- 1Department of Biological Sciences, Graduate School of Science and Technology, Kumamoto University, Kumamoto, Japan
- 2International Research Organization for Advanced Science and Technology, Kumamoto University, Kumamoto, Japan
- 3International Research Center for Agricultural and Environmental Biology, Kumamoto University, Kumamoto, Japan
- 4Center for Digital Green-innovation, Nara Institute of Science and Technology, Nara, Japan
Parasitic root-knot nematodes transform the host’s vascular cells into permanent feeding giant cells (GCs) to withdraw nutrients from the host plants. GCs are multinucleated metabolically active cells with distinctive cell wall structures; however, the genetic regulation of GC formation is largely unknown. In this study, the functions of the Arabidopsis thaliana transcription factor PUCHI during GC development were investigated. PUCHI expression was shown to be induced in early developing galls, suggesting the importance of the PUCHI gene in gall formation. Despite the puchi mutant not differing significantly from the wild type in nematode invasion and reproduction rates, puchi GC cell walls appeared to be thicker and lobate when compared to the wild type, while the cell membrane sometimes formed invaginations. In three-dimensional (3D) reconstructions of puchi GCs, they appeared to be more irregularly shaped than those in the wild type, with noticeable cell-surface protrusions and folds. Interestingly, the loss-of-function mutant of 3-KETOACYL-COA SYNTHASE 1 showed GC morphology and cell wall defects similar to those of the puchi mutant, suggesting that PUCHI may regulate GC development via very long chain fatty acid synthesis.
Introduction
Plant-parasitic nematodes, such as root-knot nematodes (RKNs, Meloidogyne spp.), are known to infect and damage a broad spectrum of crops. RKNs feed on the vascular cells of host plant roots, which impedes plant development and may even lead to plant death. Infective RKN second instar (J2) larvae enter the host plant roots and migrate to the vasculature, where they induce the formation of special feeding sites called galls. It is thought that RKNs inject various effector proteins into vascular cells, which convert these cells into specialized feeding cells known as giant cells (GCs; Williamson and Gleason, 2003).
Galls play an essential role in sustaining RKN development (Favery et al., 2016), and GCs form the majority of a gall’s volume and provide RKNs with nutrients. The development of GCs thus directly dictates gall formation and RKN development. GCs result from repeated nuclear divisions without cell division (endoduplication) and isotropic plant cell growth (Caillaud et al., 2008). The first sign that RKNs are manipulating cell development is the appearance of supernumerary nuclei. Subsequently, cell plate formation is inhibited, and these multinucleated cells continue to endoduplicate until they contain up to 100 nuclei (Wiggers et al., 1990). GCs also contain cytoplasm densely packed with organelles and often show a high level of metabolic activity; thus, they may be up to 400 times larger than normal vascular cells (Caillaud et al., 2008). Nutrient uptake into GCs from the vasculature is enhanced by the development of numerous cell wall ingrowths adjacent to the xylem contact site (Jones and Payne, 1978; Bartlem et al., 2014). Furthermore, the constant withdrawal of nutrients by RKNs converts GCs into metabolic sinks for host plants (Favery et al., 2016). However, the molecular mechanisms underlying GC formation mediated by RKNs largely remain to be elucidated.
Galls can be considered a novel organ formed from root tissues during RKN infection, with development mechanisms similar to those of lateral roots (LRs) and adventitious roots. Recent studies have suggested that the developmental processes of RKN-induced galls, LRs, and calluses have overlapping regulatory mechanisms. Hence, genes that are known to be upregulated in LRs and calluses, such as master regulatory transcription factors, have also been identified in galls (Cabrera et al., 2014; Olmo et al., 2017, 2020). In addition, plant-parasitic nematodes are known to modulate the hosts’ vascular development during gall formation (Yamaguchi et al., 2017). Currently, it remains unclear how LR development regulatory pathways are adapted to control GC development.
PUCHI encodes an APETALA2/ethylene-responsive element-binding transcription factor found exclusively in plants that regulates the differentiation of lateral organs in shoots and roots (Hirota et al., 2007; Karim et al., 2009). In shoots, PUCHI is involved in the determination of floral meristem identity and the suppression of bract growth (Karim et al., 2009), while in roots, PUCHI restricts stem cell proliferation and coordinates cell division patterns during lateral root formation (Hirota et al., 2007). Moreover, PUCHI has been shown to be induced during LR initiation (Goh et al., 2019), and recent studies have revealed that PUCHI controls LR development, at least in part, through regulation of very long chain fatty acid (VLCFA) biosynthesis (Goh et al., 2019; Trinh et al., 2019). VLCFAs have 20 or more carbon atoms in the backbone, are synthesized by the fatty acid elongase complex, and are components of various membrane, storage, and extracellular lipids (Li-Beisson et al., 2013). PUCHI activates a subset of VLCFA biosynthetic genes to regulate cell proliferation during LR development (Trinh et al., 2019; Guyomarc’h et al., 2021). Essentially, PUCHI ensures proper organogenesis through transcriptional regulation, while repressing the development of other tissues during flowering and LR development. In addition, PUCHI has been shown to function during callus formation (Trinh et al., 2019). However, the role of PUCHI during gall formation remains unclear. The results of an RNA-Seq analysis of a series of developing galls suggested that PUCHI is upregulated during gall formation (Yamaguchi et al., 2017). Here, we further examine the functions of PUCHI during RKN infection.
Materials and Methods
Plant Materials and Growth Conditions
Arabidopsis thaliana of the Columbia-0 (Col-0) ecotype, which includes puchi-1, puchi-2 (Henikoff et al., 2004; Hirota et al., 2007), kcs1-5 (SALK_200839; Shang et al., 2016), and pKCS1::GUS (Joubès et al., 2008), was used for this study. To construct ProPUCHI-GUS-tPUCHI, the PUCHI promoter (−1 to −3,892 bases upstream of the start codon) and PUCHI terminator (+1 to +1,743 bases downstream of the stop codon) sequences were cloned upstream and downstream of the beta-glucuronidase (GUS) gene, respectively. The expression cassette was then cloned into the binary vector pBIN50 (Takano et al., 2010). The construct was used to transform Col-0 and puchi-1 plants. Seeds were surface-sterilized and sown on plates with Murashige and Skoog (MS) media [0.25× MS salt mixture (Sigma-Aldrich), 0.5% (w/v) sucrose, and 0.6% (w/v) phytagel or gellan gum, pH 6.4], vernalized for 2days in the dark at 4°C, then incubated and grown under continuous light at 23°C.
Nematode Preparation and Inoculation
Root-knot nematodes were propagated as previously described (Nishiyama et al., 2015). Briefly, 6- to 7-week-old tomato (Solanum lycopersicum) cultivar Pritz plants were inoculated with 20,000 RKN J2 larvae at 3-day intervals for a total of four inoculations (80,000 J2s). The inoculated tomato plants were then transferred into hydroponics systems (Nishiyama et al., 2015), and newly hatched J2s were harvested at 2- to 4-day intervals from the hydroponic culture media and surface-sterilized before use.
For RKN-infection assays, six 5-day-old Arabidopsis seedlings were grown in a petri dish with MS media, inoculated with approximately 80 sterilized J2s per plant, and incubated under short-day conditions (8h light and 16h dark) at 25°C. The roots of the seedlings were covered with black paper to mimic nature’s dark underground environment. Galls, adult females, and egg masses were counted at 14, 28, and 42days post-infection (dpi), respectively.
Histological Analyses
For acid fuchsin staining, the roots of RKN-infected seedlings were washed in deionized water and then transferred into 1% (v/v) sodium hypochlorite for 10–30min. Roots were then washed twice with deionized water and boiled in a solution of 30-fold diluted acid fuchsin stock [25% (v/v) acetic acid and 0.035% (w/v) acid fuchsin] for 10min. Cooled samples were washed with deionized water twice and then incubated in acidified glycerol (1.2-mm hydrochloric acid in glycerol) for 2–15min at 95°C. Samples were then mounted in acidified glycerol and observed using an Axio Imager M1 microscope (Carl Zeiss) equipped with a DP71 digital camera (Olympus).
For GUS staining, roots were fixed in 90% acetone for 24h at −30°C and stained in GUS buffer [100-mm NaPO4 buffer (pH 7), 10-mm ethylenediaminetetraacetic acid (EDTA, pH 8), 0.1% Triton X-100, 3-mm K3Fe(CN)6, and 3-mm K4Fe(CN)6] with 0.5mg/ml 5-bromo-4-chloro-3-indolyl-beta-D-glucuronide (Wako) for 4h. The reaction was stopped with Carnoy’s solution [90% (v/v) methanol and 10% (v/v) acetic acid], and the roots were mounted in chloral hydrate solution [80%(w/v) chloral hydrate and 10% (v/v)glycerol]. Samples were observed using an Axio Imager M1 microscope (Carl Zeiss Microscopy) and photographed using a DP71 digital camera (Olympus).
For toluidine blue-stained sections, galls were dissected and transferred into 2% (v/v) glutaraldehyde in 20-mm cacodylate buffer (pH 7.4), vacuum-infiltrated twice for 10min each, and then incubated at 4°C overnight. Samples were dehydrated in a graded ethanol series (50% ethanol for 10min, 75% ethanol for 10min, 90% ethanol for 10min, 95% ethanol for 10min at room temperature, and anhydrous ethanol for 20min at room temperature) and embedded in Technovit 7100 (Kulzer) according to the manufacturer’s instructions. Sample blocks were then sectioned to 5μm thickness using an ultramicrotome (Leica RM2255) and stained with 0.01% (w/v) toluidine blue O (WALDECK) containing 1% (w/v) sodium borate decahydrate (Nacalai) for 2min. Samples were then rinsed in deionized water for 1min, dried, and mounted in EUKITT (O. Kindler). Samples were observed using an Axio Imager M1 microscope (Carl Zeiss) equipped with a DP71 digital camera (Olympus).
Gene Expression Analysis
Total RNA was extracted from either 40 roots or 50 galls using the RNeasy plant mini kit (Qiagen) with the RNase-Free DNase Set (Qiagen) according to the manufacturer’s instructions. First-strand cDNA was synthesized from 300ng of total RNA using the PrimeScript RT Master Mix (Takara). Transcript levels were assayed using the FastStart Essential DNA Green Master (Roche) and the LightCycler 480 system (Roche). The thermal cycler program consisted of 95°C for 5min followed by 55cycles of 95°C for 10s, 60°C for 10s, and 72°C for 10s. The glyceraldehyde 3-phosphate dehydrogenase (GAPDH) gene was used as the internal control to calculate relative expression levels. Each of the three biological replicates was performed in technical triplicate. Real-time polymerase chain-reaction (RT-PCR) analyses were performed with the specific primers (see Supplementary Table 1). In all experiments, expression levels were normalized against those of GAPDH.
Microscopy
For transmission electron microscopy (TEM) analyses, samples were fixed in 2% paraformaldehyde (w/v) and 2% glutaraldehyde (v/v) in 0.05-M cacodylate buffer (pH 7.4) at room temperature and then refrigerated at 4°C. Samples were then fixed in 2% glutaraldehyde in 0.05-M cacodylate buffer (pH 7.4) at 4°C overnight. After fixation, the samples were washed three times with 0.05-M cacodylate buffer for 30min each and post-fixed with 2% osmium tetroxide (w/v) in 0.05-M cacodylate buffer at 4°C for 3h. The samples were dehydrated in graded ethanol solutions (50% ethanol for 30min at 4°C, 70% ethanol for 30min at 4°C, 90% ethanol for 30min at room temperature, and three changes of anhydrous ethanol for 30min each at room temperature). Samples were further dehydrated in anhydrous ethanol at room temperature overnight. Then, the samples were infiltrated with propylene oxide twice for 30min each, placed into a 70:30 mixture of propylene oxide and resin (Quetol-651; Nisshin EM) for 1h, and then incubated overnight with the cap open to evaporate the excess propylene oxide. The samples were transferred to fresh 100% resin and polymerized at 60°C for 48h. The resin-embedded samples were first sectioned to 1.5μm with a glass knife using an ultramicrotome (Ultracut UCT; Leica, Vienna, Austria) and stained with 0.5% toluidine blue. Ultra-thin sections of 80nm were then prepared using a diamond knife and an ultramicrotome (Ultracut UCT; Leica, Vienna, Austria) and mounted on copper grids. The sections were stained with 2% uranyl acetate at room temperature for 15min, washed with distilled water, and stained with lead stain solution (Sigma-Aldrich) at room temperature for 3min. The grids were observed under TEM (JEM-1400Plus; JEOL Ltd., Tokyo, Japan) at an acceleration voltage of 100kV. Images were taken with a charge-coupled device camera (EM-14830RUBY2; JEOL Ltd.)
Samples imaged by confocal microscopy were first cleared with ClearSee [10% (w/v) xylitol, 15% (w/v) sodium deoxycholate, and 25% (w/v) urea] (Kurihara et al., 2015). Galls were vacuum-infiltrated twice with 4% (w/v) paraformaldehyde for 30min each at room temperature. Fixed tissues were cleared with ClearSee at room temperature for 1week or longer, depending on tissue type (Kurihara et al., 2015). For post-staining, cleared tissues were stained with Calcofluor White (100μg/ml) and 1/1,000 SYBR Green 1 in ClearSee for 1h. After staining, tissues were washed in ClearSee for 2min. Samples were imaged with a confocal laser microscope (FV3000, Olympus) with an Olympus FV 10-MCPSU (405nm and 488nm) and ×60, NA 1.35 oil objective (UPlanSApo, Olympus). All Z-stack sections were imaged at 1–1.5-μm intervals.
Image Analyses
A pretrained deep learning model based on a convolutional neural network in AIVIA (DRVision, Bellevue, WA, United States) was used to identify GCs from confocal images. A total of 17 binary images in which the cell regions were manually segmented were used as the training data set. The output images were binarized with manual intensity thresholding to determine the cell regions. The surface models for the 3D cell reconstruction were obtained using Imaris (Bitplane, Belfast, UK).
Results
PUCHI Is Expressed in RKN Feeding Sites
RNA-Seq data from developing galls showed that PUCHI was upregulated during gall formation (Supplementary Figure S1A; Yamaguchi et al., 2017). This suggests that PUCHI may play important roles related to RKN infection. Indeed, quantitative reverse transcription polymerase chain-reaction (qRT-PCR) results are consistent with the notion that RKN infection causes the accumulation of PUCHI transcripts, since PUCHI mRNA levels increased as gall formation progressed (Figure 1A). PUCHI expression peaked by 5 dpi when the transcript level increased 8-fold compared to uninfected roots (Figure 1A).
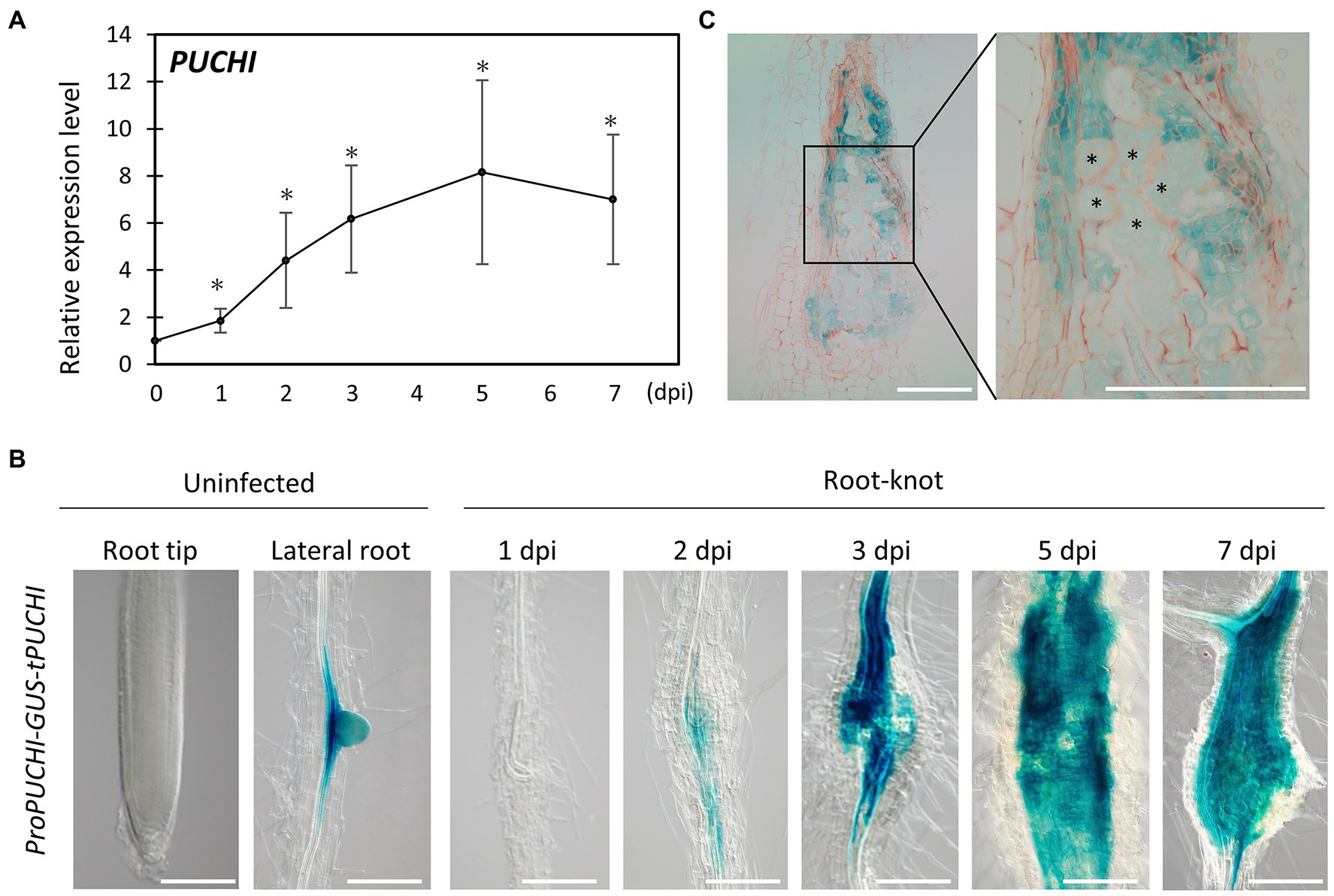
Figure 1. PUCHI expression analysis. (A) Quantitative RT-PCR results of PUCHI transcript levels in 1–7 dpi galls and uninfected roots. Means ± standard error is shown. *p<0.05 using Steel’s multiple comparison test. (B) GUS-stained ProPUCHI-GUS-tPUCHI 1–7 dpi galls and uninfected roots to denote the spatial expression patterns of PUCHI. (C) GUS-stained cross-sections of 7 dpi ProPUCHI-GUS-tPUCHI galls. * denotes GCs. Scale bars: 100μm.
To confirm the spatial expression pattern of PUCHI after RKN infection in wild-type (WT) plants, transgenic plants carrying ProPUCHI-GUS-tPUCHI construct in the WT background were examined. GUS expression was observed mostly in the lateral root primordia (LRP) of uninfected roots, which is consistent with previous work (Figure 1B; Hirota et al., 2007). In contrast, GUS expression was seen in the swelling central cylinder in the roots at 2 dpi and became more pronounced by 7 dpi (Figure 1B). Specifically, GUS expression in galls was primarily in neighboring cells surrounding the GCs, while weaker GUS staining was detected in the GCs (Figure 1C). PUCHI may function by mediating signal transductions from neighboring cells to GCs and regulating GC development non-cell autonomously. Taken together, these lines of evidence indicate that PUCHI is specifically induced during gall formation, especially in the early stages. This in turn suggests that PUCHI may play a role during gall formation.
Since PUCHI encodes a transcription factor, it is possible that PUCHI regulates its own expression. To determine whether this was the case, the ProPUCHI-GUS-tPUCHI reporter construct was also analyzed in the puchi-1 background. Similar to the situation in the WT background, GUS expression was also detected in the LRP and galls in puchi-1 plants (Supplementary Figure S1B), suggesting that PUCHI is unlikely to regulate its own expression.
PUCHI Does Not Strongly Influence RKN Infection and Development
Since RKN infections induce PUCHI expression in feeding sites, the functions of PUCHI during gall formation were investigated. Infection efficiency and gall growth were assayed in the presence of puchi-1 and puchi-2 loss-of-function mutations. Compared to the WT, these mutations did not significantly affect RKN invasion or gall formation rates (Figures 2A,B; Supplementary Figure S2A). We also measured gall diameters in these mutants and confirmed that the average puchi gall sizes were comparable to that of the WT (Figure 2C; Supplementary Figure S2B). Moreover, when compared to the WT, no significant differences in the numbers of emerging adult females and egg masses were found (Figures 2D,E). These results suggest that PUCHI alone does not regulate RKN invasion and development rates.
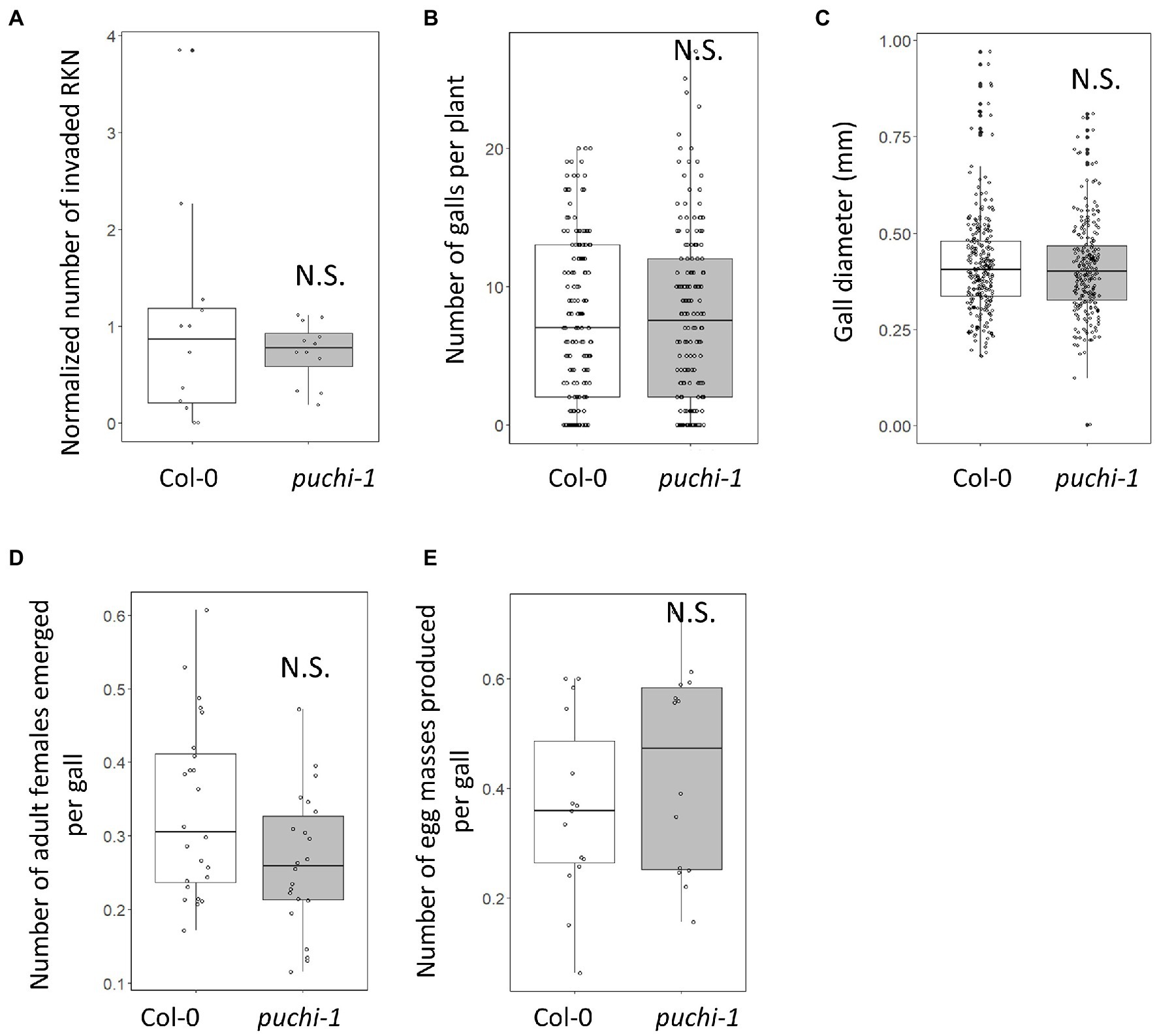
Figure 2. Effect of puchi mutation during RKN infection. Numbers of invading RKNs at 2 dpi are averages from four seedlings that were then normalized to values of WT in a root (A), galls at 14 dpi (B), gall diameters at 14 dpi (C), emerged mature females at 28 dpi (D), and egg mass at 42 dpi (E) in Col-0 and puchi-1 seedlings. Values are the means of at least three biological replicates. Statistical significance was analyzed using the Brunner-Munzel test. N.S. denotes not statistically significant.
PUCHI Plays a Role in Maintaining GC Shape During Gall Development
To further investigate the role of PUCHI in developing galls, histological analyses were performed on WT and puchi-1 galls. RKN infections cause a sophisticated transformation of root cells into GCs to nourish the RKN; this in turn results in root swellings and gall formation (Bird, 1961). Typically, 4–10 metabolically hyperactive GCs larger than other root cells were found in a single gall (Figure 3A). TEM revealed that GCs contained multiple (often endopolyploid) nuclei (Supplementary Figure S3A) and a cytoplasm densely packed with organelles (Figure 3B; Bartlem et al., 2014; Rodiuc et al., 2014).
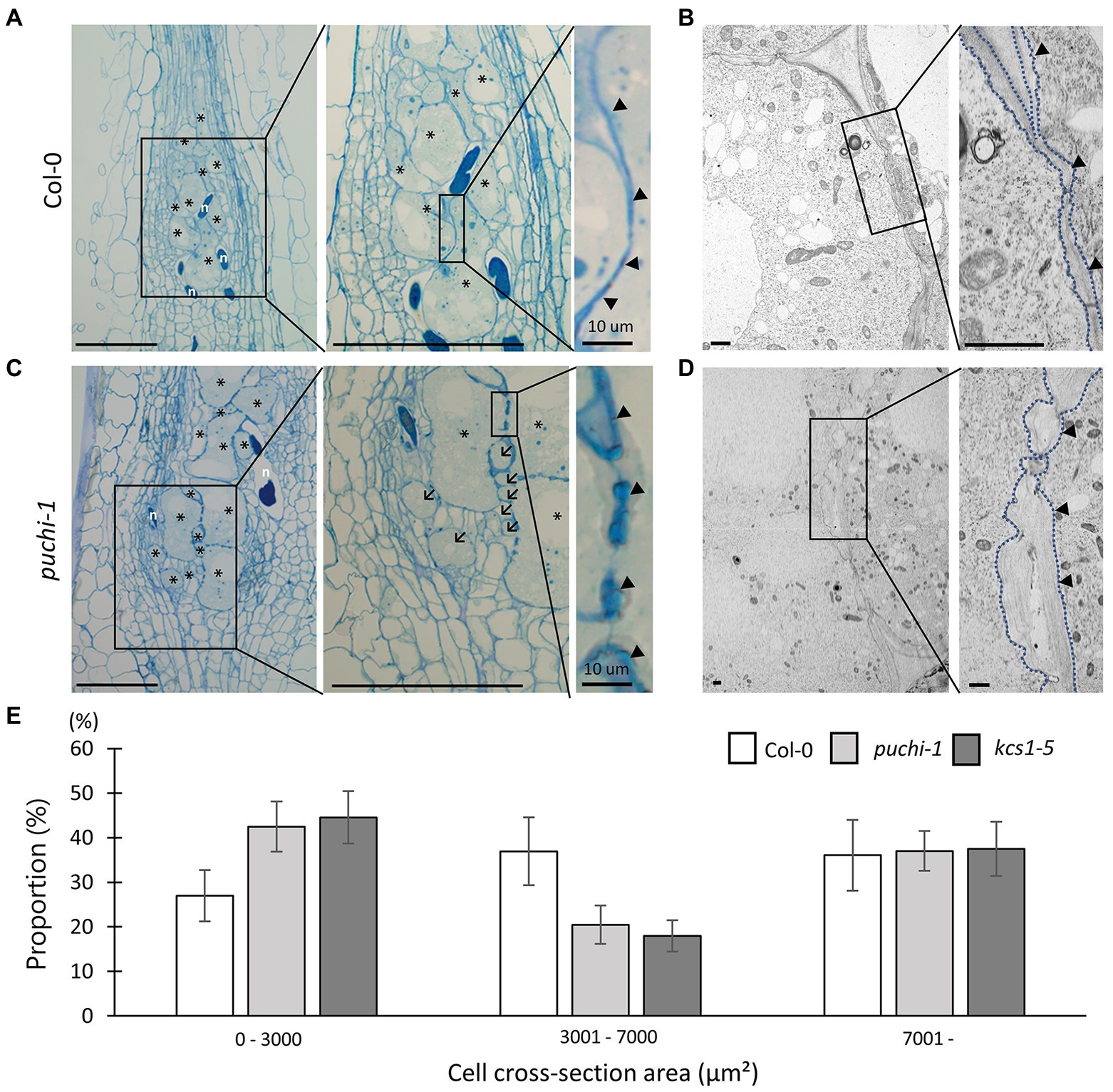
Figure 3. Observation of WT and puchi-1 GC morphology. Morphological analysis of puchi-1 and WT GCs. (A, C) Toluidine blue-stained 7 dpi WT (A) and puchi-1 (C) gall longitudinal sections. Arrows denote aberrant GCs, and arrowheads denote cell wall thickening in Col-0 and puchi-1 galls. * denotes GCs; n denotes RKNs. Scale bars: 100μm. (B, D) TEM micrographs of WT (B) and puchi-1 (D) gall cross-sections. Blue dotted lines denote GC cell walls in Col-0 and puchi-1 GCs. Arrowheads denote cell wall thickening in Col-0 and puchi-1 GCs. Scale bars: 1μm. (E) Proportions of WT, puchi-1, and kcs1-5 GCs with different cross-section area classes. Means ± standard deviation is shown. (WT: n=12, puchi-1: n=15, and kcs1-5: n=15).
In general, the cell walls between adjacent GCs appeared irregular and uneven in cross-sections compared to those of uninfected cells (Figure 3A right panel arrowheads; Figure 3B; Rodiuc et al., 2014). Cell wall thickness in the puchi-1 mutant appeared more irregular than in WT, to the point where puchi-1 GC cell walls appeared lobate in certain regions (Figure 3C right panel arrowheads; Figure 3D). Moreover, puchi-1 cell walls often formed concave invaginations that were not found in the WT (Supplementary Figure S3C).
Interestingly, small GC-like cells could be identified in puchi-1 7 dpi galls, which we tentatively named “aberrant GCs” (Figure 3C center panel arrows; Supplementary Figure S3B). The puchi-1 aberrant GCs were indeed multinucleated like normal GCs (Supplementary Figure S3B), suggesting that they were likely also products of RKN infection. To determine whether the numbers of aberrant GCs were indeed significantly higher in the puchi-1 mutant, GC distributions (based on size) were characterized in 14 dpi galls, the time at which GC differentiation is thought to be near completion. The puchi-1 galls showed an increase in the proportion of GCs with a cross-section area of 0–3,000μm2, and a decrease in those with an area of 3,001–7,000μm2 (Figure 3E). This result also suggests that the GC size distribution shifts toward smaller cells in the puchi-1 mutant, confirming that the aberrant GC phenotype is quantifiable.
At later stages of gall formation (28~42 dpi), the puchi GC defects became more pronounced (Supplementary Figure S3D). Around 42 dpi, mature females emerge from the roots to produce egg masses. Additionally, puchi aberrant GCs appeared to be more abundant during late gall development (Supplementary Figure S3D).
Puchi Produces GCs With Abnormal Morphology
To further evaluate how aberrant GCs influence gall formation in puchi mutants, GC cross-section areas and cell numbers were assayed in both WT and puchi galls. Histological analysis of cross-sections through the center of galls showed that the total area occupied by GCs at 14 dpi (Figure 4A) was significantly greater in the puchi-1 mutant than in the WT (Figure 4B). It is likely that the observed increase was due to the presence of the aberrant GCs. To address this possibility, the number of GCs per gall was counted in the WT and puchi-1 galls. The puchi-1 galls indeed contained significantly more GCs than the WT galls (Figure 4C).
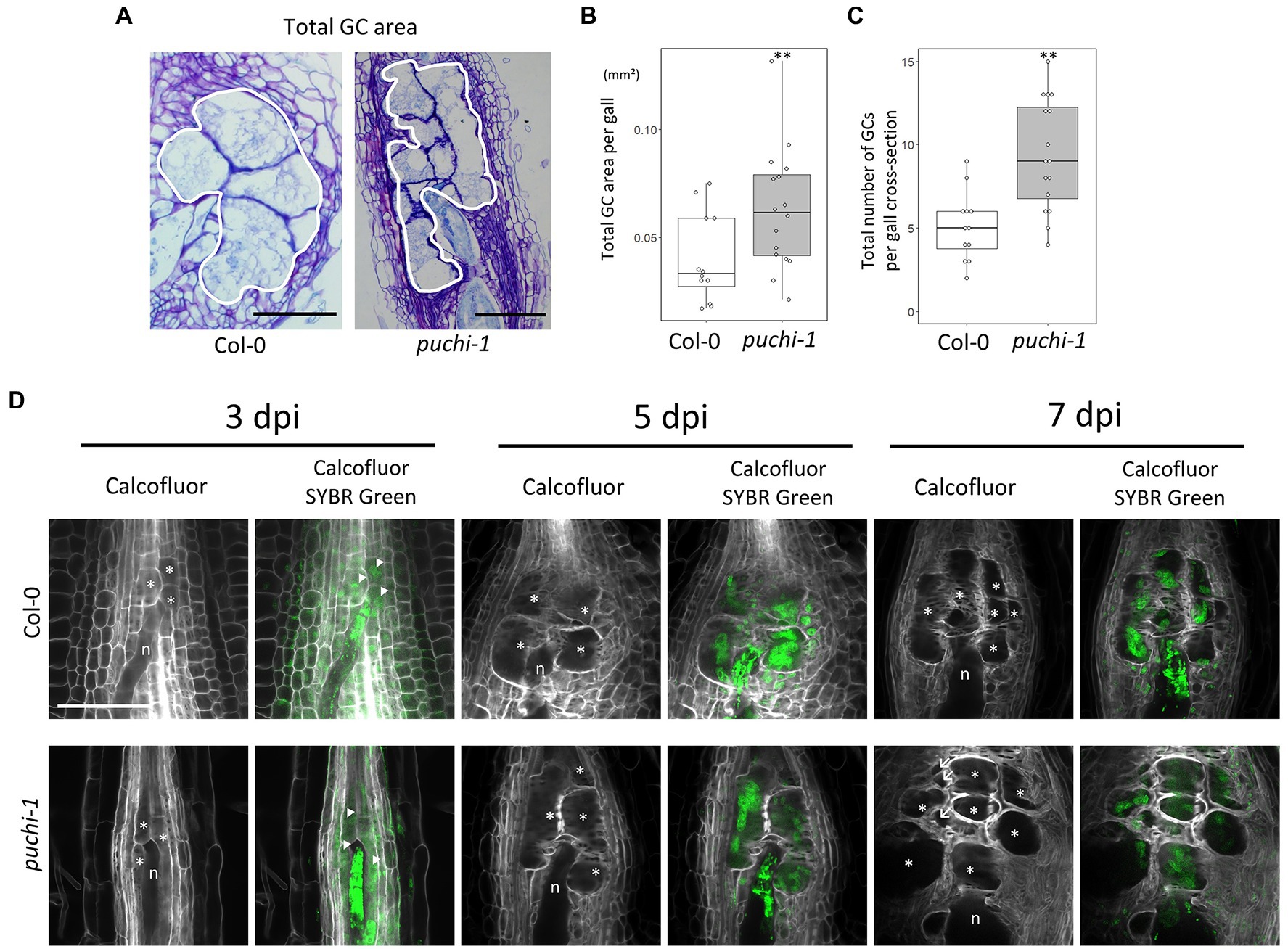
Figure 4. Development of WT and puchi-1 GCs. (A) Areas occupied by GCs in the WT and puchi-1 galls, as outlined with white borders. (B) Box plots of the total area occupied by GCs in WT and puchi-1 galls at 14 dpi. (C) Box plots of the number of GCs within the area occupied by GCs in cross-sections of WT and puchi-1 galls at 14 dpi. **p<0.01, Brunner-Munzel test. (D) Confocal microscopy images of WT and puchi-1 galls stained with Calcofluor White for cell walls and SYBR-Green for nuclei at 3, 5, and 7 dpi. n denotes RKNs. *denotes GCs. Arrow denotes aberrant GCs. Arrowheads denote nuclei.
To determine the time at which the number of GCs increased in the puchi-1 galls, the early stages of GC development in puchi galls were observed using confocal microscopy (Figure 4D). SYBR Green 1 was used to stain the nuclei, and Calcofluor White was used to stain the cell wall to visualize the GCs during the early stages of gall development (Figure 4D). puchi-1 gall cross-sections were indistinguishable from those of the WT around 3 dpi (Figure 4D). However, aberrant GCs started to appear at 7 dpi (Figure 4D). This suggests that aberrant GCs are likely to be products of defective GC formation caused by the puchi mutation, and not caused by factors present before RKN infection.
Next, to better characterize the 3D structure of aberrant GCs, Z-stack optical sections were used to build the minimum intensity projection (MinIP) of the WT and puchi GCs (Figure 5A; Supplementary Figure S4A). MinIP is used to visualize 3D structures with low intensity (Karnowski et al., 2017). Surprisingly, several puchi aberrant GCs that appeared as separate cells in one optical section were shown to be the same cells in different optical sections (Figure 5A; Supplementary Figure S4A). Next, the 3D structures of GCs in the WT and puchi-1 galls were reconstructed from Z-stack optical sections using Imaris. The 3D reconstructions clearly showed that the puchi-1 aberrant GCs were protrusions of larger GCs in other optical sections (Figure 5B; Supplementary Figure S4B). Similar results were obtained from the toluidine blue-stained sections (Supplementary Figure S4C). In WT galls, GCs did not appear to have major concavities and convexities on the surface (Figure 5B; Supplementary Figure S4B). In contrast, the puchi-1 GCs contained dramatic protrusions and invaginations (Figure 5B; Supplementary Figure S4B). When the total number of GCs was counted using 3D reconstructions instead of cross-sections, there was no significant difference between the WT and puchi-1 numbers (Figure 5C). These results suggest that although puchi does produce aberrant GCs, puchi galls do not contain more GCs. Rather, puchi produces normal numbers of normal-sized GCs with aberrant protrusions and invaginations, which may appear in single optical sections as multiple small GC-like cells.
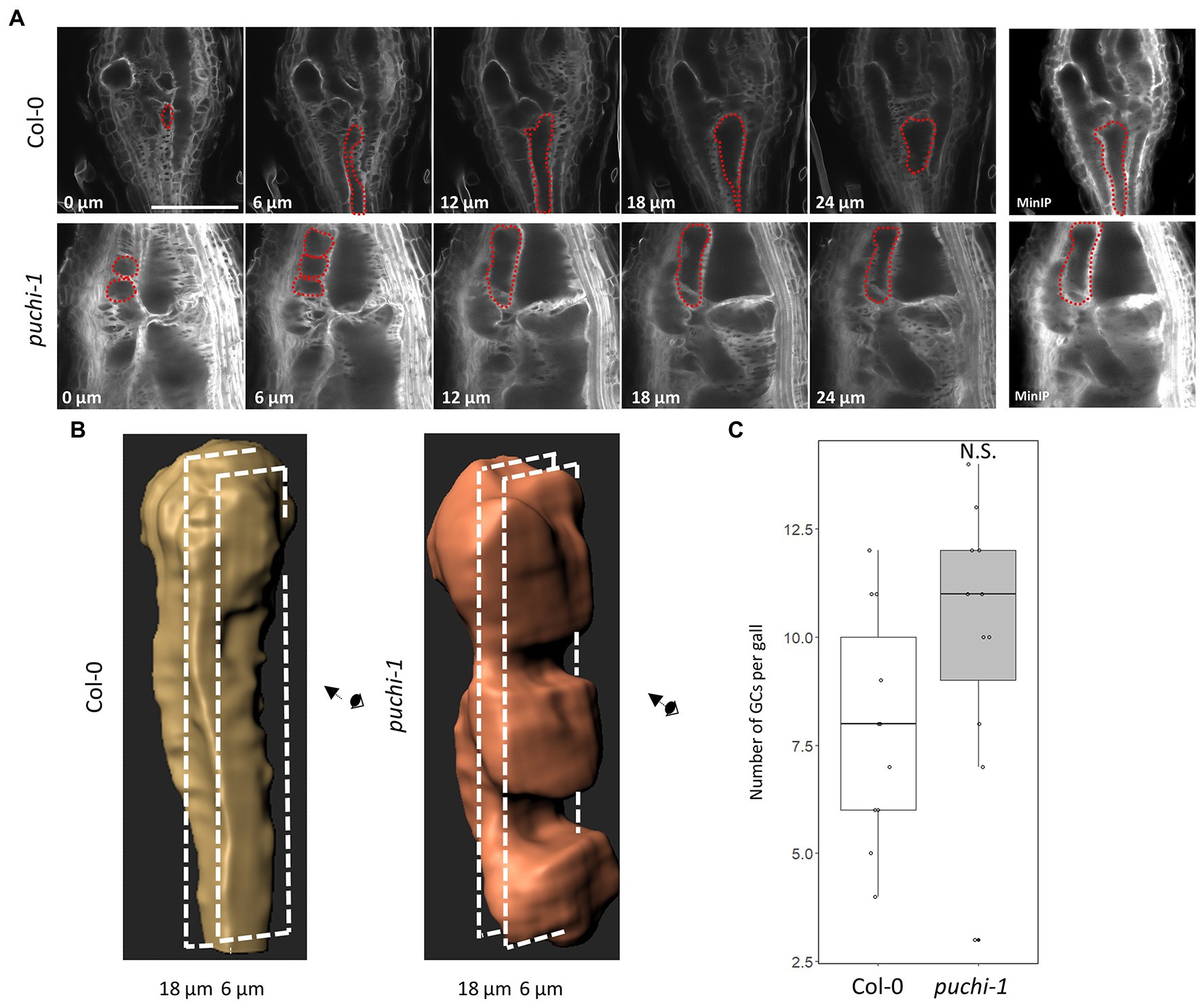
Figure 5. Reconstruction of 3D structures of WT and puchi-1 GCs. (A) Confocal optical sections of WT and puchi-1 galls stained with Calcofluor White at 7 dpi, and red dashed lines outline the same GC in different sections and the minimum intensity projection (MinIP) of the WT and puchi-1 optical section Z-stacks. Scale bars: 100μm. (B) 3D reconstructions of individual WT and puchi-1 GCs using the optical sections in (A). (C) Box plot of the number of GCs per WT and puchi-1 gall at 7 dpi calculated using GC 3D reconstructions. Statistical significance was analyzed using the Brunner-Munzel test; N.S. denotes no statistical significance.
PUCHI May Regulate Gall Formation via VLCFA Biosynthesis
A recent study revealed that PUCHI controls LR development, at least in part, through the regulation of VLCFA biosynthesis (Trinh et al., 2019). VLCFAs are synthesized by the VLCFA elongase complex through a four-step process: condensation by beta-ketoacyl-CoA synthase, reduction by beta-ketoacyl-CoA reductase, dehydration by hydroxyacyl-CoA dehydratase, and reduction by enoyl-CoA reductase (Li-Beisson et al., 2013). Interestingly, the mutants with defects in the VLCFA hydroxyacyl-CoA dehydratase PASTICCINO2 (PAS2) showed cell plate vesicles tethered to the plasma membrane, which superficially resembles the lobate GC cell wall found in puchi mutants (Bach et al., 2011; Figure 3D). It has been suggested that membrane lipids may be an important factor in mediating PUCHI’s role in gall formation. Therefore, we investigated whether PUCHI regulates the expression of VLCFA biosynthesis genes during gall formation. According to RNA-Seq data generated using developing galls (Yamaguchi et al., 2017), the expression levels of the major VLCFA elongation key enzymes fluctuated, albeit only mildly, over the course of gall formation (Supplementary Figure S5A). This suggested that these genes may be transcriptionally regulated over the course of gall formation. In galls, it was found that 3-KETOACYL-COA SYNTHASE 1 (KCS1) and KCS20 were among the most highly expressed VLCFA biosynthesis genes during gall formation and that the expression peaked at around 5dpi similar to that of PUCHI (Figure 1A; Supplementary Figure S5A).
qRT-PCR results were consistent with the notion that RKN infection induces KCS1 expression, since KCS1 transcript levels did increase significantly upon RKN infection in WT plants. However, such was not the case in puchi-1 plants, suggesting PUCHI positively regulates KCS1 expression during RKN infection (Figure 6A). The expression level of KCS20 also increased significantly after RKN infection in a PUCHI-dependent fashion; however, the magnitude of increase was much lower compared to KCS1 (Figure 6A). In contrast, KCS2 expression was significantly increased in galls of both WT and puchi-1, suggesting KCS2 is unlikely to be regulated by PUCHI (Figure 6A). Expression levels of BETA-KETOACYL REDUCTASE 1 (KCR1), PAS2, and ECERIFERUM (ECR) did not show significant increase in neither WT nor puchi-1 after RKN infection (Supplementary Figure S5B). These results suggested that among the VLCFA biosynthesis genes tested, only KCS1 and KCS20 were induced by PUCHI during gall formation. Since KCS1 showed the most pronounced upregulation, we have decided to focus on KCS1 for the subsequent analyses. To determine the converse of whether KCS1 influences PUCHI expression, PUCHI expression was examined kcs1-5 plants during RKN infection. No significant difference in PUCHI expression levels between kcs1-5 and WT in gall was observed; however, PUCHI expression was slightly, albeit significantly, increased in uninfected roots of kcs1-5 compared to the WT (Supplementary Figure S5C). This suggests that PUCHI may be transcriptionally repressed by events downstream of KCS1 as a form of negative feedback.
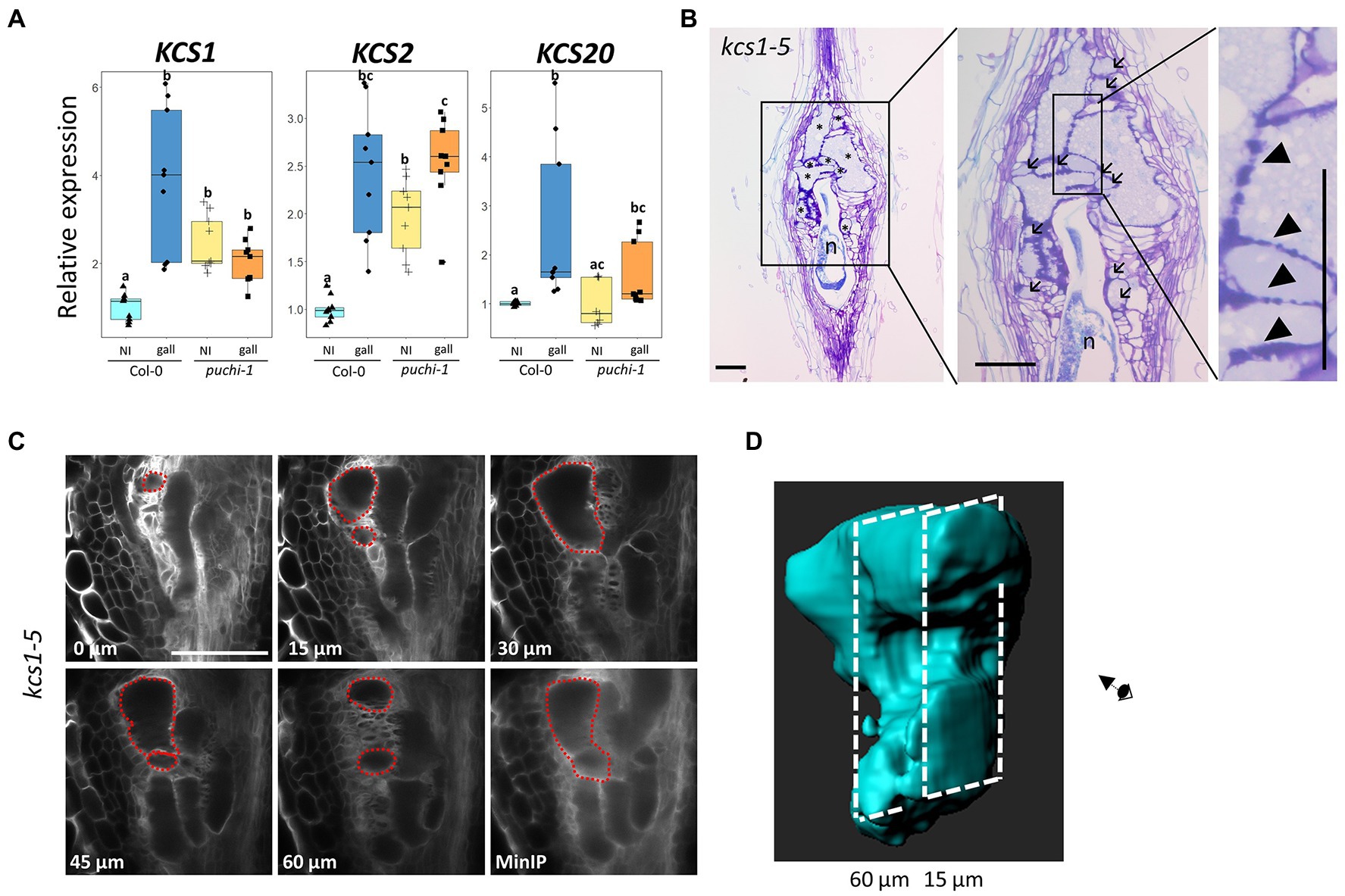
Figure 6. Expression analysis of VLCFA biosynthesis genes after RKN infection and morphological analysis of kcs1-5 galls. (A) qRT-PCR results of KCS1, KCS2, and KCS20 expression in WT (blue) and puchi-1 (orange) with uninfected roots (brighter shade) and 5 dpi galls (darker shade). Values are means of three technical replicates. Three biological replicates were performed with similar results. Alphabets denote significant differences between groups (Steel-Dwass’s multiple comparisons test, p<0.05). (B) Toluidine blue-stained longitudinal sections of kcs1-5 galls at 14 dpi. Arrows denote aberrant GCs, and arrowheads denote thickened lobate GC cell wall. * denotes GCs; n denotes RKNs. Scale bars: 100μm. (C) Confocal optical sections of kcs1-5 galls stained with Calcofluor White at 7 dpi, and red dashed line outlines the same GC in different sections and the MinIP of the kcs1-5 optical section Z-stacks. Scale bars: 100μm. (D) 3D reconstruction of individual kcs1-5 GCs using the optical sections in (C).
GUS staining in pKCS1::GUS transgenic plants was observed primarily in both uninfected main roots, LRPs and RKN-induced galls (Supplementary Figure S5D). Moreover, the puchi-1 mutation also caused a clear and consistent reduction of GUS staining in developing LRPs of pKCS1::GUS transgenic plants (Supplementary Figure S5D). In some 7 dpi puchi pKCS1::GUS galls, GUS staining was reduced to be restricted in the vascular region, whereas in the WT background, GUS staining can be found throughout the entire gall, consistent with the qRT-PCR results (Supplementary Figure S5D). Interestingly, GUS signal from pKCS1::GUS appears to be slightly stronger in the LRs of puchi-1 plants compared to the WT (Supplementary Figure S5D). This may explain the slightly increased KCS1 expression level in uninfected puchi-1 plants compared to the WT (Figure 6A). Nevertheless, the GUS-reporter line analysis validates the notion that KCS1 is positively regulated by PUCHI during gall formation.
To further investigate the roles of KCS1 during GC development, kcs1 GC morphology was observed. Accordingly, kcs1-5 galls also exhibited aberrant GCs (Figure 6B; Supplementary Figures S5E,F) and thicker lobate cell walls (Figure 6B; Supplementary Figures S5E,G), similar to those observed in puchi-1 galls, implying that KCS1 may function in the same pathway as PUCHI in GC development. To confirm whether the aberrant GCs found in the puchi and kcs1-5 galls were comparable, GC distributions (based on size) were also classified in 14 dpi WT and kcs1-5 galls. Similar to the puchi-1 gall findings, the kcs1-5 galls showed an increase in the proportion of GCs with a cross-section area of 0–3,000μm2, and a decrease in the proportion of GCs with an area of 3,001–7,000μm2 (Figure 3E). The fact that both puchi-1 and kcs1-5 galls showed a shift toward smaller GCs in cross-sections may imply that both genes utilize the same mechanism to control GC morphology. Moreover, the 3D reconstruction of kcs1-5 GCs also showed protrusions and invaginations on the surface, similar to puchi-1 GCs (Figures 6C,D). Additionally, the WT, puchi-1, and kcs1-5 galls all contained comparable proportions of GCs with a cross-section area above 7,000μm2 (Figure 3E), which is consistent with the finding that the overall GC size was comparable between the WT and puchi galls. The fact that kcs1-5 phenocopies the puchi-1 GC defects suggests that KCS1 and PUCHI may function in the same pathway to regulate GC development.
Discussion
PUCHI Is Expressed During the Early Stages of Gall Formation
The RKN infestation process can be separated into three distinct stages: invasion (~1 dpi), GC induction (~4 dpi), and nutrient acquisition (~7 dpi; Bartlem et al., 2014). GC growth occurs during the nutrient acquisition stage (Jones and Northcote, 1972; Jones and Payne, 1978). In this study, we demonstrated that the AP2/EREBP-type transcription factor PUCHI is expressed during the early stages (2~7 dpi) of gall formation (Figures 1A,B). Observation of ProPUCHI-GUS-tPUCHI roots showed that PUCHI was not detected in any root tissues except the LRP of uninfected plants. However, following RKN infection, PUCHI expression was detected in the developing galls (Figure 1B). These results suggest that PUCHI may function during gall formation. qRT-PCR results confirmed that PUCHI was most highly expressed in galls at 5 dpi (Figure 1A). Therefore, these results indicate that PUCHI is likely to be involved in early stage GC development. However, we found that the number of invading RKNs, galls formed, mature females that emerged, and egg masses produced, and the gall sizes were not significantly different between puchi and WT galls (Figure 2; Supplementary Figure S2). It has been reported that the Arabidopsis AP2 transcription factors PUCHI, DORNRÖSCHEN (DRN), and DORNRÖSCHEN-LIKE (DRNL) redundantly control floral organ initiation within the floral meristem (Chandler and Werr, 2017). Therefore, it is possible that other genes may be functionally redundant with PUCHI in RKN development regulation, such that the puchi single mutant alone does not produce dramatic defects in RKN infectivity and development. In the future, we consider it necessary to analyze higher-order mutants with these paralogs.
PUCHI Is Required for Proper GC Formation
In gall thin sections, the GCs of the puchi mutants were found to be smaller than those of the WT (Figure 3). However, 3D reconstructions of GCs generated using confocal microscopy optical sections, the aberrant puchi GCs were shown to be protrusions of larger GCs (Figure 5B; Supplementary Figure S4B). It seems that puchi galls form GCs that are more misshapen than those of WT galls and that the actual number of GCs does not change significantly. It is possible that steric constraints, such as the presence of nematodes, the rapid growth of GCs, and the presence of neighboring cells, result in the GC protrusions and indentations in WT plants (Cabrera et al., 2015, 2018). In puchi-1, these steric constraints may be more intense, as it has been reported that PUCHI represses cell divisions in LRPs (Hirota et al., 2007). Moreover, parts of the puchi-1 GC cell wall appeared to be thicker and lobate when compared to the WT GC cell wall, and sometimes formed invaginations (Figures 3A–D; Supplementary Figure S3C). Therefore, another possible explanation of how puchi-1 GCs fail to maintain their proper shape may be excessive cell membrane and cell wall material production, and we have indeed demonstrated that PUCHI may regulate VLCFA biosynthesis. Interestingly, the pas2 mutant also shows cell wall defects that are visually similar to those seen in puchi galls in the absence of RKN infection, suggesting there may be RKN-independent factors leading to these cell wall defects (Bach et al., 2011). PUCHI has been shown to control cell proliferation during LRP formation, as the puchi-1 mutant exhibits excessive cell division and produces abnormally enlarged flank cells in the early phase of LRP development (Hirota et al., 2007). Thus, it is possible that PUCHI not only suppresses mitosis in LRP formation, but also regulates cell wall formation in GCs.
PUCHI May Control the Expression of VLCFA Biosynthesis Genes During Gall Formation
Cell walls between adjacent GCs were irregularly shaped in puchi-1 galls, with some regions containing thicker walls with more elaborate lobes than found in WT galls (Figures 3A–D). This is a superficially similar result to the abortive cell plates observed in pas2-1 mutants (Bach et al., 2011). In addition, it has been reported that PUCHI regulates the VLCFA biosynthesis pathway during lateral root development (Trinh et al., 2019). These lines of evidence imply that there may be a connection between gall formation and VLCFA biosynthesis via PUCHI. We have demonstrated that KCS1 is indeed positively regulated by PUCHI during gall formation (Figure 6A), and the GCs of kcs1-5 galls also showed thicker and lobate cell walls like those of puchi-1 GCs. Furthermore, the aberrant GCs observed in kcs1-5 galls were also found to be parts of larger GCs like in puchi-1 galls (Figure 6D). This suggests that the GC defects seen in puchi mutants may be consequences of dysregulated VLCFA biosynthesis mediated by KCS1.
It should also be noted that a recent study showed that VLCFAs may play a role in the regulation of cellulose synthesis. The pas2 mutant exhibits reduced cell elongation in dark-grown hypocotyls (Zhu et al., 2020). This defect is likely due to reductions in the cellulose detected in the primary cell wall, cellulose synthase complex secretion/motility in the plasma membrane (Zhu et al., 2020). It has been reported that cellulose-deficient mutants and plants treated with the cellulose synthesis inhibitor isoxaben showed abnormal cell shapes, such as swelling and reduction of elongation in hypocotyls and roots (Arioli et al., 1998; Fagard et al., 2000; Refrégier et al., 2004; Fujita et al., 2013). These lines of evidence suggest a cross-regulatory network between VLCFAs, cell wall composition, and cell morphology. In short, the composition of the puchi-1 cell wall may have changed due to alterations in the cell membrane VLCFA composition. Both the cell wall carbohydrates and the cell membrane VLCFAs of the puchi-1 mutant may contribute to cell shape deformities, as either the altered cell wall and/or cell membrane may have buckled under the pressure exerted by the various steric hindrances in galls as described above.
Conclusion
Our findings suggest that PUCHI may be involved in the development of GCs via the regulation of VLCFA biosynthesis. Subsequent research will hopefully identify the pathway responsible for these processes, provide knowledge about how nematodes modulate GC and neighboring cell development, and further our understanding of PUCHI’s role in plant development.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: DDBJ [accession: PRJDB5797].
Author Contributions
RS, TH, MA, and MK conceived and designed the experiments. RS, MY, and TH performed the experiments. RS, AT, and SS wrote the manuscript. MK, TH, MA, AT, and SS revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the JSPS KAKENHI (grant numbers: 18H05487, 20H00422, 20KK0135, and 21K19273 to SS).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Dr. Laurent Laplaze and Dr. Soazig Guyomarc’h (IRD/Université de Montpellier) for helpful discussions, critical reading of the manuscript, and sharing materials. We are grateful to Xuehan Sun and Ryuya Okamoto (Kumamoto University) for technical support in sample preparation and nematode infection assays.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.755610/full#supplementary-material
Abbreviations
GCs, Giant Cells; RKNs, Root-Knot Nematodes; RNA-Seq, RNA Sequencing; LR, Lateral Root; MinIP, Minimum Intensity Projection; VLCFA, Very Long Chain Fatty Acid.
References
Arioli, T., Peng, L., Betzner, A. S., Burn, J., Wittke, W., Herth, W., et al. (1998). Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 279, 717–720. doi: 10.1126/science.279.5351.717
Bach, L., Gissot, L., Marion, J., Tellier, F., Moreau, P., Satiat-Jeunemaître, B., et al. (2011). Very-long-chain fatty acids are required for cell plate formation during cytokinesis in Arabidopsis thaliana. J. Cell Sci. 124, 3223–3234. doi: 10.1242/jcs.074575
Bartlem, D. G., Jones, M. G. K., and Hammes, U. Z. (2014). Vascularization and nutrient delivery at root-knot nematode feeding sites in host roots. J. Exp. Bot. 65, 1789–1798. doi: 10.1093/jxb/ert415
Bird, A. F. (1961). The ultrastructure and histochemistry of a nematode-induced giant-cell. J. Biophys. Biochem. Cytol. 11, 701–715. doi: 10.1083/jcb.11.3.701
Cabrera, J., Díaz-Manzano, F. E., Barcala, M., Arganda-Carreras, I., Almeida-Engler, J.de, Engler, G., et al. (2015). Phenotyping nematode feeding sites: three-dimensional reconstruction and volumetric measurements of giant cells induced by root-knot nematodes in Arabidopsis. New Phytol. 206, 868–880. doi: 10.1111/nph.13249
Cabrera, J., Díaz-Manzano, F. E., Sanchez, M., Rosso, M.-N., Melillo, T., Goh, T., et al. (2014). A role for LATERAL ORGAN BOUNDARIES-DOMAIN 16 during the interaction Arabidopsis–Meloidogyne spp. provides a molecular link between lateral root and root-knot nematode feeding site development. New Phytol. 203, 632–645. doi: 10.1111/nph.12826
Cabrera, J., Olmo, R., Ruiz-Ferrer, V., Abreu, I., Hermans, C., Martinez-Argudo, I., et al. (2018). A phenotyping method of giant cells from root-knot nematode feeding sites by confocal microscopy highlights a role for CHITINASE-LIKE 1 in Arabidopsis. Int. J. Mol. Sci. 19:429. doi: 10.3390/ijms19020429
Caillaud, M.-C., Dubreuil, G., Quentin, M., Perfus-Barbeoch, L., Lecomte, P., de Almeida Engler, J., et al. (2008). Root-knot nematodes manipulate plant cell functions during a compatible interaction. J. Plant Physiol. 165, 104–113. doi: 10.1016/j.jplph.2007.05.007
Chandler, J., and Werr, W. (2017). DORNRÖSCHEN, DORNRÖSCHEN-LIKE, and PUCHI redundantly control floral meristem identity and organ initiation in Arabidopsis. J. Exp. Bot. 68, 3457–3472. doi: 10.1093/jxb/erx208
Fagard, M., Desnos, T., Desprez, T., Goubet, F., Refregier, G., Mouille, G., et al. (2000). PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of Arabidopsis. Plant Cell 12, 2409–2423. doi: 10.1105/tpc.12.12.2409
Favery, B., Quentin, M., Jaubert-Possamai, S., and Abad, P. (2016). Gall-forming root-knot nematodes hijack key plant cellular functions to induce multinucleate and hypertrophied feeding cells. J. Insect Physiol. 84, 60–69. doi: 10.1016/j.jinsphys.2015.07.013
Fujita, M., Himmelspach, R., Ward, J., Whittington, A., Hasenbein, N., Liu, C., et al. (2013). The anisotropy1 D604N mutation in the Arabidopsis cellulose synthase1 catalytic domain reduces cell wall crystallinity and the velocity of cellulose synthase complexes. Plant Physiol. 162, 74–85. doi: 10.1104/pp.112.211565
Goh, T., Toyokura, K., Yamaguchi, N., Okamoto, Y., Uehara, T., Kaneko, S., et al. (2019). Lateral root initiation requires the sequential induction of transcription factors LBD16 and PUCHI in Arabidopsis thaliana. New Phytol. 224, 749–760. doi: 10.1111/nph.16065
Guyomarc’h, S., Boutté, Y., and Laplaze, L. (2021). AP2/ERF transcription factors orchestrate very long chain fatty acid biosynthesis during Arabidopsis lateral root development. Mol. Plant 14, 205–207. doi: 10.1016/j.molp.2021.01.004
Henikoff, S., Till, B. J., and Comai, L. (2004). TILLING. Traditional mutagenesis meets functional genomics. Plant Physiol. 135, 630–636. doi: 10.1104/pp.104.041061
Hirota, A., Kato, T., Fukaki, H., Aida, M., and Tasaka, M. (2007). The auxin-regulated AP2/EREBP gene PUCHI is required for morphogenesis in the early lateral root primordium of Arabidopsis. Plant Cell 19, 2156–2168. doi: 10.1105/tpc.107.050674
Jones, M. G. K., and Northcote, D. H. (1972). Multinucleate transfer cells induced in coleus roots by the root-knot nematode, Meloidogyne arenaria. Protoplasma 75, 381–395. doi: 10.1007/BF01282117
Jones, M. G. K., and Payne, H. L. (1978). Early stages of nematode-induced giant-cell formation in roots of Impatiens balsamina. J. Nematol. 10, 70–84.
Joubès, J., Raffaele, S., Bourdenx, B., Garcia, C., Laroche-Traineau, J., Moreau, P., et al. (2008). The VLCFA elongase gene family in Arabidopsis thaliana: phylogenetic analysis, 3D modelling and expression profiling. Plant Mol. Biol. 67:547. doi: 10.1007/s11103-008-9339-z
Karim, M. R., Hirota, A., Kwiatkowska, D., Tasaka, M., and Aida, M. (2009). A role for Arabidopsis PUCHI in floral meristem identity and bract suppression. Plant Cell 21, 1360–1372. doi: 10.1105/tpc.109.067025
Karnowski, K., Ajduk, A., Wieloch, B., Tamborski, S., Krawiec, K., Wojtkowski, M., et al. (2017). Optical coherence microscopy as a novel, non-invasive method for the 4D live imaging of early mammalian embryos. Sci. Rep. 7:4165. doi: 10.1038/s41598-017-04220-8
Kurihara, D., Mizuta, Y., Sato, Y., and Higashiyama, T. (2015). ClearSee: a rapid optical clearing reagent for whole-plant fluorescence imaging. Development 142, 4168–4179. doi: 10.1242/dev.127613
Li-Beisson, Y., Shorrosh, B., Beisson, F., Andersson, M. X., Arondel, V., Bates, P. D., et al. (2013). Acyl-lipid metabolism. Arabidopsis Book 11:e0161. doi: 10.1199/tab.0161
Nishiyama, H., Ngan, B. T., Nakagami, S., Ejima, C., Ishida, T., and Sawa, S. (2015). Protocol for root-knot nematode culture by a hydroponic system and nematode inoculation to Arabidopsis. Jpn. J. Nematol. 45, 45–49. doi: 10.3725/jjn.45.45
Olmo, R., Cabrera, J., Díaz-Manzano, F. E., Ruiz-Ferrer, V., Barcala, M., Ishida, T., et al. (2020). Root-knot nematodes induce gall formation by recruiting developmental pathways of post-embryonic organogenesis and regeneration to promote transient pluripotency. New Phytol. 227, 200–215. doi: 10.1111/nph.16521
Olmo, R., Cabrera, J., Moreno-Risueno, M. A., Fukaki, H., Fenoll, C., and Escobar, C. (2017). Molecular transducers from roots are triggered in Arabidopsis leaves by root-knot nematodes for successful feeding site formation: a conserved post-embryogenic de novo organogenesis program? Front. Plant Sci. 8:875. doi: 10.3389/fpls.2017.00875
Refrégier, G., Pelletier, S., Jaillard, D., and Höfte, H. (2004). Interaction between wall deposition and cell elongation in dark-grown hypocotyl cells in Arabidopsis. Plant Physiol. 135, 959–968. doi: 10.1104/pp.104.038711
Rodiuc, N., Vieira, P., Banora, M. Y., and de Almeida Engler, J. (2014). On the track of transfer cell formation by specialized plant-parasitic nematodes. Front. Plant Sci. 5:160. doi: 10.3389/fpls.2014.00160
Shang, B., Xu, C., Zhang, X., Cao, H., Xin, W., and Hu, Y. (2016). Very-long-chain fatty acids restrict regeneration capacity by confining pericycle competence for callus formation in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 113, 5101–5106. doi: 10.1073/pnas.1522466113
Takano, S., Niihama, M., Smith, H. M. S., Tasaka, M., and Aida, M. (2010). Gorgon, a novel missense mutation in the SHOOT MERISTEMLESS gene, impairs shoot meristem homeostasis in Arabidopsis. Plant Cell Physiol. 51, 621–634. doi: 10.1093/pcp/pcq028
Trinh, D.-C., Lavenus, J., Goh, T., Boutté, Y., Drogue, Q., Vaissayre, V., et al. (2019). PUCHI regulates very long chain fatty acid biosynthesis during lateral root and callus formation. Proc. Natl. Acad. Sci. U. S. A. 116, 14325–14330. doi: 10.1073/pnas.1906300116
Wiggers, R. J., Starr, J. L., and Price, H. J. (1990). DNA content and variation in chromosome number in plant cells affected by Meloidogyne incognita and M. arenaria. Phytopathology 80, 1391–1395. doi: 10.1094/Phyto-80-1391
Williamson, V. M., and Gleason, C. A. (2003). Plant–nematode interactions. Curr. Opin. Plant Biol. 6, 327–333. doi: 10.1016/S1369-5266(03)00059-1
Yamaguchi, Y. L., Suzuki, R., Cabrera, J., Nakagami, S., Sagara, T., Ejima, C., et al. (2017). Root-knot and cyst nematodes activate procambium-associated genes in Arabidopsis roots. Front. Plant Sci. 8:1995. doi: 10.3389/fpls.2017.01195
Keywords: plant-parasitic nematode, cell wall, cell morphology, VLCFA, plant-pathogen interaction
Citation: Suzuki R, Yamada M, Higaki T, Aida M, Kubo M, Tsai AY and Sawa S (2021) PUCHI Regulates Giant Cell Morphology During Root-Knot Nematode Infection in Arabidopsis thaliana . Front. Plant Sci. 12:755610. doi: 10.3389/fpls.2021.755610
Edited by:
Kengo Morohashi, Michigan State University, United StatesReviewed by:
Rosemary White, Commonwealth Scientific and Industrial Research Organisation (CSIRO), Australia Hirokazu Tanaka, Meiji University, JapanCopyright © 2021 Suzuki, Yamada, Higaki, Aida, Kubo, Tsai and Sawa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shinichiro Sawa, c2F3YUBrdW1hbW90by11LmFjLmpw
 Reira Suzuki
Reira Suzuki Mizuki Yamada2
Mizuki Yamada2 Takumi Higaki
Takumi Higaki Mitsuhiro Aida
Mitsuhiro Aida Minoru Kubo
Minoru Kubo Allen Yi-Lun Tsai
Allen Yi-Lun Tsai Shinichiro Sawa
Shinichiro Sawa