- 1State Key Laboratory for Managing Biotic and Chemical Treats to the Quality and Safety of Agro-products, Institute of Virology and Biotechnology, Zhejiang Academy of Agricultural Science, Hangzhou, China
- 2College of Chemistry and Life Sciences, Zhejiang Normal University, Jinhua, China
Grain quality is one of the key targets to be improved for rice breeders and covers cooking, eating, nutritional, appearance, milling, and sensory properties. Cooking and eating quality are mostly of concern to consumers and mainly determined by starch structure and composition. Although many starch synthesis enzymes have been identified and starch synthesis system has been established for a long time, novel functions of some starch synthesis genes have continually been found, and many important regulatory factors for seed development and grain quality control have recently been identified. Here, we summarize the progress in this field as comprehensively as possible and hopefully reveal some underlying molecular mechanisms controlling eating quality in rice. The regulatory network of amylose content (AC) determination is emphasized, as AC is the most important index for rice eating quality (REQ). Moreover, the regulatory mechanism of REQ, especially AC influenced by high temperature which is concerned as a most harmful environmental factor during grain filling is highlighted in this review.
Introduction
Rice is one of the most important staple foods, feeding more than half of the population in the world. Developing varieties with high quality is a major aim for rice breeders (James et al., 2003; Jeon et al., 2010). Starch accounts for more than 80% of the storage material in the rice endosperm and is composed of 10–30% amylose (AM) and 70–90% amylopectin (AP). AM mainly contains hundreds of glucose units with linear linkages, while AP contains thousands of glucose units and is highly branched through the α-1,6-glycosidic bond based on amylose (Takeda et al., 1990). Rice eating quality (REQ) is mainly assessed by three main physicochemical characteristics: the amylose content (AC), gel consistency (GC), and gelatinization temperature (GT; Juliano, 1985). The AC is the most important index for REQ, as it is the key determinant of the firmness and sticky nature of cooked rice (Tian et al., 2009; Tao et al., 2019). GC and GT are additional parameters representing the textural features of rice starch with the same AC (Cagampang et al., 1973; Gao et al., 2011; Zhang et al., 2020b). In recent years, certain novel functions of some starch synthesis genes have been revealed, and many genes involved in the regulation of seed development have been isolated. To obtain a comprehensive understanding of starch synthesis in rice, this review summarizes previous studies and hopefully uncovers some important regulatory mechanisms of seed development and quality control. The molecular regulation of rice quality, especially the AC, will be highlighted in this review.
Genetic Basis of Amylose Content in Rice
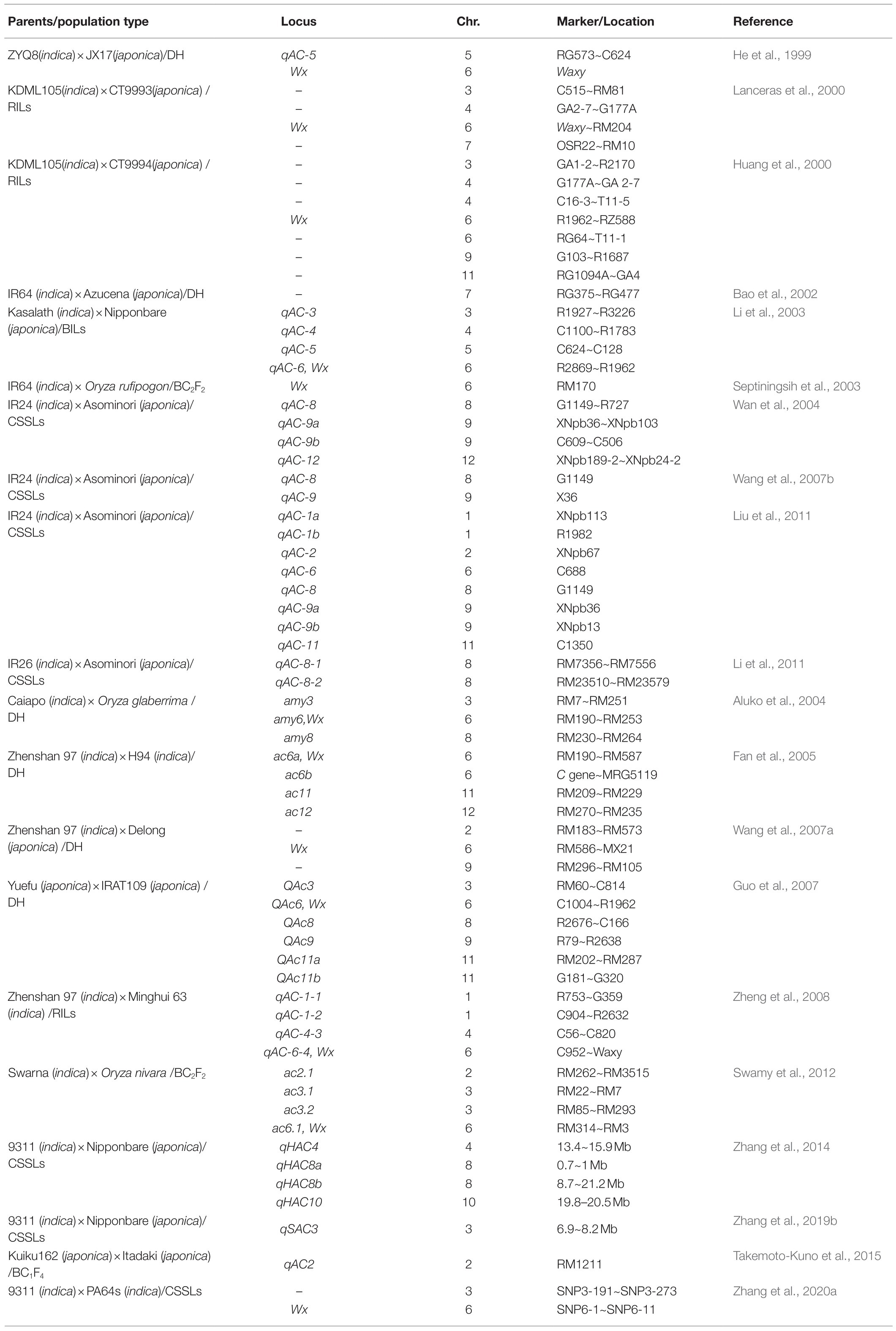
The genetic control of rice AC is relatively complex. Genetic studies using different populations, such as doubled haploid (DH), recombinant inbred lines (RILs), BCmFn, and chromosome segment substitution lines (CSSLs), have been performed (Huang et al., 2000; Lanceras et al., 2000; Li et al., 2003, 2011; Septiningsih et al., 2003; Fan et al., 2005; Guo et al., 2007; Zheng et al., 2008; Liu et al., 2011; Zhang et al., 2020a), and a series of quantitative trait loci (QTLs) and/or genes for AC have been identified in the rice genome in the past few decades (Table 1; He et al., 1999; Tan et al., 1999; Bao et al., 2002; Wang et al., 2007a; Pandey et al., 2012; Fasahat et al., 2014; Takemoto-Kuno et al., 2015; Lau et al., 2016). It is now well established that Wx on chromosome 6 is the major locus for rice AC and has been detected in almost all studies (Wang et al., 1995; Hirano and Sano, 1998). The Wx gene encodes granule-bound starch synthesis I (GBSSI), which is the key enzyme for amylose synthesis in rice (Cai et al., 1998; Huang et al., 2020b). Two alleles, Wxa and Wxb, were widely distributed in indica and japonica cultivars, respectively (Cai et al., 1998). Subsequently, more allelic variations of Wx, such as Wxop, Wxmq, Wxin, and wx, were isolated (Sato et al., 2002; Mikami et al., 2008; Liu et al., 2009; Zhang et al., 2019b, 2020c; Zhou et al., 2020). Variation in Wx can explain most of the significant alterations of rice AC in nature. QTLs on chromosome 3 have also been explored for AC modification in many different populations, such as Oryza sativa indica × Oryza sativa japonica (Lanceras et al., 2000; Zhang et al., 2019a), Oryza sativa indica (Swarna) × Oryza nivara (Swamy et al., 2012), and Oryza sativa indica (Caiapo) × Oryza glaberrima (Aluko et al., 2004). Interestingly, all the loci from indica varieties have a positive effect on AC. Their similar genetic effects and close genetic location indicate that they might represent the same locus. We named this locus qSAC3 in our previous study (Zhang et al., 2019a). Compared with the Wx allele, qSAC3 has a minor effect on rice AC. In the japonica background, introducing the indica allele of qSAC3 could mildly increase AC, and this locus was used for marker-assisted selection to improve the cooking and appearance quality in soft rice with low AC (Zhang et al., 2019a). In addition to Wx and qSAC3, two QTLs, qAC8-1 and qAC8-2, responsible for AC regulation, were also identified in multiple studies (Wan et al., 2004; Wang et al., 2007b; Li et al., 2011; Liu et al., 2011). Moreover, many other QTLs for AC were detected and distributed on all rice chromosomes, although most of them showed unstable effects across different populations or different environments. Environmental factors, such as temperature, light, and soil, were found to affect rice quality obviously, while temperature shows the greatest impact on rice AC. CSSLs with same Wx allele planted in different seasons and different locations was used to assay the AC variation (D-value) under different environments, most CSSLs showed varied D-value with their parents lines. Such results suggested that most QTLs responsible for AC determination are not stable under varying environments, and we deduced that these loci might be involved in genetic-environment interactions of AC control. Thus, AC is genetically controlled by the major locus Wx and several minor loci, such as qSAC3, qAC8-1, and qAC8-2, which could stably affect AC under multiple conditions. Fine mapping and characterization of the candidate genes of qSAC3, qAC8-1, and qAC8-2 will help us to understand their relationship with Wx and establish the exact genetic basis of AC control.
Role of Starch Biosynthesis Enzymes in Endosperm Development of Rice
Many key enzymes, such as ADP glucose pyrophosphorylase (AGPase), granule-bound starch synthase (GBSS), soluble starch synthase (SS), starch branching enzyme (SBE), and starch debranching enzyme (DBE), are involved in starch synthesis in rice seeds. Most of the enzymes that have isozymes and isoforms preferentially expressed in endosperm are responsible for starch synthesis in rice seeds, such as GBSS1 (also called Wx), SS1, SS2a (also called SSIIa/SSII-3), SS3a (also called SSIIIa/SSSIII-1), SBE1 (also called BE1), and SBE2 (also called BE2b/SBEII). Previous studies proposed that AM and AP were synthesized by different enzymes in rice. AM is mainly synthesized by GBSS1, while AP is synergistically regulated by multiple enzymes, such as SSs, SBEs, and DBEs (James et al., 2003; Jeon et al., 2010). However, recent studies have improved our understanding of the functions of SSs, which might participate in the synthesis of both AM and AP, thus affecting the rice AC.
SSs Is Essential for the AP Synthesis of Rice
It is well established that SS1, SS2a, and SS3a are responsible for AP chain elongation, while SBE1 and SBE2 control the formation of branched structures in AP (Pandey et al., 2012). The chain length distribution or degrees of polymerization (DP) in AP shows very important effects on rice quality and starch physicochemical properties (Buléon et al., 1998). The activity of rice SS1 is higher than that of SS2a and SS3a in rice endosperm. SS1 preferentially synthesizes short chains of DP 6–12. In the ss1 mutant, chains of DP 8–12 are decreased, whereas DP 6–7 chains are increased, which indicates that SS1 elongates DP 6–7 chains to DP 8–12 chains of AP (Fujita et al., 2006; Li et al., 2018). SS3a is another important enzyme for AP synthesis, and the activity of SS3a is higher than that of SS2a but lower than that of SS1. SS3a is mainly responsible for the generation of long chains (DP ≥ 30) in AP (Fujita et al., 2007). The ss3a mutant showed significantly reduced long chains of AP and abnormal starch granule morphology, which results in a floury endosperm (Ryoo et al., 2007). This result suggested that long chains catalyzed by SS3a are critical for maintaining normal structures of starch granules. In contrast, no obvious starch granule or morphological defects were observed in ss1 seeds (Fujita et al., 2006). However, the ss1/ss3a double mutant of japonica rice is sterile (Fujita et al., 2011; Hanashiro et al., 2011). These data indicated that the reduction of short chains in AP might not be enough for morphological alteration of starch granules, while the simultaneous reduction of both short and long chains could affect the formation of starch granules. SS2a was proposed to mainly produce intermediate chains (DP 13–25) of AP (Umemoto et al., 2002). The activity of SS2a is significantly different between indica and japonica. SS2a from japonica might be an inactive allele showing no or very low activity in vitro, while the indica allele has relatively higher activity (Umemoto et al., 2004). SS2a is a key gene that mainly determines rice GT, an important physiochemical property for rice eating and cooking quality (Gao et al., 2003, 2011). Introducing the indica SS2a allele into japonica rice could convert the structure of AP from the S-type (mainly in japonica cultivars) to the L-type (mostly in indica cultivars) and increase GT significantly as well (Nakamura et al., 2005).
SSs Might Play an Important Role in AM Synthesis of Rice
It was generally believed that SSs (SS1, SS2a, and SS3a) were only involved in AP synthesis. Recent studies noted that these SSs might also affect AM synthesis. The short chains of AP produced by SSs could supply substrates for the synthesis of AM (Zhu et al., 2020). SS1 is a dominant enzyme for AP synthesis, especially for the short chain of DP 6–12, as its activity accounts for approximately 70% of the total SS activity (Fujita et al., 2006). Thus, deficiency of SS1 would cause a great reduction in AP. However, the appearance of seeds and starch granules remained normal, and the AC remained unchanged in the ss1 mutant (Fujita et al., 2006). Moreover, sbe2 mutant seeds present a higher AC than wild-type seeds (Butardo et al., 2011), and knockdown of SS1 in sbe2 results in AC compensation (Abe et al., 2014). We deduced that the increase of AC in total starch was due to greatly impaired AP synthesis in sbe2, whereas defects in AM biosynthesis subtly balanced the ratio of AM to AP and returned the AC to WT level in the double mutant generated from leaky mutant of SS1crossed with sbe2 mutant. These results strongly suggested that SS1 plays important roles not only in AP synthesis but also in AM synthesis, and the short chains of AP (DP 6–12), which are mainly produced by SS1, might be important substrates for AM synthesis.
SS2a might be another SS gene involved in AM synthesis. Introducing the high activity allele SS2aInd (indica allele of SS2a) into rice plants could raise the AC whereas the effects are ecotype dependent (Yang et al., 2018; You et al., 2020). In the Wxa background, SS2aInd could increase AC dramatically (Tian et al., 2009), while in the Wxb background, SS2aInd has a minor effect on AC (Zhang et al., 2020b). The total activity of the GBSSI protein generated by Wxa is higher than that generated by Wxb (Wang et al., 1995). Therefore, we deduced that the intermediate chains of AP (DP 13–25) produced by SS2a might be substrates for AM synthesis, and Wxa may use these substrates with higher efficiency than Wxb. It will be very interesting to investigate the AC alteration in which SS2aInd is introduced into the genetic background with weaker Wx alleles, such as Wxmq and Wxhp. SS3a might not be involved in the synthesis of AM. Loss of function of SS3a caused a significant reduction in long chains (DP > 30) in AP and no obvious alteration in AM synthesis (Fujita et al., 2007), although the relative ratio of AM to AP was increased and the AC was increased in the ss3a mutant. These results indicated that long chains of AP produced by SS3a might not be used as substrates in AM synthesis. Thus, we proposed that short and intermediate chains (DP < 25) of AP might be important substrates for AM synthesis and both SS1and SS2a play important roles in this process. More evidence for this conception should be collected in the future by using other technologies, such as radio isotope tracer.
Function of SBEs in Starch Synthesis of Rice
SBE1 and SBE2 show different enzyme activities and biological functions in starch synthesis. SBE1 presents higher activity than SBE2 in rice endosperm. SBE2 has a high affinity for AP, while SBE1 is involved in branch addition in both AP and AM (Nakamura et al., 2010). Although no significant morphological defects were found in sbe1 seeds, both intermediate chains of DP 12–21 and long chains of DP ≥ 37 were reduced which resulted in a GT decrease in sbe1 seeds (Satoh et al., 2003). SBE2 seems to play a more important role in AP synthesis than SBE1 (Zhu et al., 2012; Nakata et al., 2018; Baysal et al., 2020). Short chains (DP < 17) were decreased greatly, and opacity or chalkiness appearance occurred in sbe2 (ae, amylose extender) seeds (Nishi et al., 2001; Butardo et al., 2011). This indicated that SBE2, similar to SS1, is very important for short-chain synthesis. Interestingly, in the ss1/sbe2 (ss1/ae) double mutant, similar to ss1/ss3a, normal starch granules could not be formed, and very few seeds could be produced (Abe et al., 2014). These results suggested that AP synthesis and the chain length distribution are very important for starch granule formation, which eventually affects rice yield and quality.
Other Essential Genes Regulate Seed Development and Grain Quality of Rice
Key Factors and Regulatory Network in AM Synthesis and REQ Control Related With Core Gene Wx/GBSSI
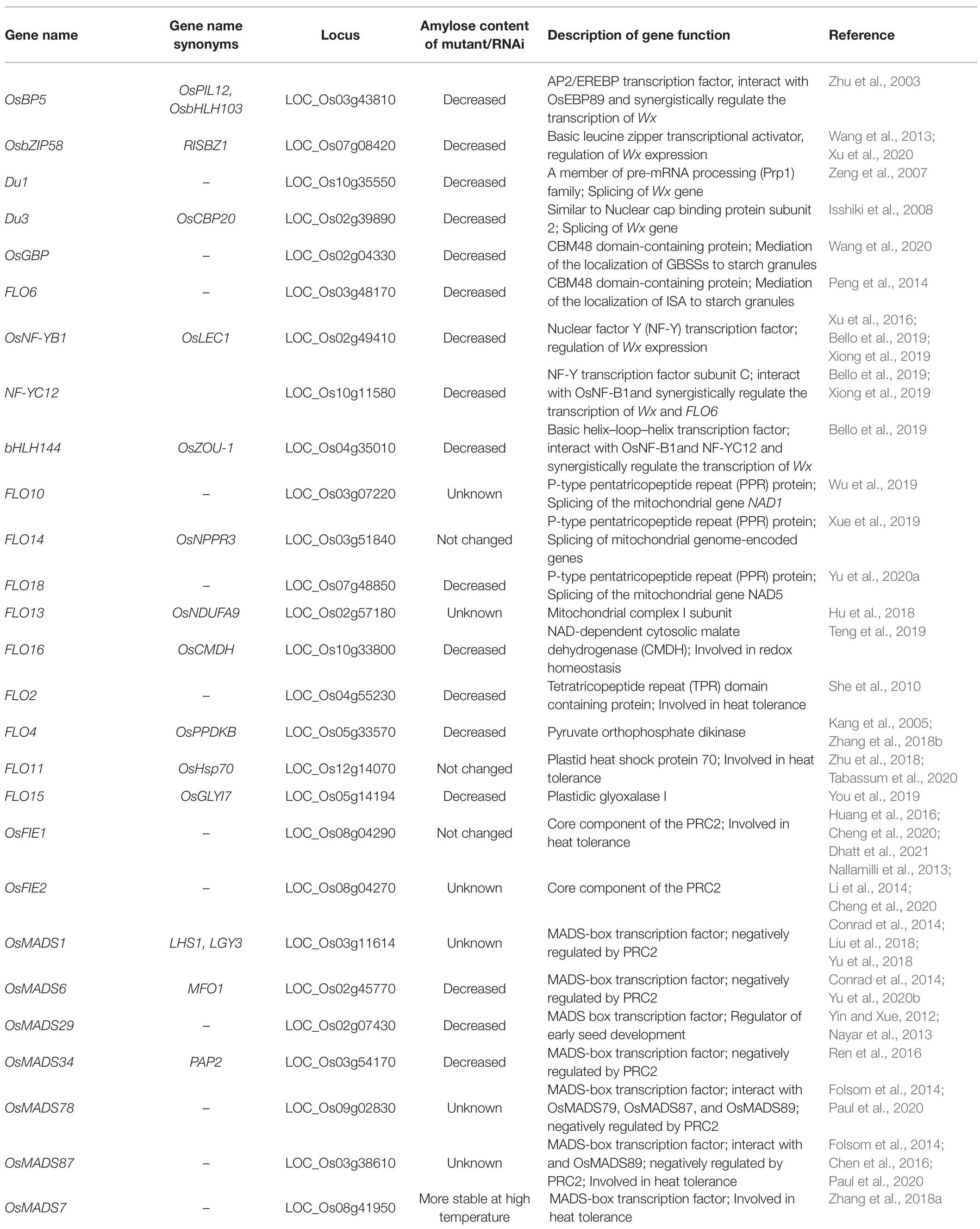
As we mentioned above, amylose synthesis is mainly controlled by the Wx gene, and many allelic Wxs, such as Wxa, Wxb, Wxin, Wxmq, Wxlv, and wx, explain a major AC variation in rice germplasm. With the development of biotechnology, more novel Wx alleles were generated and many important rice materials with different AC were produced by genetic modification or gene editing recently, and most of them occurred at coding and promoter region of Wx gene (Liu et al., 2014; Zeng et al., 2020; Huang et al., 2020a; Xu et al., 2021). Moreover, the Wx gene can be finely regulated at the transcriptional, post-transcriptional, and translational levels, and the factors involved in these processes are also important for AC modulation and rice quality control (Figure 1A).
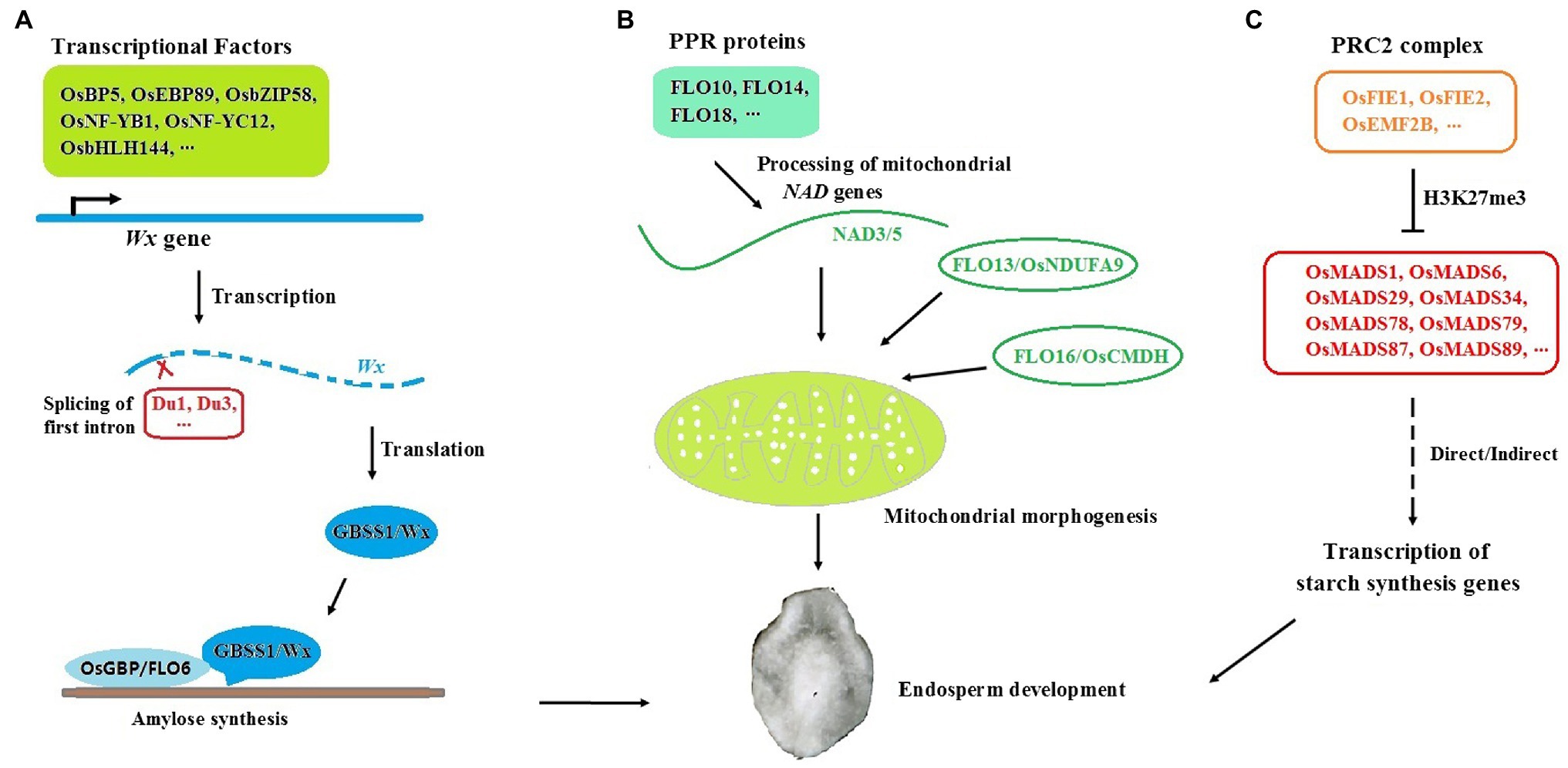
Figure 1. Regulation of amylose synthesis and endosperm development in rice seeds. (A) Wx/GBSSI is finely controlled at multiple levels. OsBP5, OsEBP89, OsbZIP58, OsNF-YB1, OsNF-YC12, and OsbHLH144 are transcriptional factors which could bind to the promoter of Wx gene and regulate its expression. Du1 and Du3 are responsible for alternative splicing of Wx. OsGBP and FLO6 might help the GBSS/Wx protein localized to starch in amylose synthesis. (B) FLO proteins (FLO10, FLO14, FLO18, FLO13, and FLO16 et al.) are involved in mitochondrial morphogenesis and endosperm development through NADH pathway. (C) The PRC2-MADS pathway for early seed development in rice. OsFIE1, OsFIE2, and OsEMF2B are important components of PRC2 complex. OsMADS1, OsMADS6, OsMADS29, OsMADS34, OsMADS78, OsMADS79, OsMADS87, and OsMADS89 are involved in the process of PRC2-mediated early endosperm development.
Many transcription factors have been reported to transactivate Wx expression by binding to cis-elements, such as 31 bp core sequences (Ge et al., 2000) and Em boxes (Cheng et al., 2002) at the upstream of Wx. For example, a bHLH transcription factor, OsBP5, together with its interacting protein, OsEBP89, binds to 31 bp and synergistically regulates the transcription of Wx (Zhu et al., 2003). OsbZIP58, a key factor in starch synthesis, was verified to directly bind to cis-elements from both Wx and SBEI and coordinately control AP and AM biosynthesis at the transcriptional level (Wang et al., 2013). Knockout or knockdown of these genes would cause alterations in AC and rice quality.
Several dull genes were isolated recently, and some of them were found to regulate Wx expression at the post-transcriptional level. Du1 encodes an mRNA splicing factor and participates in the splicing of the first intron in rice Wx (Zeng et al., 2007). Du3 encodes a protein similar to nuclear cap binding protein subunit 2 and is involved in the nuclear export of Wx mRNA (Isshiki et al., 2008). The splicing efficiency of the Wx gene was reduced and the AC decreased significantly in both du1 and du3 mutants, which indicated that post-transcriptional regulation of Wx is very important for rice quality control. In addition to the genes that regulate Wx expression and GBSSI activity, an increasing number of novel factors and pathways have been revealed to influence AM synthesis. PTST1 (Protein Targeting to Starch), which is responsible for AM synthesis in Arabidopsis leaves, was newly found to help the GBSS protein localize to starch (Seung et al., 2015). The CBM48 domain at the C-terminus of PTST1 is important for its binding activity to starch. Although GBSS itself has weak binding activity to starch, it could be recruited by PTST1 through the coiled-coil domain and subsequently bound to starch mediated by the CBM48 domain in PTST1 (Seung, 2020). OsGBP, a homolog of PTST1 in rice, could interact with both rice GBSS genes, Wx and GBSS2, in vitro. However, only AM biosynthesis in leaves but not in endosperm was greatly impaired in the osgbp mutant (Wang et al., 2020). Therefore, OsGBP may mainly function in chloroplasts, and there might be other factors involved in GBSSI locating starch in the endosperm. FLO6 (floury endosperm), another homolog of PTST1 in rice, is the most likely such gene. FLO6 contains the CBM48 domain at the C-terminus and can interact with GBSSI, GBSSII, and ISA1 to help them target starch in endosperm. Total starch and AC decreased significantly, and starch granules were abnormal in the flo6 mutant (Peng et al., 2014; Zhang et al., 2021). FLO6 might be involved in AM synthesis in rice. The transcription factor OsNF-YB1 can bind to a G-box in the Wx promoter and activate its expression (Xu et al., 2016). Moreover, OsNF-YB1 can interact with several transcription factors, such as OsNF-YC12 and OsbHLH144, and form a complex to regulate starch synthesis genes, including Wx and ISA1 (Bello et al., 2019). Mutants of osnf-yb1, osnf-yc12, and osbhlh144 displayed similar phenotypes to flo6, such as chalky endosperm, reduced grain weight, and decreased total starch and AC. Moreover, OsNF-YC12 binds to the promoter of FLO6 and directly regulates its expression (Xiong et al., 2019). Further study to reveal the biological function of FLO6 and OsGBP and their regulatory network in GBSSI activity modification will be very meaningful for elucidating starch synthesis and quality control in rice seeds.
FLO Genes Play Important Roles in Endosperm Development of Rice
In addition to flo6, many new flo mutants with floury endosperm were isolated, and most of them showed abnormal starch granules and reduced grain weight and AC (Figure 1B). Many FLO genes have been cloned and found to engage in different biological processes. FLO10, FLO14, and FLO18 encode pentatricopeptide repeat (PPR) proteins involved in RNA binding and metabolism in plant mitochondria. The processing of mitochondrial NAD genes, such as NAD1 and NAD5, was defective in flo10, flo14, or flo18 mutants (Wu et al., 2019; Xue et al., 2019; Yu et al., 2020a). NAD genes encode subunits of NADH dehydrogenase that are essential for ATP production and mitochondrial development. FLO13, known as OsNDUFA9, encodes subunit mitochondrial complex I. Loss of OsNDUFA9 changes the mitochondrial structure and greatly impairs the development of rice endosperm (Hu et al., 2018). FLO16, known as OsCMDH, encodes an NAD-dependent cytosolic malate dehydrogenase. ATP and AC were obviously reduced in the flo16 mutant (Teng et al., 2019). These reports suggested that regulators involved in the NADH pathway are essential for both mitochondrial morphogenesis and endosperm development in rice. Moreover, FLO2 was predicted to encode a tetratricopeptide repeat (TPR) domain-containing protein (She et al., 2010). The candidate gene responsible for FLO4 encodes a pyruvate orthophosphate dikinase (Kang et al., 2005; Zhang et al., 2018b). OsHsp70 is the gene responsible for the FLO11 phenotype, and FLO15 encodes glyoxalase I (Zhu et al., 2018; You et al., 2019). Clarifying the biological function of these FLO genes will be beneficial to uncover new components and pathways influencing seed development and rice quality in the future.
PRC2-MADS Cascade Is Essential for Early Seed Development of Rice
The early development of endosperm has a great influence on the quality and yield of rice. Polycomb repressive complex 2 (PRC2), which catalyzes trimethylation of histone H3 at lysine 27 (H3K27me3), is essential for the early development of endosperm (Figure 1C; Tonosaki and Kinoshita, 2015). Fertilization-independent endosperm (FIE) is an important component of PRC2. There are two FIE genes, OsFIE1 and OsFIE2, in the rice genome (Luo et al., 2009). Seed defect phenotypes, such as limited endosperm development, semisterile spikelets, and impaired grain size and quality, were obviously displayed in osfie (osfie1 or osfie2) mutants (Nallamilli et al., 2013; Li et al., 2014; Huang et al., 2016; Cheng et al., 2020). Some MADS-box genes, which are mainly responsible for floral organ identity, seem to be involved in the process of PRC2-mediated early endosperm development. For example, OsMADS6 plays an essential role in endosperm nutrient accumulation. In the osmads6 mutant, starch filling was blocked, and the relative contents of protein and soluble sugar increased, which resulted in altered grain size and quality (Yu et al., 2020b). ChIP-PCR analysis revealed that H3K27 is trimethylated in vegetative tissues where OsMADS6 is silenced (Zhang et al., 2010). Other type II MADS box genes, such as OsMADS1 (Liu et al., 2018; Yu et al., 2018), OsMADS34 (Ren et al., 2016), and OsMADS29 (Yin and Xue, 2012; Nayar et al., 2013), also contribute to early endosperm development and might be regulated by PRC2. In rice lacking OsEMF2B, another important component of PRC2, the expression of the above MADS-box genes was altered (Conrad et al., 2014; Xie et al., 2015). Moreover, several type I MADS-box genes, such as OsMADS78, OsMADS79, OsMADS87, and OsMADS89, also played essential roles in early seed development (Paul et al., 2020). OsMADS78 and OsMADS79 could interact with OsMADS87 and OsMADS89 and form a heterodimerized complex. Transgenic seeds deficient in these type I MADS-box genes exhibited accelerated endosperm cellularization and altered grain quality. The expression of these MADS genes was negatively correlated with OsFIE1 (Folsom et al., 2014). All these data suggested that PRC2-MADS might be an essential cascade for early seed development. As an increasing number of MADS genes have been found to be highly expressed in endosperm, we speculate that numerous MADS genes will function in PRC2-mediated early endosperm development and grain quality control in the future.
Regulatory Mechanisms of Rice Quality at High Temperature
In addition to genetic control, environmental factors, such as temperature, light, and soil, could affect rice quality significantly as well (Li et al., 1989; Dai et al., 1998; Yamakawa et al., 2007). For instance, GT and AC of IR661 was decreased greatly under high light condition (Li et al., 1989). Production of high-quality rice was usually associated with some specific soil (Dai et al., 1998), such as black soil in Northeast of China and distinct soil infiltrated by snow water in Niigata of Japan. However, environmental temperature might have the greatest influence on rice quality. Low total starch and AC and a highly chalky appearance were often observed in japonica cultivars under high temperature (HT; Yamakawa et al., 2007; Zhang et al., 2016). The deterioration of rice quality under HT was thought to be mainly due to the increased grain filling rate and decreased duration of grain filling (Yamakawa et al., 2007; Zhang et al., 2018a).
Expression and Splicing Efficiency of Wxb Is Important for Rice AC at HT
The reduction of the AC at HT is mostly caused by the downregulation of the Wx gene (Larkin and Park, 1999). Compared to the transcriptional inhibition of Wx by HT, post-transcriptional regulation induced by HT seems more important, especially in the Wxb background. A single nucleotide polymorphism (SNP, G to T) at the splicing site of the first intron in Wxb causes low splicing efficiency (Cai et al., 1998; Isshiki et al., 1998). The splicing efficiency is temperature-dependent. The splicing efficiency and mature transcripts of Wxb under cool temperature conditions (18°C) were much higher than those under optimal temperature conditions (25°C) and HT conditions (33°C). Two major mature transcripts could be generated from the Wxb allele under optimal temperature conditions. The large one is spliced after CT repeats (site 2), and the small one is spliced near the donor site of Wxa (site 1; Zhang et al., 2014). Two transcripts are generated almost equally from Wxb under optimal temperature conditions, while the large transcript represents the majority under HT, and the small transcript mainly exists at cool temperature (Larkin and Park, 1999; Zhang et al., 2014). These results suggested that the selection of donor sites in alternative splicing of Wxb is temperature-dependent. Alternative splicing at site 1 was suppressed by HT but promoted by cool temperature (Figure 2). We deduced that some important factors might control the selection of splicing sites and that the activity of these factors is sensitive to temperature.
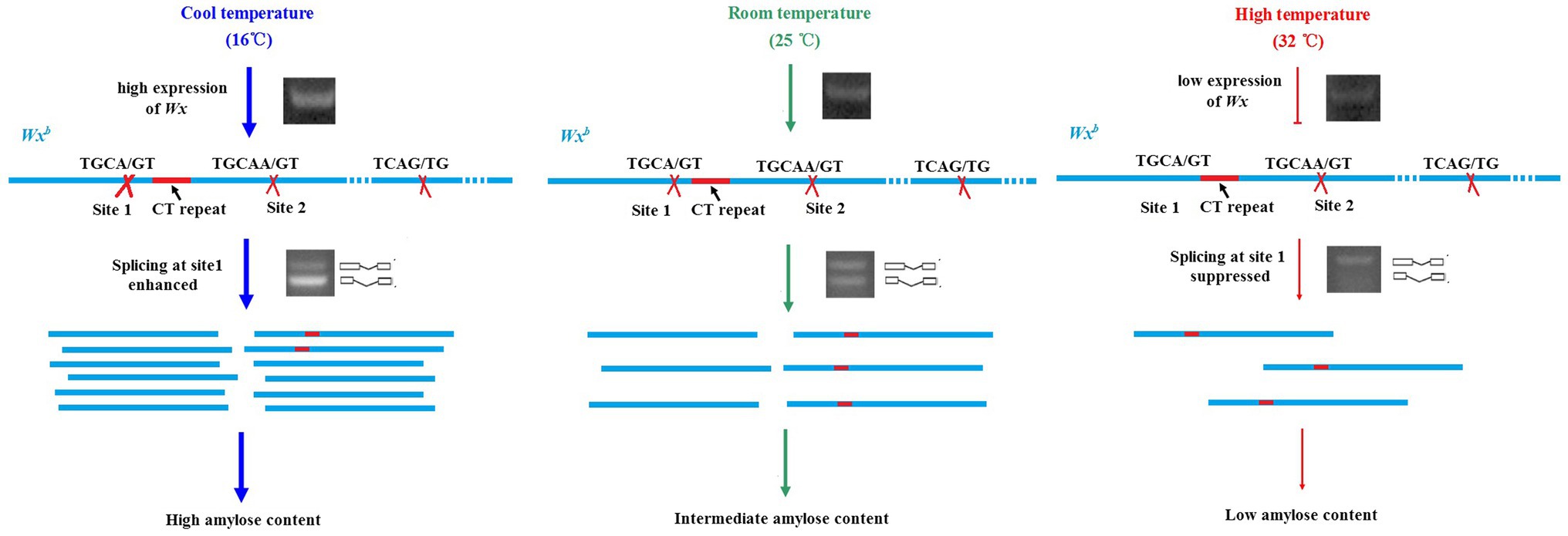
Figure 2. Transcriptional and post-transcriptional regulation of rice Wxb stimulated by different temperatures. Expression of Wxb is induced by cool temperature (16°C) but suppressed by high temperature (32°C). Alternative splicing at site 1 in the first intron of Wxb is suppressed by high temperature but promoted by cool temperature.
Indica rice is usually more tolerant to HT than japonica in terms of AC. Under the same Wxb background, the drop in AC in 9311 (indica) under HT was much smaller than that in Nipponbare (japonica). Using the CSSLs between 9311 and Nipponbare, several QTLs, qHAC8a, qHAC8b, and qHAC4, responsible for AC stabilization under HT were characterized. Introducing the indica allele of these loci into Nipponbare could enhance the splicing efficiency of Wxb, which suggested that increasing the pre-mRNA processing efficiency of the Wx gene might be an important regulatory mechanism for maintaining AC stability at HT (Figure 3A; Zhang et al., 2014). The results from MADS7-RNAi plants strongly supported this hypothesis. The floral identity gene OsMADS7 was mildly expressed in endosperm but strongly induced by HT. Suppression of OsMADS7 could improve the stability of rice AC under HT. Dynamic qRT-PCR revealed that both the expression level and the pre-mRNA processing efficiency of the Wx gene were enhanced in OsMADS7 RNAi seeds under HT during almost the entire filling stage. OsMADS7 might be the gene that can negatively regulate the expression or alternative splicing of the Wx gene under HT (Zhang et al., 2018a). Moreover, dynamic analysis revealed that grain filling rate is higher at HT than that in optimal temperature condition in both wild type ZH11 and MADS7-RNAi seeds. However, the difference in grain filling rate between HT and optimal temperature condition is smaller in MADS7-RNAi than that in ZH11, which might be another reason for relatively stable AC in MADS7-RNAi under HT condition (Zhang et al., 2018a).
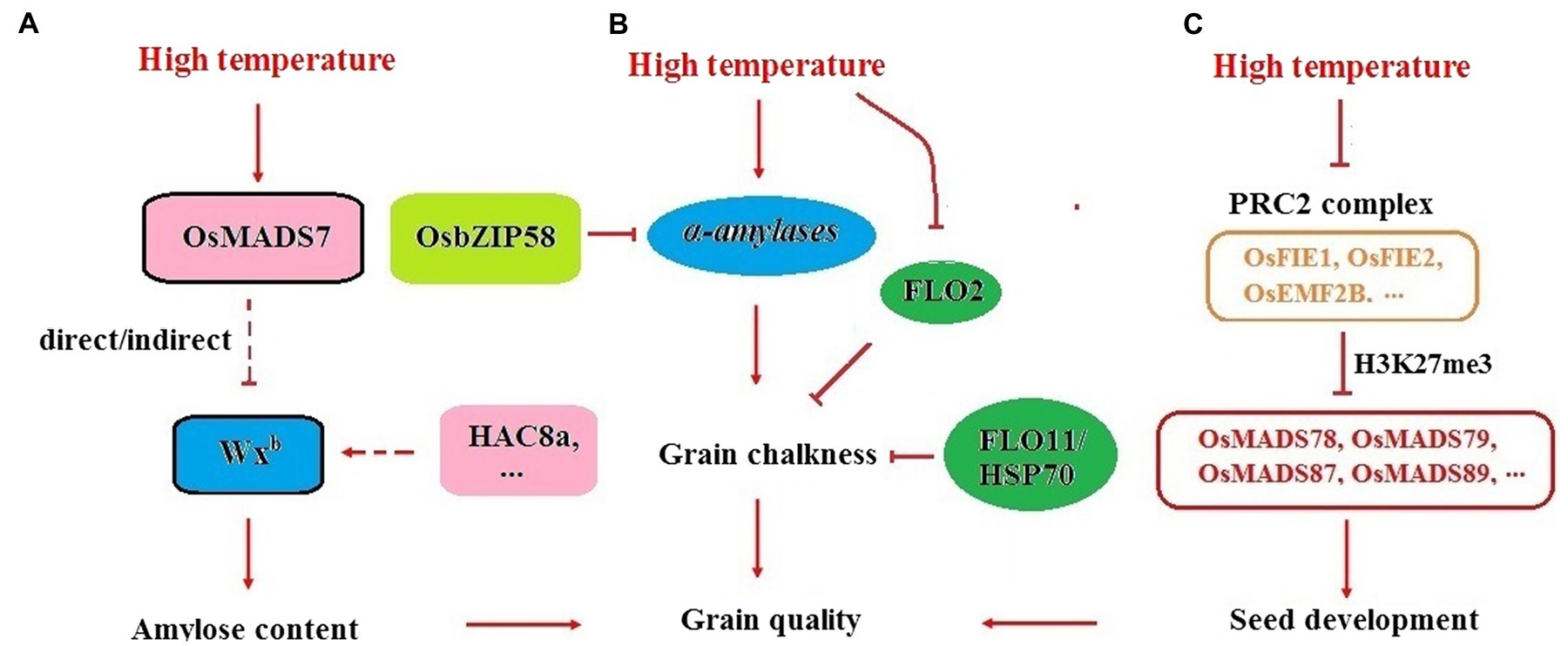
Figure 3. Seed development regulation and rice quality control under HT. (A) Increasing the efficiency of Wxb pre-mRNA processing is an important regulatory mechanism for maintaining AC stability at HT. HAC8a and OsMADS7 might be important regulators involved in this pathway. (B) α-amylases is in charge of chalky appearance under HT and OsbZIP58 seems be a key gene negatively regulating α-amylases expression. FLO2 and FLO11/HSP70 are essential genes for chalkiness production under HT. (C) PRC2-MADS pathway might be essential for the regulation of rice seed development at HT conditions. OsFIE1 might affect seed development through OsMADS87, OsMADS89, etc.
Other Starch Biosynthesis Enzymes Affect Rice AC at HT
In addition to Wx, the expression of many other starch biosynthesis genes was also changed at HT during the filling stage in rice. Overall, SS2a and SS3a were slightly downregulated, and SS1 was induced by HT. SBE1 changed slightly, whereas SBE2 decreased significantly under HT (Yamakawa et al., 2007; Liao et al., 2015). Alterations in the expression of these genes are also important for rice quality control under HT. SS3a and SBE2 are critical genes for AP synthesis. Significant downregulation of SBE2 and SS3a indicated that AP synthesis might be impaired by HT (Zhao et al., 2020). However, AM synthesis should be much more impaired than AP synthesis, since AC was reduced greatly under HT (Lin et al., 2020). The relative abilities of AM Vs AP biosynthesis under HT might be lower than those under optimal temperature conditions. As mentioned above, SS1 or SS2a is responsible for both AM and AP synthesis. Considering the great reduction in GBSSI activity (Zhang et al., 2014), the high expression of SS1 at HT might be more beneficial to the synthesis of AP than that of AM, so the reduction in AC at HT should be explained in part by the high expression of SS1. Similar to that of SS1, the high activity of the indica allele of SS2a might make AC more sensitive to HT. In contrast, knocking down SS2a might be beneficial to improve the quality of japonica rice under HT because it not only reduces rice GT but also diminishes AC effects caused by HT.
Essential Genes Regulating Seed Development and Grain Chalkiness of Rice Under HT
The PRC2-MADS pathway might also be essential for the regulation of rice endosperm development under HT conditions. Genome-wide association analysis revealed that one of the PRC2 components, OsFIE1, is a major locus for grain size regulation under HT conditions (Dhatt et al., 2021). The expression of OsFIE1 in endosperm can be suppressed by heat stress. Seed development in the osfie1 background was more sensitive to HT than that in the WT. OsMADS87 was negatively regulated by OsFIE1 but induced by HT. OsMADS87 RNAi seeds were more tolerant to HT than WT seeds by using the alteration of seed size as a trait (Figure 3C; Chen et al., 2016).
Some FLO genes might also be responsible for seed development under HT. For example, the expression of FLO2 in response to HT was different between cultivars, which indicated that FLO2 may be involved in heat tolerance during the grain filling stage (She et al., 2010). FLO11 encodes the heat shock protein OsHsp70-2, whose expression was sensitive to HT. More chalky grains were generated in the flo11 mutant than in the WT when the rice was grown at 28°C but not at 24°C, indicating that FLO11 may function under elevated temperature at the milky stage (Tabassum et al., 2020). HSPs (heat shock proteins) are molecular chaperones that delay irreversible aggregation of denatured proteins under HT condition or other stress. The expression of HSP is regulated by HSF (heat shock transcription factors) whose activity was affected by Ca2+ sensor calmodulin (CaM) in plants (Wu et al., 2012; Bourgine and Guihur, 2021). Transcripts of several HSP genes could be induced by HT (Sarkar et al., 2009), which suggested that many HSPs, HSFs and CaMs in the Ca2+–dependent heat shock signaling pathway might be essential for acquired thermotolerance of rice quality.
Dynamic analysis of gene expression in the rice endosperm revealed that α-amylases, such as Amy1A, Amy3A, and Amy3E, were greatly induced by HT. Knocking down these α-amylases significantly improved rice appearance quality under HT (Figure 3B; Hakata et al., 2012), which suggested that α-amylases might play key roles in the formation of grain quality under HT. The induced expression of starch-hydrolyzing α-amylases implied that a high speed of starch degradation might be another important cause of the increased grain chalkiness under HT conditions. The transcription factor OsbZIP58 might be an essential regulator of α-amylases. Knocking out OsbZIP58, the expression of Amy1A, Amy3A, Amy3E, and Amy1C could be increased, and osbzip58 mutants produced more chalky grains than WT at HT (Xu et al., 2020). It seems that OsbZIP58 is an effective suppressor of α-amylases in rice endosperm. It might be beneficial to increase the expression of OsbZIP58 under HT to improve the appearance quality of rice.
Conclusion and Perspective
Rice quality is a complex trait that covers biochemical, cooking, eating, nutritional, and sensory properties. Starch structure and composition largely determine rice quality, as starch is the major storage material in endosperm. Increasing consumer preference and market demand requires fine control of starch, especially the AC. Although several structural genes, chemical pathways, and regulatory networks involved in starch biosynthesis have been identified in the past few decades, the molecular mechanisms of fine control of starch metabolism remain unclear, which limits the possibility of breeding more diverse and better quality rice. It is still a major challenge for us to establish a precise genetic basis and regulatory network for grain quality, and many open questions remain to be addressed in the future. First, the AC has a decisive effect in grain quality control, and Wx is the determinant gene. Although the Wx gene has been verified to be finely regulated at multiple levels and an increasing number of essential factors have been isolated (Table 2), most regulatory mechanisms are missing. The lack of fine resolution about crystal structure and post-translational regulation of the GBSSI protein greatly limits our understanding of how to modify its activity. Moreover, in recent decades, many QTLs responsible for rice AC and many novel genes responsible for seed development have been reported. Characterization of these QTLs and genes will be immensely beneficial for clarifying the molecular mechanism of starch biosynthesis and AC control. Second, although many starch synthesis enzymes have been identified and a starch synthesis model has been established for a long time, recent research progress has provided new insights into the function of several starch synthesis enzymes, such as SSs and SBEs. Therefore, more attention should be focused on the novel functions of these starch synthesis enzymes and the physical and genetic interactions between them, which could make the model of starch synthesis more accurate. Finally, grain filling is greatly influenced by HT. The expression pattern and protein activity of many starch synthesis enzymes could be greatly altered under HT. However, only a few QTLs/genes, such as qHACs and OsMADS7, were recognized as regulatory genes involved in starch metabolism under HT. More genes and regulatory networks are expected to be explored, which will greatly contribute to breeding heat-stable rice varieties with high quality in the future.
Author Contributions
HuZ and YZ designed the manuscript. All authors listed have made a substantial, direct and intellectual contribution to the work and approved it for publication.
Funding
This work was supported by the National Key Research and Development Program of China (2016YFD0100902) and the National Natural Science Foundation of China (31401031).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We apologize to colleagues whose work is not mentioned here due to space limitation.
References
Abe, N., Asai, H., Yago, H., Oitome, N. F., Itoh, R., Crofts, N., et al. (2014). Relationships between starch synthase I and branching enzyme isozymes determined using double mutant rice lines. BMC Plant Biol. 14:80. doi: 10.1186/1471-2229-14-80
Aluko, G., Martinez, C., Tohme, J., Castano, C., Bergman, C., and Oard, J. H. (2004). QTL mapping of grain quality traits from the interspecific cross Oryza sativa×O. glaberrima. Theor. Appl. Genet. 109, 630–639. doi: 10.1007/s00122-004-1668-y
Bao, J. S., Wu, Y. R., Hu, B., Wu, P., Cui, H. R., and Shu, Q. Y. (2002). QTL for rice grain quality based on DH population derived from parents with similar apparent amylose content. Euphytica 128, 317–324. doi: 10.1023/A:1021262926145
Baysal, C., He, W., Drapal, M., Villorbina, G., Medina, V., Capell, T., et al. (2020). Inactivation of rice starch branching enzyme IIb triggers broad and unexpected changes in metabolism by transcriptional reprogramming. Proc. Natl. Acad. Sci. U. S. A. 117, 26503–26512. doi: 10.1073/pnas.2014860117
Bello, B. K., Hou, Y., Zhao, J., Jiao, G., Wu, Y., Li, Z., et al. (2019). NF-YB1-YC12-bHLH144 complex directly activates Wx to regulate grain quality in rice (Oryza sativa L.). Plant Biotechnol. J. 17, 1222–1235. doi: 10.1111/pbi.13048
Bourgine, B., and Guihur, A. (2021). Heat shock signaling in land plants: from plasma membrane sensing to the transcription of small heat shock proteins. Front. Plant Sci. 12:710801. doi: 10.3389/fpls.2021.710801
Buléon, A., Colonna, P., Planchot, V., and Ball, S. (1998). Starch granules, structure and biosynthesis. Int. J. Biol. Macromol. 23, 85–112. doi: 10.1016/S0141-8130(98)00040-3
Butardo, V. M., Fitzgerald, M. A., Bird, A. R., Gidley, M. J., Flanagan, B. M., Larroque, O., et al. (2011). Impact of down-regulation of starch branching enzyme IIb in rice by artificial microRNA- and hairpin RNA-mediated RNA silencing. J. Exp. Bot. 62, 4927–4941. doi: 10.1093/jxb/err188
Cagampang, G. B., Perez, C. M., and Juliano, B. O. (1973). A gel consistency test for eating quality of rice. J. Sci. Food Agr. 24, 1589–1594. doi: 10.1002/jsfa.2740241214
Cai, X. L., Wang, Z. Y., Xing, Y. Y., Zhang, J. L., and Hong, M. M. (1998). Aberrant splicing of intron 1 leads to the heterogeneous 5′ UTR and decreased expression of waxy gene in rice cultivars of intermediate amylose content. Plant J. 14, 459–465. doi: 10.1046/j.1365-313X.1998.00126.x
Chen, C., Begcy, K., Liu, K., Folsom, J. J., Wang, Z., Zhang, C., et al. (2016). Heat stress yields a unique MADS box transcription factor in determining seed size and thermal sensitivity. Plant Physiol. 171, 606–622. doi: 10.1104/pp.15.01992
Cheng, X., Pan, M., Zhiguo, Z., Zhou, Y., Niu, B., and Chen, C. (2020). Functional divergence of two duplicated Fertilization Independent Endosperm genes in rice with respect to seed development. Plant J. 104, 124–137. doi: 10.1111/tpj.14911
Cheng, S., Wang, Z., and Hong, M. (2002). Rice bZIP protein, REB, interacts with GCN4 motif in promoter of Waxy gene. Sci. China C Life Sci. 45, 352–359. (in Chinese). doi: 10.1360/02yc9039
Conrad, L. J., Khanday, I., Johnson, C., Guiderdoni, E., An, G., Vijayraghavan, U., et al. (2014). The polycomb group gene EMF2B is essential for maintenance of floral meristem determinacy in rice. Plant J. 80, 883–894. doi: 10.1111/tpj.12688
Dai, P., Zhou, K., Li, Y., Liu, J., Zhang, Y., Ma, G., et al. (1998). Effects of soil conditions on quality and yield of high-quality edible rice. Chin. J. Rice Sci. 12, 51–57. doi: 10.16819/j.1001-7216.1998.s1.010
Dhatt, B. K., Paul, P., Sandhu, J., Hussain, W., Irvin, L., Zhu, F., et al. (2021). Allelic variation in rice Fertilization Independent Endosperm 1 contributes to grain width under high night temperature stress. New Phytol. 229, 335–350. doi: 10.1111/nph.16897
Fan, C. C., Yu, X. Q., Xing, Y. Z., Xu, C. G., Luo, L. J., and Zhang, Q. (2005). The main effects, epistatic effects and environmental interactions of QTLs on the cooking and eating quality of rice in a doubled-haploid line population. Theor. Appl. Genet. 110, 1445–1452. doi: 10.1007/s00122-005-1975-y
Fasahat, P., Rahman, S., and Ratnam, W. (2014). Genetic controls on starch amylose content in wheat and rice grains. J. Genet. 93, 279–292. doi: 10.1007/s12041-014-0325-8
Folsom, J. J., Begcy, K., Hao, X., Wang, D., and Walia, H. (2014). Rice Fertilization-Independent Endosperm1 regulates seed size under heat stress by controlling early endosperm development. Plant Physiol. 165, 238–248. doi: 10.1104/pp.113.232413
Fujita, N., Satoh, R., Hayashi, A., Kodama, M., Itoh, R., Aihara, S., et al. (2011). Starch biosynthesis in rice endosperm requires the presence of either starch synthase I or IIIa. J. Exp. Bot. 62, 4819–4831. doi: 10.1093/jxb/err125
Fujita, N., Yoshida, M., Asakura, N., Ohdan, T., Miyao, A., Hirochika, H., et al. (2006). Function and characterization of starch synthase I using mutants in rice. Plant Physiol. 140, 1070–1084. doi: 10.1104/pp.105.071845
Fujita, N., Yoshida, M., Kondo, T., Saito, K., Utsumi, Y., Tokunaga, T., et al. (2007). Characterization of SSIIIa-deficient mutants of rice, the function of SSIIIa and pleiotropic effects by SSIIIa deficiency in the rice endosperm. Plant Physiol. 144, 2009–2023. doi: 10.1104/pp.107.102533
Gao, Z., Zeng, D., Cheng, F., Tian, Z., Guo, L., Su, Y., et al. (2011). ALK, the key gene for gelatinization temperature, is a modifier gene for gel consistency in rice. J. Integr. Plant Biol. 53, 756–765. doi: 10.1111/j.1744-7909.2011.01065.x
Gao, Z., Zeng, D., Cui, X., Zhou, Y., Yan, M., Huang, D., et al. (2003). Map-based cloning of the ALK gene, which controls the gelatinization temperature of rice. Sci. China C Life Sci. 46, 661–668. (in Chinese). doi: 10.1360/03yc0099
Ge, H. F., Wang, Z. Y., and Hong, M. M. (2000). A 31 bp fragment within the 5′ upstream region of rice Waxy gene enhances gene expression. Acta Phytophysiol. Sin. 26, 159–163. (in Chinese).
Guo, Y., Mu, P., Liu, J., Lu, Y., and Li, Z. (2007). QTL mapping and Q×E interactions of grain cooking and nutrient qualities in rice under upland and lowland environments. J. Genet. Genomics 34, 420–428. doi: 10.1016/S1673-8527(07)60046-0
Hakata, M., Kuroda, M., Miyashita, T., Yamaguchi, T., Kojima, M., Sakakibara, H., et al. (2012). Suppression of alpha-amylase genes improves quality of rice grain ripened under high temperature. Plant Biotechnol. J. 10, 1110–1117. doi: 10.1111/j.1467-7652.2012.00741.x
Hanashiro, I., Higuchi, T., Aihara, S., Nakamura, Y., and Fujita, N. (2011). Structures of starches from rice mutants deficient in the starch synthase isozyme SSI or SSIIIa. Biomacromolecules 12, 1621–1628. doi: 10.1021/bm200019q
He, P., Li, S. G., Qian, Q., Ma, Y. Q., Li, J. Z., Wang, W. M., et al. (1999). Genetic analysis of rice grain quality. Theor. Appl. Genet. 98, 502–508. doi: 10.1007/s001220051098
Hirano, H. Y., and Sano, Y. (1998). Enhancement of Wx gene expression and the accumulation of amylose in response to cool temperatures during seed development in rice. Plant Cell Physiol. 39, 807–812. doi: 10.1093/oxfordjournals.pcp.a029438
Hu, T., Tian, Y., Zhu, J., Wang, Y., Jing, R., Lei, J., et al. (2018). OsNDUFA9 encoding a mitochondrial complex I subunit is essential for embryo development and starch synthesis in rice. Plant Cell Rep. 37, 1667–1679. doi: 10.1007/s00299-018-2338-x
Huang, L., Li, Q., Zhang, C., Chu, R., Gu, Z., Tan, H., et al. (2020a). Creating novel Wx alleles with fine-tuned amylose levels and improved grain quality in rice by promoter editing using CRISPR/Cas9 system. Plant Biotechnol. J. 18, 2164–2166. doi: 10.1111/pbi.13391
Huang, X., Lu, Z., Wang, X., Ouyang, Y., Chen, W., Xie, K., et al. (2016). Imprinted gene OsFIE1 modulates rice seed development by influencing nutrient metabolism and modifying genome H3K27me3. Plant J. 87, 305–317. doi: 10.1111/tpj.13202
Huang, L., Sreenivasulu, N., and Liu, Q. (2020b). Waxy editing: old meets new. Trends Plant Sci. 25, 963–966. doi: 10.1016/j.tplants.2020.07.009
Huang, Z., Tan, X., Tragoonrung, S., and Vanavichit, A. (2000). Mapping QTLs for amylose content of grian with molecular markers in rice (Oryza sativa L.). Acta Agron. Sin. 26, 777–782. (in Chinese).
Isshiki, M., Matsuda, Y., Tasaki, A., Wong, H. L., Satoh, H., and Shimamoto, K. (2008). Du3, a mRNA cap-binding protein gene, regulates amylose content in japonica rice seeds. Plant Biotechnol. 25, 483–487. doi: 10.5511/plantbiotechnology.25.483
Isshiki, M., Morino, K., Nakajima, M., Okagaki, R. J., Wessler, S. R., Izawa, T., et al. (1998). A naturally occurring functional allele of the rice waxy locus has a GT to TT mutation at the 5′ splice site of the first intron. Plant J. 15, 133–138. doi: 10.1046/j.1365-313X.1998.00189.x
James, M. G., Denyer, K., and Myers, A. M. (2003). Starch synthesis in the cereal endosperm. Curr. Opin. Plant Biol. 6, 215–222. doi: 10.1016/S1369-5266(03)00042-6
Jeon, J. S., Ryoo, N., Hahn, T. R., Walia, H., and Nakamura, Y. (2010). Starch biosynthesis in cereal endosperm. Plant Physiol. Biochem. 48, 383–392. doi: 10.1016/j.plaphy.2010.03.006
Juliano, B. (eds.) (1985). “Criteria and test for rice grain quality,” in Rice Chemistry and Technology (Saint Paul: American Association of Cereal Chemists), 443–513.
Kang, H. G., Park, S., Matsuoka, M., and An, G. (2005). White-core endosperm floury endosperm-4 in rice is generated by knockout mutations in the C-type pyruvate orthophosphate dikinase gene (OsPPDKB). Plant J. 42, 901–911. doi: 10.1111/j.1365-313X.2005.02423.x
Lanceras, J. C., Huang, Z. L., Naivikul, O., Vanavichit, A., Ruanjaichon, V., and Tragoonrung, S. (2000). Mapping of genes for cooking and eating qualities in Thai jasmine rice (KDML105). DNA Res. 7, 93–101. doi: 10.1093/dnares/7.2.93
Larkin, P. D., and Park, W. D. (1999). Transcript accumulation and utilization of alternate and non-consensus splice sites in rice granule-bound starch synthase are temperature-sensitive and controlled by a single-nucleotide polymorphism. Plant Mol. Biol. 40, 719–727. doi: 10.1023/A:1006298608408
Lau, W. C. P., Latif, M. A., Rafii, M. Y., Ismail, M. R., and Puteh, A. (2016). Advances to improve the eating and cooking qualities of rice by marker-assisted breeding. Crit. Rev. Biotechnol. 36, 87–98. doi: 10.3109/07388551.2014.923987
Li, X., Gu, M. H., and Pan, X. B. (1989). Study on rice quality II: the influence of environmental factors on rice quality. J. Jiangsu Agr. Coll. 1, 7–12.
Li, Q., Liu, X., Zhang, C., Jiang, L., Jiang, M., Zhong, M., et al. (2018). Rice soluble starch synthase I, allelic variation, expression, function, and interaction with Waxy. Front. Plant Sci. 9:1591. doi: 10.3389/fpls.2018.01591
Li, Z., Wan, J., Xia, J., and Yano, M. (2003). Mapping of quantitative trait loci controlling physico-chemical properties of rice grains (Oryza sativa L.). Breed. Sci. 53, 209–215. doi: 10.1270/jsbbs.53.209
Li, J., Zhang, W., Wu, H., Guo, T., Liu, X., Wan, X., et al. (2011). Fine mapping of stable QTLs related to eating quality in rice (Oryza sativa L.) by CSSLs harboring small target chromosomal segments. Breed. Sci. 61, 338–346. doi: 10.1270/jsbbs.61.338
Li, S., Zhou, B., Peng, X., Kuang, Q., Huang, X., Yao, J., et al. (2014). OsFIE2 plays an essential role in the regulation of rice vegetative and reproductive development. New Phytol. 201, 66–79. doi: 10.1111/nph.12472
Liao, J. L., Zhou, H. W., Peng, Q., Zhong, P. A., Zhang, H. Y., He, C., et al. (2015). Transcriptome changes in rice (Oryza sativa L.) in response to high night temperature stress at the early milky stage. BMC Genomics 16:18. doi: 10.1186/s12864-015-1222-0
Lin, G., Yang, Y., Chen, X., Yu, X., Wu, Y., and Xiong, F. (2020). Effects of high temperature during two growth stages on caryopsis development and physicochemical properties of starch in rice. Int. J. Biol. Macromol. 145, 301–310. doi: 10.1016/j.ijbiomac.2019.12.190
Liu, Q., Han, R., Wu, K., Zhang, J., Ye, Y., Wang, S., et al. (2018). G-protein βγ subunits determine grain size through interaction with MADS-domain transcription factors in rice. Nat. Commun. 9:852. doi: 10.1038/s41467-018-03047-9
Liu, L., Ma, X., Liu, S., Zhu, C., Jiang, L., Wang, Y., et al. (2009). Identification and characterization of a novel Waxy allele from a Yunnan rice landrace. Plant Mol. Biol. 71, 609–626. doi: 10.1007/s11103-009-9544-4
Liu, X., Wan, X., Ma, X., and Wan, J. (2011). Dissecting the genetic basis for the effect of rice chalkiness, amylose content, protein content, and rapid viscosity analyzer profile characteristics on the eating quality of cooked rice using the chromosome segment substitution line population across eight environments. Genome 54, 64–80. doi: 10.1139/G10-070
Liu, D., Wang, W., and Cai, X. (2014). Modulation of amylose content by structure-based modification of OsGBSS1 activity in rice (Oryza sativa L.). Plant Biotechnol. J. 12, 1297–1307. doi: 10.1111/pbi.12228
Luo, M., Platten, D., Chaudhury, A., Peacock, W. J., and Dennis, E. S. (2009). Expression, imprinting, and evolution of rice homologs of the polycomb group genes. Mol. Plant 2, 711–723. doi: 10.1093/mp/ssp036
Mikami, I., Uwatoko, N., Ikeda, Y., Yamaguchi, J., Hirano, H. Y., Suzuki, Y., et al. (2008). Allelic diversification at the wx locus in landraces of Asian rice. Theor. Appl. Genet. 116, 979–989. doi: 10.1007/s00122-008-0729-z
Nakamura, Y., Francisco, P. B. J., Hosaka, Y., Sato, A., Sawada, T., Kubo, A., et al. (2005). Essential amino acids of starch synthase IIa differentiate amylopectin structure and starch quality between japonica and indica rice varieties. Plant Mol. Biol. 58, 213–227. doi: 10.1007/s11103-005-6507-2
Nakamura, Y., Utsumi, Y., Sawada, T., Aihara, S., Utsumi, C., Yoshida, M., et al. (2010). Characterization of the reactions of starch branching enzymes from rice endosperm. Plant Cell Physiol. 51, 776–794. doi: 10.1093/pcp/pcq035
Nakata, M., Miyashita, T., Kimura, R., Nakata, Y., Takagi, H., Kuroda, M., et al. (2018). MutMapPlus identified novel mutant alleles of a rice starch branching enzyme IIb gene for fine-tuning of cooked rice texture. Plant Biotechnol. J. 16, 111–123. doi: 10.1111/pbi.12753
Nallamilli, B. R. R., Zhang, J., Mujahid, H., Malone, B. M., Bridges, S. M., and Peng, Z. (2013). Polycomb group gene OsFIE2 regulates rice (Oryza sativa) seed development and grain filling via a mechanism distinct from Arabidopsis. PLoS Genet. 9:e1003322. doi: 10.1371/journal.pgen.1003322
Nayar, S., Sharma, R., Tyagi, A. K., and Kapoor, S. (2013). Functional delineation of rice MADS29 reveals its role in embryo and endosperm development by affecting hormone homeostasis. J. Exp. Bot. 64, 4239–4253. doi: 10.1093/jxb/ert231
Nishi, A., Nakamura, Y., Tanaka, N., and Satoh, H. (2001). Biochemical and genetic analysis of the effects of amylose-extender mutation in rice endosperm. Plant Physiol. 127, 459–472. doi: 10.1104/pp.010127
Pandey, M. K., Rani, N. S., Madhav, M. S., Sundaram, R. M., Varaprasad, G. S., Sivaranjani, A. K. P., et al. (2012). Different isoforms of starch-synthesizing enzymes controlling amylose and amylopectin content in rice (Oryza sativa L.). Biotechnol. Adv. 30, 1697–1706. doi: 10.1016/j.biotechadv.2012.08.011
Paul, P., Dhatt, B. K., Miller, M., Folsom, J. J., Wang, Z., Krassovskaya, I., et al. (2020). MADS78 and MADS79 are essential regulators of early seed development in rice. Plant Physiol. 182, 933–948. doi: 10.1104/pp.19.00917
Peng, C., Wang, Y., Liu, F., Ren, Y., Zhou, K., Lv, J., et al. (2014). FLOURY ENDOSPERM6 encodes a CBM48 domain-containing protein involved in compound granule formation and starch synthesis in rice endosperm. Plant J. 77, 917–930. doi: 10.1111/tpj.12444
Ren, D., Rao, Y., Leng, Y., Li, Z., Xu, Q., Wu, L., et al. (2016). Regulatory role of OsMADS34 in the determination of glumes fate, grain yield, and quality in rice. Front. Plant Sci. 7:1853. doi: 10.3389/fpls.2016.01853
Ryoo, N., Yu, C., Park, C. S., Baik, M. Y., Park, I. M., Cho, M. H., et al. (2007). Knockout of a starch synthase gene OsSSIIIa/Flo5 causes white-core floury endosperm in rice (Oryza sativa L.). Plant Cell Rep. 26, 1083–1095. doi: 10.1007/s00299-007-0309-8
Sarkar, N. K., Kim, Y. K., and Grover, A. (2009). Rice sHsp genes: genomic organization and expression profiling under stress and development. BMC Genomics 10:393. doi: 10.1186/1471-2164-10-393
Sato, H., Suzuki, Y., Sakai, M., and Imbe, T. (2002). Molecular characterization of Wx-mq, a novel mutant gene for low-amylose content in endosperm of rice (Oryza sativa L.). Breed. Sci. 52, 131–135. doi: 10.1270/jsbbs.52.131
Satoh, H., Nishi, A., Yamashita, K., Takemoto, Y., Tanaka, Y., Hosaka, Y., et al. (2003). Starch-branching enzyme I-deficient mutation specifically affects the structure and properties of starch in rice endosperm. Plant Physiol. 133, 1111–1121. doi: 10.1104/pp.103.021527
Septiningsih, E. M., Trijatmiko, K. R., Moeljopawiro, S., and McCouch, S. R. (2003). Identification of quantitative trait loci for grain quality in an advanced backcross population derived from the Oryza sativa variety IR64 and the wild relative O. rufipogon. Theor. Appl. Genet. 107, 1433–1441. doi: 10.1007/s00122-003-1376-z
Seung, D. (2020). Amylose in starch, towards an understanding of biosynthesis, structure and function. New Phytol. 228, 1490–1504. doi: 10.1111/nph.16858
Seung, D., Soyk, S., Coiro, M., Maier, B. A., Eicke, S., and Zeeman, S. C. (2015). PROTEIN TARGETING TO STARCH is required for localising GRANULE-BOUND STARCH SYNTHASE to starch granules and for normal amylose synthesis in Arabidopsis. PLoS Biol. 13:e1002080. doi: 10.1371/journal.pbio.1002080
She, K. C., Kusano, H., Koizumi, K., Yamakawa, H., Hakata, M., Imamura, T., et al. (2010). A novel factor FLOURY ENDOSPERM2 is involved in regulation of rice grain size and starch quality. Plant Cell 22, 3280–3294. doi: 10.1105/tpc.109.070821
Swamy, B. P. M., Kaladhar, K., Rani, N. S., Prasad, G. S. V., Viraktamath, B. C., Reddy, G. A., et al. (2012). QTL analysis for grain quality traits in 2 BC2F2 populations derived from crosses between Oryza sativa cv Swarna and 2 accessions of O. nivara. J. Hered. 103, 442–452. doi: 10.1093/jhered/esr145
Tabassum, R., Dosaka, T., Ichida, H., Morita, R., Ding, Y., Abe, T., et al. (2020). FLOURY ENDOSPERM11-2 encodes plastid HSP70-2 involved with the temperature-dependent chalkiness of rice (Oryza sativa L.) grains. Plant J. 103, 604–616. doi: 10.1111/tpj.14752
Takeda, Y., Shitaozono, T., and Hizukuri, S. (1990). Structures of sub-fractions of corn amylose. Carbohydr. Res. 199, 207–214. doi: 10.1016/0008-6215(90)84262-S
Takemoto-Kuno, Y., Mitsueda, H., Suzuki, K., Hirabayashi, H., Ideta, O., Aoki, N., et al. (2015). qAC2, a novel QTL that interacts with Wx and controls the low amylose content in rice (Oryza sativa L.). Theor. Appl. Genet. 128, 563–573. doi: 10.1007/s00122-014-2432-6
Tan, Y. F., Li, J. X., Yu, S. B., Xing, Y. Z., Xu, C. G., and Zhang, Q. (1999). The three important traits for cooking and eating quality of rice grains are controlled by a single locus in an elite rice hybrid, Shanyou 63. Theor. Appl. Genet. 99, 642–648. doi: 10.1007/s001220051279
Tao, K., Li, C., Yu, W., Gilbert, R. G., and Li, E. (2019). How amylose molecular fine structure of rice starch affects functional properties. Carbohydr. Polym. 204, 24–31. doi: 10.1016/j.carbpol.2018.09.078
Teng, X., Zhong, M., Zhu, X., Wang, C., Ren, Y., Wang, Y., et al. (2019). FLOURY ENDOSPERM16 encoding a NAD-dependent cytosolic malate dehydrogenase plays an important role in starch synthesis and seed development in rice. Plant Biotechnol. J. 17, 1914–1927. doi: 10.1111/pbi.13108
Tian, Z., Qian, Q., Liu, Q., Yan, M., Liu, X., Yan, C., et al. (2009). Allelic diversities in rice starch biosynthesis lead to a diverse array of rice eating and cooking qualities. Proc. Natl. Acad. Sci. U. S. A. 106, 21760–21765. doi: 10.1073/pnas.0912396106
Tonosaki, K., and Kinoshita, T. (2015). Possible roles for polycomb repressive complex 2 in cereal endosperm. Front. Plant Sci. 12:144. doi: 10.3389/fpls.2015.00144
Umemoto, T., Aok, N., Lin, H., Nakamura, Y., Inouchi, N., Sato, Y., et al. (2004). Natural variation in rice starch synthase IIa affects enzyme and starch properties. Funct. Plant Biol. 31, 671–684. doi: 10.1071/FP04009
Umemoto, T., Yano, M., Satoh, H., Shomura, A., and Nakamura, Y. (2002). Mapping of a gene responsible for the difference in amylopectin structure between japonica-type and indica-type rice varieties. Theor. Appl. Genet. 104, 1–8. doi: 10.1007/s001220200000
Wan, X. Y., Wan, J. M., Su, C. C., Wang, C. M., Shen, W. B., Li, J. M., et al. (2004). QTL detection for eating quality of cooked rice in a population of chromosome segment substitution lines. Theor. Appl. Genet. 110, 71–79. doi: 10.1007/s00122-004-1744-3
Wang, L. Q., Liu, W. J., Xu, Y., He, Y. Q., Luo, L. J., Xing, Y. Z., et al. (2007a). Genetic basis of 17 traits and viscosity parameters characterizing the eating and cooking quality of rice grain. Theor. Appl. Genet. 115, 463–476. doi: 10.1007/s00122-007-0580-7
Wang, J., Wan, X., Li, H., Pfeiffer, W. H., Crouch, J., and Wan, J. (2007b). Application of identified QTL-marker associations in rice quality improvement through a design-breeding approach. Theor. Appl. Genet. 115, 87–100. doi: 10.1007/s00122-007-0545-x
Wang, W., Wei, X., Jiao, G., Chen, W., Wu, Y., Sheng, Z., et al. (2020). GBSS-BINDING PROTEIN, encoding a CBM48 domain-containing protein, affects rice quality and yield. J. Integr. Plant Biol. 62, 948–966. doi: 10.1111/jipb.12866
Wang, J. C., Xu, H., Zhu, Y., Liu, Q. Q., and Cai, X. L. (2013). OsbZIP58, a basic leucine zipper transcription factor, regulates starch biosynthesis in rice endosperm. J. Exp. Bot. 64, 3453–3466. doi: 10.1093/jxb/ert187
Wang, Z. Y., Zheng, F. Q., Shen, G. Z., Gao, J. P., Snustad, D. P., Li, M. G., et al. (1995). The amylose content in rice endosperm is related to the post-transcriptional regulation of the waxy gene. Plant J. 7, 613–622. doi: 10.1046/j.1365-313X.1995.7040613.x
Wu, H. C., Luo, D. L., Vignols, F., and Jinn, T. L. (2012). Heat shock-induced biphasic Ca(2+) signature and OsCaM1-1 nuclear localization mediate downstream signalling in acquisition of thermotolerance in rice (Oryza sativa L.). Plant Cell Environ. 35, 1543–1557. doi: 10.1111/j.1365-3040.2012.02508.x
Wu, M., Ren, Y., Cai, M., Wang, Y., Zhu, S., Zhu, J., et al. (2019). Rice FLOURY ENDOSPERM10 encodes a pentatricopeptide repeat protein that is essential for the trans-splicing of mitochondrial nad1 intron 1 and endosperm development. New Phytol. 223, 736–750. doi: 10.1111/nph.15814
Xie, S., Chen, M., Pei, R., Ouyang, Y., and Yao, J. (2015). OsEMF2b acts as a regulator of flowering transition and floral organ identity by mediating H3K27me3 deposition at OsLFL1 and OsMADS4 in rice. Plant Mol. Biol. Rep. 33, 121–132. doi: 10.1007/s11105-014-0733-1
Xiong, Y., Ren, Y., Li, W., Wu, F., Yang, W., Huang, X., et al. (2019). NF-YC12 is a key multi-functional regulator of accumulation of seed storage substances in rice. J. Exp. Bot. 70, 3765–3780. doi: 10.1093/jxb/erz168
Xu, H., Li, X., Zhang, H., Wang, L., Zhu, Z., Gao, J., et al. (2020). High temperature inhibits the accumulation of storage materials by inducing alternative splicing of OsbZIP58 during filling stage in rice. Plant Cell Environ. 43, 1879–1896. doi: 10.1111/pce.13779
Xu, Y., Lin, Q., Li, X., Wang, F., Chen, Z., Wang, J., et al. (2021). Fine-tuning the amylose content of rice by precise base editing of the Wx gene. Plant Biotechnol. J. 19, 11–13. doi: 10.1111/pbi.13433
Xu, J. J., Zhang, X. F., and Xue, H. W. (2016). Rice aleurone layer specific OsNF-YB1 regulates grain filling and endosperm development by interacting with an ERF transcription factor. J. Exp. Bot. 67, 6399–6411. doi: 10.1093/jxb/erw409
Xue, M., Liu, L., Yu, Y., Zhu, J., Gao, H., Wang, Y., et al. (2019). Lose-of-function of a rice nucleolus-localized pentatricopeptide repeat protein is responsible for the floury endosperm14 mutant phenotypes. Rice 12:100. doi: 10.1186/s12284-019-0359-x
Yamakawa, H., Hirose, T., Kuroda, M., and Yamaguchi, T. (2007). Comprehensive expression profiling of rice grain filling-related genes under high temperature using DNA microarray. Plant Physiol. 144, 258–277. doi: 10.1104/pp.107.098665
Yang, B., Xu, S., Xu, L., You, H., and Xiang, X. (2018). Effects of Wx and its interaction with SSIII-2 on rice eating and cooking qualities. Front. Plant Sci. 9:456. doi: 10.3389/fpls.2018.00456
Yin, L. L., and Xue, H. W. (2012). The MADS29 transcription factor regulates the degradation of the nucellus and the nucellar projection during rice seed development. Plant Cell 24, 1049–1065. doi: 10.1105/tpc.111.094854
You, X., Zhang, W., Hu, J., Jing, R., Cai, Y., Feng, Z., et al. (2019). FLOURY ENDOSPERM15 encodes a glyoxalase I involved in compound granule formation and starch synthesis in rice endosperm. Plant Cell Rep. 38, 345–359. doi: 10.1007/s00299-019-02370-9
You, H., Zhang, O., Xu, L., Liang, C., and Xiang, X. (2020). Effects of soluble starch synthase IIa allelic variation on rice grain quality with different Waxy backgrounds. J. Sci. Food Agr. 100, 5344–5351. doi: 10.1002/jsfa.10582
Yu, J., Miao, J., Zhang, Z., Xiong, H., Zhu, X., Sun, X., et al. (2018). Alternative splicing of OsLG3b controls grain length and yield in japonica rice. Plant Biotechnol. J. 16, 1667–1678. doi: 10.1111/pbi.12903
Yu, M., Wu, M., Ren, Y., Wang, Y., Li, J., Lei, C., et al. (2020a). Rice FLOURY ENDOSPERM 18 encodes a pentatricopeptide repeat protein required for 5′ processing of mitochondrial nad5 messenger RNA and endosperm development. J. Integr. Plant Biol. 63, 834–847. doi: 10.1111/jipb.13049
Yu, X., Xia, S., Xu, Q., Cui, Y., Gong, M., Zeng, D., et al. (2020b). ABNORMAL FLOWER AND GRAIN 1 encodes OsMADS6 and determines palea identity and affects rice grain yield and quality. Sci. China Life Sci. 63, 228–238. doi: 10.1007/s11427-019-1593-0
Zeng, D., Liu, T., Ma, X., Wang, B., Zheng, Z., Zhang, Y., et al. (2020). Quantitative regulation of Waxy expression by CRISPR/Cas9-based promoter and 5′UTR-intron editing improves grain quality in rice. Plant Biotechnol. J. 18, 2385–2387. doi: 10.1111/pbi.13427
Zeng, D., Yan, M., Wang, Y., Liu, X., Qian, Q., and Li, J. (2007). Du1, encoding a novel Prp1 protein, regulates starch biosynthesis through affecting the splicing of Wxb pre-mRNAs in rice (Oryza sativa L.). Plant Mol. Biol. 65, 501–509. doi: 10.1007/s11103-007-9186-3
Zhang, H., Duan, L., Dai, J. S., Zhang, C. Q., Li, J., Gu, M. H., et al. (2014). Major QTLs reduce the deleterious effects of high temperature on rice amylose content by increasing splicing efficiency of Wx pre-mRNA. Theor. Appl. Genet. 127, 273–282. doi: 10.1007/s00122-013-2216-4
Zhang, A., Gao, Y., Li, Y., Ruan, B., Yang, S., Liu, C., et al. (2020a). Genetic analysis for cooking and eating quality of super rice and fine mapping of a novel locus qGC10 for gel consistency. Front. Plant Sci. 11:342. doi: 10.3389/fpls.2020.00342
Zhang, L., Li, N., Zhang, J., Zhao, L., Qiu, J., and Wei, C. (2021). The CBM48 domain-containing protein FLO6 regulates starch synthesis by interacting with SSIVb and GBSS in rice. Plant Mol. Biol. doi: 10.1007/s11103-021-01178-0 [Epub ahead of print].
Zhang, J., Nallamilli, B. R., Mujahid, H., and Peng, Z. (2010). OsMADS6 plays an essential role in endosperm nutrient accumulation and is subject to epigenetic regulation in rice (Oryza sativa). Plant J. 64, 604–617. doi: 10.1111/j.1365-313X.2010.04354.x
Zhang, H., Xu, H., Feng, M., and Zhu, Y. (2018a). Suppression of OsMADS7 in rice endosperm stabilizes amylose content under high temperature stress. Plant Biotechnol. J. 16, 18–26. doi: 10.1111/pbi.12745
Zhang, C., Yang, Y., Chen, Z., Chen, F., Pan, L., Lu, Y., et al. (2020b). Characteristics of grain physicochemical properties and the starch structure in rice carrying a mutated ALK/SSIIa gene. J. Agric. Food Chem. 68, 13950–13959. doi: 10.1021/acs.jafc.0c01471
Zhang, C., Yang, Y., Chen, S., Liu, X., Zhu, J., Zhou, L., et al. (2020c). A rare Waxy allele coordinately improves rice eating and cooking quality and grain transparency. J. Integr. Plant Biol. 63, 889–901. doi: 10.1111/jipb.13010
Zhang, L., Zhao, L., Lin, L., Zhao, L., Liu, Q., and Wei, C. (2018b). A novel mutation of OsPPDKB, encoding pyruvate orthophosphate dikinase, affects metabolism and structure of starch in the rice endosperm. Int. J. Mol. Sci. 19:2268. doi: 10.3390/ijms19082268
Zhang, H., Zhou, L., Xu, H., Wang, L., Liu, H., Zhang, C., et al. (2019a). The qSAC3 locus from indica rice effectively increases amylose content under a variety of conditions. BMC Plant Biol. 19:275. doi: 10.1186/s12870-019-1860-5
Zhang, C., Zhou, L., Zhu, Z., Lu, H., Zhou, X., Qian, Y., et al. (2016). Characterization of grain quality and starch fine structure of two japonica rice (Oryza sativa) cultivars with good sensory properties at different temperatures during the filling stage. J. Agric. Food Chem. 64, 4048–4057. doi: 10.1021/acs.jafc.6b00083
Zhang, C., Zhu, J., Chen, S., Fan, X., Li, Q., Lu, Y., et al. (2019b). Wxlv, the ancestral allele of rice Waxy gene. Mol. Plant 12, 1157–1166. doi: 10.1016/j.molp.2019.05.011
Zhao, Q., Ye, Y., Han, Z., Zhou, L., Guan, X., Pan, G., et al. (2020). SSIIIa-RNAi suppression associated changes in rice grain quality and starch biosynthesis metabolism in response to high temperature. Plant Sci. 294:110443. doi: 10.1016/j.plantsci.2020.110443
Zheng, X., Wu, J. G., Luo, X. Y., Xu, H. M., and Shi, C. H. (2008). The QTL analysis on maternal and endosperm genome and their environmental interactions for characters of cooking quality in rice (Oryza sativa L.). Theor. Appl. Genet. 116, 335–342. doi: 10.1007/s00122-007-0671-5
Zhou, H., Xia, D., Zhao, D., Li, Y., Li, P., Wu, B., et al. (2020). The origin of Wxla provides new insights into the improvement of grain quality in rice. J. Integr. Plant Biol. 63, 878–888. doi: 10.1111/jipb.13011
Zhu, Y., Cai, X. L., Wang, Z. Y., and Hong, M. M. (2003). An interaction between a MYC protein and an EREBP protein is involved in transcriptional regulation of the rice Wx gene. J. Biol. Chem. 278, 47803–47811. doi: 10.1074/jbc.M302806200
Zhu, L., Gu, M., Meng, X., Cheung, S. C. K., Yu, H., Huang, J., et al. (2012). High-amylose rice improves indices of animal health in normal and diabetic rats. Plant Biotechnol. J. 10, 353–362. doi: 10.1111/j.1467-7652.2011.00667.x
Zhu, X., Teng, X., Wang, Y., Hao, Y., Jing, R., Wang, Y., et al. (2018). FLOURY ENDOSPERM11 encoding a plastid heat shock protein 70 is essential for amyloplast development in rice. Plant Sci. 277, 89–99. doi: 10.1016/j.plantsci.2018.09.020
Keywords: starch biosynthesis, regulatory mechanism, rice eating quality, amylose content, high temperature
Citation: Zhang H, Xu H, Jiang Y, Zhang H, Wang S, Wang F and Zhu Y (2021) Genetic Control and High Temperature Effects on Starch Biosynthesis and Grain Quality in Rice. Front. Plant Sci. 12:757997. doi: 10.3389/fpls.2021.757997
Edited by:
Sebastien Christian Carpentier, Bioversity International (Belgium), BelgiumReviewed by:
Vesna Dragicevic, Maize Research Institute Zemun Polje, SerbiaWenqin Wang, Shanghai Normal University, China
Copyright © 2021 Zhang, Xu, Jiang, Zhang, Wang, Wang and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ying Zhu, eXpodXphYXNAMTYzLmNvbQ==
 Hua Zhang1
Hua Zhang1 Ying Zhu
Ying Zhu