- 1Faculty of Applied Biological Sciences, Gifu University, Gifu, Japan
- 2Research Faculty of Agriculture, Hokkaido University, Hokkaido, Japan
- 3Department of Biotechnology, Koneru Lakshmaiah Education Foundation, Guntur, India
- 4Experimental Plant Division, RIKEN BioResource Research Center, Tsukuba, Japan
- 5Department of Biochemistry, Central University of Rajasthan, Ajmer, India
To elucidate the unknown regulatory mechanisms involved in aluminum (Al)-induced expression of POLYGALACTURONASE-INHIBITING PROTEIN 1 (PGIP1), which is one of the downstream genes of SENSITIVE TO PROTON RHIZOTOXICITY 1 (STOP1) regulating Al-tolerance genes, we conducted a genome-wide association analysis of gene expression levels (eGWAS) of PGIP1 in the shoots under Al stress using 83 Arabidopsis thaliana accessions. The eGWAS, conducted through a mixed linear model, revealed 17 suggestive SNPs across the genome having the association with the expression level variation in PGIP1. The GWAS-detected SNPs were directly located inside transcription factors and other genes involved in stress signaling, which were expressed in response to Al. These candidate genes carried different expression level and amino acid polymorphisms. Among them, three genes encoding NAC domain-containing protein 27 (NAC027), TRX superfamily protein, and R-R-type MYB protein were associated with the suppression of PGIP1 expression in their mutants, and accordingly, the system affected Al tolerance. We also found the involvement of Al-induced endogenous nitric oxide (NO) signaling, which induces NAC027 and R-R-type MYB genes to regulate PGIP1 expression. In this study, we provide genetic evidence that STOP1-independent NO signaling pathway and STOP1-dependent regulation in phosphoinositide (PI) signaling pathway are involved in the regulation of PGIP1 expression under Al stress.
Introduction
In the last few decades, extensive studies in molecular physiological research for aluminum (Al) toxicity in acid soils (pH < 5.5) have found that activation of Al-tolerance genes governs Al resistance in plants (Delhaize and Ryan, 1995; Liu et al., 2014; Kochian et al., 2015). Several transcription factors that activate the transcription of critical Al-resistant genes (e.g., ALUMINUM-ACTIVATED MALATE TRANSPORTER 1 (ALMT1); Tokizawa et al., 2015) have been identified, including SENSITIVE TO PROTON RHIZOTOXICITY 1 (STOP1; Iuchi et al., 2007). Al-inducible expression of STOP1-regulated genes plays critical roles in Al tolerance in Arabidopsis (Sawaki et al., 2009) and is conserved in various plant species (Ohyama et al., 2013). It is important to identify the mechanisms regulating gene expression related to Al tolerance, which would be helpful in the field of breeding and the management of crops in acid soil.
Several mechanisms regulating the expression of STOP1-regulated Al-tolerance genes have been reported. For example, the expression of AtALMT1 under Al stress involves calcium signaling that includes CALCINEURIN B-LIKE PROTEIN 1 (Ligaba-Osena et al., 2017) and CALMODULIN-BINDING TRANSCRIPTION ACTIVATOR 2 (Tokizawa et al., 2015), and phosphatidylinositol signaling that includes PHOSPHATIDYLINOSITOL 4-KINASE (Wu et al., 2019). In addition, WRKY DNA-BINDING PROTEIN 46 suppresses AtALMT1 (Ding et al., 2013). These regulators have been characterized in most Al-stress root responses. However, long-term stress leads to high accumulation of Al in the shoot, which is also directly related to shoot growth inhibition (Larsen et al., 1997, 2005; Sadhukhan et al., 2020). Although most of the Al signaling mechanism in shoot is unknown, for example recently we have found Al-inducible expression of ALUMINUM SENSITIVE 3 [ALS3; encodes a bacterial-type ABC transporter-like protein that is involved in the translocation of Al (Larsen et al., 2005)] is STOP1-dependent and shows a specific response to Al in the shoots of Arabidopsis (Sawaki et al., 2016). By contrast, Al-inducible expression of ALS3 in the shoots is dependent on similar signaling mechanisms in the roots, including phosphatidylinositol signaling, although the genes involved in the pathway differ between the shoots and roots (Wu et al., 2019; Sadhukhan et al., 2020). These observations suggest that understanding the regulatory mechanisms of gene expression in shoots is critically important to explore the complexity of the Al signaling pathway.
POLYGALACTURONASE-INHIBITING PROTEIN 1 (PGIP1) gene expression is regulated by STOP1 and is strongly induced in the shoot along with ALS3 by mineral stress (especially Al) and acidic soil conditions (Sawaki et al., 2016). Although the contribution of PGIP1 to Al tolerance has not been studied yet, it has been speculated that PGIP1 plays a role in stabilizing the pectin in the cell wall under acidic conditions (Sawaki et al., 2009; Kobayashi et al., 2014). Al binds preferentially to unmethylated pectin, catalyzed by pectin methylesterase via nitric oxide (NO) signaling (Sun et al., 2016), which negatively affects cell wall structure and function by increasing rigidity and reducing cell expansion and mechanical extensibility, thus inhibiting plant growth (Tabuchi and Matsumoto, 2001; Sun et al., 2016). In contrast to ALS3, which is specifically expressed in Al, PGIP1 is also responsive to abiotic stress. Specifically, PGIP1 is induced by oligogalacturonides, a known degradation product of the cell wall in plant defense (Ferrari et al., 2003; Davidsson et al., 2017). This suggests that analysis of the response of PGIP1 expression will provide an opportunity to study Al signaling pathways in the shoot that may reveal the cross talk between stress signaling pathways, including Al stress, when compared to previous studies of ALS3 (Sadhukhan et al., 2020).
GWAS on the expression level difference of an Al-response gene is a powerful tool to identify the unknown upstream signaling pathways regulating the gene of interest (Atwell et al., 2010; Wang et al., 2020). GO enrichment and gene co-expression network analyses can add to the power of eGWAS in identifying functional candidate genes (Kobayashi et al., 2016; Sadhukhan et al., 2020; Song et al., 2021). We have conducted a genome-wide association study targeting gene expression level (eGWAS) that identified cis-mutations in the promoters of NOD26-like intrinsic protein 1; 1 (NIP1; 1), which regulates hydrogen peroxide sensitivity, and AtALMT1; multidrug and toxic compound extrusion (MATE), which encodes an Al-responsive citrate transporter, is the determinant of the expression levels of these genes in roots (Sadhukhan et al., 2017; Nakano et al., 2020a, 2020b). In addition to the cis-locus, the eGWAS of AtMATE also revealed trans-loci associated with gene expression (Nakano et al., 2020b). Through eGWAS, we also found the involvement of phosphatidylinositol and calcium signaling in the regulation of ALS3 expression under Al stress in Arabidopsis shoots (Sadhukhan et al., 2020). To identify the factors involved in Al signaling related to PGIP1 expression, we conducted an eGWAS based on the expression level of PGIP1 in Arabidopsis thaliana accessions with a reverse genetics approach. We propose both STOP1-dependent and STOP1-independent Al signalings for the transcriptional regulation of PGIP1 in the shoots of A. thaliana.
Materials and Methods
Plant Materials
Seeds of 83 worldwide natural A. thaliana accessions (Atwell et al., 2010; Cao et al., 2011; Horton et al., 2012) used in our previous GWAS (Sadhukhan et al., 2020; Nakano et al., 2020b) and T-DNA insertion lines were obtained from the Arabidopsis Biological Resource Center (ABRC, Columbus, OH, United States), the Nottingham Arabidopsis Stock Centre (NASC, Nottingham, United Kingdom), and the RIKEN BioResource Center (RIKEN BRC, Tsukuba, Japan). Prior to experimental use, the procured seeds were multiplied by a single-seed descent process. Homozygosity was confirmed in T-DNA insertion mutant line using primers from the SALK database, following their protocols.1 The sequences of the primers are given in Supplementary Table S1. The T-DNA insertion mutants used in this study were pgip1 (SALK_001662), STOP1-KO (SALK_114108), at1g64105 (SAIL_235_D03), at2g21950 (GK-707C08), at3g24480 (GABI_017A08), at5g15300 (SALK_044494C), at5g38900 (SAIL_453_G03), at5g43460 (SALK_067877C), at5g58900 (SALK_084867C), at5g58910 (SALK_064093C), at1g51070 (SALK_104253C), at2g38090 (SALK_127250C), at2g04780 (SALK_113729C) and plc9 (SALK_021982C).
Plant Growth Conditions and Stress Treatment
Seedlings were grown on nylon mesh floating on modified MGRL solution (Fujiwara et al., 1992; 2% solution with 200 μM CaCl2; initial pH 5.6) for 10 days at 22°C using a 12-h photoperiod with 37 μMol m−2 s−1 photon flux density. The culture solution was renewed every 2 days. After 10 days, the seedlings were transferred to another modified MGRL solution (without P and pH 5.0) containing 25 μM AlCl3·6H2O (Sawaki et al., 2016). The shoots were harvested after 24 h of Al treatment and immediately frozen with liquid nitrogen for RNA extraction. The same Al toxic solution containing 50 μM 2-(4-carboxyphenyl)-4,4,5,5-tetramethyl-imidazoline-1-oxyl-3-oxide (cPTIO), an NO scavenger, was used to evaluate the effect of NO (D’Alessandro et al., 2013; Sun et al., 2016). The concentration of cPTIO was based on preliminary experiments from which maximum suppressed responses were obtained without affecting the plant root (Supplementary Figures S1, S2).
Soil culture was conducted using commercial acidic soil (PROTOLEAF, Tokyo, Japan; pH 4.2; 1:2.5 w/v soil/water solution; Agrahari et al., 2020). The acidic soil was neutralized by the addition of CaCO3 (Koyama et al., 2000; Sawaki et al., 2016; Agrahari et al., 2020; 4.0 g kg−1; pH 5.1; 1:2.5 w/v soil/water solution) and used as the control soil. Plants (100 seeds) were grown for 2 weeks at 22°C during a 12-h photoperiod with 37 μMol m−2 s−1. Throughout the experiment, the plants were irrigated daily with deionized water to maintain soil moisture. The soil pH (water) and exchangeable Al were determined using the method described by Koyama et al. (2000).
RNA Extraction and Real-Time Quantitative Reverse Transcription PCR
Total RNA was isolated from the shoots using Sepasol-RNA I Super G (Nacalai Tesque, Kyoto, Japan) according to the manufacturer’s instructions. The RNA quality was analyzed using the A260/A280 ratio on a NanoVue Plus spectrophotometer (Biochrom, Holliston, United States). Total RNA was reverse-transcribed using ReverTra Ace quantitative PCR master mix with genomic DNA remover (Toyobo, Osaka, Japan) following the manufacturer’s instructions. The gene expression levels were quantified using SYBR Premix Ex Taq II (Takara Bio, Otsu, Japan) with a Dice Real Time System II MRQ thermal cycler (Takara Bio, Otsu, Japan) according to the manufacturer’s instructions. Briefly, all quantifications were carried out based on the real-time quantitative reverse transcription PCR (qRT-PCR) standard curve method of Bustin et al. (2009), as described by Kobayashi et al. (2014). For all quantifications, a standard curve was constructed using a cDNA dilution series, and the transcript levels of selected genes were quantified relative to that of the stable internal reference gene, Ubiquitin 1 (UBQ1; AT3G52590; Kobayashi et al., 2007, 2014). We have checked the invariant expression of UBQ1 for all experimental condition in used lines in this study (Supplementary Figure S3). We included a control with no reverse transcriptase to assess genomic DNA contamination, and the amplification efficiency of all primers was confirmed. The primer sequences used to amplify the selected genes are shown in Supplementary Table S2.
Expression Genome-Wide Association Study of PGIP1
The eGWAS analyses were carried out using TASSEL v3.0 software following a mixed linear model (MLM; Bradbury et al., 2007) using a total of 160,748 genome-wide single nucleotide polymorphism (SNP) information from public databases (Atwell et al., 2010; Cao et al., 2011; Horton et al., 2012),2 3 which excluded SNPs of missing data or those with less than 5% minor allele frequency, as described earlier (Nakano et al., 2020b). The heritability (h2) was estimated by the following formula h2 = (the additive genetic variance)/(the additive genetic variance + the residual variance). The suggestive SNPs were determined by quantile–quantile (Q–Q) plot analysis (Zhang et al., 2018) using a free statistics software,4 and the genes closest to the SNPs (Table 1) were identified using the TAIR 10 database.5
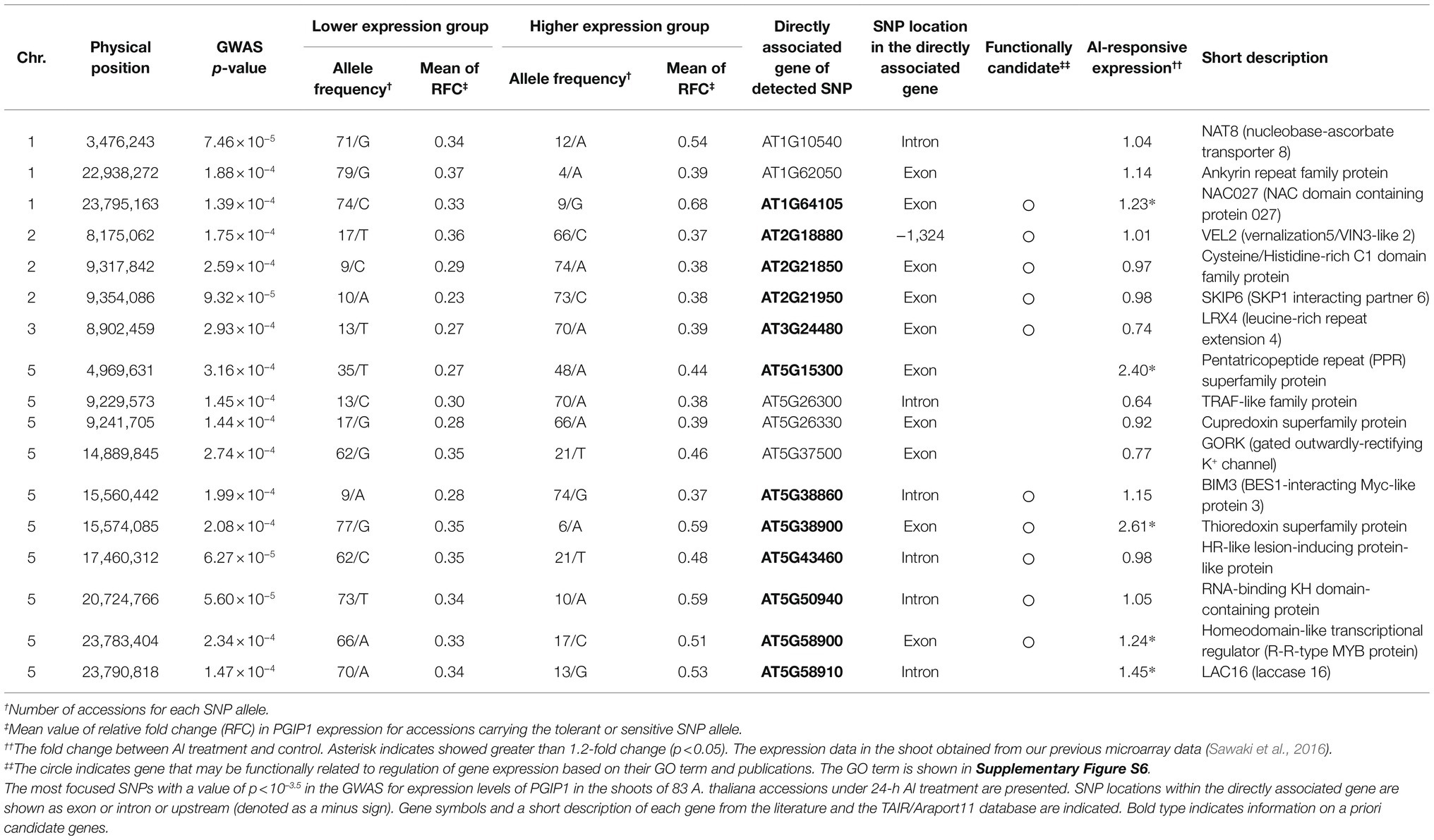
Table 1. GWAS identified SNPs with directly linked to protein-coding genes that were associated with PGIP1 expression levels in the shoots of 83 Arabidopsis thaliana accessions under Al stress.
Bioinformatics of Genes Associated With Significant SNPS
Gene ontology (GO) analysis was performed using an online tool available in the TAIR database.6 Gene polymorphisms were mined from the 1,001 Genomes database7 and POLYMORPH database.8 The genes upregulating PGIP1 expression in T87-cultured cells of Arabidopsis were identified by the Regulatory-network Research (RnR) database (Sakurai et al., 2014).9 Co-expression network analysis was conducted on the eGWAS-detected and RnR database-listed genes using the ATTED-II database (Obayashi et al., 2018).10 Cis-elements and corresponding transcription factors (TFs) of the promoters were predicted using PlantPAN 3.0 (Chang et al., 2008).11
In planta Complementation Assay of STOP1
The STOP1 complementation Arabidopsis transgenic plant was constructed as described by Ohyama et al. (2013). STOP1 genomic DNA containing the promoter (−2,848 from the first ATG) and downstream (+626 from the stop codon) regions was cloned into a binary vector (promoterless pBIG2113SF). This construct was introduced into Agrobacterium tumefaciens strain GV3101 and transformed into STOP1-KO plants by the floral dip method (Clough and Bent, 1998). A T3 homozygous line was used for the experiments.
DNA-Protein Binding Assay
The binding of STOP1 to double-stranded, synthetic promoter fragments was studied using an amplified luminescent proximity homogeneous assay (AlphaScreen™, PerkinElmer, Waltham, MA, United States) and a 276 EnSpire Multimode Plate Reader (PerkinElmer) as described by Enomoto et al. (2019). Competition assays were performed with non-biotinylated probes (450 nM) according to the manufacturer’s instructions. The competitor probes, consisting of the promoter fragments −193 to −222 bp upstream from the PGIP1 and − 2,694 to −2,723 bp upstream from the AT5G38900 start codons, respectively, were designed around the STOP1-binding site according to the Plant Cistrome Database (Sadhukhan et al., 2019).12 The mutated probes were designed following the method of Tokizawa et al. (2015). The forward and reverse probe sequences are listed in Supplementary Table S3.
Al Content in Pectin and Nuclear Magnetic Resonance Analysis
Pectin was extracted from 250 mg powdered Arabidopsis shoot tissue (500 seedlings were grown for 10 days in the hydroponics system mentioned above) in buffer containing 50 mM Tris–HCl (pH 7.2) and 50 mM cyclohexane-trans-1, 2-diamine tetra-acetate (CDTA; Bethke and Glazebrook, 2014). The extraction was continued for 15 min at 95°C with intermittent vortexing, and the sample was then centrifuged at 10,000 × g for 10 min. The supernatant containing pectin was analyzed for Al content using inductively coupled plasma mass spectrometry as described by Watanabe et al. (2015). For nuclear magnetic resonance (NMR) analysis, the supernatant containing pectin was lyophilized and dissolved in D2O. 1H-13C-Heteronuclear Single Quantum Coherence (HSQC) NMR was performed at 600.17 MHz on a JEOL ECA 600 NMR spectrometer (JEOL, Tokyo, Japan) equipped with a 5-mm FG/TH tunable probe, using the pulse sequence ‘hsqc_dec_club_pn’. NMR measurements were recorded at 70°C (Siedlecka et al., 2008). Sweep widths of 15 and 170 ppm were used to acquire the 1H and 13C spectra, respectively. For each NMR experiment, 88 scans were collected using a relaxation delay of 1.5 s.
Results
Genome-Wide Association Study to Detect Loci Associated With Expression of Arabidopsis thaliana PGIP1 Under Al Stress
We analyzed the expression of PGIP1 in the shoots of wild-type (WT) Arabidopsis, Columbia (Col-0) at different time points after root exposure to 25 μM Al. The expression of PGIP1 was induced after 12 h and was markedly induced (five-fold on average) after 24 h of treatment (Figure 1A). It is also a condition of Al accumulation in the shoots (Supplementary Figure S4). Hence, in this study, we chose 24 h as the time point for evaluating PGIP1 gene expression under Al stress to conduct the eGWAS of PGIP1 in the shoots. Next, we analyzed the expression of PGIP1 in 83 Arabidopsis accessions (Supplementary Figure S5; Supplementary Table S4) that had been treated with treated with 25 μM Al for 24 h and found that the expression range between log2 RFC −3.3 and log2 RFC 0.1 (RFC: relative fold change; compared to Col-0; Figure 1B; h2 = 88.3%). We performed a GWAS following MLM using the PGIP1 expression data, but after Bonferroni correction for multiple testing, we could not detect any significant SNPs at the genome-wide significance level. This could be due to the dependence of the statistical power of GWAS on the population size and allele frequency. Although the Q–Q plot showed only a slight deviated plot, in this study we set a suggestive threshold based on this result (p < 10–3.5) and selected the top-ranked 17 SNPs as suggestive SNPs associated with PGIP1 expression level variation (Figures 1C,D; Table 1). These potentially associated SNPs were selected for further analysis.
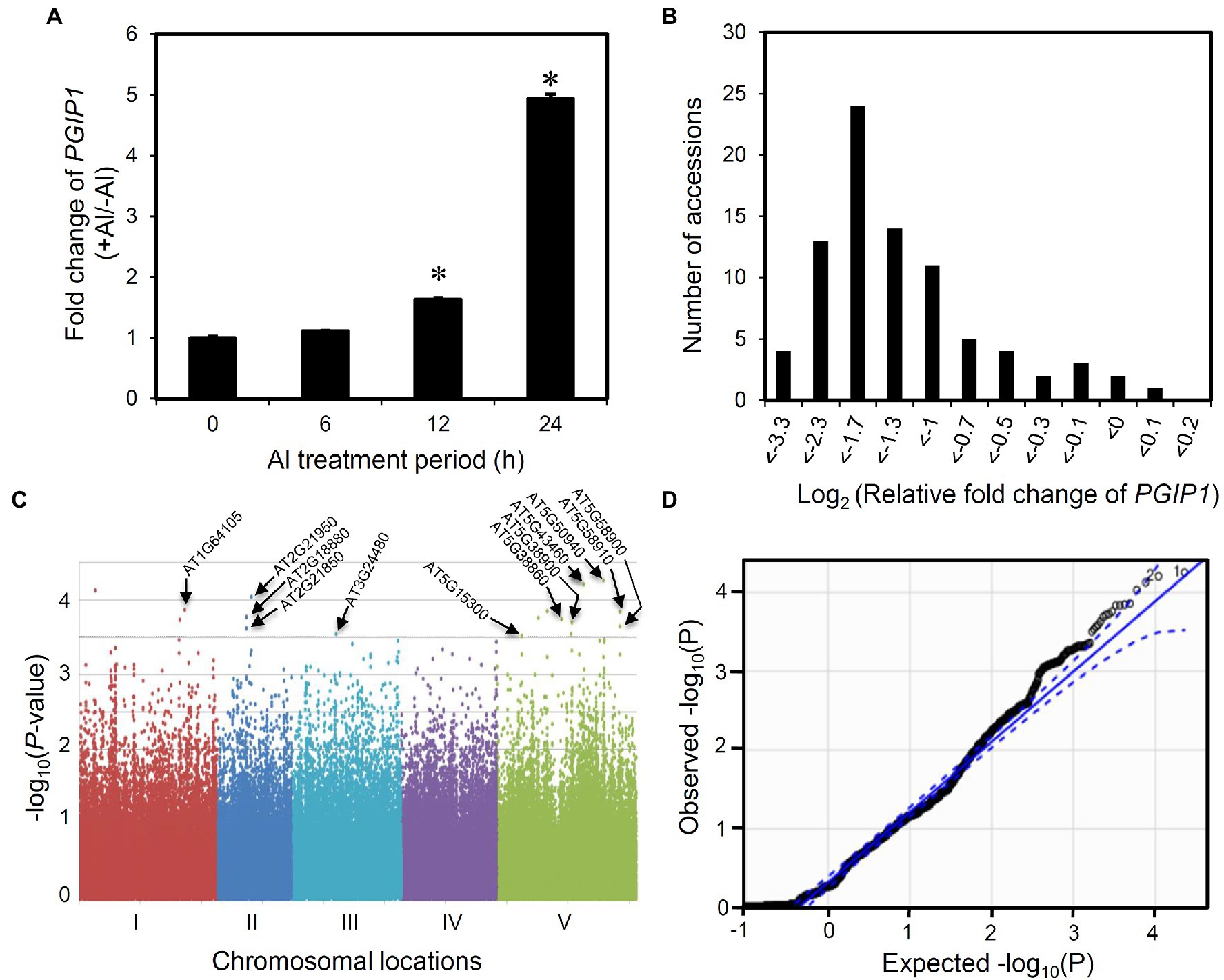
Figure 1. Expression GWAS for POLYGALACTURONASE-INHIBITING PROTEIN 1 (PGIP1) expression in Arabidopsis thaliana. (A) Fold induction of PGIP1 expression under Al stress at different time points of treatment are shown. The shoots were excised from 10-day-old approximately 100 seedlings of Arabidopsis (WT, Col-0) treated with either 0 or 25 μM Al for 0, 6, 12, and 24 h. Fold induction was calculated as the ratio of gene expression in growth media with Al to that without Al. An asterisk indicates significant difference from 0 h (Student’s t-test, p < 0.05). (B) Histogram showing the frequency distribution of PGIP1 expression levels in the shoots of 83 A. thaliana accessions. 10 days pre-grown approximately 100 seedlings of each accession were treated with 25 μM Al for 24 h for quantification of gene expression. UBQ1 was used as an internal control in the qRT-PCR experiment. The average data of three technical replicates are shown as relative fold change of PGIP1 expression compared to Col-0. (C) GWAS of PGIP1 expression levels in 83 A. thaliana accessions. The results are presented as a Manhattan plot of significance [−log10 (value of p)] versus chromosomal locations of SNPs. The horizontal dashed line indicates a value of p threshold according to the Q–Q plot of GWAS from D. Locations of the a priori candidate genes directly associated with the suggestive SNPs (p < 10–3.5; see also Table 1) based on the Q–Q plot of GWAS are shown by arrows. (D) Q–Q plot of GWAS, straight line represents expected null distribution of values of p; dots represent observed distribution of values of p; dotted line represents the 95% confidence intervals. Primers used for qRT-PCR are listed in Supplementary Table S2.
We first characterized 17 protein-coding genes carrying the 17 SNPs directly in their exons, introns, and promoters (Table 1) and then considered a priori candidate genes related to the regulation of PGIP1 expression out of these 17 genes. Out of these, seven genes belonged to the GO term of “regulation of gene expression,” “DNA-binding transcription factor activity,” “intracellular signal transduction,” and “hormone-mediated signaling pathway” and may be functionally associated with PGIP1 expression (Table 1; Supplementary Figure S6). Out of the a priori candidate genes, AT2G21950 (SKP1 interacting partner 6: SKIP6; Farrás et al., 2001) and AT3G24480 (leucine-rich repeat extension 4: LRX4; Zhao et al., 2018) are involved in hormonal signaling and cell-wall integrity, respectively. AT5G38900 (Thioredoxin superfamily protein: TRX SF) is involved in regulation of gene expression via redox signaling (Sevilla et al., 2015). On the other hand, five among the 17 genes were induced more than 1.2-fold by Al in the shoot, according to our earlier transcriptome analysis, carried out under the same experimental conditions (Sawaki et al., 2016; Table 1). Therefore, we finally selected 12 genes with functions related to transcriptional regulation and Al-responsive expression, as a priori candidate genes associated with PGIP1 expression, for subsequent analyses.
Expression Level Polymorphisms and Amino Acid Polymorphisms Caused by Detected SNP of the Candidate Genes
We examined the 12 candidate genes for expression level polymorphisms (ELPs) and amino acid polymorphisms associated with the PGIP1 expression level. The expression levels of these genes were compared between representative accession groups that carried different detected SNP alleles (Figure 2, Supplementary Figure S7). Five genes, AT1G64105 (NAC027), AT5G38900 (TRX SF) AT5G43460 (HR-like lesion-inducing protein-like protein: HR-like protein), AT5G58900 (Homeodomain-like transcriptional regulator: R-R MYB), and AT5G58910 (laccase 16: LAC16) exhibited significant differences in the level of expression between accessions carrying different SNP alleles (Figure 2). When comparing the different alleles, the minor allele group with elevated PGIP1 expression showed higher expression levels of each gene than the major allele group with low PGIP1 expression (Table 1; Figure 2). In the case of genes with SNP alleles located directly in exons, we examined the amino acid polymorphisms caused by the detected SNPs using reliable DNA sequences from the 1,001 genome database. The SNPs detected in the exons of five genes caused amino acid polymorphisms, viz. Ser29Cys in NAC027, Ala187Ser in SKIP6, Asn60Lys in LRX4, Phe134Ile in AT5G15300 (pentatricopeptide repeat superfamily protein) and His246Gln in R-R MYB. Among them, NAC027 and R-R MYB showed both ELP and amino acid polymorphism. These polymorphisms might affect the expression level variation of PGIP1 as cis-factors. In this way, eight genes were selected as the first group of possible candidate genes, which were further studied using reverse genetics.
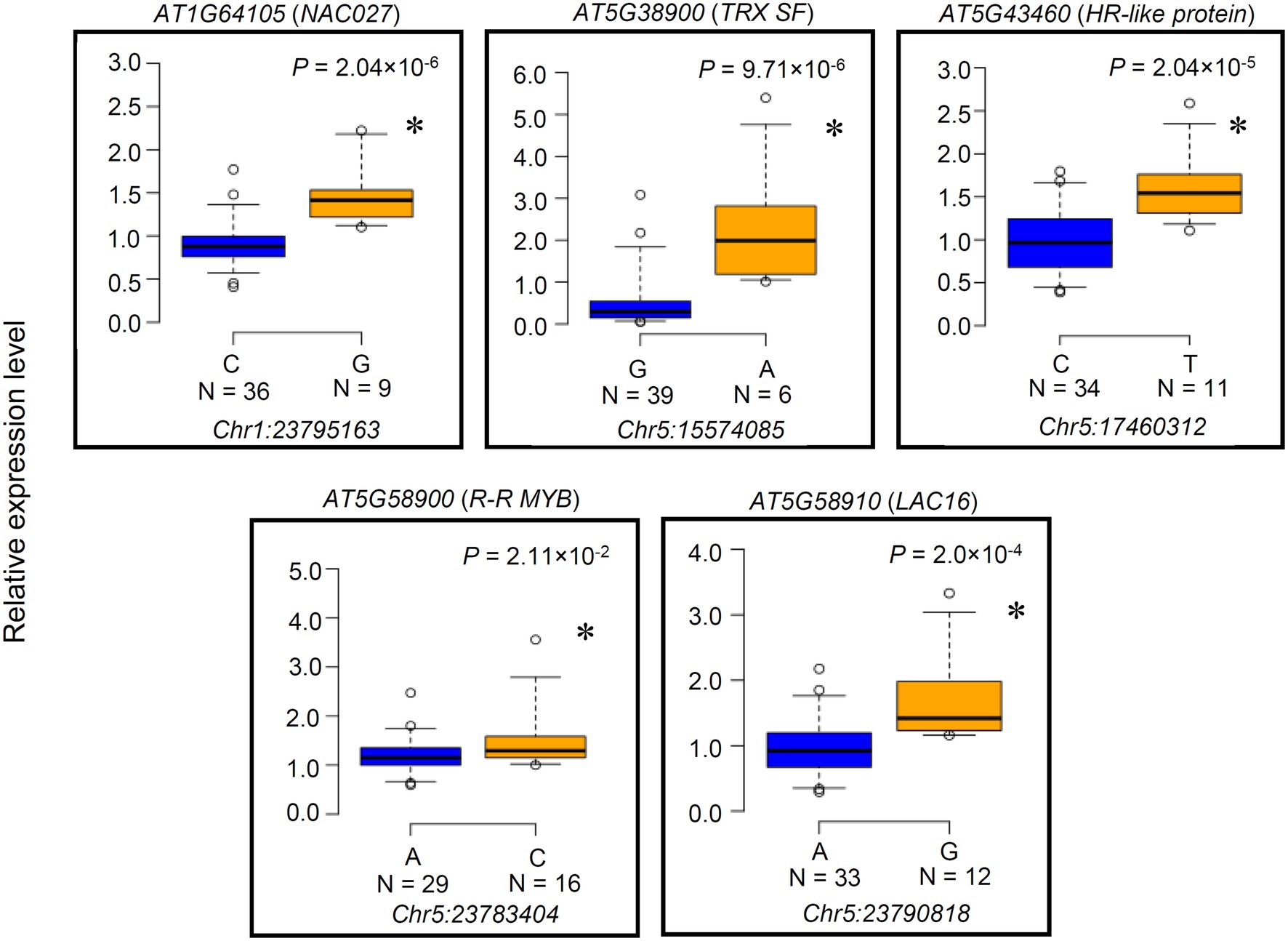
Figure 2. Expression level polymorphisms of the genes identified by expression genome-wide association study (eGWAS). The expression level variations in a priori candidate genes directly linked to SNPs (Table 1) that showed significant segregation between SNP alleles are presented as box plots. Genes whose expression levels were not significantly different between accessions are shown in Supplementary Figure S7. Expression of these genes was monitored by qRT-PCR in 45 randomly chosen accessions among the 83 accessions used in eGWAS. Accessions are grouped according to SNP alleles at a particular physical chromosome position mentioned below each box plot. Number of accessions of each SNP allele is shown under the each SNP. UBQ1 was used as an internal control. Expression level of each accession is relative to Col-0. An asterisk indicates a significant difference between the average values of group (p < 0.05, Student’s t-test).
Reverse Genetic Characterization of the Candidate Genes
To examine the effect of the candidate genes on PGIP1 expression, its expression level was quantified in the T-DNA insertion knockout (KO) or knockdown mutants (KD; Supplementary Figure S8) of the eight candidate genes. PGIP1 expression was significantly lower in the three T-DNA insertion mutants of nac027, trx sf and r-r myb than in the WT under Al treatment (Figure 3A). In particular, the reduction of PGIP1 expression in the r-r myb was as high as approximately 40%. In contrast, other mutants (at2g21950, at3g24480, at5g15300, at5g43460, at5g58910) showed no significant difference of PGIP1 expression compared to the WT under Al treatment (Figure 3A). From this analysis, we found that NAC027, TRX SF and R-R MYB are involved in the regulation of PGIP1 expression.
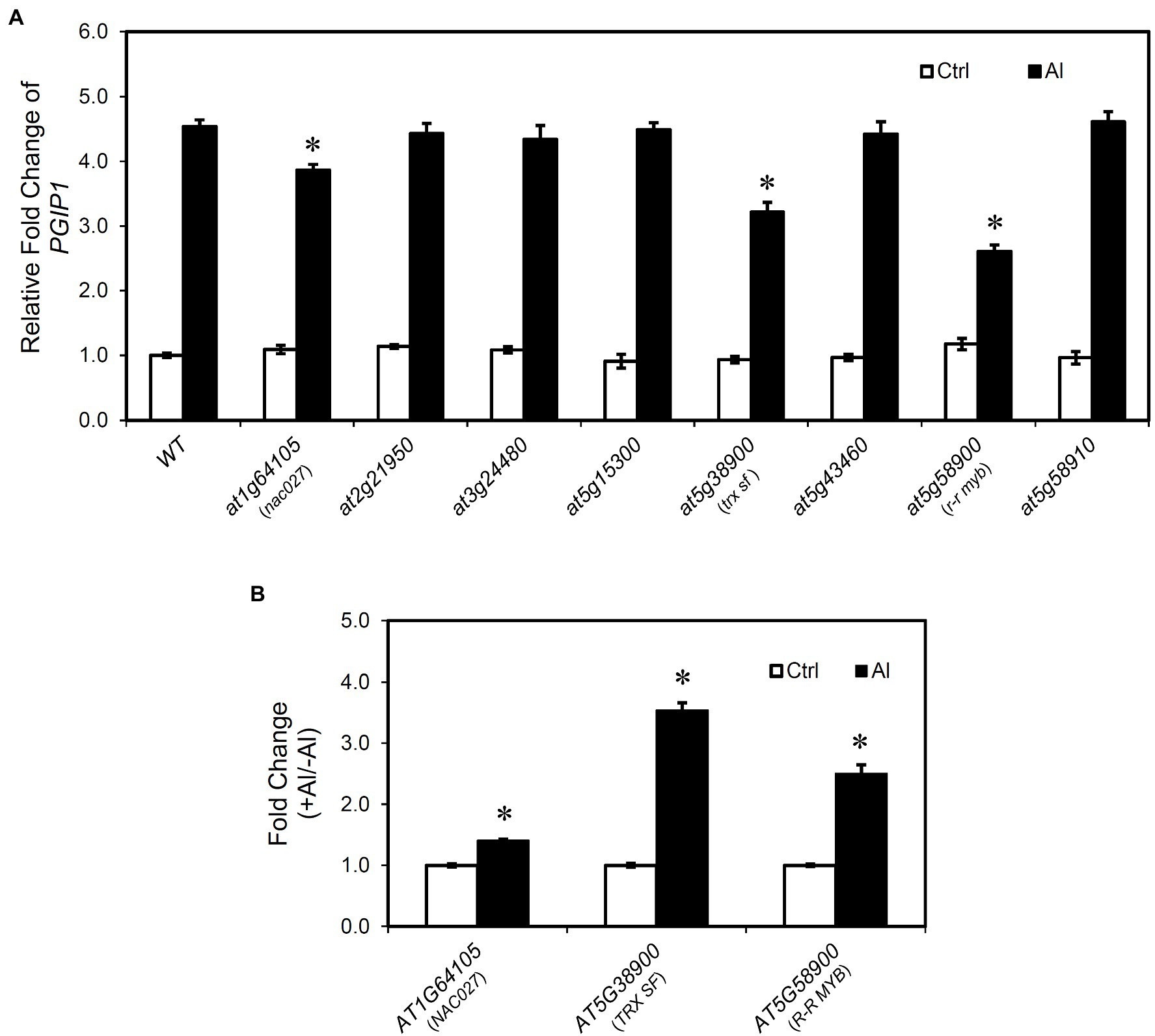
Figure 3. PGIP1 expression in the mutants of genes identified by eGWAS. (A) Relative expression levels of PGIP1 in the shoots of WT (WT, Col-0) and T-DNA insertion mutants of eight final candidate genes having expression level polymorphisms (Figure 2) and/or amino acid polymorphisms (see main text). Approximately 100 seedlings of the WT and independent homozygous T-DNA insertion mutants were grown for 10 days and treated with 0 (-Al) or 25 μM AlCl3 (+Al) for 24 h. PGIP1 expression was measured by qRT-PCR. Expression levels are expressed as relative fold changes compared to WT (-Al); significant reductions in relative fold change of PGIP1 from the Al-treated WT sample are indicated by asterisks (Student’s t-test, p < 0.05). (B) Fold changes (+Al/-Al) of AT1G64105 (NAC027), AT5G38900 (TRX SF), and AT5G58900 (R-R MYB) at 24 h of Al treatment, measured by qRT-PCR, are shown; significant fold increases from the control (-Al) samples are indicated by asterisks, *(Student’s t-test, p < 0.05). Average data of three biological replicates are presented with standard errors. UBQ1 was used as an internal control. Primers used for qRT-PCR are listed in Supplementary Table S2.
We also analyzed the patterns of expression of NAC027, TRX SF, and R-R MYB under Al stress in the shoots of WT A. thaliana after 24 h of treatment. We observed a significant induction of these genes relative to the control (Figure 3B). The Al-induced responses of NAC027 were weak, but their gene expression showed Al responses similar to PGIP1 expression. These results suggest that NAC027, TRX SF, and R-R MYB are related to the regulation of Al-induced PGIP1 expression. In addition, Al-responsive PGIP1 and its regulatory system were involved in Al tolerance. In acidic soils containing higher exchangeable Al, pgip1, like the Al-hypersensitive stop1, was much more inhibited in growth than the WT (Supplementary Figure S9). On the other hand, the growth was recovered in the neutralized soil (Supplementary Figure S9). Similarly, the growth of trx sf and r-r myb was inhibited than the WT in acidic soil, although the growth of nac027 was not severely inhibited (Supplementary Figure S9). This may be consistent with the lower degree of repression of PGIP1 in nac027 and the weaker induction of Al on the expression level of NAC027 compared to the other two genes (Figure 3).
Relationship Between STOP1 and Genes Regulating PGIP1 Identified by eGWAS
We investigated the relationship between STOP1 regulation and the eGWAS-detected genes because it has been reported that STOP1 regulates PGIP1 expression (Sawaki et al., 2009). There was no difference in expression levels of STOP1 in the nac027, trx sf, and r-r myb (Supplementary Figure S10A). In contrast, we found a significant reduction in the gene expression level of TRX SF in the STOP1-KO, whereas the other two genes showed similar expression levels compared to WT (Figure 4A). In addition, in the STOP1-complemented line, the expression of TRX SF was fully recovered similar to Col-0 (Figure 4B). These results suggest that TRX SF is regulated by the STOP1 transcription factor, while none of the three genes affect the expression level of STOP1.
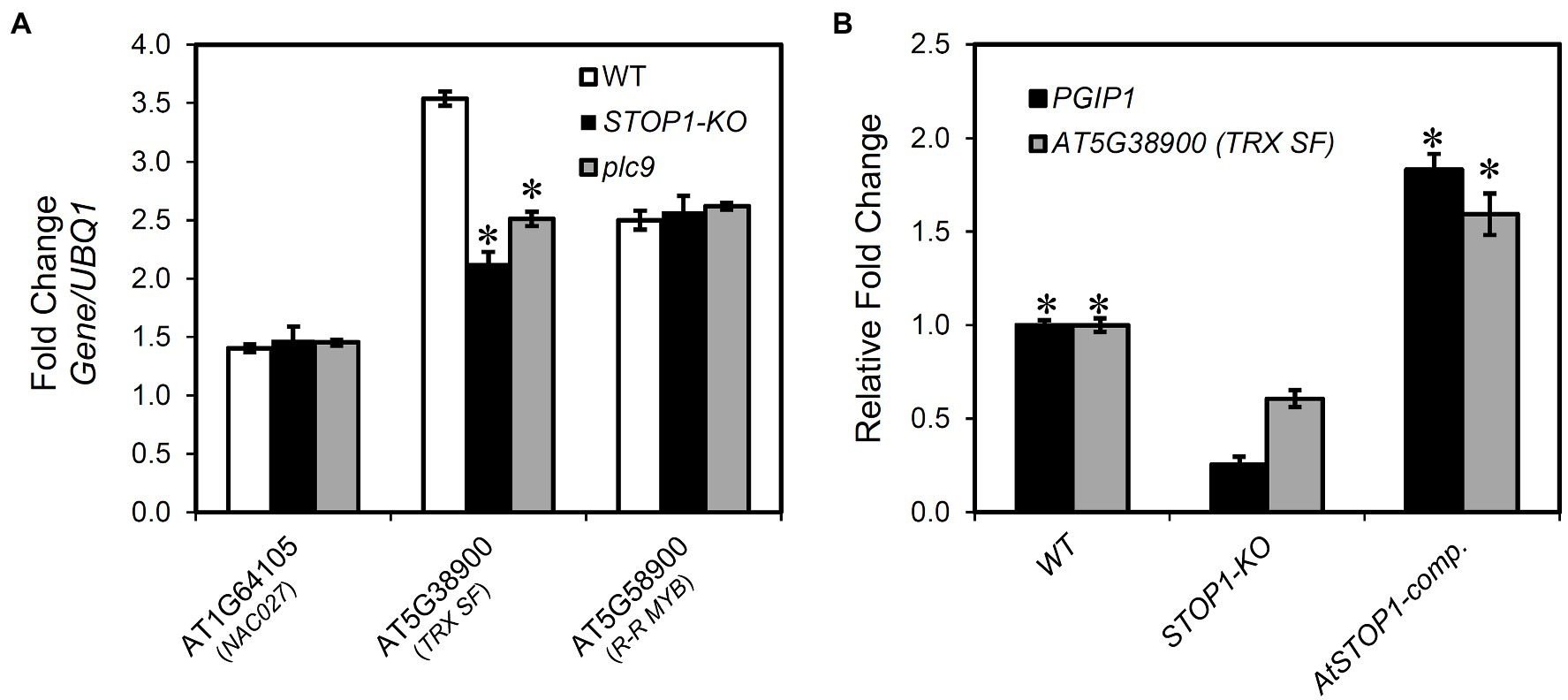
Figure 4. Relationship of SENSITIVE TO PROTON RHIZOTOXICITY 1 (STOP1) to the genes identified by eGWAS for PGIP1 expression. (A) Transcriptional regulation of the genes identified in eGWAS (NAC027, TRX SF, and R-R MYB) by STOP1 and PLC9 was studied. Seedlings of WT (WT, Col-0) and knockout (KO) lines were grown for 10 days in MGRL solution and treated with 25 μM AlCl3 for 24 h. The expression levels of NAC027, TRX SF, and R-R MYB were measured in the shoots of WT, STOP1-KO, and plc9 by qRT-PCR. An asterisk indicates a significant difference from WT (Student’s t-test, p < 0.05). (B) Recovery of the transcription of suppressed PGIP1 and TRX SF in STOP1-KO was analyzed in complemented line (AtSTOP1-comp.) after exposure to Al (25 μM AlCl3 for 24 h). Gene expression levels are relative to WT and fold changes from the control (-Al) are shown. An asterisk indicates a significant difference from STOP1-KO (Student’s t-test, p < 0.05). UBQ1 was used as an internal control. Average values of three biological replicates are presented with standard errors.
In our previous eGWAS of ALS3, we found the involvement of phosphoinositide (PI)-dependent phospholipase C9 (PLC9) signaling upstream of the STOP1 regulation system, which regulates the expression of ALS3 and PGIP1 in the shoots of A. thaliana (Sadhukhan et al., 2020). Therefore, we examined the expression of TRX SF, NAC027, and R-R MYB in the plc9. We found that the expression of TRX SF was significantly suppressed in plc9 (Figure 4A), similar to the downregulation of PGIP1 (Supplementary Figure S10B). These findings suggest that TRX SF is regulated by a PI signaling-mediated STOP1-dependent pathway. In contrast, we found that the expression of ALS3 remained unchanged in the trx sf, nac027, and r-r myb mutants (Supplementary Figure S10A). These results suggest that TRX SF differentially regulates transcription of PGIP1 and ALS3, which are co-regulated by STOP1 in Arabidopsis shoots.
In vitro Binding Analysis of STOP1 to TRX SF and PGIP1 Promoter Regions
Next, we performed a promoter binding analysis to reveal whether STOP1 directly regulates TRX SF and PGIP1 in the STOP1-dependent pathway. The STOP1-binding positions were predicted by searching enriched sequences (octamer units) in the stress-inducible promoters.13 They were identical to the binding sites provided by the Plant Cistrome Database. Therefore, we searched for putative STOP1-binding sites in the promoters of PGIP1 and TRX SF using the Plant Cistrome Database. Based on DNA affinity purification sequencing (DAP-seq), the database identified the “GGNVS” consensus sequence in the PGIP1 and TRX SF promoters for binding STOP1-like proteins (Figure 5A), as previously identified in rice by Tsutsui et al. (2011). The binding capacity of STOP1 to the sequences available from the Plant Cistrome Database was validated by an in vitro competitive binding assay using the AlphaScreen™ system (Figure 5B). A 30-bp synthetic double-stranded DNA probe, designed around the binding site in the PGIP1 (−193 to −222 bp) and TRX SF (−2,694 to −2,723) promoters, could compete for STOP1 protein binding with a known STOP1-binding site in the AtALMT1 promoter (Tokizawa et al., 2015). On the other hand, replacing the “GGNVS” consensus sequences within the PGIP1 and TRX SF probe with A/T stretches abolished STOP1 binding (Figure 5B). These results indicate that STOP1 binds directly to the PGIP1 and TRX SF promoters.
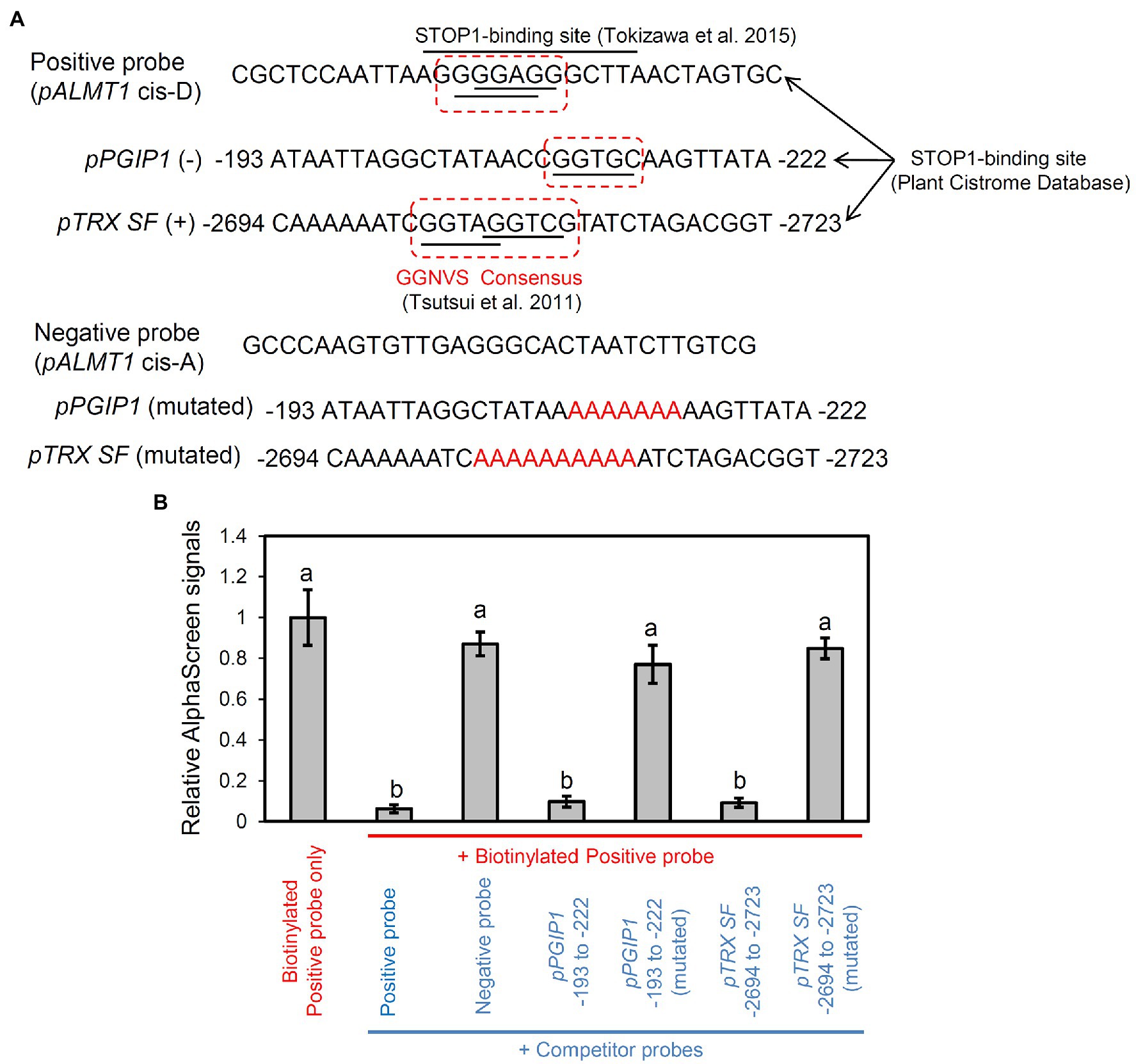
Figure 5. Binding assay of STOP1 protein with PGIP1 and TRX SF promoters. Results of competitive binding assay of synthetic double-stranded promoter fragment of PGIP1 (−193 to −222 bp) and TRX SF (−2,694 to −2,723) to in vitro translated STOP1 protein using the PerkinElmer Amplified Luminescent Proximity Homogeneous Assay (AlphaScreen™) are shown. (A) Probe sequences used in AlphaScreen are shown. The STOP1-binding sites of the PGIP1 and TRX SF promoters were chosen from the Plant Cistrome Database (http://neomorph.salk.edu/dap_web/pages/index.php). The dotted boxes indicate the GGNVS sequences that represent the STOP1-ortholog ART1 binding minimum consensus. Known binding (cis-D) and non-binding (cis-A) sites of STOP1 on the AtALMT1 promoter (Tokizawa et al., 2015) were used as positive and negative control probes, respectively. The mutated PGIP1 and TRX SF probes were designed by replacing the GGNVS consensus sequence with stretches of A/T (shown in red font). (B) Competitive binding assay where 450 nM competitor probes (shown in blue font) compete with 50 nM biotinylated positive probe (shown in red font) for binding with the STOP1 protein. The emitted light signal intensities, relative to those of the biotinylated probe without any competitor, are shown in the graph. Average values ± SD (n = 3) are presented. Lower emitted signal intensity signifies binding of the STOP1 protein to the respective competitor probe. Different letters indicate significant differences of emission intensity (Tukey’s test, p < 0.05).
Al-Inducible NO Signaling Effects Expression of Genes Regulating PGIP1
NO generation is positively correlated with cell wall pectin demethylation and alteration of cell wall metabolism under Al stress (Zhou et al., 2012; Sun et al., 2016). To establish whether Al accumulation induces NO signaling, we examined NO-inducible marker gene (AT2G06050, AT3G45140, and AT5G42650; Huang et al., 2004) expression in shoots after exposing the roots to Al or Al plus cPTIO (NO scavenger; Shi et al., 2015) for 24 h. We found that the expression of the NO marker genes was substantially induced in the Al-treated samples, whereas their expression was suppressed in the samples treated with Al plus cPTIO (Figure 6A). This suggests that Al induces NO signaling in the shoots. Next, we examined whether PGIP1 and ALS3, which are regulated by STOP1, function downstream of NO signaling by quantifying their expression levels in the shoots after exposing the roots to Al or Al plus cPTIO for 24 h. The transcript levels of PGIP1 were suppressed significantly in the Al plus cPTIO samples, whereas ALS3 expression was unchanged (Figure 6B). STOP1 expression was neither Al-induced nor affected by cPTIO.
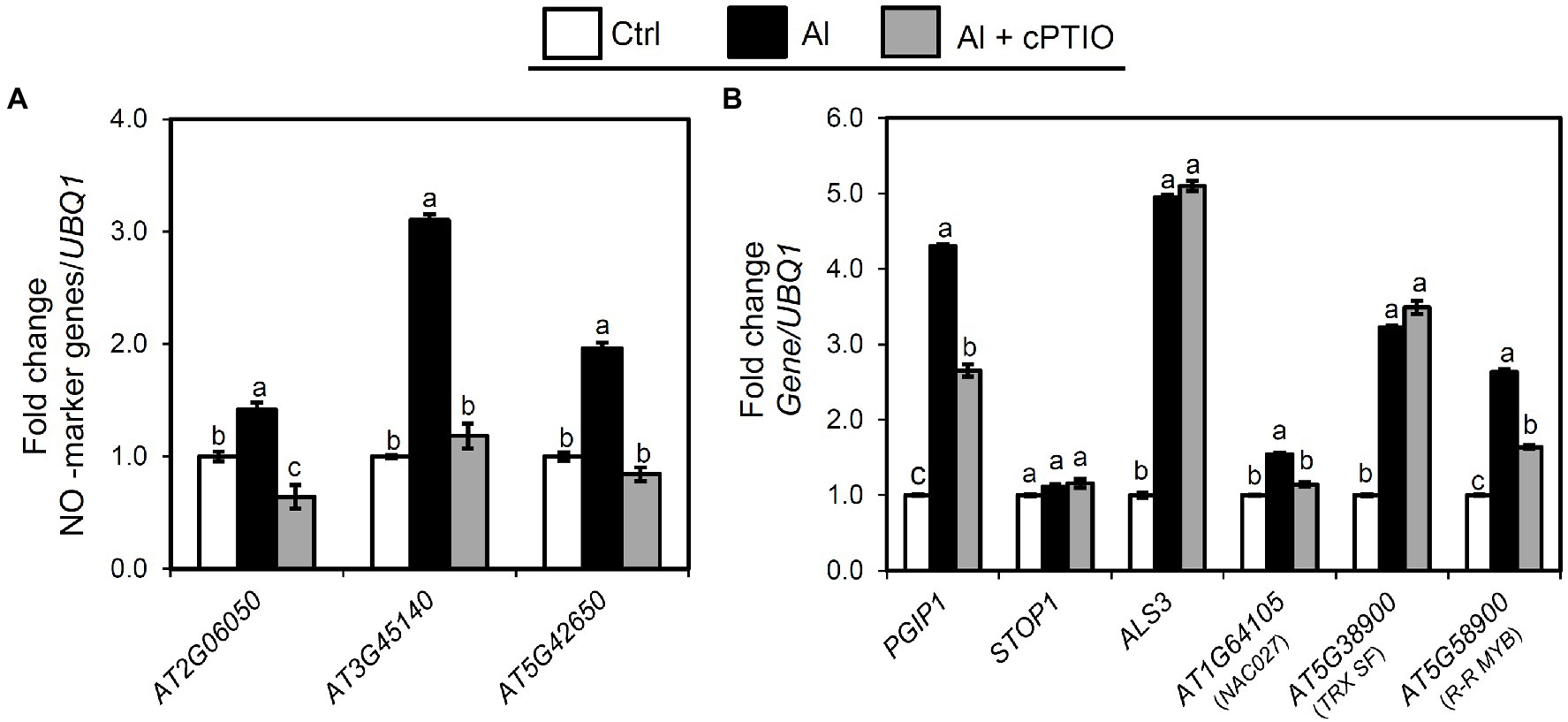
Figure 6. Relationship between Al stress and endogenous NO signaling. Ten-day-old seedlings were exposed to 25 μM Al solution containing 0 or 50 μM cPTIO (NO scavenger) for 24 h. (A) Expression analysis of NO marker genes in shoots after exposure to Al. (B) Effect of cPTIO on expression of the genes identified in eGWAS (NAC27, TRX SF, and R-R MYB), PGIP1, STOP1, and ALS3 in shoots after 24 h Al exposure. UBQ1 was used as an internal control. Average values of three biological replicates are presented with standard errors. Fold change was calculated from control (−Al). Primers used for qRT-PCR are listed in Supplementary Table S2. Different letters indicate a significant difference at p < 0.05 (Tukey test).
We also assessed whether the eGWAS-identified genes that regulate PGIP1 expression function downstream of NO signaling by quantifying their expression levels in the shoots after exposing the roots to Al or Al plus cPTIO for 24 h. We found that only NAC027 expression and R-R MYB expression were significantly suppressed in the Al plus cPTIO samples, while TRX SF remained unchanged (Figure 6B). These results clearly indicate that Al-inducible PGIP1 expression is regulated by the NO signaling pathway through NAC027 and R-R MYB, and this is not regulated by STOP1 (Figure 4A).
Transcriptional Regulation of R-R MYB and NAC027 Included in NO Signaling
The expression of NAC027 and R-R MYB was measured in each KO line. A significant reduction in the expression of NAC027 was observed in the r-r myb compared with the WT expression (Figure 7A). In contrast, the expression of R-R MYB in the nac027 showed no difference compared with wild type (Figure 7B). In addition, the PlantPAN3.0 database (provides TF binding sites in genome-wide promoters based on DAP-seq analysis of various transcription factors) identified that R-R MYB directly binds to the promoter of NAC027 (Figure 7C).
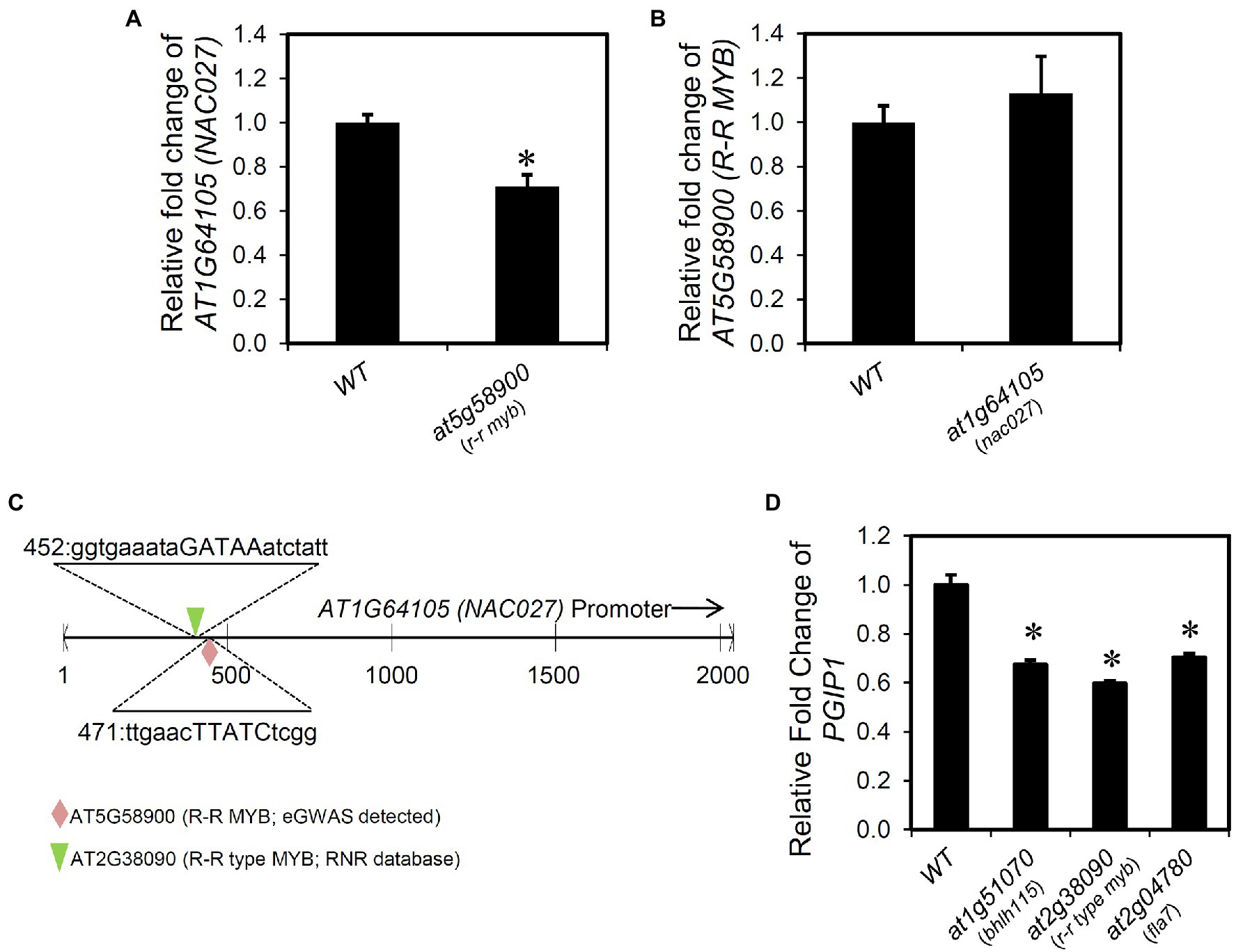
Figure 7. Transcriptional regulation of the genes regulating PGIP1. (A) Relative expression levels of NAC027 in the shoots of WT and r-r myb. (B) Relative expression levels of R–R MYB in the shoots of WT and nac027. (C) AT5G58900 (R-R MYB; eGWAS detected) and AT2G38090 (R-R type MYB; RNR database detected) binding position to the NAC027 (AT1G64105) promoter region retrieved from PlantPAN 3.0 and the corresponding sequence. (D) Relative expression levels of PGIP1 in the shoots of WT and T-DNA insertion mutants of genes (at1g51070, at2g38090, and at2g04780) reported in RNR database that regulate PGIP1 expression. Approximately 100 seedlings of WT and independent homozygous T-DNA insertion mutants were grown for 10 days and treated with 25 μM AlCl3 for 24 h. For NAC027, R-R MYB, and PGIP1, expression was measured by qRT-PCR. Expression levels are expressed as relative fold changes compared to WT. Error bars indicate standard errors across three biological replicates. UBQ1 was used as an internal control. Significant reductions compared to the Al-treated WT sample are indicated by asterisks (Student’s t-test p < 0.05). Primers used for qRT-PCR are listed in Supplementary Table S2.
Interestingly, we found regulators of PGIP1 using the RnR database that is different from the eGWAS-identified factors. The RnR database indicated that overexpression of AT1G51070 [BASIC HELIX-LOOP-HELIX 115 (BHLH115)], AT2G38090 (duplicated homeodomain-like superfamily protein), and AT2G04780 [FASCICLIN-LIKE ARABINOGALACTAN 7 (FLA7)] upregulated PGIP1 expression, which was at the approximately 99th percentile of expression regulation and about 1.3–2.0-fold expression compared with the control. In fact, we confirmed the downregulation of PGIP1 expression in the KO or KD mutants of these three genes under Al stress (Figure 7D). Among them, AT2G38090 is a member of the R-R-type MYB family (Yanhui et al., 2006) and is a close homolog of R-R MYB that we found in the eGWAS. This TF also binds to the NAC027 promoter as assessed by a PlantPAN3.0 database search, similar to R-R MYB (Figure 7C). Co-expression gene network analysis using the six genes (TRX SF, NAC027, R-R MYB, BHLH115, AT2G38090, and FLA7), whose PGIP1 expression levels were decreased in these KO or KD lines, revealed that R-R MYB was co-expressed with AT2G38090 (R-R type MYB) and FLA7 (Supplementary Figure S11). These results suggest that these genes are involved in the direct/indirect regulation of PGIP1 expression.
Discussion
The transcription of Al-tolerance genes is regulated by a complex mechanism (Delhaize et al., 2012) and involves STOP1 and cross talk with other mechanisms related to stress responses (Daspute et al., 2017). In this study, TRX SF, R-R MYB, and NAC027 involved in the regulatory mechanisms of Al-inducible PGIP1 expression were identified through eGWAS of PGIP1 expression levels under Al stress (Figure 3; Table 1). The STOP1-TRX SF pathway regulate PGIP1 expression through the PI signaling pathway via PLC9, while R-R MYB and NAC027 regulate PGIP1 expression through a STOP1-independent NO-signaling pathway (Figures 4, 6, 8). In contrast, the regulation of ALS3 expression via STOP1 in the shoots was independent of these pathways, including TRX SF, R-R MYB, and NAC027 (Figures 6, 8, Supplementary Figure S10). Taken together, the eGWAS of PGIP1 identified a portion of the complex Al signaling pathways in Arabidopsis shoots.
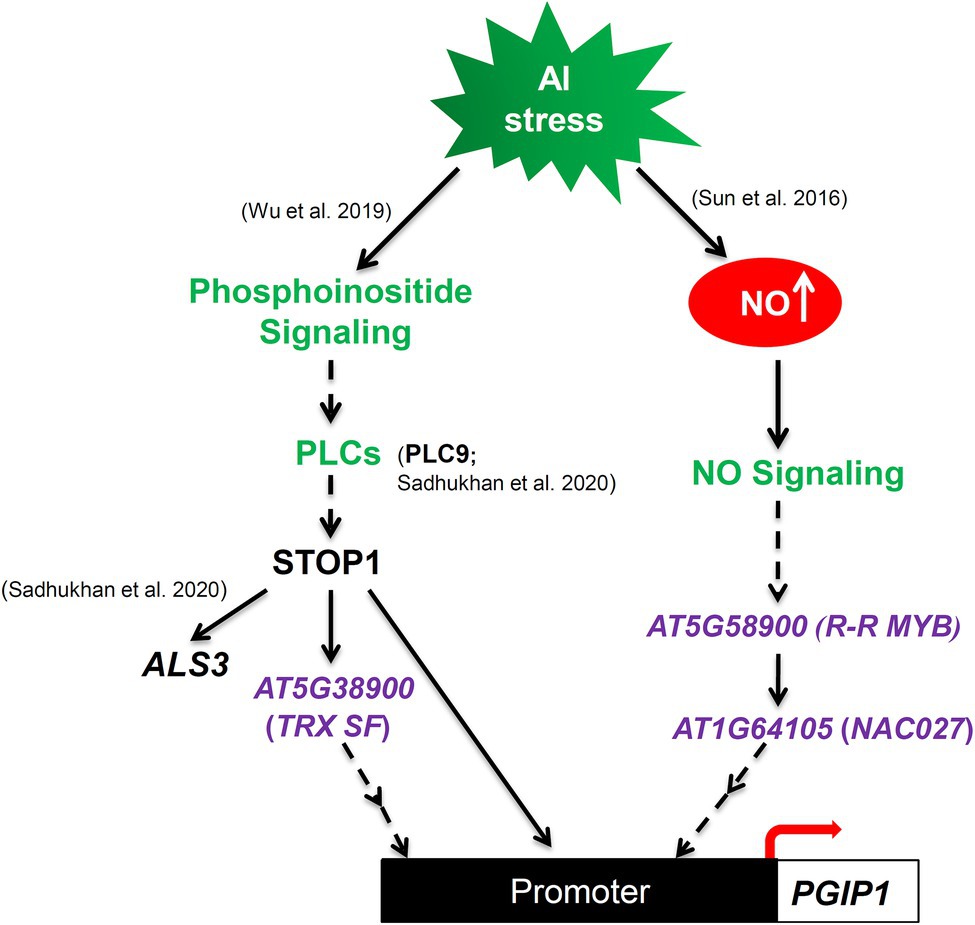
Figure 8. Schematic illustration of proposed model illustrating that Al-induced, STOP1-independent NO-signaling pathway and Al-activated STOP1 signaling cascades regulate PGIP1 expression under Al stress. TRX SF, R-R MYB, and NAC027 were identified by eGWAS for PGIP1 expression levels under Al stress in the shoots of A. thaliana. The colors green and purple represent the previously known Al signaling pathways and newly discovered genes in this study, respectively.
In the current study, we found plausible causative genes involved in the regulation of PGIP1 expression through the candidate gene-based GWAS (Table 1). Many of these genes showed genomic polymorphisms, but further reverse genetic studies clearly revealed that three genes, TRX SF, R-R MYB, and NAC027, were involved in the regulation of PGIP1 expression. The polymorphism responsible for the variation could not be determined in this study. However, using reliable DNA sequences from the 1,001 Genomes Project, we searched for polymorphisms at the three genes in several high- and low-expression accessions. In this process, we observed a haplotype containing polymorphisms in the intron sequences of TRX, associated with its expression levels. The only amino acid polymorphism caused by the detected SNPs was observed in R-R MYB. Another haplotype including a promoter deletion related to ELP was observed in NAC027 (Supplementary Figure S12). Promoter polymorphisms have been found to significantly impact gene expression level variation (Sadhukhan et al., 2017; Meloa et al., 2019; Nakano et al., 2020b; Wang et al., 2021). Functional analysis of the promoter sequence polymorphisms of NAC027 will shed light on their potential role in the differential regulation of gene expression.
A key regulatory transcription factor, STOP1, regulates the expression of various genes involved in Al tolerance along with PGIP1 (Sawaki et al., 2009; Wu et al., 2019), in which some genes are directly regulated. Our previous studies found that STOP1 directly regulates transcription of several downstream genes by binding to their promoters under not only Al but also other stress conditions: STOP1 binds to the promoter of Al-inducible AtALMT1 (Tokizawa et al., 2015) and AtMATE (Nakano et al., 2020b), low-oxygen-inducible HsfA2 (Enomoto et al., 2019), and NaCl-inducible CPK23 (Sadhukhan et al., 2019). In the present study, promoter analyses (i.e., Cistrome database and in vitro promoter assays; Figure 5) identified a functional STOP1-binding site in the promoters of PGIP1 and TRX SF. This indicates that STOP1 directly activates the Al-inducible expression of PGIP1 and TRX SF. The promoter regions of each gene commonly possessed the minimum consensus sequence of ART1/STOP1 [GGN(T/g/a/C)V(C/A/g)S(C/G); Tsutsui et al., 2011; Tokizawa et al., 2015], although the surrounding sequences of the consensus were different.
Similarly, we found that STOP1 may directly regulate TRX SF expression (Figures 4, 5), while TRX SF affected the PGIP1 expression but not STOP1 expression (Figure 3, Supplementary Figure S10), whose regulation contributed to Al tolerance (Supplementary Figure S9). One possible function of TRX SF is to control regulatory proteins by oxidative protein modification, which is a common mechanism of thioredoxin superfamily proteins (Lemaire and Miginiac-Maslow, 2004; Schmidtmann et al., 2014; Mata-Perez and Spoel, 2019). The TRX family regulates gene expression through redox activation of receptors and transcription factors under salicylic acid (SA) and brassinosteroid signaling (Ding et al., 2018; Tian et al., 2018). We previously revealed that TRX1 contributed to Al tolerance using GWAS for Al tolerance in Arabidopsis (Nakano et al., 2020a). These results suggest that TRX-mediated redox signaling is involved in gene regulation related to Al tolerance. However, ALS3 was not involved in the signaling (Supplementary Figure S10), despite the fact that ALS3 expression in shoots is regulated by STOP1 under Al stress, similar to that of PGIP1 (Sawaki et al., 2016).
We found that both NAC027 and R-R MYB (transcription factors respond to various biotic and abiotic stresses, Ascencio-Ibanez et al., 2008; Soitamo et al., 2008; Fang et al., 2018) responded to Al stress and were involved in STOP1-independent regulation of PGIP1, where they function together in the NO signaling pathway via R-R MYB binding to the NAC027 promoter (Figures 3, 4, 6, 7). However, the contribution of NAC027 to regulation and Al tolerance does not seem to be largely comparable to that of R-R MYB (Figure 3, Supplementary Figure S9), suggesting that R-R MYB, upstream of NAC027, also regulates other genes. NO signaling has been reported to be a second messenger of Al-inducible expression of several Al-tolerance genes (He et al., 2012). Additionally, PECTIN METHYLESTERASE 3, which is induced and activated by Al-dependent NO signaling (Sun et al., 2016; Ye et al., 2018), was detected as a co-expressed gene of R-R MYB and the close homolog of MYB regulating PGIP1 (Figure 7D, Supplementary Figure S11). These results suggest that the co-expressed module involved in NO signaling is related to the Al stress response. In contrast, STOP1 regulated TRX SF along with ALS3 (Figure 6), which was independent of NO signaling not included in the co-expression network.
Furthermore, several genes involved in plant cell wall biogenesis, UDP-GLUCOSE DEHYDROGENASE 4, COTTON GOLGI-RELATED 3, and FLA7, were included in the co-expression network. The cell wall plays important roles not only in the regulation of plant growth and development, but also in the perception and expression of Al toxicity (Tabuchi and Matsumoto, 2001; Eticha et al., 2005; Yang et al., 2011; Kobayashi et al., 2013; Zhu et al., 2014; Kochian et al., 2015; Sun et al., 2016). The PGIP1 was shown to be included in Al tolerance (Supplementary Figure S9), but the details of its role in Al tolerance have not been studied yet; one possibility is that it can protect the binding of Al to negatively charged ligands [e.g., polygalacturonic acid (PGA)] induced by demethylation of cell wall pectin, which is enhanced by Al (Supplementary Figures S4, S13: the degree of pectin methylesterification in Al-treated seedlings decreased to 55% of that without Al treatment). It has been reported that PGIP1 can bind to the PGA region and support the formation of a normal pectin network under biotic stress conditions (Spadoni et al., 2006). A similar alleviation was observed under proton-toxic conditions in the stop1, which showed very low expression of PGIP1 (Kobayashi et al., 2014). Under Al stress conditions, PGIP1 binding to the PGA region might reduce the formation of abnormal pectin networks, which might be caused by unusual Al binding to the PGA region. Further characterization of these events would be useful for identifying the role of PGIP1 in Al tolerance.
Conclusion
Through a candidate gene-based GWAS of PGIP1 expression, we successfully identified complex signaling of PGIP1 in response to Al stress in A. thaliana. Furthermore, we propose a model to illustrate that PGIP1 expression is regulated by a STOP1-dependent Al-induced phosphoinositide (PI) signaling through AT5G38900 (TRX superfamily protein) and STOP1-independent Al-induced endogenous NO signaling through AT1G64105 (NAC027 transcription factor) and AT5G58900 (R-R type MYB transcription factor; Figure 8). In addition, our study demonstrates the utility of an eGWAS in understanding the genetic regulation of Al signaling by exploiting the natural variation in the expression levels of key Al-responsive genes. Although a limited number of accessions were used in the current study, a future eGWAS using denser SNP information and a larger accession set, available in recent years (Togninalli et al., 2018), will open up new avenues for better understanding of Al stress signaling in plants.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
RA, YK, and HK conceived and designed the research. RA, TE, EY, TW, HI, AS, and SI performed the experiments. RA, TE, and YN analyzed the data. HK, SP, YY, and YK supervised the study. RA, YK, and HK wrote the manuscript. HK, YK, YY, and MK contributed to new reagents or analytical tools. All authors have approved the manuscript.
Funding
This work was supported by JSPS KAKENHI grant Numbers 19K05753 and 21H02088.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
RA is thankful to the Japanese Government (Monbukagakusho: MEXT) for the scholarship. YK is thankful to Gifu University Young and Mid-Career Researcher Overseas Training Program. The authors are thankful to Shiori Tai of Gifu University for assistance with phenotyping. We are grateful for the technical support provided by Mutsutomo Tokizawa at Gifu University, and Fumie Mori at RIKEN BRC. We thank the RIKEN BRC, ABRC, and NASC for providing Arabidopsis seeds. We also thankful to editage English editing service.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.774687/full#supplementary-material
Footnotes
1. ^http://signal.salk.edu/tdnaprimers.2.html
2. ^https://cynin.gmi.oeaw.ac.at/
3. ^http://1001genomes.org/index.html
5. ^http://www.arabidopsis.org
6. ^https://www.arabidopsis.org/tools/bulk/go/index.jsp
7. ^http://signal.salk.edu/atg1001/3.0/gebrowser.php
8. ^http://polymorph.weigelworld.org/cgi-bin/webapp.cgi
9. ^http://webs2.kazusa.or.jp/kagiana/rnr0912/indexff.html
11. ^http://PlantPan.mbc.nctu.edu.tw
References
Agrahari, R. K., Kobayashi, Y., Borgohain, P., Panda, S. K., and Koama, H. (2020). Aluminum-specific upregulation of GmALS3 in the shoots of soybeans: a potential biomarker for managing soybean production in acidic soil regions. Agronomy 10:1228. doi: 10.3390/agronomy10091228
Ascencio-Ibanez, J. T., Sozzani, R., Lee, T. J., Chu, T. M., Wolfinger, R. D., Cella, R., et al. (2008). Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol. 148, 436–454. doi: 10.1104/pp.108.121038
Atwell, S., Huang, Y. S., Vilhjálmsson, B. J., Willems, G., Horton, M., Li, Y., et al. (2010). Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 465, 627–631. doi: 10.1038/nature08800
Bethke, G., and Glazebrook, J. (2014). Cyclohexane diamine tetraacetic acid (CDTA) extraction of plant cell wall pectin. Bio Protoc. 4:e1357. doi: 10.21769/BioProtoc.1357
Bradbury, P. J., Zhang, Z., Kroon, D. E., Casstevens, T. M., Ramdoss, Y., and Buckler, E. S. (2007). TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23, 2633–2635. doi: 10.1093/bioinformatics/btm308
Bustin, S. A., Benes, V., Garson, J. A., Hellemans, J., Huggett, J., Kubista, M., et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622. doi: 10.1373/clinchem.2008.112797
Cao, J., Schneeberger, K., Ossowski, S., Günther, T., Bender, S., Fitz, J., et al. (2011). Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat. Genet. 43, 956–963. doi: 10.1038/ng.911
Chang, W. C., Lee, T. Y., Huang, H. D., and Pan, R. L. (2008). PlantPAN: plant promoter analysis navigator for identifying combinatorial cis-regulatory elements with distance constrain in plant gene group. BMC Genomics 9:561. doi: 10.1186/1471-2164-9-561
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for agrobacterium -mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
D’Alessandro, S., Posocco, B., Costa, A., Zahariou, G., Schiavo, F. L., Carbonera, D., et al. (2013). Limits in the use of cPTIO as nitric oxide scavenger and EPR probe in plant cells and seedlings. Front. Plant Sci. 4:340. doi: 10.3389/fpls.2013.00340
Daspute, A. A., Sadhukhan, A., Tokizawa, M., Kobayashi, Y., Panda, S. K., and Koyama, H. (2017). Transcriptional regulation of aluminum-tolerance genes in higher plants: clarifying the underlying molecular mechanisms. Front. Plant Sci. 8:1358. doi: 10.3389/fpls.2017.01358
Davidsson, P., Broberg, M., Kariola, T., Sipari, N., Pirhonen, M., and Palva, E. T. (2017). Short oligogalacturonides induce pathogen resistance-associated gene expression in Arabidopsis thaliana. BMC Plant Biol. 17:19. doi: 10.1186/s12870-016-0959-1
Delhaize, E., Ma, J. F., and Ryan, P. R. (2012). Transcriptional regulation of aluminium tolerance genes. Trends Plant Sci. 17, 341–348. doi: 10.1016/j.tplants.2012.02.008
Delhaize, E., and Ryan, P. R. (1995). Aluminium toxicity and tolerance in plants. Plant Physiol. 107, 315–321. doi: 10.1104/pp.107.2.315
Ding, Y., Sun, T., Ao, K., Peng, Y., Zhang, Y., Li, X., et al. (2018). Opposite roles of salicylic acid receptors NPR1 and NPR3/NPR4 in transcriptional regulation of plant immunity. Cell 173, 1454–1467.e15. doi: 10.1016/j.cell.2018.03.044
Ding, Z. J., Yan, J. Y., Xu, X. Y., Li, G. X., and Zheng, S. J. (2013). WRKY46 functions as a transcriptional repressor of ALMT1, regulating aluminum-induced malate secretion in Arabidopsis. Plant J. 76, 825–835. doi: 10.1111/tpj.12337
Enomoto, T., Tokizawa, M., Ito, H., Iuchi, S., Kobayashi, M., Yamamoto, Y. Y., et al. (2019). STOP1 regulates the expression of HsfA2 and GDHs that are critical for low-oxygen tolerance in Arabidopsis. J. Exp. Bot. 70, 3297–3311. doi: 10.1093/jxb/erz124
Eticha, D., Stass, A., and Horst, W. J. (2005). Cell-wall pectin and its degree of methylation in the maize root-apex: significance for genotypic differences in aluminium resistance. Plant Cell Environ. 28, 1410–1420. doi: 10.1111/j.1365-3040.2005.01375.x
Fang, Q., Wang, Q., Mao, H., Xu, J., Wang, Y., Hu, H., et al. (2018). AtDIV2, an R-R-type MYB transcription factor of Arabidopsis, negatively regulates salt stress by modulating ABA signaling. Plant Cell Rep. 37, 1499–1511. doi: 10.1007/s00299-018-2321-6
Farrás, R., Ferrando, A., Jásik, J., Kleinow, T., Ökrész, L., Tiburcio, A., et al. (2001). SKP1–SnRK protein kinase interactions mediate proteasomal binding of a plant SCF ubiquitin ligase. EMBO J. 20, 2742–2756. doi: 10.1093/emboj/20.11.2742
Ferrari, S., Vairo, D., Ausubel, F. M., Cervone, F., and De Lorenzo, G. (2003). Tandemly duplicated Arabidopsis genes that encode polygalacturonase-inhibiting proteins are regulated coordinately by different signal transduction pathways in response to fungal infection. Plant Cell 15, 93–106. doi: 10.1105/tpc.005165
Fujiwara, T., Hirai, M. Y., Chino, M., Komeda, Y., and Naito, S. (1992). Effects of sulfur nutrition on expression of the soybean seed storage protein genes in transgenic petunia. Plant Physiol. 99, 263–268. doi: 10.1104/pp.99.1.263
He, H., Zhan, J., He, L., and Gu, M. (2012). Nitric oxide signaling in aluminum stress in plants. Protoplasma 249, 483–492. doi: 10.1007/s00709-011-0310-5
Horton, M. W., Hancock, A. M., Huang, Y. S., Toomajian, C., Atwell, S., Auton, A., et al. (2012). Genome-wide patterns of genetic variation in worldwide Arabidopsis thaliana accessions from the RegMap panel. Nat. Genet. 44, 212–216. doi: 10.1038/ng.1042
Huang, X., Stettmaier, K., Michel, C., Hutzler, P., Mueller, M. J., and Durner, J. (2004). Nitric oxide is induced by wounding and influences jasmonic acid signaling in Arabidopsis thaliana. Planta 218, 938–946. doi: 10.1007/s00425-003-1178-1
Iuchi, S., Koyama, H., Iuchi, A., Kobayashi, Y., Kitabayashi, S., Kobayashi, Y., et al. (2007). Zinc finger protein STOP1 is critical for proton tolerance in Arabidopsis and coregulates a key gene in aluminum tolerance. Proc. Natl. Acad. Sci. U. S. A. 104, 9900–9905. doi: 10.1073/pnas.0700117104
Kobayashi, Y., Hoekenga, O. A., Itoh, H., Nakashima, M., Saito, S., Shaff, J. E., et al. (2007). Characterization of AtALMT1 expression in aluminum-inducible malate release and its role for rhizotoxic stress tolerance in Arabidopsis. Plant Physiol. 145, 843–852. doi: 10.1104/pp.107.102335
Kobayashi, Y., Kobayashi, Y., Watanabe, T., Shaff, J. E., Ohta, H., Kochian, L. V., et al. (2013). Molecular and physiological analysis of Al3+ and H+ rhizotoxicities at moderately acidic conditions. Plant Physiol. 163, 180–192. doi: 10.1104/pp.113.222893
Kobayashi, Y., Ohyama, Y., Kobayashi, Y., Ito, H., Iuchi, S., Fujita, M., et al. (2014). STOP2 activates transcription of several genes for Al- and low pH-tolerance that are regulated by STOP1 in Arabidopsis. Mol. Plant 7, 311–322. doi: 10.1093/mp/sst116
Kobayashi, Y., Sadhukhan, A., Tazib, T., Nakano, Y., Kusunoki, K., Kamara, M., et al. (2016). Joint genetic and network analyses identify loci associated with root growth under NaCl stress in Arabidopsis thaliana. Plant Cell Environ. 39, 918–934. doi: 10.1111/pce.12691
Kochian, L. V., Piñeros, M. A., Liu, J., and Magalhaes, J. V. (2015). Plant adaptation to acid soils: the molecular basis for crop aluminum resistance. Annu. Rev. Plant Biol. 66, 571–598. doi: 10.1146/annurev-arplant-043014-114822
Koyama, H., Kawamura, A., Kihara, T., Hara, T., Takita, E., and Shibata, D. (2000). Overexpression of mitochondrial citrate synthase in Arabidopsis thaliana improved growth on a phosphorus-limited soil. Plant Cell Physiol. 41, 1030–1037. doi: 10.1093/pcp/pcd029
Larsen, P. B., Geisler, M. J. B., Jones, C. A., Williams, K. M., and Cancel, J. D. (2005). ALS3 encodes a phloem-localized ABC transporter-like protein that is required for aluminum tolerance. Plant J. 41, 353–363. doi: 10.1111/j.1365-313X.2004.02306.x
Larsen, P. B., Kochian, L. V., and Howell, S. H. (1997). Al inhibits both shoot development and root growth in als3, an Al-sensitive Arabidopsis mutant. Plant Physiol. 114, 1207–1214. doi: 10.1104/pp.114.4.1207
Lemaire, S. D., and Miginiac-Maslow, M. (2004). The thioredoxin superfamily in Chlamydomonas reinhardtii. Photosynth. Res. 82, 203–220. doi: 10.1007/s11120-004-1091-x
Ligaba-Osena, A., Fei, Z., Liu, J., Xu, Y., Shaff, J., Lee, S. C., et al. (2017). Loss-of-function mutation of the calcium sensor CBL1 increases aluminum sensitivity in Arabidopsis. New Phytol. 214, 830–841. doi: 10.1111/nph.14420
Liu, J., Pineros, M. A., and Kochian, L. V. (2014). The role of aluminum sensing and signaling in plant aluminum resistance. J. Integr. Plant Biol. 56, 221–230. doi: 10.1111/jipb.12162
Mata-Perez, C., and Spoel, S. H. (2019). Thioredoxin-mediated redox signalling in plant immunity. Plant Sci. 279, 27–33. doi: 10.1016/j.plantsci.2018.05.001
Meloa, J. O., Martinsc, L. G. C., Barrosa, B. A., Pimentac, M. R., Lanaa, U. G. P., Duartec, C. E. M., et al. (2019). Repeat variants for the SbMATE transporter protect sorghum roots from aluminum toxicity by transcriptional interplay in cis and trans. Proc. Natl. Acad. Sci. U. S. A. 116, 313–318. doi: 10.1073/pnas.1808400115
Nakano, Y., Kusunoki, K., Hoekenga, O., Tanaka, K., Iuchi, S., Sakata, Y., et al. (2020a). Genome-wide association study and genomic prediction elucidate the distinct genetic architecture of aluminum and proton tolerance in Arabidopsis thaliana. Front. Plant Sci. 11:405. doi: 10.3389/fpls.2020.00405
Nakano, Y., Kusunoki, K., Maruyama, H., Enomoto, T., Tokizawa, M., Iuchi, S., et al. (2020b). A single-population GWAS identified AtMATE expression level polymorphism caused by promoter variants is associated with variation in aluminum tolerance in a local Arabidopsis population. Plant Direct 4:e00250. doi: 10.1002/pld3.250
Obayashi, T., Aoki, Y., Tadaka, S., Kagaya, Y., and Kinoshita, K. (2018). ATTED-II in 2018: a plant coexpression database based on investigation of the statistical property of the mutual rank index. Plant Cell Physiol. 59:e3. doi: 10.1093/pcp/pcx191
Ohyama, Y., Ito, H., Kobayashi, Y., Ikka, T., Morita, A., Kobayashi, M., et al. (2013). Characterization of AtSTOP1 orthologous genes in tobacco and other plant species. Plant Physiol. 162, 1937–1946. doi: 10.1104/pp.113.218958
Sadhukhan, A., Agrahari, R. K., Wu, L., Watanabe, T., Nakano, Y., Panda, S. K., et al. (2020). Expression genome-wide association study identifies that phosphatidylinositol-derived signalling regulates ALUMINIUM SENSITIVE3 expression under aluminium stress in the shoots of Arabidopsis thaliana. Plant Sci. 302:110711. doi: 10.1016/j.plantsci.2020.110711
Sadhukhan, A., Enomoto, T., Kobayashi, Y., Watanabe, T., Iuchi, S., Kobayashi, M., et al. (2019). Sensitive to proton rhizotoxicity1 regulates salt and drought tolerance of Arabidopsis thaliana through transcriptional regulation of CIPK23. Plant Cell Physiol. 60, 2113–2126. doi: 10.1093/pcp/pcz120
Sadhukhan, A., Kobayashi, Y., Nakano, Y., Iuchi, S., Kobayashi, M., Sahoo, L., et al. (2017). Genome-wide association study reveals that the aquaporin NIP1;1 contributes to variation in hydrogen peroxide sensitivity in Arabidopsis thaliana. Mol. Plant 10, 1082–1094. doi: 10.1016/j.molp.2017.07.003
Sakurai, N., Ara, T., Enomoto, M., Motegi, T., Morishita, Y., Kurabayashi, A., et al. (2014). Tools and databases of the komics web portal for preprocessing, mining, and dissemination of metabolomics data. Biomed. Res. Int. 2014:194812. doi: 10.1155/2014/194812
Sawaki, Y., Iuchi, S., Kobayashi, Y., Kobayashi, Y., Ikka, T., Sakurai, N., et al. (2009). STOP1 regulates multiple genes that protect Arabidopsis from proton and aluminum toxicities. Plant Physiol. 150, 281–294. doi: 10.1104/pp.108.134700
Sawaki, K., Sawaki, Y., Zhao, C. R., Kobayashi, Y., and Koyama, H. (2016). Specific transcriptomic response in the shoots of Arabidopsis thaliana after exposure to Al rhizotoxicity?: - potential gene expression biomarkers for evaluating Al toxicity in soils. Plant Soil 409, 131–142. doi: 10.1007/s11104-016-2960-8
Schmidtmann, E., König, A.-C., Orwat, A., Leister, D., Hartl, M., and Finkemeier, I. (2014). Redox regulation of Arabidopsis mitochondrial citrate synthase. Mol. Plant 7, 156–169. doi: 10.1093/mp/sst144
Sevilla, F., Camejo, D., Ortiz-Espín, A., Calderón, A., Lázaro, J. J., and Jiménez, A. (2015). The thioredoxin/peroxiredoxin/sulfiredoxin system: current overview on its redox function in plants and regulation by reactive oxygen and nitrogen species. J. Exp. Bot. 66, 2945–2955. doi: 10.1093/jxb/erv146
Shi, H., Chen, Y., Tan, D.-X., Reiter, R. J., Chan, Z., and He, C. (2015). Melatonin induces nitric oxide and the potential mechanisms relate to innate immunity against bacterial pathogen infection in Arabidopsis. J. Pineal Res. 59, 102–108. doi: 10.1111/jpi.12244
Siedlecka, A., Wiklund, S., Peronne, M. A., Micheli, F., Lesniewska, J., Sethson, I., et al. (2008). Pectin methyl esterase inhibits intrusive and symplastic cell growth in developing wood cells of Populus. Plant Physiol. 146, 323–324. doi: 10.1104/pp.107.111963
Soitamo, A. J., Piippo, M., Allahverdiyeva, Y., Battchikova, N., and Aro, E. M. (2008). Light has a specific role in modulating Arabidopsis gene expression at low temperature. BMC Plant Biol. 8:13. doi: 10.1186/1471-2229-8-13
Song, Y., Chen, P., Xuan, A., Bu, C., Liu, P., Ingvarsson, P. K., et al. (2021). Integration of genome wide association studies and co-expression networks reveal roles of PtoWRKY 42-PtoUGT76C1-1 in trans-zeatin metabolism and cytokinin sensitivity in poplar. New Phytol. 231, 1462–1477. doi: 10.1111/nph.17469
Spadoni, S., Zabotina, O., Di Matteo, A., Mikkelsen, J. D., Cervone, F., Dl, G., et al. (2006). Polygalacturonase-inhibiting protein (PGIP) interacts with pectin through a binding site formed by four clustered residues of arginine and lysine. Plant Physiol. 141, 557–564. doi: 10.1104/pp.106.076950
Sun, C., Lu, L., Yu, Y., Liu, L., Hu, Y., Ye, Y., et al. (2016). Decreasing methylation of pectin caused by nitric oxide leads to higher aluminium binding in cell walls and greater aluminium sensitivity of wheat roots. J. Exp. Bot. 67, 979–989. doi: 10.1093/jxb/erv514
Tabuchi, A., and Matsumoto, H. (2001). Changes in cell-wall properties of wheat (Triticum aestivum) roots during aluminum-induced growth inhibition. Physiol. Plant. 112, 353–358. doi: 10.1034/j.1399-3054.2001.1120308.x
Tian, Y., Fan, M., Qin, Z., Lv, H., Wang, M., Zhang, Z., et al. (2018). Hydrogen peroxide positively regulates brassinosteroid signaling through oxidation of the BRASSINAZOLE-RESISTANT1 transcription factor. Nat. Commun. 9:1063. doi: 10.1038/s41467-018-03463-x
Togninalli, M., Seren, Ü., Meng, D., Fitz, J., Nordborg, M., Weigel, D., et al. (2018). The AraGWAS catalog: a curated and standardized Arabidopsis thaliana GWAS catalog. Nucleic Acids Res. 46, D1150–D1156. doi: 10.1093/nar/gkx954
Tokizawa, M., Kobayashi, Y., Saito, T., Kobayashi, M., Iuchi, S., Nomoto, M., et al. (2015). Sensitive to proton rhizotoxicity1, calmodulin binding transcription activator2, and other transcription factors are involved in aluminum-activated malate transporter1 expression. Plant Physiol. 167, 991–1003. doi: 10.1104/pp.114.256552
Tsutsui, T., Yamaji, N., and Ma, J. (2011). Identification of a cis-acting element of ART1, a C2H2-type zinc-finger transcription factor for aluminum tolerance in rice. Plant Physiol. 156, 925–931. doi: 10.1104/pp.111.175802
Wang, Y., Tao, Z., Wang, W., Filiault, D., Qiu, C., Wang, C., et al. (2020). Molecular variation in a functionally divergent homolog of FCA regulates flowering time in Arabidopsis thaliana. Nat. Commun. 11:5830. doi: 10.1038/s41467-020-19666-0
Wang, Z., Yang, L., Wu, D., Zhang, N., and Hua, J. (2021). Polymorphisms in cis-elements confer SAUR26 gene expression difference for thermo-response natural variation in Arabidopsis. New Phytol. 229, 2751–2764. doi: 10.1111/nph.17078
Watanabe, T., Urayama, M., Shinano, T., Okada, R., and Osaki, M. (2015). Application of ionomics to plant and soil in fields under long-term fertilizer trials. Springerplus 4:781. doi: 10.1186/s40064-015-1562-x
Wu, L., Sadhukhan, A., Kobayashi, Y., Ogo, N., Tokizawa, M., Agrahari, R. K., et al. (2019). Involvement of phosphatidylinositol metabolism in aluminum-induced malate secretion in Arabidopsis. J. Exp. Bot. 70, 3329–3342. doi: 10.1093/jxb/erz179
Yang, J. L., Zhu, X. F., Peng, Y. X., Zheng, C., Li, G. X., Liu, Y., et al. (2011). Cell wall hemicellulose contributes significantly to aluminum adsorption and root growth in Arabidopsis. Plant Physiol. 155, 1885–1892. doi: 10.1104/pp.111.172221
Yanhui, C., Xiaoyuan, Y., Kun, H., Meihua, L., Jigang, L., Zhaofeng, G., et al. (2006). The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol. Biol. 60, 107–124. doi: 10.1007/s11103-005-2910-y
Ye, Y., Chunyan, D., Lanping, G., Yuan, Q., Xiaoyan, Y., Qi, C., et al. (2018). Distribution pattern of aluminum in Panax notoginseng, a native medicinal plant adapted to acidic red soils. Plant Soil 423, 375–384. doi: 10.1007/s11104-017-3510-8
Zhang, M., Ye, J., Xu, Q., Feng, Y., Yuan, X., Yu, H., et al. (2018). Genome-wide association study of cold tolerance of Chinese indica rice varieties at the bud burst stage. Plant Cell Rep. 37, 529–539. doi: 10.1007/s0029
Zhao, C., Zayed, O., Yu, Z., Jiang, W., Zhu, P., Hsu, C. C., et al. (2018). Leucine-rich repeat extensin proteins regulate plant salt tolerance in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 115, 13123–13128. doi: 10.1073/pnas.1816991115
Zhou, Y., Xu, X. Y., Chen, L. Q., Yang, J. L., and Zheng, S. J. (2012). Nitric oxide exacerbates Al-induced inhibition of root elongation in rice bean by affecting cell wall and plasma membrane properties. Phytochemistry 76, 46–51. doi: 10.1016/j.phytochem.2011.12.004
Keywords: aluminum stress, Arabidopsis thaliana, GWAS, PGIP1, phosphoinositide signaling, transcription factor, NO signaling, STOP1
Citation: Agrahari RK, Enomoto T, Ito H, Nakano Y, Yanase E, Watanabe T, Sadhukhan A, Iuchi S, Kobayashi M, Panda SK, Yamamoto YY, Koyama H and Kobayashi Y (2021) Expression GWAS of PGIP1 Identifies STOP1-Dependent and STOP1-Independent Regulation of PGIP1 in Aluminum Stress Signaling in Arabidopsis. Front. Plant Sci. 12:774687. doi: 10.3389/fpls.2021.774687
Edited by:
Marouane Baslam, Niigata University, JapanReviewed by:
Yasuhiro Sato, University of Zurich, SwitzerlandYunfeng Liu, Guangxi University, China
Copyright © 2021 Agrahari, Enomoto, Ito, Nakano, Yanase, Watanabe, Sadhukhan, Iuchi, Kobayashi, Panda, Yamamoto, Koyama and Kobayashi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuriko Kobayashi, a195dXJpa29AZ2lmdS11LmFjLmpw
 Raj Kishan Agrahari
Raj Kishan Agrahari Takuo Enomoto
Takuo Enomoto Hiroki Ito1
Hiroki Ito1 Yuki Nakano
Yuki Nakano Toshihiro Watanabe
Toshihiro Watanabe Ayan Sadhukhan
Ayan Sadhukhan Yoshiharu Y. Yamamoto
Yoshiharu Y. Yamamoto Hiroyuki Koyama
Hiroyuki Koyama Yuriko Kobayashi
Yuriko Kobayashi