- Beijing Key Laboratory of Growth and Developmental Regulation for Protected Vegetable Crops, Department of Vegetable Science, College of Horticulture, China Agricultural University, Beijing, China
Trichomes are unicellular or multicellular epidermal structures that play a defensive role against environmental stresses. Although unicellular trichomes have been extensively studied as a mechanistic model, the genes involved in multicellular trichome formation are not well understood. In this study, we first classified the trichome morphology structures in Capsicum species using 280 diverse peppers. We cloned a key gene (Hairiness) on chromosome 10, which mainly controlled the formation of multicellular non-glandular trichomes (types II, III, and V). Hairiness encodes a Cys2-His2 zinc-finger protein, and virus-induced gene silencing of the gene resulted in a hairless phenotype. Differential expression of Hairiness between the hairiness and hairless lines was due to variations in promoter sequences. Transgenic experiments verified the hypothesis that the promoter of Hairiness in the hairless line had extremely low activity causing a hairless phenotype. Hair controlled the formation of type I glandular trichomes in tomatoes, which was due to nucleotide differences. Taken together, our findings suggest that the regulation of multicellular trichome formation might have similar pathways, but the gene could perform slightly different functions in crops.
Introduction
Trichomes are specialized cell structures that commonly occur in the epidermis of terrestrial plants. Trichomes can exhibit remarkable morphological variation between plant species. Trichomes can be morphologically classified based on the number of cells, or whether they are glandular or branched (Tetsuya et al., 2008; Pattanaik et al., 2014). Due to their special structure, trichomes can serve as a model to study plant cell differentiation (Johnson, 1975; Karabourniotis et al., 2010). Trichomes protect plants against biotic and abiotic stresses, arthropod herbivores, pathogens, and UV-B radiation and prevent water loss (Werker, 2000; Kennedy, 2003; Kang et al., 2014). Secondary metabolites in glandular trichomes have been utilized as food additives, in pharmaceuticals, and as natural pesticides (Aharoni et al., 2006; Schilmiller et al., 2008; Yang et al., 2015).
In Arabidopsis, trichomes are unicellular, dendritic, and non-glandular. Transcriptional regulation of trichome initiation in Arabidopsis involves the expression of GLABRA1 (GL1), GLABRA3 (GL3), ENHANCER OF GLABRA3 (EGL3), and TRANSPARENT TESTA GLABRA1 (TTG1). GL3 and EGL3 physically interact with GL1 and TTG1 separately, forming an MYB–bHLH–WD (MBW) complex (Jürgens, 1994; Larkin, 1994; Payne et al., 2000; Szymanski et al., 2000; Bernhardt et al., 2003). GLABRA2 (GL2) encodes HD-ZIP IV transcription factors that act downstream of the complex (Ishida et al., 2007; Zhao et al., 2008). In addition, several R3 MYB regulators act as negative regulators by competing with GL1 and interacting with GL3/EGL3 to form an inactive complex. The inactive complex disrupts the activity of TTG1-GL3/EGL3-GL1, reduces the expression of GL2, and prevents neighboring cells from entering the process of trichome development (Wang et al., 2007; Wester et al., 2009; Gan et al., 2011). Furthermore, the MBW complex can be activated by Cys2-His2 (C2H2) zinc-finger proteins, such as GIS, ZFP5, ZFP6, and ZFP8, which control trichome formation on inflorescence stems and flowers (Gan et al., 2007; Zhou et al., 2011, 2013; Sun et al., 2015).
Most crops, which produce multicellular trichomes, differ from Arabidopsis in the morphological structure of trichomes. In addition, there are great differences in the morphology of trichomes among different species. For example, tobacco (Nicotiana tabacum L.) has many different types of trichomes, including glandular or non-glandular protection trichomes (Andersen et al., 1988). The protection trichomes have a single row to protect hair and fork hair. Cucumis sativus (cucumber) produces two morphologically distinct trichome types: type I glandular trichomes, which are composed of 3–15 cell bases and 4–18 celled globular glandular heads; and type II glandular trichome, which are also known as “Fruit Spines” (Chen et al., 2016). CsGL1 and CsGL3, which encode HD-ZIP I and HD-ZIP VI transcription factors, are essential for the formation of type II trichomes in cucumber (Li et al., 2015; Wang et al., 2016).
Tomato (Solanum lycopersicum L.), a model plant of Solanaceae, has seven distinct trichome types (I–VII) (Goffreda et al., 1990), of which studies focused mostly on type I and type VI glandular trichomes (Goffreda et al., 1990; Schilmiller and Charbonneau, 2012). Type I multicellular trichome formation is controlled by Wooly (Wo), which encodes an HD-ZIP IV transcription factor, and its interactors SlCycB2 and Hair (H), a B-type protein and a C2H2 zinc-finger protein (Yang et al., 2011; Gao et al., 2017; Chang et al., 2018). SlMYC1 encodes bHLH transcription factors, which when knocked down do not lead to reduced density and smaller type VI glandular trichomes, and regulation of terpene biosynthesis (Xu et al., 2018). CHALCONE ISOMERASE 1 (CHI1) encodes chalcone isomerase of flavonoid biosynthesis, which can reduce the density and flavonoids of type VI glandular trichomes (Kang et al., 2014).
The genus Capsicum produces six main trichome types (types I–VI), including three types of non-glandular (types II, III, and V) and three types of glandular trichomes (types I, IV, and VI) (Kim et al., 2012). However, there have been only a few reports on the trichome genes of pepper. The pepper trichome locus 1 (Ptl1) gene, which was reported in 2010, was mapped to an 80-kb BAC clone on chromosome 10. The mapping region included 14 open reading frames (ORFs); however, no candidate genes were identified (Kim et al., 2010).
In this study, we aimed to observe and categorize the trichome morphology of pepper trichomes. In addition, we used a map-based cloning approach to demonstrate that the Hairiness gene encodes a single C2H2 zinc-protein transcription, which controls the formation of multicellular non-glandular trichomes (types II, III, and V). Virus-induced gene silencing (VIGS) was used to determine the changes in the Hairiness gene that caused trichome loss in the hairiness lines. The CDS sequence of the Hair gene was the same between hair and hairless lines, but the promoter variation affected the Hairiness gene expression level and controlled the hairiness phenotype of pepper. This study could serve as a reference for further research on molecular mechanisms in pepper. We hope our findings can be useful for resistant pepper breeding and hybrid seed production.
Materials and Methods
Plant Materials
A total of 280 accessions used for GWAS were preserved in our laboratory (Wu et al., 2019). Two F2 populations, 18C2480 and 19Q6090, from crossing 18C2458 (hairless) × 18C3375 (hairiness) and 19Q6092 (hairiness) × 19C6093 (hairless), respectively, were used for rough mapping and fine mapping. F2:3 recombination individuals were generated from the 19Q6090 F2 populations.
Scanning Electron Microscope Observation
Samples were cut into 5 × 5 mm square and fixed with 2.5% glutaraldehyde for approximately 24 h. Thereafter, the samples were washed with 0.1 M phosphate buffer (pH 7.2) and dehydrated with 50, 70, and 90% ethanol, respectively. Finally, the samples were dried in a desiccator, and a gold film of 100–150 A was plated on the surface of the sample using an E-1010 (HITACHI) ion sputtering coater. The samples were observed using the scanning electron microscope (SEM; JSM-6390/LV; https://www.jeol.co.jp).
Trichomes Density
The young stem trichome density between the first and the second leaf and the stem between the fourth and fifth leaves, the first young leaf and the seventh leaf, calyx, and petal were dissected using a light microscope (Nikon SMZ18, https://www.nikon.com) at the 10-leaf stage. At least five individuals per line were sampled. All samples were analyzed in triplicate, and the data were expressed as the mean and SD.
Fine Mapping of Hairiness
An F2 population (18C2450) and 12 polymorphic markers were used for rough mapping. Fine mapping was performed using the F2 population (19Q6090) and eight polymorphic markers. Using the F3 recombination individuals from the F2 population (19Q6090) and five InDel markers, which were determined based on the sequenced genome sequences of parent lines to determine candidate interval. The primers used are shown in Supplementary Table 6. The physical location of the markers and genes was based on the pepper genome sequence of CM334v.1.55.
Virus-Induced Gene Silencing
Virus-induced gene silencing (VIGS) was performed according to Chung et al. (2004) with some modifications. Silence fragments were predicted using the SGN VIGS tool (https://vigs.solgenomics.net/). The constructs consisting of pTRV1, pTRV2, pTRV2-PDS, and pTRV2::21340 were transformed into the vector GV3101. A mixture of cultures containing 1:1 (v:v) of pTRV1 and pTRV2 was used as a negative control, and pTRV1 and pTRV2::PDS were used as a reporter. Approximately 1 ml syringes without needles were used to infiltrate the cotyledons of 3-week-old seedlings. The plants inoculated with Agrobacterium were grown in a growth chamber with temperatures 22°C in 16 h light and 20°C in 8 h dark cycles.
Gene Expression Analysis
The RT-PCR and real-time PCR were performed to analyze the expression levels of Hairiness from the hairiness line (19Q6092) and hairless line (19Q6093), respectively. RNA extraction and PCR were performed according to the method described by Liu et al. (2020). The primer sequences are shown in Supplementary Table 7.
Promoter Sequence Variation Analysis
The 2,200 bp sequence of the ATG upstream of the hairy gene was cloned from the hairiness line (parent line 19Q6092) and hairless line (parent line 19Q6093). Homology-based cloning primers (Supplementary Table 7) were designed according to the upstream 2,200 bp sequence in the CDS database of CM334v.1.55 (https://solgenomics.net/tools/blast/). Plant CARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) and DNA MAN (http://www.biologydir.com/dnaman-info-1940.html) were used for sequence analysis.
GUS Staining
The GUS assay was used to measure the promoter activity. Both promoters and short fragments were fused with the GUS reporter and inserted into the vector pCAMBIA1305.1. The primer sequences are shown in Supplementary Table 7. The constructs were transiently expressed in 4-week-old tobacco leaves, mediated by Agrobacterium tumefaciens strain GV3101. A. tumefaciens GV3101 and pCaMV35S::GUS were used as negative and positive controls, respectively. GUS staining solution (5-bromo-4-chloro-3-indolyl b-D-glucuronide, 37°C, 12 h) was used to evaluate GUS activity, and later incubated with 70% ethanol at 65°C to remove chlorophyll, before observing under a light microscope.
Transgenic Analysis
The full-length coding sequence of CA10g21340 from the hairiness line (19Q6092) (primers in Supplementary Table 7) was inserted into the pCAMBIA1305.1::pro21340 (hairiness) and pCAMBIA1305.1::pro21340 (hairless), which replaced GUS. The constructs were transformed into Micro Tom, mediated by the A. tumefaciens strain GV3101 (Fillatti et al., 1987).
Statistical Analysis
All samples were analyzed in triplicates, and the data are expressed as mean ± SD unless noted otherwise. Statistical significance was determined at the 0.05 (*) and 0.01 (**) levels. All experiments were repeated at least twice with three biological replicates each time.
Results
Trichome Morphology and Distribution Characteristics of Capsicum Species
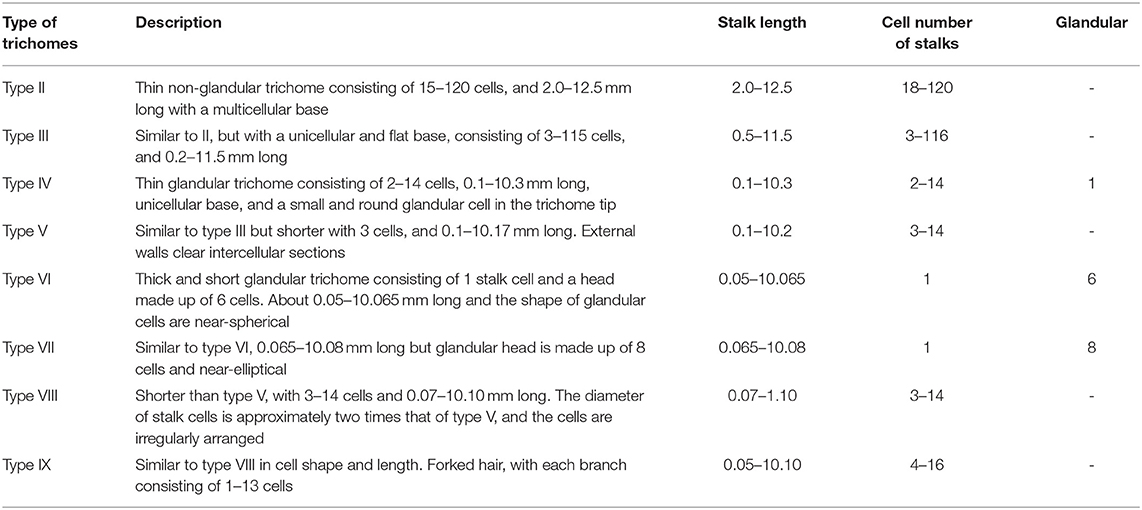
The epidermis of stems, leaves, and flowers of 280 breeding lines of 4 pepper species (Capsicum annuum, C. chinense, C. frutescens, and C. baccatum) were observed for morphological identification of trichomes. There were eight types of trichomes observed, including three types of glandular trichomes (types IV, VI, and VII) and five types of non-glandular trichomes (types II, III, V, VIII, and IX) (Figure 1). Table 1 and Figures 2A–D show the trichome morphology.
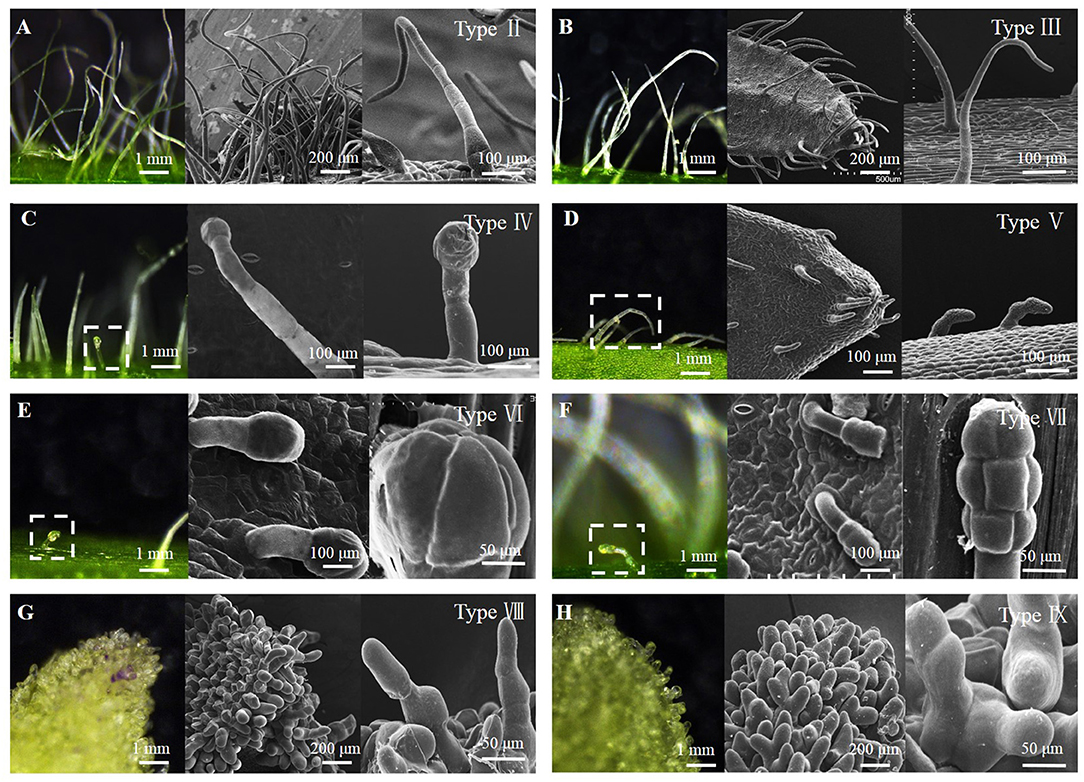
Figure 1. Morphological observation of pepper trichome types. (A) Type II, (B) Type III, (C) Type IV, (D) Type V, (E) Type VI, (F) Type VII, (G) Type VIII, and (H) Type IX. The observation was carried out under light microscopy and SEM.
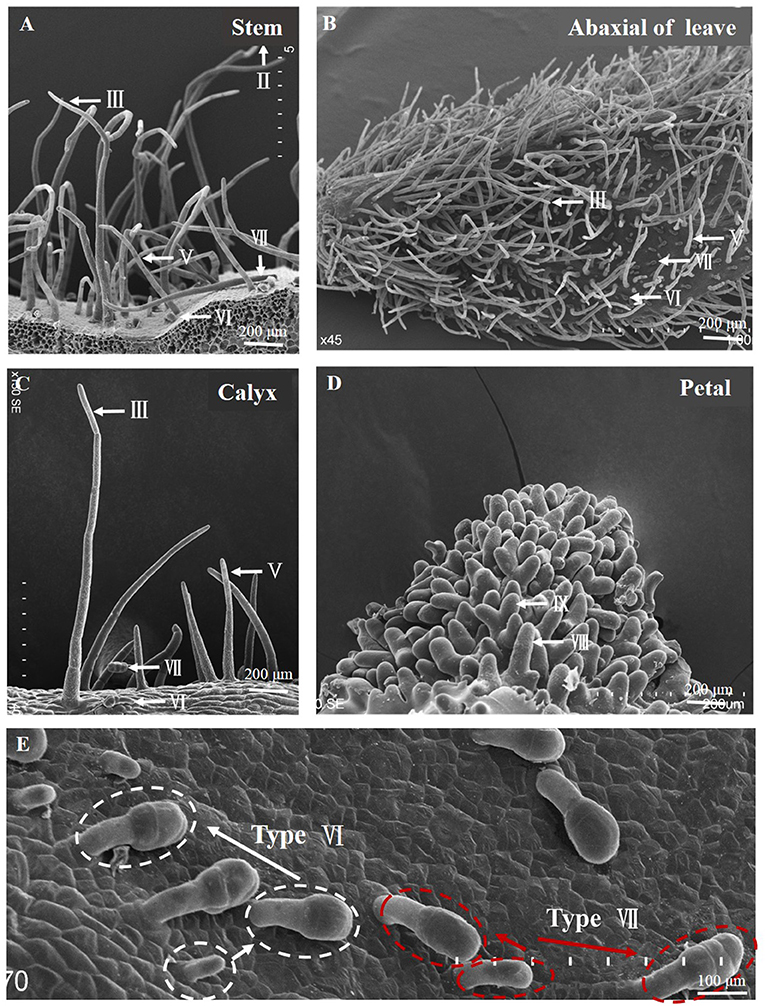
Figure 2. Morphological observation of pepper trichomes on different tissues. (A) Morphology of trichome on the stem (types II, III, V, VI, and VII). (B) Leaf adaxial (types III, V, VI, and VII), (C) Calyx (types III, V, VI, and VII), (D) Petal (types VIII and IX), and (E) Development stage of type VI and type VII.
Based on the statistics of the number of trichomes in different tissues of 280 lines, referring to Luckwill's classification of trichome density of Lycopersicon (Luckwill, 1943), pepper trichomes were divided into four grades: abundant (800–1,500 number/cm2), medium (200–800 number/cm2), sparse (1–200 number/cm2), and no (0 trichomes). Among the observed pepper species, 118 lines were hairless; 106 lines had trichomes on young stems and young leaves (8 sparse, 95 medium, and 3 abundant trichome lines); 40 lines had trichomes that occurred only on leaves (3 sparse, 30 medium, and 7 abundant trichome lines); and 16 lines showed abundant trichomes on all organs (Supplementary Table 1).
Phenotypic Analysis of Trichomes
We examined trichome phenotypes on the stems, leaves, and calyxes of the hairless inbred line “19Q6093,” hairiness inbred line “19Q6092” (CM334), and F1 progeny (Figures 3A,C,E,G,I,K). Hairiness line had four types of trichomes (types II, III, VI, and VII) on young stems, stems, and calyxes; the numbers of these trichomes were 521, 187, 78, and 63 gen/cm2; 100, 72, 4, and 5 gen/cm2; and 12, 54, 13, and 11 gen/cm2, respectively. The numbers of these trichomes (types III, V, VI, and VII) on the adaxial side of young leaves, abaxial side of young leaves, and leaves were 153, 12, 6, and 4 gen/cm2; 190, 10, 130, and 142 gen/cm2; and 78, 12, 3, and 2 gen/cm2, respectively. Hairless lines and F1 progeny were counted using the same statistical method as for the number of trichomes (Figures 3B,D,F,H,J,L).
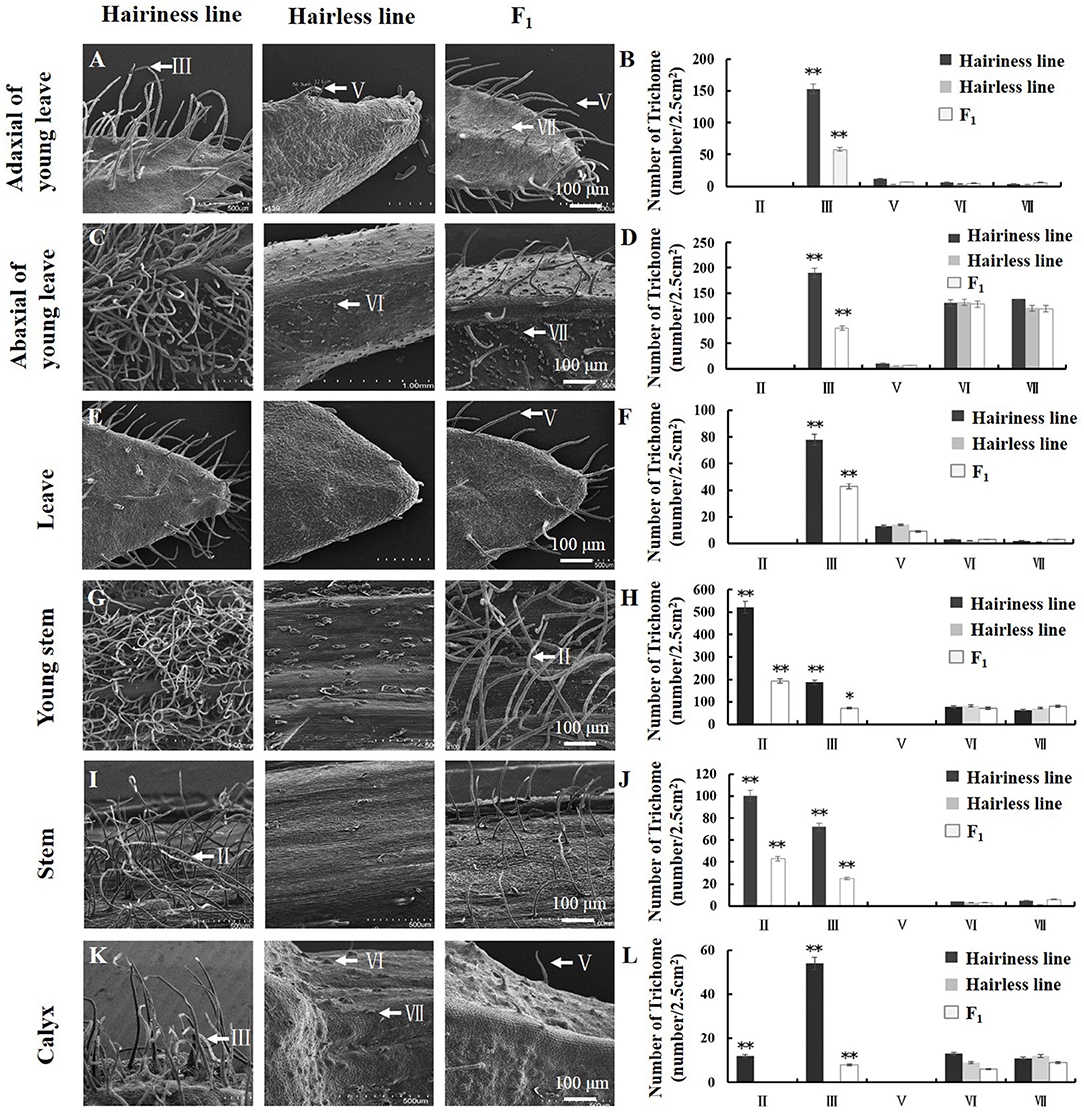
Figure 3. Trichome phenotypes and statistics of parent and F1 generation lines used for fine mapping. (A,B) Type and the number of trichomes on adaxial of young leaves. (C,D) Type and the number of trichomes on abaxial of young leaves. (E,F) Type and number of trichomes on leaves. (G,H) Type and the number of trichomes on the young stem. (I,J) Type and the number of trichomes on the stem. (K,L) Type and the number of trichomes on the calyx. SEM observation of trichomes and the arrows show types of trichomes. Error bars represent SD (n = 3). **p < 0.01, *p < 0.05.
By phenotypic comparison, type VI and VII trichomes were distributed on the hairiness line, hairless line, and F1 plants, with no significant difference in the number of trichomes among them. However, the number of type III trichomes on young leaves and leaves of hairiness line were 2.5- to 13-fold and 2-fold higher, respectively; the number of type II and III trichomes on hair line stems was 2- to 13-fold higher; and the number of type III trichomes on hair line calyxes was 7-fold higher than that on F1 progeny, whereas the hairless lines had no trichomes. These results suggest that the Hairiness gene in this study mainly controlled the formation of multicellular non-glandular trichomes (type II, III, and V) in pepper.
The phenotype of the F2 populations (18C2480 and 19Q6090) was consistent with that of the hairiness, F1 progeny, and hairless lines, with the number of plants 175, 335, and 165, respectively, for 18C2480 and 220, 429, and 220, respectively, for 19Q6090 F2 populations. Both were in good agreement with the expected 3:1 Mendelian ratio (χ2 = 0.862, p > 0.05; χ2 = 0.923, p > 0.05) (Onchiri, 2013) (Supplementary Table 2). These results suggest the presence of a single dominant locus responsible for the non-glandular trichome trait in the tested populations.
Fine Mapping of Hairiness
Hairiness parent lines (19Q6092 and 6903) were screened using 350 SSR markers, which covered the whole genome of pepper. A total of 21 polymorphic markers were obtained; with both markers SR324 and SR519 located on the same side of chromosome 10 of the Hairiness gene.
Twelve polymorphic markers were used to screen the F2 population (18C2480, n = 657) to lock the genetic interval for Hairiness (Figure 4A), and the locus was delimited by SRlj60 and SRlj1012. A total of 869 F2 individuals (19Q6090) and eight markers (HpmE031, SR711, SR724, In2423, Ind440, Ind450, InP2434, and In10-9) were used to narrow the target locus. The Hairiness gene was delimited to the region between HpmE031 and Ind440 (Figure 4B). According to the pepper reference genome sequence, the physical distance was approximately 176.62 kb. In addition, we identified five individuals with recombination near HpmE031 and Ind440. Using the F3 recombination individuals (Supplementary Table 3) and five InDel markers (Ind21340-1, In21380, Ind21430, and In21440-1) delimited the Hairiness trait for a 70-kb region, which encompassed four annotation genes (Figures 4C,D and Supplementary Table 4).
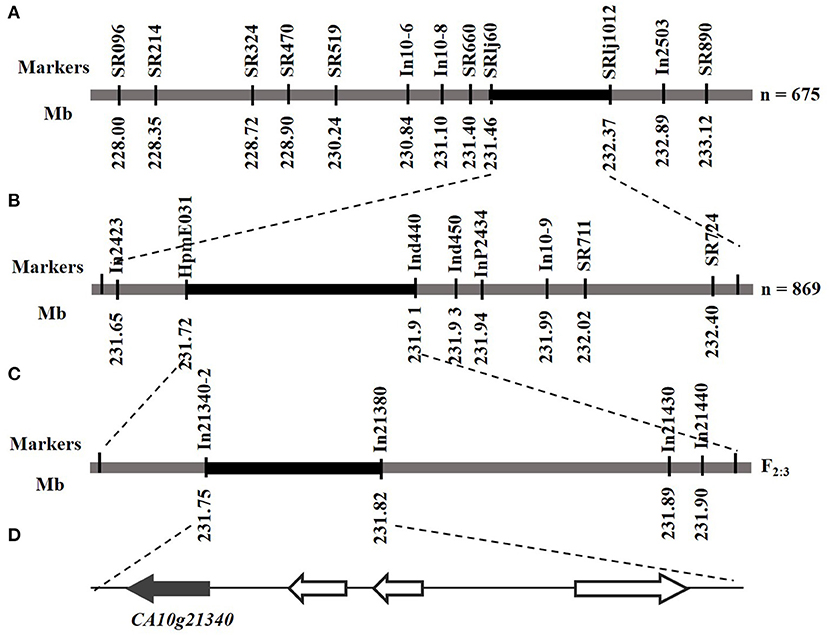
Figure 4. Genetic and physical maps of the Hairiness gene and candidate gene analysis. (A) Rough mapping of Hairiness, where n represents F2 population from the cross of 18C2458 (hairless) × 18C3375 (hairiness). Based on 12 markers and 675 F2 individuals, Hairiness was delimited between the marker SRlj60 and SRlj1012. (B) Fine mapping of Hairiness; Hairiness was mapped in a 176-kb region between markers HpmE031 and Ind440 with 869 F2 individuals (19Q6090) from the cross of 19Q6902 (hairiness) × 19Q6093 (hairless). (C) F3 exchange individuals from F2 individuals (19Q6090) to reduce the location region; positions of the markers are indicated in 70 kb region between markers In21340-2 and In21380. (D) The location of annotation genes in the target region, according to the CM334 genomics network (https://solgenomics.net/).
Identifying Candidate Genes
To determine which was Hairiness, we sequenced and analyzed the expression levels of the four annotation genes in the two parental lines. DNA sequences of four annotation genes were consistent, and only CA10g21340 showed higher expression levels in hairiness lines than in hairless lines (Supplementary Figure 1). Thus, CA10g21340 is a likely candidate gene for Hairiness.
To confirm the function of CA10g21340, VIGS was applied to pepper plants. Compared with the negative control (Figures 5A,C,E,G), the silenced plants showed a hairless phenotype (Figures 5B,D,F,H), and the number of types II and III non-glandular trichomes on the leaves and stems of TRV1 + TRV2::CA10g21340 infiltrated plants decreased significantly, and the number of type V non-glandular trichomes decreased (Figures 5G,H). The expression levels of CA10g21340 in the young leaves and young stems of TRV1 + TRV2::CA10g21340 infiltrated plants were almost 5 and 8% of that of the negative control (Figures 5I,J). Therefore, it could be presumed that CA10g21340 was the Hairiness gene that underlies the hairy phenotype.
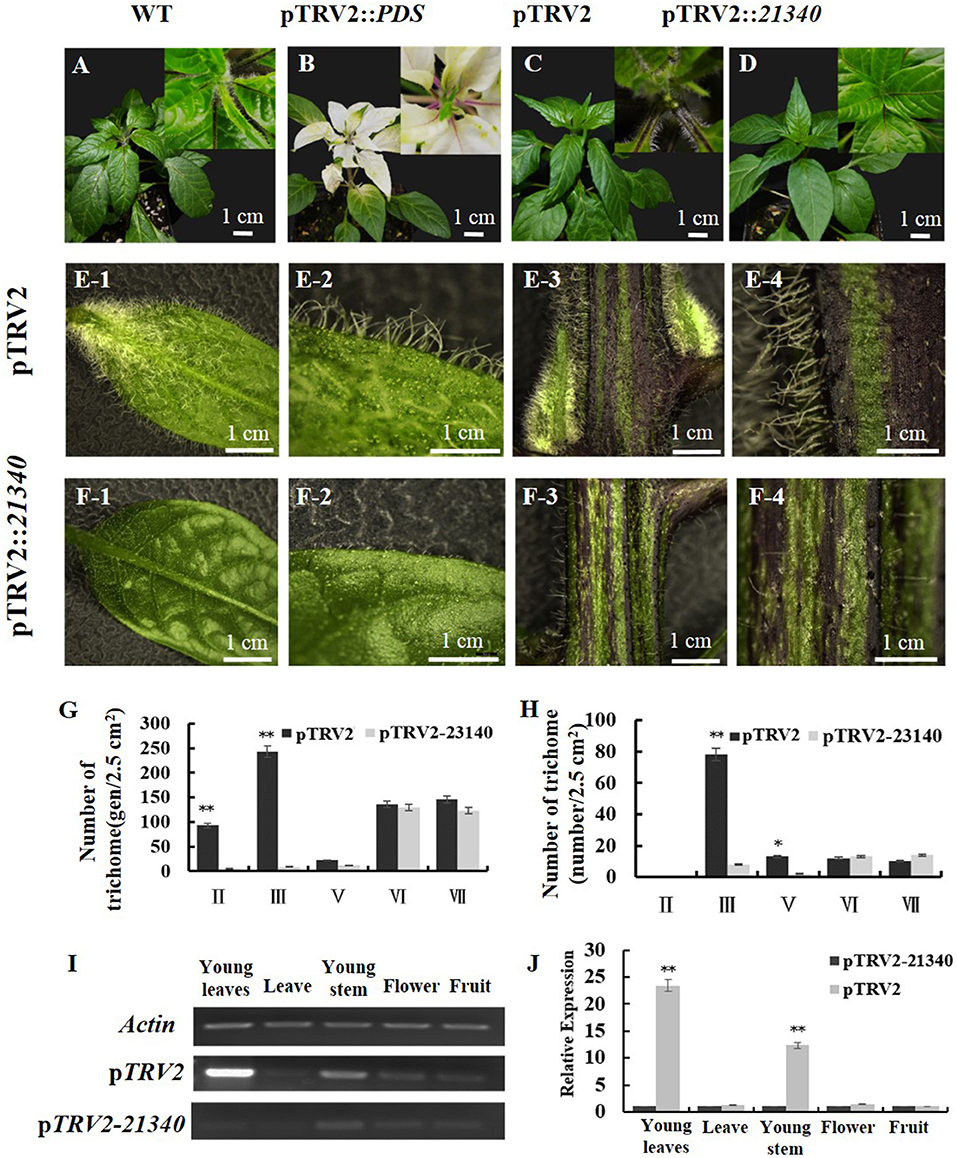
Figure 5. The phenotype of VIGS-treated plants. (A) Genotype hairiness line (CM334). (B) Expression of PDS in CM334, the plant showed photo-bleached phenotype. (C) and (E1–E4) Expression of pTRV2 in CM334, and phenotype of the plants are similar to WT. (D) and (F1–F4) Expression of pTRV2-21340 in CM334, and decreased trichomes of those plants. (G) Statistics of trichomes on leaves between pTRV2 and pTRV2-21340 infiltrated plants. (H) Statistics of trichomes on stems between WT and pTRV2-21340 infiltrated plants. (I,J) The relative expression level of CA10g21340 in differently infiltrated plants. Error bars represent the SD (n = 3). **p < 0.01, *p < 0.05. Reference gene: UIB-3 (Capana06g002873).
Hairiness Encodes a C2H2 Zinc-Finger Protein
CA10g21340 is predicted to have two exons and one intron, which encodes a putative C2H2 domain. We evaluated the phylogenetic relationship of Hairiness with several members from Arabidopsis and tomato. Phylogenetic analysis revealed that Hairiness is closely related to the H (Solyc10g078970) and GIS (AT3G58070.1) genes in tomato and Arabidopsis (Supplementary Figure 2). The H gene promoted the formation of type I multicellular trichomes, and GIS controlled the trichome formation of inflorescence stems and flowers.
Promoter Sequence Variation of Hairiness and Hairless Lines
Differential expression of Hairiness was observed in hairiness and hairless lines. Hairiness expression levels were detected in seven organs of the two parental lines (Figures 6A,B). Compared with hairless lines, the expression of Hairiness in young stems, young leaves, leaves, flowers, and fruits of hairiness lines increased significantly (Figures 6C,D).
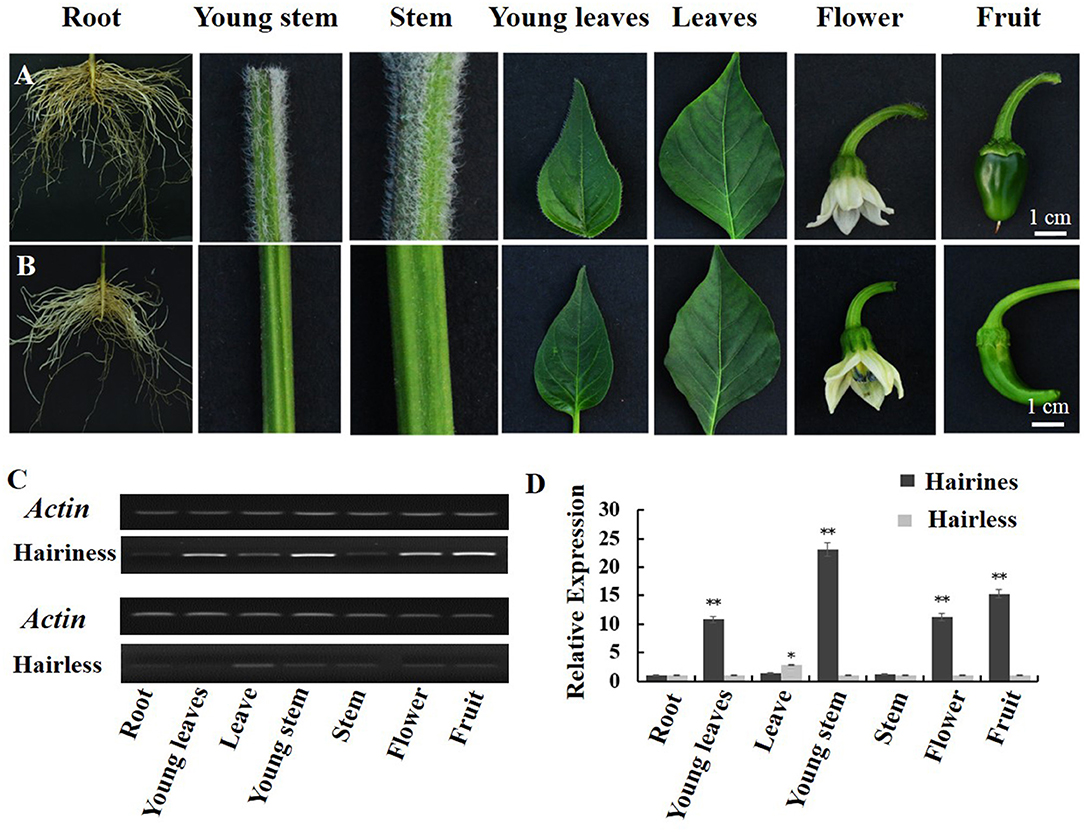
Figure 6. Relative expression levels of CA10g21340 in different tissues of hairiness and hairless lines. (A) Organ genotypes of hairiness lines (root, young stem, stem, young leaves, leaves, flower, and fruit). (B) Organ genotypes of hairless lines. (C,D) RT-PCR and real-time PCR analysis of CA10g21340. Error bars represent the SD (n = 3). **p < 0.01, *p < 0.05. Reference gene: UIB-3 (Capana06g002873).
To investigate the reason for differential expression of Hairiness in hairiness and hairless lines, an approximately 2.3 kb sequence upstream of ATG was amplified from these two lines. There were 4 large InDels, 2 small InDels, 19 SNPs, and no sequence variation in the 5 UTR (121 bp) (Figure 7A and Supplementary Figure 3).
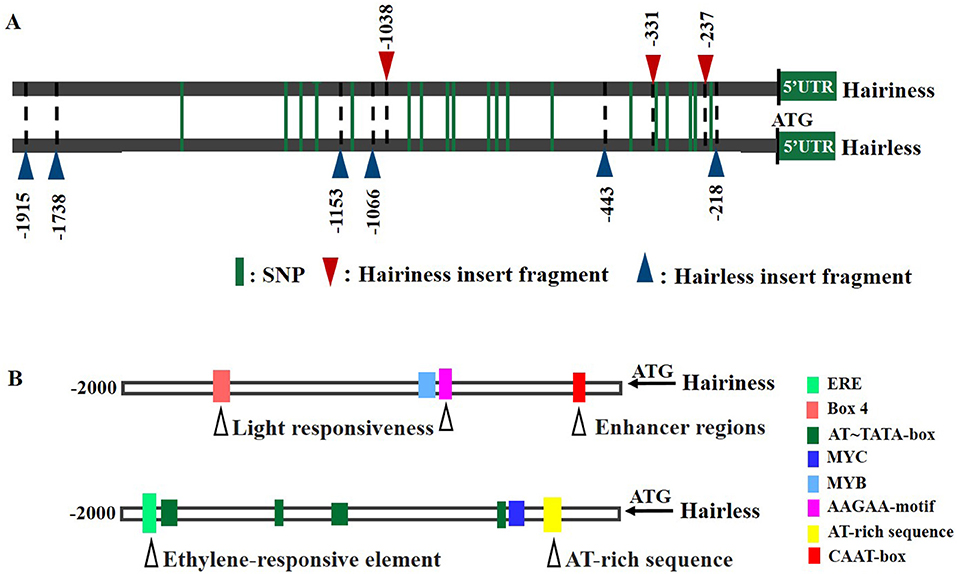
Figure 7. Promoter sequence differences of CA10g21340 between hairiness and hairless lines. (A) A representation of promoter sequences of hairiness and hairless lines. Green lines represent a single nucleotide polymorphism. The red triangle represents the hairiness line insert fragment; the Blue triangle represents the hairless line insert fragment. (B) Promoter region cis-acting element differences between hairiness and hairless lines. The black triangle represents a variation of the cis-acting element.
However, the cis-acting elements of hairiness and hairless line promoter sequences, when analyzed using the PlantCARE database (Figure 7B). The hairiness promoter had one more Box-4 (light response element), MYB (stress response element), CAAT box, and AAGAA-motif (other response elements), but lacked one ERE (hormone response element), one MYC (stress response element), one AT-rich sequence, and five AT-TATA-boxes (other response elements) (Supplementary Table 5) as compared to the hairless promoter.
Differential Expression of Hairiness Is Caused by Promoter Sequence Variation
Both promoters and their five short fragments, from ATG to upstream −1690, −1108, −582, −324, and −240 bp were separately fused with GUS to determine their activities. The GUS activity in tobacco showed that both promoters had promoter activity, but the promoter activity of Hairiness from hairless lines was significantly lower than that of hairiness lines. The GUS activity of the short fragments of promoters in the hairless line showed weak light blue, whereas, in the hairiness line, it was dark-blue to blue-black. The activity of the Hairiness gene promoter in the hairiness line was significantly higher than that in the hairless line (Figure 8A). These results suggest that promoter sequence diversity might lead to activity differences, resulting in the differential expression patterns in hairiness and hairless lines.
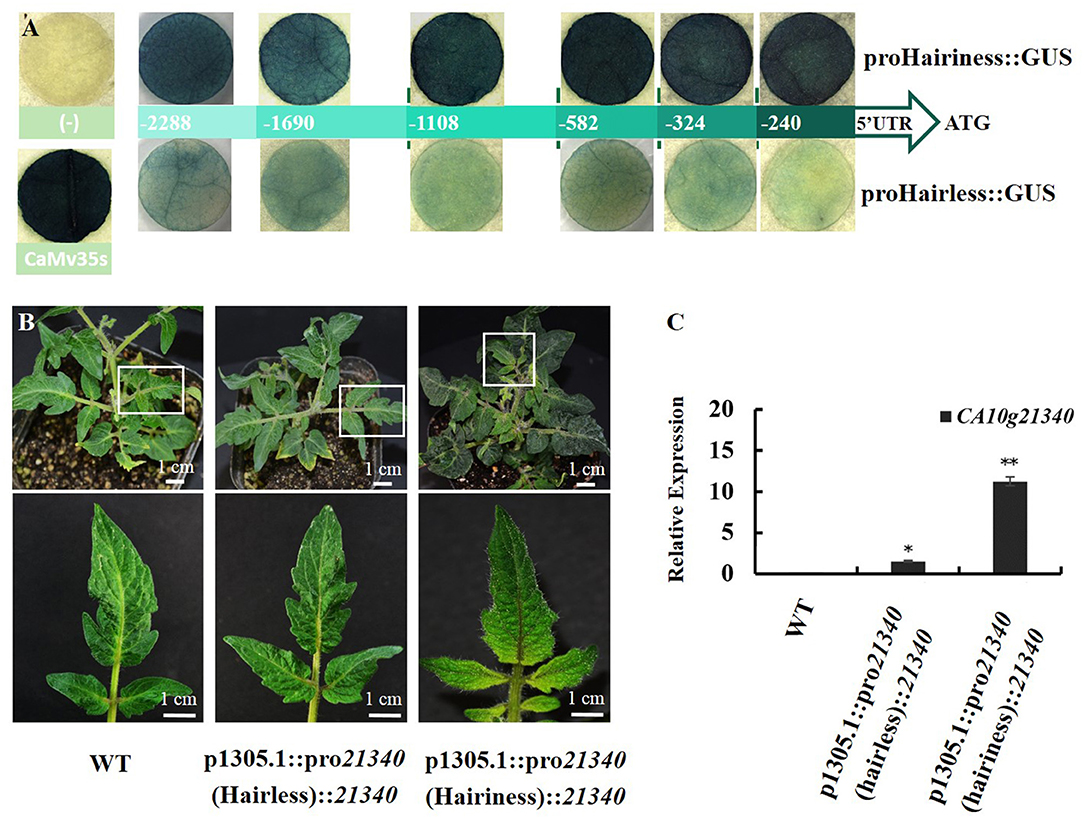
Figure 8. Comparative analysis of CA10g21340 promoter activities and genetic transformation of tomato phenotype. (A) GUS activity in leaves of Nicotiana benthamiana transiently expressing promoter short fragments between hairiness and hairless line insertion pCAMBIA1305.1. (B) Genetic transformation of tomato phenotype: CA10g21340 gene driven by pro21340 (Hairiness) and pro21340 (Hairless) in Micro Tom. (C) Expression analysis of CA10g21340 with transformation tomato. Reference gene: Actin (Solyc11g005330).
Promoter activities were compared by connecting the two promoters with CDS of Hairiness. Two constructs, proHairiness::CDS and proHairless::CDS, were developed to perform transient transformation experiments. There was no significant difference in the number of trichomes between Hairiness driven by the promoter from hairless line and control, whereas the number of trichomes was considerably higher when Hairiness was driven by the hairiness line than by the hairless line. The number of types III, V, and VI trichomes on the leaves of hairiness line promoter transgenic tomato was 3- to 4-fold higher than that in the other two (Figure 8B and Supplementary Figure 4). In addition, the expression levels of CA10g21340 in transgenic plants with hairiness promoter and hairless promoter were approximately 12 and 2 times higher, respectively, than those in control plants (Figure 8C). These results suggest that differential expression of Hairiness between hairiness and hairless lines might be caused by the variations in the gene promoter sequence, which in turn caused the hairless phenotype.
Discussion
Hairiness Triggered Non-Glandular Trichome Formation in Pepper
Morphology of unicellular trichomes of Arabidopsis thaliana (Tetsuya et al., 2008; Pattanaik et al., 2014), and multicellular trichomes of tomato (Luckwill, 1943) and cucumber (Chen et al., 2014) are well-studied. In addition, a previous study had reported on the morphology of pepper trichomes (Kim et al., 2012).
Through the observation, statistics, and morphological description of the trichomes of 280 pepper species (Figure 1, Table 1), trichomes can be divided into eight types (types II–IX) (Figure 1). According to the presence/absence of glandular cells, they could be classified into three (types IV, VI, and VII) glandular trichome and five (types II, III, V, VIII, and IX) non-glandular trichome types. Only one previous study (Kim et al., 2012) has reported on the classification of pepper trichomes, including five pepper species (C. annuum, C. chinense, C. frutescens, C. pubescens, and C. baccatum). In the present study, we investigated the trichomes of stems and leaves and reported six main trichome types (types I–V and VII), including three types of non-glandular trichomes (types II, III, and V) and three types of glandular trichomes (types I, IV, and VII).
Compared with the results of our study, the differences of glandular trichomes were types I and VII. Type I trichomes contained a long (~2 mm) multicellular stalk, a gland cell at the top, which occurred only on the stem and leaves of C. chinensis. Our investigation included four lines of C. chinensis, all of which were hairless phenotypes, and no type I was observed. Type VI and VII glandular trichomes were regarded as the same type because both had a similar structure and length (less than 0.1 mm) (Figure 1, Table 1). However, at the late stage of trichome development, the differentiation of glandular cells was significantly different in the morphology and the numbers of Type VI and VII glandular trichomes, as observed under the electron microscope (Figure 2E). Therefore, they can be classified as type VI and VII glandular cells. The differences in non-glandular trichomes, types VIII and IX, were only observed at the top edge of petals, were shorter (<0.1 mm), and the stalk cell was larger and rounder than other trichome cells. Since Kim did not observe the trichomes of flower organs, non-glandular trichomes of types VIII and IX were not reported. Overall, there were nine trichome types (types II–IX) of pepper, and based on the presence/absence of glandular cells, they can be classified into four glandular trichomes (types I, IV, VI, and VII) and five non-glandular trichomes (types II, III, V, VIII, and IX).
Phenotypic statistics are key to gene mapping. Based on the investigation of hair-type parental lines, F1 generation, and F2 population, we determined that the hairiness phenotype referred to non-glandular trichomes of types II, III, and V.
CA10g21340 Is a Strong Candidate Gene of Hairiness
Several genes have been identified as potential candidates for known multicellular trichome formation in tomato, cucumber, and tobacco through genetic mapping (Rick and Butler, 1956; Serna and Martin, 2006; Yang et al., 2014; Lashbrooke et al., 2015; Li et al., 2015; Zhao et al., 2015; Cui et al., 2016). However, genes related to trichome formation have been poorly studied in pepper plants. In 2004, Du Feng used a hairless line (02090) and hairiness line (02091) and by segregation population, genetic analysis showed that the hairiness trait of pepper was controlled by a single dominant locus. Random amplification of polymorphic DNA (RAPD) analysis of the F2 population was carried out using the BSA method. The hairiness exchange value of RAPD marker S1513630 was 3.97%, and the genetic distance was 3.98 cM; however, no follow-up tests were carried out (Du, 2004). In 2010, Kim constructed an F2 population by screening a pepper BAC library and using a hairiness line (CM334) and hairless line (Chilsungcho cultivar). The Ptl1 gene was mapped to the physical distance of 23.9 kb between markers HpmE031 and Tco on chromosome 10, in which there were 14 ORFs, but no candidate gene was identified (Kim et al., 2010). We blasted the markers HpmE031 and Tco, which are closely linked to Ptl1 in CM334v.1.55. The mapping interval was located on 231711985–231736354 of chromosome 10. In the present study, the Hairiness gene was also located by mapping-based cloning, using the marker HpmE031 to locate Hairiness in 231711985–231900189 of chromosome 10 between HpmE031 and Ind440, which included the location interval reported by Kim. We further reduced the mapping interval to 231754268–231815719 by using F3 exchange individuals and four InDel markers, which was approximately 20 kb downstream of that reported by Kim.
Four annotated genes were located in the 70-kb region of Hairiness, and we found that Ca10g21340 had the highest homology with GIS of Arabidopsis and H of tomato, which encoded C2H2 zinc-finger protein. GIS (AT3G58070) regulates the formation (An et al., 2012a) and branch cells of trichomes (An et al., 2012b) on the inflorescence of main stems, which is involved in regulating the transformation of developmental stages. H (Solyc10g078970) was involved in the multicellular trichome formation of types I and VI, which regulated the development of epidermal cells. The C2H2 zinc-finger domain is conserved in monocotyledons and dicotyledons and may have similar functions. In Arabidopsis, ZFP5 regulates the formation of trichomes on flowers, leaves, branches, and main stem inflorescences, whereas ZFP6 promotes the growth of trichomes on flowers and main stems (Gan et al., 2007; Zhou et al., 2011, 2013; Sun et al., 2015). Therefore, CA10g21340 might be a candidate gene for controlling hairiness formation in pepper plants. The expression of the VIGS suppressor gene in hairiness lines could significantly reduce the number of hairy plants, which further confirmed CA10g21340 as a strong candidate for the pepper hairiness gene.
In addition, in this study, transient expression in tobacco proved that the variation in promoter sequences led to a low expression of Hairiness in hairless lines, which could not form multicellular non-glandular trichomes. In tomato, H controlled the formation of type I glandular trichomes of tomato, which had 45% homology with Hairiness in pepper (Chang et al., 2018). Overexpression of Hairiness in tomatoes caused significant increases in trichome density. The findings of this study suggest that the regulation of multicellular trichome formation might have similar pathways, but the gene could perform slightly different functions in crops.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
JinqL, ML, and HW planned and designed the experiments. JinqL, JinkL, and SL phenotypic were observed and imaged by SEM. HW, JinqL, and ML planted and maintained field experiments. JinqL, ML, and QC evaluated the genetic transformation of tomatoes. HW, JinkL, and JinqL wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Key Research and Development Program of China (2017YFD0101903), the Beijing Fruit Vegetables Innovation Team of Modern Agricultural Industry Technology System (BAIC01-2021), and the Construction of Beijing Science and Technology Innovation and Service Capacity in Top Subjects (CEFF-PXM2019_014207_000032).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.784755/full#supplementary-material
References
Aharoni, A., Jongsma, M. A., Kim, T. Y., Ri, M. B., Giri, A. P., Verstappen, F. W. A., et al. (2006). Metabolic engineering of terpenoid biosynthesis in plants. Phytochem. Rev. 5, 49–58. doi: 10.1007/s11101-005-3747-3
An, L., Zhou, Z., Sha, S., An, Y., and Gan, Y. (2012a). GIS is required for trichome branching through GA signaling in Arabidopsis. Plant Cell Physiolo. 4, 623–631. doi: 10.1093/pcp/pcr192
An, L., Zhou, Z., Sun, L., Yan, A., and Gan, Y. (2012b). A zinc finger protein gene ZFP5 integrates phytohormone signaling to control root hair development in Arabidopsis. Plant J. 72, 474–490. doi: 10.1111/j.1365-313X.2012.05094.x
Andersen, R. A., Hamilton-Kemp, T. R., Loughrin, J. H., Hughes, C. G., Hildebrand, D. F., and Sutton, T. G. (1988). Green leaf headspace volatiles from Nicotiana tabacum lines of different trichome morphology. J. Agric. Food Chem. 36, 295–299. doi: 10.1021/jf00080a014
Bernhardt, C., Lee, M. M., Gonzalez, A., Zhang, F., Lloyd, A., and Schiefelbein, J. (2003). The bHLH genes GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) specify epidermal cell fate in the Arabidopsis root. Mol. 130, 6431–6435. doi: 10.1242/dev.00880
Chang, J., Yu, T., Yang, Q., Li, C., Xiong, C., Gao, S., et al. (2018). Hair, encoding a single C2H2 zinc finger protein, regulates multicellular trichome formation in tomato. Plant J. 67, 412–420. doi: 10.1111/tpj.14018
Chen, C., Yin, S., Liu, X., Liu, B., Yang, S., Xue, S., et al. (2016). The WD-Repeat Protein CsTTG1 Regulates Fruit Wart Formation through Interaction with the Homeodomain-Leucine Zipper I Protein Mict. Plant Physiol.171, 1156–1162. doi: 10.1104/pp.16.00112
Chen, M., Li, L, Jiang, X. F, Li, J. Y., et al. (2014). Transcriptome profiling reveals roles of meristem regulators and polarity genes during fruit trichome development in cucumber (Cucumis sativus L.). J. Exp. Bot. 17, 4943–4951. doi: 10.1093/jxb/eru258
Chung, A. E., Seong, E., Lee, S., Joung, Y. H., Oh, K., Chung, E., et al. (2004). A method of high frequency virus-induced gene silencing in chili pepper (Capsicum annuum L. cv. Bukang). Mol. Cell. 2, 377–380.
Cui, J. Y., Miao, H., Ding, L. H., Wehner, T. C., Liu, P. N., Wang, Y., et al. (2016). A new glabrous gene (csgl3) identified in trichome development in Cucumber (Cucumis sativus L.). PLoS ONE. 23, 167–173. doi: 10.1371/journal.pone.0148422
Fillatti, J. J., Kiser, J., Rose, R., and Comai, L. (1987). Efficient transfer of a glyphosate tolerance gene into Tomato using a binary agrobacterium tumefaciens vector. Nat. Biotechnol. 5, 726–730. doi: 10.1038/nbt0787-726
Gan, L., Kai, X., Chen, J. G., and Wang, S. (2011). Functional characterization of TRICHOMELESS2, a new single-repeat R3 MYB transcription factor in the regulation of trichome patterning in Arabidopsis. BMC Plant Biol. 11, 176–183. doi: 10.1186/1471-2229-11-176
Gan, Y., Liu, C., Yu, H., and Broun, P. (2007). Integration of cytokine and gibberellin signaling by Arabidopsis transcription factors GIS, ZFP8 and GIS2 in the regulation of epidermal cell fate. Development 134, 2073–2081. doi: 10.1242/dev.005017
Gao, C., Dong, L., Jin, C., Duan, S., Qi, S., Liu, K., et al. (2017). Genome-wide identification of GLABRA3 downstream genes for anthocyanin biosynthesis and trichome formation in Arabidopsis. Biochem. Bioph. Res. Co. 51, 173–181. doi: 10.1016/j.bbrc.2017.02.074
Goffreda, J. C., Szymkowiak, E. J., and Mutschler, S. (1990). Chimeric tomato plants show that aphid resistance and triacylglucose production are epidermal autonomous characters. Plant Cell. 2, 643–649. doi: 10.1105/tpc.2.7.643
Ishida, T., Hattori, S., Sano, R., Inoue, K., Shirano, Y., Hayashi, H., et al. (2007). Arabidopsis TRANSPARENT TESTA GLABRA2 is directly regulated by R2R3 MYB transcription factors and is involved in regulation of GLABRA2 transcription in epidermal differentiation. Plant Cell. 19, 2531–2543. doi: 10.1105/tpc.107.052274
Ishida, T., Kurata, T., Kiyotaka, O., and Wada, T. (2008). A genetic regulatory network in the development of trichomes and root hairs. Annu. Rev. Plant Biol. 59, 365–386. doi: 10.1146/annurev.arplant.59.032607.092949
Johnson, H. B. (1975). Plant pubescence: an ecological perspective. Bot. Rev. 41, 233–258. doi: 10.1007/BF02860838
Jürgens, M. H. A. S. (1994). Genetic dissection of trichome cell development in Arabidopsis. Cell. 3, 555–566. doi: 10.1016/0092-8674(94)90118-X
Kang, J., McRoberts, J., E J, Daniel Jones, A., and Howe, G.A. (2014). The flavonoid biosynthetic enzyme chalcone isomerase modulates terpenoid production in glandular trichomes of tomato. Plant Physiol. 34, 108–112. doi: 10.1104/pp.113.233395
Karabourniotis, G., Papadopoulos, K., Papamarkou, M., and Manetas, Y. (2010). Ultraviolet-B radiation absorbing capacity of leaf hairs. Physiol. Plantarum. 86, 414–418. doi: 10.1111/j.1399-3054.1992.tb01337.x
Kennedy, G. G. (2003). Tomato, pests, parasitoids, and predators: tritrophic interactions involving the genus Lycopersicon. Annu. Rev. Entomol. 48, 51-72. doi: 10.1146/annurev.ento.48.091801.112733
Kim, H.J., Han, J.H., Kwon, J.K., Park, M., Kim, B.D., and Choi, D. (2010). Fine mapping of pepper trichome locus 1 controlling trichome formation in Capsicum annuum L. CM334. Theor. Appl. Genet. 120, 1099–1105. doi: 10.1007/s00122-009-1237-5
Kim, H. J., Seo, E. Y., Kim, J. H., Cheong, H. J., and Choi, D. I. (2012). Morphological classification of trichomes associated with possible biotic stress resistance in the genus Capsicum. Plant Pathol. J. 28, 107–113. doi: 10.5423/PPJ.NT.12.2011.0245
Larkin, J. C. (1994). Roles of the GLABROUS1 and TRANSPARENT TESTA GLABRA genes in Arabidopsis trichome development. Plant Cell. 8, 1065–1076. doi: 10.1105/tpc.6.8.1065
Lashbrooke, J. G., Adato, A., Lotan, O., Alkan, N., Tsimbalist, T., Rechav, K., et al. (2015). The tomato MIXTA-Like transcription factor coordinates fruit epidermis conical cell development and circular lipid biosynthesis and assembly. Plant Physiol. 14,383–392. doi: 10.1104/pp.15.01145
Li, Q., Cao, C., Zhang, C., Zheng, S., and Wang, Z. (2015). The identification of Cucumis sativus Glabrous 1 (CsGL1) required for the formation of trichomes uncovers a novel function for the homeodomain-leucine zipper I gene. J. Exp. Bot. 7, 142–151. doi: 10.1093/jxb/erv046
Liu, J. Q., Ai, Y. X., Wang, Y. H., and Shen, H. L. (2020). Fine mapping of the Ca3GT gene controlling anthocyanin biosynthesis in mature unripe fruit of Capsicum annuum L. Theor. Appl. Genet. 133, 2729–2742. doi: 10.1007/s00122-020-03628-7
Luckwill, L. C. (1943). The genus Lycopersicon: a historical, biological and taxonomic survey of the wild and cultivated tomato. Aberdeen Univ. Study. 5, 120–144.
Onchiri, S. (2013). Conceptual model on application of chi-square test in education and social sciences. Educ. Res. Rev. 8, 90–98.
Pattanaik, S., Patra, B., Singh, S. K., and Yuan, L. (2014). An overview of the gene regulatory network controlling trichrome development in the model plant, Arabidopsis. Front Plant Sci. 5, 705–713. doi: 10.3389/fpls.2014.00259
Payne, C. T., Zhang, F., and Lloyd, A. M. (2000). GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1. Genetics. 156, 1349–1362. doi: 10.1093/genetics/156.3.1349
Rick, C. M., and Butler, L. (1956). Cytogenetics of the Tomato. Adv. Genet. 7, 267–382. doi: 10.1016/S0065-2660(08)60504-0
Schilmiller, A. L., and Charbonneau, R. L. (2012). Identification of a BAHD acetyltransferase that produces protective acyl sugars in tomato trichomes. Pro. Nat. Acade. Sci. 40, 16377–16382. doi: 10.1073/pnas.1207906109
Schilmiller, A. L., Last, R. L., and Pichersky, E. (2008). Harnessing plant trichome biochemistry for the production of useful compounds. The Plant J. 54, 702–711. doi: 10.1111/j.1365-313X.2008.03432.x
Serna, L., and Martin, C. (2006). Trichomes: different regulatory networks lead to convergent structures. Trends Plant Sci. 6, 274–280. doi: 10.1016/j.tplants.2006.04.008
Sun, L., Zhang, A., Zhou, Z., Zhao, Y., Yan, A., Bao, S., et al. (2015). GLABROUS INFLORESCENCE STEMS3 (GIS3) regulates trichome initiation and development in Arabidopsis. New Phytol. 206, 1202–1210. doi: 10.1111/nph.13218
Szymanski, D. B., Lloyd, A. M., and Marks, M. D. (2000). Progress in the molecular genetic analysis of trichome initiation and morphogenesis in Arabidopsis. Trends Plant Sci. 5, 214–223. doi: 10.1016/S1360-1385(00)01597-1
Wang, S., Kwak, S. H., Zeng, Q., Ellis, B. E., and Chen, J. G. (2007). TRICHOMELESS1 regulates trichome patterning by suppressing GLABRA1 in Arabidopsis. Development 134, 3873–3882. doi: 10.1242/dev.009597
Wang, Y. L., Nie, J. T., Chen, H. M., Guo, C. L., and Cai, R. (2016). Identification and mapping of Tril, a homeodomain-leucine zipper gene involved in multicellular trichome initiation in Cucumis sativus. Theor. Appl. Genet. 2, 305–312. doi: 10.1007/s00122-015-2628-4
Werker, E. (2000). Trichome diversity and development. Adv. Bot. Res. 31, 1–35. doi: 10.1016/S0065-2296(00)31005-9
Wester, K., Digiuni, S., Geier, F., Timmer, J., Fleck, C., and Hulskamp, M. (2009). Functional diversity of R3 single-repeat genes in trichome development. Development 136, 1487–1496. doi: 10.1242/dev.021733
Wu, L., Wang, P., Wang, Y., Cheng, Q., Liu, J., Sun, L., et al. (2019). Genome-wide correlation of 36 agronomic traits in the 287 pepper (Capsicum) accessions obtained from the SLAF-seq-Based GWAS. Inter. J. Mol. Sci. 20, 162–183. doi: 10.3390/ijms20225675
Xu, J., Van Herwijnen, Z. O., Dräger, D. B., Sui, C., Haring, M. A., and Schuurink, R. C. (2018). SlMYC1 regulates type VI glandular trichome formation and terpene biosynthesis in tomato glandular cells. Plant Cell. 12, 521–529. doi: 10.1105/tpc.18.00571
Yang, C., Gao, Y., Gao, S., Yu, G., Xiong, C., Chang, J., et al. (2015). Transcriptome profile analysis of cell proliferation molecular processes during multicellular trichome formation induced by tomato Wov gene in tobacco. BMC Genom. 16, 868–874. doi: 10.1186/s12864-015-2099-7
Yang, C., Li, H., Zhang, J., Wang, T., and Ye, Z. (2011). Fine-mapping of the woolly gene controlling multicellular trichome formation and embryonic development in tomato. Theor. Appl. Genet. 123, 625–633. doi: 10.1007/s00122-011-1612-x
Yang, X., Zhang, W., He, H., Nie, J., and Cai, R. (2014). Tuberculate fruit gene Tu encodes a C2H2 zinc finger protein that is required for the warty fruit phenotype in cucumber (Cucumis sativus L.). Plant J. 6, 321–329. doi: 10.1111/tpj.12531
Zhao, J. L., Pan, J. S., Guan, Y., Zhang, W. W., Bie, B. B., Wang, Y. L., et al. (2015). Micro-trichome as a class I homeodomain-leucine zipper gene regulates multicellular trichome development in Cucumis sativus. Plant Biol. 78, 832–840. doi: 10.1111/jipb.12345
Zhao, M., Morohashi, K., Hatlestad, G., Grotewold, E., and Lloyd, A. (2008). The TTG1-bHLH-MYB complex controls trichome cell fate and patterning through direct targeting of regulatory loci. Development 135, 1991–1999. doi: 10.1242/dev.016873
Zhou, Z., An, L., Sun, L., Zhu, S., Xi, W., Broun, P., et al. (2011). Zinc finger protein 5 is required for the control of trichome initiation by acting upstream of zinc finger protein8 in Arabidopsis. Plant Physiol. 157, 673–682. doi: 10.1104/pp.111.180281
Keywords: pepper, trichomes, Hairiness, zinc-finger protein, promoter
Citation: Liu J, Wang H, Liu M, Liu J, Liu S, Cheng Q and Shen H (2021) Hairiness Gene Regulated Multicellular, Non-Glandular Trichome Formation in Pepper Species. Front. Plant Sci. 12:784755. doi: 10.3389/fpls.2021.784755
Received: 28 September 2021; Accepted: 18 November 2021;
Published: 16 December 2021.
Edited by:
Kun Lu, Southwest University, ChinaReviewed by:
Xueyi Xue, University of Illinois at Urbana-Champaign, United StatesYuan-Yuan Li, Shandong Agricultural University, China
Copyright © 2021 Liu, Wang, Liu, Liu, Liu, Cheng and Shen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huolin Shen, c2hsMTYwNkBjYXUuZWR1LmNu
 Jinqiu Liu
Jinqiu Liu Haoran Wang
Haoran Wang Huolin Shen
Huolin Shen