
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 25 November 2021
Sec. Technical Advances in Plant Science
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.767522
This article is part of the Research Topic Plant Transformation View all 20 articles
Grapevine, as other woody perennials, has been considered a recalcitrant crop to produce transgenic plants. Since the production of transgenic and/or edited plants requires the ability to regenerate plants from transformed tissues, this step is often the biggest bottleneck in the process. The objective of this work is to review the state of the art technologies and strategies for the improvement of grapevine transformation and regeneration, focusing on three aspects: (i) problems associated with grapevine transformation; (ii) genes that promote grapevine regeneration; and (iii) vehicles for gene delivery. Concerning the first aspect, it is well documented that one of the main factors explaining the low success rate in obtaining transgenic plants is the regeneration process. After transgenic integration into receptor cells, tissue culture is required to regenerate transgenic seedlings from transformed cells. This process is time consuming and often requires the addition of environmentally damaging reagents (antibiotics and herbicides) to the culture medium to select transgenic plants. On the other hand, the expression of genes such as the so-called developmental regulators (DR), which induce specific development programs, can be used to avoid traditional tissue culture methods. The ectopic expression of specific combinations of DR in somatic cells has the potential to induce de novo meristems in diverse crops, including grapevine. Successful genome editing by de novo reprogramming of plant meristems in somatic tissues has been reported. Moreover, it has been shown that the expression of certain transcription factors can increase the regeneration efficiency in wheat, citrus, and rice. Finally, recent reports showed the use of nanoparticles, such as carbon dots (CDs), as an attractive alternative to Agrobacterium- and biolistic-mediated plant genetic transformation. In this way, the use of antibiotics in culture media is avoided, overcoming the loss of viability of plant tissues and accelerating the regeneration processes. It has been shown that CDs can act as a vehicle to transport plasmids to plant cells in transient transformation in several crops without negative impacts on photosynthesis or growth. Based on these advances, it is possible to combine these new available strategies and technologies to overcome the regeneration problems of species such as grapevine and other crops considered as recalcitrant.
Grapevine is one of the most widespread fruit crops in the world, with a production of about 77.1 million tons (FAOSTAT, 2019). It is cultivated both for the manufacture of wine and for its consumption as fresh fruit, and to a lesser extent to produce raisins, juices, and spirit drinks, the first use being the one for which it has more hectares allocated. Actual and future environmental conditions impose the decisions and choices regarding the management of the crop, following a path toward more sustainable alternatives. The problems related to climate change and the spread of numerous diseases require addressing solutions that can be found in the natural genetic variation of the genus Vitis (Vezzulli et al., 2019). Although grapevine can be improved through conventional breeding, it is a difficult and time-consuming process, due to the 2–3 years generation cycle and the long period of time required for the selection and testing of reliable progeny. Also, for grapevine, whose varieties are highly heterozygous, this way is even more difficult (Gray and Meredith, 1992).
Several years ago, genetic engineering emerged as an outstanding tool for the improvement of plants (Anderson et al., 2019). Genetic transformation offers the possibility of genetically modifying plants to improve agronomic traits of interest without altering the varietal identity using recombinant DNA technology, such as the transfer of resistance to diseases or herbicides to established crops. Grapevine was considered recalcitrant to genetic transformation, since one of its problems is the regeneration of plants from the tissues used for genetic transformation (Mullins et al., 1990; Nakano et al., 1994). The regeneration rate of grapevine plants after transformation and selection by antibiotics ranges between 10 and 30% of the total transformed material, and the transformation efficiency, which varies according to the genotype, down to 1%, although a 33% has been described (Torregrosa et al., 2015).
Accordingly, overcoming the problems related to tissue regeneration is one of the most essential challenges in the generation of transgenic and edited grapevine plants. In this sense, several factors have been identified that should be reviewed: the grapevine genotype; the type of tissue used to obtain the explants; the transformation methodology; the availability of regenerative transformable material, and the selection process/procedure based on antibiotics. Grapevine genetic transformation was mainly performed by infection with Agrobacterium tumefaciens (Torregrosa et al., 2015), and to a lesser extent through the biolistic techniques (Kikkert et al., 2001; Vidal et al., 2010).
The first critical factor in grapevine transformation is the production of highly regenerative transformable material, where the regeneration efficiency greatly depends on the different genotypes (Gray and Meredith, 1992). Somatic embryogenesis is the preferred regeneration procedure for the genetic transformation of grapevine. The source of starting material, the type, and the quality of the embryogenic cultures are key factors for a successful transformation (Martinelli and Gribaudo, 2001). However, despite these limitations, it has been possible to produce grapevine varieties resistant to fungal, viral, and bacterial diseases (Mauro et al., 1995; Scorza et al., 1996; Agüero et al., 2006; Nirala et al., 2010; Capriotti et al., 2020).
Beyond grapevine, the production of transgenic and edited plants requires, for most crops, the ability to regenerate plants from transformed tissues. This step is another critical factor, often reported as the biggest bottleneck in the process (Altpeter et al., 2016). After transgenic integration into recipient cells, tissue culture protocols are required to regenerate transgenic seedlings from transformed cells. This process is time consuming, usually several months, and generally requires the addition of expensive and environmentally harmful reagents (antibiotics and herbicides) to the growing medium to select the putative transgenic plants. The overexpression of certain transcription factors has been recently reported (Debernardi et al., 2020; Maher et al., 2020) as an interesting alternative to improve the transformation and regeneration processes of plants (including grapevine).
In recent years, new breeding technologies (NBT), such as gene editing via CRISPR-Cas9, have emerged as innovative genetic improvement tools for various crops of agronomic importance (Dalla Costa et al., 2017, 2019). The key elements in this system are Cas nucleases and CRISPR RNAs. The Cas9 endonuclease can cut at specific DNA target sites with the help of two small RNA molecules called CRISPR RNA (crRNA) and “trans-encoded” CRISPR RNA (tracrRNA). These two molecules can be fused artificially to form a chimeric RNA molecule called “single guide RNA” (sgRNA) (Mali et al., 2013; Qi et al., 2013). In conjunction with Cas9, sgRNA can form an “RNA-guided endonuclease,” a high precision tool capable of strategically introducing targeted mutations in the host genome (Jinek et al., 2012; Samanta et al., 2016).
On the other hand, alternative technologies are also emerging for the delivery of engineered genes into the plant cells. Carbon dots (CDs) were described as almost spherical water-soluble nanoparticles (NP) consisting of crystalline carbon domains synthesized from cheap starting materials such as peptides, carbohydrates, and, in general, a wide range of carbon sources (Li et al., 2012; Zheng et al., 2015; Peng et al., 2017). They have multiple advantages, such as being easy to obtain and for displaying an efficient plant cell uptake. It has been shown that CDs can act as a vehicle to transport plasmids to plant cells in transient transformation in several important crop species without negative impacts on photosynthesis or growth (Doyle et al., 2019; Schwartz et al., 2020).
Along with a thorough report of commonly known limitations related to grapevine transformation and regeneration, we primarily present in this review a detailed description of new alternative technologies that could provide solutions to overcome these drawbacks.
Since the genotype is one of the most influential factors in the success of a transformation protocol, the effects of the genetic background on the efficiency of plant regeneration and the corresponding culture conditions have been extensively studied (Dalla Costa et al., 2019). Thus, it has been reported that each genotype shows specific sensitivity to the infection with Agrobacterium, as well as to the antibiotics used to eliminate the bacteria, and/or to those used to select transgenic events (Zhou et al., 2014). The grapevine genetic background also influences somatic embryogenesis, the regeneration method most used in genetic engineering protocols in this crop. Several procedures have been developed for somatic embryogenesis from grapevine genotypes, and much research has been carried out using several plant tissues/organs as starting explants. The list of the plant parts widely used as suitable material for obtaining somatic embryos includes: ovaries (Yamamoto et al., 2000; Vidal et al., 2003, 2006; Gambino et al., 2005; Kikkert et al., 2005; López-Pérez et al., 2005); anthers (Franks et al., 1998; Gambino et al., 2005; Agüero et al., 2006; Prado et al., 2010); leaves (Martinelli et al., 1993; Nakano et al., 1994; Das et al., 2002); and, less frequently, stigmas and styles (Morgana et al., 2004; Carimi et al., 2005); stamen filaments (Nakajima and Matsuta, 2003; Acanda et al., 2013); nodal sections (Maillot et al., 2006, 2016); whole flowers (Gambino et al., 2007); mature seeds (Peiró et al., 2015); and tissues derived from vegetative structures, such as leaves and petioles (Martinelli et al., 1993; Das et al., 2002). The fact that most of the frequently used protocols are carried out using floral organs is indeed a strong limitation for obtaining somatic embryos, since the experiments can just be started at the flowering time. During the regeneration process, the germination of aberrant embryos and early germination can occur, which can also limit the obtaining of transformed grapevine plants since these embryos do not develop into normal plants. Among the most common aberrations, it is possible to find embryos without cotyledons, with different numbers of cotyledons, or with fused cotyledons, and trumpet-shaped or cauliflower-like cotyledons (Goebel-Tourand et al., 1993; Bornhoff et al., 2005; Li et al., 2006; Bharathy and Agrawal, 2008; Martinelli and Gribaudo, 2009; Peiró et al., 2015).
It has been proven that the synthesis and accumulation of reserve proteins during zygotic embryogenesis is regulated by abscisic acid and/or water stress (Dodeman et al., 1997). Therefore, to achieve a correct maturation of somatic embryos, two methodologies have been used: the exogenous application of abscisic acid (ABA) (Morris et al., 1990; Goebel-Tourand et al., 1993; Sholi et al., 2009) and the use of culture media with reduced water potential (Klimaszewska et al., 2000; Walker and Parrott, 2001; Kong et al., 2009; Buendía-González et al., 2012). Another strategy used has been the modulation of polyamine metabolism (Faure et al., 1991). The use of semipermeable cellulose acetate membranes has emerged as an effective alternative to improve the maturation of somatic embryos, and this is due to its ability to limit the availability of water for the embryo. In grapevines, it has been shown that the water stress produced by the membrane triggered an increase in endogenous levels of ABA, a fact that improved the maturation of somatic embryos (Acanda, 2015).
The choice of the Agrobacterium strain shows a significant effect on a successful transformation procedure of plant tissues. This factor also includes the corresponding bacterial culture conditions like density at the time of infection, coculture times, and the culture media used (Le Gall et al., 1994; Franks et al., 1998). One of the first Agrobacterium strains used for grapevine transformation was LBA4404 (Bouquet et al., 2008), showing low transformation efficiencies. To improve those transformation rates, hypervirulent Agrobacterium strains were later developed. Among them, EHA105 is currently the strain most used in grapevine transformation (Scorza et al., 1996; Franks et al., 1998; Iocco et al., 2001; Wang et al., 2005; Dutt et al., 2008; Dhekney et al., 2011; Dabauza et al., 2014; Li et al., 2015). AGL1, a hypervirulent Agrobacterium strain (Lazo et al., 1991) has also been used, verifying that agroinfiltration with this strain transformed all the tested genotypes (Urso et al., 2013).
Grapevine tissue necrosis is a regular problem that occurs during or after Agrobacterium transformation. This browning is triggered as a response to the bacteria and it was reported to be cultivar-specific (Bouquet et al., 2008). Polyphenols can be oxidized by air, peroxidases, or polyphenoloxidases. Peroxidases and polyphenoloxidases have been associated with mechanical injury and response to environmental stress. The stress response may involve the release of polyphenols from vacuoles and the de novo synthesis of phenol (Perl et al., 1996). These authors assume that the hypersensitive response could be due to the oxidation caused by high levels of peroxidase activity. Perl et al. (1996) improved plant viability and inhibited tissue necrosis in cv. “Superior Seedless” by using a combination of polyvinylpolypyrrolidone and dithiothreitol. A still unexplored alternative that could reduce this browning is the use of Agrobacterium vitis strains (A. tumefaciens biovar 3), which is a natural grapevine pathogen (Kikkert et al., 2001).
The differentiation of the positive transformation events among all the non-transformed plant tissues after infection with the Agrobacterium strain is a crucial step of the procedure. This is possible due to the action of marker genes, which are usually integrated together with the genes engineered to be expressed in the plant. There are two main types of marker genes: selection marker genes and reporter genes.
The use of marker genes allows the cells or plants carrying them to be selected in the presence of a selective agent, such as an herbicide or an antibiotic. The neomycin phosphotransferase (nptII) gene has been the most widely used in grapevine. This gene confers resistance to aminoglycoside antibiotics, such as kanamycin (Kan), paramomycin, or neomycin (Nakano et al., 1994; Scorza et al., 1995, 1996; Yamamoto et al., 2000; Iocco et al., 2001; Vidal et al., 2003, 2006; Bornhoff et al., 2005; Gambino et al., 2005; Wang et al., 2005; Agüero et al., 2006; Li et al., 2006, 2015; Dhekney et al., 2008, 2011; López-Pérez et al., 2008; Jin et al., 2009; Gago et al., 2011; Dabauza et al., 2014). Although kanamycin is the most widely selective agent used in grapevine transformation, this crop shows a high sensitivity to this antibiotic, and it is generally difficult to find a balance between the appropriate concentration for selection, without losing the viability of the embryos and plants. Another gene frequently used in grapevine transformation is hygromycin phosphotransferase (hptI), which confers resistance to the antibiotic hygromycin (Hyg) (Franks et al., 1998; Torregrosa et al., 2000; Fan et al., 2008; Nirala et al., 2010; Nookaraju and Agrawal, 2012; Dai et al., 2015). Different strategies have been assayed regarding the application timing and the optimal concentration of kanamycin (Franks et al., 1998; Iocco et al., 2001; Wang et al., 2005) and hygromycin (Franks et al., 1998; Fan et al., 2008; Nirala et al., 2010; Dai et al., 2015). Moreover, Saporta et al. (2014) made a comparison of Kan and Hyg in “Albariño” cell suspensions. Since grapevine antibiotic sensitivity showed a high genotype-specific behavior (Gray and Meredith, 1992; Torregrosa et al., 2000), sensitivity-specific assays are required for every new genetic transformation platform.
On the other hand, reporter genes give transformed plants an easily recognizable and measurable selection characteristic or phenotype. Once these genes are integrated, they allow us to know where they are expressed, in what quantity, when, and in which tissues they are transcribed. The most widely used reporter gene is the uidA gene that encodes β-glucuronidase (GUS) (Jefferson et al., 1987). This protein hydrolyzes substrates such as X-Gluc (5-bromo-4-chloro-3-indoxyl-beta-D-glucuronide) and causes a blue precipitate. The disadvantage of this method is that it is usually destructive. Alternatively, the green fluorescent protein gene (GFP), which encodes a protein that generates a chromophore emitting green fluorescence when excited by blue light or ultraviolet light, is commonly utilized as a non-destructive reporter system (Chiu et al., 1996). Both genes have been extensively used in grapevine genetic transformation (Baribault et al., 1989; Nakano et al., 1994; Scorza et al., 1995; Franks et al., 1998; Iocco et al., 2001; Das et al., 2002; Wang et al., 2005; Dutt et al., 2007, 2008; López-Pérez et al., 2008; Gago et al., 2011). More recently, He et al. (2020) reported RUBY as a non-invasive reporter that could be especially useful for monitoring gene expression in tissue culture experiments under sterile conditions in large crop plants such as fruit trees.
Alternatively to the plant transformation mediated by A. tumefaciens, several works reported the use of biolistic methodologies (Torregrosa et al., 2002; Vidal et al., 2003, 2006; Joubert et al., 2013). Biolistic is a physical transformation method, which consists of the high-speed projection of microparticles, usually gold and tungsten, impregnated with DNA (Sanford et al., 1987). The first grapevine work using biolistics was reported by Hébert et al. (1993), in which they transformed “Chancellor,” a Vitis complex interspecific hybrid. On the other hand, Scorza et al. (1995) carried out the first transformation of seedless table grapes using this technique. Nowadays, this technique is a useful transient expression tool for functional analysis in various plant materials, such as, cell suspension culture, leaf sections, and somatic embryos (Vidal et al., 2010; Jelly et al., 2014; Dalla Costa et al., 2019). A biolistic protocol based on the transient genetic transformation of “Cabernet Sauvignon” cell suspensions was developed by Torregrosa et al. (2002) to analyze the effect of anaerobiosis on the regulation of the expression of the VvAdh gene in response to anaerobiosis. Finally, biolistics transformation was also used to study the regulation of the defense gene VvPGIP in leaves sections of “Chardonnay” and ‘Thompson Seedless” somatic embryos (Joubert et al., 2013). One of the most important advantage of biolistic techniques with respect to the infection with A. tumefaciens is probably to skip the treatment with antibiotics to eliminate the bacteria, and in the case of grapevine, to avoid the before mentioned hypersensitivity reaction caused by infection. As disadvantages, low penetration depth, random integration, and putative damage to target tissue were also reported (Cunningham et al., 2018). Particle bombardment is still quite difficult to perform and requires the fine tuning of a series of critical variables such as helium pressure, particle diameter, cartridge preparation, or distance from target plant material. Additionally, purchasing a biolistic device and consumables can be expensive (Jelly et al., 2014).
The technical and biological problems mentioned above, together with the strong rejection of the consumers and the regulation of the appellations of origin, have prevented the wide development of grapevine genetic transformation. Furthermore, due to the scarce natural genetic resistance/tolerance of V. vinifera genotypes, stable transformation has been mainly oriented to the improvement of resistance to pathogens and insects (Vivier and Pretorius, 2002; Table 1). Most of the reports on fungal resistance have focused on the use of pathogenesis related proteins (PR), among which, glucanases and chitinases stand out. On the other hand, the accumulation of phytoalexins and stilbenes has been a proven strategy used to obtain resistance to fungi (Fan et al., 2008; Dabauza et al., 2014; Dai et al., 2015). The improvement of plant resistance to bacteria has been targeted by using antimicrobial genes like lytic peptides (Scorza et al., 1996; Vidal et al., 2003, 2006; Dandekar et al., 2012; Li et al., 2015). The insertion of virus capsid proteins (virus coat proteins; CP) has been used to increase the resistance to viruses such as GFLV, GVA, or GVB (Gölles et al., 1997; Tsvetkov et al., 2000; Gambino et al., 2005). Finally, the introduction of resistance to insects like root-knot nematodes or the grapevine phylloxera have been attempted by means of hairy roots transformation (Franks et al., 2006; Yang et al., 2013). In addition, the improvement of tolerance to abiotic stress has also been studied. Some of the most troublesome stress problems that have been addressed are resistance to cold (Jin et al., 2009; Tillett et al., 2012) or different sources of oxidative damage (Zok et al., 2009).
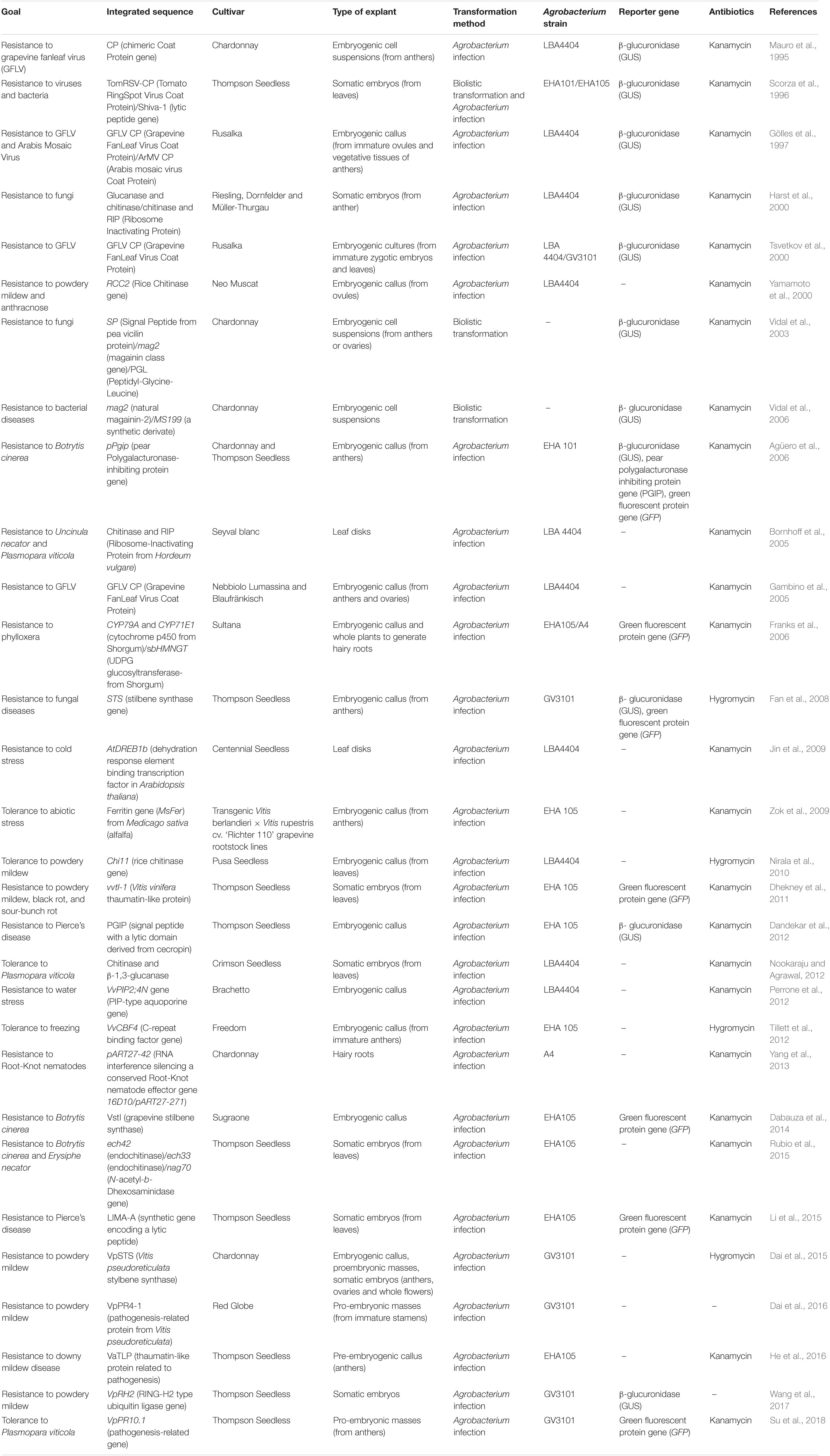
Table 1. Genetic transformation works focused on the incorporation of genes related to fungal, bacterial, viral resistance, abiotic stresses, and other pathogens in V. vinifera.
A compilation of most of the works carried out on stable transformation of V. vinifera, including the transformation methods, marker genes, and antibiotics used for selection are shown in Table 1. It is worth mentioning that most of the grapevine genetic transformation studies have used varieties such as “Thompson Seedless” or “Chardonnay,” highlighting the genotype-specificity of this procedure.
Plants are sessile organisms that are dependent on the living conditions of the environment around them, and for this reason plants have developed great plasticity to accommodate environmental effects by altering metabolism or development (Fehér et al., 2003). Pluripotency and totipotency, exceptional properties for tissue culture techniques, have contributed to several biotechnological applications. Pluripotency refers to the ability of one cell type to form another cell type, tissue, or organ, while totipotency refers to the ability of a single cell to develop, through embryogenesis, into a complete organism (Jha and Kumar, 2018). The totipotency theory was first proposed by Guttenberg (1943), but regeneration protocols were established after Skoog and Miller (1957) introduced changes in the concentration of auxins and cytokinin in the culture media. Since then, it has been possible to establish shoot regeneration for many plants (Lardon and Geelen, 2020).
The most common types of regeneration in plants are somatic embryogenesis and de novo organogenesis (Pulianmackal et al., 2014; Kareem et al., 2016). During somatic embryogenesis, dedifferentiated cells generate bipolar structures where it is possible to differentiate root and shoot meristems (Pulianmackal et al., 2014; Xu and Huang, 2014; Horstman et al., 2017; Méndez-Hernández et al., 2019). This process is achieved through abiotic stress induction or by the addition of auxins. Consequently, zygotic embryogenesis-like structures are formed, due to the action of transcription factors such as LEAFY COTYLEDON (LEC) 1 and 2, AGAMOUS- LIKE 15 (AGL15), FUSCA 3 (FUS3), BABYBOOM (BBM), and EMBRYOMAKER (EMK) (Horstman et al., 2017; Méndez-Hernández et al., 2019). On the other hand, de novo organogenesis consists of the formation of new meristems from pluripotent stem cells to build organs (Xu and Huang, 2014). This process is governed by the plant hormones auxin and cytokinin and a transcriptional cascade involving WUSCHEL-RELATED HOMEOBOX (WOX) 11 and 12, WOX5 and 7, and LATERAL ORGAN BOUNDARY DOMAIN (LBD) 16 and 29, and SHOOT MERISTEMLESS (STM) (Liu et al., 2014; Xu, 2018).
On the other hand, GROWTH-REGULATING FACTOR (GRF) genes were reported as plant-specific transcription factors involved in the establishment and maintenance of meristems and in the cellular proliferation of developing primary organs (Liebsch and Palatnik, 2020). GRF proteins interact with a transcription cofactor, GRF INTERACTION FACTOR (GIF), forming a functional transcriptional complex (Kim and Kende, 2004). GIFs can act as transcriptional coregulators enhancing the activity of GRFs. MicroRNA396 (miR396) is a conserved miRNA that recognizes a complementary sequence in GRF mRNA from seed plants. Finally, GRF expression is regulated by posttranscriptional repression by mir396 (Rodriguez et al., 2016). The combinatory action of the miR396-GRF/GIF system in regulating plant growth makes it a very valuable tool for improving crops of agronomic interest.
Grapevine, as many plant species present difficulties in transformation and regeneration. These varieties are said to be recalcitrant to being transformed and regenerated. One of the promising tools that helps reduce these difficulties is the use of genes involved in the control of plant growth and development, called developmental regulators or morphogenetic regulators. Increases in the efficiency of the transformation and regeneration of various plants using developmental regulators have been thoroughly reported (Srinivasan et al., 2007; Heidmann et al., 2011; Yang et al., 2014; Lutz et al., 2015; Lowe et al., 2016; Mookkan et al., 2017; Maher et al., 2020; Che et al., 2021).
The overexpression of the WUSCHEL (WUS) gene has been used in several models and species of crops with the aim of improving the efficiency of transformation (Lowe et al., 2016; Mookkan et al., 2017; Che et al., 2021). Lowe et al. (2016) reported in maize, a significant increase in the frequency of callus transformation with WUS2. Moreover, the combination of WUS2 and BBM led to the highest transformation frequency. Mookkan et al. (2017) reported that the coexpression of the BABY BOOM (BBM) and WUS2 maize transcription factors along with a desiccation inducible CRE/lox cleavage system allows the regeneration of inbred stable recalcitrant transgenic maize B73 and sorghum P898012 without a selectable chemical marker. An increase in the transformation frequency from 0 to 15% for the B73 genotypes and upto 6.2% for the P898012 genotypes was found without the use of selection agents. This selectable-marker-independent transformation may contribute to overcoming transformation barriers in recalcitrant species and facilitate studies using gene editing functions. More recently, Che et al. (2021) reported that the transformation of Wus2 is capable of increasing the efficiency in the regeneration of transgenic plants and the efficiency in genome editing through the CRISPR-Cas technology. In addition, the authors have developed advanced cleavage systems and transformation technology to generate high quality selectable-marker-free sorghum events and/or morphogenic genes. They conclude that Wus2-enabled genome editing may be applicable to other crops in plant transformation strategies. On the other hand, BBM was reported as a marker and an activator of a complex signaling network of different development pathways related to cell proliferation and growth (Passarinho et al., 2008). Overexpression of native and heterologous BBM genes has also been found to play a role in inducing cell proliferation and significantly improving transformation and regeneration efficiency in tobacco (Srinivasan et al., 2007), oil palm (Morcillo et al., 2007), Arabidopsis (Lutz et al., 2015), and dog rose (Yang et al., 2014).
Alternatively, to improve plant regeneration rates after gene transformation, Debernardi et al. (2020) demonstrated that the expression of a fusion protein that combines the wheat GROWTH REGULATORY FACTOR 4 (GRF4) and its cofactor INTERACTIVE FACTOR GRF 1 (GIF1) was capable of increasing regeneration when it is expressed in crops such as wheat, triticale, and rice (Table 2). The authors also evaluated the use of the GRF4–GIF1 system together with the CRISPR-Cas9 gene editing technology by designing a cassette that included the GRF4-GIF1 chimera, Cas9 and, a guide RNA (gRNA) directed to the wheat Q gene (also known as AP2L-A5). Debernardi et al. (2020) were able to recover transgenic events including seven fertile plants showing a higher number of florets per spikelet (characteristic of q-null plants). The efficiency of the GRF–GIF chimera was also tested in citrus transformation experiments by means of the generation of a citrus GRF-GIF chimera and a heterologous GRF–GIF grapevine chimera (Debernardi et al., 2020). The epicotyls transformed with both chimeras showed significant increase in the frequency of regeneration compared with those transformed with the empty vector control (Table 2).
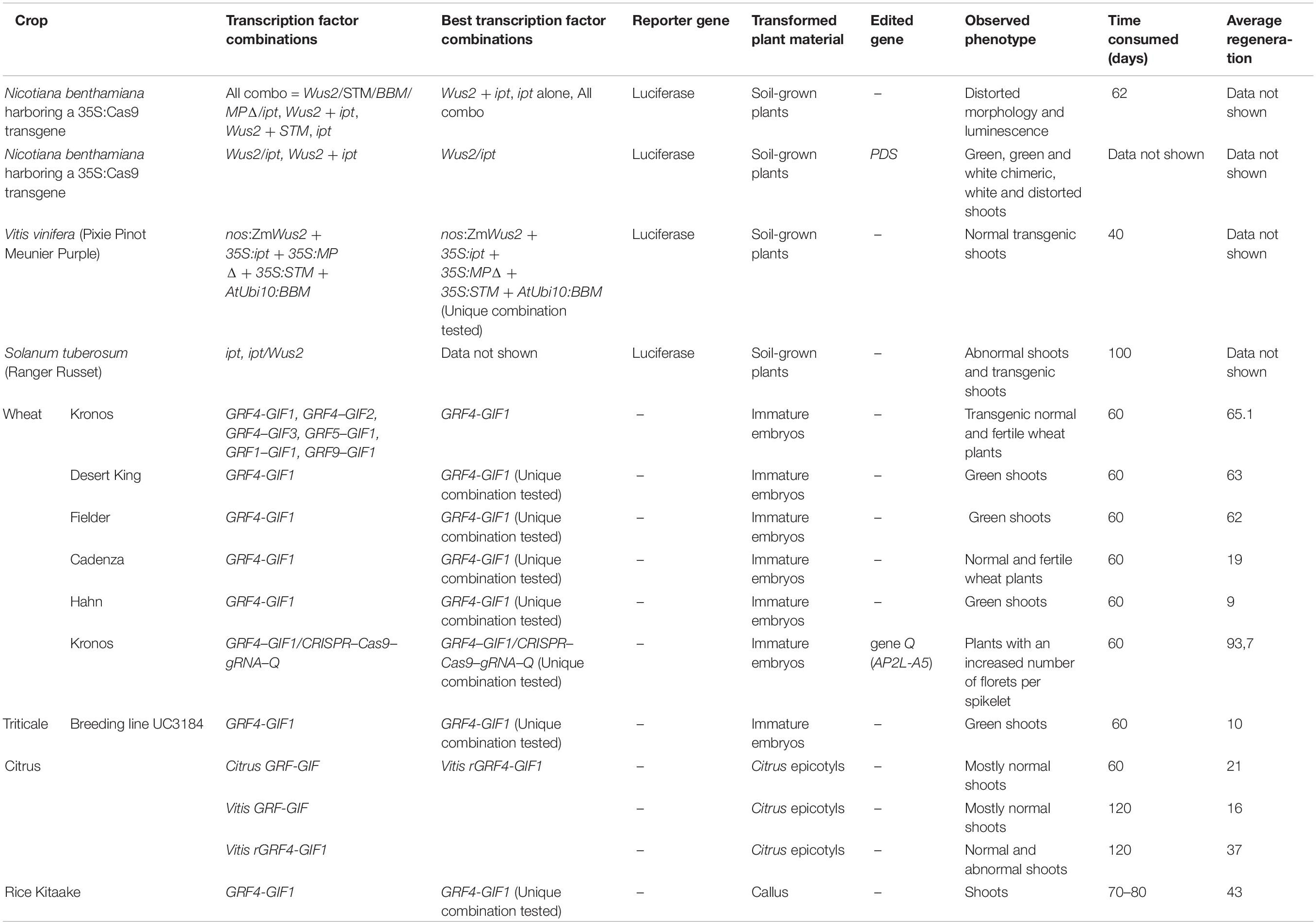
Table 2. Comparative summary of the different transcription factors used in different crops and model plants (Debernardi et al., 2020; Maher et al., 2020).
Finally, taking advantage of totipotency and pluripotency of plants, the ectopic expression of specific transcription factors called development regulators (DR) has the potential to induce meristems in somatic cells. Maher et al. (2020) presented the successful genome editing by de novo reprogramming of plant meristems in somatic tissues, which avoids tissue culture-based transformation and promises to significantly improve the utility of gene editing in plants. This innovative work proposes the induction of de novo meristems on soil-grown plants. N. benthamiana plants that constitutively expressed Cas9 were cultivated until the apical and axillary meristems were clearly differentiated, the point when they will be removed. DR combinations were delivered by A. tumefaciens at the breakpoints. Over time, de novo gene-edited shoots were formed, and editing events are passed to the next generation. Maher et al. (2020) also test the system in grapevine potato plants (Table 2).
To date, plant biotechnology lacks a method that allows passive delivery of diverse biomolecules without the aid of external force. As discussed previously, traditional methods present host-range limitations and typically target immature plant tissue (calli, meristems, or embryos), while efficient protocols have only been developed for a narrow range of plant species.
In this era, nanotechnology applications in agriculture have quickly emerged since they have little impact on environment. Due to the large surface area, tunable pore size, cargo and structure, and their tailored functionality, nanomaterials are widely used as nanoparticle-based fertilizers (Liu and Lal, 2015); antimicrobial components like silver and copper nanoparticles (Morones et al., 2005; Borkow and Gabbay, 2009; Sharon et al., 2010; Adeleye et al., 2016; Keller et al., 2017); and nanotechnology, which is being extensively applied in the genetic modification of plant DNA (Ziemienowicz et al., 2012). Nanotechnology has become a promising genetic cargo delivery toolset that is (i) plant-species independent (Su et al., 2019); and (ii) capable of high performance despite the physical barriers presented in intact plant tissues such as the plant cell wall (Etxeberria et al., 2006; Liu et al., 2009; Demirer et al., 2019a). The use of nanotechnology in gene modification enables easy operation, high efficiency (1,000 times less DNA is needed compared to conventional DNA modification techniques), versatility (nanoparticles are capable of simultaneously introducing proteins, nucleotides, and chemicals), target-specific delivery, and on-site release (Climent et al., 2009; Lin et al., 2015; Milewska-Hendel et al., 2017).
Compared with other metal-based nanomaterials, carbon-based nanomaterials show much lower environmental toxicity and higher biocompatibility due to their non-toxic carbon backbone (Chen et al., 2015; Bhattacharya et al., 2016; Mukherjee et al., 2016). Additionally, they have variety of sizes and shapes (including nanosheets, nanotubes, nanodots). Herein, carbon-based nanomaterials have become very versatile and sustainable materials, thus they have been widely applied in agriculture (Mukherjee et al., 2016; Shojaei et al., 2019; Verma et al., 2019). CDs mainly include graphene quantum dots (GQDs), carbon nanodots (CNDs), and polymer dots (PDs). These nanoparticles possess a size of less than 10 nm and have inherent photoluminescence (PL) and photostability properties, biocompatibility, abundant source, water solubility, highly tunable PL properties, easy functionalization with biomolecules, and chemical inertness (Li et al., 2012; Zheng et al., 2015; Peng et al., 2017). This kind of carbon material is much smaller than carbon nanosheets and nanotubes, allowing the CDs to pass much more easily through the biofilm of the cells of a broad range of plant phenotypes and species, including immature plant tissue and mature plants (model organisms, crop plants, and orphan crops -plants notoriously recalcitrant to transformation-) (Schwab et al., 2016; Tripathi et al., 2017). Moreover, it avoids the use of antibiotics in culture media, overcoming the loss of viability of plant tissues and accelerating the regeneration processes and has no effect on photosynthesis or growth of transformed crops and provokes no damage (Doyle et al., 2019; Li et al., 2020). Therefore, we present CD as a promising alternative that can still be used to improve the transformation and regeneration of the grapevine. Although not much work of this type has been carried out in grapevine, we consider this technique suitable to cope with more sustainable transformation protocols, which would lay the foundations for genetic improvement of grapevine in the era of food and nutrition security.
Carbon-based system for the delivery of cargo (RNA, DNA, protein, and plant protection substances) into the plant cell, is gaining relevance as it is easy, fast, and inexpensive to manufacture, requires little equipment to make, and can be adapted to a variety of application strategies to obtain genetically modified plants (Table 3). For instance, CD–plasmid nanocomplexes can act as a delivery vehicle by which plasmids can be carried into plant somatic cells, allowing transient expression. In the work of Doyle et al. (2019), plasmid-coated PEG functionalized CDs were sprayed on wheat, maize, barley, and sorghum leaves. CD-plasmid complexes containing GFP gene with a nuclear localization sequence (NLS) were successfully introduced and transiently expressed into the nucleus. The plasmid also carried the Cas9 gene and gRNA to make a ∼250 bp deletion in the wheat SPO11 genes. Importantly, spraying CD-plasmid nanocomplexes onto intact leaves can edit the genome. Similarly to Wang et al. (2020), PEI-modified CDs (CDP) with a positive charge, provides a highly efficient CD-based DNA delivery system for rapid and transient gene expression. Hygromycin resistance was achieved smearing plants leaves or soaking roots of rice with CD-plasmid complexes containing hydamycin resistance gene, whereas dipping and vacuum mature rice embryo with CD-plasmid complexes containing β-glucuronidase induced callus.
Nanosized carriers, like the oxidized multiwalled carbon nanotubes (MWCNTs) and single-walled carbon nanotubes (SWCNTs), were also studied to deliver DNA into mesophyll protoplasts, callus cells, and leaf explants. Thus, N. tabacum protoplasts were genetically transformed with the plasmid construct pGreen 0029, and a transient expression of the YFP reporter gene was shown in the protoplasts (Burlaka et al., 2015). While N. tabacum callus and leaf explants were genetically transformed by the nptII gene contained in the pGreen 0029 construct, regenerated plants were obtained on a selective medium. The investigation of Burlaka et al. (2015) demonstrated SWCNTs applicability for the transformation of protoplasts and walled plant cells. At the same time, MWCNTs demonstrated their applicability only for the transformation of protoplasts because of a limiting role of the cellulose wall against their penetration into the cells. Similarly, efficient GFP-encoding DNA plasmids or linear PCR amplicons and strong protein expression, without transgene integration, was achieved in N. benthamiana, arugula, wheat and cotton leaves, and arugula protoplasts. Demirer et al. (2019a) found that CNTs not only facilitate biomolecule transport into plant cells but also protect polynucleotides from nuclease degradation, without transgene integration. CNTs also have served as carrier of pDNA encoding yfp reporter gene for transient expression in the chloroplasts of mature arugula, watercress, tobacco and spinach plants and in isolated A. thaliana mesophyll protoplasts (Kwak et al., 2019).
The delivery of CD-based small interfering RNAs (siRNAs; double-stranded RNAs of 20–25 bp) into plant cells expands the spectrum of carbon-based NPs for molecule delivery into plant cells. RNA-induced gene silencing (also known as RNA interference) is a reliable method to study and alter the genetic form and function of plants. In Schwartz et al. (2020), PEI modified CDs (CDP) were used for delivering siRNA into the model plant N. benthamiana and tomato. Low-pressure spray application of these formulations with a spreading surfactant resulted in strong silencing of the reporter gene GFP transgenes in both species. The delivery efficacy of CD formulations was also demonstrated by the silencing of endogenous genes that encode two subunits of magnesium chelatase, an enzyme necessary for chlorophyll synthesis.
A breakthrough has been achieved in transient expression in plant somatic and embryogenic cells and protoplasts using carbon-based NPs as delivery method. This approach is very promising as it could be used to express genes and silence or increase gene expression, without gene integration, which would be particularly useful for plant developmental research. Moreover, it gives hope to the limitations of host restrictions. This kick off leads to more crops being assayed every day to test this new form of genetic cargo delivery. Particularly, for grapevine, only basic research has been carried out using NPs. However, it already stands out with interesting and eligible properties. For instance, in the work of Valletta et al. (2014), poly(lactic-co-glycolic) acid (PLGA) based CDs were demonstrated to cross the plant cell wall and membrane of V. vinifera cell cultures and grapevine-pathogenic fungi. By means of fluorescence microscopy, PLGA CDs can enter into grapevine leaf tissues through stomata openings so that they can be absorbed by the roots and transported to the shoot through vascular tissues. Viability tests demonstrated that PLGA CDs were not cytotoxic for V. vinifera-cultured cells. The cellular uptake of PLGA NPs by some important grapevine-pathogenic fungi shows promising potential for future use in agricultural applications, offering the possibility to deliver chemicals to specific targets in a controlled manner.
In the future, refinement and optimization experiments will surely lead to stable edited gene lines targeting plant germ line cells. Nevertheless, for all types of genetic modification, a comparable high efficacy with less established plant species needs to be shown.
Up to now, most of the work carried out in grapevine genetic transformation has had relative success due to the problems related to the transformation processes and the difficulty in plant regeneration. This process is generally time-consuming and involves several laborious steps (Figure 1A).
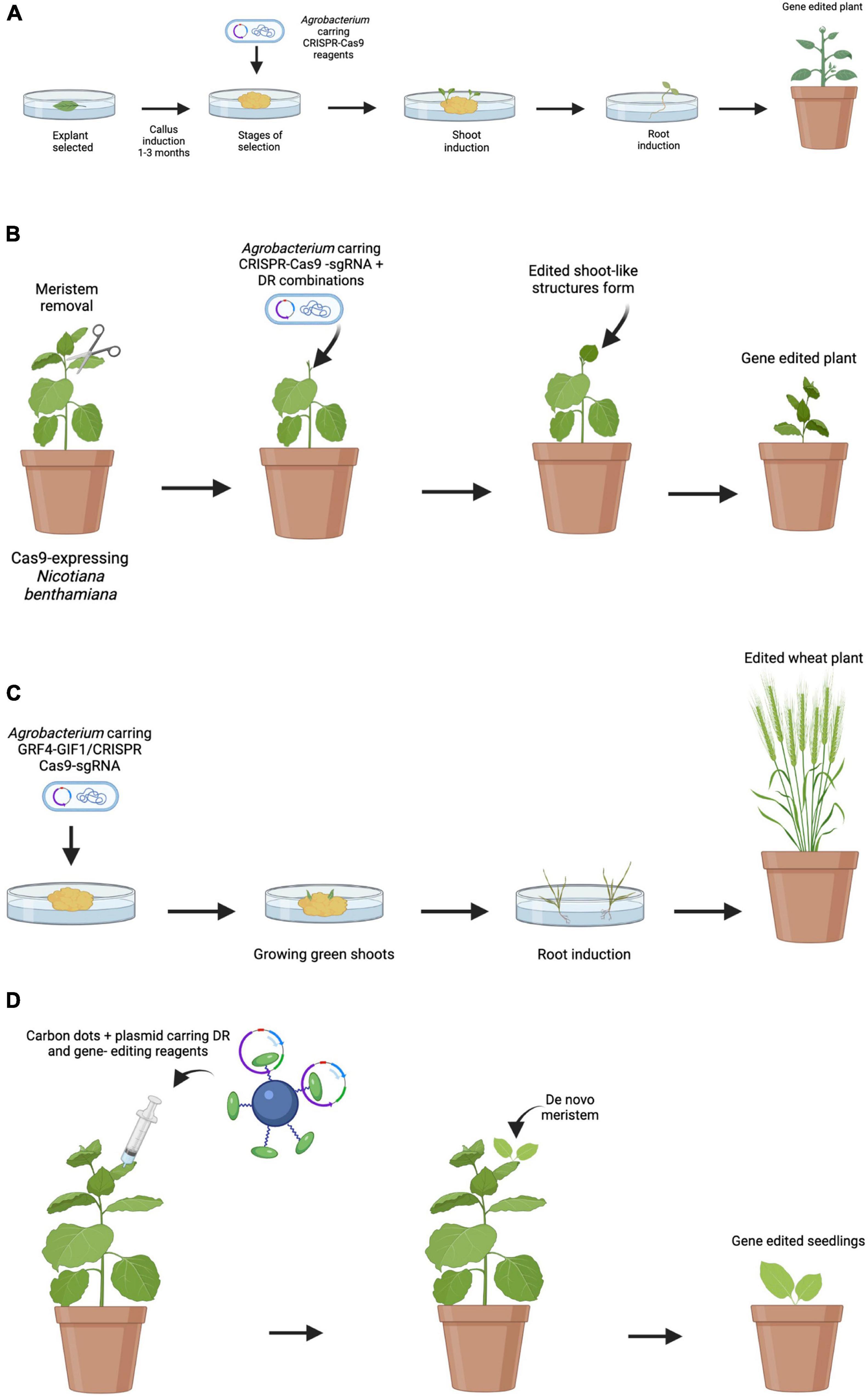
Figure 1. Comparison of traditional and emerging transformation and edition techniques. (A) Traditional Agrobacterium-mediated embryo transformation. Embryos are obtained and then incubated with A. tumefaciens. Multiple steps of selection are done to identify transgenic callus. Selected calli are transferred to shoot induction media follow by root induction media. Finally, plants are transferred to soil. (B) Induction of transgenic shoots on soil-grown plants. Meristems are removed, and DRs and gene-editing reagents are delivered by A. tumefaciens. After a while, de novo gene-edited shoots are formed and editing events are transmitted to the next generation. (C) Induction of edited shoots using the GRF–GIF chimera. GRF4–GIF1/CRISPR–Cas9–gRNA construction is delivered by A. tumefaciens. As a result, an increase in regeneration efficiency is observed. The shoots are then transferred to a medium to root and develop into whole plants. (D) Proposed model for nanoparticle mediated CRISPR/Cas9 in plant engineering. Nanoparticles can deliver DR and CRISPR/Cas9 reagents into plant cells, resulting in transgenic plants through de novo induction of meristems.
As shown in the present review, the preferred transformation method for stable grapevine transformation is Agrobacterium, and that most of the works have focused mainly on the use of few grapevine cultivars like “Chardonnay” and “Thompson Seedless.” Regarding the use of antibiotics for the selection of transformants, the determination of the optimal concentration for each genotype emerged as a necessary step since antibiotic sensitivity showed to be genotype-dependent. Accordingly, the possibility of using reporter genes such as GFP or RUBY as a control of the transformation appears as an attractive alternative. Therefore, the use of new technologies and their combination is required to facilitate the recovery of many plants in a large number of grapevine cultivars. In this sense, the application of the technologies proposed by Debernardi et al. (2020) and Maher et al. (2020) would be very useful to increase regeneration rates (Figures 1B,C). As previously mentioned, the tissues used for transformation are sensitive to infection with A. tumefaciens. For this reason, the use of nanoparticles-derived delivery systems, such as CDs, emerges as an alternative to overcome this problem with the advantage that it would allow to extend the range of hosts. This work proposes the possibility of combining the technologies developed by Debernardi et al. (2020) and Maher et al. (2020) together with the delivery of vectors mediated by nanoparticles, with the aim of overcoming problems and limitations related to the classical methodology of grapevine transformation and plant regeneration (Figure 1D).
GC and CC wrote the manuscript. SM provided critical feedback. DL conceived and performed the final edition of the manuscript. All the authors approved the final version of the manuscript.
This work was supported by the Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT): PICT2015-0822 and PICT-2018-02381. COST Action CA17111.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
GC and SM postdoctoral studies were supported by the National Scientific and Technical Research Council of Argentina (CONICET). We apologize to any researchers whose work was omitted without intention.
Acanda, Y. (2015). Embriogénesis Somática en la Vid : Control de la Maduración de los Embriones Somáticos y Generación de Nueva Variabilidad in Vitro. Doctoral thesis. Vigo: Universida de Vigo, 1–123.
Acanda, Y., Prado, M. J., González, M. V., and Rey, M. (2013). Somatic embryogenesis from stamen filaments in grapevine (Vitis vinifera L. cv. Mencía): changes in ploidy level and nuclear DNA content. Vitr. Cell. Dev. Biol. Plant 49, 276–284. doi: 10.1007/s11627-013-9499-7
Adeleye, A. S., Oranu, E. A., Tao, M., and Keller, A. A. (2016). Release and detection of nanosized copper from a commercial antifouling paint. Water Res. 102, 374–382. doi: 10.1016/j.watres.2016.06.056
Agüero, C. B., Meredith, C. P., and Dandekar, A. M. (2006). Genetic transformation of Vitis vinifera L. cvs Thompson seedless and chardonnay with the pear PGIP and GFP encoding genes. Vitis 45, 1–8.
Altpeter, F., Springer, N. M., Bartley, L. E., Blechl, A. E., Brutnell, T. P., Citovsky, V., et al. (2016). Advancing crop transformation in the era of genome editing. Plant Cell 28, 1510–1520. doi: 10.1105/tpc.16.00196
Anderson, J. A., Ellsworth, P. C., Faria, J. C., Head, G. P., Owen, M. D. K., Pilcher, C. D., et al. (2019). Genetically engineered crops: importance of diversified integrated pest management for agricultural sustainability. Front. Bioeng. Biotechnol. 7:24. doi: 10.3389/FBIOE.2019.00024
Baribault, T. J., Skene, K. G. M., and Steele Scott, N. (1989). Genetic transformation of grapevine cells. Plant Cell Rep. 8, 137–140. doi: 10.1007/BF00716825
Bharathy, P. V., and Agrawal, D. C. (2008). High frequency occurrence of single cotyledonary embryo morphotype and repetitive somatic embryogenesis in “Thompson Seedless” crossed with seven grapevine male parents. J. Grapevine Res. 47, 169–174.
Bhattacharya, K., Mukherjee, S. P., Gallud, A., Burkert, S. C., Bistarelli, S., Bellucci, S., et al. (2016). Biological interactions of carbon-based nanomaterials: from coronation to degradation. Nanomed. Nanotechnol. Biol. Med. 12, 333–351. doi: 10.1016/j.nano.2015.11.011
Borkow, G., and Gabbay, J. (2009). Copper, an ancient remedy returning to fight microbial, fungal and viral infections. Curr. Chem. Biol. 3, 272–278. doi: 10.2174/2212796810903030272
Bornhoff, B. A., Harst, M., Zyprian, E., and Töpfer, R. (2005). Transgenic plants of Vitis vinifera cv. Seyval blanc. Plant Cell Rep. 24, 433–438. doi: 10.1007/s00299-005-0959-3
Bouquet, A., Torregrosa, L., Iocco, P., and Thomas, M. R. (2008). “Grapes,” in Compendium of Transgenic Crop Plants: Transgenic Temperate Fruits and Nuts, eds C. Kole and T. C. Hall (Oxford: Blackwell Publishing), 189–232.
Buendía-González, L., Estrada-Zúñiga, M. E., Orozco-Villafuerte, J., Cruz-Sosa, F., and Vernon-Carter, E. J. (2012). Somatic embryogenesis of the heavy metal accumulator Prosopis laevigata. Plant Cell. Tissue Organ Cult. 108, 287–296. doi: 10.1007/s11240-011-0042-4
Burlaka, O. M., Pirko, Y. V., Yemets, A. I., and Blume, Y. B. (2015). Plant genetic transformation using carbon nanotubes for DNA Delivery. Cytol. Genet. 49, 349–357. doi: 10.3103/S009545271506002X
Capriotti, L., Baraldi, E., Mezzetti, B., Limera, C., and Sabbadini, S. (2020). Biotechnological approaches: gene overexpression, gene silencing, and genome editing to control fungal and oomycete diseases in grapevine. Int. J. Mol. Sci. 21:5701. doi: 10.3390/ijms21165701
Carimi, F., Barizza, E., Gardiman, M., and Schiavo Lo, F. (2005). Somatic embryogenesis from stigmas and styles of grapevine. Vitr. Cell. Dev. Biol. 41, 249–252. doi: 10.1079/IVP2004617
Che, P., Wu, E., Simon, M. K., Anand, A., Lowe, K., Gao, H., et al. (2021). Wuschel2 enables highly efficient CRISPR/Cas-targeted genome editing during rapid de novo shoot regeneration in sorghum. bioRxiv [Preprint]. doi: 10.1101/2021.06.21.449302
Chen, D., Dougherty, C. A., Zhu, K., and Hong, H. (2015). Theranostic applications of carbon nanomaterials in cancer: focus on imaging and cargo delivery. J. Control. Release 210, 230–245.
Chiu, W., Niwa, Y., Zeng, W., Hirano, T., Kobayashi, H., and Sheen, J. (1996). Engineered GFP as a vital reporter in plants. Curr. Biol. 6, 325–330. doi: 10.1016/S0960-9822(02)00483-9
Climent, E., Bernardos, A., Martínez-Máñez, R., Maquieira, A., Marcos, M. D., Pastor-Navarro, N., et al. (2009). Controlled delivery systems using antibody-capped mesoporous nanocontainers. J. Am. Chem. Soc. 131, 14075–14080. doi: 10.1021/ja904456d
Cunningham, F. J., Goh, N. S., Demirer, G. S., Matos, J. L., and Landry, M. P. (2018). Nanoparticle-mediated delivery towards advancing plant genetic engineering. Trends Biotechnol. 36, 882–897. doi: 10.1016/J.TIBTECH.2018.03.009
Dabauza, M., Velasco, L., Pazos-Navarro, M., Pérez-Benito, E., Hellín, P., Flores, P., et al. (2014). Enhanced resistance to Botrytis cinerea in genetically-modified Vitis vinifera L. plants over-expressing the grapevine stilbene synthase gene. Plant Cell. Tissue Organ Cult. 120, 229–238. doi: 10.1007/s11240-014-0598-x
Dai, L., Wang, D., Xie, X., Zhang, C., Wang, X., Xu, Y., et al. (2016). The novel gene VpPR4-1 from vitis pseudoreticulata increases powdery mildew resistance in transgenic vitis vinifera L. Front. Plant Sci. 7:695. doi: 10.3389/fpls.2016.00695
Dai, L., Zhou, Q., Li, R., Du, Y., He, J., Wang, D., et al. (2015). Establishment of a picloram-induced somatic embryogenesis system in Vitis vinifera cv. chardonnay and genetic transformation of a stilbene synthase gene from wild-growing Vitis species. Plant Cell. Tissue Organ Cult. 121, 397–412. doi: 10.1007/s11240-015-0711-9
Dalla Costa, L., Malnoy, M., and Gribaudo, I. (2017). Breeding next generation tree fruits: technical and legal challenges. Hortic. Res. 4:17067. doi: 10.1038/hortres.2017.67
Dalla Costa, L., Malnoy, M., Lecourieux, D., Deluc, L., Lecourieux, F. O., Thomas, M. R., et al. (2019). The state-of-the-art of grapevine biotechnology and new breeding technologies (NBTS). OENO One 53, 189–212. doi: 10.20870/OENO-ONE.2019.53.2.2405
Dandekar, A. M., Gouran, H., Ibáñez, A. M., Uratsu, S. L., Agüero, C. B., McFarland, S., et al. (2012). An engineered innate immune defense protects grapevines from Pierce disease. Proc. Natl. Acad. Sci. U.S.A. 109, 3721–3725. doi: 10.1073/pnas.1116027109
Das, D., Reddy, M., Upadhyaya, K., and Sopory, S. (2002). An efficient leaf-disc culture method for the regeneration via somatic embryogenesis and transformation of grape (Vitis vinifera L.). Plant Cell Rep. 20, 999–1005. doi: 10.1007/s00299-002-0441-4
Debernardi, J. M., Tricoli, D. M., Ercoli, M. F., Hayta, S., Ronald, P., Palatnik, J. F., et al. (2020). A GRF–GIF chimeric protein improves the regeneration efficiency of transgenic plants. Nat. Biotechnol. 38, 1274–1279. doi: 10.1038/s41587-020-0703-0
Demirer, G. S., Zhang, H., Goh, N. S., González-Grandío, E., and Landry, M. P. (2019a). Carbon nanotube–mediated DNA delivery without transgene integration in intact plants. Nat. Protoc. 14, 2954–2971. doi: 10.1038/s41596-019-0208-9
Demirer, G. S., Zhang, H., Matos, J. L., Goh, N. S., Cunningham, F. J., Sung, Y., et al. (2019b). High aspect ratio nanomaterials enable delivery of functional genetic material without DNA integration in mature plants. Nat. Nanotechnol. 14, 456–464. doi: 10.1038/s41565-019-0382-5
Dhekney, S. A., Li, Z. T., and Gray, D. J. (2011). Grapevines engineered to express cisgenic Vitis vinifera thaumatin-like protein exhibit fungal disease resistance. Vitr. Cell. Dev. Biol. Plant 47, 458–466. doi: 10.1007/s11627-011-9358-3
Dhekney, S. A., Li, Z. T., Dutt, M., and Gray, D. J. (2008). Agrobacterium-mediated transformation of embryogenic cultures and plant regeneration in Vitis rotundifolia Michx. (muscadine grape). Plant Cell Rep. 27, 865–872. doi: 10.1007/s00299-008-0512-2
Dodeman, V. L., Ducreux, G., and Kreis, M. (1997). Zygotic embryogenesis versus somatic embryogenesis. J. Exp. Bot. 48, 1493–1509.
Doyle, C., Higginbottom, K., Swift, T. A., Winfield, M., Bellas, C., Benito-Alifonso, D., et al. (2019). A simple method for spray-on gene editing in planta. bioRxiv [Preprint]. doi: 10.1101/805036
Dutt, M., Li, Z. T., Dhekney, S. A., and Gray, D. J. (2007). Transgenic plants from shoot apical meristems of Vitis vinifera L. “Thompson Seedless” via Agrobacterium-mediated transformation. Plant Cell Rep. 26, 2101–2110. doi: 10.1007/s00299-007-0424-6
Dutt, M., Li, Z. T., Dhekney, S. A., and Gray, D. J. (2008). A co-transformation system to produce transgenic grapevines free of marker genes. Plant Sci. 175, 423–430. doi: 10.1016/j.plantsci.2008.06.014
Etxeberria, E., Gonzalez, P., Baroja-Fernández, E., and Romero, J. P. (2006). Fluid phase endocytic uptake of artificial nano-spheres and fluorescent quantum dots by sycamore cultured cells. Plant Signal. Behav. 1, 196–200. doi: 10.4161/psb.1.4.3142
Fan, C., Pu, N., Wang, X., Wang, Y., Fang, L., Xu, W., et al. (2008). Agrobacterium-mediated genetic transformation of grapevine (Vitis vinifera L.) with a novel stilbene synthase gene from Chinese wild Vitis pseudoreticulata. Plant Cell. Tissue Organ Cult. 92, 197–206. doi: 10.1007/s11240-007-9324-2
FAOSTAT (2019). FAOSTAT. Available online at: http://www.fao.org/faostat/en/#home (accessed September 1, 2021).
Faure, O., Mengoli, M., Nougarede, A., and Bagni, N. (1991). Polyamine pattern and biosynthesis in zygotic and somatic embryo stages of Vitis vinifera. J. Plant Physiol. 138, 545–549. doi: 10.1016/S0176-1617(11)80238-5
Fehér, A., Pasternak, T. P., and Dudits, D. (2003). Transition of somatic plant cells to an embryogenic state. Plant Cell. Tissue Organ Cult. 74, 201–228. doi: 10.1023/A:1024033216561
Franks, T. K., Powell, K. S., Choimes, S., Marsh, E., Iocco, P., Sinclair, B. J., et al. (2006). Consequences of transferring three sorghum genes for secondary metabolite (cyanogenic glucoside) biosynthesis to grapevine hairy roots. Transgenic Res. 15, 181–195. doi: 10.1007/s11248-005-3737-7
Franks, T., He, D. G., and Thomas, M. (1998). Regeneration of transgenic Vitis vinifera L. Sultana plants: genotypic and phenotypic analysis. Mol. Breed. 4, 321–333. doi: 10.1023/A:1009673619456
Gago, J., Grima-Pettenati, J., and Gallego, P. P. (2011). Vascular-specific expression of GUS and GFP reporter genes in transgenic grapevine (Vitis vinifera L. cv. Albariño) conferred by the EgCCR promoter of Eucalyptus gunnii. Plant Physiol. Biochem. 49, 413–419. doi: 10.1016/j.plaphy.2011.02.005
Gambino, G., Gribaudo, I., Leopold, S., Schartl, A., and Laimer, M. (2005). Molecular characterization of grapevine plants transformed with GFLV resistance genes: I. Plant Cell Rep. 24, 655–662. doi: 10.1007/s00299-005-0006-4
Gambino, G., Ruffa, P., Vallania, R., and Gribaudo, I. (2007). Somatic embryogenesis from whole flowers, anthers and ovaries of grapevine (Vitis spp.). Plant Cell. Tissue Organ Cult. 90, 79–83. doi: 10.1007/s11240-007-9256-x
Goebel-Tourand, I., Mauro, M. C., Sossountzov, L., Miginiac, E., and Deloire, A. (1993). Arrest of somatic embryo development in grapevine: histological characterization and the effect of ABA, BAP and zeatin in stimulating plantlet development. Plant Cell. Tissue Organ Cult. 33, 91–103. doi: 10.1007/BF01997603
Gölles, R., Moser, R., Katinger, H., Laimer da Câmara Machado, M., Tsolova, V., da Câmara Machado, A., et al. (1997). Trans formation of somatic embryos of Vitis sp. With different constructs containing nucleotide sequences from nepovirus coat protein genes. Acta Hortic. 447, 265–272. doi: 10.17660/ActaHortic.1997.447.55
Gray, D. J., and Meredith, C. P. (1992). “Biotechnology of perennial fruit crops,” in Biotechnology of Perennial Fruit Crops, ed. FAO of the UN (Wallingford: CAB International), 229–262.
Guttenberg, H. v. (1943). Kulturversuche mit isolierten Pflanzenzellen. Planta 33, 576–588. doi: 10.1007/BF01916543
Harst, M., Bornhoff, B. A., Zyprian, E., and Topfer, R. (2000). Influence of culture technique and genotype on the efficiency of Agrobacterium-mediated transformation of somatic embryos (Vitis vinifera) and their conversion to transgenic plants. J. Grapevine Res. 39, 99–102.
He, R., Wu, J., Zhang, Y., Agüero, C. B., Li, X., Liu, S., et al. (2016). Overexpression of a thaumatin-like protein gene from Vitis amurensis improves downy mildew resistance in Vitis vinifera grapevine. Protoplasma 254, 1579–1589. doi: 10.1007/s00709-016-1047-y
He, Y., Zhang, T., Sun, H., Zhan, H., and Zhao, Y. (2020). A reporter for noninvasively monitoring gene expression and plant transformation. Hortic. Res. 7:152. doi: 10.1038/s41438-020-00390-1
Hébert, D., Kikkert, J. R., Smith, F. D., and Reisch, B. I. (1993). Optimization of biolistic transformation of embryogenic grape cell suspensions. Plant Cell Rep. 12, 585–589. doi: 10.1007/BF00233066
Heidmann, I., de Lange, B., Lambalk, J., Angenent, G. C., and Boutilier, K. (2011). Efficient sweet pepper transformation mediated by the BABY BOOM transcription factor. Plant Cell Rep. 30, 1107–1115. doi: 10.1007/s00299-011-1018-x
Horstman, A., Bemer, M., and Boutilier, K. (2017). A transcriptional view on somatic embryogenesis. Regeneration 4, 201–216. doi: 10.1002/reg2.91
Iocco, P., Franks, T., and Thomas, M. R. (2001). Genetic transformation of major wine grape cultivars of Vitis vinifera L. Transgenic Res. 10, 105–112. doi: 10.1023/A:1008989610340
Jefferson, R. A., Kavanagh, T. A., and Bevan, M. W. (1987). GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x
Jelly, N. S., Valat, L., Walter, B., and Maillot, P. (2014). Transient expression assays in grapevine: a step towards genetic improvement. Plant Biotechnol. J. 12, 1231–1245. doi: 10.1111/pbi.12294
Jha, P., and Kumar, V. (2018). BABY BOOM (BBM): a candidate transcription factor gene in plant biotechnology. Biotechnol. Lett. 40, 1467–1475. doi: 10.1007/s10529-018-2613-5
Jin, W., Dong, J., Hu, Y., Lin, Z., Xu, E., and Han, Z. (2009). Improved cold-resistant performance in transgenic grape (Vitis vinifera L.) overexpressing cold-inducible transcription factors AtDREB1b. HortScience 44, 35–39. doi: 10.21273/hortsci.44.1.35
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., and Charpentier, E. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–822.
Joubert, D. A., de Lorenzo, G., and Vivier, M. A. (2013). Regulation of the grapevine polygalacturonase-inhibiting protein encoding gene: expression pattern, induction profile and promoter analysis. J. Plant Res. 126, 267–281. doi: 10.1007/s10265-012-0515-5
Kareem, A., Radhakrishnan, D., Sondhi, Y., Aiyaz, M., Roy, M. V., Sugimoto, K., et al. (2016). De novo assembly of plant body plan: a step ahead of Deadpool. Regeneration 3, 182–197. doi: 10.1002/reg2.68
Keller, A. A., Adeleye, A. S., Conway, J. R., Garner, K. L., Zhao, L., Cherr, G. N., et al. (2017). Comparative environmental fate and toxicity of copper nanomaterials. NanoImpact 7, 28–40. doi: 10.1016/j.impact.2017.05.003
Kikkert, J. R., Striem, M. J., Vidal, J. R., Wallace, P. G., Barnard, J., Reisch, B. I., et al. (2005). Long-term study of somatic embryogenesis from anthers and ovaries of 12 grapevine (Vitis sp.) genotypes. Vitr. Cell. Dev. Biol. Plant 41, 232–239. doi: 10.1079/IVP2004609
Kikkert, J. R., Thomas, M. R., and Reisch, B. I. (2001). “Grapevine genetic engineering,” in Molecular Biology & Biotechnology of the Grapevine, ed. K. A. Roubelakis-Angelakis (Dordrecht: Springer), 393–410. doi: 10.1007/978-94-017-2308-4_15
Kim, J. H., and Kende, H. (2004). A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 101, 13374–13379. doi: 10.1073/pnas.0405450101
Klimaszewska, K., bernier-Cardou, M., Cyr, D. R., and Sutton, B. C. S. (2000). Influence of gelling agents on culture medium gel strength, water availability, tissue water potential, and maturation response in embryogenic cultures of Pinus strobus L. Vitr. Cell. Dev. Biol. Plant 36, 279–286. doi: 10.1007/s11627-000-0051-1
Kong, L., Dai, D., Shang, M., Li, K., and Zhang, C. X. (2009). Thidiazuron-induced somatic embryos, their multiplication, maturation, and conversion in Cinnamomum pauciflorum nees (Lauraceae). New For. 38, 131–142. doi: 10.1007/s11056-009-9135-x
Kwak, S.-Y., Lew, T. T. S., Sweeney, C. J., Koman, V. B., Wong, M. H., Bohmert-Tatarev, K., et al. (2019). Chloroplast-selective gene delivery and expression in planta using chitosan-complexed single-walled carbon nanotube carriers. Nat. Nanotechnol. 14, 447–455. doi: 10.1038/s41565-019-0375-4
Lardon, R., and Geelen, D. (2020). Natural variation in plant pluripotency and regeneration. Plants 9, 1–28. doi: 10.3390/plants9101261
Lazo, G. R., Stein, P. A., and Ludwig, R. A. (1991). A DNA transformation–competent arabidopsis genomic library in Agrobacterium. Biotechnology 9, 963–967. doi: 10.1038/nbt1091-963
Le Gall, O., Torregrosa, L., Danglot, Y., Candresse, T., and Bouquet, A. (1994). Agrobacterium-mediated genetic transformation of grapevine somatic embryos and regeneration of transgenic plants expressing the coat protein of grapevine chrome mosaic nepovirus (GCMV). Plant Sci. 102, 161–170. doi: 10.1016/0168-9452(94)90034-5
Li, Y., Xu, X., Wu, Y., Zhuang, J., Zhang, X., Zhang, H., et al. (2020). A review on the effects of carbon dots in plant systems. Mater. Chem. Front. 4, 437–448. doi: 10.1039/c9qm00614a
Li, Y., Zhao, Y., Cheng, H., Hu, Y., Shi, G., Dai, L., et al. (2012). Nitrogen-doped graphene quantum dots with oxygen-rich functional groups. J. Am. Chem. Soc. 134, 15–18. doi: 10.1021/ja206030c
Li, Z. T., Dhekney, S., Dutt, M., Van Aman, M., Tattersall, J., Kelley, K. T., et al. (2006). Optimizing Agrobacterium-mediated transformation of grapevine. Vitr. Cell. Dev. Biol. Plant 42, 220–227. doi: 10.1079/IVP2006770
Li, Z. T., Hopkins, D. L., and Gray, D. J. (2015). Overexpression of antimicrobial lytic peptides protects grapevine from Pierce’s disease under greenhouse but not field conditions. Transgenic Res. 24, 821–836. doi: 10.1007/s11248-015-9876-6
Liebsch, D., and Palatnik, J. F. (2020). MicroRNA miR396, GRF transcription factors and GIF co-regulators: a conserved plant growth regulatory module with potential for breeding and biotechnology. Curr. Opin. Plant Biol. 53, 31–42. doi: 10.1016/j.pbi.2019.09.008
Lin, G., Zhang, H., and Huang, L. (2015). Smart polymeric nanoparticles for cancer gene delivery. Mol. Pharm. 12, 314–321. doi: 10.1021/mp500656v
Liu, J., Sheng, L., Xu, Y., Li, J., Yang, Z., Huang, H., et al. (2014). WOX11 and 12 are involved in the first-step cell fate transition during de novo root organogenesis in Arabidopsis. Plant Cell 26, 1081–1093. doi: 10.1105/tpc.114.122887
Liu, Q., Chen, B., Wang, Q., Shi, X., Xiao, Z., Lin, J., et al. (2009). Carbon nanotubes as molecular transporters for walled plant cells. Nano Lett. 9, 1007–1010.
Liu, R., and Lal, R. (2015). Potentials of engineered nanoparticles as fertilizers for increasing agronomic productions. Sci. Total Environ. 514, 131–139. doi: 10.1016/j.scitotenv.2015.01.104
López-Pérez, A. J., Carreño, J., Martínez-Cutillas, A., and Dabauza, M. (2005). High embryogenic ability and plant regeneration of table grapevine cultivars (Vitis vinifera L.) induced by activated charcoal. J. Grapevine Res. 44, 79–85.
López-Pérez, A. J., Velasco, L., Pazos-Navarro, M., and Dabauza, M. (2008). Development of highly efficient genetic transformation protocols for table grape Sugraone and Crimson Seedless at low Agrobacterium density. Plant Cell. Tissue Organ Cult. 94, 189–199. doi: 10.1007/s11240-008-9404-y
Lowe, K., Wu, E., Wang, N., Hoerster, G., Hastings, C., Cho, M. J., et al. (2016). Morphogenic regulators Baby boom and Wuschel improve monocot transformation. Plant Cell 28, 1998–2015. doi: 10.1105/tpc.16.00124
Lutz, K. A., Martin, C., Khairzada, S., and Maliga, P. (2015). Steroid-inducible BABY BOOM system for development of fertile Arabidopsis thaliana plants after prolonged tissue culture. Plant Cell Rep. 34, 1849–1856. doi: 10.1007/s00299-015-1832-7
Maher, M. F., Nasti, R. A., Vollbrecht, M., Starker, C. G., Clark, M. D., and Voytas, D. F. (2020). Plant gene editing through de novo induction of meristems. Nat. Biotechnol. 38, 84–89. doi: 10.1038/s41587-019-0337-2
Maillot, P., Deglène-Benbrahim, L., and Walter, B. (2016). Efficient somatic embryogenesis from meristematic explants in grapevine (Vitis vinifera L.) cv. Chardonnay: an improved protocol. Trees Struct. Funct. 30, 1377–1387. doi: 10.1007/s00468-016-1374-9
Maillot, P., Kieffer, F., and Walter, B. (2006). Somatic embryogenesis from stem nodal sections of grapevine. J. Grapevine Res. 45, 185–189.
Mali, P., Church, G. M., Esvelt, K. M., Guell, M., Norville, J. E., Yang, L., et al. (2013). RNA-guided human genome engineering via Cas9. Science 339, 823–826.
Martinelli, L., and Gribaudo, I. (2001). “Somatic embryogenesis in grapevine,” in Molecular Biology & Biotechnology of the Grapevine, ed. K. A. Roubelakis-Angelakis (Dordrecht: Springer), 327–351. doi: 10.1007/978-94-017-2308-4_13
Martinelli, L., and Gribaudo, I. (2009). “Strategies for effective somatic embryogenesis in grapevine: an appraisal,” in Grapevine Molecular Physiology and Biotechnology, 2nd Edn, ed. K. A. Roubelakis-Angelakis (Cham: Springer), 461–493. doi: 10.1007/978-90-481-2305-6_17
Martinelli, L., Bragagna, P., Poletti, V., and Scienza, A. (1993). Somatic embryogenesis from leaf- and petiole-derived callus of Vitis rupestris. Plant Cell Rep. 12, 207–210. doi: 10.1007/BF00237055
Mauro, M. C., Toutain, S., Walter, B., Pinck, L., Otten, L., Coutos-Thevenot, P., et al. (1995). High efficiency regeneration of grapevine plants transformed with the GFLV coat protein gene. Plant Sci. 112, 97–106. doi: 10.1016/0168-9452(95)04246-Q
Méndez-Hernández, H. A., Ledezma-Rodríguez, M., Avilez-Montalvo, R. N., Juárez-Gómez, Y. L., Skeete, A., Avilez-Montalvo, J., et al. (2019). Signaling overview of plant somatic embryogenesis. Front. Plant Sci. 10:77. doi: 10.3389/fpls.2019.00077
Milewska-Hendel, A., Zubko, M., Karcz, J., Stróż, D., and Kurczyńska, E. (2017). Fate of neutral-charged gold nanoparticles in the roots of the Hordeum vulgare L. cultivar Karat. Sci Rep. 7:3014. doi: 10.1038/s41598-017-02965-w
Mookkan, M., Nelson-Vasilchik, K., Hague, J., Zhang, Z. J., and Kausch, A. P. (2017). Selectable marker independent transformation of recalcitrant maize inbred B73 and sorghum P898012 mediated by morphogenic regulators BABY BOOM and WUSCHEL2. Plant Cell Rep. 36, 1477–1491. doi: 10.1007/s00299-017-2169-1
Morcillo, F., Gallard, A., Pillot, M., Jouannic, S., Aberlenc-Bertossi, F., Collin, M., et al. (2007). EgAP2-1, an AINTEGUMENTA-like (AIL) gene expressed in meristematic and proliferating tissues of embryos in oil palm. Planta 226, 1353–1362. doi: 10.1007/s00425-007-0574-3
Morgana, C., Di Lorenzo, R., and Carimi, F. (2004). Somatic embryogenesis of Vitis vinifera L. (cv. Sugraone) from stigma and style culture. J. Grapevine Res. 43, 169–173.
Morones, J. R., Elechiguerra, J. L., Camacho, A., Holt, K., Kouri, J. B., Ram, J. T., et al. (2005). The bactericidal effect of silver nanoparticles. Nanotechnology 16, 2346–2353. doi: 10.1088/0957-4484/16/10/059
Morris, P. C., Kumar, A., Bowles, D. J., and Cuming, A. C. (1990). Osmotic stress and abscisic acid induce expression of the wheat Em genes. Eur. J. Biochem. 190, 625–630. doi: 10.1111/j.1432-1033.1990.tb15618.x
Mukherjee, A., Majumdar, S., Servin, A. D., Pagano, L., Dhankher, O. P., White, J. C., et al. (2016). Carbon nanomaterials in agriculture: a critical review. Front. Plant Sci. 7:172. doi: 10.3389/fpls.2016.00172
Mullins, M. G., Archie Tang, F. C., and Facciotti, D. (1990). Agrobacterium-mediated genetic transformation of grapevines: transgenic plants of Vitis rupestris scheele and buds of Vitis vinifera L. Nat. Biotechnol. 8, 1041–1045. doi: 10.1038/nbt1190-1041
Nakajima, I., and Matsuta, N. (2003). Somatic embryogenesis from filaments of Vitis vinifera L. and Vitis labruscana Bailey. J. Grapevine Res. 42, 53–54.
Nakano, M., Hoshino, Y., and Mii, M. (1994). Regeneration of transgenic plants of grapevine (Vitis vinifera L.) via Agrobacterium rhizogenesmediated transformation of embryogenic calli. J. Exp. Bot. 45, 649–656. doi: 10.1093/jxb/45.5.649
Nirala, N. K., Das, D. K., Srivastava, P. S., Sopory, S. K., and Upadhyaya, K. C. (2010). Expression of a rice chitinase gene enhances antifungal potential in transgenic grapevine (Vitis vinifera L.). J. Grapevine Res. 49, 181–187.
Nookaraju, A., and Agrawal, D. C. (2012). Enhanced tolerance of transgenic grapevines expressing chitinase and β-1,3-glucanase genes to downy mildew. Plant Cell. Tissue Organ Cult. 111, 15–28. doi: 10.1007/s11240-012-0166-1
Passarinho, P., Ketelaar, T., Xing, M. Van Arkel, J., Maliepaard, C., Hendriks, M. W., et al. (2008). BABY BOOM target genes provide diverse entry points into cell proliferation and cell growth pathways. Plant Mol. Biol. 68, 225–237. doi: 10.1007/s11103-008-9364-y
Peiró, R., Gammoudi, N., Yuste, A., Olmos, A., and Gisbert, C. (2015). Mature seeds for in vitro sanitation of the Grapevine leafroll associated virus (GLRaV-1 and GLRaV-3) from grape (Vitis vinifera L.). Spanish J. Agric. Res. 13:1005. doi: 10.5424/sjar/2015132-7094
Peng, Z., Han, X., Li, S., Al-Youbi, A. O., Bashammakh, A. S., El-Shahawi, M. S., et al. (2017). Carbon dots: biomacromolecule interaction, bioimaging and nanomedicine. Coord. Chem. Rev. 343, 256–277.
Perl, A., Lotan, O., Abu-Abied, M., and Holland, D. (1996). Establishment of an Agrobacterium-mediated transformation system for grape (Vitis vinifera L.): the role of antioxidants during grape–Agrobacterium interactions. Nat. Biotechnol. 14, 624–628. doi: 10.1038/nbt0596-624
Perrone, I., Gambino, G., Chitarra, W., Vitali, M., Pagliarani, C., Riccomagno, N., et al. (2012). The grapevine root-specific aquaporin VvPIP2;4N controls root hydraulic conductance and leaf gas exchange under well-watered conditions but not under water stress. Plant Physiol. 160, 965–977. doi: 10.1104/pp.112.203455
Prado, M. J., Grueiro, M. P., González, M. V., Testillano, P. S., Domínguez, C., López, M., et al. (2010). Efficient plant regeneration through somatic embryogenesis from anthers and ovaries of six autochthonous grapevine cultivars from Galicia (Spain). Sci. Hortic. (Amsterdam) 125, 342–352. doi: 10.1016/j.scienta.2010.04.019
Pulianmackal, A. J., Kareem, A. V. K., Durgaprasad, K., Trivedi, Z. B., and Prasad, K. (2014). Competence and regulatory interactions during regeneration in plants. Front. Plant Sci. 5:142. doi: 10.3389/fpls.2014.00142
Qi, L. S., Larson, M. H., Gilbert, L. A., Doudna, J. A., Weissman, J. S., Arkin, A. P., et al. (2013). Repurposing CRISPR as an RNA-γuided platform for sequence-specific control of gene expression. Cell 152, 1173–1183. doi: 10.1016/j.cell.2013.02.022
Rodriguez, R. E., Ercoli, M. F., Debernardi, J. M., and Palatnik, J. F. (2016). “Growth-regulating factors, a transcription factor family regulating more than just plant growth,” in Plant Transcription Factors: Evolutionary, Structural and Functional Aspects, ed. D. Gonzalez (San Diego, CA: Academic Press), 269–280. doi: 10.1016/B978-0-12-800854-6.00017-8
Rubio, J., Montes, C., Castro, Á, Álvarez, C., Olmedo, B., Muñoz, M., et al. (2015). Genetically engineered Thompson Seedless grapevine plants designed for fungal tolerance: selection and characterization of the best performing individuals in a field trial. Transgenic Res. 24, 43–60. doi: 10.1007/s11248-014-9811-2
Samanta, M. K., Dey, A., and Gayen, S. (2016). CRISPR/Cas9: an advanced tool for editing plant genomes. Transgenic Res. 25, 561–573. doi: 10.1007/s11248-016-9953-5
Sanford, J. C., Klein, T. M., Wolf, E. D., and Allen, N. (1987). Delivery of substances into cells and tissues using a particle bombardment process. Part. Sci. Technol. 5, 27–37. doi: 10.1080/02726358708904533
Saporta, R., De La Torre, F., Segura, A., and Vidal, J. R. (2014). Toxic effect of antibiotics in grapevine (Vitis vinifera ‘Albariño’) for embryo emergence and transgenic plant regeneration from embryogenic cell suspension. J. Grapevine Res. 53, 89–94.
Schwab, F., Zhai, G., Kern, M., Turner, A., Schnoor, J. L., and Wiesner, M. R. (2016). Barriers, pathways and processes for uptake, translocation and accumulation of nanomaterials in plants – Critical review. Nanotoxicology 10, 257–278. doi: 10.3109/17435390.2015.1048326
Schwartz, S. H., Hendrix, B., Hoffer, P., Sanders, R. A., and Zheng, W. (2020). Carbon dots for efficient small interfering RNA delivery and gene silencing in plants. Plant Physiol. 184, 647–657. doi: 10.1104/pp.20.00733
Scorza, R., Cordts, J. M., Gray, D. J., Gonsalves, D., Emershad, R. L., and Ramming, D. W. (1996). Producing transgenic “Thompson Seedless” grape (Vitis vinifera L.) plants. J. Am. Soc. Hortic. Sci. 121:4.
Scorza, R., Cordts, J. M., Ramming, D. W., and Emershad, R. L. (1995). Transformation of grape (Vitis vinifera L.) zygotic-derived somatic embryos and regeneration of transgenic plants. Plant Cell Rep. 14, 589–592. doi: 10.1007/BF00231944
Sharon, M., Choudhary, A. K., and Kumar, R. (2010). Nanotechnology in agricultural diseases and food safety. J. Phytol. 2, 83–92.
Shojaei, T. R., Salleh, M. A. M., Tabatabaei, M., Mobli, H., Aghbashlo, M., Rashid, S. A., et al. (2019). “Applications of nanotechnology and carbon nanoparticles in agriculture,” in Synthesis, Technology and Applications of Carbon Nanomaterials, eds S. A. Rashid, R. N. I. Raja Othman, and M. Z. Hussein (Amsterdam: Elsevier), 247–277.
Sholi, N. J. Y., Chaurasia, A., Agrawal, A., and Sarin, N. B. (2009). ABA enhances plant regeneration of somatic embryos derived from cell suspension cultures of plantain cv. Spambia (Musa sp.). Plant Cell. Tissue Organ Cult. 99, 133–140. doi: 10.1007/s11240-009-9585-z
Skoog, F., and Miller, C. O. (1957). Chemical regulation of growth and organ formation in plant tissues cultured in vitro. Symp. Soc. Exp. Biol. 11, 118–130.
Srinivasan, C., Liu, Z., Heidmann, I., Supena, E. D. J., Fukuoka, H., Joosen, R., et al. (2007). Heterologous expression of the BABY BOOM AP2/ERF transcription factor enhances the regeneration capacity of tobacco (Nicotiana tabacum L.). Planta 225, 341–351. doi: 10.1007/s00425-006-0358-1
Su, H., Jiao, Y. T., Wang, F. F., Liu, Y. E., Niu, W. L., Liu, G. T., et al. (2018). Overexpression of VpPR10.1 by an efficient transformation method enhances downy mildew resistance in V. vinifera. Plant Cell Rep. 37, 819–832. doi: 10.1007/s00299-018-2271-z
Su, Y., Ashworth, V., Kim, C., Adeleye, A. S., Rolshausen, P., Roper, C., et al. (2019). Delivery, uptake, fate, and transport of engineered nanoparticles in plants: a critical review and data analysis. Environ. Sci. Nano 6, 2311–2331. doi: 10.1039/c9en00461k
Tillett, R. L., Wheatley, M. D., Tattersall, E. A. R., Schlauch, K. A., Cramer, G. R., and Cushman, J. C. (2012). The Vitis vinifera C-repeat binding protein 4 (VvCBF4) transcriptional factor enhances freezing tolerance in wine grape. Plant Biotechnol. J. 10, 105–124. doi: 10.1111/J.1467-7652.2011.00648.X
Torregrosa, L., Lopez, G., and Bouquet, A. (2000). Antibiotic sensitivity of grapevine: a comparison between the effect of Hygromycin and Kanamycin on shoot development of transgenic 110 Richter rootstock (Vitis berlandieri x Vitis rupestris). S. Afr. J. Enol. Vitic. 21, 32–39. doi: 10.21548/21-1-2185
Torregrosa, L., Verriès, C., and Tesnière, C. (2002). Grapevine (Vitis vinifera L.) promoter analysis by biolistic-mediated transient transformation of cell suspensions. J. Grapevine Res. 41, 27–32.
Torregrosa, L., Vialet, S., Adivèze, A., Iocco-, C. P., and Mark, T. (2015). Grapevine (Vitis vinifera L.). Methods Mol. Biol. 344, 273–285. doi: 10.1385/1-59745-131-2:273
Tripathi, D. K., Singh, S., Singh, S., Pandey, R., Singh, V. P., Sharma, N. C., et al. (2017). An overview on manufactured nanoparticles in plants: uptake, translocation, accumulation and phytotoxicity. Plant Physiol. Biochem. 110, 2–12.
Tsvetkov, I., Tsolova, V., and Atanassov, A. (2000). Gene transfer for stress resistance in grapes. Acta Hortic. 528, 389–394.
Urso, S., Zottini, M., Ruberti, C., Lo Schiavo, F., Stanca, A. M., Cattivelli, L., et al. (2013). An Agrobacterium tumefaciens-mediated gene silencing system for functional analysis in grapevine. Plant Cell Tissue Organ Cult. 114, 49–60. doi: 10.1007/S11240-013-0305-3
Valletta, A., Chronopoulou, L., Palocci, C., Baldan, B., Donati, L., and Pasqua, G. (2014). Poly(lactic-co-glycolic) acid nanoparticles uptake by Vitis vinifera and grapevine-pathogenic fungi. J. Nanoparticle Res. 16:2744. doi: 10.1007/s11051-014-2744-0
Verma, S. K., Das, A. K., Gantait, S., Kumar, V., and Gurel, E. (2019). Applications of carbon nanomaterials in the plant system: a perspective view on the pros and cons. Sci. Total Environ. 667, 485–499.
Vezzulli, S., Zulini, L., and Stefanini, M. (2019). Genetics-assisted breeding for downy/powdery mildew and phylloxera resistance at fem. BIO Web Conf. 12:01020. doi: 10.1051/bioconf/20191201020
Vidal, J. R., Gomez, C., Cutanda, M. C., Shrestha, B. R., Bouquet, A., Thomas, M. R., et al. (2010). Use of gene transfer technology for functional studies in grapevine. Aust. J. Grape Wine Res. 16, 138–151. doi: 10.1111/j.1755-0238.2009.00086.x
Vidal, J. R., Kikkert, J. R., Donzelli, B. D., Wallace, P. G., and Reisch, B. I. (2006). Biolistic transformation of grapevine using minimal gene cassette technology. Plant Cell Rep. 25, 807–814. doi: 10.1007/s00299-006-0132-7
Vidal, J. R., Kikkert, J., Wallace, P., and Reisch, B. (2003). High-efficiency biolistic co-transformation and regeneration of “Chardonnay” (Vitis vinifera L.) containing npt-II and antimicrobial peptide genes. Plant Cell Rep. 22, 252–260. doi: 10.1007/s00299-003-0682-x
Vivier, M. A., and Pretorius, I. S. (2002). Genetically tailored grapevines for the wine industry. Trends Biotechnol. 20, 472–478. doi: 10.1016/S0167-7799(02)02058-9
Walker, D. R., and Parrott, W. A. (2001). Effect of polyethylene glycol and sugar alcohols on soybean somatic embryo germination and conversion. Plant Cell. Tissue Organ Cult. 64, 55–62. doi: 10.1023/A:1010601402098
Wang, B., Huang, J., Zhang, M., Wang, Y., Wang, H., Ma, Y., et al. (2020). Carbon dots enable efficient delivery of functional DNA in Plants. ACS Appl. Bio Mater. 3, 8857–8864. doi: 10.1021/acsabm.0c01170
Wang, L., Xie, X., Yao, W., Wang, J., Ma, F., et al. (2017). RING-H2-type E3 gene VpRH2 from Vitis pseudoreticulata improves resistance to powdery mildew by interacting with VpGRP2A. J. Exp. Bot. 68, 1669–1687. doi: 10.1093/jxb/erx033
Wang, Q., Li, P., Hanania, U., Sahar, N., Mawassi, M., Gafny, R., et al. (2005). Improvement of Agrobacterium-mediated transformation efficiency and transgenic plant regeneration of Vitis vinifera L. by optimizing selection regimes and utilizing cryopreserved cell suspensions. Plant Sci. 168, 565–571. doi: 10.1016/j.plantsci.2004.09.033
Xu, L. (2018). De novo root regeneration from leaf explants: wounding, auxin, and cell fate transition. Curr. Opin. Plant Biol. 41, 39–45. doi: 10.1016/j.pbi.2017.08.004
Xu, L., and Huang, H. (2014). Genetic and epigenetic controls of plant regeneration. Curr. Top. Dev. Biol. 108, 1–33. doi: 10.1016/B978-0-12-391498-9.00009-7
Yamamoto, T., Iketani, H., Ieki, H., Nishizawa, Y., Notsuka, K., Hibi, T., et al. (2000). Transgenic grapevine plants expressing a rice chitinase with enhanced resistance to fungal pathogens. Plant Cell Rep. 19, 639–646. doi: 10.1007/s002999900174
Yang, H. F., Kou, Y. P., Gao, B., Soliman, T. M. A., Xu, K. D., Ma, N., et al. (2014). Identification and functional analysis of BABY BOOM genes from Rosa canina. Biol. Plant. 58, 427–435. doi: 10.1007/s10535-014-0420-y
Yang, Y., Jittayasothorn, Y., Chronis, D., Wang, X., Cousins, P., and Zhong, G.-Y. (2013). Molecular characteristics and efficacy of 16D10 siRNAs in inhibiting root-knot nematode infection in transgenic grape hairy roots. PLoS One 8:e69463. doi: 10.1371/JOURNAL.PONE.0069463
Zheng, X. T., Ananthanarayanan, A., Luo, K. Q., and Chen, P. (2015). Glowing graphene quantum dots and carbon dots: properties, syntheses, and biological applications. Small 11, 1620–1636. doi: 10.1002/smll.201402648
Zhou, Q., Dai, L., Cheng, S., He, J., Wang, D., Zhang, J., et al. (2014). A circulatory system useful both for long-term somatic embryogenesis and genetic transformation in Vitis vinifera L. cv. Thompson Seedless. Plant Cell. Tissue Organ Cult. 118, 157–168. doi: 10.1007/s11240-014-0471-y
Ziemienowicz, A., Shim, Y.-S., Matsuoka, A., Eudes, F., and Kovalchuk, I. (2012). A novel method of transgene delivery into triticale plants using the Agrobacterium transferred DNA-derived nano-complex. Plant Physiol. 158, 1503–1513. doi: 10.1104/pp.111.192856
Keywords: grapevine, genetic transformation, nanotechnology, regeneration, transcription factors, development regulators, Vitis vinifera, gene editing
Citation: Campos G, Chialva C, Miras S and Lijavetzky D (2021) New Technologies and Strategies for Grapevine Breeding Through Genetic Transformation. Front. Plant Sci. 12:767522. doi: 10.3389/fpls.2021.767522
Received: 31 August 2021; Accepted: 25 October 2021;
Published: 25 November 2021.
Edited by:
Horacio Esteban Hopp, University of Buenos Aires, ArgentinaReviewed by:
Cecilia Vazquez Rovere, Institute of Agrobiotechnology and Molecular Biology (IABIMO), UEDD INTA-Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), ArgentinaCopyright © 2021 Campos, Chialva, Miras and Lijavetzky. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Diego Lijavetzky, ZGxpamF2ZXR6a3lAY29uaWNldC5nb3YuYXI=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.