- 1State Key Laboratory of Crop Stress Biology for Arid Areas, College of Life Sciences, Northwest A&F University, Yangling, China
- 2State Key Laboratory of Crop Stress Biology for Arid Areas, College of Plant Protection, Northwest A&F University, Yangling, China
- 3State Key Laboratory of Crop Stress Biology for Arid Areas, College of Innovation and Experiment, Northwest A&F University, Yangling, China
Members of the R2R3-MYB transcription factor superfamily have been implicated in plant development, improved disease resistance, and defense responses to several types of stresses. To study the function of TaMYB29 transcription factor—a member of the R2R3-MYB superfamily—in response to an avirulent race of stripe rust pathogen, Puccinia striiformis f. sp. tritici (Pst), we identified and cloned the TaMYB29 gene from wheat cultivar (cv.) AvS+Yr10 following infection with Pst. The TaMYB29 protein, comprising 261 amino acids, contains two highly conserved MYB domains. We first showed that TaMYB29 is a transcription factor, whose transcriptional levels are significantly induced by salicylic acid (SA), abscisic acid (ABA), jasmonic acid (JA), ethylene (ET), and Pst. The results showed that TaMYB29 is involved in the wheat response to stipe rust. The overexpression of the TaMYB29 gene resulted in the accumulation of reactive oxygen species (ROS) and pathogen-independent cell death in Nicotiana benthamiana leaves. The silencing of TaMYB29 gene in wheat cv. AvS+Yr10, containing the stripe rust resistance gene Yr10, promoted hyphae growth, significantly downregulated the expression of pathogenesis-related (PR) genes, and substantially reduced the wheat resistance to Pst compared with the non-silenced control. In addition, the accumulation of hydrogen peroxide (H2O2) significantly decreased, and the activity of catalase, an enzyme required for H2O2 scavenging, was elevated. Altogether, TaMYB29 positively regulates the defense response against stripe rust in wheat AvS+Yr10 by enhancing H2O2 accumulation, PR gene expression, and SA signaling pathway-induced cell death. These results provide new insights into the contribution of TaMYB29 to the defense response against rust pathogens in wheat.
Introduction
Wheat is an important food crop grown worldwide and the second most consumed crop in China (Tian et al., 2012). Puccinia striiformis f. sp. tritici (Pst), a fungal pathogen and causative agent of stripe rust disease, has severely affected the annual yield of wheat (Park et al., 2007; Chen et al., 2014). Several control measures have been implemented worldwide to control this pathogen; these aim at enhancing the tolerance of wheat to the infection (Seo and Park, 2010; Zhang et al., 2012a). The members of the Myeloblastosis (MYB) gene superfamily have been reported to actively participate in the developmental processes and defense responses of plants, attracting the worldwide attention of several plant scientists (Fujita et al., 2006; Wang et al., 2015).
Being one of the largest superfamilies of transcription factors in plants, several MYB genes have been isolated and identified from different plants (Dubos et al., 2010). The MYB protein contains a protein domain, typically 50 to 53-amino acid long, located near its N-terminus. MYB protein is divided into four categories, namely, 4R-MYB, R1R2R3-MYB, R2R3-MYB, and MYB-related, based on the number of repeats in this domain (Li et al., 2017; Zhang et al., 2018). The majority of MYB proteins belong to the R2R3-MYB gene subfamily. The R2R3-MYB proteins contain three uniformly spaced tryptophan residues within the MYB domain regions, except for R3, in which the first tryptophan residue is replaced by phenylalanine. The MYB repeat has a helix-turn-helix conformation, which forms a hydrophobic core and binds to DNA major grooves to exert their effects during transcription (Ogata and Nishimura, 1995). The first plant MYB gene COLORED1 was cloned and identified from maize (Zea mays) and is involved in anthocyanin biosynthesis (Paz-Ares et al., 1987). Since then, functions of several R2R3-MYB genes have been discovered and reported in different plant species, including fundamental metabolism, plant growth, cell apoptosis, defense response to abiotic and biotic stresses, and signal transduction (Kranz et al., 1998; Cedroni et al., 2003; Zheng et al., 2017).
The MYB gene supports a wide range of signaling crosstalks between biotic and abiotic stress signals (Grant et al., 2003; Chini et al., 2010). For example, the AtMYB96 transcription factor is involved in abscisic acid (ABA)-mediated drought response. It enhances the pathogen resistance of plants by promoting the biosynthesis of salicylic acid (SA), implying that the MYB gene affects the ABA–SA signaling crosstalk (Seo and Park, 2010). Similarly, AtMYB102 responds to salt stress, JA, ABA, wounds, and defense against herbivore attacks (De Vos et al., 2006). AtMYB108 is involved in biotic and abiotic stress crosstalks and regulated by JA (Mengiste et al., 2003). AtMYB15 is known to improve drought and salt tolerance in Arabidopsis, possibly by the ABA signaling pathway (Ding et al., 2009). TaPIMP1, an R2R3 MYB transcription factor in wheat, positively modulates defense responses to Bipolaris sorokiniana and drought stress via the ABA–SA signaling pathways in wheat (Zhang et al., 2012b). These findings indicate that several MYB genes execute their functions in a highly coordinated manner through a complex signaling network.
At the level of plant–pathogen interaction, a lot of previous studies demonstrated that the transcriptional products of MYB genes regulate plant disease resistance. In this regard, AtMYB30 is identified as a positive regulator during a hypersensitive response (HR) to pathogen attacks in plants (Vailleau et al., 2002; Raffaele et al., 2006). AtMYB46 modulates the disease susceptibility of Arabidopsis to the fungal pathogen Botrytis cinerea (Ramirez et al., 2011). Similarly, AtMYB73 of Arabidopsis thaliana is implicated in NPR1-mediated SA and JA signaling pathways against necrotrophic fungal pathogen Bipolaris oryzae (Jia et al., 2011). AtMYB108 confers plant resistance to the necrotrophic fungal pathogen Alternaria brassicicola (Mengiste et al., 2003). The tobacco MYB1 gene induced by Tobacco mosaic virus infection participates in HR reactions and systemic acquired resistance (SAR). It is located downstream of the SA regulatory pathway and regulates the expression of PR1 and other disease-related genes (Yang and Klessig, 1996). The overexpression of R2R3-MYB gene TiMYB2R-1 from Thinopyrum intermedium is known to significantly improve the resistance of transgenic wheat to the take-all disease (Liu et al., 2013). The barley MYB transcription factor gene HvMYB6 confers resistance in barley to powdery mildew. Silencing of this gene increases the sensitivity of barley to Blumeria graminis, the causative agent of powdery mildew, whereas its overexpression enhances the disease resistance of transgenic barley (Chang et al., 2013). In wheat, the overexpression of TaMYB86 significantly enhances the root rot resistance of transgenic wheat (Shan et al., 2016). TaMYB4 shows a high similarity to certain R2R3-MYB transcription factors and is implicated in the signaling pathways activated by ABA, ET, and SA hormones to induce stress defense response. In addition, it can promote programmed cell death and increase wheat tolerance to Pst in the infection phase (Ma et al., 2011; Al-Attala et al., 2014). Overexpression and underexpression experiments of the wheat R2R3 MYB gene TaPIMP1 show that TaPIMP1 provides defense against B. sorokiniana (Zhang et al., 2012b).
Although several MYB transcription factors in wheat have been functionally studied, more in-depth studies are needed to reveal the function of the MYB genes in the complex hexaploid wheat. Zhang et al. (2012a) isolated 60 wheat MYB genes containing full-length gene sequences, of which 20 genes were reported to participate in defense response to multiple abiotic stresses involving complicated signaling pathways. Although MYB is the largest transcription factor family in plants, the literature on its functions related to wheat disease resistance is scarce. To elucidate the molecular regulatory mechanisms involved in wheat–Pst interaction, we isolated a highly induced MYB gene from wheat cultivar (cv.) AvS+Yr10 after inoculating it with stripe rust fungus. The gene showed a sequence similar to that of the Chinese Spring TaMYB29. The TaMYB29 gene was induced by high salt and exogenous ABA (Zhang et al., 2012a). In addition, TaMYB29 along with TaMYB19 and TaMYB44 can co-regulate wheat plant phloem-based defense (PBD) against phloem-feeding insects; this function is executed through crosstalk with the ET signaling pathway (Zhai et al., 2017). These results imply the ability of TaMYB29 to respond to biotic and abiotic stresses and suggest that its function is mediated via different plant signaling pathways. Therefore, we further wanted to characterize the function of TaMYB29 during the wheat defense response against stripe rust. In this study, we characterized three TaMYB29 homologous genes, TaMYB29-5A, TaMYB29-5B, and TaMYB29-5D from wheat cv. AvS+Yr10 and investigated the subcellular localization and transcription activation properties of the TaMYB29-5B protein. The transcript abundance of TaMYB29 was studied using quantitative real-time PCR (qRT-PCR) in AvS and its near-isogenic lines (NILs) AvS+Yr10 seedlings inoculated with Pst pathotype CYR32, and in response to different exogenous hormone treatments. The transient expression of TaMYB29-5B confers auto active HR response to tobacco. Furthermore, we used barley stripe mosaic virus-induced gene silencing (BSMV-VIGS) to show that TaMYB29 is involved in defense against Pst. The relationships between TaMYB29 silencing and SA concentration, accumulation of reactive oxygen species (ROS), and the expression of pathogenesis-related (PR) genes were studied. In addition, we evaluated the pathogen growth and cell death. Our results suggest that TaMYB29 is a positive regulator of resistance to Pst in wheat and executes this function through the accumulation of SA and ROS. We believe that the results of this study contribute to an increased understanding of the structure and function of TaMYB29.
Materials and Methods
Plant Material
Two wheat (Triticum aestivum L.) cultivars, tobacco (Nicotiana benthamiana), and Pst pathotype CYR32 were used in this study. Wheat AvocetS (AvS) and its near-isogenic line AvSYr10NIL (AvS+Yr10) wheat seedlings carrying the Yr10 resistance gene were grown in a growth chamber at 14°C under a 16h light/8h dark photoperiod. N. benthamiana was maintained in an artificial climate room with a temperature range of 20 to 22°C, a light intensity of 20,000Lux, and a 16-h photoperiod.
Pathogen and Inoculation
The Pst race CYR32 that is compatible to wheat AvS and incompatible to AvS+Yr10 was provided by the State Key Laboratory of Crop Stress Biology for Arid Areas, NWAFU, China. The uredospores of the CYR32 pathotype were suspended in isohexadecane (IHD) and sprayed evenly on the first leaves of AvS and AvS+Yr10 wheat seedlings using a small air pump. After inoculation, these seedlings were maintained for 24h in a dark chamber with 100% relative humidity. These were subsequently returned to the original photoperiod condition. The seedlings in the control group were treated similarly with IHD containing no uredospores.
RNA Extraction From Wheat Leaves and cDNA Preparation
The wheat leaves were sampled at 0, 12, 24, 48, 72, and 120h post-inoculation (hpi) and immediately frozen in liquid nitrogen and stored at −80°C before extraction of total RNA. The total RNA of wheat leaves was extracted using the Qiagen RNeasy Plant Mini Kit (Qiagen, Valencia, CA, United States) following the manufacturer’s instructions. The quality and concentration of RNA were assessed using a 1.5% agarose gel electrophoresis and 260/280abs measurement using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, United States), respectively. Next, cDNAs were synthesized using the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, United States) following the manufacturer’s protocol.
Cloning and Sequence Analysis of TaMYB29 Gene
To clone the TaMYB29 gene, a pair of primers (M29-F/R) were designed using the Primer 5.0 software. The primers were designed based on the TaMYB29 sequence released by the Chinese Academy of Agricultural Sciences in 2012 (NCBI GenBank Accession No. JF951912.1); these were largely used to amplify the open reading frame (ORF) of the TaMYB29 gene. The PCR program was as follows: 3min at 95°C, followed by 35cycles of 30s at 95°C, 30s at 55°C, and 60s at 72°C, and finally 10min at 72°C. The PCR mixture contained the following: 0.125μl of Takara Ex Taq, 2.5μl of 10× Ex Tag buffer, 2μl of dNTP mixture, 2μl of template, and 1μl of each primer. Next, the total volume was made up to 20μl with ddH2O. The target fragments were inserted into the pGEM-T Easy vector (Promega, Madison, WI, United States) and sequenced. The TaMYB29 sequence was used to BLAST the related MYB genes at the National Center for Biotechnology Information1 and the Ensemble plant database at http://plants.ensembl.org/index.html. The DNAman program (Lynnon Biosoft, Quebec, Canada) was used to align all nucleotide sequences of these genes and corresponding deduced protein sequences.
RNA Analysis Using Quantitative Real-Time PCR
To determine the expression profiles of TaMYB29 after different treatments, a pair of primers (M29-qRT-F/R) were designed. In addition, four pairs of primers were designed to detect the expression patterns of TaPR1, TaPR2, TaPR5, and TaCAT genes by quantitative real-time PCR (qRT-PCR). Wheat housekeeping gene actin was used as an internal control. All primers used are listed in Supplementary Table S1. The qRT-PCRs were performed on a CFX96 Real-Time System (Bio-Rad, Munich, Germany). The reactions were performed in a 20μl volume containing 10μl of 2×UltraSYBR mixture, 2μl of diluted cDNA, 1μl of each primer, and 6μl of ddH2O. The amplification conditions were as follows: a pre-denaturation for 10min at 95°C, followed by 40cycles of 15s at 95°C and 60s at 60°C. A dissociation curve was generated for every reaction to ensure specific amplification. Each reaction was performed in triplicate, and reactions without templates were used as negative controls. Three independent biological replicates were used for each time point and treatment. The 2−ΔΔCt method was applied to analyze the experimental results (Livak and Schmittgen, 2001).
Determination of Endogenous SA Concentration in Wheat Leaves
To measure the endogenous SA concentration under both compatible and incompatible interactions, the wheat leaves were sampled at 0, 12, 24, 48, 72, and 120hpi with CYR32 pathotype. These were frozen in liquid nitrogen immediately. The SA concentration was analyzed using liquid chromatography-mass spectrometry (LC-MS) on an LC-30A+TripleTOF5600+ machine (AB Sciex, Singapore) according to Wang et al.’s (2017) method. In brief, 200mg of the sample was ground into a powder in liquid nitrogen and subsequently extracted with 750μl of MeOH–H2O–HOAc (90:9:1, v/v/v). The solution was centrifuged at a high speed of 10,000rpm. Next, the supernatant was collected and dried under nitrogen. The extract was dissolved in 1,000μl of HPLC-grade MeOH and filtered using a Millex-HV 0.22μm filter (Millipore, Bedford, United States). Three independent biological replicates were used for each time point.
Expression Profiles of TaMYB29 Under Hormone Treatments
To analyze the expression profiles of TaMYB29 under different hormone treatments, the one-leaf stage wheat seedlings of AvS+Yr10 were sprayed with 2μm methyl jasmonate (MeJA), 2μm ethylene (ET), 2μm abscisic acid (ABA), and 20μm salicylic acid (SA), following Wang et al.’s (2013) method. The mock control seedlings were similarly treated with an equal volume of distilled water. Subsequently, the seedlings were cut with a sterilized scissor for sampling at 0h, 2h, 6h, 12h, 24h, and 48h post-treatment (hpt). The samples were immediately frozen in liquid nitrogen and stored at −80°C. Three independent biological replicates were used for each time point and control.
Subcellular Localization of TaMYB29 Protein
To study the subcellular localization of TaMYB29 protein, a pair of primers (163-M29-F/R) with restriction enzyme HindIII and BamHI sites were designed. The PCR product of TaMYB29 was subcloned into the 16318GFP vector. Next, the recombinant plasmid was amplified in Escherichia coli strain DH5α and extracted using the OMEGA Plasmid Maxi Kit. The wheat leaves were processed with cellulase R10 (Yakult Honsha) and macerozyme R10 (Yakult Honsha) to isolate protoplasts as previously described (Yoo et al., 2007). The TaMYB29-eGFP fusion vector and only eGFP vector (control) were transformed into the wheat protoplasts separately using PEG4000. The treated wheat protoplasts were maintained in the dark at 23°C for 24h and subsequently observed under a fluorescence microscope (Olympus Corporation, Tokyo, Japan).
In addition, the gateway cloning technology was used to link the TaMYB29 gene to the pK7WGF2.0 vector carrying an enhanced green fluorescent protein (eGFP) tag. The pK7-TaMYB29-eGFP and pK7-eGFP vectors served as controls were transformed into the Agrobacterium strain GV3101 (pSoup-p19) by electroporation. Strains carrying different recombinant vectors were cultured to an optical density of 0.8 at 600nm (OD600) and injected into 4-week-old N. benthamiana leaves using a 1.0ml syringe without a needle. The transformed N. benthamiana were maintained in a greenhouse at 20 to 22°C, a light intensity of 20,000Lux, and a 16-h photoperiod. Finally, the fluorescence of eGFP was observed under a laser confocal fluorescence microscope at 48h post-infiltration.
Verification of Transcription Activation by TaMYB29 Protein
The gene of full-length TaMYB291–261 protein, its N-terminal region TaMYB291–116, and C-terminal region TaMYB29117–261 were amplified using three pairs of primers containing NdeI and EcoRI restriction sites, namely, M29-BD-F/R, M29-BD-F/R116, and M29-BD-F117/R, respectively. The PCR products were purified and linked to the pGBKT7 vectors linearized by restriction endonucleases. The three pGBKT7-TaMYB29 recombinant plasmids were directly transformed into DH5α E. coli strain and amplified. Subsequently, the pGBKT7-TaMYB29 recombinant plasmids and the pGBKT7 plasmid (negative control) were introduced into the AH109 yeast strain containing the HIS3, ADE2, MEL1, and lacZ reporter genes. The transformed yeast strains were plated on a medium lacking tryptophan (SD/-Trp) and incubated at 30°C for 2 to 3days. The positive clones on the Trp-deficient media were selected and transferred into the yeast peptone dextrose adenine (YPDA) liquid media for culturing. Next, the yeast solutions were diluted to OD600 of 1, 10−1 10−2, 10−3, and 10−4. Lastly, these yeast solutions were inoculated into a medium lacking adenine, histidine, and tryptophan (SD-His/-Ade/-Trp) as well as a medium lacking the above three amino acids (SD-His/-Ade/-Trp) and supplemented with X-α-D-galactoside (X-α-Gal). These cultures were incubated at 30°C for 3 to 4days.
Transient Expression of TaMYB29 in Nicotiana benthamiana
The TaMYB29 gene was amplified using a pair of primers (PVX-M29-F/R) containing SmaI and NotI restriction enzyme sites. The PCR product was subcloned into the pMD18-T simple vector. Afterward, the recombinant plasmid was amplified in DH5α E. coli strain and digested by SmaI and NotI restriction enzymes. Subsequently, the gene digestion product was inserted into the PVX vector which carries a 3×HA tag and was linearized by the same restriction enzymes. The PVX-TaMYB29 construct was transformed into the Agrobacterium strain GV3101 (pSoup-p19). Strains were grown in the Luria–Bertani (LB) medium containing 50mg/ml rifampicin and 25mg/ml kanamycin at 28°C for 24h. The bacteria were harvested by centrifugation and resuspended in the acetosyringone (AS) buffer containing 10mm MES (pH 5.6), 10mm MgCl2, and 150μm acetosyringone after being washed with 10mm MgCl2. Strains carrying different recombinant vectors were adjusted to a density of 0.6 at 600nm (OD600) with AS buffer and incubated for 2h in the dark at room temperature before leaf infiltration. These leaves were sampled at 1, 2, and 3days post-infiltration (dpi) and stained with 3,3′-diaminobenzidine (DAB) or trypan blue (Bai et al., 2012). The PVX-GFP construct was used as a control and treated in the same manner. These experiments were repeated at least three times and got the same result.
BSMV-VIGS-Mediated TaMYB29 Gene Silencing
A conserved region of about 200bp was selected from the cloned TaMYB29 sequence to design the primers (M29-vigs-F/R) for silencing the fragments. The PCR product was linked to the BSMV:γ modified vector to construct the BSMV:γ-TaMYB29 recombinant plasmid as previously described (Wang et al., 2020a). Next, the positive plasmid was amplified in DH5α E. coli strain. A construct carrying only the BSMV genome was used as a negative control and named BSMV:00. A construct carrying a 183-bp phytoene desaturase (PDS) gene was used as a positive control and named BSMV:PDS. The recombinant γ virus vector linearized by BssHII and RNAα, RNAγ linearized with MluI, and RNAβ with SpeI were transcribed in vitro using the mMessage mMachine T7 in vitro transcription kit (Ambion, Austin, TX, United States) following the manufacturer’s protocol. The transcription products of BSMV:α, β, γ (or γ-target) were diluted thrice with diethylpyrocarbonate (DEPC) water and mixed in a ratio of 1:1:1. Next, 70μl of FES buffer which is a kind of inoculation buffer containing a wounding agent was added to every 30μl of the mixture. The second leaves of AvS and AvS+Yr10 wheat seedlings were infected with BSMV using the method described by Holzberg et al. (2002). The BSMV-infected wheat seedlings were maintained in a 16h/8h photoperiod at 25°C. In addition, the seedlings were mock inoculated with FES buffer devoid of BSMV transcripts. At 10 to 14days after the virus inoculation, photobleaching was observed on BSMV:PDS seedlings. The third and fourth leaves were further inoculated with uredospores of Pst pathotype CYR32. Next, these plants were directly maintained in a growth chamber for 24h in the dark with 100% relative humidity at 15°C. The leaves were sampled at 0, 24, 48, and 120hpi for histological changes and analyzed by qRT-PCR.
Histological Study of Fungal Growth and Host Response
Wheat leaves sampled at 0, 24, 48, and 120hpi in control and silenced plants were observed for ROS accumulation by DAB staining and HR response areas because of the auto-fluorescence of necrotic cells using an Olympus BX-51 fluorescence microscope (Olympus Corp. Tokyo, Japan). Then, the leaf segments were stained with wheat germ agglutinin (WGA) and observed for the infection areas and hyphae development of Pst as previously described (Wang et al., 2017). At least 30 infection sites were examined on each of five randomly selected leaf segments per treatment. Infection sites with substomatal vesicles were considered successfully penetrated. Standard deviations were calculated and Tukey’s test was performed for statistical analysis using the SPSS software (SPSS, Inc. Chicago, United States).
Results
Sequence Analyses of TaMYB29 cDNA and Protein
A full-length cDNA fragment was successfully isolated due to its high expression under incompatible interaction of wheat cv. AvS+Yr10 infected with avirulent Pst CYR32. The cDNA sequence displayed 100% identity with the referred TaMYB29 sequence in the NCBI GenBank (accession no. JF951912.1). In addition, the ORF was cloned using a pair of primers to create a 786-bp fragment. The International Wheat Genomic Sequence Consortium (IWGSC) revealed homologs of TaMYB29 located on chromosomes 5A, 5B, and 5D in wheat cv. Chinese Spring. The alignment results of these nucleotide sequences also showed a high similarity ranging from 96 to 99%, referred to as TaMYB29-5A, TaMYB29-5B, and TaMYB29-5D (Supplementary Figure S1). The TaMYB29 sequence of AvS+Yr10 displayed the highest similarity to the nucleotide sequence of the Chinese Spring TaMYB29-5B.
The deduced protein of TaMYB29 had 261 amino acids, with a predicted molecular weight of 29.4 kD and an isoelectric point (pI value) of 9.05. Alignments of all wheat alleles of TaMYB29 with wheat TaMYB4 (AEG64799.1), Arabidopsis AtMYB30 (AEE77505.1), and Arabidopsis AtMYB4 (AEE86955.1) revealed two highly conserved regions R2 and R3, which contained three uniformly spaced tryptophan residues responsible for the interaction between the MYB protein and specific DNA sequences (Figure 1). The second and third tryptophan residues were conserved in all R2R3-MYB protein members, except in R3 repeats, where the first tryptophan was replaced by phenylalanine (Zhang et al., 2012a). These results revealed highly conserved amino acid sequences in these MYB proteins, indicating their similar functions in mediating stress response.

Figure 1. Alignment of the amino acid sequence of TaMYB29 with its homologous sequences. The sequences were aligned with DNAMAN. The identical residues are shaded. The black arrow line shows the MYB-binding domain. The asterisks (∗) show typical R2R3-MYB protein contains three regularly spaced tryptophan (W) residues and the first tryptophan residue was replaced by phenylalanine (F) in the R3 repeats.
Subcellular Localization of TaMYB29 Protein
To study the subcellular localization of TaMYB29 in plants, the target gene fused with the eGFP vector and only eGFP vector (control) was separately transformed into wheat protoplasts. To avoid the interference of chlorophyll, its distribution was studied individually. After analyzing the fluorescence microscopy observations, the exact localization of TaMYB29-eGFP fusion protein was found to be the nucleus of wheat protoplast, whereas green signals of only eGFP were observed both in the cytoplasm and the nucleus (Figure 2A). We subsequently used the leaves of N. benthamiana as the transformed material and repeated the subcellular localization experiment and obtained the same results (Figure 2B). Therefore, we conclude that TaMYB29 is a nuclear protein.
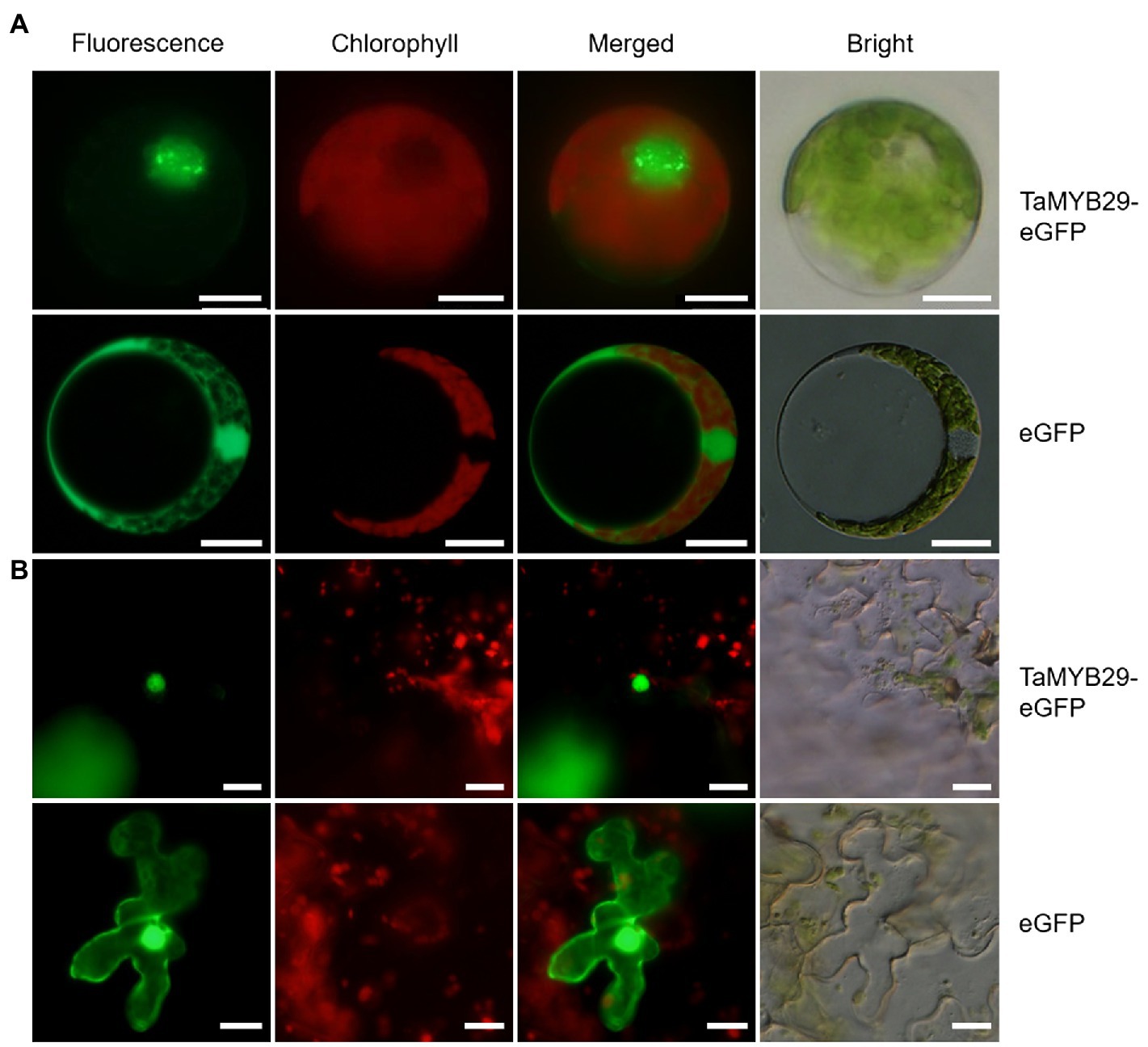
Figure 2. Subcellular localization of TaMYB29 protein. (A) TaMYB29-eGFP fusion protein and enhanced green fluorescence protein (eGFP) were expressed in wheat protoplasts; (B) TaMYB29-eGFP fusion protein and eGFP protein were expressed in the leaves of Nicotiana benthamiana. Bars=20μm.
Transcriptional Activity of TaMYB29 Protein
To identify whether the TaMYB29 protein is a transcriptional activator or repressor, pGBKT7-TaMYB291–261 and its two truncated protein plasmids pGBKT7-TaMYB291–116 and pGBKT7-TaMYB29117–261 were transformed into the AH109 yeast strain. In addition, the pGBKT7 plasmid (negative control) was transformed simultaneously. All transformants grew well on a selective synthetic defined (SD) medium lacking tryptophan (SD/-Trp); however, only pGBKT7-TaMYB291–261 and pGBKT7-TaMYB29117–261 yeast strains showed growth on the selective medium without tryptophan, histidine, and adenine (SD-Trp/-His/-Ade). Furthermore, the colonies turned blue on the same medium supplemented with X-α-Gal (SD-Trp/-His/-Ade+X-α-Gal; Figure 3). Therefore, we conclude that TaMYB29 functions as a transcriptional activator. What is more, the transactivation domain of TaMYB29 is located in the C-terminal region.
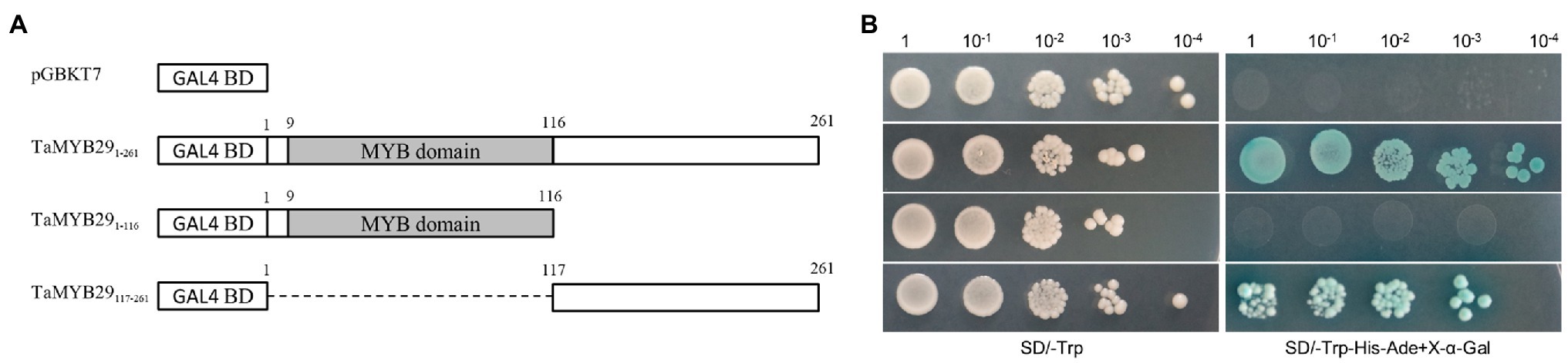
Figure 3. Transcriptional activation activity of the TaMYB29 proteins. (A) Constructs of pGBKT7-TaMYB291–261 (full-length) and pGBKT7-TaMYB291–116 (N-terminal region), pGBKT7-TaMYB29117–261 (C-terminal region), and pGBKT7 as a control were separately transformed into the yeast strain AH109. (B) The diluted yeast solutions were dropped on the SD/-Trp and SD/-Trp-His-Ade+X-α-Gal media, respectively; then, the cultures were incubated at 30°C for 3–4days.
Expression Profile of TaMYB29 in Response to Exogenous Hormones
To understand the regulation of TaMYB29 by plant hormones, the AvS+Yr10 wheat seedling leaves were treated with four different exogenous hormones (SA, ABA, JA, and ET) at the two-leaf stage. The TaMYB29 transcript levels were detected by qRT-PCR at six different time points (Figure 4). Regarding SA treatment, the relative expression of TaMYB29 showed a 2.59-fold increase at 12h and a 10-fold increase at 24h. Eventually, the increase at 48h was 11 times more than that in the control at 0h. The TaMYB29 expression of leaves treated with ABA showed a similar increasing trend. However, the increase was considerably high at 48h, 50 times as high as that in the control. With respect to JA treatment, the highest level was measured at 12h, with upregulation recorded from 12h to 48h. The upregulation was also observed in ET-treated leaves, especially at 2h, 12h, and 24h. However, the expression peak of TaMYB29 induced by JA or ET treatment was not as high as that detected with the ABA treatment. These results demonstrate that TaMYB29 exert an effect in the plant through complex hormone signal pathways.
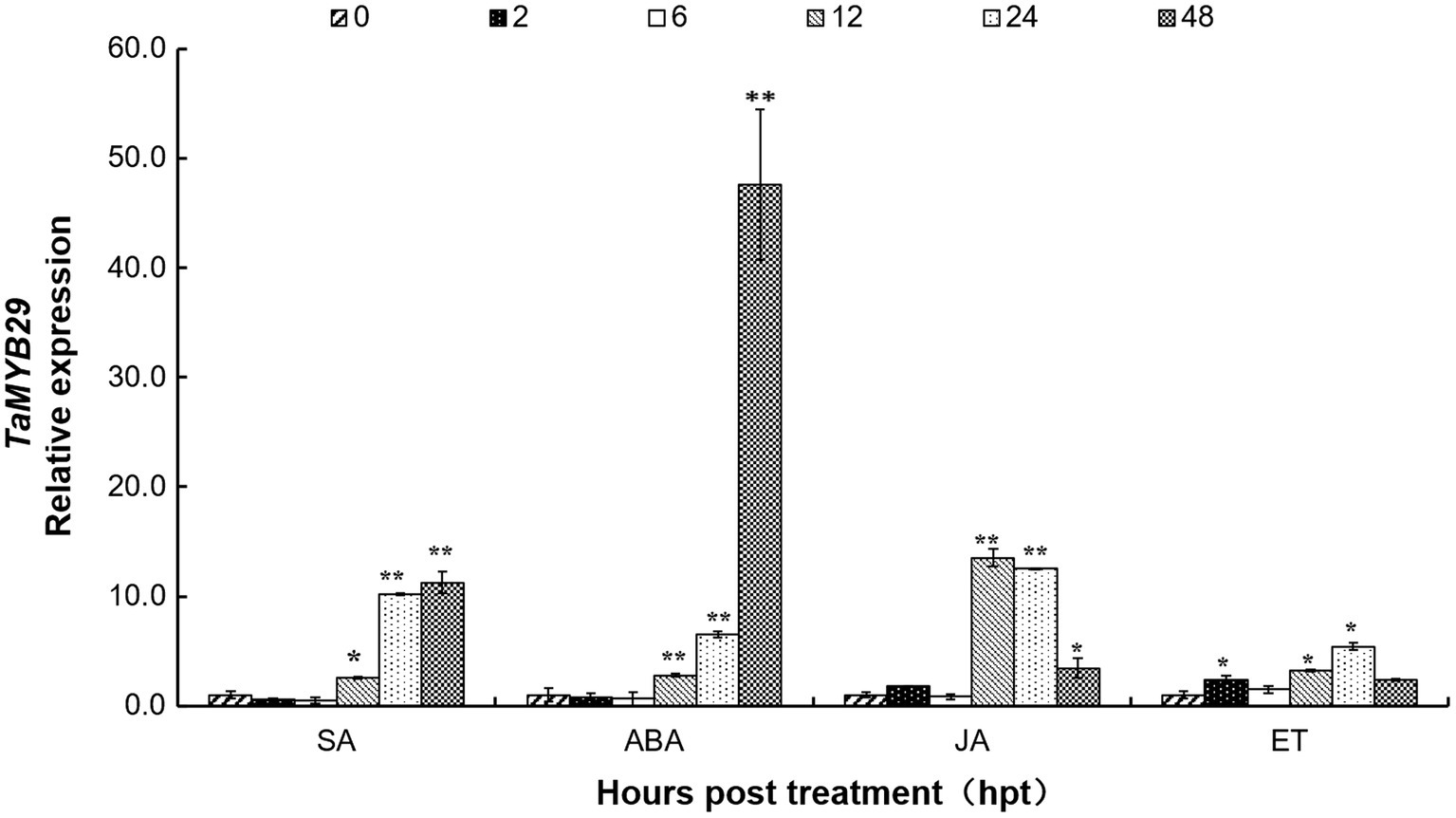
Figure 4. The expression profiles of TaMYB29 in AvS+Yr10 wheat leaves in response to exogenous hormones. The wheat leaves were sprayed with SA (salicylic acid), ABA (abscisic acid), JA (methyl jasmonate), and ET (ethylene), respectively and were sampled at 0, 2, 6, 12, 24, and 48h post-treatment (hpt). Three independent biological replications were performed. Error bars represent standard deviation among three biological replications. Student’s t-tests were used for the statistical analyses and the asterisks (∗) and (∗∗) indicate a significant difference between that time point and 0hpt with a value of p<0.05 and 0.01, respectively.
SA Levels and Expression Profiles of TaMYB29 and TaPR1 in Response to Pst
To study the expression profile of TaMYB29 during the interaction between wheat and stripe rust pathogen, the relative expression of TaMYB29 in the wheat leaves was measured by qRT-PCR at 0, 12, 24, 48, 72, and 120h in two different wheat cultivars post-inoculation with CYR32 (Figure 5A). Wheat AvocetS (AvS), which is susceptible to Pst pathotype CYR32, forms a compatible interaction, and its near-isogenic line AvSYr10NIL (AvS+Yr10), carrying the Yr10 resistance gene and highly resistant to CYR32, forms an incompatible interaction. The expression of TaMYB29 in the leaves of AvS+Yr10 plants was significantly upregulated at four different time points, except 12hpi in contrast to the 0hpi control and peaked at 48hpi, nearly 3.5 times more than that in the control. However, the expression of TaMYB29 in the leaves of AvS plants showed an opposite trend, declining slightly until 24hpi at first. Although the transcript abundance increased slightly at 48hpi compared with 0hpi, it decreased to nearly half the level at 0hpi in the end at 120 hpi. The expression of TaMYB29 in the incompatible interaction was always higher than that in the compatible interaction at five different time points. These results indicate that TaMYB29 functions during the resistance of wheat against Pst infection, especially in the incompatible interaction.
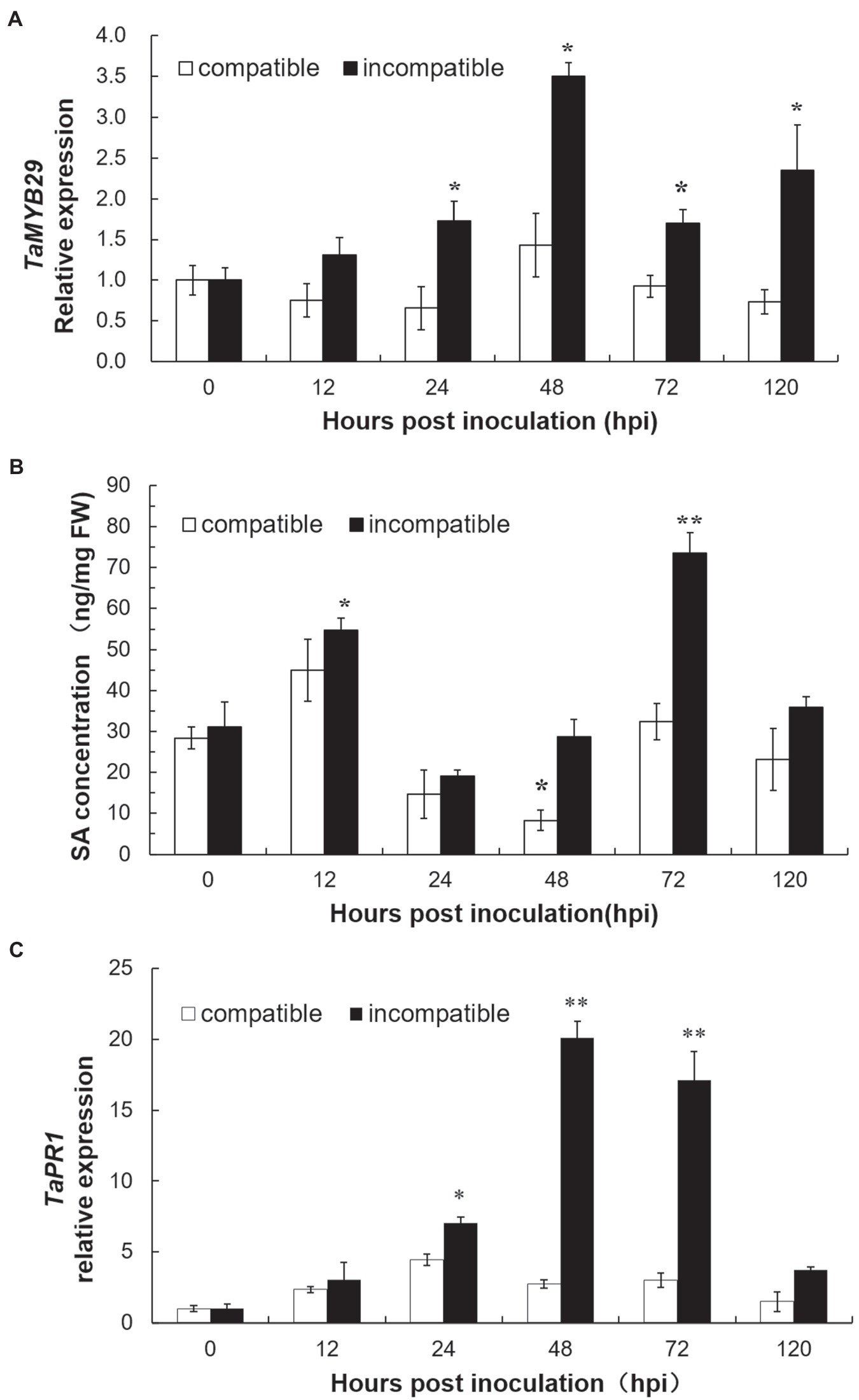
Figure 5. The expression profiles of TaMYB29 (A) and TaPR1 gene expression (C) and salicylic acid (SA) concentration (B) in AvS and AvS+Yr10 wheat leaves during the time course of stripe rust infection. The leaves were sampled at 0, 12, 24, 48, 72, and 120h post-inoculation (hpi) with three biological experiments replicates. Error bars represent standard deviation among three biological replicates. Student’s t-tests were used to for the statistical analyses and the asterisks (∗) and (∗∗) indicate a significant difference between the particular time point and 0hpi with a value of p<0.05 and 0.01.
We measured the endogenous levels of SA in both compatible and incompatible interactions at six time points (Figure 5B). In the incompatible interaction, the first significant increase in the SA levels appeared at 12 hpi, earlier than the TaMYB29 expression, and subsequently reduced at 24 and 48hpi, contrary to the observation in the TaMYB29 expression compared with the 0hpi control. The highest SA levels were obtained at 72hpi. Next, the SA levels rapidly returned to the basal levels at 120hpi. The SA levels in the compatible interaction presented a similar trend as in the incompatible system although the levels were lower than those in the incompatible system at every time point.
Furthermore, the expression of the wheat PR gene TaPR1 was measured by qRT-PCR (Figure 5C). TaPR1 was upregulated in both interactions during pathogenesis. In the incompatible interaction, the expression of TaPR1 significantly increased at 24hpi and reached the 20-fold increase at 48hpi, and maintained significantly high levels at 72hpi, which is considerably similar to the expression of TaMYB29. Next, it decreased at 120hpi, different from the expression of TaMYB29. In addition, the expression of TaPR1 in the compatible interaction was slightly upregulated; however, the level was lower as compared with that in the incompatible interaction.
TaMYB29-Induced ROS Accumulation and Pathogen-Independent Cell Death in Nicotiana benthamiana Leaves
The recognition of certain molecules released by pathogens triggers plant cell death and a subsequent defense response known as the HR response (Coll et al., 2011). To confirm whether TaMYB29 can induce ROS and cell death, the HR symptoms were detected in N. benthamiana leaves transformed by PVX:TaMYB29 and PVX:GFP constructs. After 1 to 2days post-infiltration (dpi), the accumulation of ROS was detected successfully with DAB staining (Figure 6A). The ROS accumulation in leaf areas infiltrated with PVX:TaMYB29 was apparent in contrast to that in control PVX:GFP. Subsequently, apoptosis was observed using trypan blue staining to further investigate the effects of TaMYB29 expression during ROS accumulation and pathogen-independent cell death (Figure 6B). The TaMYB29-induced cell death on N. benthamiana leaves was apparent. However, PVX:GFP control did not trigger cell death. The results showed that the expression of TaMYB29 successfully led to ROS accumulation and cell death in contrast to the GFP control, indicating that TaMYB29 played a crucial role in ROS accumulation and pathogen-independent cell death.
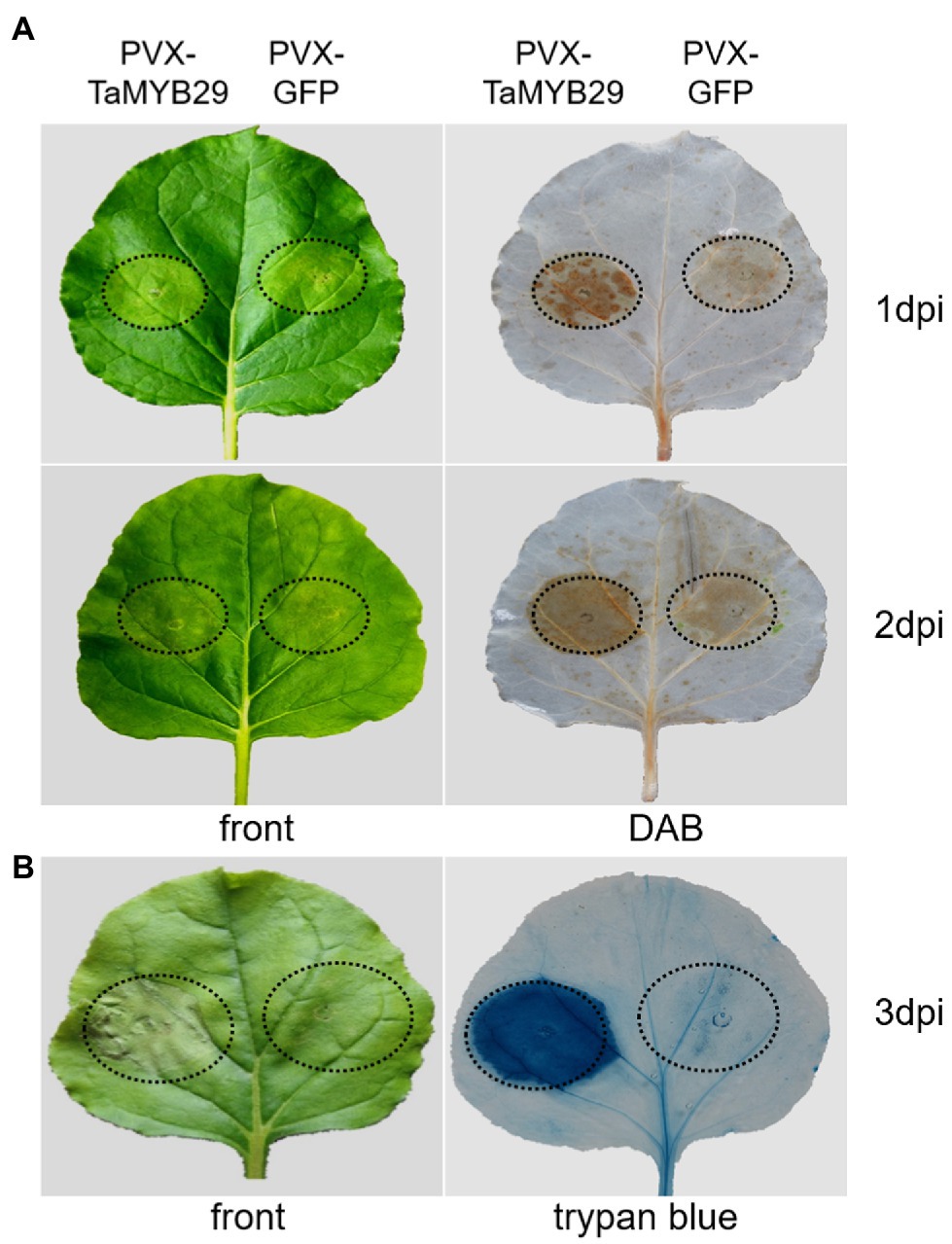
Figure 6. Expression of TaMYB29-induced ROS accumulation and pathogen-independent cell death in N. benthamiana leaves. (A) Symptoms of N. benthamiana leaf areas expressing TaMYB29 or GFP gene from 1 to 2days post-infiltration (dpi). The left and right panels represent the front and after DAB staining, respectively. (B) Symptoms of N. benthamiana leaf areas expressing TaMYB29 or GFP gene at 3 dpi. The left and right panels represent the front and after trypan blue staining, respectively. These experiments were repeated at least three times and got the same result.
TaMYB29 Gene Silencing of Wheat Reduced the Resistance to Stripe Rust
To study the effects of the TaMYB29 gene during the interaction between wheat and Pst, the target gene was silenced using the barley stripe mosaic virus-induced gene silencing (BSMV-VIGS) assay. At the beginning of the knockdown experiment, the PDS gene was used to ensure that the VIGS system worked normally and correctly. The photobleaching occurred on BSMV:PDS leaves, proving the effectiveness of the VIGS system (Figure 7A).
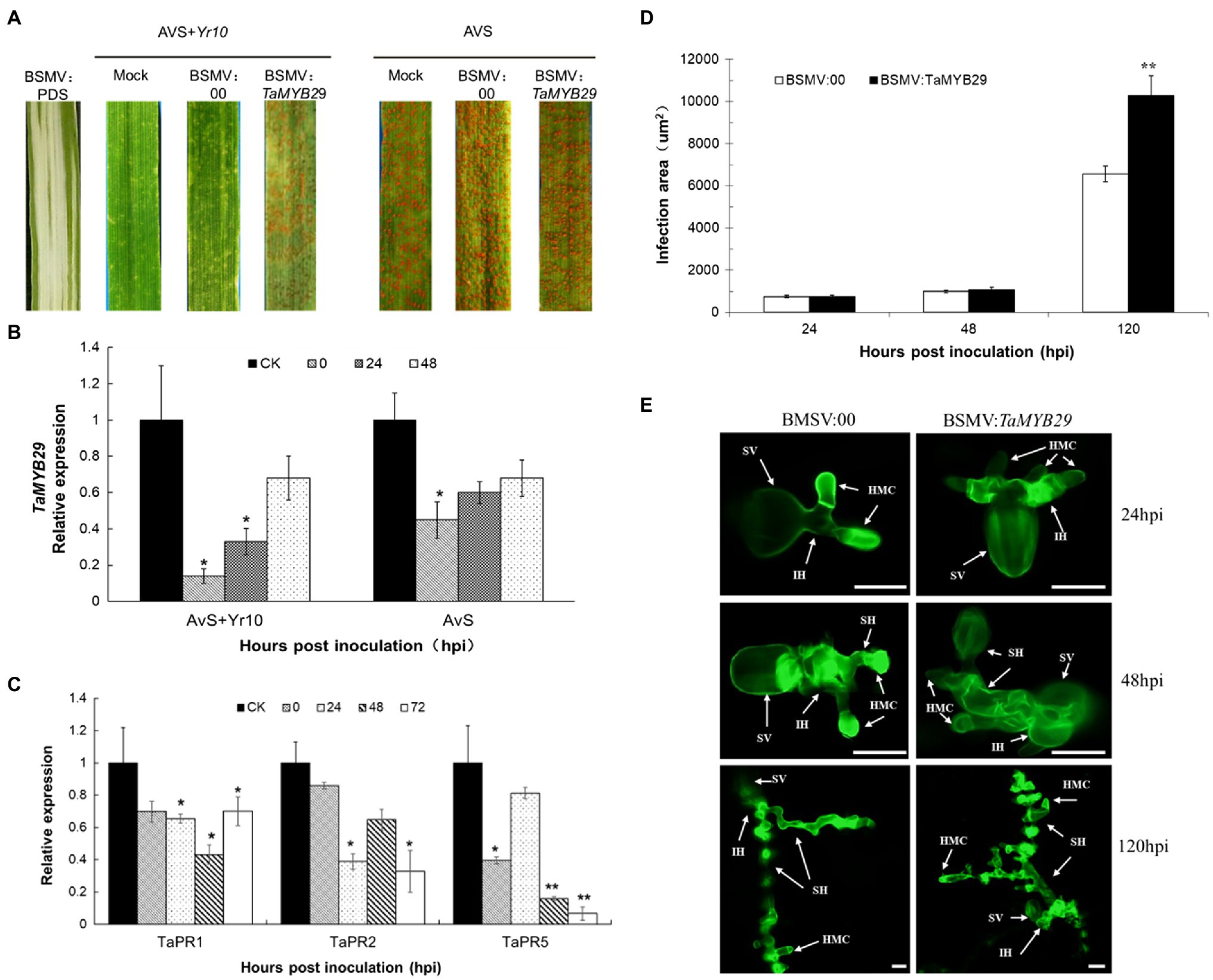
Figure 7. Functional analysis of the TaMYB29 gene by the BSMV-VIGS assay. (A) Infection type of AvS and AvS+Yr10 wheat leaves to CYR32 after inoculated with BSMV:00 or BSMV:TaMYB29. Stripe rust was inoculated 9days post-BSMV inoculation. Pictures were taken 15days post-rust inoculation. Mock: wheat leaves inoculated with FES buffer. BSMV:00 means only the BSMV genome; BSMV: TaMYB29 means a 183-bp fragment of TaMYB29 was inserted into the BSMV:γ genome. Photobleaching was evident on the leaves infected with BSMV:PDS but not on the mock-inoculated leaves. (B) Transcriptional level of TaMYB29 in AvS and AvS+Yr10 during the course of Pst infection at 0, 24, and 48hpi. CK: wheat leaves treated with BSMV:00. (C) Transcriptional level of pathogenesis-related gene TaPR1, TaPR2, and TaPR5 in TaMYB29 silenced AvS+Yr10 wheat leaves after inoculated with Pst. Leaves were sampled at 0, 24 48, and 72hpi. Three independent biological replications were performed. Error bars represent standard deviation among three biological replicates. Student’s t-tests were used for the statistical analyses and the asterisks (∗) and (∗∗) indicate a significant difference between that time point and 0hpi with a value of p<0.05 and 0.01, respectively. (D,E) Infection area (D) and hyphae development of Pst (E) in TaMYB29 knockdown AvS+Yr10 wheat leaves after infected with CYR32. Wheat leaves were sampled at 24, 48, and 120hpi and observed microscopically after stained with WGA. SV, substomatal vesicle; IH, initial hyphae; HMC, haustorial mother cell; and SH, secondary hyphae. Bars=20μm.
AvS and AvS+Yr10 wheat seedlings infected with FES buffer (mock), BSMV:00, and BSMV:TaMYB29 were further inoculated with uredospores of CYR32. Infection types were assessed based on McNeal measurement (Mcneal et al., 1971) to evaluate the differences between the phenotypes of mock, BSMV:00, and BSMV:TaMYB29. AvS+Yr10 displayed a resistant response after inoculation with CYR32 on the mock and BSMV:00 controls, characterized by a large necrosis area at the infection site. While a small number of fungal spores appeared on TaMYB29-silenced leaves. These results indicated that the resistance of wheat AvS+Yr10 with silenced TaMYB29 gene resulted in a significant decline from highly resistant to moderately susceptible. There was no susceptible phenotype change in TaMYB29-silenced AvS wheat compared with the control mock and BSMV:00 (Figure 7A). The relative expression of TaMYB29 was detected by qRT-PCR after TaMYB29 gene silencing before Pst inoculations labeled as 0hpi, and 24hpi, and 48hpi during Pst infection. The results showed that the TaMYB29 transcription level decreased in both AvS+Yr10 and AvS, which was knocked down up to 86% in incompatible interaction and 55% under compatible interaction compared with BSMV:00 labeled as CK (control check) in Figure 7B.
Pathogenesis-related genes are vital for mounting disease resistance responses in plants. TaPR1, TaPR2, and TaPR5 have been reported to be involved in systemic acquired resistance (SAR)—a type of plant immunization (Wang et al., 2020b). Therefore, we selected three PR genes as defense-related genes to investigate the effects of the knockdown of TaMYB29 before and during Pst inoculation. The results showed that the decline of TaPR1, TaPR2, and TaPR5 gene expression in BSMV:TaMYB29 compared with BSMV:00 was observed at four different time points in the incompatible interaction (Figure 7C), proving that the silencing of TaMYB29 reduced the wheat resistance to disease.
The infection area of Pst in TaMYB29-silenced AvS+Yr10 wheat leaves was examined microscopically after inoculation with CYR32. The results showed that the total infection area in TaMYB29-silenced leaves was similar to that in BSMV:00 control until 48hpi. However, significant differences between the two groups were observed at 120hpi (Figures 7D,E). Furthermore, the hyphae development of Pst in BSMV:TaMYB29 was stronger than that in BSMV:00, especially at 120hpi (Figures 7D,E).
In conclusion, TaMYB29 knockdown effectively reduced wheat resistance to stripe rust by downregulating the expression of PR genes and promoting the development of hyphae.
TaMYB29 Was Involved in ROS Accumulation and Hypersensitive Response in Wheat
We observed the accumulation of ROS and cell necrosis of AvS+Yr10 wheat leaves upon pathogen challenge. The production of H2O2 and the necrotic area in BSMV:TaMYB29 leaves were significantly less than those in BSMV:00 at 48hpi and 120hpi, creating a gap of about 2,000μm2 at 120hpi (Figures 8A,B,D). In addition, the expression patterns of TaCAT (X94352) that can eliminate H2O2 as a catalase gene in wheat were detected by qRT-PCR (Coleto et al., 2021). The results showed that the expression of TaCAT was significantly upregulated in TaMYB29-silenced leaves at 24hpi, peaked at 48hpi as compared with control non-TaMYB29-silenced leaves (Figure 8C), implying that a high expression of TaCAT reduced the ROS accumulation in TaMYB29-silenced wheat leaves. We hypothesized that the silencing of the TaMYB29 gene positively regulated the expression of the TaCAT gene, eventually decreasing the accumulation of ROS and partially eliminating the HR response in wheat mesophyll cells, thereby weakening the resistance of wheat to stripe rust.
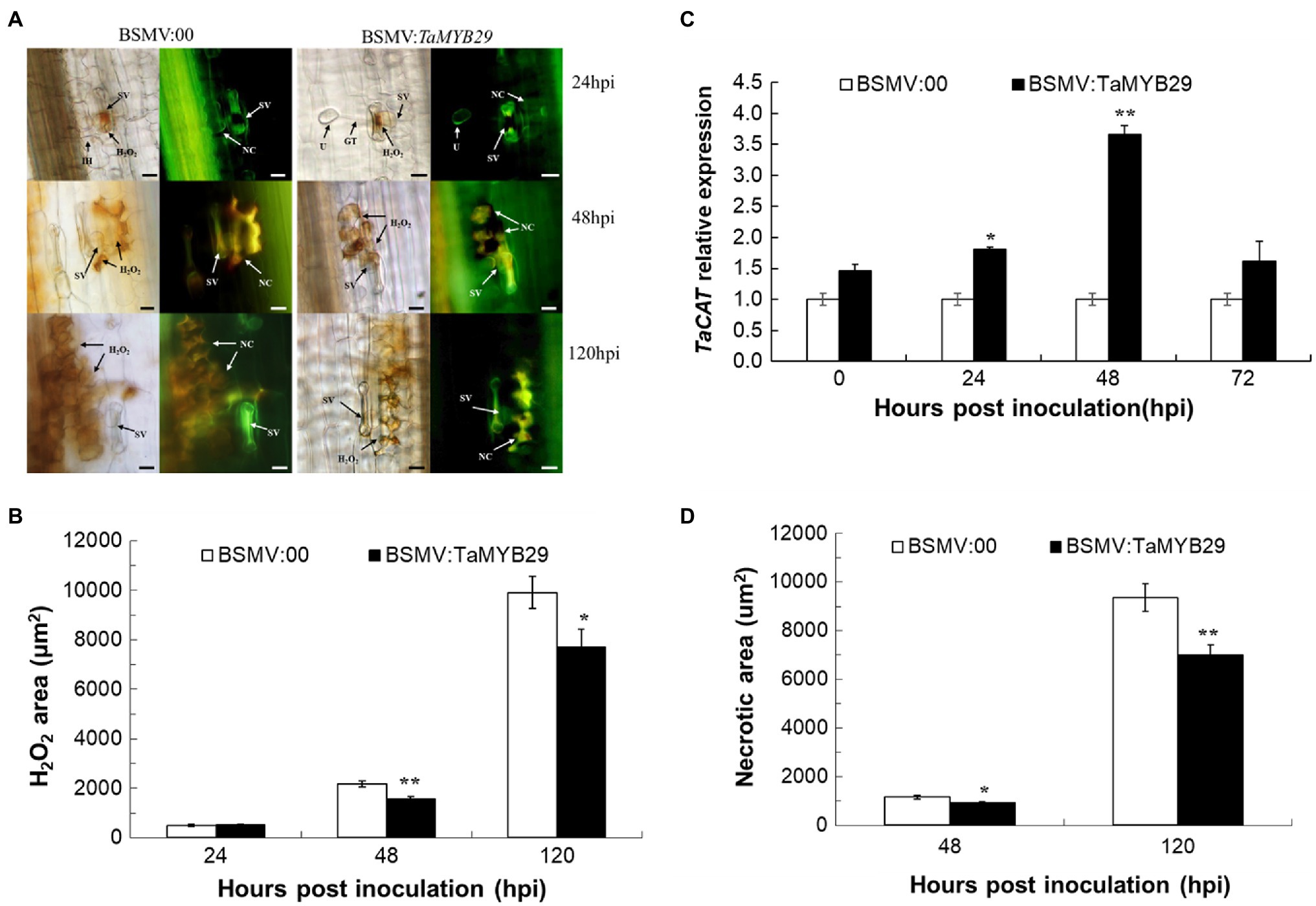
Figure 8. Reactive oxygen accumulation and related gene expression and hypersensitive response in the TaMYB29 knockdown AvS+Yr10 wheat leaves. (A) The reactive oxygen species and cell necrosis in wheat leaves in response to Pst observed under a fluorescence microscope. (B) The reactive oxygen species areas were stained with DAB and measured microscopically at 24, 48, and 120hpi. H2O2 accumulation was significantly reduced in TaMYB29 silenced leaves compared to the control. (C) Transcriptional level of ROS-related gene TaCAT in AvS+Yr10 after TaMYB29-scilencing at four time points relative to the expression in BSMV:00. (D) Cell death was measured microscopically because of the auto-fluorescence of necrotic cells at 48 and 120hpi. The area of green auto-fluorescence reduced in TaMYB29-silenced wheat plant. Pictures were taken under an epifluorescence or light microscopy at 24, 48, and 120hpi, respectively. BSMV:00 is the control group and BSMV:TaMYB29 is the experimental group in which TaMYB29 gene was silenced. U, urediniospore; GT, germ tube; NC, necrotic cell; SV, substomatal vesicle; IH, initial hyphae; HMC, haustorial mother cell; and SH, secondary hyphae. Bars=20μm. Error bars represent standard deviation among three biological replications. Student’s t-tests were used to for the statistical analyses and the asterisks (∗) and (∗∗) indicate a significant difference between particular hpi and controls with a value of p<0.05 and 0.01, respectively.
Discussion
We successfully cloned a relatively highly expressed MYB gene in response to the Pst attack in AvS+Yr10 wheat leaves. The cDNA sequence showed the same to the TaMYB29 gene (JF951912.1) searched using the NCBI GenBank database. Three homologous TaMYB29 genes obtained from the wheat genome database maintained by the IWGSC were located to wheat chromosomes 5A, 5B, and 5D separately. Multiple sequence alignment revealed that the three genes shared 96 to 99% similarity, implying that their co-silencing effectively represented the function of TaMYB29 genes in wheat. Therefore, we knocked down and measured the expression of three homologs simultaneously and used TaMYB29 to represent their functions. More in-depth research is required to determine the specific functions of each of the three homologs.
The MYB gene superfamily participates in several metabolic reactions in plants (Jin and Martin, 1999). For example, these MYB proteins interact with DNA as transcript factors to regulate the signal network of the defense response. The majority of MYB genes belong to the R2R3-MYB superfamily; proteins in this superfamily have R2 and R3 imperfect tandem repeats DNA-binding domains. A comparison of amino acid sequences with the other three MYB family genes of Arabidopsis and wheat—all of which function to resist abiotic and biotic stresses—revealed that TaMYB29 also included R2 and R3 conserved domains, suggesting its DNA-binding ability as a transcription factor (Figure 1). Its subcellular localization analysis revealed that TaMYB29 is a nuclear protein (Figure 2). Transcriptional activation assays in yeast verified the transcriptional activity of TaMYB29 and the necessity of C-terminal 117–261 amino acids for transactivation (Figure 3). Based on these results, we speculated that the TaMYB29 gene functions are similar to those of other resistance-related MYB genes previously reported as transcription factors (Morse et al., 2009; Katiyar et al., 2012; Jin et al., 2014).
The genes belonging to the MYB superfamily have been implicated in plant resistance against abiotic and biotic stresses (Saha et al., 2016). The expression of the AtMYB30 gene increased during incompatible interactions between Arabidopsis and bacterial pathogens and drought stress (Vailleau et al., 2002). The expression of AtMYB44 was upregulated following pathogen infection and treatment with defense-related SA phytohormones (Zou et al., 2013). Similarly, the expression of TiMYB2R-1 was significantly induced following G. graminis infection (Liu et al., 2013). The analysis of the expression patterns of 60 wheat TaMYB genes under different stress conditions revealed that 32 of them responded to these different stresses (Zhang et al., 2012a). Among these, TaMYB29 was induced by both high salt and exogenous ABA (Zhang et al., 2012a). We also found that TaMYB29 was induced by exogenous ABA, a finding same as that of Zhang’s study (Figure 4). In addition, we found that the transcription of TaMYB29 was significantly induced and reached the highest level at 48h post-rust inoculation in the incompatible system (Figure 5A), implicating the involvement of TaMYB29 in wheat defense response to biotic and abiotic stresses.
Plant hormones are synthesized de novo and serve as signal molecules between plants and other surrounding organisms. These are usually conserved in the plant kingdom and can regulate diverse processes including plant growth and development, and responses to abiotic and biotic stresses (Nambara and Wees, 2021). Salicylic acid is a key regulator of plant defense response against living parasitic fungi and the acquisition of SAR pathways. The primary underlying mechanism of SA is to activate the high expression of a series of transcription factors, including DNA-binding proteins containing conserved MYB domains. For example, AtMYB30, AtMYB44, and AtMYB96 participate in the resistance of Arabidopsis to Pseudomonas syringae pv. tomato strain DC3000 via the SA signaling pathway (Dong et al., 1999; Vailleau et al., 2002; Seo and Park, 2010). Wheat AvS+Yr10 containing the stripe rust resistance gene Yr10 was highly resistant to CYR32 in incompatible interaction. Real-time PCR revealed a rapid induction of TaMYB29 as early as 12h post-SA treatment (hpt) in incompatible interaction. The highest 11-fold increase was recorded at 48hpt (Figure 4). In addition, it showed significantly increased endogenous SA levels in AvS+Yr10 as early as 12hpi; the second peak appeared at 72hpi compared to the 0hpi control (Figure 5B). The expression of TaMYB29 was significantly increased at 24hpi in the incompatible interaction, which was delayed by 12h compared to the SA (Figures 5A,B). These results suggested that TaMYB29 functions downstream in the SA biosynthesis pathway to defend against Pst in the incompatible interaction.
Crosstalk between defense signaling pathways regulates the defense responses against different types of attackers (Kachroo and Kachroo, 2007; Vlot et al., 2009; Robert-Seilaniantz et al., 2011; Zou et al., 2013). Abscisic acid, an important plant hormone, is involved in plants’ response to environmental stresses, such as drought, high salinity, and extreme temperature and plant–pathogen interactions (Popko et al., 2010; Wang et al., 2017). Similarly, JA and ET are primarily involved in the interaction between plants with necrotrophic pathogens and insects as well as in the wounding process (Reymond et al., 2000). We observed that TaMYB29 was significantly induced after treatment with SA, ABA, JA, and ET exogenous hormones; the highest induced expression was detected at 48h post-treatment with ABA (Figure 4). We previously found that endogenous SA concentration increased post-ABA and JA treatments and improved the wheat defense against stripe rust in Suwon11 (Wang et al., 2017). AtMYB96-mediated ABA signals induced pathogen resistance response by promoting SA biosynthesis in Arabidopsis (Seo and Park, 2010). AtMYB44 is involved in both ET- and SA-mediated defense-related signaling pathways to regulate the plant defense response (Zou et al., 2013), implying crosstalks between SA, ABA, JA, and ET in wheat defense response to stripe rust. TaPIMP1 contributes to biotic and abiotic stress resistance by regulating defense- and stress-related genes in the ABA–SA signaling pathways in wheat (Zhang et al., 2012b). SA- and JA-mediated plant defense signaling pathways have both synergistic and antagonistic effects. The SA pathway activated by biotrophic pathogen P. syringae strongly reduced JA-mediated defenses against the attack of necrotrophic pathogen A. brassicicola in Arabidopsis (Spoel et al., 2007). On the other hand, co-treatment with low concentrations of SA and JA resulted in a synergistic effect on the transcription of PR1 in Arabidopsis explants (Mur et al., 2006). Exogenous SA and JA could induce the expression of TaMYB29 in different time points, which indicates that TaMYB29 plays a role in the communication between SA- and JA-mediated resistance signaling pathways (Figure 4). Moreover, the transcript of TaMYB29 is induced both following the treatment with SA, ABA, JA, and ET hormones, and during the defense response against stripe rust (Figures 4, 5B). In summary, these results indicate that TaMYB29 functions in wheat defenses through a complex interaction, including the ABA–SA or JA/ET–SA signaling network.
Increasing evidence shows SA as a crucial signaling molecule in plant defense against pathogens. This defense response usually causes local cell necrosis, namely, HR response and SAR as well as PR gene expression (Durrant and Dong, 2004). Pathogenesis-related genes including PR1, PR2, and PR5 are induced along with the defense response of plants to pathogens through SA signaling (Vlot et al., 2009). As an indicator gene of SA, the expression of the PR1 gene was detected during compatible and incompatible interactions. The expression of PR1 increased significantly at 24hpi, 48hpi, and 72hpi in the incompatible interaction like TaMYB29, which was 12h later than SA (Figure 5). Compared with the control, the silencing of TaMYB29 reduced the expression of three PR genes (Figure 7C), the production of H2O2 (Figure 8B), and the HR areas (Figure 8D). These findings suggested that TaMYB29 modulated the defense response in a PR gene expression-dependent manner through the signaling molecule SA. During plant defense responses, transcription factors mediate the regulation of the expression of plant host target genes, largely through the specific recognition of cis-promoter elements. Several putative MYB recognition sequences have been found in the promoter region of the PR1 gene (Abe et al., 1997). The tobacco MYB1 gene is induced during the HR response and SAR. The MYB1 protein binds to the MYB consensus-binding site in the tobacco PR1-a promoter in vitro (Yang and Klessig, 1996). In addition, AtMYB44 may regulate the PR1 gene expression by binding to its promoter region (Zou et al., 2013). Combining the expression of PR genes and TaMYB29 gene in plant defense against stripe rust with the results of TaMYB29 gene silencing, we hypothesize that TaMYB29 binds to PR cis-elements to regulate its expression. However, we cannot exclude the possibility that TaMYB29 indirectly regulates PR1 expression by regulating other target genes. These results suggest that AvS+Yr10 defense against P. striiformis depends on the expression of PR genes via the SA signaling pathway.
Yr10 gene encodes for a unique CC-NBS-LRR receptor in wheat cultivar Moro, which provides seedling or all-stage resistance (R gene-mediated resistance; Liu et al., 2014). The R gene-mediated resistance is characterized by rapid plant cell death at the infection sites, thereby hindering the fungus spread from the infection sites—a process known as plant HR response—and inducing SAR (Heath, 2000; Durrant and Dong, 2004; Ryals et al., 2013). Following the successful recognition of an avirulent gene from the pathogen by the R gene, ROS rapidly accumulate and are instantaneously released in a process termed “oxidative burst.” This is one of the earliest events of plant defense against pathogens. In our study, the transient overexpression of TaMYB29 in tobacco caused a rapid ROS increase, consequently inducing cell death at the injection sites (Figure 6). Similarly, the overexpression of AtMYB44 enhanced H2O2 accumulation and cell death via the SA signaling pathway (Zou et al., 2013). Downregulating the three homologs of TaMYB29 in AvS+Yr10 reduced the host resistance to the avirulent Pst strain CYR32, whereas no obvious phenotype difference between the TaMYB29-silenced AvS plants and controls was observed (Figure 7A), suggesting that the TaMYB29 gene is indispensable for the resistance to Pst infection mediated by Yr10. In the incompatible interaction, only extremely little accumulation of H2O2 in both TaMYB29-silenced AvS+Yr10 plants and the BSMV:00 control was observed at 24hpi. However, at 48hpi and 120hpi, the accumulation of H2O2 was reduced significantly in the TaMYB29-silenced mesophyll cells in contact with primary hyphae as compared with the control BSMV:00 (Figures 8A,B). We hypothesized that TaMYB29 affected the accumulation of ROS, which was supported by the histological observation and measurement of the production of H2O2 after knocking down TaMYB29 (Figures 8A,B). The accumulation of H2O2 was only observed in guard cells during the formation of appressorium at 24hpi (Figure 8A), which was about 12h later than that in Suwon11 inoculated with CYR23 (Wang et al., 2017). These results indicated that the wheat cultivars containing different R genes led to varying ROS production in response to different rust isolates. Catalase, a powerful antioxidant enzyme, is an H2O2 scavenger. We reported that a 3.66-fold expression of the catalase gene TaCAT was induced in TaMYB29 knocked-down leaves infected with CYR32 compared with the BSMV:00 control at 48hpi (Figure 8C). We speculated the upregulated expression of TaCAT as one of the reasons for the reduced ROS accumulation. In addition, the infection area was significantly increased after knocking down TaMYB29 at 120hpi in comparison with the BSMV:00 control wheat plants (Figures 7D,E). Altogether, TaMYB29 activated the wheat defense response against Pst by modulating H2O2 accumulation. During the defense response against pathogens, TaMYB29 is induced via the SA pathway, thus increasing the levels of ROS and the expression of PR genes, and eventually generating resistance of wheat containing the Yr10 disease resistance gene to stripe rust. However, we cannot conclude whether TaMYB29 is required in all R gene-mediated disease resistance pathways, which requires further research. The underlying mechanism of regulation of specific target genes by TaMYB29 to induce the defense response as a transcription factor requires further research. Finally, the possible use of this gene for breeding disease-resistant plant varieties needs to be assessed.
Conclusion
TaMYB29 plays an indispensable role in the wheat response against stripe rust via regulating the crosstalk between various signaling pathways. Moreover, our results indicate that TaMYB29 positively regulates the plant defense response against biological nutritional pathogens by enhancing H2O2 accumulation, PR gene expression, and cell death via the SA signaling pathway.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, and further inquiries can be directed to the corresponding authors.
Author Contributions
XW and ZK conceived the study. XW, XZ, XL, CL, and ZK advised on the experimental design and drafted the manuscript. XW, XZ, XL, QH, DG, CL, ZW, and JC performed experiments and did data analysis. XZ, XL, QH, DG, CL, ZW, and JC interpreted data. XW, XZ, XL, and ZK wrote the manuscript and other authors reviewed and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Natural Science Foundation of China General Projects (No. 31501619 and No. 32172424), 2021 College Students’ innovation and entrepreneurship training program (S202110712787), Natural Science Foundation Research Project of Shaanxi Province (2021JM-095), Shaanxi Innovation Team Project (2018TD-004).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank Yi Luo for help on the study and also thank State Key Laboratory of Crop Stress Biology for Arid Areas, Northwest A&F University, China for the equipment support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.783388/full#supplementary-material
Footnotes
References
Abe, H., Yamaguchi-Shinozaki, K., Urao, T., Iwasaki, T., Hosokawa, D., and Shinozaki, K. (1997). Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 9, 1859–1868. doi: 10.1105/tpc.9.10.1859
Al-Attala, M. N., Wang, X., Abou-Attia, M. A., Duan, X., and Kang, Z. (2014). A novel TaMYB4 transcription factor involved in the defence response against Puccinia striiformis f. sp. tritici and abiotic stresses. Plant Mol. Biol. 84, 589–603. doi: 10.1007/s11103-013-0156-7
Bai, S., Liu, J., Chang, C., Zhang, L., Maekawa, T., Wang, Q., et al. (2012). Structure-function analysis of barley NLR immune receptor MLA10 reveals its cell compartment specific activity in cell death and disease resistance. PLoS Pathog. 8:e1002752. doi: 10.1371/journal.ppat.1002752
Cedroni, M. L., Cronn, R. C., Adams, K. L., Wilkins, T. A., and Wendel, J. F. (2003). Evolution and expression of MYB genes in diploid and polyploid cotton. Plant Mol. Biol. 51, 313–325. doi: 10.1023/A:1022051100610
Chang, C., Yu, D., Jiao, J., Jing, S., Schulze-Lefert, P., and Shen, Q. H. (2013). Barley MLA immune receptors directly interfere with antagonistically acting transcription factors to initiate disease resistance signaling. Plant Cell 25, 1158–1173. doi: 10.1105/tpc.113.109942
Chen, W., Wellings, X. M., Kang, Z., and Liu, T. (2014). Wheat stripe (yellow) rust caused by Puccinia striiformis f. sp. tritici. Mol. Plant Pathol. 15, 433–446. doi: 10.1111/mpp.12116
Chini, A., Grant, J. J., Seki, M., Shinozaki, K., and Loake, G. J. (2010). Drought tolerance established by enhanced expression of the CC–NBS–LRR gene, ADR1, requires salicylic acid, EDS1 and ABI1. Plant J. 38, 810–822. doi: 10.1111/j.1365-313X.2004.02086.x
Coleto, I., Bejarano, I., Marin-Pena, A. J., Medina, J., Rioja, C., Burow, M., et al. (2021). Arabidopsis thaliana transcription factors MYB28 and MYB29 shape ammonium stress responses by regulating Fe homeostasis. New Phytol. 229, 1021–1035. doi: 10.1111/nph.16918
Coll, N. S., Epple, P., and Dangl, J. L. (2011). Programmed cell death in the plant immune system. Cell Death Differ. 18, 1247–1256. doi: 10.1038/cdd.2011.37
De Vos, M., Denekamp, M., Dicke, M., Vuylsteke, M., Van Loon, L., Smeekens, S. C., et al. (2006). The Arabidopsis thaliana transcription factor AtMYB102 functions in defense against the insect herbivore Pieris rapae. Plant Signal. Behav. 1, 305–311. doi: 10.4161/psb.1.6.3512
Ding, Z., Li, S., An, X., Liu, X., Qin, H., and Wang, D. (2009). Transgenic expression of MYB15 confers enhanced sensitivity to abscisic acid and improved drought tolerance in Arabidopsis thaliana. J. Genet. Genomics 36, 17–29. doi: 10.1016/S1673-8527(09)60003-5
Dong, H., Delaney, T. P., Bauer, D. W., and Beer, S. V. (1999). Harpin induces disease resistance in Arabidopsis through the systemic acquired resistance pathway mediated by salicylic acid and the NIM1 gene. Plant J. 20, 207–215. doi: 10.1046/j.1365-313x.1999.00595.x
Dubos, C., Stracke, R., Grotewold, E., Weisshaar, B., Martin, C., and Lepiniec, L. (2010). MYB transcription factors in Arabidopsis. Trends Plant Sci. 15, 573–581. doi: 10.1016/j.tplants.2010.06.005
Durrant, W. E., and Dong, X. (2004). Systemic acquired resistance. Annu. Rev. Phytopathol. 42, 185–209. doi: 10.1146/annurev.phyto.42.040803.140421
Fujita, M., Fujita, Y., Noutoshi, Y., Takahashi, F., Narusaka, Y., Yamaguchi-Shinozaki, K., et al. (2006). Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr. Opin. Plant Biol. 9, 436–442. doi: 10.1016/j.pbi.2006.05.014
Grant, J. J., Chini, A., Basu, D., and Loake, G. J. (2003). Targeted activation tagging of the Arabidopsis NBS-LRR gene, ADR1, conveys resistance to virulent pathogens. Mol. Plant Microbe Interact. 16, 669–680. doi: 10.1094/MPMI.2003.16.8.669
Heath, M. C. (2000). Hypersensitive response-related death. Plant Mol. Biol. 44, 321–334. doi: 10.1023/A:1026592509060
Holzberg, S., Brosio, P., Gross, C., and Pogue, G. P. (2002). Barley stripe mosaicvirus-induced gene silencing in a monocot plant. Plant J. 30, 315–327. doi: 10.1046/j.1365-313X.2002.01291.x
Jia, J., Xing, J., Dong, J., Han, J., and Liu, J. (2011). Functional analysis of MYB73 of Arabidopsis thaliana against Bipolaris oryzae. Agric. Sci. China 10, 721–727. doi: 10.1016/S1671-2927(11)60055-2
Jin, H., Cominelli, E., Bailey, P., Parr, A., Mehrtens, F., Jones, J., et al. (2014). Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J. 19, 6150–6161. doi: 10.1093/emboj/19.22.6150
Jin, H., and Martin, C. (1999). Multifunctionality and diversity within the plant MYB-gene family. Plant Mol. Biol. 41, 577–585. doi: 10.1023/A:1006319732410
Kachroo, A., and Kachroo, P. (2007). Salicylic acid-, jasmonic acid- and ethylene-mediated regulation of plant defense signaling. Genet. Eng. 28, 55–83. doi: 10.1007/978-0-387-34504-8_4
Katiyar, A., Smita, S., Lenka, S., Rajwanshi, R., Chinnusamy, V., and Bansal, K. (2012). Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis. BMC Genomics 13:544. doi: 10.1186/1471-2164-13-544
Kranz, H. D., Denekamp, M., Greco, R., Jin, H., Leyva, A., Meissner, R. C., et al. (1998). Towards functional characterisation of the members of the R2R3-MYB gene family from Arabidopsis thaliana. Plant J. 16, 263–276. doi: 10.1046/j.1365-313x.1998.00278.x
Li, X., Zhao, K., Zhang, X., and Zhou, B. (2017). Expression analysis of poplar MYB transcription factor gene family in response to salt stress. Bull. Bot. Res. 37, 424–431. doi: 10.7525/j.issn.1673-5102.2017.03.013
Liu, X., Yang, L., Zhou, X., Zhou, M., Lu, Y., Ma, L., et al. (2013). Transgenic wheat expressing Thinopyrum intermedium MYB transcription factor TiMYB2R-1 shows enhanced resistance to the take-all disease. J. Exp. Bot. 64, 2243–2253. doi: 10.1093/jxb/ert084
Liu, W., Frick, M., Huel, R., Nykiforuk, C. L., Wang, X., Gaudet, D. A., et al. (2014). The stripe rust resistance gene Yr10 encodes an evolutionary-conserved and unique CC-NBS-LRR sequence in wheat. Mol. Plant 7, 1740–1755. doi: 10.1093/mp/ssu112
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ma, Q., Wang, C., and Zhu, H. (2011). TaMYB4 cloned from wheat regulates lignin biosynthesis through negatively controlling the transcripts of both cinnamyl alcohol dehydrogenase and cinnamoyl-CoA reductase genes. Biochimie 93, 1179–1186. doi: 10.1016/j.biochi.2011.04.012
Mcneal, F. H., Konzak, C. F., Smith, E. P., Tate, W. S., and Russell, T. S. (1971). A Uniform System for Recording and Processing Cereal Research Data (Washington, DC: USDA), 34–121.
Mengiste, T., Chen, X., Salmeron, J., and Dietrich, R. (2003). The BOTRYTIS SUSCEPTIBLE1 gene encodes an R2R3MYB transcription factor protein that is required for biotic and abiotic stress responses in Arabidopsis. Plant Cell 15, 2551–2565. doi: 10.1105/tpc.014167
Morse, A. M., Whetten, R. W., Dubos, C., and Campbell, M. M. (2009). Post-translational modification of an R2R3-MYB transcription factor by a MAP kinase during xylem development. New Phytol. 183, 1001–1013. doi: 10.1111/j.1469-8137.2009.02900.x
Mur, L. A., Kenton, P., Atzorn, R., Miersch, O., and Wasternack, C. (2006). The outcomes of concentration-specific interactions between salicylate and jasmonate signaling include synergy, antagonism, and oxidative stress leading to cell death. Plant Physiol. 140, 249–262. doi: 10.1104/pp.105.072348
Nambara, E., and Wees, S. (2021). Plant hormone functions and interactions in biological systems. Plant J. 105, 287–289. doi: 10.1111/tpj.15151
Ogata, K., and Nishimura, Y. (1995). Specific DNA recognition by MYB protein. Tanpakushitsu Kakusan Koso 40, 1592–1597.
Park, R. F., Bariana, H. S., and Wellings, C. R. (2007). Preface to ‘global landscapes in cereal rust control’. Aust. J. Agric. Res. 58:469. doi: 10.1071/ARv58n6_PR
Paz-Ares, J., Ghosal, D., Wienand, U., Peterson, P. A., and Saedler, H. (1987). The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. EMBO J. 6, 3553–3558. doi: 10.1002/j.1460-2075.1987.tb02684.x
Popko, J., Hänsch, R., Mendel, R. R., Polle, A., and Teichmann, T. (2010). The role of abscisic acid and auxin in the response of poplar to abiotic stress. Plant Biol. 12, 242–258. doi: 10.1111/j.1438-8677.2009.00305.x
Raffaele, S., Rivas, S., and Roby, D. (2006). An essential role for salicylic acid in AtMYB30-mediated control of the hypersensitive cell death program in Arabidopsis. FEBS Lett. 580, 3498–3504. doi: 10.1016/j.febslet.2006.05.027
Ramirez, V., Agorio, A., Coego, A., García-Andrade, J., Hernández, M. J., Balaguer, B., et al. (2011). MYB46 modulates disease susceptibility to Botrytis cinerea in Arabidopsis. Plant Physiol. 155, 1920–1935. doi: 10.1104/pp.110.171843
Reymond, P., Weber, H., Damond, M., and Farmer, E. E. (2000). Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell 12, 707–720. doi: 10.1105/tpc.12.5.707
Robert-Seilaniantz, A., Grant, M., and Jones, J. D. (2011). Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annu. Rev. Phytopathol. 49, 317–343. doi: 10.1146/annurev-phyto-073009-114447
Ryals, J. A., Neuenschwander, U. H., Willits, M. G., Molina, A., and Hunt, M. D. (2013). Systemic acquired resistance. Plant Physiol. 1, 179–184. doi: 10.4161/psb.1.4.3221
Saha, G., Park, J. I., Ahmed, N. U., Kayum, M. A., Kang, K. K., and Nou, I. S. (2016). Characterization and expression profiling of MYB transcription factors against stresses and during male organ development in Chinese cabbage (Brassica rapa ssp. pekinensis). Plant Physiol. Biochem. 104, 200–215. doi: 10.1016/j.plaphy.2016.03.021
Seo, P. J., and Park, C. M. (2010). MYB96-mediated abscisic acid signals induce pathogen resistance response by promoting salicylic acid biosynthesis in Arabidopsis. New Phytol. 186, 471–483. doi: 10.1111/j.1469-8137.2010.03183.x
Shan, T., Hong, Y., Xu, H., Wei, X., and Zhang, Z. (2016). Development and characterization of TaMYB86-overexpressing transgenic wheat lines with resistance to common root rot. Acta Agron. Sin. 42, 1429–1436. doi: 10.3724/SP.J.1006.2016.01429
Spoel, S. H., Johnson, J. S., and Dong, X. (2007). Regulation of tradeoffs between plant defenses against pathogens with different lifestyles. Proc. Natl. Acad. Sci. U. S. A. 104, 18842–18847. doi: 10.1073/pnas.0708139104
Tian, Z., Zhong, H., Shi, R., Sun, L., Fischer, G., and Liang, Z. (2012). Estimating potential yield of wheat production in China based on cross-scale data-model fusion. Front. Earth Sci. 6, 364–372. doi: 10.1007/s11707-012-0332-0
Vailleau, F., Daniel, X., Tronchet, M., Montillet, J. L., Triantaphylidès, C., and Roby, D. (2002). A R2R3-MYB gene, AtMYB30, acts as a positive regulator of the hypersensitive cell death program in plants in response to pathogen attack. Proc. Natl. Acad. Sci. U. S. A. 99, 10179–10184. doi: 10.1073/pnas.152047199
Vlot, A. C., Dempsey, D. A., and Klessig, D. F. (2009). Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 47, 177–206. doi: 10.1146/annurev.phyto.050908.135202
Wang, X., Wang, X., Duan, Y., Yin, S., Zhang, H., Huang, L., et al. (2013). TaAbc1, a member of Abc1-like family involved in hypersensitive response against the stripe rust fungal pathogen in wheat. PLoS One 8:e58969. doi: 10.1371/journal.pone.0058969
Wang, F., Suo, Y., Wei, H., Li, M., Xie, C., Wang, L., et al. (2015). Identification and characterization of 40 isolated Rehmannia glutinosa MYB family genes and their expression profiles in response to shading and continuous cropping. Int. J. Mol. Sci. 16, 15009–15030. doi: 10.3390/ijms160715009
Wang, X., Wang, Y., Liu, P., Ding, Y., Mu, X., Liu, X., et al. (2017). TaRar1 is involved in wheat defense against stripe rust pathogen mediated by YrSu. Front. Plant Sci. 8:156. doi: 10.3389/fpls.2017.00156
Wang, X., Zhang, H., Nyamesorto, B., Luo, Y., Mu, X., Wang, F., et al. (2020a). A new mode of NPR1 action via an NB-ARC-NPR1 fusion protein negatively regulates the defence response in wheat to stem rust pathogen. New Phytol. 228, 959–972. doi: 10.1111/nph.16748
Wang, F., Yuan, S., Wu, W., Yang, Y., Cui, Z., Wang, H., et al. (2020b). TaTLP1 interacts with TaPR1 to contribute to wheat defense responses to leaf rust fungus. PLoS Genet. 16:e1008713. doi: 10.1371/journal.pgen.1008713
Yang, Y., and Klessig, D. F. (1996). Isolation and characterization of a tobacco mosaic virus-inducible myb oncogene homolog from tobacco. Proc. Natl. Acad. Sci. U. S. A. 93, 14972–14977. doi: 10.1073/pnas.93.25.14972
Yoo, S. D., Cho, Y. H., and Sheen, J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. doi: 10.1038/nprot.2007.199
Zhai, Y., Li, P., Mei, Y., Chen, M., Chen, X., Xu, H., et al. (2017). Three MYB genes co-regulate the phloem-based defence against English grain aphid in wheat. J. Exp. Bot. 68, 4153–4169. doi: 10.1093/jxb/erx204
Zhang, L., Zhao, G., Jia, J., Liu, X., and Kong, X. (2012a). Molecular characterization of 60 isolated wheat MYB genes and analysis of their expression during abiotic stress. J. Exp. Bot. 63, 203–214. doi: 10.1093/jxb/err264
Zhang, Z., Liu, X., Wang, X., Zhou, M., Zhou, X., Ye, X., et al. (2012b). An R2R3 MYB transcription factor in wheat, TaPIMP1, mediates host resistance to Bipolaris sorokiniana and drought stresses through regulation of defense- and stress-related genes. New Phytol. 196, 1155–1170. doi: 10.1111/j.1469-8137.2012.04353.x
Zhang, C., Ma, R., Xu, J., Yan, J., Guo, L., Song, J., et al. (2018). Genome-wide identification and classification of MYB superfamily genes in peach. PLoS One 13:e0199192. doi: 10.1371/journal.pone.0199192
Zheng, X., Yi, D., Shao, L., and Li, C. (2017). In silico genome-wide identification, phylogeny and expression analysis of the R2R3-MYB gene family in Medicago truncatula. J. Integr. Agric. 16, 1576–1591. doi: 10.1016/S2095-3119(16)61521-6
Keywords: TaMYB29, transcription factor, reactive oxygen species, hypersensitive response, stripe rust, VIGS
Citation: Zhu X, Li X, He Q, Guo D, Liu C, Cao J, Wu Z, Kang Z and Wang X (2021) TaMYB29: A Novel R2R3-MYB Transcription Factor Involved in Wheat Defense Against Stripe Rust. Front. Plant Sci. 12:783388. doi: 10.3389/fpls.2021.783388
Edited by:
Meixiang Zhang, Nanjing Agricultural University, ChinaReviewed by:
Zhao Zhang, China Agricultural University, ChinaGuan-Feng Wang, Shandong University, China
Qiong Zhang, University of California, Berkeley, United States
Copyright © 2021 Zhu, Li, He, Guo, Liu, Cao, Wu, Kang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhensheng Kang, a2FuZ3pzQG53c3VhZi5lZHUuY24=; Xiaojing Wang, d2FuZ3hpYW9qaW5nQG53c3VhZi5lZHUuY24=
†These authors have contributed equally to this work
 Xiaoxu Zhu
Xiaoxu Zhu Xiang Li3†
Xiang Li3† Zhensheng Kang
Zhensheng Kang Xiaojing Wang
Xiaojing Wang