- 1State Key Laboratory of Crop Stress Biology for Arid Areas, College of Horticulture, Northwest A&F University, Yangling, China
- 2Shaanxi Research Center of Apple Engineering and Technology, Yangling, China
- 3Shaanxi Key Laboratory Bio-resources, College of Bioscience and Engineering, Shaanxi University of Technology, Hanzhong, China
The red coloration of apple (Malus × domestica Borkh.) is due to the accumulation of anthocyanins in the fruit peel. Light is essential for anthocyanin biosynthesis in apple. In this study, we performed a transcriptome sequencing (RNA-seq) analysis of apple fruit exposed to light after unbagging. The identified differentially expressed genes included MdBBX21, which is homologous to Arabidopsis BBX21, suggesting it may be involved in light-induced anthocyanin biosynthesis. Additionally, MdBBX21 was localized in the nucleus and its gene was expressed earlier than MdMYB1 in apple peel treated with light. Overexpressing MdBBX21 in Arabidopsis and apple calli under light increased anthocyanin accumulation. Dual-luciferase and yeast one-hybrid assays confirmed that MdBBX21 binds to the MdHY5, MdBBX20, and MdBBX22-1/2 promoters and induces expression. At the same time, MdHY5 can also activate the expression of MdBBX21. Furthermore, bimolecular fluorescence complementation and yeast two-hybrid assays demonstrated that MdBBX21 can interact with MdHY5. This interaction can significantly enhance MdMYB1 promoter activity. These findings clarify the molecular mechanism by which MdBBX21 positively regulates light-induced anthocyanin accumulation in apple.
Introduction
Apples are one of the most widely cultivated fruits worldwide. Red-skinned apples are more popular with consumers than green-skinned or yellow-skinned apples. The redness of the peel is mainly determined by anthocyanins, which are natural water-soluble pigments (Kim et al., 2003; Zhang Y. et al., 2014). Under natural conditions, anthocyanins are stored in plant vacuoles in the form of glycosides (Passeri et al., 2016). The diversity in the colors of flowers, stems, leaves, fruits, and other plant organs and tissues is due to the type, content, and distribution of anthocyanins (Lawrence et al., 1939; Zhang H. et al., 2014).
Anthocyanins are produced by the flavonoid pathway in a process that is controlled by a series of structural genes and regulatory factors (Winkel-Shirley, 2001). The structural genes (e.g., early and late biosynthetic genes) encode various enzymes in the plant anthocyanin biosynthetic pathway. The early biosynthetic genes include those encoding chalcone synthase (CHS), chalcone isomerase (CHI), and flavanone 3-hydroxylase (F3H), whereas the late biosynthetic genes include those encoding dihydroflavonol 4-reductase (DFR), anthocyanidin synthase (ANS), and flavonoid 3-O-glycosyltransferase (UFGT) (Martin et al., 1991; Kubasek et al., 1992). Transcriptional regulators control the expression of structural genes (Quattrocchio et al., 1993, 1998; de Vetten et al., 1997). For example, MYB proteins, which are among the most important transcription factors, can combine with bHLH and WD40 proteins to form an MBW complex that binds to the promoter of structural genes to induce expression and regulate anthocyanin synthesis (Ramsay and Glover, 2005; Li, 2014). In apple, MdMYB1 and MdMYBA are responsible for apple skin coloration (Takos A. M. et al., 2006; Ban et al., 2007). Five direct tandem repeats of the MdMYB10-binding motif in the MdMYB10 promoter are associated with the accumulation of anthocyanins throughout the plant, ultimately resulting in striking phenotypes (e.g., red fruit flesh and red foliage) (Espley et al., 2007, 2009). By aligning the MdMYB1 sequence with the GDDH13 and HFTH1 genomes, a long terminal repeat retrotransposon associated with the red-skinned phenotype was detected upstream of MdMYB1 (Zhang et al., 2019). These results strongly indicate that the MdMYB1/A/10 alleles are the core transcriptional regulatory genes of the anthocyanin biosynthesis pathway.
In addition to being genetically regulated, anthocyanin production is also affected by various environmental factors, including drought (An et al., 2020), low temperatures (Xie et al., 2012), salt stress (Lotkowska et al., 2015), low nitrogen availability (Sun et al., 2018), and light (Cominelli et al., 2008). For the biosynthesis of anthocyanins in the apple peel, light is the most important environmental factor (Merzlyak and Chivkunova, 2000). The mechanism underlying light-induced anthocyanin synthesis has been well characterized as part of photomorphogenesis in the model plant Arabidopsis (Maier et al., 2013). Light signals are perceived by plants and then transmitted by photoreceptor proteins such as phytochrome (PHY) (Sharrock and Quail, 1989), cryptochrome (CRY) (Lin et al., 1995), phototropin (PHOT) (Kasahara et al., 2002), and ultraviolet light receptor (UVR8) (Rizzini et al., 2011). Different photoreceptor proteins can perceive and transmit light of different wavelengths. The blue light receptor gene MdCRY2 and the UV-B photoreceptor gene MdUVR8 have been characterized in apple (Li et al., 2013; Zhao et al., 2016). Downstream of the photoreceptors, LONG HYPOCOTYL 5 (HY5), ubiquitin E3 ligase constitutive photobiochemical enzyme 1 (COP1), and PHYTOCHROME-INTERACTING FACTORS (PIFs) are involved in the complex signaling network mediating the regulation of plant photomorphogenesis (Deng et al., 1992; Ang and Deng, 1994; Ni et al., 1998). In apple, MdHY5 binds directly to the MdMYB1 promoter to induce expression and promote the accumulation of anthocyanins under light conditions (An et al., 2017). However, under dark conditions, MdCOP1 can interact with MdMYB1 to mediate the ubiquitination and degradation of MdMYB1 (Li et al., 2012).
The B-box (BBX) protein is a zinc finger protein containing one or two B-box motifs (Reymond et al., 2001). It is a transcription factor that regulates plant photomorphogenesis along with HY5, COP1, and PIFs. Members of the BBX family have been identified in many species, including Arabidopsis (Khanna et al., 2009), rice (Huang et al., 2012), tomato (Chu et al., 2016), apple (Liu et al., 2018), and pear (Bai et al., 2019a). In Arabidopsis, the BBX protein family is divided into five subfamilies according to the protein structures. Subfamily IV comprises eight members (BBX18–25), each with two B-box zinc finger motifs (Khanna et al., 2009). The transcription factors in this subfamily are closely related to the light signaling cascade. They can interact with the COP1 and HY5 transcription factors, and can link the light signal regulatory network with other regulatory networks (e.g., hormone and temperature) (Sarmiento, 2013; Song et al., 2020). In apple, BBX22 positively regulates UV-B-induced anthocyanin synthesis, but its function depends on the synergistic effect of HY5 (Bai et al., 2014; An et al., 2019). Additionally, MdBBX20 can form a complex with MdHY5 and bind directly to the MdMYB1, MdDFR, and MdANS promoters to promote anthocyanin synthesis in apple (Fang et al., 2019b). Moreover, MdCOL4 interacts with MdHY5 to inhibit the expression of MdMYB1, whereas it can bind directly to the MdUFGT and MdANS promoters to suppress expression (Fang et al., 2019a). However, whether there are other BBX proteins involved in the synthesis of anthocyanins in apple and the relationships among these BBX proteins remain unclear.
Although the mechanism mediating light-induced anthocyanin synthesis has been partially characterized, a thorough analysis is still needed. In this study, on the basis of transcriptome sequencing data, we revealed that MdBBX21 is differentially expressed in the peel when unbagged fruit are exposed to light. Additionally, MdBBX21 responds to light and reaches peak expression levels earlier than MdBBX20, MdBBX22-1/2, and MdHY5. The overexpression of MdBBX21 in transgenic Arabidopsis plants and apple calli can induce anthocyanin accumulation under light. Further analyses indicated that MdBBX21 can activate the expression of MdBBX20, MdBBX22-1/2, and MdHY5, which subsequently promotes anthocyanin accumulation. Furthermore, MdBBX21 can interact with MdHY5 and induce the expression of MdMYB1. These findings clarify the transcriptional regulation that occurs upstream of MdMYB1 during light-induced anthocyanin biosynthesis.
Materials and Methods
Plant Materials
Six-year-old “Starkrimson Delicious” apple trees growing at the Baishui Apple Experimental Farm of Northwest A&F University, Shaanxi province, China were used as experimental materials. Fruit were covered with a paper bag (Hongtai, Shanxi, China) at 45 days after blooming and harvested at 135 days after blooming. The harvested fruit were placed in an incubator at 23°C under continuous white light (200 μmol m–2 s–1). The peels were excised at 0, 3, 6, 9, 12, 24, 48, and 72 h after initiating the light treatment. Twelve fruit were selected at each time-point, with four fruit considered as one biological replicate.
Determination of the Anthocyanin Content
Anthocyanins were extracted from the apple peels as previously described (Xie et al., 2012). Briefly, 0.5 g peel was treated with 5 mL 1% (v/v) HCl-methanol and incubated in darkness at 4°C for 24 h. After centrifuging at 13,000 × g for 10 min, the anthocyanin content in the upper liquid layer was determined using an HPLC system comprising a Waters 2998 detector (Waters, Milford, MA, United States) and a C18 column (5 μm internal diameter, 250 mm × 4.6 mm; Waters) as previously described (Liu et al., 2013). The anthocyanin content of Arabidopsis was determined as previously described (Wang et al., 2016).
RNA Extraction, Library Preparation, and RNA-seq
Total RNA was extracted using the TRIzol RNA Plant Plus Reagent (Tiangen, Beijing, China). The integrity of the RNA was checked using the 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, United States) and by agarose gel electrophoresis. The purity of the RNA was determined using the NanoPhotometer spectrophotometer (IMPLEN, Westlake Village, CA, United States). Sequencing libraries were constructed using the NEBNext® UltraTM RNA Library Prep Kit for Illumina (NE, United States). After verifying the quality of the libraries, they were sequenced using the Illumina Novaseq 6000 sequencer (150-bp paired-end sequencing) (Illumina, San Diego, CA, United States) at Novogene Bioinformatics Technology Co., Ltd., Beijing, China.
After eliminating the reads with sequencing adapters and the low-quality reads from the raw data, the remaining clean reads were aligned to the Malus Genome GDDH13 reference sequence (version 1.1) (Daccord et al., 2017) using the HISAT2 software. After the sequence alignment, the raw counts of the mapped reads for each Malus gene model in GDDH13 (version 1.1) were determined and then normalized to FPKM per million mapped reads. Using DESeq2, differentially expressed genes (DEGs) were putatively identified according to the following criteria: | log2(fold-change)| > 1 and adjusted padj < 0.05.
Subcellular Localization
The MdBBX21 coding sequence (CDS) was cloned from the cDNA derived from “Starkrimson Delicious” fruit peel. The full-length MdBBX21 open reading frame without the stop codon was inserted into the pCAMBIA2301-eGFP vector under the control of the CaMV 35S promoter to obtain the 35S:MdBBX21-eGFP construct. The primer sequences used are listed in Supplementary Table 4. Tobacco (Nicotiana benthamiana) leaves were infiltrated with Agrobacterium tumefaciens strain GV3101 cells carrying 35S:MdBBX21-eGFP or the empty vector control (pCAMBIA2301-eGFP). After 3 days, the eGFP signal in the tobacco leaves were detected using the LSM 710 confocal laser-scanning microscope (Carl Zeiss) at excitation wavelengths of 488 nm for eGFP.
Analysis of mRNA Expression
First-strand cDNA was synthesized using the HiScript® II Q RT SuperMix for qPCR (Vazyme, Nanjing, China). A quantitative real-time polymerase chain reaction (qPCR) analysis was conducted using the ChamQ Universal SYBR qPCR Master Mix (Vazyme) and the ABI StepOnePlusTM Real-Time PCR System (Applied Biosystems, Waltham, MA, United States). The MdActin gene was used as the internal control. All expression data were examined according to the delta-delta cycle threshold method (Livak and Schmittgen, 2001). The primer sequences used are listed in Supplementary Table 4.
Generation of Transgenic Plant Materials
“Orin” apple calli were transformed according to a slightly modified version of a previously reported method (Xie et al., 2012). First, calli were infected with A. tumefaciens strain LBA4404 cells carrying 35S:MdBBX21-eGFP for 20 min. The calli were then transferred to MS medium supplemented with 1 mg L–1 6-benzylaminopurine (6-BA), 1 mg L–1 2,4-dichlorophenoxyacetic acid (2,4-D), 30 g L–1 sucrose, and 8 g L–1 agar. After a 3-day incubation in darkness, the calli were transferred to screening medium (i.e., MS medium supplemented with 30 g L–1 sucrose, 1 mg L–1 2,4-D, 1 mg L–1 6-BA, 8 g L–1 agar, 250 mg L–1 carbenicillin, and 50 mg L–1 kanamycin) to screen for transformants. For the light treatment, wild-type (WT) and MdBBX21-overexpressing (MdBBX21-OX) transgenic apple calli cultivated in darkness were transferred to a light incubator and placed under constant white light (500 μmol m–2 s–1) for 3 days.
Agrobacterium tumefaciens strain GV3101 cells carrying 35S:MdBBX21-eGFP were used to infect the Arabidopsis bbx21 mutant (SALK_105390) according to the floral-dip method (Clough and Bent, 1998). Seeds of WT and T3 transgenic Arabidopsis plants were chilled at 4°C for 48 h and then placed under white light (500 μmol m–2 s–1) at 24°C under long-day conditions (16-h light/8-h dark). The anthocyanin contents of 5-day-old Arabidopsis seedlings were determined.
Yeast Two-Hybrid Assay
A yeast two-hybrid assay was performed using the MatchmakerTM Gold Yeast Two-Hybrid System (Clontech). The full-length MdBBX21 CDS was inserted into the pGADT7 (AD) vector to construct the AD-BBX21 recombinant plasmid. The MdHY5 CDS was inserted into the pGBKT7 (BD) vector to obtain the BD-HY5 recombinant plasmid. The primer sequences used are listed in Supplementary Table 4. Yeast strain Y2HGold cells were transformed with AD-BBX21 + BD-HY5, AD + BD-HY5, AD-BBX21 + BD, pGADT7-T + pGBKT7-53, or pGADT7-T + pGBKT7-Lam and then cultured on medium lacking tryptophan and leucine at 30°C. To screen for interacting proteins, the yeast cells were transferred to medium lacking leucine, tryptophan, histidine, and adenine (−L/−T/−H/−A), but supplemented with X-α-gal.
Yeast One-Hybrid Assay
The MdBBX20, MdBBX22-1/2, MdHY5, and MdMYB1 promoter fragments were inserted into the pHIS2 vector to construct the MdBBX20pro-HIS2, MdBBX22-1pro-HIS2, MdBBX22-2pro-HIS2, MdHY5pro-HIS2, and MdMYB1pro-HIS2 recombinant plasmids. The primer sequences used are listed in Supplementary Table 4. To determine the optimal 3-AT concentration, yeast strain Y187 cells containing the recombinant pHIS2 plasmids were grown on screening medium lacking histidine and tryptophan (−H/−T), but supplemented with different 3-AT concentrations. Next, Y187 yeast cells were co-transformed with MdBBX21-AD and individual recombinant pHIS2 plasmids. Interactions were detected on selection medium lacking histidine, tryptophan, and leucine (−H/−T/−L), but supplemented with the optimal 3-AT concentration.
Transient Dual-Luciferase Assay
Transient dual-luciferase assays were performed using tobacco (N. benthamiana) leaves. The MdBBX20, MdBBX22-1/2, MdHY5, and MdMYB1 promoter fragments were inserted into the pGreenII 0800-LUC vector to construct the MdBBX20pro:LUC, MdBBX22-1pro:LUC, MdBBX22-2pro:LUC, MdHY5pro:LUC, and MdMYB1pro:LUC recombinant plasmids. The full-length MdBBX21 CDS was inserted into the pGreenII 62-SK vector. The primer sequences used are listed in Supplementary Table 4. A. tumefaciens strain GV3101 cells carrying the pSoup vector were transformed with the recombinant plasmids. Leaves from 5-week-old tobacco (N. benthamiana) plants were injected according to a previously described method (Zhang et al., 2020). In order to maintain the same concentration of Agrobacterium solution, 50 ul of Agrobacterium solution MdBBX21 (OD = 0.8) and MdHY5 (OD = 0.8) were mixed with 50 ul of Agrobacterium solution MdMYB1pro:LUC for tobacco leaf injection, respectively. Also, equal volumes and concentrations of Agrobacterium solution MdBBX21 and MdHY5 were mixed, and then 50 ul of the mixture was mixed with 50 ul of Agrobacterium solution MdMYB1pro:LUC for tobacco leaf injection.
Bimolecular Fluorescence Complementation Assay
The MdBBX21 CDS was inserted into the pSPYNE vector, whereas the MdHY5 CDS was inserted into the pSPYCE vector. A. tumefaciens strain GV3101 cells were transformed with the MdBBX21-NE and MdHY5-CE recombinant plasmids. The primer sequences used are listed in Supplementary Table 4. Equal volumes of the A. tumefaciens cells carrying MdBBX21-NE and MdHY5-CE were mixed. Arabidopsis protoplast cells were infected with the A. tumefaciens cells for 15 min and then incubated at 23°C for 16–20 h. The YFP fluorescence was detected using the LSM 710 confocal laser-scanning microscope (Carl Zeiss) with an excitation wavelength of 514 nm.
Results
Transcriptome Analysis of Apple Peel Exposed to Light
To clarify the response of apple to light, we first measured the anthocyanin content in the peel of unbagged apple fruit exposed to white light for 0, 3, 6, 9, 12, 24, 48, and 72 h (Figure 1A). Anthocyanins were basically undetectable before 12 h, but they started to accumulate significantly in the peel at 24 h (Figure 1B). The peel samples at the following three time-points underwent an RNA-seq analysis: 0 h (G0), 6 h (G6), and 24 h (G24). After the strict data filtering step, the number of clean reads in each library ranged from 37.69 to 45.95 million, and the Q30 values exceeded 92% (Supplementary Table 1). The mapping rate of the clean reads to the reference genome was 91.25–92.51%. Additionally, 88.81–90.27% of the clean reads were uniquely mapped (Supplementary Table 2). All biological replicates were strongly correlated (R2 > 0.94) (Figure 1C); this correlation was confirmed by principal component analysis (Figure 1D).
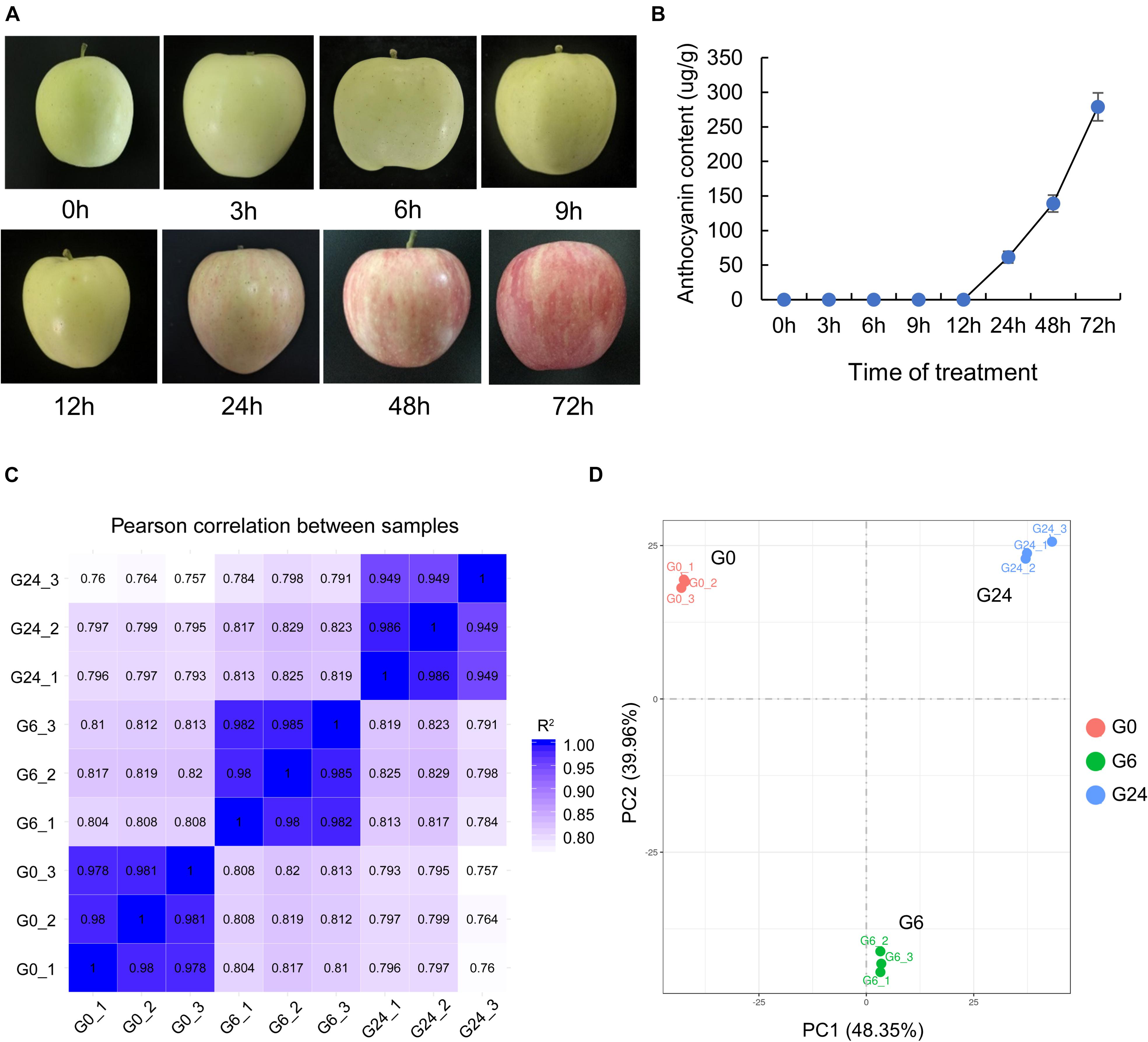
Figure 1. RNA-seq data of “Starkrimson Delicious” apple fruit exposed to light after unbagging. (A) Change in color of “Starkrimson Delicious” fruit peel under light. (B) Anthocyanin content in the peel under light. (C) Pearson correlation between samples. (D) Principal component analysis of the RNA-seq data.
Identification of Differentially Expressed Genes and Kyoto Encyclopedia of Genes and Genomes Analysis
To screen for early light-responsive genes, we compared the G0, G6, and G24 expression levels. In total, 11,941 DEGs were revealed in the G6 vs. G0, G24 vs. G6, and G24 vs. G0 pairwise comparisons. More DEGs were detected in the G6 vs. G0 comparison than in the G24 vs. G6 comparison (Figures 2A,B). These results suggest that many genes started to respond to light signals after 6 h. To further elucidate the gene expression patterns, the 11,941 DEGs were classified into eight gene expression profiles (Supplementary Figure 1). Specifically, 4,082 DEGs were classified into three significant profiles (P < 0.05), including two up-regulated profiles (profiles 6 and 7) and one down-regulated profile (profile 0) (Figure 2C). A KEGG enrichment analysis indicated genes related to flavonoid biosynthesis and phenylpropanoid biosynthesis were significantly enriched among the DEGs in profiles 0, 6, and 7 (Figure 2D).
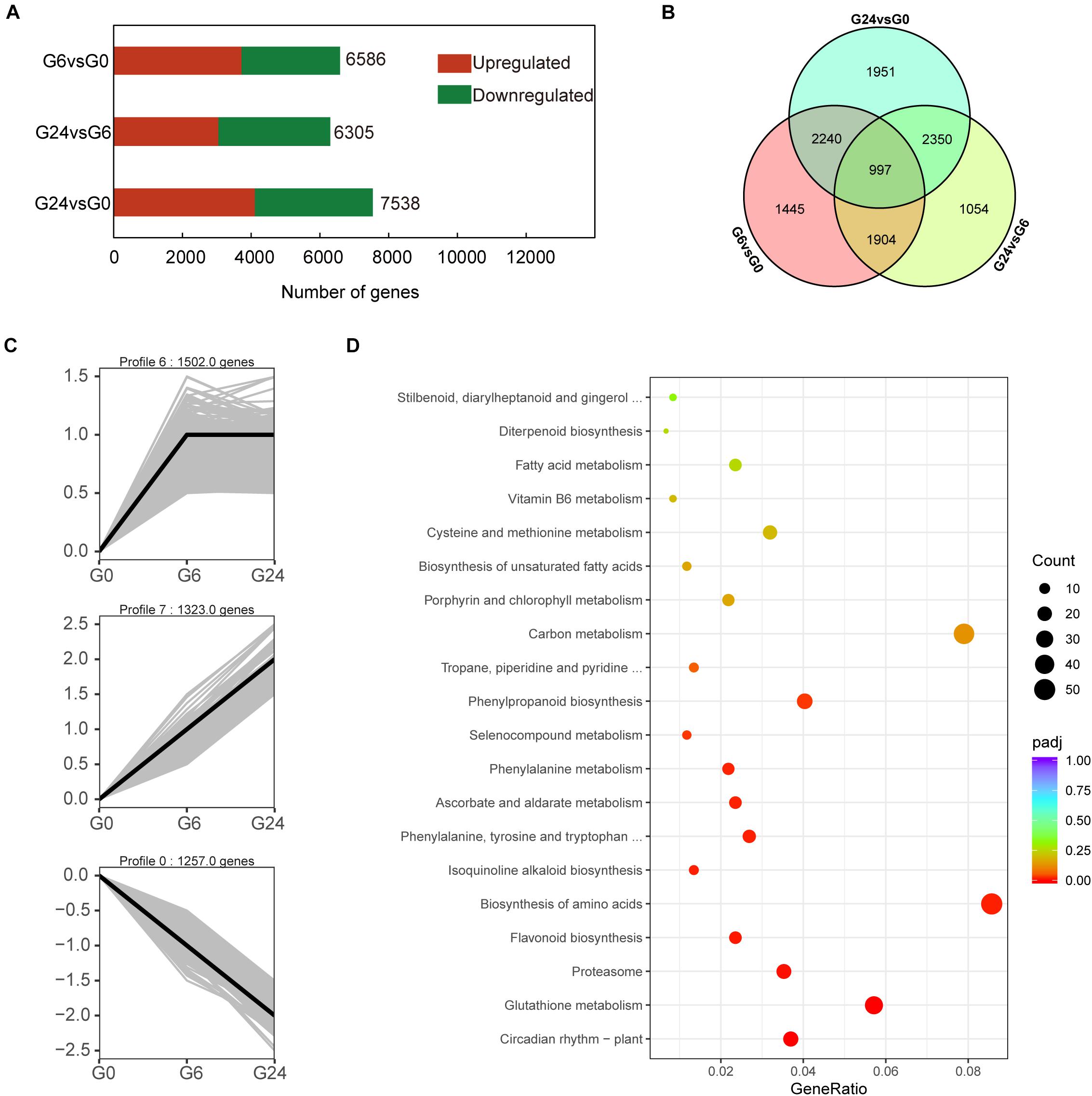
Figure 2. Identification of DEGs and KEGG analysis. (A) Number of DEGs between samples. (B) Venn diagram representation of DEGs from pairwise comparisons. (C) Three significant gene expression profiles. (D) KEGG pathway enrichment analysis of differentially expressed transcripts in profiles 0, 6, and 7.
Characterization and Expression Analysis of MdBBX21
Among the DEGs in up-regulated profile 6 (Supplementary Table 3), MD08G1021000 was revealed to be highly homologous to Arabidopsis AtBBX21. Thus, we named this gene MdBBX21. To clarify the relationship between MdBBX21 and MdBBX20, MdBBX21, and MdBBX22-1/2, we compared their amino acid sequences. The sequence identities between MdBBX21 and MdBBX20, MdBBX22-1, and MdBBX22-2 were 37.58, 31.92, and 31.63%, respectively. In the constructed phylogenetic tree, MdBBX21, MdBBX20, and MdBBX22-1/2 were clustered with Arabidopsis subfamily IV members, but they were distributed in different clades (Figure 3A).
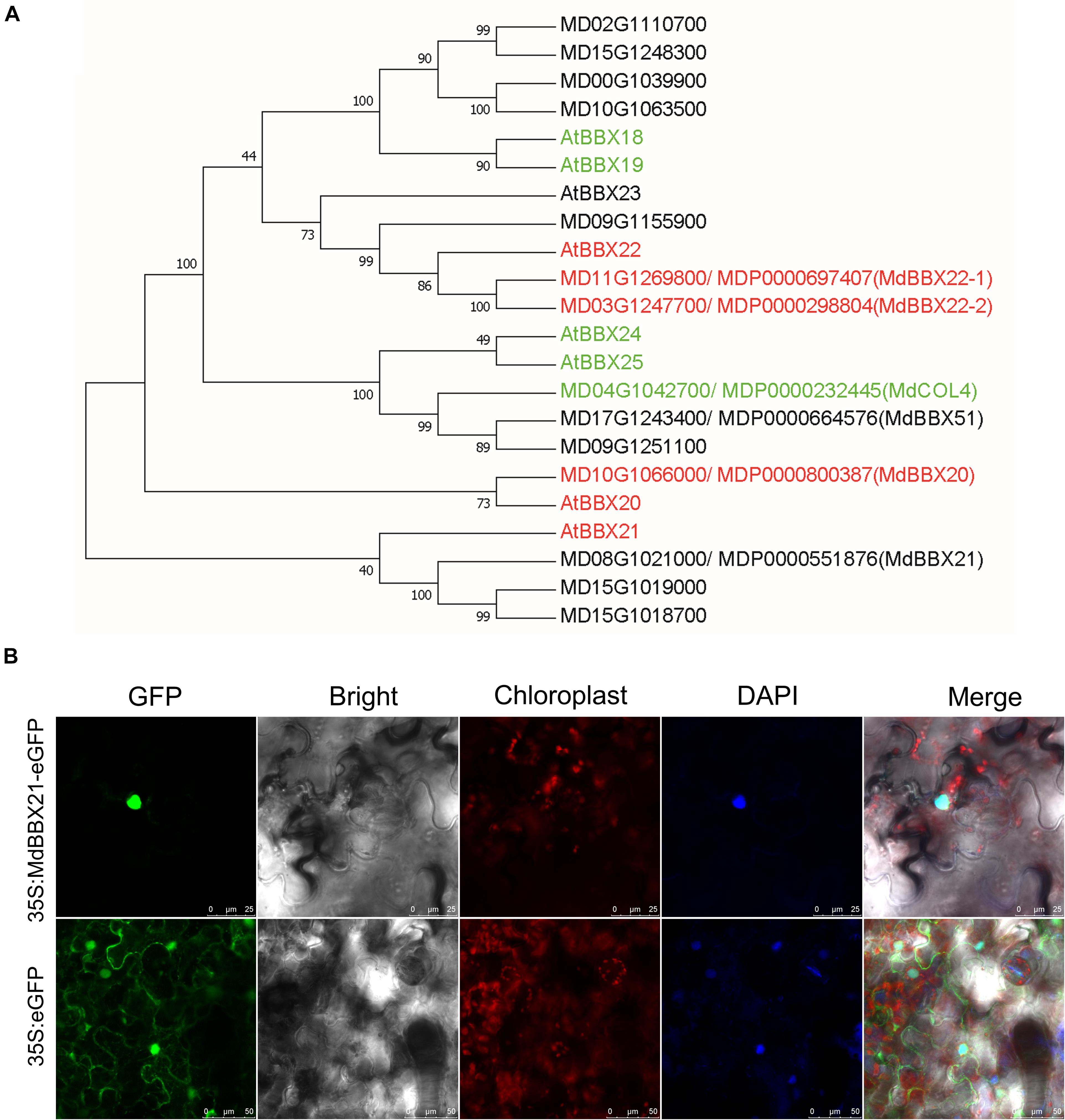
Figure 3. Characterization of MdBBX21. (A) Phylogenetic relationship of apple and Arabidopsis subfamily IV members. The red and green genes represent positive and negative regulators related to light signaling, respectively. The phylogenetic tree was constructed using the neighbor-joining method by MEGA 7.0. The bootstrap values of 1000 replicates were calculated at each node. (B) Subcellular localization of MdBBX21 expressed in tobacco leaf cells.
The correct cellular localization of a protein is critical for ensuring it functions properly. In tobacco leaf cells transiently transformed with 35S:MdBBX21-eGFP, green fluorescence was detected only in the nucleus, whereas in the control tobacco cells transiently transformed with 35S:eGFP, green fluorescence was observed throughout the cell, including in the cytoplasm and nucleus. Accordingly, MdBBX21 appears to be a nuclear protein (Figure 3B).
To determine whether MdBBX21 affects anthocyanin synthesis, we analyzed MdBBX21 expression in the apple fruit peel irradiated with white light for 0, 3, 6, 9, 12, 24, 48, and 72 h. The MdBBX21 gene was responsive to light, and its expression level started to increase at 3 h, peaking at 9 h. The expression of the regulatory genes (MdMYB1 and MdbHLH3) and structural genes (MdCHS, MdF3H, MdDFR, MdANS, and MdUFGT) related to anthocyanin synthesis peaked after 24 h. The MdWD40 expression level was essentially unchanged (Figure 4). These results imply that MdBBX21 may function upstream of MdMYB1.
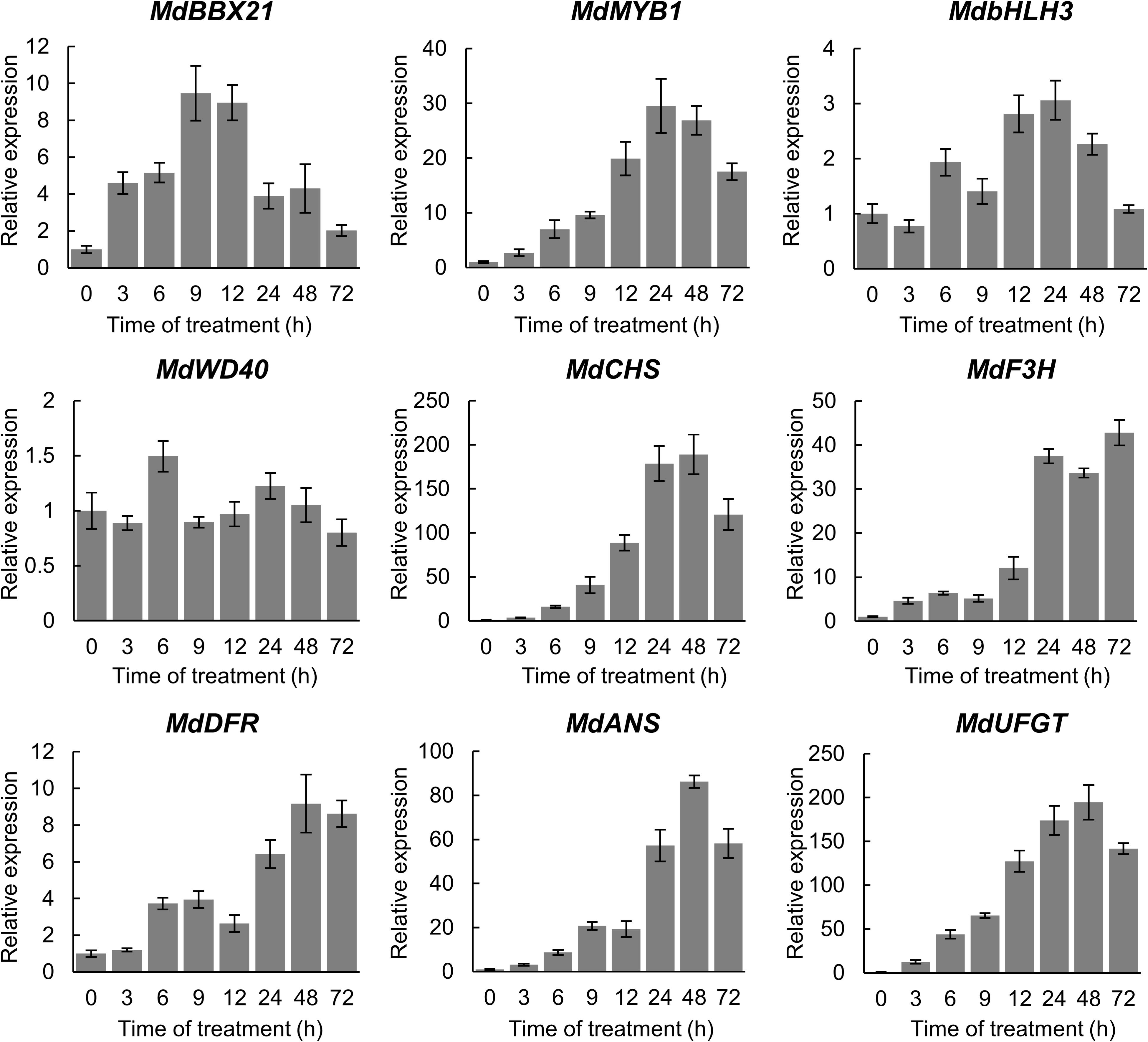
Figure 4. Relative expression levels of anthocyanin biosynthesis-related genes in the apple fruit peel irradiated with white light. The MdActin gene was used as the internal control.
Heterologous Overexpression of MdBBX21 Promotes Anthocyanin Accumulation in Arabidopsis
To confirm MdBBX21 contributes to anthocyanin biosynthesis, MdBBX21 was overexpressed in the bbx21 Arabidopsis mutant. The 35S:MdBBX21/bbx21 line accumulated more anthocyanins in the cotyledons and hypocotyls than the untransformed bbx21 Arabidopsis mutant (Figures 5A–C). The expression levels of anthocyanin-related genes (AtPAP1, AtCHS, AtF3’H, AtDFR, AtLDOX, and AtUFGT) also increased in the 35S:MdBBX21/bbx21 line, which was consistent with the changes in the anthocyanin content (Figure 5D). We also examined the expression of AtHY5 and Arabidopsis BBX subfamily IV members. The AtHY5 and AtBBX22 expression levels were higher in the 35S:MdBBX21/bbx21 seedlings than in the bbx21 Arabidopsis mutant seedlings. In contrast, there were no differences in the expression levels of the other subfamily IV BBX genes (Figure 5E and Supplementary Figure 2).
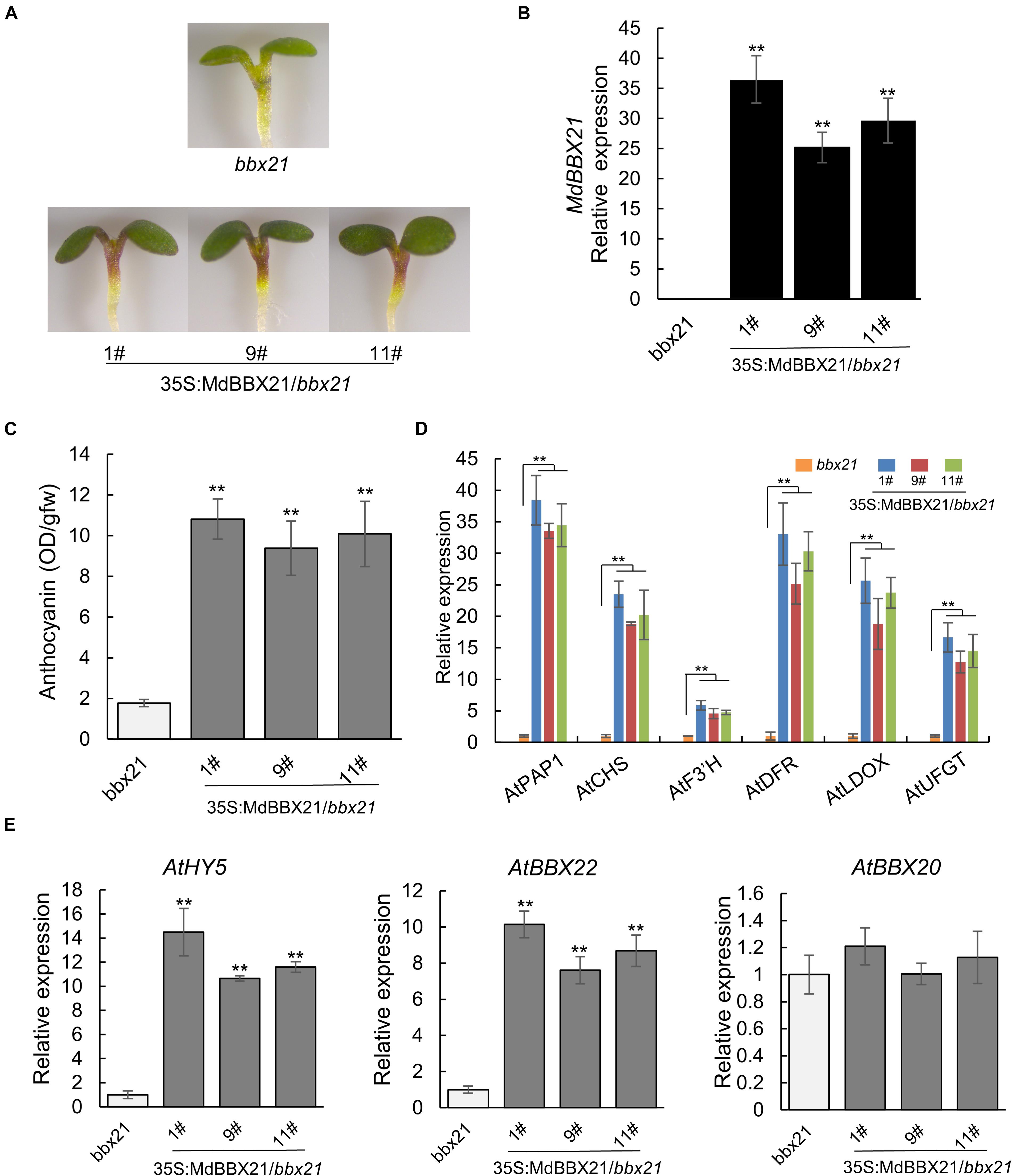
Figure 5. Overexpression of MdBBX21 promotes anthocyanin accumulation in Arabidopsis seedlings. (A) Phenotypes of 35S:MdBBX21/bbx21 and bbx21 mutant seedlings. 35S:MdBBX21/bbx21 (#1), 35S:MdBBX21/bbx21 (#9), and 35S:MdBBX21/bbx 21 (#11) represent three transgenic lines. (B) The MdBBX21’s expression level in 35S:MdBBX21/bbx21 and bbx21 mutant seedlings. (C) Anthocyanin content in 35S:MdBBX21/bbx21 and bbx21 mutant seedlings. (D) Relative expression levels of anthocyanin biosynthesis-related genes in 35S:MdBBX21/bbx21 and bbx21 mutant seedlings. (E) Relative expression levels of AtHY5, AtBBX22, and AtBBX20 in 35S:MdBBX21/bbx21 and bbx21 mutant seedlings. Error bars represent the standard deviation of three biological replicates. **P < 0.01 (Student’s t-test).
Overexpression of MdBBX21 Promotes Anthocyanin Accumulation in Apple Calli
To further confirm the MdBBX21 function related to anthocyanin biosynthesis in apple, we overexpressed MdBBX21 in apple calli (Figures 6A,B). Wild-type and MdBBX21-OX transgenic apple calli were cultured on medium in darkness and then transferred to a light incubator for a 3-day exposure to constant light. The MdBBX21-OX apple calli accumulated more anthocyanins than the WT calli (Figure 6C). As expected, the expression levels of the anthocyanin biosynthesis-related genes (MdMYB1, MdCHS, MdF3H, MdDFR, MdANS, and MdUFGT) were significantly higher in the MdBBX21-OX calli than in the WT calli (Figure 6D). These results suggest that MdBBX21 might positively regulate anthocyanin accumulation by promoting the transcription of anthocyanin biosynthesis-related genes. The transcription levels of the previously characterized genes MdBBX20, MdBBX22-1/2, MdCOL4 (MdBBX24), and MdHY5 were also analyzed by qPCR (Figure 6E and Supplementary Figure 3). The data indicated that MdHY5, MdBBX20, and MdBBX22-1/2 were significantly more highly expressed in MdBBX21-OX calli than in WT calli. The MdBBX24 expression level did not differ between the MdBBX21-OX and WT calli.
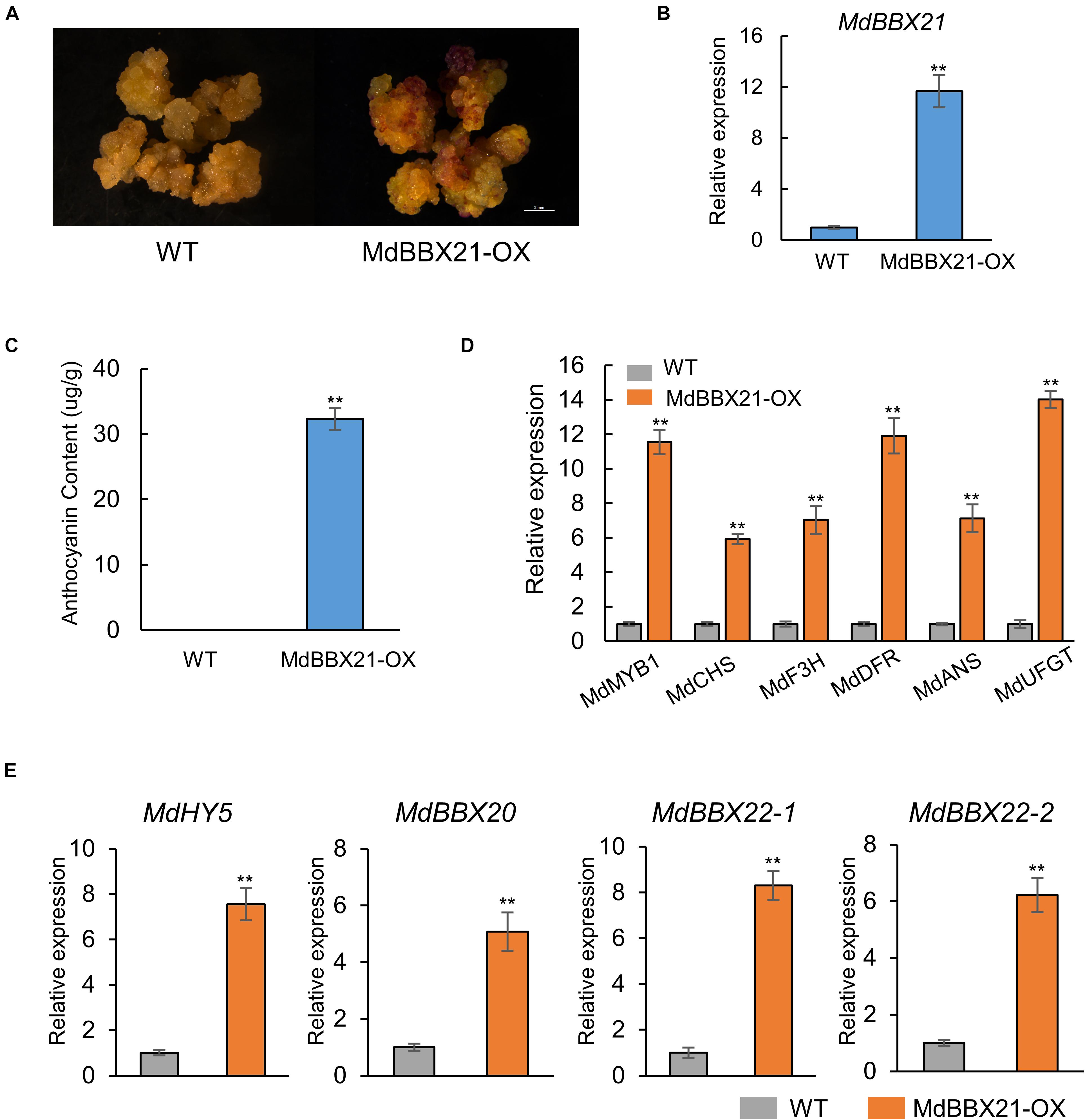
Figure 6. Overexpression of MdBBX21 promotes anthocyanin accumulation in apple calli. (A) Anthocyanin accumulation phenotypes of apple calli (WT: wild-type apple calli; MdBBX21-OX: MdBBX21 overexpression apple calli). (B) MdBBX21’s expression level in WT and MdBBX21-OX. (C) Anthocyanin contents in WT and MdBBX21-OX. (D) The expression levels of anthocyanin biosynthesis-related genes in WT and MdBBX21-OX. (E) The expression levels of MdHY5, MdBBX20, and MdBBX22-1/2 in WT and MdBBX21-OX. Error bars represent the standard deviation of three biological replicates. **P < 0.01 (Student’s t-test).
MdBBX21 Binds Directly to the MdHY5, MdBBX20, and MdBBX22-1/2 Promoters and Induces Expression
The increased MdHY5, MdBBX20, and MdBBX22-1/2 expression levels in MdBBX21-OX calli led us to speculate that MdBBX21 may directly promote the expression of these four genes. Thus, we first searched for the G-box motif, which is a BBX-binding site (Datta et al., 2008; Xu et al., 2016), in the MdHY5, MdBBX20, and MdBBX22-1/2 promoters. The G-box motif was detected in all three promoters (Figure 7A). In yeast one-hybrid assays, the Y187 yeast strains containing MdBBX21-AD + MdBBX20pro-HIS2, MdBBX21-AD + MdBBX22-1pro-HIS2, MdBBX21-AD + MdBBX22-2pro-HIS2, and MdBBX21-AD + MdHY5pro-HIS2 were able to grow on the −H/−T/−L selection medium containing 100 mM 3-AT. In contrast, the Y187 yeast strains containing AD + MdBBX20pro-HIS2, AD + MdBBX22-1pro-HIS2, AD + MdBBX22-2pro-HIS2, and AD + MdHY5pro-HIS2 did not grow under the same conditions (Figure 7B). This observation suggests that MdBBX21 can bind to the MdHY5, MdBBX20, and MdBBX22-1/2 promoters. The dual-luciferase assay results confirmed that MdBBX21 can promote the transcription of MdHY5, MdBBX20, and MdBBX22-1/2 (Figure 7C). We also analyzed the MdHY5, MdBBX20, and MdBBX22-1/2 expression patterns in the peel exposed to light. Compared with the MdBBX21 expression pattern (Figure 4), the MdBBX20, and MdBBX22-1/2 expression levels increased significantly at 6 h, and their peak expression levels occurred later than that of MdBBX21 (Supplementary Figure 4). These findings imply that MdBBX21 functions upstream of MdBBX20, and MdBBX22-1/2 and induces the expression of MdBBX20, and MdBBX22-1/2. At the same time, we found that MdHY5 was activated at 3 h. Considering HY5 being a transcription factor, we speculated that MdHY5, a master regulator of light signaling, might also control the expression of MdBBX21. Thus, we searched for the G-box motif in the MdBBX21 promoter. There were two G-box motifs present in the MdBBX21 promoter (Figure 7A). Yeast one-hybrid assay results showed that MdHY5 can bind to the MdBBX21 promoter fragment containing these two motifs (Figure 7B). The dual-luciferase assay results confirmed that MdHY5 can promote the transcription of MdBBX21 (Figure 7C).
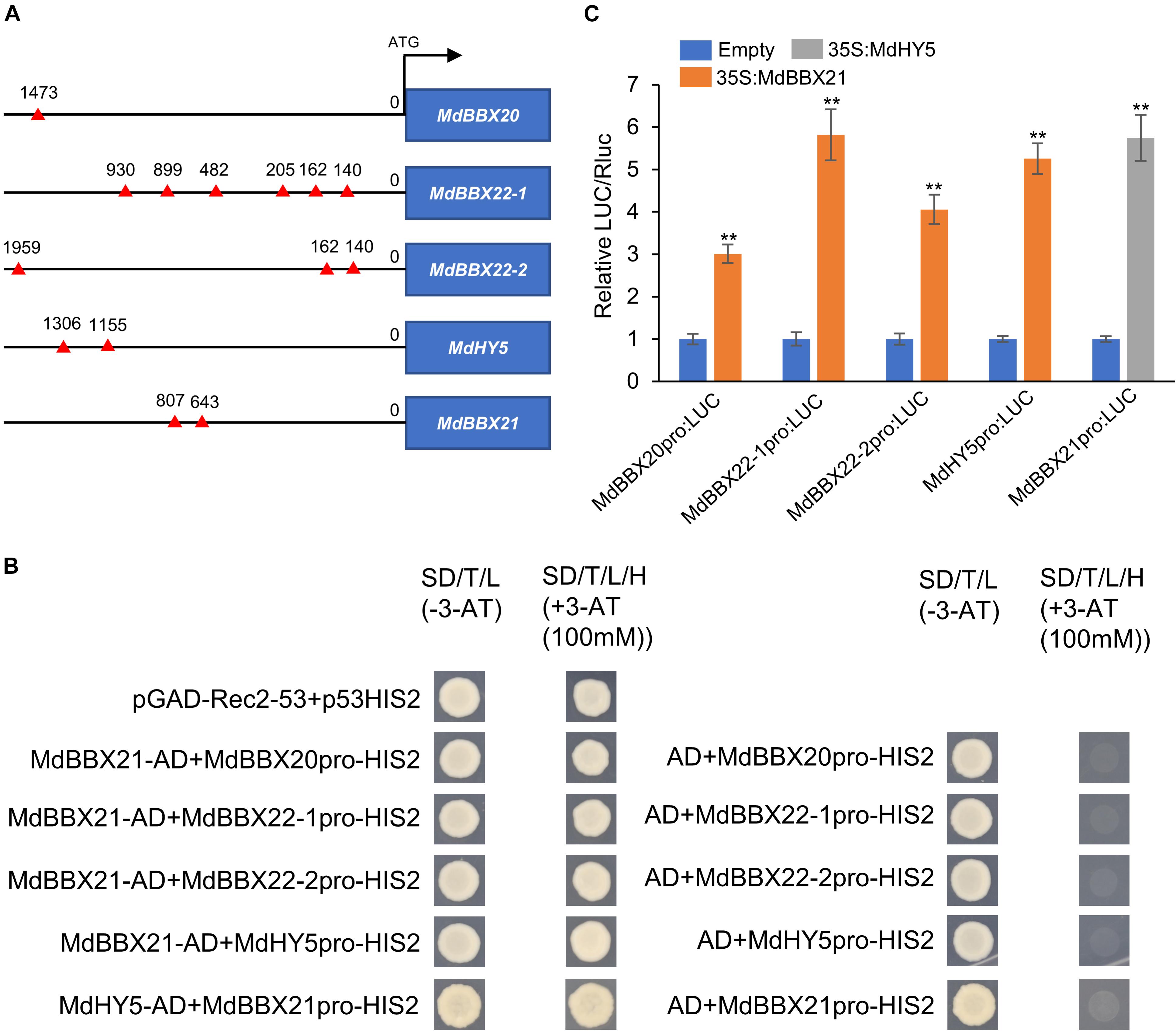
Figure 7. MdBBX21 binds directly to the MdHY5, MdBBX20, and MdBBX22-1/2 promoters and induces expression. (A) The characteristics of G-box motif in the promoters of MdHY5, MdBBX20, and MdBBX22-1/2. The red triangle represents G-box motif. (B) Yeast one-hybrid assays identified interaction of MdBBX21 with the promoters of MdHY5, MdBBX20, and MdBBX22-1/2. (C) The effects of MdBBX21 on the promoter activity of MdHY5, MdBBX20, and MdBBX22-1/2 with the dual-luciferase reporter assay. Error bars represent the standard deviation of three biological replicates. **P < 0.01 (Student’s t-test).
The Interaction Between MdBBX21 and MdHY5 Can Significantly Enhance MdMYB1 Promoter Activity
Because MdBBX20 (Fang et al., 2019b), MdBBX22-2 (An et al., 2019), PpBBX16 (Bai et al., 2019a), and PpBBX18 (Bai et al., 2019b) can interact with HY5, we speculated that MdBBX21 may also interact with MdHY5. Bimolecular fluorescence complementation assays were conducted to examine whether MdBBX21 can interact directly with MdHY5. Full-length MdBBX21 and MdHY5 CDSs were cloned into pSPYNE and pSPYCE vectors, respectively, to generate MdBBX21-NE and MdHY5-CE. Yellow fluorescence was detected in the nucleus of Arabidopsis protoplasts co-transformed with MdBBX21-NE and MdHY5-CE. However, yellow fluorescence was not observed in the BBX21-NE + CE and NE + HY5-CE controls (Figure 8A). These results indicate that MdBBX21 can interact with MdHY5. This interaction was verified in yeast two-hybrid assays (Figure 8B).
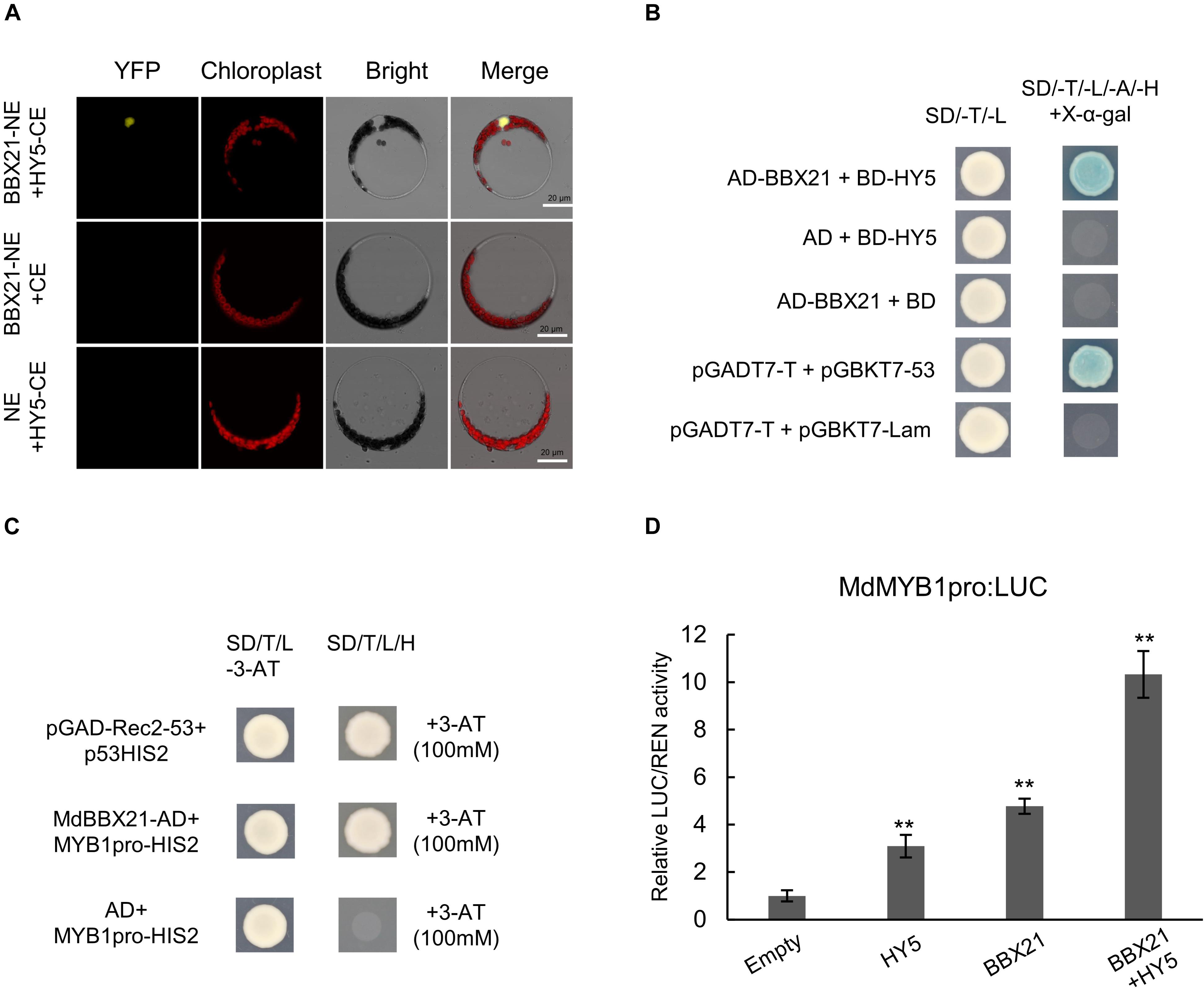
Figure 8. The interaction between MdBBX21 and MdHY5 can significantly enhance MdMYB1 promoter activity. (A) Bimolecular fluorescence complementation assay. (B) Yeast two-hybrid assay showing interaction between MdBBX21 and MdHY5. (C) Yeast one-hybrid assay showing interaction between MdBBX21 and MdMYB1 promoter. (D) Effects of MdBBX21 and MdHY5 individually and in combination on promoter activity of MdMYB1 as determined by luciferase reporter assay. Error bars represent the standard deviation of three biological replicates. **P < 0.01 (Student’s t-test).
MdBBX20 can bind directly to the MdMYB1 promoter and induce expression (Fang et al., 2019b). In this study, Y187 yeast strains containing MdBBX21-AD + MdMYB1pro-HIS2 grew on −H/−T/−L selection medium containing 100 mM 3-AT, but the Y187 yeast strains containing AD + MdMYB1pro-HIS2 did not (Figure 8C). Accordingly, MdBBX21 appears to be able to interact with the MdMYB1 promoter. On the basis of the dual-luciferase assay results, MdBBX21 can promote MdMYB1 expression (Figure 8D). Moreover, MdHY5 can also promote MdMYB1 expression, which is consistent with the results of an earlier study (An et al., 2017). We also determined that the MdBBX21–MdHY5 heterodimer enhances the MdMYB1 promoter activity more than MdBBX21 or MdHY5 alone (Figure 8D).
Discussion
MdBBX21 Responds to Light and Induces Anthocyanin Biosynthesis in Apple
Light is required for anthocyanin biosynthesis in the apple fruit peel (Saure, 1990). An exposure to light up-regulates the expression of anthocyanin biosynthesis-related structural and regulatory genes in the apple peel (Takos M. A. et al., 2006; Feng et al., 2013). However, how light signals affect the expression of these genes was unclear. There is increasing evidence that BBX proteins affect plant photomorphogenesis (Gangappa and Botto, 2014). The RNA-seq analysis in this study revealed that MdBBX21 expression in the peel of dark-grown “Starkrimson Delicious” apples is affected by the subsequent exposure to light. Additionally, MdBBX21 and Arabidopsis AtBBX21 sequences are highly similar. Earlier research demonstrated that AtBBX21 is a positive regulator of anthocyanin biosynthesis (Datta et al., 2007). Hence, we predicted that MdBBX21 and AtBBX21 might have similar functions. We analyzed MdBBX21 expression and observed that it increased in response to light. Moreover, MdBBX21 expression peaked before the expression of anthocyanin-related structural and regulatory genes peaked (Figure 4). The overexpression of MdBBX21 in the bbx21 Arabidopsis mutant resulted in a significant increase in the anthocyanin contents of hypocotyls (Figure 5). This is consistent with the fact pear PpBBX18 (homologous to AtBBX21) regulates anthocyanin accumulation in transgenic Arabidopsis plants. In the present study, the MdBBX21-OX apple calli accumulated more anthocyanins than the WT apple calli under light (Figure 6). These results indicate that MdBBX21 is responsive to light and induces anthocyanin biosynthesis.
MdBBX21 Up-Regulates MdBBX20, MdBBX22-1/2, and MdHY5 Expression
The Arabidopsis BBX subfamily IV members AtBBX20, AtBBX21, AtBBX22, and AtBBX23 positively regulate plant photomorphogenic processes in response to diverse light signals (Datta et al., 2007; Chang et al., 2008; Datta et al., 2008; Fan et al., 2012; Xu et al., 2016; Zhang et al., 2017). In pear, PpBBX16 and PpBBX18, which are subfamily IV BBX proteins, positively regulate anthocyanin synthesis by interacting with PpHY5 (Bai et al., 2019a,b). In apple, MdBBX20 and MdBBX22-1/2 reportedly promote anthocyanin accumulation in response to UV-B irradiation (Bai et al., 2014; An et al., 2019; Fang et al., 2019b). In addition, Plunkett et al. (2019) found that BBX subfamily IV member MdBBX51 and BBX subfamily I member MdBBX1 have very high expression levels during fruit development. Further, they can activate the promoter of MdMYB1 in the presence of some co-factors MYB and bHLH. However, the relationships among these BBX proteins are unknown. In this study, MdBBX21 expression increased significantly after a 3-h light treatment, and subsequently peaked at 9 h. In contrast, the MdBBX20 and MdBBX22-1/2 expression levels peaked at 12 and 24 h, respectively. The expression of structural and regulatory genes related to anthocyanin synthesis increased rapidly and peaked after 24 h (Figure 4 and Supplementary Figure 2). These findings indicate that MdBBX21 responds to light signals relatively early and likely functions upstream of MdBBX20 and MdBBX22-1/2. Dual-luciferase and yeast one-hybrid assays proved that MdBBX21 induces MdBBX20 and MdBBX22-1/2 expression after binding to their promoters. The AtBBX22, MdBBX20, and MdBBX22-1/2 expression levels increased in Arabidopsis and apple calli overexpressing MdBBX21 (Figures 5–7). Therefore, MdBBX21 can up-regulate the expression of AtBBX22, MdBBX20, and MdBBX22-1/2. However, MdBBX21 did not activate the expression of MdBBX1, MdBBX24, and MdBBX51 in apple calli (Supplementary Figure 3). In Arabidopsis, AtBBX21 can bind directly to the AtHY5 promoter and induce expression (Xu et al., 2016). In the current study, biochemical assays proved that MdBBX21 can bind directly to the MdHY5 promoter and induce expression (Figure 7). The overexpression of MdBBX21 in Arabidopsis and apple calli resulted in up-regulated HY5 expression (Figures 5, 6). These results imply that MdBBX21 and AtBBX21 have similar functions and can directly up-regulate the expression of HY5. At the same time, we found that MdHY5 can also activate the expression of MdBBX21, indicating that a positive feedback regulation mechanism exists between MdBBX21 and MdHY5.
The MdBBX21–MdHY5 Interaction Can Significantly Enhance MdMYB1 Promoter Activity Under Light
The bZIP transcription factor AtHY5 is a positive regulator of plant photomorphogenesis that functions downstream of the photoreceptors and COP1 (Lau and Deng, 2010). Additionally, AtHY5 regulates various physiological processes, including anthocyanin synthesis, lateral root formation, and hypocotyl elongation (Oyama et al., 1997). Earlier studies regarding in vitro DNA–protein interactions demonstrated that AtHY5 can bind directly and specifically to the G-box motif in the promoters of the anthocyanin-related structural genes AtF3H, AtCHS, and AtCHI and the regulatory gene AtPAP1 (Ang et al., 1998; Lee et al., 2007; Shin et al., 2013). In Arabidopsis, AtBBX21 and AtBBX22 can activate transcription. They interact with AtHY5 in vivo and induce downstream gene expression in an AtHY5-dependent and -independent manner to promote plant photomorphogenesis (Datta et al., 2007, 2008). In apples and pears, HY5 can bind directly to the MYB1 promoter (An et al., 2017; Tao et al., 2018). Both PpBBX16 and PpBBX18 cannot bind directly to the MYB1 promoter, but they can interact with HY5 and promote MYB1 expression (i.e., HY5-dependent manner) (Bai et al., 2019a,b). However, in apple, MdBBX20 can bind directly to the MdMYB1 promoter and induce expression (i.e., HY5-independent manner) (Fang et al., 2019b). In this study, we proved that MdBBX21 can bind directly to the MdMYB1 promoter and induce expression (Figure 8). Additionally, the analysis of protein–protein interactions revealed that MdBBX21 interacts with MdHY5 in yeast and plant cells. Furthermore, the interaction between MdBBX21 and MdHY5 can significantly enhance MdMYB1 promoter activity (Figure 8).
In conclusion, we systematically characterized the effect of light on MdBBX21 in the apple peel. We proved that MdBBX21 binds to the promoters of MdBBX20, MdBBX22-1/2, and MdHY5. The subsequent up-regulated expression of these genes enhances anthocyanin accumulation. Additionally, MdBBX21 can interact with MdHY5 and induce MdMYB1 expression. The results of our study will form the basis of future investigations on the functions of BBX proteins in apple.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI SRA BioProject, accession no: PRJNA767632.
Author Contributions
BZ, H-JY, and Z-YZ conceived the original screening and research plans. BZ, Z-ZZ, B-CW, N-NH, and Y-ZY supervised the experiments. BZ, H-JY, and DQ analyzed the data. BZ and Z-ZZ wrote the manuscript. H-JY revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Earmarked Fund for the National Natural Science Foundation of China (32072555), the Modern Agro-industry Technology Research System of China (CARS-27), Science and Technology Department Youth Foundation in Shaanxi province, China (2019JQ-204), and Chinese Universities Scientific Fund (2452020053).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.774446/full#supplementary-material
References
An, J. P., Qu, F. J., Yao, J. F., Wang, X. N., You, C. X., Wang, X. F., et al. (2017). The bZIP transcription factor MdHY5 regulates anthocyanin accumulation and nitrate assimilation in apple. Hortic Res. 4:17023. doi: 10.1038/hortres.2017.23
An, J. P., Wang, X. F., Zhang, X. W., Bi, S. Q., You, C. X., and Hao, Y. J. (2019). MdBBX22 regulates UV-B-induced anthocyanin biosynthesis through regulating the function of MdHY5 and is targeted by MdBT2 for 26S proteasome-mediated degradation. Plant Biotechnol. J. 17, 2231–2233. doi: 10.1111/pbi.13196
An, J. P., Zhang, X. W., Bi, S. Q., You, C. X., Wang, X. F., and Hao, Y. J. (2020). The ERF transcription factor MdERF38 promotes drought stress-induced anthocyanin biosynthesis in apple. Plant J. 101, 573–589. doi: 10.1111/tpj.14555
Ang, L. H., Chattopadhyay, S., Wei, N., Oyama, T., Okada, K., Batschauer, A., et al. (1998). Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol. Cell 1, 213–222. doi: 10.1016/s1097-2765(00)80022-2
Ang, L. H., and Deng, X. W. (1994). Regulatory hierarchy of photomorphogenic loci: allele-specific and light-dependent interaction between the HY5 and COP1 loci. Plant Cell 6, 613–628. doi: 10.1105/tpc.6.5.613
Bai, S., Saito, T., Honda, C., Hatsuyama, Y., Ito, A., and Moriguchi, T. (2014). An apple B-box protein, MdCOL11, is involved in UV-B- and temperature-induced anthocyanin biosynthesis. Planta 240, 1051–1062. doi: 10.1007/s00425-014-2129-8
Bai, S., Tao, R., Tang, Y., Yin, L., Ma, Y., Ni, J., et al. (2019a). BBX16, a B-box protein, positively regulates light-induced anthocyanin accumulation by activating MYB10 in red pear. Plant Biotechnol. J. 17, 1985–1997. doi: 10.1111/pbi.13114
Bai, S., Tao, R., Yin, L., Ni, J., Yang, Q., Yan, X., et al. (2019b). Two B-box proteins, PpBBX18 and PpBBX21, antagonistically regulate anthocyanin biosynthesis via competitive association with Pyrus pyrifolia ELONGATED HYPOCOTYL 5 in the peel of pear fruit. Plant J. 100, 1208–1223. doi: 10.1111/tpj.14510
Ban, Y., Honda, C., Hatsuyama, Y., Igarashi, M., Bessho, H., and Moriguchi, T. (2007). Isolation and functional analysis of a MYB transcription factor gene that is a key regulator for the development of red coloration in apple skin. Plant Cell Physiol. 48, 958–970. doi: 10.1093/pcp/pcm066
Chang, C. S., Li, Y. H., Chen, L. T., Chen, W. C., Hsieh, W. P., Shin, J., et al. (2008). LZF1, a HY5-regulated transcriptional factor, functions in Arabidopsis de-etiolation. Plant J. 54, 205–219. doi: 10.1111/j.1365-313X.2008.03401.x
Chu, Z., Wang, X., Li, Y., Yu, H., Li, J., Lu, Y., et al. (2016). Genomic organization, phylogenetic and expression analysis of the B-BOX gene family in tomato. Front. Plant Sci. 7:1552. doi: 10.3389/fpls.2016.01552
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Cominelli, E., Gusmaroli, G., Allegra, D., Galbiati, M., Wade, H. K., Jenkins, G. I., et al. (2008). Expression analysis of anthocyanin regulatory genes in response to different light qualities in Arabidopsis thaliana. J. Plant Physiol. 165, 886–894. doi: 10.1016/j.jplph.2007.06.010
Daccord, N., Celton, J. M., Linsmith, G., Becker, C., Choisne, N., Schijlen, E., et al. (2017). High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nat. Genet. 49, 1099–1106. doi: 10.1038/ng.3886
Datta, S., Hettiarachchi, C., Johansson, H., and Holm, M. (2007). SALT TOLERANCE HOMOLOG2, a B-box protein in Arabidopsis that activates transcription and positively regulates light-mediated development. Plant Cell 19, 3242–3255. doi: 10.1105/tpc.107.054791
Datta, S., Johansson, H., Hettiarachchi, C., Irigoyen, M. L., Desai, M., Rubio, V., et al. (2008). LZF1/SALT TOLERANCE HOMOLOG3, an Arabidopsis B-box protein involved in light-dependent development and gene expression, undergoes COP1-mediated ubiquitination. Plant Cell 20, 2324–2338. doi: 10.1105/tpc.108.061747
de Vetten, N., Quattrocchio, F., Mol, J., and Koes, R. (1997). The an11 locus controlling flower pigmentation in petunia encodes a novel WD-repeat protein conserved in yeast, plants, and animals. Genes Dev. 11, 1422–1434. doi: 10.1101/gad.11.11.1422
Deng, X. W., Matsui, M., Wei, N., Wagner, D., Chu, A. M., Feldmann, K. A., et al. (1992). COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a G beta homologous domain. Cell 71, 791–801. doi: 10.1016/0092-8674(92)90555-q
Espley, R. V., Brendolise, C., Chagné, D., Kutty-Amma, S., Green, S., Volz, R., et al. (2009). Multiple repeats of a promoter segment causes transcription factor autoregulation in red apples. Plant Cell 21, 168–183. doi: 10.1105/tpc.108.059329
Espley, R. V., Hellens, R. P., Putterill, J., Stevenson, D. E., Kutty-Amma, S., and Allan, A. C. (2007). Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 49, 414–427. doi: 10.1111/j.1365-313X.2006.02964.x
Fan, X. Y., Sun, Y., Cao, D. M., Bai, M. Y., Luo, X. M., Yang, H. J., et al. (2012). BZS1, a B-box protein, promotes photomorphogenesis downstream of both brassinosteroid and light signaling pathways. Mol. Plant 5, 591–600. doi: 10.1093/mp/sss041
Fang, H., Dong, Y., Yue, X., Hu, J., Jiang, S., Xu, H., et al. (2019b). The B-box zinc finger protein MdBBX20 integrates anthocyanin accumulation in response to ultraviolet radiation and low temperature. Plant Cell Environ. 42, 2090–2104. doi: 10.1111/pce.13552
Fang, H., Dong, Y., Yue, X., Chen, X., He, N., Hu, J., et al. (2019a). MdCOL4 interaction mediates crosstalk between UV-B and high temperature to control fruit coloration in apple. Plant Cell Physiol. 60, 1055–1066. doi: 10.1093/pcp/pcz023
Feng, F., Li, M., Ma, F., and Cheng, L. (2013). Phenylpropanoid metabolites and expression of key genes involved in anthocyanin biosynthesis in the shaded peel of apple fruit in response to sun exposure. Plant Physiol. Biochem. 69, 54–61. doi: 10.1016/j.plaphy.2013.04.020
Gangappa, S. N., and Botto, J. F. (2014). The BBX family of plant transcription factors. Trends Plant Sci. 19, 460–470. doi: 10.1016/j.tplants.2014.01.010
Huang, J., Zhao, X., Weng, X., Wang, L., and Xie, W. (2012). The rice B-box zinc finger gene family: genomic identification, characterization, expression profiling and diurnal analysis. PLoS One 7:e48242. doi: 10.1371/journal.pone.0048242
Kasahara, M., Swartz, T. E., Olney, M. A., Onodera, A., Mochizuki, N., Fukuzawa, H., et al. (2002). Photochemical properties of the flavin mononucleotide-binding domains of the phototropins from Arabidopsis, rice, and Chlamydomonas reinhardtii. Plant Physiol. 129, 762–773. doi: 10.1104/pp.002410
Khanna, R., Kronmiller, B., Maszle, D. R., Coupland, G., Holm, M., Mizuno, T., et al. (2009). The Arabidopsis B-box zinc finger family. Plant Cell 21, 3416–3420. doi: 10.1105/tpc.109.069088
Kim, S. H., Lee, J. R., Hong, S. T., Yoo, Y. K., An, G., and Kim, S. R. (2003). Molecular cloning and analysis of anthocyanin biosynthesis genes preferentially expressed in apple skin. Plant Sci. 165, 403–413. doi: 10.1016/s0168-9452(03)00201-2
Kubasek, W. L., Shirley, B. W., McKillop, A., Goodman, H. M., Briggs, W., and Ausubel, F. M. (1992). Regulation of flavonoid biosynthetic genes in germinating arabidopsis seedlings. Plant Cell 4, 1229–1236. doi: 10.1105/tpc.4.10.1229
Lau, O. S., and Deng, X. W. (2010). Plant hormone signaling lightens up: integrators of light and hormones. Curr. Opin. Plant Biol. 13, 571–577. doi: 10.1016/j.pbi.2010.07.001
Lawrence, W. J. C., Price, J. R., Robinson, G. M., and Robinson, R. (1939). The distribution of anthocyanins in flowers, fruits and leaves. Philosop. Trans. R. Soc. Lond. Series B Biol. Sci. 230, 149–178. doi: 10.1098/rstb.1939.0006
Lee, J., He, K., Stolc, V., Lee, H., Figueroa, P., Gao, Y., et al. (2007). Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19, 731–749. doi: 10.1105/tpc.106.047688
Li, S. (2014). Transcriptional control of flavonoid biosynthesis: fine-tuning of the MYB-bHLH-WD40 (MBW) complex. Plant Signal Behav. 9:e27522. doi: 10.4161/psb.27522
Li, Y. Y., Mao, K., Zhao, C., Zhao, X. Y., Zhang, H. L., Shu, H. R., et al. (2012). MdCOP1 ubiquitin E3 ligases interact with MdMYB1 to regulate light-induced anthocyanin biosynthesis and red fruit coloration in apple. Plant Physiol. 160, 1011–1022. doi: 10.1104/pp.112.199703
Li, Y. Y., Mao, K., Zhao, C., Zhao, X. Y., Zhang, R. F., Zhang, H. L., et al. (2013). Molecular cloning and functional analysis of a blue light receptor gene MdCRY2 from apple (Malus domestica). Plant Cell Rep. 32, 555–566. doi: 10.1007/s00299-013-1387-4
Lin, C., Robertson, D. E., Ahmad, M., Raibekas, A. A., Jorns, M. S., Dutton, P. L., et al. (1995). Association of flavin adenine dinucleotide with the Arabidopsis blue light receptor CRY1. Science 269, 968–970. doi: 10.1126/science.7638620
Liu, X., Li, R., Dai, Y., Chen, X., and Wang, X. (2018). Genome-wide identification and expression analysis of the B-box gene family in the Apple (Malus domestica Borkh.) genome. Mol. Genet. Genom. 293, 303–315. doi: 10.1007/s00438-017-1386-1
Liu, Y., Che, F., Wang, L., Meng, R., Zhang, X., and Zhao, Z. (2013). Fruit coloration and anthocyanin biosynthesis after bag removal in non-red and red apples (Malus × domestica Borkh.). Molecules 18, 1549–1563. doi: 10.3390/molecules18021549
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lotkowska, M. E., Tohge, T., Fernie, A. R., Xue, G. P., Balazadeh, S., and Mueller-Roeber, B. (2015). The arabidopsis transcription factor MYB112 promotes anthocyanin formation during salinity and under high light stress. Plant Physiol. 169, 1862–1880. doi: 10.1104/pp.15.00605
Maier, A., Schrader, A., Kokkelink, L., Falke, C., Welter, B., Iniesto, E., et al. (2013). Light and the E3 ubiquitin ligase COP1/SPA control the protein stability of the MYB transcription factors PAP1 and PAP2 involved in anthocyanin accumulation in Arabidopsis. Plant J. 74, 638–651. doi: 10.1111/tpj.12153
Martin, C., Prescott, A., Mackay, S., Bartlett, J., and Vrijlandt, E. (1991). Control of anthocyanin biosynthesis in flowers of Antirrhinum majus. Plant J. 1, 37–49. doi: 10.1111/j.1365-313x.1991.00037.x
Merzlyak, M. N., and Chivkunova, O. B. (2000). Light-stress-induced pigment changes and evidence for anthocyanin photoprotection in apples. J. Photochem. Photobiol. B 55, 155–163. doi: 10.1016/s1011-1344(00)00042-7
Ni, M., Tepperman, J. M., and Quail, P. H. (1998). PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95, 657–667. doi: 10.1016/s0092-8674(00)81636-0
Oyama, T., Shimura, Y., and Okada, K. (1997). The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 11, 2983–2995. doi: 10.1101/gad.11.22.2983
Passeri, V., Koes, R., and Quattrocchio, F. M. (2016). New challenges for the design of high value plant products: stabilization of anthocyanins in plant vacuoles. Front. Plant Sci. 7:153. doi: 10.3389/fpls.2016.00153
Plunkett, B. J., Henry-Kirk, R., Friend, A., Diack, R., Helbig, S., Mouhu, K., et al. (2019). Apple B-box factors regulate light-responsive anthocyanin biosynthesis genes. Sci. Rep. 9:17762. doi: 10.1038/s41598-019-54166-2
Quattrocchio, F., Wing, J. F., Leppen, H., Mol, J., and Koes, R. E. (1993). Regulatory genes controlling anthocyanin pigmentation are functionally conserved among plant species and have distinct sets of target genes. Plant Cell 5, 1497–1512. doi: 10.1105/tpc.5.11.1497
Quattrocchio, F., Wing, J. F., van der Woude, K., Mol, J. N., and Koes, R. (1998). Analysis of bHLH and MYB domain proteins: species-specific regulatory differences are caused by divergent evolution of target anthocyanin genes. Plant J. 13, 475–488. doi: 10.1046/j.1365-313x.1998.00046.x
Ramsay, N. A., and Glover, B. J. (2005). MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 10, 63–70. doi: 10.1016/j.tplants.2004.12.011
Reymond, A., Meroni, G., Fantozzi, A., Merla, G., Cairo, S., Luzi, L., et al. (2001). The tripartite motif family identifies cell compartments. Embo J. 20, 2140–2151. doi: 10.1093/emboj/20.9.2140
Rizzini, L., Favory, J. J., Cloix, C., Faggionato, D., O’Hara, A., Kaiserli, E., et al. (2011). Perception of UV-B by the Arabidopsis UVR8 protein. Science 332, 103–106. doi: 10.1126/science.1200660
Sarmiento, F. (2013). The BBX subfamily IV: additional cogs and sprockets to fine-tune light-dependent development. Plant Signal Behav. 8:e23831. doi: 10.4161/psb.23831
Saure, M. C. (1990). External control of anthocyanin formation in apple. Scientia Horticulturae 42, 181–218. doi: 10.1016/0304-4238(90)90082-P
Sharrock, R. A., and Quail, P. H. (1989). Novel phytochrome sequences in Arabidopsis thaliana: structure, evolution, and differential expression of a plant regulatory photoreceptor family. Genes Dev. 3, 1745–1757. doi: 10.1101/gad.3.11.1745
Shin, D. H., Choi, M., Kim, K., Bang, G., Cho, M., Choi, S. B., et al. (2013). HY5 regulates anthocyanin biosynthesis by inducing the transcriptional activation of the MYB75/PAP1 transcription factor in Arabidopsis. FEBS Lett. 587, 1543–1547. doi: 10.1016/j.febslet.2013.03.037
Song, Z., Bian, Y., Liu, J., Sun, Y., and Xu, D. (2020). B-box proteins: pivotal players in light-mediated development in plants. J. Integr. Plant Biol. 62, 1293–1309. doi: 10.1111/jipb.12935
Sun, X., Jia, X., Huo, L., Che, R., Gong, X., Wang, P., et al. (2018). MdATG18a overexpression improves tolerance to nitrogen deficiency and regulates anthocyanin accumulation through increased autophagy in transgenic apple. Plant Cell Environ. 41, 469–480. doi: 10.1111/pce.13110
Takos, A. M., Jaffé, F. W., Jacob, S. R., Bogs, J., Robinson, S. P., and Walker, A. R. (2006). Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 142, 1216–1232. doi: 10.1104/pp.106.088104
Takos, M. A., Robinson, P. S., and Walker, R. A. (2006). Transcriptional regulation of the flavonoid pathway in the skin of dark-grown ‘Cripps’ Red’ apples in response to sunlight. J. Horticultural Sci. Biotechnol. 81, 735–744. doi: 10.1080/14620316.2006.11512131
Tao, R., Bai, S., Ni, J., Yang, Q., Zhao, Y., and Teng, Y. (2018). The blue light signal transduction pathway is involved in anthocyanin accumulation in ‘Red Zaosu’ pear. Planta 248, 37–48. doi: 10.1007/s00425-018-2877-y
Wang, Y., Wang, Y., Song, Z., and Zhang, H. (2016). Repression of MYBL2 by both microRNA858a and HY5 leads to the activation of anthocyanin biosynthetic pathway in arabidopsis. Mol. Plant 9, 1395–1405. doi: 10.1016/j.molp.2016.07.003
Winkel-Shirley, B. (2001). Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 126, 485–493. doi: 10.1104/pp.126.2.485
Xie, X. B., Li, S., Zhang, R. F., Zhao, J., Chen, Y. C., Zhao, Q., et al. (2012). The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit colouration in response to low temperature in apples. Plant Cell Environ. 35, 1884–1897. doi: 10.1111/j.1365-3040.2012.02523.x
Xu, D., Jiang, Y., Li, J., Lin, F., Holm, M., and Deng, X. W. (2016). BBX21, an Arabidopsis B-box protein, directly activates HY5 and is targeted by COP1 for 26S proteasome-mediated degradation. Proc. Natl. Acad. Sci. U.S.A. 113, 7655–7660. doi: 10.1073/pnas.1607687113
Zhang, B., Yang, H. J., Yang, Y. Z., Zhu, Z. Z., Li, Y. N., Qu, D., et al. (2020). mdm-miR828 participates in the feedback loop to regulate anthocyanin accumulation in apple peel. Front. Plant Sci. 11:608109. doi: 10.3389/fpls.2020.608109
Zhang, H., Jordheim, M., Lewis, D. H., Arathoon, S., Andersen, O. M., and Davies, K. M. (2014). Anthocyanins and their differential accumulation in the floral and vegetative tissues of a shrub species (Rhabdothamnus solandri A. Cunn). Scientia Horticulturae 165, 29–35. doi: 10.1016/j.scienta.2013.10.032
Zhang, Y., Butelli, E., and Martin, C. (2014). Engineering anthocyanin biosynthesis in plants. Curr. Opin. Plant Biol. 19, 81–90. doi: 10.1016/j.pbi.2014.05.011
Zhang, L., Hu, J., Han, X., Li, J., Gao, Y., Richards, C. M., et al. (2019). A high-quality apple genome assembly reveals the association of a retrotransposon and red fruit colour. Nat. Commun. 10:1494. doi: 10.1038/s41467-019-09518-x
Zhang, X., Huai, J., Shang, F., Xu, G., Tang, W., Jing, Y., et al. (2017). A PIF1/PIF3-HY5-BBX23 transcription factor cascade affects photomorphogenesis. Plant Physiol. 174, 2487–2500. doi: 10.1104/pp.17.00418
Keywords: apple, light, anthocyanin, MdBBX21, MdMYB1
Citation: Zhang B, Zhu Z-Z, Qu D, Wang B-C, Hao N-N, Yang Y-Z, Yang H-J and Zhao Z-Y (2021) MdBBX21, a B-Box Protein, Positively Regulates Light-Induced Anthocyanin Accumulation in Apple Peel. Front. Plant Sci. 12:774446. doi: 10.3389/fpls.2021.774446
Received: 12 September 2021; Accepted: 26 October 2021;
Published: 12 November 2021.
Edited by:
Yongliang Liu, University of Kentucky, United StatesReviewed by:
Shaohua Zeng, South China Botanical Garden, Chinese Academy of Sciences (CAS), ChinaRichard Victor Espley, The New Zealand Institute for Plant and Food Research Ltd., New Zealand
Copyright © 2021 Zhang, Zhu, Qu, Wang, Hao, Yang, Yang and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hui-Juan Yang, aHVpanVhbnlhbmdAbndzdWFmLmVkdS5jbg==; Zheng-Yang Zhao, emhhb3p5QG53c3VhZi5lZHUuY24=
†These authors have contributed equally to this work
 Bo Zhang
Bo Zhang Zhen-Zhen Zhu
Zhen-Zhen Zhu Dong Qu
Dong Qu Bo-Chen Wang1,2
Bo-Chen Wang1,2 Hui-Juan Yang
Hui-Juan Yang Zheng-Yang Zhao
Zheng-Yang Zhao