- 1College of Horticulture Science and Engineering, Shandong Agricultural University, Tai’an, China
- 2State Key Laboratory of Crop Biology, Shandong Agricultural University, Tai’an, China
- 3Shandong Collaborative Innovation Center for Fruit and Vegetable Production With High Quality and Efficiency, Tai’an, China
- 4College of Life Sciences, Shandong Agricultural University, Tai’an, China
- 5Shandong Huayu University of Technology, Dezhou, China
The OVATE family protein (OFP) genes (OFPs) have been shown to respond to salt stress in plants. However, the regulatory mechanism for salt tolerance of the peach (Prunus persica) OFP gene PpOFP1 has not been elucidated. In this study, using yeast two-hybrid screening, we isolated a nucleus-localized ZF-HD_dimer domain protein PpZFHD1, which interacts with the PpOFP1 protein in the peach cultivar “Zhongnongpan No.10”. A segmentation experiment further suggested that the interaction happens more specifically between the N-terminal, contains ZF-HD_dimer domain, of PpZFHD1 and the C-terminal, consists of OVATE domain, of PpOFP1. Additionally, quantitative real-time polymerase chain reaction (qRT-PCR) experiments indicate that transcription of these two genes are induced by 200 mmol/L (mM) NaCl treatment. Heterogeneous transformation experiments suggested that the growth status of transformed yeast strain over-expressing each of these two genes was more robust than that of control (CK). Furthermore, transgenic tomato plants over-expressing PpOFP1 were also more robust. They had a higher content of chlorophyll, soluble proteins, soluble sugars, and proline. Activities of the superoxide dismutase (SOD), peroxidase (POD), and catalase (CAT) in these plants were higher, and tissues from these plants exhibited a lower relative conductivity and malondialdehyde (MDA) content. These results suggest that PpOFP1 physically interacts with PpZFHD1 and confers salt tolerance to tomato and yeast, thus revealing a novel mechanism for regulating salt tolerance in peach and other perennial deciduous trees.
Introduction
Research Progress of Fruit Trees in Response to Salt Stress
The extremity of agricultural salt damage has increased significantly in the 21st century. The extent of salinization has approached 900 million hm2 globally, and includes some 100 million hm2 in China alone (Fang et al., 2021). The effects of salt damage on fruit trees vary with climate conditions, light intensity, soil conditions, and plant species. Osmotic regulation, chlorophyll content, antioxidant defense system, and cell membrane stability are all affected by salt stress (Singh and Sharma, 2018). Under salt stress the chlorophyll content in grape leaves decreased differentially among varieties (Sivritepe and Eriş, 1999). Yuan et al. (2016) found that VaPAT1, a transcription factor of the GRAS family from Vitis amurensis, can respond to salt stress. Furthermore, over-expressing VaPAT1 in Arabidopsis increase the salt tolerance of transgenic lines by increasing the proline and soluble sugar content. Transgenic lemon plants over-expressing FcWRKY40 exhibit increased salt tolerance, increased proline content, and improved ion transport (Dai et al., 2018). When stressed with salt, the soluble sugar content of papaya varieties with strong salt tolerance increased. In salt sensitive varieties it initially increased, but decreased over time (Chander and Palaniappan, 2016). Al-Shorafa et al. (2014) reported that after salt treatment proline accumulated in strawberry leaves and membrane permeability increased.
Plant OVATE Family Protein Transcription Factors
The OFP transcription factor containing an OVATE domain composing of 70 amino acids, was first identified in pear-shaped tomato (Liu et al., 2002). Zhang et al. (2020) found that AtOFP2 and AtOFP5 could interact directly with the tubule regulatory protein TONNEU2 (TON2), and alter the normal development and cell morphology of the embryo sac. An interaction of AtOFP4, AtKNATs, and AtBLHs proteins in Arabidopsis regulates the formation of secondary cell walls (Liu and Douglas, 2015). The interaction between AtOFP1 and AtKu of Arabidopsis can regulate the DNA repair process. In rice, OsOFP8 protein can interact with OsGSK2 and be phosphorylated, which helps to regulate the feedback circuit of BR signaling, thus affecting hormone response in tissues (Yang et al., 2016). CaOvate inhibited the expression of CaGA20ox1 gene, thus changing the shape of pepper fruit (Tsaballa et al., 2011). The co-expression of MaMADS1 and MaOFP1 proteins in banana can alter fruit shape and improve quality related to hardness, soluble solid, and soluble sugar content (Liu et al., 2017).
In addition to the regulation of plant growth and development, the OFPs gene play key roles in the process of coping with abiotic stress. Drought stress can induce the expression of AtOFP8, and the drought resistance of Arabidopsis overexpressing the AtOFP8 gene is enhanced (Tang et al., 2018). Rice plants over-expressing OsOFP6 showed stronger drought and cold resistance, while the inhibition of OsOFP6 expression resulted in hypersensitivity, indicating that it played an important role in coping with abiotic stress (Ma et al., 2017). It is suggested that MdOFPs in apple may participate in the regulation of salt stress responses (Li et al., 2019).
Zinc Finger Homologous Domain Proteins in Plants
Zinc finger homologous domain proteins are plant-specific transcription factors which were first identified in the C4 plant chrysanthemum (Windhövel et al., 2001). They have been implicated in the regulation of the gene coding process of PEPCase (Windhövel et al., 2001). They can specifically identify and bind nucleic acids or proteins (Takatsuji, 1999; Krishna et al., 2003; Wang et al., 2016), regulate gene expression, and play an important role in stress response and defense activation of plants (Mackay and Crossley, 1998). Many studies have identified ZF-HD gene families in Arabidopsis, peach, apple, land cotton, and coconut (Xu et al., 2014; Abdullah et al., 2018; Shalmani et al., 2019; Sun et al., 2020). AtZFHD5, a ZF-HD gene in Arabidopsis, plays an important role in bud regeneration and participates in ABA and cytokinin response (Wang et al., 2011; Kim et al., 2019). Shin-Young et al. (2011) found that the leaves of Arabidopsis expressing AtZFHD5 were larger and grew faster. Moreover, AtMIF1 may interact with a ZF-HD protein and interfere with the normal function of ZF-HD protein. Fifteen known ZFHD genes in upland cotton are involved in the early development and pigment synthesis of fibers (Abdullah et al., 2018). Overexpression of rice OsZFHD1 and OsZFHD2 resulted in the back curling of leaves (Xu et al., 2014). CoZFHD16 participates in the regulation of coconut fruit growth and development (Sun et al., 2020).
Arabidopsis AtZFHD1 can respond to salt, ABA and drought treatment, and can specifically bind to the ERD1 promoter (Tran et al., 2007; Wang et al., 2014). Most of the ZF-HD genes in tomato are expressed in flower buds, and several of them respond to abiotic stress and hormone treatment (Khadiza et al., 2017). There are 31 ZF-HD genes in the genome of Chinese cabbage, including seven BraMIF genes and 24 BraZFHD genes. Their expression is regulated by abiotic stress, vernalization, and photoperiod induction (Wang et al., 2016). In barley, HvZFHD1 is regulated by dehydration, salt stress and heat stress. All increased the expression of HvZFHD1 (Abu-Romman and Al-Hadid, 2017).
Recently, OFP genes have been implicated in abiotic stress response in Arabidopsis, rice (Ma et al., 2017; Tang et al., 2018). To date, there have been few studies of the OFP transcription factors in fruit trees, including peach where salt stress restricts the healthy development of the peach industry. Salt stress/resistance in peach trees would greatly benefit the industrial goal of safe production, ecological cultivation, and quality improvement. This study addresses the function of the PpOFP1 gene in peach, and explores its biological role in response to salt stress. We analyzed both protein function and gene expression in transgenic plants in attempt to provide a new genetic improvement of important characters of peach.
Materials and Methods
Plant Materials
In this study, the expression pattern of PpOFP1 and PpZFHD1 was detected in various tissues including root, stem, leaf, flower, seed, pericarp, and mesocarp of the peach cultivar “Zhongnongpan No. 10” at normal growth condition. The peach plants which have grown for 40 days after seeds germination were treated with 200 mM NaCl, and within 24 h at unequal time interval, the leaf was collected for qRT-PCR. Transgenic tomato over-expressing PpOFP1 and wild-type which have grown for 50 days after seeds germination were treated with 150 mM NaCl, and within 21 days at every 7 days, the leaf was collected for qRT-PCR. Tissues were collected and frozen in liquid nitrogen, and stored at −80°C for later use.
Bioinformatics Analysis
The proteome data and annotation files for peach were download from GDR1. The whole proteome data and annotation files of Arabidopsis thaliana were downloaded from phytozome v12.1 database2. The alignment file including 2588 ZF-HD_dimer domain protein sequences (PF04770) was downloaded from the pfam database3. The hmmer model was established from the alignment file and was used to search the peach and Arabidopsis proteome databases using Hmmer 3.1b software4. Candidate genes with e value less than 1 × 10–10 were selected and validated with the NCBI-CDD online tool5. The ZF-HD_dimer domain proteins in peach and Arabidopsis were aligned using the MUSCLE program6. The neighbor joining (NJ) method in Mega 77 was used to construct a phylogenetic tree where a bootstrap value was set at 1,000 times. The evolutionary tree was embellished by Evolview software8. The orthologous gene of PpZFHD1 in Arabidopsis thaliana was identified using the MCScanX package9 with default setting. Subcellular localization was predicted by Cell-PLoc 2.010. The promoter cis-elements were analyzed using the online software PlantCARE11.
RNA Extraction and Quantitative Real-Time Polymerase Chain Reaction
Total RNA was extracted from ∼0.3 g of each tissue with the RNAprep Pure Plant Kit (TianGen, Beijing, China) according to the manufacturer’s instructions. First-strand cDNA was synthetized with HIScript® III RT SuperMix for qPCR (+gDNA wiper) (Vazyme, Nanjing, China) according to the manufacturer’s instructions. qRT-PCR was conducted using ChamQTM Universal SYBR® qPCR Master Mix (Without ROX) (Vazyme, Nanjing, China) on a CFX96 real-time PCR detection system (Bio-Rad). Each assay was done in triplicate or greater. The relative expressions were estimated using the 2–ΔΔCT method (Livak and Schmittgen, 2001) with the Ppactin and SIactin genes as the internal reference. The qPCR primers are listed in Supplementary Table 1.
Production of Yeast Lines Over-Expressing PpOFP1 and PpZFHD1
The full-length coding sequence (CDS) of PpOFP1 and PpZFHD1 were downloaded from GDR (see text footnote 1) and amplified using the leaf cDNA. The primers are listed in Supplementary Table 2. The fragment was inserted into the pYES2 vector, and the positive over-expression colonies were propagated, and the plasmids were extracted and transferred into INVSc1 yeast. The OD value of yeast solution was adjusted to 1.2 at the wavelength of 600 nm, and then diluted to 10–1, 10–2, 10–3, 10–4, and 10–5. They were plated on -U solid medium with NaCl concentrations of 0, 200, and 400 mM, and cultured at 30°C for 72 h for observation of growth status.
Production of Transgenic Tomato Lines Over-Expressing PpOFP1
The PpOFP1 CDS was amplified using the leaf cDNA and spliced into the pZp211-GFP to form 35S::PpOFP1-GFP construct. The primers are listed in Supplementary Table 2. The resultant construct was introduced into Agrobacterium tumefaciens GV3101. Tomato sections were then transformed with Agrobacterium and transferred to selective medium containing kanamycin. Identification of transgenic plants was performed by PCR and qRT-PCR analysis. To verify that the tomato was an over-expression line, DNA was isolated from ∼0.3 g leaf tissue by the DNAsecure Plant Kit (TianGen, Beijing, China) according to the manufacturer’s instruction. The PCR products were visualized using agarose gel electrophoresis. qRT-PCR analysis was performed as above.
Morphological Analysis of Leaf and Stem
Microscopic observation of leaf and stem morphology were performed by paraffin sectioning according to Wang et al. (2020). For the scanning electron microscopy, the samples were treated, and micrographs were taken as described by Li et al. (2002).
Determination of Physiological Indexes
Physiological indexes including chlorophyll content, electrical conductivity, MDA content, soluble sugar content, soluble protein content, proline content, and antioxidant enzyme (SOD, POD, and CAT) activity of plant leaves were determined according to Zhao et al. (2002). At each sampling time point, at least three biological replicates were performed for each sample (over-expressor and wild-type lines). We totally sampled four times (0, 7, 14, and 21 days) for over-expressor and wild-type lines, respectively. The Analysis of Variance (ANOVA) was performed based on comparison among the four sampling time points for over-expressor and wild-type lines, respectively.
Subcellular Localization of PpOFP1 and PpZFHD1 Proteins
The full-length CDS of PpOFP1 and PpZFHD1 without stop codons were amplified and inserted into the PRI-GFP (35S::GFP) vector for detection of subcellular localization. The primers are listed in Supplementary Table 2. 35S::PpOFP1-GFP, 35S::PpZFHD1-GFP and the control 35S::GFP construct was used to infect onion epidermal cells via Agrobacterium tumefaciens strain GV3101. After 72 h incubation, the GFP fluorescence signals in the transformed onion cells were observed using a Zeiss LSM880 fluorescence microscope, and the images were analyzed using the ZEN lite software (Zeiss).
Yeast Two-Hybrid Assays
The coding region of the PpOFP1 gene and various PpOFP1 deletion derivatives were individually cloned into the pGBKT7 vector (Clontech) as bait, and the coding region of the PpZFHD1 gene, and various PpZFHD1 deletion derivatives were cloned into the pGADT7 vector (Clontech) as prey. The primers are listed in Supplementary Table 2. Different combinations of bait and prey vectors were transformed into the yeast strain Y2H cell and initially selected on selective medium (SD/–Trp/–Leu) at 30°C for ∼72 h. After the yeast cells had grown, the putative transformants were transferred to selective medium SD/-Leu/-Trp/-His/-Ade and SD/–Leu/–Trp/–His/–Ade with X-α-gal.
Bimolecular Fluorescence Complementary Assays
The coding regions of PpOFP1 and PpZFHD1 without stop codon were amplified using PCR with appropriate primers in Supplementary Table 1 and cloned into pSPYNE-35S and pSPYCE-35S, respectively. The primers are listed in Supplementary Table 2. The resulting constructs were transformed into the Agrobacterium tumefaciens LBA4404 strain. The PpOFP1-pSPYNE and PpZFHD1-pSPYCE plasmids were mixed together (1:1), after which onion epidermal cells were transfected with the mixture for 30 min at 28°C; a mixture of PpOFP1-pSPYNE and pSPYCE was used as a control. The onion epidermal cells were then transferred to solid medium. After 48 h at 28°C, the onion epidermal cells were observed at an excitation wavelength of 488 nm under a laser scanning confocal microscope (LSM880) (Carl Zeiss, Oberkochen, Germany).
Statistical Analysis
Statistical analysis was performed with IBM SPSS Statistics 25 software, and Duncan’s test was used to analyze significance (P < 0.05).
Results and Analysis
Expression Pattern, Subcellular Localization and Transgenic Validation of PpOFP1
The expression pattern of PpOFP1 was detected in various tissues (root, stem, leaf, flower, seed, exocarp, and mesocarp) under normal growth conditions (Figure 1A). The results showed that the PpOFP1 was expressed in all tissues, and that higher expression was found in mesocarp and stem, followed by root and flower. The expression of PpOFP1 in leaf was induced by ∼3 times after 1 h treatment with 200 mM NaCl, and decreased sharply after 12 h (Figure 1B), indicating that PpOFP1 could respond to high salt stress.
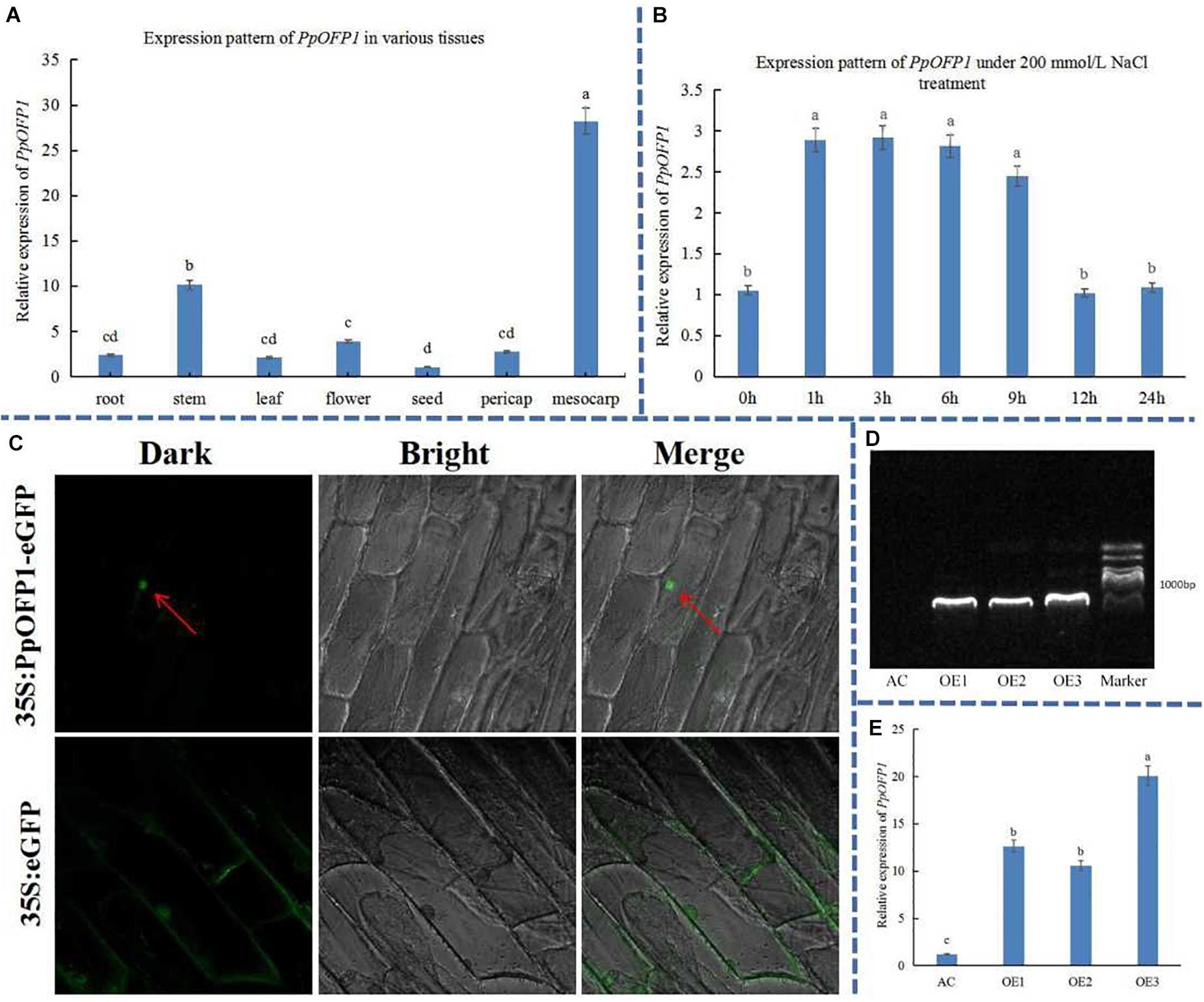
Figure 1. Expression pattern, subcellular localization, and transgenic validation of PpOFP1. (A) Expression pattern of PpOFP1 in various tissues. (B) Expression pattern of PpOFP1 under 200 mM NaCl treatment. (C) Detection of subcellular localization of PpOFP1. Red arrows indicate where a green fluorescence signal was detected in the nucleus. (D) Transgenic validation of PpOFP1 by DNA electrophoresis. (E) Transgenic validation of PpOFP1 by qRT-PCR. AC is wild-type, OE1, OE2, and OE3 are over-expressor lines of PpOFP1. DNA maker is 2,000 bp. The primer pairs used for cloning and qRT-PCR of PpOFP1 is listed in Supplementary Tables 1, 2. Different lower-case letters indicate significant differences between means as measured by ANOVA followed by Duncan’s multiple range test (P < 0.05, the same below).
The PpOFP1 protein was predicted to be located in the nucleus (Supplementary Figure 1) by Cell-PLoc 2.0 tool (see text footnote 10). To validate the subcellular localization of PpOFP1, onion epidermal cells were transfected with Agrobacterium tumefaciens carrying the PpOFP1-GFP construct. The green fluorescence signal of cells transfected with the 35S::PpOFP1-GFP construct was only detected in the nucleus (Figure 1C), while that of 35S::GFP empty vector was dispersed throughout the whole cell, indicating that the protein of PpOFP1 was localized in the nucleus.
Three independent transgenic tomato plants over-expressing PpOFP1 (OE1, OE2, and OE3) were obtained. DNA electrophoresis showed that the size of bands of these PpOFP1 over-expressors were expected (Figure 1D), and sequencing confirmed that they were PpOFP1. qRT-PCR results showed that the expression of PpOFP1 was upregulated by 10–20 fold in the over-expressor lines (Figure 1E).
Phenotypic Analysis of Transgenic Tomato Over-Expressing PpOFP1
Under normal growth conditions the transgenic lines had smaller and curled leaves, thicker stems, and thinner branches than the wild-type plants (Figures 2A,B). Scanning electron microscopy (Figure 2C) bright field images (Figure 2D) show that the structure of leaf epidermal cells of transgenic lines was more compact, flatter, longer, and had a significantly increased stomata density (Figure 2C). The length and width of stem cells were reduced, the shape was irregular, and the edge was wrinkled (Figure 2D). These results suggest that PpOFP1 may play a regulatory role in growth and development of leaf and stem.

Figure 2. Phenotypic analysis of transgenic tomato over-expressing PpOFP1. (A) Phenotypes of whole transgenic tomato under normal growth conditions. (B) Leaf phenotypes of transgenic tomato under normal growth conditions. (C) Scanning electron microscopy of leaf epidermal cell structure under normal growth conditions. (D) Cross-section of paraffin embedded stem cell tissue under normal growth conditions. (E) Phenotype of transgenic tomato under high salt stress.
Before salt stress (0 day), both wild-type and transgenic tomato grew vigorously (Figure 2E). After 7 days treatment with 150 mM NaCl, tomato plants appeared somewhat wilted, but the leaf curling and wilting of wild-type was more pronounced (Figure 2E). On the 14th day, the wild-type showed severe wilting and partial necrosis, and the lateral branches drooped. The growth state of transgenic tomato leaves, however, was better than that of wild-type plants, and the leaf wilting was less (Figure 2E). The tomato plants were rewatered without salt after 14 days, and both wild-type and transgenic tomato basically returned to normal status (Figure 2E). This indicated that the salt tolerance of transgenic tomato was better than that of wild-type.
Assays of Physiological Indexes Change of Tomato Over-Expressing PpOFP1 Under Salt Stress
To further explore the physiological mechanism of PpOFP1 to salt stress, we measured a variety of physiological indexes of wild-type and transgenic tomato after salt stress. The results showed that the chlorophyll content in leaf increased at first, but then decreased under salt stress. After rehydration with normal water, the chlorophyll content returned to the initial level. Throughout whole treatment process, the chlorophyll content of wild-type plants was always lower than that of transgenic plants (Figure 3A). The relative conductivity and MDA content of leaves increased with stress time, and those values for wild-type were always higher than for transgenic plants (Figures 3B,C). The content of soluble sugar and soluble protein also increased with stress time, the accumulation in transgenic tomato was more rapid than in wild-type. After rewatering, the content of soluble sugar and soluble protein in transgenic tomato leaves decreased significantly (Figures 3D,E). The content of proline increased significantly during the 14 days of salt stress, and decreased to normal level after 7 days of rehydration. Throughout the whole salt stress, the proline accumulation of transgenic tomato was higher than that of wild-type (Figure 3F). Figures 3G–I show that the enzyme activity increased with salt stress time, and the enzyme activity of transgenic plants increased significantly. But after re-watering, the enzyme activity decreased close to the normal level, indicating that the ability to remove reactive oxygen species was higher in transgenic plants than that of wild-type. These results further confirmed that the salt tolerance of transgenic lines was significantly higher than that of wild type.
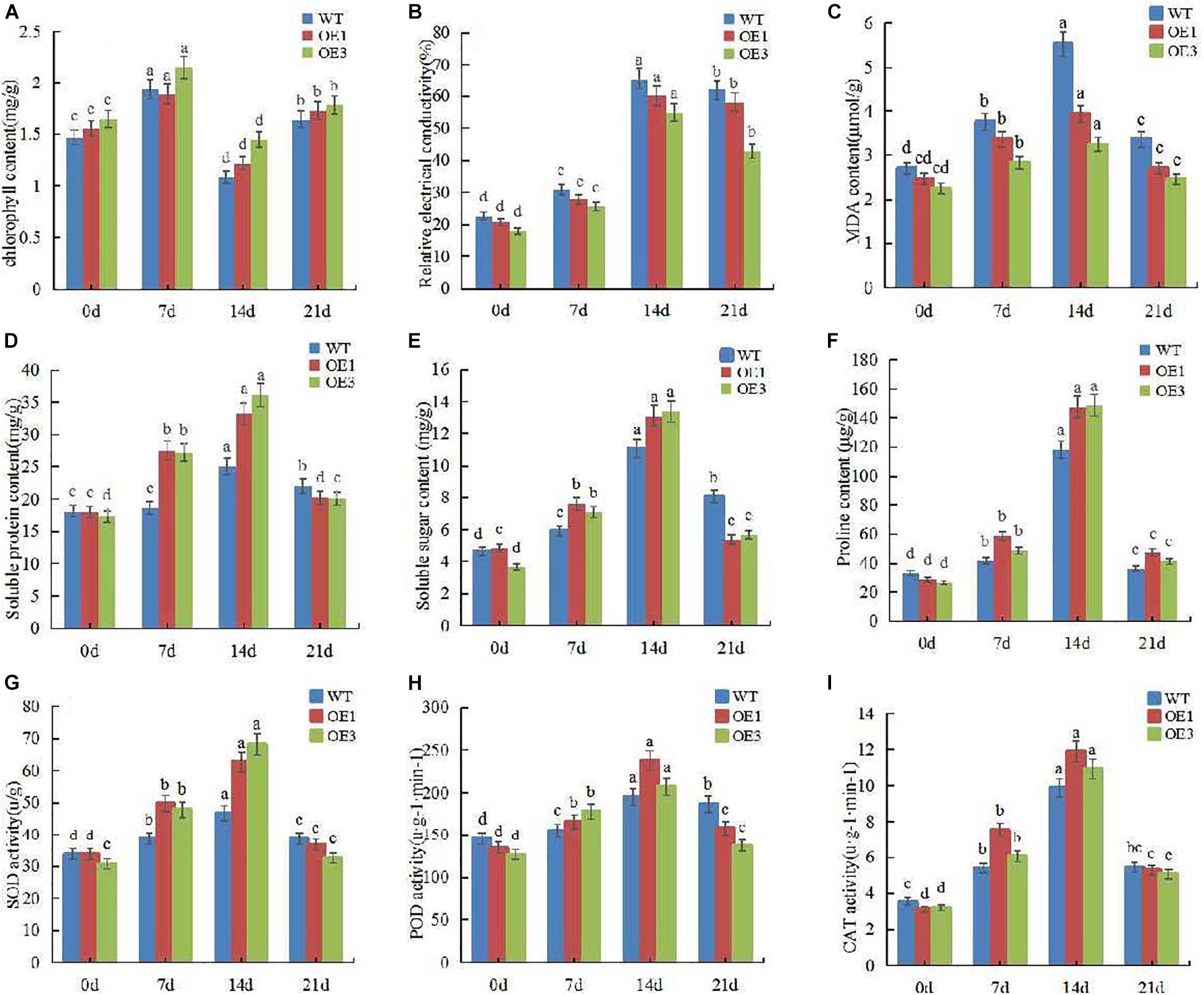
Figure 3. Determination of various physiological indexes of tomato leaf receiving 150 mM NaCl followed by no-salt watering after 14 days. (A) Chlorophyll content of leaf under 150 mM NaCl and rewatering. (B) Relative conductivity of leaf under 150 mM NaCl and rewatering. (C) MDA content of leaf under 150 mM NaCl and rewatering. (D) Soluble protein content of leaf under 150 mM NaCl and rewatering. (E) Soluble sugar content of leaf under 150 mM NaCl and rewatering. (F) Proline content of leaf under 150 mM NaCl and rewatering. (G) SOD activity of leaf under 150 mM NaCl and rewatering. (H) POD activity of leaf under 150 mM NaCl and rewatering. (I) CAT activity of leaf under 150 mM NaCl and rewatering. Different lower-case letters indicate significant differences between means as measured by ANOVA followed by Duncan’s multiple range test (P < 0.05, the same below).
Verification of Interaction Between PpOFP1 and PpZFHD1 Protein
The full-length CDS of PpZFHD1 was amplified and ligated to the pGADT7 vector, and transformed to yeast cell line Y2H along with the PpOFP1-pGBKT7 construct. Interaction was verified using a SD/-T/-L, SD/-T/-L/-H/-A, and SD/-T/-L/-H/-A + X-α-gal plate. Figure 4A shows the pink yeast growing normally on both SD/-T/-L and SD/-T/-L/-H/-A plates, while yeast colonies on the SD/-T/-L/-H/-A + X-α-gal plate turned blue, indicating an interaction between the PpOFP1 and PpZFHD1 proteins.
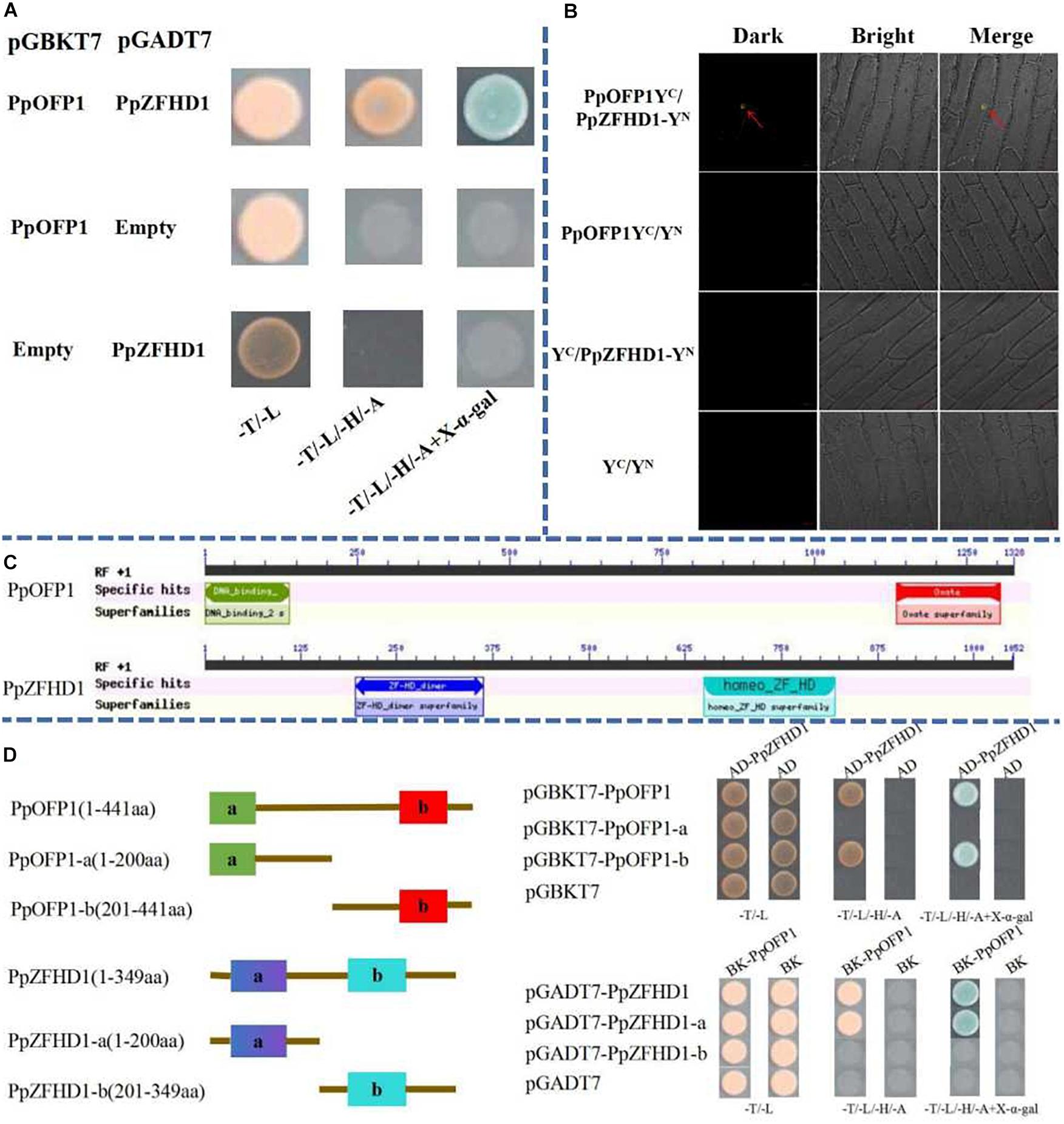
Figure 4. Interaction between PpOFP1 and PpZFHD1 proteins. (A) Interaction between PpOFP1 and PpZFHD1 proteins was verified by yeast double hybridization assay. (B) Interaction between PpOFP1 and PpZFHD1 proteins was verified by BiFC. Red arrows indicate where a green fluorescence signal was detected in the nucleus. (C) Conserved domains in PpOFP1 and PpZFHD1 proteins were predicted by the NCBI CDD online tool (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). PpOFP1 has two conserved domains: the N-terminal DNA-binding domain (PF13724) and the C-terminal OVATE domain (PF04844). There are also two domains in PpZFHD1, the ZF-HD_dimer domain (PF04770) at the N-terminal and the homeo_ZF_HD domain (TIGR01565) at the C-terminal. (D) Identification of the interaction region between PpOFP1 and PpZFHD1. a and b of PpOFP1 protein in (D) indicate the N-terminal DNA-binding domain (PF13724) and the C-terminal OVATE domain (PF04844), respectively. a and b of PpZFHD1 protein in (D) indicate the ZF-HD_dimer domain (PF04770) at the N-terminal and the homeo_ZF_HD domain (TIGR01565) at the C-terminal, respectively.
To verify that PpOFP1 and PpZFHD1 can interact in vivo, a double molecule fluorescence complementary experiment was conducted. The results showed that the onion epidermal cells co-infected by PpZFHD1-YN and PpOFP1-YC produced the yellow fluorescence signal, while no fluorescence was seen the in other co-infections: PpZFHD1-YN and YC; YN and PpOFP1-YC; and YC and YN (Figure 4B).
To further explore which domains of PpOFP1 and PpZFHD1 interact, we conducted a domains segmentation experiment. As depicted in Figure 4C, PpOFP1 has two conserved domains: the N-terminal DNA-binding domain (PF13724) and the C-terminal OVATE domain (PF04844). There are also two domains in PpZFHD1, the ZF-HD_dimer domain (PF04770) at the N-terminal and the homeo_ZF_HD domain (TIGR01565) at the C-terminal. Different combinations of functional domains were co-transformed into Y2H yeast cells, and interactions were detected with SD/-Trp/-Leu and SD/-Trp/-His/-Trp/-Ade plates. The results show that the combination of the OVATE domain of PpOFP1 and the ZF-HD_dimer domain of PpZFHD1 can grow on SD/-Trp/-His/-Trp/-Ade plates and become blue (Figure 4D). It is suggested that the OVATE domain of PpOFP1 and ZF-HD_dimer domain of PpZFHD1 are very important in the interaction between PpOFP1 and PpZFHD1, while the N-terminal DNA-binding domain of PpOFP1 and the C-terminal homeo_ZF_HD domain of PpZFHD1 do not affect the interaction between PpOFP1 and PpZFHD1.
Expression Pattern, Subcellular Localization and Transgenic Validation of PpZFHD1
We have analyzed the domain organization of ZF-HD family members, which included ZF-HD_dimer domain (PF04770). We identified 17 and 10 family members from the whole genomes of Arabidopsis and peach, respectively. Searching for protein conserved domains through NCBI-CDD online tool, the above ZF-HD proteins all contain ZF-HD_dimer domain. As the phylogenetic tree shown (Supplementary Figure 2), PpZFHD1 (Prupe.1G274700.1) closer clustered with AtZFHD1 (At1G69600.1) than other members. Collinearity analysis flanking PpZFHD1 (Prupe.1G274700.1) between Arabidopsis and peach suggested that orthologous gene of PpZFHD1 in Arabidopsis is AtZFHD1 (Supplementary Table 3).
The expression pattern of PpZFHD1 was investigated in various tissues (root, stem, leaf, flower, seed, exocarp, and mesocarp) under normal growth conditions (Figure 5A). The results showed that PpZFHD1 was expressed in all tissues, and strongest in seed and mesocarp. The results of qRT-PCR showed that after 200 mM NaCl treatment, the expression of PpZFHD1 in leaf was strongly induced, with a peak value of 50-fold (Figure 5B), indicating that PpZFHD1 responds to high salt stress.

Figure 5. Expression pattern, subcellular localization of PpZFHD1, and growth status of yeast over-expressing PpZFHD1 responsive to high salt stress. (A) Expression pattern of PpZFHD1 in various tissues. (B) Expression pattern of PpZFHD1 under 200 mM NaCl. (C) Detection of subcellular localization of PpZFHD1. (D) Growth status of yeast over-expressing PpZFHD1 responsive to high salt stress. Red arrows indicate where a green fluorescence signal was detected in the nucleus. Different lower-case letters indicate significant differences between means as measured by ANOVA followed by Duncan’s multiple range test (P < 0.05, the same below).
The PpZFHD1 protein was predicted to be localized into the nucleus (Supplementary Figure 3) by the Cell-PLoc 2.0 tool (see text footnote 10). To accurately locate the subcellular localization of PpZFHD1, we transfected onion epidermal cells with Agrobacterium tumefaciens which carried the PpZFHD1-GFP construct. As shown in Figure 5C, the green fluorescence of cells transfected with 35S::PpZFHD1-GFP fusion protein fluoresced only in the nucleus, while that of cells transfected with empty vector dispersed throughout the whole cell. These results indicate that the PpZFHD1 protein was localized into the nucleus.
To further verify whether over-expression of PpZFHD1 can confer salt tolerance to yeast, we transformed the pYES2-PpZFHD1 construct, and the empty vector pYES2 (CK) into yeast strain INVSc1. The OD value of yeast solution was adjusted to 1.2 at the wavelength of 600 nm, and then diluted to 10–1, 10–2, 10–3, 10–4, and 10–5. They were plated on -U solid medium with NaCl concentrations of 0, 200, and 400 mM, and cultured at 30°C for 72 h. Figure 5D indicates that the yeast plaque over-expressing PpZFHD1 grew better than CK under high salt stress. This is consistent with that the yeast plaque over-expressing PpOFP1 grew better than CK under high salt stress (Supplementary Figure 4).
Discussion
Response of PpOFP1 and PpZFHD1 to Salt Stress
If there are many stress response-related cis-elements in the gene promoter, the gene may participate in the responsive to abiotic stress process. In this study, we analyzed the cis-elements of the promoters of PpOFP1 (Supplementary Table 4) and PpZFHD1 (Supplementary Table 5), and found that there was a binding site (MBS) of MYB involved in drought induction in the both promoters. MYB transcription factors play a wide range of regulatory roles in stress response (Li et al., 2015). Many ZFHD genes have been found to be involved in abiotic stress responses. Sun et al. (2021) found that most ZFHD genes in tobacco could respond to drought and salt stress, and NtZFHD21 was highly expressed in response to the drought treatments. Furthermore, gene silencing of the NtZFHD21 gene reduced the drought resistance of tobacco. In this study, we found that the expression levels of PpOFP1 and PpZFHD1 were significantly increased under NaCl stress (Figures 1B, 5B). These results suggest that PpOFP1 and PpZFHD1 can respond to salt stress, which indicates that they may cooperate in the regulation of salt tolerance.
PpOFP1 Promotes the Growth of Transgenic Tomato Plants
Photosynthesis is a necessary process for the growth and development of plants, which has a great impact on the yield and quality of fruit trees. Chlorophyll is the basis of plant photosynthesis, and the change of chlorophyll content is the main basis to measure plant growth. The change of chlorophyll content in this experiment indicates that stress had less damage to tomato overexpressing PpOFP1. We considered that the increase of stomatal density on the leaf epidermis of overexpression plants enhances photosynthesis and makes plants more resistant to adversity, and then grow more vigorously. We found that the growth of transgenic PpOFP1 plants was significantly better than that of wild-type plants under salt stress, and the plant state recovered faster after rewatering. In this study, the leaves of transgenic tomato progenies were obviously curled and shrunk, and the arrangement of leaf epidermis cells was more compact, stomata increased significantly, which indicated that the overexpression of PpOFP1 in tomato plants might increase drought resistance by changing leaf morphology and structure and increasing stomatal density. The salt tolerance of plants is closely related to drought tolerance (Li et al., 2019). This may be because the curled and shrunken leaves of transgenic plants reduce the water loss of leaves and increase the light area of the whole plant, so the plant grows better.
PpOFP1 Promotes Antioxidant Enzyme Activity
Plant salt stress can damage the lipid membrane system, and malondialdehyde (MDA) is the main product of membrane lipid peroxidation. It can break many macromolecular substances that perform biological functions, such as protein, nucleic acid and enzyme (Iskender et al., 2013). Therefore, the relative conductivity and MDA content of leaves are important indicators to measure the degree of plant damage. By measuring these two indicators, we found that high salt stress caused more damage to wild-type tomato than to our transgenic plants, and that the damage degree of PpOFP1-OE3 was lower than that of PpOFP1-OE1. This may be because the expression level of PpOFP1 in PpOFP1-OE3 was higher than that of PpOFP1-OE1, which indicated that the plants over-expressing PpOFP1 have stronger resistance to salt stress. Further analysis of the antioxidant enzymes POD, SOD, and CAT showed that their activity of under salt stress was highest in PpOFP1-OE3, followed by PpOFP1-OE1 and finally wild type. We speculated that PpOFP1 could protect transgenic tomato from salt stress injury by regulating the activity of antioxidant enzymes.
PpOFP1 Promotes the Accumulation of Osmotic Adjustment Substances
Osmotic adjustment ability is one of the most basic characteristics of plant resistance to salt, and proline is the most common osmotic regulator. Many plants accumulate proline under stress conditions. The increase of proline in stress-resistant varieties is greater than that of stress-sensitive varieties (Hayat et al., 2012). In this study, the proline content of overexpression PpOFP1-OE3 lines treated with salt was higher than that of wild type. In addition, soluble sugar and soluble protein also participate in osmotic regulation. Our study found that soluble sugar and soluble protein content of wild-type tomato under salt stress were lower than that of PpOFP1 transgenic plants. PpOFP1 may enhance salt tolerance of transgenic plants by increasing both the content of osmotic substances, and the activity of antioxidant enzymes. Overexpression of the kumquat gene FcWRKY40 in both tobacco and lemon can promote high levels of proline and regulate ion transport, improving the tolerance of transgenic plants to salt (Dai et al., 2018). Studies with Malus halliana have shown that an increased accumulation of sucrose, amino acids, alkaloids, carotenoids, and other metabolites can eliminate excessive reactive oxygen species in cells and improve the salt tolerance of plants (Jia et al., 2020).
Interaction Between PpOFP1 and PpZFHD1 Is Involved in Salt Stress
Transcription factors mostly play roles in the nucleus and regulate the normal progress of various plant reactions. The localization results of this study showed that both PpOFP1 and PpZFHD1 were located in the nucleus. This is consistent with results in other species (Liu et al., 2002; Xu et al., 2014). It indicates that both PpOFP1 and PpZFHD1 can function as transcription factors. In order to further study the regulatory network of PpOFP1 in plants, we confirmed the interaction between PpZFHD1 and PpOFP1 in the nucleus by the yeast two hybrid test and bimolecular fluorescence complementary analysis (BiFC). Moreover, the N-terminal ZF-HD_dimer domain of PpZFHD1 is rich in cysteine and histidine residues. This is the region that interacts with C-terminal OVATE domain in PpOFP1 (Figure 4D), indicating that the ZF-HD_dimer domain is an indispensable part of stable inheritance and function of ZF-HD genes. ZF-HD_dimer domains can be used as novel dimer domains to form homodimers and heterodimers, and highly conserved cysteine is essential for protein-protein interactions. Moreover, potential homodimerization and heterodimerization may enhance the transcriptional activity of the ZF-HD protein. If so, this would have a significant impact on the expression of target genes and may therefore affect the stress response (Nakashima and Yamaguchi-Shinozaki, 2010). The interaction between the ZF-HD_dimer domain of TsZFHD1 and a domain of TsNAC1 can competitively inhibit the formation of homodimer, so the co-expression of TsZFHD1 and TsNAC1 can more effectively regulate the expression of target genes (Liu et al., 2019). We speculate that this mode of action may also exist between PpZFHD1 and PpOFP1. PpOFP1 binds to ZF-HD_dimer domain through OVATE domain to inhibit the formation of self homodimer of PpZFHD1, and then more effectively regulates the expression of downstream target genes.
Conclusion
In summary, our study demonstrates that PpOFP1 physically interacts with PpZFHD1 and confers salt tolerance to tomato and yeast. Our findings characterize the molecular mechanisms relating to PpOFP1 in peach and revealing a novel mechanism for regulating salt tolerance in peach and other perennial deciduous trees.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
LL, WX, QT, and SJ designed and performed the experiments and analyzed the data. QT and SJ wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was partially supported by the National Key Research and Development Plan (2018YFD1000104); National Natural Science Foundation of China (31872041); Provincial Natural Science Foundation of Shandong (ZR2018MC023); Shandong Province Agricultural Good Seed Project grants, 2020LZGC007 and 2020LZGC00702; and Funding for Major Agricultural Application Technology Innovation Projects in Shandong Province.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.759955/full#supplementary-material
Footnotes
- ^ https://www.rosaceae.org/species/prunus_persica/genome_v2.0.a1
- ^ https://phytozome.jgi.doe.gov/pz/portal.html
- ^ http://pfam.xfam.org/family/PF04770#tabview=
- ^ http://hmmer.janelia.org
- ^ https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi
- ^ http://www.drive5.com/muscle/
- ^ https://megasoftware.net/
- ^ https://www.evolgenius.info/evolview/#login
- ^ https://github.com/wyp1125/MCScanx
- ^ http://www.csbio.sjtu.edu.cn/bioinf/Cell-PLoc-2
- ^ http://bioinformatics.psb.ugent.be/webtools/plantcare/html/
References
Abdullah, M., Cheng, X., Cao, Y. P., Su, X. Q., Manzoor, M. A., Gao, J. S., et al. (2018). Zinc finger-homeodomain transcriptional factors (ZHDs) in upland cotton (Gossypium hirsutum): genome-wide identification and expression analysis in fiber development. Front. Genet. 9:357. doi: 10.3389/fgene.2018.00357
Abu-Romman, S., and Al-Hadid, K. (2017). Novel zinc finger-homeodomain gene from barley (HvZFHD1) is differentially regulated during spike development and under hormonal treatments and abiotic stresses. Not. Bot. Horti Agrobot. 45, 89–96. doi: 10.15835/nbha45110612
Al-Shorafa, W., Mahadeen, A., and Al-Absi, K. (2014). Evaluation for salt stress tolerance in two strawberry cultivars. Am. J. Agric. Biol. Sci. 9, 334–341. doi: 10.3844/ajabssp.2014.334.341
Chander, M. S., and Palaniappan, R. (2016). Oxidative stress and changes in antioxidant and biochemical constituents in papaya (Carica papaya L.) under salt stress. J. Hortic. Sci. 2, 134–138.
Dai, W. S., Wang, M., Gong, X. Q., and Liu, J. H. (2018). The transcription factor FcWRKY40 of Fortunella crassifolia functions positively in salt tolerance through modulation of ion homeostasis and proline biosynthesis by directly regulating SOS2 and P5CS1 homologs. New Phytol. 219, 972–989. doi: 10.1111/nph.15240
Fang, S., Hou, X., and Liang, X. (2021). Response mechanisms of plants under saline-alkali stress. Front. Plant Sci. 12:1049. doi: 10.3389/fpls.2021.667458
Hayat, S., Hayat, Q., Alyemeni, M. N., Wani, A. S., Pichtel, J. T., and Ahmad, A. (2012). Role of proline under changing environments: a review. Plant Signal. Behav. 7, 1456–1466. doi: 10.4161/psb.21949
Iskender, E., Kayis, T., Coskun, M., Dursun, O., and Cogun, H. Y. (2013). Changes in antioxidative enzyme activity, glycogen, lipid, protein, and malondialdehyde content in cadmium-treated Galleria mellonella larvae. Ann. Entomol. Soc. Am. 106, 371–377. doi: 10.1603/an12137
Jia, X., Zhu, Y., Zhang, R., Zhu, Z., Zhao, T., Cheng, L. R., et al. (2020). Ionomic and metabolomic analyses reveal the resistance response mechanism to saline-alkali stress in Malus halliana seedlings. Plant Physiol. Biochem. 147, 77–90. doi: 10.1016/j.plaphy.2019.12.001
Khadiza, K., Nath, U. K., Robin, A. H. K., Park, J. I., Lee, D. J., Kim, M. B., et al. (2017). Genome-wide analysis and expression profiling of zinc finger homeodomain (ZHD) family genes reveal likely roles in organ development and stress responses in tomato. BMC Genomics 18:695. doi: 10.1186/s12864-017-4082-y
Kim, J. B., Kang, J. Y., Park, M. Y., Song, M., Kim, Y. C., and Kim, S. Y. (2019). Arabidopsis zinc fnger homeodomain protein ZHD5 promotes shoot regeneration and confers other cytokinin-related phenotypes when overexpressed. Plant Cell Tissue Organ Cult. 137, 181–185. doi: 10.1007/s11240-018-01546-7
Krishna, S. S., Majumdar, I., and Grishin, N. V. (2003). SURVEY AND SUMMARY: structural classification of zinc fingers. Nucleic Acids Res. 31, 532–550. doi: 10.1093/nar/gkg161
Li, C., Ng, C. K. Y., and Fan, L. M. (2015). MYB transcription factors, active players in abiotic stress signaling. Environ. Exp. Bot. 114, 80–91. doi: 10.1016/j.envexpbot.2014.06.014
Li, H., Dong, Q., Zhao, Q., and Ran, K. (2019). Genome-wide identification, expression profiling, and protein-protein interaction properties of ovate family proteins in apple. Tree Genet. Genomes 15, 1–11.
Li, Q., Xing, L., Bai, S., Lu, W., and Xian, Z. (2002). Isolation of hag1 and its regulation by plant hormones during in vitro floral organogenesis in Hyacinthus orientalis l. Planta 215, 533–540. doi: 10.1007/s00425-002-0796-3
Liu, F., Jiyi, G., Yuke, L., Yubin, Z., and Yin, Y. (2019). Excavation of drought and salt responsive functional genes and their application in cotton breeding. Mol. Plant Breed. 17, 7395–7400.
Liu, J. H., Zhang, J., Wang, J. Y., Zhang, J. B., Miao, H. X., and Jia, C. H. (2017). MuMADS1 and MaOFP1 regulate fruit quality in a tomato ovate mutant. Plant Biotechnol. J. 16, 989–1001. doi: 10.1111/pbi.12843
Liu, J., van Eck, J., Cong, B., and Tanksley, S. D. (2002). A new class of regulatory genes underlying the cause of pearshaped tomato fruit. Proc. Natl. Acad. Sci. U.S.A. 99, 13302–13306. doi: 10.1073/pnas.162485999
Liu, Y. Y., and Douglas, C. J. (2015). A role for OVATE FAMILY PROTEIN1 (OFP1) and OFP4 in a BLH6-KNAT7 multi-protein complex regulating secondary cell wall formation in Arabidopsis thaliana. Plant Signal. Behav. 10:e1033126. doi: 10.1080/15592324.2015.1033126
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative pcr and the 2-ΔΔct method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ma, Y., Yang, C., and He, Y. (2017). Rice OVATE family protein 6 regulates plant development and confers resistance to drought and cold stresses. J. Exp. Bot. 68, 4885–4898. doi: 10.1093/jxb/erx309
Mackay, J. P., and Crossley, M. (1998). Zinc fingers are sticking together. Trends Biochem. Sci. 23, 1–4. doi: 10.1016/s0968-0004(97)01168-7
Nakashima, K., and Yamaguchi-Shinozaki, K. (2010). Regulons involved in osmotic stress-responsive and cold stress-responsive gene expression in plants. Physiol. Plant. 126, 62–71. doi: 10.1111/j.1399-3054.2005.00592.x
Shalmani, A., Muhammad, I., Sharif, R., Zhao, C., Ullah, U., and Zhang, D. (2019). Zinc fnger-homeodomain genes: evolution, functional diferentiation, and expression profling under fowering-related treatments and abiotic stresses in plants. Evol. Bioinform. 15, 1–16.
Shin-Young, H., Ok-Kyoung, K., Sang-Gyu, K., Moon-Sik, Y., and Chung-Mo, P. (2011). Nuclear import and DNA binding of the ZHD5 transcription factor is modulated by a competitive peptide inhibitor in Arabidopsis. J. Biol. Chem. 286, 1659–1668. doi: 10.1074/jbc.m110.167692
Singh, A., and Sharma, P. C. (2018). Recent insights into physiological and molecular regulation of salt stress in fruit crops. Adv. Plants Agric. Res. 8, 171–183.
Sivritepe, N., and Eriş, A. (1999). Determination of salt tolerance in some grapevine cultivars (Vitis vinifera L.) under in vitro conditions. Turk. J. Biol. 23, 473–486.
Sun, J., Xie, M., Li, X., Li, Z., Wang, Q., Ding, A., et al. (2021). Systematic investigations of the ZF-HD gene family in tobacco reveal their multiple roles in abiotic stresses. Agronomy 11:406. doi: 10.3390/agronomy11030406
Sun, X., Fan, H., Shufang, G., Rui, L., Fengfeng, J., and Yong, X. (2020). Identification and bioinformatics analysis of coconut ZF-HD gene family. Chin. J. Trop. Crops 41:284.
Takatsuji, H. (1999). Zinc-finger proteins: the classical zinc finger emerges in contemporary plant science. Plant Mol. Biol. 39, 1073–1078.
Tang, Y., Zhang, W., Yin, Y. L., Feng, P., Li, H., and Chang, Y. (2018). Expression of ovate family protein 8 affects epicuticular waxes accumulation in Arabidopsis thaliana. Bot. Stud. 59, 1–9. doi: 10.1186/s40529-018-0228-8
Tran, L. S. P., Nakashima, K., Sakuma, Y., Osakabe, Y., Qin, F., Simpson, S. D., et al. (2007). Co-expression of the stress-inducible zinc finger homeodomain ZFHD1 and NAC transcription factors enhances expression of the ERD1 gene in Arabidopsis. Plant J. 49, 46–63. doi: 10.1111/j.1365-313x.2006.02932.x
Tsaballa, A., Pasentsis, K., Darzentas, N., and Tsaftaris, A. S. (2011). Multiple evidence for the role of an Ovate-like gene in determining fruit shape in pepper. BMC Plant Biol. 11:46. doi: 10.1186/1471-2229-11-46
Wang, H., Yin, X. J., Li, X. Q., Wang, L., Zheng, Y., Xu, X. Z., et al. (2014). Genome-wide identification, evolution and expression analysis of the grape (Vitis vinifera L.) zinc finger-homeodomain gene family. Int. J. Mol. Sci. 15, 5730–5748. doi: 10.3390/ijms15045730
Wang, L., Hua, D., He, J., Duan, Y., Chen, Z., and Hong, X. (2011). Auxin Response Factor2 (ARF2) and its regulated homeodomain gene HB33 mediate abscisic acid response in Arabidopsis. PLoS Genet. 7:e1002172. doi: 10.1371/journal.pgen.1002172
Wang, Q., Xu, G., Zhao, X., Zhang, Z., Wang, X., Liu, X., et al. (2020). Transcription factor TCP20 regulates peach bud endodormancy by inhibiting DAM5/DAM6 and interacting with ABF2. J. Exp. Bot. 71, 1585–1597. doi: 10.1093/jxb/erz516
Wang, W. L., Wu, P., Li, Y., and Hou, X. L. (2016). Genome-wide analysis and expression patterns of ZF-HD transcription factors under different developmental tissues and abiotic stresses in Chinese cabbage. Mol. Genet. Genomics 291, 1451–1464. doi: 10.1007/s00438-015-1136-1
Windhövel, A., Hein, I., Dabrowa, R., and Stockhaus, J. (2001). Characterization of a novel class of plant homeodomain proteins that bind to the C4 phosphoenolpyruvate carboxylase gene of Flaveria trinervia. Plant Mol. Biol. 45, 201–214.
Xu, Y., Wang, Y., Long, Q., Huang, J., Wang, Y., and Zhou, K. (2014). Overexpression of OsZHD1, a zinc fnger homeodomain class homeobox transcription factor, induces abaxially curled and drooping leaf in rice. Planta 239, 803–816. doi: 10.1007/s00425-013-2009-7
Yang, C., Shen, W., He, Y., Tian, Z., and Li, J. (2016). OVATE family protein 8 positively mediates brassinosteroid signaling through interacting with the GSK3-like kinase in rice. PLoS Genet. 12:e1006118. doi: 10.1371/journal.pgen.1006118
Yuan, Y. Y., Fang, L. C., Karungo, S. K., Zhang, L. L., Gao, Y. Y., Li, S. H., et al. (2016). Overexpression of VaPAT1, a GRAS transcription factor from Vitis amurensis, confers abiotic stress tolerance in Arabidopsis. Plant Cell Reports 35, 655–666. doi: 10.1007/s00299-015-1910-x
Zhang, X. W., Wu, J. L., Yu, Q., Liu, R. Y., Wang, Z. Y., and Sun, Y. (2020). AtOFPs regulate cell elongation by modulating microtubule orientation via direct interaction with TONNEAU2. Plant Science 292, 110405. doi: 10.1016/j.plantsci.2020.110405
Keywords: Prunus persica, salt tolerance, OVATE Family Protein PpOFP1, ZF-HD_dimer domain protein PpZFHD1, protein interaction
Citation: Tan Q, Jiang S, Wang N, Liu X, Zhang X, Wen B, Fang Y, He H, Chen X, Fu X, Li D, Xiao W and Li L (2021) OVATE Family Protein PpOFP1 Physically Interacts With PpZFHD1 and Confers Salt Tolerance to Tomato and Yeast. Front. Plant Sci. 12:759955. doi: 10.3389/fpls.2021.759955
Received: 17 August 2021; Accepted: 04 October 2021;
Published: 12 November 2021.
Edited by:
Abidur Rahman, Iwate University, JapanReviewed by:
Arif Ashraf, University of Massachusetts Amherst, United StatesZhisheng Xu, Nanjing Agricultural University, China
Copyright © 2021 Tan, Jiang, Wang, Liu, Zhang, Wen, Fang, He, Chen, Fu, Li, Xiao and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ling Li, bGlsaW5nc2RhdUAxNjMuY29t; Wei Xiao, Z3VsaWdodDk4NjkxOEAxNjMuY29t
†These authors have contributed equally to this work
 Qiuping Tan
Qiuping Tan Shan Jiang5†
Shan Jiang5†