- 1State Key Laboratory of Plant Physiology and Biochemistry, College of Life Sciences, Zhejiang University, Hangzhou, China
- 2State Key Laboratory of Subtropical Silviculture, Zhejiang A & F University, Hangzhou, China
- 3Research Centre for Plant RNA Signaling and Zhejiang Provincial Key Laboratory for Genetic Improvement and Quality Control of Medicinal Plants, College of Life and Environmental Sciences, Hangzhou Normal University, Hangzhou, China
In response to changing environments, plants regulate gene expression and subsequent metabolism to acclimate and survive. A superfamily of acyl-activating enzymes (AAEs) has been observed in every class of creatures on planet. Some of plant AAE genes have been identified and functionally characterized to be involved in growth, development, biotic, and abiotic stresses via mediating diverse metabolic pathways. However, less information is available about AAEs superfamily in tomato (Solanum lycopersicum), the highest value fruit and vegetable crop globally. In this study, we aimed to identify tomato AAEs superfamily and investigate potential functions with respect to aluminum (Al) stress that represents one of the major factors limiting crop productivity on acid soils worldwide. Fifty-three AAE genes of tomato were identified and named on the basis of phylogenetic relationships between Arabidopsis and tomato. The phylogenetic analysis showed that AAEs could be classified into six clades; however, clade III contains no AAE genes of tomato. Synteny analyses revealed tomato vegetable paralogs and Arabidopsis orthologs. The RNA-seq and quantitative reverse-transcriptase PCR (qRT-PCR) analysis indicated that 9 out of 53 AAEs genes were significantly up- or downregulated by Al stress. Numerous cis-acting elements implicated in biotic and abiotic stresses were detected in the promoter regions of SlAAEs. As the most abundantly expressed gene in root apex and highly induced by Al, there are many potential STOP1 cis-acting elements present in the promoter of SlAAE3-1, and its expression in root apex was specific to Al. Finally, transgenic tobacco lines overexpressing SlAAE3-1 displayed increased tolerance to Al. Altogether, our results pave the way for further studies on the functional characterization of SlAAE genes in tomato with a wish of improvement in tomato crop in the future.
Introduction
Aluminum (Al) toxicity is one of the major limiting factors affecting the crop productivity in acidic soils, which occupy nearly 50% of the potential arable lands of the world (von Uexküll and Mutert, 1995). When soil pH is lower than 5.5, ionic Al, mainly Al3+, predominates in soil solution, which is highly toxic to plants. The initial and most visible symptom of Al toxicity is inhibition of root elongation by ravaging cell structure of the root apex and thus limiting the mineral nutrient and water uptake and, consequently, hindering the plant growth and development (Kochian, 1995; Ma, 2007; Ryan et al., 2011; Liu et al., 2014). To adapt to Al-toxic environment, plants have evolved two major types of Al-tolerance mechanisms, namely, external exclusion (preventing Al from entering cells of root apex) and internal tolerance mechanisms (detoxifying Al via complexation and sequestration) (Kochian et al., 2004, 2015; Liu et al., 2014). Substantial advances have been made toward elucidating the physiological and molecular mechanisms by which plants cope with Al stress (Kochian et al., 2015; Yang et al., 2019). Recently, it has been shown that metabolic change might play important roles in response to Al stress (Lou et al., 2016a,b; Xian et al., 2020). However, the molecular basis of the role of metabolic alterations in Al stress response still needs further elucidation.
The activation of carboxylic acids provides the precursors for pathways that lead to the metabolism of a diverse variety of metabolites, including lipids, amino acids (aa), sugars, and secondary metabolites. In plants, the acyl-activating enzymes (AAEs) superfamily consists of acyl-coenzyme A synthetases (ACSs), 4-coumarate:coenzyme A ligases (4CLs), luciferases, and non-ribosomal peptide synthetases, which are involved in many primary and secondary metabolic pathways. All members of the AAE family have low sequence similarity to each other but share many highly conserved motifs, such as the AMP-binding domain (Shockey and Browse, 2011). As the members of the Arabidopsis AAE superfamily were systematically analyzed and identified, more and more metabolic functions of plant AAE superfamily genes have been reported. For example, the Arabidopsis peroxisomal-localized OPCL1 (OPDA-CoA ligase) gene, At1g20510, is involved in the biosynthesis of jasmonic acid in Arabidopsis (Koo et al., 2006; Kienow et al., 2008). The rice fatty acyl-CoA synthase gene, OsACOS12, is involved in regulating lipid metabolism-mediated tapetum-programmed cell death, which ultimately affects the male fertility of rice (Yang et al., 2017). The petunia malonyl-CoA synthase gene, PhAAE13, is specifically involved in anthocyanin biosynthesis in flowers (Chen et al., 2017). Recently, a rice 4CL4 belonging to 4-coumarate:coenzyme A ligases was reported to be involved in Al resistance (Liu et al., 2020).
Al-induced secretion of organic acids, including citrate, malate, and oxalate, has been well-documented as a very important mechanism by which plants resist the Al toxicity (Yang et al., 2019). Although transporters responsible for Al-induced citrate and malate secretion, respectively, have been characterized in a variety of plant species, genes encoding oxalate transporter remain unclear (Yang et al., 2019). Accumulating evidence suggests that oxalic acid has an important role in plant responses to both biotic (Molano-Flores, 2001; Jang et al., 2016) and abiotic stresses, including calcium regulation, ion homeostasis, metal stress, and other pathways (Palmieri et al., 2019). It has been reported that one of the AAE family members, AAE3 (acyl-activating enzyme3), is involved in oxalic acid metabolism–mediated plant growth and development and in resistance to biotic and abiotic stresses. For example, Foster et al. (2012) identified an AAE3 gene encoding an oxalyl-CoA synthase in Arabidopsis and found that it participated in seed development and fungal pathogen defense by catalyzing CoA-dependent oxalate metabolism (Foster et al., 2012). Lou et al. (2016a) found that rice bean (Vigna umbellata) VuAAE3 is involved in oxalate degradation and Al tolerance. Recently, Xian et al. (2020) also found that wild soybean GsAAE3 similarly influences its tolerance to Cd and Al stress by catalyzing the oxalate metabolism. Therefore, it appears that AAE family proteins might have important roles in Al stress responses by regulating metabolic pathways.
In this study, we identified 53 AAE genes from tomato genome and found 9 differentially expressed AAE genes, including SlAAE3-1 and SlAAE3-2, under Al stress. The expression pattern analysis of SlAAE3-1 suggested that its expression is specific to Al stress. Therefore, our results contribute not only to enrich the molecular mechanism of Al stress response in tomato but also to provide a theoretical basis for improving tomato Al tolerance through genetic improvement and molecular breeding techniques.
Materials and Methods
Plant Materials and Grown Conditions
Tomato (Solanum lycopersicum) cultivar Ailsa Craig (AC) was used in this study (Horticulture Research International, Warwick, United Kingdom). The seeds were sterilized with 10% NaClO (v/v) for 15 min, washed thoroughly with sterile water, and soaking in sterilized water overnight. After that, the seeds were sown on Petri dishes containing 1/5 strength Hoagland nutrient solution (pH 5.5). The nutrient solution consisted of KNO3 (1.0 mM), Ca(NO3)2 (1.0 mM), MgSO4 (0.4 mM), and (NH4)H2PO4 (0.2 mM) and the micronutrients NaFeEDTA (20 μM), H3BO3 (3.0 μM), MnCl2 (0.5 μM), CuSO4 (0.2 μM), ZnSO4 (0.4 μM), and (NH4)6Mo7O24 (1 μM), with 0.8% agar (Sigma-Aldrich, Saint Louis, United States). Perti dishes were kept in the dark at 4°C for 2 days and then germinated in a plant growth room with a daytime 16 h/24°C and 8 h/22°C night regime. Germinated seedlings with uniform primary root length (4 cm) were transferred to 1/5 Hoagland nutrient solution (pH 5.5) with (NH4)H2PO4 concentration of 10 μM.
Identification of Acyl-Activating Enzyme Superfamily in Tomato
The Hidden Markov Model (HMM) file corresponding to the AMP-bind domain (PF00501) was downloaded from the Pfam protein family database1 (El-Gebali et al., 2019). HMMER 3.2 was used to search against the AAE superfamily genes from the annotated tomato genome obtained from Phytozome version 12.12 (Finn et al., 2011; Goodstein et al., 2012). All candidate genes that may contain AMP-binding domain based on HMMER results were further examined by confirming the existence of the AMP-binding core sequence using the PFAM and the SMART program3 (Letunic and Bork, 2018). The length of aa sequences, protein molecular weights (MWs), and isoelectric point of identified tomato AAE superfamily proteins were obtained by using tools from the ExPasy website.4
Phylogenetic Analysis of Acyl-Activating Enzyme Supfamily Members
The sequences of 53 identified tomato AAEs and Arabidopsis all 60 AAE sequences according to two studies previously reported (Shockey et al., 2002, 2003; De Azevedo Souza et al., 2008) were used to create multiple protein sequence alignments using ClustalW in MEGA 7.05 (Kumar et al., 2016) with default parameters. The alignment results were used to construct a phylogenetic tree using the neighbor-joining method with 1,000 bootstrap replicates. The phylogenetic tree was displayed using the R package ggtree (Yu et al., 2017).
Gene Structure and Conserved Motif Analysis
The exon-intron distribution of each tomato AAE superfamily genes (SlAAEs) was analyzed by comparing predicted coding sequences with their corresponding genomic sequences using TBtools program (Chen et al., 2020). Conserved motifs of tomato AAE protein sequences were investigated using the online software MEME5.0.46 (Bailey et al., 2009) with the following motif parameters: number of repetitions (any), maximum number of motif (20), and the optimum width of each motif (between 6 and 100 residues).
Chromosomal Distribution and Gene Duplication Analysis
All SlAAEs were mapped to 12 tomato chromosomes based on physical location information from the database of tomato genome using TBtools (Chen et al., 2020). Multiple Collinearity Scan Toolkit, McScanX (Wang et al., 2012) with the default parameters was used to analyze the tandem repeats and segmental duplication events of SlAAEs superfamily in the tomato genome and synteny of AAE superfamily genes between tomato and Arabidopsis.
Expression Analysis of Aluminum-Responsive SlAAE Genes
To investigate Al-responsive SlAAEs, the analysis of RNA-seq data (Jin et al., 2020) and qRT-PCR were performed. For qRT-PCR, seedlings were subjected to the modified 1/5 Hoagland nutrient solution (pH 5.0; 10 μM NH4H2PO4) containing 5 μM Al or 5 μM of CdCl2, or LaCl3, or 3 μM of CuCl2 for 6 h. RNA samples were extracted from both root tips (1 cm in length) after treatment. One microgram of DNA-free RNA was transcribed into first strand cDNA by PrimeScript RT Master Mix (TaKaRa). The qRT-PCR was carried out with the Roche LightCyler 480 instrument using SYBR Green chemistry (Toyobo, Osaka, Japan). The reaction conditions were 40 cycles at 95°C for 15 s, 60°C for 10 s, and 72°C for 15 s. The primer sequences used in this study are listed in Supplementary Table 1. Expression data of target genes were normalized with tomato GAPDH (Wang et al., 2016) by the ΔΔCt method. Each reaction was performed with three repeats from different biological samples.
Promoter Analysis
The promoter data were obtained from Phytozome version 12.1 (see text footnote 2). The promoter analysis was conducted by searching 2.0 kb upstream sequences of the coding sequences against the PlantCARE database7 to identify their related cis-elements (Lescot et al., 2002). After sorting the cis-elements obtained from PlantCARE, the results were visualized and mapped to the AAE promoter using the TBtools software (Chen et al., 2020).
Overexpression of SlAAE3-1 in Tobacco and Aluminum Tolerance Evaluation
The open read frame of SlAAE3-1 was amplified by PCR using gene-specific primer pair TGGTACCATGGAGAGTATGACGCTC and CCGGGATCCCTACGCTCCAAATTTAGG, cloned into pCAMBIA1300 vector driven by Cauliflower mosaic virus 35S promoter and transformed into Agrobacterium tumefaciens (strain GV1301). Tobacco plants were transformed as described by Horsch et al. (1985). Transgenic lines carrying SlAAE3-1 were selected using PCR with the primers described above. For evaluating Al tolerance of SlAAE3-1-overexpressing lines, seeds from T2 homozygous and wild-type lines were first sterilized, soaked, and germinated as described above. When the length of the primary root had reached about 4 cm, the seedlings were transferred to the modified 1/5 Hoagland nutrient solution (pH 5.0; 10 μM NH4H2PO4) containing 5 μM Al for 4 days. The solution was renewed every 2 days. The Al sensitivity was evaluated by relative root elongation expressed as (root elongation with Al treatment/root elongation without Al) × 100. After the treatment, root apex was stained with propidium iodide (PI) solution (5 μg/ml) for 1 min, washed with deionized water for 30 s, then observed, and captured using confocal laser scanning microscopy (Zeiss LSM710, Jena, Germany).
Statistical Analysis
The Student’s t-test was performed in Microsoft Excel (version 2016, Microsoft Corp., Redmond, WA, United States). Data are given as means ± standard deviation (SD) of three independent biological replicates. A p-value < 0.05 was considered to be statistically significant.
Results
Identification of the Acyl-Activating Enzymes Superfamily in Tomato
We identified 53 members of AAEs superfamily in S. lycopersicum by searching the AAE consensus motif (PF00501) equal to PROSITE PS00455 (Shockey et al., 2003) following a previously described analysis pipeline (Jin et al., 2020). Gene characteristics, including the length of the coding sequence, the length of the protein sequence, the protein MW, isoelectric point (pI), and protein sequence, were analyzed (Supplementary Table 2). Among the 53 SlAAE proteins, Solyc07g043650 was identified to be the smallest protein with 455 aa, whereas the largest one was Solyc01g006640 (2,320 aa). The MW of SlAAE proteins ranged from 50.4 to 256.8 kDa, and the pI varied from 5.13 (Solyc08g076300) to 9.43 (Solyc03g005090).
Phylogenetic Analysis and Classification of SlAAE Genes
To probe the phylogenetic relationships among these 53 SlAAEs, we constructed an unrooted phylogenetic tree for SlAAEs together with 60 AtAAEs retrieved from previously published data (Shockey et al., 2002, 2003; De Azevedo Souza et al., 2008) by using neighbor-joining method. In consistent with previous result (Shockey et al., 2003), AAE superfamily could be separated into six distinct subfamilies (Figure 1). Among the 53 SlAAE proteins, 15 belong to clade I, 4 to clade II, 19 to clade IV (the largest clade), 14 to clade V, and 1 to clade VI (the smallest clade). Notably, clade III, a special subfamily, contains only 19 AtAAEs (Figure 1).
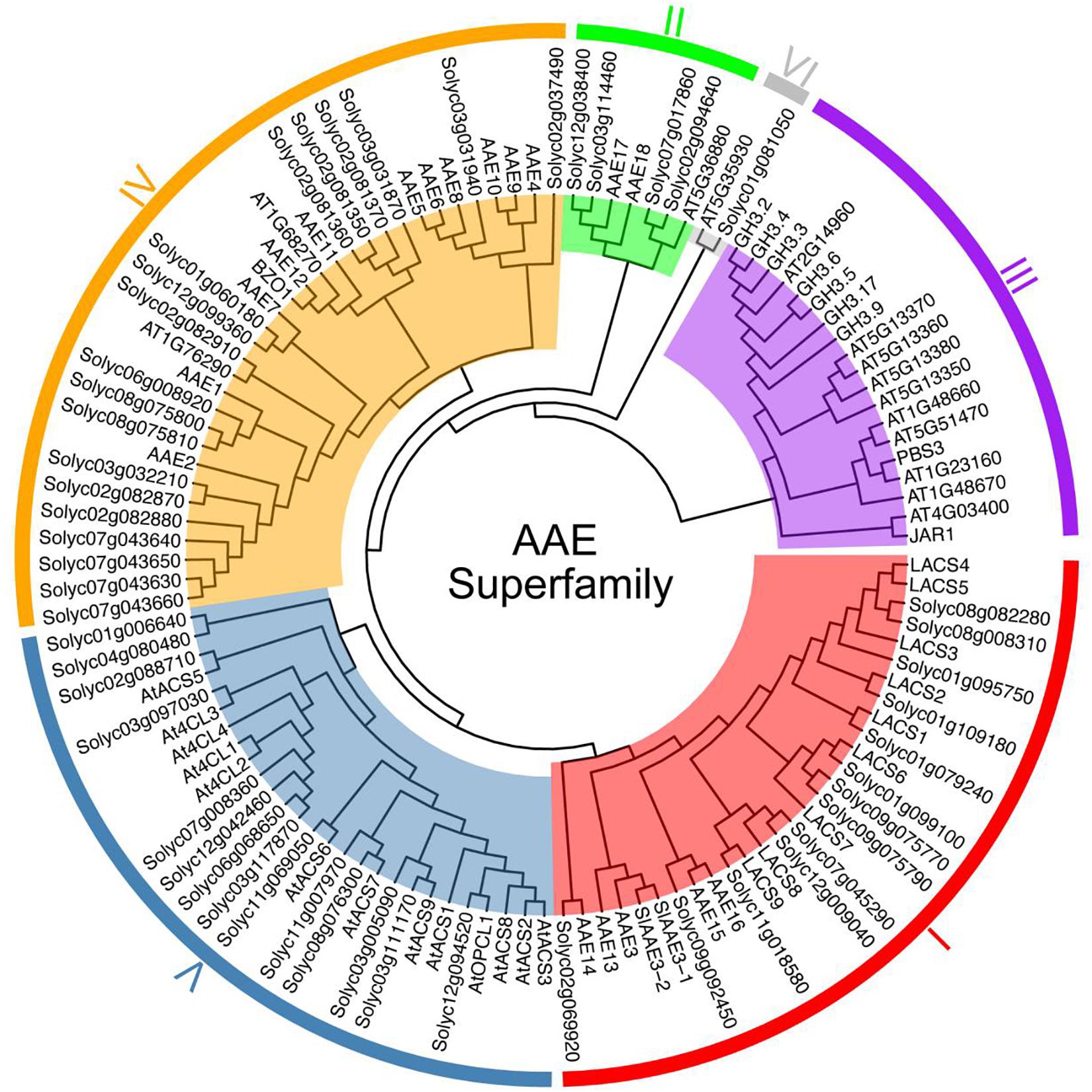
Figure 1. The phylogenetic analysis of tomato (S. lycopersicum) acyl-activating enzymes (AAEs) (SlAAEs). The phylogenetic analysis of AAEs from tomato and Arabidopsis using the complete protein sequences. The neighbor-joining (NJ) tree was constructed using the MEGA 7.0 software with the pairwise deletion option, and 1,000 bootstrap replicates were used to assess tree reliability. AAEs from tomato and Arabidopsis fell in six separate subfamilies as I–VI.
According to the feature of known long-chain acyl-CoA synthetase (LACS) proteins (Iijima et al., 1996; Fulda et al., 1997), 11 SlAAE members from clade I contain the eukaryote-type linker domain, a motif of between 30 and 70 aa residues (Figure 2), and may be active against long-chain fatty acids. However, similar to previously characterized AtLACSs (Shockey et al., 2002), the other four members (i.e., SlAAE3-1, SlAAE3-2, Solyc02g069920, and Solyc09g092450) probably do not produce the activity of the LACS enzyme even though they showed highly sequence similarity to 11 members. So far, the biological functions of 13 AAEs from clade I remain unknown except Solyc01g079240 (SlLACS1) and Solyc01g109180 (SlLACS2), both of which are reported to be involved in wound-induced suberization of tomato fruit (Han et al., 2018).
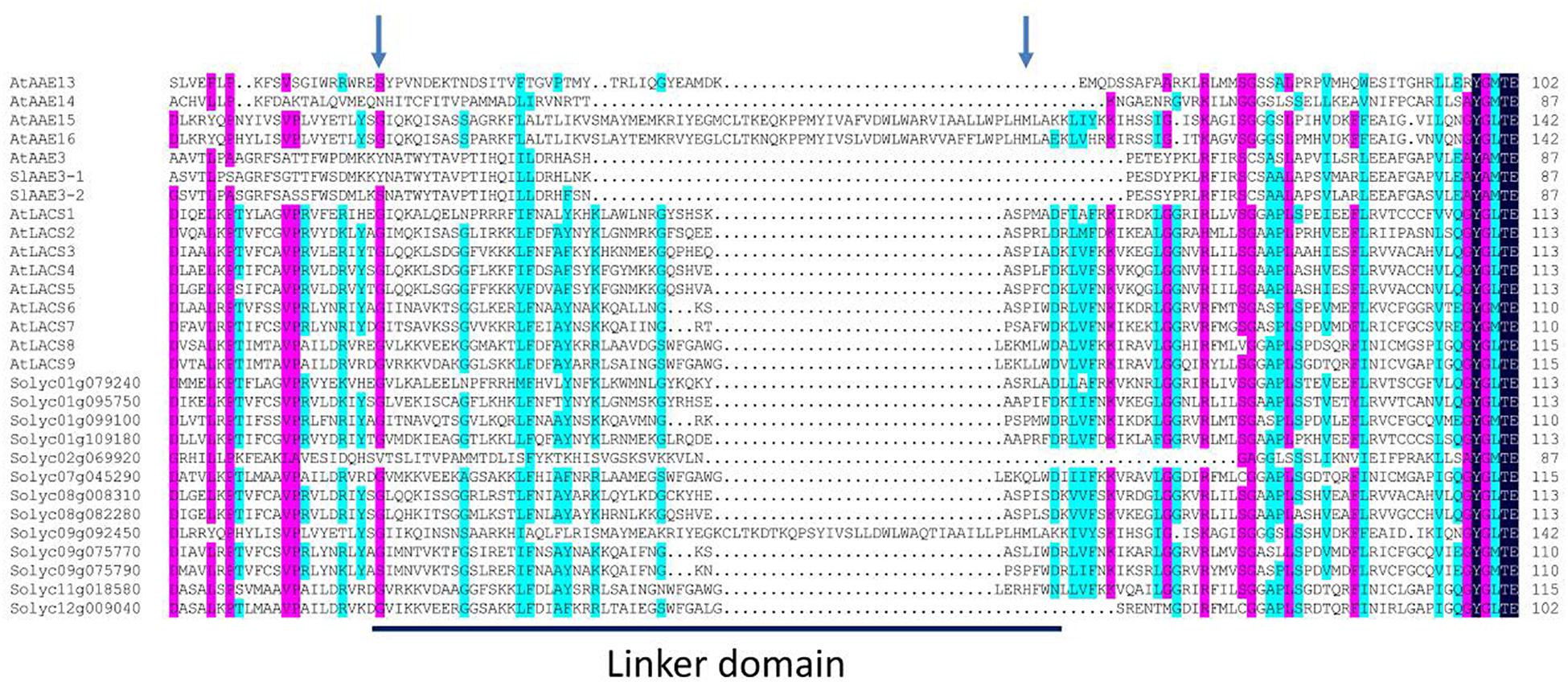
Figure 2. Multiple sequence alignment and linker domain structure of clade I proteins. The sequences of all acyl-activating enzyme (AAE) subfamily I proteins were aligned by DNAMAN software, and the sequence alignment and linker domain annotation were performed according to the reported sequence alignment results of Arabidopsis (Shockey et al., 2002). The blue arrows indicate the left and right boundaries of the linker domains, and the amino acid sequences in the horizontal line indicate the linker domains.
Six members of clade II could be further separated into two subgroups. Solyc02g094640 and Solyc07g017860 from clade II share 78% sequence similarity with AT5G63880 that was reported functioning as acetyl-CoA synthetase involved in lipid synthesis in seeds (Ke et al., 2000). The results of multiple sequence alignment among Solyc03g114460, Solyc12g038400, AAE17, and AAE18 revealed 69% sequence identity. However, all proteins from clade II share only 51% sequence similarity on average. These results suggest that these two subgroups might have distinct functions and require further biochemical assays to elucidate their functions.
Clade III consisted of 19 Arabidopsis AAE family members, termed adenylases, which are considered to participate in multiple important plant hormones (e.g., JA, IAA, and SA) signaling pathways through ATP-dependent adenylation of these hormones (Staswick et al., 2002; Shockey et al., 2003). Notably, no tomato AAEs were included in this clade. Among 19 tomato AAE genes in clade IV, only Solyc07g043630 (SlAACS1) has been reported to be involved in biosynthesis of acylsugars in tomato trichomes (Fan et al., 2020), while the function of the remaining genes has not been characterized yet.
Clade V contains 13 putative Arabidopsis 4-coumarate CoA ligases (At4Cls) (Shockey et al., 2003) and 14 putative tomato 4CLs. The 4CLs play a vital role in enhancing the mechanical support of plants and protecting plants from biotic and abiotic stresses depended on biosynthesis of lignins, flavonoids, and other compounds (Lavhale et al., 2018). For example, 4CL gene involving in biosynthesis of lignin is upregulated in tomato (S. lycopersicum) upon Alternaria solani inoculation (Shinde et al., 2017). In rice, Al repressed the expression of 4CL4, resulting in less lignin accumulation and more 4-coumaric acid and ferulic acid accumulation (Liu et al., 2020). In Arabidopsis, At4CL1/2 required for biosynthesis of lignin was upregulated, while At4CL3 involved in flavonoid was downregulated by wounding (Lee and Douglas, 1996; Ehlting et al., 1999; Soltani et al., 2006).
Gene Structure and Protein Motif Analysis of SlAAE Genes
During the evolution of multigene families, diversification of gene structure may facilitate evolutionary co-option of genes for new functions to adapt to changing environments (Shang et al., 2013; Hu et al., 2015). To better understand the structural diversity of SlAAE genes, the conserved motifs and exon-intron organizations were analyzed according to the phylogenetic relationships (Figure 3).
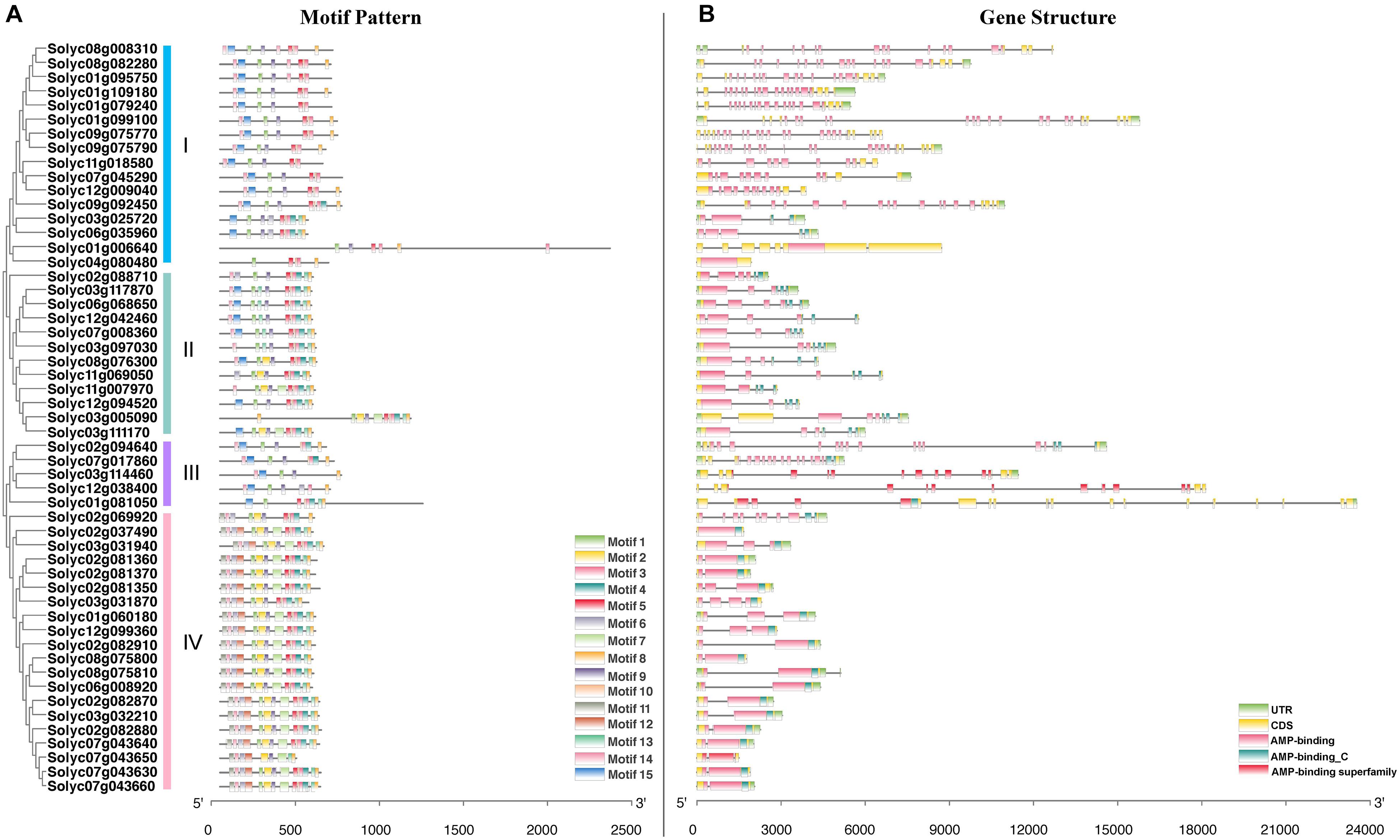
Figure 3. Phylogenetic relationships, gene structure, and architecture of conserved protein motifs in AAE genes from tomato. (A) The motif composition of tomato acyl-activating enzyme (AAE) proteins in accordance to the phylogenetic relationship. Fifteen motifs are shown in different colored boxes. The length of motifs in each AAE protein was displayed proportionally. (B) The exon-intron structure of SlAAE genes. Each exon-intron structure analysis of tomato AAE genes was completed using the online tool GSDS. The length of gene can be estimated using the scale at the bottom.
The potential conserved motifs of all SlAAE proteins were presented according to the MEME motif analysis as described (Xie et al., 2018). As a result, 15 different motifs were identified from all SlAAEs, which were successively named as motifs 1–15 (Figure 3A). The sequence information of each motif is displayed in Figure 4. Among these motifs, motifs 3, 5, and 9 belong to the AMP-binding domain, which are widely distributed on all SlAAEs. As expected, SlAAEs members within the same cluster in the phylogenetic tree commonly share a similar motif compositions (Figure 3A), indicating that they might have functional similarities.

Figure 4. The sequence information of each conserved motif (1–15). Conserved motifs of acyl-activating enzyme (AAE) superfamily members were found using the MEME online tool with parameters: number of repetitions (any), maximum number of motif (20), and the optimum width of each motif (between 6 and 100 residues).
As shown in Figure 3B and Supplementary Table 2, SlAAEs genes contain 0–22 introns. For instance, while Solyc02g037490 and Solyc04g080480 have no intron, Solyc01g099100, Solyc09g075770, and Solyc09g075790 have 22 introns. Others have at least one intron, but genes with 8, 12, 14, 15, 20, and 21 introns were not observed. Most of SlAAE members in the same group have a similar gene structure. For example, most of group IV members contained only a single intron. These results suggest that SlAAEs possessing similar gene structures and motifs were clustered in the same group and might have evolved similar functions in tomato.
Chromosomal Localization of SlAAE Genes
To probe the chromosomal distribution of all SlAAE genes, all members were mapped to tomato chromosomes based on physical location information derived from the database of tomato genome. The result showed that all of the 53 SlAAEs could be mapped onto 10 out of 12 tomato chromosomes (except Chr05 and Chr10) in an increasing order from short-arm to long-arm telomere (Figure 5), and most of SlAAE genes were distributed in the chromosome ends. In addition, bias change in gene number was inspected. Among 10 chromosomes, Chr02 contained largest number of AAE genes, while Chr04 had least.
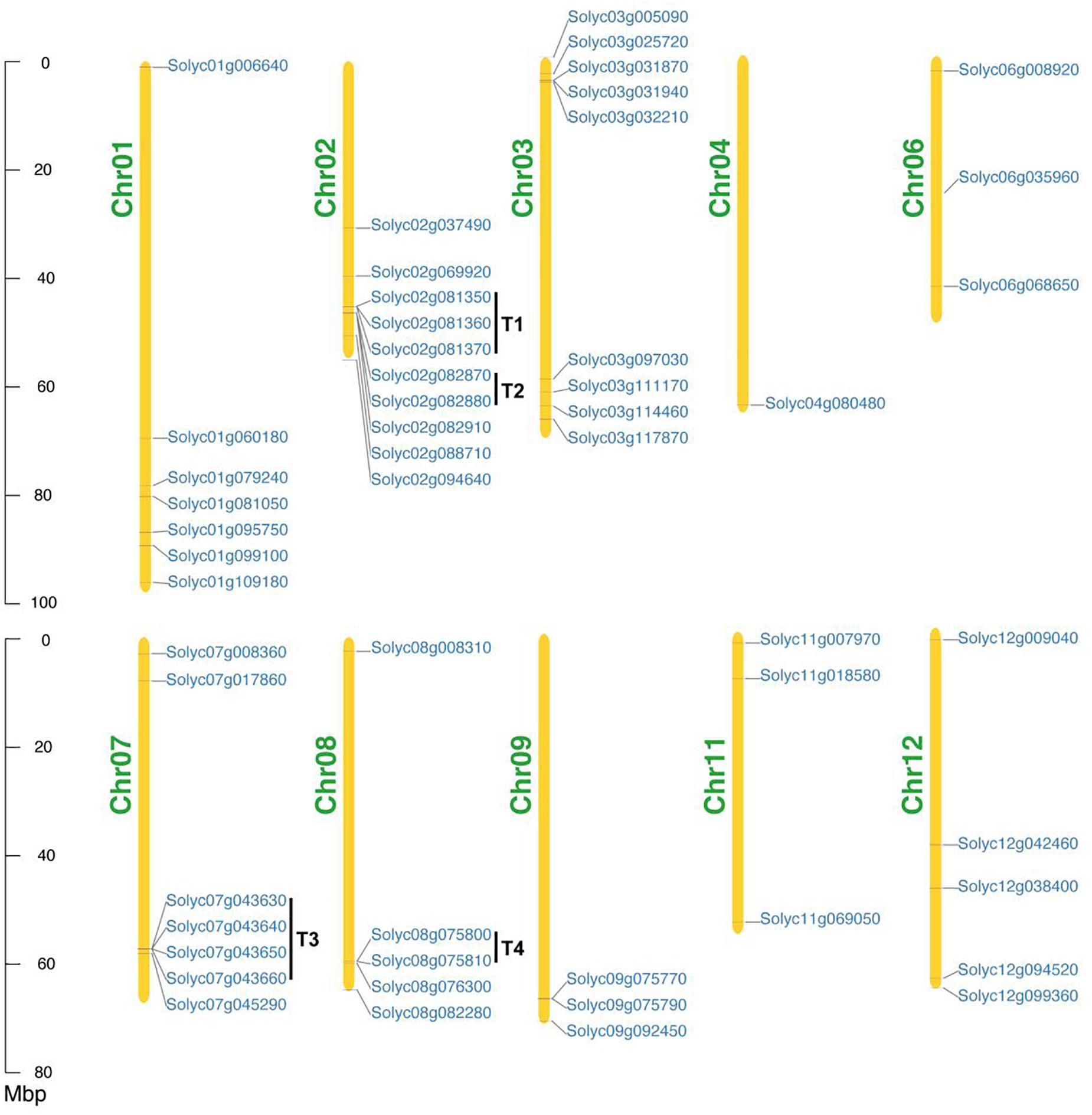
Figure 5. Chromosomal locations of SlAAE genes. In total, 53 SlAAE genes are distributed on 10 of 12 tomato chromosomes. Mbp indicates the scale. T, tandem duplication; Chr, chromosome.
According to a previous study (Holub, 2001), 200 kb of a chromosomal area containing two or more genes is defined as a tandem duplication event. As shown in Figure 5, four gene pairs present as tandem duplication (T) observed on three chromosomes, i.e., T1 (Solyc02g081350, Solyc02g081360, and Solyc02g081370) and T2 (Solyc02g082870 and Solyc02g082880) on Chr02, T3 (Solyc07g043630, Solyc07g043640, Solyc07g043650, and Solyc07g043660) on Chr07, and T4 (Solyc08g075800 and Solyc08g075810) on Chr08. Therefore, in-tandem AAE duplicates comprise 21% of the whole tomato AAE superfamily.
Syntenic Analysis of AAE Genes in Tomato Genome
Besides the tandem duplication events, we also investigated the segmental duplication in the tomato genome relating to the recurring polyploidization events, which generated gene duplicates that have usually been retained in extant tomato genome (Wang and Paterson, 2011). In this study, Eight pairs (named pair 1–8) of syntenic AAE paralogs were observed within the tomato genome (Figure 6A). According to the phylogenetic tree (Figure 1), the tomato paralogs belong to clade I (syntenic pairs 1 and 8), clade IV (syntenic pairs 2, 3, and 4), as well as clade V (syntenic pairs 5, 6, and 7), thus allowing us to propose the conserved functions between the syntenic pairs.
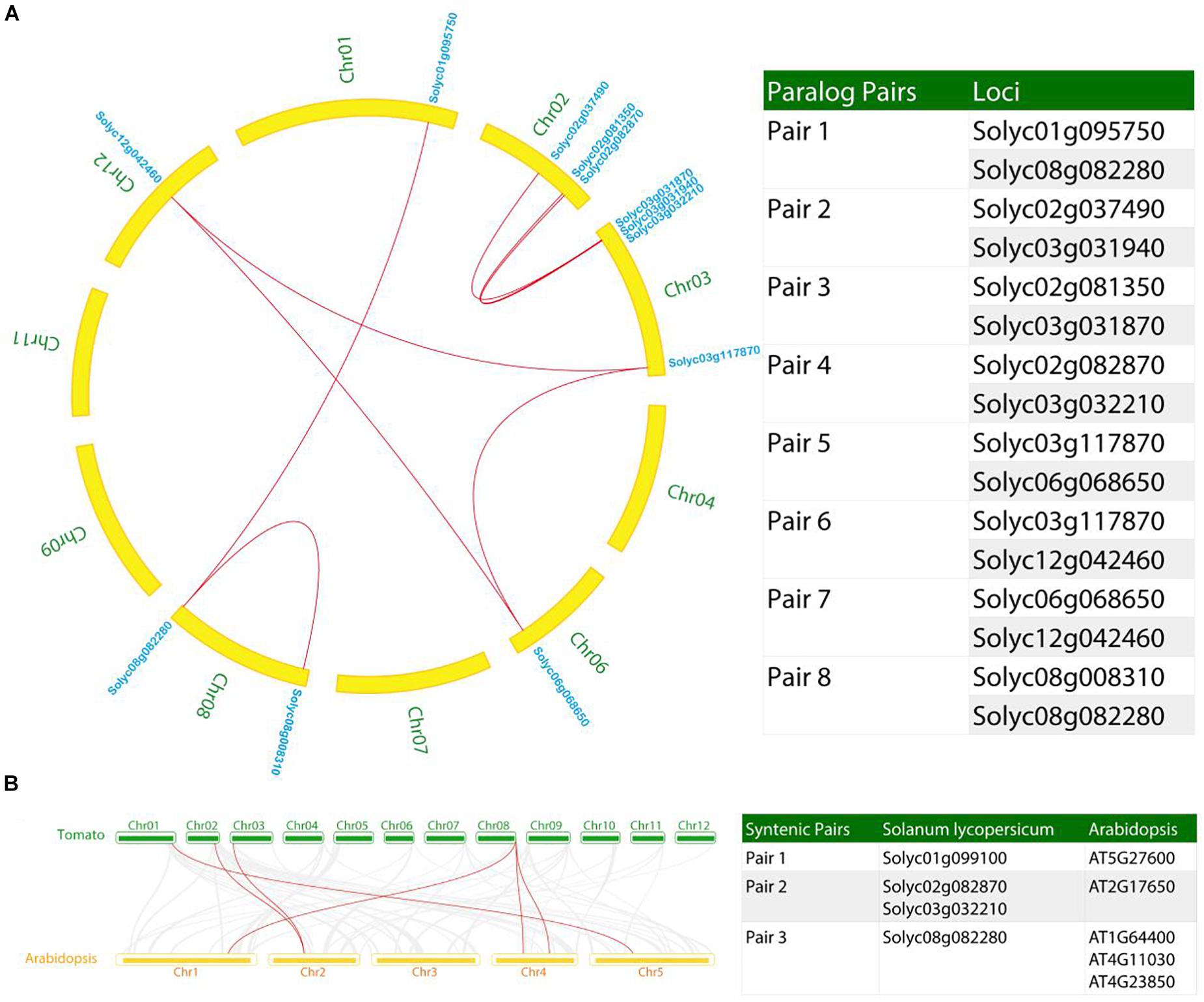
Figure 6. Syntenic analysis of AAE genes in the tomato genome. (A) Identification of paralog pairs in syntenic blocks within the tomato genome. Ten paralog pairs were identified. (B) Synteny analysis of AAE genes between tomato and Arabidopsis. Three syntenic pairs were found in this analysis. Gray lines in the background indicate the collinear blocks within tomato and Arabidopsis, while the red lines highlight the syntenic AAE gene pairs.
Furthermore, we constructed a comparative syntenic map between tomato and Arabidopsis (Figure 6B). Three syntenic pairs comprising four SlAAE genes were identified in syntenic blocks. We also found that duplicated AAE genes exhibited conserved synteny with Arabidopsis genes. For example, Solyc02g082870 is microsyntenic to Solyc03g032210 (paralog pair 4), both of which are syntenic to AT2G17650 (ortholog pair 2, Figure 6B). Solyc08g082280 is microsyntenic to Solyc01g095750 (paralog pair 1) and syntenic to three Arabidopsis genes (ortholog pair 3). Based on these information, we could have inferred the functions of these tomato genes based on the functions of Arabidopsis genes, though the functions of these genes in Arabidopsis are yet to be investigated. Nevertheless, the biological functions of these ortholog genes could have been conservatively evolved since the last common ancestor of tomato and Arabidopsis, which is estimated to have existed approximately 150 million years ago (Ku et al., 2000).
Expression Profiles of SlAAEs Under Aluminum Stress
We have previously demonstrated that rice bean (V. umbellata) VuAAE3, a gene showing a high sequence identity to SlAAE3-1 (Solyc03g025720) and SlAAE3-2 (Solyc06g035960), was involved in Al tolerance (Lou et al., 2016a). This prompted us to investigate the potential function of SlAAE genes in Al stress response after a systemic analysis of the SlAAE gene family. On the basis of tomato root tip Al stress-responsive expressed genes identified from the results of RNA-seq (SRP227103) (Jin et al., 2020), we found that 50 out of 53 SlAAE genes could be detected by RNA-seq in the tomato root apex (Supplementary Table 3). However, six AAE genes showed FPKM value lower than six and were hardly expressed either without or with Al stress. In addition, the expression of 35 AAE genes was not induced by Al stress in tomato root apexes (Supplementary Table 3). Finally, only 9 out of 53 SlAAEs were identified to be differentially regulated, 8 were upregulated, and 1 were downregulated by 5 μM of Al (Supplementary Table 4 and Figure 7A). Notably, Solyc03g025720 (SlAAE3-1) was highly expressed and greatly induced by Al compared with others. We then examined the specificity of SlAAE3-1 expression by exposing tomato seedlings to various metals, including Al, Cd, La, and Cu. The expression of SlAAE3-1 was greatly induced by Al but not by other metals (Figure 7B). To verify the reliability of the RNA-seq data, 14 SlAAE genes were selected for the qRT-PCR analysis. As shown in Figure 7C, all 14 SlAAE genes displayed similar expression patterns to that obtained using RNA-seq. A good correlation (R2 = 0.8415) was observed for their expression in plot qRT-PCR results against that of RNA-seq, indicating that the RNA-seq data accurately reflected the transcriptional changes induced by Al stress.
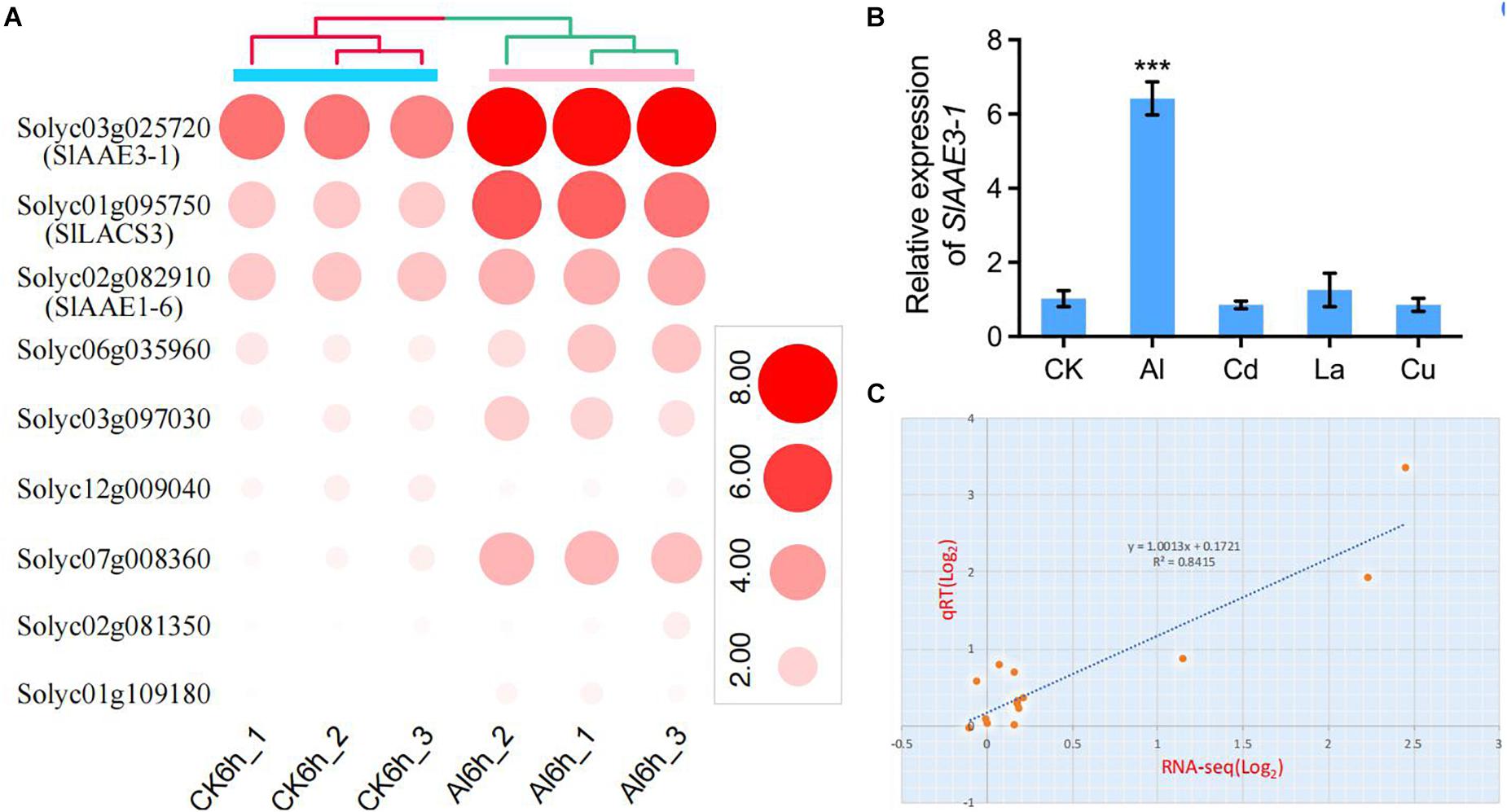
Figure 7. Expression profiles of SlAAEs under Al stress. (A) Heatmap of nine differentially expressed SlAAE genes during Al stress. (B) Expression specificity of SlAAE3-1 gene. Seedlings were subjected to 1/5 Hoagland solution containing 5 μM AlCl3, or 5 μM CdCl2, 5 μM LaCl3, or 3 μM CuCl2 for 6 h. Data are the means ± SD (n = 3); the asterisk indicates significant differences between control and treatment at P < 0.001 using one-way ANOVA. (C) Correlation of gene expression levels between the RNA-seq data and quantitative reverse-transcriptase (qRT-PCR) analysis. Fourteen SlAAEs potentially responding to Al were selected and subjected to the qRT-PCR analysis using the same RNA as for RNA-seq. Both x- and y-axes are shown in Log2 scale.
Identification of Stress-Responsive cis-Acting Elements of SlAAEs
To explore the cis-acting elements probably implicated in the expression regulation of SlAAEs, 2.0 kb sequences upstream of the start codon of SlAAEs were analyzed using the PlantCARE database (Lescot et al., 2002). The results showed that most of the predicted cis-acting elements were associated with phytohormone responses. In addition, there were MYB-binding site, light-responsive element, 60K protein site, ATBP-1 binding site, and endosperm-specific negative expression. However, we did not find Al-responsive element predicted from the PlantCARE (Figure 8). Sensitive to proton rhizotoxicity 1 (STOP1) is a C2H2-type zinc finger transcription factor that regulates expression of many downstream genes involved in Al tolerance by binding to STOP1 cis-acting elements GGN(T/g/a/C)V(C/A/g)S(C/G) present in their promoters (Tsutsui et al., 2011; Liu et al., 2016). Therefore, we analyzed the 2 kb length promoter sequence of SlAAE3-1 and identified 28 cis-acting elements that have potentials to interact with STOP1 (Table 1), suggesting that the STOP1 regulatory module may also be present in tomato.

Figure 8. Phylogenetic and cis-element analysis of SlAAE family promoters. A total of 53 of promoter sequences from tomato genome were scanned using PlantCARE. Sorted cis-elements were then mapped on promoters of the corresponding AAEs and visualized using TBtools. Colored rectangles represent different cis-element.
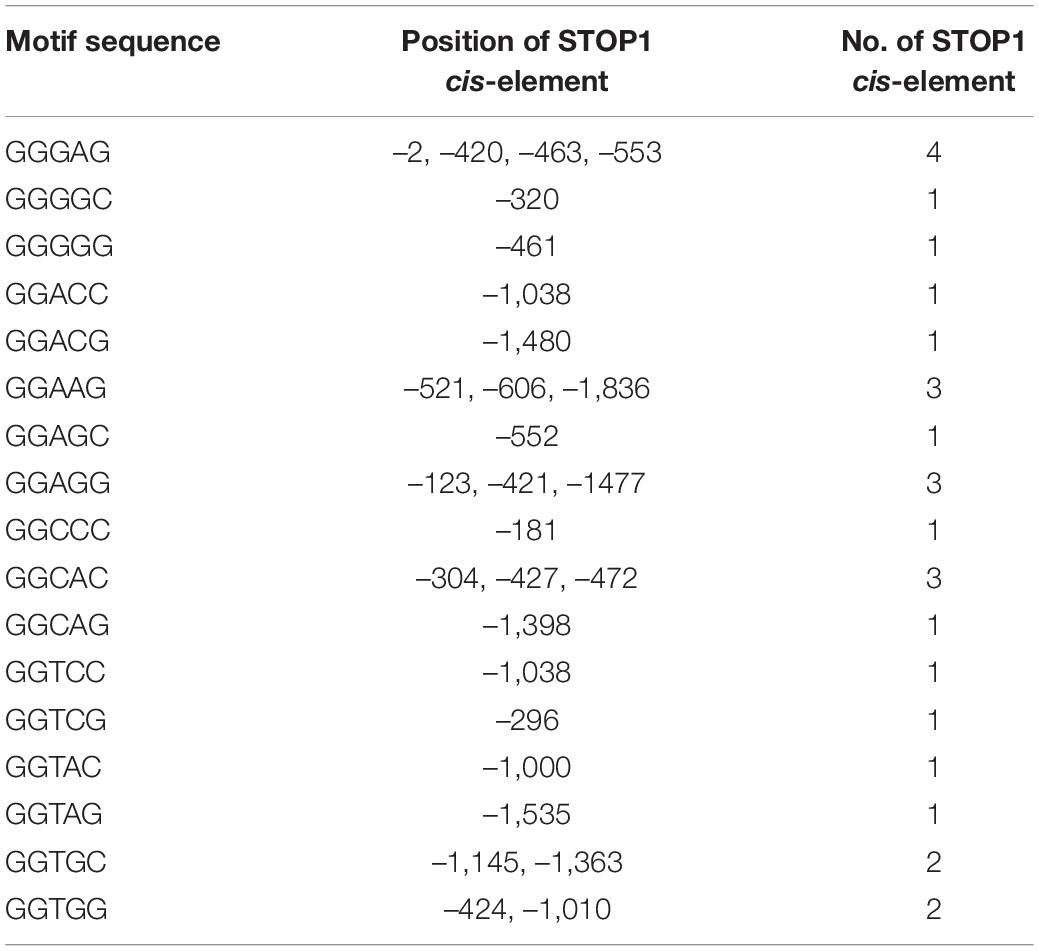
Table 1. The number and position of sensitive to proton rhizotoxicity 1 (STOP1) cis-acting elements of SlAAE3-1 promoter.
Effect of SlAAE3-1 Overexpression in Tobacco on the Tolerance to Aluminum
To confirm that the identified differentially expressed SlAAE genes are exactly involved in tolerance to stresses, we developed transgenic tobacco lines overexpressing SlAAE3-1 that is most significantly induced by Al in the tomato root apex. Three independent SlAAE3-1 overexpressing tobacco lines (i.e., OE1, OE2, and OE3) were selected for examining their tolerance to Al stress. Under normal condition, the wild-type (WT) and transgenic lines showed no difference in root elongation. However, in the presence of 5 μM of Al, the elongation of the primary root of transgenic lines was significantly greater than WT lines (Figures 9A,B). In addition, the PI staining was used to check cell damage. In the absence of Al, PI was hardly stained both in the WT roots and transgenic lines (Figure 9C). However, Al stress resulted in the red fluorescence signals to be more severely accumulated in the WT root apex than in the transgenic lines (Figure 9C), suggesting that transgenic tobacco lines were more tolerant to Al stress than WT plants. Therefore, SlAAE3-1 plays roles in the tolerance to Al stress, consisting with its transcriptional regulation by Al.
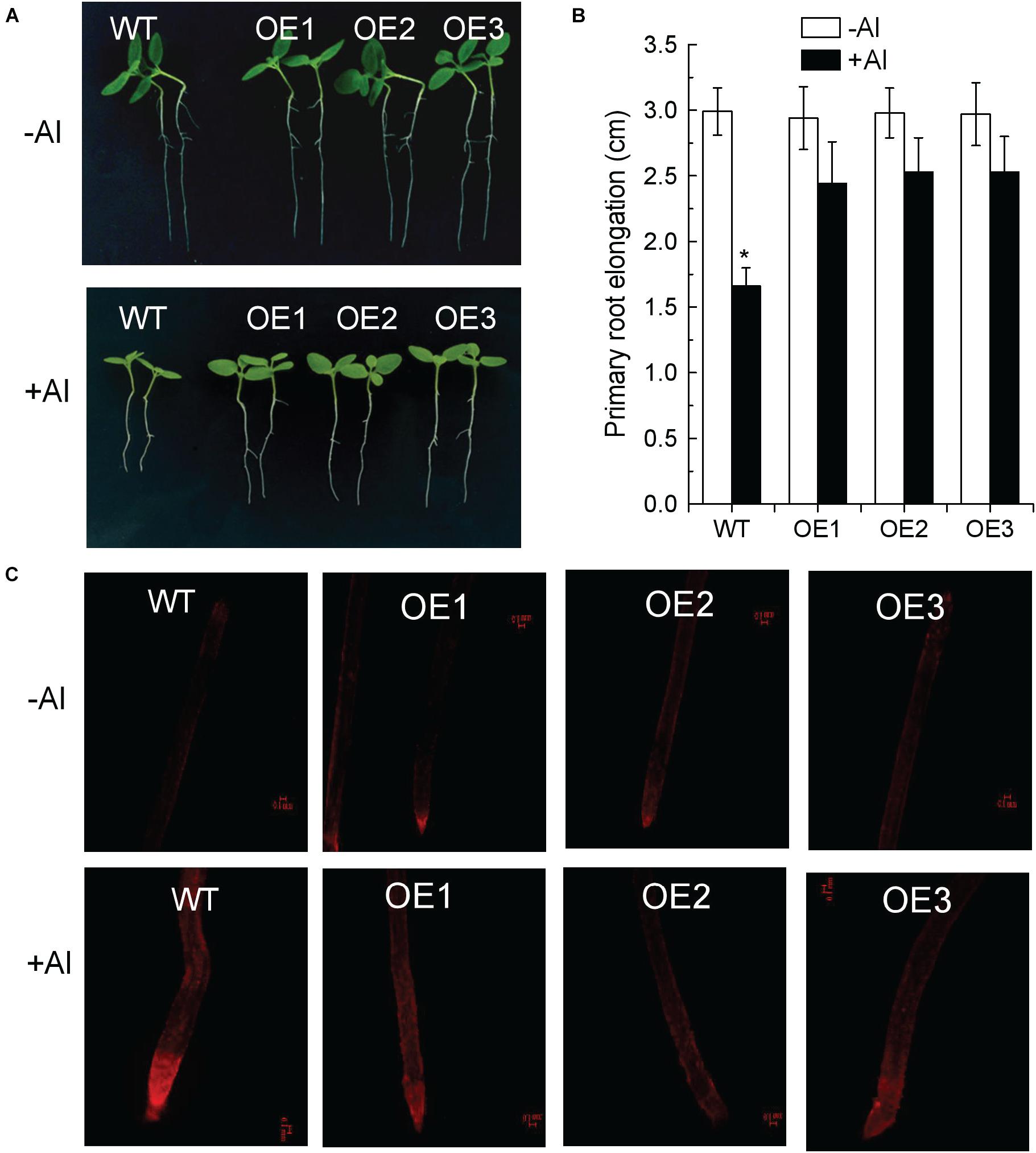
Figure 9. Overexpression of SlAAE3-1 in tobacco improved Al tolerance. (A) Al tolerance phenotype of SlAAE3-1 overexpressing tobacco lines. (B) Primary root elongation. Data are the means ± standard deviation (SD) (n = 10); the asterisk indicates significant differences between control and treatment at P < 0.001 using one-way ANOVA. (C) Propidium iodide staining, fluorescence signals were analyzed using confocal microscopy, bars = 100 μm.
Discussion
In higher plants, AAEs, also called acyl adenylate-forming (Conti et al., 1996; Chang et al., 1997) or AMP-binding proteins (Fulda et al., 1997), are involved in numerous metabolic pathways, such as fatty acid β-oxidation (Schnurr et al., 2004), oxalate catabolism (Foster et al., 2016), and malonic acid degradation (Chen et al., 2011). In the current study, we systemically analyzed the AAE superfamily in tomato and identified a total of 53 members (Supplementary Table 2). We further divided tomato AAE superfamily into five distinct clades based on the phylogenetic analysis (Figure 1). SlAAEs and AtAAEs from clades I, II, IV, V, and VI showed that these genes were not only homologous but could be evolved from a common ancestor. However, AAEs from clade III indicated that this clade of AtAAE genes had a different ancestor with tomato (Figure 1). It is also interesting to note that clade III AAEs encompass 19 Arabidopsis plant hormone adenylases including JAR1. Substrate-dependent ATP-32P-PPi isotope exchange experiment demonstrated that JAR1 is specifically active on JA, while some members from this clade are active on auxin (Staswick et al., 2002). Here, we found that all 53 tomato AAE superfamily members had hardly been reported functionally, except SlLACS1 (Solyc01g079240) and SlLACS2 (Solyc01g109180) (Han et al., 2018). Therefore, the systematic analysis and identification of the tomato AAE superfamily in this study facilitate further studying on the biological function of the superfamily.
As one of the most important metabolisms in plants, phenylpropanoid metabolism provides precursors for more than 8,000 metabolites contributing to plant development and plant-environment interplay (Lavhale et al., 2018; Dong and Lin, 2021). The reaction catalyzed by 4-coumarate-CoA ligase (4CL) is the third step of the first three shared common steps of the general phenylpropanoid pathway, which is responsible for channelizing precursors for various phenylpropanoids (Fraser and Chapple, 2011; Lavhale et al., 2018; Dong and Lin, 2021). In plants, 4CL enzymes belong to AAE superfamily and catalyze the reaction that converts methoxy or hydroxycinnamic acid derivatives to corresponding CoA thioesters (Shockey et al., 2003; Lavhale et al., 2018). In addition, the 4CL enzymes play vital roles in plant physiology or in responses to biotic and abiotic stresses (Fritzemeier et al., 1987; Lee and Douglas, 1996; Sun et al., 2013; Abdollahi Mandoulakani et al., 2017; Blanco-Ulate et al., 2017). In rice, 4CL4-knockout mutants increase the Al tolerance by reducing the binding of Al to the cell walls caused by increased accumulation of 4-coumaric acid and ferulic acid that strengthens the cross-linking of the hemicellulose (Liu et al., 2020). In this study, we also identified 14 putative tomato 4CL and 4CL-like enzymes in clade V, but their biological functions have to be characterized in future. According to the RNA-seq data, we found that the expression level of 2 4CL genes, Solyc03g097030 and Solyc06g035960, increased under Al stress for 6 h (Figure 7A). 4CLs in dicots, such as tomato and Arabidopsis, could be grouped into two clusters, namely, type I and type II (Supplementary Figure 1). Type I is mainly involved in lignin biosynthesis, whereas type II cluster is involved in phenylpropanoid biosynthesis other than lignin (Gui et al., 2011; Lavhale et al., 2018). However, in monocot plants, such as rice, five Os4CLs were mainly categorized into type III except Os4CL2 that belongs to type II (Gui et al., 2011; Sun et al., 2013). As shown in Supplementary Figure 1, Solyc03g097030 belongs to type I, indicating that Solyc03g097030 responds to Al stress possibly by regulating lignin biosynthesis. However, it remains possible that Solyc03g097030 might be involved in other biosynthesis pathways, not just lignin biosynthesis. This proposition is supported by a recent report that although Os4CL4 belongs to type III, it does participate in the regulation of lignin biosynthesis (Liu et al., 2020).
In general, the presence of multiple paralogs in multigene families may relate to the recurring polyploidization events of the angiosperm lineage, which generated gene duplicates that have often retained in extant plant genomes (Wang and Paterson, 2011). It has been shown that a genome-wide duplication event happened in tomato about 83–123 Myr (Sato et al., 2012). Over time, these gene duplicates may have culminated in sub- or neofunctionalization and, subsequently, acquired new functions that are occasionally retained, thus resulting in functional diversity and proliferation of genes derived from a common ancestor gene (Veitia, 2005). In this study, we revealed four tandem duplication segments (Figure 5), which may result in an intensification of gene expression. For example, maize with in-tandem MATE genes (three-copy allele) show a greater Al tolerance as enhanced overall expression of these genes (Maron et al., 2013). Orthologs and paralogs are two essentially different types of homologous genes that are associated with speciation or duplication (Koonin, 2005; Gabaldón and Koonin, 2013). In current study, eight pairs of syntenic AAE paralogs were found within the tomato genome (Figure 6A), and three ortholog pairs were identified in syntenic blocks between tomato and Arabidopsis (Figure 6B). According to the phylogenetic tree, we found that tomato paralog pair 4 belong to the clade IV (Figures 1, 6A), and this paralog pair showed synteny with Arabidopsis AT2G17650 (ortholog pair 2 in Figure 6B), suggesting that these genes could share conserved functions. These results suggested that the analysis of synteny of genes contributes to inferring novel gene functions based on known genes.
Long-chain acyl-CoA synthetases represent a subgroup of AAE superfamily that activates free fatty acids to acyl-CoA and as such play vital roles in long-chain or very-long-chain fatty acids metabolism (Shockey et al., 2002; Shockey and Browse, 2011; Zhao et al., 2021). The loss of catalytic activity of LACS often causes pleiotropic phenotypes such as organ fusion (Weng et al., 2010), male sterility (Jessen et al., 2011), deficient cuticle (Ingram and Nawrath, 2017), delayed seed germination (Shockey et al., 2002), and plant ability to respond to various environmental stresses including drought (Bessire et al., 2007; Lü et al., 2009; Zhao et al., 2019), hypoxia (Licausi et al., 2011; Schmidt et al., 2018; Schmidt and van Dongen, 2019; Xie et al., 2020), and biotic stress (Bessire et al., 2007; Tang et al., 2007; Lü et al., 2009). It has been shown that eukaryotic-type LACSs usually contain the linker domain (Shockey et al., 2002). Combining the phylogenetic tree (Figure 1) and the result of multiple sequence alignment between clade I members (Supplementary Figure 1), We identified 11 tomato LACS members containing a linker domain and nine known AtLACS (1–9) (Shockey et al., 2002). Interestingly, although four tomato members (i.e., SlAAE3-1, SlAAE3-2, Solyc02g069920, and Solyc09g092450) and five Arabidopsis members (i.e., AtAAE3, AtAAE13/14/15/16) belong to clade I-like LACSs, these enzymes do not contain a linker domain. AAE3 has been reported to be involved in oxalate degradation (Foster et al., 2016); however, whether it is involved in other metabolic pathways remains unknown. The above analysis suggests that AAE3 may be involved in fatty acid metabolism in addition to long-chain (i.e., LCFAs; C16–C18) or very-long-chain fatty acids (i.e., VLCFAs; ≥C20), which preferentially activated by LACSs (Shockey et al., 2002; Lü et al., 2009; Shockey and Browse, 2011; Zhao et al., 2021). But, the function of AAE3 still needs further study.
We identified nine tomato SlAAE genes that rapidly responded to Al stress in the tomato root apex, among which SlAAE3-1 was most abundantly expressed and dramatically upregulated (Figure 7 and Supplementary Table 4). Previous studies have shown that rice bean VuAAE3 and wild soybean GsAAE3 are implicated in Al tolerance by regulating oxalate acetylation (Lou et al., 2016a; Xian et al., 2020). Here, we demonstrated that tomato SlAAE3-1 plays the same role with respect to Al tolerance (Figure 9). However, the role of other Al-responsive SlAAE genes in Al tolerance has to be investigated. There is considerable evidence that AAE genes play critical roles in the plant growth and development (Schnurr et al., 2004; Lü et al., 2009; Chen et al., 2011; Yang et al., 2017) and improving tolerance to abiotic stresses, including salinity (Zhou et al., 2020), drought (Bessire et al., 2007; Zhao et al., 2019), and metal stress (e.g., Cd and Al) (Lou et al., 2016a; Xian et al., 2020). Therefore, the functional roles of these SlAAEs in various stress responses could be inferred. In accordance with this supposition, many cis-acting elements related to biotic and abiotic stresses have been identified to be present in their promoters (Figure 8).
In summary, we provided the first integrated bioinformatic information of SlAAE superfamily in tomato including gene identification, structure, chromosomal location, duplication, and expression regulation by Al stress. A total of 53 SlAAE genes were identified, which is essential for the functional characterization of SlAAE3 genes in tomato in future. Furthermore, the RNA-seq data and qRT-PCR analysis have identified nine Al-responsive SlAAE genes and characterized one of them, SlAAE3-1, to be implicated in Al tolerance, which pave the way for identifying novel genes involved in Al tolerance. In addition, the central role of SlAAE members in diverse metabolisms shed light on the importance of this family in responding to both biotic and abiotic stresses.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
JY and WC conceived the research. JJ, QH, and PL performed the experiments. HL provided the technical assistance. JJ and QH analyzed the data. JJ, WC, and JY wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was financially supported by grants from the Zhejiang Provincial Natural Science Foundation (LY19C150006), the National Natural Science Foundation of China (31601765), the China Postdoctoral Science Foundation (to WC), and the China Scholarship Council.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.754147/full#supplementary-material
Footnotes
- ^ http://pfam.xfam.org/
- ^ https://phytozome.jgi.doe.gov/pz/portal.html
- ^ http://smart.embl-heidelberg.de/smart/batch.pl
- ^ https://www.expasy.org/
- ^ https://www.megasoftware.net/
- ^ http://meme-suite.org/tools/meme
- ^ http://bioinformatics.psb.ugent.be/webtools/plantcare/html/
References
Abdollahi Mandoulakani, B., Eyvazpour, E., and Ghadimzadeh, M. (2017). The effect of drought stress on the expression of key genes involved in the biosynthesis of phenylpropanoids and essential oil components in basil (Ocimum basilicum L.). Phytochemistry 139, 1–7. doi: 10.1016/j.phytochem.2017.03.006
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, 202–208. doi: 10.1093/nar/gkp335
Bessire, M., Chassot, C., Jacquat, A. C., Humphry, M., Borel, S., Petétot, J. M. C., et al. (2007). A permeable cuticle in Arabidopsis leads to a strong resistance to Botrytis cinerea. EMBO J. 26, 2158–2168. doi: 10.1038/sj.emboj.7601658
Blanco-Ulate, B., Hopfer, H., Figueroa-Balderas, R., Ye, Z., Rivero, R. M., Albacete, A., et al. (2017). Red blotch disease alters grape berry development and metabolism by interfering with the transcriptional and hormonal regulation of ripening. J. Exp. Bot. 68, 1225–1238. doi: 10.1093/jxb/erw506
Chang, K. H., Xiang, H., and Dunaway-Mariano, D. (1997). Acyl-adenylate motif of the acyl-adenylate/thioester-forming enzyme superfamily: a site-directed mutagenesis study with the pseudomonas sp. strain CBS3 4-chlorobenzoate:coenzyme A ligase. Biochemistry 36, 15650–15659. doi: 10.1021/bi971262p
Chen, C. J., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y. H., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, G., Liu, H., Wei, Q., Zhao, H., Liu, J., and Yu, Y. (2017). The acyl-activating enzyme PhAAE13 is an alternative enzymatic source of precursors for anthocyanin biosynthesis in petunia flowers. J. Exp. Bot. 68, 457–467. doi: 10.1093/jxb/erw426
Chen, H., Kim, H. U., Weng, H., and Browse, J. (2011). Malonyl-CoA synthetase, encoded by ACYL ACTIVATING ENZYME13, is essential for growth and development of Arabidopsis. Plant Cell 23, 2247–2262. doi: 10.1105/tpc.111.086140
Conti, E., Franks, N. P., and Brick, P. (1996). Crystal structure of firefly luciferase throws light on a superfamily of adenylate-forming enzymes. Structure 4, 287–298. doi: 10.1016/S0969-2126(96)00033-0
De Azevedo Souza, C., Barbazuk, B., Ralph, S. G., Bohlmann, J., Hamberger, B., and Douglas, C. J. (2008). Genome-wide analysis of a land plant-specific acyl:coenzymeA synthetase (ACS) gene family in Arabidopsis, poplar, rice and Physcomitrella. New Phytol. 179, 987–1003. doi: 10.1111/j.1469-8137.2008.02534.x
Dong, N. Q., and Lin, H. X. (2021). Contribution of phenylpropanoid metabolism to plant development and plant–environment interactions. J. Integr. Plant Biol. 63, 180–209. doi: 10.1111/jipb.13054
Ehlting, J., Büttner, D., Wang, Q., Douglas, C. J., Somssich, I. E., and Kombrink, E. (1999). Three 4-coumarate:coenzyme A ligases in Arabidopsis thaliana represent two evolutionarily divergent classes in angiosperms. Plant J. 19, 9–20. doi: 10.1046/j.1365-313X.1999.00491.x
El-Gebali, S., Mistry, J., Bateman, A., Eddy, S. R., Luciani, A., Potter, S. C., et al. (2019). The Pfam protein families database in 2019. Nucleic Acids Res. 47, 427–432. doi: 10.1093/nar/gky995
Fan, P. X., Wang, P. P., Lou, Y. R., Leong, B. J., Moore, B. M., Schenck, C. A., et al. (2020). Evolution of a plant gene cluster in Solanaceae and emergence of metabolic diversity. eLife 9:e56717. doi: 10.7554/eLife.56717
Finn, R. D., Clements, J., and Eddy, S. R. (2011). HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39, 29–37. doi: 10.1093/nar/gkr367
Foster, J., Kim, H. U., Nakata, P. A., and Browse, J. (2012). A previously unknown oxalyl-CoA synthetase is important for oxalate catabolism in Arabidopsis. Plant Cell 24, 1217–1229. doi: 10.1105/tpc.112.096032
Foster, J., Luo, B., and Nakata, P. A. (2016). An oxalyl-CoA dependent pathway of oxalate catabolism plays a role in regulating calcium oxalate crystal accumulation and defending against oxalate-secreting phytopathogens in Medicago truncatula. PLoS One 11:e0149850. doi: 10.1371/journal.pone.0149850
Fraser, C. M., and Chapple, C. (2011). The phenylpropanoid pathway in Arabidopsis. Arab. Book 9:e0152. doi: 10.1199/tab.0152
Fritzemeier, K. H., Cretin, C., Kombrink, E., Rohwer, F., Taylor, J., Scheel, D., et al. (1987). Transient induction of phenylalanine ammonia-lyase and 4-coumarate: CoA ligase mRNAs in potato leaves infected with virulent or avirulent races of Phytophthora infestans. Plant Physiol. 85, 34–41. doi: 10.1104/pp.85.1.34
Fulda, M., Heinz, E., and Wolter, F. P. (1997). Brassica napus cDNAs encoding fatty acyl-CoA synthetase. Plant Mol. Biol. 33:12. doi: 10.1023/A:1005780529307
Gabaldón, T., and Koonin, E. V. (2013). Functional and evolutionary implications of gene orthology. Nat. Rev. Genet. 14, 360–366. doi: 10.1038/nrg3456
Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., et al. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, 1178–1186. doi: 10.1093/nar/gkr944
Gui, J., Shen, J., and Li, L. (2011). Functional characterization of evolutionarily divergent 4-coumarate:coenzyme A ligases in rice. Plant Physiol. 157, 574–586. doi: 10.1104/pp.111.178301
Han, X. Y., Mao, L., Lu, W. J., Tao, X. Y., Wei, X. P., and Luo, Z. S. (2018). Abscisic acid induces differential expression of genes involved in wound-induced suberization in postharvest tomato fruit. J. Integr. Agric. 17, 2670–2682. doi: 10.1016/s2095-3119(18)62142-2
Holub, E. B. (2001). The arms race is ancient history in Arabidopsis, the wildflower. Nat. Rev. Genet. 2, 516–527. doi: 10.1038/35080508
Horsch, R. B., Fry, J. E., Hoffmann, N. L., Eichholtz, D., Rogers, S. G., and Fraley, R. T. (1985). A simple and general method for transferring genes into plants. Science 227, 1229–1231. doi: 10.1126/science.227.4691.1229
Hu, W., Wei, Y. X., Xia, Z. Q., Yan, Y., Hou, X. W., Zou, M. L., et al. (2015). Genome-wide identification and expression analysis of the NAC transcription factor family in cassava. PLoS One 10:e0136993. doi: 10.1371/journal.pone.0136993
Iijima, H., Fujino, T., Minekura, H., Suzuki, H., Kang, M. J., and Yamamoto, T. (1996). Biochemical studies of two rat acyl-CoA synthetases, ACS1 and ACS2. Eur. J. Biochem. 242, 186–190. doi: 10.1111/j.1432-1033.1996.0186r.x
Ingram, G., and Nawrath, C. (2017). The roles of the cuticle in plant development: organ adhesions and beyond. J. Exp. Bot. 68, 5307–5321. doi: 10.1093/jxb/erx313
Jang, J. Y., Choi, Y. H., Shin, T. S., Kim, T. H., Shin, K. S., Park, H. W., et al. (2016). Biological control of Meloidogyne incognita by Aspergillus niger F22 producing oxalic acid. PLoS One 11:e0156230. doi: 10.1371/journal.pone.0156230
Jessen, D., Olbrich, A., Knüfer, J., Krüger, A., Hoppert, M., Polle, A., et al. (2011). Combined activity of LACS1 and LACS4 is required for proper pollen coat formation in Arabidopsis. Plant J. 68, 715–726. doi: 10.1111/j.1365-313x.2011.04722.x
Jin, J. F., Wang, Z. Q., He, Q. Y., Wang, J. Y., Li, P. F., Xu, J. M., et al. (2020). Genome-wide identification and expression analysis of the NAC transcription factor family in tomato (Solanum lycopersicum) during aluminum stress. BMC Genomics 21:288. doi: 10.1186/s12864-020-6689-7
Ke, J. S., Behal, R. H., Back, S. L., Nikolau, B. J., Wurtele, E. S., and Oliver, D. J. (2000). The role of pyruvate dehydrogenase and acetyl-coenzyme A synthetase in fatty acid synthesis in developing Arabidopsis seeds. Plant Physiol. 123, 497–508. doi: 10.1104/pp.123.2.497
Kienow, L., Schneider, K., Bartsch, M., Stuible, H. P., Weng, H., Miersch, O., et al. (2008). Jasmonates meet fatty acids: functional analysis of a new acyl-coenzyme A synthetase family from Arabidopsis thaliana. J. Exp. Bot. 59, 403–419. doi: 10.1093/jxb/erm325
Kochian, L. V. (1995). Cellular mechanisms of aluminum toxicity and resistance in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 46, 237–260. doi: 10.1146/annurev.pp.46.060195.001321
Kochian, L. V., Hoekenga, O. A., and Piñeros, M. A. (2004). How do crop plants tolerate acid soils? Mechanisms of aluminum tolerance and phosphorous efficiency. Annu. Rev. Plant Biol. 55, 459–493. doi: 10.1146/annurev.arplant.55.031903.141655
Kochian, L. V., Piñeros, M. A., Liu, J. P., and Magalhaes, J. V. (2015). Plant adaptation to acid soils: the molecular basis for crop aluminum resistance. Annu. Rev. Plant Biol. 66, 571–598. doi: 10.1146/annurev-arplant-043014-114822
Koo, A. J. K., Chung, H. S., Kobayashi, Y., and Howe, G. A. (2006). Identification of a peroxisomal acyl-activating enzyme involved in the biosynthesis of jasmonic acid in Arabidopsis. J. Biol. Chem. 281, 33511–33520. doi: 10.1074/jbc.M607854200
Koonin, E. V. (2005). Orthologs, paralogs, and evolutionary genomics. Annu. Rev. Genet. 39, 309–338. doi: 10.1146/annurev.genet.39.073003.114725
Ku, H. M., Vision, T., Liu, J. P., and Tanksley, S. D. (2000). Comparing sequenced segments of the tomato and Arabidopsis genomes: large-scale duplication followed by selective gene loss creates a network of synteny. Proc. Natl. Acad. Sci. U.S.A. 97, 9121–9126. doi: 10.1073/pnas.160271297
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lavhale, S. G., Kalunke, R. M., and Giri, A. P. (2018). Structural, functional and evolutionary diversity of 4-coumarate-CoA ligase in plants. Planta 248, 1063–1078. doi: 10.1007/s00425-018-2965-z
Lee, D., and Douglas, C. J. (1996). Two divergent members of a tobacco 4-coumarate:coenzyme A ligase (4CL) gene family (cDNA structure, gene inheritance and expression, and properties of recombinant proteins). Plant Physiol. 112, 193–205. doi: 10.1104/pp.112.1.193
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327.
Letunic, I., and Bork, P. (2018). 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 46, 493–496. doi: 10.1093/nar/gkx922
Licausi, F., Kosmacz, M., Weits, D. A., Giuntoli, B., Giorgi, F. M., Voesenek, L. A. C. J., et al. (2011). Oxygen sensing in plants is mediated by an N-end rule pathway for protein destabilization. Nature 479, 419–422. doi: 10.1038/nature10536
Liu, J. P., Piñeros, M. A., and Kochian, L. V. (2014). The role of aluminum sensing and signaling in plant aluminum resistance. J. Integr. Plant Biol. 56, 221–230. doi: 10.1111/jipb.12162
Liu, M., Xu, J., Lou, H., Fan, W., Yang, J., and Zheng, S. (2016). Characterization of VuMATE1 expression in response to iron nutrition and aluminum stress reveals adaptation of rice bean (Vigna umbellata) to acid soils through Cis regulation. Front. Plant Sci. 7:511. doi: 10.3389/fpls.2016.00511
Liu, S., Zhao, L., Liao, Y., Luo, Z., Wang, H., Wang, P., et al. (2020). Dysfunction of the 4-coumarate:coenzyme A ligase 4CL4 impacts aluminum resistance and lignin accumulation in rice. Plant J. 104, 1233–1250. doi: 10.1111/tpj.14995
Lou, H. Q., Fan, W., Xu, J. M., Gong, Y. L., Jin, J. F., Chen, W. W., et al. (2016a). An oxalyl-CoA synthetase is involved in oxalate degradation and aluminum tolerance. Plant Physiol. 172, 1679–1690. doi: 10.1104/pp.16.01106
Lou, H. Q., Gong, Y. L., Fan, W., Xu, J. M., Liu, Y., Cao, M. J., et al. (2016b). A formate dehydrogenase confers tolerance to aluminum and low pH. Plant Physiol. 171, 294–305. doi: 10.1104/pp.16.01105
Lü, S. Y., Song, T., Kosma, D. K., Parsons, E. P., Rowland, O., and Jenks, M. A. (2009). Arabidopsis CER8 encodes LONG-CHAIN ACYL-COA SYNTHETASE 1 (LACS1) that has overlapping functions with LACS2 in plant wax and cutin synthesis. Plant J. 59, 553–564. doi: 10.1111/j.1365-313x.2009.03892.x
Ma, J. F. (2007). Syndrome of aluminum toxicity and diversity of aluminum resistance in higher plants. Int. Rev. Cytol. 264, 225–252. doi: 10.1016/S0074-7696(07)64005-4
Maron, L. G., Guimarães, C. T., Kirst, M., Albert, P. S., Birchler, J. A., Bradbury, P. J., et al. (2013). Aluminum tolerance in maize is associated with higher MATE1 gene copy number. Proc. Natl. Acad. Sci. U.S.A. 110, 5241–5246. doi: 10.1073/pnas.1220766110
Molano-Flores, B. (2001). Herbivory and calcium concentrations affect calcium oxalate crystal formation in Leaves of Sida (Malvaceae). Ann. Bot. 88, 387–391. doi: 10.1006/anbo.2001.1492
Palmieri, F., Estoppey, A., House, G. L., Lohberger, A., Bindschedler, S., Chain, P. S. G., et al. (2019). Oxalic acid, a molecule at the crossroads of bacterial-fungal interactions. Adv. Appl. Microbiol. 106, 49–77. doi: 10.1016/bs.aambs.2018.10.001
Ryan, P. R., Tyerman, S. D., Sasaki, T., Furuichi, T., Yamamoto, Y., Zhang, W. H., et al. (2011). The identification of aluminium-resistance genes provides opportunities for enhancing crop production on acid soils. J. Exp. Bot. 62, 9–20. doi: 10.1093/jxb/erq272
Sato, S., Tabata, S., Hirakawa, H., Asamizu, E., Shirasawa, K., Isobe, S., et al. (2012). The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485, 635–641. doi: 10.1038/nature11119
Schmidt, R. R., and van Dongen, J. T. (2019). The ACBP1-RAP2.12 signalling hub: a new perspective on integrative signalling during hypoxia in plants. Plant Signal. Behav. 14:e1651184. doi: 10.1080/15592324.2019.1651184
Schmidt, R. R., Fulda, M., Paul, M. V., Anders, M., Plum, F., Weits, D. A., et al. (2018). Low-oxygen response is triggered by an ATP-dependent shift in oleoyl-CoA in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 115, E12101–E12110. doi: 10.1073/pnas.1809429115
Schnurr, J., Shockey, J., and Browse, J. (2004). The acyl-CoA synthetase encoded by LACS2 is essential for normal cuticle development in Arabidopsis. Plant Cell 16, 629–642. doi: 10.1105/tpc.017608
Shang, H. H., Li, W., Zou, C. S., and Yuan, Y. L. (2013). Analyses of the NAC transcription factor gene family in Gossypium raimondii Ulbr.: chromosomal location, structure, phylogeny, and expression patterns. J. Integr. Plant Biol. 55, 663–676. doi: 10.1111/jipb.12085
Shinde, B. A., Dholakia, B. B., Hussain, K., Panda, S., Meir, S., Rogachev, I., et al. (2017). Dynamic metabolic reprogramming of steroidal glycol-alkaloid and phenylpropanoid biosynthesis may impart early blight resistance in wild tomato (Solanum arcanum Peralta). Plant Mol. Biol. 95, 411–423. doi: 10.1007/s11103-017-0660-2
Shockey, J. M., Fulda, M. S., and Browse, J. (2003). Arabidopsis contains a large superfamily of acyl-activating enzymes. phylogenetic and biochemical analysis reveals a new class of acyl-coenzyme A synthetases. Plant Physiol. 132, 1065–1076. doi: 10.1104/pp.103.020552
Shockey, J. M., Fulda, M. S., and Browse, J. A. (2002). Arabidopsis contains nine long-chain acyl-coenzyme A synthetase genes that participate in fatty acid and glycerolipid metabolism. Plant Physiol. 129, 1710–1722. doi: 10.1104/pp.003269
Shockey, J., and Browse, J. (2011). Genome-level and biochemical diversity of the acyl-activating enzyme superfamily in plants: biochemistry and evolution of plant AAE proteins. Plant J. 66, 143–160. doi: 10.1111/j.1365-313X.2011.04512.x
Soltani, B. M., Ehlting, J., Hamberger, B., and Douglas, C. J. (2006). Multiple cis-regulatory elements regulate distinct and complex patterns of developmental and wound-induced expression of Arabidopsis thaliana 4CL gene family members. Planta 224, 1226–1238. doi: 10.1007/s00425-006-0296-y
Staswick, P. E., Tiryaki, I., and Rowe, M. L. (2002). Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation. Plant Cell 14, 1405–1415. doi: 10.1105/tpc.000885
Sun, H., Li, Y., Feng, S., Zou, W., Guo, K., Fan, C., et al. (2013). Analysis of five rice 4-coumarate:coenzyme A ligase enzyme activity and stress response for potential roles in lignin and flavonoid biosynthesis in rice. Biochem. Biophys. Res. Commun. 430, 1151–1156. doi: 10.1016/j.bbrc.2012.12.019
Tang, D. Z., Simonich, M. T., and Innes, R. W. (2007). Mutations in LACS2, a long-chain acyl-coenzyme A synthetase, enhance susceptibility to avirulent Pseudomonas syringae but confer resistance to Botrytis cinerea in Arabidopsis. Plant Physiol. 144, 1093–1103. doi: 10.1104/pp.106.094318
Tsutsui, T., Yamaji, N., and Feng Ma, J. (2011). Identification of a Cis-Acting element of ART1, a C2H2-Type zinc-finger transcription factor for aluminum tolerance in rice. Plant Physiol. 156, 925–931. doi: 10.1104/pp.111.175802
Veitia, R. A. (2005). Paralogs in polyploids: one for all and all for one? Plant Cell 17, 4–11. doi: 10.1105/tpc.104.170130
von Uexküll, H. R., and Mutert, E. (1995). Global extent, development and economic impact of acid soils. Plant Soil 171, 1–15. doi: 10.1007/BF00009558
Wang, G. D., Zhang, S., Ma, X. C., Wang, Y., Kong, F. Y., and Meng, Q. W. (2016). A stress-associated NAC transcription factor (SlNAC35) from tomato plays a positive role in biotic and abiotic stresses. Physiol. Plant. 158, 45–64. doi: 10.1111/ppl.12444
Wang, X. Y., and Paterson, A. H. (2011). Gene conversion in angiosperm genomes with an emphasis on genes duplicated by polyploidization. Genes 2, 1–20. doi: 10.3390/genes2010001
Wang, Y. P., Tang, H. B., Debarry, J. D., Tan, X., Li, J., Wang, X. Y., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, 49–63. doi: 10.1093/nar/gkr1293
Weng, H., Molina, I., Shockey, J., and Browse, J. (2010). Organ fusion and defective cuticle function in a lacs1 lacs2 double mutant of Arabidopsis. Planta 231, 1089–1100. doi: 10.1007/s00425-010-1110-4
Xian, P. Q., Cai, Z. D., Cheng, Y. B., Lin, R. B., Lian, T. X., Ma, Q. B., et al. (2020). Wild soybean oxalyl-CoA synthetase degrades oxalate and affects the tolerance to cadmium and aluminum stresses. Int. J. Mol. Sci. 21:8869. doi: 10.3390/ijms21228869
Xie, L. J., Tan, W. J., Yang, Y. C., Tan, Y. F., Zhou, Y., Zhou, D. M., et al. (2020). Long-chain acyl-CoA synthetase LACS2 contributes to submergence tolerance by modulating cuticle permeability in Arabidopsis. Plants 9:262. doi: 10.3390/plants9020262
Xie, T., Chen, C. J., Li, C. H., Liu, J. R., Liu, C. Y., and He, Y. H. (2018). Genome-wide investigation of WRKY gene family in pineapple: evolution and expression profiles during development and stress. BMC Genomics 19:490. doi: 10.1186/s12864-018-4880-x
Yang, J. L., Fan, W., and Zheng, S. J. (2019). Mechanisms and regulation of aluminum-induced secretion of organic acid anions from plant roots. J. Zhejiang Univ. Sci. B 20, 513–527. doi: 10.1631/jzus.b1900188
Yang, X. J., Liang, W. Q., Chen, M. J., Zhang, D. B., Zhao, X. X., and Shi, J. X. (2017). Rice fatty acyl-CoA synthetase OsACOS12 is required for tapetum programmed cell death and male fertility. Planta 246, 105–122. doi: 10.1007/s00425-017-2691-y
Yu, G. C., Smith, D. K., Zhu, H. C., Guan, Y., and Lam, T. T. Y. (2017). GGTREE: an r package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 8, 28–36. doi: 10.1111/2041-210x.12628
Zhao, H. Y., Kosma, D. K., and Lü, S. Y. (2021). Functional role of long-chain acyl-CoA synthetases in plant development and stress responses. Front. Plant Sci. 12:640996. doi: 10.3389/fpls.2021.640996
Zhao, L. F., Haslam, T. M., Sonntag, A., Molina, I., and Kunst, L. (2019). Functional overlap of long-chain acyl-CoA synthetases in Arabidopsis. Plant Cell Physiol. 60, 1041–1054. doi: 10.1093/pcp/pcz019
Keywords: AAEs superfamily, abiotic stress, Al stress, carboxylic acid, organic acid, oxalate, tomato
Citation: Jin JF, He QY, Li PF, Lou HQ, Chen WW and Yang JL (2021) Genome-Wide Identification and Gene Expression Analysis of Acyl-Activating Enzymes Superfamily in Tomato (Solanum lycopersicum) Under Aluminum Stress. Front. Plant Sci. 12:754147. doi: 10.3389/fpls.2021.754147
Received: 09 August 2021; Accepted: 04 November 2021;
Published: 02 December 2021.
Edited by:
Miguel A. Pineros, Robert W. Holley Center for Agriculture & Health, United States Department of Agriculture-Agricultural Research Service (USDA-ARS), United StatesReviewed by:
Luqing Zheng, Nanjing Agricultural University, ChinaPeijian Cao, Zhengzhou Tobacco Research Institute of CNTC, China
Suzhi Zhang, Maize Research Institute of Sichuan Agricultural University, China
Copyright © 2021 Jin, He, Li, Lou, Chen and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Wei Chen, MTU4NTgyMjM4MDdAMTYzLmNvbQ==; Jian Li Yang, eWFuZ2ppYW5saUB6anUuZWR1LmNu
 Jian Feng Jin
Jian Feng Jin Qi Yu He
Qi Yu He Peng Fei Li
Peng Fei Li He Qiang Lou
He Qiang Lou Wei Wei Chen
Wei Wei Chen Jian Li Yang
Jian Li Yang