- 1The Cancer Hospital of the University of Chinese Academy of Sciences (Zhejiang Cancer Hospital), Institute of Basic Medicine and Cancer, Chinese Academy of Sciences, Hangzhou, China
- 2College of Biomedical Engineering and Instrument Science, Zhejiang University, Hangzhou, China
Mahonia bealei (M. bealei) is a traditional Chinese medicine containing a high alkaloid content used to treat various diseases. Generally, only dried root and stem are used as medicines, considering that the alkaloid content in M. bealei leaves is lower than in the stems and roots. Some previous research found that alkaloid and flavonoid contents in the M. bealei leaves may increase when exposed to ultraviolet B (UV-B) radiation. However, the underlying mechanism of action is still unclear. In this study, we used titanium dioxide material enrichment and mass-based label-free quantitative proteomics techniques to explore the effect and mechanism of M. bealei leaves when exposed to UV-B treatment. Our data suggest that UV-B radiation increases the ATP content, photosynthetic pigment content, and some enzymatic/nonenzymatic indicators in the leaves of M. bealei. Moreover, phosphoproteomics suggests phosphoproteins related to mitogen-activated protein kinase (MAPK) signal transduction and the plant hormone brassinosteroid signaling pathway as well as phosphoproteins related to photosynthesis, glycolysis, the tricarboxylic acid cycle, and the amino acid synthesis/metabolism pathway are all affected by UV-B radiation. These results suggest that the UV-B radiation activates the oxidative stress response, MAPK signal transduction pathway, and photosynthetic energy metabolism pathway, which may lead to the accumulation of secondary metabolites in M. bealei leaves.
Introduction
Mahonia Bealei is a kind of evergreen shrub widely grown in China and some other countries in Asia, Europe, and America. Its main active ingredients are berberine, jatrorrhizine, and palmatine, which have heat-clearing, detoxifying, and antiphlogistic effects and may be used to treat stomach disorders, inflammation, infections like hepatitis, and skin-related diseases. In addition, a previous study (Deng et al., 2013) points out that, besides berberine, jatrorrhizine, and palmatine, Alishan Mahonia contains columbamine, and its 1-diphenyl-2-trinitrophenylhydrazine (DPPH) radical scavenging activity is five times higher than the activity of vitamin C. Alishan Mahonia may exert a certain inhibitory effect on pain (Hu et al., 2011). Moreover, studies also find that jateorhizine, berberine, and palmatine from Mahonia can inhibit the activity of fungus (Wang et al., 2018). So far, only dried roots and stems named Gonglaomu were officially used as medicines in China, considering that the alkaloid contents in M. bealei leaves are lower than those in stems and roots (Zhu et al., 2015). Therefore, studies on increasing the alkaloid contents in M. bealei leaves could be valuable for improving medicinal material utilization.
As one of the environmental factors, UV-B radiation can alter the primary and secondary metabolism in plants. Studies show that UV-B stimulation can significantly increase the content of phenols and the activity of antioxidant enzymes in Salvia Miltiorrhiza leaves (Liu et al., 2013). Wang et al. (2016) find that UV-B radiation can stimulate tanshinone production in hairy root cultures of Salvia Miltiorrhiza, and when combining UV-B radiation with methyl jasmonate treatment, the content of tanshinone can be increased by 4.9 times over the control. Moreover, an integrative omic study indicates that the NAC1 gene is positively involved in the increase of salvianolic acid in S. Miltiorrhiza under UV-B radiation (Ying et al., 2020). Chen et al. (2020) investigated the metabolic variations in Dendrobium officinal Kimura et Migo stem under UV-B radiation and found that the treatment may increase flavonoids, polysaccharides, and alkaloids. Furthermore, in Chrysanthemum morifolium leaves, caffeoylquinic acid, fatty acid, and flavonoid contents all increased when the leaves were exposed to UV-B radiation (Yang et al., 2018). In previous studies, we also find that the contents of benzylisoquinoline alkaloids, such as berberine, jateorhizine, palmatine, and columbamine, in M. bealei leaves increased under UV-B radiation and dark treatment (Zhang L. et al., 2014; Zhu et al., 2021). We also discovered that the enhanced tricarboxylic acid (TCA) cycle under UV-B radiation might regulate the energy metabolism to provide extra power for the biosynthesis of secondary metabolites. However, the mechanism of induction is not fully understood.
Protein post-translational modifications (PTMs) include protein phosphorylation, ubiquitylation, glycosylation, methylation, acetylation, and so on. Protein phosphorylation is one of the most well-studied modification types involving the regulation of transcription, translation, protein degradation, and cell signaling and communication. For instance, sucrose nondependent protein kinase 2.10 (SnRK2.10) protein phosphorylation in Arabidopsis thaliana under salt stress is regarded as the response to dehydration pressure (Maszkowska et al., 2019), and phosphorylation increased in proteins related to secondary metabolism in sugarcane under drought stress (Vital et al., 2017). In C. roseus, UV-B radiation activates calcium-dependent protein phosphorylation, the glycolysis pathway, and the TCA-cycle proteins phosphorylation changes as well as an oxidative stress response affecting secondary metabolic pathways, such as aromatic amino acids and phenylpropanoids (Zhong et al., 2019). Yet the protein phosphorylation levels in M. bealei under UV-B radiation might be related to metabolic changes, especially for alkaloids.
In this study, we used titanium dioxide (TiO2) material enrichment and mass-based label-free quantitative proteomics techniques to explore the effect and mechanism of M. bealei leaves when exposed to UV-B treatment. Label-free proteomic and mass spectrum detection techniques were applied for phosphopeptide identification. Furthermore, phosphoproteomic results were confirmed by qRT-PCR analyses.
Materials and Methods
Plant Material and Treatment
Seedlings of Mahonia bealei (Fort.) Carr. were provided by the College of Pharmacy Zhejiang University (Hangzhou, China). Plants were 3 years old and grown in the greenhouse. UV-B treatment was then carried out in special UV-B chambers with four UV-B lamps (Philips 20 W ultraviolet B TL20W/12RS, Philips). The intensity of UV-B irradiation on the leaf surface was 104.4 kJ m –2 d –1, which was measured from 275 to 320 nm with a UV radiometer (Beijing Normal University photoelectric instrument factory, Beijing, China). The following conditions were applied: 26°C and humidity of 58%. The plants were divided into two groups: plants exposed to UV-B radiation for 6 h were labeled as the treatment or UV-B group and the control group comprised those growing under regular light for 6 h. After the parallel treatment, the leaves were collected and snap-frozen in liquid nitrogen and then stored in a refrigerator at −80°C for subsequent analysis. Three independent biological replicates were conducted in our study (Supplementary Figure 1).
Measurement of ATP Content
ATP-content determination was carried out using an ATP Content Assay kit (Solarbio, Beijing, China). In simple terms, fresh leaves of M. bealei were fully ground on ice with 1 mL extraction buffer and then centrifuged by 8,000 × g at 4°C for 10 min. The supernatant was collected and mixed with 500 μL of chloroform, followed by centrifugation of 10,000 × g at 4°C for 3 min, after which the supernatant was collected. The water bath reaction was performed according to the requirements of the kit. The ATP content was obtained by comparing the absorbance increase at 340 nm before and after the reaction with the absorbance increase of ATP standard substance.
Analysis of Physiological Changes
The peroxidase activity (POD), superoxide dismutase activity (SOD), total antioxidant capacity (T-AOC), plant malondialdehyde (MDA) content, and hydrogen peroxide (H2O2) content were measured by different kits following the manufacturer’s protocols (Nanjingjiancheng, Jiangsu, China). Phenylalanine ammonia-lyase (PAL) activity was determined as previously described (Huang et al., 2010) with minor modifications. Leaf samples were homogenized within 1 mL of extracting solution on ice and centrifuged by 12,000 × g at 4°C for 30 min. The supernatant was used in the final reaction mixture. PAL activity was measured by recording the absorbance at 290 nm.
Contents of carotenoids (Car), chlorophyll a (Chl a), and chlorophyll b (Chl b) were determined according to Wellburn and Lichtenthaler (1984). The content of brassinosteroid was determined by a Plant Brassinosteroids (BR) ELISA Kit (Boshen Biotechnology, Jiangsu, China).
For the total flavonoid assay, 0.5 mL of 5% NaNO2 was added into 1 mL methanol extraction of leaves. Next, 0.5 mL of 10% Al (NO3)3 was added to the reaction mixture and mixed with 1.5 mL of 2 M NaOH. After incubation for 15 min, the absorbance was measured at 510 nm (Li et al., 2015). Total anthocyanin content was measured as described by Neff and Chory (1998). For total alkaloid analysis, the obtained residue was dissolved in 1 mL of phosphate buffer solution (pH 4.5) after which the solution was transferred to a glass tube, and 1 mL of bromocresol green solution (0.03%) was added. After 30 min, 1 mL of chloroform was added, followed by vortexing for 2 min. The lower layer was separated after 5 min. The extraction was repeated once, and the lower phase was combined. The extracts were analyzed by using a UV–Vis spectrophotometer at 418 nm. The data were expressed as mg berberine equivalents (BE)/g fresh weight (FW) with berberine as a reference standard for the total alkaloid assay (Zhu et al., 2021). Next, the Pearson correlation coefficient was used to analyze the correlation between BR, total alkaloids, total flavonoids, and anthocyanins.
Protein Extraction
A portion (0.5 g) of the samples were ground into powder in liquid nitrogen and then transferred to acetone solution containing 10% trichloroacetic acid and 0.07% 2-mercaptoethanol. After vortexing, the samples were incubated at −20°C for 1 h, and the supernatant was discarded after centrifugation at 9,000 × g for 20 min at 4°C. The precipitation was then cleaned twice with acetone solution containing 0.07% 2-mercaptoethanol. After drying, the precipitation was lysed at room temperature for 1 h with lysis buffer (7 M urea, 2 M thiourea, 5% CHAPS, and 2 mM tributylphosphine). The resulting suspension was centrifuged at 20,000 × g for 20 min at 25°C, the supernatant was collected as total protein, and the protein concentration was determined by the Bradford method with bovine serum albumin as the standard.
Protein Purification and Digestion
One hundred microgram proteins were adjusted to the final volume of 100 μL. Next, 400 μL of methanol, 100 μL of chloroform, and 300 μL of double distilled water were added successively. After mixing, the sample protein was centrifuged at 20,000 × g for 10 min. The upper phase was discarded, and 300 μL of methanol was added to the lower phase and centrifuged at 20,000 × g for 10 min again. Solid particles were collected for subsequent enzymatic hydrolysis.
The purified protein was suspended in 50 mM ammonium bicarbonate, then reduced with 50 mM dithiothreitol at 56°C for 30 min in the dark, and subsequently alkylated with 50 mM iodoacetamide at 37°C for 30 min in the dark. Finally, the proteins were digested with trypsin at a 1:100 enzyme/protein ratio at 37°C for 16 h in the dark and preliminary desalted with Strata X Column (Phenomenex, United States).
Phosphopeptide Enrichment
The phosphorylated peptide enrichment kit (PTM BIO, Hangzhou, China) was used to enrich phosphopeptides. Peptides (1 mg) were resuspended in 1 mL of binding buffer. After mixing, 50 μL of TiO2 materials (8 mg) were added and lightly shaken for 30 min. They were centrifuged by 2,000 × g for 3 min to discard the supernatant, and TiO2 materials were washed using washing buffer. Finally, phosphopeptides were eluted, dried, and dissolved in 0.1% formic acid for nanoflow liquid chromatography-mass spectrometry.
Analysis Using Nanoliquid Chromatography-Mass Spectrometry
The peptides dissolved in 0.1% (v/v) formic acid were subjected to a homemade reverse analysis column (15 cm length, 75 μm i.d.). Buffer phase A was the 0.1% formic acid and 2% acetonitrile, and buffer phase B contained 90% acetonitrile and 0.1% formic acid. On an EASY nLC 1000 UPLC System, the following linear separation gradient was set: 0–86 min, 17% B. 86–110 min, 17–28% B, 110–115 min, 28–80% B, 115–120 min at 80% B, flow rate 500 nL/min.
The peptides were subjected to an NSI source followed by tandem mass spectrometry (MS/MS) in Q Exactive™ Plus (Thermo) coupled online to the UPLC. The full scanning m/z scan range was 350–1,800 with a voltage of 2.0 kV. An Orbitrap detects the complete peptide at 70,000 resolution. For selecting the peptides for MS/MS, HCD was set to 28% to detect fragments at 35,000 resolution in the Orbitrap A data-dependent program that scans 20,000 ions/s after one MS scan, dynamically excludes 15.0 s. Automatic gain control (AGC) was set to 1E5. The fixed first mass was set to 100 m/z.
Protein Identification
Maxquant (V1.5.2.8) was used to retrieve the secondary mass spectrometry data and set the retrieval parameters: identified proteins were searched against the UniProtKB database (uniprot-reviewed viridiplantae with 40,254 entries, downloaded 2020.04.22). A reverse database was added to calculate the false positive rate (FPR) caused by random matching, and a common pollution database was added to the database to eliminate the influence of contaminated proteins in the identification results. Trypsin was used for digestion. The number of missing bits was set to two. The first search and the main search level mother ion quality error tolerance were set to 20 and 5 ppm, respectively. The quality of secondary ions had an error tolerance of 0.02 Da. The cysteine alkylation was set to fixed, variable modifiers for oxidation of methionine, protein N acetylation, deamidation (NQ), and protein tyrosine phosphorylation of serine-threonine identification PSM of FDR was set to 1%.
Functional Analysis of Phosphoproteins
The submitted proteins were annotated for subcellular localization using Wolfpsort,1 a software that predicts subcellular localization. Heatmap Illustrator (heml.1.0.3.7) was used to provide an overall representation of intensities of the phosphorylation site of only phosphorylated proteins in the control or UV-B group. MapMan bin codes were used to categorize phosphoproteins. DAVID functional annotation analysis has phosphoproteins that were classified into molecular function, cellular component, and the biological process by GO terms. Then, the results were presented by the micro-bioinformation platform.2 The Kyoto Encyclopedia of Genes and Genomes (KEGG) database3 was used to annotate protein pathways, and the pathways were enriched by Metascape4 and performed by the micro-bioinformation platform. Phosphoproteins’ protein--protein interactions were analyzed in STRING.5 The required confidence score was set as >0.700 for highly confident interactions. A K-means algorithm in STRING clusters the network.
Motif Analysis of Phosphorylation Modification Sites
The Motif-X algorithm was used to extract significantly enriched amino acid motifs surrounding the identified phosphosites. The sequence window was limited to 13 amino acids, and foreground peptides were prealigned with the phosphosite centered. The proteome data set from the Uniprot database were used as the background database. The occurrence threshold was set at the minimum of 20 peptides, and the P-value threshold was set at < 10–6.
RNA Extraction and Quantitative Real-Time PCR
About 0.1 g of fresh leaf samples were taken from M. bealei and ground into powder in a sterilized mortar with liquid nitrogen. RNA was extracted from leaf tissue using a Quick RNA Isolation kit (Huayueyang Biotechnology, Beijing, China). After that, the sample RNA was reverse transcribed using a Reverse Transcription System (5 × All-In-OneRT MasterMix, abmgood, Canada). Primers were designed using the Primer Premier 5.0, and qRT-PCR was performed with MasterMiX-No Dyekit on a CFX real-time PCR detection system (Bio-rad, Hercules, CA, United States). Finally, quantitative differences between groups were evaluated by the relative quantitative method (2–ΔΔCT). Actin was used as a single reference gene (Zhu et al., 2019).
Results
ATP-Content Change in Mahonia bealei Leaves Exposed to Ultraviolet B Radiation
ATP from the control and UV-B groups in M. bealei leaves was extracted and analyzed. The results show that ATP contents in the control and UV-B groups were 2.33 and 3.20 μmol g–1, respectively (Figure 1 and Supplementary Table 1), which indicates that UV-B radiation increased the ATP content of M. bealei leaves. Because ATP can provide phosphate groups for the phosphorylation process, this pre-experiment result also reflects the difference in phosphorylation levels between groups to some extent (Zhong et al., 2019).
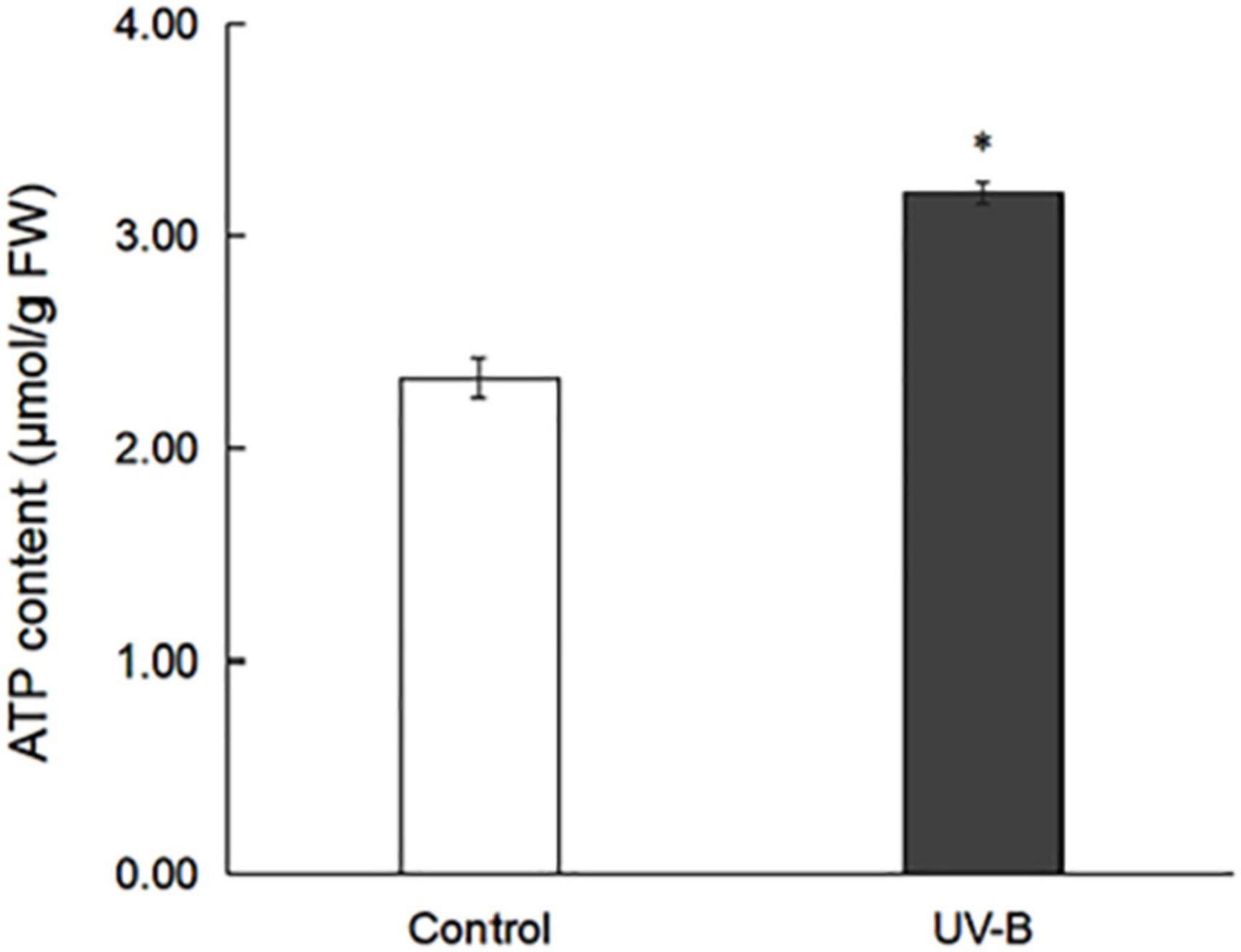
Figure 1. ATP content in M. bealei leaves exposed to UV-B radiation. ATP content was determined by analyzing the increase in absorbance at 340 nm before and after the reaction. ATP content was calculated as μmol per fresh weight (g). Data are shown as means ± SD from three independent biological replicates. Student’s t-test was used to compare values between the control and UV-B groups. *p < 0.05.
Physiological and Chemical Changes in Mahonia bealei Leaves Exposed to Ultraviolet B Radiation
To study oxidative stress, MDA content, H2O2 content, POD activity, SOD activity, T-AOC, and PAL activity in M. bealei leaves exposed to UV-B radiation were determined (Figures 2A–F and Supplementary Table 1). The results show that MDA and H2O2 contents were increased, and these antioxidant enzyme activities were significantly increased after UV-B radiation. The contents of photosynthetic pigments (chlorophyll and carotenoids) were also increased after UV-B radiation (Figures 2G,H and Supplementary Table 1).
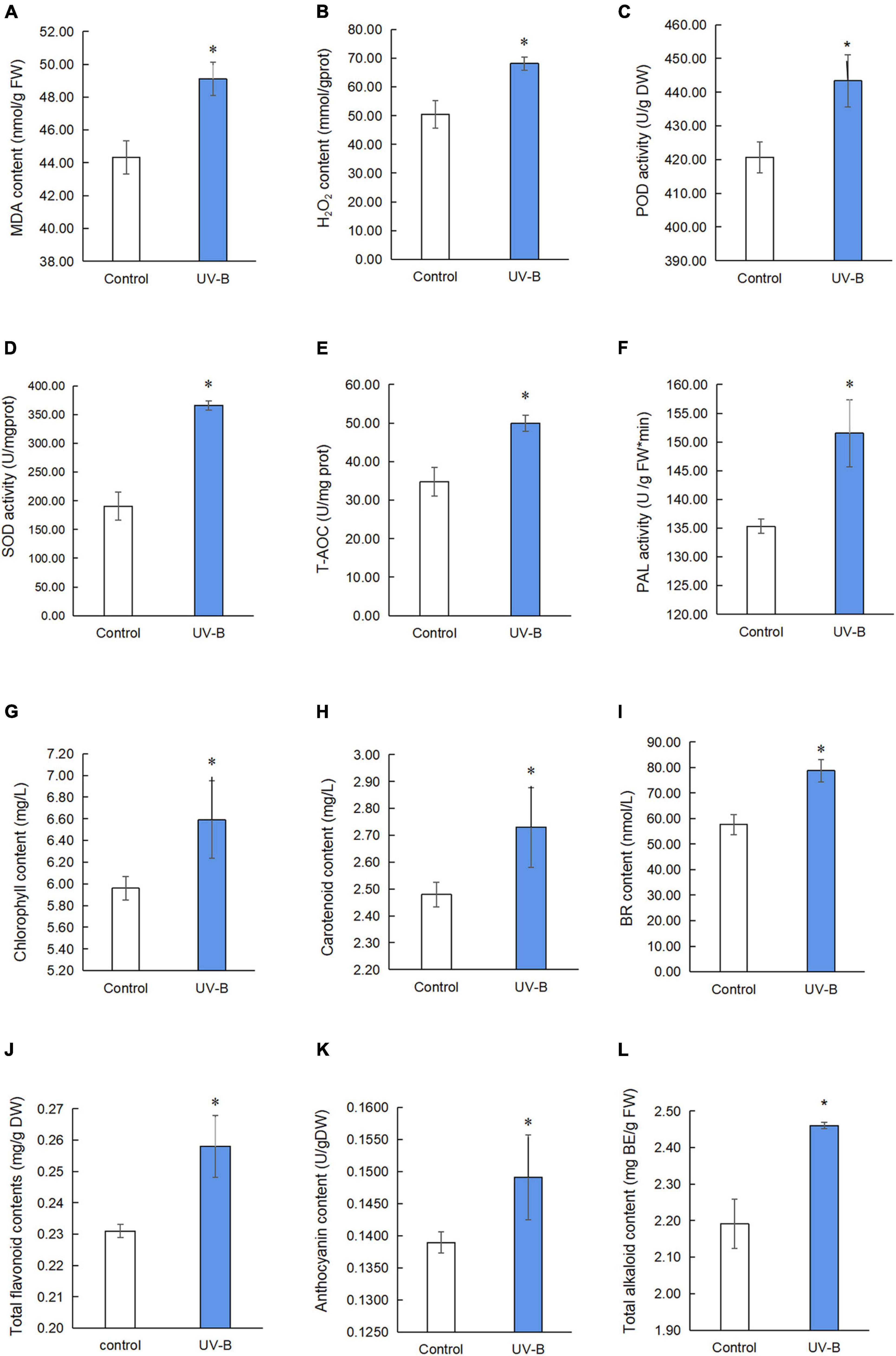
Figure 2. Physiological and chemical analyses of M. bealei leaves exposed to UV-B radiation. (A) MDA content; (B) hydrogen peroxide (H2O2); (C) POD activity; (D) SOD activity; (E) T-AOC; (F) PAL activity; (G) chlorophyll content; (H) carotenoid content; (I) BR content; (J) total flavonoid contents; (K) anthocyanin content; (L) total alkaloids content. Data are presented as mean ± SD from three independent biological replicates. POD, peroxidase; SOD, superoxide dismutase; T-AOC, total antioxidant capacity; MDA, malondialdehyde; PAL, phenylalanine ammonia-lyase; BR, brassinosteroid; DW, dry weight; FW, fresh weight; UV-B, ultraviolet B treatment for 6 h. *p < 0.05.
Next, the content of brassinosteroid, a plant hormone, was determined by an ELISA kit. The results showed that the content of brassinosteroids in M. bealei leaves was significantly increased after UV-B induction (Figure 2I and Supplementary Table 1). In addition, secondary metabolites, such as total flavonoid, anthocyanin, and total alkaloid content, were also increased (Figures 2J–L and Supplementary Table 1). Moreover, the correlation analysis of BR, total alkaloids, total flavonoids, and anthocyanins showed that BR had a significant correlation with total alkaloids and total flavonoids (Supplementary Figure 2).
Identification and Functional Categories of Phosphoproteins in Mahonia bealei Leaves Exposed to Ultraviolet B Radiation
To display the phosphorylation situation of proteins in M. bealei under UV-B radiation, phosphoproteomic analysis was performed (Supplementary Figure 2). One hundred ninety-two phosphorylation sites on 148 proteins were identified, of which 128 proteins were successfully quantified (Supplementary Table 2). The data were used for the subsequently differential site analysis; 54 significantly changed phosphoproteins were identified in the UV-B group, of which 31 and 14 phosphoproteins were increased and decreased, respectively (p < 0.05, fold change > 1.2) (Figures 3A,B). Subcellular localization of the identified phosphorylated proteins shows that phosphorylated proteins are mainly located in chloroplast, cytoplasm, and nucleus (Figure 3C). In addition, nine proteins were phosphorylated only in the control or UV-B groups (Figure 3D).
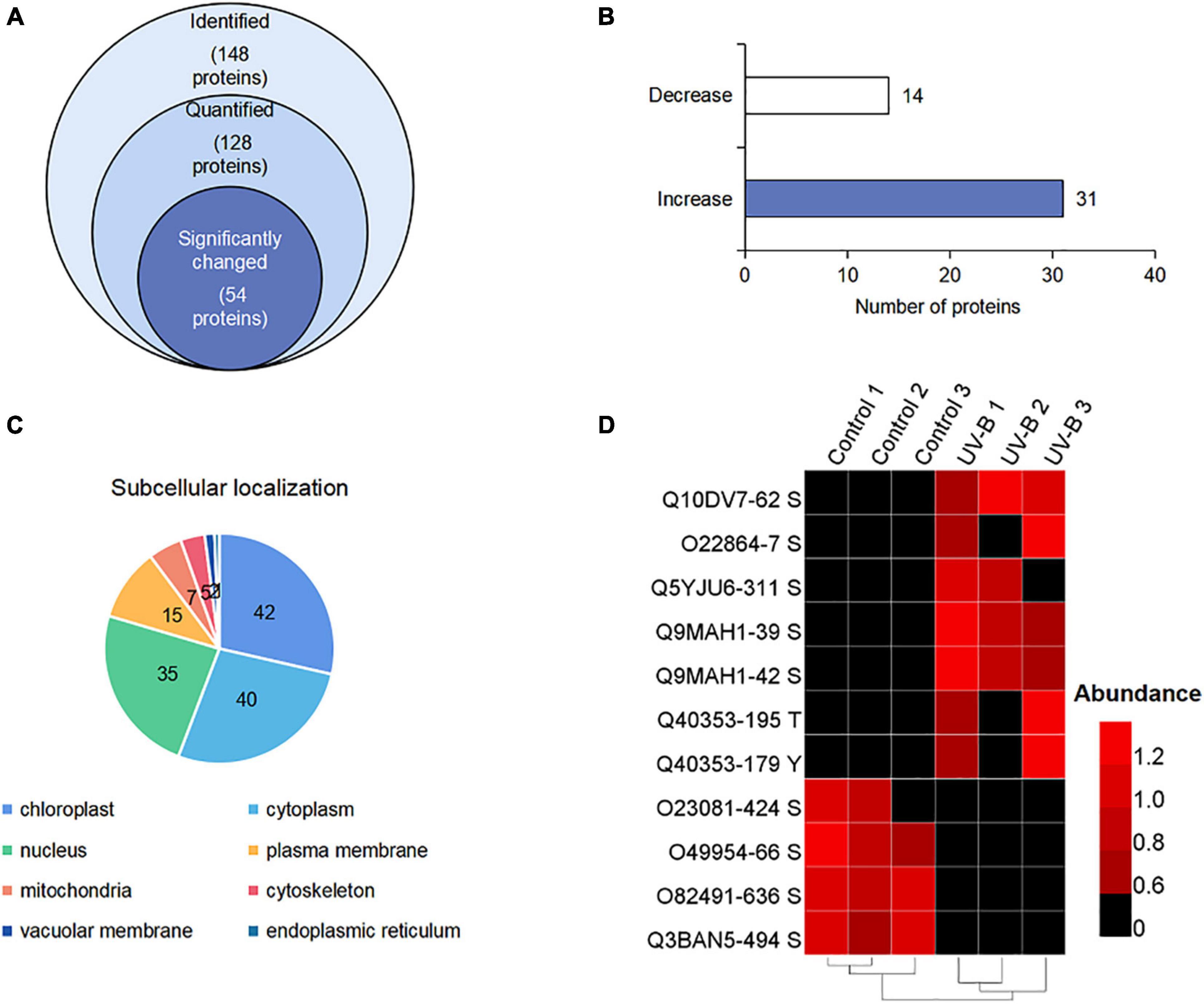
Figure 3. (Phospho) proteome response upon UV-B stress in M. bealei leaves. (A) Venn diagram of all identified phosphoproteins; (B) bar chart of significantly changed phosphoproteins; (C) subcellular localization analysis of phosphorylated proteins; (D) heat map showing values of phosphosite intensities of phosphorylated and absolutely dephosphorylated proteins under UV-B radiation.
MapMan bin codes were used for functional category analysis of phosphoproteins. One hundred forty-eight phosphoproteins were classified into 24 categories. They were mainly related to protein synthesis/post-translational modification/degradation, signaling, RNA transcriptional regulation, cell organization/vesicle transport, and photosynthesis (Supplementary Figure 4). GO term classification was involved in protein phosphorylation and photosynthesis of biological process (BP); ATP binding and protein binding of cellular component (CC); plasma membrane and cytoplasm of molecular function (MF) (Supplementary Figure 5).
KEGG pathway enrichment found that phosphoproteins were mainly involved in MAPK signaling pathways, photosynthesis, glycolysis, plant hormone signal transduction, and biosynthesis of amino acids. Moreover, many phosphoproteins were also involved in the secondary metabolic pathways (Supplementary Figure 6). Combined with KEGG pathway enrichment results, KEGG mapping and KEGG color were used to match the differentially phosphorylated proteins into the KEGG pathway. Using qRT-PCR verification results, three pathways were obtained through integrated analysis: MAPK signal pathway and plant hormone signaling (Figure 4), photosynthesis-glycolysis-TCA cycle pathway (Figure 5), and amino acid biosynthesis pathway (Figure 6).
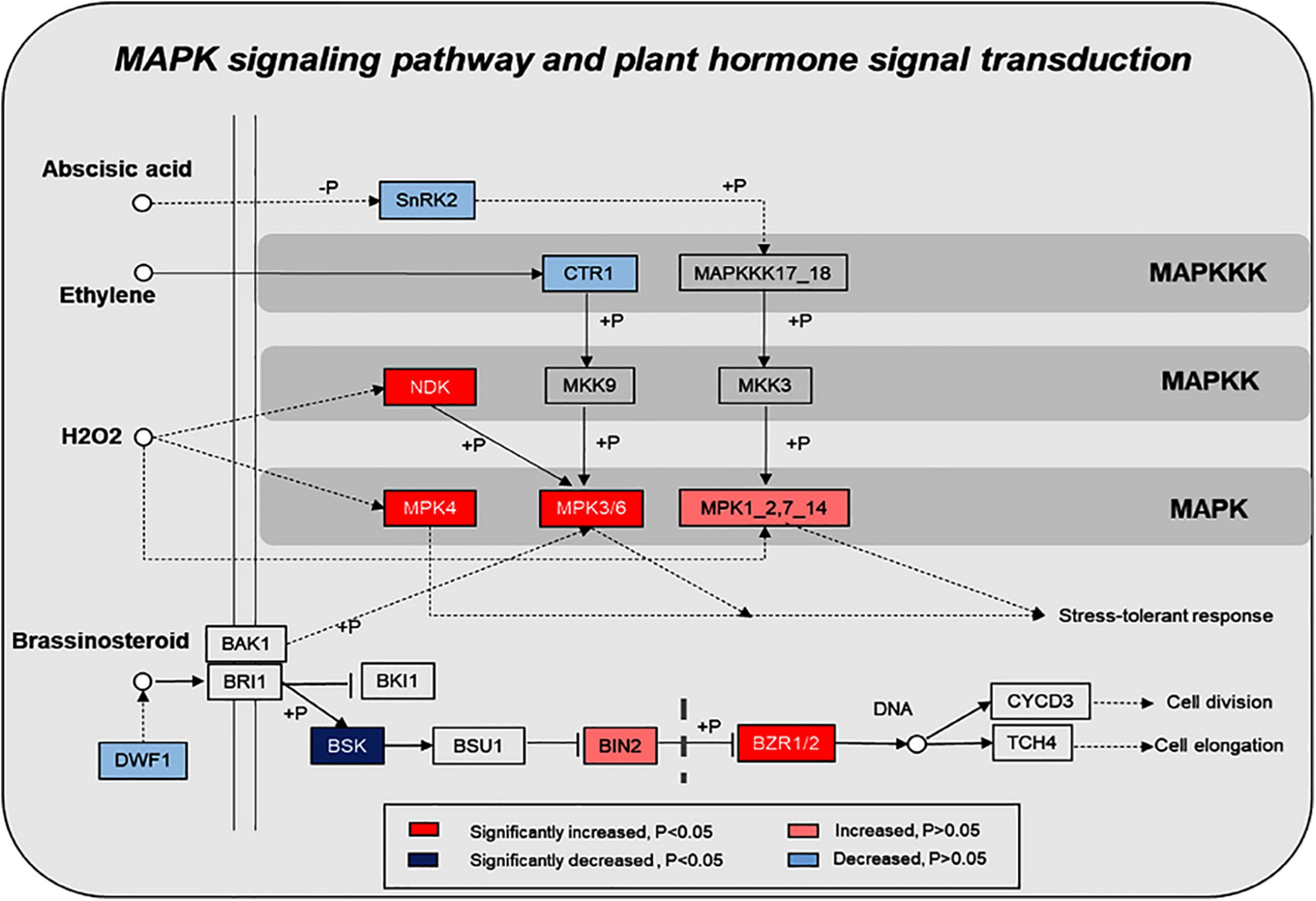
Figure 4. Changes of proteins in plant hormone signal transduction and MAPK signaling pathway in response to the UV-B treatments. The pathways are drawn based on the KEGG database. SNRK2, serine/threonine-protein kinase SRK2; CTR, serine/threonine-protein kinase CTR1; NDK, nucleoside-diphosphate kinase; MPK: mitogen-activated protein kinase; DWF1, Delta24-sterol reductase; BSK, BR-signaling kinase; BIN2, protein brassinosteroid insensitive 2; BZR1/2, brassinosteroid resistant 1/2.
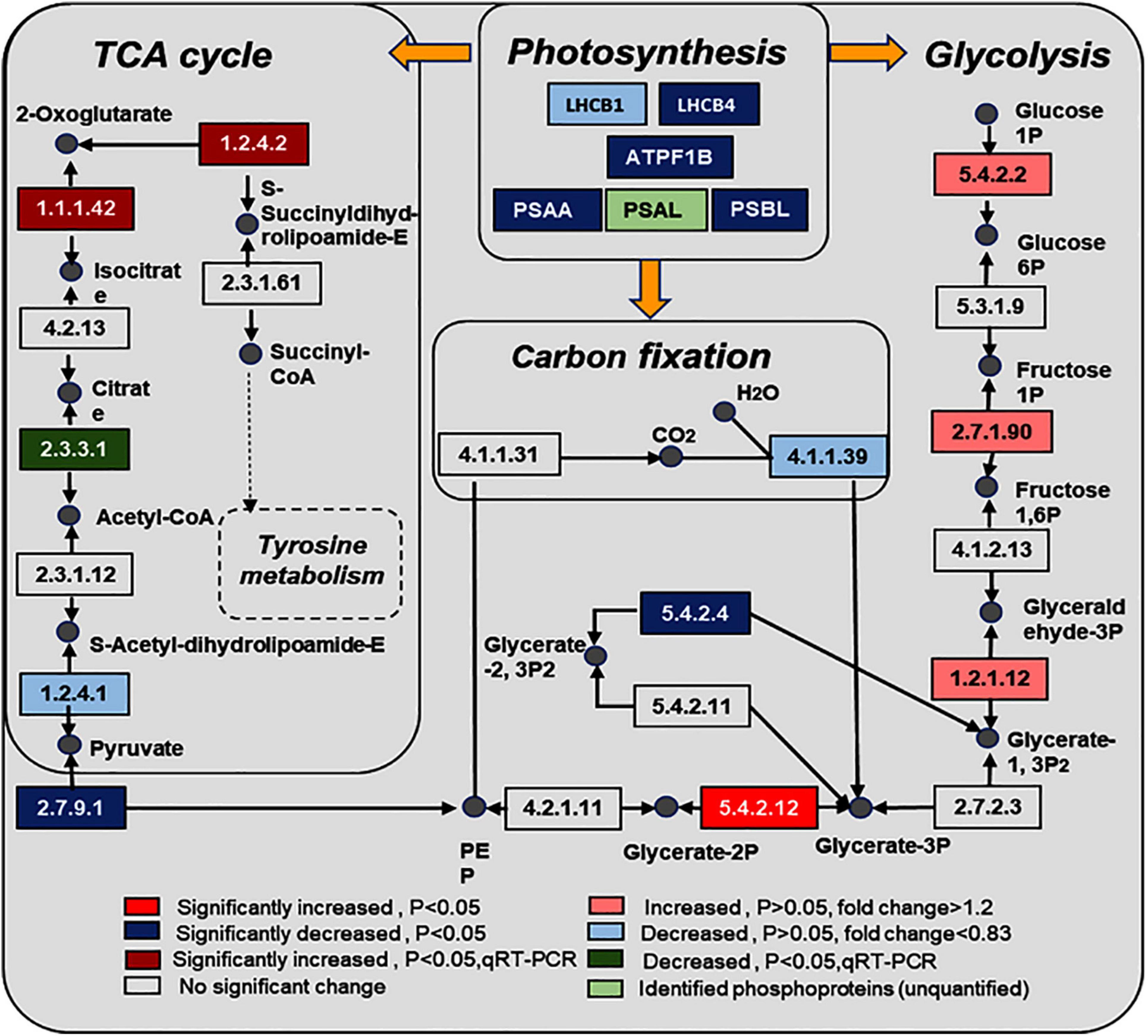
Figure 5. Changes of proteins in photosynthesis, glycolysis, carbon fixation, and citrate cycle in response to the UV-B treatments. The pathways are drawn based on the KEGG database. LHCB1, light-harvesting complex II chlorophyll a/b binding protein 1; LHCB4, light-harvesting complex II chlorophyll a/b binding protein 4; PSAA, photosystem I P700 chlorophyll a apoprotein A1; PSAL, photosystem I subunit XI; PSBL, photosystem II PsbL protein; ATPF1B, F-type H+/Na+-transporting ATPase subunit beta; EC numbers: 5.4.2.2, phosphoglucomutase (PGM); 2.7.1.90, diphosphate dependent phosphofructokinase; 1.2.1.12, glyceraldehyde 3-phosphate dehydrogenase (GAPDH); 5.4.2.12:2, 3-bisphosphoglycerate-independent phosphoglycerate mutase; 2.7.9.1, pyruvate, orthophosphate dikinase (PPSD); 1.2.4.1, pyruvate dehydrogenase E1 component (OSJ); 1.1.1.42, isocitrate dehydrogenase (CICDH); 1.2.4.2, 2-oxoglutarate dehydrogenase E1 component (odhA); 4.1.1.39, ribulose-bisphosphate carboxylase large chain; 5.4.2.4, bisphosphoglycerate/phosphoglycerate mutase; 2.3.3.1, citrate synthase (ACLA).
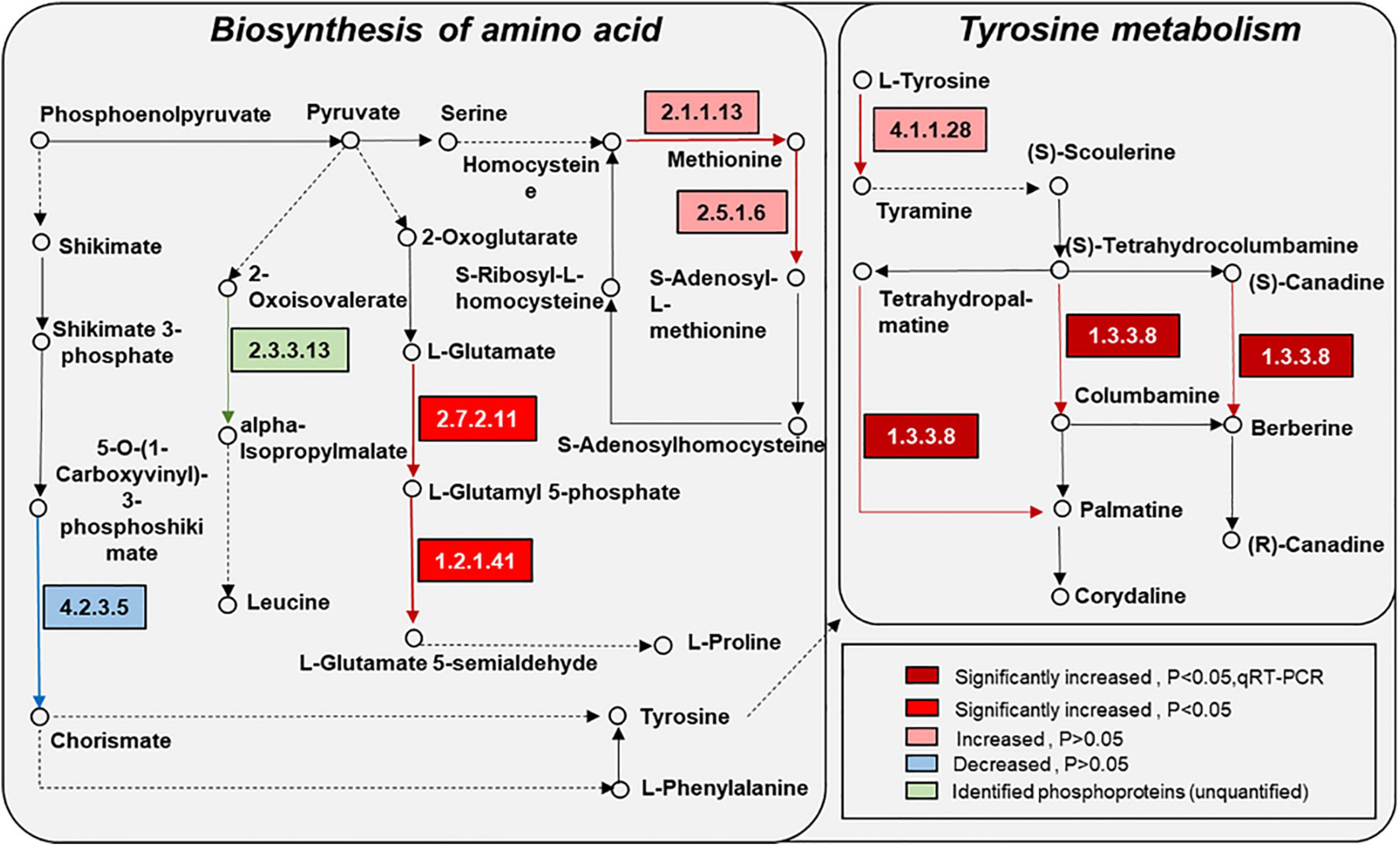
Figure 6. Changes of proteins in amino acid synthesis and metabolism in response to the UV-B treatments. The pathways are drawn based on the KEGG database. EC numbers: 4.2.3.5, chorismate synthase; 2.3.3.13, 2-isopropylmalate synthase; 2.1.1.13, 5-methyltetrahydropteroyltriglutamate-homocysteine methyltransferase; 2.7.2.11, glutamate 5-kinase; 1.2.1.41, glutamate-5-semialdehyde dehydrogenase; 2.5.1.6, S-adenosylmethionine synthetase; 4.1.1.28, aromatic-L-amino-acid/L-tryptophan decarboxylase; 1.3.3.8, tetrahydroprotoberberine oxidase (STOX).
Motif Analysis of Protein Phosphorylation Modifications
Protein motif analysis calculated the trend of amino acid sequences in the phosphorylation site region by counting the regularity of amino acid sequences before and after all phosphorylation sites in the sample. Motif-X analysis results revealed that, for S-phosphorylation, the [SP], were potential substrates of MAPK, cyclin-dependent kinase, and cyclin-dependent kinase-like, and that [RxxS], which can be identified by mitogen-activated protein kinase kinase (MAPKK), calmodulin-dependent protein kinase (CaMK)II, and protein kinase A, were the most enriched motif (Wang K. et al., 2014; Zhang et al., 2014; Figure 7A).
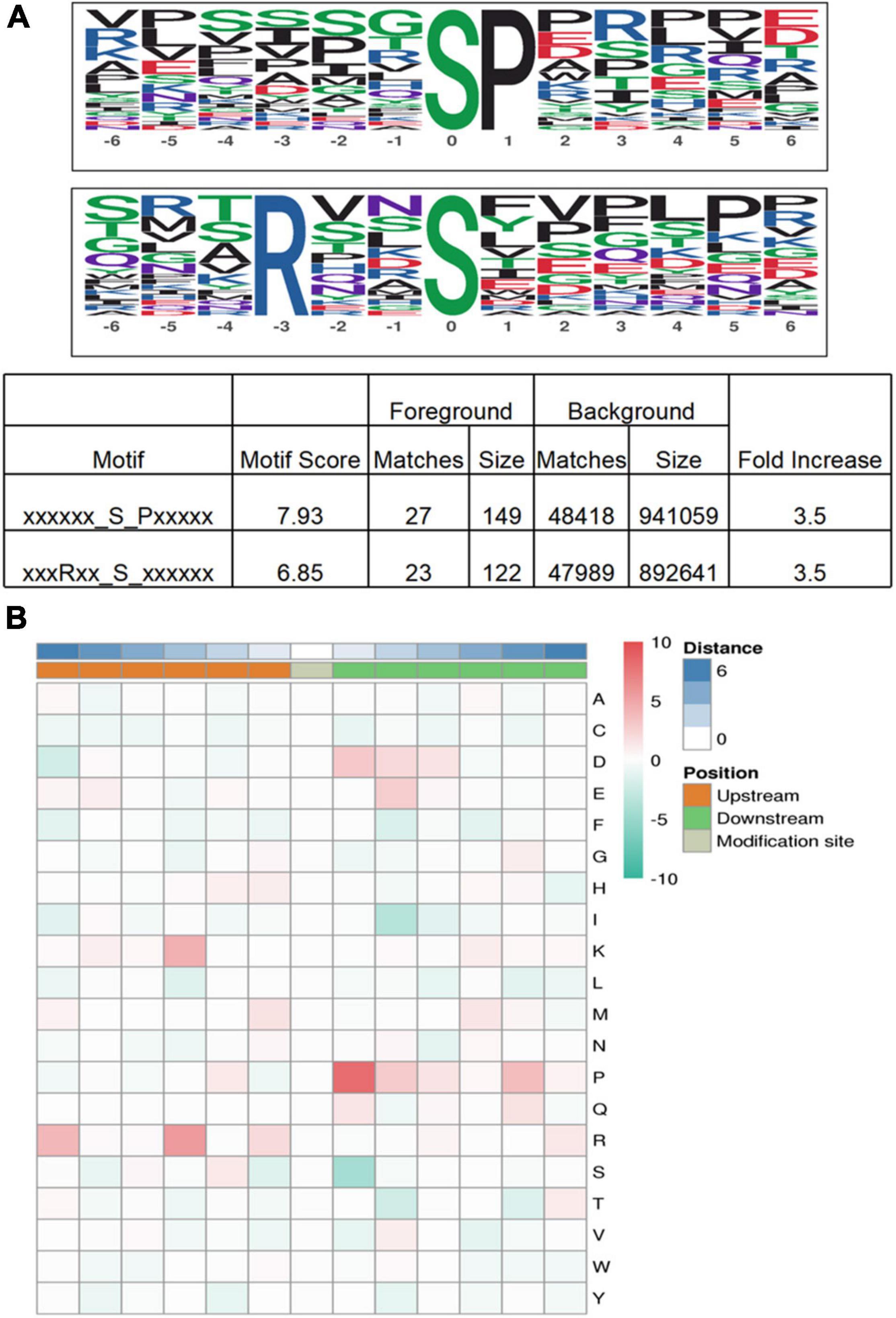
Figure 7. Motif analysis of phosphoproteins in M. bealei leaves exposed to UV-B radiation. (A) This modified site feature sequence and its enrichment statistics are from MoMo software and Motif-X analysis for overrepresented phosphorylation motifs of all identified phosphosites in M. bealei leaves; (B) The motif enrichment heat map of upstream and downstream amino acids of all identified modification sites. Red indicates that this amino acid is significantly enriched near the modification site, and green indicates that this amino acid is significantly reduced near the modification site.
The motif enrichment heat map of upstream and downstream amino acids of all identified modification sites shows that arginine (R) and proline (P) near the serine residue (S) were the most conserved amino acids upstream and downstream of modified serine sites, and arginine (R) and proline (P) were significantly overrepresented at positions 3 and −1, respectively. In addition, lysine (K) frequently occurred at position 3, whereas aspartic acid (D) and glutamic acid (E) were overrepresented at positions −1 and −2, respectively (Figure 7B).
Protein–Protein Interaction Analysis of Differential Phosphoproteins
Protein–protein interaction networks of the phosphoproteins were obtained using the open-access STRING software. One hundred forty-two nodes connected by 127 interactions in phosphoproteins, 51 nodes connected by 19 interactions in significantly changed phosphoproteins were found. Cluster networks analysis in the STRING software showed that most compacts were constituted by phosphoproteins involved in carbon metabolism, ribosome, photosynthesis, oxidative phosphorylation, brassinosteroid, and MAPK cascade pathways (Supplementary Figure 7A). Most compacts were constituted by significantly changed phosphoproteins involved in ribosome, photosynthesis, and oxidative phosphorylation pathways (Supplementary Figure 7B).
Quantitative Real-Time PCR Analyses
qRT-PCR analysis of several genes involved in the light reaction, glycolysis, TCA cycle, and secondary metabolism was performed. Early light-induced protein 2 (elip), chlorophyll a-b binding protein4.1 (lhcb), cytosolic isocitrate dehydrogenase (cicdh), 2-oxoglutarate dehydrogenase E1/E2 component (odha), (s)-tetrahydroprotoberberine oxidase (stox), pyruvate dehydrogenase E1 component subunit alpha (osj) and s-adenosylmethionine synthase1 (sam) expression increased in the UV-B group, but quantitative results of 2,3-bisphosphoglycerate-independent phosphoglycerate mutase (pgm) and citrate synthase (acla) were reduced compared to the control group (Figure 8 and Supplementary Table 3).
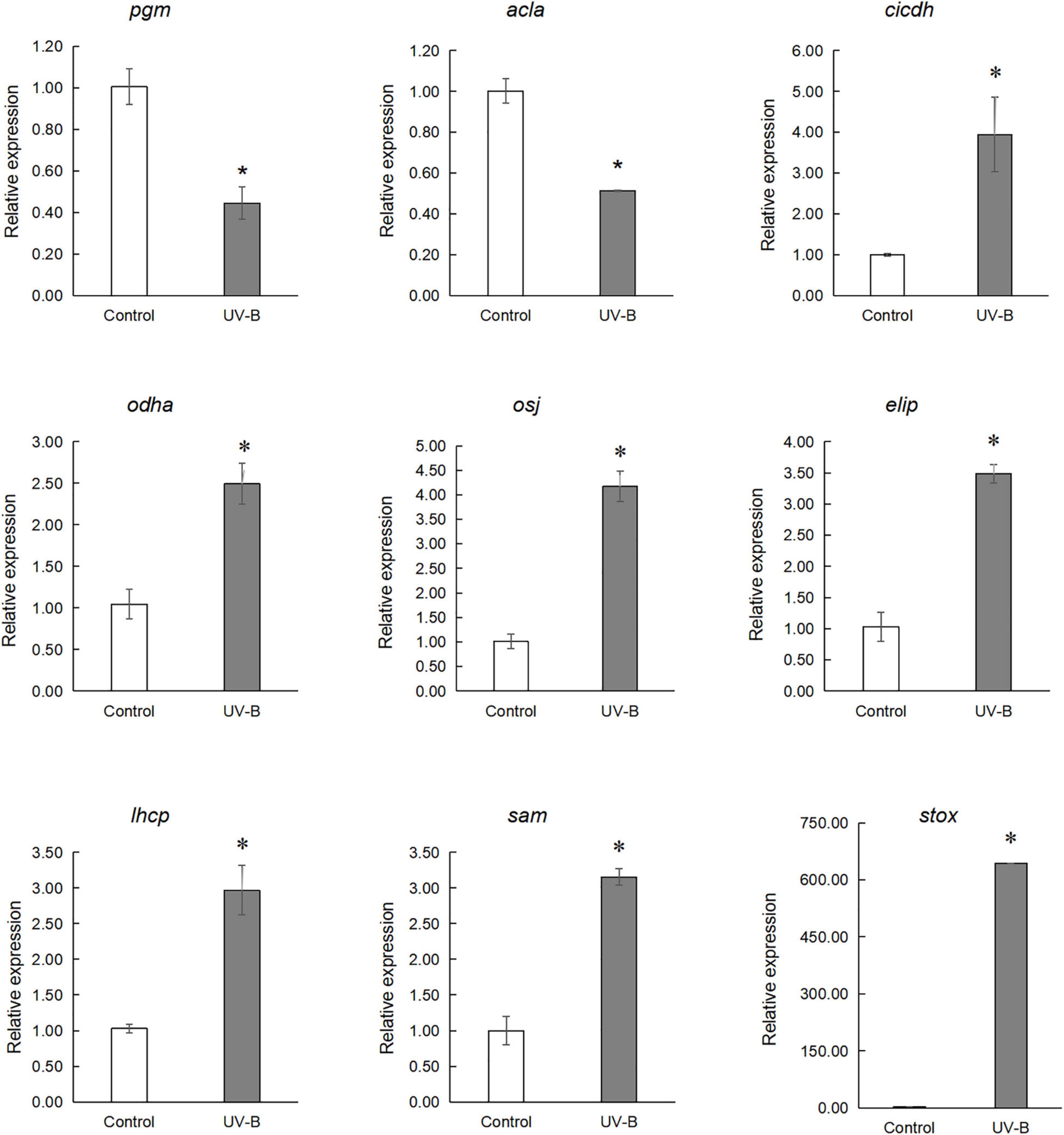
Figure 8. qRT-PCR analysis of genes involved in the light reaction, glycolysis, TCA cycling, and secondary metabolism. Early light-induced protein 2 (elip), chlorophyll a-b binding protein 4.1 (lhcb), cytosolic isocitrate dehydrogenase (cicdh), 2-oxoglutarate dehydrogenase E1/E2 component (odha), (S)-tetrahydroprotoberberine oxidase (stox), pyruvate dehydrogenase E1 component subunit alpha (osj) and S-adenosylmethionine synthase 1 (sam) expression increased in UV-B group, but quantitative results of 2,3-bisphosphoglycerate-independent phosphoglycerate mutase (pgm) and Citrate synthase (acla) are reduced in UV-B group (*p < 0.05).
Discussion
Oxidative Stress Induces Secondary Metabolite Biosynthesis in Mahonia bealei Leaves Exposed to Ultraviolet B Radiation
UV-B radiation can induce oxidative stress, which produces reactive oxygen species (Ortega-Hernández et al., 2019) and then activates the plant ROS scavenging system (Das and Roychoudhury, 2014). Our study results show that the contents of MDA and H2O2 in M. bealei leaves were increased after exposure to UV-B radiation, and correspondingly, the activity of POD and SOD and T-AOC under the UV-B radiation was significantly improved (p < 0.05), which was significant for the detoxification of oxidants produced by UV-B (Czégény et al., 2016; Köhler et al., 2017). Kasote et al. (2015) suggest that the compounds involved in the phenylpropanoid pathway and the phenolics are important for antioxidation, which is consistent with our results. In the present study, PAL activity, total flavonoids, and anthocyanin contents were significantly increased in M. bealei leaves exposed to UV-B radiation (Figures 2F,J,K). UV-B radiation promotes the accumulation of flavonoids and anthocyanins, which can also be interpreted as a way for plants to prevent cell damage. Previous studies show that flavonoids and anthocyanins, as UV-B absorbing compounds and antioxidants, have a significant role in reducing the adverse effect of UV radiation on plant cells (Julkunen-Tiitto et al., 2015; Su et al., 2016). This study suggests that the oxidative damage of M. bealei leaves could be alleviated by the accumulation of flavonoids and anthocyanins under UV-B radiation.
Ultraviolet B Induces Secondary-Metabolite Biosynthesis by Activating the Mitogen-Activated Protein Kinase Signal Pathway and Regulating Brassinosteroids
Functional categorization of identified phosphoproteins indicates that these proteins are mainly involved in protein metabolism, signaling, RNA, cell, and photosynthesis. Furthermore, the pathway enrichment analysis shows that MAPK signaling and plant hormone signaling transduction pathways are significantly enriched in response to UV-B radiation. The identified phosphoproteins are mapped into the KEGG database; the results show that eight and six phosphoproteins were significantly changed in the plant MAPK signaling pathway and plant hormone signaling transduction pathway, respectively (Figure 4).
MAPK cascades are the common feature in eukaryotic cells. As a general feature, there are three kinases, including MAP3K (Mitogen-Activated Protein Kinase Kinase Kinase, MAPKKK), MAP2K (Mitogen-activated protein kinase, MAPKK), and MAPK. Among them, MAPK cascades are important for plant growth (Zhang et al., 2018), development (Chen et al., 2018; Sun et al., 2018; Xue et al., 2020), and abiotic stress (de Zelicourt et al., 2016; Ding et al., 2018). Moreover, MAPK cascades are more likely involved in secondary metabolisms, such as regulation of camalexin (Thulasi Devendrakumar et al., 2018), nicotine (De Boer et al., 2011), anthocyanin (Lei et al., 2014), and phytoalexin biosynthesis (Genot et al., 2017). Recent studies show that phytohormones also participated in the regulation of secondary metabolite biosynthesis under environmental stresses in plants (Chu et al., 2011; Yang et al., 2012; Li et al., 2017). In this study, KEGG pathway analysis shows that some phosphorylated proteins, such as BR-signaling kinase (BSK), brassinosteroids resistant 1/2 (BZR1/2), and protein brassinosteroid insensitive 2 (BIN2), are associated with the brassinosteroid regulation signaling pathway, and the phosphorylated protein Delta24-sterol reductase (DWF1) is associated with brassinosteroid biosynthetic changes after exposure to UV-B induction. Therefore, we speculated that UV-B radiation activates the brassinosteroid biosynthesis pathway and the related signal transduction. Next, we determined the content of brassinosteroid in the samples, and our results show that UV-B stimulates brassinosteroid content, which was significantly increased in M. bealei leaves (Figure 2J). In Arabidopsis thaliana, the BR signal from the cell membrane receptor BRI1 (brassinosteroid insensitive 1) passed through the phosphorylation-mediated cascade BSK1-BSU1-BIN2/GSK1 (BSK1—BRI1 suppressor 1—BIN2/glycogen synthase kinase-1), eventually reaching the transcription factor BZR1, which can trigger BR response (Wang et al., 2012, 2014). Some studies show that inhibition of BR biosynthesis may weaken plant photosynthesis and affect plant tolerance to stress (Cui et al., 2011; Jiang et al., 2012; Ahammed et al., 2014). Also, Cui et al. (2012) suggest that BR utilizes hydrogen peroxide (H2O2)- and nitric oxide (NO)-mediated mechanisms to provide stress tolerance. Additionally, brassinosteroids have a certain role in stimulating secondary metabolic synthesis in plants. For example, Ahammed et al. (2012) find that brassinosteroids stimulate the accumulation of flavonoids in tomato leaves. Similarly, Xi et al. (2013) find that the applications of BR can promote anthocyanin biosynthesis in grapes.
We conducted a simple correlation statistical analysis on the contents of BR, total alkaloids, total flavonoids, and anthocyanins. Our results show that BR had a significant correlation with total alkaloids and total flavonoids (Supplementary Figure 2). Furthermore, crosstalk was observed in the MAPK signaling pathway and brassinosteroid signaling pathway: BR could reduce the activation of BIN2 to inhibit mitogen-activated protein kinase kinase 4, 5, 7, and 9 (MKK4/5/7/9), which induce the phosphorylation of mitogen-activated protein kinase 3 and 6 (MPK3/6) in negatively regulated stomatal cell formation (Kim et al., 2012). Therefore, our data suggest that the MAPK signal pathway and plant hormone signal transduction pathway, especially the BR signaling pathway, are activated by UV-B radiation and can affect the synthesis mechanism of subsequent secondary metabolites.
Ultraviolet Radiation Stimulates the Combined Action Mechanism of Phosphoproteins in the Photosynthetic–Glycolysis–Tricarboxylic Acid Cycle Pathway
Phosphoprotein KEGG pathway enrichment analysis shows that carbon metabolism–related pathways were significantly enriched, such as photosynthesis, glycolysis, TCA cycle, and carbon fixation, and proteins involved in these pathways were significantly changed. The phosphorylation level of light-harvesting complex II chlorophyll a/b binding protein 4 (LHCB4), photosystem I P700 chlorophyll an apoprotein A1 (PSAA), and photosystem II PsbL protein (PSBL) were significantly decreased in photosynthesis. At the same time, phosphorylated 2,3-bisphosphoglycerate-independent phosphoglycerate mutase was upregulated, and bisphosphoglycerate/phosphoglycerate mutase was downregulated in glycolysis. Studies show that UV-B radiation can affect membrane-binding proteins, such as enzymes involved in pigment synthesis, and lead to degradation damage of plant thylakoid membranes (Pradhan et al., 2006). Our experimental results show that the contents of chlorophyll and carotenoids were significantly increased, which may be related to photosynthesis’s function of photosynthetic pigments in absorbing and transferring light energy or causing primary photochemical reactions. Glycolysis and the TCA cycle were the fundamental pathways to produce energy for plant survival, and the phosphorylation level of proteins related to glycolysis and TCA pathways were significantly changed. Furthermore, the ATP content in the leaves under UV-B radiation was significantly increased (Figure 1). We know that photosynthesis and secondary metabolites are intrinsically related (Maeda, 2019), and secondary metabolites were induced by UV-B radiation to absorb and/or dissipate solar energy, which might alleviate the UV-B damage to plants (Liu et al., 2012). These results suggest that UV-B radiation, which stimulates the combined action mechanism of phosphoproteins in the photosynthetic–glycolysis–TCA cycle pathway, may consequently provide the substrate and energy for the synthesis of secondary metabolites.
Amino Acid Metabolism Was Enhanced by Ultraviolet B Radiation to Provide the Precursors for Flavonoid and Alkaloid Biosynthesis
Pathway enrichment analysis showed that the biosynthesis of the amino acid pathway was significantly enriched. Also, the identified phosphoproteins were mainly involved in amino acid biosynthesis and metabolism (Figure 6). Phosphorylated delta-1-pyrroline-5-carboxylate synthetase, which is involved in the proline synthesis pathway, was significantly increased in the UV-B group compared with the control group. Phosphorylated chorismate synthase involved in the biosynthesis of tyrosine and phenylalanine and 5-methyltetrahydropteroyltri glutamate-homocysteine methyltransferase and s-adenosylmethionine synthetase involved in the methionine biosynthesis were also altered. Amino acids and their derivatives have an important role in protein synthesis, growth and development, and stress of plants (Hildebrandt et al., 2015). Moreover, the metabolism of amino acids is closely related to energy metabolism, carbon and nitrogen balance, and secondary metabolism (Pratelli and Pilot, 2014). Specifically, plant amino acids, such as phenylalanine, tyrosine, and tryptophan, help protein synthesis and affect the biosynthesis of many growth hormones and secondary metabolites, which often could resist biotic and abiotic stress (Tzin and Galili, 2010). In this study, the contents of total flavonoid and anthocyanin in M. bealei leaves were increased under UV-B radiation because of the enhanced PAL activity, which may result from changes of phosphorylated proteins in phenylalanine biosynthesis and metabolism-related pathways. Tyrosine was the central hub to myriad specialized metabolic pathways, and phenylalanine and tyrosine were also the initial substrates for isoquinoline alkaloid biosynthesis. The phosphorylated aromatic L-amino acid/L-tryptophan decarboxylase, which was involved in the preliminary metabolism of tyrosine, was upregulated. Moreover, phosphorylated PAL, a key enzyme in the flavonoid biosynthesis pathway, and phosphorylated S-adenosylmethionine synthetase, an important enzyme in the alkaloid biosynthesis pathway, also significantly changed. Additionally, gene expression of stox and sam, which were involved in the biosynthesis of the berberine, columbamine, and palmatine, were significantly upregulated. Previous literature also shows that UV-B induced total flavones and alkaloids’ biosynthesis (Zhu et al., 2021), which is consistent with our data. In addition, the total alkaloid content in samples of the control and UV-B treatment groups was verified in this study, and it was found that the alkaloid content in leaves of the UV-B group was increased (Figure 2L). Accordingly, we conclude that the amino acid biosynthesis and metabolic pathways activated by UV-B are induced to promote some secondary metabolites’ biosynthesis in M. bealei leaves.
Conclusion
In the present study, we analyze the phosphoproteomics of M. bealei leaves that were exposed to UV-B radiation for 6 h. The results show that ultraviolet-B radiation stimulates the oxidative stress system, MAPK signaling, amino acid synthesis, and metabolism pathway leaves and promotes the accumulation of secondary metabolites, total flavonoids, and total alkaloids in M. bealei. The analysis of phosphoproteins in these pathways led to the following conclusions: (1) oxidative stress induces secondary metabolite biosynthesis in M. bealei leaves under UV-B radiation; (2) UV-B induces secondary metabolite biosynthesis by activating the MAPK signal pathway and regulating brassinosteroid (BR); (3) UV-B radiation stimulates the combined action mechanism of phosphoproteins in the photosynthetic–glycolysis–TCA cycle pathway; (4) UV-B-activates amino acid synthesis and the metabolism pathway in leaves promoting the synthesis of secondary metabolites flavonoid and alkaloid. This study provides a new perspective for exploring the ultraviolet-induced mechanism of increasing alkaloid content of the secondary metabolites of M. bealei leaves. However, due to the imperfection of the transcriptome database and insufficient abundance of the phosphoproteomics data, this mechanism needs to be further analyzed.
Data Availability Statement
The data presented in the study are deposited in the ProteomeXchange repository, accession number PXD028873.
Author Contributions
AL and WZ conceived the idea, designed the research, and drafted the manuscript. AL performed the experiments and analyzed the data. SL, ZZ, and JT supervised the study. MT, YL, and HH provided suggestions for the revision of the manuscript. All authors read and approved the final manuscript.
Funding
This work was financially supported by the National Science Foundation of China (Nos. 81603078 and 81872973).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.794906/full#supplementary-material
Supplementary Figure 1 | Map of the experimental design process.
Supplementary Figure 2 | Correlation analysis of brassinosteroid with total alkaloids and flavonoids of secondary metabolites.
Supplementary Figure 3 | The quality control analysis. (A) Pearson’s correlation coefficient. (B) The relative standard deviation (RSD). (C) Three methods of statistical analysis to evaluate quantitative repeatability.
Supplementary Figure 4 | Functional categories of phosphoproteins in M. bealei leaves exposed to UV-B radiation. The functional category of phosphoproteins was determined using MapMan bin codes. One hundred forty-eight phosphoproteins were mapped and classified into 24 categories. The top four categories are presented in detail with pie charts. “Not assigned” indicates protein without ontology.
Supplementary Figure 5 | The top classified enriched GO terms involved in UV-B radiation.
Supplementary Figure 6 | The KEGG pathway enrichment bubble map of phosphoproteins under UV-B induced. KEGG pathway analysis shows that phosphoproteins are mainly involved in MAPK signaling pathways, photosynthesis, glycolysis, plant hormone signal transduction, and biosynthesis of amino acids. Moreover, many phosphoproteins have been identified in the secondary metabolic pathways.
Supplementary Figure 7 | Protein–protein interaction analysis of identified phosphoproteins in M. bealei leaves under UV-B radiation.
Footnotes
- ^ http://www.genscript.com/psort/wolf_psort.html
- ^ http://www.bioinformatics.com.cn
- ^ http://www.kegg.jp/
- ^ http://metascape.org/gp/
- ^ http://string-db.org/
References
Ahammed, G. J., Xia, X. J., Li, X., Shi, K., Yu, J. Q., and Zhou, Y. H. (2014). Role of brassinosteroid in plant adaptation to abiotic stresses and its interplay with other hormones. Curr. Protein Pept. Sci. 16, 462–473. doi: 10.2174/1389203716666150330141427
Ahammed, G. J., Zhou, Y. H., Xia, X. J., Mao, W. H., Shi, K., and Yu, J. Q. (2012). Brassinosteroid regulates secondary metabolism in tomato towards enhanced tolerance to phenanthrene. Biol. Plant. 7, 154–158. doi: 10.1007/s10535-012-0128-9
Chen, L. F., Yang, D. D., Zhang, Y. W., Wu, L. M., Zhang, Y. Y., Ye, L., et al. (2018). Evidence for a specific and critical role of mitogen-activated protein kinase 20 in uni-to-binucleate transition of microgametogenesis in tomato. New Phytol. 219, 176–194. doi: 10.1111/nph.15150
Chen, Y., Shen, Q., Lv, P., and Sun, C. B. (2020). Comparative metabolomic analyses of dendrobium officinale kimura et migo responding to UV-B radiation reveal variations in the metabolisms associated with its bioactive ingredients. PeerJ 8:e9107. doi: 10.7717/peerj.9107
Chu, H. Y., Wegel, E., and Osbourn, A. (2011). From hormones to secondary metabolism: the emergence of metabolic gene clusters in plants. Plant J. 66, 66–79. doi: 10.1111/j.1365-313X.2011.04503.x
Cui, F., Liu, L., Zhao, Q., Zhang, Z., Li, Q., Lin, B., et al. (2012). Arabidopsis ubiquitin conjugase UBC32 is an ERAD component that functions in brassinosteroid-mediated salt stress tolerance. Plant Cell 24, 233–244. doi: 10.1105/tpc.111.093062
Cui, J. X., Zhou, Y. H., Ding, J. G., Xia, X. J., Shi, K., Chen, S. C., et al. (2011). Role of nitric oxide in hydrogen peroxide-dependent induction of abiotic stress tolerance by brassinosteroids in cucumber. Plant Cell Environ. 34, 347–358. doi: 10.1111/j.1365-3040.2010.02248.x
Czégény, G., Le Martret, B., Pávkovics, D., Dix, P. J., and Hideg, É (2016). Elevated ROS scavenging enzymes contribute to acclimation to UV-B exposure in transplastomic tobacco plants, reducing the role of plastid peroxidases. J. Plant Physiol. 201, 95–100. doi: 10.1016/j.jplph.2016.05.026
Das, K., and Roychoudhury, A. (2014). Reactive oxygen species (ROS) and response of antioxidants as ROS-scavengers during environmental stress in plants. Front. Environ. Sci. 2:53. doi: 10.3389/fenvs.2014.00053
De Boer, K., Tilleman, S., Pauwels, L., Vanden Bossche, R., De Sutter, V., Vanderhaeghen, R., et al. (2011). APETALA2/ETHYLENE response factor and basic helix-loop-helix tobacco transcription factors cooperatively mediate jasmonate-elicited nicotine biosynthesis. Plant J. 66, 1053–1065. doi: 10.1111/j.1365-313X.2011.04566.x
de Zelicourt, A., Colcombet, J., and Hirt, H. (2016). The role of MAPK modules and ABA during abiotic stress signaling. Trends Plant Sci. 21, 677–685. doi: 10.1016/j.tplants.2016.04.004
Deng, S., May, B. H., Zhang, A. L., Lu, C., and Xue, C. C. L. (2013). Plant extracts for the topical management of psoriasis: a systematic review and meta-analysis. Br. J. Dermatol. 169, 769–782. doi: 10.1111/bjd.12557
Ding, H., He, J., Wu, Y., Wu, X., Ge, C., Wang, Y., et al. (2018). The tomato mitogen-activated protein kinase SlMPK1 is as a negative regulator of the high-temperature stress response. Plant Physiol. 177, 633–651. doi: 10.1104/pp.18.00067
Genot, B., Lang, J., Berriri, S., Garmier, M., Gilard, F., Pateyron, S., et al. (2017). Colcombet J. constitutively active arabidopsis MAP kinase 3 triggers defense responses involving salicylic acid and SUMM2 resistance protein. Plant Physiol. 174, 1238–1249. doi: 10.1104/pp.17.00378
Hildebrandt, T. M., Nesi, A. N., Araujo, W. L., and Braun, H. P. (2015). Amino acid catabolism in plants. Mol. Plant. 8, 1563–1579. doi: 10.1016/j.molp.2015.09.005
Hu, W. C., Yu, L. L., and Wang, M. H. (2011). Antioxidant and antiproliferative properties of water extract from Mahonia bealei (fort.) carr. Leaves. Food Chem. Toxicol. 49, 799–806. doi: 10.1016/j.fct.2010.12.001
Huang, J., Gu, M., Lai, Z., Fan, B., Shi, K., Zhou, Y. H., et al. (2010). Functional analysis of the arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol. 153, 1526–1538. doi: 10.1104/pp.110.157370
Jiang, Y. P., Cheng, F., Zhou, Y. H., Xia, X. J., Mao, W. H., Shi, K., et al. (2012). Cellular glutathione redox homeostasis plays an important role in the brassinosteroid induced increase in CO2 assimilation in Cucumis sativus. New Phytol. 194, 932–943. doi: 10.1111/j.1469-8137.2012.04111.x
Julkunen-Tiitto, R., Nenadis, N., Neugart, S., Robson, M., Agati, G., Vepsäläinen, J., et al. (2015). Assessing the response of plant flavonoids to UV radiation: an overview of appropriate techniques. Phytochem. Rev. 14, 273–297. doi: 10.1007/s11101-014-9362-4
Kasote, D. M., Katyare, S. S., Hegde, M. V., and Bae, H. (2015). Significance of antioxidant potential of plants and its relevance to therapeutic applications. Int. J. Biol. Sci. 11, 982–991. doi: 10.7150/ijbs.12096
Kim, T. W., Michniewicz, M., Bergmann, D. C., and Wang, Z. Y. (2012). Brassinosteroid regulates stomatal development by GSK3-mediated inhibition of a MAPK pathway. Nature 482, 419–422. doi: 10.1038/nature10794
Köhler, H., Contreras, R. A., Pizarro, M., Cortés-Antíquera, R., and Zúñiga, G. E. (2017). Antioxidant responses induced by UVB radiation in deschampsia antarctica desv. Front. Plant Sci. 8:1–9. doi: 10.3389/fpls.2017.00921
Lei, L., Li, Y., Wang, Q., Xu, J., Chen, Y., Yang, H., et al. (2014). Activation of MKK9-MPK3/MPK6 enhances phosphate acquisition in Arabidopsis thaliana. New Phytol. 203, 1146–1160. doi: 10.1111/nph.12872
Li, D., Xu, G., Ren, G., Sun, Y., Huang, Y., and Liu, C. (2017). The application of ultra-high performance liquid chromatography coupled with a LTQ-orbitrap mass technique to reveal the dynamic accumulation of secondary metabolites in licorice under ABA stress. Molecules 22:1742. doi: 10.3390/molecules22101742
Li, L. Y., Long, W. F., Wan, X. L., Ding, Q., Zhang, F., and Wan, D. (2015). Studies on quantitative determination of total alkaloids and berberine in five origins of crude medicine “Sankezhen”. J. Chromatogr. Sci. 53, 307–311. doi: 10.1093/chromsci/bmu060
Liu, J. L., Shu, Z. M., Liang, Z. S., Shi, X. L., and Zhang, Y. M. (2013). UV-B radiation effects on phenolic changes and antioxidant activity in Salvia miltiorrhiza bunge leaf. J. Food Agric. Environ. 11, 2788–2791.
Liu, M. L., Cao, B., Zhou, S., and Liu, Y. (2012). Responses of the flavonoid pathway to UV-B radiation stress and the correlation with the lipid antioxidant characteristics in the desert plant Caryopteris mongolica. Acta Ecol. Sin. 32, 150–155. doi: 10.1016/j.chnaes.2012.04.004
Maeda, H. A. (2019). Evolutionary diversification of primary metabolism and its contribution to plant chemical diversity. Front. Plant Sci. 10:881. doi: 10.3389/fpls.2019.00881
Maszkowska, J., Dębski, J., Kulik, A., Kistowski, M., Bucholc, M., Lichocka, M., et al. (2019). Phosphoproteomic analysis reveals that dehydrins ERD10 and ERD14 are phosphorylated by SNF1-related protein kinase 2.10 in response to osmotic stress. Plant Cell Environ. 42, 931–946. doi: 10.1111/pce.13465
Neff, M. M., and Chory, J. (1998). Genetic interactions between phytochrome A, phytochrome B, and cryptochrome 1 during Arabidopsis development. Plant Physiol. 118, 27–35. doi: 10.1104/pp.118.1.27
Ortega-Hernández, E., Nair, V., Welti-Chanes, J., Cisneros-Zevallos, L., and Jacobo-Velázquez, D. A. (2019). Wounding and UVB light synergistically induce the biosynthesis of phenolic compounds and ascorbic acid in red prickly pears (opuntia ficus-indica cv. Rojo Vigor). Int. J. Mol. Sci. 20:5327. doi: 10.3390/ijms20215327
Pradhan, M., Joshi, P., Nair, J., Ramaswamy, N., Iyer, R., Biswal, B., et al. (2006). UVB exposure enhances senescence of wheat leaves: modulation by photosynthetically active radiation. Radiat. Environ. Biophys. 45, 221–229. doi: 10.1007/s00411-006-0055-2
Pratelli, R., and Pilot, G. (2014). Regulation of amino acid metabolic enzymes and transporters in plants. J. Exp. Bot. 65, 5535–5556. doi: 10.1093/jxb/eru320
Su, N. N., Lu, Y. W., Wu, Q., Liu, Y., Xia, Y. Y., Xia, K., et al. (2016). UV-B-induced anthocyanin accumulation in hypocotyls of radish sprouts continues in the dark after irradiation. Sci. Food Agric. 96, 886–892. doi: 10.1002/jsfa.7161
Sun, T. J., Nitta, Y., Zhang, Q., Wu, D., Tian, H., Lee, J. S., et al. (2018). Antagonistic interactions between two MAP kinase cascades in plant development and immune signaling. EMBO Rep. 19:e45324. doi: 10.15252/embr.201745324
Thulasi Devendrakumar, K., Li, X., and Zhang, Y. (2018). MAP kinase signalling: interplays between plant PAMP- and effector-triggered immunity. Cell Mol. Life Sci. 75, 2981–2989. doi: 10.1007/s00018-018-2839-3
Tzin, V., and Galili, G. (2010). New insights into the shikimate and aromatic amino acids biosynthesis pathways in plants. Mol. Plant. 3, 956–972. doi: 10.1093/mp/ssq048
Vital, C. E., Giordano, A., de Almeida Soares, E., Williams, T. C. R., Mesquita, R. O., Vidigal, P. M. P., et al. (2017). An integrative overview of the molecular and physiological responses of sugarcane under drought conditions. Plant Mol. Biol. 94, 577–594. doi: 10.1001/s11103-017-0611-y
Wang, C. H., Zheng, L. P., Tian, H., and Wang, J. W. (2016). Synergistic effects of ultraviolet-B and methyl jasmonate on tanshinone biosynthesis in Salvia miltiorrhiza hairy roots. J. Photochem. Photobiol. B Biol. 159, 93–100. doi: 10.1016/j.jphotobiol.2016.01.012
Wang, K., Zhao, Y., Li, M., Gao, F., Yang, M. K., Wang, X., et al. (2014). Analysis of phosphoproteome in rice pistil. Proteomics 14, 2319–2334. doi: 10.1002/pmic.201400004
Wang, W., Bai, M. Y., and Wang, Z. Y. (2014). The brassinosteroid signaling network-a paradigm of signal integration. Curr. Opin. Plant Biol. 21, 147–153. doi: 10.1016/j.pbj.2014.07.012
Wang, Z. Y., Bai, M. Y., Oh, E., and Zhu, J. Y. (2012). Brassinosteroid signaling network and regulation of photomorphogenesis. Annu. Rev. Genet. 46, 701–724. doi: 10.1146/annurev-genet-102209-163450
Wang, Z. Y., Wang, N., Liu, X., Wang, Q., Xu, B., Liu, P., et al. (2018). Broadleaf mahonia attenuates granulomatous lobular mastitis-associated inflammation by inhibiting CCL-5 expression in macrophages. Int. J. Mol. Med. 41, 340–352. doi: 10.3892/ijmm.2017.3246
Wellburn, A. R., and Lichtenthaler, H. (1984). “Formulae and program to determine total carotenoids and chlorophylls a and b of leaf extracts in different solvents,” in Advances in Photosynthesis Research. Advances in Agricultural Biotechnology, Vol. 2, ed. C. Sybesma (Dordrecht: Springer), doi: 10.1007/978-94-017-6368-4_3
Xi, Z. M., Zhang, Z. W., Huo, S. S., Luan, L. Y., Gao, X., Ma, L. N., et al. (2013). Regulating the secondary metabolism in grape berry using exogenous 24-epibrassinolide for enhanced phenolics content and antioxidant capacity. Food Chem. 141, 3056–3065. doi: 10.1016/j.foodchem.2013.05.137
Xue, X. Y., Bian, C., Guo, X. Y., Di, R., and Dong, J. (2020). The MAPK substrate MASS proteins regulate stomatal development in Arabidopsis. PLoS Genet. 16:e1008706. doi: 10.1371/journal.pgen.1008706
Yang, D., Ma, P., Liang, X., Wei, Z., Liang, Z., Liu, Y., et al. (2012). PEG and ABA trigger methyl jasmonate accumulation to induce the MEP pathway and increase tanshinone production in salvia miltiorrhiza hairy roots. Physiol. Plant. 146, 173–183. doi: 10.1111/j.1399-3054.2012.01603.x
Yang, Y. J., Jiang, Z. F., Guo, J., Yang, X., Xu, N., Chen, Z. H., et al. (2018). Transcriptomic analyses of chrysanthemum morifolium ramat under UV-B radiation treatment reveal variations in the metabolisms associated with bioactive components. Ind. Crop Prod. 124, 475–486. doi: 10.1016/j.indcrop.2018.08.011
Ying, X. J., Fan, H., Chen, Y., Li, L. Z., Song, W., Fan, Y. M., et al. (2020). Integrative omic and transgenic analyses reveal the positive effect of ultraviolet-B irradiation on salvianolic acid biosynthesis through upregulation of SmNAC1. Plant J. 104, 781–799. doi: 10.1111/tpj.14952
Zhang, L., Zhu, W., Zhang, Y. Z., Yang, B. X., Fu, Z. R., Li, X. M., et al. (2014). Proteomics analysis of Mahonia bealei leaves with induction of alkaloids via combinatorial peptide ligand libraries. J. Proteomics 110, 59–71. doi: 10.1016/j.jprot.2014.07.036
Zhang, M. M., Su, J. B., Zhang, Y., Xu, J., and Zhang, S. Q. (2018). Conveying endogenous and exogenous signals: MAPK cascades in plant growth and defense. Curr. Opin. Plant Biol. 45, 1–10. doi: 10.1016/j.pbi.2018.04.012
Zhang, M., Lv, D. W., Ge, P., Bian, Y. W., Chen, G. X., Zhu, G. R., et al. (2014). Phosphoproteome analysis reveals new drought response and defense mechanisms of seedling leaves in bread wheat (Triticum aestivum L.). J. Proteom. 109, 290–308. doi: 10.1016/j.jprot.2014.07.010
Zhong, Z. H., Liu, S. Z., Zhu, W., Ou, Y. T., Hisateru, Y., Keisuke, H., et al. (2019). Phosphoproteomics reveals the biosynthesis of secondary metabolites in catharanthus roseus under ultraviolet B radiation. J. Proteome Res. 18, 3328–3341. doi: 10.1021/acs.jproteome.9b00267
Zhu, W., Han, H. T., Liu, A. M., Guan, Q. J., Kang, J. N., David, L., et al. (2021). Combined ultraviolet and darkness regulation of medicinal metabolites in mahonia bealei revealed by proteomics and metabolomics. J. Proteomics. 233:104081. doi: 10.1016/j.jprot.2020.104081
Zhu, W., Hu, J., Wang, X., Tian, J. K., and Komatsu, S. (2015). Organ-specific analysis of mahonia using gel-free/label-free proteomic technique. J. Proteome Res. 14, 2669–2685. doi: 10.1021/acs.jproteome.5b00208
Keywords: phosphoproteomics, Mahonia bealei, ultraviolet-B radiation, abiotic stress, secondary metabolism
Citation: Liu A, Liu S, Li Y, Tao M, Han H, Zhong Z, Zhu W and Tian J (2022) Phosphoproteomics Reveals Regulation of Secondary Metabolites in Mahonia bealei Exposed to Ultraviolet-B Radiation. Front. Plant Sci. 12:794906. doi: 10.3389/fpls.2021.794906
Received: 15 October 2021; Accepted: 29 November 2021;
Published: 11 January 2022.
Edited by:
Wenyan Han, Tea Research Institute, Chinese Academy of Agricultural Sciences (CAAS), ChinaReviewed by:
Xiaojian Yin, China Pharmaceutical University, ChinaJinwei Suo, Zhejiang Agriculture and Forestry University, China
Copyright © 2022 Liu, Liu, Li, Tao, Han, Zhong, Zhu and Tian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Zhu, emh1d2VpMjAwNzEwMEAxNjMuY29t; Jingkui Tian, dGprQHpqdS5lZHUuY24=
 Amin Liu1,2
Amin Liu1,2 Yaohan Li
Yaohan Li Haote Han
Haote Han Wei Zhu
Wei Zhu Jingkui Tian
Jingkui Tian