
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 06 January 2022
Sec. Plant Development and EvoDevo
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.787826
Chinese cabbage is an important leaf heading vegetable crop. At the heading stage, its leaves across inner to outer show significant morphological differentiation. However, the genetic control of this complex leaf morphological differentiation remains unclear. Here, we reported the transcriptome profiling of Chinese cabbage plant at the heading stage using 24 spatially dissected tissues representing different regions of the inner to outer leaves. Genome-wide transcriptome analysis clearly separated the inner leaf tissues from the outer leaf tissues. In particular, we identified the key transition leaf by the spatial expression analysis of key genes for leaf development and sugar metabolism. We observed that the key transition leaves were the first inwardly curved ones. Surprisingly, most of the heading candidate genes identified by domestication selection analysis obviously showed a corresponding expression transition, supporting that key transition leaves are related to leafy head formation. The key transition leaves were controlled by a complex signal network, including not only internal hormones and protein kinases but also external light and other stimuli. Our findings provide new insights and the rich resource to unravel the genetic control of heading traits.
Chinese cabbage (Brassica rapa ssp. pekinensis) is a widely cultivated vegetable crop in East Asia. It has a unique product organ, a leafy head, which is a distinct type of plant architecture shared by several vegetable crops, including cabbage, mustard, endive, and lettuce (Wang et al., 2014; Yu et al., 2020). The vegetative growth period of Chinese cabbage can be divided into three stages: seedling, rosette and heading stage (Yu et al., 2013). At the seedling stage, the leaves are round and flat with long petioles, while at the rosette stage, the inner new leaves are continually formed at the shoot apical meristem (SAM) and subsequently become large with short petioles, which start to grow more upright surrounding the axis of the enlarged, but compressed stem (Opena et al., 1998; Sun et al., 2019).
When it comes to the heading stage, some rosette leaves begin to incurve and fold upward. As more new leaves form and expand around the SAM, they become increasingly entrapped until they remain folded in the center to form leafy heads as nutrient storage organs. When leafy heads are ready for harvest, the leaves are wrinkled with broad midveins and broad fleshy petioles (Sun et al., 2019), and different regions of the blade and petiole also show varying degrees of curvature. In addition, various types of leaf morphology can be distinguished from the inner to outer leaves. The inner head leaves (HLs) curve inward, are wrapped by outer HLs, and are not exposed to the sunlight. They can be white, orange, or yellow, while outer HLs are green, grow upright, and curve outward (Li et al., 2012; Li Y. et al., 2019). In contrast, the outermost leaves are non-heading and close to the soil surface and easily become senescent and rotting. Generally, the outer leaves are considered to promote head formation by supplying photosynthates to the inner leaves for head formation, as well as providing shade to the head (He et al., 2000; Wang et al., 2014). Leaf morphological and functional differentiation with the growth stage and spatial position indicate that leafy head formation is a very complex process.
As for the leafy head domestication trait, some genes involved in adaxial–abaxial polarity pathway and hormones related genes, were found to be under selection in leaf-heading accessions, indicating their contribution to leafy head formation (Cheng et al., 2016; Liang et al., 2016; Cai et al., 2021). Genetic analysis has demonstrated that the ad-ab polarity pathway gene BcpLH affected the inward curvature of leaves (Yu et al., 2000; Ren et al., 2020). Transcription factors, such as BrpTCP4 and BrpSPL9, have been proven to be involved in leafy head formation by affecting head shape and heading time, respectively (Mao et al., 2014; Wang et al., 2014). In addition, gibberellin biosynthesis related gene BrKS1 was identified for leafy head formation by screening the EMS mutant library of Chinese cabbage (Gao et al., 2020). Auxin biosynthesis genes (AUXs) have also been investigated and demonstrated to be involved in leafy head formation, possibly by regulating the concentration of auxin (He et al., 2000).
In addition to the differences in leaf morphology, the leaf functions are also different; i.e., the rosette leaves act as photosynthetic organs, while leafy head serves as a nutrient sink organ (He et al., 2000). Carbohydrate accumulation in the leaves was also considered to be closely related to leafy head formation (Ito and Kato, 1957; Wang et al., 2012). Sucrose, fructose and glucose are the main soluble sugars, most of which in edible organs come from the photosynthate of source leaves. Sucrose is the main form of photosynthate output, which can be decomposed into fructose and glucose in sink cells (Park et al., 2008). The distribution and composition of carbohydrates in sink tissues depends on the relative gene expression of enzymes in carbohydrate metabolism. A recent study investigated differing sugar accumulation and expression patterns of sugar-metabolizing enzyme genes in heading leaves of Chinese cabbage, and then identified the important role of sucrose synthase (SUS1) in leafy head formation (Liu et al., 2021).
To reveal the molecular mechanism underlying leafy head formation, a range of transcriptomic studies were performed based on the comparison of changes in transcript abundance of only certain leaves at different development stages, or between heading Chinese cabbage or non-heading pak-choi (Wang et al., 2012; Li J. et al., 2019; Sun et al., 2019). Wang et al. (2012) discovered that some stimuli (such as carbohydrate levels, light, and hormones), and transcription factors, protein kinases and calcium, may play important roles in leafy head development. In addition, hormones such as auxin, cytokinin, abscisic acid, gibberellin, and brassinosteriods (BR), were found to perhaps play important roles in leafy head formation (Gao et al., 2017; Gu et al., 2017; Li J. et al., 2019). However, previous transcriptome analyses to date have focused on only the inner leaves between different stages or simply separated the blade and midrib of internal and external leaves of leafy heads, which ignores the various leaf morphologies at heading stages that may play diverse and potentially fundamental roles in leafy head formation. To date, the genetic basis and molecular mechanism of leafy head formation remain largely unclear, including the roles of leaf blades and petioles.
Considering the significant differences in leaf morphology, including age, color, size, and curvature degree, the inner to outer leaves of leafy head at the heading stage reflect their functional diversification and specialization; i.e., the outer leaves might act as source tissues, while the inner leaves act as sink tissues. In addition, even on the same leaf, the different regions show significantly different structures and curvature degrees, indicating their different contributions to the leaf heading process. Here, we investigated the transcriptome profiles of continuous head leaves from inside to outside at the heading stage, including inner SAM, inner leaves (covered and not exposed to sunlight), and outer leaves (exposed to sunlight), which represented different leaf morphology. Moreover, five different regions in the blade and petiole of head leaves were further separated considering that different regions on the same head leaf show different degrees of curvature. The series-spatial profiles of genome-wide gene expression were presented in a total of 24 different leaf tissues, identifying the key transition head leaves that likely have an important role in leaf-heading. Overall, our study provided new insights into leafy head formation.
The leafy head formation of Chinese cabbage is attributed to significantly spatial leaf morphological differentiation. The head leaves presented two regions, the blade and petiole, which show different structures and curvature degrees. The head leaves from the inside to the outside also showed significant morphological variations (Figure 1A). In addition to the gradually increasing leaf size, the color of the leaf blades changed from yellow (L1–L4) to yellow–green (L5–L6), then became light green (L7) and dark green (L8–L9). Moreover, the SAM and inner leaves (L1–L6) curved inward, while the outer leaves (L8–L9) curved slightly outward. Of all the leaves, L7 was positioned at the boundary between the inner incurved leaves and outer outward-curved leaves, and they covered all the inner leaves and provided shade to form the head. Here, L7 was defined as an outer leaf.
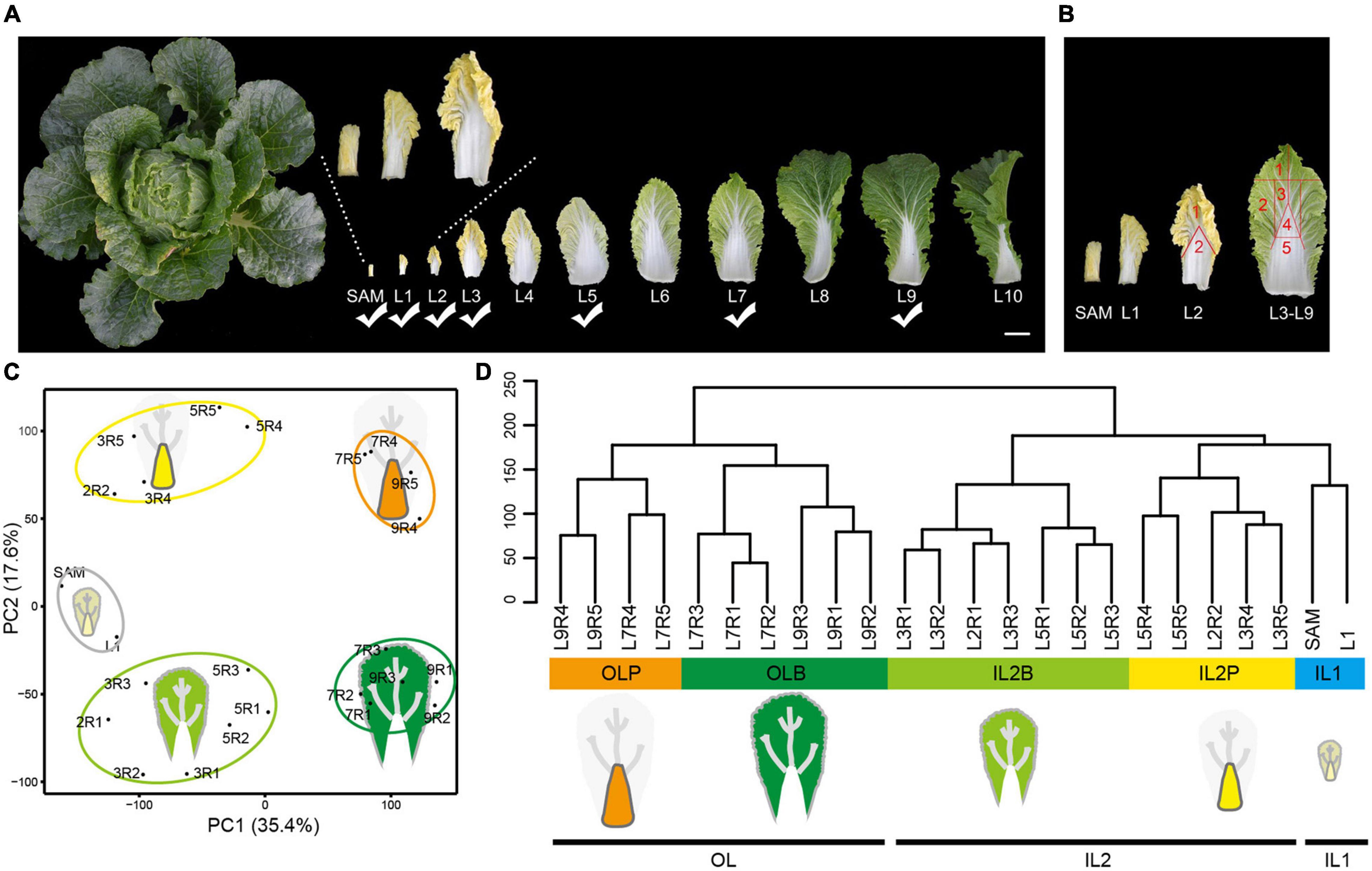
Figure 1. Transcriptome profiling of Chinese cabbage leaves in different leaf layers at the heading stage. (A) Representative pictures of leaves spanning 11 leaf layers. The leaf layers marked with a check mark were used for RNA-seq. Scale bar is 5 cm. (B) L2 was divided into two regions (including leaf blades and petioles) for sampling. L3, L5, L7, and L9 were divided into five regions (named R1, R2, R3, R4, and R5, respectively) for sampling. (C) PCA of the transcriptomes of the 24 different tissues. The first PC (PC1) distinguished leaf positioning from SAM and explained 35.4% of the total sample variation. The second PC (PC2; 17.6% of sample variance) clearly distinguished the petioles from the leaf blades. (D) The cluster dendrogram for total samples. IL1 inner leaves 1; IL2B, blade of inner leaves 2; IL2P, petiole of inner leaves 2; OLB, outer leaf blade; OLP, outer leaf petiole.
To explore the molecular processes underlying the leafy head formation, comprehensive transcriptome analysis was conducted on 24 leaf tissue samples with two biological replicates for each tissue, including the shoot apical meristem with the incipient youngest leaves (SAM), whole inner leaves (L1), and dissected blade and petiole tissues from three inner leaves (L2, L3, and L5), and two outer leaves (L7, L9), which represented significant differences in leaf morphology (Figures 1A,B). Transcriptome sequencing yielded 7.43 × 108 high-quality clean reads from 48 samples (1.549 × 107 reads per sample, on average; Supplementary Table 1). All biological replicates showed a high correlation, with a Pearson correlation coefficient of 0.92–0.99 (Supplementary Figure 1). After mapping clean reads to the Chinese cabbage reference genome, transcripts per million (TPM) was calculated for each gene. A total of 27,876 expressed genes were identified (Supplementary Figure 2A and Supplementary Table 2), and 75.08% of genes (20929/27876) were commonly expressed in all samples (Supplementary Figure 2B).
Principal component analysis (PCA) showed a clear separation along principal components (PC) 1 and 2. PC1 separated the leaf samples according to their distance from the SAM, whereas PC2 separated the blade and petiole tissues. In combination with hierarchical clustering analysis, the transcriptomes were divided into five distinct leaf tissue groups. Samples from SAM and L1 formed inner leaves 1 (IL1) and represented the tissues that were closest to the shoot apex. Blades of inner leaves 2 (IL2B) and petioles of inner leaves 2 (IL2P) represented the blade and petiole tissues of L2–L5, whereas the outer leaf blade (OLB) and outer leaf petiole (OLP) represented the blade and petiole tissues of outer leaves (L7 and L9), respectively (Figures 1C,D). These groups were in accordance with our observation of leaf morphological differentiation in these selected leaf samples, indicating that our sampling strategy would identify tissue-specific genes from different head leaves.
Differentially expressed genes (DEGs) were identified in different leaves at the whole-genome level to explore the diverse and potential functions among these leaves. A total of 9,133 DEGs were found, including 5,186 DEGs among leaf blades and 6,136 DEGs among petioles, as well as 5,514 DEGs between leaf blades and petioles (Supplementary Figure 3). A display of DEGs (Figure 2A) showed that they were divided into 14 co-expression clusters (CEC1–CEC14; Supplementary Figure 4) using the k-means clustering, and these CECs were closely related to the five groups of HL samples in Figures 1C,D.
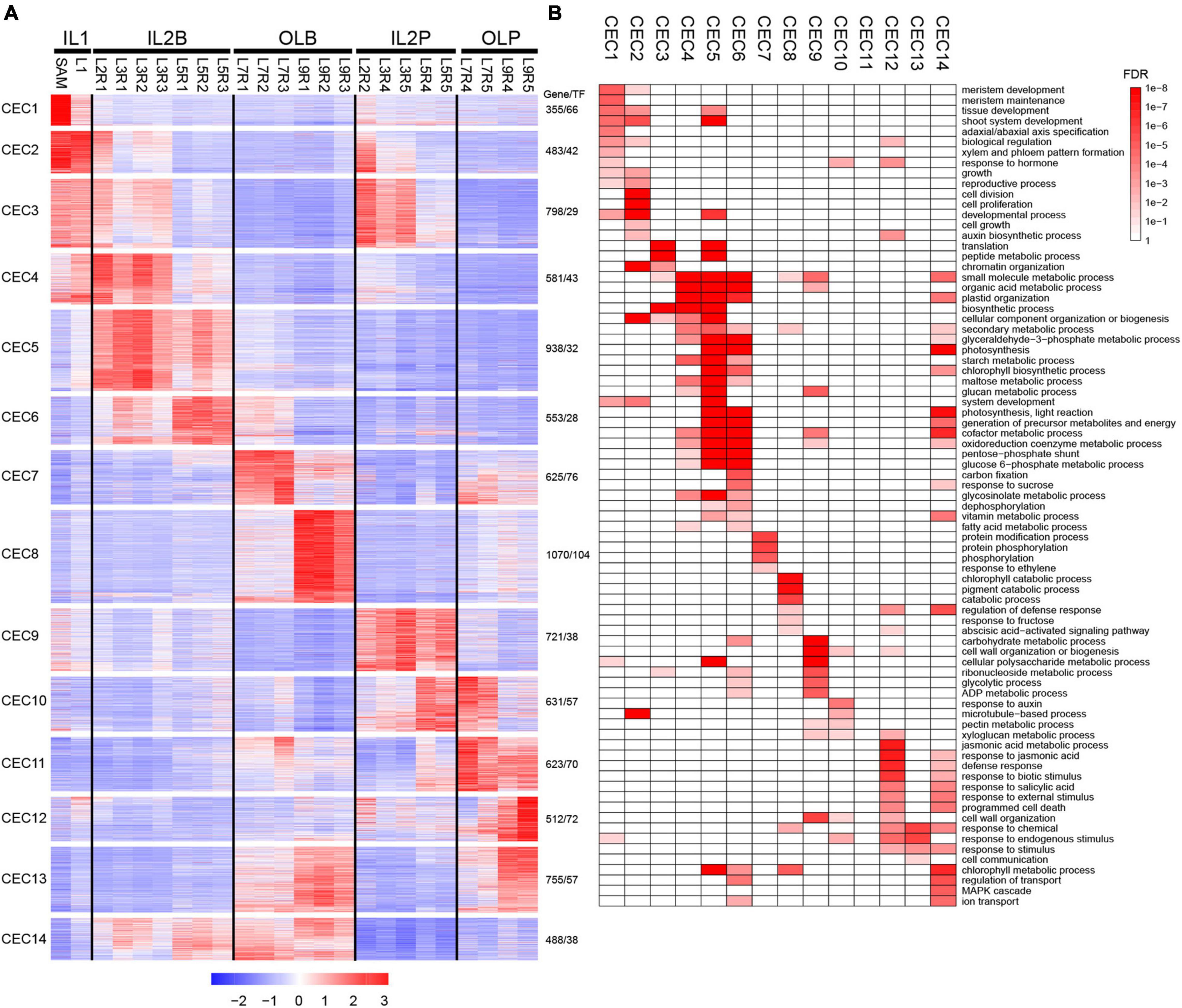
Figure 2. Expression profiles and functional transition of 9,133 DEGs in different leaves. (A) K-means clustering of DEGs. A total of 9,133 DEGs were grouped into 14 clusters. For each gene, expression data were Z-score standardized. (B) Gene ontology (GO) slim categories of each co-expressed cluster were presented as a heatmap with a color gradient based on the false discovery rate (FDR) values. Only significant GO categories (FDR < 0.05) were displayed.
The high expression level of DEGs in IL1 was best represented by CEC 1 and 2 (Figure 2A and Supplementary Table 3). CEC 1 was highly expressed in SAM and was represented by genes related to meristem development, tissue development, and adaxial/abaxial polarity (Figure 2B). For example, STM, KNAT2, and KNAT6 in SAM are required to maintain indeterminate cell fate and to prevent cell differentiation in the meristem (Carles and Fletcher, 2003). In addition, BOP1 and BOP2 are expressed in the proximal region to directly activate the adaxially expressed AS2 and the boundary-expressed LOB (Jun et al., 2010). CEC 2 was highly expressed at SAM and L1 and contained a set of genes related to cell division, cell proliferation, cell growth, and auxin biosynthetic process (Figure 2B), such as CDKB1, CDKB2, CYCA1;1, AN3, ANT, FKD1, and PIN1. Previous studies showed that CDKBs, CYCAs, ANT, and AN3 were involved in cell proliferation during leaf development (Powell and Lenhard, 2012; Kalve et al., 2014). In developing leaf veins, FORKED1 is required for localization of PIN1, contributing to organ positioning, separation, and outgrowth (Vernoux et al., 2000; Hou et al., 2010).
The high expression level of DEGs in IL2B was best represented by CEC 4–6 (Figure 2A and Supplementary Table 3). CEC 4, 5, and 6 were all enriched in the organic acid metabolic process, starch metabolic process, plastid organization, and glycosinolate metabolic process. Additionally, both CEC 5 and 6 contained substantial expression of genes related to photosynthetic light reactions, as well as several key genes of the Calvin cycle, including PSAE2, PSBQ2, CAB, PSAK, and PSAL (Figure 2B). The high expression level of DEGs in IL2P was best represented by CEC 9 (Figure 2A and Supplementary Table 3). The CEC 9 were enriched in plant cell wall-related process, including wall organization or biogenesis, and pectin metabolic processes (Figure 2B). For example, IRX9, IRX3, and IRX15 are essential for normal xylan synthesis and deposition in the secondary cell wall (Doering et al., 2012). In addition, the genes encoding expansins (EXPA3, EXPA4, EXPA9, and EXP12) were also enriched in CEC 9 (Kalve et al., 2014).
The high expression level of DEGs in OLB was best represented by CEC 7 and 8 (Figure 2A and Supplementary Table 3). CEC 7 were highly expressed in leaf blade of L7 and were represented by genes related to protein modification process, protein phosphorylation, and protein serine/threonine kinase activity (Figure 2A), such as BRI1, CPK5, and CRK39. These results reflected that the L7 was characterized by obvious signal transduction and gene regulation characteristics. CEC 8, with a high expression level in the leaf blade of L9, contained a set of genes related to chlorophyll catabolic process, cellular catabolic process, regulation of defense response, response to fructose, and abscisic acid-activated signaling pathway (Figure 2B). For example, MCCA, ACD1, ACD2, and HGO were involved in the catabolic process (Ding et al., 2012; Pattanayak et al., 2012; Han et al., 2013). Abscisic acid (ABA) regulates various developmental processes and adaptive stress responses in plants (Cutler et al., 2010). In addition, many abscisic acid-responsive genes were also enriched in CEC 8, including ABI5, ABI1, SNRK2, CIPK15, NAC019, ANAC2, PYL7, and MYB74.
The high expression level of DEGs in OLP was best represented by CECs 11 and 12 (Figure 2A and Supplementary Table 3). The genes in CEC 11 were highly expressed in the petioles of L7–L9, but they were not enriched in any Gene ontology (GO) term. The genes in CEC 12 were highly expressed in the petiole of L9. These genes might be mainly involved in the abscisic acid-activated signaling pathway, jasmonic acid (JA) metabolic process, JA responsion, defense responsion, biotic stimulus responsion, salicylic acid responsion, and external stimulus responsion. Many JA pathway genes were enriched in CEC 12, including JA biosynthetic genes (AOC3, OPR3, JASSY, AOS, and LOX4) and JA response genes (MYBR1, ILL6, TAT3, JAZ1, JAZ5, JAZ6, MYC2, JAZ10, JAZ8, and JAZ3) (Huang et al., 2017; Wasternack and Song, 2017). These results suggest that L9 may play a role in protecting the leafy head from external damage.
Although whole-genome transcriptomic profiling suggested five distinct groups for leaf samples (Figures 1C,D), a total of 3,051 highly expressed genes, including 195 TFs, in CECs 3 and 4, CECs 6 and 14, and CEC 10 may have similar functional processes in IL1-IL2B, IL2B-OLB, and IL2P-OLP, respectively, as these genes represented similar expression patterns between groups (Figure 2A and Supplementary Table 3). These shared genes may have important roles in the functional transition from one group to another. The discovery of group-specific and group-shared highly expressed genes indicated the dynamic expression profiling existing in heading leaves.
Cell proliferation and elongation rates along different axes directions shape leaf morphology (Tsukaya, 2003; Horiguchi et al., 2006; Nakata and Okada, 2013). We identified previous characterized cell proliferation-related genes and cell expansion-related genes (Powell and Lenhard, 2012; Du et al., 2018; Supplementary Table 4 and Supplementary Figure 5), to investigate the role of cell proliferation and expansion genes in morphology differentiation of head leaves. We found that most cell proliferation genes peaked in IL1 HLs, including SAM and L1, but were rapidly downregulated in IL2 HLs and outer HLs (Figure 3A), implying their important role in the initiation of the constantly growing inner leaves. Unlike cell proliferation genes, most cell expansion genes were highly expressed in the blade regions of IL2 HLs and outer HLs, and they displayed more diverse expression patterns and were divided into four clusters (Figure 3A). Notably, several genes in cluster VI had higher expression levels in R1 and R2 than in R3, such as BrTCP5.1, BrTCP5.2, BrNGA1.1, and BrNGA1.2, indicating their potential role in leaf marginal growth of HLs. The genes from cluster V were dominantly expressed in the petioles, indicating that they have a specific role in petiole growth. Furthermore, genes from clusters V, VI, and VII showed significantly different patterns between IL2 HLs and outer HLs. Cluster VI genes showed significant downregulation in outer HLs compared with IL2 HLs, while cluster VII genes dominantly expressed in outer HLs than IL2 HLs and cluster V genes showed the highest expression in the outer HL L7. These results indicated that L7 are special transition leaves, which was in accordance with our observation that L7 were positioned as key boundary leaves between the outer and inner leaves. Overall, our results suggested that leafy head growth included a developmental transition of cell proliferation-dominant in IL1 HLs to cell expansion-dominant in IL2 and outer leaves, and cell division/cell expansion of the leaf blade and petiole were coordinated by different genes. In addition, most cell expansion genes showed different expression pattern between IL2 HLs and outer HLs, resulting in a significant change in the regulation of cell division and cell expansion in L7 compared with IL2 HLs and outer leaves (Figure 3B).
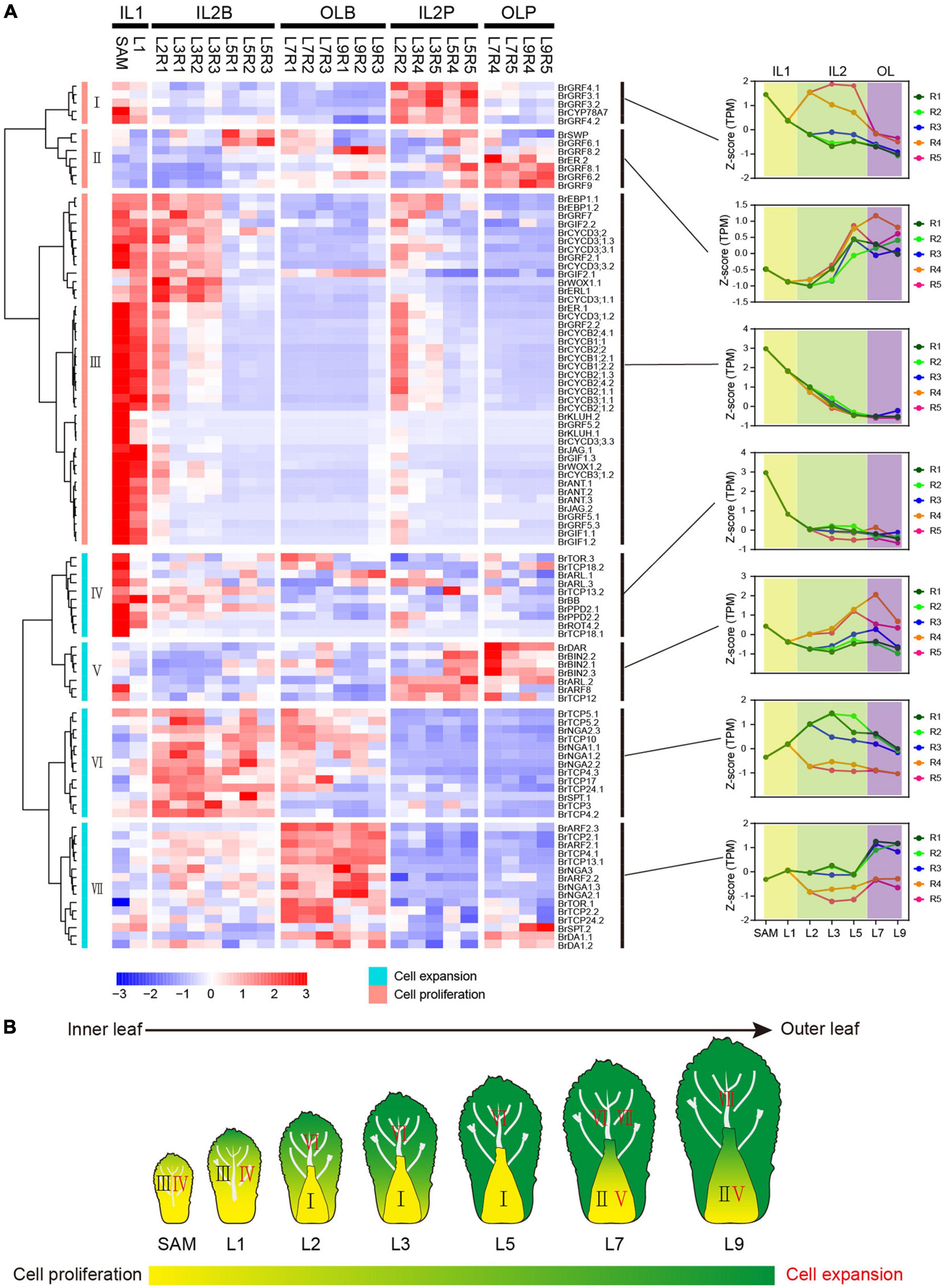
Figure 3. Expression pattern of cell proliferation and cell expansion genes. (A) Hierarchical clustering analysis of cell proliferation genes and cell expansion genes. For each gene, expression data were Z-score standardized. (B) Expression model of cell proliferation genes and cell expansion genes in Chinese cabbage at the heading stage.
The curvature of Chinese cabbage HLs is generally considered to be caused by the asymmetric cell growth between the adaxial and abaxial axis of the leaf (Cheng et al., 2016; Liang et al., 2016; Yu et al., 2020). To explore the role of ad-ab polarity genes in the curvature of head leaves, their expression patterns were analyzed (Supplementary Table 5). Almost all adaxial genes, except BrAS1.1, showing a significantly low expression level in the outer HLs (Figures 4A,B). For the abaxial gene, two major expression patterns were observed. Many abaxial genes, except BrKAN2.1 and BrKAN2.3, showed a similar expression pattern with adaxial genes; they were predominantly expressed in inner HLs, downregulated from the inner to the outer leaves, and significantly repressed in the outer HLs (Figures 4A,C). Conversely, there were still many abaxial genes were upregulated from the inner to the outer leaves, and significantly upregulated in the outer HL L7, such as BrARF2.1, BrARF2.2, and BrARF2.3 (Figures 4D–F). Interestingly, the adaxial gene BrAS1.1 and two abaxial genes BrKAN2.1 and BrKAN2.3 were specifically upregulated in L7. A homologous gene of AS1 (LsAS1) in lettuce has been confirmed to be involved in leafy head formation (Yu et al., 2020), whereas BrKAN2.1 and BrKAN2.3 were demonstrated to be under strong selection in heading Chinese cabbage (Cheng et al., 2016). Altogether, most ad-ab polarity genes showed significantly different expression patterns in the outer HLs compared with inner HLs.
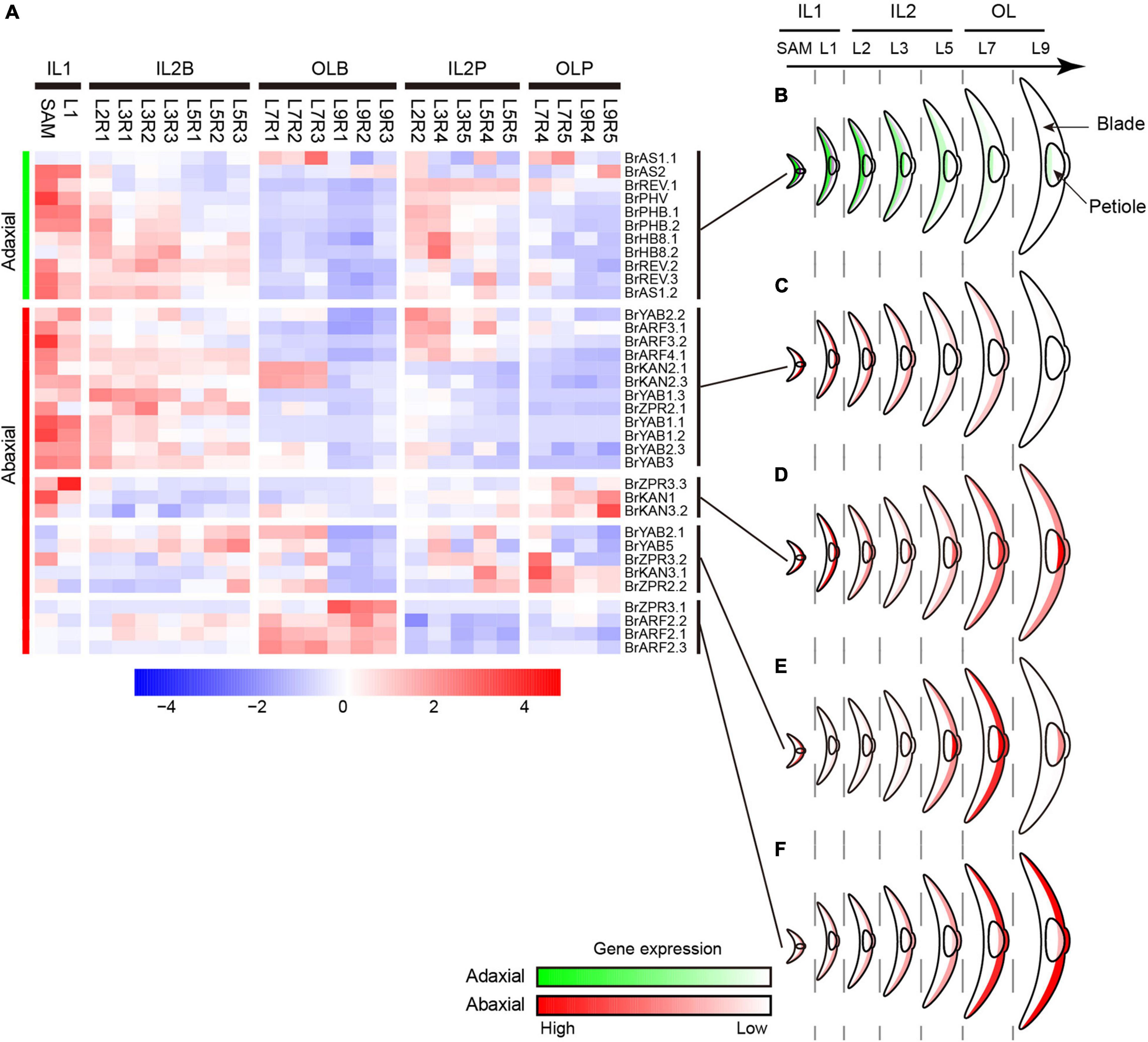
Figure 4. Expression profiles of ad-ab polarity genes. (A) Expression characteristics of ad-ab polarity genes in leafy head. For each gene, expression data were Z-score standardized. (B) Expression pattern of adaxial genes. (C–F) Four expression patterns of abaxial genes. Crescent indicated transverse section of a leaf.
Considering the morphological observations, PCA and clustering analysis, and expression of genes related to cell division/expansion and ad-ab polarity, L7 were positioned at the boundary between the inner leaves and outer leaves. L7 curved inward, thus showing a similar characteristic to inner leaves. However, L7 covered all inner leaves, providing shade for inner leaves, but the top of the blades exposed to light were light-green, thus showed similar to outer leaves. Therefore, L7 were classified into the outer leaves group by PCA and clustering. In agreement with this observation, expression analysis of genes related to cell division/expansion and ad-ab polarity revealed that L7 were a transition state showing three patterns. First, L7 showed a similar expression pattern to adjacent inner leaves L5 but were different from adjacent outer L9. Second, L7 showed a similar pattern to the outer L9 but were different from the inner L5. Third, L7 showed a specific pattern compared to the adjacent inner L5 and outer L9. In summary, our results confirmed that L7 and adjacent leaves L5 showed consecutive transition states and acted as transition leaves, but L7 were the key transition leaves.
In addition to the differences in leaf morphology between the inner and outer leaves at the heading stage, at this stage, the inner leaves are shielded from light, acting as storage organs, whereas the outer leaves are green and exposed to light, implicating their ability to photosynthesize to provide a carbon source for head development (He et al., 2000). Sucrose is the main form of photoassimilate transportation from the photosynthetic “sources” to the heterotrophic “sinks” (Fu et al., 2011; Chen et al., 2012). Here, we further identified and analyzed the accumulation of soluble sugar in HLs and expression characteristics of related gene (Supplementary Table 6).
Almost all genes encoding sucrose-phosphate synthase (SPS), a key enzyme of sucrose synthesis and its activity dictating the identities of source tissues (Ruan, 2014; Rodrigues et al., 2020), were predominantly expressed in outer leaves L7 and L9 (Figure 5A). Consistent with this, several key sucrose transporters (BrSWEET11s, BrSWEET12s, and two BrSUT1) were also preferentially expressed in outer leaves L7 and L9 (Figure 5A). In Arabidopsis, AtSWEET11 and AtSWEET12 are highly expressed in source leaves and are co-expressed with genes involved in sucrose biosynthesis and phloem loading (Gottwald et al., 2000; Chen et al., 2012). These results indicated that the outer leaves from L7 to L9 were the sites of sucrose synthesis. Interestingly, four genes encoding sucrose transporters (BrSUT2.1, BrSUT2.2, BrSUT4.1, and BrSUT4.2) were highly expressed in the petioles of HLs (Figure 5A). According to previous reports, SUT2 and SUT4 showed higher expression in sink cells and tissues (Meyer et al., 2004; Schulz et al., 2011; Schneider et al., 2012). These seemed to indicate that the petioles may be sink tissues. Almost all genes encoding SUS, which is an important sucrose-degrading enzyme hydrolyzing sucrose to fructose and UDP-glucose and considered a marker for sink strength (Ruan, 2014), were preferentially expressed in the inner HLs (SAM to L5) (Figure 5A), while other sucrose degrading genes such as BrCIN3, BrCIN7.2, and BrCIN1, showed significantly higher expression in the outer HLs than inner HLs. In addition, starch is an important storage substance in sink organs, and its important synthetic genes were highly expressed in inner HLs (SAM to L5), while lowly expressed in outer HLs. These results also supported that heading leaves from SAM to L5, as well as all petioles, could be storage organs for storing nutrients.
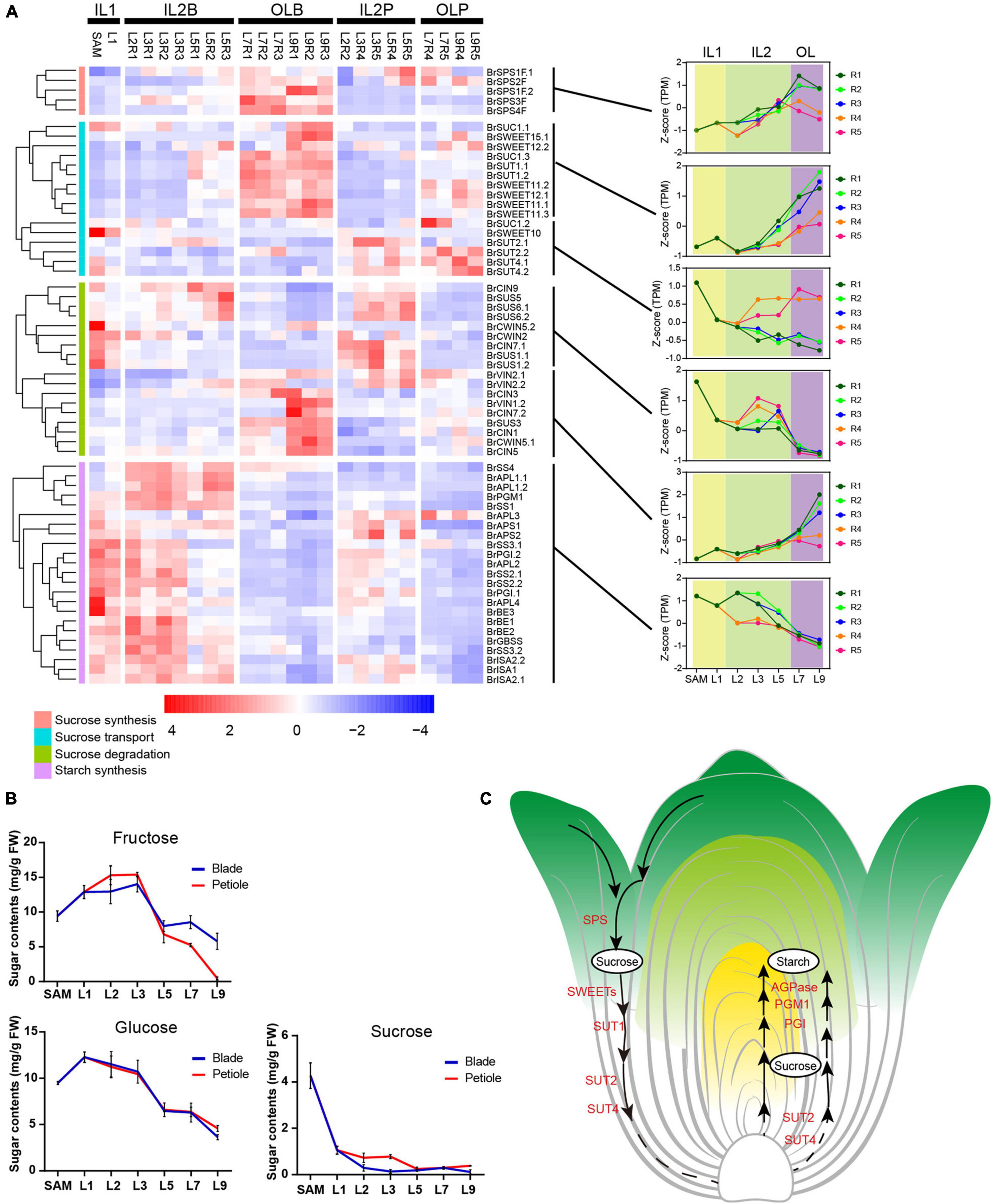
Figure 5. The expression pattern of starch, sucrose and transport genes in leafy head. (A) Expression profiles of the genes related to sucrose synthesis, transport, degradation, and starch synthesis. For each gene, expression data were Z-score standardized. (B) Soluble sugar content in different leaves. The X axis depicts 12 samples, and the y axis depicts the sugar content. Error bars represent the SD (n = 2). (C) A model proposed for sucrose synthesis, transport, degradation and starch synthesis in leafy head.
We further determined the content of sucrose, fructose and glucose in different head leaves. In accordance with the expression pattern of SUSs, the trend curves of fructose and glucose content in different head leaves were shown as inverse V shapes (Figure 5B). The content of fructose increased dramatically from L1 to L2, peaked at L3, and tapered off from L5–L9, and glucose content was maintained at a high level in the inner HLs (SAM to L3), decreased rapidly from L5 to L9. While the content of sucrose was constantly decreased from inner to outer HLs (Figure 5B). Taken together, our results demonstrated that outer leaves were source tissues that provide energy, while inner leaves the sink tissues for storing nutrients, and the L7 were the key transition leaves for leafy head development (Figure 5C).
To explore the contribution of the key transition leaves in leafy head formation, we next analyzed the expression characteristics of the previously identified candidate genes for leafy head formation. Since heading is the result of artificial domestication (Cheng et al., 2016; Zhang et al., 2019), the genes under strong selection in heading B. rapa might be candidate genes involved in leafy head formation. In this study, besides ten reported heading-related genes, 13 other leaf-heading candidate genes were further discovered in the domesticated selection region collected from our previously published data (Supplementary Table 7; Cheng et al., 2016; Cai et al., 2021). Surprisingly, nine of the ten reported heading-candidate genes were obviously activated or inhibited in the key transition leaves L7, including two downregulated genes (ARF3.1 and ARF4.1) and seven upregulated genes (KAN2.1, KAN2.3, BRX.1, BRX.2, BrPIN3.3, BrFL5.1, and BrSAL4.2) (Figure 6A and Supplementary Table 7). Moreover, eight of the 13 newly identified candidate genes also showed specific expression trends (Figure 6B and Supplementary Table 7). The expression level of BrKS (a reported heading-related gene) in the key transition leaves and inner leaves was significantly higher than that of the outer L9, and peaked at the petiole R4 region of the key transition leaves (Figure 6A). The significantly different expression of the heading candidate genes in the key transition leaves compared with other leaves confirmed that these genes will no doubt be the targets of future functional studies, and suggested that the transition leaves were related to leafy head formation.
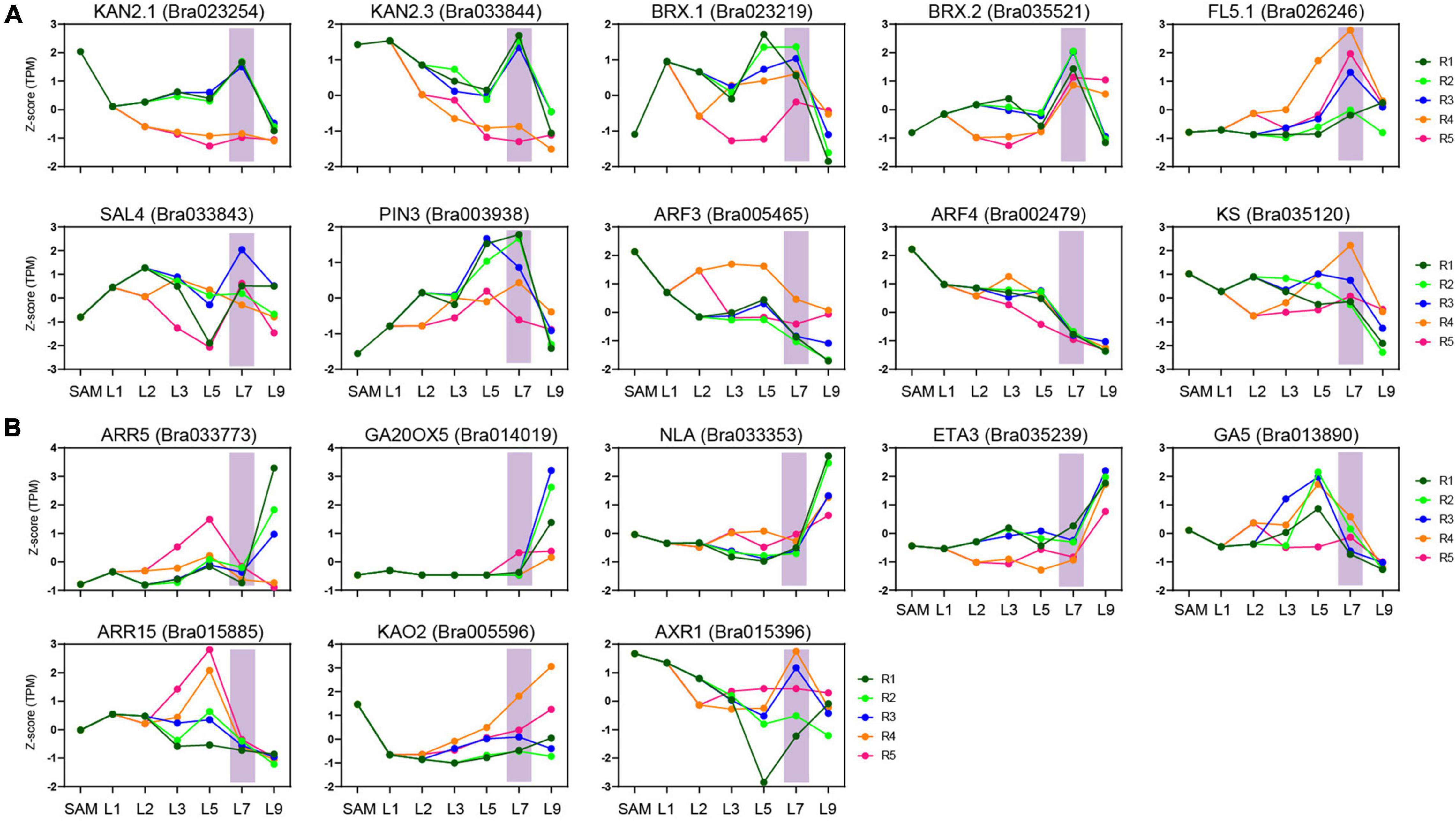
Figure 6. Expression characteristics of heading candidate genes. (A) The expression characteristics of reported heading candidate genes in key transition leaves. (B) The expression characteristics of heading candidate genes newly identified by domesticated selection analysis.
To identify the pathways contributed to the special state of the transition leaf, weighted gene coexpression network analysis (WGCNA) were performed. A total of 17 modules were identified by WGCNA (Supplementary Figure 6). Among them, five modules (greenyellow, magenta, purple, turquoise, and yellow) displayed close associations with transition leaves. Two modules, greenyellow and magenta, were specifically related to the transition leaves and contained many genes encoding protein kinase, including mitogen-activated protein kinases (MAPK), calcium-dependent protein kinases (CDPKs), and cysteine-rich receptor-like kinases (CRKs) (Figure 7A, Supplementary Figures 7, 8, and Supplementary Table 8). These protein kinases are key actors in plant signaling and play an important role in plant development, and external stimulus and phytohormone responses (Xu and Zhang, 2015; Baba et al., 2018; Shi et al., 2018). For example, MPK12 is a negative regulator of auxin signaling, and its kinase activity is increased following auxin treatment (Lee et al., 2009). In addition, the activity of MAPKs is also regulated by BR signals (Kim et al., 2012). In Arabidopsis, AtCRK5 can phosphorylate the hydrophilic loops of PIN3, impacting auxin distribution (Ding et al., 2011; Baba et al., 2019). AtCRK1 regulates the light response, and the Atcrk1-1 mutant shows serious growth defects under continuous illumination (Baba et al., 2018). Here, from the inner to outer leaves, CRK1, CPK5, MPK12, and CML4 displayed increased expression in the blades of L7 but rapidly decreased in L9, indicating that key transition leaves L7 might be regulated by complex signal interactions, not only light and other external stimuli, but also internal hormones.
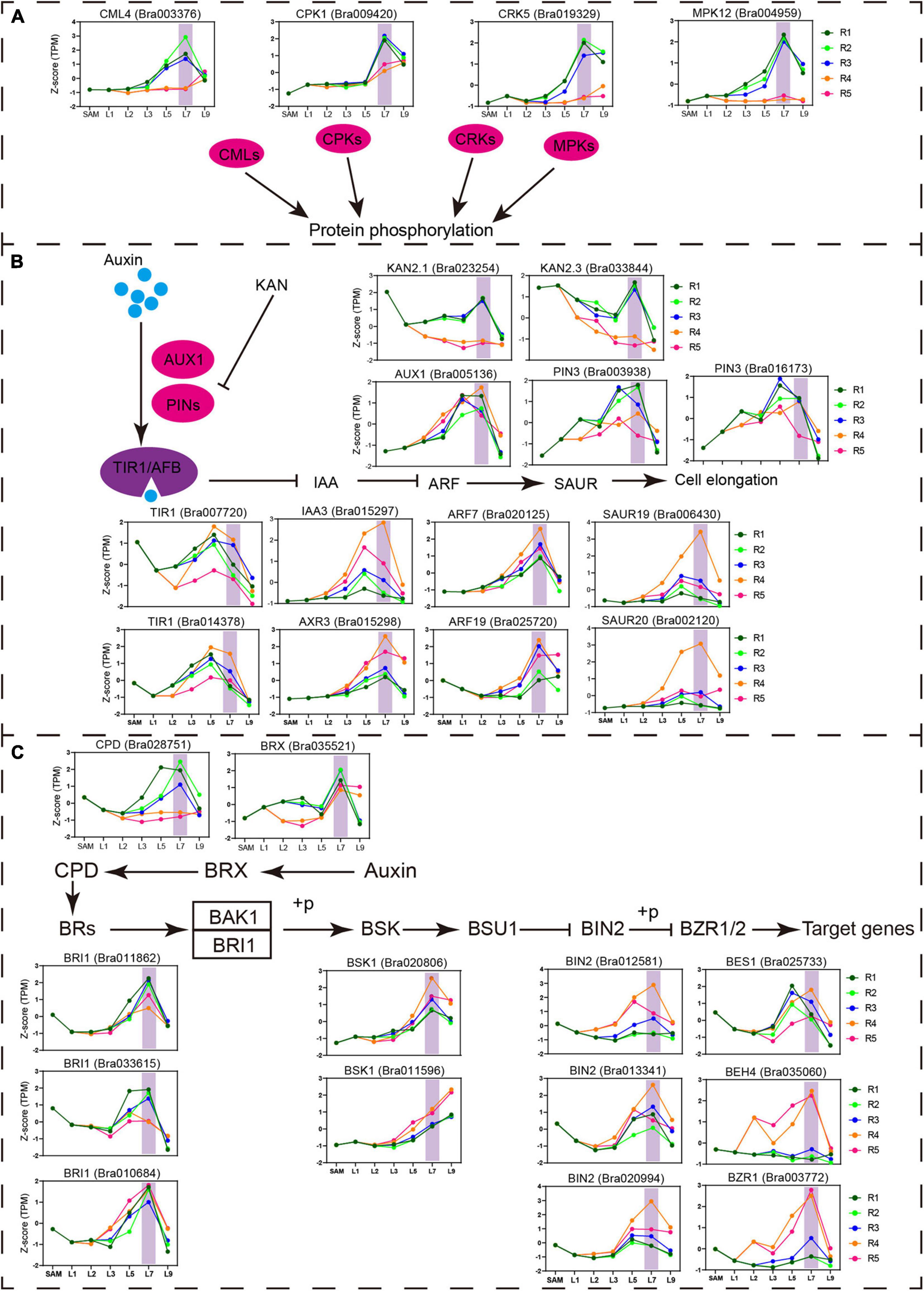
Figure 7. Important pathway identified in the key transition leaves. (A) Expression profiles of the genes related to protein phosphorylation and calcium ion binding identified in key transition leaves. Vertical shade boxes indicate transition leaves position. (B) Expression profiles of auxin signaling genes upregulated in key transition leaves. Vertical shade boxes indicate transition leaves position. (C) Expression profiles of BR signaling genes upregulated in key transition leaves. Vertical shade boxes indicate transition leaves position.
As expected, multiple phytohormonal pathways were activated in the petioles or blades of the transition leaves (Figures 7B,C and Supplementary Figure 7). Many auxin signaling pathway genes PIN3, TIR1, AXR3, ARF7, ARF19, SAUR20, and SAUR19 were highly expressed in key transition leaves L7 (Figure 7B). As reported, ARF7, ARF19, and SAUR19 constituted a module that mediates the bending growth of hypocotyls in response to light and gravity stimulation (Wang et al., 2020). The icu6, semi-dominant mutation of AXR3, results in leaf incurvature (Pérez-Pérez et al., 2010). Meanwhile, AXR3 could be induced by exogenous BR and is significantly downregulated in bri1 plants (Kim et al., 2006; Nakamura et al., 2006). Many BR signaling pathway genes showed predominant expression in transition leaves, including synthesis gene CPD, receptor gene BRI, and signal transduction genes BSK1, BIN2, BES1, BZR1, and BEH4 (Figure 7C). The mutation of BRI1 resulted in shorter petioles and curled leaves in Arabidopsis (Yang et al., 2011). BRX mediates the feedback regulation loop between BR synthesis and auxin signaling (Mouchel et al., 2006; Scacchi et al., 2009; Marhava et al., 2018). Overexpression of Chinese cabbage BrBRX.2 in Arabidopsis caused curling leaves with changes in leaf ad–ab polarity patterning (Zhang et al., 2021). Those results indicated that the crosstalk between auxin and BR may play an important role in key transition leaves.
In addition, a lot of light-responsive genes were also activated in transition leaves, such as FHY3, DAG2, RAX2, COP1, PHOT1, and PHYB (Supplementary Figure 8). COP1 is a central repressor of photomorphogenesis and directly interacts with BIN2 and modulates its kinase activity (Ling et al., 2017). PHOT1 is a blue-light photoreceptor that regulates leaf development and morphology by mediating auxin efflux (Jenness et al., 2020). Some important transcription factors that regulate leaf development were also identified, including PIF4, TCP24, TCP2, MYB30, HB12, BLH2, BrKAN2.1, and BrAS1.1, were identified (Supplementary Figure 8). MYB30 is closely integrated with the phytochrome-PIF4/PIF5 signaling module to participate in photomorphogenesis (Yan et al., 2020). Collectively, our data show that a complex signal network integrates plant hormones and external stimuli to maintain the special state of transition leaves.
To explore the mechanism of leafy head formation, many transcriptome studies of Chinese cabbage leaves have been conducted (Wang et al., 2012; Li J. et al., 2019; Sun et al., 2019). However, most heading candidate genes discovered through domestication selection analysis, genetic analysis, or molecular biology have not been identified by these studies. The reason may be that previous transcriptome studies only focused on a certain leaf between different stages, or analyzed mixed-leaf samples, thereby ignoring the diverse and potentially fundamental roles of complex head leaves with different morphology during leaf heading and limiting the value of transcriptome data to understand the process of leafy head formation. In this study, we constructed a spatial transcriptome landscape of leafy head using 24 dissected leaf tissues at the heading stage. Our samples contained continuous head leaves representing very different leaf morphologies. For example, the outer leaves wee yellow-green or green, and curved outward, while the inner leaves were covered by outer leaves, showing yellow and inwardly-curved. And the different part of the leaves by dissecting different regions from blades and petioles due to their structure or curvature degree is different. Our spatial sampling approach enabled the detection of tissue-specific or low abundance genes and their spatial expression changes in a leafy head. For example, the previously identified leaf head related genes such as BrKAN2.1 showed enrichment in blades, while BrARF3.1 was predominantly expressed in petioles. More importantly, it enabled the identification of the key transition leaves, because most of the genes involved in leaf development, including the genes related to cell division/expansion and ad-ab polarity, and the genes involved in sucrose metabolism showed significant expression changes in the key transition leaves (L7) or in both the key transition and their adjacent leaves (L5).
Surprisingly, most heading candidate genes (17/23), including those previously identified and those newly identified by domesticated selection analysis, indeed showed very different expression in the key transition leaves. In addition, an independent experiment was conducted to observe the morphological changes of transition leaves from the rosette stage to the heading stage (Figure 8). At the rosette stage, the new leaves were continually formed at the SAM, and the rosette leaves started to grow more upright. The leaves were arranged as a spiral path. As plants gradually grew up, the fifth whorls of leaves from outside to inside began to curve inward gradually until folding upward, thus covering all the inner leaves and finally forming a leafy head (Figure 8), which were similar to the key transition leaves (L7, the fourth whorls of leaves from outside to inside) identified from the transcriptome analysis. Conversely, the first fourth whorls of leaves, including the outermost leaves that were totally covered, did not curve inward and kept growing upright, resulting in the gentle outward curved in the top of their blades. Our observations showed the key transition leaves as the first inwardly-curved leaves that subsequently folded upward, which covered all the inner leaves and supported a frame for leafy head formation. These results strongly support the existence of special transition leaves, that play very important roles in leafy head formation, suggesting that the key transition leaves are worthy of attention in future heading research.

Figure 8. Morphological changes of Chinese cabbage (Brassica rapa cv Chiifu-401-42) from rosette stage to heading stage. The edges of key transition leaves were marked by colored dotted lines.
Accordingly, we further analyzed the specific transcriptional characteristics of the transition leaves through WGCNA. The transition leaves showed the enrichment of transcripts associated with protein kinases, auxin and BR pathways, and the light-responsive pathway, which was consistent with a previous study (Wang et al., 2012). Moreover, many of the previously identified heading-related genes were involved in the auxin and BR signaling pathways, including ARF3, ARF4, PIN3, and BRX. Interestingly, the protein kinases that play an important role in cell signal transduction (Xu and Zhang, 2015; Shin et al., 2016) were preferentially highly expressed in the leaf blade of the key transition leaves. Comparatively, many auxin and BR signaling genes were enriched at the R4 position of the key transition leaves, which were located at the top of the petiole and connected to the leaf blade and the petiole (Figure 7). Gao et al. (2017) found an uneven distribution of auxin, and the level of auxin peaked at the lower area of the leaf blades in the HLs in the early folding stage, which was caused by the polar transport of auxin. These results suggested that the transition leaves first curved inward, resulting from complex signal network regulation, including internal hormones, such as auxin and BR, as well as external stimuli, such as light. In addition, the leaf blade and leaf petiole showed different contributions to these signal integrations, and the region R4 is a site worthy of future attention for hormone content analysis.
In summary, the spatial transcriptome of the Chinese cabbage leafy head enabled the identification of the key transition leaves and provided a new perspective for leaf heading in Chinese cabbage. The values of the key transition leaves were illustrated to elucidate the genetic control of the complex leaf morphological and functional differentiation underlying leaf head formation. It also provides a valuable reference for leaf-heading research in other vegetables with leafy head, such as cabbage, mustard, lettuce, and endive, and the identification of the key transition leaves may be a valuable starting point to understand leaf head formation. However, the transition leaves may not be the exact leaves L7 identified by our study due to different species, different growth conditions, and even different heading types of the same species. In addition, our comprehensive and spatial transcriptome data cover almost all the aerial parts of Chinese cabbage at the heading stage. These data can be used to study leaf morphology and development, as well as to analyze the synthesis and accumulation of nutrient substances in leafy heads, such as glucosinolate, and explore the differentiation between homeologs after whole genome triplication.
Chinese cabbage (B. rapa cv Chiifu-401-42) was sown in potted trays in a greenhouse at August. After 3 weeks, seedlings were transplanted in the field with plastic sheds at the Chinese Academy of Agricultural Sciences. Eleven-week-old plants were used for sampling. Two normal growing Chinese cabbages were randomly selected. Since all head leaves arise on the axis of the enlarged but compressed stem, the spiral path of leaves may have five leaves in two whorls or eight leaves in three whorls (Zhang et al., 2021). The leaves in the same whorl showed similar size and similar morphology. Accordingly, all head leaves (leaf length > 2 cm) from the outer leaves to inner leaves were separated and collected in order, while the young and small inner leaves (leaf length < 2 cm) with the SAM were not separated and were collected as a mixed sample. A total of 30 leaves were obtained from each head. From the inside to the outside, every three leaves with similar morphology and similar size were taken as a whorl of leaves and a total of 10 whorls of leaves were obtained (Figure 1A). One leaf from every whorl was selected, named L1–L10, but only SAM, L1, L2, L3, L5, L7, and L9 were chosen for transcriptome analysis. L2 was dissected into two parts, leaf petiole (L2R2) and leaf blade (L2R1) for RNA-seq, while for L3, L5, L7, and L9, five regions, including the top (R1), outer margin (R2), and middle region (R3) of the blade and the top (R4) and middle (R5) regions of the petiole, were sampled. After sampling, a total of 48 samples from two biological replicates (two leafy heads) were ultimately used for transcriptome sequencing via the Illumina platform.
Total RNA was extracted using the TRIzol reagent (Invitrogen, CA, United States). RNA integrity was confirmed using the 2100 Bioanalyzer. RNA-seq libraries were constructed according to the manufacturer’s protocol of the Vazyme mRNA-seq library preparation kit (Vazyme) and were sequenced to generate 150-nucleotide paired-end reads on a HiSeq platform (Illumina).
The Chiifu reference genome (Brapa_v1.5; Cheng et al., 2016) was downloaded from http://brassicadb.org/brad/. After removing low-quality reads using the NGS QC Toolkit_v2.3.3 (Patel and Jain, 2012), clean reads were mapped to the Brapa_v1.5 reference genome using Hisat2-2.1.0 (Kim et al., 2015) with default settings for parameters. The bam files of uniquely mapped reads were used as inputs for the StringTie v1.3.4 software (Pertea et al., 2016), and TPM values were calculated to measure the expression levels of genes. To reduce transcription noise, only the genes with an average TPM ≥ 1 between two biological replicates were considered to be expressed and used for subsequent analysis.
Based on TPM values, DEGs were detected with a Fisher exact test (P-value cutoff < 0.05 and log2 fold change > 1 or <−1). By comparing the expression levels of genes at the corresponding positions of adjacent leaf blades (R1 vs R1, R2 vs R2, and R3 vs R3); that is, if a gene was upregulated or downregulated in multiple comparisons, it was considered a differential gene among leaf blades. SAM or HL1 were also used to compare adjacent blades. A similar method was also used to identify DEGs among petioles. The DEGs were identified between leaf blades and petioles by comparing the expression levels of genes in leaf blades and petioles from the same leaf; that is, if the expression level of a gene in any leaf blade sample was upregulated or downregulated relative to any petiole samples, it was considered to be a differential gene between leaf blades and petioles. Pearson correlation coefficient was calculated between biological replicates with the normalized expression levels of log2 (TPM value +1). Hierarchical clustering and PCA analysis were performed using the prcomp and hclust functions in R software (R Core Team, 2013) with default settings. The syntenic genes between A. thaliana and B. rapa were identified as previously described (Liang et al., 2016). An orthologous gene dataset was established for obtaining functional annotations of B. rapa genes based on A. thaliana genes.
Co-expression analysis was performed on all samples using MeV (V 4.9) with the k-means method (Gasch and Eisen, 2002). The normalized expression values of genes with Z-scores were used for k-Means clustering. Gene ontology enrichment analysis of each co-expression cluster was performed using agriGO with the singular enrichment analysis method (FDR < 0.05) (Tian et al., 2017).
The leaf blade or petiole samples from the same leaf layer were mixed together and ground in liquid nitrogen. Subsequently, 600 μl water and 800 μl internal standard (10 mg/ml arabinose water) were added to 200 mg ground powder, vortexed and mixed well. After sonication and of centrifugation for 10 min each, the mixture was filtered through a 0.22 μm filter membrane. Then, 895 μl of acetonitrile and 5 μl 20% ammonia water were added to 100 μl of filtrate, and the mixture was shaken to mix. The samples were maintained at in 4°C for 10 h and then filtered with a 0.22 μm organic membrane filter. The sugar content was then determined by UPLC-MS/MS (ACQUITY UPLC I-Class-Xevo TQ-S Micro, Waters).
To identify modules of highly correlated genes, those genes with an average TPM of more than 1 were used to perform the co-expression network analysis by WGCNA. The one-step network construction and module detection were performed using the function blockwiseModules, with the following settings: the soft threshold power was set to 16, the maxBlockSize was set to 30,000, the minModuleSize was set to 30, and a dynamic tree cutoff was 0.25.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI with the accession number PRJNA778186 (https://www.ncbi.nlm.nih.gov/sra?linkname=bioproject_sra_all&from_uid=778186).
XW, JW, and JL designed the study. JL and XG grew and collected the plant materials and performed the content assay of soluble sugars and morphological observation. XG and RL analyzed the RNA-seq data. XG, JL, and XW wrote the manuscript, with the help from JW and RL. All authors have discussed the results and approved the manuscript.
This work was supported by the National Natural Science Foundation of China (31630068 and 32072594) and the Central Public-Interest Scientific Institution Basal Research Fund (Y2020PT21), Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences. This research was carried out at the Key Laboratory of Biology and Genetic Improvement of Horticultural Crops, Ministry of Agriculture, People’s Republic of China.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Xia Cui and Haijing Wang for experimental assistance from Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.787826/full#supplementary-material
Supplementary Figure 1 | Pearson correlation coefficients of transcriptome data and distribution of TPM normalized expression. (A) Pearson correlation coefficients of transcriptome data from biological replicates for 24 leaf tissues. (B) Distribution of TPM normalized expression among 24 leaf tissues.
Supplementary Figure 2 | Distribution of expressed genes. (A) The numbers and proportion of expressed genes in 24 leaf tissues. (B) Distribution of expressed genes among different leaves.
Supplementary Figure 3 | Distribution of DEGs among different leaves/tissue-types. (A) DEGs overlap for the blades of neighboring leaf. (B) DEGs overlap for the petioles of neighboring leaf. (C) Overlap of DEGs between leaf blades and petioles.
Supplementary Figure 4 | K-means clustering grouped 9133 DEGs into 14 clusters. The X axis depicts 24 samples, and the Y axis represents the expression data per gene. For each gene, expression data were Z-score standardized.
Supplementary Figure 5 | Cell proliferation and cell expansion genes constitute a regulatory network that controls leaf size.
Supplementary Figure 6 | WGCNA of all expressed genes. (A) Hierarchical cluster tree showing co-expression modules identified by WGCNA. (B) Module–sample association. Each row corresponds to a module, labeled with a color as in panel (A). Each column corresponds to a specific tissue. (C) Expression profiles of the 17 module eigengenes in all tissues. The X axis indicates the tissues; The Y axis indicates the value of the module eigengene.
Supplementary Figure 7 | Expression pattern of some important genes identified in key transition leaves. (A) Expression profiles of the genes related to protein phosphorylation identified in KTLs. Vertical shade boxes indicate the position of key transition leaves. (B) Many auxin signaling genes were up-regulated in key transition leaves. Vertical shade boxes indicate the position of key transition leaves.
Supplementary Figure 8 | Expression pattern of some important genes in transition leaves. (A) Many light-responsive genes were identified in key transition leaves. Vertical shade boxes indicate the position of key transition leaves. (B) Many genes encoding transcription factors were up-regulated in key transition leaves. Vertical shade boxes indicate the position of key transition leaves.
Baba, A. I., Andrási, N., Valkai, I., Gorcsa, T., Koczka, L., Darula, Z., et al. (2019). AtCRK5 protein kinase exhibits a regulatory role in hypocotyl hook development during skotomorphogenesis. Int. J. Mol. Sci. 20:3432.
Baba, A. I., Rigó, G., Ayaydin, F., Rehman, A. U., Andrási, N., Zsigmond, L., et al. (2018). Functional analysis of the Arabidopsis thaliana CDPK-related kinase family: AtCRK1 regulates responses to continuous light. Int. J. Mol. Sci. 19:1282. doi: 10.3390/ijms19051282
Cai, X., Chang, L., Zhang, T., Chen, H., Zhang, L., Lin, R., et al. (2021). Impacts of allopolyploidization and structural variation on intraspecific diversification in Brassica rapa. Genome Biol. 22:166. doi: 10.1186/s13059-021-02383-2
Carles, C. C., and Fletcher, J. C. (2003). Shoot apical meristem maintenance: the art of a dynamic balance. Trends Plant Sci. 8, 394–401. doi: 10.1016/S1360-1385(03)00164-X
Chen, L. Q., Qu, X. Q., Hou, B. H., Sosso, D., Osorio, S., Fernie, A. R., et al. (2012). Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science 335, 207–211. doi: 10.1126/science.1213351
Cheng, F., Sun, R., Hou, X., Zheng, H., Zhang, F., Zhang, Y., et al. (2016). Subgenome parallel selection is associated with morphotype diversification and convergent crop domestication in Brassica rapa and Brassica oleracea. Nat. Genet. 48, 1218–1224. doi: 10.1038/ng.3634
Cutler, S. R., Rodriguez, P. L., Finkelstein, R. R., and Abrams, S. R. (2010). Abscisic acid: emergence of a core signaling network. Annu. Rev. Plant Biol. 61, 651–679. doi: 10.1146/annurev-arplant-042809-112122
Ding, G., Che, P., Ilarslan, H., Wurtele, E. S., and Nikolau, B. J. (2012). Genetic dissection of methylcrotonyl CoA carboxylase indicates a complex role for mitochondrial leucine catabolism during seed development and germination. Plant J. 70, 562–577. doi: 10.1111/j.1365-313X.2011.04893.x
Ding, Z., Galván-Ampudia, C. S., Demarsy, E., Łangowski, Ł, Kleine-Vehn, J., Fan, Y., et al. (2011). Light-mediated polarization of the PIN3 auxin transporter for the phototropic response in Arabidopsis. Nat. Cell Biol. 13, 447–452. doi: 10.1038/ncb2208
Doering, A., Lathe, R., and Persson, S. (2012). An update on xylan synthesis. Mol. Plant 5, 769–771. doi: 10.1093/mp/sss049
Du, F., Guan, C., and Jiao, Y. (2018). Molecular mechanisms of leaf morphogenesis. Mol. Plant 11, 1117–1134. doi: 10.1016/j.molp.2018.06.006
Fu, Q., Cheng, L., Guo, Y., and Turgeon, G. R. (2011). Phloem loading strategies in relation to water relations in trees and herbaceous plants. Plant Physiol. 157, 1518–1527. doi: 10.1104/pp.111.184820
Gao, L., Lyu, S. W., Tang, J., Zhou, D., Bonnema, G., Xiao, D., et al. (2017). Genome-wide analysis of auxin transport genes identifies the hormone responsive patterns associated with leafy head formation in Chinese cabbage. Sci. Rep. 7:42229. doi: 10.1038/srep42229
Gao, Y., Huang, S., Qu, G., Fu, W., Zhang, M., Liu, Z., et al. (2020). The mutation of ent-kaurene synthase, a key enzyme involved in gibberellin biosynthesis, confers a non-heading phenotype to Chinese cabbage (Brassica rapa L. ssp. pekinensis). Hortic. Res. 7:178. doi: 10.1038/s41438-020-00399-6
Gasch, A. P., and Eisen, M. B. (2002). Exploring the conditional coregulation of yeast gene expression through fuzzy k-means clustering. Genome Biol. 3:research0059. doi: 10.1186/gb-2002-3-11-research0059
Gottwald, J. R., Krysan, P. J., Young, J. C., Evert, R. F., and Sussman, M. R. (2000). Genetic evidence for the in planta role of phloem-specific plasma membrane sucrose transporters. Proc. Natl. Acad. Sci. U. S. A. 97, 13979–13984. doi: 10.1073/pnas.250473797
Gu, A., Meng, C., Chen, Y., Wei, L., Dong, H., Lu, Y., et al. (2017). Coupling Seq-BSA and RNA-Seq analyses reveal the molecular pathway and genes associated with heading type in Chinese cabbage. Front. Genet. 8:176. doi: 10.3389/fgene.2017.00176
Han, C., Ren, C., Zhi, T., Zhou, Z., Liu, Y., Chen, F., et al. (2013). Disruption of fumarylacetoacetate hydrolase causes spontaneous cell death under short-day conditions in Arabidopsis. Plant Physiol. 162, 1956–1964. doi: 10.1104/pp.113.216804
He, Y. K., Xue, W. X., Sun, Y. D., Yu, X. H., and Liu, P. L. (2000). Leafy head formation of the progenies of transgenic plants of Chinese cabbage with exogenous auxin genes. Cell Res. 10, 151–160. doi: 10.1038/sj.cr.7290044
Horiguchi, G., Ferjani, A., Fujikura, U., and Tsukaya, H. (2006). Coordination of cell proliferation and cell expansion in the control of leaf size in Arabidopsis thaliana. J. Plant Res. 119, 37–42. doi: 10.1007/s10265-005-0232-4
Hou, H., Erickson, J., Meservy, J., and Schultz, E. A. (2010). FORKED1 encodes a PH domain protein that is required for PIN1 localization in developing leaf veins. Plant J. 63, 960–973. doi: 10.1111/j.1365-313X.2010.04291.x
Huang, H., Liu, B., Liu, L., and Song, S. (2017). Jasmonate action in plant growth and development. J. Exp. Bot. 68, 1349–1359. doi: 10.1093/jxb/erw495
Ito, H., and Kato, T. (1957). Studies on the head formation of Chinese cabbage: histological and physiological studies of head formation. J. Jpn. Soc. Hortic. Sci. 26, 154–162. doi: 10.2503/jjshs.26.154
Jenness, M. K., Tayengwa, R., and Murphy, A. S. (2020). An ATP-binding cassette transporter, ABCB19, regulates leaf position and morphology during phototropin1-mediated blue light responses. Plant Physiol. 184, 1601–1612. doi: 10.1104/pp.20.00223
Jun, J. H., Ha, C. M., and Fletcher, J. C. (2010). BLADE-ON-PETIOLE1 coordinates organ determinacy and axial polarity in Arabidopsis by directly activating ASYMMETRIC LEAVES2. Plant Cell 22, 62–76. doi: 10.1105/tpc.109.070763
Kalve, S., De Vos, D., and Beemster, G. T. (2014). Leaf development: a cellular perspective. Front. Plant Sci. 5:362. doi: 10.3389/fpls.2014.00362
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Kim, H., Park, P. J., Hwang, H. J., Lee, S. Y., Oh, M. H., and Kim, S. G. (2006). Brassinosteroid signals control expression of the AXR3/IAA17 gene in the cross-talk point with auxin in root development. Biosci. Biotechnol. Biochem. 70, 768–773. doi: 10.1271/bbb.70.768
Kim, T. W., Michniewicz, M., Bergmann, D. C., and Wang, Z. (2012). Brassinosteroid regulates stomatal development by GSK3-mediated inhibition of a MAPK pathway. Nature 482, 419–422. doi: 10.1038/nature10794
Lee, J. S., Wang, S., Sritubtim, S., Chen, J. G., and Ellis, B. E. (2009). Arabidopsis mitogen-activated protein kinase MPK12 interacts with the MAPK phosphatase IBR5 and regulates auxin signaling. Plant J. 57, 975–985. doi: 10.1111/j.1365-313X.2008.03741.x
Li, J., Zhang, X., Lu, Y., Feng, D., Gu, A., Wang, S., et al. (2019). Characterization of non-heading mutation in heading Chinese cabbage (Brassica rapa L. ssp. pekinensis). Front. Plant Sci. 10:112. doi: 10.3389/fpls.2019.00112
Li, Y., Fan, Y., Jiao, Y., Wu, J., Zhang, Z., Yu, X., et al. (2019). Transcriptome profiling of yellow leafy head development during the heading stage in Chinese cabbage (Brassica rapa subsp. pekinensis). Physiol. Plant 165, 800–813. doi: 10.1111/ppl.12784
Li, Y., Liu, Z., Wang, Y., Ning, Y., Xin, X., Yang, S., et al. (2012). Identification of quantitative trait loci for yellow inner leaves in Chinese cabbage (Brassica rapa L. ssp. pekinensis) based on SSR and SRAP markers. Sci. Hortic. 133, 10–17. doi: 10.1016/j.scienta.2011.10.023
Liang, J., Liu, B., Wu, J., Cheng, F., and Wang, X. (2016). Genetic variation and divergence of genes involved in leaf adaxial-abaxial polarity establishment in Brassica rapa. Front. Plant Sci. 7:94. doi: 10.3389/fpls.2016.00094
Ling, J. J., Li, J., Zhu, D., and Deng, X. W. (2017). Noncanonical role of Arabidopsis COP1/SPA complex in repressing BIN2-mediated PIF3 phosphorylation and degradation in darkness. Proc. Natl. Acad. Sci. U. S. A. 114, 3539–3544. doi: 10.1073/pnas.1700850114
Liu, Q., Li, J., and Liu, W. (2021). Sugar accumulation and characterization of metabolizing enzyme genes in leafy head of Chinese cabbage (Brassica campestris L. ssp. pekinensis). Hortic. Environ. Biotechnol. 62, 17–29. doi: 10.1007/s13580-020-00294-y
Mao, Y., Wu, F., Yu, X., Bai, J., Zhong, W., and He, Y. (2014). MicroRNA319a-targeted Brassica rapa ssp. pekinensis TCP genes modulate head shape in chinese cabbage by differential cell division arrest in leaf regions. Plant Physiol. 164, 710–720. doi: 10.1104/pp.113.228007
Marhava, P., Bassukas, A. E. L., Zourelidou, M., Kolb, M., Moret, B., Fastner, A., et al. (2018). A molecular rheostat adjusts auxin flux to promote root protophloem differentiation. Nature 558, 297–300. doi: 10.1038/s41586-018-0186-z
Meyer, S., Lauterbach, C., Niedermeier, M., Barth, I., Sjolund, D. R., and Sauer, N. (2004). Wounding enhances expression of AtSUC3, a sucrose transporter from Arabidopsis sieve elements and sink tissues. Plant Physiol. 134, 684–693. doi: 10.1104/pp.103.033399
Mouchel, C. F., Osmont, K. S., and Hardtke, C. S. (2006). BRX mediates feedback between brassinosteroid levels and auxin signalling in root growth. Nature 443, 458–461. doi: 10.1038/nature05130
Nakamura, A., Nakajima, N., Goda, H., Shimada, Y., Hayashi, K., Nozaki, H., et al. (2006). Arabidopsis Aux/IAA genes are involved in brassinosteroid-mediated growth responses in a manner dependent on organ type. Plant J. 45, 193–205. doi: 10.1111/j.1365-313X.2005.02582.x
Nakata, M., and Okada, K. (2013). The leaf adaxial-abaxial boundary and lamina growth. Plants 2, 174–202. doi: 10.3390/plants2020174
Opena, R. T., Kuo, C. G., and Voon, J. Y. (1998). Breeding and seed production of Chinese cabbage in the tropics and subtropics. Taiwan: Asian Vegetable Research and Development Centre
Park, J. Y., Canam, T., Kang, K. Y., Ellis, D. D., and Mansfield, S. D. (2008). Over-expression of an arabidopsis family A sucrose phosphate synthase (SPS) gene alters plant growth and fibre development. Transgenic Res. 17, 181–192. doi: 10.1007/s11248-007-9090-2
Patel, R. K., and Jain, M. (2012). NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One 7:e30619. doi: 10.1371/journal.pone.0030619
Pattanayak, G. K., Venkataramani, S., Hortensteiner, S., Kunz, L., Christ, B., Moulin, M., et al. (2012). Accelerated cell death 2 suppresses mitochondrial oxidative bursts and modulates cell death in Arabidopsis. Plant J. 69, 589–600. doi: 10.1111/j.1365-313X.2011.04814.x
Pérez-Pérez, J. M., Candela, H., Robles, P., López-Torrejón, G., del Pozo, J. C., and Micol, J. L. (2010). A role for AUXIN RESISTANT3 in the coordination of leaf growth. Plant Cell Physiol. 51, 1661–1673. doi: 10.1093/pcp/pcq123
Pertea, M., Kim, D., Pertea, G. M., Leek, J. T., and Salzberg, S. L. (2016). Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11, 1650–1667. doi: 10.1038/nprot.2016.095
R Core Team (2013). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. Available online at: http://www.R-project.org
Ren, W., Wu, F., Bai, J., Li, X., Yang, X., Xue, W., et al. (2020). BcpLH organizes a specific subset of microRNAs to form a leafy head in Chinese cabbage (Brassica rapa ssp. pekinensis). Hortic. Res. 7:1. doi: 10.1038/s41438-019-0222-7
Rodrigues, C. M., Müdsam, C., Keller, I., Zierer, W., Czarnecki, O., Corral, J. M., et al. (2020). Vernalization alters sink and source identities and reverses phloem translocation from taproots to shoots in sugar beet. Plant Cell 32, 3206–3223. doi: 10.1105/tpc.20.00072
Ruan, Y. (2014). Sucrose metabolism: gateway to diverse carbon use and sugar signaling. Annu. Rev. Plant Biol. 65, 33–67. doi: 10.1146/annurev-arplant-050213-040251
Scacchi, E., Osmont, K. S., Beuchat, J., Salinas, P., Navarrete-Gómez, M., Trigueros, M., et al. (2009). Dynamic, auxin-responsive plasma membrane-to-nucleus movement of Arabidopsis BRX. Development 136, 2059–2067. doi: 10.1242/dev.035444
Schneider, S., Hulpke, S., Schulz, A., Yaron, I., Höll, J., Imlau, A., et al. (2012). Vacuoles release sucrose via tonoplast-localised SUC4-type transporters. Plant Biol. 14, 325–336. doi: 10.1111/j.1438-8677.2011.00506.x
Schulz, A., Beyhl, D., Marten, I., Wormit, A., Neuhaus, E., Poschet, G., et al. (2011). Proton-driven sucrose symport and antiport are provided by the vacuolar transporters SUC4 and TMT1/2. Plant J. 68, 129–136. doi: 10.1111/j.1365-313X.2011.04672.x
Shi, S., Li, S., Asim, M., Mao, J., Xu, D., Ullah, Z., et al. (2018). The Arabidopsis calcium-dependent protein kinases (CDPKs) and their roles in plant growth regulation and abiotic stress responses. Int. J. Mol. Sci. 19:1900. doi: 10.3390/ijms19071900
Shin, A. Y., Han, Y. J., Baek, A., Ahn, T., Kim, S. Y., Nguyen, T. S., et al. (2016). Evidence that phytochrome functions as a protein kinase in plant light signalling. Nat. Commun. 7:11545. doi: 10.1038/ncomms11545
Sun, X., Basnet, R. K., Yan, Z., Bucher, J., Cai, C., Zhao, J., et al. (2019). Genome-wide transcriptome analysis reveals molecular pathways involved in leafy head formation of Chinese cabbage (Brassica rapa). Hortic. Res. 6:130. doi: 10.1038/s41438-019-0212-9
Tian, T., Liu, Y., Yan, H., You, Q., Yi, X., Du, Z., et al. (2017). agriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 45, W122–W129. doi: 10.1093/nar/gkx382
Tsukaya, H. (2003). Organ shape and size: a lesson from studies of leaf morphogenesis. Curr. Opin. Plant Biol. 6, 57–62. doi: 10.1016/s1369526602000055
Vernoux, T., Kronenberger, J., Grandjean, O., Laufs, P., and Traas, J. (2000). PIN-FORMED 1 regulates cell fate at the periphery of the shoot apical meristem. Development 127, 5157–5165.
Wang, F., Li, L., Li, H., Liu, L., Zhang, Y., Gao, J., et al. (2012). Transcriptome analysis of rosette and folding leaves in Chinese cabbage using high-throughput RNA sequencing. Genomics 99, 299–307. doi: 10.1016/j.ygeno.2012.02.005
Wang, X., Yu, R., Wang, J., Lin, Z., Han, X., Deng, Z., et al. (2020). The Asymmetric Expression of SAUR genes mediated by ARF7/19 promotes the gravitropism and phototropism of plant hypocotyls. Cell Rep. 31:107529. doi: 10.1016/j.celrep.2020.107529
Wang, Y., Wu, F., Bai, J., and He, Y. (2014). BrpSPL9 (Brassica rapa ssp. pekinensis SPL9) controls the earliness of heading time in Chinese cabbage. Plant Biotechnol. J. 12, 312–321. doi: 10.1111/pbi.12138
Wasternack, C., and Song, S. (2017). Jasmonates: biosynthesis, metabolism, and signaling by proteins activating and repressing transcription. J. Exp. Bot. 68, 1303–1321. doi: 10.1093/jxb/erw443
Xu, J., and Zhang, S. (2015). Mitogen-activated protein kinase cascades in signaling plant growth and development. Trends Plant Sci. 20, 56–64. doi: 10.1016/j.tplants.2014.10.001
Yan, Y., Li, C., Dong, X., Li, H., Zhang, D., Zhou, Y., et al. (2020). MYB30 is a key negative regulator of Arabidopsis photomorphogenic development that promotes PIF4 and PIF5 protein accumulation in the light. Plant Cell 32, 2196–2215. doi: 10.1105/tpc.19.00645
Yang, C., Zhang, C., Lu, Y., Jin, J., and Wang, X. (2011). The mechanisms of brassinosteroids’ action: from signal transduction to plant development. Mol. Plant 4, 588–600. doi: 10.1093/mp/ssr020
Yu, C., Yan, C., Liu, Y., Liu, Y., Jia, Y., Lavelle, D., et al. (2020). Upregulation of a KN1 homolog by transposon insertion promotes leafy head development in lettuce. Proc. Natl. Acad. Sci. U. S. A. 117, 33668–33678. doi: 10.1073/pnas.2019698117
Yu, X., Peng, J., Feng, X., Yang, S., Zheng, Z., Tang, X., et al. (2000). Cloning and structural and expressional characterization of BcpLH gene preferentially expressed in folding leaf of Chinese cabbage. Sci. China C Life Sci. 43, 321–329. doi: 10.1007/BF02879292
Yu, X., Wang, H., Zhong, W., Bai, J., Liu, P., and He, Y. (2013). QTL mapping of leafy heads by genome resequencing in the RIL population of Brassica rapa. PLoS One 8:e76059. doi: 10.1371/journal.pone.0076059
Zhang, K., Wang, X., and Cheng, F. (2019). Plant polyploidy: origin, evolution, and its influence on crop domestication. Hortic. Plant J. 5, 231–239. doi: 10.1016/j.hpj.2019.11.003
Keywords: leafy head, transcriptome, key transition leaves, hormone, Chinese cabbage
Citation: Guo X, Liang J, Lin R, Zhang L, Wu J and Wang X (2022) Series-Spatial Transcriptome Profiling of Leafy Head Reveals the Key Transition Leaves for Head Formation in Chinese Cabbage. Front. Plant Sci. 12:787826. doi: 10.3389/fpls.2021.787826
Received: 01 October 2021; Accepted: 08 December 2021;
Published: 06 January 2022.
Edited by:
Hirokazu Tsukaya, The University of Tokyo, JapanReviewed by:
Hui Feng, Shenyang Agricultural University, ChinaCopyright © 2022 Guo, Liang, Lin, Zhang, Wu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaowu Wang, d2FuZ3hpYW93dUBjYWFzLmNu
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.