- 1Swift Current Research and Development Center, Agriculture and Agri-Food Canada (AAFC), Swift Current, SK, Canada
- 2Morden Research and Development Centre, Agriculture and Agri-Food Canada, Morden, MB, Canada
- 3Department of Plant Sciences, University of Saskatchewan, Saskatoon, SK, Canada
The hexaploid spring wheat cultivar, Carberry, was registered in Canada in 2009, and has since been grown over an extensive area on the Canadian Prairies. Carberry has maintained a very high level of leaf rust (Puccinia triticina Eriks.) resistance since its release. To understand the genetic basis of Carberry’s leaf rust resistance, Carberry was crossed with the susceptible cultivar, Thatcher, and a doubled haploid (DH) population of 297 lines was generated. The DH population was evaluated for leaf rust in seven field environments at the adult plant stage. Seedling and adult plant resistance (APR) to multiple virulence phenotypes of P. triticina was evaluated on the parents and the progeny population in controlled greenhouse studies. The population was genotyped with the wheat 90 K iSelect single nucleotide polymorphism (SNP) array, and quantitative trait loci (QTL) analysis was performed. The analysis using field leaf rust response indicated that Carberry contributed nine QTL located on chromosomes 1B, 2B (2 loci), 2D, 4A, 4B, 5A, 5B, and 7D. The QTL located on 1B, 2B, 5B, and 7D chromosomes were observed in two or more environments, whereas the remainder were detected in single environments. The resistance on 1B, detected in five environments, was attributed to Lr46 and on 7D, detected in seven environments to Lr34. The first 2B QTL corresponded with the adult plant gene, Lr13, while the second QTL corresponded with Lr16. The seedling analysis showed that Carberry carries Lr2a, Lr16, and Lr23. Five epistatic effects were identified in the population, with synergistic interactions being observed for Lr34 with Lr46, Lr16, and Lr2a. The durable rust resistance of Carberry is attributed to Lr34 and Lr46 in combination with these other resistance genes, because the resistance has remained effective even though the P. triticina population has evolved virulent to Lr2a, Lr13, Lr16, and Lr23.
Introduction
Wheat (Triticum aestivum L.) is the most widely cultivated crop globally, and it is a major source of calories and protein for the world population (Shiferaw et al., 2013; Shewry and Hey, 2015). Significant constraints to increased wheat production in Canada and internationally are the rust diseases, such as leaf rust (Puccinia triticina Eriks.), stem rust (Puccinia graminis Pers.: Pers. f. sp. Tritici Eriks. & E. Henn.), and stripe rust (Puccinia striiformis Westend. f. sp. tritici) (Mcintosh et al., 1995; Kolmer, 2005; McCallum et al., 2007). Historically, leaf rust had caused major crop losses in North America (Peturson, 1958; Kolmer, 2005; McCallum et al., 2007). Leaf rust was abundant in the Prairie Provinces of Canada in 1953, 1954, and 1955, and it was prevalent and well established in the region somewhat later than stem rust (Peturson, 1958). Considerable rust damage was caused in all 3 years but particularly in 1954, when both leaf and stem rusts were heavy throughout most of Manitoba and Saskatchewan and in a considerable area in the east-central Alberta. Corresponding wheat yield reduction in the Western Canada due to leaf rust and stem rust was estimated at about 45 million bushels in 1953, 150 million bushels in 1954, and 9 million bushels in 1955. Leaf rust was also a production problem in the 1980s when the cultivar AC Barrie was the dominant wheat cultivar in the western Canada (McCallum and DePauw, 2008). Leaf rust can still pose a serious threat to wheat production if breeding for resistance and other management practices are relaxed (Aboukhaddour et al., 2020).
Over time, improved resistance to leaf rust was achieved in western Canada by developing cultivars with additional genes for resistance (McCallum and DePauw, 2008). For example, a hard red spring wheat cultivar, Thatcher was susceptible to leaf rust, although it was the first significant stem rust resistant cultivar grown in Canada extensively over a long period of time extending from 1939 to 1968 (McCallum et al., 2007; McCallum and DePauw, 2008). Thatcher was selected from a double cross Marquis/Iumillo//Marquis/Kanred wheat in 1925 and released in 1935 (Hayes et al., 1936). The resistant durum variety, Iumillo, and the winter wheat variety, Kanred are the ancestors of Thatcher, from which it inherited some of its resistance (Hayes et al., 1936). However, Thatcher is generally very susceptible to leaf rust, except to a few races, and has been used as a universal susceptible genetic background to develop the Thatcher near isogenic wheat lines (McCallum et al., 2016). Using a cross of a Romanian wheat line Fundulea 900 and Thatcher, Zhang et al. (2017) reported a minor effect of leaf rust resistance QTL on chromosome 2DS, QLr.hebau-2DS contributed by Thatcher. Zhang et al. (2017) additionally indicated that Lr22b may confer residual resistance in field nurseries when challenged with isolates virulent on Lr22b, or another gene linked to Lr22b confers this resistance from Thatcher. Previously, Dyck (1979) indicated that Thatcher carries Lr22b which confers adult plant resistance (APR) to leaf rust to only a few virulence phenotypes.
Carberry is a semi-dwarf doubled haploid (DH), hard red spring wheat variety that is derived from the cross, Alsen by Superb made in 2000 at the Swift Current Research and Development Centre, AAFC, SK, Canada and registered in 2009 (DePauw et al., 2011). It was grown over 2.3 Mha in the years 2011--20191. Carberry was resistant to both leaf rust and stem rust at the time of its release (DePauw et al., 2011) and currently, it still has resistance to both rust diseases and moderate resistance to stripe rust. Superb has the resistance genes, Lr2a and Lr10 (McCallum and Seto-Goh, 2010), and Alsen is reported to have genes, Lr2a, Lr10, Lr13, Lr23, and Lr34 (Oelke and Kolmer, 2005).
Resistance has been and will continue to be the major means for controlling cereal rusts (Roelfs, 1988). Two classes of genes, namely all stage (seedling) resistance (ASR) and APR are known. ASR is expressed throughout the life of the plant, whereas APR is expressed only at later stages in the plant’s development (Ellis et al., 2014; da Silva et al., 2018). Most of the designated genes confer ASR against leaf rust and some QTL, reported in different genetic backgrounds, provide APR (McIntosh et al., 2014). Lr34 and Sr2 are APR genes that have been deployed in conjunction with other ASR and APR genes providing resistance in wheat cultivars widely grown over many years, therefore, demonstrating durable resistance (Ellis et al., 2014). ASR is governed by major or race-specific genes and is often characterized by its short longevity as compared to some APR genes. For example, the ASR gene, Lr10 became ineffective and Lr16 became partially ineffective within a few years of their deployment in Canadian wheat cultivar, Selkirk (da Silva et al., 2018). In Canada, improved leaf resistance has been achieved primarily due to the use of genes, such as Lr13, Lr14a, Lr16, Lr21, and Lr34 (McCallum and DePauw, 2008; McCallum et al., 2016).
Breeding for resistance has evolved with the advent of molecular mapping technologies and the development of markers linked with resistance genes. Identifying and mapping genes, and developing genetic markers for marker-assisted breeding is helpful to develop wheat varieties with an acceptable level of resistance. This study was conducted to understand the genetic basis of leaf rust resistance in Carberry through a cross with the susceptible variety, Thatcher.
Materials and Methods
Plant Material
A DH population of 297 lines generated from F1 plants of a cross Carberry/Thatcher (CT) was used in this study. Carberry is resistant to leaf rust (DePauw et al., 2011), whereas Thatcher is susceptible (Dyck et al., 1966). The population was evaluated for ASR in the greenhouse and APR in the greenhouse and in the field.
Disease Evaluation
Seedling Leaf Rust Analysis
To determine the number and identity of the all stage (seedling) leaf rust resistance genes in Carberry, the parents and the CT population were inoculated at the seedling stage with multiple virulent phenotypes of P. triticina. Urediniospores of single purified isolates of P. triticina were used to inoculate this population at the two-leaf growth stage as described by McCallum et al. (2020). The isolates used included 1-1 BBBD, 96-12-3 MBDS, 128-1 MBRJ, 74-2 MGBJ, 11-180-1 TDBG, 06-1-1 TDBG, 77-2 TJBJ, 9-1 SBDG, 161-1 FBDS, 08-5-1 TDBB,19-129-1 TBTN, and 18-10-1 TBBS (McCallum et al., 2020). The race letter codes used in the isolate names define the virulence/avirulence formula of each isolate as described by Long and Kolmer (1989). Plants were rated to determine the infection type 12–14 days post-inoculation (McCallum et al., 2020). Host lines that produced infection types “;” (hypersensitive flecks), “1” (small uredinia with necrosis), and “2” (small to medium sized uredinia with chlorosis) were considered resistant, and those that produced infection types “3” (medium sized uredinia without chlorosis or necrosis) and “4” (large uredinia without chlorosis or necrosis) were considered susceptible.
Determination of Leaf Rust Resistance in Superb and Alsen, the Parental Lines of Carberry
From the initial seedling tests, Carberry appeared to have the resistance gene, Lr16, which was not reported in either of its parents, Superb and Alsen, and appeared to lack Lr10 that was reported to be present in both the parental lines (Oelke and Kolmer, 2005; McCallum and Seto-Goh, 2010). Seeds of the parental lines which were used to make the cross that resulted in Carberry were tested for seedling leaf rust resistance and the presence or absence of molecular markers associated with Lr16. Sixteen seeds of each parental line (Superb and Alsen) were planted in individual root trainers. The check lines, Carberry, Thatcher, the Thatcher near isogenic lines with Lr2a, Lr10, Lr16, and Lr23 and the standard set of 16 North American leaf rust differential lines were also planted. These plants were then inoculated with the isolate, 20-140-1 TBRD, which is virulent to Lr2a and Lr23 but avirulent to Lr10 and Lr16. Then a second set of plants, as described above, was inoculated with the isolate, 19-123-2 TBGJ which is virulent to Lr2a, Lr23, and Lr10, but avirulent to Lr16.
Leaf tissue was sampled from Superb, Alsen, and check lines, and the DNA was extracted. Carberry, Superb, Alsen, and check lines were genotyped with two Kompetitive Allele-Specific PCR (KASP) markers, kwm677 and kwm849 that are diagnostic of Lr16 (Kassa et al., 2017). KASP assays were performed as described by Kassa et al. (2017).
Lr13 Adult Plant Resistance Evaluation
To determine the presence or absence of Lr13 in Carberry, it was grown along with Thatcher and the CT population in square pots (15 cm) in the greenhouse, and one pot was inoculated with each of the Lr13 avirulent isolates 16-284-1 TGBQ and 17-358-1 TBBJ at the flag leaf stage, as described by McCallum et al. (2020). Both isolates were virulent to Lr2a and Lr23, but had intermediate (16-284-1 TGBQ) or avirulent (17-358-1 TBBJ) reactions to Lr16. Plants were rated 14 days later for the infection type produced and classified as resistant or susceptible as described above. Although both Lr34 and Lr46 APR genes were present in this population, the infection type for Lr13 was lower or more resistant than that produced by either of these genes, allowing for the identification of Lr13 in the presence of Lr34 and/or Lr46.
Hybrid Necrosis Test to Determine the Presence of Lr13 in Carberry
Leaf rust resistance Lr13 and progressive necrosis Ne2m are conditioned by a single pleiotropic gene (Zhang et al., 2016). When Lr13/Ne2m carriers are crossed with Ne1 carriers, the F1 progeny show the distinctive phenotype of progressive necrosis where leaves, starting with the first leaf, undergo necrosis and die-off as the plant develops. To confirm the presence of Lr13 in Carberry, Carberry was crossed with Kubanka, tetraploid wheat that is a carrier of Ne1. The F1 progeny were grown in conditions as described above and were observed for progressive necrosis once the third to the fourth leaves fully emerged. Zhang et al. (2016) mapped Lr13 and Ne2 using a DH population from the cross Thatcher/Thatcher-Lr13. DH lines were crossed with Kubanka in their study to confirm the cosegregation of Lr13 and Ne2. F1 progeny from these crosses with Kubanka were used as positive (Lr13 present) and negative (Lr13 absent) controls for the presence of progressive necrosis.
Field Trials
The field trials were conducted near Swift Current, SK in 2014, 2015, 2016, and 2018 and at Morden, MB in 2016, 2019, and 2020. Entries were planted in single 1 m rows in groups of five flanked by susceptible spreader rows. At Morden, the experiments consisted of two replications in a randomized complete block design, whereas the trials at Swift Current were planted as single entries with repeated parents and checks. Given the population size of nearly 300 lines, each allele at each locus is replicated roughly 150 times in the population that behaves as a diploid. The spreader rows were inoculated with a mixture of leaf rust races for ease of disease development and infection of entries as previously explained in Bokore et al. (2020). Briefly, the inoculum of P. triticina was generated by increasing the urediniospores of all races in the proportions they were found in the western Canada in the year prior to the field trial. Urediniospores of these multi-race mixtures were used to inoculate spreader rows susceptible to leaf rust at both Swift Current and Morden. For each year, all the isolates generated during the virulence survey of Manitoba and Saskatchewan were combined to generate this field inoculum. In each year, the same P. triticina race composition was used in Morden and Swift Current trials.
At the Morden location, urediniospores were suspended in light mineral oil (Soltrol, Chevron Phillips Chemical Company) and sprayed on the leaves of the spreader rows at early tillering. Subsequently, leaf rust developed on the spreader rows and urediniospores were windblown to the test lines to provide infection. At Swift Current, spreader rows of susceptible genotypes were needle inoculated with leaf rust urediniospores (Bokore et al., 2020). Irrigation misting was used to provide conditions suitable for the development and spread of leaf rust. Leaf rust severity was scored from 0 to 100% using the modified Cobb Scale (Peterson et al., 1948). Infection response (IR) was recorded as resistant (R), resistant to moderately resistant (RMR), moderately resistant (MR), mesothetic (X), moderately resistant to moderately susceptible (MRMS), moderately susceptible (MS), moderately susceptible to susceptible (MSS), and susceptible (S). To utilize the data for the analysis of main effect QTL, the epistatic effects, and the infection response scores were converted to numerical values as R = 1, RMR = 2, MR = 3, X = 4, MRMS = 5, MS = 6, MSS = 7, and S = 8. Simple means of two replications of each experiment were used for the QTL analysis at Morden, whereas single plot data was used for the QTL analysis at Swift Current.
Genotyping and Linkage Mapping
The DNAs of the parents and 297 lines were extracted from young leaves using the DNeasy 96 Plant Kit (QIAGEN Science, MD, United States). The lines and parents were genotyped with the wheat 90K iSelect single nucleotide polymorphism (SNP) genotyping array (Illumina Inc., San Diego, CA). The raw data were processed using GenomeStudio v2.0 software (Illumina). Of the 81,587 SNPs contained on the 90K iSelect SNP genotyping array, 8,360 high quality polymorphic SNPs were identified. The SNPs were identified by filtering to include only those with two major cluster frequencies displaying near 1:1 segregation expected for a DH population (each cluster containing >35 and <65% of total lines) with the third cluster containing a maximum 5% of total lines. SNPs were further filtered to include only cluster plots with a high (>0.6) GenTrain Score (the GenomeStudio clustering algorithm measuring SNP calling quality ranging from 0 to 1). Finally, only SNPs with a high (>90%) call frequency were accepted. The resulting SNP calls were then exported to MS Excel and converted to a binary mapping matrix by phasing the SNP calls corresponding to Thatcher as “A” and the SNP calls corresponding to Carberry as “B” for each SNP.
The genetic map was built using a two-step strategy as previously described by Fowler et al. (2016) and Perez-Lara et al. (2016). First, markers were clustered into linkage groups with a stringent cut off p-value of 1–10 and a maximum distance between markers of 15 cM, using the minimum spanning tree map (MSTMap) software (Wu et al., 2008). Next, the linkage groups were refined using the MapDisto version 1.7.5 software (Lorieux, 2012) using a cut off recombination value of 0.35, a minimum logarithm of odds (LOD) score of 3.0, and a Kosambi mapping function (Kosambi, 1944). The best order of markers was generated using both “AutoCheckInversions” and “AutoRipple” commands. Linkage groups were assigned to their belonging chromosomes based on the existing high density SNP maps of wheat (Cavanagh et al., 2013; Maccaferri et al., 2014, Wang et al., 2014).
Statistical Analysis
Pearson’s correlation coefficients, among disease data of different environments, were calculated using the CORR procedure of SAS v.9.3 (SAS Institute, Cary, NC, United States). Broad-sense heritability and narrow sense heritability of the disease resistance were calculated by QTLNetwork 2.0 (Yang et al., 2005, 2008). As the population used in the present study was DH, dominance gene effects were absent. Broad-sense heritability was calculated as the variance of genetic main effects divided by phenotypic variance [V(G)/V(P)], whereas narrow sense heritability was calculated as the variance of additive genetic effect divided by phenotypic variance [V(A)/V(P)]. The epistasis heritability was calculated as the variance of additive × additive divided by phenotypic variance [V(AA)/V(P)]. To compare single gene effects with combined gene effects, we performed the analysis of variance and Duncan’s Multiple Range Test (DMRT) with SAS software (SAS Institute, NC v.9.3). The comparison among gene combination effects focused on a pool of four genes Lr2a, Lr16, Lr34 and Lr46, present in Carberry. To perform the combined gene effect analysis, the DH lines were sorted into different classes based on markers that were associated with each gene. Lr13, while not effective on its own in the field, may have some degree of interaction and a background effect through its interaction with the many other resistance genes present in this population. As the number of interactions gets to be large, when these many effective genes are involved, Lr13 was not included in the combined analysis.
Detection of Main and Epistatic Quantitative Trait Loci Effects Using Field Data
The analysis of the main effect of QTL was carried out for each environment on DS and IR data, and on seedling infection response data by MapQTL 6 software, Kyazma (Van Ooijen, 2009). The QTL analysis performed based on the seedling infection response was used to compare with the QTL identified using the field data (results not presented). Simple interval mapping followed by multiple QTL mapping (MQM) approaches were conducted to detect the main effect of QTL. Cofactor markers were selected using automatic cofactor selection based on the backward elimination of markers and/or adjusted by selecting a set of markers manually. To determine the significant threshold of LOD values, a permutation test of 1,000 iterations was performed. The significance of each QTL was declared at 5% probability.
To determine the epistatic interactions between the main effect QTL, an epistasis analysis was performed by QTLNetwork 2.0, which was used to detect single-locus and epistatic QTL simultaneously (Yang et al., 2005, 2008). Mixed-model-based composite interval mapping (MCIM) within QTLNetwork 2.0 was selected for a one-dimensional (1D) genome scan to search for single-locus QTL. To determine epistatic effects, a two-dimensional (2D) genome scan procedure was used. The main effect QTL and epistatic interaction were declared significant at 5% probability.
Results
Greenhouse Leaf Rust Reaction Analysis
All isolates tested were virulent on Thatcher and avirulent on Carberry (Table 1). The segregation ratio was consistent with a single resistance gene when the progeny lines were inoculated with 11-180-1 TDBG, 06-1-1 TDBG, 95-77-2 TJBJ, and 18-10-1 TBBS (Table 2). This resistance gene gave a ‘1+’ infection type, characteristic of the reaction of these isolates to the Thatcher-Lr16 line (Table 1). Each of the progeny lines was scored as having the resistant or susceptible allele for this gene and the results for each of these virulent phenotypes were mapped as QTL for seedling leaf rust resistance in the Lr16 region of chromosome 2BS. This gene was also effective against all the other virulent phenotypes consistent with their avirulent response to Lr16 [though both 95-74-2 MGBJ and 95-77-2 TJBJ had intermediate pustule types on lines with Lr16 (Table 1)].
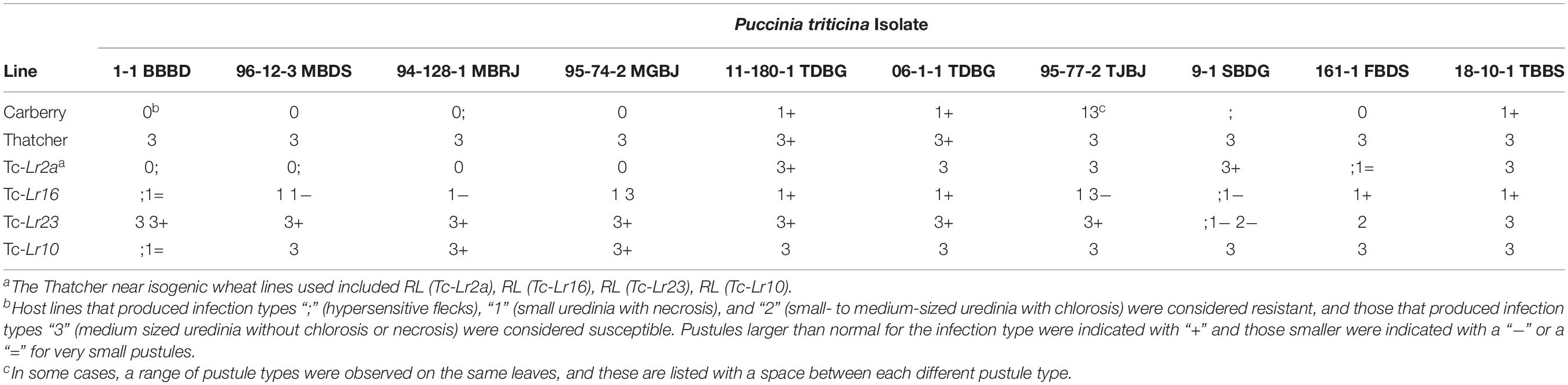
Table 1. Puccinia triticina isolates used for seedling test and the reaction of known wheat lines against each isolate.
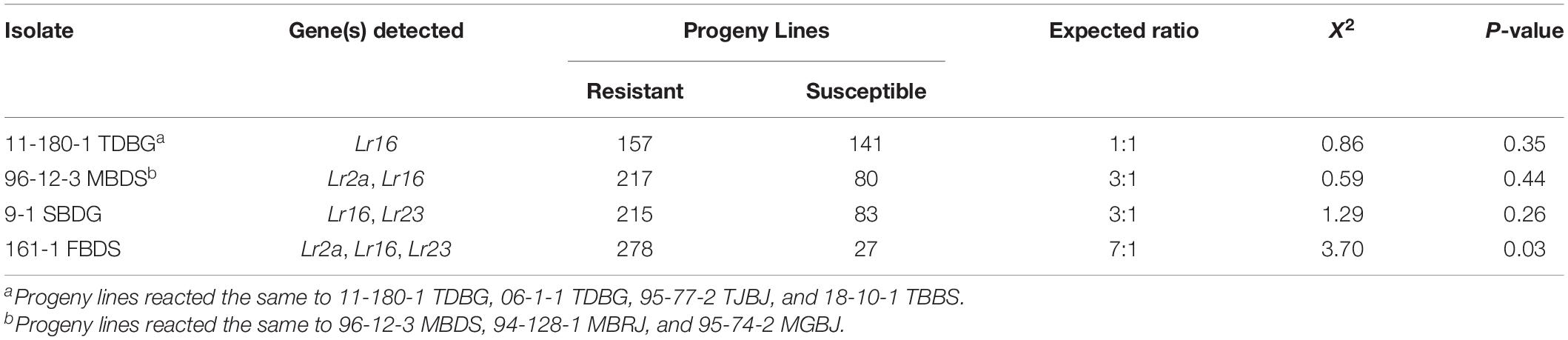
Table 2. Reaction of the Carberry/Thatcher population against P. triticina isolates at the seedling stage.
When 96-12-3 MBDS, 94-128-1 MBRJ, and 95-74-2 MGBJ were inoculated onto the progeny lines, the segregation ratios were consistent with two effective resistance genes, Lr16 and a second resistance gene thought to be Lr2a since it was effective against isolates avirulent to Lr2a and ineffective to isolates virulent to Lr2a (Table 2). This second gene had a very resistant infection type consistent with Lr2a (Table 1) which made the determination of the presence or absence of this gene possible in the presence of Lr16. All the progeny lines were scored for the presence or absence of this resistance gene and it was mapped as a QTL for seedling leaf rust resistance to the Lr2 region of chromosome 2DS.
The segregation ratio of the progeny lines to 9-1 SBDG also indicated the presence of two resistance genes, one of which was Lr16; however, this isolate is virulent to Lr2a, so a third seedling resistance gene was also present. This third resistance gene was ineffective against all the isolates tested except 9-1 SBDG and 161-FBDS, which were the only isolates avirulent to Lr23. When progeny lines were scored for the presence or absence of this resistance gene, the results also mapped to chromosome 2BS and is thought to be Lr23, which was reported to be present in Alsen, one of the parents of Carberry (Oelke and Kolmer, 2005). The population appeared to segregate three resistance genes to 161-1 FBDS, Lr16, Lr2a, and Lr23, fitting both a three and four gene ratio. This third gene (Lr23) also appeared to have some effect on 1-1 BBBD but this reaction could not be determined as consistently as it could when these lines were inoculated with 9-1 SBDG or 161-FBDS.
The seedling resistance gene, Lr10 was reported to be present in both the parents of Carberry (Oelke and Kolmer, 2005; McCallum and Seto-Goh, 2010). When isolates, 08-5-1 TDBB and 19-129-1 TBTN, both of which were avirulent on Lr10, were inoculated onto the progeny, the reactions were the same as to 11-180-1 TDBG, 06-1-1 TDBG, 95-77-2 TJBJ, and 18-10-1 TBBS, indicating the presence of Lr16, but with no additional resistance gene. From this, it appears that Lr10 is not present in Carberry.
Determination of Leaf Rust Resistance in Superb and Alsen, the Parental Lines for Carberry
When plants of the parental lines of Carberry (Superb and Alsen) were inoculated with the isolate 20-140-1 TBRD, which was virulent to Lr2a and Lr23 but avirulent to Lr10 and Lr16, all 16 Superb plants were resistant with a consistent infection type of “11+ 2−“ characteristic of Lr10. The Alsen plants, however, varied in their response to this isolate with 14 resistant plants and two susceptible plants. This demonstrates that at least some plants of Alsen did not have Lr10. When a second set of plants was inoculated with the isolate, 19-123-2 TBGJ, which was virulent to Lr2a, Lr23, and Lr10, but avirulent to Lr16, all the Superb plants were uniformly susceptible, but the Alsen plants again varied for their infection types with nine resistant and seven susceptible plants.
When these same plants were marker tested, marker alleles corresponded with resistance to races, 20-140-1 TBRD and 19-123-2 TBGJ. The data showed that Carberry carries Lr16, Superb lacks Lr16, and Alsen is heterogeneous for Lr16. The presence of Lr16 in the Alsen plants corresponded with a resistant infection type when inoculated with the P. triticina isolate, 19-123-2 TBGJ. The phenotypic and genotypic data confirm that Carberry carries Lr16 inherited from Alsen (Supplementary Table 1).
Lr13 Adult Plant Resistance Evaluation
When adult plants of Carberry, Thatcher, and each line in the CT population were inoculated at the adult plant stage with each of the isolates, 16-284-1 TGBQ and 17-358-1 TBBJ, Carberry was resistant and Thatcher was susceptible. The progeny lines segregated for resistance with similar reactions to both isolates. All progeny lines with Lr16 were resistant to both the isolates. Even though 16-284-1 TGBQ is classified as virulent on Lr16, the reaction is intermediate, and host plants were more resistant at the adult plant stage than at the seedling stage. For the progeny lines without Lr16, 50 were resistant and 87 were susceptible, which indicated the presence of another resistance gene, likely Lr13, but the results did not fit a single gene segregation ratio. Even though both Lr34 and Lr46 were present in this population, they did not affect the detection of Lr13 since plants with Lr13 had uniform infection types of “1” or ‘1–2,” whereas Lr34 and Lr46 produce infection types with mixtures of pustule types, including “3” or susceptible pustules.
Hybrid Necrosis Test to Determine the Presence of Lr13 in Carberry
Seedlings of the parental lines of Carberry, Thatcher, Thatcher-Lr13, and Kubanka grew normally with healthy green leaves all the way to early tillering. Similarly, hybrids between Thatcher/Thatcher-Lr13 DH lines that lacked Lr13 and Kubanka showed the same healthy growth pattern. Hybrids between Carberry and Kubanka and between Thatcher/Thatcher-Lr13 DH lines carrying Lr13 and Kubanka showed strong progressive necrosis (Figure 1). Necrosis of the first leaf became evident once the third leaf had fully emerged.

Figure 1. Seedlings of Kubanka, Carberry, Thatcher Lr13 (Tc-Lr13) and F1 of Carberry × Kubanka, Tc-Lr13 × Kubanka and Thatcher (Tc) × Kubanka. Hybrid necrosis in the Carberry × Kubanka F1 generation lines confirms the presence of Lr13 in Carberry.
Disease Evaluation in the Field
The resistant parent Carberry conferred high resistance to leaf rust with low disease severity (DS) and IR ranging from R to MR across environments, whereas the susceptible parent Thatcher had high DS and IR ranging from MSS to S (Table 3). The disease severity of the CT population ranged from 0.0 to 90% across environments at Morden and from 0.5 to 80% across environments at Swift Current. Although Swift Current across years had lower disease scores than Morden, the disease pressure observed in each environment was sufficient for discriminating among the lines and mapping loci associated with quantitative resistance. A wide range of heritability values of disease response was observed in the population (Table 3). Broad sense heritability of DS ranged from 0.25 to 0.63, and of IR from 0.19 to 0.54 across environments. Furthermore, narrow sense heritability ranged from 0.22 to 0.61 for the DS and from 0.18 to 0.51 for the IR.
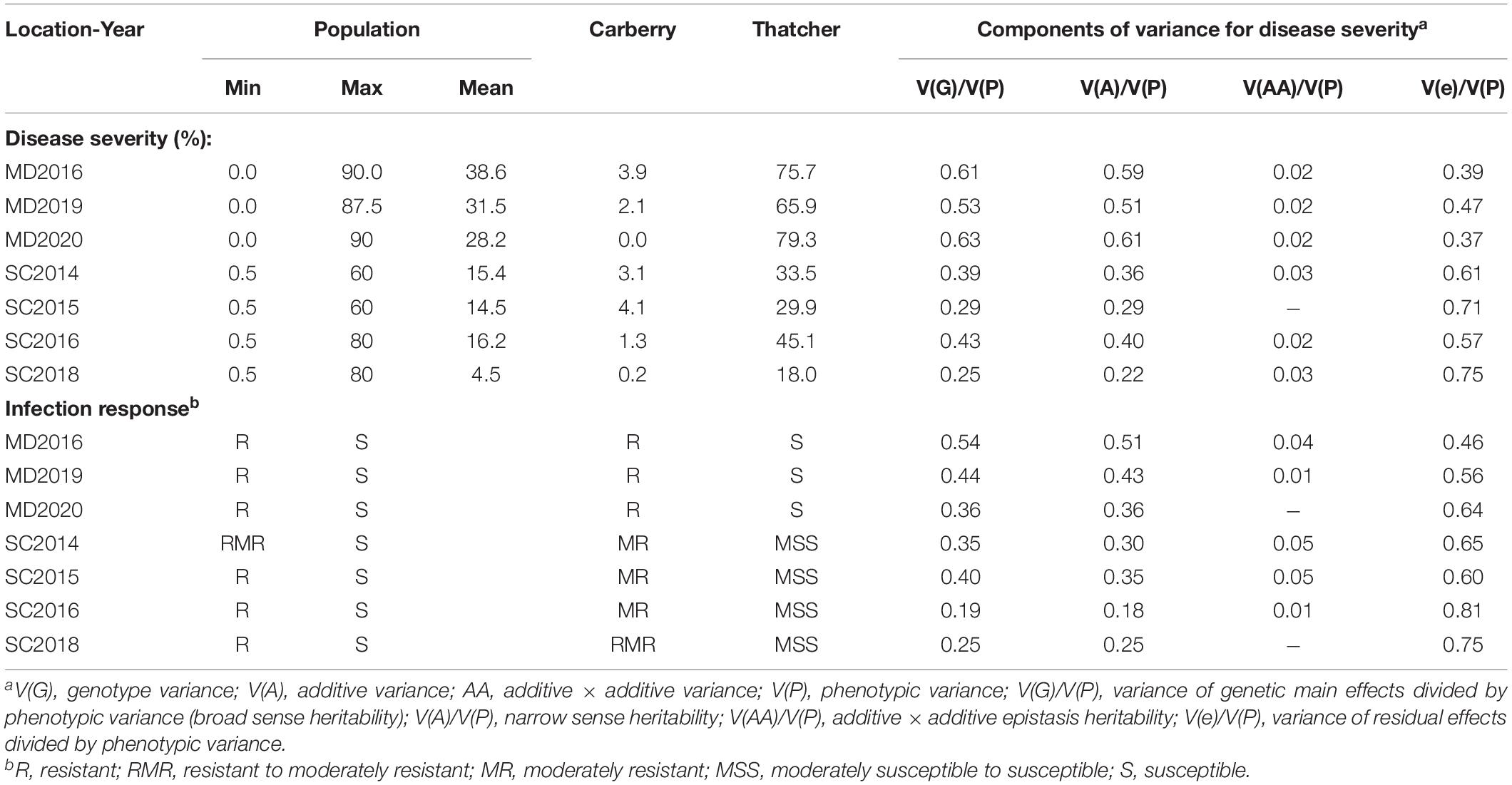
Table 3. Mean and range of the leaf rust severity and infection response scores for the Carberry/Thatcher doubled haploid (DH) population and parents and heritabilities from field nurseries near Morden (MD), MB for 3 years, and Swift Current (SC), SK for 5 years.
Except at Morden in 2016 and 2019, which displayed bimodal distributions, the DS of the population was continuous with a preponderance of lines showing low-disease scores (Figure 2). When the most resistant (0 to <20%) and susceptible (>60 to <90%) portions of the distribution tails for leaf rust severity were considered, markers showed a disproportionate representation for Lr34 for the Morden 2019 and 2016 environments. In the Morden 2019 environment, out of 120 lines in the resistant mode, 99 of them carried the Lr34 resistance allele compared with 21 of the lines which did not carry the resistance allele. In the susceptible mode, only 6 out of 39 lines possessed the Lr34 resistance allele. The same trend was observed in the Morden 2016 environment in which 84 out of 103 lines in the resistant mode had the Lr34 resistance allele, while 19 lines did not. In the susceptible mode, from a total of 39 lines, only 6 lines had the Lr34 resistance allele. Correlation coefficients (Table 4) among environments for disease severity were highly significant and ranged from moderate to high (r = 0.54 to 0.90, P < 0.0001).
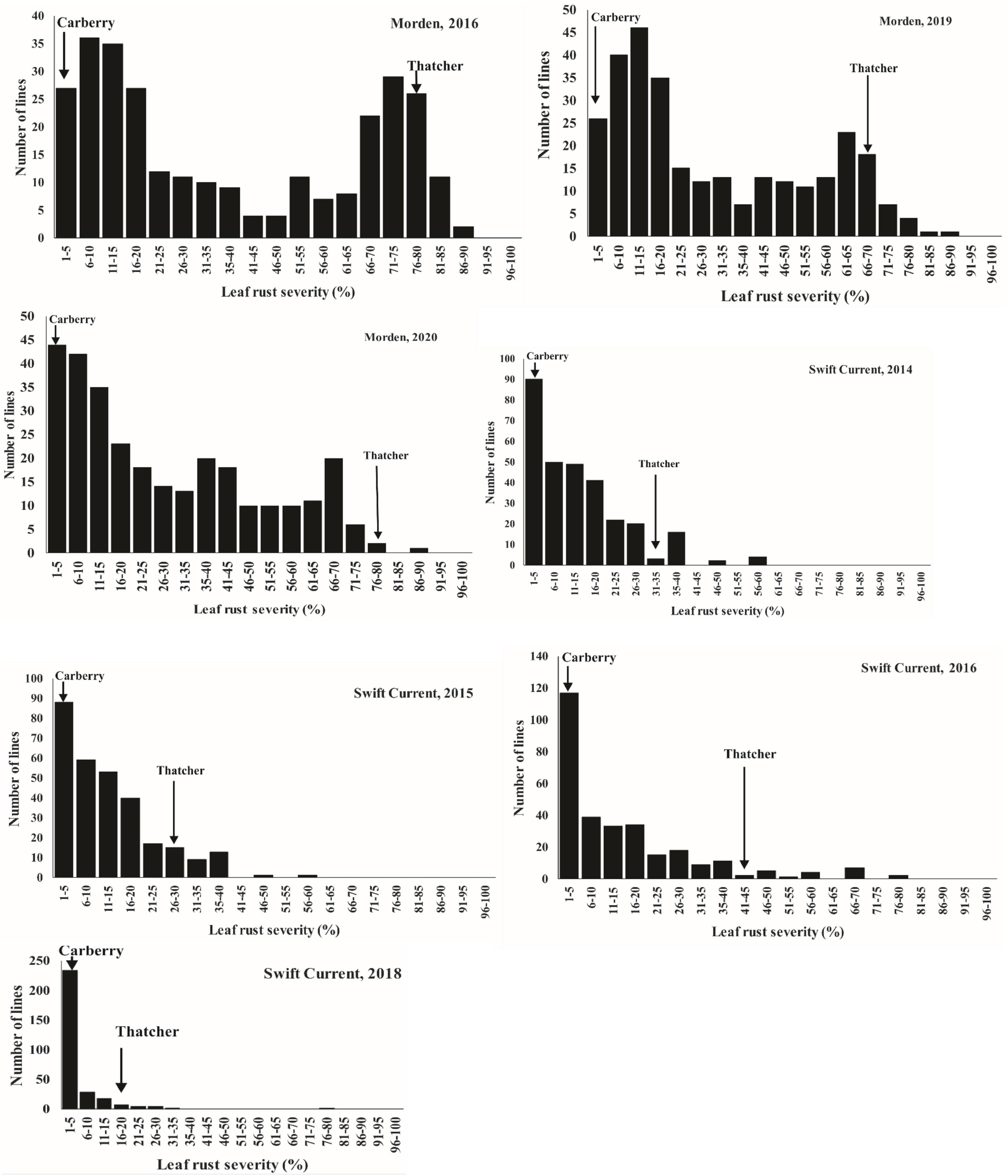
Figure 2. Distributions of leaf rust severity for Carberry/Thatcher lines evaluated over multiple years in Morden, MB and Swift Current, SK locations. Arrows indicate the severity of the parents.
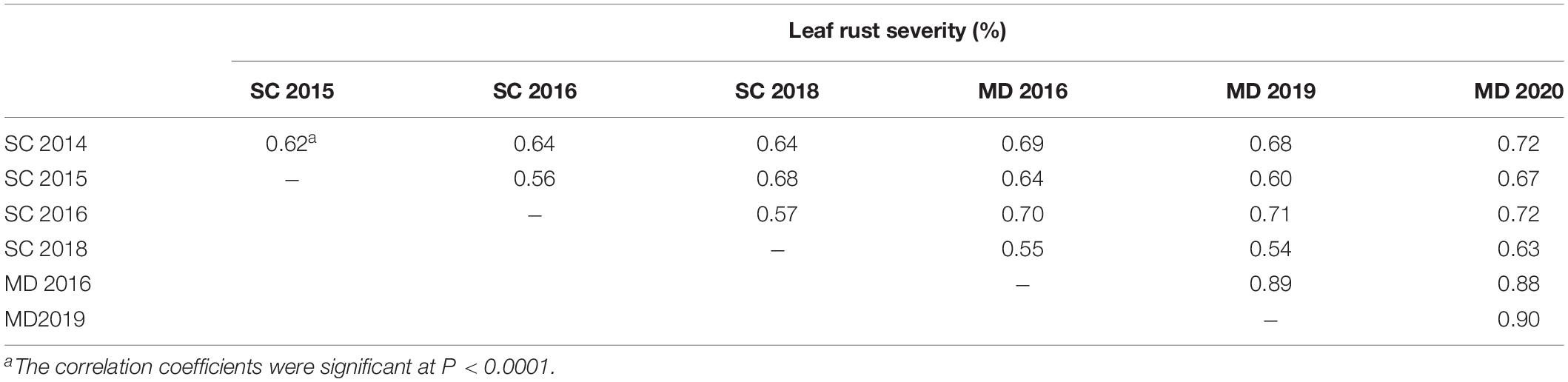
Table 4. Pearson correlation coefficients (r) of the relation between leaf rust severities of the DH lines of the Carberry/Thatcher population evaluated near Morden (MD) and Swift Current (SC) from 2014 to 2020.
Linkage Map
The genetic map of the CT population consisted of 8,360 polymorphic SNP markers (Supplementary Table 2). The map covered 3645.8 cM of the wheat genome, corresponding to an average density of 0.44 cM per marker. All of the 21 wheat chromosomes, except 4D, were represented in 28 linkage groups.
Leaf Rust Resistance Quantitative Trait Loci Identified
Overall, nine QTL associated with field data were detected on chromosomes, 1B (designated QLr.spa-1B), 2B [2 loci (QLr.spa-2B.1; QLr.spa-2B.2)], 2D (QLr.spa-2D), 4A (QLr.spa-4A), 4B (QLr.spa-4B), 5A (QLr.spa-5A), 5B (QLr.spa-5B), and 7D (QLr.spa-7D) (Table 5 and Figure 3). Besides their effectiveness at the adult plant stage, QLr.spa-2B.1 and QLr.spa-2D were significant at the seedling stage. Four of the QTL located on 1B, 2B (2 loci), and 7D were detected in multiple environments, whereas the remaining were detected in one or a maximum of two out of seven test environments. Carberry contributed to the desirable alleles at all the detected loci, and no QTL was obtained from Thatcher. Based on the position of QTL- associated markers in a hexaploid wheat high density 90K SNP map by Wang et al. (2014), the identified QTLs were assigned with the chromosome arms, 1BL, 2BS (2 loci), 2DS, 4AS, 4BS, 5AL, 5BS, and 7DS.
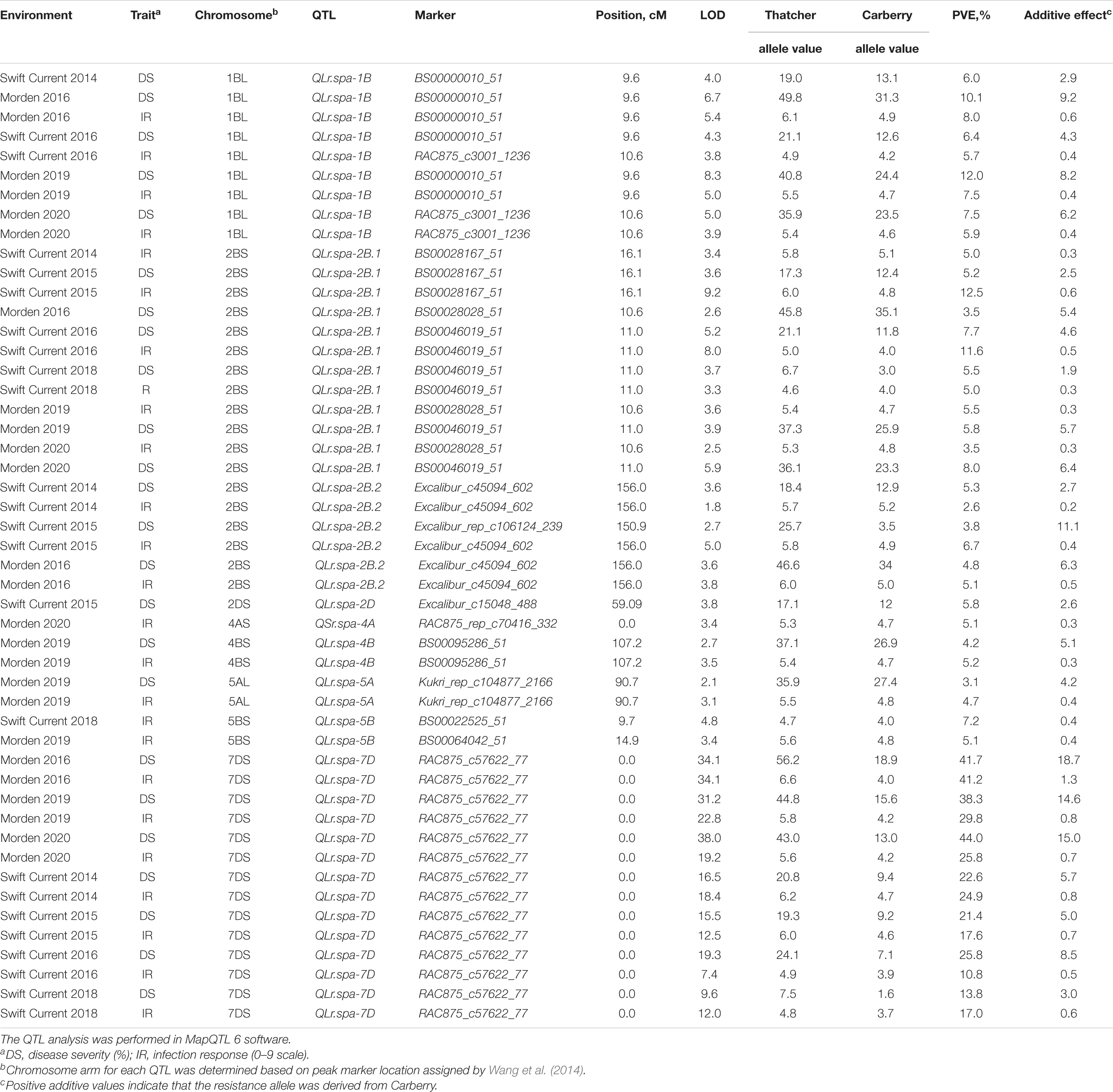
Table 5. Quantitative trait loci (QTL), their chromosome arm location, peak associated marker and LOD, position on the chromosome in centiMorgans (cM), phenotypic value associated with the parental type allele, percent of phenotypic variation explained (PVE%), and additive effect associated with response to leaf rust disease severity and infection response detected in the Carberry/Thatcher population evaluated in field nurseries near Morden, MB, and Swift Current, SK.
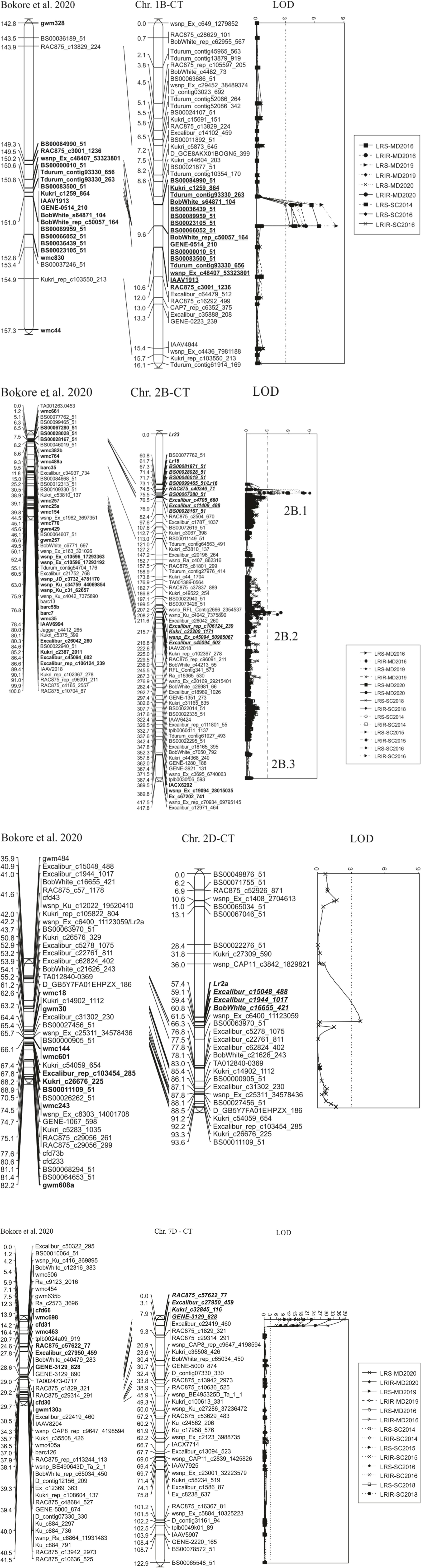
Figure 3. Quantitative trait loci (QTL) for leaf rust resistance identified in Carberry/Thatcher (CT) population evaluated in seven field environments at the adult plant stage near Morden (MD), MB and Swift Current (SC), SK, Canada. QTL detected in the population on chromosome 4A, 4B, and 5B were not presented in graphs. Note that 2B.1 stands for QLr.spa-2B.1, 2B.2 for QLr.spa-2B.2, and 2B.3 for QLr.spa-2B.3. QLr.spa-2B.3 was not detected by MapQTL and QTLNetwork as the main effect QTL, but it was revealed by epistasis analysis interacting with QLr.spa-2D. Disease traits, leaf rust severity (LRS) and leaf rust infection response (LRIR), and test locations Morden (MD) and Swift Current (SC) and the year of field evaluation. The Carberry/Thatcher (CT) population map was aligned with a hexaploid high density consensus map published by Bokore et al. (2020).
The QLr.spa-1B accounted for the percent of phenotypic variation explained (PVE) of up to 12.0% for DS and 8% PVE for IR and was associated with peak markers BS00000010_51 and RAC875_c3001_1236, and with a maximum LOD of 8.3 (Table 5). QLr.spa-1B was consistently detected in five of the seven environments. The QLr.spa-2B.1 QTL was detected in all test environments compared to QLr.spa-2B.2 which was detected in three out of seven environments. Based on the position of QTL-associated markers in the CT population linkage map, QLr.spa-2B.1 and QLr.spa-2B.2 were about 145.0 cM distance from each other. QLr.spa-2B.1 accounted for a PVE of 3.5 to 12.5% for DS and IR, whereas QLr.spa-2B.2 accounted for 2.6 to 6.7% PVE for both traits. A QTL analysis performed on the seedling data of avirulent races against Lr16 (11-180-1 TDBG, 06-1-1 TDBG, 95-77-2 TJBJ, and 18-10-1 TBBS) produced a significant QTL at the same location as QLr.spa-2B.1 identified with field data.
QLr.spa-7D was a major QTL detected at all environments and was associated with the peak marker, RAC875_c57622_77 with LOD values ranging from 7.4 to 28.0. The QTL accounted for PVE values of up to 44% for DS and 30% for IR. Minor QTL, such as QLr.spa-2D, QLr.spa-4B, and QLr.spa-5A were confined to single environments. QLr.spa-4B and QLr.spa-5A were significant for both DS and IR, whereas QLr.spa-2D was associated only with DS. A QTL from the analysis of phenotypic data of Lr2a avirulent races, 96-12-3 MBDS, 94-128-1 MBRJ, and 95-74-2 MGBJ coincided with QLr.spa-2D. The other minor QTL, QLr.spa-5B was detected at two environments associated with IR, but not with severity.
Epistatic Effect Quantitative Trait Loci
A total of five additive by additive epistatic interactions were detected by QTLNetwork analysis (Table 6). The interactions involved QTL that was also identified by MapQTL and an additional QTL on chromosome 2B, designated as QLr.spa-2B.3, which was not identified by MapQTL. Of the five interactions detected, positive epistatic effects with an enhanced level of disease resistance were observed between QLr.spa-1B (Lr46) and QLr.spa-7D (Lr34), QLr.spa-2B.1 (Lr16) and QLr.spa-7D (Lr34), and QLr.spa-2D (Lr2a) and QLr.spa-7D (Lr34). The QLr.spa-7D (Lr34) QTL showed the greatest epistatic effects with QLr.spa-1B (Lr46) and QLr.spa-2B.1 (Lr16) and less so with QLr.spa-2D (Lr2a). Negative epistatic effects were detected between QLr.spa-1B (Lr46) and QLr.spa-2B.1 (Lr16), and QLr.spa-2B.3 and QLr.spa-2D (Lr2a). The negative association between the alleles at QLr.spa-1B (Lr46) and QLr.spa-2B.1 (Lr16), revealed that the Lr16 interaction with Lr46 did not act as synergistically as expected. Furthermore, the heritability values for the additive × additive epistasis were very low from 0.01 to 0.05 (Table 6). The most common interaction was between QLr.spa-1B and QLr.spa-7D.
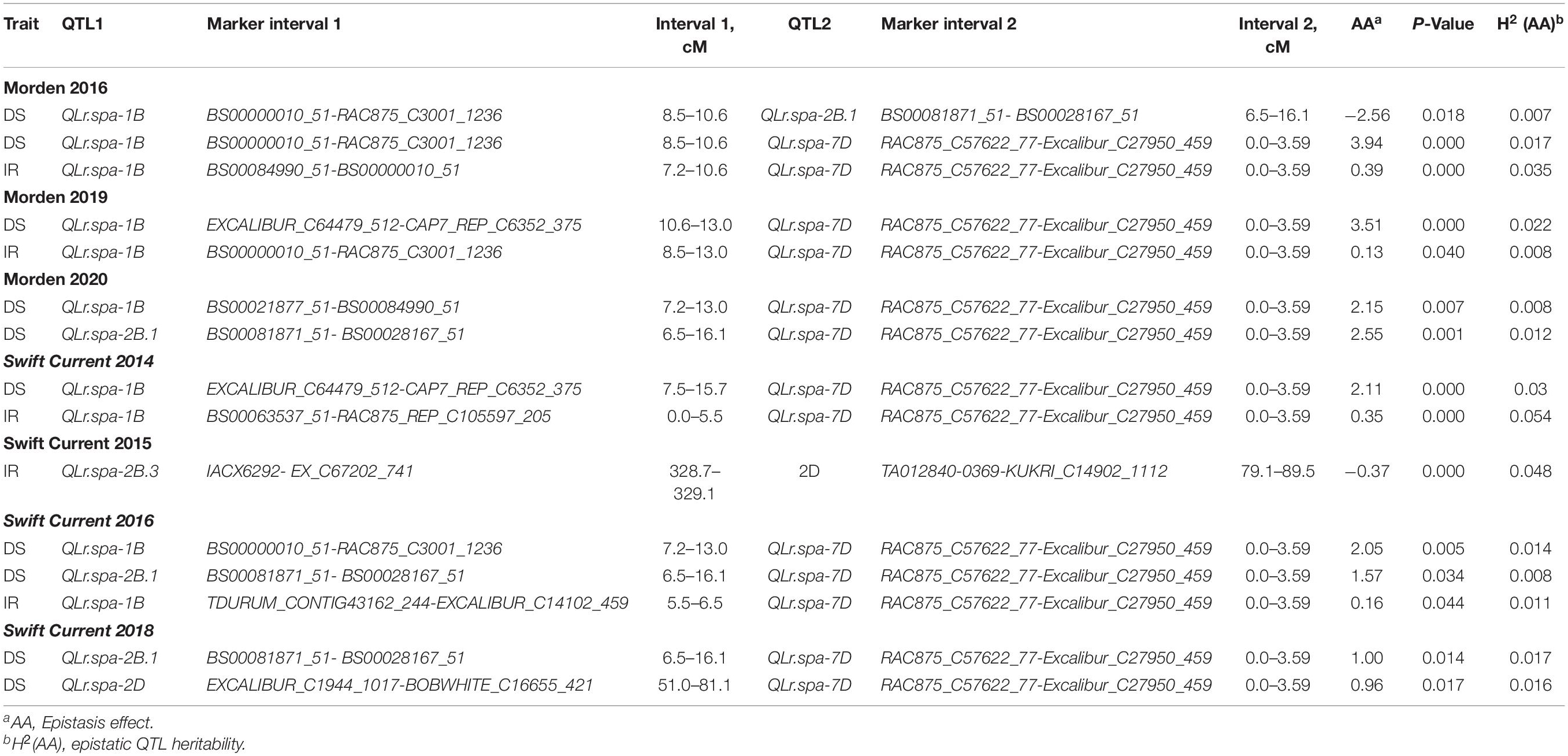
Table 6. Epistatic interactions between Carberry leaf rust resistance genes detected by QTLNetwork in the Carberry/Thatcher population evaluated at Morden, MB for 3 years and Swift Current, SK for 4 years.
The ANOVA results for gene combination analysis using SAS aimed at investigating the effects of various gene combinations can be seen in Figure 4 and Supplementary Table 3. The results showed that the association of the Lr34 gene with Lr46, Lr16, or Lr2a generally increased the level of disease resistance. At the Morden location in both 2019 and 2020, the effect of Lr2a or Lr16 alone was marginal, whereas Lr46 was moderately effective and Lr34 was more effective (Figure 4). When Lr46 and Lr34 were combined in the same lines, these had a very good level of resistance, particularly when combined with Lr2a and/or Lr16.
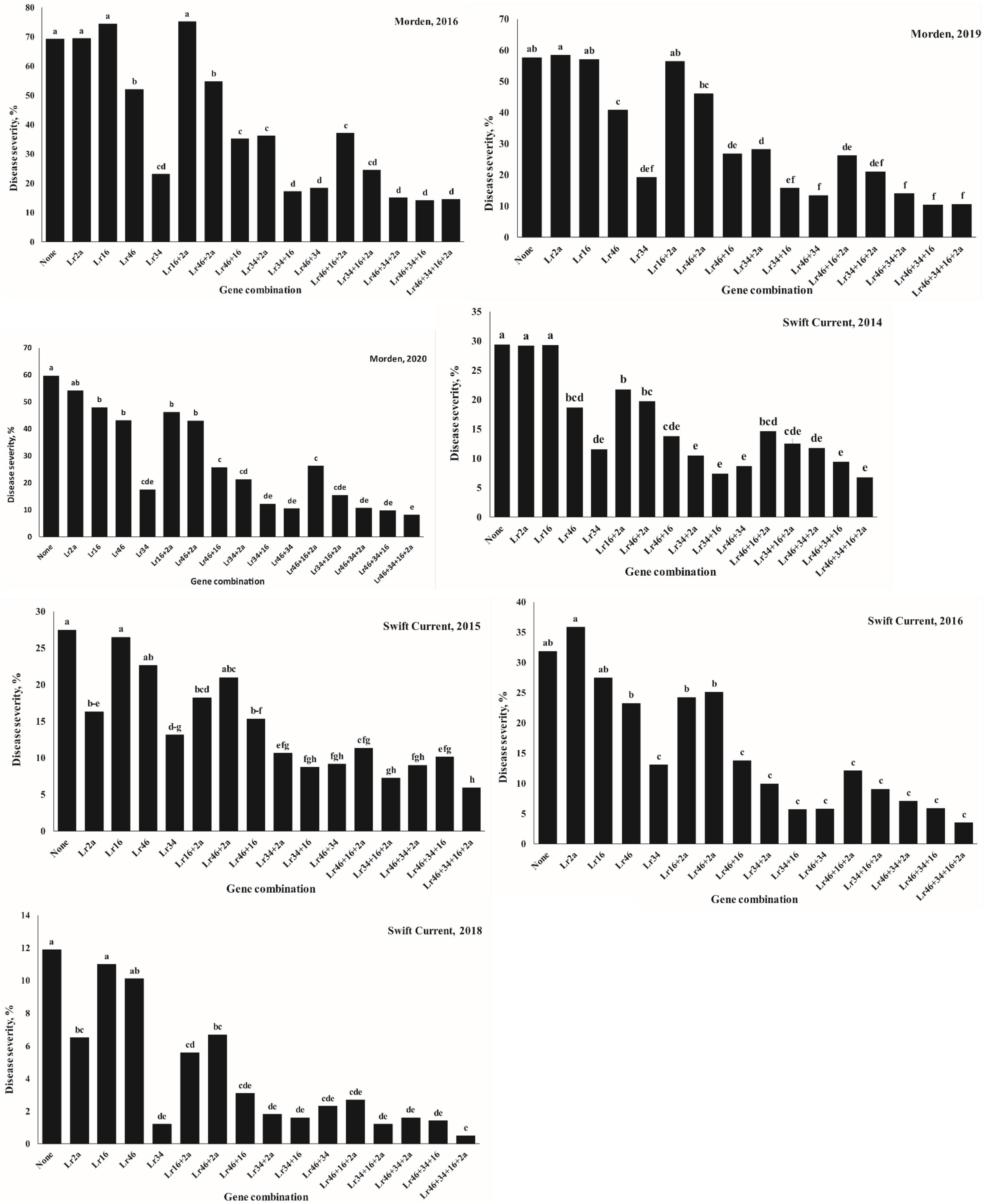
Figure 4. Effects of gene combinations on adult plant leaf rust response in the Carberry/Thatcher population evaluated near Morden, AB and Swift Current, SK in different years.
Discussion
The variation from moderate to high correlation values observed in the leaf rust severity was consistent with the plotted distributions of the CT population across environments and suggested a partial differential response to the environment. These results were suggestive of a leaf rust resistance complex of genes in Carberry, which is supported by low to moderate heritability values. Genetic analysis confirmed the complex nature of leaf rust resistance in Carberry that has been expressed at a high level since its commercial release with the identification of three seedling genes (Lr2a, Lr16, Lr23) and three APR genes (Lr13, Lr34, Lr46). We also detected minor QTL on chromosomes, 2B, 4A, 4B, 5A, and 5B that contributed to Carberry’s leaf rust resistance. The analysis of two-way epistatic effects revealed interactions between Lr34 and Lr46, and Lr34 and Lr16, which significantly boosted the resistance of Carberry.
The roughly bimodal distributions of the population observed at Morden 2016 and 2019, could be due to the high expression of the 7D QTL (Lr34) behaving like a qualitative gene explaining up to 42% of the phenotypic variation in 2016, and 38% in 2019, and low or insignificant expression by other Carberry genes/QTL. The first mode of the DH lines expressed a low-disease severity similar to the resistant parent, Carberry, whereas the other group had high-disease severity which was similar to the susceptible parent, Thatcher. Besides having a large effect by itself, Lr34 also interacted with other genes forming two large groups, those with Lr34 and those without. However, there were also DH lines that could not be categorized into Carberry type or Thatcher type due to the presence of the minor effect QTL which was segregating in the population. For example, besides the 7D QTL, three other QTL with a cumulative phenotypic effect of 18% segregated in 2016, and four QTL which cumulatively explained 25% of the phenotypic variation segregated in 2019 (Table 5).
Of the two resistance QTL detected using the field data on the chromosome arm 2BS, QLr.spa-2B.1 corresponded with Lr16. The consistent expression of Lr16 in most of the field tests in the current study was irrespective of the presence of races partially virulent on the gene in Canada (Samborski, 1984; McCallum et al., 2016, 2020). Predictive markers for Lr16, such as SSR markers, wmc661, wmc764, and gwm210 were located nearby QLr.spa-2B.1 in a high-density consensus map published by Bokore et al. (2020), while SNP marker, BS00099465_51 mapped within the QLr.spa-2B.1 QTL interval (Kassa et al., 2017; Figure 3). The presence of Lr16 in Carberry was puzzling based on the gene determinations in the parental lines, Alsen and Superb, which did not detect Lr16 in either of the parents, Superb (McCallum and Seto-Goh, 2010) and Alsen (Oelke and Kolmer, 2005). However, our seedling leaf rust assays and the use of predictive KASP markers supported the QTL analysis that Carberry carries Lr16. The puzzle was solved with the testing of multiple parental plants and although all the Superb plants tested were negative for Lr16, the Alsen plants tested were heterogeneous for Lr16. Thus, Carberry inherited Lr16 from Alsen. Bokore et al. (2020) mapped the Lr16 in Carberry using different mapping populations. Many other Canadian wheat varieties are known to have Lr16 (McCartney et al., 2005; McCallum et al., 2016; Kassa et al., 2017; Toth et al., 2018).
Both of the parents of Carberry, Superb and Alsen, were reported to have the seedling leaf rust resistance gene, Lr10 (Oelke and Kolmer, 2005; McCallum and Seto-Goh, 2010). However, no QTL was detected for Lr10, and seedling testing with two Lr10 avirulent isolates indicated that the population did not segregate for Lr10 and that Carberry did not have Lr10. When seed stocks of the parents that were used to make Carberry were tested, Superb appeared to be uniformly resistant to an Lr10 avirulent isolate, but Alsen had a small proportion of susceptible plants demonstrating that these plants did not have Lr10. Therefore, it is likely that the Alsen plant that was crossed to produce Carberry had Lr16 but lacked Lr10.
The detection of the third 2BS gene, Lr23, with the seedling test, but not in field trials, suggested that the ineffectiveness of the gene in the field was attributed to virulent races or a weak expressivity of the gene on adult plants. The frequency of virulence to Lr23 varied from 37.5 to 56.1% from 2015–2019 in Canada (McCallum et al., 2021). Oelke and Kolmer (2005) reported one of Carberry’s parents, Alsen possesses Lr23. McIntosh and Dyck (1975) indicated the presence of an unknown gene in Thatcher that inhibits the expression of Lr23 under Canadian field conditions, and partially in Australian conditions. Originally believed to be derived from a durum wheat cultivar Gaza, Lr23 was later introgressed into hexaploid wheat (McIntosh and Dyck, 1975).
A third seedling gene identified in Carberry, Lr2a was determined by the seedling test and revealed by field data. Carberry was expected to carry Lr2a as both its parents, Alsen (Oelke and Kolmer, 2005) and Superb (McCallum and Seto-Goh, 2010) are reported to possess this gene. Lr2a is present in a wide range of North American wheat germplasm (Oelke and Kolmer, 2004; McCallum et al., 2016). Lr2a which corresponded with the field QTL, QLr.spa-2D showed a minor effect on DS and it was significant only in one environment. The effectiveness of the Lr2a gene in only one out of seven environments is consistent with the frequency of isolates virulent on the gene in the P. triticina population in western Canada dramatically keep increasing after the year 2000 (McCallum and Seto-Goh, 2010; McCallum et al., 2020). As the gene sometimes synergistically interacts with Lr34, it could still be valuable in resistance breeding.
Superb was reported not to have any APR genes (McCallum and Seto-Goh, 2010), and Alsen was reported to have Lr13 and Lr34 (Oelke and Kolmer, 2005). However, the current analysis demonstrated that Lr46 was an important component of the resistance in Carberry. Carberry’s QLr.spa-1B QTL is the same as the slow rusting or APR and the pleiotropic gene, Lr46, located on the chromosome arm 1BL (Singh et al., 1998; William et al., 2003; Lillemo et al., 2013). The QTL at QLr.spa-1B was also effective against stem rust (data not shown). Studies are limited in showing the presence of Lr46 in Canadian wheat germplasm. It was recently demonstrated that Superb has Lr46 (Lewarne, 2021), which would have been the donor parent for Lr46 in Carberry. Furthermore, recently published studies indicated a QTL on 1BL that was associated with multiple disease resistance in Carberry and Vesper that could be Lr46 (Bokore et al., 2017, Bokore et al., 2020). The detection of the QLr.spa-1B (Lr46) in five out of seven environments with the current study indicates the importance of this gene in resistance breeding.
The QLr.spa-7D QTL on 7DS represented the major slow rusting pleiotropic gene Lr34. As opposed to Lr46, Lr34 is common in many wheat varieties in Canada (McCallum and DePauw, 2008; McCallum et al., 2012, 2016) and several other countries (Singh et al., 2011; Lillemo et al., 2013). The presence of the Lr34 gene in Carberry has been documented in various reports (McCallum et al., 2012; Randhawa et al., 2013; Bokore et al., 2020), and it was transferred to Carberry from Alsen (Oelke and Kolmer, 2005).
The QTL on chromosome 2B, QLr.spa-2B.2, corresponding with APR gene Lr13. The detection of this gene in Carberry was expected as one of its parents, Alsen, possesses the gene (Oelke and Kolmer, 2005). Lr13 linked markers, Excalibur_c26042_260 and wsnp_Ku_c4042_7375890, reported by Zhang et al. (2016), were placed within 5.15 and 8.58 cM distance from the QLr.spa-2B.2 Carberry QTL peak marker, Excalibur_c45094_602, on the genetic map of the CT population (Figure 3). The gene is generally effective at the early postseedling stage (Dyck et al., 1966; Zhang et al., 2016). Lr13 is a recessive gene as described by Dyck et al. (1966) in a study made using a cross of Thatcher/Manitou, the Canadian variety Manitou, possessing the gene, was originally transferred from Frontana in which it behaved partially dominant. This gene has been widely deployed in the Canadian and American wheat germplasm (McCallum et al., 2016), including Alsen, the immediate parent of Carberry’s immediate parent Alsen (Oelke and Kolmer, 2005). Zhang et al. (2016) reported that Lr13 is the same gene as Ne2m, a gene known to govern hybrid necrosis in wheat. In indoor adult plant testing, we established the presence of Lr13 in the progeny of the population of Carberry/Thatcher. Given that hybrids between Carberry and Kubanka (Ne1 carrier) exhibited progressive necrosis, the presence of Lr13 in Carberry was confirmed. The gene, Lr13 is an example of an APR gene that has a race-specific response to leaf rust.
Carberry additionally has genes with relatively small effects on disease resistance identified on the chromosome arms, 4AS, 4BS, 5AL, and 5BS, which were significant in only one or a maximum of two out of seven environments. Despite the detection of several QTL in the resistant parent Carberry, we found that no QTL was contributed by the susceptible parent, Thatcher. The minor genes identified in Carberry may play a cumulative role in the resistance by acting in the synergy that is at a level that we could not detect in our epistasis analysis. Some of these minor QTLs were found to be close to genomic regions that are associated with other rust species on a hexaploid wheat consensus map (Bokore et al., 2020), making them useful regions to consider in resistance breeding. For example, the 4AS leaf rust QTL marker RAC875_rep_c70416_332 was placed only at 0.25 cM proximal to BobWhite_c20163_456, a marker for stripe rust resistance similarly reported in Carberry (Bokore et al., 2020). Additionally, BS00095286_51 associated with the 4BS leaf rust QTL was located at 31.5 cM proximal to wmc617, an SSR marker for resistance to stem rust race Ug99 in Carberry (Singh et al., 2013) and 37.2 cM proximal to Tdurum_contig27799_114, a marker for a stripe rust resistance QTL which was similarly reported in Carberry (Bokore et al., 2020).
Understanding the epistatic genetic effects of multiple leaf rust resistance genes is useful to develop wheat varieties with appropriate gene combinations and durable resistance. Results of the present study demonstrated that the strength of Carberry’s years of leaf rust resistance in the farmer’s fields in Canada may, in part, be attributable to the synergistic additive by additive epistatic effects of Lr34 with Lr46, Lr16, and/or Lr2a. For over 100 years, Lr34 has remained effective against the leaf rust pathogen. Lr34 has been cloned and encoded an ABC transporter protein unlike cloned all stage resistance genes, but how this protein confers resistance to rust pathogen is not known (Krattinger et al., 2009). Singh et al. (2011) indicated that gene combinations could minimize the development of virulent races on race-specific resistance genes. Also, wheat varieties having Lr34 in combination with other genes are more durable compared with varieties that lack Lr34 (Oelke and Kolmer, 2004; Singh et al., 2006, 2011). Singh et al. (2011) reported that combinations of 4 to 5 APR genes usually result in “near immunity” or high level resistance.
In conclusion, since its registration in Canada in 2009, the hexaploid spring wheat cultivar, Carberry, was grown over an extensive area in Canada for several years and maintained a high level of leaf rust (Puccinia triticina Eriks.) resistance. The present study characterized the genetic basis of what has turned out to be durable resistance in Carberry through several race-specific and non-specific race-resistance genes. Using the adult plant leaf rust response data, we identified nine QTLs located on chromosomes 1B, 2B.1, 2B.2, 2D, 4A, 4B, 5A, 5B, and 7D some of which represented previously documented genes. For example, the resistance on 1B corresponded with Lr46, 7D with Lr34, one of the QTL on 2B with Lr13, the other QTL on 2B with Lr16, and 2D with Lr2a. In addition to field studies, our seedling tests revealed race-specific genes, Lr2a, Lr16, and Lr23. Although Lr2a and Lr16 were also revealed using adult plant response data, Lr23 did not show any significant effect in adult plants due to the presence of gene-specific virulent races in recent years. Synergistic epistatic effects were revealed for Lr34 with Lr46, Lr16, or Lr2a with a combination of these genes contributing to higher resistance. Single events of negative interactions were detected between Lr16 and Lr46, and between Lr2a and an unknown QTL on chromosome 2B. However, these gene combinations had reduced the disease symptoms compared to each gene alone. Generally, the durability of leaf rust resistance in Carberry could be attributed to Lr34 and Lr46, because the resistance has remained effective even though the P. triticina population has evolved virulence to Lr2a, Lr13, Lr16, and Lr23.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
RK, RC, and RD developed the mapping population. RK, RC, BDM, CH, and FB conceived, designed, and supervised the study. BDM, FB, RK, and RC performed the field trials and phenotyping. BDM performed seedling resistance trials. CH performed the hybrid necrosis test and predictive marker tests. FB, BM, CP, and AN’D performed genotyping, genetic data mining, and genetic mapping. FB analyzed the data and drafted the manuscript. FB, BDM, RK, CH, RC, and RD revised the manuscript. All authors reviewed the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are grateful for the financial support of AAFC, the Canadian Wheat Research Coalition through the Canadian Agricultural Partnership AgriScience Program, and CTAG2 through Genome Canada with the additional support of the Western Grains Research Foundation, Government of Saskatchewan, Saskatchewan Wheat Development Commission, Alberta Wheat Commission, Manitoba Agriculture, DuPont Pioneer, Viterra, Manitoba Wheat and Barley Growers Association, and SeCan.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.775383/full#supplementary-material
Footnotes
References
Aboukhaddour, R., Fetch, T., McCallum, B. D., Harding, M. W., Beres, B. L., and Graf, R. J. (2020). Wheat diseases on the prairies: A Canadian story. Plant Pathol. 69, 418–432. doi: 10.1111/ppa.13147
Bokore, F. E., Cuthbert, R. D., Knox, R. E., Randhawa, H. S., Hiebert, C., DePauw, W., et al. (2017). Quantitative trait loci for resistance to stripe rust of wheat revealed using global field nurseries and opportunities for stacking resistance genes. Theor. Appl. Genet. 130, 2617–2635. doi: 10.1007/s00122-017-2980-7
Bokore, F. E., Knox, R. E., Cuthbert, R. D., Pozniak, C. J., McCallum, B. D., and N’Diaye (2020). Mapping quantitative trait loci associated with leaf rust resistance in five spring wheat populations using single nucleotide polymorphism markers. PLoS One 15:e0230855. doi: 10.1371/journal.pone.0230855
Cavanagh, C. R., Chao, S., Wang, S., Huang, B. E., Stephen, S., Kiani, S., et al. (2013). Genome-wide comparative diversity uncovers multiple targets of selection for improvement in hexaploid wheat landraces and cultivars. Proc. Natl. Acad. Sci. USA 110, 8057–8062. doi: 10.1073/pnas.1217133110
da Silva, G. B. P., Zanella, C. M., Martinelli, J. A., Chaves, M. S., Hiebert, C. W., McCallum, B. D., et al. (2018). Quantitative trait loci conferring leaf rust resistance in hexaploid wheat. Phytopathology 108, 1344–1354. doi: 10.1094/phyto-06-18-0208-rvw
DePauw, R. M., Knox, R. E., McCaig, T. N., Clarke, F. R., and Clarke, J. M. (2011). Carberry hard red spring wheat. Can. J. Plant Sci. 91, 529–534. doi: 10.4141/cjps10187
Dyck, P. L. (1979). Identificatin of the gene for adult plant leas rust resistance in Thatcher. Can. J. Plant Sci. 59, 499–501. doi: 10.4141/cjps79-078
Dyck, P. L., Samborski, D. J., and Anderson, R. G. (1966). Inheritance of adult-plant leaf rust resistance derived from the common wheat varieties Exchange and Frontana. Can. J. Genet. Cytol. 8, 665–671. doi: 10.1139/g66-082
Ellis, J. G., Lagudah, E. S., Spielmeyer, W., and Dodds, P. N. (2014). The past, present and future of breeding rust resistant wheat. Front. Plant Sci. 5:641. doi: 10.3389/fpls.2014.00641
Fowler, D. B., N’Diaye, A., Laudencia-Chingcuanco, D., and Pozniak, C. J. (2016). Quantitative trait loci associated with phenological development, low-temperature tolerance, grain quality, and agronomic characters in wheat (Triticum aestivum L.). PLoS One 11:e0152185.
Hayes, H., Ausemus, E., Stakman, E., Bailey, C., Wilson, H., Bamberg, R., et al. (1936). Thatcher wheat. Stn Bull— Minn. Agric Exp. Stn. 1936:325.
Kassa, M. T., You, F. M., Hiebert, C. W., Pozniak, C. J., Fobert, P. R., and Sharpe (2017). Highly predictive SNP markers for efficient selection of the wheat leaf rust resistance gene Lr16. BMC Plant Biol. 17:45. doi: 10.1186/s12870-017-0993-7:45
Kolmer, J. A. (2005). Tracking wheat rust on a continental scale. Curr. Opin. Plant Biol. 8, 441–449. doi: 10.1016/j.pbi.2005.05.001
Kosambi, D. D. (1944). The estimation of map distances from recombination values. Ann. Eugen. 12, 172–175. doi: 10.1007/978-81-322-3676-4_16
Krattinger, S. G., Lagudah, E. S., Spielmeyer, W., Singh, R. P., Huerta-Espino, J., McFadden, H., et al. (2009). A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 323, 1360–1363. doi: 10.1126/science.1166453
Lewarne, M. (2021). Characterization and genetic mapping of leaf rust (Puccinia triticina) resistance genes Lr2a and Lr46 in Canadian spring wheat (Triticum aestivum) germplasm. M.Sc. thesis. Manitoba: University of Manitoba, 156.
Lillemo, M., Joshi, A. K., Prasad, R., Chand, R., and Singh, R. P. (2013). QTL for spot blotch resistance in bread wheat line Saar co-locate to the biotrophic disease resistance loci Lr34 and Lr46. Theor. Appl. Genet. 126, 711–719. doi: 10.1007/s00122-012-2012-6
Long, D. L., and Kolmer, J. A. (1989). A North American system of nomenclature for Puccinia recondita f. sp. tritici. Phytopathology 79, 525–529.
Lorieux, M. (2012). MapDisto: Fast and efficient computation of genetic linkage maps. Mol. Breed. 30, 1231–1235.
Maccaferri, M., Cane, M. A., Sanguineti, M. C., Salvi, S., Colalongo, M. C., Massi, A., et al. (2014). A consensus framework map of durum wheat (Triticum durum Desf.) suitable for linkage disequilibrium analysis and genome-wide association mapping. BMC Genomics 15:873. doi: 10.1186/1471-2164-15-873
McCallum, B., Fetch, T., and Chong, J. (2007). Cereal rust control in Canada. Aust. J. Agric. Res. 58:639. doi: 10.1071/AR06145
McCallum, B. D., and DePauw, R. M. (2008). A review of wheat cultivars grown in the Canadian prairies. Can. J. Plant Sci. 88, 649–677. doi: 10.4141/CJPS07159
McCallum, B. D., Hiebert, C. W., Cloutier, S., Bakkeren, G., Rosa, S. B., Humphreys, D. G., et al. (2016). A review of wheat leaf rust research and the development of resistant cultivars in Canada. Can. J. Plant Pathol. 38, 1–18. doi: 10.1080/07060661.2016.1145598
McCallum, B. D., Humphreys, D. G., Somers, D. J., Dakouri, A., and Cloutier, S. (2012). Allelic variation for the rust resistance gene Lr34/Yr18 in Canadian wheat cultivars. Euphytica 183, 261–274.
McCallum, B. D., Reimer, E., McNabb, W., Foster, A., Rosa, S., and Xue, A. (2021). Physiologic specialization of Puccinia triticina, the causal agent of wheat leaf rust, in Canada in 2015-2019. Can. J. Plant Pathol. 2021:1888156. doi: 10.1080/07060661.2021.1888156
McCallum, B. D., Reimer, E., McNabb, W., Foster, A., and Xue, A. (2020). Physiological specialization of Puccinia triticina, the causal agent of wheat leaf rust, in Canada in 2014. Can. J. Plant Pathol. 42, 520–526. doi: 10.1080/07060661.2020.1723705
McCallum, B. D., and Seto-Goh, P. (2010). The inheritance of leaf rust resistance in the wheat cultivars ‘Superb’, ‘McKenzie’ and ‘HY644’. Can. J. Plant Pathol. 32, 387–395. doi: 10.1080/07060661.2010.499266
McCartney, C. A., Somers, D. J., McCallum, B. D., Thomas, J., Humphreys, D. G., Menzies, J. G., et al. (2005). Microsatellite tagging of the leaf rust resistance gene Lr16 on wheat chromosome 2BSc. Mol. Breed 15, 329–337. doi: 10.1007/s11032-004-5948-7
McIntosh, R., Dubcovsky, J., Rogers, J., Morris, C., Appels, R., and Xia, X. (2014). Catalogue of gene symbols for wheat: 2013-14 Supplement. Available online at: http://www.shigen.nig.ac.jp/wheat/komugi/genes/symbolClassList.jsp (accessed July 28, 2021).
McIntosh, R., and Dyck, P. (1975). Cytogenetical studies in wheat. VII Gene Lr23 for reaction to Puccinia recondita in Gabo and related cultivars. Aust. J. Biol. Sci. 28, 201–212. doi: 10.1071/BI9750201
Mcintosh, R. A., Wellings, C. R., and Park, R. F. (1995). Wheat rusts: An atlas of resistance genes. Australia: CSIRO Publishing, doi: 10.1071/AR07117
Oelke, L. M., and Kolmer, J. A. (2004). Characterization of leaf rust resistance in hard red spring wheat cultivars. Plant Dis. 88, 1127–1133. doi: 10.1094/PDIS.2004.88.10.1127
Oelke, L. M., and Kolmer, J. A. (2005). Genetics of leaf rust resistance in spring wheat cultivars Alsen and Norm. Phytopathology 95, 773–778. doi: 10.1094/phyto-95-0773
Perez-Lara, E., Semagn, K., Chen, H., Iqbal, M., N’Diaye, A., Kamran, A., et al. (2016). QTLs associated with agronomic traits in the Cutler × AC Barrie spring wheat mapping population using single nucleotide polymorphic markers. PLoS One 11:e0160623.
Peterson, R. F., Campbell, A. B., and Hannah, A. E. (1948). A diagrammatic scale for estimating rust intensity on leaves and stems of cereals. Can. J. Res. 26, 496–500. doi: 10.1139/cjr48c-033
Peturson, B. (1958). Wheat rust epidemics in western Canada in 1953, 1954 and 1955. Can. J. Plant Sci. 38, 16–28. doi: 10.4141/cjps58-004
Randhawa, H. S., Asif, M., Pozniak, C., Clarke, J. M., Graf, R. J., Fox, S. L., et al. (2013). Application of molecular markers to wheat breeding in Canada. Plant Breed. 132, 458–471. doi: 10.1111/pbr.12057
Roelfs, A. (1988). “Control of stem and leaf rusts of wheat,” in Fifth Regional Wheat Workshop for Eastern, Central, and Southern Africa and the Indian Ocean, eds M. Van Ginkel DG Tanner (Mexico, DF: CIMMYT), 244–250.
Samborski, D. J. (1984). Occurrence and virulence of Puccinia recondita in Canada in 1983. Can. J. Plant Patholol. 6, 238–242. doi: 10.1080/07060668409501560
Shewry, P. R., and Hey, S. J. (2015). The contribution of wheat to human diet and health. Food Energy Secur. 4, 178–202. doi: 10.1002/fes3.64
Shiferaw, B., Smale, M., Braun, H. J., Duveiller, E., Reynolds, M., and Muricho, G. (2013). Crops that feed the world 10. Past successes and future challenges to the role played by wheat in global food security. Food Sec. 5, 291–317. doi: 10.1007/s12571-013-0263-y
Singh, A., Knox, R. E., DePauw, R. M., Singh, A. K., Cuthbert, R. D., Campbell, H. L., et al. (2013). Identification and mapping in spring wheat of genetic factors controlling stem rust resistance and the study of their epistatic interactions across multiple environments. Theor. Appl. Genet. 126, 1951–1964. doi: 10.1007/s00122-013-2109-6
Singh, R. P., Huerta-Espino, J., Bhavani, S., Herrera-Foessel, S. A., Singh, D., Singh, P. K., et al. (2011). Race non-specific resistance to rust diseases in CIMMYT spring wheats. Euphytica 179, 175–186. doi: 10.1007/s10681-010-0322-9
Singh, R. P., Huerta-Espino, J., and William, M. (2006). Breeding for resistance to biotic stresses. Plant Breed. 2006, 310–322.
Singh, R. P., Mujeeb-Kazi, A., and Huerta-Espino, J. (1998). Lr46: A gene conferring slow-rusting resistance to leaf rust in wheat. Phytopathology 88, 890–894. doi: 10.1094/phyto.1998.88.9.890
Toth, J., Pandurangan, S., Burt, A., Mitchell, F. J., and Kumar, S. (2018). Marker-assisted breeding of hexaploid spring wheat in the Canadian prairies. Can. J. Plant Sci. 99, 111–127. doi: 10.1139/cjps-2018-0183
Van Ooijen, J. (2009). MapQTL 6: Software for the mapping of quantitative trait loci in experimental populations of diploid species. Netherlands: Wageningen.
Wang, S., Wong, D., Forrest, K., Allen, A., Chao, S., Huang, B. E., et al. (2014). Characterization of polyploid wheat genomic diversity using a high-density 90,000 single nucleotide polymorphism array. Plant biotechnol. J. 12, 787–796. doi: 10.1111/pbi.12183
William, M., Singh, R. P., Huerta-Espino, J., Islas, S. O., and Hoisington, D. (2003). Molecular marker mapping of leaf rust resistance gene Lr46 and its association with stripe rust resistance gene Yr29 in wheat. Phytopathology 93, 153–159. doi: 10.1094/phyto.2003.93.2.153
Wu, Y., Bhat, P. R., Close, T. J., and Lonardi, S. (2008). Efficient and accurate construction of genetic linkage maps from the minimum spanning tree of a graph. PLoS Genet. 4:e1000212.
Yang, J., Hu, C., Hu, H., Yu, R., Xia, Z., Ye, X., et al. (2008). QTLNetwork: mapping and visualizing genetic architecture of complex traits in experimental populations. Bioinformatics 24, 721–723. doi: 10.1093/bioinformatics/btm494
Yang, J., Hu, C. C., Ye, X. Z., and Zhu, J. (2005). QTLNetwork 2.0. Institute of Bioinformatics. Hangzhou: Zhejiang University.
Zhang, P., Hiebert, C. W., McIntosh, R. A., McCallum, B. D., Thomas, J. B., Hoxha, S., et al. (2016). The relationship of leaf rust resistance gene Lr13 and hybrid necrosis gene Ne2m on wheat chromosome 2BS. Theor. Appl. Genet. 129, 485–493. doi: 10.1007/s00122-015-2642-6
Keywords: Triticum aestivum, leaf rust, QTL mapping, SNP markers, gene combination
Citation: Bokore FE, Knox RE, Hiebert CW, Cuthbert RD, DePauw RM, Meyer B, N’Diaye A, Pozniak CJ and McCallum BD (2022) A Combination of Leaf Rust Resistance Genes, Including Lr34 and Lr46, Is the Key to the Durable Resistance of the Canadian Wheat Cultivar, Carberry. Front. Plant Sci. 12:775383. doi: 10.3389/fpls.2021.775383
Received: 13 September 2021; Accepted: 17 November 2021;
Published: 06 January 2022.
Edited by:
Morten Lillemo, Norwegian University of Life Sciences, NorwayReviewed by:
Karansher Singh Sandhu, Washington State University, United StatesJian Ma, Sichuan Agricultural University, China
Copyright © 2022 Amidou N’Diaye, Curtis J. Pozniak and Her Majesty the Queen in Right of Canada, as represented by the Minister of Agriculture and Agri-Food Canada for the contribution of Firdissa E. Bokore, Ron E. Knox, Colin W. Hiebert, Richard D. Cuthbert, Ron M. DePauw, Brad Meyer, and Brent D. McCallum. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Firdissa E. Bokore, ZmlyZGlzc2EuYm9rb3JlQGFnci5nYy5jYQ==; Brent D. McCallum, YnJlbnQubWNjYWxsdW1AYWdyLmdjLmNh
†Present address: Ron M. DePauw, Advancing Wheat Technologies, Calgary, AB, Canada
 Firdissa E. Bokore
Firdissa E. Bokore Ron E. Knox1
Ron E. Knox1 Colin W. Hiebert
Colin W. Hiebert Ron M. DePauw
Ron M. DePauw Amidou N’Diaye
Amidou N’Diaye Curtis J. Pozniak
Curtis J. Pozniak Brent D. McCallum
Brent D. McCallum