
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 20 January 2022
Sec. Crop and Product Physiology
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.800625
This article is part of the Research Topic Dissection of the Molecular Basis of Fatty Acid Composition in Oil Crops and Molecular Breeding of Oil Crops With Improved Fatty Acid Composition View all 13 articles
 Xiaorong Xiao1,2,3†
Xiaorong Xiao1,2,3† Rui Wang1†
Rui Wang1† Shahneela Khaskhali1†
Shahneela Khaskhali1† Zhiliang Gao1
Zhiliang Gao1 Wenya Guo1,2
Wenya Guo1,2 Honggang Wang1
Honggang Wang1 Xiaolei Niu1
Xiaolei Niu1 Chaoze He1
Chaoze He1 Xiaohui Yu1*
Xiaohui Yu1* Yinhua Chen1,2*
Yinhua Chen1,2*Glycerol-induced resistance to various pathogens has been reported in different plants. Glycerol kinase (GK), a vital rate-limiting enzyme that catalyzes glycerol conversion to glycerol-3-phosphate (G3P), participates in responses to both abiotic and biotic stresses. However, its physiological importance in rice defenses against pathogens remains unclear. In this research, quantification analysis revealed that GK levels were significantly induced in rice leaves infected by Xanthomonas oryzae pv. oryzae (Xoo) strain PXO99. A typical GK-encoding gene OsNHO1 was cloned in rice. The transcriptional levels of OsNHO1 were significantly induced by salicylic acid, jasmonic acid, and Xoo-PXO99. Ectopic expression of OsNHO1 partially rescued the resistance to P. s. pv. phaseolicola in the Arabidopsis nho1 mutant. In the overexpressing transgenic rice lines (OsNHO1-OE), the content of GK and the transcriptional level of OsNHO1 were increased and the resistance to bacterial blight and blast was improved, while reduced OsNHO1 expression impaired the resistance in OsNHO1-RNAi lines. The wax contents and expression of the wax synthesis regulatory genes were significantly increased in the overexpression lines but decreased in the OsNHO1-RNAi lines. We then confirmed the interaction partner of OsNHO1 using yeast two-hybrid and bimolecular fluorescence complementation assays. The transcription of the interaction partner-encoding genes OsSRC2 and OsPRs in OsNHO1-RNAi lines was downregulated but upregulated in OsNHO1-OE lines. Thus, we concluded that OsNHO1 provided disease resistance by affecting the wax content and modulating the transcription levels of PR genes.
Plants are threatened by many pathogens through environmental exposure. However, they have evolved a series of complex defense mechanisms. Constitutive resistance and inducible resistance are two typical defense mechanisms by which plants deal with pathogen invasion (Mysore and Ryu, 2004; Ellis, 2006). The constitutive defenses are provided by plant cell walls, cytoskeleton, obstacles, and a variety of secondary metabolites (Yun et al., 2003). Induced defenses include the accumulation of active antimicrobial substances, the activation of plant defense signal transduction pathways, calcium influx, the accumulation of reactive oxygen species (ROS), the production of nitric oxide, the occurrence of hypersensitivity reactions, the expression of defense-related genes, etc. (Lipka, 2005; Lee et al., 2017). In recent decades, significant progress has been made in understanding inducible defense mechanisms, ranging from the pathogen-associated molecular pattern (PAMP)-induced basal resistance to effector-induced cultivar-specific resistance (Jones and Dangl, 2006; Delventhal et al., 2017).
Recent evidence has suggested that primary metabolic pathways and metabolic signaling in both plants and pathogens can interface with disease-related signaling (Rolland and Sheen, 2002). The components of primary metabolism can act as signals regulating plant defense (Schaaf and Hess, 1995; Chandra-Shekara et al., 2007). Both the fatty acid and carbohydrate metabolism play important roles in plant defense and are involved in cross-talk with various phytohormones, namely, salicylic acid (SA), jasmonic acid (JA), and abscisic acid (ABA) (Scheideler et al., 2002; Kachroo et al., 2003, 2004, 2005). Vitamin B1 and sucrose also induce resistance to pathogens in Arabidopsis and rice (Oryza sativa), respectively (Ahn et al., 2005; Gómez-Ariza et al., 2007).
Glycerol is a common cellular metabolite present in a wide range of organisms. Glycerol metabolism is initiated upon its conversion to glycerol-3-phosphate (G3P), which can be derived via glycerol kinase (GK)-mediated phosphorylation of glycerol or G3P dehydrogenase (G3Pdh)-mediated reduction of dihydroxyacetone phosphate (DHAP) (Chanda et al., 2008). The participation of glycerol and its metabolites in host defense has been reported in Arabidopsis, wheat, pepper, and soybean (Kachroo et al., 2004, 2005, 2008; Chandra-Shekara et al., 2007). AtNHO1, which encodes flagellin-induced GK in Arabidopsis, is an essential factor in gene-for-gene resistance against Pst DC3000 and basal resistance against Colletotrichum higginsianum (Kang et al., 2003; Chanda et al., 2008). G3P levels in Arabidopsis are associated with defense against the hemibiotrophic fungal pathogen Colletotrichum higginsianum. Transgenic plants that are impaired in the utilization of plastidial G3P, accumulate elevated levels of pathogen-induced G3P and display enhanced resistance. TaGLI1, which encodes GK, contributes to systemic acquired resistance against Puccinia striiformis f. sp. Tritici in wheat (Yang et al., 2013). All these previous findings suggested that regulating glycerol metabolism could enhance the immune response in plants.
Glycerol-3-phosphate can be transported between the cytosol and the plastidial stroma. In the plastids, G3P is acylated with oleic acid (18:1) by the ACT1-encoded G3P acyltransferase. This ACT1-utilized 18:1 is derived from stearoyl-acyl carrier protein (ACP)-desaturase (SSI2)-catalyzed desaturation of stearic acid (18:0). The 18:1-ACP generated by ACT1 either enters the prokaryotic lipid biosynthetic pathway through acylation of G3P or is exported from the plastids as a CoA-thioester to enter the eukaryotic lipid biosynthetic pathway (Brisson et al., 2001). C16 and C18 chain fatty acids are important precursors for wax synthesis in the endoplasmic reticulum (Wang et al., 2018). As a type of secondary metabolite, wax is widely involved in many physiological resistance processes, namely, stress defenses and resistance to pests and diseases. Wax and cutin, the main components of the cuticle, form the first line of defense against pathogen infection in plants and they play a critical role in physical resistance (physical barrier) and chemical resistance (bacteriostasis) as constitutive defense components (Ye et al., 2009). The inducible wax component can also act as a signal molecule or inducer to activate downstream resistance reactions and exert its chemical resistance function (He et al., 2018).
As a model monocot plant, rice (Oryza sativa) is one of the staple crops in many countries and has significant economic significance. However, the yield is adversely impacted by bacterial blight and rice blast caused by Xanthomonas oryzae pv. oryzae (Xoo) and Magnaporthe oryzae (M. oryzae), respectively. There is an urgent need to identify broad-spectrum resistance genes to the diseases. In this study, we isolated a rice GK gene OsNHO1 and found that OsNHO1 can contribute to the non-host resistance in Arabidopsis. Moreover, it acts as a positive regulator in resistance to Xoo-PXO99 and M. oryzae Y34. Overexpression of the OsNHO1 gene significantly increased the wax content of transgenic plants and regulated the expression of the downstream PR genes. In this study, we provided evidence that modifying glycerol metabolism may also regulate the resistance of rice by affecting wax synthesis.
Bacterial strains Xanthomonas oryzae pv. oryzae (Xoo)-PXO99, M. oryzae Y34, P. s. phaseolicola, E. coli DH5α, and A. tumefaciens were preserved and routinely cultured in our laboratory.
Arabidopsis ecotype Columbia [Col-0, wild type (WT)], nho1 mutant (Col-0 background), and rice (Oryza sativa L.) var. Nippobare, DJ, and TP309 were provided by the Institute of Microbiology, the Chinese Academy of Sciences. All the Arabidopsis plants were grown in growth chambers at 20°C at night and 22°C during the day with a 10-h/day photoperiod. Rice plants were grown at 30°C/28°C with a 14 h/10 h day/night cycle.
The cDNA sequence of OsNHO1 (LOC_Os04g55410) was obtained from the Rice Annotation Project Database (http://rice.plantbiology.msu.edu/) by BLAST with the amino acid sequence of AtNHO1 gene (accession number: AT1G80460). We designed specific primers tailed with BamH I and Sac I (OsNHO1-F/R, Supplementary Table S1) to amplify the full-length OsNHO1. The identification of the amplified OsNHO1 was verified by sequencing. Multiple sequence alignment of OsNHO1 with other GK proteins was conducted using DNAMAN Version 6.0 (Lynnon Corporation, Canada). A phylogenetic tree including the OsNHO1 and other GK proteins was constructed using the MEGA7 (Tamura, Stecher, and Kumar 2016) program. The accession numbers of proteins used in multiple sequence alignment and phylogenetic analysis are listed in Supplementary Table S2.
We designed specific primers (Ppronho1-62F/R, Supplementary Table S1) to clone the promoter sequence of OsNHO1 by chromosome walking. The fragment was sequenced, and the promoter sequence was analyzed with the PlantCARE online website (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) and visualized by TBtools software (Chen et al., 2020) (Supplementary Figure S1).
Germinant WT (TP309) rice seedlings were grown in pots at 30°C/28°C with a 14 h/10 h day/night cycle. For hormone treatments, rice plants with 4 leaves were sprayed with 1 mmol/l SA and 0.1 mmol/l methyl jasmonate (MeJA), and the control plants were sprayed with deionized water. Leaves were sampled at 0, 1, 4, 8, 12, and 24 h after treatment. For pathogen infection, rice leaves were detached from plants and infected with Xoo-PXO99 (OD 0.4–0.6), and the control was infected with water. The leaves were sampled at 0, 1, 4, 8, 12, and 24 h. All the samples were immediately frozen in liquid nitrogen and stored at −80°C.
The roots, stems, and leaves of TP309 rice plants were sampled and ground in liquid nitrogen. Total RNA was extracted using the TRIzol method. DNase I (Invitrogen) was employed to digest the genomic DNA. Total RNA (1 μg) was subsequently used for first-strand cDNA synthesis catalyzed by M-MLV reverse transcriptase with Oligo dT18 primer. cDNA was stored at −20°C. Quantitative real-time PCR (qRT-PCR) was performed using the SYBR Green Mix PCR Kit, with GAPDH as an internal reference gene (Supplementary Table S1). The gene-specific primers used for qPCR analysis are given in Supplementary Table S1.
The OsNHO1 full-length fragment was digested with BamH I and Sac I, and then inserted into the binary vector pTCK303 (Wang et al., 2004). The fragment for RNAi vector construction was amplified using specific primers (OsNHO1i-F/R, Supplementary Table S1) with Spe I/BamH I and Sac I/Kpn I restriction enzyme sites. The purified fragments of approximately 600 bp were digested with Spe I and Sac I, inserted into pTCK303 in the sense orientation and then digested with BamH I and Kpn I in the antisense orientation. The recombinant plasmids pTCK303-NHO1 full and pTCK303-NHO1i were transferred into the Agrobacterium LBA4404 strain. Then, the two positive strains were preserved and used for rice plant transformation.
Transgenic rice plants were acquired via the Agrobacterium-mediated transformation method (Wang et al., 2004). Homozygous transgenic rice offspring were obtained after three generations of self-crossing, as detected by qRT-PCR and Western blotting.
Xoo-PXO99 was cultivated on a solid pressure-sensitive adhesive (PSA) medium at 28°C and then a single colony was grown in a liquid PSA medium at 180 revolutions per minute (rpm) for 24 h. Subsequently, the bacteria were cultivated on solid PSA medium again at 28°C in the dark for 2 d. Then, the bacterial culture was suspended in sterile water, and the solution was adjusted to OD600 = 0.5. Then, rice leaves were treated with the bacteria. Ten to twenty rice plants for each line at the four-leaf stage were included in the experiment. Three fully expanded leaves of each plant were cut for further observation and analysis. The lengths of disease speckles and diseased leaves were surveyed after 14 days.
M. oryzae isolate Y34 was cultivated on oatmeal agar containing 30 g/l oatmeal and 15 g/l agar. After 15 min of sterilization at 115°C, 100 μg/ml carbenicillin, 50 μg/ml kanamycin, and 50 μg/ml streptomycin were added. Rice leaves at the four-leaf stage were cut into fragments of approximately 6 cm and washed with sterile water. The leaves were placed on filter paper wetted with 100 mg/l 6-benzylaminopurine (6-BA). The two ends of each leaf were fixed with cotton to ensure that the leaf remained close to the filter paper. Bacterial colonies of the same size were selected and inoculated into each leaf using a 0.5-cm diameter perforator. The bacterial incidence in the leaves was surveyed 1 week later.
The content of GK in the offspring of transgenic rice was determined by the double-antibody sandwich method of ELISA, and the specific operation was carried out according to the instructions of the kit (Shanghai Jining, China, Cat.no.JN19516).
For wax quantification, we measured the wax content by the hot chloroform extraction method (Zhou et al., 2013). Leaves of WT and transgenic rice were cut into pieces and dipped into 60°C chloroform. The leaves were removed immediately after 30 s of oscillation. After the extraction volatilized at room temperature, the wax was weighed.
To identify proteins interacting with OsNHO1, yeast transformation, and library screening were performed using the Match-MakerTM GAL4 Two Hybrid System 3 (Clontech, Mountain View, California, USA). A cDNA library was generated from rice samples subjected to abiotic stress and biotic stress and cloned into pGADT7, which was used as a prey protein. The coding region of OsNHO1 was cloned into pGBKT7 as a bait protein. The resultant vectors were used to transform yeast strain AH109. Positive clones were selected on a medium lacking leucine, threonine, and histidine, and positivity was ensured by culturing a medium lacking leucine, threonine, histidine, and adenine but containing X-α-Gal. Then, the samples were sequenced and verified using phytozome and rice annotation databases.
For the bimolecular fluorescence complementation (BiFC) assay, the full-length coding sequences of OsNHO1 and OsSRC2 were cloned into the binary BiFC vectors pSPYNE-35S and pSPYCE-35S using a gateway system (Walter et al., 2004). The combination vectors OsNHO1-YFPC and OsSRC2-YFPN were coinfiltrated into Nicotiana benthamiana via an Agrobacterium-mediated method. Yellow fluorescent protein (YFP) signals were detected with a confocal microscope.
Data are presented as the mean ± SD. Significant differences between transgenic lines and WT plants were analyzed using a t-test (p < 0.05). The displayed measured values are the means of three biological replicates.
Glycerol kinase (GK) is the critical enzyme in the production of glycerol metabolites and belongs to the FGGY superfamily. A BLASTp search showed that only one gene locus (LOC_Os04g55410) in rice shared high homology with GKs from monocot and dicot plants. The coding sequence of this locus was cloned from rice and named the OsNHO1 gene. Multiple sequence alignment analysis showed that OsNHO1 contained one conserved ATP-binding motif (DQGTTSTR) and two FGGY signature motifs (Figure 1A). The phylogenetic tree of GK proteins from several species showed that OsNHO1 had a close relationship with GKs from the grass family (Figure 1B). The evolutionary relationships were consistent with the relatedness of the species.
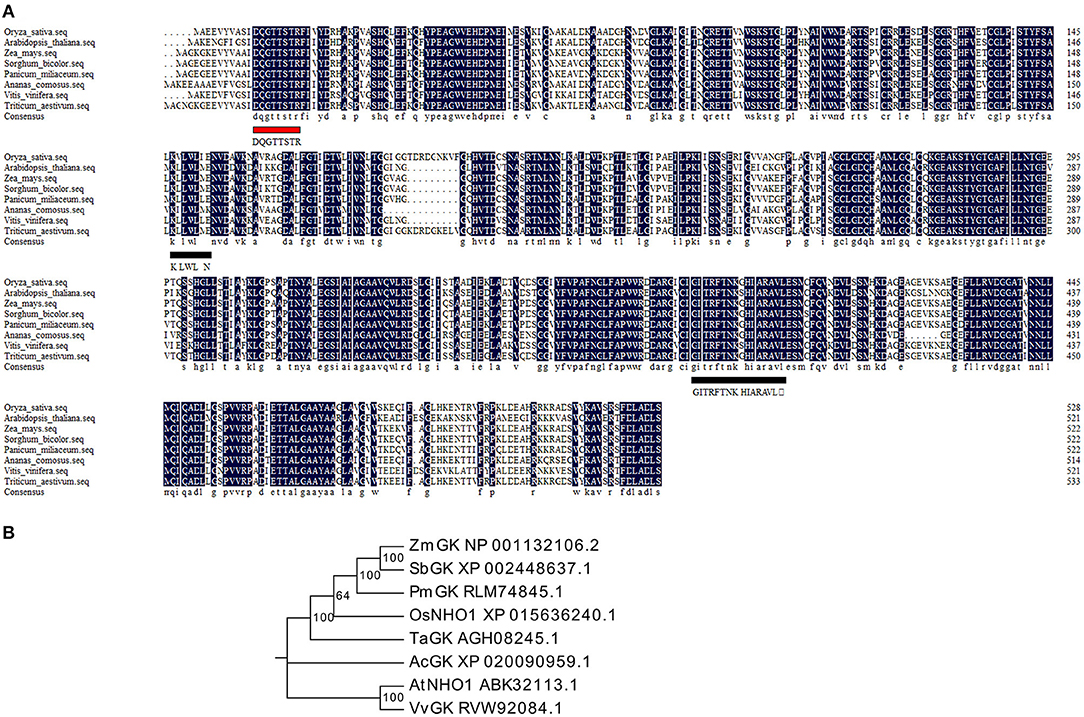
Figure 1. Sequence and phylogenetic analysis of OsNHO1 and GK proteins from other species. (A) Protein sequence alignment using DNAMAN software. The ATP-binding motif is underlined with a red solid line. The FGGY signature motives are underlined with a black solid line. (B) Phylogenetic analysis of GK proteins constructed using Mega6 based on the neighbor-joining method. Oryza sativa (accession number: XP_015636240.1), Arabidopsis thaliana (accession number: ABK32113.1), Triticum aestivum (accession number: AGH08245.1), Zea mays (accession number: NP_001132106.2), Sorghum bicolor (accession number: XP_002448637.1), Panicum miliaceum (accession number: RLM74845.1), Ananas comosus (accession number: XP_020090959.1), and Vitis vinifera (accession number: RVW92084.1). GK, glycerol kinase.
To determine whether GK is involved in the resistance of rice against Xoo, we inoculated the leaves with Xoo-PXO99, and compared the GK content in the leaves at different time points. The GK content was strongly increased at 12 h postinoculation (hpi) compared with the control (Figure 2A). The results indicated that GK is involved in response to Xoo.
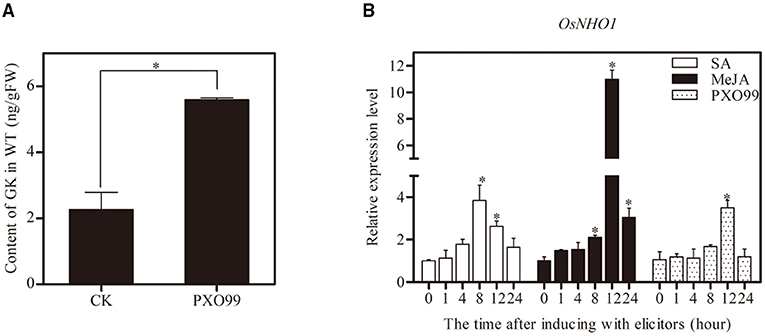
Figure 2. Determination of GK content and expression profiles of OsNHO1 treated with Xoo-PXO99. (A) The GK level was increased by Xoo infection in rice leaves after inoculation with the Xoo-PXO99. (B) Transcriptional levels of OsNHO1 in plants treated with JA, SA, and Xoo-PXO99. Each value represents the mean ± SE of three biological replicates. GK, glycerol kinase; JA, jasmonic acid; SA, salicylic acid. * indicates a significant difference at the 0.05 level.
The expression profile of OsNHO1 was analyzed by qPCR in rice cultivar TP309 seedlings treated with Xoo-PXO99, JA, and SA (Figure 2B). In the presence of JA, the relative expression of OsNHO1 slowly increased before peaking at 12 h, with a 10-fold increase compared with that of the control. OsNHO1 was also induced by SA. The relative expression level of OsNHO1 increased nearly 4-fold compared with the control at 8 h and then gradually declined. When inoculated with Xoo-PXO99, OsNHO1 was significantly induced only at 12 h. These results showed that OsNHO1 could be induced by JA, SA, and Xoo-PXO99.
AtNHO1 is required for both the general and specific resistance against bacteria and fungi and is involved in flagellin-induced non-host resistance to Pseudomonas in Arabidopsis. To explore whether OsNHO1 functions in a similar way to the homologous gene AtNHO1 in Arabidopsis, we obtained Arabidopsis nho1 mutant plants with OsNHO1 ectopic expressed. The transgenic progeny was inoculated with the pathogen P. s. pv. phaseolicola and the bacterial counts were conducted at 0 d and 3 days. The bacterial number in Col-0 at 3 days was 1.6 times that at 0 day and in the nho1 mutant, it was 5.5 times that at 0 day (Figure 3). However, in the presence of OsNHO1, the bacterial number at 3 days decreased to 1.8 times higher than that at 0 day. In addition, bacterial fecundity in Col-0 and nho1 + OsNHO1 was significantly lower than that in the nho1 mutant (Figure 3), indicating that OsNHO1 functions similarly to AtNHO1 in resisting P. s. pv. phaseolicola.
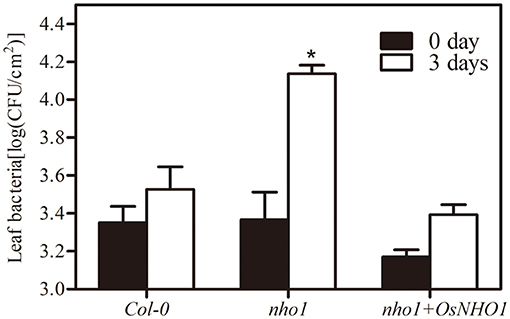
Figure 3. Bacterial growth of P. s. phaseolicola in Col-0, nho1 mutant, and nho1 + OsNHO1 lines was measured at 0 and 3 days after inoculation. Asterisks indicate significant differences compared with the other two varieties at 3 days (p < 0.05) by one-way ANOVA followed by the Tukey's highest significant difference (HSD) analysis. The error bars indicate the SEs. Three independent experiments were performed with similar results.
The AtNHO1 was required for defense against Pseudomonas bacteria in Arabidopsis (Lu et al., 2001). To test the function of the OsNHO1 gene in rice defense, we obtained OsNHO1 overexpression (OE) and knockdown transgenic plants using the Agrobacterium-mediated genetic transformation method. After four generations of screening, homozygous transgenic lines (T4), namely, two OsNHO1-OE lines and two OsNHO1-RNAi lines, were used for subsequent research. The relative expression level of OsNHO1 in the knockdown transgenic lines RNAi 72 and RNAi 85 was reduced by 50%, while it was increased approximately 3-fold in the OE lines OsNHO1-OE1 and OsNHO1-OE2 (Figure 4A). Western blotting results also showed that OsNHO1 was expressed in the OsNHO1-OE lines (Figure 4B).
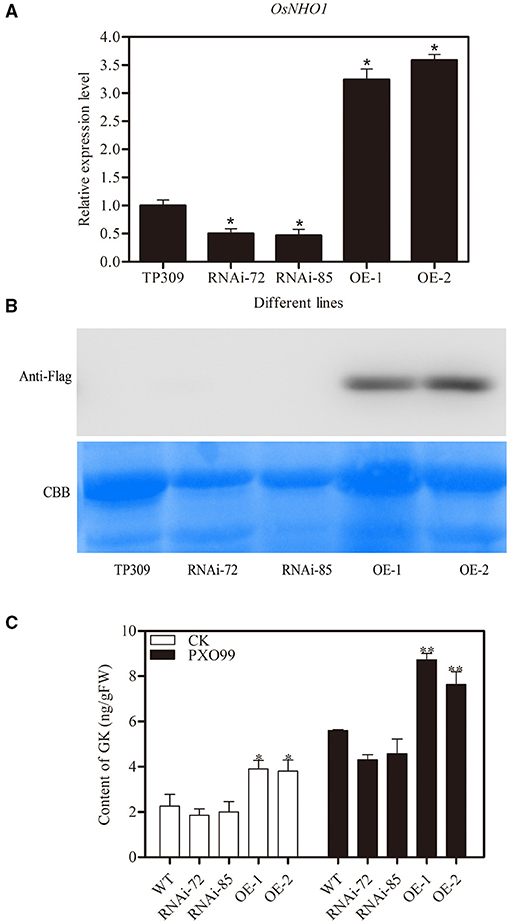
Figure 4. Identification of transgenic materials. (A) OsNHO1 transcript levels in RNAi, OE, and WT (TP309) plants by qPCR. (B) Protein-level determination in different plants. (C) Content of GK in different transgenic plants. GK, glycerol kinase; OE, overexpression; WT, wild type. * indicates a significant difference at the 0.05 level; ** indicates a significant difference at the 0.01 level.
To confirm whether OsNHO1 could affect GK accumulation, we quantified the GK contents in different transgenic plants. The content of GK in OsNHO1-OE plants was significantly higher than that in WT plants but was only marginally reduced in OsNHO1-RNAi plants compared with WT plants (Figure 4C). Meanwhile, to further verify that GK is involved in rice responses to pathogens, we measured the changes in GK contents in different materials after pathogen treatments. We found that the GK level was strongly increased in both the OsNHO1-OE and OsNHO1-RNAi lines after inoculation with Xoo-PXO99 at 12 hpi compared with 0 hpi (Figure 4C).
The obtained transgenic plants were used for Xoo and blast resistance test. We infected rice leaves with Xoo-PXO99 and measured the lesion length produced by Xoo-PXO99 after 13–15 days (Figures 5A,B), and the lesion area produced by Y34 was measured after 7–8 days (Figures 5C,D). We observed that the lesion length in the WT line was approximately 8.5 cm, while it was 11 cm in the OsNHO1-RNAi lines and 6 cm in the OsNHO1-OE lines. In addition, the lesion areas in the two overexpression lines were significantly less than those in the WT line, while the lesion areas in the two OsNHO1-RNAi lines were increased approximately 2-fold compared with those in the WT line.
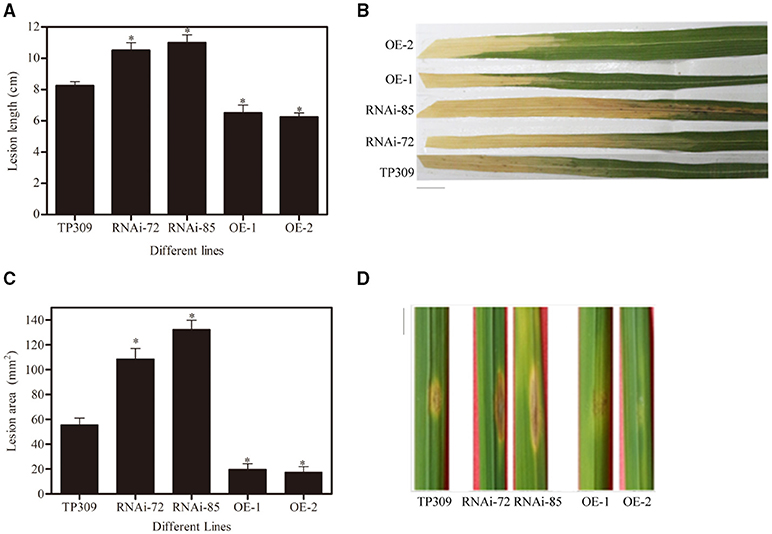
Figure 5. Disease resistance identification of transgenic materials. (A) Lesion lengths in transgenic plants were measured at 14 days postinoculation with Xoo-PXO99. (B) A photograph depicting disease symptom development in leaves 14 d postinfection with Xoo-PXO99. Scale bars = 1 cm. (C) Lesion areas of transgenic plants were measured using ImageJ software at 7 days postinoculation with Y34. (D) A photograph depicting disease symptom development in leaves 7 d postinfection with Y34. Scale bars = 1 cm. TP309: wild-type; RNAi-72 and RNAi-85: two RNAi-OsNHO1 lines; OE1 and OE2, two OsNHO1 overexpression lines. Asterisks indicate significant differences compared to TP309 (p < 0.05) by one-way ANOVA followed by Tukey's HSD analysis. Error bars indicate SEs. Three independent experiments were performed with similar results.
These results showed that the defects in OsNHO1 led to increased susceptibility to Xoo-PXO99 and M. oryzae Y34 while overexpressing OsNHO1 inhibited the spread of bacterial blight and blast. Thus, we concluded that OsNHO1 positively regulates rice immunity against pathogens.
Glycerol metabolism is initiated upon its conversion to G3P via the GK-mediated phosphorylation of glycerol. G3P is involved in the synthesizing of C16 and C18 chain fatty acids, which are essential precursors for wax synthesis. To analyze whether GK is related to wax synthesis, we measured the wax contents in different OsNHO1 transgenic lines. The wax content in the OsNHO1-RNAi lines was reduced in comparison with that in TP309. However, the wax content in the OsNHO1-OE lines was higher than that in the WT (Figure 6A).
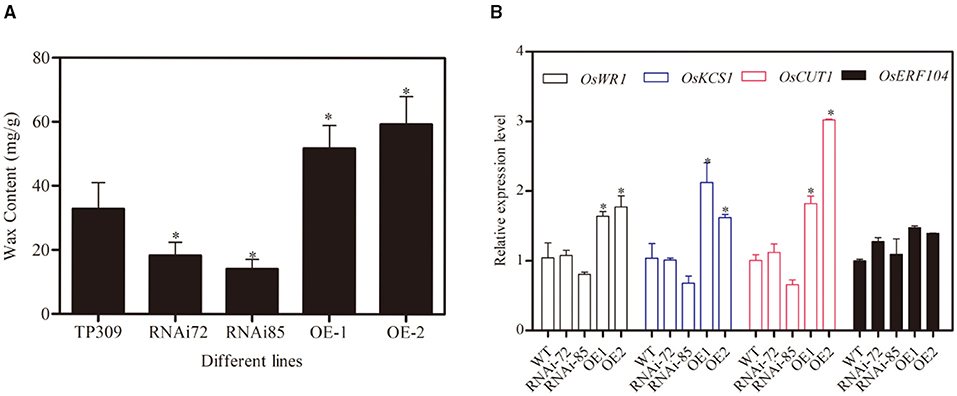
Figure 6. Contents of wax and relative expression of OsWRs genes in the WT (TP309), OsNHO1-RNAi, and OsNHO1-OE lines. (A) Contents of wax were measured by the hot chloroform extraction method. (B) Relative expression of OsWRs genes. Each value represents the mean ± SE of three biological replicates. Asterisks indicate significant differences compared to TP309 (p < 0.05) by one-way ANOVA followed by the Tukey's HSD analysis. OE, overexpression; WT, wild type.
Rice wax synthesis regulatory genes (OsWRs) modulate wax synthesis by the alteration of long-chain fatty acids and alkanes (Wang et al., 2012; Zhou et al., 2013). In this study, we analyzed the relative transcript levels of OsWR1, OsKCS1, OsCUT1, and OsERF104 in the transgenic lines (Figure 6B). These four genes were upregulated in OsNHO1-OE lines and showed no significant difference in the OsNHO1-RNAi lines compared with the WT. These results implied that overexpression of the OsNHO1 gene increased the content of wax.
Glycerol-induced resistance was reported in non-host resistance with upregulation of several PR genes (Jiang et al., 2009; Zhang et al., 2015). Pathogenesis-related proteins (PRs) are important components in plant responses to biotic and abiotic stresses. In this study, we analyzed the relative transcript levels of OsPRs in the transgenic lines (Figure 7). We found that the expression of OsPR10, OsNH1, and OsICS1 genes was no significant difference, while OsPR5, OsAOS1, and OsWRKY6 showed different transcription levels in different materials. Silencing of OsNHO1 decreased the relative expression of OsPR5 by 85%, while overexpression of OsNHO1 increased the expression of OsPR5.
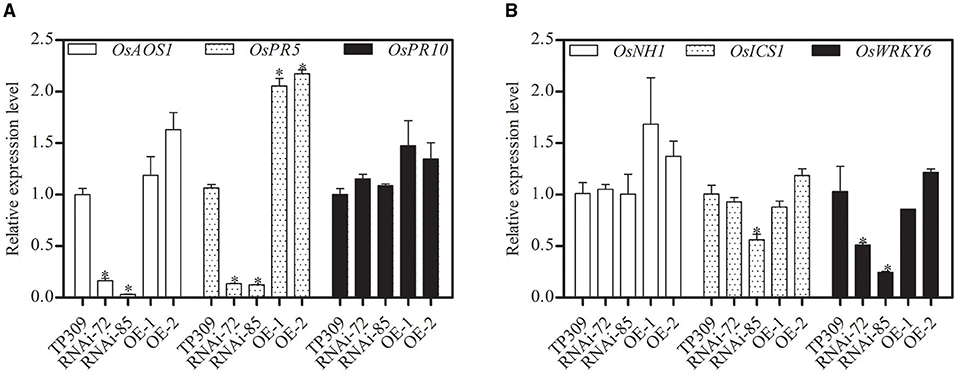
Figure 7. Relative expression of OsPRs in transgenic lines determined by qPCR with an SYBR® Premix Ex Taq TM II Kit. The transcript levels of PR genes in WT were considered 100%. Expression levels of the target gene were normalized to the ACTIN reference gene. (A) Relative expression of OsAOS1, OsPR5, and OsPR10. (B) Relative expression of OsNH1, OsICS1, and OsWRKY6. Each value represents the mean ± SE of three biological replicates. Asterisks indicate significant differences compared to TP309 (P < 0.05) by one-way ANOVA followed by Tukey's HSD analysis. The error bars indicate the standard errors. Three independent experiments were performed with similar results. qPCR, quantitative PCR; WT, wild type.
To identify the components in the OsNHO1-mediated signaling pathway, we screened the interaction partners of OsNHO1 by the yeast two-hybrid method using a rice cDNA library prepared from rice leaf tissue treated with biotic stresses. We obtained 13 potential candidate proteins (Table 1). Among these candidate proteins, the protein LOC_Os01g70790 contains a C2 domain (calcium-dependent lipid-binding domain, CaLB) and exhibits highly homologous with the protein SRC2 in soybeans, which is induced by cold stress. In vivo interaction between OsNHO1 and OsSRC2 indicated by BiFC analysis showed that these two proteins colocalized in the nucleus (Figure 8).
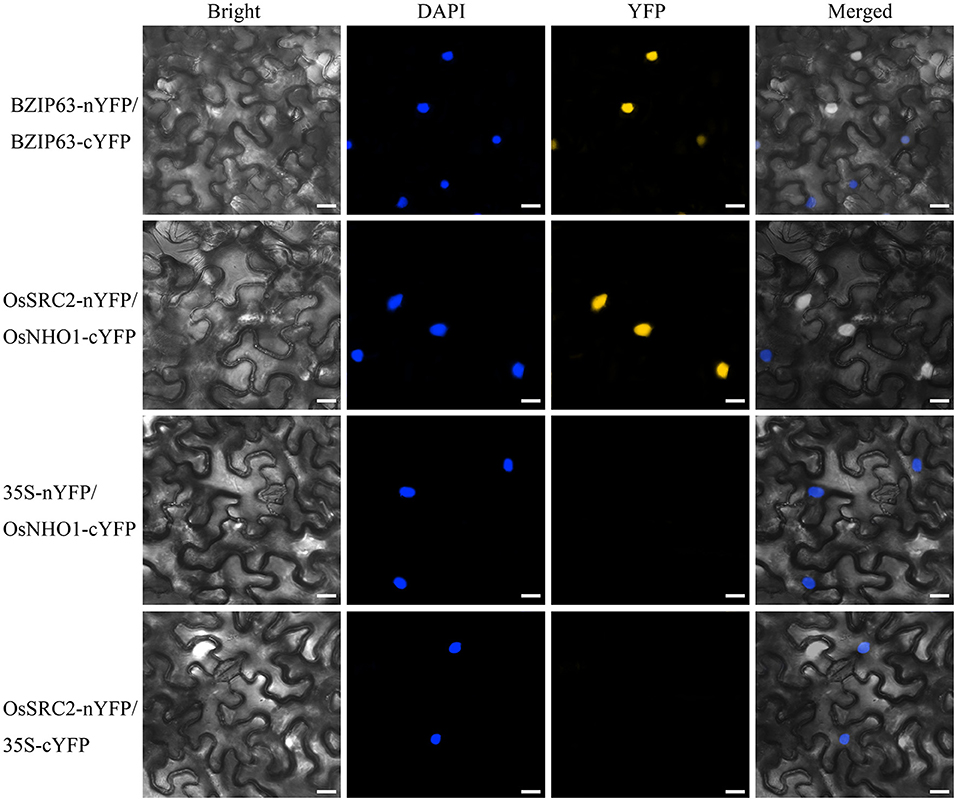
Figure 8. In vivo interaction between OsNHO1 and OsSRC2 by BiFC analysis. N. benthamiana leaves were co-infiltrated with Agrobacterium carrying 35S::OsSRC2-nYFP and 35S::OsNHO1-cYFP. BZIP63 was used as a positive control. 35S-nYFP/OsNHO1-cYFP and OsSRC2-nYFP/35S-cYFP were used as negative controls. Fluorescence [yellow fluorescent protein (YFP)] and bright and merged field confocal images were acquired at 72 h. DAPI was used for nuclear staining. Scale bars = 20 μm.
C2 domain-containing proteins play important roles in plant immunity, and the OsSRC2 protein contains a C2 domain. Previous results show that OsSRC2 functions as the interacting partner of OsNHO1. To analyze whether SRC2 is regulated by NHO1, we detected the transcription levels of OsSRC2 in different OsNHO1 transgenic lines by qPCR (Supplementary Figure S2). OsSRC2 was upregulated in the OsNHO1-OE lines but downregulated in the OsNHO1-RNAi lines. These results implied that OsNHO1 may interact with the OsSRC2 protein to modulate responses to pathogens.
Glycerol can be utilized as a sole carbon and energy source for both bacteria and fungi (Wei et al., 2004). This study found that glycerol may work as a significant transferred metabolite from plant to pathogen. The way to assimilate glycerol is the phosphorylative catabolic pathway, the key enzyme of glycerol metabolism is GK. Early study found that glycerol-insensitive mutants gli1 seedings lacking glycerol kinase are more resistant to abiotic stress (Eastmond, 2004), but NHO1 is needed for resistance to the fungal pathogen Botrytis cinerea and to the bacterial resistance. Consistent with results in Arabidopsis, we found that OsNHO1 was significantly induced by Xoo-PXO99 (Figure 2). Moreover, OsNHO1-OE plants showed increased resistance to Xoo-PXO99 and Y34, and OsNHO1-RNAi plants were more susceptible to pathogens (Figure 5). The correlation of OsNHO1 transcriptional levels with different reactions implies an essential role of these biochemical processes in disease resistance. A possible explanation might be that GK can alter plant glycerol pools then affect nutrient availability for pathogens.
Cutin is an extracellular lipid polymer that contributes to protective cuticle barrier functions against biotic and abiotic stresses in land plants. The cuticle wax is chemically composed of lipids. GK deficiency alters the expression of genes involved in lipid and carbohydrate metabolism (Rahib et al., 2007). In contrast, the wax biosynthetic genes OWR1, OsKCS1, and OsCUT1 in OsNHO1-OE plants are upregulated (Figure 6B). Corresponding to waxy gene expression, the wax content in OsNHO1-OE materials increases accordingly (Figure 5).
In addition, cuticle wax in plants is considered to contribute to drought, insect (Lee et al., 2014), and pathogen resistance (Jenks et al., 1994; Özer et al., 2017). Studies have shown that the content of wax in plants may also be correlated with disease resistance. For example, the wax contents of maize disease-resistant varieties were significantly higher than those of susceptible varieties (Russin et al., 1997), and the wax contents of leaves of resistant cassava varieties were higher than those of susceptible varieties (Zinsou et al., 2006). Similarly, in this study, we found that the wax content in the OsNHO1-OE plants, which showed increased resistance to Xoo-PXO99 and Y34, was far greater than that in OsNHO1-RNAi plants (Figure 5). This indicated that abnormal OsNHO1 expression may disturb rice pathogen resistance by altering the wax content.
Plant hormones can work as signal molecules and then affect plant responses to stress by mediating the deposition of cuticle wax. Ethylene (ETH) could increase the content and change the structure of wax to protect citrus plants from Penicillium digitatum invasion (Cajuste et al., 2010). Under water stress, ABA promoted the expression of several wax-synthesis genes in Arabidopsis by upregulating the expression of the transcription factor MYB96 and eventually promoted the accumulation of wax (Seo et al., 2011). Exogenous SA, MeJA, and 1-aminocyclopropane-1-carboxylate (ACC) could induce the deposition of cuticle wax in Brassica napus. SA is essential for sugars and glycerol-mediated disease resistance (Qian et al., 2015). Glycerol-induced resistance was reported in non-host resistance with upregulation of several PR genes and ROS accumulation. OsPR5 and OsICS1 are the key genes of the SA signaling pathway. JA and cutin wax belong to the fatty acid metabolism pathway and have a common synthetic precursor (Zhang et al., 2020). OsAOS1 is involved in the JA signal pathway. The expression of PR genes was significantly reduced in Arabidopsis waxy epidermis mutants cer6 and cer2 (Garbay et al., 2007), suggesting that the transcription level of PR genes is closely related to waxy components. Remarkably, the transcriptional level of the OsPR5 gene in OsNHO1-OE plants was higher than those in WT and OsNHO1-RNAi plants (Figure 7A), indicating that OsNHO1 have an additive influence on cuticular wax biosynthesis that is SA-dependent.
Glycerol metabolism is initiated upon its conversion to G3P via the GK-mediated phosphorylation of glycerol. G3P is involved in the synthesis of C16 and C18 chain fatty acids, which are important precursors for wax synthesis. In addition, cuticle wax in plants is considered to contribute to drought, insect (Lee et al., 2014), and pathogen resistance (Jenks et al., 1994; Özer et al., 2017). Studies have shown that the content of wax in plants may also be correlated with disease resistance. For example, the wax contents of maize disease-resistant varieties were significantly higher than those of susceptible varieties (Russin et al., 1997), and the wax contents of leaves of resistant cassava varieties were higher than those of susceptible varieties (Zinsou et al., 2006). Similarly, in our research, we found that the wax content in the OsNHO1-OE plants, which showed increased resistance to Xoo-PXO99 and Y34, was far greater than that in OsNHO1-RNAi plants (Figure 7). This indicated that abnormal OsNHO1 expression may disturb rice pathogen resistance by altering the wax content.
The cuticle wax is chemically composed of lipids. In mice, SRC2 and SRC3 regulate epidermis-specific sphingolipid production (Oda et al., 2009). In plants, SRC2 is a C2 domain-containing protein or calcium-dependent lipid-binding protein. C2 domains are found in over 100 different proteins with functions ranging from signal transduction to vesicular trafficking. The C2 domain of CaSRC2-1 is crucial for plasma membrane targeting, and the PcINF1-SRC2-1 complex is required in PcINF1-induced pepper immunity (Liu et al., 2015). In addition, the transcript level of OsSRC2 was upregulated by M. oryzae according to the Rice MetaSysB database (Sureshkumar et al., 2019). Other C2 domain-containing proteins, such as SS52 in pepper (Kim et al., 2008; Sakamoto et al., 2009); OsERG1, OsERG3, and GTPase-activating protein (GAP) in rice (Kim et al., 2003; Cheung et al., 2008); and BON1/CPN1, BAP1, and BAP2 in Arabidopsis (Liu et al., 2005; Yang et al., 2006, 2007), play important roles in plant immunity. Here, we found that the OsSCR2 protein, which is one of the interaction partners of OsNHO1 (Table 1), contains a C2 domain. BiFC results showed that these two proteins co-localized in the nucleus (Figure 7). Our results showed that the transcriptional levels of the OsSRC2 gene in OsNHO1-OE plants were higher than those in WT and OsNHO1-RNAi plants (Supplementary Figure S2).
Thus, we concluded that OsNHO1 participates in a novel pathway regulating plant defense. OsNHO1 significantly contributed to pathogen resistance to bacterial blight and rice blast by interacting with OsSRC2 protein upon pathogen infection, affecting the content of wax and modulating the expression of PR genes. OsNHO1 was a potential candidate gene for disease resistance engineering. Accordingly, further investigations may focus on the following important areas.
The regulation between OsNHO1 and its partner OsSRC2 should be further investigated. The roles of OsSRC2 in innate immunity and their relationships with wax generation and PR gene activation will be of great interest in future explorations.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
YC, XY, XN, and CH designed the research. HW, WG, and ZG performed the research. XX, RW, and SK wrote the paper. All authors read and approved the final manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank the National Key Research and Development Program of China (2018YFD1000500), the Key R&D Program of Hainan Province (ZDYF2019063), and PHD CJ for providing TBtools software. We are particularly grateful to Xiaofei Zhang from Alliance of Bioversity International and CIAT for his revised recomendation.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.800625/full#supplementary-material
Ahn, I. P., Kim, S., and Lee, Y. H. (2005). Vitamin B1 functions as an activator of plant disease resistance. Plant Physiol. 138, 1505–1515. doi: 10.1104/pp.104.058693
Brisson, D., Vohl, M. C., St-Pierre, J., Hudson, T., and Gaudet, D. (2001). Glycerol: a neglected variable in metabolic processes? Bioessays 23, 534–542 doi: 10.1002/bies.1073
Cajuste, J. F., González-Candelas, L., Veyrat, A., García-Breijo, F. J., and Reig-Armiñana, J. (2010). Epicuticular wax content and morphology as related to ethylene and storage performance of ‘Navelate’ orange fruit. Postharvest Biol. Technol. 1, 29–35. doi: 10.1016/j.postharvbio.2009.07.005
Chanda, B., Venugopal, S. C., Kulshrestha, S., Navarre, D. A., Downie, B., Vaillancourt, L., et al. (2008). Glycerol-3-phosphate levels are associated with basal resistance to the hemibiotrophic fungus colletotrichum higginsianum in Arabidopsis. Plant Physiol. 147, 2017–2029. doi: 10.1104/pp.108.121335
Chandra-Shekara, A.C., Venugopal, S.C., Barman, S.R., Kachroo, A., and Kachroo, P. (2007). Plastidial fatty acid levels regulate resistance gene-dependent defense signaling in Arabidopsis. In: Proceedings of the National Academy of Sciences of the United States of America. doi: 10.1073/pnas.0609259104
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). Tbtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Cheung, M. Y., Zeng, N. Y., Tong, S. W., Li, W. F., Xue, Y., Zhao, K. J., et al. (2008). Constitutive expression of a rice GTPase-activating protein induces defense responses. New Phytol. 179, 530–545. doi: 10.1111/j.1469-8137.2008.02473.x
Delventhal, R., Rajaraman, J., Stefanato, F. L., Rehman, S., Aghnoum, R., Mcgrann, G., et al. (2017). A comparative analysis of nonhost resistance across the two Triticeae crop species wheat and barley. BMC Plant Biol. 17, 1–17. doi: 10.1186/s12870-017-1178-0
Eastmond, P. J. (2004). Glycerol-insensitive Arabidopsis mutants: gli1seedlings lack glycerol kinase, accumulate glycerol and are more resistant to abiotic stress. Plant J. 37, 617–625. doi: 10.1111/j.1365-313X.2003.01989.x
Ellis, J. (2006). Insights into nonhost disease resistance: can they assist disease control in agriculture? Plant Cell. 18, 523–528. doi: 10.1105/tpc.105.040584
Garbay, B., Tautu, M. T., and Costaglioli, P. (2007). Low level of pathogenesis-related protein 1 mRNA expression in 15-day-old Arabidopsis cer6-2 and cer2 eceriferum mutants. Plant Sci. 2, 299–305.
Gómez-Ariza, J., Campo, S., Rufat, M., Estopà, M, Messeguer, J., Segundo, B. S., et al. (2007). Sucrose-mediated priming of plant defense responses and broad-spectrum disease resistance by overexpression of the maize pathogenesis-related PRs protein in rice plants. Mol. Plant Microbe. Interact. 20, 832–842. doi: 10.1094/MPMI-20-7-0832
He, Y., Han, J., Liu, R., Ding, Y., Wang, J., Sun, L., et al. (2018). Integrated transcriptomic and metabolomic analyses of a wax deficient citrus mutant exhibiting jasmonic acid-mediated defense against fungal pathogens. Hortic. Res. 5, 43. doi: 10.1038/s41438-018-0051-0
Jenks, M. A., Joly, R. J., Peters, P. J., Rich, P. J., Axtell, J. D., Ashworth, E. N. (1994). Chemically Induced Cuticle Mutation Affecting Epidermal Conductance to Water Vapor and Disease Susceptibility in Sorghum bicolor (L.) Moench. Plant Physiol. 105, 1239–1245. doi: 10.1104/pp.105.4.1239
Jiang, C., Hasegawa, M., Shimono, M., Sugano, S., Maeda, S., Inoue, H., et al. (2009). Suppression of the rice fatty-acid desaturase gene OsSSI2 enhances resistance to blast and leaf blight diseases in rice. Molec. Plant-Microbe Interac. 22:820–829. doi: 10.1094/MPMI-22-7-0820
Jones, J., and Dangl, J. (2006). The plant immune system. Nature 444, p. 323–329. doi: 10.1038/nature05286
Kachroo, A., Fu, D. Q., Havens, W., Navarre, D. R., Kachroo, P., and Ghabrial, S. A. (2008). An oleic acid–mediated pathway induces constitutive defense signaling and enhanced resistance to multiple pathogens in soybean. Mol. Plant Microbe. Interact. 21, 564–575. doi: 10.1094/MPMI-21-5-0564
Kachroo, A., Lapchyk, L., Fukushige, H., Hildebrand, D., Klessig, D., and Kachroo, P. (2003). Plastidial fatty acid signaling modulates salicylic acid- and jasmonic acid-mediated defense pathways in the Arabidopsis ssi2 mutant. Plant Cell. 15, 2952–2965. doi: 10.1105/tpc.017301
Kachroo, A., Venugopal, S. C., Lapchyk, L., Falcone, D., and Hildebrand, D., Kachroo (2004). Oleic acid levels regulated by glycerolipid metabolism modulate defense gene expression in Arabidopsis. Proc. Nat. Acad. Sci. US Am. 101, 5152–5157. doi: 10.1073/pnas.0401315101
Kachroo, P., Venugopal, S. C., Navarre, D. A., Lapchyk, L., and Kachroo, A. (2005). Role of salicylic acid and fatty acid desaturation pathways in ssi2-mediated signaling. Plant Physiol. 139, 1717–1735. doi: 10.1104/pp.105.071662
Kang, L., Li, J., Zhao, T., Xiao, F., Tang, X., Thilmony, R., et al. (2003). Interplay of the Arabidopsis nonhost resistance gene NHO1 with bacterial virulence. Proc. Nat. Acad. Sci. 100, 3519–3524. doi: 10.1073/pnas.0637377100
Kim, C., Koo, Y., Jin, J., and Moon, B. (2003). Rice C2 domain proteins are induced and translocated to the plasma membrane in response to a fungal elicitor. Biochemistry. 42, 11625–11631. doi: 10.1021/bi034576n
Kim, Y.-C., Kim, S.-Y., Choi, D., Ryu, C.-M., and Park, J. M. (2008). Molecular characterization of a pepper C2 domain-containing SRC2 protein implicated in resistance against host and non-host pathogens and abiotic stresses. Planta 227, 1169–1179. doi: 10.1007/s00425-007-0680-2
Lee, H.-A., Lee, H.-Y., Seo, E., Lee, J., Kim, S.-B., Oh, S., et al. (2017). Current understandings of plant nonhost resistance. Molec. Plant-Microbe Interac. 30, 5–15. doi: 10.1094/MPMI-10-16-0213-CR
Lee, S. B., Kim, H., Kim, R. J., and Suh, M. C. (2014). Overexpre Arabidopsis MYB96 confers drought resistance in Camelina sativa via cuticular wax accumulation. Plant cell Rep. 33, 1535–1546. doi: 10.1007/s00299-014-1636-1
Lipka, V. (2005). Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science. 310, 1180–1183. doi: 10.1126/science.1119409
Liu, J., Jambunathan, N., and McNellis, T. W. (2005). Transgenic expression of the von Willebrand A domain of the BONZAI 1/COPINE 1 protein triggers a lesion-mimic phenotype in Arabidopsis. Planta. 221, 85–94. doi: 10.1007/s00425-004-1413-4
Liu, Z., Qiu, A., Shi, L., Cai, J., Huang, X., Yang, S., et al. (2015). SRC2-1 is required in PcINF1-induced pepper immunity by acting as an interacting partner of PcINF1. J. Exp.Bot. 13, 3683–3698. doi: 10.1093/jxb/erv161
Lu, M., Tang, X., and Zhou, J. M. (2001). Arabidopsis NHO1 is required for general resistance against pseudomonas bacteria. Plant Cell. 13, 437–447. doi: 10.2307/3871287
Mysore, K. S., and Ryu, C. M. (2004). Nonhost resistance: how much do we know? Trends Plant Sci. 9, 97–104. doi: 10.1016/j.tplants.2003.12.005
Oda, Y., Uchida, Y., Moradian, S., Crumrine, D., Elias, P. M., and Bikle, D. D. (2009). Vitamin D receptor and coactivators SRC2 and 3 regulate epidermis-specific sphingolipid production and permeability barrier formation. J. Invest. Dermatol. 129, 1367–1378. doi: 10.1038/jid.2008.380
Özer, N., Şabudak, T., Özer, C., Gindro, K., Schnee, S., and Solak, E. (2017). Investigations on the role of cuticular wax in resistance to powdery mildew in grapevine. J. Gen Plant Pathol. 5, 316–328. doi: 10.1007/s10327-017-0728-5
Qian, Y., Tan, D. X., Reiter, R. J., and Shi, H. (2015). Comparative metabolomic analysis highlights the involvement of sugars and glycerol in melatonin-mediated innate immunity against bacterial pathogen in Arabidopsis. Sci. Rep. 5, 15815. doi: 10.1038/srep15815
Rahib, L., MacLennan, N. K., Horvath, S., Liao, J. C., and Dipple, K. M. (2007). Glycerol kinase deficiency alters expression of genes involved in lipid metabolism, carbohydrate metabolism, and insulin signaling. Eur. J. Hum. Genet. 15, 646–657. doi: 10.1038/sj.ejhg.5201801
Rolland, F., and Sheen, M. J. (2002). Supplement: signal transduction || sugar sensing and signaling in plants. Plant Cell. 14, S185–S205. doi: 10.1105/tpc.010455
Russin, J. S., Guo, B. Z., Tubajika, K. M., Brown, R. L., Cleveland, T. E., and Widstrom, N. W. (1997). Comparison of Kernel Wax from corn genotypes resistant or susceptible to aspergillus flavus. Phytopathology 87, 529–533. doi: 10.1094/PHYTO.1997.87.5.529
Sakamoto, M., Tomita, R., and Kobayashi, K. (2009). A protein containing an XYPPX repeat and a C2 domain is associated with virally induced hypersensitive cell death in plants. FEBS Lett. 583, 2552–2556. doi: 10.1016/j.febslet.2009.07.020
Schaaf, J., and Hess, W. D. (1995). Primary metabolism in plant defense. Plant Physiol. 108, 949–960. doi: 10.1104/pp.108.3.949
Scheideler, M., Schlaich, N. L., Fellenberg, K., Beissbarth, T., Hauser, N. C., Vingron, M., et al. (2002). Monitoring the switch from housekeeping to pathogen defense metabolism in Arabidopsis thaliana using cDNA arrays. J. Biol. Chem. 277, 10556–10561. doi: 10.1074/jbc.M104863200
Seo, P. J., Lee, S. B., Suh, M. C., Park, M. J., Go, Y. S., and Park, C. M. (2011). The MYB96 transcription factor regulates cuticular wax biosynthesis under drought conditions in Arabidopsis. Plant cell 23, 1138–1152 doi: 10.1105/tpc.111.083485
Sureshkumar, V., Dutta, B., Kumar, V., Prakash, G., Mishra, D. C., and Chathurvedi, K. K., et al. (2019). RiceMetaSysB: a database of blast and bacterial blight responsive genes in rice and its utilization in identifying key blast-resistant WRKY genes. J. Biol. Databaaases 2019. doi: 10.1093/database/baz015
Walter, M., Chaban, C., Schütze, K., Batistic, O., Weckermann, K., Näke, C., et al. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40, 428–438. doi: 10.1111/j.1365-313X.2004.02219.x
Wang, T., Xing, J., Liu, X., Yao, Y., Hu, Z., Peng, H., et al. (2018). GCN5 contributes to stem cuticular wax biosynthesis by histone acetylation of CER3 in Arabidopsis. J. Exper. Botany. 69, 2911–2922. doi: 10.1093/jxb/ery077
Wang, Y., Wan, L., Zhang, L., Zhang, Z., Zhang, H., Quan, R., et al. (2012). An ethylene response factor OsWR1 responsive to drought stress transcriptionally activates wax synthesis related genes and increases wax production in rice. Plant Mol. Biol. 78, 275–288. doi: 10.1007/s11103-011-9861-2
Wang, Z., Chen, C., Xu, Y., Jiang, R., and Xu, Z. (2004). A practical vector for efficient knockdown of gene expression in rice (Oryza sativa L.). Plant Mol. Biol. Rep. 22, 409–417. doi: 10.1007/BF02772683
Wei, Y., Shen, W., Dauk, M., Wang, F., Selvaraj, G., and Zou, J. (2004). Targeted gene disruption of glycerol-3-phosphate dehydrogenase in Colletotrichum gloeosporioides reveals evidence that glycerol is a significant transferred nutrient from host plant to fungal pathogen. J. Biol. Chem. 1, 429–435. doi: 10.1074/jbc.M308363200
Yang, H., Li, Y., and Hua, J. (2006). The C2 domain protein BAP1 negatively regulates defense responses in Arabidopsis. Plant J. 48, 238–248. doi: 10.1111/j.1365-313X.2006.02869.x
Yang, H., Yang, S., Li, Y., and Hua, J. (2007). The Arabidopsis BAP1 and BAP2 genes are general inhibitors of programmed cell death. Plant Physiol. 145, 135–146. doi: 10.1104/pp.107.100800
Yang, Y., Jing, Z., Peng, L., Xing, H., Li, C., Wei, G., et al. (2013). Glycerol-3-phosphate metabolism in wheat contributes to systemic acquired resistance against puccinia striiformis f. sp. tritici. PLoS ONE. 8, e81756. doi: 10.1371/journal.pone.0081756
Ye, X., Gao, Q. M., Yu, K., Lapchyk, L., and Kachroo, P. (2009). An intact cuticle in distal tissues is essential for the induction of systemic acquired resistance in plants. Cell Host and Microbe. 5, 151–165. doi: 10.1016/j.chom.2009.01.001
Yun, B. W., Atkinson, H. A., Gaborit, C., Greenland, A., Read, N. D., Pallas, J. A., et al. (2003). Loss of actin cytoskeletal function and EDS1 activity, in combination, severely compromises non-host resistance in Arabidopsis against wheat powdery mildew. Plant J. 34, 768–777. doi: 10.1046/j.1365-313X.2003.01773.x
Zhang, Q., Li, X., Long, X., Hu, B., Xiao, X., Zhang, X., et al. (2020). Metabolism of the cutin and wax of plants and their disease resistance mechanisms (in Chinese). J. Zhejiang AandF Univ. 37, 1207–1215. doi: 10.11833/j.issn.2095-0756.20190745
Zhang, Y., Smith, P., Maximova, S. N., and Guiltinan, M. J. (2015). Application of glycerol as a foliar spray activates the defence response and enhances disease resistance of Theobroma cacao. Mol. Plant Pathol. 16, 27–37. doi: 10.1111/mpp.12158
Zhou, X., Jenks, M. A., Liu, J., Liu, A., Zhang, X., Xiang, J., et al. (2013). Overexpression of transcription factor OsWR2 regulates wax and cutin biosynthesis in rice and enhances its tolerance to water deficit. Plant Molec. Biol. Repor. 32, 719–731. doi: 10.1007/s11105-013-0687-8
Keywords: rice, glycerol kinase, non-host resistance, bacterial blight, pathogen, wax
Citation: Xiao X, Wang R, Khaskhali S, Gao Z, Guo W, Wang H, Niu X, He C, Yu X and Chen Y (2022) A Novel Glycerol Kinase Gene OsNHO1 Regulates Resistance to Bacterial Blight and Blast Diseases in Rice. Front. Plant Sci. 12:800625. doi: 10.3389/fpls.2021.800625
Received: 23 October 2021; Accepted: 26 November 2021;
Published: 20 January 2022.
Edited by:
Yong Xiao, Chinese Academy of Tropical Agricultural Sciences, ChinaReviewed by:
Huanbin Zhou, Institute of Plant Protection, Chinese Academy of Agricultural Sciences (CAAS), ChinaCopyright © 2022 Xiao, Wang, Khaskhali, Gao, Guo, Wang, Niu, He, Yu and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaohui Yu, eGlhb2h1aXl1QGhhaW5hbnUuZWR1LmNu; Yinhua Chen, eWhjaGVuQGhhaW5hbnUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.