- 1Institute of Botany, Jiangsu Province and Chinese Academy of Sciences (Nanjing Botanical Garden Mem. Sun Yat-Sen), Nanjing, China
- 2College of Forestry, Nanjing Forestry University, Nanjing, China
Glehnia littoralis is a medicinal halophyte that inhabits sandy beaches and has high ecological and commercial value. However, the molecular mechanism of salt adaptation in G. littoralis remains largely unknown. Here, we cloned and identified a non-specific phospholipase C gene (GlNPC3) from G. littoralis, which conferred lipid-mediated signaling during the salt stress response. The expression of GlNPC3 was induced continuously by salt treatment. Overexpression of GlNPC3 in Arabidopsis thaliana increased salt tolerance compared to wild-type (WT) plants. GlNPC3-overexpressing plants had longer roots and higher fresh and dry masses under the salt treatment. The GlNPC3 expression pattern revealed that the gene was expressed in most G. littoralis tissues, particularly in roots. The subcellular localization of GlNPC3 was mainly at the plasma membrane, and partially at the tonoplast. GlNPC3 hydrolyzed common membrane phospholipids, such as phosphotidylserine (PS), phosphoethanolamine (PE), and phosphocholine (PC). In vitro enzymatic assay showed salt-induced total non-specific phospholipase C (NPC) activation in A. thaliana GlNPC3-overexpressing plants. Plant lipid profiling showed a significant change in the membrane-lipid composition of A. thaliana GlNPC3-overexpressing plants compared to WT after the salt treatment. Furthermore, downregulation of GlNPC3 expression by virus-induced gene silencing in G. littoralis reduced the expression levels of some stress-related genes, such as SnRK2, P5SC5, TPC1, and SOS1. Together, these results indicated that GlNPC3 and GlNPC3-mediated membrane lipid change played a positive role in the response of G. littoralis to a saline environment.
Introduction
Glehnia littoralis Fr. Schmidt ex Miq. is a medicinal and edible plant in the Umbelliferae family. Glehnia littoralis is rich in coumarins, coumarin glycosides, phospholipids, and polysaccharides (Yuan et al., 2002). The peeled and dried roots and rhizomes of G. littoralis are commonly used as a traditional Chinese herbal medicine for moistening the lungs, removing phlegm, relieving cough, curing gastrointestinal disorders, recovering from surgery, and immunoregulation, as well as for their anti-inflammatory properties (Huang et al., 2012; Li et al., 2020). Moreover, the tender leaves of G. littoralis are edible as a vegetable. Therefore, the demand for G. littoralis as a clinical medication and health care product is always high. Glehnia littoralis is a perennial halophyte that grows on sandy beaches in Northern Pacific countries and regions, such as eastern China, Japan, the Korean Peninsula, Russia, and the United States (Li et al., 2018). A previous study elucidated how G. littoralis adapts to high-salinity environments by analyzing its anatomical and morphological characteristics, such as the secretory trichomes and thick cuticle cover on leaves (Voronková et al., 2011), but the molecular mechanism of salt adaptation in G. littoralis remains largely unknown.
The phospholipid bilayer of the plasma membrane is the first barrier to the external environment. Phospholipases, including phospholipase A (PLA1, sPLA2, and pPLA), phospholipase C (PLC), and phospholipase D (PLD) hydrolyze membrane phospholipids, and affect the structure and stability of the cellular membrane and signaling responses to environmental stimuli (Wang, 2005; Scherer et al., 2010; Hong et al., 2016). The products of phospholipases, such as phosphatidic acid (PA) and inositol 1,4,5-trisphosphate, are cellular messengers that regulate various biological processes (Testerink and Munnik, 2005; Wang et al., 2006; Tang et al., 2007). PLD and PLC/diacylglycerol kinases (DGK) play a major role in the stress-induced generation of PA (Munnik and Testerink, 2009). The activities of the phospholipases are induced rapidly, coupled with a highly dynamic level of PA, in response to hormones, as well as abiotic and biotic stimuli (Bargmann and Munnik, 2006; Li et al., 2006; Peters et al., 2010). PA acts as a cellular mediator by binding with some regulatory proteins, anchoring these proteins to the membrane, modulating their catalytic activities, and altering membrane structure (Wang et al., 2006; Testerink and Munnik, 2011). PA-interacting proteins include various kinases, transcription factors, phosphatases, and other target proteins during cellular processes (Zhang et al., 2004, 2012; Yu et al., 2010; Kim et al., 2019; Shen et al., 2019, 2020).
Non-specific phospholipase C (NPC) is a subtype of PLC that hydrolyzes common membrane phospholipids, such as phosphatidylcholine (PC), phosphatidylethanolamine (PE), and phosphatidylserine (PS), to produce sn-1,2-diacylglycerol (DAG) and a corresponding phosphorylated headgroup. DAG is phosphorylated by DGK to generate PA. Unlike PI-PLC (another PLC subtype), which is found widely in animals, plants, and bacteria, NPC is found only in bacteria and plants, and has distinct evolutionary features (Nakamura, 2014; Nakamura and Ngo, 2020). In the model plant Arabidopsis thaliana, six NPCs have been identified, most of which have been reported to be involved in multiple biological processes. NPC4 is associated with the plasma membrane, and the recombinant protein of NPC4 shows enzymatic activity toward PC and PE (Nakamura et al., 2005). NPC4 is involved in the plant response to abscisic acid (ABA), auxin, phosphate deficiency, hyperosmotic conditions, and salt and Al stresses (Nakamura et al., 2005; Gaude et al., 2008; Peters et al., 2010; Wimalasekera et al., 2010; Kocourková et al., 2011; Yang et al., 2021). NPC4 and NPC3 are important in brassinolide-mediated signaling in root development (Wimalasekera et al., 2010). NPC4 is also involved in phosphosphingolipid hydrolysis and remodeling in the presence of a phosphate deficiency during root growth (Yang et al., 2021). NPC1 plays a vital role in plant heat tolerance (Krčková et al., 2015). NPC1 is localized at secretory pathway compartments, such as the endoplasmic reticulum or Golgi apparatus, so is NPC2. NPC2 expression is suppressed after Pseudomonas syringae attack, and NPC2 is involved in plant immune responses, such as PTI, ETI, and SA (Krčková et al., 2018). The double-mutant npc2npc6 displays a lethal homozygous phenotype and a defect in the heterozygous gametophyte (Ngo et al., 2018). NPC5 is a cytosolic protein. NPC5 and its derived DAG mediate lateral root development under salt stress, and NPC5 is involved in galactolipid accumulation during phosphate limitation (Gaude et al., 2008; Peters et al., 2014). The localization of NPC6 is present in both chloroplast and microsomal fractions, but not in cytosolic or nuclear fractions (Cai et al., 2020). NPC6 promotes seed oil content and enhances PC and galactolipid turnover to TAG (Cai et al., 2020).
The important roles of NPCs in other plant species have been gradually revealed following the NPC findings in A. thaliana. For example, Cai et al. (2020) reported that BnNPCs affect seed mass and yield, and that BnNPC6.C01 is positively associated with seed oil content in oilseed rape. In rice, five OsNPCs are identified. The cytosolic and membrane-associated OsNPC1 modulates silicon distribution and secondary cell wall deposition in nodes and grains, affecting mechanical strength and seed shattering (Cao et al., 2016). OsNPC6 is involved in mesocotyl elongation in rice. OsNPC6-overexpressing plants exhibit a shorter mesocotyl than the wild-type (WT) and npc6 mutant (Yang et al., 2021). Yang et al. (2021) also indicated that all five OsNPCs showed plasma membrane localization. These studies suggest that plant NPCs are involved in numerous biological processes, although the functional properties of most plant NPCs await exploration. In the current study, we cloned and identified GlNPC3 from G. littoralis, which played a positive role in the salt stress response.
Materials and Methods
Plant Materials and Growth Conditions
The G. littoralis plants used in this study were obtained and grown as described by Li et al. (2018). The seedlings were subjected to salt (200mM NaCl), drought (20% PEG 6000), or hormone [100μM ABA or 100μM methyl jasmonate (MeJA)] stress treatments for 0, 6, or 24h. The shoots and roots were sampled separately at different time points in each treatment.
Arabidopsis thaliana Columbia (Col-0, WT) and its transgenic seeds were sown on Murashige and Skoog (MS) medium with vitamins (1% sucrose and 1% agar) and supplemented with 75mM NaCl for the salt stress treatment. The plants were grown in a vertical position (for root growth) in a growth chamber under a 14-h-light (23°C)/10-h-dark (20°C) photoperiod. The roots were measured using the ImageJ software (NIH, Bethesda, MD, United States).
In silico Characterization of GlNPC3
The coding sequence (CDS) of GlNPC3 was amplified from G. littoralis cDNA using the GlNPC3-F1 and GlNPC3-R1 primers. The locations of the exons and introns were defined using GSDS V2.0.1 The protein domains were identified with Pfam database.2 The GlNPC3 upstream promoter sequence was amplified using the Genome Walking Kit (Takara, Dalian, China) with the SP1, SP2, and SP3 primers. The plant regulatory elements in the GlNPC3 promoter region were obtained from the PlantCARE database.3 All the primers are listed in Supplementary Table S1. The gene, protein, CDS, and a 2,070-bp promoter sequence of GlNPC3 are shown in Data Sheet 1.
Vector Construction and Plant Transformation
To generate the GlNPC3-overexpressing construct (pSuper::GlNPC3), the CDS was amplified using the GlNPC3-F2/GlNPC3-R2 primers. The product was cloned into the pSuper1300 binary vector, which was pCAMBIA1300 containing a Super promoter (Ni et al., 1995; Cao et al., 2013), and digested with XbaI/SalI. To generate the PromoterGlNPC3::GUS construct, a 2,070-bp promoter sequence was amplified using the Pro-GlNPC3-F/Pro-GlNPC3-R primers and cloned into the PMV plant binary vector (Hua et al., 2021). The primers are listed in Supplementary Table S1.
Arabidopsis thaliana transformation was performed according to the floral dip method (Clough and Bent, 1998). The harvested seeds were selected on MS medium containing 25mg/L hygromycin until the T3 generation. The homozygous lines were used for the experiments.
Subcellular Localization of GlNPC3
The CDS sequence of GlNPC3 was inserted into the pSuper1300GFP vector using the GlNPC3-F3 and GlNPC3-R3 primers to generate pSuper::GFP-GlNPC3 recombinant plasmid. The N-terminus of GlNPC3 was translationally fused to GFP under the control of Super promoter. A plasma membrane (PM) marker (AtCBL1n-OFP) and tonoplast marker (AtCBL6-OFP) were used as described by Batistič et al. (2010). The plasmids were introduced into Agrobacterium tumefaciens (GV3101) for transient expression in tobacco (Nicotiana benthamiana), and the tobacco leaves were infiltrated as described previously (Li et al., 2017). Fluorescence was observed using an LSM 780 confocal microscope (Zeiss, Jena, Germany).
GUS Staining
Histochemical staining for GUS expression in transgenic A. thaliana was performed according to the method of Jefferson (1987).
Protein Expression and Enzyme Activity Assays
The GlNPC3 CDS was cloned into the pCold I vector (Takara) with a His-tag at its N-terminus and transformed into E. coli BL21 (DE3). Isopropyl β-D-thiogalactopyranoside (0.1mM) was added and the culture was incubated at 15°C for 24h to induce expression of the His-GlNPC3 fusion protein. The protein was purified with TALON® Metal Affinity Resin (Clontech, Palo Alto, CA, United States) according to the manufacturer’s protocol.
GlNPC3 activity was assayed as described previously (Nakamura et al., 2005; Peters et al., 2010) with some modifications. The purified His-GlNPC3 protein or total plant protein was incubated for 1h at 37°C in a 500μl reaction mixture (50mM Tris-HCl, pH 7.3, 50mM NaCl, and 5% glycerol) in the presence of a sonicated micellar suspension of 200μM of the fluorescent head-group-labeled lipid substrate. The lipid substrates 18:1 NBD-PS (810198C), 18:1 NBD-PE (810145P), and 18:1 Cy5-PC (850483C) were obtained from Avanti Polar Lipids, Inc. (Alabaster, AL, United States). The reaction was stopped by vigorous vortex with 1ml of ethyl acetate and 0.75ml of 0.45% NaCl. Centrifuge the reaction at 1,500g for 10min and remove supernatant (water-soluble phase) to a new tube carefully, do not suck middle layer (protein) and lower organic phase (lipid). The water-soluble phase was used to determine fluorescence using a microplate reader (SpectraMax ID5; Molecular Devices, San José, CA, United States) with the following parameters: NBD, excitation at 460nm and emission at 534nm; Cy5, excitation at 648nm and emission at 662nm.
RNA Extraction and Quantitative Real-Time PCR
RNA extraction and quantitative real-time PCR (RT-qPCR) were performed according to protocols described previously (Li et al., 2020). The primers for RT-qPCR are listed in Supplementary Table S1.
Lipid Analysis
The lipid extraction and electrospray ionization tandem mass spectrometry (ESI-MS/MS) analyses were performed as described previously (Welti et al., 2002; Devaiah et al., 2006).
Virus-Induced Gene Silencing in G. littoralis
Virus-induced gene silencing (VIGS) was performed as described previously (Liu et al., 2002). A 409-bp fragment of GlPDS (phytoene desaturase gene) was amplified from the G. littoralis CDS using the PDS-F/PDS-R primers and constructed using pTRV II as the positive control. Similarly, 199- and 200-bp fragments of the CDS within GlNPC3 were cloned into pTRVII using the vigs-F1/vigs-R1 and vigs-F2/vigs-R2 primers. The fragments were designed using the SGN VIGS tool4 (Fernandez-Pozo et al., 2015). The recombinant vectors were transferred into A. tumefaciens strain GV3101. Suspensions of A. tumefaciens containing recombinant vectors were mixed with suspension of A. tumefaciens containing pTRV I (helper vector) for infiltration. The silencing effect was monitored in the filtered leaves after 14days by RT-qPCR. The silencing phenotype (photobleaching phenotype) of VIGS-PDS was used as an indicator of VIGS efficiency. All primers are listed in Supplementary Table S1. The fragments used for the VIGS assay are listed in Data Sheet 2.
Results
Cloning and Identification of GlNPC3
The phospholipase-mediated lipid signaling pathway plays an important role in the abiotic stress response in plants. Based on the G. littoralis salt-related transcriptome data obtained in our previous study (Li et al., 2018), we cloned a NPC that showed a positive response to salt treatment in G. littoralis. This gene contained an open reading frame of 1,563bp encoding a protein of 520 amino acids (Figure 1A). Phylogenetic analysis of this NPC revealed a close relationship to Daucus carota L. DcNPC3; therefore, we named this gene GlNPC3 (Supplementary Figure S1). GlNPC3 contained a phosphoesterase domain with three motifs commonly present in plant NPCs (Figures 1B,C). To better understand the function of GlNPC3, the 2,070-bp GlNPC3 promoter sequence was cloned. We analyzed the main regulatory cis-elements in the GlNPC3 promoter region using the PlantCARE database (Figure 1C; Data Sheet 1). Some regulatory elements are involved in plant hormone and abiotic stress responses, such as the light-responsive G-box, GT1-motif, TCT-motif, and GATA-motif; hormones responsive to AREB, the CGTCA-motif, and the TGA-motif; and the stress-responsive MYB, MYC, and STRE.
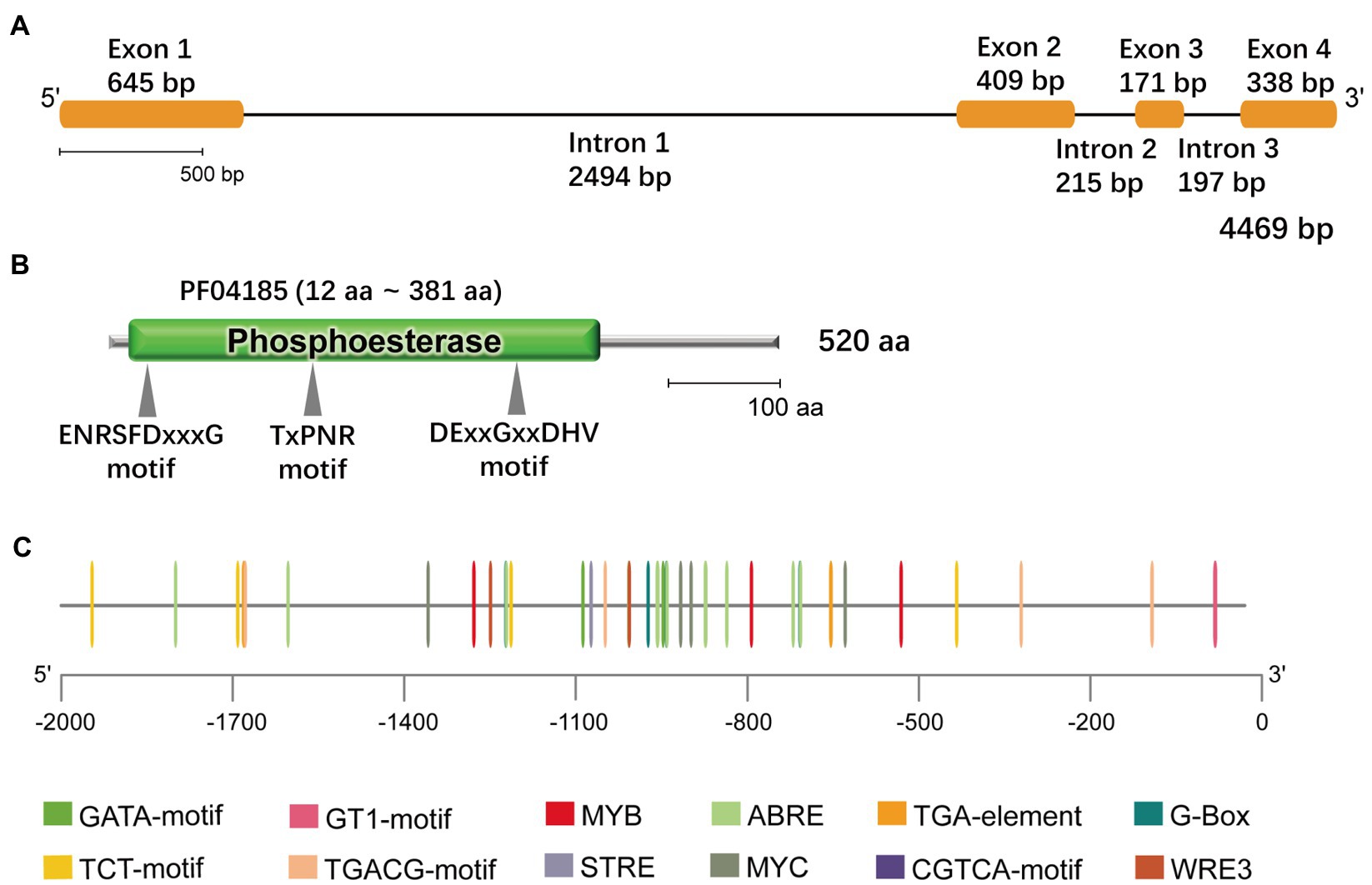
Figure 1. Bioinformatics features of GlNPC3. (A) GlNPC3 exons and introns obtained through GSDS V2.0. The yellow rectangles represent exons, and the black lines represent introns. (B) Schematic representation of the GlNPC3 protein; the phosphoesterase domain (Pfam entry PF04185) and conserved domains are indicated. (C) Plant regulatory elements in the GlNPC3 promoter region were obtained from the PlantCARE database.
GlNPC3 Plays a Positive Role in the Salt Stress Response
We performed RT-qPCR to reveal changes in GlNPC3 gene expression when the G. littoralis seedlings were exposed to ABA, MeJA, NaCl, or osmotic stress (PEG). The results showed that GlNPC3 positively responded to these treatments in shoots and roots, and the expression of GlNPC3 increased continuously within 24h after the salt treatment (Figure 2). The trends in GlNPC3 expression determined by RT-qPCR were consistent with those shown in our previous RNA-seq data (Li et al., 2018). As a stable G. littoralis genetic transformation system has not been established, we transformed recombinant plasmid (pSuper::GlNPC3) into A. thaliana (Col-0) to verify the biological function of GlNPC3 in the salt stress response. Homozygous transgenic lines were isolated and exogenous GlNPC3 expression was evaluated in the transgenic lines by reverse transcription PCR (Supplementary Figure S2). It was found that exogenous GlNPC3 was over-expressed in transgenic lines OE1 and OE2 (Supplementary Figure S2). The OE lines showed no obvious phenotype when grown under normal conditions compared to WT plants. However, when plants were grown in medium supplemented with 75mM NaCl, the main root length of the OE lines were significantly longer than that of the WT (Figure 3). Moreover, soil-cultivated plants were exposed to the salt treatment. Four-week-old plants were watered with Hoagland’s culture solution containing 100mM NaCl, and the concentration was maintained for 10days. The fresh and dry masses were higher in OE lines than the WT under the salt treatment (Supplementary Figure S3). The leaf size of the WT decreased compared to that of the OE lines. Thus, these results suggest a positive role for GlNPC3 in G. littoralis salt tolerance.
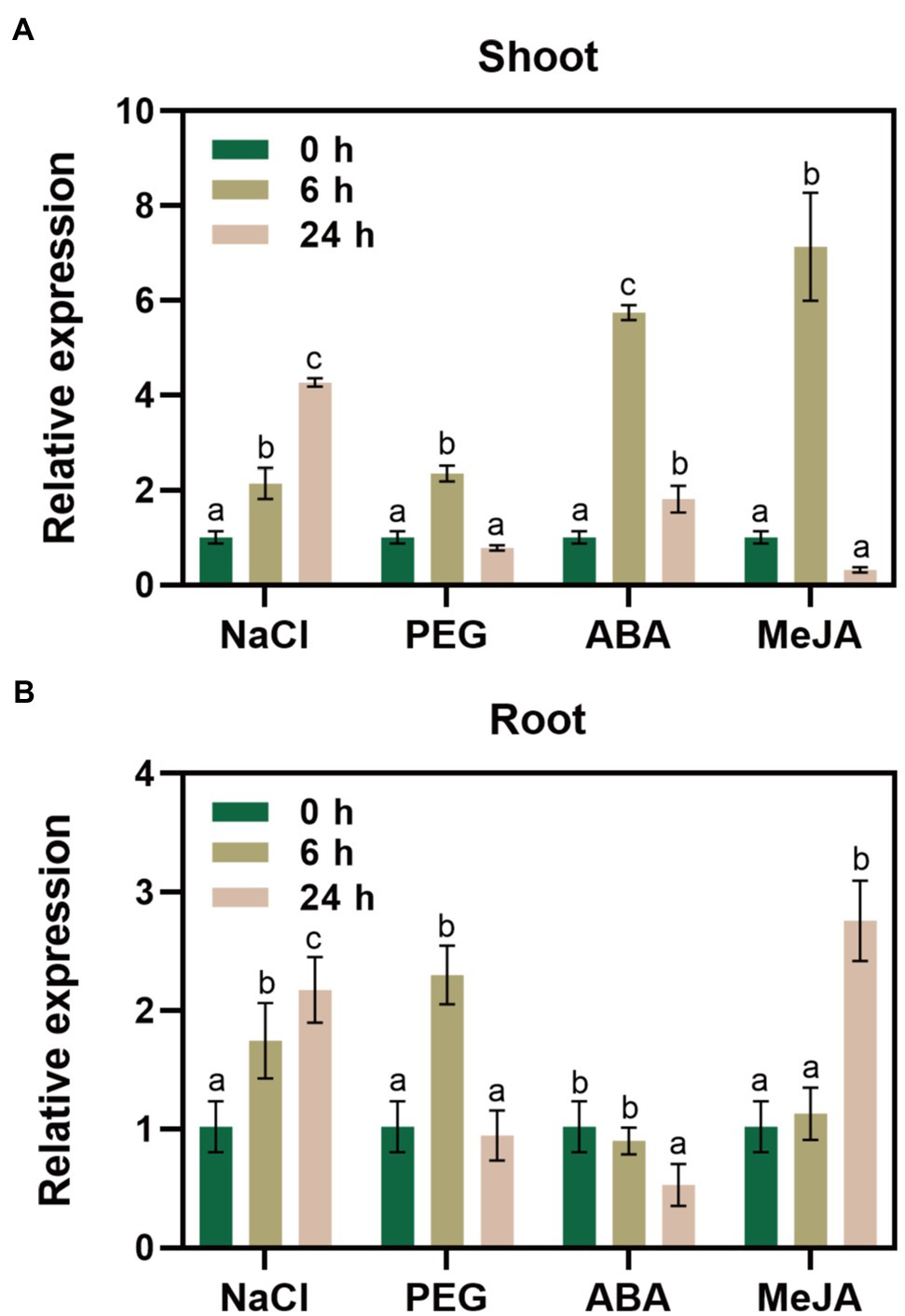
Figure 2. Relative expression levels of GlNPC3 under different stress conditions. GlNPC3 expression levels in shoots (A) and roots (B) under different stress conditions. Glehnia littoralis seedlings were subjected to salt (200mM NaCl), drought (20% PEG 6000), or hormone (100μM ABA or 100μM MeJA) treatments for 0, 6, or 24h. The shoots and roots of G. littoralis were harvested at the indicated time intervals for RNA extraction. GlCYP2 was used as the internal control. Data represent means three biological replicates with three pooled plants each ± SD (three technical replicates per biological replicate). Different letters indicate statistically significant difference in each treatment (p<0.05, Duncan’s multiple range test).
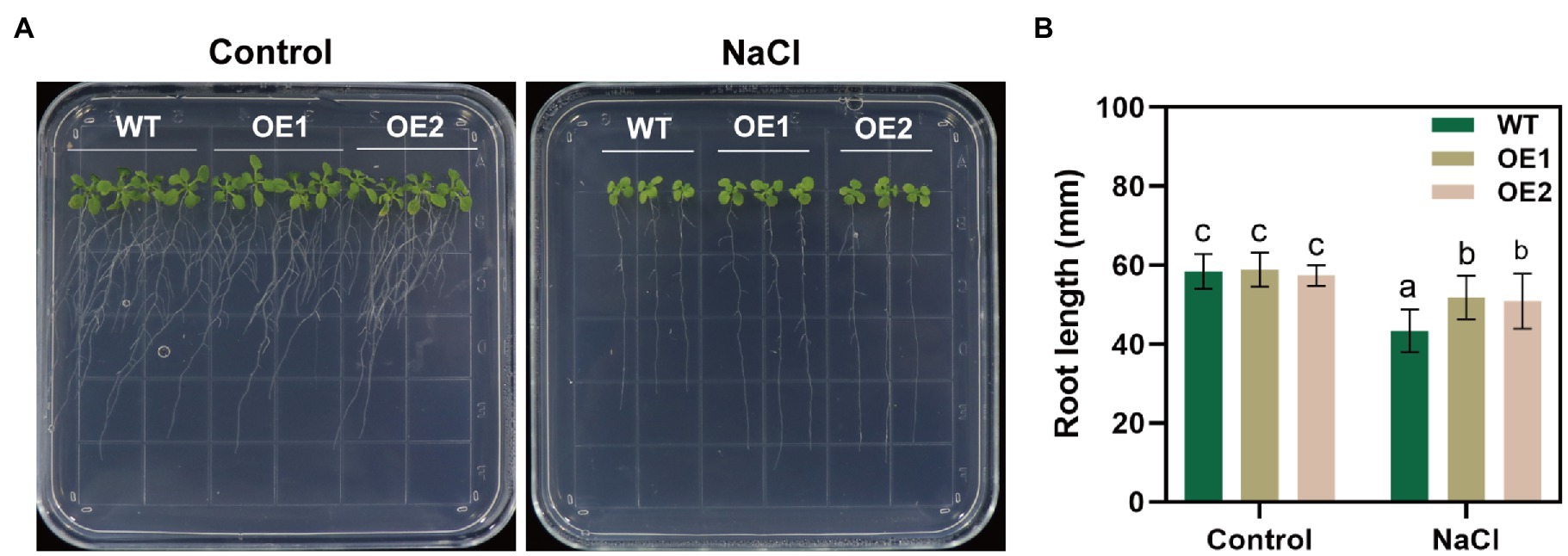
Figure 3. Effect of NaCl on root length in the GlNPC3-overexpressing plants. (A) Phenotypes of the wild-type (WT) and transgenic Arabidopsis thaliana during salt stress. The seeds were grown on agar plates supplemented with 0mM (control) or 75mM NaCl for 14days. Representative photographs of the plants are shown. (B) Root length of 14-day-old seedlings. Data represent mean±SD, n=48. Different letters above each bar indicate a significant difference (p<0.05, Duncan’s multiple range test).
GlNPC3 Expression Pattern and Localization
We examined the tissue expression pattern and subcellular localization to further understand the function of GlNPC3 in G. littoralis. RT-qPCR revealed that GlNPC3 was expressed in most tissues, including roots, rhizomes, leaves, flowers, pedicels, leaf sheath, the rachis, and the petioles (Figure 4A). GlNPC3 was highly expressed in roots, at a level more than three times higher than in other tissues (Figure 4A). A GUS construct driven by the GlNPC3 native promoter (PromoterGlNPC3::GUS) was transformed into A. thaliana. As shown in Figures 4B–H, GUS staining occurred in the seminal and lateral roots, hypocotyl, leaves, stamens, and stems. PromoterGlNPC3::GUS displayed a high level of expression in leaves at the A. thaliana eight-leaf stage; however, little expression was detected at the four-leaf stage and in old leaves (Figures 4B,C,E). The expression of GlNPC3 varied spatiotemporally. Moreover, the expression of PromoterGlNPC3::GUS also responded to exogenous NaCl, mannitol or the hormones treatments in the roots of transgenic A. thaliana seedlings (Supplementary Figure S4). To explore the subcellular localization of GlNPC3, GFP-GlNPC3 fusion protein was co-expressed with a PM marker or tonoplast marker tagged with orange fluorescent protein (OFP) in N. benthamiana leaves (Batistič et al., 2010). The distribution of green and orange fluorescence indicated that GlNPC3 was predominantly localized at the plasma membrane, with some staining of the tonoplast (Figure 5).
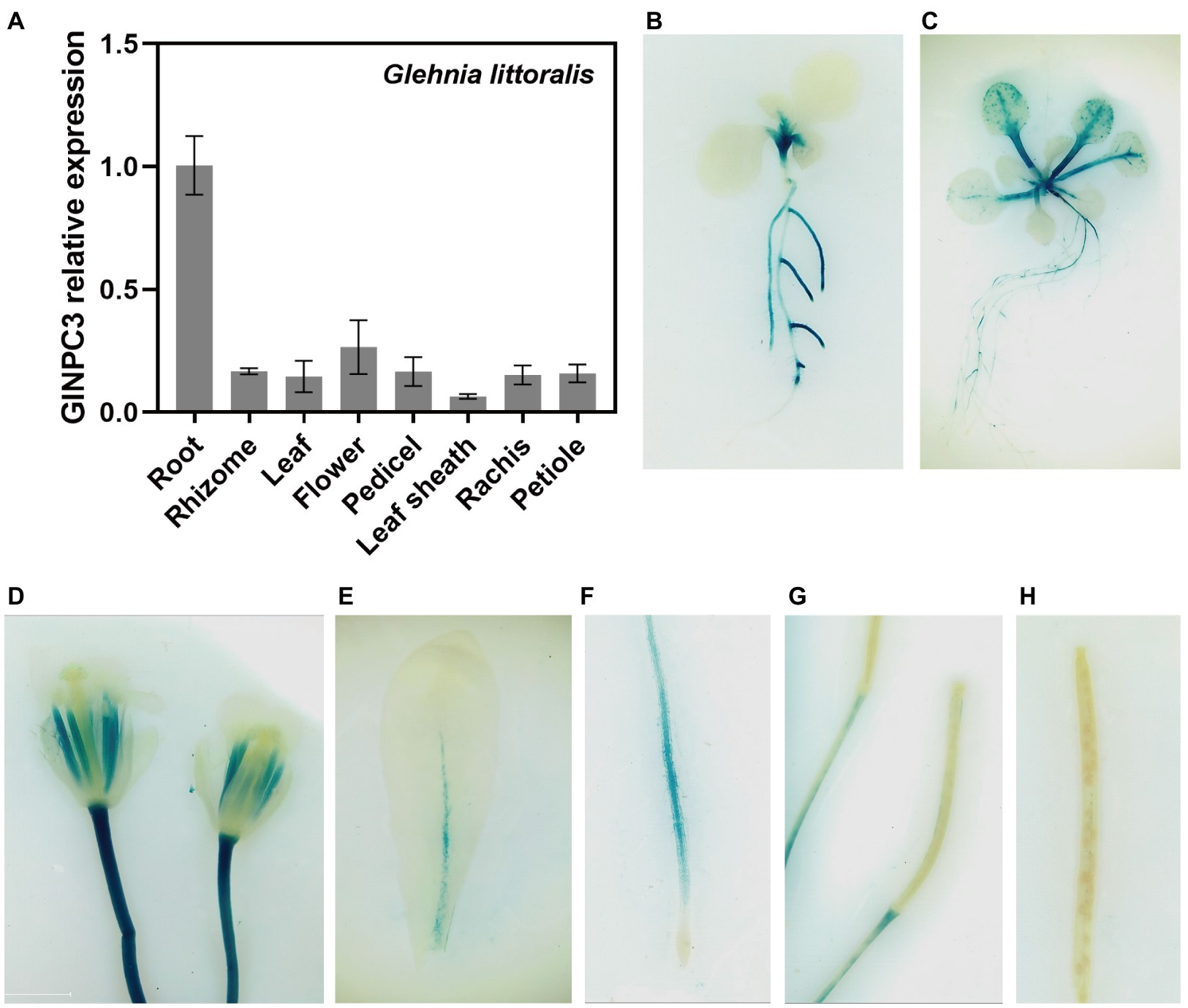
Figure 4. Expression pattern of GlNPC3 in plants. (A) Tissue-specific expression analysis of GlNPC3 in G. littoralis. Data represent means±SD of three biological replicates with three pooled tissues (plants) each. GlEXP1 was used as the internal control. (B–H) Histochemical GUS-staining of transgenic A. thaliana (PromoterGlNPC3::GUS). Histochemical GUS-stained 4-day-old seedling (B), 10-day-old seedling (C), flower (D), old leaf (E), root (F), and siliques (G,H) are shown.
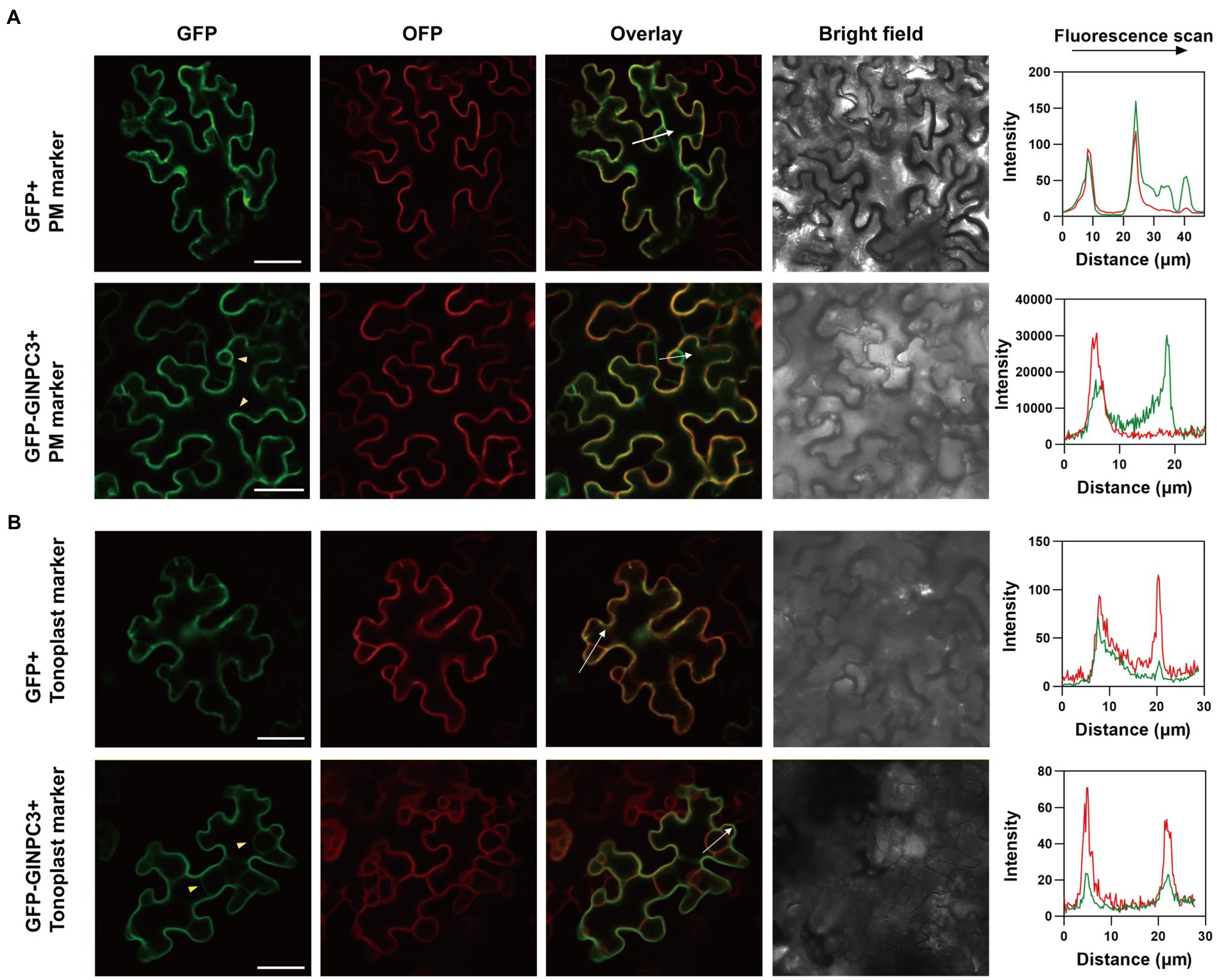
Figure 5. Subcellular localization of GlNPC3 in leaf epidermal cells of N. benthamiana. (A) Upper row, GFP empty vector was co-expressed with a plasma membrane (PM) marker. Lower row, GFP-GlNPC3 fusion protein was co-expressed with the PM marker. PM marker, AtCBL1n-OFP. Bars, 40μm. The fluorescence intensity profiles along the direction of white arrows were obtained using Zen (blue edition). The yellow arrow heads indicate the PM and tonoplast localization of GlNPC3. (B) Upper row, GFP empty vector was co-expressed with the tonoplast marker. Lower row, GFP-GlNPC3 fusion protein was co-expressed with the tonoplast marker. Tonoplast marker, AtCBL6-OFP. Bars, 40μm. The fluorescence intensity profiles along the direction of white arrows were obtained using Zen (blue edition). The yellow arrow heads indicate the tonoplast localization of GlNPC3.
Biochemical Characterization of GlNPC3
To determine the biochemical characteristics of GlNPC3, the GlNPC3 protein fused with a His-tag at its N-terminus (His-GlNPC3) was expressed in E. coli and subjected to an in vitro enzymatic assay (Figure 6A, inset). GlNPC3 activity was assayed using various fluorescent head group-labeled phospholipid substrates, including NBD-PS, NBD-PE, and Cy5-PC. Phospholipases hydrolyze lipid substrates and produce a soluble group with fluorescence, which is easy to detect with a microplate reader. In our results, His-GlNPC3 fusion protein displayed twice the hydrolyzing activity toward the PS substrate compared to PC, and five times the activity compared to PE (Figure 6A). As a negative control, His-tag protein (empty vector) displayed little enzymatic activity toward these substrates (Figure 6A). Next, we extracted total protein from WT and GlNPC3-overexpressing lines (OE1 and OE2) for enzymatic activity assay, respectively. Total NPC activity was assayed using the PS substrate. The WT and OE lines showed similar NPC activity under the control condition. NPC activity was significantly induced by salt treatment (Figure 6B). Total NPC activity was higher in the OE lines after the salt treatment compared to the WT. These results suggest that GlNPC3 contributes to the overall NPC activity in plants during the salt stress response.
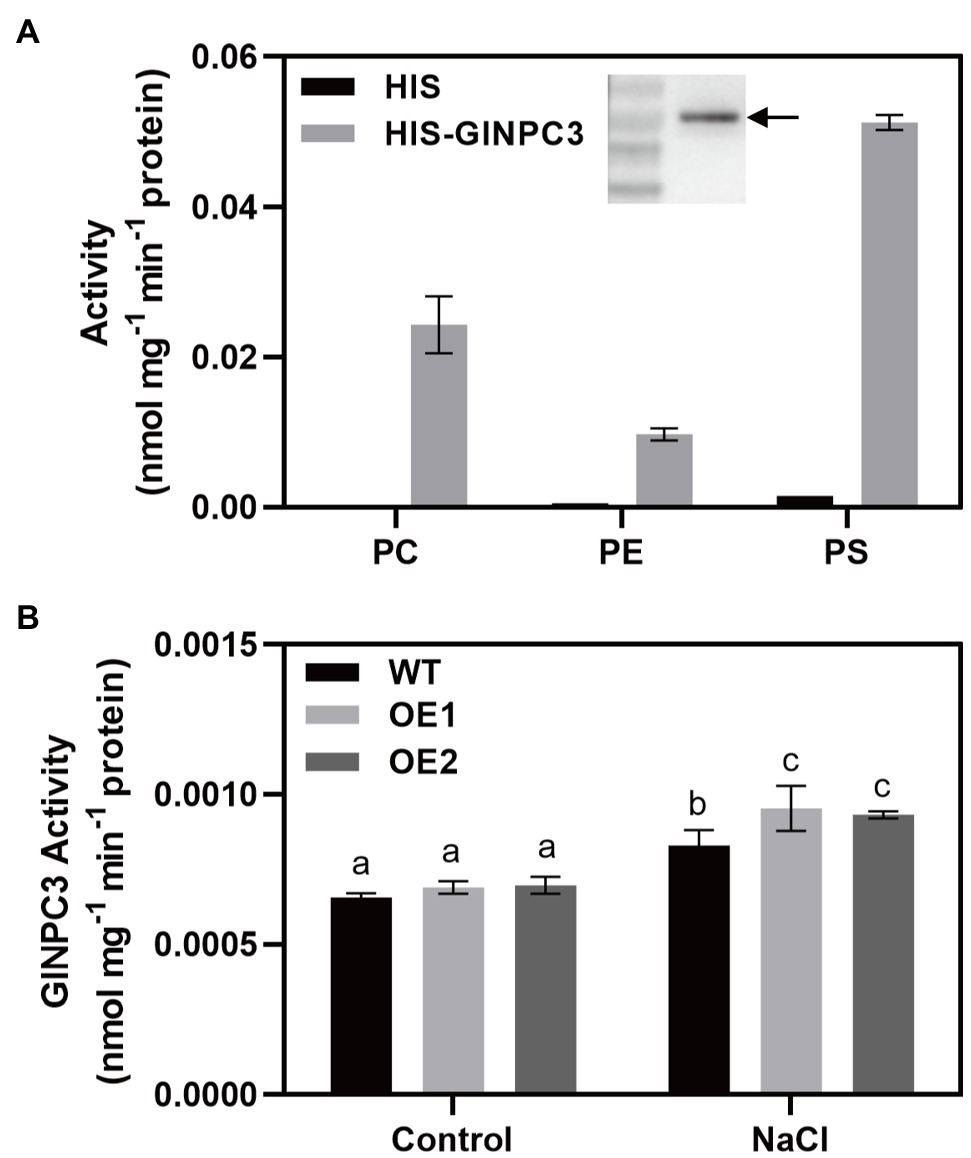
Figure 6. GlNPC3 activity assay in vitro. (A) His-GlNPC3 activity assay in the presence of different fluorescent head-group-labeled lipid substrates. Data are mean±SD of three measurements. Inset, immunoblotting assay of His-GlNPC3 fusion protein with the His antibody. Arrow indicates the protein. (B) Total non-specific phospholipase C (NPC) activities of WT and transgenic A. thaliana. Total protein was extracted from A. thaliana seedlings exposed or not to 75mM NaCl for 24days. Fluorescent head group-labeled phosphotidylserine (PS; 200μM) was used as the substrate. Data represent means±SD three biological replicates. Different letters above each bar indicate significant differences (p<0.05, Duncan’s multiple range test).
Overexpression of GlNPC3 Alters Membrane-Lipid Composition in Transgenic Plants
Membrane lipids affect the stability of membranes, and are involved in various cellular responses to abiotic and biotic stresses. To investigate the mechanism underlying GlNPC3 lipid metabolism in response to salt stress, we determined the lipid composition of transgenic A. thaliana using ESI-MS/MS-based lipid profiling (Figure 7). Under normal condition, there was no significant difference in the levels of PC, PE, phosphatidylinositol (PI), PS, phosphatidylglycerol (PG), DAG, and PA between WT and GlNPC3-overexpressing seedlings (OE1; Figures 7A–H). After 45min of salt treatment, the lipid composition changed to varying degrees (Figure 7). The levels of PC, PS, and PG in OE1 decreased obviously compared to that of WT. DAG content increased significantly in WT after exposure to the salt treatment, however, no significant difference occurred in OE1 (Figure 7G). Generally, NPC hydrolyzed glycerolipids to generate DAG, and DAG simultaneously generated PA through DGK kinases. Then, we analyzed PA content. In our result, there was no significant difference in the level of total PA between salt-treated and non-treated plants. Furthermore, a molecular species analysis revealed the lipid composition of the plants (Supplementary Figure S5). The molecular species in the seedlings with a higher mass spectral signal were 34:3, 34:2, 36:4, 36:5, and 36:6 for PC, PE, PI, DAG, and PA; 42:2, 42:3, 40:2, and 40:3 for PS; and 34:4, 34:3, and 32:1 for PG. Among them, both 36:4 PA and 34:6 PA in OE1 were significantly higher than that in WT, and the same DAG species were increased in OE1 under normal condition (Supplementary Figure S5). The changes in major lipid molecular species (Supplementary Figure S5) were almost consistent with those shown in sum (Figure 7).
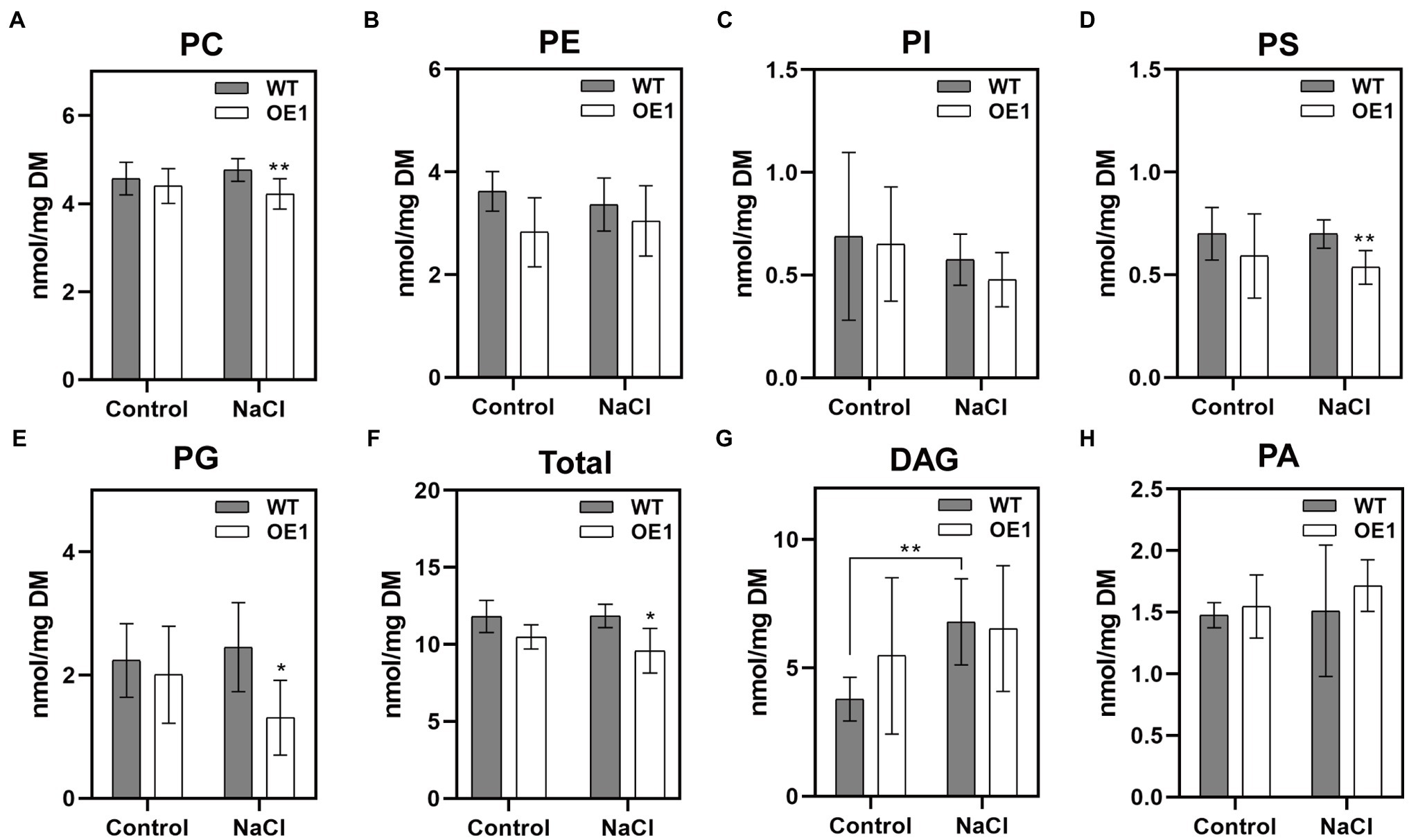
Figure 7. Effect of GlNPC3 overexpression on membrane lipid contents in A. thaliana. (A–H) Comparison of lipid changes between the WT and OE1. Total lipids were extracted from 10-day-old medium-grown A. thaliana seedlings that were exposed or not to 150mM NaCl for 45min. Lipids were quantified by electrospray ionization tandem mass spectrometry (ESI-MS/MS). Total lipids refer to the amounts of PC, PE, PI, PS, and PG. Data represent means±SD of five replicates (each replicate contained 15~20 plants). Asterisks indicate significant differences from the WT: *p≤0.05; **p≤0.01, Duncan’s multiple range test. DM, dry mass.
Downregulation of GlNPC3 Affects the Transcript Levels of Some Stress-Related Genes
It was difficult to obtain transgenic G. littoralis plants, so we used VIGS to transiently silence the GlNPC3 gene in G. littoralis leaves for rapid functional analysis. First, the endogenous G. littoralis phytoene desaturase gene (GlPDS), which causes photobleaching, was used as a positive control to assess VIGS efficiency (Supplementary Figure S6). The photobleaching phenotype was different from senescence (Supplementary Figure S6A). Photobleaching was confined to infiltrated leaves, perhaps because of the unique morphology of G. littoralis, which has extremely short rhizomes and a long petiole, and a basal petiole that expands into the sheath. Thus, only infiltrated leaves were used for the RNA extraction and RT-qPCR analysis. Then, we selected two interfering fragments (VIGSNPC3-1 and VIGSNPC3-2) of the GlNPC3 CDS for VIGS-silencing. The specificity of gene silencing was detected in infiltrated leaves (Supplementary Figures S6B–D). GlNPC3 expression was almost 50 and 70% reduced in the injected leaves of VIGSNPC3-1 and VIGSNPC3-2, respectively (Supplementary Figure S6B). We also checked two of the GlNPC3 homologous genes. Their expression levels were not reduced as much as that of GlNPC3 (Supplementary Figures S6C,D). RT-qPCR analysis was performed to investigate whether the GlNPC3-mediated lipid changes were accompanied by a gene expression response. According to the RT-qPCR results, most of the stress-related genes were significantly induced by the NaCl treatment (Figure 8). In particular, transcription of GlSnRK2 (comp37685_c0_seq1), GlP5CS (comp33363_c0_seq1), GlWRKY (comp25557_c0_seq2), GlSOS1 (comp30905_c0_seq3), and GlCIPK (comp35199_c0_seq4) increased more than 3-fold in response to the salt stress, while the transcription of GlTPC1 (comp35393_c0_seq6) increased more than 10-fold. The induction of most of the tested genes was significantly inhibited in the VIGSNPC3-1 and VIGSNPC3-2 leaves (Figure 8).
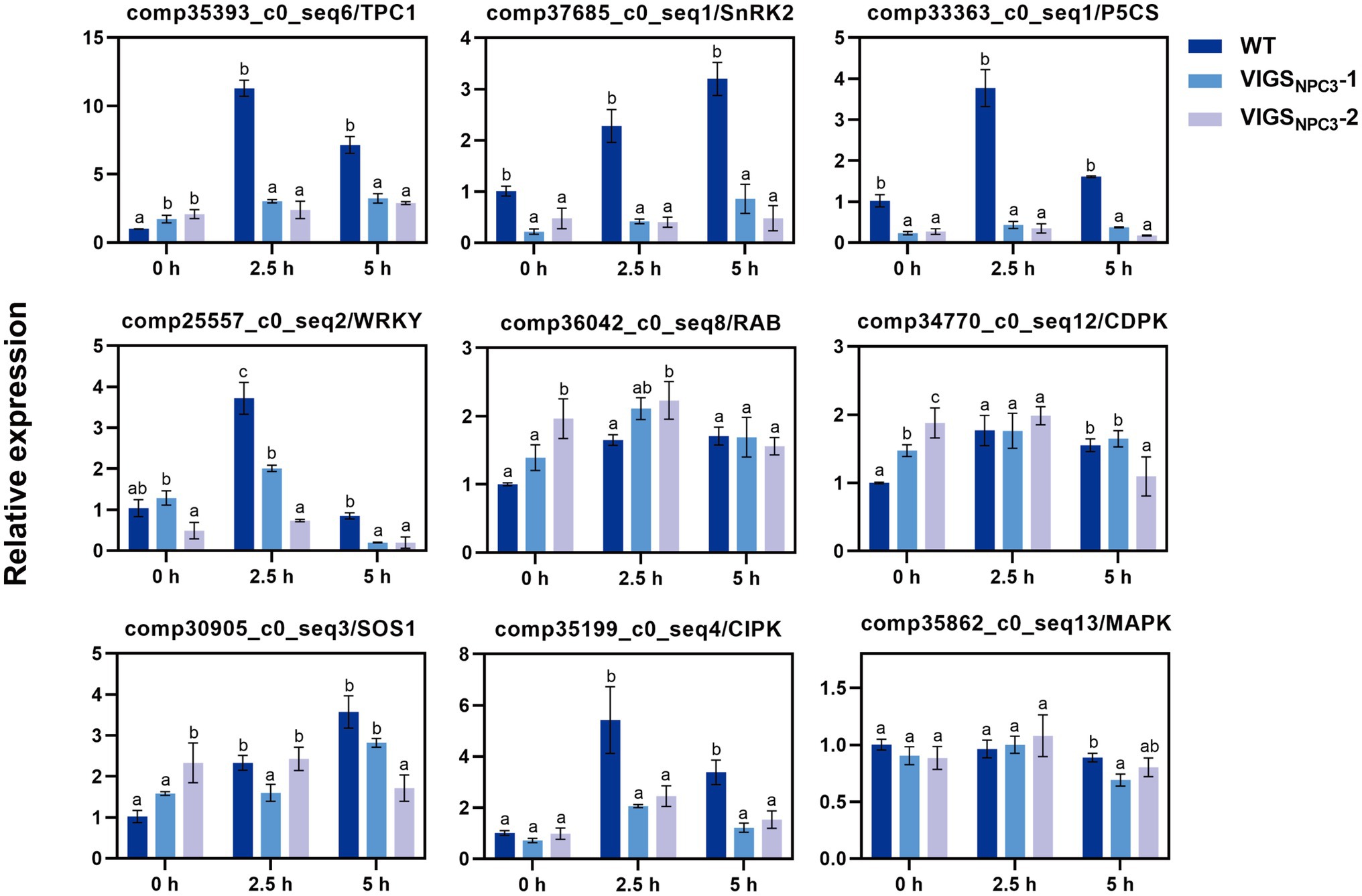
Figure 8. Relative expression of stress-related genes in G. littoralis WT, VIGSNPC3-1, and VIGSNPC3-2 leaves. For NaCl treatment, the pot-cultured G. littoralis plants were watered with Hoagland’s culture solution containing 200mM NaCl. Data represent means±SD three biological replicates. Different letters above each bar indicate significant differences (p<0.05, Duncan’s multiple range test). GlEXP1 was used as the internal control.
Discussion
Glehnia littoralis is a medicinal halophyte that grows in coastal habitats. There is an inevitable association between saline adaptability and the accumulation of active medicinal ingredients in G. littoralis. Shu et al. (2019) indicated that the accumulation of furocoumarins was increased by NaCl stress in G. littoralis. Few studies have been reported on the salt tolerance mechanism of G. littoralis. In our previous study, we performed a comprehensive transcriptome analysis of the response of G. littoralis to salt stress, and obtained a large number of differentially expressed genes involved in basic metabolism, secondary metabolism, transportation, and signal transduction (Li et al., 2018). We cloned and identified a differentially expressed NPC gene named GlNPC3 (Figure 1). RT-qPCR showed that the expression level of GlNPC3 increased continuously upon salt treatment (Figure 2). The characterization of A. thaliana GlNPC3-overexpressing plants showed that overexpression of GlNPC3 enhanced the salt tolerance of A. thaliana. Therefore, we performed a series of detailed experiments to elucidate the involvement of GlNPC3 in the salt stress response in G. littoralis.
Similar to most NPCs, GlNPC3 contained a conserved phosphoesterase domain necessary for esterase activity and NPC catalytic activity (Figure 1 and Supplementary Figure S1). In A. thaliana, AtNPC1, AtNPC2, and AtNPC6 contain a signal peptide at the C-terminus, but AtNPC3, AtNPC4, and AtNPC5 lack it (Pokotylo et al., 2013). GlNPC3 contains no signal peptide and was clustered with AtNPC3, AtNPC4, and AtNPC5 in the phylogenetic analysis. The 50–100 amino acids at the C-terminus of NPCs are the most variable regions among NPC subfamilies. These regions may be responsible for various molecular functions and subcellular localization of different NPCs (Pokotylo et al., 2013). The gene expression changes under the different treatments showed that GlNPC3 responds to various stresses (Figure 2). Our analysis of the GlNPC3 promoter sequence also showed the variety of stress-related regulatory elements in the GlNPC3 promoter sequence, suggesting that GlNPC3 might be involved in a variety of transcriptionally regulatory processes. Moreover, we found that the expression of the GUS reporter driven by the GlNPC3 promoter in the roots of A. thaliana was upregulated after NaCl, mannitol or the hormone treatments during 2h (Supplementary Figure S4). However, the expression of GlNPC3 did not increase at 6 and 24h after ABA treatment in the root of G. littoralis (Figure 1B). We supposed that the response of GlNPC3 to ABA might be earlier or later than the time points we analyzed in G. littoralis.
In previous studies, AtNPCs were expressed at different levels in the roots (Peters et al., 2010), and some AtNPCs were involved in the regulation of root development. For example, AtNPC2 and AtNPC6 are required for root growth in Arabidopsis (Ngo et al., 2019). AtNPC4 is involved in the root response to salt stress (Kocourková et al., 2011), and AtNPC5 mediates lateral root development during salt stress (Peters et al., 2014). Our results showed that GlNPC3 was expressed at the highest levels in the roots of G. littoralis, followed by flowers (Figure 4). Expression of the GUS reporter driven by the native GlNPC3 promoter in A. thaliana exhibited a similar pattern with that of G. littoralis. However, there were some differences compared to G. littoralis. For example, PromoterGlNPC3::GUS was highly expressed in the stem of A. thaliana, while G. littoralis had extremely short rhizomes and the expression level in rhizomes was similar to that in the petiole. Moreover, the roots of A. thaliana GlNPC3-overexpressing plants were longer than that of the WT under the salt treatment (Figure 3). Based on the growth characteristics of G. littoralis, the roots of G. littoralis, which are deeply embedded in the beach, are not only the main medicinal parts but also play a positive role in salt tolerance. Therefore, GlNPC3 may be related to the response of roots to salt stress.
The subcellular localization of GlNPC3 was mainly at the plasma membrane and partially at the tonoplast, different from other NPCs (Figure 5). Both AtNPC1 and AtNPC2 are localized at the endoplasmic reticulum and Golgi apparatus (Krčková et al., 2015; Ngo et al., 2018), and AtNPC2 is localized at the plastid (Ngo et al., 2018). AtNPC4 is associated with the plasma membrane, whereas AtNPC5 localizes to the cytosol (Gaude et al., 2008; Peters et al., 2014). The subcellular localization of the NPC isoforms is generally related to enzyme activity and biological function (Nakamura and Ngo, 2020). The localization of GlNPC3 suggests that it may hydrolyze phospholipids at the plasma membrane and tonoplast, and may be involved in the lipid-mediated signaling pathway or in the lipid metabolism.
Previous studies have reported that NPCs hydrolyze a variety of common lipids, such as PC, PE, and PS (Pokotylo et al., 2013). Changes in NPC substrates and products affect intracellular lipid signaling messengers, and the composition and remodeling of membrane phospholipids, and are involved in a series of cellular responses (Nakamura et al., 2005; Gaude et al., 2008; Krčková et al., 2015; Ngo et al., 2018). We analyzed GlNPC3 catalytic activity using fluorescent head group-labeled lipids as substrates. Compared to previously reported NPCs, the hydrolytic activity of GlNPC3 in the presence of PS was higher than that in the presence of PC in vitro (Figure 6A); however, the PS content in the plants was significantly lower than that of PC and PE (Figures 7A,B,D). Because of the limitation associated with the use of a single molecular species substrate (18:1) in the in vitro assay, detailed analysis of phospholipids was needed in the plants. In this study, NPC enzyme activity increased in GlNPC3-overexpressing plants after the in vitro salt treatment (Figure 6B). Then, the changes in phospholipid content of the WT and OE1 were determined under salt stress (Figure 7). The common phospholipids, such as PC, PS, and PG, decreased significantly in OE1 compared to the WT under NaCl treatment, suggesting that these lipids may be hydrolyzed by GlNPC3 in plant. Direct measurement of the DAG product showed that the DAG contents increased significantly in the WT at 45min after the salt treatment. However, no significant difference was observed between WT and OE1. Previous studies showed that DAG was phosphorylated to PA under hyperosmotic stress conditions (Arisz et al., 2009; Munnik and Testerink, 2009; Nakamura, 2014). We then analyzed PA content; however, the increase in PA did not occur at 45min after the salt treatment. Why OE plants had less substrates during NaCl treatment, and no significant increase in DAG and PA levels? On one hand, the lipid molecules involved in hydrolysis and synthesis are not limited to these classes that we analyzed. For example, PC also contributes to DAG-mediated triacylglycerol (TAG) synthesis (Bates et al., 2009; Lu et al., 2009). Moreover, Cai et al. (2020) found that AtNPC6 promoted polar glycerolipid and galactolipid [monogalactosyldiacylglycerol (MGDG) and digalactosyldiacylglycerol (DGDG)] conversion to TAG production. In contrast, MGDG and DGDG were converted to DAG by monogalactosyldiacylglycerol synthase (MGD) and digalactosyldiacylglycerol synthase (DGD). TAG and galactolipid were not measured in this study. So, we speculated that OE plants may promote polar glycerolipid turnover to DAG to form TAG during salt treatment, and affect DAG and PA levels. Under normal condition, although no significant difference was observed in DAG content between WT and OE1, OE1 still displayed more DAG content than WT (average 5.47 vs. 3.78nmol/mg DM), and less total polar lipids than WT (average 10.49 vs. 11.82nmol/mg DM; Figures 7F,G). This data suggested that GlNPC3 overexpression contributed the generation of DAG under the normal condition. Together, lipid profiles suggested over-accumulation of GlNPC3 affected the proportion of different membrane lipids and the spatial and electronic properties of membrane, conferring salt tolerance to plants.
On the other hand, the “time limitation” might affect the determination of PA and DAG content. A significantly salt-induced increase in PA and DAG between WT and OE line may occur in the other time points during salt treatment. PA is metabolized rapidly in plants, requiring appropriate time points and rigorous operation, and the extraction and determination of PA are complicated and costly (Welti et al., 2002; Shadyro et al., 2004; Nguyen et al., 2017). Because of the spatiotemporal complexity of monitoring lipid in living cells and tissues of plants, a real-time quantitative and visualized method is required. For example, Potocky et al. (2014) constructed a PA biosensor with a Spo20p-PA binding domain fusing YFP protein, which reflected PA dynamics in transiently transformed tobacco pollen tubes as shown by confocal laser scanning microscopy. Subsequently, Li et al. (2019) developed a PA-specific biosensor, PAleon, which monitors and visualizes the concentration and dynamics of PA in plant cells under abiotic stress based on Förster resonance energy transfer (FRET) technology. PA biosensors have revealed the spatiotemporal dynamics of PA in plants. Similarly, DAG, PS, PtdIns(4,5)P2 (phosphatidylinositol-4,5-biphosphates), PtdIns4P (phosphatidylinositol-4-phosphate), and PtdIns3P (phosphatidylinositol-3-phosphate) biosensors have been successfully used in plant cells (Vermeer et al., 2006, 2009; van Leeuwen et al., 2007; Thole et al., 2008; Yamaoka et al., 2011; Simon et al., 2014; Vermeer et al., 2017). These biosensors could be used for real-time monitoring of lipid dynamics and provide new insight into lipid metabolism in G. littoralis.
Finally, we analyzed the expression of some stress-related genes in WT and VIGS-silenced G. littoralis plants to investigate whether the GlNPC3-mediated lipid changes were associated with gene expression. The expression of SOS1 (salt overly sensitive 1), TPC1 (two-pore channel 1), SnRK2 (Ser/Thr protein kinase), P5CS (∆1-pyrroline-5-carboxylate synthetase), CIPK (CBL-interacting protein kinase), and the WRKY transcription factor genes in G. littoralis was significantly upregulated under the salt treatment, but little changed after VIGS-induced GlNPC3 silencing of the plants (Figure 8). These genes play pivotal roles in responses to multiple stresses in model plants. Among them, the vacuolar ion channel TPC1 is responsible for a reactive oxygen species-assisted Ca2+ wave during the salt stress response (Evans et al., 2016). The SOS signaling pathway gene SOS1 plays a key role in Na+ export (Zhu, 2001, 2016). Therefore, combined with the membrane localization of GlNPC3, these findings suggest that a GlNPC3-mediated lipid change may be accompanied by the response of stress-related genes affecting G. littoralis salt tolerance.
In conclusion, we cloned and identified a NPC (GlNPC3) from the medicinal halophyte G. littoralis, and uncovered a potentially functional connection between GlNPC3-mediated phospholipid signaling and the salt-stress response in G. littoralis. Further investigation is needed to determine the role of GlNPC3 in lipid remodeling, and how it is regulated by upstream mediators during the stress response. These results will provide insights into studies on plants living in special environments.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
LL and YZ designed the experiments. LL performed the experiments, analyzed the data, and wrote the manuscript. YB provided support for protein expression and enzyme activity assays. NL and QC provided support for plant growth. XQ helped in bioinformatics analysis. HF, XY, and DL provided the materials and technical support. CL and YZ revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by grants from the National Natural Science Foundation of China (No. 31800272).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Sheng Xu at Institute of Botany, Jiangsu Province and Chinese Academy of Sciences for providing the pTRVI and pTRVII vector. We also thank Qun Zhang at Nanjing Agricultural University for providing valuable advices in manuscript writing.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.769599/full#supplementary-material
Footnotes
1. ^http://gsds.gao-lab.org/index.php
3. ^http://bioinformatics.psb.ugent.be/webtools/plantcare/html/
References
Arisz, S. A., Testerink, C., and Munnik, T. (2009). Plant PA signaling via diacylglycerol kinase. Biochim. Biophys. Acta 1791, 869–875. doi: 10.1016/j.bbalip.2009.04.006
Bargmann, B. O., and Munnik, T. (2006). The role of phospholipase D in plant stress responses. Curr. Opin. Plant Biol. 9, 515–522. doi: 10.1016/j.pbi.2006.07.011
Bates, P. D., Durrett, T. P., Ohlrogge, J. B., and Pollard, M. (2009). Analysis of acyl fluxes through multiple pathways of triacylglycerol synthesis in developing soybean embryos. Plant Physiol. 150, 55–72. doi: 10.1104/pp.109.137737
Batistič, O., Waadt, R., Steinhorst, L., Held, K., and Kudla, J. (2010). CBL-mediated targeting of CIPKs facilitates the decoding of calcium signals emanating from distinct cellular stores. Plant J. 61, 211–222. doi: 10.1111/j.1365-313X.2009.04045.x
Cai, G., Fan, C., Liu, S., Yang, Q., Liu, D., Wu, J., et al. (2020). Nonspecific phospholipase C6 increases seed oil production in oilseed Brassicaceae plants. New Phytol. 226, 1055–1073. doi: 10.1111/nph.16473
Cao, L., Wang, L., Zheng, M., Cao, H., Ding, L., Zhang, X., et al. (2013). Arabidopsis AUGMIN subunit8 is a microtubule plus-end binding protein that promotes microtubule reorientation in hypocotyls. Plant Cell 25, 2187–2201. doi: 10.1105/tpc.113.113472
Cao, H., Zhuo, L., Su, Y., Sun, L., and Wang, X. (2016). Non-specific phospholipase C1 affects silicon distribution and mechanical strength in stem nodes of rice. Plant J. 86, 308–321. doi: 10.1111/tpj.13165
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Devaiah, S. P., Roth, M. R., Baughman, E., Li, M., Tamura, P., Jeannotte, R., et al. (2006). Quantitative profiling of polar glycerolipid species from organs of wild-type Arabidopsis and a phospholipase Dalpha1 knockout mutant. Phytochemistry 67, 1907–1924. doi: 10.1016/j.phytochem.2006.06.005
Evans, M. J., Choi, W.-G., Gilroy, S., and Morris, R. J. (2016). A ROS-assisted calcium wave dependent on the AtRBOHD NADPH oxidase and TPC1 cation channel propagates the systemic response to salt stress. Plant Physiol. 171, 1771–1784. doi: 10.1104/pp.16.00215
Fernandez-Pozo, N., Rosli, H. G., Martin, G. B., and Mueller, L. A. (2015). The SGN VIGS tool: user-friendly software to design virus-induced gene silencing (VIGS) constructs for functional genomics. Mol. Plant 8, 486–488. doi: 10.1016/j.molp.2014.11.024
Gaude, N., Nakamura, Y., Scheible, W. R., Ohta, H., and Dormann, P. (2008). Phospholipase C5 (NPC5) is involved in galactolipid accumulation during phosphate limitation in leaves of Arabidopsis. Plant J. 56, 28–39. doi: 10.1111/j.1365-313X.2008.03582.x
Hong, Y., Zhao, J., Guo, L., Kim, S. C., Deng, X., Wang, G., et al. (2016). Plant phospholipases D and C and their diverse functions in stress responses. Prog. Lipid Res. 62, 55–74. doi: 10.1016/j.plipres.2016.01.002
Hua, B., Chang, J., Wu, M., Xu, Z., Zhang, F., Yang, M., et al. (2021). Mediation of JA signalling in glandular trichomes by the woolly/SlMYC1 regulatory module improves pest resistance in tomato. Plant Biotechnol. J. 19, 375–393. doi: 10.1111/pbi.13473
Huang, G. J., Deng, J. S., Liao, J. C., Hou, W. C., Wang, S. Y., Sung, P. J., et al. (2012). Inducible nitric oxide synthase and cyclooxygenase-2 participate in anti-inflammatory activity of imperatorin from Glehnia littoralis. J. Agric. Food Chem. 60, 1673–1681. doi: 10.1021/jf204297e
Jefferson, R. A. (1987). Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol. Biol. Report. 5, 387–405. doi: 10.1007/BF02667740
Kim, S. C., Nusinow, D. A., Sorkin, M. L., Pruneda-Paz, J., and Wang, X. (2019). Interaction and regulation between lipid mediator phosphatidic acid and circadian clock regulators. Plant Cell 31, 399–416. doi: 10.1105/tpc.18.00675
Kocourková, D., Krčková, Z., Pejchar, P., Veselková, S., Valentová, O., Wimalasekera, R., et al. (2011). The phosphatidylcholine-hydrolysing phospholipase C NPC4 plays a role in response of Arabidopsis roots to salt stress. J. Exp. Bot. 62, 3753–3763. doi: 10.1093/jxb/err039
Krčková, Z., Brouzdová, J., Daněk, M., Kocourková, D., Rainteau, D., Ruelland, E., et al. (2015). Arabidopsis non-specific phospholipase C1: characterization and its involvement in response to heat stress. Front. Plant Sci. 6:928. doi: 10.3389/fpls.2015.00928
Krčková, Z., Kocourková, D., Daněk, M., Brouzdová, J., Pejchar, P., Janda, M., et al. (2018). The Arabidopsis thaliana non-specific phospholipase C2 is involved in the response to pseudomonas syringae attack. Ann. Bot. 121, 297–310. doi: 10.1093/aob/mcx160
Li, L., Li, N., Fang, H., Qi, X., and Zhou, Y. (2020). Selection and validation of reference genes for normalisation of gene expression in Glehnia littoralis. Sci. Rep. 10:7374. doi: 10.1038/s41598-020-63917-5
Li, L., Li, M., Qi, X., Tang, X., and Zhou, Y. (2018). De novo transcriptome sequencing and analysis of genes related to salt stress response in Glehnia littoralis. Peer J. 6:e5681. doi: 10.7717/peerj.5681
Li, W., Song, T., Wallrad, L., Kudla, J., Wang, X., and Zhang, W. (2019). Tissue-specific accumulation of pH-sensing phosphatidic acid determines plant stress tolerance. Nat. Plants 5, 1012–1021. doi: 10.1038/s41477-019-0497-6
Li, L., Wang, F., Yan, P., Jing, W., Zhang, C., Kudla, J., et al. (2017). A phosphoinositide-specific phospholipase C pathway elicits stress-induced Ca2+ signals and confers salt tolerance to rice. New Phytol. 214, 1172–1187. doi: 10.1111/nph.14426
Li, M., Welti, R., and Wang, X. (2006). Quantitative profiling of Arabidopsis polar glycerolipids in response to phosphorus starvation. Roles of phospholipases D zeta1 and D zeta2 in phosphatidylcholine hydrolysis and digalactosyldiacylglycerol accumulation in phosphorus-starved plants. Plant Physiol. 142, 750–761. doi: 10.1104/pp.106.085647
Liu, Y., Schiff, M., and Dinesh-Kumar, S. P. (2002). Virus-induced gene silencing in tomato. Plant J. 31, 777–786. doi: 10.1046/j.1365-313X.2002.01394.x
Lu, C., Xin, Z., Ren, Z., and Miquel, M. (2009). An enzyme regulating triacylglycerol composition is encoded by the ROD1 gene of Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 106, 18837–18842. doi: 10.1073/pnas.0908848106
Munnik, T., and Testerink, C. (2009). Plant phospholipid signaling: “in a nutshell.” J. Lipid Res. 50, S260–S265. doi: 10.1194/jlr.R800098-JLR200
Nakamura, Y. (2014). “NPC: nonspecific phospholipase Cs in plant functions,” in Phospholipases in Plant Signaling. ed. X. Wang (Berlin Heidelberg: Springer-Verlag), 55–67.
Nakamura, Y., Awai, K., Masuda, T., Yoshioka, Y., Takamiya, K., and Ohta, H. (2005). A novel phosphatidylcholine-hydrolyzing phospholipase C induced by phosphate starvation in Arabidopsis. J. Biol. Chem. 280, 7469–7476. doi: 10.1074/jbc.M408799200
Nakamura, Y., and Ngo, A. H. (2020). Non-specific phospholipase C (NPC): an emerging class of phospholipase C in plant growth and development. J. Plant Res. 133, 489–497. doi: 10.1007/s10265-020-01199-8
Ngo, A. H., Kanehara, K., and Nakamura, Y. (2019). Non-specific phospholipases C, NPC2 and NPC6, are required for root growth in Arabidopsis. Plant J. 100, 825–835. doi: 10.1111/tpj.14494
Ngo, A. H., Lin, Y. C., Liu, Y. C., Gutbrod, K., Peisker, H., Dormann, P., et al. (2018). A pair of nonspecific phospholipases C, NPC2 and NPC6, are involved in gametophyte development and glycerolipid metabolism in Arabidopsis. New Phytol. 219, 163–175. doi: 10.1111/nph.15147
Nguyen, A., Rudge, S. A., Zhang, Q., and Wakelam, M. J. O. (2017). Using lipidomics analysis to determine signalling and metabolic changes in cells. Curr. Opin. Biotechnol. 43, 96–103. doi: 10.1016/j.copbio.2016.10.003
Ni, M., Cui, D., Einstein, J., Narasimhulu, S., Vergara, C. E., and Gelvin, S. B. (1995). Strength and tissue specificity of chimeric promoters derived from the octopine and mannopine synthase genes. Plant J. 7, 661–676. doi: 10.1046/j.1365-313X.1995.7040661.x
Peters, C., Kim, S.-C., Devaiah, S., Li, M., and Wang, X. (2014). Non-specific phospholipase C5 and diacylglycerol promote lateral root development under mild salt stress in Arabidopsis. Plant Cell Environ. 37, 2002–2013. doi: 10.1111/pce.12334
Peters, C., Li, M., Narasimhan, R., Roth, M., Welti, R., and Wang, X. (2010). Nonspecific phospholipase C NPC4 promotes responses to abscisic acid and tolerance to hyperosmotic stress in Arabidopsis. Plant Cell 22, 2642–2659. doi: 10.1105/tpc.109.071720
Pokotylo, I., Pejchar, P., Potocky, M., Kocourková, D., Krčková, Z., Ruelland, E., et al. (2013). The plant non-specific phospholipase C gene family. Novel competitors in lipid signalling. Prog. Lipid Res. 52, 62–79. doi: 10.1016/j.plipres.2012.09.001
Potocky, M., Pleskot, R., Pejchar, P., Vitale, N., Kost, B., and Zarsky, V. (2014). Live-cell imaging of phosphatidic acid dynamics in pollen tubes visualized by Spo20p-derived biosensor. New Phytol. 203, 483–494. doi: 10.1111/nph.12814
Scherer, G. F., Ryu, S. B., Wang, X., Matos, A. R., and Heitz, T. (2010). Patatin-related phospholipase A: nomenclature, subfamilies and functions in plants. Trends Plant Sci. 15, 693–700. doi: 10.1016/j.tplants.2010.09.005
Shadyro, O., Yurkova, I., Kisel, M., Brede, O., and Arnhold, J. (2004). Formation of phosphatidic acid, ceramide, and diglyceride on radiolysis of lipids: identification by MALDI-TOF mass spectrometry. Free Radic. Biol. Med. 36, 1612–1624. doi: 10.1016/j.freeradbiomed.2004.03.013
Shen, L., Tian, Q., Yang, L., Zhang, H., Shi, Y., Shen, Y., et al. (2020). Phosphatidic acid directly binds with rice potassium channel OsAKT2 to inhibit its activity. Plant J. 102, 649–665. doi: 10.1111/tpj.14731
Shen, L., Zhuang, B., Wu, Q., Zhang, H., Nie, J., Jing, W., et al. (2019). Phosphatidic acid promotes the activation and plasma membrane localization of MKK7 and MKK9 in response to salt stress. Plant Sci. 287:110190. doi: 10.1016/j.plantsci.2019.110190
Shu, X., Li, N., Tang, X., and Zhou, Y. (2019). Effects of NaCl stress on photosynthetic physiology and active component of different Glehnia littoralis provenance. Jiangsu J. Agric. Sci. 4, 790–797. doi: 10.3969/j.issn.1000-4440.2019.04.006
Simon, M. L., Platre, M. P., Assil, S., van Wijk, R., Chen, W. Y., Chory, J., et al. (2014). A multi-colour/multi-affinity marker set to visualize phosphoinositide dynamics in Arabidopsis. Plant J. 77, 322–337. doi: 10.1111/tpj.12358
Tang, R.-H., Han, S., Zheng, H., Cook, C. W., Choi, C. S., Woerner, T. E., et al. (2007). Coupling diurnal cytosolic Ca2+ oscillations to the CAS-IP3 pathway in Arabidopsis. Science 315, 1423–1426. doi: 10.1126/science.1134457
Testerink, C., and Munnik, T. (2005). Phosphatidic acid: a multifunctional stress signaling lipid in plants. Trends Plant Sci. 10, 368–375. doi: 10.1016/j.tplants.2005.06.002
Testerink, C., and Munnik, T. (2011). Molecular, cellular, and physiological responses to phosphatidic acid formation in plants. J. Exp. Bot. 62, 2349–2361. doi: 10.1093/jxb/err079
Thole, J. M., Vermeer, J. E., Zhang, Y., Gadella, T. W. Jr., and Nielsen, E. (2008). Root hair defective 4 encodes a phosphatidylinositol-4-phosphate phosphatase required for proper root hair development in Arabidopsis thaliana. Plant Cell 20, 381–395. doi: 10.1105/tpc.107.054304
van Leeuwen, W., Vermeer, J. E., Gadella, T. W. Jr., and Munnik, T. (2007). Visualization of phosphatidylinositol 4,5-bisphosphate in the plasma membrane of suspension-cultured tobacco BY-2 cells and whole Arabidopsis seedlings. Plant J. 52, 1014–1026. doi: 10.1111/j.1365-313X.2007.03292.x
Vermeer, J. E., Thole, J. M., Goedhart, J., Nielsen, E., Munnik, T., and Gadella, T. W. Jr. (2009). Imaging phosphatidylinositol 4-phosphate dynamics in living plant cells. Plant J. 57, 356–372. doi: 10.1111/j.1365-313X.2008.03679.x
Vermeer, J. E., van Leeuwen, W., Tobeña-Santamaria, R., Laxalt, A. M., Jones, D. R., Divecha, N., et al. (2006). Visualization of PtdIns3P dynamics in living plant cells. Plant J. 47, 687–700. doi: 10.1111/j.1365-313X.2006.02830.x
Vermeer, J. E. M., van Wijk, R., Goedhart, J., Geldner, N., Chory, J., Gadella, T. W. J. J., et al. (2017). In vivo imaging of diacylglycerol at the cytoplasmic leaflet of plant membranes. Plant Cell Physiol. 58, 1196–1207. doi: 10.1093/pcp/pcx012
Voronkova, N. M., Burkovskaya, E. V., Bezdeleva, T. A., and Burundukova, O. L. (2011). Morphological and biological features of plants related to their adaptation to coastal habitats. Russ. J. Ecol. 39, 1–7. doi: 10.1134/s1067413608010013
Wang, X. (2005). Regulatory functions of phospholipase D and phosphatidic acid in plant growth, development, and stress responses. Plant Physiol. 139, 566–573. doi: 10.1104/pp.105.068809
Wang, X., Devaiah, S. P., Zhang, W., and Welti, R. (2006). Signaling functions of phosphatidic acid. Prog. Lipid Res. 45, 250–278. doi: 10.1016/j.plipres.2006.01.005
Welti, R., Li, W., Li, M., Sang, Y., Biesiada, H., Zhou, H. E., et al. (2002). Profiling membrane lipids in plant stress responses. Role of phospholipase D alpha in freezing-induced lipid changes in Arabidopsis. J. Biol. Chem. 277, 31994–32002. doi: 10.1074/jbc.M205375200
Wimalasekera, R., Pejchar, P., Holk, A., Martinec, J., and Scherer, G. F. (2010). Plant phosphatidylcholine-hydrolyzing phospholipases C NPC3 and NPC4 with roles in root development and brassinolide signaling in Arabidopsis thaliana. Mol. Plant 3, 610–625. doi: 10.1093/mp/ssq005
Yamaoka, Y., Yu, Y., Mizoi, J., Fujiki, Y., Saito, K., Nishijima, M., et al. (2011). PHOSPHATIDYLSERINE SYNTHASE1 is required for microspore development in Arabidopsis thaliana. Plant J. 67, 648–661. doi: 10.1111/j.1365-313X.2011.04624.x
Yang, D., Liu, X., Yin, X., Dong, T., Yu, M., and Wu, Y. (2021). Rice non-specific phospholipase C6 is involved in mesocotyl elongation. Plant Cell Physiol. 62, 985–1000. doi: 10.1093/pcp/pcab069
Yu, L., Nie, J., Cao, C., Jin, Y., Yan, M., Wang, F., et al. (2010). Phosphatidic acid mediates salt stress response by regulation of MPK6 in Arabidopsis thaliana. New Phytol. 188, 762–773. doi: 10.1111/j.1469-8137.2010.03422.x
Yuan, Z., Tezuka, Y., Fan, W., Kadota, S., and Li, X. (2002). Constituents of the underground parts of Glehnia littoralis. Chem. Pharm. Bull. 50, 73–77. doi: 10.1248/cpb.50.73
Zhang, Q., Lin, F., Mao, T., Nie, J., Yan, M., Yuan, M., et al. (2012). Phosphatidic acid regulates microtubule organization by interacting with MAP65-1 in response to salt stress in Arabidopsis. Plant Cell 24, 4555–4576. doi: 10.1105/tpc.112.104182
Zhang, W., Qin, C., Zhao, J., and Wang, X. (2004). Phospholipase D alpha 1-derived phosphatidic acid interacts with ABI1 phosphatase 2C and regulates abscisic acid signaling. Proc. Natl. Acad. Sci. U. S. A. 101, 9508–9513. doi: 10.1073/pnas.0402112101
Zhu, J.-K. (2001). Plant salt tolerance. Trends Plant Sci. 6, 66–71. doi: 10.1016/s1360-1385(00)01838-0
Keywords: Glehnia littoralis, halophyte, non-specific phospholipase C, salt stress, lipid
Citation: Li L, Li N, Qi X, Bai Y, Chen Q, Fang H, Yu X, Liu D, Liang C and Zhou Y (2021) Characterization of the Glehnia littoralis Non-specific Phospholipase C Gene GlNPC3 and Its Involvement in the Salt Stress Response. Front. Plant Sci. 12:769599. doi: 10.3389/fpls.2021.769599
Edited by:
Elisabeth Jamet, Université Toulouse III Paul Sabatier, FranceReviewed by:
Ana María Laxalt, National University of Mar del Plata, ArgentinaVolodymyr Kravets, Institute of Bioorganic Chemistry and Petrochemistry (NAN Ukraine), Ukraine
Copyright © 2021 Li, Li, Qi, Bai, Chen, Fang, Yu, Liu, Liang and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yifeng Zhou, bmpnemhvdUAxNjMuY29t; Chengyuan Liang, bGlhbmdjeTYxOEBjbmJnLm5ldA==
 Li Li
Li Li Naiwei Li1
Naiwei Li1 Yang Bai
Yang Bai Hailing Fang
Hailing Fang Xu Yu
Xu Yu