
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 04 November 2021
Sec. Functional Plant Ecology
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.766550
This article is part of the Research Topic Genetic Diversity and Genomic Insights into Physiological Adaptations of Grasses to Diverse Ecological Niches View all 8 articles
Parts of this article's content have been modified or rectified in:
Erratum: Genome-Wide Investigation of the NAC Transcription Factor Family in Miscanthus sinensis and Expression Analysis Under Various Abiotic Stress
 Gang Nie†
Gang Nie† Zhongfu Yang†
Zhongfu Yang† Jie He†
Jie He† Aiyu Liu
Aiyu Liu Jiayi Chen
Jiayi Chen Shuan Wang
Shuan Wang Xia Wang
Xia Wang Guangyan Feng
Guangyan Feng Dandan Li
Dandan Li Yan Peng
Yan Peng Linkai Huang
Linkai Huang Xinquan Zhang*
Xinquan Zhang*The NAC transcription factor family is deemed to be a large plant-specific gene family that plays important roles in plant development and stress response. Miscanthus sinensis is commonly planted in vast areas of marginal lang as forage, ornamental grass, or bioenergy crop which demand a relatively high resistance to abiotic stresses. The recent release of a draft chromosome-scale assembly genome of M. sinensis provided a basic platform for the genome-wide investigation of NAC proteins. In this study, a total of 261 M. sinensis NAC genes were identified and a complete overview of the gene family was presented, including gene structure, conserved motif compositions, chromosomal distribution, and gene duplications. Results showed that gene length, molecular weights (MW), and theoretical isoelectric points (pI) of NAC family were varied, while gene structure and motifs were relatively conserved. Chromosomal mapping analysis found that the M. sinensis NAC genes were unevenly distributed on 19 M. sinensis chromosomes, and the interchromosomal evolutionary analysis showed that nine pairs of tandem duplicate genes and 121 segmental duplications were identified, suggesting that gene duplication, especially segmental duplication, is possibly associated with the amplification of M. sinensis NAC gene family. The expression patterns of 14 genes from M. sinensis SNAC subgroup were analyzed under high salinity, PEG, and heavy metals, and multiple NAC genes could be induced by the treatment. These results will provide a very useful reference for follow-up study of the functional characteristics of NAC genes in the mechanism of stress-response and potential roles in the development of M. sinensis.
Miscanthus sinensis is a rhizomatous C4 perennial grass which is widely applied in paper production and cultivated as forage, ornamental grass, and a bioenergy crop (Clifton-Brown and Lewandowski, 2002; Heaton et al., 2004; Nie et al., 2021). Generally, M. sinensis tend to be planted in marginal land with varied kinds of abiotic stress, such as limited water supply and heavy metal pollution (Yu et al., 2014), so as to maximize the avoidance competition of arable land (Mohr and Raman, 2013). Miscanthus sinensis can produce high biomass yields and potentially increase soil carbon when compared with other C4 grasses like Zea mays and Saccharum officinarum, which is enabled by a relatively highly efficient photosynthesis process and a great resistance to abiotic stress (Greef et al., 1997; Ye et al., 2005; Ezaki et al., 2008; Long and Spence, 2013; Nie et al., 2017) owing to its extensive root system and perennial growth habit (Byrt et al., 2011; Van der Weijde et al., 2013). However, the molecular regulation network and genetic components of the characteristics are still unclear due to the absence of genomic information of the species owing to the complexity of its genomes, which hinders the application of modern breeding approaches (Mitros et al., 2020).
Continued genetic improvement of bioenergy plants for their excellent features is important, especially to enhance productivity and resistance when exposed to abiotic stress. Recently, a draft chromosome-scale assembly of M. sinensis genome was published, which promoted the process of future genetic improvement of perennial biomass crops (Mitros et al., 2020). NAC proteins have shown significant functions in controlling plant growth and development in active response to abiotic and/or biotic stress (Aida et al., 1997; Nakashima et al., 2012; Kou et al., 2014). NAC domain protein family is deemed a transcription factor (TF) family containing a highly conserved N-terminal DNA-binding domain and a variable C-terminal domain. Commonly, it is divided into five subdomains (A–E) and was originally identified from three consensus sequences: NAM (no apical meristem), Arabidopsis ATAF1/2, and CUC2 (cup-shaped cotyledon; Ooka et al., 2003; Nakashima et al., 2007; Jensen et al., 2010). Although NAC gene family has been widely studied and the identified genes could potentially be used for plant improvement, there is no systematic concern in M. sinensis referring to limited availability of genome resources.
In Arabidopsis, AtNAC1 and AtNAC2 were reported to be involved in lateral root development by downregulating auxin signals (Guo et al., 2005), while NAP is related to plant leaf senescence (Guo and Gan, 2006) and floral morphogenesis (Sablowski and Meyerowitz, 1998). In Medicago truncatula, loss of MtNST1 resulted in the decrease of lignin content (Zhao et al., 2010). The overexpression of TaNAC69 enhanced the transcript levels of stress-induced genes and improved the drought tolerance of Triticum aestivum (Xue et al., 2011), while the overexpression of TaNAC29 increased salt tolerance by reducing H2O2 accumulation and membrane damage (Xu et al., 2015). OsNAC6 was induced by low temperature, salinity, drought, and ABA damage (Ohnishi et al., 2005; Nakashima et al., 2007). In soybean (Glycine max), 31 genes were identified to contain complete open reading frames encoding GmNAC protein. Among them, nine of the GmNAC genes were induced by drought, salinity, cold, and ABA treatments (Tran et al., 2009). In addition, the SsNAC23 gene in sugarcane homologous to OsNAC6 is associated with cold and PEG-mediated water stress (Nogueira et al., 2005). In Tartary buckwheat (Fagopyrum tataricum), eight FtNAC genes were identified as potentially participating in fruit development (Liu et al., 2019). In addition, stress-responsive NAC (SNAC) proteins were also reported to have critical roles in regulating plant abiotic stress tolerance, which would be useful for further genetic improvement (Nakashima et al., 2012).
The genome-wide identification and analysis of the NAC gene family will provide an important foundation to understand how abiotic stress response and signaling has evolved in plant development. In this study, a complete overview was reported on gene structure, duplication, and phylogenetic characteristics of M. sinensis NAC genes at the genome level. Also, an expression analysis of NAC genes that suffered from various abiotic stresses was comprehensively analyzed. The result will be helpful for further study of the functional characteristics of NAC genes in the mechanism of M. sinensis stress-responsive.
The M. sinensis genome resource was available in the PHYTOZOME database1 (Mitros et al., 2020). The HMM profile for the NAC transcription factor (PF02365) was downloaded from the Pfam database2 (Eddy, 2011). The NAC proteins from M. sinensis genome were identified from the annotated protein database and were used by HMMER 3.0 software (default parameters) on the similarity in sequence network with Hidden Markov Model (the cutoff value was set to 0.01; Finn et al., 2014; Lozano et al., 2015; Moturu et al., 2018). Then, the candidate NAC genes were further confirmed by Pfam and NCBI CDD. All candidate protein sequences through the batch sequence search of Pfam3 were uploaded to search for domain NAM structure (default parameters). Similarly, we uploaded all candidate protein sequences to NCBI batch CD-search4 for the structural domain search. Finally, the purified protein model5 was used to analyze the physicochemical parameters, including the CDs (Coding Sequence) length, molecular weight (MW), and theoretical isoelectric points (PI; Wilkins et al., 1999). Cis-acting regulatory elements were analyzed using PlantCARE software (Lescot et al., 2002). The online ProtComp 9.06 was used to predict the subcellular localization of the protein. The membrane-bound NAC proteins were predicted by TMHMM server v.2.07 (Zhou et al., 2018b).
The NAC protein sequences of Arabidopsis were retrieved from the TAIR 11 (Arabidopsis genome database; Rhee et al., 2003).8 The amino acid sequence alignment of M. sinensis and Arabidopsis thaliana NAC genes was performed by the combination of MEGA version 6.0 and Genedoc software (Nicholas et al., 1997), and the phylogenetic tree was constructed by using the MEGA 6.0 with Poisson model, using the NJ method with pairwise gaps deletion and 1,000 bootstraps (Tamura et al., 2011). According to the classification of AtNAC, the 261 M. sinensis NAC genes were divided into different subgroups.
To explore the exon/intron structure of M. sinensis NAC genes, the online Gene Structure Display Server 2.0 tools was used. The conserved motif of NAC proteins of M. sinensis was predicted by using the Multiple Em for Motif Elicitation (MEME)9 program with the default settings (Bailey et al., 2015).
The chromosomal positions information of NAC genes was acquired from the M. sinensis genome annotations. The chromosomal map of M. sinensis NAC genes was drafted by MG2C.10 Segmental and tandem duplications have been suggested to represent two of the main causes of gene family expansion in plants (Cannon et al., 2004). Segmental duplications of multiple genes through polyploidy followed by chromosome rearrangements were carried out (Yu et al., 2005). Tandem duplications were characterized as multiple members of one family occurring within the same intergenic region or in neighboring intergenic regions (Ramamoorthy et al., 2008). In this study, MCScanX (default parameters) was used to examine segmental and tandem duplications (Wang et al., 2012). The corresponding plot was created by Dual Synteny Plot for MCscanx in TBtools platform11 (Chen et al., 2020), and the NAC gene ID of M. sinensis was highlighted to obtain homologous pairs. Thus, the syntenic analysis maps between M. sinensis and other 10 plants (Brachypodium distachyon, Panicum hallii, Saccharum spontaneum, Setaria italic, Sorghum bicolor, Z. mays, M. truncatula, G. max, Trifolium repens, and A. thaliana) were constructed.
The M. sinensis plant seeds “0819” were collected from 29°57′30.1″, 102°24′36.4″, and 1,388 m asl. The plastic pots with a matrix of quartz sand were used to plant the M. sinensis seedlings, and 1X Hoagland’s nutrient solution was used to cultivate. Plant materials were grown under a constant temperature (28/25°C with cycles of 12/12 h for light and darkness), respectively, in the incubator. When the plants had 5–6 leaves, five different stress treatments started. These stress conditions were set to evaluate the gene expression pattern, including drought stress (20% PEG 6000), arsenic stress (As5+, 200 mg/L Na2HAsO4·7H2O), chromium stress (Cr6+, 200 mg/L K2Cr2O7), and cadmium stress (Cd2+, 200 mg/L CdCl2·2.5H2O), and salt stress (300 mmol/L NaCl). The plants were separately treated by five stresses for 6 days. At 0, 1, 3, and 6 days, samples under each treatment were obtained with three independent biological replicates. After that, all samples were immediately frozen in liquid nitrogen and then were kept at −80°C for RNA extraction.
Direct-zol™ RNA MiniPrep Kit (Zymo Research, China) was used to extract total RNA according to the manufacturer’s instructions. The genomic DNA was eliminated by DNase I (6 U/μl; Zymo Research, China). A NanoDrop ND-2000 (Thermo Scientific, United States) spectrophotometer and 1% agarose gel electrophoresis were used to detect the RNA quality of all samples. The cDNA was synthesized with The 5X All-In-One RT MasterMix (with AccuRT Genomic DNA Removal Kit) by following instructions, including total RNA 2 μg, 4X AccuRT Reaction Mix 2 μl, 5X AccuRT Reaction Stopper 2 μl, and 5X All-In-One RT MasterMix 4 μl. All cDNA was stored in reserve at −20°C. The qRT-PCR technique was performed to validate the expression of M. sinensis NAC genes selected from SNAC subgroup, and Premier 3.0 was used to design the specific primers (Table 1). A total of 10 μl volume consists of 5 μl abm® EvaGreen 2X qPCR Master Mix (Applied Biological Materials, Inc., Canada), 2.9 μl ddH2O, 1.5 μl synthesized cDNA product, and 0.3 μl each primer. Real-time RT-PCR reaction procedures are as follows: one cycle of 95°C for 10 min to activate enzyme, 95°C for 15 s for denaturation, and 60°C for 60 s for anneal/extension, for a total of 35 cycles. Finally, Tm and melting curve analysis were obtained to verify the specificity of each primer. The relative gene expression values were analyzed using the 2−ΔΔCt method (Schmittgen and Livak, 2008). The reference gene (Unigene33024) was set to standardize the expression data (Zhong et al., 2020).
To identify the putative NAC proteins in M. sinensis genome, a HMM (hidden Markov model) search was performed (NAM domain, PF02365) against the M. sinensis genome downloaded from Phytozome database.1 Subsequently, 261 putative M. sinensis NAC genes were identified after removing redundant sequences preliminarily. The protein sequence length of all M. sinensis NAC proteins ranged from 71 (MisinT039500.1) to 691 (Misin18G098700.1) amino acids with an average of 357. The molecular weights (MW) of the proteins ranged from 7575.7 to 77657.7 Da, and the theoretical isoelectric points (pI) ranged from 4.14 (Misin14G114600.1) to 10.88 (Misin13G160800.1), with an average of 6.92. The subcellular localization of 261 M. sinensis NAC proteins was predicted as nuclear (56.3%) or extracellular (43.7%). The basic information of all identified M. sinensis NAC genes, including the coding sequence length (CDs), protein sequence length, and genomic location, was mentioned in Supplementary Table S1. A further 12 M. sinensis NAC proteins were identified, which contained α-helical trans-membrane (TM) domain (Supplementary Table S1). They may respond to abiotic stress (Zhou et al., 2018b), and are highly homologous to Arabidopsis NAC2 and TIP, respectively.
An unrooted neighbor-joining (NJ) tree (with 1,000 bootstraps) was constructed by using the amino acid sequence alignment of NAC proteins from M. sinensis and A. thaliana (Figure 1). The results showed that 261 M. sinensis NAC genes could be divided into 15 subgroups, ANAC063, SENU5, ONAC003, AtNAC3, ANAC001, ATAF, ANAC011, NAP, OsNAC7, NAC1, NAC2, ONAC022, TIP, NAM, and a M. sinensis-specific subgroup named MisinNAC together with A. thaliana. The ANAC001 subgroup contained only two M. sinensis NAC proteins, and the ANAC063 subgroup contained the most members of M. sinensis NAC proteins. However, no NAC members in M. sinensis were detected from the OsNAC8. Besides, many stress-responsive NAC genes that regulate gene expression associated with plant abiotic stress as factors were assigned to the SNAC subgroup, including AtNAC3, NAP, and ATAF in the present study. Cis-elements analysis of SNAC genes in M. sinensis showed that most of genes may be associated in a variety of plant stress response pathways and hormonal regulation, such as auxin, abscisic acid, and MeJA (Supplementary Table S2).
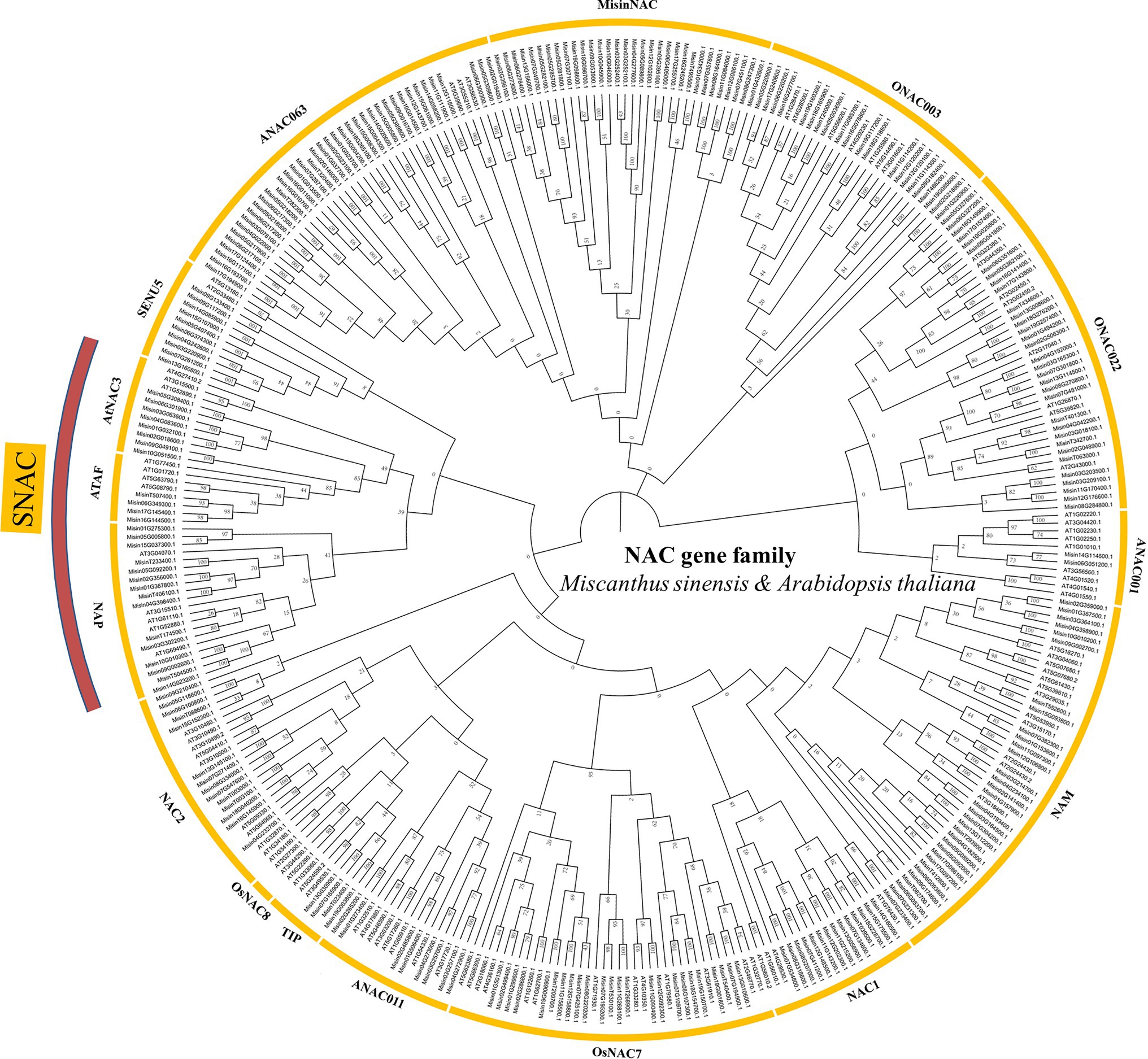
Figure 1. Unrooted phylogenetic tree representing relationships among the NAC proteins of Miscanthus sinensis and Arabidopsis thaliana. NAC genes were used to construct the Neighbor-Joining tree with the parameters including a Blosum62 cost matrix, the Jukes-Cantor model, global alignment, and bootstrap value of 1,000.
Ten conserved motifs were identified in the MEME tool to reveal the structural similarity and diversification of M. sinensis NAC proteins (Figure 2). The length of these conserved motifs was varied (11–57 amino acids) and showed a highly diverse distribution. And the details of the sequence logos of each motif were listed in Supplementary Figure S1. Misin10G045900.1 only contains one motif, whereas eight types motifs were present in Misin19G098600.1 (which contained the highest number). In the NAC2, AtNAC3, ATAF, and NAP subgroups, motif 1–6 and motif 8 were the most conserved parts. Motif 10 only appeared in ONAC003, ANAC063, SENU5, and MisinNAC, which may reveal that it was a typical part of these subgroups. Similar motif compositions and similar positions were exhibited in M. sinensis NAC genes (which located in the same subgroup), indicating that genes in the same subgroup may have similar functions. However, the specific biological function of most of these motifs is not classified and requires further investigation. The structural features of all identified NAC genes in M. sinensis were analyzed to estimate the evolution of the NAC gene family. As shown in Figure 2, among the coding sequences of the entire NAC genes, 18 (approximately 6.90%) were intronless, and over half (132, approximately 50.57%) had three exons. Only three genes (Misin11G114300.1, Misin09G174600.1, and Misin01G432600.1) had more than seven exons. Most genes from the same subgroups had a similar exon/intron structure, and Misin18G098700.1 had more than 20 kb gene structure, while MisinT039500.1 had the shortest structure with 216 bp.
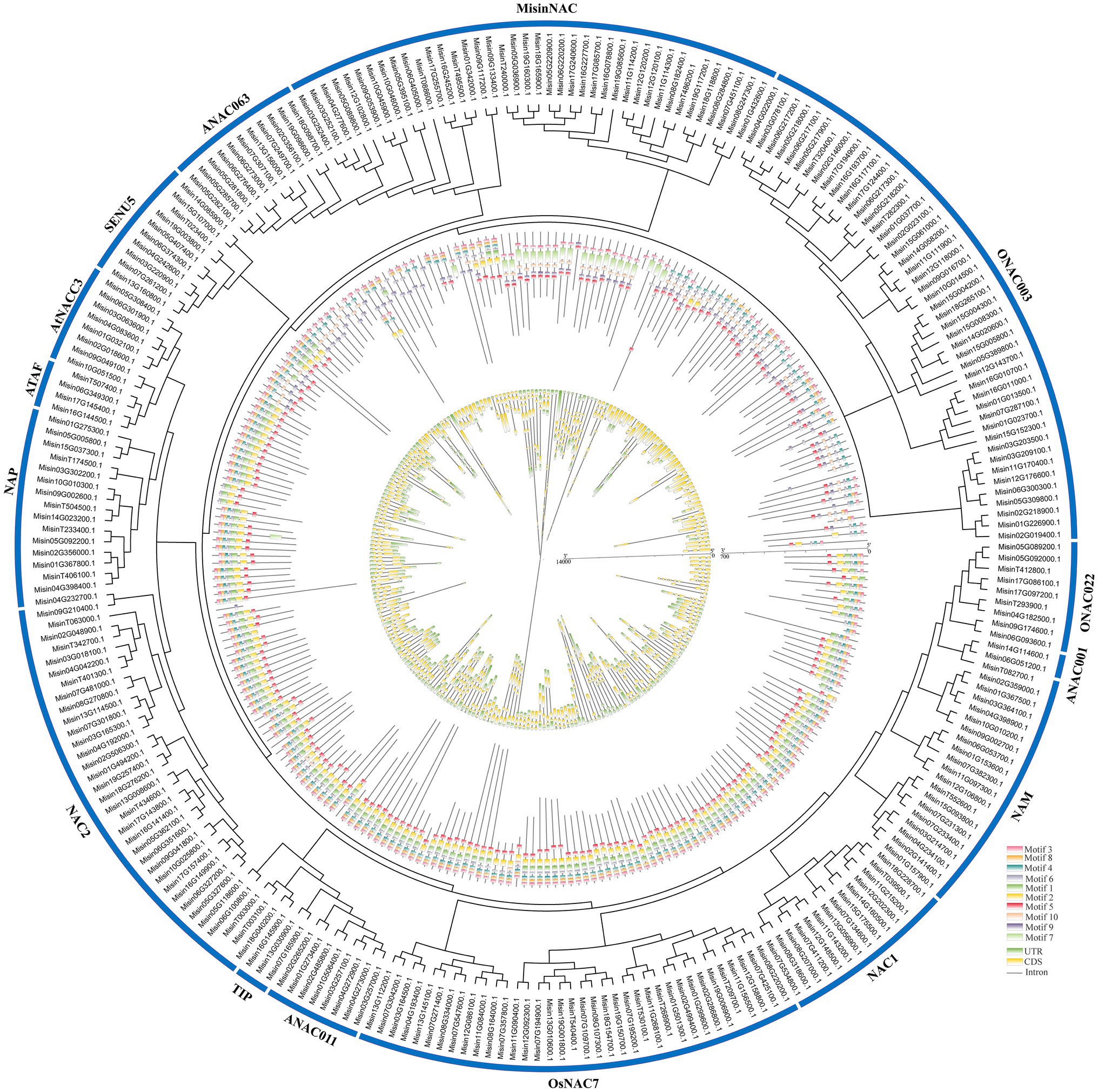
Figure 2. Phylogenetic relationships, motifs pattern, and gene structure of NAC genes from M. sinensis. The unrooted phylogenetic tree was constructed based on the full-length sequences of NAC proteins. The motifs composition of the NAC proteins was identified by MEME, and grey lines represent the non-conserved sequences. Each motif is indicated by a colored box numbered at the bottom, and the sequence information for each motif is provided in Supplementary Figure S1. The exon-intron structure of NAC genes was performed by using the online tool GSDS. Lengths of exons and introns of each M. sinensis NAC gene were exhibited proportionally.
The genomic locations of M. sinensis NAC are shown in Figure 3 (Supplementary Table S1). A total of 234 NAC genes were unevenly distributed on 19 chromosomes. The maximum number of NAC genes exhibited in Chromosome 7 (22, ~9.40%), while the least number of NAC genes (7, ~2.99%) were located on chromosome 10. Additionally, the distribution of NAC genes on chromosomes 10, 11, and 12 was relatively concentrated. There were 27 NAC genes just mapping to the scaffolds of M. sinensis genome. Tandem duplication is an essential source for the origin and evolution of multigene families. In this study, only nine pairs of tandem duplicate genes in M. sinensis NAC gene family were identified after the duplication events of NAC genes were examined, which were linked with the red line (Figure 4). There were 121 segmental duplication gene pairs identified in M. sinensis NAC gene family in all subpopulations. The results suggested that gene duplication, especially segmental duplication, may provide the primary driving force of evolution and may be possibly associated with the amplification of M. sinensis NAC gene family.
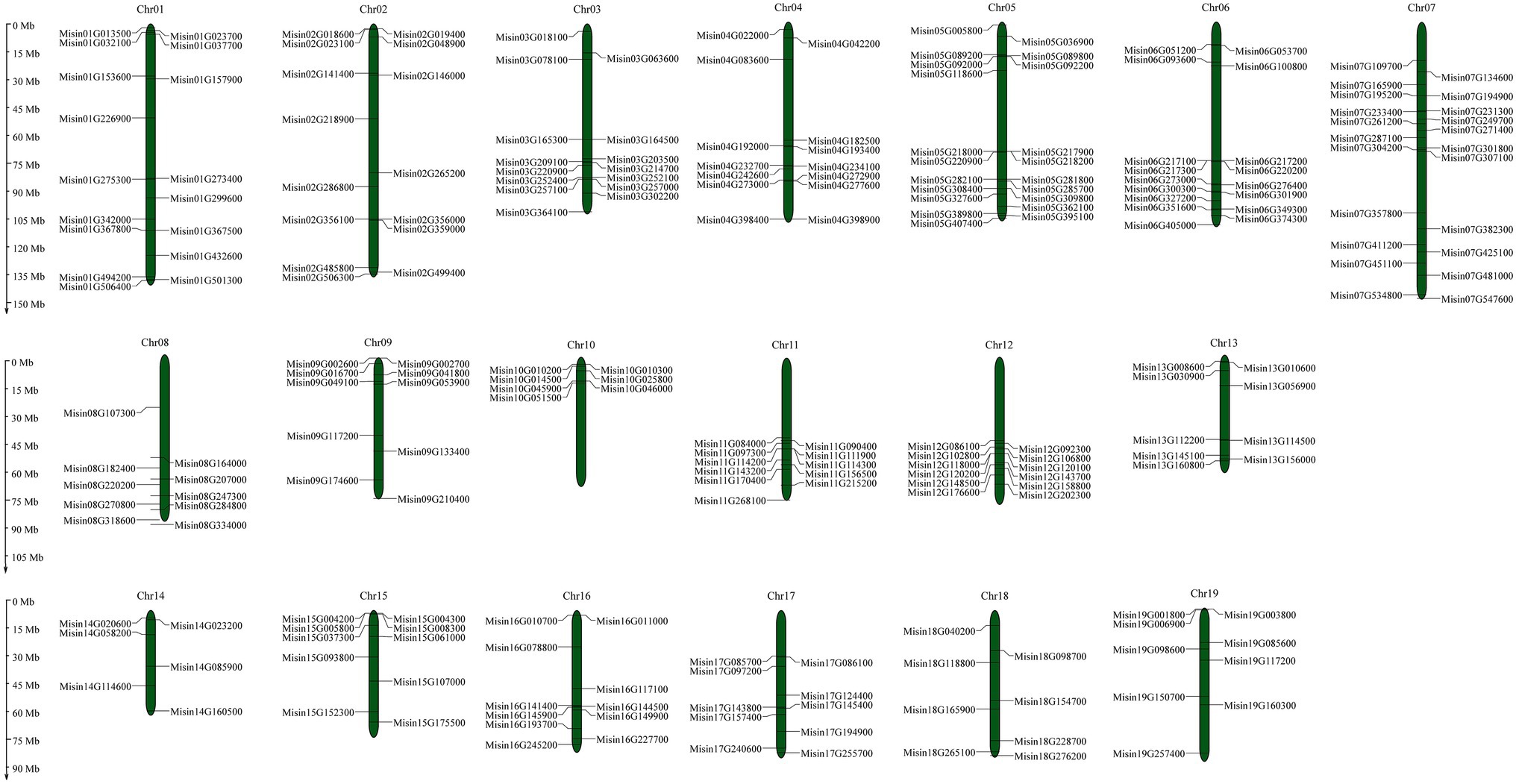
Figure 3. Distribution of 234 M. sinensis NAC genes on 19 chromosomes. Vertical bars represent the chromosomes, and the chromosome number is to the top of each chromosome. The scale on the left represents chromosome length (Mb).
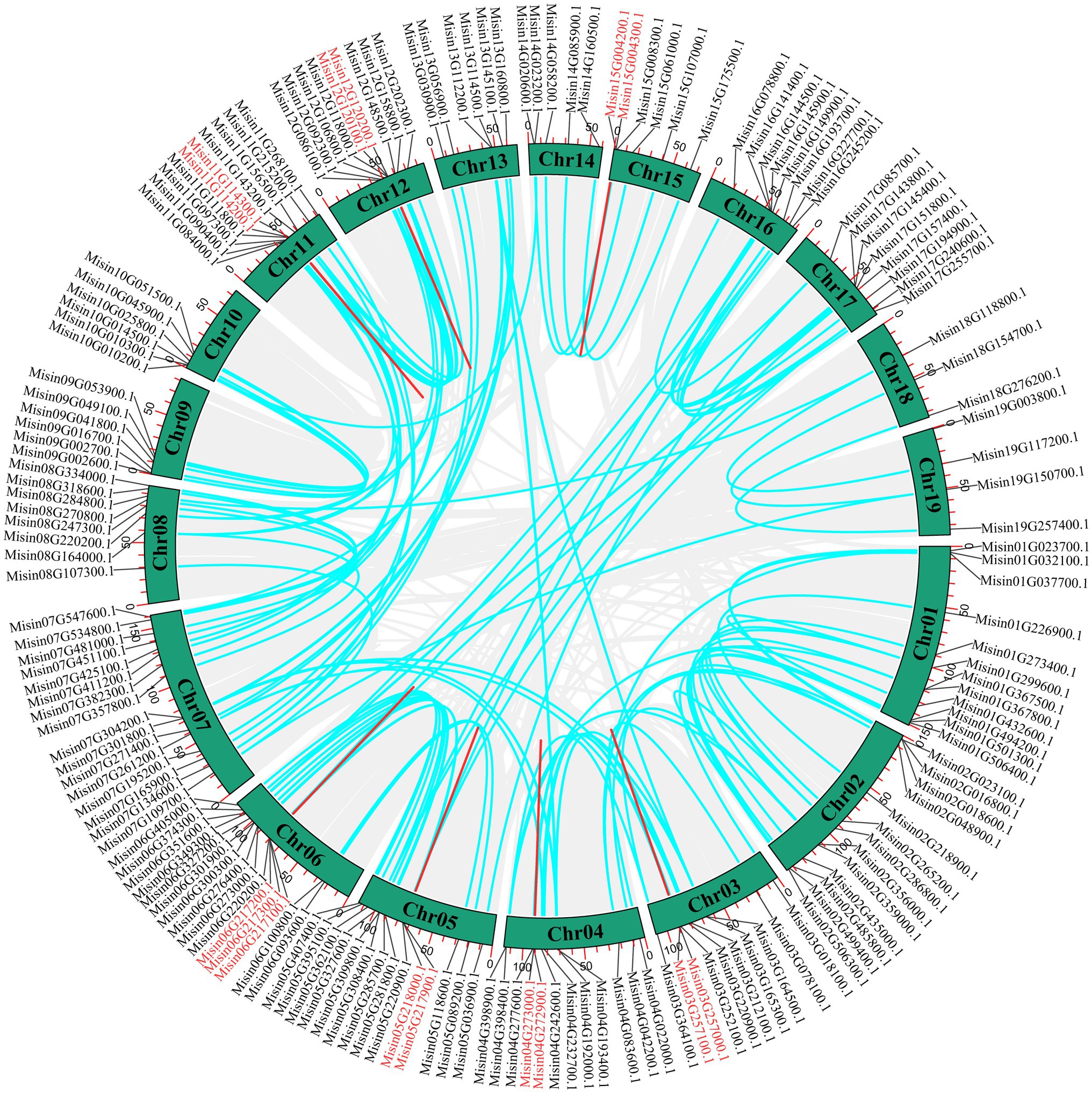
Figure 4. Schematic representations of the interchromosomal relationships of M. sinensis NAC genes. Gray lines indicate all synteny blocks in the genome, and the blue lines indicate segmental duplicates of NAC gene pairs, and the red lines indicate tandem duplications of NAC gene pairs.
In order to further explore the evolutionary relationship of NAC gene family in M. sinensis, 10 comparative syntenic maps consisting of four dicotyledonous plant (A. thaliana, M. truncatula, G. max, and T. repens) and six monocotyledonous plants (B. distachyon, P. hallii, S. spontaneum, S. italic, S. bicolor, and Z. mays) were constructed (Figure 5). The number of homologous pairs between M. sinensis and 10 other species was 2, 8, 25, 55, 148, 202, 205, 210, 211, and 215, respectively (Supplementary Table S3). By comparison, it was found that there are more homologous genes between M. sinensis and monocotyledons, indicating that these species were associated with the phylogenetic relationship.
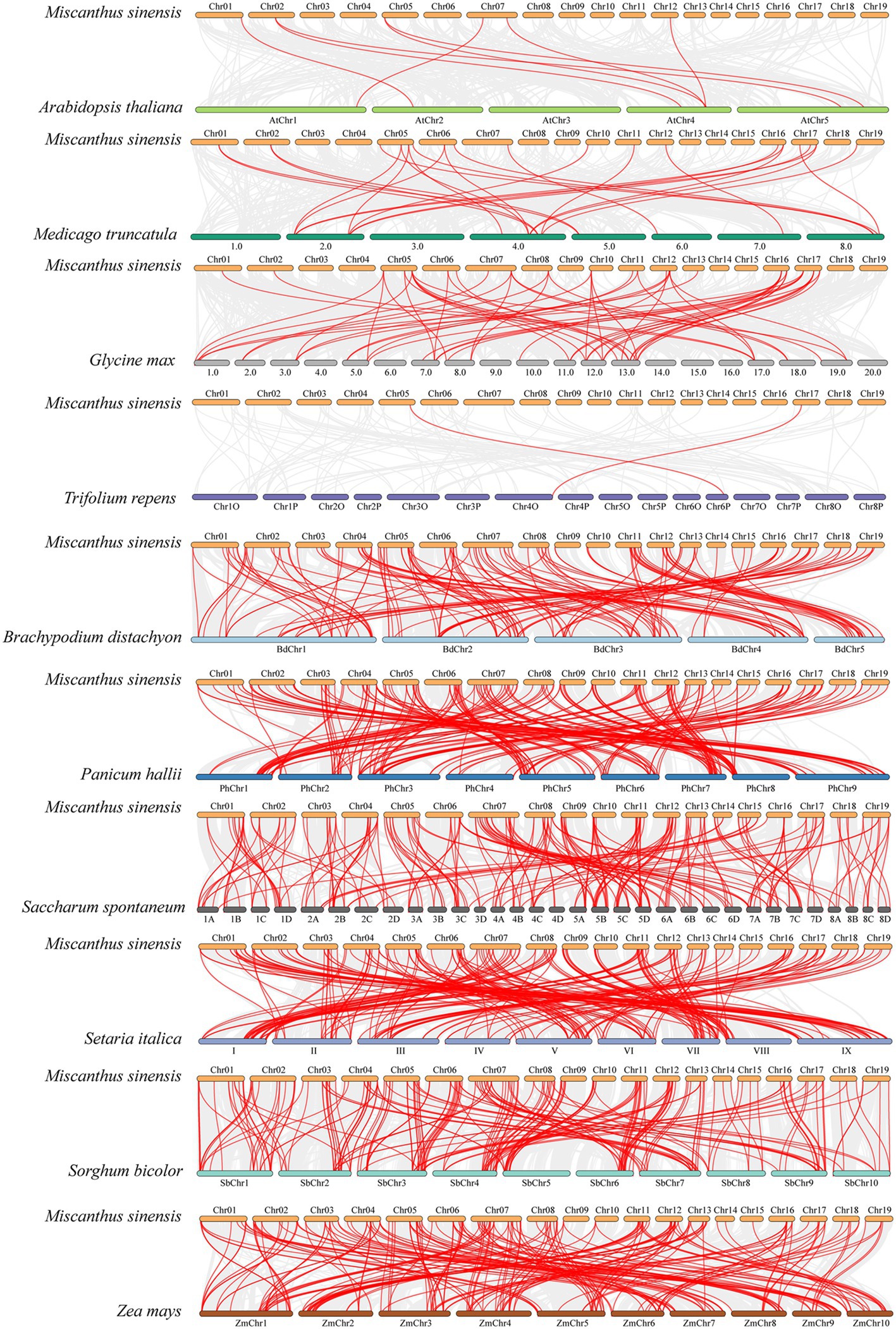
Figure 5. Synteny analysis of NAC genes between M. sinensis and 10 representative plant species. Gray lines in the background indicate the collinear blocks within the M. sinensis and other plant genomes, whereas the red lines highlight the syntenic NAC gene pairs.
It is evident that SNAC plays a vitally important role in the regulation of plant stress tolerance. To investigate the expression pattern of NAC genes under abiotic stress, 14 M. sinensis NAC genes were selected based on the phylogenetic tree in SNAC subgroup. Under salt treatment (Figure 6), most genes were shown to be significantly upregulated and obviously occurred at 1d (Misin03G063600.1 and Misin09G049100.1), 3d (Misin05G005800.1, Misin01G367800.1, Misin04G398400.1, Misin09G002600.1, and MisinT504500.1), or 6d (Misin06G301900.1, Misin06G349300.1, Misin17G145400.1, Misin16G144500.1, and Misin05G092200.1), which indicates an emergency rescue for plant response to stress (Figure 6).
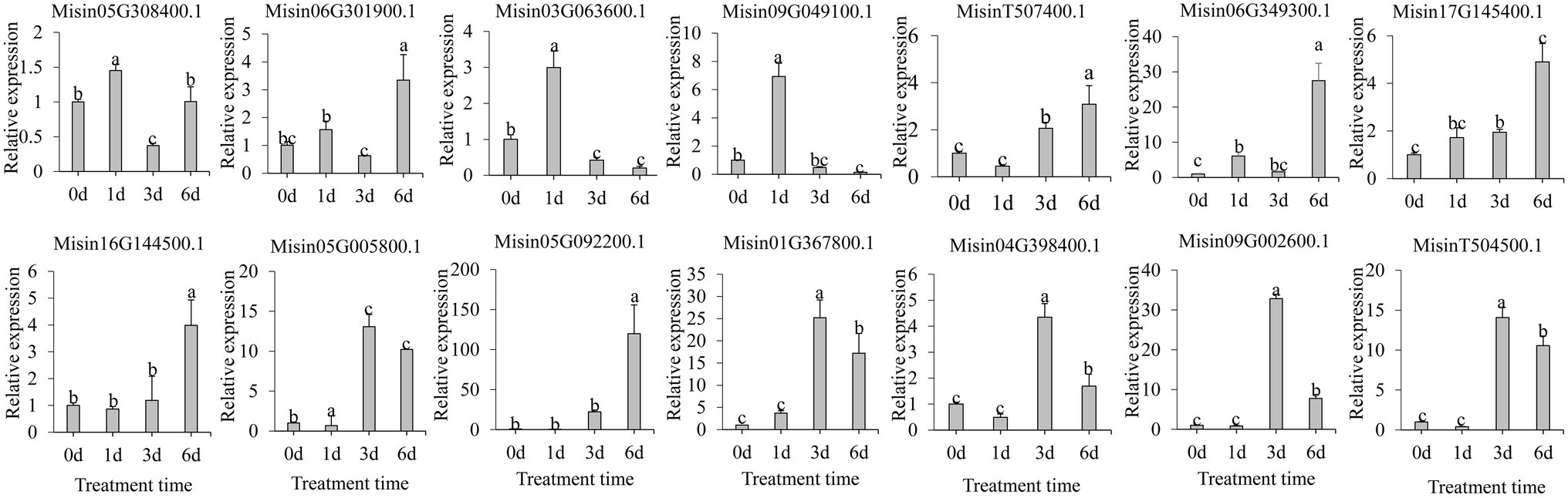
Figure 6. Expression profiles of 14 selected M. sinensis NAC genes in response to salinity treatment. Data were normalized to reference gene Unigene33024, and vertical bars indicate SD. No common letter above bar indicates a significant difference by least significant difference (LSD; p < 0.05).
In contrast, the gene expression profile was very different between drought and salt treatments. The expression of most selected candidate genes did not result in significant fold changes, and some genes were shown to be downregulated (MisinT507400.1, Misin17G145400.1, Misin16G144500.1, and Misin04G398400.1) after exposure to PEG treatment (Figure 7).
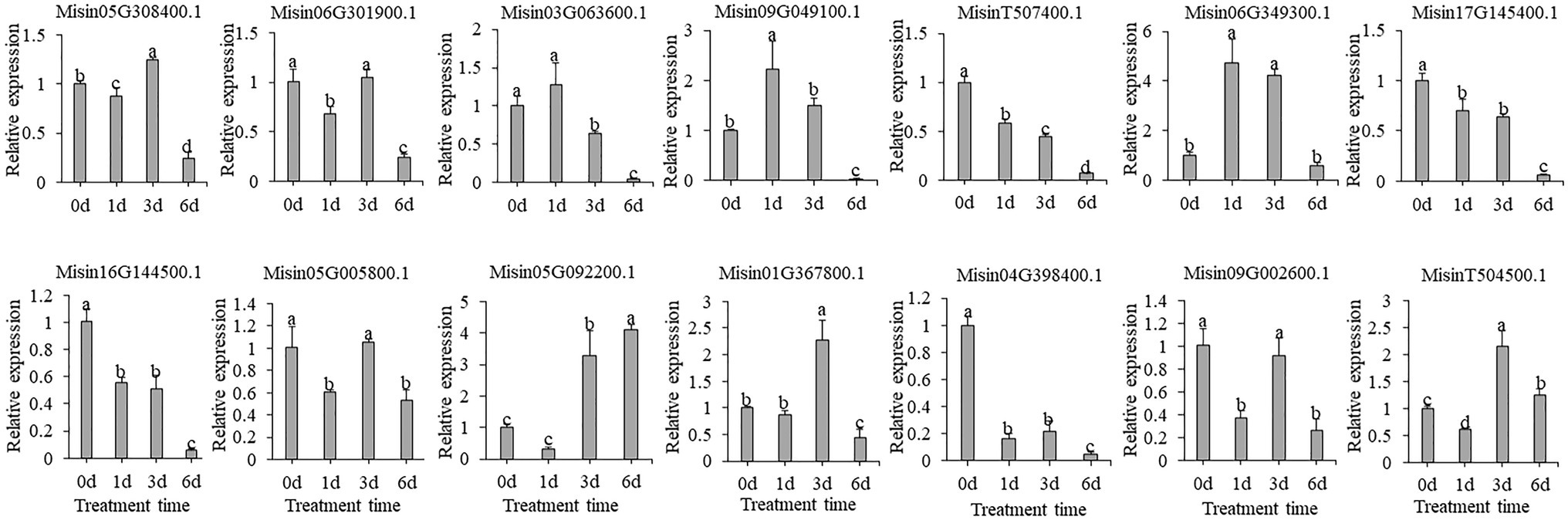
Figure 7. Expression profiles of 14 selected M. sinensis NAC genes in response to PEG treatment. Data were normalized to reference gene Unigene33024, and vertical bars indicate SD. No common letter above bar indicates a significant difference by least significant difference (LSD; p < 0.05).
Multiple NAC genes could be induced by heavy metal treatment. In Cr treatment, most genes had significantly increased expression, especially for Misin06G349300.1, Misin05G092200.1, and Misin01G367800.1. However, two NAC genes were downregulated during treatment time (Figure 8A). Consistently, there were also intense changes of gene expression under Cd treatment, and most genes upregulated continuously except for Misin05G308400.1 and Misin06G301900.1, for which peak values of relative expression appeared at 1d (Figure 8B). A similar gene expression pattern was found during As treatment. For instance, four NAC genes (Misin06G349300.1, Misin05G092200.1, Misin01G367800.1, and Misin09G002600.1) were highly induced and showed continuously increasing expression (Figure 8C). To determine gene function prediction, the gene expression pattern could provide crucial information.

Figure 8. Expression profiles of 14 selected M. sinensis NAC genes in response to (A) Cr (B) Cd and (C) As treatment. Data were normalized to reference gene Unigene33024, and vertical bars indicate SD. No common letter above bar indicates a significant difference by least significant difference (LSD; p < 0.05).
Miscanthus genus is one of the most promising non-food bioenergy crops belonging to the Tribe Andropogoneae (Poaceae), which contains many important C4 crops, including sugarcane (S. officinarum L.), sorghum (S. bicolor L.), and maize (Z. mays L.; Nie et al., 2016). However, owing to the complexity of Miscanthus genome, the lack of reliable genome information and unavailable genetic databases limited related research within the genus. Recently, chromosome-scale assembly of Miscanthus lutarioriparius genome (Miao et al., 2020), Miscanthus floridulus genome (Zhang et al., 2021), and M. sinensis genome (Mitros et al., 2020) was published, which would provide a valuable genetic resource for Miscanthus breeding improvement and molecular regulation network of stress resistance. The NAC gene family is a crucial plant-specific transcription factor family that is well known for its involvement in plant growth and developmental activities along with resistance to biotic and abiotic stresses (Wang et al., 2014; Dalman et al., 2017).
Attributing to the differences in genome size for each species, members of the NAC gene family identified in multiple plant species were varied. In A. thaliana, there were 117 NAC genes obtained from the genome database (Ooka et al., 2003), while 72 NAC genes have been identified in Lolium perenne (Nie et al., 2020), 80 in F. tataricum (Liu et al., 2019), 108 in Dactylis glomerata (Yang et al., 2021), 152 in G. max (Le et al., 2011), 151 in Oryza sativa (Nuruzzaman et al., 2010), 152 in Z. mays (Shiriga et al., 2014), and 288 in T. aestivum (Guérin et al., 2019). In this study, a total of 261 NAC genes were identified based on the M. sinensis genome (size of 1.88 G, Mitros et al., 2020), which is higher than most other plant species, indicating they contribute to the varied genome size (such as A. thaliana, 125 Mb; Korbinian et al., 2011): L. perenne, 2.04 Gb (Byrne et al., 2015); F. tataricum, 489.3 Mb (Zhang et al., 2017); D. glomerata, 1.78 Gb (Huang et al., 2020); G. max, 1.025 Gb (Shen et al., 2018); O. sativa, 466 Mb (Guillaume et al., 2003); and T. aestivum, 14.5 Gb (Appels et al., 2018). Continued gene duplication events occurred even after differentiation from their earliest ancestors. Besides, a collinearity analysis showed that there were nine groups of tandem duplication and 121 segmental duplication events in M. sinensis NAC gene family were identified. The duplication event of M. sinensis increases the genome size rather than increasing many NAC gene members, while tandem replication of NAC genes has been observed in many species. The result may be related to the expansion of long terminal repeat retrotransposons. Moreover, the structure of the genes was relatively conserved, while the length, MW, and pI of the NAC genes varied. The motifs of transcription factors are often related to DNA binding, protein interaction, and transcriptional activity (Liu et al., 2019). Owing to the function of NAC genes being predicted via alignment with model plants, such as Arabidopsis, the conserved motifs and gene structures may provide fundamental information for classification in M. sinensis (Nie et al., 2020).
Some NAC proteins have a TM motif in the C-terminus and show subcellular localization at the plasma membrane or endoplasmic reticulum. These proteins may be regulated by post-translational regulation under specific conditions (Seo et al., 2010; Kim et al., 2012; Lee et al., 2012). In this research, 12 M. sinensis NAC proteins were identified, which contained α-helical TM domain. In line with that, several species have membrane-bound NAC transcription factors, such as 13 identified in Arabidopsis, 13 in tomato (Solanum Lycopersicum L.), 11 in soybean, and five in rice, and most are induced by abiotic stresses (Kim et al., 2007; Le et al., 2011; Bhattacharjee et al., 2017). A previous study confirmed that the gene expression patterns can provide crucial information for determining gene function prediction (Zhou et al., 2018a). In this study, results showed that the NAP subgroup members Misin05G092200.1 and Misin01G367800.1 had increased expression under all five stress conditions, which was perhaps related to the abiotic stress resistance of M. sinensis. Interestingly, a previous study revealed that NAC genes in the NAP subgroup were involved in plant leaf senescence and floral morphogenesis, as well as associated to abscisic acid hormones regulating (Sablowski and Meyerowitz, 1998; Guo and Gan, 2006), which could be an important reason indicating stress resistance function of these kinds of genes. Besides, it was proved that AtSOG1ΔNAC (AT1G25580) was induced by salinity and the stability of SOG1 protein under salinity stress could be regulated by N-terminal NAC domain and the C-terminal (Mahapatra and Roy, 2019). Based on phylogenetic and conserved gene structure analysis, it was speculated that Misin11G114300.1 (together with AT1G25580, which were assigned into ONAC003 subgroup) had possible relevance to salinity stress. Arabidopsis resistance to drought, high salinity, and abscisic acid was significantly enhanced through the overexpression of AtNAC3 (Tran et al., 2004), which was similar to these homologous genes in M. sinensis from expression results. However, although Misin05G308400.1 and Misin06G301900.1 were duplicated genes within the same subgroup (AtNAC3), the expression pattern of Misin05G308400.1 was different from that of Misin06G301900.1 in salinity treatment, which might be caused by variation in gene regulation after duplication events. The result was consistent with a previous study in F. tataricum that showed the expression pattern between FtNAC15 and FtNAC38. An Arabidopsis NAC transcription factor family gene (AtNAP) functions as a negative regulator via transcriptional repression of AREB1 under salt stress (Seok et al., 2017). Expression of the SNAC gene Misin09G002600.1 in M. sinensis significantly reached its peak value at 6d, which was over 100 times higher than the control in salinity stress. In general, these findings supported the view that the NAC proteins in SNAC subgroup actively participated in plant development and stress response (Takasaki et al., 2010, 2015), and these candidate genes with putative functions could be further validated in future.
In addition, a previous study had shown that MsSND1 (annotated Misin19G001800.1 in the genome belonging to the subgroup OsNAC7) is an NAC transcriptional master regulator orchestrating secondary cell wall biosynthesis in Miscanthus (Golfier et al., 2017), and the biosynthesis of lignin could directly reduce the entry of heavy metals into the root system (Cheng et al., 2014). Consequently, the predicted function of Misin19G001800.1 may play a role in the tolerance of M. sinensis to heavy metals, which would provide a new insight to further verify the function of M. sinensis NAC genes.
The release of M. sinensis genome provided a basic platform for the genome-wide investigation of NAC proteins. A total of 261 M. sinensis NAC genes were identified, and a comprehensive analysis of the gene family was presented in this study, including chromosomal distribution, conserved motif compositions, gene structure, and gene duplications. Results showed that gene length, MW, and pI of NAC family were varied, while gene structure and motifs were relatively conserved. Chromosomal mapping analysis found that the M. sinensis NAC genes were unevenly distributed on 19 M. sinensis chromosomes, and the interchromosomal evolutionary analysis showed that nine pairs of tandem duplicate genes and 121 segmental duplications were identified. The expression patterns of 14 genes from M. sinensis SNAC subgroup were analyzed under high salinity, PEG, and heavy metals, and multiple NAC genes could be induced by the treatment, indicating that abiotic stress-responsive NAC-type transcription factors play important roles for improving the stress tolerance of M. sinensis. However, further investigation of extensive functional characterization of the NAC gene family in M. sinensis is needed. Consistently, current results will provide very useful information for a follow-up study of NAC genes in stress-response and potential genetic improvement for better adaptivity in marginal land.
Publicly available data sets were analyzed in this study. These data can be found at: https://phytozome-next.jgi.doe.gov/info/Msinensis_v7_1.
XZ, GN, YP, and LH contributed to experiment design and funding acquisition. Material preparation and data collection were performed by ZY, JH, AL, and JC. DL, ZY, JH, GF, and XW analyzed data. The first draft of the manuscript was written by GN, ZY, SW, and JH. All authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
This work was supported by China Agriculture Research System of MOF and MARA, Grassland Basic Resources Investigation Research in China (2017FY100602), and the National Natural Science Foundation of China (NSFC 31801432).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.766550/full#supplementary-material
Supplementary Figure S1 | The sequence information for each motif.
1. ^https://phytozome-next.jgi.doe.gov/info/Msinensis_v7_1
3. ^http://pfam.xfam.org/search#tabview=tab1
4. ^https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi
5. ^https://web.expasy.org/protparam/
6. ^http://linux1.softberry.com/berry.phtml
7. ^http://www.cbs.dtu.dk/services/TMHMM/
8. ^https://www.arabidopsis.org/
9. ^http://meme-suite.org/tools/meme
Aida, M., Ishida, T., Fukaki, H., Fujisawa, H., and Tasaka, M. (1997). Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell 9, 841–857. doi: 10.1105/tpc.9.6.841
Appels, R., Eversole, K., Stein, N., Feuillet, C., Keller, B., Rogers, J., et al. (2018). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361:eaar7191. doi: 10.1126/science.aar7191
Bailey, T. L., Johnson, J., Grant, C. E., and Noble, W. S. (2015). The MEME suite. Nucleic Acids Res. 43, W39–W49. doi: 10.1093/nar/gkv416
Bhattacharjee, P., Das, R., Mandal, A., and Kundu, P. (2017). Functional characterization of tomato membrane-bound NAC transcription factors. Plant Mol. Biol. 93, 511–532. doi: 10.1007/s11103-016-0579-z
Byrne, S. L., Nagy, I., Pfeifer, M., Armstead, I., Swain, S., Studer, B., et al. (2015). A synteny-based draft genome sequence of the forage grass. Plant J. 84, 816–826. doi: 10.1111/tpj.13037
Byrt, C., Grof, C., and Furbank, R. (2011). C4 plants as biofuel feedstocks: optimising biomass production and feedstock quality from a lignocellulosic perspective. J. Integr. Plant Biol. 53, 120–135. doi: 10.1111/j.1744-7909.2010.01023.x
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D., and May, G. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 4:10. doi: 10.1186/1471-2229-4-10
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, H. M., He, Y., et al. (2020). Tbtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Cheng, H., Zhao, Y. J., Liu, Y., Ye, Z. H., Wu, M. L., Sun, C. C., et al. (2014). Metal (Pb, Zn and cu) uptake and tolerance by mangroves in relation to root anatomy and lignification/suberization. Tree Physiol. 34, 646–656. doi: 10.1093/treephys/tpu042
Clifton-Brown, J. C., and Lewandowski, I. (2002). Screening Miscanthus genotypes in field trials to optimise biomass yield and quality in southern Germany. Eur. J. Agron. 16, 97–110. doi: 10.1016/S1161-0301(01)00120-4
Dalman, K., Wind, J., Nemesio-Gorriz, M., Hammerbacher, A., Lundén, K., Ezcurra, I., et al. (2017). Overexpression of PaNAC03, a stress induced NAC gene family transcription factor in Norway spruce leads to reduced flavonol biosynthesis and aberrant embryo development. BMC Plant Biol. 17:6. doi: 10.1186/s12870-016-0952-8
Eddy, S. R. (2011). Accelerated profile HMM searches. PLoS Comput. Biol. 7:e1002195. doi: 10.1371/journal.pcbi.1002195
Ezaki, B., Nagao, E., Yamamoto, Y., Nakashima, S., and Enomoto, T. (2008). Wild plants, Andropogon virginicus L. and Miscanthus sinensis Anders, are tolerant to multiple stresses including aluminum, heavy metals and oxidative stresses. Plant Cell Rep. 27, 951–961. doi: 10.1007/s00299-007-0503-8
Finn, R. D., Bateman, A., Clements, J., Coggill, P., Eberhardt, R. Y., Eddy, S. R., et al. (2014). Pfam: the protein families database. Nucleic Acids Res. 42, D222–D230. doi: 10.1093/nar/gkt1223
Guillaume, B., Karsten, H., and Kenneth, H. W. (2003). A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res. 13, 137–144. doi: 10.1101/gr.751803
Golfier, P., Volkert, C., He, F., Rausch, T., and Wolf, S. (2017). Regulation of secondary cell wall biosynthesis by a nac transcription factor from miscanthus. Plant Direct 1:e00024. doi: 10.1002/pld3.24
Greef, J. M., Deuter, M., Jung, C., and Schondelmaier, J. (1997). Genetic diversity of European Miscanthus species revealed by AFLP fingerprinting. Genet. Resour. Crop. Evol. 44, 185–195. doi: 10.1023/A:1008693214629
Guérin, C., Roche, J., Allard, V., Ravel, C., Mouzeyar, S., and Bouzidi, M. F. (2019). Genome-wide analysis, expansion and expression of the NAC family under drought and heat stresses in bread wheat (T. aestivum L.). PLoS One 14:e213390. doi: 10.1371/journal.pone.0213390
Guo, Y., and Gan, S. (2006). AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J. 46, 601–612. doi: 10.1111/j.1365-313X.2006.02723.x
Guo, H. S., Xie, Q., Fei, J. F., and Chua, N. H. (2005). MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. Plant Cell 17, 1376–1386. doi: 10.1105/tpc.105.030841
Heaton, E. A., Long, S. P., Voigt, T. B., Jones, M. B., and Clifton-Brown, J. (2004). Miscanthus for renewable energy generation: European Union experience and projections for Illinois. Mitig. Adapt. Strateg. Glob. Chang. 9, 433–451. doi: 10.1023/B:MITI.0000038848.94134.be
Huang, L., Feng, G., Yan, H., Zhang, Z., Bushman, B. S., Wang, J., et al. (2020). Genome assembly provides insights into the genome evolution and flowering regulation of orchardgrass. Plant Biotechnol. J. 18, 373–388. doi: 10.1111/pbi.13205
Jensen, M. K., Kjaersgaard, T., Nielsen, M. M., Galberg, P., Petersen, K., O’shea, C., et al. (2010). The Arabidopsis thaliana NAC transcription factor family: structure–function relationships and determinants of ANAC019 stress signalling. Biochem. J. 426, 183–196. doi: 10.1042/BJ20091234
Kim, S. Y., Kim, S. G., Kim, Y. S., Seo, P. J., Bae, M., Yoon, H. K., et al. (2007). Exploring membrane-associated NAC transcription factors in Arabidopsis: implications for membrane biology in genome regulation. Nucleic Acids Res. 35, 203–213. doi: 10.1093/nar/gkl1068
Kim, M. J., Park, M. J., Seo, P. J., Song, J. S., Kim, H. J., and Park, C. M. (2012). Controlled nuclear import of the transcription factor NTL6 reveals a cytoplasmic role of SnRK2.8 in the drought-stress response. Biochem. J. 448, 353–363. doi: 10.1042/BJ20120244
Korbinian, S., Stephan, O., Felix, O., Klein, J. D., Xi, W., Christa, L., et al. (2011). Reference-guided assembly of four diverse Arabidopsis thaliana genomes. Proc. Natl. Acad. Sci. U. S. A. 108, 10249–10254. doi: 10.1073/pnas.1107739108
Kou, X., Wang, S., Wu, M., Guo, R., Xue, Z., Meng, N., et al. (2014). Molecular characterization and expression analysis of NAC family transcription factors in tomato. Plant Mol. Biol. Report. 32, 501–516. doi: 10.1007/s11105-013-0655-3
Le, D. T., Nishiyama, R., Watanabe, Y., Mochida, K., Yamaguchi-Shinozaki, K., Shinozaki, K., et al. (2011). Genome-wide survey and expression analysis of the plant-specific NAC transcription factor family in soybean during development and dehydration stress. DNA Res. 18, 263–276. doi: 10.1093/dnares/dsr015
Lee, S., Seo, P. J., Lee, H. J., and Park, C. M. (2012). A NAC transcription factor NTL4 promotes reactive oxygen species production during drought-induced leaf senescence in Arabidopsis. Plant J. 70, 831–844. doi: 10.1111/j.1365-313X.2012.04932.x
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., VandePeer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Liu, M., Ma, Z., Sun, W., Huang, L., Wu, Q., Tang, Z., et al. (2019). Genome-wide analysis of the NAC transcription factor family in Tartary buckwheat (Fagopyrum tataricum). BMC Genomics 20:113. doi: 10.1186/s12864-019-6331-8
Long, S. P., and Spence, A. K. (2013). Toward cool C4 crops. Annu. Rev. Plant Biol. 64, 701–722. doi: 10.1146/annurev-arplant-050312-120033
Lozano, R., Hamblin, M. T., Prochnik, S., and Jannink, J. L. (2015). Identification and distribution of the NBS-LRR gene family in the cassava genome. BMC Genomics 16:360. doi: 10.1186/s12864-015-1554-9
Mahapatra, K., and Roy, S. (2019). An insight into the folding and stability of arabidopsis thaliana sog1 transcription factor under salinity stress in vitro. Biochem. Biophys. Res. Commun. 515, 531–537. doi: 10.1016/j.bbrc.2019.05.183
Miao, J., Feng, Q., Li, Y., Zhao, Q., Zhou, C., Lu, H., et al. (2020). Chromosome-scale assembly and analysis of biomass crop Miscanthus lutarioriparius genome. Nat. Commun. 12:2458. doi: 10.1038/s41467-021-22738-4
Mitros, T., Session, A. M., James, B. T., Wu, G. A., and Rokhsar, D. S. (2020). Genome biology of the paleotetraploid perennial biomass crop Miscanthus. Nat. Commun. 11:5442. doi: 10.1038/s41467-020-18923-6
Mohr, A., and Raman, S. (2013). Lessons from first generation biofuels and implications for the sustainability appraisal of second generation biofuels. Energy Policy 63, 114–122. doi: 10.1016/j.enpol.2013.08.033
Moturu, T. R., Thula, S., Singh, R. K., Nodzyński, T., Vařeková, R. S., Friml, J., et al. (2018). Molecular evolution and diversification of the SMXL gene family. J. Exp. Bot. 69, 2367–2378. doi: 10.1093/jxb/ery097
Nakashima, K., Takasaki, H., Mizoi, J., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2012). NAC transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 1819, 97–103. doi: 10.1016/j.bbagrm.2011.10.005
Nakashima, K., Tran, L. S., Van Nguyen, D., Fujita, M., Maruyama, K., Todaka, D., et al. (2007). Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J. 51, 617–630. doi: 10.1111/j.1365-313X.2007.03168.x
Nicholas, K. B., Nicholas, H. B., and Deerfield, D. W. (1997). GeneDoc: analysis and visualization of genetic variation. Eb News 4:14.
Nie, G., Huang, L., Zhang, X., Taylor, M., Jiang, Y., Yu, X., et al. (2016). Marker-trait association for biomass yield of potential bio-fuel feedstock Miscanthus sinensis from Southwest China. Front. Plant Sci. 7:802. doi: 10.3389/fpls.2016.00802
Nie, G., Liao, Z., Zhong, M., Zhou, J., Cai, J., Liu, A., et al. (2021). MicroRNA-mediated responses to chromium stress provide insight into tolerance characteristics of Miscanthus sinensis. Front. Plant Sci. 12:1019. doi: 10.3389/fpls.2021.666117
Nie, G., Tang, L., Zhang, Y., Huang, L., Ma, X., Cao, X., et al. (2017). Development of SSR markers based on transcriptome sequencing and association analysis with drought tolerance in perennial grass Miscanthus from China. Front. Plant Sci. 8:801. doi: 10.3389/fpls.2017.00801
Nie, G., Yang, X., Yang, Z., Zhong, M., Zhu, Y., Zhou, J., et al. (2020). Genome-wide investigation of the NAC transcript factor family in perennial ryegrass (Lolium perenne L.) and expression analysis under various abiotic stressor. Genomics 112, 4224–4231. doi: 10.1016/j.ygeno.2020.06.033
Nogueira, F. T., Schlögl, P. S., Camargo, S. R., Fernandez, J. H., Rosa, V. E., Pompermayer, P., et al. (2005). SsNAC23, a member of the NAC domain protein family, is associated with cold, herbivory and water stress in sugarcane. Plant Sci. 169, 93–106. doi: 10.1016/j.plantsci.2005.03.008
Nuruzzaman, M., Manimekalai, R., Sharoni, A. M., Satoh, K., Kondoh, H., Ooka, H., et al. (2010). Genome-wide analysis of NAC transcription factor family in rice. Gene 465, 30–44. doi: 10.1016/j.gene.2010.06.008
Ohnishi, T., Sugahara, S., Yamada, T., Kikuchi, K., Yoshiba, Y., Hirano, H. Y., et al. (2005). OsNAC6, a member of the NAC gene family, is induced by various stresses in rice. Genes Genet. Syst. 80, 135–139. doi: 10.1266/ggs.80.135
Ooka, H., Satoh, K., Doi, K., Nagata, T., Otomo, Y., Murakami, K., et al. (2003). Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 10, 239–247. doi: 10.1093/dnares/10.6.239
Ramamoorthy, R., Jiang, S. Y., Kumar, N., Venkatesh, P. N., and Ramachandran, S. (2008). A comprehensive transcriptional profiling of the WRKY gene family in rice under various abiotic and phytohormone treatments. Plant Cell Physiol. 49, 865–879. doi: 10.1093/pcp/pcn061
Rhee, S. Y., Beavis, W., Berardini, T. Z., Chen, G., Dixon, D., Doyle, A., et al. (2003). The Arabidopsis information resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res. 31, 224–228. doi: 10.1093/nar/gkg076
Sablowski, R. W., and Meyerowitz, E. M. (1998). A homolog of NO APICAL MERISTEM is an immediate target of the floral homeotic genes APETALA3/PISTILLATA. Cell 92, 93–103. doi: 10.1016/S0092-8674(00)80902-2
Schmittgen, T. D., and Livak, K. J. (2008). Analyzing real-time PCR data by the comparative C T method. Nat. Protoc. 3:1101. doi: 10.1038/nprot.2008.73
Seo, P. J., Kim, M. J., Park, J. Y., Kim, S. Y., Jeon, J., Lee, Y. H., et al. (2010). Cold activation of a plasma membrane-tethered NAC transcription factor induces a pathogen resistance response in Arabidopsis. Plant J. 61, 661–671. doi: 10.1111/j.1365-313X.2009.04091.x
Seok, H. Y., Woo, D. H., Nguyen, L. V., Tran, H. T., Tarte, V. N., Mehdi, S. M., et al. (2017). Arabidopsis AtNAP functions as a negative regulator via repression of AREB1 in salt stress response. Planta 245, 329–341. doi: 10.1007/s00425-016-2609-0
Shen, Y., Liu, J., Geng, H., Zhang, J., Liu, Y., Zhang, H., et al. (2018). De novo assembly of a Chinese soybean genome. Sci. China Life Sci. 61, 1–14. doi: 10.1007/s11427-018-9360-0
Shiriga, K., Sharma, R., Kumar, K., Yadav, S. K., Hossain, F., and Thirunavukkarasu, N. (2014). Genome-wide identification and expression pattern of drought-responsive members of the NAC family in maize. Meta Gene 2, 407–417. doi: 10.1016/j.mgene.2014.05.001
Takasaki, H., Maruyama, K., Kidokoro, S., Ito, Y., Fujita, Y., Shinozaki, K., et al. (2010). The abiotic stress-responsive NAC-type transcription factor OsNAC5 regulates stress-inducible genes and stress tolerance in rice. Mol. Gen. Genomics. 284, 173–183. doi: 10.1007/s00438-010-0557-0
Takasaki, H., Maruyama, K., Takahashi, F., Fujita, M., Yoshida, T., Nakashima, K., et al. (2015). SNAC-as, stress-responsive NAC transcription factors, mediate ABA-inducible leaf senescence. Plant J. 84, 1114–1123. doi: 10.1111/tpj.13067
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Tran, L. S., Nakashima, K., Sakuma, Y., Simpson, S. D., Fujita, Y., Maruyama, K., et al. (2004). Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16, 2481–2498. doi: 10.1105/tpc.104.022699
Tran, L. S., Quach, T. N., Guttikonda, S. K., Aldrich, D. L., Kumar, R., Neelakandan, A., et al. (2009). Molecular characterization of stress-inducible GmNAC genes in soybean. Mol. Genet. Genom. 281, 647–664. doi: 10.1007/s00438-009-0436-8
Van der Weijde, T., Alvim Kamei, C. L., Torres, A. F., Vermerris, W., Dolstra, O., Visser, R. G., et al. (2013). The potential of C4 grasses for cellulosic biofuel production. Front. Plant Sci. 4:107. doi: 10.3389/fpls.2013.00107
Wang, Z., Rashotte, A., Moss, A., and Dane, F. (2014). Two NAC transcription factors from Citrullus colocynthis, CcNAC1, CcNAC2 implicated in multiple stress responses. Acta Physiol. Plant. 36, 621–634. doi: 10.1007/s11738-013-1440-5
Wang, Y., Tang, H., DeBarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40:e49. doi: 10.1093/nar/gkr1293
Wilkins, M. R., Gasteiger, E., Bairoch, A., Sanchez, J. C., and Hochstrasser, D. F. (1999). Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 112, 531–552. doi: 10.1385/1-59259-584-7:531
Xu, Z., Wang, C., Xue, F., Zhang, H., and Ji, W. (2015). Wheat NAC transcription factor TaNAC29 is involved in response to salt stress. Plant Physiol. Biochem. 96, 356–363. doi: 10.1016/j.plaphy.2015.08.013
Xue, G. P., Way, H. M., Richardson, T., Drenth, J., Joyce, P. A., and McIntyre, C. L. (2011). Overexpression of TaNAC69 leads to enhanced transcript levels of stress up-regulated genes and dehydration tolerance in bread wheat. Mol. Plant 4, 697–712. doi: 10.1093/mp/ssr013
Yang, Z., Nie, G., Feng, G., Han, J., Huang, L., Zhang, X., et al. (2021). Genome-wide identification, characterization, and expression analysis of the NAC transcription factor family in orchardgrass (Dactylis glomerata L.). BMC Genomics 22:178. doi: 10.1186/s12864-021-07485-6
Ye, B., Saito, A., and Minamisawa, K. (2005). Effect of inoculation with anaerobic nitrogen-fixing consortium on salt tolerance of Miscanthus sinensis. Soil Sci. Plant Nutr. 51, 243–249. doi: 10.1111/j.1747-0765.2005.tb00028.x
Yu, J., Wang, J., Lin, W., Li, S., Li, H., Zhou, J., et al. (2005). The genomes of Oryza sativa: a history of duplications. PLoS Biol. 3:e38. doi: 10.1371/journal.pbio.0030038
Yu, Y., Yi, Z., and Zhou, G. (2014). Research progress and comprehensive utilization of Miscanthus. Chin. Bull. Life Sci. 26, 474–480. doi: 10.13376/j.cbls/2014070
Zhang, G., Ge, C., Xu, P., Wang, S., Chen, S., Han, Y., et al. (2021). The reference genome of Miscanthus floridulus illuminates the evolution of saccharinae. Nat. Plants 7, 1–11. doi: 10.1038/s41477-021-00908-y
Zhang, L., Li, X., Ma, B., Gao, Q., Du, H., Han, Y., et al. (2017). The Tartary buckwheat genome provides insights into Rutin biosynthesis and abiotic stress tolerance. Mol. Plant 10, 1224–1237. doi: 10.1016/j.molp.2017.08.013
Zhao, Q., Gallego-Giraldo, L., Wang, H., Zeng, Y., Ding, S. Y., Chen, F., et al. (2010). An NAC transcription factor orchestrates multiple features of cell wall development in Medicago truncatula. Plant J. 63, 100–114. doi: 10.1111/j.1365-313X.2010.04223.x
Zhong, M., Yang, X., Hu, Y., Huang, L., Peng, Y., Li, Z., et al. (2020). Identification of candidate reference genes for quantitative RT-PCR in Miscanthus sinensis subjected to various abiotic stresses. Mol. Biol. Rep. 47, 2913–2927. doi: 10.1007/s11033-020-05392-9
Zhou, Q., Zhang, S., Chen, F., Liu, B., Wu, L., Li, F., et al. (2018a). Genome-wide identification and characterization of the SBP-box gene family in petunia. BMC Genomics 19:193. doi: 10.1186/s12864-018-4537-9
Keywords: Miscanthus sinensis, NAC transcription factor, gene family, abiotic stress, gene expression
Citation: Nie G, Yang Z, He J, Liu A, Chen J, Wang S, Wang X, Feng G, Li D, Peng Y, Huang L and Zhang X (2021) Genome-Wide Investigation of the NAC Transcription Factor Family in Miscanthus sinensis and Expression Analysis Under Various Abiotic Stresses. Front. Plant Sci. 12:766550. doi: 10.3389/fpls.2021.766550
Received: 29 August 2021; Accepted: 11 October 2021;
Published: 04 November 2021.
Edited by:
Devendra Ram Malaviya, Indian Grassland and Fodder Research Institute (ICAR), IndiaReviewed by:
Vishal Acharya, Institute of Himalayan Bioresource Technology (CSIR), IndiaCopyright © 2021 Nie, Yang, He, Liu, Chen, Wang, Wang, Feng, Li, Peng, Huang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xinquan Zhang, emhhbmd4cUBzaWNhdS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.