- 1Engineering Research Center for Breeding of New Varieties of Tropical Crops, Ministry of Education, Haikou, China
- 2College of Horticulture, Hainan University, Haikou, China
The comparison of chloroplast genome (cpDNA) sequences among different plant species is an important source of plant molecular phylogenetic data. In this paper, the cpDNA sequences of 13 different oil-tea camellia samples were compared to identify an undetermined oil-tea camellia species from Hainan Province. The cpDNA of the samples was sequenced and resequenced, and divergence hotspots and simple sequence repeat (SSR) variations were analyzed. Bayesian inference (BI) and maximum-likelihood (ML) phylogenetic trees were constructed based on the full cpDNA sequences. The cpDNA sequences were 156512∼157089 bp in length and had the circular tetrad structure typical of angiosperms. The inverted repeats (IRs) of different species included varying contractions and expansions. The cpDNA sequences of the samples of the undetermined species of oil-tea camellia from Hainan Province and Camellia gauchowensis from Xuwen County were identical. In total, 136 genes were annotated, including 91 protein-coding genes (PCGs), 37 tRNA genes and 8 rRNA genes. The GC content of the cpDNA was 37.3%. The small single-copy (SSC)/IR boundary was rich in variation. Divergence hotspots were mainly located in the intergenic space (IGS) and coding sequences (CDSs), and there were obvious differences in divergence hotspots among species. The same divergence hotspots were found in Camellia vietnamensis, Camellia gauchowensis and the undetermined species of oil-tea camellia from Hainan Province. A total of 191∼198 SSR loci were detected. Most of the SSRs included A or T, and the distribution of SSRs in the cpDNA was uneven. Different species shared common SSRs and exhibited unique SSRs. Based on the full cpDNA sequences, the evolutionary relationships of different species of Camellia were well identified. The thirteen samples were classified into 2 clades and 6 subclades, and the different sections of Camellia clustered on the same branch in 2 clades and 2 subclades. Camellia vietnamensis was more closely related to the undetermined species of oil-tea camellia from Hainan Province and the sample of Camellia gauchowensis from Xuwen County than to the sample of Camellia gauchowensis from Luchuan County. Camellia osmantha was closely related to Camellia gauchowensis and Camellia vietnamensis. In conclusion, the cpDNA of different oil-tea camellia species has a conserved tetrad structure with certain length polymorphisms. SSRs are expected to be developed as “barcodes” or “identity cards” for species identification. SSR variations and other factors result in abundant divergence hotspots in the CDSs and IGS (one non-CDS region), indicating that full cpDNA sequences can be used for the species identification and phylogenetic analysis of Camellia. Accordingly, the undetermined species of oil-tea camellia from Hainan Province is likely Camellia vietnamensis, Camellia vietnamensis and Camellia gauchowensis may be the same species, and additional genetic evidence is needed to determine whether Camellia osmantha is a new independent species. The previous division of related sections of Camellia may need readjustment based on full cpDNA sequences.
Introduction
Oil-tea camellia trees, as one of the four largest woody oil plants in the world, are endemic in China and have a long history of cultivation. The group consists of nearly 20 species of Camellia, such as Camellia oleifera, Camellia meiocarpa, and Camellia vietnamensis, and approximately 30 common subspecific taxa. Camellia oil is rich in tea polyphenols, tea saponins and other health components and free of cholesterol, erucic acid and other harmful components. The oil has extremely high nutritional and health-beneficial value and thus has strong market competitiveness and wide market prospects (Zhu et al., 2010). Oil-tea camellia trees, with the characteristics of strong resistance, wide adaptability and good tolerance of typhoons (Chen et al., 2012), are suitable for afforestation in low-yielding or desolated woodlands and can be expected to provide immense ecological benefits.
Oil-tea camellia trees have been planted in Hainan Province for approximately 2000 years. Four 600-year-old trees were discovered as the most ancient individuals, and the area of oil-tea camellia afforestation reached approximately 6,000 hectares in the 1950s to 1960s. As people in Hainan Province have always had a special preference for camellia oil, regarding it as “magic oil,” camellia oil has always been in short supply; thus, the selection and breeding of afforestation varieties is urgently needed to promote the rapid development of the Hainan oil-tea camellia industry (Dai and Zhong, 2017). Since 2007, C. oleifera cultivars from outside Hainan Island have been introduced to Wuzhishan city in Hainan Province for afforestation, as local oil-tea camellia tree seedlings planted for afforestation show weak growth, low survival rates and poor economic performance. New cultivars bred from native germplasm resources have been increasingly realized as crucial to the development of the oil-tea camellia industry in Hainan Province. Furthermore, the lack of afforestation cultivars of local species of oil-tea camellia trees is the primary bottleneck in the development of the oil-tea camellia industry in Hainan Province (Chen et al., 2017).
The oil-tea camellia trees in Hainan Province have been identified as Camellia oleifera (Yuan et al., 2014; Ye et al., 2015), and the features of native oil-tea camellia trees, such as large fruits with thick pericarps and the unique oil scent, are different from those of C. oleifera grown outside Hainan Island, which is attributed to the influence of the tropical environment (Zheng D. J. et al., 2016; Zheng W. W. et al., 2016). However, C. oleifera cultivars introduced from outside Hainan Island presented poor growth, harvest properties and survival rates (Chen et al., 2017). Overall, the above results indicate that native oil-tea camellia trees from Hainan Province might not belong to C. oleifera.
Traditional plant identification methods based on morphological characteristics have difficulties eliminating interference factors such as environmental factors and tree ages, and DNA barcoding does not differentiate closely related species because of resolution limitations (Hebert et al., 2003; Zhang et al., 2015). The chloroplast genome (cpDNA), which has uniparental maternal inheritance, is small, and its variations provide much more information than that obtained from a single DNA barcode without recombination. cpDNA can be used to identify different species and even different populations of the same species; therefore, it is called a “super barcode” (Li et al., 2015). The coding sequences (CDSs) and non-coding sequences (non-CDSs) of cpDNA differ greatly in evolutionary rates and have low sequencing costs and small splicing errors. cpDNA has the advantages of convenience, accuracy and low cost for exploring the systematic evolution, classification and identification of plant species (Semerikova and Semerikov, 2014). Therefore, in the past 30 years, researchers have increasingly preferred to use cpDNA comparisons for the identification, classification and evolutionary relationship determination of plant species (Leigh et al., 2013). Camellia plants easily hybridize and cross-pollinate and may also have intraspecific polyploids, making identification at the species level difficult (Liu et al., 2012). Therefore, identification studies need to be carried out from multiple perspectives, among which cpDNA comparative analysis is important for studying species identification and evolutionary relationships. At the time of writing this paper, the NCBI genome database1 contained 7556 complete cpDNA sequences, 45 of which were from Camellia (10 cpDNA sequences assembled by this research group have not yet been published), providing a foundation for discussing the evolutionary relationships and species identification of Camellia and other plants.
In brief, to breed appropriate afforestation cultivars and guide the introduction and collection of suitable germplasm resources, it is necessary to compare the cpDNA sequences of different oil-tea camellia samples and identify the undetermined oil-tea camellia species from Hainan Province. Furthermore, the results of cpDNA analyses also have important significance in brand creation and cultural value development of camellia oil.
Materials and Methods
Plant Materials
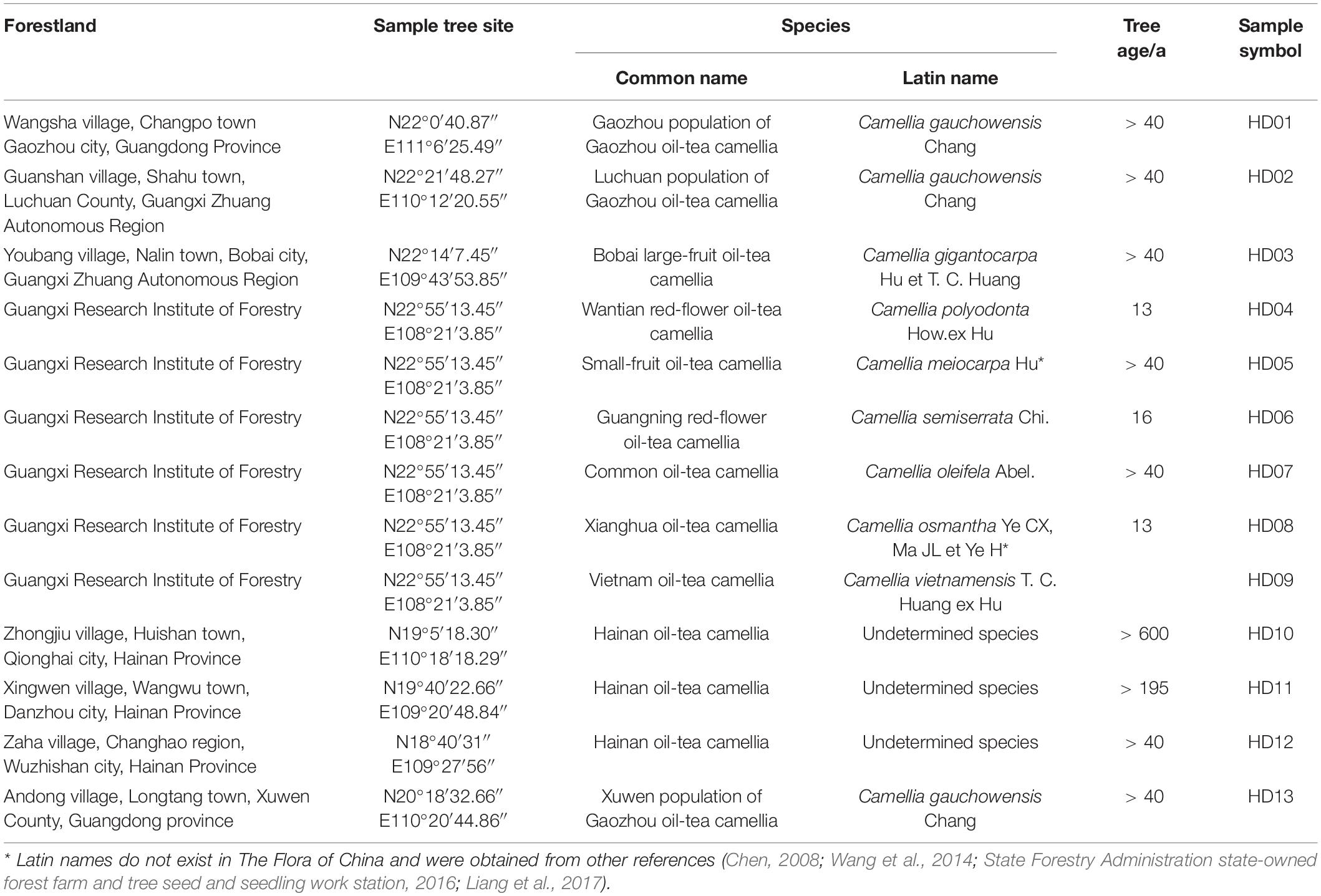
Information on the leaf sampling of forestland and the location, species, age and number of each plant sampled is shown in Table 1. From October 15 to November 5, 2017, 30 leaves were collected from each plant, immediately frozen in liquid nitrogen in the field, and brought back to the laboratory for storage in an ultralow-temperature (−80°C) freezer.
Chloroplast Genome Extraction, Sequencing and Genome Library Construction
Whole-genome DNA was extracted from 10 g samples of fresh leaves using an E.Z.N.A.® XXX DNA Kit. An Illumina TruSeq™ Nano DNA Sample Prep Kit was used to construct a PE library, and an 8-cycle enriched library was amplified by PCR. The target band was recovered in 2% Certified Low Range Ultra Agarose. PicoGreen nucleic acid dye was quantitatively detected with a TBS380 microfluorometer and mixed in proportion to the obtained data. A TruSeq PE Cluster Kit V3-BOT-HS was used to amplify and generate DNA clusters by bridging PCR with the cBot System. Finally, the DNA clusters were sequenced on the Illumina HiSeq 4000-PE150 sequencing platform to produce the original sequences (raw read length of 150 bp). The original sequences were subjected to quality control, whereby adapter sequences and bases containing non-AGCT at the 5′ end were removed, reads with sequencing quality values less than Q20 were trimmed, reads with N proportions more than or equal to 10% were removed, and joint sequences and small segments with lengths less than 75 bp were discarded after pruning. As a result, high-quality read sequences (clean reads) were obtained. The NT library was randomly selected to detect whether the sequencing results were contaminated.
Chloroplast Genome Splicing, Annotation, and Submission of Camellia oleifera
SOAPdenovo (version: 2.04) short-sequence assembly software was used to assemble the clean data from Camellia oleifera. The optimal assembly results were obtained after the parameters were adjusted several times. Then, the following two methods were used to screen cpDNA contigs: a homologous sequence searching method based on the cpDNA sequences of related species and a screening method based on cpDNA characteristics such as larger copy numbers, lower GC contents and unique kmer frequencies. Then, the reads were mapped to the assembled contigs, and local assembly and the optimization of the assembly results were performed according to paired ends and read overlaps. GapCloser (version 1.12) software was used to repair inner gaps in the assembly results, and redundant sequences were removed to obtain the final assembly results. Homologous alignment prediction and de novo prediction were combined to predict the genome of Camellia oleifera. Homologous alignment prediction was performed using the protein-coding genes (PCGs) of reference genomes. The PCGs were rapidly aligned to the sample genome sequence, poor alignment results were filtered to remove redundancy, and then GeneWise was used to produce exact alignment. AUGUSTUS software was used to predict de novo genes in plant mitochondrial/chloroplast genomes. Finally, EVidenceModeler V1.1.1 software was used to integrate the gene set and obtain the Camellia oleifera coding genes. DOGMA, RNAmmer-1.2 and TRNAscan-SE V1.3.1 were used to predict ncRNA in the genome. After the amino acid sequences of Camellia oleifera were predicted based on the identified genes, they were compared with the known protein database, and the Camellia oleifera genes were annotated with corresponding functional information. The optimal comparison result of each gene was retained as the annotation result. The amino acid sequences of the samples were compared with the non-redundant protein (NR), Swiss-Prot, eggNOG, Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) databases to obtain the functional annotation information of the PCGs of Camellia oleifera. After sequence annotation, the genome sequence was edited by Sequin and submitted to GenBank under accession number MN078090.
Chloroplast Genome Splicing, Annotation and Submission of the Other 12 Oil-Tea Camellia Trees
First, SPAdes software was used to preliminarily splice the clean data. Based on the above cpDNA data from Camellia oleifera and PCG sequences, blastn and Exonerate comparisons were performed (the criteria were an e-value of 1e-10 and a protein similarity threshold of 70%). The scaffold that matched each gene was selected, and splicing coverage was determined to remove fragments that were obviously not part of the target genome. PRICE and MITObim were used to carry out extended merging and splicing of the collected fragmented target sequence, and this process was iterated 50 times. With the results of iteration splicing, Bowtie2 was used to examine the original sequencing reads, paired reads were selected, and SPAdes was used for resplicing. The path was examined, and an obvious ring graph was selected. Otherwise, the iterative stitching and comparison steps were repeated until the ring graph was assembled successfully. The comparison of the obtained cpDNA of all oil-tea camellia species with the PCGs of Camellia oleifera described above was performed by blastn, and comparisons between PCGs and nucleic acids were performed to confirm the existence and boundaries of genes. If the predicted amino acid sequence was too long or too short, the starting codon was adjusted, other variable codons were used, or the gene was checked for introns. Exonerate software was used to compare the amino acid sequences of Camellia oleifera genes to determine intron boundaries and lengths. Chloroplast tRNA annotations were submitted to tRNAscan-SE online for annotation. For rRNA annotation, the sequences were submitted to the RNAmmer 1.2 server for prediction and supplemented by homologous sequence alignment to correct the boundary ranges. The tRNA annotation and ribonuclease rnpB were submitted to ARAGORN and Bcheck, respectively, for annotation. After sequence annotation of the HD01∼HD06 and HD08∼HD13 samples, the sequences were edited by Sequin and submitted to GenBank under accession numbers MN078084∼MN078089 and MN078091∼MN078093 (the sequences of the HD10∼HD13 samples were the same), respectively.
Characterization of Chloroplast Genome
Bitmap Graph Creation
The physical map of cpDNA was drafted by submitting the edited GenBank annotation file to OGDRAW. According to the assembled results of the 13 samples, non-CDSs were extracted by scripts, and the distribution regions [large single-copy (LSC), small single-copy (SSC), and inverted repeat (IR)] and sizes were determined.
Expansion and Contraction Analysis of Inverted Repeat Boundaries
First, a script was used to identify IR region A (IRA) and IR region B (IRB) sequences to determine the boundary positions of IRs. Then, according to the genome annotation results, the genes that crossed or were closest to the IR boundaries were located. Finally, the distances from the gene boundaries to the IR boundaries were extracted, and the results were plotted using AI.
Analysis of the Divergence Hotspots of Genomic Systems
Based on the literature (Yang et al., 2013), mVISTA software was used to analyze the evolutionary divergence hotspots of the genomic system, with the cpDNA of Panax ginseng C. A. Mey used as a reference. MAFFT software was used for multisequence alignment, and then a script was used to obtain the input file required by mVISTA. mVISTA was used to obtain the original output results. After overlapping gene names were adjusted and intron annotations were added, a divergence hotspot diagram was obtained.
Simple Sequence Repeat Analysis
According to the method reported in the literature (Zheng et al., 2016), the SSR sequences were analyzed with MISA software, with the parameters set as follows: 1-8, 2-4, 3-4, 4-3, 5-3 and 6-3.
Construction of a Chloroplast Genome Phylogenetic Tree
The complete cpDNAs of all Camellia plants were downloaded from the NCBI database, and 64 sequences were obtained. Hartia laotica (LAOstipa camellia) (NC_041509.1) was chosen as an outgroup species. The above 65 sequences were combined with 10 sample sequences (the HD10∼HD13 sequences were identical, and HD10 was used to represent these 4 samples) for phylogenetic analysis. The LSC, SSC and IR regions of 75 sequences were extracted, and LSC + IR + SSC data was used for analysis. The data was examined using MAFFT software (default parameters), sequence pruning was performed using Gblocks (parameters: -t, D, -b5, h), and a phylogenetic tree was constructed using MrBayes and IQ-TREE software.
Bayesian Inference
The outgroup was again set as Hartia laotica (NC_041509.1). The model parameters were LSET NST = 6 and rates = invgamma, which denoted a nucleic acid molecule replacement model of GTR. The rate variation across sites followed an inverse gamma model. The prior probability model parameters were set to the default values. The parameters of Markov chain Monte Carlo (MCMC) sampling were Nruns = 2, Nchain = 4, Ngen = 1000000, Samplefreq = 500 and Temp = 0.05, indicating that two groups of analyses were run simultaneously. One cold chain and three hot chains were set in each group to run 1000000 generations, and the Markov chain was sampled once every 500 generations. When the original tree results were obtained from MrBayes software, branches unrelated to the sample were cut off to obtain the final phylogenetic tree.
Maximum Likelihood
For model checking, the optimal model was selected via the IQ-TREE model finder. The optimal model of the full sequence was K3Pu + F + R2.
To build the Maximum likelihood (ML) tree, the outgroup was set as Hartia laotica (NC_041509.1), and the parameters were set as -BB 1000 and -ALRT 1000. When the original tree results were obtained, branches unrelated to the sample were cut off to obtain the final phylogenetic tree.
Results and Analysis
Sequencing and Assembly of Chloroplast Genome
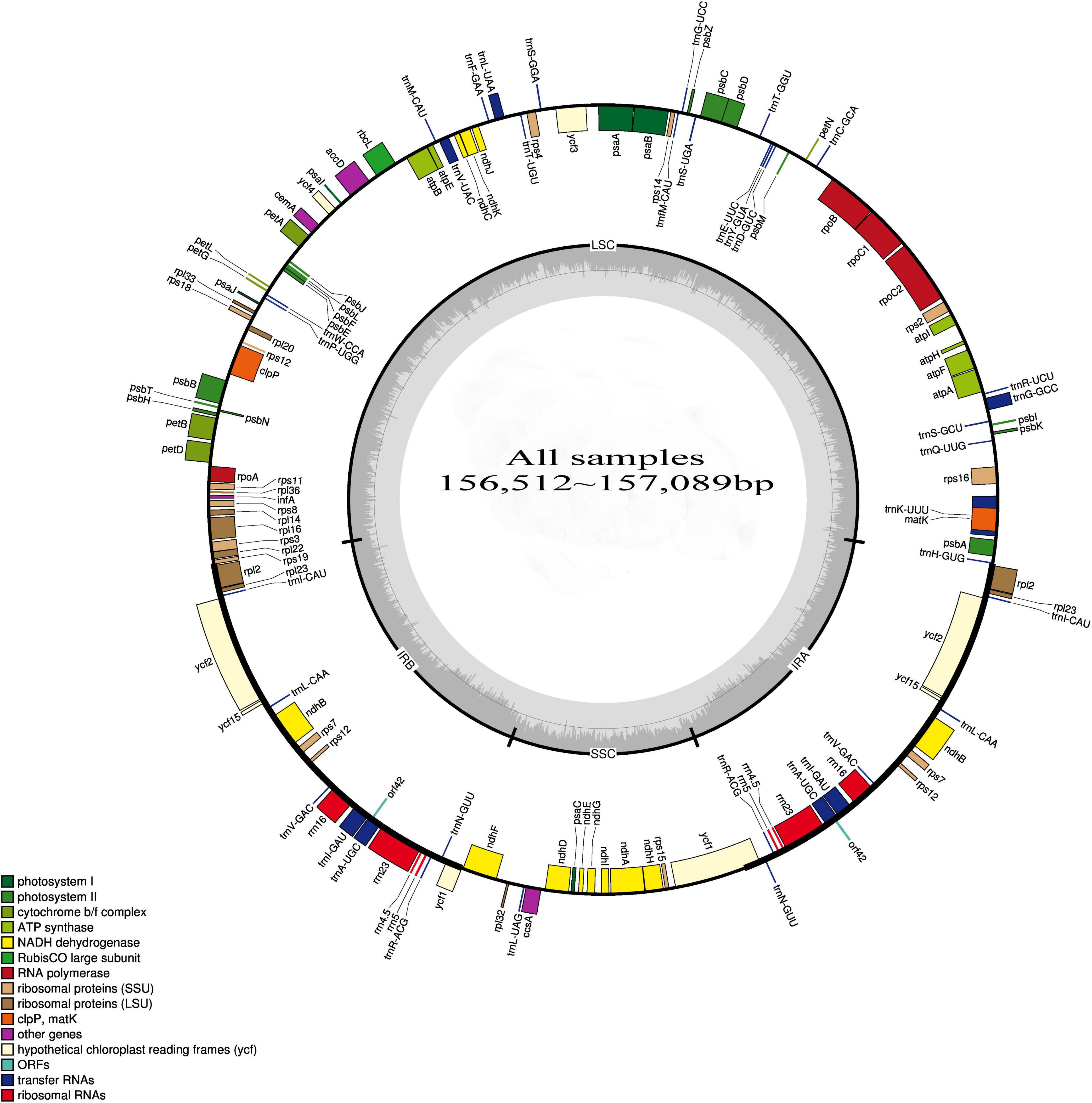
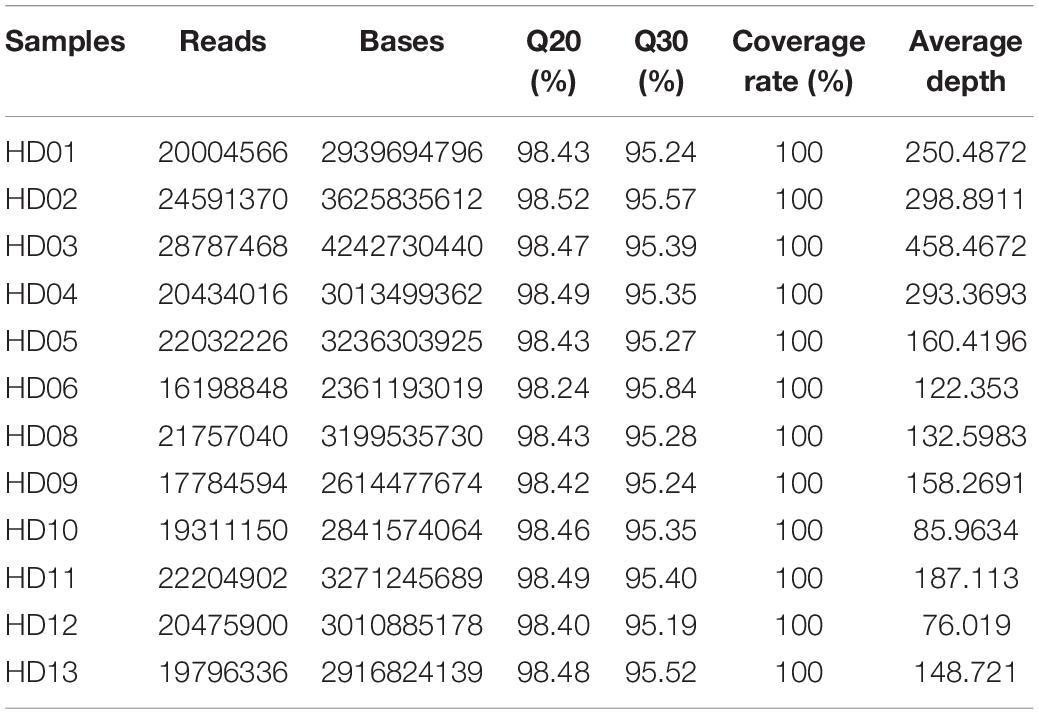
In the cpDNA sequencing of HD07 (Camellia oleifera), the raw data totaled 3235 MB, and the clean data totaled 2875 MB after quality control processing. The GC content of the clean data was 40.67%, the Q20-value was 97.95%, and the Q30 value was 94.02%. The cpDNA map is shown in Figure 1. The GC content of the genome was 37.29%, there were no unknown bases in the assembly sequence, and the sequencing coverage rate reached 100%. These results indicated that the quality of the cpDNA sequencing and assembly results was very high. cDNA resequencing of the other 12 samples was performed, and the cpDNA map of Camellia oleifera was used as a reference. The statistical results of the sequencing data are shown in Table 2. The numbers of reads ranged from 161988448 to 28787468, the base numbers ranged from 2361193019 to 4242730440, the Q20 value was above 98.24%, the Q30 value was above 95.19%, the sequencing coverage rate reached 100%, and the average sequencing depth ranged from 76.019 to 458.4672 times, all indicating that the sequencing results were relatively reliable. The clean reads of these samples were mapped to create Figure 1, and then the cpDNA map of each sample was assembled.
Organization of Camellia Chloroplast Genome
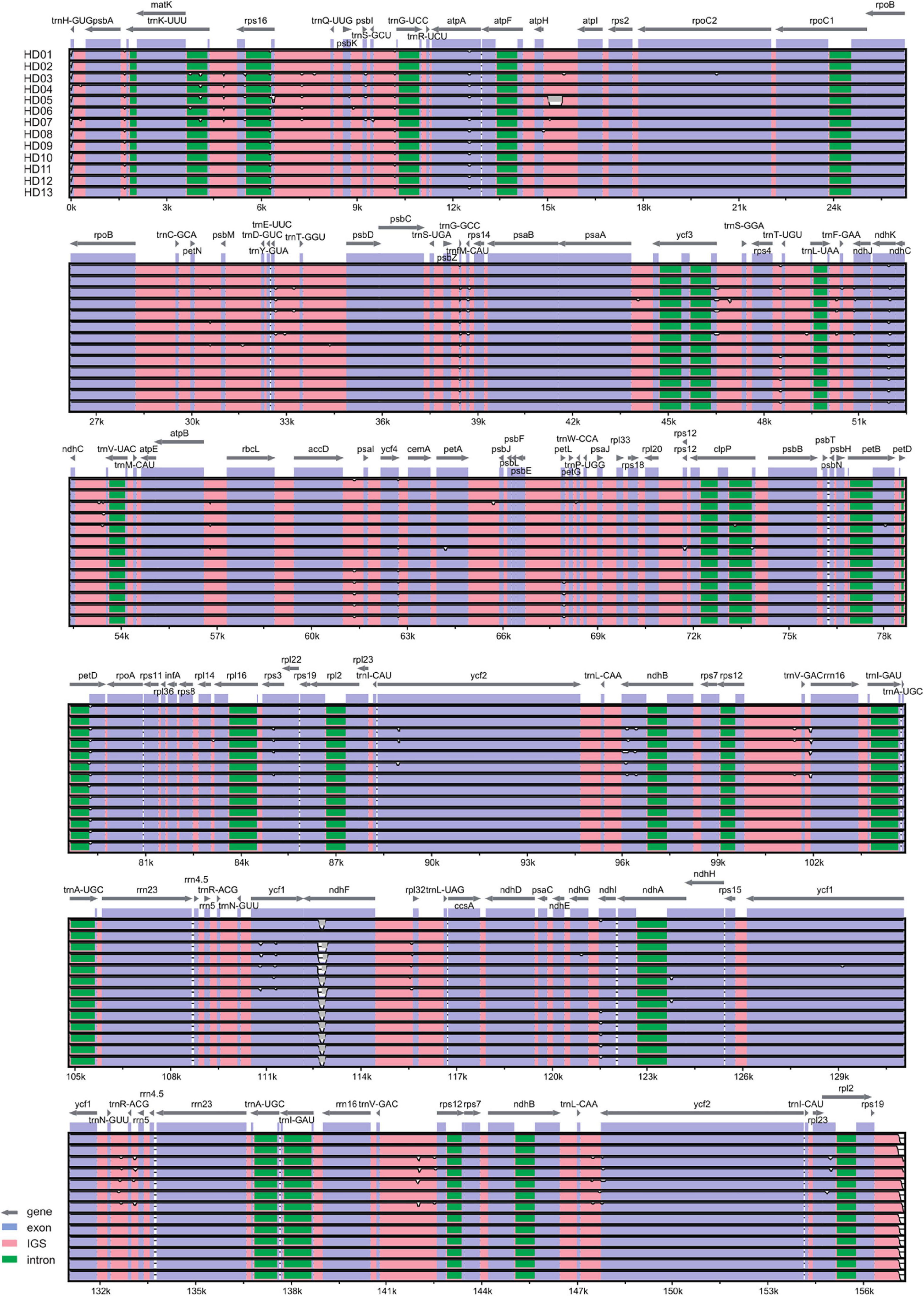
The cpDNA structures and sequence variations of 13 different oil-tea camellia samples were compared. The cpDNA of oil-tea camellia samples was highly conserved, and no inversion or translocation variations were observed. The full-length cpDNA sequences of all samples were 156512∼157089 bp, with a circular tetrad structure containing LSC, SSC, IRA, and IRB regions (Figure 1).
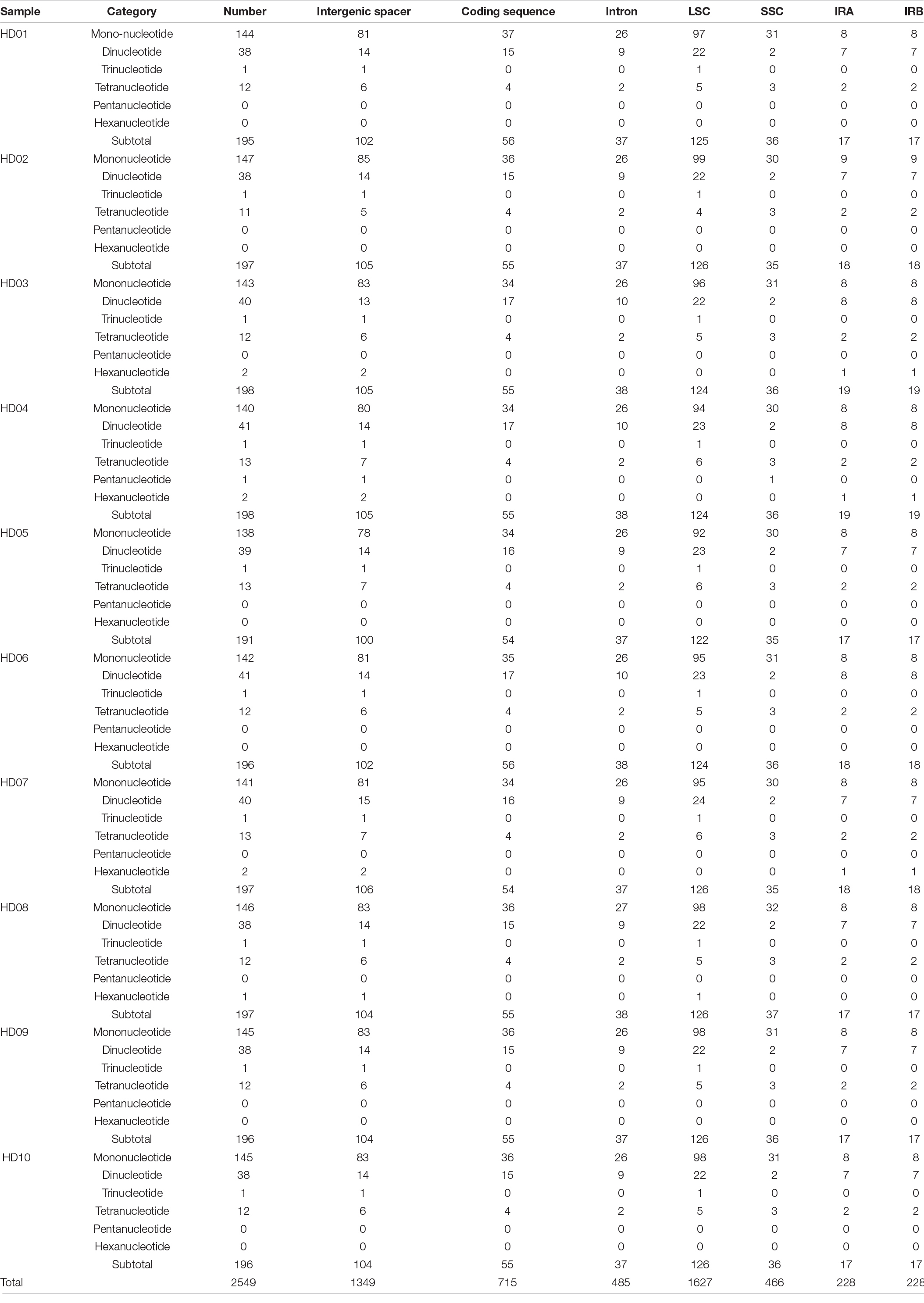
The structural information of each part of the cpDNA tetrad of the 13 samples is shown in Table 3. The total lengths of the cpDNA sequences ranged from 156512 bp in HD05 (C. meiocarpa) to 157089 bp in HD02 (C. gauchowensis from Luchuan County). The IR lengths ranged from 25943 bp in HD05 (C. meiocarpa) to 26165 bp in HD02 (C. gauchowensis from Luchuan County). The LSC lengths ranged from 86224 bp in HD05 (C. meiocarpa) to 86657 bp in HD04 (C. polyodonta) and HD09 (C. vietnamensis), and the SSC lengths ranged from 18132 bp in HD02 (C. gauchowensis from Luchuan County) to 18902 bp in HD05 (C. meiocarpa). The variation in SSC length was the main factor affecting the cpDNA length, while the length variations in the two IRs were less important. The GC content of the cpDNA of the 13 samples was 37.29% and very stable. These results indicated that the cpDNA of the different known species had significant polymorphisms in total length and tetrad region length. The cpDNA sequences of HD10∼HD12 (the undetermined species of oil-tea camellia from Hainan Province) and HD13 (C. gauchowensis from Xuwen County) were identical, indicating that HD10∼HD12 (the undetermined species of oil-tea camellia from Hainan Province) and HD13 (C. gauchowensis from Xuwen County) were closely related. The cpDNA of HD10, which represented the undetermined species (HD10-HD12), HD13 (C. gauchowensis from Xuwen County) and the other 9 samples was analyzed as reported below.
As shown in Figure 1 and Table 4, the cpDNA of all samples contained 136 genes, including 84 genes in the LSC region, 12 genes in the SSC region, and 20 genes each in the IRA and IRB regions. The gene sequences, gene contents, and length ratios of CDSs and non-CDSs of genes were consistent, reflecting good collinearity of the gene distribution. Thus, a circle diagram was used to annotate the cpDNA of the 13 samples (Figure 1). According to the statistics of translation products, 90 PCGs, 8 rRNA genes and 37 tRNA genes were detected.
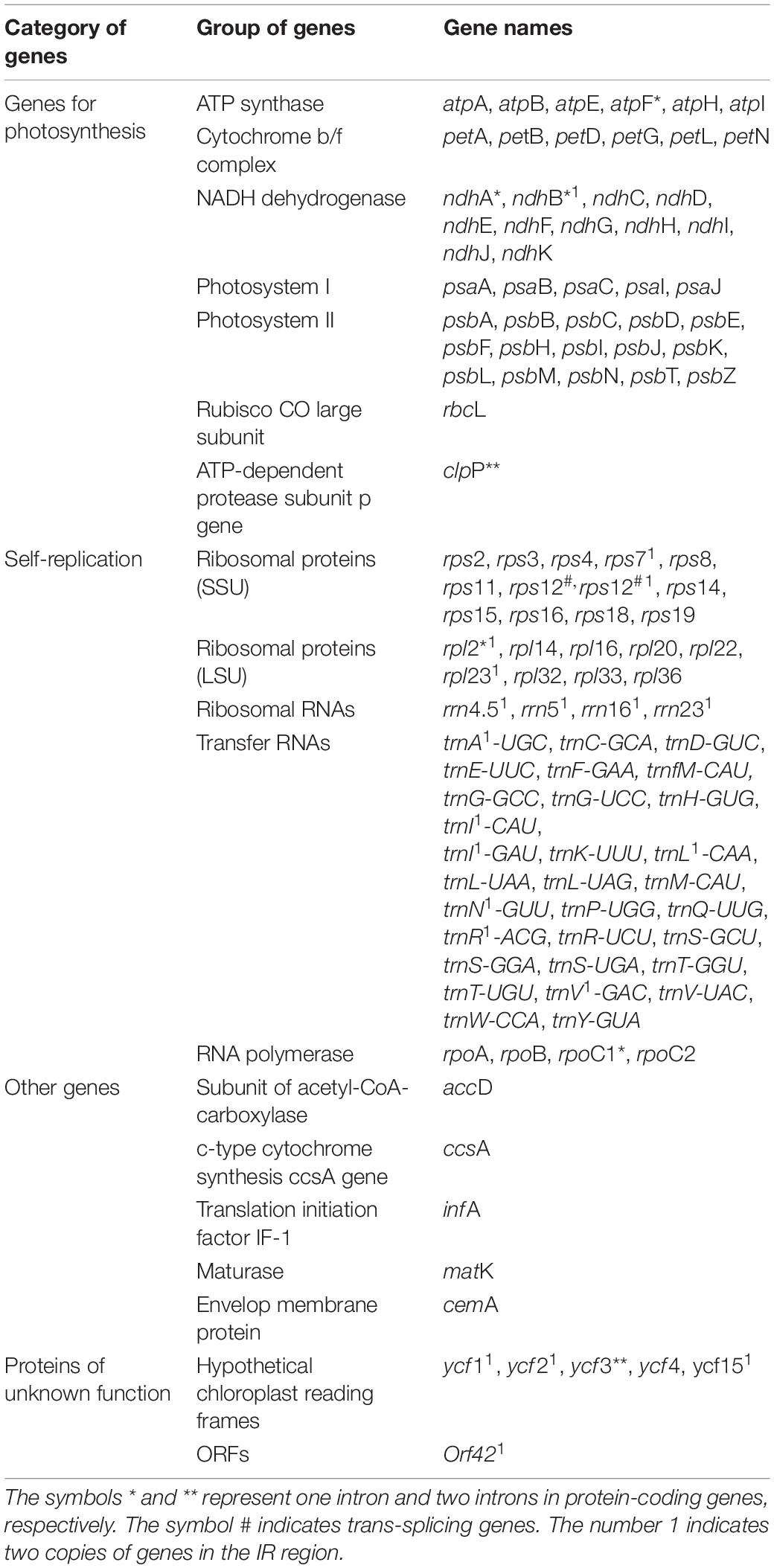
The list of genes classified according to function is shown in Table 4. All genes were divided into 4 categories and 19 groups, including genes for photosynthesis (7 groups), self-replication genes (5 groups), other genes (5 groups) and unknown-protein function genes (2 groups). Further analysis of Figure 1 and Tables 3, 4 shows that all genes in the cpDNA tetrad were not evenly distributed in each region. PCGs were distributed in all regions of the tetrad, including 60 genes in the LSC region, 18 genes in the IR regions and 12 genes in the SSC region. All 8 rRNA genes were distributed in the IR regions. Twenty-two tRNA genes were located in the LSC region, 14 were distributed in the IR regions, and 1 was located in the SSC region. There were 19 genes with 2 copies, all of which were in the IRA and IRB regions, including 4 PCGs (ndhB, rps7, rpl2, and rpl23), all 4 rRNA genes (rrn4.5, rrn5, rrn16, and rrn23), 7 tRNA genes (trnA-UGC, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG and trnV-GAC) and 4 genes of unknown function (ycf1, ycf2, ycf15, and orf42). The rps12 gene had 3 copies, one of which was located in the LSC region, while the others were located in the IRA and IRB regions, and this gene was the only trans-splicing gene. There were 5 genes with one intron, including the genes atpF, ndhA, ndhB, rpl2 and rpoC1, and 2 genes with 2 introns, including the genes ycf3 and clpP.
Inverted Repeat Contraction and Expansion
Inverted repeat expansion and contraction analysis of the cpDNA of the 13 samples was performed to investigate the gene variation in the boundaries of IRs, LSCs and SSCs, as shown in Figure 2. The cpDNA of the 13 samples was highly conserved at the boundaries on both sides of the LSC region, the rpl2 gene was 106 bp from the boundary of the IRA region, and the trnH-GUG gene was 2 bp from the boundary of the LSC region. The rps19 gene straddled the boundary of the LSC and IRB regions, and it contained 46 bp of the IRB. The ycf1 gene crossed the boundary between the SSC and IRA regions, and it extended 963 bp∼1209 bp into the SSC region. That is, in the 8 samples, including HD01 (C. gauchowensis from Gaozhou city), HD06 (C. semiserrrata), HD08 (C. osmantha), HD09 (C. vietnamensis), HD10 (the undetermined species of oil-tea camellia from Hainan Province) and HD13 (C. gauchowensis from Xuwen County), the gene was located at 1069 bp. In HD02 (C. gauchowensis from Luchuan County), the gene was located at 1209 bp. In HD03 (C. gigantocarpa), the gene was is located at 967 bp, and in HD04 (C. polyodonta), HD05 (C. meiocarpa) and HD07 (C. oleifera), it was located at 963 bp. The genes ycf1 and ndhF were located on both sides of the SSC and IRB boundary of HD04 (C. polyodonta), HD05 (C. meiocarpa) and HD07 (C. oleifera) and at 140 bp and 34 bp, respectively, on both sides of the SSC and IRB boundary in the population of HD02 (C. gauchowensis from Luchuan County). The genes were located at 106 bp and 57 bp on both sides of the SSC and IRB boundary in the 7 samples of HD06 (C. semiserrrata), HD08 (C. osmantha), HD09 (C. vietnamensis), HD10-HD12 (the undetermined species of oil-tea camellia from Hainan Province) and HD13 (C. gauchowensis from Xuwen County), respectively. The ycf1 gene extended into the IRB region by 2 bp and 26 bp at the SSC and IRB boundary in the samples of HD01 (C. gauchowensis from Gaozhou city) and HD03 (C. gigantocarpa), respectively. The ndhF gene was located on the SSC side of the boundary between the SSC and IRB regions in the populations of HD01 (C. gauchowensis from Gaozhou city) and HD03 (C. gigantocarpa). In conclusion, the IRs of different oil-tea camellia species contracted and expanded differently, resulting in variations in the relative lengths of tetrads and the full length of cpDNA.
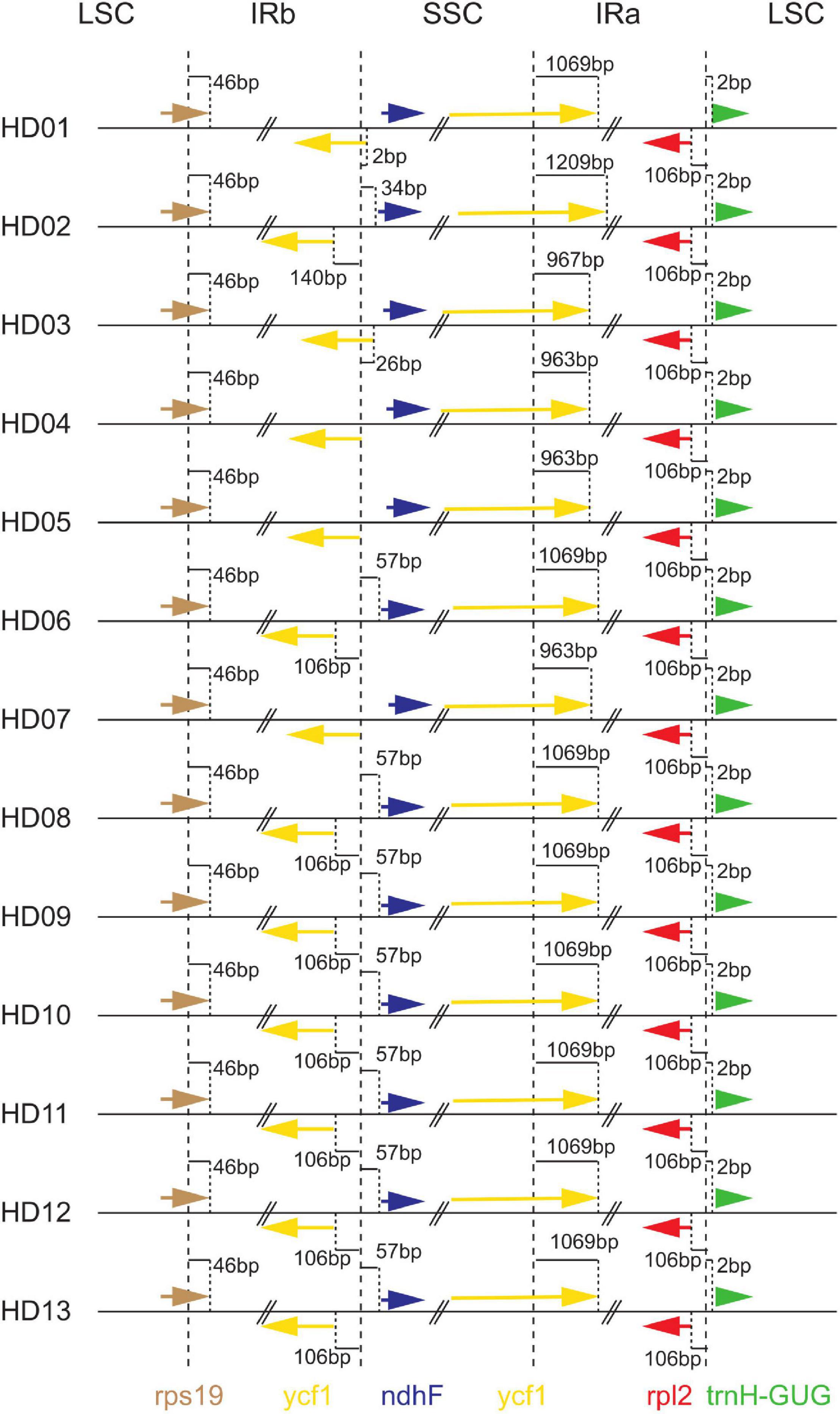
Figure 2. Comparison of the border positions of large single-copy (LSC), inverted repeat (IR) and small single-copy (SSC) among the thirteen chloroplast genome samples.
Synteny Analysis and Divergence Hotspots
The results of the phylogenetic divergence hotspot analysis of cpDNA from the 13 samples are shown in Figure 3. The depressed gaps in the figure are the divergence hotspots. Based on the pairwise genomic synteny of the cpDNA, the distribution characteristics of the divergence hotspots are shown in Table 5. There were 42 divergence hotspots in the intergenic space (IGS), accounting for 60% of the total. There were 23 divergence hotspots in exons (CDSs), accounting for 32.86% of the total. Only 5 divergence hotspots were located in introns, accounting for 7.14% of the total. There were 44 divergence hotspots in the LSC region, accounting for 62.86% of the total. Only 6 divergence hotspots were located in the SSC region, accounting for 8.57% of the total. The frequency of divergence hotspots in the IRs was intermediate but asymmetrical between the IRA and IRB regions, accounting for 12.86% and 15.71% of the total, respectively. The LSC and IGS regions were the main regions containing variations in the latent sequence. Among the 23 PCGs, the exons of trnH-GUC, atpA, ndhK, trnfM-CAU, petD, ndhB, ycf1, and ndhF were very important divergence hotspots. Among the 42 IGSs, rps19∼trnH-GUC, ycf2∼TrnL-CAA, rps12∼TrnV-GAC, rrn5∼trnR-ACG, TrnR-ACG∼trnN-GUU, rpl32∼ndhF, trnG-UCC∼trnS-GCU, trnQ-UUG∼rps16 and rps16∼trnKUUU were especially rich in divergence hotspots. The introns of the rps16 and trnK-UUU genes also contained important divergence hotspots. The IGS and a few exons were the main divergence hotspots, and the introns contained few divergence hotspots. As shown in Figure 3, the divergence hotspots of HD01 (C. gauchowensis from Gaozhou city), HD02 (C. gauchowensis from Luchuan County), HD09 (C. vietnamensis), HD10-HD12 (the undetermined species of oil-tea camellia from Hainan Province), and HD13 (C. gauchowensis from Xuwen County) were identical, indicating that the undetermined species of oil-tea camellia from Hainan Province was closely related to C. gauchowensis and C. vietnamensis.
Simple Sequence Repeats of Chloroplast Genome
The characteristics of SSR polymorphisms and distributions in different samples are shown in Table 6. The total numbers of SSRs in different samples ranged from 191 to 198, and the numbers of monobase to hexabase SSRs were 138 to 147, 38 to 41, 1, 12 to 13, 0 to 1, and 0 to 2, respectively; thus, polymorphisms were found in the number of SSRs and the sequence of repeat units. Tribase SSRs were located in the IGS of the LSC region. Tetrabase SSRs were mainly distributed in the IGS of the LSC region, and the numbers of SSRs differed by 1 at most, while the numbers of SSRs in the SSC, IRA and IRB regions were stable at 3, 2 and 2, respectively. There were only 2 dibase SSRs in the SSC regions of different samples, and a number of polymorphisms were found in the IGS and exons of other regions, which were mainly distributed in the IGS and exons of the LSC region, accounting for more than half of the total. There were a number of monobase SSR polymorphisms in all regions of the tetrad. The number of monobase SSRs was the highest in the LSC and IGS regions, and the number of single-base SSRs was the highest in exons. None of the samples except HD04 (C. polyodonta) contained pentabase SSRs. Hexabase SSRs were found in the IGS of the IRs of HD03 (C. gigantocarpa), HD04 (C. polyodonta) and HD07 (C. oleifera), and one was found in the IGS of the LSC region of HD08 (C. osmantha). The distributions of SSRs in the IGS, exons and introns of the tetrad of cpDNA were uneven, and the numbers of mono- and dibase SSR polymorphisms varied among species or samples. Tetrabase SSRs were represented by the most types of repeated-unit sequences, at up to 12 types, although this group of SSRs was not the largest. Among the 12 kinds of tetrabase SSRs, 11 were present in the same number in different samples. The loci of the LSC regions of HD04 (C. polyodonta) and HD08 (C. osmantha) were identified by a pentabase SSR and a hexabase SSR, which might be useful as identification markers for the respective species.
The SSRs of different samples were classified according to sequence differences in repeat units and could be divided into SSRs with the same repeat unit in all samples (Table 7) and SSRs with specific repeat units in different samples (Table 8). As shown in the two tables, the majority of SSR repeat units were mainly composed of A and T, and SSRs containing C or G were rarely observed, indicating that the SSRs of different samples had an obvious bias in the base types of repeat units. Comprehensive analysis of the two tables shows that few SSRs of specific repeat units were found in different oil-tea camellia species, and most of those identified were the same. As shown in Table 8, the only tribase SSR was TTC, and tetrabase SSRs included AAAT, AATA, AGAT, ATAG, CCCT, GAGG, GACT, TCTA, TCTT and TTTC. These tribase and tetrabase SSRs in different samples not only contained the same repeat units but were also present in the same number. The numbers of monobases A, C, and T and dibases AT, CT, GA, and TC were inconsistent among samples, indicating polymorphisms of the same mono- and dibase SSRs.
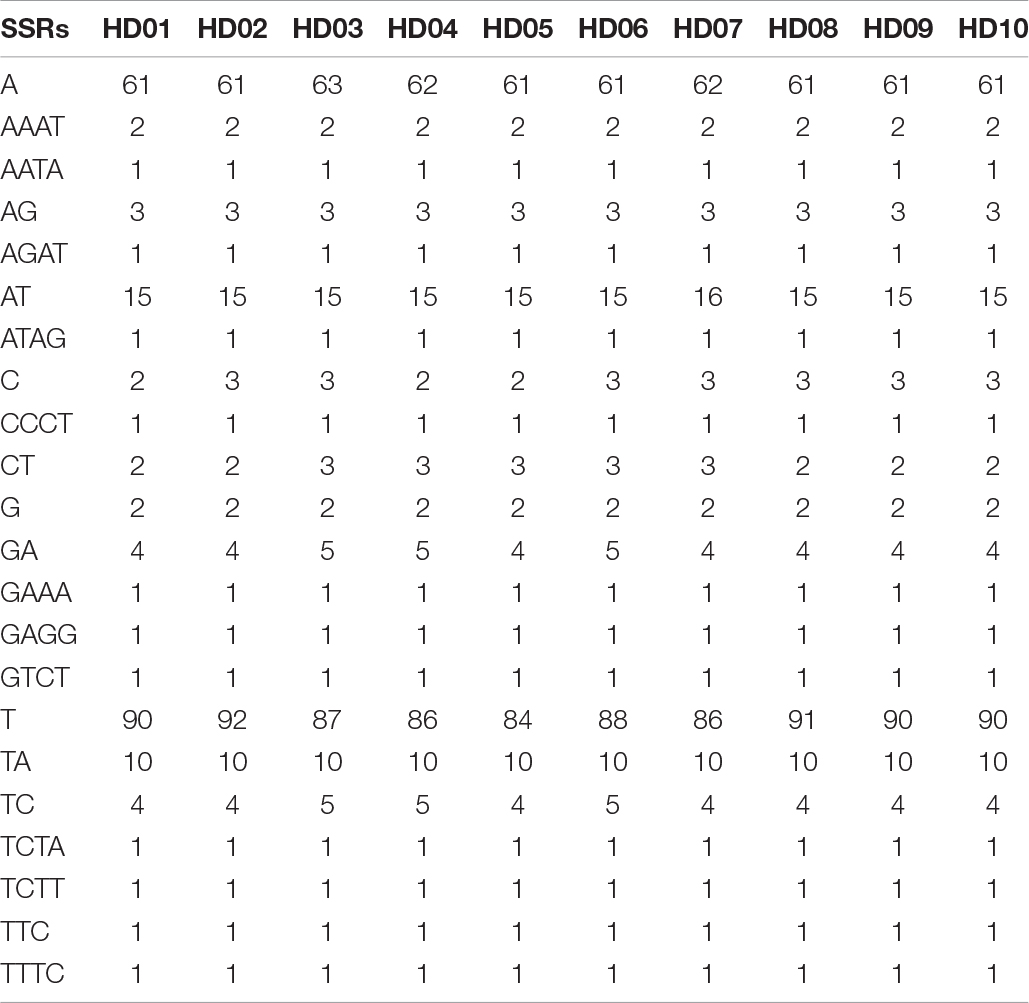
Table 7. Common Simple sequence repeats (SSRs) in the chloroplast genome (cpDNA) of different samples.
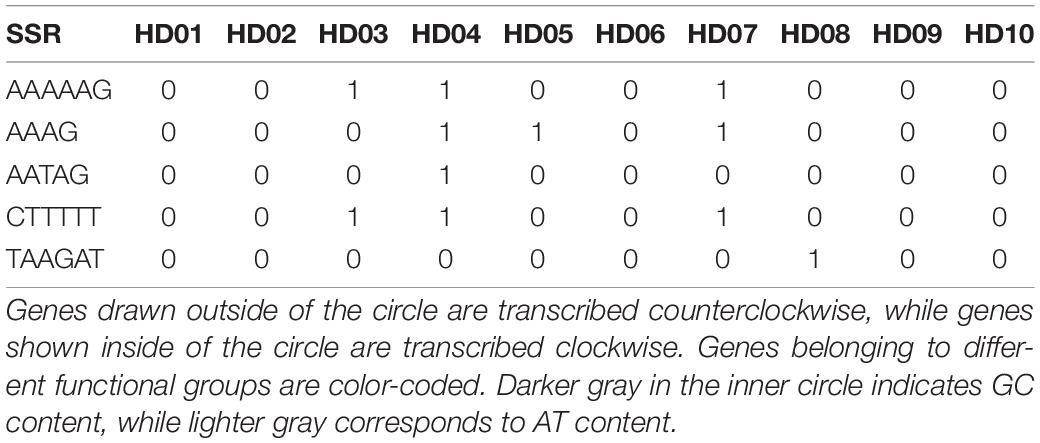
Table 8. Unique Simple sequence repeats (SSRs) in the chloroplast genome (cpDNA) of different samples.
As shown in Tables 6, 8, HD04 (C. polyodonta) uniquely contained one pentabase SSR, AATAG, in the IGS of the SSC region, and HD08 (C. osmantha) uniquely contained one TAAGAT hexabase SSR in the IGS of the LSC region. One AAAAAG SSR and one CTTTTT SSR were found in the IRs of HD03 (C. gigantocarpa), HD04 (C. polyodonta) and HD07 (C. oleifera), but the other samples did not contain these hexabase SSRs. HD04 (C. polyodonta), HD05 (C. meiocarpa) and HD07 (C. oleifera) all contained the tetrabase SSR AAAG in the IGS of the LSC region, but the other samples did not. The SSRs of HD09 (C. vietnamensis), HD01 (C. gauchowensis from Gaozhou city), HD02 (C. gauchowensis from Luchuan County), HD13 (C. gauchowensis from Xuwen County) and HD10∼HD12 (the undetermined species of oil-tea camellia from Hainan Province) contained the same repeat units and differed from those of other oil-tea camellia species. Therefore, these SSR combinations may be used for species identification, and HD04 (C. polyodonta) and HD08 (C. osmantha) also have unique SSR markers. The undetermined species of oil-tea camellia from Hainan Province may be closely related to C. vietnamensis and C. gauchowensis.
In addition, according to the sequencing results, the SSRs of different samples also contained 27-30 interval SSRs, in which the TTC tribase SSR was distributed. All types, intermediate sequence lengths, compositions of base pairs and lengths of SSRs in the samples showed polymorphism. The maximum interval SSR was 484 bp in length, containing three copies each of T(9) and T(8) and one copy each of T(12) and A(12). HD04 (C. polyodonta) had a specific SSR complex, namely, A(10)(AAAG)3*, wherein (AAAG)3* was AAAGAAAGA. Therefore, the intermediate sequence and the SSR complex can be used to infer genetic diversity, and A(10)(AAAG)3* may be a unique marker of HD04 (C. polyodonta).
Phylogenetic Inference
Hartia laotica was taken as the outgroup, and Bayesian inference (BI) and ML phylogenetic analyses of the whole cp-DNA of HD01-HD10 and 7 other Camellia species were performed. The results are shown in Figures 4, 5.
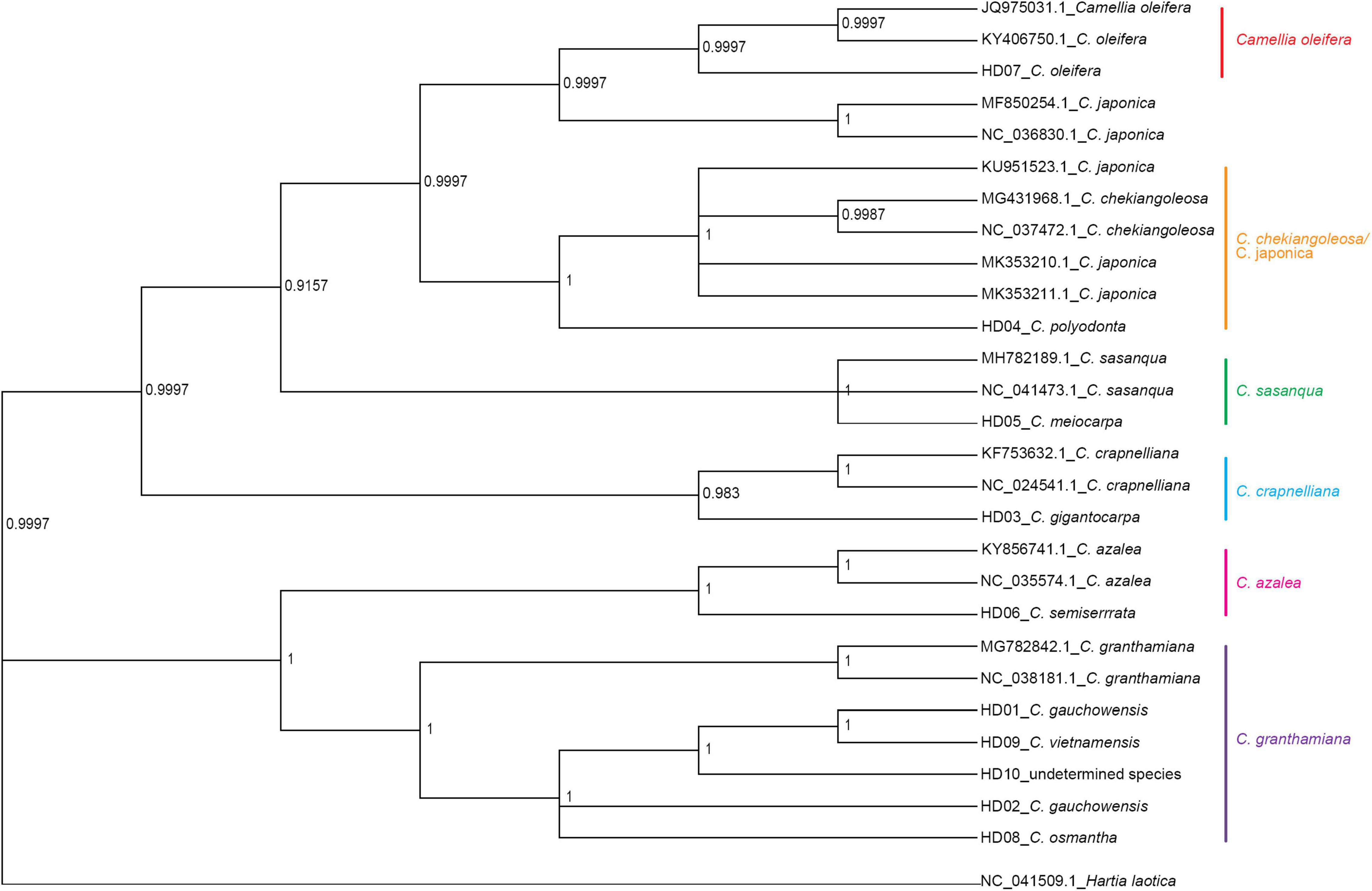
Figure 4. Phylogenetic tree of 13 samples based on chloroplast genome (cpDNA) constructed using the Bayes inference method (BI).
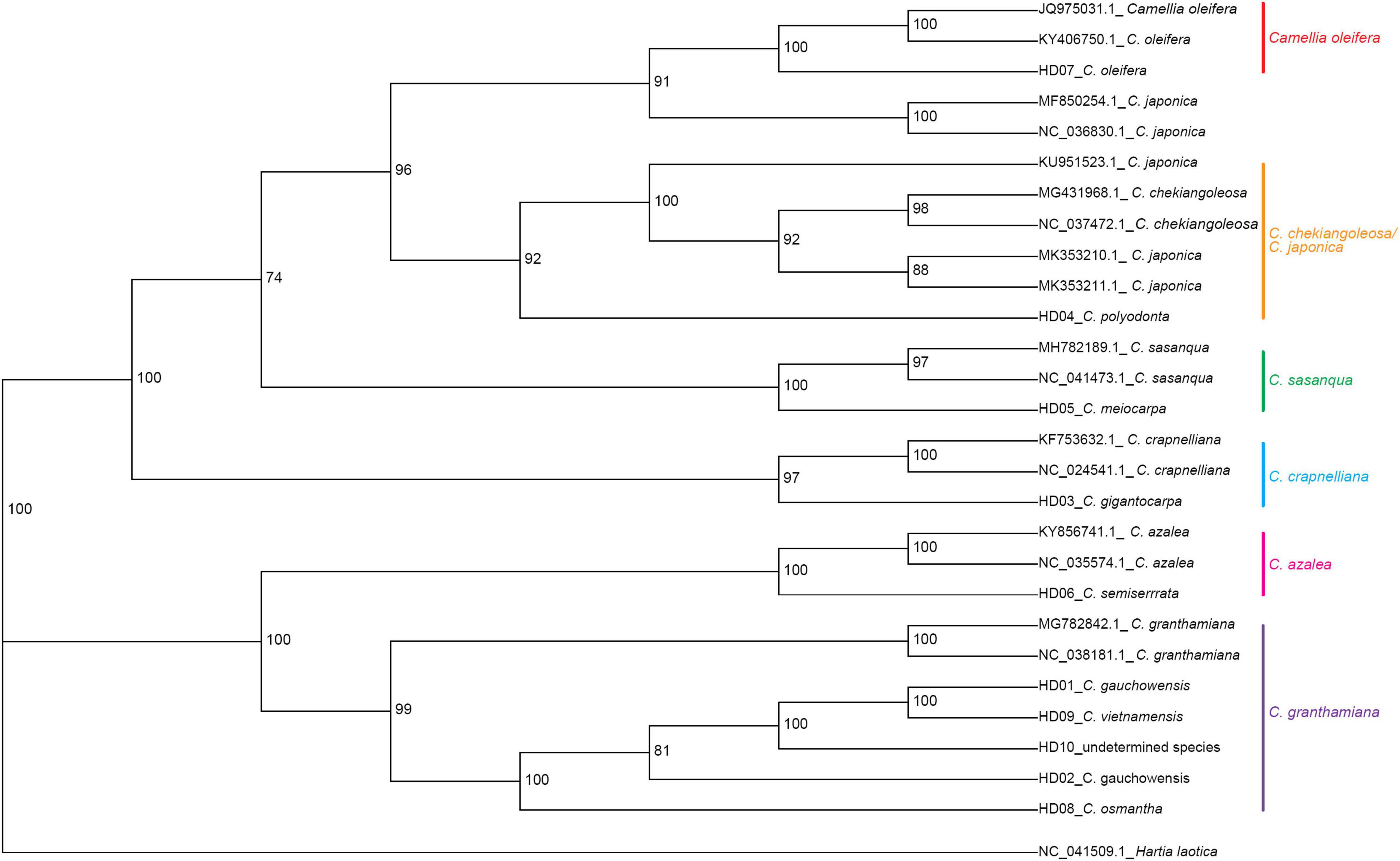
Figure 5. Phylogenetic tree of 13 samples based on chloroplast genome (cpDNA) constructed using the maximum likelihood method (ML).
The samples belonged to the same two clades. HD03 (C. gigantocarpa), HD04 (C. polyodonta), HD05 (C. meiocarpa) and HD07 (C. oleifera) belonged to one major clade. Four samples belonging to four different subclades, namely, HD07 (C. oleifera), were identified as C. oleifera. HD03 (C. gigantocarpa) was associated with C. crapaelliana, HD04 (C. polyodonta) was associated with C. chekiangoleosa and C. japonica, and HD05 (C. meiocarpa) and C. sasanque belonged to different subclades. The rest of the samples belonged to another major clade, and HD06 (C. semiserrrata) and C. aztea were clustered into one subclade. The samples of HD01 (C. gauchowensis from Gaozhou city), HD02 (C. gauchowensis from Luchuan County), HD09 (C. vietnamensis), HD10 (the undetermined species of oil-tea camellia from Hainan Province), HD08 (C. osmantha) and C. granthamiana belonged to another subclade. HD01 (C. gauchowensis from Gaozhou city), HD02 (C. gauchowensis from Luchuan County), HD10 (the undetermined species of oil-tea camellia from Hainan Province), HD09 (C. vietnamensis) and HD08 (C. osmantha) were clustered on the same branch of this subclade. In other words, two samples (HD11∼HD12) of oil-tea camellia from Hainan Province and HD13 (C. gauchowensis from Xuwen County) were also located on this branch. Therefore, the 13 samples were clustered into 6 subclades, namely, the first major clade of 4 subclades, including the C. oleifera subclade, C. chekiangoleosa and C. japonica subclade, C. sasanque subclade, and C. crapaelliana subclade. The second major clade of the two subclades included the C. aztea subclade and the C. granthamiana subclade.
Although the topological maps constructed by the two methods were different in terms of the end nodes of the subclades, such as C. chekiangoleosa and C. japonica, C. sasanque and C. granthamiana, and the genetic distances among the different samples within each subclade were inconsistent, the results of clustering analysis of the 13 samples, including the major clades and subclades, were not affected.
In conclusion, HD01 (C. gauchowensis from Gaozhou city), HD02 (C. gauchowensis from Luchuan County), HD09 (C. vietnamensis), HD10-HD12 (the undetermined species of oil-tea camellia from Hainan Province) and HD13 (C. gauchowensis from Xuwen County) may be different ecotypes of C. vietnamensis or C. gauchowensis. HD08 (C. osmantha) was closely related to C. vietnamensis and C. gauchowensis. Therefore, the phylogenetic relationships of different species and populations of Camellia could be well identified based on whole cpDNA sequences.
The branch nodes of HD09 (C. vietnamensis) and HD01 (C. gaochowensis from Gaozhou city) were the outermost nodes, and the branch node of HD10 (the undetermined species of oil-tea camellia from Hainan Province) was one level inward. The phylogenetic relationship between HD10-HD12 (the undetermined species of oil-tea camellia from Hainan Province) and HD01 (C. gaochowensis from Gaozhou city) or HD09 (C. vietnamensis) was closer than that between HD02 (C. gauchowensis from Luchuan County) and HD01 (C. gaochowensis from Gaozhou city) or HD09 (C. vietnamensis). These results indicate that the three samples of undetermined species of oil-tea camellia from Hainan Province and C. gaochowensis from Xuwen County were closely related to C. gaochowensis from Gaozhou city and C. vietnamensis. The results also illustrate that the undetermined species of oil-tea camellia from Hainan Province may be C. vietnamensis, and C. gaochowensisu may be merged with C. vietnamensis.
Discussion
Chloroplast Genome Variation and Evolution
The total length of plant cpDNA is generally between 107 and 218 kb and consists of LSC and SSC regions and two IRs in a tetrad structure (Henry et al., 2016). Changes in the total length are mainly caused by the boundary contraction and expansion of IRs (Wang et al., 2008). The cpDNA of the 13 samples also showed a tetrad structure, and the maximum differences in total, IR, LSC and SSC lengths were 577 kb, 222 kb, 433 kb, and 770 kb, respectively. The results also showed that the IRs of different samples exhibited different boundary contractions and expansion phenomena on the SSC sides, which resulted in great differences in the SSC and IR sequences among different samples. Therefore, the results in this paper are basically consistent with previous reports of plants of Camellia with cpDNA lengths below 600 kb, and the different cpDNA samples had a consistent type, quantity, and order (Wang et al., 2008), showing that the Camellia cpDNA structure is highly conserved and confirming the conservatism of the plant cpDNA structure. The results in this paper also confirmed that the boundary contraction and expansion of IRs are the main reasons for differences in the size of cpDNA and indicated that IRs play an important role in stabilizing the structure of cpDNA (Chen et al., 2014). The results in this paper showed a large number of different SSR variants in the cpDNA of different samples, which also led to changes in cpDNA size. Therefore, SSR variants in Camellia may be another important reason for small variations in cpDNA size.
Application Prospect of Simple Sequence Repeats in Chloroplast Genome
Simple Sequence Repeats have the advantages of high polymorphism, codominance, and a wide distribution, and SSRs of cpDNA share these advantages as well as the characteristics of conserved sequences, a simple structure, and uniparental inheritance (Palmer and Thompson, 1982). Therefore, SSRs have been widely applied in studies on species evolution (Kaundun and Matsumoto, 2011) and genetic diversity analyses (Flannery et al., 2006; Allender et al., 2007). They are often developed as “barcodes” or “identity cards” for plant species identification. The results in this paper show that the SRs of oil-tea camellia are SSRs with a preference for A/T, and they have polymorphisms in composition, number and length, which is consistent with previous sequencing results in tea plants (Wang et al., 2008). The results also show that C. ployodonta contains a unique SSR (AATAG) in the IGS of the SSC region, and C. gigantocapa, C. polyodonta and C. oleifera each contain 3 SSRs (AAAAAG and CTTTTT, respectively) in the IGS of IRs. The existence of pentabase SSRs was not previously reported in cpDNA sequences of Camellia plants (Wang et al., 2008), making this the first discovery of such SSRs in Camellia plants. These specific SSRs may have important application significance in the species identification of C. gigantocapa, C. polyodonta, and C. oleifera. In this paper, 13 samples of 7 oil-tea camellia species were examined. The abundant cpSSR variation could be divided into two categories: SSRs shared among all samples and SSRs with inconsistencies in different samples. Pairs of species always showed differences in SSRs. The former category may reflect the specificity of kinship above the genus level, while the latter may reflect differences in species within Camellia. In short, if various codes are established for SSRs with differences in grading units, each oil-tea camellia species will differ from others in its code combination. The similarity of different codes also directly indicates the kinship and phylogenetic relatedness among species. Consistent with previous predictions for oil-tea camellia (Leigh, 2013), with the accumulation of cpSSR sequence information from different oil-tea camellia species, it is entirely possible to successfully establish “barcodes” or “identity cards” for individual oil-tea camellia species.
Genetic Relationships Between Different Species of Camellia and Identification of the Undetermined Species of Oil-Tea Camellia From Hainan Province
The phylogenetic trees of 7 ornamental species of Camellia were constructed by ML and BI methods based on cpDNA, LSC, SSC, CDS, intron, IGS and IR sequences. The topological structures of different sequence data were highly similar. The analysis method had no effects on the topological maps; specifically, the same topological maps were obtained based on the cpDNA, IGS, SSC and IR sequences (Leigh et al., 2013). With the development of high-throughput sequencing technology, it is becoming increasingly common to construct phylogenetic trees based on full cpDNA sequences (Nie et al., 2012; Chen et al., 2014; Huang et al., 2014; Song et al., 2017; Yang et al., 2019). The results in this paper showed that divergence hotspots and SSR variants were distributed mostly in the IGS (one kind of non-CDS), somewhat distributed in CDSs, and less common in introns (another kind of non-CDS). Therefore, in this paper, based on full chloroplast genome sequences, phylogenetic trees were constructed to explore the evolutionary relationships between different oil-tea camellia species. C. oleifera, C. meiocarpa, C. vietnamensis, C. gigantocarpa, C semiserrrata, and C. polyodonta were located on different branches, and a consensus regarding these species divisions in terms of taxonomy of oil-tea camellia has gradually been reached (Chen, 2008; State Forestry Administration state-owned forest farm and tree seed and seedling work station, 2016). Furthermore, it is feasible to identify different species of Camellia plants based on the clustering analysis results in this paper.
HD10∼HD12 (the undetermined species of oil-tea camellia from Hainan Province) and HD13 (C. gauchowensis from Xuwen County) were clustered on the same branch as HD09 (C. vietnamensis), and their genetic relationship with HD09 (C. vietnamensis) was closer than that with HD02 (C. gauchowensis from Luchuan County), which indicated that the undetermined species of oil-tea camellia from Hainan Province may be C. vietnamensis. Based on these results combined, C. vietnamensis, C. gauchowensis and the undetermined species of oil-tea camellia from Hainan Province had the same divergence hotspots, while the sample of C. gauchowensis from Xuwen County (located at the southernmost tip of Leizhou Peninsula and on the northern Qiongzhou Strait) had the same cpDNA sequence as the undetermined species of oil-tea camellia from Hainan Province, indicating that C. gaochowensis and C. vietnamensis may be merged into the same species.
Although previous studies have attempted to prove that C. osmantha is a new oil-tea camellia species based on agronomic and economic traits, inter SSR (ISSR) molecular markers of nuclear DNA, and other evidence (Wang et al., 2014; Liang et al., 2017), the results in this paper show that HD08 (C. osmantha) is closely related to C. gauchowensis and C. vietnamensis. Therefore, additional genetic evidence needs to be collected to resolve the controversial issue of whether C. osmantha is a new species of Camellia.
Phylogenetic trees were constructed in this paper, and 3 species of Section Heterogenea Sealy (C. gigantocarpa, C. granthamiana, and C. crapnelliana), 5 species of Section Camellia (C. semiserrrata, C. polyodonta, C. japonica, C. azalea, and C. chekiangoleosa) and 5 species of Section Oleifera H.T. Chang (C. oleifera, C. meiocarpa, C. vietnamensis, C. gauchowensis, C. sasanqua, and C. osmantha, which is a new species undergoing confirmation but not included in this citation) were used (Zhang and Min, 1999). Based on the topological structures of the phylogenetic trees obtained with different sequence regions and different methods, the species of Section Heterogenea Sealy, Section Camellia and Section Oleifera H.T. Chang belong to two clades. In addition, one subclade contained the species from the different sections of Camellia in each of the two clades, and one subclade contained C. granthamiana (Section Heterogenea Sealy) as well as C. vietnamensis and C. gauchowensis (Section Oleifera H.T. Chang). Another subclade included C. oleifera (Section Oleifera H.T. Chang) and C. japonica (Section Camellia), and the other 4 subclades contained the species from the same section. The results show that the previous classification of Camellia plant sections may not be accurate enough. Section division is based on the morphological characteristics of different species, which are affected by environmental conditions, tree ages and other factors, producing a risk of unreasonable division (Luo et al., 1999). In this paper, cpDNA sequence differences were used to distinguish and cluster species, as such differences are not affected by the environment and may be more suitable for classification (Zhang and Min, 1999; Leigh et al., 2013). It is possible that the results in this paper can provide important information on the classification of sections of Camellia and how to adjust them.
Conclusion
Based on whole-genome high-throughput sequencing, the full cpDNA of seven Camellia species was successfully assembled. The tetrad structure was conserved with good collinearity, but certain polymorphisms in length were found due to the contraction and expansion of IRs, SSRs and other variations. The largest numbers of divergence hotspots and SSR variants were observed in the IGS and CDSs, respectively. Different species showed specific SSRs, and these SSRs can be developed as “barcodes” or “ID cards” for species identification. Therefore, full cpDNA sequences can be used for the identification and phylogenetic analysis of Camellia plants. The phylogenetic trees based on full cpDNA sequences showed that HD10∼HD12 (the undetermined species of oil-tea camellia from Hainan Province) probably belonged to Camellia vietnamensis, while Camellia vietnamensis and Camellia gauchowensis may be merged into the same species. Camellia osmantha is closely related to Camellia gauchowensis and Camellia vietnamensis, and additional genetic evidence is needed to determine whether it is an independent new species. The current section-level division of Camellia plants based on morphology may need to be adjusted based on cpDNA sequence differences.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm. nih.gov/genbank/, MN078084; https://www.ncbi.nlm.nih.gov/genbank/, MN078085; https://www.ncbi.nlm.nih.gov/genbank/, MN078086; https://www.ncbi.nlm.nih.gov/genbank/, MN078087; https://www.ncbi.nlm.nih.gov/genbank/, MN078088; https://www.ncbi.nlm.nih.gov/genbank/, MN078089; https://www.ncbi. nlm.nih.gov/genbank/, MN078090; https://www.ncbi.nlm. nih.gov/genbank/, MN078091; https://www.ncbi.nlm.nih.gov/genbank/, MN078092; https://www.ncbi.nlm.nih.gov/gen bank/, MN078093.
Author Contributions
JC was the lead author, and she completed the main research work of this manuscript. YG was the second author, and she assisted the lead author in the experiment. KZ was the assistant supervisor of JC, a Ph.D. student, and he is responsible for specific guidance work. XH was the supervisor of JC, a Ph.D. student. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
Allender, C. J., Allainguillaume, J., Lynn, J., and King, G. J. (2007). Simple sequence repeats reveal uneven distribution of genetic diversity in chloroplast genomes of Brassica oleracea L. and (n=9) wild relatives. Theor. Appl. Genet. 114, 609–618. doi: 10.1007/s00122-006-0461-5
Chen, C. M., Ma, C. L., Ma, J. Q., Liu, S. C., and Chen, L. (2014). Sequencing of Chloroplast Genome of Camellia sinensis and Genetic Relationship for Camellia Plants Based on Chloroplast DNA Sequences. J. Tea Sci. 34, 371–380.
Chen, L. Q., Yang, W. B., Li, Y., Wang, Y., Fu, D. Q., Niu, X. Q., et al. (2012). The development course and prospect of camellia oil industry in Hainan Island. Mod. Agricult. Sci. Technol. 1:374.
Chen, Y. Z. (2008). Excellent germplasm resources of oil-tea camellia. Beijing: China Forestry Press, 159.
Chen, Y. Z., Chen, L. S., Li, R. F., and Ma, L. (2017). Survey and the industry development proposals of Camellia sp. Resources in Haina n. Trop. Forest. 45, 49–52.
Dai, J., and Zhong, S. J. (2017). Development Status of Camellia Oil Industry in Hainan and Suggestions. Trop. Agricult. Eng. 41, 61–64.
Flannery, M. L., Mitchel, F. J. G., Coyne, S., Kavanagh, T. A., Burke, J. I., Salamin, N., et al. (2006). Plastid genome characterisation in Brassica and Brassicaceae using a new set of nine SSRs [J]. Theor. Appl. Genet. 113, 1221–1231. doi: 10.1007/s00122-006-0377-0
Hebert, P. D. N., Cywinska, A., Ball, S. L., and Dewaard, J. R. (2003). Biological identifications through DNA barcodes[J]. Proc. R. Soc. B-Biol. Sci. 270, 313–321. doi: 10.1098/rspb.2002.2218
Henry, D., Lin, C. S., Yu, M., and Chang, W. J. (2016). Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genom. Biol. 17, 134–163. doi: 10.1186/s13059-016-1004-2
Huang, H., Shi, C., Liu, Y., Mao, S. Y., and Gao, L. Z. (2014). Thirteen Camellia chloroplast genome sequences determined by high-throughput sequencing:genome structure and phylogenetic relationships. BMC Evol. Biol. 14:151. doi: 10.1186/1471-2148-14-151
Kaundun, S. S., and Matsumoto, S. (2011). Molecular evidence for maternal inheritance of the chloroplast genome in tea,Camellia sinensis (L.) O. Kuntze. J. Sci. Food Agricult. 91, 2660–2663. doi: 10.1002/jsfa.4508
Leigh, F. J., Mackay, I., Oliveira, H. R., Nicholas, E., Richard, G. A., Huw, H. J., et al. (2013). Using diversity of the chloroplast genome to examine evolutionary history of wheat species. Genet. Resour. Crop Evol. 60, 1831–1842. doi: 10.1007/s10722-013-9957-4
Li, X., Yang, Y., Henry, R. J., Rossetto, M., Wang, Y. T., and Chen, S. (2015). Plant DNA barcoding:from genes to genome. Biol. Rev. 90, 157–166. doi: 10.1111/brv.12104
Liang, G. X., Liu, K., Ma, J. L., Chen, G. C., Ye, H., and Jiang, Z. P. (2017). Molecular taxonomy and identifi cation of Camellia osmantha. Nonwood Forest Res. 35, 26–29.
Liu, Y., Yang, S. X., Ji, P. Z., and Gao, L. Z. (2012). Phylogeography of Camellia taliensis (Theaceae) inferred from chloroplast and nuclear DNA: insights into evolutionary history and conservation[J]. BMC Evol. Biol. 12:92. doi: 10.1186/1471-2148-12-92
Luo, C. Q., Tan, X. F., and Qi, L. L. (1999). A classification summary on plant of Genus Camellia. J. Cent. South Forest. Coll. 19, 78–81.
Nie, X. J., Lu, S. Z., Zhang, Y. X., Du, X. H., Wang, L., Biradar, S. S., et al. (2012). Complete chloroplast genome sequence of a major invasive species, Crofton Weed (Ageratina adenophora). PLoS One 7:e36869. doi: 10.1371/journal.pone.0036869
Palmer, J. D., and Thompson, W. F. (1982). Chloroplast DNA rearrangements are more frequent when a large inverted repeat sequence is lost. Cell 29, 537–550. doi: 10.1016/0092-8674(82)90170-2
Semerikova, S. A., and Semerikov, V. L. (2014). Molecular phylogenetic analysis of the genus Abies (Pinaceae) based on the nucleotide sequence of chloroplast DNA. Russ J. Genet. 50, 7–19. doi: 10.1134/S1022795414010104
Song, J., Long, Y. H., Lin, L. M., Yin, F., and Xing, C. B. (2017). Analysis on structure and phylogeny of chloroplast genomes in Araliaceae species. Chin. Trad. Herb. Drugs 48, 5070–5075.
State Forestry Administration state-owned forest farm and tree seed and seedling work station (2016). Chinese oil tea cultivars. Beijing: China Forestry Press, 11–15.
Wang, D. X., Ye, H., Ma, J. L., and Zhou, Z. D. (2014). Evaluation and selection of Camellia osmantha germplasm resources. Nonwood Forest Res. 32, 159–162.
Wang, R. J., Cheng, C. L., Chang, C. C., Wu, C. L., Su, T. M., and Chaw, S. M. (2008). Dynamics and evolution of the inverted repeat-large single copy junctions in the chloroplast genomes of monocots[J]. BMC Evol. Biol. 8, 36–51. doi: 10.1186/1471-2148-8-36
Yang, J. B., Yang, S. X., Li, H. T., Yang, J., and Li, D. Z. (2013). Comparative Chloroplast Genomes of Camellia Species[J]. PLoS One 8, 1–12. doi: 10.1371/journal.pone.0073053
Yang, Z., Wang, G. X., Ma, Q. H., Ma, W. X., Liang, L. S., and Zhao, T. T. (2019). The complete chloroplast genomes of three Betulaceae species: implications for molecular phylogeny and historical biogeography. PeerJ. 7, 1–25. doi: 10.7717/peerj.6320
Ye, Z. C., Wu, Y. G., Dai, J., Zhou, K. B., and Hu, X. W. (2015). Distribution of Duckweed Community and the Environmental Factors that Affect the Community Structure. J. Trop. Biol. 2015, 310–314.
Yuan, J., Han, Z. Q., He, S. Y., Huang, L. Y., and Zhou, N. F. (2014). Investigation and Cluster Analysis of Main Morphological and Economical Characters for Oiltea Resource in Hainan Province. J. Plant Genet. Resour. 15, 1380–1384.
Zhang, J., Chen, M., Dong, X., Lin, R. Z., Fan, J. H., and Chen, Z. D. (2015). Evaluation of fpur commonly used DNA barcoding loci for Chinese medicinal plants of the family Schisandraceae[J]. PLoS One 10:e0125574. doi: 10.1371/journal.pone.0125574
Zhang, W. J., and Min, T. L. (1999). A Cytogeological Study of Genus Camellia[J]. Acta Bot. Yunnan. 21, 184–196.
Zheng, D. J., Pan, X. Z., Zhang, D. M., Xie, L. S., Zeng, J. H., Zhang, Z. L., et al. (2016). Survey and analysis on Oli-tea camellia resource in Hainan. J. Northw. Forest. Univ. 31, 130–135.
Zheng, W. W., Chen, J. H., Hao, Z. D., and Shi, J. S. (2016). Comparative Analysis of the Chloroplast Genomic Information of Cunninghamia lanceolata (Lamb.) Hook with Sibling Species from the Genera Cryptomeria D. Don, Taiwania Hayata, and Calocedrus Kurz. Internat. J. Mole. Sci. 17, 1–16. doi: 10.3390/ijms17071084
Keywords: oil-tea camellia, cpDNA, divergence hotspots, SSRs, phylogenetic tree
Citation: Chen J, Guo Y, Hu X and Zhou K (2022) Comparison of the Chloroplast Genome Sequences of 13 Oil-Tea Camellia Samples and Identification of an Undetermined Oil-Tea Camellia Species From Hainan Province. Front. Plant Sci. 12:798581. doi: 10.3389/fpls.2021.798581
Received: 20 October 2021; Accepted: 17 December 2021;
Published: 07 February 2022.
Edited by:
Jacqueline Batley, University of Western Australia, AustraliaCopyright © 2022 Chen, Guo, Hu and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xinwen Hu, MTI2MzcxNDk5NUBxcS5jb20=; Kaibing Zhou, emtiQGhhaW5hbnUuZWR1LmNu
 Jing Chen
Jing Chen Yujian Guo
Yujian Guo Xinwen Hu
Xinwen Hu Kaibing Zhou
Kaibing Zhou