
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 17 January 2022
Sec. Plant Abiotic Stress
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.801239
This article is part of the Research Topic Identification and Functional Dissection of Stress-responsive Genes in Cotton View all 26 articles
 Xiaokang Fu1†
Xiaokang Fu1† Yonglin Yang2†
Yonglin Yang2† Meng Kang1
Meng Kang1 Hengling Wei1
Hengling Wei1 Boying Lian1
Boying Lian1 Baoquan Wang1
Baoquan Wang1 Liang Ma1
Liang Ma1 Pengbo Hao1
Pengbo Hao1 Jianhua Lu1
Jianhua Lu1 Shuxun Yu1*
Shuxun Yu1* Hantao Wang1*
Hantao Wang1*
The caleosin (CLO) protein family displays calcium-binding properties and plays an important role in the abiotic stress response. Here, a total of 107 CLO genes were identified in 15 plant species, while no CLO genes were detected in two green algal species. Evolutionary analysis revealed that the CLO gene family may have evolved mainly in terrestrial plants and that biological functional differentiation between species and functional expansion within species have occurred. Of these, 56 CLO genes were identified in four cotton species. Collinearity analysis showed that CLO gene family expansion mainly occurred through segmental duplication and whole-genome duplication in cotton. Sequence alignment and phylogenetic analysis showed that the CLO proteins of the four cotton species were mainly divided into two types: H-caleosins (class I) and L-caleosins (class II). Cis-acting element analysis and quantitative RT–PCR (qRT–PCR) suggested that GhCLOs might be regulated by abscisic acid (ABA) and methyl jasmonate (MeJA). Moreover, transcriptome data and qRT–PCR results revealed that GhCLO genes responded to salt and drought stresses. Under salt stress, gene-silenced plants (TRV: GhCLO06) showed obvious yellowing and wilting, higher malondialdehyde (MDA) content accumulation, and significantly lower activities of superoxide dismutase (SOD) and peroxidase (POD), indicating that GhCLO06 plays a positive regulatory role in cotton salt tolerance. In gene-silenced plants (TRV: GhCLO06), ABA-related genes (GhABF2, GhABI5, and GhNAC4) were significantly upregulated after salt stress, suggesting that the regulation of salt tolerance may be related to the ABA signaling pathway. This research provides an important reference for further understanding and analyzing the molecular regulatory mechanism of CLOs for salt tolerance.
Caleosins (CLOs) are calcium-binding proteins encoded by small gene families, sometimes called peroxygenases (PXGs) in databases, and are widely distributed in terrestrial plants (Khalil et al., 2014; Shen et al., 2016; Rahman et al., 2018). Caleosins (Pfam PF05042) are members of the EC: 1.11.2.3 class of oxidoreductases, and they also have a wide range of biological functions (Rahman et al., 2018). CLO proteins usually contain a highly conserved single calcium-binding EF hand motif, a lipid-binding domain and two invariant heme-coordinating histidine residues (Hanano et al., 2006; Kim et al., 2011; Shen et al., 2014; Rahman et al., 2018). Additionally, there is a region containing several predicted kinase sites proximal to the C-terminus (Shen et al., 2014; Song et al., 2014; Charuchinda et al., 2015), and these structures are usually important features for identifying the caleosin family and its classification. In general, there are two different CLO isomers in angiosperms, labeled H (high) and L (low), where H-forms contain an additional C-terminal motif of approximately 30–50 residues that is absent from L-forms, and L-caleosins evolve from H-caleosins (Khalil et al., 2014; Shen et al., 2014, 2016; Rahman et al., 2018). Among the eight CLO proteins found in Arabidopsis, AtCLO1-3, and AtCLO8 are H-forms, and AtCLO4-7 is an L-form (Shen et al., 2016). Caleosin is considered a structural stabilizer of lipid droplets and is named for its ability to combine with calcium (Shen et al., 2016). If caleosin has heme groups coordinated by two invariant histidine residues, it will have specific types of lipid peroxygenase activity (Hanano et al., 2006; Blée et al., 2012; Benaragama et al., 2017). Some CLO subtypes can bind to a variety of cellular bilayer membranes, such as the endoplasmic reticulum (ER) and plasmalemma, through a single transmembrane domain (Partridge and Murphy, 2009; Hanano et al., 2015; Purkrtová et al., 2015).
Soil salinity seriously affects world agricultural production (Munns and Gilliham, 2015). Salt stress is an abiotic stress factor that seriously affects the growth, development and survival of plants (Ganie et al., 2019; Xu et al., 2020). In China, saline-alkali soils account for 25% of farmland and are underutilized (Liu and Wang, 2021). Cultivating salt-tolerant plants and deeply understanding the salt tolerance mechanism of plants play an important role in agricultural production and sustainable development of the environment (Deinlein et al., 2014; Liu and Wang, 2021). Some studies have confirmed that the CLO gene family may be related to signal transduction and a variety of abiotic stress responses (Kim et al., 2011; Khalil et al., 2014). In Arabidopsis, AtCLO1 (ATS1; At4g26740) has been found to actively participate in the degradation of storage lipids in oil bodies (OBs) (Poxleitner et al., 2006) and to have Ca2+-dependent peroxygenase activity, which may be related to oxylipin signaling pathways and plant defense responses (Hanano et al., 2006). AtCLO3 (RD20; At2g33380) was significantly induced under salt, drought, and abscisic acid (ABA) stresses, and the tolerance of its mutant rd20 to these stresses was significantly reduced (Takahashi et al., 2000; Partridge and Murphy, 2009; Aubert et al., 2010). In Arabidopsis overexpressing RD20, 13-hydroxy-9,11,15-octadecaterinoic acid (a linolenate-derived hydroxide) was enriched; the level of reactive oxygen species (ROS) increased in plants with early gibberellin-dependent flowering and ABA hypersensitivity at seed germination, indicating that RD20 is directly related to abiotic stress (Blée et al., 2014). Compared with the wild type, Arabidopsis plants with high AtCLO4 expression were less sensitive to exogenous ABA, salt and mannitol stresses, but a loss-of-function mutant (atclo4) was hypersensitive (Kim et al., 2011). In OsEFA27, the first OB calcium protein identified in rice, experimental results showed that the protein was induced by exogenous ABA (Frandsen et al., 1996). Wei et al. (2011) identified 6 OsCLO genes, 3 (OsCLO-2, OsCLO-3, and OsCLO-6), that can be induced by drought stress. In wheat, CLO3 plays an important role in low-temperature stress, stomatal regulation and G (GTP-binding protein) protein signal transduction (Khalil et al., 2011).
Cotton is an important cash crop and plays an important role in the world’s textile industry (Du et al., 2018; Zhang J. et al., 2020). The yield and quality of cotton are severely impaired under exposure to various external stresses, such as salinity and drought (Zhou et al., 2014; Abdelraheem et al., 2019). The caleosin (CLO) gene family has been identified in Arabidopsis and some other species, and it has been found that it has an important relationship with signal transduction and a variety of abiotic stresses (Kim et al., 2011; Blée et al., 2014; Khalil et al., 2014). However, there are few studies on the function of CLO genes in cotton, and the regulatory mechanism is not clear. Therefore, it is necessary to explore the potential function of the CLO gene family in cotton. In this work, the members of the CLO gene family were identified in 15 plant species, and phylogenetic analysis was performed. The expression profiles and preliminary functions of GhCLOs in response to salt stress were analyzed. Virus-induced gene silencing verified that GhCLO06 has a positive regulatory effect on salt tolerance in cotton, and this positive function may be related to the ABA signaling pathway. The results provide an important reference for further exploring the potential roles of CLO genes in cotton stress resistance.
Genome and protein sequence data for Gossypium arboreum (CRI), Gossypium raimondii (JGI) and Gossypium barbadense (HAU) were downloaded from CottonFGD (Zhu et al., 2017), and those for Gossypium hirsutum (ZJU) were obtained from CottonGen (Yu et al., 2014). Eight Arabidopsis AtCLO1-8 sequences (Shen et al., 2014) were obtained from the Arabidopsis thaliana TAIR website1. The hidden Markov model (HMM) profile (PF05042) of the conserved caleosin domain was downloaded from the Pfam database2. The HMMER 3.0 program (Finn et al., 2011) was used to identify all CLO gene family members based on the published genomes of species [e-value (E) < 10–20]. Searches were also performed against ten other species, namely, Micromonas pusilla, Physcomitrella patens, Azolla filiculoides, Oryza sativa, Eucalyptus grandis, Glycine max, Populus trichocarpa, and Theobroma cacao (data downloaded from phytozome_V13), Picea abies (data downloaded from the PlantGenIE.org website3), and Ostreococcus lucimarinus (data downloaded from the NCBI genome website4). Furthermore, the conserved domains of all the candidate CLO protein sequences were identified using the online Simple Modular Architecture Research Tool (SMART) (Letunic et al., 2021). The CLO genes (except AtCLOs) were named based on gene positions on the chromosomes.
Phosphorylation sites of CLO were predicted using NetPhos 3.1 (Blom et al., 1999, 2004). The isoelectric point (pI) and molecular weight (MW) of CLO proteins were analyzed by the ExPASy Proteomics Server5. Transmembrane domain analysis of CLO protein sequences was performed using TMHMM6.
To study the phylogenetic relationships among different species, multiple sequence alignments of CLO protein sequences were carried out using the Clustal X program (Larkin et al., 2007) and imaged using ESPript 3.0 (Robert and Gouet, 2014). The alignment result was employed to construct a neighbor-joining (NJ) tree by MEGA 7.0 software, 1,000 bootstrap repetitions were used to increase the reliability of interior branches, and the default values were used for other parameters (Kumar et al., 2016).
To better understand the conservation of the CLO genes, the GSDS 2.0 program was used to analyze the structures of the CLO gene family (Hu et al., 2015). The gene loci of four cotton species were confirmed according to the genome annotation data and drew by TBtools software (Chen C. et al., 2020).
This study used a BLASTp search (E-value < 1e-10) to align protein sequences in three cotton species, and the MCScanX program in TBtools was employed to perform genome collinearity analysis based on the BLASTp results (Wang et al., 2012; Chen C. et al., 2020). The circular maps of identified CLO gene pairs in three cotton species were displayed using TBtools software (Chen C. et al., 2020). The coding sequences of CLO homologous gene pairs were used to calculate the ratios of non-synonymous (Ka) substitutions and synonymous (Ks) substitutions by the NG methods of TBtools to evaluate the selection pressure on these gene pairs (Hurst, 2002; Chen C. et al., 2020). Normally, Ka/Ks < 1 indicates purifying selection, Ka/Ks = 1 indicates neutral selection, and Ka/Ks > 1 indicates positive selection. The divergence times of the homologous gene pairs were estimated using the formula t = Ks/2r, with r = 2.6 × 10–9 representing neutral substitution (Sun et al., 2019).
The conserved domains of GhCLO proteins were analyzed using the online software MEME 5.1.0 with the following optimized parameters: the maximum number of motifs was set to 6, and other parameters were set to default values (Bailey et al., 2009). The GhCLO promoter regions containing 2,000 bp of DNA upstream of the initiation codon (ATG) were extracted from the G. hirsutum genome database (Yu et al., 2014). The 2,000-bp upstream regions were analyzed by PlantCARE software to detect cis-acting elements in the promoter regions (Lescot et al., 2002).
The upland cotton material TM-1 was planted in greenhouse with a suitable environment (light/dark cycle: 28°C for 16 h and dark for 8 h) to explore the reaction to NaCl, PEG, methyl jasmonate (MeJA), and ABA treatment. When the seedlings reached the stage with two flat true leaves, their roots were soaked in 200 mM NaCl and 30% PEG6000, respectively. And the leaf samples were collected after 0, 1, 3, 6, 12, and 24 h of treatment. The leaves were sprayed with 100 mM MeJA and 200 mM ABA, respectively, and the leaves of three seedlings were collected from every treatment at 0, 1, 3, 6, 9, 12, and 24 h after the stress treatments. Three biological replicates were collected from each plant and immediately frozen in liquid nitrogen.
RNA-Seq data were obtained from the SRA database (PRJNA490626) (Hu et al., 2019). Raw RNA-seq reads were filtered using the SRAToolkit (v 2.9.2) (Leinonen et al., 2011) and trimmed by Trimmomatic (v 0.3.9) (Bolger et al., 2014) to generate clean reads, and the filtered clean RNA-seq reads were analyzed by HISAT2 (v 2.1.0) (Kim et al., 2015), SAMtools (v 1.9) (Li et al., 2009), and StringTie (v 2.0) (Pertea et al., 2015). Gene expression was measured in fragments per kilobase per million (FPKM) values, and expression levels were expressed as log2 (FPKM + 1) values (Chen P. et al., 2020). HemI 1.0.3.7 software was used to visualize the results (Deng et al., 2014).
A Polysaccharides and Polyphenolics-rich RNAprep Pure Plant Kit (TIANGEN, Beijing, China) was used to extract total RNA from collected samples, and the RNA samples were reverse transcribed into complementary DNA (cDNA) using the Prime Script RT Reagent Kit (TaKaRa, Japan). An ABI 7500 real-time PCR system (Applied Biosystems, United States) was used to perform qRT–PCR (Promega, Madison, WI, United States) with three biological replicates. The qRT–PCR primers of GhCLOs were listed in Supplementary Table 1, and GhACTIN was used as a constituent expression control in qRT–PCR experiments. The results were calculated using the 2–Δ Δ Ct relative quantitative method (Livak and Schmittgen, 2001).
Virus-induced gene silencing (VIGS) assays were carried out by using tobacco rattle virus (TRV) vectors (Burch-Smith et al., 2004). The TRV system contains two vectors, pTRV1 (pYL192) and pTRV2 (pYL156), and the gene was silenced by inserting the target gene fragment of the pTRV2 vector. The web-based SGN VIGS Tool7 was used to design the silenced fragment of GhCLO06. The fragment was PCR-amplified and cloned into the pTRV2 vector to produce pTRV2:GhCLO06 constructs. Oligo 7 software was used to design primers (Supplementary Table 1) (Rychlik, 2007). The pTRV2:00, pTRV2:GhCLO06, pTRV2:GhPDS, and pTRV1 (pYL192) constructs were transformed into Agrobacterium tumefaciens strain LBA4404. The above cultures were collected by centrifugation and resuspended in infiltration buffer (10 mM MgCl2, 10 mM MES, and 200 μM acetosyringone) to a 1.5 OD600 value. After incubation at room temperature for 3 h, the first three kinds of Agrobacterium suspensions containing vectors of pTRV2:00, pTRV2:GhCLO06, and pTRV2:GhPDS were mixed with the same amount of Agrobacterium suspension holding the vector of pTRV1 (pYL192). Cotton seedlings were grown at 16 h/8 h (light/dark) at 25°C. After the cotyledons of cotton were flattened, the seedlings were infiltrated with mixed culture using a 1 ml syringe. Leaves were collected for RNA extraction and interference efficiency detection. At the three-leaf stage, silenced plant roots were soaked in 200 mM NaCl solution, and deionized water was used as a control. The treatments were repeated three times. Cotton plant wilting rates were calculated as the percentage of wilted plants to the total stressed plants. Malondialdehyde (MDA), superoxide dismutase (SOD), and peroxidase (POD) were extracted and identified according to standard methods (Solarbio, Beijing, China). The error bars represent the standard deviations of three biological replicates.
To identify the CLO genes in green plants, 107 CLO genes were identified in G. hirsutum, G. barbadense, G. raimondii, GossypiumArboretum, and 11 other species (Figure 1 and Supplementary Table 2) including green algae (O. lucimarinus, M. pusilla), a bryophyte (P. patens), a pteridophyte (A. filiculoides), a gymnosperm (P. abies), a monocot (O. sativa), and eudicots (E. grandis, G. max, P. trichocarpa, A. thaliana, and T. cacao). The CLO genes identified in these species were named with a species-specific letter as a prefix and a numerical suffix, which was based on the chromosomal position of the gene (Supplementary Table 2). The evolutionary relationships of these 15 species and the number of corresponding CLO genes were determined (Figure 1). No CLO genes were identified in the green algae (O. lucimarinus and M. pusilla). Among the six plant evolutionary lineages from lower plants to higher plants, the size of the CLO gene family varied from 0 to 19 members (Supplementary Table 2). Three species had more than 10 members, five species had 7–10 members, and the other species had fewer than 5 members.
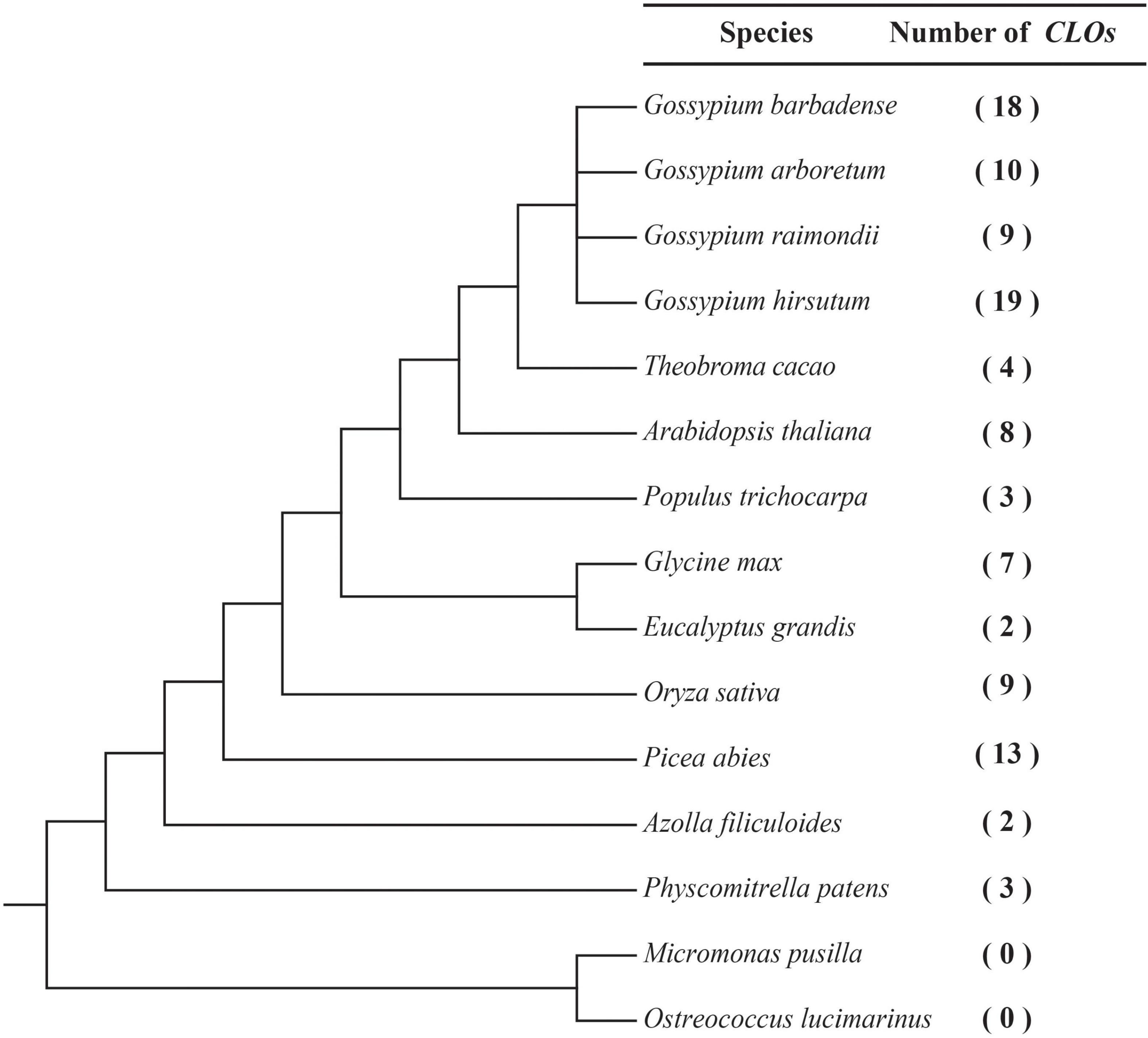
Figure 1. Inferred phylogenetic relationships among 15 species. The number of CLO genes detected in each genome is indicated on the right.
In total, 19, 18, 9, and 10 CLO genes were identified in G. hirsutum, G. barbadense, G. raimondii, and G. arboreum, respectively. The protein lengths of GhCLO, GbCLO, GaCLO, and GrCLO members varied from 209 to 236 (aa), 143 to 296 (aa), 202 to 285 (aa), and 202 to 287 (aa), respectively. The physicochemical properties of CLOs showed that the pI of the protein was between 5.812 and 9.268, and the molecular weight was between 16.16 and 33.369 kDa. Subcellular localization results showed that CLO proteins were mainly located in the cytoplasm and periplasmic region (Supplementary Table 3).
To study the evolutionary relationships of the CLO gene family, referring to a study in Arabidopsis (Shen et al., 2014), 107 CLO proteins were classified into three categories (named class I, class II, and class III) (Supplementary Figure 1). Among the 3 types of CLO proteins, class I was clustered with H (high)-caleosins of Arabidopsis, which was also the largest category, with 66 CLO proteins (11 species). Class II contained 37 proteins (11 species), which were clustered with L (low)-caleosins of A. thaliana. In addition to these 2 categories, a unique class, class III, was formed, which contained 4 CLO genes, including all bryophyte (P. patents) CLO genes (3) and one pteridophyte (A. filiculoides) CLO gene (Supplementary Figure 1). Except for those in gymnosperms (P. abies), the CLO genes were distributed in the 2 clusters (class I and class II). These results showed that in the long-term evolutionary process, the CLO gene family formed certain species differences from lower plants to higher plants, but it remained highly conserved within species.
To further investigate the phylogenetic relationships and understand the structural diversity and structural characteristics of CLO genes, the intron/exon structures of each CLO from G. hirsutum, G. barbadense, G. arboreum, and G. raimondii were analyzed (Supplementary Figure 2). The numbers of exons and introns in CLO genes in cotton ranged from 5 to 9 and 4 to 8, respectively. Eighty percent (45/56) of the CLO genes contained 6 exons and 5 introns, except for GaCLO04 and GbCLO16 (5 exons and 4 introns); GrCLO01, GrCLO05, GaCLO08, GaCLO10, GhCLO05, GbCLO01, GbCLO05, and GbCLO10 (6 exons and 5 introns); and GbCLO01 (9 exons and 8 introns). The number of exons/introns is related to the organism’s ability to adapt to adverse environmental conditions, structural divergence and functional differentiation (Xu et al., 2012; Shang et al., 2017). Motif analysis of 56 CLO amino acid sequences of 4 cotton genomes (G. hirsutum, G. barbadense, G. arboreum, and G. raimondii) was carried out by the MEME program. Six motifs (motifs 1–6) were identified in the CLO proteins (Supplementary Figure 2): 68% (38/56) contained motif 4; class I contained motif 1, motif 2 and motif 6; and class II contained motif 4, motif 1, motif 6 and motif 3 but not the GbCLO15. These results showed that the CLO gene family was highly conserved in terms of protein sequence and gene structure, but the structural differences of some genes might also lead to functional differentiation.
Multiple alignments were performed using 56 caleosin protein sequences from 4 cotton species and 8 caleosin protein sequences from A. thaliana. The alignments of H-form insertions and EF-hand Ca2+-binding motifs are shown in Figure 2. Among the 64 CLO proteins, 42 contained H-form insertions and were called H-caleosins, and the others were called L-caleosins (Supplementary Table 3). The N-terminus was the main difference between L- and H-isoform caleosins. An insertion in the N-terminus of the middle hydrophobic region of the H-isoform made its N-terminus larger (Naested et al., 2000; Hanano et al., 2006; Shen et al., 2014). The EF-hand Ca2+-binding motifs of GaCLO04 were partially lost, resulting in the incompleteness of the domain, which might cause it to lose its ability to bind calcium (Shen et al., 2016).
The chromosomal distributions of GrCLO, GaCLO, GbCLO, and GhCLO genes were visualized according to the genomic positions of 56 cotton CLO genes (Supplementary Figure 3). Ten GaCLO genes were distributed on scaffolds A02, A09, A10, A12 and 2, and 9 GrCLO genes of G. raimondii were distributed on chromosomes D02, D06, D08, D11, and D12. Among the 19 CLO genes in G. hirsutum, 10 came from the At subgenome and 9 from the Dt subgenome. Nine CLO genes were identified in the At and Dt genomes of G. barbadense. The number (18) of CLOs identified in allotetraploid G. barbadense was one less than the sum of the numbers in the two diploid cotton species (G. raimondii and G. arboreum). The distributions of CLO genes in tetraploid G. hirsutum and G. barbadense were similar, but there were differences in chromosome distribution corresponding to the diploid species (G. arboreum and G. raimondii).
The allotetraploid cotton species G. hirsutum is derived from the hybridization of two diploid cotton species (G. arboreum and G. raimondii) (Zhang Q. et al., 2020). The types of gene replication mainly include tandem duplication, segmental duplication and whole-genome duplication (WGD) (Cannon et al., 2004). BLASTp and MCScanX software were used for homologous sequence alignment and collinearity analysis of CLO genes in four cotton species, and the results were used to identify the duplication types of tetraploid cotton species (G. hirsutum and G. barbadense) (Wang et al., 2012). The analysis of the gene duplication types of G. hirsutum and G. barbadense showed that CLO family genes mainly came from segmental duplication or WGD (Supplementary Table 4), which indicated that segmental duplication or WGD played an important role in the evolution of the CLO gene family. Homologous gene pairs were determined from the results of the gene sequence comparison program BLASTp, the genomes of G. hirsutum, G. arboreum, and G. raimondii were analyzed for collinearity, and the results were visualized (Figure 3). Most of the non-synonymous (Ka)/synonymous (Ks) values of all identified CLO homologous gene pairs were less than 1, and only 3 pairs [(GhCLO05/GaCLO08), (GhCLO10/GaCLO05), and (GhCLO10/GhCLO19)] showed values greater than 1, indicating that these 3 gene pairs might undergo positive selection, resulting in gene differentiation and new biological functions, while other genes were under strong purifying selection (Supplementary Table 5). The divergence time of CLO genes in G. raimondii, G. arboreum and the two subgenomes of G. hirsutum was predicted by the formula “t = Ks/2r” (r = 2.6 × 10–9) (Zhang et al., 2015). The results showed that except for the two gene pairs [(GhCLO05/GaCLO08) and (GhCLO07/GaCLO10)], the divergence time of CLOs of three cotton species might have occurred 1.203 to 50.848 million years ago (MYA) (Supplementary Table 5).
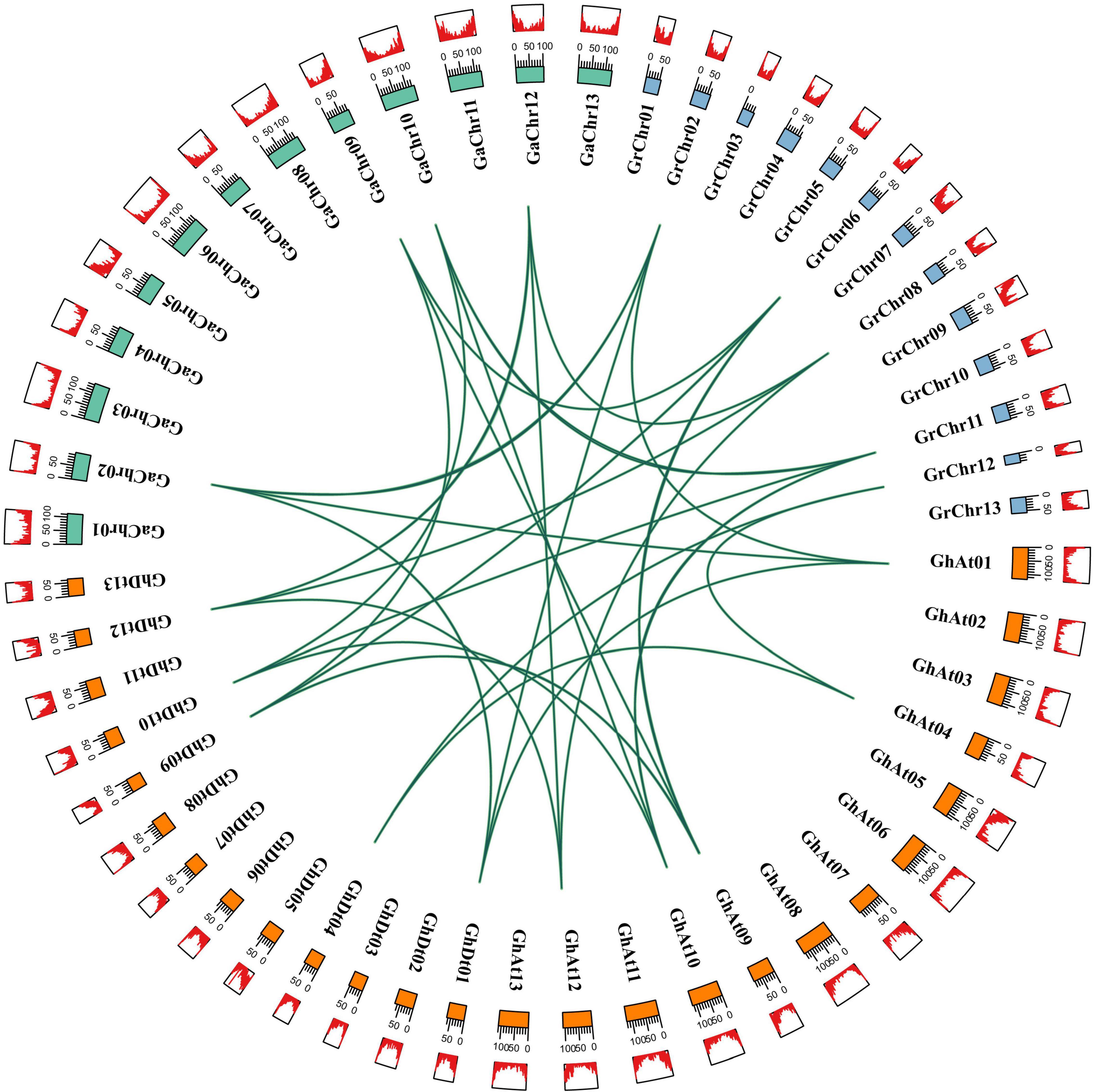
Figure 3. CLO homologous gene pairs among Gossypium arboreum, Gossypium raimondii, and Gossypium hirsutum. Orange, blue and green represent chromosomes of G. hirsutum, G. raimondii and G. arboreum, respectively; red represents the density of genes on chromosomes.
To better study the possible functions of GhCLOs in abiotic stress and hormone regulation, the 2,000-bp promoter regions of 19 GhCLO genes were analyzed by PlantCARE (Figure 4 and Supplementary Table 6). Among the 5 hormones, the number of cis-elements related to ABA hormone was the largest (38), distributed in 13 CLO promoters, and followed by MeJA, including 34 cis-elements. In addition, there were 3 stress-related elements, namely, defense and stress (TC-rich repeats), drought (MBS) and low temperature (LTR), with numbers of 13, 12, and 10, respectively. Low-temperature stress elements were distributed in the promoters of 8 genes, and the other 2 types of elements were distributed in the promoters of 10 CLO genes. In addition, there were 16 elements (W-box) in the GhCLO family and distributed in the promoters of 10 GhCLO genes (Supplementary Table 6). Studies have shown that the W-box plays an important role in the response to salt stress (Xu et al., 2018; Yao et al., 2020). These results revealed that GhCLOs might be related to hormones and multiple abiotic stresses.
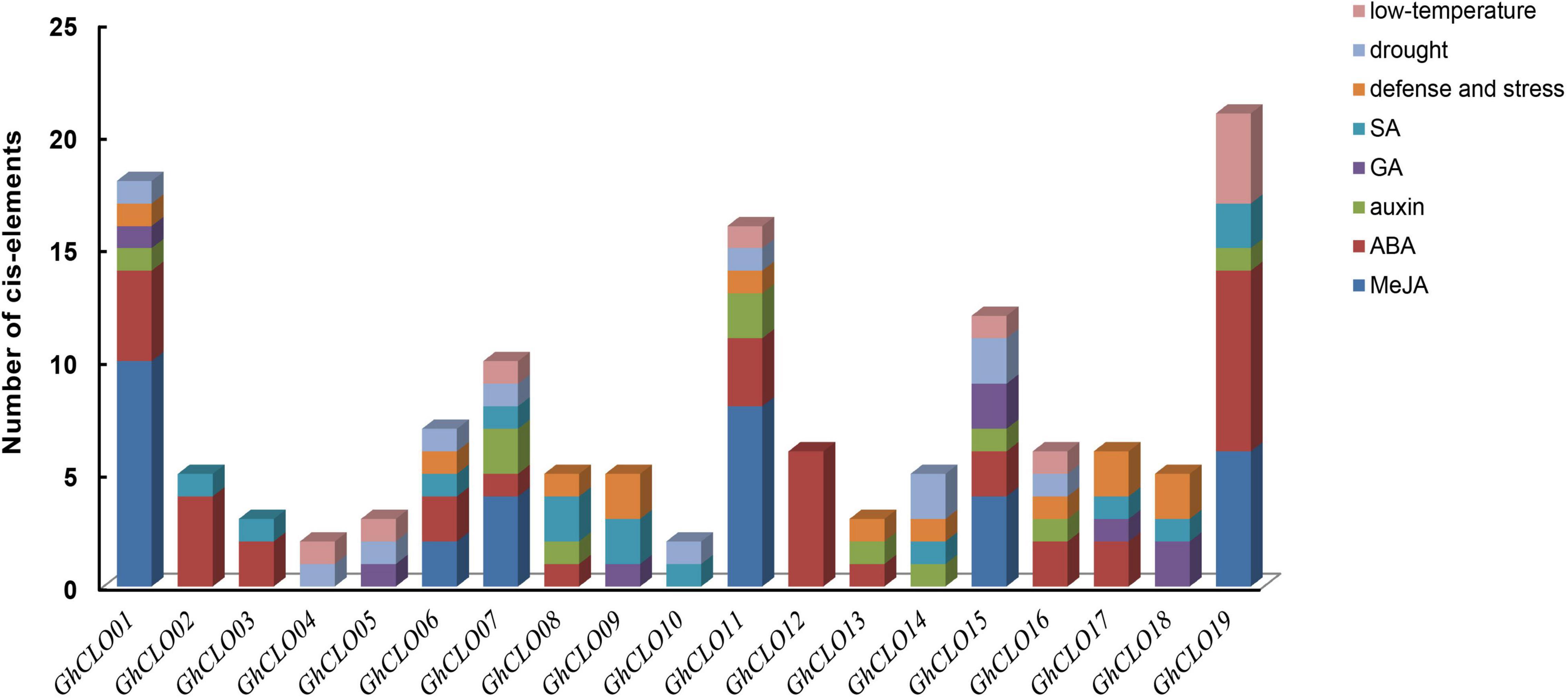
Figure 4. Cis-elements of GhCLO genes in promoter regions. The numbers of different cis-elements are presented as bars, with similar cis-elements shown by the same colors.
To deeply study the potential biological functions of the GhCLO gene family, their tissue specificity in cotton was analyzed, including root, steam, leaf, torus, petal, pistil, sepal, and bract tissues (Figure 5A and Supplementary Table 7). GhCLO06 was highly expressed in many tissues; GhCLO02, GhCLO07, GhCLO16, and GhCLO12 were highly expressed in individual tissues, but the expressions of 9 GhCLO genes, including GhCLO04, GhCLO05, GhCLO08, GhCLO09, GhCLO13, GhCLO14, GhCLO18, GhCLO17, and GhCLO15, were very low in the eight tissues. The differences of tissue-specific expression indicated that the functions of the GhCLOs might have been differentiated in the long-term evolutionary process, and its specific biological functions might diverge among tissues.
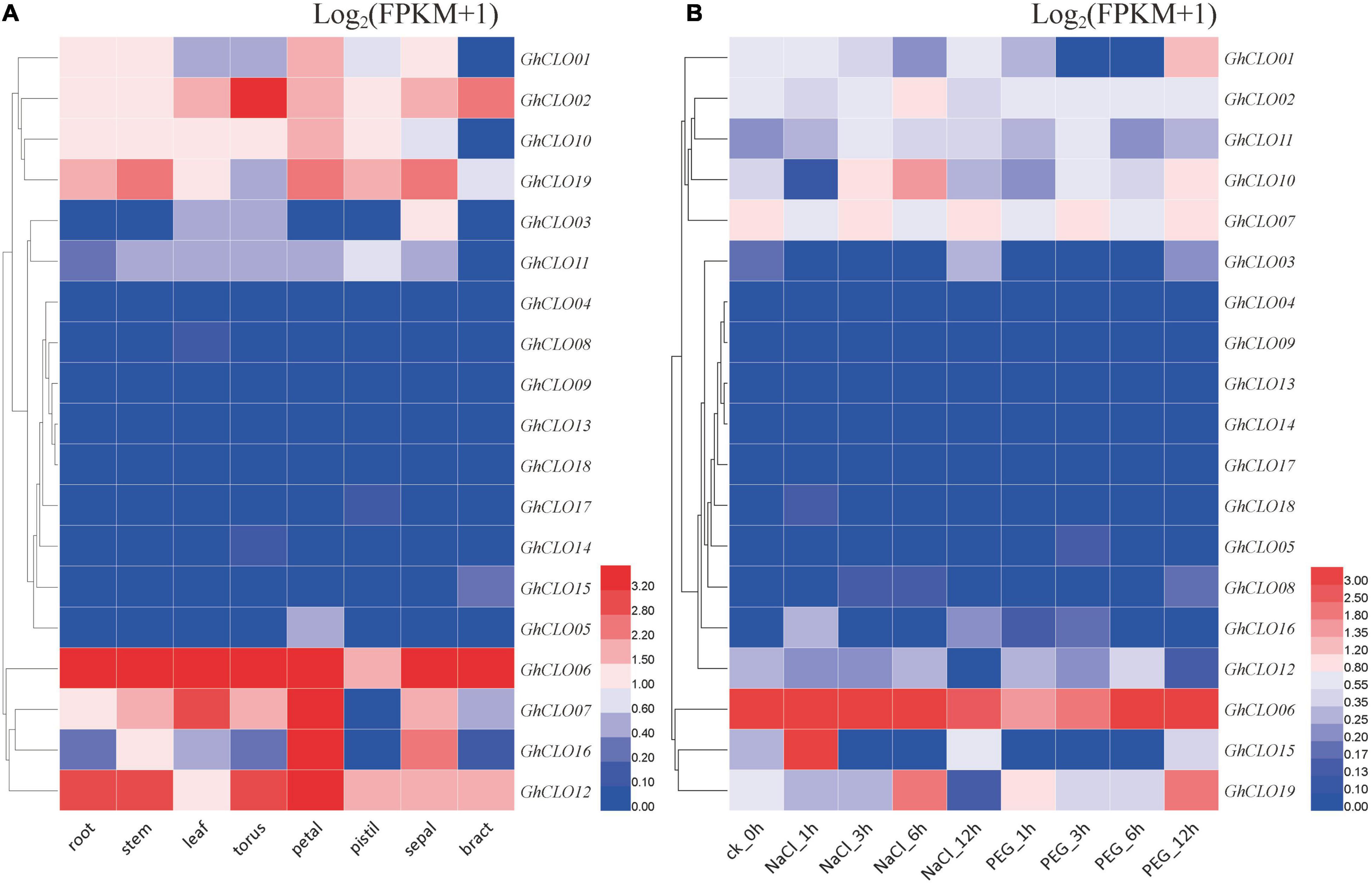
Figure 5. Expression profiles of GhCLOs in different tissues (A) and responses to different stresses (B). The tissues or treatments are shown at the bottom, the genes are shown on the right, and the phylogenetic relationships are shown on the left. The color scale in the upper right corner of the heatmap represents the FPKM values, which were standardized by log2(FPKM + 1). FPKM, fragments per kilobase per million.
Transcriptome data were also used to analyze the GhCLO gene family under salt and drought stresses (Figure 5B and Supplementary Table 7). There were significant differences in the expression trends of GhCLO genes after salt and drought treatments. GhCLO01, GhCLO02, GhCLO07, GhCLO10, and GhCLO11 showed varying trends after treatment, and the expression levels were relatively low in different treatment periods. The expression levels of GhCLO03, GhCLO04, GhCLO05, GhCLO08, GhCLO09, GhCLO13, GhCLO14, GhCLO16, GhCLO17, and GhCLO18 were very low at different stages after treatment. GhCLO06 was induced after salt and drought treatments, and its expression was higher at different stages after treatment. These results suggested that GhCLOs might have functional differences in response to abiotic stress.
Based on the cis-elements in the promoter of the GhCLO genes and the results of previous studies, 6 GhCLO genes were selected for qRT–PCR experiments under salt treatment (Figure 6A). The expressions of GhCLO10 and GhCLO11 first increased and then decreased after treatment, and the expression levels were the highest at 6 h. GhCLO02 and GhCLO16 were significantly induced at 24 and 12 h after treatment, respectively. The expression of GhCLO06 was higher than that of the control (0 h) in different treatment periods. GhCLO09, GhCLO13, GhCLO14, GhCLO16, GhCLO17, and GhCLO18 were selected for drought treatment analysis (Figure 6B). The expression levels of GhCLO01 and GhCLO11 were higher at 3 and 24 h after treatment, respectively. The expression levels of GhCLO03 and GhCLO12 in different periods after drought stress treatment were lower than those in the control (0 h). GhCLO06 and GhCLO19 were induced at 12 h after treatment. These results showed that GhCLO genes had different expression patterns in response to salt and drought stresses.
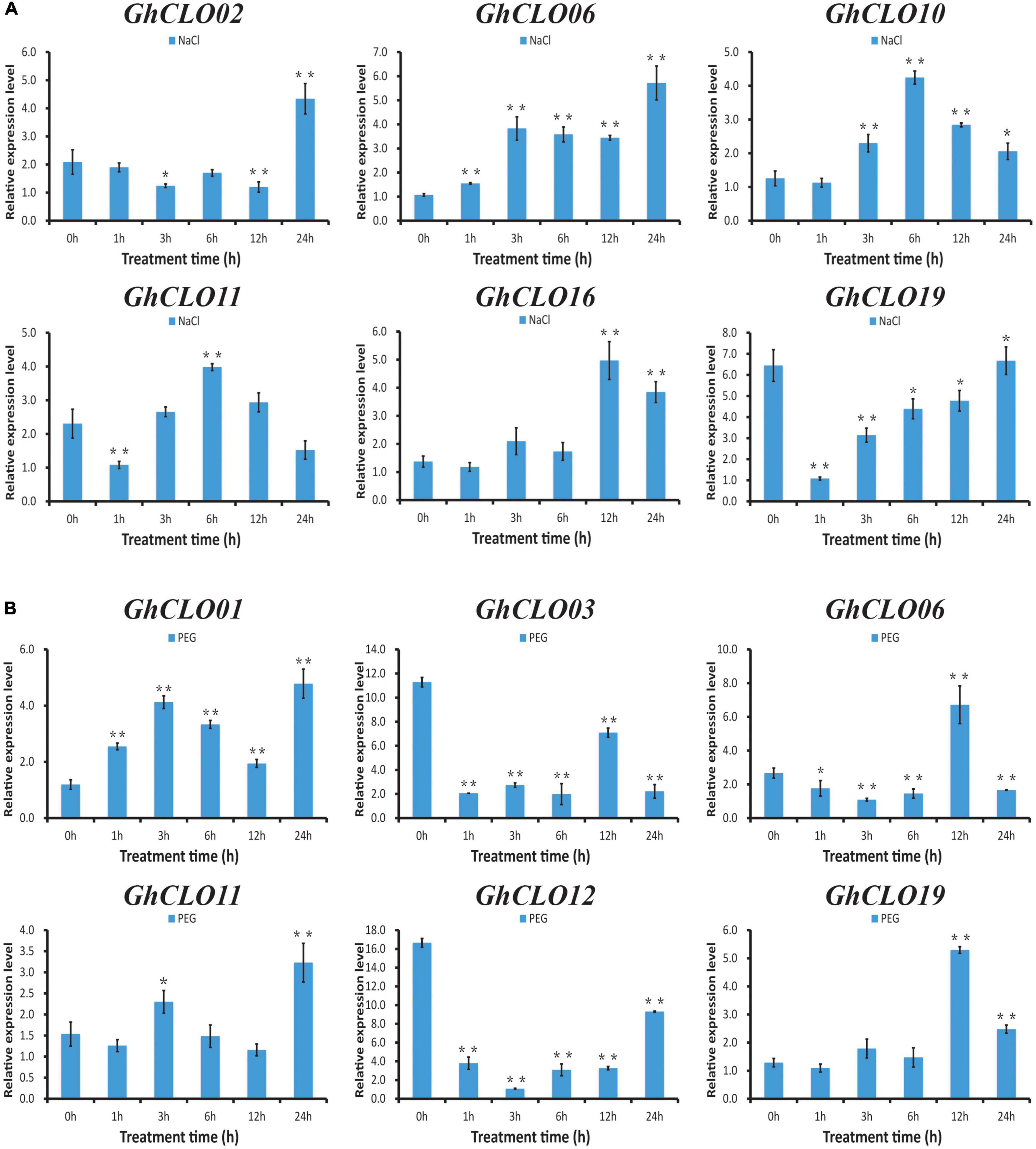
Figure 6. Relative expression levels of GhCLO genes under salt and PEG treatments detected by qRT-PCR. (A) The relative expression levels of selected genes under control conditions (water) and salt treatment. (B) The relative expression levels of selected genes under control conditions (water) and PEG treatment. Error bars show the standard deviations of three biological replicates. *P < 0.05, **P < 0.01 (t-test).
In the prediction of cis-elements in the promoter, it was found that the numbers of cis-elements related to ABA and MeJA hormones were larger (Supplementary Table 6). Studies have shown that ABA and MeJA in plants play critical roles in the response to a variety of abiotic stresses, such as high salinity, drought stress and cold (Reyes and Chua, 2007; Su et al., 2011; Chen et al., 2019; Tavallali and Karimi, 2019). Six GhCLO genes were selected for exogenous ABA and MeJA stress analysis (Figure 7). After exogenous ABA treatment, the expression levels of GhCLO01, GhCLO06, and GhCLO19 were the highest at 9 h. The expression levels of GhCLO03, GhCLO07, and GhCLO16 peaked at 3 h (Figure 7A). After treatment with exogenous MeJA, the expression levels of GhCLO06, GhCLO07, and GhCLO15 were significantly lower than those in the control (0 h). The relative expression level of GhCLO19 increased gradually 3–12 h after treatment (Figure 7B). These results suggested that GhCLOs might be regulated by MeJA or ABA.
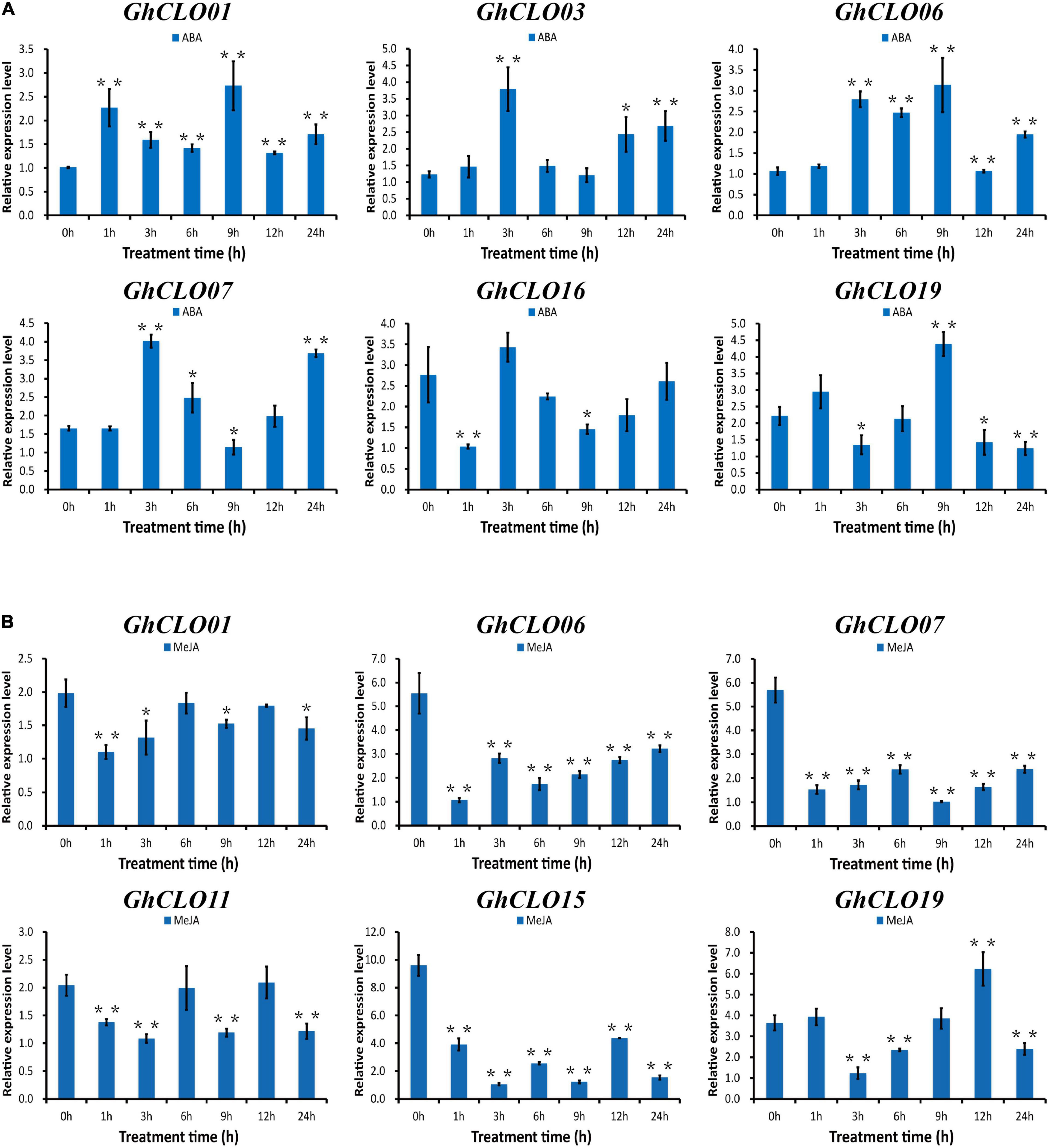
Figure 7. Relative expression levels of GhCLO genes under ABA and MeJA treatments. Two true leaves were sprayed with 200 mM ABA and 100 mM MeJA, and water was used as a blank control. (A) The relative expression levels of selected genes under control conditions (water) and ABA treatment. (B) The relative expression levels of selected genes under control conditions (water) and MeJA treatment. Error bars show the standard deviations of three biological replicates. *P < 0.05, **P < 0.01 (t-test).
By combining transcriptome data and qRT–PCR analysis, it was found that the GhCLO06 was responded positively to salt and drought stresses (Figures 5B, 6A). Previous studies have shown that a CLO gene AtCLO3 (RD20) is closely related to salt stress and ABA (Takahashi et al., 2000; Aubert et al., 2010; Blée et al., 2014). These results suggested that GhCLO06 might play an important role in response to salt stress.
To further verify our prediction, we carried out a VIGS experiment to verify the role of the GhCLO06 gene in cotton under salt stress. Ten days after infection, plants in which the GhPDS gene was silenced (positive control) exhibited the albino phenotype (Figure 8A), which indicated the effectiveness of the experiment. qRT–PCR showed that the relative expression level of TRV:GhCLO06 decreased significantly compared with that of the control plant TRV:00, indicating that the gene was significantly inhibited (Figures 8B,C). To estimate the salt resistance of the target gene-silenced cotton plants, TRV:00 and TRV:GhCLO06 plants were treated with 200 mM NaCl and deionized water (control) for 4 days. Compared with the control plants (TRV: 00), the leaves of TRV:GhCLO06 cotton plants under salt treatment displayed considerable damage, including yellowing and wilting (Figure 8B). The wilting rate of TRV:GhCLO06-silenced plants was significantly higher than that of control plants (TRV:00) (Figure 8D). In addition, physiological indexes such as MDA content and SOD and POD activity in the leaves of gene-silenced (TRV:GhCLO06) plants and control (TRV:00) plants were investigated (Figures 8E–G). No significant differences in the MDA content or SOD and POD activities were observed between control (TRV:00) plants and target gene-silenced (TRV:GhCLO06) plants under normal conditions. Under salt stress for 4 days, compared with the control (TRV:00) plants, the MDA content of the target gene-silenced (TRV:GhCLO06) plants increased significantly, but the activities of SOD and POD decreased significantly. Under salt stress, the expressions of the GhABF2, GhABI5, and GhNAC4 genes related to ABA signal regulation were significantly upregulated, and the transcriptions of three genes in the target gene-silenced (TRV:GhCLO06) plants were 6 times higher than that before treatment (Figure 9).

Figure 8. Silencing of GhCLO06 decreased resistance to salt stress in cotton. (A) Leaf whitening of TRV:GhPDS (positive control). (B) Phenotype of the control plants (TRV:00) and gene-silenced plants with GhCLO06 gene silencing (TRV:GhCLO06) under normal conditions (Mock) and salt stress for 4 days. (C) Relative expression of GhCLO06 in the control plants (TRV:00) and gene-silenced plants (TRV:GhCLO06) determined via qPCR analysis. (D–G) Physiological parameters were quantified in plants cultivated under normal (mock) and salt stress conditions. (D) Wilting rate; (E) malondialdehyde (MDA) content; (F) superoxide dismutase (SOD) activity; (G) peroxidase (POD) activity. Error bars indicate standard deviations estimated by three independent experiments. **P < 0.01 (t-test).
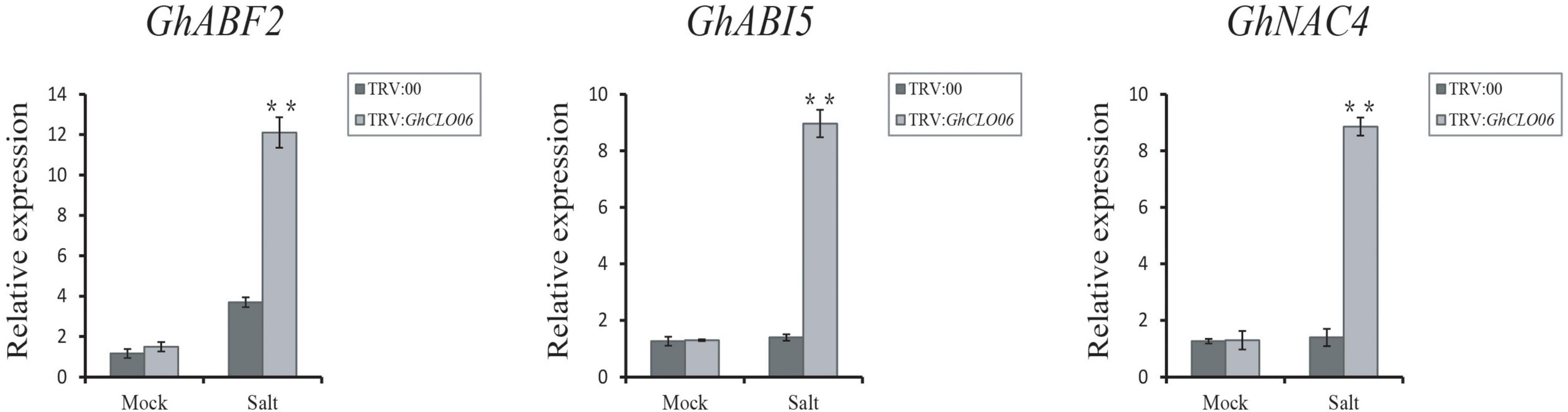
Figure 9. Abscisic acid-responsive genes GhABF2, GhABI5, and GhNAC4 in control plants and TRV:GhCLO06 plants in normal (mock) and salt environments; ABA, abscisic acid. Error bars indicate standard deviations estimated by three independent experiments. **P < 0.01 (t-test).
The completion of whole-genome sequencing provides support for the whole-genome identification and evolutionary analysis of gene families in many plants. In this study, the CLO families of 13 terrestrial plants and 2 green algal species were analyzed, and a total of 107 CLO genes were identified. Interestingly, CLO genes were detected in terrestrial plants but not in the two tested green algal species (Figure 1 and Supplementary Table 2). We speculated that the current CLO gene family may have evolved mainly in terrestrial plants. During the evolution of terrestrial plants, the size of the CLO gene family varied from 2 to 19 members. Three species had more than 10 members, five species had 7–10 members, and the other species had fewer than 5 members (Figure 1). Studies have found that the CLO gene family is widespread in terrestrial organisms and plays a role in a variety of stress responses (Wei et al., 2011; Hanano et al., 2015; Purkrtová et al., 2015). The size of the gene family in species evolution might be closely related to its species-specific function. These results indicated that the CLO gene family may have experienced specific biological functional differentiation during evolution in terrestrial organisms, and species-specific expansion has occurred after the evolution of these species. This phenomenon also occurred in other gene families, such as the HMGS gene family (Liu et al., 2019).
A total of 19, 18, 10, and 9 CLO genes were identified in G. hirsutum, G. barbadense, G. arboreum, and G. raimondii, respectively (Supplementary Table 2). Allotetraploid cotton is the result of genomic hybridization and doubling approximately 1–1.5 MYA (Wendel and Cronn, 2003; Li et al., 2015), and gene loss is most likely an ongoing process in allotetraploid cotton (Zhang et al., 2015), which may have resulted in G. barbadense (18) lacking a CLO gene compared with G. hirsutum (19) and provides strong support for the study of cotton polyploidy.
To investigate the conservation of the CLO gene sequences in cotton, exon–intron structures and conserved motifs were analyzed (Supplementary Table 3). The numbers of exons and introns of CLO genes in cotton ranged from 5 to 9 and from 4 to 8, respectively. Eighty percent (45/56) of CLO proteins contained 6 exons and 5 introns. In addition, among the six types of motifs identified, the motifs on the same branch in the four cotton species showed a high degree of conservation (Supplementary Figure 2). The structural similarity of the CLO gene family reveals structural conservation in evolution, and the structural differences among individual genes also reflect diversity in evolution.
Polyploidy is a manifestation of plant adaptability to the environment, and it is also an important mechanism of new species formation (Ramsey and Schemske, 1998). To further study the evolutionary relationship between two diploid species (G. arboreum and G. raimondii) and allotetraploid species (G. hirsutum and G. barbadense), we analyzed the chromosomal distribution of CLO genes and gene duplication events (Supplementary Figure 3 and Supplementary Table 4). The results showed that the chromosomal distributions of CLO genes in G. arboreum and the corresponding At subgenome of allotetraploid cotton were not identical, which may be caused by chromosome translocation in the process of tetraploid cotton speciation (Hu et al., 2019). The chromosomal locations of CLO genes in G. raimondii and the corresponding Dt subgenome of allotetraploid cotton were highly consistent (Supplementary Figure 3), illustrating that CLO genes in the allotetraploid cotton Dt subgenome were highly conserved in the process of evolution. A similar chromosome evolutionary distribution was found in the RPD3 gene family (Zhang J. et al., 2020). The replication of a single gene, chromosome or genome is the main force of plant genome evolution (Paterson et al., 2012). The duplication types of 37 genes in G. hirsutum and G. barbadense showed that 35 CLO genes were formed by WGD or segmental duplication based on collinearity analysis. The cotton NF-YA, NHX, and GT47 families showed enlargement as a result of WGD and segmental duplication (Wu et al., 2019; Fu et al., 2020; Zhang Q. et al., 2020). Studies have reported that cotton has a decaploid ancestor, which has experienced an extremely complex polyploid process (Wang et al., 2016). A- and D-genome diploid cotton began to differentiate from a common ancestor 5–10 MYA (Hu et al., 2019). Subsequently, G. hirsutum evolved from the hybridization of two diploid cotton plants approximately 1–2 MYA (Zhang et al., 2015). In G. hirsutum, the deduced divergence times of most CLO homologous gene pairs are between 4.37 and 14.56 MYA (Supplementary Table 5), accompanied by the differentiation of ancestral genomes of A and D, which is similar to the pattern observed for the RPD3 family (Zhang J. et al., 2020). The predicted divergence time of some genes ranged from 16.1 to 50.8 MYA, and the divergence of these genes might have gone through the diploid ancestor period of cotton or even occurred in the early decaploid ancestor period of cotton. In addition, through analysis of selective stress in the evolution of the CLO gene family, the results showed that the Ka/Ks ratios of almost all gene pairs were less than 1 (Supplementary Table 5), indicating that the CLO gene family was under strong purifying selection during long-term evolution and is functionally conserved.
CLO family genes are closely related to abiotic stress and signal transduction (Khalil et al., 2011; Kim et al., 2011). After salt treatment, the expression trend of GhCLO10 and GhCLO11 first increased and then decreased, GhCLO06 was induced, and the expression level in each period was higher than that in the control (water). After drought treatment, compared with those in the control, the expression levels of GhCLO03 and GhCLO12 decreased, and GhCLO06 and GhCLO19 were significantly induced at 12 h (Figure 6). The above results suggest that these GhCLO genes respond to salt and drought stress, and the difference in gene expression levels may be related to the importance of participating in the stress response.
In eukaryotes, transcriptional regulation is the main mechanism of gene expression regulation, and cis-acting elements are involved in the transcriptional regulation of genes (Ding et al., 2018; Chen P. et al., 2020; Yang et al., 2020). In general, gene expression depends on the presence or absence of these elements (Priest et al., 2009). In the prediction of cis-elements in the promoter, it was found that the numbers of cis-elements related to ABA and MeJA were the larger (Figure 4 and Supplementary Table 6). MeJA and ABA not only regulate plant growth and development but also participate in plant defense responses to environmental stress, such as mechanical injury and osmotic stress (Ellis and Turner, 2001; Anderson et al., 2004). After ABA stress treatment, the six genes were induced to varying degrees; after MeJA stress, the expression levels of GhCLO06, GhCLO07, and GhCLO15 were significantly lower than those of the control (0 h) (Figure 7). The differences in the expression levels of GhCLO genes revealed that GhCLO genes may adopt different response patterns under the stimulation of exogenous ABA and MeJA. In addition, through the analysis of transcriptome data under salt and PEG stress, it was found that the relative expression level of GhCLO06 in each period was significantly higher than that of other genes (Figure 5B), indicating that GhCLO06 may play a more important role in salt and drought stress responses than other GhCLO genes.
VIGS-TRV is an important technology for studying the gene function of cotton. The TRV vector is widely used in the study of functional genes related to the abiotic stress response of cotton (Cai et al., 2019; Zhang Q. et al., 2020). After salt stress, the target gene-silenced plants (TRV:GhCLO06) exhibited obvious yellowing and wilting (Figure 8B). Malondialdehyde (MDA), superoxide dismutase (SOD), and peroxidase (POD) are important indicators of cell oxidative damage (Mittler, 2006; Li et al., 2020). A large number of studies have shown that salt stress and other factors can lead to the accumulation of reactive oxygen species (ROS), and superoxide dismutase play an important role in the clearance of ROS (Miller et al., 2008, 2010). After salt stress, the MDA content, SOD and POD activities of gene silenced plants (TRV:GhCLO06) increased. Compared with that in the control plants (TRV:00), the MDA content in gene silenced plants (TRV:GhCLO06) showed higher accumulation, but the activities of SOD and POD decreased significantly in gene silenced plants (TRV:GhCLO06) (Figures 8E–G). Taken together, these findings demonstrated that GhCLO06 was a positive regulator of salt tolerance in plant.
In addition to ROS, gene tolerance to salt stress may also involve other physiological and biochemical mechanisms (Yu et al., 2020), among which ABA related to salt stress has been widely studied (Jia et al., 2002; Perin et al., 2019; Zhang Q. et al., 2020). In Arabidopsis, AtCLO3 and AtCLO4 have been confirmed to be related to ABA signal transduction (Kim et al., 2011; Blée et al., 2014). ABF2 (Liang et al., 2016), ABI5 (Skubacz et al., 2016), and NAC4 (Trishla and Kirti, 2021) are considered to be important genes for ABA signal regulation. After salt stress, the expression levels of GhABF2, GhABI5, and GhNAC4 in gene silenced plants were significantly upregulated (Figure 9). These results suggested that GhCLO06 might regulate salt tolerance by activating the ABA signaling pathway after salt stress.
In this work, a total of 107 CLO genes were obtained from the whole-genome identification of 15 plant species. CLO genes are ubiquitous in terrestrial plants but may be lacking in some green algal species. In addition, the gene structure, phylogeny and biological characteristics of CLO family members of four cotton species were systematically analyzed. qRT–PCR analysis suggested that some CLO genes might play important roles in the cotton response to salt stress. This research also revealed that the GhCLO06 gene might play a positive role of salt tolerance and might be regulated by ABA signaling pathway in cotton. Further studies on the role of ABA homeostasis under salt stress will help clarify the comprehensive effect of the cotton CLO gene on salt tolerance.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
XF and HTW conceived and designed the study and prepared the manuscript. XF, YY, MK, HLW, and BL performed the experiments. BW, LM, JL, and PH assisted with the analysis and interpretation of the data. SY participated in the design of the experiments and provided a critical review. All authors have read, edited, and approved the current version of the manuscript.
This research was financed by the National Key R&D Program of China (2020YFD1001004) and the China Agriculture Research System (Grant No. CARS-15-06).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.801239/full#supplementary-material
Supplementary Figure 1 | Phylogenetic tree of the CLO proteins in 13 species. Three categories of CLO proteins are indicated using different colors by iTOL.
Supplementary Figure 2 | Phylogenetically aligned conserved motif and gene structure analysis of CLO genes in four cotton species.
Supplementary Figure 3 | Distribution of CLOs among the chromosomes of four cotton species. Panels (A–D) represent the chromosomal locations of CLO genes from Gossypium hirsutum (A), Gossypium barbadense (B), Gossypium arboreum (C), and Gossypium raimondii (D), respectively. The chromosome numbers are shown on the left side of each vertical bar.
Supplementary Table 1 | A list of the primers used in this study.
Supplementary Table 2 | Nomenclature and sequencing of CLO genes.
Supplementary Table 3 | Detailed physicochemical characteristics of CLO proteins in G. hirsutum, G. barbadense, G. arboreum, and G. raimondii.
Supplementary Table 4 | Duplicate type of CLOs in G. hirsutum and G. barbadense.
Supplementary Table 5 | Ka/Ks ratios and occurrence times of segmentally duplicated CLO gene pairs in three cotton species.
Supplementary Table 6 | Statistical results of phytohormones-responsive cis-acting elements in the promoter segments of CLOs.
Supplementary Table 7 | The FPKM value of GhCLO genes in different tissues and under two different abiotic stresses.
Abdelraheem, A., Esmaeili, N., O’connell, M., and Zhang, J. (2019). Progress and perspective on drought and salt stress tolerance in cotton. Industrial Crops Products 130, 118–129. doi: 10.1016/j.indcrop.2018.12.070
Anderson, J. P., Badruzsaufari, E., Schenk, P. M., Manners, J. M., Desmond, O. J., Ehlert, C., et al. (2004). Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell 16, 3460–3479. doi: 10.1105/tpc.104.025833
Aubert, Y., Vile, D., Pervent, M., Aldon, D., Ranty, B., Simonneau, T., et al. (2010). RD20, a stress-inducible caleosin, participates in stomatal control, transpiration and drought tolerance in Arabidopsis thaliana. Plant Cell Physiol. 51, 1975–1987. doi: 10.1093/pcp/pcq155
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Benaragama, I., Meesapyodsuk, D., Beattie, A. D., and Qiu, X. (2017). Identification and functional analysis of new peroxygenases in oat. Planta 246, 711–719. doi: 10.1007/s00425-017-2729-2721
Blée, E., Boachon, B., Burcklen, M., Le Guédard, M., Hanano, A., Heintz, D., et al. (2014). The reductase activity of the Arabidopsis caleosin RESPONSIVE TO DESSICATION20 mediates gibberellin-dependent flowering time, abscisic acid sensitivity, and tolerance to oxidative stress. Plant Physiol. 166, 109–124. doi: 10.1104/pp.114.245316
Blée, E., Flenet, M., Boachon, B., and Fauconnier, M.-L. (2012). A non-canonical caleosin from Arabidopsis efficiently epoxidizes physiological unsaturated fatty acids with complete stereoselectivity. FEBS J. 279, 3981–3995. doi: 10.1111/j.1742-4658.2012.08757.x
Blom, N., Gammeltoft, S., and Brunak, S. (1999). Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 294, 1351–1362. doi: 10.1006/jmbi.1999.3310
Blom, N., Sicheritz-Pontén, T., Gupta, R., Gammeltoft, S., and Brunak, S. (2004). Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 4, 1633–1649. doi: 10.1002/pmic.200300771
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Burch-Smith, T. M., Anderson, J. C., Martin, G. B., and Dinesh-Kumar, S. P. (2004). Applications and advantages of virus-induced gene silencing for gene function studies in plants. Plant J. 39, 734–746. doi: 10.1111/j.1365-313X.2004.02158.x
Cai, C., Wang, X., Zhang, B., and Guo, W. (2019). Tobacco rattle virus-induced gene silencing in cotton. Methods Mol. Biol. 1902, 105–119. doi: 10.1007/978-1-4939-8952-2_9
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D., and May, G. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 4:10. doi: 10.1186/1471-2229-4-10
Charuchinda, P., Waditee-Sirisattha, R., Kageyama, H., Yamada, D., Sirisattha, S., Tanaka, Y., et al. (2015). Caleosin from Chlorella vulgaris TISTR 8580 is salt-induced and heme-containing protein. Biosci. Biotechnol. Biochem. 79, 1119–1124. doi: 10.1080/09168451.2015.1010480
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, P., Wei, F., Cheng, S., Ma, L., Wang, H., Zhang, M., et al. (2020). A comprehensive analysis of cotton VQ gene superfamily reveals their potential and extensive roles in regulating cotton abiotic stress. BMC Genomics 21:795. doi: 10.1186/s12864-020-07171-z
Chen, R., Ma, J., Luo, D., Hou, X., Ma, F., Zhang, Y., et al. (2019). CaMADS, a MADS-box transcription factor from pepper, plays an important role in the response to cold, salt, and osmotic stress. Plant Sci. 280, 164–174. doi: 10.1016/j.plantsci.2018.11.020
Deinlein, U., Stephan, A. B., Horie, T., Luo, W., Xu, G., and Schroeder, J. I. (2014). Plant salt-tolerance mechanisms. Trends Plant Sci. 19, 371–379. doi: 10.1016/j.tplants.2014.02.001
Deng, W., Wang, Y., Liu, Z., Cheng, H., and Xue, Y. (2014). HemI: a toolkit for illustrating heatmaps. PLoS One 9:e111988. doi: 10.1371/journal.pone.0111988
Ding, X., Li, J., Pan, Y., Zhang, Y., Ni, L., Wang, Y., et al. (2018). Genome-Wide identification and expression analysis of the UGlcAE gene family in tomato. Int. J. Mol. Sci. 19:1583. doi: 10.3390/ijms19061583
Du, X., Huang, G., He, S., Yang, Z., Sun, G., Ma, X., et al. (2018). Resequencing of 243 diploid cotton accessions based on an updated a genome identifies the genetic basis of key agronomic traits. Nat. Genet. 50, 796–802. doi: 10.1038/s41588-018-0116-x
Ellis, C., and Turner, J. G. (2001). The Arabidopsis mutant cev1 has constitutively active jasmonate and ethylene signal pathways and enhanced resistance to pathogens. Plant Cell 13, 1025–1033. doi: 10.1105/tpc.13.5.1025
Finn, R. D., Clements, J., and Eddy, S. R. (2011). HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39, W29–W37. doi: 10.1093/nar/gkr367
Frandsen, G., Müller-Uri, F., Nielsen, M., Mundy, J., and Skriver, K. (1996). Novel plant Ca(2+)-binding protein expressed in response to abscisic acid and osmotic stress. J. Biol. Chem. 271, 343–348. doi: 10.1074/jbc.271.1.343
Fu, X., Lu, Z., Wei, H., Zhang, J., Yang, X., Wu, A., et al. (2020). Genome-Wide identification and expression analysis of the NHX (Sodium/Hydrogen Antiporter) gene family in cotton. Front. Genet. 11:964. doi: 10.3389/fgene.2020.00964
Ganie, S. A., Molla, K. A., Henry, R. J., Bhat, K. V., and Mondal, T. K. (2019). Advances in understanding salt tolerance in rice. Theor. Appl. Genet. 132, 851–870. doi: 10.1007/s00122-019-03301-3308
Hanano, A., Bessoule, J.-J., Heitz, T., and Blée, E. (2015). Involvement of the caleosin/peroxygenase RD20 in the control of cell death during Arabidopsis responses to pathogens. Plant Signal. Behav. 10:e991574. doi: 10.4161/15592324.2014.991574
Hanano, A., Burcklen, M., Flenet, M., Ivancich, A., Louwagie, M., Garin, J., et al. (2006). Plant seed peroxygenase is an original heme-oxygenase with an EF-hand calcium binding motif. J. Biol. Chem. 281, 33140–33151. doi: 10.1074/jbc.M605395200
Hu, B., Jin, J., Guo, A., Zhang, H., Luo, J., and Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Hu, Y., Chen, J., Fang, L., Zhang, Z., Ma, W., Niu, Y., et al. (2019). Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat. Genet. 51, 739–748. doi: 10.1038/s41588-019-0371-375
Hurst, L. D. (2002). The Ka/Ks ratio: diagnosing the form of sequence evolution. Trends Genet. 18:486. doi: 10.1016/s0168-9525(02)02722-2721
Jia, W., Wang, Y., Zhang, S., and Zhang, J. (2002). Salt-stress-induced ABA accumulation is more sensitively triggered in roots than in shoots. J. Exp. Bot. 53, 2201–2206. doi: 10.1093/jxb/erf079
Khalil, H. B., Brunetti, S. C., Pham, U. M., Maret, D., Laroche, A., and Gulick, P. J. (2014). Characterization of the caleosin gene family in the Triticeae. BMC Genomics 15:239. doi: 10.1186/1471-2164-15-239
Khalil, H. B., Wang, Z., Wright, J. A., Ralevski, A., Donayo, A. O., and Gulick, P. J. (2011). Heterotrimeric Gα subunit from wheat (Triticum aestivum), GA3, interacts with the calcium-binding protein, Clo3, and the phosphoinositide-specific phospholipase C, PI-PLC1. Plant Mol. Biol. 77, 145–158. doi: 10.1007/s11103-011-9801-9801
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Kim, Y. Y., Jung, K. W., Yoo, K. S., Jeung, J. U., and Shin, J. S. (2011). A stress-responsive caleosin-like protein, AtCLO4, acts as a negative regulator of ABA responses in Arabidopsis. Plant Cell Physiol. 52, 874–884. doi: 10.1093/pcp/pcr039
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., Mcgettigan, P. A., Mcwilliam, H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Leinonen, R., Sugawara, H., and Shumway, M. (2011). The sequence read archive. Nucleic Acids Res. 39, D19–D21. doi: 10.1093/nar/gkq1019
Lescot, M., Dehais, P., Thijs, G., Marchal, K., Moreau, Y., De Peer, Y. V., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Letunic, I., Khedkar, S., and Bork, P. (2021). SMART: recent updates, new developments and status in 2020. Nucleic Acids Res. 49, D458–D460. doi: 10.1093/nar/gkaa937
Li, B., Zheng, J.-C., Wang, T.-T., Min, D.-H., Wei, W.-L., Chen, J., et al. (2020). Expression analyses of soybean VOZ transcription factors and the role of in drought and salt stress tolerance. Int. J. Mol. Sci. 21:2177. doi: 10.3390/ijms21062177
Li, F., Fan, G., Lu, C., Xiao, G., Zou, C., Kohel, R. J., et al. (2015). Genome sequence of cultivated upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat. Biotechnol. 33, 524–530. doi: 10.1038/nbt.3208
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). Genome project data processing S: the sequence alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Liang, C., Meng, Z., Meng, Z., Malik, W., Yan, R., Lwin, K. M., et al. (2016). GhABF2, a bZIP transcription factor, confers drought and salinity tolerance in cotton (Gossypium hirsutum L.). Sci. Rep. 6:35040. doi: 10.1038/srep35040
Liu, L., and Wang, B. (2021). Protection of halophytes and their uses for cultivation of saline-alkali soil in China. Biology 10:353. doi: 10.3390/biology10050353
Liu, W., Zhang, Z., Zhu, W., Ren, Z., Jia, L., Li, W., et al. (2019). Evolutionary conservation and divergence of genes encoding 3-Hydroxy-3-methylglutaryl coenzyme a synthase in the allotetraploid cotton species Gossypium hirsutum. Cells 8:412. doi: 10.3390/cells8050412
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Miller, G., Shulaev, V., and Mittler, R. (2008). Reactive oxygen signaling and abiotic stress. Physiol. Plant. 133, 481–489. doi: 10.1111/j.1399-3054.2008.01090.x
Miller, G., Suzuki, N., Ciftci-Yilmaz, S., and Mittler, R. (2010). Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 33, 453–467. doi: 10.1111/j.1365-3040.2009.02041.x
Mittler, R. (2006). Abiotic stress, the field environment and stress combination. Trends Plant Sci. 11, 15–19. doi: 10.1016/j.tplants.2005.11.002
Munns, R., and Gilliham, M. (2015). Salinity tolerance of crops - what is the cost? New Phytol. 208, 668–673. doi: 10.1111/nph.13519
Naested, H., Frandsen, G. I., Jauh, G. Y., Hernandez-Pinzon, I., Nielsen, H. B., Murphy, D. J., et al. (2000). Caleosins: Ca2+-binding proteins associated with lipid bodies. Plant Mol. Biol. 44, 463–476. doi: 10.1023/a:1026564411918
Partridge, M., and Murphy, D. J. (2009). Roles of a membrane-bound caleosin and putative peroxygenase in biotic and abiotic stress responses in Arabidopsis. Plant Physiol. Biochem. 47, 796–806. doi: 10.1016/j.plaphy.2009.04.005
Paterson, A. H., Wendel, J. F., Gundlach, H., Guo, H., Jenkins, J., Jin, D., et al. (2012). Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492, 423–427. doi: 10.1038/nature11798
Perin, E. C., Da Silva, Messias, R., Borowski, J. M., Crizel, R. L., Schott, I. B., et al. (2019). ABA-dependent salt and drought stress improve strawberry fruit quality. Food Chem. 271, 516–526. doi: 10.1016/j.foodchem.2018.07.213
Pertea, M., Pertea, G. M., Antonescu, C. M., Chang, T.-C., Mendell, J. T., and Salzberg, S. L. (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295. doi: 10.1038/nbt.3122
Poxleitner, M., Rogers, S. W., Lacey Samuels, A., Browse, J., and Rogers, J. C. (2006). A role for caleosin in degradation of oil-body storage lipid during seed germination. Plant J. 47, 917–933. doi: 10.1111/j.1365-313X.2006.02845.x
Priest, H. D., Filichkin, S. A., and Mockler, T. C. (2009). Cis-regulatory elements in plant cell signaling. Curr. Opin. Plant Biol. 12, 643–649. doi: 10.1016/j.pbi.2009.07.016
Purkrtová, Z., Chardot, T., and Froissard, M. (2015). N-terminus of seed caleosins is essential for lipid droplet sorting but not for lipid accumulation. Arch. Biochem. Biophys. 579, 47–54. doi: 10.1016/j.abb.2015.05.008
Rahman, F., Hassan, M., Rosli, R., Almousally, I., Hanano, A., and Murphy, D. J. (2018). Evolutionary and genomic analysis of the caleosin/peroxygenase (CLO/PXG) gene/protein families in the Viridiplantae. PLoS One 13:e0196669. doi: 10.1371/journal.pone.0196669
Ramsey, J., and Schemske, D. W. (1998). Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu. Rev. Ecol. Syst. 29, 467–501. doi: 10.1146/annurev.ecolsys.29.1.467
Reyes, J. L., and Chua, N.-H. (2007). ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. Plant J. 49, 592–606. doi: 10.1111/j.1365-313X.2006.02980.x
Robert, X., and Gouet, P. (2014). Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324. doi: 10.1093/nar/gku316
Rychlik, W. (2007). OLIGO 7 primer analysis software. Methods Mol. Biol. 402, 35–60. doi: 10.1007/978-1-59745-528-2_2
Shang, X., Cao, Y., and Ma, L. (2017). Alternative splicing in plant genes: a means of regulating the environmental fitness of plants. Int. J. Mol. Sci. 18:432. doi: 10.3390/ijms18020432
Shen, Y., Liu, M., Wang, L., Li, Z., Taylor, D. C., Li, Z., et al. (2016). Identification, duplication, evolution and expression analyses of caleosins in Brassica plants and Arabidopsis subspecies. Mol. Genet. Genomics 291, 971–988. doi: 10.1007/s00438-015-1156-x
Shen, Y., Xie, J., Liu, R. D., Ni, X. F., Wang, X. H., Li, Z. X., et al. (2014). Genomic analysis and expression investigation of caleosin gene family in Arabidopsis. Biochem. Biophys. Res. Commun. 448, 365–371. doi: 10.1016/j.bbrc.2014.04.115
Skubacz, A., Daszkowska-Golec, A., and Szarejko, I. (2016). The role and regulation of ABI5 (ABA-Insensitive 5) in plant development, abiotic stress responses and phytohormone crosstalk. Front. Plant Sci. 7:1884. doi: 10.3389/fpls.2016.01884
Song, W., Qin, Y., Zhu, Y., Yin, G., Wu, N., Li, Y., et al. (2014). Delineation of plant caleosin residues critical for functional divergence, positive selection and coevolution. BMC Evol. Biol. 14:124. doi: 10.1186/1471-2148-14-124
Su, M., Li, X.-F., Ma, X.-Y., Peng, X.-J., Zhao, A.-G., Cheng, L.-Q., et al. (2011). Cloning two P5CS genes from bioenergy sorghum and their expression profiles under abiotic stresses and MeJA treatment. Plant Sci. 181, 652–659. doi: 10.1016/j.plantsci.2011.03.002
Sun, H., Wei, H., Wang, H., Hao, P., Gu, L., Liu, G., et al. (2019). Genome-wide identification and expression analysis of the BURP domain-containing genes in Gossypium hirsutum. BMC Genomics 20:558. doi: 10.1186/s12864-019-5948-y
Takahashi, S., Katagiri, T., Yamaguchi-Shinozaki, K., and Shinozaki, K. (2000). An Arabidopsis gene encoding a Ca2+-binding protein is induced by abscisic acid during dehydration. Plant Cell Physiol. 41, 898–903. doi: 10.1093/pcp/pcd010
Tavallali, V., and Karimi, S. (2019). Methyl jasmonate enhances salt tolerance of almond rootstocks by regulating endogenous phytohormones, antioxidant activity and gas-exchange. J. Plant Physiol. 23, 98–105. doi: 10.1016/j.jplph.2019.02.001
Trishla, V. S., and Kirti, P. B. (2021). Structure-function relationship of Gossypium hirsutum NAC transcription factor, GhNAC4 with regard to ABA and abiotic stress responses. Plant Sci. 302:110718. doi: 10.1016/j.plantsci.2020.110718
Wang, X., Guo, H., Wang, J., Lei, T., Liu, T., Wang, Z., et al. (2016). Comparative genomic de-convolution of the cotton genome revealed a decaploid ancestor and widespread chromosomal fractionation. New Phytol. 209, 1252–1263. doi: 10.1111/nph.13689
Wang, Y., Tang, H., Debarry, J., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40:e49. doi: 10.1093/nar/gkr1293
Wei, Z., Ma, H., and Ge, X. (2011). Phylogenetic analysis and drought-responsive expression of the rice caleosin gene family (in Chinese). Chinese Sci. Bull. (Chinese Ver) 56, 1612–1621. doi: 10.1360/972011-972422
Wendel, J. F., and Cronn, R. C. (2003). Polyploidy and the evolutionary history of cotton. Adv. Agronomy 78, 139–186. doi: 10.1016/s0065-2113(02)78004-78008
Wu, A., Hao, P., Wei, H., Sun, H., Cheng, S., Chen, P., et al. (2019). Genome-Wide identification and characterization of glycosyltransferase family 47 in cotton. Front. Genet. 10:824. doi: 10.3389/fgene.2019.00824
Xu, G., Guo, C., Shan, H., and Kong, H. (2012). Divergence of duplicate genes in exon-intron structure. Proc. Natl. Acad. Sci. U S A. 109, 1187–1192. doi: 10.1073/pnas.1109047109
Xu, H.-S., Guo, S.-M., Zhu, L., and Xing, J.-C. (2020). Growth, physiological and transcriptomic analysis of the perennial ryegrass in response to saline stress. R. Soc. Open Sci. 7:200637. doi: 10.1098/rsos.200637
Xu, Z., Raza, Q., Xu, L., He, X., Huang, Y., Yi, J., et al. (2018). GmWRKY49, a salt-responsive nuclear protein, improved root length and governed better salinity tolerance in transgenic. Front. Plant Sci. 9:809. doi: 10.3389/fpls.2018.00809
Yang, X., Zhang, J., Wu, A., Wei, H., Fu, X., Tian, M., et al. (2020). Genome-Wide identification and expression pattern analysis of the gene family of cotton in fiber development and under stresses. Front. Genet. 11:566469. doi: 10.3389/fgene.2020.566469
Yao, J., Shen, Z., Zhang, Y., Wu, X., Wang, J., Sa, G., et al. (2020). Populus euphratica WRKY1 binds the promoter of H+-ATPase gene to enhance gene expression and salt tolerance. J. Exp. Bot. 71, 1527–1539. doi: 10.1093/jxb/erz493
Yu, J., Jung, S., Cheng, C.-H., Ficklin, S. P., Lee, T., Zheng, P., et al. (2014). CottonGen: a genomics, genetics and breeding database for cotton research. Nucleic Acids Res. 42, D1229–D1236. doi: 10.1093/nar/gkt1064
Yu, Z., Duan, X., Luo, L., Dai, S., Ding, Z., and Xia, G. (2020). How plant hormones mediate salt stress responses. Trends Plant Sci. 25, 1117–1130. doi: 10.1016/j.tplants.2020.06.008
Zhang, J., Wu, A., Wei, H., Hao, P., Zhang, Q., Tian, M., et al. (2020). Genome-wide identification and expression patterns analysis of the RPD3/HDA1 gene family in cotton. BMC Genomics 21:643. doi: 10.1186/s12864-020-07069-w
Zhang, Q., Zhang, J., Wei, H., Fu, X., Ma, L., Lu, J., et al. (2020). Genome-wide identification of NF-YA gene family in cotton and the positive role of GhNF-YA10 and GhNF-YA23 in salt tolerance. Int. J. Biol. Macromol. 165, 2103–2115. doi: 10.1016/j.ijbiomac.2020.10.064
Zhang, T., Hu, Y., Jiang, W., Fang, L., Guan, X., Chen, J., et al. (2015). Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 33, 531–537. doi: 10.1038/nbt.3207
Zhou, M., Sun, G., Sun, Z., Tang, Y., and Wu, Y. (2014). Cotton proteomics for deciphering the mechanism of environment stress response and fiber development. J. Proteom. 105, 74–84. doi: 10.1016/j.jprot.2014.03.017
Keywords: cotton, caleosin protein, salt tolerance, abscisic acid, GhCLO06
Citation: Fu X, Yang Y, Kang M, Wei H, Lian B, Wang B, Ma L, Hao P, Lu J, Yu S and Wang H (2022) Evolution and Stress Responses of CLO Genes and Potential Function of the GhCLO06 Gene in Salt Resistance of Cotton. Front. Plant Sci. 12:801239. doi: 10.3389/fpls.2021.801239
Received: 25 October 2021; Accepted: 23 December 2021;
Published: 17 January 2022.
Edited by:
Jinfa Zhang, New Mexico State University, United StatesReviewed by:
Jinpeng Wang, Institute of Botany, Chinese Academy of Sciences (CAS), ChinaCopyright © 2022 Fu, Yang, Kang, Wei, Lian, Wang, Ma, Hao, Lu, Yu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuxun Yu, eXN4MTk1MzExQDE2My5jb20=; Hantao Wang, dy53YW5naGFudGFvQDE2My5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.