- Department of Microbiology and Plant Pathology, Institute for Integrative Genome Biology, University of California, Riverside, Riverside, CA, United States
Plants have evolved variable phenotypic plasticity to counteract different pathogens and pests during immobile life. Microbial infection invokes multiple layers of host immune responses, and plant gene expression is swiftly and precisely reprogramed at both the transcriptional level and post-transcriptional level. Recently, the importance of epigenetic regulation in response to biotic stresses has been recognized. Changes in DNA methylation, histone modification, and chromatin structures have been observed after microbial infection. In addition, epigenetic modifications may be preserved as transgenerational memories to allow the progeny to better adapt to similar environments. Epigenetic regulation involves various regulatory components, including non-coding small RNAs, DNA methylation, histone modification, and chromatin remodelers. The crosstalk between these components allows precise fine-tuning of gene expression, giving plants the capability to fight infections and tolerant drastic environmental changes in nature. Fully unraveling epigenetic regulatory mechanisms could aid in the development of more efficient and eco-friendly strategies for crop protection in agricultural systems. In this review, we discuss the recent advances on the roles of epigenetic regulation in plant biotic stress responses.
Introduction
Epigenetic modification is a regulatory mechanism of gene expression caused by changes in chromatin structure and function without alteration of the DNA sequence. Epigenetic regulation of both mammalian and plant genomes has been intensely studied over the last two decades. Unlike animals, plants are sessile and unable to escape from variable environmental extremes or biotic stressors like herbivores and pathogens. To withstand pathogens and pests, plants have evolved highly sophisticated defense regulatory mechanisms (Jones et al., 2016; Arnold et al., 2019; Bakhtiari et al., 2019; Zhou and Zhang, 2020). A growing number of studies have unveiled that epigenetic regulation is crucial for shaping plant immunity and phenotypic variations during plant-microbe interaction.
Plants lack an adaptive immune system or specialized cells for immune response. Plants detect conserved pathogen-associated molecular patterns (PAMPs) via the host plasma membrane-associated pattern recognition receptors (PRRs). This recognition triggers general nonspecific immune responses, named PAMP-triggered immunity (PTI; Bigeard et al., 2015; Nürnberger and Kemmerling, 2018). To suppress PTI and promote a successful invasion, pathogens evolved the ability to send effectors into plant cells to modulate host gene expression and suppress plant immunity (Irieda et al., 2019). In turn, host cells have evolved the ability to recognize pathogen effectors using intracellular nucleotide-binding domain and leucine-rich repeat receptors (NLRs), which activate innate immune responses against pathogen infection (Jones et al., 2016; Zhou and Zhang, 2020). The recognition of effectors by NLRs induces rapid and robust effector-triggered immunity (ETI; Cui et al., 2015; Lolle et al., 2020). Expression and activation of NLRs are precisely regulated at many different levels, including transcriptional regulation, alternative transcript splicing, non-coding small RNA (sRNA)-mediated post-transcriptional regulation, translational regulation, post-translational modification, NLR dimerization or oligomerization, and proteasome-mediated degradation (Li et al., 2015; Lai and Eulgem, 2018). Failure of NLR activation or over accumulation of NLRs can lead to serious diseases or autoimmune responses, respectively. In plants, the local defense response triggers subsequent systemic acquired resistance (SAR) in distal leaves to prevent or reduce future infection (Fu and Dong, 2013). PTI, ETI, and SAR are associated with elevated levels of the phytohormone salicylic acid (SA), a phenolic compound produced by a wide range of prokaryotic and eukaryotic organisms. Besides SA, jasmonic acid (JA) and ethylene are also important phytohormones in biotic stress signaling (Burger and Chory, 2019). In addition, chemicals, such as microbe or host-derived molecules, can act as priming stimuli by inducing disease resistance, which enables a more robust response by the host defense system to future attacks. Further, this ability can sometimes be transmitted to progeny (Conrath et al., 2015; Mauch-Mani et al., 2017; Lopez Sanchez et al., 2021). This transgenerational priming is highly correlated to inherited epigenetic modifications.
Epigenetic regulation of gene expression involves various components, including enzymes that catalyze or remove DNA methylation and histone post-translational modification (PTM), sRNAs, and chromatin remodelers. Together, these components contribute to the precise integrated transcriptional regulation of gene expression. Molecular interactions, including protein-protein, protein-DNA, protein-RNA, and RNA-DNA-protein complexes, establish, erase, or edit epigenetic marks on both genomic DNA or histones to alter chromatin structures and accessibility. The major roles of DNA methylation include the maintenance of plant genome stability by inhibiting the movement of transposable elements (TEs), and the repression of gene expression by recruiting gene expression suppressors or preventing the binding of transcription factors to the methylated DNA. Heterochromatic siRNAs (hc-siRNA) are associated with RNA-directed DNA methylation (RdDM) involved in the deposition of DNA methylation and transcriptional gene silencing (TGS; Kim and Zilberman, 2014; Matzke and Mosher, 2014; Bond and Baulcombe, 2015; Erdmann and Picard, 2020). PTMs at the N-terminal tails of core histones (H2A, H2B, H3, and H4) impact the interaction of histones with DNA, transcription factors, and chromatin remodelers to regulate gene expression (Liu et al., 2010; Deal and Henikoff, 2011). In addition, DNA methylation and histone H1 jointly maintain transcriptional homeostasis by silencing TEs and aberrant intragenic transcripts (Choi et al., 2020). In plants, emerging evidence shows that various epigenetic regulatory mechanisms including DNA methylation dynamics, changes in histone density and variants, and histone PTMs play important roles in regulating plant defense responses (Deleris et al., 2016; Ramirez-Prado et al., 2018; Huang et al., 2019). Different histone marks, together with their specific writers, readers, and erasers coordinate the transcription of defense-related genes (Ramirez-Prado et al., 2018; Hu et al., 2019). While the role of chromatin remodelers and their crosstalk with DNA methylation and histone modification in regulating plant growth and development has been extensively studied (Han et al., 2015), their potential roles in regulating plant immune responses require further investigation. Here, we review recent discoveries on epigenetic regulation of plant immune responses, including the responses of the Arabidopsis mutant plants of DNA methylation components, histone modification readers, and chromatin remodelers to non-viral pathogens (Table 1), and in crop diseases (Table 2).
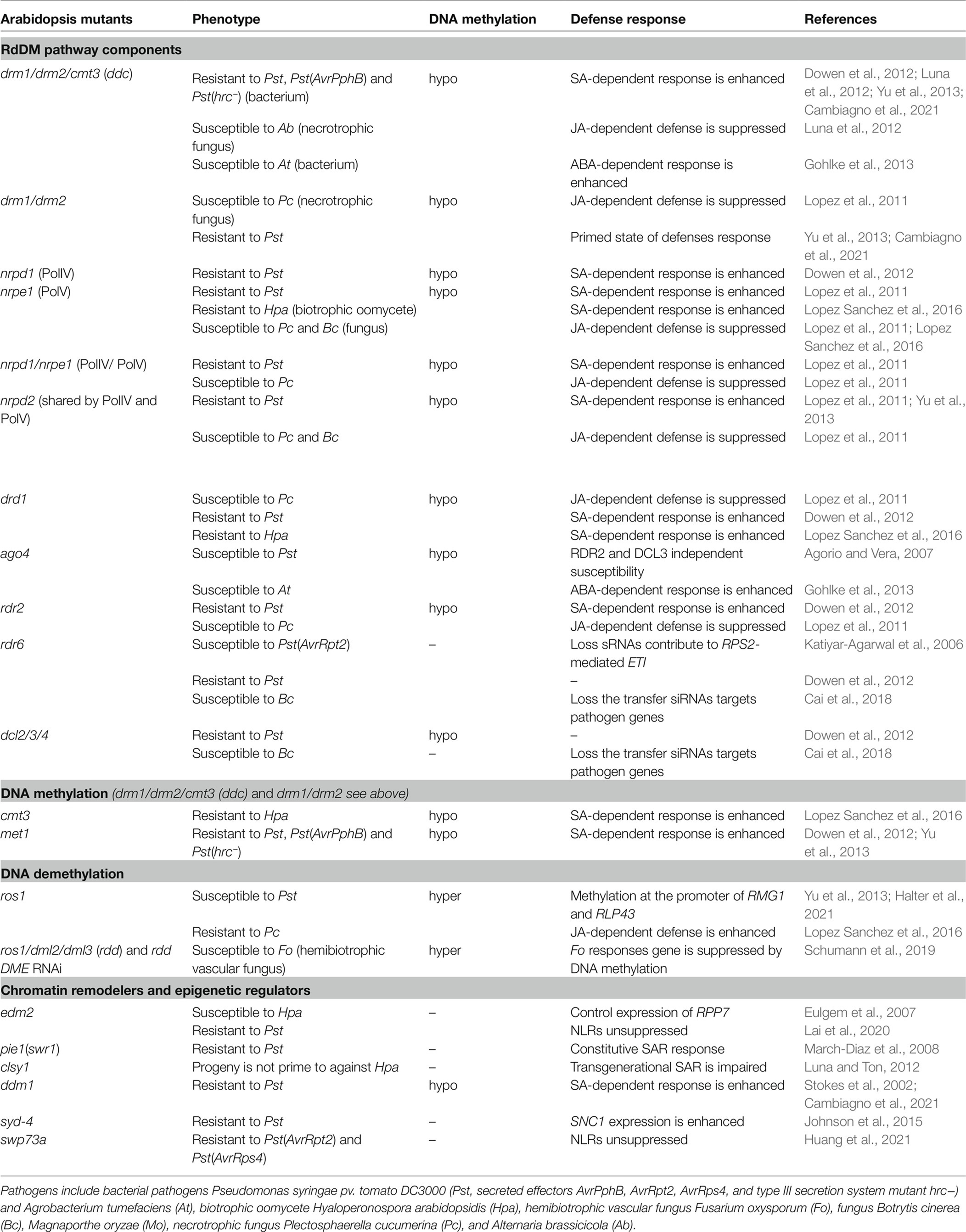
Table 1. The responses of the Arabidopsis mutants plants of DNA methylation components, histone readers, and chromatin remodelers to different pathogens.
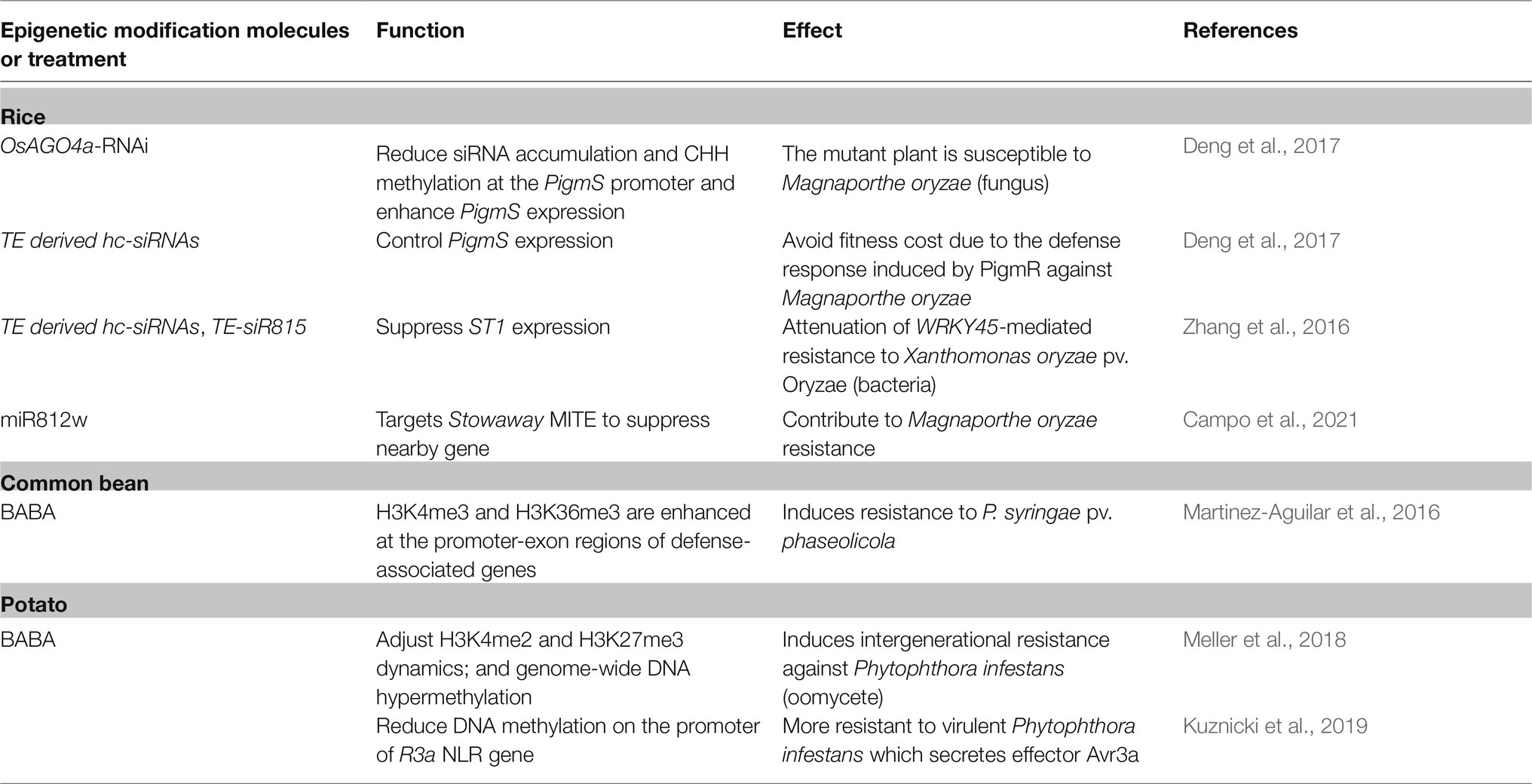
Table 2. Examples of molecular regulators or treatments which cause epigenetic modification and regulate crop disease response.
Small Rna-Mediated Epigenetic Modification Regulates Plant Defense
In plants, sRNAs, including microRNAs (miRNAs) and small-interfering RNAs (siRNAs), are generated by the type III ribonuclease Dicer or Dicer-like (DCL) proteins and are incorporated into Argonaute (AGO) proteins to induce gene silencing in a sequence-specific manner (Baulcombe, 2004). An sRNA is loaded into an AGO protein and then induces TGS or post-transcriptional gene silencing (PTGS) of their target genes endogenously or even in interacting organisms (Baulcombe, 2004; Weiberg et al., 2013; Cai et al., 2018; Huang et al., 2019). In general, miRNAs are processed from single-stranded primary RNA precursors with stem-loop structures, whereas siRNAs are generated from double-stranded RNAs (dsRNAs) that are derived from invert repeats, sense-antisense transcript pairs, or products of RNA-dependent RNA polymerases (RDRs). Plant siRNAs can be further divided into trans-acting siRNAs (ta-siRNAs; Allen et al., 2005) or secondary phased siRNAs (phasiRNAs; Fei et al., 2013), hc-siRNAs, natural antisense transcripts-derived siRNAs (nat-siRNAs; Katiyar-Agarwal et al., 2006), and long siRNAs (lsiRNAs; Katiyar-Agarwal et al., 2007) based on their biogenesis pathways (Borges and Martienssen, 2015; Huang et al., 2019). Different types of sRNAs were reported to precisely regulate the expression of NLRs and plant defense signaling genes to activate plant immune responses and to balance the trade-off between plant growth and defense (Huang et al., 2019). Here, we mainly focus on the functions of some sRNAs that play a direct role in epigenetic regulation in the following sections.
Heterochromatic siRNAs, which are typically 24–30 nt in length, play a central role in the canonical RdDM pathway (Figure 1A). The biogenesis of hc-siRNAs is dependent on the RDR2-DCL3-AGO4/6/9 pathway and also requires plant-specific RNA polymerase IV and V (PolIV and PolV; Figure 1A). They are derived from TEs, repeats, and heterochromatic regions and act to direct de novo DNA methylation and/or histone modifications at the target region (Matzke and Mosher, 2014). Many PRR/NLR loci or clusters are associated with TEs or repeats (Cambiagno et al., 2018) and, thus, are regulated by hc-siRNA-mediated epigenetic regulation. One example of this is the rice NLR Pigm locus, which confers durable resistance to the rice blast fungus Magnaporthe oryzae (Deng et al., 2017; Table 2). The Pigm locus encodes a cluster of NLRs and NLR pseudogenes, including PigmR and PigmS. Constitutive expression of PigmR confers resistance to M. oryzae but causes yield losses. PigmS, which is highly expressed in pollen and panicles, forms a heterodimer with PigmR to suppress PigmR-mediated resistance to avoid fitness costs. The PigmS promoter contains two tandem miniature transposons (MITEs), which associate with hc-siRNAs. Thus, the precise control of PigmS expression is mediated by hc-siRNA at the epigenetic level.
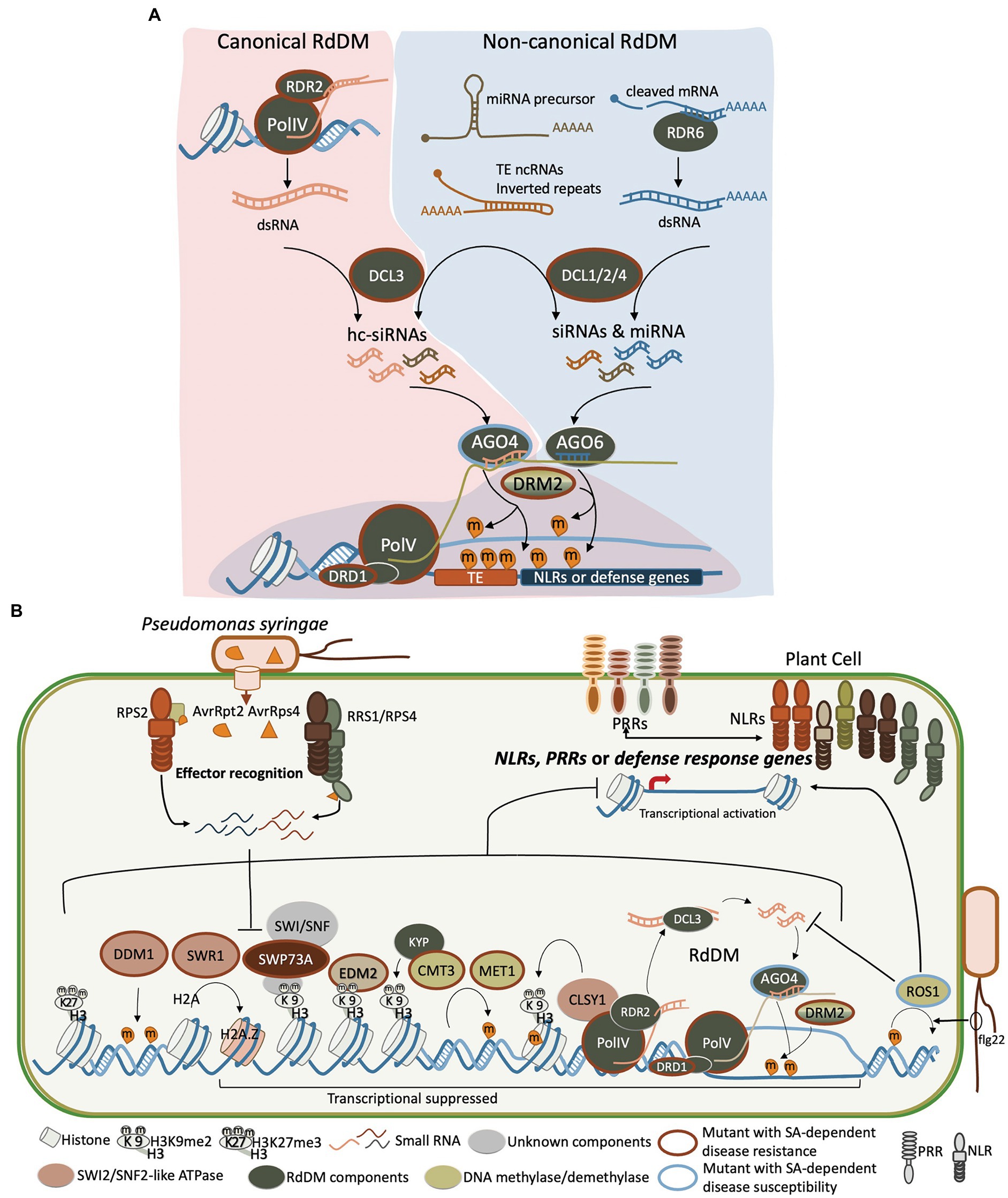
Figure 1. The epigenetic regulatory mechanisms act coordinately in reprogramming gene expression when plants encounter biotic stress. The known factors in Arabidopsis that regulate the biotic stress involve small RNAs, RdDM, methylation and demethylation of DNA, histone modification, and chromatin remodelers, which are present here. (A) Small RNAs participate in conical and non-canonical RdDM pathways to regulate DNA methylation on TEs and defense-related genes. The RDR2- and DCL3-dependent hc-siRNAs are key components in PolIV-RDR2-DCL3-AGO4-PolV-DRM2, the conical RdDM pathway, which establish and reinforce DNA methylation at TEs and regulate the nearby defense response genes. The siRNAs derived from mRNA precursor, TE non-coding RNAs (TE ncRNAs), inverted repeats, or dsRNAs produced by RDR6 are processed by DCL2/4 also participate in establishing the DNA methylation and regulate the expression of defense-related genes. (B) Deposition of DNA methylation and on the NLRs, PRRs, or defense response genes leads to a transcriptionally suppressed status. Other components acting with the chromatin remodeler SWP73A (dark red oval) are not clear (unknown component is shown in gray oval). RdDM components including PolIV and PolV, DCLs, RDRs, AGO4/6, and DRD1 are shown in a dark green oval. DNA methyltransferases DRM2, MET1, and CMT3 are shown in a light green oval. SWI2/SNF2-like ATPases Decreased DNA Methylation 1 (DDM1), Swi2/Snf2-related 1 (SWR1), and Classy1 (CLSY1) are shown in an orange oval. The H3K9me2 reader EDM2 is shown in the light brown oval. Mutant plants that display a resistant phenotype to pathogen (such as Pst) relying on an SA-dependent response are surrounded with a red outline, whereas the blue outline indicates a more susceptible phenotype.
In addition to NLRs, hc-siRNAs also regulate signaling components of plant defense. For example, a rice hc-siRNA, TE-siR815, derived from a MITE in the first intron of the transcription factor WRKY45-1 allele, induces TGS of a leucine-rich repeat receptor kinase-type protein gene, ST1. ST1 is a key component in the WRKY45 signaling pathway and this suppression leads to attenuation of WRKY45-mediated resistance to bacterial blight of rice caused by Xanthomonas oryzae pv. Oryzae (Xoo; Zhang et al., 2016; Table 2). Unlike the WRKY45-1 allele, the WRKY45-2 allele does not contain the TE-siR815-generating MITE, which allows signaling pathway activation against Xoo. Other than hc-siRNA, miRNA can participate in non-canonical RdDM (Cuerda-Gil and Slotkin, 2016). A rice-specific miR812w, which originates from the Stowaway MITE, targets Stowaway MITE to suppress the expression of nearby genes through miRNA-directed DNA methylation, contributing to M. oryzae resistance (Campo et al., 2021; Table 2). Taken together, these studies demonstrate that sRNAs can regulate plant defense response through RdDM.
Dynamics of Dna Methylation in Response To Biotic Stresses
In plants, DNA methylation is observed on cytosine in the context of symmetric CG and CHG, and asymmetric CHH (where H = A, C, or T). CHH methylation is primarily established by de novo DNA methylation through RdDM by the RNA scaffolds produced by PolV, which recruit DNA (cytosine-5)-methyltransferase DRM2 and hc-siRNAs that are produced by RDR2 and DCL3 and associated with AGO4/6/9 in Arabidopsis. The PolIV-RDR2-DCL3-AGO4-PolV-DRM2 pathway forms a feedback loop to reinforce DNA methylation at heterochromatin regions and TEs (Figure 1). In addition to this canonical RdDM, DNA methylation can also be established by the RDR6-DCL2-derived 21 and 22 nt siRNA pathway, which is also dependent on AGO4 and AGO6 (Nuthikattu et al., 2013; Cuerda-Gil and Slotkin, 2016). Afterward, spreading of CG and CHG methylation is maintained by DNA (cytosine-5) Methyltransferase 1 (MET1) and a plant-specific Chromomethylase 3 (CMT3), respectively (Zhang et al., 2018). DNA methylation is reversible and the process of DNA demethylation can be passive or active. The passive process occurs in the absence of DNA methylation on newly synthesized DNA strands. Active DNA demethylation requires the direct removal of a methyl group from DNA by DNA demethylases. Arabidopsis has four DNA demethylases, including DEMETER (DME), DME-Like 1/Repressor of Silencing 1 (DML1/ROS1), DML2, and DML3 (Kumar et al., 2018).
DNA methylation dynamics are dependent on the equilibrium between methylation and demethylation pathways. In genome-wide regulation, the DNA methylation within repetitive sequences or TEs are altered in response to infection of the bacterial pathogen Pseudomonas syringae pv. tomato DC3000 (Pst) and SA treatment and subsequently regulate the transcription of neighboring genes (Agorio and Vera, 2007; Dowen et al., 2012). In general, Arabidopsis mutant plants with DNA hypomethylation are more resistant to disease and exhibit an elevated SA-dependent response. For example, met1, drm1/drm2 and drm1/drm2/cmt3 (ddc), nrpd1 (PolIV mutant), nrpe1 (PolV mutant), nrpd1/nrpe1, nrpd2 (subunit shared by PolIV and PolV), drd1 (defective in RNA-directed DNA methylation), rdr2, and dcl2/3/4 are more resistant to the bacterial pathogen Pst; cmt3, drd1, and nrpe1 are more resistant to the obligate biotrophic oomycete pathogen Hyaloperonospora arabidopsidis (Hpa; Lopez et al., 2011; Dowen et al., 2012; Yu et al., 2013; Lopez Sanchez et al., 2016; Cambiagno et al., 2021; Table 1 and Figure 1A).
Conversely, some mutations in sRNA biogenesis and RdDM-dependent pathways are more susceptible to pathogen infection. For example, mutation in RDR6, which is required for phasing siRNA and nat-siRNA biogenesis and also mediates non-canonical RdDM, does not have an obvious change on global DNA methylation and is more susceptible to Pst strain that secrets effector AvrRpt2, Pst (AvrRpt2), but displays minor changes in response to Pst infection (Katiyar-Agarwal et al., 2006; Dowen et al., 2012; Nuthikattu et al., 2013). AvrRpt2 triggers NLR RPS2-mediated ETI so this suggested that RDR6-mediated siRNAs play a crucial role for RPS2-mediated ETI. The rdr6 and dcl2/3/4 mutants, which greatly reduce the biogenesis of siRNAs, are more susceptible to fungal pathogen Botrytis cinerea (Bc). This susceptible phenotype is caused by the fact that the host plant has lost the siRNAs that move into fungal cells to suppress fungal virulence-related genes (Cai et al., 2018). Drd1, nrpe1, nrpd1/nrpe1, and nrpd2 also display enhanced susceptibility to the necrotrophic fungus Plectosphaerella cucumerina (Lopez Sanchez et al., 2016) and the ddc mutant is more susceptible to necrotrophic fungus Alternaria brassicicola (Lopez et al., 2011; Luna et al., 2012). This is due to the fact that the defensive signaling against P. cucumerina and A. brassicicola is JA-dependent, which is suppressed in nrpe1 and ddc mutants. RdDM mutant ago4 is more susceptible to Pst but this phenotype is independent of other upstream components of the RdDM pathway including RDR2 and DCL3 (Agorio and Vera, 2007). Thus, this response could be caused by other regulatory mechanism of AGO4, which is suggested from the following study. The ddc and ago4 mutants were found to be more susceptible to Agrobacterium tumefaciens, the bacterium that causes crown gall tumors. This enhanced susceptibility phenotype was abscisic acid (ABA) dependent, which is also regulated by DNA methylation (Gohlke et al., 2013). ABA plays a pivotal role in abiotic stress responses and has negative impacts on plant immunity against diverse pathogens (Robert-Seilaniantz et al., 2011). This impact is attributed to ABA-mediated suppression of plant immune responses induced by immune hormones SA, JA, and ethylene (Mine et al., 2017). Thus, dynamic changes in DNA methylation in response to pathogen infection play a pivotal role in plant immune responses.
Active demethylation also shapes transcriptional reprogramming of immune response genes upon infection of different pathogens (Table 1). In Arabidopsis, the bacterial PAMP, flagellin-derived peptide flg22, derepresses RdDM targeted genes, such as an NLR gene, RMG1, and a PRR gene, RLP43, through ROS1-directed demethylation on their promoters (Yu et al., 2013; Halter et al., 2021; Figure 1B). In addition, ROS1 antagonizes RdDM-dependent methylation at RMG1 locus, which may also contribute to anti-bacterial response (Halter et al., 2021). The ros1 mutant shows hypermethylation and is more susceptible to Hpa. Conversely, ros1 is more resistant to P. cucumerina, which is associated with JA-dependent defense pathways (Lopez Sanchez et al., 2016). Furthermore, the triple mutant of DNA demethylases ros1/dml2/dml3 (rdd) and the quadruple mutant rdd DME RNAi lines display enhanced susceptibility to a hemibiotrophic vascular fungal pathogen, Fusarium oxysporum, that causes disease in many important crops. The tissue-specific expression of four DNA demethylases DME, ROS1, DML2, and DML3 act cooperatively to construct resistance against F. oxysporum (Schumann et al., 2019). It was also found that some Arabidopsis NLRs can be demethylated by ROS1, DML2, and DML3 within their promoters and transcribed regions (Kong et al., 2020). Thus, active demethylation processed by DNA demethylases regulates defense response genes upon pathogen infection (Table 1).
Histone Modification and Crosstalk with Dna Methylation Modulate Plant Defense Responses
Post-translational modifications on histone proteins have direct impacts on the chromatin structure and contribute to the transcriptional regulation of gene expression. Histone modification is a reversible process and is modulated by specific writers that add the modification, erasers that remove the modification, or readers that sense the modification and transduce the downstream signaling pathways. Some histone PTMs are associated with specific transcriptional states. In general, H3K4me (methylation of Histone 3 at Lys4), H3K36me, H3K9ac (acetylation of H3 at Lys9), and H3K27ac are markers for transcriptional activation, whereas H3K27me3 is mainly linked to transcriptional silencing of genes (Xiao et al., 2016). H3K9me2 and H3K9me3 are enriched in heterochromatic regions with a high density of TEs or repeats, where they have a constitutive repressive function (Liu et al., 2010; He et al., 2011). In euchromatic regions, H3K9me2 has been observed to span the entire gene and is correlated with low expression levels (Zhou et al., 2010; West et al., 2014).
The epigenetic regulation of defense-related genes mediated by histone modification was comprehensively discussed in recent reviews (Ramirez-Prado et al., 2018; Hu et al., 2019). Here, we emphasize the crosstalk between histone modification and DNA methylation on plant biotic stress as methylation of DNA and H3K9 is highly correlated with gene silencing in eukaryotes. This link between DNA methylation and H3K9 modification was revealed by binding of CMT3 with the histone methyltransferase Kryptonite/SUVH4 (KYP). The interaction of CMT3 and KYP/SUVH4 constitutes a self-reinforcing loop between histone and DNA methylation in plants which is important for TE silencing (Du et al., 2015; Figure 1B). In addition to DNA methylation on TEs regulating the transcription of neighboring NLR genes (Dowen et al., 2012), some NLR loci or clusters associated with TEs are also controlled by histone marks, such as H3K9me2 (Lai and Eulgem, 2018). Through Arabidopsis mutant screening, the methylation of DNA and H3K9 was observed to regulate resistance against Pst infection (Cambiagno et al., 2021) and infestation by the pest, green peach aphid Myzus persicae (Annacondia et al., 2021). The study revealed that nrpd1 and kyp mutant plants are more resistant to aphids (Annacondia et al., 2021). Whether and how does the crosstalk between DNA and H3K9 methylation contribute to aphid resistance is still largely unknown. The drm1/drm2, ddc, and suvh4/5/6 mutant plants do not exhibit constitutive expression of the defense gene marker PR1 (PATHOGENESIS-RELATED GENE 1) but are more resistant to Pst due to a faster and stronger PR1 induction after Pst infection compared to wild-type plants (Cambiagno et al., 2021). This suggest that drm1/drm2, ddc, and suvh4/5/6 mutant plants acquired a primed state of defense against Pst, which is regulated by both DNA and H3K9 methylation levels. Another example of genes participating in the crosstalk of DNA and H3K9 methylation is Increase in Bonsai Methylation 1 (IBM1), which encodes a histone demethylase and directly associates with the gene body that has the repressive mark, H3K9me2. IBM1 removes mono- and dimethylation of histone lysines and negatively regulates DNA methylation at CHG loci in the genic regions. IBM1 positively regulates Arabidopsis defense responses against Pst at the chromatic level by derepressing the defense marker genes PR1, PR2, and the PTI marker FRK1 (Chan and Zimmerli, 2019). Thus, the crosstalk between histone modification and DNA methylation contributes to the epigenetic regulation of gene expression in response to pathogen infection.
Chromatin Remodelers and Epigenetic Regulators Modulate Plant Immunity
While defense-related genes are regulated by covalent DNA and histone modifications, chromatin remodeling proteins also play an important role in regulating NLRs, plant defense signaling components, SA-, and JA-pathway genes. Conserved chromatin remodeling complexes are composed of multiple subunits which regulate gene expression by altering nucleosome composition and interactions at the chromatin structure level. The SWI/SNF chromatin remodeling complexes were initially identified from Saccharomyces cerevisiae. They have been broadly studied in many different organisms and can either “read” or “shape” the chromatin landscapes to regulate gene transcription (Raab et al., 2015; Pulice and Kadoch, 2016). SWI/SNF complexes facilitate the activation or repression of the target gene transcription by binding to the DNA or interacting with histones and transcription factors (Zhu et al., 2013; Grossi et al., 2020).
In Arabidopsis, the SWI/SNF complex has a well-established role in gene expression regulation in plant growth and development (Han et al., 2015), but only a few studies link the function of SWI/SNF complex subunits to plant immunity (Figure 1B). Most studies on the role of SWI/SNF complexes in plant defense response focus on the SWI2/SNF2-like ATPase subunits. For instance, Swi2/Snf2-related 1 (SWR1) complex replaces the histone H2A with the histone variant H2A.Z to maintain the suppression of several SA-dependent defense genes (March-Diaz et al., 2008). These genes include CLASSY1 (CLSY1), which is implicated in the RdDM pathway, is required for accumulation of hc-siRNA, and interacts with H3K9 methylation (Luna and Ton, 2012; Zhang et al., 2013; Zhou et al., 2018), as well as DDM1, which is required for DNA methylation and regulates expression of NLRs (Jeddeloh et al., 1998, 1999; Stokes et al., 2002; Li et al., 2010; Cambiagno et al., 2021). In addition, the expression of NLR SNC1 is suppressed by SPLAYED (SYD), another SWI2/SNF2-like ATPase, which is confirmed by the elevated transcription in the syd mutant. However, no direct SYD binding site or DNA region has been identified (Walley et al., 2008; Johnson et al., 2015). This could be a result of indirect regulation mediated by SYD.
Other than the SWI2/SNF2-like ATPase subunits, a recent study revealed that Arabidopsis SWP73A, a SWI/SNF2 non-ATPase subunit and an ortholog of the mammalian BRG1-Associated Factor 60 (BAF60), acts as a negative regulator of a group of NLRs to prevent autoimmunity in the absence of pathogens (Huang et al., 2021). Upon infection of Pst (AvrRpt2) or Pst (AvrRPS4), SWP73A is silenced by two bacterial-induced sRNAs post-transcriptionally, which allows rapid induction of these NLRs to activate plant immune responses (Figure 1B). For some NLRs, such as RPS2 and ZAR1, SWP73A binds with H3K9me2 at their transcription starting site and promoter regions directly to potentiate its suppression function on the expression of these NLRs. For some other NLRs, such as RPS4 and RRS1, SWP73 does not bind to their promoters and transcription starting sites, but instead suppresses their expression indirectly by suppressing Cell Division Cycle 5 (CDC5), a key regulator of RNA splicing, which subsequently interferes with the alternative splicing of these NLRs (Huang et al., 2021). This finding uncovers a new layer of epigenetic control over the precise regulation of NLRs. Potential roles of other SWI/SNF complex subunits in plant immunity remain to be explored.
Other epigenetic regulators, such as ENHANCED DOWNY MILDEW 2 (EDM2), also help modulate the expression of Arabidopsis NLRs (Eulgem et al., 2007; Lai et al., 2020). EDM2 binds to the H3K9me2 at the proximal polyadenylation sites of RPP7 and RPP4, which suppresses the maturation of the short transcripts and promotes the accumulation of full length functional RPP7 and RPP4 (Tsuchiya and Eulgem, 2013). EDM2 binds to H3K9me2 at TEs inside or near NLR genes and plays a role in balancing transcript levels of these NLRs. While the expression level of full length RPP7 mRNA increases, EDM2 also represses the expression of other NLRs, which is evidenced by the fact that the edm2 mutant is more resistant to Pst.
Epigenetic Memories and Defense Priming
Biotic stress-induced epigenetic changes, triggered by bacteria, fungi, or insect herbivory, can sometimes be transmitted to the progeny, leading to transgenerational priming (Luna et al., 2012; Rasmann et al., 2012; Lopez Sanchez et al., 2021; Moran-Diez et al., 2021). A global clustering DNA methylation study revealed transgenerational acquired resistance-related patterns, which were identified after three generations of Pst exposure. The major change to DNA methylated regions occurred at the CG context in gene bodies (Stassen et al., 2018). A screening for Hpa-resistant Arabidopsis was performed using epigenetic recombinant inbred lines (epiRILs) generated from ddm1 mutant, which has reduced DNA methylation in all sequence contexts, crossing to wild-type plants. The selected Hpa-resistant lines had no growth defect and a stronger PR1 induction after Hpa infection compared to wild-type plants which revealed a priming of SA-inducible defenses. Through transcriptome and DNA methylome analysis of these Hpa-resistant epiRILs lines, it was found that genome-wide priming of defense-related genes is sufficient to provide quantitative disease resistance and is heritable (Furci et al., 2019). Therefore, Arabidopsis epigenomic responses at the DNA methylation level in previous generations could contribute to transgenerational acquired resistance.
In addition, several studies have revealed that chemical treatment can lead to epigenetic adjustment for enhanced plant disease resistance. For instance, β-aminobutyric acid (BABA) treatment primes Arabidopsis PTI against the necrotrophic bacteria, Pectobacterium carotovorum. This priming is mediated by H3K9K14ac and H3K4me2 (Po-Wen et al., 2013). Additionally, treatment with BABA or 2,6-dichloroisonicotinic acid in the common bean leads to enhancement in H3K4me3 and H3K36me3 at the promoter-exon regions of defense-associated genes (Martinez-Aguilar et al., 2016; Table 2). BABA treatment also has priming effects in potatoes and induces intergenerational resistance against oomycete Phytophthora infestans through epigenetic adjustment of H3K4me2 and H3K27me3 dynamics. After BABA treatment, H3K4me2 was shown to be transiently induced in NPR1 (Non-expressor of PR genes) and SNI1 (Suppressor of NPR1 Inducible 1) resulting in tuning of the SA-responsive gene and enhanced occupancy on the gene body of defense response genes WRKY1, PR1, and PR2 in primed plants and their descendants (Meller et al., 2018; Table 2). Progeny of the BABA-primed potato was shown to carry lower DNA methylation on the promoter of R3a NLR gene with a higher transcription level of R3a and activate to virulent P. infestans which secretes effector Avr3a (Kuznicki et al., 2019; Table 2). Thus, the priming response from chemical-primed treatment is highly regulated at the DNA and histone methylation levels.
Conclusion and Perspectives
Here, we reviewed recent advances on the regulatory role of epigenetic mechanisms, including RdDM, DNA methylation dynamics, histone modifications, and chromatin remodeling, in plant immune responses. The role of hc-siRNAs in guiding DNA methylation endogenously in the RdDM pathway has been well demonstrated, but whether hc-siRNAs can also direct cross-kingdom DNA methylation of target genes in interacting pathogens and organisms remains to be investigated. Epigenetic modifications, including DNA methylation and histone modification, could be heritable without the need to introduce an initial trigger for targeted manipulation, making it an attractive approach to modify a locus for the desired disease-resistant trait. Notably, the components needed to manipulate methylation can be delivered by direct application of RNAs, protein regulators, or priming molecules to plant cells (Mauch-Mani et al., 2017; Que et al., 2019; Gallego-Bartolome, 2020; Watanabe et al., 2021). Therefore, it is possible to bypass genetic transformation to manipulate gene expression through epigenetic modification in economically important crops.
A major challenge in crop management lies in the multiple biotic and abiotic stresses occurring concurrently in the field. The limited information of crosstalk between abiotic stress and biotic stress responses in crop plants makes it difficult to develop strategies to trigger an efficient broad-spectrum resistance response. Though studies probing the relationship between epigenetic regulation and plant biotic stress are emerging, the connection between epigenetic modification at gene loci and disease-resistant traits in different varieties of crops still needs further exploration. In the future, this could become even more important to deal with unpredictable effects due to climate change.
Author Contributions
C-YH and HJ wrote the manuscript and organized the table and figure. All authors contributed to the article and approved the submitted version.
Funding
We appreciate the support for Dr. Hailing Jin’s laboratory from the National Institute of Health (R35 GM136379), the National Science Foundation (IOS2017314), the United States Department of Agriculture National Institute of Food and Agriculture (2021-67013-34258 and 2019-70016-29067), the Australian Research Council Industrial Transformation Research Hub (IH190100022), and the CIFAR Fungal Kingdom fellowship to HJ.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We sincerely apologize that we could not include many related interesting studies due to space limitations. We thank Rachael Hamby and Angela Chen for editing the manuscript.
References
Agorio, A., and Vera, P. (2007). ARGONAUTE4 is required for resistance to Pseudomonas syringae in Arabidopsis. Plant Cell 19, 3778–3790. doi: 10.1105/tpc.107.054494
Allen, E., Xie, Z., Gustafson, A. M., and Carrington, J. C. (2005). microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121, 207–221. doi: 10.1016/j.cell.2005.04.004
Annacondia, M. L., Markovic, D., Reig-Valiente, J. L., Scaltsoyiannes, V., Pieterse, C. M. J., Ninkovic, V., et al. (2021). Aphid feeding induces the relaxation of epigenetic control and the associated regulation of the defense response in Arabidopsis. New Phytol. 230, 1185–1200. doi: 10.1111/nph.17226
Arnold, P. A., Kruuk, L. E. B., and Nicotra, A. B. (2019). How to analyse plant phenotypic plasticity in response to a changing climate. New Phytol. 222, 1235–1241. doi: 10.1111/nph.15656
Bakhtiari, M., Formenti, L., Caggia, V., Glauser, G., and Rasmann, S. (2019). Variable effects on growth and defense traits for plant ecotypic differentiation and phenotypic plasticity along elevation gradients. Ecol. Evol. 9, 3740–3755. doi: 10.1002/ece3.4999
Bigeard, J., Colcombet, J., and Hirt, H. (2015). Signaling mechanisms in pattern-triggered immunity (PTI). Mol. Plant 8, 521–539. doi: 10.1016/j.molp.2014.12.022
Bond, D. M., and Baulcombe, D. C. (2015). Epigenetic transitions leading to heritable, RNA-mediated de novo silencing in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U. S. A. 112, 917–922. doi: 10.1073/pnas.1413053112
Borges, F., and Martienssen, R. A. (2015). The expanding world of small RNAs in plants. Nat. Rev. Mol. Cell Biol. 16, 727–741. doi: 10.1038/nrm4085
Burger, M., and Chory, J. (2019). Stressed out about hormones: how plants orchestrate immunity. Cell Host Microbe 26, 163–172. doi: 10.1016/j.chom.2019.07.006
Cai, Q., Qiao, L., Wang, M., He, B., Lin, F. M., Palmquist, J., et al. (2018). Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science 360, 1126–1129. doi: 10.1126/science.aar4142
Cambiagno, D. A., Nota, F., Zavallo, D., Rius, S., Casati, P., Asurmendi, S., et al. (2018). Immune receptor genes and pericentromeric transposons as targets of common epigenetic regulatory elements. Plant J. 96, 1178–1190. doi: 10.1111/tpj.14098
Cambiagno, D. A., Torres, J. R., and Alvarez, M. E. (2021). Convergent epigenetic mechanisms avoid constitutive expression of immune receptor gene subsets. Front. Plant Sci. 12:703667. doi: 10.3389/fpls.2021.703667
Campo, S., Sanchez-Sanuy, F., Camargo-Ramirez, R., Gomez-Ariza, J., Baldrich, P., Campos-Soriano, L., et al. (2021). A novel transposable element-derived microRNA participates in plant immunity to rice blast disease. Plant Biotechnol. J. 19, 1798–1811. doi: 10.1111/pbi.13592
Chan, C., and Zimmerli, L. (2019). The histone demethylase IBM1 positively regulates arabidopsis immunity by control of defense gene expression. Front. Plant Sci. 10:1587. doi: 10.3389/fpls.2019.01587
Choi, J., Lyons, D. B., Kim, M. Y., Moore, J. D., and Zilberman, D. (2020). DNA methylation and histone H1 jointly repress transposable elements and aberrant intragenic transcripts. Mol. Cell 77, 310–323.e7. doi: 10.1016/j.molcel.2019.10.011
Conrath, U., Beckers, G. J., Langenbach, C. J., and Jaskiewicz, M. R. (2015). Priming for enhanced defense. Annu. Rev. Phytopathol. 53, 97–119. doi: 10.1146/annurev-phyto-080614-120132
Cuerda-Gil, D., and Slotkin, R. K. (2016). Non-canonical RNA-directed DNA methylation. Nat. Plants 2:16163. doi: 10.1038/nplants.2016.163
Cui, H., Tsuda, K., and Parker, J. E. (2015). Effector-triggered immunity: from pathogen perception to robust defense. Annu. Rev. Plant Biol. 66, 487–511. doi: 10.1146/annurev-arplant-050213-040012
Deal, R. B., and Henikoff, S. (2011). Histone variants and modifications in plant gene regulation. Curr. Opin. Plant Biol. 14, 116–122. doi: 10.1016/j.pbi.2010.11.005
Deleris, A., Halter, T., and Navarro, L. (2016). DNA methylation and demethylation in plant immunity. Annu. Rev. Phytopathol. 54, 579–603. doi: 10.1146/annurev-phyto-080615-100308
Deng, Y., Zhai, K., Xie, Z., Yang, D., Zhu, X., Liu, J., et al. (2017). Epigenetic regulation of antagonistic receptors confers rice blast resistance with yield balance. Science 355, 962–965. doi: 10.1126/science.aai8898
Dowen, R. H., Pelizzola, M., Schmitz, R. J., Lister, R., Dowen, J. M., Nery, J. R., et al. (2012). Widespread dynamic DNA methylation in response to biotic stress. Proc. Natl. Acad. Sci. U. S. A. 109, E2183–E2191. doi: 10.1073/pnas.1209329109
Du, J., Johnson, L. M., Jacobsen, S. E., and Patel, D. J. (2015). DNA methylation pathways and their crosstalk with histone methylation. Nat. Rev. Mol. Cell Biol. 16, 519–532. doi: 10.1038/nrm4043
Erdmann, R. M., and Picard, C. L. (2020). RNA-directed DNA methylation. PLoS Genet. 16:e1009034. doi: 10.1371/journal.pgen.1009034
Eulgem, T., Tsuchiya, T., Wang, X. J., Beasley, B., Cuzick, A., Tor, M., et al. (2007). EDM2 is required for RPP7-dependent disease resistance in Arabidopsis and affects RPP7 transcript levels. Plant J. 49, 829–839. doi: 10.1111/j.1365-313X.2006.02999.x
Fei, Q., Xia, R., and Meyers, B. C. (2013). Phased, secondary, small interfering RNAs in posttranscriptional regulatory networks. Plant Cell 25, 2400–2415. doi: 10.1105/tpc.113.114652
Fu, Z. Q., and Dong, X. (2013). Systemic acquired resistance: turning local infection into global defense. Annu. Rev. Plant Biol. 64, 839–863. doi: 10.1146/annurev-arplant-042811-105606
Furci, L., Jain, R., Stassen, J., Berkowitz, O., Whelan, J., Roquis, D., et al. (2019). Identification and characterisation of hypomethylated DNA loci controlling quantitative resistance in Arabidopsis. eLife 8:e40655. doi: 10.7554/eLife.40655
Gallego-Bartolome, J. (2020). DNA methylation in plants: mechanisms and tools for targeted manipulation. New Phytol. 227, 38–44. doi: 10.1111/nph.16529
Gohlke, J., Scholz, C. J., Kneitz, S., Weber, D., Fuchs, J., Hedrich, R., et al. (2013). DNA methylation mediated control of gene expression is critical for development of crown gall tumors. PLoS Genet. 9:e1003267. doi: 10.1371/journal.pgen.1003267
Grossi, E., Raimondi, I., Goni, E., Gonzalez, J., Marchese, F. P., Chapaprieta, V., et al. (2020). A lncRNA-SWI/SNF complex crosstalk controls transcriptional activation at specific promoter regions. Nat. Commun. 11:936. doi: 10.1038/s41467-020-14623-3
Halter, T., Wang, J., Amesefe, D., Lastrucci, E., Charvin, M., Singla Rastogi, M., et al. (2021). The Arabidopsis active demethylase ROS1 cis-regulates defence genes by erasing DNA methylation at promoter-regulatory regions. eLife 10:e62994. doi: 10.7554/eLife.62994
Han, S. K., Wu, M. F., Cui, S., and Wagner, D. (2015). Roles and activities of chromatin remodeling ATPases in plants. Plant J. 83, 62–77. doi: 10.1111/tpj.12877
He, G., Elling, A. A., and Deng, X. W. (2011). The epigenome and plant development. Annu. Rev. Plant Biol. 62, 411–435. doi: 10.1146/annurev-arplant-042110-103806
Hu, Y., Lu, Y., Zhao, Y., and Zhou, D. X. (2019). Histone acetylation dynamics integrates metabolic activity to regulate plant response to stress. Front. Plant Sci. 10:1236. doi: 10.3389/fpls.2019.01236
Huang, C. Y., Rangel, D. S., Qin, X., Bui, C., Li, R., Jia, Z., et al. (2021). The chromatin-remodeling protein BAF60/SWP73A regulates the plant immune receptor NLRs. Cell Host Microbe 29, 425–434.e4. doi: 10.1016/j.chom.2021.01.005
Huang, C. Y., Wang, H., Hu, P., Hamby, R., and Jin, H. (2019). Small RNAs - big players in plant-microbe interactions. Cell Host Microbe 26, 173–182. doi: 10.1016/j.chom.2019.07.021
Irieda, H., Inoue, Y., Mori, M., Yamada, K., Oshikawa, Y., Saitoh, H., et al. (2019). Conserved fungal effector suppresses PAMP-triggered immunity by targeting plant immune kinases. Proc. Natl. Acad. Sci. U. S. A. 116, 496–505. doi: 10.1073/pnas.1807297116
Jeddeloh, J. A., Bender, J., and Richards, E. J. (1998). The DNA methylation locus DDM1 is required for maintenance of gene silencing in Arabidopsis. Genes Dev. 12, 1714–1725. doi: 10.1101/gad.12.11.1714
Jeddeloh, J. A., Stokes, T. L., and Richards, E. J. (1999). Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat. Genet. 22, 94–97. doi: 10.1038/8803
Johnson, K. C., Xia, S., Feng, X., and Li, X. (2015). The chromatin remodeler SPLAYED negatively regulates SNC1-mediated immunity. Plant Cell Physiol. 56, 1616–1623. doi: 10.1093/pcp/pcv087
Jones, J. D., Vance, R. E., and Dangl, J. L. (2016). Intracellular innate immune surveillance devices in plants and animals. Science 354:aaf6395. doi: 10.1126/science.aaf6395
Katiyar-Agarwal, S., Gao, S., Vivian-Smith, A., and Jin, H. (2007). A novel class of bacteria-induced small RNAs in Arabidopsis. Genes Dev. 21, 3123–3134. doi: 10.1101/gad.1595107
Katiyar-Agarwal, S., Morgan, R., Dahlbeck, D., Borsani, O., Villegas, A. Jr., Zhu, J. K., et al. (2006). A pathogen-inducible endogenous siRNA in plant immunity. Proc. Natl. Acad. Sci. U. S. A. 103, 18002–18007. doi: 10.1073/pnas.0608258103
Kim, M. Y., and Zilberman, D. (2014). DNA methylation as a system of plant genomic immunity. Trends Plant Sci. 19, 320–326. doi: 10.1016/j.tplants.2014.01.014
Kong, W., Xia, X., Wang, Q., Liu, L. W., Zhang, S., Ding, L., et al. (2020). Impact of DNA demethylases on the DNA methylation and transcription of Arabidopsis NLR genes. Front. Genet. 11:460. doi: 10.3389/fgene.2020.00460
Kumar, S., Chinnusamy, V., and Mohapatra, T. (2018). Epigenetics of modified DNA bases: 5-methylcytosine and beyond. Front. Genet. 9:640. doi: 10.3389/fgene.2018.00640
Kuznicki, D., Meller, B., Arasimowicz-Jelonek, M., Braszewska-Zalewska, A., Drozda, A., and Floryszak-Wieczorek, J. (2019). BABA-induced DNA methylome adjustment to intergenerational defense priming in potato to Phytophthora infestans. Front. Plant Sci. 10:650. doi: 10.3389/fpls.2019.00650
Lai, Y., and Eulgem, T. (2018). Transcript-level expression control of plant NLR genes. Mol. Plant Pathol. 19, 1267–1281. doi: 10.1111/mpp.12607
Lai, Y., Lu, X. M., Daron, J., Pan, S., Wang, J., Wang, W., et al. (2020). The Arabidopsis PHD-finger protein EDM2 has multiple roles in balancing NLR immune receptor gene expression. PLoS Genet. 16:e1008993. doi: 10.1371/journal.pgen.1008993
Li, X., Kapos, P., and Zhang, Y. (2015). NLRs in plants. Curr. Opin. Immunol. 32, 114–121. doi: 10.1016/j.coi.2015.01.014
Li, Y., Tessaro, M. J., Li, X., and Zhang, Y. (2010). Regulation of the expression of plant resistance gene SNC1 by a protein with a conserved BAT2 domain. Plant Physiol. 153, 1425–1434. doi: 10.1104/pp.110.156240
Liu, C., Lu, F., Cui, X., and Cao, X. (2010). Histone methylation in higher plants. Annu. Rev. Plant Biol. 61, 395–420. doi: 10.1146/annurev.arplant.043008.091939
Lolle, S., Stevens, D., and Coaker, G. (2020). Plant NLR-triggered immunity: from receptor activation to downstream signaling. Curr. Opin. Immunol. 62, 99–105. doi: 10.1016/j.coi.2019.12.007
Lopez, A., Ramirez, V., Garcia-Andrade, J., Flors, V., and Vera, P. (2011). The RNA silencing enzyme RNA polymerase v is required for plant immunity. PLoS Genet. 7:e1002434. doi: 10.1371/journal.pgen.1002434
Lopez Sanchez, A., Pascual-Pardo, D., Furci, L., Roberts, M. R., and Ton, J. (2021). Costs and benefits of transgenerational induced resistance in Arabidopsis. Front. Plant Sci. 12:644999. doi: 10.3389/fpls.2021.644999
Lopez Sanchez, A., Stassen, J. H., Furci, L., Smith, L. M., and Ton, J. (2016). The role of DNA (de)methylation in immune responsiveness of Arabidopsis. Plant J. 88, 361–374. doi: 10.1111/tpj.13252
Luna, E., Bruce, T. J., Roberts, M. R., Flors, V., and Ton, J. (2012). Next-generation systemic acquired resistance. Plant Physiol. 158, 844–853. doi: 10.1104/pp.111.187468
Luna, E., and Ton, J. (2012). The epigenetic machinery controlling transgenerational systemic acquired resistance. Plant Signal. Behav. 7, 615–618. doi: 10.4161/psb.20155
March-Diaz, R., Garcia-Dominguez, M., Lozano-Juste, J., Leon, J., Florencio, F. J., and Reyes, J. C. (2008). Histone H2A.Z and homologues of components of the SWR1 complex are required to control immunity in Arabidopsis. Plant J. 53, 475–487. doi: 10.1111/j.1365-313X.2007.03361.x
Martinez-Aguilar, K., Ramirez-Carrasco, G., Hernandez-Chavez, J. L., Barraza, A., and Alvarez-Venegas, R. (2016). Use of BABA and INA as activators of a primed state in the common bean (Phaseolus vulgaris L.). Front. Plant Sci. 7:653. doi: 10.3389/fpls.2016.00653
Matzke, M. A., and Mosher, R. A. (2014). RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat. Rev. Genet. 15, 394–408. doi: 10.1038/nrg3683
Mauch-Mani, B., Baccelli, I., Luna, E., and Flors, V. (2017). Defense priming: an adaptive part of induced resistance. Annu. Rev. Plant Biol. 68, 485–512. doi: 10.1146/annurev-arplant-042916-041132
Meller, B., Kuznicki, D., Arasimowicz-Jelonek, M., Deckert, J., and Floryszak-Wieczorek, J. (2018). BABA-primed histone modifications in potato for intergenerational resistance to Phytophthora infestans. Front. Plant Sci. 9:1228. doi: 10.3389/fpls.2018.01228
Mine, A., Berens, M. L., Nobori, T., Anver, S., Fukumoto, K., Winkelmuller, T. M., et al. (2017). Pathogen exploitation of an abscisic acid- and jasmonate-inducible MAPK phosphatase and its interception by Arabidopsis immunity. Proc. Natl. Acad. Sci. U. S. A. 114, 7456–7461. doi: 10.1073/pnas.1702613114
Moran-Diez, M. E., Martinez de Alba, A. E., Rubio, M. B., Hermosa, R., and Monte, E. (2021). Trichoderma and the plant heritable priming responses. J. Fungi 7:318. doi: 10.3390/jof7040318
Nürnberger, T., and Kemmerling, B. (2018). “Pathogen-associated molecular patterns (PAMP) and PAMP-triggered immunity,” in Annual Plant Reviews Online. ed. J. A. Roberts (Wiley), 16–47.
Nuthikattu, S., McCue, A. D., Panda, K., Fultz, D., DeFraia, C., Thomas, E. N., et al. (2013). The initiation of epigenetic silencing of active transposable elements is triggered by RDR6 and 21-22 nucleotide small interfering RNAs. Plant Physiol. 162, 116–131. doi: 10.1104/pp.113.216481
Po-Wen, C., Singh, P., and Zimmerli, L. (2013). Priming of the Arabidopsis pattern-triggered immunity response upon infection by necrotrophic Pectobacterium carotovorum bacteria. Mol. Plant Pathol. 14, 58–70. doi: 10.1111/j.1364-3703.2012.00827.x
Pulice, J. L., and Kadoch, C. (2016). Composition and function of mammalian SWI/SNF chromatin remodeling complexes in human disease. Cold Spring Harb. Symp. Quant. Biol. 81, 53–60. doi: 10.1101/sqb.2016.81.031021
Que, Q., Chilton, M. M., Elumalai, S., Zhong, H., Dong, S., and Shi, L. (2019). Repurposing macromolecule delivery tools for plant genetic modification in the era of precision genome engineering. Methods Mol. Biol. 1864, 3–18. doi: 10.1007/978-1-4939-8778-8_1
Raab, J. R., Resnick, S., and Magnuson, T. (2015). Genome-wide transcriptional regulation mediated by biochemically distinct SWI/SNF complexes. PLoS Genet. 11:e1005748. doi: 10.1371/journal.pgen.1005748
Ramirez-Prado, J. S., Piquerez, S. J. M., Bendahmane, A., Hirt, H., Raynaud, C., and Benhamed, M. (2018). Modify the histone to win the battle: chromatin dynamics in plant-pathogen interactions. Front. Plant Sci. 9:355. doi: 10.3389/fpls.2018.00355
Rasmann, S., De Vos, M., Casteel, C. L., Tian, D., Halitschke, R., Sun, J. Y., et al. (2012). Herbivory in the previous generation primes plants for enhanced insect resistance. Plant Physiol. 158, 854–863. doi: 10.1104/pp.111.187831
Robert-Seilaniantz, A., Grant, M., and Jones, J. D. (2011). Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annu. Rev. Phytopathol. 49, 317–343. doi: 10.1146/annurev-phyto-073009-114447
Schumann, U., Lee, J. M., Smith, N. A., Zhong, C., Zhu, J. K., Dennis, E. S., et al. (2019). DEMETER plays a role in DNA demethylation and disease response in somatic tissues of Arabidopsis. Epigenetics 14, 1074–1087. doi: 10.1080/15592294.2019.1631113
Stassen, J. H. M., Lopez, A., Jain, R., Pascual-Pardo, D., Luna, E., Smith, L. M., et al. (2018). The relationship between transgenerational acquired resistance and global DNA methylation in Arabidopsis. Sci. Rep. 8:14761. doi: 10.1038/s41598-018-32448-5
Stokes, T. L., Kunkel, B. N., and Richards, E. J. (2002). Epigenetic variation in Arabidopsis disease resistance. Genes Dev. 16, 171–182. doi: 10.1101/gad.952102
Tsuchiya, T., and Eulgem, T. (2013). An alternative polyadenylation mechanism coopted to the Arabidopsis RPP7 gene through intronic retrotransposon domestication. Proc. Natl. Acad. Sci. U. S. A. 110, E3535–E3543. doi: 10.1073/pnas.1312545110
Walley, J. W., Rowe, H. C., Xiao, Y., Chehab, E. W., Kliebenstein, D. J., Wagner, D., et al. (2008). The chromatin remodeler SPLAYED regulates specific stress signaling pathways. PLoS Pathog. 4:e1000237. doi: 10.1371/journal.ppat.1000237
Watanabe, K., Odahara, M., Miyamoto, T., and Numata, K. (2021). Fusion peptide-based biomacromolecule delivery system for plant cells. ACS Biomater Sci. Eng. 7, 2246–2254. doi: 10.1021/acsbiomaterials.1c00227
Weiberg, A., Wang, M., Lin, F. M., Zhao, H., Zhang, Z., Kaloshian, I., et al. (2013). Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 342, 118–123. doi: 10.1126/science.1239705
West, P. T., Li, Q., Ji, L., Eichten, S. R., Song, J., Vaughn, M. W., et al. (2014). Genomic distribution of H3K9me2 and DNA methylation in a maize genome. PLoS One 9:e105267. doi: 10.1371/journal.pone.0105267
Xiao, J., Lee, U. S., and Wagner, D. (2016). Tug of war: adding and removing histone lysine methylation in Arabidopsis. Curr. Opin. Plant Biol. 34, 41–53. doi: 10.1016/j.pbi.2016.08.002
Yu, A., Lepere, G., Jay, F., Wang, J., Bapaume, L., Wang, Y., et al. (2013). Dynamics and biological relevance of DNA demethylation in Arabidopsis antibacterial defense. Proc. Natl. Acad. Sci. U. S. A. 110, 2389–2394. doi: 10.1073/pnas.1211757110
Zhang, H., Lang, Z., and Zhu, J. K. (2018). Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell Biol. 19, 489–506. doi: 10.1038/s41580-018-0016-z
Zhang, H., Ma, Z. Y., Zeng, L., Tanaka, K., Zhang, C. J., Ma, J., et al. (2013). DTF1 is a core component of RNA-directed DNA methylation and may assist in the recruitment of pol IV. Proc. Natl. Acad. Sci. U. S. A. 110, 8290–8295. doi: 10.1073/pnas.1300585110
Zhang, H., Tao, Z., Hong, H., Chen, Z., Wu, C., Li, X., et al. (2016). Transposon-derived small RNA is responsible for modified function of WRKY45 locus. Nat. Plants 2:16016. doi: 10.1038/nplants.2016.16
Zhou, M., Palanca, A. M. S., and Law, J. A. (2018). Locus-specific control of the de novo DNA methylation pathway in Arabidopsis by the CLASSY family. Nat. Genet. 50, 865–873. doi: 10.1038/s41588-018-0115-y
Zhou, J., Wang, X., He, K., Charron, J. B., Elling, A. A., and Deng, X. W. (2010). Genome-wide profiling of histone H3 lysine 9 acetylation and dimethylation in Arabidopsis reveals correlation between multiple histone marks and gene expression. Plant Mol. Biol. 72, 585–595. doi: 10.1007/s11103-009-9594-7
Zhou, J. M., and Zhang, Y. (2020). Plant immunity: danger perception and signaling. Cell 181, 978–989. doi: 10.1016/j.cell.2020.04.028
Keywords: epigenetics, DNA methylation, histone modification, small RNAs, chromatin remodelers, plant defense, plant-microbes interaction
Citation: Huang C-Y and Jin H (2022) Coordinated Epigenetic Regulation in Plants: A Potent Managerial Tool to Conquer Biotic Stress. Front. Plant Sci. 12:795274. doi: 10.3389/fpls.2021.795274
Edited by:
Stefan Simm, Universitätsmedizin Greifswald, GermanyReviewed by:
Daisuke Miki, Shanghai Center for Plant Stress Biology, Shanghai Institute for Biological Sciences (CAS), ChinaCopyright © 2022 Huang and Jin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hailing Jin, aGFpbGluZ2pAdWNyLmVkdQ==
 Chien-Yu Huang
Chien-Yu Huang Hailing Jin
Hailing Jin