
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 05 November 2021
Sec. Technical Advances in Plant Science
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.768393
This article is part of the Research Topic Plant Transformation View all 20 articles
The available methods for plant transformation and expansion beyond its limits remain especially critical for crop improvement. For grass species, this is even more critical, mainly due to drawbacks in in vitro regeneration. Despite the existence of many protocols in grasses to achieve genetic transformation through Agrobacterium or biolistic gene delivery, their efficiencies are genotype-dependent and still very low due to the recalcitrance of these species to in vitro regeneration. Many plant transformation facilities for cereals and other important crops may be found around the world in universities and enterprises, but this is not the case for apomictic species, many of which are C4 grasses. Moreover, apomixis (asexual reproduction by seeds) represents an additional constraint for breeding. However, the transformation of an apomictic clone is an attractive strategy, as the transgene is immediately fixed in a highly adapted genetic background, capable of large-scale clonal propagation. With the exception of some species like Brachiaria brizantha which is planted in approximately 100 M ha in Brazil, apomixis is almost non-present in economically important crops. However, as it is sometimes present in their wild relatives, the main goal is to transfer this trait to crops to fix heterosis. Until now this has been a difficult task, mainly because many aspects of apomixis are unknown. Over the last few years, many candidate genes have been identified and attempts have been made to characterize them functionally in Arabidopsis and rice. However, functional analysis in true apomictic species lags far behind, mainly due to the complexity of its genomes, of the trait itself, and the lack of efficient genetic transformation protocols. In this study, we review the current status of the in vitro culture and genetic transformation methods focusing on apomictic grasses, and the prospects for the application of new tools assayed in other related species, with two aims: to pave the way for discovering the molecular pathways involved in apomixis and to develop new capacities for breeding purposes because many of these grasses are important forage or biofuel resources.
Apomixis in plants is defined as an asexual type of reproduction by seeds where the maternal plant produces clonal offsprings, combining the advantages of propagation by seeds and those of propagation by clone (Kandemir and Saygili, 2015). This reproductive mode, although rare among the major crop families, is a common trait in the polyploid C4 grasses (Poaceae family), many of which are important forage species. Some genera included in this group are Panicum, Pennisetum, Urochloa(Brachiaria), Eragrostis, Tripsacum, Paspalum, Cenchrus, Brachipodium, Bothriochloa, Bouteloua, Callipedium, Dischantium, Hyparrhenia, Melinis, Setaria, Chloris (Quero-Carrillo et al., 2010). For many years our group has been working on deciphering the genetic pathways involved in apomixis in Eragrostiscurvula or “weeping lovegrass,” a facultative apomictic grass native to Southern Africa and well-adapted to grow in EEUU, Australia, and the semiarid regions of Argentina. We recently demonstrated that apomixis in this grass is a gene-regulated trait linked to a chromosome region inherited in a Mendelian way (Zappacosta et al., 2019), which is consistent with other previously reported findings (Voigt and Bashaw, 1972). Additionally, we were able to sequence and assemble the genome of “Victoria,” a diploid accession with high resolution (Carballo et al., 2019), which demonstrated a strong epigenetic mechanism controlling the trait (Selva et al., 2020; Carballo et al., 2021). This complex epigenetic landscape makes it essential to analyze the trait directly in an apomictic context.
Plants are organisms with a plastic development and as such have the potential to differentiate new organs from the stem cell niche (pluripotency). Also, plant cells can be “reprogrammed” to produce an entirely identical plant (clone) through embryogenesis. This phenomenon is called totipotency and is exploited in biotechnology to induce somatic embryogenesis and produce transgenic plants. In grasses, success in genetic transformation is strongly dependent on the tissue culture procedures and on the ability of plant cells to regenerate a whole plant (Figure 1A). This usually involves the induction of embryogenic callus in a medium supplemented with auxins and the development of embryos in a medium with cytokinins, such as 6-benzylaminopurine (BAP) or thidiazuron (TDZ) or metatopolines at low concentrations (Sánchez-Romero, 2021). Several morphogenic regulators are able to modify calli responsiveness to embryogenesis, transformation, and regeneration (Fehér, 2019; Gordon-Kamm et al., 2019). Also, it is commonly observed that even the genotype and explant source might condition these callus responses (Echenique et al., 1996; Colomba et al., 2006; Yadav et al., 2009; Seo et al., 2010; Cabral et al., 2011; Kumar and Bhat, 2012; Takamori et al., 2015). As genotype effects are unavoidable, different explants, culture conditions, and morphogenic regulators should be assayed to improve the transformation and regeneration efficiencies. Several plant tissues have been used as explants to induce embryogenic calli, such as zygotic embryos, scutella, apical shoots, inflorescences, anthers, and basal parts of the leaves, among others. The actively dividing cells present in the callus phase provide a suitable target for transformation (Delporte et al., 2012).
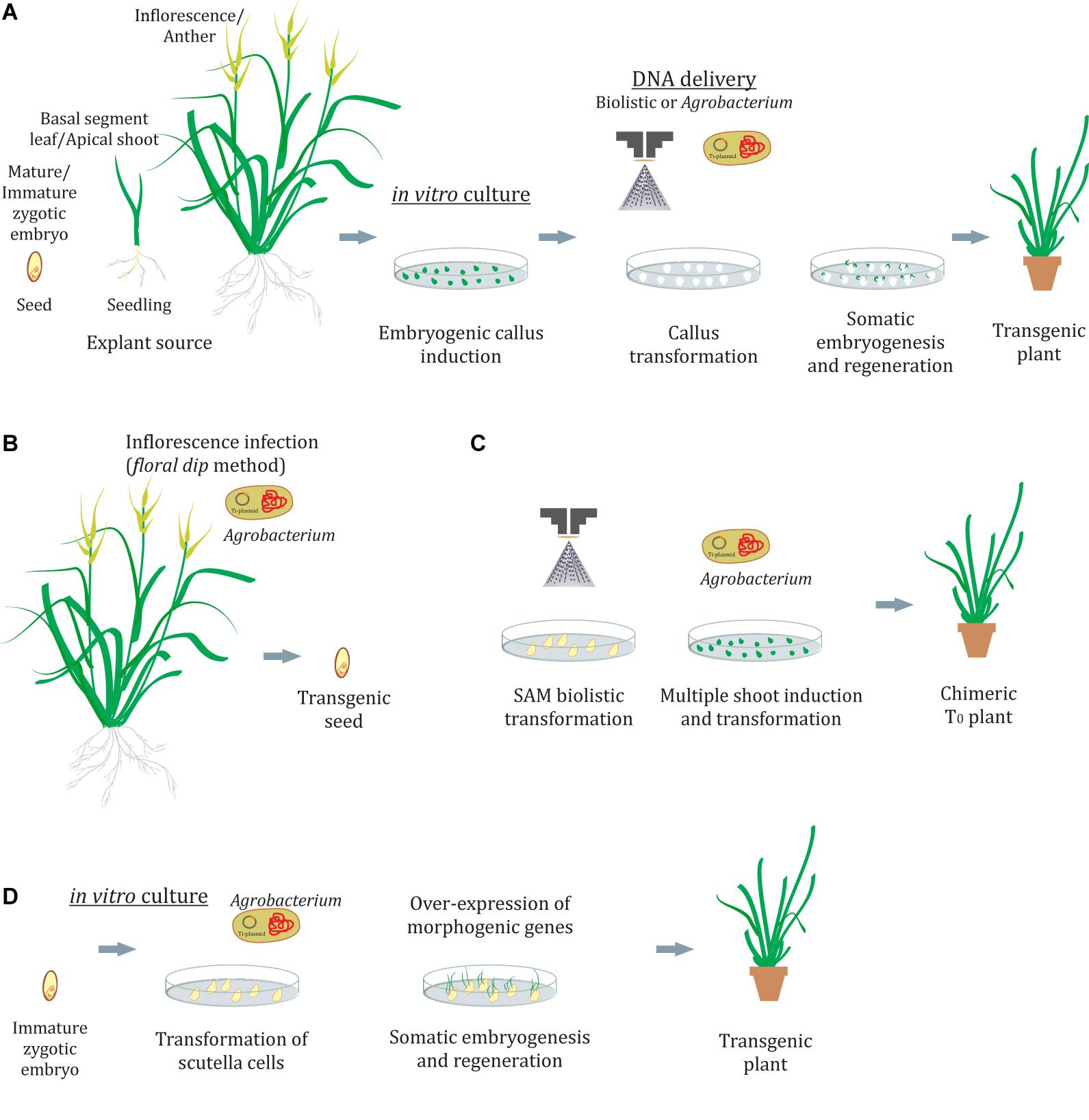
Figure 1. Genetic transformation in grasses. (A) Transformation of calli and regeneration by indirect somatic embryogenesis. (B,C) In planta methods by floral dip (B) or shoot apical meristem transformation (C). (D) Transformation of scutella cells with morphogenic regulators and regeneration without the callus phase by direct somatic embryogenesis. SAM, shoots apical meristem.
To achieve a successful genetic transformation not only is it critical to establish a suitable tissue culture system but also determine an effective DNA delivery method. The most commonly used methods in grasses are biolistic- and Agrobacterium-mediated transformation. The former, in theory, can be applied to any species or plant tissue owing to its physical character but requires trained personnel and also specialized and expensive instruments such as a gun device (Ozyigit and YucebilgiliKurtoglu, 2020). Furthermore, the DNA delivered by particle bombardment could result in variable expression levels in different transformed clones owing to the random integration of the expression cassettes in the host genome. In addition, if either several integrations of the same gene or integrations in methylated zones occur, it could result in transgene silencing (Delporte et al., 2012; Anami et al., 2013; Dong and Ronald, 2021). Also, the sequences of the expression cassettes introduced are prone to be truncated reducing the transformation efficiency (Shou et al., 2004). On the other hand, Agrobacterium-mediated transformation appears to be more suitable since it produces low transgene copy number insertions and confers more stability over generations with reduced gene silencing (Singh and Prasad, 2016). However, as Agrobacterium-mediated transformation is a biological process it is still dependent on the interaction of the Agrobacterium strain with the host plant genome, which is known to be influenced by several factors. Phenolic compounds produced by the host plant are necessary to induce the expression of Agrobacterium vir genes. In monocots, which are not natural hosts and are considered recalcitrant to Agrobacterium infection, these compounds are not synthesized and the addition of acetosyringone during plant and bacterial interaction supports the transference of the gene (Jha et al., 2011). Furthermore, 20 min of vacuum treatment and cocultivation for 2–3 days have been necessary to enhance this interaction (Li and Qu, 2011; Lin et al., 2017). Another important issue in plant transformation is the selection agent used to detect and isolate the primary transformed cells. Cells subjected to repetitive stresses related to transformation, antibiotic selection, and the more-or-less long-term in vitro cultures are exposed to increased risk of somaclonal variation (Lu et al., 2006; Carloni et al., 2014; Ishigaki et al., 2014). This variation refers to the genetic changes in either the DNA content (ploidy changes) or the DNA structure (chromosome rearrangements and point mutations) that are common in regenerated plants. For example, albinism in regenerated seedlings is usually indicative of a somaclonal event in the chloroplast genome (Mozgova et al., 2012). A reduction of time in culture, inducing direct regeneration of apical shoots without a callus phase, and employing non-antibiotic selection methods (i.e., capacity to metabolize mannose) offer new alternatives to alleviate concerns about somaclonal variation (O’Kennedy et al., 2004; Jha et al., 2011).
Owing to their efficient use of energy and high adaptability to harsh conditions, C4 grasses and their wild relatives constitute a source of genes that can be useful to genetically improve crop plants for agronomic traits such as high yield and stress resistance. Even when the transformation of grasses is a difficult task, this technology has been used to improve forage and turf in different aspects, such as digestibility, drought, cold, freezing, and salt tolerance, virus-resistance, among other traits in species like Bahiagrass, Tall fescue, Switchgrass, and Kentucky bluegrass (Wang and Brummer, 2012). Additionally, there have been many efforts focused on the use of these grasses for the production of biofuels with encouraging progress (Weijde et al., 2013; Mazarei et al., 2020). On the other hand, apomixis is a promising alternative that can provide a direct mechanism for the exploitation of heterosis. This is an agronomically important feature in breeding programs that would dramatically reduce the costs of hybrid seed production, especially for crops like maize, rice, or wheat. Alternatively, the transformation of an apomictic clone is an attractive strategy as the transgene is immediately fixed in a highly adapted genetic background, capable of large-scale clonal propagation. Although not providing complete transgene containment, gene transfer between apomictic species occurs at low frequency and over short distances, as has been shown in Kentucky bluegrass (Poa pratensis) and Bahiagrass (Paspalum notatum) (Johnson et al., 2006; Sandhu et al., 2010). Many efforts have been taken to understand the nature of apomixis, and several apomictic related genes have been identified by differential expression analyses (reviewed in Hojsgaard and Hörandl, 2019; Kaushal et al., 2019; Schmidt, 2020). Although it was suggested that the asexual mode of reproduction could have evolved from the altered expression of genes in the sexual pathway (Carman, 1997), only partial engineering of apomixis could be obtained by transgenesis in Arabidopsis thaliana (Ravi et al., 2008; d’Erfurth et al., 2009; Ravi and Chan, 2010) and rice (Khanday et al., 2019; Wang et al., 2019) due to the complexity of the trait. Although all the above efforts represent significant progress in the possibility of transferring the trait, an understanding of the mechanism that operates in true apomicts is crucial to getting efficient synthetic apomixis. This knowledge could be obtained by altering the genes or genetic pathways involved using genome-editing approaches in tissues or using single cells (protoplasts) in automated platforms (Dlugosz et al., 2016). Automation would enable large-scale screens, such as those performed by Wang et al. (2015), where CRISPR-mediated mutations were used to determine the essential genes required for human cell proliferation. By using an automated cell screen, every gene could be knocked out sequentially in the crop cells for a massive functional analysis (Altpeter et al., 2016). Regardless of the approach selected, this functional analysis and also the exploitation of traits with agronomic relevance present in apomictic grasses is still dependent on genetic transformation and plant regeneration, which are the main bottlenecks in the process. In this study, we review the current status of the in vitro culture and genetic transformation methods focusing on apomictic grasses (Table 1), and the prospects for the application of new tools assayed in other related species, with two aims: to pave the way for discovering the molecular pathways involved in apomixis and to develop new capacities for breeding purposes because many of these grasses are important forage or biofuel resources.
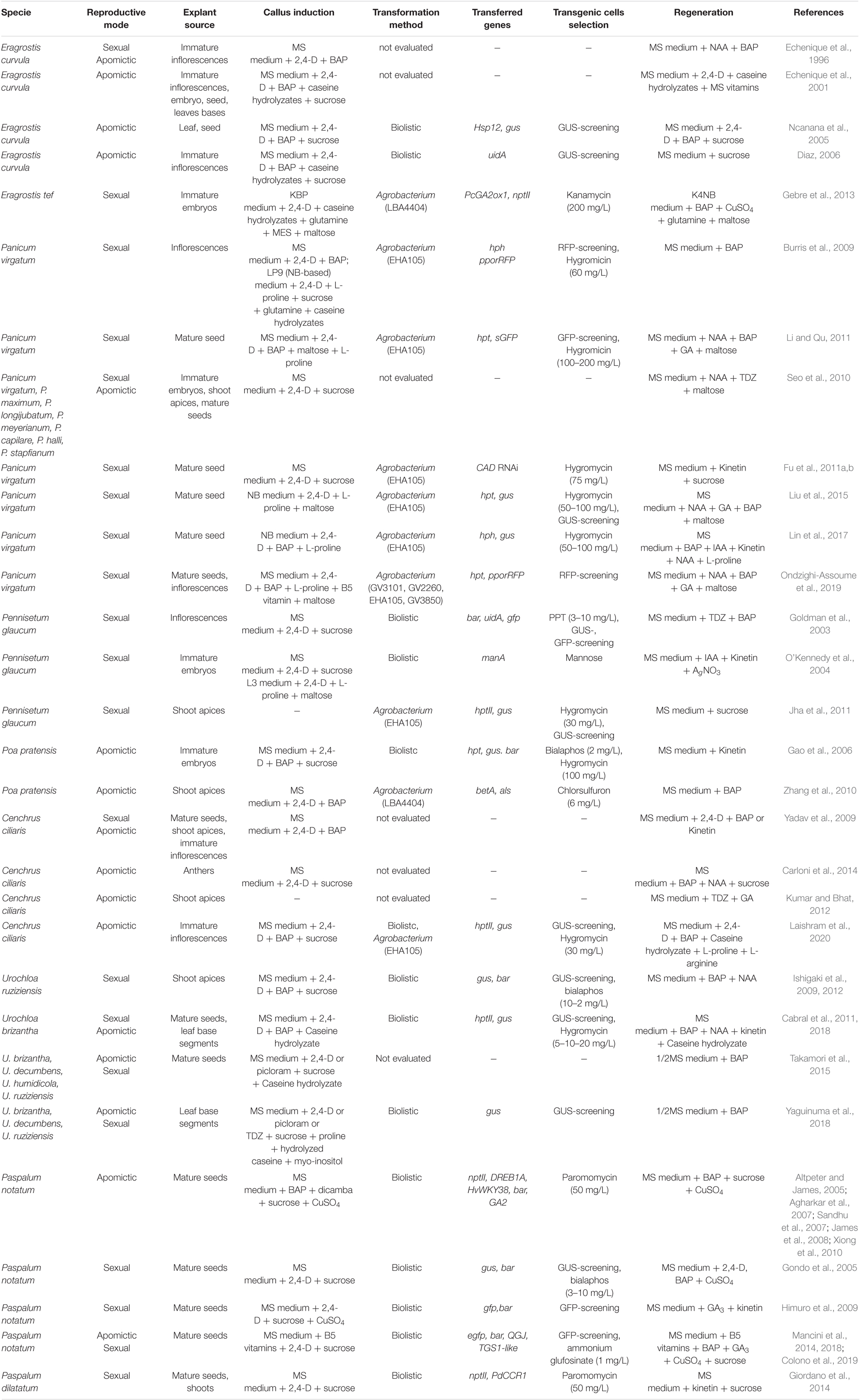
Table 1. Summary of methods for in vitro culture, transformation and regeneration of apomictic grasses and their relatives.
Eragrostis curvula (Schrad.) Nees is a polymorphic grass native to Southern Africa, naturalized in the semiarid regions of Argentina. Lovegrasses were introduced to Australia, the United States, and Argentina as soil conservation cultivars or forage grasses, having important agronomic traits such as drought resistance and a perennial habit, with forage quality being the most limiting factor. Most E. curvula cultivars are polyploids and reproduce by diplosporous apomixis. Diploids are always sexual and are very infrequent in nature, with tetraploid apomicts being the most frequently used cultivars for forage. In recent years our group has been working on different aspects of the reproductive biology of this grass, and different genetic and genomic resources have been developed, such as more than 40 floral and leaf transcriptomes (Cervigni et al., 2008a; Garbus et al., 2017), a genome assembly (Carballo et al., 2019), more than 4,520 SSRs (Cervigni et al., 2008b; Carballo et al., 2019), the first linkage map for the species highly saturated with GBS-SNPs (Zappacosta et al., 2019), genes and/or alleles conferring resistance to abiotic stresses (Carballo et al., 2019; Selva et al., 2020), and genes involved in lignin pathways (Díaz et al., 2010; Carballo et al., 2019).
Tissue culture protocols for E. curvula have been established using panicles as explant donors obtaining regeneration by embryogenesis and organogenesis (Echenique et al., 1996). Inflorescences just emerging from the flag leaf were cultured on Murashige and Skoog medium (1962) supplemented with different concentrations of 2,4-D and BAP, the most suitable concentrations for calli growth and development being 9 and 18 μM 2,4-D combined with 0.044 μM BAP. Fertile plants were obtained from four facultative apomictic genotypes (Morpa, Tanganyika, Don Pablo, and Kromdraii) with different efficiencies, as the regeneration capacity is influenced by the genotype since the two sexual materials included in the study did not regenerate (Echenique et al., 1996). Later Echenique et al. (2001) analyzed four different explant sources (immature inflorescences, embryos, seeds, and leaf bases) from three out of the four genotypes previously mentioned (Morpa, Don Pablo, and Kromdraai). Immature inflorescences were the most suitable explants for inducing embryogenic calli of the three cultivars. More recently, we have started to use isolated mature embryos as explants to establish a new in vitro regeneration protocol. Unlike their immature counterparts, mature embryos are available in large quantities throughout the year, being easily stored in the form of dried seeds (Delporte et al., 2005). These embryos are cultured with the scutellum facing upward on MS basal supplemented medium, and they produce structures like a callus and/or somatic embryos that arise from the shoot apical meristem (SAM) and then develop seedlings. Callus formation and/or somatic embryogenesis from the scutella of immature embryos were observed in Eragrostis tef (Zucc.) Trotter (Gugsa and Kumlehn, 2011).
Some efforts have also been made to develop a genetic transformation protocol in this genus. Ncanana et al. (2005) reported the development of plant regeneration and transformation protocols for E. curvula cv. Ermelo. Calli were generated from leaf and seed tissues and were transformed by biolistic bombardment with the yeast Saccharomyces cerevisiaeHsp12 gene under the maize ubiquitin promoter. Although successful transformation and transcription of the Hsp12 gene occurred, no Hsp12 protein was found in the transformed plants. Diaz (2006) evaluated the transient expression of the uidA gene introduced by biolistic bombardment in embryogenic calli obtained from immature inflorescences. Three different promoters were analyzed, and the maize ubiquitin gene promoter gave the best response. It was also possible to get transient transformation by inoculating mature seeds with two Agrobacterium tumefaciens strains, AGL0 and AGL1, both containing the binary vector Ppzp201BUGI (Terenti Romero, 2015).
Another member of the genus is Eragrostis tef (Zucc.) Trotter, a staple food crop in Ethiopia and cultivated in several countries for grain and forage production. It is not an apomictic species but it can also grow on marginal soils and under climate conditions not suitable for major cereals such as maize, wheat, and rice. It is an extremely low-yielding crop, mainly due to lodging and prolonged drought during the growing season (Numan et al., 2021). To increase the yield by introducing dwarfism, Gebre et al. (2013) transformed embryogenic calli obtained from immature embryos with the gibberellic acid (GA) inactivating gene PcGA2ox under the control of a triple CaMV 35S promoter using Agrobacterium. Calli were induced in a culture medium supplemented with KBP minerals and 2,4-D. The selection was performed in KBP medium with kanamycin, and regeneration in K4NB, where fully viable transformed plants carrying the transgene were obtained (detected by PCR and then sequenced). Some of them showed the expected phenotype. According to Numan et al. (2021), there must be a well-established transformation and regeneration system in E. tef to adopt advanced genetic engineering technologies. One of the major obstacles for the targeted breeding of E. tef is the presence of genes in two genomes (AA and BB: 2n = 4x = 40 chromosomes), as was shown in the recently released genome assembly (VanBuren et al., 2020). Target genes affecting key traits for this crop that can be modified by CRISPR-Cas have recently been selected from the genome assembly (Numan et al., 2021).
Panicum is a genus with approximately 450 species that grows in tropical and subtropical regions (Zuloaga and Soderstrom, 1985). The genus includes important forage species, such as Guineagrass (Panicum maximum Jacq.) and Switchgrass (Panicum virgatum L.), the former reproducing by apomixis. Attention to this group was particularly focused on Switchgrass owing to its inclusion in developmental programs as a model to produce lignocellulosic ethanol or second-generation biofuels (Merrick and Fei, 2015; Lin et al., 2017). It is self-incompatible and has varied ploidy levels with tetraploid and octoploid being the most common. As a perennial grass with a deep root system, Switchgrass is effective for soil conservation and could be helpful for removing the herbicide Atrazine, which is used in field crops for broadleaf weed control. In addition, this crop presents pest- and disease-tolerance traits has a high biomass yielding potential, and the ability to grow in marginal lands with low nutrient requirements (Lin et al., 2017).
Seo et al. (2010) reported an optimized protocol for shoot regeneration in these grasses starting from mature seed-derived calli of two species, P. longijubatum, and P. meyerianum. The effects of different carbohydrates and the optimal hormonal combination in the culture medium were determined. The maximum shoot regeneration was obtained using maltose (30 g/L; a twofold increase compared with sucrose) and TDZ (1 mg/L) without naphthalene acetic acid (NAA). This optimal regeneration medium was used on nine Panicum spp., and the efficiency of shoot regeneration resulted in being genotype-dependent, ranging from 69.9% in P. meyerianum to 0% in Panicum maximum. However, using calli derived from immature embryos in spite of mature seeds, higher regeneration frequencies (41–75%) were observed for three cultivars of Guineagrass, showing the influence of the explant source on shoot regeneration. The high capacity of immature embryos to regenerate shoots in plant tissue culture has also been reported previously in barley (Chang et al., 2003) and maize (Frame et al., 2000).
Genetic transformation has been an important tool for studying gene function and for germplasm improvement in Switchgrass. For instance, the downregulation of the cinnamyl alcohol dehydrogenase (CAD) gene or the caffeic acid O-methyltransferase (COMT) gene decreased lignin content and improved sugar release, giving Switchgrass the potential to increase digestibility and biofuel production during the fermentation processes (Fu et al., 2011a, b). In this context, several research projects have been focused on overcoming the recalcitrance of some cultivars to genetic transformation. The first report on obtaining highly regenerable type II calli used inflorescences as explants and a modified NB-based medium (LP9), containing 2,4-D and 100 mg/L–1 of L-proline (Burris et al., 2009). The maximum transformation efficiency (TE: transgenic plants obtained/total of calli exposed to Agrobacterium) of 4.4% was obtained using the EHA105 Agrobacterium strain. However, type II and type I calli were not separated during the transformation process and only type II callus yielded transgenic plants, suggesting that this TE can probably be increased by only selecting type II calli for transformation. Afterward, the identification and selection of type II calli from mature caryopses resulted in transformation efficiency increases of 50–90% in three different cultivars using Murashige and Skoog (1962) based medium for all the culture stages, supplemented with 2g/L–1 of L-proline (Li and Qu, 2011). In this study, the regeneration rate was higher than 80% in all cultivars, also showing the high competence of type II calli for the regeneration process. Importantly, vacuum application during Agrobacterium infection, desiccation at the co-cultivation stage, and resting after infection also facilitated Switchgrass transformation and selection (Li and Qu, 2011; Lin et al., 2017). On the other hand, histological analysis of calli derived from seeds revealed the occurrence of a “shell-core” structure being the “core” that was able to develop on type II calli (Liu et al., 2015; Lin et al., 2017). Following the dissection of this “core,” it was possible to recover transgenic plants of a cultivar previously reported as highly recalcitrant (Liu et al., 2015). In addition, it has been noticed that optimization of the seed sterilization step increased callus induction by 20% (Lin et al., 2017). Finally, the development of embryogenic cell suspension cultures derived from type II calli accelerated the regeneration of transgenic plants (Ondzighi-Assoume et al., 2019). However, somaclonal variation has been reported to occur during embryogenic callus suspension culture (Lu et al., 2006; Liu et al., 2018).
This genus includes the apomictic Pennisetum squamulatum and also the sexual relatives, pearl millet (Pennisetum glaucum L.), and Napier grass (Pennisetum purpureum Schumach.). The latter two are important forage crops in tropical and subtropical areas of the world, pearl millet also being a staple food in Africa and India, and Napier grass having great potential as an energy crop, and especially as a source for biofuel production. Thus, the development of a reliable transformation protocol for these crops will complement the classical breeding programs, and the effect of putative apomixis gene(s) from P. squamulatum could also be tested. Apomixis in P. squamulatum is transmitted by the apospory-specific genomic region (ASGR), a physically large, hemizygous, and non-recombining chromosomal region (Ozias-Akins et al., 1998). Multiple copies of the PsASGR-BABY BOOM-like (PsASGR-BBML) gene were found within the ASGR, and it has been proposed as a candidate gene related to parthenogenesis. BBM transcripts were originally identified in microspore cultures of Brassica napus (BnBBM) undergoing somatic embryogenesis (Boutilier et al., 2002). More recently, Conner et al. (2015) attempted to analyze the function of the PsASGR-BBML gene in P. squamulatum by creating an RNAi apomictic line to knock down the expression of this gene. Since the direct transformation and regeneration of P. squamulatum was not possible, it was decided to clone the ASGR in the sexual tetraploid pearl millet and the transgenic plants obtained exhibited parthenogenesis (Conner et al., 2015). In addition, sexual lines with an RNAi construct to silence BBML genes were generated and then these lines were fertilized with pollen from P. squamulatum. The resulting F1 hybrids inheriting the RNAi construct and harboring the ASGR showed a reduced number of parthenogenetic embryo sacs owing to the silencing of the BBML gene (Conner et al., 2015). The protocol employed above for the genetic transformation of pearl millet was previously described by Goldman et al. (2003), who optimized the conditions for biolistic-mediated transformation using embryogenic callus derived from inflorescences as explants. Shaving the spikelet primordia of a single inflorescence proved to be an excellent way to increase the amount of tissue capable of embryogenic calli production. By rotating the axis of the rachis, inflorescences could be shaved multiple times, allowing large quantities of responsive spikelets. Callus induction was performed in the dark in an MS medium containing sucrose (30 g/L) and a maximum of 5 mg/l 2,4-D. For plant regeneration, BAP (0.1 mg/L) and TDZ (0.1 mg/L) were used. TDZ was beneficial due to reducing in vitro culture and the overall time required for regenerating plants (Goldman et al., 2003). Delaying herbicide selection by 10–14 days after transformation resulted in greater effectiveness in the recovery of resistant plants.
Herbicide selection could either allow the regeneration of escapes (even at high selection pressure) or be deleterious to the regeneration process. It is commonly observed that the necrotic tissue surrounding the positive transgenic cells may prevent their growth. However, mannose-positive selection promoted regeneration and growth of the transgenic cells while non-transgenic cells were starved but not killed (O’Kennedy et al., 2004). Transgenic phosphomannose isomerase (PMI)-expressing cells acquired the ability to convert mannose-6-phosphate to fructose-6-phosphate, whereas the non-transgenic cells accumulated the former (Reed et al., 2001). The accumulation of mannose-6-phosphate in cells inhibits phosphoglucose isomerase, thereby causing a block in glycolysis (Goldsworthy and Street, 1965). The manA gene was shown to be a superior selectable marker in maize and wheat when compared with antibiotic or herbicide marker genes (Reed et al., 2001; Wright et al., 2001). The manA gene from E. coli has been used to produce transgenic plants of P. glaucum (O’Kennedy et al., 2004). With this purpose, embryogenic calli derived from immature zygotic embryos were bombarded and regenerated in a medium with IAA (0.2 mg/L), kinetin (0.5 mg/L), and AgNO3 (10 mg/L). Although the transformation efficiency was low, the system was effective in selecting almost only transgenic tissue, eliminating the labor-intensive tissue culture selection and molecular analysis of putative transgenic plants (O’Kennedy et al., 2004).
Transformation of shoot apical meristems (SAMs) either through Agrobacterium or biolistics has already been reported (McCabe et al., 1988; Gould and Magallanes-Cedeno, 1998; Zapata et al., 1999; Cho et al., 2003; Goldman et al., 2003; Zhang et al., 2010). In the absence of tissue dedifferentiation steps, less somaclonal variation and genotype regeneration dependence were observed. Nevertheless, the direct regeneration of transformants in this method always generates chimeric plants. It was proposed that more stable transformants could be generated by the multiplication of transgenic apical meristem cells by treatment with the phenylurea-type cytokinin TDZ (Zhong et al., 1996; Srivatanakul et al., 2000; Goldman et al., 2003; Gairi and Rashid, 2004). Kinetin was also reported to induce multiple shoots in pearl millet (Jha et al., 2009), and the addition of 50 μM CuSO4 could enhance their proliferation in Napiergrass (Umami et al., 2012). Following this method, Jha et al. (2011) generated a multiple shoot regeneration system in P. glaucum (without an intervening callus phase) for Agrobacterium-mediated transformation. Briefly, emerging shoot apices consisting of SAMs were cocultivated with the Agrobacterium EHA105 strain for 3 days in an MS medium containing acetosyringone (400 μM, essential for the successful transformation of pearl millet). After the recovery phase, shoot apices were transferred to the selection medium to allow the growth of the transformed shoots. The surviving shoots were transferred onto a preregeneration medium with BAP (17.6 μM) for 4–6 weeks to stimulate the production of transgenic multiple shoots that were then separated individually and regenerated on a medium with no growth regulators for 2–3 weeks. Stable transgenics were obtained, and the highest transformation frequency observed was 5.8% (number of transgenic plants/total number of explants inoculated) (Jha et al., 2011). Previously, Yookongkaew et al. (2007) reported a related protocol for the transformation and regeneration of several Thai rice cultivars showing that this technique is efficient and genotype-independent. This technique is also an alternative for species that are recalcitrant in producing embryogenic calli and it was used efficiently for P. purpureum. However, after bombardment and selection, the transformation efficiency was very low (0.64%) because the regeneration capacity was compromised by the aged callus (4–12 months) (Gondo et al., 2017).
Kentucky Bluegrass (Poa pratensis L.) is aC3turf grass adapted to a wide range of soils and climates (Van Wijk, 1997). It is sensitive to high salinity and drought (McDonnell and Conger, 1984) and reproduces by facultative apomixis (Bashaw, 1980; Albertini et al., 2001). Gao et al. (2006) reported a protocol that made it possible to obtain a large number of transgenic plants via particle bombardment of embryogenic calli derived from immature embryos. Callus induction was performed in an MS-based medium supplemented with 2,4-D (2 mg/L), BAP (0.1 mg/L), and gelrite as a gelling agent. Kinetin (0.2 mg/L) was added to the regeneration medium. In this study, the efficiency of two selectable marker genes, hpt, and bar, was tested and compared during the obtention of transgenic lines. No escapes were found under selection with 100 mg/L of hygromycin or 2 mg/L of bialaphos, hygromycin being superior to bialaphos since 77.8% of the hygromycin-resistant calli regenerated plants, whereas only 34.3% of the bialaphos resistant ones were able to regenerate. Moreover, the selection of transformed calli with hygromycin was faster and required only 4–8 weeks in culture (vs. 14–16 weeks for bialaphos), and the transformation efficiency (transgenic plants/bombarded callus) was higher (TEHygromycin 22% and TEBialaphos7.5%). In addition, bialaphos-mediated selection resulted in the regeneration of some albino plants, probably related to the time in culture. Overall, a long in vitro culture period showed a decline of 68% in the regeneration capacity (Gao et al., 2006). Despite the high regeneration capacity observed in this study, it is important to mention that only one genotype was considered. Additionally, all the recovered transgenic plants showed a complex integration pattern (with rearrangements and multiple transgene insertions), showing inactivation of the uidA gene after 3 months in the soil, probably as a consequence of gene silencing by multiple insertions. An Agrobacterium-mediated protocol was described by Zhang et al. (2010) for the transformation of three Kentucky bluegrass cultivars using a multiple shoot induction approach. The betA gene from E. coli was introduced together with the als selectable marker gene, which encodes a choline dehydrogenase enzyme and catalyzes the synthesis of the plant stress-protectant betaine (Sakamoto and Murata, 2001). In brief, shoot apices were cultured in an MS medium supplemented with 2,4-D (0.2 mg/L) plus BAP (2 mg/L). After 20 days in culture, meristematic cell clumps were subcultured for 8–12 days in an MS medium with BAP (2 mg/L) for the formation of multiple shoot clumps (MSCs) and were then divided and proliferated for 15 days in a medium supplemented with 2,4-D (0.07 mg/L) and BAP (2 mg/L). The MSCs were then cocultured for 2–4 days with the Agrobacterium strain LBA4404 followed by 10 days on rest medium with cefotaxime (10 mg/L). A chlorsulfuron selection (6 mg/L) was performed for 45 days with subcultures every 15 days. The highest transformation frequency (number of PCR-positive plants/number of shoot tips evaluated) was 1.42% with 87.5% of the surviving shoots being escapes (Zhang et al., 2010).
Buffelgrass (Cenchrus ciliaris L. syn. Pennisetum ciliare) is a warm-season C4 perennial forage grass native to Africa and India, reproducing predominantly through apomixis (Fisher et al., 1954; Bhat et al., 2001). It is highly drought-tolerant and is mainly used as fodder owing to its high biomass productivity (Martin et al., 1995; Rao et al., 1996). Some cultivars have shown recalcitrance to somatic embryogenesis (Colomba et al., 2006), and it was proposed that the understanding of the developmental events during this process could help in determining the role of phytohormones during the induction of somatic embryos (Yadav et al., 2009). These authors reported a histological analysis of regenerating calli of apomictic and sexual plants obtained from mature seeds, shoot tips, and immature inflorescences. Only the latter explant was able to produce embryogenic calli (with a maximum of 33%) in an MS medium supplemented with BAP (0.5 mg/L) and 2,4-D (with a maximum concentration of 5 mg/L) (Yadav et al., 2009). Regeneration of somatic embryos was obtained in the same medium supplemented with 2,4-D (0.25 mg/L) and BAP or kinetin (1–5 mg/L), with both cytokinins yielding comparative results. In the same way (Carloni et al., 2014), somatic embryos were induced from three apomictic genotypes of Buffel grass. Anthers were placed on MS-based induction medium containing 2,4-D (6 mg/L) for 90 days (subcultured after 45 days), and the regeneration was achieved in a MS medium supplemented with BAP (1 mg/L) and NAA (0.5 mg/L). Although anthers have been proposed as an optimal tissue for somatic embryogenesis (Kruczkowska et al., 2002), between 8 and 12 months were spent following this protocol for the regeneration. After this time several chromosomal rearrangements were found in the regenerated plants (Carloni et al., 2014).
Using a different approach, Kumar and Bhat (2012) tested the regeneration of five genotypes of Buffelgrass (including the apomictic one studied by Yadav et al., 2009) via multiple shoot induction. Briefly, seeds were germinated in an MS medium with TDZ (3 mg/L) to induce a high concentration of endogenous cytokinins (according to Yookongkaew et al., 2007). Shoot tips from 4-day old seedlings were excised and subcultured for 3–5 weeks with the same concentration of TDZ (3 mg/L). Shoots regenerated on medium containing TDZ (≥3 mg/L) were found to be stunted. To overcome this problem, GA3 (2 mg/L) was added to the shoot elongation medium. After 2 weeks, shoots were divided and transferred to MS plus indole-3-acetic acid (IAA, 3 mg/L) for rooting. All the genotypes were shown to be highly responsive to multiple shoot induction in a relatively short period of time (10–11 weeks). Based on these results, Laishram et al. (2020) performed an assessment of biolistic and Agrobacterium-mediated genetic transformation methods in C. ciliaris. Immature inflorescences of an apomictic cultivar were cultured in an MS medium supplemented with 2,4-D (3 mg/L) and BAP (0.5 mg/L) for embryogenic callus induction. Calli were maintained in an MS medium with casein hydrolyzate (0.3 g/L), L-proline (0.4 g/L), and L-glutamine (0.4 g/L) (according to Shashi, 2013), and subcultured every 21 days. It was observed that the regeneration ability declined after the third round of subcultures, and therefore earlier calli were used for genetic transformation. The marker gene GUS was used to screen transgenic events and hygromycin (30 mg/L) was employed during its selection. Stable transgenic plants were obtained only by biolistics with the transformation efficiency (transgenic plants confirmed by PCR/calli bombarded) of 0.2%. Although the regeneration rate was high (40 plants could be regenerated from the 100 resistant calluses obtained), there were a lot of escapes (90%), suggesting that further optimization of the transformation protocol is needed instead of the regeneration one.
Urochloa P. Beauv. (syn. Brachiaria Griseb. spp.) species are tropical forage grasses native to Africa (Keller-Grein et al., 1996), well adapted to poor soils, and resistant to long dry periods (Pinheiro et al., 2000). These grasses were introduced into South America in the eighteenth century (Miles et al., 1996), and are cultivated in millions of hectares in Brazil for cattle grazing (Montanari, 2013). Most of this cultivated area is covered by only two cultivars, both apomictic and tetraploid, U. brizantha cv. Marandu and U. decumbens cv. Basilisk (Simioni and Valle, 2009). Another species, U. humidicola, also apomictic and usually hexaploid, was introduced into Brazil from Australia. The only cultivated diploid and sexual species is U. ruziziensis, native to the African savannas (Valle and Savidan, 1996). For the latter species (U. ruziziensis), Ishigaki et al. (2009) established protocols for the development of MSCs and embryogenic calli using SAMs as explants. The addition of 2,4-D (0.5 mg/L) and BAP (2 mg/L) to an MS medium was the most effective treatment for the development of MSCs (21.4%). The highest regeneration capacity (53.6%) was observed in the same basal medium supplemented with BAP (1 mg/L) or with kinetin and GA3 (2 mg/L both). The highest rate of embryogenic callus induction (16.7%) was recorded in an MS medium with the addition of 2,4-D (4 mg/L) and BAP (0.2 mg/L), whereas the most effective treatment for plant regeneration was the use of solid MS medium supplemented with BAP (2 mg/L) and NAA (0.1 mg/L) (47.6%). MSCs and calli were maintained by subculturing onto fresh media every 30 or 14–21 days, respectively. Embryogenic calli maintained its shoot regeneration capacity for more than 1 year. Although both MSCs and embryogenic calli showed a high regenerative potential, some albino plants were obtained from the calli (Ishigaki et al., 2009). Afterward, in a transient GUS expression assay, Ishigaki et al. (2012) determined that embryogenic calli were more suitable targets than MSCs for particle bombardment transformation. These authors bombarded 9-month-old embryogenic calli obtaining four bialaphos resistant, but sterile, plants (TE: 1.4%). Later, by using flow cytometry it was observed that embryogenic calli older than 2 months were polyploydized and these events progressed over time (Ishigaki et al., 2014). The authors hypothesized that this response in the culture could be triggered by the accumulation of 2,4-D in old calli.
Cabral et al. (2011) evaluated the natural tetraploid apomictic genotype U.brizantha cv. Marandu and the diploid sexual U. brizantha for their capacities to produce somatic embryos and regenerate plants. When mature seeds of the apomictic cv. were used as explants, the highest induction of embryogenic calli (77%) and plant regeneration (54%) were observed in MS media with 2,4-D (3 mg/L) and BAP (0.2 mg/L) or BAP (1 mg/L), NAA (0.5 mg/L), and kinetin (2.5 mg/L), respectively. Calli were subcultured monthly on fresh medium and maintained for 5 months, but It was observed that 4-month-old cultures only produced albino plants. Interestingly, a decrease in the pH of the medium (from 5.8 to 4.0) not only reduced contamination but also increased the number of regenerating buds/shoots per clump and the total of regenerated plants. The same basal medium and growth regulator combination gave the best results regarding somatic embryogenesis and plant regeneration using leaf basal segments as explants (Cabral et al., 2011). These authors also observed that the apomictic genotype showed higher percentages of somatic embryogenesis and regeneration (77 and 64%, respectively) than the sexual one (45 and 21%), highlighting a genotype dependent response in U. brizantha. Afterward, with the optimal culture conditions for somatic embryogenesis described above, embryogenic calli, and cell suspensions were bombarded with plasmids containing GUS and hptII cassettes and tested for transient and stable expression (Cabral et al., 2018). The phased selection on hygromycin (5–10–20 mg/L) only delivered one transgenic plant, and it was negative for GUS histochemical assay and the amplification of the hptII gene by PCR. On the other hand, bombarded cell suspensions produced hygromycin-resistant calli that also showed stable GUS expression (both GUS and hptII genes were detected by PCR). These calli maintained their proliferative capacity for 10 months but were not able to regenerate shoots.
Takamori et al. (2015) tested two synthetic auxins (2,4-D and picloram) at different concentrations for callus induction, somatic embryogenesis, and plant regeneration in U. brizantha cv. Marandu using mature seeds as explants, obtaining more regenerated plants with picloram (1 mg/L). In the same study, 2,4-D at a concentration of 8 mg/L showed the lowest shoot conversion and the production of albino plantlets (4 and 8 mg/L). No albinos were observed with picloram. The regeneration was performed initially on MS with no growth regulators after maintaining the embryogenic calli for 72 days in callus induction media (subcultured every 14 days). Indirect light (1.7 μMm–2s–1) for 14 days was essential to avoid purple pigmentation (anthocyanin production) in regenerating calli. Finally, the calli were transferred to MS (half-strength) supplemented with BAP (2 mg/L) for 14 days. The effect of picloram was also tested for the embryogenic potential of other Urochloa species (U. decumbens, U. humidicola, and U. ruziziensis) with 1 mg/L being the most effective concentration. U. brizanthaand U. decumbens were the genotypes that were most prone to somatic embryogenesis, whereas U. humidicola and U. ruziziensis showed a minimal response. Similar results were obtained by Yaguinuma et al. (2018) using leaf base segments as explants. In U. decumbens, 1 mg/L of picloram was more effective in regenerating plants than 1 mg/L of 2,4-D (49 vs. 14%), and U. ruziziensis tended to be more recalcitrant for plant regeneration. Furthermore, the authors proposed cytokinin (TDZ) as an alternative callus inductor to reduce somaclonal variation linked to auxins. In this way, although fewer calli were obtained, plantlets of all genotypes were regenerated with 4 mg/L of TDZ, but the best response came from U. brizantha. Finally, picloram also showed better results than 2,4-D in a transient GUS expression experiment.
Bahiagrass (Paspalum notatum Flugge) is native to South America and used as turf and forage grass in tropical and subtropical regions from the United States to Argentina (Burton, 1967). This grass tolerates marginal soil fertility and has excellent resilience against drought, heat, and insect and nematode invasion. Two cytotypes, sexual diploids and apomictic tetraploids of Bahiagrass are grown (Forbes and Burton, 1961). The apomictic tetraploid cultivar “Argentine” is superior to the sexual diploid “Pensacola” owing to its darker green color, higher density, and seed production in shorter periods. Hence, with its asexual seed production and uniform seed progeny, the “Argentine” cultivar is an attractive target for genetic engineering of stress tolerance. Altpeter and James (2005) established a transformation protocol using embryogenic calli derived from mature seeds of this cultivar that were induced in an MS medium supplemented with BAP (5 μM) and dicamba (13.5 μM). After 6 weeks in culture, the calli were bombarded with a constitutive nptII expression cassette. The selection was initiated 1 week later on paromomycin (50 mg/L) and performed for 4 weeks (with subcultures every 2 weeks) on an MS-based shoot regeneration medium supplemented with BAP (5 μM). With this protocol, 2–10% resistant calli were obtained. Out of these, 1.5–4% regenerated plants and 60% of these plants were confirmed by PCR to be transgenic. Based on this protocol, James et al. (2008) obtained apomictic transgenic plants of Bahiagrass expressing the transcription factor DREB1A from barley, regulated by the abiotic stress-inducible HVA1 promoter. Transgenic lines showed faster recovery and increased survival under repeated dehydration–rehydration cycles than wild-type plants. Moreover, the transgene did not affect normal plant growth under non-stress conditions. The same protocol was used for the constitutive expression of HvWKY38 in transgenic plants of Bahiagrass (Xiong et al., 2010), obtaining a regeneration efficiency (resistant calli/calli bombarded) of 3.7% and a TE (transgenic lines/calli bombarded) of 2.7%. Although some lines were sterile and others had reduced seed set, three lines (out of 17 obtained) showed wild-type phenotypes and produced a similar number of inflorescences and seeds. The transgenic plants showed improved survival and biomass accumulation following dehydration and were superior to the wild-type plants in water retention, regeneration of new roots, and photosynthetic efficiency.
Uniform transgene transmission and expression could be a challenge during biolistic-mediated transformation, especially when large amounts of DNA are delivered into cells. Furthermore, transgenic plants frequently show complex integration patterns and silencing of transgenes (Gao et al., 2006). For this reason, Sandhu et al. (2007) attempted to optimize the process using two minimal transgene expression cassettes for particle bombardment-mediated cotransformation in Bahiagrass. Two unlinked nptIIand bar genes were excised from plasmids and cointroduced into embryogenic calli derived from mature seeds of the apomictic cv “Argentine.” After selection on medium containing paramomycin as selection agent, a TE of 10% was obtained with 95% of cotransformation frequency. Bar gene showed more complex integration patterns, probably owing to the presence of recombination hotspots at the CaMV35S sequence. The presence of recombinogenic spots within the expression cassettes and larger quantities of DNA during transformation seems to increase the complexity of transgenic loci (Sandhu and Altpeter, 2008). Despite the complex integration of these minimal cassettes, high expression, and coexpression levels were observed. Probably, this could be due to insertions on different loci whereas plasmid sequences tend to become linked. With a similar approach, Agharkar et al. (2007) achieved the constitutive expression of the AtGA2 gene in Bahiagrass (cultivar “Argentine”). Minimal cassettes of nptII and AtGA2 were transferred through biolistics and it resulted in 100% of cointegration and coexpression together with a simple integration pattern. No escapes were observed during the selection process and a TE of 1.3% was reached (eight independent transgenic lines were obtained from 600 bombarded calli). The AtGA2 expressing lines hydroxylated and degraded the GAs showing very low levels of GA, which was reflected in shorter stems and leaves, delayed flowering, and an increase in tillering. So, the semidwarf phenotype showed an enhanced turf quality.
Antibiotic selection through the constitutive expression of gene-conferring resistance is not always desirable in plant genetic engineering because it may hamper the normal growth or recovery of transgenic plants. The use of a visual marker without chemical dependence has made it possible to increase the transformation efficiency and reduce the screening time (Ahlandsberg et al., 1999; Zhu et al., 2004; Baranski et al., 2006). Accordingly, Himuro et al. (2009) proposed visual screening and selection of transgenic diploid Bahiagrass plants using the GFP reporter gene. Embryogenic calli for particle-bombardment transformation were induced from mature seeds in a liquid MS medium supplemented with 2,4-D (2 mg/L). After 2 weeks, primary calli were transferred to solid MS with the same concentration of 2,4-D. Then, microcalli (not larger than 2 mm in diameter) were divided and subcultured every 2 weeks in a medium containing 2,4-D (2 mg/L), BAP (0.1 mg/L), and CuSO4 (50 μM) at 31°C. Microcalli were transformed and the GFP expressing sectors were separated using fine-tipped forceps and transferred to a fresh medium. All these visually selected calli were able to regenerate in an MS medium with GA3 (1 mg/L) and kinetin (1 mg/L), obtaining an overall TE of 2.4%. Microcalli production resulted in the exposure of a larger surface area for the approaching particles during the bombardment and also for the culture conditions. Previously, it had been reported that high Cu content and high temperature during in vitro culture could probably enhance the regeneration of transgenic plants (Gondo et al., 2005). Furthermore, these conditions were adopted by Mancini et al. (2014) who observed a regeneration efficiency of 78% during the genetic transformation of tetraploid apomictic Bahiagrass. In this study, mature seeds were used as explants for embryogenic calli induction in an MS medium with B5 vitamins and 2,4-D (0.25 mg/L), and calli were cotransformed with plasmids containing egfp and bar genes. Regeneration was performed in an MS medium with B5 vitamins supplemented with BAP (5 μM), GA3 (1 μM), and CuSO4 (50 μM). A cotransformation frequency of 40.7% and a TE (independent transgenic lines/calli bombarded) of 8% were observed. The transgenic plants obtained were phenotypically normal and fertile after 4 weeks of selection in ammonium glufosinate (1 mg/L). Afterward, this protocol was successfully applied in Paspalum notatum for the functional characterization of the apomixis-related genes QUI-GON JINN (QGJ) and trimethylguanosine synthase 1 (TGS1)-like (Mancini et al., 2018; Colono et al., 2019). The high Cu content in the culture medium as a regeneration enhancer, firstly reported by Gondo et al. (2005) and later validated, appears to be a promising approach that could be replicated in other warm-season recalcitrant species. On the other hand, Giordano et al. (2014) obtained transgenic Paspalum dilatatum plants in which a sense-suppression gene cassette, delivered free of vector backbone and integrated separately to the selectable marker, reduced the CCR1 transcript levels and, as a consequence, the lignin content.
The success of the induction of embryogenic callus, somatic embryos, and the resulting recovery of viable plants is not always achievable for many species. Furthermore, tissue culture may demand long and complex treatments or procedures, and it often causes undesired and unpredictable changes in plant genomes. The in planta transformation methods avoid callus culture and plant regeneration (Figures 1B,C). The most widely used of these methods is a floral dip, originally developed for Arabidopsis (Clough and Bent, 1998), where the flower buds are dipped into an Agrobacterium suspension leading to the transformation of the ovule cells (Figure 1B). Floral dip methods have also been described for grass species including wheat (Zale et al., 2009), maize (Mu et al., 2012), Setariaviridis (Martins et al., 2015; Saha and Blumwald, 2016), and rice (Ratanasut et al., 2017). However, a broader application remains elusive mainly because the method is dependent not only on successful Agrobacterium infection but also on the inflorescence anatomy and the capacity of the plant to produce large amounts of seeds. Direct DNA delivery into pollen cells by ultrasonication or magnetofection has also been described as an alternative to creating transgenic seeds through pollination of plants with transformed pollen (Yang et al., 2017; Zhao et al., 2017). In addition to gametophyte cells, SAMs were used as targets for in planta biolistic transformation in wheat (Hamada et al., 2017; Figure 1C). Although primary T0 plants were chimeric, it was possible to recover T1 plants carrying the transgene, suggesting that genome modifications occurring at the SAMs are able to pass to the next generation. Even when the transformation efficiency (transgenic T1 plants/embryos bombarded) was lower than that of conventional tissue culture-based methods (0.8%), stable transformed lines of the recalcitrant cultivar Haruyokoi were obtained.
With regard to the limited host range of Agrobacterium tumefaciens, the compelling need to identify or generate non-Agrobacterium delivery systems emerges to increase the scope of genetic transformation. Furthermore, Agrobacterium-engineered crops for commercialization are restricted by licensing (Chi-Ham et al., 2012). It has long been recognized that the machinery required for DNA delivery to plant cells is not exclusive of Agrobacterium, since it is also present in species of the Rhizobium genus (Broothaerts et al., 2005; Rudder et al., 2014; Lacroix et al., 2016; Lacroix and Citovsky, 2019). It was observed that R. etli can successfully transform N. benthamiana and N. tabacum with a plasmid containing a T-DNA segment (Lacroix and Citovsky, 2016). However, the expression levels are about 10 times lower than those obtained with A. tumefaciens. Nonetheless, R. etli might be more efficient with other plant species, such as its specific hosts. Alternatively, Zuniga-Soto et al. (2015) obtained transgenic rice with Ensifer adhaerens-mediated infection of two Japonica cultivars with transformation efficiencies comparable with the Agrobacterium-mediated ones. Moreover, with this method, these authors were able to obtain a transgenic line from the recalcitrant Indica variety IR64. The genome of E. adhaerens (syn. Sinorhizobium adherens) not only has the same essential genes as Agrobacterium T-DNA but also non-essential ones, which could exert a positive impact on the virulence and ability to transform host tissues (Rudder et al., 2014). Although E. adharens-based protocols are still at the early stages, there are sustained efforts to expand them (Rathore et al., 2019) and indeed, they have a high potential in crop biotechnology.
On the other hand, virus-based vectors provide an alternative for delivering genome engineering reagents to plant cells (Liu and Zhang, 2020; Wang et al., 2020). Among these are the RNA viruses, which for monocots include wheat streak mosaic virus (WSMV) and barley stripe mosaic virus (BSMV) (Lee et al., 2012). Single-stranded (ss) DNA viruses, such as geminiviruses, are able to infect a wide range of host plant species like wheat, barley, corn, oat, and rye, and have been widely adopted as vectors (Choi et al., 2000). Furthermore, they replicate inside the host cells and produce a high amount of replicons enhancing the targeting cell response (Hanley-Bowdoin et al., 2013). Although the cargo capacity of geminiviruses is quite restricted, these vectors have been engineered for the expression of heterologous proteins in plants (Lozano-Durán, 2016). BSMV-based vectors were used for gene silencing in barley (Hordeum vulgare) and could be a suitable option in grasses as well as the RNAi approach (Holzberg et al., 2002; Scofield and Nelson, 2009). The virus-induced gene silencing exploits the RNA-based plant antiviral defense response, which degrades the RNA produced by infecting viruses. In this way, by inserting a plant gene sequence into the viral vector, gene transcripts become targets for degradation (Kumagai et al., 1995; Ratcliff et al., 1997; Baulcombe, 1999).
Genome editing (GE) is a new powerful technique and is considered a precision plant breeding tool (Kausch et al., 2019). Thus, the CRISPR/Cas technology used in its multiple versions (via transgenesis or by delivering ribonucleoproteins, base, and prime editing, etc.) offers new opportunities for improving different traits in important crops, sometimes combined with synthetic biology (for a review, see Chen et al., 2019; Zhu et al., 2020). Furthermore, nanotechnology has shown higher transformation efficiency than conventional methods, and in combination with GE, it has a great potential for engineering plants (Ahmar et al., 2021). The above considerations that influence the response of grasses concerning tissue culture, plant regeneration, and plant transformation methods are also important for GE. The knowledge generated from functional studies in apomictic grasses through transgenesis could support the induction of apomixis via GE in sexual plants to preserve the hybrid vigor for multiple generations in economically important crops (Scheben and Hojsgaard, 2020; Fiaz et al., 2021). However, the genomic information in apomictic grasses needed to perform GE is scarce and often unavailable. Although no genome-edited apomictic grasses have yet been reported, targeted mutations have been achieved in related grasses, such as Panicum virgatum (Liu et al., 2018) and Setariaviridis (Basso et al., 2021), using the CRISPR/Cas technology.
To exploit Agrobacterium as a routine biological engineering tool, it was necessary to develop the “binary vector” system from the large tumor inducing (Ti) plasmid (Hoekema et al., 1983; Bevan, 1984). This system allowed the separation of transfer DNA (T-DNA) in a small episome from the virulence genes encoded by a disarmed Ti plasmid. Afterward, the superbinary vector pSB1 with extra vir genes (B, G, part of C and D) was developed and resulted in the expansion of the host range of plants amenable to transformation (Komari, 1990; Hiei et al., 1994; Ishida et al., 1996; Komari et al., 1996; Cheng et al., 1997; Tingay et al., 1997). Recently, an improved pVIR ternary vector system was able to enhance the transformation frequencies of recalcitrant maize inbreds (Anand et al., 2018). Moreover, transformed calli grow faster, reducing the transformation process by 2–3 weeks. However, a high non-quality events frequency (i.e., events with backbone integration and/or multicopy events) was observed, probably due to the increased T-strand delivery. Similar results were described using the hypervirulent strain of Agrobacterium AGL0 (Zhi et al., 2015).
Agrobacterium elicits a wide plant defense response and modifies cell metabolism during infection, which in turn could affect the transformation efficiency (Ditt et al., 2006; Torres et al., 2006; Zipfel et al., 2006). Myo-inositol has several functions in eukaryotes, affecting a variety of developmental and physiological processes (Boss et al., 2006; York, 2006; Michell, 2008; Munnik and Nielsen, 2011), and it is a common constituent of standard plant culture media, as its addition is believed to improve plant regeneration (Murashige and Skoog, 1962; Gamborg et al., 1968; Loewus and Loewus, 1983; Schenk and Hildebrandt, 2011). However, a report in Ryegrass (Lolium perenne L.) revealed that the removal of myo-inositol from the callus culture media, in combination with a cold shock pretreatment and the addition of L-glutamine (L-Gln) prior to and during Agrobacterium-infection, resulted in about 84% of the treated calli being stably transformed (Zhang et al., 2013). Additionally, nearly 60% of the stable-transformed calli regenerated into green plants. Furthermore, the authors showed that under these conditions, the expression of pathogenesis-related (PRs) genes is modified, and probably plant defense response is attenuated, leading to an enhancement in the transformation efficiency. Similar results were obtained when the protocol was applied to cells, a rice cells suspension, suggesting that a broad application of this protocol can improve the transformation of recalcitrant monocot species (Zhang et al., 2013).
An alternative way to improve transformation and/or minimize callus culture concerns in grasses emerges from a genetic perspective. The ectopic expression of a few transcription factors (TFs) can induce the development of spontaneous somatic embryos in transformed tissues (Lotan et al., 1998; Stone et al., 2001; Boutilier et al., 2002; Figure 1D). Studies on its role in 2,4-D-mediated somatic embryogenesis have revealed an interacting network that acts on hormone pathways (see Horstman et al., 2017 and reference within). In the light of these facts, Lowe et al. (2016) successfully enhanced somatic embryogenesis and transformation in recalcitrant maize, rice, sorghum, and sugarcane varieties through the constitutive coexpression of TFs as BBM and WUS2. In this report, immature embryos were directly transformed by Agrobacterium-mediated infection, resulting in growth stimulation of embryogenic tissues. However, once embryogenic callus is formed the BBM and WUS2 expression cassettes must be eliminated to avoid deleterious pleiotropic effects in the regenerating plants. Although the excision method employed was efficient in the recovery of healthy and fertile, male and female plants, 3 months of calli culture was still required. An alternative scenario was described later by restricting the expression of BBM and WUS2 to the transformation targets (scutella and young leaves) using specific regulatory elements (Lowe et al., 2018). In this way, somatic embryos were developed on immature embryos within the first week after Agrobacterium infection. Furthermore, these embryos were able to germinate into healthy fertile plants without any intervening callus phase. This direct somatic embryogenesis response appeared to be genotype independent, and often dozens of somatic embryos could be observed on the surface of the developing scutella at similar rates as that of the zygotic embryos. This method requires less than half the normal time for traditional callus-based transformation protocols and eliminates the somaclonal issue. In another genetic approach, Debernardi et al. (2020) demonstrated that the overexpression in calli of a sequence encoding a chimera, composed of the GRF4 (growth-regulating factor 4) transcription factor and its GIF1 (GRF-interacting factor 1) cofactor, substantially increased the regeneration efficiency in wheat, rice, and citrus, including recalcitrant species. Also, the GRF4–GIF1 chimera resulted in high transformation and regeneration efficiencies, even in shorter protocols and contingent upon diverse conditions. On this basis, a protocol to select transgenic shoots without using antibiotic-based markers was formulated with promising results. The addition of the GRF4–GIF1 chimera in wheat also allowed the regeneration of transgenic lines in a cytokinin-free medium. Furthermore, transgene excision was not necessary to recover normal and fertile plants.
Owing to the efficient use of energy and the high adaptability to harsh conditions, apomictic grasses, many of which are C4, constitute a source of genes that can be useful for improving agronomic traits in crop plants, such as high yield and resistance to biotic and abiotic stresses, especially in the current climate change scenario. Also, this group of grasses represents very useful resources either for second-generation biofuel production or forage. Furthermore, transferring apomixis into economically important crops would be one of the major achievements of modern agriculture. A prerequisite to doing this is to understand how apomixis works. The way to acquire this knowledge is by modifying candidate genes directly in apomictic species that provide the correct epigenetic landscape for the expression of these genes. In this way, it would be possible to characterize their correct function. However, an efficient transformation protocol is still needed to engineer these grasses. Recently, several advances that contributed to enhancing the embryogenic and regeneration responses of cultured tissues have been obtained. Among these, high Cu content, the use of maltose instead of sucrose, a decrease in pH (from 5.8 to 4.0), and the removal of myo-inositol from the culture media have shown to be effective for enhancing the regeneration response. In addition, the use of alternative auxins and the expression of morphogenic genes during in vitro culture, the in planta genetic transformation methods, and genome editing emerge as promising approaches to increase the transformation efficiency of apomictic grasses.
AB and VE: conceptualization and writing original draft. VE: supervision. AB, ES, HP, and VE: writing, review, and editing. All authors have read and agreed to the published version of the manuscript.
This project has received funding from the Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT, PICT Raíces 2017-0879), Universidad Nacional del Sur (PGI 24/A199), and Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), Argentina.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
2,4-D, 2,4-dichlorophenoxiyacetic acid; BAP, 6-benzylaminopurine; Dicamba, 3,6-Dichloro-2-methoxybenzoic acid; GA, Gibberellic acid; IAA, Indole acetic acid; NAA, Naphthalene acetic acid; TE, Transformation efficiency; TDZ, Thidiazuron.
Agharkar, M., Lomba, P., Altpeter, F., Zhang, H., Kenworthy, K., and Lange, T. (2007). Stable expression of AtGA2ox1 in a low-input turfgrass (Paspalum notatum Flugge) reduces bioactive gibberellin levels and improves turf quality under field conditions. Plant Biotechnol. J. 5, 791–801. doi: 10.1111/j.1467-7652.2007.00284.x
Ahlandsberg, S., Sathish, P., Sun, C., and Jansson, C. (1999). Green fluorescent protein as a reporter system in the transformation of barley cultivars. Physiologia Plantarum 107, 194–200. doi: 10.1034/j.1399-3054.1999.100207.x
Ahmar, S., Mahmood, T., Fiaz, S., Mora-Poblete, F., Shafique, M. S., Chattha, M. S., et al. (2021). Advantage of nanotechnology-based genome editing system and its application in crop improvement. Front. Plant Sci. 12:663849. doi: 10.3389/fpls.2021.663849
Albertini, E., Barcaccia, G., Porceddu, A., Sorbolini, S., and Falcinelli, M. (2001). Mode of reproduction is detected by Parth1 and Sex1 SCAR markers in a wide range of facultative apomictic Kentucky bluegrass varieties. Mol. Breed. 7, 293–300. doi: 10.1023/A:1011673112747
Altpeter, F., and James, V. (2005). Genetic transformation of turf-type bahiagrass (Paspalum notatum Flugge) by biolistic gene transfer. Int. Turfgrass Soc. Res. J. 10:485.
Altpeter, F., Springer, N. M., Bartley, L. E., Blechl, A. E., Brutnell, T. P., Citovsky, V., et al. (2016). Advancing crop transformation in the era of genome editing. Plant Cell 28, 1510–1520. doi: 10.1105/tpc.16.00196
Anami, S., Njuguna, E., Coussens, G., Aesaert, S., and Van Lijsebettens, M. (2013). Higher plant transformation: principles and molecular tools. Int. J. Dev. Biol. 57, 483–494. doi: 10.1387/ijdb.130232mv
Anand, A., Bass, S. H., Wu, E., Wang, N., McBride, K. E., Annaluru, N., et al. (2018). An improved ternary vector system for Agrobacterium-mediated rapid maize transformation. Plant Mol. Biol. 97, 187–200. doi: 10.1007/s11103-018-0732-y
Baranski, R., Klocke, E., and Schumann, G. (2006). Green fluorescent protein as an efficient selection marker for Agrobacterium rhizogenes mediated carrot transformation. Plant Cell Rep. 25, 190–197. doi: 10.1007/s00299-005-0040-2
Bashaw, E. C. (1980). “Apomixis and its application in crop improvement,” in Hybridization of Crop Plants, eds W. R. Fehr and H. H. Hadley (Madison, WI: American Society of Agronomy), 45–63.
Basso, M. F., Duarte, K. E., Santiago, T. R., de Souza, W. R., Garcia, B. D. O., da Cunha, B. D. B., et al. (2021). Efficient genome editing and gene knockout in Setaria viridis with CRISPR/Cas9 directed gene editing by the non-homologous end-joining pathway. Plant Biotechnol. (Tokyo, Japan) 38, 227–238. doi: 10.5511/plantbiotechnology.21.0407a
Baulcombe, D. C. (1999). Fast forward genetics based on virus-induced gene silencing. Curr. Opin. Plant Biol. 2, 109–113. doi: 10.1016/s1369-5266(99)80022-3
Bevan, M. (1984). Binary Agrobacterium vectors for plant transformation. Nucleic Acids Res. 12, 8711–8721. doi: 10.1093/nar/12.22.8711
Bhat, V., Dalton, S. J., Kumar, S., Bhat, B. V., Gupta, M. G., and Morris, P. (2001). Particle-inflow-gun-mediated genetic transformation of buffel grass (Cenchrus ciliaris L.): optimizing biological and physical parameters. J. Appl. Genet. 42, 405–412.
Boss, W. F., Davis, A. J., Im, Y. J., Galvão, R. M., and Perera, I. Y. (2006). Phosphoinositide metabolism: towards an understanding of subcellular signaling. Subcell Biochem. 39, 181–205. doi: 10.1007/0-387-27600-9_8
Boutilier, K., Offringa, R., Sharma, V. K., Kieft, H., Ouellet, T., Zhang, L., et al. (2002). Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 14, 1737–1749. doi: 10.1105/tpc.001941
Broothaerts, W., Mitchell, H. J., Weir, B., Kaines, S., Smith, L. M. A., Yang, W., et al. (2005). Gene transfer to plants by diverse species of bacteria. Nature 433, 629–633. doi: 10.1038/nature03309
Burris, J. N., Mann, D. G. J., Joyce, B. L., and Stewart, C. N. (2009). An improved tissue culture system for embryogenic callus production and plant regeneration in switchgrass (Panicum virgatum L.). BioEnergy Res. 2, 267–274. doi: 10.1007/s12155-009-9048-8
Cabral, G., Carneiro, V., Lacerda, A. L., Valle, C., Martinelli, A., and Dusi, D. (2011). Somatic embryogenesis and organogenesis in apomictic and sexual Brachiariabrizantha. Plant Cell Tissue Organ Cult. 107, 271–282. doi: 10.1007/s11240-011-9978-7
Cabral, G. B., Carneiro, V., Gomes, A. C., Lacerda, A. L., Martinelli, A., and Dusi, D. (2018). GenetictransformationofBrachiariabrizanthacv. Marandubybiolistics. Anais Academia Brasileira Ciencias 90, 1789–1797.
Carballo, J., Santos, B., Zappacosta, D., Garbus, I., Selva, J. P., Gallo, C. A., et al. (2019). A high-quality genome of Eragrostiscurvula grass provides insights into Poaceae evolution and supports new strategies to enhance forage quality. Sci. Rep. 9:10250. doi: 10.1038/s41598-019-46610-0
Carballo, J., Zappacosta, D., Marconi, G., Gallardo, J., Di Marsico, M., Gallo, C. A., et al. (2021). Differential methylation patterns in apomictic vs. sexual genotypes of the diplosporous grass eragrostiscurvula. Plants 10:946. doi: 10.3390/plants10050946
Carloni, E., Ribotta, A. N., Lopez, E., Griffa, S., Quiroga, M., Tommasino, E., et al. (2014). Somatic embryogenesis from in vitro anther culture of apomictic buffel grass genotypes and analysis of regenerated plants using flow cytometry. Plant Cell Tissue Organ. Culture (PCTOC) 117, 311–322. doi: 10.1007/s11240-014-0441-4
Carman, J. G. (1997). Asynchronous expression of duplicate genes in angiosperms may cause apomixis, bispory, tetraspory, and polyembryony. Biol. J. Linnean Soc. 61, 51–94. doi: 10.1111/j.1095-8312.1997.tb01778.x
Cervigni, G. D. L., Paniego, N., Díaz, M., Selva, J. P., Zappacosta, D., Zanazzi, D., et al. (2008a). Expressed sequence tag analysis and development of gene associated markers in a near-isogenic plant system of Eragrostiscurvula. Plant Mol. Biol. 67, 1–10. doi: 10.1007/s11103-007-9282-4
Cervigni, G. D.L., Paniego, N., Pessino, S., Selva, J. P., Díaz, M., Spangenberg, G., et al. (2008b). Gene expression in diplosporous and sexual Eragrostiscurvula genotypes with differing ploidy levels. Plant Mol. Biol. 67, 11–23. doi: 10.1007/s11103-008-9305-9
Chang, Y., von Zitzewitz, J., Hayes, P. M., and Chen, T. H. H. (2003). High frequency plant regeneration from immature embryos of an elite barley cultivar (Hordeum vulgare L. cv. Morex). Plant Cell Rep. 21, 733–738. doi: 10.1007/s00299-003-0607-8
Chen, K., Wang, Y., Zhang, R., Zhang, H., and Gao, C. (2019). CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu. Rev. Plant Biol. 70, 667–697. doi: 10.1146/annurev-arplant-050718-100049
Cheng, M., Fry, J. E., Pang, S., Zhou, H., Hironaka, C. M., Duncan, D. R., et al. (1997). Genetic transformation of wheat mediated by Agrobacterium tumefaciens. Plant Physiol. 115, 971–980. doi: 10.1104/pp.115.3.971
Chi-Ham, C. L., Boettiger, S., Figueroa-Balderas, R., Bird, S., Geoola, J. N., Zamora, P., et al. (2012). An intellectual property sharing initiative in agricultural biotechnology: development of broadly accessible technologies for plant transformation. Plant Biotechnol. J. 10, 501–510. doi: 10.1111/j.1467-7652.2011.00674.x
Cho, M. J., Choi, H. W., Okamoto, D., Zhang, S., and Lemaux, P. G. (2003). Expression of green fluorescent protein and its inheritance in transgenic oat plants generated from shoot meristematic cultures. Plant Cell Rep. 21, 467–474. doi: 10.1007/s00299-002-0542-0
Choi, I. R., Stenger, D. C., Morris, T. J., and French, R. (2000). A plant virus vector for systemic expression of foreign genes in cereals. Plant J. 23, 547–555. doi: 10.1046/j.1365-313x.2000.00820.x
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Colomba, E. L., Grunberg, K., Griffa, S., Ribotta, A., Mroginski, L., and Biderbost, E. (2006). The effect of genotype and culture medium on somatic embryogenesis and plant regeneration from mature embryos of fourteen apomictic cultivars of buffel grass (Cenchrus ciliaris L.). Grass Forage Sci. 61, 2–8.
Colono, C., Ortiz, J. P. A., Permingeat, H. R., Souza Canada, E. D., Siena, L. A., Spoto, N., et al. (2019). A Plant-Specific TGS1 homolog influences gametophyte development in sexual tetraploid paspalum notatum ovules. Front. Plant Sci. 10:1566. doi: 10.3389/fpls.2019.01566
Conner, J. A., Mookkan, M., Huo, H., Chae, K., and Ozias-Akins, P. (2015). A parthenogenesis gene of apomict origin elicits embryo formation from unfertilized eggs in a sexual plant. Proc. Natl. Acad. Sci. U.S.A. 112, 11205–11210. doi: 10.1073/pnas.1505856112
Debernardi, J. M., Tricoli, D. M., Ercoli, M. F., Hayta, S., Ronald, P., Palatnik, J. F., et al. (2020). A GRF-GIF chimeric protein improves the regeneration efficiency of transgenic plants. Nat. Biotechnol. 38, 1274–1279. doi: 10.1038/s41587-020-0703-0
Delporte, F., Jacquemin, J.-M., Masson, P., and Watillon, B. (2012). Insights into the regenerative property of plant cells and their receptivity to transgenesis: wheat as a research case study. Plant Signal. Behav. 7, 1608–1620. doi: 10.4161/psb.22424
Delporte, F., Li, S., and Jacquemin, J.-M. (2005). Calluses initiated from thin mature embryo fragments are suitable targets for wheat transformation as assessed by long-term GUS expression studies. Plant Cell Tissue Organ. Cult. 80, 139–149. doi: 10.1007/s11240-004-9221-x
d’Erfurth, I., Jolivet, S., Froger, N., Catrice, O., Novatchkova, M., and Mercier, R. (2009). Turning meiosis into mitosis. PLoS Biol. 7:e1000124. doi: 10.1371/journal.pbio.1000124
Diaz, M. L. (2006). Biotecnología Para el Mejoramiento de la Calidad Nutritiva en Pasto Llorón, Eragrostiscurvula (Schrad.) Nees. Ph.D. thesis. Bahía Blanca: Universidad Nacional del Sur.
Díaz, M. L., Garbus, I., and Echenique, V. (2010). Allele-specific expression of a weeping lovegrass gene from the lignin biosynthetic pathway, caffeoyl-coenzyme A 3-O-methyltransferase. Mol. Breed. 26, 627–637. doi: 10.1007/s11032-010-9399-z
Ditt, R. F., Kerr, K. F., de Figueiredo, P., Delrow, J., Comai, L., and Nester, E. W. (2006). The Arabidopsis thaliana transcriptome in response to Agrobacterium tumefaciens. Mol. Plant Microbe Interact. 19, 665–681. doi: 10.1094/mpmi-19-0665
Dlugosz, E. M., Lenaghan, S. C., and Stewart, C. N. Jr. (2016). A robotic platform for high-throughput protoplast isolation and transformation. J. Vis. Exp. e54300. doi: 10.3791/54300
Dong, O. X., and Ronald, P. C. (2021). Targeted DNA insertion in plants. Proc. Natl. Acad. Sci. U.S.A. 118:e2004834117. doi: 10.1073/pnas.2004834117
Echenique, C. V., Díaz, M., Polci, P. A., and Mroginski, L. A. (2001). Embryogenic cell suspensions from different explants and cultivars of Eragrostiscurvula (Schrad.) Nees. Biocell 25, 131–138.
Echenique, V., Polci, P., and Mroginski, L. (1996). Plant regeneration in weeping lovegrass, (Eragrostiscurvula) through inflorescence culture. Plant Cell Tissue Organ. Culture 46, 123–130. doi: 10.1007/BF00034845
Fehér, A. (2019). Callus, dedifferentiation, totipotency, somatic embryogenesis: what these terms mean in the era of molecular plant biology? Front. Plant Sci. 10:536. doi: 10.3389/fpls.2019.00536
Fiaz, S., Wang, X., Younas, A., Alharthi, B., Riaz, A., and Ali, H. (2021). Apomixis and strategies to induce apomixis to preserve hybrid vigor for multiple generations. GM Crops Food 12, 57–70. doi: 10.1080/21645698.2020.1808423
Fisher, W. D., Bashaw, E. C., and Holt, E. C. (1954). Evidence of apomixis in Pennisetum ciliare and Cenchrus setigerus. Agron J. 46, 401–404.
Forbes, I. Jr., and Burton, G. W. (1961). Cytology of diploids, natural and induced tetraploids, and intra-species hybrids of bahiagrass, paspalum notatum Flugge1. Crop. Sci. 1:crosci1961.0011183X000100060006x. doi: 10.2135/cropsci1961.0011183X000100060006x
Frame, B. R., Zhang, H., Cocciolone, S. M., Sidorenko, L. V., Dietrich, C. R., Pegg, S. E., et al. (2000). Production of transgenic maize from bombarded type II callus: effect of gold particle size and callus morphology on transformation efficiency. In Vitro Cell. Dev. Biol. - Plant 36, 21–29. doi: 10.1007/s11627-000-0007-5
Fu, C., Mielenz, J. R., Xiao, X., Ge, Y., Hamilton, C. Y., Rodriguez, M., et al. (2011a). Genetic manipulation of lignin reduces recalcitrance and improves ethanol production from switchgrass. Proc. Natl. Acad. Sci. U.S.A. 108:3803. doi: 10.1073/pnas.1100310108
Fu, C., Xiao, X., Xi, Y., Ge, Y., Chen, F., Bouton, J., et al. (2011b). Downregulation of Cinnamyl Alcohol Dehydrogenase (CAD) leads to improved saccharification efficiency in switchgrass. BioEnergy Res. 4, 153–164. doi: 10.1007/s12155-010-9109-z
Gairi, A., and Rashid, A. (2004). TDZ-induced somatic embryogenesis in non-responsive caryopses of rice using a short treatment with 2,4-D. Plant Cell Tissue Organ. Culture 76, 29–33. doi: 10.1023/A:1025864605846
Gamborg, O. L., Miller, R. A., and Ojima, K. (1968). Nutrient requirements of suspension cultures of soybean root cells. Exp. Cell Res. 50, 151–158. doi: 10.1016/0014-4827(68)90403-5
Gao, C., Jiang, L., Folling, M., Han, L., and Nielsen, K. K. (2006). Generation of large numbers of transgenic Kentucky bluegrass (Poa pratensis L.) plants following biolistic gene transfer. Plant Cell Rep. 25, 19–25. doi: 10.1007/s00299-005-0005-5
Garbus, I., Romero, J. R., Selva, J. P., Pasten, M. C., Chinestra, C., Carballo, J., et al. (2017). De novo transcriptome sequencing and assembly from apomictic and sexual Eragrostis curvula genotypes. PLoS One 12:e0185595. doi: 10.1371/journal.pone.0185595
Gebre, E., Gugsa, L., Schlüter, U., and Kunert, K. (2013). Transformation of tef (Eragrostis tef) by Agrobacterium through immature embryo regeneration system for inducing semi-dwarfism. South Afr. J. Bot. 87, 9–17. doi: 10.1016/j.sajb.2013.03.004
Giordano, A., Liu, Z., Panter, S. N., Dimech, A. M., Shang, Y., Wijesinghe, H., et al. (2014). Reduced lignin content and altered lignin composition in the warm season forage grass Paspalum dilatatum by down-regulation of a Cinnamoyl CoA reductase gene. Transgenic Res. 23, 503–517. doi: 10.1007/s11248-014-9784-1
Goldman, J. J., Hanna, W. W., Fleming, G., and Ozias-Akins, P. (2003). Fertile transgenic pearl millet [Pennisetum glaucum (L.) R. Br.] plants recovered through microprojectile bombardment and phosphinothricin selection of apical meristem-, inflorescence-, and immature embryo-derived embryogenic tissues. Plant Cell Rep. 21, 999–1009. doi: 10.1007/s00299-003-0615-8
Goldsworthy, A., and Street, H. E. (1965). The carbohydrate nutrition of tomato roots: VIII. The mechanism of the inhibition by D-Mannose of the respiration of excised roots. Ann. Bot. 29, 45–58. doi: 10.1093/oxfordjournals.aob.a083936
Gondo, T., Tsuruta, S.-I., Akashi, R., Kawamura, O., and Hoffmann, F. (2005). Green, herbicide-resistant plants by particle inflow gun-mediated gene transfer to diploid bahiagrass (Paspalum notatum). J. Plant Physiol. 162, 1367–1375. doi: 10.1016/j.jplph.2005.03.005
Gondo, T., Umami, N., Muguerza, M., and Akashi, R. (2017). Plant regeneration from embryogenic callus derived from shoot apices and production of transgenic plants by particle inflow gun in dwarf napier grass (Pennisetum purpureum Schumach.). Plant Biotechnol. (Tokyo, Japan) 34, 143–150. doi: 10.5511/plantbiotechnology.17.0623a
Gordon-Kamm, B., Sardesai, N., Arling, M., Lowe, K., Hoerster, G., Betts, S., et al. (2019). Using morphogenic genes to improve recovery and regeneration of transgenic plants. Plants (Basel) 8:38. doi: 10.3390/plants8020038
Gould, J. H., and Magallanes-Cedeno, M. (1998). Adaptation of cotton shoot apex culture to agrobacterium-mediated transformation. Plant Mol. Biol. Rep. 16, 1–10.
Gugsa, L., and Kumlehn, J. (2011). Somatic embryogenesis and massive shoot regeneration from immature embryo explants of tef. Biotechnol. Res. Int. 2011:309731. doi: 10.4061/2011/309731
Hamada, H., Linghu, Q., Nagira, Y., Miki, R., Taoka, N., and Imai, R. (2017). An in planta biolistic method for stable wheat transformation. Sci. Rep. 7:11443. doi: 10.1038/s41598-017-11936-0
Hanley-Bowdoin, L., Bejarano, E. R., Robertson, D., and Mansoor, S. (2013). Geminiviruses: masters at redirecting and reprogramming plant processes. Nat. Rev. Microbiol. 11, 777–788. doi: 10.1038/nrmicro3117
Hiei, Y., Ohta, S., Komari, T., and Kumashiro, T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6, 271–282. doi: 10.1046/j.1365-313x.1994.6020271.x
Himuro, Y., Gondo, T., Yamakawa, K., and Akashi, R. (2009). Genetic transformation of bahiagrass (Paspalum notatum Flügge) by visually screening cells expressing green fluorescent protein. Grassland Sci. 55, 216–220. doi: 10.1111/j.1744-697X.2009.00162.x
Hoekema, A., Hirsch, P. R., Hooykaas, P. J. J., and Schilperoort, R. A. (1983). A binary plant vector strategy based on separation of vir- and T-region of the Agrobacterium tumefaciens Ti-plasmid. Nature 303, 179–180. doi: 10.1038/303179a0
Hojsgaard, D., and Hörandl, E. (2019). The rise of apomixis in natural plant populations. Front. Plant Sci. 10:358. doi: 10.3389/fpls.2019.00358
Holzberg, S., Brosio, P., Gross, C., and Pogue, G. P. (2002). Barley stripe mosaic virus-induced gene silencing in a monocot plant. Plant J. 30, 315–327. doi: 10.1046/j.1365-313x.2002.01291.x
Horstman, A., Bemer, M., and Boutilier, K. (2017). A transcriptional view on somatic embryogenesis. Regeneration (Oxford, England) 4, 201–216. doi: 10.1002/reg2.91
Ishida, Y., Saito, H., Ohta, S., Hiei, Y., Komari, T., and Kumashiro, T. (1996). High efficiency transformation of maize (Zea mays L.) mediated by Agrobacterium tumefaciens. Nat. Biotechnol. 14, 745–750. doi: 10.1038/nbt0696-745
Ishigaki, G., Gondo, T., Rahman, M. M., Umami, N., and Akashi, R. (2014). Spontaneous appearance of polyploids in plants regenerated from embryogenic calli derived from seedling-meristems of ruzigrass (Brachiariaruziziensis Germain et Everard). Grassland Sci. 60, 24–30. doi: 10.1111/grs.12040
Ishigaki, G., Gondo, T., Suenaga, K., and Akashi, R. (2009). Multiple shoot formation, somatic embryogenesis and plant regeneration from seed-derived shoot apical meristems in ruzigrass (Brachiariaruziziensis). Grassland Sci. 55, 46–51. doi: 10.1111/j.1744-697X.2009.00137.x
Ishigaki, G., Gondo, T., Suenaga, K., and Akashi, R. (2012). Fertile transgenic Brachiariaruziziensis (ruzigrass) plants by particle bombardment of tetraploidized callus. J. Plant Physiol. 169, 546–549. doi: 10.1016/j.jplph.2011.11.013
James, V. A., Neibaur, I., and Altpeter, F. (2008). Stress inducible expression of the DREB1A transcription factor from xeric, Hordeum spontaneum L. in turf and forage grass (Paspalum notatum Flugge) enhances abiotic stress tolerance. Transgenic Res. 17, 93–104. doi: 10.1007/s11248-007-9086-y
Jha, P., Shashi, Rustagi, A., Agnihotri, P., Kulkarni, V. M., and Bhat, V. (2011). Efficient Agrobacterium-mediated transformation of Pennisetum glaucum (L.) R. Br. using shoot apices as explant source. Plant Cell Tissue Organ. Culture (PCTOC) 107, 501–512.
Jha, P., Yadav, C., Anjaiah, V., and Bhat, V. (2009). In vitro plant regeneration through somatic embryogenesis and direct shoot organogenesis in Pennisetum glaucum (L.) R. Br. In Vitro Cell. Dev. Biol. - Plant 45, 145–154.
Johnson, P. G., Larson, S. R., Anderton, A. L., Patterson, J. T., Cattani, D. J., and Nelson, E. K. (2006). Pollen-Mediated gene flow from kentucky bluegrass under cultivated field conditions. Crop Sci. 46, 1990–1997. doi: 10.2135/cropsci2005.09.0316
Kandemir, N., and Saygili, I. (2015). Apomixis: new horizons in plant breeding. Turkish J. Agric. For. 39, 549–556. doi: 10.3906/tar-1409-74
Kausch, A. P., Nelson-Vasilchik, K., Hague, J., Mookkan, M., Quemada, H., Dellaporta, S., et al. (2019). Edit at will: genotype independent plant transformation in the era of advanced genomics and genome editing. Plant Sci. 281, 186–205. doi: 10.1016/j.plantsci.2019.01.006
Kaushal, P., Dwivedi, K. K., Radhakrishna, A., Srivastava, M. K., Kumar, V., Roy, A. K., et al. (2019). Partitioning apomixis components to understand and utilize gametophytic apomixis. Front. Plant Sci. 10:256. doi: 10.3389/fpls.2019.00256
Keller-Grein, G., Maass, B., and Hanson, J. (1996). “Natural variation in Brachiaria and existing germplasm collections,” in Brachiaria: Biology, Agronomy, and Improvement, eds J. W. Miles, B. L. Maass, C. B. Valle, and V. Kumble (Cali: Centro Internacional de Agricultura Tropical (CIAT)), 16–42.
Khanday, I., Skinner, D., Yang, B., Mercier, R., and Sundaresan, V. (2019). A male-expressed rice embryogenic trigger redirected for asexual propagation through seeds. Nature 565, 91–95. doi: 10.1038/s41586-018-0785-8
Komari, T. (1990). Transformation of cultured cells of Chenopodium quinoa by binary vectors that carry a fragment of DNA from the virulence region of pTiBo542. Plant Cell Rep. 9, 303–306. doi: 10.1007/BF00232856
Komari, T., Hiei, Y., Saito, Y., Murai, N., and Kumashiro, T. (1996). Vectors carrying two separate T-DNAs for co-transformation of higher plants mediated by Agrobacterium tumefaciens and segregation of transformants free from selection markers. Plant J. 10, 165–174. doi: 10.1046/j.1365-313x.1996.10010165.x
Kruczkowska, H., Pawłowska, H., and Skucińska, B. (2002). Influence of anther pretreatment on the efficiency of androgenesis in barley. J. Appl. Genet. 43, 287–296.
Kumagai, M. H., Donson, J., della-Cioppa, G., Harvey, D., Hanley, K., and Grill, L. K. (1995). Cytoplasmic inhibition of carotenoid biosynthesis with virus-derived RNA. Proc. Natl. Acad. Sci. U.S.A. 92, 1679–1683. doi: 10.1073/pnas.92.5.1679
Kumar, S., and Bhat, V. (2012). High-frequency direct plant regeneration via multiple shoot induction in the apomictic forage grass Cenchrus ciliaris L. In Vitro Cell. Dev. Biol. Plant 48, 241–248.
Lacroix, B., and Citovsky, V. (2016). A Functional Bacterium-to-Plant DNA transfer machinery of rhizobium etli. PLoS Pathog 12:e1005502. doi: 10.1371/journal.ppat.1005502
Lacroix, B., and Citovsky, V. (2019). Pathways of DNA transfer to plants from Agrobacterium tumefaciens and related bacterial species. Ann. Rev. Phytopathol. 57, 231–251. doi: 10.1146/annurev-phyto-082718-100101
Lacroix, B., Citovsky, V., Winans, S. C., and Collier, R. J. (2016). Transfer of DNA from Bacteria to Eukaryotes. mBio 7, e863–e816. doi: 10.1128/mBio.00863-16
Laishram, S. D., Goyal, S., Shashi, Kulkarni, V. M., Kumar, S., and Bhat, V. (2020). Assessment of biolistic and Agrobacterium-mediated genetic transformation methods in Cenchrus ciliaris. Nucleus 63, 303–312. doi: 10.1007/s13237-020-00332-1
Lee, W. S., Hammond-Kosack, K. E., and Kanyuka, K. (2012). Barley stripe mosaic virus-mediated tools for investigating gene function in cereal plants and their pathogens: virus-induced gene silencing, host-mediated gene silencing, and virus-mediated overexpression of heterologous protein. Plant Physiol. 160, 582–590. doi: 10.1104/pp.112.203489
Li, R., and Qu, R. (2011). High throughput Agrobacterium-mediated switchgrass transformation. Biomass Bioenergy 35, 1046–1054. doi: 10.1016/j.biombioe.2010.11.025
Lin, C.-Y., Donohoe, B. S., Ahuja, N., Garrity, D. M., Qu, R., Tucker, M. P., et al. (2017). Evaluation of parameters affecting switchgrass tissue culture: toward a consolidated procedure for Agrobacterium-mediated transformation of switchgrass (Panicum virgatum). Plant Methods 13:113. doi: 10.1186/s13007-017-0263-6
Liu, H., and Zhang, B. (2020). Virus-Based CRISPR/Cas9 genome editing in plants. Trends Genet. 36, 810–813. doi: 10.1016/j.tig.2020.08.002
Liu, Y., Merrick, P., Zhang, Z., Ji, C., Yang, B., and Fei, S. Z. (2018). Targeted mutagenesis in tetraploid switchgrass (Panicum virgatum L.) using CRISPR/Cas9. Plant Biotechnol. J. 16, 381–393. doi: 10.1111/pbi.12778
Liu, Y.-R., Cen, H.-F., Yan, J.-P., Zhang, Y.-W., and Zhang, W.-J. (2015). Inside out: high-efficiency plant regeneration and Agrobacterium-mediated transformation of upland and lowland switchgrass cultivars. Plant Cell Rep. 34, 1099–1108. doi: 10.1007/s00299-015-1769-x
Loewus, F. A., and Loewus, M. W. (1983). myo-Inositol: its biosynthesis and metabolism. Ann. Rev. Plant Physiol. 34, 137–161. doi: 10.1146/annurev.pp.34.060183.001033
Lotan, T., Ohto, M., Yee, K. M., West, M. A., Lo, R., Kwong, R. W., et al. (1998). Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93, 1195–1205. doi: 10.1016/s0092-8674(00)81463-4
Lowe, K., La Rota, M., Hoerster, G., Hastings, C., Wang, N., Chamberlin, M., et al. (2018). Rapid genotype “independent” Zea mays L. (maize) transformation via direct somatic embryogenesis. In Vitro Cell Dev. Biol. Plant 54, 240–252. doi: 10.1007/s11627-018-9905-2
Lowe, K., Wu, E., Wang, N., Hoerster, G., Hastings, C., Cho, M.-J., et al. (2016). Morphogenic regulators baby boom and wuschel improve monocot transformation. Plant Cell 28, 1998–2015. doi: 10.1105/tpc.16.00124
Lozano-Durán, R. (2016). Geminiviruses for biotechnology: the art of parasite taming. New Phytol. 210, 58–64. doi: 10.1111/nph.13564
Lu, S., Wang, Z., Peng, X., Guo, Z., Zhang, G., and Han, L. (2006). An efficient callus suspension culture system for triploid bermudagrass (Cynodon transvaalensis × C. dactylon) and somaclonal variations. Plant Cell, Tissue Organ. Culture 87, 77–84. doi: 10.1007/s11240-006-9138-7
Mancini, M., Permingeat, H., Colono, C., Siena, L., Pupilli, F., Azzaro, C., et al. (2018). The MAP3K-Coding QUI-GON JINN (QGJ) gene is essential to the formation of unreduced embryo sacs in paspalum. Front. Plant Sci. 9:1547. doi: 10.3389/fpls.2018.01547
Mancini, M., Woitovich, N., Permingeat, H. R., Podio, M., Siena, L. A., Ortiz, J. P. A., et al. (2014). Development of a modified transformation platform for apomixis candidate genes research in Paspalum notatum (bahiagrass). In Vitro Cell. Dev. Biol. - Plant 50, 412–424. doi: 10.1007/s11627-014-9596-2
Martin, M. H., Cox, J. R., and Ibarra-F, F. (1995). Climatic effects on buffelgrass productivity in the Sonoran Desert. J. Range Manag. 48, 60–63.
Martins, P. K., Ribeiro, A. P., Cunha, B. A. D., Kobayashi, A. K., and Molinari, H. B. C. (2015). A Simple and Highly Efficient Agrobacterium -Mediated Transformation Protocol for Setariaviridis</i>. Biotechnology reports (Amsterdam, Netherlands) [Online], Vol. 6. Available online at: https://doi.org/10.1016/j.btre.2015.02.002 (accessed June, 2015).
Mazarei, M., Baxter, H. L., Srivastava, A., Li, G., Xie, H., Dumitrache, A., et al. (2020). Silencing folylpolyglutamate Synthetase1 (FPGS1) in switchgrass (Panicum virgatum L.) improves lignocellulosic biofuel production. Front. Plant Sci. 11:843. doi: 10.3389/fpls.2020.00843
McCabe, D. E., Swain, W. F., Martinell, B. J., and Christou, P. (1988). Stable transformation of soybean (Glycine Max) by particle acceleration. Biotechnology (Nature Publishing Company) 6, 923–926. doi: 10.1038/nbt0888-923
McDonnell, R. E., and Conger, B. V. (1984). Callus induction and plantlet formation from mature embryo explants of Kentucky bluegrass. Crop Sci. 24, 573–578.
Merrick, P., and Fei, S. (2015). Plant regeneration and genetic transformation in switchgrass — A review. J. Integr. Agric. 14, 483–493. doi: 10.1016/S2095-3119(14)60921-7
Michell, R. H. (2008). Inositol derivatives: evolution and functions. Nat. Rev. Mol. Cell Biol. 9, 151–161. doi: 10.1038/nrm2334
Miles, J. W., Maass, B. L., Do Valle, C. B., and Kumble, V. (1996). “Centro Internacional de Agricultura Tropical; Empresa Brasileira de Pesquisa Agropecuária),” in Brachiaria: Biology, Agronomy, and Improvement, eds J. W. Miles, B. L. Maass, C. B. Do Valle, and V. Kumble (Cali: Centro Nacional de Pesquisa de Gado de Corte (CNPGC)).
Montanari, R. E. A. (2013). Correlación de la productividad de un forraje con las propiedades físicas de un Ultisol en Aquidauana. Revista Ceres [online] 60, 102–110. doi: 10.1590/S0034-737X2013000100015
Mozgova, G. V., Zaitseva, O. I., and Lemesh, V. A. (2012). Structural changes in chloroplast genome accompanying albinism in anther culture of wheat and triticale. Cereal Res. Commun. 40, 467–475.
Mu, G., Chang, N., Xiang, K., Sheng, Y., Zhang, Z., and Pan, G. (2012). Genetic transformation of maize female inflorescence following floral dip method mediated by agrobacterium. Biotechnology(Faisalabad) 11, 178–183. doi: 10.3923/biotech.2012.178.183
Munnik, T., and Nielsen, E. (2011). Green light for polyphosphoinositide signals in plants. Curr. Opin. Plant Biol. 14, 489–497. doi: 10.1016/j.pbi.2011.06.007
Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plantarum 15, 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Ncanana, S., Brandt, W., Lindsey, G., and Farrant, J. (2005). Development of plant regeneration and transformation protocols for the desiccation-sensitive weeping lovegrass Eragrostiscurvula. Plant Cell Rep. 24, 335–340. doi: 10.1007/s00299-005-0940-1
Numan, M., Khan, A. L., Asaf, S., Salehin, M., Beyene, G., Tadele, Z., et al. (2021). From traditional breeding to genome editing for boosting productivity of the ancient grain tef [Eragrostis tef (Zucc.) Trotter]. Plants 10:628. doi: 10.3390/plants10040628
O’Kennedy, M. M., Burger, J. T., and Botha, F. C. (2004). Pearl millet transformation system using the positive selectable marker gene phosphomannose isomerase. Plant Cell Rep. 22, 684–690. doi: 10.1007/s00299-003-0746-y
Ondzighi-Assoume, C. A., Willis, J. D., Ouma, W. K., Allen, S. M., King, Z., Parrott, W. A., et al. (2019). Embryogenic cell suspensions for high-capacity genetic transformation and regeneration of switchgrass (Panicum virgatum L.). Biotechnol. Biofuels 12:290. doi: 10.1186/s13068-019-1632-3
Ozias-Akins, P., Roche, D., and Hanna, W. W. (1998). Tight clustering and hemizygosity of apomixis-linked molecular markers in Pennisetum squamulatum implies genetic control of apospory by a divergent locus that may have no allelic form in sexual genotypes. Proc. Natl. Acad. Sci. U.S.A. 95, 5127–5132. doi: 10.1073/pnas.95.9.5127
Ozyigit, I. I., and YucebilgiliKurtoglu, K. (2020). Particle bombardment technology and its applications in plants. Mol. Biol. Rep. 47, 9831–9847. doi: 10.1007/s11033-020-06001-5
Pinheiro, A. A., Pozzobon, M. T., do Valle, C. B., Penteado, M. I. O., and Carneiro, V. T. C. (2000). Duplication of the chromosome number of diploid Brachiariabrizantha plants using colchicine. Plant Cell Rep. 19, 274–278. doi: 10.1007/s002990050011
Quero Carrillo, A. R., Enríquez Quiroz, J. F., Morales Nieto, C. R., and Miranda Jiménez, L. (2010). Apomixis y su importancia en la selección y mejoramiento de gramineas forrajeras tropicales. Revisión. Rev. Mex. Ciencias Pecuarias 1, 25–42.
Rao, A. S., Singh, K. C., and Wight, J. R. (1996). Productivity of Cenchrus ciliaris in relation to rain-fall and fertilization. J. Range Manag. 49, 143–146.
Ratanasut, K., Rod-In, W., and Sujipuli, K. (2017). In planta agrobacterium-mediated transformation of rice. Rice Sci. 24, 181–186. doi: 10.1016/j.rsci.2016.11.001
Ratcliff, F., Harrison, B. D., and Baulcombe, D. C. (1997). A similarity between viral defense and gene silencing in plants. Science 276, 1558–1560. doi: 10.1126/science.276.5318.1558
Rathore, D. S., Zuniga-Soto, E., and Mullins, E. (2019). Ensifer-Mediated Transformation (EMT) of rice (Monocot) and oilseed rape (Dicot). Methods Mol. Biol. 1864, 37–48. doi: 10.1007/978-1-4939-8778-8_3
Ravi, M., and Chan, S. W. L. (2010). Haploid plants produced by centromeremediated genome elimination. Nature 464, 615–618. doi: 10.1038/nature0884
Ravi, M., Marimuthu, M. P. A., and Siddiqi, I. (2008). Gamete formation without meiosis in Arabidopsis. Nature 451, 1121–1124. doi: 10.1038/nature06557
Reed, J., Privalle, L., Powell, M. L., Meghji, M., Dawson, J., Dunder, E., et al. (2001). Phosphomannose isomerase: an efficient selectable marker for plant transformation. In Vitro Cell. Dev. Biol. - Plant 37, 127–132. doi: 10.1007/s11627-001-0024-z
Rudder, S., Doohan, F., Creevey, C. J., Wendt, T., and Mullins, E. (2014). Genome sequence of Ensiferadhaerens OV14 provides insights into its ability as a novel vector for the genetic transformation of plant genomes. BMC Genomics 15:268. doi: 10.1186/1471-2164-15-268
Saha, P., and Blumwald, E. (2016). Spike-dip transformation of Setariaviridis. Plant J. 86, 89–101. doi: 10.1111/tpj.13148
Sakamoto, A., and Murata, N. (2001). The use of bacterial choline oxidase, a glycinebetaine-synthesizing enzyme, to create stress-resistant transgenic plants. Plant Physiol. 125, 180–188. doi: 10.1104/pp.125.1.180
Sánchez-Romero, C. (2021). “Use of meta-topolin in somatic embryogenesis,” in Meta-topolin: A Growth Regulator for Plant Biotechnology and Agriculture, eds N. Ahmad and M. Strnad (Singapore: Springer Singapore), 187–202.
Sandhu, S., and Altpeter, F. (2008). Co-integration, co-expression and inheritance of unlinked minimal transgene expression cassettes in an apomictic turf and forage grass (Paspalum notatum Flugge). Plant Cell Rep. 27, 1755–1765. doi: 10.1007/s00299-008-0599-5
Sandhu, S., Altpeter, F., and Blount, A. R. (2007). Apomictic bahiagrass expressing the bar gene is highly resistant to glufosinate under field conditions. Crop Sci. 47, 1691–1697. doi: 10.2135/cropsci2006.11.0699
Sandhu, S., Blount, A. R., Quesenberry, K. H., and Altpeter, F. (2010). Apomixis and ploidy barrier suppress pollen-mediated gene flow in field grown transgenic turf and forage grass (Paspalum notatum Flüggé). Theor. Appl. Genet. 121, 919–929. doi: 10.1007/s00122-010-1360-3
Scheben, A., and Hojsgaard, D. (2020). Can we use gene-editing to induce apomixis in sexual plants? Genes 11:781. doi: 10.3390/genes11070781
Schenk, R., and Hildebrandt, A. C. (2011). Medium and techniques for induction and growth of monocotyledonous and dicotyledonous plant cell cultures. Canadian J. Bot. 50, 199–204. doi: 10.1139/b72-026
Schmidt, A. (2020). Controlling apomixis: shared features and distinct characteristics of gene regulation. Genes (Basel) 11:329. doi: 10.3390/genes11030329
Scofield, S. R., and Nelson, R. S. (2009). Resources for virus-induced gene silencing in the grasses. Plant Physiol. 149, 152–157. doi: 10.1104/pp.108.128702
Selva, J. P., Zappacosta, D., Carballo, J., Rodrigo, J. M., Bellido, A., Gallo, C. A., et al. (2020). Genes modulating the increase in sexuality in the facultative diplosporous grass eragrostiscurvula under water stress conditions. Genes 11:969. doi: 10.3390/genes11090969
Seo, M.-S., Takahara, M., and Takamizo, T. (2010). Optimization of culture conditions for plant regeneration of Panicum spp. through somatic embryogenesis. Grassland Sci. 56, 6–12. doi: 10.1111/j.1744-697X.2009.00166.x
Shashi (2013). Developmental Morphogenesis and in vitro Genetic Manipulation of Cenchrus ciliaris L, Vol. VI. New Delhi: University of Delhi, 234.
Shou, H., Frame, B. R., Whitham, S. A., and Wang, K. (2004). Assessment of transgenic maize events produced by particle bombardment or Agrobacterium-mediated transformation. Mol. Breed. 13, 201–208. doi: 10.1023/B:MOLB.0000018767.64586.53
Simioni, C., and Valle, C. (2009). Chromosome duplication in Brachiaria (A. Rich.) Stapf allows intraspecific crosses. Crop Breed. Appl. Biotechnol. 9, 328–334.
Singh, R. K., and Prasad, M. (2016). Advances in Agrobacterium tumefaciens-mediated genetic transformation of graminaceous crops. Protoplasma 253, 691–707. doi: 10.1007/s00709-015-0905-3
Srivatanakul, M., Park, S. H., Sanders, J. R., Salas, M. G., and Smith, R. H. (2000). Multiple shoot regeneration of kenaf (Hibiscus cannabinus L.) from a shoot apex culture system. Plant Cell Rep. 19, 1165–1170. doi: 10.1007/s002990000256
Stone, S. L., Kwong, L. W., Yee, K. M., Pelletier, J., Lepiniec, L., Fischer, R. L., et al. (2001). LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc. Natl. Acad. Sci. U.S.A. 98, 11806–11811. doi: 10.1073/pnas.201413498
Takamori, L. M., Neto, N. B. M., Vieira, L., and Ribas, A. (2015). Optimization of somatic embryogenesis and in vitro plant regeneration of Urochloa species using picloram. In Vitro Cell. Dev. Biol. - Plant 51, 554–563.
Terenti Romero, C. M. (2015). Obtención y Evaluación de Nuevos Materiales de Pasto Llorón (Eragrostiscurvula - Schrad. Nees) y Desarrollo de Protocolos Biotecnológicos Para su Utilización en Programas de Mejoramiento de la Especie. Doctor en Agronomía thesis. Bahía Blanca: Universidad Nacional del Sur.
Tingay, S., McElroy, D., Kalla, R., Fieg, S., Wang, M., Thornton, S., et al. (1997). Agrobacterium tumefaciens-mediated barley transformation. Plant J. 11, 1369–1376. doi: 10.1046/j.1365-313X.1997.11061369.x
Torres, M. A., Jones, J. D., and Dangl, J. L. (2006). Reactive oxygen species signaling in response to pathogens. Plant Physiol. 141, 373–378. doi: 10.1104/pp.106.079467
Umami, N., Gondo, T., Ishigaki, G., Rahman, M. M., and Akashi, R. (2012). Efficient nursery production and multiple-shoot clumps formation from shoot tiller-derived shoot apices of dwarf napiergrass (Pennisetum purpureum Schumach). J. Warm Regional Soc. Anim. Sci. 55, 121–127.
Valle, C. B. D., and Savidan, Y. H. (1996). “Genetics, cytogenetics, and reproductive biology of Brachiaria,” in Brachiaria: Biology, Agronomy, and Improvement, eds J. W. M. Miles, L. Brigitte, C. B. Valle, and V. Kumble (Campo Grande, BR: Centro Internacional de Agricultura Tropical (CIAT)).
Van Wijk, A. J. P. (1997). “Breeding amenity grasses,” in Ecological Aspects of Breeding Fodder Crops and Amenity Grasses, eds Z. Staszewski, W. Mlyniec, and R. Osinski (Radzikow: Plant Breeding and Acclimatization Institute).
VanBuren, R., Man Wai, C., Wang, X., Pardo, J., Yocca, A. E., Wang, H., et al. (2020). Exceptional subgenome stability and functional divergence in the allotetraploid Ethiopian cereal teff. Nat. Commun. 11:884. doi: 10.1038/s41467-020-14724-z
Voigt, P. W., and Bashaw, E. C. (1972). Apomixis and sexuality in eragrostis curvula1. Crop Sci. 12:crosci1972.0011183X001200060039x. doi: 10.2135/cropsci1972.0011183X001200060039x
Wang, C., Liu, Q., Shen, Y., Hua, Y., Wang, J., Lin, J., et al. (2019). Clonal seeds from hybrid rice by simultaneous genome engineering of meiosis and fertilization genes. Nat. Biotechnol. 37, 283–286. doi: 10.1038/s41587-018-0003-0
Wang, M., Gao, S., Zeng, W., Yang, Y., Ma, J., and Wang, Y. (2020). Plant virology delivers diverse toolsets for biotechnology. Viruses 12:1338. doi: 10.3390/v12111338
Wang, T., Birsoy, K., Hughes, N. W., Krupczak, K. M., Post, Y., Wei, J. J., et al. (2015). Identification and characterization of essential genes in the human genome. Science 350, 1096–1101.
Wang, Z.-Y., and Brummer, E. C. (2012). Is genetic engineering ever going to take off in forage, turf and bioenergy crop breeding? Ann. Bot. 110, 1317–1325. doi: 10.1093/aob/mcs027
Weijde, T., Alvim Kamei, C., Torres, A., Vermerris, W., Dolstra, O., Visser, R., et al. (2013). The potential of C4 grasses for cellulosic biofuel production. Front. Plant Sci. 4:107. doi: 10.3389/fpls.2013.00107
Wright, M., Dawson, J., Dunder, E., Suttie, J., Reed, J., Kramer, C., et al. (2001). Efficient biolistic transformation of maize (Zea mays L.) and wheat (Triticum aestivum L.) using the phosphomannose isomerase gene, pmi, as the selectable marker. Plant Cell Rep. 20, 429–436. doi: 10.1007/s002990100318
Xiong, X., James, V. A., Zhang, H., and Altpeter, F. (2010). Constitutive expression of the barley HvWRKY38 transcription factor enhances drought tolerance in turf and forage grass (Paspalum notatum Flugge). Mol. Breed. 25, 419–432.
Yadav, C. B., Jha, P., Mahalakshmi, C., Anjaiah, V., and Bhat, V. (2009). Somatic embryogenesis and regeneration of Cenchrus ciliaris genotypes from immature inflorescence explants. Biol. Plantarum 53, 603–609. doi: 10.1007/s10535-009-0111-2
Yaguinuma, D. H., Takamori, L. M., de Oliveira, A. M., Vieira, L. G. E., and Ribas, A. F. (2018). In vitro regeneration from leaf-base segments in three genotypes of Urochloa spp. Crop Pasture Sci. 69, 527–534.
Yang, L., Cui, G., Wang, Y., Hao, Y., Du, J., Zhang, H., et al. (2017). Expression of foreign genes demonstrates the effectiveness of pollen-mediated transformation in zea mays. Front. Plant Sci. 8:383. doi: 10.3389/fpls.2017.00383
Yookongkaew, N., Srivatanakul, M., and Narangajavana, J. (2007). Development of genotype-independent regeneration system for transformation of rice (Oryza sativa ssp. indica). J. Plant Res. 120, 237–245. doi: 10.1007/s10265-006-0046-z
York, J. D. (2006). Regulation of nuclear processes by inositol polyphosphates. Biochim. Biophys. Acta 1761, 552–559. doi: 10.1016/j.bbalip.2006.04.014
Zale, J. M., Agarwal, S., Loar, S., and Steber, C. M. (2009). Evidence for stable transformation of wheat by floral dip in Agrobacterium tumefaciens. Plant Cell Rep. 28, 903–913. doi: 10.1007/s00299-009-0696-0
Zapata, C., Park, S. H., El-Zik, K. M., and Smith, R. H. (1999). Transformation of a Texas cotton cultivar by using Agrobacterium and the shoot apex. Theoretical Appl. Genet. 98, 252–256. doi: 10.1007/s001220051065
Zappacosta, D., Gallardo, J., Carballo, J., Meier, M., Rodrigo, J. M., Gallo, C. A., et al. (2019). A high-density linkage map of the forage grass eragrostiscurvula and localization of the diplospory locus. Front. Plant Sci. 10:918. doi: 10.3389/fpls.2019.00918
Zhang, K., Wang, J., Hu, X., Yang, A., and Zhang, J. (2010). Agrobacterium-mediated transformation of shoot apices of Kentucky bluegrass (Poa pratensis L.) and production of transgenic plants carrying a betA gene. Plant Cell Tissue Organ. Culture (PCTOC) 102, 135–143. doi: 10.1007/s11240-010-9713-9
Zhang, W. J., Dewey, R. E., Boss, W., Phillippy, B. Q., and Qu, R. (2013). Enhanced Agrobacterium-mediated transformation efficiencies in monocot cells is associated with attenuated defense responses. Plant Mol. Biol. 81, 273–286. doi: 10.1007/s11103-012-9997-8
Zhao, X., Meng, Z., Wang, Y., Chen, W., Sun, C., Cui, B., et al. (2017). Pollen magnetofection for genetic modification with magnetic nanoparticles as gene carriers. Nat. Plants 3, 956–964. doi: 10.1038/s41477-017-0063-z
Zhi, L., TeRonde, S., Meyer, S., Arling, M. L., Register, J. C. 3rd, Zhao, Z. Y., et al. (2015). Effect of Agrobacterium strain and plasmid copy number on transformation frequency, event quality and usable event quality in an elite maize cultivar. Plant Cell Rep. 34, 745–754. doi: 10.1007/s00299-014-1734-0
Zhong, H., Sun, B., Warkentin, D., Zhang, S., Wu, R., Wu, T., et al. (1996). The competence of maize shoot meristems for integrative transformation and inherited expression of transgenes. Plant Physiol. 110, 1097–1107. doi: 10.1104/pp.110.4.1097
Zhu, H., Li, C., and Gao, C. (2020). Applications of CRISPR–Cas in agriculture and plant biotechnology. Nat. Rev. Mol. Cell Biol. 21, 661–677. doi: 10.1038/s41580-020-00288-9
Zhu, Y. J., Agbayani, R., and Moore, P. H. (2004). Green fluorescent protein as a visual selection marker for papaya (Carica papaya L.) transformation. Plant Cell Rep. 22, 660–667. doi: 10.1007/s00299-004-0755-5
Zipfel, C., Kunze, G., Chinchilla, D., Caniard, A., Jones, J. D., Boller, T., et al. (2006). Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125, 749–760. doi: 10.1016/j.cell.2006.03.037
Zuloaga, F., and Soderstrom, T. (1985). Classification of the Outlying Species of New World Panicum (Poaceae: Paniceae). Washington, DC: Smithsonian Institution Press.
Keywords: genetic transformation, apomictic grasses, plant regeneration, DNA-delivery methods, editing, morphogenic regulators
Citation: Bellido AM, Souza Canadá ED, Permingeat HR and Echenique V (2021) Genetic Transformation of Apomictic Grasses: Progress and Constraints. Front. Plant Sci. 12:768393. doi: 10.3389/fpls.2021.768393
Received: 31 August 2021; Accepted: 05 October 2021;
Published: 05 November 2021.
Edited by:
Horacio Esteban Hopp, University of Buenos Aires, ArgentinaReviewed by:
Marco Dalla Rizza, National Institute for Agricultural Research (INIA), UruguayCopyright © 2021 Bellido, Souza Canadá, Permingeat and Echenique. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Viviana Echenique, ZWNoZW5pcUBjcmliYS5lZHUuYXI=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.