
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 16 December 2021
Sec. Plant Breeding
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.784105
This article is part of the Research Topic Soybean Molecular Breeding and Genetics View all 25 articles
Early leaf senescence phenotype in soybean could be helpful to shorten the maturation period and prevent green stem disorder. From a high-density mutation library, we identified two early leaf senescence soybean mutant lines, els1-1 (early leaf senescence 1) and els1-2. The chlorophyll contents of both els1-1 and els1-2 were low in pre-senescent leaves. They degraded rapidly in senescent leaves, revealing that ELS1 is involved in chlorophyll biosynthesis during leaf development and chlorophyll degradation during leaf senescence. The causal mutations in els1 were identified by next-generation sequencing-based bulked segregant analysis. ELS1 encodes the ortholog of the Arabidopsis CaaX-like protease BCM1, which is localized in chloroplasts. Soybean ELS1 was highly expressed in green tissue, especially in mature leaves. The accumulation of photosystem I core proteins and light-harvesting proteins in els1 was low even in pre-senescent leaves, and their degradation was accelerated during leaf senescence. These results suggest that soybean ELS1 is involved in both chlorophyll synthesis and degradation, consistent with the findings in Arabidopsis BCM1. The gene els1, characterized by early leaf senescence and subsequent early maturation, does not affect the flowering time. Hence, the early leaf senescence trait regulated by els1 helps shorten the harvesting period because of early maturation characteristics. The els1-1 allele with weakly impaired function of ELS1 has only a small effect on agricultural traits and could contribute to practical breeding.
Soybean (Glycine max) is an essential crop for oil production, human consumption, and livestock feed. As with many crops, adjusting the flowering and maturation time in soybean breeding is very important for the target adaptation area. Early soybean harvesting is one of the critical agricultural processes that might increase yields, other factors being double-cropping systems, crop rotation, and cultivation at high latitudes. However, most of the early maturation genes that have been isolated so far shorten the maturation by accelerating the flowering time (Liu et al., 2008; Watanabe et al., 2009, 2011; Xia et al., 2012; Lu et al., 2020). To diversify soybean harvest time, early maturing genetic resources with causal genes that do not affect flowering time are required, but there have been no reports so far. In addition, soybean harvest is often affected by the green stem disorder (GSD), in which chlorophyll degradation is suppressed without leaf senescence during harvest (Phillips et al., 1984; Hobbs et al., 2006). GSD reduces seed quality because mechanical harvesting contaminates seeds with leaf and stem juices (Hill et al., 2006). Since early chlorophyll degradation accelerated early leaf senescence in Arabidopsis (Ono et al., 2019), controlling chlorophyll degradation might help adjust soybean harvest time without affecting the flowering time and prevent GSD. However, little about the effects of changes in chlorophyll metabolism on agricultural traits has been known.
Chlorophyll is an essential molecule for capturing light during photosynthesis and is synthesized during leaf development and degraded during leaf senescence. Higher plants have two types of chlorophyll: chlorophyll a (Chl a) and chlorophyll b (Chl b). Chl a consists of photosystem I (PSI), photosystem II (PSII), and cytochrome b6f. Chl b is present only in the PSI-associated light-harvesting complex II (LHCI) and the PSII-associated light-harvesting complex II (LHCII). Chlorophyll-binding proteins such as PSI and PSII convert light energy into chemical energy during light reactions. The subunits of PSI, PSII, LHCI, and LHCII are encoded by Psa, Psb, Lhca, and Lhcb, respectively.
Free chlorophyll and its intermediate forms react with light to produce harmful reactive oxygen species, ultimately leading to cell death (Apel and Hirt, 2004). Hence, both chlorophyll synthesis and degradation are strictly regulated at the genetic level. The first step of chlorophyll synthesis begins with synthesizing protoporphyrin IX, a chlorophyll precursor, from glutamyl-tRNAGlu via several steps. Then, protoporphyrin IX is converted to Mg-protoporphyrin IX (MgP) by magnesium chelatase (MgCh). MgCh is composed of three subunits: GENEMOS UNCOUPLED 5 (GUN5)/CHLH, CHLD, and CHLI, and is activated by GUN4 (Gibson et al., 1995; Larkin et al., 2003; Davison et al., 2005; Verdecia et al., 2005). Finally, MgP is converted to Chl a via several steps. Chl b is converted from Chl a by the chlorophyll a oxygenase (CAO; Tanaka et al., 1998).
The first step of chlorophyll degradation begins with the conversion of Chl a to pheophytin a by magnesium dechelatase STAY-GENE/NON-YELLOWING 1 (SGR/NYE1; Armstead et al., 2007; Kusaba et al., 2007; Park et al., 2007; Ren et al., 2007; Sato et al., 2007; Shimoda et al., 2016). Subsequently, pheophytin a undergoes several steps to open the porphyrin ring and finally becomes colorless. Chl b is converted to Chl a by the Chl b-degrading enzymes NON-YELLOW COLORING 1 (NYC1) and NYC1-LIKE (NOL) and gets degraded via the Chl a degradation pathway (Kusaba et al., 2007; Sato et al., 2009). In addition, PSII subunit proteins NON-YELLOW COLORING 4/THYLAKOID FORMATION 1 (NYC4/THF1) and cytG/PsbM control the degradation of chlorophyll-binding proteins (Huang et al., 2013; Yamatani et al., 2013; Kohzuma et al., 2017).
Recently, a chloroplast-localized CaaX-like protease BALANCE of CHLOROPHYLL METABOLISM (BCM), which regulates chlorophyll synthesis and degradation, was isolated from Arabidopsis thaliana (Wang et al., 2020). BCM1 and its paralog BCM2 are also present in Arabidopsis. The functions of BCM1 and BCM2 overlap, and their double mutants show phenotypes of pale green leaves during leaf development and early chlorophyll degradation during leaf senescence. BCM1 is localized in thylakoid membranes and interacts with GUN4 and SGR to control chlorophyll synthesis and degradation (Wang et al., 2020). BCM1 has also been reported as an Mg2+ transporter and controls chlorophyll synthesis (Zhang et al., 2020). BCM1 is considered a factor that regulates chlorophyll synthesis and degradation at the protein level. Soybean has two copies of the Arabidopsis BCM ortholog. One of them has been reported to be the classical G gene that determines the color of the green seed coat in soybeans (Wang et al., 2018). The G gene has lost its function during domestication from wild soybean (G. soja) to cultivated soybean. Modern soybeans with yellow seed coat are the g mutant, and the yellow seed coat is assumed to be caused by reduced chlorophyll synthesis and/or increased chlorophyll degradation due to the loss of function of the G gene. In the present study, we isolated and physiologically characterized novel early leaf senescence mutants els1 ( early leaf senescence 1) in soybean and confirmed that ELS1 encodes a paralog of the G gene, an ortholog of Arabidopsis BCM1, by using next-generation sequencing (NGS)-based bulk DNA analysis.
Two mutant lines els1-1 and els1-2 that exhibit an early chlorophyll degradation phenotype were identified in the high-density mutant library by ethyl methanesulfonate (EMS) treatment twice (Tsuda et al., 2015). The mutants els1-1 and els1-2 were backcrossed with the wild-type (WT) cultivar Enrei, and els1-1, els1-2, and WTs were used in the experiment. The plants were cultivated in a field or a greenhouse 2018–2021 at NARO (36°20' N, 140°110' E) in Tsukuba City, Ibaraki Prefecture, Japan. Field cultivation was carried out in row-plots with 80 cm row spacing and 15 or 30 cm inter-plant spacing. Pot cultivation was carried out using a 30 cm diameter pot of Nippi Horticultural Land No. 1 soil (Kumiai Nippi Engeibaido No. 1. Nihon Hiryo, Japan). Dark treatment of soybean was performed as described by Kohzuma et al. (2017). The soybean stage was determined according to Fehr et al. (1971).
Leaf chlorophyll content was non-destructively measured using a SPAD-502 Plus instrument (Konica-Minolta, Japan). The photosynthetic pigment was extracted with 80% acetone after crushing the leaves with liquid nitrogen. Chl a and Chl b contents were determined using the method described by Porra et al. (1989). The carbon assimilation rate was measured in sunlight using the Rapid Photosynthesis Measuring System MIC-100 (Masa International, Japan; Tanaka et al., 2021).
Genomic DNA was extracted from fresh leaves according to the method described by Tsuda et al. (2015). DNA bulks for next-generation sequencing (NGS)-based bulk DNA analyses were prepared from the els1-2 × WT F3 population. DNA from 20 WT and 20 mutant individuals from each F3 family of WT and mutant types were bulked. The mutant and WT DNA bulks were sequenced on an Illumina HiSeq X10 platform (Illumina Inc., San Diego, CA, USA) at Macrogen Inc. (Seoul, Republic of Korea). A 150 bp paired-end library was constructed using genomic DNA following the TruSeq™ DNA PCR Free protocol for a 350 bp insert (Illumina). NGS-based bulked segregant analysis was conducted following Dougherty et al. (2018) protocol with slight modifications. The obtained data were trimmed and mapped to the reference genome Gmax_275_v2.0 using CLC Genomics Workbench ver.11 (CLC Bio, Denmark) with the following parameters: adaptor trim, ambiguous limit two, quality limit 0.01, removal of three 5'- and 3'-terminal nucleotides, discard read pairs with a minimum number of nucleotides less than 50 bp, no global alignment, no masking mode, linear gap cost, no auto-detect paired distances, match score one, mismatch cost two, deletion cost three, insertion cost three, length fraction 0.9, and similarity fraction 0.96. Variants against the reference genome were identified in the aligned reads with basic variant detection module ver.2.0 with the following parameters: ignore non-specific matches and broken read pairs, minimum read coverage ten, minimum count three, base quality filter, neighborhood radius five, minimum central quality 20, and minimum neighborhood quality 15. A shared variant track was prepared from the variant tracks for WT and mutant bulks by using the identified shared variants module ver to collect all variations between bulks and reference. 1.2 with a frequency parameter of 1%. Then, the variant frequency in the aligned reads of each bulk was re-calculated using the identified known mutations from mappings module ver.1.1 against the shared variant track with the following parameters: minimum coverage one, detection frequency 0.05, ignore broken pairs, and ignore non-specific matches. In addition, all positions overlapping with known gene annotations and the resulting amino acid changes or splicing site changes were searched using the amino acid changes module ver. 2.5 and the splice site effect predicted by module ver.1.4. Finally, variant filtering was performed according to the variant frequency of more than 75% for mutant bulk, less than 25% for WT bulk, and a variant with amino acid change.
The mutation site of ELS1 in els1-1 was determined by Sanger sequencing using a SupreDye v3.1 Cycle Sequencing Kit (EdgeBioSystems, USA) and an ABI 3500xl genetic analyzer (Thermo Fisher Scientific, United States) according to the manufacturers’ instructions. Primers used for Sanger sequencing are shown in Supplemental Table 1. To confirm co-segregation between the causative mutations of els1-1 and els1-2 and their phenotype in the segregating populations, derived cleaved-amplified polymorphic sequence (dCAPS) markers with amplified fragment lengths of approximately 200 bp were designed. Primer pairs and restriction enzymes for the dCAPS markers are shown in Supplemental Table 2. We investigated the genotype and phenotype co-separation of 120 individuals in the els1-1 × Enrei F2 population and 40 individuals in the els1-2 × Enrei F3 population.
Total RNA was isolated from soybean tissues using TRI Reagent (MOR, United States) or RNeasy (Qiagen, Netherlands). According to the manufacturer’s protocol, first-strand cDNA was synthesized from 500 ng total RNA using ReverTra ACE qPCR RT Master Mix with gDNA Remover (TOYOBO, Japan). The synthesized cDNA was diluted tenfold and used as a template for qRT-PCR. The qRT-PCR was performed using Kapa SYBR Fast qPCR Kit (Kapa Biosystems, USA) and the ViiA7 real-time PCR system (Thermo Fisher Scientific). PCR conditions were performed according to the protocol. The primer pairs used for qRT-PCR are listed in Supplemental Table 3.
For bule native PAGE analysis, fresh soybean leaves were crushed with liquid nitrogen and then suspended in homogenizing buffer (50 mM HEPES-KOH, pH 7.8, 400 mM sucrose, 10 mM NaCl, and 2 mM MgCl2). Homogenates were filtered through a two-layer Miracloth and centrifuged at 5000 × g for 10 min at 4°C. The obtained thylakoid membrane pellets were suspended in 25BTH20G buffer (50 mM Bis-Tris–HCl, pH 7.0, 20% glycerol) and centrifuged at 14000 rpm for 1 min at 4°C. The thylakoid membranes corresponding to 100 mg fresh weight (FW) were dissolved in 200 μl of 1% β-dodecyl-maltoside in the dark and on ice. Solubilized thylakoid membrane proteins were electrophoresed using Native PAGE ™ 4 to 16%, Bis-Tris, 1.0 mm, Mini Protein Gel (Thermo Fisher Scientific), according to Yamatani et al. (2018). For 2D-SDS PAGE, the excised strip-shaped gel was heat-denatured in equilibration buffer (250 mM Tris, pH 6.8, 4% SDS, 1% dithiothreitol [DTT], 0.1% bromophenol blue [BPB], and 10% glycerol) at 70°C for 5 min and then shaken for 20 min at room temperature. Electrophoresis was performed using an acrylamide gel containing 6 M urea.
For western blot and SDS-PAGE analysis, total protein was extracted from 100 mg FW soybean leaves with 400 μl of 2× SDS buffer (0.125 M Tris, pH 6.8, 4% SDS, 4% mercaptoethanol, 1% BPB, and 20% glycerol) followed by heat denaturation at 100°C for 5 min and then 10-fold diluted with 1× SDS buffer (62.5 mM Tris, pH 6.8, 2% SDS, 2% mercaptoethanol, 0.5% BPB, and 10% glycerol). The extracted proteins were electrophoresed on an acrylamide gel. Protein transfer was performed using the Trans-Blot® Turbo ™ Transfer System (Bio-Rad, Hercules, CAUSA) or Mini Trans-Blot® Cell (BIO-RAD). Antibodies against D2, Lhca1-Lhca4, Lhcb1, Lhcb3, Lhcb4, PsaH, and PsaL for western blotting were purchased from Agrisera (Agrisera, Sweden). Antibodies against D1 (Kato et al., 2012), VAR2 (Sakamoto et al., 2003), and TIC110 (Kikuchi et al., 2013) were also used. Protein detection was performed using an ECL Prime Western Blotting Detection System (Cytiva, USA) and ImageQuant LAS 4000 mini (Cytiva). SDS-PAGE gels were stained with CBB Stain One Super (Ready to Use) (Nacalai, Japan).
The following soybean genes were used: GmACTIN (Glyma.08G182200), ELS1/GmBCM1 (Glyma.11G043400), GmBCM2 (Glyma.01G198500), GmSGR1 (Glyma.11G027400), GmNYC1 (Glyma.09G191200), GmNAC01 (Glyma.15G254000), GmSAG15 (Glyma.06G162200), GmLhca1 (Glyma.02G064700), GmLhca2 (Glyma.16G016100), GmLhcb1 (Glyma.16G165500), GmLhcb2 (Glyma.02G305400), GmPsaH (Glyma.07G019700), and GmPsaL (Glyma.18G241700).
The two early leaf senescence mutants, els1-1 and els1-2, were isolated from a mutant library in which high-density mutations were induced by EMS treatments (Tsuda et al., 2015). Unlike the WT cultivar., Enrei, the upper 4th leaves of els1-1 at 5 weeks after flowering (5 WAF) exhibited an early yellowing phenotype (Figure 1A). The chlorophyll content of the upper 4th leaves of els1-1 and els1-2 had lower chlorophyll content than the WT cultivar Enrei at 0 WAF (SPAD: Enrei, 37.92 ± 0.69; els1-1, 30.30 ± 1.05; els1-2, 28.50 ± 0.91) (Figure 1B). Chlorophyll in both els1 mutants was degraded earlier than Enrei at 6 WAF (SPAD value: Enrei, 47.52 ± 1.35; els1-1, 19.88 ± 1.41; els1-2, 12.75 ± 1.05). Since the chlorophyll content of els1-2 diminished faster than els1-1, the els1-2 phenotype was more severe than the els1-1 phenotype. Aerial photographs acquired by a drone at 3 and 5 WAF also showed that the leaves of els1 exhibited an early chlorophyll degradation phenotype (Figure 1B). These results indicate that els1 shows phenotypes of pale green in pre-senescent leaves and early chlorophyll degradation in senescent leaves.
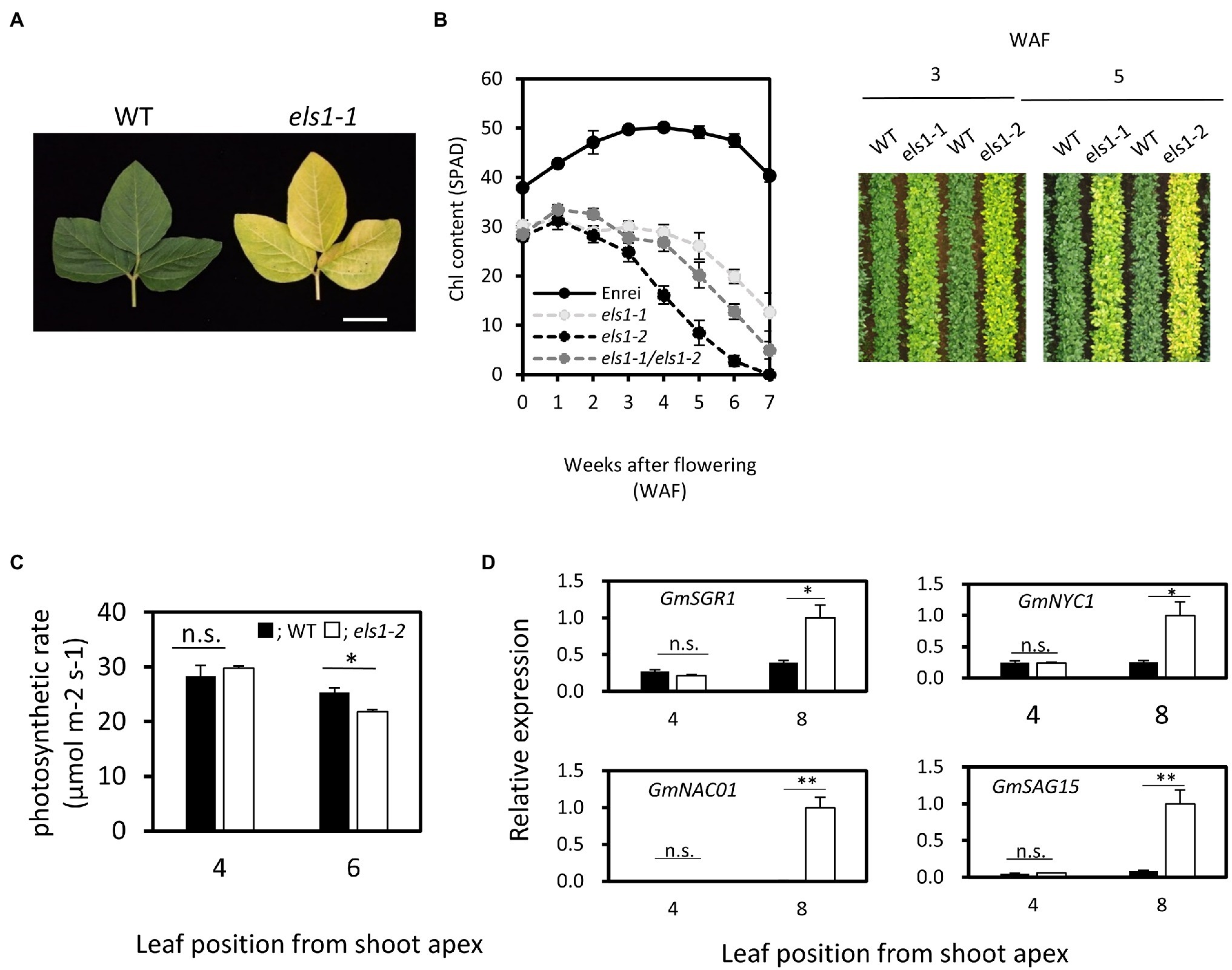
Figure 1. Physiological characterization of els1 during natural leaf senescence. (A) els1-1 showed an early yellowing phenotype compared to WT during natural leaf senescence. The photo shows the upper 4th leaves at 5 weeks after flowering. The scale bar indicates 5 cm. (B) Time course of chlorophyll content in els1 during natural during natural leaf senescence. The chlorophyll content (SPAD) of upper 4th leaves was measured from the flowering. Solid lines; Enrei, light gray dotted lines; els1-1, dotted lines; els1-2, dark gray dotted lines; heterozygote lines between els1-1 and els1-2. The error bars indicate the standard error (SE) (n = 6 biological replicates). The right panels were aerial photographs taken by a drone at 3 and 5 weeks after flowering. (C) The carbon assimilation rates of pre-senescent (the upper 4th leaves) and senescent (the upper 6th leaves) leaves (n = 5 biological replicates) of WT and els1-2. Black bars indicate WT, white bars indicate els1-2. Error bars indicate standard error (SE). *p < 0.05, **p < 0.01, n.s., not significant (Student’s t-test). (D) Expression pattern of senescence-inducible genes in WT and els1-2 during natural leaf senescence. Total RNA from pre-senescent (the upper 4th leaves) and senescent (the upper 8th leaf) leaves of WT and els1-2 were examined. Expression levels were standardized using GmACTIN (n = 6 biological replicates).
To determine if the causative genes of els1-1 and els1-2 are at the same locus, a complementation test was performed by crossing these plants. Heterozygote plants between els1-1 and els1-2 showed the identical phenotypes of pale green and early chlorophyll degradation as both parents, suggesting that mutations of els1-1 and els1-2 occurred in the same gene (Figure 1B).
The content of both Chl a and Chl b in els1-2 was lower than that in the WT at 0 WAF (Chl a: WT, 2.35 ± 0.18 nmol mg−1 FW; els1-2, 1.57 ± 0.07 nmol mg−1 FW; Chl b: WT, 0.58 ± 0.04 nmol mg−1 FW; els1-2, 0.32 ± 0.02 nmol mg−1 FW; Supplemental Figure 1). The Chl a/b ratio of els1-2 was higher than that of the WT at 0WAF (WT, 4.06 ± 0.02; els1-2, 4.79 ± 0.06, p < 0.01), indicating that the accumulation of Chl b was more significantly reduced in els1-2. In addition, since the decreasing rate of chlorophyll content of els1-2 at 4 WAF was higher than that of the WT, early chlorophyll degradation occurred in els1-2 (Chl a: WT, 3.39 ± 0.11 nmol mg−1 FW; els1-2, 0.88 ± 0.09 nmol mg−1 FW; Chl b: WT, 0.96 ± 0.03 nmol mg−1 FW; els1-2, 0.14 ± 0.01 nmol mg−1 FW).
Next, we examined the physiological functions of the leaves in els1-2 during natural leaf senescence. Carbon assimilation rate of pre-senescent leaves (the upper 4th leaves) did not differ between the WT and els1-2 (WT, 28.31 ± 1.99 μmol m−2 s−1; els1-2, 29.79 ± 0.41 μmol m−2 s−1). However, the carbon assimilation rate in the senescent leaves of els1-2 (the upper 6th leaves) decreased faster than in the WT (WT, 25.34 ± 0.87 μmol m−2 s−1; els1-2, 21.78 ± 1.13 μmol m−2 s−1; Figure 1C). Additionally, the expression levels of senescence-inducible genes (GmSGR1, magnesium dechelatase gene; GmNYC1, Chl b -degrading enzyme gene; GmNAC01, senescence-inducible transcription factor gene; GmSAG15, senescence-inducible marker gene) in pre-senescent leaves (the upper 4th leaves) of els1-2 during natural leaf senescence was not significantly different from that of the WT, but that of els1-2 in senescent leaves (the upper 8th leaves) were significantly increased compared to the WT (Figure 1D). These results also physiologically confirmed that els1 exhibits an early leaf senescence phenotype during natural leaf senescence.
Moreover, we investigated dark-induced leaf senescence using primary leaves in els1-2. The results showed that els1-2 exhibited an early chlorophyll degradation phenotype 4 days after dark incubation (4 DAD; Supplemental Figure 2A). In addition, the chlorophyll content of els1-2 was lower than that of the WT in pre-senescent leaves (0 DAD) and decreased faster than that of the WT in senescent leaves (4 DAD; Supplemental Figure 2B). During dark treatment, the expression level of senescence-inducible genes in els1-2 at 0 DAD was not significantly different from that of the WT, but that of els1-2 at 4 DAD was considerably higher than that of the WT (Supplemental Figure 2C). Taken together, these results indicate that els1 shows the phenotype of pale green leaves and early leaf senescence not only during natural leaf senescence but also during dark-induced leaf senescence.
The causative gene of the els1-2 mutant was determined by NGS-based bulked segregant analysis using the F3 population for els1-2. Twenty individual plants from each F3 family of WT and mutant types were bulked. As a result of variant filtering of single nucleotide polymorphisms (SNPs) and indels obtained from NGS-based bulk segregant analysis in els1-2 under three conditions (variant frequency of more than 75% in mutant bulk, variant frequency of less than 25% in WT bulk, and a variant with amino acid change), seven genes were identified as candidates. Among them, Glyma.11G043400 was considered the candidate gene for ELS1 because the variant frequency was 100% in the mutant bulk (Supplemental Table 4). Glyma.11G043400 of els1-2 had a base substitution from cytosine to adenine at position 1,027 bp from the start codon in the 8th exon, resulting in the formation of a premature stop codon at the 349th tyrosine (Figure 2A). As the complementation test above suggested that els1-1 and els1-2 are mutants at the same locus, we examined the coding region sequence of Glyma.11G043400 in els1-1 by Sanger sequencing. We found a base substitution from thymine to cytosine in the 4th exon which resulted in an amino acid substitution from serine to proline at position 233aa (Figure 2A). Since the serine 233aa is conserved in eudicots (A. thaliana) to monocots (O. sativa), this amino acid substitution would be expected to affect protein function (Figure 2B). We analyzed genotypes of 120 individuals in the F2 population of els1-1 × Enrei and 40 individuals in the F3 population of els1-2 × Enrei to examine whether els1-1 and els1-2 mutations in Glyma.11G043400 were consistent with the early leaf senescence phenotype. The mutant alleles of els1-1 and els1-2 were completely co-segregated with the early leaf senescence phenotype (Supplemental Figures 3, 4), indicating that ELS1 encodes Glyma.11G043400, the Arabidopsis ortholog GmBCM1 of the chloroplast-localized CaaX-like protease, BCM1. els1-2 is considered a null allele because it forms a premature stop codon, whereas els1-1 is a weak allele because it has an amino acid substitution and a milder phenotype of early leaf senescence than els1-2.
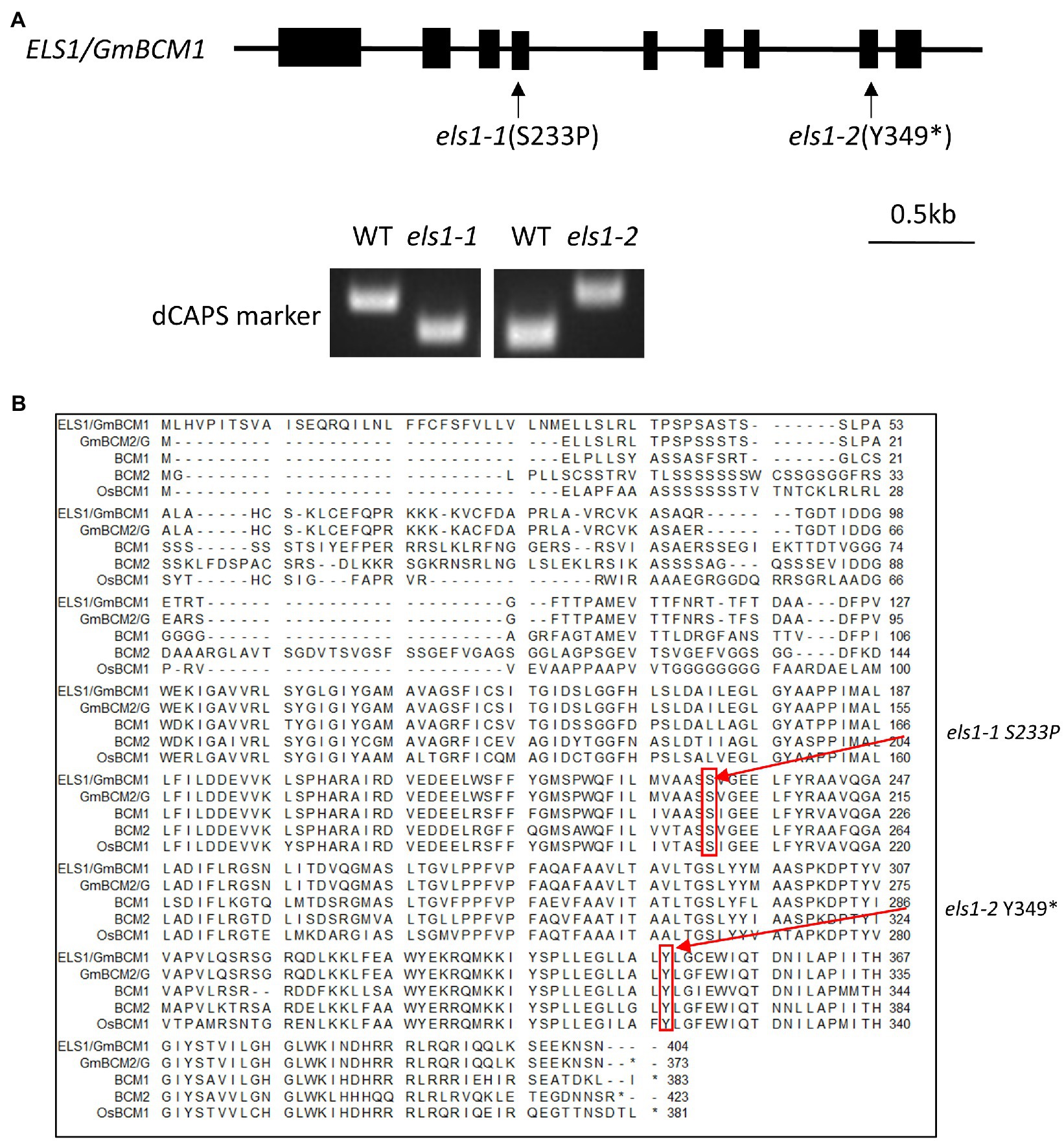
Figure 2. Isolation of ELS1/GmBCM1. (A) Gene structure of ELS1/GmBCM1. Black boxes indicate exons. Arrows indicate the positions of mutations. els1-1 had an amino acid substitution from serine to proline at position 233, and els1-2 had a nonsense mutation at position 349. The dCAPS marker was developed at the position of the base substitution. (B) Alignment diagram of ELS1/GmBCM1. The red arrows indicate the positions of mutations. els1-1 and els1-2 have single-base-substitution causing amino acid substitution in ELS1/GmBCM1. BCM1; AT2G35260 (A. thaliana), BCM2; AT4G17840 (A. thaliana), OsBCM1; Os03g0100030 (O. sativa). The asterisk “*” indicates a single nucleotide substitution resulting in a premature termination at the amino acid position.
Among the various tissues of Enrei, ELS1/GmBCM1 was highly expressed in mature leaves, and its expression level in different tissues was similar to that in GmBCM2 (Glyma.01G198500, classical locus G), GmBCM1 paralog (Figure 3A). Since Arabidopsis BCM1 is induced by light and BCM2 is induced by leaf senescence, we investigated the expression pattern of ELS1/GmBCM1 and GmBCM2 in soybean during dark treatment. The expression levels of both ELS1/GmBCM1 and GmBCM2 rapidly decreased at 1 DAD and remained low after 2 DAD (Figure 3B). Expression of the antenna protein genes GmLhca1 and GmLhcb1 is induced by light, and that of the chlorophyll-degrading enzyme genes GmSGR1 and GmNYC1 are influenced by leaf senescence (Figure 3B). Taken together, ELS1/GmBCM1 and GmBCM2, unlike Arabidopsis BCM2, are genes induced by light and not by leaf senescence.
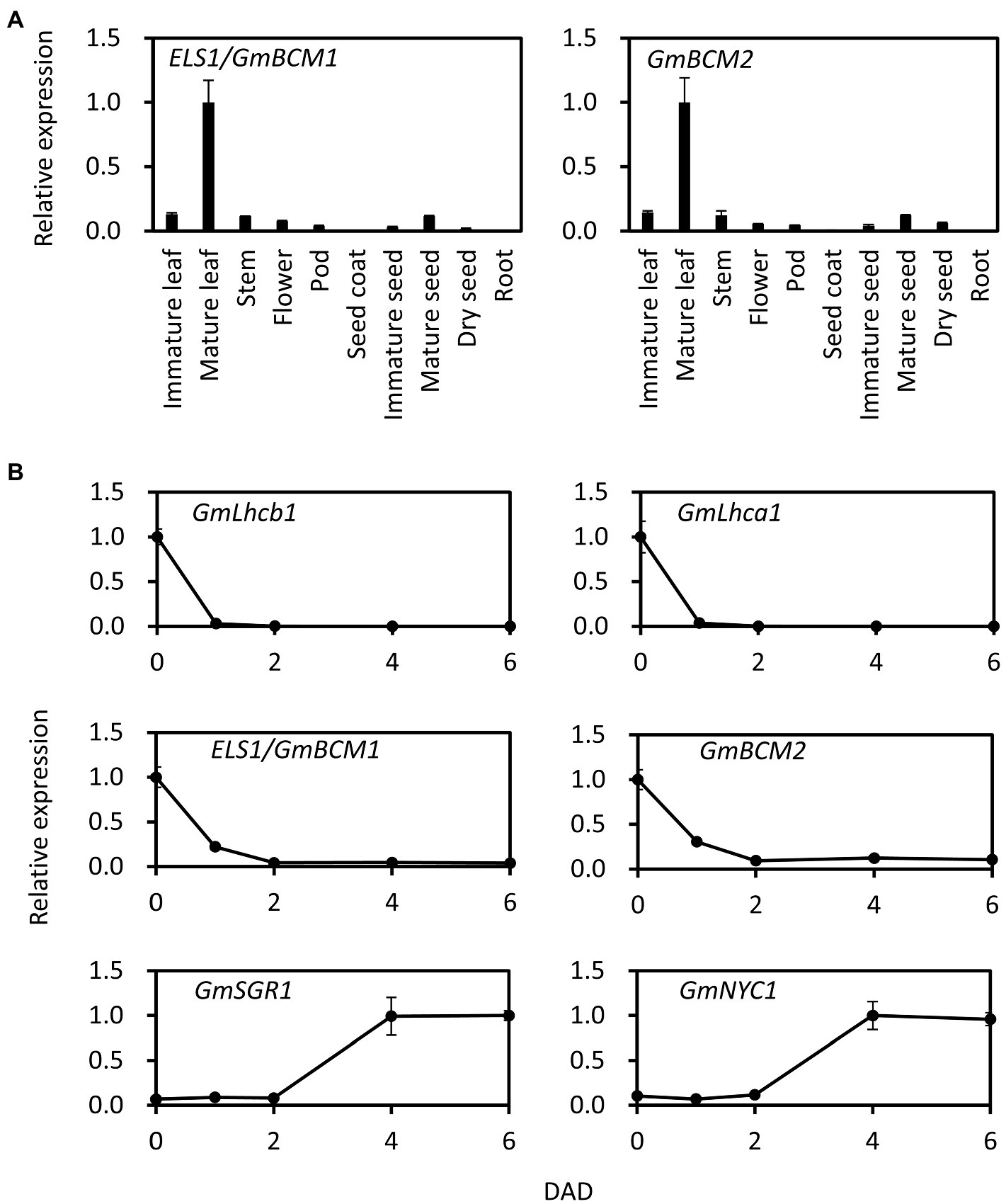
Figure 3. Expression pattern of ELS1/GmBCM1 and GmBCM2 in tissue and during dark treatment. (A) Total RNA from each tissue of WT soybean cultivar Enrei was used for qRT-PCR. (B) Total RNA from primary leaves of WT soybean cultivar Enrei during dark treatment was used for qRT-PCR. Expression levels were standardized using GmACTIN. Error bars indicate the standard error (SE; n = 5 biological replicates).
As els1-2 reveals a pale green phenotype at 0 WAF (SPAD value of the upper 4th pre-senescent leaves: WT, 36.65 ± 0.73; els1-2, 27.95 ± 0.91, p < 0.01), the composition of the photosystem proteins may have changed. The accumulation of PSII-LHCII super-complex and LHCII trimer in pre-senescent leaves of els1-2 was lower than in the WT (Figures 4A,B). The composition of chloroplast proteins of D1 and D2 of the PSII core subunits in pre-senescent leaves did not differ between the WT and els1-2 (Figure 4C). In contrast, the amount of PSI core subunits PsaH and PsaL, LHCI subunits Lhca1-4 and LHCII subunits Lhcb1, Lhcb3, and Lhcb4 of els1-2 was lower than that of WT. The reduced amount of Chl b-binding protein of LHCI and LHCII in pre-senescent leaves in els1-2 was consistent with increment of the Chl a/b ratio as mentioned above (Supplemental Figure 1). The chloroplast proteins that are not chlorophyll-binding proteins, YELLOW VARIEGATED2 (VAR2), translocon at the inner envelope membrane of chloroplasts 110 (TIC110), and Rubisco large subunit (RBSCL), were not different between the WT and els1-2 in pre-senescent leaves. These results indicate that the loss of function of ELS1 altered the composition of the chlorophyll-binding proteins PSI, LHCI and LHCII in pre-senescent leaves.
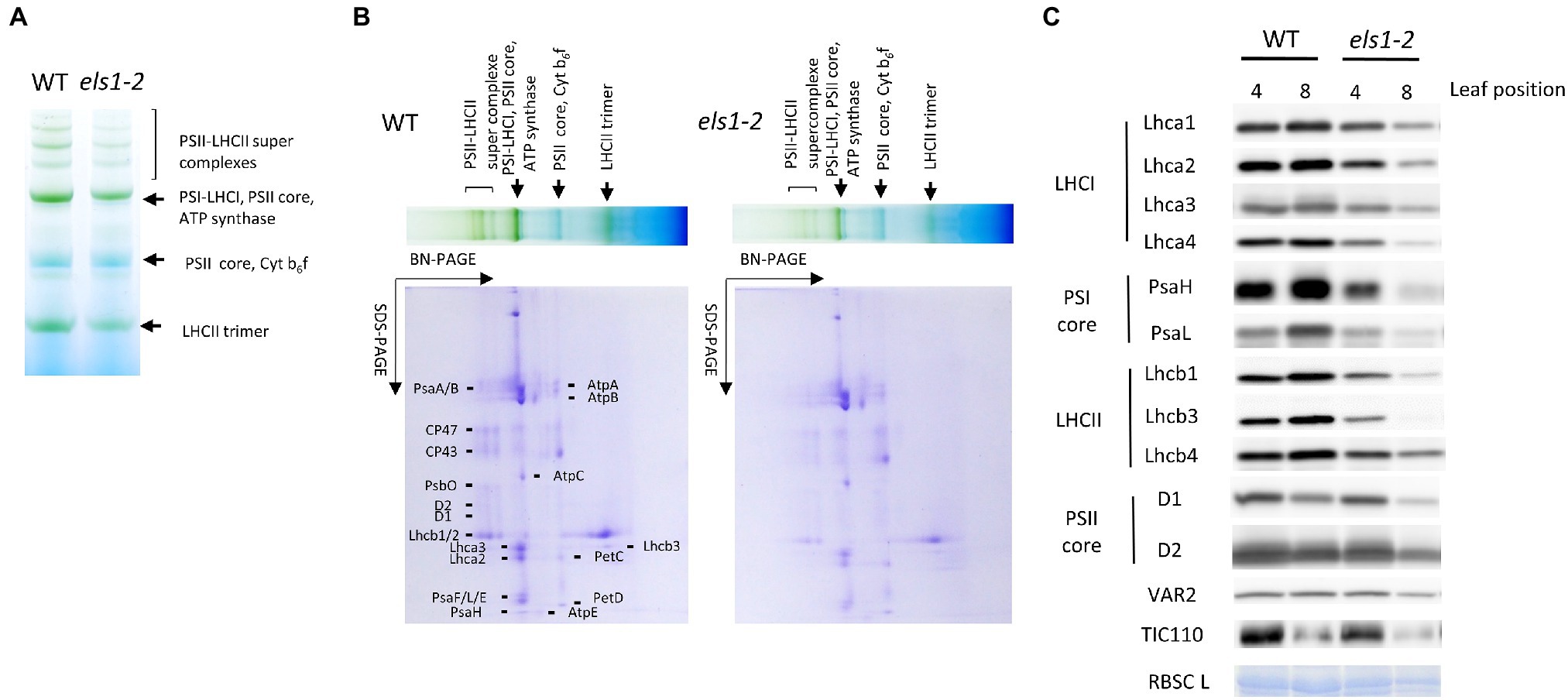
Figure 4. Thylakoid protein accumulation in els1-2. To examine the thylakoid membrane proteins and pigment complexes from pre-senescent leaves (the upper 4th leaves) of WT and els1-2, we performed (A) blue native PAGE analysis and (B) two-dimensional electrophoresis. SDS-PAGE was visualized by CBB staining. (C) Western blot of chloroplast proteins from pre-senescent (the upper 4th leaves) and senescent (the upper 8th leaves) leaves of WT and els1-2. Lhca1-4 are LHCI apoproteins; PsaH and L are photosystem I reaction center subunits; Lhcb1, 3, and 4 are LHCII apoproteins; D1 and D2 are photosystem II reaction center subunits; VAR2, TIC110, and RBSC L represent thylakoid membrane-localized protein, envelope-localized translocon protein, and stroma-localized protein, respectively. RBSC L was visualized by CBB staining.
In senescent leaves (the upper 8th leaves) at 0 WAF (SPAD value: WT, 34.80 ± 1.04; els1-2, 12.88 ± 1.12, p < 0.01), the amount of PSI, PSII, LHCI and LHCII of els1-2 was reduced earlier than that of WT. In senescent leaves, the accumulation of RBSCLs in els1-2 was reduced faster than in the WT, consistent with the physiological loss of leaf function in els1-2 as described above.
The expression levels of Lhca1, Lhca2, Lhcb1, Lhcb2, PsaH, and PsaL in pre-senescent leaves of els1-2 were slightly higher than those in the WT, but the difference was not significant (Supplemental Figure 5). These results indicate that the decreased accumulation of PSI core protein and LHC in els1 was not due to reduced gene expression.
The els1 showed an early maturation phenotype than the WTs, especially els1-2 matured earlier relative to the els1-1 (Figure 5A). To examine in more detail, the number of days to reach each growth stage from R1 to R8 of els1-1 and els1-2 was compared with those of their WTs. The counterparts of WTs for els1-1 and els1-2 in the segregating population were named WTels1-1 and WTels1-2, respectively. The flowering initiation stage R1 and the pod elongation stage R4 of els1-1 and els1-2 were not significantly different from those in WTs (days to each stage, R1: WTels1-1, 36.00 ± 0.26 day; els1-1, 35.50 ± 0.43 day; WTels1-2, 35.33 ± 0.49 day; els1-2, 35.00 ± 0.37 day; R4: WTels1-1, 48.50 ± 1.15 day; els1-1, 46.50 ± 1.06 day; WTels1-2, 46.67 ± 0.99 day; els1-2, 46.67 ± 0.21 day; Figure 5B). In contrast, the days to full ripening stage R8 in els1-1 and els1-2 were significantly earlier than those of the WT (R8: WTels1-1, 115.83 ± 0.40 day; els1-1, 100.33 ± 1.74 day; WTels1-2, 107.17 ± 2.97 day; els1-2, 85.33 ± 1.02 day; Figure 5B).
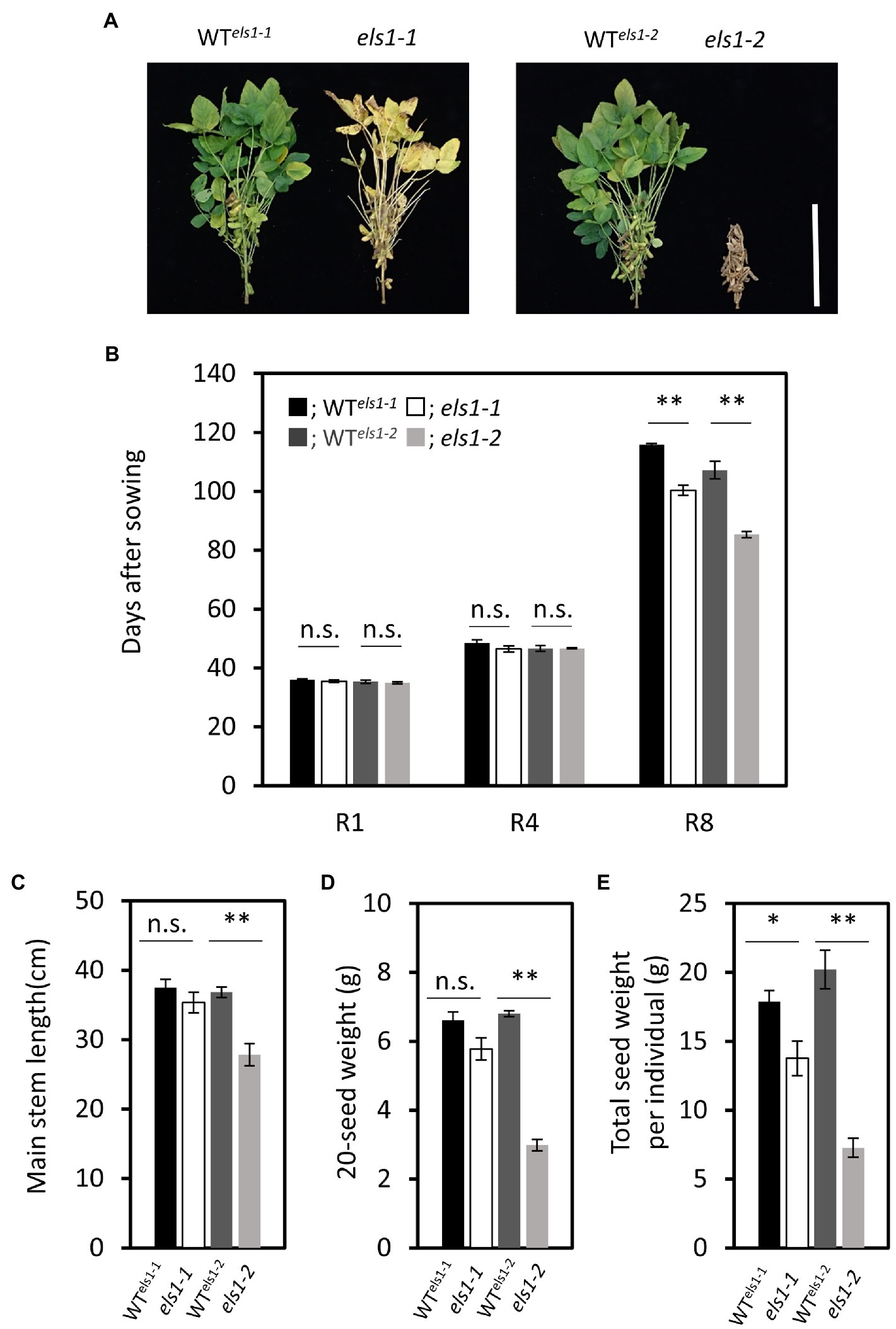
Figure 5. The differences of agronomic traits between els1 and their WT. (A) Phenotype of WT els1-1, els1-1, WT els1-2 and els1-2 at about 80 days after sowing. The scale bar indicates 30 cm. (B) Days to each growth stage in els1 (n = 6 biological replicates). Black bars indicate WT of els1-1, white bars indicate els1-1. Dark gray bars indicate WT of els1-2, light gray bars indicate els1-2. Error bars indicate the standard error (SE). *p < 0.05, **p < 0.01, n.s., not significant (Student’s t-test). (C) Main stem length in els1 (n = 6 biological replicates). (D) 20-seed weight of els1 (n = 6 biological replicates). (E) The total seed weight per plant in els1 (n = 6 biological replicates).
The main stem length of els1-2 was significantly shorter than that of the WT (WTels1-2, 36.83 ± 0.75 cm; els1-2, 27.83 ± 1.60 cm) while that of els1-1 was slightly shorter than that of the WT, but there was no significant difference (WTels1-1, 37.50 ± 1.20 cm; els1-1, 35.33 ± 1.48 cm; Figure 5C). The 20-seed weight of els1-2 was significantly lower than that of the WT (WTels1-2, 6.81 ± 0.09 g; els1-2, 2.99 ± 0.17 g) while that of els1-1 was slightly lower than that of the WT, but there was no significant difference (WTels1-1, 6.61 ± 0.24 g; els1-1, 5.78 ± 0.32 g; Figure 5D). Total seed weight per individual of els1-2 was significantly lower than that of the WT (WTels1-2, 20.21 ± 1.40 g; els1-2, 7.27 ± 0.70 g) while that of els1-1 was lower than the WT (WTels1-1, 17.87 ± 0.82 g; els1-1, 13.77 ± 1.25 g) and intermediate between WT and els1-2 (Figure 5E).
In recent years, the causative genes of mutants and QTLs have been efficiently and quickly isolated using NGS analysis. The MutMap method developed in rice (Abe et al., 2012) was subsequently applied to soybean to characterize the causative genes of several mutants. Recently, the causative gene of the spl-1 mutant induced by EMS was isolated by NGS-based bulk segregant analysis (Al Amin et al., 2019). NGS analysis was performed using 20 WT and 20 mutant bulks and they were narrowed down to seven candidate genes. Similarly, in the present study, we also efficiently narrowed down to one candidate gene of els1-2 by NGS-based bulked segregant analysis and succeeded in identifying the causative genes of both els1-1 and els1-2 by genetic analysis (Figure 2).
Bulk DNA analysis using NGS revealed that ELS1 encodes the ortholog of Arabidopsis BCM1, a chloroplast-localized CaaX-like protease (Figure 2). Other CaaX proteases such as Rce1 and Ste24 localize to the ER membrane (Bracha-Drori et al., 2008), regulate membrane localization of substrates, and clogging of the ER translocon (Ast et al., 2016). It has been reported that BCM1 localized in chloroplasts does not have CaaX protease activity and interacts with GUN4 and SGR to control chlorophyll synthesis and degradation (Wang et al., 2020). The bcm1bcm2 double mutant in Arabidopsis exhibits phenotypes of pale green and early leaf senescence (Wang et al., 2020; Zhang et al., 2020). Similarly, els1 revealed the phenotypes of pale green and early leaf senescence (Figure 1; Supplemental Figure 2), suggesting that its function is conserved among higher plants. Since soybean experienced two whole-genome duplications about 59 million years ago and 13 million years ago and has a paleopolyploid genome (Schmutz et al., 2010), nearly 75% of the genes are present in multiple copies. There are two copies of GmBCM, GmBCM1, and GmBCM2, in the soybean genome. GmBCM2, the paralog of ELS1/GmBCM1, has been reported as the causative gene that controls the green seed coat in soybean (Wang et al., 2018). The GmBCM2 allele in the wild species (G. soja) is the function allele, whereas the loss-of-function mutant allele, gmbcm2, is common in cultivated soybean. Cultivated soybeans with yellow seed coats are caused by a loss of GmBCM2 function, resulting in decreased chlorophyll synthesis and/or increased chlorophyll degradation. The GmBCM2 allele of the WT donor plant of the mutant library is gmbcm2; therefore, els1-1 and els1-2 are the gmbcm1gmbcm2 double mutants. Duplicated genes may undergo pseudogenization, sub-functionalization, or neofunctionalization (Innan and Kondrashov, 2010). Since cultivated yellow soybeans with the gmbcm2 mutation do not reduce green leaf color, the two GmBCMs are likely working redundantly in soybean leaves. The strong expression levels of both GmBCM1 and GmBCM2 in the leaves support this hypothesis (Figure 3A). Phenotypes of pale green and early chlorophyll degradation in els1 were more pronounced due to the loss of both GmBCM gene functions.
In Arabidopsis, BCM1 expression is induced by light, whereas BCM2 expression is induced by leaf senescence (Wang et al., 2020; Zhang et al., 2020). Interestingly, ELS1/GmBCM1 and GmBCM2 expression in soybean rapidly decrease at 1DAD, suggesting that both GmBCMs regulate light responses during photosynthesis (Figure 3B). Since many genes responsible for the light reaction during photosynthesis are strongly expressed in green tissues such as leaves and are induced by light, these findings were consistent with GmBCM1 and GmBCM2. In contrast, ELS1/GmBCM1 and GmBCM2 expression were not induced by dark-induced leaf senescence at 6DAD, suggesting that their expression regulation might differ from Arabidopsis BCM2 (Figure 3B).
Blue native PAGE analysis and western blot analyses showed that the amount of PSI, LHCI and LHCII was reduced in els1-2 compared to the WT in the pre-senescent leaves (Figure 4). In contrast, the accumulation of PSII core protein and other chloroplast-localized proteins was not notably different between the WT and els1-2. The Chl a/b ratio of els1-2 was higher in the pre-senescent leaves than in the WT (Supplemental Figure 1). This was consistent with the protein analysis that the Chl a/b ratio increased with decreasing accumulation of LHC (the only Chl b binding protein). Thus, the pale green phenotype in els1 could be due to the reduced amounts of PSI and LHC (Figure 4). The expression levels of PSI and LHC genes were not reduced in els1-2 as compared with the WT (Supplemental Figure 5). These results suggest that ELS1 regulates the light reaction of photosynthesis by controlling the accumulation of PSI and LHC at the protein level.
BCM1 in Arabidopsis destabilizes the magnesium dechelatase SGR, and the bcm1bcm2 double mutant accumulates SGR even before leaf senescence and shows an early chlorophyll degradation phenotype (Wang et al., 2020). An early leaf senescence phenotype in els1 might be due to the high accumulation of SGR, similar to the Arabidopsis bcm1bcm2 double mutant. Since there was no significant difference in SGR expression before leaf senescence between the WT and els1-2, the regulation of SGR accumulation by BCM is likely to occur at the protein level in soybean (Figure 1D; Supplemental Figure 2C). In senescent leaves of els1-2, the amount of PSI and LHC subunits decreased faster, and the Chl a/b ratio gradually increased (Figure 4). Previous studies have proposed that the degradation of PSI and LHC is regulated by SGR-mediated chlorophyll degradation, and the degradation of PS II is regulated by NYC4/THF1 (Huang et al., 2013; Yamatani et al., 2013). BCM1 inhibits chlorophyll degradation via SGR, and the pronounced degradation of PSI and LHC in senescent leaves of els1-2 was consistent with previous results. In this study, els1 showed early chlorophyll degradation during leaf senescence and decreased carbon assimilation rate and induction of senescence-related genes, indicating an early leaf senescence phenotype (Figure 1D; Supplemental Figure 2C). The Arabidopsis bcm1bcm2 double mutant exhibited premature chlorophyll degradation during senescence, but decreased leaf functionality was similar to that of the WT (Wang et al., 2020). In contrast, SGR overexpression lines showed early chlorophyll degradation and decreased leaf functionality (Park et al., 2007). Since BCM1 destabilizes SGR and inhibits chlorophyll degradation, we speculated that reduced leaf functionality in els1 might occur after early chlorophyll degradation due to the high accumulation of SGR.
In this study, we successfully isolated two els1 alleles. Mutations in els1-1 and els1-2 caused the conserved amino acid substitution from serine to proline at position 233aa and nonsense mutation at position 349aa, respectively (Figure 2). The position of the amino acid substitution in els1-1 is conserved between Arabidopsis and rice (Wang et al., 2018). Although there are no reports on the functional domain of the region, this amino acid residue is expected to be important for the function of the GmBCM1 protein because els1-1 showed a pale green and early senescence phenotype. Both els1-1 and els1-2 mutants revealed a pale green phenotype and early leaf senescence, but the extent differed (Figure 1B). These results suggest that els1-1 is a weak allele with impaired protein function, whereas els1-2 is a strong allele with complete loss of function. Recently, natural variations of gmbcm1/yl2 and gmgcm2/yl1 have been isolated (Liu et al., 2020). The double mutant of yl1yl2 exhibited phenotypes of pale green and early chlorophyll degradation, similar to els1. yl1 has been reported to have a 1 bp deletion resulting in a frameshift and null mutation in GmBCM2. Although the effect of yl1 variation on agricultural traits has not been reported yet, there may be a significant effect due to early chlorophyll degradation, such as els1-2.
The days to flowering (R1) and pod elongation (R4) of els1-1 and els1-2 were almost the same as those of the WT. On the other hand, the days to the full ripening of pod (R8) of els1-1 and els1-2 were significantly less than those of the WT (Figure 5B). For example, the number of days until the full ripening stage (R8) of els1-2 was 20 days shorter than the WT. The plant height of els1-1 was slightly lower than the WT, but the difference was not significant (Figure 5C). In addition, the plant height of els1-1 decreased mildly compared to that of els1-2. The total seed weight per individual of els1-1 was slightly lower than that of the WT, but its extent was intermediate between the WT and els1-2 (Figure 5D). In other words, the weak allele of els1-1 is expected to accelerate the harvest time without significantly affecting agronomic traits, such as flowering time and yield.
Moreover, early chlorophyll degradation in els1 is expected to suppress green stem disorders. Interestingly, the heterozygous line between els1-1 and els1-2 showed an intermediate chlorophyll degradation phenotype (Figure 1B). This indicates that the phenotype of els1 might be able to quantitatively manipulate early chlorophyll degradation depending on the strength of the allele. Isolation of novel alleles of different strengths of ELS1 might allow for fine-tuning of the harvesting period without affecting agronomic traits.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
HY and AK designed the study. HY, TH, and AK conducted the study. TY and MK provided information and advice regarding gene function analysis. HY, TY, MK, and AK wrote the manuscript. All authors contributed to the article and approved the submitted version.
This study was supported by JSPS KAKENHI (Grant Number 19 J00195) to HY and a grant from the Ministry of Agriculture, Forestry, and Fisheries of Japan [Genomics-Based Technology for Agricultural Improvement (IVG1005)] to AK.
The authors declare that the research was conducted without any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Prof. Wataru Sakamoto (Okayama University) and Prof. Masato Nakai (Osaka University) for providing anti-D1, anti-VAR2, and anti-TIC110. We thank the National Institute of Agricultural and Biological Sciences (NIAS) staff members for supporting the cultivation management in the field.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.784105/full#supplementary-material
Chl a, Chlorophyll a; Chl b, Chlorophyll b; DAD, Days after dark incubation; els1, Early leaf senescence 1; FW, Fresh weight; GSD, Green stem disorder; LHCI, Light-harvesting complex II; LHCII, Light-harvesting complex II; NGS, Next-generation sequencing; PSI, Photosystem I; PSII, Photosystem II; qRT-PCR, Quantitative RT-PCR; SNP, Single nucleotide polymorphism; WAH, Weeks after flowering; WT, Wild-type.
Abe, A., Kosugi, S., Yoshida, K., Natsume, S., Takagi, H., Kanzaki, H., et al. (2012). Genome sequencing reveals agronomically important loci in rice using MutMap. Nat. Biotechnol. 30, 174–178. doi: 10.1038/nbt.2095
Al Amin, G. M., Kong, K., Sharmin, R. A., Kong, J., Bhat, J. A., and Zhao, T. (2019). Characterization and rapid gene-mapping of leaf lesion mimic phenotype of spl-1 mutant in soybean (Glycine max (L.) merr.). Int. J. Mol. Sci. 20, 2193. doi: 10.3390/ijms20092193
Apel, K., and Hirt, H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399. doi: 10.1146/annurev.arplant.55.031903.141701
Armstead, I., Donnison, I., Aubry, S., Harper, J., Hörtensteiner, S., James, C., et al. (2007). Cross-species identification of Mendel’s I locus. Science 315, 73. doi: 10.1126/science.1132912
Ast, T., Michaelis, S., and Schuldiner, M. (2016). The protease Ste24 clears clogged translocons. Cell 164, 103–114. doi: 10.1016/j.cell.2015.11.053
Bracha-Drori, K., Shichrur, K., Lubetzky, T. C., and Yalovsky, S. (2008). Functional analysis of Arabidopsis postprenylation CaaX processing enzymes and their function in subcellular protein targeting. Plant Physiol. 148, 119–131. doi: 10.1104/pp.108.120477
Davison, P. A., Schubert, H. L., Reid, J. D., Iorg, C. D., Heroux, A., Hill, C. P., et al. (2005). Structural and biochemical characterization of Gun4 suggests a mechanism for its role in chlorophyll biosynthesis. Biochemistry 44, 7603–7612. doi: 10.1021/bi050240x
Dougherty, L., Singh, R., Brown, S., Dardick, C., and Xu, K. (2018). Exploring DNA variant segregation types in pooled genome sequencing enables effective mapping of weeping trait in Malus. J. Exp. Bot. 69, 1499–1516. doi: 10.1093/jxb/erx490
Fehr, W. R., Caviness, C. E., Burmood, D. T., and Pennington, J. S. (1971). Stage of development descriptions for soybeans, glycine max (L.) Merrill. Crop Sci. 11, 929–931. doi: 10.2135/cropsci1971.0011183X001100060051x
Gibson, L. C. D., Willows, R. D., Kannangara, C. G., von Wettstein, D., and Hunter, C. N. (1995). Magnesium-protoporphyrin chelatase of Rhodobacter sphaeroides: reconstitution of activity by combining the products of the bchH, -I, and -D genes expressed in Escherichia coli. Proc. Natl. Acad. Sci. 92, 1941–1944. doi: 10.1073/pnas.92.6.1941
Hill, C. B., Hartman, G. L., Esgar, R., and Hobbs, H. A. (2006). Field evaluation of green stem disorder in soybean cultivars. Crop Sci. 46, 879–885. doi: 10.2135/cropsci2005.0207
Hobbs, H. A., Hill, C. B., Grau, C. R., Koval, N. C., Wang, Y., Pedersen, W. L., et al. (2006). Green stem disorder of soybean. Plant Dis. 90, 513–518. doi: 10.1094/PD-90-0513
Huang, W., Chen, Q., Zhu, Y., Hu, F., Zhang, L., Ma, Z., et al. (2013). Arabidopsis thylakoid formation 1 is a critical regulator for dynamics of PSII-LHCII complexes in leaf senescence and excess light. Mol. Plant 6, 1673–1691. doi: 10.1093/mp/sst069
Innan, H., and Kondrashov, F. (2010). The evolution of gene duplications: classifying and distinguishing between models. Nat. Rev. Genet. 11, 97–108. doi: 10.1038/nrg2689
Kato, Y., Sun, X., Zhang, L., and Sakamoto, W. (2012). Cooperative D1 degradation in the photosystem II repair mediated by chloroplastic proteases in Arabidopsis. Plant Physiol. 159, 1428–1439. doi: 10.1104/pp.112.199042
Kikuchi, S., Bédard, J., Hirano, M., Hirabayashi, Y., Oishi, M., Imai, M., et al. (2013). Uncovering the protein translocon at the chloroplast inner envelope membrane. Science 339, 571–574. doi: 10.1126/science.1229262
Kohzuma, K., Sato, Y., Ito, H., Okuzaki, A., Watanabe, M., Kobayashi, H., et al. (2017). The non-mendelian green cotyledon gene in soybean encodes a small subunit of photosystem II. Plant Physiol. 173, 2138–2147. doi: 10.1104/pp.16.01589
Kusaba, M., Ito, H., Morita, R., Iida, S., Sato, Y., Fujimoto, M., et al. (2007). Rice NON-YELLOW COLORING1 is involved in light-harvesting complex II and grana degradation during leaf senescence. Plant Cell 19, 1362–1375. doi: 10.1105/tpc.106.042911
Larkin, R. M., Alonso, J. M., Ecker, J. R., and Chory, J. (2003). GUN4, a regulator of chlorophyll synthesis and intracellular signaling. Science 299, 902–906. doi: 10.1126/science.1079978
Liu, B., Kanazawa, A., Matsumura, H., Takahashi, R., Harada, K., and Abe, J. (2008). Genetic redundancy in soybean photoresponses associated with duplication of the phytochrome A gene. Genetics 180, 995–1007. doi: 10.1534/genetics.108.092742
Liu, M., Wang, Y., Nie, Z., Gai, J., Bhat, J. A., Kong, J., et al. (2020). Double mutation of two homologous genes YL1 and YL2 results in a leaf yellowing phenotype in soybean [Glycine max (L.) Merr]. Plant Mol. Biol. 103, 527–543. doi: 10.1007/s11103-020-01008-9
Lu, S., Dong, L., Fang, C., Liu, S., Kong, L., Cheng, Q., et al. (2020). Stepwise selection on homeologous PRR genes controlling flowering and maturity during soybean domestication. Nat. Genet. 52, 428–436. doi: 10.1038/s41588-020-0604-7
Ono, K., Kimura, M., Matsuura, H., Tanaka, A., and Ito, H. (2019). Jasmonate production through chlorophyll a degradation by stay-green in Arabidopsis thaliana. J. Plant Physiol. 238, 53–62. doi: 10.1016/j.jplph.2019.05.004
Park, S. Y., Yu, J. W., Park, J. S., Li, J., Yoo, S. C., Lee, N. Y., et al. (2007). The senescence-induced staygreen protein regulates chlorophyll degradation. Plant Cell 19, 1649–1664. doi: 10.1105/tpc.106.044891
Phillips, D. A., Pierce, R. O., Edie, S. A., Foster, K. A., and Knowles, P. F. (1984). Delayed leaf senescence in soybean. Crop Sci. 24, 518–522. doi: 10.2135/cropsci1984.0011183X002400030022x
Porra, R. J., Thompson, W. A., and Kriedemann, P. E. (1989). Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta Bioenerg. 975, 384–394. doi: 10.1016/S0005-2728(89)80347-0
Ren, G., An, K., Liao, Y., Zhou, X., Cao, Y., Zhao, H., et al. (2007). Identification of a novel chloroplast protein AtNYE1 regulating chlorophyll degradation during leaf senescence in Arabidopsis. Plant Physiol. 144, 1429–1441. doi: 10.1104/pp.107.100172
Sakamoto, W., Zaltsman, A., Adam, Z., and Takahashi, Y. (2003). Coordinated regulation and complex formation of YELLOW VARIEGATED1 and YELLOW VARIEGATED2, chloroplastic FtsH metalloproteases involved in the repair cycle of photosystem II in Arabidopsis thylakoid membranes. Plant Cell 15, 2843–2855. doi: 10.1105/tpc.017319
Sato, Y., Morita, R., Katsuma, S., Nishimura, M., Tanaka, A., and Kusaba, M. (2009). Two short-chain dehydrogenase/reductases, NON-YELLOW COLORING 1 and NYC1-LIKE, are required for chlorophyll b and light-harvesting complex II degradation during senescence in rice. Plant J. 57, 120–131. doi: 10.1111/j.1365-313X.2008.03670.x
Sato, Y., Morita, R., Nishimura, M., Yamaguchi, H., and Kusaba, M. (2007). Mendel’s green cotyledon gene encodes a positive regulator of the chlorophyll-degrading pathway. Proc. Natl. Acad. Sci. 104, 14169–14174. doi: 10.1073/pnas.0705521104
Schmutz, J., Cannon, S. B., Schlueter, J., Ma, J., Mitros, T., Nelson, W., et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463, 178–183. doi: 10.1038/nature08670
Shimoda, Y., Ito, H., and Tanaka, A. (2016). Arabidopsis STAY-GREEN, Mendel’s green cotyledon gene, encodes magnesium-dechelatase. Plant Cell 28, 2147–2160. doi: 10.1105/tpc.16.00428
Tanaka, A., Ito, H., Tanaka, R., Tanaka, N. K., Yoshida, K., and Okada, K. (1998). Chlorophyll a oxygenase (CAO) is involved in chlorophyll b formation from chlorophyll a. Proc. Natl. Acad. Sci. 95, 12719–12723. doi: 10.1073/pnas.95.21.12719
Tanaka, Y., Taniyoshi, K., Imamura, A., Mukai, R., Sukemura, S., Sakoda, K., et al. (2021). MIC-100, a new system for high-throughput phenotyping of instantaneous leaf photosynthetic rate in the field. Funct. Plant Biol. doi: 10.1071/FP21029
Tsuda, M., Kaga, A., Anai, T., Shimizu, T., Sayama, T., Takagi, K., et al. (2015). Construction of a high-density mutant library in soybean and development of a mutant retrieval method using amplicon sequencing. BMC Genomics 16, 1014. doi: 10.1186/s12864-015-2079-y
Verdecia, M. A., Larkin, R. M., Ferrer, J.-L., Riek, R., Chory, J., and Noel, J. P. (2005). Structure of the mg-chelatase cofactor GUN4 reveals a novel hand-shaped fold for porphyrin binding. PLoS Biol. 3:e151. doi: 10.1371/journal.pbio.0030151
Wang, M., Li, W., Fang, C., Xu, F., Liu, Y., Wang, Z., et al. (2018). Parallel selection on a dormancy gene during domestication of crops from multiple families. Nat. Genet. 50, 1435–1441. doi: 10.1038/s41588-018-0229-2
Wang, P., Richter, A. S., Kleeberg, J. R. W., Geimer, S., and Grimm, B. (2020). Post-translational coordination of chlorophyll biosynthesis and breakdown by BCMs maintains chlorophyll homeostasis during leaf development. Nat. Commun. 11, 1254. doi: 10.1038/s41467-020-14992-9
Watanabe, S., Hideshima, R., Xia, Z., Tsubokura, Y., Sato, S., Nakamoto, Y., et al. (2009). Map-based cloning of the gene associated with the soybean maturity locus E3. Genetics 182, 1251–1262. doi: 10.1534/genetics.108.098772
Watanabe, S., Xia, Z., Hideshima, R., Tsubokura, Y., Sato, S., Yamanaka, N., et al. (2011). A map-based cloning strategy employing a residual heterozygous line reveals that the GIGANTEA gene is involved in soybean maturity and flowering. Genetics 188, 395–407. doi: 10.1534/genetics.110.125062
Xia, Z., Watanabe, S., Yamada, T., Tsubokura, Y., Nakashima, H., Zhai, H., et al. (2012). Positional cloning and characterization reveal the molecular basis for soybean maturity locus E1 that regulates photoperiodic flowering. Proc. Natl. Acad. Sci. 109, E2155–E2164. doi: 10.1073/pnas.1117982109
Yamatani, H., Kohzuma, K., Nakano, M., Takami, T., Kato, Y., Hayashi, Y., et al. (2018). Impairment of Lhca4, a subunit of LHCI, causes high accumulation of chlorophyll and the stay-green phenotype in rice. J. Exp. Bot. 69, 1027–1035. doi: 10.1093/jxb/erx468
Yamatani, H., Sato, Y., Masuda, Y., Kato, Y., Morita, R., Fukunaga, K., et al. (2013). NYC4, the rice ortholog of Arabidopsis THF1, is involved in the degradation of chlorophyll-protein complexes during leaf senescence. Plant J. 74, 652–662. doi: 10.1111/tpj.12154
Keywords: soybean, chlorophyll, CaaX-like protease, NGS-based bulked segregant analysis, GmBCM1
Citation: Yamatani H, Heng T, Yamada T, Kusaba M and Kaga A (2021) Identification and Characterization of an Early Leaf Senescence Gene ELS1 in Soybean. Front. Plant Sci. 12:784105. doi: 10.3389/fpls.2021.784105
Received: 27 September 2021; Accepted: 22 November 2021;
Published: 16 December 2021.
Edited by:
Guo-Liang Jiang, Virginia State University, United StatesCopyright © 2021 Yamatani, Heng, Yamada, Kusaba and Kaga. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Akito Kaga, a2FnYUBhZmZyYy5nby5qcA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.