- 1Department of Horticulture, Washington State University, Pullman, WA, United States
- 2Department of Plant Pathology, Washington State University, Pullman, WA, United States
Calcium (Ca2+) signaling in plant cells is an essential and early event during plant-microbe interactions. The recognition of microbe-derived molecules activates Ca2+ channels or Ca2+ pumps that trigger a transient increase in Ca2+ in the cytoplasm. The Ca2+ binding proteins (such as CBL, CPK, CaM, and CML), known as Ca2+ sensors, relay the Ca2+ signal into down-stream signaling events, e.g., activating transcription factors in the nucleus. For example, CaM and CML decode the Ca2+ signals to the CaM/CML-binding protein, especially CaM-binding transcription factors (AtSRs/CAMTAs), to induce the expressions of immune-related genes. In this review, we discuss the recent breakthroughs in down-stream Ca2+ signaling as a dynamic process, subjected to continuous variation and gradual change. AtSR1/CAMTA3 is a CaM-mediated transcription factor that represses plant immunity in non-stressful environments. Stress-triggered Ca2+ spikes impact the Ca2+-CaM-AtSR1 complex to control plant immune response. We also discuss other regulatory mechanisms in which Ca2+ signaling activates CPKs and MAPKs cascades followed by regulating the function of AtSR1 by changing its stability, phosphorylation status, and subcellular localization during plant defense.
Introduction—Calcium Signaling Cascades Control Plant Defense Responses
Plant immune systems rely on multiple layers of recognition systems to confer full protection to pathogen attack. For example, pattern recognition receptors (PRRs) on cell surfaces recognize pathogen-associated molecular patterns (PAMPs) or damage-associated molecular patterns (DAMPs), each of which is derived from pathogenic microbes or damaged plants themselves (Jones and Dangl, 2006; Tanaka and Heil, 2021). This leads to PRR-mediated immunity, or so-called pattern-triggered immunity or PTI (Denancé et al., 2013; Yuan et al., 2017). Pathogens secrete virulence determinants referred to as effectors to inhibit PTI or other plant physiological responses. However, some effectors are recognized by intracellular nucleotide-binding domains and leucine-rich repeat proteins (NLRs), which result in NLR-mediated immunity, or so-called effector-triggered immunity or ETI (Jones et al., 2016). Notably, cellular responses during both PTI and ETI involve dynamic changes in cytosolic Ca2+ concentrations (Zhivotovsky and Orrenius, 2011; Yuan et al., 2020, 2021). Changes in cytosolic Ca2+ concentrations are sensed by the Ca2+-signaling toolkit (Marcec et al., 2019) e.g., Ca2+ sensors and/or decoders [calmodulin (CaM), CaM-like proteins (CML), “calcineurin B-like protein” (CBL)-“CBL-interacting protein kinases” (CBL-CIPK), and calcium-dependent protein kinases (CPKs or CDPKs)], which, together with mitogen-activated protein kinases (MAPKs) activation, coordinate the transcriptional reprogramming of defense genes through activation of various transcript factors (TFs) (Figure 1).
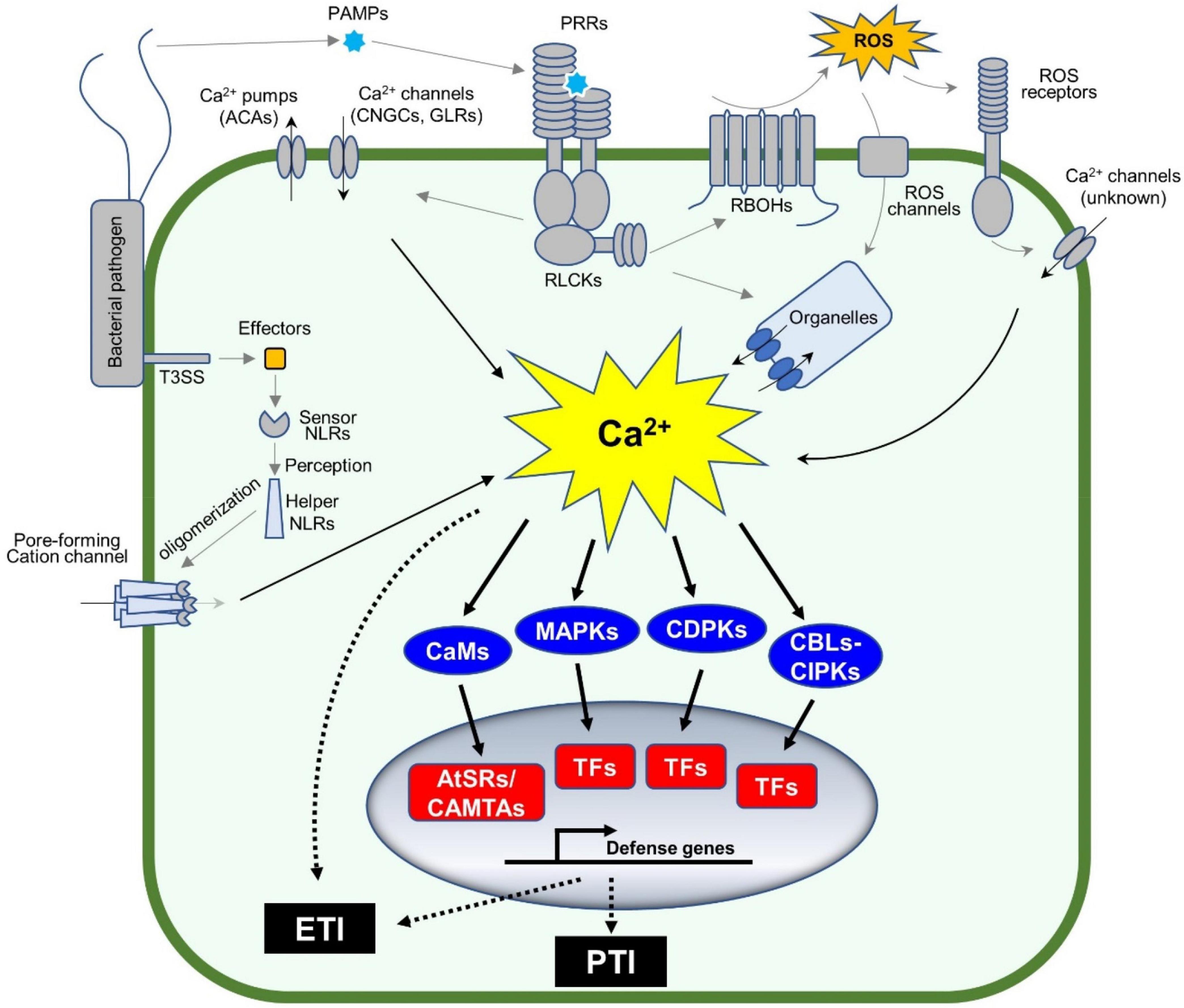
Figure 1. Calcium signaling cascades control plant defense gene expression. Plants sense pathogens (e.g., bacteria in the figure) through pattern recognition receptors (PRRs) by recognizing the pathogen-associated molecular patterns (PAMPs). The activated PRRs phosphorylate themselves and other proteins including receptor-like cytoplasmic kinases (RLCKs). The phosphorylated RLCKs activate Ca2+ channels or Ca2+ pumps, leading to the dynamic changes in cytosolic Ca2+ concentration, while the activated RLCKs phosphorylate RBOHs to produce apoplastic ROS accumulation, which indirectly induces the Ca2+ influx in the plant cell. Ca2+ sensors, such as CaM/CML, CBLs, and CDPKs, together with MAPKs, decode the pattern-triggered Ca2+ spiking into downstream signaling. CBL-activated CIPKs and CDPKs phosphorylate a variety of transcription factors (TFs), such as WRKYs, to induce the expression of defense-related genes. In addition, the TFs (e.g., AtSRs/CAMTAs) activated by Ca2+-bound CaMs are involved in Ca2+-mediated immune response. The entire process is important to activate pattern-triggered immunity (PTI). On the other hand, pathogens secrete proteins, so-called effectors, into the plant cell to repress the plant immune system, also known as ETS. However, the effectors are recognized by intracellular nucleotide-binding domain and leucine-rich repeat proteins (NLRs) to activate a strong and rapid immune response, known as effector-triggered immunity (ETI). The activated NLRs oligomerize and form a non-canonical Ca2+-permeable channel in the plasma membrane, to induce a strong Ca2+ influx (Ca2+ burst) followed by PCD.
The dynamic changes in cytosolic Ca2+ concentrations are an early event during immune responses, where the Ca2+ channels and Ca2+ pumps are activated to form specific Ca2+ signatures to each stimulus (Marcec et al., 2019; Marcec and Tanaka, 2022). For example, cyclic nucleotide-gated ion channel 2 (CNGC2) forms a heteromeric cation channel with CNGC4 playing an important role in the flg22-induced rise of Ca2+ in plant cells. In this event, PRR-activated receptor-like cytoplasmic kinases (RLCKs), e.g., botrytis-induced kinase 1 (BIK1), activate the Ca2+ channel through the phosphorylation of CNGC4, but not CNGC2. In addition, BIK1 also phosphorylates CaM7 that, in turn, binds to the IQ motif in CNGC2 and CNGC4 to suppress the activity of the heteromeric Ca2+ channel (Tian et al., 2019), which could be a desensitization mechanism to regulate the immune response. The application of H2O2 induces Ca2+ influx, where the hydrogen peroxide-induced Ca2+ increase 1 (HPCA1), which functions as an extracellular H2O2 receptor and is required for H2O2-induced Ca2+ rise (Figure 1). Recently, the NLR receptor hopz-activated resistance 1 (ZAR1) resistome was revealed to form a Ca2+-permeable channel to trigger the programmed cell death (PCD) (Wang et al., 2019; Bi et al., 2021). Another study revealed that the active NLR, N requirement gene 1 (NRG1), also forms puncta in the plasma membrane, which is a non-selective cation channel leading to permeability for Mg2+ and Ca2+, but not Cl– (Jacob et al., 2021). These channels based on ZAR1 or NRG1 likely cause a strong, prolonged Ca2+ signature as a Ca2+ burst, which plays a central role during ETI-mediated PCD (Jacob et al., 2021). Most studies on Ca2+ channels focus on the plasma membrane-localized channels, since Ca2+ channel blocker, La3+ or Gd3+, can suppress the NLRs-forming Ca2+ channels. It is not clear how the organelle membrane localized Ca2+ channels are involved in plant immune response. Currently, there are many unanswered questions, for example, how Ca2+ burst induces PCD and other ETI. Ca2+/CaM-binding transcription factors (CAMTAs) or Arabidopsis thaliana signal responsives (AtSRs) could be a sensor of the Ca2+ burst (Yuan et al., 2021) as described in the Section 2 below.
AtSR1/CAMTA3 is a transcriptional regulator in response to biotic stress-induced Ca2+ changes, and plays a suppressor role in the plant immune system (Du et al., 2009) since atsr1/camta3 mutants show an autoimmune phenotype, including elevated salicylic acid (SA) and reactive oxygen species (ROS) concentrations, and enhanced resistance to bacterial and fungal pathogens (Galon et al., 2008; Du et al., 2009). AtSR1/CAMTA3 requires CaM binding for its activation, suggesting a role for Ca2+ in repressing its function in plant immunity. However, accumulating evidence suggests that AtSR1/CAMTA3 functions as more than a negative regulator in the plant immune system. In this review, we summarize the recent progress related to studies on AtSRs/CAMTAs during plant defense responses that could help in our understanding of their unique roles in the plant immune system.
AtSR1/CAMTA3 Is a Central Signaling Component in Plant Immune Responses
CaM-Mediated Regulation of AtSR1/CAMTA3
AtSR1/CAMTA3 is known as a Ca2+/CaM-regulated transcription factor involved in transcriptional reprogramming during plant immune response. AtSR1 binds to the CGCG cis-regulatory element in the promoter of enhanced disease susceptibility 1 (EDS1), non-race-specific disease resistance1 (NDR1), and non-expresser of PR genes1 (NPR1) to modulate their expression in SA-mediated plant immunity (Yuan et al., 2018b,2021). AtSR1 also regulates isochorismate synthase 1 (ICS1) to suppress plant immunity at both 20oC and 28oC (Du et al., 2009). AtSR1 contributes to systemic acquired resistance (SAR) through the regulation of NDR1 expression (Nie et al., 2012). A recent study revealed that AtSR1 is required to establish a proper plant immune response to basal resistance or ETI-triggered PCD, also known as hypersensitive response (HR) cell death (Yuan et al., 2021; Figure 2).
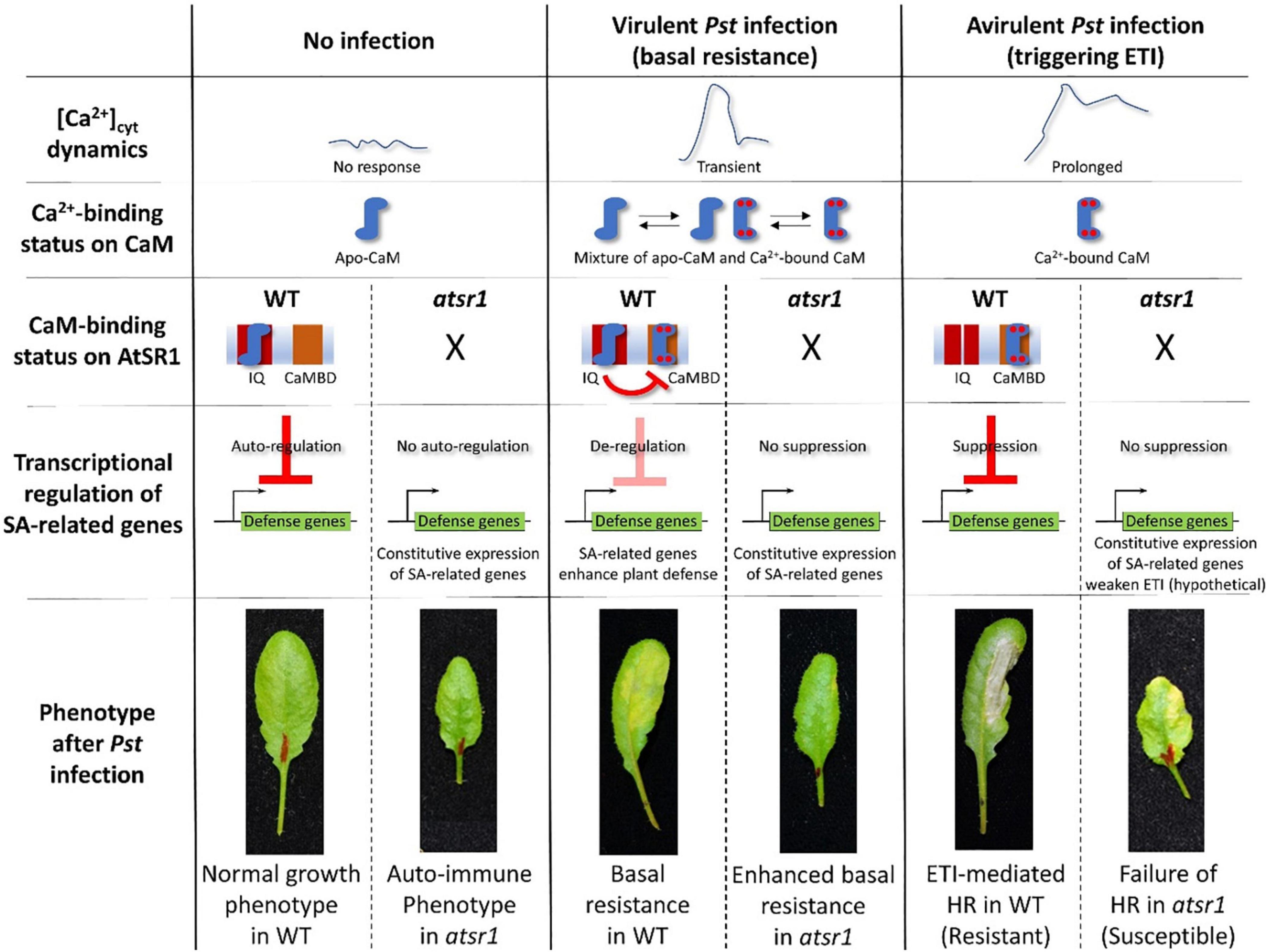
Figure 2. AtSR1/CAMTA3 acts as a signaling switch with dependence on Ca2+-binding status of CaM during plant immune response. In non-stressful environments, there is no [Ca2+]cyt dynamic. Hence, the status of CaM is not in a saturated phase at four Ca2+ binding sites, designated as apo-CaM. Apo-CaM interacts with the IQ motifs of AtSR1. This CaM-AtSR1 complex may have an effect on the stability of AtSR1, which suppresses the transcriptional expression of defense-related genes (e.g., EDS1, ICS1, and NPR1) as an autoregulator. Thus, suppression of plant immunity maintains normal plant growth and development. Under the virulent pathogen infection, [Ca2+]cyt is transiently increased, which confers a status of mixture of Apo-CaM and Ca2+-saturated CaM. Ca2+-saturated CaM interacts with the CaMBD of AtSR1, while Apo-CaM interacts with the IQ motifs. This CaM-AtSR1 complex loses its binding activity to the cis-regulatory element “CGCG,” and de-regulates the transcriptional expression of the defense-related genes. This eventually activates the basal resistance or PTI. Under avirulent pathogen infection, [Ca2+]cyt is constitutively increased, which leads to stable binding of Ca2+ ions to CaM. The Ca2+-saturated CaM interacts only with the CaMBD of AtSR1. This CaM-AtSR1 complex regulates the ETI-triggered PCD. In the atsr1 mutant, due to the loss of negative regulation of the NPR1 transcription by AtSR1, the plants constitutively activate basal immune response, resulting in the autoimmune phenotype. Under pathogen infection in the atsr1 mutant due to the lack of fine-tuning for transcriptional regulation of defense-related genes, the immune response is disturbed in the mutants, i.e., enhanced basal resistance and failure of ETI-induced PCD to the virulent and avirulent bacterial pathogens, respectively. Refer to Yuan et al. (2021) for further details.
The AtSRs family contains different types of CaM-binding domains (CaMBDs). CaM binding to AtSR1 is essential for the function of AtSR1 (Reddy et al., 2011). Yuan et al. (2021) demonstrated that a loss-of-function mutation on CaMBD did not complement the atsr1 mutant phenotype, i.e., failed to suppress the EDS1 expression, suggesting that CaMBD is essential for the AtSR1 function. In contrast, a gain-of-function mutant, atsr1-4D, in which a mutation was located at the first IQ motif, displayed constitutive down-regulation of transcriptional expressions of EDS1 and NDR1 (Nie et al., 2012). Given that the first IQ motif binds to apo-CaM (Ca2+ free CaM), and the CaMBD binds to Ca2+-bound CaM (Yuan et al., 2021), AtSR1 acts as a signaling switch with dependence on the level of cytosolic Ca2+ concentration. The complementation with double mutation at the IQ motif and CaMBD restored the plant phenotype similar to WT, which indicated that the IQ motif and the CaMBD in AtSR1 interact during the plant immune response (Kim et al., 2017; Yuan et al., 2018c; Figure 2).
Recently, AtSRs were also reported to negatively regulate the pipecolic acid (Pip)-mediated plant immune response (Kim et al., 2020; Sun et al., 2020). The biosynthesis of Pip was activated in camta1/2/3 and the transcriptional expressions of agd2-like defense response protein 1 (ALD1) and flavin-dependent monooxygenase 1 (FMO1), which both encode two pip biosynthesis enzymes, were greatly induced in camta1/2/3. Moreover, CBP60g and SARD1 were identified to regulate Pip production by ALD1 and FMO1; also, AtSR1 protein interacted with the CGCG box in the promoter of calmodulin binding protein 60g (CBP60g), but not that of SAR deficient 1 (SARD1), to repress the CBP60g expression (Sun et al., 2020). In addition, the atsr1 auto-immunity phenotype was compromised by not only sard1 cbp60g double mutant, but ald1 fmo1 double mutant as well (Sun et al., 2020). These observations suggest that SA and N-Hydroxypipecolic acid (NHP) crosstalk to mediate plant immune response. This idea is supported by the observation that the application of Pip to plant leaf promoted NPR1 stability (Kim et al., 2020). How AtSR1 with or without Ca2+-bound CaM regulates pip biosynthesis remains to be determined.
A recent study revealed that AtSR1/CAMTA3 is more than a transcriptional repressor in the plant immune response (Figure 3). For example, AtSR1 was found to mediate DAMP-induced signaling, whereas AtPep1-induced reprograming of JA-responsive genes requires functional AtSR1 (Yuan et al., 2020). Extracellular ATP (eATP) induces defense-related transcriptomes in which the CGCG cis-regulatory element was highly enriched in the eATP-responsive promoters (Jewell and Tanaka, 2019; Jewell et al., 2019). Interestingly, AtSR1/CAMTA3 is required for defense gene induction in response to eATP treatment. A recent report demonstrated that CaM-AtSR1 interaction regulates RNAi-mediated immune response against viral infection, where CaM3- and CaM6-bound AtSR1 positively regulate the RNAi system (Wang Y. et al., 2021). AtSR1 interacts with the promoter of RNA-dependent RNA polymerase 6 (RDR6) and bifunctional nuclease-2 (BN2) to induce the transcriptional expression of RDR6 and BN2, respectively. RDR6 is known to convert single-stranded (ss) RNA into double-stranded (ds) RNA to induce RNA silencing (Harmoko et al., 2013), while BN2 is a ribonuclease that degrades microRNAs to activate RNAi. Given that the virus enters into plant cells through natural wound sites and herbivory of insects, wound-induced Ca2+ influx in plant cells may promote the interaction between CaMs and AtSR1 to activate the antiviral RNAi system. In contrast, the virus (a geminivirus CLCuMuV in the reported case) employs an effector protein, V2, to impair the interaction between CaM3 and AtSR1 to suppress plant defense (Wang Y. et al., 2021), which provides corroborative evidence of AtSR1-mediated immune response against the virus.
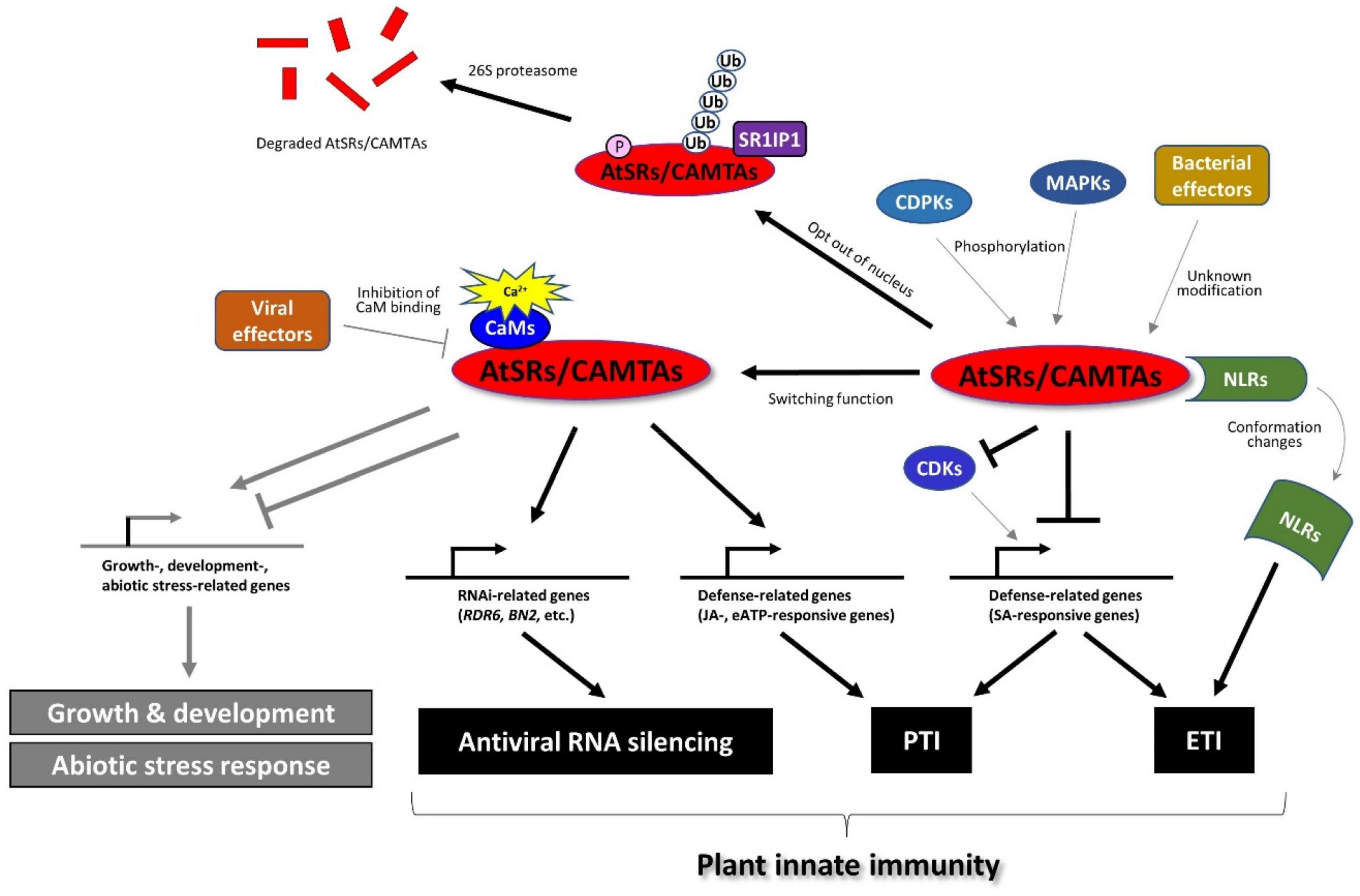
Figure 3. AtSRs/CAMTAs play the role of a hub in plant immune response. AtSRs are the suppressor of plant immunity and repress defense-related gene expressions through the interaction with the “CGCG” box in their promoter. Pathogen infection induces Ca2+ influx as described in Figure 1, which impacts the Ca2+-CaM-AtSRs complex and subsequently removes the suppressor of AtSRs in SA-based plant immunity to induce the defense-related gene expression (e.g., EDS1, and NPR1, etc.). However, effector proteins derived from the bacterial pathogens (e.g., Pst carrying AvrRpt2 or AvrRps4) modify AtSRs, which is sensed by NLRs, e.g., DSC1 and DSC2, followed by inducing ETI-based plant defense response. In addition, the rise of cytoplasmic Ca2+ activates MPKs and CPKs, which phosphorylate AtSRs, resulting in the export of AtSRs from the nucleus and eventually the degradation of AtSRs. In contrast, AtSRs act as transcriptional activators for plant immune response by reprograming the transcription of JA- and eATP-responsive genes. In addition, CaM-AtSRs interaction is targeted by viral effectors, e.g., V2 derived from the geminivirus CLCuMuV, which regulates RNAi-mediated immune response against viral infection. In other respects, AtSRs also play an important role in plant growth, development, and abiotic stress responses, which are not discussed in this review.
Phosphorylation-Based Regulation of AtSR1/CAMTA3
Facing pathogen challenge, it is necessary for the activation and establishment of plant immune response to overcome the suppressor of AtSR1 (Yuan et al., 2018b). The pathogen infection promotes the degradation of AtSR1 through proteasome-mediated pathway where AtSR1 interaction protein 1 (SR1IP1) is a substrate-adaptor for cullin3-based E3 ubiquitin ligase that likely regulates the AtSR1 degradation (Zhang et al., 2014). A recent study revealed that MAPKs cascade contribute to proteasome-mediated turnover of AtSR1 (Jiang et al., 2020). The application of flg22-induced phosphorylation of AtSR1 (mediated by mitogen-activated protein kinases 3 (MPK3) and MPK6, but not MPK4) further results in destabilization of the AtSR1 protein. Recently, the CPKs or CDPKs are also revealed to be involved in pathogen-induced phosphorylation events of AtSR1 (Jiang et al., 2020). The Arabidopsis genomic DNA encodes a large number of the CPK gene family, which contains 34 members and is grouped into four subgroups. The stability of the AtSR1 protein was greatly disrupted in the co-expression of CPK1, CPK2, and CPK5, where only CPK5 was reported to be activated by flg22 (Jiang et al., 2020). Taken together, MAPKs and CDPKs appear to be essential for the phosphorylation and degradation of AtSR1 during plant-microbe interactions, although AtSR1 is not phosphorylated directly by CPK5 in vitro (Jiang et al., 2020).
Controlling the subcellular localization is another way to sequester the negative function of AtSR1 in plant immunity. AtSR1 contains two nuclear localization sequences (NLSs) (Yang and Poovaiah, 2002). The flg22 triggers the subcellular re-localization of the AtSR1 from the nucleus to the cytoplasm, where MAPKs regulate AtSR1 phosphorylation to export AtSR1 out of the nucleus (Jiang et al., 2020), although the MAPK-independent regulation remains to be studied. In addition, the protein phosphatase, such as protein phosphatase 2C (PP2C), is a major regulator of plant immune response. However, the role of phosphatase in AtSR1-mediated plant defense remains to be studied.
AtSR1/CAMTA3 as a Guardee
RIN4 is a well-studied guarded effector target, or so-called “guardee.” However, there are more potential guardees. For example, EXO70B1, a subunit of the exocyst complex, can be a guardee that can activate ETI via the truncated NLR TN2, where CPK5 is required for this TN-mediated immunity (Liu et al., 2017). Also, AtSR1/CAMTA3 was reported as a guardee based on accumulating evidence (Liu et al., 2017). Two toll/interleukin-1 receptor (TIR)-NLR proteins, called dominant suppressor of camta3 number 1 (DSC1) and DSC2 directly interacts with AtSR1, where DSC1 and DSC2 guard the guardee AtSR1. Upon infection of avirulent Pst strains carrying avrRps4 and avrRpt2, a rapid degradation of AtSR1 is induced probably following phosphorylation and/or ubiquitination of AtSR1. The degradation of AtSR1 contributes to the activation of downstream immune responses by de-repression of defense-related genes, e.g., EDS1 and NDR1 (Figure 3). This notion explains how the auto-immunity phenotype in atsr1 requires DSC1 and DSC2 (Lolle et al., 2017). AtSR1 degradation leads to the activation of NLRs-mediated plant immunity. Thus, it is reasonable to hypothesize that a modification of AtSR1 by an unidentified pathogen effector, such as phosphorylation and proteolysis, is sensed by the DSC1 and DSC2, or other unknown NLRs. It would be interesting to know if DSC1 and/or DSC2 might sense the phosphorylated AtSR1, by MPK3, MPK6 and CPK5, as mentioned above. In other respects, DSC1 and DSC2 were identified to be localized in the nucleus; hence, they may also sense the subcellular re-localization of the AtSR1.
Mediator-Associated Regulation of AtSR1/CAMTA3
In eukaryotic cells, RNA polymerase II (Pol II) is required for most transcriptions of general protein-coding genes and several non-coding RNA (ncRNAs) genes (Palazzo and Lee, 2015). The mediator is the highly conserved, large multi-subunit regulator, which works together with RNA Pol II, TFs, and co-TFs to activate the gene expression upon perception of environmental and developmental stimuli. The genetic analysis revealed that cyclin-dependent kinase 8 (CDK8), a mediator subunit, modulates the AtSR1-regulated SA signaling pathway in plant immune response (Huang et al., 2019), where a strong autoimmune phenotype of the triple mutant camta1/2/3 (atsr2/4/1) is partially suppressed in the cdk8 null mutant. Further study revealed that CDK8 regulates SA biosynthesis genes, such as ICS1 and EDS5, probably via AtSRs-mediated transcriptional reprogramming, although a detailed mechanism of how CDK8 regulates AtSRs remains to be studied (Huang et al., 2019). Another report revealed that CDK8 recruits NPR1 and WRKY18 to promote defense gene expression, where SA further facilitates the interaction between CDK8 and NPR1. In addition, CDK8 interacts with other TFs, TGA5 and TGA7, together with NPR1, to induce PR1 expression (Chen et al., 2019). It is interesting to speculate that AtSR1 is the key regulator of CDKs during plant immune responses, but the mechanisms remain to be investigated further.
AtSR1 as Positive Regulator of Plant Growth and Development
Facing pathogen infection, a plant reduces the resource and energy for growth and development, and switches to activate and establish plant immune response to restrict the invading pathogen (Denancé et al., 2013). During the resting stage, AtSR1 represses the plant defense to maintain plant growth and development (Yuan et al., 2018a). Hence, AtSR1 is involved in promoting growth. AtSR1 was found to be involve in IAA and BR signal transduction. AtSR1 interacts with the “CGCG” box in the promoter of IAA1 and IAA19 to regulate their expressions, and regulates DWF4 expression through binding to its promoter (Yuan et al., 2018a). However, the mechanisms involved are not clearly understood.
Conclusion and Future Perspectives
In recent years, a great deal of progress has been made in understanding Ca2+ channels and the associated down-stream signaling. However, there are still several key questions which remain to be addressed. Although many Ca2+ sensors have been identified that regulate plant immune response, the role of CMLs is still unclear and remains to be studied. In addition, the pathogen-triggered modifications (such as phosphorylation and ubiquitination) of AtSR1 suppresses its negative function in plant immunity, however, the molecular mechanism of recovery of AtSR1 after successful prevention of pathogen infection needs to be addressed. There are several major questions: (1) The role of AtSR1 in plant growth remains unclear (Yuan et al., 2018a). (2) Further studies need to be carried out on AtSR1-mediated plant defense against herbivory. (3) The molecular mechanism of AtSR1 decoding Ca2+ signaling through IQ motif and/or CaMBD still needs to be addressed. (4) How AtSR1 crosstalks with hormonal pathways, such as gibberellins (GAs), brassinosteroids (BRs), ethylene (ET) and auxin is not understood.
Since the focus of this special issue is on signaling in plant biotic interactions, it is appropriate to point out that Ca2+/CaM-mediated signaling plays a unique role in both pathogenic (e.g., AtSR1 discussed above) and symbiotic interactions [Ca2 +/CaM-dependent protein kinase (CCaMK)] in plants. It is well recognized that CCaMK, a Ca2+/CaM-binding protein (Patil et al., 1995), plays a key role in fungal and bacterial symbioses (Patil et al., 1995; Gleason et al., 2006; Routray et al., 2013; Wang T. et al., 2021). However, the focus of this review is on plant immune response, hence any discussion on symbioses is beyond the scope of this review.
Author Contributions
PY, KT, and BP were involved in writing this review. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by National Science Foundation grants (1021344 and 1557813 to BP and 1557813 to KT) as well as USDA NIFA (Hatch project no. 1015621 to KT and BP).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors would like to appreciate the help of Lorie Mochel in preparing this review.
References
Bi, G., Su, M., Li, N., Liang, Y., Dang, S., Xu, J., et al. (2021). The ZAR1 resistosome is a calcium-permeable channel triggering plant immune signaling. Cell 184, 3528–3541.e12. doi: 10.1016/j.cell.2021.05.003
Chen, J., Mohan, R., Zhang, Y., Li, M., Chen, H., Palmer, I. A., et al. (2019). NPR1 Promotes Its Own and Target Gene Expression in Plant Defense by Recruiting CDK8. Plant Physiol. 181, 289–304. doi: 10.1104/pp.19.00124
Denancé, N., Sánchez-Vallet, A., Goffner, D., and Molina, A. (2013). Disease resistance or growth: the role of plant hormones in balancing immune responses and fitness costs. Front. Plant Sci. 4:155. doi: 10.3389/fpls.2013.00155
Du, L., Ali, G. S., Simons, K. A., Hou, J., Yang, T., Reddy, A. S. N., et al. (2009). Ca2+/calmodulin regulates salicylic-acid-mediated plant immunity. Nature 457, 1154–1158. doi: 10.1038/nature07612
Galon, Y., Nave, R., Boyce, J. M., Nachmias, D., Knight, M. R., and Fromm, H. (2008). Calmodulin-binding transcription activator (CAMTA) 3 mediates biotic defense responses in Arabidopsis. FEBS Lett. 582, 943–948. doi: 10.1016/j.febslet.2008.02.037
Gleason, C., Chaudhuri, S., Yang, T., Munoz, A., Poovaiah, B. W., and Oldroyd, G. E. D. (2006). Nodulation independent of rhizobia induced by a calcium-activated kinase lacking autoinhibition. Nature 441, 1149–1152. doi: 10.1038/nature04812
Harmoko, R., Fanata, W. I. D., Yoo, J. Y., Ko, K. S., Rim, Y. G., Uddin, M. N., et al. (2013). RNA-dependent RNA polymerase 6 is required for efficient hpRNA-induced gene silencing in plants. Mol. Cells 35, 202–209. doi: 10.1007/s10059-013-2203-2
Huang, J., Sun, Y., Orduna, A. R., Jetter, R., and Li, X. (2019). The Mediator kinase module serves as a positive regulator of salicylic acid accumulation and systemic acquired resistance. Plant J. 98, 842–852. doi: 10.1111/tpj.14278
Jacob, P., Kim, N. H., Wu, F., El-Kasmi, F., Chi, Y., Walton, W. G., et al. (2021). Plant “helper” immune receptors are Ca2+-permeable nonselective cation channels. Science 373:420. doi: 10.1126/science.abg7917
Jewell, J. B., Sowders, J. M., He, R., Willis, M. A., Gang, D. R., and Tanaka, K. (2019). Extracellular ATP Shapes a Defense-Related Transcriptome Both Independently and along with Other Defense Signaling Pathways. Plant Physiol. 179, 1144–1158. doi: 10.1104/pp.18.01301
Jewell, J. B., and Tanaka, K. (2019). Transcriptomic perspective on extracellular ATP signaling: a few curious trifles. Plant Signal. Behav. 14:1659079. doi: 10.1080/15592324.2019.1659079
Jiang, X., Hoehenwarter, W., Scheel, D., and Lee, J. (2020). Phosphorylation of the CAMTA3 Transcription Factor Triggers Its Destabilization and Nuclear Export. Plant Physiol. 184, 1056–1071. doi: 10.1104/pp.20.00795
Jones, J. D. G., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Jones, J. D. G., Vance, R. E., and Dangl, J. L. (2016). Intracellular innate immune surveillance devices in plants and animals. Science 354:aaf6395. doi: 10.1126/science.aaf6395
Kim, Y., Gilmour, S. J., Chao, L., Park, S., and Thomashow, M. F. (2020). Arabidopsis CAMTA Transcription Factors Regulate Pipecolic Acid Biosynthesis and Priming of Immunity Genes. Mol Plant 13, 157–168. doi: 10.1016/j.molp.2019.11.001
Kim, Y. S., An, C., Park, S., Gilmour, S. J., Wang, L., Renna, L., et al. (2017). CAMTA-Mediated Regulation of Salicylic Acid Immunity Pathway Genes in Arabidopsis Exposed to Low Temperature and Pathogen Infection. Plant Cell 29, 2465–2477. doi: 10.1105/tpc.16.00865
Liu, N., Hake, K., Wang, W., Zhao, T., Romeis, T., and Tang, D. (2017). CALCIUM-DEPENDENT PROTEIN KINASE5 Associates with the Truncated NLR Protein TIR-NBS2 to Contribute to exo70B1-Mediated Immunity. Plant Cell 29, 746–759. doi: 10.1105/tpc.16.00822
Lolle, S., Greeff, C., Petersen, K., Roux, M., Jensen, M. K., Bressendorff, S., et al. (2017). Matching NLR Immune Receptors to Autoimmunity in camta3 Mutants Using Antimorphic NLR Alleles. Cell Host Microbe 21, 518–529.e4. doi: 10.1016/j.chom.2017.03.005
Marcec, M. J., Gilroy, S., Poovaiah, B. W., and Tanaka, K. (2019). Mutual interplay of Ca2+ and ROS signaling in plant immune response. Plant Sci. 283, 343–354. doi: 10.1016/j.plantsci.2019.03.004
Marcec, M. J., and Tanaka, K. (2022). Crosstalk between calcium and ROS signaling during flg22-triggered immune response in Arabidopsis leaves. Plants 11:14. doi: 10.3390/plants11010014
Nie, H., Zhao, C., Wu, G., Wu, Y., Chen, Y., and Tang, D. (2012). SR1, a Calmodulin-Binding Transcription Factor, Modulates Plant Defense and Ethylene-Induced Senescence by Directly Regulating NDR1 and EIN3. Plant Physiol. 158, 1847–1859. doi: 10.1104/pp.111.192310
Palazzo, A. F., and Lee, E. S. (2015). Non-coding RNA: what is functional and what is junk? Front. Genet. 6:2. doi: 10.3389/fgene.2015.00002
Patil, S., Takezawa, D., and Poovaiah, B. W. (1995). Chimeric plant calcium/calmodulin-dependent protein kinase gene with a neural visinin-like calcium-binding domain. Proc. Natl. Acad. Sci. U. S. A. 92, 4897–4901. doi: 10.1073/pnas.92.11.4897
Reddy, A. S. N., Ali, G. S., Celesnik, H., and Day, I. S. (2011). Coping with Stresses: roles of Calcium- and Calcium/Calmodulin-Regulated Gene Expression. Plant Cell 23, 2010–2032. doi: 10.1105/tpc.111.084988
Routray, P., Miller, J. B., Du, L., Oldroyd, G., and Poovaiah, B. W. (2013). Phosphorylation of S344 in the calmodulin-binding domain negatively affects CCaMK function during bacterial and fungal symbioses. Plant J. 76, 287–296. doi: 10.1111/tpj.12288
Sun, T., Huang, J., Xu, Y., Verma, V., Jing, B., Sun, Y., et al. (2020). Redundant CAMTA Transcription Factors Negatively Regulate the Biosynthesis of Salicylic Acid and N-Hydroxypipecolic Acid by Modulating the Expression of SARD1 and CBP60g. Mol. Plant 13, 144–156. doi: 10.1016/j.molp.2019.10.016
Tanaka, K., and Heil, M. (2021). Damage-Associated Molecular Patterns (DAMPs) in Plant Innate Immunity: applying the Danger Model and Evolutionary Perspectives. Annu. Rev. Phytopathol. 59, 53–75. doi: 10.1146/annurev-phyto-082718-100146
Tian, W., Hou, C., Ren, Z., Wang, C., Zhao, F., Dahlbeck, D., et al. (2019). A calmodulin-gated calcium channel links pathogen patterns to plant immunity. Nature 572, 131–135. doi: 10.1038/s41586-019-1413-y
Wang, J., Hu, M., Wang, J., Qi, J., Han, Z., Wang, G., et al. (2019). Reconstitution and structure of a plant NLR resistosome conferring immunity. Science 364:eaav5870. doi: 10.1126/science.aav5870
Wang, T., Guo, J., Peng, Y., Lyu, X., Liu, B., Sun, S., et al. (2021). Light-induced mobile factors from shoots regulate rhizobium-triggered soybean root nodulation. Science 374, 65–71. doi: 10.1126/science.abh2890
Wang, Y., Gong, Q., Wu, Y., Huang, F., Ismayil, A., Zhang, D., et al. (2021). A calmodulin-binding transcription factor links calcium signaling to antiviral RNAi defense in plants. Cell Host Microbe 29, 1393–1406.e7. doi: 10.1016/j.chom.2021.07.003
Yang, T., and Poovaiah, B. W. (2002). A Calmodulin-binding/CGCG Box DNA-binding Protein Family Involved in Multiple Signaling Pathways in Plants. J. Biol. Chem. 277, 45049–45058. doi: 10.1074/jbc.M207941200
Yuan, P., Tanaka, K., Du, L., and Poovaiah, B. W. (2018b). Calcium Signaling in Plant Autoimmunity: a Guard Model for AtSR1/CAMTA3-Mediated Immune Response. Mol. Plant 11, 637–639. doi: 10.1016/j.molp.2018.02.014
Yuan, P., Yang, T., and Poovaiah, B. W. (2018c). Calcium Signaling-Mediated Plant Response to Cold Stress. Int. J. Mol. Sci. 19:3896. doi: 10.3390/ijms19123896
Yuan, P., Du, L., and Poovaiah, B. (2018a). Ca2+/Calmodulin-Dependent AtSR1/CAMTA3 Plays Critical Roles in Balancing Plant Growth and Immunity. Int. J. Mol. Sci. 19:1764. doi: 10.3390/ijms19061764
Yuan, P., Jauregui, E., Du, L., Tanaka, K., and Poovaiah, B. W. (2017). Calcium signatures and signaling events orchestrate plant–microbe interactions. Curr. Opin. Plant Biol. 38, 173–183. doi: 10.1016/j.pbi.2017.06.003
Yuan, P., Jewell, J. B., Behera, S., Tanaka, K., and Poovaiah, B. W. (2020). Distinct Molecular Pattern-Induced Calcium Signatures Lead to Different Downstream Transcriptional Regulations via AtSR1/CAMTA3. Int. J. Mol. Sci. 21:8163. doi: 10.3390/ijms21218163
Yuan, P., Tanaka, K., and Poovaiah, B. W. (2021). Calmodulin-binding transcription activator AtSR1/CAMTA3 fine-tunes plant immune response by transcriptional regulation of the salicylate receptor NPR1. Plant Cell Environ. 44, 3140–3154. doi: 10.1111/pce.14123
Zhang, L., Du, L., Shen, C., Yang, Y., and Poovaiah, B. W. (2014). Regulation of plant immunity through ubiquitin-mediated modulation of Ca2+–calmodulin–AtSR1/CAMTA3 signaling. Plant J. 78, 269–281. doi: 10.1111/tpj.12473
Keywords: AtSR1/CAMTA3, Ca2+ signaling, CaMs/CMLs, CBL-CIPK, CPKs, MAPKs, plant immune response
Citation: Yuan P, Tanaka K and Poovaiah BW (2022) Calcium/Calmodulin-Mediated Defense Signaling: What Is Looming on the Horizon for AtSR1/CAMTA3-Mediated Signaling in Plant Immunity. Front. Plant Sci. 12:795353. doi: 10.3389/fpls.2021.795353
Received: 14 October 2021; Accepted: 15 December 2021;
Published: 11 January 2022.
Edited by:
Keiko Yoshioka, University of Toronto, CanadaReviewed by:
Justin Lee, Leibniz-Institut für Pflanzenbiochemie (IPB), GermanySung Un Huh, Kunsan National University, South Korea
Copyright © 2022 Yuan, Tanaka and Poovaiah. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: B. W. Poovaiah, cG9vdmFpYWhAd3N1LmVkdQ==
 Peiguo Yuan
Peiguo Yuan Kiwamu Tanaka
Kiwamu Tanaka B. W. Poovaiah
B. W. Poovaiah