- 1Department of Biological Sciences, Michigan Technological University, Houghton, MI, United States
- 2Department of Horticultural Sciences, University of Florida, Gainesville, FL, United States
- 3Kaveri Seed Company Limited, Secunderabad, India
- 4Renewable Resources and Enabling Sciences Center, National Renewable Energy Laboratory, Golden, CO, United States
- 5Department of Biochemistry and Molecular Biology, University of Georgia, Athens, GA, United States
- 6Complex Carbohydrate Research Center, University of Georgia, Athens, GA, United States
The precise role of KNAT7 transcription factors (TFs) in regulating secondary cell wall (SCW) biosynthesis in poplars has remained unknown, while our understanding of KNAT7 functions in other plants is continuously evolving. To study the impact of genetic modifications of homologous and heterologous KNAT7 gene expression on SCW formation in transgenic poplars, we prepared poplar KNAT7 (PtKNAT7) overexpression (PtKNAT7-OE) and antisense suppression (PtKNAT7-AS) vector constructs for the generation of transgenic poplar lines via Agrobacterium-mediated transformation. Since the overexpression of homologous genes can sometimes result in co-suppression, we also overexpressed Arabidopsis KNAT7 (AtKNAT7-OE) in transgenic poplars. In all these constructs, the expression of KNAT7 transgenes was driven by developing xylem (DX)-specific promoter, DX15. Compared to wild-type (WT) controls, many SCW biosynthesis genes downstream of KNAT7 were highly expressed in poplar PtKNAT7-OE and AtKNAT7-OE lines. Yet, no significant increase in lignin content of woody biomass of these transgenic lines was observed. PtKNAT7-AS lines, however, showed reduced expression of many SCW biosynthesis genes downstream of KNAT7 accompanied by a reduction in lignin content of wood compared to WT controls. Syringyl to Guaiacyl lignin (S/G) ratios were significantly increased in all three KNAT7 knockdown and overexpression transgenic lines than WT controls. These transgenic lines were essentially indistinguishable from WT controls in terms of their growth phenotype. Saccharification efficiency of woody biomass was significantly increased in all transgenic lines than WT controls. Overall, our results demonstrated that developing xylem-specific alteration of KNAT7 expression affects the expression of SCW biosynthesis genes, impacting at least the lignification process and improving saccharification efficiency, hence providing one of the powerful tools for improving bioethanol production from woody biomass of bioenergy crops and trees.
Introduction
Plant cell walls serve as significant sinks for the irreversible sequestering of fixed atmospheric carbon (Pauly and Keegstra, 2008). Lignocellulosic biomass from secondary cell walls (SCW) is one of the most promising bioenergy feedstocks for producing second-generation bioethanol (Demura and Ye, 2010; Nookaraju et al., 2013; Zhong et al., 2019). The unraveling of the poplar genome sequence by Tuskan et al. (2006) has opened up many new avenues into the production of transgenic poplars with altered expression of cell wall genes for enhanced saccharification and biomass growth assisting in an efficient bioconversion of cell wall biomass to bioethanol (Sahoo and Maiti, 2014; Busov, 2018; Cho et al., 2019; Bryant et al., 2020). A battery of transcription factors (TFs) regulates SCW formation (Zhong and Ye, 2014). For example, Zhong et al. (2008) suggested that NAC TFs are the top-tier master regulators directly activating the expression of several lower-level TFs, including MYBs and KNAT7. Second-tier regulators like MYB46 also directly regulate the expression of third-tier targets like KNAT7, impacting the biosynthesis of SCW components, namely, cellulose, xylan, and lignin (Ko et al., 2012a; Rao and Dixon, 2018). Thus, KNAT7 TF expression is directly or indirectly regulated by upstream regulators, but KNAT7 also plays an essential role in SCW formation. Genetic manipulation of KNAT7 gene expression in poplars has not yet been reported, but such knowledge will assist in using transgenic woody biomass for bioethanol production in the future.
In Arabidopsis, AtKNAT7 (KNOTTED ARABIDOPSIS THALIANA) is one of the KNOX (KNOTTED1-like homeodomain) TF gene family members, which is highly conserved across the angiosperms (Rao and Dixon, 2018; Ma et al., 2019). There are at least two main classes of KNOX genes. Class I KNOX genes play an essential role in meristem function, leaf development, and hormone homeostasis (Hake et al., 2004). In contrast, the functions of Class II KNOX genes have primarily remained unclear until recently, although some members were reported to be involved in root development (e.g., Hay and Tsiantis, 2010). AtKNAT7, belonging to the AtKNOX II gene family, recently came into the limelight because of its paradoxical role in regulating SCW biosynthesis (Zhong et al., 2008; Li et al., 2012). It was suggested that AtKNAT7 is a negative regulator (or transcriptional repressor) of the downstream genes involved in SCW formation in Arabidopsis because, in the knockout mutant Atknat7, significant thickening in the secondary walls of interfascicular fibers was observed than the wild type plants. However, SCWs of vessels of Atknat7 mutant remained thin-walled and showed irregular xylem (irx) or collapsed xylem phenomenon due to weak SCWs. Thus, AtKNAT7 appears to be differentially regulating SCW formation in different cells/tissues of the same plant. AtKNAT7 was also shown to be a functional ortholog of poplar KNAT7 (PtKNAT7) because PtKNAT7 complemented the Atknat7 mutant (Li et al., 2012). Overexpression of AtKNAT7 in wild-type Arabidopsis showed thinning in the interfascicular fiber walls, and RT-PCR results showed upregulation of many SCW biosynthetic genes in Atknat7 mutant (Li et al., 2012). In contrast, primary cell wall gene expression remained unaffected. Collectively, these results suggested that AtKNAT7 TF is a negative regulator of SCW formation in fiber walls (but perhaps a positive regulator of SCW synthesis in vessel walls). However, contrasting results were earlier reported by Zhong et al. (2008) in Arabidopsis expressing engineered dominant repression variant of AtKNAT7. Dominant repression of AtKNAT7 caused a severe reduction in secondary wall thickening in both interfascicular fibers and intrafascicular xylem fibers in inflorescence stems, suggesting that AtKNAT7 TF is a positive regulator of SCW formation in fibers. The severity of reduction in cell wall thickness correlated with the degree of repression. Similarly, Pandey et al. (2016) identified NbKNAT7 as a positive regulator of SCW formation in tobacco by using virus-induced gene silencing (VIGS) experiments. The NbKNAT7 overexpression lines had thicker cell walls, suggesting that KNAT7 TF is a positive regulator of the SCW biosynthesis in tobacco. Also, the saccharification from SCW biomass of the VIGS NbKNAT7 lines was 40% higher than the controls. It was recently reported that KNAT7 also positively controls the xylan biosynthesis pathway by binding to the promoter of the IRX9 gene, and expression studies showed that the transcript levels of IRX9, IRX10, and FRA8 genes were lower in the Atknat7 mutant (He et al., 2018). So, the data reported so far regarding the role of KNAT7 in SCW regulation appears to be confusing and contrasting. No studies have been reported in woody plant species like poplars regarding the precise role of KNAT7 during SCW formation.
The main goal of this research was to understand how genetic modifications in the expression of the PtKNAT7 gene affect the SCW formation and study its impact on the saccharification efficiency of lignocellulosic biomass from such transgenic poplars. Here, we investigated the role of PtKNAT7 in poplars by creating overexpression and antisense suppression lines of transgenic poplar. In addition, we present results of heterologous overexpression of AtKNAT7 in transgenic poplars to examine the impact of such genetic manipulations on SCW formation and saccharification properties in transgenic poplars. These experiments were set to circumvent the co-suppression that commonly occurs during the overexpression of native genes due to high sequence homology.
Materials and Methods
Vector Construction and Plant Transformation
The PtKNAT7 gene overexpression (sense) and suppression (antisense) vector constructs were designated as PtKNAT7-OE and PtKNAT7-AS. The full-length KNAT7 cDNA from poplar stems was amplified using gene-specific primers (see Supplementary Table 1) with integrated XbaI and SacI Restriction Enzyme sites and inserted into the DX15pBI101 vector by replacing the GUS gene (Kindly provided by Dr. K-H Han, Michigan State University). The PtKNAT7 insert was flipped over in the opposite direction for the antisense construct from the sense constructs. For the second part of this study, we overexpressed AtKNAT7 in poplars using the same DX15 promoter and designated this construct as AtKNAT7-OE. For this vector construct, AtKNAT7 cDNA was amplified using gene-specific primers and ligated into XbaI/SacI digested DX15pBI101 vector to replace the GUS gene. All the three constructs, PtKNAT7-OE, PtKNAT7-AS, and AtKNAT7-OE, were used for the transformation of hybrid poplar (Populus tremula × Populus alba clone 717-1 B4) explants using Agrobacterium C58 strain by leaf disc infiltration method (Liu et al., 2012).
RNA Extraction and Gene Expression Studies
Total RNA was isolated from Stem Differentiating Xylem (SDX) scraped from 20 to 50 aerial internodes of hybrid poplar plants using the TRIZOL (Ambion, Life Technologies) method as described earlier (Liu et al., 2012). Extracted RNA was treated with DNase (Turbo DNA free, Thermo-fisher). First-strand cDNA synthesis was performed using 1 μg of total RNA using reverse transcription kit (Applied Bio-systems). qRT-PCR was performed using PowerUp SYBR green master mix (Applied Biosystems). Real-time primers were designed for poplar KNAT7 and Arabidopsis KNAT7 using Integrated DNA Technologies (IDT) software. The total reaction was 12 μl containing 6 μl of SYBR green, 1 μl of each primer (1 μM), 1 μl of cDNA template, and 3 μl of RNase-free water. The reaction for each gene was performed in triplicates with thermocycler conditions as follows: 95°C for 10 min followed by 45 cycles for 95°C for 30 s, and 60°C for 30 s. Relative gene expression was calculated by the ΔCT method. Actin7 was used as an internal control (Zhang et al., 2010).
Microscopic Studies
For histology, sections were cut at the seventh internode from the apex of poplar plants (with about 50 internodes above the soil) and preserved in ice-cold FAA (37% formaldehyde, glacial acetic acid, and 95% ethanol). The internodes were infiltrated under a vacuum for about 10–15 min and kept overnight at 4°C. Dehydration and embedding in wax were done for fixing the sections on slides. Dewaxing was performed using two washes of xylene and decreasing concentrations of ethanol followed by washing with water. Autofluorescence was observed from dewaxed sections using fluorescent light imaging.
Growth Measurements
The growth characteristics of the greenhouse-grown plants were measured every week. All the independent lines for each gene construct were grown under identical conditions in the greenhouse as described by Liu et al. (2012). Three parameters, namely, stem height, stem thickness, and the number of leaves were measured for one-month-old plants from week 1 until the day of harvest in week 9 in the greenhouse.
Lignin Analysis Using Pyrolysis-Molecular Mass Beam Spectrometry
After scraping the developing xylem from transgenic plants, the wood was dried at room temperature for 2 weeks and then milled through a 20-gage mesh filter. One gram of wood powder was stored in falcon tubes, destarched with amylase and ethanol extracted (described in the following section), and 4 mg of destarched-extracted material was used for lignin analysis. Three samples from each gene construct were sent to National Renewable Energy Laboratory (NREL) for analysis by Pyrolysis-Molecular Mass Beam Spectrometry (Py-MBMS) to estimate lignin content and for S/G ratio measurement (Decker et al., 2018). Duplicate samples were analyzed for each line.
Saccharification Assays
Pretreatment and enzymatic hydrolysis of biomass was carried out as described previously with some modifications (Biswal et al., 2015, 2018). Before the analysis, biomass was treated with alpha-amylase (0.47 U per mg biomass, Sigma Cat # A6255) in 100 mM ammonium formate (pH 5.0) buffer at 25°C for 48 h to remove starch, followed by three water and two acetone washes. Then biomass samples were kept under the hood for 72 h for drying. This was followed by an ethanol Soxhlet extraction for an additional 24 h to remove extractives. After drying overnight, pretreatment was carried out with 5 mg of dry biomass at 180°C for 17.5 min. About 40 μl of buffer-enzyme stock, 8% CTec2 (Novozymes) in 1.0 M sodium citrate buffer, was added to the pretreated biomass. The samples were incubated at 50°C for 70 h. After 70 h of incubation, the hydrolysate was analyzed using Megazyme’s GOPOD and XDH assays as described earlier (Biswal et al., 2015, 2018).
Results
Gene Expression Studies in KNAT7 Transgenic Poplar Lines
The relative expression of the KNAT7 gene in wild-type control (WT) and transgenic KNAT7 poplars were quantified using qRT-PCR. The PtKNAT7 gene transcripts were significantly increased in PtKNAT7-OE lines compared to the WT plants (Figure 1A), whereas significant suppression of PtKNAT7 transcript formation was observed in PtKNAT7-AS transgenic trees (Figure 1B). Figure 1C shows the substantial upregulation in AtKNAT7 transcripts in AtKNAT7-OE transgenic poplar lines. We used the same PtKNAT7-OE, PtKNAT7-AS, and AtKNAT7-OE lines for further analyses. We selected four representative poplar SCW genes, namely CesA8, IRX9, PAL, and CCR for the expression studies in these transgenic poplar lines. In the case of PtKNAT7-OE lines, all four SCW genes were upregulated and the same genes were significantly downregulated in PtKNAT7-AS lines (Figure 2A). We also observed that the same four SCW biosynthesis genes were also upregulated in the AtKNAT7-OE lines of transgenic poplar lines (Figure 2B). These results suggest that KNAT7 TF is involved in the expression of downstream SCW genes and transgenic plants show expected altered expression of SCW genes.
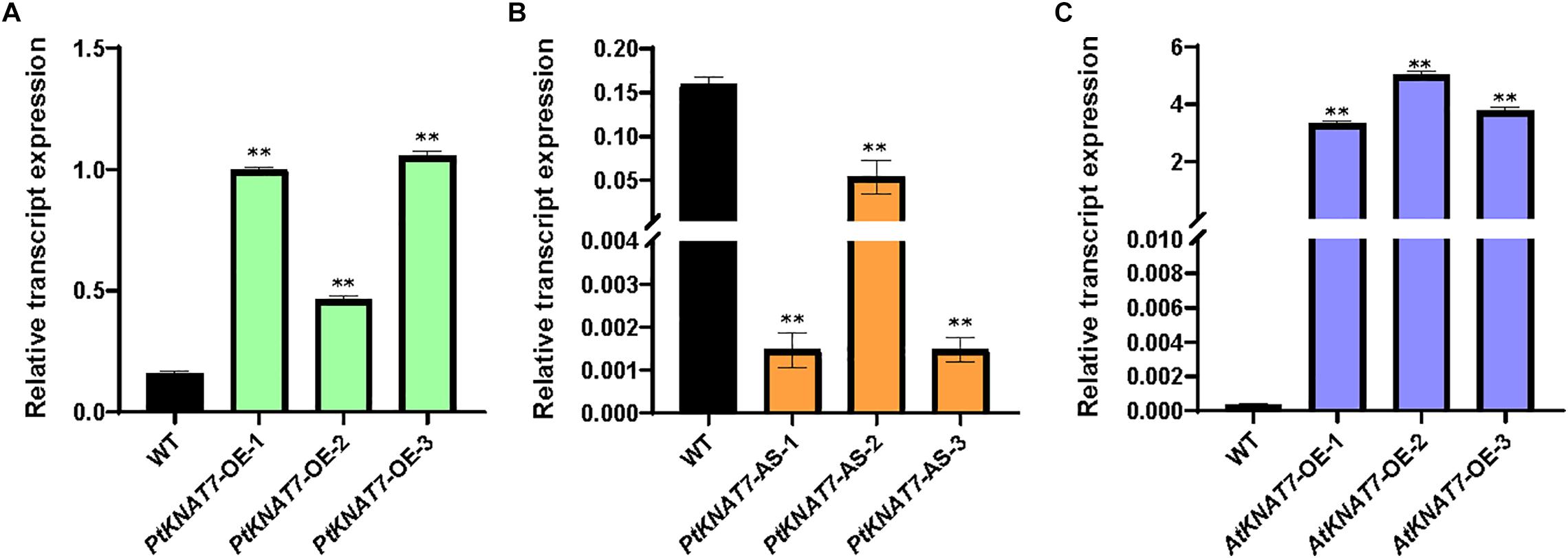
Figure 1. Relative transcript expression was estimated through quantitative RT-PCR using three independent transgenic lines in each of the poplar gene constructs: PtKNAT7-OE (A), PtKNAT7-AS (B), and AtKNAT7-OE (C). Sampling was done in triplicate of each sample of the transgenic line.
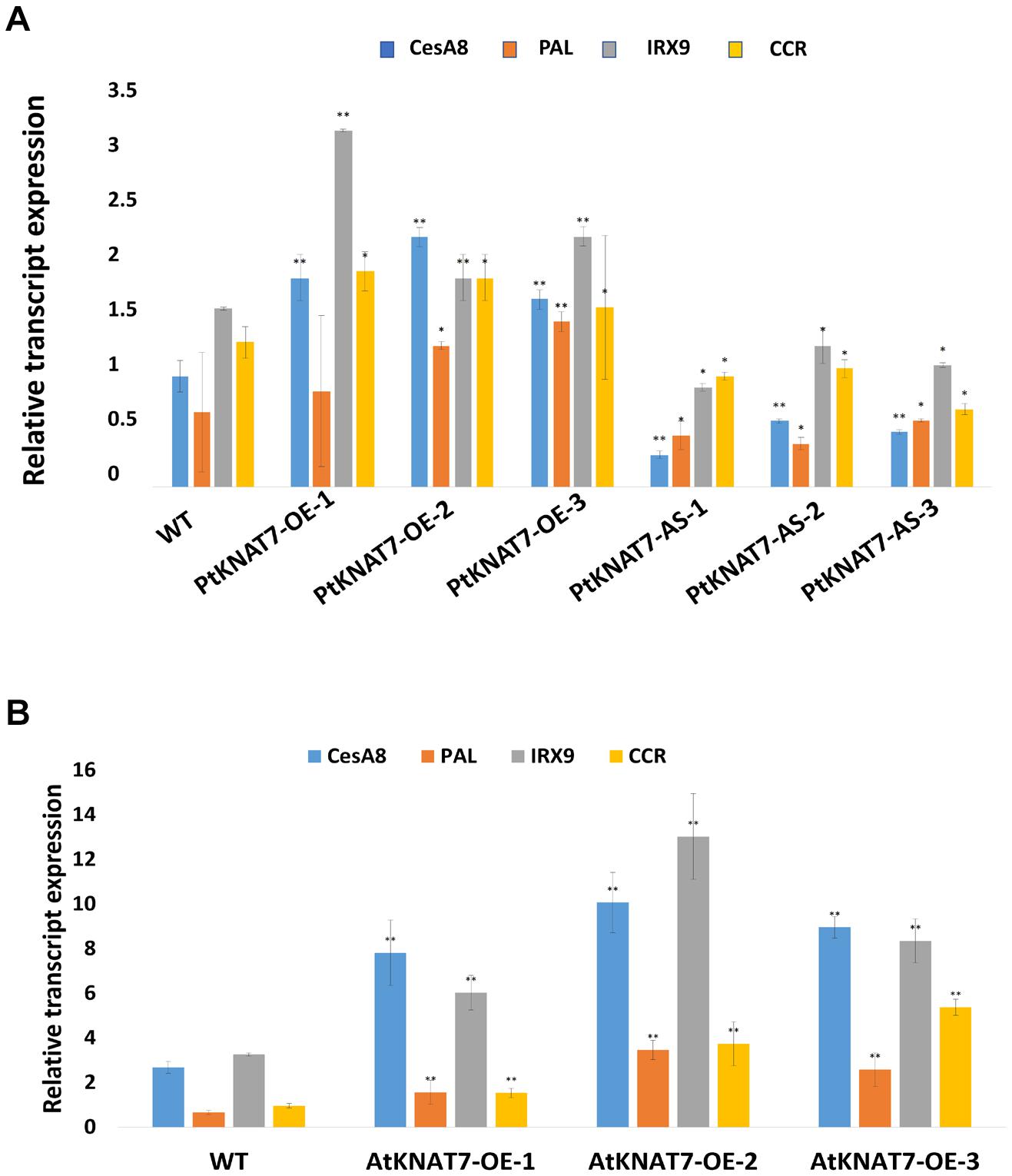
Figure 2. Effect of KNAT7 transgene expression on four selected SCW biosynthetic genes in poplars, namely, CesA8, PAL, IRX9, and CCR. (A) SCW gene expressions patterns in PtKNAT7-OE and PtKNAT7-AS transgenic lines. (B) SCW gene expressions patterns in AtKNAT7-OE transgenic lines. Bars represent the average of three replicates of three transgenic lines. All differences were significant when compared to WT controls.
Growth Measurements of Transgenic KNAT7 Poplar Plants
Once the homologous or heterologous KNAT7 gene overexpression or silencing was confirmed in a variety of transgenic poplar KNAT7 lines using gene expression studies, the same three independent transgenic lines for each construct were chosen for measuring the growth parameters. The plant height, stem thickness, and the number of leaves measurements were recorded every week until the day of harvest in about 9 weeks (Figures 3A–C). The height of most of the three transgenic lines was not significantly greater than WT plants, except for the PtKNAT7-OE-3 line, and PtKNAT7-AS-3 line (Figure 3A) by week 9 but eight of the nine transgenic lines examined had significantly increased stem diameter (Figure 3B) than the WT plants by week 9. In terms of the number of leaves in all three types of transgenic lines, there was a significant increase as compared to WT plants by week 9 (Figure 3C). Thus, growth phenotypes of most of the transgenic KNAT7 lines examined were either similar or a little better than WT lines by the time of harvest (Supplementary Figure 1).
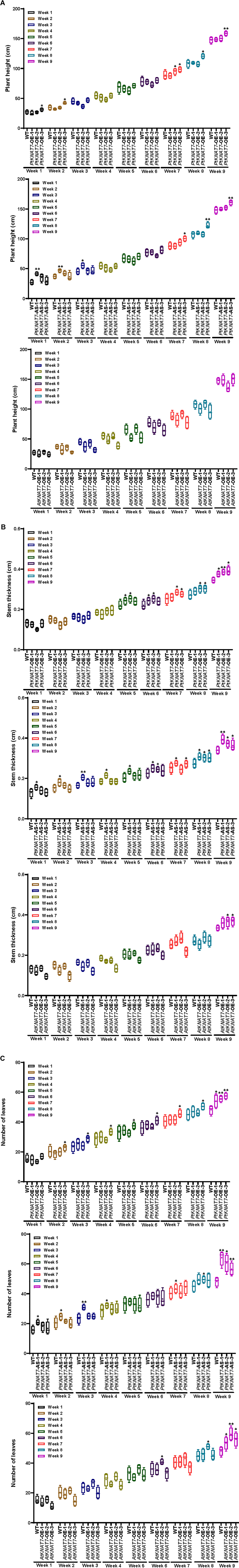
Figure 3. Phenotypic measurements were performed every week in WT control and transgenic poplars until the day of harvest in the 9th week. (A) Plant Height, (B) stem thickness, and (C) the number of leaves in PtKNAT7-OE, PtKNAT7-AS, and AtKNAT7-OE were measured for 9 weeks until the day of harvest. The data is represented as a means ± SD (n = 3).
Transgenic KNAT7 Poplar Lines Show Altered Xylem Growth Phenotype
Pandey et al. (2016) reported that genetic modifications of the KNAT7 gene expression in tobacco showed the altered amount of xylem occupying stem sections. VIGS and RNAi-suppression lines for the NbKNAT7 gene showed about 50% increase in xylem area with reduced xylem fiber wall thickness whereas NbKNAT7 overexpression lines showed increased xylem cell wall thickness but did not show such increase in xylem area. Here, we used autofluorescence of lignin in the dewaxed stem sections from the top 7th internode to estimate the xylem area in transgenic KNAT7 poplars compared to WT plants (Figures 4A–E). In the case of PtKNAT7-OE overexpression lines (Figures 4B,E), we observed a 15% decrease in the xylem area as compared to controls (Figures 4A,E) whereas a 26% increase in the xylem area was observed in the PtKNAT7-AS lines (Figures 4C,E) over the WT control plants (Figures 4A,E). About a 13% increase in the xylem area was observed in AtKNAT7-OE sections (Figures 4D,E) over control. Qualitatively, a slight increase in the lignin autofluorescence signal was also observed in the KNAT7 overexpression lines of poplars as compared to the stem sections of WT control plants (Figures 4A,B). However, no changes in autofluorescence levels were observed in the case of the AtKNAT7-OE lines than controls (Figure 4D). There were no obvious differences in the fiber or vessel wall thickness in any of the KNAT7 transgenic lines and these observations are similar to a previous report by Zhong et al. (2008).
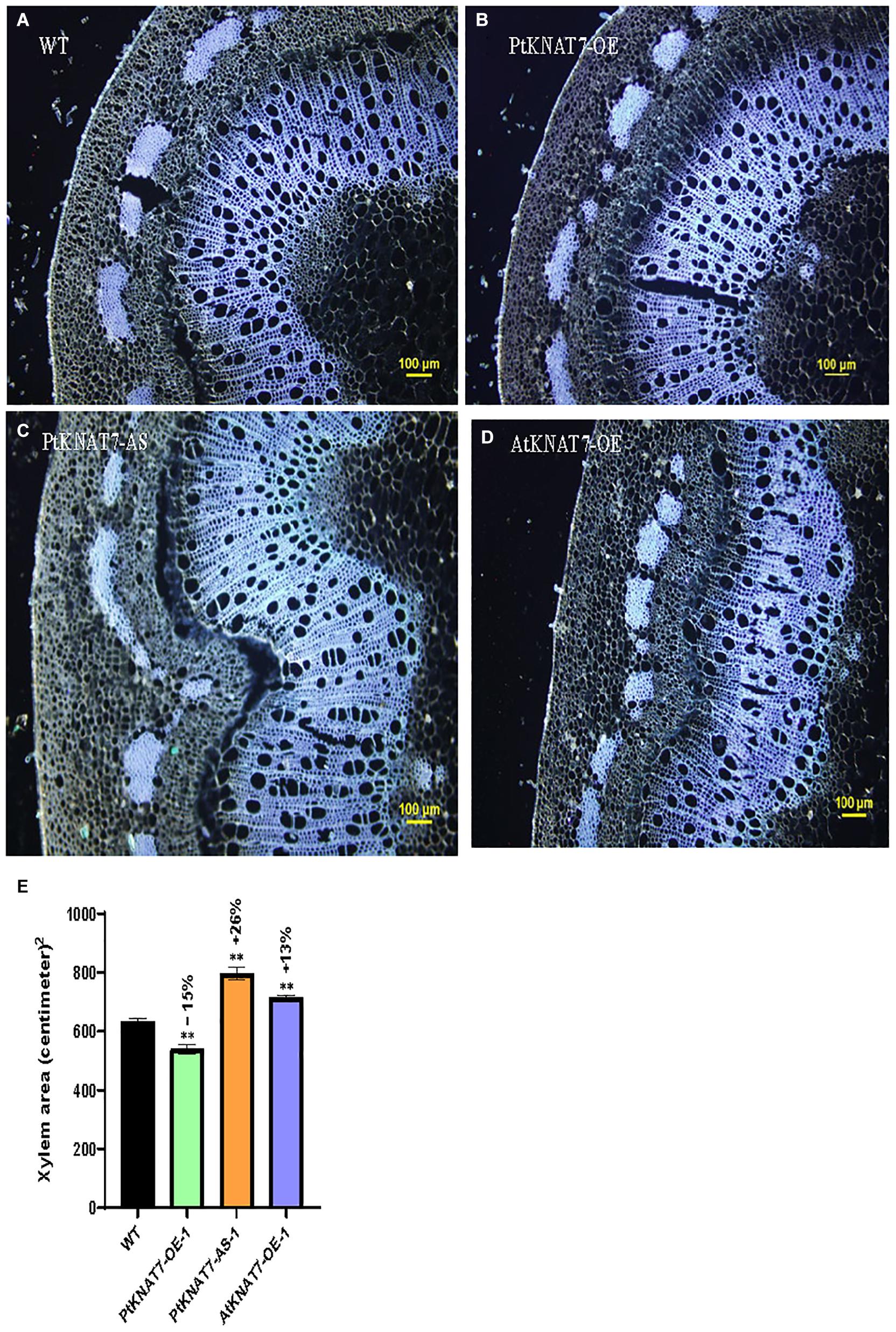
Figure 4. Autofluorescence images of cross-sections and measurements of xylem area. Tissue sections for histological studies were performed at the 7th internode from the top. Cross-section of wild type (A) compared from PtKNAT7-OE (B), PtKNAT7-AS (C), AtKNAT7-OE (D), Scale bars-100 micrometers. (E) Xylem area measurements in WT and three KNAT7 transgenic lines in poplars. Significant differences were observed as compared to WT controls. Measurements were calculated from the xylem area observed at three separate points and averaged (n = 3).
Changes in KNAT7 Gene Expression Leads to Altered Lignin Content and Constitution in Some Transgenic Lines
Lignin analysis was performed using py-MBMS. No significant changes in lignin content were observed for PtKNAT7-OE lines, but there was a significant decrease of about 6% lignin in the case of PtKNAT7-AS lines as compared to the controls (Figure 5A). AtKNAT7-OE expression lines also did not show any observable changes in lignin content. However, the S/G lignin ratios were significantly increased in all three KNAT7 transgenic lines (Figure 5B). Overall, a 5–7% increase in PtKNAT7-OE, 8–12% increase in PtKNAT7-AS lines, and 7% increase in AtKNAT7-OE lines were observed for the S/G lignin ratios as compared to controls. A change in the S/G lignin ratio could bring about structural and other property changes in the cell walls. Therefore, we further explored how these changes in the cell wall composition resulted in changes in the saccharification efficiency.
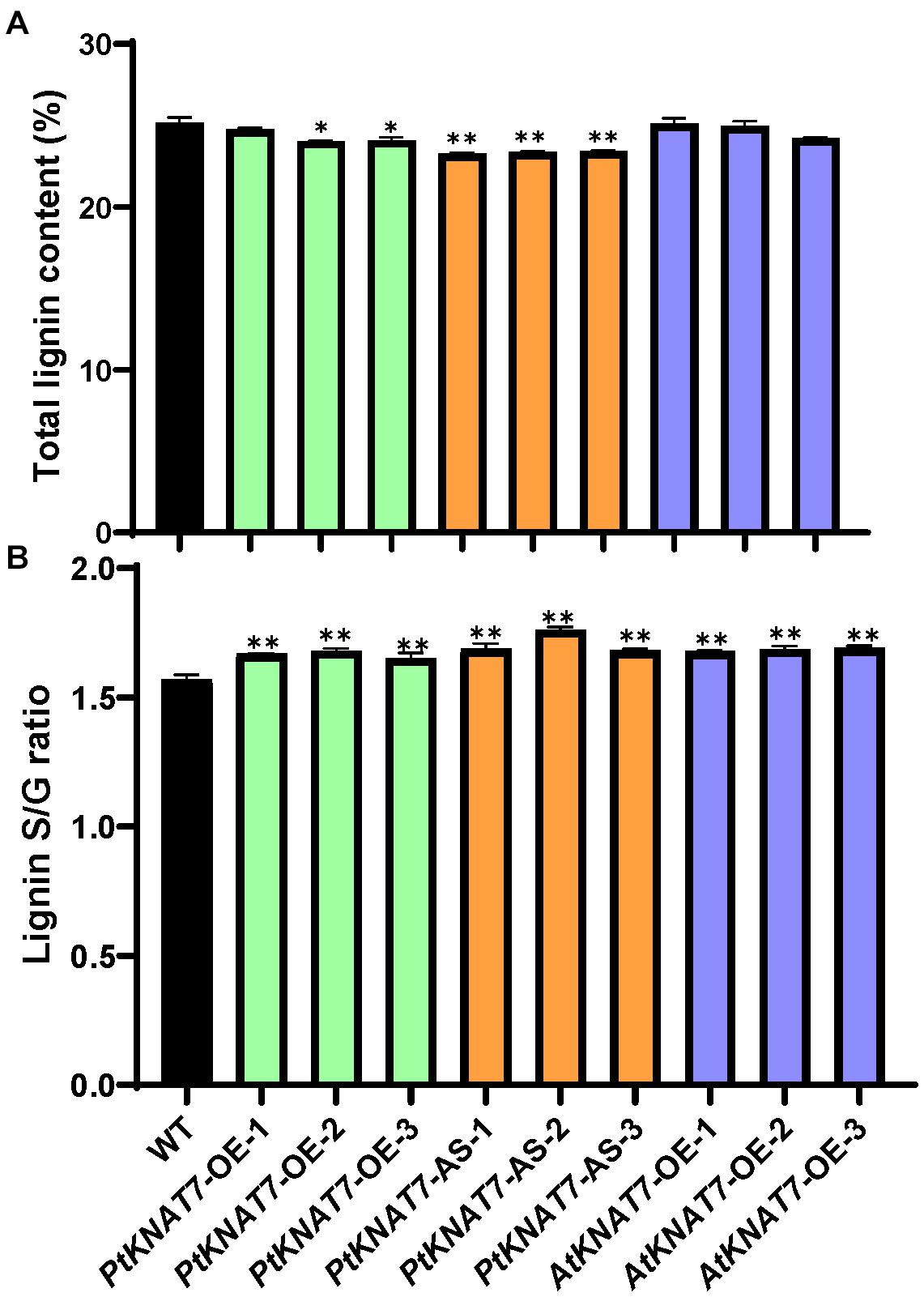
Figure 5. Lignin content and S/G lignin ratio measurements in poplar transgenic lines: (A) Lignin contents in three PtKNAT7-OE, PtKNAT7-AS, and AtKNAT7-OE transgenic lines in poplar. (B) S/G lignin ratio measurements in three PtKNAT7-OE, PtKNAT7-AS, and AtKNAT7-OE transgenic lines in poplar. Data analyses were performed in duplicate for each sample. Significant observations are marked by * when compared to WT controls (P-value < 0.05).
Saccharification Efficiency Was Significantly Improved in the Genetically Manipulated KNAT7 Transgenic Lines of Poplars
Saccharification efficiency measurements were performed to estimate simple sugar release from the wood samples obtained from greenhouse-grown transgenic and WT poplar plants. Glucose release was increased in all KNAT7 transgenic lines as compared to WT controls. PtKNAT7-OE lines showed an overall 22–26% increase in glucose release whereas PtKNAT7-AS lines showed a 44–53% increase in glucose release over the WT controls (Figure 6A). There was also a 24–30% increase in glucose release for AtKNAT7-OE expression lines (Figure 6A). The data on xylose released also showed a similar trend of sugar release with a 28–34% increase in PtKNAT7-OE lines, 55–67% in PtKNAT7-AS lines, and 37–44% increase in AtKNAT7-OE lines (Figure 6B). The total increase in sugar release varied from 24 to 58% in these KNAT7 transgenic lines and that could be mainly attributed to the alteration in the SCW formation that occurred as a result of changes in the expression of KNAT7 genes (Figure 6C). Also, the increase in the S/G ratio and changes in lignin content might have contributed to the reduced cell wall recalcitrance.
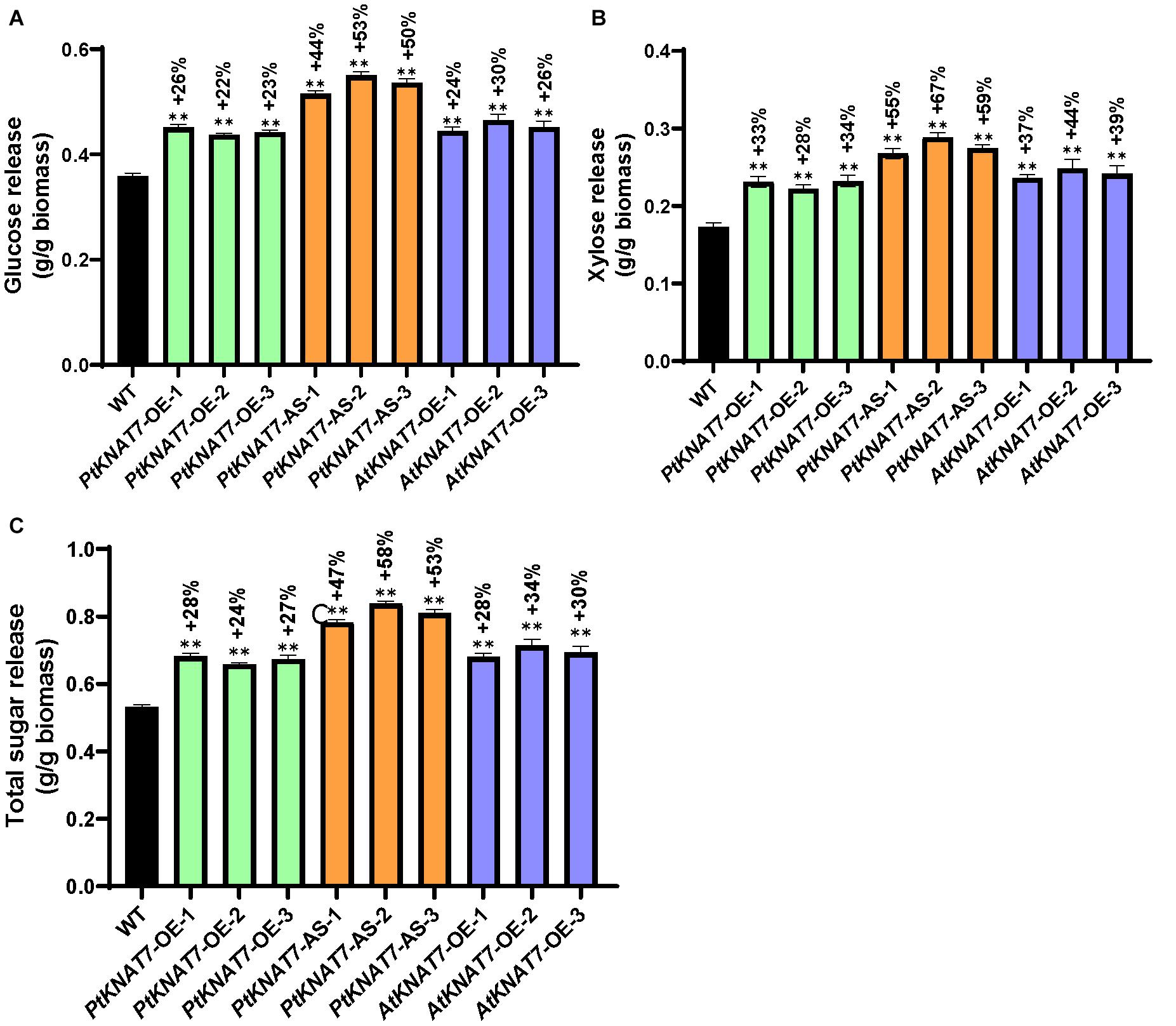
Figure 6. Saccharification yield of three PtKNAT7-OE, PtKNAT7-AS, and AtKNAT7-OE lines. (A) Glucose released, (B) xylose released, and (C) total sugars released from all the three independent KNAT7 transgenic lines of poplars. Significant differences were observed when compared to WT controls, P-value < 0.01.
Discussion
Investigations into the role of KNAT7, one of the members of the plant KNOX II TF family, began with the Arabidopsis microarray data mining studies where the KNAT7 gene was highly co-expressed with Cellulose synthase A (CESA) genes involved in the SCW formation (Brown et al., 2005; Ehlting et al., 2005; Persson et al., 2005). To understand the role of KNAT7 in SCW synthesis, Zhong et al. (2008) reported that dominant repression of AtKNAT7 TF exhibited reduced SCW thickening in interfascicular and xylary fibers and vessels (but no irx phenomenon) suggesting a positive regulation of SCW biosynthesis by KNAT7 TF. Overexpression of the KNAT7 gene did not change the SCW thickness of vessels and fibers in transgenics compared to wild-type control plants (Zhong et al., 2008). Similarly, in another study by Pandey et al. (2016), overexpression of NbKNAT7 in tobacco resulted in thickening of the SCW in the xylem while suppression of NbKNAT7 in tobacco VIGS and RNAi plants resulted in thinner xylem SCWs coupled with increased xylem area. Furthermore, the positive regulation of xylan biosynthesis by KNAT7 TF has been recently reported in Arabidopsis inflorescence stems (He et al., 2018) and seed mucilage formation (Wang Y. et al., 2020). The use of constitutive promoters like CaMV 35S driving the expression of transgenes for such studies is known to produce several undesirable growth outcomes. In order to circumvent such growth phenotypes, Ko et al. (2012b) described the discovery of utility promoters, such as DX15 that drive strong developing xylem-specific expression of transgenes. A number of papers have described the use of DX15 promoters for biomass engineering (e.g., Cho et al., 2019). We have, therefore, used DX15-promoter for driving the expression of KNAT7 transgenes in this work. In the present study, we overexpressed poplar and Arabidopsis KNAT7 genes in transgenic poplars under the regulation of DX15 promoters and our observations suggest that KNAT7 acts like a positive regulator in poplars. It is also possible that there are species-specific differences in KNAT7 functions in SCW biosynthesis. Overall, the xylem area in PtKNAT7-AS lines increased by 26% while the xylem area was decreased in PtKNAT7-OE lines by 15% as compared to WT stems confirming the earlier observations in NbKNAT7 transgenic lines of tobacco (Pandey et al., 2016), where the increased xylem proliferation in antisense lines could be partially attributed as a compensatory mechanism in response to reduced fiber cell wall thickness. However, in the present study, we did not observe any significant changes in the cell wall thickness in stems of PtKNAT7 transgenic lines. It has not escaped our attention that AtKNAT7-OE transgenics showed xylem area phenotype similar to PtKNAT7-AS and not like PtKNAT7-OE. We are still far away from understanding the impact of such manipulations on plant phenotypes and it appears that at least, in this case, specific-specific variations in the KNAT7 gene source might be responsible for this phenotype. But further investigations are warranted into this phenomenon in the future.
On the contrary, Li et al. (2012) initially claimed KNAT7 TF to be a negative regulator of SCW formation because Atknat7 mutants had thicker interfascicular fibers but curiously also had irx phenotype in the vessels. However, these studies could not explain the irx phenotype in vessels when interfascicular fiber cells displayed SCW thickening at the same time. Recently, Qin et al. (2020) and Wang S. et al. (2020) elegantly reconciled these paradoxical observations by suggesting that KNAT3 and KNAT7 work synergistically in fibers but antagonistically in vessels in the regulation of SCW biosynthesis. As suggested by He et al. (2018), KNAT7 may not simply activate or repress every component of the secondary wall equally. Rather, it may differentially activate and suppress the deposition of various SCW components in specific cell types. Also, it is possible that KNAT7 functions either as an activator or repressor, depending upon the composition of the TF network active in different tissues or cell types of plants. Lastly, one cannot ignore species-specific variations in the transgenes that may result in different phenotypes.
We observed that overexpression of the PtKNAT7 and AtKNAT7 genes did not significantly increase the total lignin content although some monolignol biosynthesis genes, as well as some other representative genes involved in SCW formation, were upregulated. This observation is similar to Zhong et al. (2008) who also did not see significant changes in lignin quantities when the AtKNAT7 gene was overexpressed. However, antisense PtKNAT7 suppressed lines in poplars had lower lignin content and suppressed expression of SCW genes. This observation is again different than the earlier findings by Li et al. (2012), where they reported increased lignin deposition in Arabidopsis knat7 mutants. We also observed that S/G lignin ratios were significantly increased in both PtKNAT7-OE, AtKNAT7-OE, and PtKNAT7-AS lines. Considering the potential importance of KNAT7 TF in syringyl lignin biosynthesis (Qin et al., 2020; Wang S. et al., 2020), it is expected that overexpression of KNAT7 may increase S lignin synthesis but it is puzzling why antisense suppression of KNAT7 also increased S lignin content. It is possible that all the components of the TF network that interact with KNAT7 are not yet completely identified and there might be some complex regulation of S lignin synthesis or suppression of G lignin production that might be occurring in such transgenic lines.
Recently, Yoo et al. (2018) reported a negative correlation between lignin content and saccharification efficiency of poplar woody tissues and a positive correlation between S/G ratio and saccharification efficiency of SCW biomass. The current report is the first time when transgenic KNAT7 poplar lines have been examined for their lignin quantities and content as well as saccharification efficiencies. We have observed increased saccharification efficiencies in all the transgenic lines of poplar altered in KNAT7 gene expression compared to WT controls. The decrease in lignin content in PtKNAT7-AS lines and an increase in S/G ratios in all transgenic lines are positively associated with increased saccharification efficiencies. Similar to present findings, Pandey et al. (2016) also observed that tobacco NbKNAT7 VIGS lines had increased saccharification efficiency than control plants.
While the focus of this study is on the biotechnological application of genetic manipulation of KNAT7, it is a common observation that in transgenic plants manipulated in lignin biosynthesis genes show abnormal growth phenotypes (e.g., Ko et al., 2012a). We observed that KNAT7 transgenic poplar lines show almost similar or slightly better growth phenotypes compared to WT control plants. These observations are similar to an earlier report in tobacco KNAT7 transgenic plants where normal-looking transgenic KNAT7 plants were produced (Pandey et al., 2016). An increase in saccharification efficiencies, indicative of increased bioethanol production potential, without altering growth is a desirable characteristic of these KNAT7 transgenic poplar lines from a bioenergy production perspective. In conclusion, the genetic alteration of KNAT7 could be one of the powerful strategies for increasing saccharification efficiency in transgenic bioenergy plants and has a great potential for improving bioethanol production.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
YA and CJ planned the experiments and interpreted the results. AN initiated the KNAT7 experiments in poplars. AB performed saccharification assays. AH-W and CD did lignin analysis. All authors contributed to manuscript preparation.
Funding
CJ acknowledges funding from the National Science Foundation to the “Sustainable Forest-Based Biofuel Pathways to Hydrocarbon Transportation Fuels” project at Michigan Technological University (Grant number #1230803). CJ and AN’s work was also partially supported by the World Class University project of the Ministry of Science and Technology of South Korea (R31-2009-000-20025-0). YA thank the Government of India for the National Overseas Scholarship and the Department of Biological Sciences, Michigan Technological University for sponsoring his doctoral research. This work was authored in part by Alliance for Sustainable Energy, LLC, the manager and operator of the National Renewable Energy Laboratory for the United States DOE under Contract No. DE-AC36-08GO28308. AB acknowledges the Center for Bioenergy Innovation (Oak Ridge National Laboratory), a US Department of Energy (DOE) Bioenergy Research Center supported by the Office of Biological and Environmental Research in the DOE Office of Science.
Author Disclaimer
The views expressed in the article do not necessarily represent the views of the DOE or the United States Government. The United States Government retains and the publisher, by accepting the article for publication, acknowledges that the United States Government retains a non-exclusive, paid-up, irrevocable, worldwide license to publish or reproduce the published form of this work or allow others to do so, for the United States Government purposes.
Conflict of Interest
AN is currently employed by Kaveri Seeds.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.762067/full#supplementary-material
References
Biswal, A. K., Atmodjo, M. A., Li, M., Baxter, H. L., Yoo, C. G., Pu, Y., et al. (2018). Sugar release and growth of biofuel crops are improved by downregulation of pectin biosynthesis. Nat. Biotechnol. 36, 249–257. doi: 10.1038/nbt.4067
Biswal, A. K., Hao, Z., Pattathil, S., Yang, X., Winkeler, K., Collins, C., et al. (2015). Downregulation of GAUT12 in Populus deltoides by RNA silencing results in reduced recalcitrance, increased growth and reduced xylan and pectin in a woody biofuel feedstock. Biotechnol. Biofuels 8:41. doi: 10.1186/s13068-015-0218-y
Brown, D. M., Zeef, L. A. H., Ellis, J., Goodacre, R., and Turner, S. R. (2005). Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 17, 2281–2295. doi: 10.1105/tpc.105.031542
Bryant, N. D., Pu, Y., Tschaplinski, T. J., Tuskan, G. A., Muchero, W., Kalluri, U. C., et al. (2020). Transgenic poplar designed for biofuels. Trends Plant Sci. 25, 881–896. doi: 10.1016/j.tplants.2020.03.008
Busov, V. B. (2018). Manipulation of growth and architectural characteristics in trees for increased woody biomass production. Front. Plant Sci. 9:1505. doi: 10.3389/fpls.2018.01505
Cho, J. S., Jeon, H. W., Kim, M. H., Vo, T. K., Kim, J., Park, E. J., et al. (2019). Wood forming tissue−specific bicistronic expression of Pd GA 20ox1 and Ptr MYB 221 improves both the quality and quantity of woody biomass production in a hybrid poplar. Plant Biotechnol. J. 17, 1048–1057. doi: 10.1111/pbi.13036
Decker, S. R., Harman-Ware, A. E., Happs, R. M., Wolfrum, E. J., Tuskan, G. A., Kainer, D., et al. (2018). High Throughput Screening Technologies in Biomass Characterization. Front. Energy Res. 6:120. doi: 10.3389/fenrg.2018.00120
Demura, T., and Ye, Z.-H. (2010). Regulation of plant biomass production. Curr. Opin. Plant Biol. 13, 298–303. doi: 10.1016/j.pbi.2010.03.002
Ehlting, J., Mattheus, N., Aeschliman, D. S., Li, E., Hamberger, B., Cullis, I. F., et al. (2005). Global transcript profiling of primary stems from Arabidopsis thaliana identifies candidate genes for missing links in lignin biosynthesis and transcriptional regulators of fiber differentiation: global transcript profiling of stems. Plant J. 42, 618–640. doi: 10.1111/j.1365-313X.2005.02403.x
Hake, S., Smith, H. M. S., Holtan, H., Magnani, E., Mele, G., and Ramirez, J. (2004). The role of knox genes in plant development. Annu. Rev. Cell Dev. Biol. 20, 125–151. doi: 10.1146/annurev.cellbio.20.031803.093824
Hay, A., and Tsiantis, M. (2010). KNOX genes: versatile regulators of plant development and diversity. Development 137, 3153–3165. doi: 10.1242/dev.030049
He, J.-B., Zhao, X.-H., Du, P.-Z., Zeng, W., Beahan, C. T., Wang, Y.-Q., et al. (2018). KNAT7 positively regulates xylan biosynthesis by directly activating IRX9 expression in Arabidopsis: KNAT7 positively regulates xylan biosynthesis. J. Integr. Plant Biol. 60, 514–528. doi: 10.1111/jipb.12638
Ko, J. H., Kim, W. C., Kim, J. Y., Ahn, S. J., and Han, K. H. (2012a). MYB46-mediated transcriptional regulation of secondary wall biosynthesis. Mol. Plant 5, 961–963. doi: 10.1093/mp/sss076
Ko, J. H., Kim, H. T., Hwang, I., and Han, K. H. (2012b). Tissue-type-specific transcriptome analysis identifies developing xylem-specific promoters in poplars. Plant Biotechnol. J. 10, 587–596. doi: 10.1111/j.1467-7652.2012.00690.x
Li, E., Bhargava, A., Qiang, W., Friedmann, M. C., Forneris, N., Savidge, R. A., et al. (2012). The Class II KNOX gene KNAT7 negatively regulates secondary wall formation in Arabidopsis and is functionally conserved in Populus. New Phytologist 194, 102–115. doi: 10.1111/j.1469-8137.2011.04016.x
Liu, Y., Xu, F., Gou, J., Al-Haddad, J., Telewski, F. W., Bae, H.-J., et al. (2012). Importance of two consecutive methionines at the N-terminus of a cellulose synthase (PtdCesA8A) for normal wood cellulose synthesis in aspen. Tree Physiol. 32, 1403–1412. doi: 10.1093/treephys/tps096
Ma, Q., Wang, N., Hao, P., Sun, H., Wang, C., Ma, L., et al. (2019). Genome-wide identification and characterization of TALE superfamily genes in cotton reveals their functions in regulating secondary cell wall biosynthesis. BMC Plant Biol. 19:432. doi: 10.1186/s12870-019-2026-1
Nookaraju, A., Pandey, S. K., Bae, H.-J., and Joshi, C. P. (2013). Designing cell walls for improved bioenergy production. Mol. Plant 6, 8–10. doi: 10.1093/mp/sss111
Pandey, S. K., Nookaraju, A., Fujino, T., Pattathil, S., and Joshi, C. P. (2016). Virus-induced gene silencing (VIGS)-mediated functional characterization of two genes involved in lignocellulosic secondary cell wall formation. Plant Cell Rep. 35, 2353–2367. doi: 10.1007/s00299-016-2039-2
Pauly, M., and Keegstra, K. (2008). Cell-wall carbohydrates and their modification as a resource for biofuels. Plant J. 54, 559–568. doi: 10.1111/j.1365-313X.2008.03463.x
Persson, S., Wei, H., Milne, J., Page, G. P., and Somerville, C. R. (2005). Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proc. Natl. Acad. Sci. U. S. A. 102, 8633–8638. doi: 10.1073/pnas.0503392102
Qin, W., Yin, Q., Chen, J., Zhao, X., Yue, F., He, J., et al. (2020). The class II KNOX transcription factors KNAT3 and KNAT7 synergistically regulate monolignol biosynthesis in Arabidopsis. J. Exp. Bot. 71, 5469–5483. doi: 10.1093/jxb/eraa266
Rao, X., and Dixon, R. A. (2018). Current models for transcriptional regulation of secondary cell wall biosynthesis in grasses. Front. Plant Sci. 9:399. doi: 10.3389/fpls.2018.00399
Sahoo, D., and Maiti, I. (2014). Biomass derived from transgenic tobacco expressing the Arabidopsis CESA3ixr1-2 gene exhibits improved saccharification. Acta Biol. Hung. 65, 189–204. doi: 10.1556/abiol.65.2014.2.7
Tuskan, G. A., DiFazio, S., Jansson, S., Bohlmann, J., Grigoriev, I., Hellsten, U., et al. (2006). The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313, 1596–1604. doi: 10.1126/science.1128691
Wang, S., Yamaguchi, M., Grienenberger, E., Martone, P. T., Samuels, A. L., and Mansfield, S. D. (2020). The Class II KNOX genes KNAT3 and KNAT7 work cooperatively to influence deposition of secondary cell walls that provide mechanical support to Arabidopsis stems. Plant J. 101, 293–309. doi: 10.1111/tpj.14541
Wang, Y., Xu, Y., Pei, S., Lu, M., Kong, Y., Zhou, G., et al. (2020). KNAT7 regulates xylan biosynthesis in Arabidopsis seed-coat mucilage. J. Exp. Bot. 71, 4125–4139. doi: 10.1093/jxb/eraa189
Yoo, C. G., Dumitrache, A., Muchero, W., Natzke, J., Akinosho, H., Li, M., et al. (2018). Significance of lignin S/G ratio in biomass recalcitrance of Populus trichocarpa variants for bioethanol production. ACS Sustain. Chem. Eng. 6, 2162–2168. doi: 10.1021/acssuschemeng.7b03586
Zhang, D., Du, Q., Xu, B., Zhang, Z., and Li, B. (2010). The actin multigene family in Populus: organization, expression and phylogenetic analysis. Mol. Genet. Genomics 284, 105–119. doi: 10.1007/s00438-010-0552-5
Zhong, R., Cui, D., and Ye, Z. (2019). Secondary cell wall biosynthesis. New Phytol. 221, 1703–1723. doi: 10.1111/nph.15537
Zhong, R., Lee, C., Zhou, J., McCarthy, R. L., and Ye, Z.-H. (2008). A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 20, 2763–2782. doi: 10.1105/tpc.108.061325
Keywords: antisense, developing xylem, overexpression, saccharification, secondary cell wall biosynthesis
Citation: Ahlawat YK, Nookaraju A, Harman-Ware AE, Doeppke C, Biswal AK and Joshi CP (2021) Genetic Modification of KNAT7 Transcription Factor Expression Enhances Saccharification and Reduces Recalcitrance of Woody Biomass in Poplars. Front. Plant Sci. 12:762067. doi: 10.3389/fpls.2021.762067
Received: 20 August 2021; Accepted: 06 October 2021;
Published: 26 October 2021.
Edited by:
Wusirika Ramakrishna, Central University of Punjab, IndiaReviewed by:
Kyung-Hwan Han, Michigan State University, United StatesAimin Wu, South China Agricultural University, China
Dipak Kumar Sahoo, Iowa State University, United States
Copyright © 2021 Ahlawat, Nookaraju, Harman-Ware, Doeppke, Biswal and Joshi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chandrashekhar P. Joshi, Y3Bqb3NoaUBtdHUuZWR1
 Yogesh Kumar Ahlawat1,2
Yogesh Kumar Ahlawat1,2 Anne E. Harman-Ware
Anne E. Harman-Ware Ajaya K. Biswal
Ajaya K. Biswal Chandrashekhar P. Joshi
Chandrashekhar P. Joshi