- Department of Grassland Science, College of Animal Science and Technology, Northeast Agricultural University, Harbin, China
Alfalfa is widely grown worldwide as a perennial high-quality legume forage and as a good ecological landcover. The cysteine synthase (CSase) gene family is actively involved in plant growth and development and abiotic stress resistance but has not been systematically investigated in alfalfa. We identified 39 MsCSase genes on 4 chromosomes of the alfalfa genome. Phylogenetic analysis demonstrated that these genes were clustered into six subfamilies, and members of the same subfamily had similar physicochemical properties and sequence structures. Overexpression of the CSase gene in alfalfa increased alkali tolerance. Compared with control plants, the overexpression lines presented higher proline, soluble sugars, and cysteine and reduced glutathione contents and superoxide dismutase and peroxidase activities as well as lower hydrogen peroxide and superoxide anion contents after alkali stress. The relative expression of γ-glutamyl cysteine synthetase gene (a downstream gene of CSase) in the overexpression lines was much higher than that in the control line. The CSase gene enhanced alkalinity tolerance by regulating osmoregulatory substances and improving antioxidant capacity. These results provide a reference for studying the CSase gene family in alfalfa and expanding the alkali tolerance gene resources of forage plants.
Introduction
Cysteine is the first organic substance in plants found to contain both sulfur and nitrogen and is a precursor of sulfur-containing metabolites such as methionine (Takahashi et al., 2011), glutathione (GSH) and Fe-S clusters, which play an important role in plant development and metabolic processes (Droux, 2004; Van Hoewyk et al., 2008). The synthesis of cysteine can be roughly divided into the absorption and reduction of elemental sulfur (Kopriva, 2006; Davidian and Kopriva, 2010). Cysteine synthase (CSase) is involved in the final step of cysteine synthesis; this enzyme catalyzes the synthesis of cysteine from H2S and O-acetylserine (OAS) (Jez and Dey, 2013; Romero et al., 2014).
The CSase gene is often referred to as the O-acetylserine(thiol)lyase gene (OAS-TL) and belongs to the CSase gene family (Droux et al., 1998; Wirtz and Hell, 2006; Alvarez et al., 2010a). The CSase gene family was previously identified in Arabidopsis (Arabidopsis thaliana) and was found to comprise nine genes divided into five subfamilies, all of whose members contain PLP-binding sites (PXXSVKDR) that are highly conserved across species (Yamaguchi et al., 2000). However, cytosolic OAS-A1, plastidial OAS-B, and mitochondrial OAS-C were identified as the three OASTLs that were also expressed at relatively high levels and interacted with ser acetyltransferase (SAT) (Bonner et al., 2005; Heeg et al., 2008; Jez and Dey, 2013). In addition, CYS-D1 and CYS-D2 also have weak cysteine synthesis functions in mitochondria (Yamaguchi et al., 2000). CSases compose a protein family whose members have multiple functions, and CSase genes in different tissue sites may have different functions. For example, L-cysteine desulfhydrase 1 (DES1) in the cytoplasm has L-cysteine desulfhydrase activity, sulfocysteine synthase (SCS) in the chloroplast encodes S-thiocysteine synthase, and the mitochondrial enzyme CAS-C1 has β-cyanoalanine synthase (CAS) activity (Alvarez et al., 2010a; Bermúdez et al., 2010). Recently, it has also been shown that CSase genes are involved in environmental stress responses such as responses to high-salt conditions and heavy metals, and overexpression of CSase genes has been shown to increase the ability of plants to adapt to oxidative stress (Ning et al., 2009; Xie et al., 2012). Moreover, a wide range of defense compounds that can respond to adverse environments use cysteine as a precursor (Alvarez et al., 2010b). Taking the GSH metabolic pathway as an example, the synthesis of GSH as an antioxidant molecule is restricted by cysteine, and in turn GSH is a precursor for the synthesis of phytochelatins (PCs), thiolated peptides involved in the detoxification of heavy metals (Cui et al., 2012, 2014). When plants need to enhance GSH biosynthesis under heavy metal stress, CSase can increase cysteine production and subsequently affect the synthesis of downstream substances and achieve improved plant tolerance.
The earliest report of the CYS-C1 gene in Arabidopsis involved a CAS that converts cyanide and cysteine to β-cyanoalanine and H2S in mitochondria. CYS-C1 and Cys-C act together to complete the cyclic pathway of cyanide detoxification (García et al., 2010; Álvarez et al., 2012b). However, the activity of CYS-C1 during cysteine synthesis is also relatively high (Hatzfeld et al., 2000), and CYS-C1 is considered a member of the CSase gene family (Watanabe et al., 2008). Since SAT and CSase interact for efficient synthesis of cysteine, authentic CSase can interact with SAT (Droux et al., 1998; Romero et al., 2014). Moreover, the SlOAS7 gene in the CYS-C subfamily was found to interact with SAT in tomato (Liu et al., 2018). Based on the above information, it is speculated that CYS-C subfamily members may also be true CSases.
Alfalfa (Medicago sativa L.), which is widely grown in Asia, Europe, and America, is a high-quality perennial forage plant of the legume family; alfalfa is high yielding and rich in nutrients and is one of the most important forage species for healthy and efficient livestock breeding. The CSase gene family has been extensively studied in many species, and genome-wide analyses have identified members of the CSase gene family in Arabidopsis (Yamaguchi et al., 2000), tomato (Solanum lycopersicum L.) (Liu et al., 2018), foxtail millet (Setaria italica (L.) P. Beauvois) (Liu et al., 2019), and sorghum (Sorghum bicolor) (Akbudak et al., 2018), but our knowledge of the CSase gene family in forage crop species such as alfalfa is still limited. The recently published genome of alfalfa (cultivar Xinjiangdaye) provides an important resource for further molecular studies of this species (Chen et al., 2020). Previous work by our group found that this gene responds to alkali stress (Song et al., 2017, 2021). Based on this information, a total of 39 CSase genes were identified and classified into 6 subfamilies in this study, and bioinformatic analyses including phylogenetic analysis, motif composition analysis, and gene duplication analysis were performed to provide a theoretical basis for clarifying the evolutionary history and biological functions of the members of this gene family. In addition, we successfully cloned a CSase gene (belonging to the CYS-C1 subfamily) from alfalfa, transferred it into alfalfa, and analyzed its potential function. The results showed that this gene encodes a protein that promotes cysteine synthesis and improves the alkalinity tolerance of overexpression lines by increasing the antioxidant capacity of the plant.
Materials and Methods
Identification of Cysteine Synthase Gene Family Members in Alfalfa
The sequences of the nine identified AtCSase genes were obtained from the NCBI database.1 The M. sativa Xinjiangdaye genome sequence was downloaded from a website.2 MsCSases were identified by two rounds of BLASTP. A hidden Markov model (HMM) was used by Pfam 31.13 to ensure that the PF00291 domain was retained, and DNAMAN was used for sequence comparison searches for the PLP-binding site (PXXSVKDR) in alfalfa.
Phylogenetic Analysis and Multiple Sequence Alignment
A phylogenetic tree was generated by MEGA 5 using the NJ method, with 1,000 bootstrap replicates. Multiple sequence alignments of CSases were created with ClustalX.
Analysis of Conserved Motifs and Conserved Domains
The conserved motif structures within the CSase sequences were identified by MEME Suite Version 5.2.04 with the following parameters: zero or one occurrence per sequence of site distribution, a maximum of 10 misfits and a maximum width of motif between 6 and 50. NCBI Batch CD-Search5 was used to analyze the conserved domains of the CSase proteins, after which the domains were visualized by TBtools.
Vector Construction and Plant Transformation
We used the CSase gene of M. truncatula (Medtr7g078070.1) as a reference sequence to clone the CSase gene of M. sativa. Transient expression vectors for tobacco and overexpression vectors for alfalfa were constructed by the one-step cloning method. Then, the expression vectors were transformed into Agrobacterium rhizogenes by using the freeze-thaw method. The CSase gene was transformed into alfalfa via Agrobacterium mediation using the cotyledon method, and regenerated alfalfa plants were obtained. The bar gene detection method and fluorescence quantitative analysis technology were used to screen overexpression plants. The sequences of the primers used are shown in Supplementary Table 1.
Plant Growth Conditions and Treatments
Nicotiana benthamiana plants were grown in plastic pots filled with vermiculite. Approximately 1-month-old seedlings were used for transient expression. M. sativa Longmu 801 was used in this study. Softwood cuttings from the CK line and overexpression lines OV#L11, OV#L12, and OV#L13 were transplanted into plastic pots containing vermiculite (one plant per pot). All the plants were grown under a 16 h light/8 h dark photoperiod under a day/night temperature cycle of 22°C/18°C. Hoagland solution (1/10 strength) was applied to the plants every 3 days.
For NaHCO3 treatment, 150 mM NaHCO3 was applied for 5 d, and a 5 d recovery period was selected as the best condition for identifying stress phenotypes. Samples were taken at 0, 1, 6, 12, 24, 48 h, and 5 d after the beginning of the treatment. Three biological replicates were included per line.
Subcellular Localization Analysis
To explore the subcellular localization of CSase proteins, we constructed a transient expression vector. CSase gene was inserted downstream from the double CaMV 35S promoter in the pCAMBIA1300 vector. The pCAMBIA1300 vector carries GFP gene. The sequences of the primers used are listed in Supplementary Table 1. The resulting vector was introduced into the Agrobacterium tumefaciens strain GV3101. We used a syringe to infiltrate Agrobacterium tumefaciens strain GV3101 containing a tobacco transient expression vector into 1-month-old tobacco leaves. After infiltration, the plants were cultivated for 72 h under dark conditions. The fluorescence signal in the infested tobacco leaves was subsequently observed by confocal microscopy.
Determination of Physiological Indicators and Expression Analysis of Related Genes
Physiological traits including GSH, cysteine, Pro, and MDA contents and SOD and POD activities were measured using reagent kits (Nanjing Jiancheng Bioengineering Institute, Nanjing, China). The instructions of the kits were followed for specific test procedures.
For qRT-PCR, total RNA was isolated from alfalfa samples using an RNeasy Plant Mini Kit (CWBIO, Jiangsu, China), and cDNA was synthesized using a kit (Vazyme). qRT-PCR was used to analyze the relative expression levels of alfalfa CSase-responsive genes. The GADPH gene was used as a reference. The sequences of the primers used are shown in Supplementary Table 1.
Statistical Analyses
To determine significance, all statistical analyses were performed by using Microsoft Excel.
Results
Identification of MsCSase Genes in Alfalfa
First, a total of 39 MsCSase gene sequences were retrieved from alfalfa using BLAST, PF00291 domain and PLP-binding site (PXXSVKDR) searches and named MsCSase01 to MsCSase39 according to their chromosome locations (Supplementary Table 2). They were unevenly mapped onto chromosomes 1, 4, 5, and 7, which contained 14, 9, 7, and 9 genes, respectively (Supplementary Table 2). Information about their coding DNA sequences (CDS) and resulting protein sequences are presented in Supplementary Table 3. Then, characterization of the proteins revealed that the predicted isoelectric points (pIs) of the MsCSase proteins ranged from 5.17 to 9.08 (Supplementary Table 2). Except for MsCSase01, MsCSase31 and MsCSase36, the length and molecular mass did not widely vary (Supplementary Table 2). The phylogenetic tree results demonstrated that the MsCSase proteins could be divided into 6 subfamilies according to the clades and classification from Arabidopsis, including 14, 9, 5, 4, and 4 members in the CysA subfamily, CysB subfamily, SCS subfamily, CysD subfamily and CysC subfamily, respectively (Figure 1). Similar to that which occurred in a study in tomato (Liu et al., 2018), MsCSase25, MsCSase27, and MsCSase28 were separated into a separate family and did not belong to the other five subfamilies. These results indicated that the characteristics and patterns of evolution in various species are more likely to differ.
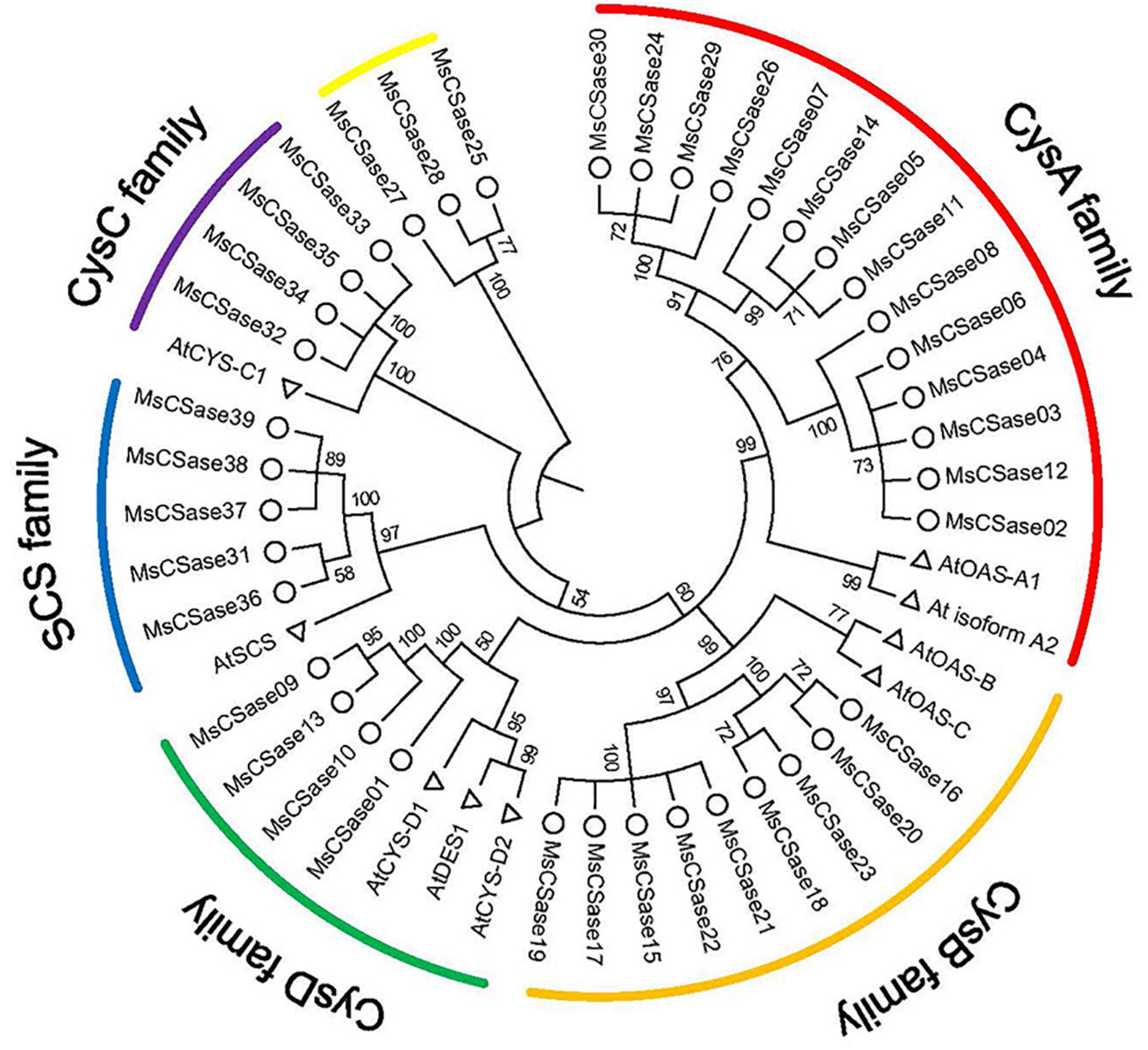
Figure 1. Phylogenetic tree generated using the NJ method. The tree represents relationships among CSases of alfalfa and Arabidopsis. The different colored arcs indicate different CSase subfamilies. Δ and ° represent CSase genes in alfalfa and Arabidopsis, respectively. The number represents the bootstrap value of the node. Larger node values imply a higher confidence level.
Structural Features and Synteny Analysis
Genetic structural diversity supported the phylogenetic groupings to some extent (Wei et al., 2016). Therefore, we analyzed the relationship between gene structure and phylogenetic clustering to gain insight into the evolution of the MsCSase gene family in alfalfa (Figures 2A,B). Gene structure analysis showed that genes within the same subfamily presented similar structures; for instance, the CysA subfamily members contained 10 exons, and the exon distribution of genes on the same branch was largely similar. In addition, the protein motif analysis (the conserved motifs via sequence logo are shown in Supplementary Figure 1) by MEME found a similar pattern and the order and distribution of the motifs were roughly similar among the members of the subfamilies (Figure 2C). These results supported the close evolutionary relationship of the classification of these MsCSase subfamilies. Moreover, the MsCSase genes had five characterized domains, including PLN02565, the PLN02565 superfamily, Trp-synth-beta_II superfamily and PLN02556 domains, all of which are related to cysteine synthesis (Figure 2D). Overall, the domain similarity suggested that these genes may have similar functions, but the differences in the activity of their encoded enzymes may be related to differences in gene structure and motifs between subfamily members.
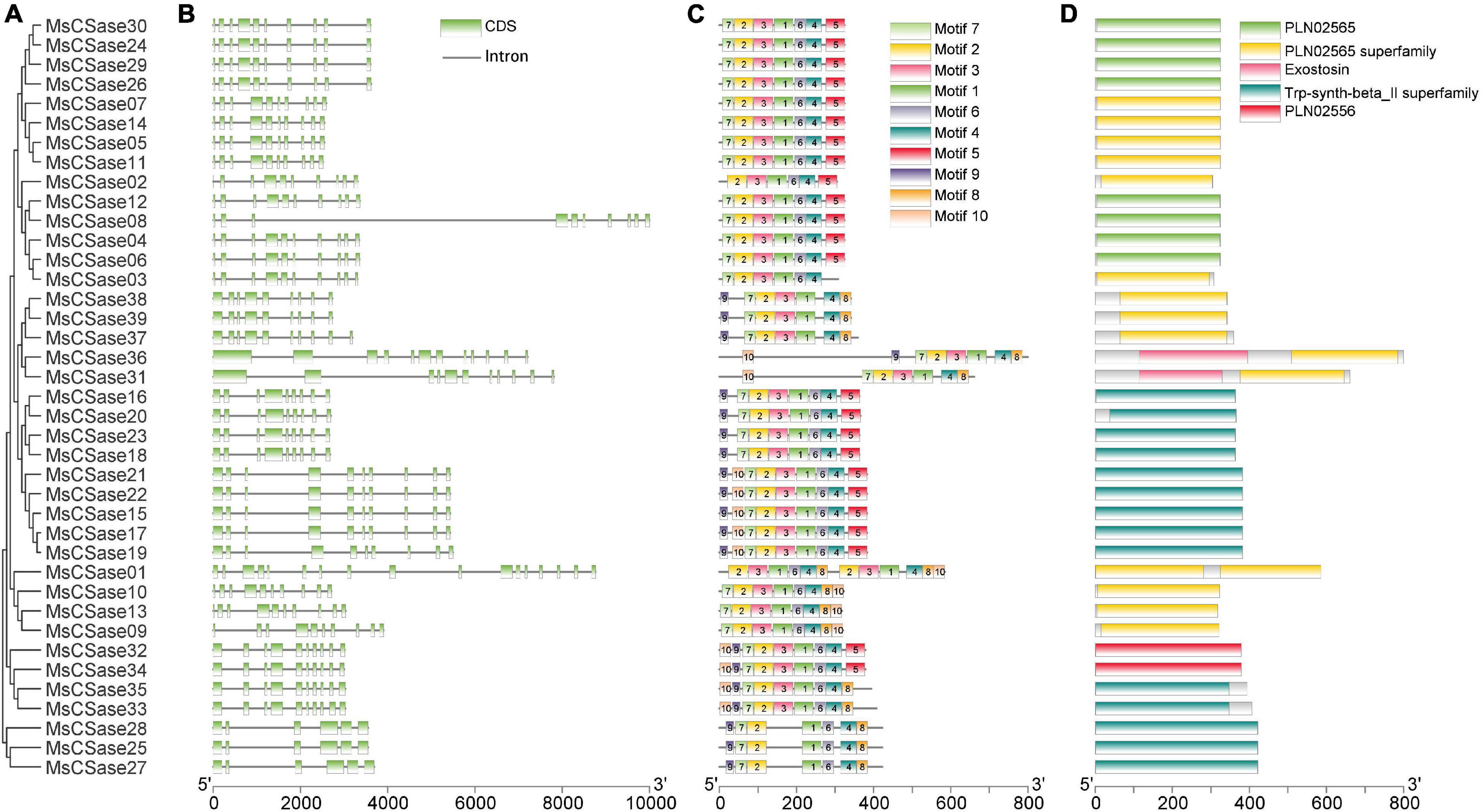
Figure 2. Architecture of conserved protein motifs, domains and gene structures among the MsCSase gene family members in alfalfa. (A) Phylogenetic tree of alfalfa MsCSase proteins constructed using MEGA 5 software. (B) Gene structure of the MsCSase gene family members. The green boxes indicate exons; the black lines indicate introns. (C) Motif composition of MsCSase proteins. The motifs, numbered 1–10, are displayed in different colored boxes. The results of a sequence logo analysis of the conserved motifs are provided in Supplementary Figure 1. The length of the protein can be estimated using the scale at the bottom. (D) The characterized domains of the MsCSase genes were identified. The different colors represent different domains.
To elucidate the mechanism through which the MsCSase gene family members in alfalfa expanded, gene duplication events were identified. Two pairs of tandem duplication genes (MsCSase9/10, MsCSase22/23) and 35 groups of synteny gene pairs in which MsCSase07/24 were segmentally duplicated genes were identified by TBtools and MCScanX software (Supplementary Table 4). Duplicated genes were located on chromosomes 1, 5, and 10 (Supplementary Figure 2). Taken together, the results indicated that there was no obvious relationship between chromosome length and the number of genes. Some MsCSase genes may have been generated by gene duplications and tandem and segmental duplications contributed to the evolution of MsCSases in alfalfa.
Molecular Cloning and Subcellular Localization of CSase
Using the Medtr7g078070.1 gene of Medicago truncatula as a probe, we cloned the gene with accession number MK334208 named CSase from alfalfa (cultivar Longmu 801). Sequence analysis showed that the amino acid sequences of MsCSas32, MsCSase34, and CSase were nearly identical (Supplementary Figure 3); they were 99.91% similar at the nucleotide level, and only one nucleotide differed between MsCSase34 and CSase (Supplementary Figure 4). Combining the results of the phylogenetic evolutionary tree analysis with these results, we determined that the CSase gene belonged to the CYS-C1 subfamily and was highly conserved in two different alfalfa varieties.
qRT-PCR analysis of different alfalfa tissue parts revealed that the relative expression of CSase was higher in the leaves than in the other tissues (Figure 3A). Moreover, the relative expression in mature leaves was much higher than that in other tissues. To further clarify the location of gene activity, we evaluated the subcellular localization of CSase. We fused its ORF sequence without the terminal codon to GFP at the N-terminus under the control of the CaMV 35S promoter and ultimately transiently expressed it in tobacco (N. benthamiana) leaf epidermal cells. The results showed that the presence of the CSase–GFP fusion protein in the chloroplasts of the cells (Figure 3B). This finding also directly validates the accuracy of the qRT-PCR results.
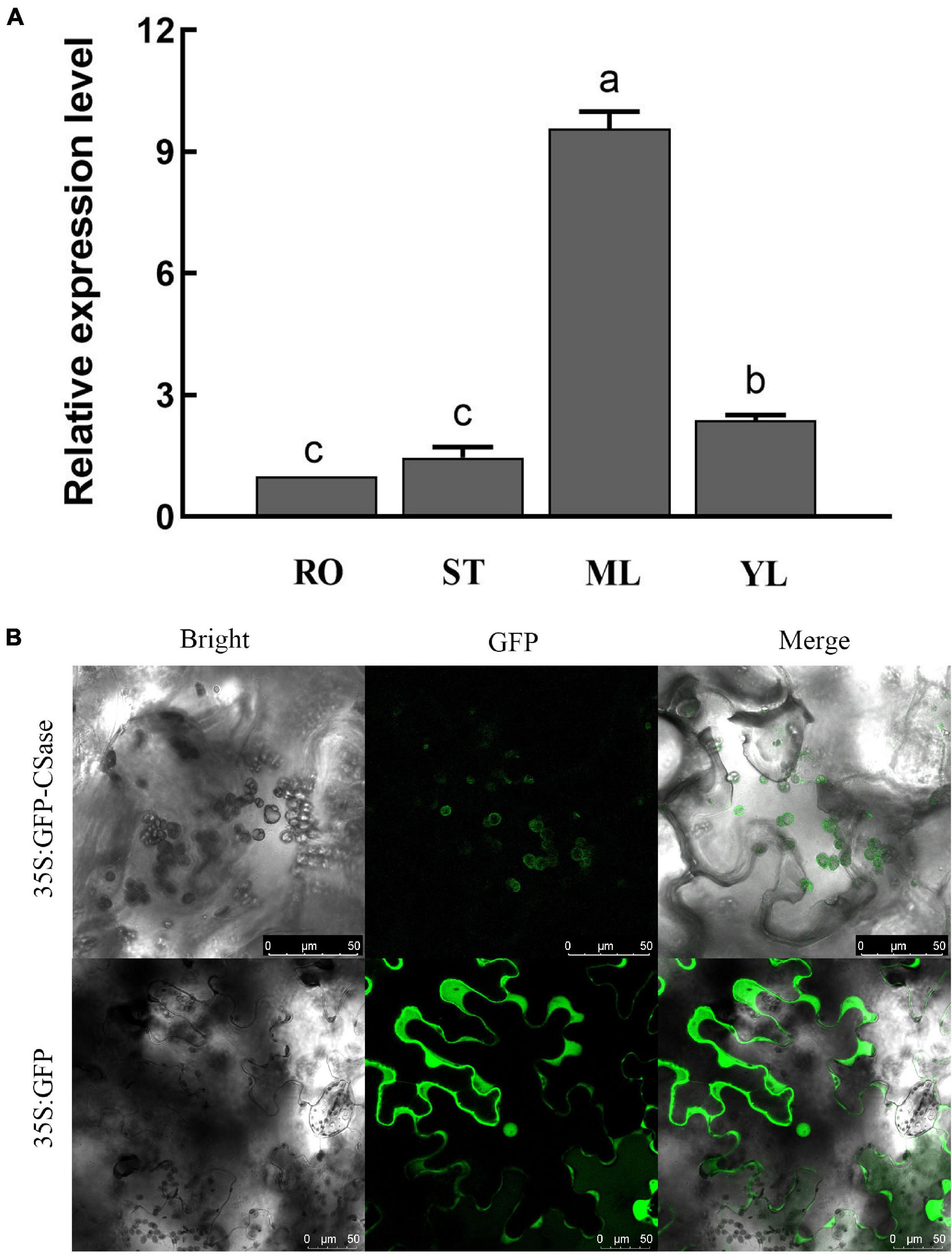
Figure 3. Expression pattern and subcellular localization of CSase. (A) Differential expression of representative CSases in different tissues according to qRT-PCR. RO, root; ST, stem; ML, mature leaf; and YL, young leaf. (B) Confocal laser scanning microscopy images of tobacco leaf cells expressing the CSase protein (35S:GFP-CSase) and GFP protein (35S:GFP). Scale bars = 50 μm.
Overexpression of Cysteine Synthase Enhances Alkali Tolerance in Transgenic Alfalfa
qRT-PCR analysis under different abiotic stresses revealed that CSase could respond positively to salt, alkali and drought stresses in alfalfa, and the response to alkali stress was more pronounced than that to the other stressors (Figure 4A). To determine whether alfalfa alkalinity tolerance was altered by up-regulation of CSase, we constructed alfalfa overexpression vectors (Supplementary Figure 5) and transformed the CDS of CSase into the alfalfa cultivar Longmu 801 to obtain CSase-overexpressing transgenic lines. The glucosamine gene is a marker gene that is present only in the vector itself and not in alfalfa. Bar gene detection revealed successful infestation of alfalfa in response to a bacterial solution (Supplementary Figure 6). qRT-PCR analysis showed that the relative expression of the MsCSase gene in the control (CK) lines were much lower than that in the OV# L11-, OV# L12-, and OV#L13-overexpressed lines (Figure 4B). This indicates that the CSase gene was successfully overexpressed in alfalfa. We ultimately selected the CK line and overexpression lines OV#L11, OV#L12, and OV#L13 for subsequent experiments.
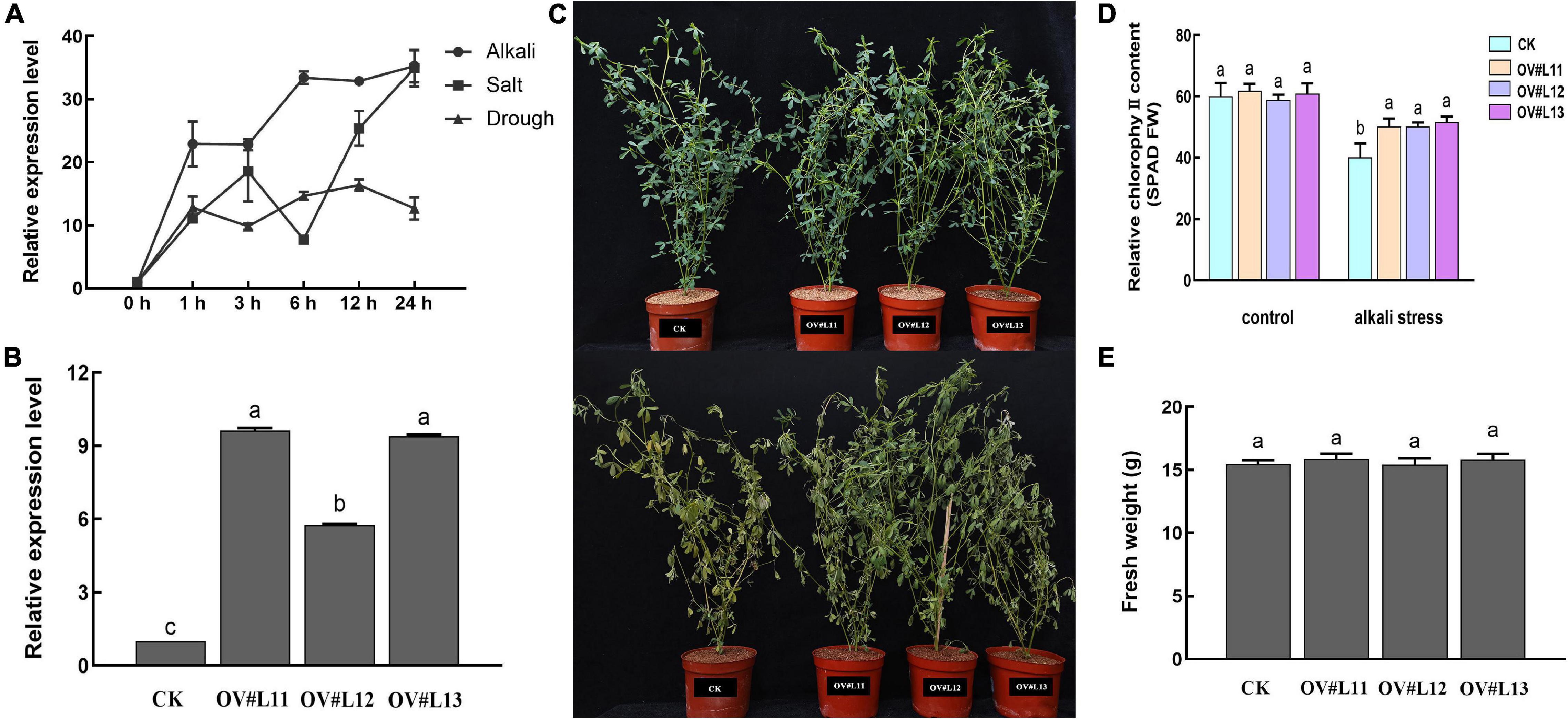
Figure 4. Overexpression of CSase confers alkalinity tolerance to alfalfa. (A) Relative expression of CSase under salt, alkali and drought stress. (B) Relative expression of Bar in different lines. (C) Phenotypic plots of different lines under normal growth and alkali conditions. (D) Tissue SPAD value of different lines under normal and alkali stress conditions. (E) Fresh weight of each line under normal conditions. The average of three independent samples was calculated and is shown, together with the standard error of the mean (bars) between samples. Different lowercase letters under the same conditions indicate that the difference is significant (P < 0.05).
Under normal growth conditions, there was no significant difference in phenotype between the CK and overexpression lines, and the differences between the aboveground biomass fresh weight and the relative chlorophyll content were not significant. However, after 5 d of alkali stress, the leaf wilting of the overexpression lines was much lower than that of the CK (Figure 4C). The alkali treatment led to degradation of the relative chlorophyll content (Figure 4D), but the chlorophyll contents were still higher in the overexpression lines than in the CK line. Moreover, the difference in aboveground biomass between the lines was not significant (Figure 4E). Overall, overexpressing CSase provided increased tolerance to alkali stress in alfalfa.
To clarify the reason for the increased alkali tolerance of the CSase overexpression lines, we also measured the contents of malondialdehyde (MDA), proline (Pro) and soluble sugars (SSs) in the CSase overexpression lines and CK plants grown under normal and alkaline conditions. However, after alkali stress, the contents of MDA, Pro and SSs increased within each line, and the increase in MDA content in the overexpression line was significantly lower than that in CK, while the contents of osmoregulatory substances containing Pro and SSs were significantly higher than those in CK (Figures 5A–C). It can be hypothesized that, compared with the CK plants, the overexpression lines are alkaline tolerant due to their lower degree of membrane damage and higher accumulation of osmoregulatory substances.
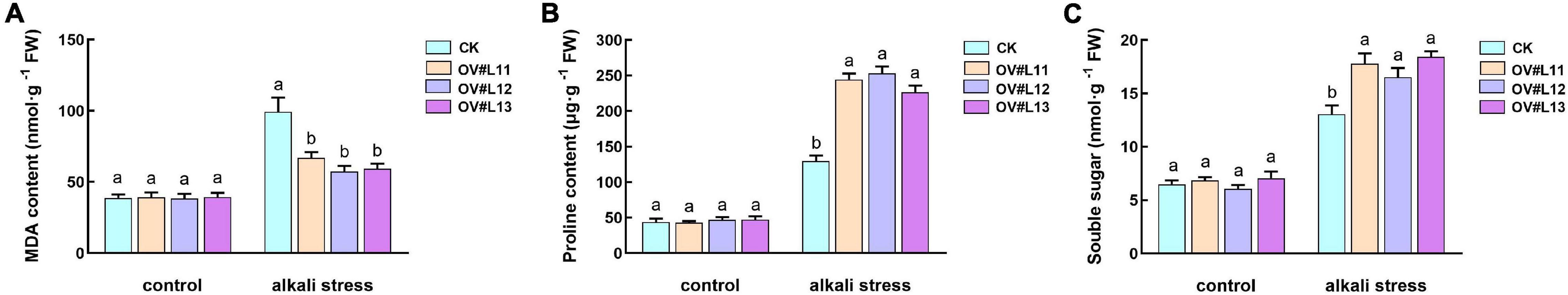
Figure 5. Contents of MDA (A), Pro (B), and SSs (C) in each line under normal conditions and after alkali stress. One-month-old alfalfa seedlings were subjected to 150 mM NaHCO3 solution for 5 days. The average of three independent samples was calculated and is shown, together with the standard error of the mean (bars) between samples. Different lowercase letters under the same conditions indicate that the difference is significant (P < 0.05).
Cysteine Synthase Overexpression Increased the Cysteine and Glutathione Contents to Improve Alkali Tolerance
CSases have been shown to encode CSase proteins (Hatzfeld et al., 2000). The increase in CSase content promotes an increase in cysteine content in plants. In the present study, the cysteine content in the overexpression lines was significantly higher than that in CK under normal growth conditions, and the cysteine content under alkali stress was higher than that in the CK and tended to increase (Figure 6A). Accordingly, we speculated that cysteine plays a role in improving alkali tolerance in plants.
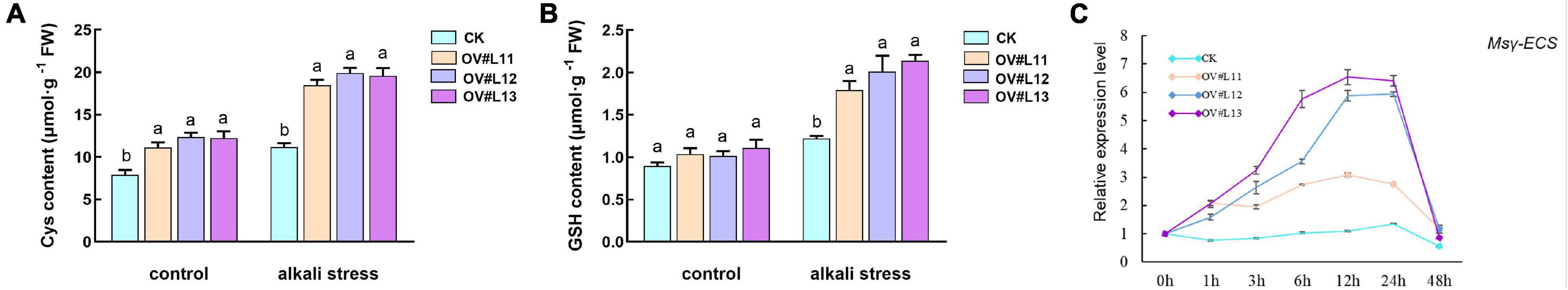
Figure 6. Contents of cysteine (A) and GSH (B) and the relative expression of the Msγ-ECS gene (C) in each line under normal conditions and after alkali stress. One-month-old alfalfa seedlings were subjected to 150 mM NaHCO3 solution for 5 days. The average of three independent samples was calculated and is shown, together with the standard error of the mean (bars) between samples. Different lowercase letters under the same conditions indicate that the difference is significant (P < 0.05).
Based on our preliminary research, we found that cysteine is a precursor for the synthesis of antioxidant substances such as GSH (Droux, 2004; Wirtz and Droux, 2005; Kopriva, 2006) and that GSH plays an important role in enhancing antioxidant capacity and transducing redox-sensitive signals in plants (Cnubben et al., 2001; Matés et al., 2002; Pastori and Foyer, 2002). Therefore, we determined the GSH content and the relative expression of γ-glutamyl cysteine synthetase (γ-ECS) in each line. The results showed that the GSH content in the plants increased in response to alkali stress and that the overexpression lines contained more GSH (Figure 6B). The trends of the relative expression of γ-ECS in the overexpression and CK lines were similar, but the relative expression of γ-ECS was higher in the overexpression line than in the CK line (Figure 6C). Overexpression of CSase and SAT in tobacco also significantly increased the relative expression of γ-ECSs as well as the GSH content in the plants (Nakamura et al., 2014). Thus, the overexpression of the CSase gene could regulate downstream metabolic pathways, which led to an increase in the relative expression of the downstream γ-ECS gene, and the increase in cysteine content provided the possibility of an increase in GSH content. This also laid the foundation for the improvement in alkali tolerance in the overexpression line.
Cysteine Synthase Overexpression Increased the Antioxidant Capacity of Transgenic Alfalfa
Studies on DES1 and OAS-A1 in Arabidopsis showed that cysteine is an important determinant of antioxidant capacity in the cytoplasm (Loìpez-Martiìn et al., 2008; Alvarez et al., 2010a), while SCS plays an important role in chloroplast redox (Bermúdez et al., 2010). In addition, cysteine and GSH are also associated with plant antioxidants. Therefore, we measured the superoxide anion (OFR) and hydrogen peroxide (H2O2) contents and the enzymatic activity of several antioxidant enzymes. Under normal growth conditions, although the H2O2 and OFR contents in the overexpression line were lower than those in the CK line, only the H2O2 content differed significantly (Figures 7A,B). The levels in both lines increased significantly after alkali stress, but the levels in the overexpression lines were significantly lower than those in the CK line. Thus, we speculated that the overexpression lines may be superior to the CK line in terms of the scavenging ability of reactive oxygen species (ROS).
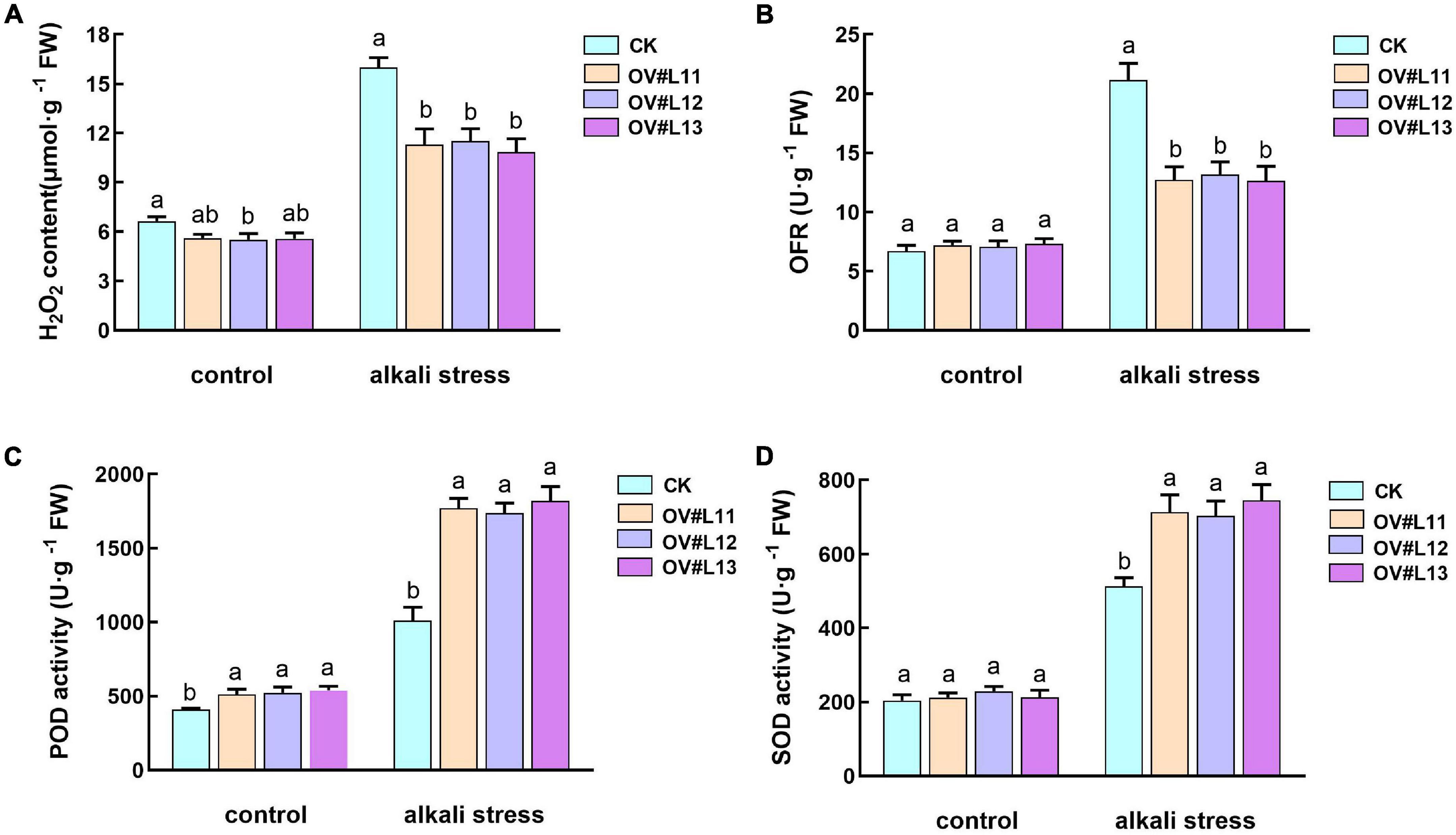
Figure 7. Content of H2O2 (A) and OFR (B) and the activities of POD (C) and SOD (D) in each line under normal conditions and after alkali stress. One-month-old alfalfa seedlings were subjected to 150 mM NaHCO3 solution for 5 days. The average of three independent samples was calculated and is shown, together with the standard error of the mean (bars) between samples. Different lowercase letters under the same conditions indicate that the difference is significant (P < 0.05).
Based on the above speculation, we also measured the activities of peroxidase (POD) and superoxide dismutase (SOD). The results showed that the activities of both enzymes were much higher in the overexpression lines than in the CK plants under alkali stress, and the activities of POD in the former were also significantly higher than those in the CK plants under normal growth conditions (Figures 7C,D). In conclusion, it was clear that the overexpression lines could reduce the accumulation of ROS by increasing the activity of antioxidant enzymes under alkali stress to enhance the alkali tolerance of the overexpression lines.
Discussion
The CSase gene family is widespread in plants, and its members play an important role in cysteine synthesis, cyanide metabolism and other pathways. The synthesis of cysteine is the main function of the members of this gene family, and the CSase they encode is involved in the final step of cysteine synthesis. Cysteine is the first reduced sulfur donor organic molecule synthesized in plants and is involved in the synthesis of a variety of compounds involved in defense, redox, and other functions and occupies a central position in plant metabolism. Most studies on the classification and functional determination of the CSase gene family have focused on Arabidopsis (Barroso et al., 1995; Domínguez-Solís et al., 2004; Alvarez et al., 2010a; Álvarez et al., 2012a; Birke et al., 2012). However, studies on the identification, classification and related functions of CSase gene family members in specific species are lacking. For this reason, we conducted the present study on the MsCSase gene family in alfalfa.
The 39 MsCSase genes that were identified were unevenly distributed across only a few chromosomes. Although previous studies have also shown that CSase genes are unevenly distributed on only a few specific chromosomes, they have not shown that the genes are subject to fragment duplication or tandem duplication events (Yamaguchi et al., 2000; Akbudak et al., 2018; Liu et al., 2018, 2019). However, we identified two pairs of tandem repeat genes and one pair of gene-generating fragment duplication genes in alfalfa (Bennetzen et al., 2005; Zhu et al., 2014). These results suggested that tandem and fragment duplication events play a role in expansion of the alfalfa MsCSase gene family.
Based on phylogenetic analysis and previous Arabidopsis studies, we divided the alfalfa MsCSase gene family members (excluding MsCSase25, MsCSase27, and MsCSase28) into 5 subfamilies. The gene structures, motifs and domains of each subfamily member were somewhat similar. The results of the structural analysis support the reliability of the phylogenetic analysis. Differences in the physicochemical properties and structures of different subfamilies may result in differences in the activity of the enzymes or the function of the genes (Liszewska et al., 2007; Noda et al., 2016). The identified MsCSase25, MsCSase27, and MsCSase28, which do not belong to other subfamilies, although they have PLP-binding sites, still need to be verified whether they encode active cysteine synthases.
In terms of gene function validation, we successfully cloned the CSase gene (MsCSase32, MsCSase34) and produced CSase-overexpressing alfalfa. Stress tests showed that overexpression of CSase significantly improved alkali tolerance in alfalfa. Alkali stress signals induce the biosynthesis and accumulation of compatible osmotic solutes, including SSs and Pro, to improve tolerance. When the plants were subjected to alkali stress, compared with the CK plants, the overexpression lines accumulated more of these substances to reduce the intracellular osmotic potential, and the tolerance of the overexpression plants was improved by the accumulation of these substances.
CSase activity and the cysteine content increase under metal and salt stresses and that overexpression of CSase improves the antioxidant capacity and tolerance of plants (Domìnguez-Solìs et al., 2001; Youssefian et al., 2001; Domínguez-Solís et al., 2004; Fediuc et al., 2005; Pajuelo et al., 2007; Gotor et al., 2015). GSH content increases in overexpression lines under environmental stresses (Sabetta et al., 2017) and GSH plays important roles in scavenging ROS and transducing stress signals (Meyer, 2008; Zagorchev et al., 2013; Leng et al., 2015). In addition, redox- and ROS-dependent regulatory networks are important for photosynthesis in chloroplasts (Strand et al., 2015; Gütle et al., 2016). In the present study, we found that the increased resistance of the transgenic lines are due to the overexpression of the CSase gene leading to an increase in the content of antioxidants such as cysteine and downstream GSH, which in turn leads to enhanced antioxidant capacity. This was also evidenced by the increase in SOD and POD activities and the decrease in H2O2 and OFR contents in overexpression plants under alkali stress. Moreover, the enhanced antioxidant capacity of the transgenic lines may make their photosynthesis less affected by alkali stress and subsequently have higher SPAD values. The enhanced stress resistance resulted in good phenotypes of overexpression lines under alkali stress. In conclusion, CSase overexpression lines enhance plant tolerance by increasing the antioxidant capacity of plants. This conclusion is also consistent with the results of the previous group, which showed that CSase can respond to alkali stress and that increased cysteine content can improve the antioxidant capacity of the plants (Song et al., 2017, 2021).
Interestingly, numerous studies have shown that CYS-C1 has dual functions in synthesizing cysteine and β-cyanoalanine (Ikegami et al., 1988; Watanabe et al., 2008; Marrero-Degro et al., 2010). However, CYS-C1 currently synthesizes CAS only in mitochondria. The encoded CSase product (CAS) is localized in the mitochondria of Arabidopsis and is involved in the detoxification of HCN (Álvarez et al., 2012b). In the present study, this protein was localized in chloroplasts and enhanced the alkalinity tolerance of plants by synthesizing cysteine. From this, we hypothesized that CSase could have the ability to encode both CSase and CAS but selectively encodes one of the enzymes depending on the expression location. The above hypothesis needs to be further investigated. Whether CSase in alfalfa can also encode CAS also needs to be further investigated.
Conclusion
In this study, we focused on 39 alfalfa MsCSase family members and classified them into 6 subfamilies first on the basis of the results of a phylogenetic analysis followed by a gene structure analysis, conserved domain characterization and a synteny analysis and on the basis of the high similarity in these aspects of members within the same subfamily. Subsequently, we cloned CSase and successfully overexpressed it in alfalfa. Evidence from both physiological experiments and the determination of the relative expression of downstream genes indicated that the overexpression lines can significantly improve alkali stress tolerance in alfalfa by increasing oxidative stress protection and the levels of osmoregulatory substances. These findings set the stage for the study of the CSase gene family. We will focus our future work on the associated metabolic pathways to further clarify the molecular mechanism of basal tolerance.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
GC and YY designed the experiments. YY wrote the first draft of the article and GC revised it. All authors participated in the experiments and read and approved the final manuscript.
Funding
This study was supported in part by the National Natural Science Foundation of China (31872998).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors thank to lab members for assistance. And we extend our thanks to the reviewers and editor for their careful reading and helpful comments on this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.792862/full#supplementary-material
Footnotes
- ^ http://ncbi.nlm.nih.gov/
- ^ https://figshare.com/projects/whole_genome_sequencing_and_assembly_of_Medicago_sativa/66380
- ^ https://pfam.xfam.org/
- ^ http://meme-suite.org/tools/meme
- ^ https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi
References
Akbudak, M. A., Filiz, E., and Uylas, S. (2018). Identification of O-acetylserine(thiol)lyase (OASTL) genes in sorghum (Sorghum bicolor) and gene expression analysis under cadmium stress. Mol. Biol. Rep. 46, 343–354. doi: 10.1007/s11033-018-4477-0
Alvarez, C., Calo, L., Romero, L. C., García, I., and Gotor, C. (2010a). An O-acetylserine(thiol)lyase homolog with l-cysteine desulfhydrase activity regulates cysteine homeostasis in Arabidopsis. Plant Physiol. 152, 656–669. doi: 10.1104/pp.109.147975
Álvarez, C., García, I., Moreno, I., Pérez-Pérez, M. E., Crespo, J. L., Romero, L. C., et al. (2012a). Cysteine-generated sulfide in the cytosol negatively regulates autophagy and modulates the transcriptional profile in Arabidopsis. Plant Cell 24, 4621–4634. doi: 10.1105/tpc.112.105403
Álvarez, C., García, I., Romero, L. C., and Gotor, C. (2012b). Mitochondrial sulfide detoxification requires a functional isoform o-acetylserine(thiol)lyase c in Arabidopsis thaliana. Mol. Plant 5, 1217–1226.
Alvarez, C., Lozano-Juste, J., Romero, L. C., García, I., Gotor, C., León, J., et al. (2010b). Inhibition of Arabidopsis O-acetylserine(thiol)lyase A1 by tyrosine nitration. J. Biol. Chem. 286, 578–586.
Barroso, C., Vega, J., and Gotor, C. (1995). A new member of the cytosolic O-acetylserine(thiol)lyase gene family in Arabidopsis thaliana. FEBS Lett. 363:1.
Bennetzen, J., Yu, J., Wang, J., Lin, W., Li, S., Li, H., et al. (2005). The genomes of Oryza sativa: a history of duplications. PLoS Biol. 3:e38. doi: 10.1371/journal.pbio.0030038
Bermúdez, M. A., Páez-Ochoa, M. A., Gotor, C., and Romero, L. C. (2010). Arabidopsis S-sulfocysteine synthase activity is essential for chloroplast function and long-day light-dependent redox control. Plant Cell 22, 403–416. doi: 10.1105/tpc.109.071985
Birke, H., Haas, Florian, H., De Kok, Luit, J., Balk, J., et al. (2012). Cysteine biosynthesis, in concert with a novel mechanism, contributes to sulfide detoxification in mitochondria of Arabidopsis thaliana. Biochem. J. 445, 275–283. doi: 10.1042/bj20120038
Bonner, E. R., Cahoon, R. E., Knapke, S. M., and Jez, J. M. (2005). Molecular basis of cysteine biosynthesis in plants. J. Biol. Chem. 280, 38803–38813. doi: 10.1074/jbc.M505313200
Chen, H., Zeng, Y., Yang, Y., Huang, L., Tang, B., Zhang, H., et al. (2020). Allele-aware chromosome-level genome assembly and efficient transgene-free genome editing for the autotetraploid cultivated alfalfa. Nat. Commun. 11:2494. doi: 10.1038/s41467-020-16338-x
Cnubben, N., Rietjens, I., Wortelboer, H., Zanden, J. V., and Bladeren, P. (2001). The interplay of glutathione-related processes in antioxidant defense. Environ. Toxicol. Pharmacol. 10, 141–152.
Cui, W., Chen, H., Zhu, K., Jin, Q., Xie, Y., Jin, C., et al. (2014). Cadmium-induced hydrogen sulfide synthesis is involved in cadmium tolerance in Medicago sativa by reestablishment of reduced (Homo)glutathione and reactive oxygen species Homeostases. PLoS One 9:e109669.
Cui, W., Li, L., Gao, Z., H, Wu, Xie, Y., and Shen, W. (2012). Haem oxygenase-1 is involved in salicylic acid-induced alleviation of oxidative stress due to cadmium stress in Medicago sativa. J. Exp. Bot. 63, 5521–5534.
Davidian, J.-C., and Kopriva, S. (2010). Regulation of sulfate uptake and assimilation – the same or not the same? Mol. Plant 3, 314–325. doi: 10.1093/mp/ssq001
Domìnguez-Solìs, J. R., Gutiérrez-Alcalá, G., Romero, L. C., and Gotor, C. (2001). The cytosolic O-acetylserine(thiol)lyase gene is regulated by heavy metals and can function in cadmium tolerance. J. Biol. Chem. 276, 9297–9302. doi: 10.1074/jbc.M009574200
Domínguez-Solís, J. R., López-Martín, M. C., Ager, F. J., Ynsa, M. D., Romero, L. C., and Gotor, C. (2004). Increased cysteine availability is essential for cadmium tolerance and accumulation in Arabidopsis thaliana. Plant Biotechnol. J. 2, 469–476. doi: 10.1111/j.1467-7652.2004.00092.x
Droux, M. (2004). Sulfur assimilation and the role of sulfur in plant metabolism: a survey. Photosynthesis Res. 79, 331–348. doi: 10.1023/b:pres.0000017196.95499.11
Droux, M., Ruffet, M. L., Douce, R., and Job, D. (1998). Interactions between serine acetyltransferase and O-acetylserine (thiol) lyase in higher plants–structural and kinetic properties of the free and bound enzymes. Eur. J. Biochem. 255, 235–245.
Fediuc, E., Lips, S. H., and Erdei, L. (2005). O-acetylserine (thiol) lyase activity in Phragmites and Typha plants under cadmium and NaCl stress conditions and the involvement of ABA in the stress response. J. Plant Physiol. 162, 865–872. doi: 10.1016/j.jplph.2004.11.015
García, I., Castellano, J. M., Vioque, B., Solano, R., Gotor, C., and Romero, L. C. (2010). Mitochondrial β-cyanoalanine synthase is essential for root hair formation in Arabidopsis thaliana. Plant Cell 22, 3268–3279. doi: 10.1105/tpc.110.076828
Gotor, C., Laureano-Marín, A. M., Moreno, I., Aroca, Á., García, I., and Romero, L. C. (2015). Signaling in the plant cytosol: cysteine or sulfide? Amino Acids 47, 2155–2164.
Gütle, D. D., Roret, T., Müller, S. J., Couturier, J., Lemaire, S. D., Hecker, A., et al. (2016). Chloroplast FBPase and SBPase are thioredoxin-linked enzymes with similar architecture but different evolutionary histories. Proc. Natl. Acad. Sci. U.S.A. 113, 6779–6784. doi: 10.1073/pnas.1606241113
Hatzfeld, Y., Maruyama, A., Schmidt, A., Noji, M., Ishizawa, K., and Saito, K. (2000). β-cyanoalanine synthase is a mitochondrial cysteine synthase-like protein in spinach and Arabidopsis1. Plant Physiol. 123, 1163–1172. doi: 10.1104/pp.123.3.1163
Heeg, C., Kruse, C., Jost, R., Gutensohn, M., Ruppert, T., Wirtz, M., et al. (2008). Analysis of the Arabidopsis O-Acetylserine(thiol)lyase gene family demonstrates compartment-specific differences in the regulation of cysteine synthesis. Plant Cell 20, 168–185. doi: 10.1105/tpc.107.056747
Ikegami, F., Takayama, K., and Murakoshi, I. (1988). Purification and properties of β-cyano-l-alanine synthase fromVicia angustifolia. Phytochemistry 27, 2011–2016.
Jez, J. M., and Dey, S. (2013). The cysteine regulatory complex from plants and microbes: what was old is new again. Curr. Opin. Struct. Biol. 23, 302–310. doi: 10.1016/j.sbi.2013.02.011
Kopriva, S. (2006). Regulation of sulfate assimilation in Arabidopsis and beyond. Ann. Bot. 97, 479–495. doi: 10.1093/aob/mcl006
Leng, X., Jia, H., Sun, X., Shangguan, L., Mu, Q., Wang, B., et al. (2015). Comparative transcriptome analysis of grapevine in response to copper stress. Sci. Rep. 5:17749. doi: 10.1038/srep17749
Liszewska, F., Lewandowska, M., Pochocka, D., and Sirko, A. (2007). Mutational analysis of O-acetylserine (thiol) lyase conducted in yeast two-hybrid system. Biochim. Biophys. Acta 1774, 450–455.
Liu, D., Li, J., Lu, J., Tian, B., Liu, X., Yang, G., et al. (2019). Cloning and functional analysis of four O-Acetylserine (thiol) lyase family genes from foxtail millet. Plant Physiol. Biochem. 139, 325–332. doi: 10.1016/j.plaphy.2019.03.032
Liu, D., Lu, J., Li, H., Wang, J., and Pei, Y. (2018). Characterization of the O-acetylserine(thiol)lyase gene family in Solanum lycopersicum L. Plant Mol. Biol. 99, 123–134. doi: 10.1007/s11103-018-0807-9
Loìpez-Martiìn, M. C., Becana, M., Romero, L. C., and Gotor, C. (2008). Knocking out cytosolic cysteine synthesis compromises the antioxidant capacity of the cytosol to maintain discrete concentrations of hydrogen peroxide in Arabidopsis. Physiology 147, 562–572. doi: 10.1104/pp.108.117408
Marrero-Degro, J., Marcano-Velázquez, J., and Siritunga, D. (2010). Isolation and characterization of novel β-cyanoalanine synthase and cysteine synthase genes from cassava. Plant Mol. Biol. Rep. 29, 514–524. doi: 10.1007/s11105-010-0255-4
Matés, J. M., Pérez-Gómez, C., Núñez de Castro, I., Asenjo, M., and Márquez, J. (2002). Glutamine and its relationship with intracellular redox status, oxidative stress and cell proliferation/death. Int. J. Biochem. Cell Biol. 34, 439–458.
Meyer, A. J. (2008). The integration of glutathione homeostasis and redox signaling. J. Plant Physiol. 165, 1390–1403. doi: 10.1016/j.jplph.2007.10.015
Nakamura, M., Ochiai, T., Noji, M., Ogura, Y., and Saito, K. (2014). An improved tolerance to cadmium by overexpression of two genes for cysteine synthesis in tobacco. Plant Biotechnol. 31, 141–147.
Ning, H., Zhang, C., Yao, Y., and Yu, D. (2009). Overexpression of a soybean O-acetylserine (thiol) lyase-encoding gene GmOASTL4 in tobacco increases cysteine levels and enhances tolerance to cadmium stress. Biotechnol. Lett. 32, 557–564. doi: 10.1007/s10529-009-0178-z
Noda, M., Nakamura, M., Takamiya, R., Tamura, T., Ito, T., and Kodama, H. (2016). A spinach O-acetylserine(thiol)lyase homologue, SoCSaseLP, suppresses cysteine biosynthesis catalysed by other enzyme isoforms. Biochim. Open 2, 24–32.
Pajuelo, E., Carrasco, J. A., Romero, L. C., Chamber, M. A., and Gotor, C. (2007). Evaluation of the metal phytoextraction potential of crop legumes. Regulation of the expression of O-Acetylserine (Thiol)lyase under metal stress. Plant Biol. 9, 672–681. doi: 10.1055/s-2007-965439
Pastori, G. M., and Foyer, C. H. (2002). common components, networks, and pathways of cross-tolerance to stress. The central role of “redox” and abscisic acid-mediated controls. Plant Physiol. 129, 460–468. doi: 10.1104/pp.011021
Romero, L. C., Aroca, M. A., Laureano-Marín, A., Moreno, I., García, I., and Gotor, C. (2014). Cysteine and cysteine-related signaling pathways in Arabidopsis thaliana. Mol. Plant 7, 264–276.
Sabetta, W., Paradiso, A., Paciolla, C., and de Pinto, M. C. (2017). “Chemistry, biosynthesis, and antioxidative function of glutathione in plants,” in Glutathione in Plant Growth, Development, and Stress Tolerance, eds M. Hossain, M. Mostofa, P. Diaz-Vivancos, D. Burritt, M. Fujita, and L. S. Tran (Cham: Springer), 1–27. doi: 10.1007/978-3-319-66682-2_1
Song, T., Sun, N., Dong, L., and Cai, H. (2021). Enhanced alkali tolerance of rhizobia-inoculated alfalfa correlates with altered proteins and metabolic processes as well as decreased oxidative damage. Plant Physiol. Biochem. 159, 301–311.
Song, T., Xu, H., Sun, N., Jiang, L., Tian, P., Yong, Y., et al. (2017). Metabolomic analysis of alfalfa (Medicago sativa L.) root-symbiotic rhizobia responses under alkali stress. Front. Plant Sci. 8:1208. doi: 10.3389/fpls.2017.01208
Strand, D. D., Livingston, A. K., Satoh-Cruz, M., Froehlich, J. E., Maurino, V. G., and Kramer, D. M. (2015). Activation of cyclic electron flow by hydrogen peroxide in vivo. Proc. Natl. Acad. Sci. U.S.A. 112, 5539–5544. doi: 10.1073/pnas.1418223112
Takahashi, H., Kopriva, S., Giordano, M., Saito, K., and Hell, R. (2011). Sulfur assimilation in photosynthetic organisms: molecular functions and regulations of transporters and assimilatory enzymes. Annu. Rev. Plant Biol. 62, 157–184. doi: 10.1146/annurev-arplant-042110-103921
Van Hoewyk, D., Pilon, M., and Pilon-Smits, E. A. H. (2008). The functions of NifS-like proteins in plant sulfur and selenium metabolism. Plant Sci. 174, 117–123. doi: 10.1016/j.plantsci.2007.10.004
Watanabe, M., Kusano, M., Oikawa, A., Fukushima, A., Noji, M., and Saito, K. (2008). Physiological roles of the β-substituted alanine synthase gene family in Arabidopsis. Plant Physiol. 146, 310–320. doi: 10.1104/pp.107.106831
Wei, Y., Shi, H., Xia, Z., Tie, W., Ding, Z., Yan, Y., et al. (2016). Genome-wide identification and expression analysis of the WRKY gene family in cassava. Front. Plant Sci. 7:25. doi: 10.3389/fpls.2016.00025
Wirtz, M., and Droux, M. (2005). Synthesis of the sulfur amino acids: cysteine and methionine. Photosynthesis Res. 86, 345–362. doi: 10.1007/s11120-005-8810-9
Wirtz, M., and Hell, R. (2006). Functional analysis of the cysteine synthase protein complex from plants: structural, biochemical and regulatory properties. J. Plant Physiol. 163, 273–286. doi: 10.1016/j.jplph.2005.11.013
Xie, Y., Lai, D., Mao, Y., Zhang, W., Shen, W., and Guan, R. (2012). Molecular cloning, characterization, and expression analysis of a novel gene encoding l-cysteine desulfhydrase from Brassica napus. Mol. Biotechnol. 54, 737–746. doi: 10.1007/s12033-012-9621-9
Yamaguchi, Y., Nakamura, T., Kusano, T., and Sano, H. (2000). Three Arabidopsis genes encoding proteins with differential activities for cysteine synthase and β-cyanoalanine synthase. Plant Cell Physiol. 41, 465–476.
Youssefian, S., Nakamura, M., Orudgev, E., and Kondo, N. (2001). Increased cysteine biosynthesis capacity of transgenic tobacco overexpressing an O-acetylserine(thiol) lyase modifies plant responses to oxidative stress. Plant Physiol. 126, 1001–1011.
Zagorchev, L., Seal, C., Kranner, I., and Odjakova, M. (2013). A central role for thiols in plant tolerance to abiotic stress. Int. J. Mol. Sci. 14, 7405–7432. doi: 10.3390/ijms14047405
Keywords: alfalfa, CSase gene family, genome-wide analysis, gene overexpression, alkali stress
Citation: Yuan Y, Song T, Yu J, Zhang W, Hou X, Kong Ling Z and Cui G (2022) Genome-Wide Investigation of the Cysteine Synthase Gene Family Shows That Overexpression of CSase Confers Alkali Tolerance to Alfalfa (Medicago sativa L.). Front. Plant Sci. 12:792862. doi: 10.3389/fpls.2021.792862
Received: 11 October 2021; Accepted: 13 December 2021;
Published: 04 January 2022.
Edited by:
Rajeev K. Varshney, International Crops Research Institute for the Semi-Arid Tropics (ICRISAT), IndiaCopyright © 2022 Yuan, Song, Yu, Zhang, Hou, Kong Ling and Cui. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guowen Cui, Y2d3NjAzQDE2My5jb20=
 Yuying Yuan
Yuying Yuan Tingting Song
Tingting Song Jinqiu Yu
Jinqiu Yu Wenkai Zhang
Wenkai Zhang Zelai Kong Ling
Zelai Kong Ling Guowen Cui
Guowen Cui