- State Key Laboratory of Crop Stress Biology for Arid Areas, College of Plant Protection, Northwest A&F University, Yangling, China
Wheat powdery mildew, caused by the obligate biotrophic ascomycete fungal pathogen Blumeria graminis f. sp. tritici (Bgt), is a major threat to wheat production worldwide. It is known that Arabidopsis thaliana glucan synthase-like 5 (AtGSL5) improves the resistance of wheat to powdery mildew by increasing its anti-penetration abilities. However, the function of glucan synthase-like (GSL) orthologs in crop species remains largely unknown. In this study, TaGSL22, a novel functional ortholog of AtGSL5, was isolated as the only Bgt-induced GSL gene in wheat. Phylogenetic analysis indicated that TaGSL22 was conserved within the group of Gramineae and showed a closer relationship to GSL orthologs from monocots than to those from dicots. The TaGSL22 transcript was highest in the wheat leaves, followed by stems then roots. TaGSL22 was localized in the cell membrane and cytoplasm of wheat protoplasts, as predicted by transmembrane structure analysis. In addition, expression of TaGSL22 was induced by the plant hormones ethylene (ETH) and salicylic acid (SA), but down-regulated by jasmonate (JA) and abscisic acid (ABA). The transcript level of TaGSL22 was up-regulated in the incompatible interaction between Bgt and wheat, whereas it remained relatively unchanged in the compatible interaction. Knocking down of TaGSL22 by virus-induced gene silencing (VIGS) induced a higher infection type in the wheat–Bgt interaction. The TaGSL22-silenced plants exhibited reduced resistance to Bgt, accompanied by decreased callose accumulation. Our study shows a conserved function of GSL genes in plant immunity associated with penetration resistance, and it indicates that TaGSL22 can be used to improve papilla composition and enhance resistance to wheat powdery mildew.
Introduction
Wheat is one of the earliest cultivated crops in history. More than 10,000 years ago, humans began to plant Triticum monococcum for use as food (Araus et al., 2007). Today it remains the main source of calories and protein globally, and cultivating wheat varieties with high nutrition, yield, and resistance to biotic and abiotic stresses is key to ensuring food security (Mondal et al., 2016). Wheat powdery mildew is a fungal disease caused by Blumeria graminis f. sp. tritici (Bgt) (Belanger et al., 2003). The disease occurs in all wheat-growing regions in the world, and its distribution is very extensive. The reproduction of Bgt mainly depends on the conidia and ascospores of the pathogen, and the disease often occurs in a large area following the spread of airflow. Typically, wheat powdery mildew will cause a 5–19% yield loss of the crop, while in serious epidemic years that number may reach to more than 30%. As incidences of the disease increase, the safety of global wheat production is seriously threatened (Li et al., 2019). Planting disease-resistant wheat cultivars is the most effective and cost-effective strategy for the control of powdery mildew. At present, more than 60 loci of powdery mildew resistance genes have been identified, of which only a few have been cloned successfully, such as Pm5e and Pm41 (Li et al., 2020; Xie et al., 2020). However, because most of these resistance genes are based on a gene-for-gene system and are race-specific, they are at risk of being overcome by new emerging races of the pathogen (McDonald and Linde, 2002). Genes that contribute to disease resistance with a broad spectrum are needed to provide durable resistance.
The cell wall of plants is the first barrier against the invasion of pathogens (Kacprzak et al., 2011). In the resistance of plants to pathogenic fungi and oomycetes, the plant cell wall will self-modify where the pathogens attempt to invade. This results in a complex structure known as a papilla, which is essentially a physical barrier created to slow down or even prevent the invasion of pathogens. The papilla contains a variety of chemical components with clear anti-microbial functions, such as thionins and phenolic compounds (McLusky et al., 1999). Callose, the most prominent component of the papillae, is likely indispensable in papillae disease resistance. The prevention of pathogen invasion has been observed to be associated with the deposit of relatively high amounts of callose at sites of attempted penetration, forming typical papillae (Voigt, 2016).
The glucan synthase-like (GSL) gene family exists in many plants and is responsible for the production of callose, which is present not only at sites of pathogen attack and fungal penetration, but also in the pollen cell wall and in callus plugs in wounds (Voigt et al., 2006). For example, Arabidopsis thaliana glucan synthase-like 5 (AtGSL5) (also known as the powdery mildew resistance gene 4, PMR4) in Arabidopsis thaliana is involved in the formation of the callose wall that separates tetrads, making it essential for the formation of pollen (Enns et al., 2005). In addition, AtGSL5 improves the resistance of A. thaliana to powdery mildew by increasing its anti-penetration abilities (Blümke et al., 2013). It was also found that CRISPR/Cas-9-targeted mutagenesis of the PMR4 gene in tomato plants produced mutants that displayed a reduction (but not a complete loss) of susceptibility to the tomato powdery mildew pathogen Oidium neolycopersici (Martinez et al., 2020). The functional ortholog of AtGSL5 in barley was identified as HvGSL6, whose down-regulation led to reduced glucan deposition and increased cell wall penetration by powdery mildew (Chowdhury et al., 2016). In wheat, eight different TaGSL genes were identified and ascertained to mediate the synthesis and regulation of callose under different environmental conditions in various tissues (Voigt et al., 2006). Functional analysis of members of the GSL gene family in wheat revealed that either TaGSL8 or TaGSL10 are involved in plant regeneration, while the TaGSL3 and TaGSL8 RNAi transgenic lines showed slightly reduced resistance against Fusarium graminearum (Rana et al., 2014). However, the GSL genes associated with papillary callose accumulation in common wheat have not been identified, and their organ- and tissue-specific expressions and involvement in disease resistance remain unknown.
A number of plant hormones play critical roles in the defense systems of plants (Shigenaga and Argueso, 2016). For example, salicylic acid (SA) is considered to be an important signaling molecule that induces an immune reaction in plants. It is involved in the hypersensitive response and the systemic acquired resistance response in a plant, both of which can regulate reactive oxygen species and antioxidants to improve plant resistance (Ryals et al., 1994). It has also been reported that the SA pathway in A. thaliana is negatively regulated by callose synthase, the loss of which can result in SA-dependent disease resistance to powdery mildew (Nishimura et al., 2003). Jasmonate (JA) is a plant hormone produced by the metabolism of unsaturated fatty acids, and it is involved in the regulation of plant resistance to pathogens, insects, and mechanical damage (Koo and Howe, 2009). In Arabidopsis, AtGSL5 was found to negatively regulate JA and SA production or signaling to enhance disease resistance to powdery mildew, and the negative regulation of defense signaling by this gene could be independent of callose production (Wawrzynska et al., 2010). Ethylene (ETH) is a gaseous plant hormone derived from methionine, which is abundant in plants. The ETH response factor subfamily plays an important role in the establishment and regulation of defense systems of plants by balancing positive and negative transcriptional regulation (Bari and Jones, 2009). In fact, in most cases, these plant hormones are more effective when they act in relation to each other, making them indispensable members of the plant disease resistance response network. However, the regulatory function of these plant hormones in the defense of common wheat triggered by a GSL gene is not clear.
In the present study, the wheat ortholog gene TaGSL22 of the Arabidopsis gene AtGSL5 was identified in wheat. Transcript profiling of TaGSL22 was analyzed in wheat seedlings inoculated with virulent and avirulent Bgt races, and in response to various plant hormones. Tissue-specific expression and subcellular localization of TaGSL22 were determined. The BMSV-VIGS technique was performed to demonstrate how TaGSL22 was involved in callose regulation and disease resistance to Bgt. Taken together, our results indicated that the TaGSL22 gene plays a role in callose synthesis and resistance to Bgt in wheat.
Materials and Methods
Plant Material and Pathogen Preparation
The common wheat variety Asosan/8Cc, which is highly resistant to Bgt E09 and susceptible to Bgt A13, was used throughout the study. The plants were placed under a transparent cover in a greenhouse with a temperature of 17°C, 75% humidity, and a 16 h photoperiod of light (18 μmol/m2/s). Inoculation with Bgt was carried out after about 7–10 days, when the wheat seedlings grew to one leaf (Zeng et al., 2010). Before inoculation, the seedlings were misted with water. Fresh conidia were then inoculated on the wheat leaves by the shaking powder method. About 7 days later, when the susceptible control “Jingshuang 16” reached infection type (IT) 4, the ITs of the tested materials were rated using a scale of 0–4 (Supplementary Table 1).
Identification and Phylogenetic Analysis of TaGSL22
The protein sequence of AtGSL5 in Arabidopsis was used to search the UniProt protein database for its ortholog gene in wheat. A 423 bp DNA sequence of TaGSL22 was found in the linked NCBI database using the amino acid sequence of the TaGSL22 protein obtained in the UniProt protein database. Using this sequence, ortholog genes of TaGSL22 in other plant species were identified by a BlastX search. A phylogenetic tree was constructed with sequences of the seven homologous genes obtained in BlastX using the molecular evolutionary genetic analysis software MEGA7. The signal peptide and transmembrane domain of the protein were predicted by online software TMHMM-2.0, and its subcellular localization was analyzed and predicted by online software cNLS Mapper.
RNA Extraction and Quantitative Real-Time Polymerase Chain Reaction
Samples measuring about 100 mg were collected from leaves, stems, and roots at the seedling stage (7–10 days of growth), then frozen immediately in liquid nitrogen and stored at −80°C for RNA extraction using a plant RNA small extraction kit (Magen, Guangzhou, China). First-strand cDNA was synthesized in accordance with the instructions of the Hiscript III 1st Strand cDNA Synthesis Kit (+gDNA wiper) (Vazyme, Nanjing, China). For quantitative real-time polymerase chain reaction (qRT-PCR) analysis, ChamQ Universal SYBR qPCR Master Mix (Vazyme) was used on a Quant Studio 5 fluorescence quantitative instrument (Thermo Fisher, Waltham, MA, United States). Gene encoding wheat elongation factor (EF-1α) was used as the internal reference gene (Supplementary Table 1). After qRT-PCR analysis, the 2–Δ Δ CT method was used to calculate the relative expression of the target gene (Livak and Schmittgen, 2001). The experiments consisted of three biological repeats and three technical repeats. The sequences of quantitative primers are shown in Supplementary Table 2.
Subcellular Localization of TaGSL22::GFP Fusion Protein
The pJIT163-GFP vector was used to construct a TaGSL22 expression vector (pJIT163-TaGSL22::GFP) for subcellular localization (Wang et al., 2013). Endonuclease BamHI and XbaI were used to digest the pJIT163 plasmid, and the reaction was carried out on a Bio-Rad MyCycler PCR (Bio-Rad, Hercules, CA, United States) at 37°C for 4 h. After the reaction was complete, the product was recovered using a Gel Extraction Kit (Omega, Norcross, GA, United States). The TaGSL22, along with the recognition sequences and homologous arms, was amplified and ligated with the linear vector using a ClonExpress Ultra One Step Cloning Kit (Vazyme) with two CaMV 35S promoters. The ligated product was then transformed into E. coli DH5α and sequenced to confirm the successful construction of the TaGSL22 expression vector. Wheat protoplasts were prepared using a wheat protoplast preparation and transformation kit (Coolaber, Beijing, China). The protoplasts were observed with a FV3000 confocal laser scanning microscope (Olympus Corp., Tokyo, Japan) using a GFP channel (Alexa Flour 488, green) and an SV2 channel (Alexa Flour 565, red).
Transcript Profiling of TaGSL22 Induced by Different Plant Hormones
The seedlings were treated with methyl jasmonate (MeJA) (0.1 mM), ETH (0.2 mM), abscisic acid (ABA) (0.2 mM), and SA (1 mM), respectively. Leaf samples were collected at 0, 0.5, 3, 6, 12, and 24 h after each treatment to conduct qRT-PCR analysis as described above. The mock control was treated with water. Data were normalized to the expression level of the wheat elongation factor TaEF-1α. The transcript level of genes in control plants at time 0 was standardized as 1. The qRT-PCR analysis for each respective experiment was repeated three times.
Production of TaGSL22-Silenced Wheat Plants by Virus-Induced Gene Silencing
The viral vectors, including BMSV: α, BMSV: β, BMSV: γ, and BMSV: γ-phytoene desaturase gene (PDS), were donated by Professor Jun Guo at Northwest A&F University. The BSMV-VIGS system was performed as previously described (Yuan et al., 2011). TaGSL22 was connected to BMSV: γ-PDS plasmid by homologous recombination to form γ-TaGSL22 using a ClonExpress Ultra One Step Cloning Kit (Vazyme). BMSV: α and BMSV: γ were digested with endonuclease Mlu, BMSV: γ-PDS and BMSV: γ-TaGSL22 were digested with BssHII, BMSV: β was digested with SpeI, and the linearized vector was transcribed in vitro with an in vitro transcription kit (Promega, Madison, WI, United States). The transcripts of linearized plasmids α, β, and γ were mixed with appropriate amounts of 1 × FES buffer (0.1 M glycine, 0.06 M K2HPO4, 1% w/v tetrasodium pyrophosphate, 1% w/v bentonite, and 1% w/v celite, pH 8.5) (Pogue et al., 1998). When the seedlings reached the stage of two leaves and one heart, the second leaves were inoculated with the virus, and one pot of seedlings was kept uninoculated to serve as control. The wheat leaves were treated in darkness at 25°C for 24 h. After phenotypic observation, the virus-infected wheat seedlings were selected and inoculated with Bgt E09, then transferred to the growth chamber under the same conditions as described above. Samples were taken at 0, 48, and 120 h post infection (hpi) for qRT-PCR, and the silencing efficiency was measured. The primers used are listed in Supplementary Table 2.
Histopathological Observation of the Powdery Mildew Pathogen in the TaGSL22-Silenced Plant
During virus-induced gene silencing (VIGS), histopathological observation was performed on samples taken at 24, 48, and 96 hpi with Bgt E09. The collected leaves were cut with scissors into 2 cm segments and placed in a decolorizing solution of anhydrous ethanol and glacial acetic acid for 2 days. The leaf segments were then transferred into a saturated chloral hydrate solution for transparency, and finally into a 50% glycerol solution. After wheat germ agglutinin (WGA) staining, the samples were then observed under an Olympus BX53 microscope (Olympus Corp., Tokyo, Japan). Representative colonies from the control and silenced groups were selected for imaging, and the colony areas, number of mycelial branches, and mycelium lengths were recorded.
Microscopic Analysis of Callose Accumulation in the TaGSL22-Silenced Plant
During VIGS, freshly cut leaf tissues collected at 24 hpi with Bgt E09 were decolorized and washed twice with 50% (v/v) ethanol for 15 min, rinsed with water, then stained with aniline blue solution [67 mM K2HPO4, 0.05% (w/v) aniline blue]. The degree of callose deposition was determined in fields of 1 mm2 using a BX-51 microscope (Olympus Corp., Tokyo, Japan) (Xu et al., 2020). Representative pictures of the callose accumulation in the control and silenced groups, with scale bars measuring 100 μm, were selected for imaging. The average number of callose foci per field of view (1 mm2) was calculated from 30 fields.
Statistical Analysis
The data processing system SPSS Statistics 25 (IBM, Armonk, NY, United States) was used for statistical analysis. The Student’s t-test method was used, wherein * and ** represented significant differences at P < 0.05 and P < 0.01, respectively. Relative gene quantification was calculated by the comparative ΔΔCT method. All experiments were performed and analyzed separately with three biological replicates.
Results
TaGSL22 Gene Was Identified as a Wheat Ortholog of Arabidopsis thaliana Glucan Synthase-Like 5
To identify the putative ortholog of AtGSL5 in common wheat, the UniProt protein database1 was searched using the amino acid sequence of the AtGSL5 protein as the query sequence. A wheat ortholog gene with the highest sequence identity, TaGSL22, was extracted (UniProtKB: Q4JHU1).
The phylogenetic relationships of GSL genes in seven other plant species, Aegilops tauschii, Hordeum vulgare, Setaria viridis, Zea mays, Oryza sativa, Gossypium hirsutum, and A. thaliana, were investigated to identify their orthologs. A phylogenetic tree was then constructed using Mega 7 (Supplementary Figure 1A). It was inferred that the GSL gene was relatively conservative in Gramineae (in the same clade as Hordeum and Aegilops). The average hydrophobicity of the TaGSL22 protein was 0.289, indicating it was a hydrophobic protein. It was predicted that the TaGSL22 presented in cytoplasm and had transmembrane domains (Supplementary Figure 1B). According to the structure analysis, no signal peptide or nuclear localization were predicted.
TaGSL22 Expression Increased in the Resistance Reaction to Bgt E09
To characterize the expression pattern of TaGSL22 in wheat after a fungal infection, its transcript profiles with races E09 (avirulent) and A13 (virulent) were examined using RT-qPCR (Figure 1). Expression levels were recorded in a time course at 12/24 h intervals across the first 96 h after inoculation. It was found that the expression of TaGSL22 was increased at 48 hpi, and subsequently decreased with Bgt E09 infection. The peak at 48 hpi was about 3.8 times that of the control (0 hpi), indicating that gene expression was significantly induced. After 48 h, the expression of TaGSL22 decreased slowly, but its value was always higher than during the first 24 h. In contrast, inoculation of Bgt A13 suppressed the overall expression of the GSL gene, except at 24 hpi when a 1.5 times expression was noted. These results indicated that TaGSL22 was highly induced in the wheat powdery mildew resistance response, but not in the susceptible reaction.
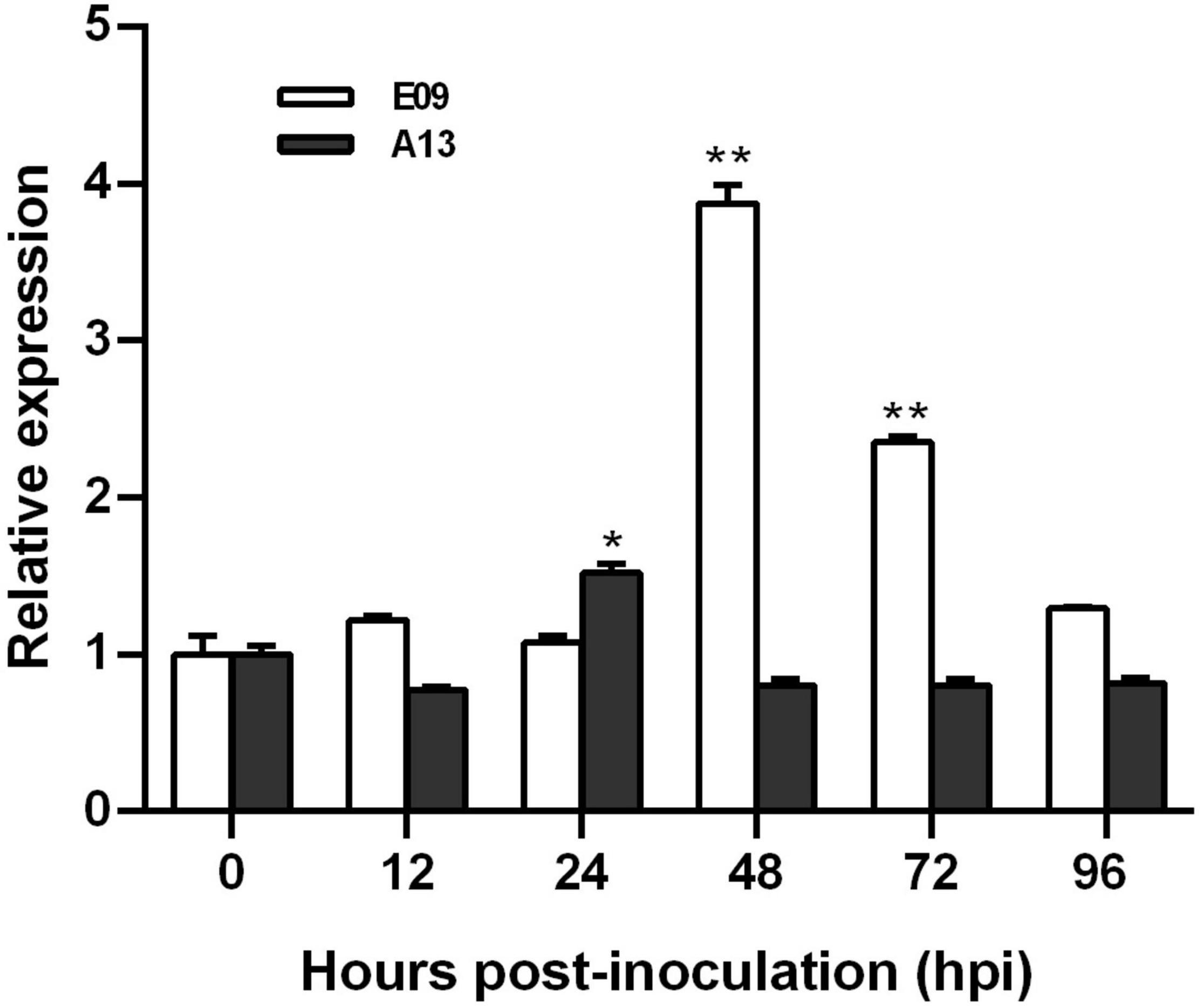
Figure 1. Transcript profiles of TaGSL22 after inoculation with avirulent (E09) and virulent (A13) Bgt races. Relative gene quantification was calculated by the comparative ΔΔCT method. Data were normalized to the expression level of the wheat elongation factor TaEF-1α. * and ** indicate significant differences (P < 0.05 and P < 0.01) from the mock-inoculated control at 0 hpi using Student’s t-test. Error bars represent SEM of three biological replicates.
Expression of TaGSL22 Was Highest in Leaves
A qRT-PCR assay was used to assess the expression patterns of TaGSL22 in different tissues, including root, stem, and leaf. The results showed that TaGSL22 was detected in all tested plant tissues (Figure 2). TaGSL22 had the highest expression in leaves and the lowest expression in roots. If the expression level in root tissues was defined as 1, then its expression in the stem and leaf tissues were 1.41 and 1.85, respectively.
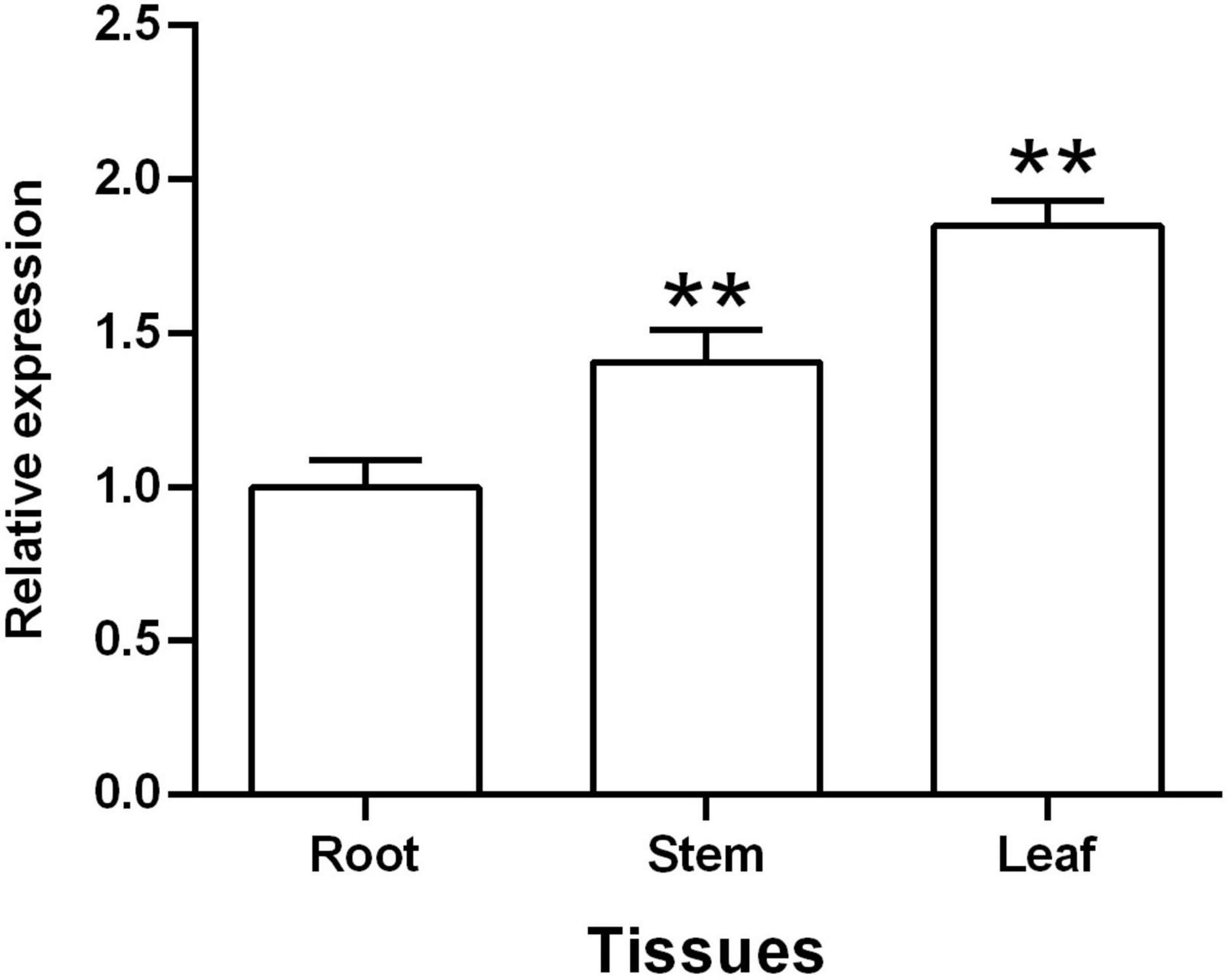
Figure 2. Tissue-specific expression of TaGSL22. Relative gene quantification was calculated by the comparative ΔΔCT method. Data were normalized to the expression level of the wheat elongation factor TaEF-1α. ** indicates a significant difference (P < 0.01) from the root by Student’s t-test. Error bars represent SEM of three biological replicates.
TaGSL22 Was Localized in the Cell Membrane and Cytoplasm of Wheat
To verify the subcellular localization predicted by the cNLS Mapper online software based on the amino acid sequence of the TaGSL22 protein, the pJIT163 empty plasmid (pJIT163:00) and pJIT163 recombinant plasmid with green fluorescent protein (GFP) fusion (pJIT163:TaGSL22) were transformed into wheat protoplasts. It was found that the protoplasts with pJIT163:00 emitted green fluorescence in the cell membrane, cytoplasm, and nucleus under the GFP channel, and under the SV2 channel, red fluorescence was observed only in the region where the organelles were located (Figure 3). In the protoplasts transformed with pJIT163:TaGSL22, the cell membrane and cytoplasm emitted green fluorescence under the GFP channel. These results suggested that the TaGSL22 protein was located in the cell membrane and cytoplasm, which was consistent with the initial prediction based on the amino acid sequence.
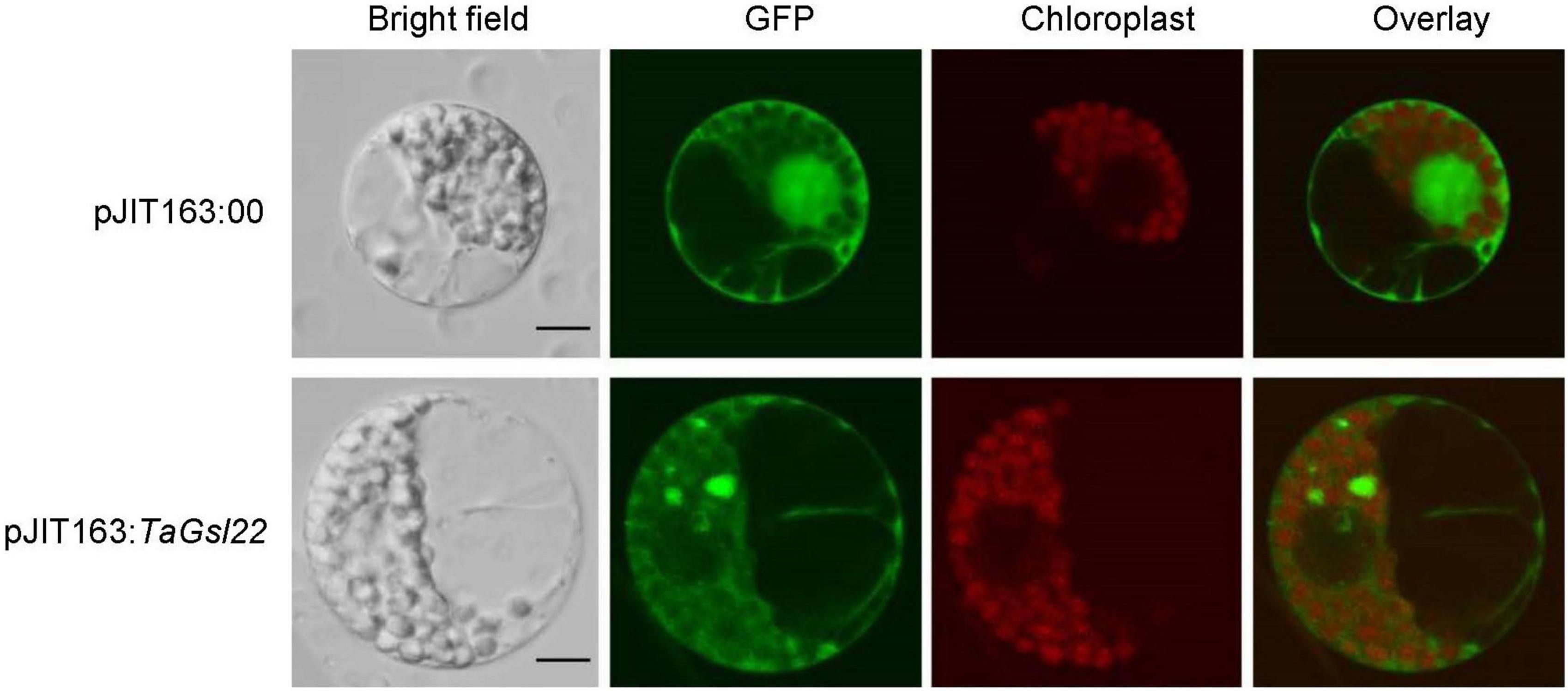
Figure 3. Subcellular localization of the TaGSL22 protein. pJIT163:00, pJIT163 empty plasmid; pJIT163:TaGSL22, pJIT163 recombinant plasmid with TaGSL22; overlay, superposition of green fluorescent protein (GFP) channel and chloroplast channel; bright field images show the equivalent field observed under white light. Scale bars: 10 μm.
TaGSL22 Was Induced by Plant Hormones Ethylene and Salicylic Acid
To understand the regulatory effect of plant hormones on TaGSL22 in wheat, the seedlings of variety Asosan/8Cc were treated with MeJA (0.1 mM), ETH (0.2 mM), ABA (0.2 mM), and SA (1 mM), respectively (Figure 4). After MeJA or ABA treatment, expression of TaGSL22 was down-regulated. In contrast, the TaGSL22 transcript level dropped to its minimum at 6 h post treatment (hpt) with ETH, then increased dramatically at 12 hpt until reaching its maximum at 24 hpt, at about 2.8 times that of the control (0 hpt). When treated with SA, expression of the gene decreased initially, then increased gradually to reach its maximum at 12 hpt, then decreased again. It was therefore inferred that TaGSL22 may be involved in the ETH- and SA-mediated signaling pathways.
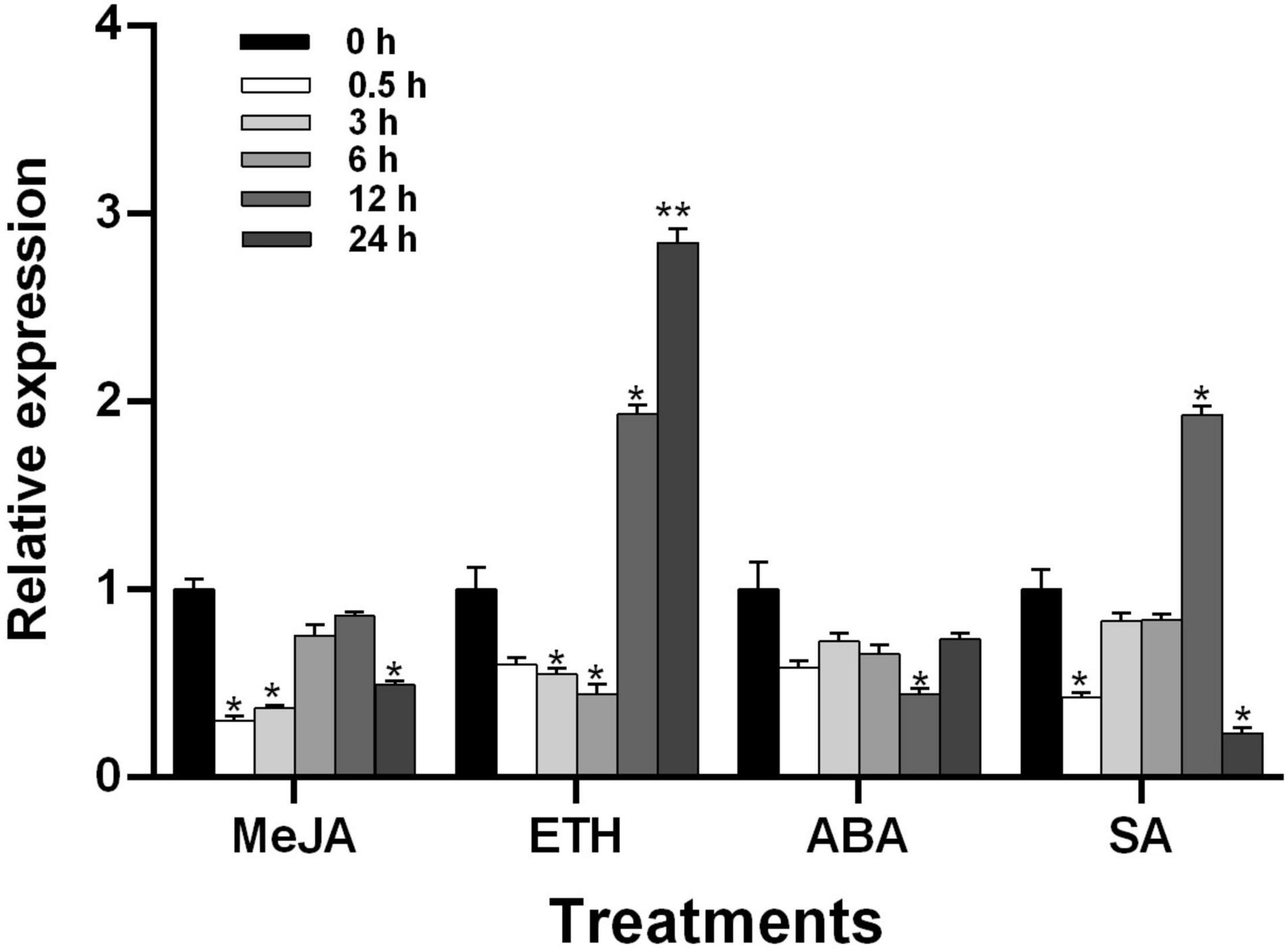
Figure 4. Transcript profiles of TaGSL22 under different hormone treatments. MeJA, methyl jasmonate; ETH, ethylene; ABA, abscisic acid; SA, salicylic acid. The mock control was treated with water. Data were normalized to the expression level of the wheat elongation factor TaEF-1α. Relative gene quantification was calculated by the comparative ΔΔCT method. * and ** indicate a significant difference (P < 0.05 and P < 0.01) from the control at 0 hpt using Student’s t-test. Error bars represent SEM of three biological replicates.
Silencing of TaGSL22 Reduced Resistance to Powdery Mildew in Wheat
To further assess the function of TaGSL22 in wheat, the gene was silenced via a BSMV-based VIGS approach (Singh et al., 2012). Ten days after inoculation with γ-PDS as a control for VIGS, an efficient silencing of the PDS gene was indicated by the observation of photobleaching symptoms on the leaves (Figure 5A). The leaves inoculated with both the γ virus and the γ-TaGSL22 virus exhibited chlorotic stripes, and no symptoms were observed on the leaves without virus inoculation. To evaluate the role of TaGSL22 in resistance against the powdery mildew pathogen Bgt, the fourth leaf with evident virus virulence was inoculated with Bgt E09 and the disease IT was scored at 10 dpi. The gene-silenced group was found to have significantly more spores on its leaves than on leaves in the control groups (Figure 5B).
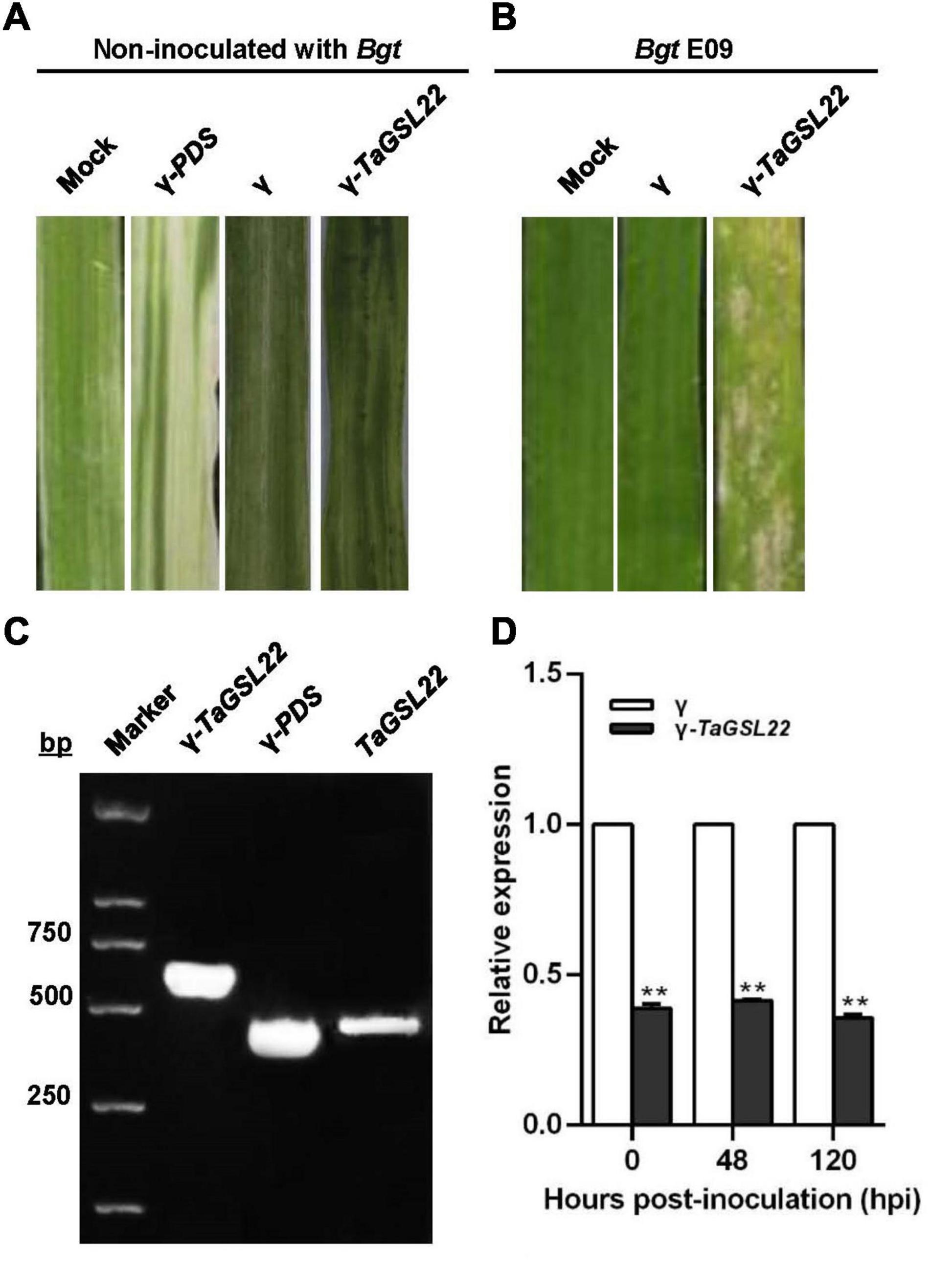
Figure 5. Functional assessment of TaGSL22 in the wheat–Bgt interaction by gene silencing. (A) The leaf phenotype of successful virus infection; photographs were captured 12 days post-inoculation (dpi); mock: leaves inoculated with water; γ-PDS: transcripts of α, β, γ-PDS plasmids inoculated on leaves; γ: transcripts of α, β, γ plasmids inoculated on leaves; γ-TaGSL22: transcripts of α, β, γ-TaGSL22 plasmids inoculated on leaves. (B) Disease phenotypes of the leaves pre-inoculated with BSMV: γ then the avirulent E09 Bgt race; mock: the leaves were only inoculated with Bgt. Photos were taken at 10 dpi. (C) PCR amplification of γ-TaGSL22, γ-PDS, and TaGSL22 from corresponding leaves. (D) Assessment of TaGSL22 silencing efficiency. Relative gene quantification was calculated by the comparative ΔΔCT method, and data were normalized to the expression level of the wheat elongation factor TaEF-1α. ** indicates a significant difference at P < 0.01 from the control group (γ) by Student’s t-test. Error bars represent SEM for three biological replicates.
The involvement of TaGSL22 in the plant resistance reaction was verified by PCR amplification of γ-TaGSL22, γ-PDS, and TaGSL22 from corresponding leaves. qRT-PCR analysis was carried out to determine whether the gene was effectively silenced at the transcript level (Figure 5C). At three time points, the endogenous TaGSL22 transcript levels were significantly reduced in leaves inoculated with the γ-TaGSL22 virus compared with those in control plants (Figure 5D). The results indicated that the wheat resistance to Bgt was reduced after down-regulation of TaGSL22.
Down-Regulation of TaGSL22 Led to Significantly More Virulent Fungal Infections
To further clarify the role of TaGSL22 during the fungal colonization on leaves, samples were taken at three time points and observed under a fluorescence microscope (Figure 6A). At 24 hpi, the appressoria were two times longer in the TaGSL22 down-regulated leaves (γ-TaGSL22) compared to those in the control plants (γ). Secondary hypha were observed at 48 hpi in the TaGSL22-silenced leaves, while in the control plants, the primary germ tube was only just germinated from the conidium. At 96 hpi, the hypha mass was significantly larger in the gene-silenced plants than in the control plants. The leaves of the gene-silenced group displayed a significant increase in the number of mycelia branches and mycelia lengths compared to those in the control group, and the powdery mildew colony areas were also larger (Figures 6B–D). Over time, all three measured parameters increased dramatically at the histological level. A considerably faster growth of the fungus in the TaGSL22-silenced plants suggested that down-regulation of TaGSL22 increased the Bgt fungal infection.
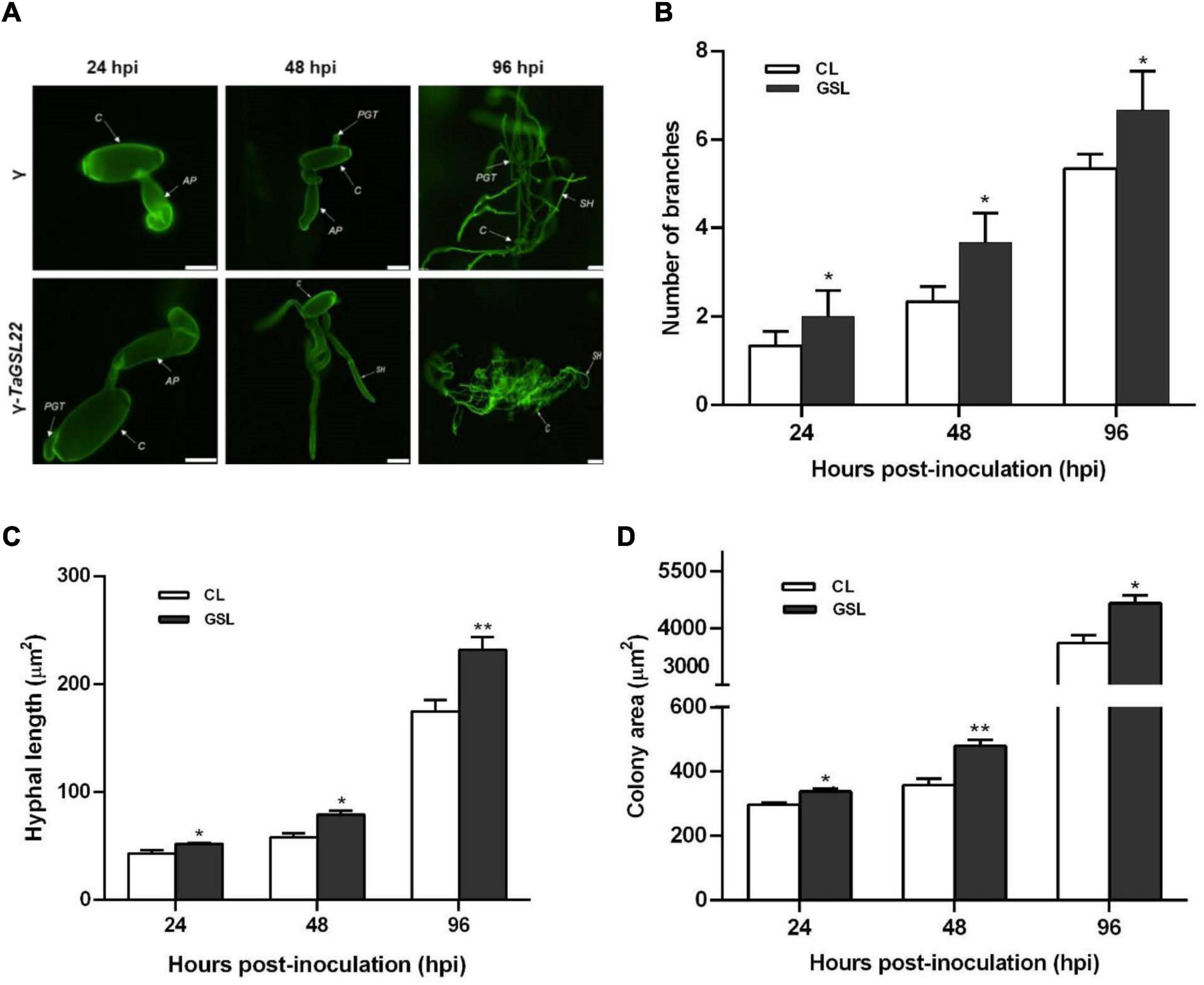
Figure 6. (A) Histopathological observation of Bgt infection on leaves after gene silencing. Samples of control plants (γ) and the gene-silenced plants (γ-TaGSL22) were taken at 24, 48, and 96 hpi. The fungal structures were stained with wheat germ agglutinin (WGA). C, conidia; AP, appressorium; PGT, primary germ tube; SH, secondary hyphae. Scale bars: 5 μm. (B) Number of branches, (C) hyphal length, and (D) colony area of Bgt after gene silencing. The leave samples were collected at 24, 48, and 96 hpi, and all results were obtained from 50 infection sites. CL, control leaf; GSL, gene-silenced leaf. Error bars represent SEM of three biological replicates. * and ** represent significant differences at P < 0.05 and P < 0.01, respectively, from the control by Student’s t-test.
Silencing of TaGSL22 Resulted in Reduced Levels of Callose
In order to determine whether the silencing of the TaGSL22 gene affected the deposition of callose in epidermal cells during Bgt infection, infected wheat leaves were stained with aniline blue to examine callose accumulation (Figure 7A). Samples of control plants (wild-type and γ) and the gene-silenced plants (γ-TaGSL22) were collected at 24 hpi with Bgt E09 and stained, then microscopic observation was carried out. It was observed that the fluorescent signals detected in the gene-silenced plants were significantly reduced compared to signals in the control plants, whereas the wild-type and empty vector plants showed similar intensities of callose accumulation signals (Figure 7B).
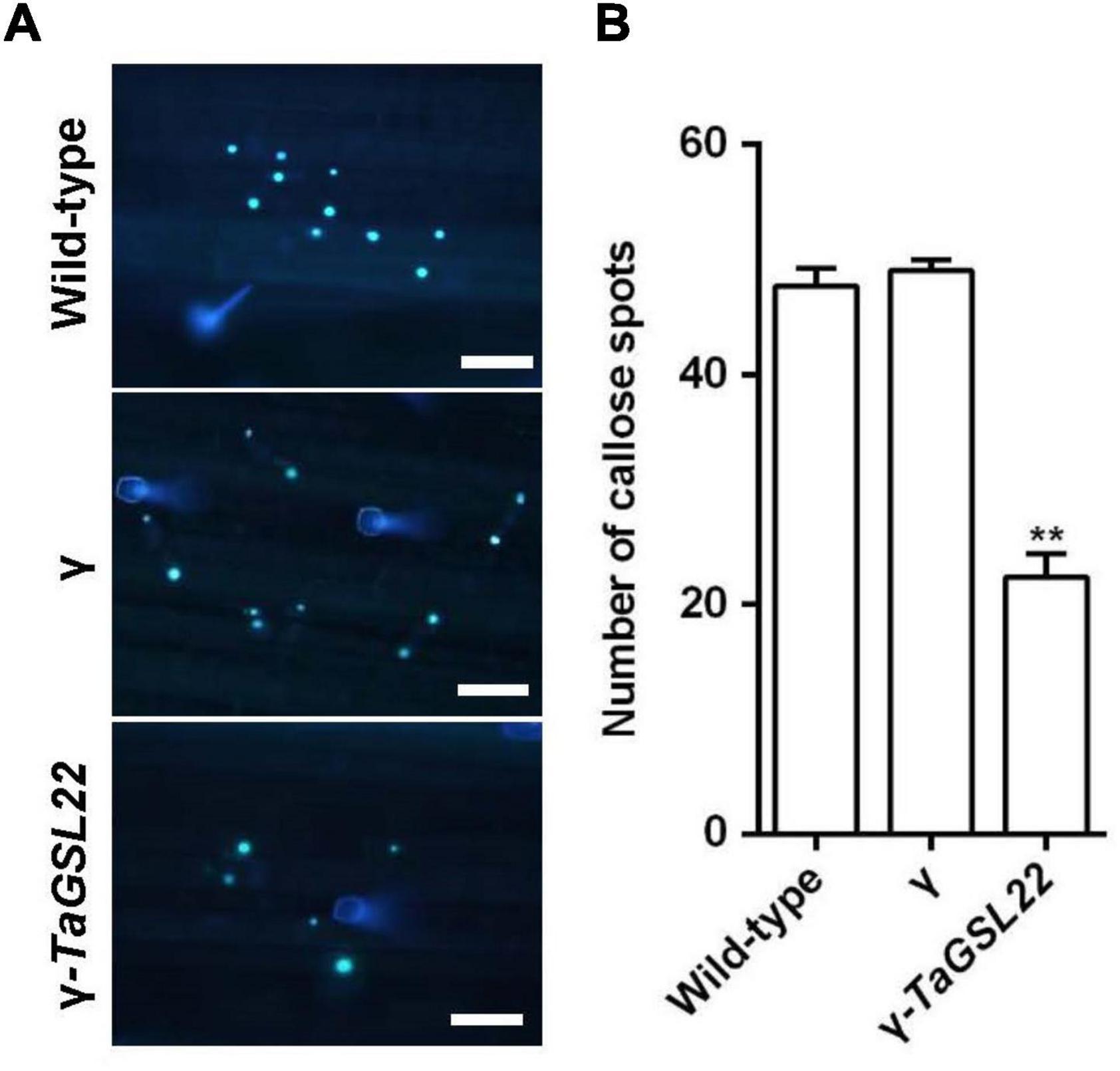
Figure 7. Down-regulation of TaGSL22 expression reduced the callose deposition in host plant. (A) Representative wheat leaves stained with aniline blue for callose deposition (bar = 100 μm). Pictures of control plants (wild-type and γ) and the gene-silenced plants (γ-TaGSL22) were taken at 24 hpi with Bgt avirulent race E09 at ×10 magnification with an Olympus BX-51 microscope. (B) The average number of callose foci per field of view (1 mm2) was calculated from 30 fields for each sample. Error bars represent SEM of three biological replicates. ** represent significant differences at P < 0.01 (Student’s t-test).
Discussion
Callose Synthase Glucan Synthase-Likes Play Different Roles in Various Plant Species and Tissues
Callose, a β-1,3-glucan, plays critical regulatory roles in plants, such as in sieve tube metabolism and gametophyte development (Legentil et al., 2015; Shi et al., 2015). It has long been observed that the deposition of relatively high amounts of callose and cellulose at sites of attempted penetration induces the formation of effective papillae, preventing the invasion of pathogens (Voigt, 2016). The most prominent cell wall polymer of papillae is β-1,3-glucan callose (Mangin, 1895). It has been reported that callose can enhance the resistance of melon to aphids (Shinoda, 1993), and early callose deposition increased the resistance of Arabidopsis to powdery mildew (Ellinger et al., 2013). Furthermore, overexpression of the Arabidopsis callose synthase gene AtGSL5 in barley induced penetration resistance to the barley powdery mildew pathogen Blumeria graminis f. sp. hordei (Bgh) (Blümke et al., 2013). Silencing of HvGSL6, encoding the barley callose synthase that falls into the AtGSL5 clade, also caused a decline in callose accumulation in powdery mildew-induced papillae and resulted in an increased penetration rate of Bgh (Chowdhury et al., 2016).
To date, a number of TaGSL genes have been identified in different wheat tissues with various regulatory functions under different environmental conditions (Voigt et al., 2006). TaGSL8 or TaGSL10 are involved in plant regeneration, and the knocking down of TaGSL3 and TaGSL8 slightly reduced the resistance against F. graminearum (Rana et al., 2014). In the present study, phylogenetic analysis revealed that TaGSL22 had the closest sequence similarity with AtGSL5. Transcript profile analysis at different time points revealed a significant up-regulation of the TaGSL22 gene following infection by Bgt. As AtGSL5 was the only GSL induced during pathogen infection (Dong et al., 2008), our results suggested that TaGSL22 was likely the ortholog of AtGSL5. The tissue-specific expression analysis of TaGSL22 showed that its expression was highest in the leaves compared to the roots and stems, which is consistent with the finding by Voigt et al. (2006). Silencing of TaGSL22 reduced the accumulation of callose significantly and increased the Bgt growth rate. Our study is the first report on the GSL genes associated with papillary callose accumulation and powdery mildew resistance in common wheat.
Pathogen-Induced Callose Synthase in Poaceae May Be Involved in Different Plant Defense Regulating Pathways Than Dicots
Unlike in barley and wheat, it was found that silencing the pathogen-induced callose synthase AtGSL5 in Arabidopsis results in a significant reduction of callose deposition, leading to an unexpected increase in fungal resistance (Jacobs et al., 2003). Similarly, silencing of the callose synthase gene SlPMR4, the ortholog of AtGSL5 in tomato, resulted in increased resistance to the adapted powdery mildew pathogen O. neolycopersici (Huibers et al., 2013). Interestingly, the increased resistance in Arabidopsis has previously been explained to result from a hyperactivation of the SA-dependent defense pathway due to the loss of callose (Nishimura et al., 2003). It has also been suggested that SA may be involved in the regulation of stress response genes, since similar results were found in a study of induced plant defense responses against chewing insects in Arabidopsis (Stotz et al., 2000). Although reciprocal phenomena were observed from the down-regulation of GSL in Arabidopsis and barley, the function of callose deposition was verified by the overexpression of AtGSL5 in barley and Arabidopsis, resulting in complete penetration resistance to Bgh and Golovinomyces cichoracearum, respectively (Blümke et al., 2013; Ellinger et al., 2013). Combining this result with the fact that silencing the HvGSL6 gene leads to a loss-of-resistance phenotype that is not clouded by an off-target increase in the SA-dependent defense pathway, Chowdhury et al. (2016) proposed that the pathogen-induced callose synthase observed in barley may not be involved in regulating other plant defense pathways, as compared to observations in the dicotyledonous plants Arabidopsis and tomato.
In the present study, it was indicated that the expression of TaGSL22 increased at 12 hpt with ETH and SA, respectively. Interestingly, reciprocal trends were observed at 24 hpt. While the transcript level of TaGSL22 kept increasing under ETH treatment, the TaGSL22 expression was suppressed by SA. Considering the loss of callose induced SA-dependent defense pathways in Arabidopsis, further investigation is needed to understand the relationship between GSL regulation and SA-dependent defense pathways in wheat and barley.
Utilization of TaGSL22 in Wheat Resistance Breeding
The nucleotide sequence of TaGSL22 had the highest similarity with those of A. tauschii and H. vulgare, indicating that the gene is relatively conservative in Gramineae plants and evolves slowly. This gives new opportunities for investigating orthologous genes in relative species and provides new targets for increasing crop resistance against penetration. Our study identified a novel wheat ortholog gene, TaGSL22, that may contribute to improved resistance in wheat breeding. In future studies, we plan to search the wheat whole-genome resequencing database for any natural variations in TaGSL22 that can be identified for breeding applications. Molecular markers can be developed and tested in a natural wheat accession to verify the association between Bgt resistance and the TaGSL22 allele. The identified polymorphic markers could then be used for marker-assisted selection for introgression of TaGSL22 in wheat resistance breeding.
It is noteworthy that although HvGSL6 has been identified in barley and its RNAi transgenic lines resulted in increased penetration by Bgh (Chowdhury et al., 2016), studies on the overexpression of HvGSL6 have not been reported. We are working on developing TaGSL22 overexpression wheat lines to further investigate its positive regulation of resistance to wheat powdery mildew. However, transgenic crops are not widely accepted for agricultural production. Recently, A donor-DNA-free CRISPR/Cas-based approach was reported to increase expression of target genes by creating structural variations in rice (Lu et al., 2021). This cutting-edge technology knocks out the sequence between the target gene and the promotor of a neighboring high-expressed gene, resulting in gene knock-up by ligating the high-expressed promotor with the coding region of the target gene. It is believed that this advanced gene knock-up technology sheds light on the potential applications of an identified resistance gene in crop resistance enhancement without introducing alien DNA residues by plant transformation.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
PC and ZW designed the experiments and wrote the manuscript. ZW, YR, and PC performed the experiments. ZW, PC, PJ, KM, and QL analyzed the data. PC acquired the funding. BW supervised the project. All authors read and approved the final manuscript.
Funding
This study was financially supported by the grants from the National Natural Science Foundation of China (Grant Nos. 32072410 and 31701745), Key Research and Development Project of Shaanxi Province (Grant No. 2021ZDLNY01-01), Natural Science Basic Research Program of Shaanxi Province (Grant No. 2020JQ-257), and Scientific Startup Foundation for Doctors of Northwest A&F University (Grant No. 2452019582).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Jun Guo, Qiao Wang, and Chang Su for providing reagents, materials, and analysis tools. We also thank reviewers for their constructive comments.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.800077/full#supplementary-material
Footnotes
References
Araus, J. L., Ferrio, J. P., Buxo, R., and Voltas, J. (2007). The historical perspective of dryland agriculture: Lessons learned from 10,000 years of wheat cultivation. J. Exp. Bot. 58, 131–145. doi: 10.1093/jxb/erl133
Bari, R., and Jones, J. D. (2009). Role of plant hormones in plant defence responses. Plant. Mol. Biol. 69, 473–488. doi: 10.1007/s11103-008-9435-0
Belanger, R. R., Benhamou, N., and Menzies, J. G. (2003). Cytological Evidence of an Active Role of Silicon in Wheat Resistance to Powdery Mildew (Blumeria graminis f. sp. tritici). Phytopathology 93, 402–412. doi: 10.1094/PHYTO.2003.93.4.402
Blümke, A., Somerville, S. C., and Voigt, C. A. (2013). Transient expression of the Arabidopsis thaliana callose synthase PMR4 increases penetration resistance to powdery mildew in barley. Adv. Biosci. Biotechnol. 4, 810–813. doi: 10.4236/abb.2013.48106
Chowdhury, J., Schober, M. S., Shirley, N. J., Singh, R. R., Jacobs, A. K., Douchkov, D., et al. (2016). Down-regulation of the glucan synthase-like 6 gene (HvGsl6) in barley leads to decreased callose accumulation and increased cell wall penetration by Blumeria graminis f. sp. hordei. New Phytol. 212, 434–443. doi: 10.1111/nph.14086
Dong, X., Hong, Z., Chatterjee, J., Kim, S., and Verma, D. P. S. (2008). Expression of callose synthase genes and its connection with Npr1 signaling pathway during pathogen infection. Planta 229, 87–98. doi: 10.1007/s00425-008-0812-3
Ellinger, D., Naumann, M., Falter, C., Zwikowics, C., Jamrow, T., Manisseri, C., et al. (2013). Elevated early callose deposition results in complete penetration resistance to powdery mildew in Arabidopsis. Plant. Physiol. 161, 1433–1444. doi: 10.1104/pp.112.211011
Enns, L. C., Kanaoka, M. M., Torii, K. U., Comai, L., Okada, K., and Cleland, R. E. (2005). Two callose synthases, GSL1 and GSL5, play an essential and redundant role in plant and pollen development and in fertility. Plant. Mol. Biol. 58, 333–349. doi: 10.1007/s11103-005-4526-7
Huibers, R. P., Loonen, A. E., Gao, D., Van den Ackerveken, G., Visser, R. G., and Bai, Y. (2013). Powdery mildew resistance in tomato by impairment of SlPMR4 and SlDMR1. PLoS One 8:e67467. doi: 10.1371/journal.pone.0067467
Jacobs, A. K., Lipka, V., Burton, R. A., Panstruga, R., Strizhov, N., Schulze-Lefert, P., et al. (2003). An Arabidopsis callose synthase, GSL5, is required for wound and papillary callose formation. Plant Cell 15, 2503–2515. doi: 10.1105/tpc.016097
Kacprzak, P., Macioszek, V. K., and Kononowicz, A. K. (2011). Induced systemic resistance (isr) in the protection of plants against pathogenic fungi. Postepy. Biol. Komorki. 38, 129–142.
Koo, A. J., and Howe, G. A. (2009). The wound hormone jasmonate. Phytochemistry 70, 1571–1580. doi: 10.1016/j.phytochem.2009.07.018
Legentil, L., Paris, F., Ballet, C., Trouvelot, S., Daire, X., Vetvicka, V., et al. (2015). Molecular interactions of β-(1→ 3)-glucans with their receptors. Molecules 20, 9745–9766. doi: 10.3390/molecules20069745
Li, G., Cowger, C., Wang, X., Carver, B. F., and Xu, X. (2019). Characterization of Pm65, a new powdery mildew resistance gene on chromosome 2AL of a facultative wheat cultivar. Theor. Appl. Genet. 132, 2625–2632. doi: 10.1007/s00122-019-03377-2
Li, M. M., Dong, L. L., Li, B. B., Wang, Z. Z., Xie, J. Z., Qiu, D., et al. (2020). A CNL protein in wild emmer wheat confers powdery mildew resistance. New Phytol. 228, 1027–1037. doi: 10.1111/nph.16761
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2–Δ Δ CT Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, Y., Wang, J., Chen, B., et al. (2021). A donor-DNA-free CRISPR/Cas-based approach to gene knock-up in rice. Nat. Plants 7, 1445–1452. doi: 10.1038/s41477-021-01019-4
Martinez, M. I. S., Bracuto, V., Koseoglou, E., Appiano, M., Jacobsen, E., Visser, R. G. F., et al. (2020). CRISPR/Cas9-targeted mutagenesis of the tomato susceptibility gene PMR4 for resistance against powdery mildew. BMC Plant. Biol. 20:284–287. doi: 10.1186/s12870-020-02497-y
McDonald, B. A., and Linde, C. (2002). The population genetics of plant pathogens and breeding strategies for durable resistance. Euphytica 124, 163–180. doi: 10.1023/A:1015678432355
McLusky, S. R., Bennett, M. H., Beale, M. H., Lewis, M. J., Gaskin, P., and Mansfield, J. W. (1999). Cell wall alterations and localized accumulation of feruloyl-3’-methoxytyramine in onion epidermis at sites of attempted penetration by Botrytis allii are associated with actin polarisation, peroxidase activity and suppression of flavonoid biosynthesis. Plant. J. 17, 523–534. doi: 10.1046/j.1365-313X.1999.00403.x
Mondal, S., Singh, R. P., Mason, E. R., Huerta-Espino, J., Autrique, E., and Joshi, A. K. (2016). Grain yield, adaptation and progress in breeding for early-maturing and heat-tolerant wheat lines in South Asia. Field Crops Res. 192, 78–85. doi: 10.1016/j.fcr.2016.04.017
Nishimura, M. T., Stein, M., Hou, B. H., Vogel, J. P., Edwards, H., and Somerville, S. C. (2003). Loss of a callose synthase results in salicylic acid-dependent disease resistance. Science 301, 969–972. doi: 10.1126/science.1086716
Pogue, G. P., Lindbo, J. A., Dawson, W. O., and Turpen, T. H. (1998). “Tobamovirus transient expression vectors: tools for plant biology and high level expression of foreign proteins in plants,” in Plant Molecular Biology Manual, eds S. B. Gelvin and R. A. Schilperoot (Dordrecht: Kluwer Academic Publishers), 1–27. doi: 10.1385/0-89603-321-x:1
Rana, I. A., Salomon, S., Schafer, W., and Becker, D. (2014). Downregulation of Glucan Synthase-Like (TaGSL) genes in wheat leads to inhibition of transgenic plant regeneration. In Vitro Cell. Dev. Biol. Plant. 50, 696–706. doi: 10.1007/s11627-014-9636-y
Ryals, J., Uknes, S., and Ward, E. (1994). Systemic Acquired-Resistance. Plant. Physiol. 104, 1109–1112. doi: 10.4161/psb.1.4.3221
Shi, X., Sun, X., Zhang, Z., Feng, D., Zhang, Q., Han, L., et al. (2015). GLUCAN SYNTHASE-LIKE 5 (GSL5) plays an essential role in male fertility by regulating callose metabolism during microsporogenesis in rice. Plant. Cell Physiol. 56, 497–509. doi: 10.1093/pcp/pcu193
Shigenaga, A. M., and Argueso, C. T. (2016). No hormone to rule them all: Interactions of plant hormones during the responses of plants to pathogens. Semin. Cell Dev. Biol. 56, 174–189. doi: 10.1016/j.semcdb.2016.06.005
Shinoda, T. (1993). Callose reaction induced in melon leaves by feeding of melon aphid, Aphis gossypii Glover as possible aphid-resistant factor. Jpn. J. Appl. Entomol. Zool. 37, 145–152. doi: 10.1303/jjaez.37.145
Singh, A., Liang, Y. C., Kumar, P., Jiang, C. Z., and Reid, M. S. (2012). Co-silencing of the Mirabilis antiviral protein (MAP) permits virus-induced gene silencing (VIGS) of other genes in Four O’Clock plants (Mirabilis jalapa). J. Horticult. Sci. Biotechnol. 87, 334–340. doi: 10.1080/14620316.2012.11512873
Stotz, H. U., Pittendrigh, B. R., Kroymann, J., Weniger, K., Fritsche, J., Bauke, A., et al. (2000). Induced plant defense responses against chewing insects. Ethylene signaling reduces resistance of Arabidopsis against Egyptian cotton worm but not diamondback moth. Plant. Physiol. 124, 1007–1018. doi: 10.1104/pp.124.3.1007
Voigt, C. A. (2016). Cellulose/callose glucan networks: The key to powdery mildew resistance in plants? New Phytol. 212, 303–305. doi: 10.1111/nph.14198
Voigt, C. A., Schafer, W., and Salomon, S. (2006). A comprehensive view on organ-specific callose synthesis in wheat (Triticum aestivum L.): Glucan synthase-like gene expression, callose synthase activity, callose quantification and deposition. Plant. Physiol. Biochem. 44, 242–247. doi: 10.1016/j.plaphy.2006.05.001
Wang, X., Bai, J., Liu, H., Sun, Y., Shi, X., and Ren, Z. (2013). Overexpression of a maize transcription factor ZmPHR1 improves shoot inorganic phosphate content and growth of Arabidopsis under low-phosphate conditions. Plant. Mol. Biol. Rep. 31, 665–677. doi: 10.1007/s11105-012-0534-3
Wawrzynska, A., Rodibaugh, N. L., and Innes, R. W. (2010). Synergistic activation of defense responses in Arabidopsis by simultaneous loss of the GSL5 callose synthase and the EDR1 protein kinase. Mol. Plant. Microbe Interact. 23, 578–584. doi: 10.1094/MPMI-23-5-0578
Xie, J., Guo, G., Wang, Y., Hu, T., Wang, L., Li, J., et al. (2020). A rare single nucleotide variant in Pm5e confers powdery mildew resistance in common wheat. New Phytol. 228, 1011–1026. doi: 10.1111/nph.16762
Xu, Q., Tang, C. L., Wang, L. K., Zhao, C. C., Kang, Z. S., and Wang, X. J. (2020). Haustoria - arsenals during the interaction between wheat and Puccinia striiformis f. sp. tritici. Mol. Plant Pathol. 21, 83–94.
Yuan, C., Li, C., Yan, L., Jackson, A. O., Liu, Z., Han, C., et al. (2011). A high throughput barley stripe mosaic virus vector for virus induced gene silencing in monocots and dicots. PLoS One 6:e26468. doi: 10.1371/journal.pone.0026468
Keywords: wheat powdery mildew, BMSV-VIGS, disease resistance, callose, glucan synthase-like, TaGSL22
Citation: Cheng P, Wang Z, Ren Y, Jin P, Ma K, Li Q and Wang B (2021) Silencing of a Wheat Ortholog of Glucan Synthase-Like Gene Reduced Resistance to Blumeria graminis f. sp. tritici. Front. Plant Sci. 12:800077. doi: 10.3389/fpls.2021.800077
Received: 22 October 2021; Accepted: 06 December 2021;
Published: 23 December 2021.
Edited by:
Guotian Li, Huazhong Agricultural University, ChinaCopyright © 2021 Cheng, Wang, Ren, Jin, Ma, Li and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peng Cheng, cGVuZ2NoZW5nQG53YWZ1LmVkdS5jbg==; Baotong Wang, d2FuZ2J0QG53YWZ1LmVkdS5jbg==
 Peng Cheng
Peng Cheng Zihao Wang
Zihao Wang Yanyan Ren
Yanyan Ren Pengfei Jin
Pengfei Jin Kangjie Ma
Kangjie Ma Qiang Li
Qiang Li Baotong Wang
Baotong Wang