
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 30 November 2021
Sec. Technical Advances in Plant Science
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.786328
This article is part of the Research Topic Plant Transformation View all 20 articles
Transgenic technology is increasingly used in forest-tree breeding to overcome the disadvantages of traditional breeding methods, such as a long breeding cycle, complex cultivation environment, and complicated procedures. By introducing exogenous DNA, genes tightly related or contributed to ideal traits—including insect, disease, and herbicide resistance—were transferred into diverse forest trees, and genetically modified (GM) trees including poplars were cultivated. It is beneficial to develop new varieties of GM trees of high quality and promote the genetic improvement of forests. However, the low transformation efficiency has hampered the cultivation of GM trees and the identification of the molecular genetic mechanism in forest trees compared to annual herbaceous plants such as Oryza sativa. In this study, we reviewed advances in transgenic technology of forest trees, including the principles, advantages and disadvantages of diverse genetic transformation methods, and their application for trait improvement. The review provides insight into the establishment and improvement of genetic transformation systems for forest tree species. Challenges and perspectives pertaining to the genetic transformation of forest trees are also discussed.
As important plant materials, forest trees are crucial for ecological preservation, climate regulation, building materials, road greening, and energy supply (Trumbore et al., 2015). However, breeding forest trees take a long time, and advanced-generation breeding populations are needed due to their long life span. Molecular breeding methods based on genetic transformation facilitate breeding and overcome the disadvantages of directional improvement, breeding, and the difficulty of distant hybridization and inter-specific hybridization that characterizes traditional breeding (Fang and Han, 2019). Furthermore, two or more traits can be simultaneously improved in genetically modified (GM) trees, enabling the improvement of the adaptability and productivity of forest trees (Martínez-Gómez, 2019). Genetic transformation is complicated by the complexity and diversity of forest genomes. Compared to annual crops, transgenic research on forestry trees started late and efficient transformation systems for many tree species have not been successfully established. Thus, it is important and indispensable to understand the parameters of genetic transformation of forest trees and elucidate challenges in transgenic technology, accelerating the application of GM trees for sustainable development.
To date, transgenic investigation of trees has focused on the transformation efficiency using marker genes, the regeneration of the transgenic plant tissues into complete plants, the introduction of introducing exogenous genes into receptor genomes, and gene function and regulation mechanism (Yu et al., 2021). Genetic engineering has been successfully applied to forest trees including Populus, used as a model plant for gene function research based on the establishment of a complete genetic transformation system (Zhou Y. et al., 2020; Guo et al., 2021). To generate GM forest trees, the most widely used transgenic method involves Agrobacterium tumefaciens-mediated. Gene gun-mediated, pollen tube pathway, and protoplast transformation methods can also be used. Mobile genetic techniques, such as clustered regularly interspaced short palindromic repeats (CRISPR)-associated (CRISPR-Cas) systems, have recently been applied to forest tree breeding (Shivram et al., 2021). Transgenic technology has been used to modify insect, herbicide, abiotic stress, disease resistance, wood properties, flowering regulation, and phytoremediation (Liao and An, 2013; Ding et al., 2016; Xu and Zhai, 2021; Yu et al., 2021).
In this study, we reviewed the advantages and disadvantages of diverse genetic transformation methods and their application to forest trees for enhancing insect, disease, and abiotic-stress resistance. Also, the challenges in transgenic engineering of forest trees are presented, and potential future work is discussed. This review provides insight into the establishment and improvement of genetic transformation systems for forest tree species.
Agrobacterium tumefaciens-mediated transformation is the most commonly used method of genetic transformation of forest trees. A. tumefaciens could deliver DNA molecules into plant cells for integration of exogenous genes into the host genome (Chilton et al., 1977). A. tumefaciens-mediated genetic transformation of plants is a rare example of naturally occurring trans-kingdom DNA transfer (Lacroix and Citovsky, 2013). A. tumefaciens-mediated genetic transformation system has been widely applied in poplars, such as Populus tomentosa, Populus alba × Populus glandulosa, Populus simonii × Populus nigra, and Cinnamomum camphora (Table 1) (Du et al., 2008; Li et al., 2020a,b; Ma et al., 2020; Guo et al., 2021). Furthermore, it has also been successfully established in some other forest trees, such as Betula platyphylla, Eucalyptus urophylla, and Juglans (Wang, 2015; Li et al., 2021a; Zhong et al., 2021).
Due to its simplicity and high repetition rate, leaf disk transformation is the most widely used method for plant transforming using A. tumefaciens. The transformation receptor, infection and coculture time, of genetic transformation might differ among forest tree species (Table 2). However, as explants, young leaves (top 3–5 leaves of tissue culture plantlets) are amenable to genetic transformation, and the infection and coculture times depend on the secondary metabolites produced by plants. Additionally, bacterial density in the logarithmic growth phase (optical density at 600 nm = 0.6–0.8) is suitable for the genetic transformation of most forest trees. Moreover, callus induction from plant organs and infection with A. tumefaciens can be conducted to generate transgenic plants. The stability of callus germination can be tested by using beta-glucuronidase (GUS)-labeled vectors to infect calli of Hevea brasiliensis (Lardet et al., 2011). A large number of transgenic plants can be produced in a relatively short period by A. tumefaciens-mediated transformation, contributing to the acquisition and rapid renewal of transgenic trees.
Agrobacterium tumefaciens-mediated transformation is simple, economical, and efficient. It is also important for investigating gene function and the cultivation of transgenic plants. However, A. tumefaciens infection is limited to certain species and genotypes. A. tumefaciens residues may form crown galls, resulting in the yield reduction of transgenic plants (Guo et al., 2019). Further consideration should be given to the field applications of A. tumefaciens-mediated transgenic trees.
The principle of the gene gun method is to use accelerators to transfer particles coated with exogenous genes into receptor cells, tissues, or organs so that the exogenous genes can be integrated into the receptor genome and expressed (Zhang et al., 2013). This method is mainly applied to crops and some fruit trees such as wheat, corn, bean, and citrus (Ozyigit and Yucebilgili Kurtoglu, 2020). Compared to A. tumefaciens-mediated transformation, the gene gun method is not limited by genotype (Ozyigit and Yucebilgili Kurtoglu, 2020). The applications of gene gun-mediated transformation in forest trees are listed in Table 3; gene gun technology has considerable potential in forest tree research.
The efficiency of gene gun-mediated transformation is affected by receptor types, culture conditions, and transformation parameters (Wang B. et al., 2018). The transformation efficiency of plant cells or tissues with strong regeneration ability and strong physiological activity is high. For example, a highly efficient transformation system involving particle bombardment of the callus of date palm was reported (Mousavi et al., 2014). Gene gun transformation is also unrestricted in terms of the materials and cells to which it can be applied. The gene gun method could overcome the drawbacks of A. tumefaciens-mediated transformation and improve transformation efficiency. For example, the drought-related genes JERF36, ZxZF, AREB, and GST were cotransformed into Populus euramericana by particle bombardment to generate transgenic poplar with drought tolerance (Cui, 2012). However, gene gun-mediated transformation has low transformation efficiency, inserts multiple gene copies, and can inactivate or silence the transformed genes. Additionally, the exogenous genes are expressed unstably and easily lost on bombardment. There is an increased likelihood of non-transformants or chimerism, possibly leading to abnormal gene expression and coinhibition.
The pollen tube pathway uses pollen tubes naturally formed after plant pollination to carry out genetic transformation and typically comprises three steps, namely, foreign gene injection into the pollen tube, integration into the plant genome, and selection of transgenic plants (Wang M. et al., 2018). Compared to other transformation methods, the pollen tube pathway undergoes a short period of application in transgenic plants, and there are few reports of its use in forest trees, so further research should be needed in this field. The pollen tube method was used to introduce the total DNA of P. alba into Populus liaoningensis sp. nov × Populus deltoids cv. “N001,” and the phenotypic characteristics of the donor poplar were evident in four transgenic lines (Zhao, 2016). Intriguingly, Ve gene transfer into the walnut genome by the pollen tube pathway resulted in a lower malformed fruit rate than stigma-cutting addition or microinjection (Hou et al., 2004). The method has been applied in crops, e.g., Oryza sativa, and Glycine max, but in few forest tree species (Guo and Zhou, 2018; Zhang H. et al., 2021; Table 4).
Although the pollen tube pathway is less frequently used than A. tumefaciens-mediated transformation, it overcomes the genotype restriction of the latter. For example, the genetic transformation of cotton is restricted by genotype, and transgenic cotton lines can be generated by the pollen tube pathway to enhance insect and herbicide resistance (Showalter et al., 2009). This has the advantage of simplicity and is also inexpensive but has low transformation efficiency (Zhang, 2010; Cui et al., 2013; Wang M. et al., 2018). However, the method is limited by flowering time and is not applicable to gymnosperms, as it is dependent on naturally formed pollen tubes (Jian et al., 2012).
Genetic transformation of protoplasts refers to the transfer of exogenous genes into plants, using protoplasts as receptors to generate transgenic plants with stable expression of exogenous genes. Protoplasts, as single-cell systems, are not (or less) affected by the surrounding cells and microenvironment. Compared to protoplast transformation in annual herbaceous plants, such as Arabidopsis thaliana, tobacco, and O. sativa (Jiang et al., 2006; Sun et al., 2013; Zhao et al., 2014), the separation and regeneration of protoplasts in forest trees is difficult, although advances have been made (Table 5). For example, protoplasts were isolated from petals and leaves of Camellia sinensis (Liu et al., 2017; Peng et al., 2018; Ye et al., 2021); however, large-scale analysis is still under way. Intriguingly, the addition of aminophosphoric acid inhibitors degraded the cell wall, as verified in elm (Chang et al., 2018b). Additionally, green fluorescent protein (GFP) was transformed into Elaeis guineensis protoplast by a polyethylene glycol (PEG)-mediated method, and a protoplast transformation system of this species was established for the first time (Masani et al., 2014).
Protoplasts can be extracted from almost all organs and tissues and show intrinsic developmental and spatial characteristics. The regenerated plants develop from single-cell systems, which are easy to purify and stable. Therefore, the introduction of exogenous genes into protoplasts has advantages compared to other exosomes. PEG-mediated transformation, shock perforation transformation, liposome-mediated transformation, and A. tumefaciens coculture transformation are commonly used to construct protoplast-based genetic transformation systems (Zhao and Chen, 2004). The PEG-mediated method is the most widely used type of protoplast transformation and can be combined with the electroshock method to improve transformation efficiency (Lenaghan and Neal Stewart, 2019). Since there is no cell wall, this method overcomes the obstacles of poor hybrid compatibility and low cell-transformation efficiency. In addition, protoplasts can be isolated from uniform cell suspension cultures, mainly from calli. A system for protoplast regeneration to whole plants has been established in A. thaliana, which showed that WUS and DRN were necessary for protoplast regeneration and greatly facilitated this process (Xu et al., 2021). Cell-wall regeneration is a key step in protoplast regeneration to whole plants (Zhang Q. et al., 2021). Early screening of molecular targets by protoplasts enabled the establishment of efficient and automatic protoplast isolation, transformation, and screening methods in crops. However, protoplast separation and regeneration in forest trees are more difficult than in annual crops and have not been well-developed, hampering the development and application of protoplast transformation in forest trees.
Instantaneous genetic transformation enables the investigation of gene function and comparison of genetic constructs of recombined genes (Canto, 2016). Instantaneous transformation can be mediated by particle bombardment, PEG, plant virus vector, and A. tumefaciens. Due to the cost of particle bombardment equipment, the low success rate of protoplast culture, and scarcity of viral vectors, A. tumefaciens-mediated instantaneous transformation is typically used (Li et al., 2020c).
Leaf osmosis is the most commonly used instantaneous gene expression method in A. tumefaciens infection. For example, a method was established to reduce individual differences in the instantaneous transformation of Camptotheca acuminata (Wang B. et al., 2018). The transcription factor LoNAC18 was transferred into larch by A. tumefaciens instantaneous transformation, demonstrating that LoNAC18 is involved in the regulation of PEG-mediated simulated drought stress in larch (Zhang et al., 2020). Besides leaves, it could also be used in stems and roots. An instantaneous transformation system was established for vacuum osmotic infection of poplar stem segments, enabling identification of the functions of genes involved in vascular tissue differentiation and regulation of xylem development (Li et al., 2021b). Instantaneous transformation of roots has been applied in medicinal plants and soybeans, but there are few reports in forest trees (Meng et al., 2019; Xia et al., 2020; Table 6). Intriguingly, a simple and efficient A. tumefaciens-mediated instantaneous gene expression system was developed for diverse trees—including birch, poplar, and Tamarix—in which the whole plantlet, leaf, and stem are used as explants for instantaneous expression (Zheng et al., 2012). As genetic information on forest trees accumulates, the instantaneous transformation will enable the exploration of metabolic pathways and subcellular localization of forest tree genes. Therefore, considering the low transformation efficiency and non-availability of genetic transform systems, it is necessary to improve A. tumefaciens-mediated instantaneous transformation of forest trees for transgenic research.
Virus-induced gene silencing (VIGS) is a transcription suppression technique that facilitates the functional analysis of genes. VIGS has been applied in diverse plants, including herbs and fruit trees (Dommes et al., 2019) but few forest trees (Cui and Wang, 2017; Dommes et al., 2019). VIGS technology based on tobacco rattle virus (TRV) was successfully applied in Populus euphratica, Populus canescens (Shen et al., 2015), H. brasiliensis (Li et al., 2021c), and Olea europaea (Koudounas et al., 2020). Since VIGS can rapidly reduce the expression of target genes, it facilitates molecular function research in plants, including forest trees. Therefore, it is necessary to determine the optimal conditions for VIGS to silence target genes in forest trees, including the viral vector, ambient temperature, plant age or development stages, and inoculation method (Shi et al., 2021). Overall, VIGS enables gene function analysis of trees.
Agrobacterium tumefaciens-mediated transformation is affected by genotype and secondary metabolites. It is difficult to establish the A. tumefaciens-mediated genetic transformation system in some plants, but the method is important for investigating gene function in forest trees. In dicotyledonous plants, A. tumefaciens-mediated transformation is the first choice due to its high transformation efficiency. The gene gun method compensates for the genotype limitation of A. tumefaciens-mediated transformation. Additionally, gene gun-mediated transformation is important for research on gymnosperms such as Pinus, but its application is limited by cost. The pollen tube pathway and protoplast transformation methods may be preferred for some forest trees. Instantaneous transformation enables the establishment of stable genetic transformation systems and expression of the genes of Populus, Pinus, and other forest trees in tobacco or other easily transformed plants. For most investigations of gene function, A. tumefaciens-mediated transformation is used for multiple forest trees. However, consideration should be given to other methods, particularly transformation from scratch in forest trees because A. tumefaciens residues can lead to crown gall development and yield reduction (Stanton, 2018).
Multiple insect-resistant genes—including Bacillus thuringiensis (Bt), protease inhibitor (PI), Androctonus australis hector insect toxin (AaIT), and chitinase genes—have been identified and applied in trees (Ren et al., 2021). Among them, Bt is the most widely used in insect resistance. Stable transfer of Bt into forest trees was first reported in transgenic poplar (McCown et al., 1991). Intriguingly, the simultaneous application of two Bt genes expanded the scope of insect resistance in transgenic forest trees (Wang et al., 2012; Dong et al., 2015). Consequently, means of enhancing forest tree resistance to insects by transforming two or more Bt genes warrant further research.
Overexpression of PI genes, including serine protease inhibitors (SPIs) and Kunitz trypsin inhibitor (KTI), resulted in insect death and prevented resistance development (Major and Constabel, 2008; Clemente et al., 2019). Bivalent resistance genes (CryIAc and API) were introduced into poplar, and the mortality rate of larvae was 60–80% (Li et al., 2007). In addition, genetic transformation with PI and Bt genes enhanced the insect resistance of transgenic plants. Transgenic poplar with API and dual Bt genes were toxic to Lepidoptera and Coleoptera and showed greater insect resistance than plants transformed with a single Bt gene (Wang G. et al., 2018).
The GM improvement of insect resistance has been realized in diverse forest trees, including Populus (Ren et al., 2021), Eucalyptus (Shao et al., 2002), Picea (Hammerbacher et al., 2014), Ulmus (Newhouse et al., 2007), Pinus (Grace et al., 2005), and Tsuga (Merkle et al., 2014). Transgenic forest trees were first used commercially in China (Chang et al., 2018a). Exogenous genes were expressed stably in 8- and 10-year-old transgenic poplar trees, and there was no significant developmental difference between 10-year-old transgenic and non-transgenic poplars (Ren et al., 2017). The current investigations suggested that the additive effect existed in transgenic forest trees with the same or different kinds of insect-resistant genes, which presented broad-spectrum insect resistance. The stability of exogenous insect resistance genes in transgenic forest trees was verified in 10-year-old transgenic poplars. However, the stability and effectiveness of insect resistance require validation in transgenic forest trees as perennials. Additionally, whether insects will develop tolerance warrants further investigation.
It is necessary to control weeds during the early stages of tree growth. Mechanical herbicides are inefficient and costly and affect the normal growth and development of forest trees. Therefore, it is preferable to cultivate herbicide-resistant tree varieties. Bialaphos resistance (bar) is the most widely used herbicide resistance selective marker gene; it is derived from the soil bacterium Streptomyces hygroscopicus and induces resistance to phosphate-based broad-spectrum herbicides, such as Liberty Basta, and Finale (Lebedev et al., 2016). Bar has been inserted into diverse species and hybrids of P. alba, Eucalyptus, Picea abies, oak, and various conifers (Brukhin et al., 2000; Confalonieri et al., 2000; Harcourt et al., 2000; Bishop-Hurley et al., 2001; Zhang et al., 2005; Álvarez et al., 2009). These investigations indicated the broad application of bar in herbicide-resistant transgenic trees.
In addition, glutathione S-transferase (GST) genes encoding specific herbicide resistance to acetylchloroaniline were introduced into poplar hybrids, enhancing herbicide resistance (Gullner et al., 2001). The poplar clones INRA 353-38 (Populus tremula × Populus tremuloides) and 717-1B4 (P. tremula × P. alba) transformed with bar, and the male sterility gene BARNASE showed stable herbicide resistance within 8 years (Li et al., 2008). The selection of herbicide-resistant trees provides an alternative to non-chemical weed control. In future, gene-editing technology may be used to improve the herbicide resistance of forest trees.
Disease resistance genes are mainly used in the molecular breeding of forest trees to improve plant antiviral and antibacterial defenses. Trichosanthin (TCS), a broad-spectrum antiviral gene, was transformed into Paulownia by A. tumefaciens-mediated method, and transgenic Paulownia lines with strong disease resistance were screened out (Liu et al., 2011). HbLFG1, a negative regulator of plant immunity, promoted infection by Erysiphe quercicola of H. brasiliensis (Li et al., 2021d). Poplar is threatened by Melampsora species, which cause poplar leaf rusts. Overexpression of A. thaliana GALACTINOL SYNTHASE3 (AtGolS) and Cucumber sativus RAFFINOSE SYNTHASE (CsRFS) in hybrid poplar (P. alba × Populus grandidentata) increased susceptibility to Melampsora aecidiodes infection (La Mantia et al., 2018). Additionally, constitutive overexpression of PtrWRKY18 and PtrWRKY35 in poplar activated disease-related genes and increased the resistance of poplar to Melampsora, suggesting functional redundancy (Jiang et al., 2017). Besides, miRNA can promote plant disease resistance by participating in hormone signaling and regulating resistance (R) genes. In transgenic poplar, miR472a positively regulates resistance to Colletotrichum gloeosporioides infection by targeting NBS-LRR and negatively regulates resistance to Cytospora chrysosperma infection (Su et al., 2018). At present, there are many studies on miRNA and disease resistance in rice, potato, and other crops, but there are few reports in forest trees (Natarajan et al., 2018; Zhang et al., 2018). Further investigation of the roles of miRNAs in pathogen infection of trees is needed.
Studies of tree disease resistance and genetic engineering have promoted the breeding and improvement of tree varieties. Future studies should focus on the regulatory networks of tree responses to pathogens to reduce disease susceptibility.
Plant abiotic stresses include cold, freezing, drought, salt, nutrient deficiency, and heavy metals (Gong et al., 2020). Investigation of gene function in response to abiotic stress could improve the environments of trees and so, expand their ranges in specific ecosystems and increase species richness (Xu and Zhai, 2021). Therefore, breeding new varieties of trees with strong resistance to stress is warranted.
Tree genetic engineering research has focused on salt and drought tolerance. Overexpression of WOX11/12A and ThNAC12 in poplar increased salt tolerance, reactive oxygen species (ROS) scavenging, and the antioxidant enzyme activity of transgenic plants (Wang et al., 2021a,b). Interference with FDL expression enhanced the drought resistance of transgenic poplar (Yu et al., 2019). The K+/Na+ homeostasis of root cells and tolerance to salt stress were improved in transgenic poplar overexpressing JERF36s (Ding et al., 2020). These studies provided insight into the mechanism of salt tolerance improvement in plants and will facilitate breeding strategies to improve salt tolerance. The introduction of BpMBF1 into poplar significantly improved cold resistance (Wang, 2020). Instantaneous overexpression of JrGRAS2 in walnut enhanced the tolerance to high temperature (Yang et al., 2018). Overexpression of PsnICE1 significantly enhanced the cold stress tolerance and antioxidant enzyme activity of transgenic poplars (Wang et al., 2021d).
Many other adverse environmental conditions also affect plant growth. The transcriptomic profiles of poplar under stresses suggest candidate genes of breeding (Yao et al., 2018; Chen et al., 2020), enabling investigation of plant regulatory networks. The correlations among stress response regulatory signals need further investigation.
Wood structure and quality are critical traits for genetic improvement. Lignin content can be reduced by introducing genes that inhibit key enzymes in the lignin synthesis pathway. Downregulation of coumaroyl shikimate 3′-hydroxylase (C3′H), cinnamate 4-hydroxylase (C4H), and 4-coumarate-CoA ligase gene (4CL) reduces the lignin content in transgenic hybrid eucalyptus (Eucalyptus urophylla × Eucalyptus grandis) (Sykes et al., 2015). Cell-specific downregulation of 4CL decreased the lignin content of transgenic poplars (Cao et al., 2020). In transgenic poplars with suppressed C3H and hydroxycinnamoyl transferase (HCT), the fiber cell diameter, vessel molecular diameter, and cell wall thickness were smaller, leading to decreased lignin content (Zhou et al., 2018). Therefore, suppression of lignin biosynthesis-related genes in transgenic forest trees decreased the lignin content, thus improving wood properties and biomass utilization.
Overexpression of GAlactUronosylTransferase12 (GAUT12) in poplar increased xylan and galacturonic acid production and decreased growth (Biswal et al., 2018b). Accordingly, downregulation of GAUT12 significantly improved saccharification efficiency and promoted the growth of transgenic poplars (Biswal et al., 2015). In addition, downregulation of GAUT4 by RNA interference (RNAi) decreased the homogalacturonan (HG) and rhamnogalacturonan II (RG-II) contents and increased the biomass yield (Biswal et al., 2018a). Therefore, the GAUT gene family negatively regulates plant growth by regulating xylan biosynthesis. The suppression of ACAULIS5 expression reduced the stem cytokinin level in hybrid aspen (P. tremula × P. tremuloides) and reduced secondary stem growth (Milhinhos et al., 2020). In P. tomentosa, PtSS3 is important in sucrose metabolism and growth and participates in wood formation (Li et al., 2020a). Brassinosteroid (BR) signaling plays an important role in secondary growth and wood formation. The BR signaling pathway affects xylem development synergetic with PdC3H17, a positive regulator of auxin-mediated xylem formation (Tang et al., 2020). However, to overcome the influence of the environment and obtain stable traits during the growth of transgenic plants, further improvement of the technology and accumulation of genes related to wood properties is needed.
Plants undergo the transition from infancy to reproductive maturity before flowering. Furthermore, trees experience a longer vegetative period than crops, prolonging the breeding cycle (Liao and An, 2013). However, genetic engineering can shorten infancy and alter flowering time in forest trees. FLOWERING LOCUS T (FT) is a floral hormone that affects plant flowering, growth, and development (Wigge, 2011). Overexpression of FT-induced flowering of Eucalyptus, and early flowering trees were found to be vigorous, showing a high branching phenotype (Klocko et al., 2016).
Transformation of poplars with HSP:AtFT and PsEND1:barnase-barstar vectors resulted in disturbed pollen development and the formation of male-sterile plants (Briones et al., 2020). Additionally, LEAFY (LFY) is necessary for the induction of flower organ-recognition genes. It endows root explant cells with the fate of flowers and allows callus to form flowers and flower organs without producing leaves (Wagner et al., 2004). In sweetgum, RNAi was used to inhibit LEAFY gene expression, generating sterile transgenic plants (Qiao et al., 2007). A vector with the RNAi-LFY cassette was transferred into P. alba, which markedly altered flower morphology and led to female flower sterility (Klocko et al., 2021). However, in asexual forest trees, sterility associated with LFY expression inhibition can alleviate the gene flow of seeds and pollen, although the effects on tree shape and wood production are unclear (Klocko et al., 2021). However, the use of LFY suppressor genes could be costly, and further research is needed.
RNA-based approaches, including RNAi and CRISPR system, enable highly targeted modifications to enhance yield and stress resistance. These methods are typically based on A. tumefaciens-mediated genetic transformation. In RNAi, small interfering RNAs downregulate target gene expression without affecting the expression of other genes and are important for plant improvement (Rajput et al., 2021). GM agroforestry poplars obtained by RNAi exhibited reduced plantation isoprene emissions without compromising woody biomass production (Monson et al., 2020). Transgenic poplars carrying PTRARF2.1-RNAi showed severe leaf phenotypes, such as irregular shape and small size, and stimulated expression of auxin-response genes (Fu et al., 2019). In addition, RNAi allows the targeting of specific plant pathogens to control plant diseases (Kuo and Falk, 2020). By silencing CYP33C9 by RNAi in vitro, the feeding, reproduction, oviposition, hatchability, and pathogenicity of Bursaphelenchus xylophilus nematode were inhibited (Qiu et al., 2019). The effects of RNAi should be studied and applied in other tree species.
The CRISPR system for precision breeding has been applied in poplar, Eucalyptus, and other forest tree species (Müller et al., 2020; Elorriaga et al., 2021). For example, the knockout of CSE by CRISPR-Cas9 improved lignocellulosic biomass without growth retardation in GM poplar (Jang et al., 2021). Knockout of the root growth transcription factor PDNF-YB21 by CRISPR-Cas repressed the root growth and drought resistance in poplar (Zhou Y. et al., 2020). These studies aimed to improve sustainable production, induce DNA-free targeted mutations, and alter plant architecture, sex, and floral development. CRISPR-Cas technology does not introduce exogenous genes into the genomes of forest trees and so, has higher biosafety than other transgenic techniques. CRISPR-Cas is the most promising gene-editing technology developed to date (Bewg et al., 2018).
Genetic engineering can improve the traits of forest trees, shorten the breeding period, and enable the cultivation of new varieties with high commercial value by introducing exogenous genes (Figure 1). Genetic transformation also enables the exploration of gene function in forest trees. However, there are many difficulties and problems to overcome in forest trees. One of them is the genetic transformation of vectors with multiple foreign genes. The introduction of multiple exogenous genes concurrently could improve the traits of forest trees, but the construction of vectors carrying multiple genes is more difficult and some may not play the expected roles in transgenic trees. For example, transgenic poplar with two insect-resistance genes (Cry1Ac and Cry3A) and two salt-tolerant genes (mtlD and BADH) did not show improved salt tolerance (Zhou X. et al., 2020). Additionally, the balance between the expression of exogenous genes and growth/development requires investigation—whether increased resistance weakens other traits in transgenic forest trees is unclear.
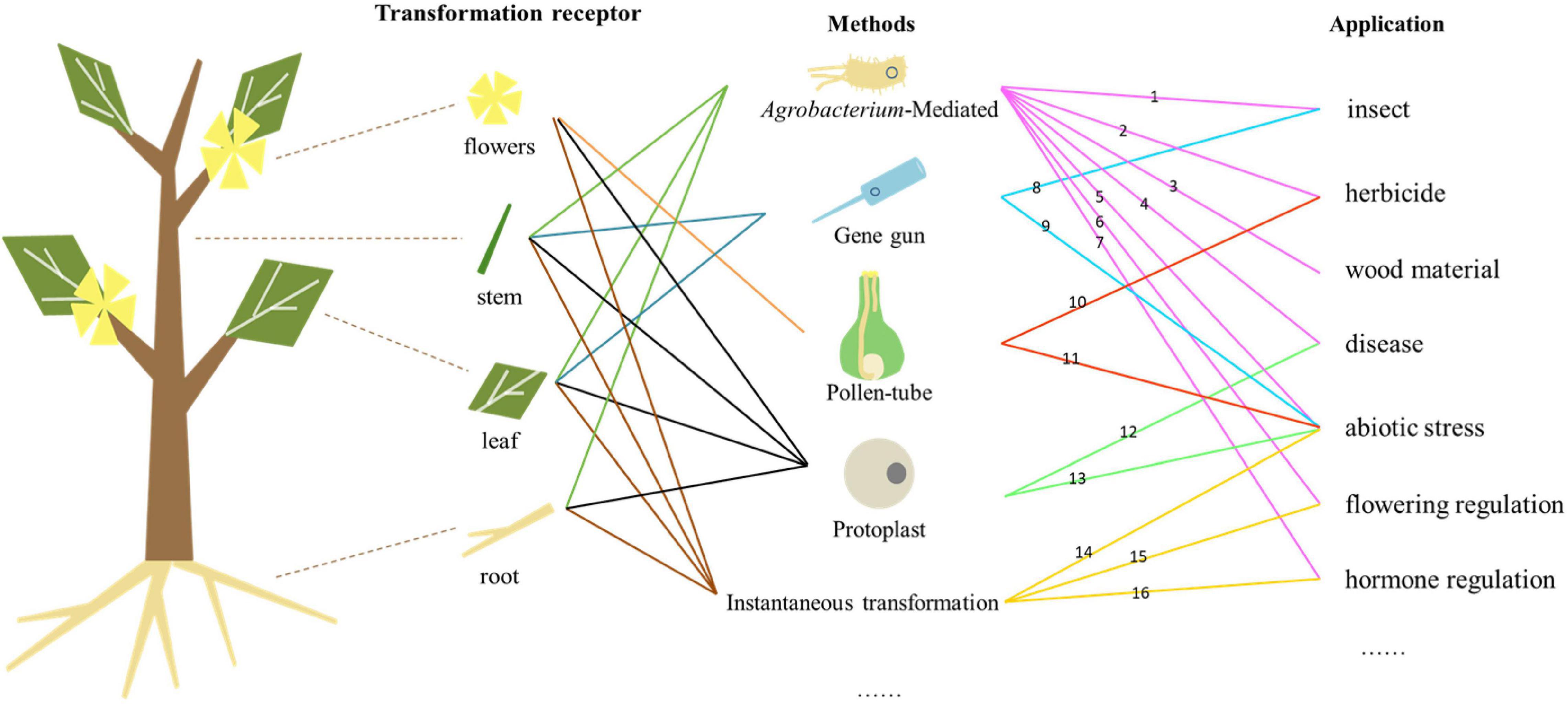
Figure 1. Relationships among genetic transformation receptors, methods, and applications. Numbers represent different gene types. (1) Bt, API, etc., (2) Bar, aroA, GST, etc., (3) 4CL, C3H, C4H, etc., (4) TCS, PPV, WRKY, etc., (5) MBF, BADH, etc., (6) FT, LFY, etc., (7) GAUT, BAK, RGL, etc., (8) Bt, etc., (9) GERF, AREB, etc., (10) Bar, etc., (11) AFP, etc., (12) hph, etc., (13) FLA, etc., (14) DREBRB, ALDH21, etc., (15) SAG, etc., and (16) pBI121, etc.
Due to the uncertainty over insertion sites, genetic transformation inevitably generates chimerism, for instance, in stems and young leaves. For example, in peach (Prunus persica), A. tumefaciens-mediated transformation is inefficient, with a low level of correspondence between transformed and regenerative cells, and a high rate of chimerism in the buds produced during transformation (Ricci et al., 2020). Although chimeras enhance the cultivation of some ornamental plants, the purification and stable inheritance of target types can be problematic. Conversely, the protoplast is a single-cell system that can develop into a complete plant, enabling stable inheritance of traits. Several genes enhance protoplast regeneration, in particular, callus formation, which might promote the use of protoplast-mediated gene transformation (Xu et al., 2021). A strict screening system is needed for protoplast transformation, identifying transgenic plants with improvements in the desired traits.
Genetic engineering is controversial due to the potential for harm to the environment. The long-term stability of transgenic forest trees needs to be investigated, and the environmental impact of GM trees is still debated. A 5-year field trial showed no effect of Bt transgenic 741 poplar on arthropods or soil bacterial diversity (Zuo et al., 2018). A robust biosafety framework is necessary, with precautions followed for domesticated trees. An international group of researchers in silviculture, forest tree breeding, forest biotechnology, and environmental risk assessment examined how the environmental risk assessment paradigm used for genetic engineering crop plants could be applied to the genetic engineering of trees for plantation. It is also important to differentiate between environmental risk assessment for confined field trials of genetic engineering trees and unconfined or commercial-scale release (Häggman et al., 2013).
It is important to establish a rapid and reliable transformation system for forest trees, considering their long growth cycle and low transformation efficiency. Multi-omics techniques and modern biotechnology will facilitate the molecular breeding of forest trees. Transgenic research on trees will improve transformation efficiency and enable the safety evaluation of transgenic plants for commercial application. Leveraging the genetic transformation of forest trees for ecosystem restoration, energy supply, and sustainable production is a major challenge.
YY and CW were involved in planning and drafting the manuscript. DX modified the manuscript. YL collated the table contents. YW conceived of the presented idea and supervised this study. All authors discussed the results and commented on the manuscript.
This study was supported by the National Natural Science Foundation of China (32071504 and 31670671).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Álvarez, R., Álvarez, J. M., Humara, J. M., Revilla, Á, and Ordás, R. J. (2009). Genetic transformation of cork oak (Quercus suber L.) for herbicide resistance. Biotechnol. Lett. 31, 1477–1483. doi: 10.1007/s10529-009-0033-2
Bewg, W. P., Ci, D., and Tsai, C. (2018). Genome editing in trees: from multiple repair pathways to long-term stability. Front. Plant Sci. 9:1732. doi: 10.3389/fpls.2018.01732
Bishop-Hurley, S. L., Zabkiewicz, R. J., Grace, L., Gardner, R. C., Wagner, A., and Walter, C. (2001). Conifer genetic engineering: transgenic Pinus radiata (D. Don) and Picea abies (Karst) plants are resistant to the herbicide buster. Plant Cell Rep. 20, 235–243. doi: 10.1007/s002990100317
Biswal, A. K., Atmodjo, M. A., Pattathil, S., Amos, R. A., Yang, X., Winkeler, K., et al. (2018b). Working towards recalcitrance mechanisms: increased xylan and homogalacturonan production by overexpression of GAlactUronosylTransferase12 (GAUT12) causes increased recalcitrance and decreased growth in Populus. Biotechnol. Biofuels 11:9. doi: 10.1186/s13068-017-1002-y
Biswal, A. K., Atmodjo, M. A., Li, M., Baxter, H. L., Yoo, C. G., Pu, Y., et al. (2018a). Sugar release and growth of biofuel crops are improved by down regulation of pectin biosynthesis. Nat. Biotechnol. 36, 249–257. doi: 10.1038/nbt.4067
Biswal, A. K., Hao, Z., Pattathil, S., Yang, X., Winkeler, K., Collins, C., et al. (2015). Downregulation of GAUT12 in Populus deltoides by RNA silencing results in reduced recalcitrance, increased growth and reduced xylan and pectin in a woody biofuel feedstock. Biotechnol. Biofuels 8:41. doi: 10.1186/s13068-015-0218-y
Briones, M. V., Hoenicka, H., Canas, L. A., Beltran, J. P., Hanelt, D., Sharry, S., et al. (2020). Efficient evaluation of a gene containment system for poplar through early flowering induction. Plant Cell Rep. 39, 577–587. doi: 10.1007/s00299-020-02515-1
Bruegmann, T., Wetzel, H., Hettrich, K., Smeds, A., Willför, S., Kersten, B., et al. (2019). Knockdown of PCBER1, a gene of neolignan biosynthesis, resulted in increased poplar growth. Planta 249, 515–525. doi: 10.1007/s00425-018-3021-8
Brukhin, V., Clapham, D., Elfstrand, M., and von Arnold, S. (2000). Basta tolerance as a selectable and screening marker for transgenic plants of Norway spruce. Plant Cell Rep. 19, 899–903. doi: 10.1007/s002990000217
Canto, T. (2016). “Transient expression systems in plants: potentialities and constraints,” in Advanced Technologies for Protein Complex Production and Characterization, Vol. 896, ed. M. Cristina Vega (Cham: Springer International Publishing), 287–301. doi: 10.1007/978-3-319-27216-0_18
Cao, S., Huang, C., Luo, L., Zheng, S., Zhong, Y., Sun, J., et al. (2020). Cell-specific suppression of 4-Coumarate-CoA ligase gene reveals differential effect of lignin on cell physiological function in Populus. Front. Plant Sci. 11:589729. doi: 10.3389/fpls.2020.589729
Chang, S., Sun, W., Xu, G., Wang, Z., Yin, M., Han, Q., et al. (2018b). Isolating method of plant protoplast and its research advances of application. Mol. Plant Breed. 16, 1271–1277.
Chang, S., Mahon, E. L., MacKay, H. A., Rottmann, W. H., Strauss, S. H., Pijut, P. M., et al. (2018a). Genetic engineering of trees: progress and new horizons. In Vitro Cell. Dev. Biol. Plant 54, 341–376. doi: 10.1007/s11627-018-9914-1
Chen, H. (2008). Introduction of Exogenous DNA of Populus euphratica into White Poplars via Pollen-Tube Pathway. Beijing: Beijing Forest University.
Chen, Y., Jiang, Y., Chen, Y., Feng, W., Liu, G., Yu, C., et al. (2020). Uncovering candidate genes responsive to salt stress in Salix matsudana (Koidz) by transcriptomic analysis. PLoS One 15:e236129. doi: 10.1371/journal.pone.0236129
Chilton, M. D., Drummond, M. H., Merio, D. J., Sciaky, D., Montoya, A. L., Gordon, M. P., et al. (1977). Stable incorporation of plasmid DNA into higher plant cells: the molecular basis of crown gall tumorigenesis. Cell 11, 263–271. doi: 10.1016/0092-8674(77)90043-5
Cho, J. S., Jeon, H. W., Kim, M. H., Vo, T. K., Kim, J., Park, E. J., et al. (2019). Wood forming tissue-specific bicistronic expression of PdGA20ox1 and PtrMYB221 improves both the quality and quantity of woody biomass production in a hybrid poplar. Plant Biotechnol. J. 17, 1048–1057. doi: 10.1111/pbi.13036
Clemente, M., Corigliano, M., Pariani, S., Sánchez-López, E., Sander, V., and Ramos-Duarte, V. (2019). Plant serine protease inhibitors: biotechnology application in agriculture and molecular farming. Int. J. Mol. Sci. 20:1345. doi: 10.3390/ijms20061345
Coleman, H. D., Cánovas, F. M., Man, H., Kirby, E. G., and Mansfield, S. D. (2012). Enhanced expression of glutamine synthetase (GS1a) confers altered fibre and wood chemistry in field grown hybrid poplar (Populus tremula × alba) (717-1B4). Plant Biotechnol. J. 10, 883–889. doi: 10.1111/j.1467-7652.2012.00714.x
Confalonieri, M., Belenghi, B., Balestrazzi, A., Negri, S., Facciotto, G., Schenone, G., et al. (2000). Transformation of elite white poplar (Populus alba L.) cv. ‘Villafranca’ and evaluation of herbicide resistance. Plant Cell Rep. 19, 978–982. doi: 10.1007/s002990000230
Cui, H., and Wang, A. (2017). An efficient viral vector for functional genomic studies of Prunus fruit trees and its induced resistance to Plum pox virus via silencing of a host factor gene. Plant Biotechnol. J. 15, 344–356. doi: 10.1111/pbi.12629
Cui, X. (2012). Multi-Drought Resistance Genes Transformation and Integration Mechanism Poplar. Beijing: Chinese Academy of Forestry.
Cui, X., Zhang, B., Ding, C., and Su, X. (2013). Plant transgenic technology and its application to genetic improvement of forest trees. World For. Res. 26, 41–46.
Ding, C., Zhang, W., Li, D., Dong, Y., Liu, J., Huang, Q., et al. (2020). Effect of overexpression of JERFs on intracellular K+/Na+ balance in transgenic poplar (Populus alba × P. berolinensis) under salt stress. Front. Plant Sci. 11:1192. doi: 10.3389/fpls.2020.01192
Ding, L., Chen, Y., Wei, X., Ni, M., Zhang, J., Wang, H., et al. (2017). Laboratory evaluation of transgenic Populus davidiana× Populus bolleana expressing Cry1Ac + SCK, Cry1Ah3, and Cry9Aa3 genes against gypsy moth and fall webworm. PLoS One 12:e178754. doi: 10.1371/journal.pone.0178754
Ding, L., Zhi, W., and Hua, W. (2016). Progress and prospect of research in transgenic poplar. For. Res. 29, 124–132.
Dommes, A. B., Gross, T., Herbert, D. B., Kivivirta, K. I., and Becker, A. (2019). Virus-induced gene silencing: empowering genetics in non-model organisms. J. Exp. Bot. 70, 757–770. doi: 10.1093/jxb/ery411
Dong, Y., Du, S., Zhang, J., Yang, M., and Wang, J. (2015). Differential expression of dual Bt genes in transgene poplar Juba (Populus deltoides cv. ‘Juba’) transformed by two different transformation vectors. Can. J. For. Res. 45, 60–67. doi: 10.1139/cjfr-2014-0335
Du, L., Zeng, X., Zhou, S., and Bao, M. (2008). Transfer of green fluorescent protein gene into embryogenic calluses of Cinnamomum camphora. Acta Bot. Boreali Occidentalia Sin. 28, 465–469.
Elorriaga, E., Klocko, A. L., Ma, C., Plessis, M., An, X., Myburg, A. A., et al. (2021). Genetic containment in vegetatively propagated forest trees: CRISPR disruption of LEAFY function in Eucalyptus gives sterile indeterminate inflorescences and normal juvenile development. Plant Biotechnol. J. 19, 1743–1755. doi: 10.1111/pbi.13588
Fan, D., Ran, L., Hu, J., Ye, X., Xu, D., Li, J., et al. (2020). MiR319a/TCP module and DELLA protein regulate trichome initiation synergistically and improve insect defenses in Populus tomentosa. New Phytol. 227, 867–883. doi: 10.1111/nph.16585
Fu, Y., Win, P., Zhang, H., Li, C., Shen, Y., He, F., et al. (2019). PtrARF2.1 is involved in regulation of leaf development and lignin biosynthesis in poplar trees. Int. J. Mol. Sci. 20:4141. doi: 10.3390/ijms20174141
Génissel, A., Leplé, J., Millet, N., Augustin, S., Jouanin, L., and Pilate, G. (2003). High tolerance against Chrysomela tremulae of transgenic poplar plants expressing a synthetic cry3Aa gene from Bacillus thuringiensis ssp tenebrionis. Mol. Breed. 11, 103–110. doi: 10.1023/A:1022453220496
Gong, Z., Xiong, L., Shi, H., Yang, S., Herrera-Estrella, L. R., Xu, G., et al. (2020). Plant abiotic stress response and nutrient use efficiency. Sci. China Life Sci. 63, 635–674. doi: 10.1007/s11427-020-1683-x
Grace, L. J., Charity, J. A., Gresham, B., Kay, N., and Walter, C. (2005). Insect-resistant transgenic Pinus radiata. Plant Cell Rep. 24, 103–111. doi: 10.1007/s00299-004-0912-x
Gullner, G., Kömives, T., and Rennenberg, H. (2001). Enhanced tolerance of transgenic poplar plants overexpressing γ-glutamylcysteine synthetase towards chloroacetanilide herbicides. J. Exp. Bot. 52, 971–979. doi: 10.1093/jexbot/52.358.971
Guo, M., Ye, J., Gao, D., Xu, N., and Yang, J. (2019). Agrobacterium-mediated horizontal gene transfer: mechanism, biotechnological application, potential risk and forestalling strategy. Biotechnol. Adv. 37, 259–270. doi: 10.1016/j.biotechadv.2018.12.008
Guo, Q., Jiang, J., Yao, W., Li, L., Zhao, K., Cheng, Z., et al. (2021). Genome-wide analysis of poplar HD-Zip family and over-expression of PsnHDZ63 confers salt tolerance in transgenic Populus simonii × P. nigra. Plant Sci. 311:111021. doi: 10.1016/j.plantsci.2021.111021
Guo, X., and Zhou, X. (2018). Transgenic wheat was obtained by pollen tube mediated capsicum total DNA. Jiangsu Agric. Sci. 46, 36–38.
Häggman, H., Raybould, A., Borem, A., Fox, T., Handley, L., Hertzberg, M., et al. (2013). Genetically engineered trees for plantation forests: key considerations for environmental risk assessment. Plant Biotechnol. J. 11, 785–798. doi: 10.1111/pbi.12100
Hammerbacher, A., Paetz, C., Wright, L. P., Fischer, T. C., Bohlmann, J., Davis, A. J., et al. (2014). Flavan-3-ols in Norway spruce: biosynthesis, accumulation, and function in response to attack by the bark beetle-associated fungus Ceratocystis polonica. Plant Physiol. 164, 2107–2122. doi: 10.1104/pp.113.232389
Hao, Z., Dai, Y., Zhang, S., and Wang, P. (2021). Genetic transformation method of hairy roots mediated by Agrobacterium rhizogenes for Pyrus betulaefolia. Hubei Agric. Sci. 60, 151–154.
Harcourt, R. L., Kyozuka, J., Floyd, R. B., Bateman, K. S., Tanaka, H., Decroocq, V., et al. (2000). Insect- and herbicide-resistant transgenic eucalypts. Mol. Breed. 6, 307–315. doi: 10.1023/A:1009676214328
Hou, L., Li, X., Cui, G., Li, J., and Qiao, Y. (2004). Application of several genetic transformation techniques in transgenic breeding of Walnut. Shandong For. Sci. Technol. 1, 8–9.
Huang, Y., Liu, H., Jia, Z., Fang, Q., and Luo, K. (2012). Combined expression of antimicrobial genes (Bbchit1 and LJAMP2) in transgenic poplar enhances resistance to fungal pathogens. Tree Physiol. 32, 1313–1320. doi: 10.1093/treephys/tps079
Jang, H., Bae, E., Kim, M., Park, S., Choi, N., Pyo, S., et al. (2021). CRISPR-knockout of CSE gene improves saccharification efficiency by reducing lignin content in hybrid poplar. Int. J. Mol. Sci. 22:9750. doi: 10.3390/ijms22189750
Jeon, H., Cho, J., Park, E., Han, K., Choi, Y., and Ko, J. (2016). Developing xylem-preferential expression of PdGA20ox1, a gibberellin 20-oxidase 1 from Pinus densiflora, improves woody biomass production in a hybrid poplar. Plant Biotechnol. J. 14, 1161–1170. doi: 10.1111/pbi.12484
Jian, C., Li, K., and Ou, W. (2012). Research progress in pollen-tube pathway method in transgenic plants. Chin. J. Trop. Crops 33, 956–961. doi: 10.1104/pp.108.125229
Jiang, L., Kong, X., Wu, X., and Li, S. (2006). Isolation and purification of protoplasts from tobacoo leaves. J. Anhui Univ. Nat. Sci. Ed. 30, 91–94.
Jiang, Y., Guo, L., Ma, X., Zhao, X., Jiao, B., Li, C., et al. (2017). The WRKY transcription factors PtrWRKY18 and PtrWRKY35 promote Melampsora resistance in Populus. Tree Physiol. 37, 665–675. doi: 10.1093/treephys/tpx008
Jin, Y., Tang, R., Wang, H., Jiang, C., Bao, Y., Yang, Y., et al. (2017). Overexpression of Populus trichocarpa CYP85A3 promotes growth and biomass production in transgenic trees. Plant Biotechnol. J. 15, 1309–1321. doi: 10.1111/pbi.12717
Ke, Q., Wang, Z., Ji, C. Y., Jeong, J. C., Lee, H. S., Li, H., et al. (2016). Transgenic poplar expressing codA exhibits enhanced growth and abiotic stress tolerance. Plant Physiol. Biochem. 100, 75–84. doi: 10.1016/j.plaphy.2016.01.004
Klocko, A. L., Goddard, A. L., Jacobson, J. R., Magnuson, A. C., and Strauss, S. H. (2021). RNAi suppression of LEAFY gives stable floral sterility, and reduced growth rate and leaf size, in field-Grown poplars. Plants 10:1594. doi: 10.3390/plants10081594
Klocko, A. L., Ma, C., Robertson, S., Esfandiari, E., Nilsson, O., and Strauss, S. H. (2016). FT overexpression induces precocious flowering and normal reproductive development in Eucalyptus. Plant Biotechnol. J. 14, 808–819. doi: 10.1111/pbi.12431
Koudounas, K., Thomopoulou, M., Angeli, E., Tsitsekian, D., Rigas, S., and Hatzopoulos, P. (2020). Virus-Induced gene silencing in olive tree (Oleaceae). Methods Mol. Biol. 2172, 165–182. doi: 10.1007/978-1-0716-0751-0_13
Kourosh, M., Ali, M., Samaneh, S. M., Sun, W., Zhang, J., Amir, A. Z. Y., et al. (2018). Functional analysis of overexpressed PtDRS1 involved in abiotic stresses enhances growth in transgenic poplar. Plant Physiol. Biochem. 126, 22–31. doi: 10.1016/j.plaphy.2018.01.023
Kuo, Y., and Falk, B. W. (2020). RNA interference approaches for plant disease control. BioTechniques 69, 469–477. doi: 10.2144/btn-2020-0098
La Mantia, J., Unda, F., Douglas, C. J., Mansfield, S. D., and Hamelin, R. (2018). Overexpression of AtGolS3 and CsRFS in poplar enhances ROS tolerance and represses defense response to leaf rust disease. Tree Physiol. 38, 457–470. doi: 10.1093/treephys/tpx100
Lacroix, B., and Citovsky, V. (2013). The roles of bacterial and host plant factors in Agrobacterium-mediated genetic transformation. Int. J. Dev. Biol. 57, 467–481. doi: 10.1387/ijdb.130199bl
Lardet, L., Leclercq, J., Bénistan, E., Dessailly, F., Oliver, G., Martin, F., et al. (2011). Variation in GUS activity in vegetatively propagated Hevea brasiliensis transgenic plants. Plant Cell Rep. 30, 1847–1856. doi: 10.1007/s00299-011-1092-0
Lebedev, V. G., Faskhiev, V. N., Kovalenko, N. P., Shestibratov, K. A., and Miroshnikov, A. I. (2016). Testing transgenic aspen plants with bar gene for herbicide resistance under semi-natural conditions. Acta Nat. 8, 92–106.
Lenaghan, S. C., and Neal Stewart, C. (2019). An automated protoplast transformation system. Methods Mol. Biol. 1917, 355–363. doi: 10.1007/978-1-4939-8991-1_26
Levee, V., Major, I., Levasseur, C., Tremblay, L., MacKay, J., and Seguin, A. (2009). Expression profiling and functional analysis of Populus WRKY23 reveals a regulatory role in defense. New Phytol. 184, 48–70. doi: 10.1111/j.1469-8137.2009.02955.x
Li, J., Gao, K., Lei, B., Zhou, J., Guo, T., and An, X. (2020a). Altered sucrose metabolism and plant growth in transgenic Populus tomentosa with altered sucrose synthase PtSS3. Transgenic Res. 29, 125–134. doi: 10.1007/s11248-019-00184-9
Li, J., Jia, H., Sun, P., Zhang, J., Xia, Y., Hu, J., et al. (2020b). The WUSCHELa (PtoWUSa) is involved in developmental plasticity of adventitious root in poplar. Genes 11:176. doi: 10.3390/genes11020176
Li, J., Meilan, R., Ma, C., Barish, M., and Strauss, S. H. (2008). Stability of herbicide resistance over 8 years of coppice in field-grown, genetically engineered poplars. West. J. Appl. For. 23, 89–93. doi: 10.1093/wjaf/23.2.89
Li, K., Fan, J., Zhao, Z., Li, L., and Zhu, H. (2007). Transformation of CryIAc and API two insect-resistant genes to poplar (Popular alba × P. Glandulosa). For. Res. 20, 699–704.
Li, L., Gu, H., Yue, Y., Yang, X., and Wang, L. (2020c). Advances in transient transformation systems of woody plants. Mol. Plant Breed. 18, 7784–7794. doi: 10.13271/j.mpb.018.007784
Li, L., Qi, L., Han, Y., Wang, Y., and Li, W. (2000). A study on the introduction of male sterility of antiinsect transgenic Populus nigra by TA29-Barnase gene. Sci. Silvae Sin. 36, 28–32.
Li, L., Zhou, Y., Cheng, X., Sun, J., Marita, J. M., Ralph, J., et al. (2003). Combinatorial modification of multiple lignin traits in trees through multigene cotransformation. Proc. Natl. Acad. Sci. U.S.A. 100, 4939–4944. doi: 10.1073/pnas.0831166100
Li, Q., Yan, S., and Chen, S. (2021a). Genetic transformation of BpERF98 gene and abiotic stress response of transgenic plant in Betula platyphylla. Bull. Bot. Res. 1–11. doi: 10.1007/978-3-319-23534-9_1
Li, H., Guo, D., Wang, Y., Zhu, J., Qu, L., and Peng, S. (2021c). Tobacco rattle virus-induced gene silencing in Hevea brasiliensis. Biosci. Biotechnol. Biochem. 85, 562–567. doi: 10.1093/bbb/zbaa085
Li, X., An, Y., Huang, L., Zeng, W., and Lu, M. (2021b). Establishment of a transient transformation system for stem segments of poplar 84K. Sci. Silvae Sin. 57, 82–89.
Li, X., Li, S., Liu, Y., He, Q., Liu, W., Lin, C., et al. (2021d). HbLFG1, a rubber tree (Hevea brasiliensis) lifeguard protein, can facilitate powdery mildew infection by regulating plant immunity. Mol. Physiol. Plant Pathol. doi: 10.1094/PHYTO-08-20-0362-R
Liao, W., and An, X. (2013). Research status and development trend of transgenic trees. China Biotechnol. 33, 148–160.
Liu, H. (2012). The Establishment of Walnut Genetic Transformation System by Pollen Tube Pathway. Beijing: Chinese Academy Forestry.
Liu, J., Huang, Y., Weng, M., Luo, L., Zhang, H., Wang, C., et al. (2011). Transformation of TCS gene to paulownia elongata and its virus resistance. Sci. Silvae Sin. 47, 171–176.
Liu, M., Xu, J., Ye, J., Li, Z., Fan, R., and Li, L. (2020). Agrobacterium tumefaciens-mediated transformation of leaf callus in Eucommia ulmoides. Sci. Silvae Sin. 56, 79–88.
Liu, X., Du, X., Chen, S., Hu, X., Huang, H., and Tong, Z. (2021). Agrobacterium rhizogenes mediated high frequency hairy root induction system and genetic transformation in Betula luminifera. J. Agric. Biotechnol. 29, 495–505.
Liu, Y., Jin, X., Ma, L., Cao, D., Gong, Z., and Ling, W. (2017). Isolation and purification of mesophyll protoplasts from the leaves of Camellia sinensis. Plant Sci. J. 35, 908–911.
Lu, Y., Su, W., Bao, Y., Wang, S., He, F., Wang, D., et al. (2020). Poplar PdPTP1 gene negatively regulates salt tolerance by affecting ion and ROS homeostasis in Populus. Int. J. Mol. Sci. 21:1065. doi: 10.3390/ijms21031065
Ma, X., Liang, X., Lv, S., Guan, T., Jiang, T., and Cheng, Y. (2020). Histone deacetylase gene PtHDT902 modifies adventitious root formation and negatively regulates salt stress tolerance in poplar. Plant Sci. 290:110301. doi: 10.1016/j.plantsci.2019.110301
Major, I. T., and Constabel, C. P. (2008). Functional analysis of the kunitz trypsin inhibitor family in poplar reveals biochemical diversity and multiplicity in defense against herbivores. Plant Physiol. 146, 888–903. doi: 10.1104/pp.107.106229
Martínez-Gómez, P. (2019). Editorial for special issue “plant genetics and molecular breeding”. Int. J. Mol. Sci. 20:2659. doi: 10.3390/ijms20112659
Masani, M. Y., Noll, G. A., Parveez, G. K., Sambanthamurthi, R., and Prufer, D. (2014). Efficient transformation of oil palm protoplasts by PEG-mediated transfection and DNA microinjection. PLoS One 9:e96831. doi: 10.1371/journal.pone.0096831
McCown, B. H., McCabe, D. E., Russell, D. R., Robison, D. J., Barton, K. A., and Raffa, K. F. (1991). Stable transformation of Populus and incorporation of pest resistance by electric discharge particle acceleration. Plant Cell Rep. 9, 590–594. doi: 10.1007/BF00232339
Meng, D., Yang, Q., Dong, B., Song, Z., Niu, L., Wang, L., et al. (2019). Development of an efficient root transgenic system for pigeon pea and its application to other important economically plants. Plant Biotechnol. J. 17, 1804–1813. doi: 10.1111/pbi.13101
Meng, L., Li, H., Jin, D., Cui, D., and Wang, B. (2004). Transformation of Populus deltoids with CH5B gene. Biotechnol. Bull. 3, 48–51.
Merkle, S. A., Montello, P. M., Reece, H. M., and Kong, L. (2014). Somatic embryogenesis and cryostorage of eastern hemlock and Carolina hemlock for conservation and restoration. Trees 28, 1767–1776. doi: 10.1007/s00468-014-1084-0
Meyermans, H., Morreel, K., Lapierre, C., Pollet, B., De Bruyn, A., Busson, R., et al. (2000). Modifications in lignin and accumulation of phenolic glucosides in poplar xylem upon down-regulation of caffeoyl-coenzyme a O-methyltransferase, an enzyme involved in lignin biosynthesis. J. Biol. Chem. 275, 36899–36909. doi: 10.1074/jbc.M006915200
Milhinhos, A., Bollhöner, B., Blazquez, M. A., Novák, O., Miguel, C. M., and Tuominen, H. (2020). ACAULIS5 is required for cytokinin accumulation and function during secondary growth of Populus trees. Front. Plant Sci. 11:601858. doi: 10.3389/fpls.2020.601858
Monson, R. K., Winkler, B., Rosenstiel, T. N., Block, K., Merl-Pham, J., Strauss, S. H., et al. (2020). High productivity in hybrid-poplar plantations without isoprene emission to the atmosphere. Proc. Natl. Acad. Sci. U.S.A. 117, 1596–1605. doi: 10.1073/pnas.1912327117
Mousavi, M., Mousavi, A., Habashi, A. A., and Dehsara, B. (2014). Genetic transformation of date palm (Phoenix dactylifera L. Cv. ‘Estamaran’) via particle bombardment. Mol. Biol. Rep. 41, 8185–8194. doi: 10.1007/s11033-014-3720-6
Müller, N. A., Kersten, B., Leite Montalvão, A. P., Mähler, N., Bernhardsson, C., Bräutigam, K., et al. (2020). A single gene underlies the dynamic evolution of poplar sex determination. Nat. Plants 6, 630–637. doi: 10.1038/s41477-020-0672-9
Natarajan, B., Kalsi, H. S., Godbole, P., Malankar, N., Thiagarayaselvam, A., Siddappa, S., et al. (2018). MiRNA160 is associated with local defense and systemic acquired resistance against Phytophthora infestans infection in potato. J. Exp. Bot. 69, 2023–2036. doi: 10.1093/jxb/ery025
Newhouse, A. E., Schrodt, F., Liang, H., Maynard, C. A., and Powell, W. A. (2007). Transgenic American elm shows reduced Dutch elm disease symptoms and normal mycorrhizal colonization. Plant Cell Rep. 26, 977–987. doi: 10.1007/s00299-007-0313-z
Ozyigit, I. I., and Yucebilgili Kurtoglu, K. (2020). Particle bombardment technology and its applications in plants. Mol. Biol. Rep. 47, 9831–9847. doi: 10.1007/s11033-020-06001-5
Park, E., Kim, H., Choi, Y., Lee, C., Nguyen, V. P., Jeon, H., et al. (2015). Overexpression of gibberellin 20-oxidase1 from Pinus densiflora results in enhanced wood formation with gelatinous fiber development in a transgenic hybrid poplar. Tree Physiol. 35, 1264–1277. doi: 10.1093/treephys/tpv099
Peng, Z., Tong, H., Liang, G., Shi, Y., and Yuan, L. (2018). Protoplast isolation and fusion induced by PEG with leaves and roots of tea plant (Camellia sinensis l. O. Kuntze). Acta Agron. Sin. 44, 463–470. doi: 10.3724/sp.j.1006.2018.00463
Qiao, G., Luan, W., Pan, H., and Zhou, R. (2007). The LEAFY gene in RNA interference (RNAi) transgenic Liquidambar formosana mediated by Agrobacterium tumefaciems. J. Zhejiang Agric. For. Univ. 24, 140–144.
Qiu, X., Yang, L., Ye, J., Wang, W., Zhao, T., Hu, H., et al. (2019). Silencing of cyp-33C9 gene affects the reproduction and pathogenicity of the pine wood nematode, Bursaphelenchus xylophilus. Int. J. Mol. Sci. 20:4520. doi: 10.3390/ijms20184520
Rajput, M., Choudhary, K., Kumar, M., Vivekanand, V., Chawade, A., Ortiz, R., et al. (2021). RNA interference and CRISPR/Cas gene editing for crop improvement: paradigm shift towards sustainable agriculture. Plants 10:1914. doi: 10.3390/plants10091914
Rao, H., Chen, Y., Huang, M., Wang, M., and Wu, N. (2000). Genetic transformation of poplar NL-80106 transferred by Bt gene and its insect-resistance. J. Plant Resour. Environ. 9, 1–5.
Ren, Y., Zhang, J., Liang, H., Wang, J., and Yang, M. (2017). Inheritance and expression stability of exogenous genes in insect-resistant transgenic poplar. Plant Cell Tissue Organ Cult. 130, 567–576. doi: 10.1007/s11240-017-1247-y
Ren, Y., Zhou, X., Dong, Y., Zhang, J., Wang, J., and Yang, M. (2021). Exogenous gene expression and insect resistance in dual Bt toxin Populus × euramericana ‘neva’ transgenic plants. Front. Plant Sci. 12:660226. doi: 10.3389/fpls.2021.660226
Ricci, A., Sabbadini, S., Prieto, H., Padilla, I. M., Dardick, C., Li, Z., et al. (2020). Genetic transformation in peach (Prunus persica L.): challenges and ways forward. Plants 9:971. doi: 10.3390/plants9080971
Shang, A., Tian, C., Zhao, L., and Tian, Y. (2008). Establishment of genetic transformation system of Euonymus japonicus ‘Cu Zhi’ mediated by Agrobacterium tumefaciens. Acta Hortic. Sin. 35, 409–414.
Shao, Z., Chen, W., Luo, H., Ye, X., and Zhang, J. (2002). Studies on the introduction of the cecrpin d gene into Eucalyptus urophylla to breed the resistant varieties to Pseudomonas solanacearum. Sci. Silvae Sin. 38, 92–97.
Shen, Z., Sun, J., Yao, J., Wang, S., Ding, M., Zhang, H., et al. (2015). High rates of virus-induced gene silencing by tobacco rattle virus in Populus. Tree Physiol. 35, 1016–1029. doi: 10.1093/treephys/tpv064
Shi, G., Hao, M., Tian, B., Cao, G., Wei, F., and Xie, Z. (2021). A methodological advance of tobacco rattle virus-induced gene silencing for functional genomics in plants. Front. Plant Sci. 12:671091. doi: 10.3389/fpls.2021.671091
Shivram, H., Cress, B. F., Knott, G. J., and Doudna, J. A. (2021). Controlling and enhancing CRISPR systems. Nat. Chem. Biol. 17, 10–19. doi: 10.1038/s41589-020-00700-7
Showalter, A. M., Heuberger, S., Tabashnik, B. E., and Carrière, Y. (2009). A primer for using transgenic insecticidal cotton in developing countries. J. Insect Sci. 9, 1–39. doi: 10.1673/031.009.2201
Shu, W., Zhou, H., Jiang, C., Zhao, S., Wang, L., Li, Q., et al. (2019). The auxin receptor TIR1 homolog (PagFBL 1) regulates adventitious rooting through interactions with Aux/IAA28 in Populus. Plant Biotechnol. J. 17, 338–349. doi: 10.1111/pbi.12980
Su, Y., Li, H., Wang, Y., Li, S., Wang, H., Yu, L., et al. (2018). Poplar miR472a targeting NBS-LRRs is involved in effective defense response to necrotrophic fungus Cytospora chrysosperma. J. Exp. Bot. 69, 5519–5530. doi: 10.1093/jxb/ery304/5075698
Sun, H., Lang, Z., Zhu, L., and Huang, D. (2013). Optimized condition for protoplast isolation from maize, wheat and rice leaves. Chin. J. Biotechnol. 29, 224–234.
Sun, Z., Feng, D., Liu, J., Zhao, J., Wang, Y., Zhang, L., et al. (2005). The study of effect the transformed AFP gene on development of apricot fruits. J. Shandong Agric. Univ. 36, 161–166.
Sykes, R. W., Gjersing, E. L., Foutz, K., Rottmann, W. H., Kuhn, S. A., Foster, C. E., et al. (2015). Down-regulation of p-coumaroyl quinate/shikimate 3′-hydroxylase (C3′H) and cinnamate 4-hydroxylase (C4H) genes in the lignin biosynthetic pathway of Eucalyptus urophylla × E. Grandis leads to improved sugar release. Biotechnol. Biofuels 8:128. doi: 10.1186/s13068-015-0316-x
Tang, H., Wallbraun, M., Ren, Z., Reustle, G. M., and Krczal, G. (2001). Genetic transformation of the trichoderma endochitinase gene ThEn-42 to somatic embryos of English walnut. Acta Hortic. Sin. 28, 12–18.
Tang, X., Wang, C., Liu, Y., He, G., Ma, N., Chai, G., et al. (2020). Brassinosteroid signaling converges with auxin-mediated C3H17 to regulate xylem formation in Populus. Front. Plant Sci. 11:586014. doi: 10.3389/fpls.2020.586014
Thakur, A. K., Aggarwal, G., and Srivastava, D. K. (2012). Genetic modification of lignin biosynthetic pathway in Populus ciliata wall. Via Agrobacterium-mediated antisense CAD gene transfer for quality paper production. Nat. Acad. Sci. Lett. 35, 79–84. doi: 10.1007/s40009-012-0018-x
Trumbore, S., Brando, P., and Hartmann, H. (2015). Forest health and global change. Science 349, 814–818. doi: 10.1126/science.aac6759
Wagner, D., Wellmer, F., Dilks, K., William, D., Smith, M. R., Kumar, P. P., et al. (2004). Floral induction in tissue culture: a system for the analysis of LEAFY-dependent gene regulation. Plant J. 39, 273–282. doi: 10.1111/j.1365-313X.2004.02127.x
Wang, B. (2020). Genetic Transformation and Cold Resistance of BpMBF1 in Populus leucopyrami-Dalis 1, L1. Shandong: Shandong Agricultural University.
Wang, B., Gu, J., Huo, C., Chang, C., and Yu, F. (2018). Establishment of Agrobacterium tumefaciens mediated transient transformation system in Camptotheca acuminata leaves. Mol. Plant Breed. 16, 5624–5630.
Wang, G., Dong, Y., Liu, X., Yao, G., Yu, X., and Yang, M. (2018). The current status and development of insect-resistant genetically engineered poplar in china. Front. Plant Sci. 9:1408. doi: 10.3389/fpls.2018.01408
Wang, G., Xia, X., Zhong, W., Fang, H., and Jiang, M. (2003). Regeneration and antibacterial peptide gene transformation of cherry leaves. Acta Hortic. Sin. 30, 209–211.
Wang, G., Yang, M., Huo, X., and Liu, X. (2012). Comparison of exogenous gene expression and insect resistance ability of transgenic 741 poplars with single and double Bt genes. Acta Entomol. Sin. 55, 798–803.
Wang, J., Tang, J., Cui, Y., Chen, Y., Zhao, H., and Yan, X. (2020). Establishment of Agrobacterium-mediated genetic transformation system in Lycium ruhenicium Murr. North. Hortic. 1, 104–110.
Wang, K. (2015). Preface. Agrobacterium protocols. Methods Mol. Biol. 1224, vii–viii. doi: 10.1007/978-1-4939-1658-0_26
Wang, L., Chen, W., Liu, Y., Yang, R., Yin, X., Ma, N., et al. (2014). Optimization of Agrobacterium- mediated transient gene expression system and its utilization in RNAi based gene silencing of rose (Rosa hybrida) petals. J. Agric. Biotechnol. 22, 133–140.
Wang, M., Sun, R., Zhang, B., and Wang, Q. (2018). Pollen tube pathway-mediated cotton transformation. Methods Mol. Biol. 1902, 67–73. doi: 10.1007/978-1-4939-8952-2_6
Wang, L., Wen, S., Wang, R., Wang, C., Gao, B., and Lu, M. (2021a). PagWOX11/12a activates PagCYP736A12 gene that facilitates salt tolerance in poplar. Plant Biotechnol. J. 11, 2249–2260. doi: 10.1111/pbi.13653
Wang, R., Zhang, Y., Wang, C., Wang, Y., and Wang, L. (2021b). ThNAC12 from Tamarix hispida directly regulates ThPIP2;5 to enhance salt tolerance by modulating reactive oxygen species. Plant Physiol. Biochem. 163, 27–35. doi: 10.1016/j.plaphy.2021.03.042
Wang, Y., Zhang, Y., Zhang, X., Zhao, X., Zhang, Y., Wang, C., et al. (2021d). Poplar PsnICE1 enhances cold tolerance by binding to different cis-acting elements to improve reactive oxygen species-scavenging capability. Tree Physiol. doi: 10.1093/treephys/tpab084
Wang, Y., Liu, X., Zhang, Q., Zheng, Y., and Li, D. (2021c). Isolation of palm mesophyII protoplasts and establishment of transient transformation system. J. Huazhong Agric. Univ. 40, 154–159.
Wei, H., Movahedi, A., Xu, C., Sun, W., Li, L., Wang, P., et al. (2020). Overexpression of PtHMGR enhances drought and salt tolerance of poplar. Ann. Bot. 125, 785–803. doi: 10.1093/aob/mcz158
Wei, W., Zhang, Q., Wu, J., Ma, X., and Gu, L. (2021). Establishment of high-efficiency callus induction and transient transformation system of Chinese fir. Mol. Plant Breed. 1–15.
Wigge, P. A. (2011). FT, a mobile developmental signal in plants. Curr. Biol. 21, R374–R378. doi: 10.1016/j.cub.2011.03.038
Wu, N., Sun, Q., Yao, B., Fan, Y., Rao, H., Hang, M., et al. (2000). Acquisition of insect-resistant transgenic AaIT poplar. Chin. J. Biotechnol. 16, 13–17.
Xia, P., Hu, W., Liang, T., Yang, D., and Liang, Z. (2020). An attempt to establish an Agrobacterium-mediated transient expression system in medicinal plants. Protoplasma 257, 1497–1505. doi: 10.1007/s00709-020-01524-x
Xu, M., Du, Q., Tian, C., Wang, Y., and Jiao, Y. (2021). Stochastic gene expression drives mesophyll protoplast regeneration. Sci. Adv. 7:eabg8466. doi: 10.1126/sciadv.abg8466
Xu, S., and Zhai, Q. (2021). Advances of genetic transformation in woody plants. J. Henan For. Sci. Technol. 41, 10–13.
Xu, S., Zhang, Y., Li, S., Chang, L., Wu, Y., and Zhang, J. (2020). Plastid-expressed Bacillus thuringiensis (Bt) cry3Bb confers high mortality to a leaf eating beetle in poplar. Plant Cell Rep. 39, 317–323. doi: 10.1007/s00299-019-02492-0
Yang, G., Gao, X., Ma, K., Li, D., Jia, C., Zhai, M., et al. (2018). The walnut transcription factor JrGRAS2 contributes to high temperature stress tolerance involving in Dof transcriptional regulation and HSP protein expression. BMC Plant Biol. 18:367. doi: 10.1186/s12870-018-1568-y
Yang, R., Wang, A., Zhang, J., Dong, Y., Yang, M., and Wang, J. (2016). Genetic transformation and expression of transgenic lines of Populus × euramericana with insect-resistance and salt-tolerance genes. Genet. Mol. Res. 15:gmr8635. doi: 10.4238/gmr.15028635
Yang, Y., Tang, R., Jiang, C., Li, B., Kang, T., Liu, H., et al. (2015). Overexpression of the PtSOS2 gene improves tolerance to salt stress in transgenic poplar plants. Plant Biotechnol. J. 13, 962–973. doi: 10.1111/pbi.12335
Yao, W., Wang, S., Zhou, B., and Jiang, T. (2016). Transgenic poplar overexpressing the endogenous transcription factor ERF76 gene improves salinity tolerance. Tree Physiol. 36, 896–908. doi: 10.1093/treephys/tpw004
Yao, W., Zhao, K., Cheng, Z., Li, X., Zhou, B., and Jiang, T. (2018). Transcriptome analysis of poplar under salt stress and over-expression of transcription factor NAC57 gene confers salt tolerance in transgenic Arabidopsis. Front. Plant Sci. 9:1121. doi: 10.3389/fpls.2018.01121
Ye, J., Zhao, D., Liang, Y., Lu, J., and Zheng, X. (2021). Research progress on protoplast preparation of tea plants. J. Tea 47, 75–79.
Yu, D., Wildhagen, H., Tylewicz, S., Miskolczi, P. C., Bhalerao, R. P., and Polle, A. (2019). Abscisic acid signalling mediates biomass trade-off and allocation in poplar. New Phytol. 223, 1192–1203. doi: 10.1111/nph.15878
Yu, M., Yu, Y., Li, H., Yang, J., and Wang, C. (2020). Protoplast isolation from the leaves of Populus trichocarpa and subcellular localization of BpFLA20 gene. J. Northeast For. Univ. 48, 9–14.
Yu, Y., Yin, Q., and Yu, S. (2021). Advances in genetic engineering of forests. For. Prod. Special China 87-88:90. doi: 10.13268/j.cnki.fbsic.2021.03.035
Zhan, Y., Liu, Z., Wang, Y., Wang, Z., Yang, C., and Liu, G. (2001). Transformation of insect resistant gene into birch. J. Northeast For. Univ. 29, 4–6.
Zhang, C., Hampp, R., and Nehls, U. (2005). Investigation of horizontal gene transfer in poplar/Amanita muscaria ectomycorrhizas. Environ. Biosafety Res. 4, 235–242. doi: 10.1051/ebr:2006004
Zhang, D., Das, D. B., and Rielly, C. D. (2013). Potential of microneedle-assisted micro-particle delivery by gene guns: a review. Drug Deliv. 21, 571–587. doi: 10.3109/10717544.2013.864345
Zhang, H., Zhang, Y., Du, Y., and Song, Y. (2021). The effects on the soybean root system of transgenic GmXTH gene by two transgenic methods. Mol. Plant Breed. 1–10.
Zhang, L. (2010). Research of indentifying transformed hemp with pollen tube pathway method by GISH. Heilongjiang Agric. Sci. 8, 14–15.
Zhang, L., Xiong, H., Cao, Q., Zhao, J., and Zhang, H. (2020). Drought resistance of larch NAC gene by transient genetic transformation. Bull. Bot. Res. 40, 394–400.
Zhang, Q., Wei, W., Zhang, H., and Gu, L. (2021). Research progress on regeneration of cell wall from forest tree protoplasts. Mol. Plant Breed. 1–20. Available online at: http://kns.cnki.net/kcms/detail/46.1068.S.20210430.1320.006.html
Zhang, X., Bao, Y., Shan, D., Wang, Z., Song, X., Wang, Z., et al. (2018). Magnaporthe oryzae induces the expression of a MicroRNA to suppress the immune response in rice. Plant Physiol. 177, 352–368. doi: 10.1104/pp.17.01665
Zhao, H., and Chen, F. (2004). Genetic transformation of protoplasts from somatic cells of plants. Acta Bot. Boreali Occidentalia Sin. 24, 1329–1341.
Zhao, H., Jiang, J., Li, K., and Liu, G. (2017). Populus simonii × Populus nigra WRKY70 is involved in salt stress and leaf blight disease responses. Tree Physiol. 37, 827–844. doi: 10.1093/treephys/tpx020
Zhao, H., Wei, J., Lu, J., Shi, C., Wang, H., and Song, Y. (2004). Studies on the cultivation of low lignin content of Populus tomentosa with antisense CCoAOMT gene. Prog. Nat. Sci. 14, 108–112.
Zhao, S., Lu, Y., Zhang, Z., Zhao, Y., and Wang, L. (2014). Protoplast preparation and transient expression system in maize and Arabidopsis. J. Anhui Agric. Sci. 42, 3479–3482.
Zhao, X. (2016). Introduction of exogenous DNA of Populus alba into sect. Aigeiros via pollen-tube pathway. Chin. Bull. Bot. 51, 533–541.
Zhao, Y., Song, D., Sun, J., and Li, L. (2013). Populus endo-beta-mannanase PtrMAN6 plays a role in coordinating cell wall remodeling with suppression of secondary wall thickening through generation of oligosaccharide signals. Plant J. 74, 473–485. doi: 10.1111/tpj.12137
Zheng, L., Liu, G., Meng, X., Li, Y., and Wang, Y. (2012). A versatile Agrobacterium-mediated transient gene expression system for herbaceous plants and trees. Biochem. Genet. 50, 761–769. doi: 10.1007/s10528-012-9518-0
Zhong, L., Xiao, Y., Li, J., Zhang, Y., Zhang, X., Wei, Q., et al. (2021). Establishment of genetic transformation system of Eucalyptus urophylla clone GLU4. Mol. Plant Breed. 1–16.
Zhou, C., Guo, W., Wang, D., Yu, C., Lu, M., and Li, Y. (2005). Exploration of the transformation parameters of the leaf discs of bergamot by particle bombardment in virtue of transient expression of GUS gene. Acta Bot. Boreali Occidentalia Sin. 25, 7–12.
Zhou, X., Dong, Y., Zhang, Q., Xiao, D., Yang, M., and Wang, J. (2020). Expression of multiple exogenous insect resistance and salt tolerance genes in Populus nigra L. Front. Plant Sci. 11:1123. doi: 10.3389/fpls.2020.01123
Zhou, X., Ren, S., Lu, M., Zhao, S., Chen, Z., Zhao, R., et al. (2018). Preliminary study of cell wall structure and its mechanical properties of C3H and HCT RNAi transgenic poplar sapling. Sci. Rep. 8:10508. doi: 10.1038/s41598-018-28675-5
Zhou, Y., Zhang, Y., Wang, X., Han, X., An, Y., Lin, W., et al. (2020). Root-specific NF-Y family transcription factor, PdNF-YB21, positively regulates root growth and drought resistance by abscisic acid-mediated indoylacetic acid transport in Populus. New Phytol. 2, 407–426. doi: 10.1111/nph.16524
Keywords: forest trees, transgenic technology, genetically modified trees, trait improvement, application
Citation: Yin Y, Wang C, Xiao D, Liang Y and Wang Y (2021) Advances and Perspectives of Transgenic Technology and Biotechnological Application in Forest Trees. Front. Plant Sci. 12:786328. doi: 10.3389/fpls.2021.786328
Received: 30 September 2021; Accepted: 25 October 2021;
Published: 30 November 2021.
Edited by:
Horacio Esteban Hopp, University of Buenos Aires, ArgentinaReviewed by:
Minsheng Yang, Agricultural University of Hebei, ChinaCopyright © 2021 Yin, Wang, Xiao, Liang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanwei Wang, eXd3YW5nQGJqZnUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.