- 1Biotechnology Research Institute, Chinese Academy of Agricultural Sciences, Beijing, China
- 2State Key Laboratory for Conservation and Utilization of Subtropical Agro-Bioresources, South China Agricultural University, Guangzhou, China
- 3Guangdong Laboratory for Lingnan Modern Agriculture, Guangzhou, China
Leaf senescence is the terminal stage of leaf development. Both light and the plant hormone ethylene play important roles in regulating leaf senescence. However, how they coordinately regulate leaf senescence during leaf development remains largely unclear. In this study, we show that FHY3 and FAR1, two homologous proteins essential for phytochrome A-mediated light signaling, physically interact with and repress the DNA binding activity of EIN3 (a key transcription factor essential for ethylene signaling) and PIF5 (a bHLH transcription factor negatively regulating light signaling), and interfere with their DNA binding to the promoter of ORE1, which encodes a key NAC transcription factor promoting leaf senescence. In addition, we show that FHY3, PIF5, and EIN3 form a tri-protein complex(es) and that they coordinately regulate the progression of leaf senescence. We show that during aging or under dark conditions, accumulation of FHY3 protein decreases, thus lifting its repression on DNA binding of EIN3 and PIF5, leading to the increase of ORE1 expression and onset of leaf senescence. Our combined results suggest that FHY3 and FAR1 act in an age gating mechanism to prevent precocious leaf senescence by integrating light and ethylene signaling with developmental aging.
Introduction
Leaf senescence is the last stage of leaf development, during which macromolecules (such as nucleic acids and proteins) are degraded in an orderly fashion, and the resulting nutrients are mobilized from old leaves to actively growing tissues or storage organs, thus increasing plant fitness (Lim et al., 2007). Leaf senescence can be conceptually divided into three phases: initiation (onset), reorganization (progression) and termination (completion), with each process being tightly regulated by genetic, developmental and environmental factors (Gan and Amasino, 1997; Nam, 1997; Li et al., 2018).
Over the past few decades, the utilization of genetic and molecular biology approaches, and more recently, the use of multi-omics technologies together with computational biology tools have greatly aided in the identification of key players and the associated gene regulatory networks (GRNs) regulating the various processes of leaf senescence (Woo et al., 2019). Particularly illuminating, several GRNs of NAC (NAM/ATAF/CUC) and WRKY transcription factors have been shown to change dynamically as leaf senescence progresses (Kim et al., 2016). For instance, it has been shown that the GRNs involving the NAC transcription factor ORE1 (ORESARA1, means “long living” in Korean) play an essential role in promoting leaf senescence (Oh et al., 1997; Park et al., 2019). ORE1 protein promotes leaf senescence by directly activating the expression of numerous chlorophyll catabolic genes (CCGs), such as NYE1 (NON-YELLOWING 1), NYC1 (NON-YELLOW COLORING 1), and PAO (PHEOPHORBIDE A OXYGENASE), and senescence associated genes (SAGs) on one hand (Qiu et al., 2015), and on the other hand, inhibits the function of the chloroplast maintenance factor GLK1 (GOLDEN-LIKE 1) via protein-protein interaction (Rauf et al., 2013). Expression of ORE1 in young leaves is repressed at the posttranscriptional level, and during aging, the repression of ORE1 expression is alleviated due to age-dependent down-regulation of MIR164 expression by EIN2 (ETHYLENE INSENSITIVE 2, a key regulator essential for ethylene signaling) (Kim et al., 2009). It has also been shown that expression of ORE1 is positively regulated by several transcription factors, including EIN3/EIL1 (ETHYLENE INSENSITIVE 3/EIN3-LIKE 1), ATAF1 (ARABIDOPSIS TRANSCRIPTION ACTIVATION FACTOR 1), ABI5/EEL (ABA INSENSITIVE 5/ENHANCED EM LEVEL), and PIF4/5 (PHYTOCHROME-INTERACTING FACTOR 4/5) (Li et al., 2013; Sakuraba et al., 2014; Song et al., 2014). Thus, ORE1 acts in multiple coherent feed-forward loops to promote leaf senescence by integrating signals from ethylene, abscisic acid (ABA), salinity and light/dark into developmental aging.
Light is a key environmental factor influencing the onset and progress of leaf senescence. Darkness (light deprivation), low intensity of light or shade (low Red: Far-Red ratios) are known to induce leaf senescence (Lim et al., 2007; Brouwer et al., 2012, 2014; Liebsch and Keech, 2016). Recent studies have shown that in Arabidopsis, the red light photoreceptor phyB plays a role in inhibiting leaf senescence (Sakuraba et al., 2014), whereas in far red light enriched environment, phyA represses but phyB induces leaf senescence (Lim et al., 2018). In addition, recent studies showed that a group of bHLH proteins named phytochrome-interacting factors (PIFs) also promote leaf senescence. PIF4 and PIF5, whose protein accumulation is stimulated by darkness or shade, can directly activate the expression of EIN3 and ORE1 to promote leaf senescence (Sakuraba et al., 2014). In another study, it was shown that PIF4 regulates chlorophyll degradation, chloroplast activity, dark-induced ethylene biosynthesis and ethylene-induced leaf senescence (Song et al., 2014). These studies suggest that light and ethylene signaling pathways converge on EIN3 and ORE1 to regulate leaf senescence. Furthermore, recent studies showed that leaf senescence is also regulated by the circadian clock. For example, it was shown that the evening complex (EC) can directly regulate the expression of MYELOCYTOMATOSIS-RELATED PROTEIN 2 (MYC2), a key transcription factor mediating jasmonates (JA)-induced leaf senescence (Zhang et al., 2018). In another study, it was reported that PRR9 (PSEUDO-RESPONSE REGULATOR 9), a key component of the circadian clock, directly regulates the expression of ORE1 and MIR164, thus forming a feed-forward loop regulating leaf senescence (Kim et al., 2018). Despite the progress made in this field, the detailed molecular mechanisms of light signaling regulating leaf senescence, particularly how light signaling integrates with ethylene signaling and developmental aging to coordinately regulate the onset of leaf senescence, still remain largely unclear.
Arabidopsis FHY3 (FAR-RED ELONGATED HYPOCOTYL 3) and FAR1 (FAR-RED IMPAIRED RESPONSE 1) were initially identified as two positive regulators of phytochrome A signaling and far-red light mediated photomorphogenic development (Hudson et al., 1999; Wang and Deng, 2002). They encode two homologous transcription factors derived from transposase and they regulate phyA signaling by direct activating the expression of FHY1 (FAR-RED ELONGATED HYPOCOTYL 1) and FHL (FHY1-LIKE), whose gene products encodes two homologous chaperone proteins required for light-induced phyA nuclear translocation (Lin et al., 2007). Follow-up studies have demonstrated that FHY3 and FAR1 also play a wide range of biological roles, including UV-B signaling, circadian clock entrainment and flowering, chloroplast biogenesis and chlorophyll biosynthesis, ABA signaling and branching (Wang and Wang, 2015). Recently, it was reported that FHY3 and FAR1 also regulate leaf senescence by directly repressing the expression of WRKY28 and salicylic acid (SA) biosynthesis (Tian et al., 2020).
In this study, we demonstrate that FHY3 and FAR1 repress leaf senescence by physically interacting with EIN3 and PIF5, and inhibiting their transcription activation activity on ORE1 and other SAGs. Our results expand the functional roles of FHY3 and FAR1, and deepen our understanding of the molecular mechanisms regulating leaf senescence through integration of the light and ethylene signaling pathways. Our results suggest that FHY3 and FAR1 act in an age gating mechanism to prevent precocious leaf senescence.
Results
FHY3 and FAR1 Repress Leaf Senescence and Depend on EIN3 and EIL1
We previously showed that FHY3 and FAR1 proteins physically interact with both EIN3 and PIF5 transcription factors (Liu et al., 2017, 2020), while both EIN3 and PIF5 were reported to up-regulate the expression of ORE1, a key NAC transcription factor promoting leaf senescence (Sakuraba et al., 2014; Qiu et al., 2015). Thus, we hypothesized that FHY3 and FAR1 may regulate leaf senescence through modulating the functionality of the EIN3-ORE1 and PIF5-ORE1 transcriptional modules. To test our hypothesis, we first compared the leaf phenotype of the fhy3 far1 double mutant and wild type plant (Col-0) grown under long-day (16 h light/8 h darkness) conditions. The result showed that the fhy3 far1 plants indeed showed an obvious early leaf senescence phenotype, with lower chlorophyll contents and higher expression levels of several well-known senescence-induced genes (SEN4, SAG12, SAG13 and SAG29) in the fourth leaves of 30 and 36 day-old plants compared with those in the same-aged wild type plants (Supplementary Figures 1A–D). Consistent with this, the leaves of FHY3 overexpressors (FHY3OE) senesced later than the wild type plants (Supplementary Figure 2). Since dark treatment is known to induce rapid and synchronous leaf senescence and is adopted to simulate natural senescence (Weaver et al., 1998; Weaver and Amasino, 2001), thus we exposed the third and fourth rosette leaves detached from 4-week-old wild type (Col-0) and fhy3 far1 mutant plants to darkness. We found that the detached leaves from fhy3 far1 mutants showed significantly faster senescence than the wild type, with significantly lower chlorophyll content (Figures 1A,B), consistent with their effects on age-dependent senescence. The expression levels of SEN4 and SAG12 were also much higher in the fhy3 far1 mutant than those in wild type plants (Figure 1C). Moreover, with 4-day darkness treatment (4 DDI), 32-day-old fhy3-11 and fhy3 far1 plants exhibited early leaf senescence when compared with the same-aged wild type plants, while the FHY3OE plants showed late leaf senescence (Supplementary Figure 2). Collectively, these results indicate that FHY3 and FAR1 negatively regulate leaf senescence with or without darkness induction.
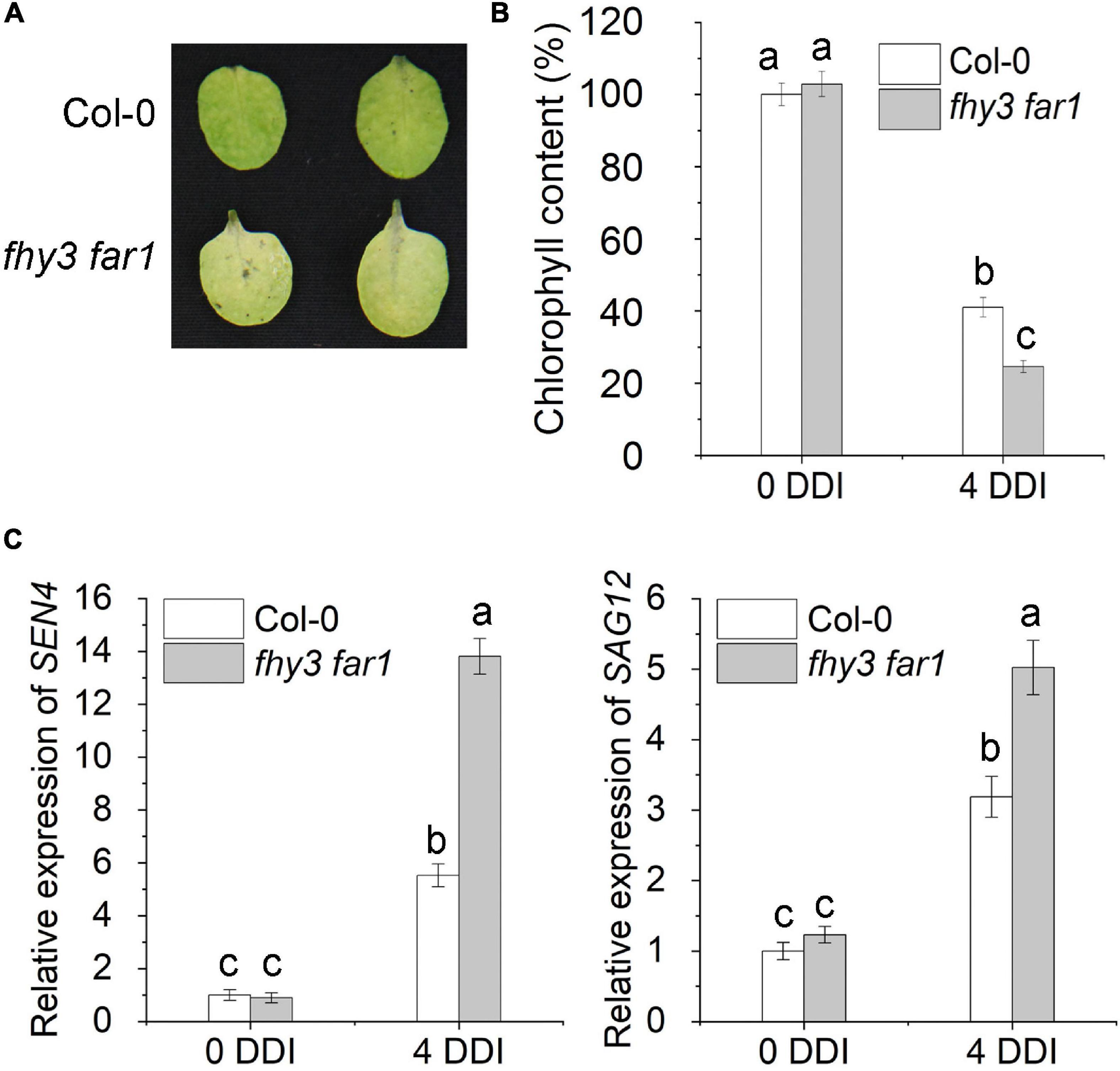
Figure 1. FHY3/FAR1 negatively regulates leaf senescence upon darkness induction. (A) The senescence phenotypes of the detached leaves of 4-week-old Col-0 wild type and fhy3 far1 double mutant plants incubated under darkness for 4 days (4 DDI). (B) The chlorophyll content of the fourth leaves in (A). Error bars represent SD (n = 6). Letters indicate significant differences by two-sided LSD test (p < 0.05). (C) Quantitative RT-PCR analysis of SEN4 (left) and SAG12 (right) gene expression in the fourth leaves of Col-0 and fhy3 far1 without (0 DDI) or with darkness induction for 4 days (4 DDI). Error bars represent SD (n = 3). Letters indicate significant differences by two-sided LSD test (p < 0.05).
It is well known that ethylene promotes leaf senescence, and two closely related transcription factors, EIN3 and EIL1, are essential for ethylene signaling (Chao et al., 1997). It has been shown that EIN3 and EIL1 can directly activate the expression of several senescence-associated genes, such as ORE1, NAP, and WRKY75 (Qiu et al., 2015; Guo et al., 2017; Zhang et al., 2017), to promote leaf senescence. We previously showed that FHY3 and FAR1 can directly interact with EIN3 and EIL1 in vivo and in vitro to coordinately regulate the phosphate starvation response in Arabidopsis (Liu et al., 2017). Thus, we examined the available databases of EIN3 and FHY3 target genes (Ouyang et al., 2011; Chang et al., 2013; Wang et al., 2016) and identified several senescence associated genes among the co-regulated genes by FHY3 and EIN3, including WRKY75, ORE1, NAP, SAG20, and SAG21. qRT-PCR analysis verified that the expression patterns of these genes indeed changed in the fhy3 far1 mutant compared with the wild type (Supplementary Figure 3).
To investigate the genetic interaction between FHY3/FAR1 and EIN3/EIL1 in regulating leaf senescence, we constructed the fhy3 ein3 eil1 triple mutant and compared its leaf senescence phenotype with that of ein3 eil1, and fhy3-11. We found that after 4 days in darkness, the detached leaves of the fhy3 ein3 eil1 mutant, but not fhy3-11, maintained green just like the ein3 eil1 parental mutant (Figure 2A). In addition, their distinct leaf yellowing phenotypes of the detached 4-day-old leaves were consistent with the results of a quantitative assay of chlorophyll contents in these mutants (Figure 2B). We also observed a similar senescence pattern for naturally senescence plants with or without darkness treatment (Supplementary Figure 4). Moreover, expression of the senescence associated genes SAG12, ORE1, and WRKY75 at the indicated leaf ages were also in consistent with the senescence phenotypes of the dark-treated leaves (Figures 2C–E). Taking together, these results indicate that FHY3 and FAR1 act upstream of EIN3/EIL1 to regulate leaf senescence.
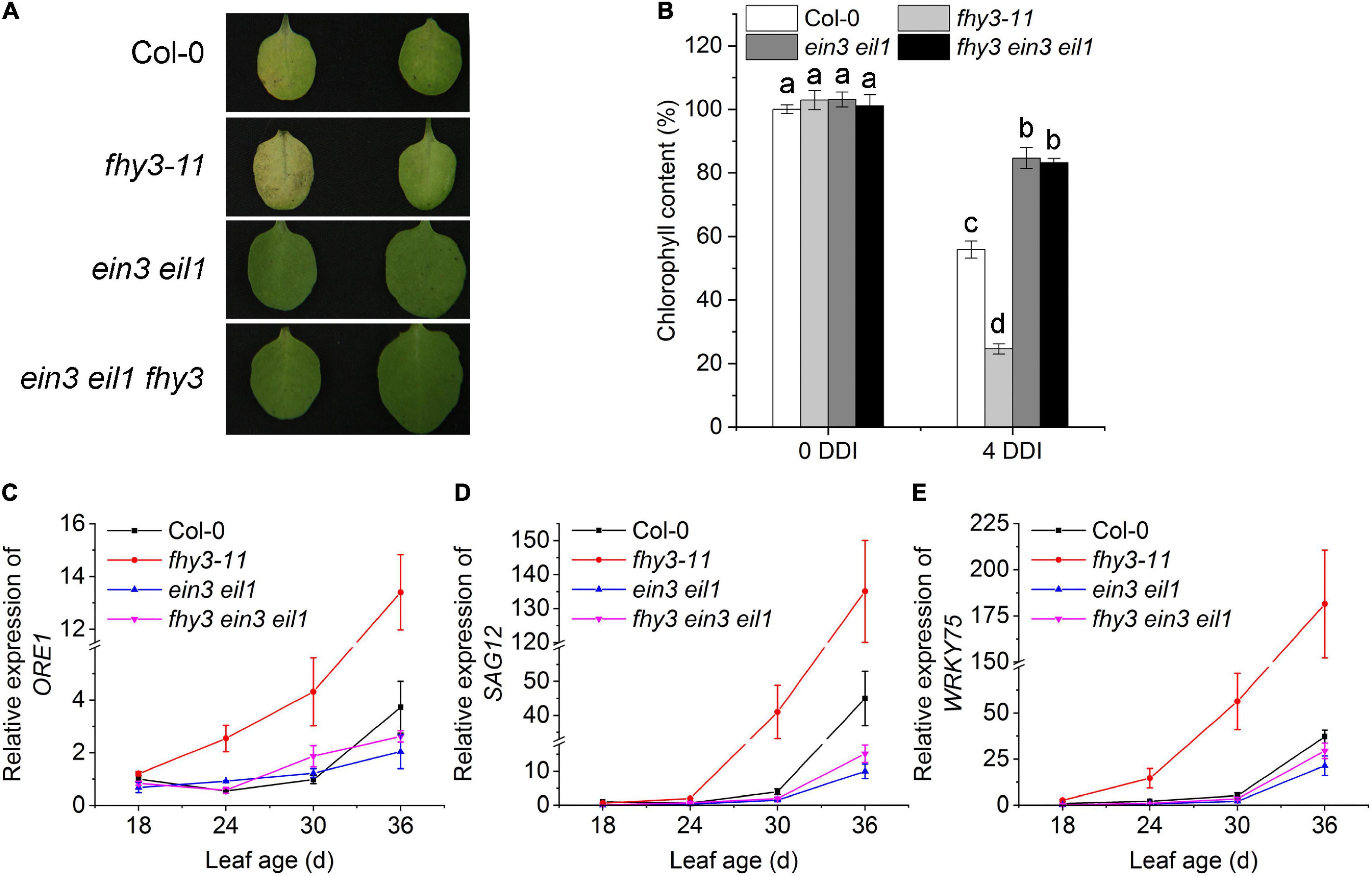
Figure 2. Function of FHY3/FAR1 in regulating leaf senescence depends on EIN3/EIL1. (A) The senescence phenotypes of the detached leaves of 4-week-old Col-0, fhy3-11, ein3 eil1, and fhy3 ein3 eil1 plants incubated under darkness for 4 days. (B) The chlorophyll content of the fourth leaves in (A). Letters indicate significant differences by two-sided LSD test (p < 0.05). (C–E) Quantitative RT-PCR analysis of the expression of ORE1 (C), SAG12 (D), and WRKY75 (E) in the fourth leaves of Col-0, fhy3-11, ein3 eil1, fhy3 ein3 eil1 at the indicated leaf age. Error bars represent SD (n = 3).
ORE1 Acts Downstream of FHY3 and FAR1
Previous studies have shown that ORE1, a key regulator of leaf senescence, is a direct target gene of EIN3 (Qiu et al., 2015). To elucidate the genetic relationship between ORE1 and FHY3/FAR1 in controlling leaf senescence, we generated ore1 single mutant using CRISPR/Cas9 technology (Supplementary Figures 5A,B) and constructed ore1 fhy3 far1 triple mutant. The fourth detached leaves from two independent ore1 single mutants, ore1-3 and ore1-4, showed later leaf senescence with darkness treatment than those from wild type plants (Supplementary Figure 5C). The detached leaves of the fhy3 far1 mutant became yellow after dark treatment for 4 days, whereas the detached leaves of ore1 and ore1 fhy3 far1 remained green (Figure 3A and Supplementary Figure 5C). The distinct leaf yellowing phenotypes were consistent with the results of chlorophyll content assay (Figure 3B). Similarly, the 32-day-old plants with or without darkness induction also showed that the ore1 fhy3 far1 triple mutant exhibited a slower senescence phenotype than the fhy3 far1 double mutant (Supplementary Figure 5D). Consistently, the expression of SAG12 and SEN4 was induced much slower and lower in the ore1 fhy3 far1 triple mutant compared with the fhy3 far1 double mutant, similar to the ore1 single mutant (Figures 3C,D). These observations indicate that ORE1 acts downstream of FHY3 and FAR1 in regulating leaf senescence.
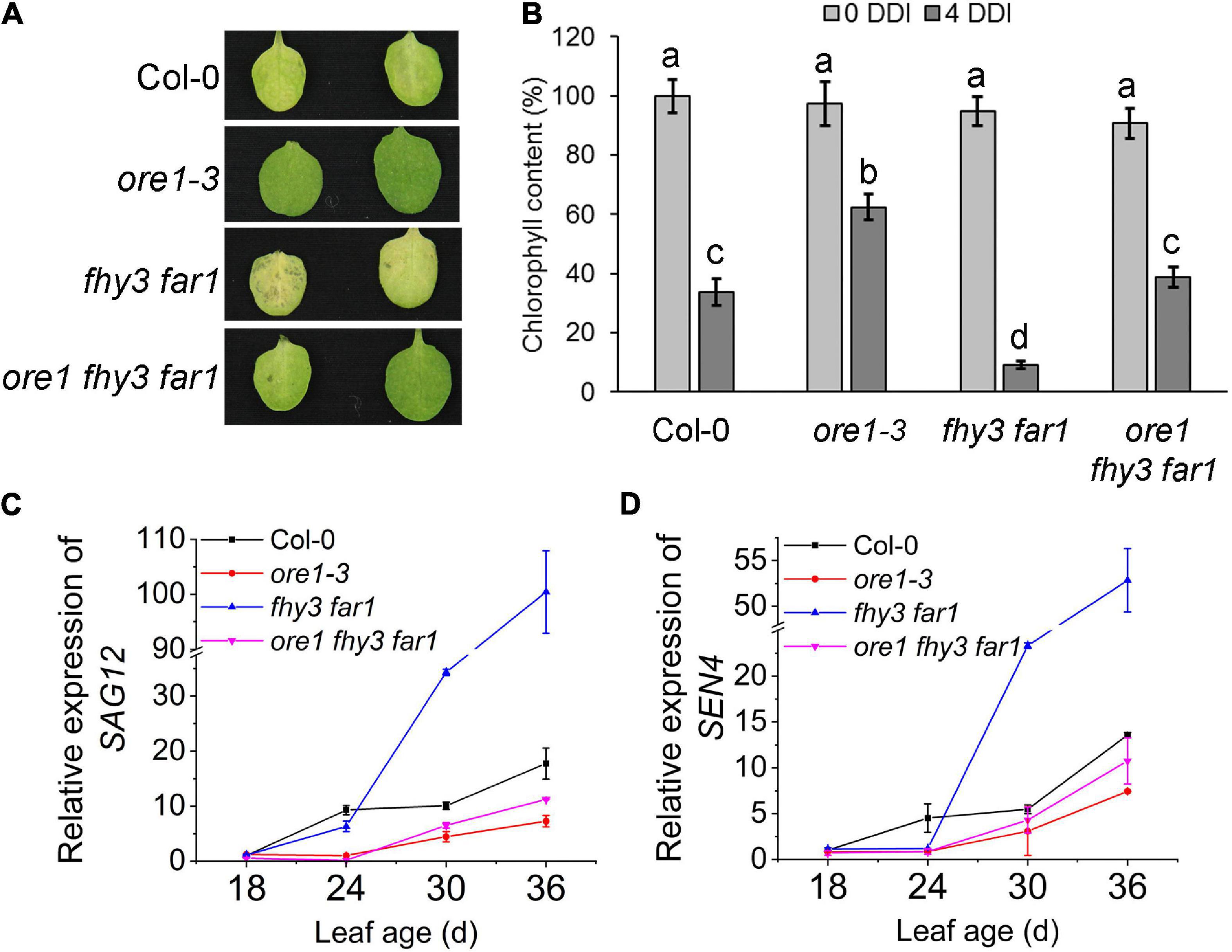
Figure 3. ORE1 acts downstream of FHY3 and FAR1. (A) The senescence phenotypes of the detached leaves of 4-week-old Col-0, ore1-3, fhy3 far1, and ore1 fhy3 far1 plants incubated under darkness for 4 days. The ore1-3 mutant was described in Supplementary Figure 5. (B) The chlorophyll content of the fourth leaves in (A). Error bars represent SD (n = 6). Letters indicate significant differences by two-sided LSD test (p < 0.05). (C,D) Quantitative RT-PCR analysis of SAG12 (C) and SEN4 (D) expression in the fourth leaves of Col-0 and fhy3 far1 at the indicated leaf age. Error bars represent SD (n = 3).
Considering that WRKY75 is another direct target of EIN3 (Chang et al., 2013), we also constructed wrky75 fhy3 far1 triple mutant and observed its senescence phenotype. The leaf senescence phenotype and chlorophyll content showed that the mutation of WRKY75 partially rescued the early senescence phenotype of fhy3 far1 (Supplementary Figures 6A,B). Consistent with this, the expression of SAG12 and SEN4 was much higher in the 30 and 36-day wrky75 fhy3 far1 triple mutant compared with those in the same age wrky75 or wild type, and was close to that in the fhy3 far1 double mutant (Supplementary Figures 6C,D), suggesting that WRKY75 may also play a minor role in FHY3/FAR1-regulated leaf senescence.
FHY3 Represses the DNA Binding Activity of EIN3 to the ORE1 Promoter
We previously reported that FHY3 and FAR1 can directly interact with the DNA binding domain of EIN3/EIL1 proteins (Liu et al., 2017), we thus speculated whether FHY3 and FAR1 can regulate the function of EIN3/EIL1 during leaf senescence. Results from a dual-luciferase reporter (DLR) system with a 3.5-kb ORE1 promoter sequence in Nicotiana benthamiana leaves (Figures 4A,B) showed that EIN3 alone promoted the transcription of ORE1, whereas FHY3 alone seemed have no obvious effect on ORE1 transcription (Figures 4A,B). However, when FHY3 was co-expressed with EIN3 in N. benthamiana leaves, the induction of ORE1 expression by EIN3 was significantly repressed (Figures 4A,B), suggesting that FHY3 represses the transcriptional activation activity of EIN3 on ORE1. We next examined whether the physical interaction between FHY3 and EIN3 may affect the DNA binding activity of EIN3. We first used yeast one-hybrid assay to test the effect of FHY3 protein on binding of EIN3 to the EIN3 binding site (EBS, 5′-ATGAACCT-3′, 5x EBS was used here) in the ORE1 promoter. The results showed that there was no obvious binding between the FHY3 protein and ORE1 promoter or the 5xEBS fragment (Figure 4C). However, we found that when FHY3 protein was added (construct AD-FHY3), the ability of EIN3 binding to the ORE1 promoter or the 5xEBS fragment was drastically decreased (Figure 4C). To confirm this, we detected the direct binding activity of EIN3 by electrophoretic mobility shift assay (EMSA). Since the N-terminal fragment of EIN3 (EIN3N, amino acids 141–352) contains the DNA binding domain and has been verified to possess the binding ability (Li et al., 2013), thus we produced the EIN3N proteins and tested its binding activity to the biotin-labeled 60-bp ORE1 promoter fragment (containing the EBS, designed as Biotin-ORE1 EBS). We found that when FHY3 protein was added, the ability of EIN3 binding to the ORE1 promoter was drastically decreased (Figure 4D). Since no binding of FHY3 to the ORE1 promoter was detected (Figure 4C), these results suggest that FHY3 likely regulates ORE1 expression via the FHY3-EIN3 interaction rather than through direct binding to the ORE1 promoter. To verify this, we generated transgenic plants expressing HA-tagged EIN3 (EIN3-HA) in the wild type (EIN3-HA/Col-0) and fhy3 far1 double mutant backgrounds (EIN3-HA/fhy3 far1). Chromatin immunoprecipitation combined with quantitative PCR (ChIP-qPCR) showed more enrichment of the ORE1 promoter fragment containing the EBS (Solano et al., 1998) in the EIN3-HA/fhy3 far1 seedlings, compared to the EIN3-HA/Col-0 seedlings (Figure 4E). These results indicate that FHY3 represses the DNA binding activity of EIN3 to the ORE1 promoter.
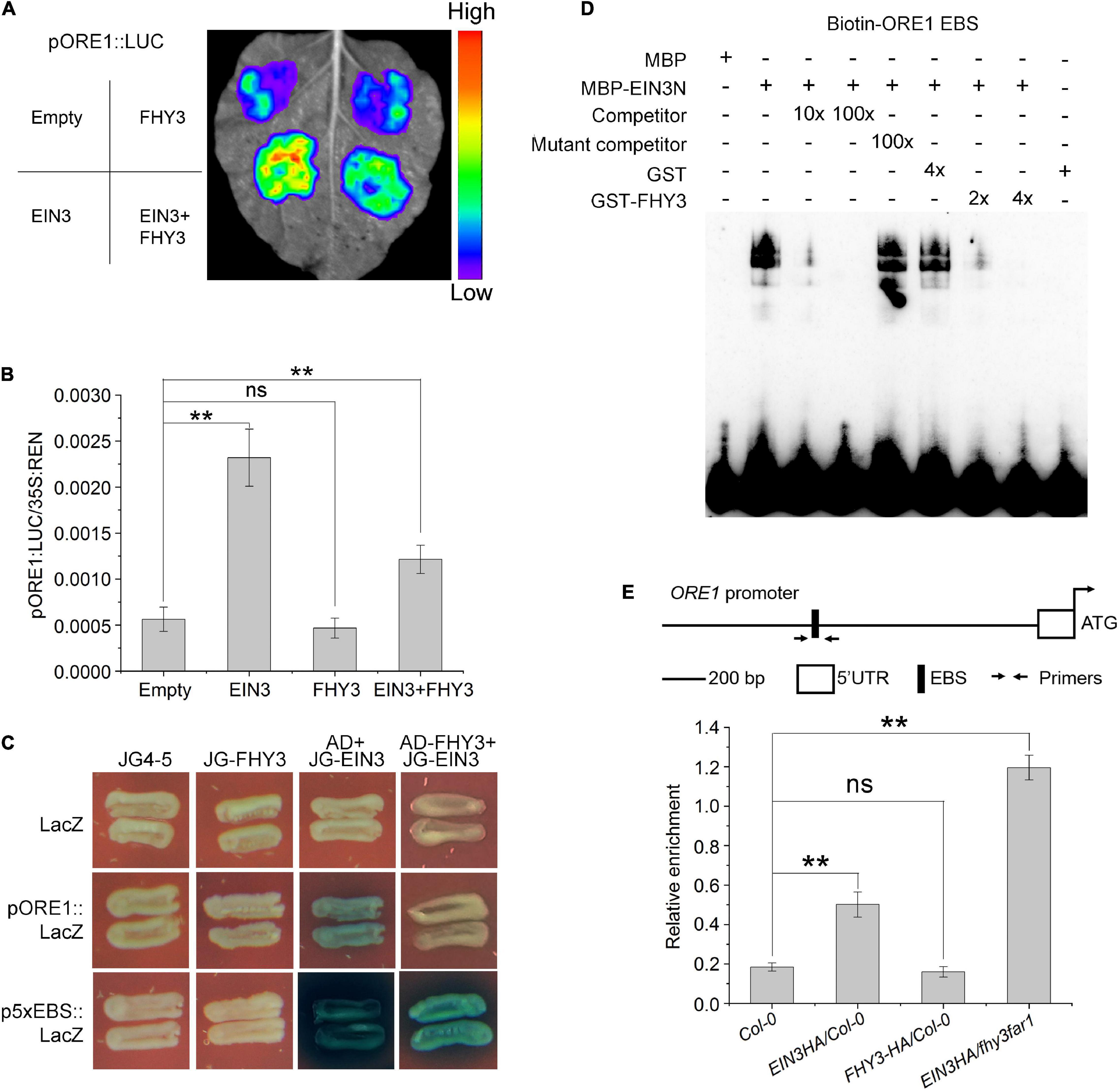
Figure 4. FHY3 repress the DNA binding activity of EIN3. (A) Transient expression assay shows that the addition of FHY3 leads to down-regulation of the ORE1 expression. (B) Dual-luciferase assay of LUC expression in (A). The expression of REN was used as an internal control. LUC/REN ration represents the relative activity of the promoters. Values are means ± SD (n = 3). Significant difference was identified with Student’s t-test. ns, no significance; **p < 0.05. The pSPYNE-35S and pCAMBIA1307 empty vectors were used as controls. (C) Yeast one-hybrid assay shows that EIN3 binds to the ORE1 promoter through the EIN3-binding site (EBS) and FHY3 partially disrupted the interaction between EIN3 and the ORE1 promoter. 5x EBS, 5 repeats of the EBS in the ORE1 promoter. (D) EMSA results show that FHY3 affects the binding of EIN3 (EIN3N, aa 141–352) to the ORE1 promoter. Different concentration of GST-FHY3 protein (aa 186–839) was applied (4 μg for lane 7, 8 μg for lane 8). Eight μg of GST was added to lane 6 as a negative control. (E) ChIP-qPCR shows the in vivo binding of EIN3 to EBS in the ORE1 promoter. Cross-linked chromatins from EIN3-HA were precipitated with anti-HA antibodies. Eluted DNA was subjected to amplification of the neighboring sequences of EBS by qRT-PCR. Col-0 and FHY3-HA plants were used as negative controls. Error bars represent SD (n = 3). Significant difference was identified with Student’s t-test. ns, no significance; **p < 0.05.
FHY3 Represses the DNA Binding Activity of PIF5
Previous studies have shown PIF4 and PIF5 promote leaf senescence by activating the expression of EIN3, ORE1, ABI5, and EEL (Sakuraba et al., 2014). Our previous work showed that FHY3 and FAR1 interact with PIF5 both in vivo and in vitro, and that the bHLH domain of PIF5, which is necessary for DNA binding, is responsible for the interaction with FHY3 (Liu et al., 2020). We thus speculated that FHY3 and FAR1 may also regulate PIF5 function in inducing leaf senescence. To test this possibility, we constructed fhy3 pif5 double mutant and phenotypic assay showed that, compared with wild type, pif5-3 leaves displayed a little delayed senescence and the fhy3 pif5 double mutant exhibited an intermediate phenotype between the fhy3-11 and pif5-3 single mutants after dark treatment. Consistent with this, chlorophyll content in the fhy3 pif5 double mutant was lower than the pif5-3 single mutant but higher than the fhy3-11 single mutant (Figures 5A,B). Moreover, the expression patterns of ORE1, SAG12, and SEN4 in leaves of these mutants were consistent with their senescence phenotypes (Figures 5C–E). These results suggest that FHY3 acts antagonistically with PIF5 in regulating leaf senescence. In support of this, we found that overexpression of FHY3 partially repressed the early senescence phenotype of the PIF5OE plants (Supplementary Figures 7A,B).
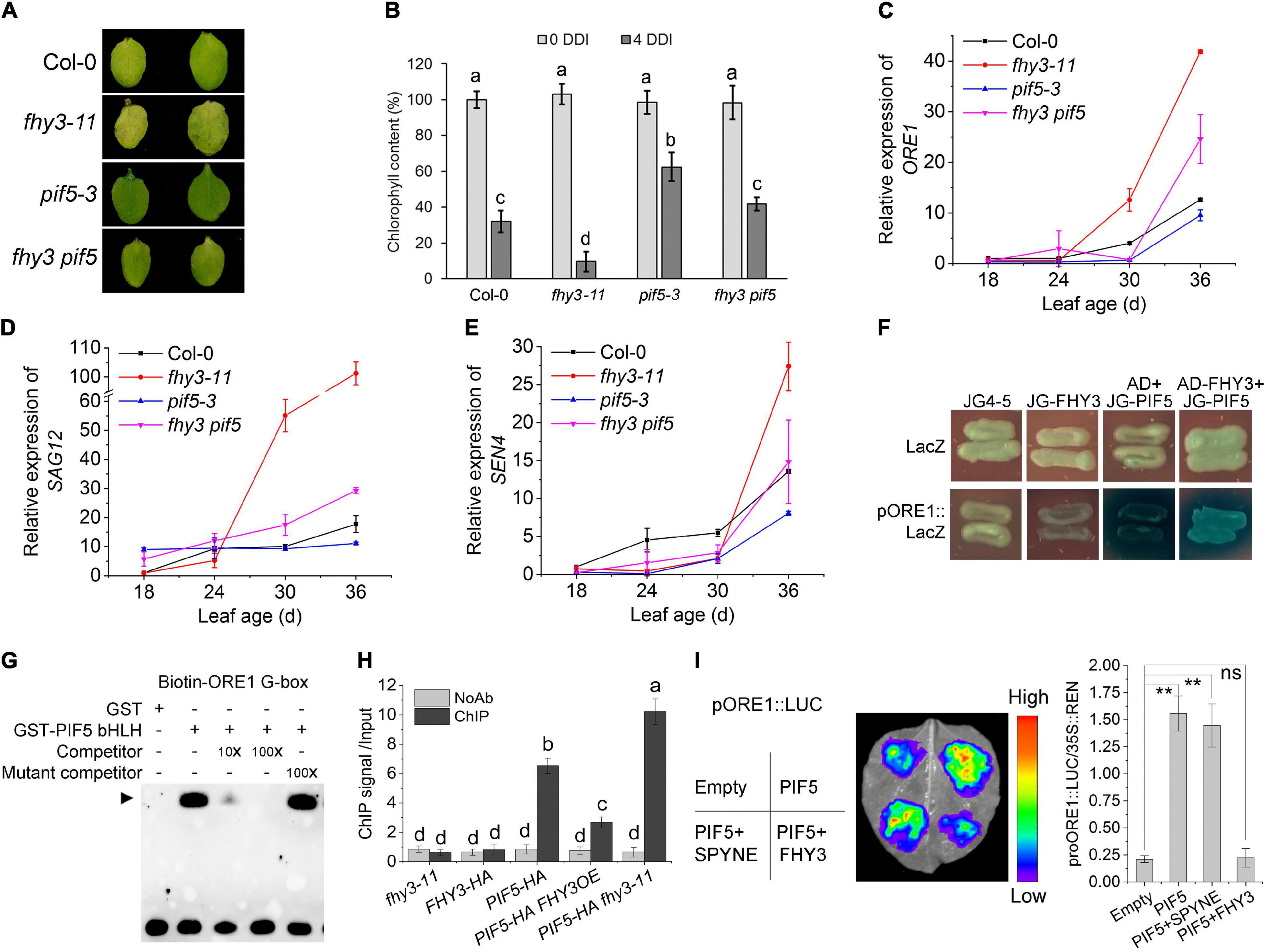
Figure 5. FHY3/FAR1 represses the transcriptional activity of PIF5. (A) The senescence phenotypes of the detached leaves of 4-week-old Col-0, pif5-3, fhy3-11, and fhy3 pif5 plants incubated under darkness for 4 days. (B) The chlorophyll content of the fourth leaves in (A). Error bars represent SD (n = 6). Letters indicate significant differences by two-side LSD test (p < 0.05). (C–E) Quantitative RT-PCR analysis of ORE1 (C), SAG12 (D), and SEN4 (E) expression in the fourth leaves of Col-0, fhy3-11, pif5-3, and fhy3 pif5 at the indicated leaf age. Error bars represent SD (n = 3). (F) Yeast one-hybrid assay shows that PIF5 binds to the ORE1 promoter and that FHY3 interferes with its binding. (G) EMSA shows that PIF5 directly binds to the G-box motif in the ORE1 promoter. (H) ChIP-qPCR assay using p35S:PIF5-HA transgenic seedlings shows that the enrichment of ORE1 promoter fragment by PIF5 was inhibited in the presence of FHY3. Leaf tissues from 4-week-old plants were harvested. Values are mean ± SD (n = 3). Letters indicate significant differences by two-sided LSD test (p < 0.05). (I) Transient expression assay using N. benthamiana leaves. Tobacco leaves were infiltrated with A. tumefaciens transformed with the indicated reporter and effector constructs. The relative LUC activity normalized to REN activity (LUC/REN) are shown. Error bars represent SD (n = 3). Significant difference was identified with Student’s t-test. ns, no significance; **p < 0.05. The pSPYNE-35S empty vector was used as the control.
We next tested whether FHY3 can repress PIF5’s DNA binding activity to the ORE1 promoter. Our yeast one-hybrid assay and EMSA experiments showed that PIF5 could directly bind to the ORE1 promoter fragment (Figures 5F,G), which is consistent with previous report by Sakuraba et al. (2014), and the addition of FHY3 proteins repressed or interfered with the binding activity of PIF5 (Figure 5F). ChIP-qPCR and DLR assays also verified that PIF5 could directly bind to the ORE1 promoter and induce expression of ORE1 in vivo, while addition of FHY3 repressed PIF5-induced expression of ORE1 (Figures 5H,I), suggesting that the interaction between FHY3 and PIF5 represses PIF5 binding to the ORE1 promoter.
FHY3 Mediates the Formation of a Tri-Protein Complex to Regulate Leaf Senescence
We previously showed that FHY3 protein directly interacts with both EIN3 and PIF5 (Liu et al., 2017, 2020), thus we wondered whether FHY3, EIN3, and PIF5 can form a tri-protein complex. To test this hypothesis, we conducted yeast three-hybrid experiment and luciferase complementation imaging (LCI) assay in tobacco. Yeast three-hybrid result showed that the interaction between EIN3 and PIF5 could hardly be detected in yeast, but addition of FHY3 obviously increased the interaction between EIN3 and PIF5 (Figure 6A), suggesting that FHY3 may bridge the interaction between EIN3 and PIF5. Similarly, the LCI results showed that the interaction between EIN3 and PIF5 was very weak, but when FHY3 was co-expressed, the luciferase activity was strongly induced (Figure 6B).
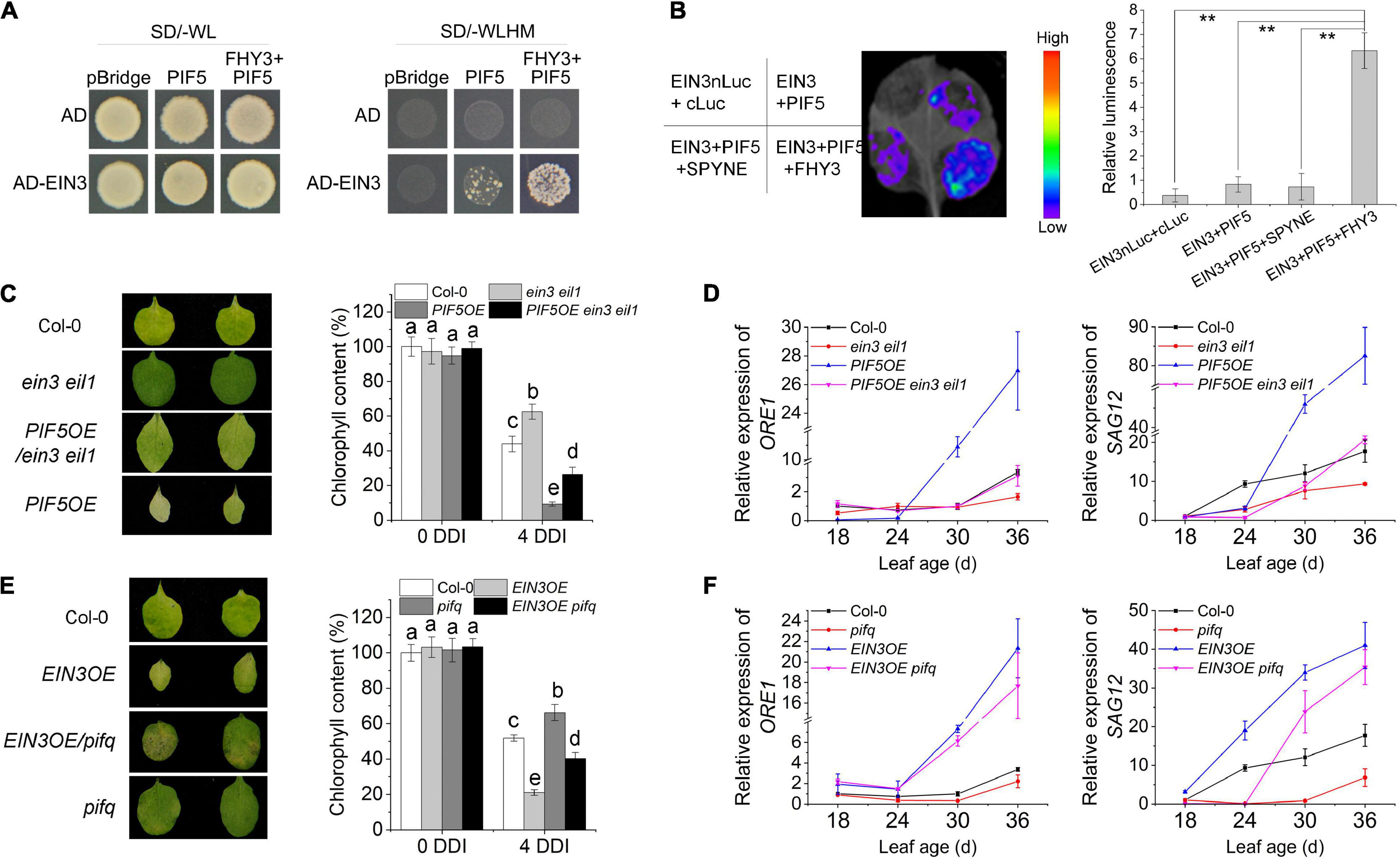
Figure 6. FHY3 and FAR1 mediate the formation of a tri-protein complex to regulate leaf senescence. (A) Yeast three-hybrid assay demonstrates that FHY3 bridges a tri-protein complex containing EIN3, FHY3, and PIF5. (B) Luciferase complementation imaging (LCI) assay and relative luminescence intensity quantification show that FHY3 enhances the interaction between EIN3 and PIF5 in vivo. Error bars represent SD (n = 3). Letters indicate significant differences by two-sided LSD test (p < 0.05). (C) The senescence phenotypes and the chlorophyll content of detached leaves of 4-week-old ein3 eil1, PIF5OE/ein3 eil1, and PIF5OE plants incubated under darkness for 4 days. Letters indicate significant differences by two-sided LSD test (p < 0.05). (D) Quantitative RT-PCR analysis of ORE1 and SAG12 expression in the fourth leaves of ein3 eil1, PIF5OE/ein3 eil1 and PIF5OE at the indicated leaf age. Error bars represent SD (n = 3). (E) The senescence phenotypes and the chlorophyll content of detached leaves of 4-week-old EIN3OE, EIN3OE/pifq and pifq plants incubated under darkness for 4 days. Letters indicate significant differences by two-sided LSD test (p < 0.05). (F) Quantitative RT-PCR analysis of ORE1 and SAG12 expression in the fourth leaves of EIN3OE, EIN3OE/pifq and pifq at the indicated leaf age. Error bars represent SD (n = 3).
To further investigate the genetic relationship between PIF5 and EIN3/EIL1, we generated the PIF5OE/ein3 eil1 mutant combination by genetic crosses. Leaf phenotyping, chlorophyll content measurement and downstream senescence-associated genes (ORE1 and SAG12) expression assay all revealed that mutation of EIN3 and EIL1 partially rescued the early senescence phenotype of PIF5OE (Figures 6C,D). We also generated the EIN3OE/pifq mutant. Phenotypic assay showed that its leaves senesced earlier than pifq but later than EIN3OE (Figure 6E). The expression levels of their downstream targets ORE1 and SAG12 in the EIN3OE/pifq plants were also intermediate between their parents EIN3OE and pifq mutants (Figure 6F). These results suggest that PIF5 likely works in parallel with EIN3/EIL1, but downstream of FHY3.
Regulation of FHY3 Expression by Developmental Age and Darkness
To investigate how the expression and protein levels of FHY3 and FAR1 were regulated during the aging process, we analyzed the expression of FHY3 and FAR1 at the four indicated age in wild type (Col-0) leaves. qRT-PCR analysis showed that the expression of FHY3 and FAR1 was up-regulated in leaves of about 24-day-old and then down-regulated afterward (Figure 7A). To examine developmental regulation of the FHY3 protein level, we generated transgenic plants expressing GFP-FHY3 fusion protein driven by the 35S promoter (GFP-FHY3/Col-0). The functionality of the over-expressed GFP-FHY3 fusion protein was verified by phenotyping under mimicked shade conditions (Supplementary Figure 8). Total protein was extracted from the fourth leaf at the indicated leaf ages and we found that the levels of GFP-FHY3 fusion protein rapidly decreased in leaves older than 24 days (Figure 7B). This result was verified in wild type plants using anti-FHY3 antibodies (Supplementary Figure 9). By contrast, most of the senescence-associated genes (including ORE1) were sharply up-regulated in leaves older than 24 days, as previously shown (Figure 7C and Supplementary Figures 1D,E). These results indicate that FHY3 possibly functions at the early stage to repress leaf senescence.
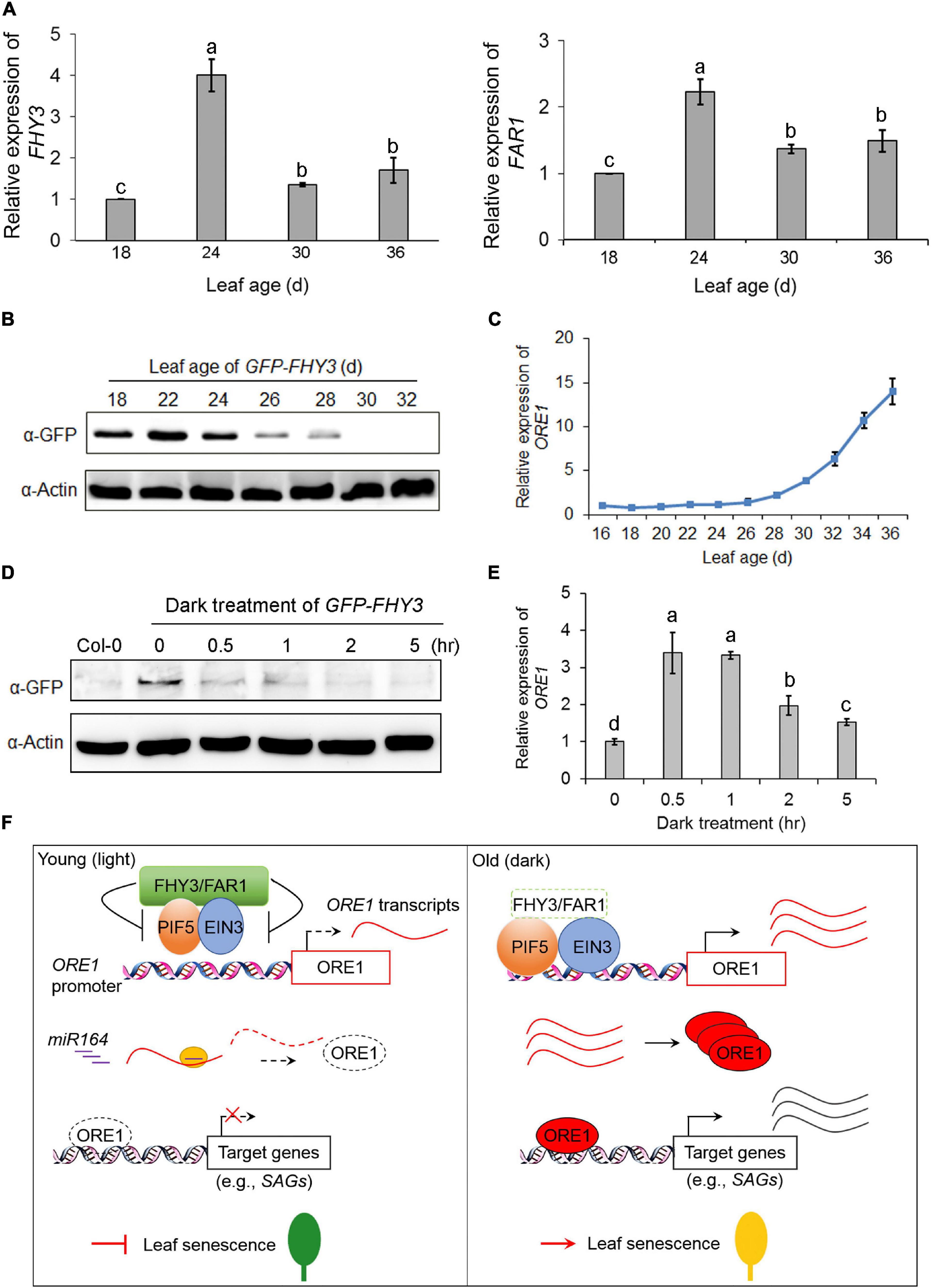
Figure 7. qRT-PCR analysis of FHY3 and FAR1 transcripts and immunoblot analysis of FHY3 protein accumulation. (A) qRT-PCR analysis of FHY3 (left) and FAR1 (right) expression in the fourth leaves of Col-0 at the indicated leaf age. Error bars represent SD (n = 3). Letters indicate significant differences by LSD test (p < 0.05). (B) Immunoblot assays showing GFP-FHY3 fusion protein levels in the fourth leaves of the GFP-FHY3 overexpressors at the indicated leaf age. Actin was used as the internal control. (C) qRT-PCR analysis of ORE1 expression in the fourth leaves of the GFP-FHY3 overexpressors at the indicated leaf age. Error bars represent SD (n = 3). (D) Western blotting assay showing that FHY3 protein level rapidly declined in seedlings treated with darkness. The GFP-FHY3 overexpressor seedlings were grown in continuous light for 7 days and then incubated in darkness for the indicated time lengths and then harvested for immunoblot analysis. Anti-GFP antibodies were used to detect FHY3 proteins and actin was adopted as a loading control. (E) qRT-PCR analysis of ORE1 expression in the fourth leaves of the GFP-FHY3 overexpressors at the indicated points of dark treatment. Error bars represent SD (n = 3). Letters indicate significant differences by LSD test (p < 0.05). (F) A proposed age gating model of FHY3 and FAR1 in regulating leaf senescence. In young, green leaves (or under light conditions), FHY3 protein is accumulated above a threshold level, and it represses the DNA binding activity of EIN3 and PIF5 to the ORE1 promoter, thus repressing ORE1 expression. In addition, ORE1 transcript is targeted for degradation by miR164 at a posttranscriptional level. As a result, leaf senescence is inhibited (left). In old leaves (or under darkness), FHY3 and FAR1 protein levels decrease, thus lifting their repression on EIN3 and PIF5, leading to ORE1 expression. Meanwhile, ORE1 transcript accumulates due to reduced expression of MIR164, allowing translation of ORE1 protein to promote leaf senescence (right).
Next, we examined the changes of FHY3 protein levels during dark treatment. The GFP-FHY3 transgenic seedlings were grown under continuous white light for 7 days and then transferred to darkness for the indicated times. Western blot analysis showed that the FHY3 protein levels decreased quickly upon dark treatment (Figure 7D). By contrast, we observed a rapid up-regulation of ORE1 gene expression in dark-treated seedlings (Figure 7E).
Discussion
One effective way to increase crop productivity is to increase planting density. However, high-density planting could trigger shade avoidance syndrome, including exaggerated stem elongation, less branching, early flowering and premature leaf senescence, thus lowering crop productivity (Brouwer et al., 2014). Our previous studies showed that FHY3 and FAR1 play important roles in regulating multiple aspects of shade avoidance response, including hypocotyl elongation, branch number, flowering time, and plant defense (Liu et al., 2019; Xie et al., 2020a,b).
In this study, we collected several lines of evidence to show that FHY3 and FAR1 act as negative regulators of leaf senescence. We showed that fhy3 far1 mutant senesced earlier (turn yellow earlier, more rapid loss of chlorophyll, and earlier induction of senescence-associated gene expression) than wild type under normal long-day conditions, as well as in dark-treated detached leaves (Figure 1 and Supplementary Figure 1). Genetic epistasis assay showed that FHY3 and FAR1 act upstream of EIN3, EIL1, PIF5 and ORE1 (Figures 2–5). We further showed that FHY3 and FAR1 directly interact with EIN3, EIL1, and PIF5 and repress their binding to the ORE1 promoter (Figures 4, 5; Liu et al., 2020). Moreover, we accumulated evidence suggesting that FHY3, EIN3 and PIF5 can form a tri-protein complex(es) to coordinately regulate leaf senescence (Figure 6). Further, we showed that the protein level of FHY3 is rapidly down-regulated in leaves older than 24 days/or is rapidly down-regulated by dark treatment, concomitant with the observed rapid induction of ORE1 and other SAG genes (Figure 7). Based on these results, we propose a model that FHY3 and FAR1 act as an age gating mechanism to prevent precocious leaf senescence. In young leaves (less than 24 days old) or plants under normal light conditions, FHY3/FAR1 proteins accumulate and they inhibit the DNA binding activities of EIN3 and PIF5 to the ORE1 promoter through direct physical interaction. As a result, ORE1 expression is reduced. In addition, the expression of ORE1 is further negatively regulated by miR164 at a posttranscriptional level in young leaves (Kim et al., 2009). Thus, both transcriptional and posttranscriptional repression of ORE1 expression may constitute a “double-secure” mechanism to prevent precocious leaf senescence in young leaves. During aging or under dark conditions, FHY3 protein level decreases so that its inhibitory effect on EIN3 and PIF5 is lifted. Meanwhile, the expression of EIN3 and PIF5 is up-regulated, while expression of MIR164 is down-regulated (Kim et al., 2009; Li et al., 2013; Song et al., 2014). These molecular events collectively lead to rapid induction of ORE1 expression, thus promoting leaf senescence (Figure 7F). Our model is consistent with and provides a mechanistic explanation for the earlier reports that activation of ethylene signaling can only trigger leaf senescence in leaves that have reached a defined age (Buchanan-Wollaston et al., 2005; Jing et al., 2005).
Our previous study demonstrated that both FHY3 and FAR1 directly bind to the promoter of CIRCADIAN CLOCK ASSOCIATED1 (CCA1), a key component of the core oscillator of the circadian clock, and activate its expression, while PIF5 could also directly bind to the CCA1 promoter but repress its expression (Liu et al., 2020). Furthermore, we showed that PIF5 physically interacts with FHY3 and suppresses its transcriptional activation activity on CCA1 expression (Liu et al., 2020). On the other hand, it has been demonstrated that CCA1 directly suppresses ORE1 expression to counteract leaf senescence (Song et al., 2018). In this study we found that fhy3 pif5 double mutant exhibited an intermediate level of leaf senescence phenotype between the fhy3-11 and pif5-3 single mutants and that overexpression of FHY3 partially repressed the early senescence of the PIF5OE plants. Thus there is a probability that the compromised phenotype of the fhy3 pif5 double mutant is due to the negative effect of PIF5 on transcriptional activity of FHY3 and therefore reduction of CCA1 expression.
It is also worth noting that earlier studies have found that in the fhy3 far1 mutant, the levels of both SA and reactive oxygen species (ROS) increased (Wang et al., 2016). Both SA and ROS are known to act as positive regulators of leaf senescence (Buchanan-Wollaston et al., 2005; Rivas-San and Plasencia, 2011). Thus, it is possible that FHY3 and FAR1 may also regulate leaf senescence through the SA and ROS signaling pathways. Interestingly, these studies have shown that FHY3 and FAR1 can directly regulate the expression of myo-Inositol-1-phosphate synthase1 (MIPS1) and HEMB1 (which encodes a 5-aminolevulinic acid dehydratase in the chlorophyll biosynthetic pathway), and that constitutive expression of MIPS1 or HEMB1 can partially or largely rescued the cell death phenotype and oxidative stress in fhy3 far1 (Ma et al., 2016; Wang et al., 2016). Interestingly, a recent study reported that the transcription factor WRKY75 can promotes SA production by inducing the transcription of SA INDUCTION-DEFICIENT2 (SID2) and suppresses H2O2 scavenging, partly by repressing the transcription of CATALASE2 (CAT2) (Guo et al., 2017). Similarly, a recent study reported that FHY3 and FAR1 regulate leaf senescence by repressing the expression of WRKY28 and thus suppressing SA biosynthesis (Tian et al., 2020). The detailed molecular mechanism interconnecting FHY3/FAR1-mediated transcriptional regulation of ORE1 with the SA and ROS signaling pathways in coordinating leaf senescence will be an interesting avenue for future research.
Besides ethylene and SA, other phytohormones, including cytokinins, auxins, ABA, and JA are also known to regulate leaf senescence (Lim et al., 2007; Jibran et al., 2013). Particularly worth mentioning, FHY3 and FAR1 have been previously shown to regulate multifaceted developmental processes by integrating light signaling with multiple hormone signaling pathways (Wang and Wang, 2015). For example, we previously showed that FHY3 and FAR1 can directly activate the expression of ABI5 and regulate ABA responses in plants (Tang et al., 2013). We also showed that FHY3 directly interacts with EIN3 and that both of them can directly bind to distinct cis-elements on the promoter of PHOSPHOATE STARVATION RESPONSE1 (PHR1) to coordinately regulate light- and ethylene-mediated phosphate starvation response (Liu et al., 2017). We additionally showed that FHY3 can also physically with multiple JASMONATE ZIM-DOMAIN (JAZ) proteins and MYC2, a group of key regulators of JA responses, to coordinately regulate JA-mediated growth and defense responses (Liu et al., 2019). Thus, it is expected that FHY3 and FAR1 may regulate leaf senescence through cross talking with these hormone signaling pathways as well. These results on one hand, suggest that FHY3 and FAR1 may indeed act as a signaling hub regulating leaf senescence via integrating various internal and external signals, and on the other hand, call for more detailed research to fully elucidate the detailed molecular mechanisms of FHY3 and FAR1 in regulating leaf senescence.
Materials and Methods
Plant Materials and Growth Conditions
The Arabidopsis thaliana ecotype Columbia (Col-0) is the parent line for all mutants and transgenic plants used in this study. Transgenic lines in different genetic backgrounds and multiple mutants are constructed by genetic crosses. fhy3-11 (SALK_002711) and far1-4 (SALK_031652) were obtained from the ABRC, ein3 eil1 (Alonso et al., 2003), pif5-3, pifq (Zhong et al., 2012) were described previously. FHY3 overexpressors (FHY3OE) and FAR1 overexpressors (FAR1OE) have been described in Ma et al. (2017) and Liu et al. (2019), respectively. The wrky75 mutant has been described in Guo et al. (2017). Double or triple mutants were generated by genetic crosses.
Arabidopsis seeds were surface-sterilized and plated on Murashige and Skoog (MS) medium (4.4 g/L MS salts, 1% [w/v] sucrose, pH 5.8, and 8 g/L agar). After stratification at 4°C for 3 days, the seedlings were transferred to soil and grown at 22°C under long-day conditions (16-h light/8-h dark). The white light source was provided by LED (PAR = 100 μmol m–2 s–1).
Construction of Plasmids and Generation of Transgenic Plants
For JG-EIN3, JG-PIF5, and JG-FHY3 constructs, the individual full-length coding sequences of EIN3, PIF5, and FHY3 were ligated to the vector pB42AD and designed as JG-EIN3, JG-PIF5, and JG-FHY3, respectively. For AD-FHY3 construct, the coding sequences of FHY3 was ligated to the pEG202 vector (Clontech) and designed as AD-FHY3. To create pORE1:LacZ, the ORE1 promoter was amplified from genomic DNA and inserted into pLacZ2μ vector (Lin et al., 2007) digested with EcoRI and XhoI. To construct p5x EBS:LacZ, 5 repeats of the ORE1 promoter fragment containing the EIN3 binding site (5′-aatatactttacaaggttcatgcatgcatacattgttttc-3′) was amplified and inserted into SalI digested pLacZi2μ vector (Lin et al., 2007). The five tandem repeats of EBS (5x EBS) were designed in the primer pairs P03 and P04. Two subfragments, 5x EBS-1 (amplified with primer pair P03) and 5xEBS-2 (amplified with primer pair P04), together with pLacZi2μ vector (digested with EcoRI/XhoI) were incubated in 2x Gibson Assembly Master Mix (New England Biolabs) to generate the construct p5xEBS:LacZ for yeast one hybrid.
For yeast three-hybrid assay, the coding sequence of EIN3 was cloned from cDNA into EcoRI-digested pGADT7 vector to generate the AD-EIN3 construct. PIF5 coding sequence was amplified from cDNA and cloned into EcoRI-digested pBridge vector to generate the pBridge-PIF5 construct. Then FHY3 coding sequence was amplified from cDNA and inserted into BglII-digested pBridge-PIF5 to generate the pBridge-PIF5-FHY3 construct.
To generate GST-FHY3 (aa 186-TAA), the FHY3 fragment (aa 186-TAA) was amplified from cDNA and inserted into EcoRI digested pGEX-5x-1.
Plasmids of the 35S promoter-driven effectors for dual Luc reporter system were described previously (Liu et al., 2017, 2019). To generate pORE1:Luc, a 3.5-kb genomic promoter sequence upstream of the coding region of ORE1 was amplified, and inserted into SalI digested pGreenII-0800 vector (Hellens et al., 2005).
The oligonucleotide primers for the constructs above are summarized in Supplementary Table 1. The constructs were verified by DNA sequencing analysis.
FHY3OE (35S:FLAG-FHY3-HA), PIF5OE (35S:PIF5-HA) were lab stock (Liu et al., 2017; Xie et al., 2017). FHY3OE PIF5OE transgenic plants were generated by crossing FHY3OE and PIF5OE. PIF5OE/ein3 eil1 transgenic plants were generated by crossing PIF5OE and ein3 eil1. GFP-FHY3 (35S:GFP-FHY3) transgenic plants were obtained by cloning FHY3 coding sequences into the pEGAD vector and transforming the construct into Col-0 background. Homozygotes were characterized by hygromycin resistance in the T3 population. EIN3-HA (35S:EIN3-HA) transgenic plants were obtained by cloning EIN3 coding sequences into the pCAMBIA1307 vector and transforming the construct into Col-0 background. Homozygotes were characterized by hygromycin resistance in the T3 population. EIN3HA/fhy3 far1 transgenic plants were obtained by crossing EIN3-HA transgenic plants with fhy3 far1, and the homozygotes were characterized by PCR-based genotyping of the F2 population.
Measurement of Chlorophyll Content
Chlorophyll contents were measured in the third and fourth leaves using a SPAD Chlorophyll Meter (SPAD-502 Plus, Konica Minolta). Each leaf was evenly divided into 5–6 spots, and one measurement was taken per spot. The average value of the 5–6 measurements (SPAD Unit) represents a single data point and one biological replicate. Six individual leaves of each genotype are measured, and three biological replicates were performed.
RNA Extraction, Reverse Transcription, and Real-Time PCR
Total RNA was extracted from the fourth leaf of the indicated leaf ages using Trizol reagent (Invitrogen). Reverse transcription was performed using reverse transcriptase (Tiangen). cDNA was diluted 1:10 and subjected to quantitative PCR using SuperReal PreMix Plus (Tiangen) and a 7500 Real Time PCR System (Applied Biosystems, United States) cycler according to the manufacturer’s manual. The level of ACT2 transcript was adopted as an internal control. The oligonucleotide primers for Real-time PCR are summarized in Supplementary Table 1.
Yeast One-Hybrid Assay
To detect the binding of EIN3, FHY3 or PIF5 proteins to the ORE1 promoter, plasmids of indicated JG-fusion proteins (such as JG-FHY3, JG-EIN3 or JG-PIF5) were cotransformed with the indicated ORE1 promoter reporter plasmids into the yeast strain EGY48. Transformants grown on the SD/-Trp/-Ura medium (Clontech, United States) were transferred to the selection medium containing raffinose, galactose, and 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (Amresco, United States) for blue color development. To test the effect of FHY3 on the binding of PIF5 or EIN3 to ORE1 promoter, AD-FHY3 and JG-EIN3 or JG-PIF5 were cotransformed with the indicated ORE1 promoter reporter plasmids into the yeast strain EGY48. Transformants grown on the SD/-Ura/-Trp/-His medium (Clontech, United States) were transferred to the selection medium for blue color development.
Yeast Three-Hybrid Assay
Vectors were cotransfected into the AH109 yeast strain according to the manufacturer’s protocol (Clontech, United States). Yeast were grown on selection plate (SD/-Trp/-Leu) for 3–4 days and then transferred to selection plate (SD/-Trp/-Leu/-Met/-His). Positive interactions were recognized by growth on the SD/-Trp/-Leu/-Met/-His plate.
Electrophoretic Mobility Shift Assay
Biotin-labeled/unlabeled or mutant ORE1 and EIN3 promoter oligonucleotide probes were listed in Supplementary Table 1. MBP-EIN3N and GST-PIF5 bHLH vectors were constructed as described previously (Liu et al., 2017; Xie et al., 2017). GST, GST-PIF5 bHLH, GST-FHY3 (aa 186-TAA), MBP, and MBP-EIN3N fusion proteins were expressed in the Escherichia coli strain BL21. The recombinant proteins were purified using either GST-agarose or amylose resin affinity chromatography. EMSA was performed using a LightShift Chemiluminescent EMSA kit (Pierce, United States) according to the manufacturer’s instructions. Briefly, synthetic DNA oligonucleotide probes labeled with biotin were incubated with the indicated recombinant proteins in the presence or absence of excess amounts of unlabeled competitors for 10 min at room temperature. The DNA-protein complexes were separated on 6% native polyacrylamide gels. To analyze FHY3 protein function, 1, 2, and 4 μg of GST-FHY3 were used.
Chromatin Immunoprecipitation Combined With Quantitative PCR
Chromatin immunoprecipitation was performed as described previously (Saleh et al., 2008). Briefly, 2 g of leaf tissues from 4-week-old Col-0 (used as a negative control, set to a value of 1), EIN3-HA, EIN3-HA/fhy3 far1 were collected and fixed in 1% formaldehyde for 20 min under a vacuum, followed by neutralization using 0.125 M glycine for additional 5 min. The leaves were then washed for three times with water followed by chromatin isolation. Anti-HA antibodies were added to the sonicated chromatin followed by incubation overnight to precipitate the bound DNA fragments. After salmon sperm-sheared DNA/protein A agarose beads, the bound DNA was eluted and amplified with primers corresponding to sequences in the ORE1 promoter. Each experiment was performed three times using different pools of seedlings. The oligonucleotide primers for qPCR are summarized in Supplementary Table 1.
Transient Dual-Luciferase Reporter System
Transient expression in N. benthamiana was performed as described previously (Sparkes et al., 2006). A. tumefaciens strain GV3101 carrying the reporter plasmid (pORE1:LacZ or p5x EBS:LacZ) and effector plasmids were cultured in liquid Luria-Bertani medium overnight. The dense cultures were incubated into fresh medium by 1:100 dilution and incubated for 6–8 h. The bacteria were then pelleted at 4,000 rpm for 15 min, and resuspended in an infiltration buffer (5 g/L glucose, 10 mM MgCl2, 10 mM MES-KOH, pH 5.7; adding 150 μM acetosyringone right before use) to and OD600 of 0.6. The resuspended agrobacteria containing different constructs were mixed equally and then infiltrated into tobacco leaves using 1 mL syringes without needles. Plants were incubated for 2 or 3 days. Firefly luciferase and Renilla luciferase activities were assayed as described previously (Li et al., 2010).
Luciferase Complementation Imaging
The firefly LCI assays were performed using N. benthamiana leaves. Plasmids for LCI were described previously (Liu et al., 2017, 2019). Both the nLUC- (N-terminal luciferase) and cLUC- (C-terminal luciferase) fusion constructs with or without pSPYNE-FHY3 (empty vector pSPYNE as control) were co-infiltrated into N. benthamiana leaves via A. tumefaciens-mediated co-infiltration. The infiltrated plants were incubated for 2 or 3 days before examining using the Night SHADE LB 985 Plant Imaging System (Berthold, German).
Accession Numbers
Sequences of all genes analyzed in this work are available at TAIR under the flowing AGI codes: ORE1 (AT5G39610), FHY3 (AT3G22170), FAR1 (AT4G15090), PIF5 (AT3G59060), NAP (AT1G69490), WRKY75 (AT5G13080), SEN4 (AT4G30270), EIN3 (AT3G20770), SAG12 (AT5G45890), SAG13 (AT2G29350), SAG21 (AT4G02380), SAG20 (AT3G10985), SAG29 (AT5G13170), UBQ10 (AT4G05320), ACT2 (AT3G18780).
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
HYW, YX, and MM designed the research and wrote the manuscript. MM, YL, BW, YX, HBW, and DK performed the experiments and analyzed the data. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the grant from the National Natural Science Foundation of China (31770210).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Hongwei Guo (South University of Science and Technology) for kindly providing wrky75 mutant and Jigang Li (China Agricultural University) for providing the anti-FHY3 antibodies.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.770060/full#supplementary-material
References
Alonso, J. M., Stepanova, A. N., Solano, R., Wisman, E., Ferrari, S., Ausubel, F. M., et al. (2003). Five components of the ethylene-response pathway identified in a screen for weak ethylene-insensitive mutants in Arabidopsis. Proc. Natl. Acad. Sci. U S A 100, 2992–2997. doi: 10.1073/pnas.0438070100
Brouwer, B., Gardestrom, P., and Keech, O. (2014). In response to partial plant shading, the lack of phytochrome A does not directly induce leaf senescence but alters the fine-tuning of chlorophyll biosynthesis. J. Exp. Bot. 65, 4037–4049. doi: 10.1093/jxb/eru060
Brouwer, B., Ziolkowska, A., Bagard, M., Keech, O., and Gardestrom, P. (2012). The impact of light intensity on shade-induced leaf senescence. Plant Cell Environ. 35, 1084–1098. doi: 10.1111/j.1365-3040.2011.02474.x
Buchanan-Wollaston, V., Page, T., Harrison, E., Breeze, E., Lim, P. O., Nam, H. G., et al. (2005). Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J. 42, 567–585. doi: 10.1111/j.1365-313X.2005.02399.x
Chang, K. N., Zhong, S., Weirauch, M. T., Hon, G., Pelizzola, M., Li, H., et al. (2013). Temporal transcriptional response to ethylene gas drives growth hormone cross-regulation in Arabidopsis. Elife 2:e00675. doi: 10.7554/eLife.00675.022
Chao, Q., Rothenberg, M., Solano, R., Roman, G., Terzaghi, W., and Ecker, J. R. (1997). Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 89, 1133–1144. doi: 10.1016/S0092-8674(00)80300-1
Gan, S., and Amasino, R. M. (1997). Making sense of senescence (molecular genetic regulation and manipulation of leaf senescence). Plant Physiol. 113, 313–319. doi: 10.1104/pp.113.2.313
Guo, P., Li, Z., Huang, P., Li, B., Fang, S., Chu, J., et al. (2017). A tripartite amplification loop involving the transcription factor WRKY75, salicylic acid, and reactive oxygen species accelerates leaf senescence. Plant Cell 29, 2854–2870. doi: 10.1105/tpc.17.00438
Hellens, R. P., Allan, A. C., Friel, E. N., Bolitho, K., Grafton, K., Templeton, M. D., et al. (2005). Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1:13. doi: 10.1186/1746-4811-1-13
Hudson, M., Ringli, C., Boylan, M. T., and Quail, P. H. (1999). The FAR1 locus encodes a novel nuclear protein specific to phytochrome A signaling. Genes Dev. 13, 2017–2027. doi: 10.1101/gad.13.15.2017
Jibran, R., Hunter, D. A., and Dijkwel, P. P. (2013). Hormonal regulation of leaf senescence through integration of developmental and stress signals. Plant Mol. Biol. 82, 547–561. doi: 10.1007/s11103-013-0043-2
Jing, H. C., Schippers, J. H., Hille, J., and Dijkwel, P. P. (2005). Ethylene-induced leaf senescence depends on age-related changes and OLD genes in Arabidopsis. J. Exp. Bot. 56, 2915–2923. doi: 10.1093/jxb/eri287
Kim, H., Kim, H. J., Vu, Q. T., Jung, S., McClung, C. R., Hong, S., et al. (2018). Circadian control of ORE1 by PRR9 positively regulates leaf senescence in Arabidopsis. Proc. Natl. Acad. Sci. U S A 115, 8448–8453. doi: 10.1073/pnas.1722407115
Kim, J. H., Nam, H. G., and Lim, P. O. (2016). Regulatory network of NAC transcription factors in leaf senescence. Curr. Opin. Plant Biol. 33, 48–56. doi: 10.1016/j.pbi.2016.06.002
Kim, J. H., Woo, H. R., Kim, J., Lim, P. O., Lee, I. C., Choi, S. H., et al. (2009). Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science 323, 1053–1057. doi: 10.1126/science.1166386
Li, J., Li, G., Gao, S., Martinez, C., He, G., Zhou, Z., et al. (2010). Arabidopsis transcription factor ELONGATED HYPOCOTYL5 plays a role in the feedback regulation of phytochrome A signaling. Plant Cell 22, 3634–3649. doi: 10.1105/tpc.110.075788
Li, Z., Peng, J., Wen, X., and Guo, H. (2013). Ethylene-insensitive3 is a senescence-associated gene that accelerates age-dependent leaf senescence by directly repressing miR164 transcription in Arabidopsis. Plant Cell 25, 3311–3328. doi: 10.1105/tpc.113.113340
Li, Z., Woo, H. R., and Guo, H. (2018). Genetic redundancy of senescence -associated transcription factors in Arabidopsis. J. Exp. Bot. 69, 811–823. doi: 10.1093/jxb/erx345
Liebsch, D., and Keech, O. (2016). Dark-induced leaf senescence: new insights into a complex light-dependent regulatory pathway. New Phytol. 212, 563–570. doi: 10.1111/nph.14217
Lim, J., Park, J. H., Jung, S., Hwang, D., Nam, H. G., and Hong, S. (2018). Antagonistic roles of phyA and phyB in far-red light-dependent leaf senescence in Arabidopsis thaliana. Plant Cell Physiol. 59, 1753–1764. doi: 10.1093/pcp/pcy153
Lim, P. O., Kim, H. J., and Nam, H. G. (2007). Leaf senescence. Annu. Rev. Plant Biol. 58, 115–136. doi: 10.1146/annurev.arplant.57.032905.105316
Lin, R., Ding, L., Casola, C., Ripoll, D. R., Feschotte, C., and Wang, H. (2007). Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science 318, 1302–1305. doi: 10.1126/science.1146281
Liu, Y., Ma, M., Li, G., Yuan, L., Xie, Y., Wei, H., et al. (2020). Transcription factors FHY3 and FAR1 regulate light-induced CIRCADIAN CLOCK ASSOCIATED1 gene expression in Arabidopsis. Plant Cell 32, 1464–1478. doi: 10.1105/tpc.19.00981
Liu, Y., Wei, H., Ma, M., Li, Q., Kong, D., Sun, J., et al. (2019). Arabidopsis FHY3 and FAR1 regulate the balance between growth and defense responses under shade conditions. Plant Cell 31, 2089–2106. doi: 10.1105/tpc.18.00991
Liu, Y., Xie, Y., Wang, H., Ma, X., Yao, W., and Wang, H. (2017). Light and ethylene coordinately regulate the phosphate starvation response through transcriptional regulation of PHOSPHATE STARVATION RESPONSE1. Plant Cell 29, 2269–2284. doi: 10.1105/tpc.17.00268
Ma, L., Tian, T., Lin, R., Deng, X. W., Wang, H., and Li, G. (2016). Arabidopsis FHY3 and FAR1 regulate light-induced myo-inositol biosynthesis and oxidative stress responses by transcriptional activation of MIPS1. Mol. Plant 9, 541–557. doi: 10.1016/j.molp.2015.12.013
Ma, L., Xue, N., Fu, X., Zhang, H., and Li, G. (2017). Arabidopsis thaliana FAR-RED ELONGATED HYPOCOTYLS3 (FHY3) and FAR-RED-IMPAIRED RESPONSE1 FAR1) modulate starch synthesis in response to light and sugar. New Phytol. 213, 1682–1696. doi: 10.1111/nph.14300
Nam, H. G. (1997). The molecular genetic analysis of leaf senescence. Curr. Opin. Biotechnol. 8, 200–207. doi: 10.1016/S0958-1669(97)80103-6
Oh, S. A., Park, J. H., Lee, G. I., Paek, K. H., Park, S. K., and Nam, H. G. (1997). Identification of three genetic loci controlling leaf senescence in Arabidopsis thaliana. Plant J. 12, 527–535. doi: 10.1111/j.0960-7412.1997.00527.x
Ouyang, X., Li, J., Li, G., Li, B., Chen, B., Shen, H., et al. (2011). Genome-wide binding site analysis of FAR-RED ELONGATED HYPOCOTYL3 reveals its novel function in Arabidopsis development. Plant Cell 23, 2514–2535. doi: 10.1105/tpc.111.085126
Park, S. H., Jeong, J. S., Seo, J. S., Park, B. S., and Chua, N. H. (2019). Arabidopsis ubiquitin-specific proteases UBP12 and UBP13 shape ORE1 levels during leaf senescence induced by nitrogen deficiency. New Phytol. 223, 1447–1460. doi: 10.1111/nph.15879
Qiu, K., Li, Z., Yang, Z., Chen, J., Wu, S., Zhu, X., et al. (2015). EIN3 and ORE1 accelerate degreening during ethylene-mediated leaf senescence by directly activating chlorophyll catabolic genes in Arabidopsis. PLoS Genet. 11:e1005399. doi: 10.1371/journal.pgen.1005399
Rauf, M., Arif, M., Dortay, H., Matallana-Ramírez, L. P., Waters, M. T., Nam, H. G., et al. (2013). ORE1 balances leaf senescence against maintenance by antagonizing G2-like-mediated transcription. EMBO Rep. 14, 382–388. doi: 10.1038/embor.2013.24
Rivas-San, V. M., and Plasencia, J. (2011). Salicylic acid beyond defence: its role in plant growth and development. J. Exp. Bot. 62, 3321–3338. doi: 10.1093/jxb/err031
Sakuraba, Y., Jeong, J., Kang, M. Y., Kim, J., Paek, N. C., and Choi, G. (2014). Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nat. Commun. 5:4636. doi: 10.1038/ncomms5636
Saleh, A., Alvarez-Venegas, R., and Avramova, Z. (2008). An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat. Protoc. 3, 1018–1025. doi: 10.1038/nprot.2008.66
Solano, R., Stepanova, A., Chao, Q., and Ecker, J. R. (1998). Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE- INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 12, 3703–3714. doi: 10.1101/gad.12.23.3703
Song, Y., Jiang, Y., Kuai, B., and Li, L. (2018). CIRCADIAN CLOCK- ASSOCIATED 1 inhibits leaf senescence in Arabidopsis. Front. Plant Sci. 9:280. doi: 10.3389/fpls.2018.00280
Song, Y., Yang, C., Gao, S., Zhang, W., Li, L., and Kuai, B. (2014). Age-triggered and dark-induced leaf senescence require the bHLH transcription factors PIF3, 4, and 5. Mol. Plant 7, 1776–1787. doi: 10.1093/mp/ssu109
Sparkes, I. A., Runions, J., Kearns, A., and Hawes, C. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1, 2019–2025. doi: 10.1038/nprot.2006.286
Tang, W., Ji, Q., Huang, Y., Jiang, Z., Bao, M., and Wang, H. (2013). FAR-RED ELONGATED HYPOCOTYL3 and FAR-RED IMPAIRED RESPONSE1 transcription factors integrate light and abscisic acid signaling in Arabidopsis. Plant Physiol. 163, 857–866. doi: 10.1104/pp.113.224386
Tian, T., Ma, L., Liu, Y., Xu, D., Chen, Q., and Li, G. (2020). Arabidopsis FAR-RED ELONGATED HYPOCOTYL3 integrates age and light signals to negatively regulated leaf senescence. Plant Cell 32, 1574–1588. doi: 10.1105/tpc.20.00021
Wang, H., and Deng, X. W. (2002). Arabidopsis FHY3 defines a key phytochrome A signaling component directly interacting with its homologous partner FAR1. EMBO J. 21, 1339–1349. doi: 10.1093/emboj/21.6.1339
Wang, H., and Wang, H. (2015). Multifaceted roles of FHY3 and FAR1 in light signaling and beyond. Trends Plant Sci. 20, 453–461. doi: 10.1016/j.tplants.2015.04.003
Wang, W., Tang, W., Ma, T., Niu, D., Jin, J. B., Wang, H., et al. (2016). A pair of light signaling factors FHY3 and FAR1 regulates plant immunity by modulating chlorophyll biosynthesis. J. Integr. Plant Biol. 58, 91–103. doi: 10.1111/jipb.12369
Weaver, L. M., and Amasino, R. M. (2001). Senescence is induced in individually darkened Arabidopsis leaves, but inhibited in whole darkened plants. Plant Physiol. 127, 876–886. doi: 10.1104/pp.010312
Weaver, L. M., Gan, S., Quirino, B., and Amasino, R. M. (1998). A comparison of the expression patterns of several senescence-associated genes in response to stress and hormone treatment. Plant Mol. Biol. 37, 455–469. doi: 10.1023/A:1005934428906
Woo, H. R., Kim, H. J., Lim, P. O., and Nam, H. G. (2019). Leaf senescence: systems and dynamics aspects. Annu. Rev. Plant Biol. 70, 347–376. doi: 10.1146/annurev-arplant-050718-095859
Xie, Y., Liu, Y., Ma, M., Zhou, Q., Zhao, Y., Zhao, B., et al. (2020a). Arabidopsis FHY3 and FAR1 integrate light and strigolactone signaling to regulate branching. Nat. Commun. 11:1955. doi: 10.1038/s41467-020-15893-7
Xie, Y., Zhou, Q., Zhao, Y., Li, Q., Liu, Y., Ma, M., et al. (2020b). FHY3 and FAR1 integrate light signals with the miR156-SPL module-mediated aging pathway to regulate arabidopsis flowering. Mol. Plant 13, 483–498.
Xie, Y., Liu, Y., Wang, H., Ma, X., Wang, B., Wu, G., et al. (2017). Phytochrome-interacting factors directly suppress MIR156 expression to enhance shade-avoidance syndrome in Arabidopsis. Nat. Commun. 8:348. doi: 10.1038/s41467-017-00404-y
Zhang, S., Li, C., Wang, R., Chen, Y., Shu, S., Huang, R., et al. (2017). The Arabidopsis mitochondrial protease FtSH4 is involved in leaf senescence via regulation of WRKY-dependent salicylic acid accumulation and signaling. Plant Physiol. 173, 2294–2307. doi: 10.1104/pp.16.00008
Zhang, Y., Wang, Y., Wei, H., Li, N., Tian, W., and Chong, K. (2018). Circadian evening complex represses jasmonate-induced leaf senescence in Arabidopsis. Mol. Plant 11, 326–337. doi: 10.1016/j.molp.2017.12.017
Keywords: Arabidopsis, leaf senescence, FHY3/FAR1, EIN3, PIF5, ORE1
Citation: Xie Y, Ma M, Liu Y, Wang B, Wei H, Kong D and Wang H (2021) Arabidopsis FHY3 and FAR1 Function in Age Gating of Leaf Senescence. Front. Plant Sci. 12:770060. doi: 10.3389/fpls.2021.770060
Received: 07 September 2021; Accepted: 07 October 2021;
Published: 28 October 2021.
Edited by:
Ute Hoecker, University of Cologne, GermanyReviewed by:
Cornelia Klose, University of Freiburg, GermanyHongwei Guo, Southern University of Science and Technology, China
Copyright © 2021 Xie, Ma, Liu, Wang, Wei, Kong and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haiyang Wang, d2h5YW5nQHNjYXUuZWR1LmNu
†These authors share first authorship
 Yurong Xie
Yurong Xie Mengdi Ma
Mengdi Ma Yang Liu
Yang Liu Baobao Wang1
Baobao Wang1 Hongbin Wei
Hongbin Wei Haiyang Wang
Haiyang Wang