
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 17 January 2022
Sec. Plant Development and EvoDevo
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.803341
This article is part of the Research Topic Environmental and molecular control of bud dormancy and bud break in woody perennials: An integrative approach View all 23 articles
Parts of this article's content have been modified or rectified in:
Erratum: Time-Resolved Analysis of Candidate Gene Expression and Ambient Temperature During Bud Dormancy in Apple
Winter dormancy – a period of low metabolic activity and no visible growth – appears as an adaptation to harsh winter conditions and can be divided into different phases. It is tightly controlled by environmental cues, with ambient temperature playing a major role. During endodormancy, a cultivar-specific amount of cold needs to be perceived, and during ecodormancy, heat hours accumulate before bud burst and anthesis in spring. Expression analysis, performed in several key fruit tree species, proved to be very useful in elucidating the molecular control of onset and release of dormancy. However, the time resolution of these experiments has been limited. Therefore, in this study, dense time-series expression analysis was conducted for 40 candidate genes involved in dormancy control, under the cool-temperate climate conditions in Dresden. Samples were taken from the cultivars ‘Pinova’ and ‘Gala,’ which differ in flowering time. The set of candidate genes included well-established dormancy genes such as DAM genes, MdFLC-like, MdICE1, MdPRE 1, and MdPIF4. Furthermore, we tested genes from dormancy-associated pathways including the brassinosteroid, gibberellic acid, abscisic acid (ABA), cytokinin response, and respiratory stress pathways. The expression patterns of well-established dormancy genes were confirmed and could be associated with specific dormancy phases. In addition, less well-known transcription factors and genes of the ABA signaling pathway showed associations with dormancy progression. The three ABA signaling genes HAB1_chr15, HAI3, and ABF2 showed a local minimum of gene expression in proximity of the endodormancy to ecodormancy transition. The number of sampling points allowed us to correlate expression values with temperature data, which revealed significant correlations of ambient temperature with the expression of the Malus domestica genes MdICE1, MdPIF4, MdFLC-like, HAB1chr15, and the type-B cytokinin response regulator BRR9. Interestingly, the slope of the linear correlation of temperature with the expression of MdPIF4 differed between cultivars. Whether the strength of inducibility of MdPIF4 expression by low temperature differs between the ‘Pinova’ and ‘Gala’ alleles needs to be tested further.
In deciduous fruit trees such as apple (Malus × domestica Borkh.), phases of high metabolic activity, growth, and reproduction alternate with phases of no growth and low levels of activity, called dormancy. During dormancy, all vulnerable tissues are either removed or protected by very robust bud scales, which allows surviving harsh winter conditions. The cycles of activity and dormancy need to be synchronized well with environmental conditions in order to maximize survival during winter, optimize the timing of reproduction and yield, and maximize the length of growing seasons (Cooke et al., 2012). Which environmental cues regulate onset, progression, and release of dormancy in deciduous fruit trees is not entirely resolved, but ambient temperature and photoperiod appear to play major roles (Heide, 2008; Cooke et al., 2012; Maurya and Bhalerao, 2017). In contrast to most fruit tree species, M. × domestica is not sensitive to photoperiod and thus uses ambient temperature as a major environmental cue to control the timing of dormancy (Heide and Prestrud, 2005; Tanino et al., 2010; Cooke et al., 2012; Singh et al., 2017). Whether light quality serves as an additional cue needs to be determined.
Winter dormancy consists of three distinct phases, namely, paradormancy, endodormancy, and ecodormancy (Lang, 1987). During paradormancy, the termination of shoot growth is induced by other parts of the growing plant (Singh et al., 2017). However, dormant buds are still competent to leaf out, when disconnected from internal inhibitory signals from the tree and when grown under suitable conditions (Saure, 1985). During endodormancy, internal conditions prevent buds from leafing out, even if disconnected from the tree and placed under suitable growth conditions. In order to transit from endodormancy to ecodormancy, a genotype-specific amount of cold needs to be perceived, which is called chilling requirement (CR) (Coville, 1920). Freezing tolerance is highest during endodormancy, where water is bound and unfreeze (Erez et al., 1990; Faust et al., 1991). After the transition to ecodormancy, bud behavior is controlled predominantly by external conditions. If temperature accumulation is sufficiently high, buds will leaf out. For the phase transition from ecodormancy to bud break, it is heated, which needs to accumulate (heat requirement, HR). Several temperature-based models have been developed to measure the amounts of cold and heat that are required to progress through dormancy and allow to forecast the timing of bud break (Fadon et al., 2020). The most commonly used models are the chilling hours/Weinberger-Eggert model (Weinberger, 1950), the Utah model (Richardson et al., 1974), and the dynamic model (Fishman et al., 1987a,b; Erez et al., 1990). Model comparisons suggest that the goodness of performance of each model depends on the climatic region and the plant species it is applied to Luedeling et al. (2009); Chmielewski et al. (2012). The most commonly used model for heat quantification is the growing degree hour (GDH) model by Anderson et al. (1986). How chill and heat units are perceived and how their accumulation is resolved molecularly are still open questions.
In regions of the mild climate, chilling accumulates very slowly, leading to a prolonged phase of endodormancy, which may overlap with ecodormancy (Saure, 1985; Malagi et al., 2015). In temperate climate zones, there is sufficient chilling, and the phase transition to ecodormancy occurs at winter times when low temperature alone is sufficient to inhibit further growth. Global warming poses significant challenges to the progression of dormancy that are not universal but rather specific to the perceived environmental conditions. In mild climates, not enough chill units accumulate, which leads to irregular budburst and to lower crop yield (Erez, 2000). In the temperate zone, warm springs cause a shift to earlier bud break, with the consequence of the high risk of flower damage by early spring frost (Chmielewski et al., 2004; Estrella et al., 2007). Therefore, it is of high interest to unravel the molecular mechanisms of winter dormancy, which will allow developing tools for breeding apple cultivars, which are resilient to climate change.
In recent years, progress has been made on resolving the molecular mechanisms of dormancy control (Falavigna et al., 2019). Dormancy-Associated MADS-Box (DAM) genes were the first genes identified from the evergrowing mutant in peach (Bielenberg et al., 2008), and later also in other fruit-bearing species of the Rosaceae, such as plum, cherry, Japanese apricot, apple, and pear (Mimida et al., 2015; Rothkegel et al., 2017; Zhao et al., 2018; Vimont et al., 2019; Quesada-Traver et al., 2020).
Short Vegetative Phase (SVP) genes are structurally similar to DAM genes and are also important for dormancy control. Lines with transgenically downregulated levels of MdDAM and MdSVP genes are not able to get into the dormant state (Wu et al., 2017, 2021; Moser et al., 2020). Interestingly, MdDAM, MdSVP, and Md Flowering Locus C-like (MdFLC-like) genes form multimeric complexes that bind to specific DNA regions (Falavigna et al., 2021).
Since functional verification by transgenic lines is difficult, gene expression analysis has been very important to get insight into genes and pathways involved in dormancy control. Several RNAseq studies have been conducted in apple, peach, cherry, and apricot (Falavigna et al., 2014; Takeuchi et al., 2018; Artlip et al., 2019; Vimont et al., 2019; Moser et al., 2020; Yu et al., 2020; Zhu et al., 2020). The strict filtering strategy of Takeuchi et al. (2018) revealed a small set of genes being involved in endodormancy phase transition and showed expression correlation with ambient temperature under laboratory conditions. Among them were MdFLC-like and PHYTOCHROME-INTERACTING FACTOR 4 (MdPIF4), both have been associated with a temperature-sensitive expression before. In Arabidopsis thaliana, PIFs interact with light receptors and mediate growth responses by modifying the chromatin organization (Kumar et al., 2012; Willige et al., 2021). MdFLC-like in apple has been shown to be upregulated on cold treatment and may confer a growth-inhibiting effect (Porto et al., 2015; Nishiyama et al., 2021).
Although whole-genome expression analysis has revealed important insight, the time resolution of sampling dates is very limited due to the long time span that needs to be covered. A common strategy has been to sample once a month or to sample after plants have perceived a defined amount of chill hours (CH), which results in a total of four to eight sampling dates across winter (Moser et al., 2020; Yu et al., 2020; Falavigna et al., 2021). Vimont et al. (2019) also sampled every month; however, they improved the study considerably by selectively increasing sampling density in the weeks before phase transition (Vimont et al., 2019). By doing so, they increased the sampling density to biweekly for a few weeks. Yet, higher sampling densities are required to adequately capture the dynamic process of dormancy and the transitions between phases. Therefore, to unravel the molecular mechanisms of winter dormancy, a gene expression analysis of selected dormancy candidate genes was carried out. In this study, the high sampling density allowed capturing the dynamic process of dormancy and the transition between its individual phases. Buds of the two apple cultivars ‘Gala’ and ‘Pinova’ were collected from a commercial orchard in the Southeast of Germany, under the cool-temperate climate conditions in Dresden (Germany), which is located in the transition zone to the continental climate. The expression patterns of these genes were determined in densely spaced samples, which were taken weekly, covering the complete time span of winter dormancy. The dense sampling allowed us to identify specific expression patterns that were associated either with endodormancy or ecodormancy or patterns that reached across both phases. Correlations of gene expression with temperature under natural conditions provided further evidence for temperature-dependent expression.
Spur buds of the two M.×domestica Borkh. cultivars ‘Pinova’ and ‘Gala’ grafted on M9 rootstocks were collected at the Orchard Görnitz in Neusörnewitz, located near Dresden, Germany (latitude 51.14908600295051, longitude 13.54123977860281). Sampling took place every week from September 20, 2017 to April 18, 2018. Per time point and cultivar, four biological replicates were collected with five spur buds for each replicate.
The timing of the phase transition from endodormancy to ecodormancy was determined by scion cuttings. Therefore, extension shoots with a swollen terminal bud were cut every week and placed in a growth chamber at 20°C long days (16 h light) and relative humidity of 60%. Scions were placed in jars containing tap water, which was renewed every week. The developmental stages of generative terminal buds were determined for three replicates. Scions were cut every week from December 12, 2017 to February 14, 2018. The date of transition was defined as the cutting date of scions that under bud break forcing conditions reached the developmental stage BBCH 59 (ballon) after 5 weeks. Bud break and flowering in the field were scored every week during spring 2018 on 10 trees per cultivar.
Hourly temperature data were obtained from the meteorological station located in Coswig at a distance of 3.38 km from the orchard. The accumulation of chilling hours was calculated by counting any hour as one unit if its temperature lies between 0 and 7.2°C (Weinberger, 1950). Chill portions (CP) were calculated by the dynamic model by Fishman et al. (1987a,b); Erez et al. (1990). Heat accumulation was calculated using the ASYMCUR GDH model by Anderson with the base, optimal, and critical temperatures of 4°C, 25°C, and 36°C, respectively (Anderson et al., 1986; Milyaev et al., 2021).
Immediately after harvesting, bud scales were removed, and samples were frozen on dry ice. Frozen buds were homogenized with the Retsch Mixer Mill MM400 (Retsch, Germany), total RNA was extracted using the InviTrap Spin Plant RNA Mini Kit (Stratec Molecular GmbH, Berlin, Germany), and DNA was removed with the DNA-Free Kit (Life Technologies GmbH, Darmstadt, Germany). cDNA was synthesized with oligodT primer and 1 μg RNA as template and diluted 1:20 for gene expression analysis.
Primers were designed with the NCBI Tool Primer-BLAST, combining Primer3 and BLAST with the standard settings and M. domestica taxid 3750 as reference organism. To assure specificity to the gene of interest, suggested primer sequences were only considered if they did not target any other sequence with zero to two mismatches. All mRNA target sequences were extracted from the GDDH1.3 Genome (Daccord et al., 2017). To identify M. domestica genes that are homologous to the selected Prunus persica genes of interest, nucleotide BLAST against the M. domestica GDDH1.3 genome was used. Exceptions were sequences for Md Flowering locus T 2 (MdFT2). The sequence of MdFT2 (HQ424012.1 ‘Pinova’ and HF31588-RA ‘Hanfu line’) was not found in the GDDH1.3 genome by BLAST, and only the sequence of MdFT1 (HQ424013.1 ‘Pinova,’ MD12G1262000 ‘Golden Delicious’, HF38537-RA ‘Hanfu line’) was found in the GDDH1.3 genome. Therefore, the primers MdFT2-qRT-PCR of Kotoda et al. (2010) were used for PCR. All primer pairs were tested on ‘Pinova’ and ‘Gala’ cDNA by qRT-PCR (95°C for 15 s, 60°C for 30 s, 72°C for 30 s, for 40 cycles), and specificity was confirmed by melting curve analysis. For PIF4, primer efficiency was determined for each cultivar via concentration series and was very similar for both cultivars (Supplementary Figure 1). A full list of primers is given in Supplementary Table 1. The presence of MdDAM1 splice variants was tested by PCR on cDNA of four replicates of ‘Pinova’ samples collected between November 1, 2017 and December 13, 2017 at an annealing temperature of 57°C, an extension time of 1 min, and with 40 PCR cycles.
Expression analysis was performed using the BioMark HD high-throughput system with the 96.96 Dynamic Array (Fluidigm, South San Francisco, California, United States). For the winter period of 2017/2018, three biological and two technical replicates of two cultivars at 28 were analyzed with 44 primer pairs (Supplementary Table 1). The 96.96 Dynamic Array Integrated Fluidic Chip assay was performed according to the instructions of the manufacturer and as described by Reim et al. (2020). Expression data were analyzed with the Fluidigm Real-Time polymerase chain reaction (PCR) Analysis 3.1.3 software (Fluidigm, South San Francisco, California, United States). The amplification curve quality check was performed, and three primer pairs and three samples (i.e., 114_rep1_Pinova, 120_rep3_Pinova, and 123_rep1_Gala) were removed from further analysis as their quality score was below the set threshold of 0.65. The linear baseline correction and auto cycle threshold (Ct) determination were used for retrieving CT values. The expression stability of reference genes was validated using RefFinder (Xie et al., 2012). Relative expression values were calculated using the 2–ΔΔCT method, using sample 101 as the calibrator (Pfaffl, 2001). Graphical representation of expression data was performed with ggplot2 using R (Wickham, 2016, R foundation, Vienna, Austria).
Mean expression values per time point and cultivar were correlated with temperature data. Therefore, the following temperature values were used: mean daily temperature at the sampling date, and the sum of CH or CP of all days within 2 weeks before sampling was calculated. The correlation matrix was calculated and plotted using the Pearson correlation method with the R package corrplot (Wei and Simko, 2021). Scatter plots and linear regressions were performed using R and ggplot2 (Wickham, 2016).
k-means clustering was performed using the R package “cluster,” and the R package “factoextra” was used to visualize the clusters (Kassambara, 2017; Maechler et al., 2021). The optimal cluster number was determined by considering the within the sum of squares per number of clusters. For iterations, 25 random starting assignments were used, which revealed 25 different clustering results. Of these results, the best one with the smallest sum of squares was chosen.
A set of 40 candidate genes was selected to determine specific expression patterns during winter dormancy in buds of the two apple cultivars ‘Gala’ and ‘Pinova’ (Supplementary Table 2). These include transcription factors and metabolic enzymes that are clearly or less explicitly associated with dormancy.
The MADS-box transcription factor genes MdDAM1, MdDAM2, MdDAM4, MdSVPa, and MdSVPb are well known to control the progression of dormancy in fruit trees of the Rosacea (Falavigna et al., 2019). The gene MdFLC-like also belongs to MADS-box transcription factors and is the closest paralog of the A. thaliana flowering time gene Flowering Locus C (FLC) (Kumar et al., 2016). It was chosen because MdFLC-like was upregulated after cold exposure, and evidence exists for a growth-inhibiting effect, which suggests that MdFLC-like is a good candidate for playing a role in dormancy (Porto et al., 2015; Takeuchi et al., 2018; Nishiyama et al., 2021). Additionally, it was located on top of chromosome 9, where quantitative trait loci (QTL) - and genome-wide association studies identified loci controlling bud break in apple (Vimont et al., 2019; Celton et al., 2011; Allard et al., 2016; Urrestarazu et al., 2017). Additionally, the four AP2/ethylene-responsive transcription factor genes Md Early Bud Break1 (MdEBB1), APETALA 2 on chromosome 7(AP2chr7), APETALA 2/Ethylene-Responsive Factor 113 (AP2_ERF113), and Ethylene-Responsive Factor on chromosome 4 (ERFchr4) described below were also included. EBB1 has been identified in poplar to play a role in dormancy release (Yordanov et al., 2014) and was associated with bud break in apples (Wisniewski et al., 2015). In pear, correlations of PpEBB1 with cyclin D3 were observed, and the hypothesis was put forward that EBB1 could regulate bud break by activating cell cycle regulator genes to reinitiate cell division (Tuan et al., 2016). AP2chr7 and ERFchr4 were differentially expressed in MdDAM1-silenced transgenic apple trees and changed expression during the course of dormancy (Moser et al., 2020). AP2_ERF113 showed temperature-sensitive expression variation during dormancy, similar to the gene PIF4, the MYC2-like transcription factor MD14G1126900 (MYC2-like), F-box transcription factor MD11G1009500 (MdFbox), Peroxidase 42-like MD10G1321200 (Md_peroxidase), and 4-Coumarate coenzyme A ligase 5 MD13G1257800 (CoA_ligase) (Takeuchi et al., 2018). MdbZIP and the NAC-like transcription factor NAM were included because both are differentially expressed in MdDAM1-silenced trees and varied expression during dormancy progression (Moser et al., 2020). The transcription factor gene Inducer of CBF Expression1 (ICE1) was found in a pear to be associated with endodormancy (Takemura et al., 2015) and was identified as the candidate gene in previous QTL studies investigating the timing of bud break, similar to PACLOBUTRAZOL RESISTANCE 1 (MdPRE1) (Miotto et al., 2019). Additional to MdPRE1, two more members of the Brassinosteroid pathway were selected, namely, Brassinosteroid-6-Oxidase 2 (BR6OX) and the receptor Kinase inhibitor BKI1. BR6Ox and BKI1 were chosen because both showed differential expression in dormancy-related RNA-seq experiments in cherry, peach, and apricot (Vimont et al., 2019; Yu et al., 2020). Of the gibberellic acid pathway, two gibberellin 3-oxidases 1 (GA3Ox1, MD08G1138400, MD15G1116300) and one gibberellin 2-oxidase 8 (GA2Ox8, MD11G1225400) were included because close homologs were differentially expressed in cherry, peach, and apricot during dormancy progression (Vimont et al., 2019; Yu et al., 2020). From the abscisic acid (ABA) pathway, the following four transcription factor genes were included: ABSCISIC ACID RESPONSIVE ELEMENTS-BINDING FACTOR 2 (ABF2, MD15G1081800), ABF2 on chromosome 8 (ABFchr8), (MD08G1099600), ABA-Insensitive 5 (ABI5) on chromosome 12,(ABI5chr12, MD12G1024300), and ABI5 on chromosome 14 (ABI5chr14, MD14G1021600). These were included because they were found to control a cluster of differentially expressed genes during dormancy in cherry (Vimont et al., 2019). Further members of the ABA pathway that were included in this study were the genes Group A protein type 2C phosphatases HAB1chr15 (MD15G1212000), HAB1chr2 (MD02G1084600), and the HIGHLY ABA-INDUCED PP2C Gene 3 (HAI3, MD06G1106000) and the epoxidase ABA1, which catalyzes the first step of ABA biosynthesis (MD02G1318300, MD07G1003000). This group of genes was differentially expressed during dormancy progression in peach and apricot (Yu et al., 2020). From the cytokinin signaling pathway, the type-B cytokinin response regulators BRR1, 7, 8, 9, and 10 were included in this study, as they were found to play a role in the endodormancy to ecodormancy transition in apple previously (Cattani et al., 2020). Not only hormones but also respiratory stress has been proposed to play a role in dormancy release (Vimont et al., 2019); therefore, the gene Respiratory burst oxidase protein F (RBOH, MD14G1211700) was included, which is differentially expressed in cherry, peach, and apricot, as well as Md_peroxidase, mentioned above).
To be able to associate expression patterns with the timing of dormancy, the transition of endodormancy to ecodormancy was phenologically determined by scion cuttings and the end of dormancy by scoring bud break in the field (Lang, 1987). During winter 2017/2018, the transition of endodormancy to ecodormancy occurred on January 17 for ‘Pinova’ and on January 31 for ‘Gala’ (Supplementary Figure 2). Until January 17, 2018, trees had received 72.9 CP or 1,344 CH (Supplementary Figure 3). Therefore, the CR for ‘Pinova’ 2018 was 72.9 CP or 1,344 CH. Slightly higher was the CR for ‘Gala,’ 83.9 CP or 1,552 CH, which was reached on January 31. Bud break occurred between April 4 and April 11, 2018 in both cultivars, after an accumulation of 2,619–2,915 GDH.
The time points of sample collections were chosen to cover all dormancy phases from the end of summer (September 20) to spring (April 18). Dense sampling allows monitoring expression changes that occur during short time spans. The expression pattern of 40 selected genes was surveyed, and for 35 genes, distinct patterns of expression were determined, and five genes did show no expression. To group genes by overall expression pattern across all time points, k-means clustering analyses were performed. Expression patterns were clustered into six different clusters (Figure 1). Cluster 1 and cluster 6 are distinct with good separation, whereas clusters 2 and 3, as well as clusters 4 and 5, are less distant from each other.
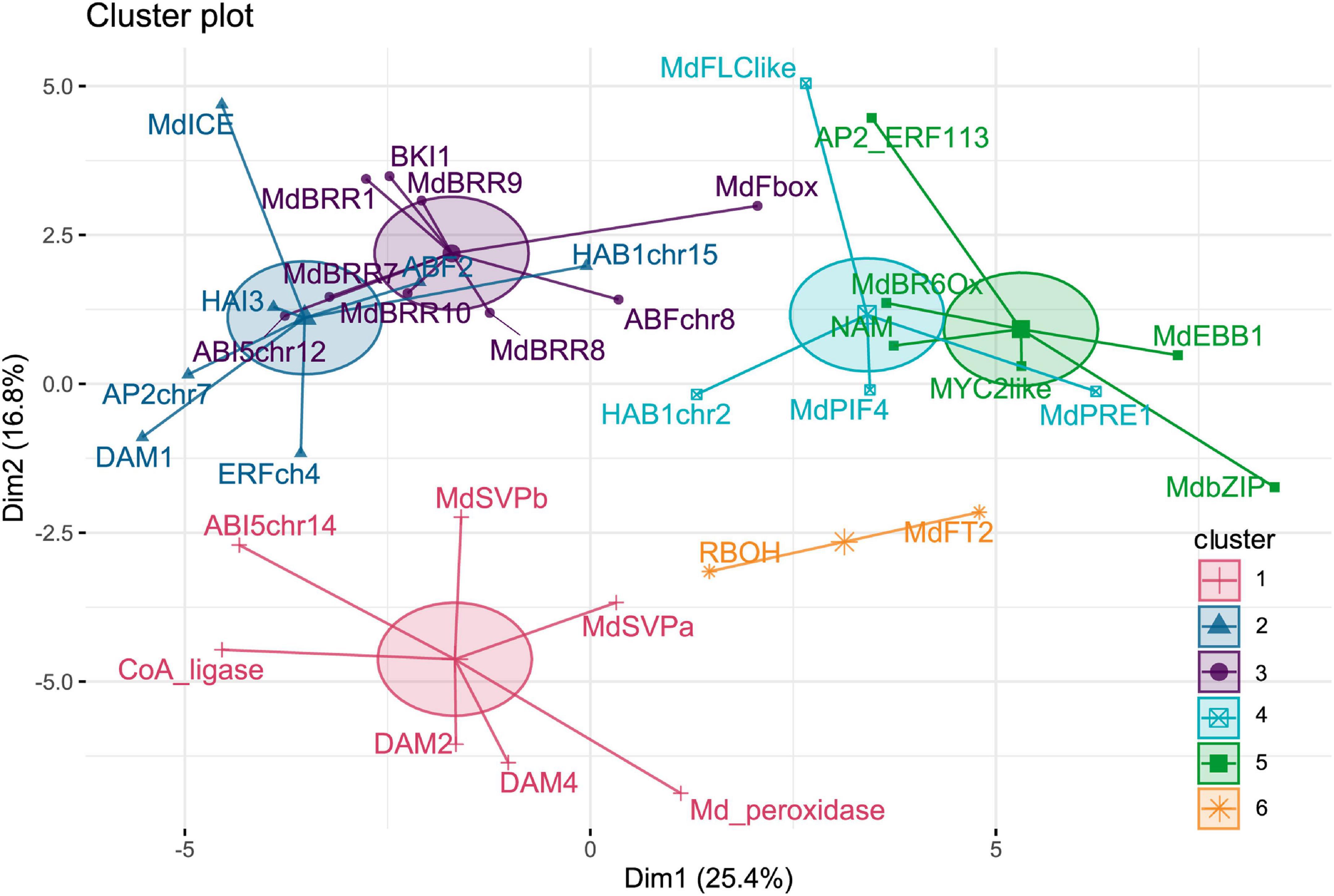
Figure 1. Clusters of genes determined by k-means clustering of gene expression patterns. A total of 35 genes were clustered into six clusters according to their gene expression patterns across all samples.
Genes that belong to cluster 1 were highly expressed at the very beginning of dormancy (Figures 2A–E. See Supplementary Table 3 for collection dates). The two known dormancy-related SVP genes MdSVPa and MdSVPb belong to this first cluster (Figures 2A,B). Their expression levels were high at the beginning of dormancy, dropped to a local minimum during the ecodormancy to endodormancy transition, and increased again to reach a second maximum of expression during mid of ecodormancy. The genes CoA_ligase and Md_peroxidase showed a pattern of expression that is very similar to MdSVPa and MdSVPb (Figures 2C,D). Difference in expression between the two cultivars ‘Gala’ and ‘Pinova’ was observed for MdSVPa, which showed stronger expression in ‘Gala’ compared with ‘Pinova’, especially during the phase transition and during ecodormancy. The known dormancy-associated MADS-box gene MdDAM2 also belongs to cluster 1, with high expression levels at the beginning of dormancy (Figure 2E). Less specific was the expression pattern of ABI5chr14, which also belongs to cluster 1 (Supplementary Figure 4). Genes of cluster 2 increased expression levels at first and showed a local maximum of expression during endodormancy (Figures 2F–L, 3A). MdDAM4 and MdDAM1 belong to cluster 2 and both show a single peak of expression during endodormancy. The peaks of expression occurred progressively, earlier in MdDAM4 than in MdDAM1. A similar pattern was observed for AP2chr7 and ERFchr4. Both genes are members of the ERF/AP2 transcription factor family that are involved in transcriptional control of stress responses and are less well known for playing a role in apple dormancy. ‘Pinova’ expression levels were higher in MdDAM4 and AP2chr7 and lower in MdDAM1 compared with ‘Gala’; however, no shift in the timing of expression was observed. The MADS-box transcription factors MdSVPa, MdSVPb, MdDAM1, MdDAM4, and MdFLC-like (Figure 3I) have been shown to form multimeric complexes (Falavigna et al., 2021). In this study, we observed overlapping expression patterns of these genes, which may indicate that multimeric complexes including MdSVPa/b-MdDAM1 or MdDAM4, and complexes including MdSVPa/b and MdFLC-like could potentially be built during time spans of overlapping expression. For DAM1 in pear, it was shown that alternative splicing variants exist (Li et al., 2021). Since alternative splicing is another mechanism that occurs in relation to temperature-dependent regulation, we also assessed whether splice variants of MdDAM1 were present in our samples between November 1 and December 22, 2017; however, we did not find evidence for any splice variant of MdDAM1 in our samples (Supplementary Figure 5). The expression pattern of MdICE1 (Figure 2J) showed a broader single peak of expression. Expression levels started to rise during endodormancy, were highly expressed during phase transition, declined continuously during ecodormancy, and reached low levels before bud break. The expression peaks occurred approximately 1 week later in ‘Pinova’ compared with ‘Gala.’ In A. thaliana, ICE1 is an inducer of the cold acclimation pathway that leads to freezing tolerance. Although freezing tolerance overlaps with endodormancy, it appears to be independent of endodormancy (Bilavcik et al., 2015). Furthermore, evidence exists that ICE1 can activate FLC expression (Lee et al., 2015). Since the expression curve of MdICE1 peaked earlier than that of MdFLC-like, MdICE1 may also induce expression of MdFLC-like. The genes HAI3, ABF2, and HAB1chr15 also belong to cluster 2 (Figures 2K,L, 3A). These three genes showed an expression peak during endodormancy, reduced expression, and a local minimum of expression 2–3 weeks before the phase transition to ecodormancy. A second local maximum of expression occurred during ecodormancy. The expression of ABF2 showed high variation between sampling weeks and was strongly expressed in ‘Gala’ compared with ‘Pinova’ (Figure 2K). Since HAB1chr15, HAI3, and ABF2 are all parts of the ABA signaling pathway, these results clearly suggest involvement of ABA signaling during endodormancy and ecodormancy and during phase transition, when expression levels were low. Less distinct were the expression patterns of genes belonging to cluster 3 (Figures 3B,C and Supplementary Figures 4B–H). MdBRR1 and MdBRR9 show a broad expression curve across dormancy. ‘Gala’ samples showed higher expression compared with ‘Pinova’ in both genes. Less specific expression patterns, however, certainly expressed during dormancy and with reduced levels before bud break was observed for the genes ABFchr8, MdFbox, BKI1, MdBRR7, MdBRR8, MdBRR10, and ABI5chr12 (Supplementary Figures 4B–H). Genes that belong to cluster 4 show predominant expression during ecodormancy. The genes MdPIF4, HAB1chr2, MdPRE1, and MdEBB1 belong to this cluster (Figures 3D–G). MdPIF4 and HAB1chr2 showed a small peak of expression during endodormancy and a larger expression peak during ecodormancy. For both genes, expression levels were higher in ‘Pinova’ compared with ‘Gala’. MdPRE1 and MdEBB1 had a single peak in their expression pattern during ecodormancy. Cluster 4 comprises expression patterns with a single broader peak that is not lowered during phase transition. MdbZIP and MdFLC-like belong to this cluster (Figures 3H,I), as well as the following genes with less clear expression patterns: AP2_ERF113, NAM, MYC2like, and MdBR6Ox (Supplementary Figures 4I,L). The expression levels of NAM were much higher in ‘Gala’ compared with ‘Pinova’ throughout dormancy progression. Cluster 6 is a distinct cluster and comprises the two genes MdFT2 and RBOH (Figures 3J,K). Both genes were expressed after dormancy and were associated with bud break.
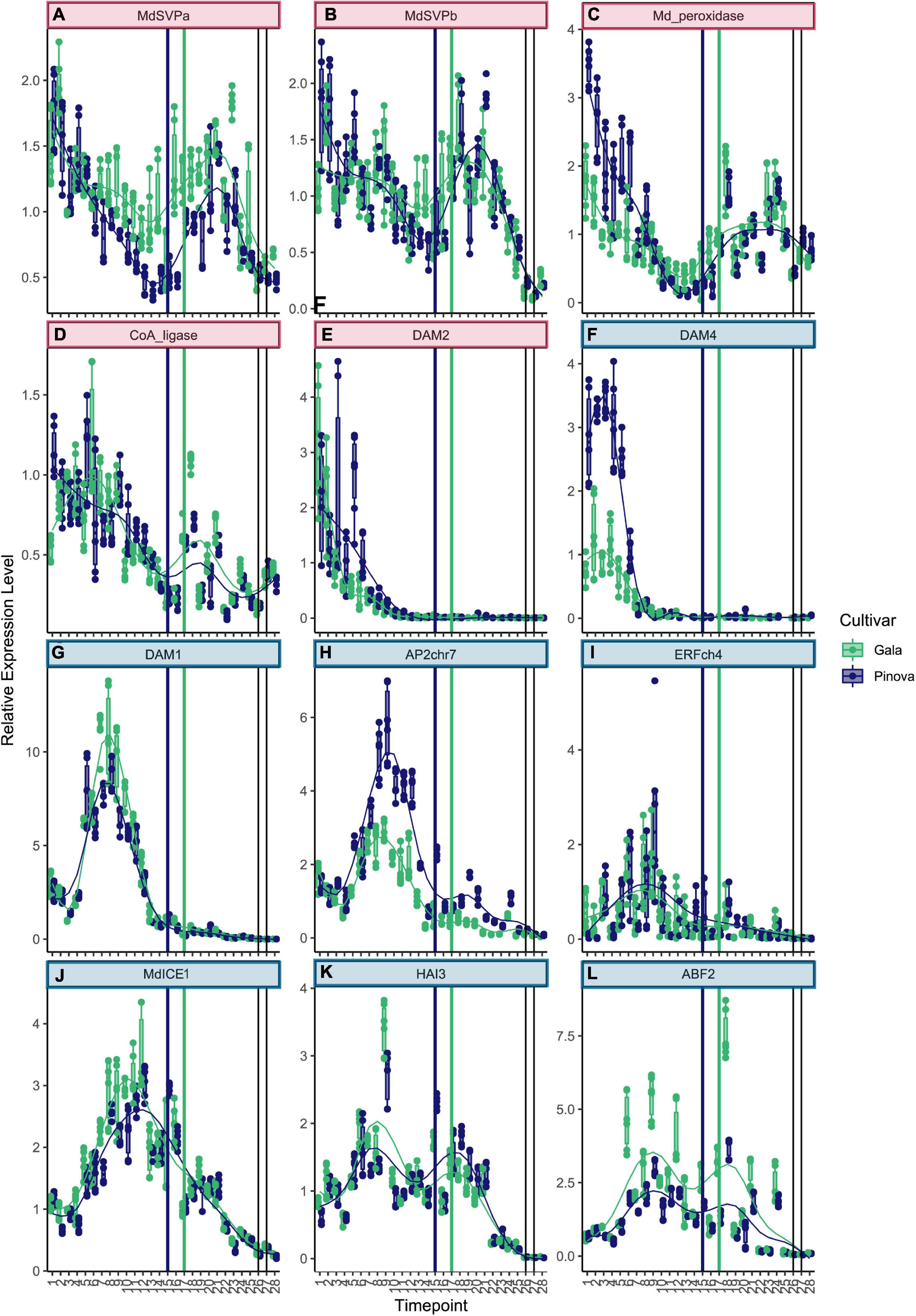
Figure 2. Expression patterns of genes belonging to clusters 1 and 2. (A) MdSVPa, MdSVPb (B), and Md_peroxidase (C) were highly expressed at the beginning of dormancy, expression levels dropped to a local minimum shortly before the endodormancy to ecodormancy phase transition, and raised again to peak during ecodormancy. (D) CoA-ligase showed a similar pattern. (E) MdDAM2 expression levels were highest at the first three sampling dates during early fall. (F) Expression of MdDAM4 peaked on October 18 in both cultivars, levels were higher in ‘Pinova’ (blue) compared with ‘Gala’ (green). (G) MdDAM1 peaked on November 23, similar to AP2chr7 (H) and ERFchr4 (I). (J) The expression curve of MdICE was broader. (K) The expression pattern of HAI3 and ABF2 (L) showed two peaks. Cluster number is indicated by colored box harboring the gene name. The colors of the boxes match the cluster colors in Figure 1. Blue and green dots represent ‘Pinova’ and ‘Gala’ samples, respectively. Vertical lines mark the transition from ecodormancy to endodormancy of ‘Pinova’ (blue, January 17) and ‘Gala’ (green, January 31, 2018). Gray vertical lines indicate the time of bud break. Numbers on the x-axis indicate sample collection dates specified in Supplementary Table 3.
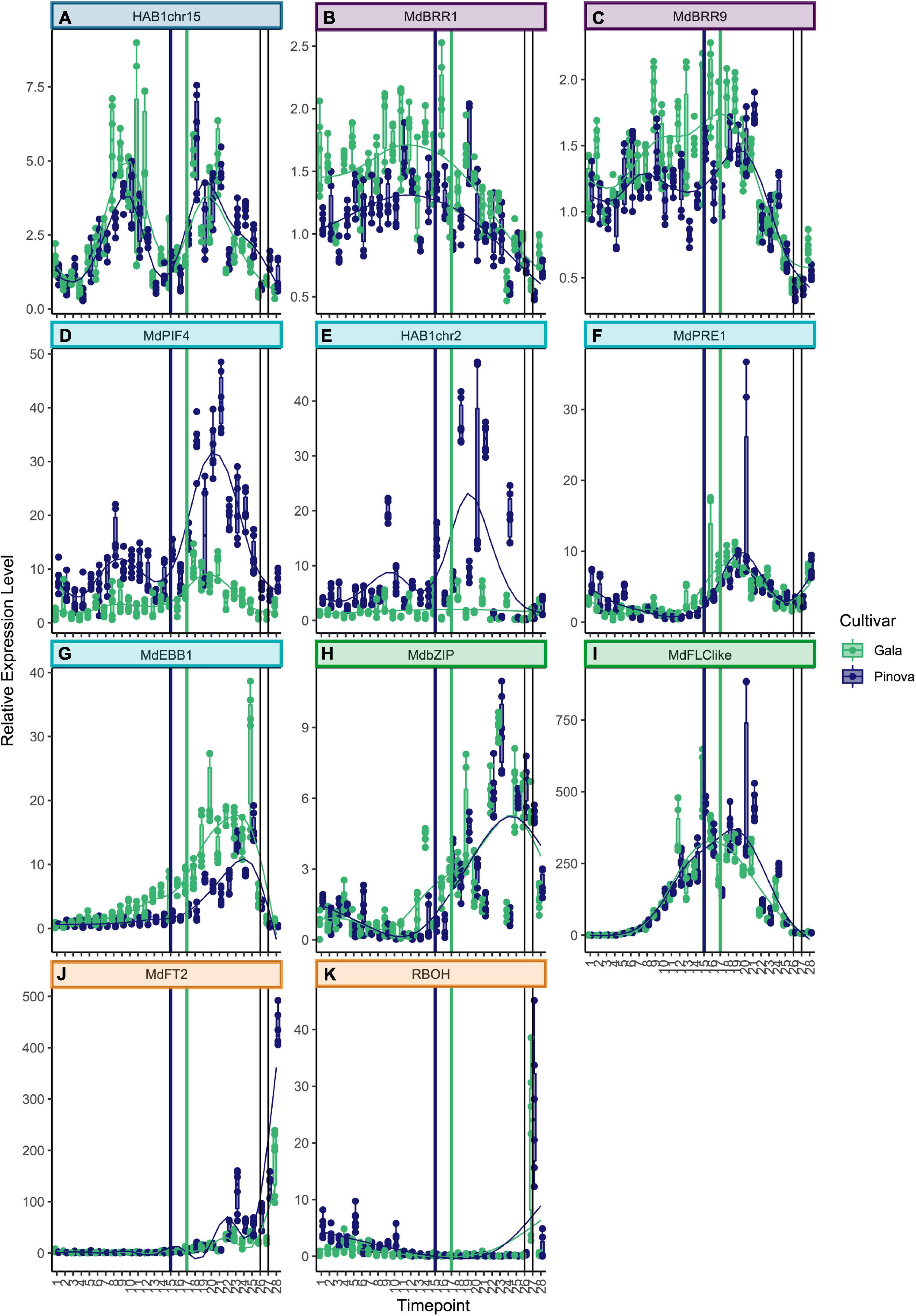
Figure 3. Expression patterns of genes belonging to clusters 2–6. (A) HAB1chr15 showed two similarly strong peaks of expression, on December 6 and February 14, and the local minimum was on January 10, 1 week before the ecodormancy to endodormancy transition. (B) MdBRR1 and MdBRR9 (C) were expressed broadly across dormancy and declined before bud break. (D) Expression of MdPIF4 as well as for HAB1chr2 (E) showed two peaks of which the first peak was lower than at the second peak, and ‘Pinova’ expression levels were higher compared to ‘Gala.’ (F) The expression of MdPRE1 was low during endodormancy and showed a maximum of expression on February 7 and 14. (G) The expression levels of MdEBB1 and MdbZIP peaked slightly later, on March 7 and March 14 (H). (I) MdFLC-like was expressed broadly and it peaked later in ‘Pinova’ compared with ‘Gala.’ (J) RBOH and MdFT2 (K) were not expressed during dormancy; however, they were strongly induced on bud break. The color scheme is identical to the one in Figure 2.
No expression could be detected for the three genes involved in GA metabolism, two GA3 oxidases, one GA2 oxidase, and for the two copies of the ABA biosynthesis genes ABA1 (MD02G1318300, MD07G1003000).
These results allow us to speculate on the expression differences between ‘Gala’ and ‘Pinova’ that could potentially cause the difference in dormancy phase transition between these cultivars. Prominent differences between cultivars can be found in the amplitude of expression levels rather than in a time shift. The observed expression pattern also indicates a reduction of expression during endodormancy to ecodormancy transition in several genes. Whether the incomplete reduction of MdSVPa expression in Gala during phase transition has an effect on the timing of phase transition could be tested in further experiments.
Dormancy progression highly depends on ambient temperatures – on chill unit accumulation during endodormancy and on the accumulation of heat units during ecodormancy. To gain insight into the mechanism that allows a temperature-dependent progression of the dormancy cycle, it was tested whether mean expression values of any gene in our field-collected samples correlate with temperature values (Supplementary Figure 6). Strong correlations with environmental temperature indicate the influence of temperature on the control of gene expression.
When using accumulated temperature data – accumulated GDH, accumulated CH, or CP – as an environmental variable, correlation analysis did not reveal any meaningful correlations. In order to be able to still use chilling as an environmental variable, we reduced the interval of temperature accumulation to shorter intervals. The most meaningful observed correlations were correlations with the sum of chill units that accumulated 2 weeks before sampling, as well as with mean daily temperature at sampling date (Figure 4A). Using mean daily temperature at the date of sampling for correlation analysis will likely not capture processes that rely on longer time spans. In this study, we expected to identify genes that are able to react to temperature change rather promptly. We imagined dormancy control to occur at multiple time scales and also that the successive progression of several short-term temperature-sensitive events could potentially underlie the observed long-term dormancy progression. Expression of MdICE1 was positively correlated with CH and showed the highest correlation coefficient of R = 0.617 (p < 0.05) (Figure 4B). This finding is in line with the knowledge of ICE1 (A. thaliana) being an upstream transcription factor that targets cold-responsive genes conferring freezing tolerance (Gilmour et al., 1998). The highest correlation of expression with mean daily temperature showed HAB1chr15, followed by MdFLC-like, MdBRR9, and MdPIF4. All four genes are negatively correlated with higher expression at lower temperatures (Figures 4C–F). The expression pattern of HAB1chr15 showed two peaks, which interestingly coincided very well with the lowest mean daily temperatures, resulting in the highest correlation coefficient with a mean daily temperature of all investigated genes (R = −0.754, p < 0.05). The expression of MdFLC-like was also highly correlated with mean daily temperature (R = −0.680, p < 0.05). Interestingly, MdFLC-like was only expressed when the mean daily temperature was below 9.2°C in these field-collected samples. These findings fit an increased MdFLC-like expression after cold treatment in chambers (Porto et al., 2015). The third highest correlation coefficient in correlations with mean temperature was revealed with the expression levels MdBRR9 (R = −0.634, p < 0.05). The strongest correlation with temperature showed the expression of MdPIF4, however, only after separating the two cultivars (R = −0.845 for ‘Gala’, R = −0.943 for ‘Pinova’, and R = −0.525 all data). This correlation confirms the temperature-sensitive expression variation observed previously (Takeuchi et al., 2018). Also, PIF4 is well known for its thermoregulatory role in A. thaliana, however, mostly for its activation by heat (Kumar et al., 2012). Further study is required to establish, whether allelic differences between ‘Pinova’ and ‘Gala’ MdPIF4 can confer variation in temperature sensitivity between the two cultivars and whether this difference underlies phenological variation between cultivars.
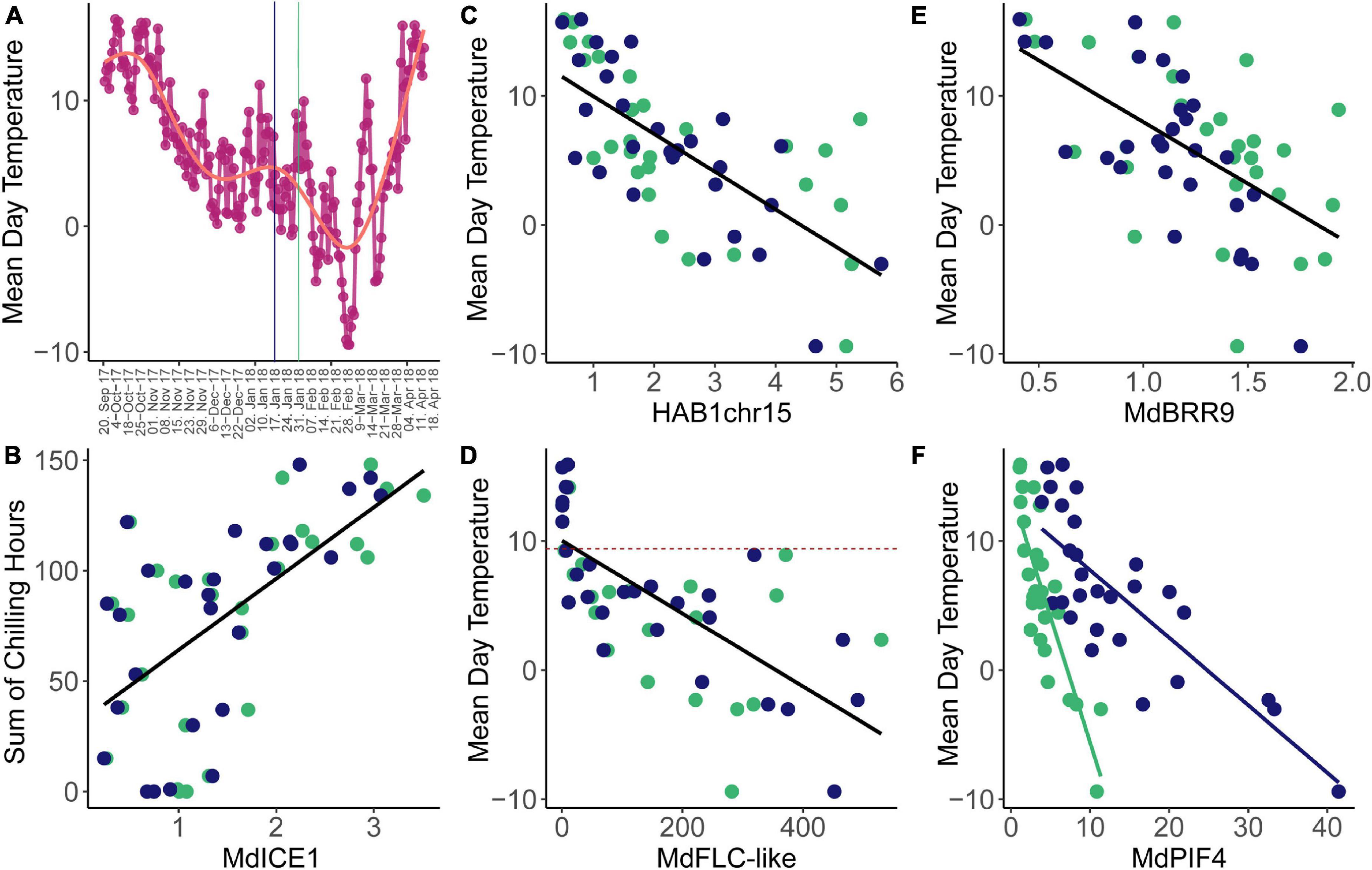
Figure 4. Correlations observed between expression levels and temperature traits. (A) Mean daily temperature varied from the first to the last time point of sampling. Vertical lines indicate the timing of the transition from endodormancy to ecodormancy as in Figures 1–3. (B) MdICE1 mean expression levels per time point correlated significantly with the amount of CH that accumulated during the 2 weeks before sampling. (C) HAB1chr16 mean expression levels were highly correlated with mean daily temperature, similar to MdFLC-like (D) and MdBRR9 (E). The dashed lines indicate the mean daily temperature of 9.2°C (D). (F) The slopes of the linear correlation differ between the two genotypes in MdPIF4 differed. Blue and green dots represent ‘Pinova’ and ‘Gala’ samples, respectively.
Gene expression data of dormancy candidate genes were presented, covering the seasons fall, winter, and spring with collected samples every week of two cultivars. In this study, the genes surveyed revealed a set of genes that showed continuous progression of overlapping expression patterns, from paradormancy via endodormancy and ecodormancy to bud break (Figure 5). The association of well-established dormancy genes with the distinct phases of dormancy was confirmed; however, evidence was also provided for genes associated with winter dormancy that are less well known for dormancy control. AP2chr7 and ERFchr4 expression levels associated with endodormancy, and several genes of the ABA signaling pathway (HAB1, HAI3, and ABF3) showed a distinct two-peak expression pattern that was revealed through dense sampling. Furthermore, high correlations of expression level with temperature values for MdICE1, HAB1chr15, MdFLC-like, MdBRR9, and MdPIF4 were identified. Interestingly, the slope of the linear correlation of temperature with the expression of MdPIF4 differed between cultivars. However, whether the two MdPIF4 alleles differ in their temperature sensitivity needs to be tested further.
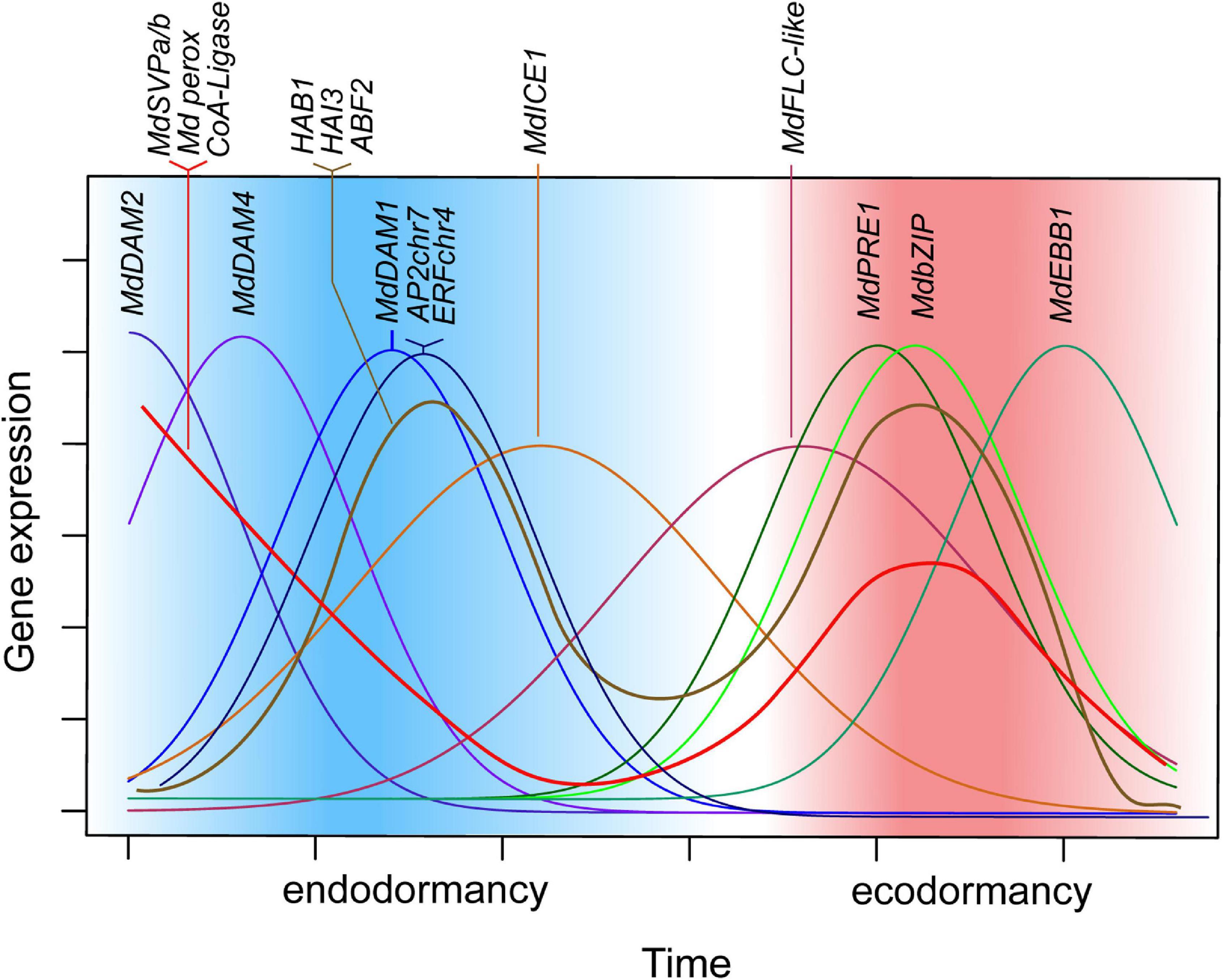
Figure 5. Schematic representation of candidate-gene expression patterns during dormancy progression. Expression of MdDAM2, MdDAM4, and MdDAM1, as well as AP2chr7 and ERFchr4, occurs in successive order with a single peak during endodormancy. Similarly, during ecodormancy, expression of MdPRE1, MdbZIP, and MdEBB1 show single, consecutive peaks of expression. Expression of MdSVPa, MdSVPb, MdPeroxidase, and CoA-Ligase is high at the beginning of dormancy and during mid ecodormancy and drops toward phase transitions of endodormancy to ecodormancy and of ecodormancy to bud break. Thus, it appears that multimeric complexes of MdSVPa/b with MdDAM1, MdDAM2, MdDAM4, and MdFLC-like can be formed all through dormancy but phase transitions. MdICE1 and MdFLC-like show broader expression curves that also occur in successive order; however, they are both highly expressed during endodormancy to ecodormancy transition.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
JL, AP, and HF designed the research. JL performed the experiments and analyzed the data. JL and HF wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the Federal Office for Agriculture and Food via project 2819BINZ01.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We are grateful to M. Görnitz for allowing us to sample apple buds in his orchard. We want to thank A. Lauber (JKI Dresden) for technical help, E. Zyprian and M. Schneider (JKI Siebeldingen, Germany) for making available and technical assistance with the Fluidigm BioMark HD system, and M. Kraatz (Saxon State Office for Environment, Agriculture and Geology, Nossen, Germany) for kindly providing temperature data.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.803341/full#supplementary-material
Allard, A., Bink, M. C., Martinez, S., Kelner, J. J., Legave, J. M., di Guardo, M., et al. (2016). Detecting QTLs and putative candidate genes involved in budbreak and flowering time in an apple multiparental population. J. Exp. Bot. 67, 2875–2888. doi: 10.1093/jxb/erw130
Anderson, J. L., Richardson, E. A., and Kesner, C. D. (1986). Validation of chill unit and flower bud phenology models for ‘Montmorency’ sour cherry. Acta Hortic. 184, 71–78.
Artlip, T., McDermaid, A., Ma, Q., and Wisniewski, M. (2019). Differential gene expression in nontransgenic and transgenic “M.26” apple overexpressing a peach CBF gene during the transition from eco-dormancy to bud break. Hortic. Res. 6:86. doi: 10.1038/s41438-019-0168-9
Bielenberg, D. G., Wang, Y., Li, Z. G., Zhebentyayeva, T., Fan, S. H., Reighard, G. L., et al. (2008). Sequencing and annotation of the evergrowing locus in peach [Prunus persica (L.) Batsch] reveals a cluster of six MADS-box transcription factors as candidate genes for regulation of terminal bud formation. Tree Genet. Genomes 4, 495–507. doi: 10.1007/s11295-007-0126-9
Bilavcik, A., Zamecnik, J., and Faltus, M. (2015). Cryotolerance of apple tree bud is independent of endodormancy. Front. Plant Sci. 6:695. doi: 10.3389/fpls.2015.00695
Cattani, A. M., da Silveira Falavigna, V., Silveira, C. P., Buffon, V., Dos Santos Maraschin, F., Pasquali, G., et al. (2020). Type-B cytokinin response regulators link hormonal stimuli and molecular responses during the transition from endo- to ecodormancy in apple buds. Plant Cell Rep. 39, 1687–1703. doi: 10.1007/s00299-020-02595-z
Celton, J. M., Martinez, S., Jammes, M. J., Bechti, A., Salvi, S., Legave, J. M., et al. (2011). Deciphering the genetic determinism of bud phenology in apple progenies: a new insight into chilling and heat requirement effects on flowering dates and positional candidate genes. New Phytol. 192, 378–392. doi: 10.1111/j.1469-8137.2011.03823.x
Chmielewski, F.-M., Blümel, K., and Pálešová, I. (2012). Climate change and shifts in dormancy release fordeciduous fruit crops in Germany. Clim. Res. 54, 209–219.
Chmielewski, F. M., Muller, A., and Bruns, E. (2004). Climate changes and trends in phenology of fruit trees and field crops in Germany, 1961-2000. Agric. For. Meteorol. 121, 69–78. doi: 10.1016/S0168-1923(03)00161-8
Cooke, J. E., Eriksson, M. E., and Junttila, O. (2012). The dynamic nature of bud dormancy in trees: environmental control and molecular mechanisms. Plant Cell Environ. 35, 1707–1728. doi: 10.1111/j.1365-3040.2012.02552.x
Coville, F. V. (1920). The influence of cold in stimulating the growth of plants. Proc. Natl. Acad. Sci. U.S.A. 6, 434–435. doi: 10.1073/pnas.6.7.434
Daccord, N., Celton, J. M., Linsmith, G., Becker, C., Choisne, N., Schijlen, E., et al. (2017). Highquality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nat. Genet. 49, 1099–1106. doi: 10.1038/ng.3886
Erez, A. (2000). “Bud dormancy; phenomenon, problems and solutions in the tropics and subtropics,” in Temperate Fruit Crops in Warm Climates, 1 Edn, ed. A. Erez (Dordrecht: Springer Netherlands), 17–48.
Erez, A., Fishman, S., Linsley-Noakes, G. C., and Allan, P. (1990). The dynamicmodel for rest completion in peach buds. Acta Hortic. 276, 165–174.
Estrella, N., Sparks, T. H., and Menzel, A. (2007). Trends and temperature response in the phenology of crops in Germany. Glob. Change Biol. 13, 1737–1747. doi: 10.1111/j.1365-2486.2007.01374.x
Fadon, E., Fernandez, E., Behn, H., and Luedeling, E. (2020). A conceptual framework for winter dormancy in deciduous trees. Agronomy 10:241. doi: 10.3390/agronomy10020241
Falavigna, V. D., Porto, D. D., Buffon, V., Margis-Pinheiro, M., Pasquali, G., and Revers, L. F. (2014). Differential transcriptional profiles of dormancy-related genes in apple buds. Plant Mol. Biol. Report. 32, 796–813. doi: 10.1007/s11105-013-0690-0
Falavigna, V. D. S., Guitton, B., Costes, E., and Andrés, F. (2019). I want to (bud) break free: the potential role of DAM and SVP-like genes in regulating dormancy cycle in temperate fruit trees. Front. Plant Sci. 9:1990. doi: 10.3389/fpls.2018.01990
Falavigna, V. D. S., Severing, E., Lai, X., Estevan, J., Farrera, I., Hugouvieux, V., et al. (2021). Unraveling the role of MADS transcription factor complexes in apple tree dormancy. New Phytol. 232, 2071–2088. doi: 10.1111/nph.17710
Faust, M., Liu, D. H., Millard, M. M., and Stutte, G. W. (1991). Bound versus free-water in dormant apple buds – a theory for endodormancy. Hortscience 26, 887–890. doi: 10.21273/Hortsci.26.7.887
Fishman, S., Erez, A., and Couvillon, G. A. (1987a). The temperature dependence of dormancy breaking in plants—computer simulation of processes studied under controlled temperatures. J. Theor. Biol. 126, 309–321.
Fishman, S., Erez, A., and Couvillon, G. A. (1987b). The temperature dependence of dormancy breaking in plants—mathematical analysis of a two-step model involving a cooperative transition. J. Theor. Biol. 124, 473–483.
Gilmour, S. J., Zarka, D. G., Stockinger, E. J., Salazar, M. P., Houghton, J. M., and Thomashow, M. F. (1998). Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J. 16, 433–442. doi: 10.1046/j.1365-313x.1998.00310.x
Heide, O. M. (2008). Interaction of photoperiod and temperature in the control of growth and dormancy of Prunus species. Sci. Hortic. 115, 309–314. doi: 10.1016/j.scienta.2007.10.005
Heide, O. M., and Prestrud, A. K. (2005). Low temperature, but not photoperiod, controls growth cessation and dormancy induction and release in apple and pea. Tree Physiol. 25, 109–114.
Kassambara, A. (2017). Practical Guide to Cluster Analysis in R. Available online at: http://www.sthda.com: STHDA (accessed November 5, 2021).
Kotoda, N., Hayashi, H., Suzuki, M., Igarashi, M., Hatsuyama, Y., Kidou, S., et al. (2010). Molecular characterization of FLOWERING LOCUS T-like genes of apple (Malusx domestica Borkh.). Plant Cell Physiol. 51, 561–575. doi: 10.1093/pcp/pcq021
Kumar, G., Arya, P., Gupta, K., Randhawa, V., Acharya, V., and Singh, A. K. (2016). Comparative phylogenetic analysis and transcriptional profiling of MADS-box gene family identified DAM and FLC-like genes in apple (Malusx domestica). Sci. Rep. 6:20695. doi: 10.1038/srep20695
Kumar, S. V., Lucyshyn, D., Jaeger, K. E., Alós, E., Alvey, E., Harberd, N. P., et al. (2012). Transcription factor PIF4 controls the thermosensory activation of flowering. Nature 484, 242–245. doi: 10.1038/nature10928
Lee, J. H., Jung, J. H., and Park, C. M. (2015). INDUCER OF CBF EXPRESSION 1 integrates cold signals into FLOWERING LOCUS C-mediated flowering pathways in Arabidopsis. Plant J. 84, 29–40. doi: 10.1111/tpj.12956
Li, J. Z., Yan, X. H., Ahmad, M., Yu, W. J., Song, Z. Z., Ni, J. B., et al. (2021). Alternative splicing of the dormancy-associated MADS-box transcription factor gene PpDAM1 is associated with flower bud dormancy in ‘Dangshansu’ pear (Pyrus pyrifolia white pear group). Plant Physiol. Biochem. 166, 1096–1108. doi: 10.1016/j.plaphy.2021.07.017
Luedeling, E., Zhang, M. H., and Girvetz, E. H. (2009). Climatic changes lead to declining winter chill for fruit and nut trees in California during 1950-2099. PLoS One 4:e6166. doi: 10.1371/journal.pone.0006166
Maechler, M., Rousseeuw, P., Struyf, A., Hubert, M., and Hornik, K. (2021). cluster: Cluster Analysis Basics and Extensions [Online]. Available online at: https://CRAN.R-project.org/package=cluster (accessed November 5, 2021).
Malagi, G., Sachet, M. R., Citadin, I., Herter, F. G., Bonhomme, M., Regnard, J. L., et al. (2015). The comparison of dormancy dynamics in apple trees grown under temperate and mild winter climates imposes a renewal of classical approaches. Trees Struct. Funct. 29, 1365–1380. doi: 10.1007/s00468-015-1214-3
Maurya, J. P., and Bhalerao, R. P. (2017). Photoperiod- and temperature-mediated control of growth cessation and dormancy in trees: a molecular perspective. Ann. Bot. 120, 351–360. doi: 10.1093/aob/mcx061
Milyaev, A., Kofler, J., Klaiber, I., Czemmel, S., Pfannstiel, J., Flachowsky, H., et al. (2021). Toward systematic understanding of flower bud induction in apple: a multi-omics approach. Front. Plant Sci. 12:604810. doi: 10.3389/fpls.2021.604810
Mimida, N., Saito, T., Moriguchi, T., Suzuki, A., Komori, S., and Wada, M. (2015). Expression of DORMANCY-ASSOCIATED MADS-BOX (DAM)-like genes in apple. Biol. Plant. 59, 237–244. doi: 10.1007/s10535-015-0503-4
Miotto, Y. E., Tessele, C., Czermainski, A. B. C., Porto, D. D., Falavigna, V. D. S., Sartor, T., et al. (2019). Spring is coming: genetic analyses of the bud break date locus reveal candidate genes from the cold perception pathway to dormancy release in apple (Malusx domestica Borkh.). Front. Plant Sci. 10:33. doi: 10.3389/fpls.2019.00033
Moser, M., Asquini, E., Miolli, G. V., Weigl, K., Hanke, M. V., Flachowsky, H., et al. (2020). The MADS-box gene MdDAM1 controls growth cessation and bud dormancy in apple. Front. Plant Sci. 11:1003. doi: 10.3389/fpls.2020.01003
Nishiyama, S., Matsushita, M. C., Yamane, H., Honda, C., Okada, K., Tamada, Y., et al. (2021). Functional and expressional analyses of apple FLC-like in relation to dormancy progress and flower bud development. Tree Physiol. 41, 562–570. doi: 10.1093/treephys/tpz111
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. doi: 10.1093/nar/29.9.e45
Porto, D. D., Bruneau, M., Perini, P., Anzanello, R., Renou, J. P., dos Santos, H. P., et al. (2015). Transcription profiling of the chilling requirement for bud break in apples: a putative role for FLC-like genes. J. Exp. Bot. 66, 2659–2672. doi: 10.1093/jxb/erv061
Quesada-Traver, C., Guerrero, B. I., Badenes, M. L., Rodrigo, J., Rios, G., and Lloret, A. (2020). Structure and expression of bud dormancy-associated MADS-box genes (DAM) in European Plum. Front. Plant Sci. 11:1288. doi: 10.3389/fpls.2020.01288
Reim, S., Rohr, A. D., Winkelmann, T., Weiss, S., Liu, B. Y., Beerhues, L., et al. (2020). Genes involved in stress response and especially in phytoalexin biosynthesis are upregulated in four malus genotypes in response to apple replant disease. Front. Plant Sci. 10:1724. doi: 10.3389/fpls.2019.01724
Richardson, E. A., Seeley, S. D., and Walker, D. R. (1974). A model for estimating the completion of rest for “Redhaven” and “Elberta” peach trees. Hortic Sci. 9, 331–332.
Rothkegel, K., Sanchez, E., Montes, C., Greve, M., Tapia, S., Bravo, S., et al. (2017). DNA methylation and small interference RNAs participate in the regulation of MADS-box genes involved in dormancy in sweet cherry (Prunus avium L.). Tree Physiol. 37, 1739–1751. doi: 10.1093/treephys/tpx055
Saure, M. C. (1985). “Dormancy release in desciduous fruit trees,” in Horticultural Reviews, ed. J. Janick (Westport, CT: Wiley).
Singh, R. K., Svystun, T., AlDahmash, B., Jönsson, A. M., and Bhalerao, R. P. (2017). Photoperiod- and temperature-mediated control of phenology in trees - a molecular perspective. New Phytol. 213, 511–524. doi: 10.1111/nph.14346
Takemura, Y., Kuroki, K., Shida, Y., Araki, S., Takeuchi, Y., Tanaka, K., et al. (2015). Comparative transcriptome analysis of the less-dormant Taiwanese pear and the dormant Japanese pear during winter season. PLoS One 10:e0139595. doi: 10.1371/journal.pone.0139595
Takeuchi, T., Matsushita, M. C., Nishiyama, S., Yamane, H., Banno, K., and Tao, R. (2018). RNA-sequencing analysis identifies genes associated with chilling-mediated endodormancy release in apple. J. Am. Soc. Hort. Sci. 143:194. doi: 10.21273/jashs04345-18
Tanino, K. K., Kalcsits, L., Silim, S., Kendall, E., and Gray, G. R. (2010). Temperature-driven plasticity in growth cessation and dormancy development in deciduous woody plants: a working hypothesis suggesting how molecular and cellular function is affected by temperature during dormancy induction. Plant Mol. Biol. 73, 49–65. doi: 10.1007/s11103-010-9610-y
Tuan, P. A., Bai, S. L., Saito, T., Imai, T., Ito, A., and Moriguchi, T. (2016). Involvement of EARLY BUD-BREAK, an AP2/ERF transcription factor gene, in bud break in Japanese pear (Pyrus pyrifolia Nakai) lateral flower buds: expression, histone modifications and possible target genes. Plant Cell Physiol. 57, 1038–1047. doi: 10.1093/pcp/pcw041
Urrestarazu, J., Muranty, H., Denancé, C., Leforestier, D., Ravon, E., Guyader, A., et al. (2017). Genome-wide association mapping of flowering and ripening periods in apple. Front. Plant Sci. 8:1923. doi: 10.3389/fpls.2017.01923
van Dyk, M. M., Soeker, M. K., Labuschagne, I. F., and Rees, D. J. G. (2010). Identification of a major QTL for time of initial vegetative budbreak in apple (Malus x domestica Borkh.). Tree Genet. Genomes 6, 489–502.
Vimont, N., Fouché, M., Campoy, J. A., Tong, M., Arkoun, M., Yvin, J. C., et al. (2019). From bud formation to flowering: transcriptomic state defines the cherry developmental phases of sweet cherry bud dormancy. BMC Genomics 20:974. doi: 10.1186/s12864-019-6348-z
Wei, T., and Simko, V. (2021). R Package ‘Corrplot’: Visualization of a Correlation Matrix [Online]. Available online at: https://github.com/taiyun/corrplot (accessed April 10, 2021).
Weinberger, J. H. (1950). Chilling requirements of peach varieties. J. Am. Soc. Hort. Sci. 56, 11–16.
Willige, B. C., Zander, M., Yoo, C. Y., Phan, A., Garza, R. M., Trigg, S. A., et al. (2021). PHYTOCHROME-INTERACTING FACTORs trigger environmentally responsive chromatin dynamics in plants. Nat. Genet. 53, 955–961. doi: 10.1038/s41588-021-00882-3
Wisniewski, M., Norelli, J., and Artlip, T. (2015). Overexpression of a peach CBF gene in apple: a model for understanding the integration of growth, dormancy, and cold hardiness in woody plants. Front. Plant Sci. 6:85. doi: 10.3389/fpls.2015.00085
Wu, R., Cooney, J., Tomes, S., Rebstock, R., Karunairetnam, S., Allan, A. C., et al. (2021). RNAimediated repression of dormancy-related genes results in evergrowing apple trees. Tree Physiol. 41, 1510–1523. doi: 10.1093/treephys/tpab007
Wu, R., Tomes, S., Karunairetnam, S., Tustin, S. D., Hellens, R. P., Allan, A. C., et al. (2017). SVP-like MADS box genes control dormancy and budbreak in apple. Front. Plant Sci. 8:477. doi: 10.3389/fpls.2017.00477
Xie, F., Xiao, P., Chen, D., Xu, L., and Zhang, B. (2012). miRDeepFinder: a miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. doi: 10.1007/s11103-012-9885-2
Yordanov, Y. S., Ma, C., Strauss, S. H., and Busov, V. B. (2014). EARLY BUD-BREAK 1 (EBB1) is a regulator of release from seasonal dormancy in poplar trees. Proc. Natl. Acad. Sci. U.S.A. 111, 10001–10006. doi: 10.1073/pnas.1405621111
Yu, J., Conrad, A. O., Decroocq, V., Zhebentyayeva, T., Williams, D. E., Bennett, D., et al. (2020). Distinctive gene expression patterns define endodormancy to ecodormancy transition in apricot and peach. Front. Plant Sci. 11:180. doi: 10.3389/fpls.2020.00180
Zhao, K., Zhou, Y. Z., Ahmad, S., Xu, Z. D., Li, Y. S., Yang, W. R., et al. (2018). Comprehensive cloning of Prunus mume dormancy associated MADS-box genes and their response in flower bud development and dormancy. Front. Plant Sci. 9:17. doi: 10.3389/fpls.2018.00017
Keywords: apple, dormancy, temperature, DAM genes, endodormancy, ecodormancy
Citation: Lempe J, Peil A and Flachowsky H (2022) Time-Resolved Analysis of Candidate Gene Expression and Ambient Temperature During Bud Dormancy in Apple. Front. Plant Sci. 12:803341. doi: 10.3389/fpls.2021.803341
Received: 27 October 2021; Accepted: 24 November 2021;
Published: 17 January 2022.
Edited by:
Fernando Andrés, INRA UMR Amélioration Génétique et Adaptation des Plantes Méditerranéennes et Tropicales, FranceReviewed by:
Timothy Artlip, Appalachian Fruit Research Station (ARS-USDA), United StatesCopyright © 2022 Lempe, Peil and Flachowsky. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Janne Lempe, amFubmUubGVtcGVAanVsaXVzLWt1ZWhuLmRl
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.