
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 17 November 2021
Sec. Plant Breeding
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.777494
This article is part of the Research TopicAdvances in Breeding for Quantitative Disease ResistanceView all 28 articles
 Yike Liu1,2
Yike Liu1,2 Guang Zhu1
Guang Zhu1 Zhangwang Zhu1
Zhangwang Zhu1 Lin Chen1
Lin Chen1 Hongli Niu3,4
Hongli Niu3,4 Weijie He1
Weijie He1 Hanwen Tong1
Hanwen Tong1 Jinghan Song1
Jinghan Song1 Yuqing Zhang1
Yuqing Zhang1 Dongfang Ma2,3,4*
Dongfang Ma2,3,4* Chunbao Gao1,2*
Chunbao Gao1,2*Fusarium seedling blight (FSB) is an important disease of wheat occurring as part of the Fusarium disease complex consisting also of Fusarium head blight (FHB). 240 Chinese elite cultivars and lines were evaluated in greenhouse experiments for FSB resistance and genotyped using the wheat 90 K single nucleotide polymorphism arrays. Among them, 23 accessions had an average lesion length of less than 0.6 cm, exhibiting potential for breeding for FSB resistance in wheat. Jingfumai 1 and Yangmai 11 had a relatively high resistance to both FSB and FHB simultaneously. Six relatively stable quantitative trait loci (QTLs) were detected on chromosome arms 1DL, 3AS, 3BL, 6BL, 7AL, and Un using the mixed linear model approach, interpreting 4.83–7.53% of phenotypic variation. There was a negative correlation between the average FSB lesion length and the BLUE FHB index with a low coefficient, and resistance to both diseases appeared to be conferred by different QTLs across the same population. Four KASP markers were detected on 1DL, 3AS, 3BL, and 6BL in QTLs to facilitate marker-assisted selection. Combined with transcriptome data analysis, eight defense-related genes were considered as candidates for mapping QTLs. The resistant elite germplasm, mapped QTLs, and KASP markers developed in this study are useful resources for enhancing Fusarium seedling blight in wheat breeding.
Fusarium seedling blight (FSB) and Fusarium head blight (FHB), primarily caused by Fusarium pathogens, refer to are economically devastating diseases in wheat (Triticum aestivum L.) as well as other small grain cereals across the world (Bai and Shaner, 2004; Li X. et al., 2010; Ren et al., 2016). Fusarium seedling blight can cause extensive damage to growing seedlings or foot rot later during the growing season, leading to reduced emergence and crop establishment and consequently yield losses in wheat (Wiese, 1987; Antalová et al., 2020). Moreover, FSB can provide a pathogen source for following FHB infection, creating reddish scabby spikes (Haigh et al., 2009; Li X. et al., 2010). Due to the global climate change and tillage management, FSB and FHB usually reach epidemic levels, causing huge yield losses across millions of hectares in global wheat production regions (Cheng et al., 2015; Liu et al., 2016). In addition, both FSB and FHB produce various mycotoxins during infection, with high toxicity, posing a threat to people as well as livestock (Pestka and Smolinski, 2005; Liu et al., 2012).
China is the largest producer and consumer of wheat (Shi and Ling, 2018). Cultivars play a major role in national wheat production, and developing and using resistant cultivars can confer protection to Fusarium pathogens. The analysis of the probable association between FSB and FHB can help develop new strategies to combat the Fusarium disease complex. Twelve Polish spring wheat cultivars and 18 spring wheat accessions from CIMMYT were examined for resistance to FSB and FHB by applying a highly aggressive fungal isolate, and no correlation was found between the two resistance types (Wisniewska and Busko, 2005). No significant correlation was also detected between FSB infection and FHB index and between FSB infection and DON content in a Wuhan/Nyubai doubled haploid (DH) wheat population (Somers et al., 2003; Tamburic-Ilincic et al., 2009). In subsequent research, QTLs for Fusarium resistance at seedling and spike stages were different, but further verification was required for various wheat populations (Tamburic-Ilincic et al., 2009). Comparatively, there have also been reports concerning the positive association between FSB and FHB resistance (Mesterhazy, 1987; Wu et al., 2005; Shin et al., 2014). Mesterhazy (1987) found a significant correlation between FSB and FHB resistance, and the most resistant genotypes at the seedling stage could yield the FHB resistant material with a large probability. By inoculation of wheat coleoptiles with Fusarium graminearum isolates, Wu et al. (2005) found a significant correlation between FSB and FHB resistance in the same genotype in the field. Using the clip-dipping inoculation method, Shin et al. (2014) found the remarkable correlation between the lesion length and Type II FHB resistance and suggested that the method for the evaluation of FSB resistance may provide a simple and feasible way for the early screening of FHB resistance in wheat.
Using linkage analysis, quite a few QTLs associated with FHB resistance were detected in 21 wheat chromosomes reported (Buerstmayr et al., 2009; Ma et al., 2020), with 7 FHB genes (Fhb1-Fhb7) being formally cataloged (Zhu et al., 2020). But FSB has not received much attention so far, and there have been very few studies on the QTLs for FSB resistance. A QTL on chromosome 5B controlling FSB resistance was identified in a DH population, and its linked marker WMC75 interpreted 13.8% of the phenotypic variation (Tamburic-Ilincic et al., 2009). Single major QTLs for FSB resistance, caused by Microdochium nivale and Microdochium majus, were detected on the chromosomes 1AL and 2BS, respectively (Ren et al., 2016).
Genome-wide association studies (GWAS) on the basis of linkage disequilibrium (LD) offer several advantages over linkage mapping, which has gained success in the analysis of different quantitative characteristics in wheat (Sapkota et al., 2019; Hu et al., 2020). For example, using 166 elite wheat varieties from Yellow and Huai River Valleys Wheat District in China, 120 common loci were detected for their associations with grain yield, among which 78 were potentially new (Li et al., 2019). In our previous studies, five QTLs were identified for their consistent associations with FHB resistance in a natural population, among which the QTLs on 5AS, 5AL, and 7DS were possibly new (Zhu et al., 2020). However, GWAS to identify FSB in wheat has not been reported yet, and the molecular mechanisms for FSB remain poorly understood.
In the present study, we evaluated FSB resistance in Chinese elite wheat lines and then performed GWAS and QTL analyses. The study aimed to (1) identify wheat germplasms with FSB resistance that could be used as resistance donors in breeding and confirm the relationship between FSB and FHB resistance caused by Fusarium pathogens, (2) uncover novel FSB-resistant loci that could be used in molecular marker-assisted breeding. The findings provide an insight into the genetics of FSB response in Chinese cultivars, and the developed markers associated with the mapped QTLs may be used for breeding FSB resistance wheat.
A total of 240 common wheat cultivars or elite lines (Supplementary Table 1) were selected as the natural population to evaluate FSB resistance and perform GWAS analysis, as described in our previous study (Zhu et al., 2020). The population included 229 elite wheat cultivars (lines) developed in the main wheat-growing areas of China, covering 12 provinces with five agroecological systems, and could represent the current situation of breeding in China. The remaining 11 genotypes belonged to CIMMYT (10) and Australian (1). Seeds were harvested in the Wuhan Nanhu farm of Hubei Academy of Agricultural Sciences (N 30.28°, E 114.19°) during the cropping seasons in 2018–2019.
Wheat coleoptiles at the seedling stage were inoculated with conidiospores using the previously described method (Li X. et al., 2010; Cheng et al., 2015) with minor alterations. For seedling inoculation, the concentration of the macroconidia suspension for the aggressive isolate F. graminearum Huanggang 1 (Zhu et al., 2020) was regulated to 5 × 105 spores/mL with sterilized distilled water. Forty full wheat seeds per cultivars (lines) were disinfected using 0.1% HgCl2 for 1 min and then rinsed twice using sterilized distilled water. Sterilized seeds were placed on wet filter paper in Petri dishes and incubated at 20°C in the dark for 2 days. Then 20 seeds with steady growth were picked, transferred to a sterilized germination box (length, width, and height of 11.5, 11.5, and 9.8 cm, respectively) with three layers of wet filter papers, and kept in the dark at 20°C for 1 day. Top coleoptiles (2–3 mm) were dissected, and a 3-μL aliquot of the macroconidial suspension was injected into the slant side of the dissected seedlings. Inoculated seedlings were stored in the germination box in the dark at 20°C, in dark as previously mentioned. The brown lesions of diseased seedlings were measured at 7 day post inoculation, and the lesion length was determined as previously described (Li X. et al., 2010). For each genotype, 20 wheat seedlings were examined each time, and the average value was used for subsequent analysis. The experiments were performed independently with triple replications.
The t-tests, as well as Pearson’s correlation analysis for independent samples, were conducted using IBM SPSS Statistics version 19.0 (IBM Corporation, Armonk, NY, United States). Histograms showing the distribution of the lesion length (cm) of 240 cultivars (lines) were made for each replicate with a script executed in R version 3.5.1.1
Illumina 90 K SNP array genotyping was performed on 240 wheat accessions (Wang et al., 2014). Calling and filtering for SNPs, kinship, and population structure analysis have all been all elaborately explained in the previous research (Zhu et al., 2020). A total of 19,803 with MAF of >5 and <20% missing data of 22,922 polymorphic SNPS were employed for subsequent analysis (Zhu et al., 2020). Population structure analysis was performed via ADMIXTURE.2 The population fell into three subgroups, basically based on geographic origin and pedigree (Zhu et al., 2020).
Associations between genotypic and phenotypic data were analyzed in Tassel v5.0. A kinship (K) + PCA model was used to perform the MLM analysis for controlling the background variation and eliminating spurious marker-trait associations (MTAs). R2 exhibiting the variation explained by SNP was documented (Bradbury et al., 2007). SNPs with an adjusted-log10 (P-value) of ≥3.0 were considered associated with FSB resistance. The remarkable loci in a minimum of two repetitions detected in the research were stable QTLs. Remarkable SNP markers in one linkage disequilibrium on the same chromosome represented one locus.
The SNP markers remarkably associated with FSB resistance were identified using GWAS and transformed into Kompetitive Allele-Specific PCR (KASP) markers to facilitate their application in MAS. The SNP contextual sequences were obtained at GrainGenes3 and the primers were designed by PolyMarker4 or the primer premier 5.0 (PREMIER Biosoft International, Palo Alto, CA, United States). Amplification was performed initially at 95°C for 15 min, 10 cycles of touchdown PCR (at 95°C for 20 s; an initial touchdown at 65°C, followed by a reduction of −1°C per cycle for 25 s), and the final 30 additional cycles for annealing (95°C for 10 s; 60°C for 60 s). Fluorescence signals were inspected under the multifunctional microplate reader PHERAstarPlus (BMG LABTECH, Ortenberg, Germany) and determined via KlusterCaller (LGC Genomics, Teddington, United Kingdom).
To identify the candidate genes associated with typical SNPs, physical positions of markers before the chromosome name were introduced into Ensembl,5 and genes within a 2 Mb distance from typical SNPs were detected to assess their candidacy for FSB resistance. The transcript IDs of all these genes were obtained from wheat sequences (Alaux et al., 2018). We used another publicly available database, expVIP,6 to obtain the expression profiles of all these genes in wheat seedling coleoptile organs infected by Fusarium spp. (Ma et al., 2014; Powell et al., 2017). To visualize the expression profiles, heat maps were drawn using TBtools (Chen et al., 2018) from differently expressed genes, with the absolute value of log2 fold change of ≥1 or ≤–1 at either time point. Up-regulated genes in resistant varieties associated with disease resistance annotated by RefSeq Annotation v1.1 (Appels et al., 2018) were identified as candidate genes. The candidate genes were further used for searing the sequences with high similarity via NCBI, combined with a basic local alignment search tool (BLAST).7
The assessment of FSB resistance in 240 wheat accessions showed a lesion length within the range of 0.075–2.896, with a normal distribution in three replications (Table 1 and Figure 1). Pearson’s correlation coefficients among three replications ranged from 0.535 to 0.577 with a significant difference (P < 0.01), and the average values were significantly associated with repeats, with correlation coefficients of 0.826, 0.857, and 0.828 for each repeat (Table 1). Further analysis indicated that 23 accessions, including the elite cultivars Zhoumai 17, Yanzhan 4110, Yunong 035, Jimai 38, Yumai 69, Jingfumai 1, Zhoumai 16, Yannong 24, and Yangmai 11 showed an average lesion length of less than 0.6, with a potential for breeding for FSB resistance in wheat. There were 105, 71, 29, and 12 accessions showing the average lesion lengths within the ranges of 0.6–1.0, 1.01–1.40, 1.41–1.80, and >1.80, respectively. Representative accessions with different grades of resistance to FSB are represented in Table 2.
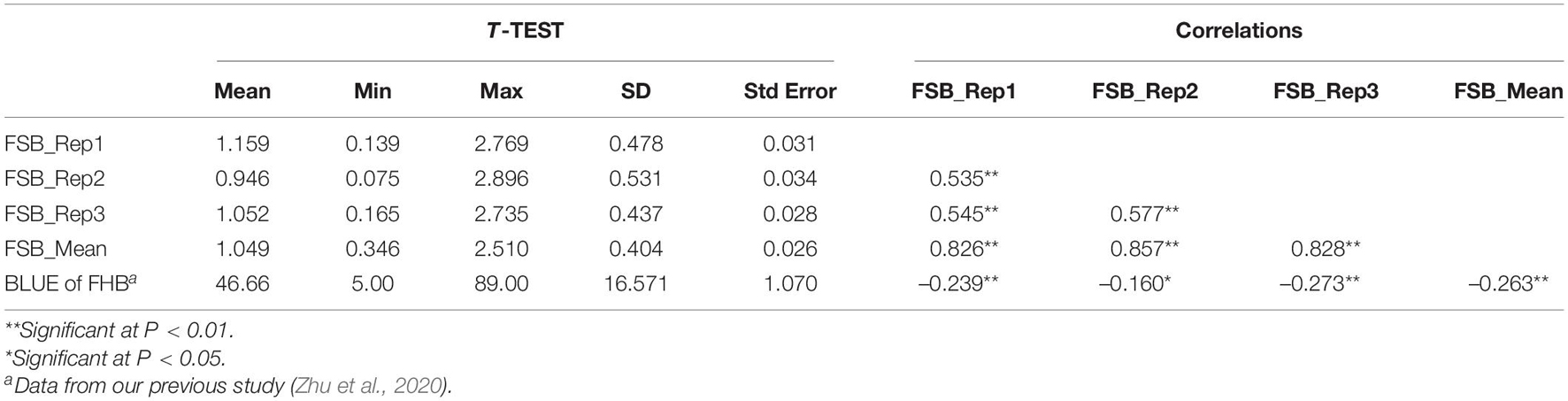
Table 1. Descriptive statistics and correlation coefficients of Fusarium seeding blight and Fusarium head blight of 240 wheat cultivars (lines).
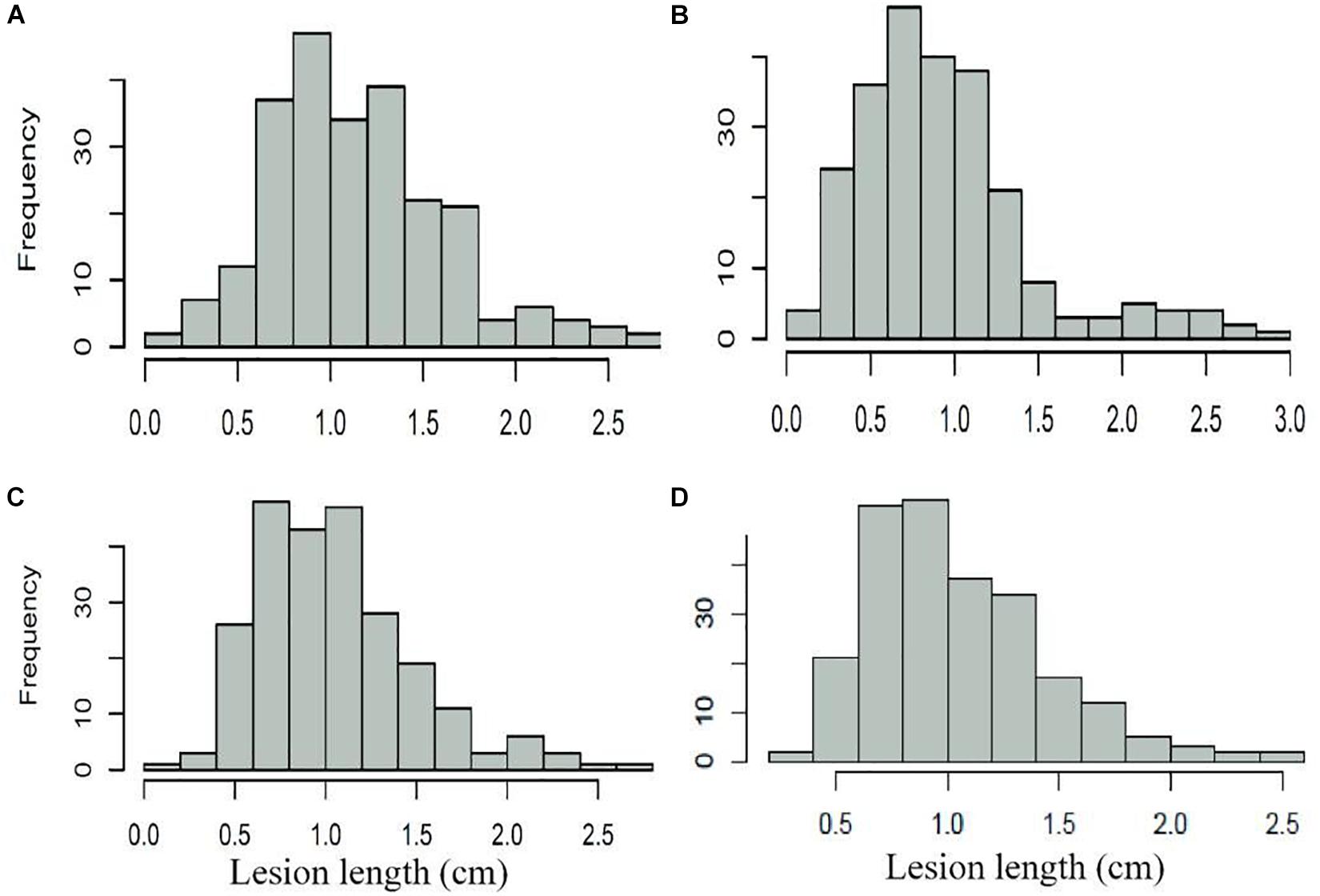
Figure 1. Frequency distribution of Fusarium seeding blight of 240 wheat cultivars (lines). (A) FSB_Rep1; (B) FSB_Rep2; (C) FSB_Rep3; and (D) FSB_mean.

Table 2. Materials with different resistance levels to Fusarium seedling blight (FSB) (only representative materials are shown).
Correlation coefficients were determined based on FSB infection values in the research and BLUE values of FHB indices, calculated within 4 years, according to the results of our previous study (Zhu et al., 2020) using the same population. The average FSB lesion length was negatively correlated with the BLUE FHB index across the population, although a low coefficient of R = –0.263 was determined (Table 1). The most notable cultivar Sumai3 and its derivative Ning7840 with a high FHB resistance showed quite low resistance to FSB in this assay. Conversely, the FHB susceptible cultivars Zhengyumai 9987 and Zhoumai 17 (Zhu et al., 2020) showed relatively high resistance to FSB. However, some accessions such as Jingfumai 1 and Yangmai 11 had relatively high resistance to both FSB and FHB simultaneously (Zhu et al., 2020).
Six QTLs on chromosome arms 1DL, 3AS, 3BL, 6BL, 7AL, and Un, designated as Qfsb.hbaas-1DL, Qfsb.hbaas-3AS, Qfsb.hbaas-3BL, Qfsb.hbaas-6BL, Qfsb.hbaas-7AL, and Qfsb.hbaas-un, respectively, were significant in a minimum of two repetitions, interpreting phenotypic variation of 4.83–7.53% (Table 3 and Figure 2). Representative significant markers for these QTLs were IWB41243, IWB64668, IWB3107, IWA3221, IWB41907, and IWB36312, respectively. For the goal of identifying minor QTL, this study was underpowered because of small population size which results in not seeing high signals.
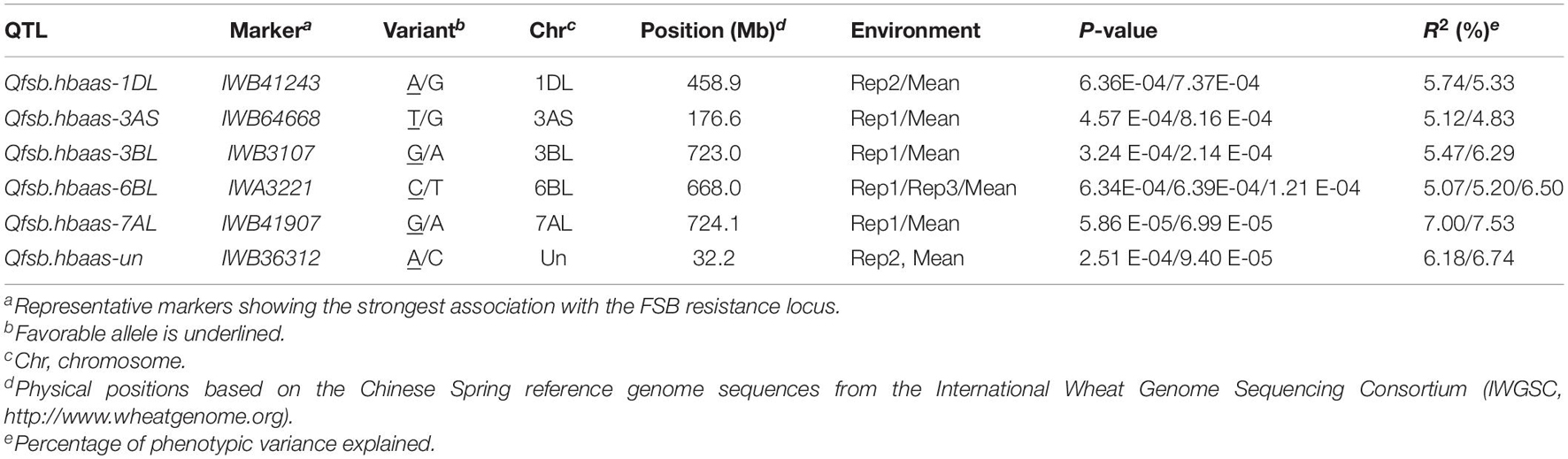
Table 3. Loci significantly associated with FSB resistance in at least two environments in the 240 wheat cultivars (lines) using the mixed linear model (MLM) model in Tassel v5.0.

Figure 2. Manhattan plots from genome-wide association scan for Fusarium seedling blight (FSB) severities among 240 wheat accessions in (A) FSB_Rep1, (B) FSB_Rep2, (C) FSB_Rep3, and (D) FSB_mean. Dashed red horizontal line is the significant threshold level.
Of the 240 genotypes, 12, 180, 130, 204, 220, and 217 possessed the resistance alleles Qfsb.hbaas-1DL, Qfsb.hbaas-3AS, Qfsb.hbaas-3BL, Qfsb.hbaas-6BL, Qfsb.hbaas-7AL, and Qfsb.hbaas-un, respectively, on the basis of marker analysis (Table 4 and Supplementary Table 1). The mean FSB lesion length in accessions with favorable Qfsb.hbaas-1DL alleles was 21.4% shorter than with unfavorable alleles. Discrepancies between Qfsb.hbaas-6BL and Qfsb.hbaas-7AL were much greater (31.7 and 36.3%, respectively). In Qfsb.hbaas-3AS, Qfsb.hbaas-3BL, and Qfsb.hbaas-un, the FSB lesion lengths were reduced by 17.4, 13.8, and 8.8%, respectively (Table 4).
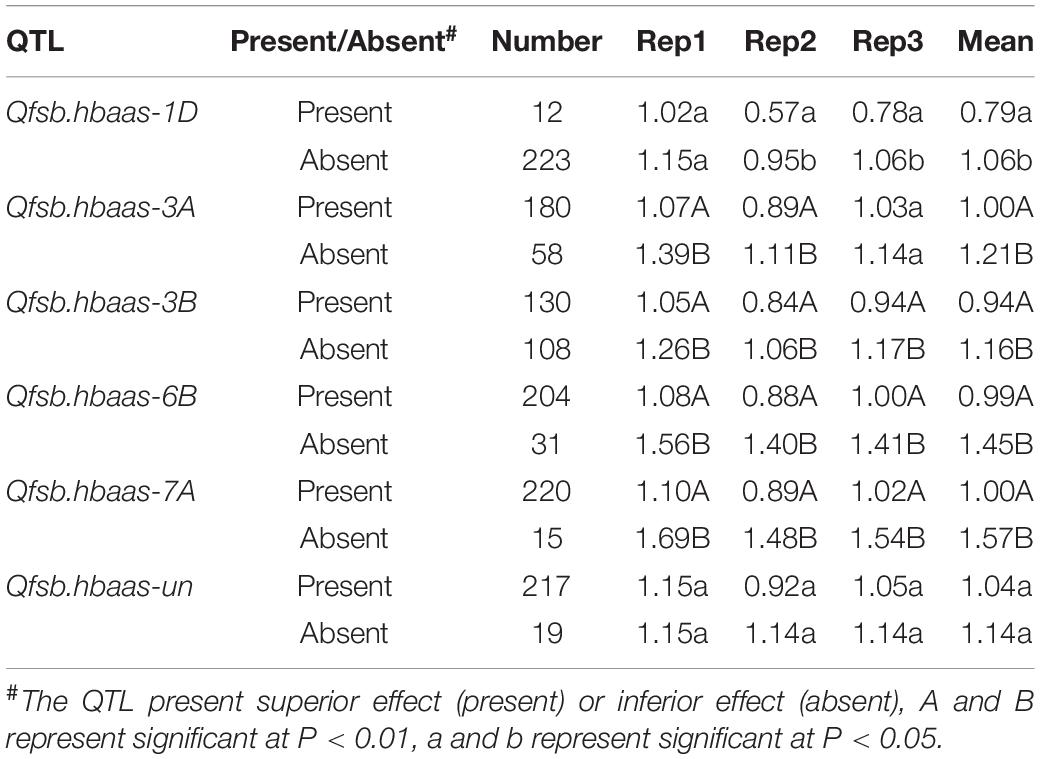
Table 4. T-tests for differences in Fusarium seedling blight between two groups of wheat accessions with contrasting resistance or susceptibility alleles for quantitative trait loci (QTL) on chromosomes 1D, 3A, 3B, 6B, 7A, and Un.
To examine the pyramiding effects of favorable alleles of various QTLs, we analyzed the number of favorable alleles in 6 mapped loci per accession. Favorable alleles were 0–5. Linear regression (r2 = 0.872) revealed the correlation between disease severity and the number of favorable alleles (Figure 3 and Supplementary Table 2). Accessions including a larger number of favorable alleles, such as Zhoumai16 (5), Xikemai4 (4), and Zhoumai17 (4), exhibited strong FSB resistance. Conversely, Yang 07–15, with no favorable alleles, exhibited low FSB resistance (Supplementary Table 1).
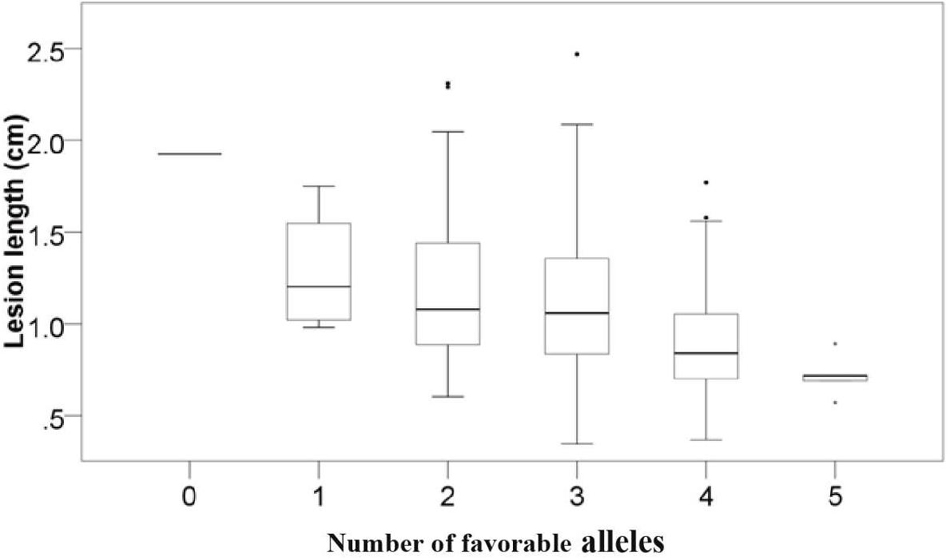
Figure 3. Relationship between the number of favorable quantitative trait locis (QTLs) and the mean FSB severity. Lesion length indicates the FSB severity.
The SNPs (IWB41243, IWB64668, IWB3107, and IWA3221), associated with Qfsb.hbaas-1DL, Qfsb.hbaas-3AS, Qfsb.hbaas-3BL, and Qfsb.hbaas-6BL, respectively, were successfully used to develop KASP markers (Table 5). All 240 wheat accessions were genotyped by these KASP markers. The results demonstrated that the genotypes from the KASP test were identical to the chip assay with low-frequency oscillations (2.5, 5.0, 3.3, and 3.8% for each marker, respectively).

Table 5. Primer sequences of single nucleotide polymorphism (SNP) markers for validation in wheat lines by Kompetitive Allele-Specific PCR (KASP) assay.
A total of 291 candidate genes were located within the candidate regions. Combined with transcriptome data from public databases (Ma et al., 2014; Powell et al., 2017), 57 genes were differently expressed in wheat seedling coleoptile organs after infection by Fusarium spp. (Figure 4 and Supplementary Table 3). Among them, eight unique annotated genes involved in plant disease resistance were considered as candidates for mapping QTLs (Table 6). Two genes encoding the disease resistance protein RPM1 and receptor-like protein kinase were identified as candidates for Qfsb.hbaas-1DL. A gene encoding L-type lectin receptor kinase might contribute to FSB resistance for Qfsb.hbaas-3AS. A gene encoding MADS-box protein was considered as a candidate for Qfsb.hbaas-6BL. For Qfsb.hbaas-7AL, a gene encoding NAC domain-containing protein was identified. Three genes encoding serine/threonine kinase-like protein, HCBT-like defense response protein, and subtilisin-like protease might contribute to FSB resistance for Qfsb.hbaas-un.
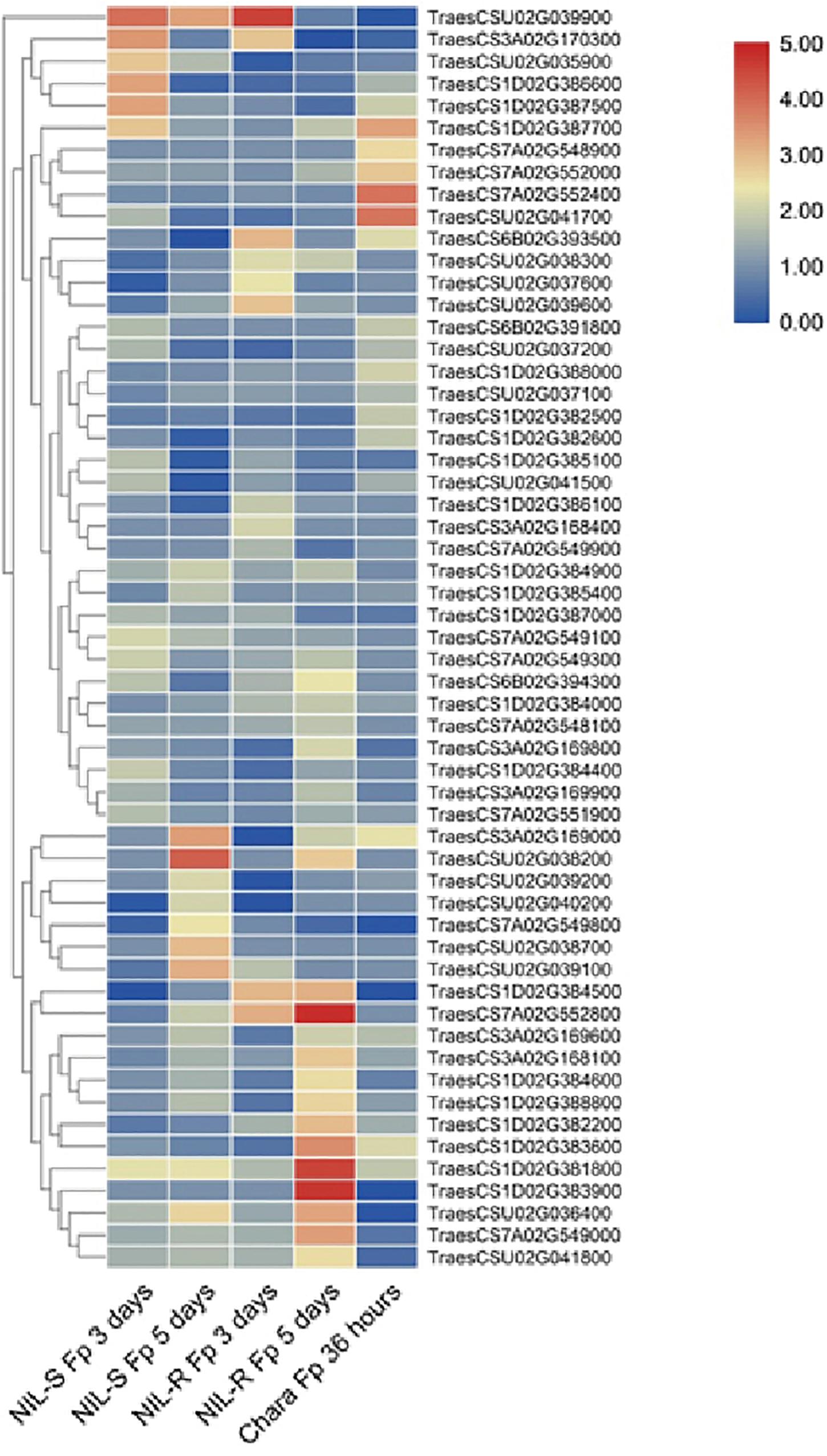
Figure 4. Clustering heatmap for differentially expressed genes infected by Fusarium spp. compared to those in the mock. S-NIL1-Fp 3 days, susceptible isolines infected by Fusarium spp. after 3 days; S-NIL1-Fp 5 days, susceptible isolines infected by Fusarium spp. after 5 days; R-NIL1-Fp 3 days, resistant isolines infected by Fusarium spp. after 3 days; R-NIL1-Fp 5 days, resistant isolines infected by Fusarium spp. after 3 days; Chara-Fp 36 h, wheat culture “Chara” infected by Fusarium spp. after 36 h.
Breeding the cultivars with resistance to FSB and FHB offers an efficient way to control complex diseases and decrease yield losses or mycotoxin occurrence in agricultural products. Combining the two resistance traits in one elite wheat cultivar is challenging due to its exposure to regulated independent genetic loci and also the restricted size of resistant germplasm in the natural environment (Tamburic-Ilincic et al., 2009). In this study, 229 elite Chinese wheat cultivars and lines, which represent the genetic diversity in newly assembled accessions in China (Jia et al., 2020), were investigated. A total of 54 cultivars reached the maximum annual acreage of 1 × 105 ha during 2000–2016, and quite a few cultivars such as Liangxing 99, Zhoumai 18, Jimai 22, Zhoumai 22, Zhengmai 9023, and Aikang 58, have been used as founder parents in breeding programs (Jia et al., 2020). Among them, 23 accessions showed a relatively high-level resistance with an average lesion length of less than 0.6 cm, exhibiting the potential for breeding for FSB resistance in wheat. Despite the negative correlation between FSB infection and FHB index in the population (Table 1), combined with our previous research results (Zhu et al., 2020), we also found that some cultivars such as Jingfumai 1 and Yangmai 11 had relatively great resistance to both FSB and FHB simultaneously. Jingfumai 1 and Yangmai 11 both bred in the Middle and Lower Yangtze River Valleys were red-grained spring wheat and high resistant to pre-harvest sprouting. The spike length of the two lines was 8.0 and 8.4 cm, and the spikelet number was 18.3 and 17.2, respectively. The research conformed to the findings reported by Ren et al. (2015), who found that the FSB-resistant cultivar Petrus was simultaneously resistant to FHB. These lines were good parent candidates for future crosses in breeding for Fusarium seedling resistance and head blight resistance in wheat.
The correlation analysis between FSB and FHB resistance has been reported in previous studies (Mesterhazy, 1987; Gosman et al., 2005; Ruckenbauer et al., 2001; Tamburic-Ilincic et al., 2009; Shin et al., 2014). Few studies revealed a positive association between FSB and FHB resistance. Shin et al. (2014) reported the significant correlation coefficients between the lesion lengths and Type II resistance to FSB and FHB, but the number of samples was not very large and only 29 Korean winter wheat cultivars were chosen in trials. The CIMMYT spring wheat line LSP2 was proved to have a high susceptibility to FSB and resistance to FHB, caused by Fusarium spp. (Ren et al., 2016). The widely planted British winter wheat cultivar Rialto was highly resistant to FSB, caused by Microdochium spp., while some reports revealed its high susceptibility to FHB (Srinivasachary et al., 2008). Some wheat cultivars, including Chinese local cultivars Wangshuibai and Sumai3, were highly resistant to FHB, and high susceptibility to FSB was also found (Mesterhazy, 1987; Wu et al., 2005; Li X. et al., 2010). Using Sumai3 and Falat as the cultivars resistant and susceptible to FHB, respectively, Sorahinobar et al. (2016) observed little correspondence between wheat seedling tolerance to F. graminearum crude extract and resistance to FHB. In both our previous (Zhu et al., 2020) and present studies, Sumai3 showed an FSB-susceptible reaction while exhibiting FHB resistance in response to Fusarium spp. Negative correlations, albeit low, between FSB and FHB resistance were observed in the present study. These results also agreed with the findings published by Bruins et al. (1993); Ruckenbauer et al. (2001), Gosman et al. (2005), and Tamburic-Ilincic et al. (2009), who discovered that greenhouse experiments in seedling cannot be used when selecting for FHB resistance.
Two transgenic wheat lines expressing two anti-fungal peptides exhibited enhanced resistance to FSB and FHB, while FHB resistance could be detected in the other five lines (Liu et al., 2012). Transgenic wheat overexpressing an A. thaliana NPR1 gene could increase the severity of FSB, although FHB resistance increased simultaneously (Gao et al., 2013). Li X. et al. (2010) firstly reported a close association between FHB and FSB resistance in wheat using distinct molecular profiles for disease-associated gene expression and suggested that there may be two resistance mechanisms in wheat spikes and seedlings in response to FHB pathogens. Some studies have also shown different QTLs for resistance to FSB and FHB (Tamburic-Ilincic et al., 2009; Ren et al., 2016). In our previous study, five QTL on chromosome arms 1AS, 2DL, 5AS, 5AL, and 7DS were associated with FHB resistance, explaining 5.4–10.3% of phenotypic variation (Zhu et al., 2020). Using the same population, we identified six entirely different QTL on chromosome arms 1DL, 3AS, 3BL, 6BL, 7AL, and Un, interpreting phenotypic variation of 4.83–7.53% (Table 3 and Figure 2). Different regions suggested the differences in QTLs for resistance to FSB and FHB and resistance to FSB and FHB is probably independent. Thus, due to gene recombination, a few accessions in this research exhibited Fusarium resistance in seedling and head.
There might be two main reasons why discrepant alterations in genes or QTLs could be associated with resistance to both FSB and FHB. The first reason is the infection time; FSB infection occurs during the seedling growth, whereas FHB infection occurs during the flowering stage. It is well known that different genes can be involved in the resistance of host plants to one disease in various stages of plant development (Li H. B. et al., 2010). The second reason is the infection of different organs by the two diseases. Miedaner (1997) put forward the idea that complex interactions can occur between the resistance to diseases across different plant growth stages, plant organs, or host genotypes. We suggested that the mechanisms and genes involved in resistance to Fusarium during seedling growth and spike formation are possibly different, and separate screening is essential to evaluate the resistance to FSB and FHB, caused by Fusarium in breeding programs.
The discovery of novel genes or QTLs is a constant challenge and extremely important in wheat breeding. Many QTLs associated with FHB resistance have been detected (Zhu et al., 2020), whereas there have been very few studies on QTLs for resistance to FSB. Using a Wuhan/Nyubai doubled haploid (DH) wheat population, merely one QTL, controlling FSB resistance, detected on chromosome 5B and marker WMC75, could interpret 13.8% phenotypic variation in the trait (Tamburic-Ilincic et al., 2009). Using the Rialto/LSP2 DH population, a single major QTL conferring FSB resistance to Microdochium majus was found on the chromosome 1AL in all four experiments, accounting for 32.5–56.6% of the phenotypic variation; a significant QTL conferring FSB resistance to Microdochium nivale was discovered on chromosomes 2BS and explained 29.3–55.0% of the phenotypic variation (Ren et al., 2016). In this research, we detected six QTLs on chromosome arms 1DL, 3AS, 3BL, 6BL, 7AL, and Un that were significant for resistance to FSB and previously uncharacterized in wheat, and therefore they were likely to be novel QTLs for FSB resistance. The average lesion length dramatically decreased when the number of favorable alleles increased (Figure 3). This finding suggested the prospective role of these QTLs in FSB resistance. But some QTls such as Qfsb.hbaas-1D and Qfsb.hbaas-7A have very tiny minor allele frequencies, they may not be real signals. To validate the real effects of these 6 mapped QTLs, typical significant markers should be examined using various bi-parental and natural populations.
Combined with the analysis of transcriptome data, we identified eight unique annotated genes involved in plant disease resistance in wheat in IWGSC RefSeq v1.1, which were linked to the six QTLs (Table 6). The RPM1 is a CC-NB-LRR protein that conferred resistance against Pseudomonas syringae pv. maculicola 1 (Mackey et al., 2002; Su et al., 2017), and TaRPM1 might play a key part in the wheat innate immune response to the infection caused by the powdery mildew pathogen (Nie and Ji, 2019). Receptor-like protein kinases, which are the largest gene family in plants, play essential roles in combating infection caused by pathogens (Liu et al., 2017). TaCRK2, a novel receptor-like kinase gene, plays a positive role in resistance to leaf rust in wheat through the regulation of the HR cell death process induced by P. triticina (Gu et al., 2020). L-type lectin receptor kinases are omnipresent in plants and play an important role in the initiation of innate immunity (Wang and Bouwmeester, 2017). An L-type lectin receptor kinase in Haynaldia villosa conferred powdery mildew resistance in wheat (Wang et al., 2018). Moreover, MIKC-type MADS-box genes exhibited new expression patterns in response to biotic stress (Schilling et al., 2020). The NAC protein constituting the most important plant transcription factors could enhance resistance to Fusarium head blight, as well as stripe rust (Ning et al., 2010; Perochon et al., 2019). Serine/threonine kinase, one of the largest protein kinase gene families, could confer resistance to powdery mildew and stripe rust in wheat (Cao et al., 2011; Gou et al., 2015). HCBT-like defense response protein, which was rapidly and transiently expressed after being induced by the pathogen, plays an essential role in fungal pathogen resistance (Brooks et al., 2002). The subtilisin-like protease is associated with pathogenicity in fungi and plays an important role in resistance to leaf rust in wheat (Fan et al., 2016).
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
YL, DM, and CG guided the design of the experiment. YL, GZ, ZZ, LC, HN, WH, HT, JS, and YZ directed the data analysis. YL and GZ conducted data analysis and wrote the manuscript. DM and CG supervised the experiment and confirmed the manuscript. YL was the guarantor of this work, so she could have full access to all the data in the research and responsible for the integrity of the data and the accuracy of the data analysis. All authors contributed to the article and approved the submitted version.
This research was supported by Hubei Provincial Special Project of Central Government Guides Local Science and Technology Development (2020ZYYD011) and the Hubei Key Laboratory of Food Crop Germplasm and Genetic Improvement (2018zjj09).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.777494/full#supplementary-material
Alaux, M., Rogers, J., Letellier, T., Flores, R., Alfama, F., Pommier, C., et al. (2018). Linking the International Wheat Genome Sequencing Consortium bread wheat reference genome sequence to wheat genetic and phonemic data. Genome Biol. 19:111. doi: 10.1186/s13059-018-1491-4
Antalová, Z., Bleša, D., Martinek, P., and Matušinsky, P. (2020). Transcriptional analysis of wheat seedlings inoculated with Fusarium culmorum under continual exposure to disease defence inductors. PLoS One 15:e0224413. doi: 10.1371/journal.pone.0224413
Appels, R., Eversole, K., Feuillet, C., Keller, B., Rogers, J., Stein, N., et al. (2018). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361:eaar7191. doi: 10.1126/science.aar7191
Bai, G., and Shaner, G. (2004). Management and resistance in wheat and barley to Fusarium head blight. Annu. Rev. Phytopathol. 42, 135–161. doi: 10.1146/annurev.phyto.42.040803.140340
Bradbury, P. J., Zhang, Z., Kroon, D. E., Casstevens, T. M., Ramdoss, Y., and Buckler, E. S. (2007). TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23, 2633–2635. doi: 10.1093/bioinformatics/btm308
Brooks, S. A., Huang, L., Gill, B. S., and Fellers, J. P. (2002). Analysis of 106 kb of contiguous DNA sequence from the D genome of wheat reveals high gene density and a complex arrangement of genes related to disease resistance. Genome 45, 963–972. doi: 10.1139/g02-049
Bruins, M. B., Karsai, M., Schepers, I. J., and Snijders, C. H. A. (1993). Phytotoxicity of deoxynivalenol to wheat tissue with regard to in vitro selection for Fusarium head blight resistance. Plant Sci. 94, 195–206. doi: 10.1016/0168-9452(93)90020-Z
Buerstmayr, H., Ban, T., Anderson, J. A., Buerstmayr, H., Ban, T., and Anderson, J. A. (2009). QTL mapping and marker-assisted selection for Fusarium head blight resistance in wheat: a review. Plant Breed. 128, 1–26. doi: 10.1556/crc.36.2008.suppl.b.1
Cao, A., Xing, L., Wang, X., Yang, X., Wang, W., Sun, Y., et al. (2011). Serine/threonine kinase gene Stpk-V, a key member of powdery mildew resistance gene Pm21, confers powdery mildew resistance in wheat. Proc. Natl. Acad. Sci. U.S.A. 108, 7727–7732. doi: 10.1073/pnas.1016981108
Chen, C., Rui, X., Hao, C., and He, Y. (2018). TBtools, a toolkit for biologists integrating various biological data handling tools with a user-friendly interface. BioRxiv [preprint] 289660. doi: 10.1101/289660
Cheng, W., Song, X. S., Li, H. P., Cao, L. H., Sun, K., and Qiu, X. L. (2015). Host-induced gene silencing of an essential chitin synthase gene confers durable resistance to Fusarium head blight and seedling blight in wheat. Plant Biotechnol. J. 13, 1335–1345. doi: 10.1111/pbi.12352
Fan, T., Bykova, N. V., Rampitsch, C., and Xing, T. (2016). Identification and characterization of a serine protease from wheat leaves. Eur. J. Plant Pathol. 146, 293–304. doi: 10.1007/s10658-016-0914-x
Gao, C. S., Kou, X. J., Li, H. P., Zhang, J. B., Saad, A. S. I., and Liao, Y. C. (2013). Inverse effects of Arabidopsis NPR1 gene on fusarium seedling blight and fusarium head blight in transgenic wheat. Plant Pathol. 62, 383–392. doi: 10.1111/j.1365-3059.2012.02656.x
Gosman, N., Chandler, E., Thomsett, M., Draeger, R., and Nicholson, P. (2005). Analysis of the relationship between parameters of resistance to Fusarium head blight and in vitro tolerance to deoxynivalenol of the winter wheat cultivar WEK0609H. Eur. J. Plant Pathol. 111, 57–66. doi: 10.1007/s10658-004-2733-8
Gou, J. Y., Li, K., Wu, K., Wang, X. D., Lin, H. Q., Cantu, D., et al. (2015). Wheat stripe rust resistance protein WKS1 reduces the ability of the thylakoid-associated ascorbate peroxidase to detoxify reactive oxygen species. Plant Cell. 27, 1755–1770. doi: 10.1105/TPC.114.134296
Gu, J., Sun, J., Liu, N., Sun, X., Liu, C., Wu, L., et al. (2020). A novel cysteine-rich receptor-like kinase gene, TaCRK2, contributes to leaf rust resistance in wheat. Mol. Plant Pathol. 21, 732–746. doi: 10.1111/mpp.12929
Haigh, I. M., Jenkinson, P., and Hare, M. C. (2009). The effect of a mixture of seed-borne Microdochium nivale var. majus and Microdochium nivale var. nivale infection on Fusarium seedling blight severity and subsequent stem colonisation and growth of winter wheat in pot experiments. Eur. J. Plant Pathol. 124, 65–73. doi: 10.1007/s10658-008-9393-z
Hu, W., Gao, D., Wu, H., and Liu, J. (2020). Genome-wide association mapping revealed syntenic loci QFhb-4AL and QFhb-5DL for Fusarium head blight resistance in common wheat (Triticum aestivum L.). BMC Plant Biol. 20:29. doi: 10.1186/s12870-019-2177-0
Jia, M., Yang, L., Zhang, W., Rosewarne, G., Li, J., Yang, E., et al. (2020). Genome-wide association analysis of stripe rust resistance in modern Chinese wheat. BMC Plant Biol. 20:491. doi: 10.1186/s12870-020-02693-w
Li, F., Wen, W., Liu, J., Zhang, Y., Cao, S., He, Z., et al. (2019). Genetic architecture of grain yield in bread wheat based on genome-wide association studies. BMC Plant Biol. 19:168. doi: 10.1186/s12870-019-1781-3
Li, H. B., Xie, G. Q., Ma, J., Liu, G. R., Wen, S. M., Ban, T., et al. (2010). Genetic relationships between resistances to Fusarium head blight and crown rot in bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 121, 941–950. doi: 10.1007/s00122-010-1363-0
Li, X., Zhang, J. B., Song, B., Li, H. P., Xu, H. Q., Qu, B., et al. (2010). Resistance to Fusarium head blight and seedling blight in wheat is associated with activation of a cytochrome P450 gene. Phytopathology 100, 183–191. doi: 10.1094/PHYTO-100-2-0183
Liu, P. L., Du, L., Huang, Y., Gao, S. M., and Meng, Y. (2017). Origin and diversification of leucine-rich repeat receptor-like protein kinase (LRR-RLK) genes in plants. BMC Evol. Biol. 17:47. doi: 10.1186/s12862-017-0891-5
Liu, Y., Tong, H., Zhu, Z., Chen, L., Zou, J., Zhang, Y., et al. (2016). Progress in research on mechanism of resistance to fusarium head blight in wheat. Sci. Agric. Sin. 49, 1476–1488. doi: 10.3864/j.issn.0578-1752.2016.08.005
Liu, Z. W., Li, H. P., Cheng, W., Peng, Y., Zhang, J. B., Gong, A. D., et al. (2012). Enhanced overall resistance to Fusarium seedling blight and Fusarium head blight in transgenic wheat by co-expression of anti-fungal peptides. Eur. J. Plant Pathol. 134, 721–732. doi: 10.1007/s10658-012-0048-8
Ma, J., Stiller, J., Zhao, Q., Jian, M., Stiller, J., Qiang, Z., et al. (2014). Transcriptome and allele specificity associated with a 3bl locus for fusarium crown rot resistance in bread wheat. PLoS One 9:e113309. doi: 10.1371/journal.pone.0113309
Ma, Z., Xie, Q., Li, G., Jia, H., Zhou, J., Kong, Z., et al. (2020). Germplasms, genetics and genomics for better control of disastrous wheat Fusarium head blight. Theor. Appl. Genet. 133, 1541–1568. doi: 10.1007/s00122-019-03525-8
Mackey, D., Holt, B. F., Wiig, A., and Dangl, J. L. (2002). RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108, 743–754. doi: 10.1016/S0092-8674(02)00661-X
Mesterhazy, A. (1987). Selection of head blight resistant wheats through improved seedling resistance. Plant Breed. 98, 25–36. doi: 10.1111/j.1439-0523.1987.tb01086.x
Miedaner, T. (1997). Breeding wheat and rye for resistance to Fusarium diseases. Plant Breed. 116, 201–220. doi: 10.1111/j.1439-0523.1997.tb00985.x
Nie, Y. B., and Ji, W. Q. (2019). Cloning and characterization of disease resistance protein RPM1 genes against powdery mildew in wheat line N9134. Cereal. Res. Commun. 47, 473–483. doi: 10.1556/0806.47.2019.27
Ning, X., Zhang, G., Liu, X. Y., Deng, L., Cai, G. L., Zhang, Y., et al. (2010). Characterization of a novel wheat NAC transcription factor gene involved in defense response against stripe rust pathogen infection and abiotic stresses. Mol. Biol. Rep. 37, 3703–3712. doi: 10.1007/s11033-010-0023-4
Perochon, A., Kahla, A., Vranić, M., Jia, J., Malla, K. B., Craze, M., et al. (2019). A wheat NAC interacts with an orphan protein and enhances resistance to Fusarium head blight disease. Plant Biotechnol. J. 17, 1892–1904. doi: 10.1111/pbi.13105
Pestka, J. J., and Smolinski, A. T. (2005). Deoxynivalenol: toxicology and potential effects on humans. J. Toxicol. Env. Health, Part B. 8, 39–69. doi: 10.1080/10937400590889458
Powell, J. J., Carere, J., Fitzgerald, T. L., Stiller, J., Covarelli, L., Xu, Q., et al. (2017). The Fusarium crown rot pathogen Fusarium pseudograminearum triggers a suite of transcriptional and metabolic changes in bread wheat (Triticum aestivum L.). Ann. Bot. 119, 853–867. doi: 10.1093/aob/mcw207
Ren, R., Foulkes, J., Mayes, S., Yang, X., and Ray, R. V. (2016). Identification of novel quantitative trait loci for resistance to Fusarium seedling blight caused by Microdochium majus and M. nivale in wheat. Field Crop Res. 191, 1–12. doi: 10.1016/j.fcr.2016.03.011
Ren, R., Yang, X., and Ray, R. V. (2015). Comparative aggressiveness of Microdochium nivale and M. majus and evaluation of screening methods for Fusarium seedling blight resistance in wheat cultivars. Eur. J. Plant Pathol. 141, 281–294. doi: 10.1007/s10658-014-0541-3
Ruckenbauer, P., Buerstmayr, H., and Lemmens, M. (2001). Present strategies in resistance breeding against scab (Fusarium spp.). Euphytica 119, 123–129. doi: 10.1023/A:1017598523085
Sapkota, S., Hao, Y., Johnson, J., Buck, J., Aoun, M., and Mergoum, M. (2019). Genome-wide association study of a worldwide collection of wheat genotypes reveals novel quantitative trait loci for leaf rust resistance. Plant Genome 12:190033. doi: 10.3835/plantgenome2019.05.0033
Schilling, S., Kennedy, A., Pan, S. R., Jermiin, L. S., and Melzer, R. (2020). Genome-wide analysis of MIKC-type MADS-box genes in wheat: pervasive duplications, functional conservation and putative neofunctionalization. New Phytol. 225, 511–529. doi: 10.1101/585232
Shi, X. L., and Ling, H. Q. (2018). Current advances in genome sequencing of common wheat and its ancestral species. Crop J. 6, 15–21. doi: 10.1016/j.cj.2017.11.001
Shin, S., Kim, K. H., Kang, C. S., Cho, K. M., Park, C. S., Okagaki, R., et al. (2014). A simple method for the assessment of Fusarium head blight resistance in korean wheat seedlings inoculated with Fusarium graminearum. Plant Pathol. J. 30, 25–32. doi: 10.5423/PPJ.OA.06.2013.0059
Somers, D. J., Fedak, G., and Savard, M. (2003). Molecular mapping of novel genes controlling Fusarium head blight resistance and deoxynivalenol accumulation in spring wheat. Genome 46, 555–564. doi: 10.1139/g03-033
Sorahinobar, M., Niknam, V., Ebrahimzadeh, H., Soltanloo, H., Moradi, B., and Bahram, M. (2016). Lack of association between Fusarium graminearum resistance in spike and crude extract tolerance in seedling of wheat. Eur. J. Plant Pathol. 144, 525–538. doi: 10.1007/s10658-015-0792-7
Srinivasachary, Gosman, N., Steed, A., Simmonds, J., Leverington-Waite, M., Wang, Y., et al. (2008). Susceptibility to Fusarium head blight isassociated with the Rht-D1b semi-dwarfing allele in wheat. Theor. Appl. Genet. 116, 1145–1153. doi: 10.1007/s00122-008-0742-2
Su, J., Spears, B. J., Kim, S. H., and Gassmann, W. (2017). Constant vigilance: plant functions guarded by resistance proteins. Plant J. 93, 637–650. doi: 10.1111/tpj.13798
Tamburic-Ilincic, L., Somers, D., Fedak, G., and Schaafsma, A. (2009). Different quantitative trait loci for Fusarium resistance in wheat seedlings and adult stage in the Wuhan/Nyubai wheat population. Euphytica 165, 453–458. doi: 10.1007/s10681-008-9747-9
Wang, S., Wong, D., Forrest, K., Allen, A., Chao, S., Huang, B. E., et al. (2014). Characterization of polyploid wheat genomic diversity using a high-density 90000 single nucleotide polymorphism array. Plant Biotechnol. J. 12, 787–796. doi: 10.1111/pbi.12183
Wang, Y., and Bouwmeester, K. (2017). L-type lectin receptor kinases: new forces in plant immunity. PLoS Pathog. 13:e1006433. doi: 10.1371/journal.ppat.1006433
Wang, Z., Cheng, J., Fan, A., and Zhao, J. (2018). LecRK-V, an L-type lectin receptor kinase in Haynaldia villosa, plays positive role in resistance to wheat powdery mildew. Plant Biotechnol. J. 16, 50–62. doi: 10.1111/pbi.12748
Wiese, M. V. (1987). Compendium of Wheat Diseases, 2nd Edn. Saint Paul, MN: The American Phytopathological Society.
Wisniewska, H., and Busko, M. (2005). Evaluation of spring wheat resistance to Fusarium seedling blight and head blight. Biologia 60, 287–293.
Wu, A. B., Li, H. P., Zhao, C. S., and Liao, Y. C. (2005). Comparative pathogenicity of fusarium graminearum isolates from China revealed by wheat coleoptile and floret inoculations. Mycopathologia 160, 75–83. doi: 10.1007/s11046-005-1153-4
Keywords: common wheat, Fusarium seedling blight, Fusarium head blight, GWAS, QTL
Citation: Liu Y, Zhu G, Zhu Z, Chen L, Niu H, He W, Tong H, Song J, Zhang Y, Ma D and Gao C (2021) Investigation and Genome-Wide Association Analysis of Fusarium Seedling Blight Resistance in Chinese Elite Wheat Lines. Front. Plant Sci. 12:777494. doi: 10.3389/fpls.2021.777494
Received: 15 September 2021; Accepted: 18 October 2021;
Published: 17 November 2021.
Edited by:
Jianjun Chen, University of Florida, United StatesReviewed by:
Mohsen Mohammadi, Purdue University, United StatesCopyright © 2021 Liu, Zhu, Zhu, Chen, Niu, He, Tong, Song, Zhang, Ma and Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dongfang Ma, bWFkb25nZmFuZzE5ODRAMTYzLmNvbQ==; Chunbao Gao, Z2NiZ3lid2pAMTYzLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.