- 1College of Traditional Chinese Medicine, Yunnan University of Chinese Medicine, Kunming, China
- 2Key Laboratory of Economic Plants and Biotechnology, Yunnan Key Laboratory for Wild Plant Resources, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, China
Genome size is known to influence phenotypic traits in leaves and seeds. Although genome size is closely related to cellular and developmental traits across biological kingdoms, floral longevity is a floral trait with important fitness consequence, but less is known about the link between floral longevity and sizes of genomes and cells. In this study, we examined evolutionary coordination between genome size, floral longevity, and epidermal cell size in flowers and leaves in 13 Paphiopedilum species. We found that, across all the study species, the genome size was positively correlated with floral longevity but negatively associated with labellum epidermal cell size, and a negative relationship was found between floral longevity and labellum epidermal cell size. This suggested that genome size is potentially correlated with floral longevity, and genome size has an important impact on life-history trait. In addition, genome size was positively correlated with leaf epidermal cell size, which was different from the relationship in flower due to different selective pressures they experienced or different functions they performed. Therefore, genome size constraints floral longevity, and it is a strong predictor of cell size. The impact of genome size on reproduction might have more implications for the evolution of flowering plants and pollination ecology.
Introduction
It is well known that genome size varies significantly within plants, even in very closely related plant species (Šmarda and Bureš, 2010; Pellicer et al., 2018). However, the ecological adaptation and evolutionary significance of genome size remains unclear. Phylogenetic studies have revealed that increases and decreases of genome size have taken place many times within lineages (Soltis et al., 2003; Leitch et al., 2005; Simonin and Roddy, 2018). Genome size variation may suffer from strong selection pressures (Petrov, 2002), which suggests that some cost may exist to associate large genomes, or benefits from small genome size (Knight et al., 2005; Beaulieu et al., 2007). Therefore, there has been a growing interest in identifying the phenotypic consequences from species with different genome sizes (Knight et al., 2005; Beaulieu et al., 2007; Simonin and Roddy, 2018; Théroux-Rancourt et al., 2021).
Variations in genome sizes may be closely related to cell sizes across biological kingdoms (Cavalier-Smith, 2005; Simonin and Roddy, 2018; Théroux-Rancourt et al., 2021). Such coordination is indirectly and/or directly linked with whole-organism fitness characters (Jockusch, 1997). Previous studies have found that the cell size is positively correlated with genome size (Bennett, 1972; Edwards and Endrizzi, 1975; Sugiyama, 2005). In addition, there is a positive relationship between genome size and cell cycle duration (Rees et al., 1966; Baetcke et al., 1967; Bennett et al., 1983; Lawrence, 1985). In view of these cellular associations, it is assumed that many physiological and functional traits may link with genome size.
Observations have shown that there are positive and negative relationships between genome size and the structural characteristics in leaves, such as cell size, densities of veins and stomata, stomatal size, leaf mass per unit area, and leaf specific area (Beaulieu et al., 2007; Simonin and Roddy, 2018). In addition, changes in genome size are also associated with variation in many functional physiological traits, such as maximum rate of photosynthesis, minimum generation time, seed mass, flower size, and environmental variables (Bennett, 1972; Knight and Ackerly, 2002; Knight et al., 2005; Meagher et al., 2005; Beaulieu et al., 2007; Simonin and Roddy, 2018; Roddy et al., 2020; Faizulah et al., 2021; Théroux-Rancourt et al., 2021). In turn, the variation of plant traits may impact the evolution of genome size (Simonin and Roddy, 2018; Roddy et al., 2020; Théroux-Rancourt et al., 2021). It has been proposed that genome size is a predictor of cell sizes and photosynthetic rates in terrestrial vascular plants (Roddy et al., 2020). In addition, seed mass is strongly positively correlated with genome size (Beaulieu et al., 2008). Environmentally, genome size has been reported to vary with latitude, precipitation, altitude, and temperature (Sims and Price, 1985; Wakamiya et al., 1993; Knight and Ackerly, 2002; Knight et al., 2005). These suggest that variations in genome size may be adaptive.
Flower is a sexual reproductive organ of flowering plants, which has an important impact on its reproduction success (Rosas-Guerrero et al., 2014). Floral longevity is the length of duration that flowers remain open and functional, which is a key floral characteristic that impacts the reproductive fitness of plants (Primack, 1985). Floral longevity varies considerably from a day or less (e.g., Ipomoea purpurea) up to 2 months (e.g., some species from Orchidaceae) among flowering plants (Kerner von Marilaum, 1895; Gori, 1983; Primack, 1985; Ashman and Schoen, 1994; Zhang et al., 2017, 2021). Some studies have indicated that the duration of the life history in leaves was correlated with genome size within vascular plants (Finch, 1990; Morgan and Westoby, 2005; Beaulieu et al., 2007, 2008; Théroux-Rancourt et al., 2021). However, a previous study found a weak positive correlation between leaf lifespan, leaf mass per area, and genome size, suggesting that among angiosperms, genome size and leaf strategy are not related (Morgan and Westoby, 2005). Variation of floral longevity may imply different ecological strategies (Zhang et al., 2021). Therefore, understanding the genetic basis of variation in floral longevity is of great value. However, the potential link between floral longevity and the sizes of genome and cell is still lacking. We speculated that genome size may be related to floral longevity through cell size effect on physiological function within floral organs. Species with larger genome and longer floral longevity might need longer developmental duration (Feng et al., 2021).
The family Orchidaceae is an important group of flowering plants with great values in ornamental, medical, conservation, and evolutionary research (Zhang et al., 2018). The high diversity in morphology, floral longevity, growth form, habitat, and genome size mean that orchid species exhibit various biological and ecological consequences (Cox et al., 1998; Chase, 2005; Leitch et al., 2009; Zhang et al., 2017, 2018, 2021). The genus Paphiopedilum species are commonly named as “Lady's or Venus's Slipper” orchids, and these species are among the most cultivated and horticulturally important plants (Liu et al., 2009; Parveen et al., 2012). This genus Paphiopedilum was divided into three subgenera as follows: Parvisepalum, Brachypetalum, and Paphiopedilum (Guo et al., 2015; Tsai et al., 2020). Species of this genus are characterized by their exotic flowers with a deep shoe-shaped labellum, a structure unique to orchids that is a strongly modified petal, which is the main classification characteristics and pollination channel (Liu et al., 2009), and the droop and wilting of labellum were regarded as the end of floral longevity (Sugiura et al., 2001). Their flowers have long life span, and they can remain on the plant for 62 days (Zhang et al., 2017). However, there are few studies on factors influencing floral longevity of Paphiopedilum species (Zhang et al., 2017, 2021). In this study, we examined the potential link between floral longevity, sizes of genomes, and cells in 13 Paphiopedilum species. Specifically, we hypothesized that floral longevity is positively correlated with genome size, such that species with longer floral longevity may have larger genome. We also examined whether the scaling of genome size and cell size is similar between flower and leaf, or such relationship is different as a result of distinct selective pressures they experienced.
Materials and Methods
Plant Materials
A total of 13 Paphiopedilum species from three subgenera, namely, Parvisepalum, Brachypetalum, and Paphiopedilum were used in this study (Supplementary Figure 1). These plants are grown and kept in the greenhouse at Kunming Institute of Botany, Chinese Academy of Sciences (102°41′ 25°10′ N; elevation 1,990 m). It is helpful to eliminate the effect of environmental divergence on their growth by applying similar culturing conditions. Therefore, variations may mirror the effect of a genetic consequence.
Genome Size
Genome size (1C, pg) (the DNA content of cells in the unreplicated phase from leaf tissue) was measured by flow cytometry in Kunming Institute of Botany, Chinese Academy of Sciences. Three heart leaves of the studied plant tops of species have not been expanded, which were collected and fixed in precooled mGb dissociation solution (45 mM MgCl2·6H2O, 20 mM MOPS, 30 mM sodium citrate, 1% (W/V) PVP 40, 0.2% (v/v) Triton X-100, 10 mM Na2EDTA, 20μl/ml β-mercaptoethanol, pH 7.5). The tissues immediately are chopped in the buffer with a new razor blade, and then mixed the homogenate by pipetting up and down for several times. The homogenate was filtered using the 42-μm nylon mesh and incubated with DNA fluorochrome propidium iodide (PI). PI is used at 50 mg/ml simultaneously with RNase (Dolezel and Bartos, 2005; Dolezel et al., 2007; Tian et al., 2011).
The stained nuclear suspension samples were detected by BD FACScaliburTM flow cytometry (Becton, Dickinson and Company, San Jose, CA 95131, USA). The cytometer was equipped with an argon ion laser operating at 488 nm. The PI fluorescence was collected using the 620 nm fluorescence-2 (FL2) filter. The sample flow rate was set at ~100 nuclei/s, and at least 6,000 nuclei were acquired for each sample. The results were analyzed using BD CellQuestTM Pro software (BD, USA). The results with coefficient of variation values (CV) < 5% were considered as reliable (Supplementary Figure 2). The emitted light fluorescence intensity of PI was detected by 488 nm blue light excitation.
By observing the fluorescence peaks of PI-DNA complex of the tested sample and the internal standard (Zea mays, 1C = 2.41 pg; Schnable et al., 2009), the ratio of DNA content of the two individuals can be obtained, and then multiplied by the C value of internal standard plant, the C value of the tested plant can be tested and calculated by the following equation: DNA content of tested the sample = internal standard DNA content × fluorescence intensity of tested/fluorescence intensity of internal standard (Tian et al., 2011). Orchids are known to differ in cellular ploidy levels (endopolyploidy) in different tissues (Teixeira da Silva et al., 2014; Bateman et al., 2018; Li et al., 2020). In this study, to test the differences of genome size between leaves and other tissues, tissues from labellum and root of Paphiopedilum villosum were used to measure the genome size. We have found that values of genome sizes between leaf and labellum and root tissues are insignificantly different (Supplementary Figure 3). The measure method of genome size (1C, pg) from labellum (the even central part of labellum, Supplementary Figure 4) and root (root tip) was the same as leaf.
Cell Size
Six labellum from newly opened flowers from six plants of each species were fixed in a formalin acetic acid-alcohol solution (37% formaldehyde, glacial acetic acid, 95% ethanol, and deionized water in a 10:5:50:35 mixture) for microscopic analysis. The even central part of labellum was cut into a section to ensure they are flattened, which was used to measure the cell size of labellum (Supplementary Figure 4), and the sampled part of labellum in all plants was the same. Six mature leaves from six different plants per species were sampled. Abaxial epidermis from the middle part of a mature leaf was coated with a thin layer without color, transparent nail polish. These films were gently torn away from the leaf surface with tweezers. These films were mounted on a microscope slide, and images were taken using the light microscope. All samples were obtained to observe epidermal cell and measured using the ImageJ software (National Institutes of Health, Bethesda, MD, USA). Epidermal cell size was subsequently calculated as suggested by Carins Murphy et al. (2016) using the following equation: Epidermal cell size = [1 – (mean stomatal size × stomatal density)]/epidermal cell density.
Floral Longevity
The 10–20 newly emerged floral buds from different plants per species were marked. An individual flower was regarded as “opening” when the dorsal sepal upward and any floral visitor could enter labellum. A flower was identified as “wilting” when perianth was discolored or when labellum began to wilt, thereby ending its function (Sugiura et al., 2001).
Statistical Analysis
Correlations between variables were analyzed using both Pearson's correlation and phylogenetically independent contrasts (PIC). The evolutionary correlations were tested with PIC analysis by using the “ape” package, combining molecular phylogenetic relationships (Cox et al., 1997; Guo et al., 2015). Statistical analyses were performed using R v.4.0.0 (R Core Team, 2020).
Results
Across all studied species, the genome size from the leaf tissue, floral longevity, and epidermal cell size of labellum and leaf varied considerably (Figure 1; Table 1). Specially, genome size (mean genome size ranging from 17.83 to 29.41 pg in the largest species) and floral longevity (mean floral longevity ranging from 26 to 62.13 days in the longest species) varied by up to 1.65 and 2.60 times, respectively. The epidermal cell size of labellum and leaf also varied substantially between species, with a 1.90-fold and a 2.35-fold range, respectively (Table 1).
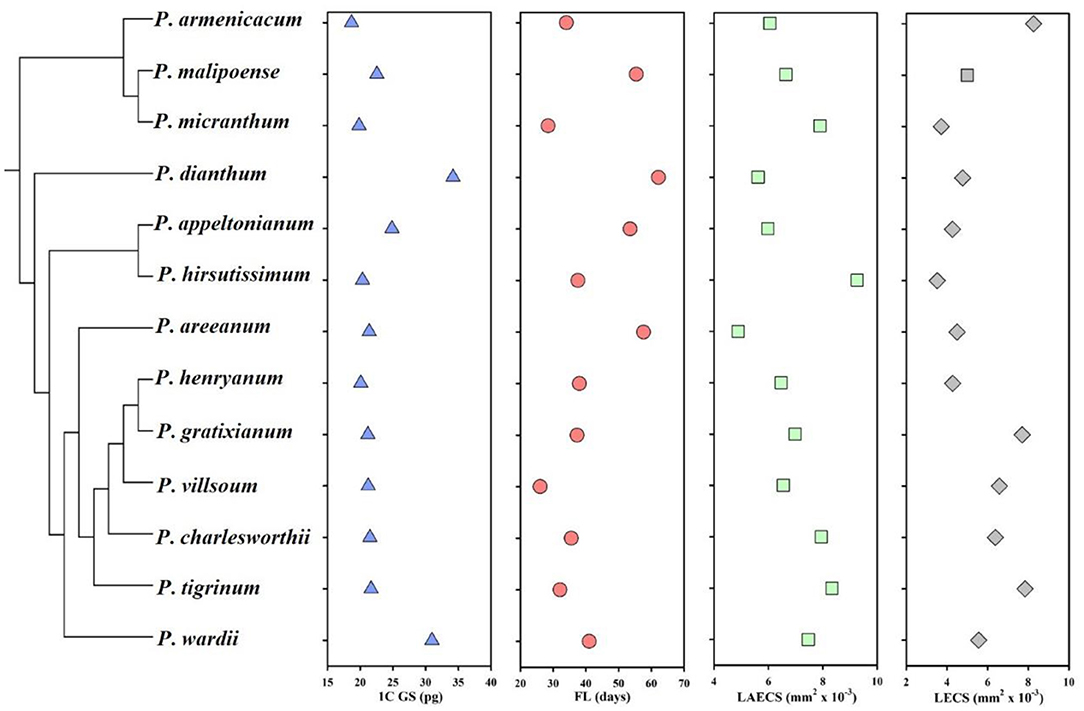
Figure 1. Mean of raw values in genome size, floral longevity, and epidermal cell size of labellum and leaf from 13 Paphiopedilum species. GS, genome size; FL, floral longevity; LAECS, labellum epidermal cell size; LECS, leaf epidermal cell size. The phylogenetic relationships of the studied Paphiopedilum species in this study are obtained from the study by Cox et al. (1997) and Guo et al. (2015).
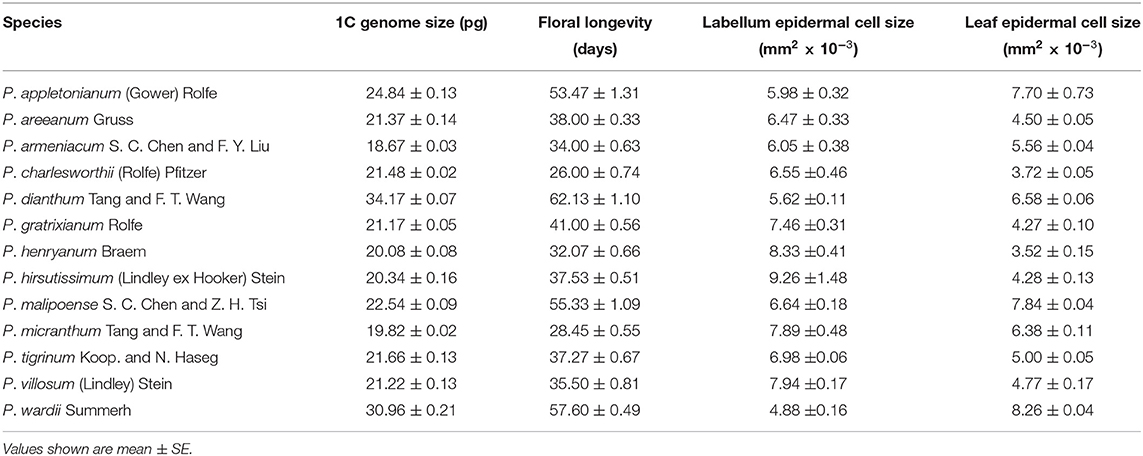
Table 1. Mean values of genome size from leaf tissue (n = 3), floral longevity (10–20 flowers from different plants), epidermal cell size of labellum (n = 6), and leaf (n = 6) from 13 Paphiopedilum species.
Genome size had a significant positive correlation with floral longevity among all species whether or not phylogeny was considered (Figure 2). The epidermal cell size in labellum had a significant negative correlation with genome size and floral longevity even after phylogeny was considered (Figure 2). In addition, genome size was positively correlated with leaf epidermal cell size in both non-phylogenetic and phylogenetic analyses (Figure 3).
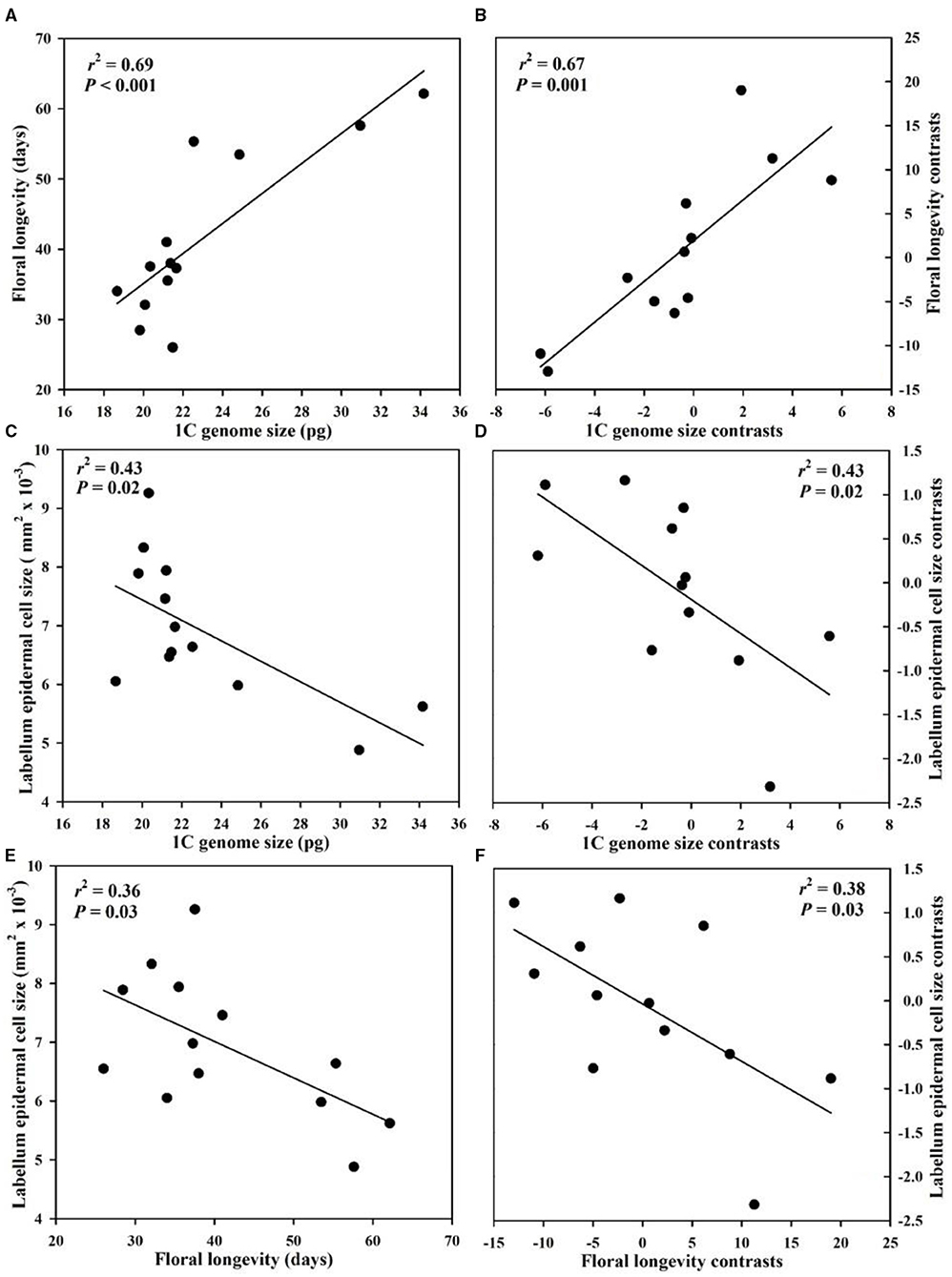
Figure 2. Correlations among genome size, floral longevity, and labellum epidermal cell size from 13 Paphiopedilum species. (A,C,E) Pearson's regressions; (B,D,F) phylogenetically independent contrast correlations.
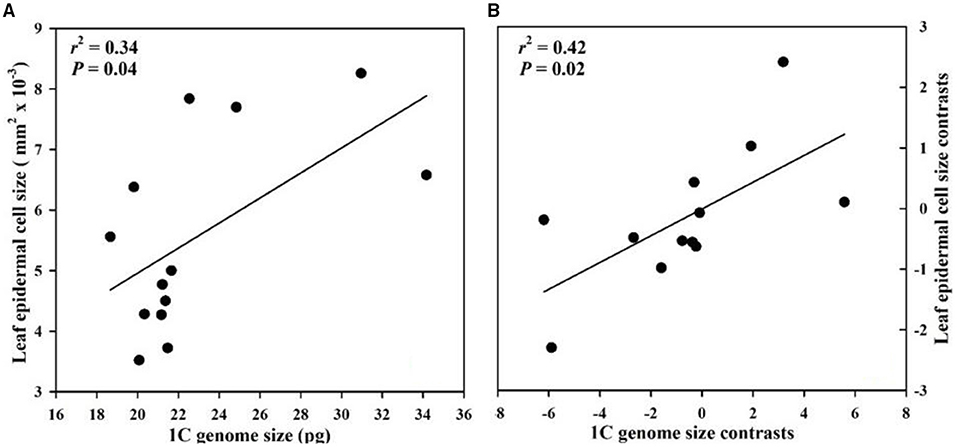
Figure 3. Correlations between genome size and leaf epidermal cell size from 13 Paphiopedilum species. (A) Pearson's regressions; (B) phylogenetically independent contrast correlations.
Discussion
To determine whether floral longevity was driven by genome size and cell size, we obtained data for genome size, floral longevity, and the epidermal cell size of flower and leaf for 13 Paphiopedilum species. We first tested whether floral longevity is associated with the sizes of genomes and epidermal cells. Our results showed that coordinated changes in genome size and cell size to potentially constraint floral longevity in flower (labellum) and leaf of Paphiopedilum. Our results provide new insights into the significance of genome size variation in maintaining floral longevity and evolution of floral traits in orchid plants.
Variation in genome size has aroused studies for seeking evidences as to its phenotypic consequences in plants and animals (Chung et al., 1998; Monaghan and Metcalfe, 2000; Morgan and Westoby, 2005; Beaulieu et al., 2006, 2007; Faizulah et al., 2021; Théroux-Rancourt et al., 2021). Genome size may be closely linked to cell size in biological kingdoms (Cavalier-Smith, 2005). The results may indirectly and/or directly link with whole-organism fitness values (Jockusch, 1997). Longevity is a biological trait with important fitness consequences (Monaghan and Metcalfe, 2000). Differences of longevity between species are likely to be linked to divergences in genome sizes and tissue maintenances (Kirkwood, 1985; Monaghan and Metcalfe, 2000). Our results showed that floral longevity was evolutionarily correlated with genome size and floral epidermal cell size (labellum). Species with longer floral longevity had a large genome and a small labellum cell. Our findings implied that the sizes of genome and cell played an important role in regulating floral longevity. The correlation between genome size and floral longevity that we have shown is very interesting. Whether it may be a causal link and what mechanisms might underlie it remain to be further studied. Endopolyploidy is regarded as the simultaneous existence of multiple ploidy nuclei in adjacent somatic cells of the same individual, tissue, or organ, and it is caused by normal DNA replication without cell division (Barlow, 1978). Recent studies showed that endopolyploidization commonly appeared, and levels from different tissues were different in Orchidaceae (Yang and Loh, 2004; Teixeira da Silva et al., 2014; Bateman et al., 2018; Li et al., 2020). For example, in Ophyrys, the endoreplication was frequent and (Bateman et al., 2018), while there was no evidence of endoreplication in Dactylorhiza (Bateman et al., 2018). In Spathoglottis plicata, the pedicel at the development stage showed the high level of endoploidy, but no endoploidy was found in calyx, petals, and ovary (Yang and Loh, 2004). However, the polyploidy from floral tissues of Paphiopedilum delenatii Guillaumin has not been demonstrated (Teixeira da Silva et al., 2014). In our study, we tried to avoid the interference of internal polyploidy by selecting young leaves. Moreover, we have measured genome sizes from labellum and root tissues, and our results showed that genome sizes from leaves and from other tissues were consistent (Supplementary Figure 3).
Floral longevity can influence the visiting frequency of pollinators, thus affecting plant reproductive rate. Meanwhile, the length of floral longevity is likely to reflect a trade-off between maintenance costs and plant fitness consequences (Kerner von Marilaum, 1895). Therefore, a trade-off should exist between investment and return in flowering plants. Time taken for DNA synthesis, and developmental duration in organs, is longer with larger genomes (Finch, 1990; Monaghan and Metcalfe, 2000; Feng et al., 2021). Within the angiosperms, the genome size of the orchids is relatively large (Leitch et al., 2009), and the reason for this is not clear. But a significant negative correlation between metabolic rate (photosynthetic rate, Amass) and genome size was found (Beaulieu et al., 2007), and the sizes of genomes and cells constraint photosynthetic rate (Roddy et al., 2020). The large genome is thought to be correlated with the low metabolic rate (Beaulieu et al., 2007).
The floral longevity in orchids is generally much longer than other flowering plant. Thus, large genome size would be possibly correlated with floral longevity. Probably, exceeding a certain value, increases of genome size may need to pay a phenotypic cost in biological function. The cell division duration in organs may be an important life history traits (e.g., floral longevity), and Paphiopedilum dianthum with longer floral longevity has longer duration of floral bud differentiation than that of P. micrantum and P. henryanum with shorter floral longevity (Feng et al., 2021). In this study, the strong positive correlation is between floral longevity and genome size in 13 Paphiopedilum species. This result contradicts previous studies showing insignificant relationship between leaf longevity and genome size (Morgan and Westoby, 2005), which suggests that flower and leaf traits of Paphiopedilum have evolved independently, and this possible explanation is that different selective pressures flower and leaf experienced and they acted as different functions (Roddy et al., 2013; Zhang et al., 2017).
In this study, genome size was a strong predictor of leaf epidermal cell size whether or not phylogenetic relatedness of species was considered. Genome size constrains cell size in leaves, so that cell size varies widely as cell grow and differentiate to influence its structure and function in the context of various factors, and then define phenotype (Bennett, 1987; Franks et al., 2012; Simova and Herben, 2012; Théroux-Rancourt et al., 2021), which potentially affect rates of leaf water loss (transpiration) and photosynthetic rate (Beaulieu et al., 2007; Théroux-Rancourt et al., 2021). In our study, we found that the correlation between genome size and cell size from flowers and leaves was different, implying that selective pressures they experienced may be different due to their structures and functions.
Conclusion
We uncovered significant evolutionary correlations between floral longevity and sizes of genomes and cells in Paphiopedilum using a phylogenetic comparative method. These relationships between the sizes of genomes and cell and floral longevity perhaps represent a genotype and phenotype selection relationships. In addition, our results showed that genome size is a strong predictor of cell size. These findings provide novels insights into floral physiological and developmental traits and genome sizes of evolutionary correlations among flowering plant species. Therefore, sampling more species with different genome sizes in orchids and/or angiosperms should be necessary. Further investigation should pay close attention to test for a more direct influence of genome size on floral functional traits. Besides, answering the primary questions needs a continued effort to combine floral functional trait studies with the evolution of plant genome size studies. The endeavor of joining results is critical to reveal evolutionary relationships between genome sizes with floral functional traits in flowering plants.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
F-PZ and S-BZ designed the study and wrote and revised the manuscript. F-PZ collected the samples and data, carried out the experiments, and analyzed the data. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Natural Science Foundation of China (31960224), the Young Top Talents of the Ten Thousand Talents Plan in Yunnan Province (YNWR-QNBJ-2018-337), the Project for Innovation Team of Yunnan Province (202105AE160012), the Yunnan Provincial Science and Technology Department-Applied Basic Research Joint Special Funds of Yunnan University of Chinese Medicine (202001AZ070001-041).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors thank Jing-Qiu Feng, Wei Zhang, and Xue-Wei Fu for providing the images of Paphiopedilum species. They also thank the editors and reviewers for valuable suggestions that greatly improve the quality of the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.793516/full#supplementary-material
References
Ashman, T. L., and Schoen, D. J. (1994). How long should flowers live? Nature 371, 788–791. doi: 10.1038/371788a0
Baetcke, K., Sparrow, A., Nauman, C., and Schwemmer, S. S. (1967). The relationship of DNA content to nuclear and chromosome volumes and to radiosensitivity (LD50). Proc. Natl. Acad. Sci. USA 58, 533–540. doi: 10.1073/pnas.58.2.533
Barlow, P. W (1978). Endopolyploidy: Towards an understanding of its biological significance. Acta Biotheoretica. 27, 1–18. doi: 10.1007/BF00048400
Bateman, M. R., Guy, J. J., Rudall, P. J., Leitch, I. J., Pellicer, J., and Leitch, A. R. (2018). Evolutionary and functional potential of ploidy increase within individual plants: somatic ploidy mapping of the complex labellum of sexually deceptive bee orchids. Ann. Bot. 122, 133–150. doi: 10.1093/aob/mcy048
Beaulieu, J. M., Leitch, I. J., and Knight, C. A. (2007). Genome size evolution in relation to leaf strategy and metabolic rates revisited. Ann. Bot. 99, 495–505. doi: 10.1093/aob/mcl271
Beaulieu, J. M., Leitch, I. J., Patel, S., Pendharkar, A., and Knight, C. A. (2008). Genome size is a strong predictor of cell size and stomatal density in angiosperms. New Phytol. 179, 975–986. doi: 10.1111/j.1469-8137.2008.02528.x
Beaulieu, J. M., Moles, A. T., Leitch, I. J., Bennett, M. D., Dickie, J. B., and Knight, C. A. (2006). Correlated evolution of genome size and seed mass. New Phytol. 173, 422–437. doi: 10.1111/j.1469-8137.2006.01919.x
Bennett, M. D (1972). Nuclear DNA content and minimum generation time in herbaceous plants. Proc. R. Soc. B 181, 109–135. doi: 10.1098/rspb.1972.0042
Bennett, M. D (1987). Variation in genomic form in plants and its ecological implications. New Phytol. 106, 177–200. doi: 10.1111/j.1469-8137.1987.tb04689.x
Bennett, M. D., Heslop-Harrison, J. S., Smith, J. B., and Ward, J. P. (1983). DNA density in mitotic and meiotic metaphase chromosomes of plants and animals. J. Cell Sci. 63, 173–179. doi: 10.1242/jcs.63.1.173
Carins Murphy, M. R., Jordan, G. J., and Brodribb, T. J. (2016). Cell expansion not cell differentiation predominantly co-ordinates veins and stomata within and among herbs and woody angiosperms grown under sun and shade. Ann. Bot. 118, 1127–1138. doi: 10.1093/aob/mcw167
Cavalier-Smith, T (2005). Economy, speed and size matter: evolutionary forces driving nuclear genome miniaturization and expansion. Ann. Bot. 95, 147–175. doi: 10.1093/aob/mci010
Chase, M. W (2005). Classification of Orchidaceae in the age of DNA data. Curtis's Bot. Mag. 22, 2–7. doi: 10.1111/j.1355-4905.2005.00466.x
Chung, J., Lee, J. H., Arumuganathan, K., Graef, G. L., and Specht, J. E. (1998). Relationships between nuclear DNA content and seed and leaf size in soybean. Theor. Appl. Genet. 96, 1064–1068. doi: 10.1007/s001220050840
Cox, A. V., Abdelnour, G. J., Bennett, M. D., and Leitch, I. J. (1998). Genome size and karyotype evolution in the slipper orchids (Cypripedioideae: Orchidaceae). Am. J. Bot. 85, 681–687. doi: 10.2307/2446538
Cox, A. V., Pridgeon, A. M., Albert, V. A., and Chase, M. W. (1997). Phylogenetics of the slipper orchids (Cypripedioideae, Orchidaceae): nuclear rDNA ITS sequences. Plant Syst. Evol. 208, 197–223. doi: 10.1007/BF00985442
Dolezel, J., and Bartos, J. (2005). Plant DNA flow cytometry and estimation of nuclear genome size. Ann. Bot. 95, 99–110. doi: 10.1093/aob/mci005
Dolezel, J., Greilhuber, J., and Suda, J. (2007). Estimation of nuclear DNA content in plants using flow cytometry. Nat. Protoc. 2, 2233–2244. doi: 10.1038/nprot.2007.310
Edwards, G. A., and Endrizzi, J. L. (1975). Cell size nuclear size and DNA content relationships in Gossypium. Can. J. Genet. Cytol. 17, 181–186. doi: 10.1139/g75-024
Faizulah, L., Morton, J., Hersch-Green, E. I., Walczyk, A. M., Leitch, A. R., and Leitch, I. J. (2021). Exploring environmental selection on genome size in angiosperms. Trends Plant Sci. 26, 1039–1049. doi: 10.1016/j.tplants.2021.06.001
Feng, J. Q., Xia, Q., Zhang, F. P., Wang, J. W., and Zhang, S. B. (2021). Is seasonal flowering time of Paphiopedilum species caused by differences in initial time of floral bud differentiation? AoB Plants 13: plab 053. doi: 10.1093/aobpla/plab053
Finch, C. E (1990). Longevity, Senescence, and the Genome University of Chicago Press. Chicago, IL: University of Chicago Press
Franks, P. J., Leitch, I. J., Ruszala, E. M., Hetherington, A. M., and Beerling, D. J. (2012). Physiological framework for adaptation of stomata to CO2 from glacial to future concentrations. Philos. T. R. Soc. B 367, 537–546. doi: 10.1098/rstb.2011.0270
Gori, D. F (1983). “Post-pollination phenomena and adaptive floral changes,” in Handbook of Experimental Pollination Biology, eds Jones, C.E., Little, R.J. (New York, NY: Scientific and Academic), 31–49.
Guo, Y. Y., Luo, Y. B., Liu, Z. J., and Wang, X. Q. (2015). Reticulate evolution and sea-level fluctuations together drove species diversification of slipper orchids in South-East Asia. Mol. Ecol. 24, 2838–2855. doi: 10.1111/mec.13189
Jockusch, E. L (1997). An evolutionary correlate of genome size change in plethodontid salamanders. Proc. R. Soc. B 264, 597–604. doi: 10.1098/rspb.1997.0085
Kerner von Marilaum, A (1895). The Natural History of Plants, Their Forms, Growth, Reproduction and Distribution. New York, NY: Henry Holt.
Kirkwood, T. B. L (1985). “Comparative aspects of longevity,” in Handbook of the Biology of Aging, eds Finch, C. E. and Schneider, E. L. New York, NY: Van Nostrand Reinhold, 27–45.
Knight, C. A., and Ackerly, D. D. (2002). Variation in nuclear DNA content across environmental gradients: a quantile regression analysis. Ecol. Lett. 5, 66–76. doi: 10.1046/j.1461-0248.2002.00283.x
Knight, C. A., Molinari, N. A., and Petrov, D. A. (2005). The large genome constraint hypothesis: evolution, ecology and phenotype. Ann. Bot. 95, 177–190. doi: 10.1093/aob/mci011
Lawrence, M. E (1985). Senecio L. (Asteraceae) in Australia: nuclear DNA amounts. Aust. J. Bot. 33, 221–232. doi: 10.1071/BT9850221
Leitch, I. J., Kahandawala, I., Suda, J., Hanson, L., Ingrouille, M. J., Chase, M. W., et al. (2009). Genome size diversity in orchids: consequences and evolution. Ann. Bot. 104, 469–481. doi: 10.1093/aob/mcp003
Leitch, I. J., Soltis, D. E., Soltis, P. S., and Bennett, M. D. (2005). Evolution of DNA amounts across land plants (Embryophyta). Ann. Bot. 95, 207–217. doi: 10.1093/aob/mci014
Li, C. N., Fu, Q. J., Shen, G. Z., Zhao, F. K., Zhang, X. Y., and Ruan, R. X. (2020). Applications of flow cytometry in orchidaceae. J. Nucl. Agricult. Sci. 34, 0973–0981. doi: 10.11869/j.issn.100-8551.2020.05.0973
Liu, Z. J., Chen, X. Q., Chen, L. J., and Lei, S. P. (2009). The Genus Paphiopedilum in China. Beijing: Science Press.
Meagher, T. R., Gillies, A. C. M., and Costich, D. E. (2005). Genome size, quantitative genetics and the genomic basis for flower size evolution in Silene latifolia. Ann. Bot. 95, 247–254. doi: 10.1093/aob/mci018
Monaghan, P., and Metcalfe, N.B. (2000). Genome size and longevity. Trends Genet. 16, 331–332. doi: 10.1016/S0168-9525(00)02051-5
Morgan, H. M., and Westoby, M. (2005). The relationship between Nuclear DNA content and leaf strategy in seed plant. Ann. Bot. 96, 1321–1330. doi: 10.1093/aob/mci284
Parveen, I., Singh, H. K., Raghuvanshi, S., and Pradhan, U. C. (2012). DNA barcoding of endangered Indian Paphiopedilum species. Mol. Ecol. 12, 82–90. doi: 10.1111/j.1755-0998.2011.03071.x
Pellicer, J., Hidalgo, O., Dodsworth, S., and Leitch, I. J. (2018). Genome size diversity and its impact on the evolution of land plants. Genes 9:88. doi: 10.3390/genes9020088
Petrov, D. A (2002). Mutational equilibrium model of genome size evolution. Theor. Popul. Biol. 61, 531–543. doi: 10.1006/tpbi.2002.1605
Primack, R. B (1985). Longevity of individual of flowers. Annu. Rev. Ecol. Syst. 16, 15–37. doi: 10.1146/annurev.es.16.110185.000311
R Core Team (2020). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. Available online at: www.R-project.org/
Rees, H., Cameron, F. M., Hararika, M. H., and Jones, G. H. (1966). Nuclear variation between diploid angiosperms. Nature 211, 828–830. doi: 10.1038/211828a0
Roddy, A. B., Guilliams, C. M., Lilittham, T., Farmer, J., Wormser, V., Pham, T., et al. (2013). Uncorrelated evolution of leaf and petal venation patterns across the angiosperm phylogeny. J. Exp. Bot. 64, 4081–4088. doi: 10.1093/jxb/ert247
Roddy, A. B., Theroux-Rancourt, G., Abbo, T., Brodersen, C. R., and Simonin, K. A. (2020). The scaling of genome size and cell size limits maximum rates of photosynthesis with implications for ecological strategies. Int. J. Plant Sci. 181, 75–87. doi: 10.1086/706186
Rosas-Guerrero, V., Aguila, R., Martén-Rodríguez, S., Ashworth, L., Lopezaraiza-Mikel, M., Bastida, J. M., et al. (2014). A quantitative review of pollination syndromes: do floral traits predict effective pollinators? Ecol. Lett. 17, 388–400. doi: 10.1111/ele.12224
Schnable, P. S., Ware, D., Fulton, R. S., Stein, J. C., Wei, F. S., Pasternak, S., et al. (2009). The B73 Maize genome: complexity, diversity, and dynamics. Science 326:1112. doi: 10.1126/science.1178534
Simonin, K. A., and Roddy, A. B. (2018). Genome downsizing, physiological novelty, and the global dominance of flowering plants. PLoS Biol. 16:e2003706. doi: 10.1371/journal.pbio.2003706
Simova, I., and Herben, T. (2012). Geometrical constraints in the scaling relationships between genome size, cell size and cell cycle length in herbaceous plants. Proc. R. Soc. B 279, 867–875. doi: 10.1098/rspb.2011.1284
Sims, L. E., and Price, H. J. (1985). Nuclear DNA content variation in Helianthus (Asteraceae).Am. J. Bot. 72, 1213–1219. doi: 10.1002/j.1537-2197.1985.tb08374.x
Šmarda, P., and Bureš, P. (2010). Understanding intraspecific variation in genome size in plants. Preslia 82, 41–61. doi: 10.1055/s-0029-1240632
Soltis, D. E., Soltis, P. S., Bennett, M. D., and Leitch, I. J. (2003). Evolution of genome size in the angiosperms. Am. J. Bot. 90, 1596–1603. doi: 10.3732/ajb.90.11.1596
Sugiura, N., Fujie, T., Inoue, K., and Kitamura, K. (2001). Flowering phenology, pollination, and fruit set of Cypripedium macranthos var. rebunense, a threatened lady's slipper (Orchidaceae). J. Plant Res. 114, 171–178. doi: 10.1007/PL00013980
Sugiyama, S (2005). Developmental basis of interspecific differences in leaf size and specific leaf area among C3 grass species. Funct. Ecol. 19, 916–924. doi: 10.1111/j.1365-2435.2005.01044.x
Teixeira da Silva, J., Giang, D., Dobránszki, J., Zeng, S., and Tanaka, M. (2014). Ploidy analysis of Cymbidium, Phalaenopsis, Dendrobium and Paphiopedillum (Orchidaceae), and Spathiphyllum and Syngonium (Araceae). Biologia 69, 750–755. doi: 10.2478/s11756-014-0370-z
Théroux-Rancourt, G., Roddy, A. B., Earles, M. J., Gilbert, M. E., Maciej, A., Zwieniecki, M. A., et al. (2021). Maximum CO2 diffusion inside leaves is limited by the scaling of cell size and genome size. Proc. R. Soc. B 288:20203145. doi: 10.1098/rspb.2020.3145
Tian, X. M., Zhou, X. Y., and Gong, N. (2011). Application of flow cytometry in plant research—analysis of nuclear DNA content and ploidy level in plant cells. Chinese Agricult. Sci. Bull. 9, 21–27. doi: 10.1093/mp/ssq070
Tsai, C. C., Liao, P. C., Ko, Y. Z., Chen, C. H., and Chiang, Y. C. (2020). Phylogeny and Historical Biogeography of Paphiopedilum Pfitzer (Orchidaceae) Based on Nuclear and Plastid DNA. Front. Plant Sci. 11:126. doi: 10.3389/fpls.2020.00126
Wakamiya, I., Newton, R. J., Johnston, S. J., and Price, J. H. (1993). Genome size and environmental factors in the genus Pinus. Am. J. Bot. 80, 1235–1241. doi: 10.1002/j.1537-2197.1993.tb15360.x
Yang, M., and Loh, C. S. (2004). Systemic endopolyploidy in Spathoglottis plicata (orchidaceae) development. BMC Cell Biol. 5:33. doi: 10.1186/1471-2121-5-33
Zhang, F. P., Feng, J. Q., Huang, J. L., Huang, W., Fu, X. W., Hu, H., et al. (2021). Floral Longevity of Paphiopedilum and Cypripedium is associated with floral morphology. Front. Plant Sci. 12:637236. doi: 10.3389/fpls.2021.637236
Zhang, F. P., Yang, Y. J., Yang, Q. Y., Zhang, W., Brodribb, T. J., Hao, G. Y., et al. (2017). Floral mass per area and water maintenance traits are correlated with floral longevity in Paphiopedilum (Orchidaceae). Front. Plant Sci. 8:501. doi: 10.3389/fpls.2017.00501
Keywords: genome size, floral longevity, cell size, flower, leaf, slipper orchids
Citation: Zhang F-P and Zhang S-B (2021) Genome Size and Labellum Epidermal Cell Size Are Evolutionarily Correlated With Floral Longevity in Paphiopedilum Species. Front. Plant Sci. 12:793516. doi: 10.3389/fpls.2021.793516
Received: 12 October 2021; Accepted: 19 November 2021;
Published: 16 December 2021.
Edited by:
Elena M. Kramer, Harvard University, United StatesReviewed by:
Philipp M. Schlüter, University of Hohenheim, GermanyXiaohua Jin, Institute of Botany, Chinese Academy of Sciences (CAS), China
Copyright © 2021 Zhang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shi-Bao Zhang, c2J6aGFuZ0BtYWlsLmtpYi5hYy5jbg==
 Feng-Ping Zhang
Feng-Ping Zhang Shi-Bao Zhang2*
Shi-Bao Zhang2*