
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 02 December 2021
Sec. Plant Breeding
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.756741
 Ling Qiao1,2†
Ling Qiao1,2† Justin Wheeler2†
Justin Wheeler2† Rui Wang2
Rui Wang2 Kyle Isham2
Kyle Isham2 Natalie Klassen2
Natalie Klassen2 Weidong Zhao2
Weidong Zhao2 Meng Su2
Meng Su2 Junli Zhang3
Junli Zhang3 Jun Zheng1*
Jun Zheng1* Jianli Chen2*
Jianli Chen2*Cadmium (Cd) is a heavy metal that can cause a variety of adverse effects on human health, including cancer. Wheat comprises approximately 20% of the human diet worldwide; therefore, reducing the concentrations of Cd in wheat grain will have significant impacts on the intake of Cd in food products. The tests for measuring the Cd content in grain are costly, and the content is affected significantly by soil pH. To facilitate breeding for low Cd content, this study sought to identify quantitative trait loci (QTL) and associated molecular markers that can be used in molecular breeding. One spring wheat population of 181 doubled haploid lines (DHLs), which was derived from a cross between two hard white spring wheat cultivars “UI Platinum” (UIP) and “LCS Star” (LCS), was assessed for the Cd content in grain in multiple field trials in Southeast Idaho, United States. Three major QTL regions, namely, QCd.uia2-5B, QCd.uia2-7B, and QCd.uia2-7D, were identified on chromosomes 5B, 7B, and 7D, respectively. All genes in these three QTL regions were identified from the NCBI database. However, three genes related to the uptake and transport of Cd were used in the candidate gene analysis. The sequences of TraesCS5B02G388000 (TaHMA3) in the QCd.uia2-5B region and TraesCS7B02G320900 (TaHMA2) and TraesCS7B02G322900 (TaMSRMK3) in the QCd.uia2-7B region were compared between UIP and LCS. TaHMA2 on 7B is proposed for the first time as a candidate gene for grain Cd content in wheat. A KASP marker associated with this gene was developed and it will be further validated in near-isogenic lines via a gene-editing system in future studies.
Wheat (Triticum aestivum L.) is a critically important food crop, providing 20% of the calories consumed by the population worldwide. Due to environmental pollution and climate change, wheat faces many challenges, including biological and abiotic stress (Bowne et al., 2012; Wegulo, 2012; Guzmán et al., 2016). Recently, heavy metal stress has attracted increased attention (Rizwan et al., 2016). Compared to other heavy metals, cadmium (Cd) is more toxic, has higher bioavailability, and is more easily accumulated in crops. Therefore, Cd pollution is an important risk factor for the environment and human health (Shi et al., 2019).
Cd causes serious problems for both crop production and the human diet (Wagner and Donald, 1993; Prasad, 1995). A long-term intake of food with high levels of Cd causes human health risks, including itai-itai disease, cardiovascular disease, cancer, chronic kidney disease, and bone disease. The maximum allowable Cd concentration in wheat grain is 0.2 mg kg–1, but only 30 μg kg–1 is the maximum allowable amount in baby food products (FAO/WHO, 2010). Two methods can be used to prevent Cd from entering the human food chain. The first method is to decrease plant availability by changing the form of Cd in soil. Soil acidification alters the form of Cd present in soil, increases the presence of Cd2+ and the bioavailability of Cd, and ultimately leads to increased Cd absorption and accumulation by plants (Naidu et al., 1994; Zeng et al., 2011). The second method is to breed crop cultivars that take up and accumulate less Cd. Studying the genetic basis of Cd uptake and transport in crops will contribute to the breeding approach.
The mechanism of Cd absorption and transport by plants has been described in rice. There are two mechanisms for Cd to enter plant root cells. First, Cd can enter plants via the same mechanisms used for the absorption of essential mineral elements, such as Mn, Zn, and Fe (Nakanishi et al., 2006; Lu et al., 2009; Takahashi et al., 2011a,b; Ishimaru et al., 2012; Sasaki et al., 2012; Song et al., 2014). Second, Cd can enter plants via chelation with small molecules such as plant-chelating peptides and enter the root cells in the form of Cd phytochelatins and other binding states (Clemens, 2006). OsNRAMP5 is a transshipment protein gene involved in the absorption of external Mn2+, Cd2+, and Fe3+ by rice root cells (Ishimaru et al., 2012; Yang et al., 2014). The ability of xylem-mediated Cd transfer from roots to aerial parts determines the Cd content in rice stems and grains (Clemens and Ma, 2016). OsHMA3 can transport Cd2+ into vacuoles to isolate and reduce Cd transport to the aboveground parts, thereby reducing the Cd toxicity (Ueno et al., 2010; Miyadate et al., 2011; Sasaki et al., 2014; Lu et al., 2019). Cd is transported from roots to the aboveground parts and then from phloem to various tissues and organs. Uraguchi et al. (2011) isolated the Cd transport protein gene OsLCT1, which was mainly expressed in leaves and stem nodes at the reproductive stage of rice. The CAL1 gene played an important regulatory role in the process of Cd distribution in the aboveground parts of rice (Luo et al., 2018). In durum wheat, a single dominant gene, Cdu1-B located on chromosome 5B, was associated with low Cd concentration (Penner et al., 1995; Clarke et al., 1997; Knox et al., 2009; Abuhammad et al., 2016; Oladzad et al., 2018; Salsman et al., 2018), accounting for more than 80% of variation in the accumulation of Cd in grain (Wiebe et al., 2010; Harris and Taylor, 2013). Maccaferri et al. (2019) discovered a metal transporter gene (TdHMA3-B1) on chromosome 5BL, with a non-functional variant causing high accumulation of Cd in grain.
Genetic studies of Cd in common wheat lag that of rice and durum wheat due to the large genome size. Two quantitative trait loci (QTL) for the accumulation of Cd were identified on wheat chromosomes 4A and 5D, explaining up to 17% of phenotypic variation (Ci et al., 2012). Ban et al. (2020) identified additional two QTL for low Cd content in grain on chromosomes 4BS and 6BL. Using genome-wide association scans, Guttieri et al. (2015) identified Cd-associated single-nucleotide polymorphisms (SNPs) on 5AL in a region homologous to Cdu1 locus on 5BL in durum wheat. Zhang et al. (2020) found three TaHMA3 genes (i.e., TaHMA3-A1, TaHMA3-B1, and TaHMA3-D1) in common wheat, all of which encode transporters located in the vacuolar membrane. The absolute expression level of these genes was very low in all three wheat cultivars compared with that of OsHMA3 in rice, especially in the roots.
Genetic research on the uptake and accumulation of Cd in crops is generally lacking. The QTL identification is the foundation of gene cloning and molecular marker-assisted breeding. Therefore, research to discover QTL for Cd content and associated molecular markers under different soil pH and Cd conditions will have both theoretical and practical values.
The objectives of this study were to identify QTL and to analyze potential candidate genes for grain Cd content in spring wheat in relation to the genes controlling the grain Cd content in durum and rice.
This study used 181 doubled haploid lines (DHLs) which were developed using a wheat × maize hybridization system (Laurie and Bennett, 1986) through the services of Heartland Plant Innovation in Kansas, United States. The DHLs were derived from the F1 generation of a cross between two high yielding hard white spring wheat cultivars, namely, UI Platinum (UIP) and LCS Star (LCS). UIP was developed by the University of Idaho Agricultural Experiment Station and released in 2014 (Chen et al., 2016). LCS was developed and released by Limagrain Cereal Seeds. Both parents have a semi-dwarfing allele at the Rht-B1 locus and similar plant height but have alternative alleles for the two major photoperiod response genes. UIP has the photoperiod insensitive alleles at loci for both PPD-B1b and PPD-D1b, while LCS has the sensitive alleles. As a result, UIP flowers earlier than LCS when grown under short-day conditions.
This study also used 127 spring wheat cultivars and elite lines to validate QTL identified in the DHLs. These lines were from multiple wheat breeding programs in the Pacific Northwest of the United States and the International Maize and Wheat Improvement Center (CIMMYT) in Mexico, as described by Wang et al. (2017).
The parents and DHLs were planted and assessed in four-field trials, with two dryland trials in Soda Springs (SS), ID (42°43′ N, 111°35′ W, altitude 1,760 m) in 2017 and 2018 (17SS and 18SS), and two irrigated trials in Ashton (AS), ID (44°4′ N, 111°23′ W, altitude 1,603 m) in 2017 and 2018 (17AS and 18AS). The 127 spring wheat cultivars and elite lines were planted in the same field as DHLs in SS in 2017. The soil in both the locations had pH < 6, but AS had lower pH and lower Cd content than that in SS (Table 1).
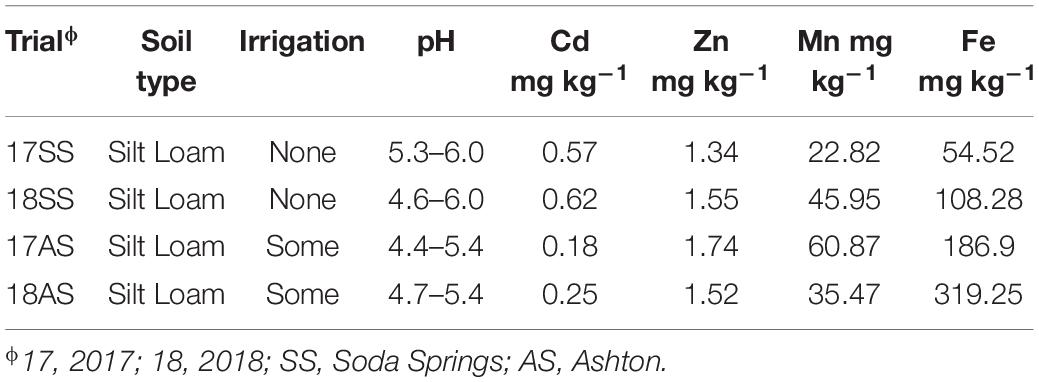
Table 1. Content of four metals and pH in 0–30 cm soil in two irrigated and two non-irrigated field trials.
The two 2018 trials used seven row plots with 3.0 m in length, 1.5 m in width, and 0.25 m between the rows. The DH and parental lines were arranged in a randomized complete block design with two replications. Because of limited seed, the two 2017 trials had one replicate of four row plots with 1.5 m in width, 1.5 m in length, and 0.5 m between the rows. Field management in both SS and AS used common field practices, and the plots were managed by cooperating growers.
Composite core soil samples were taken from each location to establish a baseline profile for N, P, K, Zn, Fe, Cu, Mn, S, Cd, Cr, Ni, Pb, soil type, organic matter, pH, and salinity. Soil was sampled by splitting the field into two parts based on the environmental layout of the field (i.e., slopes and dips in the field). Several core samples for each part were drawn at 15 and 30 cm. Samples were submitted as 0–15 and 15–30 cm to determine the depth profile of the elements in the soil. Full elemental analysis for macronutrients and micronutrients, including Cd levels, was conducted at the Utah State University analytical lab.1
Plots were harvested using a Wintersteiger Classic small plot combine (2002 Wintersteiger Elite, Wintersteiger Seedmech) equipped with a Harvest Master weighing system (HM-400, Juniper Systems, United States). From each plot, 300 g samples of grain were milled using a Perten 3100 Laboratory Mill (2012 Perten Instruments, United States). For each line, a 15 g subsample of milled whole grain flour was sent to the University of Idaho Analytical Sciences Laboratory for elemental analysis.
Milled grain samples were digested in 30% nitric acid and Cd content measured by using inductively coupled plasma mass spectrometer collision/reaction (ICP-MScx). Samples were prepared by using the SMM.57.070.05 protocol maintained by the University of Idaho Analytical Sciences Laboratory. The concentrations of Cd in the milled samples were determined using an Agilent 7800 inductively coupled plasma mass spectrometer (ICP-MS) (Agilent 7800 ICP-MScx, United States).
The content of Cd in each grain sample from all trials was used in the subsequent analysis. The best linear unbiased prediction (BLUP) and the broad-sense heritability (H2) were calculated from data sets across years and locations by using SAS V8.0 (SAS Institute, Cary, NC, United States) (Smith et al., 1998). The rate of decrease (DR) of Cd content in grain across the range of soil pH from the trials was calculated as follows: (AS - SS)/AS × 100, where AS is the mean Cd content in grain from all AS trails and SS is the mean for SS. The Spearman’s correlations of Cd content across four trials were calculated, and BLUP data were derived from multiple trials.
A genetic map of the mapping population was generated with 14,236 polymorphic SNPs from the wheat Illumina 90k SNP assay, representing all 21 hexaploid wheat chromosomes (Isham et al., 2021). The 7DS linkage map also included additional seven KASP markers published by Isham et al. (2021). All linkage details were used in QTL analysis in this study.
QTL analysis was conducted using individual and BLUP data sets for grain Cd content by using the composite interval mapping (CIM) method in JMP Genomics 9.0.2 Significant QTL were determined with the expectation maximization algorithm at a threshold of 2.5 [logarithm of the odds (LOD) ≥ 2.5] (Lin et al., 1996). The names for QTL followed the International Rules of Genetic Nomenclature.3 The software output provided a proportion of phenotypic variance (R2) and the additive effects for each marker. The source of the allelic effect of the parent UIP or LCS was indicated by negative or positive estimates of the additive effects, respectively. The LOD threshold of 2.5 was set for entry and retention in the model. Epistatic analysis was performed with the IciMappingVer.4.1 EPI module (LOD = 5, step = 1 cM, PIN = 0.0001).
To determine the physical positions for identified QTL regions, a BLAST search4 was performed to align the QTL-associated peak and flanking SNP marker sequences with the reference wheat genome assembly constructed in the cv. Chinese Spring (CS) sequence (RefSeq v1.0, the International Wheat Genome Consortium).
Genes within the target region were identified using the genome browser (JBrowse) on the triticeae multiomics website (Triticeae Multi-omics).5 The sequences of common wheat genes were retrieved based on the intervals of major QTL identified from https://urgi.versailles.inra.fr/download/iwgsc/IWGSC_RefSeq_Annotations/v1.0/ (Zheng et al., 2019). The sequences were used to perform a BLAST search against the genome sequence databases of rice6 and durum wheat7 to identify orthologous gene pairs. The collinearity of these genes was analyzed using MCscan (Python version).8 Functional annotation and enrichment analysis of genes in segments were carried out in the Gene Ontology (GO) database using the R package cluster Profiler.
The candidate genes that were related to the uptake and transport of Cd were used in comparative analysis between the two parental lines using the resequencing data generated by the program of the corresponding author (Chen, personal communication). Gene-specific markers were designed based on the sequence differences between UIP and LCS. The markers were genotyped in the original mapping population and in 127 diverse lines in the validation panel. The contribution of the candidate genes to the Cd content in grain was validated based on the association analysis between marker data and the grain Cd content, and the allelic effect of the candidate gene was analyzed with a t-test in SAS V8.0.
The Cd content in LCS was generally higher than that in UIP, except in 18SS. Based on the BLUP value, the Cd content in grain was 0.108 mg kg–1 for LCS, 0.099 mg kg–1 for UIP, and 0.083–0.126 mg kg–1 for the DHLs (Table 2). The Cd content of the parents and DHLs were lower than the maximum level of grain Cd proposed by FAO/WHO (0.200 mg kg–1) (FAO/WHO, 2010). The estimated H2 for the Cd content was 0.68, which was high (H2 > 0.50), indicating that it was affected more by genetic vs. environmental factors (Table 2). However, the Spearman’s correlations ranged from 0.313 to 0.414 across different trials, indicating that the Cd content of the DHLs is affected by environment (Table 3). The Cd content of the DHLs showed continuous variation, suggesting multigene genetic control.
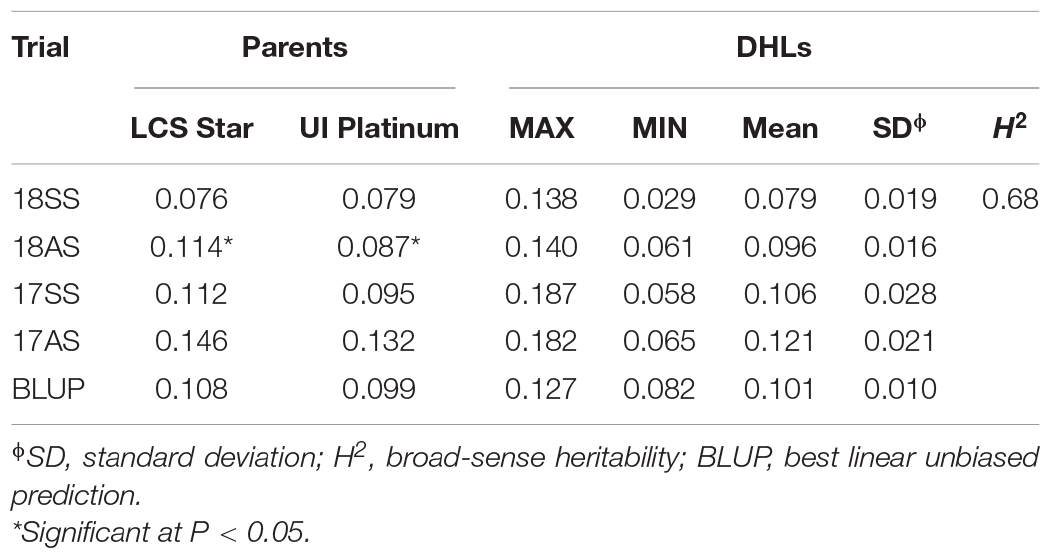
Table 2. Phenotypic performance and distribution of Cd content (mg kg–1) in parents and the doubled haploid lines in four-field trials.
A total of 10 QTL explaining 6.37–15.34% of the phenotypic variance were detected (Table 4). Three major QTL regions, namely, QCd.uia2-5B, QCd.uia2-7B, and QCd.uia2-7D, were detected in more than three environments (Table 4 and Figure 1). LCS had alleles for higher Cd content at QCd.uia2-5B and QCd.uia2-7B, and UIP had an allele for higher Cd content at QCd.uia2-7D (Table 4). QCd.uia2-5B was detected in three data sets, namely, 18SS, 18AS, and BLUP, explaining 7.2–11.1% of grain Cd content. It was physically mapped in a 558.41–585.75 Mb interval on 5BL. QCd.uia2-5B is near Cdu1-B on chromosome 5BL (Penner et al., 1995; Clarke et al., 1997; Abuhammad et al., 2016; Oladzad et al., 2018; Salsman et al., 2018). QCd.uia2-7B was detected in three data sets, namely, 18AS, 17AS, and BLUP, explaining 7.2–10.6% of grain Cd content. It was physically mapped in a 559.14–601.17 Mb interval on 7BL. QCd.uia2-7D was detected in four data sets, namely, 17AS, 18AS, 18SS, and BLUP, explaining 7.63–12.29% of grain Cd content. It was physically mapped in a 59.74–68.42 Mb interval on 7DS. No Cd content-related QTL have been reported earlier in these two intervals; therefore, QCd.uia2-7B and QCd.uia2-7D are likely novel. The other seven minor QTL were only detected in one to two data sets, explaining 6–15% of phenotypic variation (Table 4).
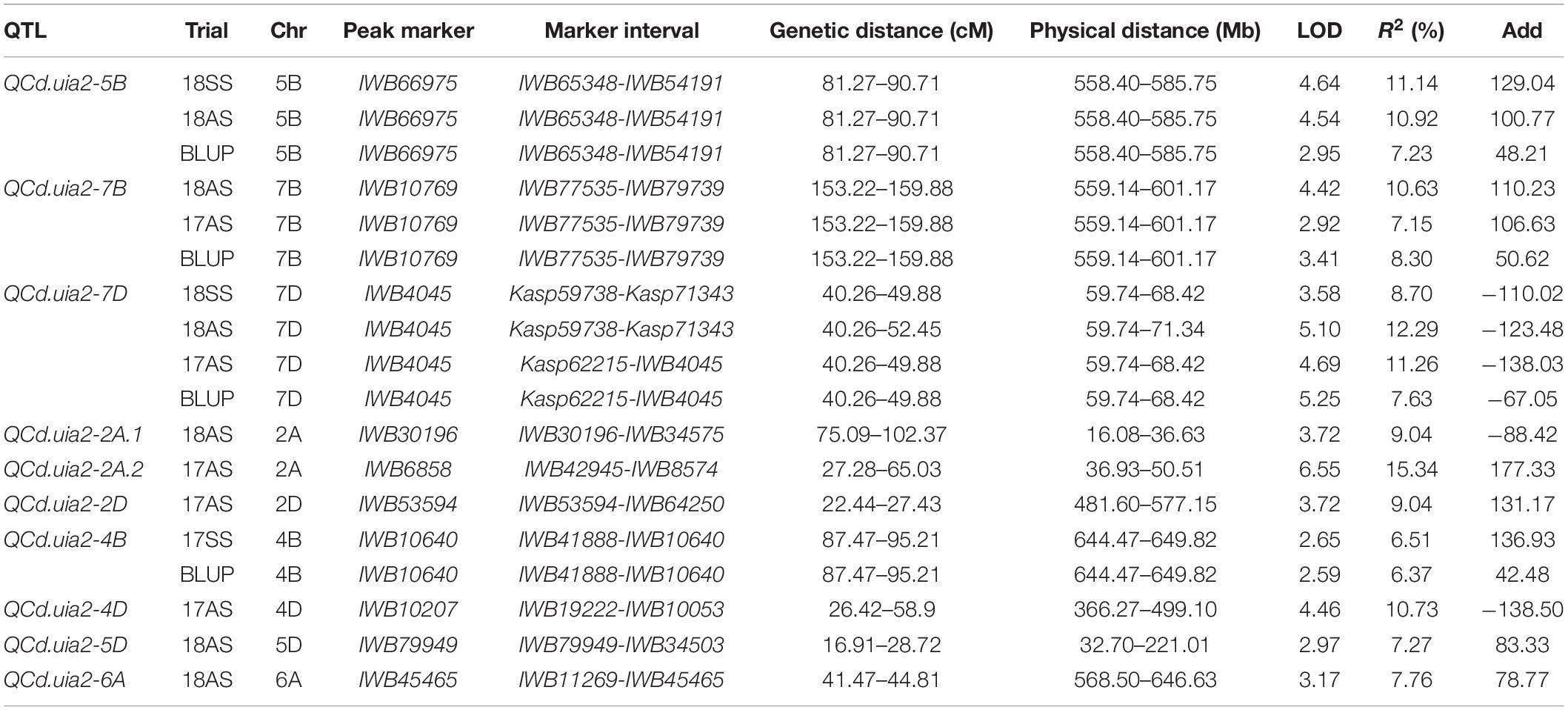
Table 4. Major quantitative trait loci (QTL) for Cd content (mg kg–1) in grain detected in the DH population.
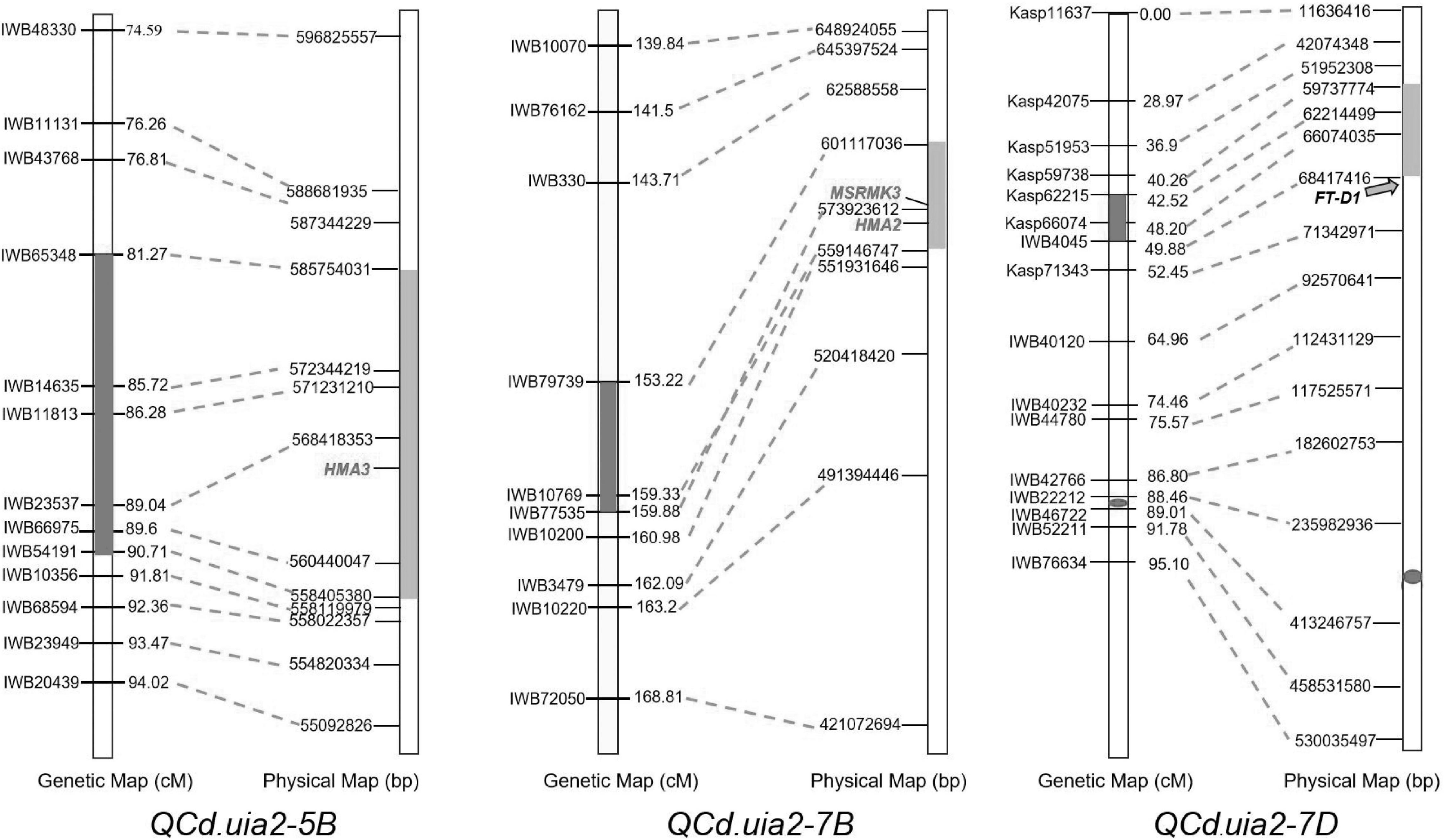
Figure 1. Comparative genetic linkage and physical maps of QCd.uia2-5B, QCd.uia2-7B, and QCd.uia2-7D.
LCS contributed the Cd-increasing alleles for both QCd.uia2-5B and QCd.uia2-7B and UIP contributed the Cd-increasing allele for QCd.uia2-7D, and these three QTL regions have additive effects toward increasing Cd content in grain (Table 4 and Figure 1). There was no epistatic effect observed for the three major QTL regions (Supplementary Table 1). The average Cd content increased as the number of alleles increased (Table 5 and Figure 2). The DHLs with low Cd alleles at all the three QTL regions had 0.0161 mg kg–1 less Cd content compared to those with contrasting alleles. The combination of negative alleles from QCd.uia2-5B, QCd.uia2-7B, and QCd.uia2-7D had the largest effect on the Cd content.
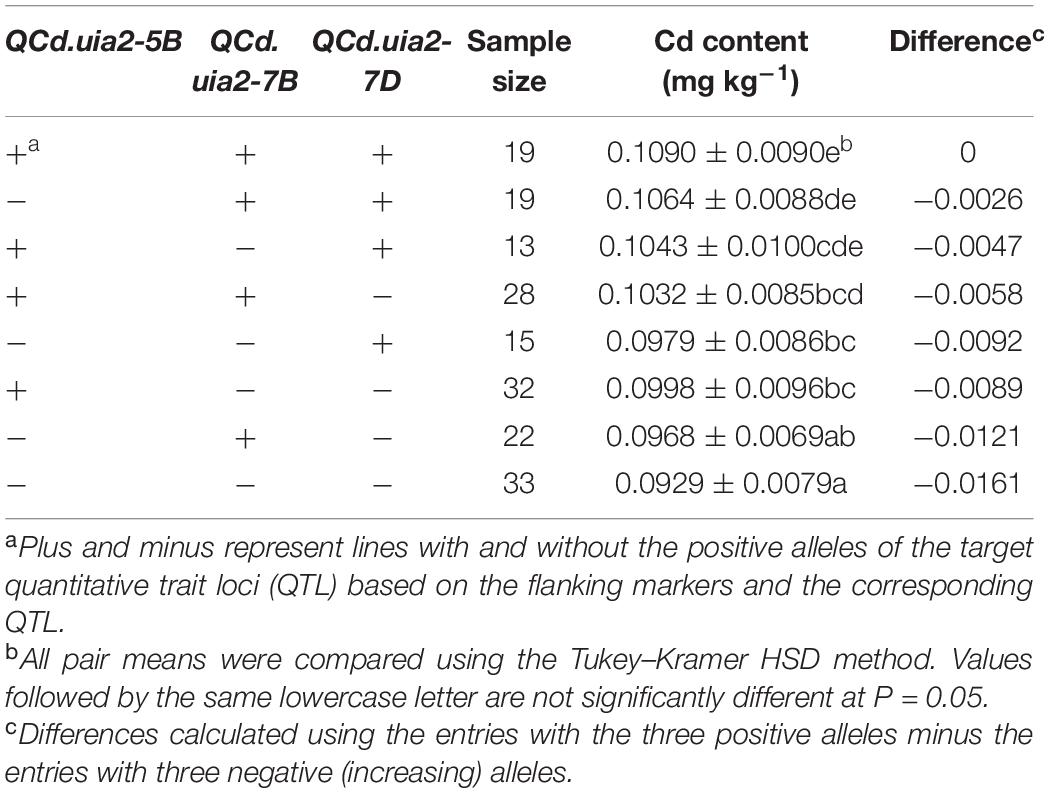
Table 5. Additive effects of the QTL on 5B, 7B, and 7D for Cd content (mg kg–1) in grain across sites in the UI Platinum × LCS Star-derived double haploid population.
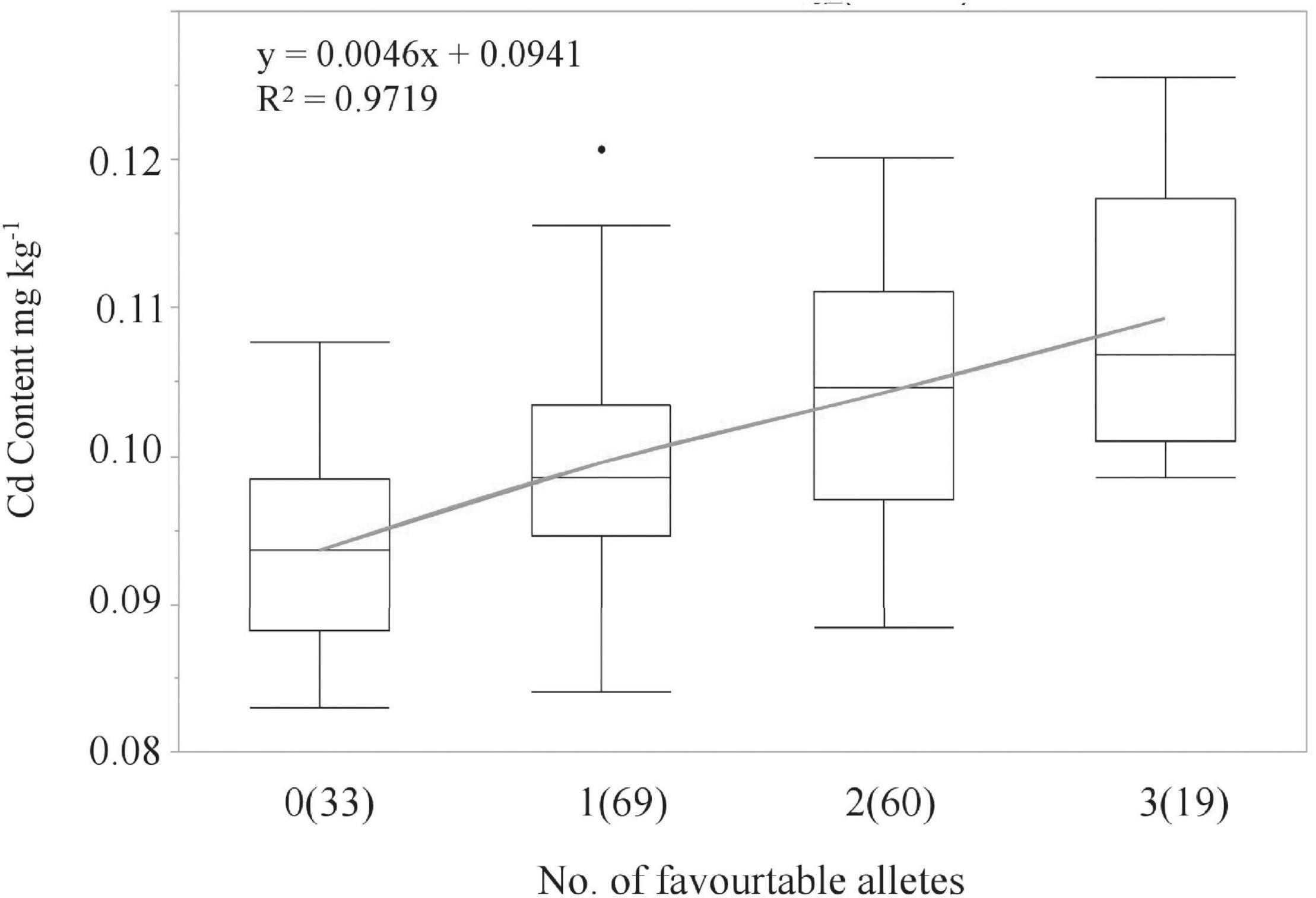
Figure 2. Linear regressions between the number of high Cd alleles (number of lines) and BLUP Cd content in the DH population. Numbers of lines carrying the corresponding number of favorable alleles are shown in brackets. X and Y in the equation represent the number of high Cd alleles and BLUP Cd content, respectively.
QCd.uia2-5B was physically mapped in a 558.41–585.75 Mb interval on 5BL, and 293 genes were found in this interval in CS. Nine of these genes were involved in metal ion transport according to gene functional annotations in the GO public database (Table 6). The functional annotation of TraesCS5B02G388000 was for the transport of Zn and Cd. The corresponding gene TraesCS5B02G388000 in wheat was TRITD5Bv1G197380 (TdHMA3) in durum wheat and Os07g0232900 (OsHMA3) in rice. Therefore, TraesCS5B02G388000 was named TaHMA3 in wheat.
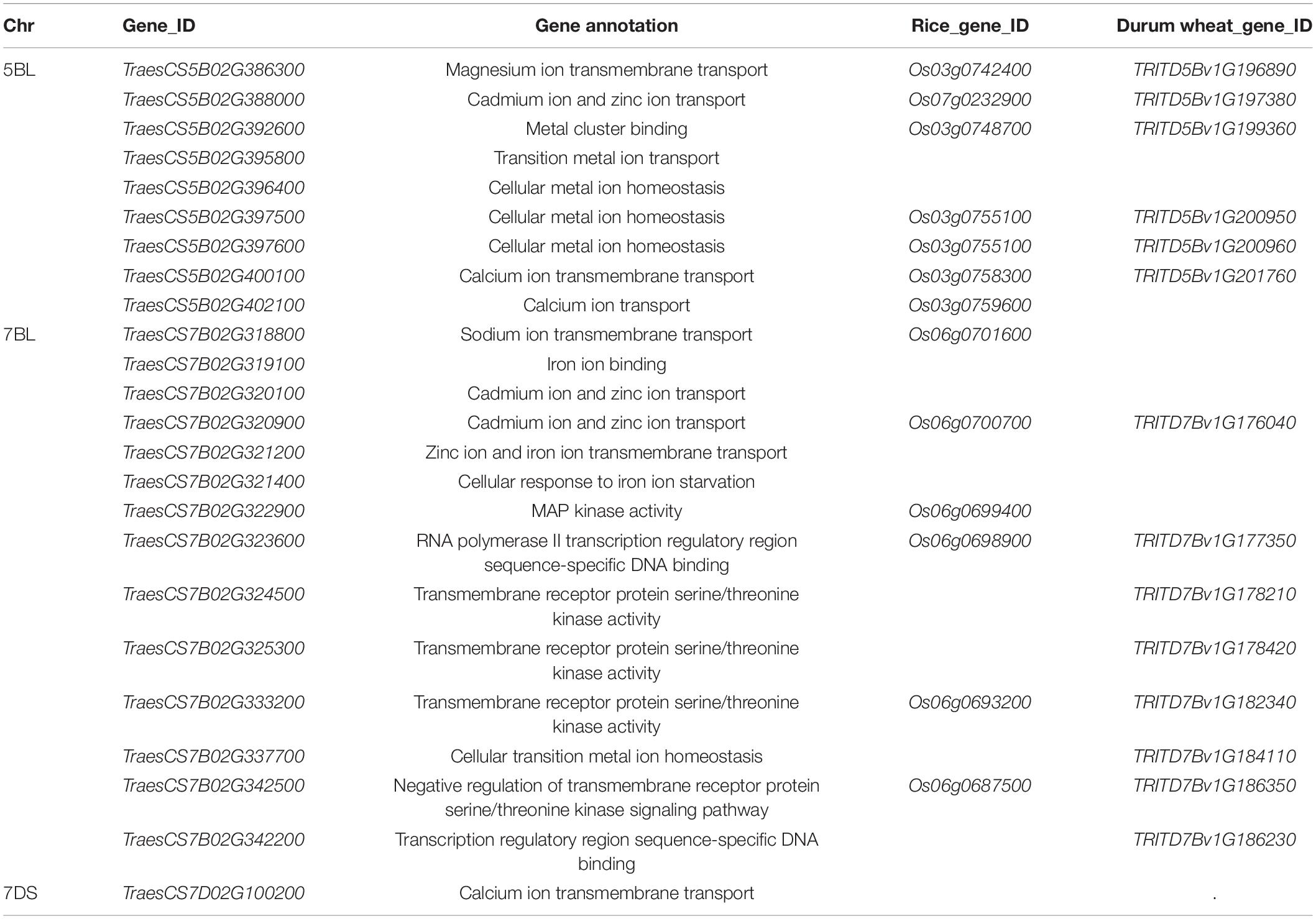
Table 6. Candidate genes significantly associated with Cd and either metal uptake or transport in the three major QTL regions identified in this study.
The QCd.uia2-7B was physically mapped in a 559.14–601.17 Mb interval on 7BL and 307 genes were found in this interval in CS. Out of these genes, 12 were involved in the transmembrane transport of metal ions, such as Zn, Fe, and Cd (Table 6). TraesCS7B02G320900 is homologous to OsHMA2 (Os06g0700700) and TraesCS7B02G322900 is homologous to OsMSRMK3 (Os06g0699400) in rice. TraesCS7B02G320900 and TraesCS7B02G322900 were named TaHMA2 and TaMSRMK3, respectively.
The interval of QCd.uia2-7D has 128 genes in CS. Only TraesCS7D02G100200 participates in the transmembrane transport of Ca and other divalent cations (Table 6).
We analyzed the coding and promoter regions of TaHMA3, TaHMA2, TaMSRMK3, and TraesCS7D02G100200 from the resequencing data of UIP and LCS. The three genes contain 2,487, 2,298, and 1,134 nucleotides and encode 829, 766, and 378 amino acids in coding sequence, respectively. In TaHMA3 gene sequence of LCS, one SNP (at 1,974 bp G/A) was detected, which resulted in the exchange of amino acids between arginine and glutamine. By predicting the protein structure, the amino acid variation of TaHMA3 did not change the three-dimensional (3D) structure of protein (Supplementary Figure 1). Four SNPs were detected in TaHMA2 gene sequence, one SNP (at 3,633 bp A/G) resulted in a synonymous mutation of glycine, and the other three SNPs were non-synonymous mutations (Figure 3). One SNP at 3,094 bp C/A resulted in the exchange of amino acids between leucine and methionine. One SNP at 3,893 bp G/C resulted in the exchange of amino acids between glycine and alanine. The other SNP at 3,963 bp C/G resulted in the exchange of amino acids between isoleucine and methionine. The exchange of a single amino acid at the 338 site of UIP predicted to increase an α-helix on the 3D structure (Supplementary Figure 1). No sequence polymorphism was found in the gene sequences of TaMSRMK3 and TraesCS7D02G100200. The KASP marker for TaHMA2 shown in Figure 3 was significantly associated with grain Cd content in all environments, except for 17SS (Table 7). The effect of TaHMA2 was also significantly associated with grain Cd content in 127 diverse spring wheat cultivars and elite lines (Table 8).
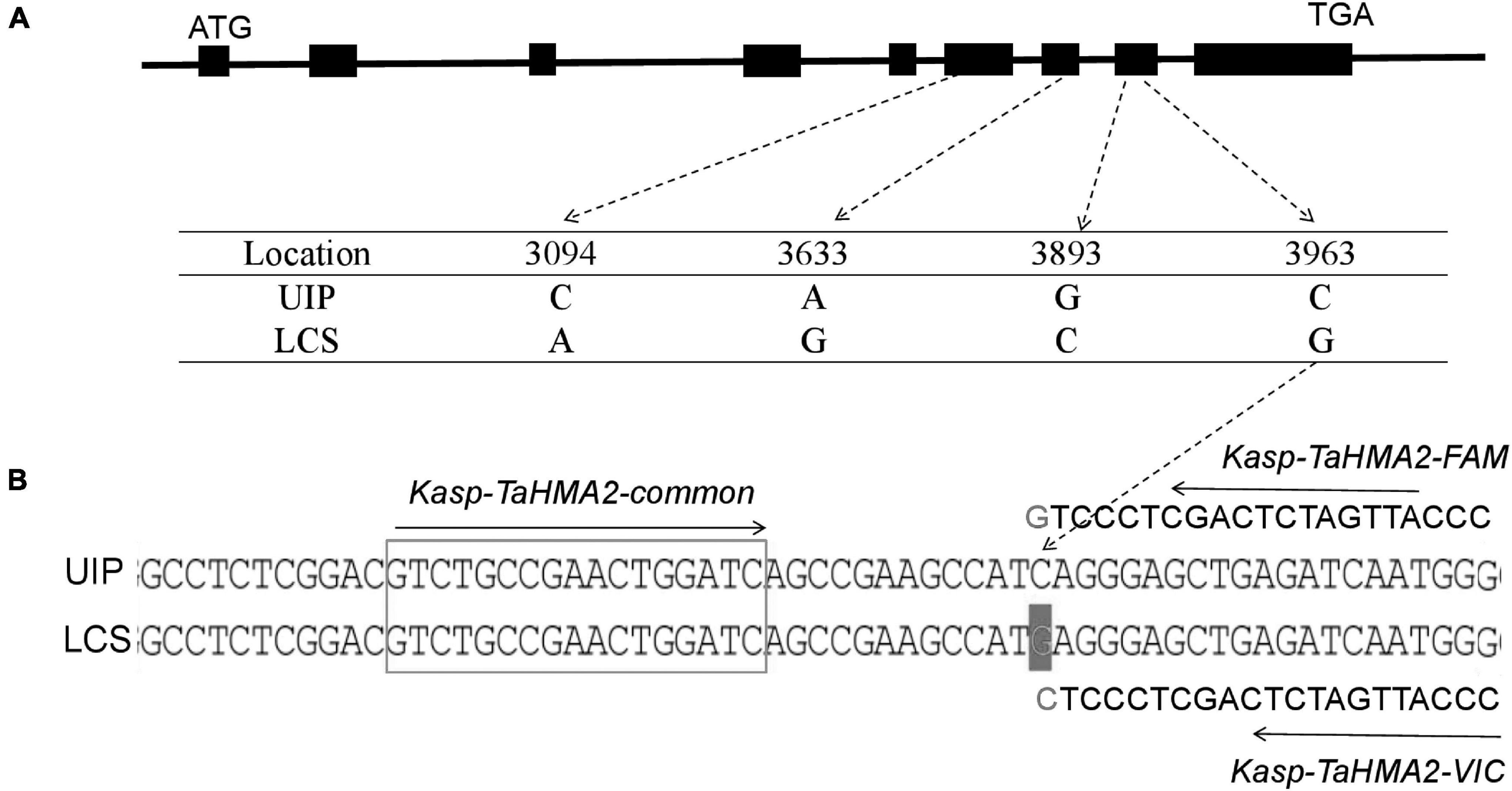
Figure 3. Sequence comparison of TaHMA2 in two parents and KASP marker associated with the candidate gene. (A) Schematic diagram of nucleotide polymorphism for TaHMA2. The polymorphic site and relative positions are indicated on the genomic sequence of TaHMA2. Exons are indicated by black boxes, flanking regions and introns are indicated by solid black lines. (B) A KASP marker was designed using the nucleotide polymorphism of TaHMA2.
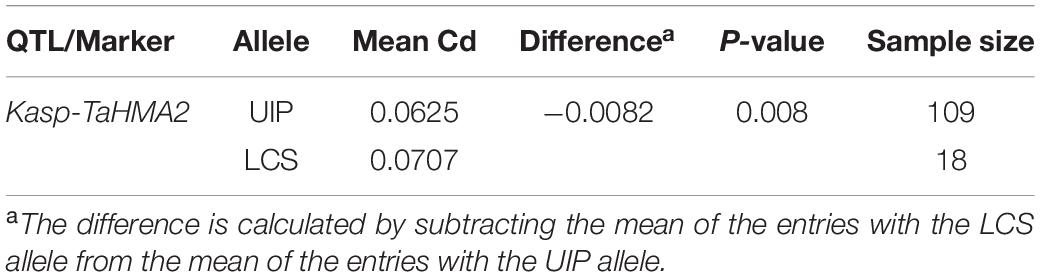
Table 8. Allelic effect of TaHMA2 on grain Cd content in 127 diverse spring wheat lines grown in 17SS.
The uptake of Cd in plants depends on the plant itself, the concentration of Cd in soil, and the soil properties, such as soil pH, organic matter content, and cation exchange capacity (Eriksson et al., 1996; Benavides et al., 2005; Kim et al., 2016; Zhuang et al., 2021). Soil pH is negatively correlated with Cd content in grain (Kirkham, 2006; Baize et al., 2009). In this study, the soil pH changed from acidic to neutral across the two experimental sites AS and SS. The grain Cd content of the two parents and the population means decreased, although the Cd content in soil was lower in acidic location AS than in the neutral location SS (Table 2). This result supports the conclusion that the soil pH is the most important factor contributing to Cd uptake in wheat (Nan et al., 2002; Liu et al., 2015). In acid soil, Cd is mainly free Cd2+, and at neutral or alkaline pH, Cd forms CdCl, CdHCO3, and hydrated CdCO3, which increases the adsorption capacity of Cd and reduces the accumulation of Cd in plants (Reddy and Patrick, 1977; Sebastian and Prasad, 2014; Volpe et al., 2015; Ismael et al., 2019). Therefore, avoiding soil acidification will reduce the bioavailability of Cd in soil. We also observed a year effect of Cd content in grain. The grain Cd content in parents and in the DHLs in 2018 was lower than in 2017 in the same location. The Cd and Fe content in soil was higher in 2018 than in 2017, which might be the cause of the year effect.
Three QTL regions, namely, QCd.uia2-5B, QCd.uia2-7B, and QCd.uia2-7D, were identified in 2–3 location-year trials. QCd.uia2-7B and QCd.uia2-7D are novel QTL in common wheat. The three QTL regions have additive effects that can be used in breeding low grain Cd cultivars. However, none of QTL regions was detected in all four trials. QCd.uia2-5B was only detected in the two 2018 trials (18SS and 18AS), QCd.uia2-7B only in AS trials 17AS and 18AS, and QCd.uia2-7D was detected in three of the four trials. The effects of the three QTL regions were generally small, explaining up to 12% of total phenotypic variation (Table 4). The Cd content of grain in common wheat is generally much lower than that in durum and rice, and the two parents did not differ greatly in grain Cd content, which possibly explains the small effect of the three QTL identified in this study. To improve the power of QTL detection for grain Cd content, it is essential to do Cd screening of grain in controlled environments and using near-isogenic lines.
Based on the physical location, annotation of candidate gene function, and comparison of homologous genes, we found three genes that regulate the uptake and transport of Cd in durum and/or rice and also identified three orthologous genes in wheat, namely, TaHMA3 (TraesCS 5B02G388000), TaHMA2 (TraesCS7B02G320900), and TaMSRMK3 (TraesCS7B02G322 900). TaHMA3 encodes a P1B-type heavy metal ATPase 3 (HMA3) that is orthologous to OsHMA3. OsHMA3 is a transporter protein located on the vacuolar membrane of the root, which can transport Cd absorbed by root to the vacuole, thus limiting the transport of Cd to the aboveground plant parts (Miyadate et al., 2011; Sasaki et al., 2014; Maccaferri et al., 2019; Lei et al., 2020). TaHMA2 is orthologous to rice HMA2 (OsHMA2). OsHMA2, a type of efflux metal transporter expressed on the cell membrane, is involved in root-to-shoot transport and plays a role in Zn and Cd loading into the xylem (Satoh-Nagasawa et al., 2012; Takahashi et al., 2012). TaMSRMK3 is orthologous to rice OsMSRMK3. The expression of OsMSRMK3 is upregulated by heavy metal stress (Agrawal et al., 2003).
Based on the sequence comparison of the three candidate genes, TaHMA2 was a candidate gene validated using the gene-specific KASP marker. The function of TaHMA2 is being sought via gene-editing technology in an ongoing project. This finding is an important starting point for understanding the molecular mechanism of Cd absorption, transport, and accumulation in wheat and provides a theoretical basis for breeding low cadmium varieties using molecular technology.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
JC and LQ designed the experiment and developed the original manuscript. LQ, JZe, MS, and JZa did sequence analysis and genotyping of the candidate genes. JC, LQ, JW, RW, KI, NK, and WZ did the field experiments. LQ, RW, and JZa performed the phenotypic data analysis and QTL detection. JC, LQ, JZe, and JZa revised the manuscript. All authors approved the submitted version of the manuscript.
This study was supported by the Research Project Supported by Shanxi Scholarship Council of China (2020-159), the Agricultural Science Research of Shanxi Academy of Agricultural Sciences (YCX2020BH2, YCX2020YQ47, YCX2020YQ34 and YZGC013), the National Research Initiative Competitive Grant 2017-67007-25939 from the USDA NIFA, the Idaho Agricultural Experimental Station Project IDA01627, and the Idaho Wheat Commission Projects.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We acknowledge Zhongfu Ni and Weilong Guo and their student Wenxin Wang at China Agricultural University for assistance in sequence analysis. We also want to thank Juliet Marshall at University of Idaho for her assistance in planting field trials. In addition, we thank John Bonman, a USDA-ARS retired scientist for his editing comments.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.756741/full#supplementary-material
Abuhammad, W. A., Mamidi, S., Kumar, A., Pireseyedi, S., Manthey, F. A., Kianian, S. F., et al. (2016). Identification and validation of a major cadmium accumulation locus and closely associated SNP markers in North Dakota durum wheat cultivars. Mol. Breed. 36:112. doi: 10.1007/s11032-016-0536-1
Agrawal, G. K., Agrawal, S. K., Shibato, J., Iwahashi, H., and Rakwal, R. (2003). Novel rice MAP kinases OsMSRMK3 and OsWJUMK1 involved in encountering diverse environmental stresses and developmental regulation. Biochem. Biophys. Res. Commun. 300, 775–783. doi: 10.1016/S0006-291X(02)02868-1
Baize, D., Bellanger, L., and Tomassone, R. (2009). Relationships between concentrations of trace metals in wheat grains and soil. Agron. Sustain Dev. 29, 297–312. doi: 10.1051/agro:2008057
Ban, Y., Ishikawa, G., Ueda, H., Ishikawa, N., and Yanaka, M. (2020). Novel quantitative trait loci for low grain cadmium concentration in common wheat (Triticum aestivum L.). Breeding Sci. 70, 331–341. doi: 10.1270/jsbbs.19150
Benavides, M. P., Gallego, S. M., and Tomaro, M. L. (2005). Cadmium toxicity in plants. Braz. J. Plant Physiol. 17, 21–34. doi: 10.1590/S1677-04202005000100003
Bowne, J. B., Erwin, T. A., Juttner, J., Schnurbusch, T., Langridge, P., Bacic, A., et al. (2012). Drought responses of leaf tissues from wheat cultivars of differing drought tolerance at the metabolite level. Mol. Plant. 5, 418–429. doi: 10.1093/mp/ssr114
Chen, J., Wheeler, J., O’Brien, K., Zhao, W., Klassen, N., Zhang, J., et al. (2016). Registration of ‘UI Platinum’ hard white spring wheat. J. Plant Regist. 10, 36–40. doi: 10.3198/jpr2015.06.0037crc
Ci, D., Jiang, D., Li, S., Wollenweber, B., Dai, T., and Cao, W. (2012). Identification of quantitative trait loci for cadmium tolerance and accumulation in wheat. Acta Physiol. Plant. 34, 191–202. doi: 10.1007/s11738-011-0818-5
Clarke, J., Leisle, D., and Kopytko, G. (1997). Inheritance of cadmium concentration in five durum wheat crosses. Crop Sci. 37, 1722–1726. doi: 10.2135/cropsci1997.0011183X003700060008x
Clemens, S. (2006). Evolution and function of phytochelatin synthases. J. Plant Physiol. 163, 319–332. doi: 10.1016/j.jplph.2005.11.010
Clemens, S., and Ma, J. F. (2016). Toxic heavy metal and metalloid accumulation in crop plants and foods. Annu Rev. Plant Biol. 67, 489–512. doi: 10.1146/annurev-arplant-043015-112301
Eriksson, J., Oborn, I., Jansson, G., and Andersson, A. (1996). Factors influencing Cd-content in crops: results from Swedish field investigations. Swed. J. Agric. Res. 26, 125–133. doi: 10.1016/0921-4488(95)00737-7
FAO/WHO (2010). Evaluation of certain food additives and contaminants (Seventy- third report of the joint FAO/WHO expert committee on food additives). WHO Techn. Rep. Ser. 2010:960.
Guttieri, M. J., Baenziger, P. S., Frels, K., Carver, B., and Waters, B. M. (2015). Prospects for selecting wheat with increased zinc and decreased cadmium concentration in grain. Crop Sci. 55, 1712–1728. doi: 10.2135/cropsci2014.08.0559
Guzmán, C., Autrique, J. E., Mondal, S., Singh, R. P., Govindan, V., Morales-Dorantes, A., et al. (2016). Response to drought and heat stress on wheat quality, with special emphasis on bread-making quality, in durum wheat. Field Crop Res. 186, 157–165. doi: 10.1016/j.fcr.2015.12.002
Harris, N. S., and Taylor, G. J. (2013). Cadmium uptake and partitioning in durum wheat during grain filling. BMC Plant Bio. 13:103. doi: 10.1186/1471-2229-13-103
Isham, K., Wang, R., Zhao, W., Wheeler, J., Klassen, N., Akhunov, E., et al. (2021). QTL mapping for grain yield and three yield components in a population derived from two high-yielding spring wheat cultivars. Theor. Appl. Genet. 134, 2079–2095. doi: 10.1007/s00122-021-03806-1
Ishimaru, Y., Takahashi, R., Bashir, K., Shimo, H., Senoura, T., Sugimoto, K., et al. (2012). Characterizing the role of rice NRAMP5 in manganese, iron and cadmium transport. Sci. Rep. 2:286. doi: 10.1038/srep00286
Ismael, M. A., Elyamine, A. M., Moussa, M. G., Cai, M., Zhao, X., and Hu, C. (2019). Cadmium in plants: Uptake, toxicity, and its interactions with selenium fertilizers. Metallomics 11, 255–277. doi: 10.1039/c8mt00247a
Kim, S. C., Kim, H. S., Seo, B. H., Owens, G., and Kim, K. R. (2016). Phytoavailability control based management for paddy soil contaminated with Cd and Pb: Implications for safer rice production. Geoderma 270, 83–88. doi: 10.1016/j.geoderma.2015.11.031
Kirkham, M. B. (2006). Cadmium in plants on polluted soils: Effects of soil factors, hyperaccumulation and amendments. Geoderma 137, 19–32. doi: 10.1016/j.geoderma.2006.08.024
Knox, R., Pozniak, C. J., Clarke, F. R., Clarke, J. M., Houshmand, S., and Singh, A. K. (2009). Chromosomal location of the cadmium uptake gene (Cdu1-B) in durum wheat. Genome 52, 741–747. doi: 10.1139/g09-042
Laurie, D., and Bennett, M. (1986). Wheat x maize hybridization. Can. J. Genet. Cytol. 28, 313–316. doi: 10.1139/g86-046
Lei, G. J., Fujii-Kashino, M., Wu, D. Z., Hisamo, H., Saisho, D., Deng, F. L., et al. (2020). Breeding for low cadmium barley by introgression of a Sukkula-like transposable element. Nat. Food. 1, 489–499. doi: 10.1038/s43016-020-0130-x
Lin, H., Qian, H., Zhuang, J., Lu, J., Min, S., Xiong, Z., et al. (1996). RFLP mapping of QTLs for yield and related characters in rice (Oryza sativa L.). Theor. Appl. Genet. 92, 920–927. doi: 10.1007/BF00224031
Liu, K., Lv, J., He, W., Zhang, H., Cao, Y., and Dai, Y. (2015). Major factors influencing cadmium uptake from the soil into wheat plants. Ecotox Environ. Saf. 113, 207–213. doi: 10.1016/j.ecoenv.2014.12.005
Lu, C., Zhang, L. X., Tang, Z., Huang, X. Y., Ma, J. F., and Zhao, F. J. (2019). Producing cadmium-free Indica rice by overexpressing OsHMA3. Environ. Int. 126, 619–626. doi: 10.1016/j.envint.2019.03.004
Lu, L. L., Tian, S. K., Yang, X. E., Li, T. Q., and He, Z. L. (2009). Cadmium uptake and xylem loading are active processes in the hyperaccumulator Sedum alfredii. J. Plant Physiol. 166, 579–587. doi: 10.1016/j.jplph.2008.09.001
Luo, J. S., Huang, J., Zeng, D. L., Peng, J. S., Zhang, G. B., Ma, H. L., et al. (2018). A defensin-like protein drives cadmium efflux and allocation in rice. Nat. Commun. 9:645. doi: 10.1038/s41467-018-03088-0
Maccaferri, M., Harris, N. S., Twardziok, S. O. Pasam, R. K., Gundlach, H., Spannagl, M., et al. (2019). Durum wheat genome highlights past domestication signatures and future improvement targets. Nat. Genet. 51, 885–895. doi: 10.1038/s41588-019-0381-3
Miyadate, H., Adachi, S., Hiraizumi, A., Tezuka, K., Nakazawa, N., Kawamoto, T., et al. (2011). OsHMA3, a P1B-type of ATPase affects root-to-shoot cadmium translocation in rice by mediating efflux into vacuoles. New Phytol. 189, 190–199. doi: 10.1111/j.1469-8137.2010.03459.x
Naidu, R., Bolan, N. S., Kookana, R. S., and Tiller, K. G. (1994). Ionic-strength and pH effects on the sorption of cadmium and the surface charge of soils. Eur. J. Soil Sci. 45, 419–429. doi: 10.1111/j.1365-2389.1994.tb00527.x
Nakanishi, H., Ogawa, I., Ishimaru, Y., Mori, S., and Nishizawa, N. K. (2006). Iron deficiency enhances cadmium uptake and translocation mediated by the Fe2+ transporters OsIRT1 and OsIRT2 in rice. Soil Sci. Plant Nutr. 52, 464–469. doi: 10.1111/j.1747-0765.2006.00055.x
Nan, Z., Li, J., Zhang, J., and Cheng, G. (2002). Cadmium and zinc interactions and their transfer in soil-crop system under actual field conditions. Sci. Total Environ. 285, 187–195. doi: 10.1016/S0048-9697(01)00919-6
Oladzad, A. A., Kumar, A., Pirseyedi, S., Salsman, E., Dobrydina, M., Sharma, P. R., et al. (2018). Identification and validation of a new source of low grain cadmium accumulation in durum wheat. G3 8, 923–932. doi: 10.1534/g3.117.300370
Penner, G. A., Bezte, L. J., Leisle, D., and Clarke, J. (1995). Identification of RAPD markers linked to a gene governing cadmium uptake in durum wheat. Genome 38, 543–547. doi: 10.1139/g95-070
Prasad, M. N. V. (1995). Cadmium toxicity and tolerance in vascular plants. Environ. Exp. Bot. 35, 525–545. doi: 10.1016/0098-8472(95)00024-0
Reddy, C. N., and Patrick, W. H. (1977). Effect of redox potential and pH on the uptake of cadmium and lead by rice plants. J. Environ. Qual. 6, 259–262. doi: 10.2134/jeq1977.00472425000600030005x
Rizwan, M., Ali, S., Abbas, T., Rehman, M. Z., Hannan, F., Keller, C., et al. (2016). Cadmium minimization in wheat: a critical review. Ecotoxicol. Environ. Saf. 130, 43–53. doi: 10.1016/j.ecoenv.2016.04.001
Salsman, E., Kumar, A., AbuHammad, W., Abbasabadi, A. O., Dobrydina, M., Chao, S. M., et al. (2018). Development and validation of molecular markers for grain cadmium in durum wheat. Mol. Breeding 38:28. doi: 10.1007/s11032-018-0788-z
Sasaki, A., Yamaji, N., and Ma, J. F. (2014). Overexpression of OsHMA3 enhances Cd tolerance and expression of Zn transporter genes in rice. J. Exp. Bot. 65, 6013–6021. doi: 10.1093/jxb/eru340
Sasaki, A., Yamaji, N., Yokosho, K., and Ma, J. F. (2012). Nramp5 is a major transporter responsible for manganese and cadmium uptake in rice. Plant Cell 24, 2155–2167. doi: 10.1105/tpc.112.096925
Satoh-Nagasawa, N., Mori, M., Nakazawa, N., Kawamoto, T., Nagato, Y., Sakurai, K., et al. (2012). Mutations in Rice (Oryza sativa) Heavy metal ATPase 2 (OsHMA2) restrict the translocation of zinc and cadmium. Plant Cell Physiol. 53, 213–224. doi: 10.1093/pcp/pcr166
Sebastian, A., and Prasad, M. N. V. (2014). Cadmium minimization in rice. A review. Agron Sustain Dev. 34, 155–173. doi: 10.1007/s13593-013-0152-y
Shi, G. L., Li, D. J., Wang, Y. F., Liu, C. H., Hu, Z. B., Lou, L. Q., et al. (2019). Accumulation and distribution of arsenic and cadmium in winter wheat (Triticum aestivum L.) at different developmental stages. Sci. Total Environ. 667, 532–539. doi: 10.1016/j.scitotenv.2019.02.394
Smith, S. E., Kuehl, R., Ray, I., Hui, R., and Soleri, D. (1998). Evaluation of simple methods for estimating broad-sense heritability in stands of randomly planted genotypes. Crop Sci. 38, 1125–1129. doi: 10.2135/cropsci1998.0011183X003800050003x
Song, W. Y., Mendoza-Cózatl, D. G., Lee, Y., Schroeder, J. I., Ahn, S. N., Lee, H. S., et al. (2014). Phytochelatin-metal(loid) transport into vacuoles shows different substrate preferences in barley and Arabidopsis. Plant Cell Environ. 37, 1192–1201. doi: 10.1111/pce.12227
Takahashi, R., Bashir, K., Ishimaru, Y., Nishizawa, N. K., and Nakanishi, H. (2012). The role of heavy-metal ATPases, HMAs, in zinc and cadmium transport in rice. Plant Signal Behav. 7, 1605–1607. doi: 10.4161/psb.22454
Takahashi, R., Ishimaru, Y., Nakanishi, H., and Nishizawa, N. K. (2011a). Role of the iron transporter OsNRAMP1 in cadmium uptake and accumulation in rice. Plant Signal Behav. 6, 1813–1816. doi: 10.4161/psb.6.11.17587
Takahashi, R., Ishimaru, Y., Senoura, T., Shimo, H., Ishikawa, S., Arao, T., et al. (2011b). The OsNRAMP1 iron transporter is involved in Cd accumulation in rice. J. Exp. Bot. 62, 4843–4850. doi: 10.1093/jxb/err136
Ueno, D., Yamaji, N., Kono, I., Huang, C. F., Ando, T., Yano, M., et al. (2010). Gene limiting cadmium accumulation in rice. P. Natl. Acad. Sci. USA. 107, 16500–16505. doi: 10.1073/pnas.1005396107
Uraguchi, S., Kamiya, T., Sakamoto, T., Kasai, K., Sato, Y., Nagamura, Y., et al. (2011). Low-affinity cation transporter (OsLCT1) regulates cadmium transport into rice grains. Proc. Natl. Acad. Sci. USA. 108, 20959–20964.
Volpe, M. G., Nazzaro, M., Stasio, M. D., Siano, F., Coppola, R., and Marco, A. D. (2015). Content of micronutrients, mineral and trace elements in some Mediterranean spontaneous edible herbs. Chem. Cent. J. 9:57. doi: 10.1186/s13065-015-0137-9
Wagner, G. J., and Donald, L. S. (1993). Accumulation of cadmium in crop plants and its consequences to human health. Adv. Agron. 51, 173–212. doi: 10.1016/S0065-2113(08)60593-3
Wang, R., Chen, J., Anderson, J. A., Zhang, J., Zhao, W., Wheeler, J., et al. (2017). Genome-wide association mapping of Fusarium head blight resistance in spring wheat lines devel- oped in the Pacific Northwest and CIMMYT. Phytopathology 107, 1486–1495. doi: 10.1094/PHYTO-02-17-0073-R
Wegulo, S. N. (2012). Factors influencing deoxynivalenol accumulation in small grain cereals. Toxins. 4, 1157–1180. doi: 10.3390/toxins4111157
Wiebe, K., Harris, N. S., Faris, J. D., Clarke, J. M., Knox, R. E., Taylor, G. J., et al. (2010). Targeted mapping of Cdu1, a major locus regulating grain cadmium concentration in durum wheat (Triticum turgidum L. var durum). Theor. Appl. Genet. 121, 1047–1058. doi: 10.1007/s00122-010-1370-1
Yang, M., Zhang, Y., Zhang, L., Hu, J., Zhang, X., Lu, K., et al. (2014). OsNRAMP5 contributes to manganese translocation and distribution in rice shoots. J. Exp. Bot. 65, 4849–4861. doi: 10.1093/jxb/eru259
Zeng, F. R., Ali, S., Zhang, H., Quyang, Y., Qiu, B. Y., Wu, F. B., et al. (2011). The influence of pH and organic matter content in paddy soil on heavy metal availability and their uptake by rice plants. Environ. Pollut. 159, 84–91. doi: 10.1016/j.envpol.2010.09.019
Zhang, L., Gao, C., Chen, C., Zhang, W., and Zhao, F. J. (2020). Overexpression of rice OsHMA3 in wheat greatly decreases cadmium accumulation in wheat grain. Environ. Sci. Technol. 54, 10100–10108. doi: 10.1021/acs.est.0c02877
Zheng, X. W., Wen, X. J., Qiao, L., Zhao, J. J., Zhang, X. J., Li, X., et al. (2019). A novel QTL QTrl.saw-2D.2 associated with the total root length identified by linkage and association analyses in wheat (Triticum aestivum L.). Planta 250, 129–143. doi: 10.1094/PDIS-01-19-0001-RE
Keywords: cadmium, grain, spring wheat, doubled haploid population, quantitative trait locus
Citation: Qiao L, Wheeler J, Wang R, Isham K, Klassen N, Zhao W, Su M, Zhang J, Zheng J and Chen J (2021) Novel Quantitative Trait Loci for Grain Cadmium Content Identified in Hard White Spring Wheat. Front. Plant Sci. 12:756741. doi: 10.3389/fpls.2021.756741
Received: 11 August 2021; Accepted: 28 October 2021;
Published: 02 December 2021.
Edited by:
Kun Lu, Southwest University, ChinaReviewed by:
Dongcheng Liu, Chinese Academy of Sciences (CAS), ChinaCopyright © 2021 Qiao, Wheeler, Wang, Isham, Klassen, Zhao, Su, Zhang, Zheng and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jun Zheng, c3hua3l6akAxMjYuY29t; Jianli Chen, amNoZW5AdWlkYWhvLmVkdQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.