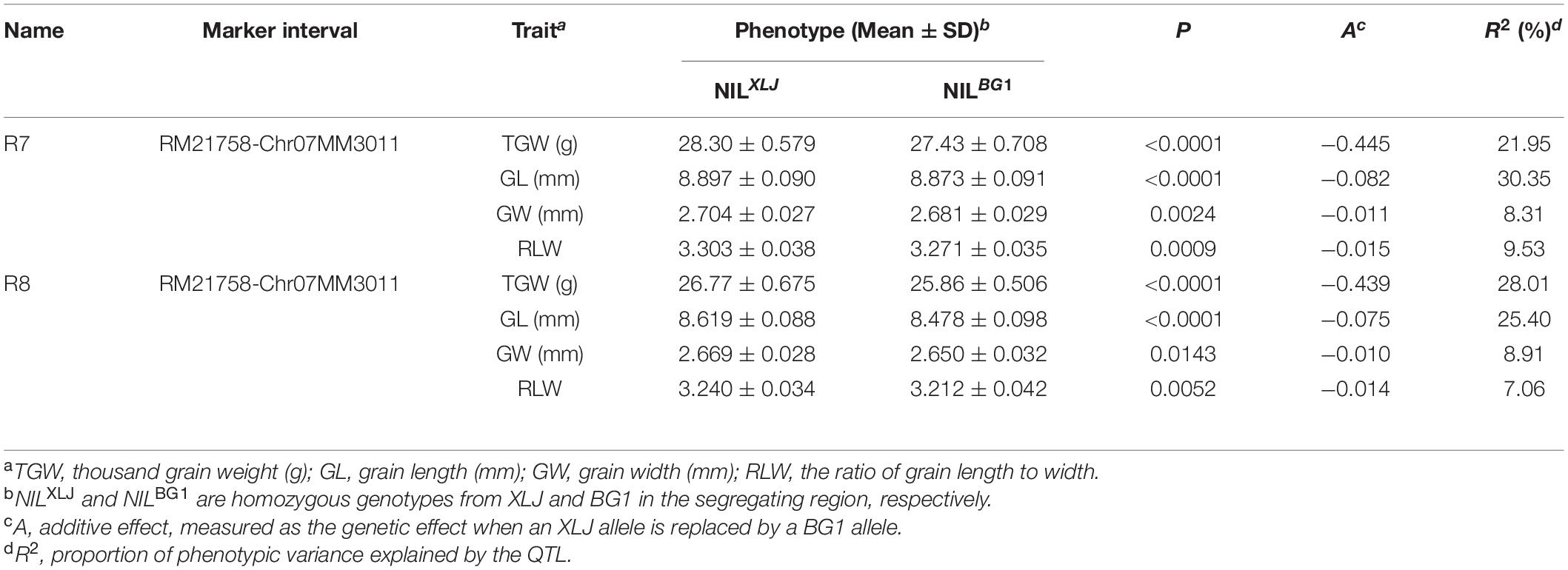
We have identified a grain size QTL, the qGS7.1, in the X7-9-RM351 interval on chromosome 7 (Figure 1A). For further validation and fine-mapping of the target region, 12 polymorphic markers were designed based on the sequence differences between BG1 and XLJ. Consequently, the RM21758 became the new boundary when all plants were homozygous for it. Meanwhile, the R7 and R8 populations were derived from two segregated single plants selected from the R1 population (Figure 1B) to validate the qGS7.1, and to exclude the non-target interval in BC4F7 populations. Both showed a significant enhancement of GW, GL, and TGW between the two parental genotypic groups. These results showed that the qGS7.1 with the XLJ allele have increased TGW, GL, and GW by 0.439–0.445 g, 0.075–0.082 mm, and.010–0.011 mm, and had R2 values of 21.95–28.01, 25.40–30.35, and 8.31–8.91%, respectively (Table 1). The effects detected in the two populations were comparable and indicated that the qGS7.1 was located in the region between RM21758 and Chr07MM3011.
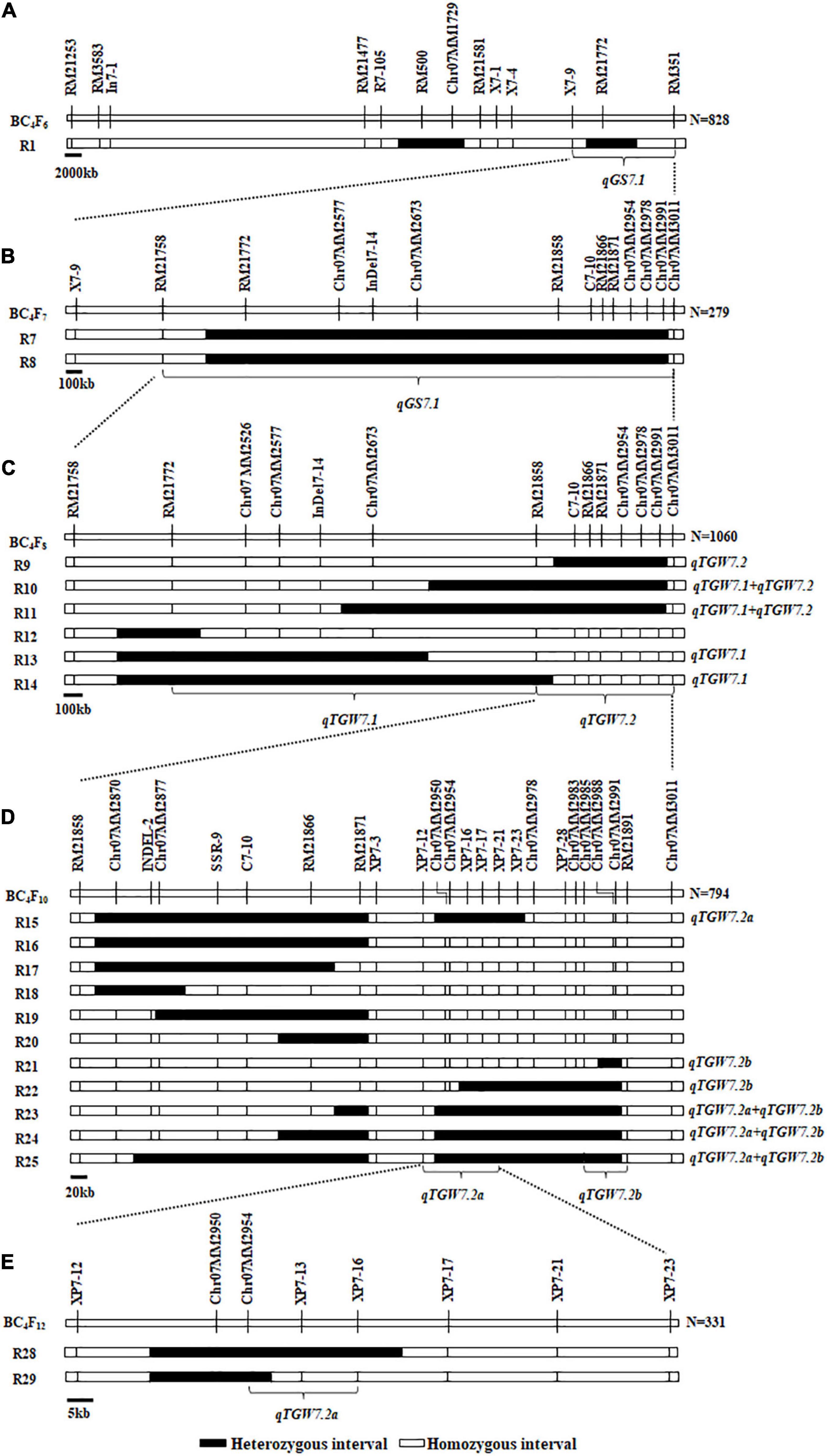
Figure 1. Genotypic compositions of residual heterozygous populations in the target regions. (A) Composition of R1 population in previous study. (B) Two residual heterozygous populations in BC4F7. (C) Six residual heterozygous populations in BC4F8. (D) Eleven residual heterozygous populations in BC4F10. (E) Two residual heterozygous populations in BC4F12.
Dissection of qGS7.1 Into Three Quantitative Trait Loci
Six progeny populations with sequential segregating regions jointly covering the qGS7.1 region (Figure 1C) were developed in BC4F8 populations. There were no significant differences in the R12 population (Table 2). In the remaining five populations, significant enhancements were discovered in XLJ alleles for TGW. The additive effects were −0.477, −0.670, −0.531, −0.265, and −0.180 g in R9, R10, R11, R13, and R14, respectively (Table 2). These results indicated that there were two hypotheses for the consistent allelic direction. Despite this, there were also significantly different effects among the five populations. Firstly, there were two QTLs for the TGW located in these populations. One was mapped in R9 but not in R13 and R14, while the other was segregated in R13 and R14 but not in R9. Both were located in R10 and R11, thus, the additive effects were higher in the two populations than those in the other populations. Secondly, only one QTL was segregated in these populations with highly unstable effects.
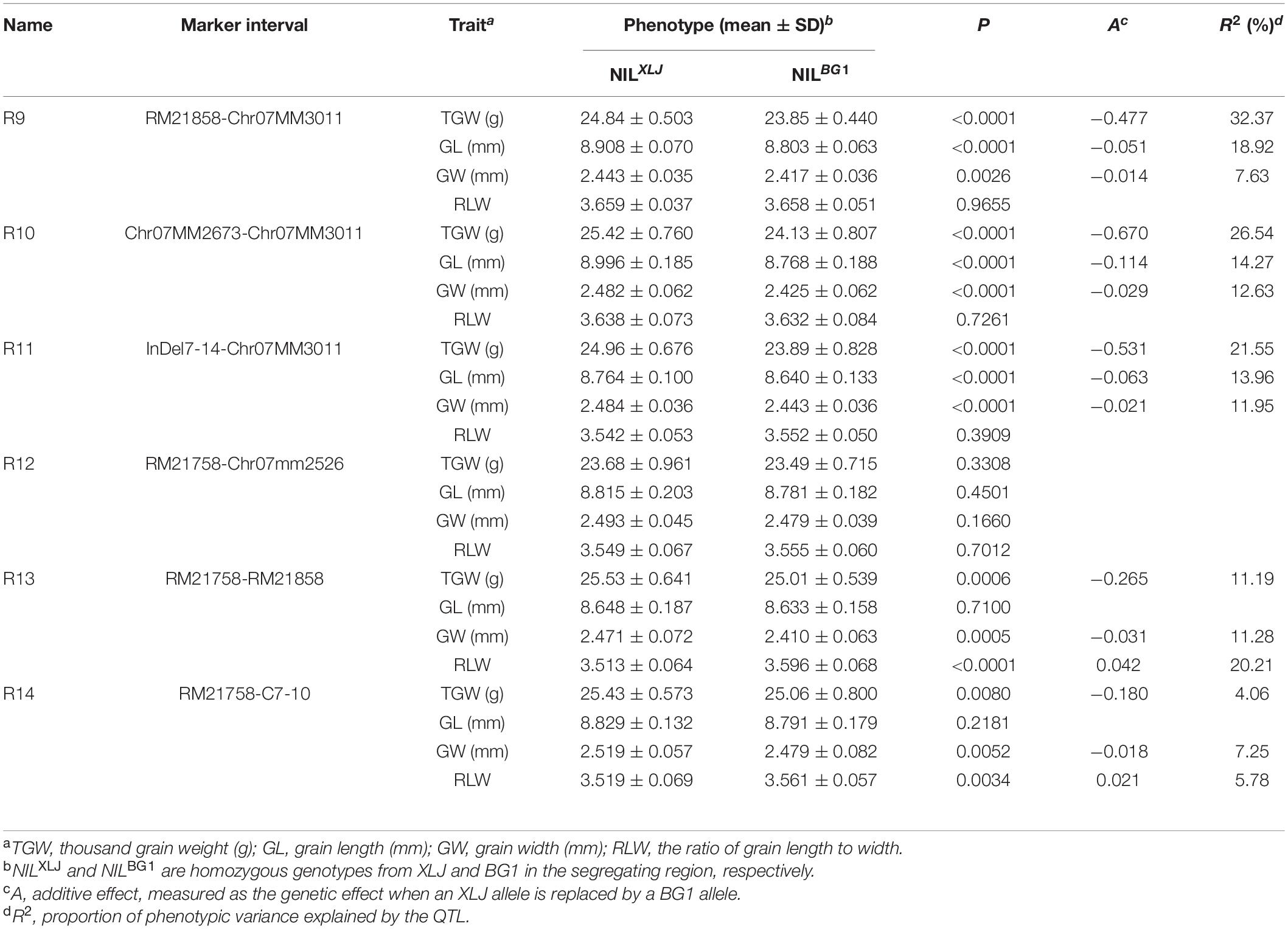
Table 2. Dissection of qGS7.1 into two quantitative trait loci (QTLs) using six residual heterozygous populations in BC4F8.
For GL, significant effects were detected in three populations (Table 2) with the enhancing alleles always derived from XLJ. However, there were no significant differences between the two parental genotypic groups in R13 and R14 populations. To sum up, it could be concluded that the qGS7.1 was a composite of two independent QTLs (Figure 1C). The first QTL, named qTGW7.1, was located in the common segregating region of R13 and R14, with the allele from XLJ increasing the TGW and GW. The second QTL, named qTGW7.2, was located in the heterozygous interval in R9, with the allele from XLJ increasing the TGW, GL, and GW.
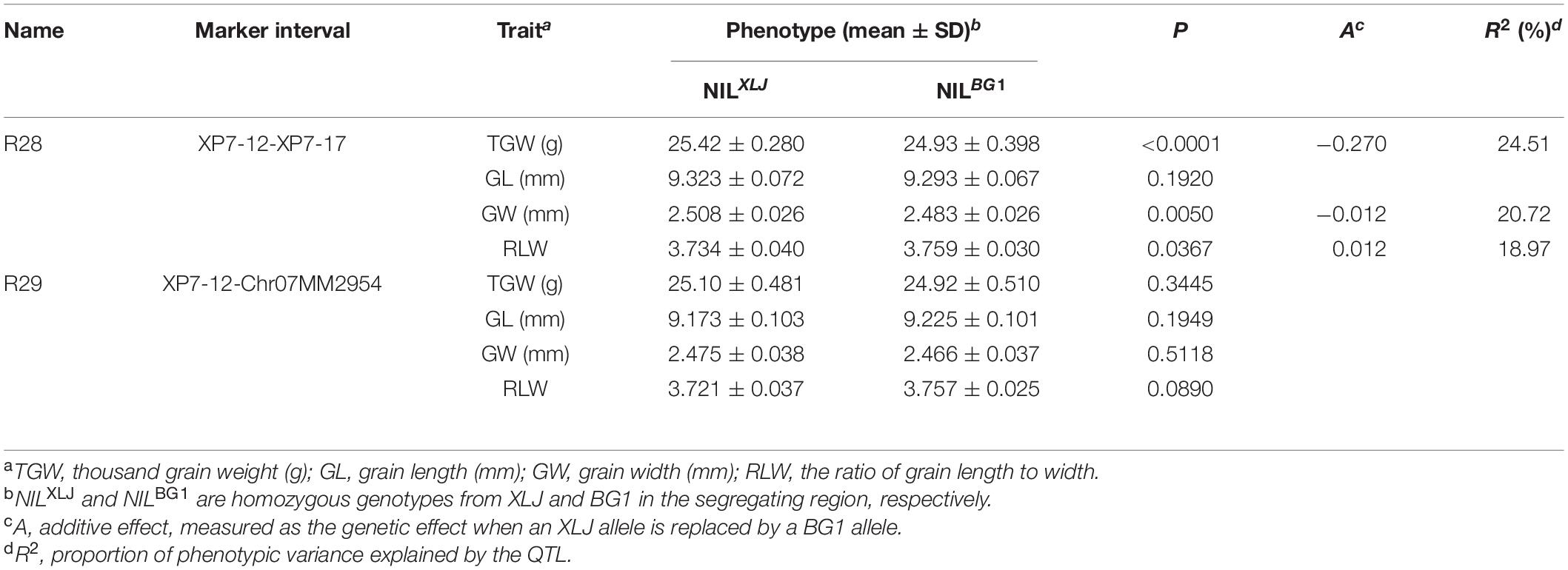
Finally, the qTGW7.2 was selected for further analysis. Eleven populations (R15-R25) were developed from 11 heterozygous individuals in BC4F9 populations (Figure 1D). Significant differences were found in the TGW, GL, and GW with the positive allele from XLJ in the R23, R24, and R25 populations. The genotypic effects were similar to the qTGW7.2 (Table 3). In the R15 population, significant genotypic effects were detected in TGW and GW, but not in GL. In R21 and R22 populations, similar additive effects for GL were −0.050 and −0.053 mm, respectively explaining the 8.32 and 20.81% of the genotypic variance in both populations. There was no significant difference in R16, R17, R18, R19, and R20 between the two parental genotypic groups (Table 3).
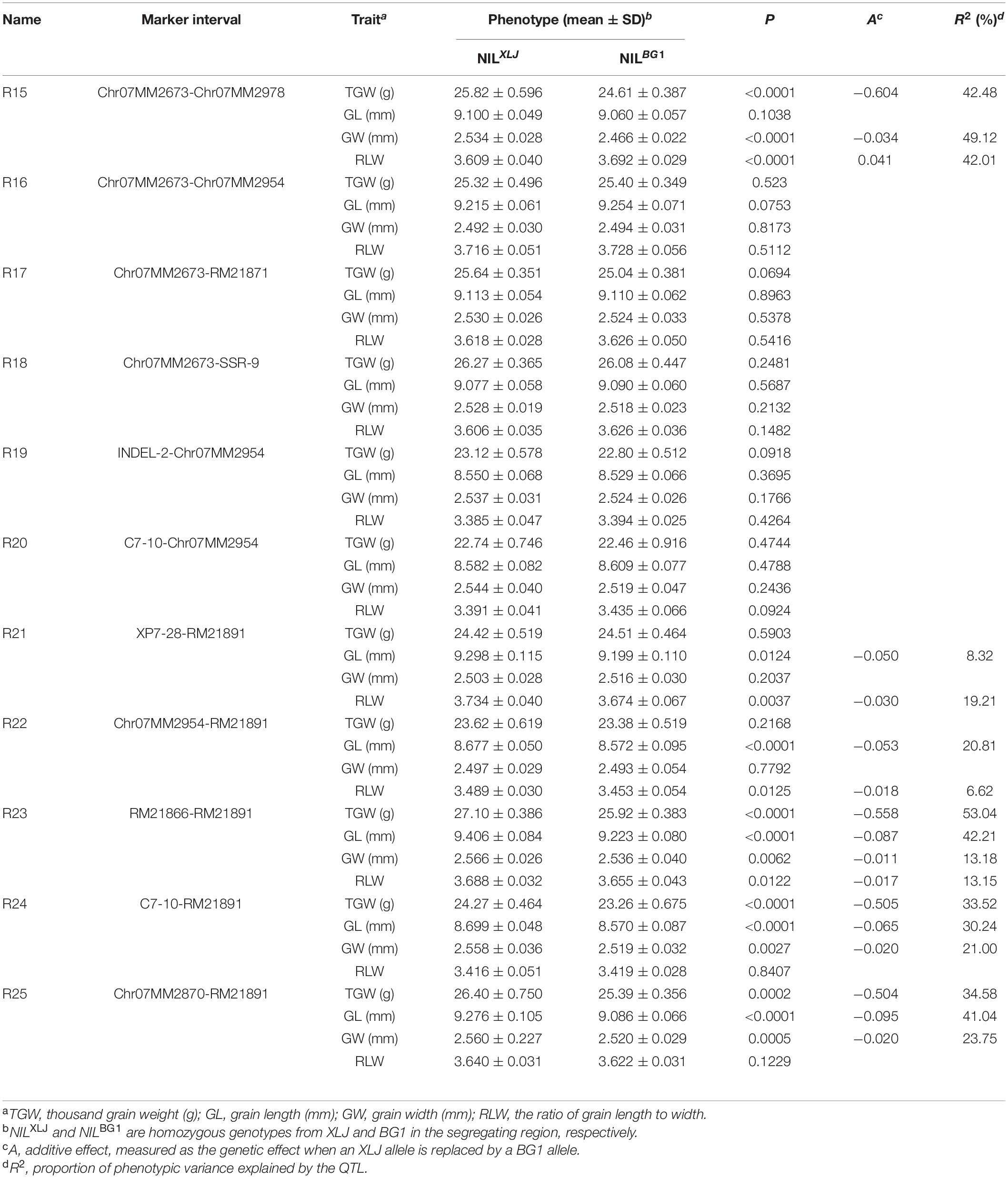
Table 3. Dissection of qTGW7.2 into two QTLs using eleven residual heterozygous populations in BC4F10.
Combined with the separated graphical genotypes of R15 and R21 populations, the qTGW7.2 was dissected into two separate QTLs (Figure 1D). The first QTL, the qTGW7.2a, had considerable effects on TGW and GW within a 53.96 kb region spanning XP7-12 to XP7-16. The second QTL, the qTGW7.2b, was located between Chr07MM2985 and RM21891, a 52.71 kb interval, and had affected the GL and RLW but had little effect on the TGW.
Three NIL populations (N1-N3) derived from the R15, R16, and R21 populations were developed to validate the function of the qTGW7.2a and the qTGW7.2b (Supplementary Table 2). Meanwhile, two progeny populations, R26 and R27, derived from two recombinants only containing a qTGW7.2a, were used in this study (Supplementary Figure 2). In the N1 population, significant genotypic effects were detected for TGW and GW, which was coincident with R26 and R27 populations. Highly significant genotypic effects were detected for GL, GW, and RLW in the N3 population. There was no significant difference in the N2 population (Supplementary Table 2).
As described above, the qGS7.1 was dissected into three independent QTLs. Primarily, the qTGW7.1 was located in a region flanked by RM21772 and RM21858. The XLJ allele increased the TGW by.018g and.265g, and also the GW by.018mm and.031mm (Figure 1C and Table 2). The qTGW7.2a mainly controls TGW through regulating GW with enhancing alleles derived from XLJ. Simultaneously, the qTGW7.2b has affected the GL and GW in opposite ways with no significant effect on the TGW (Supplementary Figure 3). For further analysis, the qTGW7.2a was selected for its stable function and considerable effect.
Fine-Mapping qTGW7.2a Into a 21.10-kb Region
For further mapping of qTGW7.2a, we constructed a BC4F11 population consisting of 6,128 individuals. Two recombinants in the RM21871-XP7-23 interval were utilized to develop two progeny populations, R28 and R29. Highly significant phenotypic effects were detected in TGW and GW in the R28 population. The additive effects were 0.213 g and 0.013 mm, having R2 of 11.29 and 9.70%, respectively. There were no significant differences in the R29 population (Table 4). According to the mapping results of the BC4F12 population, we mapped the qTGW7.2a to the 21.10 kb interval between Chr07MM2954 and XP7-16 (Figure 1E).
The qTGW7.2a increased the TGW and GW with the enhancing allele from XLJ as compared to grain size and weight between NIL-qTGW7.2aXLJ and NIL-qTGW7.2aBG1 (Figures 2A–E), which were derived from the R28 population. Additionally, compared with NIL-qTGW7.2aBG1, besides the flag leaf width, the NIL-qTGW7.2aXLJ showed comparable effects in the heading date, plant height, panicle number and length, grain number per panicle, and other agronomic traits (Figures 2F–P). Grain size is restricted by the size of the spikelet hull in rice, which is determined by both cell proliferation and expansion. Therefore, we compared the cell number and the cell size of the outer glume epidermal cells between NIL-qTGW7.2aXLJ and NIL-qTGW7.2aBG1 (Figure 3A). There was no significant difference in cell number or cell length between NIL-qTGW7.2aXLJ and NIL-qTGW7.2aBG1 (Figures 3B,C). However, the cell width of NIL-qTGW7.2aXLJ was significantly greater than NIL-qTGW7.2aBG1 (Figure 3D). These findings suggest that the grain size increase in NIL-qTGW7.2aXLJ is predominantly due to the cell width expansion.
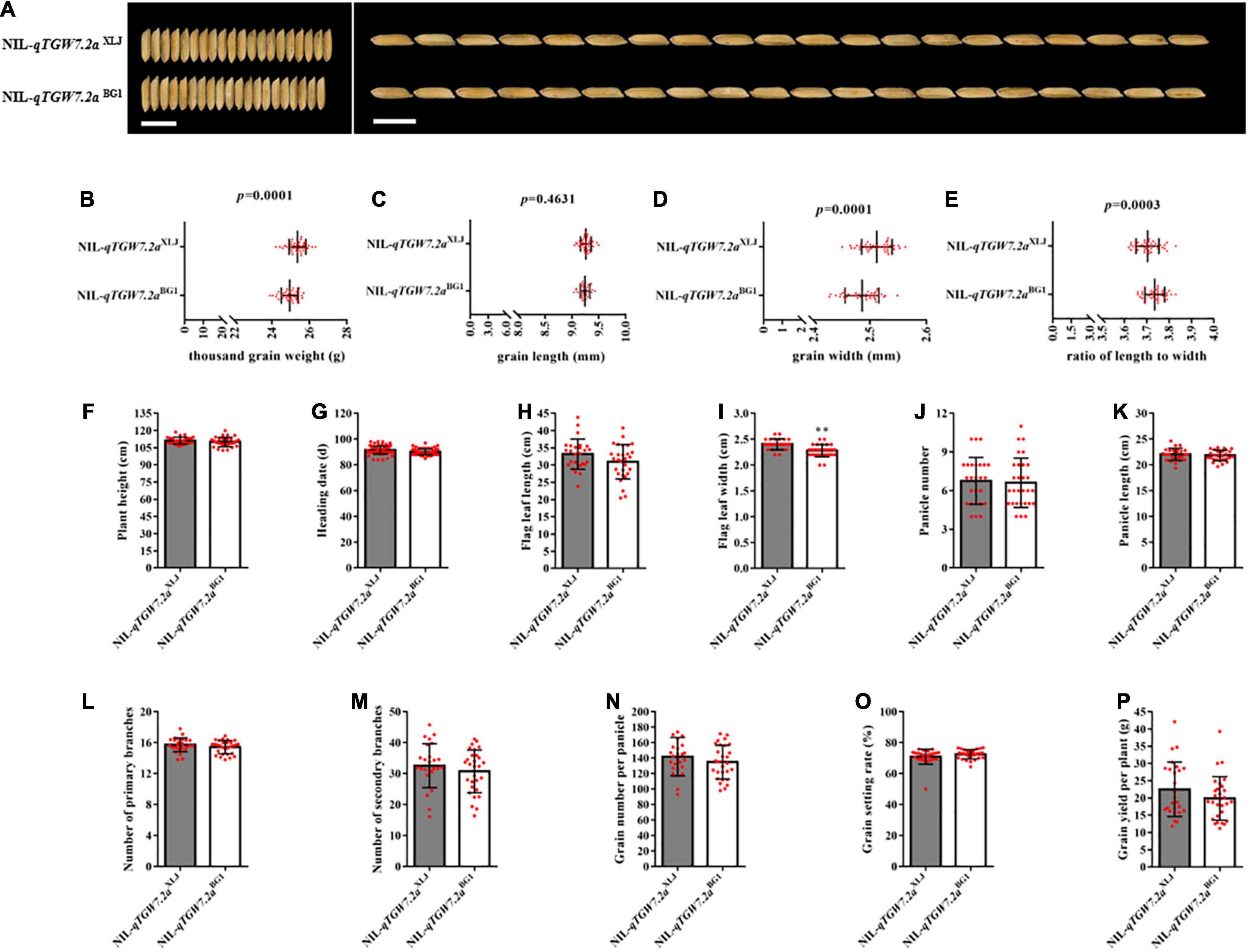
Figure 2. qTGW7.2a regulates grain width and weight. (A) Grain phenotypes of rice NIL plants. Bar = 1 cm. (B) Comparison of thousand grain weight. (C) Comparison of grain length. (D) Comparison of grain width. (E) Comparison of the ratio of grain length to width. (F) Comparison of plant height. (G) Comparison of heading date. (H) Comparison of flag leaf length. (I) Comparison of flag leaf width. (J) Comparison of panicle number. (K) Comparison of panicle length. (L) Comparison of number of primary branches. (M) Comparison of secondary branches. (N) Comparison of grain number per panicle. (O) Comparison of grain setting rate. (P) Comparison of grain yield per plant. Data are given as mean ± SD. Student’s t-test was used to generate P value; **P < 0.01; red dot, the number of plants for traits measurement.
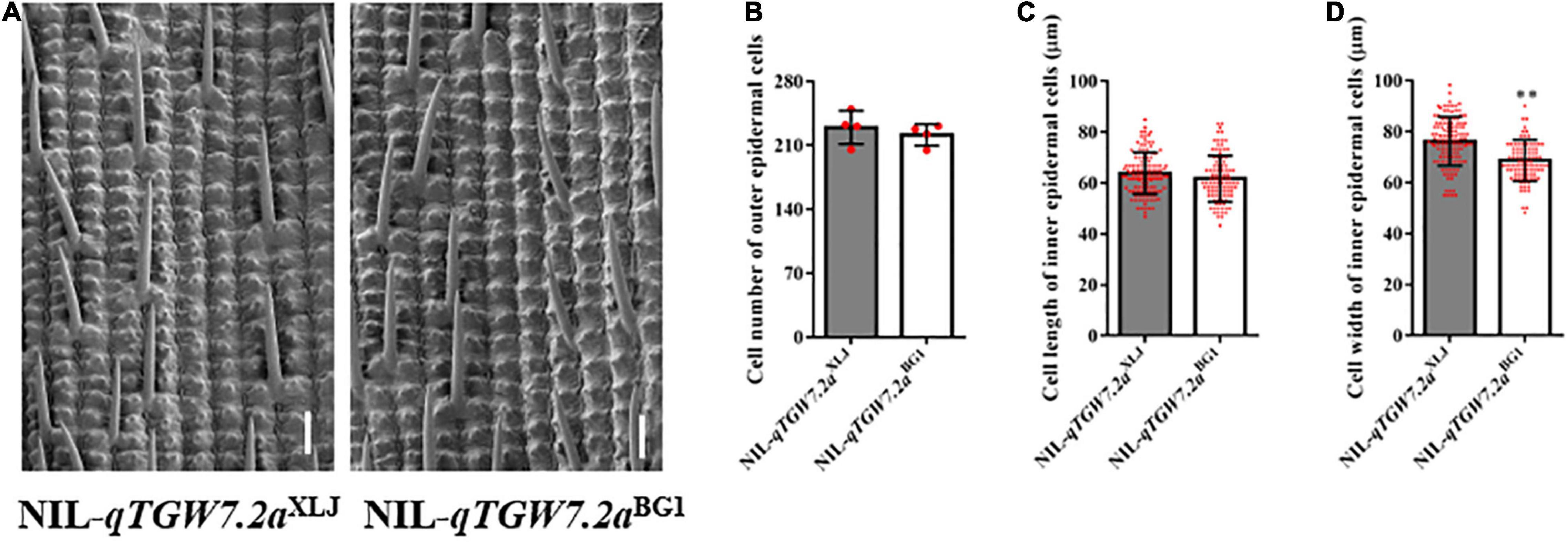
Figure 3. Scanning electron microscopic observation and analysis of the glume. (A) Scanning electron micrograph of the outer glume epidermal cells between NIL-qTGW7.2aXLJ and NIL-qTGW7.2aBG1. Bar = 100 μm. (B) Cell number of outer epidermal cells. (C) Cell length of outer epidermal cells. (D) Cell width of outer epidermal cells. Data are given as mean ± SD. Student’s t-test was used to generate P value; **P < 0.01. Red dot, the number of grains used to calculate cell number in (B), and cell numbers to measure cell length (C) and cell width (D).
Candidate Genes of qTGW7.2a and qTGW7.2b
There are four open reading frames (ORFs) located in the region spanning qTGW7.2a. The LOC_Os07g39470 encodes a rice GRAS family protein, CIGR2, which suppresses cell death in rice inoculated with a rice blast via activation of a Heat Shock Transcription Factor, OsHsf23 (Tanabe et al., 2016). The LOC_Os07g39480 encodes WRKY78, a transcriptional factor that is involved in regulating plant height and seed size (Zhang et al., 2011). Lastly, the LOC_Os07g39490 and LOC_Os07g39500 are unknown functional proteins (Supplementary Table 3).
Sequences of the genome sequence in four genes between the NIL-qTGW7.2aXLJ and NIL-qTGW7.2aBG1 were compared. Mutations were found at 61, 47, 43, and 77 sites of the four genes, respectively (Supplementary Tables 4–7). Two synonymous SNPs were detected in LOC_Os07g39470, indicating that there were no differences between the two alleles. For LOC_Os07g39480, there were four polymorphic sites, three of which were synonymous and one 3-bp deletion in the XLJ allele, resulting in a serine deletion. For LOC_Os07g39490, three SNPs include one synonymous and two non-synonymous were detected between two NILs, resulting in two amino acids substituted; especially, a 2-bp deletion in NIL-qTGW7.2aBG1 led to the terminal 62 residues were truncated. Finally, there were 16 SNP variations in LOC_Os07g39500 between two NILs, including twelve non-synonymous mutations and a premature stop codon at T784C in the BG1 allele. However, there were three fragment deletions/insertions in the promoter and one at the 5′-UTR region of LOC_Os07g39470. One fragment deletions/insertion of LOC_Os07g39480 was at its promoter (Supplementary Tables 6, 7).
Subsequently, the expression levels of four candidates in the panicles of NIL-qTGW7.2aXLJ and NIL-qTGW7.2aBG1 were analyzed (Figure 4). The expression levels of the LOC_Os07g39490 and the LOC_Os07g39500 were significantly higher in NIL-qTGW7.2aXLJ than in NIL-qTGW7.2aBG1, while there were no significant differences between the two NILs in the LOC_Os07g39470 and the LOC_Os07g39480.
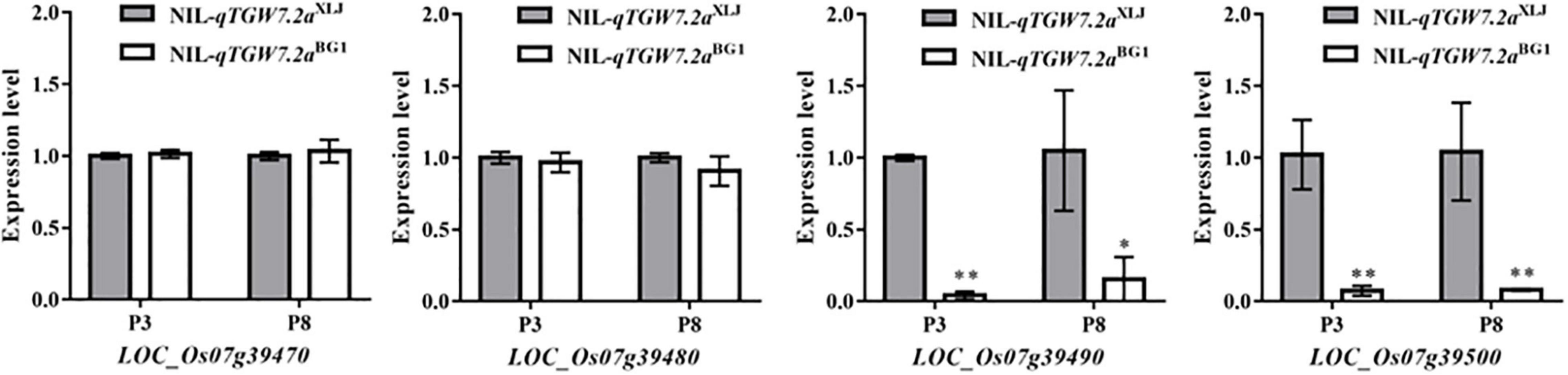
Figure 4. The transcript levels of annotated genes between NIL-qTGW7.2aXLJ and NIL-qTGW7.2aBG1. The experiment was performed using panicles of 1 ≤ P < 3 cm (P3) and 5 ≤ P < 8 cm (P8) collected from NIL-qTGW7.2aXLJ and NIL-qTGW7.2aBG1. Data are given as mean ± SD. Student’s t-test was used to generate P value; *P < 0.05; **P < 0.01.
According to the Gramene Database1, there are eight candidate genes in the qTGW7.2b region (Supplementary Table 8). One of them encodes proteins with known functional domains. The LOC_Os07g39750 encodes acetylcholinesterase relating to the positive regulation of gravitropic response in rice seedlings (Yamamoto et al., 2015). Four of them encode putative functional proteins, including LOC_Os07g39740, which encodes a GDSL-like lipase/acyl hydrolase, LOC_Os07g39780 for a SUMO-activating enzyme subunit 2, LOC_Os07g39800 for a transcription repressor HOTR, and LOC_Os07g39810, which encodes a protein that belongs to the lipase class 3 family. The remaining annotated genes LOC_Os07g39760, LOC_Os07g39770, and LOC_Os07g39790 are for unknown functional proteins.
Sequence comparisons of the eight annotated genes between NIL-qTGW7.2bXLJ and NIL-qTGW7.2bBG1 were performed. Thirty-eight variants were found of the LOC_Os07g39810 (Supplementary Table 9). No variants were detected in the sequences of LOC_Os07g39760 and LOC_Os07g39770 and in the 2-6 sites of the other five genes (Supplementary Table 10), but few of them occurred in the coding sequence (CDS). For LOC_Os07g39790, three mutations were identified, including the C2088G and the T2233C synonymous substitution, as well as a three-nucleotide insertion in NIL-qTGW7.2bXLJ. For the other four genes, most SNPs were detected in the promoter and terminator regions.
Discussion
Grain size is jointly regulated by large-effect and small-effect QTLs. Remarkable progress has been achieved by the discovery of large-effect QTLs affecting the yield and quality in recent years. Notably, rarely have small-effect QTLs been cloned in rice (Chan et al., 2020; Du et al., 2021). It is recognized that the complex traits, especially as grain yield and its component traits, were controlled by many genes. However, in recent years, the number of the cloned large-effect QTLs is decreasing. Small-effect QTL does not have the same effect as the large-effect QTL, but compared with the large-effect QTL, the small-effect QTL usually affects a single trait rather than the multiple agronomic traits or, even, the adverse changes. So, the small-effect QTLs could be selected and be used to fine-tune the function of a large-effect QTL, thereby making the varieties fitter to the market demand. Aggregation of small-effect QTLs with the same genetic direction may even show greater phenotypic changes. Moreover, molecular characterization of these small-effect QTLs, which have a direct and consistent influence on grain yield, would be beneficial to establish a regulatory network for rice grain development. These indicate that if we pay more attention to the small-effect QTLs, it is possible to detect some dominant alleles that have not been selected, and they will have more potential applications in breeding.
In the present study, the qGS7.1 has increased the TGW, GL, and GW by 3.17, 0.27, and 0.86%, respectively, with the enhancing allele derived from XLJ (Table 1). Then, the qGS7.1 is dissected into three closely linked QTLs, qTGW7.1, qTGW7.2a, and qTGW7.2b. The latter two small-effect QTLs, regulating grain size and grain shape, were identified and fine-mapped. Grain size and grain shape are both determined by grain length and width. While grain size is the main factor affecting the grain weight, grain shape mainly influences grain preference and may not be related to grain weight (Dong et al., 2018). qTGW7.2a was limited between Chr07MM2954 and XP7-16 with a 21.10 kb interval, affecting grain width and weight. qTGW7.2b, which inversely affects gra in length and width, was mapped into the 52.71 kb region between Chr07MM2985 and RM21891.
Small-effect QTLs were susceptible to environmental and genetic background. However, the effects of a genetic context could be counteracted under a highly homozygous genetic context. All the populations used in this study were derived from a single plant of high generation BC4F6 populations with the same background and were cultivated in Fuyang and Lingshui following the chronological order. The qTGW7.2a could be detected in both environments, but the effects on TGW and GW were not stable. The additive effects on TGW and GW increased by XLJ allele were in the range of 0.213 to 0.604 g and 0.013 to 0.034 mm in the two environments, respectively (Tables 3, 4 and Supplementary Table 2). Especially for qTGW7.2b, in R21 and R22 populations, the qTGW7.2b regulates the grain length and has little influence on the grain width and grain weight. Despite this, in the N3 population, the qTGW7.2b was detected affecting the grain length and grain width with opposite allelic directions and had little effect on grain weight (Table 3 and Supplementary Table 2). These results suggested that small-effect QTLs could be steadily detected using the residual heterozygous method, but the effects of QTL could be affected by environmental interaction.
Among QTLs with large effects, GW2, GS5, GW5/GSE5, and GW6 are those which regulate grain weight through controlling grain width (Song et al., 2007; Li et al., 2011; Xu et al., 2015; Duan et al., 2017; Liu et al., 2017; Shi et al., 2020). In our study, the qTGW7.2a has increased the grain width and weight but did not influence the other agronomic traits. These suggest that the qTGW7.2a could be used for yield improvement.
For large-effect QTLs, the following have similar effects with qTGW7.2b on grain length and width that regulate grain shape: GL7/GW7, GW8, and GS9 (Wang et al., 2012; Wang S. K. et al., 2015; Wang Y. X. et al., 2015; Zhao et al., 2018). In the XLJ allele, the qTGW7.2b increased the grain length but decreased the grain width. This result has little effect on grain weight and, therefore, indicated that the qTGW7.2b could be used to fine-tune the grain size.
Four annotated genes were found in the 21.1 kb interval covering the qTGW7.2a. Firstly, the LOC_Os07g39470 encodes CIGR2 belonging to the rice GRAS family, and members of this family encode transcriptional regulators with functions in a wide range of signaling mechanisms, such as growth and development, hormone signaling, and plant defense (Tanabe et al., 2016). However, there were only two synonymous SNPs between the CIGR2 alleles. Secondly, the LOC_Os07g39480 encodes a transcriptional factor, the WRKY78, which was involved in regulating the plant height and seed size. Knocking down of the WRKY78 led to a semi-dwarf and small seed phenotype by reducing the cell length (Zhang et al., 2011). However, in addition to the three SNPs showing synonymous mutation, there was just one serine deletion in the CDS of NIL-qTGW7.2aXLJ. The expression level of the WRKY78 was comparable between the two NILs. Combined with the fragment deletions/insertion of the LOC_Os07g39470 and the LOC_Os07g39480, these results may be due to how the deletions/insertion at the promoter region were not the cis-acting element and does not affect their mRNA expression level. Previous studies showed that the coding region introduced a premature stop codon resulting in premature termination of translation could influence grain size, such as GW2, GS3, qLGY3/GW3p6, WTG1, OsMAPK6, TGW6, and GL6 (Fan et al., 2006; Song et al., 2007; Ishimaru et al., 2013; Liu S. Y. et al., 2015; Huang et al., 2017; Liu et al., 2018; Wang A. H. et al., 2019; Wang C. S. et al., 2019). In our study, the LOC_Os07g39490 and the LOC_Os07g39500 encode hypothetical proteins. In its coding region, a non-synonymous mutation existed as a premature stop codon and prevents the transcription of a mature protein in NIL-qTGW7.2aBG1. Therefore, more studies in the gene editing, such as CRISPR/Cas9-targeted mutagenesis and gene overexpression, need to be done to confirm the gene for the qTGW7.2a.
The 52.71 kb region surrounding the qTGW7.2b contained eight annotated genes. For two annotated genes, LOC_Os07g39760 and LOC_Os07g39770, no variants between NIL-qTGW7.2bXLJ and NIL-qTGW7.2bBG1 were detected. For four other annotated genes, LOC_Os07g39740, LOC_Os07g39750, LOC_Os07g39780, and LOC_Os07g39800, only a few SNPs were detected at the beginning of promoter regions and the terminator regions, and no variants were detected in the coding domain sequence. For the LOC_Os07g39780, there were two synonymous mutations and a Ser insertion detected in the NIL-qTGW7.2bXLJ. For the remaining annotated gene, the LOC_Os07g39810, which encodes an expression protein, three long fragment insertions in the promoter region, and an insertion of nine nucleotides or three amino acids were detected in the NIL-qTGW7.2bXLJ. Work is underway to test whether LOC_Os07g39780 or LOC_Os07g39810 is the gene underlying the QTL qTGW7.2b for grain shape.
Conclusion
Two small-effect QTLs for grain size and grain shape, qTGW7.2a and qTGW7.2b, were fine-mapped in this study. qTGW7.2a was limited to a 21.10 kb region containing four genes. This QTL regulates grain width and weight, which has potential for yield improvement. The qTGW7.2b, which inversely regulates grain length and width, was within a 52.71 kb interval. This QTL has potential for fine-tuning the grain shape and the grain appearance. These results provide a basis for QTL cloning and offer new resources for the yield and quality improvement.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
PX performed most of the experiments, analyzed the data, and wrote the manuscript. Y-YC contributed to sequencing and constructed the populations. X-XW, B-FW, and Q-QY analyzed the data collection the phenotypes and revised the manuscript. KG and Y-WK conducted the field trials. L-PS, PY, L-YC, Y-XZ, X-DZ, and S-HC designed the experiments, supervised and completed the writing, and reviewed the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by Projects of International Cooperation NSFC (31961143016), Science and Technology Innovation Project of Chinese Academy of Agricultural Sciences (CAAS-ASTIP-2013-CNRRI), and Fundamental Research Funds for Central Public Welfare Research Institutes of Chinese Rice Research Institute (CPSIBRF-CNRRI-202102).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.804444/full#supplementary-material
Footnotes
References
Chan, A. N., Wang, L. L., Zhu, Y. J., Fan, Y. Y., Zhuang, J. Y., and Zhang, Z. H. (2020). Identification through fine mapping and verification using CRISPR/Cas9-targeted mutagenesis for a minor QTL controlling grain weight in rice. Theoret. Appl. Genet. 1, 327–337. doi: 10.1007/s00122-020-03699-6
Dong, N. Q., Sun, Y., Guo, T., Shi, C. L., Zhang, Y. M., Kan, Y., et al. (2020). UDP-glucosyltransferase regulates grain size and abiotic stress tolerance associated with metabolic flux redirection in rice. Nat. Commun. 11:2629. doi: 10.1038/s41467-020-16403-5
Dong, Q., Zhang, Z. H., Wang, L. L., Zhu, Y. J., Fan, Y. Y., Mou, T. M., et al. (2018). Dissection and fine-mapping of two QTL for grain size linked in a 460-kb region on chromosome 1 of rice. Rice 11:44. doi: 10.1186/s12284-018-0236-z
Du, J. H., Fan, Y. Y., Wang, L., and Zhuang, J. Y. (2008). Dissection of QTLs for yield traits by using near isogenic lines derived from residual heterozygous lines in rice. Chin. J. Rice Sci. 1, 1–7.
Du, Z. X., Huang, Z., Li, J. B., Bao, J. Z., Tu, H., Zeng, C. H., et al. (2021). QTGW12a, a naturally varying QTL, regulates grain weight in rice. Theoret. Appl. Genet. 134, 2767–2776. doi: 10.1007/s00122-021-03857-4
Duan, P. G., Rao, Y. C., Zeng, D. L., Yang, Y. L., Xu, R., Zhang, B. L., et al. (2014). SMALL GRAIN 1, which encodes a mitogen-activated protein kinase kinase 4, influences grain size in rice. Plant J. 77, 547–557. doi: 10.1111/tpj.12405
Duan, P. G., Xu, J. S., Zeng, D. L., Zhang, B. L., Geng, M. F., Zhang, G. Z., et al. (2017). Natural variation in the promoter of GSE5 contributes to grain size diversity in rice. Mol. Plant 10, 685–694. doi: 10.1016/j.molp.2017.03.009
Fan, C. C., Xing, Y. Z., Mao, H. L., Lu, T. T., Han, B., Xu, C. G., et al. (2006). GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theoret. Appl. Genet. 112, 1164–1171. doi: 10.1007/s00122-006-0218-1
Fan, Y. W., and Li, Y. B. (2019). Molecular, cellular and Yin-Yang regulation of grain size and number in rice. Mol. Breed. 39, 1–25. doi: 10.1007/s11032-019-1078-0
Guo, T., Chen, K., Dong, N. Q., Shi, C. L., Ye, W. W., Gao, J. P., et al. (2018). GRAIN SIZE and NUMBER1 negatively regulates the OsMKKK10-OsMKK4-OsMPK6 cascade to coordinate the trade-off between grain number per panicle and grain size in rice. Plant Cell 30, 871–888. doi: 10.1105/tpc.17.00959
Guo, T., Lu, Z. Q., Shan, J. X., Ye, W. W., Dong, N. Q., and Lin, H. X. (2020). ERECTA1 acts upstream of the OsMKKK10-OsMKK4-OsMPK6 cascade to control spikelet number by regulating cytokinin metabolism in rice. Plant Cell 32, 2763–2779. doi: 10.1105/tpc.20.00351
Hu, J., Wang, Y. X., Fang, Y. X., Zeng, L. J., Xu, J., Yu, H. P., et al. (2015). A rare allele of GS2 enhances grain size and grain yield in rice. Mol. Plant 8, 1455–1465. doi: 10.1016/j.molp.2015.07.002
Hu, Z. J., Lu, S. J., Wang, M. J., He, H. H., Sun, L., Wang, H. R., et al. (2018). A novel QTL qTGW3 encodes the GSK3/SHAGGY-Like Kinase OsGSK5/OsSK41 that interacts with OsARF4 to negatively regulate grain size and weight in rice. Mol. Plant 11, 736–749. doi: 10.1016/j.molp.2018.03.005
Huang, K., Wang, D. K., Duan, P. G., Zhang, B. L., Xu, R., Li, N., et al. (2017). WIDE and THICK GRAIN 1, which encodes an otubain-like protease with deubiquitination activity, influences grain size and shape in rice. Plant J. 91, 849–860. doi: 10.1111/tpj.13613
Huang, R. Y., Jiang, L. R., Zheng, J. S., Wang, T. S., Wang, H. C., Huang, Y. M., et al. (2013). Genetic bases of rice grain shape: So many genes, so little known. Trends Plant Sci. 18, 218–226. doi: 10.1016/j.tplants.2012.11.001
Huang, X. Z., Qian, Q., Liu, Z. B., Sun, H. Y., He, S. Y., Luo, D., et al. (2009). Natural variation at the DEP1 locus enhances grain yield in rice. Nat. Genet. 41, 494–497. doi: 10.1038/ng.352
Ishimaru, K., Hirotsu, N., Madoka, Y., Murakami, N., Hara, N., Onodera, H., et al. (2013). Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield. Nat. Genet. 45, 707–711. doi: 10.1038/ng.2612
Kinoshita, N., Kato, M., Koyasaki, K., Kawashima, T., Nishimura, T., Hirayama, Y., et al. (2017). Identification of quantitative trait loci for rice grain quality and yield-related traits in two closely related Oryza sativa L. Subsp. japonica cultivars grown near the northernmost limit for rice paddy cultivation. Breed. Sci. 67, 191–206. doi: 10.1270/jsbbs.16155
Li, J., Chu, H. W., Zhang, Y. H., Mou, T. M., Wu, C. Y., Zhang, Q. F., et al. (2012). The rice HGW gene encodes a ubiquitin-associated (UBA) domain protein that regulates heading date and grain weight. PLoS One 7:e34231. doi: 10.1371/journal.pone.0034231
Li, N., and Li, Y. H. (2016). Signaling pathways of seed size control in plants. Curr. Opin. Plant Biol. 33, 23–32. doi: 10.1016/j.pbi.2016.05.008
Li, Q. P., Lu, L., Liu, H. Y., Bai, X. F., Zhou, X., Wu, B., et al. (2020). A minor QTL, SG3, encoding an R2R3-MYB protein, negatively controls grain length in rice. Theoret. Appl. Genet. 133, 2387–2399. doi: 10.1007/s00122-020-03606-z
Li, Y. B., Fan, C. C., Xing, Y. Z., Jiang, Y. H., Luo, L. J., Sun, L., et al. (2011). Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat. Genet. 43, 1266–1269. doi: 10.1038/ng.977
Liu, J. F., Chen, J., Zheng, X. M., Wu, F. Q., Lin, Q. B., Heng, Y. Q., et al. (2017). GW5 acts in the brassinosteroid signalling pathway to regulate grain width and weight in rice. Nat. Plants 3:17043. doi: 10.1038/nplants.2017.43
Liu, L. C., Tong, H. N., Xiao, Y. H., Che, R. H., Xu, F., Hu, B., et al. (2015). Activation of Big Grain1 significantly improves grain size by regulating auxin transport in rice. Proc. Natl. Acad. Sci. U S A. 112, 11102–11107. doi: 10.1073/pnas.1512748112
Liu, Q., Han, R. X., Wu, K., Zhang, J. Q., Ye, Y. F., Wang, S. S., et al. (2018). G-protein βγ subunits determine grain size through interaction with MADS-domain transcription factors in rice. Nat. Commun. 9:852. doi: 10.1038/s41467-018-03047-9
Liu, S. Y., Hua, L., Dong, S. J., Chen, H. Q., Zhu, X. D., Jiang, J. E., et al. (2015). OsMAPK6, a mitogen-activated protein kinase, influences rice grain size and biomass production. Plant J. 84, 672–681. doi: 10.1111/tpj.13025
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lo, S. F., Cheng, M. L., Hsing, Y. I. C., Chen, Y. S., Lee, K. W., Hong, Y. F., et al. (2020). Rice Big Grain 1 promotes cell division to enhance organ development, stress tolerance and grain yield. Plant Biotechnol. J. 18, 1969–1983. doi: 10.1111/pbi.13357
Mackay, T. F. C., Stone, E. A., and Ayroles, J. F. (2009). The genetics of quantitative traits: Challenges and prospects. Nat. Rev. Genet. 10, 565–577. doi: 10.1038/nrg2612
Murray, M. G., and Thompson, W. F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 19, 4321–4325.
Qi, P., Lin, Y. S., Song, X. J., Shen, J. B., Huang, W., Shan, J. X., et al. (2012). The novel quantitative trait locus GL3.1 controls rice grain size and yield by regulating Cyclin-T1;3. Cell Res. 22, 1666–1680.
Ruan, B. P., Shang, L. U., Zhang, B., Hu, J., Wang, Y. X., Lin, H., et al. (2020). Natural variation in the promoter of TGW2 determines grain width and weight in rice. New Phytol. 227, 629–640. doi: 10.1111/nph.16540
Shi, C. L., Dong, N. Q., Guo, T., Ye, W. W., Shan, J. X., and Lin, H. X. (2020). A quantitative trait locus GW6 controls rice grain size and yield through the gibberellin pathway. Plant J. 103, 1174–1188. doi: 10.1111/tpj.14793
Shi, C. L., Ren, Y. L., Liu, L. L., Wang, F., Zhang, H., Tian, P., et al. (2019). Ubiquitin specific protease 15 has an important role in regulating grain width and size in rice. Plant Physiol. 180, 381–391. doi: 10.1104/pp.19.00065
Si, L. Z., Chen, J. Y., Huang, X. H., Gong, H., Luo, J. H., Hou, Q. Q., et al. (2016). OsSPL13 controls grain size in cultivated rice. Nat. Genet. 48, 447–456. doi: 10.1038/ng.3518
Song, X. J., Huang, W., Shi, M., Zhu, M. Z., and Lin, H. X. (2007). A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 39, 623–630. doi: 10.1038/ng2014
Song, X. J., Kuroha, T., Ayano, M., Furuta, T., Nagai, K., Komeda, N., et al. (2015). Rare allele of a previously unidentified histone H4 acetyltransferase enhances grain weight, yield, and plant biomass in rice. Proc. Natl. Acad. Sci. U S A. 112, 76–81. doi: 10.1073/pnas.1421127112
Tanabe, S., Onodera, H., Hara, N., Ishii-Minami, N., Day, B., Fujisawa, Y., et al. (2016). The elicitor-responsive gene for a GRAS family protein, CIGR2, suppresses cell death in rice inoculated with rice blast fungus via activation of a heat shock transcription factor, OsHsf23. Biosci. Biotech. Bioch. 80, 145–151. doi: 10.1080/09168451.2015.1075866
Wang, A. H., Hou, Q. Q., Si, L. Z., Huang, X. H., Luo, J. H., Lu, D. F., et al. (2019). The PLATZ transcription factor GL6 affects grain length and number in Rice. Plant Physiol. 180, 2077–2090. doi: 10.1104/pp.18.01574
Wang, C. S., Tang, S. C., Zhan, Q. L., Hou, Q. Q., Zhao, Y., Zhao, Q., et al. (2019). Dissecting a heterotic gene through GradedPool-Seq mapping informs a rice-improvement strategy. Nat. Commun. 10:2982. doi: 10.1038/s41467-019-11017-y
Wang, S. K., Li, S., Liu, Q., Wu, K., Zhang, J. Q., Wang, S. S., et al. (2015). The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nat. Genet. 47, 949–954. doi: 10.1038/ng.3352
Wang, S. K., Wu, K., Yuan, Q. B., Liu, X. Y., Liu, Z. B., Lin, X. Y., et al. (2012). Control of grain size, shape and quality by OsSPL16 in rice. Nat. Genet. 44, 950–954. doi: 10.1038/ng.2327
Wang, W. H., Wang, L. L., Zhu, Y. J., Fan, Y. Y., and Zhuang, J. Y. (2019). Fine-mapping of qTGW1.2a, a quantitative trait Locus for 1000-grain weight in rice. Rice Sci. 4, 220–228.
Wang, Y. X., Xiong, G. S., Hu, J., Jiang, L., Yu, H., Xu, J., et al. (2015). Copy number variation at the GL7 locus contributes to grain size diversity in rice. Nat. Genet. 47, 944–948. doi: 10.1038/ng.3346
Wu, W. G., Liu, X. Y., Wang, M. H., Meyer, R. S., Luo, X. J., Ndjiondjop, M., et al. (2017). A single-nucleotide polymorphism causes smaller grain size and loss of seed shattering during African rice domestication. Nat. Plants 3:17064. doi: 10.1038/nplants.2017.64
Wu, W. X., Zheng, X. M., Lu, G. W., Zhong, Z. Z., Gao, H., Chen, L. P., et al. (2013). Association of functional nucleotide polymorphisms at DTH2 with the northward expansion of rice cultivation in Asia. Proc. Natl. Acad. Sci. U S A. 8, 2775–2780. doi: 10.1073/pnas.1213962110
Wu, Y., Wang, Y., Mi, X. F., Shan, J. X., Li, X. M., Xu, J. L., et al. (2016). The QTL GNP1 encodes GA20ox1, which increases grain number and yield by increasing cytokinin activity in rice panicle meristems. PLoS Genet. 12:e1006386. doi: 10.1371/journal.pgen.1006386
Xia, D., Zhou, H., Liu, R. J., Dan, W. H., Li, P. B., Wu, B., et al. (2018). GL3.3, a novel QTL encoding a GSK3/SHAGGY-like Kinase, epistatically interacts with GS3 to produce extra-long grains in Rice. Mol. Plant 11, 754–756. doi: 10.1016/j.molp.2018.03.006
Xiong, H. Y., Yu, J. P., Miao, J. L., Li, J. J., Zhang, H. L., Wang, X., et al. (2018). Natural variation in OsLG3 increases drought tolerance in Rice by inducing ROS scavenging. Plant Physiol. 178, 451–467. doi: 10.1104/pp.17.01492
Xu, C. J., Liu, Y., Li, Y. B., Xu, X. D., Xu, C. G., Li, X. H., et al. (2015). Differential expression of GS5 regulates grain size in rice. J. Exp. Bot. 66, 2611–2623. doi: 10.1093/jxb/erv058
Xue, P., Zhang, Y. X., Lou, X. Y., Zhu, A. K., Chen, Y. Y., Sun, B., et al. (2019). Mapping and genetic validation of a grain size QTL qGS7.1 in rice (Oryza sativa L.). J. Integrat. Agricult. 8, 1838–1850. doi: 10.1016/S2095-3119(18)62113-6
Yamamoto, K., Shida, S., Honda, Y., Shono, M., Miyake, H., Oguri, S., et al. (2015). Overexpression of acetylcholinesterase gene in rice results in enhancement of shoot gravitropism. Biochem. Biophys. Res. Commun. 465, 488–493. doi: 10.1016/j.bbrc.2015.08.044
Ying, J. Z., Ma, M., Bai, C., Huang, X. H., Liu, J. L., Fan, Y. Y., et al. (2018). TGW3, a major QTL that negatively modulates grain length and weight in rice. Mol. Plant 11, 750–753. doi: 10.1016/j.molp.2018.03.007
Yu, J. P., Miao, J. L., Zhang, Z. Y., Xiong, H. Y., Zhu, X. Y., Sun, X. M., et al. (2018). Alternative splicing of OsLG3b controls grain length and yield in japonica rice. Plant Biotechnol. J. 16, 1667–1678. doi: 10.1111/pbi.12903
Yu, J. P., Xiong, H. Y., Zhu, X. Y., Zhang, H. L., Li, H. H., Miao, J. L., et al. (2017). OsLG3 contributing to rice grain length and yield was mined by Ho-LAMap. BMC Biol. 15:28. doi: 10.1186/s12915-017-0365-7
Zhang, C. Q., Xu, Y., Lu, Y., Yu, H. X., Gu, M. H., and Liu, Q. Q. (2011). The WRKY transcription factor OsWRKY78 regulates stem elongation and seed development in rice. Planta 234, 541–554. doi: 10.1007/s00425-011-1423-y
Zhang, H., Zhu, Y. J., Zhu, A. D., Fan, Y. Y., Huang, T. X., Zhang, J. F., et al. (2020). Fine-mapping of qTGW2, a quantitative trait locus for grain weight in rice (Oryza sativa L.). PeerJ 8:e8679. doi: 10.7717/peerj.8679
Zhang, Y., Xiong, Y., Liu, R. Y., Xue, H. W., and Yang, Z. B. (2019). The Rho-family GTPase OsRac1 controls rice grain size and yield by regulating cell division. Proc. Natl. Acad. Sci. U S A. 116, 16121–16126. doi: 10.1073/pnas.1902321116
Zhao, D. S., Li, Q. F., Zhang, C. Q., Zhang, C., Yang, Q. Q., Pan, L. X., et al. (2018). GS9 acts as a transcriptional activator to regulate rice grain shape and appearance quality. Nat. Commun. 9:1240. doi: 10.1038/s41467-018-03616-y
Zhu, Y. J., Zhang, Z. H., Chen, J. Y., Fan, Y. Y., Mou, T. M., Tang, S. Q., et al. (2019). Fine mapping of qTGW10-20.8, a QTL having important contribution to grain weight variation in rice. Crop J. 7, 587–597. doi: 10.1016/j.cj.2019.01.006
Keywords: rice, quantitative trait locus, grain size, small-effect, residual heterozygous population
Citation: Xue P, Chen Y-y, Wen X-x, Wang B-f, Yang Q-q, Gong K, Kang Y-w, Sun L-p, Yu P, Cao L-y, Zhang Y-x, Zhan X-d and Cheng S-h (2022) Dissection of Closely Linked Quantitative Trait Locis Controlling Grain Size in Rice. Front. Plant Sci. 12:804444. doi: 10.3389/fpls.2021.804444
Received: 29 October 2021; Accepted: 06 December 2021;
Published: 20 January 2022.
Edited by:
Marcelino Perez De La Vega, Universidad de León, SpainReviewed by:
Sarla Neelamraju, Indian Institute of Rice Research (ICAR), IndiaJauhar Ali, International Rice Research Institute (IRRI), Philippines
Copyright © 2022 Xue, Chen, Wen, Wang, Yang, Gong, Kang, Sun, Yu, Cao, Zhang, Zhan and Cheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ying-xin Zhang, emhhbmd5aW5neGluQGNhYXMuY24=; Xiao-deng Zhan, emhhbnhpYW9kZW5nQGNhYXMuY24=; Shi-hua Cheng, Y2hlbmdzaGlodWFAY2Fhcy5jbg==
 Pao Xue
Pao Xue Yu-yu Chen
Yu-yu Chen Xiao-xia Wen1
Xiao-xia Wen1 Bei-fang Wang
Bei-fang Wang Yi-wei Kang
Yi-wei Kang Lian-ping Sun
Lian-ping Sun Xiao-deng Zhan
Xiao-deng Zhan Shi-hua Cheng
Shi-hua Cheng