- 1School of Life Sciences and Food Engineering, Hebei University of Engineering, Handan, China
- 2Institute of Environment and Sustainable Development in Agriculture, Chinese Academy of Agricultural Sciences, Beijing, China
- 3College of Natural Resources and Environment, Northwest Agriculture and Forestry University, Yangling, China
- 4Key Laboratory of Beijing for Identification and Safety Evaluation of Chinese Medicine, Institute of Chinese Material Medica, China Academy of Chinese Medical Sciences, Beijing, China
- 5State Key Laboratory for Biology of Plant Diseases and Insect Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing, China
In this study, the capacity to tune root morphogenesis by a plant growth-promoting rhizobacterium, Streptomyces lincolnensis L4, was investigated from various aspects including microbial physiology, root development, and root endophytic microbial community. Strain L4 was isolated from the root-associated soil of 7-year plantation of Artemisia annua. Aiming at revealing the promotion mechanism of Streptomyces on root growth and development, this study first evaluated the growth promotion characters of S. lincolnensis L4, followed by investigation in the effect of L4 inoculation on root morphology, endophytic microbiota of root system, and expression of genes involved in root development in Arabidopsis thaliana. Streptomyces lincolnensis L4 is able to hydrolyze organic and inorganic phosphorus, fix nitrogen, and produce IAA, ACC deaminase, and siderophore, which shaped specific structure of endophytic bacterial community with dominant Streptomyces in roots and promoted the development of roots. From the observation of root development characteristics, root length, root diameter, and the number of root hairs were increased by inoculation of strain L4, which were verified by the differential expression of root development-related genes in A. thaliana. Genomic traits of S. lincolnensis L4 which further revealed its capacity for plant growth promotion in which genes involved in phosphorus solubilization, ACC deamination, iron transportation, and IAA production were identified. This root growth-promoting strain has the potential to develop green method for regulating plant development. These findings provide us ecological knowledge of microenvironment around root system and a new approach for regulating root development.
Introduction
Rhizosphere is the soil zone surrounding plant root, which derive but differ from bulk soil (Hartmann et al., 2007). The interaction between root and rhizosphere soil shaped specific physical and chemical character of rhizosphere soil, and also the structure and function of microbial community (Fan et al., 2020). As a consequence, rhizosphere microbes also take effect on the growth and development of host plant. It has been intensively reported that microbes associated with root systems are diverse and usually demonstrate beneficial activity to host plant by stimulating plant growth, reducing pathogenesis, and alleviating abiotic stresses, which are assigned as plant growth-promoting rhizobacteria (PGPR). These PGPR are crucial component of phytomicrobiome that is associated with physiological regulation in plants (Smith et al., 2015). The function of rhizosphere microbes in plant phenotype regulation has important implications for plant phenology in various climates and for increasing agricultural production (Singh et al., 2010).
During the interaction between rhizosphere microbes and host plant, auxins synthesized by rhizosphere microorganisms play a critical role (Reyes, 2006; Li et al., 2018; Treesubsuntorn et al., 2018). The function of auxins from microorganisms as phytohormone raised the intriguing possibility for regulating plant growth and development by utilizing rhizosphere microbes or synthetic microbial consortium (Chen et al., 2017). According to the study of isolated PGPR and their physiological trait, the function of PGPR has been reported as inhibiting phytopathogens (Rai and Nabti, 2017), improving plant nutrient supply, producing phytohormones, and modifying physicochemical properties of soil (Khan et al., 2020). On the other hand, these PGPR strains are also able to enhance the disease resistance of plants through changing soil microbial community (Bharti et al., 2015; Wang et al., 2020).
Streptomyces spp. are ubiquitous actinobateria in soils, rhizosphere, and endophytic microbiota. At present, the applications of Streptomyces in agriculture are mainly as biocontrol agents (Yuan and Crawford, 1995). However, for application as a PGPR, the promotion mechanism of Streptomyces still needs further investigation. It has been reported in some studies that Streptomyces is able to promote plant growth by the producing 1-aminocyclopropane-1-carboxylate deaminase (ACC), indole-3-acetic acid (IAA), and siderophores to cope with abiotic stress (El-Tarabily, 2008; Rungin et al., 2012). However, little is known about the role of Streptomyces in host root development (Wu et al., 2019).
In our previous study, the increased abundance of actinobacteria was observed in root-associated soil with 7-year plantation of Artemisia annua (Shi et al., 2021). It is the only raw material for the production of artemisinin, which is the most effective drug to treat malaria so far. The enrichment of actinobacteria implied that they may have connection with the growth of A. annua. Thus, in this study, the actinobacterial strains that harbor plant growth-promoting character were isolated from the root-associated soil with 7-year plantation of A. annua. Specifically, the function in plant growth promotion and root development of actinobacterial isolate was further investigated.
Materials and Methods
Soil Sampling and Bacteria Isolation
The root-associated soils of A. annua YQ2 were sampled from the field with 7-year plantation history at Rong’an in China. Due to the annual plantation of A. annua, the soils that have been plowed every year were actually the mixture of 7-year accumulated rhizosphere soil. Five rhizosphere sample replicates were collected by shaking off the soil which tightly adhered to root. The replicate that showed the highest enrichment of actinobacteria in our previous study (Shi et al., 2021) was selected for bacterial isolation in this study.
For the isolation of actinobacteria, one gram of homogenized rhizosphere soil was inoculated to LB liquid medium and incubated in 28°C with 150 rpm rotary shaking for 24 h. The enriched soil microbial culture was diluted for 10, 100 and 1,000 times gradient. These diluted liquid cultures (200 μL for each sample) were spread on actinobacteria selecting Gause’s agar medium (20 g L–1 soluble starch, 1 g–1 KNO3, 0.5 g L–1 K2HPO4, 0.5 g L–1 MgSO4 ⋅ 7H2O, 0.5 g L–1 NaCl, 0.01 g L–1 FeSO4 ⋅ 7H2O, 20 g L–1 agar, 1 L of distilled water, pH = 7.4–7.6) and incubated in 28°C for 5 days.
Identification of Actinomyces
The colonies from selective medium were picked and cultured rotationally in 5 ml of liquid LB medium for 48 h at 30°C. Genomic DNA of each culture was isolated by bacterial DNA extraction kit (OMEGA Bacterial DNA Kit). The purified DNA was resuspended in TE buffer and stored at -20°C for further analysis.
For identifying taxonomy of isolated strains, the sequence of 16S rRNA gene was amplified using the bacterial-specific primers, 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′). Polymerase chain reaction (PCR) amplifications were performed with 2 × Taq PCR Mix (TIANGEN BIOTECH, China), 1.0 μM of each primer, genomic DNA as template and ddH2O. Three independent PCR amplifications for each isolate were performed for 30 cycles with denaturation at 94°C (60 s), annealing at 55°C (30 s), and extension at 72°C (60 s), followed by a final extension at 72°C (10 min). The resulted PCR products were sequenced with platform (SinoGenoMax, China). The taxonomy of each isolate was confirmed by the alignment of these amplified sequences with NR database in GenBank with BLAST and the comparison with nucleotide sequence homology of 16S rRNA gene for bacteria. Highly homologous sequences were aligned, and neighbor joining trees were generated using Molecular Evolutionary Genetics Analysis X.
Microbial Physiological Assay of Bacterial Isolates
Phosphate Solubilization
Phosphate solubilization abilities of all isolates were screened according to the method of Surange et al. (1997). The tests were carried out in triplicates. The presence of lysis circle around the bacterial colony was observed as indicator of phosphate solubilization. The diameters of the lysis circle and colony were measured digitally. The ability of phosphate solubilization was represented by phosphorus solubilizing index (SI) based on the definition: SI = (diameter of halo zone + colony diameter)/colony diameter.
Nitrogen Fixation
The capability of autotrophic nitrogen fixation in isolated strains was characterized by detecting their growth in nitrogen-free medium. The isolated bacteria were inoculated in nitrogen-free medium (5.0 g mannitol or glucose L–1, 0.2 g KH2PO4 L–1, 0.2 g MgSO4 ⋅ 7H2O L–1, 0.2 g NaCl L–1, 0.2 g CaSO4 ⋅ 2H2O L–1, 5 g CaCO3 L–1, 15 g agar L–1, 1 L of distilled water, pH = 7.0–7.2). These cultures were incubated on the nitrogen-free medium at 30°C for 5 days. The formation of bacterial colony was defined as the ability of nitrogen fixation.
Siderophore Production
According to the method of Schwyn and Nelands (1987), the blue agar chrome azurol S (CAS) medium was used to characterize the siderophore production potential of all bacterial isolates (Alexander and Zuberer, 1991). Quantification of siderophore produced from each isolated strain was performed in liquid medium supplemented with CAS. These isolates with a yellowish orange halo were further inoculated into liquid MKB medium and incubated at 30°C for 3 days. Then, cell-free supernatants from the culture were collected by centrifugation (10,000 × g for 5 min), and 3 mL of the bacterial supernatant was added into the same amount of CAS solution, followed by dark incubation for 1 h. The uninoculated supernatant was used as blank control for comparison and calculation. All treatments were performed in triplicates. Absorbance of the isolates was measured at a wave length of 630 nm using spectrophotometer. The ratio, A/Ar (OD630 of treatments/OD630 of blank control), represented the capacity of producing siderophores.
Indole-3-Acetic Acid Production
The IAA production capacities of the strains were determined according to the method of Patten and Glick (2002). The IAA concentration was calculated from a calibration curve of IAA standard solution ranging from 0 to 20 mg/L (Supplementary Figure 1).
1-Aminocyclopropane-1-Carboxylate Deaminase Activity Assay
For investigating the activity of ACC deaminase, each isolated strain was inoculated on ADF solid medium and cultured at 30°C for 7 days. If the strain could grow, it was transferred to the liquid medium with ACC as the sole nitrogen source (Jaemsaeng et al., 2018). The strain that kept growing after three generations was defined as ACC deaminase active strain.
Genome Sequencing and Annotation
Genome Sequencing
Genomic DNA was extracted with the SDS method (Lim et al., 2016). Sequencing libraries were generated using NEBNext® Ultra™ DNA Library Prep Kit for Illumina (NEB, United States) referring to the manufacturer’s recommendations. Purified libraries (AMPure XP system) were analyzed for size distribution by Agilent 2100 Bioanalyzer and quantified using real-time PCR. Library sequencing was performed with Illumina HiSeq/NovaSeq PE150 platform at the Beijing Allwegene Technology Co., Ltd. Quality filtered paired reads were assembled by the SPAdes (v3.13.0) (Bankevich et al., 2012) software into a number of scaffolds. Finally, scaffolds with larger than 500 bp were selected for subsequent analysis.
Genome Annotation
The genome was annotated using Kyoto Encyclopedia of Genes and Genomes (KEGG; Kanehisa et al., 2021). Genome component including the coding gene, repetitive sequences, noncoding RNA, protein signal peptide, secretory protein, pseudogene, prophage, genomics islands, and clustered regularly interspaced short palindromic repeat (CRISPR) sequences were comprehensively predicted. Coding genes involved in plant growth promoting were screened manually.
Measurement of Plant Growth and Root Development
The gnotobiotic root elongation assay was applied for assessing the effect of bacterial isolates on the growth of Arabidopsis thaliana Col-0 seedlings. Col-0 seeds were surface sterilized before cultivation. The seeds (approximately 0.2 g per treatment) were washed in 70% ethanol for 1 min followed by washing with 1% sodium hypochlorite for 8 min. These sterilized seeds were further rinsed with sterile distilled water at least five times for removing sterilizing reagent. After sterilization, 15 seeds were placed in half-cut MS agar medium with sterilized scalpel and tweezer. The supernatant of 100 μL bacterial solution that was precultured in liquid LB (after 3 days of activation in LB liquid medium, stand for about 2 h, wait for the cell to precipitate naturally, and measure the number of supernatant spores by coating method, 400–600 mL–1) was inoculated on the surface of seeds (liquid LB and sterile water were used as control). Each plate was placed vertically in a 28°C greenhouse, with 12-h light/dark cycles (18 μmol m–2 s–1). The primary root lengths were measured after 21 days.
Bacterial Communities Associated With the Roots of Arabidopsis thaliana
Root Sampling
The Arabidopsis plants in plate were sampled and washed with sterile water to clean the medium residues. The roots were collected for further endophytic microbial community analysis. All treatments were conducted in triplicates.
DNA Extraction, Amplification, and Sequencing
Total genomic DNA was extracted using DNA isolation kit (MoBio Laboratories, Carlsbad, CA, United States) following the manual. Purity and quality of the genomic DNA were checked on 1% agarose gels and NanoDrop spectrophotometer (Thermo Scientific). The bacterial 16S rRNA gene (V3–V4) was amplified by primer pairs 338F (5’-ACTCCTACGGGAGGCAGCAG-3’)/806R (5’-GGACTACHVGGGTWTCTAAT-3’) (Xu et al., 2016). The PCR was carried out on a Mastercycler Gradient (Eppendorf, Germany) (Yan et al., 2022). Amplicon sequencing was performed on Miseq platform at Allwegene, Beijing. Index and barcode sequences were removed from resulted reads, and quality was controlled using Illumina Analysis Pipeline version 2.6.
All raw data sequenced were deposited in the NCBI Sequence Read Archive (SRA) with the accession number PRJNA761252.
Sequence Analysis and Annotation
The raw data were first screened by removing the sequences shorter than 230 bp or with a low-quality score (≤20). These filtered sequences were separated using the sample-specific barcode sequences. Qualified reads were clustered into operational taxonomic units (OTUs) at a similarity level of 97% (Edgar, 2013) using Uparse algorithm of Vsearch (v2.7.1) software.
QIIME (v1.8.0) was used to generate rarefaction curves and to calculate the richness and diversity indices based on the OTU information. To compare the number of communities in samples of different components and structures, stacked bar chart was built (Jami et al., 2013). Based on the results of taxonomic annotation and relative abundance, R (v3.6.0) software was used for bar-plot diagram analysis.
Quantitative PCR of Root Development-Related Genes in Arabidopsis thaliana
The treated and control A. thaliana was sampled after 21 days of incubation and washed with sterile water to clean off the attached medium. The root samples were cut and grinded directly in the Total RNA Extractor (Sangon Biotech) with a proportion as 100 mg samples per 1mL solution. Total RNA was extracted according to the instructions (Sangon Biotech). Qualified RNA was reversely transcribed into cDNA by reverse transcription kit, and then the genes controlling root length and root hair were quantified.
The cDNA template was diluted 10–20 times, according to the instructions of SYBR Premix Ex Taq™ (perfect real time) (Takara) kit, and PCR (CFX96TOUCH) experiments were performed. PCR amplifications were performed with 2 × Taq SYBR Green qPCR Premix (Takara, Japan), 1.0 μM of each primer, genomic DNA as template and ddH2O. Three independent PCR amplifications for each isolate were performed for 45 cycles with denaturation at 94°C (30 s), annealing at 57°C (30 s), and extension at 72°C (30 s). Each experiment was performed in triplicate. The primer sequence for quantitative PCR was shown in Supplementary Table 1. Gene encoding 18S rRNA was used as internal reference for calibration and comparison.
Results
Streptomyces spp. Isolated From Root-Associated Soil in Artemisia annua
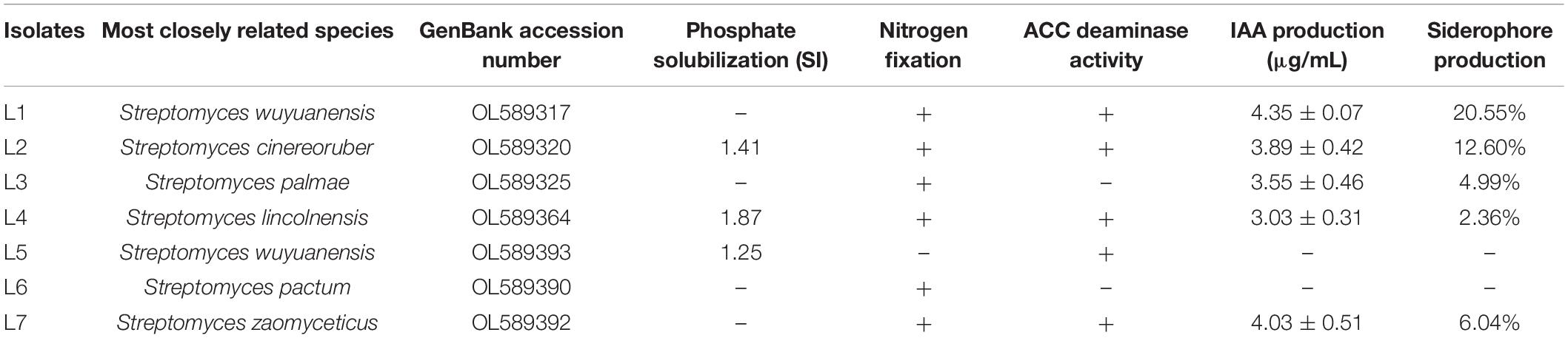
According to the full-length sequences of 16S rRNA genes, seven isolated actinobacterial strains in total were identified. All of these strains were assigned in Streptomyces genus with the identity of 16S rRNA gene > 97% as criteria. Based on the closest species in the sequences of 16S rRNA gene, the seven Streptomyces isolates were assigned to species level and named as Streptomyces wuyuanensis L1, Streptomyces cinereoruber L2, Streptomyces palmae L3, Streptomyces lincolnensis L4, Streptomyces wuyuanensis L5, Streptomyces pactum L6, and Streptomyces zaomyceticus L7 (Table 1). The phylogenetic relationship based on full sequences of 16S rRNA genes of all seven Streptomyces with corresponding type strains confirmed their taxonomy (Figure 1).
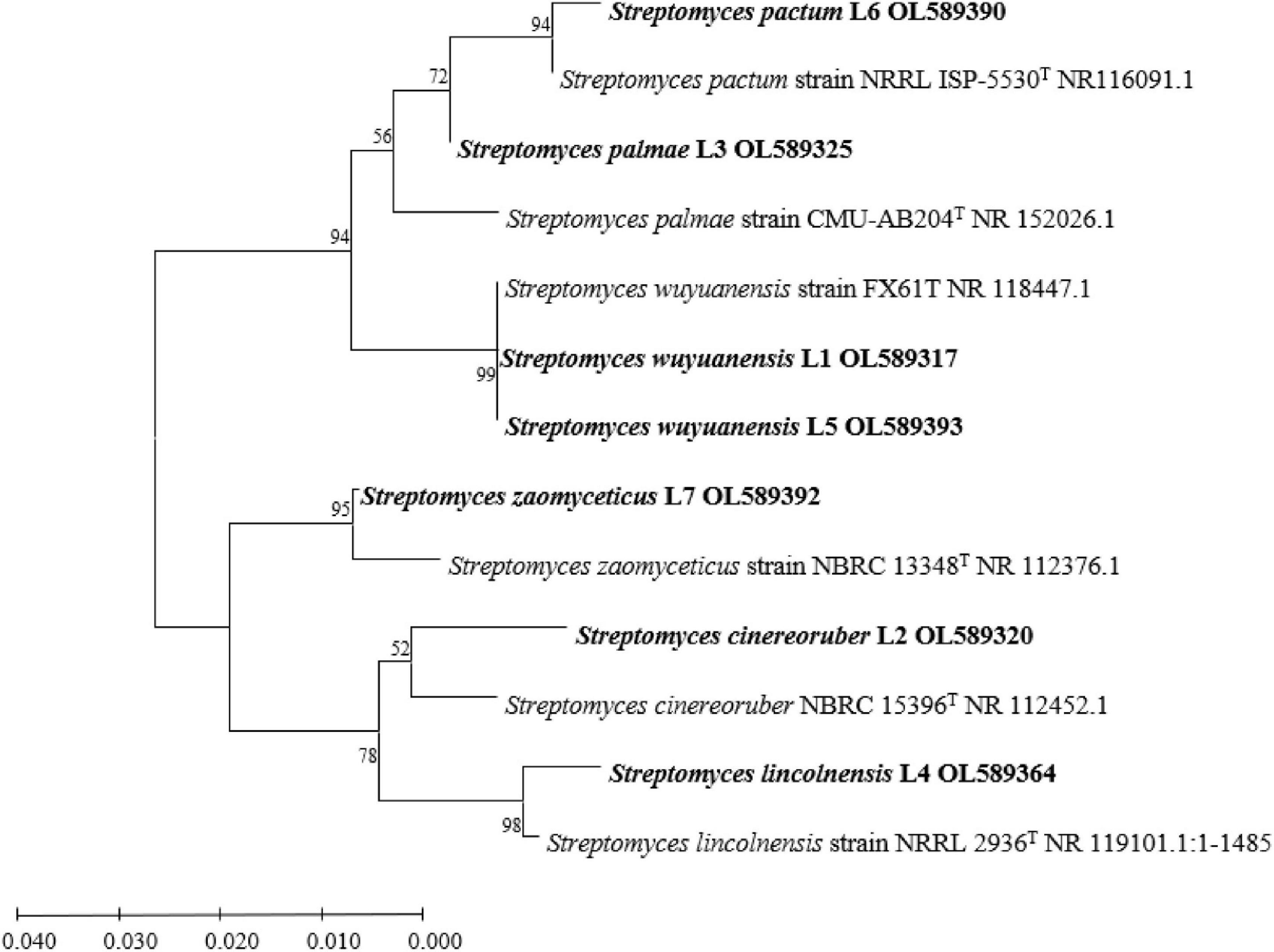
Figure 1. Phylogenetics tree based on16S rRNA gene sequence of isolates L1–L7 and Streptomyces type strains.
Plant Growth Promoting Potential of Isolated Streptomyces
The plant growth promoting potential of seven isolated Streptomyces was characterized in vitro by determining qualitatively and/or quantitatively the ability of phosphate solubilization, nitrogen fixation, ACC deaminase, IAA, and iron carrier production (Table 1).
Phosphate Solubilization
Strains L2, L4, and L5 among these seven isolated strains were identified as phosphate solubilizing microbes according to their ability to solubilize inorganic phosphorus with the ratio between halo plus colony diameter and colony diameter of 1.41, 1.87, and 1.25, respectively. In addition, strain L4 has the ability to dissolve organic phosphorus (Supplementary Figure 2).
Nitrogen Fixation
Strains L1, L2, L3, L4, L6, and L7, except for strain L5, exhibited their nitrogen fixation ability judged by their apparent growth on nitrogen-free medium after at least three continuous transferring (Supplementary Figure 3).
Indole-3-Acetic Acid Production
The concentration of IAA in the culture of strain L1 kept increased after inoculation and reached to the highest value of 4.35 ± 0.07 μg/mL on the 10th day. The IAA concentration of strain L2 increased to the highest on the sixth day and stabilized with the value of 3.89 ± 0.42 μg/mL. Exceptionally, due to that strain L3 began to produce pigment on the fifth day and interfered the measurement of IAA, the highest value of IAA was observed on the fifth day as 3.55 ± 0.46 μg/mL. The production of IAA in strain L4 reached to the highest on the sixth day and decreased on the ninth day, with the highest value of 3.03 ± 0.31 μg/mL. Strain L7 also began to produce pigment on the seventh day, so the highest value of IAA, 4.03 ± 0.51 μg/mL, was observed on the fifth day. There was no IAA detected in strains L5 and L6 (Figure 2).
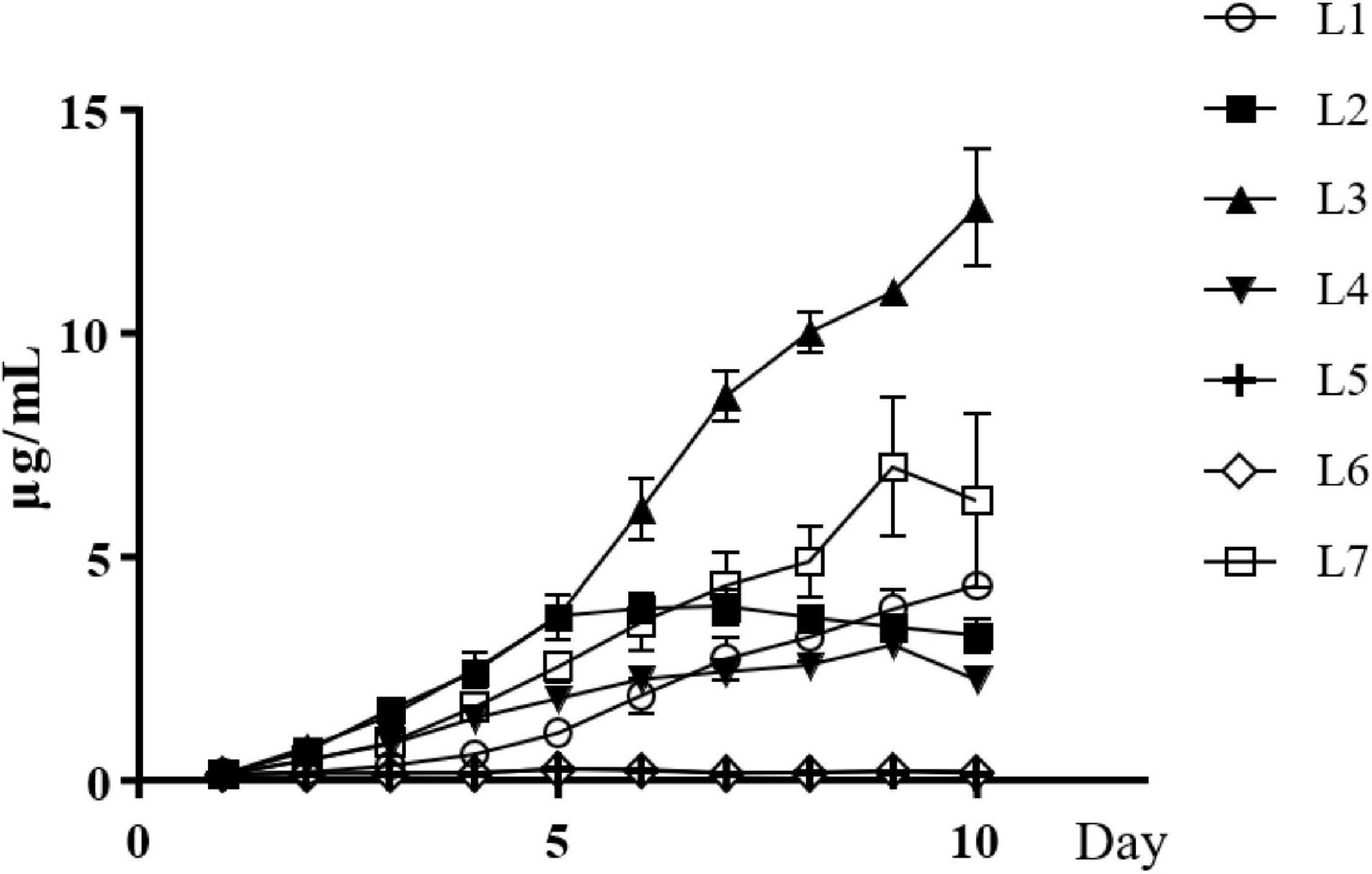
Figure 2. IAA production by seven Streptomyces isolates, Streptomyces wuyuanensis L1, Streptomyces cinereoruber L2, Streptomyces palmae L3, Streptomyces lincolnensis L4, and Streptomyces zaomyceticus L7.
Siderophore Activity
The concentration of siderophore (iron carrier) in Streptomyces cultures was changed with incubation time when these strains were grown in iron carrier medium. The highest concentrations of iron carrier for each strain were 20.55% at 48 h for L1 and 12.6%, 4.99%, 2.36%, and 6.04% at 36 h for L2, L3, L4, and L7, respectively. There was no iron carrier activity in strains L5 and L6.
1-Aminocyclopropane-1-Carboxylate Deaminase Activity
Strains L1, L2, L4, L5, and L7 were grown well after three continuous transferring on ADF medium supplemented with ACC as the sole nitrogen source, indicating their ACC deaminase activity for decomposing ACC into ACC butanoic acid and ammonia that could be utilized as direct nitrogen source by plants (Table 1).
Root Growth Parameters
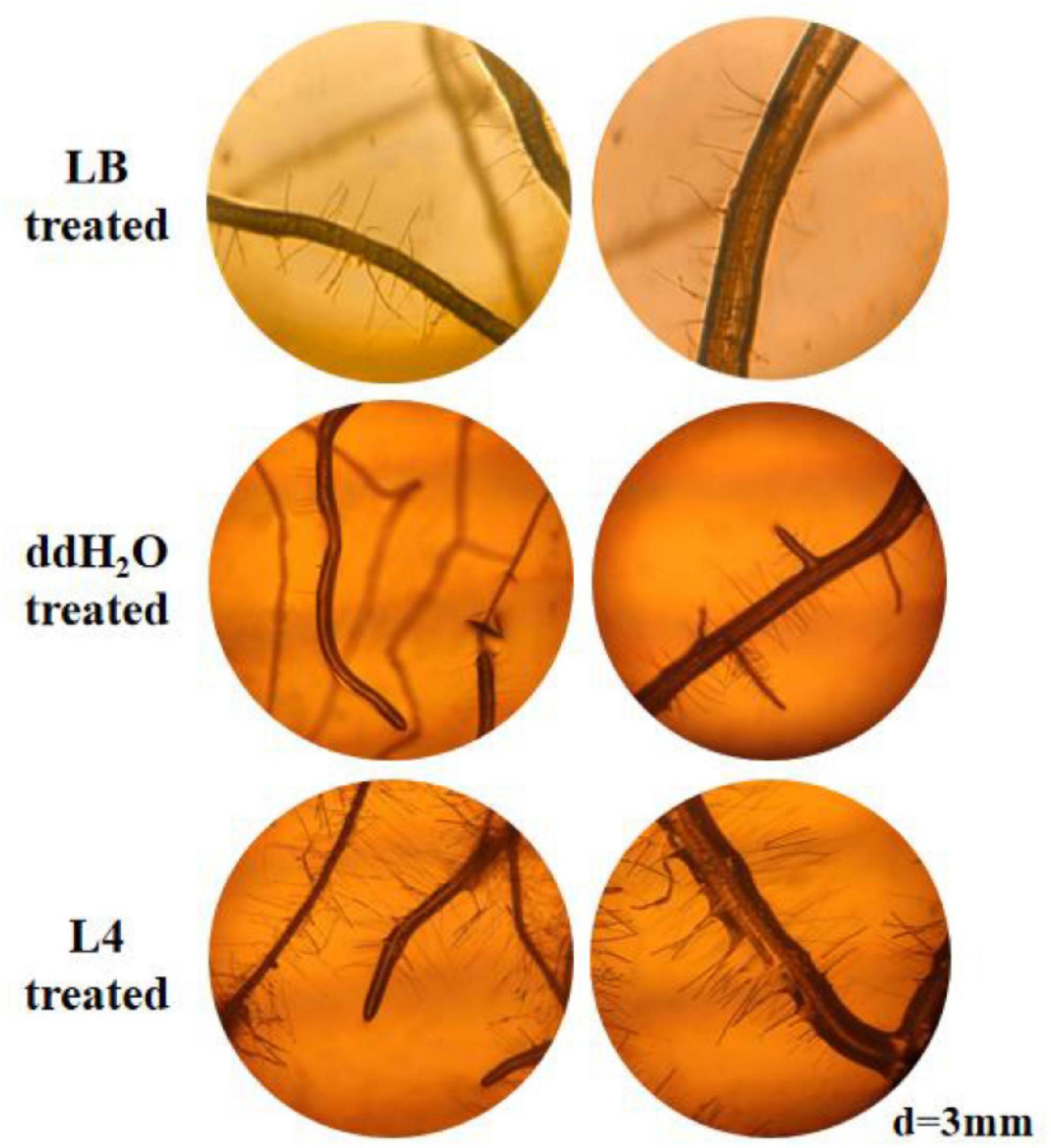
As a primary indicator of root growth, the taproot lengths of A. thaliana treated by all seven Streptomyces isolates, LB medium and ddH2O, were measured. The taproot length treated by strains L1, L2, L4, L6, and L7 was increased in comparison with LB and ddH2O-treated control (Figures 3A,B). Strain S. lincolnensis L4 demonstrated the highest taproot length (32 mm) among all treatments. More than changes in taproots, the average length (Figure 3C), and number (Figures 3D, 4) of root hairs in A. thaliana treated by strain L4 were also promoted, which indicated the potential of strain L4 to change root morphogenesis. Thus, S. lincolnensis L4 was further investigated for its function in endophytic microbiota and its genomic trait.
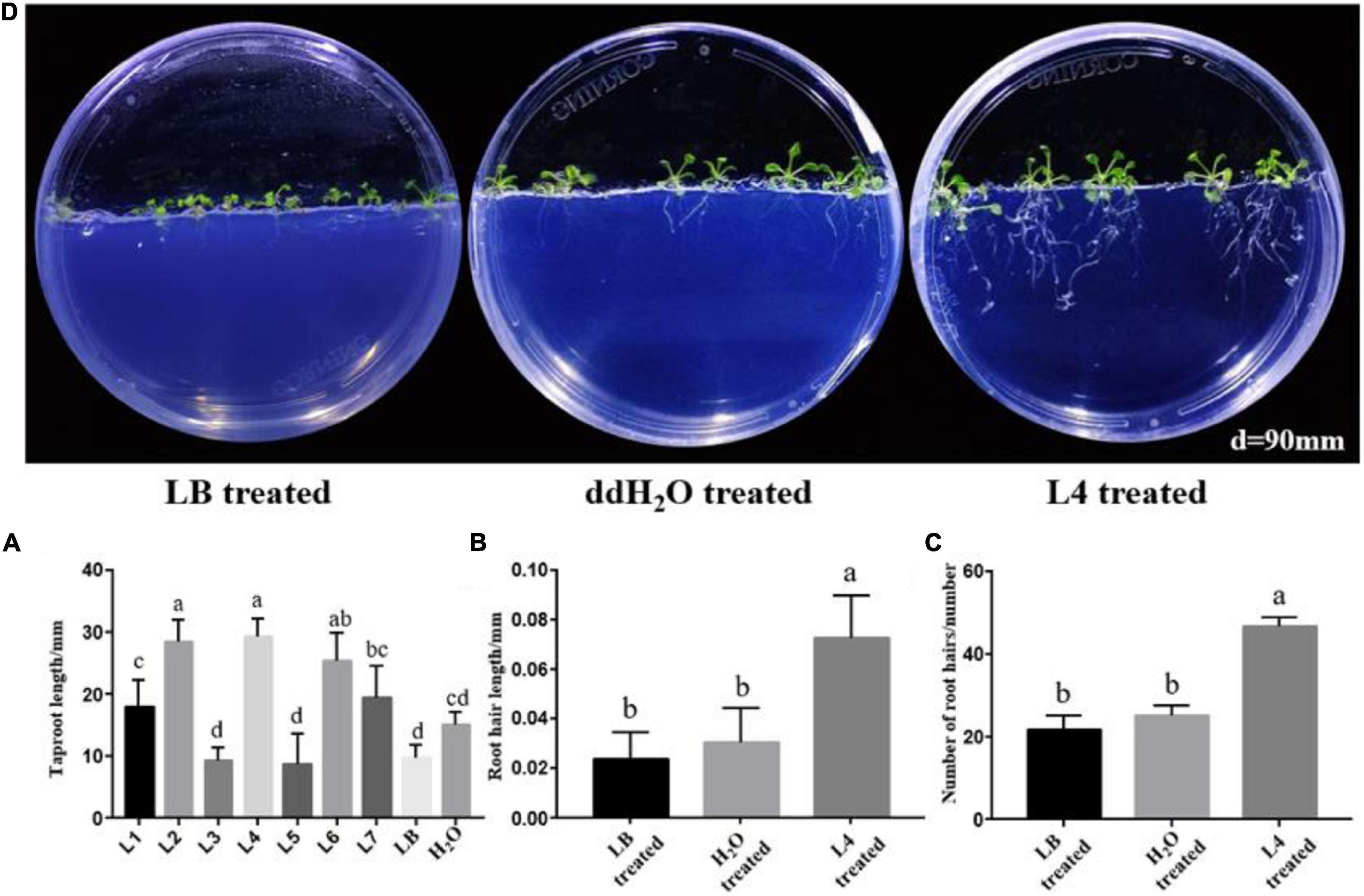
Figure 3. Root morphology influenced by the colonization of PGPR isolates. (A) The growth of Arabidopsis thaliana treated with S. lincolnensis L4; (B) changes in taproot length of A. thaliana after colonization by strain Streptomyces wuyuanensis L1, Streptomyces cinereoruber L2, Streptomyces palmae L3, S. lincolnensis L4, Streptomyces wuyuanensis L5, Streptomyces pactum L6, and Streptomyces zaomyceticus L7. It can be seen from the figure that L4 strain has the best effect on promoting the root length; (C) changes in root hair length after treatment by S. lincolnensis L4; (D) changes in number of root hairs per 3 mm taproot after treatment by S. lincolnensis L4.
Effect of Streptomyces lincolnensis L4 on Root Endophytic Bacterial Community in Arabidopsis thaliana
From Illumina sequencing of 16S rRNA gene, 833578 sequences in total were obtained from three treatments including three root samples from L4-treated and three root samples from ddH2O-treated A. thaliana. After removing sequences from mitochondria and chloroplasts, an average of 14,295 reads per sample was generated, with a total of 382 bacterial OTUs, representing composition of endophytic bacterial community in A. thaliana. OTUs with abundance greater than 1% were defined as dominant.
The richness and diversity of endophytic bacterial communities were calculated. In comparison with ddH2O-treated control, the abundance of Streptomyces as dominant genus in the group treated with S. lincolnensis L4 was much higher than that in the ddH2O-treated control (Figure 5). These results indicated that S. lincolnensis L4 had been colonized into the root of A. thaliana. However, the bacterial alpha diversity, represented by Shannon index, was significantly reduced in S. lincolnensis L4 treatments (Supplementary Figure 4).
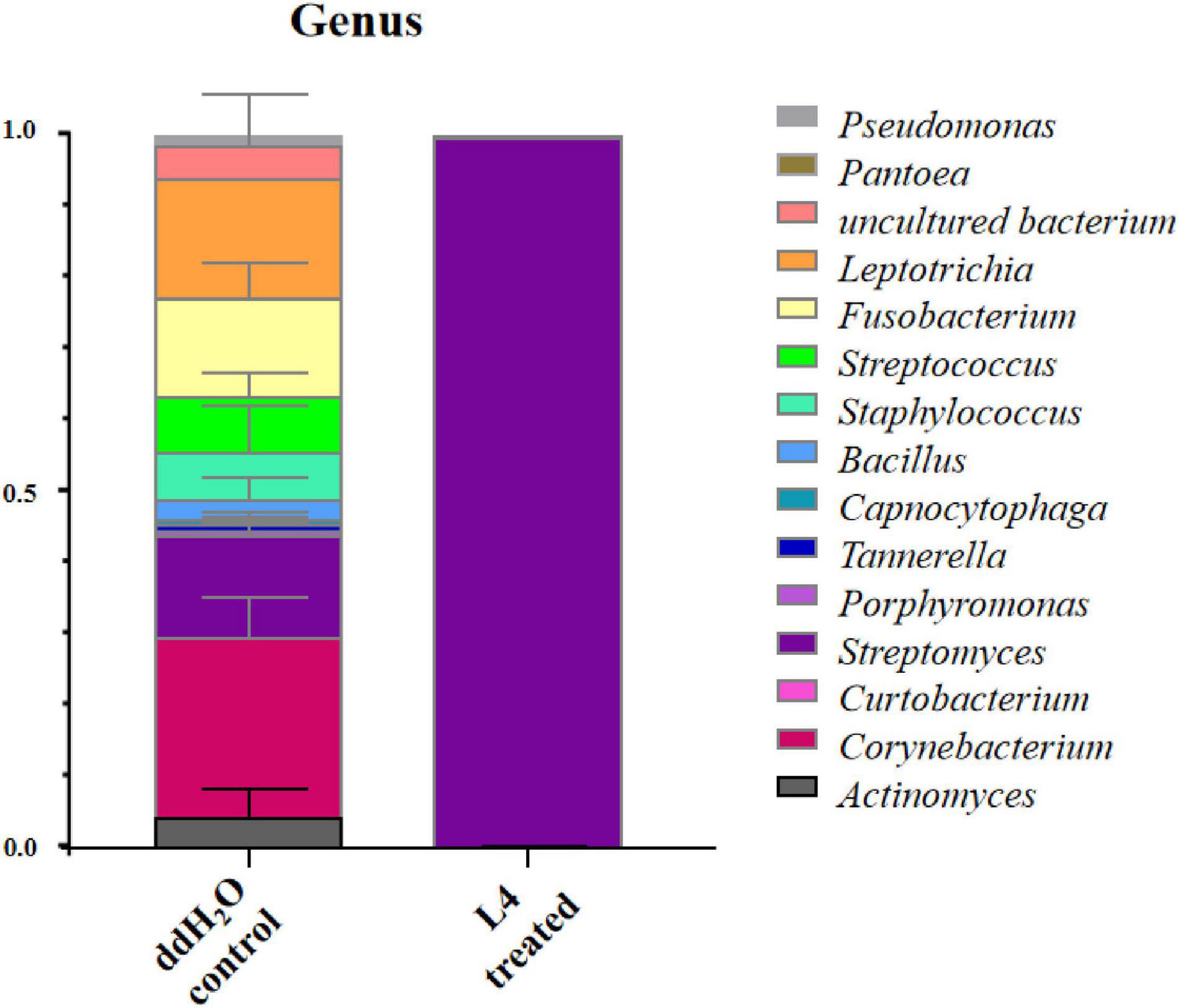
Figure 5. Endophytic bacterial community at genus level in the root of A. thaliana after treatment with S. lincolnensis L4.
Genome of Streptomyces lincolnensis L4
The assembled draft genome of S. lincolnensis L4 is 10.54 Mb in estimated length, comprising 82 scaffolds with an overall G+C content of 71.20%. A total of 9,334 protein-coding DNA sequences (CDS) and 100 predicted RNAs (92 tRNAs, 12 rRNAs, and 1 tmRNA) were identified from the draft genome. The genome sequence of S. lincolnensis L4 had been submitted NCBI GenBank with accession number PRJNA753994.
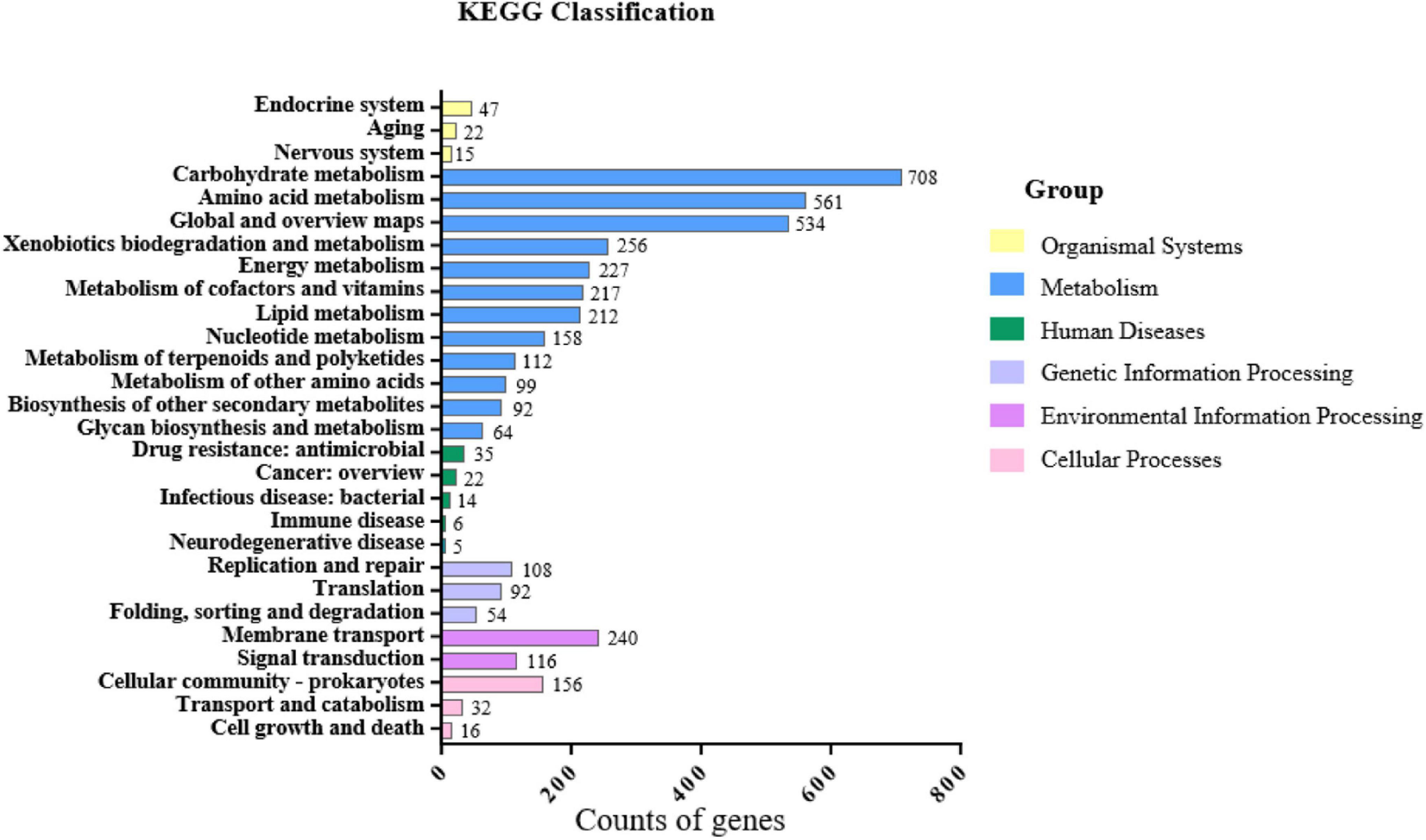
From the function annotation of KEGG, besides two major categories of central carbon metabolism “carbohydrate metabolism” (708 CDSs) and “amino acid metabolism” (561 CDSs), these characterized categories including “xenobiotics biodegradation and metabolism” (256 CDSs), “metabolism of terpenoids and polyketides” (112 CDSs), and “biosynthesis of other secondary metabolites” (92 CDSs) may be involved in plant growth promotion (Figure 6).
Specifically, genes encoding function of ACC deamination, iron transportation, and IAA synthesis were identified from genome in S. lincolnensis L4. The presence of gene encoding ACC deaminase (1-aminocyclopropane-1-carboxylate deaminase, EC 3.5.99.7) in L4 genome indicates its potential to promote plant growth by decreasing ethylene level resulting in extensive root growth (Sheehy et al., 1991). Furthermore, IucA/IucC family protein including aerobactin synthase (EC 6.3.2.39) (Lorenzo and Neilands, 1986) and N(2)-citryl-N(6)-acetyl-N(6)-hydroxylysine (EC 6.3.2.38) (Hickey and Cianciotto, 1997) functioned in siderophore biosynthesis was identified in L4 genome. In addition, several genes involved in IAA production were identified including genes encoding tryptophan 2-monooxygenase (EC 1.13.12.3), and aldehyde dehydrogenase AldA (EC 1.2.1.3), which play a role in the early stage of IAA synthesis (Krause et al., 2015; Mcclerklin et al., 2018). However, other encoding gene(s) involved in IAA synthesis was not annotated in the draft genome of S. lincolnensis L4 (Table 2).
Expression Changes of Root Development-Related Genes After Colonization of Streptomyces lincolnensis L4
To further explore the role of S. lincolnensis L4 in the process of root hair elongation of A. thaliana, we selected seven genes related to root hair growth and development (akt1, kojak, rop2, lrl3, rsl4, rhd6, and rhd2) and quantified their expression in the root of L4-treated A. thaliana by performing real-time quantitative PCR with primers reported in the previous study (Forman and Torres, 2002; Sieberer et al., 2005; Dias and Sá-Correia, 2013; Nakamura et al., 2018; Wang et al., 2019; Supplementary Table 1). Expression of four genes lrl3 and rhd6 was upregulated in treatment group; expression of three genes akt1, kojak, and rop2 was decreased; rsl4 and rhd2 have no significant difference compared with the control group (Figure 7). These upregulated genes implied their function in root development induced by the colonization of S. lincolnensis L4, in which gene lrl3 encodes AtLRL3 that mediate the development of root hair cells in Arabidopsis by acting downstream from the regulatory complexes that specify pattern formation in the epidermis (Karas et al., 2009), and gene rhd6 controls the number of root hairs (Menand et al., 2007).
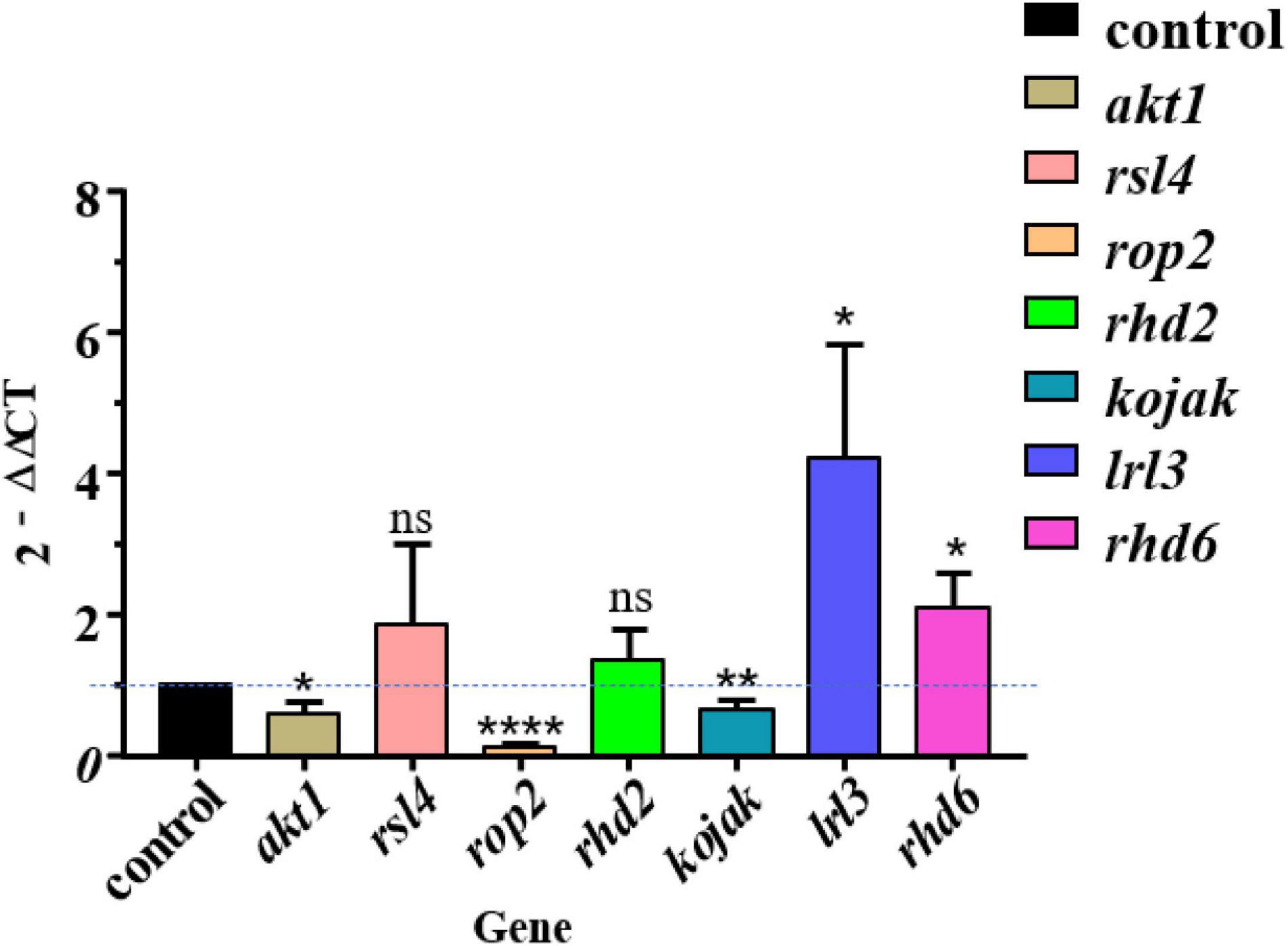
Figure 7. Quantitative PCR revealed the differential expression of genes involved in root development in A. thaliana after colonization of S. lincolnensis L4. Expression of four genes lrl3 and rhd6 were upregulated in the L4 treatment group compared with the control group; expression of three genes akt1, kojak, and rop2 were decreased; rsl4 and rhd2 have no significant difference compared with the control group. The “2–ΔΔCT” method was used for calculating the expression change of target genes with 18S rRNA gene as housekeeping gene, in which 2–ΔΔCT> 1 or <1 represents the upregulation or downregulation of target genes. The significances were presented by *P < 0.05; **P < 0.01; ****P < 0.0001.
On the contrary, expression of Rop2 GTPase-encoding gene (rop2) was downregulated by the inoculation of S. lincolnensis L4 at late stage of root development in our treatment, probably due to that it is a positive regulator of root hair initiation (Jones et al., 2002). Similarly, both gene akt1 and gene kojak were downregulated due to the inoculation of strain L4. Gene akt1 is an important gene for K+ transport in roots (Wang et al., 2016). Gene kojak encoded KOJAK/AtCSLD3 (KOJAK encodes a cellulose synthase-like protein, AtCSLD3) function in the biosynthesis of β-glucan-containing polysaccharides that are required during root hair elongation (Favery et al., 2001). This may be also because they function in the earliest stage but the analysis was conducted on the late stage of root hair development. Gene rhd2 facilitates Ca2+ influx for growth (Foreman et al., 2003) and rsl4 encodes RSL4 protein involved in signaling, secretion, and cell wall modification (Vijayakumar et al., 2016). Expression of the two genes was significant, which may be related to the stabilization of gene expression in the later stage of root growth and development.
Discussion
The plant growth promotion could be the result of the beneficial functions of applied PGPR isolates, such as plant growth hormone production, nitrogen fixation, and P solubilization, and these microorganisms with the function of promoting plant growth and development will be optimal substitute of chemical fertilizers (Hassen et al., 2018). In this study, a plant growth-promoting bacterium, S. lincolnensis L4, was isolated from the rhizosphere soil of A. annua. In the aseptic seedling experiment, S. lincolnensis L4 had a positive effect on the root growth and development of Arabidopsis. Compared with control, root length, number of root hairs, and length of root hair were improved after the colonization of L4 in roots. The results of real-time quantitative PCR showed that the expression of genes involved in root development in Arabidopsis was regulated by the inoculation of L4 culture which verified the interaction between S. lincolnensis L4 and host plant. The analysis of community composition and structure of endophytic bacteria showed that the relative abundance of Streptomyces reached more than 99.8% in the root of A. thaliana treated by S. lincolnensis L4, which had become the dominant endophytes compared with other genera, indicating that S. lincolnensis L4 as PGPR bacteria had actual interaction with A. thaliana by colonization in its root. These results, combined with IAA production, ACC deamination, and iron carrier production, revealed that S. lincolnensis L4 harbors the ability to stimulate plant root growth by tuning its root morphogenesis. On the other hand, the domination of Streptomyces spp. in roots reduced the alpha diversity, which may have risks to break the ecological balance of endophytic community even though negative effect to root growth and development was not observed in this study (Sagita et al., 2021). The changes in endophytic microbial diversity which cause by alien microbial inoculation should be considered during PGPR strategic design, which could be optimized in the proper inoculating dosage or synthetic consortium.
It has been well studied that siderophore production and IAA synthesis are related to efficient nutrient uptake and significant enhancement of the root system, respectively (Patten and Glick, 2002; Etesami et al., 2015; Abbamondi et al., 2016). Siderophore-producing bacteria indirectly promoted plant growth by preventing the proliferation of pathogens by decreasing the amount of available iron (Olanrewaju et al., 2017; Compant et al., 2019). The majority of the isolated bacteria in this study, including S. lincolnensis L4, were able to synthesize IAA, indicating its potential of PGPR trait similar to the previous studies (Spaepen and Vanderleyden, 2011). However, the observed concentration of IAA changed probably due to the various pathways for biosynthesis of IAA in different species (Majeed et al., 2015). High levels of IAA promote the formation of lateral roots (Gupta and Pandey, 2019) and increase root length (Olanrewaju et al., 2017). Even though the IAA synthesis pathway was not completely annotated in the draft genome of strain L4, the apparent IAA production and promoted root development demonstrated its function to regulated root growth via IAA accumulation. On the other hand, ACC deaminase activity that produces certain amount of α-ketobutyrate was observed in S. lincolnensis L4. As a product of ACC deamination, ammonia could be utilized by plant as direct nitrogen, which has been proved in other PGPR strains (Penrose and Glick, 2003; Checcucci et al., 2017; Jaemsaeng et al., 2018). These above-described PGPR traits in S. lincolnensis L4 were further verified in genomic analysis and quantitative assay of root growth-related genes in Arabidopsis, which conclusively implied its role in plant growth promotion through regulating root development.
Conclusion (Final Remark)
From the aspects of microbial physiology, genomic analysis, endophytic bacterial community, root phenotype, and gene expression of host plant, we have demonstrated that isolated Streptomyces strains harbor various traits including N2 fixation, P-solubilizing, siderophore activity, ACC deaminase activity, and IAA-producing bacteria which present among the natural population. Through these characteristics, strain L4 is specifically capable of regulating the root length and root hair number of Arabidopsis which was verified by the expression of gene related to root development. Thus, this Streptomyces strain offers potential in field applications as root growth-promoting agents. Further studies should be focused on the detailed molecular and functional characterization of these PGPR for unraveling the mechanism of root development driven by associated microbes.
Plant growth-promoting rhizobacteria have been proved on their ability of nutrient acquisition, phosphorus solubilization and phytohormones synthesis in crops. It has been considered as a feasible alternative to chemical approaches (Olanrewaju et al., 2017). These metabolic potentials of Streptomyces in plant growth promotion have received increasing attentions (Olanrewaju and Babalola, 2019). We believe that these microorganisms with the function of promoting plant growth and development will be optimal substitute of chemical fertilizers, and also ensure the sustainable development of agriculture.
Data Availability Statement
The datasets generated for this study can be found in the NCBI under accession numbers PRJNA753994 and PRJNA761252. The genome of Streptomyces lincolnensis L4 is available at: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA753994. The community data are available at: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA761252.
Author Contributions
WF: conceptualization, methodology, formal analysis, investigation, data curation, and writing—original draft. YP and YS: conceptualization, validation, resources, and investigation. JC: writing—review and editing and visualization. DG and YL: writing—reviewing and editing. GH: writing—reviewing and editing and supervision. DH: writing—reviewing and editing, supervision, project administration, and funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by Central Public Interest Scientific Institution Basal Research Fund (Y2019LM02-02) and the Agricultural Science and Technology Innovation Program (ASTIP).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.802737/full#supplementary-material
References
Abbamondi, G. R., Tommonaro, G., Weyens, N., Thijs, S., Sillen, W., Gkorezis, P., et al. (2016). Plant growth-promoting effects of rhizospheric and endophytic bacteria associated with different tomato cultivars and new tomato hybrids. Chem. Biol. Technol. Agric. 3:1. doi: 10.1186/s40538-015-0051-3
Alexander, D. B., and Zuberer, D. A. (1991). Use of chrome azurol S reagents to evaluate siderophore production by rhizosphere bacteria. Biol. Fertil. Soils 12, 39–45. doi: 10.1007/BF00369386
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Bharti, N., Barnawal, D., Maji, D., and Kalra, A. (2015). Halotolerant PGPRs prevent major shifts in indigenous microbial community structure under salinity stress. Microb. Ecol. 70, 196–208. doi: 10.1007/s00248-014-0557-4
Checcucci, A., Azzarello, E., Bazzicalupo, M., De Carlo, A., Emiliani, G., Mancuso, S., et al. (2017). Role and regulation of ACC deaminase gene in Sinorhizobium meliloti: is it a symbiotic, rhizospheric or endophytic gene? Front. Genet. 8:6. doi: 10.3389/fgene.2017.00006
Chen, S., Li, X., Lavoie, M., Jin, Y., Xu, J., Fu, Z., et al. (2017). Diclofop-methyl affects microbial rhizosphere community and induces systemic acquired resistance in rice. J. Environ. Sci. (China) 51, 352–360. doi: 10.1016/j.jes.2016.06.027
Compant, S., Samad, A., Faist, H., and Sessitsch, A. (2019). A review on the plant microbiome: ecology, functions, and emerging trends in microbial application. J. Adv. Res. 19, 29–37. doi: 10.1016/j.jare.2019.03.004
Dias, P. J., and Sá-Correia, I. (2013). The drug:H+ antiporters of family 2 (DHA2), siderophore transporters (ARN) and glutathione: H+ antiporters (GEX) have a common evolutionary origin in hemiascomycete yeasts. Dias SáCorreia BMC Genomics 14:901. doi: 10.1186/1471-2164-14-901
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
El-Tarabily, K. A. (2008). Promotion of tomato (Lycopersicon esculentum Mill.) plant growth by rhizosphere competent 1-aminocyclopropane-1-carboxylic acid deaminase-producing streptomycete actinomycetes. Plant Soil 308, 161–174. doi: 10.1007/s11104-008-9616-2
Etesami, H., Alikhani, H. A., and Hosseini, H. M. (2015). Indole-3-acetic acid (IAA) production trait, a useful screening to select endophytic and rhizosphere competent bacteria for rice growth promoting agents. MethodsX 2, 72–78. doi: 10.1016/j.mex.2015.02.008
Fan, D., Subramanian, S., and Smith, D. L. (2020). Plant endophytes promote growth and alleviate salt stress in Arabidopsis thaliana. Sci. Rep. 10:12740. doi: 10.1038/s41598-020-69713-5
Favery, B., Ryan, E., Foreman, J., Linstead, P., Boudonck, K., Steer, M., et al. (2001). KOJAK encodes a cellulose synthase-like protein required for root hair cell morphogenesis in Arabidopsis. Genes Dev. 15, 79–89. doi: 10.1101/gad.188801
Foreman, J., Demidchik, V., Bothwell, J. H. F., Mylona, P., Miedema, H., Torresk, M. A., et al. (2003). Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 422, 442–446. doi: 10.1038/nature01485
Forman, H. J., and Torres, M. (2002). Reactive oxygen species and cell signaling: respiratory burst in macrophage signaling. Am. J. Respir. Crit. Care Med. 166, S4–S8. doi: 10.1164/rccm.2206007
Gupta, S., and Pandey, S. (2019). Unravelling the biochemistry and genetics of ACC deaminase-An enzyme alleviating the biotic and abiotic stress in plants. Plant Gene 18:100175. doi: 10.1016/j.plgene.2019.100175
Hartmann, A., Rothballer, M., and Schmid, M. (2007). Lorenz Hiltner, a pioneer in rhizosphere microbial ecology and soil bacteriology research. Plant Soil 312, 7–14. doi: 10.1007/s11104-007-9514-z
Hassen, W., Neifar, M., Cherif, H., Najjari, A., Chouchane, H., Driouich, R. C., et al. (2018). Pseudomonas rhizophila S211, a new plant growth-promoting rhizobacterium with potential in pesticide-bioremediation. Front. Microbiol. 9:34. doi: 10.3389/fmicb.2018.00034
Hickey, E. K., and Cianciotto, N. P. (1997). An iron- and fur-repressed Legionella pneumophila gene that promotes intracellular infection and encodes a protein with similarity to the Escherichia coli aerobactin synthetases. Infect. Immunity 65, 133–143. doi: 10.1128/iai.65.1.133-143.1997
Jaemsaeng, R., Jantasuriyarat, C., and Thamchaipenet, A. (2018). Molecular interaction of 1-aminocyclopropane-1-carboxylate deaminase (ACCD)-producing endophytic Streptomyces sp. GMKU 336 towards salt-stress resistance of Oryza sativa L. cv. KDML105. Sci Rep 8, 1950. doi: 10.1038/s41598-018-19799-9
Jami, E., Israel, A., Kotser, A., and Mizrahi, I. (2013). Exploring the bovine rumen bacterial community from birth to adulthood. ISME J. 7, 1069–1079. doi: 10.1038/ismej.2013.2
Jones, M. A., Shen, J. J., Fu, Y., Li, H., Yang, Z., and Grierson, C. S. (2002). The Arabidopsis Rop2 GTPase is a positive regulator of both root hair initiation and tip growth. Plant Cell 14, 763–776. doi: 10.1105/tpc.010359
Kanehisa, M., Sato, Y., and Kawashima, M. (2021). KEGG mapping tools for uncovering hidden features in biological data. Protein Sci. doi: 10.1002/pro.4172
Karas, B., Amyot, L., Johansen, C., Sato, S., Tabata, S., Kawaguchi, M., et al. (2009). Conservation of lotus and Arabidopsis basic helix-loop-helix proteins reveals new players in root hair development. Plant Physiol. 151, 1175–1185. doi: 10.1104/pp.109.143867
Khan, N., Ali, S., Tariq, H., Latif, S., Yasmin, H., Mehmood, A., et al. (2020). Water conservation and plant survival strategies of rhizobacteria under drought stress. Agronomy 10:1683. doi: 10.3390/agronomy10111683
Krause, K., Henke, C., Asiimwe, T., Ulbricht, A., Klemmer, S., Schachtschabel, D., et al. (2015). Biosynthesis and secretion of indole-3-acetic acid and its morphological effects on Tricholoma vaccinum-spruce ectomycorrhiza. Appl. Environ. Microbiol. 81, 7003–7011. doi: 10.1128/AEM.01991-15
Li, M., Guo, R., Yu, F., Chen, X., Zhao, H., Li, H., et al. (2018). Indole-3-acetic acid biosynthesis pathways in the plant-beneficial bacterium Arthrobacter pascens ZZ21. Int. J. Mol. Sci. 19:443. doi: 10.3390/ijms19020443
Lim, H. J., Lee, E. H., Yoon, Y., Chua, B., and Son, A. (2016). Portable lysis apparatus for rapid single-step DNA extraction of Bacillus subtilis. J. Appl. Microbiol. 120, 379–387. doi: 10.1111/jam.13011
Lorenzo, V. D., and Neilands, J. B. (1986). Characterization of iucA and iucC genes of the aerobactin system of plasmid Co1V-K30 in Escherichia coli. J. Bacteriol. 167, 350–355. doi: 10.1128/jb.167.1.350-355.1986
Majeed, A., Abbasi, M. K., Hameed, S., Imran, A., and Rahim, N. (2015). Isolation and characterization of plant growth-promoting rhizobacteria from wheat rhizosphere and their effect on plant growth promotion. Front. Microbiol. 6:198. doi: 10.3389/fmicb.2015.00198
Mcclerklin, S. A., Lee, S. G., Harper, C. P., Nwumeh, R., Jez, J. M., and Kunkel, B. N. (2018). Indole-3-acetaldehyde dehydrogenase-dependent auxin synthesis contributes to virulence of Pseudomonas syringae strain DC3000. PLoS Pathog. 14:e1006811. doi: 10.1371/journal.ppat.1006811
Menand, B., Yi, K., Jouannic, S., Hoffmann, L., Ryan, E., Linstead, P., et al. (2007). An ancient mechanism controls the development of cells with a rooting function in land plants. Science 316, 1477–1480. doi: 10.1126/science.1142618
Nakamura, M., Claes, A. R., Grebe, T., Hermkes, R., Viotti, C., Ikeda, Y., et al. (2018). Auxin and ROP GTPase signaling of polar nuclear migration in root epidermal hair cells. Plant Physiol. 176, 378–391. doi: 10.1104/pp.17.00713
Olanrewaju, O. S., and Babalola, O. O. (2019). Streptomyces: implications and interactions in plant growth promotion. Appl. Microbiol. Biotechnol. 103, 1179–1188. doi: 10.1007/s00253-018-09577-y
Olanrewaju, O. S., Glick, B. R., and Babalola, O. O. (2017). Mechanisms of action of plant growth promoting bacteria. World J. Microbiol. Biotechnol. 33:197. doi: 10.1007/s11274-017-2364-9
Patten, C. L., and Glick, B. R. (2002). Role of Pseudomonas putida indoleacetic acid in development of the host plant root system. Appl. Environ. Microbiol. 68, 3795–3801. doi: 10.1128/AEM.68.8.3795-3801.2002
Penrose, D. M., and Glick, B. R. (2003). Methods for isolating and characterizing ACC deaminase-containing plant growth-promoting rhizobacteria. Physiol. Plant 118, 10–15. doi: 10.1034/j.1399-3054.2003.00086
Rai, A., and Nabti, E. (2017). “Plant growth-promoting bacteria: importance in vegetable production,” in Microbial Strategies for Vegetable Production, eds A. Zaidi and M. Khan (Cham: Springer), 23–48.
Reyes, J. C. (2006). Chromatin modifiers that control plant development. Curr. Opin. Plant Biol. 9, 21–27. doi: 10.1016/j.pbi.2005.11.010
Rungin, S., Indananda, C., Suttiviriya, P., Kruasuwan, W., Jaemsaeng, R., and Thamchaipenet, A. (2012). Plant growth enhancing effects by a siderophore-producing endophytic streptomycete isolated from a Thai jasmine rice plant (Oryza sativa L. cv. KDML105). Antonie Van Leeuwenhoek 102, 463–472. doi: 10.1007/s10482-012-9778-z
Sagita, R., Quax, W. J., and Haslinger, K. (2021). Current state and future directions of genetics and genomics of endophytic fungi for bioprospecting efforts. Front. Bioeng. Biotechnol. 9:649906. doi: 10.3389/fbioe.2021.649906
Schwyn, B., and Nelands, J. B. (1987). Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160, 47–56. doi: 10.1016/0003-2697(87)90612-9
Sheehy, R. E., Honma, M., Yamada, M., Sasaki, T., Martineau, B., and Hiatti, W. R. (1991). Isolation, sequence, and expression in Escherichia coli of the Pseudomonas sp. strain ACP gene encoding 1-aminocyclopropane1-carboxylate deaminase. J. Bacteriol. 173, 5260–5265. doi: 10.1128/jb.173.17.5260-5265.1991
Shi, Y., Pan, Y., Xiang, L., Zhu, Z., and Fu, W. (2021). Assembly of rhizosphere microbial communities in Artemisia annua: recruitment ofplant growth-promoting microorganisms and inter-kingdom interactions between bacteria and fungi. Plant Soil doi: 10.1007/s11104-021-04829-9
Sieberer, B. J., Ketelaar, T., Esseling, J. J., and Emons, A. M. (2005). Microtubules guide root hair tip growth. New Phytol. 167, 711–719. doi: 10.1111/j.1469-8137.2005.01506.x
Singh, B. K., Bardgett, R. D., Smith, P., and Reay, D. S. (2010). Microorganisms and climate change: terrestrial feedbacks and mitigation options. Nat. Rev. Microbiol. 8, 779–790. doi: 10.1038/nrmicro2439
Smith, D. L., Praslickova, D., and Ilangumaran, G. (2015). Inter-organismal signaling and management of the phytomicrobiome. Front. Plant Sci. 6:722. doi: 10.3389/fpls.2015.00722
Spaepen, S., and Vanderleyden, J. (2011). Auxin and plant-microbe interactions. Cold Spring Harb. Perspect. Biol. 3:a001438. doi: 10.1101/cshperspect.a001438
Surange, S., Wollum, A. G., Kumar, N., and Nautiyal, C. S. (1997). Characterization of Rhizobium from root nodules of leguminous trees growing in alkaline soils. Can. J. Microbiol. 43, 891–894. doi: 10.1139/m97-130
Treesubsuntorn, C., Dhurakit, P., Khaksar, G., and Thiravetyan, P. (2018). Effect of microorganisms on reducing cadmium uptake and toxicity in rice (Oryza sativa L.). Environ. Sci. Pollut. Res. Int. 25, 25690–25701. doi: 10.1007/s11356-017-9058-6
Vijayakumar, P., Datta, S., and Dolan, L. (2016). Root hair defective six-like4 (RSL4) promotes root hair elongation by transcriptionally regulating the expression of genes required for cell growth. New Phytol. 212, 944–953. doi: 10.1111/nph.14095
Wang, C. X., Qi, C. Y., Luo, J. H., Liu, L., He, Y., and Chen, L. Q. (2019). Characterization of LRL5 as a key regulator of root hair growth in maize. Plant J. 98, 71–82. doi: 10.1111/tpj.14200
Wang, M., Xue, J., Ma, J., Feng, X., Ying, H., and Xu, H. (2020). Streptomyces lydicus M01 regulates soil microbial community and alleviates foliar disease caused by Alternaria alternata on cucumbers. Front. Microbiol. 11:942. doi: 10.3389/fmicb.2020.00942
Wang, X. P., Chen, L. M., Liu, W. X., Shen, L. K., Wang, F. L., Zhou, Y., et al. (2016). AtKC1 and CIPK23 synergistically modulate AKT1-mediated low-potassium stress responses in Arabidopsis. Plant Physiol. 170, 2264–2277. doi: 10.1104/pp.15.01493
Wu, Q., Lu, C., Ni, M., Wang, H., Liu, W., and Chen, J. (2019). Streptomyces lydicus A01 affects soil microbial diversity, improving growth and resilience in tomato. J. Integr. Plant Biol. 61, 182–196. doi: 10.1111/jipb.12724
Xu, N., Tan, G., Wang, H., and Gai, X. (2016). Effect of biochar additions to soil on nitrogen leaching, microbial biomass and bacterial community structure. Eur. J. Soil Biol. 74, 1–8. doi: 10.1016/j.ejsobi.2016.02.004
Yan, X., An, J., Yin, Y., Gao, C., Wang, B., and Wei, S. (2022). Heavy metals uptake and translocation of typical wetland plants and their ecological effects on the coastal soil of a contaminated bay in Northeast China. Sci. Total Environ. 803:149871. doi: 10.1016/j.scitotenv.2021.149871
Keywords: Streptomyces lincolnensis, PGPR, indole-3-acetic acid (IAA), root morphogenesis, endophytic microbes
Citation: Fu W, Pan Y, Shi Y, Chen J, Gong D, Li Y, Hao G and Han D (2022) Root Morphogenesis of Arabidopsis thaliana Tuned by Plant Growth-Promoting Streptomyces Isolated From Root-Associated Soil of Artemisia annua. Front. Plant Sci. 12:802737. doi: 10.3389/fpls.2021.802737
Received: 27 October 2021; Accepted: 25 November 2021;
Published: 10 January 2022.
Edited by:
Sheng Qin, Jiangsu Normal University, ChinaReviewed by:
Chunxu Song, China Agricultural University, ChinaKe Zhao, Sichuan Agricultural University, China
Copyright © 2022 Fu, Pan, Shi, Chen, Gong, Li, Hao and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guangfei Hao, d3V5dWNvb2xAMTI2LmNvbQ==; Dongfei Han, aGFuZG9uZ2ZlaUBjYWFzLmNu
 Wenbo Fu
Wenbo Fu Yanshuo Pan2,3
Yanshuo Pan2,3 Yuhua Shi
Yuhua Shi Daozhi Gong
Daozhi Gong Guangfei Hao
Guangfei Hao Dongfei Han
Dongfei Han