- 1Beijing Academy of Agriculture and Forestry Sciences, Beijing, China
- 2College of Grassland Science, Beijing Forestry University, Beijing, China
Pheophytinase (PPH), the phytol hydrolase, plays important roles in chlorophyll degradation. Nevertheless, little attention has been paid to the PPHs in warm-season grass species; neither its detailed function in photosynthesis has been systematically explored to date. In this study, we isolated ZjPPH from Zoysia japonica, an excellent warm-season turfgrass species. Quantitative real-time PCR analysis and promoter activity characterization revealed that the expression of ZjPPH could be induced by senescence, ABA, and dark induction. Subcellular localization observation proved that ZjPPH was localized in the chloroplasts. Overexpression of ZjPPH accelerated the chlorophyll degradation and rescued the stay-green phenotype of the Arabidopsis pph mutant. Moreover, ZjPPH promoted senescence with the accumulation of ABA and soluble sugar contents, as well as the increased transcriptional level of SAG12 and SAG14. Transmission electron microscopy investigation revealed that ZjPPH caused the decomposition of chloroplasts ultrastructure in stable transformed Arabidopsis. Furthermore, chlorophyll a fluorescence transient measurement analysis suggested that ZjPPH suppressed photosynthesis efficiency by mainly suppressing both photosystem II (PSII) and photosystem I (PSI). In conclusion, ZjPPH plays an important role in chlorophyll degradation and senescence. It could be a valuable target for genetic editing to cultivate new germplasms with stay-green performance and improved photosynthetic efficiency.
Introduction
Chlorophyll degradation is the phenotype landmark of leaf senescence processes in plants. The biochemistry and topology of chlorophyll degradation have been explored systematically during the recent decades (Hortensteiner, 2013; Kuai et al., 2018). Specifically, the identification of important stay-green mutants unmasked the key catalyzing steps in the chlorophyll breakdown pathway. In brief, the breakdown of chlorophyll was started with the conversion of chlorophyll b (Chl b) to Chl a catalyzed by two Chl b reductase paralogous, NYC1 and NOL (Yu et al., 2021). Next, magnesium was released from Chl a by the STAY-GREEN protein (SGR) to yield pheophytin a (Shimoda et al., 2016). Then, the pheophytin a was converted to pheophorbide a with the action of pheophytinase (PPH) (Zhang et al., 2016). Finally, the pheophorbide a was subsequently catalyzed by PAO, RCCR, and TIC55, generating red chlorophyll catabolite (RCC), phyllobilin (pFCC), and hydroxy-pFCC, respectively (Kuai et al., 2018). The above steps were finished inside the chloroplasts and were termed as the first part of the chlorophyll degradation pathway. The other part of the pathway resulted in a diversity of phyllobilins and took place outside the chloroplasts (Guo et al., 2021).
The PPH gene was first reported to be involved in chlorophyll breakdown machinery during leaf senescence in Arabidopsis (Schelbert et al., 2009) and rice (Morita et al., 2009). Bamboo (Bambusa emeiensis) PPH (also called BeCRN1) is localized in the chloroplasts, and its constitutive overexpression rescued the stay-green phenotype of Arabidopsis crn1 mutants (Wei et al., 2013). Knockdown of SlPPH delayed chlorophyll breakdown and accumulated pheophytin during leaf senescence in tomato (Solanum lycopersicum) (Guyer et al., 2014). Perennial ryegrass (Lolium perenne) PPH was reported to accelerate chlorophyll degradation and might be a direct downstream target gene of abscisic acid and cytokinin-related transcription factors (Zhang et al., 2016). Apple (Malus domestica) MdPPH, a target of MdERF17, was required for peel Chl degradation and pigment accumulation (Han et al., 2018). Warm-season grasses are important for both agricultural production and ecosystem services (Lee et al., 2019). Nevertheless, little attention has been paid to the PPH gene in warm-season grass species; neither its function has been explored to date.
The popularization of Chl a fluorescence transient measurements (JIP test) provides us a convenient and reliable tool to evaluate the status and performance of photosystem II (PSII) reaction centers, photosynthetic electron transport, and the attribute of both the donor and acceptor sides of PSII (Hu et al., 2016; Kalaji et al., 2016, 2018; Zagorchev et al., 2021). JIP test revealed that the overexpression of the γ-tocopherol methyl transferase gene improved the efficiency of photosystem and consequently enhanced the drought adaptability of transgenic Brassica juncea plants (Yusuf et al., 2010). Analysis of Chl a fluorescence indicated that the heterologous expression of wheat TaFBA1 alleviated the damage to the photosynthetic apparatus of transgenic tobacco under high salinity (Zhao et al., 2017). JIP test demonstrated that the overexpression of Fld preserved the integrity and functionality of the photosynthetic machinery in tobacco leaves infected with Botrytis cinereal (Rossi et al., 2017). However, the detailed influence of PPH, the catalyzer of the primary electron acceptor in PSII (pheophytin a), on photosystem remains poorly explored. The question on the specific functional machinery of PPH in photosynthesis still has unfilled gaps.
Zoysia grass is an excellent warm-season turfgrass species and is widely utilized in football pitches and landscaping (Xuan et al., 2013; Chen et al., 2015). Compared with the cool-season turfgrasses, the most limiting factor preventing the popularization of Zoysia japonica is the shorter green period (Teng et al., 2016). Consequently, getting a better comprehension of the chlorophyll degradation and photosynthetic mechanisms is important. The objectives of this study were to characterize ZjPPH in controlling chlorophyll degradation and to investigate its detailed regulatory mechanism in photosynthesis. It will contribute to breeding new Z. japonica cultivar with improved quality in the future.
Materials and Methods
Plant Materials and Growth Conditions
Zoysia japonica cultivar “Zenith” seeds, purchased from the Patten Seed Company (Lakeland, GA, United States), were cultivated in the TS1 nutrient medium (Klasmann-Deilmann, Germany). The seedlings were kept in an RXZ-380D-LED growth chamber (Ningbo Jiang Nan Instrument Factory, Ningbo, China), which is set at 28/25 oC (day/night), 65% humidity, and approximately 400 μmol m–2s–1photosynthetic active radiation (PAR). Arabidopsis thaliana “Columbia” (wild-type background) and pph mutant (SALK_000095) were kept in a growth chamber set at 24/22 oC (day/night) with 65% humidity and 400 μmol m–2s–1 PAR. The photoperiod of the growth chambers was set to 16 h per 24 h. Nicotiana benthamiana was cultivated under identical growth conditions as Z. japonica. The plants were irrigated by 1/2 strength Hoagland’s solution weekly (Hoagland and Arnon, 1950).
Cloning of ZjPPH and Its Promoter
Total RNA was extracted from “Zenith” leaves using Trizol and then it was used to generate cDNA using PrimeScript RT reagent kit (TaKaRa, Dalian, China). Genomic DNA was extracted using the CTAB methods from fresh leaves of Zenith. BLAST analysis was carried out using homologous proteins as queries in the Zoysia grass genome sequence to design primers used for cloning ZjPPH (Table 1). PCR amplification was performed to amplify the ZjPPH gene and its promoter sequence, respectively. The PCR products were then purified and inserted into the pMD-19T cloning vector (TaKaRa, Dalian, China) with ampicillin resistance. After antibiotic selection and PCR verification, the plasmids containing the complete coding sequence (CDS) (pMD-ZjPPH) and the promoter sequence (pMD-ZjPPHpro) from the base 0 were generated, respectively. The correct plasmids examined by Sanger sequencing and alignment were selected and used in the subsequent experiments.
Quantitative Real-Time PCR
Total RNA was isolated from different organs of Z. japonica, including roots, stems, and leaves, separately. Total RNA samples were also extracted from leaves at different senescent stages as described previously (Teng et al., 2016). Phytohormone treatments, including 10 μmol ABA, 10 μmol MeJA, and 0.5 mM SA, and dark induction were applied to investigate the expression characteristics following a protocol described by Teng et al. (2017). qRT-PCR data calculation was performed using the ΔΔCt method (Livak and Schmittgen, 2001). The Z. japonica β-actin gene (GenBank accession No. GU290546) was used as the housekeeping gene (Teng et al., 2017).
Bioinformatic Analysis of ZjPPH and Its Promoter Sequence
The NCBI BLAST analysis was carried out to search for the homologs. Phylogenetic tree was built with the neighbor-joining method using MEGA v.5 software (Tamura et al., 2011). The Ks/Ka ratio was generated using DnaSP6 software (Rozas et al., 2017). Based on the PlantCARE online database, the cis-elements in the promoter sequence were identified (Lescot et al., 2002). The molecular weight (MW) and the theoretical isoelectric point (PI) were calculated using the compute pI/Mw tool1. The subcellular localization pattern was predicted using the online program TargetP 12.
Plasmid Construction
The ZjPPH CDS was inserted into the plant binary vector 3302Y3 (Jia et al., 2016) to generate the 35S:ZjPPH:YFP construct used for subcellular localization investigation. The ZjPPH CDS was infused into the pTA7002 vector used for generating transgenic Arabidopsis lines. The promoter sequence of ZjPPH was inserted into the pCambia1391Z vector to generate the ZjPPHpro:GUS construct used for the GUS staining determination.
Subcellular Localization Observation
For the subcellular localization observation, Agrobacterium tumefaciens EHA105 harboring 35S:ZjPPH:YFP was transformed into the lower epidermal cells of N. benthamiana leaves using an injection syringe according to the transient expression protocol (Sparkes et al., 2006). After 48-h incubation, the fluorescence in the tobacco cells were checked and photographed using a SP8 laser confocal scanning microscope (Leica, Mannheim, Germany).
Generation of Transgenic Arabidopsis thaliana Lines and Physiological Determinations
The A. tumefaciens GV3101 containing the pTA7002-ZjPPH construct, and pTA7002 empty vector were used to generate transgenic and control A. thaliana lines using the floral dip method, respectively (Clough and Bent, 1998). The seeds harvested were screened on MS medium supplemented with 30 mg L–1 hygromycin. After PCR verification, T3 lines with 100% hygromycin resistance were selected for further morphological assessment. Three-week-old healthy lines growing in MS medium were transplanted to filter paper immersed in 30 μM dexamethasone (DEX) (Sigma-Aldrich, Munich, Germany) and 0.01% Tween-20 solution diluted with ddH2O. After 4 days of induction, the plants were photographed in a photostudio using an EOS 80D digital camera (Canon, Tokyo, Japan).
The chlorophyll content was extracted using 95% ethanol, and total chlorophyll contents were then quantified based on the absorbance recorded (Teng et al., 2016). ABA content was measured using an ELISA kit (H251, Jiancheng Bioengineering Institute, Nanjing, China) based on the protocol provided by the manufacturer. Soluble sugar content was determined using the 3,5-dinitrosalicylic acid method (Sun et al., 2020). Leaf fresh weight was adopted in assessing these indicators above.
A Handy PEA analyzer (Hansatech, Kings Lynn, United Kingdom) was used for the determination of chlorophyll fluorescence parameters. For dark incubation, fresh leaves were covered by a leaf clip at the middle of the leaf blades for 30 min. Then, the clips were removed and chlorophyll fluorescence was immediately measured by exposing the leaves to a two-second saturating light pulse of 3,500 μmol photons m–2 s–1. Photosynthetic parameters were recorded and calculated according to the protocol provided by the manufacture. Ten replicates were utilized for each of the CK, line-3, and line-7. The parameters were processed using Handy PEA v.1.3 software. Origin Pro v.2019b (OriginLab Corporation, Northampton, MA, United States) was used for data presentation.
Transmission Electron Microscopy Observation
Transmission electron microscopy observation was performed based on the protocol of Park et al. (2007) with moderate modifications. After DEX incubation, leaf tissues were soaked in the fixative buffer with 2% paraformaldehyde, 2% glutaraldehyde, and 50 mM sodium cacodylate (pH 7.2). Next, the samples were washed with 50 mM sodium cacodylate buffer (pH 7.2) three times at 4°C. Then, the samples were treated in the 50 mM sodium cacodylate buffer containing 1% osmium tetroxide (pH 7.2) at 4°C for 2 h. After being washed three times with distilled water at 25°C, the samples were incubated in 0.5% uranyl acetate for at least 30 min at 4°C. A gradient series of propylene oxide and ethanol was used for sample dehydration. Finally, the samples were embedded in resin for generating ultrathin sections using an ultra-microtome. The samples were then placed on copper grids after being polymerized at 70°C for 24 h. Finally, they were treated with 2% uranyl acetate for 5 min and with Reynolds’ lead citrate for 2 min at 25°C. The ultrastructure of chloroplasts was then observed and photographed using a HT7700 transmission electron microscope (Hitachi, Tokyo, Japan). We examined 10 cells in each sample and photographed the representative chloroplast structure in well-cut sections.
Statistical Analysis
Data were analyzed through Student’s t-test or one-way ANOVA using SPSS version 15.0 (IBM, Chicago, IL, United States). *p < 0.05 were recognized statistically significant. All data are presented as the mean ± SD (n = 3 at least).
Results
Cloning and Bioinformatic Analysis of ZjPPH
The ZjPPH (GenBank accession No.:MW882937) was successfully cloned by RT-PCR based on the genome sequence database of Z. japonica. The coding sequence of ZjPPH is 1479 bp in length, encoding 492 amino acids. Its theoretical pI and MW are 8.19 and 54.67 kD, respectively. A phylogenetic tree was constructed and indicated that ZjPPH was most closely related to the PPH orthologs in Zea mays and Sorghum bicolor (Figure 1A). Conserved domain analysis revealed that ZjPPH contained a conserved domain of α/β hydrolases and a PPH motif (PVYIVGNSLGG) containing a catalytic Ser residue (Figure 1B). The synonymous and non-synonymous substitution rates (Ka/Ks) were calculated to classify the evolutionary selection types. It showed that the Ka/Ks values of ZjPPH were less than one, suggesting that ZjPPH is under purifying selection (Figure 1C).
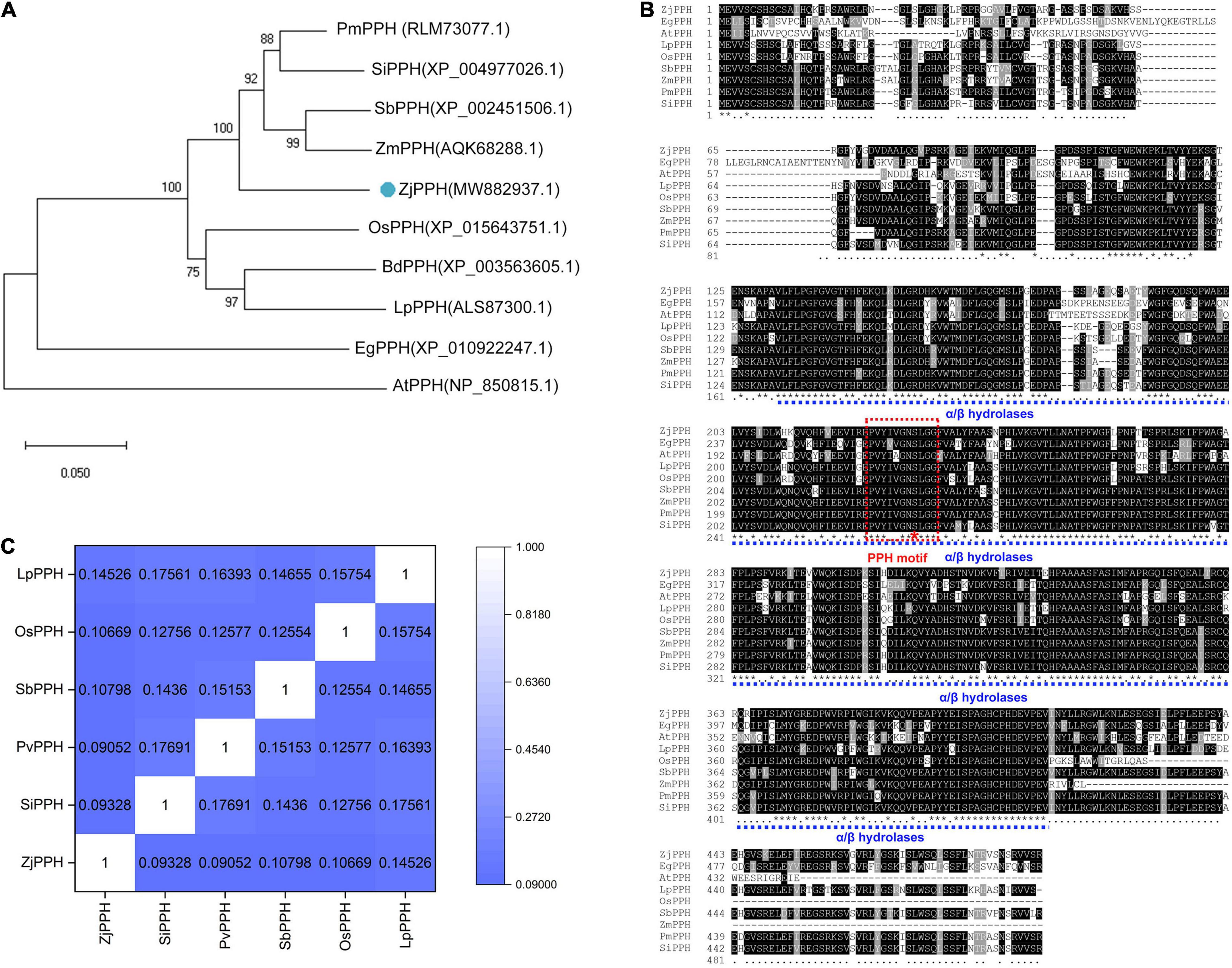
Figure 1. Bioinformatic analysis of ZjPPH. (A) Phylogenetic tree: orthologous proteins in Panicum miliaceum (PmPPH), Setaria italica (SiPPH), Sorghum bicolor (SbPPH), Zea mays (ZmPPH), Zoysia japonica (ZjPPH), Oryza sativa (OsPPH), Brachypodium distachyon (BdPPH), Lolium perenne (LpPPH), Elaeis guineensis (EgPPH), and Arabidopsis thaliana (AtPPH) were used for analysis. (B) Multiple sequences alignment and conserved domain prediction. (C) Ka/Ks values.
Isolation of Promoter and GUS Staining Assay
A 1500-bp nucleotide promoter sequence was obtained by PCR amplification. Cis-elements prediction results demonstrated that the ZjPPH promoter contained two ABRE elements involved in ABA responsiveness, one CGTCA motif and one TGACG motif involved in MeJA-responsiveness, two TCA elements participant in salicylic acid responsiveness, and one ERE elements correlated with ethylene-responsive element (Figure 2A).
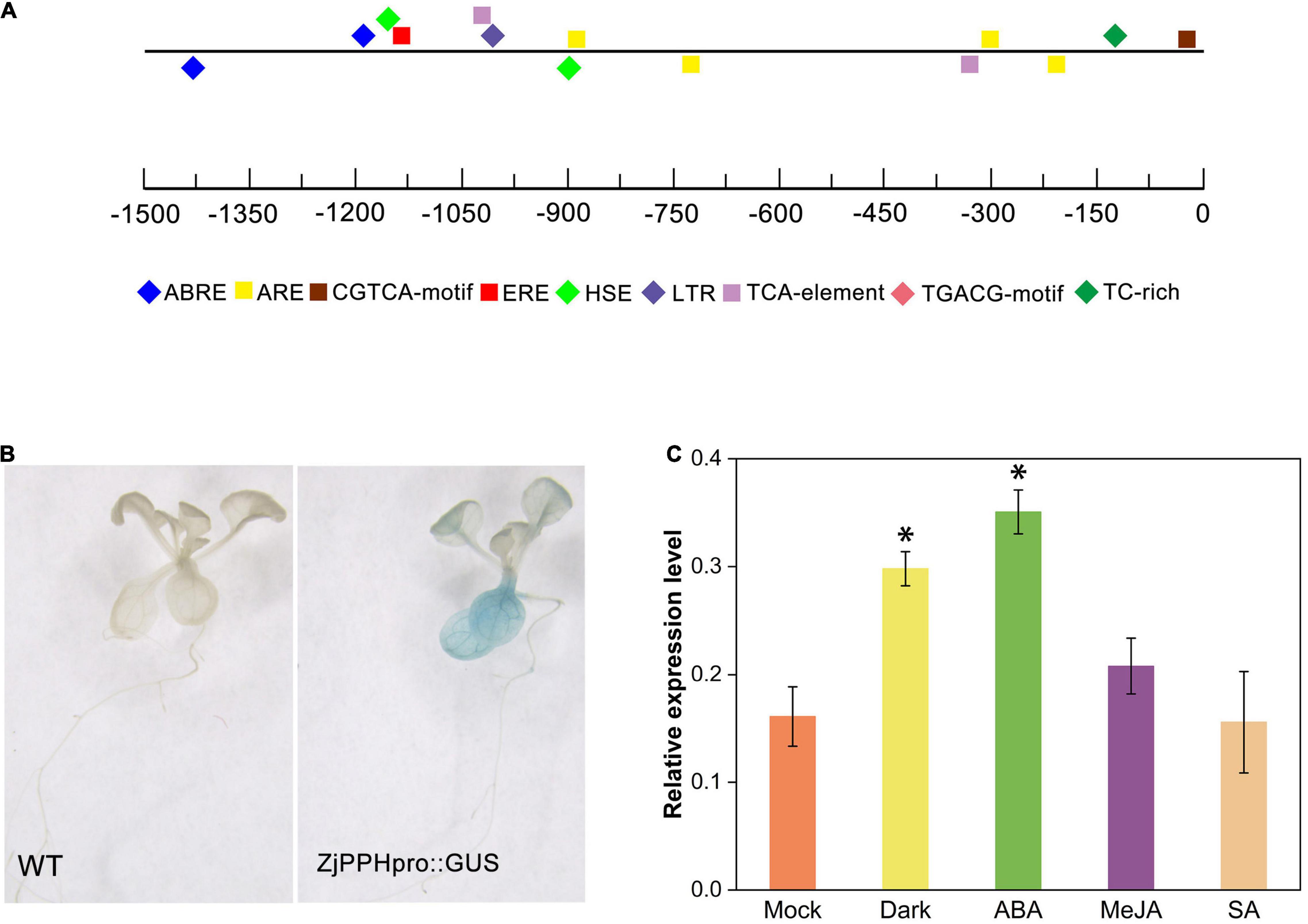
Figure 2. Promoter analysis and GUS assay. (A) cis-elements prediction; (B) GUS staining results. (C) GUS expression level under different treatments. *Indicate significant differences at p ≤ 0.05 based on Student’s t-test.
Using transgenic Arabidopsis seedlings, GUS histochemical staining was performed to further characterize the promoter. It displayed that the promoter sequence could drive GUS expression in transgenic plants. Specifically, blue pattern was abundantly found in the leaves and root (Figure 2B). The GUS expression level showed that the 48 h of dark and ABA treatment could induce the expression of GUS gene, whereas the MeJA and SA treatment caused no significant changes, indicating the role of ABA and dark treatment in regulating the promoter activity (Figure 2C).
Transcriptional Expression Characteristics of ZjPPH
The expression of ZjPPH exhibited a tissue-specific character with a higher level in the leaf than in the root and stem (Figure 3A). The expression level of ZjPPH was found more abundantly in senescent leaves than in mature and young leaves (Figure 3B). Hormone inducement analysis revealed that the transcriptional activity of ZjPPH could be activated by ABA, MeJA, SA, and dark treatment within 24 h of treatment (Figures 3C–F). Specifically, ABA and dark treatments upregulated the expression level of ZjPPH from 6 h and maintained a higher level at 24 h. MeJA treatment induced the expression of ZjPPH from 3 h and continuously kept the highest level from 6 h to the end of the experiment. SA increased the expression of ZjPPH from 1 h and reached the highest level at 6 h, and then it decreased gradually from 6 to 24 h.
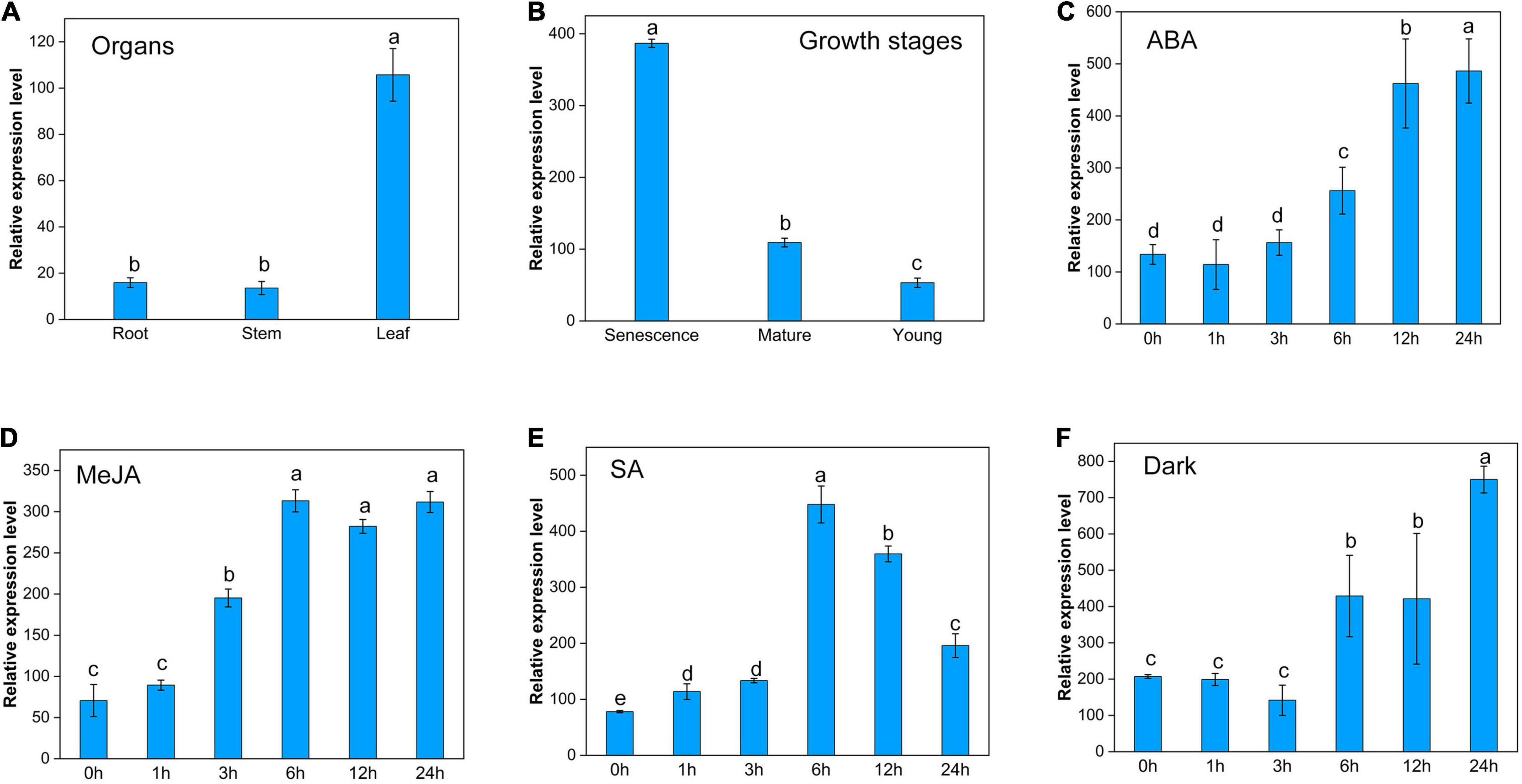
Figure 3. Transcriptional expression characters of ZjPPH. (A) Different organs. (B) Leaves at different growth stages. Continuous expression tendency under different treatments: ABA (C), MeJA (D), SA (E), and dark (F). Data were expressed as mean ± SD (n = 4). Different letters indicate significant differences at p ≤ 0.05 based on Fisher’s protected least significant difference (LSD) test.
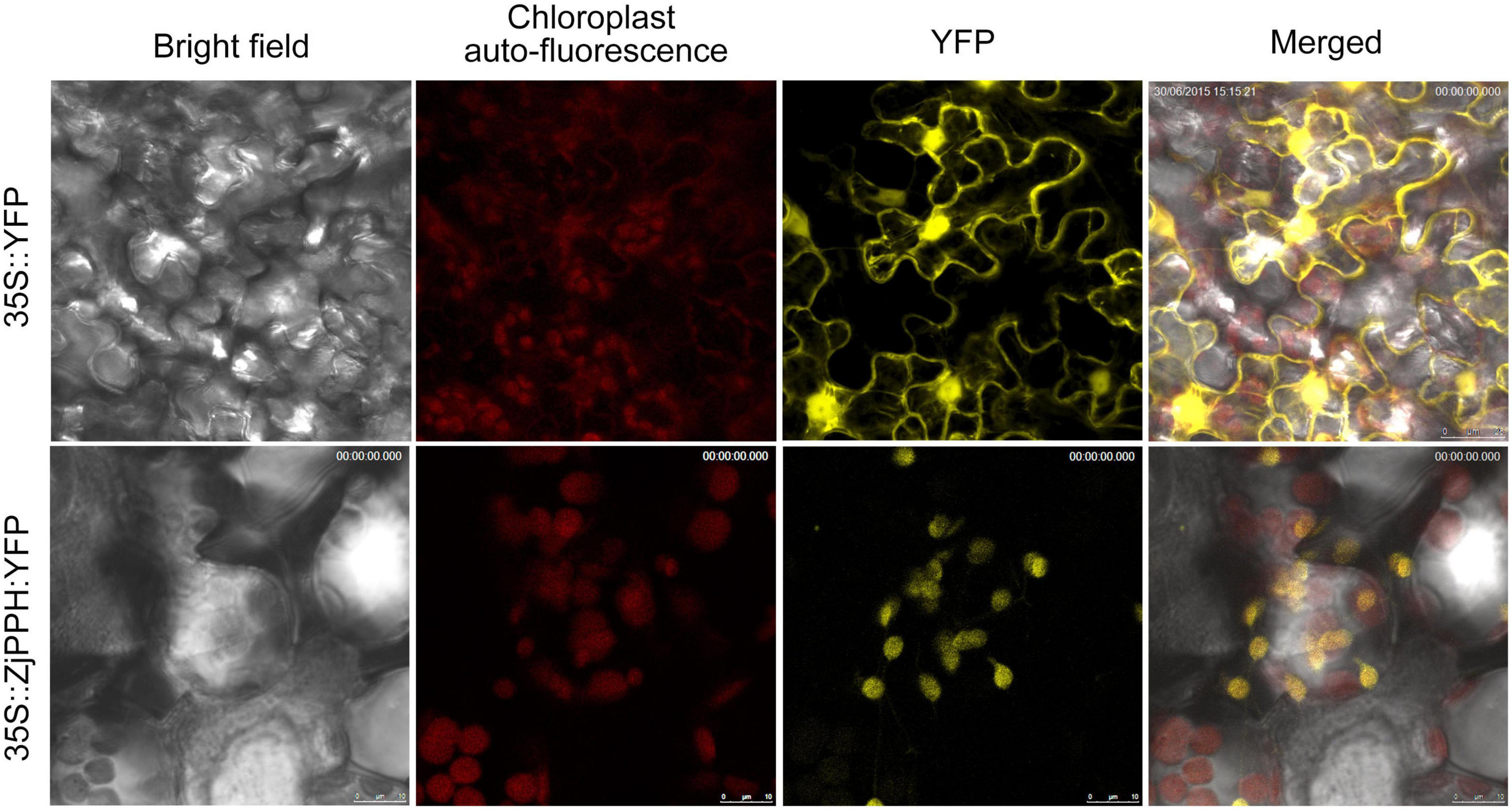
Subcellular Localization of ZjPPH
The TargetP 1.1 online program demonstrated that the ZjPPH was a chloroplast-specific localized protein. To further verify the subcellular localization pattern of ZjPPH, transient expression analysis was employed. As shown in Figure 4, the tobacco lower epidermis cells transformed with 35S:YFP were full of YFP signal. In contrast, the YFP signal was only detected in the chloroplast of the tobacco cells transformed with 35S:ZjPPH:YFP construct. The results proved that ZjPPH was a chloroplast-specific localized protein.
Overexpression of ZjPPH Accelerated Chlorophyll Degradation and Senescence
Transgenic Arabidopsis lines overexpressing ZjPPH were generated to explore the function of ZjPPH. The ZjPPH-overexpressing Arabidopsis lines showed an obvious yellowing phenotype compared with control (Figure 5A). The qRT-PCR analysis proved that the ZjPPH was efficiently expressed in the transgenic lines (Figure 5B). Correspondingly, chlorophyll content determination results showed that the chlorophyll contents was higher in the control than in the transgenics (Figure 5C). Moreover, ZjPPH also rescued the stay-green phenotype of the pph mutant (Figure 5A). ABA content determination showed that overexpression of ZjPPH increased the ABA content in transgenic lines compared with control (Figure 5D). The soluble sugar content was also found to be higher in the transgenic lines (Figure 5E).
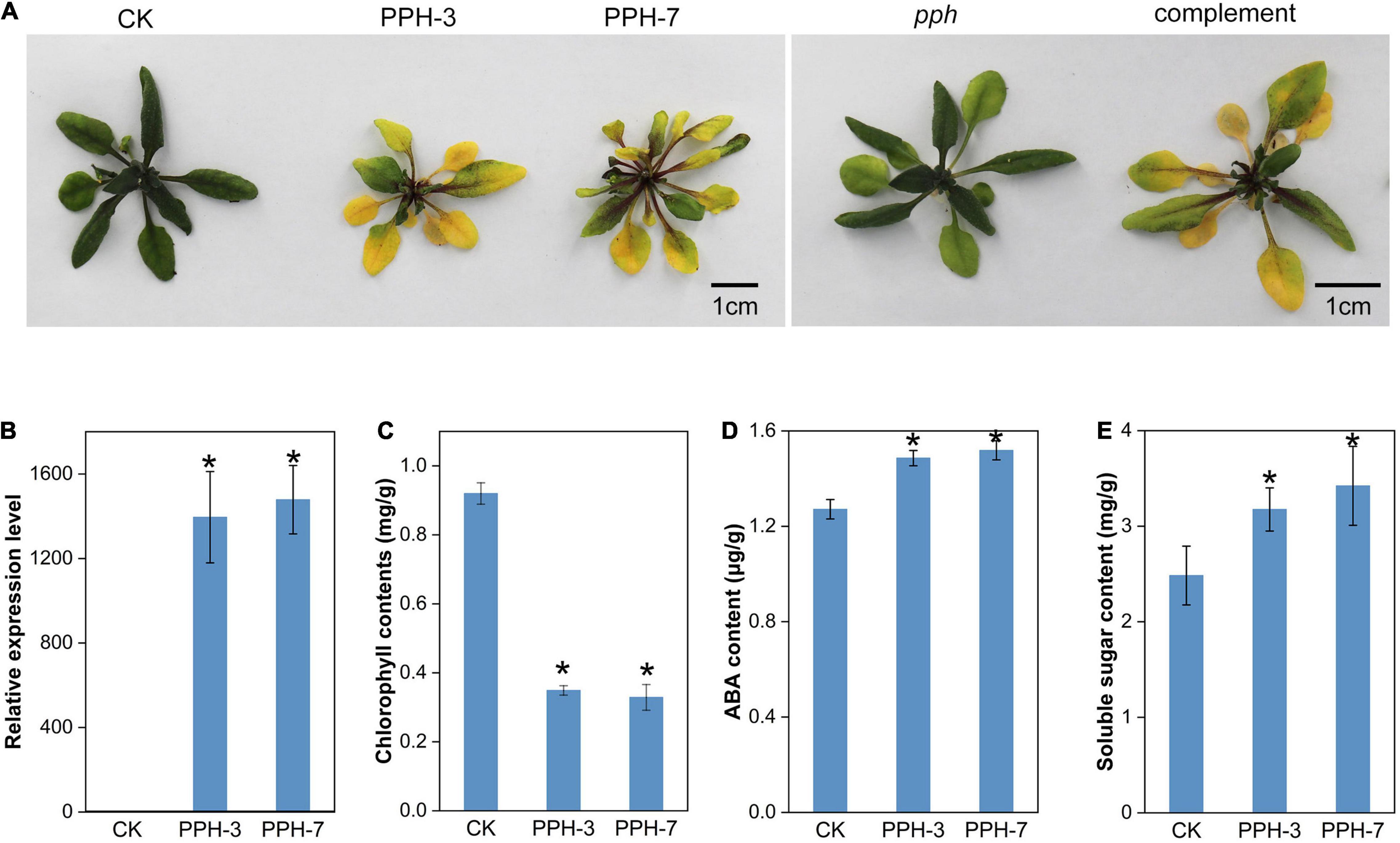
Figure 5. Phenotype observation and physiological determination of ZjPPH-overexpressing lines. (A) Picture of ZjPPH-overexpressing lines and complementary line. (B) Expression-level assessment of ZjPPH in transgenic lines. (C) Chlorophyll contents determination. (D) ABA content. (E) Soluble sugar content. Data were expressed as mean ± SD (n = 4). *Indicate significant differences at p ≤ 0.05 based on Student’s t-test.
Two senescence marker genes, SAG12 and SAG14, were selected to assess the impact of ZjPPH in senescence processes. It turned out that the expression levels of the two senescence marker genes were upregulated significantly (Figures 6A,B). It indicated that the overexpression of ZjPPH promoted senescence. In addition, the expression of four photosynthetic efficiency marker genes was monitored to evaluate the photosynthetic rate. The results revealed that the overexpression of ZjPPH significantly downregulated the expression of CAB1 (Chl a/b binding protein 1), PsaF (PSI component), RbcL (rubisco large subunit), and RCA (rubisco activase) (Figures 6C–F). It is rational to propose that ZjPPH could decrease the photosynthetic efficiency, which should be further evidenced by the determination of ultrastructure characteristics of chloroplasts and the detailed photosynthetic parameters.
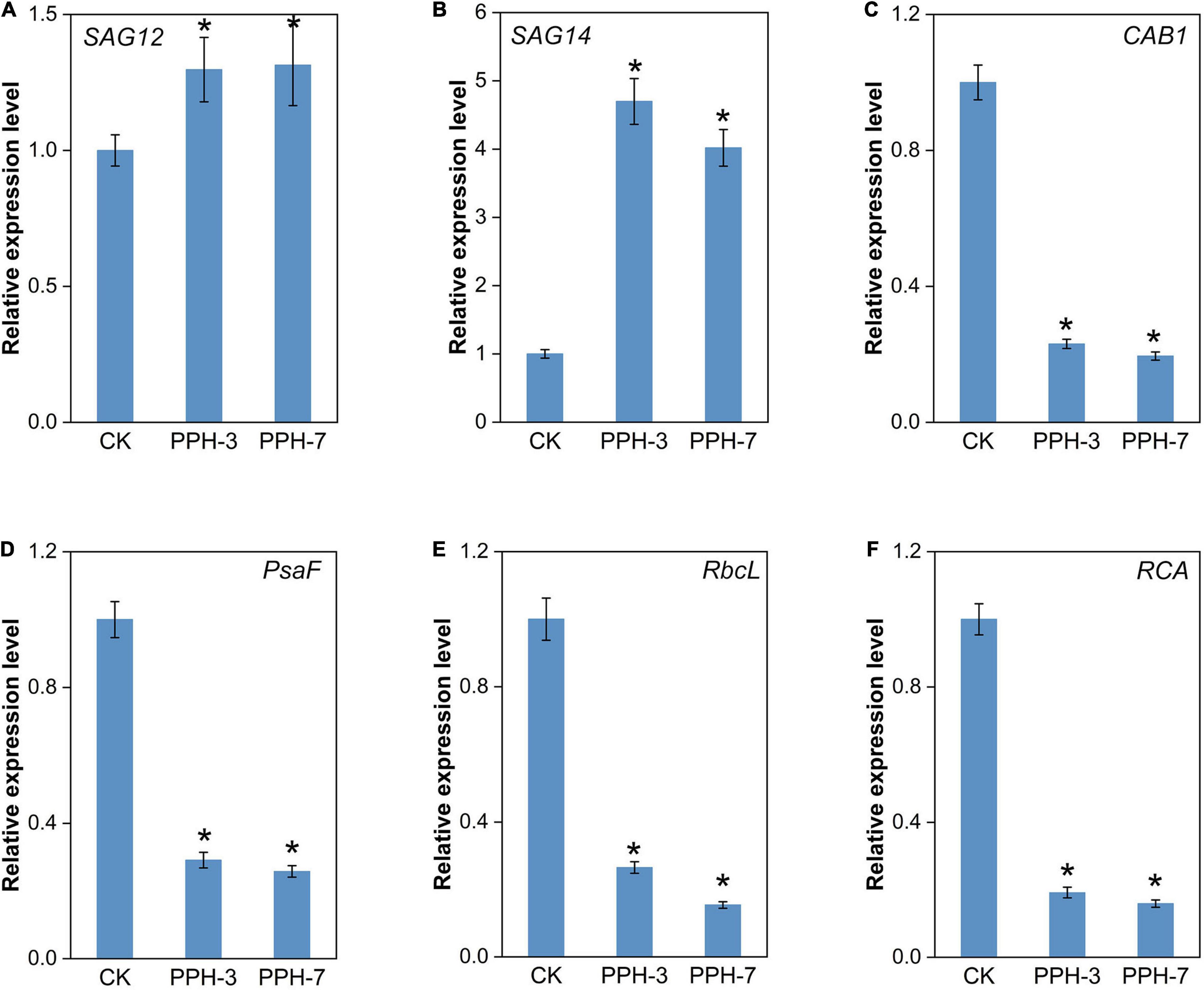
Figure 6. Expression analysis of senescence markers and photosynthesis-related genes. (A) SAG12. (B) SAG14. (C) CAB1. (D) PsaF. (E) RbcL. (F) RCA. Data were expressed as mean ± SD (n = 4). *Indicate significant differences at p ≤ 0.05 based on Student’s t-test.
Overexpression of ZjPPH Caused Ultrastructure Changes of Chloroplasts
Transmission electron microscopic analysis was carried out to reveal the ultrastructure changes of chloroplasts caused by the overexpression of ZjPPH. The results revealed that the overexpression of ZjPPH caused drastic changes to the ultrastructure of chloroplasts. In detail, the chloroplasts in the control maintained a high stability status with normal grana stacks, starch particles, and thylakoid membrane (Figure 7A). In contrast, the chloroplasts showed a decomposition tendency in the ZjPPH-overexpressing plants with fewer grana stacks and looser thylakoid membrane (Figure 7B).
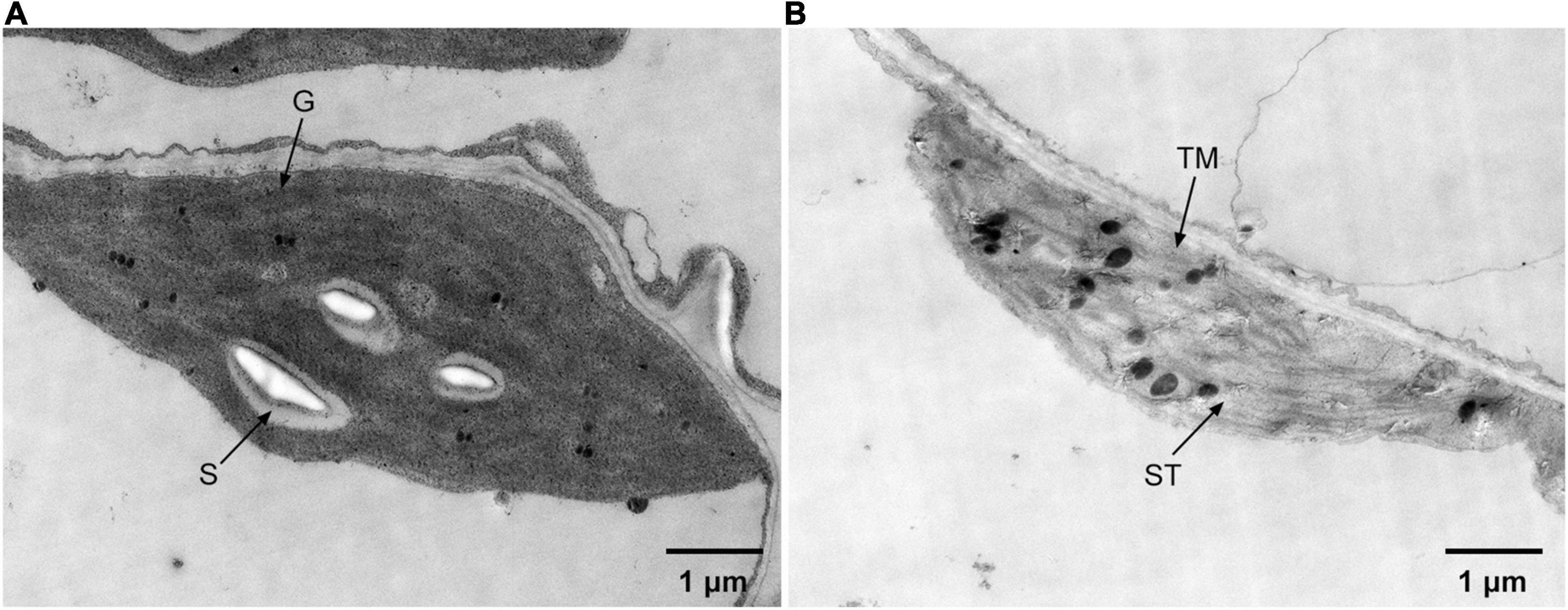
Figure 7. Ultrastructure of chloroplasts in panel (A). Control (B). ZjPPH-overexpressing plants. ST, stroma; S. starch; G. grana stacks; TM, thylakoid membrane.
The JIP Test Revealed That the Overexpression of ZjPPH Suppressed Photosynthetic Efficiency
The JIP test revealed that FO/FV and FV/FM were higher in the control than in the transgenic lines, indicating less of active photosynthesizing structures and photosynthetic membranes in the transgenic lines (Supplementary Table 1). Comparison of normalized OJIP curves provides detailed information of structural and functional differences of the photosystem (Hu et al., 2016). As shown in Figure 8A, the O-J step values were higher in the transgenic lines than in the control. It demonstrated that the expression of ZjPPH retarded the electron flow in the acceptor side of PSII.
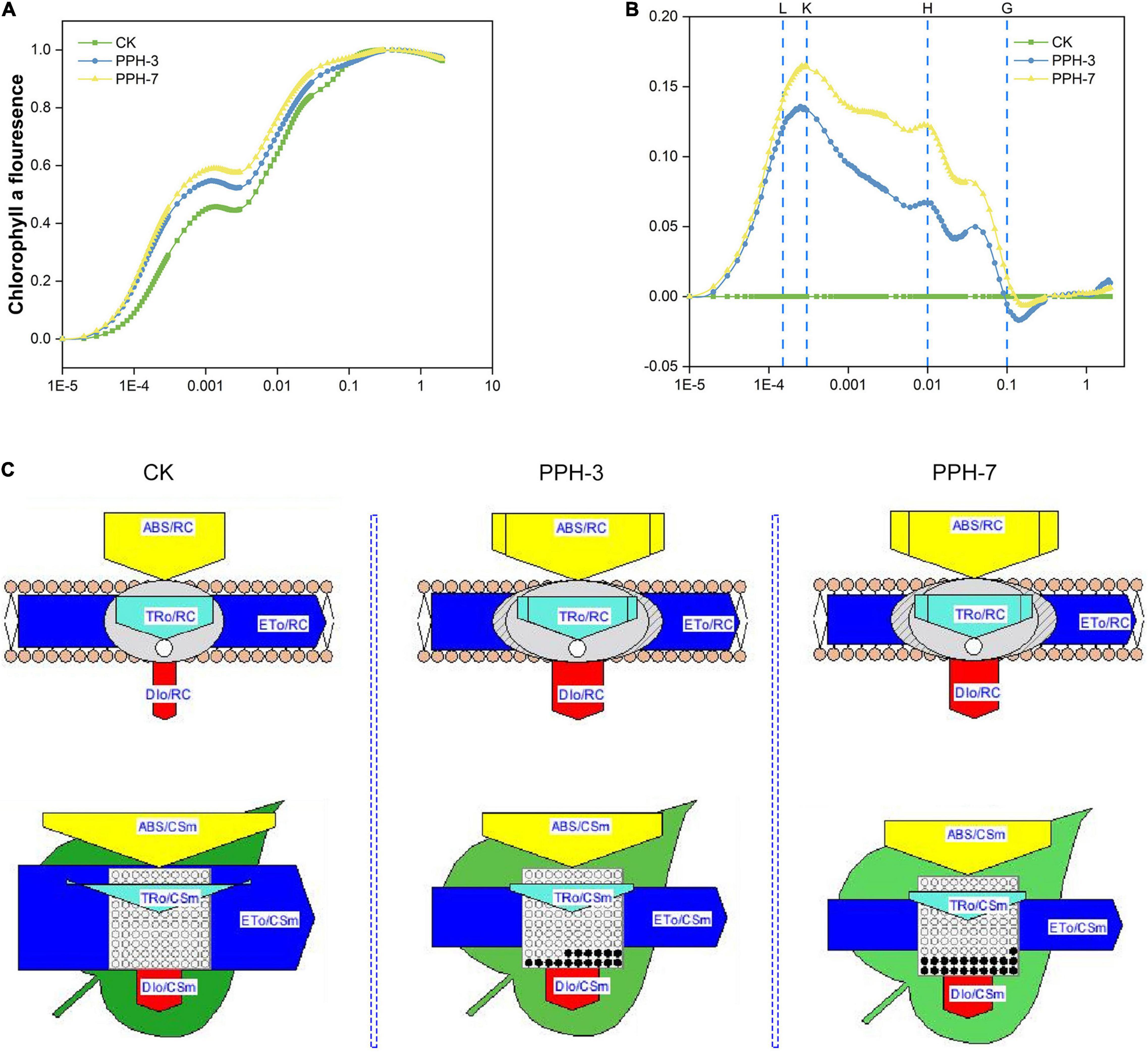
Figure 8. Prompt chlorophyll a fluorescence transient analysis of the ZjPPH-overexpressing lines. (A) The standard OJIP curves. (B) The relative variable fluorescence Vt of the transient chlorophyll a fluorescence. (C) The proportion of phenomenological energy flux parameters of the transgenic lines and the control displayed by the leaf pipeline model. The fluorescence parameters were displayed as means of ten replicates. The L, K, H, and G bands between the transgenic lines and control were significantly different at p ≤ 0.05 based on Student’s t-test. The width of the arrow represents the relative values of the associated parameters; dark circle represents non-active reaction centers.
The L-band is an indicator of the PSII status in the thylakoid grana membrane. A positive L-band was observed, suggesting the ungrouping of the antenna complexes and attenuated connectivity between LHCII and PSII reaction centers (Figure 8B and Supplementary Figure 1A). The positive K-band revealed the inactivation of OEC, resulting in the slower electron transport toward PSII reaction center (Supplementary Figure 1B). In this study, no obvious H-band was identified, indicating that the expression of ZjPPH caused no significant changes of the PQ pool volume and electron transduction between the two PS (Supplementary Figure 1C). The negative G-band implied that the PSI acceptors pool is bigger (Supplementary Figure 1D). Because the intersystem electron transduction did not show significant difference, it is rational to conclude that the PSI was suppressed in the transgenic lines. The lower RE0/CSm of the transgenic lines also supported this assumption (Supplementary Table 1).
Phenomenological energy pipeline models help to visualize the influence caused by the overexpression of ZjPPH. It showed that membrane model parameters, including ABS/RC, TR0/RC, and DI0/RC, were increased in the transgenic lines (Figure 8C). However, the ET0/RC was showed no significant changes. It indicated the decreased efficiency of PSII reaction center in electron transport (Janeeshma et al., 2021). Cross-section (CS)-level indicators, including ABS/CSm, TR0/CSm, and ET0/CSm, were lower in the transgenic lines, while the DI0/CSm increased (Figure 8C). In addition, less active reaction centers were identified in the ZjPPH-overexpressing lines. The decrease of ABS/CSm, TR0/CSm, and ET0/CSm was related to the increased inactive reaction centers and suppressed PSII efficiency (Janeeshma et al., 2021). The increase of the DI0/CSm reflected the inefficiency in the utilization of light energy.
Discussion
Interpreting the chlorophyll degradation and photosynthetic mechanism is of vital importance for prolonging the green period of Z. japonica. In this study, the PPH gene was successfully isolated, and bioinformatic analysis proved that it is a member of α/β hydrolases. The conserved PPH motif with a Ser residue confirmed its role as a serine-type hydrolase to PPH belongs (Guyer et al., 2018). The indicator of gene function prediction, Ka/Ks ratio (Gaut et al., 1996), demonstrated that ZjPPH is functionally conserved. Transient expression analysis demonstrated that ZjPPH was a chloroplast-specific localized protein, and it is in consistent with the homologous proteins in Arabidopsis (Schelbert et al., 2009), bamboo (Wei et al., 2013), and perennial ryegrass (Zhang et al., 2016).
The promoter activity characterization and gene transcriptional expression analysis parallelly revealed that ZjPPH was expressed in both leaves and roots and could be induced by ABA and dark treatments. The expression of rice PPH was detected in the root, supporting our assumption that ZjPPH might be involved in other activities besides photosynthesis (Morita et al., 2009). LpPPH was reported to be upregulated by exogenous ABA, and the expression of bamboo PPH increased after darkness. As revealed by previous works, the expression of PPH increased to a rather high level during senescence (Schelbert et al., 2009; Zhang et al., 2016). In this study, ZjPPH expression level is positively associated with senescence, suggesting its role in senescence process. MeJA and SA were important hormones actively involved in senescence (Schippers et al., 2015). Different from the promoter activity analysis, the qRT-PCR results revealed that ZjPPH could also be upregulated by MeJA and SA, indicating the role of ZjPPH in MeJA and SA-induced senescence process. The different sampling time might be responsible for this result. In general, promoter activity characterization with the GUS reporting system usually needs longer incubating time than gene expression-level test. PPH is involved in chlorophyll degradation during leaf senescence in Arabidopsis (Schelbert et al., 2009). The heterologous expression of bamboo PPH in Arabidopsis caused fast yellowing phenotype (Wei et al., 2013). Transient expression of LpPPH caused chlorophyll breakdown in N. benthamiana (Zhang et al., 2016). In agreement with the previous reports, the functionality of ZjPPH was confirmed in accelerating chlorophyll degradation in this study. The ABA and soluble sugar contents determination revealed the expression of ZjPPH led to the accumulation of ABA and soluble sugar. ABA and sugar accumulation play positive roles in promoting senescence (Wingler et al., 2005; Schippers et al., 2015). SAG12 and SAG14 were senescence markers (Kim et al., 2008; Schelbert et al., 2009). The transcript level of SAG12 and SAG14 was increased, indicating that the senescence processes were activated in the transgenic lines. It was in accordance with the findings in perennial ryegrass (Zhang et al., 2016). The transcript abundance of the photosynthetic markers, such as CAB1, PsaF, RbcL, and RCA, was decreased, leaving a hint of photosynthesis retardation in the transgenic lines (Sun et al., 2016). The transcript abundance of the photosynthetic marker was decreased, leaving a hint of photosynthesis retardation in the transgenic lines.
Arabidopsis pph-1 mutant showed nearly unchanged thylakoid stacks and unreduced membrane density compared with wild type after dark-induced senescence (Schelbert et al., 2009). The same is true of rice pph mutant in which the decomposition of the chloroplast ultrastructure was retarded during dark incubation (Morita et al., 2009). Our TEM observation revealed that the overexpression of ZjPPH promoted the decomposition of photosynthesis system, proving the important roles of ZjPPH in adjusting photosynthetic systems of chloroplast. JIP test results supported the TEM observation that the antenna complexes were ungrouped and the connectivity between LHCII and PSII reaction centers was attenuated. It has been reported that PPH accelerates PSI reaction center and dimeric PSII protein degradation (Morita et al., 2009; Ren et al., 2010). Similar results have been obtained for the JIP test determination in this study that ZjPPH suppressed the efficiency both PSII and PSI. PPH was responsible for catalyzing pheophytin a, the primary electron acceptor in PSII. Surprisingly, the intersystem electron transport chain between PSII and PSI showed no significant differences in the transgenic lines. The decreased efficiency of energy transformation of PSII reaction center as well as the inefficiency of light energy utilization was responsible for the insensitivity of the intersystem electron transduction in the transgenic lines. It seems reasonable to speculate that ZjPPH suppressed photosynthesis efficiency by mainly retarding PSII and PSI, without significantly altering the intersystem electron transduction.
Conclusion
In summary, the proposed working model of the molecular regulating machinery of ZjPPH in regulating senescence and photosynthesis is summarized in Figure 9. The expression of ZjPPH could be induced by ABA, dark, and senescence. ZjPPH was localized in the chloroplasts and its overexpression accelerated chlorophyll degradation. ZjPPH accelerated senescence with the accumulation of ABA and soluble sugar contents, as well as the increased transcriptional level of SAG12 and SAG14. JIP test analysis revealed that ZjPPH suppressed photosynthesis efficiency by mainly suppressing both PSII and PSI, and photosynthesis-related genes. Consequently, ZjPPH plays important roles in chlorophyll degradation and photosynthesis. It could be a valuable gene for genetic editing to cultivate new cultivars with stay-green trait and improved photosynthetic efficiency.
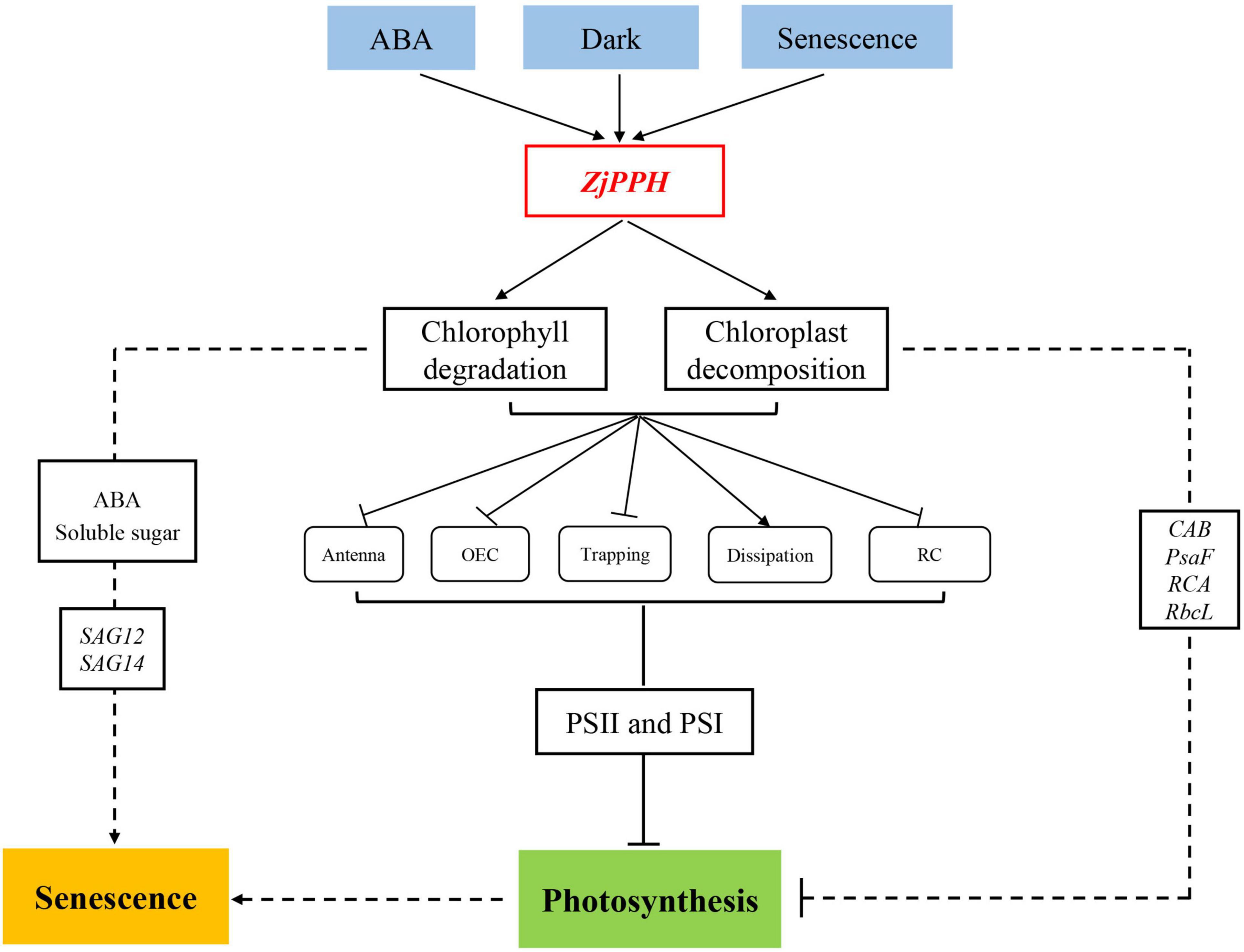
Figure 9. Proposed model of the molecular regulating machinery of ZjPPH in senescence and photosynthesis.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
XF and JW designed the study. KT, CH, and YY performed the experiments. KT, HZ, HL, and LX wrote the manuscript. All authors have read and approved the manuscript.
Funding
This study was supported by the Beijing Natural Science Foundation (No. 6204039), National Natural Science Foundation of China (No. 31901397), Scientific Funds of Beijing Academy of Agriculture and Forestry Sciences (KJCX20210431), and National Key Research and Development Program of China (2019YFF03032043).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank Kai Wang from the Hansha Scientific Instruments Ltd., for the Handy PEA parameter analysis. We would also like to thank Duowei Li from the Northwest University for providing the pTA7002 vector.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.786570/full#supplementary-material
Supplementary Figure 1 | Different normalizations of the fluorescence rise kinetics OJIP curves. (A) L-band. (B) K-band. (C) H-band. (D) G-band.
Footnotes
References
Chen, Y., Li, L. L., Zong, J. Q., Chen, J. B., Guo, H. L., Guo, A. G., et al. (2015). Heterologous expression of the halophyte Zoysia matrella H+-pyrophosphatase gene improved salt tolerance in Arabidopsis thaliana. Plant Physiol. Biochem. 91, 49–55. doi: 10.1016/j.plaphy.2015.04.004
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Gaut, B. S., Morton, B. R., McCaig, B. C., and Clegg, M. T. (1996). Substitution rate comparisons between grasses and palms: synonymous rate differences at the nuclear gene Adh parallel rate differences at the plastid gene rbcL. Proc. Natl. Acad. Sci. U. S. A. 93, 10274–10279. doi: 10.1073/pnas.93.19.10274
Guo, Y., Ren, G., Zhang, K., Li, Z., and Guo, H. (2021). Leaf senescence: progression, regulation, and application. Mol. Hortic. 1:5.
Guyer, L., Hofstetter, S. S., Christ, B., Lira, B. S., Rossi, M., and Hortensteiner, S. (2014). Different mechanisms are responsible for chlorophyll dephytylation during fruit ripening and leaf senescence in tomato. Plant Physiol. 166, 44–56. doi: 10.1104/pp.114.239541
Guyer, L., Salinger, K., Krugel, U., and Hortensteiner, S. (2018). Catalytic and structural properties of pheophytinase, the phytol esterase involved in chlorophyll breakdown. J. Exp. Bot. 69, 879–889. doi: 10.1093/jxb/erx326
Han, Z., Hu, Y., Lv, Y., Rose, J. K. C., Sun, Y., Shen, F., et al. (2018). Natural variation underlies differences in ETHYLENE RESPONSE FACTOR17 activity in fruit peel degreening. Plant Physiol. 176, 2292–2304. doi: 10.1104/pp.17.01320
Hoagland, D. R., and Arnon, D. I. (1950). The water-culture method for growing plants without soil. Calif. Agric. Exp. Stn. Circ. 347, 357–359.
Hortensteiner, S. (2013). Update on the biochemistry of chlorophyll breakdown. Plant Mol. Biol. 82, 505–517. doi: 10.1007/s11103-012-9940-z
Hu, Z., Fan, J., Xie, Y., Amombo, E., Liu, A., Gitau, M. M., et al. (2016). Comparative photosynthetic and metabolic analyses reveal mechanism of improved cold stress tolerance in bermudagrass by exogenous melatonin. Plant Physiol. Biochem. 100, 94–104. doi: 10.1016/j.plaphy.2016.01.008
Janeeshma, E., Kalaji, H. M., and Puthur, J. T. (2021). Differential responses in the photosynthetic efficiency of Oryza sativa and Zea mays on exposure to Cd and Zn toxicity. Acta Physiol. Plant. 43:12.
Jia, N., Liu, X. M., and Gao, H. B. (2016). A DNA2 homolog is required for DNA damage repair, cell cycle regulation, and meristem maintenance in plants. Plant Physiol. 171, 318–333. doi: 10.1104/pp.16.00312
Kalaji, H. M., Jajoo, A., Oukarroum, A., Brestic, M., Zivcak, M., Samborska, I. A., et al. (2016). Chlorophyll a fluorescence as a tool to monitor physiological status of plants under abiotic stress conditions. Acta Physiol. Plant. 38:102.
Kalaji, H. M., Rackova, L., Paganova, V., Swoczyna, T., Rusinowski, S., and Sitko, K. (2018). Can chlorophyll-a fluorescence parameters be used as bio-indicators to distinguish between drought and salinity stress in Tilia cordata Mill?. Environ. Exp. Bot. 152, 149–157. doi: 10.1016/j.envexpbot.2017.11.001
Kim, C. Y., Bove, J., and Assmann, S. M. (2008). Overexpression of wound-responsive RNA-binding proteins induces leaf senescence and hypersensitive-like cell death. New Phytol. 180, 57–70. doi: 10.1111/j.1469-8137.2008.02557.x
Kuai, B., Chen, J. Y., and Hortensteiner, S. (2018). The biochemistry and molecular biology of chlorophyll breakdown. J. Exp. Bot. 69, 751–767. doi: 10.1093/jxb/erx322
Lee, M. S., Mitchell, R., Heaton, E., Zumpf, C., and Lee, D. K. (2019). Warm-season grass monocultures and mixtures for sustainable bioenergy feedstock production in the Midwest, USA. Bioenerg. Res. 12, 43–54.
Lescot, M., Dehais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–Δ Δ CT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Morita, R., Sato, Y., Masuda, Y., Nishimura, M., and Kusaba, M. (2009). Defect in non-yellow coloring 3, an α/β hydrolase-fold family protein, causes a stay-green phenotype during leaf senescence in rice. Plant J. 59, 940–952. doi: 10.1111/j.1365-313X.2009.03919.x
Park, S. Y., Yu, J. W., Park, J. S., Li, J., Yoo, S. C., Lee, N. Y., et al. (2007). The senescence-induced staygreen protein regulates chlorophyll degradation. Plant Cell 19, 1649–1664. doi: 10.1105/tpc.106.044891
Ren, G. D., Zhou, Q., Wu, S. X., Zhang, Y. F., Zhang, L. G., Huang, J. R., et al. (2010). Reverse genetic identification of CRN1 and its distinctive role in chlorophyll degradation in Arabidopsis. J. Integr. Plant Biol. 52, 496–504. doi: 10.1111/j.1744-7909.2010.00945.x
Rossi, F. R., Krapp, A. R., Bisaro, F., Maiale, S. J., Pieckenstain, F. L., and Carrillo, N. (2017). Reactive oxygen species generated in chloroplasts contribute to tobacco leaf infection by the necrotrophic fungus Botrytis cinerea. Plant J. 92, 761–773. doi: 10.1111/tpj.13718
Rozas, J., Ferrer-Mata, A., Sanchez-DelBarrio, J. C., Guirao-Rico, S., Librado, P., Ramos-Onsins, S. E., et al. (2017). DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 34, 3299–3302. doi: 10.1093/molbev/msx248
Schelbert, S., Aubry, S., Burla, B., Agne, B., Kessler, F., Krupinska, K., et al. (2009). Pheophytin pheophorbide hydrolase (Pheophytinase) is involved in chlorophyll breakdown during leaf senescence in Arabidopsis. Plant Cell 21, 767–785. doi: 10.1105/tpc.108.064089
Schippers, J. H. M., Schmidt, R., Wagstaff, C., and Jing, H. C. (2015). Living to die and dying to live: the survival strategy behind leaf senescence. Plant Physiol. 169, 914–930. doi: 10.1104/pp.15.00498
Shimoda, Y., Ito, H., and Tanaka, A. (2016). Arabidopsis STAY-GREEN, mendel’s green cotyledon gene, encodes magnesium-dechelatase. Plant Cell 28, 2147–2160. doi: 10.1105/tpc.16.00428
Sparkes, I. A., Runions, J., Kearns, A., and Hawes, C. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1, 2019–2025. doi: 10.1038/nprot.2006.286
Sun, T. X., Shao, K., Huang, Y., Lei, Y. Y., Tan, L. Y., and Chan, Z. L. (2020). Natural variation analysis of perennial ryegrass in response to abiotic stress highlights LpHSFC1b as a positive regulator of heat stress. Environ. Exp. Bot. 179: 104192.
Sun, X. B., Sun, C. Y., Li, Z. G., Hu, Q., Han, L. B., and Luo, H. (2016). AsHSP17, a creeping bentgrass small heat shock protein modulates plant photosynthesis and ABA-dependent and independent signalling to attenuate plant response to abiotic stress. Plant Cell Environ. 39, 1320–1337. doi: 10.1111/pce.12683
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739.
Teng, K., Chang, Z. H., Li, X., Sun, X. B., Liang, X. H., Xu, L. X., et al. (2016). Functional and RNA-sequencing analysis revealed expression of a novel Stay-Green gene from Zoysia japonica (ZjSGR) caused chlorophyll degradation and accelerated senescence in Arabidopsis. Front. Plant Sci. 7:1894. doi: 10.3389/fpls.2016.01894
Teng, K., Tan, P. H., Xiao, G. Z., Han, L. B., Chang, Z. H., and Chao, Y. H. (2017). Heterologous expression of a novel Zoysia japonica salt-induced glycine-rich RNA-binding protein gene, ZjGRP, caused salt sensitivity in Arabidopsis. Plant Cell Rep. 36, 179–191. doi: 10.1007/s00299-016-2068-x
Wei, Q., Cao, H. M., Li, Z. R., Kuai, B. K., and Ding, Y. L. (2013). Identification of an AtCRN1-like chloroplast protein BeCRN1 and its distinctive role in chlorophyll breakdown during leaf senescence in bamboo (Bambusa emeiensis ‘Viridiflavus’). Plant Cell Tiss. Org. 114, 1–10. doi: 10.1007/s11240-013-0298-y
Wingler, A., Purdy, S., MacLean, J. A., and Pourtau, N. (2005). The role of sugars in integrating environmental signals during the regulation of leaf senescence. J. Exp. Bot. 57, 391–399. doi: 10.1093/jxb/eri279
Xuan, J., Song, Y., Zhang, H., Liu, J., Guo, Z., and Hua, Y. (2013). Comparative proteomic analysis of the stolon cold stress response between the C4 Perennial grass species Zoysia japonica and Zoysia metrella. PLoS One 8:e75705. doi: 10.1371/journal.pone.0075705
Yu, G. H., Xie, Z. I., Zhang, J., Lei, S. S., Lin, W. J., Xu, B., et al. (2021). NOL-mediated functional stay-green traits in perennial ryegrass (Lolium perenne L.) involving multifaceted molecular factors and metabolic pathways regulating leaf senescence. Plant J. 106, 1219–1232. doi: 10.1111/tpj.15204
Yusuf, M. A., Kumar, D., Rajwanshi, R., Strasser, R. J., Tsimilli-Michael, M., and Govindjee, et al. (2010). Overexpression of γ-tocopherol methyl transferase gene in transgenic Brassica juncea plants alleviates abiotic stress: physiological and chlorophyll a fluorescence measurements. Biochim. Biophys. Acta Bioenerg. 1797, 1428–1438. doi: 10.1016/j.bbabio.2010.02.002
Zagorchev, L., Atanasova, A., Albanova, I., Traianova, A., Mladenov, P., Kouzmanova, M., et al. (2021). Functional characterization of the photosynthetic machinery in Smicronix galls on the parasitic plant cuscuta campestris by JIP-Test. Cells 10:1399. doi: 10.3390/cells10061399
Zhang, J., Yu, G. H., Wen, W. W., Ma, X. Q., Xu, B., and Huang, B. R. (2016). Functional characterization and hormonal regulation of the PHEOPHYTINASE gene LpPPH controlling leaf senescence in perennial ryegrass. J. Exp. Bot. 67, 935–945. doi: 10.1093/jxb/erv509
Keywords: pheophytinase, chlorophyll degradation, photosynthesis, chlorophyll a fluorescence, senescence, Zoysia japonica
Citation: Teng K, Yue Y, Zhang H, Li H, Xu L, Han C, Fan X and Wu J (2021) Functional Characterization of the Pheophytinase Gene, ZjPPH, From Zoysia japonica in Regulating Chlorophyll Degradation and Photosynthesis. Front. Plant Sci. 12:786570. doi: 10.3389/fpls.2021.786570
Received: 19 October 2021; Accepted: 16 November 2021;
Published: 23 December 2021.
Edited by:
Magda Pál, ELKH Agricultural Institute, Centre for Agricultural Research, HungaryReviewed by:
Zhulong Chan, Huazhong Agricultural University, ChinaJing Zhang, Nanjing Agricultural University, China
Copyright © 2021 Teng, Yue, Zhang, Li, Xu, Han, Fan and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xifeng Fan, ZmFueGlmZW5nY2F1QDE2My5jb20=; Juying Wu, d3VqdXlpbmdAZ3Jhc3MtZW52LmNvbQ==
 Ke Teng
Ke Teng Yuesen Yue
Yuesen Yue Hui Zhang1
Hui Zhang1 Hui Li
Hui Li Lixin Xu
Lixin Xu